- 1Department of Orthopedics, Orthopaedic Research Institute, West China Hospital, Sichuan University, Chengdu, China
- 2Boston University Henry M. Goldman School of Dental Medicine, Boston, MA, United States
- 3West China School of Public Health, Sichuan University, Chengdu, China
- 4Department of Preventive Dentistry, West China Hospital of Stomatology, Sichuan University, Chengdu, China
- 5College of Electronics and Information Engineering, Sichuan University, Chengdu, China
Background: Staphylococcus aureus (S. aureus) is the leading cause of various infective diseases, including topical soft tissue infections. The goals of this study were to investigate the roles of YycF and CodY in the regulation of biofilm formation and pathogenicity.
Methods: Electrophoretic mobility shift assay (EMSA) was conducted to validate the bound promoter regions of YycF protein. We constructed the codY up-regulated or down-regulated S. aureus mutants. The biofilm biomass was determined by crystal violet microtiter assay and scanning electron microscopy (SEM). Quantitative RT-PCR analysis was used to detect the transcripts of biofilm-related genes. The live and dead cells of S. aureus biofilm were also investigated by confocal laser scanning microscopy (CLSM). We constructed an abscess infection in Sprague Dawley (SD) rat models to determine the effect of CodY on bacterial pathogenicity. We further used the RAW264.7, which were cocultured with S. aureus, to evaluate the effect of CodY on macrophages apoptosis.
Result: Quantitative RT-PCR analyses reveled that YycF negatively regulates codY expression. EMSA assays indicated that YycF protein directly binds to the promoter regions of codY gene. Quantitative RT-PCR confirmed the construction of dual- mutant stains codY + ASyycF and codY-ASyycF. The SEM results showed that the biofilm formation in the codY + ASyycF group was sparser than those in the other groups. The crystal violet assays indicated that the codY + ASyycF group formed less biofilms, which was consistent with the immunofluorescence results of the lowest live cell ration in the codY + ASyycF group. The expression levels of biofilm-associated icaA gene were significantly reduced in the codY + strain, indicating codY negatively regulates the biofilm formation. Furthermore, CodY impedes the pathogenicity in a rat-infection model. After cocultured with bacteria or 4-h in vitro, the apoptosis rates of macrophage cells were lowest in the codY + group.
Conclusions: YycF negatively regulate the expression of codY. By interaction with codY, YycF could modulate S. aureus biofilm formation via both eDNA- dependent and PIA- dependent pathways, which can be a significant target for antibiofilm. CodY not only impedes the pathogenicity but also has a role on immunoregulation. Thus, the current evidence may provide a supplementary strategy for managing biofilm infections.
Introduction
Staphylococcus aureus (S. aureus), a Gram-positive opportunistic pathogen, is the leading cause of various infectious diseases, including topical soft tissue infections, osteomyelitis, and endocarditis (Wertheim et al., 2004). Due to biofilm formation, S. aureus infection has become more challenging to treat in recent years (Schilcher and Horswill, 2020). Biofilm formation is responsible for persistent infections, difficult to eradicate, and much more resistant to environmental stimuli. Thus, biofilms are an essential target for infection treatment, and many strategies targeting biofilms have been developed to attenuate the pathogenicity of bacteria. In S. aureus, approximately 16 TCSs (Two-component systems) play a role in adaptation to environmental changes. YycFG is the only TCS essential for the viability of bacteria and significantly modulates gene expression, which is associated with biofilm formation and pathogenicity (Villanueva et al., 2018; Jenul and Horswill, 2019; Rapun-Araiz et al., 2020). YycFG TCS, also known as the VicRK/WalRK TCS, consists of the sensor histidine protein kinase YycG and its cognate response regulator YycF. By phosphorylation, YycG activates YycF and controls downstream gene expression (Wang et al., 2021).
YycFG has a major role in controlling biofilm formation in low-G + C Gram-positive bacteria, including S. aureus (Dubrac et al., 2007). By directly binding to the promoter region of the ica operon, activated YycF can positively trigger extracellular polysaccharide (EPS) synthesis, which is associated with biofilm construction (Xu et al., 2017). The ica operon is a chromosomal gene locus that comprises the intercellular adhesion gene icaA and regulates the production of polysaccharide intercellular adhesin (PIA). Additionally, YycF can indirectly stimulate the expression of ica operon by controlling the expression of the global transcriptional regulator SarA, resulting in biofilm aggregation (Wu et al., 2021b). Hence, the YycFG TCS represents a promising target to modulate S. aureus biofilms.
Research has shown that CodY [control of dciA (decoyinine induced operon) Y] is a global repressor regulator in Gram-positive bacteria (Chapeton-Montes et al., 2020). In response to environmental signals such as the amount of branched-chain amino acids (BCAAs) [isoleucine, leucine, and valine (ILV)] and GTP, CodY adjusts metabolism and virulence gene regulation (Pohl et al., 2009). Via recognition of a conserved sequence motif (AATTTTCWGAAAATT) (Brinsmade, 2017), CodY competes with RNA polymerase for binding to a promoter and primarily represses the target genes. In S. aureus, biofilm development is thought to occur mainly via PIA-dependent and PIA-independent biofilm formation pathways. CodY can act as a repressor of ica and modulates PIA-dependent biofilm formation (Majerczyk et al., 2008). PIA-independent biofilms are mainly based on the aggregation of extracellular DNA (eDNA) and/or protein. CodY also contributes to PIA-independent formation by repressing the expression of secreted proteases and nucleases (Nuc) (Mlynek et al., 2020). Both eDNA and PIA can work synergistically in biofilm organization.
CodY-targeted biofilm genes have been extensively studied in S. aureus, but the regulatory relationship between CodY and YycFG TCS is largely unknown (Augagneur et al., 2020). In this study, we used electrophoretic mobility shift assays (EMSAs) to verify the binding of YycF to codY promoters and identify negative regulation of YycF on CodY by RT–PCR to gain insight into the relationship between CodY and YycFG and their coordinating adjustments to S. aureus biofilm formation and pathogenicity. We showed that S. aureus YycF acts as a repressor to control the activity of CodY, thus contributing to biofilm formation and pathogenesis in infectious diseases.
Methods and materials
Bacterial strains and biofilm growth conditions
As previously described, S. aureus strain ATCC29213 was cultured in tryptic soy broth (TSB) at 37°C and 5% CO2. Briefly, 500 μL of S. aureus suspension was inoculated into 10 mL fresh TSB medium to mid-logarithmic phase (optical density at 600 nm [OD600] = 0.5), and a log-phase suspension was prepared for further investigation. For biofilm formation, sterilized glass disks (10-mm diameter) were dropped into 24-well microtiter plates and cocultured with log-phased suspension for 24 h.
Electrophoretic mobility shift assay to detect bound codY promoter regions of YycF protein
We performed electrophoretic mobility shift assays to determine whether the YycF protein could directly bind to the promoter regions of codY. To generate YycF His-Tag fusion proteins, pET-22b (Novagen) was applied to yield pET-yycF at Huabio Biotech (Hangzhou, China). Then, the above plasmids were transformed into E. coli BL21 for recombinant proteins. We isolated recombinant proteins from bacterial suspension culture after a 3-h induction with 1 mM IPTG. The acquired recombinant proteins were purified through affinity chromatography on Ni2+ NTA agarose (Qiagen). The purified YycF protein was visualized via Coomassie staining after SDS–PAGE.
The PCR amplicon of the codY promoter region was generated from the S. aureus ATCC29213 genomic DNA sample using primers labeled with the 5′ FAM (Roche) (see Table 1). The amplified DNA fragments were purified according to the manufacturer’s instructions (Tiangen Biotech, Beijing, China). After purification, labeled DNA fragments (0.02 pmol) were incubated with recombinant YycF protein at various concentrations from 0 to 60 pmol. After 30 min of incubation on ice, the samples were loaded on native PAGE gels in 0.5 × TBE buffer (44.5 mM Tris-HCl, 44.5 mM boric acid, 1 mM EDTA, pH 8.0). Native PAGE was prepared with 5 × TBE (445 mM Tris-HCl, 445 mM boric acid, 10 mM EDTA, pH 8.0), 30% Acr-Bis (29:1), 50% glycerinum, 10% ammonium persulfate (APS), and N,N,N′,N′-tetramethylethylenediamine (TEMED). Gel electrophoresis was performed at 110 V for 90 min on ice, according to our previous study (Lei et al., 2018).
Construction of codY-upregulated or -downregulated Staphylococcus aureus mutants
We constructed the S. aureus yycF downregulating strain (ASyycF) as previously described (Wu et al., 2021a). To investigate the subsequent effects of codY, we constructed codY-upregulated or -downregulated expression mutants. To downregulate codY expression, antisense sequences were applied to construct a codY-downregulated expression mutant by transformation of a plasmid expressing antisense codY (AScodY) into S. aureus ATCC29213. AScodY was engineered by Sangon Biotech (Shanghai, China) by inserting the antisense sequences of codY into restriction sites between BamHI and EcoRI. In addition, a codY-upregulated expression mutant (codY +) was constructed by transformation of the codY-encoding sequences inserted into the pDL278 plasmid in ATCC29213.
