- 1Jiangsu Key Laboratory of Zoonosis, Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou, China
- 2Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agrifood Safety and Quality, Ministry of Agriculture of China, Yangzhou University, Yangzhou, China
This study aimed to investigate the prevalence and characterization of tet(X4) in Escherichia coli isolates from a pig farm in Shanghai, China, and to elucidate tet(X4) dissemination mechanism in this swine farm. Forty-nine (80.33%) E. coli strains were isolated from 61 samples from a pig farm and were screened for the presence of tet(X). Among them, six (12.24%) strains were positive for tet(X4) and exhibited resistance to tigecycline (MIC ≥ 16 mg/L). They were further sequenced by Illumina Hiseq. Six tet(X4)-positive strains belonged to ST761 with identical resistance genes, resistance profiles, plasmid replicons, and cgMLST type except that additional ColE10 plasmid was present in isolate SH21PTE35. Isolate SH21PTE31, as a representative ST761 E. coli strain, was further sequenced using Nanopore MinION. The tet(X4) in SH21PTE31 was located on IncFIA18/IncFIB(K)/IncX1 hybrid plasmid pYUSHP31-1, highly similar to other tet(X4)-carrying IncFIA18/IncFIB(K)/IncX1 plasmids from ST761 E. coli and other E. coli lineages in China. These IncFIA18/IncFIB(K)/IncX1 plasmids shared closely related multidrug resistance regions, and could reorganize, acquire or lose resistance modules mediated by mobile elements such as ISCR2 and IS26. Phylogenetic analysis were performed including all tet(X4)-positive isolates obtained in this pig farm combined with 43 tet(X4)-positive E. coli from pigs, cow, pork, wastewater, and patients with the same ST from NCBI. The 50 tet(X4)-carrying E. coli ST761 isolates from different areas in China shared a close phylogenetic relationship (0-49 SNPs). In conclusion, clonal transmission of tet(X4)-positive E. coli ST761 has occurred in this swine farm. E. coli ST761 has the potential to become a high-risk clone for tet(X4) dissemination in China.
Introduction
Tigecycline is considered as a last-resort antimicrobial agent to treat serious infections caused by multidrug-resistant bacteria, particularly carbapenem-resistant Enterobacteriaceae (Yaghoubi et al., 2021). However, the recent identification of novel plasmid-borne tigecycline resistance genes tet(X3) in Acinetobacter baumannii and tet(X4) in Escherichia coli from animals in China significantly impairs the clinical efficacy of tigecycline (He et al., 2019). Thus far, tet(X) and its variants [tet(X1)∼tet(X47)] have been identified in Gram-negative pathogens and encode flavin-dependent monooxygenase that modify tigecycline (Aminov, 2021; Li R. et al., 2021; Umar et al., 2021; Zhang et al., 2021). Among them, the mobile tet(X4) gene has been increasingly identified in E. coli from various sources including food-producing animals, wild birds, food products, humans, and the environment, mainly in China (He et al., 2019; Fang et al., 2020; Li et al., 2020; Li Y. et al., 2021; Dong et al., 2022; Liu et al., 2022). It has sporadically reported in countries outside of China, e.g., Singapore, Pakistan, Vietnam, United Kingdom, and Norway (Ding et al., 2020; Marathe et al., 2021; Mohsin et al., 2021; Dao et al., 2022; Martelli et al., 2022). The tet(X4) has subsequently detected in various Enterobacteriaceae species, such as Proteus, A. baumannii, Aeromonas caviae, Citrobacter freundii, Enterobacter cloacae, E. hormaechei, Klebsiella pneumoniae, and Shewanella xiamenensis (Chen et al., 2019; He et al., 2019; Zeng et al., 2021; Dao et al., 2022; Li et al., 2022; Wu et al., 2022; Zhai et al., 2022).
Although tigecycline is not applied in livestock, the tet(X4) gene and tigecycline resistance are frequently described in E. coli from food-producing animals (mainly pigs) in China (He et al., 2019; Fang et al., 2020; Li Y. et al., 2021; Liu et al., 2022). The heavy use of tetracyclines in animal production might facilitate the emergence and spread of tet(X) in livestock (He et al., 2019). In addition, conjugative/mobilizable plasmids and mobile elements play an essential role in the dissemination of tet(X4) in Enterobacteriaceae (Aminov, 2021). In this study, we aimed to investigate the prevalence and characterization of tet(X4) in E. coli isolates from one pig farm in Shanghai, China, to provide insights into the spread of tet(X4) in this swine farm.
Materials and methods
Sample collection and tet(X) detection
On 15 July 2021, 61 non-duplicate samples from pig feces (n = 41) and pig feed (n = 20) were collected from a pig farm in Shanghai, China. Samples were incubated in LB broth for 18∼24 h and then cultured on the MacConkey agar with and without 2 mg/L tigecycline. One E. coli isolate per plate was selected and identified by 16S rRNA gene sequencing (Kim et al., 2010). The presence of tet(X) were detected by PCR and sequencing (Wang et al., 2019).
