
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 26 August 2022
Sec. Microbiotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.968439
This article is part of the Research TopicMicrobial Proteases: Biochemical Studies, Immobilization and Biotechnological ApplicationView all 7 articles
 Hao Chen1,2,3,4
Hao Chen1,2,3,4 Jie Wu1,2
Jie Wu1,2 Xiaodan Huang3,4
Xiaodan Huang3,4 Xuzhong Feng5
Xuzhong Feng5 Hongwu Ji3,4
Hongwu Ji3,4 Liangzhong Zhao1,2*
Liangzhong Zhao1,2* Jianrong Wang1,2,6*
Jianrong Wang1,2,6*Proteases are important for decomposition of proteins to generate peptides or amino acids and have a broad range of applications in different industries. Herein, a gene encoding an alkaline protease (AprBcp) from Bacillus circulans R1 was cloned and bioinformatics analyzed. In addition, a series of strategies were applied to achieve high-level expression of AprBcp in Bacillus subtilis. The maximum activity of AprBcp reached 165,870 U/ml after 60 h fed-batch cultivation in 50 l bioreactor. The purified recombinant AprBcp exhibited maximum activity at 60°C and pH 10.0, and remained stable in the range from pH 8.0 to 11.0 and 30 to 45°C. Metal ions Ca2+, Mn2+, and Mg2+ could improve the stability of AprBcp. Furthermore, the recombinant AprBcp displayed great potential application on the recovery of protein from soybean dregs. The results of this study will provide an effective method to prepare AprBcp in B. subtilis and its potential application on utilization of soybean dregs.
Proteases could decompose proteins to form small peptides or amino acids by cleaving the peptide bonds (Razzaq et al., 2019; Cheng et al., 2021). Proteases are found widely in different sources and mainly from animals, plants, and microorganisms (Ding et al., 2020). Compared with animals and plants, microorganisms are more suitable for production of proteases due to their easy cultivation, extracellular secretion, and excellent properties for industrial application (Contesini et al., 2018). Among different microorganisms, Bacillus sp. is the prominent producer for preparation of different proteases, some of which have been commercially applied in food, detergent, feed, and leather industry (Baweja et al., 2016; Contesini et al., 2018). According to the previous studies, proteases from Bacillus sp. are mainly composed of alkaline and neutral protease, especially alkaline protease which exhibits maximum relative activity in the range from pH 8.0 to 11.0 (Solanki et al., 2021). Owning to high activity and stability in the range of pH 8.0–11.0, alkaline protease is suitable for application in areas with alkaline conditions such as detergent, leather, and recovery of protein from food-byproduct (Barzkar, 2020; Matkawala et al., 2021). Therefore, many researchers are dedicated to isolation and characterization of alkaline protease from different Bacillus. Until now, many alkaline proteases have been isolated and characterized from Bacillus (Contesini et al., 2018). However, the expression level of some alkaline protease from wild-type strain is beneath the requirements of industry application. Thus, it is crucial to develop an effective way to obtain high production of alkaline protease.
As a mature expression host, B. subtilis is widely used to produce a variety of enzymes and proteins due to its extracellular secretion of target recombinant protein (Song et al., 2015; Cui et al., 2018). Compared with E. coli and Pichia pastoris, B. subtilis exhibits several advantages such as ease of genetic manipulation, being classified as generally recognized as safe (GRAS) and easy cultivation, and short fermentation time (Gu et al., 2018; Wang et al., 2021b). Recently, several proteases are heterologously expressed in B. subtilis including alkaline and neutral proteases from Bacillus clausii TCCC1004, Geobacillus stearothermophilus (G. stearothermophilus), Bacillus licheniformis, and Idiomarina sp. C9-1 (Liu et al., 2013, 2019; Zhou et al., 2018; Chang et al., 2021). However, the extracellular production levels of heterologous proteases in B. subtilis vary greatly with respect to proteases from different sources. It is particularly noteworthy that promoter and signal peptides play a pivotal role on the expression levels of heterologous protein in B. subtilis (Zhang et al., 2017; Cai et al., 2019; Guo et al., 2021). A series of previous researches demonstrated that the expression level of recombinant protein is improved by 1.1- to 15.2-fold through promoter and signal peptide optimization (Song et al., 2017; Zhang et al., 2017; Fu et al., 2018; Liu et al., 2019; Yao et al., 2019; Kang et al., 2020; Guo et al., 2021). Interestingly, the results of those studies indicated that the strength of promoter and secretion efficiency of signal peptide vary greatly in accordance with the recombinant protein from different microbial sources. For instance, a promoter Pshuttle-09 from B. licheniformis exhibits 8-fold strength than promoter P43 for expression of galactosidase in B. subtilis (Yang et al., 2013). Nevertheless, the production levels of alkaline proteases from B. clausii TCCC1004 in B. subtilis are almost same in B. subtilis when driving by promoter P43 and Pshuttle-09 (Liu et al., 2019). Similarly, the secretion efficiency of the same signal peptide varies differently according to different recombinant protein in B. subtilis (Guan et al., 2016; Zhang et al., 2016b; Liu et al., 2019; Kang et al., 2020). Based on the above-mentioned results, it is concluded that it is necessary to optimize promoter or signal peptide to achieve high-level expression of each target protein. Analysis of previous studies we found that plasmid-based expression is the most favorite choice for heterologous expression of recombinant protein in B. subtilis. However, plasmid-based expression has some disadvantages such as unstable in medium without antibiotic, and vectors with antibiotic resistances, which limit its further industrial application (Yang et al., 2016; Chen et al., 2017). Compared with plasmid-based expression, integrative approach expression of recombinant protein seems more suitable for industrial application due to its stable and no antibiotic resistance-encoding gene (Zhou et al., 2021). Additionally, integrative approach expression is convenient for further genetic manipulation of recombinant strain (Zhou et al., 2020, 2021).
Soybean dregs, also known as okara, are the by-product of soybean products, with about 2.8 million tones annually produced in China (Li et al., 2012). Although the soybean products mainly make use of the protein in soybean, there is still a lot left in soybean dregs. Many studies have shown that protein accounts for 15.2–37.5% of dry weight of soybean dregs and contains all essential amino acids necessary for good health (Li et al., 2012; Fayaz et al., 2019). So far, soybean dregs mainly are discarded as waste, with a small amount used as animal feed or fertilizers, resulting in environmental pollution and waste resources. Therefore, soybean dregs can be consumed as an underutilized and cheap plant protein resource for human consumption. To date, several methods have been used to recover protein from soybean dregs such as fermentation, chemical, and enzymatic methods (Cao et al., 2019; Wang et al., 2021a; Heng et al., 2022). Among the above-mentioned methods, the enzymatic method has attracted more attention due to its mild reaction conditions and environmental friendliness. As the key factor of enzymatic method, protease could hydrolyze peptide bonds to produce small peptides, and then, recover most of the protein from soybean dregs.
In this study, the gene encodes an alkaline protease from B. circulans R1 (named AprBcp) was cloned and bioinformatics analyzed. Meanwhile, the promoters and signal peptides were optimized to drive the heterologous expression of AprBcp in B. subtilis. Moreover, integrative approach and large-scale cultivation were carried out to achieve high-level production of AprBcp in B. subtilis. Additionally, the recombinant AprBcp was purified and characterized. Finally, the application of recombinant AprBcp for recovery of protein from soybean dregs was investigated. The results of this study will provide an effective method to produce AprBcp in B. subtilis and its potential application on utilization of soybean dregs.
The Escherichia coli strain top 10 was used to propagate plasmids. B. circulans strain R1 was isolated from Okara and conserved in our laboratory. The B. subtilis strains RIK1285, pMD20T, and in-fusion cloning kit were purchased from Takara Biotechnology (Dalian, China). Q5® high-fidelity DNA polymerase and protein marker were purchased from New England Biolabs (NEB, Ipswich, MA, United States). Different inhibitors, such as pepstatin A, iodoacetamide, ethylenedia minetetraacetic acid (EDTA), and phenylmethanesulfonyl fluoride (PMSF), and substrates, such as casein, bovine serum albumin (BSA), keratin, soybean protein isolate, and chicken egg albumin, were purchased from Yuanye Biotechnology (Shanghai, China). Soybean dregs were by-products of manufacturing fermented soybean whey-based tofu, dried by microwave drying, ground, and sieved with 60 mesh. The soybean dregs comprised 22.03% protein and 10.23% water.
LBK (LB with 30 μg/ml kanamycin) was used to screen E. coli top 10 and B. subtilis RIK1285 contained plasmids with kanamycin-resistant gene (KanR). Fermentation medium 1 was used for the production of AprBcp in 24-well plates and shake flask, which was composed of 1.5% peptone, 2.5% yeast extract, 1% citrate sodium, 0.3% K2HPO4, and 4% maltose. Fermentation medium 2 was used for large-scale production of AprBcp in 7 and 50 l bioreactor, which contained 7% maltose, 2.5% soybean meal, 2% corn steep liquor, 0.3% citrate sodium, 1% K2HPO4, 2% wheat bran, and 0.3% CaCl2.
