- Centre for Mechanical Engineering, Materials and Processes, Department of Life Sciences, University of Coimbra, Coimbra, Portugal
Gallium (Ga) is considered a high-tech Critical Metal, used in the manufacture of several microelectronic components containing either gallium arsenide (GaAs) or gallium nitride (GaN). The current high demand for this critical metal urges the development of effective recovery processes from secondary resources such as mine tailings or electronic recycling material. The importance of bioleaching as a biotechnological process to recover metals prompted this study, where an integrative approach combining experimental and genomic analysis was undertaken to identify potential mechanisms involved in bioleaching ability and strategies to cope with high metal(loid)s concentrations in five mine isolates. The Clusters of Orthologous Group (COG) annotation showed that the “amino acid transport and metabolism” [E] was the most predominant functional category in all genomes. In addition, the KEEG pathways analysis also showed predicted genes for the biosynthetic pathways of most amino acids, indicating that amino acids could have an important role in the Ga leaching mechanism. The presence of effective resistance mechanisms to Ga and arsenic (As) was particularly important in GaAs bioleaching batch assays, and might explain the divergence in bioleaching efficiency among the bacterial strains. Rhodanobacter sp. B2A1Ga4 and Sphingomonas sp. A2-49 with higher resistance, mainly to As, were the most efficient bioleaching strains under these conditions. In bioleaching assays using cell-free spent medium Arthrobacter silviterrae A2-55 with lower As resistance outperformed all the other stains. Overall, higher efficiency in Ga leaching was obtained in bioleaching assays using GaAs when compared to GaN.
Introduction
Gallium (Ga) is a rare element with an abundance of about 16 ppm in the Earth’s crust (Moskalyk, 2003) and due to its semiconducting properties the industrial usage of this element has gained great interest (Gray et al., 2013). Nowadays, Ga is widely used in a variety of industrial applications and primarily in electronics. This high-tech metal is used in the manufacture of several microelectronic components containing either gallium arsenide (GaAs) or gallium nitride (GaN). Due to the technological advances and a modern society increasingly reliant on electronic devices, Ga has been classified as a critical metal, as being of high economic importance, and simultaneously subject to high supply risks (Hayes and McCullough, 2018; Girtan et al., 2021). In a circular economy concept, Ga and other critical metals can be recovered from secondary sources such as mine tailing and end-of-life electronic equipment (e-waste) (Hagelüken et al., 2016; Işıldar et al., 2019). E-wastes contain high-tech critical metals with a concentration higher than that present in the primary ores, which turns them a remarkable resource for metal recovery (Mishra et al., 2021).
Biotechnology processes such as the bioleaching that take advantage of microbial activity or metabolites produced by them, to solubilize valuable metals from secondary sources have developed into a successful and expanding area (Olson et al., 2003; Sedlakova-Kadukova et al., 2020). Bioleaching offers a more efficient and ecological biotechnology, with lower costs when compared to traditional physicochemical methods (Gao et al., 2020). A diverse group of bacteria found ubiquitously, or isolated from metal contaminated sites such as mines, is able to solubilize metals due to the production of metabolic products (Işıldar et al., 2019; Mishra et al., 2021). Several Pseudomonas species (Li et al., 2015; Potysz et al., 2016; Biswal et al., 2019), Chromobacterium violaceum (Li et al., 2015), and Bacillus megaterium (Motaghed et al., 2014), are cyanogenic bacteria able to bioleach e-waste, targeting valuable metals and the platinum group of metals (PGM), i.e., Au, Ag, Pt, Pd, Rh (Işıldar et al., 2019). These cyanogenic bacteria produce hydrogen cyanide and form water-soluble metal cyanide complexes by reacting with metals containing solids such as e-waste (Faramarzi et al., 2004; Brandl et al., 2008). Heterotrophic bacteria can also contribute to the bioleaching of critical metals through the production of organic acids solubilizing metals, namely, acetic acid, lactic acid, formic acid, oxalic acid, citric acid, succinic acid, and gluconic acid (Bosecker, 1997; Işıldar et al., 2019). Bioleaching of Rare Earth Elements (REE) from waste materials by biogenic gluconic acid was carried out by members of Acinetobacter, Pseudomonas, and Gluconobacter genera (Işıldar et al., 2019). Studies on bioleaching of Ga by heterotrophic bacteria are scanty, however, Maneesuwannarat and co-workers reported the bioleach of Ga from GaAs by Cellulosimicrobium funkei (Maneesuwannarat et al., 2016a) and from GaN by Arthrobacter creatinolyticus (Maneesuwannarat et al., 2016b), respectively.
Bioleaching bacteria show a biphasic response to metals in the environment (Pourhossein and Mousavi, 2018). They have the ability to solubilize and mobilize metals. However, when metal concentration increases beyond certain levels, it becomes toxic and has a negative effect on bioleaching efficiency. These bacteria have to rely on metal resistance mechanisms to survive, since high metal concentrations disrupt cellular function by damaging vital enzymatic functions, by directly affecting DNA structure, membrane lipids and proteins, and by disturbing ion balance (Chandrangsu et al., 2017; Igiri et al., 2018). Several bacterial are able to cope with metals and resist their toxicity using a diversity of strategies such as: modification of the metal redox state, metals precipitation or sorption to the cell surface, uptake and intracellular chelation, efflux mediated by specific transporters or the secretion of metal chelating agents to the environment (Srivastava and Kowshik, 2013).
The advances in high-quality and high-throughput sequencing technologies are massively increasing the number of bacterial genomes available, providing an opportunity to gain insights into several biological processes, such as the prediction of microbial interactions, the genetic diversity and the metal resistance mechanisms (Cárdenas et al., 2016; Zhang et al., 2018). Genome analysis can also improve our knowledge of bioleaching microorganisms by predicting their metabolic potential to mobilize and bioleach valuable metals.
In this work, the genomic sequence of five heterotrophic bacteria, isolated from different Portuguese mines was obtained (Rugamonas sp. A1-17, Sphingomonas sp. A2-49, Arthrobacter silviterrae A2-55, Rhodanobacter sp. B2A1Ga4, and Undibacterium sp. Jales W-56), and a comprehensive comparative genomic analysis based on several bioinformatics tools was performed to identify potential mechanisms involved in bioleaching ability and strategies to cope with high metal(loid)s concentrations. The ability of these five strains to bioleach Ga from GaAs and GaN was also determined in different experimental conditions.
Materials and methods
Bacterial strains isolation and growth
In a survey for bacteria with leaching ability, five bacterial strains showed the ability to mobilize Gallium (Ga) from GaAs and GaN and were selected for this study. These strains were isolated from different Portuguese mines and were deposited in the University of Coimbra Bacteria Culture Collection (UCCCB). Rugamonas sp. A1-17 (UCCCB48), Sphingomonas sp. A2-49 (UCCCB49), and Arthrobacter sp. A2-55 (UCCCB146), were isolated from the water of an uranium mine (Urgeiriça), Rhodanobacter sp. B2A1Ga4 (UCCCB 112) was isolated from sediments of a tungsten mine (Panasqueira) and Undibacterium sp. Jales W-56 (UCCCB147) was isolated from sediments from a gold mine (Jales). The bacterial strains were cultured in modified Reasoner’s 2A broth medium (mR2Ab), containing per liter: 0.25 g of yeast extract, 0.5 g of tryptone, 1.0 g of glucose, 0.3 g of K2HPO4, 0.024 g of MgSO4, and 0.3 g of sodium pyruvate. The pH of the medium was adjusted to pH 6.0. Cultures were grown at 25°C with orbital shaking at 140 rpm for 8 h (late exponential phase of growth), 24 h (stationary phase of growth) and 48 h (late stationary phase of growth), respectively.
Genome sequencing, annotation, and strain identification
Total bacterial DNA extraction was performed using the E.Z.N.A.® Bacterial DNA Kit (Omega Bio-Tek) according to manufacturer instructions. Libraries of total genomic DNA were prepared using Nextera XT Preparation Kit (Illumina, San Diego, CA, United States) following the manufacturer’s instructions. Libraries were purified using HighPrep PCR Clean-up beads (MagBio Genomics, Inc.). Fragment analyzer 5200 (Agilent NGS Fragment 1-6000 pb methods) was used to check the fragment size distribution and molarity of each library. Nine-picomolar libraries were sequenced on an Illumina MiSeq System with 2 × 300 bp chemistry (MiSeq Reagent Kit v3). Pairing, trimming, and assembly based on Bruijn graphs were performed using CLC Genomics Workbench v9.5.4 (Qiagen) using default parameters. Genome sequences were annotated using RAST server (Aziz et al., 2008) and the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al., 2016). The draft genomes of Rugamonas sp. A1-17, Sphingomonas sp. A2-49, Arthrobacter silviterrae A2-55, and Undibacterium sp. Jales W-56 were deposited at DDBJ/ENA/GenBank under the accessions, JAJLPB000000000, JAJLPA000000000, JAJLOZ000000000, and JAJLQW000000000, respectively. The draft genome of Rhodanobacter sp. B2A1Ga-4 (JADBJR000000000) was already deposited at DDBJ/ENA/GenBank (Caldeira et al., 2021) and was used in this work. The taxonomic identification of the five mine isolates was based on the 16S rRNA gene by searching in the EzBioCloud Database the closest relative type strains (Yoon et al., 2017), and also by a genome-scale taxonomic analysis using the JSpeciesWS online services1 for calculation of the Average Nucleotide Identity based on the BLAST algorithm (ANIb) (Goris et al., 2007) and Tetranucleotide frequency correlation coefficient (TETRA) (Teeling et al., 2004) between the sequenced genomes and closely related type strains genomes. The type (strain) genome server (TYGS) (Meier-Kolthoff and Göker, 2019) was also used to confirm the taxonomic identities of the strains.
Structural and functional analyses of bacterial genomes
The sequenced genomes were functionally annotated in terms of cluster of orthologous groups (COGs) using online eggNOG-mapper v2 (Cantalapiedra et al., 2021). Additionally, genomes were also annotated in terms of Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology identifiers (KO) and mapped to KEGG pathways by the KEGG Automatic Annotation Server (KAAS) (Moriya et al., 2007), to gain insight about metabolic traits of the bacterial strains. The Venn diagram with shared and unique genes (COGs) among the 5 genomes was constructed using the Venn-Diagram free web tool of Bioinformatics and Evolutionary Genomics2.
The RAST Server (Aziz et al., 2008) and the COG functional annotation (Cantalapiedra et al., 2021) were used for identification of genes potentially involved in arsenic resistance and in iron transport. The resulting protein sequences were BLASTp searched against the non-redundant NCBI protein database for confirmation. Only those sequences that reported high hits to the correct functions were considered in our analyses and all of them were also checked in the NCBI Conserved Domains database.