Antimicrobial susceptibility testing
The MICs of tigecycline were determined in all E. coli strains using the broth microdilution method and interpreted according to EUCAST clinical breakpoint (MIC ≥ 1 mg/L)1. The tet(X4)-positive isolates were further tested susceptibility to other 13 antimicrobial agents including ampicillin, cefotaxime, meropenem, gentamicin, amikacin, streptomycin, tetracycline, chloramphenicol, florfenicol, nalidixic acid, ciprofloxacin, colistin, and sulfamethoazole/trimethoprim by using the broth microdilution method. The results were interpreted according to Clinical Laboratory Standards Institute (CLSI) M100, 30th edition. Florfenicol (> 16 mg/L) and streptomycin (> 16 mg/L) were interpreted according to the epidemiological cut-off values for E. coli set by EUCAST (see Text Footnote 1). The E. coli strain ATCC 25922 was used for quality control.
Conjugation experiments
Conjugation experiments were conducted according to a previously described protocol (Chen et al., 2007) using E. coli C600 (streptomycin-resistant) as the recipient strain. Transconjugants were selected on MacConkey agar plates supplemented with 2 mg/L tigecycline and 3,000 mg/L streptomycin.
Whole genome sequencing and analysis
The tet(X4)-positive E. coli strains were sequenced on the Illumina Hiseq platform, and the quality-trimmed raw sequence data were assembled into contigs using SPAdes v.3.8.2 with -careful and -cov cut-off auto options. One representative E. coli isolate SH21PTE31 was sequenced using Nanopore MinION, assembling with Unicycler version 0.4.9. The genome sequences of them were analyzed multilocus sequence typing (MLST), resistance genes, and plasmid replicons by using the Center for Genomic Epidemiology (CGE) pipeline2. The tet(X4)-carrying plasmid pYUSHP31-1 in strain SH21PTE31 was analyzed by ISfinder3, BLAST4 and the Gene Construction Kit 4.5 (Textco BioSoftware, Inc., Raleigh, NC, United States). pYUSHP31-1 was compared with other similar plasmids using BLASTn and BRIG.
Phylogenetic analysis of tet(X4)-Positive ST761 Escherichia coli strains
The genome sequences of 43 tet(X4)-positive ST761 E. coli strains in the NCBI database were downloaded (data collected on July 7th, 2022) (Supplementary Table 1). The phylogenetic tree of all the tet(X4)-carrying ST761 E. coli strains obtained from this pig farm and NCBI was constructed using Parsnp5 and visualized by iTOL6. Core genome MLST (cgMLST) profiles based on 2,513 alleles were analyzed using cgMLSTFinder 1.27.
Nucleotide sequence accession number
The whole genome sequences of tet(X4)-positive E. coli isolates have been deposited in the GenBank under accession number PRJNA836295.
Results and discussion
Characterization of tet(X4)-positive Escherichia coli isolates
A total of 49 E. coli strains were obtained from 61 samples. Among them, six strains (12.24%) from different fecal samples were positive for tet(X4), including five strains isolated under selection with tigecycline and one strain isolate without selection. The tet(X4)-positive isolates exhibited resistance to tigecycline (MIC ≥ 16 mg/L), and the remaining isolates showed susceptibility to tigecycline with MICs of 0.125 to 0.5 mg/L. These tet(X4)-positive isolates were also resistant to ampicillin, tetracycline, chloramphenicol, florfenicol, and sulfamethoazole/trimethoprim, but susceptible to cefotaxime, meropenem, gentamicin, amikacin, streptomycin, colistin, nalidixic acid, and ciprofloxacin (Supplementary Table 2). However, all tigecycline-resistant isolates failed to transfer tet(X4) to E. coli C600 via conjugation.
The draft genome sequences of six tet(X4)-positive E. coli strains were obtained by Illumina (Supplementary Table 3). All six tet(X4)-positive E. coli strains belonged to ST761 with identical resistance genes [blaTEM–1, tet(A), tet(M), floR, qnrS1, sul3, dfrA5 and mef(B)] and plasmid replicons [IncFIA, IncFIB(K), IncX1, IncR], except that additional ColE10 plasmid was present in isolate SH21PTE35 (Figure 1).
