- Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China
Lysobacter enzymogenes has been applied as an abundant beneficial microorganism to control plant disease; however, most L. enzymogenes strains have been mainly reported to control fungal diseases, not bacterial diseases. In this study, two L. enzymogenes strains were characterized, of which CX03 displayed a broad spectrum of antagonistic activities toward multiple bacteria, while CX06 exhibited a broad spectrum of antagonistic activities toward diverse fungi and oomycete, and the whole genomes of the two strains were sequenced and compared. The genome annotation showed that the CX03 genome comprised a 5,947,018 bp circular chromosome, while strain CX06 comprised a circular 6,206,196 bp chromosome. Phylogenetic analysis revealed that CX03 had a closer genetic relationship with L. enzymogenes ATCC29487T and M497-1, while CX06 was highly similar to L. enzymogenes C3. Functional gene annotation analyses of the two L. enzymogenes strains showed that many genes or gene clusters associated with the biosynthesis of different secondary metabolites were found in strains CX03 and CX06, which may be responsible for the different antagonistic activities against diverse plant pathogens. Moreover, comparative genomic analysis revealed the difference in bacterial secretory systems between L. enzymogenes strains CX03 and CX06. In addition, numerous conserved genes related to siderophore biosynthesis, quorum sensing, two-component systems, flagellar biosynthesis and chemotaxis were also identified in the genomes of strains CX03 and CX06. Most reported L. enzymogenes strains were proven mainly to suppress fungi, while CX03 exhibited direct inhibitory activities toward plant bacterial pathogens and showed an obvious role in managing bacterial disease. This study provides a novel understanding of the biocontrol mechanisms of L. enzymogenes, and reveals great potential for its application in plant disease control.
Introduction
Lysobacter species are widely distributed in nature, including in soil, fresh water, and even some extreme environments (Ramamoorthy et al., 2001; Folman et al., 2003). The Lysobacter genus was first proposed in 1978 and was named for its ability to produce multiple antibiotics that dissolve the cell walls or membranes of other microorganisms (Panthee et al., 2016). Lysobacter strains are characterized as sliding and glutinous on agar medium plate (de Bruijn et al., 2015). Lysobacter belongs to the Xanthomonadaceae family of Gammaproteobacteria, and was once indistinguishable from Polyangium and Sorangium or confused with Xanthomonas and Stenotrophomonas. At the time it was identified as a separate genus, Lysobacter contained four species: L. antibioticus, L. brunescens, L. enzymogenes, and L. gummosus (Hayward et al., 2010). To date, approximately 25 Lysobacter species have been identified, but only a few strains have been subjected to genome-wide. Recently, Lysobacter has been described as a new source for bioactive natural products, and has been used to control plant disease. For example, L. enzymogenes OH11 produced heat-stable antifungal factor (HSAF) that showed degrading activity against the chitinous hyphae of Fusarium graminearum (Odhiambo et al., 2017). A 2,5-diketopiperazine produced by L. capsica AZ78 showed significant antagonistic activities toward Phytophthora infestans and Plasmopara viticola (Puopolo et al., 2014). L. antibiobioticus HS124 produced 4-hydroxyphenylacetic acid (4-HPAA), an antibiotic that was able to control the root-knot nematode in tomato (Lee et al., 2013).
Lysobacter enzymogenes has been reported as a potential biocontrol agent to control diverse plant diseases due to its capacity to generate secondary metabolites with antibiotic activity or generate a variety of extracellular enzymes with degrading ability toward the cell walls of fungal pathogens (Hayward et al., 2010; Xie et al., 2012). L. enzymogenes C3 was reported to inhibit wheat root rot caused by Bipolaris sorokiniana (Zhang and Yuen, 1999), lawn brown spot triggered by Rhizoctonia solani (Giesler and Yuen, 1998), and soybean rust resulting from Uromyces appendiculatus (Yuen et al., 2001). In addition, strain C3 had strong antagonistic activity against Pythium ultimum and various nematodes (Kobayashi and Yuen, 2005; Li et al., 2008). L. enzymogenes OH11 effectively antagonized the growth of many plant pathogenic fungi, including R. solani, P. capsica and P. aphanidermatum (Qian et al., 2009). L. enzymogenes 3.1T8 was isolated from the inter-root of cucumber and showed an antagonistic effect against P. aphanidermatum and exhibited a significant effect in controlling Corynespora cassiicola and P. capsica disease (Folman et al., 2003). L. enzymogenes M497-1 was reported to have broad bacteriolytic specificity and was used to lyse anaerobic Gram-positive rods, including Micrococcus radiodurans, Staphylococcus aureus and Peptococcus saccharolyticus (Takami et al., 2017). L. enzymogenes N4-7 was used as a biocontrol agent to control plant fungal diseases and nematodes for the production of β-1,3-glucanase, which has hydrolysis activity on fungal cell walls (Palumbo et al., 2003). In addition, L. enzymogenes LE16 reduced the organic phosphorus and increased the soluble P in the soil, thus promoting plant growth (Chen et al., 2019). However, most reported L. enzymogenes strains possess the ability to suppress plant pathogenic fungi, oomycete or nematodes, while very few L. enzymogenes strains have been reported to control bacterial disease.
It was reported that the reason Lysobacter spp. can control plant disease involved in their ability to generate a variety of lytic enzymes and antibiotics. Previous studies showed that Lysobacter strains harbored large numbers of genes related to antagonistic activities, and whole-genome annotation indicated that each Lysobacter strain shared 12–16 secondary metabolites (Panthee et al., 2016). L. enzymogenes was proven to suppress fungi with multiple secondary metabolites and lytic antibiotics. The cyclic lipodepsipeptide HSAF derived from L. enzymogenes C3 inhibited the activity of fungal neuro phthalamide synthase and altered the composition of sphingolipids in the cell membrane, resulting in depolarization of the target fungal hyphae (hyphae expansion, numerous branches, etc.) and thus exhibited potent antifungal activity (Kobayashi and Yuen, 2005; Yuen et al., 2018). L. enzymogenes OH11 could also produce HSAF, which showed significant antagonistic activities toward diverse pathogenic fungi, and was reported to be a highly effective and broad-spectrum antibiotic of microbial origin. HSAF was not toxic to plants or animals, and maintained high antagonistic activity at high temperatures and under acidic or alkaline conditions, demonstrating good potential for development and application (Yu et al., 2007; Olson et al., 2015). In addition, many other functional genes were closely related to the biosynthesis of HSAF. Overexpression of the quorum sensing related gene lesR significantly impaired HSAF biosynthesis levels and antifungal activities (Qian et al., 2014). Deletion of letR (a TetR-family protein gene) in L. enzymogenes OH11 could significantly increase HSAF levels and the transcription of key biosynthetic genes (Wang et al., 2017). Other non-ribosomal peptides produced by L. enzymogenes were also reported to have antipathogenic activities. WAP-8294A, a cyclic lipodepsipeptide isolated from L. enzymogenes OH11, was reported to have potent activity against Gram-positive bacterial pathogens, especially S. aureus (Hashizume et al., 2001). However, most studies have focused on the antifungal compounds produced by L. enzymogenes, especially HSAF, while only a few reports of secondary metabolites have been related to antibacterial activity. Previous studies showed that L. enzymogenes degraded phytopathogenic (micro)organism cell walls by secreting a variety of highly active extracellular hydrolases, such as protease cellulases, glucanases and chitinases. L. enzymogenes strains C3, OH11 and N4–7 synthesized three extracellular β-1,3-glucanases encoded by gluABC genes (Palumbo et al., 2003, 2005). The capacity of the strain to generate β-1,3-glucanases and control brown patches in tall fescue was reduced when all three genes of L. enzymogenes C3 were mutated simultaneously (Palumbo et al., 2005). L. enzymogenes OH11 secreted proteases that significantly inhibited the growth of R. solani. The α-hydrolytic protease secreted by L. enzymogenes 3.1T8 could degrade proteins in the nematode body wall, and effectively antagonize nematode-induced plant diseases (Folman et al., 2004). In addition, L. enzymogenes OH11 used the type IV secretion system (T4SS) as the main contact dependent weapon to control Pectobacterium carotovorum (Shen et al., 2021). Type IV pilus-driven twitching motility, which was proven to promote bacterial colonization on host fungal surfaces in L. enzymogenes OH11, was also recognized as one of the antifungal mechanisms (Xia et al., 2018). Moreover, induced resistance was also described as a mechanism for disease control of L. enzymogenes strains. Resistance elicited by L. enzymogenes C3 suppressed the conidial germination of B. sorokiniana on the phylloplane, thus reducing the severity of leaf spot (Kilic-Ekici and Yuen, 2003). In addition, other Lysobacter species applied to biological control have similar antagonistic mechanisms. L. antibioticus HS124 showed a significant control effect on Phytophthora Blight for the generation of 4-hydroxyphenylacetic acid and many lytic enzymes (Ko et al., 2009). Notably, L. antibioticus OH13 produced phenazine antibiotics that showed antagonistic activities against several bacteria (Zhao et al., 2016). L. capsica AZ78 produced cyclo (L-Pro-L-Tyr), a 2,5-diketopiperazine that was effective in inhibiting the growth of the sporangia of P. infestans (Puopolo et al., 2014).
In this study, two L. enzymogenes strains CX03 and CX06 exhibited distinct antagonistic activities toward different plant pathogens. To reveal the possible different biocontrol mechanisms of the two strains, the genomes of CX03 and CX06 were sequenced and compared with those of the typical Lysobacter strains C3, M497-1, 55 and 76, which were reported to be beneficial to plant growth. Phylogenetic analysis was used to demonstrate the taxonomic positions of CX03 and CX06, and their relationships with other Lysobacter strains. Core genes related to the biosynthesis of known and unknown secondary metabolites of the different Lysobacter strains were predicted and compared. Comparative genome analysis was performed to mine the common and unique functional genes involved in other biocontrol factors, such as the bacterial secretion system and the biosynthesis of lyase. These data will provide novel insights into the biocontrol mechanisms of L. enzymogenes and provide new antibiotic resources for disease control.
Materials and methods
Strain isolation, culture conditions, microscopic analysis and bacterial DNA extraction
Strains CX03 and CX06 were isolated from rhizosphere soil in Guizhou Province and swamp soil in Sichuan Province, China, according to a standard 10-fold dilution plating assay as described by previous study, respectively (Wang et al., 2013). Strains CX03 and CX06 grew stably on nutrient broth (NB) agar medium at 28°C. Gliding motility of strains CX03 and CX06 were evaluated on 5% tryptic soy agar (TSA) medium according to the reported method (Zhou et al., 2015). Strains CX03 and CX06 were inoculated to NB media (1/1000 inoculation proportion) with shaking at 28°C, and the colony number of CX03 and CX06 and the pH value of CX03 and CX06 culture were measured every four hours. The genomic DNA of strains CX03 and CX06 was extracted from the cultured bacterial colonies (OD600 = 1.0) according to the manufacturer’s instructions of the TIANamp Bacteria DNA kit (Tiangen Biotech (Beijing, China) Co., Ltd.). Scanning electron microscopy (SEM) and transmission electron microscopy (TEM; SU8010, Hitachi, Japan, 10.0 kV) were used to observe the micromorphology of the two strains.
Antagonistic and biocontrol assays
The antagonistic abilities of strains CX03 and CX06 against different plant pathogens were measured by plate bioassays, and the control efficacies of strains CX03 and CX06 against cabbage black rot and the stem rot of Chinese cabbage were tested in pot experiments. Briefly, the cabbage was inoculated with X. campestris pv. campestris (1 × 108cfu/ml) by spray method, 1 day later, the culture of CX03 and CX06 (1 × 108cfu/ml) were inoculated to cabbage (inoculated with X. campestris pv. campestris) by spray method, respectively, and kasugamycin was used as chemical control. The Chinese cabbage was inoculated with R. solani by root irrigation, 1 day later, the culture of CX03 and CX06 (1 × 108cfu/ml) were inoculated to Chinese cabbage (inoculated with R. solani), respectively, and validamycin was used as chemical control. Plate tests were used to evaluate the ability to produce siderophores, proteases (Prts), and chitinases according to previously reported methods (Murata, 1991; Chatterjee et al., 1995).
Biosynthesis of selenium nanoparticles
The selenium (Se) nanoparticle (SeNP) biosynthesis capacities of strains CX03 and CX06 were measured based on previously described method (Zhu et al., 2018) with slight adjustment. The selenium standard curve was plotted according to the previously reported method (Biswas et al., 2011). The activated strains CX03 and CX06 were inoculated in LB medium containing 2.5, 5, 10, 15, 45, and 75 mM Se, and the samples were taken after 24 h of continuous shaking cultivation. The culture was centrifuged at 10,000 rpm for 20 min, the sediment bacteria were resuspended in ddH2O, and the above steps were repeated three times. One milliliter of suspension was placed in a 4 ml centrifuge tube, and 1 ml of Na2S (1 M) solution was added and mixed well for 1 h. Then, the absorbance value at 500 nm was measured by ultraviolet spectrophotometry and each sample was repeated three times. The measured absorbance was used to calculate the concentration of selenium nanoparticles in the samples according to the standard curve.
Whole-genome sequencing and annotation
The genomes of strains CX03 and CX06 were sequenced using a PacBio Sequel platform by Allwegene Technologies Corporation, Beijing, China. Circular genome visualization of the two strains were created by CGView (Tatusova et al., 2016). The functional genes were predicted using GeneMarkS software (version 4.17). Transfer RNA (tRNA) and ribosomal RNA (rRNA) were predicted through tRNAscan-SE version 2.0 (Lowe and Chan, 2016) and RNAmmer version 1.2 (Lagesen et al., 2007), respectively. Gene Ontology database (GO) (Ashburner et al., 2000), Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2006), RAST (Rapid Annotation using Subsystem Technology) (Aziz et al., 2008), Pfam1, Swiss-Prot2 (Bairoch and Apweiler, 2000), Carbohydrate-Active enZYmes Database (CAZy) (Cantarel et al., 2009), Transporter Classification Database (TCDB) (Saier et al., 2009) and the enhanced COG database (Bairoch and Apweiler, 2000; Tatusov et al., 2003; El-Gebali et al., 2019).
