- 1School of Life Sciences, Anhui Agricultural University, Hefei, China
- 2State Key Laboratory of Tea Plant Biology and Utilization, Anhui Agricultural University, Hefei, China
Insect-associated fungi are a potentially rich source of novel natural products with antibacterial activity. Here, we investigated the community composition and phylogenetic diversity of gut-associated fungi of the dragonfly (Crocothemis Servilia) using a combination of culture-dependent and culture-independent methods. A total of 42 fungal isolates were obtained from the guts of the dragonfly, which belonged to four classes and thirteen different genera. Amplicon sequencing analyses revealed that the fungal communities were more diverse, and a total of 136 genera were identified and dominated by the genera Wojnowiciella and Phoma. The antibacterial bioassay showed that five fungal crude extracts of representative isolates have shown antibacterial activities. Among them, the extract of Phoma sp. QTH17 showed the best antibacterial activities against Escherichia coli, Micrococcus tetragenus, and Staphylococcus aureus with the disc diameter of inhibition zone diameter (IZD) of 6.50, 10.80, and 8.70 mm, respectively. Chemical analysis of Phoma sp. QTH17 led to the discovery of five known compounds, including ergosterol (1), 3-Chlorogentisyl alcohol (2), epoxydon (3), epoxydon 6-methylsalicylate ester (4) and mannitol (5). Among them, the compound 3 exhibited potent antibacterial activities against E. coli, M. tetragenus, and S. aureus with the IZD of 7.00, 14.00, and 12.50 mm, respectively, which were slightly weaker than those of the positive gentamicin sulfate with the IZD of 11.13, 18.30, and 12.13 mm, respectively. In conclusion, our results confirmed that the diversity of gut-associated fungi of C. Servilia could be expected to explore the resource of new species and antibacterial substances.
Introduction
Insects are the most abundant group of animals and coexist with various microorganisms (Park et al., 2018). Due to the complexity and diversity of their habitats and diets of their insect partners, the insects-associated microorganisms are also diverse (Long et al., 2022). Fungi are one of the largest groups of eukaryotic organisms on Earth and play a significant role in insect development and fitness (Gibson and Hunter, 2010; Sarikaya Bayram et al., 2010). The gut microbiome in insects has been a hot research topic in microbiology, and which diversity determines their host functions (Xia et al., 2020). The culture-dependent and culture-independent approaches have recently been used to evaluate the fungal composition and diversity of insects’ guts. For example, two approaches were used to assess gut fungal diversity in termites, and characterize gut fungal communities of the wild and laboratory-reared ghost moth larvae (Makonde et al., 2013; Liu et al., 2021). Although there are many studies on the diversity of fungi associated with insects, most are only focused on social insects.
Insect-associated fungi have been isolated from various insects, including termites, honeybee, leaf-cutting ants, Dactylopius, caterpillars, and black soldier fly (Pereira et al., 2016; Vera-Ponce de León et al., 2016; Tauber et al., 2019; Višňovská et al., 2020; Nagam et al., 2021; Tegtmeier et al., 2021). Yeast species were the most common fungi in the insect gut, and have beneficial effects on their host, such as immune system enhancement, antimicrobial activity, and nutrient supplementation (Stefanini, 2018; Meriggi et al., 2020). Some filamentous fungi were also frequently isolated, including Trichoderma, Pyrenochaeta, Fusarium, Acremonium, Chaetomium, and Penicillium (Xu et al., 2020; Nagam et al., 2021). Those insect gut-associated fungi were the resource of new species and novel bioactive products, and have attracted the wide attention of scholars (Zhang et al., 2008; Moubasher et al., 2017). For instance, two novel yeast species isolated from the guts of termites play a symbiotic role in the host concerning xylan degradation and production of vitamins (Handel et al., 2016). Two novel compounds were purified and identified from Trichoderma harzianum isolated from the gut of Pantala flavescens, one of the compounds has displayed moderate antibacterial activity against Bacillus subtilis (Zhang et al., 2020). Some active compounds with antibacterial activity against important human pathogens have been found from insect-associated fungi, including long-horned grasshoppers, termites, and black soldier fly (Li et al., 2014; Correa et al., 2019; Nagam et al., 2021). Thus, there is an enormous potential for the production of secondary metabolites with biological activities by insect gut-associated fungi.
As a flying invertebrate, the dragonfly belongs to the class Insecta and the order Odonata in the phylum Arthropoda. Additionally, the dragonfly is a flesh-eating insect, and beneficial for agriculture and forestry (Maoka et al., 2020). Dragonfly guts microbiome has an important impact on host ecology and evolution and is related to dietary differences (Nair and Agashe, 2016; Nobles and Jackson, 2020). Gut-associated fungi of dragonfly were highly diverse and discovered as the resource of bioactive compounds (Shao et al., 2015; Lu et al., 2017; Zhang et al., 2020). Crocothemis servilia is one of Asia’s most widely distributed dragonfly species, mature males and most females are scarlet-red and yellow, respectively (Joshi et al., 2020). However, no study reported the gut fungal diversity of the dragonfly. Our understanding of the species range, biological activity, and secondary metabolites of these gut-associated fungi of the dragonfly is still limited. Here, we investigated the gut fungal community of the dragonfly by using both culture-dependent and culture-independent methods. The antibacterial activity and secondary metabolites of the gut-associated fungi were also evaluated.
Materials and methods
Sample collection
Dragonflies (C. servilia), which were medium-sized and authenticated by assistant professor Xin Yu (College of Life Sciences, Chongqing Normal University), were collected from a pond in Anhui Agricultural University, Hefei, China (GPS: 31°86′N, 117°26′E) in September 2018. The collected samples were rapidly transported to the laboratory and starved for 24 h, then some samples were stored at –20°C for fungal isolation and at –80°C for DNA extraction, respectively.
Fungal isolation
Two dragonfly individuals (C. servilia) were surface-sterilized in 75% ethanol for 3 min, followed by rinsing three times with sterilized water. Subsequently, the guts of samples were dissected and homogenized with 5 mL of sterilized water. Finally, the homogenates were diluted in a 10-fold series (i.e., 10–1, 10–2, 10–3), and an aliquot of 100 μL was spread onto three different isolation media: Potato dextrose agar (PDA) (Xu et al., 2020), malt extract agar (MEA) (Shao et al., 2015), ISP medium no. 2 (ISP 2) agar (Xu et al., 2020). All isolation media were added with ampicillin (10 mg/mL) and streptomycin (10 mg/mL) to suppress the growth of bacteria. Pure colonies of fungi from the appropriate dilution were transferred into a new PDA medium and incubated aerobically at 28°C. All these isolated fungal strains were maintained on PDA slants at 4°C until use. The fungi were also preserved on glycerol suspensions (25%, v/v) at –80°C.