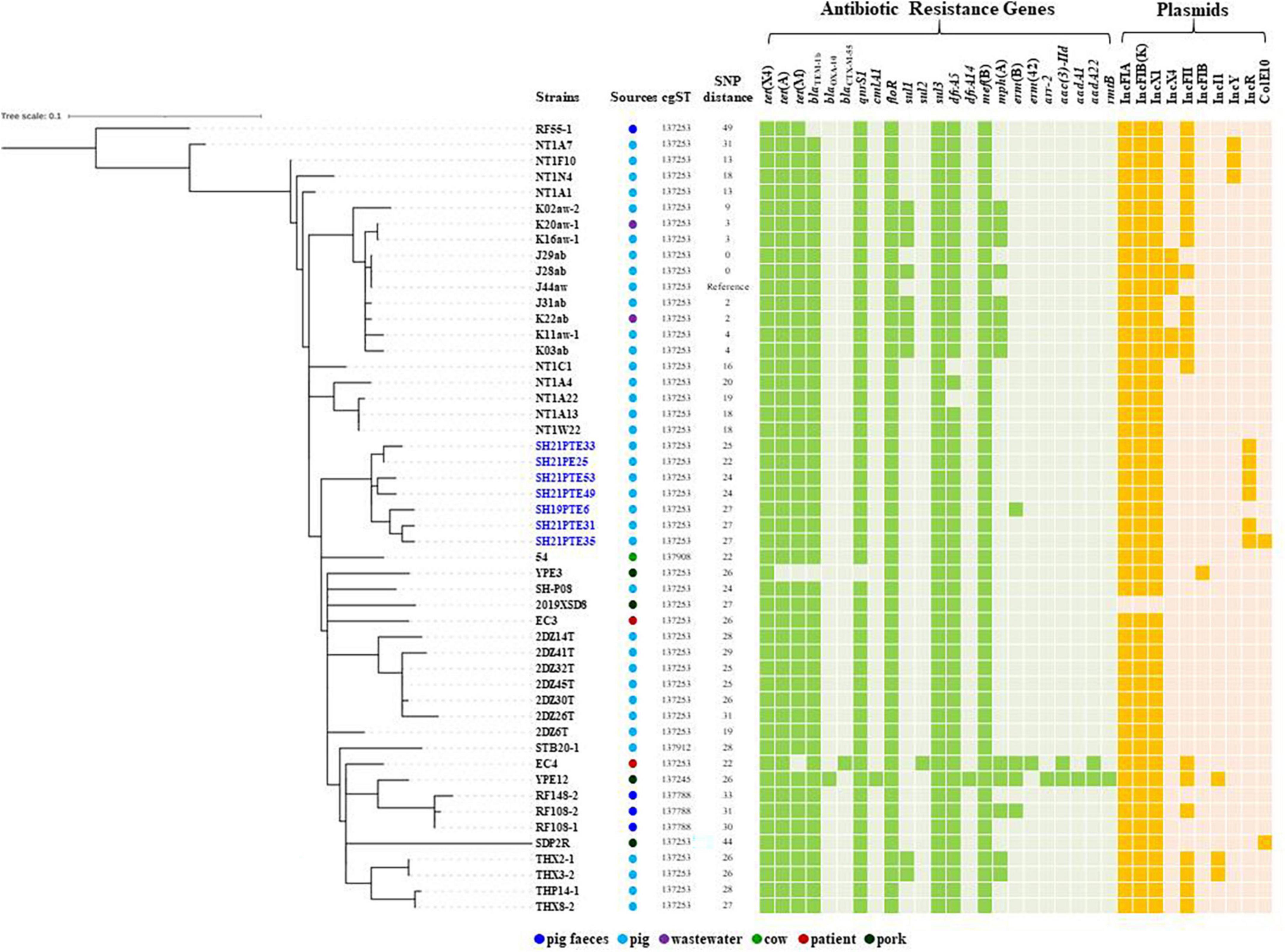
Figure 1. The maximum likelihood tree of tet(X4)-positive E. coli ST761 isolates in this study compared with tet(X4)-positive E. coli ST761 isolates from NCBI based on cgSNP analysis. Antibiotic resistance genes and plasmid replicons with >95% sequence homology and >60% coverage are shown. The isolates obtained in this study and in the same pig farm were indicated in blue.
tet(X4)-Carrying plasmid pYUSHP31-1
The complete sequences of isolate SH21PTE31, as a representative ST761 E. coli strain, was obtained. A total of 43,674 reads were obtained, and the sequencing data volume was approximately 1,000 Mbp. The minimal, maximum and average read lengths were 8,260 bp, 150,801 bp and 22,897.3 bp, respectively. The read length N50 of the total sequencing data were 28,637 bp. The isolate SH21PTE31 consisted of one chromosome (4,706,168 bp) and four plasmids (Supplementary Table 3). Among them, tet(X4) and another eight resistance genes were co-located on the largest plasmid, designated as pYUSHP31-1. This plasmid had a size of 104,163 bp, and belonged to the hybrid IncFIA18/IncFIB(K)/IncX1 plasmid. It was highly similar to our previously reported plasmid pYUSHP6-tetX (GenBank accession no. MW423609) from ST761 E. coli isolate SH19PTE6 collected from the same pig farm in 2019 (Wang et al., 2021), and also showed high identity (> 99.7%) to multiple tet(X4)-carrying IncFIA18/IncFIB(K)/IncX1 plasmids from ST761 E. coli strains in China, such as pNT1W22-tetX4 (pig, CP075470), pRF108-2_97k_tetX (pig, MT219820), pSTB20-1T (pig, CP050174), p54-tetX (cow, CP041286), pYPE12-101k-tetX4 (pork, CP041443), and pYPE3-92k-tetX4 (pork, CP041453) (Figure 2). Similar IncFIA18/IncFIB(K)/IncX1 plasmids harboring tet(X4) were also present among other E. coli lineages obtained from a pig farm in Jiangsu province, China (Li Y. et al., 2021), e.g., pNT1N31-tetX4 (ST716, CP075481), pNT1F25-tetX4 (ST1421, CP075471), pNT1F31-tetX4 (ST206, CP045188), pNT1N25-tetX4 (ST641, CP075485), and pNT1F34-tetX (ST10115, CP075486) (Figure 2), highlighting the importance role of horizontal transfer of plasmids in the tet(X4) dissemination between different bacteria.
