
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 September 2022
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.973046
Roseburia intestinalis is an anaerobic bacterium that produces butyric acid and belongs to the phylum Firmicutes. There is increasing evidence that this bacterium has positive effects on several diseases, including inflammatory bowel disease, atherosclerosis, alcoholic fatty liver, colorectal cancer, and metabolic syndrome, making it a potential “Next Generation Probiotic.” We investigated the genomic characteristics, probiotic properties, cytotoxicity, oral toxicity, colonization characteristics of the bacterium, and its effect on the gut microbiota. The genome contains few genes encoding virulence factors, three clustered regularly interspaced short palindromic repeat (CRISPR) sequences, two Cas genes, no toxic biogenic amine synthesis genes, and several essential amino acid and vitamin synthesis genes. Seven prophages and 41 genomic islands were predicted. In addition to a bacteriocin (Zoocin A), the bacterium encodes four metabolic gene clusters that synthesize short-chain fatty acids and 222 carbohydrate-active enzyme modules. This bacterium is sensitive to antibiotics specified by the European Food Safety Authority, does not exhibit hemolytic or gelatinase activity, and exhibits some acid resistance. R. intestinalis adheres to intestinal epithelial cells and inhibits the invasion of certain pathogens. In vitro experiments showed that the bacterium was not cytotoxic. R. intestinalis did not affect the diversity or abundance of the gut flora. Using the fluorescent labelling method, we discovered that R. intestinalis colonizes the cecum and mucus of the colon. An oral toxicity study did not reveal any obvious adverse effects. The lethal dose (LD)50 of R. intestinalis exceeded 1.9 × 109 colony forming units (CFU)/kg, whereas the no observed adverse effect level (NOAEL) derived from this study was 1.32 × 109 CFU/kg/day for 28 days. The current research shows that, R. intestinalis is a suitable next-generation probiotic considering its probiotic properties and safety.
Probiotics are defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (Salminen et al., 2021). The global market for probiotics is expected to reach $91.1 billion by 2026, up significantly from $61.1 billion in 2021.1 Currently, most probiotic strains are derived from a limited number of bacterial species. These include Lactobacillus and Bifidobacterium species. Large-scale applications of metagenomic sequencing and bacterial genome editing methods have greatly expanded the range of bacteria with potential health benefits, and these bacteria are termed “Next Generation Probiotics” (NGPs; O’Toole et al., 2017). The development of NGPs requires by preclinical research, safety testing, pharmacokinetics, pharmacodynamics, and Phase 1–3 clinical trials.
Roseburia intestinalis is an anaerobic bacterium that colonizes in the intestine and belongs to the phylum Firmicutes (Duncan et al., 2002; Nie et al., 2021). During colitis, R. intestinalis modulates the immune response and maintains tight junction integrity (Luo et al., 2019). Its flagellin regulates the long noncoding RNA HIF1A-AS2 (Quan et al., 2018), inhibits the activation of the NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3) inflammasome (Wu et al., 2020), and stimulates the differentiation of regulatory T (Treg) cells to suppress colitis (Zhu et al., 2018). R. intestinalis inhibits intestinal inflammation by increasing the secretion of anti-inflammatory cytokines such as thymic stromal lymphopoietin (TSLP), transforming growth factor (TGF)-β, and interleukin (IL)-10 (Shen et al., 2018). Moreover, R. intestinalis alleviates colitis by affecting the brain-gut axis (Xu et al., 2021).
Roseburia intestinalis can also improve atherosclerosis (Kasahara et al., 2018; Liu et al., 2020) and alcohol-related liver diseases (Seo et al., 2020). There is evidence that the amount of R. intestinalis declines in patients with colorectal cancer (Montalban-Arques et al., 2021) and that treatment with the bacterium alone can prevent or even treat the disease (Liang et al., 2017). The abundance of R. intestinalis decreased in individuals suffering from amyotrophic lateral sclerosis (Nicholson et al., 2021), nonalcoholic steatohepatitis (Pan et al., 2021), pancreatic ductal adenocarcinoma (Zhou et al., 2021), and HIV infection (Dillon et al., 2017). R. intestinalis abundance was negatively correlated with waist circumference in patients with metabolic syndrome (Qin et al., 2021). Moreover, the bacterium has also been associated with graft-versus-host disease (Devaux et al., 2020) and insulin sensitivity in patients with type 2 diabetes (Lee et al., 2019). In fact, a recent study demonstrated R. intestinalis is associated with coronavirus disease 2019 (COVID-19), as the amount of the bacterium decreased significantly in severe patients (Xu et al., 2022). The relationship between R. intestinalis and various diseases is shown in Figure 1.
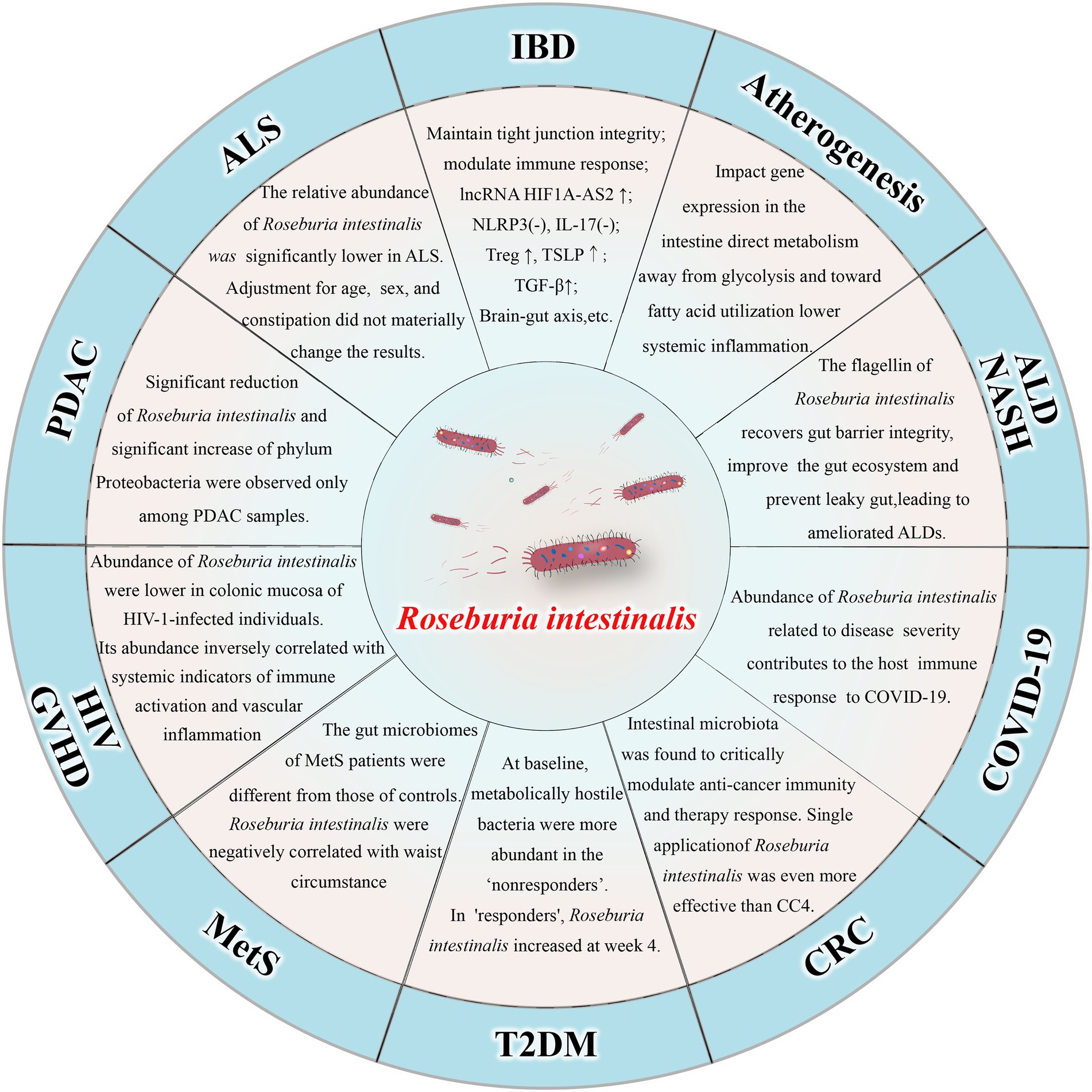
Figure 1. Relationship between Roseburia intestinalis and various diseases. IBD: inflammatory bowel disease; ALD: alcohol-related liver diseases; NASH: nonalcoholic steatohepatitis; COVID-19: coronavirus disease 2019; CRC: colorectal cancer; T2DM: type 2 diabetes mellitus; MetS: metabolic syndrome; HIV: human immunodeficiency virus; GVHD: Graft-versus-host disease; PDAC: pancreatic ductal adenocarcinoma; ALS: amyotrophic lateral sclerosis.
Currently, there are many guidelines related to probiotics, including those issued by the United States Food and Drug Administration (FDA), the World Gastroenterology Organization (WGO), the Product Safety Enforcement Forum of Europe (EU-PROSAFE; Vankerckhoven et al., 2008), and the International Scientific Association for Probiotics and Prebiotics (ISAPP; Hill et al., 2014). This study examined the safety and probiotic properties of R. intestinalis as well as its possible application in healthcare, in accordance with the above guidelines.
Roseburia intestinalis L1-82 (DSMZ14610) was purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). Bacteroides fragilis (ATCC 25285) and Bacteroides vulgatus were obtained from Ningbo Mingzhou Biotechnology Co., Ltd. (Zhejiang, China). The above bacteria were cultured under anaerobic conditions at 37°C. An anaerobic culture environment was created using Atmosphere Generation Systems (AnaeroJar™ ASSEMBLY and AnaeroGen™, OXOID, Thermo Fisher Scientific). The culture medium (Miquel et al., 2015) was brain heart infusion (BHI) containing 0.5% yeast extract (OXOID), 1 mg/ml cellobiose (Macklin, China), 1 mg/ml maltose (Solarbio, China), 0.5 mg/ml cellobiose (Macklin, China), and 0.5 mg/ml cysteine (Solarbio, China). We established a standard curve to convert the absorbance values at 600 nm (OD600) to CFU values (Supplementary Figure S1). Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 25923, Salmonella typhimurium CMCC 50115, Bacillus subtilis DSM 1088, and Bacillus cereus ATCC 11778 were used as controls. These strains were cultured in nutrient broth at 37°C in an aerobic environment.
Genome data for R. intestinalis were obtained from the National Center for Biotechnology Information (NCBI) database (GCF_900537995.1). The Comprehensive Antibiotic Resistance Database (CARD; Alcock et al., 2020) and Virulence Factor Database (VFDB; Liu et al., 2022) were used to predict virulence and antibiotic resistance genes. Genes related to biogenic amines, essential amino acids, and vitamin synthesis were identified in the bacterial genome using the Rapid Annotations using Subsystems Technology (RAST) server (Overbeek et al., 2014). The Carbohydrate-Active enZYmes (CAZy; Drula et al., 2021) database was used to analyze the carbohydrate metabolism ability of R. intestinalis.
CRISPRCasFinder was used to identify the CRISPR loci and associated Cas genes (Couvin et al., 2018). PHAge Search Tool Enhanced Release (PHASTER; Arndt et al., 2016) was used to identify and characterize the prophages within the genome. The bacteriocin operons were identified and visualized using BAGEL4 (van Heel et al., 2018). IslandViewer (Bertelli et al., 2017) was used to predict genomic islands (GIs) in the genome. All the above predictions were made using default parameters. GutSMASH (Pascal Andreu et al., 2021) was used to identify and analyze specific gene clusters associated with primary metabolism and energy capture.
Several bacterial strains considered potential probiotics or pathogens in inflammatory bowel disease (IBD) were selected, and their genomes were compared with that of R. intestinalis. This included strains of R. intestinalis and other species of the genus Roseburia. GenBank accession numbers for these strains are listed in Supplementary Table S1. Genes involved in acid or bile salt tolerance, intestinal adhesion colonization, and antioxidant activity were retrieved from a previous study (Hussein et al., 2020) and aligned using the NCBI Basic Local Alignment Search Tool (BLAST) tools. We aligned the query genome with that of R. intestinalis using the BLAST Ring Image Generator (BRIG; Alikhan et al., 2011).
Acid tolerance was tested as previously described (Anandharaj et al., 2015). The bacterium was grown anaerobically at 37°C for 24 h, centrifuged at 4000 rpm for 10 min, and the suspension was washed twice with phosphate-buffered saline (PBS). We used 1 M HCL to adjust the pH of the medium in order to simulate the acidic environment of the stomach. R. intestinalis was added to the medium at varying pH values and cultured at 37°C for 24 h. Viable cells were counted on BHI agar plates, and the results were expressed as log10 colony-forming units (CFU)/ml.
CSH was measured as described previously (Sharma et al., 2019). R. intestinalis in logarithmic growth phase was washed twice and resuspended in PBS, with the density adjusted to between 0.8 and 1.0. The n-hexadecane and bacterial solution were mixed at a ratio of 1:5 (v:v), and the mixture was vortexed for 2 min. The mixture was kept at room temperature for 45 min, and the aqueous phase was used to determine the OD600. CSH was calculated using the following formula:
CSH (%) = × 100.
A0 and A represent the OD600 values measured before and after mixing with n-hexadecane, respectively.
Auto-aggregation ability was identified based on the method described by Reuben et al. (2020), with slight modifications. The bacterium was rinsed twice and resuspended in PBS, and the OD600 was adjusted to 1.0 (A0). The bacterial suspension was incubated at 37°C for 2–24 h, and the OD600 of the supernatant was measured. Auto-aggregation capacity was calculated using to the following formula:
Auto-aggregation (%) = (1 - ) × 100.
Co-aggregation ability was examined using a previously described method (Solieri et al., 2014). Equal volumes of R. intestinalis and pathogenic bacterial suspensions were mixed and vortexed for 10 s. A suspension of each bacterium alone was used as a control. A series of OD600 measurements were taken at different time points. The co-aggregation was calculated as follows:
Co-aggregation (%) = [1 - ] × 100.
For each time point, Ax represents the OD600 of R. intestinalis, and Ay represents the absorbance of each pathogenic bacterium.
The bile salt tolerance test was performed as previously described (Hussein et al., 2020). R. intestinalis at 108 CFU/ml was inoculated into BHI medium containing 0.3% porcine bile salts (Solarbio, China). To simulate human intestinal conditions, 200 μl of bacterial broth was seeded in a 96-well microplate and incubated in an anaerobic incubator at 37°C for 0–8 h. Bacterial growth was monitored hourly by measuring OD600.
The ability to adhere to intestinal epithelial cells was evaluated in a manner similar to that reported by Dudík et al. (2020). Caco-2 and HT-29 cells were cultured in Roswell Park Memorial Institute (RPMI)-1,640 medium containing 10% fetal bovine serum (Procell, China), seeded in six-well plates, and the medium was changed every 48 h. The cells were cultured continuously for 21 days. Bacteria were washed and resuspended in PBS (OD600 = 1.0). The plate was placed in a cell incubator at 37°C for 1 h with 1 ml of bacterial solution added to each well. The plate was rinsed three times with PBS, and 1 ml of 0.05% trypsin was added to digest the cells. The digested cell suspension was serially diluted in PBS and colonies were counted on BHI agar plates.