Taxonomic identification of fungal isolates
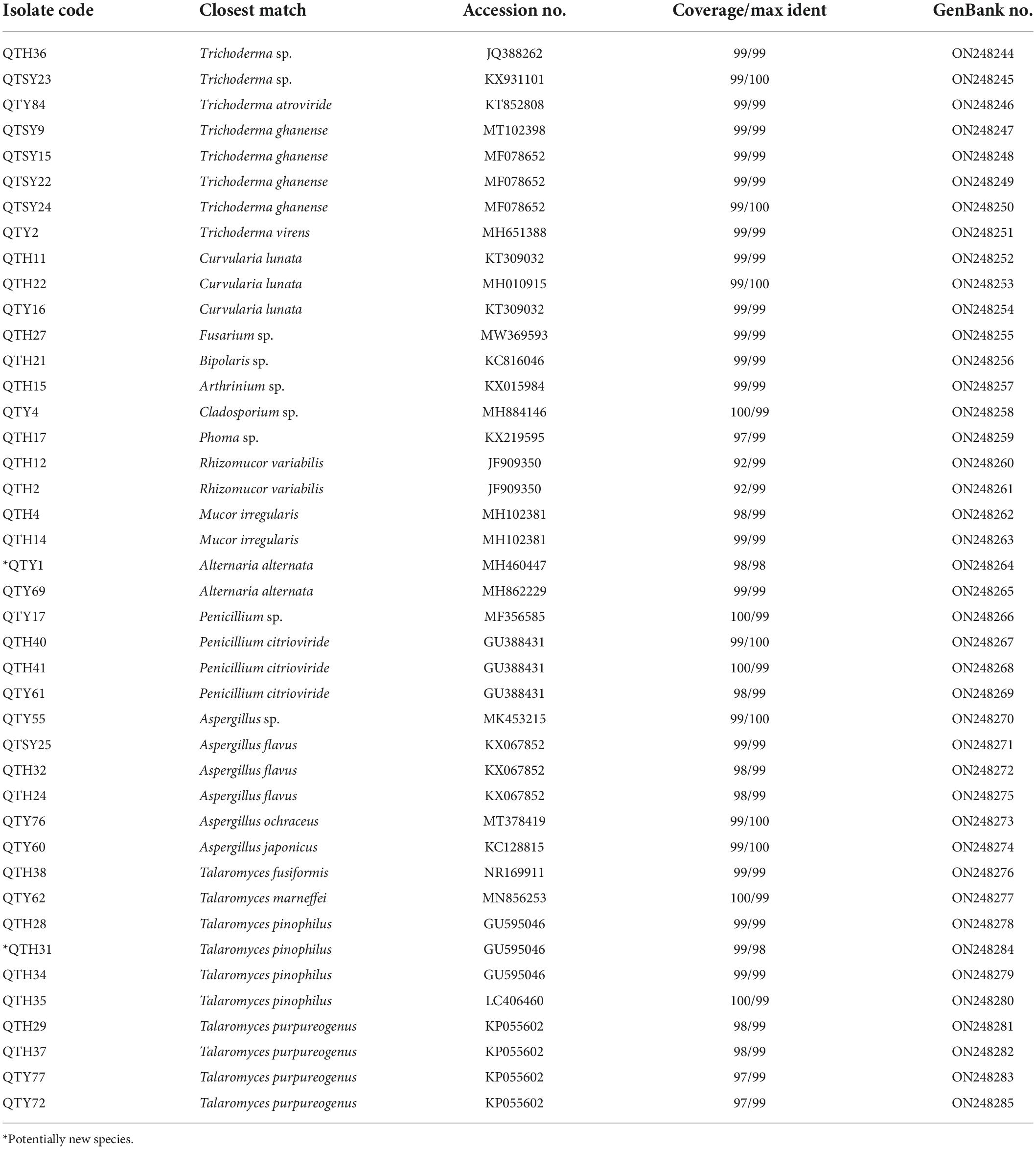
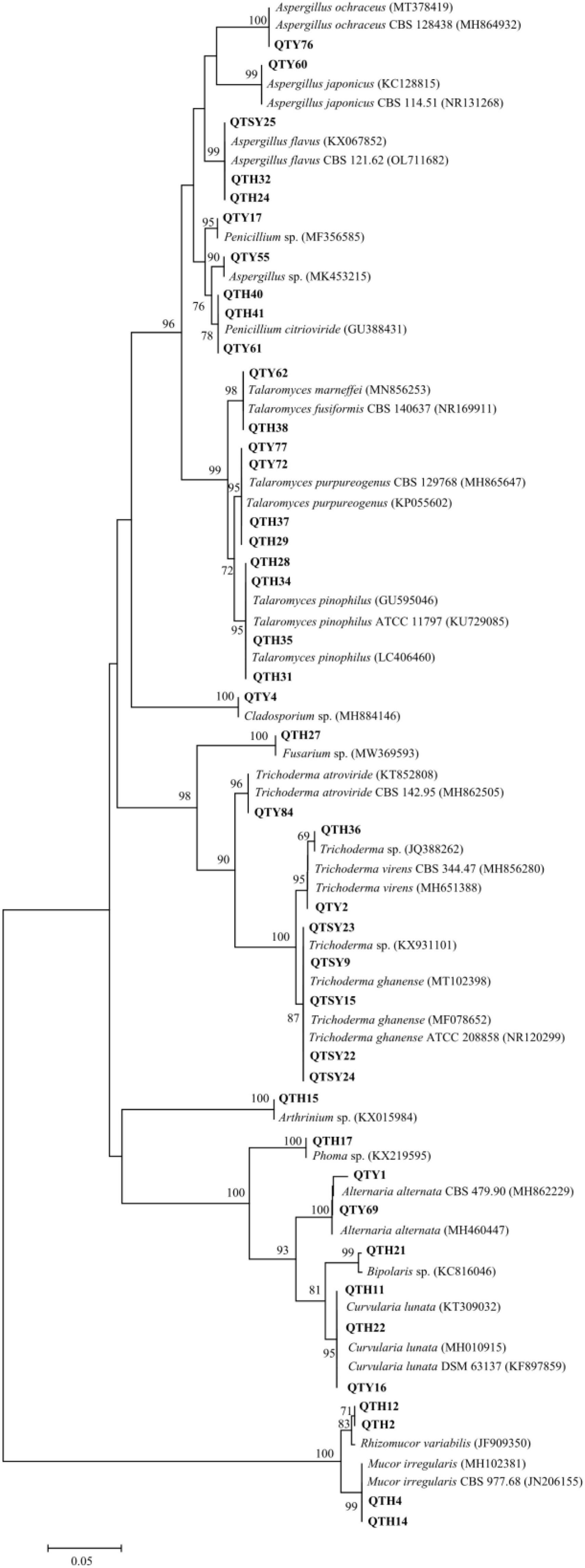
All fungal isolates were identified by molecular techniques and morphological characteristics (Li et al., 2014; Xu et al., 2020). Each fungus was cultured in PDA plate at 28°C for 3–4 days. Then the fungal genomic DNA was purified using the Fast DNA Extraction Kit [Tiangen Biotech (Beijing) Co., Ltd., Beijing, China] according to the manufacturer’s instructions. As mentioned before (Shao et al., 2015), PCR amplification for the 5.8S rDNA gene sequence was performed using the universal fungal primers ITS1 and ITS4. The quality was visualized on 1% agarose gel by electrophoresis. Qualified products amplified from PCR were sent to General Biosystems (Anhui) Co., Ltd. for sequencing. All achieved sequences’ affiliations were compared with that of available data in the NCBI database using the BLAST program. Sequence alignment and Neighbor-joining phylogenetic tree were constructed using MEGA software version 5.0. Bootstrap replication (1,000 replications) was used to assess the topology of the phylogenetic tree (Felsenstein, 1985). The ITS sequence data were submitted and deposited in GenBank under accession numbers ON248244-ON248285.