Comparative genomic analysis
The taxonomic position of two strains CX03 and CX06 were confirmed by multilocus gene sequence analysis (MLSA) according to the sequences of conserved genes (16S rRNA, gyrB, atpD and rpoD) (Supplementary Table 1). Sequence alignment of strains CX03 and CX06 with other Lysobacter strains was performed using MUSCLE and the phylogenetic tree was constructed through the maximum likelihood method in MEGA 6.0 (Tamura et al., 2013). In addition, average nucleotide identity (ANI) and in silico DNA–DNA hybridization (DDH) were executed as described previously (Goris et al., 2007). According to the phylogenetic analysis, four representative strains with released complete genomes, including L. enzymogenes M497-1 (AP014940.1), L. enzymogenes C3 (CP013140.1), L. capsici 55 (CP011130.1) and L. antibioticus 76 (CP011129.1) were chosen for comparative genomic analysis. Genome pairwise alignment was conducted using MAUVE comparison software, and Venn diagrams were generated using R package (Richter and Rosselló-Móra, 2009).
Functional genes related secondary metabolites and microbe–plant interactions
Gene clusters involved in the biosynthesis of secondary metabolites were predicted using antiSMASH 2.03, and compared among the L. enzymogenes CX03, L. enzymogenes CX06, L. enzymogenes M497-1, L. enzymogenes C3, L. capsici 55, and L. antibioticus 76 strains. Functional genes involved in the bacterial secretion system, quorum sensing, two-component system and seleno compound metabolism, biosynthesis of siderophores, flagella and chemotaxis, polyketone, lipopolysaccharide, and peptidoglycan biosynthesis were retrieved according to NCBI databases. The homology analysis of sequences for various functional genes in L. enzymogenes CX03, L. enzymogenes CX06, L. enzymogenes M497-1, L. enzymogenes C3, L. capsici 55 and L. antibioticus 76 were compared based on the KEGG database at the amino acid level.
Results
Antagonistic activity and biochemical characteristics
Antagonistic activity and pot experiment
Antagonistic spectrum assays revealed that the two strains CX03 and CX06 displayed the opposite results. Strain CX03 exhibited broad, strong antagonistic activities toward multiple plant-pathogenic bacteria, including X. campestris pv. campestris, Clavibacter michiganensis subsp. sepedonicum, Ralstonia solanacearum, Pseudomonas syringae pv. tomato, and P. syringae pv. lachrymans, and slight inhibitory activities toward plant-pathogenic fungi F. oxysporum and C. cassiicola, while strain CX06 showed strong inhibitory activities toward many plant-pathogenic fungi including Verticillium dahlia, C. cassiicola, Botrytis cinerea, F. oxysporum, Colletotrichum spp., R. solani and the oomycete P. capsici (Figure 1). Furthermore, strain CX03 showed a notable effect in controlling black rot on cabbage (Figure 2), while CX06 exhibited an obvious influence in controlling stem rot on Chinese cabbage (Figure 2). Lysobacter sp. was reported to produce siderophores which were associated with the biocontrol mechanisms. Such as L. enzymogenes strains C3 and LE16 were all reported to produce siderophores (de Bruijn et al., 2015; Chen et al., 2020). In the study, orange halos appeared around colonies of strains CX03 and CX06 on CAS agar plates, indicating that the two strains produced siderophores (Supplementary Figure 1). L. enzymogenes secreted a variety of extracellular hydrolases and were related to its antagonistic activity (Xie et al., 2012). In our study, enzyme activity assays showed that strains CX03 and CX06 produced chitinase (Supplementary Figure 1) and protease (Supplementary Figure 1).
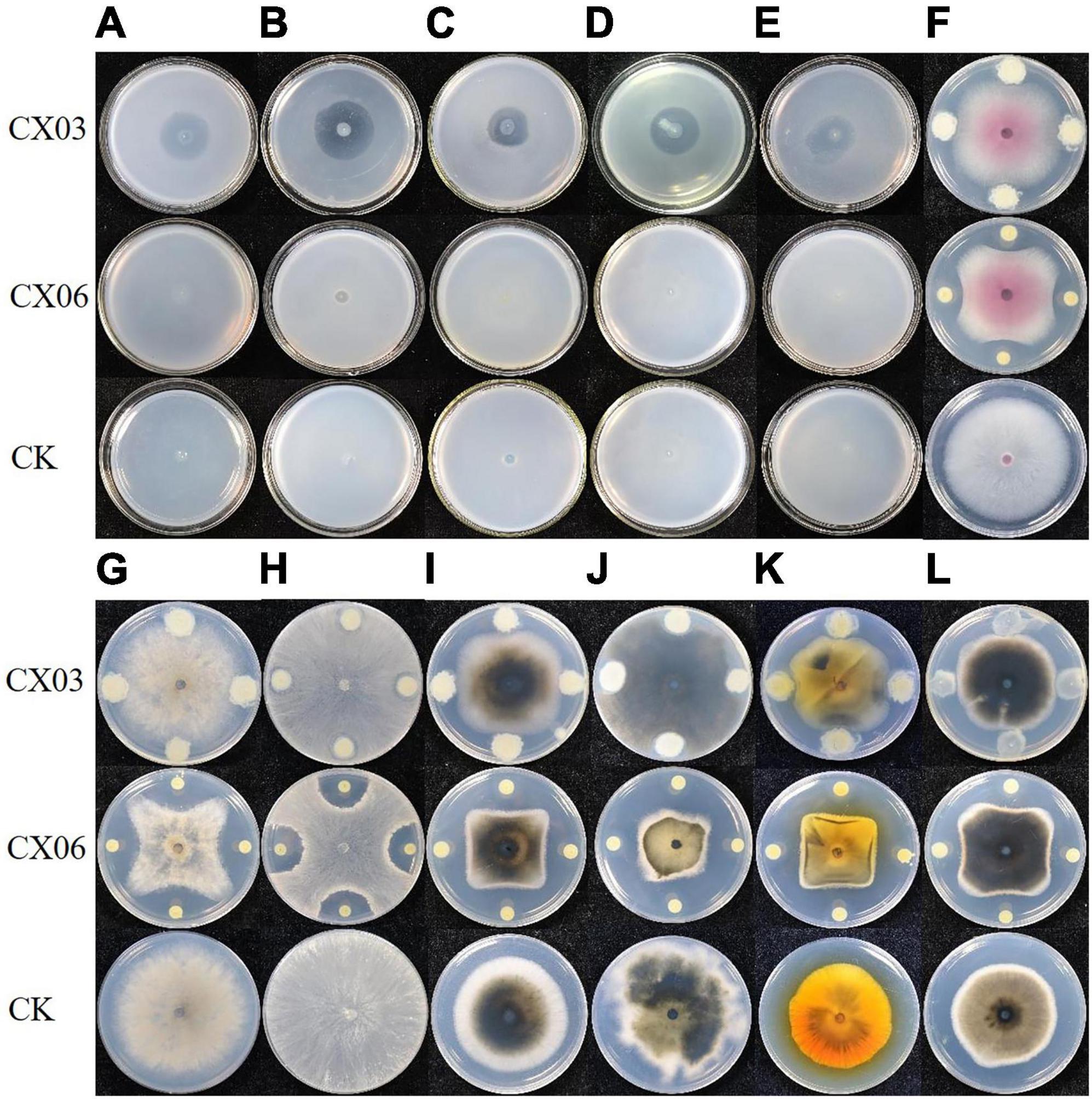
Figure 1. Antagonistic assay of strains CX03 and CX06 against various plant pathogens. (A) Clavibacter michiganensis subsp. sepedonicus, (B) C. michiganensis subsp. michiganensis; (C) Agrobacterium vitis, (D) Xanthomonas campestris pv. campestris, (E) Pseudomonas syringae pv. lachrymans, (F) Fusarium oxysporum, (G) Botrytis cinerea, (H) Rhizoctonia solani, (I) Corynespora cassiicola, (J) Diaporthe batatas, (K) Stemphylium solani, (L) Phytophthora capsica. CK, control.

Figure 2. Control efficiency of L. enzymogenes CX03 and CX06 against black rot on cabbage (A) and stem rot on Chinese cabbage (B). The white frames indicated the pathogenesis of stem rot on Chinese cabbage. Kasugamycin and validamycin were used as chemical control, respectively.
Organism information
The colonies of strains CX03 and CX06 were creamy-yellow, with irregular, flat to convex forms with undulated edges (Supplementary Figure 1). Strains CX03 and CX06 were measured as rod-shaped bacteria, and displayed gliding motility (Supplementary Figure 1). Strains CX03 and CX06 could grow in 1% NaCl (w/v), at an optimum temperature 28–30°C, and an optimum pH of 8. CX03 and CX06 were positive for citrate and gelatin hydrolysis tests. Biology assays showed that strain CX03 could utilize D-maltose, D-trehalose, D-fructose, D-galactose, and D-galacturonic acid, while CX06 could utilize D-maltose, gentiobiose, D-melibiose, D-mannose, and L-fucose. The growth curve showed that strains CX03 and CX06 were in the rapid growth stage between 16 and 20 h after inoculation, and reached a stable period at 32 h after incubation (OD600 = 2.8) (Supplementary Figure 2A). The pH of the fermentation broth of biocontrol agents may be continuously change during the fermentation. In our study, the pH of strains CX03 and CX06 fermentation rose approximately from 6.5 to 8.3 within 48 h of incubation and tended to be stable (Supplementary Figure 2B).
Comparison of CX03, CX06 and other sequenced Lysobacter strains
General genomic features of Lysobacter enzymogenes CX03 and CX06
The complete genome of L. enzymogenes CX03 comprised a 5,947,018 bp chromosome with an average G+C content of 69.67%, and the complete genome of L. enzymogenes CX06 comprised a circular 6,206,196 bp chromosome with an average G+C content of 69.91% (Supplementary Table 2). The genome structure and functions of strains CX03 and CX06 were shown in the graphical circular genome map (Figure 3). In total, 5,065 genes were predicted in the genome of strain CX03, including 4,957 protein coding genes, 69 RNA genes, and 39 pseudogenes. Using the KEGG, Swiss-Prot, GO, Pfam, NR, TCDB, PHI, and VFDB databases, 4.689 (92.58%), 1,631 (32.20%), 2,958 (58.40%), 2,958 (58.40%), 4,755 (93.88%), 319 (6.30%), 464 (9.20%), and 273 (5.40%) of the ORFs in the CX03 genome were classified into different groups, respectively (Supplementary Table 2). For strain CX06, 5,158 ORFs were predicted in the genome, including 5,059 protein coding genes, 70 RNA genes, and 29 pseudogenes. Similarly, according the KEGG, Swiss-Prot, GO, Pfam, NR, TCDB, PHI, and VFDB databases, 4.867 (94.36%), 1,642 (31.83%), 2,965 (57.48%), 2,965 (57.48%), 4,908 (95.15%), 323 (6.26%), 466 (9.03%), and 328 (6.36%) of the ORFs in CX06 genomes were classified into different groups, respectively (Supplementary Table 2).
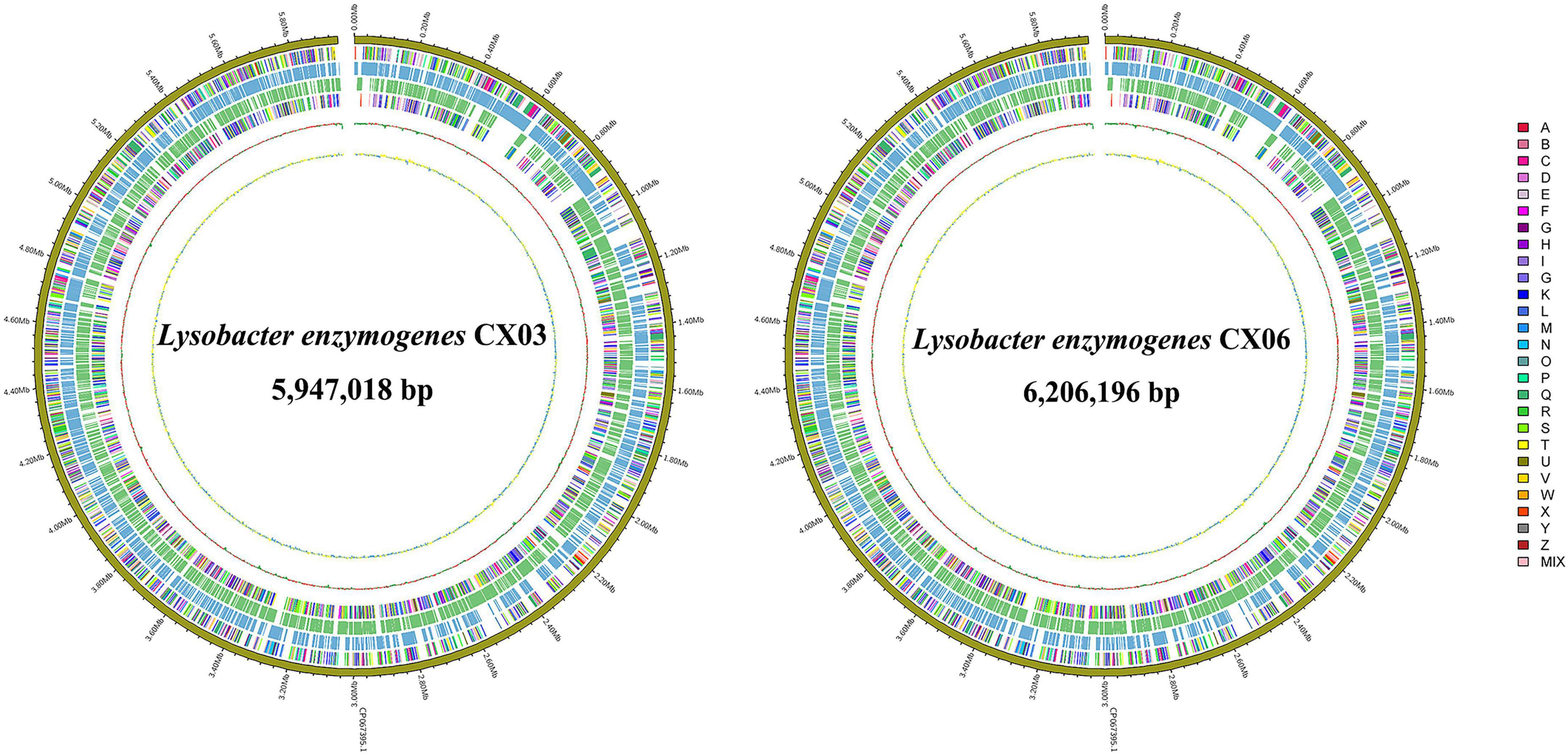
Figure 3. Graphical circular maps of the L. enzymogenes CX03 and CX06 chromosome genomes generated using the CGview server. From outside to center, ring 1 showed the genome sequence position, rings 2 and 5 showed protein-coding genes oriented in the forward (colored by COG categories) and reverse (colored by COG categories) directions, respectively. Rings 3 and 4 showed coding genes in the forward (blue) and reverse (green) directions, respectively. Ring 6 showed the G+C% content and the inner most ring showed the GC skews, where blue indicated positive values and yellow indicated negative values.