The hemolytic activity experiments were based on Sonakshi et al. (Rastogi et al., 2020). R. intestinalis was inoculated onto BHI agar containing 5% (v/v) sterile defibrillated sheep blood, and cultured at 37°C under anaerobic conditions for 72 h. A clear halo around the colony indicates β-hemolysis, a green halo indicates α-hemolysis, and no halo indicates γ-hemolysis. S. aureus ATCC 25923 was used as a positive control.
DNase activity was measured using a previously described method (Rastogi et al., 2020). R. intestinalis was inoculated onto DNase agar medium (Hopebio, China) for 72 h. A solution of 1 M HCl was then added to the plate and the colonies were observed after a few minutes to determine whether a clear area was present around them. The positive control bacteria were S. aureus ATCC 25923 and the negative control bacteria were E. coli ATCC 25922.
Approximately 50 μl of bacterial suspension (109 CFU/ml) was added to a gelatin biochemical detection tube (Hopebio, China) and incubated at 37°C for 48 h. The tube was placed in a refrigerator for 20 min, and tilted to observe whether the medium could solidify. If the medium is liquid, gelatin is hydrolyzed and gelatinase activity is positive. Bacillus cereus ATCC 11778 was used as a positive control and E. coli ATCC 25922 was used as a negative control.
Roseburia intestinalis was resuspended in the BHI medium and adjusted to a concentration of 107 CFU/ml. Next, 200 μl of the bacterial suspension was added to a 96-well plate, and the plate was incubated at 37°C for 72 h in an anaerobic jar. The wells were rinsed thrice with PBS. Then, 200 μl of methanol was added to each well, and the methanol was removed after fixing for 15 min. Subsequently, 200 μl of 1% crystal violet was added, and the wells were washed three times with PBS after 15 min. After air-drying for 30 min, 200 μl of 30% glacial acetic acid was added to the plate. The liquid in the well was transferred to a new plate and the OD550 was measured. A 30% glacial acetic acid solution was used as blank control. As controls, we selected two anaerobic (Bacteroides fragilis and Bacteroides vulgatus) and one aerobic probiotic (B. subtilis), in addition to three other pathogens (E. coli, S. aureus, S. typhimurium). Using the average OD (ODc) of the negative control group as a cut-off value, the bacteria were categorized into those without biofilm (OD ≤ ODc), weak (ODc < OD ≤ 2ODc), moderate (2 ODc < OD ≤ 4ODc) and strong (OD > 4ODc) biofilm formation (Cozzolino et al., 2020).
According to European Food Safety Authority, probiotics must be tested for resistance to the following 9 antibiotics: ampicillin, vancomycin, gentamicin, kanamycin, streptomycin, erythromycin, clindamycin, tetracycline, and chloramphenicol. The disk diffusion method (Rocha-Mendoza et al., 2020) was used to determine the susceptibility of R. intestinalis to 17 antibiotics. By measuring the zone of inhibition according to Clinical and Laboratory Standards Institute (CLSI) criteria (CLSI, 2016), we were able to determine the susceptibility of the bacteria to antibiotics.
Three different methods (Fan et al., 2019) were used to examine the ability of R. intestinalis to defend against pathogen invasion of epithelial cells. HT-29 cells infected with 1 × 107 CFU/ml of E. coli (multiplicity of infection [MOI] = 100) and incubated for 2 h served as a control group.
For the competition test, 1 × 108 CFU/ml of R. intestinalis and 1 × 107 CFU/ml of E. coli were added simultaneously to the cells and incubated for 2 h. For the displacement test, cells were first incubated with 1 × 107 CFU/ml of E. coli for 1 h, and then 1 × 108 CFU/ml of R. intestinalis was added and incubated for 1 h. For the exclusion test, HT-29 cells were first pre-incubated with 1 × 108 CFU/ml of R. intestinalis for 1 h, followed by the addition of 1 × 107 CFU/ml of E. coli for 1 h at 37°C. All groups were washed with PBS and the medium was changed to RPMI-1640 containing 1% penicillin/streptomycin/gentamicin (Biosharp, China) and incubated for 2 h at 37°C. To count intracellular E. coli, the cells were dissociated with 0.05% trypsin and treated with 0.1% Triton X-100 for 8 min. Results are expressed as the percentage decrease in E. coli invasion into cells in the experimental group compared with the control group.
Two human colorectal cancer cell lines, HT-29 and Caco-2, as well as the normal human colon epithelial cell line NCM460, were cultured at 37°C in a humidified environment containing 5% CO2 in RPMI-1640 medium containing 10% fetal bovine serum (Procell, China). After the cells had grown to 80–90% confluency, they were digested with 0.05% trypsin and seeded into appropriate cell culture plates according to the experimental procedure.
The effect of R. intestinalis on cell viability was evaluated using a cell counting kit (CCK-8 assay). R. intestinalis was co-cultured with the cells for 4 h at an MOI of 100. The bacteria-containing medium was replaced with medium supplemented with 1% penicillin/streptomycin/gentamicin, followed by incubation at 37°C for 2 h to eliminate all extracellular bacteria (Li et al., 2021). For further experiments, the medium was changed to normal RPMI-1640 medium and cell viability was evaluated at different time points. The control group consisted of cells treated with E. coli.
Roseburia intestinalis (108 CFU/ml) was co-cultured with the NCM460 cells at 37°C for 4 h. Lactate dehydrogenase (LDH) released by the damaged cells was measured using an LDH assay kit (Beyotime, China). The results were expressed as a percentage of LDH activity in each group relative to the positive control reagent provided by the kit (Hussein et al., 2020).
HT-29 cells were co-cultured with bacteria (108 CFU/ml) for 4 h, followed by a change to media containing 1% penicillin/streptomycin/gentamicin, and cultured for two additional hours at 37°C. The tests were conducted according to the manufacturer’s instructions (Beyotime, China), and the relative fluorescence values (RFU) were compared between the experimental and control groups.
HT-29 cells were cultured in 6-well plates. As soon as the cells reached 80% confluence, 100 μl of R. intestinalis or E. coli (108 CFU/ml) was added to the plate, and 30 μM of DNA synthesis inhibitor (hydroxyurea, Beyotime) served as the positive control. An EdU Cell Proliferation Kit (Beyotime) was used to detect cell proliferation after 24 h. The FUJI-ImageJ software was used to quantify the percentage of EdU-positive cells (Lu et al., 2019).
The bacteria were centrifuged at 12000 rpm for 5 min and washed thrice with PBS. An equal volume of 1× CFDA-SE staining solution (Beyotime) was added to the bacterial suspension, and the mixture was incubated for 30 min at 37°C in the dark. The samples were then washed thrice with PBS to remove the unbound dye. Fluorescence was observed using a fluorescence microscope at an excitation wavelength of 488 nm.
To investigate the characteristics of R. intestinalis colonization of the GIT, we used a previously described method (Zhao et al., 2022). R. intestinalis was collected during the logarithmic growth phase, stained with CFDA-SE, and resuspended in PBS at a concentration of 1 × 1010 CFU/ml. The mice were then gavaged with 200 μl of bacterial solution, whereas the control group was administered PBS. Mice were euthanized 2 to 72 h after gavage, and 1 cm long sections of the duodenum, jejunum, ileum, cecum, colon, and rectum were removed. After thoroughly rinsing the intestinal tube with PBS, the particulate matter was filtered using a 40 μm cell strainer. Flow cytometry (Cytek AthenaTM) was used to analyze the ratio of CFDA-SE labelled R. intestinalis in the intestine. Mice gavaged with non-fluorescently labelled R. intestinalis were used as blank controls.
For frozen sections, mice were euthanized 12 h after administration of R. intestinalis, cecal and colonic specimens were collected and washed in sterile PBS, embedded with Optimal cutting temperature (OCT) reagent (Sakura, Japan), and frozen sections were obtained immediately (Leica CM1950).
To analyze the effect of R. intestinalis on the structure of the intestinal flora, mice were gavaged with R. intestinalis at a concentration of 1 × 109 CFU/ml for 14 days. On days 7 and 14, mouse feces were harvested for 16S rRNA sequencing.
A MagPure Stool DNA kit (Magen, China) was used to extract DNA from the microbial communities. DNA was quantified using a Qubit Fluorometer with the Qubit dsDNA Kit (Invitrogen, United States), and quality was checked using a 1% agarose gel. Polymerase chain reaction (PCR) primers 341F (5’-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5’-GGACTACHVGGGTWTCTAAT-3′) were used to amplify the variable regions V3–V4 of the bacterial 16S rRNA genes. The primers were tagged with Illumina adapter, pad, and linker sequences. PCR enrichment was performed using a 50 μl reaction containing 30 ng template, fusion PCR primer, and master mix. The PCR cycling conditions were 94°C for 3 min, 30 cycles of 94°C for 30 s, 56°C for 45 s, 72°C for 45 s, and 10 min at 72°C for extension. PCR products were purified using AmpureXP beads and eluted with elution buffer. The libraries were qualified using an Agilent 2,100 bioanalyzer (Agilent, United States). The validated libraries were sequenced on an Illumina MiSeq platform (BGI, Shenzhen, China) following the standard Illumina pipeline and 2 × 300 bp paired-end reads were generated.
Oral toxicity studies were performed using male C57BL/6 J mice (6–8 weeks) and all animal experiments were approved by the Central South University Animal Ethics Committee (XMSB-2022-0198). The animals were housed under standard conditions with alternating periods of light and dark (12 h each) at a temperature of 25 ± 2°C and with free access to food and water. The animals were acclimated for 7 days before the experiment. R. intestinalis was cultured in an anaerobic environment at 37°C for 24 h in the BHI medium. Bacteria were centrifuged for 10 min at 4000 rpm, rinsed, and resuspended in PBS.
The acute oral toxicity test was conducted according to the method described by Anantha et al. (Metlakunta and Soman, 2020) and the Organisation for Economic Co-operation and Development (OECD) guideline No.423 with certain modifications. This study was conducted in a stepwise manner. As a first step, three mice (group A1) were gavaged with R. intestinalis at a dose of 1.9 × 109 CFU/kg. It represents 133–1,330 times the empirical dose level of oral probiotics in humans (i.e., 108–109 CFU or 1.43 × 106–1.43 × 107 CFU/kg/day in a 70 kg individual; Steppe et al., 2014). In the following 48 h, there were no mortality, morbidity, or abnormal clinical signs. To confirm this, the same dosage was administered to another group (A2), while a control group of mice (A0) was administered PBS. The animals were monitored for 14 consecutive days for clinical manifestations, such as changes in the skin and fur, mucous membranes, respiration, and behavioral changes. The animals were euthanized at the end of the observation period, and the gross appearance of vital organs, organ weights, and histopathology were assessed. Blood was collected from the animals for biochemical and hematological analyses.
According to OECD No.407, this part of the study evaluated the toxicity of R. intestinalis after repeated administration for 28 days to establish dose–response relationships and determine the NOAEL. Each group consisted of six mice, and the control group (S0) was administered PBS via gavage. The experimental groups were divided into three dose levels corresponding to 66–657 times (S1), 77–769 times (S2), and 92–923 times (S3) the empirical dose of oral probiotics in humans. The mice were monitored daily for changes in clinical signs, mortality, fur and skin, respiration, behavioral patterns, body weight, food intake, and water consumption. On the 29th day, the mice were euthanized and blood samples were collected for analysis of hematology and biochemical indicators. The gross appearance of vital organs, organ weights, and histopathology were assessed.
Statistical analyses were performed using SPSS version 25.0. To ensure homogeneity of variance and normality of the data, the Kolmogorov–Smirnov test was used. The values are presented as the mean ± standard deviation for normally distributed data or as the median and interquartile range for data that were not normally distributed. When comparing two groups, p values were derived from the Student’s t-test for normally distributed data, and the Mann–Whitney was used for data with other distributions. For multi-group comparisons, p values were determined using one-way ANOVA. We considered p < 0.05 statistically significant in all comparisons.
The RAST annotation results indicated that the genome contained 4,340 coding sequences and 81 RNA genes (Supplementary Figure S2). In total 252 subsystems were functionally annotated. Carbohydrates accounted for the largest number of genes (200), followed by amino acids and derivatives (192). Pathosystems Resource Integration Center (PATRIC; Davis et al., 2020) was used to generate a circular genome map (Figure 2).
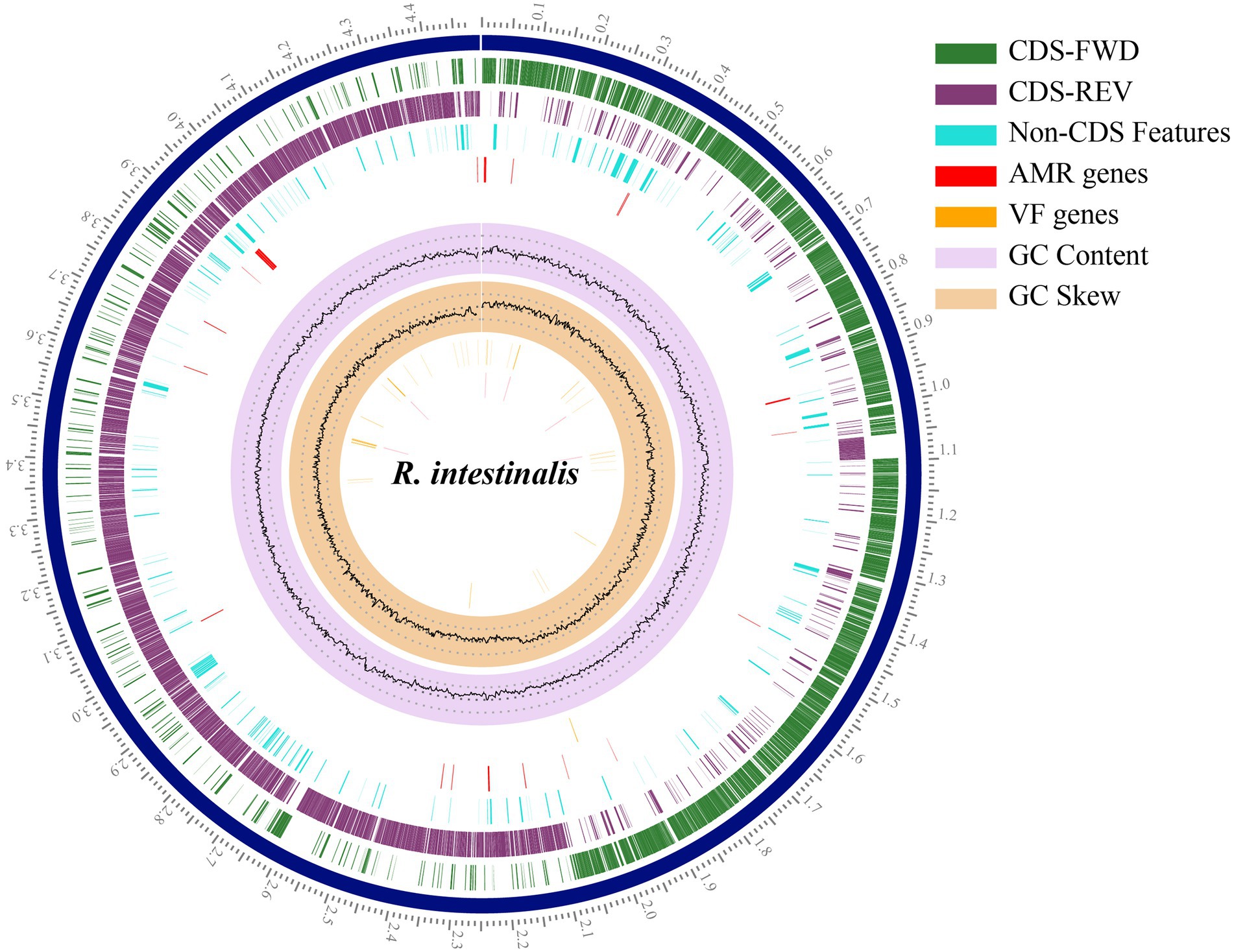
Figure 2. Circular genome map of Roseburia intestinalis. The outermost scales indicate the base location in Mbp. From the innermost circles: GC skew, GC content, virulence factor genes, AMR genes, Non-CDS features. The last two circles illustrate the coding sequence, the green circle represents the forward strand, and the purple circle represents the backward strand.