Culture-independent community analysis
Six dragonfly individuals (C. servilia) were surface-sterilized in 75% ethanol for 3 min, followed by rinsing three times with sterilized water. Then, the entire guts were removed with sterile forceps, and the gut total genome DNA of the two samples was extracted using the CTAB method (Doyle, 1987). The concentration and purity of DNA were monitored on 2% agarose gels. We generated three samples for amplicon sequencing in this study. Libraries were prepared and sequenced using specific primers ITS5-1737F and ITS2-2043R to amplify region ITS1 of the ITS gene of fungi. At last, the libraries were sequenced on an Illumina NovaSeq platform to generate approximately 250 bp paired-end reads.
Paired-end reads were merged using FLASH V1.2.7 and analyzed using QIIME V1.9.1. We used the QIIME V1.9.1 and UCHIME Algorithm for quality control, filtering of chimeric sequences, and obtaining effective tags (Caporaso et al., 2010; Edgar et al., 2011; Haas et al., 2011). Sequences having greater than 97% similarity were assigned to the same operational taxonomic units (OTUs) using the software Uparse v7.0.1001 (Edgar, 2013). The taxonomy of each representative sequence was assessed by using the MUSCLE software V 3.8.31, and comparison against the Unite database based on the blast algorithm (Edgar, 2004). Alpha diversity and species annotation were based on phylogenetic relationship construction. Raw data is accessible from NCBI’s short read archive under accession number PRJNA827522.
Antibacterial activities
The filter paper dispersion method was used to evaluate the antibacterial activity of some representative crude extracts (Xu et al., 2020). Each fungus was inoculated in a 250 mL Erlenmeyer flask containing 150 mL of ME liquid medium and cultured for 7 d at 180 rpm and 28 ± 0.5°C. The culture broth filtrate was exhaustively extracted three times with ethyl acetate (EtOAc, 1:1, v/v). Then, the ethyl acetate extract was evaporated under vacuum to yield fungal crude extract. The fungal crude extract was dissolved in acetone to get a concentration of 6 mg/mL, and 5 μL of the tested solution was pipetted onto a sterile filter paper disc (6 mm in diameter), which was placed onto pre-prepared LBA medium (1% tryptone, 1% NaCl, 0.5% yeast extract, 2% agar) plates with a suspension of the pathogenic bacteria. The pathogenic bacteria were Escherichia coli (ATCC8739), Micrococcus tetragenus (ATCC35098), and Staphylococcus aureus (ATCC6538). The sterile paper disc treated with acetone alone and gentamicin sulfate were served as a negative and positive control, respectively. Each inhibition assay was repeated three times and evaluated by measuring the diameters of inhibition zone diameter (IZD) (in mm).
Secondary metabolite characterization
One fungal strain QTH17 was selected for purification and identification of compounds in this study. Strain QTH17 was grown on PDA medium plates for 3–4 days at 28°C. Then the fresh mycelia (3 plugs of 5 mm) were inoculated into 250 mL Erlenmeyer flasks containing 150 mL of ME liquid medium and cultivated for 4 days at 28°C with shaking at 180 rpm. After that, aliquots (15 mL) of the culture were transferred into 1,000 mL Erlenmeyer flasks filled with 400 mL of the same medium and cultured at 28°C for 7 days with shaking at 180 rpm. A total of 16 L of the fermentation broth of QTH17 was filtered and extracted with EtOAc three times (3 × 16 L). The EtOAc phase was subsequently concentrated in vacuo to obtain 3.0 g of crude extract.
The crude extract (3.0 g) was dissolved in 10 mL solvent of methanol/methylene chloride (2/8, v/v) and mixed with 6.0 g dry silica gel powder (100–200 mesh, Qingdao Haiyang Chemical Co., Ltd., Qingdao, China). Then, the mixture was passed through silica-gel column chromatography (60 g, 200–300 mesh, Qingdao Haiyang Chemical Co., Ltd., Qingdao, China; Φ: 60 mm, the height of the silica-gel column: 1,000 mm), eluting with a gradient of CH2Cl2/MeOH (100:0, 100:1, 100:2, 100:4, 100:8, 100:16, v/v), to yield six fractions (Frs. 1–6). Fr6 was recrystallized from methanol, yielding a white crystal (compound 5, 3 mg). Fr1 and Fr2 were further purified by crystallization from CH2Cl2/MeOH to give the compounds 1 (7 mg) and 2 (3 mg), respectively. Fr4 was subjected to a Sephadex LH-20 column with MeOH to yield compounds 3 (5 mg) and 4 (2 mg).
The structures of all compounds were determined using spectroscopic analysis. High Resolution-Mass Spectrometry (HR-ESI-MS) spectra were measured on a TripeTOF 4600 mass analyzer (Bruker, United States). 1H/13C-nuclear magnetic resonance (NMR) data were acquired using Agilent DD2 600Hz spectrometer (Agilent, United States) with tetramethylsilane (TMS) as an internal standard, and chemical shifts (δ) were reported as parts per million (ppm) values.
Antibacterial assay of compounds
The antibacterial activity of the pure compounds isolated from QTH17 were determined using previously described the method of filter paper dispersion (Xu et al., 2020) and the microdilution method of the minimum inhibitory concentrations (MICs) (Li et al., 2014). MICs of compounds were measured and determined at five different concentration gradients in disposable 96-well microtiter dishes. A stock solution of each compound (200 μg/mL) was further twofold diluted in LB broth and added separately into individual microplate wells (100 μL/well). Then, a standard amount of the tested microbes (1.0 × 106 CFU/mL) were added per well. Thus, the compounds were tested in the final concentration range of 100–3.125 μg/mL. The 96-well plates were incubated at 37°C for 12–14 h. The lowest concentration of compounds at which the bacterial growth was inhibited, was defined as MIC, as shown by no turbidity. Gentamicin sulfate was used as a positive control. Each determination was performed three times.
Results
Diversity of cultivable fungi
In this study, a total of 42 fungi were isolated from the guts of C. Servilia using the culture-dependent method (Table 1 and Figure 1). All fungal isolates were identified by the ITS1/IST4 region of the 5.8S rDNA gene and comparison with available sequences in GenBank. Sequence analysis revealed that all fungal isolates were classified into 2 fungal phyla, 4 classes, 6 orders, 9 families, 13 genera. Ascomycota was the predominant fungi with a proportion of 90.48%. Thirty-eight isolates were grouped into three classes [Eurotiomycetes (47.62%), Sordariomycetes (23.81%), and Dothideomycetes (19.05%)] within the phylum Ascomycota. The other 4 strains (9.52%) were distributed in the Zygomycete within the phylum Zygomycota.
The largest number (20) of the strains belonged to the class Eurotiomycetes and was only distributed in Eurotiales order. The most common genera identified was Talaromyces in this study. In total, 10 strains were in the genus Talaromyces and identified as T. fusiformis (1 isolate), T. marneffei (1 isolate), T. pinophilus (4 isolates), and T. purpureogenus (4 isolates). Among them, the strain QTH31 showed only 98% similarity to T. pinophilus, which suggested a potential new species. Six isolates of the family Aspergillaceae were mainly concentrated in the genus Aspergillus. Three strains showed highly similar to A. flavus with an identity of 99%. Another three strains belonged to the genera Aspergillus and had 100% similar sequences to Aspergillus sp., A. japonicus, and A. ochraceus, respectively. The last four strains belonged to the genus Penicillium, and three strains of them were identified as P. citrioviride.