Minimum inhibitory concentration
The Minimum Inhibitory Concentration (MIC) for Ga and arsenite [As(III)] was determined in 96 multiwell plates using the standard broth micro dilution method in mR2Ab medium. The Ga concentration in the assays ranged between 10 and 0.125 mM and As(III) concentration ranged between 2.0 and 0.125 mM. Metal stock solutions were prepared in a concentration of 0.2 M gallium (III) nitrate (GaN3O9, Alfa Aesar) and 0.5 M sodium arsenite [NaAsO2, Merck)] and were sterilized by filtration. The assays were performed in duplicate to ensure the reproducibility of the experiments. Bacterial growths were analyzed after 48 h of incubation at 25°C. MIC values were the lowest concentration of each tested metal that inhibited growth of the microorganism after the incubation time in the appropriate growth conditions (Andrews, 2002). The inhibition of growth was considered as the absence of difference between the assay and its respective non-inoculated control, in terms of absorbance.
Bioleaching experiments
The bioleaching experiments were performed in 20 ml of leaching medium containing either 10 mg of GaAs (Alfa Aesar) manually ground in a mortar, or 10 mg of GaN powder (Sigma Aldrich), at 25°C and 150 rpm in an orbital shaking incubator for 21 days. Three different types of bioleaching experiments were performed: (i) batch growth experiments, where the different strains were inoculated into mR2Ab medium (leaching medium) with an initial OD 600 of 0.1; (ii) pre cultivated bacteria cultures at different growth phases (late exponential, 8 h; stationary, 24 h, and late stationary, 48 h) were used as leaching medium; and (iii) cell-free spent medium of cultures from stationary (24 h) and late stationary (48 h) phases of growth, which were obtained by centrifugation (9,000 rpm, 15 min.), followed by filtering through 0.2 μm filters, were used as leaching medium. Control experiments with mR2Ab medium without bacteria and in presence of GaAs or GaN were run in parallel and in the same conditions. All bioleaching experiments were run in duplicate or triplicate.
Analytical methods
The pH was monitored at the beginning of each bioleach experiment and every 7 days until the end of the bioleaching period. pH variations were measured in a pH® meter, pHenomenal pH 1100L (VWR Chemicals). Soluble Ga amount in liquid medium was quantified spectrophotometrically by a modification of the bromopyrogallol red (BPR) method (Huang et al., 1997), optimized for Ga. Briefly, samples were prepared by adding 200 μl of buffer solution (1.25 ml of 0.2 M KCl; 2.65 ml of 0.2 M HCL in 50 ml deionized water), 70 μl of 0.2% SDS (sodium dodecyl sulfate), 100 μl of leaching medium and 200 μl of 0.01% BPR in deionized water to make up a final volume of 1 ml. Calibration curve was prepared using standard Ga concentrations from 0 to 300 μM diluted from a gallium(III) nitrate (GaN3O9, Alfa Aesar) stock solution (1 mM). Samples were quantified spectrophotometrically at 540 nm.
Statistical analysis
The Ga leaching capacities between all strains, incubation times, mineral substrate (GaAs, GaN) and growth phases of the biochemically active cells/cell-free spent media were analyzed by performing a multifactorial Permutational Multivariate Analysis of Variance (PERMANOVA) in order to determine significant differences between the factors analyzed (Bray-Curtis; Monte-Carlo test; PRIMER6 (v6.1.13) and PERMANOVA + (v1.0.3), PRIMER-E Ltd). Significant differences of leaching abilities between assays and controls were evaluated with a one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparison test using the software GraphPad Prism version 6.00 for Windows (GraphPad Software, San Diego, CA, United States)3.
Results
Taxonomic identification and genomic features of the assembled bacterial genomes
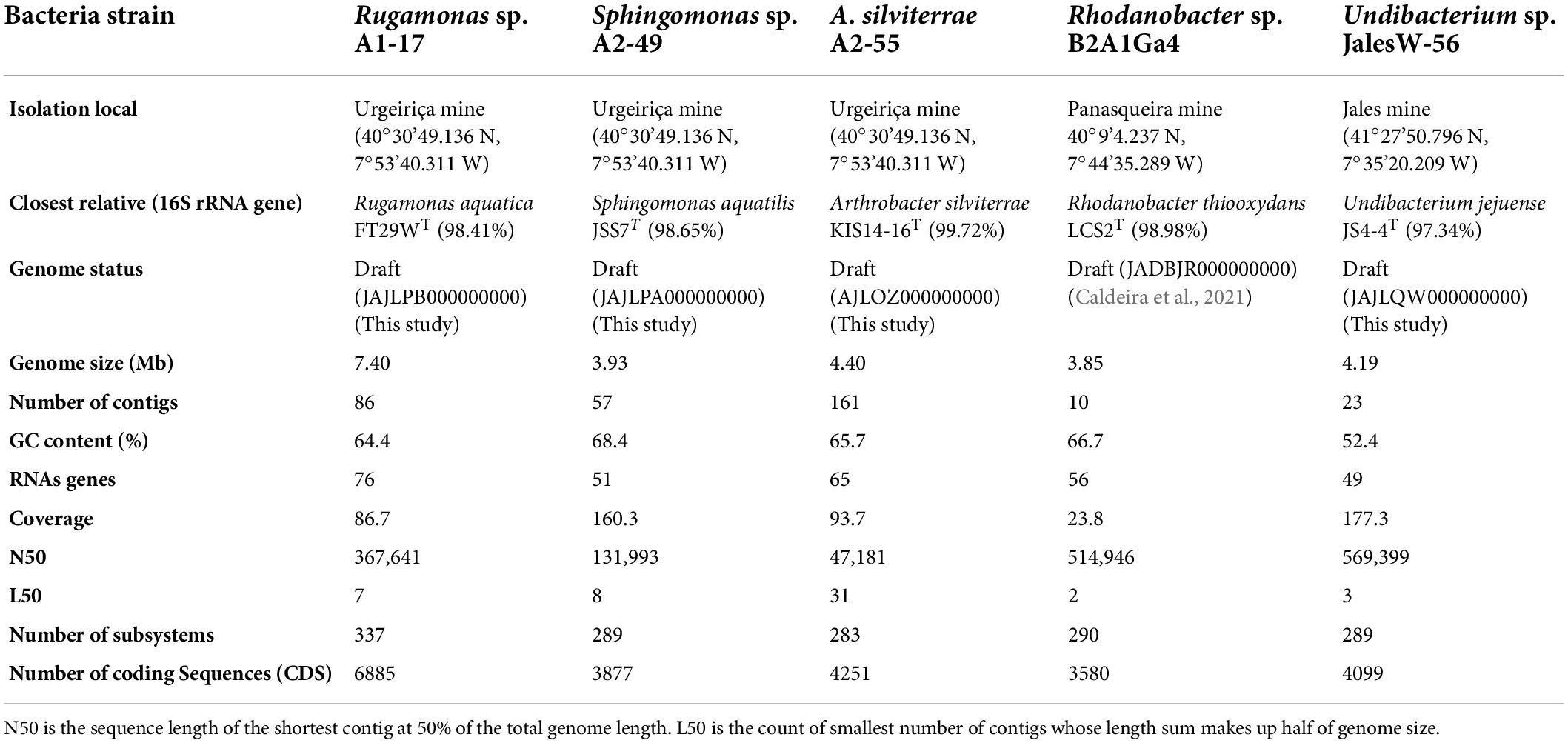
The taxonomic identification of the bacterial strains based on the 16S rRNA gene showed that the isolates from Urgeiriça mine A1-17, A2-49, and A2-55, were closely related to Rugamonas aquatica FT29WT with 98.41% of sequence similarity, Sphingomonas aquatilis JSS7T with 98.65% sequence similarity and Arthrobacter silviterrae KIS14-16T with 99.72% sequence similarity, respectively (Table 1). Strain Jales W-56 showed the highest 16S rRNA sequence similarity (97.34%) with Undibacterium jejuense JS4-4T. Strain B2A1Ga4 was already identified as a Rhodanobacter sp. (Caldeira et al., 2021) having as closest relative Rhodanobacter thiooxydans LCS2T with 98.98% of sequence similarity (Table 1). However, the values below the thresholds of 95.0% for ANIb and 0.989 for TETRA between the sequenced genomes and their closely related type strains genomes strongly indicates that they belong to different species (Supplementary Data 1). The only exception was obtained with strain A2-55, which have values of ANIb and TETRA with Arthrobacter silviterrae KIS14-16T above the threshold of 95.0% and 0.999, respectively, suggesting its inclusion in this species (Supplementary results). These results were further supported by the TYGS analysis, in particular by the digital DNA-DNA hybridization (dDDH) value of 80.3% between the genomes of strain A2-55 and Arthrobacter silviterrae KIS14-16T, higher than the threshold of 70.0% used to delineate species. All the other sequenced genomes showed dDDH values below the threshold 70.0% with their selected type strains genomes (Supplementary Data 2).
The genome features of the four genomes obtained in this study by Illumina sequencing, as well as the genome features of Rhodanobacter sp. B2A1Ga4 previously sequenced (Caldeira et al., 2021) are summarized in Table 1. Rugamonas sp. A1-17 has a larger genome with 7.4 Mb compared to the other genomes which sized between 3.85 and 4.40 Mb. The Undibacterium Jales W-56 genome has the lowest G + C content (52.4%); all the other genomes have G + C contents higher than 64% (Table 1).
Comparative genome analysis
Analysis of orthologous genes
The Clusters of Orthologous Group (COG) annotation of the five genomes showed that 78.5 to 83.2% of the coding sequences (CDS) were matched to putative proteins with known functions and assigned to 21 of the COG categories (Figure 1A). The most abundant COG broad functional category in all genomes was “Metabolism” with 45.0 to 52.8% of genes involved in this function. Among this COG category, the “amino acid transport and metabolism” [E] was the most predominant functional category (8.5–13.2%), followed by “carbohydrate transport and metabolism” [G] (4.4 to 11.2%) and “energy production and conversion” [C] (6.7 to 8.5%), which were also well-represented functional categories (Figure 1A). The other two COG broad functional categories “Information Storage and Processing” represented about 20.2 to 35.6% and “Cellular Process and Signaling” represented 19.2 to 29.3% of the COG categories, respectively (Figure 1A). A comparative analysis of the genomes showed that 772 orthologous genes were shared by all the strains (Figure 1B). The COG functional categories determined for these shared genes were very similar to those assigned for each individual genome. The “amino acid transport and metabolism” [E] was the most predominant functional category (10.4%) among the “Metabolism” function. However, the shared genes were enriched in the functional category “translation, ribosomal structure and biogenesis” [J] representing 14.9%, while in individual genomes this category represented only 4.2 to 7.2% (Figures 1A,B).