In Roseburia intestinalis, we detected three CRISPR sequences and two Cas genes. The CRISPR sequences were located at 1020388–1020441 bp, 1,222,931–1,223,608 bp, and 1,225,355–1,226,103 bp, while the Cas gene sites were located at 1015665–1019051 bp and 1,216,581–1,217,327 bp. The genome contained no genes involved in biogenic amine biosynthesis other than spermidine synthase (speE) and carboxynorspermidine decarboxylase (nspC). The genome contains essential amino acid synthesis genes (tryptophan, methionine, threonine, arginine, and cysteine), as well as vitamin synthesis genes (i.e., Thiamine, Riboflavin, Folate, Pantothenate, and Biotin; Table 1).
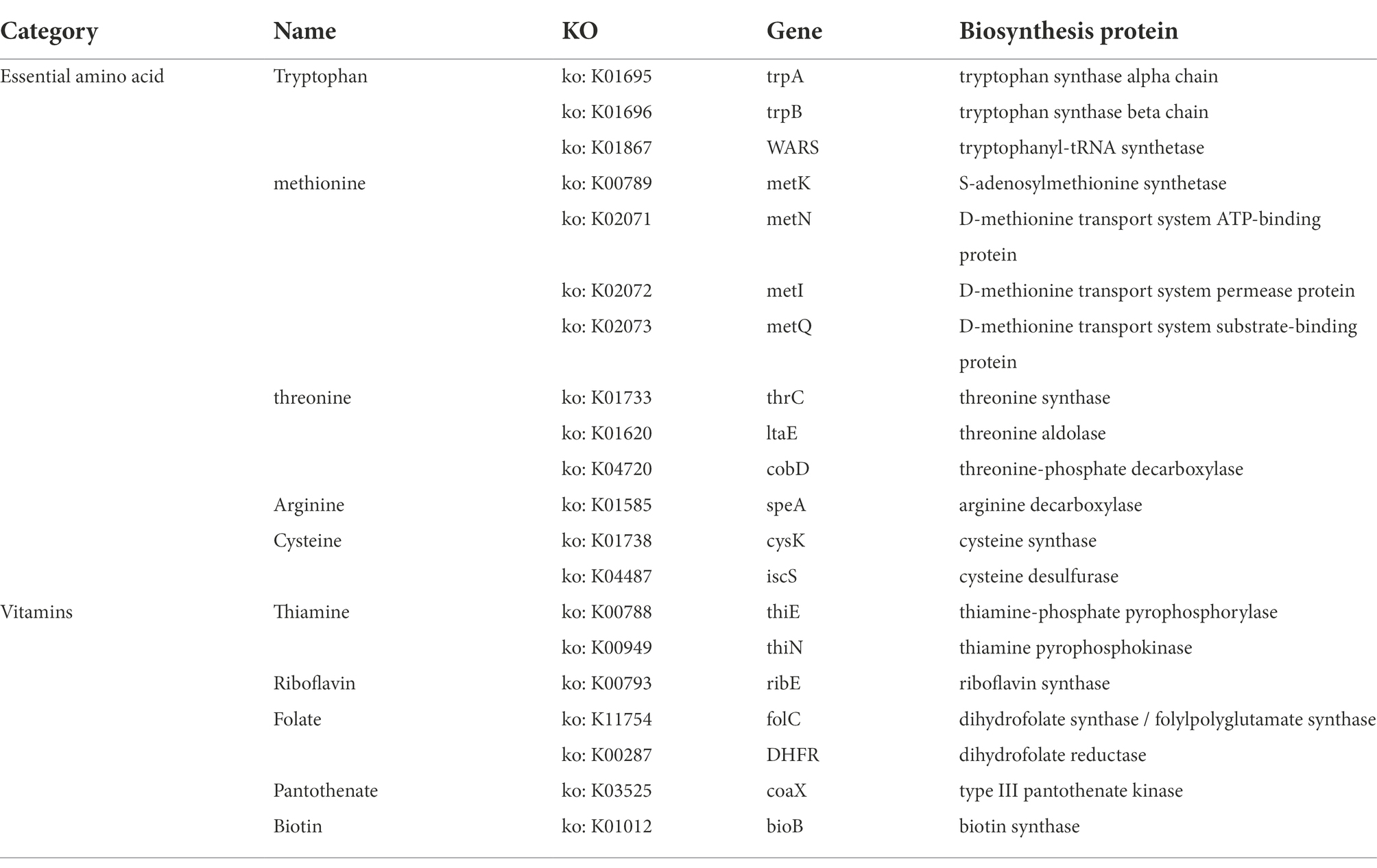
Table 1. Associated biosynthetic genes detected in R. intestinalis genome about essential amino acids or vitamins.
PHASTER identified seven prophages within the genome, of which three are incomplete and four are questionable. The details of the prophages are provided in Supplementary Table S2. Using the BAGEL4 webserver, we detected that the bacterium harbored a bacteriocin cluster (Figure 3C) of Zoocin A class within contig 93.3 (3723994–3,744,393 bp). AOI consisted of RNA pseudouridine synthase, a transcriptional regulatory protein (LanR), a putative lantibiotic resistance two-component sensor kinase precursor (LanK), and multiple open reading frames (ORFs). Using gutSMASH, we discovered that the genome contained six metabolism-related gene clusters (Table 2). Four of these clusters are associated with short-chain fatty acid metabolism, including butyric acid synthesis.
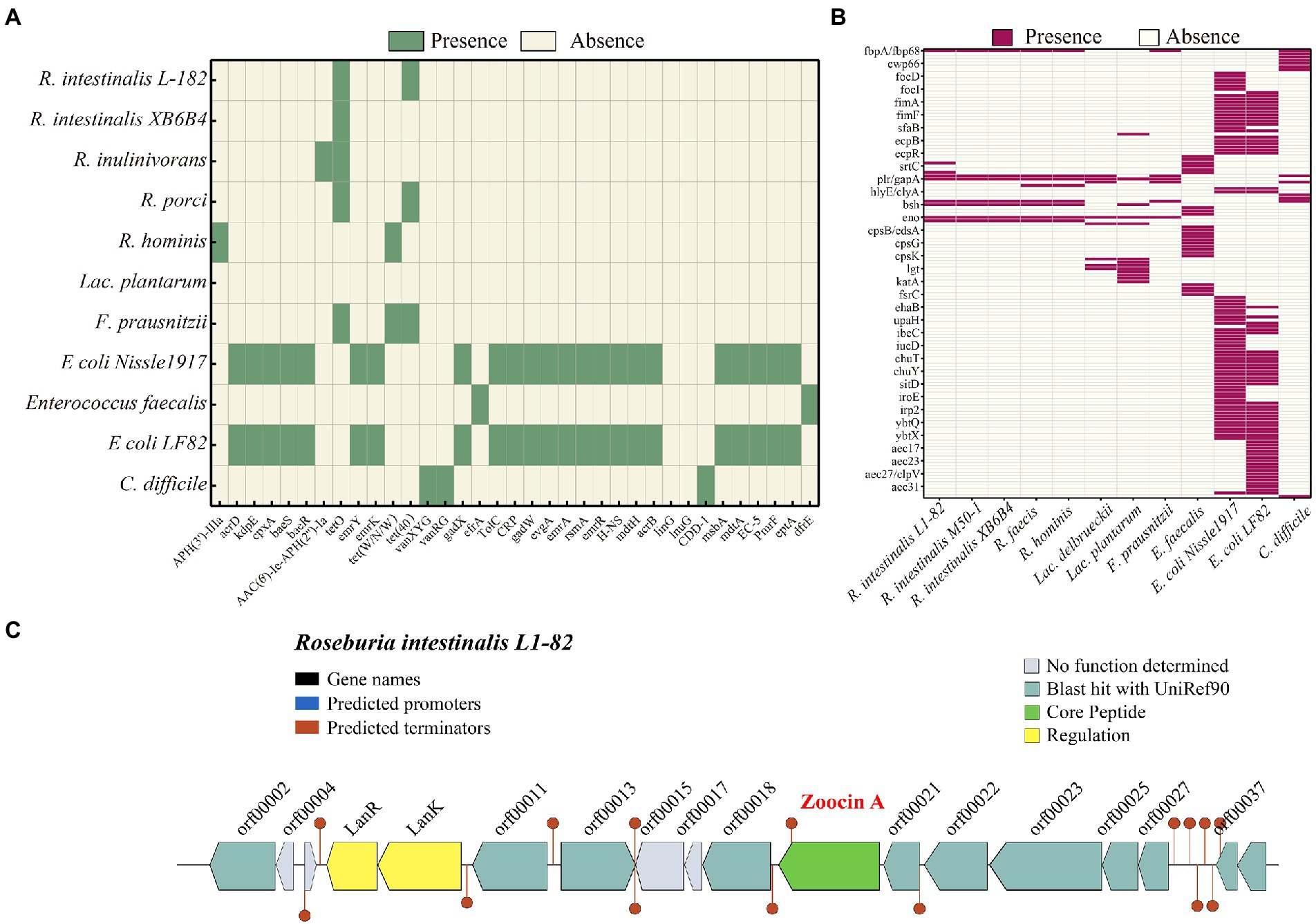
Figure 3. Heatmap showing the clustering of the compared R. intestinalis genomes based on the presence of antibiotic resistance genes (A) and virulence genes (B). (C) Organization of bacteriocin cluster genes. orf00002, RNA pseudouridine synthase; orf00011, S-adenosylmethionine synthase; orf00013, Cyclic di-GMP phosphodiesterase; orf00018, UDP-N-acetylglucosamine 1-carboxyvinyltransferase; orf00021, Shikimate kinase; orf00022, Foldase protein PrsA; orf00023, Transcription-repair-coupling factor; orf00025, Peptidyl-tRNA hydrolase; orf00027, Bifunctional protein GlmU; orf00037, Putative septation protein SpoVG; orf00039, Glucose-1-phosphate adenylyltransferase.
The CAZy database identified 222 carbohydrate-active enzyme modules in the genome. A total of 136 glycoside hydrolase families, 48 glycosyltransferases, 14 carbohydrate esterase families, and 24 carbohydrate-binding module families were identified. Using IslandViewer, we combined two prediction algorithms, islandPath-DIMOB and SIGI-HMM, and identified 41 GIs (Supplementary Figure S3). In the predicted GIs, no annotated virulence, antibiotic resistance, or pathogenicity genes were identified.
The genome contains tetO and Tet(40), which are associated with tetracycline resistance, whereas other Roseburia genera and other probiotic/pathogenic bacteria contain more antibiotic resistance genes (Figure 3A). According to VFDB, the R. intestinalis genome contains genes related to adhesion (fbpA/fbp68, ebpC, efaA, plr/gapA), and lipid and fatty acid metabolism (panD; Figure 3B). Compared to other probiotics (all belonging to Firmicutes), R. intestinalis contains more genes that encode cell adhesion-related proteins and genes related to gastrointestinal survival (Supplementary Figure S4). The genome contains seven genes related to antioxidant activity, suggesting a strong ability to survive in the host environment. Furthermore, we compared its genome with the genomes of six probiotics and found that F. prausnitzii, a probiotic used in the treatment of IBD (Machiels et al., 2014), possessed the highest degree of similarity (Supplementary Figure S5).
R. intestinalis was tested for its resistance to 17 antibiotics (Table 3). The bacterium was sensitive to most antibiotics, had intermediate resistance to streptomycin and kanamycin, and was only resistant to amikacin.
As shown in Figure 4, R. intestinalis exhibited no gelatinase, (Figure 4A) DNase (Figure 4E), or hemolytic activity. As a positive control, S. aureus ATCC 25923 exhibited β-hemolysis (Figure 4F). R. intestinalis survived in an environment with a pH of 4 (Figure 4B). The proliferation of bacteria was inhibited in the presence of 0.3% bile salt, whereas bacteria survived in the artificial intestinal fluid (Figures 4C,K). R. intestinalis was able to adhere effectively to HT-29 and Caco-2 cells (Figures 4H,I) and exhibited a hydrophobicity of 12.25 ± 1.01% in n-hexadecane. At 24 h, R. intestinalis had a maximum auto-aggregation ability of 80% (Figure 4L), and the co-aggregation experiment (Figure 4J) showed a strong co-aggregation ability (69–80%). Competition, displacement, and exclusion experiments showed that R. intestinalis inhibited the internalization of E. coli into epithelial cells (Figure 4D). R. intestinalis had a higher biofilm-forming ability than several probiotics and pathogens (Figure 4G). This may allow it to gain a competitive growth advantage and enhance its ability to adhere to the intestinal epithelium.