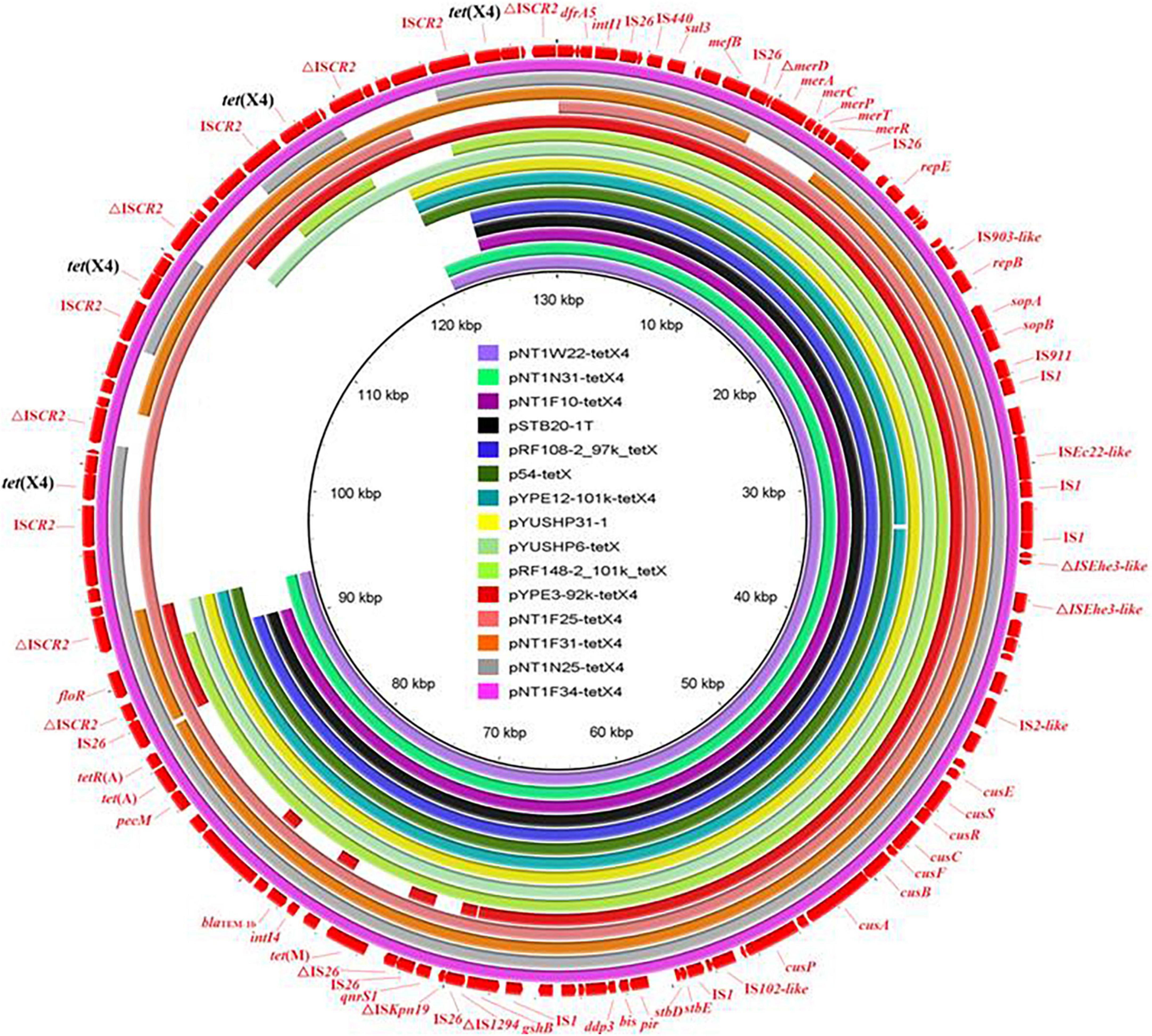
Figure 2. Sequence comparison of tet(X4)-carrying plasmid pYUSHP31-1 from E. coli isolate SH21PTE31 in this study with other similar IncFIA18/IncFIB(K)/IncX1 plasmids using BRIG. The reference sequence pNT1F34-tetX (CP075486) is indicated in red in the outer circle.
As shown in Figure 3, these IncFIA18/IncFIB(K)/IncX1 plasmids shared closely related multidrug resistance regions (MRRs). The MRRs in all were bounded by one copy of IS26 and IS1, respectively. The pYUSHP31-1 MRR (53,134 bp) contained nine resistance genes and consisted of five regions bounded by IS26 or ISCR2 (Figure 3A). The first of these (2,813 bp) comprised one copy of IS26 and a putative open reading frame encoding recombinase family protein, which was absent in other similar plasmids.