The methods of construction for dual mutants were modified according to our previous study (Zhang et al., 2022). To generate overexpression strain codY + ASyycF (yycF low-expression and codY overexpression mutant), codY–ASyycF (codY low-expression and yycF low-expression mutant), anti-sense sequences of yycF were obtained by oligonucleotides synthesis and connected with cody coding region or antisense codY cloned into the pDL278 shuttle vector (Sangon Biotech, Shanghai, China), generating recombinant plasmid pDL278 codY + ASyycF or pDL278 codY–ASyycF.
Analysis of gene expression using quantitative real-time PCR
To investigate the interactions between yycF and codY and the effect on biofilm-associated gene expression, quantitative real-time PCR (qRT–PCR) was performed. The codY–yycF–, codY–, codY+yycF–, codY+, and ATCC29213 (as a control) strains were cultured to the mid-logarithmic phase. Total RNA was extracted and purified from each strain with the MasterPure™ RNA Purification Kit (Epicenter Technologies, Madison, WI, USA). The purified RNA was reverse transcribed to cDNA with the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). Quantitative real-time PCR assays were performed with a LightCycler 480 system (Roche, Basel, Switzerland) with the primers listed in Table 1 and the 16S rRNA gene as an internal control. Threshold cycle values (CT) were determined, and the abundance of each gene was expressed relative to that of the 16S rRNA gene. Each sample was analyzed in triplicate, and the data were analyzed according to the 2–ΔΔCT method.
Crystal violet assay and epifluorescence staining for biofilm biomass
A crystal violet assay was performed to evaluate the biomass of biofilms, including codY–ASyycF, codY–, codY+ASyycF, codY+, and ATCC29213 (as a control). After 24 h of culture in TSB medium, the biofilm samples were stained with 0.1% (w/v) crystal violet for 15 min. The dye bound to the biofilms was transferred into a new plate, and the absorbance was measured with a microplate reader (ELX800, Gene) at 595 nm (Wu et al., 2021b). In addition, the biofilms were labeled with SYTO9 and PI for epifluorescence observation. Live strains were stained green, while dead strains appeared red. Three random fields in each specimen were visualized using epifluorescence microscopy (Nikon Eclipse TE-2000S, Melville, NY).
Characterizing biofilm morphologies
To observe the biofilm structure of each group, scanning electron microscopy (SEM) (Inspect, Hillsboro, OR, USA; SEM) was conducted. The 24 h biofilm samples were washed with PBS twice and fixed with 2.5% glutaraldehyde for 4 h. Then, the biofilm samples were dehydrated and dried in a critical point dryer. After being coated with gold powder, micrographs of the biofilm samples were evaluated.
Abscess model for evaluation of pathogenicity
The ability of S. aureus to form biofilms contributes to major microbial infections. To determine the effect of codY on biofilm infection, which can be modulated by YycF, we constructed an abscess infection in Sprague Dawley (SD) rat models. The animal experiments were approved by West China Hospital Animal Welfare Committee (NO. 20220606004). The rats were randomized into four experimental groups: S. aureus as a positive control, SSN group, AScodY, codY + and normal control group (n = 5 rats per group). After anesthetization with ketamine (60 μg/g) and xylazine (6 μg/g), we injected 0.1 mL of a bacterial suspension (1.2 × 109CFU/mL) into deep calf muscle and observed after 36 h. A palpable fluctuant mass in the calf muscle was identified for model establishment (Wyss et al., 2004). For histopathological analysis, the muscle tissue was excised and fixed in 10% neutral-buffered formalin for 48 h. Tissue sections were processed and stained with hematoxylin-eosin (HE) according to standard protocols (Cardiff et al., 2014).
The effect of CodY on macrophages
We also used RAW264.7 cells to evaluate the effect of CodY on macrophage apoptosis. RAW264.7 cells were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS). Bacterial suspensions of S. aureus ATCC29213, codY- and codY + strains at the log phase were diluted to achieve a multiplicity of infection (MOI) of 100:1. The number of S. aureus was determined by serial dilution with the plate counting method. Cells were inoculated into a 6-well plate at 3.0 × 105 cells/well. After the cells grew for 12 h and formed a monolayer, 200 μL of S. aureus (MOI = 100:1) was added to each well for 4 h and treated with lysostaphin (10 μg/mL) for 12 min to kill extracellular S. aureus. To detect RAW264.7 cell apoptosis, we used the Annexin V-FITC/propidium iodide (AV/PI) dual staining AP-101-100-kit (Multisciences, China) to test the apoptosis rate of RAW264.7 cells following the manufacturer’s instructions. Briefly, after coculture with S. aureus, the cells were digested with trypsin, collected by centrifugation, washed with PBS, stained with Annexin V-FITC and PI, and analyzed by FCM (Becton CytoFLEX) (Xu et al., 2020). The cell concentration for FCM was modulated to 1.0 × 107/mL. Each sample was added with 5μL Annexin V-FITC and PI for 30 min at 4°C. After centrifugation at 300 g for 5min, the supernatant was removed, resuspended in 500 mL PBS and analyzed for cell apoptosis.
Data analysis
All statistical data were analyzed in SPSS 16.0 (SPSS Inc., Chicago, IL, USA). The Shapiro–Wilk test was used to analyze the distribution of data, and the Bartlett test was used to determine the homogeneity of variances. For parametric testing, we adopted one-way ANOVA to assess the statistical significance of variables followed by the Tukey test. Differences in the data were considered significant at P < 0.05.
Results
YycF negatively regulates CodY expression
The methods of construction for dual mutants were modified according to our previous study (Zhang et al., 2022). To generate overexpression strain codY + ASyycF (yycF low-expression and codY overexpression mutant), codY-ASyycF (codY low-expression and yycF low-expression mutant), anti-sense sequences of yycF were obtained by oligonucleotides synthesis and connected with codY coding region or antisense codY cloned into the pDL278 shuttle vector (Sangon Biotech, Shanghai, China), generating recombinant plasmid pDL278 codY + ASyycF or pDL278 codY-ASyycF. Quantitative RT–PCR analyses revealed that the transcription of codY was elevated in ASyycF strains (Figure 1A). To reveal the possible interactions between YycF and candidate targeted genes, EMSA was performed on the promoter regions of the codY gene. As demonstrated in Figure 1B, the promoter region of codY contained a putative YycF-binding consensus motif. The YycF protein directly binds to the promoter regions of the codY gene.
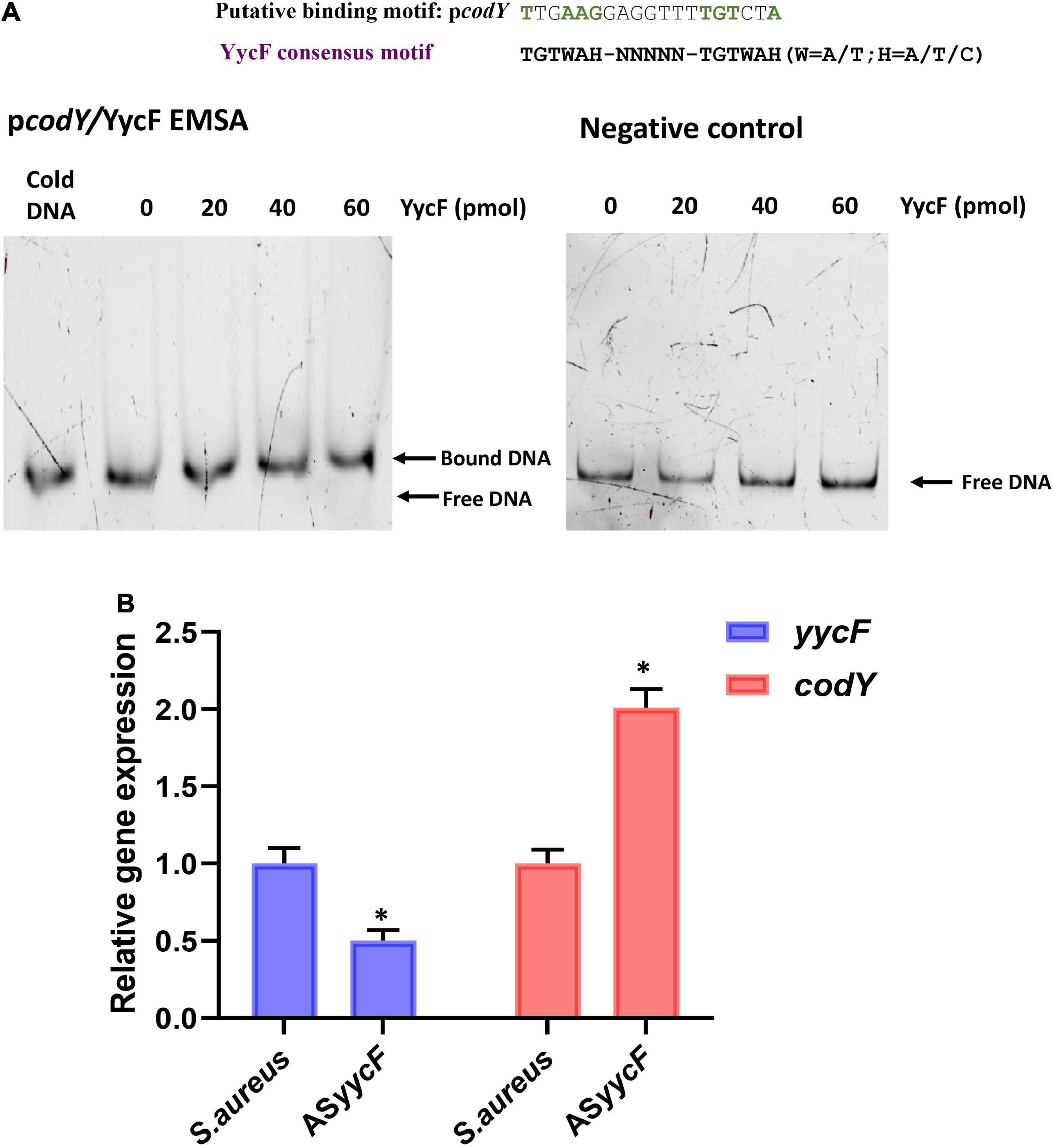
Figure 1. YycF negatively regulates codY expression. (A) Consensus YycF binding motif and candidate sequences in promoters of codY. TGTWAH-NNNNN-TGTWAH, where W is A/T and H is A/T/C; EMSA in which promoter regions were obtained by PCR and FAM-labeled. As the negative control, a DNA fragment the same size as the promoter region and similar AT: GC mole ratio, but missing the YycF consensus binding sequence, was used to rule out non-specific binding. (B) Quantitative RT-PCR analysis showed the gene transcripts in S. aureus, and ASyycF strains. S. aureus gene expression was relatively quantified by RT-PCR using 16S as an internal control (n = 5, *P < 0.05).