Meanwhile, 4,101 functional genes in the CX03 genome and 4,044 protein-coding genes in the CX06 genome were annotated to COG categories, including transcription, cell wall/membrane/envelope biogenesis, amino acid transport and metabolism, lipid transport and metabolism, translation, ribosomal structure and biogenesis (Supplementary Table 3). In addition, 895 genes in the CX03 genome and 1,044 genes in the CX06 genome were not annotated in the COG database, and the functions need to be further verified.
Comparison of the CX03 and CX06 genomes with other typical Lysobacter strains
Four publicly available typical Lysobacter strains with released complete genome sequences, L. enzymogenes M497-1, L. enzymogenes C3, L. capsici 55 and L. antibioticus 76 were selected for comparative genomic analysis. The results showed that the whole genome sizes of the six Lysobacter strains ranged from 5.92 to 6.39 Mb, with G+C contents of 66.60–69.90%, and the predicted CDSs ranged from 4,801 to 5,198. Moreover, the genome size of strain CX03 was larger than that of strain 76, but smaller than those of strains CX06, M497-1, C3 and 55. In addition, all the six Lysobacter strains contained one circular chromosome without a plasmid (Supplementary Table 4).
A phylogenetic tree was constructed according to four conserved genes (16S rRNA, gyrB, atpD and rpoB) to reveal the genetic relationships of L. enzymogenes CX03 and CX06 with other representative Lysobacter strains. The phylogenetic analysis indicated that the Lysobacter species could be clustered into two primary clades. The first clade consisted of three groups including L. enzymogenes, L. capsici, and L. antibioticus species, and strains CX03 and CX06 were clearly classified as L. enzymogenes (Supplementary Figure 3). Based on the observed distance relationships, the two strains CX03 and CX06 were clustered in different subclades of L. enzymogenes, and L. enzymogenes CX03 was closely related to L. enzymogenes ATCC 29487T, and M497-1, while L. enzymogenes CX06 was closely related to L. enzymogenes YC36, C3, and OH11.
Average nucleotide identity (ANI) and in silico DNA–DNA hybridization (DDH) were used to calculate the similarity of bacteria, and strains exhibiting ANI values ≥96% and DDH values ≥70% were usually regarded as the same species (Zhang et al., 2016). However, low ANI and DDH values were calculated between CX03 and other Lysobacter strains, even the L. enzymogenes strains (Supplementary Figure 4). The ANI values between CX03 and other L. enzymogenes strains CX06, M497-1, YC36, C3, and OH11 were approximately 88%, and the DDH values were approximately 35%. Lower ANI and DDH values were obtained when other Lysobacter species were regarded as references. Only the ANI and DDH values between L. enzymogenes CX06 and L. enzymogenes strains YC36, C3 exceeded 96% and 70%, respectively.
The whole genome sequences including L. enzymogenes strains CX03, CX06, M497-1, C3, OH11, L. capsici strains 55 and AZ78, L. antibioticus strains 76 and ATCC29479 were compared using Mauve. The alignments demonstrated that obvious local collinear block (LCB) inversion was detected among the five L. enzymogenes strains CX03, CX06, M497-1, C3, and OH11, but the CX06 genome was highly syntenic with M497-1. At the species level, more gene insertion or deletion occurred between L. enzymogenes strains CX03, CX06 and L. capsici strains 55, AZ78 or L. antibioticus strains 76, ATCC29479, which indicated the far genetic distances of the different Lysobacter species (Figures 4A–C).
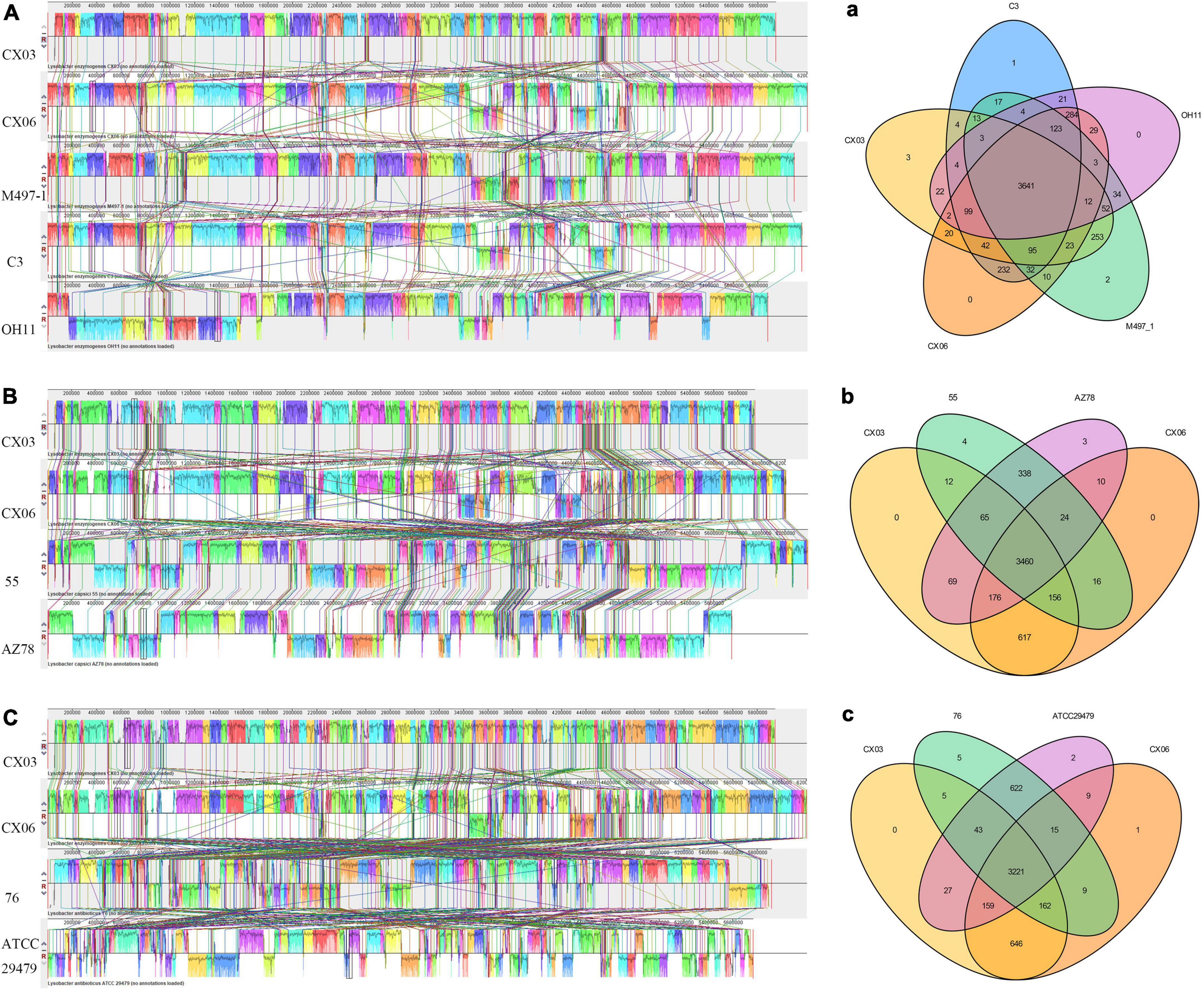
Figure 4. Global alignment of the genome sequences of completely sequenced L. enzymogenes CX03 and CX06 against other representative Lysobacter genome sequences. (A) Mauve progressive alignment of the genomes of CX03, CX06 and other representative L. enzymogenes strains M497-1, C3 and OH11. (B) Mauve progressive alignment of the genomes of CX03, CX06 and representative L. capsici strains 55 and AZ78. (C) Mauve progressive alignment of the genomes of CX03, CX06 and representative L. antibioticus 76, ATCC29479. Boxes with the same color indicate syntenic regions. Boxes below the horizontal strain line indicate inverted regions. Rearrangements are indicated with the colored lines. The scale is in nucleotides. (a) Venn diagram showing the numbers of shared and unique Clusters of Orthologous Genes among the genomes of CX03, CX06 and other representative L. enzymogenes strains M497-1, C3 and OH11. (b) Venn diagram showing the numbers of shared and unique Clusters of Orthologous Genes among the genomes of CX03, CX06 and the representative L. capsici strains 55 and AZ78. (c) Venn diagram showing the numbers of shared and unique Clusters of Orthologous Genes among the genomes of CX03, CX06 and the representative L. antibioticus strains 76, ATCC29479.
A Venn diagram indicating the specific genes among L. enzymogenes strains CX03, CX06, M497-1, C3, OH11, L. capsici strains 55, AZ78 and L. antibioticus strains 76, ATCC29479 was constructed (Figure 4). In total, 3,641 homologous genes existed in L. enzymogenes strains CX03, CX06, M497-1, C3, and OH11. CX03 shared 4,092 orthologous genes with M497-1, 3,934 orthologous genes with CX06, 3,901 orthologous genes with C3, and only 3,835 orthologous genes with OH11. Among the five L. enzymogenes strains, three unique genes were found in the CX03 genome, two unique genes were found in the M497-1 genome, one unique gene was found in the C3 genome, and no unique genes was observed in the genomes of strains CX06 and OH11. Moreover, 3,460 orthologous genes were found among the genomes of strains CX03, CX06, and L. capsici strains 55, AZ78. CX03 shared 3,693 orthologous genes with 55, and shared 3,770 orthologous genes with AZ78. Intriguingly, four unique genes were found in strain 55, three unique genes were found in strain AZ78, and no unique genes were found in strains CX03 and CX06. Furthermore, 3,221 orthologous genes were found among the genomes of strains CX03, CX06 and L. antibioticus strains 76, ATCC29479. CX03 shared 3,431 orthologous genes with 76, and shared 3,450 orthologous genes with ATCC29479. In addition, five unique genes were found in the 76 genome, two unique genes were found in the ATCC29479 genome, one unique gene was found in the CX06 genome, but no unique genes were found in the CX03 genome.
Functional genes related secondary metabolites and microbe–plant interactions
Gene clusters involved in the biosynthesis of antibiotics
To compare the gene clusters involved in antibiotic synthesis in L. enzymogenes CX03 and CX06, four Lysobacter strains including L. enzymogenes M497-1, L. enzymogenes C3, L. capsici 55, and L. antibioticus 76 were selected. For strain CX03, 11 gene clusters associated with secondary metabolite biosynthesis were detected, including six gene clusters encoding NRPSs (non-ribosomal peptide synthetases), two gene clusters encoding lanthipeptide, one gene cluster encoding arylpolyene, one gene cluster encoding RiPP-like and one redox-cofactor (Table 1). Moreover, 16 gene clusters linked with secondary metabolite biosynthesis were searched in the CX06 genome, including ten gene clusters encoding NRPSs, three gene clusters encoding lanthipeptide, one encoding RiPP-like protein, one arylpolyene and one redox-cofactor. According to the anti-SMASH database, seven secondary metabolites including xanthomonadin 1, Le-pyrrolopyrazines, lankacidin C, and four unknown antibiotics existed in strains CX03, CX06 and C3 with high homology (Supplementary Figure 5) (Table 1). Four secondary metabolites including thailanstatin A and three unknown antibiotics were detected in strain CX03, but not in strain CX06 (Figure 5A). Nine secondary metabolites including HSAF, WAP-8294A2, chlorotonil A, BE-43547A1, and three unknown antibiotics existed in strain CX06, but not in strain CX03 (Figure 5B). Interestingly, the gene cluster encoding thailanstatin A was detected only in the CX03 genome, not in the genomes of strains CX06, C3, M497-1, 55, 76 (Table 1). An unknown secondary metabolite with core coding genes entF and entE was found in the genomes of L. enzymogenes strains CX03, M497-1 and L. antibioticus 76, but not in L. enzymogenes strains CX06, C3 and L. capsici 55. Another unknown secondary metabolite with the core coding gene lanKC was only detected in L. enzymogenes CX03 and L. capsica 55, but not in other Lysobacter strains (Figure 5A). In addition, many gene clusters associated with the biosynthesis of different secondary metabolites were only found in L. enzymogenes strains, not in L. antibioticus or L. capsici strains, such as Le-pyrrolopyrazines, thailanstatin A, WAP-8294A2, chlorotonil A, BE-43547A1 and four unknown antibiotics (Figure 5 and Supplementary Figure 5). Notably, two secondary metabolites (HSAF and WAP-8294A2) that have been widely studied were not detected in the genomes of L. enzymogenes CX03 and L. antibioticus 76 (Figure 5).