Another representative class was Sordariomycetes, including Hypocreales, and Xylariales orders. Eight strains were in the genus Trichoderma and were identified as Trichoderma sp. (2 isolates), T. atroviride (1 isolate), T. ghanense (4 isolates), and T. virens (1 isolate). The other one strain belonging to the family Hypocreaceae was identified as Fusarium sp. with an identity of 99%. The last one strain belonging to Arthrinium genus showed similar to Arthrinium sp. with an identity of 99%.
The strains of Dothideomycetes (8 isolates) were assigned to two orders, including the Pleosporales (7 isolates) and Cladosporiales (1 isolate). The three strains in Pleosporaceae showed highly similar to Curvularia lunata with more than 99% identity. Both strains belonging to Pleosporaceae were identified as Alternaria alternata with more than 99% identity. The strain QTY1 showed only 98% similarity to A. alternata, which was regarded as a potential new species. In addition, two strains of Pleosporales were identified as Bipolaris sp. and Phoma sp., respectively. Only one strain belonging to the order Cladosporiales was grouped into the genus Cladosporium with an identity of 99% match to Cladosporium sp.
Finally, the last four strains of class Zygomycete belonging to the phylum Zygomycota were distributed in the same order Mucorales. Both strains belonging to Mucoraceae were identified as Rhizomucor variabilis with an identity of 99%. The other two strains exhibited a sequence match of 99% to Mucor irregularis.
Culture-independent community analysis
To investigate the gut fungal community composition of C. Servilia, the culture-independent method was conducted. In total, 244,994 final reads with an average length of 290 bp were obtained from three samples, and 376 OTUs were acquired (QTC1, QTC2, QTC3) (Supplementary Table 1). Rarefaction curves showed that all samples reached stable values, indicating that the sequencing depth was sufficient (Supplementary Figure 1). In terms of alpha diversity, all samples displayed similar levels of diversity, including the Ace, Chao1, Shannon, and Simpson diversity indexes (Supplementary Table 1). The fungal community composition of different samples was estimated at the phylum and genus levels. The OTUs from all three samples were primarily assigned to the phyla Ascomycota (99.19%) and Basidiomycota (0.43%), and Ascomycota was the most abundant phylum (Figure 2A). At the genus level, 136 genera were identified across investigated gut samples (Supplementary Table 2). Among which, Wojnowiciella (52.51%) and Phoma (45.45%) were the main dominant fungal genera in gut fungi (Figure 2B). Moreover, several genii of cultivable filamentous fungi were also found in the culture-independent method, such as the genus Trichoderma, Curvularia, Fusarium, Bipolaris, Cladosporium, Rhizomucor, Mucor, Alternaria, Penicillium, Aspergillus, and Talaromyces, etc. (Supplementary Table 2).
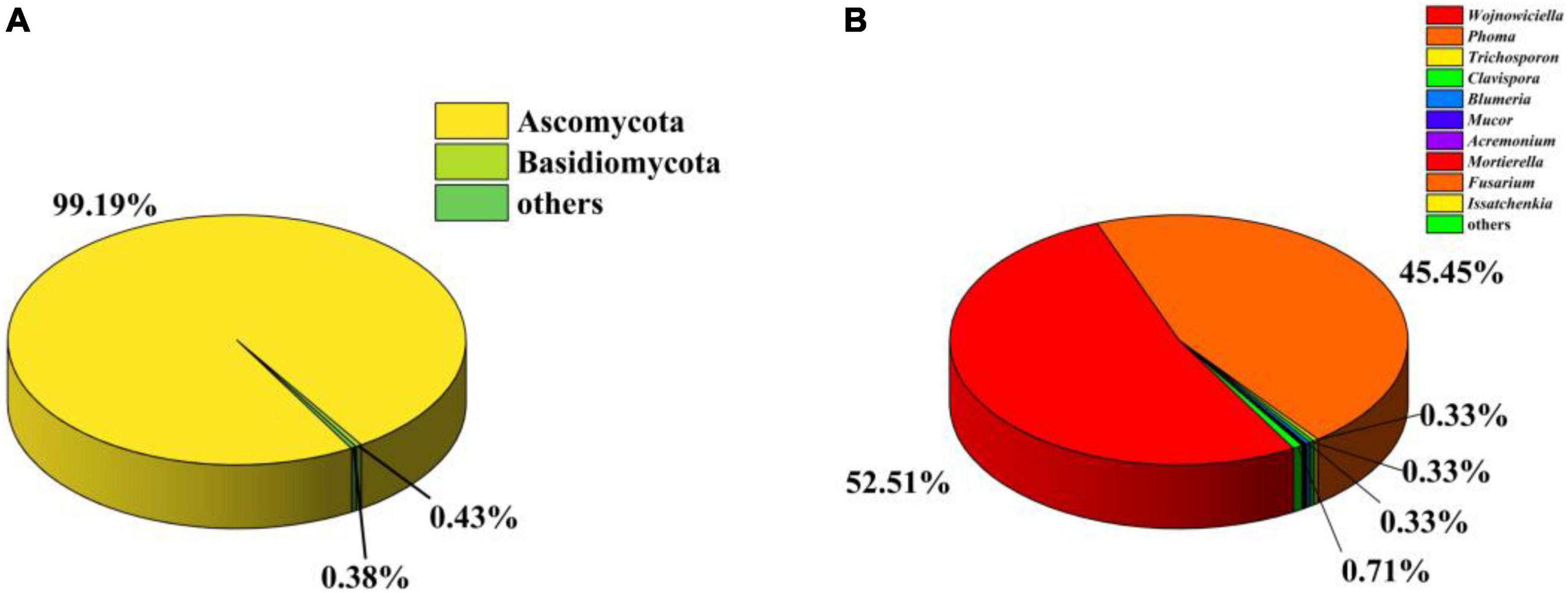
Figure 2. Analysis of culture-independent microbial communities. Relative abundance of phylum (A) and genus (B) within gut fungal communities.
Antibacterial activities of cultivable fungi
The antibacterial results of crude extracts of representative strains from 11 different genera were shown in Table 2. The results showed that five fungal crude extracts (QTH14, QTH15, QTH17, QTH21, and QTH27) exhibited inhibitory activities against at least one of the bacterial strains under the concentration of 30 μg/filter paper. Among them, QTH17 displayed potent antibacterial activities against E. coli, M. tetragenus and S. aureus with the disc diameter of IZD of 6.50, 10.80, and 8.70 mm, respectively, which were slightly weaker than those of the positive gentamicin sulfate with the IZD of 13.60, 16.50, and 12.30 mm, respectively.