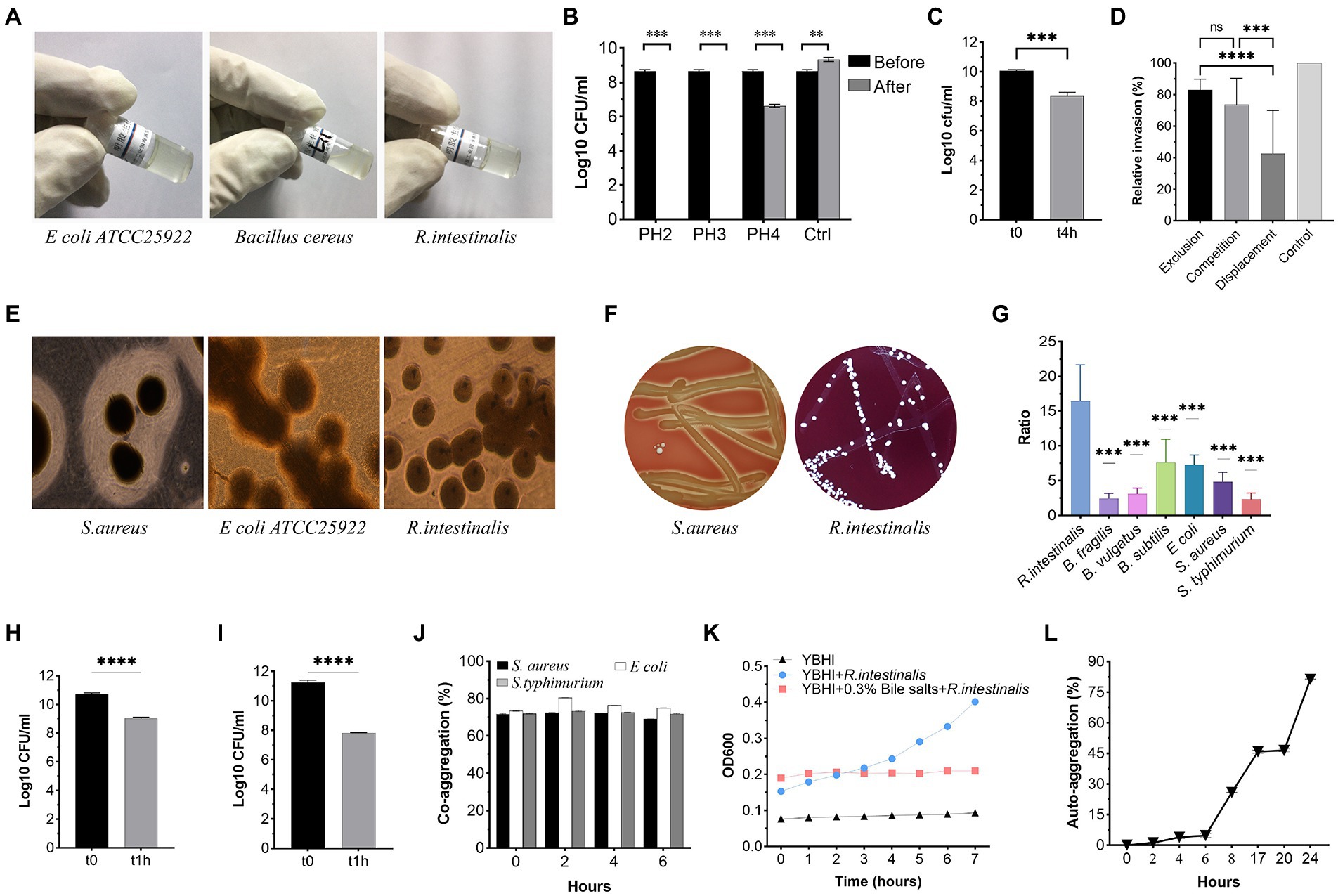
Figure 4. Phenotypic safety assessment of probiotic Characteristics. (A) Gelatinase activity of R. intestinalis compared to Bacillus cereus ATCC 14579 (positive control) and E coli ATCC 25922 (negative control). (B) Acid tolerance. (C,K) Bile salt and artificial intestinal fluid tolerance. (D) Inhibition of internalization of pathogens. (E) DNase activity. A clear zone around the colony represents DNase activity. (F) Hemolytic activities. (G) Biofilm formation. The y-axis represents the ratio of the OD550 of the bacteria to the ODc. (H,I) Adhesion of R. intestinalis on Caco-2 and HT-29 cells. (J,L) Co-aggregation and auto-aggregation abilities. * indicates p < 0.05, ns indicates no significance.
After treatment with R. intestinalis, there was no statistically significant difference in cell viability between the experimental and control groups (p > 0.05); however, cell viability was significantly reduced after treatment with E. coli (Figures 5A,B). As shown in Figure 5C, the RFU of the experimental group was not significantly different from that of the control group, whereas that of the E. coli treatment group was significantly different (p < 0.05). Compared to the positive control, there was no significant difference in LDH activity between the R. intestinalis treatment and control groups. This indicates that this probiotic bacterium did not cause notable cell damage (Figure 5F). The proportion of EdU-positive cells in the treatment group did not differ significantly from that in the control group but differed significantly from that in the positive control group (p < 0.05). This indicated that R. intestinalis treatment did not affect DNA replication during cellular proliferation (Figures 5D,E).
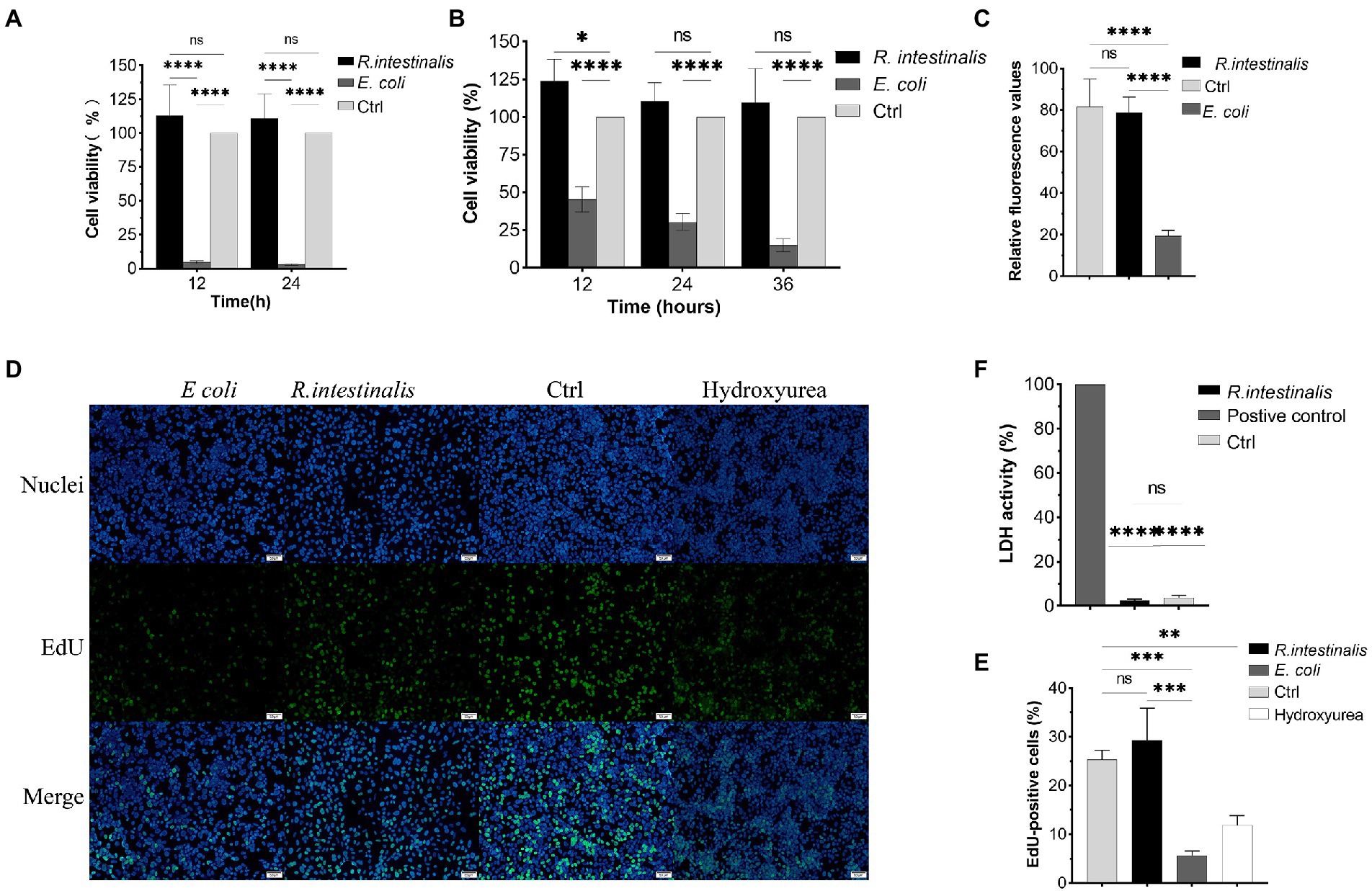
Figure 5. Cytotoxic effects of R. intestinalis. (A,B) Cytotoxic effects of R. intestinalis on HT-29 and NCM460 cells after 12–36 h of treatment. (C) Calcein-PI staining in live and dead cell detection test. (D,E) EdU cellular proliferation test. The hydroxyurea treatment group served as the positive control. (F) LDH activity test. * indicates p < 0.05, ns indicates no significance.
There was no statistically significant difference among the groups in any of the α-diversity indices (Figures 6A–C), indicating that oral administration of R. intestinalis did not modify gut microbiota diversity and abundance. Non-metric multidimensional scaling (NMDS) analysis using a weighted UniFrac analysis revealed distinct differences in microbiota structure (Figures 6D–H). We found that the highest relative abundance of R. intestinalis occurred 7 days after oral administration (Figures 7A-C). Species with core effects were analyzed using the LEfSe method. The core strains after 7 days of administration were Firmicutes, Clostridiales, Clostridia, Lachnospiraceae among others. After 14 days, the core species were Barnesiella and Porphyromonadaceae (Figure 7D). The PICRUSt2 tool based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to predict pathway enrichment, and we observed differences in pathway enrichment after the administration of R. intestinalis (Supplementary Figure S6).
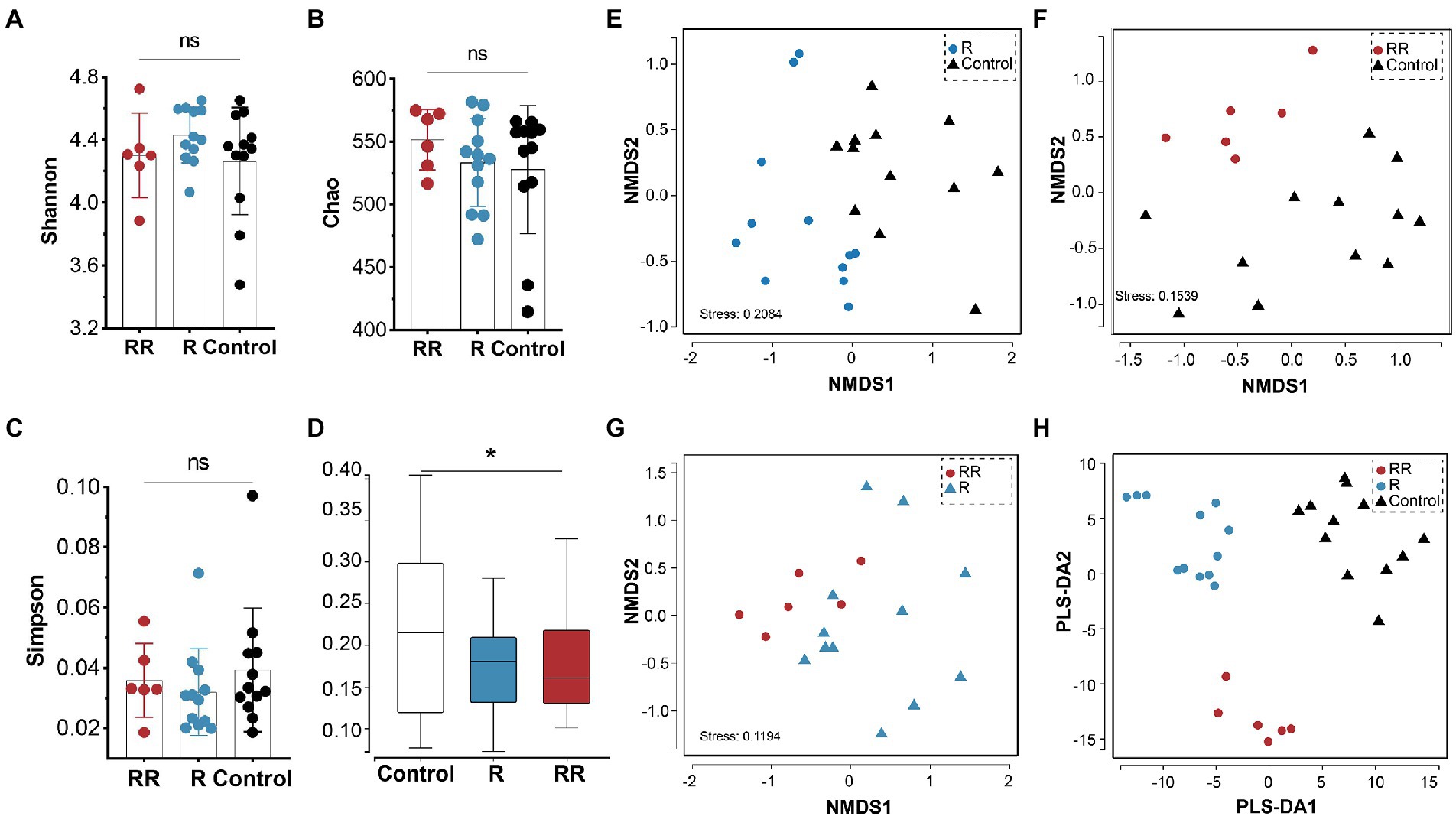
Figure 6. Cecal microbiota α-diversity and β-diversity following administration of R. intestinalis for 7 days (R group) or 14 days (RR group). (A) Shannon’s index. (B) Chao’s index. (C) Simpson’s index. (D) Weighted UniFrac distance of the cecal microbiota. (E–G) NMDS analysis among groups. (H) Partial least squares discrimination analysis (PLS-DA) analysis. * indicates p < 0.05, ns indicates no significance.