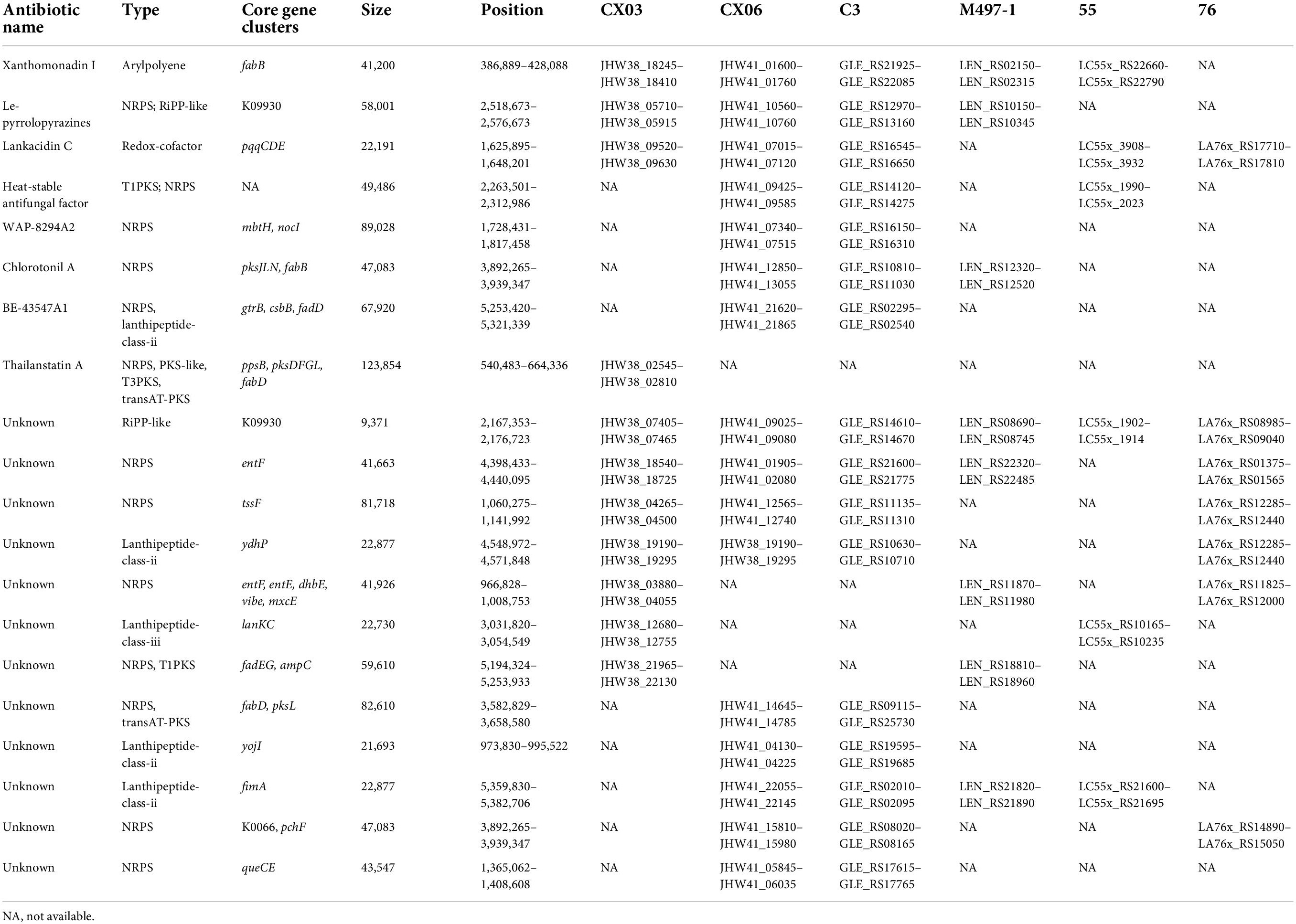
Table 1. Comparison of core gene clusters involved in secondary metabolites biosynthesis in strains CX03, CX06, MA97-1, C3, 55 and 76.
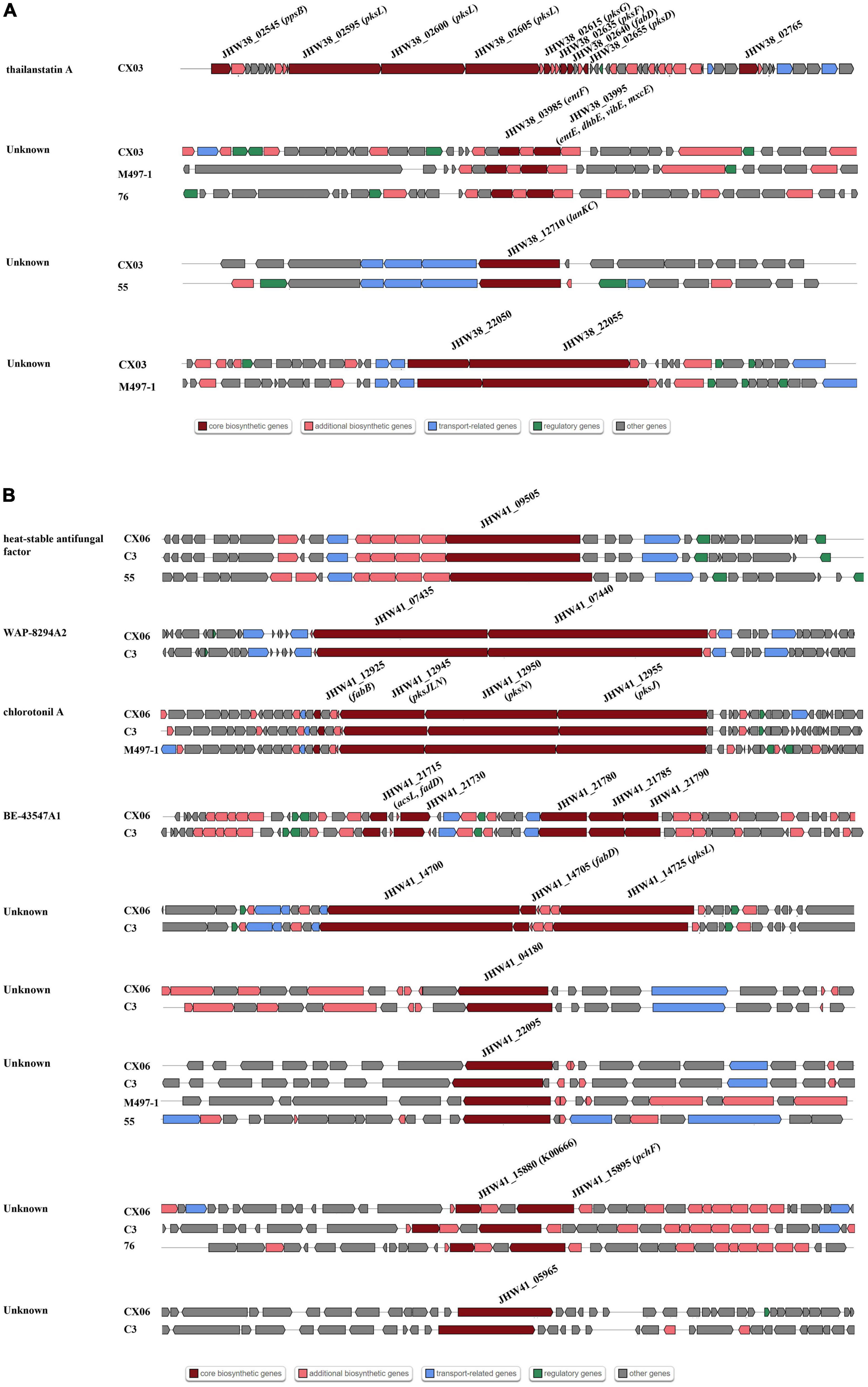
Figure 5. Comparisons of gene clusters for secondary metabolites synthesis with other representative Lysobacter strains C3, M497-1, 55 and 76. (A) Gene clusters existed in strain CX03, not in strain CX06. (B) Gene clusters existed in strain CX06, not in strain CX03. Dark red indicated the core biosynthetic genes in different gene clusters. The core biosynthesis genes were marked in the diverse gene clusters.
Bacterial secretion system
Comparative genomic analysis of the bacterial secretion systems among strains CX03, CX06, M497-1, C3, 55 and 76 showed that L. enzymogenes CX03 harbored a highly conserved T2SS (type II secretion system) gene cluster (gspDEFGHIJKLM) (Supplementary Figure 6), covering 11,589 bp with 10 ORFs. All the core genes were found in Lysobacter strains CX06, M497-1, C3, 55 and 76, and shared a high consistency at the amino acid level (exceeding 80%) (Supplementary Table 5).
A large sct/hrp gene cluster of ten genes (sctJLQSTUV, yscN, hrpAB) was searched in the genome of L. enzymogenes CX03, and shared high similarities with other Lysobacter strains (exceeding 80%), except the gene sctQ encoding cytoplasmic ring protein SctQ of the type III secretion system, which showed a low sequence identity with strains 55 and 76 (74% and 64%, respectively) (Supplementary Table 5 and Supplementary Figure 6).
Moreover, ten key genes related to T4SS (virB1,2,3,4,6,8,9,10,11 and virD4) were all found in strains CX03, CX06, M497-1, C3, 55 and 76 (Supplementary Figure 6). However, only four genes (virB4, virB9, virB11, and virD4) retrieved from the CX03 genome shared a high similarity (identity > 80%) with other four Lysobacter strains, while other genes shared a low sequence identity (below <80%). In particular, the sequence identity of the virB6 gene was less than 70% between CX03 and M497-1, C3, 55, 76, and was only 37% between CX03 and CX06 (Supplementary Table 5).
T6SS widely existed in many Gram-negative bacteria. In this study, 28 genes linked with the T6SS were only found in the genomes of strains CX06 and C3, not in CX03, M407-1, 55 and 76 (Supplementary Figure 6). The 28 genes were detected to encode different putative proteins, including 12 core genes (tssABCHEFGMLK, tagFH) for type VI secretion system proteins, and two genes (vgrG and hcp) related to extracellular structural components of the secretion machine and specific effectors. Furthermore, most T6SS genes were multicopy in strains CX06 and C3 (Supplementary Table 5).
In addition, 11 genes associated with the Sec system were detected in the CX03 genome, and possessed high sequence identities with those of the other five Lysobacter strains, except for the secG gene, which only showed 66% and 63% identities between CX03 and strains 55, 76. Three genes (tolC, hlyB and hlyD) associated with the T1SS were found in the genome of CX03, but the genes hylB and hlyD were absent in the genomes of CX06, C3 and 55. Three genes (tatABC) involved in twin arginine targeting were all present in the genomes of CX03, CX06, M497-1, C3, 55 and 76; however the identities of the corresponding genes between CX03 and other L. enzymogenes strains were higher than those between CX03 and L. capsici 55, L. antibioticus 76 strains (Supplementary Table 5 and Supplementary Figure 6).
Flagella, type IV pilus and chemotaxis
The comparative genomic analysis showed that sixteen key genes (mainly the fli and flg gene clusters) associated with flagella biosynthesis were searched in the L. enzymogenes CX03 genome. Most genes encoding flagella-associated proteins were also detected in L. enzymogenes CX06, M497-1, C3 and L. capsici 55, while only three genes (fliH, fliI and fliO) were detected in the genome of L. antibioticus 76, and the gene flgD was not detected in the L. enzymogenes CX06 genome. The identities of these genes between CX03 and CX06, M497-1, and C3 were higher than those between CX03 and 55, 76. Most genes detected in the CX03 genome shared higher amino acid similarities with L. enzymogenes CX06 and C3, but showed a low similarity with that in M497-1. Only one gene fliI shared a high sequence identity (exceeding 90%) among the Lysobacter strains CX03, CX06, M497-1, C3, 55 and 76 (Supplementary Table 6). In addition, 15 genes (pilWECVGABDUMOPQT and fimT) involved in type IV pilus were found in the genomes of strains CX03, CX06, MA97-1, C3, 55 and 76 (Supplementary Table 6). Most of these genes from strain CX03 shared high homology with those in other Lysobacter stains at the amino acid level (identities > 80%). However, the identities of pilE, pilC, pilV, pilA and fimT genes between CX03 and other Lysobacter strains were less than 70%. Similarly, pilW in strain CX03 shared higher identities with L. enzymogenes strains CX06, M497-1 and C3, but showed a low similarity with L. capsici 55 and L. antibioticus 76. Moreover, 7 genes (cheW, pilJ, pilI, cheB, wspD, wspB, wspA) involved in chemotaxis were sought in the genome of L. enzymogenes CX03, and shared a high homology with the other three L. enzymogenes strains (>80%) at the amino acid level. However, the wspD gene encoding purine-binding chemotaxis protein CheW retrieved from the genome of strain CX03 did not exist in strain CX06, and exhibited very low identities with those in strains 55 and 76 (Supplementary Table 6).