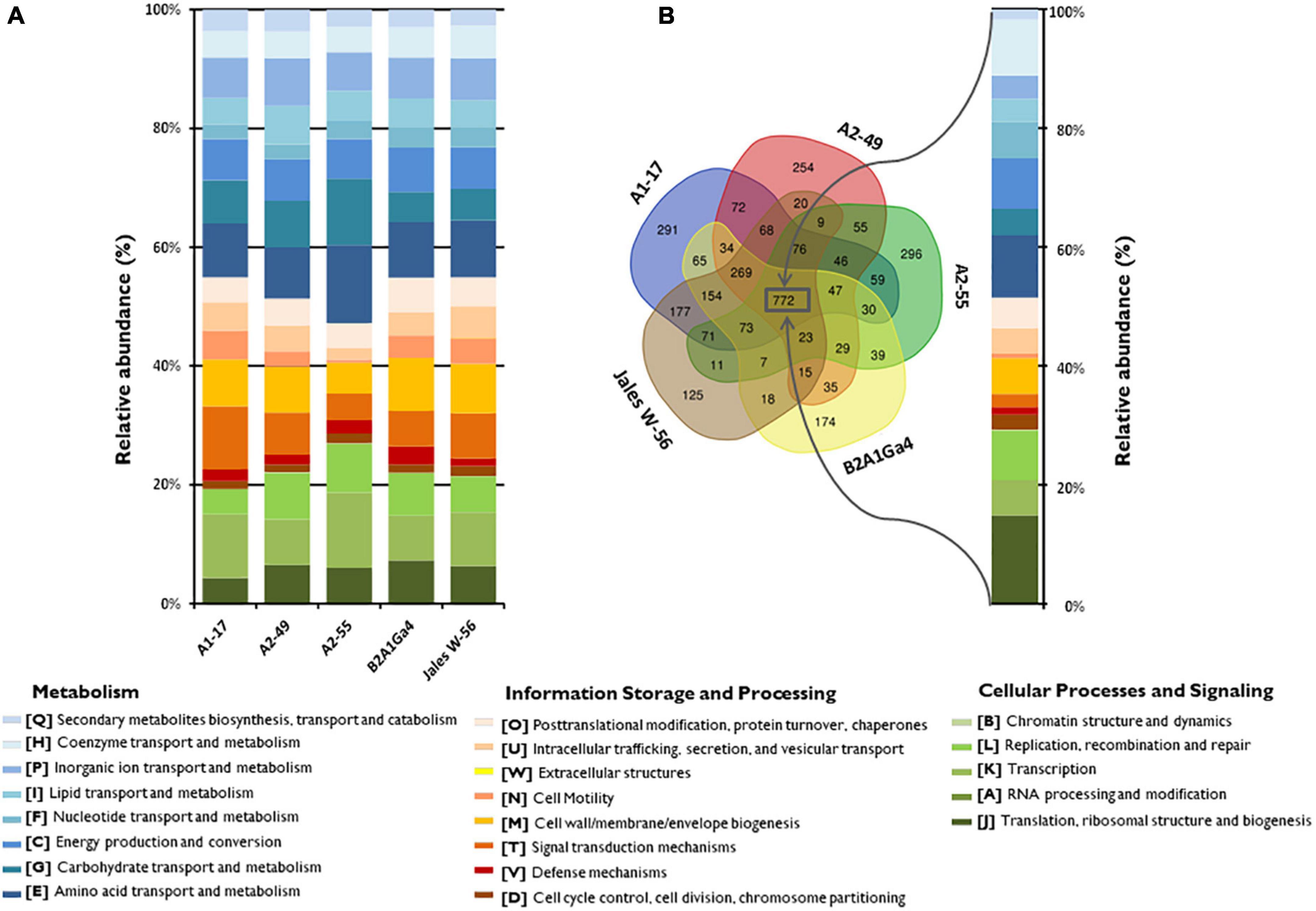
Figure 1. Distribution and comparison of COG functional categories in the analyzed genomes. (A) Relative abundances (%) of the 21 different COG functional categories in each genome according to three broad functional groups (Metabolism; Information Storage and Processing; Cellular Processes and Signaling). (B) Venn diagram showing the shared and unique gene clusters (COGs) among the genomes and the relative abundances (%) of the shared COG categories.
Kyoto encyclopedia of genes and genomes pathway enrichment analysis
The metabolic potential of the genomes under study was also investigated by functional annotation and pathway mapping in the KEGG Automatic Annotation Server (KAAS). In all genomes, the majority of the genes was related to metabolism pathways (64.0–71.6%), and the gene families related to carbohydrate metabolism (15.3–19.3%) and amino acid metabolism (13.0–16.4%) were the most abundant among the metabolic genes (Supplementary Data 3). These results corroborate the ones obtained by the COG functional categories distributions. Since amino acids metabolism was predominant in these bacterial strains the biosynthetic pathways of each amino acid were further characterized and compared among these organisms. KEEG modules for the biosynthesis of amino acids threonine (M00018), tryptophan (M00023), arginine (M00844, M00845), lysine (M00016), proline (M00015), valine (M00019), isoleucine (M00019, M00570), leucine (M00432), and also the biosynthesis pathway for chorismate (M00022) the precursor of aromatic amino acids were present in all genomes (Table 2). None of the strains had the complete pathway for the biosynthesis of methionine from aspartate (M00017). Although the biosynthesis of glutamate, aspartate, asparagine and alanine were not represented in KEEG modules since they are derived from intermediates of central metabolism in short pathways, most of the strains had the potential to synthesize these amino acids. Exceptions were observed with strain Sphingomonas sp. A2-49 where the pyruvate to alanine conversion pathway was absent and strain Undibacterium sp. Jales W-56 where the oxaloacetate conversion pathway to aspartate is also absent. Glutamine and glycine conversion pathways from glutamate and threonine, respectively, were also present in all genomes.
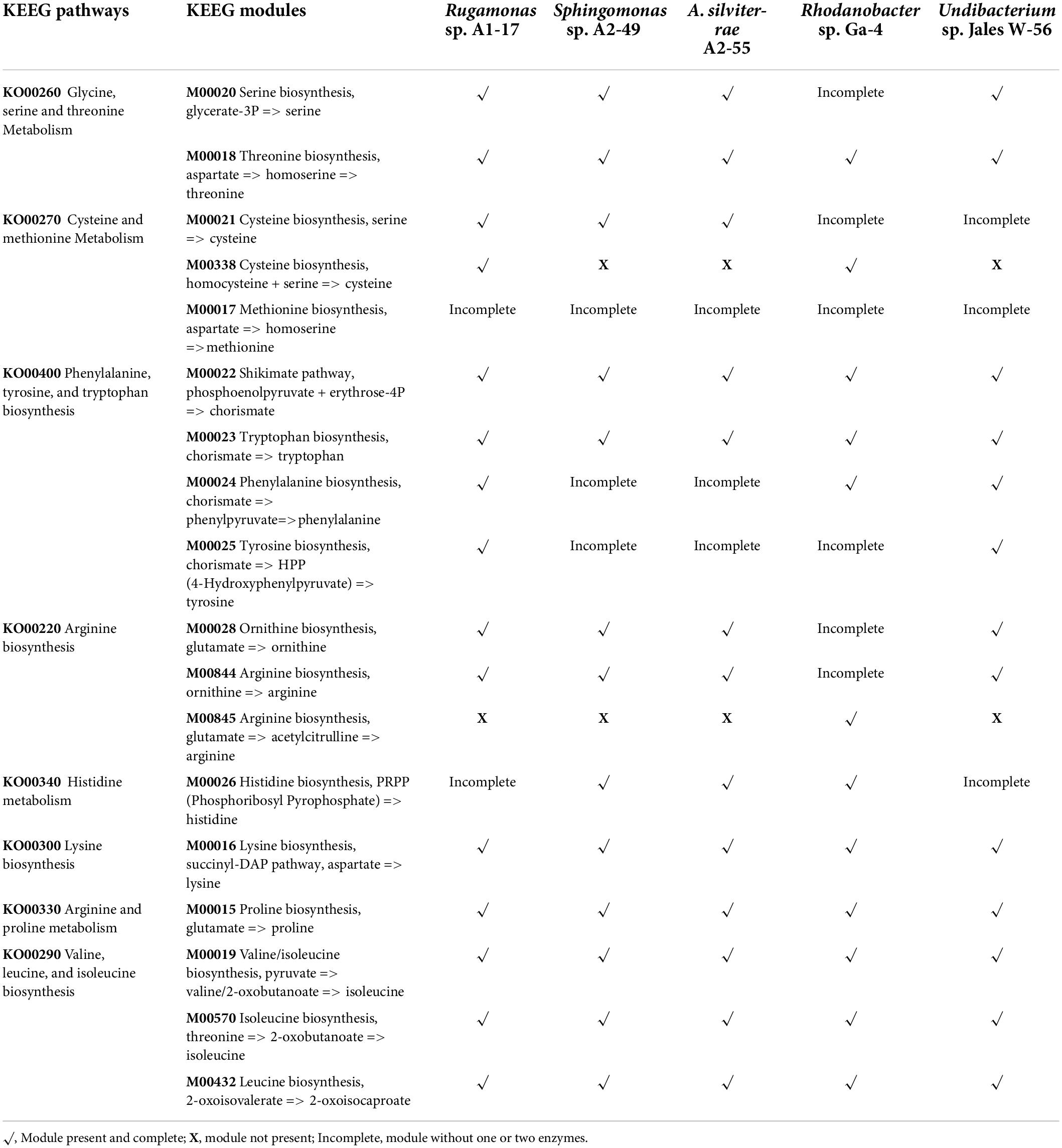
Table 2. KEEG pathways and modules for amino acids biosynthesis and metabolism determined by KAAS (KEGG Automatic Annotation Server).
Arsenic resistance gene analysis
The five genomes were analyzed to identify genes involved in arsenic resistance. The genomes of A. silviterrae A2-55 and Undibacterium sp. Jales W-56 showed simpler ars gene clusters, with an arsR gene encoding a metalloregulatory protein, an acr3 or aquaporin Z gene encoding an arsenite efflux pump and an arsC gene encoding an enzyme able of reducing arsenate to arsenite (Figure 2). The arsenic gene cluster arsRCBH was identified in the genomes of Rugamonas sp. A1-17 and Sphingomonas sp. A2-49. This late strain was the only one to have arsN2 genes in the ars gene clusters, however, the precise function of this gene has not yet been elucidated (Figure 2). The genome of Rhodanobacter sp. B2A1Ga4 showed a higher number and more complex ars gene clusters with a wide variety of gene configurations involved in arsenic resistance. This strain is the only one to present an arsRCDAB cluster, that besides the arsR and arsC, have arsD, arsA, and arsB genes adjacent, encoding an ArsA ATPase, an ArsD chaperone, and an arsenite efflux pump (Figure 2).