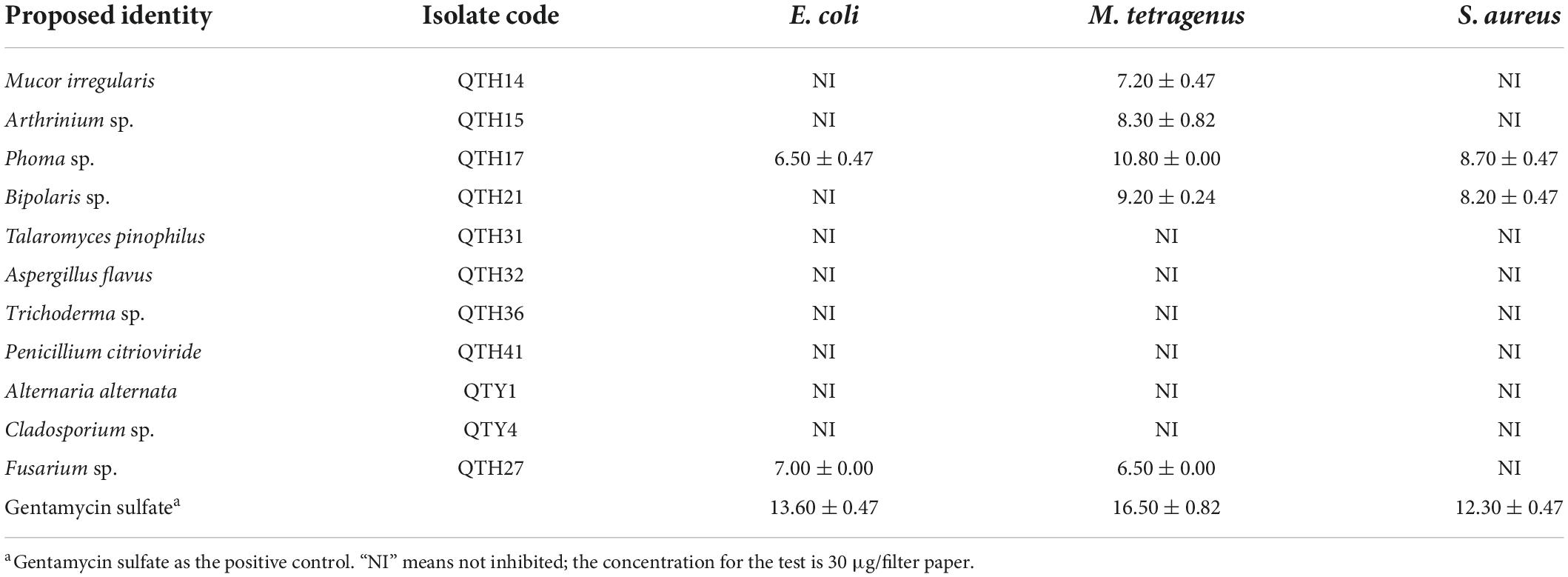
Table 2. Inhibition zone diameters of inhibiting effects of fungal extracts on three tested bacteria (mm).
Separation and identification of the compounds of QTH17
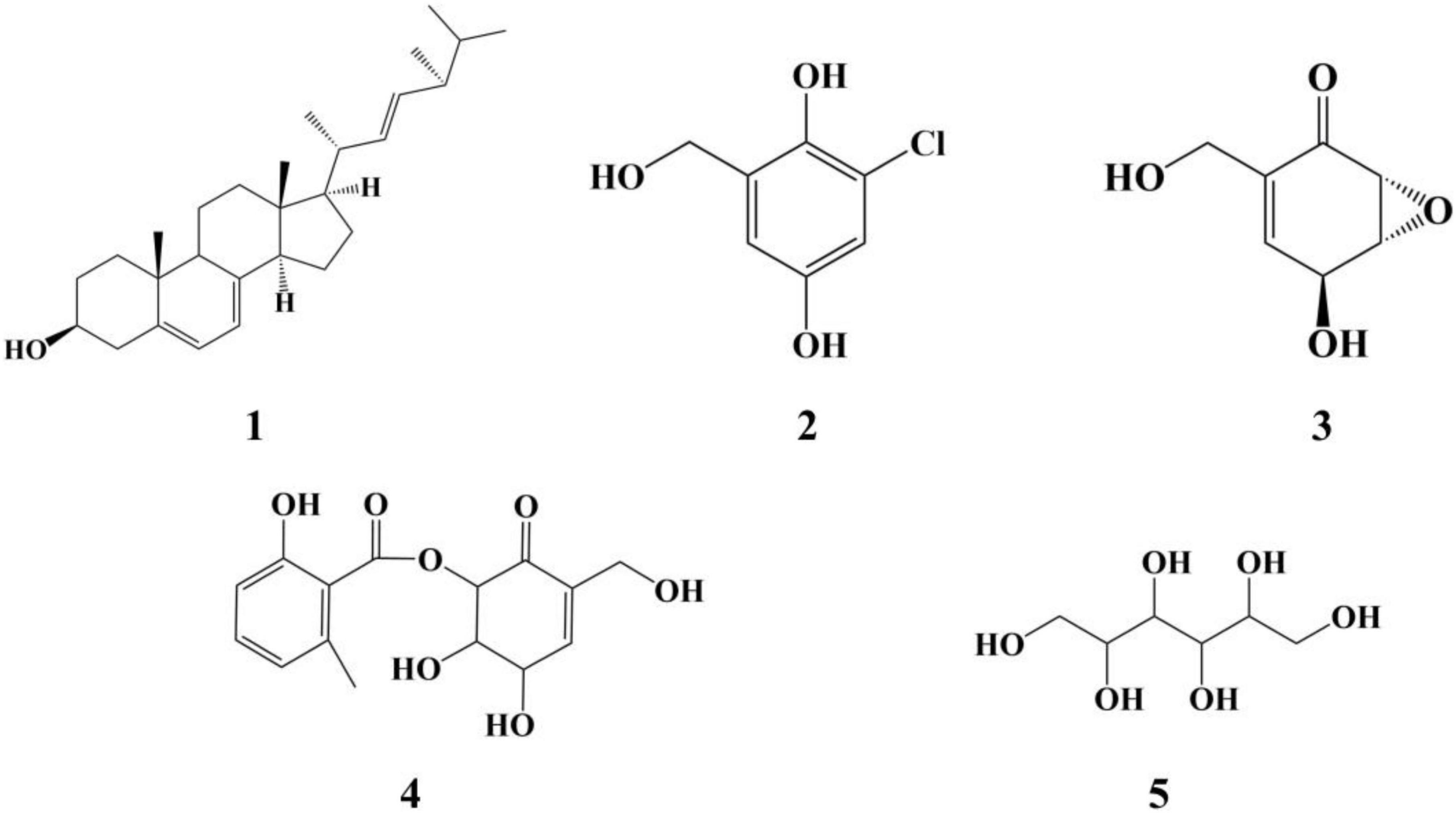
Five compounds were purified from the ME liquid fermentation product of Phoma sp. QTH17 (Figure 3), and their structures were determined to be ergosterol (1) (Huang et al., 2011), 3-chlorogentisyl alcohol (2) (Li et al., 2005), epoxydon (3) (Venkatasubbaiah et al., 1992a), epoxydon 6-methylsalicylate ester (4) (Venkatasubbaiah and Chilton, 1992b), and mannitol (5) (Branco et al., 2010) by spectroscopic data (Supplementary Figures 2–9) analyses, and the comparison of their derivative data in the literature.
Antibacterial activities of compounds
The antibacterial activities of the compounds isolated from QTH17 were shown in Table 3. The results revealed that three compounds (2–4) exhibited inhibitory activities against at least one of the bacterial strains under the concentration of 30 μg/filter paper. Specifically, compound 3 exhibited potent antibacterial activities against E. coli, M. tetragenus, and S. aureus with the IZD of 7.00, 14.00, and 12.50 mm, respectively, which were slightly weaker than those of the positive gentamicin sulfate with the IZD of 11.13, 18.30, and 12.13 mm. Compound 2 exhibited moderate inhibitory activities against M. tetragenus with the IZD of 9.00 mm, while it had no inhibition effect on E. coli and S. aureus. Compound 4 only displayed potent inhibitory activity against S. aureus with the IZD of 11.00 mm. However, compounds 3 and 4 lacked activities against three tested strains with MIC values of more than 100 μg/mL.