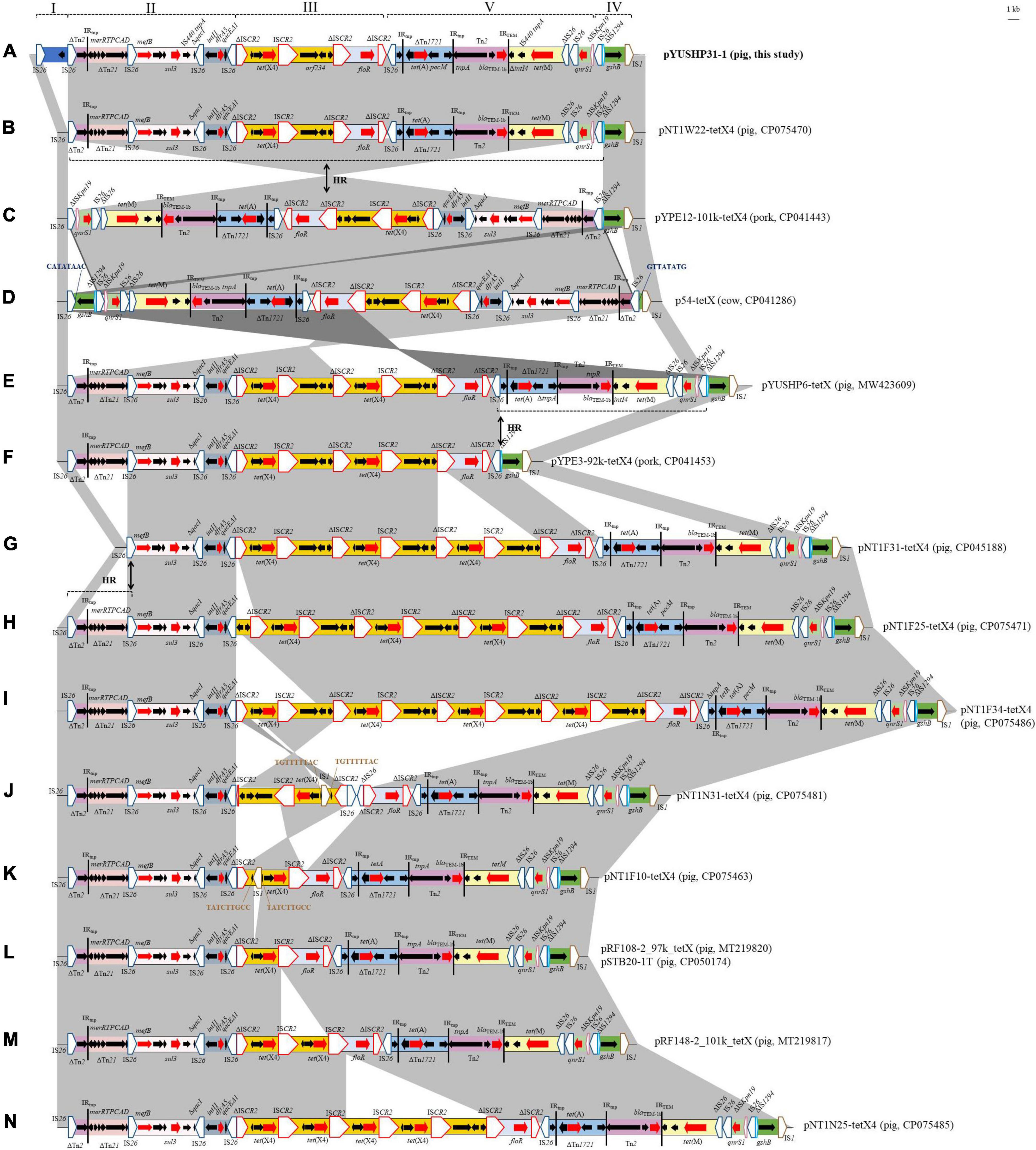
Figure 3. Genetic organization of the multidrug resistance region of plasmid pYUSHP31-1 and structural comparison with other IncFIA18/IncFIB(K)/IncX1 plasmids. I to IV indicate five regions bounded by IS26 or ISCR2 in pYUSHP31-1. The extents and directions of orientation of resistance genes (thick red arrow) and other genes are indicated by arrows. Regions with >99% identity are shaded in gray. 1 indicates a truncated gene or mobile element. Insertion sequences (ISs) are shown as boxes labeled with the IS name. Labeled vertical arrows with IS boxes denote the insertion position of IS elements. Direct repeats are indicated by arrows and sequences. Tall bars represent the 38-bp IR of transposons (Tn). Arrows labeled with “HR” and dotted lines indicate where homologous recombination could explain differences between structures.
The second part (∼14.8 kb) contained three resistance genes mefB, sul3, and dfrA5; four copies of IS26 and incomplete transposon Tn2 and Tn21. This fragment was also present in other IncFIA18/IncFIB(K)/IncX1 plasmids, but differed by 46-bp shorter (limited to pNT1N25-tetX4) or 126-bp longer Tn2 except pYUSHP6-tetX (identical to pYUSHP31-1, obtained from the same pig farm); deletion of a 5,198-bp structure (IS26-ΔTn2-ΔTn21) in pNT1F31-tetX4 (Figure 3G).