Two-component system
In addition, 25 TCSs (two-component system) were searched in the genome of strain CX03, most of which were highly similar to those of strains CX06, M497-1, C3, 55 and 76. Based on the topological characteristics of sensor histidine kinase (HK) and response regulator (RR), the 31 TCSs were assigned to diverse subfamilies. For, example, phosphate regulation TCSs such as phoR/B and phoQ/P were detected in the genomes of strains CX03, CX06, M497-1, C3, 55 and 76, with amino acid sequence identities exceeding 93% (Supplementary Table 7). The TCSs kdpD/E, algR/Z, and pilH/G retrieved from CX03 shared high similarity with those in the other five Lysobacter strains (>90 identity). Nevertheless, the identities of wspA/B, wspD/E and wspF/R which were associated with chemotaxis between CX03 and CX06, M497-1, and C3 (>80%), were higher than those between CX03 and 55, 76 (<70%) (Supplementary Table 7). Interestingly, the rpf C/F TCS in strain CX03 owned a high similarity with those in strains CX06 and C3 (92% and 91%, respectively), but exhibited a low identity to strain M497-1 (47% and 48%, respectively). Furthermore, the mdt TCS was only found in the genomes of L. enzymogenes strains CX03 and M497-1 with the sequence identities exceeding 80% and most were multicopy genes. Although two genes mdtAC were also found in the genome of L. capsici 55, their identities with those in L. enzymogenes CX03 at the amino acid level were very low (41% and 77%, respectively). The rcs TCSs were not detected in strains L. enzymogenes CX06 and L. antibioticus 76, and low homology of genes rcsC and rcsB was found between CX03 and C3, M497-1, 55 strains, especially the sequence identity of rcsC gene was less than 80% (Supplementary Table 7).
Quorum sensing
The comparative genomic analysis showed that 40 genes associated with quorum sensing (QS) were retrieved in the CX03 genome. In this study, the key gene expR, which was related to N-acyl homoserine lactones (AHLs), was observed in strain CX03 and shared a high homology with other Lysobacter strains. In addition, the QS gene clusters, tox, rpf, phn and the gene hfq were highly conserved among strains CX03, CX06, and the other four Lysobacter strains (Supplementary Table 8). However, many genes shared a higher sequence identity between CX03 and CX06, M497-1, C3 than that between CX03 and 55, 76 (Supplementary Table 8).
Selenium metabolism
In this study, L. enzymogenes CX03 and CX06 were tested to generate elemental Se nanoparticles (SeNPs) from Na2SeO3. The highest concentration of Na2SeO3 tolerated for strains CX03 and CX06 was 75 mM, and the production of SeNPs achieved a maximum when the concentration of Na2SeO3 was 5 mM (Supplementary Figure 7). Whole genome annotation demonstrated that many genes linked with selenium metabolism were found in the genomes of L. enzymogenes CX03 and CX06, and shared high similarity with Lysobacter strains M497-1, C3, 55 and 76. Among these genes, eight genes (metB, metC, metE, metG, metH, mdeA, and trxB) were related to selenium association, and two genes (cysN and cysD) were involved in selenium transportation. However, the identities of these genes between CX03 and L. enzymogenes strains CX06, M497-1, C3 (90–98%) were higher than those between CX03 and L. capsici 55, L. antibioticus 76 (83–96%) (Supplementary Table 9).
Biosynthesis of siderophores
In addition, the result showed that eight key genes (entACDFE, dhbA, mcyF, and racD) associated with the biosynthesis of siderophore group non-ribosomal peptides were found in the genomes of strains CX03, CX06, M497-1, C3, 55 and 76. Most genes in strain CX03 shared high sequence identities (exceeding 70%) between strains CX06, M497-1, C3 at the amino acid level, except the gene entD encoding 4′-phosphopantetheinyl, which showed low homology between CX03 and CX06, M497-1, C3, 55 and 76 (<50% identity) (Supplementary Table 10).
Polyketone, lipopolysaccharide, peptidoglycan biosynthesis
Moreover, 42 putative protein coding genes involved in the biosynthesis of polyketone, lipopolysaccharide and peptidoglycan were searched in the genome of L. enzymogenes CX03 and CX06, including four genes (rfbABCD) related to polyketide sugar unit biosynthesis, 17 genes (mainly the lpx gene cluster) related to lipopolysaccharide biosynthesis, 19 genes (mainly the mur gene cluster) related to peptidoglycan biosynthesis, and two genes proA, proB related to carbapenem biosynthesis. All these genes possessed high homology with those of the other five Lysobacter strains at the amino acid level, except the gene rfbD, which showed low homology between CX03 and 55, 76 (69% and 71% identities, respectively) (Supplementary Table 11).
Discussion
Lysobacter enzymogenes has been widely reported to have antagonistic abilities against diverse plant-pathogenic fungi due to its ability to produce various antifungal compounds and multiple lytic enzymes. In this study, two L. enzymogenes strains exhibited distinct antipathogenic activities; strain CX03 showed broad-spectrum antagonistic capacities against various plant-pathogenic bacteria, while strain CX06 showed broad-spectrum inhibiting abilities toward multiple plant-pathogenic fungi. To explore the possible biocontrol mechanisms for the different antipathogenic capacities of the two L. enzymogenes strains, especially the potential biocontrol mechanism of strain CX03 inhibiting bacteria, the complete genomes of the two strains were sequenced and comparative genomic analysis among different Lysobacter strains was performed.
Based on the phylogenetic analysis, L. enzymogenes strains clustered into two subgroups indicating the high diversity of the species, which was consistent with a previous study (Gómez Expósito et al., 2015). ANI and DDH were widely used to calculate the similarity of bacteria, however, the ANI and DDH values between strain CX03 and other L. enzymogenes strains CX06, M497-1, YC36, C3, and OH11 were below 95% and 70%, respectively. Only the ANI and DDH values between CX06 and L. enzymogenes strains YC36, C3 were greater than 95% and 70%, respectively. Even though the ANI and DDH values between strains M497-1 and YC36, C3, OH11 which had been classified as L. enzymogenes, were still below 95% and 70%, respectively. These results may indicate that ANI and DDH was not suitable for the classification in Lysobacter strains. Pairwise alignment of whole genomes played an important role in the diversity analysis of lineages in different species, especially for the evolution of species and subspecies (Nowell et al., 2014). Thus, the obvious gene transfer or large local collinear block inversion between CX03, CX06 and 55, AZ78, 76, ATCC29479 indicated that these strains belonged to different Lysobacter spp., supporting the phylogenetic result that these strains were in different clades. In addition, more orthologous genes were detected between CX03, CX06 and L. enzymogenes strains than between CX03, CX06 and L. capsici or L. antibioticus strains, which proved the above result of the phylogenetic analysis that strains CX03 and CX06 were classified as L. enzymogenes, followed by L. antibioticus strains, L. capsici strains successively.
Lysobacter was a bacterial genus known to generate a number of antibiotics (Hashizume et al., 2016). However, the antimicrobial activities of different Lysobacter spp. were significantly different. Previous studies showed that most L. enzymogenes, L. antibiotics, L. capsica and L. gummosus strains had strong antagonistic abilities toward many fungi and oomycetes, but only a few L. antibioticus strains exhibited antagonistic activity against the bacteria X. campestris pv. campestris (Gómez Expósito et al., 2015). However, in our study, L. enzymogenes CX03 showed broad-spectrum antagonistic capacities toward plant-pathogenic bacteria and an obvious influence in controlling black rot on cabbage. In fact, most reported secondary metabolites synthesized by L. enzymogenes were reported to show antagonistic activities against plant fungal pathogen. L. enzymogenes C3 was demonstrated to suppress diseases caused by multiple fungal pathogens, including B. sorokiniana (Zhang and Yuen, 1999), F. graminearum (Jin et al., 2003), R. solani (Giesler and Yuen, 1998), U. appendiculatus (Yuen et al., 2001), P. ultimum (Kobayashi et al., 2005), and Magnaporthe poae (Kobayashi and Yuen, 2005). The main antifungal factor in strain C3 was HSAF, which had a novel structural features and inhibited against many fungal species (Yu et al., 2007). HSAF could disrupt the polarized growth of fungi by targeting the biosynthesis of sphingolipids, which were essential components of eukaryotic cell membranes and signaling molecules associated with numerous cellular process (Li et al., 2006). Genes related to hybrid polyketide synthase-non-ribosomal peptide synthetase (PKS-NRPS) were reported as key genes for the production of HSAF. The destruction of the PKS-NRPS gene in strain C3 lost the capacity to produce HSAF and reduce the ability to restrain fungal growth (Yu et al., 2007; Li et al., 2012). HSAF production was also proved to be a crucial mechanism to control cyst nematodes in L. enzymogenes C3 (Yuen et al., 2018). The comparative genomic analysis showed that the genes and gene clusters involved in secondary metabolite synthesis were significantly different among the six Lysobacter strains. As expected, a gene cluster for the biosynthesis of HSAF was found in the CX06 and C3 genomes, but not in CX03 genome, corresponding to the antifungal activity of L. enzymogenes CX06. The WAP-8294A compound was reported to antagonize Gram-positive organisms, and the NRPSs comprised 45 functional domains, which were related to the assembly of the 12 modules of the WAP-8294A antibiotic (Chen et al., 2015). L. enzymogenes OH11 was reported to produce WAP-8294A2, a cyclic lipodepsipeptide with potent anti-S. aureus ability (Zhang et al., 2011). However, very few studies reported its antagonistic activity against plant pathogenic bacteria. The WAP biosynthetic gene cluster in the genome of L. enzymogenes OH11 was identified with two large NRPS genes, WAPS1 and WAPS2 (Chen et al., 2015), and further knockdown of ORF3, ORF4, and ORF8 genes reduced the antagonistic ability of the mutant strain against Gram-positive bacteria (Zhang et al., 2011). L. enzymogenes CX03 showed antagonistic activities against C. michiganensis, but no genes or gene clusters responsible for the biosynthesis of WAP-8294A2 were found in the genome of strain CX03. Moreover, although genes or gene clusters associated with WAP-8294A2 were found in the CX06 genome, strain CX06 did not show antagonistic activity against plant pathogenic bacteria. These results might indicate that the inhibitory on C. michiganensis of L. enzymogenes CX03 was not related to WAP-8294A2. The antiSMASH analysis showed that four secondary metabolites (thailanstatin A and three unknown compounds) were found in strain CX03, not in strain CX06, especially thailanstatin A, which was only detected in strain CX03 and not in other Lysobacter strains. Thailanstatin A (TST-A) was first reported as an effective antiproliferative natural product generated from Burkholderia thailandensis MSMB43 (Liu et al., 2016). Thailanstatins were biosynthesized by a hybrid polyketide synthase-non-ribosomal peptide synthetase pathway, and the biosynthetic gene cluster was similar to that of FR901464, a prototype spliceosome inhibitor produced by Pseudomonas sp. 2663. It was also proven to inhibit pre-mRNA splicing (a new mechanism of anti-neoplastic action) as potently as FR901464 (Liu et al., 2013). Moreover, several pks genes involved in the biosynthesis of bacillaene polyketides were found in the genome of strain CX03. Polyketides were reported as a large family of secondary metabolites with antibacterial, immunosuppressive and antitumor activities (Chen et al., 2006). Interestingly, only strain CX03 exhibited direct antagonistic activities against many bacteria, while the other L. enzymogenes strains CX06, C3 and M497-1 did not exhibit antibacterial activities. Thus, thailanstatin A may be an unusual antibiotic that showed antibacterial activities, though the specific biological function of thailanstatin A remained to be confirmed. In addition, a core gene lanKC related to unknown secondary metabolite encoding class III lanthionine synthetase was found in the genome of strain CX03. Previous study showed that lanthionine synthetase complex was involved in the modification and transport of lantibiotics which displayed potent antimicrobial activity (Lu et al., 2012). Notably, phenazine antibiotics produced by L. antibioticus were reported to have antagonistic activities against several bacteria (Puopolo et al., 2014), however, no definite genes or gene clusters associated with the biosynthesis of phenazine were retrieved from the genome of L. enzymogenes CX03.
In addition, five other secondary metabolites (Chlorotonil A, BE-43547A1 and five unknown antibiotics) were found in strain CX06, but not in strain CX03. Chlorotonil A, a tricyclic macrolide generated by S. cellulosum, was first described in 2008 (Gerth et al., 2008), and was tested to exhibit possible antimalarial activity (Held et al., 2014). BE-43547A1 was isolated as a member of seven macrocyclic dipeptides families from Streptomyces strain A43547 in 1998 with different congeners A1, A2, B1, B2, C1, and C2. The BE-4357 members were found to exhibit obvious hypoxia-selective growth-inhibitory ability toward pancreatic cancer cells (Villadsen et al., 2017), and had structural similarities to the cytotoxic natural products rakicidins A and B (McBrien et al., 1995). Given the potential fact that HSAF was the main antibiotic for antagonizing plant pathogenic fungi, the specific functions of these five secondary metabolites need to be further verified.
Furthermore, seven secondary metabolites (xanthomonadin I, Le-pyrrolopyrazines, Lankacidin C and four unknown compounds) were found in strains CX03 and CX06. Xanthomonadin was first reported in Xanthomonas, and was unimportant during pathogenesis (Rajagopal et al., 1997). To date, the function of xanthomonadin was unknown, but it was proven to provide protection against photodynamic lipid peroxidation in liposomes and could be used for protection against photodamage (Poplawsky et al., 2000). Lankacidin C, also named bundling A or T-2636 C, was described as an antitumor antibiotic that was isolated independently from diverse Streptomyces species. Lankacidins have been reported to have powerful antimicrobial abilities against many Gram-positive bacteria, including some strains resistant to conventional macrolide antibiotics (Kende et al., 1995). Given that CX03 and CX06 showed distinct antagonistic activities toward plant pathogens, the seven secondary metabolites played dispensable roles in antimicrobial activity. Worth mentioning, many gene clusters related to unknown secondary metabolites were identified in the L. enzymogenes genomes, and these secondary metabolites could be a potential resource for novel antibiotics; however, in-depth chemical and functional analyses are needed to verify this possibility.