CodY interaction with YycF affects biofilm morphology
Quantitative RT–PCR demonstrated that in the dual-mutant stains codY + ASyycF and codY-ASyycF, the expression levels of yycF genes were significantly reduced. Furthermore, the expression levels of the biofilm-associated icaA gene were significantly reduced in the codY + ASyycF strain compared with the S. aureus and codY-ASyycF strains (P < 0.05; Figure 2A), which can be attributed to the reduced biomass of the codY + ASyycF strain. The SEM results showed that the biofilm formation in the codY + ASyycF group was sparser than those in the other groups (Figure 2B), and that codY interacted with yycF in regulating biofilm formation. Quantitatively, we evaluated the ability of the S. aureus strains to form biofilms in the TSB culture. The biomass was quantified via the crystal violet assay, and the codY + ASyycF group formed fewer biofilms than the S. aureus group, as demonstrated by the reduction in OD595 values from 1.9 to 1.0 (Figure 3A). Similarly, the immunofluorescence density of the live cells in the codY + ASyycF group was the lowest compared with the S. aureus and codY-ASyycF groups (Figure 3B).
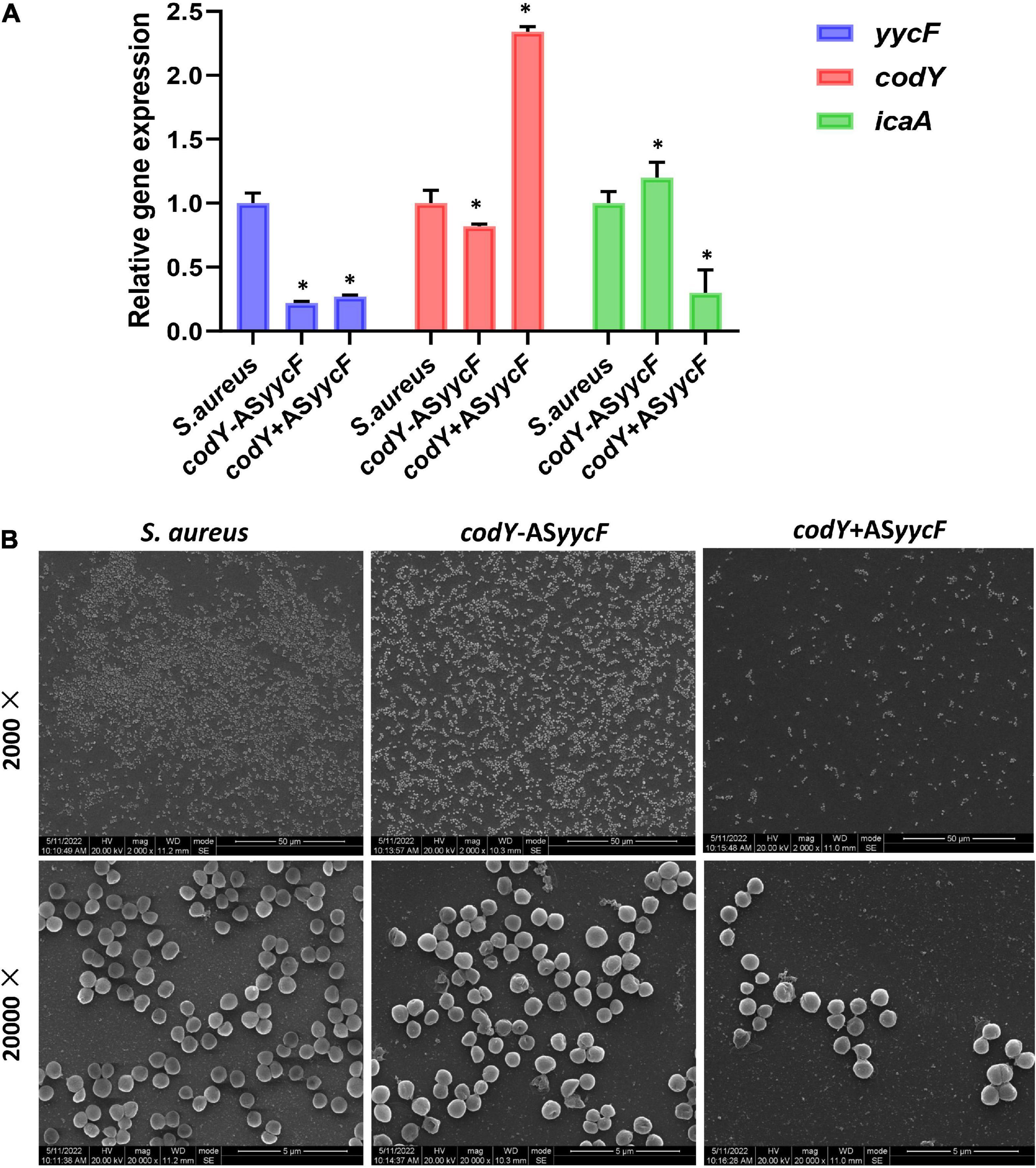
Figure 2. CodY interaction with yycF affects biofilm morphology. (A) Quantitative RT-PCR analysis showed the gene expressions in S. aureus, codY + ASyycF, and codY-ASyycF dual- mutant stains. S. aureus gene expression was relatively quantified by RT-PCR using 16S as an internal control (n = 5, *P < 0.05). (B) SEM images of S. aureus, codY + ASyycF, and codY-ASyycF dual- mutant stains.