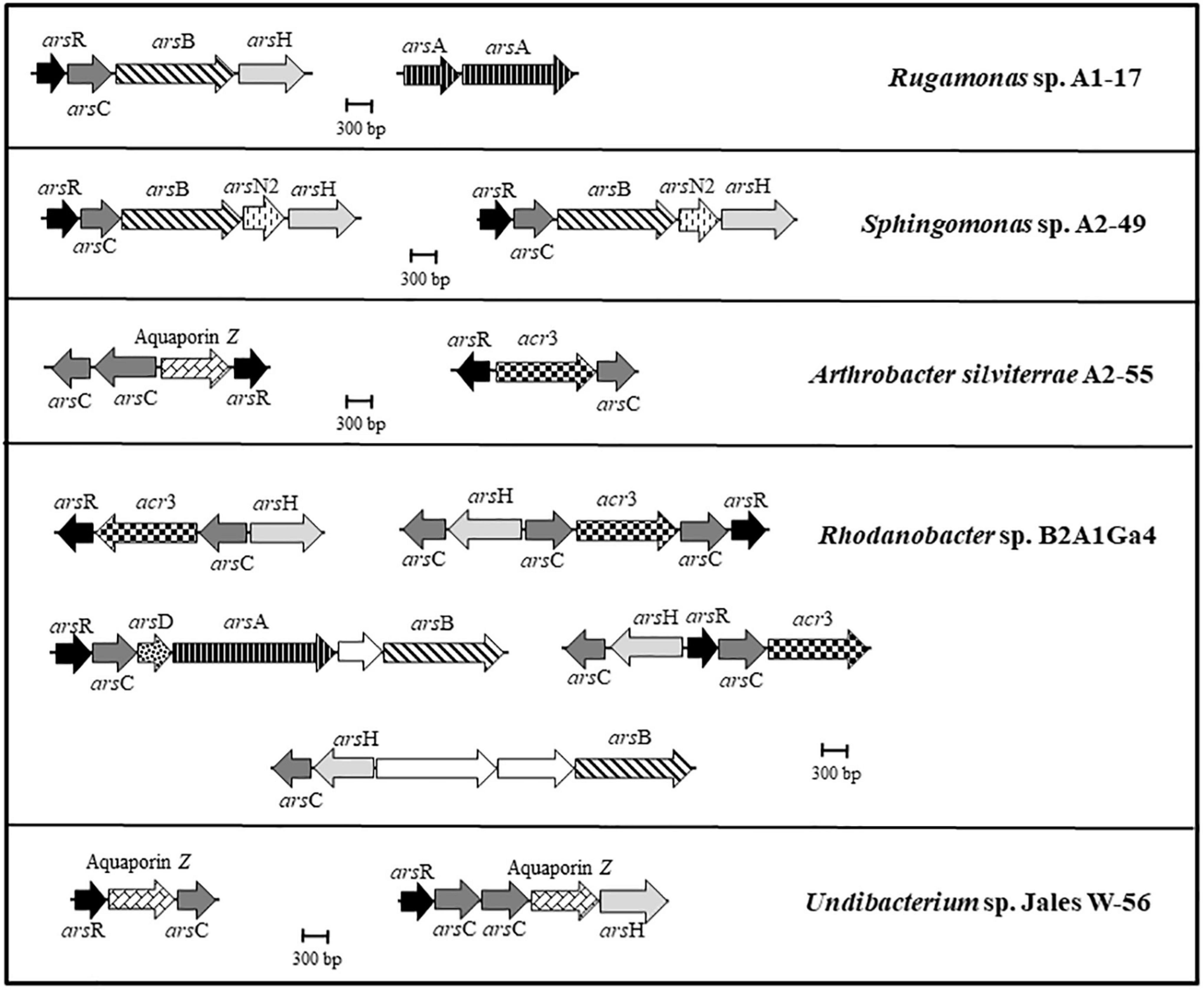
Figure 2. Genomic organization of the ars gene clusters in the analyzed genomes. Arrows represent different genes, and homologous genes are represented by the same color and pattern, except for the white arrows that represent genes identified as not being related to arsenic resistance.
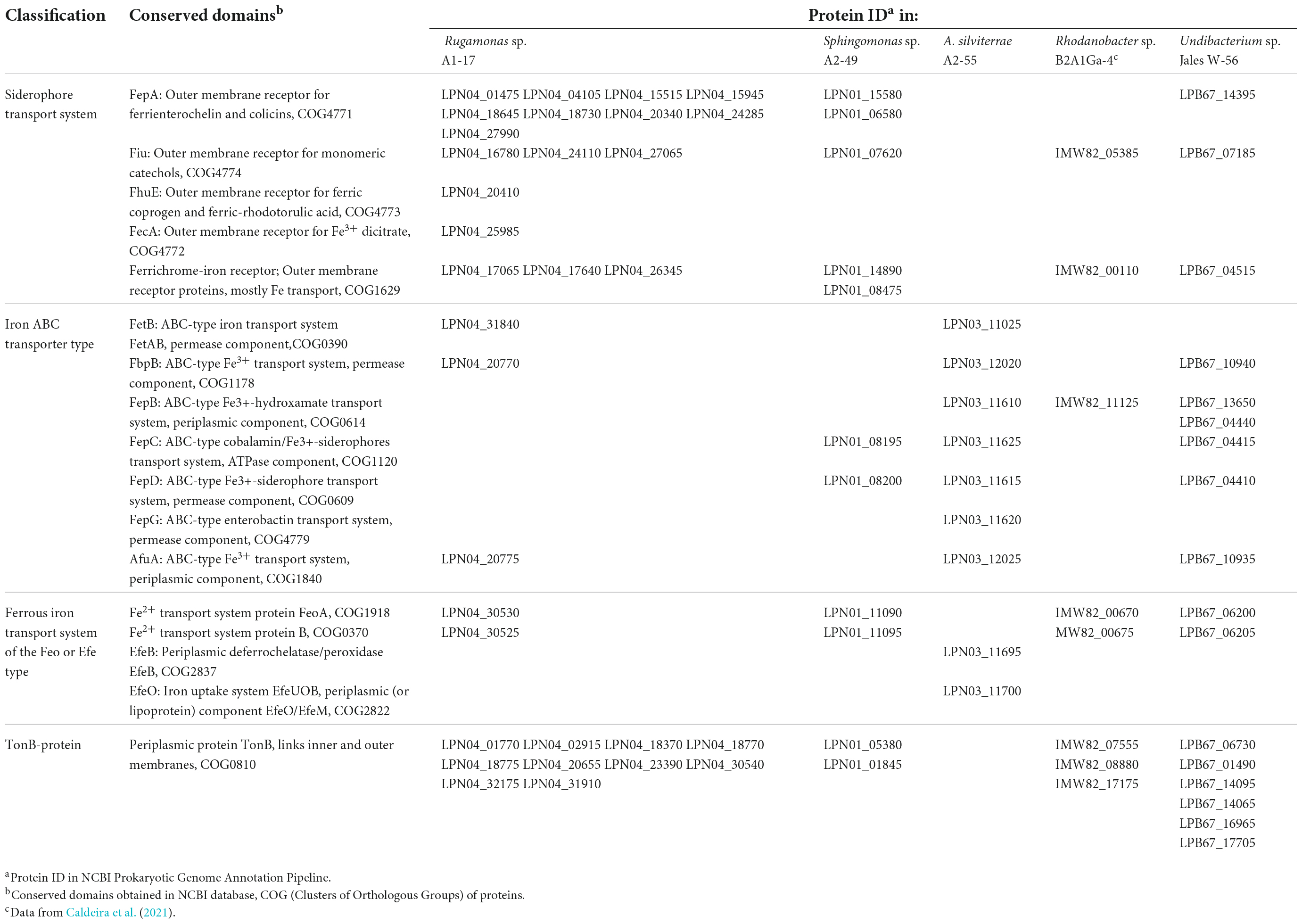
Iron transport systems
The mechanisms involved in Ga resistance are not well known, however, iron transport systems have an important role in this situation. The sequenced bacterial genomes showed the presence of iron uptake systems for both ferrous and ferric forms of elemental iron. In the genome of Rugamonas sp. A1-17 it was identified a larger number of putative outer membrane receptors for high-affinity uptake of siderophore-bound Fe3+ complexes when compared to the other Gram-negative bacterial genomes (Sphingomonas sp. A2-49, Rhodanobacter sp. B2A1Ga4, and Undibacterium sp. Jales W-56) (Table 3). The transport of iron-siderophore complexes through the outer membrane requires energy provided by the energy-transducing protein complex composed by TonB protein and the accessory proteins ExbB and ExbD (TonB-ExbB-ExbD). TonB proteins were predicted in all Gram-negative genomes and were present in a high number in the genomes of Rugamonas sp. A1-17 and Undibacterium sp. Jales W-56 (Table 3). Moreover, accessory proteins ExbB and ExbD were also present in all of these genomes, and in strains Rugamonas sp. A1-17, Rhodanobacter sp. B2A1Ga4, and Undibacterium sp. Jales W-56, in more than one set of ExbB-ExbD protein complex. A. silviterrae A2-55 as Gram-positive bacteria lacks an outer membrane and requires neither outer membrane receptors nor TonB-ExbB-ExbD systems. This Gram-positive strain has diverse ABC transporters analogous to those of the Gram-negative to transport iron-siderophore complexes across the cell membrane (Table 3). Among the Gram-negative bacteria, strains Rugamonas sp. A1-17 and Undibacterium sp. Jales W-56 were the only ones that presented putative periplasmic ABC transporters (AfuA: ABC-type Fe3+ transport system, FepB: ABC-type Fe3+-hydroxamate transport system) that are able to shuttle iron-siderophore complexes from outer- membrane receptors to the cytoplasmic membrane ABC transporters (FbpB: ABC-type Fe3+ transport system, FepC: ABC-type cobalamin/Fe3+-siderophores transport system, and FepD: ABC-type Fe3+-siderophore transport system). The genome of Rhodanobacter sp. B2A1Ga4 only showed a putative periplasmic ABC transporter and the genome of Sphingomonas sp. A2-49 only showed cytoplasmic membrane ABC transporters (Table 3). A putative iron export permease protein (FetB: ABC-type iron transport system) was only identified in the genomes of Rugamonas sp. A1-17 and A. silviterrae A2-55. Uptake systems for ferrous iron were also identified, the FeoAB uptake system was present in all Gram negative genomes and the EfeOB uptake system was present in the genome of A. silviterrae A2-55 (Table 3).
Minimum inhibitory concentration
The Minimum Inhibitory Concentration (MIC) for Ga and As(III) was determined in liquid assays in 96-multiwell plates. Rhodanobacter sp. B2A1Ga4 showed the highest MIC values for both metal(loid)s, 7.5 mM for As(III) and 2 mM for Ga, respectively. The lowest MIC values for As(III) were obtained with strains A. silviterrae A2-55 and Undibacterium sp. Jales W-56, while the strain Sphingomonas sp. A2-49 exhibited the lowest MIC value for Ga (Table 4).