The 24-well plate cultivation was used to screen the recombinant strains with different promoters and signal peptides. The shake flask cultivation was used to isolate the recombinant strains with higher alkaline protease activity. Large-scale production of AprBcp was carried out in 7- and 50-L bioreactor. The detailed protocol is provided in Supplementary Methods.
Based on the sequence of alkaline protease gene from B. circulans strain NCTC2610 (GenBank: UAPZ01000012.1: 120935-122077), two primers, bcp-fw and bcp-rev, were designed for PCR amplification. The gene encoding AprBcp was cloned by PCR amplification using genomic DNA of B. circulans R1 as template. The PCR product was ligated to pMD20T and sequenced. The primers used in this study are shown in Supplementary Table 1.
Sequence alignment was performed by blastn or blastx provided by the National Center for Biotechnology Information (NCBI). Amino acid sequences and signal peptide prediction of AprBcp were analyzed by Expasy.1 The sequences were analyzed for identity using ClustalW2 and ENDscript/ESPript.3 Homologous models were generated on the Swiss-model online server. PyMOL was used to analyze the obtained model. The alkaline serine protease from B. lentus was chosen as a template (Protein Data Bank code: 1st3.1.A).
The vector pHY was constructed as template to construct different plasmids (Figure 1). The vector pHY was composed of temperature-sensitive replicon PE194 and a kanamycin-resistant gene (KanR) which functions in B. subtilis and E. coli, as well as the pUC origin of replication from vector pBluescript IISK(+). Additionally, it included B. subtilis-derived promoter P43, which located upstream from the multi-cloning site (MCS), the 5′-upstream repair, and 3′-downstream repair template of α-amylase from B. subtilis 168.
The gene bcp was ligated to pHY to form expression vector pHY-bcp. For promoter optimization, nine highly active promoters reported by previous studies were selected and synthesized for heterologous expression of AprBcp. The fragment pHY without promoter P43 was obtained from pHY-bcp digested by Eco52I and MluI. And then, each different promoter flanked by Eco52I-MluI was separately ligated to fragment pHY to form nine expression plasmids. For dual promoter plasmids, the second promoter was inserted into the upstream of the single promoter expression plasmid (Figure 1). The sequences of nine highly active promoters are shown in Supplementary Table 1 and the primers used in this part are shown in Supplementary Table 1.
For signal peptide optimization, the expression vector with the highly active dual promoters was used to screen the efficient signal peptide. Nine signal peptides were selected and synthesized by over-lapping PCR, and ligated to dual promoter expression plasmid by in-fusion cloning. The process for construction of plasmids with signal peptides is shown in Figure 1. Plasmids with different promoters and signal peptides used in this study are shown in Table 1. The sequences of nine signal peptides are shown in Supplementary Table 3 and the primers used in this part are also shown in Supplementary Table 1.
The bcp expression cassette, with highly active dual promoter, efficient signal peptide and bcp, was integrated to α-amylase locus of B. subtilis 1285. The method for chromosomal integration of bcp expression cassette was similar to previous studies and the detailed process is provided in Supplementary Methods and Figure 2 (Zakataeva et al., 2010; Zhang et al., 2016a; Zhou et al., 2021). The recombinant strains with bcp expression cassette integrated to α-amylase locus were cultivated in a 500 ml flask for 48 h. The recombinant strain with the highest activity at 500 ml shake flask was further cultivated in 7 and 50 l bioreactor, respectively.
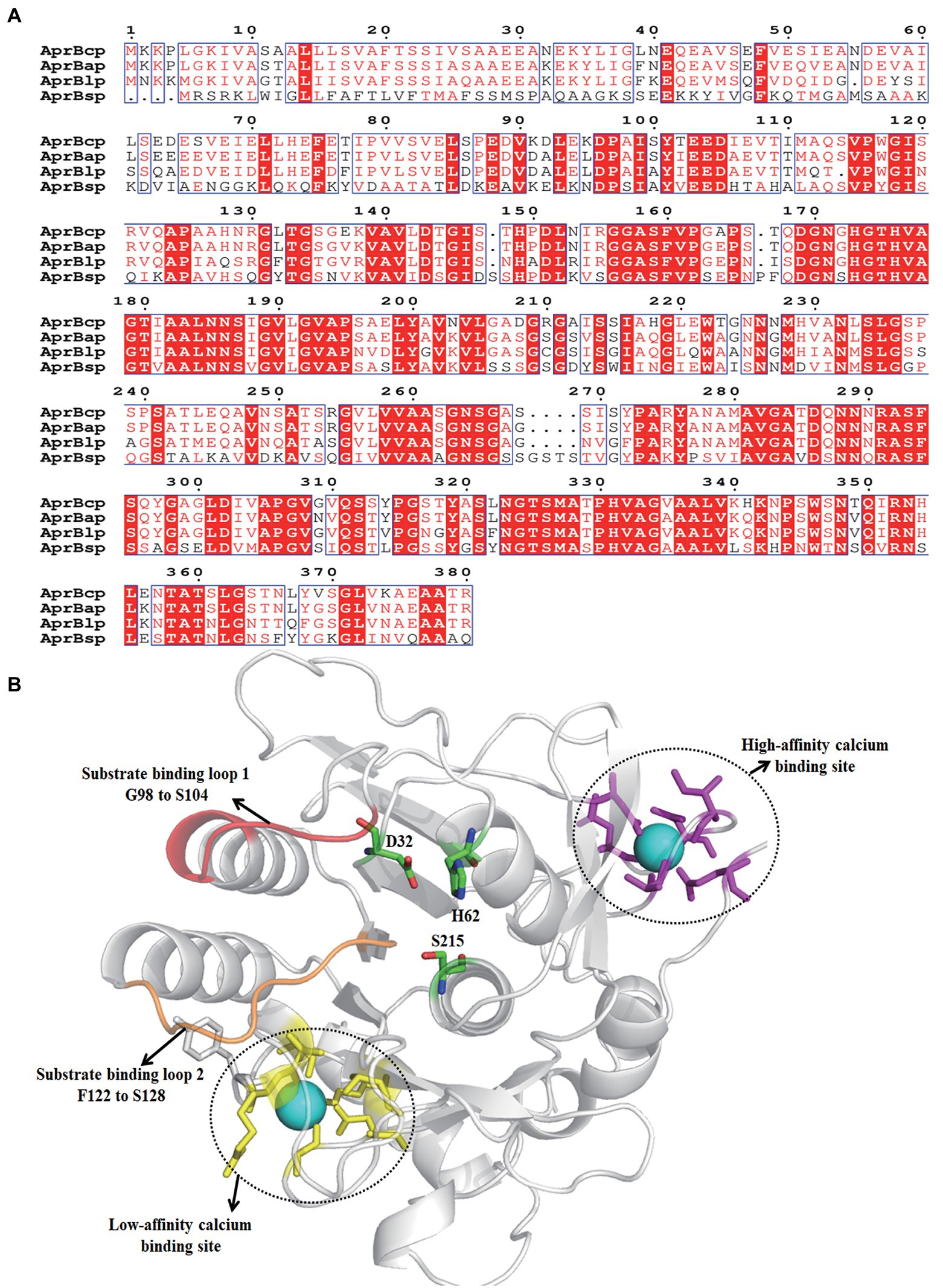
Figure 2. Sequence analysis of AprBcp. (A) Sequence alignment of AprBcp with other alkaline proteases. The listed sequences included the alkaline protease AprBlp from Bacillus lehensis (AFK0897 0.1), AprBap from Bacillus alcalophilus (FJ940727.1) and AprBsp from B. subtilis (WP_003327717.1). (B) Homology modeling structure of AprBcp.
The fermentation supernatant after centrifugation was filtered through with a 0.22 μm membrane, then concentrated and buffer-exchanged to Tris–HCl buffer (20 mM, pH 7.5) by ultrafiltration with a membrane of 10 kDa cutoff. The AprBcp was purified by HiTrap SP Fast Flow chromatography. The purified AprBcp was collected and analyzed by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
The protease activity was detected according to the Folin’s phenol method (Wang et al., 2019). A quantity of 1 g casein was dissolved in 100 ml boric acid buffer (pH 10.0, 50 mM) and used as substrate. Firstly, 200 μl of diluted enzyme was added to 200 μl of 1% (w/v) casein solution and incubated at 60°C for 10 min. Secondly, 400 μl of 400 mM trichloroacetic acid (TCA) was added to end the reaction, and then centrifuged at 11,000 × g for 5 min. Finally, 200 μl of the supernatant was added into 1 ml of 400 mM Na2CO3 and 200 μl of Folin’s phenol reagent and incubated at 40°C for 20 min. The reactions with enzymes added after TCA were used as controls. One unit of alkaline protease activity was defined as the amount of enzyme that releases 1 μg of tyrosine from casein at 60°C and pH 10.0.
The optimal pH of AprBcp was assayed in different 50 mM buffer from pH 6.0 to 11.0 (Na2HPO4–NaH2PO4 for pH 6.0–7.0, Tris–HCl for pH 8.0–9.0, and boric acid-NaOH for pH 10.0–11.0). The relative activity at different pH was calculated by setting pH 10.0 as 100%. For the pH stability, AprBcp was incubated at 30°C in 50 mM buffer with different pH from 6.0 to 11.0 for 6 h, and then the residual activity was determined. The highest enzyme activity was considered as 100%, and other values were recorded as a percentage of the highest value.