The third region corresponded to the core tet(X4) structure [ΔISCR2-orf1-abh-tet(X4)-ISCR2-orf2-orf3-orf4-ΔISCR2] and downstream floR-ΔISCR2 module, as observed in other IncFIA18/IncFIB(K)/IncX1 plasmids with one to four copies of tet(X4) structure (Figures 3A–J). Compared with that of pYUSHP31-1, partial tet(X4) structure [ΔISCR2-orf1-abh-tet(X4)-ISCR2] with varied copies was identified in plasmids pNT1F10-tetX4, pRF108-2_97k_tetX, pSTB20-1T, pRF148-2_101k_tetX, and NT1N25-tetX4 (Figures 3K–N); one copy of IS1 was inserted into orf1 within the tet(X4) structure with 9-bp direct repeats in plasmids pNT1F10-tetX4 and pNT1N31-tetX4, and the latter plasmid carried the tet(X4) fragment in the opposite orientation and additional two copies of IS26 upstream of floR-ΔISCR2 module (Figure 3J). As previously described (Liu et al., 2022), ISCR2 is associated with tet(X4) transmission by forming an rolling-cycle transposable unit, thus generating tandem copies of tet(X4)-harboring structures in different IncFIA18/IncFIB(K)/IncX1 plasmids.
The fourth segment (∼18.4 kb) included one copy of IS26, an incomplete Tn1721 carrying tetracycline resistance gene tet(A) and an intact Tn2 (tnpA-tnpR-blaTEM–1b), followed by 5,391-bp module [ΔintI4-IS440 tnpA-tet(M)-ΔIS26] and qnrS1 structure (IS26-qnrS1-ΔISKpn19). This region was also found in other IncFIA18/IncFIB(K)/IncX1 plasmids with the same ΔISKpn19/IS26 boundary except pYPE3-92k-tetX4 (Figure 3F). IS26-mediated homologous recombination could explain the loss or acquisition of this region.
The last segment comprising a 3,507-bp structure (IS26-ΔIS1294-gshB-IS1) was identical to segments in other plasmids except p54-tetX (Figure 3D). Insertion of an extra copy of IS26 downstream of gshB, followed by homologous recombination between it and the first IS26 of MRR, may explain the opposite location of an approximately 50.2-kb fragment within MRR in p54-tetX compared to pYUSHP31-1. Similar recombination between two IS26 elements located in inverse orientations may also occur in pYPE12-101k-tetX4, leading to the presence of ∼47.8 kb fragment with the opposite orientation within MRR (Figure 3C).
These tet(X4)-carrying IncFIA18/IncFIB(K)/IncX1 plasmids may evolve from the same ancestor, and form variable but related MRRs by insertions, deletions, or rearrangements of different resistance modules mediated by mobile elements such as IS26 and ISCR2.
Phylogenomic analysis of tet(X4)-Positive ST761 Escherichia coli strains
Escherichia coli ST761 has been increasingly reported in different sources associated with tet(X4) in China, particularly from pigs (Supplementary Table 1). To further compare the genetic differences between tet(X4)-positive E. coli isolates of the same ST, we performed a phylogenomic analysis based on cgSNP. The results revealed a relatively close genetic relationship (0-49 SNPs) among 50 tet(X4)-positive ST761 E. coli isolates (Figure 1). Among them, cgST 137253 (n = 44) was the most prevalent type, and it contained two isolates from patients, two from wastewater, three from pork, and 37 from pigs including six strains obtained in this study and SH19PTE6 from the same pig farm (Figure 1). It indicates that clonal transmission has occurred in this swine farm. The plasmid replicons [IncFIA, IncFIB(K), IncX1] possibly associated with tet(X4) were present in all isolates, and the core resistance genes [blaTEM–1, tet(A), tet(M), floR, qnrS1, sul3, dfrA5 and mef(B)] within tet(X4)-carrying IncFIA/IncFIB(K)/IncX1 plasmid pYUSHP31-1 were shared by 45 strains (Figure 1).
Although horizontal transfer mediated by plasmids (e.g., IncQ, and IncX1) and insertion sequences (e.g., ISCR2, IS26, and IS1) is the main mechanism for tet(X4) transmission (Aminov, 2021; Liu et al., 2022; Yu et al., 2022), clonal spread of tet(X4)-carrying strains, such as E. coli ST877, ST10, and ST48 clones is also responsible for tet(X4) dissemination between animals and humans (Cui et al., 2022). The E. hormaechei co-harboring tet(X4) and blaNDM could also clonally spread from the slaughterhouse to the retail market (Li et al., 2022). E. coli ST761 isolates carrying tet(X4) has been detected in pigs, cow, pork, wastewater, and patients in different areas from China sharing a close phylogenetic relationship, suggesting that the ST761 lineage has the potential to be a successful clone to transfer tet(X4) and other resistance genes as well in China.