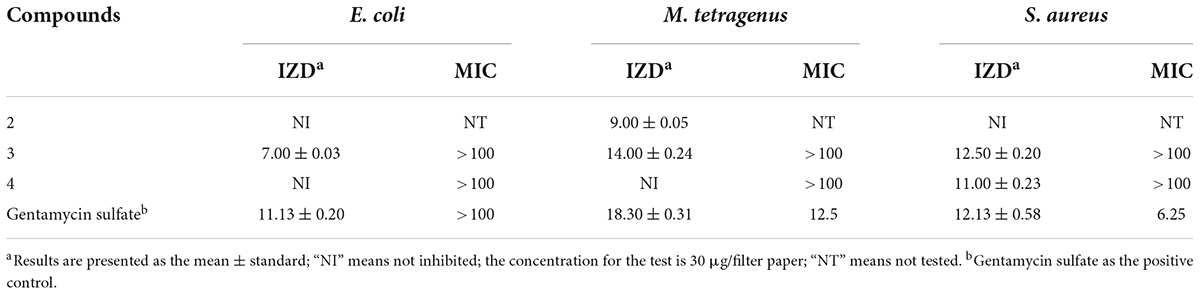
Table 3. Inhibition zone diameter (mm) and minimum inhibitory concentration (MIC) values (μg/mL) of compounds against three tested bacteria.
Discussion
The insect gut contains a vast number of microbes, including bacteria, fungi, and viruses, many of which influence the health and development of their hosts (Muratore et al., 2020). However, the fungal component of the microbiota is frequently overlooked compared to bacteria (Engel and Moran, 2013). Insect-associated fungi have been an essential source of new microbial resources and bioactive compounds (Zhang et al., 2011; Nagam et al., 2021; Tong et al., 2021). Culture-dependent and -independent methods are commonly used to investigate the fungal community composition of insect gut (Xu et al., 2020; Callegari et al., 2021). Forty-two fungi, including two potentially new species, were isolated and characterized by the culture-dependent method and molecular biological identification in this study. Concomitantly, the fungal community composition of the gut was further analyzed by the culture-independent method. Furthermore, five compounds were purified and characterized from Phoma sp. QTH17 has antibacterial activities. To our knowledge, this is the first report on understanding fungal community composition and screening of biologically active secondary metabolites of gut-associated fungi isolated from the C. Servilia, which will provide a resource for microbial diversity and antibiotics.
Thirteen different genera were found in the guts of C. Servilia by the culture-dependent method, which was highly similar to the gut-associated fungi of Pantala flavescens larvae (Shao et al., 2015). Moreover, Penicillium, Aspergillus, and Cladosporium have been associated with other insects’ guts, such as termites, honeybees, beetles, and palm weevils (Moubasher et al., 2017; Xu et al., 2020). Trichoderma, Cladosporium, Penicillium, and Talaromyces were also found in the guts of aquatic insect larvae, which have cellulolytic potential (Belmont-Montefusco et al., 2020). Besides, C. lunata and A. alternata isolated from the gut have been reported as plant pathogens (Fahim Abbas et al., 2021). However, some culturable fungi are the first reported fungi of dragonfly gut, such as Bipolaris sp., Arthrinium sp., M. irregularis, T. marneffei, T. pinophilus, and T. purpureogenus. This study further confirmed the association between filamentous fungi and dragonflies.
However, isolation methods of culturable fungi still have some limitations. The composition of C. Servilia gut fungal community was accurately evaluated by the culture-independent method in this study. A total of 136 genus-level OTUs were detected in the dragonfly gut sample, whereas only thirteen genera were isolated by culture. This result provides the impetus for strategies to obtain more fungi from the dragonfly gut in future studies, for instance, media-fungus pairings (Oberhardt et al., 2015). Notably, the most frequently isolated cultivable fungi correspond with fungal OTUs in the culture-independent method. Nevertheless, only Phoma sp. was isolated from the main dominant fungal genera, Wojnowiciella was not isolated. Synthetic low-nutrient agar (SNA) medium at 25°C was used to isolate Wojnowiciella, which remains to be explored further (Marin-Felix et al., 2019). Additionally, due to the taxonomic resolution of fungal community has remained relatively low, some fungal OTUs were unidentified (Muratore et al., 2020).
To validate that gut-associated fungi represent a source of antimicrobials with activity against pathogenic bacteria, 11 different genera were tested in vitro assays using three different bacteria. However, there are only a few strains that have shown activity against pathogenic bacteria. For example, Fusarium sp. QTH27, Mucor irregularis QTH14, and Bipolaris sp. QTH21 exhibited weak antibacterial activity against M. tetragenus. Similarly, most fungi isolated from the gut of Pantala flavescens larvae had weak or no activity against pathogenic bacteria (Shao et al., 2015). Notably, Phoma sp. QTH17 has shown higher activities against both Gram-positive and Gram-negative bacteria. Moreover, Phoma spp. was a significant source of bioactive compounds, which have demonstrated a range of antibacterial, antifungal, and cytotoxic activity (Rai et al., 2020). Ergosterol (1) and mannitol (5) are common fungal metabolites and act as fungal cell membrane growth factors and metabolic stores, respectively (de Lira Mota et al., 2012; Meena et al., 2015). 3-Chlorogentisyl alcohol (2), epoxydon (3), and epoxydon 6-methylsalicylate ester (4) have been isolated from Phoma spp. (Venkatasubbaiah et al., 1992a; Nenkep et al., 2010).
The antibacterial bioassay showed that compound 3 showed the moderate antibacterial activities against E. coli, M. tetragenus, and S. aureus using the filter paper dispersion method, but no inhibition was found in the MIC test with the maximum concentration of 100 μg/mL. The result was consistent with previous report that compound 3 gave the MIC value of 128 μg/mL against S. aureus (Trisuwan et al., 2008). Additionally, three bacterial strains were not susceptible to compound 4 using the microdilution method. Therefore, MIC values might better mirror the antibacterial activity of compound against the tested strains (Di Lodovico et al., 2020).
Conclusion
Here, the diversity of the gut-associated fungi isolated from the C. Servilia was analyzed using both culture-dependent and culture-independent methods. This study improved our understanding of gut-associated fungi and further increase fungal species from C. Servilia. Antibacterial activity assays showed that several of these fungi have antibacterial activities, among which the fungus Phoma sp. QTH17 was the most prominent. In addition, five known compounds were identified. These results suggest that dragonfly gut-associated fungi represent a promising and underexplored resource for exploring antibiotics.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/ (ON248244–ON248285 and PRJNA827522).