The optimal temperature of AprBcp was measured at different temperatures from 40°C to 75°C. The relative activity at different temperatures was calculated by setting 60°C as 100%. The thermal stability of AprBcp was investigated by incubating at temperatures from 40°C to 70°C for 10 min, and the residual activity was determined at pH 10.0 and 60°C. Furthermore, thermal stabilities of AprBcp at 60°C in presence of 2, 5, and 10 mM Ca2+ were determined by incubation at different time intervals. The residual activity was calculated by taking the non-heated AprBcp activity as 100%.
Several inhibitors, such as pepstatin A, PMSF, EDTA, and iodoacetamide, were used to investigate the inhibition effects on the activity of AprBcp. The residual activity of AprBcp was detected in presence of these inhibitors and calculated by taking the non-treated AprBcp activity as 100%.
The effect of different metal ions on the activity of AprBcp was analyzed by incubating enzyme samples for 6 h at room temperature in 50 mM Tris–HCl buffer (pH 8.0), containing 1 and 5 mM of Ca2+, Mg2+, Na+, K+, Li+, Zn2+, Mn2+, Co2+, Hg2+, Ag+, and Cu2+. The residual activity was determined as described previously.
For substrate specificity, casein, BSA, keratin, soy protein isolate, and chicken egg albumin were used as substrate, respectively. The relative activity on each substrate was expressed as the percentage of that on casein.
The soybean dregs powders were mixed with deionized water at a ratio of 3% (w/w), and homogenized using a homogenizer at a speed of 11,000 × g for 3 min before it was adjusted pH to 9.0 with 1 M NaOH. The mixtures were gradually raised to 50°C prior to the addition of proteinase AprBcp, and then continuously shook at that temperature for 3 h. The resulting suspensions were incubated in boiling water for 10 min to inactivate the proteinase, and subsequently centrifuged at 11,000 × g for 30 min to remove the insoluble residue. The collected supernatant was acidified with 3 M HCl to pH 7.0.
Different ratios of enzyme to substrate (500, 1,000, 1,500, 2,000, and 2,500 U/g), ratio of substrate to water (1, 2, 3, 4, and 5%, w/w), hydrolysis time (2, 3, 4, 5, and 6 h), hydrolysis temperature (40, 45, 50, 55, and 60°C), and pH (7, 8, 9, 10, and 11) were selected successively for optimization.
The protein recovery (RP, %) was used as a measurement of the soybean dregs’ protein extraction efficiency and defined as the ratio of the protein mass in the supernatant to that in the soybean dregs. The protein content was determined using the GB 5009.5-2016 of the Chinese standard.
All measurements were done in triplicate. The data were analyzed by the Statistical Package for the Social Sciences (SPSS) software (version 18.0, SPSS Inc., United States) and were expressed as mean ± standard deviation (SD).
The results of NCBI-blastn and open reading frame finder exhibited that the open reading frame of bcp was 1,143 bp, which encoded a polypeptide with 381 amino acid residues. The sequence of bcp was deposited in the GenBank of NCBI (accession no. ON733249). Based on the results of NCBI-blastp, we deduced that AprBcp is an alkaline serine protease, which belonged to S8 family peptidase. In addition, AprBcp displayed 92.3, 89.4, and 78.7% identity against alkaline serine protease from B. circulans NCTC2610 (accession no. SPU21234.1), Alkalihalobacillus clausii (accession no. WP_094423791), and Alkalihalobacillus patagoniensis (accession no. WP_09419032 9.1), respectively. Multiple alignment analyses of different alkaline serine protease with AprBcp displayed that alkaline proteases from bacillus are composed of three different domains, which include signal peptide, pro-peptide, and mature peptide domain (Figure 2A). For AprBcp, the first 27 amino acid residues, the sequence from Ala28 to Met111, and the sequence from Ala112 to Arg380 were signal peptide, pro-peptide, and mature peptide domain, respectively.
The tertiary structure of AprBcp with mature peptide domain was obtained by homology modeling using the alkaline proteases from Bacillus lentus as a template (PDB deposition: 1st3.1.A). As shown in Figure 2B, the catalytic triad consisted of D32, H62, and S215. In addition, two surface loops, located from position G98 to S104 and F122 to S128, were responsible for substrate binding. Similar to already reported alkaline proteases (Zhang et al., 2019), AprBcp also has two calcium-binding sites. The first calcium-binding site with high affinity is involved of a loop from position A72 to L80 and side chains of Gln2 and Asp40. The second calcium-binding site, a low-affinity binding site, was formed by two bound water and residues Tyr165, Ala168 Asp191, and Arg241.
In this study, 10 promoters were chosen to drive the expression of AprBcp in B. subtilis 1285 (Table 1). The process for construction of expression vectors with different promoters was depicted in Figure 1. The obtained 10 expression vectors were transformed to B. subtilis 1285 and the strengths of those 10 promoters were evaluated by extracellular activity of AprBcp. As exhibited in Supplementary Figure 2 and Figure 3A, the recombinant strain Bs7 containing plasmid pHY7 with promoter Pcry3A exhibited the highest activity at both 24-well plate and shake flask cultivation, followed by Bs8 (PBsapr), Bs3 (PBsamy), Bs4 (PBaamy), and Bs6 (PHpaII). For cultivated 500 ml shake flask, the highest activity of Bs7 was 5,580 U/ml, which is 1.13- to 2.73-fold higher than that of another recombinant strain containing different promoters (Figure 3A). Until now, there are few reports showed that promoter Pcry3A from Bacillus thuringiensis could drive high-level expression of alkaline protease in B. subtilis (Widner et al., 2000). The results of this study demonstrated that promoter Pcry3A was more suitable for heterologous expression of recombinant AprBcp in B. subtilis 1285 than another nine promoters. Our results were similar to previous studies, which demonstrated that optimization of promoter could effectively improve the production of recombinant protein in B. subtilis (Song et al., 2015; Song et al., 2017; Zhang et al., 2017; Liu et al., 2019; Kang et al., 2020).
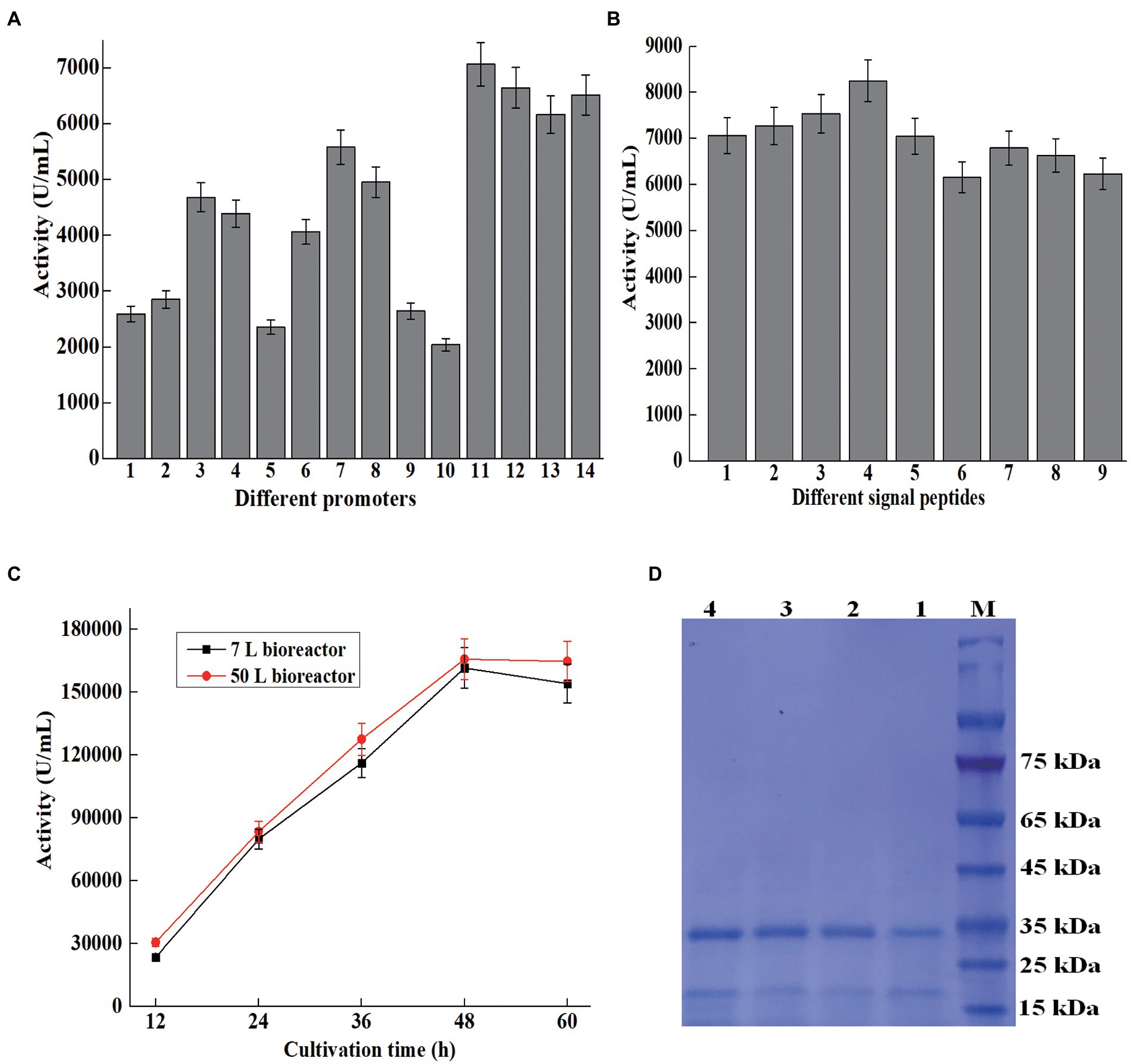
Figure 3. Overexpression of AprBcp in Bacillus subtilis. (A) Promoter optimization. Numbers 1–14 represent promoter P43, PgsiB, PBsamy, PBaamy, PBsnpr, PHpaII, Pcry3A, PBsapr, PBlapr, P09, PBsapr-cry3A, PBaamy-cry3A, PBsamy-cry3A, and PHpaII-cry3A. (B) Signal peptide optimization. Numbers 1–9 represent signal peptide SPbcp, SPBsamy, SPBsapr, SPBschi, SPBsnpr, SPDacB, SPVpr, SPYncM, and SPBsap. (C) Large-scale production of AprBcp in 7 and 50 l bioreactor. (D) SDS-PAGE analysis of supernatant from different cultivation time in 50 l bioreactor. Numbers 1–4 represent supernatant from 12 to 60 h.