Siderophores, which were mostly encoded by NRPS gene clusters, have been shown to play a crucial role in iron competition with other microorganisms, and could be synthesized by Lysobacter strains. For example, L. enzymogenes C3 exhibited iron-chelating ability on CAS plate, and the iron availability was related to the antibacterial capacity (de Bruijn et al., 2015). The potential biocontrol agent L. enzymogenes LE16, which showed broad antagonistic activities against many pathogenic bacteria, could produce siderophores, proteases, and phosphatases in pure culture (Chen et al., 2020). L. capsica AZ78, which has considerable potential for the biocontrol of phytopathogenic microorganisms, harbored specific genes involved in the production of siderophores (Gerardo et al., 2016). L. antibioticus HS124, which showed antifungal activity against P. capsici was reported to produce catechol type siderophores, and the antifungal ability when Fe (III) was added was approximately 1.5 times higher than that in the absence of Fe (III) (Ko et al., 2011). Many siderophores have been widely described, such as enterobactin, a triscatechol derivative of a cyclic triserine lactone, which was achieved by non-ribosomal peptide synthetases (Raymond et al., 2003). Previous studies showed that six Ent proteins, EntA-F were related to the synthesis of enterobactin. For instance, EntF, an N-terminal elongation/condensation domain was reported as a catalyst in enterobactin assembly. EntD was known as a posttranslational modification catalyst and the EntD-mediated conversion of apo-forms of EntB and EntF to phosphopantetheinylated played important roles in the acyl activation and transfer of enterobactin assembly (Gehring et al., 1998). Pyochelin was described as a type of thiazolines siderophore which was also assembled by NRPS. Among the two gene clusters pchEF and pchDCBA which related to the pyochelin assembly, pchF was required for production of the entire pyochelin molecule (Quadri et al., 1999). In this study, CX03 and CX06 could produce siderophores on CAS agar plates, and ent gene cluster related to the production of enterobactin were found in the CX03 and CX06 genomes. So, enterobactin might be one of the siderophores produced by strains CX03 and CX06. In addition, the pchF gene was also found in strain CX06, considering its important role in the siderophore assembly, pyochelin might be another siderophore produced by strain CX06. However, these guesses need further experiments to verify.
Lysobacter members were well-known for their ability to produce a variety of extracellular enzymes, such as chitinases, elastases, glucanases, endonucleases, proteases and lipases (Zhang et al., 2001; Gökçen et al., 2014; Gómez Expósito et al., 2015). Chitinolytic activity has been proven to be one of the mechanisms related to biological control in L. enzymogenes. For example, the chitinolytic in L. enzymogenes C3 could cause conidial deformation and abnormal germ tube formation in fungi (Zhang and Yuen, 2000). L. enzymogenes C3 was reported to generate three extracellular β-1,3-glucanases, which were encoded by the gluABC genes, and the inhibiting ability of strain C3 against Pythium damping-off and Bipolaris leaf spot decreased when the three genes were deleted (Palumbo et al., 2005). Proteolytic ability in Lysobacter strains has attracted most attention for its potential application in industrial processes (Gökçen et al., 2014) and was reported to inhibit phytopathogenic bacteria such as Erwinia carotovora (Vasilyeva et al., 2014). In this study, strains CX03 and CX06 were tested to produce cellulase and phosphatases, which might indicate their potential applications in biological control.
Previous studies have shown that various secretion systems exist in Gram-negative bacteria, and most have been identified in Lysobacter strains (Tseng et al., 2009). The secretion systems ranged from single transporters to multi-component complexes and were divided into six types in Gram-negative bacteria, type I to type VI secretion systems (Costa et al., 2015). T2SS was well-conserved and harbored 12–15 components calling Gsp proteins (GspA to GspO and GspS), which was crucial for bacterial survival and growth in a host or in an environment niche (Filloux, 2004; Korotkov et al., 2012). T2SS in Lysobacter genomes was reported to secrete cell wall degrading enzymes (Cianciotto, 2005), which may be responsible for the behavior of Lysobacter strains to lyse the cells of many microorganisms (Christensen and Cook, 1978). In this study, CX03 and CX06 harbored 10 gsp genes encoding T2SS components, which were highly conserved in Lysobacter strains; thus, Gsps may be linked to protein secretion and transportation, but the role of T2SS in L. enzymogenes CX03 and CX06 needs to be confirmed in the future.
The type III secretion system (T3SS) had been reported to translocate bacterial proteins into host cells (Galan and Wolf-Watz, 2006). T3SS was found in various pathogenic Gram-negative bacteria such as Salmonella, Shigella, Yersinia and Pseudomonas, and was responsible for protein transportation across the inner bacterial membrane (Dale et al., 2002; Ghosh, 2004). Recent studies have shown that the distribution of T3SS was not limited to pathogens, but also existed in certain endosymbiotic bacteria or biocontrol bacteria (Dale et al., 2001). For example, L. enzymogenes C3, a bacterium demonstrated earlier to be used in biological control of fungi, was identified to contain a T3SS, which was an essential part in bacterial-fungal interactions (Reedy, 2004).
T4SS was characterized as a multiprotein complex that may deliver DNA, effectors and protein-DNA complexes to the extracellular milieu or into the eukaryotic and prokaryotic target cells (Sgro et al., 2019). The VirB/D4 T4SS in S. maltophilia and X. citri were found to be associated with the transfer of effectors lethal to bacterial competitors, playing important roles in hosts and environmental colonization (Souza et al., 2015; Bayer-Santos et al., 2019). A recent study showed that L. enzymogenes OH11 employed T4SS as the main contact dependent weapon to suppress other soilborne bacteria. The T4SS-mediated killing behavior of strain OH11 was proven to be essential to inhibit the plant pathogenic bacterium P. carotovorum (Shen et al., 2021). In this study, the virB/D T4SS was present in the conserved genetic organization in L. enzymogenes strains CX03, CX06, M497-1 and C3, which indicated that the T4SS may play a vital role in the antagonistic activities of the bacterium; however, further studies need to explore the function of T4SS in L. enzymogenes.
T6SS was functionally defined in 2006, and existed in various Gram-negative bacteria (Pukatzki et al., 2006); it has been reported to be cell envelope spanning machine that can translocate effector proteins into eukaryotic and prokaryotic cells (Ho et al., 2014). T6SS consisted of 13 conserved core genes (mainly tss and tag gene clusters) and several accessory genes including the imp gene cluster, and multiple copies genes vgrG and hcp, which were hypothesized to be minimally necessary for function and the conserved genes varied in different species (Chang et al., 2014). In this study, T6SS was only detected in L. enzymogenes strains CX06 and C3, not in strains CX03, M497-1, 55 and 76, which was consistent with the previous report that genes encoding T6SS were only searched in L. enzymogenes strain C3 and L. gummosus strain 3.2.11 (de Bruijn et al., 2015). The variation in secretion system may reveal that T6SS corresponded to the diversity of different Lysobacter strains, although the specific function needs further confirmation.
Selenium was a significant element in environmental remediation and human and animal health; most beneficial microorganisms have been reported to produce extracellular selenium, such as Bacillus and Rahnella strains (Zhang et al., 2018). R. aquatilis ZF7 and HX2 were reported to produce elemental Se nanoparticles (SeNPs) (Yuan et al., 2020), and the potential beneficial bacterium R. aceris ZF458 harbored abundant genes involved in selenium metabolism (Xu et al., 2022). The cys and met gene clusters were reported to be closely related to selenium biosynthesis (Zhu et al., 2018). In this study, L. enzymogenes CX03 and CX06 showed strong ability to produce SeNPs from Na2SeO3; many genes involved in selenium metabolism were detected in the genomes of strains CX03 and CX06, which shared a high similarity with those in the genomes of other Lysobacter strains, and most homologous genes were also found in the R. aceris ZF458 genome (Xu et al., 2022). Considering the ability to synthesize SeNPs, L. enzymogenes CX03 and CX06 will have wide application prospects in nano-agriculture.
Lysobacter was reported as non-motile cells, with gliding motility or twitching motility replacing flagellar motility as a common form of movement in Lysobacter strains (Siddiqi and Im, 2016). For example, L. capsica strains L31 and L. enzymogenes strains L30 showed no motility after 4 days of incubation on soft SSM agar medium, while these strains spread from the point of inoculation, similar to gliding motility after 12 days of incubation (Gómez Expósito et al., 2015). Previous studies showed that no genes related to flagella synthesis were present in the genomes of L. antibioticus strains, and flagellar biosynthesis in many Lysobacter strains was non-functional due to the lack of genes encoding essential flagellar components (de Bruijn et al., 2015; Tomada et al., 2016). For example, L. capsica AZ78 did not contain genes for flagellar filaments, such as fliC, leading to the phenotypic description that the strain was non-motile (Park et al., 2008). In addition, only genes associated with the components of the flagellar apparatus were detected in L. enzymogenes C3, and the genes have been proven non-functional previously (Hayashi et al., 2001). In fact, L. enzymogenes strains exhibited a twitching behavior that was powered by type IV pilus (T4P) (Fulano et al., 2020a). For example, in the model strain L. enzymogenes OH11, many pilus structural component proteins including the major pilus subunit PilA, the motor proteins PilB and the outer membrane secretin PilQ were essential for the biogenesis of T4P and the function of twitching motility (Xia et al., 2018). Other minor pilins such as PiLVWE were also reported to play important roles in the initiation of pilus assembly, and PilD was a bi-functional enzyme which performed the N-terminal methylation of mature pilins (Nguyen et al., 2015). In addition, several flagellar type III secretion system (FT3SS) components including FlhA, FlhB, FliI, and FliR in L. enzymogenes OH11 were reported to acquire a distinct function to control T4P-driven twitching motility (Fulano et al., 2020b). In this study, flagellar apparatus genes and many T4P related genes were found in strains CX03 and CX06, which might reveal that twitching motility was the main form of movement in strains CX03 and CX06.
Previous studies showed that the biosynthesis of many antibiotic compounds in Lysobacter strains was regulated by intercellular signaling or quorum sensing mediated by the diffusible signal factor (DSF)-dependent system and the Clp (cyclic AMP receptor (CRP)-like protein) regulator (He and Zhang, 2008; Deng et al., 2011). For example, the small molecule metabolite (LeDSF3) in L. enzymogenes OH11 was proven to regulate the biosynthesis of HSAF. Furthermore, LeDSF3 upregulated the expression of the global regulator Clp, and the knock-out of clp reduced HSAF production (Han et al., 2015). WspR was reported as an important diguanylate cyclase (DGC) involved in HSAF regulation in L. enzymogenes OH11. Phosphorylation activated DGC activity of WspR and weakened the WspR-CdgL interaction, thus contributing to the accumulation of the cdi-GMP-bound CdgL, which in turn decreased HSAF biosynthesis operon transcription (Lin et al., 2021). In addition, LeDSF3-regulated HSAF production was dependent on the two-component regulatory system RpfC/RpfG, which has been served as the sensor/response regulator of DSF (Slater et al., 2000). Besides, DSF and diffusible factor (DF), were two chemically distinct autoinducers (Barber et al., 1997), and both Rpf/DSF and DF signaling systems were related to the modulation of HSAF biosynthesis in L. enzymogenes (Qian et al., 2013). Furthermore, Clp signaling in L. enzymogenes OH11 played a positive role in regulating the biosynthesis of HSAF and WAP-8294A2 (Wang et al., 2014), and the mutation of the clp gene in L. enzymogenes C3 reduced the antimicrobial activity in vitro, and the control efficiency to pythium damping-off in sugar beet (Kobayashi et al., 2005). In this study, the RpfC/RpfG TCS and clp gene were both found in L. enzymogenes strains CX03 and CX06, and shared a high homology with that in C3, indicating that the genes could be associated with the biosynthesis of antibiotics, which need to be further confirmed. AHL is the most studied autoinducer in Gram-negative bacteria controlling infections of various pathogens and expR encodes receptor for AHL. In the study, the gene expR was detected in the genomes of many L. enzymogenes strains, however, almost all the reported members of Lysobacter do not produce AHL. For example, the Le0959 of L. enzymogenes OH11 failed to directly degard AHL, but could bind with Pcol, and this was functioned as a new quorum quenching protein (Liao et al., 2021).
Conclusion
This study described two L. enzymogenes strains CX03 and CX06, with significantly different antagonistic activities, and provided a comprehensive comparative genomic analysis for the potential biocontrol mechanisms of the two strains. Strain CX03 presented a broad-spectrum antagonistic activity toward different bacteria and showed a prominent effect in controlling black rot on cabbage, while strain CX06 exhibited a broad-spectrum inhibitory ability against diverse fungi and oomycete, and showed a remarkable effect in controlling stem rot on Chinese cabbage. Whole genomic and phylogenetic analysis demonstrated that the two strains CX03 and CX06 belonged to L. enzymogenes. Comparative genomic analysis showed that diverse secondary metabolites with different antagonistic activities were found in L. enzymogenes strains CX03 and CX06. Moreover, large numbers of genes involved in siderophore biosynthesis, bacterial secretion system, quorum sensing, selenium metabolism, two-component system were also detected in the CX03 and CX06 genomes. All these features of strains CX03 and CX06 revealed new insight into the comprehensive metabolic pathways. To our knowledge, strain CX03 is one of the few L. enzymogenes strains with the ability to directly inhibit bacteria, and this was the first study to systematically analyze the functional genes responsible for the biosynthesis of potential antibiotics with antibacterial activity in L. enzymogenes strains. Overall, all these features of L. enzymogenes strains CX03 and CX06 indicated that the two strains would be potential biocontrol agents for plant disease control, and the study provides detailed insights for the understanding of biocontrol mechanisms in L. enzymogenes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/ (CP067395.1 and CP067396.1); https://www.ncbi.nlm.nih.gov/ (SRR19560894 and SRR19568259).
Author contributions
SX, LL, and BL conceived and designed the experiments. SX and ZZ performed the experiments and analyzed the data. SX and ZZ wrote the manuscript. LL, XX, YS, TF, and AC revised the manuscript. All authors have read and approved of the final version of the manuscript.