Author contributions
YZ designed the research and supervised the study. PC, LL, SS, ZH, and KK performed the experiments and analyzed the data. PC and LL analyzed and wrote the manuscript. All authors revised the manuscript and approved the final version for submission.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) (32270015), the Anhui Province Natural Science Funds for Distinguished Young Scholars (2108085J18), and the Open Fund of State Key Laboratory of Tea Plant Biology and Utilization (SKLTOF20200104).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.970990/full#supplementary-material
References
Belmont-Montefusco, E. L., Nacif-Marçal, L., Assunção, E. N., Hamada, N., and Nunes-Silva, C. G. (2020). Cultivable cellulolytic fungi isolated from the gut of amazonian aquatic insects. Acta Amazonica 50, 346–354. doi: 10.1590/1809-4392202000902
Branco, A., Santos, J. D. G., Pimentel, M. M. A. M., Osuna, J. T. A., Lima, L. S., and David, J. M. (2010). D-mannitol from Agave sisalana biomass waste. Ind. Crop Prod. 32, 507–510. doi: 10.1016/j.indcrop.2010.06.025
Callegari, M., Crotti, E., Fusi, M., Marasco, R., Gonella, E., Noni, I. D., et al. (2021). Compartmentalization of bacterial and fungal microbiomes in the gut of adult honeybees. npj Biofilms Microbiom. 7:42. doi: 10.1038/s41522-021-00212-9
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittingeret, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Correa, Y., Cabanillas, B., Jullian, V., Álvarez, D., Castillo, D., Dufloer, C., et al. (2019). Identification and characterization of compounds from Chrysosporium multifidum, a fungus with moderate antimicrobial activity isolated from Hermetia illucens gut microbiota. PLoS One 14:e0218837. doi: 10.1371/journal.pone.0218837
de Lira Mota, K. S., de Oliveira Pereira, F., de Oliveira, W. A., Lima, I. O., and de Oliveira Lima, E. (2012). Antifungal activity of Thymus vulgaris L. Essential Oil and its constituent phytochemicals against Rhizopus oryzae: Interaction with ergosterol. Molecules 17, 14418–14433. doi: 10.3390/molecules171214418
Di Lodovico, S., Menghini, L., Ferrante, C., Recchia, E., Castro-Amorim, J., Gameiro, P., et al. (2020). Hop extract: An efficacious antimicrobial and anti-biofilm agent against multidrug-resistant staphylococci strains and cutibacterium acnes. Front. Microbiol. 11:1852. doi: 10.3389/fmicb.2020.01852
Doyle, J. J. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15. doi: 10.2307/4119796
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgar, R. C. (2013). UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects-diversity in structure and function. Fems. Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Fahim Abbas, M., Batool, S., Khaliq, S., Mubeen, S., Azziz-ud-Din, Ullah, N., et al. (2021). Diversity of fungal pathogens associated with loquat and development of novel virulence scales. PLoS One 16:e0257951. doi: 10.1371/journal.pone.0257951
Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using thebootstrap. Evolution 39, 783–791.
Gibson, C. M., and Hunter, M. S. (2010). Extraordinarily widespread and fantastically complex: Comparative biology of endosymbiotic bacterial and fungal mutualists of insects. Ecol. Lett. 13, 223–234. doi: 10.1111/j.1461-0248.2009.01416.x
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Handel, S., Wang, T. F., Yurkov, A. M., and König, H. (2016). Sugiyamaella mastotermitis sp. nov. And Papiliotrema odontotermitis f.a., sp. nov. from the gut of the termites Mastotermes darwiniensis and Odontotermes obesus. Int. J. Syst. Evol. Micr. 66, 4600–4608. doi: 10.1099/ijsem.0.001397
Huang, L. Y., Cao, Y. Y., Xu, H., and Chen, G. N. (2011). Separation and purification of ergosterol and stigmasterol in Anoectochilus roxburghii (wall) Lindl by high-speed counter-current chromatography. J. Sep. Sci 34, 385–392. doi: 10.1002/jssc.201000577
Joshi, S., Poorten, N. V. D., Sumanapala, A., Nielsen, E., Patel, J., Nielsen, B., et al. (2020). New records of polymorphism in Asian Libellulid dragonflies (Insecta: Odonata). Int. J. Odonatol. 2020, 337–356. doi: 10.1080/13887890.2020.1788999
Li, S., Shao, M. W., Lu, Y. H., Kong, L. C., Jiang, D. H., and Zhang, Y. L. (2014). Phytotoxic and antibacterial metabolites from Fusarium proliferatum ZS07 isolated from the gut of long-horned grasshoppers. J. Agric. Food Chem. 62, 8997–9001. doi: 10.1021/jf502484n
Li, Y., Li, X., and Son, B. W. (2005). Antibacterial and radical scavenging epoxycyclohexenones and aromatic polyols from a marine isolate of the fungus Aspergillus. Nat. Prod. Sci. 11, 136–138.
Liu, G., Zheng, X., Long, H., Rao, Z., Cao, L., and Han, R. (2021). Gut bacterial and fungal communities of the wild and laboratory-teared thitarodes larvae, host of the chinese medicinal fungus ophiocordyceps sinensis on tibetan plateau. Insects 12:327. doi: 10.3390/insects12040327
Long, Y. H., Zhang, Y., Huang, F., Liu, S., Gao, T., and Zhang, Y. L. (2022). Diversity and antimicrobial activities of culturable actinomycetes from Odontotermes formosanus (Blattaria: Termitidae). BMC Microbiol. 22:80. doi: 10.1186/s12866-022-02501-5
Lu, Y. H., Jin, L. P., Kong, L. C., and Zhang, Y. L. (2017). Phytotoxic, antifungal and immunosuppressive metabolites from Aspergillus terreus QT122 isolated from the gut of dragonfly. Curr. Microbiol. 74, 1–6. doi: 10.1007/s00284-016-1157-y
Makonde, H. M., Boga, H. I., Osiemo, Z., Mwirichia, R., Stielow, J. B., Göker, M., et al. (2013). Diversity of termitomyces associated with fungus-farming termites assessed by cultural and culture-independent methods. PLoS One 8:e56464. doi: 10.1371/journal.pone.0056464
Maoka, T., Kawase, N., Ueda, T., and Nishida, R. (2020). Carotenoids of dragonflies, from the perspective of comparative biochemical and chemical ecological studies. Biochem. Syst. Ecol. 89:104001. doi: 10.1016/j.bse.2020.104001
Marin-Felix, Y., Hernández-Restrepo, M., Wingfield, M. J., Akulov, A., Carnegie, A. J., Cheewangkoon, R., et al. (2019). Genera of phytopathogenic fungi: GOPHY 2. Stud. Mycol. 92, 47–133. doi: 10.1016/j.simyco.2018.04.002
Meena, M., Prasad, V., Zehra, A., Gupta, V. K., and Upadhyay, R. S. (2015). Mannitol metabolism during pathogenic fungal–host interactions under stressed conditions. Front. Microbiol. 6:1019. doi: 10.3389/fmicb.2015.01019
Meriggi, N., Paola, M. D., Cavalieri, D., and Stefanini, I. (2020). Saccharomyces cerevisiae-insects association: Impacts, biogeography, and extent. Front. Microbiol. 11:1629. doi: 10.3389/fmicb.2020.01629
Moubasher, A. H., Abdel-Sater, M. A., and Zeinab, S. (2017). Yeasts and filamentous fungi inhabiting guts of three insect species in Assiut. Egypt. Mycosphere 8, 1297–1316. doi: 10.5943/mycosphere/8/9/4