Conclusion
Our findings suggest that tet(X4)-positive ST761 E. coli was the main reason for spread and persistence of tet(X4) in this pig farm. Importantly, E. coli ST761 has the potential to become a high-risk clone for tet(X4) dissemination in China. On the other hand, the tet(X4)-carrying IncFIA18/IncFIB(K)/IncX1 hybrid plasmids within ST761 E. coli lineage could reorganize, acquire or lose resistance modules mediated by mobile elements such as ISCR2 and IS26. The horizontal transfer of similar IncFIA18/IncFIB(K)/IncX1 plasmids further facilitates the tet(X4) dissemination in distinct lineages.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
XJ and JW conceived the study. M-JL, HW, Z-YW, and YJ carried out the experiments. JW, Z-YW, and YJ analyzed the data. JW wrote the manuscript. Z-MP and XJ revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the fifth phase of “333 Project” Scientific Research project in Jiangsu Province (BRA2020002) and Key Research and Development Program (Modern Agriculture) project of Jiangsu Province (grant no. BE2021331).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.967313/full#supplementary-material
Footnotes
- ^ www.eucast.org
- ^ http://www.genomicepidemiology.org/
- ^ https://www-is.biotoul.fr/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ https://harvest.readthedocs.io/en/latest/content/parsnp.html
- ^ https://itol.embl.de/
- ^ https://cge.food.dtu.dk/services/cgMLSTFinder/
References
Aminov, R. (2021). Acquisition and spread of antimicrobial resistance: A tet(X) case study. Int. J. Mol. Sci. 22:3905. doi: 10.3390/ijms22083905
Chen, C., Chen, L., Zhang, Y., Cui, C. Y., Wu, X. T., He, Q., et al. (2019). Detection of chromosome-mediated tet(X4)-carrying Aeromonas caviae in a sewage sample from a chicken farm. J. Antimicrob. Chemother. 74, 3628–3630. doi: 10.1093/jac/dkz387
Chen, L., Chen, Z. L., Liu, J. H., Zeng, Z. L., Ma, J. Y., and Jiang, H. X. (2007). Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J. Antimicrob. Chemother. 59, 880–885. doi: 10.1093/jac/dkm065
Cui, C. Y., Li, X. J., Chen, C., Wu, X. T., He, Q., Jia, Q. L., et al. (2022). Comprehensive analysis of plasmid-mediated tet(X4)-positive Escherichia coli isolates from clinical settings revealed a high correlation with animals and environments-derived strains. Sci. Total Environ. 806:150687. doi: 10.1016/j.scitotenv.2021.150687
Dao, T. D., Kasuga, I., Hirabayashi, A., Nguyen, D. T., Tran, H. T., Vu, H., et al. (2022). Emergence of mobile tigecycline resistance gene tet(X4)-harbouring Shewanella xiamenensis in a water environment. J. Glob. Antimicrob. Resist. 28, 140–142. doi: 10.1016/j.jgar.2021.12.022
Ding, Y., Saw, W. Y., Tan, L., Moong, D., Nagarajan, N., Teo, Y. Y., et al. (2020). Emergence of tigecycline- and eravacycline-resistant Tet(X4)-producing Enterobacteriaceae in the gut microbiota of healthy Singaporeans. J. Antimicrob. Chemother. 75, 3480–3484. doi: 10.1093/jac/dkaa372
Dong, N., Zeng, Y., Cai, C., Sun, C., Lu, J., Liu, C., et al. (2022). Prevalence, transmission, and molecular epidemiology of tet(X)-positive bacteria among humans, animals, and environmental niches in China: An epidemiological, and genomic-based study. Sci. Total Environ. 818:151767. doi: 10.1016/j.scitotenv.2021.151767
Fang, L. X., Chen, C., Cui, C. Y., Li, X. P., Zhang, Y., Liao, X. P., et al. (2020). Emerging high-level tigecycline resistance: Novel tetracycline destructases spread via the mobile Tet(X). Bioessays 42:e2000014. doi: 10.1002/bies.202000014
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
Kim, T. W., Kim, Y. H., Kim, S. E., Lee, J. H., Park, C. S., and Kim, H. Y. (2010). Identification and distribution of Bacillus species in doenjang by whole-cell protein patterns and 16S rRNA gene sequence analysis. J. Microbiol. Biotechnol. 20, 1210–1214. doi: 10.4014/jmb.1002.02008
Li, R., Liu, Z., Li, Y., Xiao, X., and Wang, Z. (2022). Characterization of blaNDM-positive Enterobacteriaceae reveals the clonal dissemination of Enterobacter hormaechei coharboring blaNDM and tet(X4) along the pork production chain. Int. J. Food Microbiol. 372:109692. doi: 10.1016/j.ijfoodmicro.2022.109692
Li, R., Lu, X., Peng, K., Liu, Z., Li, Y., Liu, Y., et al. (2020). Deciphering the structural diversity and classification of the mobile tigecycline resistance gene tet(X)-bearing plasmidome among bacteria. mSystems 5:e00134–20. doi: 10.1128/mSystems.00134-20