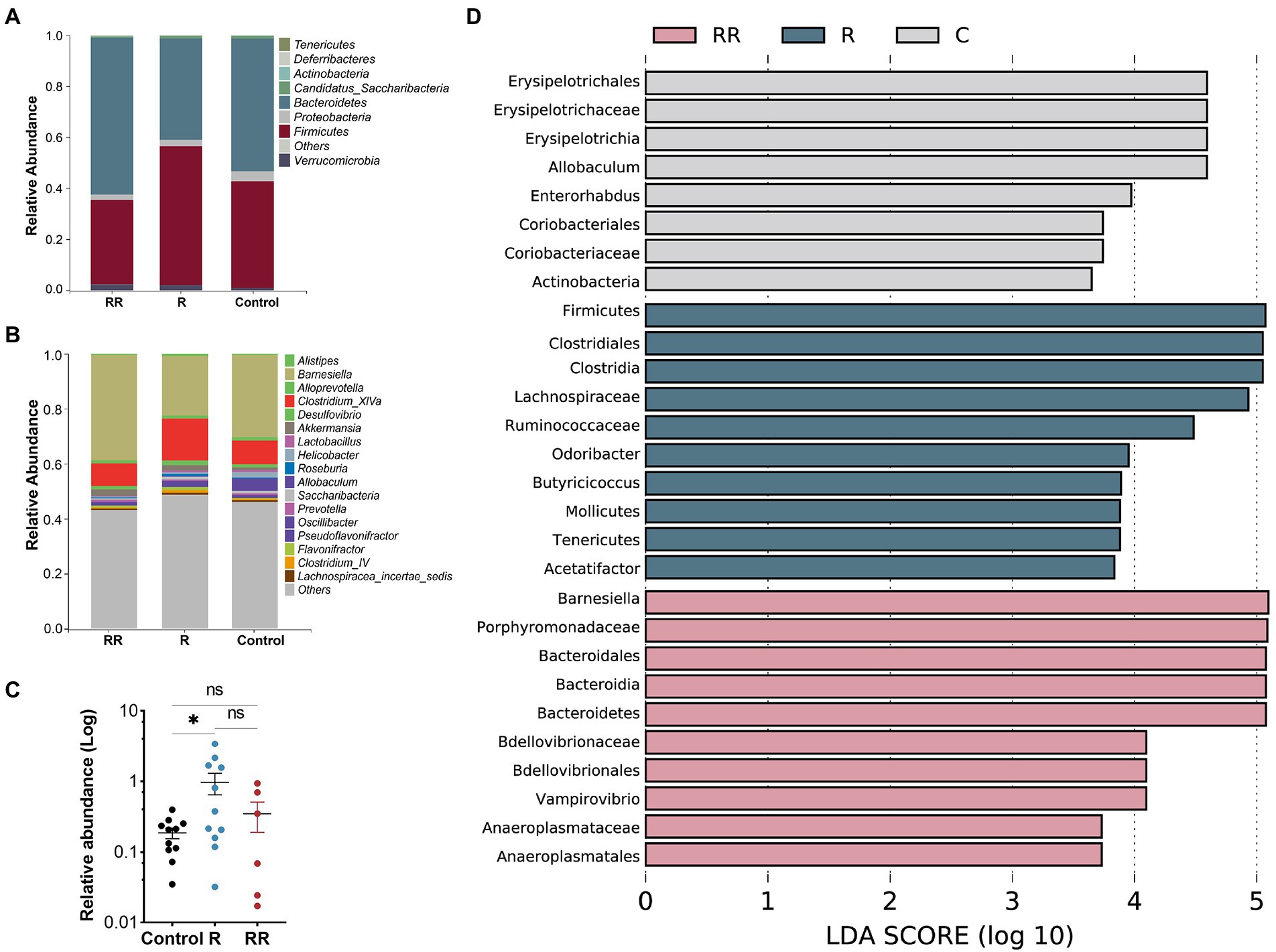
Figure 7. Classification of microflora at the phylum (A), and genus (B) levels. (C) Relative abundance of Roseburia intestinalis among different groups. R: 7 days group; RR: 14 days group. (D) LEfSe analysis between different groups, displaying only genera with absolute values of linear discriminant analysis (LDA) scores greater than 2.0. * indicates p < 0.05, ns indicates no significance.
Two hours after oral administration, fluorescently labelled bacteria (Figure 8A) were detected in all intestinal segments, with the highest concentration in the colon. After 48 h, the content of R. intestinalis in the cecum exceeded that in the colon. After 72 h, the bacterium primarily colonized the cecum and colon, whereas its distribution in the duodenum, jejunum, ileum, and rectum was scarce (Figure 8B; Supplementary Figure S7). The results of the frozen section demonstrated that the spatial colonization sites of R. intestinalis were primarily located in the cecum and colonic mucus layers (Figure 8C).
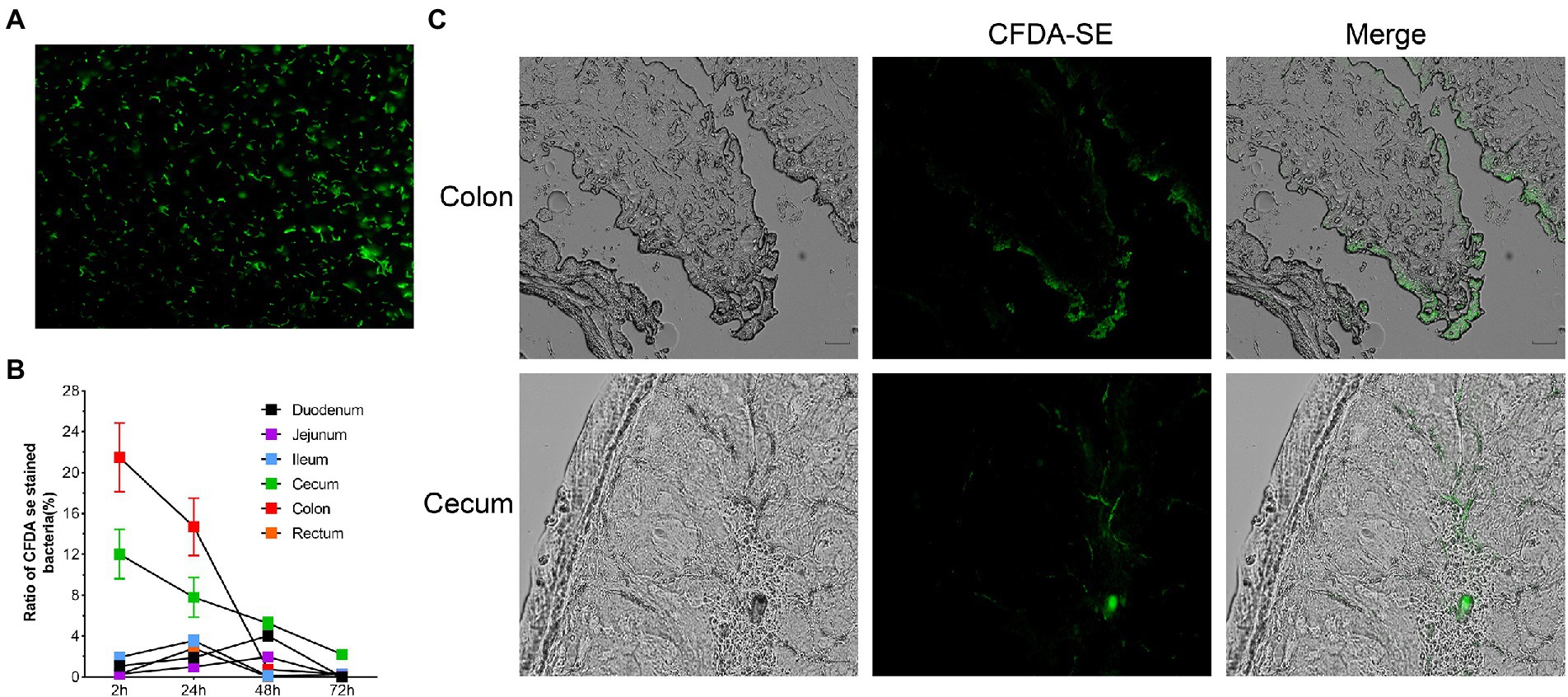
Figure 8. Using flow cytometry and frozen sections to characterize the colonization of Roseburia intestinalis in the GIT. (A) CFDA-SE labelled R. intestinalis investigated under fluorescence microscopy. (B) R. intestinalis fluorescence at different time points and in different areas of the intestine. (C) Frozen sections were used to observe the distribution of R. intestinalis in the colon and cecum.
During observation period, no mortality, morbidity, or abnormal clinical manifestations were observed. The average daily food and water intake is presented in Supplementary Tables S3, S4. Body weight, organ weight, and colon length were not significantly different between the experimental and control groups (Figures 9A–D). The organs exhibited no obvious pathological damage such as necrosis, inflammation, or proliferation (Figure 9E). All hematological parameters in the mice were comparable between the groups (Supplementary Table S5). There were no significant differences between the experimental and control groups in terms of alanine aminotransferase, aspartate aminotransferase, γ-glutamyltransferase, total bile acid, and creatinine (Table 4). Notably, serum uric acid levels in the experimental group were significantly lower than those in the control group, suggesting that R. intestinalis may affect purine and uric acid metabolism. Based on these results, we determined that oral R. intestinalis has an LD50 exceeding 1.9 × 109 CFU/kg.
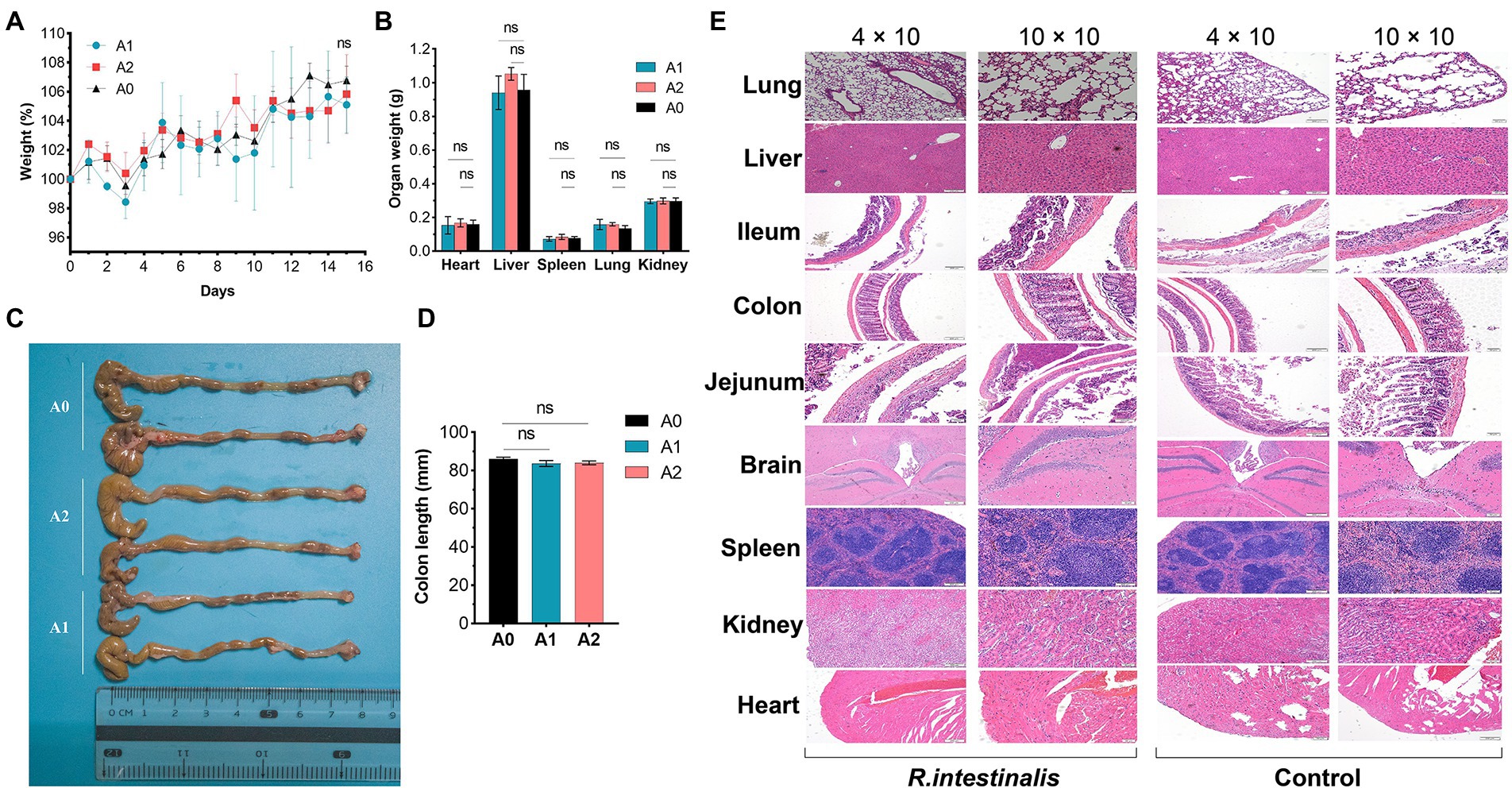
Figure 9. Acute toxicity study. (A) Body weight. (B) Weight of vital organs (heart, lung, liver, spleen, kidney). (C,D) Colon length. (E) Hematoxylin–eosin staining pathological examination results of vital organs. * indicates p < 0.05, ns indicates no significance.
No deaths or dosing-related adverse reactions were observed at any dose level during the 28-day experimental period. The average daily food and water intake is presented in Supplementary Tables S6, S7. Body weight, organ weight, and colon length did not differ significantly between groups (Figures 10A–D). No obvious pathological changes, such as necrosis, inflammation, or proliferation, were observed in the organs (Figure 10E). The hematological parameters did not differ significantly among the groups (Supplementary Table S8). In terms of alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), γ-glutamyltransferase (γ-GT), total bile acid (TBA), and urea levels (Table 5), no significant differences were observed between the groups. Notably, the serum uric acid level in the experimental group was significantly lower than that in the control group, in agreement with the results of the acute oral toxicity test. Based on these data, the NOAEL for the oral administration of R. intestinalis was estimated at 1.32 × 109 CFU/kg/day for 28 days.
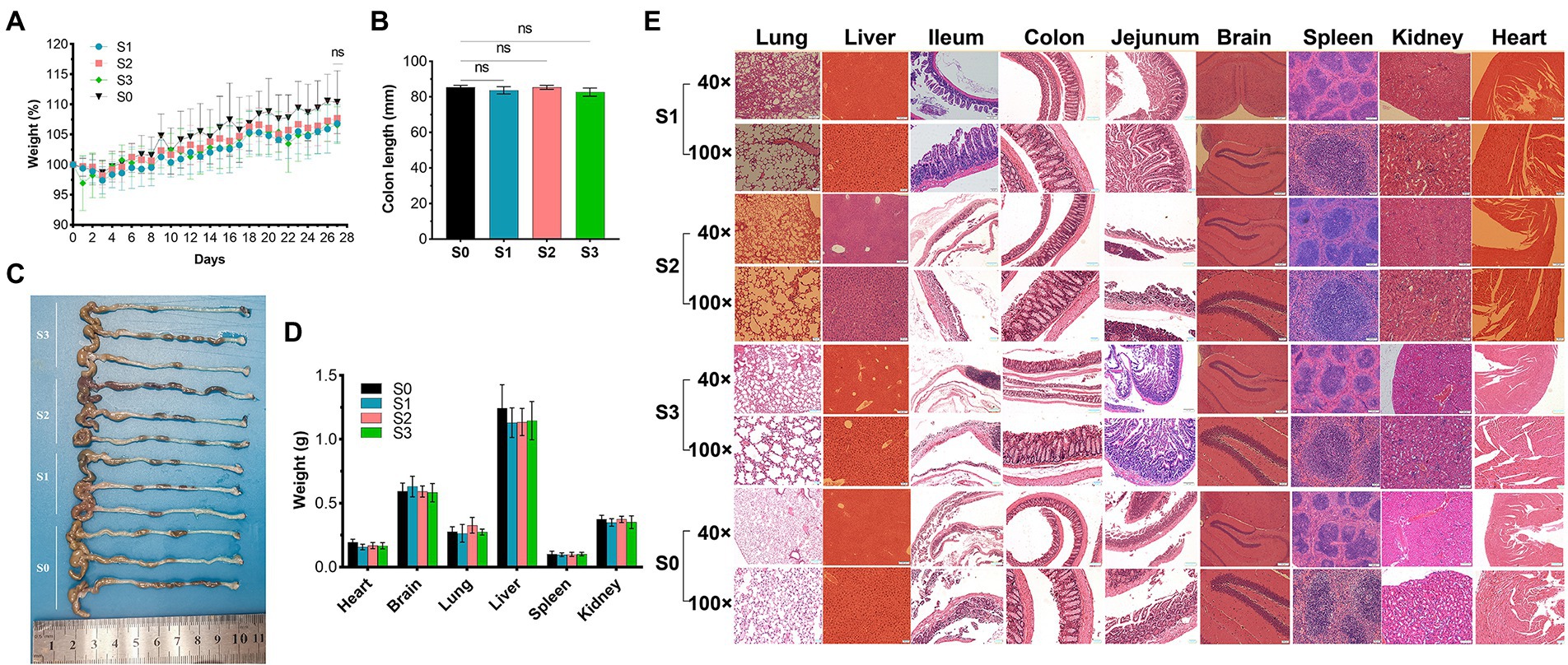
Figure 10. 28-Day repeated dose study. (A) Body weight. (B,C) Colon length. (D) Weight of vital organs (heart, lung, brain, liver, spleen, kidney). (E) Hematoxylin–eosin staining pathological examination of organs. * indicates p < 0.05, ns indicates no significance.