As a key regulatory element, promoter plays an important role on the expression level of heterologous gene in B. subtilis. Many studies demonstrated that the strength of dual promoter was normally higher than the single in B. subtilis (Guan et al., 2016; Zhang et al., 2017; Liu et al., 2019; Kang et al., 2020). For further improvement of the production of AprBcp, expression vectors with dual promoter were constructed (Table 1; Figure 1). According to previous work, the transcriptional strength of a dual promoter depends on the promoter adjacent to the gene, so promoter Pcry3A was located adjacent to bcp. Promoter PBsapr, PBsamy, PBaamy, and PHpaII were inserted to pHY7 upstream Pcry3A, generating plasmids with dual promoter such as PBsapr-cry3A, PBaamy-cry3A, PBsamy-cry3A, and PHpaII-cry3A, which were in accordance with plasmids pHY11, pHY12, pHY13, and pHY14, respectively. As exhibited in Supplementary Figure2 and Figure 3A, the extracellular activities of recombinant strains with dual promoter were higher than Bs7 (Pcry3A), which demonstrated that dual promoter could further improve the production of recombinant AprBcp. Among those recombinant strains, Bs11 (PBsapr-cry3A) with highest extracellular activity (7,068 U/ml), which was 1.26-fold higher than that of Bs7 (Pcry3A). According to the previous literatures, two tandem promoters can improve the production of recombinant protein in B. subtilis. As reported by Kang et al. (2020), the production of recombinant amidase drove by dual promoter PamyE-cdd was 1.37-fold higher than that of the control. Similarly, the activity of extracellular BcaPRO from recombinant strain containing dual promoter PBsamy-Baamy was 1.5-fold of the activity of strain containing single promoter PBaamy (Liu et al., 2019). In recent years, different dual tandem promoters have been reported to drive expression of recombinant protein in B. subtilis such as PBsamy-Baamy, PamyE-cdd, PspoVG-spoVG142, PHpaII-amyQ, and PgsiB-HpaII (Guan et al., 2016; Zhang et al., 2017; Liu et al., 2019; Kang et al., 2020). To our best knowledge, there are no reports about dual promoter PBsapr-cry3A. The results of this study revealed that dual tandem promoter PBsapr-cry3A with great potential for heterologous expression of recombinant protein in B. subtilis.
As a key regulatory element, signal peptide plays an important role on efficient extracellular expression of target protein in B. subtilis (Song et al., 2017; Fu et al., 2018). To further improve the production of AprBcp in B. subtilis, eight efficient sec-type signal peptides from B. subtilis were selected to replace the native signal peptide of AprBcp (Table 1; Figure 1). As shown in Supplementary Figure 3 and Figure 3B, the recombinant strain BS17 (containing signal peptide SPBschi) exhibited the highest activity among those eight recombinant strains containing different signal peptides. The maximum extracellular activity of recombinant strain BS17 was 8,253 U/ml, which was almost about 1.09- to 1.33-fold higher than that of the other strain with different signal peptides. Until now, there are few studies reported the signal peptide SPBschi of chitinase from B. subtilis. Generally, signal peptides of amylase and protease from B. subtilis, B. licheniformis, or Bacillus amyloliquefaciens are usually used to the secretion of recombinant protein in B. subtilis. The result of this study revealed that signal peptide SPBschi of chitinase is more efficient than that from α-amylase, alkaline, and neutral protease.
Generally, the signal peptide is composed of positively charged N-region, hydrophobic H-region, and C-region. The positively charged N-region is responsible for the interaction with the translocation machinery. Additionally, the hydrophobic H-region, following the charged N-region, functions on the process of insertion of signal peptide into the membrane. Previous studies indicated that the efficiency of signal peptide is related to its positively charged N-region and hydrophobic H-region (Guo et al., 2021). In this study, the N-domain charge and the ratio of hydrophobic amino acids of eight efficient Sec-type signal peptides used in this study were from 2 to 4, and 54.5 to 72.7%, respectively (Supplementary Table 4). The most efficient signal peptide SPBschi with the highest N-domain charge, but lowest ratio of hydrophobic amino acids, which indicated that the efficiency of secretion may not direct relative to the properties described above. Based on the results of previous (Liu et al., 2019; Kang et al., 2020) and this study, we concluded that each target protein has its preference for signal peptide and it is necessary to screen a suitable signal peptide for efficient extracellular production of target protein.
During the processes of promoter and signal peptide optimization, the expression of AprBcp was mediated by free plasmids, and the kanamycin was required to maintain expression vectors. However, cultivation added with kanamycin and the plasmid with kanamycin resistance gene marker are both prohibited in many application industries (Yang et al., 2016). Therefore, it is necessary to develop a mark-free expression way to produce AprBcp. The expression vector pHY17 contains temperature-sensitive replicon PE194, which will be inactivated when cultivation temperature is up to 45°C. Then, the fragment with dual promoter PBsapr-cry3A, signal peptide SPBschi, bcp, 5′-upstream and 3′-downstream homologous template of α-amylase will be integrated to α-amylase locus of B. subtilis1285. A total of 10 mark-free strains named Bs23 to Bs32 were selected and cultivated in shake flask. The extracellular activities of these 10 mark-free strains were from 8,123 to 8,365 U/ml and the strain Bs27 with highest activity (8,365 U/ml) was chosen for large-scale fermentation.
The large-scale fermentation of Bs27 was carried in 7- and 50-L bioreactor. The maximum activity produced by recombinant strain Bs27 in 7- and 50-L bioreactor were 161,661 and 165,870 U/ml, respectively (Figure 3C). In recent years, several engineered Bacillus strains, such as B. subtilis, B. licheniformis, and B. amyloliquefaciens, were cultivated in a bioreactor to prepare alkaline protease (Liu et al., 2019; Wang et al., 2019; Zhou et al., 2020, 2021). Previous studies reported that the extracellular alkaline protease activity of engineered B. licheniformis 2709, B. amyloliquefaciens K11, and B. subtilis WB600 reached 57,763, 30,200, and 27,860 U/ml, respectively, in 5 or 15 l fermenter (Wang et al., 2016, 2019; Liu et al., 2019; Zhou et al., 2021). However, for most of the studies, the method for alkaline protease activity assay was based on the method of the People’s Republic of China GB/T 23527-2009, in which one unit of alkaline protease activity was defined as the amount of enzyme that releases 1 μg of tyrosine from casein at 40°C and pH 10.5. Thus, our results were not fully comparable with these values since activity assays with different conditions have been used. SDS-PAGE analysis of fermented supernatant from different cultivation times in 50-L bioreactor revealed that AprBcp was approximately free from contaminating proteins, which facilitates downstream processing (Figure 3D).
The process for purification of AprBcp is shown in Supplementary Table 5. After ultrafiltration and affinity chromatography, AprBcp was purified 2.09-fold to homogeneity and specific activity reached 58,621 U/mg, which was higher than many previous reported alkaline proteases but lower than those from Idiomarina sp. C9-1 (99,511.9 U/mg), Termitomyces albuminosus (180,000 U/mg), Trametes cingulata CTM10101 (94,000 U/mg), and Vibrio metschnikovii J1 (120,000 U/mg; Jellouli et al., 2009; Zheng et al., 2011; Omrane Benmrad et al., 2016; Zhou et al., 2018). SDS-PAGE analysis showed that the purified AprBcp was about 30 kDa corresponding to molecular mass of mature peptide of AprBcp, which is similar to other alkaline proteases from Bacillus (Contesini et al., 2018). It has been reported that alkaline proteases from Bacillus species almost consist of a signal peptide, pro-peptide, and subtilisin domain. Theoretically, alkaline proteases from Bacillus are activated by three steps: (1) folding of alkaline protease containing different domains, (2) auto-processing between the pro-peptide and subtilisin domain, and (3) degradation of the pro-peptide by activated subtilisin (Uehara et al., 2013).