Muratore, M., Sun, Y., and Prather, C. (2020). Environmental nutrients alter bacterial and fungal gut microbiomes in the common Meadow Katydid. Orchelimum vulgare. Front. Microbiol. 11:557980. doi: 10.3389/fmicb.2020.557980
Nagam, V., Aluru, R., Shoaib, M., Dong, G. R., Li, Z., Pallaval, V. B., et al. (2021). Diversity of fungal isolates from fungus-growing termite Macrotermes barneyi and characterization of bioactive compound from Xylaria escharoidea. Insect Sci. 28, 392–402. doi: 10.1111/1744-7917.12799
Nair, A., and Agashe, D. (2016). Host-specific spatial and temporal variation in culturable gut bacterial communities of dragonflies. Curr. Sci. 110, 1513–1523. doi: 10.18520/cs/v110/i8/1513-1523
Nenkep, V. N., Yun, K., Li, Y., Choi, H. D., Kang, J. S., and So, B. W. (2010). New production of haloquinones, bromochlorogentisylquinones A and B, by a halide salt from a marine isolate of the fungus Phoma herbarum. J. Antibiot. 63, 199–201. doi: 10.1038/ja.2010.15
Nobles, S., and Jackson, C. R. (2020). Effects of life stage, site, and species on the dragonfly gut microbiome. Microorganisms 8:183. doi: 10.3390/microorganisms8020183
Oberhardt, M. A., Zarecki, R., Gronow, S., Lang, E., Klenk, H.-P., Gophna, U., et al. (2015). Harnessing the landscape of microbial culture media to predict new organism–media pairings. Nat. Commun. 6:8493. doi: 10.1038/ncomms9493
Park, J. M., You, Y. H., Back, C. G., Kim, H. H., Ghim, S. Y., and Park, J. H. (2018). Fungal load in Bradysia agrestis, a phytopathogen-transmitting insect vector. Symbiosis 74, 145–158. doi: 10.1007/s13199-017-0494-3
Pereira, S. J., Costa, R. R., Nagamoto, N. S., Forti, L. C., Pagnocca, F. C., and Rodrigues, A. (2016). Comparative analysis of fungal communities in colonies of two leaf-cutting ant species with different substratum preferences. Fungal. Ecol. 21, 65–78. doi: 10.1016/j.funeco.2016.03.004
Rai, M., Gade, A., Zimowska, B., Ingle, A. P., and Ingle, P. (2020). Harnessing the potential of novel bioactive compounds produced by endophytic phoma spp. - biomedical and agricultural applications. Acta Sci. Pol Hortoru 19, 31–45. doi: 10.24326/asphc.2020.6.3
Sarikaya Bayram, Ö, Bayram, Ö, Valerius, O., Park, H. S., Irniger, S., Gerke, J., et al. (2010). LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet. 6:e1001226. doi: 10.1371/journal.pgen.1001226
Shao, M. W., Lu, Y. H., Miao, S., Zhang, Y., Chen, T. T., and Zhang, Y. L. (2015). Diversity, bacterial symbionts and antibacterial potential of gut-associated fungiisolated from the Pantala flavescens Larvae in China. PLoS One 10:e0134542. doi: 10.1371/journal.pone.0134542
Stefanini, I. (2018). Yeast-insect associations: It takes guts. Yeast 35, 315–330. doi: 10.1002/yea.3309
Tauber, J. P., Nguyen, V., Lopez, D., and Evans, J. D. (2019). Effects of a resident yeast from the honeybee gut on immunity, microbiota, and nosema disease. Insects 10:296. doi: 10.3390/insects10090296
Tegtmeier, D., Hurka, S., Mihajlovic, S., Bodenschatz, M., Schlimbach, S., and Vilcinskas, A. (2021). Culture-independent and culture-dependent characterization of the black soldier fly gut microbiome reveals a large proportion of culturable bacteria with potential for industrial applications. Microorganisms 9:1642. doi: 10.3390/microorganisms9081642
Tong, Z. W., Xie, X. H., Wang, T. T., Lu, M., Jiao, R. H., and Ge, H. M. (2021). Acautalides A-C, neuroprotective diels-alder adducts from solid-state cultivated Acaulium sp. H-JQSF. Org. Lett. 23, 5587–5591. doi: 10.1021/acs.orglett.1c02089
Trisuwan, K., Rukachaisirikul, V., Sukpondma, Y., Preedanon, S., Phongpaichit, S., Rungjindamai, N., et al. (2008). Epoxydons and a pyrone from the marine-derived fungus Nigrospora sp. PSU-F5. J. Nat. Prod. 71, 1323–1326. doi: 10.1021/np8002595
Venkatasubbaiah, P., and Chilton, W. S. (1992b). An epoxydon-derived ester from a Phoma sp. pathogenic to rhubarb. J. Nat Prod. 1992, 639–643. doi: 10.1021/np50083a013
Venkatasubbaiah, P., Dyke, C. G. V., and Chilton, W. S. (1992a). Phytotoxic metabolites of phoma sorghina, a new foliar pathogen of pokeweed. Mycologia 84, 715–723. doi: 10.1080/00275514.1992.12026197
Vera-Ponce de León, A., Sanchez-Flores, A., Rosenblueth, M., and Martínez-Romero, E. (2016). Fungal community associated with Dactylopius (Hemiptera: Coccoidea: Dactylopiidae) and its role in uric acid metabolism. Front. Microbiol. 7:954. doi: 10.3389/fmicb.2016.00954
Višňovská, D., Petr, P., Šigut, M., Kostovčík, M., Kolařík, M., Kotásková, N., et al. (2020). Caterpillar gut and host plant phylloplane mycobiomes differ: A new perspective on fungal involvement in insect guts. FEMS Microbiol. Ecol. 96:116. doi: 10.1093/femsec/fiaa116
Xia, X., Lan, B., Tao, X., Lin, J., and You, M. (2020). Characterization of Spodoptera litura gut bacteria and their role in feeding and growth of the host. Front. Microbiol. 11:1492. doi: 10.3389/fmicb.2020.01492
Xu, X., Shao, M. W., Yin, C. P., Mao, Z. C., Shi, Z. C., Yu, X. Y., et al. (2020). Diversity, bacterial symbionts, and antimicrobial potential of termite-associated fungi. Front. Microbiol. 11:300. doi: 10.3389/fmicb.2020.00300
Zhang, S. X., Sun, F. F., Liu, L. J., Bao, L. Y., Fang, W., Yin, C. P., et al. (2020). Dragonfly-associated Trichoderma harzianum QTYC77 is not only a potential biological control agent of Fusarium oxysporum f. sp. cucumerinum but also a source of new antibacterial agents. J. Agric. Food Chem. 68, 14161–14167. doi: 10.1021/acs.jafc.0c05760
Zhang, Y. L., Ge, H. M., Zhao, W., Dong, H., Xu, Q., Li, S. H., et al. (2008). Unprecedented immunosuppressive polyketides from Daldinia eschscholzii, a mantis-associated fungus. Angew. Chem. Int. Ed. Engl. 47, 5823–5826. doi: 10.1002/anie.200801284
Keywords: Crocothemis servilia, fungal diversity, insect-associated fungi, antibacterial activity, secondary metabolites
Citation: Cui P, Liu L, Huang Z, Shi S, Kong K and Zhang Y (2022) Diversity, antibacterial activity and chemical analyses of gut-associated fungi isolated from the Crocothemis servilia. Front. Microbiol. 13:970990. doi: 10.3389/fmicb.2022.970990
Received: 16 June 2022; Accepted: 15 August 2022;
Published: 16 September 2022.
Edited by:
Crisalejandra Rivera-Perez, Centro de Investigación Biológica del Noroeste (CIBNOR), MexicoReviewed by:
Cesar Salvador Cardona-Felix, Centro Interdisciplinario de Ciencias Marinas (IPN), MexicoSujogya Kumar Panda, KU Leuven, Belgium
Copyright © 2022 Cui, Liu, Huang, Shi, Kong and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinglao Zhang, emhhbmd5bEBhaGF1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Pu Cui1†
Pu Cui1† Yinglao Zhang
Yinglao Zhang