Genomic, phenotypic, and oral safety studies should be conducted on potential probiotic strains to assess their safety and feasibility for industrial use.
A few virulence factors (VFs) have also been found in the genomes of nonpathogenic bacteria (Niu et al., 2013). Certain VFs are involved in host–microbe interactions, while others are involved in cell adhesion and host defense (Li et al., 2018). VF genes found in the R. intestinalis genome were related to adhesion (fbpA/fbp68, ebpC, efaA, plr/gapA) and lipid and fatty acid metabolism (panD). FbpA may play a role in the prevention of pathogenic colonization by inhibiting biofilm formation (Wang et al., 2017). EfaA is associated with adhesion of probiotics bacteria to both biotic and abiotic surfaces (Choeisoongnern et al., 2021). GroEL may inhibit colitis by inhibiting pro-inflammatory M1 macrophages and promoting the secretion of anti-inflammatory cytokines (Dias et al., 2021). The genome contains two tetracycline resistance genes, but a subsequent antibiotic susceptibility test showed that R. intestinalis was susceptible to tetracycline. Gene modifications or pseudogenes may have causes this seemingly paradoxical result. Additionally, the expression of these genes may usually be low or they are induced only under specific circumstances, such as stimuli or signals from the environment (Li et al., 2018). Therefore, the presence of these resistance genes does not hinder the safety of R. intestinalis as a probiotic.
The CRISPR-Cas system is considered an defense mechanism against mobile genetic elements (Holý et al., 2020). This system can protect against re-invasion by capturing and integrating foreign nucleic acid fragments from the initial invasion. CRISPR-Cas genes have also been found in some probiotics, such as Bifidobacterium (Pan et al., 2020) and Lactobacillus reuteri (Alayande et al., 2020). Amino acids can be converted to biogenic amines via microbial decarboxylation. Histamine and tyramine (Zhu et al., 2020) are prevalent in fermented foods, and the excessive intake of biogenic amines can be harmful. The R. intestinalis genome contained genes involved in spermidine biosynthesis. Spermidine improves gut barrier integrity (Ma et al., 2020b), reduces obesity and cancer mortality, and provides anti-inflammatory and stem cell senescence protection (Madeo et al., 2018). Probiotics, including Akkermansia muciniphila, may have anti-aging and anti-obesity effects, that are linked to increased levels of spermidine in the host (Grajeda-Iglesias et al., 2021).
Microbes in the gut synthesize several vitamins and essential amino acids that contribute to the host amino acid homeostasis (Gill et al., 2006; Lin et al., 2017). R. intestinalis contains genes for the biosynthesis of five essential amino acids and five vitamins. Prophages are generally considered beneficial to their bacterial hosts, especially in the gastrointestinal environment (Manrique et al., 2017). The presence of prophages not only increases genetic variability but may also allow bacteria to cope with adverse environmental conditions (Casjens, 2003). Several probiotics contain prophages, including Lactococcus, Bifidobacterium, Lactobacillus (Pei et al., 2021), and L. rhamnosus (Brandt et al., 2001). Additionally, studies have suggested that prophages are present in more than 92% of Lactobacillus genomes (Sun et al., 2015). Jeffrey et al. (Cornuault et al., 2020) identified two active prophages in the genome of R. intestinalis and demonstrated that these prophages can influence short-term changes in the gut microbiota composition.
Bacteriocins are products synthesized by bacterial ribosomes that possess bacteriostatic properties (Deraz et al., 2005). Increasing antibiotic resistance has prompted researchers to focus on bacteriocins, since these may serve as alternatives to antibiotics (Cotter et al., 2013). R. intestinalis can produce Zoocin A, a 30 kDa D-alanyl-L-alanine endopeptidase bacteriocin that inhibits streptococcal growth by binding and cleaving bacterial peptidoglycan (Gargis et al., 2009). L. plantarum (Seddik et al., 2017) and Enterococcus spp. (Ben Braïek and Smaoui, 2019) are two probiotics microbes known to produce bacteriocins. R. intestinalis contains four short-chain fatty acid (SCFA) synthesis gene clusters, including a butyrate synthesis gene cluster. Butyrate has several beneficial effects, including as an energy source for colonic epithelial cells and exerting anti-inflammatory properties by inhibiting nuclear factor (NF)-κB (Segain et al., 2000). Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor growth by modulating the Wnt pathway and gut microbiota (Chen et al., 2020). Moreover, VSL#3 modulates the gut microbiota-SCFA-hormone axis to combat obesity and diabetes (Yadav et al., 2013).
As the human genome encodes only approximately 17 carbohydrate-degrading enzymes, those produced by gut bacteria play a critical role in carbohydrate degradation (Bhattacharya et al., 2015). R. intestinalis contains 222 carbohydrate metabolism-related genes that likely play important roles in host carbohydrate metabolism. Maria et al. (Leth et al., 2020) demonstrated that R. intestinalis expresses RiCBM86, an enzyme that binds and absorbs xylan in the gut, reduces inflammation, and improves atherosclerosis (Kasahara et al., 2018). This bacterium is also a major degrader of β-mannans (La Rosa et al., 2019) and can produce xylanase to degrade xylan (Mirande et al., 2010). GIs are an important aspect of horizontal gene transfer and can transmit antibiotic resistance and virulence genes (Juhas et al., 2009). R. intestinalis contains 41 GIs, none of which contain virulence, antibiotic resistance, or pathogenicity genes.
Although the gastric juice pH is 1–3, it increases to 6 after food ingestion (De Angelis et al., 2006). The pH values of the duodenum, terminal ileum, cecum, and rectum are 6.0, 7.4, 5.7, and 6.7, respectively (Fallingborg, 1999). Several methods have been reported for enhancing the tolerance of probiotics to bile salts and acids. Gou et al. (2021) used soybean lecithin and whey protein concentrate to treat Lacticaseibacillus paracasei and successfully increased its tolerance to bile salts and acids. Evidence suggests that 5% lactose can enhance the bile salt tolerance of L. bulgaricus (Mena and Aryana, 2018), whereas soy lecithin (Hu et al., 2015) can enhance the bile salt resistance of L. plantarum and whey protein (Vargas et al., 2015) can enhance the acid and bile salt tolerance of Streptococcus thermophilus. Biofilm formation by probiotics can inhibit pathogens from colonizing the mucosa (Deng et al., 2020). R. intestinalis forms a higher percentage of biofilms than many common probiotics and pathogenic bacteria, further highlighting its probiotic properties.
Following the administration of R. intestinalis, the serum uric acid level decreased significantly, suggesting that the bacterium may influence the host’s purine and uric acid metabolism. Our in vitro study indicates that R. intestinalis can degrade 60% of the uric acid in the surrounding environment within 24 h (data not shown), although further investigation is required to determine the underlying mechanism.
R. intestinalis did not adversely affect the abundance or variety of the gut flora. After 14 days of continuous administration, the relative abundance of R. intestinalis did not increase proportionally. However, the structure of the flora changed, and the predominant flora consisted of Barnesiella and porphyromonadaceae. The relative abundance of R. intestinalis was determined by 16S rRNA sequencing of stool samples. Owing to their ease of collection, feces are often used to study the gut microbiota. Nonetheless, this method has a limited ability to detect the mucosal-associated microbiota. Mucosal-associated microbiota and luminal microbiota have only a partial association (Zmora et al., 2018). Due to their biofilm-like structure, mucosal-associated microbiota promote beneficial functions in intestinal epithelial cells (Sonnenburg et al., 2004), and conventional sampling methods (feces or intestinal tissue) may underestimate their levels (Donaldson et al., 2016). R. intestinalis adheres to the intestinal mucosa and colonizes the cecum and colonic mucosa. Flow cytometry results showed that there was a low abundance of R. intestinalis in the intestinal contents (Figure 8B). Similarly, some probiotics, such as B. lactis, showed a decreasing trend in its levels after prolonged administration (Ma et al., 2020a). Therefore, we speculated that the variation in the gut flora is stratified rather than continuous, and a relatively balanced state of host-microbe symbiosis exists in the gut microbiota (Arumugam et al., 2011). Studies indicate that the gut microbiota is partially stable, with approximately 40 species of bacteria forming a core microbiota that persists in humans for at least 1 year (Donaldson et al., 2016). Additionally, mucosal flora analysis revealed an increase in Enterobacteriaceae and a decrease in Clostridium in patients with Crohn’s disease. However, these features disappeared when stool samples were examined (Gevers et al., 2014).
Probiotics may have an impact on the host even though they do not necessarily interact with indigenous microbiota (Hill et al., 2014). Instead, they may normalize the disturbed microbiota and modulate it in a beneficial manner. It is difficult to identify patterns of change in commonly altered microbes among studies that report probiotic-associated microbiome alterations. The average relative abundance of R. intestinalis in healthy individuals is estimated at only 0.09377% according to the Gmrepo database (Dai et al., 2022). Thus, R. intestinalis is not predominant in the entire intestinal flora composition. This may explain why, after taking R. intestinalis for a short period, it remains difficult for the bacterium to become the predominant. A clinical study also found that after administration L. paracasei DG, the α-diversity of the intestinal flora did not change, but the β-diversity and structure changed (Ferrario et al., 2014) with an increase in Brucella and Faecalis. Administration of R. intestinalis increased the levels of certain beneficial gut bacteria. Porphyromonadaceae abundance increased in long-lived populations, and the proportion of R. intestinalis also increased (Ren et al., 2021). The abundance of Barnesiella, Porphyromonadaceae, and Roseburia also increased after vitamin D1 administration in patients with Crohn’s disease (Schäffler et al., 2018), indicating that these bacteria may function synergistically. In addition to its resistance to IBD (Weiss et al., 2014), Barnesiella is a valuable “oncomicrobiotic” for antitumor immunomodulator therapy (Daillère et al., 2016). Barnesiella has also been associated with remission from obesity and hepatic steatosis (Rodriguez et al., 2020) and with the clearance of vancomycin-resistant Enterococcus in the gut (Ubeda et al., 2013). Porphyromonadaceae has been associated with reduced visceral fat and a healthier metabolic profile in the elderly (Tavella et al., 2021). Therefore, the probiotic effect of R. intestinalis may be that it constitutes an interaction network with several beneficial gut microbes, such as Barnesiella and Porphyromonadaceae.
CFDA-SE is a fluorescent dye with many advantages such as strong fluorescence, low toxicity, and good stability. Several researchers (Lee et al., 2004; Wang et al., 2019, 2021; Zhao et al., 2022) have used this method to tag and track bacteria. R. intestinalis was detected in all intestinal sites 2 h after administration, whereas after 48 h, R. intestinalis was predominantly located in the cecum, and not present in the colon. This distribution may be related to the physiology of the different parts of the gut (Donaldson et al., 2016; Martinez-Guryn et al., 2019). Using frozen section analysis, we observed that R. intestinalis colonizing the cecum and mucus layer of the colon. Abbeele et al. demonstrated that R. intestinalis colonized the mucin layer using in vitro intestinal models (Van den Abbeele et al., 2013), which corresponds with our observations in vivo.
In conclusion, we performed a comprehensive safety assessment of R. intestinalis. An in vitro study and genomic analysis revealed that R. intestinalis was not cytotoxic and caused no safety concerns associated with antibiotic resistance genes, virulence factors, biogenic amine production, gelatinase, or DNase activity. In vivo experiments showed that orally administrated R. intestinalis mainly colonizes the cecum and colonic mucus layers without altering the abundance and diversity of the gut microbiota. An oral toxicity study conducted in mice revealed that R. intestinalis was not toxic and could reduce serum uric acid levels. This study highlights R. intestinalis as a non-pathogenic strain suitable for use as a “Next Generation Probiotic.”
The raw sequences generated for this study can be found in the NCBI Short Read Archive under BioProject no. PRJNA850191.
The animal study was reviewed and approved by Central South University Animal Ethics Committee (XMSB-2022-0198).
CZ analyzed the strain phenotypically and genetically and drafted the manuscript. KM and KN helped with genomic and bioinformatic analyses. WL and XinW contributed to the phenotypic experiments. YH helped to carry out the experiments on mice and fluorescently label of bacteria. MD contributed to revising the manuscript. XiaW oversaw the project and reviewed and revised the manuscript. All authors contributed to the article and approved the submitted version.