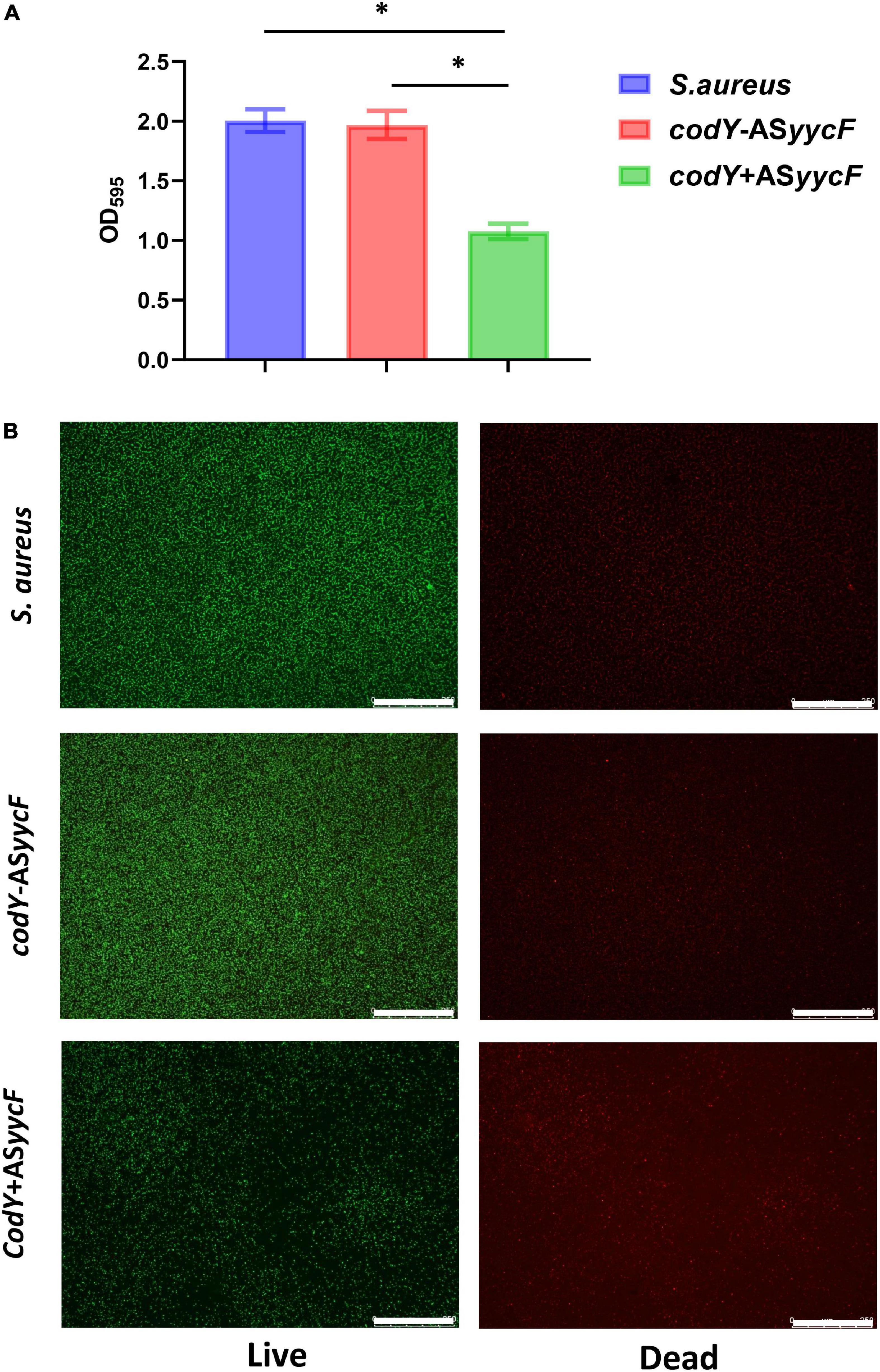
Figure 3. CodY interaction with yycF regulates the biofilm biomass. (A) Biomass of S. aureus, codY + ASyycF, and codY-ASyycF dual- mutant stains were quantified by crystal violet staining. Optical densities at 595 nm were measured (n = 5, *P < 0.05). (B) The CLSM observations of S. aureus, codY + ASyycF and codY-ASyycF dual- mutant stains. Green, viable bacteria (SYTO 9); red, dead bacteria (PI); scale bars, 250 μm.
CodY suppressed biofilm morphology and biofilm-associated genes
To further explore the potential roles of the codY gene in biofilm formation, we constructed the codY + strain (codY overexpression strain) and AScodY strain (codY low-expression strain). Quantitative RT–PCR analyses demonstrated the construction of the codY + strain and AScodY strain (Figure 4A, blue column). Furthermore, the expression levels of the biofilm-associated icaA gene were significantly reduced in the codY + strain but increased in the AScodY strain (P < 0.05; Figure 4A, red column), indicating that codY negatively regulates biofilm-associated genes. The SEM results showed that the biofilm formation in the codY + group was sparser than that in the other groups (Figure 4B). In particular, the AScodY strains presented dense biofilms. The biomass was quantitively measured by crystal violet staining (Figure 5A). The codY + group formed the lowest biomass, while the AScodY strain presented the highest biomass, as demonstrated by the reduction in OD595 values from 2.5 to 1.0 (Figure 5A). Accordingly, the immunofluorescence density of the live cells in the codY + group was the lowest compared with the S. aureus and AScodY groups (Figure 5B).
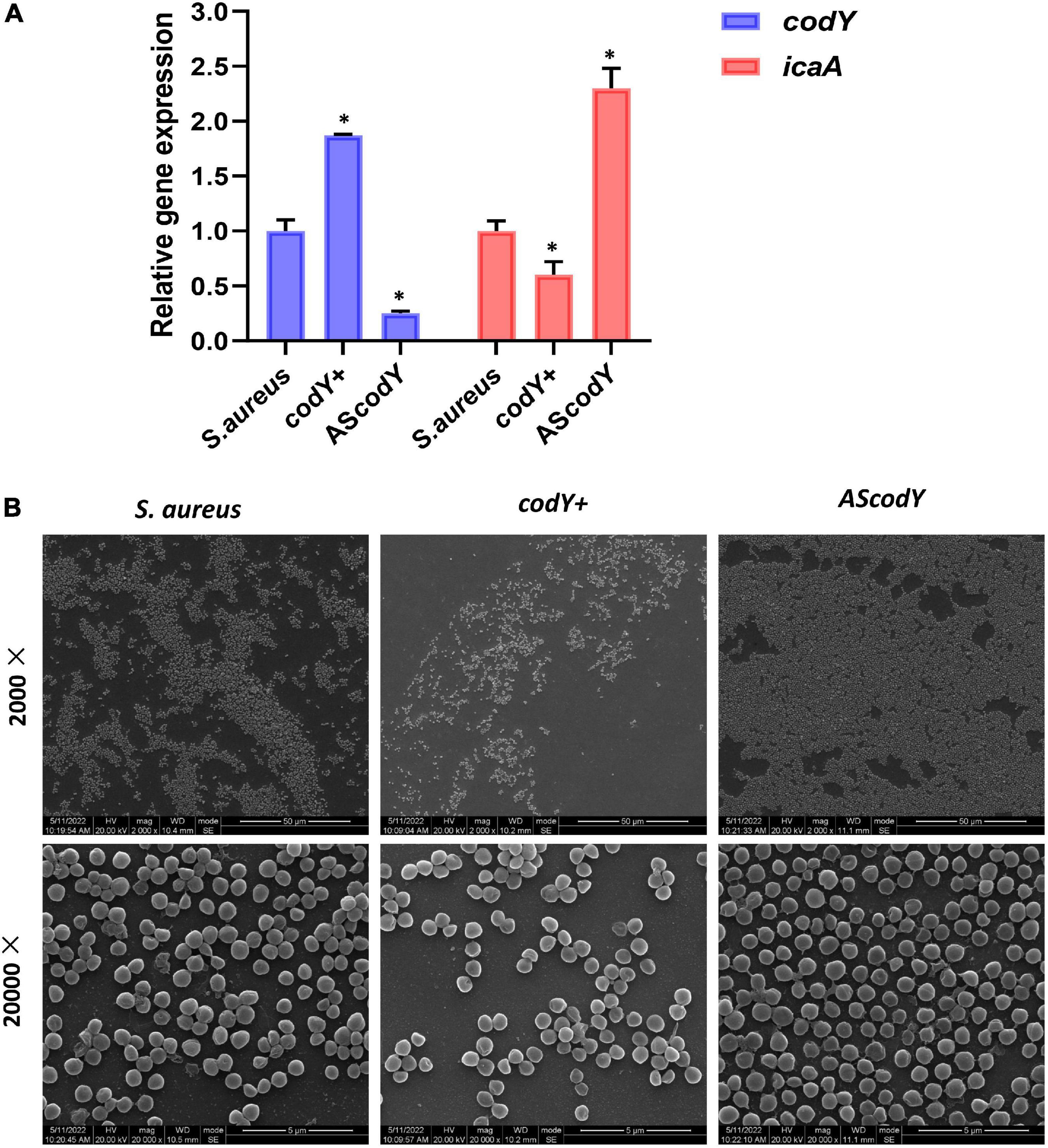
Figure 4. CodY suppressed biofilm morphology and biofilm-associated genes. (A) Quantitative RT-PCR analysis showed the gene expressions in S. aureus, codY + and codY- mutant stains. S. aureus gene expression was relatively quantified by RT-PCR using 16S as an internal control (n = 5, *P < 0.05). (B) SEM images of S. aureus, codY + and codY- mutant stains.