Li, R., Peng, K., Xiao, X., Wang, Y., and Wang, Z. (2021). Characterization of novel ISAba1-bounded tet(X15)-bearing composite transposon Tn6866 in Acinetobacter variabilis. J. Antimicrob. Chemother. 76, 2481–2483. doi: 10.1093/jac/dkab182
Li, Y., Wang, Q., Peng, K., Liu, Y., Xiao, X., Mohsin, M., et al. (2021). Distribution and genomic characterization of tigecycline-resistant tet(X4)-positive Escherichia coli of swine farm origin. Microb. Genom. 7:000667. doi: 10.1099/mgen.0.000667
Liu, D., Wang, T., Shao, D., Song, H., Zhai, W., Sun, C., et al. (2022). Structural diversity of the ISCR2-mediated rolling-cycle transferable unit carrying tet(X4). Sci. Total Environ. 826:154010. doi: 10.1016/j.scitotenv.2022.154010
Marathe, N. P., Svanevik, C. S., Ghavidel, F. Z., and Grevskott, D. H. (2021). First report of mobile tigecycline resistance gene tet(X4)-harbouring multidrug-resistant Escherichia coli from wastewater in Norway. J. Glob. Antimicrob. Resist. 27, 37–40. doi: 10.1016/j.jgar.2021.07.019
Martelli, F., AbuOun, M., Cawthraw, S., Storey, N., Turner, O., Ellington, M., et al. (2022). Detection of the transferable tigecycline resistance gene tet(X4) in Escherichia coli from pigs in the United Kingdom. J. Antimicrob. Chemother. 77, 846–848. doi: 10.1093/jac/dkab439
Mohsin, M., Hassan, B., Martins, W., Li, R., Abdullah, S., Sands, K., et al. (2021). Emergence of plasmid-mediated tigecycline resistance tet(X4) gene in Escherichia coli isolated from poultry, food and the environment in South Asia. Sci. Total Environ. 787:147613. doi: 10.1016/j.scitotenv.2021.147613
Umar, Z., Chen, Q., Tang, B., Xu, Y., Wang, J., Zhang, H., et al. (2021). The poultry pathogen Riemerella anatipestifer appears as a reservoir for Tet(X) tigecycline resistance. Environ. Microbiol. 23, 7465–7482. doi: 10.1111/1462-2920.15632
Wang, J., Wu, H., Mei, C. Y., Wang, Y., Wang, Z. Y., Lu, M. J., et al. (2021). Multiple mechanisms of tigecycline resistance in Enterobacteriaceae from a pig farm. China. Microbiol. Spectr. 9:e0041621. doi: 10.1128/Spectrum.00416-21
Wang, L., Liu, D., Lv, Y., Cui, L., Li, Y., Li, T., et al. (2019). Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, eravacycline, and omadacycline in a clinical Acinetobacter baumannii isolate. Antimicrob. Agents Chemother. 64:e01326–19. doi: 10.1128/AAC.01326-19
Wu, Y., He, R., Qin, M., Yang, Y., Chen, J., Feng, Y., et al. (2022). Identification of plasmid-mediated tigecycline-resistant gene tet(X4) in Enterobacter cloacae from pigs in China. Microbiol. Spectr. 10:e0206421. doi: 10.1128/spectrum.02064-21
Yaghoubi, S., Zekiy, A. O., Krutova, M., Gholami, M., Kouhsari, E., Sholeh, M., et al. (2021). Tigecycline antibacterial activity, clinical effectiveness, and mechanisms and epidemiology of resistance: Narrative review. Eur. J. Clin. Microbiol. Infect. Dis. 5, 1–20. doi: 10.1007/s10096-020-04121-1
Yu, R., Chen, Z., Schwarz, S., Yao, H., and Du, X. D. (2022). Mobilization of tet(X4) by IS1 family elements in porcine Escherichia coli isolates. Antimicrob. Agents Chemother. 66:e0159721. doi: 10.1128/AAC.01597-21
Zeng, Y., Dong, N., Liu, C., Lu, J., and Zhang, R. (2021). Presence of tet(X4)-positive Citrobacter freundii in cancer patient with chemotherapy-induced persistent diarrhea. J. Glob. Antimicrob. Resist. 24, 88–89. doi: 10.1016/j.jgar.2020.11.007
Zhai, W., Tian, Y., Lu, M., Zhang, M., Song, H., Fu, Y., et al. (2022). Presence of mobile tigecycline resistance gene tet(X4) in clinical Klebsiella pneumoniae. Microbiol. Spectr. 10:e0108121. doi: 10.1128/spectrum.01081-21
Keywords: plasmids, ST761, tigecycline resistance, tet(X4), Escherichia coli
Citation: Wang J, Lu M-J, Wang Z-Y, Jiang Y, Wu H, Pan Z-M and Jiao X (2022) Tigecycline-resistant Escherichia coli ST761 carrying tet(X4) in a pig farm, China. Front. Microbiol. 13:967313. doi: 10.3389/fmicb.2022.967313
Received: 12 June 2022; Accepted: 19 July 2022;
Published: 09 August 2022.
Edited by:
Kristina Kadlec, Independent Researcher, Wunstorf, GermanyReviewed by:
Jianmin Zhang, South China Agricultural University, ChinaPo-Xing Zheng, Academia Sinica, Taiwan
Copyright © 2022 Wang, Lu, Wang, Jiang, Wu, Pan and Jiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinan Jiao, amlhb0B5enUuZWR1LmNu
 Jing Wang
Jing Wang Meng-Jun Lu1,2
Meng-Jun Lu1,2 Zhen-Yu Wang
Zhen-Yu Wang Zhi-Ming Pan
Zhi-Ming Pan Xinan Jiao
Xinan Jiao