Bioleaching experiments
Batch growth
In batch growth assays where the bacterial strains were inoculated at the beginning of the bioleaching experiments, the efficiency of Ga leaching increased over time (factor “incubation time” was statistically relevant, p < 0.05) (Supplementary Data 4), and was maximum at 21 days, for most of the bacterial strains in both Ga mineral substrates, GaAs and GaN (Figure 3). The only exceptions were obtained with Sphingomonas sp. A2-49 and A. silviterrae A2-55 that reached the highest values of Ga leaching at 14 days of incubation from GaAs (53.0%) and from GaN (25.0%), respectively. Among the bacterial mine isolates, Rhodanobacter sp. B2A1Ga4 showed the highest efficiency to leach Ga from GaAs (56.0%) and from GaN (40%) (Figure 3). Together with Sphingomonas sp. A2-49 (53.0% leaching), for GaAs, the high leaching capacities were statistically significant when compared to the lowest performing strains: A. silviterrae A2-55 and Undibacterium sp. Jales W-56, which reached, respectively, only 33.6 and 29.0% of Ga leaching. Effectively, the later strains did not exhibit significant differences comparing leaching on days 7 and 21, unlike the other strains. All bacterial strains performed better in presence of GaAs (29.0 to 56.0%) than in GaN (24.4 to 40.4%) except Undibacterium sp. Jales W-56, with statistical significance (p < 0.05) after 21 days of experiment (Figure 3).
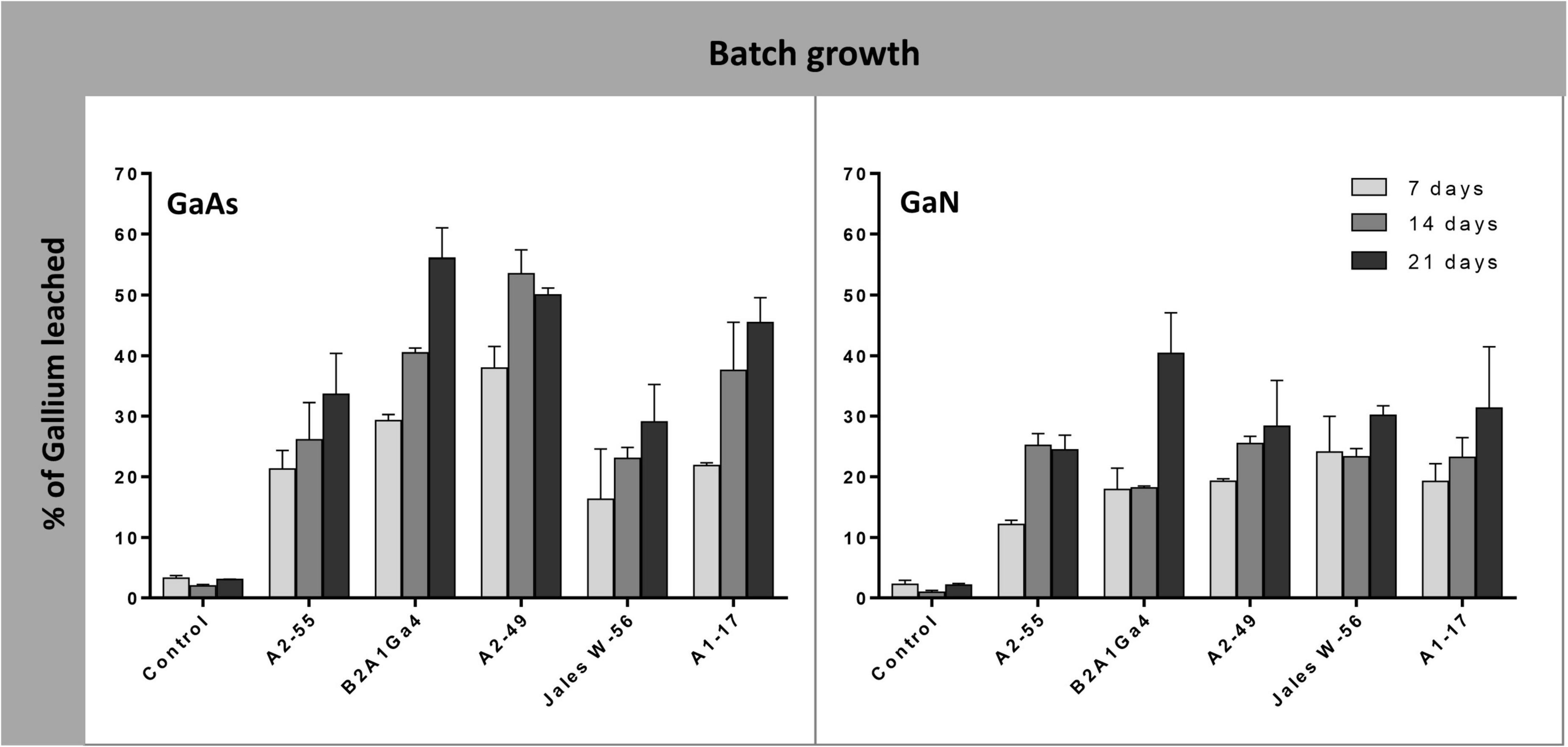
Figure 3. Gallium leaching from GaAs and GaN in batch growth of Arthrobacter silviterrae A2-55, Rhodanobacter sp. B2A1Ga4, Sphingomonas sp. A2-49, Undibacterium sp. Jales W-56, and Rugamonas sp. A1-17, with mR2Ab medium, pH = 6.0. Ga in leaching medium was determined at day 7, 14 and 21 of the experiments. Data are the mean values (± standard deviations) obtained from two or three independent experiments.
Cultures in different phases of growth
In these bioleaching experiments, bacterial strains were first grown under their optimum conditions until they reached the desired phase of growth (late exponential, 8 h; stationary, 24 h, and late stationary, 48 h (Supplementary Data 4). The cultures at different phases of growth were then used as leaching media in the assays. For all bacteria strains, the efficiency of Ga leaching of cultures at the three different phases of growth was similar for GaAs, with marginal differences of 1.0 to 12.0% in Ga leaching capacity between them, and no statistical significance (Figure 4). This trend was observed at all incubation times (7, 14, and 21 days). The factor “growth phase” was statistically relevant when analyzing the results obtained with GaN. The use of cultures, in the stationary phase or in late stationary phase, significantly increased the Ga leaching by 5.0–9.0% after 21 days, for both strains Undibacterium sp. Jales W-56 and Rugamonas sp. A1-17 comparatively to the Ga leaching using cultures in the late exponential phase (Figure 4). The results obtained with GaN separated these two strains, with a higher leaching ability (p < 0.05), from strains A. silviterrae A2-55, Rhodanobacter sp. B2A1Ga4, and Sphingomonas sp. A2-49 with a lower leaching capacity on days 14 and 21. Significantly higher efficiency of Ga leaching was always obtained at 21 days for both GaN and GaAs.
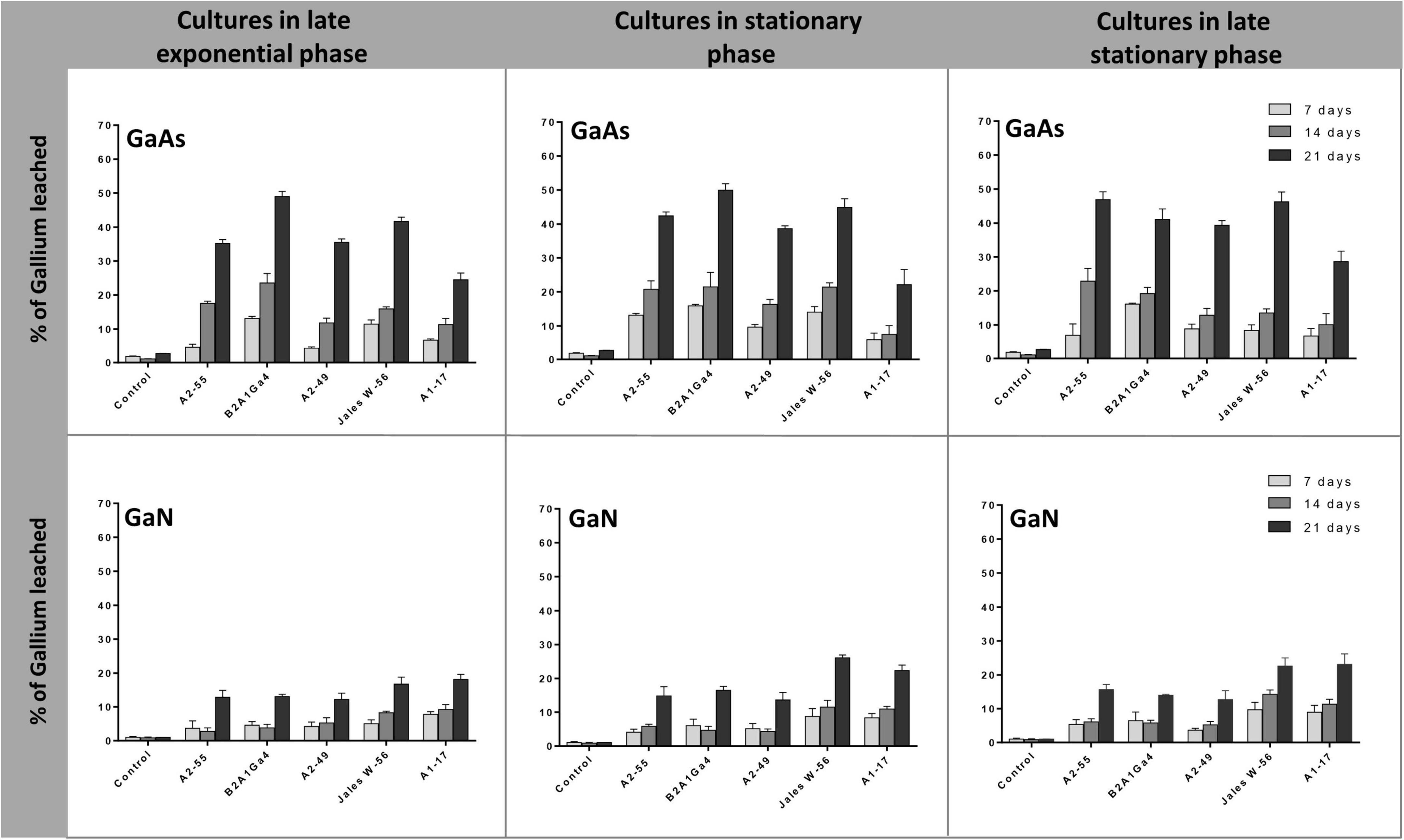
Figure 4. Gallium leaching from GaAs and GaN by bacterial cultures at different growth phases (late exponential, 8 h; stationary, 24 h, and late stationary, 48 h). Strains: Arthrobacter silviterrae A2-55, Rhodanobacter sp. B2A1Ga4, Sphingomonas sp. A2-49, Undibacterium sp. Jales W-56, and Rugamonas sp. A1-17. Ga in leaching medium was determined at day 7, 14, and 21 of the experiments. Data are the mean values (± standard deviations) obtained from two or three independent experiments.