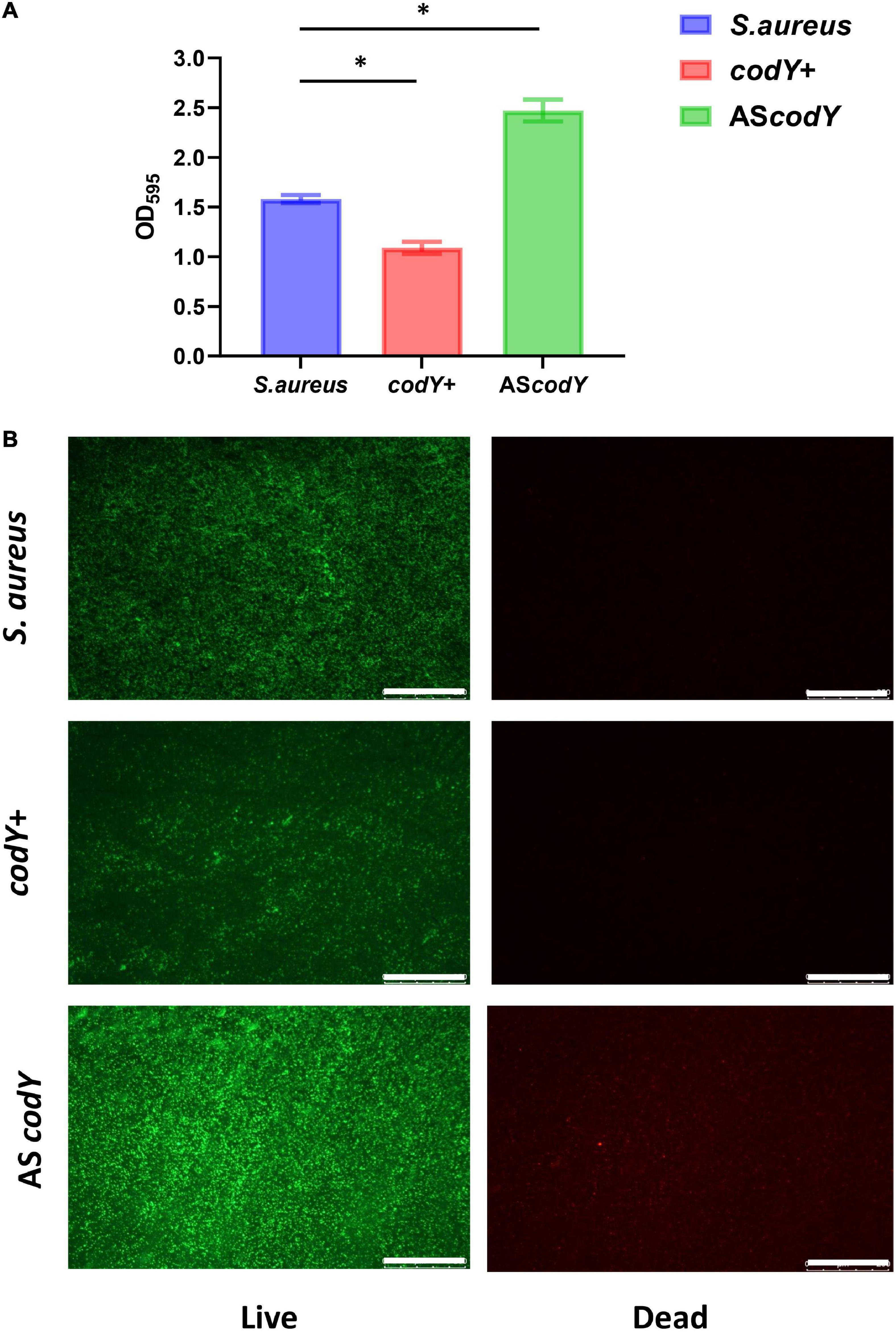
Figure 5. CodY decreases the biomass of S. aureus biofilm. (A) Biomass of S. aureus, codY + and codY- mutant stains were quantified by crystal violet staining. Optical densities at 595 nm were measured (n = 5, *P < 0.05). (B) The CLSM observations of S. aureus, codY + and codY- mutant stains. Green, viable bacteria (SYTO 9); red, dead bacteria (PI); scale bars, 250 μm.
CodY impeded pathogenicity in a rat infection model
Thirty-six hours after muscle injection of S. aureus, codY+, and AScodY strains, the rats were sacrificed by euthanasia under deep anesthesia (ketamine/xylazine) by cervical dislocation. The infection sites were dissected under macroscopic observation. Among all groups, infectious lesions with diameters of approximately 3 mm and 1.5 mm were observed in the S. aureus and AScodY groups, respectively. However, in the codY-overexpressing group (codY +), the abscess in muscle was obscure, and there were only unhealthy tissues with a diameter of less than 2 mm (Figure 6A). Correspondingly, the percentage of inflammatory cell infiltration was measured by ImageJ, and there were approximately 13% inflammatory cells in the S. aureus group. The percentages of inflammatory cell infiltration were approximately 9 and 6% for the AScodY and codY + groups, respectively (Figure 6B). After coculture with bacteria for 4 h in vitro, the apoptosis rates of macrophages were measured by flow cytometry. The total apoptosis rate in S. aureus (ATCC29213) was 86.61%. In the AScodY group, the total apoptosis rate was 72.60%, which was higher than that of 66.24% in the codY + group (Figure 6C).
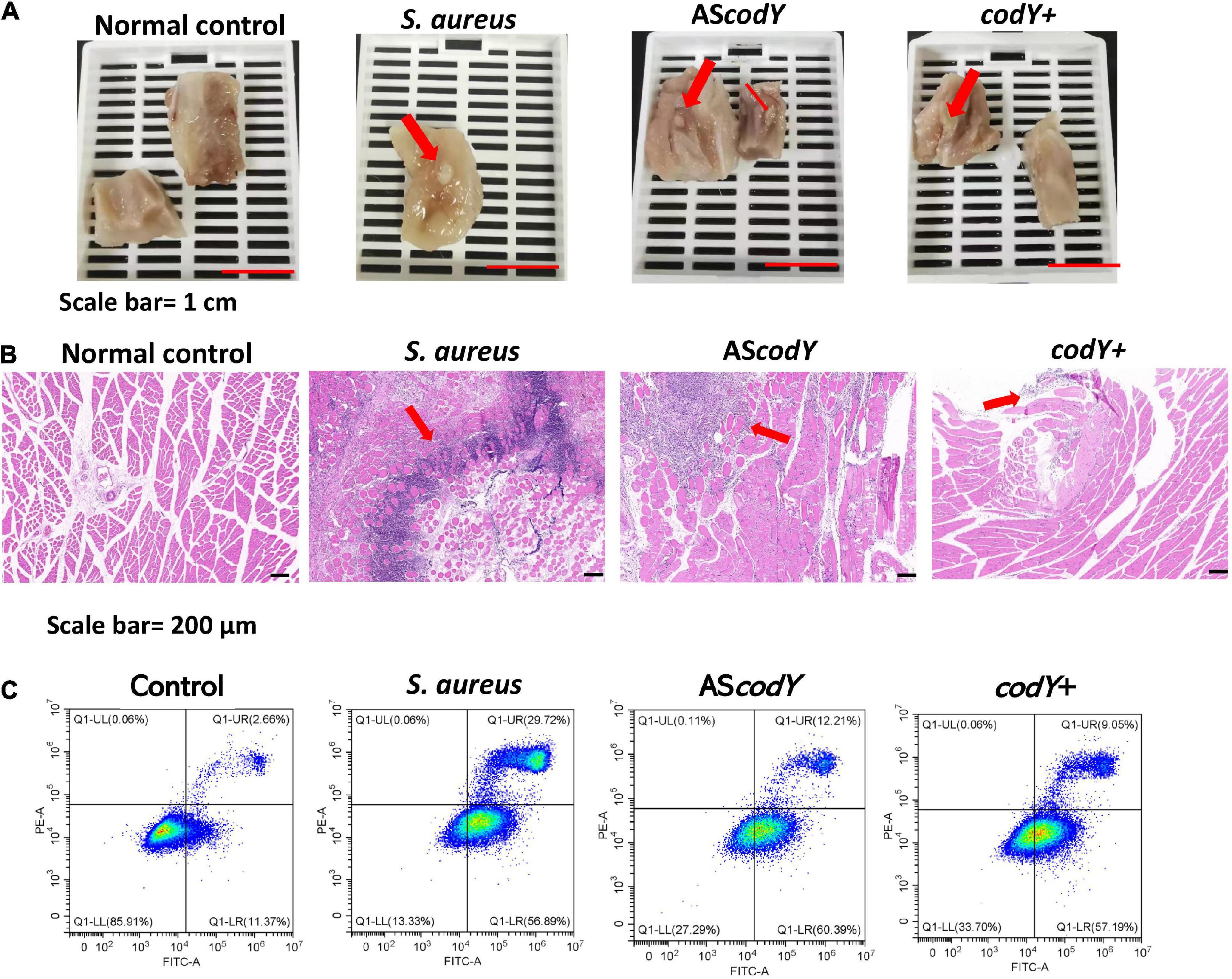
Figure 6. CodY impedes pathogenicity in a rat-infection model. (A) Gross observation of the abscess in S. aureus, codY + and codY- infected group; scale bars, 1 cm. (B) HE staining of the abscess lesions in S. aureus, codY + and codY- infected group; scale bars, 200 μm. (C) The apoptosis rates of macrophage cells when co-cultured with S. aureus, codY + and codY- respectively were measured by the flow cytometer.