The values of Km and Vmax of purified AprBcp were 2.35 mg/ml and 63,038 μM min−1 ml−1, respectively. Kinetic parameters of purified AprBcp indicated that it has high substrate affinity and catalytic efficiency toward casein. Based on previous studies, we found that Km values of Bacillus alkaline proteases vary differently. The Km values of alkaline proteases from B. cereus-S6-3, Bacillus stearothermophilus, Bacillus safensis RH12, and B. circulans M34 are 3.07, 0.62, 2.2, and 0.96 mg casein/mL, respectively (Sari et al., 2015; Abdel-Naby et al., 2017, 2020; Rekik et al., 2019).
The optimum pH for maximum activity of AprBcp was 10.0, and the relative activities were above 65% in the range from pH 7.0 to 11.0 (Figure 4A). Generally, the most reported alkaline proteases exhibit higher activity in the range of 8.0–11.0. The maximum activities of alkaline proteases from B. pumilus AR57 and Bacillus megaterium RRM2 are detected at 9.0 and 10.0, respectively (Rajkumar et al., 2011; Jagadeesan et al., 2020). While, the optimal pH of an alkaline protease from Bacillus halodurans C-125 is 12.0 (Tekin et al., 2021). For pH stability, AprBcp exhibited high residual activity (above 80%) in the range from pH 8.0 to 11.0 after 5 h of incubating at 25°C (Figure 4B), which is similar to previous reported alkaline protease (Rajkumar et al., 2011; Jagadeesan et al., 2020; Tekin et al., 2021).
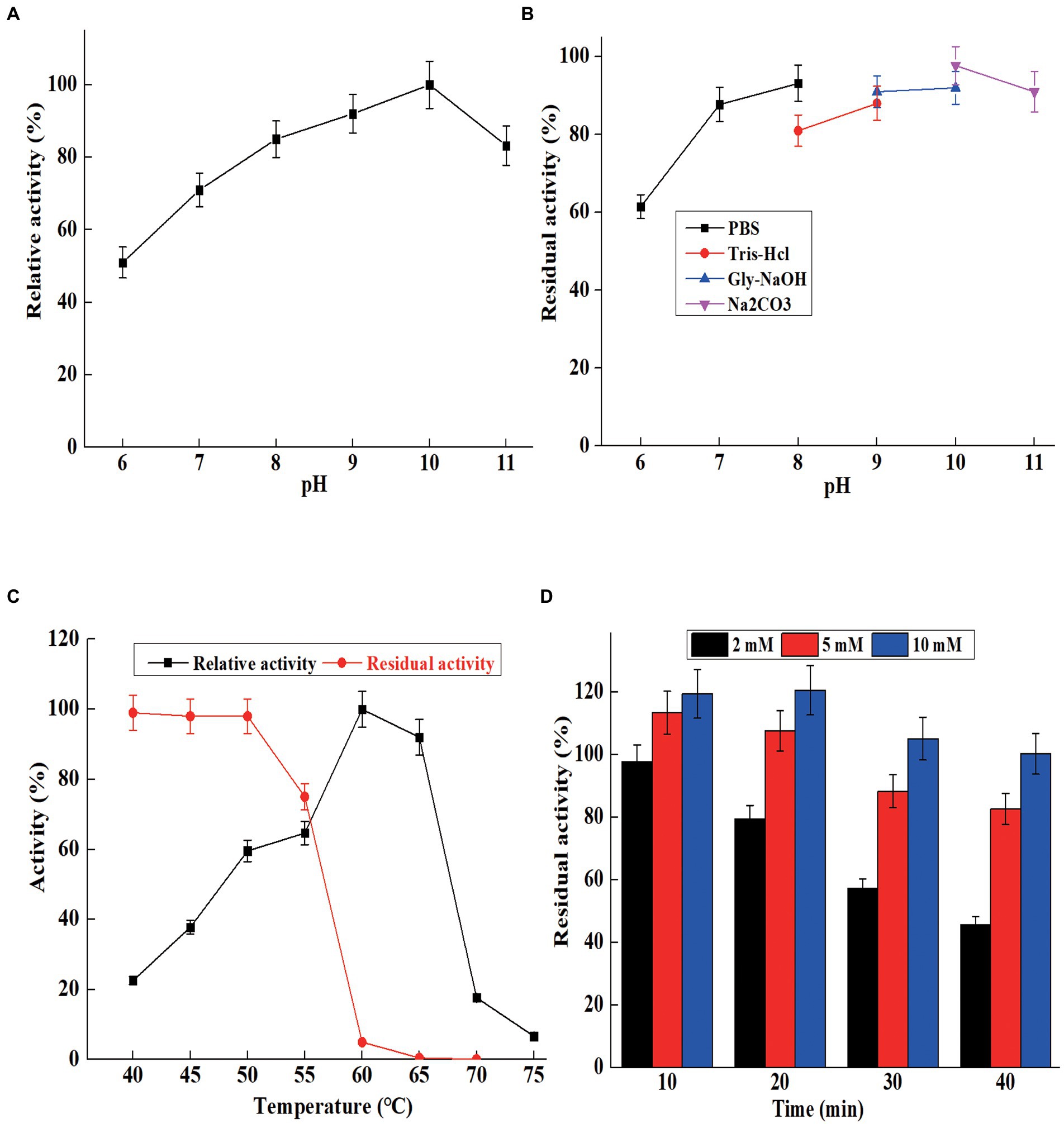
Figure 4. The characterization of purified AprBcp. (A) Optimum pH of purified AprBcp. (B) pH stability of purified AprBcp. (C) Optimum temperature and thermal stability of purified AprBcp. (D) The effects of different concentration of Ca2+ on the stability of AprBcp incubating at 60°C.
For optimum temperature, AprBcp exhibited maximum relative activity at 60°C and remained highly active from 50 to 65°C (Figure 4C). The optimum temperature of already studied alkaline proteases verifies differently according to their origin. Normally, alkaline proteases from Bacillus showed high activity in the range from 50 to 80°C (Contesini et al., 2018). As far as thermal stability analysis is concerned, the purified AprBcp was incubated in buffers with and without Ca2+. The residual activities of AprBcp in the absence of Ca2+ at 50, 55, 60, and 65°C were 98.2, 75.3, 5.2, and 0.5%, respectively, after heat treatment for 10 min (Figure 4C). As shown in Figure 4D, it was obvious that addition with Ca2+ could effectively improve the thermal stability of AprBcp. The residual activities of AprBcp in the presence of 2, 5, and 10 mM Ca2+ were 45.8, 82.3, and 100.3%, respectively, after heat treatment at 60°C for 40 min. It has been reported that Ca2+ plays an important role on the stability of alkaline protease. For instance, the stability of alkaline protease from Trametes cingulata CTM10101, Nocardiopsis alba OM-5, and B. safensis S406 was enhanced by 1.24-to 4.52-fold, respectively (Omrane Benmrad et al., 2016; Mhamdi et al., 2017; Chauhan et al., 2021). According to the results from Zhang et al. (2019), Ca2+ could as an activator to improve the stability and activity of alkaline protease from Bacillus species is attributed to its electrostatic interaction with alkaline protease. In this study, there were two calcium-binding sites in AprBcp to interact with Ca2+, which improved the stability of AprBcp in extreme conditions.
It has been reported that proteases from different sources are sensitive to various inhibitors (Barzkar, 2020). Usually, serine proteases are inhibited by PMSF, diisopropyl fluorophosphate, and 3,4-dichloroisocoumarin. The relative activities of AprBcp treated with different inhibitors are shown in Table 2. AprBcp was strongly inhibited by PMSF and the relative activity was only 27.2 and 0.5%, respectively, in the presence of 1 and 5 mM PMSF, suggesting that serine is relative to the catalytic activity. Based on previous studies, we found that the activities of most serine proteases were completely inhibited by PMSF at a concentration of 5 mM (Barzkar, 2020; Jagadeesan et al., 2020; Chang et al., 2021). However, for an alkaline serine protease from Pseudoalteromonas sp. 129–1, the maximum activity inhibition is detected in the presence of 15 mM PMSF (Wu et al., 2015). In addition, the relative activities of AprBcp were 86.3 and 73.2%, respectively, in presence of 1 and 5 mM EDTA, revealing that some metallic ions are important for catalytic activity of AprBcp. The inhibitory effect of EDTA on AprBcp is similar to previous studies (Omrane Benmrad et al., 2018; Sharma et al., 2020; Chang et al., 2021). The alkaline proteases from Nocardiopsis dassonvillei OK-18, B. stearothermophilus CAU209, and Penicillium chrysogenum X5 are inhibited by 30.8, 40.5, and 22.2%, respectively, in presence of 1 to 10 mM EDTA (Omrane Benmrad et al., 2018; Sharma et al., 2020; Chang et al., 2021). Compared with PMSF and EDTA, pepstatin A and iodoacetamide exhibited little inhibitory effects on the activity of AprBcp, which indicated that-SH group and aspartate were not involved in the catalytic activity of AprBcp.