Cell-free spent medium
Cell-free spent medium from cultures at stationary phase of growth (24 h) and late stationary phase of growth (48 h) was also used to evaluate the efficiency of Ga leaching from GaAs and GaN. The results showed that spent medium from A. silviterrae A2-55 at stationary phase had the highest efficiency to leach Ga from GaAs, reaching 35.9 and 57.0% after 14 and 21 days, respectively, significantly outperforming all other strains (Figure 5). However, the use of the spent medium at late stationary phase from all the other 4 strains improved their Ga leaching performance from GaAs. This enhancement was clearly evident (p < 0.05) for Rhodanobacter sp. B2A1Ga4, that showed an increase of soluble Ga amounts of 13.0 and 27.0% using the cell-free spent medium from cultures at late stationary growth phase, after 14 and 21 days, respectively. Cell free-spent medium from all strains showed low efficiency of Ga leaching from GaN. The maximum Ga leaching amounts from GaN, 21.7 and 23.9%, were obtained using cell free-spent medium from cultures of Undibacterium sp. Jales W-56 and Rugamonas sp. A1-17 at stationary phase, respectively, after 21 days. Both strains performance differed significantly from the other three strains. However, the use of medium of late stationary phase improved significantly the Ga leaching efficiency of the three other strains, particularly on day 21.
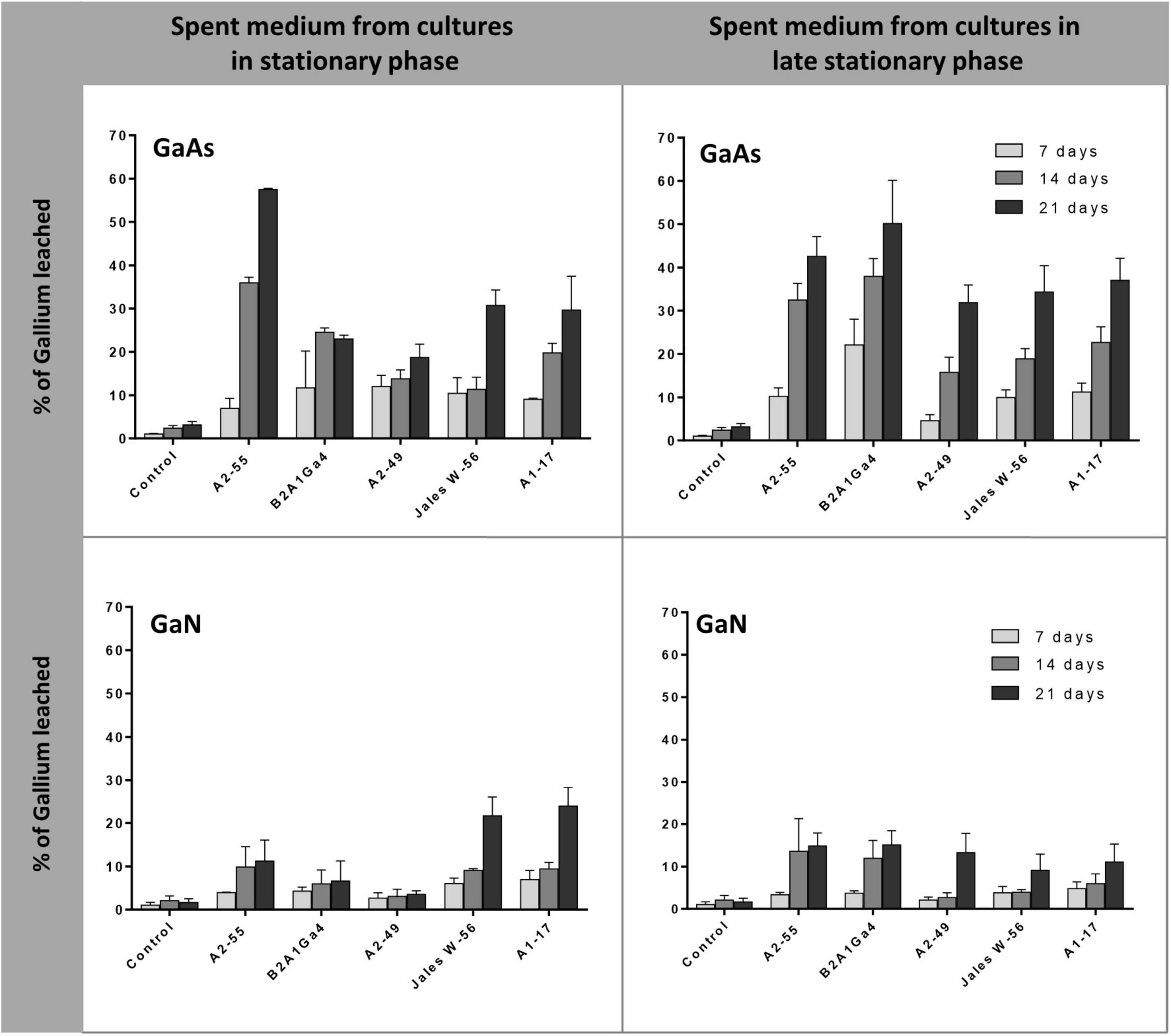
Figure 5. Gallium leaching from GaAs and GaN by cell-free spent medium of cultures at different phases (stationary, 24 h, and late stationary, 48 h). Strains: Arthrobacter silviterrae A2-55, Rhodanobacter sp. B2A1Ga4, Sphingomonas sp. A2-49, Undibacterium sp. Jales W-56, and Rugamonas sp. A1-17. Ga in leaching medium was determined at day 7, 14, and 21 of the experiments. Data are the mean values (± standard deviations) obtained from two or three independent experiments.
pH variation
pH values were measured in the beginning and during the bioleaching experiments after 7, 14, and 21 days. pH changes were observed in all the different bioleaching experiments used in this study. In batch experiments the initial pH of mR2Ab was adjusted to 6.0 and by day 7 the pH value was between 7.0 and 7.8, and at the end of the experiments, the pH values reached 8.0 or 8.5. In the bioleaching experiments with cultures or spent medium in different phases of growth as leaching medium, the initial pH was higher between 6.8 and 7.9, since the bacterial strains have previously grown in this medium. pH changes throughout the assays were also observed, and after 21 days the pH reached values of between 8.0 and 9.0. However, the pH values of the inoculated controls did not have major changes and remained between 6.0 and 6.5 throughout the assays.
Discussion
Bacteria that inhabit metal-contaminated environments, such as mines, play unique roles in the biochemical cycling of metals, including mineral dissolution by leaching processes and metal recovery. The present study combines experimental and whole-genome analysis approaches to investigate the ability of five distinct bacterial mine isolates to leach and mobilize Ga from GaAs and GaN.
All these bacterial genomes shared a large number of homologous genes that were assigned to COG functional categories, of which the “amino acid transport and metabolism” [E], “translation, ribosomal structure and biogenesis” [J], and “replication, recombination, and repair” [L] were the most abundant. These findings show that these heterotrophic bacteria have the ability to synthesize amino acids as energy supply [E], and that the enrichment in genes coding for structures involved in protein synthesis [J] and DNA repair [L] might provide an adaptive strategy to cope with high metal concentrations of their natural environment. High concentrations of heavy metals can cause damage to bacterial cells (Chandrangsu et al., 2017; Igiri et al., 2018) and mechanisms of defense such as the synthesis of proteins involved in transport and detoxification, and also proteins involved in DNA repair can be trigged as rescue systems (Zhang et al., 2018; Mathivanan et al., 2021).
Genome analysis of these five strains showed predicted genes for the biosynthetic pathways of most amino acids, indicating that they potentially can synthesize most of the amino acids. The presence of these amino acids in the growth supernatant of cultures of these strains could have an important role in the Ga leaching mechanism (Supplementary Data 4). Previous studies have already pointed out that amino acids/peptides/proteins produced by heterotrophic bacteria were involved in Ga leaching from GaAs and GaN (Maneesuwannarat et al., 2016a,b). More recently, Kudpeng et al. (2021) reported that amino acids/peptides/proteins produced by Macrococcus caseolyticus and Acinetobacter calcoaceticus were capable of gold bioleaching from silicate ore. The Ga leaching ability of these molecules is related to the presence of charges in their structure, which varies with different pHs due to the pK values of the carboxylic and amino groups. It was shown that basic pH conditions promoted deprotonation of the carboxylic and amino groups of amino acids/peptides/proteins, resulting in a higher number of negative charges and interaction with positively charged Ga (Maneesuwannarat et al., 2016a,b, 2019). In the bioleaching experiments of this study, the progressive increase of Ga in the leachate medium was always accompanied by an increase of pH values, reaching values between 8 and 9, after 21 days of incubation. This pH range also seemed to favor the bioleaching process, as the highest values of Ga leaching were obtained at 21 days in most bioleaching assays. These alkaline pH values also showed that these bacterial strains do not produce organic acids as a strategy to mobilize Ga. The comparison of the different bioleaching assays demonstrated a divergence in bioleaching efficiency among the bacterial strains, which could be correlated with the different metabolic features presented in their genomes. Thus, the bacterial genomes were analyzed to unveil the mechanisms of resistance to arsenic (As) and Ga, the two metal(loid)s used in the bioleaching assays. All bacterial genomes harbored several clusters of ars genes, with a different variety and combination of genes involved in the As resistance. Strain Rhodanobacter sp. B2A1Ga4, with the highest As resistance, exhibited multiple and redundant ars genes in complex ars gene clusters in its genome in comparison to all other genomes. Furthermore, the presence of an arsA gene within the arsRCDAB cluster enhances the efflux activity of ArsB, by coupling the ATP-hydrolyzing activity of an ATPase, increasing As (III) extrusion and resistance to this metalloid (Firrincieli et al., 2019). Strains Rugamonas sp. A1-17 and Sphingomonas sp. A2-49 with MICs values of 3.75 and 1 mM, respectively, harbored arsRCBH gene clusters. The presence of an additional arsN2 gene in the ars gene clusters of Sphingomonas sp. A2-49 was an interesting finding. This gene is often found in ars operons, suggesting a role of the ArsN proteins in arsenic resistance. However, the exact functions of ArsN proteins are not yet known (Chauhan et al., 2009; Chen et al., 2016; Ben-Fekih et al., 2018). Strains A. silviterrae A2-55 and Undibacterium sp. Jales W-56 exhibited low As resistance (MIC = 0.5 mM). Genomic evidences showed simpler ars gene clusters with variations of the canonical arsRBC cluster, where the arsB gene was replaced by acr3 or aquaporin Z gene. Although the role of ACR3 protein as an efflux pump is well known (Fu et al., 2009; Ben-Fekih et al., 2018), the role of aquaporins in As resistance is more recent. The aquaporin Z genes identified in these genomes encode for aquaglyceroporins GlpF, which usually transport water and organic solutes such as glycerol (Borgnia et al., 1999), but can also function as an As (III) efflux pump replacing the ArsB transporter (Yang et al., 2012; Mukhopadhyay et al., 2014; Yang et al., 2015). Literature reports strains, such as Ochrobactrum tritici (Branco et al., 2008; Sousa et al., 2015) or Herminiimonas arsenicoxydans (Muller et al., 2006), as having a high number of ars genes, which confer high resistance to arsenic. Multiple and redundant ars genes in Prokaryotes commonly give rise to higher levels of resistance to arsenic (Li and Krumholz, 2007). Additionally, these redundant ars genes may be expressed differentially depending on the environmental conditions, which may constitute an advantage when facing arsenic stress (Ben-Fekih et al., 2018).