Discussion
Staphylococcus aureus is a major human pathogen that is responsible for a wide range of infectious diseases. The propensity of bacteria to form biofilms is one of most crucial factors contributing to pathogenesis and resistance (McCarthy et al., 2015). In S. aureus, biofilm organization is thought to occur mainly via two mechanisms, polysaccharide intercellular adhesin (PIA)-based and eDNA/protein-based pathways. Both pathways contribute to the construction of a self-produced extracellular matrix, which is primarily comprised of exopolysaccharides, proteins, and extracellular DNA (eDNA) for cell-to-cell or cell-to-host attachment. The potential mechanisms of biofilm formation are critical for developing strategies to control biofilms and biofilm-related infections.
PIA synthesis is modulated by the ica locus. According to a previous study, the expression of ica is positively controlled by YycFG, which is the only essential TCS in S. aureus that regulates bacterial metabolism, including virulence and biofilm formation (Clausen et al., 2003; Wu et al., 2019b). YycFG is reported to modulate Staphylococcus epidermidis biofilm formation in an ica-dependent manner (Xu et al., 2017). In Bacillus subtilis, YycFG is directly involved in regulation of cell wall synthesis and modification (Wu et al., 2019). The YycG protein acts as a sensor to respond to environmental signals, and YycF can directly regulate different sets of vital functional genes by binding to promoter regions (Winkler and Hoch, 2008). The putative recognition sequence of YycF is composed of two hexanucleotide direct repeats separated by five nucleotides [5′-TGT(A/T)A(A/T/C)-N5-TGT(A/T)A(A/T/C)-3′]. However, CodY, as a global regulator, can negatively regulate ica expression and inhibit biofilm formation. In Clostridium difficile, the variability of CodY-dependent regulation is an important contributor to the bacterial virulence and sporulation (Nawrocki et al., 2016). In Bacillus subtilis, CodY can be seen to regulate the entire protein utilization pathway (Barbieri et al., 2015). Additionally, in major gram-positive pathogens, several virulence factors are regulated by CodY (Stenz et al., 2011). To identify the interaction of YycF and codY, we analyzed the promoter of codY, and the consensus motif of YycF was found (Figure 1A). The EMSA results revealed that YycF can bind to the promoter of codY and potentially regulate its expression (Figure 1A).
The mutation in yycF reduced biofilm formation and led to decreased transcripts in the ica operon (Howden et al., 2011). However, our RT–qPCR assays showed downregulation in yycF combined with subsequent elevated expression in codY (Figure 1B). Thus, we speculated that YycF negatively modulated the expression of codY. To further explore the interactions of YycF with codY, we constructed dual mutants expressing codY and yycF (Figure 2). In the codY-ASyycF group (indicating that both codY and yycF expression were downregulated), the PCR results indicated that the expression of codY decreased while ica expression significantly increased. Consistent with previous work, ica (PIA synthesis) is negatively regulated by CodY (Majerczyk et al., 2008, 2010). With the relatively higher ica expression in the codY-ASyycF group, the biofilm biomass increased (Figures 2B, 3A). Whereas the codY + ASyycF group presented lower ica expression, the biofilm biomass significantly decreased (Figures 2B, 3A).
In addition, YycFG TCS (also known as WalRK, VicRK, and MicAB TCS) plays a central role in bacterial viability (Haag and Bagnoli, 2017; Villanueva et al., 2018). In the codY + ASyycF group, the density of strains was significantly downregulated (Figure 3). However, both yycF and codY expression decreased in the codY-ASyycF group, and the accumulation of strains was similar to that in the S. aureus group (Figure 3). Therefore, the downregulation of yycF can inhibit S. aureus viability, and this viability alteration can be partially complemented by repressing codY. CodY, as a global transcription factor, typically represses gene expression and regulates physiology for growth and survival under various levels of nutrient depletion (King et al., 2018). YycF as an essential and global regulator responds to various physiological metabolic processes in S. aureus (Wu et al., 2021a). Therefore, multiple reasons including eDNA- dependent and PIA- dependent pathways as well as slow growth rate and reduced viability will inhibit biofilm formation. In addition to CodY/YycF regulated eDNA- dependent and PIA- dependent pathways, we also found reduced viability in Figures 3, 5 and slow growth rate in Supplementary Material.
In addition to bacterial growth and biofilm formation, YycF also regulates the expression of genes involved in cell wall metabolism and virulence (Bleul et al., 2021). Subsequently, to observe the specific effect of codY on biofilm organization, which can be modulated by YycF, we constructed codY overexpression (codY +) and downregulation (codY-) mutants. The decrease in CodY activity promotes cell aggregation and biofilm formation (Brinsmade, 2017). Our SEM result in Figure 4B indicated a significant increase in the AScodY biofilm, which is consistent with this conclusion. By utilizing available eDNA and PIA, CodY demonstrates a synergistic effect combining a DNA-dependent strategy with a PIA-based strategy for biofilm formation (Mlynek et al., 2020). Considering the present findings, YycF negatively modulates codY expression while positively participating in an eDNA/PIA dual-dependent manner for biofilm organization in S. aureus (Figure 7).
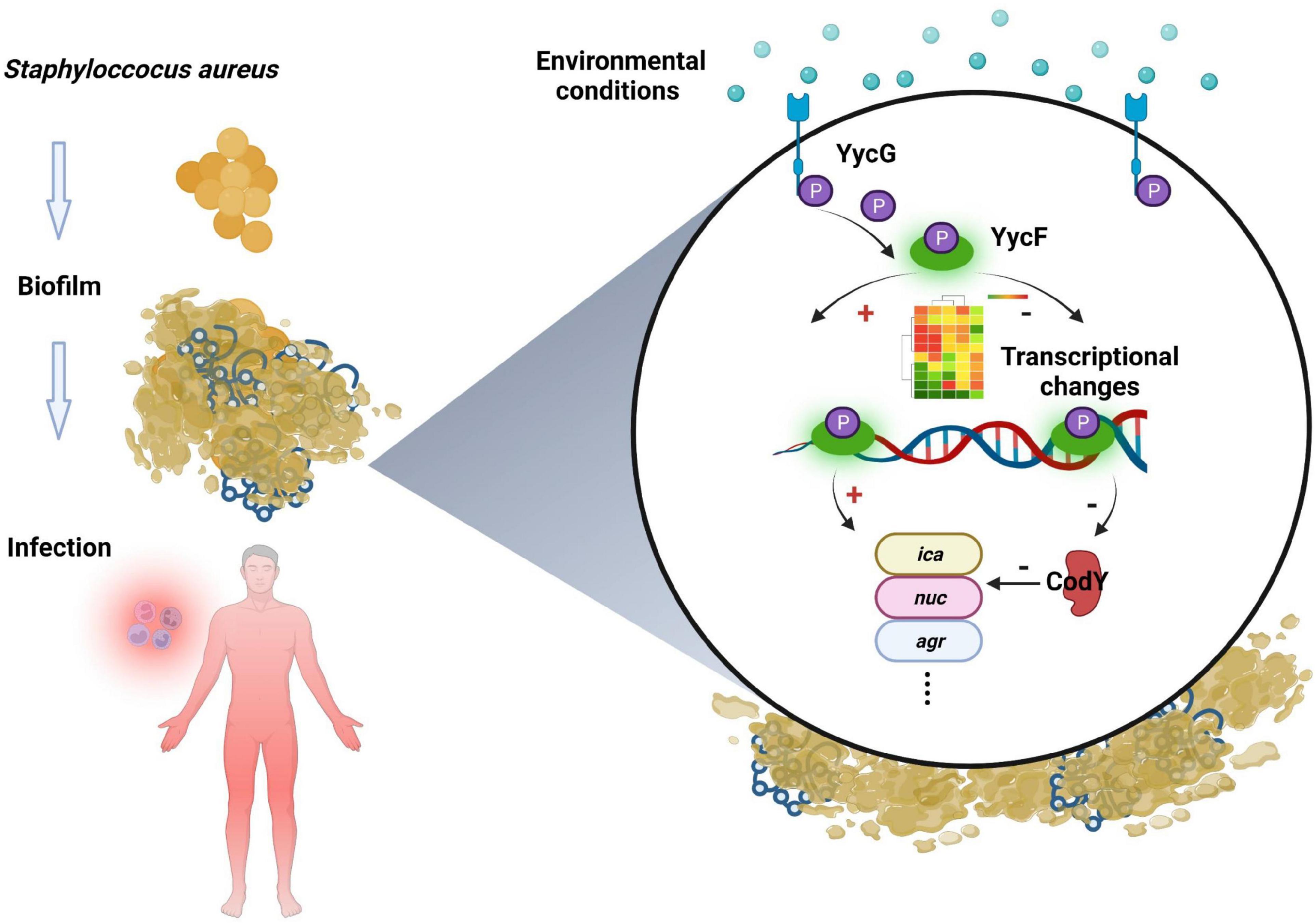
Figure 7. Working model of CodY interactions with YycF affects biofilm formation in Staphylococcus aureus.