This project was supported by the National Natural Science Foundation of China (NSFC no. 81970494).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.973046/full#supplementary-material
Alayande, K. A., Aiyegoro, O. A., Nengwekhulu, T. M., Katata-Seru, L., and Ateba, C. N. (2020). Integrated genome-based probiotic relevance and safety evaluation of lactobacillus reuteri PNW1. PLoS One 15:e0235873. doi: 10.1371/journal.pone.0235873
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–d525. doi: 10.1093/nar/gkz935
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402. doi: 10.1186/1471-2164-12-402
Anandharaj, M., Sivasankari, B., Santhanakaruppu, R., Manimaran, M., Rani, R. P., and Sivakumar, S. (2015). Determining the probiotic potential of cholesterol-reducing lactobacillus and Weissella strains isolated from gherkins (fermented cucumber) and south Indian fermented koozh. Res. Microbiol. 166, 428–439. doi: 10.1016/j.resmic.2015.03.002
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Arumugam, M., Raes, J., Pelletier, E., Le Paslier, D., Yamada, T., Mende, D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180. doi: 10.1038/nature09944
Ben Braïek, O., and Smaoui, S. (2019). Enterococci: Between emerging pathogens and potential probiotics. Biomed. Res. Int. 2019, 1–13. doi: 10.1155/2019/5938210
Bertelli, C., Laird, M. R., Williams, K. P., Lau, B. Y., Hoad, G., Winsor, G. L., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–w35. doi: 10.1093/nar/gkx343
Bhattacharya, T., Ghosh, T. S., and Mande, S. S. (2015). Global profiling of carbohydrate active enzymes in human gut microbiome. PLoS One 10:e0142038. doi: 10.1371/journal.pone.0142038
Brandt, K., Tilsala-Timisjärvi, A., and Alatossava, T. (2001). Phage-related DNA polymorphism in dairy and probiotic lactobacillus. Micron 32, 59–65. doi: 10.1016/S0968-4328(00)00030-5
Casjens, S. (2003). Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49, 277–300. doi: 10.1046/j.1365-2958.2003.03580.x
Chen, D., Jin, D., Huang, S., Wu, J., Xu, M., Liu, T., et al. (2020). Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 469, 456–467. doi: 10.1016/j.canlet.2019.11.019
Choeisoongnern, T., Sirilun, S., Waditee-Sirisattha, R., Pintha, K., Peerajan, S., and Chaiyasut, C. (2021). Potential probiotic enterococcus faecium OV3-6 and its bioactive peptide as alternative bio-preservation. Foods 10:2264. doi: 10.3390/foods10102264
CLSI. (2016). “Performance standards for antimicrobial susceptibility testing,” in CLSI Document M100S. 26th Edn. (Wayne, PA: Clinical and Laboratory Standards Institute).
Cornuault, J. K., Moncaut, E., Loux, V., Mathieu, A., Sokol, H., Petit, M. A., et al. (2020). The enemy from within: a prophage of Roseburia intestinalis systematically turns lytic in the mouse gut, driving bacterial adaptation by CRISPR spacer acquisition. ISME J. 14, 771–787. doi: 10.1038/s41396-019-0566-x
Cotter, P. D., Ross, R. P., and Hill, C. (2013). Bacteriocins - a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105. doi: 10.1038/nrmicro2937
Couvin, D., Bernheim, A., Toffano-Nioche, C., Touchon, M., Michalik, J., Néron, B., et al. (2018). CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 46, W246–w251. doi: 10.1093/nar/gky425
Cozzolino, A., Vergalito, F., Tremonte, P., Iorizzo, M., Lombardi, S. J., Sorrentino, E., et al. (2020). Preliminary evaluation of the safety and probiotic potential of Akkermansia muciniphila DSM 22959 in comparison with lactobacillus rhamnosus GG. Microorganisms 8:189. doi: 10.3390/microorganisms8020189
Dai, D., Zhu, J., Sun, C., Li, M., Liu, J., Wu, S., et al. (2022). GMrepo v2: a curated human gut microbiome database with special focus on disease markers and cross-dataset comparison. Nucleic Acids Res. 50, D777–d784. doi: 10.1093/nar/gkab1019
Daillère, R., Vétizou, M., Waldschmitt, N., Yamazaki, T., Isnard, C., Poirier-Colame, V., et al. (2016). Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic Immunomodulatory effects. Immunity 45, 931–943. doi: 10.1016/j.immuni.2016.09.009
Davis, J. J., Wattam, A. R., Aziz, R. K., Brettin, T., Butler, R., Butler, R. M., et al. (2020). The PATRIC bioinformatics resource center: expanding data and analysis capabilities. Nucleic Acids Res. 48, D606–d612. doi: 10.1093/nar/gkz943
De Angelis, M., Rizzello, C. G., Fasano, A., Clemente, M. G., De Simone, C., Silano, M., et al. (2006). VSL#3 probiotic preparation has the capacity to hydrolyze gliadin polypeptides responsible for celiac Sprue. Biochim. Biophys. Acta 1762, 80–93. doi: 10.1016/j.bbadis.2005.09.008
Deng, Z., Luo, X. M., Liu, J., and Wang, H. (2020). Quorum sensing, biofilm, and intestinal mucosal barrier: involvement the role of probiotic. Front. Cell. Infect. Microbiol. 10:538077. doi: 10.3389/fcimb.2020.538077
Deraz, S. F., Karlsson, E. N., Hedström, M., Andersson, M. M., and Mattiasson, B. (2005). Purification and characterisation of acidocin D20079, a bacteriocin produced by lactobacillus acidophilus DSM 20079. J. Biotechnol. 117, 343–354. doi: 10.1016/j.jbiotec.2005.02.005
Devaux, C. A., Million, M., and Raoult, D. (2020). The Butyrogenic and lactic bacteria of the gut microbiota determine the outcome of Allogenic hematopoietic cell transplant. Front. Microbiol. 11:1642. doi: 10.3389/fmicb.2020.01642
Dias, A. M. M., Douhard, R., Hermetet, F., Regimbeau, M., Lopez, T. E., Gonzalez, D., et al. (2021). Lactobacillus stress protein GroEL prevents colonic inflammation. J. Gastroenterol. 56, 442–455. doi: 10.1007/s00535-021-01774-3
Dillon, S. M., Kibbie, J., Lee, E. J., Guo, K., Santiago, M. L., Austin, G. L., et al. (2017). Low abundance of colonic butyrate-producing bacteria in HIV infection is associated with microbial translocation and immune activation. AIDS 31, 511–521. doi: 10.1097/QAD.0000000000001366
Donaldson, G. P., Lee, S. M., and Mazmanian, S. K. (2016). Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32. doi: 10.1038/nrmicro3552
Drula, E., Garron, M.-L., Dogan, S., Lombard, V., Henrissat, B., and Terrapon, N. (2021). The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 50, D571–D577. doi: 10.1093/nar/gkab1045
Dudík, B., Kiňová Sepová, H., Bilka, F., Pašková, Ľ., and Bilková, A. (2020). Mucin pre-cultivated lactobacillus reuteri E shows enhanced adhesion and increases mucin expression in HT-29 cells. Antonie Van Leeuwenhoek 113, 1191–1200. doi: 10.1007/s10482-020-01426-1
Duncan, S. H., Hold, G. L., Barcenilla, A., Stewart, C. S., and Flint, H. J. (2002). Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int. J. Syst. Evol. Microbiol. 52, 1615–1620. doi: 10.1099/00207713-52-5-1615
Fallingborg, J. (1999). Intraluminal pH of the human gastrointestinal tract. Dan. Med. Bull. 46, 183–196.
Fan, H., Chen, Z., Lin, R., Liu, Y., Wu, X., Puthiyakunnon, S., et al. (2019). Bacteroides fragilis strain ZY-312 defense against Cronobacter sakazakii-induced necrotizing Enterocolitis In vitro and in a neonatal rat model. mSystems 4:19. doi: 10.1128/mSystems.00305-19
Ferrario, C., Taverniti, V., Milani, C., Fiore, W., Laureati, M., De Noni, I., et al. (2014). Modulation of fecal Clostridiales bacteria and butyrate by probiotic intervention with lactobacillus paracasei DG varies among healthy adults. J. Nutr. 144, 1787–1796. doi: 10.3945/jn.114.197723
Gargis, S. R., Heath, H. E., Heath, L. S., Leblanc, P. A., Simmonds, R. S., Abbott, B. D., et al. (2009). Use of 4-sulfophenyl isothiocyanate labeling and mass spectrometry to determine the site of action of the streptococcolytic peptidoglycan hydrolase zoocin A. Appl. Environ. Microbiol. 75, 72–77. doi: 10.1128/AEM.01647-08
Gevers, D., Kugathasan, S., Denson, L. A., Vázquez-Baeza, Y., Van Treuren, W., Ren, B., et al. (2014). The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392. doi: 10.1016/j.chom.2014.02.005
Gill, S. R., Pop, M., Deboy, R. T., Eckburg, P. B., Turnbaugh, P. J., Samuel, B. S., et al. (2006). Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359. doi: 10.1126/science.1124234
Gou, X., Zhang, L., Zhao, S., Ma, W., and Yang, Z. (2021). Application of the combination of soybean lecithin and whey protein concentrate 80 to improve the bile salt and acid tolerance of probiotics. J. Microbiol. Biotechnol. 31, 840–846. doi: 10.4014/jmb.2103.03017
Grajeda-Iglesias, C., Durand, S., Daillère, R., Iribarren, K., Lemaitre, F., Derosa, L., et al. (2021). Oral administration of Akkermansia muciniphila elevates systemic antiaging and anticancer metabolites. Aging (Albany NY) 13, 6375–6405. doi: 10.18632/aging.202739
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document. The international scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Holý, O., Parra-Flores, J., Lepuschitz, S., Alarcón-Lavín, M. P., Cruz-Córdova, A., Xicohtencatl-Cortes, J., et al. (2020). Molecular characterization of Cronobacter sakazakii strains isolated from powdered Milk. Foods 10:20. doi: 10.3390/foods10010020
Hu, B., Tian, F., Wang, G., Zhang, Q., Zhao, J., Zhang, H., et al. (2015). Enhancement of bile resistance in lactobacillus plantarum strains by soy lecithin. Lett. Appl. Microbiol. 61, 13–19. doi: 10.1111/lam.12418
Hussein, W. E., Abdelhamid, A. G., Rocha-Mendoza, D., García-Cano, I., and Yousef, A. E. (2020). Assessment of safety and probiotic traits of enterococcus durans OSY-EGY, isolated From Egyptian artisanal cheese, using comparative genomics and phenotypic analyses. Front. Microbiol. 11:608314. doi: 10.3389/fmicb.2020.608314
Juhas, M., van der Meer, J. R., Gaillard, M., Harding, R. M., Hood, D. W., and Crook, D. W. (2009). Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33, 376–393. doi: 10.1111/j.1574-6976.2008.00136.x
Kasahara, K., Krautkramer, K. A., Org, E., Romano, K. A., Kerby, R. L., Vivas, E. I., et al. (2018). Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat. Microbiol. 3, 1461–1471. doi: 10.1038/s41564-018-0272-x
La Rosa, S. L., Leth, M. L., Michalak, L., Hansen, M. E., Pudlo, N. A., Glowacki, R., et al. (2019). The human gut Firmicute Roseburia intestinalis is a primary degrader of dietary β-mannans. Nat. Commun. 10, 905. doi: 10.1038/s41467-019-08812-y
Lee, S. E., Choi, Y., Jun, J. E., Lee, Y. B., Jin, S. M., Hur, K. Y., et al. (2019). Additional effect of dietary fiber in patients with type 2 diabetes mellitus using metformin and sulfonylurea: An open-label. Pilot Trial. Diabetes Metab J 43, 422–431. doi: 10.4093/dmj.2018.0090
Lee, Y. K., Ho, P. S., Low, C. S., Arvilommi, H., and Salminen, S. (2004). Permanent colonization by lactobacillus casei is hindered by the low rate of cell division in mouse gut. Appl. Environ. Microbiol. 70, 670–674. doi: 10.1128/AEM.70.2.670-674.2004
Leth, M. L., Ejby, M., Madland, E., Kitaoku, Y., Slotboom, D. J., Guskov, A., et al. (2020). Molecular insight into a new low-affinity xylan binding module from the xylanolytic gut symbiont Roseburia intestinalis. FEBS J. 287, 2105–2117. doi: 10.1111/febs.15117
Li, Q., Hu, W., Liu, W. X., Zhao, L. Y., Huang, D., Liu, X. D., et al. (2021). Streptococcus thermophilus inhibits colorectal tumorigenesis Through secreting β-Galactosidase. Gastroenterology 160, 1179–1193.e14. doi: 10.1053/j.gastro.2020.09.003
Li, B., Zhan, M., Evivie, S. E., Jin, D., Zhao, L., Chowdhury, S., et al. (2018). Evaluating the safety of potential probiotic enterococcus durans KLDS6.0930 using whole genome sequencing and Oral toxicity study. Front. Microbiol. 9:1943. doi: 10.3389/fmicb.2018.01943
Liang, Q., Chiu, J., Chen, Y., Huang, Y., Higashimori, A., Fang, J., et al. (2017). Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin. Cancer Res. 23, 2061–2070. doi: 10.1158/1078-0432.CCR-16-1599
Lin, R., Liu, W., Piao, M., and Zhu, H. (2017). A review of the relationship between the gut microbiota and amino acid metabolism. Amino Acids 49, 2083–2090. doi: 10.1007/s00726-017-2493-3
Liu, S., Zhao, W., Liu, X., and Cheng, L. (2020). Metagenomic analysis of the gut microbiome in atherosclerosis patients identify cross-cohort microbial signatures and potential therapeutic target. FASEB J. 34, 14166–14181. doi: 10.1096/fj.202000622R
Liu, B., Zheng, D., Zhou, S., Chen, L., and Yang, J. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–d917. doi: 10.1093/nar/gkab1107
Lu, W., Gao, B., Fan, J., Cheng, P., Hu, Y., Jie, Q., et al. (2019). Mesenchymal progenitors derived from different locations in long bones display diverse characteristics. Stem Cells Int. 2019, 1–11. doi: 10.1155/2019/5037578
Luo, W., Shen, Z., Deng, M., Li, X., Tan, B., Xiao, M., et al. (2019). Roseburia intestinalis supernatant ameliorates colitis induced in mice by regulating the immune response. Mol. Med. Rep. 20, 1007–1016. doi: 10.3892/mmr.2019.10327
Ma, C., Huo, D., You, Z., Peng, Q., Jiang, S., Chang, H., et al. (2020a). Differential pattern of indigenous microbiome responses to probiotic Bifidobacterium lactis V9 consumption across subjects. Food Res. Int. 136:109496. doi: 10.1016/j.foodres.2020.109496
Ma, L., Ni, Y., Wang, Z., Tu, W., Ni, L., Zhuge, F., et al. (2020b). Spermidine improves gut barrier integrity and gut microbiota function in diet-induced obese mice. Gut Microbes 12, 1–19. doi: 10.1080/19490976.2020.1832857
Machiels, K., Joossens, M., Sabino, J., De Preter, V., Arijs, I., Eeckhaut, V., et al. (2014). A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63, 1275–1283. doi: 10.1136/gutjnl-2013-304833