Currently, little information is available on the mechanisms of Ga resistance. Nonetheless, studies indicate that iron metabolism plays an important role in Ga resistance and cellular protection against the potential toxicity of this metal (Gugala et al., 2019; Caldeira et al., 2021). It is predominantly assumed that Ga crosses cell membranes using the iron transport systems, in particular the Fe-siderophore transport system, as Ga is considered an iron mimetic (Gugala et al., 2019; Li et al., 2022). Among the bacterial strains tested in the current study, the highest Ga resistance was demonstrated by Rhodanobacter sp. B2A1Ga4 and A. silviterrae A2-55 (MIC = 2 mM). A lower number of genes encoding iron transporter systems were identified in Rhodanobacter sp. B2A1Ga4 genome in comparison to other genomes, might prevent Ga import and accumulation, resulting in higher Ga resistance. A noteworthy finding was the presence of a gene encoding for a putative FetB protein, in the genome of A. silviterrae A2-55, which is reported as an iron efflux system (Nicolaou et al., 2013). This system can be used by these cells to extrude Ga, increasing the Ga resistance. Strains Rugamonas sp. A1-17 and Undibacterium sp. Jales W-56 showed an intermediate Ga resistance (MICs = 1.5–1.0 mM), and a high number of proteins involved in iron transport were predicted in these genomes. However, in the genome of Rugamonas sp. A1-17 is also predicted a putative FetB protein, as in genome of A. silviterrae A2-55, which may contribute to the Ga resistance of this strain. The lowest resistance to Ga was presented by Sphingomonas sp. A2-49 (MIC 0.25 mM) which carries a large number of putative outer membrane receptors for Fe-siderophores complexes, providing a high number of potential Ga targets. Ga resistance related to iron transport systems, has also been reported in other microorganisms. Ga resistance in P. aeruginosa increased after the inactivation of the hitA gene encoding an iron transporter (García-Contreras et al., 2013). In E. coli, the deletion of genes encoding proteins involved in Fe-siderophores import complexes, such as the FepG, FecA and the accessory TonB proteins, reduced Ga import and intracellular accumulation of this metal (Graves et al., 2019; Gugala et al., 2019). Additionally, a previous study with a mutant of Rhodanobacter sp. B2A1Ga4, in which the ferrous iron FeoAB uptake system was inactivated, showed that acquisition of iron by this system is critical to control the oxidative stress in presence of indium and Ga, enhancing the resistance to both metals (Caldeira et al., 2021).
The presence of effective resistance mechanisms to Ga and As was particularly important in GaAs bioleaching batch assays where bacterial growth occur simultaneously with the mobilization of Ga and As into the leaching medium. Actually, under this condition, a correlation could be established: strains with higher resistance, mainly to As, performed better in this bioleaching assays, such as Rhodanobacter sp. B2A1Ga4 and Sphingomonas sp. A2-49. On the other hand, strains with lower resistance, such as A. silviterrae A2-55 and Undibacterium sp. Jales W-56, were not so efficient in the bioleaching process. However, in GaAs bioleaching assays, where cultures were first grown in absence of GaAs and then used either as it is or as cell-free spent medium, both strains clearly improved their bioleaching efficiency. In fact, A. silviterrae A2-55 outperformed all the other stains (57.0% Ga leached) with cell-free spent medium from the stationary phase of growth.
In this work, the genes involved in nitrogen metabolism were not deeply explored, since these heterotrophic bacteria naturally have mechanisms to deal with the nitrogen released from the bioleaching assays with GaN. In Ga bioleaching assays from GaN, the presence of active bacterial cells seems to be important, since free-cell spent medium assays presented the lowest values of Ga leaching (2.0 to 24.0%). Moreover, older cultures favored the bioleaching process from GaN, indicating that the metabolic by-products produced in the late stages of growth and the pH provided more efficient conditions for Ga mobilization. Overall, the efficiency of Ga bioleaching from GaN was lower when compared to GaAs. The refractory property of GaN generated difficulties in the process of bioleaching, and it was shown that pretreatment with high temperatures improved the efficiency of the bioleaching process (Maneesuwannarat et al., 2016b).
The integrative approach that combines the experimental and genomic analysis undertaken in this work showed that the genetic features related to amino acids metabolism and genetic mechanisms that potentially account for Ga and As resistance in these bacterial mine strains are related to their different ability to mobilize and leach Ga from GaAs and GaN. Genomic analyses to identify metabolic traits linked to the bioleaching process as presented here improve our current knowledge and further promote the industrial applications of bioleaching technologies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, for JAJLPB000000000 (Rugamonas sp. A1-17); for JAJLPA000000000 (Sphingomonas sp. A2-49); for JAJLOZ000000000 (Arthrobacter silviterrae A2-55); for JAJLQW000000000 (Undibacterium sp. Jales W-56); and for JADBJR000000000 (Rhodanobacter sp. B2A1Ga4).
Author contributions
AC and RB conceived and designed the experiments and wrote the manuscript. AC performed the genome analysis and the experiments. AC, RF, PM, and RB analyzed the data. RF performed the statistical analysis. RB and PM contributed with reagents, materials, and analysis tools. All authors revised and approved the manuscript.
Funding
This study was supported by the project MicroMineR–PTDC/CTA-AMB/31820/2017 funded by Fundação para a Ciência e Tecnologia (FCT) and Programa Operacional da Região Centro, by EU Horizon 2020 Research and Innovation Program (Grant no 821096) Project Biorecover, EU program ERA-MIN2 (Grant no 67), FEDER funds through the program COMPETE–Programa Operacional Factores de Competitividade–and through FCT, under the project UIDB/00285/20.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.970147/full#supplementary-material
Footnotes
- ^ http://jspecies.ribohost.com/jspeciesws
- ^ http://bioinformatics.psb.ugent.be/webtools/Venn/
- ^ www.graphpad.com
References
Andrews, J. M. (2002). Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 49, 1049. doi: 10.1093/jac/dkf083
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: Rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Ben-Fekih, I., Zhang, C., Li, Y. P., Zhao, Y., Alwathnani, H. A., Saquib, Q., et al. (2018). Distribution of arsenic resistance genes in Prokaryotes. Front. Microbiol. 9:2473. doi: 10.3389/fmicb.2018.02473
Biswal, B. K., Chen, Z., and Yang, E.-H. (2019). Hydrothermal process reduced Pseudomonas aeruginosa PAO1-driven bioleaching of heavy metals in a novel aerated concrete synthesized using municipal solid waste incineration bottom ash. Chem. Eng. J. 360, 1082–1091. doi: 10.1016/j.conbuildmat.2020.118005
Borgnia, M., Nielsen, S., Engel, A., and Agre, P. (1999). Cellular and molecular biology of the aquaporin water channels. Annu. Rev. Biochem. 68, 425–458. doi: 10.1146/annurev.biochem.68.1.425
Bosecker, K. (1997). Bioleaching: Metal solubilization by microorganisms. FEMS Microbiol. Rev. 20, 591–604. doi: 10.1111/j.1574-6976.1997.tb00340.x
Branco, R., Chung, A. P., and Morais, P. V. (2008). Sequencing and expression of two arsenic resistance operons with different functions in the highly arsenic-resistant strain Ochrobactrum tritici SCII24T. BMC Microbiol. 8:95. doi: 10.1186/1471-2180-8-95
Brandl, H., Lehmann, S., Faramarzi, M. A., and Martinelli, D. (2008). Biomobilization of silver, gold, and platinum from solid waste materials by HCN-forming microorganisms. Hydrometallurgy 94, 14–17. doi: 10.1016/j.hydromet.2008.05.016
Caldeira, J. B., Chung, A. P., Morais, P. V., and Branco, R. (2021). Relevance of FeoAB system in Rhodanobacter sp. B2A1Ga4 resistance to heavy metals, aluminium, gallium, and indium. Appl. Microbiol. Biotechnol. 105, 3301–3314. doi: 10.1007/s00253-021-11254-6
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829. doi: 10.1093/molbev/msab293
Cárdenas, J. P., Quatrini, R., and Holmes, D. S. (2016). Genomic and metagenomic challenges and opportunities for bioleaching: A mini-review. Res. Microbiol. 167, 529–538. doi: 10.1016/j.resmic.2016.06.007
Chandrangsu, P., Rensing, C., and Helmann, J. D. (2017). Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 15, 338–350. doi: 10.1038/nrmicro.2017.15
Chauhan, N. S., Ranjan, R., Purohit, H. J., Kalia, V. C., and Sharma, R. (2009). Identification of genes conferring arsenic resistance to Escherichia coli from an effluent treatment plant sludge metagenomic library. FEMS Microbiol. Ecol. 67, 130–139. doi: 10.1111/j.1574-6941.2008.00613.x
Chen, J., Yoshinaga, M., Garbinski, L. D., and Rosen, B. P. (2016). Synergistic interaction of glyceraldehydes-3-phosphate dehydrogenase and ArsJ, a novel organoarsenical efflux permease, confers arsenate resistance. Mol. Microbiol. 100, 945–953. doi: 10.1111/mmi.13371
Faramarzi, M. A., Stagars, M., Pensini, E., Krebs, W., and Brandl, H. (2004). Metal solubilization from metal-containing solid materials by cyanogenic Chromobacterium violaceum. J. Biotechnol. 113, 321–326. doi: 10.1016/j.jbiotec.2004.03.031
Firrincieli, A., Presentato, A., Favoino, G., Marabottini, R., Allevato, E., Stazi, S. R., et al. (2019). Identification of resistance genes and response to arsenic in Rhodococcus aetherivorans BCP1. Front. Microbiol. 10:888. doi: 10.3389/fmicb.2019.00888