In S. epidermidis, YycF is bound to the promoter of ica and increases ica expression (Xu et al., 2017). Similarly, YycF was shown to interact with the ica promoter region and contribute to PIA-based biofilms in our previous study (Wu et al., 2021b). In the present study, YycF was speculated to modulate PIA/eDNA-based biofilm formation by repressing codY. Therefore, YycF negatively modulates Cody for PIA/eDNA-based biofilms and also affects ica for PIA biofilms. Notably, the biofilm disassembly of the dual mutant (codY + ASyycF) was more obvious than that of codY + (Figures 2B, 4B). As S. aureus in biofilms is 1,000 times more tolerant to antibiotics and recalcitrance than planktonic cells, the susceptibility of the pathogen was reversible without the shelter of the biofilm (Shenkutie et al., 2020; Gimza and Cassat, 2021).
A potential mechanism of CodY limits the host damage of S. aureus, in which it transitions from a commensal bacterium to an invasive pathogen. The decreased CodY activity promotes a more invasive lifestyle of S. aureus (Waters et al., 2016). Similarly, our animal experimental results indicated that the ability of AScodY strains to invade was higher than that of codY + and they formed a larger abscess (Figure 6A). Similar with Montgomery et al., we found CodY can represses virulence in vivo. In Montgomery study, Cody as a global regulator can decrease expression of agr and saeRS, as well as the gene encoding the toxin alpha-hemolysin (hla). Also, Cody can restrain the expression of the lukF-PV gene, encoding part of the Panton-Valentine leukocidin (PVL) (Montgomery et al., 2012). By multiple pathways, Cody can mediate the virulence of USA300. In our study, CodY-mediated repression was focus on CodY/YycF interaction and biofilm formation. And our results indicated CodY can impede the pathogenicity of S. aureus by biofilm inhibition which has a potential role on immunoregulation. By histological examination, we observed that invasive AScodY stains could recruit more immune cell infiltration surrounding the infectious region than codY + strains (Figure 6B). Therefore, YycF could indirectly enhance bacterial aggregation by repressing CodY (Figure 7). According to this mechanism, our previous antisense yycF (ASyycF) is base-paired with yycF and downregulates yycF expression, which indicates therapeutic potential for infectious diseases (Wu et al., 2021a). One previous study indicated that CodY repression of sae expression (an exoprotein expression TCS SaeRS to secrete virulence factors) delays immune evasion and reduces immune cell death (Mlynek et al., 2018). In the present study, the AScodY group also had a higher apoptosis rate of macrophage cells than that of the codY + group (Figure 6C). However, in the S. aureus ATCC29213 group, the apoptosis rate was even higher than that in the AScodY group. This may be induced by the positive regulation of CodY on genes such as fnbA and spa, which encode the microbial surface components recognizing adhesive matrix molecule (MSCRAMM) proteins (Brinsmade, 2017). It could be speculated that CodY as a repressor of target can also positively regulate bacterial redox balance and protease induced biofilm formation (Shivers et al., 2006; Roux et al., 2014). In addition, the construction of AScodY including the introduction of an exogenous plasmid vector may interfere intracellular homeostasis (Senadheera et al., 2009; Lei et al., 2015). All those items will affect bacterial metabolism and indirectly interaction between AScodY and macrophages, which may attribute to a lower apoptosis rate in AScodY group instead of S. aureus.
Conclusion
In summary, YycF binds to the promoter regions of codY and negatively regulates the expression of codY. By interacting with codY, YycF could modulate S. aureus biofilm formation via both eDNA-dependent and PIA-dependent pathways, which can be a significant target for anti-biofilms. CodY impedes pathogenicity and also has a role in immunoregulation. CodY not only impedes the pathogenicity but also has a role on immunoregulation. By interacting with CodY, YycF plays essential roles in host-pathogen interactions and pathogenesis. Thus, the current evidence may provide a supplementary strategy for managing biofilm infections.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the corresponding authors.
Ethics statement
The animal study was reviewed and approved by West China Hospital Animal Welfare Committee (No. 20220606004).
Author contributions
SW, BQ, LL, and GF: conceptualization (equal), data curation (equal), formal analysis (equal), validation (equal), writing—original draft and review, and editing (equal). SD, YL, and HZ: conceptualization (supporting), formal analysis (supporting), funding acquisition (lead), writing—review, and editing (equal). All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by the Sichuan Provincial Natural Science Foundation of China (Grant Nos. 2022YFS0161 and 2021YJ0455) and the Central Guiding Local Science and Technology Development Fund project in 2022 (XZ202201YD0018C).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.967567/full#supplementary-material
References
Augagneur, Y., King, A. N., Germain-Amiot, N., Sassi, M., Fitzgerald, J. W., Sahukhal, G. S., et al. (2020). Analysis of the CodY RNome reveals RsaD as a stress-responsive riboregulator of overflow metabolism in Staphylococcus aureus. Mol. Microbiol. 113, 309–325. doi: 10.1111/mmi.14418
Barbieri, G., Voigt, B., Albrecht, D., Hecker, M., Albertini, A. M., Sonenshein, A. L., et al. (2015). CodY regulates expression of the Bacillus subtilis extracellular proteases Vpr and Mpr. J. Bacteriol. 197, 1423–1432. doi: 10.1128/JB.02588-14
Bleul, L., Francois, P., and Wolz, C. (2021). Two-component systems of S. aureus: Signaling and sensing mechanisms. Genes 13:34.
Brinsmade, S. R. (2017). CodY, a master integrator of metabolism and virulence in Gram-positive bacteria. Curr. Genet. 63, 417–425. doi: 10.1007/s00294-016-0656-5
Cardiff, R. D., Miller, C. H., and Munn, R. J. (2014). Manual hematoxylin and eosin staining of mouse tissue sections. Cold Spring Harb. Protoc. 2014, 655–658. doi: 10.1101/pdb.prot073411
Chapeton-Montes, D., Plourde, L., Deneve, C., Garnier, D., Barbirato, F., Colombie, V., et al. (2020). Tetanus toxin synthesis is under the control of a complex network of regulatory genes in Clostridium tetani. Toxins 12:328. doi: 10.3390/toxins12050328
Clausen, V. A., Bae, W., Throup, J., Burnham, M. K., Rosenberg, M., and Wallis, N. G. (2003). Biochemical characterization of the first essential two-component signal transduction system from Staphylococcus aureus and Streptococcus pneumoniae. J. Mol. Microbiol. Biotechnol. 5, 252–260. doi: 10.1159/000071077
Dubrac, S., Boneca, I. G., Poupel, O., and Msadek, T. (2007). New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 189, 8257–8269. doi: 10.1128/JB.00645-07
Gimza, B. D., and Cassat, J. E. (2021). Mechanisms of antibiotic failure during Staphylococcus aureus osteomyelitis. Front. Immunol. 12:638085. doi: 10.3389/fimmu.2021.638085
Haag, A. F., and Bagnoli, F. (2017). The role of two-component signal transduction systems in Staphylococcus aureus virulence regulation. Curr. Top. Microbiol. Immunol. 409, 145–198. doi: 10.1007/82_2015_5019
Howden, B. P., McEvoy, C. R., Allen, D. L., Chua, K., Gao, W., Harrison, P. F., et al. (2011). Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog. 7:e1002359. doi: 10.1371/journal.ppat.1002359
Jenul, C., and Horswill, A. R. (2019). Regulation of Staphylococcus aureus virulence. Microbiol Spectr 7:10.1128/microbiolspec.GPP3-0031-2018. doi: 10.1128/microbiolspec.GPP3-0031-2018
King, A. N., Borkar, S. A., Samuels, D. J., Batz, Z., Bulock, L. L., Sadykov, M. R., et al. (2018). Guanine limitation results in CodY-dependent and -independent alteration of Staphylococcus aureus physiology and gene expression. J. Bacteriol. 200:e00136-18. doi: 10.1128/JB.00136-18