Madeo, F., Eisenberg, T., Pietrocola, F., and Kroemer, G. (2018). Spermidine in health and disease. Science 359:2788. doi: 10.1126/science.aan2788
Manrique, P., Dills, M., and Young, M. J. (2017). The human gut phage community and its implications for health and disease. Viruses 9:141. doi: 10.3390/v9060141
Martinez-Guryn, K., Leone, V., and Chang, E. B. (2019). Regional diversity of the gastrointestinal microbiome. Cell Host Microbe 26, 314–324. doi: 10.1016/j.chom.2019.08.011
Mena, B., and Aryana, K. (2018). Short communication: lactose enhances bile tolerance of yogurt culture bacteria. J. Dairy Sci. 101, 1957–1959. doi: 10.3168/jds.2017-13919
Metlakunta, A. S., and Soman, R. J. (2020). Safety evaluation of Bacillus coagulans SNZ 1969 in Wistar rats. Regul. Toxicol. Pharmacol. 110:104538. doi: 10.1016/j.yrtph.2019.104538
Miquel, S., Leclerc, M., Martin, R., Chain, F., Lenoir, M., Raguideau, S., et al. (2015). Identification of metabolic signatures linked to anti-inflammatory effects of Faecalibacterium prausnitzii. MBio 6:15. doi: 10.1128/mBio.00300-15
Mirande, C., Kadlecikova, E., Matulova, M., Capek, P., Bernalier-Donadille, A., Forano, E., et al. (2010). Dietary fibre degradation and fermentation by two xylanolytic bacteria Bacteroides xylanisolvens XB1A and Roseburia intestinalis XB6B4 from the human intestine. J. Appl. Microbiol. 109, 451–460. doi: 10.1111/j.1365-2672.2010.04671.x
Montalban-Arques, A., Katkeviciute, E., Busenhart, P., Bircher, A., Wirbel, J., Zeller, G., et al. (2021). Commensal Clostridiales strains mediate effective anti-cancer immune response against solid tumors. Cell Host Microbe 29, 1573–1588.e7. doi: 10.1016/j.chom.2021.08.001
Nicholson, K., Bjornevik, K., Abu-Ali, G., Chan, J., Cortese, M., Dedi, B., et al. (2021). The human gut microbiota in people with amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener 22, 186–194. doi: 10.1080/21678421.2020.1828475
Nie, K., Ma, K., Luo, W., Shen, Z., Yang, Z., Xiao, M., et al. (2021). Roseburia intestinalis: A beneficial gut organism From the discoveries in genus and species. Front. Cell. Infect. Microbiol. 11:757718. doi: 10.3389/fcimb.2021.757718
Niu, C., Yu, D., Wang, Y., Ren, H., Jin, Y., Zhou, W., et al. (2013). Common and pathogen-specific virulence factors are different in function and structure. Virulence 4, 473–482. doi: 10.4161/viru.25730
O’Toole, P. W., Marchesi, J. R., and Hill, C. (2017). Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2, 17057. doi: 10.1038/nmicrobiol.2017.57
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Pan, X., Kaminga, A. C., Liu, A., Wen, S. W., Luo, M., and Luo, J. (2021). Gut microbiota, glucose, lipid, and water-electrolyte metabolism in children With nonalcoholic fatty liver disease. Front. Cell. Infect. Microbiol. 11:683743. doi: 10.3389/fcimb.2021.683743
Pan, M., Nethery, M. A., Hidalgo-Cantabrana, C., and Barrangou, R. (2020). Comprehensive mining and characterization of CRISPR-Cas Systems in Bifidobacterium. Microorganisms 8:720. doi: 10.3390/microorganisms8050720
Pascal Andreu, V., Roel-Touris, J., Dodd, D., Fischbach, M. A., and Medema, M. H. (2021). The gut SMASH web server: automated identification of primary metabolic gene clusters from the gut microbiota. Nucleic Acids Res. 49, W263–W270. doi: 10.1093/nar/gkab353
Pei, Z., Sadiq, F. A., Han, X., Zhao, J., Zhang, H., Ross, R. P., et al. (2021). Comprehensive scanning of Prophages in lactobacillus: distribution, diversity, antibiotic resistance genes, and linkages with CRISPR-Cas systems. mSystems 6:e0121120. doi: 10.1128/mSystems.01211-20
Qin, Q., Yan, S., Yang, Y., Chen, J., Li, T., Gao, X., et al. (2021). A Metagenome-wide association study of the gut microbiome and metabolic syndrome. Front. Microbiol. 12:682721. doi: 10.3389/fmicb.2021.682721
Quan, Y., Song, K., Zhang, Y., Zhu, C., Shen, Z., Wu, S., et al. (2018). Roseburia intestinalis-derived flagellin is a negative regulator of intestinal inflammation. Biochem. Biophys. Res. Commun. 501, 791–799. doi: 10.1016/j.bbrc.2018.05.075
Rastogi, S., Mittal, V., and Singh, A. (2020). In vitro evaluation of probiotic potential and safety assessment of lactobacillus mucosae strains isolated from Donkey’s lactation. Probiotics Antimicrob Proteins 12, 1045–1056. doi: 10.1007/s12602-019-09610-0
Ren, M., Li, H., Fu, Z., and Li, Q. (2021). Succession analysis of gut microbiota structure of participants from long-lived families in Hechi, Guangxi, China. Microorganisms 9:2524. doi: 10.3390/microorganisms9122524
Reuben, R. C., Roy, P. C., Sarkar, S. L., Rubayet Ul Alam, A. S. M., and Jahid, I. K. (2020). Characterization and evaluation of lactic acid bacteria from indigenous raw milk for potential probiotic properties. J. Dairy Sci. 103, 1223–1237. doi: 10.3168/jds.2019-17092
Rocha-Mendoza, D., Kosmerl, E., Miyagusuku-Cruzado, G., Giusti, M. M., Jiménez-Flores, R., and García-Cano, I. (2020). Growth of lactic acid bacteria in milk phospholipids enhances their adhesion to Caco-2 cells. J. Dairy Sci. 103, 7707–7718. doi: 10.3168/jds.2020-18271
Rodriguez, J., Hiel, S., Neyrinck, A. M., Le Roy, T., Pötgens, S. A., Leyrolle, Q., et al. (2020). Discovery of the gut microbial signature driving the efficacy of prebiotic intervention in obese patients. Gut 69, 1975–1987. doi: 10.1136/gutjnl-2019-319726
Salminen, S., Collado, M. C., Endo, A., Hill, C., Lebeer, S., Quigley, E. M. M., et al. (2021). The international scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 18, 649–667. doi: 10.1038/s41575-021-00440-6
Schäffler, H., Herlemann, D. P., Klinitzke, P., Berlin, P., Kreikemeyer, B., Jaster, R., et al. (2018). Vitamin D administration leads to a shift of the intestinal bacterial composition in Crohn’s disease patients, but not in healthy controls. J. Dig. Dis. 19, 225–234. doi: 10.1111/1751-2980.12591
Seddik, H. A., Bendali, F., Gancel, F., Fliss, I., Spano, G., and Drider, D. (2017). Lactobacillus plantarum and its probiotic and food potentialities. Probiotics Antimicrob Proteins 9, 111–122. doi: 10.1007/s12602-017-9264-z
Segain, J. P., Raingeard de la Blétière, D., Bourreille, A., Leray, V., Gervois, N., Rosales, C., et al. (2000). Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn’s disease. Gut 47, 397–403. doi: 10.1136/gut.47.3.397
Seo, B., Jeon, K., Moon, S., Lee, K., Kim, W. K., Jeong, H., et al. (2020). Roseburia spp. abundance associates with alcohol consumption in humans and its administration ameliorates alcoholic fatty liver in mice. Cell Host Microbe 27, 25–40.e6. doi: 10.1016/j.chom.2019.11.001
Sharma, K., Attri, S., and Goel, G. (2019). Selection and evaluation of probiotic and functional characteristics of autochthonous lactic acid bacteria isolated from fermented wheat flour dough Babroo. Probiotics Antimicrob Proteins 11, 774–784. doi: 10.1007/s12602-018-9466-z
Shen, Z., Zhu, C., Quan, Y., Yang, J., Yuan, W., Yang, Z., et al. (2018). Insights into Roseburia intestinalis which alleviates experimental colitis pathology by inducing anti-inflammatory responses. J. Gastroenterol. Hepatol. 33, 1751–1760. doi: 10.1111/jgh.14144
Solieri, L., Bianchi, A., Mottolese, G., Lemmetti, F., and Giudici, P. (2014). Tailoring the probiotic potential of non-starter lactobacillus strains from ripened Parmigiano Reggiano cheese by in vitro screening and principal component analysis. Food Microbiol. 38, 240–249. doi: 10.1016/j.fm.2013.10.003
Sonnenburg, J. L., Angenent, L. T., and Gordon, J. I. (2004). Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 5, 569–573. doi: 10.1038/ni1079
Steppe, M., Van Nieuwerburgh, F., Vercauteren, G., Boyen, F., Eeckhaut, V., Deforce, D., et al. (2014). Safety assessment of the butyrate-producing Butyricicoccus pullicaecorum strain 25-3(T), a potential probiotic for patients with inflammatory bowel disease, based on oral toxicity tests and whole genome sequencing. Food Chem. Toxicol. 72, 129–137. doi: 10.1016/j.fct.2014.06.024
Sun, Z., Harris, H. M., McCann, A., Guo, C., Argimón, S., Zhang, W., et al. (2015). Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 6, 8322. doi: 10.1038/ncomms9322
Tavella, T., Rampelli, S., Guidarelli, G., Bazzocchi, A., Gasperini, C., Pujos-Guillot, E., et al. (2021). Elevated gut microbiome abundance of Christensenellaceae, Porphyromonadaceae and Rikenellaceae is associated with reduced visceral adipose tissue and healthier metabolic profile in Italian elderly. Gut Microbes 13, 1–19. doi: 10.1080/19490976.2021.1880221
Ubeda, C., Bucci, V., Caballero, S., Djukovic, A., Toussaint, N. C., Equinda, M., et al. (2013). Intestinal microbiota containing Barnesiella species cures vancomycin-resistant enterococcus faecium colonization. Infect. Immun. 81, 965–973. doi: 10.1128/IAI.01197-12
Van den Abbeele, P., Belzer, C., Goossens, M., Kleerebezem, M., De Vos, W. M., Thas, O., et al. (2013). Butyrate-producing clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 7, 949–961. doi: 10.1038/ismej.2012.158
van Heel, A. J., de Jong, A., Song, C., Viel, J. H., Kok, J., and Kuipers, O. P. (2018). BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 46, W278–w281. doi: 10.1093/nar/gky383
Vankerckhoven, V., Huys, G., Vancanneyt, M., Vael, C., Klare, I., Romond, M.-B., et al. (2008). Biosafety assessment of probiotics used for human consumption: recommendations from the EU-PROSAFE project. Trends Food Sci. Technol. 19, 102–114. doi: 10.1016/j.tifs.2007.07.013
Vargas, L. A., Olson, D. W., and Aryana, K. J. (2015). Whey protein isolate improves acid and bile tolerances of Streptococcus thermophilus ST-M5 and lactobacillus delbrueckii ssp. bulgaricus LB-12. J. Dairy Sci. 98, 2215–2221. doi: 10.3168/jds.2014-8869
Wang, G., Chen, Y., Fei, S., Xie, C., Xia, Y., and Ai, L. (2021). Colonisation with endogenous lactobacillus reuteri R28 and exogenous lactobacillus plantarum AR17-1 and the effects on intestinal inflammation in mice. Food Funct. 12, 2481–2488. doi: 10.1039/D0FO02624G
Wang, G., Liu, Y., Lu, Z., Yang, Y., Xia, Y., Lai, P. F., et al. (2019). The ameliorative effect of a lactobacillus strain with good adhesion ability against dextran sulfate sodium-induced murine colitis. Food Funct. 10, 397–409. doi: 10.1039/C8FO01453A
Wang, L., Si, W., Xue, H., and Zhao, X. (2017). A fibronectin-binding protein (FbpA) of Weissella cibaria inhibits colonization and infection of Staphylococcus aureus in mammary glands. Cell. Microbiol. 19:2731. doi: 10.1111/cmi.12731
Weiss, G. A., Chassard, C., and Hennet, T. (2014). Selective proliferation of intestinal Barnesiella under fucosyllactose supplementation in mice. Br. J. Nutr. 111, 1602–1610. doi: 10.1017/S0007114513004200
Wu, X., Pan, S., Luo, W., Shen, Z., Meng, X., Xiao, M., et al. (2020). Roseburia intestinalis-derived flagellin ameliorates colitis by targeting miR-223-3p-mediated activation of NLRP3 inflammasome and pyroptosis. Mol. Med. Rep. 22, 2695–2704. doi: 10.3892/mmr.2020.11351
Xu, F., Cheng, Y., Ruan, G., Fan, L., Tian, Y., Xiao, Z., et al. (2021). New pathway ameliorating ulcerative colitis: focus on Roseburia intestinalis and the gut-brain axis. Ther. Adv. Gastroenterol. 14, 175628482110044. doi: 10.1177/17562848211004469
Xu, X., Zhang, W., Guo, M., Xiao, C., Fu, Z., Yu, S., et al. (2022). Integrated analysis of gut microbiome and host immune responses in COVID-19. Front. Med. 16, 263–275. doi: 10.1007/s11684-022-0921-6
Yadav, H., Lee, J. H., Lloyd, J., Walter, P., and Rane, S. G. (2013). Beneficial metabolic effects of a probiotic via butyrate-induced GLP-1 hormone secretion. J. Biol. Chem. 288, 25088–25097. doi: 10.1074/jbc.M113.452516
Zhao, X., Zhao, C., Yang, L., Jiang, L., Zhang, J., Yu, X., et al. (2022). Spatial and temporal persistence of fluorescent Lactiplantibacillus plantarum RS-09 in intestinal tract. Front. Microbiol. 13:843650. doi: 10.3389/fmicb.2022.843650
Zhou, W., Zhang, D., Li, Z., Jiang, H., Li, J., Ren, R., et al. (2021). The fecal microbiota of patients with pancreatic ductal adenocarcinoma and autoimmune pancreatitis characterized by metagenomic sequencing. J. Transl. Med. 19, 215. doi: 10.1186/s12967-021-02882-7
Zhu, Y., Guo, L., and Yang, Q. (2020). Partial replacement of nitrite with a novel probiotic lactobacillus plantarum on nitrate, color, biogenic amines and gel properties of Chinese fermented sausages. Food Res. Int. 137:109351. doi: 10.1016/j.foodres.2020.109351
Zhu, C., Song, K., Shen, Z., Quan, Y., Tan, B., Luo, W., et al. (2018). Roseburia intestinalis inhibits interleukin-17 excretion and promotes regulatory T cells differentiation in colitis. Mol. Med. Rep. 17, 7567–7574. doi: 10.3892/mmr.2018.8833
Keywords: probiotics, Roseburia intestinalis, safety, genome mining, oral toxicity, cytotoxicity, intestinal colonization
Citation: Zhang C, Ma K, Nie K, Deng M, Luo W, Wu X, Huang Y and Wang X (2022) Assessment of the safety and probiotic properties of Roseburia intestinalis: A potential “Next Generation Probiotic”. Front. Microbiol. 13:973046. doi: 10.3389/fmicb.2022.973046
Received: 19 June 2022; Accepted: 05 August 2022;
Published: 08 September 2022.
Edited by:
Hui Ni, Jimei University, ChinaReviewed by:
Santosh Kumar Tiwari, Maharshi Dayanand University, IndiaCopyright © 2022 Zhang, Ma, Nie, Deng, Luo, Wu, Huang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyan Wang, d3h5MjIwMDExQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.