Fu, H. L., Meng, Y., Ordóñez, E., Villadangos, A. F., Bhattacharjee, H., Gil, J. A., et al. (2009). Properties of arsenite efflux permeases (Acr3) from Alkaliphilus metalliredigens and Corynebacterium glutamicum. J. Bio. Chem. 284, 19887–19895. doi: 10.1074/jbc.M109.011882
Gao, X.-Y., Liu, X.-J., Fu, C.-A., Gu, X.-F., Lin, J.-Q., Liu, X.-M., et al. (2020). Novel strategy for improvement of the bioleaching efficiency of Acidithiobacillus ferrooxidans based on the AfeI/R quorum sensing system. Minerals 10:222. doi: 10.3390/min10030222
García-Contreras, R., Lira-Silva, E., Jasso-Chávez, R., Hernández-González, I. L., Maeda, T., Hashimoto, T., et al. (2013). Isolation and characterization of gallium resistant Pseudomonas aeruginosa mutants. Int. J. Med. Microbiol. 303, 574–582. doi: 10.1016/j.ijmm.2013.07.009
Girtan, M., Wittenberg, A., Grilli, M. L., de Oliveira, D. P. S., Giosuè, C., and Ruello, M. L. (2021). The critical raw materials issue between scarcity, supply risk, and unique properties. Materials 14:1826. doi: 10.3390/ma14081826
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Graves, J. L. Jr., Ewunkem, A. J., Ward, J., Staley, C., Thomas, M. D., Rhinehardt, K., et al. (2019). Experimental evolution of gallium resistance in Escherichia coli. Evol. Med. Public Health 2019, 169–180. doi: 10.1093/emph/eoz025
Gray, F., Kramer, D. A., and Bliss, J. D. (2013). “Gallium and gallium compounds,” in Kirk-othmer encyclopedia of chemical technology, (Hoboken, NJ: Wiley Online Library). doi: 10.1016/j.wasman.2016.12.035
Gugala, N., Chatfield-Reed, K., Turner, R. J., and Chua, G. (2019). Using a chemical genetic screen to enhance our understanding of the antimicrobial properties of gallium against Escherichia coli. Genes (Basel) 10:34. doi: 10.3390/genes10010034
Hagelüken, C., Lee-Shin, J. U., Carpentier, A., and Heron, C. (2016). The EU circular economy and its relevance to metal recycling. Recycling 1, 242–253. doi: 10.3390/recycling1020242
Hayes, S. M., and McCullough, E. A. (2018). Critical minerals: A review of elemental trends in comprehensive criticality studies. Resour. Policy 59, 192–199. doi: 10.1016/j.resourpol.2018.06.015
Huang, X., Zhang, W., Han, S., Yin, Y., Xu, G., and Wang, X. (1997). Spectrophotometric determination of Sb(III) in Sb(III)/Sb(V) binary mixtures using sodium dodecylsulfate/nonylphenoxy polyethoxyethanol mixed micellar media. Talanta 45, 127–135. doi: 10.1016/S0039-9140(97)00114-8
Igiri, B. E., Okoduwa, S. I. R., Idoko, G. O., Akabuogu, E. P., Adeyi, A. O., and Ejiogu, I. K. (2018). Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: A review. J. Toxicol. 2018:2568038. doi: 10.1155/2018/2568038
Işıldar, A., van Hullebusch, E. D., Lenz, M., Du Laing, G., Marra, A., Cesaro, A., et al. (2019). Biotechnological strategies for the recovery of valuable and critical raw materials from waste electrical and electronic equipment (WEEE) – A review. J. Hazard. Mater. 362, 467–481. doi: 10.1016/j.jhazmat.2018.08.050
Kudpeng, K., Thayanukul, P., and Thiravetyan, P. (2021). Bioleaching of gold from silicate ore by Macrococcus caseolyticus and Acinetobacter calcoaceticus: Effect of medium, amino acids and growth supernatant. Minerals 11:580. doi: 10.3390/min11060580
Li, F., Liu, F., Huang, K., and Yang, S. (2022). Advancement of gallium and gallium-based compounds as antimicrobial agents. Front. Bioeng. Biotechnol. 10:827960. doi: 10.3389/fbioe.2022.827960
Li, J., Liang, C., and Ma, C. (2015). Bioleaching of gold from waste printed circuit boards by Chromobacterium violaceum. J. Mater. Cycles Waste Manag. 17, 529–539. doi: 10.1007/s10163-014-0276-4
Li, X., and Krumholz, L. R. (2007). Regulation of arsenate resistance in Desulfovibrio desulfuricans G20 by an arsRBCC operon and an arsC gene. J. Bacteriol. 189, 3705–3711. doi: 10.1128/JB.01913-06
Maneesuwannarat, S., Kudpeng, K., Yingchutrakul, Y., Roytrakul, S., Vangnai, A. S., Yamashita, M., et al. (2019). A possible protein model involved in gallium arsenide leaching by Cellulosimicrobium funkei. Miner. Eng. 137, 207–216.
Maneesuwannarat, S., Teamkao, P., Vangnai, A. S., Yamashita, M., and Thiravetyan, P. (2016b). Possible mechanism of gallium bioleaching from gallium nitride (GAN) by Arthrobacter creatinolyticus: Role of amino acids/peptides/proteins bindings with gallium. Process Saf. Environ. Prot. 103, 36–45. doi: 10.1016/j.psep.2016.06.036
Maneesuwannarat, S., Vangnai, A. S., Yamashita, M., and Thiravetyan, P. (2016a). Bioleaching of gallium from gallium arsenide by Cellulosimicrobium funkei and its application to semiconductor/electronic wastes. Process Saf. Environ. Prot. 99, 80–87. doi: 10.1016/j.psep.2015.10.008
Mathivanan, K., Chandirika, J. U., Vinothkanna, A., Yin, H., Liu, X., and Meng, D. (2021). Bacterial adaptive strategies to cope with metal toxicity in the contaminated environment – A review. Ecot. Environ. Safety. 226:112863. doi: 10.1016/j.ecoenv.2021.112863
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. doi: 10.1038/s41467-019-10210-3
Mishra, S., Panda, S., Akcil, A., Dembele, S., and Agcasulu, I. (2021). A Review on chemical versus microbial leaching of electronic wastes with emphasis on base metals dissolution. Minerals 1:1255. doi: 10.3390/min11111255
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. doi: 10.1093/nar/gkm321
Moskalyk, R. R. (2003). Gallium: The backbone of the electronics industry. Miner. Eng. 16, 921–929. doi: 10.1016/j.mineng.2003.08.003
Motaghed, M., Mousavi, S. M., Rastegar, S. O., and Shojaosadati, S. A. (2014). Platinum and rhenium extraction from a spent refinery catalyst using Bacillus megaterium as a cyanogenic bacterium: Statistical modeling and process optimization. Bioresour. Technol. 171, 401–409. doi: 10.1016/j.biortech.2014.08.032
Mukhopadhyay, R., Bhattacharjee, H., and Rosen, B. P. (2014). Aquaglyceroporins: Generalized metalloid channels. Biochim. Biophys. Acta 1840, 1583–1591. doi: 10.1016/j.bbagen.2013.11.021
Muller, D., Simeonova, D. D., Riegel, P., Mangenot, S., Koechler, S., Lievremont, D., et al. (2006). Herminiimonas arsenicoxydans sp. nov., a metalloresistant bacterium. Int. J. Syst. Evol. Microbiol. 56, 1765–1769. doi: 10.1099/ijs.0.64308-0
Nicolaou, S. A., Fast, A. G., Nakamaru-Ogiso, E., and Papoutsakis, E. T. (2013). Overexpression of fetA (ybbL) and fetB (ybbM), encoding an iron exporter, enhances resistance to oxidative stress in Escherichia coli. Appl. Environ. Microbiol. 79, 7210–7219. doi: 10.1128/AEM.02322-13
Olson, G. J., Brierley, J. A., and Brierley, C. L. (2003). Bioleaching review part B: Progress in bioleaching: Applications of microbial processes by the minerals industries. Appl. Microbiol. Biotechnol. 63, 249–257. doi: 10.1007/s00253-003-1404-6
Potysz, A., Lens, P. N. L., van de Vossenberg, J., Rene, E. R., Grybos, M., Guibaud, G., et al. (2016). Comparison of Cu, Zn and Fe bioleaching from Cu-metallurgical slags in the presence of Pseudomonas fluorescens and Acidithiobacillus thiooxidans. Appl. Geochem. 68, 39–52. doi: 10.1016/j.apgeochem.2016.03.006
Pourhossein, F., and Mousavi, S. M. (2018). Enhancement of copper, nickel, and gallium recovery from LED waste by adaptation of Acidithiobacillus ferrooxidans. Waste Manag. 79, 98–108. doi: 10.1016/j.wasman.2018.07.010
Sedlakova-Kadukova, J., Marcincakova, R., Luptakova, A., Vojtko, M., Fujda, M., and Pristas, P. (2020). Comparison of three different bioleaching systems for Li recovery from lepidolite. Sci. Rep. 10:14594. doi: 10.1038/s41598-020-71596-5
Sousa, T., Branco, R., Piedade, A. P., and Morais, P. V. (2015). Hyper Accumulation of Arsenic in Mutants of Ochrobactrum tritici Silenced for Arsenite Efflux Pumps. PLoS One 10:e0131317. doi: 10.1371/journal.pone.0131317
Srivastava, P., and Kowshik, M. (2013). Review Article: Mechanisms of metal resistance and homeostasis in Haloarchaea. Archea 2013:732864. doi: 10.1155/2013/732864
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Teeling, H., Waldmann, J., Lombardot, T., Bauer, M., and Glöckner, F. O. (2004). TETRA: A web-service and a stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences. BMC Bioinformatics 5:163. doi: 10.1186/1471-2105-5-163
Yang, H. C., Fu, H. L., Lin, Y. F., and Rosen, B. P. (2012). Pathways of arsenic uptake and efflux. Curr. Top. Membr. 69, 325–358. doi: 10.1016/B978-0-12-394390-3.00012-4
Yang, Y., Wu, S., Lilley, R. M., and Zhang, R. (2015). The diversity of membrane transporters encoded in bacterial arsenic-resistance operons. PeerJ 3:e943. doi: 10.7717/peerj.943
Yoon, S.-H., Ha, S.-M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Keywords: mine isolates, genome analysis, metabolic features, gallium nitride (GaN), gallium arsenide (GaAs)
Citation: Chung AP, Francisco R, Morais PV and Branco R (2022) Genome mining to unravel potential metabolic pathways linked to gallium bioleaching ability of bacterial mine isolates. Front. Microbiol. 13:970147. doi: 10.3389/fmicb.2022.970147
Received: 15 June 2022; Accepted: 15 August 2022;
Published: 13 September 2022.
Edited by:
Hongzhi Tang, Shanghai Jiao Tong University, ChinaReviewed by:
Ricardo Jasso-Chávez, Instituto Nacional de Cardiología Ignacio Chávez, MexicoHaiyang Hu, Shanghai Jiao Tong University, China
Qiang Li, University of Jinan, China
Copyright © 2022 Chung, Francisco, Morais and Branco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Paula Chung, YW5hLmNodW5nQHVjLnB0
 Ana Paula Chung
Ana Paula Chung Romeu Francisco
Romeu Francisco Paula V. Morais
Paula V. Morais Rita Branco
Rita Branco