Several metal ions were assessed for their effects on the stability of AprBcp (Table 3). As activator, metal ions Ca2+, Mn2+, and Mg2+ could improve the residual activity of AprBcp by 115.2, 112.3, and 108.5%, respectively, at a concentration of 5 mM. Besides, AprBcp was inhibited by Cu2+ and the residual activities were 40.7 and 25.9%, respectively, in the presence of 1 and 5 mM Cu2+. Other metal ions, such as K+, Na+, Co2+, Zn2+, and Al3+ exhibited little effects on the stability of AprBcp. According to the previous studies, metal ions act as activator or inhibitor mainly depend on the specific target alkaline proteases (Contesini et al., 2018; Barzkar, 2020). The result of this study and some previous research found that Cu2+ is an inhibitor. Otherwise, the alkaline proteases from B. safensis RH12 and Penicillium chrysogenum X5 are activated by Cu2+ and the residual activities are upregulated by 34.2 and 5.3%, respectively, when treated with 1 or 5 mM Cu2+ (Omrane Benmrad et al., 2018; Rekik et al., 2019).
The substrate preferences of AprBcp were characterized with various natural proteinaceous substrates (Table 4). AprBcp exhibited the highest activity toward casein, followed by soybean protein isolate (61.2%) and bovine serum albumin (46.2%). Furthermore, AprBcp also displayed little activity against keratin and chicken egg albumin.
To search for the effect of the proteinase AprBcp enzymatic hydrolysis of soybean dregs on the protein recovery, single-factor experiments were carried out (Figure 5). The protein recovery (RP) gradually increased with the increase in the ratio of enzyme to substrate and leveled off after 2,000 U/g (Figure 5A). Therefore, the selected ratio of enzyme to substrate was 2,000 U/g. In addition, as ratio of substrate to water increased, the RP first increased and then decreased, reaching the maximum value at 2%, however, with no significant difference between 2 and 3% (Figure 5B). Considering the improvement of industrial production efficiency, ratio of substrate to water = 3% was selected. As shown in Figure 5C, when the hydrolysis time reached 4 h, the RP was the highest (69.64%). But, in industrial production, shorter enzymatic digestion time can prevent the growth of stray bacteria and reduce reaction costs. Thus, 3 h was selected as the hydrolysis time. Temperature is an important index affecting protease activity. The optimal temperature for enzymolysis of soybean dregs by proteinase AprBcp was 45°C, which was close to our above study (Figure 5D). When pH was 9.0, proteinase AprBcp exhibited the best hydrolysis efficiency in soybean dregs with a protein recovery of 71.81% (Figure 5E), which was higher than the results reported by Figueiredo et al. (2018).
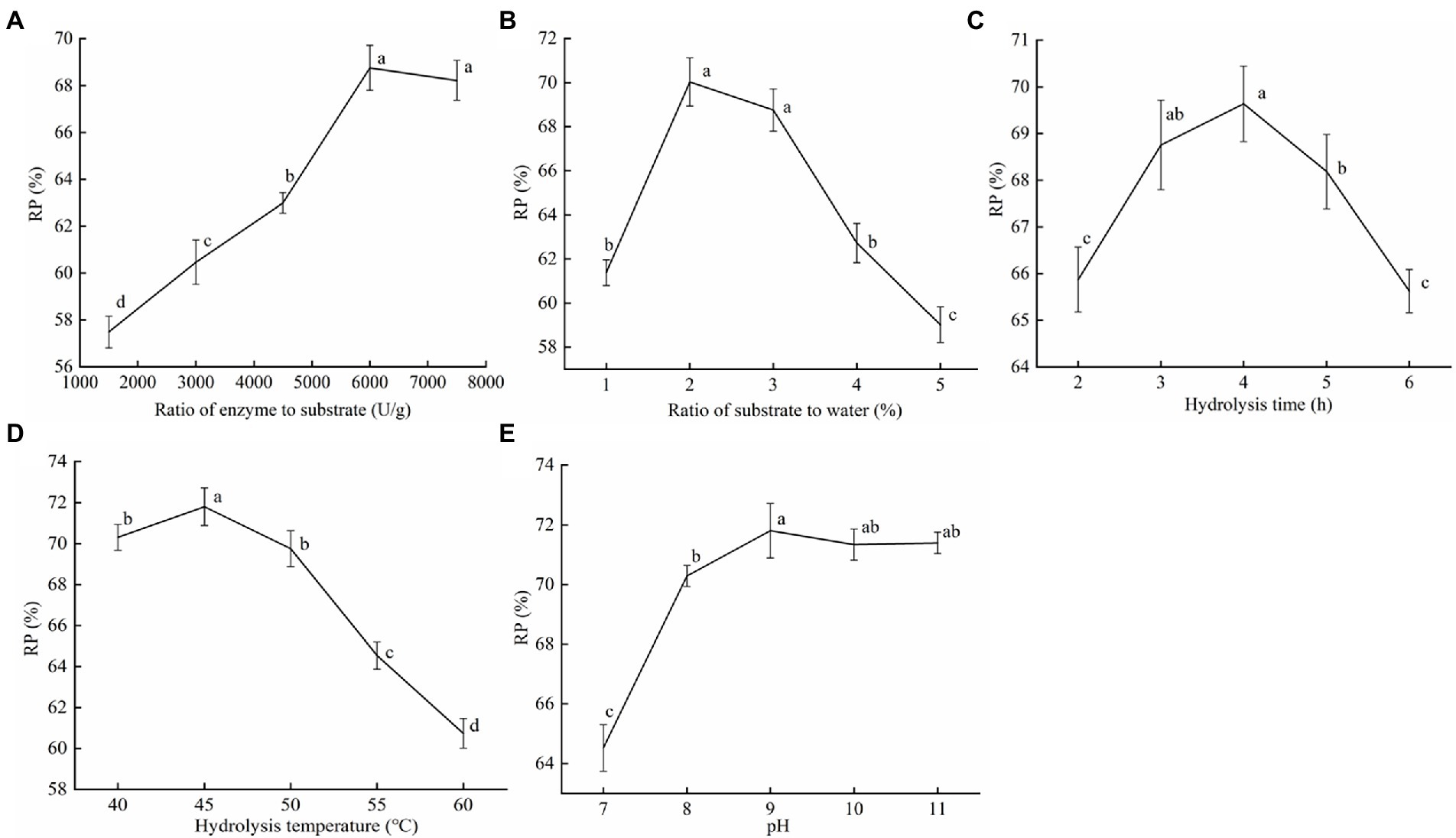
Figure 5. The effect of the proteinase AprBcp enzymatic hydrolysis of soybean dregs on the protein recovery. (A) Ratio of enzyme to substrate. (B) Ratio of substrate to water. (C) Hydrolysis time. (D) Hydrolysis temperature. (E) pH. Different lower case letters indicate significant differences (p < 0.05).
In conclusion, an alkaline protease (AprBcp) from B. circulans R1 was overexpressed, purified, and characterized. AprBcp belonged to S8 family peptidase. The maximum activity of recombinant strain Bs27 reached 165,870 U/ml. The purified AprBcp was most active at pH 10.0 and 60°C. Additionally, Ca2+ could as an activator to improve the stability of AprBcp. Furthermore, the recombinant AprBcp exhibited great potential application on recovery protein from soybean dregs. The excellent properties and overexpression of AprBcp will provide a basis for its further application on recovery protein from by-products from food and agriculture industry.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material. GenBank ON733249.
HC contributed to gene clone, construct recombinant strain, and bioinformatics analysis of AprBcp and writing. JWu contributed to characterization, purification, and kinetic parameters of AprBcp. XH contributed to recovery of protein from soybean dregs by AprBcp. XF contributed to large-scale production of AprBcp. HJ contributed to project administration. LZ contributed to the funding acquisition. JWa contributed to promoter optimization, signal peptide optimization, and experiment planning. All authors contributed to the article and approved the submitted version.
This work was supported by the Science and Technology Innovation Program of Hunan Province (2019TP1028, 2019SK2122, 2019NK4229, and 2022NK2039), Postgraduate Scientific Research Innovation Project of Hunan Province (CX20211281, CX20211282), the Natural Science Foundation of Hunan Province (2021JJ40514), Guangdong Provincial Key Laboratory of Aquatic Product Processing and Safety (GDPKLAPPS2203), the National Key R&D Program (2019YFD0902000), and the Guangdong Innovation Team of Seafood Green Processing Technology (2019KCXTD011).
XF was employed by Shenzhen Shanggutang Food Development Co., Ltd. JWa was employed by Shenzhen Raink Ecology & Environment Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.968439/full#supplementary-material
1. ^https://www.expasy.org/ (Accessed July 25, 2022).
2. ^https://www.genome.jp/tools-bin/clustalw (Accessed July 25, 2022).
3. ^https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi(Accessed July 25, 2022).