Funding
This work was performed with the support of the China Agriculture Research System of MOF and MARA; the Science and Technology Innovation Program if the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS); and the Key Laboratory of Horticultural Crops Genetic Improvement, Ministry of Agriculture in China (IVF2020).
Acknowledgments
We would like to acknowledge Yang Cao (Allwegene Technologies Corporation) for the genome data analysis. Finally, we thank the editor and reviewers for their kind suggestions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.966986/full#supplementary-material
Supplementary Figure 1 | Production of siderophores, extracellular enzymes and general morphological characteristics, gliding motility of L. enzymogenes CX03 and CX06.
Supplementary Figure 2 | Growth and pH curves of L. enzymogenes CX03 and CX06. (A) Colony concentrations of CX03 and CX06 at different culture times. (B) pH values of CX03 and CX06 suspensions at different culture times.
Supplementary Figure 3 | Phylogenetic tree highlighting the relative positions of L. enzymogenes CX03 and CX06 among other Lysobacter strains. The phylogenetic tree was constructed based on four housekeeping genes (16S rRNA, gyrB, atpD, rpoD) according to the aligned gene sequences using maximum likelihoods derived from MEGA 6.0 software. Bootstrap values (1,000 replicates) were shown at the branch points. The scale bar indicates 0.05 nucleotide substitutions pernucleotide position.
Supplementary Figure 4 | Percentage of the average nucleotide identities (ANI) and in silico DNA-DNA hybridization (DDH) among the selected Lysobacter strains. ANI values were computed for a pairwise genome comparison using the OrthoANIu algorithm. The percentage of ANI was shown on the top right. DDH values were calculated by using the Genome-to-Genome Distance Calculator (GGDC). The percentage of DDH was shown on the bottom left.
Supplementary Figure 5 | Comparisons of gene clusters for secondary metabolites (both existed in CX03 and CX06) with other representative Lysobacter strains C3, M497-1, 55 and 76. Dark red indicated the core biosynthetic genes in different gene clusters. The core biosynthesis genes were marked in the diverse gene clusters.
Supplementary Figure 6 | Comparison of the bacterial secretion system gene clusters of L. enzymogenes CX03 and CX06 against four previous fully sequenced Lysobacter genomes. (A) Type I secretion system. (B) Type II secretion system. (C) Type III secretion system. (D) Type IV secretion system. (E) Sec (secretion) system. (F) Type VI secretion system. The same color represented genes with the same or similar biological function. Arrows denoted putative transcriptional units. The length of blocks represented the size of genes (1 cm = 1000 bp).
Supplementary Figure 7 | Determination of selenium synthesis capabilities of L. enzymogenes CX03 and CX06. (A) Nanoselenium formation by strains CX03 and CX06 grown in LB broth supplemented with different concentrations of Na2SO3. Error bars represent the range of standard errors. Three biological replicates were performed for each treatment, and the experiments were independently repeated three times. (B) The survival of strains CX03 and CX06 in different concentrations of Na2SO3.
Footnotes
- ^ https://pfam.xfam.org/
- ^ http://www.ebi.ac.uk/uniprot/
- ^ https://antismash.secondarymetabolites.org
References
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: Rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bairoch, A., and Apweiler, R. (2000). The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28, 45–48. doi: 10.1093/nar/28.1.45
Barber, C. E., Tang, J. L., Feng, J. X., Pan, M. Q., Wilson, T. J., Slater, H., et al. (1997). A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24, 555–566. doi: 10.1046/j.1365-2958.1997.3721736.x
Bayer-Santos, E., Cenens, W., Matsuyama, B. Y., Oka, G. U., Di, S. G., and Mininel, I. D. V. (2019). The opportunistic pathogen Stenotrophomonas maltophilia utilizes a type IV secretion system for interbacterial killing. PLoS Pathog. 15:e1007651. doi: 10.1371/journal.ppat.1007651
Biswas, K. C., Barton, L. L., Tsui, W. L., Shuman, K., Gillespie, J., and Eze, C. S. (2011). A novel method for the measurement of elemental selenium produced by bacterial reduction of selenite. J. Microbiol. Methods. 86, 140–144. doi: 10.1016/j.mimet.2011.04.009
Cantarel, B. L., Coutinho, P. M., Corinne, R., Thomas, B., Vincent, L., and Bernard, H. (2009). The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 37, 233–238. doi: 10.1093/nar/gkn663
Chang, J. H., Desveaux, D., and Creason, A. L. (2014). The ABCs and 123s of bacterial secretion systems in plant pathogenesis. Annu. Rev. Phytopathol. 52, 317–345. doi: 10.1146/annurev-phyto-011014-015624
Chatterjee, A., Cui, Y., Liu, Y., Dumenyo, C. K., and Chatterjee, A. K. (1995). Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-L-homoserine lactone. Environ. Microbiol.Appl. Environ. Microbiol. 61, 1959–1967. doi: 10.1128/aem.61.5.1959-1967.1995
Chen, D., Huang, J., and Yuan, L. (2019). A new function of the biocontrol bacterium Lysobacter enzymogenes LE16 in the mineralization of soil organic phosphorus. Plant Soil 442, 299–309. doi: 10.1007/s11104-019-04175-x
Chen, D. M., Yang, H. J., Huang, J. G., and Yuan, L. (2020). Lysobacter enzymogenes LE16 autolysates have potential as biocontrol agents—Lysobacter sp. autolysates as biofungicide. J. Appl. Microbiol. 129, 1684–1692. doi: 10.1111/jam.14752
Chen, H., Olson, A. S., Su, W., Dussault, P. H., and Du, L. (2015). Fatty Acyl Incorporation in the Biosynthesis of WAP-8294A, a Group of Potent Anti-MRSA Cyclic Lipodepsipeptides. RSC Adv. 5, 105753–105759. doi: 10.1039/c5ra20784c
Chen, X. H., Vater, J., Piel, J., Franke, P., Scholz, R., Schneider, K., et al. (2006). Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 188, 4024–4036. doi: 10.1128/JB.00052-06
Christensen, P., and Cook, F. D. (1978). Lysobacter, a New Genus of Nonfruiting, Gliding Bacteria with a High Base Ratio. Int. J. Syst. Bacteriol. 28, 367–393. doi: 10.1099/00207713-28-3-367
Cianciotto, N. P. (2005). Type II secretion: A protein secretion system for all seasons. Trends Microbiol. 13, 581–588. doi: 10.1016/j.tim.2005.09.005
Costa, T. R., Felisberto-Rodrigues, C., Meir, A., Prevost, M. S., Redzej, A., Trokter, M., et al. (2015). Secretion systems in Gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 13, 343–359. doi: 10.1038/nrmicro3456
Dale, C., Plague, G. R., Wang, B., Ochman, H., and Moran, N. A. (2002). Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc. Natl. Acad. Sci. U.S.A. 99, 12397–12402. doi: 10.1073/pnas.182213299
Dale, C., Young, S. A., Haydon, D. T., and Welburn, S. C. (2001). The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc. Natl. Acad. Sci. U.S.A. 98, 1883–1888. doi: 10.1073/pnas.98.4.1883
de Bruijn, I., Cheng, X., de Jager, V., Expósito, R. G., Watrous, J., Patel, N., et al. (2015). Comparative genomics and metabolic profiling of the genus Lysobacter. BMC Genomics 16:991. doi: 10.1186/s12864-015-2191-z
Deng, Y., Wu, J., Tao, F., and Zhang, L. H. (2011). Listening to a new language: DSF-based quorum sensing in Gram-negative bacteria. Chem. Rev. 111, 160–173. doi: 10.1021/cr100354f
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, 427–432. doi: 10.1093/nar/gky995
Filloux, A. (2004). The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta 1694, 163–179. doi: 10.1016/j.bbamcr.2004.05.003
Folman, L. B., Klein, M., Postma, J., and Van Veen, J. A. (2004). Production of antifungal compounds by Lysobacter enzymogenes isolate 3.1T8 under different conditions in relation to its efficacy as a biocontrol agent of Pythium aphanidermatum in cucumber. Biol. Control. 31, 145–154. doi: 10.1016/j.biocontrol.2004.03.008
Folman, L. B., Postma, J., and Van Veen, J. A. (2003). Characterisation of Lysobacter enzymogenes (Christensen and Cook, 1978) strain 3.1T8, a powerful antagonist of fungal diseases of cucumber. Microbiol. Res. 158, 107–115. doi: 10.1078/0944-5013-00185
Fulano, A. M., Shen, D., Kinoshita, M., Chou, S. H., and Qian, G. (2020a). The homologous components of flagellar type III protein apparatus have acquired a novel function to control twitching motility in a non-flagellated biocontrol bacterium. Biomolecules 10:733. doi: 10.3390/biom10050733
Fulano, A. M., Shen, D., Zhang, E. H., Shen, X., Chou, S. H., Minamino, T., et al. (2020b). Functional divergence of flagellar type III secretion system: A case study in a non-flagellated, predatory bacterium. Comput. Struct. Biotechnol. J. 18, 3368–3376. doi: 10.1016/j.csbj.2020.10.029
Galan, J., and Wolf-Watz, H. (2006). Protein delivery into eukaryotic cells by type III secretion machines. Nature 444, 567–573. doi: 10.1038/nature05272
Gehring, A. M., Mori, I., and Walsh, C. T. (1998). Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry 37, 2648–2659. doi: 10.1021/bi9726584
Gerardo, P., Selena, T., Paolo, S., Marco, M., Kristof, E., Michele, P., et al. (2016). The Lysobacter capsici AZ78 Genome Has a Gene Pool Enabling it to Interact Successfully with Phytopathogenic Microorganisms and Environmental Factors. Front. Microbiol. 7:96. doi: 10.3389/fmicb.2016.00096
Gerth, K., Steinmetz, H., Höfle, G., and Jansen, R. (2008). Chlorotonil A, a macrolide with a unique gem-dichloro-1,3-dione functionality from Sorangium cellulosum, So ce1525. Angew. Chem. Int. Ed. Engl. 47, 600–602. doi: 10.1002/anie.200703993
Ghosh, P. (2004). Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68, 771–795. doi: 10.1128/MMBR.68.4.771-795.2004
Giesler, L. J., and Yuen, G. Y. (1998). Evaluation of Stenotrophomonas maltophilia strain C3 for biocontrol of brown patch disease. Crop Prot. 17, 509–513. doi: 10.1016/S0261-2194(98)00049-0
Gökçen, A., Vilcinskas, A., and Wiesner, J. (2014). Biofilm-degrading enzymes from Lysobacter gummosus. Virulence 5, 378–387. doi: 10.4161/viru.27919
Gómez Expósito, R., Postma, J., Raaijmakers, J. M., and De Bruijn, I. (2015). Diversity and Activity of Lysobacter Species from Disease Suppressive Soils. Front. Microbiol. 6:1243. doi: 10.3389/fmicb.2015.01243
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Han, Y., Wang, Y., Tombosa, S., Wright, S., Huffman, J., Yuen, G., et al. (2015). Identification of a small molecule signaling factor that regulates the biosynthesis of the antifungal polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes. Appl. Microbiol. Biotechnol. 99, 801–811. doi: 10.1007/s00253-014-6120-x
Hashizume, H., Hirosawa, S., Sawa, R., Muraoka, Y., Ikeda, D., Naganawa, H., et al. (2016). Corrigendum: Tripropeptins, novel antimicrobial agents produced by Lysobacter sp. II. structure elucidation. J. Antibiot. 69, 889–891. doi: 10.1038/ja.2016.104
Hashizume, H., Igarashi, M., Hattori, S., Hori, M., Hamada, M., and Takeuchi, T. (2001). Tripropeptins, novel antimicrobial agents produced by Lysobacter sp. I. Taxonomy, isolation and biological activities. J. Antibiot. 54, 1054–1059. doi: 10.7164/antibiotics.54.1054
Hayashi, F., Smith, K. D., Ozinsky, A., Hawn, T. R., Yi, E. C., Goodlett, D. R., et al. (2001). The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103. doi: 10.1038/35074106
Hayward, A. C., Fegan, N., Fegan, M., and Stirling, G. R. (2010). Stenotrophomonas and Lysobacter: Ubiquitous plant-associated gamma-proteobacteria of developing significance in applied microbiology. J. Appl. Microbiol. 108, 756–770. doi: 10.1111/j.1365-2672.2009.04471.x
He, Y. W., and Zhang, L. H. (2008). Quorum sensing and virulence regulation in Xanthomonas campestris. FEMS. Microbiol. Rev. 32, 842–857. doi: 10.1111/j.1574-6976.2008.00120.x
Held, J., Gebru, T., Kalesse, M., Jansen, R., Gerth, K., Müller, R., et al. (2014). Antimalarial activity of the myxobacterial macrolide chlorotonil a. Antimicrob. Agents Chemother. 58, 6378–6384. doi: 10.1128/aac.03326-14
Ho, B. T., Dong, T. G., and Mekalanos, J. J. (2014). A view to a kill: The bacterial type VI secretion system. Cell Host Microbe 15, 9–21. doi: 10.1016/j.chom.2013.11.008
Jin, Y., Osborne, L., Jochum, C., and Yuen, G. (2003). Biocontrol of Fusarium Head Blight in Wheat by Lysobacter enzymogenes C3. Phytopathology 93:S93.