Lei, L., Stipp, R. N., Chen, T., Wu, S. Z., Hu, T., and Duncan, M. J. (2018). Activity of Streptococcus mutans VicR is modulated by antisense RNA. J. Dent. Res. 97, 1477–1484. doi: 10.1177/0022034518781765
Lei, L., Yang, Y., Mao, M., Li, H., Li, M., Yang, Y., et al. (2015). Modulation of biofilm exopolysaccharides by the Streptococcus mutans vicX gene. Front. Microbiol. 6:1432. doi: 10.3389/fmicb.2015.01432
Majerczyk, C. D., Dunman, P. M., Luong, T. T., Lee, C. Y., Sadykov, M. R., Somerville, G. A., et al. (2010). Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192, 2861–2877. doi: 10.1128/JB.00220-10
Majerczyk, C. D., Sadykov, M. R., Luong, T. T., Lee, C., Somerville, G. A., and Sonenshein, A. L. (2008). Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190, 2257–2265. doi: 10.1128/JB.01545-07
McCarthy, H., Rudkin, J. K., Black, N. S., Gallagher, L., O’Neill, E., and O’Gara, J. P. (2015). Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 5:1. doi: 10.3389/fcimb.2015.00001
Mlynek, K. D., Bulock, L. L., Stone, C. J., Curran, L. J., Sadykov, M. R., Bayles, K. W., et al. (2020). Genetic and biochemical analysis of CodY-mediated cell aggregation in Staphylococcus aureus reveals an interaction between extracellular DNA and polysaccharide in the extracellular matrix. J. Bacteriol. 202:e00593-19. doi: 10.1128/JB.00593-19
Mlynek, K. D., Sause, W. E., Moormeier, D. E., Sadykov, M. R., Hill, K. R., Torres, V. J., et al. (2018). Nutritional regulation of the sae two-component system by CodY in Staphylococcus aureus. J. Bacteriol. 200:e00012-18. doi: 10.1128/JB.00012-18
Montgomery, C. P., Boyle-Vavra, S., Roux, A., Ebine, K., Sonenshein, A. L., and Daum, R. S. (2012). CodY deletion enhances in vivo virulence of community-associated methicillin-resistant Staphylococcus aureus clone USA300. Infect. Immun. 80, 2382–2389. doi: 10.1128/IAI.06172-11
Nawrocki, K. L., Edwards, A. N., Daou, N., Bouillaut, L., and McBride, S. M. (2016). CodY-dependent regulation of sporulation in Clostridium difficile. J. Bacteriol. 198, 2113–2130. doi: 10.1128/JB.00220-16
Pohl, K., Francois, P., Stenz, L., Schlink, F., Geiger, T., Herbert, S., et al. (2009). CodY in Staphylococcus aureus: A regulatory link between metabolism and virulence gene expression. J. Bacteriol. 191, 2953–2963. doi: 10.1128/JB.01492-08
Rapun-Araiz, B., Haag, A. F., De Cesare, V., Gil, C., Dorado-Morales, P., Penades, J. R., et al. (2020). Systematic reconstruction of the complete two-component sensorial network in Staphylococcus aureus. mSystems 5:e00511-20. doi: 10.1128/mSystems.00511-20
Roux, A., Todd, D. A., Velázquez, J. V., Cech, N. B., and Sonenshein, A. L. (2014). CodY-mediated regulation of the Staphylococcus aureus Agr system integrates nutritional and population density signals. J. Bacteriol. 196, 1184–1196. doi: 10.1128/JB.00128-13
Schilcher, K., and Horswill, A. R. (2020). Staphylococcal biofilm development: Structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev. 84:e00026-19. doi: 10.1128/MMBR.00026-19
Senadheera, D., Krastel, K., Mair, R., Persadmehr, A., Abranches, J., Burne, R. A., et al. (2009). Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J. Bacteriol. 191, 6415–6424. doi: 10.1128/JB.00793-09
Shenkutie, A. M., Yao, M. Z., Siu, G. K., Wong, B. K. C., and Leung, P. H. (2020). Biofilm-induced antibiotic resistance in clinical Acinetobacter baumannii isolates. Antibiotics 9:817. doi: 10.3390/antibiotics9110817
Shivers, R. P., Dineen, S. S., and Sonenshein, A. L. (2006). Positive regulation of Bacillus subtilis ackA by CodY and CcpA: Establishing a potential hierarchy in carbon flow. Mol. Microbiol. 62, 811–822. doi: 10.1111/j.1365-2958.2006.05410.x
Stenz, L., Francois, P., Whiteson, K., Wolz, C., Linder, P., and Schrenzel, J. (2011). The CodY pleiotropic repressor controls virulence in gram-positive pathogens. FEMS Immunol. Med. Microbiol. 62, 123–139. doi: 10.1111/j.1574-695X.2011.00812.x
Villanueva, M., Garcia, B., Valle, J., Rapun, B., Ruiz de Los Mozos, I., Solano, C., et al. (2018). Sensory deprivation in Staphylococcus aureus. Nat. Commun. 9:523. doi: 10.1038/s41467-018-02949-y
Wang, S., Long, L., Yang, X., Qiu, Y., Tao, T., Peng, X., et al. (2021). Dissecting the role of VicK phosphatase in aggregation and biofilm formation of Streptococcus mutans. J. Dent. Res. 100, 631–638. doi: 10.1177/0022034520979798
Waters, N. R., Samuels, D. J., Behera, R. K., Livny, J., Rhee, K. Y., Sadykov, M. R., et al. (2016). A spectrum of CodY activities drives metabolic reorganization and virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 101, 495–514. doi: 10.1111/mmi.13404
Wertheim, H. F., Vos, M. C., Ott, A., van Belkum, A., Voss, A., Kluytmans, J. A., et al. (2004). Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364, 703–705. doi: 10.1016/S0140-6736(04)16897-9
Winkler, M. E., and Hoch, J. A. (2008). Essentiality, bypass, and targeting of the YycFG (VicRK) two-component regulatory system in gram-positive bacteria. J. Bacteriol. 190, 2645–2648. doi: 10.1128/JB.01682-07
Wu, S., Liu, Y., Lei, L., and Zhang, H. (2019a). Antisense yycG regulation of antibiotic sensitivity of methicillin-resistant Staphylococcus aureus in chronic osteomyelitis. Surg. Infect. 20, 472–479. doi: 10.1089/sur.2019.016
Wu, S., Liu, Y., Lei, L., and Zhang, H. (2019b). Virulence of methicillin-resistant Staphylococcus aureus modulated by the YycFG two-component pathway in a rat model of osteomyelitis. J. Orthop. Surg. Res. 14:433. doi: 10.1186/s13018-019-1508-z
Wu, S., Liu, Y., Lei, L., and Zhang, H. (2021a). An antisense yycF RNA modulates biofilm organization of methicillin-resistant Staphylococcus aureus and pathogenicity in a rat model of osteomyelitis. Antibiotics 10:603. doi: 10.3390/antibiotics10050603
Wu, S., Liu, Y., Lei, L., and Zhang, H. (2021b). Antisense yycG modulates the susceptibility of Staphylococcus aureus to hydrogen peroxide via the sarA. BMC Microbiol. 21:160. doi: 10.1186/s12866-021-02218-x
Wu, X., Song, Q., and Han, A. (2019). Interacting proteins of the essential two-component system YycFG in Bacillus subtilis. J. Basic Microbiol. 59, 950–959. doi: 10.1002/jobm.201800701
Wyss, M. T., Honer, M., Spath, N., Gottschalk, J., Ametamey, S. M., Weber, B., et al. (2004). Influence of ceftriaxone treatment on FDG uptake–an in vivo [18F]-fluorodeoxyglucose imaging study in soft tissue infections in rats. Nucl. Med. Biol. 31, 875–882. doi: 10.1016/j.nucmedbio.2004.06.001
Xu, J., Yao, H., Wang, S., Li, H., and Hou, X. (2020). Mangiferin inhibits apoptosis and autophagy induced by Staphylococcus aureus in RAW264.7 cells. J. Inflamm. Res. 13, 847–857. doi: 10.2147/JIR.S280091
Xu, T., Wu, Y., Lin, Z., Bertram, R., Gotz, F., Zhang, Y., et al. (2017). Identification of genes controlled by the essential YycFG two-component system reveals a role for biofilm modulation in Staphylococcus epidermidis. Front. Microbiol. 8:724. doi: 10.3389/fmicb.2017.00724
Keywords: antisense, biofilm formation, Staphylococcus aureus, YycFG, CodY
Citation: Wu S, Qin B, Deng S, Liu Y, Zhang H, Lei L and Feng G (2022) CodY is modulated by YycF and affects biofilm formation in Staphylococcus aureus. Front. Microbiol. 13:967567. doi: 10.3389/fmicb.2022.967567
Received: 13 June 2022; Accepted: 30 August 2022;
Published: 11 October 2022.
Edited by:
Gowrishankar Muthukrishnan, University of Rochester Medical Center, United StatesReviewed by:
Asad U. Khan, Aligarh Muslim University, IndiaYichao Wu, Huazhong Agricultural University, China
Steven Gill, University of Rochester, United States
Copyright © 2022 Wu, Qin, Deng, Liu, Zhang, Lei and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Lei, bGVpbGVpQHNjdS5lZHUuY24=; Guoying Feng, Z3VvaW5nX2ZlbmdAc2N1LmVkdS5jbg==
†These authors share first authorship
 Shizhou Wu1†
Shizhou Wu1† Shu Deng
Shu Deng Lei Lei
Lei Lei