Abdel-Naby, M. A., Ahmed, S. A., Wehaidy, H. R., and El-Mahdy, S. A. (2017). Catalytic, kinetic and thermodynamic properties of stabilized Bacillus stearothermophilus alkaline protease. Int. J. Biol. Macromol. 96, 265–271. doi: 10.1016/j.ijbiomac.2016.11.094
Abdel-Naby, M. A., El-Wafa, W. M. A., and Salem, G. E. M. (2020). Molecular characterization, catalytic, kinetic and thermodynamic properties of protease produced by a mutant of Bacillus cereus-S6-3. Int. J. Biol. Macromol. 160, 695–702. doi: 10.1016/j.ijbiomac.2020.05.241
Barzkar, N. (2020). Marine microbial alkaline protease: an efficient and essential tool for various industrial applications. Int. J. Biol. Macromol. 161, 1216–1229. doi: 10.1016/j.ijbiomac.2020.06.072
Baweja, M., Tiwari, R., Singh, P. K., Nain, L., and Shukla, P. (2016). An alkaline protease from Bacillus pumilus MP 27: functional analysis of its binding model toward its applications as detergent additive. Front. Microbiol. 7:1195. doi: 10.3389/fmicb.2016.01195
Cai, D., Rao, Y., Zhan, Y., Wang, Q., and Chen, S. (2019). Engineering Bacillus for efficient production of heterologous protein: current progress, challenge and prospect. J. Appl. Microbiol. 126, 1632–1642. doi: 10.1111/jam.14192
Cao, Z. H., Green-Johnson, J. M., Buckley, N. D., and Lin, Q. Y. (2019). Bioactivity of soy-based fermented foods: a review. Biotechnol. Adv. 37, 223–238. doi: 10.1016/j.biotechadv.2018.12.001
Chang, C., Gong, S., Liu, Z., Yan, Q., and Jiang, Z. (2021). High level expression and biochemical characterization of an alkaline serine protease from Geobacillus stearothermophilus to prepare antihypertensive whey protein hydrolysate. BMC Biotechnol. 21:21. doi: 10.1186/s12896-021-00678-7
Chauhan, J. V., Mathukiya, R. P., Singh, S. P., and Gohel, S. D. (2021). Two steps purification, biochemical characterization, thermodynamics and structure elucidation of thermostable alkaline serine protease from Nocardiopsis alba strain OM-5. Int. J. Biol. Macromol. 169, 39–50. doi: 10.1016/j.ijbiomac.2020.12.061
Chen, J., Jin, Z., Gai, Y., Sun, J., and Zhang, D. (2017). A food-grade expression system for d-psicose 3-epimerase production in Bacillus subtilis using an alanine racemase-encoding selection marker. Bioresour. Bioprocess. 4:9. doi: 10.1186/s40643-017-0139-7
Cheng, J. H., Wang, Y., Zhang, X. Y., Sun, M. L., Zhang, X., Song, X. Y., et al. (2021). Characterization and diversity analysis of the extracellular proteases of Thermophilic Anoxybacillus caldiproteolyticus 1A02591 from deep-sea hydrothermal vent sediment. Front. Microbiol. 12:643508. doi: 10.3389/fmicb.2021.643508
Contesini, F. J., Melo, R. R., and Sato, H. H. (2018). An overview of Bacillus proteases: from production to application. Crit. Rev. Biotechnol. 38, 321–334. doi: 10.1080/07388551.2017.1354354
Cui, W., Han, L., Suo, F., Liu, Z., Zhou, L., and Zhou, Z. (2018). Exploitation of Bacillus subtilis as a robust workhorse for production of heterologous proteins and beyond. World J. Microbiol. Biotechnol. 34:145. doi: 10.1007/s11274-018-2531-7
Ding, Y., Yang, Y., Ren, Y., Xia, J., Liu, F., Li, Y., et al. (2020). Extracellular production, characterization, and engineering of a Polyextremotolerant Subtilisin-like protease from feather-degrading Thermoactinomyces vulgaris strain CDF. Front. Microbiol. 11:605771. doi: 10.3389/fmicb.2020.605771
Fayaz, G., Plazzotta, S., Calligaris, S., Manzocco, L., and Nicoli, M. C. (2019). Impact of high pressure homogenization on physical properties, extraction yield and biopolymer structure of soybean okara. LWT-Food Sci. Technol. 113:108324. doi: 10.1016/j.lwt.2019.108324
Figueiredo, V. D., Yamashita, F., Vanzela, A., Ida, E. I., and Kurozawa, L. E. (2018). Action of multi-enzyme complex on protein extraction to obtain a protein concentrate from okara. J. Food Sci. Technol. 55, 1508–1517. doi: 10.1007/s13197-018-3067-4
Fu, G., Liu, J., Li, J., Zhu, B., and Zhang, D. (2018). Systematic screening of optimal signal peptides for secretory production of heterologous proteins in Bacillus subtilis. J. Agric. Food Chem. 66, 13141–13151. doi: 10.1021/acs.jafc.8b04183
Gu, Y., Xu, X., Wu, Y., Niu, T., Liu, Y., Li, J., et al. (2018). Advances and prospects of Bacillus subtilis cellular factories: from rational design to industrial applications. Metab. Eng. 50, 109–121. doi: 10.1016/j.ymben.2018.05.006
Guan, C., Cui, W., Cheng, J., Liu, R., Liu, Z., Zhou, L., et al. (2016). Construction of a highly active secretory expression system via an engineered dual promoter and a highly efficient signal peptide in Bacillus subtilis. New Biotechnol. 33, 372–379. doi: 10.1016/j.nbt.2016.01.005
Guo, X., Chai, C., An, Y., Peng, C., Shi, N., Wang, W., et al. (2021). Rational design of signal peptides for improved MtC1LPMO production in Bacillus amyloliquefaciens. Int. J. Biol. Macromol. 175, 262–269. doi: 10.1016/j.ijbiomac.2021.02.034
Heng, X., Chen, H., Lu, C., Feng, T., Li, K., and Gao, E. (2022). Study on synergistic fermentation of bean dregs and soybean meal by multiple strains and proteases. LWT-Food Sci. Technol. 154:112626. doi: 10.1016/j.lwt.2021.112626
Jagadeesan, Y., Meenakshisundaram, S., Saravanan, V., and Balaiah, A. (2020). Sustainable production, biochemical and molecular characterization of thermo-and-solvent stable alkaline serine keratinase from novel Bacillus pumilus AR57 for promising poultry solid waste management. Int. J. Biol. Macromol. 163, 135–146. doi: 10.1016/j.ijbiomac.2020.06.219
Jellouli, K., Bougatef, A., Manni, L., Agrebi, R., Siala, R., Younes, I., et al. (2009). Molecular and biochemical characterization of an extracellular serine-protease from Vibrio metschnikovii J1. J. Ind. Microbiol. Biotechnol. 36, 939–948. doi: 10.1007/s10295-009-0572-5
Kang, X. M., Cai, X., Huang, Z. H., Liu, Z. Q., and Zheng, Y. G. (2020). Construction of a highly active secretory expression system in Bacillus subtilis of a recombinant amidase by promoter and signal peptide engineering. Int. J. Biol. Macromol. 143, 833–841. doi: 10.1016/j.ijbiomac.2019.09.144
Li, B., Qiao, M., and Lu, F. (2012). Composition, nutrition, and utilization of okara (soybean residue). Food Rev. Int. 28, 231–252. doi: 10.1080/87559129.2011.595023
Liu, Y., Shi, C., Li, D., Chen, X., Li, J., Zhang, Y., et al. (2019). Engineering a highly efficient expression system to produce BcaPRO protease in Bacillus subtilis by an optimized promoter and signal peptide. Int. J. Biol. Macromol. 138, 903–911. doi: 10.1016/j.ijbiomac.2019.07.175
Liu, B., Zhang, J., Fang, Z., Gu, L., Liao, X., Du, G., et al. (2013). Enhanced thermostability of keratinase by computational design and empirical mutation. J. Ind. Microbiol. Biotechnol. 40, 697–704. doi: 10.1007/s10295-013-1268-4
Matkawala, F., Nighojkar, S., Kumar, A., and Nighojkar, A. (2021). Microbial alkaline serine proteases: production, properties and applications. World J. Microbiol. Biotechnol. 37:63. doi: 10.1007/s11274-021-03036-z
Mhamdi, S., Bkhairia, I., Nasri, R., Mechichi, T., Nasri, M., and Kamoun, A. S. (2017). Evaluation of the biotechnological potential of a novel purified protease BS1 from Bacillus safensis S406 on the chitin extraction and detergent formulation. Int. J. Biol. Macromol. 104, 739–747. doi: 10.1016/j.ijbiomac.2017.06.062
Omrane Benmrad, M., Moujehed, E., Ben Elhoul, M., Mechri, S., Bejar, S., Zouari, R., et al. (2018). Production, purification, and biochemical characterization of serine alkaline protease from Penicillium chrysogenum strain X5 used as excellent bio-additive for textile processing. Int. J. Biol. Macromol. 119, 1002–1016. doi: 10.1016/j.ijbiomac.2018.07.194
Omrane Benmrad, M., Moujehed, E., Ben Elhoul, M., Zarai Jaouadi, N., Mechri, S., Rekik, H., et al. (2016). A novel organic solvent-and detergent-stable serine alkaline protease from Trametes cingulata strain CTM10101. Int. J. Biol. Macromol. 91, 961–972. doi: 10.1016/j.ijbiomac.2016.06.025
Rajkumar, R., Jayappriyan, K. R., and Rengasamy, R. (2011). Purification and characterization of a protease produced by Bacillus megaterium RRM2: application in detergent and dehairing industries. J. Basic Microbiol. 51, 614–624. doi: 10.1002/jobm.201000517
Razzaq, A., Shamsi, S., Ali, A., Ali, Q., Sajjad, M., Malik, A., et al. (2019). Microbial proteases applications. Front. Bioeng. Biotechnol. 7:110. doi: 10.3389/fbioe.2019.00110
Rekik, H., Zarai Jaouadi, N., Gargouri, F., Bejar, W., Frikha, F., Jmal, N., et al. (2019). Production, purification and biochemical characterization of a novel detergent-stable serine alkaline protease from Bacillus safensis strain RH12. Int. J. Biol. Macromol. 121, 1227–1239. doi: 10.1016/j.ijbiomac.2018.10.139
Sari, E., Logoglu, E., and Oktemer, A. (2015). Purification and characterization of organic solvent stable serine alkaline protease from newly isolated Bacillus circulans M34. Biomed. Chromatogr. 29, 1356–1363. doi: 10.1002/bmc.3431
Sharma, A. K., Kikani, B. A., and Singh, S. P. (2020). Biochemical, thermodynamic and structural characteristics of a biotechnologically compatible alkaline protease from a haloalkaliphilic, Nocardiopsis dassonvillei OK-18. Int. J. Biol. Macromol. 153, 680–696. doi: 10.1016/j.ijbiomac.2020.03.006
Solanki, P., Putatunda, C., Kumar, A., Bhatia, R., and Walia, A. (2021). Microbial proteases: ubiquitous enzymes with innumerable uses. 3 Biotech 11:428. doi: 10.1007/s13205-021-02928-z
Song, Y., Fu, G., Dong, H., Li, J., Du, Y., and Zhang, D. (2017). High-efficiency secretion of beta-Mannanase in Bacillus subtilis through protein synthesis and secretion optimization. J. Agric. Food Chem. 65, 2540–2548. doi: 10.1021/acs.jafc.6b05528
Song, Y., Nikoloff, J. M., and Zhang, D. (2015). Improving protein production on the level of regulation of both expression and secretion pathways in Bacillus subtilis. J. Microbiol. Biotechnol. 25, 963–977. doi: 10.4014/jmb.1501.01028
Tekin, A., Uzuner, U., and Sezen, K. (2021). Homology modeling and heterologous expression of highly alkaline subtilisin-like serine protease from Bacillus halodurans C-125. Biotechnol. Lett. 43, 479–494. doi: 10.1007/s10529-020-03025-6
Uehara, R., Angkawidjaja, C., Koga, Y., and Kanaya, S. (2013). Formation of the high-affinity calcium binding site in pro-subtilisin E with the insertion sequence IS1 of pro-Tk-subtilisin. Biochemistry 52, 9080–9088. doi: 10.1021/bi401342k
Wang, J., Li, X., Chen, H., Lin, B., and Zhao, L. (2021b). Heterologous expression and characterization of a high-efficiency Chitosanase from Bacillus mojavensis SY1 suitable for production of chitosan oligosaccharides. Front. Microbiol. 12:781138. doi: 10.3389/fmicb.2021.781138
Wang, H., Yang, L., Ping, Y., Bai, Y., Luo, H., Huang, H., et al. (2016). Engineering of a Bacillus amyloliquefaciens strain with high neutral protease producing capacity and optimization of its fermentation conditions. PLoS One 11:e0146373. doi: 10.1371/journal.pone.0146373
Wang, F., Zeng, J., Gao, H., and Sukmanov, J. (2021a). Effects of different physical technology on compositions and characteristics of bean dregs. Innov. Food Sci. Emerg. 73:102789. doi: 10.1016/j.ifset.2021.102789
Wang, H., Zhang, X., Qiu, J., Wang, K., Meng, K., Luo, H., et al. (2019). Development of Bacillus amyloliquefaciens as a high-level recombinant protein expression system. J. Ind. Microbiol. Biotechnol. 46, 113–123. doi: 10.1007/s10295-018-2089-2
Widner, B., Thomas, M., Sternberg, D., Lammon, D., Behr, R., and Sloma, A. (2000). Development of marker-free strains of Bacillus subtilis capable of secreting high levels of industrial enzymes. J. Ind. Microbiol. Biot. 25, 204–212. doi: 10.1038/sj.jim.7000051
Wu, S., Liu, G., Zhang, D., Li, C., and Sun, C. (2015). Purification and biochemical characterization of an alkaline protease from marine bacteria Pseudoalteromonas sp. 129-1. J. Basic Microbiol. 55, 1427–1434. doi: 10.1002/jobm.201500327
Yang, S., Kang, Z., Cao, W., Du, G., and Chen, J. (2016). Construction of a novel, stable, food-grade expression system by engineering the endogenous toxin-antitoxin system in Bacillus subtilis. J. Biotechnol. 219, 40–47. doi: 10.1016/j.jbiotec.2015.12.029
Yang, M., Zhang, W., Ji, S., Cao, P., Chen, Y., and Zhao, X. (2013). Generation of an artificial double promoter for protein expression in Bacillus subtilis through a promoter trap system. PLoS One 8:e56321. doi: 10.1371/journal.pone.0056321
Yao, D., Su, L., Li, N., and Wu, J. (2019). Enhanced extracellular expression of Bacillus stearothermophilus alpha-amylase in Bacillus subtilis through signal peptide optimization, chaperone overexpression and alpha-amylase mutant selection. Microb. Cell Factories 18:69. doi: 10.1186/s12934-019-1119-8
Zakataeva, N. P., Nikitina, O. V., Gronskiy, S. V., Romanenkov, D. V., and Livshits, V. A. (2010). A simple method to introduce marker-free genetic modifications into the chromosome of naturally nontransformable Bacillus amyloliquefaciens strains. Appl. Microbiol. Biotechnol. 85, 1201–1209. doi: 10.1007/s00253-009-2276-1
Zhang, K., Duan, X., and Wu, J. (2016a). Multigene disruption in undomesticated Bacillus subtilis ATCC 6051a using the CRISPR/Cas 9 system. Sci. Rep. 6:27943. doi: 10.1038/srep27943
Zhang, K., Su, L., Duan, X., Liu, L., and Wu, J. (2017). High-level extracellular protein production in Bacillus subtilis using an optimized dual-promoter expression system. Microb. Cell Factories 16:32. doi: 10.1186/s12934-017-0649-1
Zhang, J., Wang, J., Zhao, Y., Li, J., and Liu, Y. (2019). Study on the interaction between calcium ions and alkaline protease of bacillus. Int. J. Biol. Macromol. 124, 121–130. doi: 10.1016/j.ijbiomac.2018.11.198
Zhang, W., Yang, M., Yang, Y., Zhan, J., Zhou, Y., and Zhao, X. (2016b). Optimal secretion of alkali-tolerant xylanase in Bacillus subtilis by signal peptide screening. Appl. Microbiol. Biotechnol. 100, 8745–8756. doi: 10.1007/s00253-016-7615-4
Zheng, S., Wang, H., and Zhang, G. (2011). A novel alkaline protease from wild edible mushroom Termitomyces albuminosus. Acta Biochim. Pol. 58, 269–273.
Zhou, C., Qin, H., Chen, X., Zhang, Y., Xue, Y., and Ma, Y. (2018). A novel alkaline protease from alkaliphilic Idiomarina sp. C9-1 with potential application for eco-friendly enzymatic dehairing in the leather industry. Sci. Rep. 8:16467. doi: 10.1038/s41598-018-34416-5
Zhou, C., Yang, G., Zhang, L., Zhang, H., Zhou, H., and Lu, F. (2021). Construction of an alkaline protease overproducer strain based on Bacillus licheniformis 2709 using an integrative approach. Int. J. Biol. Macromol. 193, 1449–1456. doi: 10.1016/j.ijbiomac.2021.10.208
Keywords: alkaline protease, overexpression, Bacillus circulans, Bacillus subtilis, soybean dregs
Citation: Chen H, Wu J, Huang X, Feng X, Ji H, Zhao L and Wang J (2022) Overexpression of Bacillus circulans alkaline protease in Bacillus subtilis and its potential application for recovery of protein from soybean dregs. Front. Microbiol. 13:968439. doi: 10.3389/fmicb.2022.968439
Received: 14 June 2022; Accepted: 21 July 2022;
Published: 26 August 2022.
Edited by:
Benevides Costa Pessela, Spanish National Research Council (CSIC), SpainReviewed by:
Tianwen Wang, Xinyang Normal University, ChinaCopyright © 2022 Chen, Wu, Huang, Feng, Ji, Zhao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangzhong Zhao, c3lzMTY5QDE2My5jb20=; Jianrong Wang, d2FuZ3lpcWluZzA3MjhAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.