Kanehisa, M., Goto, S., Hattori, M., Aoki-Kinoshita, K. F., and Hirakawa, M. (2006). From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 34, 354–357. doi: 10.1093/nar/gkj102
Kende, A. S., Kaldor, I., Dorey, G., Koch, K., and Liu, K. (1995). Total synthesis of the macrolide antitumor antibiotic lankacidin C. J. Am. Chem. Soc. 117, 8258–8270. doi: 10.1021/ja00136a025
Kilic-Ekici, O., and Yuen, G. Y. (2003). Induced resistance as a mechanism of biological control by Lysobacter enzymogenes Strain C3. Phytopathology 93, 1103–1110. doi: 10.1094/PHYTO.2003.93.9.1103
Ko, H. S., Jin, R. D., Krishnan, H. B., Lee, S. B., and Kim, K. Y. (2009). Biocontrol ability of Lysobacter antibioticus HS124 against Phytophthora blight is mediated by the production of 4-hydroxyphenylacetic acid and several lytic enzymes. Curr. Microbiol. 59, 608–615. doi: 10.1007/s00284-009-9481-0
Ko, H. S., Tindwa, H., Jin, R. D., Yong, S. L., Hong, S. H., Hyun, H. N., et al. (2011). Investigation of Siderophore production and Antifungal activity against Phytophthora capsici as related to Iron (III) nutrition by Lysobacter antibioticus HS124. Korean J. Soil Sci. Fert. 44, 650–656. doi: 10.7745/KJSSF.2011.44.4.650
Kobayashi, D. Y., Reedy, R. M., Palumbo, J. D., Zhou, J. M., and Yuen, G. Y. (2005). A clp gene homologue belonging to the Crp gene family globally regulates lytic enzyme production, antimicrobial activity, and biological control activity expressed by Lysobacter enzymogenes strain C3. Appl. Environ. Microbiol. 71, 261–269. doi: 10.1128/aem.71.1.261-269.2005
Kobayashi, D. Y., and Yuen, G. Y. (2005). The role of clp-regulated factors in antagonism against Magnaporthe poae and biological control of summer patch disease of Kentucky bluegrass by Lysobacter enzymogenes C3. Can. J. Microbiol. 51, 719–723. doi: 10.1139/w05-056
Korotkov, K. V., Sandkvist, M., and Hol, W. G. (2012). The type II secretion system: Biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 10, 336–351. doi: 10.1038/nrmicro2762
Lagesen, K., Hallin, P., Rødland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Lee, Y. S., Anees, M., Hyun, H. N., and Kim, K. Y. (2013). Biocontrol potential of Lysobacter antibioticus HS124 against the root-knot nematode, Meloidogyne incognita, causing disease in tomato. Nematology 15, 545–555. doi: 10.1163/15685411-00002700
Li, S., Du, L., Yuen, G., and Harris, S. D. (2006). Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 17, 1218–1227. doi: 10.1091/mbc.e05-06-0533
Li, S., Jochum, C. C., Yu, F., Zaleta-Rivera, K., Du, L., Harris, S. D., et al. (2008). An antibiotic complex from Lysobacter enzymogenes strain C3: Antimicrobial activity and role in plant disease control. Phytopathology 98, 695–701. doi: 10.1094/phyto-98-6-0695
Li, Y., Huffman, J., Li, Y., Du, L., and Shen, Y. (2012). 3-Hydroxylation of the polycyclic tetramate macrolactam in the biosynthesis of antifungal HSAF from Lysobacter enzymogenes C3. Med. Chem. Commun. 3, 982–986. doi: 10.1039/c2md20026k
Liao, J., Shen, D., Lin, L., Chen, H., Jin, Y., Chou, S. H., et al. (2021). Bacterial quorum sensing quenching activity of Lysobacter leucyl aminopeptidase acts by interacting with autoinducer synthase. Comput. Struct. Biotechnol. J. 19, 6179–6190. doi: 10.1016/j.csbj.2021.11.017
Lin, L., Xu, K., Shen, D., Chou, S. H., Gomelsky, M., and Qian, G. (2021). Antifungal weapons of Lysobacter, a mighty biocontrol agent. Environ. Microbiol. 23, 5704–5715. doi: 10.1111/1462-2920.15674
Liu, X., Biswas, S., Berg, M. G., Antapli, C. M., and Cheng, Y. Q. (2013). Genomics-Guided Discovery of Thailanstatins A, B, and C As Pre-mRNA Splicing Inhibitors and Antiproliferative Agents from Burkholderia thailandensis MSMB43. J. Nat. Prod. 76, 685–693. doi: 10.1021/np300913h
Liu, X., Zhu, H., Biswas, S., Cheng, Y. Q., and Biotechnology, S. (2016). Improved production of cytotoxic thailanstatins A and D through metabolic engineering of Burkholderia thailandensis MSMB43 and pilot scale fermentation. Synth. Syst. Biotechnol. 1, 34–38. doi: 10.1016/j.synbio.2016.02.002
Lowe, T. M., and Chan, P. P. (2016). tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, 54–57. doi: 10.1093/nar/gkw413
Lu, P., Hontecillas, R., Horne, W. T., Carbo, A., Viladomiu, M., Pedragosa, M., et al. (2012). Computational modeling-based discovery of novel classes of anti-inflammatory drugs that target lanthionine synthetase C-like protein 2. PLoS One 7:e34643. doi: 10.1371/journal.pone.0034643
McBrien, K. D., Berry, R. L., Lowe, S. E., Neddermann, K. M., Bursuker, I., Huang, S., et al. (1995). Rakicidins, new cytotoxic lipopeptides from Micromonospora sp. fermentation, isolation and characterization. J. Antibiot. 48, 1446–1452. doi: 10.7164/antibiotics.48.1446
Murata, H. J. (1991). Molecular Cloning of an aepA Gene that Activates Production of Extracellular Pectolytic, Cellulolytic, and Proteolytic Enzymes in Erwinia carotovora subsp. carotovora. Mol. Plant Microbe Interact. 4, 239–246. doi: 10.1094/MPMI-4-239
Nguyen, Y., Sugiman-Marangos, S., Harvey, H., Bell, S. D., Charlton, C. L., Junop, M. S., et al. (2015). Pseudomonas aeruginosa minor pilins prime type IVa pilus assembly and promote surface display of the PilY1 adhesin. J. Biol. Chem. 290, 601–611. doi: 10.1074/jbc.M114.616904
Nowell, R. W., Green, S., Laue, B. E., and Sharp, P. M. (2014). The extent of genome flux and its role in the differentiation of bacterial lineages. Genome Biol. Evol. 6, 1514–1529. doi: 10.1093/gbe/evu123
Odhiambo, B. O., Xu, G., Qian, G., and Liu, F. (2017). Evidence of an Unidentified Extracellular Heat-Stable Factor Produced by Lysobacter enzymogenes (OH11) that Degrade Fusarium graminearum PH1 Hyphae. Curr. Microbiol. 74, 437–448. doi: 10.1007/s00284-017-1206-1
Olson, A. S., Chen, H., Du, L., and Dussault, P. (2015). Synthesis of a 2,4,6,8,10-dodecapentanoic acid thioester as a substrate for biosynthesis of Heat Stable Antifungal Factor (HSAF). RSC Adv. 5, 11644–11648. doi: 10.1039/c4ra14829k
Palumbo, J. D., Sullivan, R. F., and Kobayashi, D. Y. (2003). Molecular characterization and expression in Escherichia coli of three beta-1,3-glucanase genes from Lysobacter enzymogenes strain N4-7. J. Bacteriol. 185, 4362–4370. doi: 10.1128/jb.185.15.4362-4370.2003
Palumbo, J. D., Yuen, G. Y., Jochum, C. C., Tatum, K., and Kobayashi, D. Y. (2005). Mutagenesis of beta-1,3-Glucanase Genes in Lysobacter enzymogenes Strain C3 Results in Reduced Biological Control Activity Toward Bipolaris Leaf Spot of Tall Fescue and Pythium Damping-Off of Sugar Beet. Phytopathology 95, 701–707. doi: 10.1094/phyto-95-0701
Panthee, S., Hamamoto, H., Paudel, A., and Sekimizu, K. (2016). Lysobacter species: A potential source of novel antibiotics. Arch. Microbiol. 198, 839–845. doi: 10.1007/s00203-016-1278-5
Park, J. H., Kim, R., Aslam, Z., Jeon, C. O., and Chung, Y. R. (2008). Lysobacter capsici sp. nov., with antimicrobial activity, isolated from the rhizosphere of pepper, and emended description of the genus Lysobacter. Int. J. Syst. Evol. Microbiol. 58, 387–392. doi: 10.1099/ijs.0.65290-0
Poplawsky, A. R., Urban, S. C., and Chun, W. (2000). Biological Role of Xanthomonadin Pigments in Xanthomonas campestris pv. campestris. Environ. Microbiol.Appl. Environ. Microbiol. 66, 5123–5127. doi: 10.1128/AEM.66.12.5123-5127.2000
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Puopolo, G., Cimmino, A., Palmieri, M. C., Giovannini, O., Evidente, A., and Pertot, I. (2014). Lysobacter capsici AZ78 produces cyclo(L-Pro-L-Tyr), a 2,5-diketopiperazine with toxic activity against sporangia of Phytophthora infestans and Plasmopara viticola. J. Appl. Microbiol. 117, 1168–1180. doi: 10.1111/jam.12611
Qian, G., Wang, Y., Liu, Y., Xu, F., He, Y. W., Du, L., et al. (2013). Lysobacter enzymogenes uses two distinct cell-cell signaling systems for differential regulation of secondary-metabolite biosynthesis and colony morphology. Appl. Environ. Microbiol. 79, 6604–6616. doi: 10.1128/aem.01841-13
Qian, G., Xu, F., Venturi, V., Du, L., and Liu, F. (2014). Roles of a solo LuxR in the biological control agent Lysobacter enzymogenes strain OH11. Phytopathology 104, 224–231. doi: 10.1094/PHYTO-07-13-0188-R
Qian, G. L., Hu, B. S., Jiang, Y. H., and Liu, F. Q. (2009). Identification and characterization of Lysobacter enzymogenes as a biological control agent against some fungal pathogens. Agric. Sci. China 8, 68–75. doi: 10.1016/S1671-2927(09)60010-9
Quadri, L. E., Keating, T. A., Patel, H. M., and Walsh, C. T. (1999). Assembly of the Pseudomonas aeruginosa nonribosomal peptide siderophore pyochelin: In vitro reconstitution of aryl-4, 2-bisthiazoline synthetase activity from PchD, PchE, and PchF. Biochemistry 38, 14941–14954. doi: 10.1021/bi991787c
Rajagopal, L., Sundari, C. S., Balasubramanian, D., and Sonti, R. V. (1997). The bacterial pigment xanthomonadin offers protection against photodamage. FEBS. Lett. 415, 125–128. doi: 10.1016/s0014-5793(97)01109-5
Ramamoorthy, V., Viswanathan, R., Raguchander, T., Prakasam, V., and Samiyappan, R. (2001). Induction of systemic resistance by plant growth promoting rhizobacteria in crop plants against pests and diseases. Crop Prot. 20, 1–11. doi: 10.1016/S0261-2194(00)00056-9
Raymond, K. N., Dertz, E. A., and Kim, S. S. (2003). Enterobactin: An archetype for microbial iron transport. Proc. Natl. Acad. Sci. U.S.A. 100, 3584–3588. doi: 10.1073/pnas.0630018100
Reedy, R. M. (2004). Identification of a type III secretion system in Lysobacter enzymogenes strain C3: Molecular characterization and functional analysis of the pathway as an integral part in bacterial-fungal interactions. Piscataway, NJ: The State University of New Jersey New Brunswick.
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Saier, M. H., Reddy, V. S., Tamang, D. G., and Västermark, A. (2009). The transporter classification database: Recent advances. Nucleic Acids Res. 1, 251–258. doi: 10.1093/nar/gkn862
Sgro, G. G., Oka, G. U., Souza, D. P., Cenens, W., Bayer-Santos, E., Matsuyama, B. Y., et al. (2019). Bacteria-Killing Type IV secretion systems. Front. Microbiol. 10:1078. doi: 10.3389/fmicb.2019.01078
Shen, X., Wang, B., Yang, N., Zhang, L., Shen, D., Wu, H., et al. (2021). Lysobacter enzymogenes antagonizes soilborne bacteria using the type IV secretion system. Environ. Microbiol. 23, 4673–4688. doi: 10.1111/1462-2920.15662
Siddiqi, M. Z., and Im, W. T. (2016). Lysobacter hankyongensis sp. nov., isolated from activated sludge and Lysobacter sediminicola sp. nov., isolated from freshwater sediment. Int. J. Syst. Evol. Microbiol. 66, 212–218. doi: 10.1099/ijsem.0.000699
Slater, H., Alvarez-Morales, A., Barber, C. E., Daniels, M. J., and Dow, J. M. (2000). A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol. Microbiol. 38, 986–1003. doi: 10.1046/j.1365-2958.2000.02196.x
Souza, D. P., Oka, G. U., Alvarez-Martinez, C. E., Bisson-Filho, A. W., Dunger, G., Hobeika, L., et al. (2015). Bacterial killing via a type IV secretion system. Nat. Commun. 6:6453. doi: 10.1038/ncomms7453
Takami, H., Toyoda, A., Uchiyama, I., Itoh, T., Takaki, Y., Arai, W., et al. (2017). Complete genome sequence and expression profile of the commercial lytic enzyme producer Lysobacter enzymogenes M497-1. DNA Res. 24, 169–177. doi: 10.1093/dnares/dsw055
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tatusov, R. L., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Kiryutin, B., Koonin, E. V., et al. (2003). The COG database: An updated version includes eukaryotes. BMC Bioinformatics 4:41. doi: 10.1186/1471-2105-4-41
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Tomada, S., Puopolo, G., Perazzolli, M., Musetti, R., Loi, N., and Pertot, I. (2016). Pea broth enhances the biocontrol efficacy of Lysobacter capsici AZ78 by triggering cell motility associated with biogenesis of type IV Pilus. Front. Microbiol. 7:1136. doi: 10.3389/fmicb.2016.01136
Tseng, T. T., Tyler, B., and Setubal, J. (2009). Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 9(Suppl. 1):S2. doi: 10.1186/1471-2180-9-s1-s2
Vasilyeva, N. V., Shishkova, N. A., Marinin, L. I., Ledova, L. A., Tsfasman, I. M., Muranova, T. A., et al. (2014). Lytic peptidase L5 of Lysobacter sp. XL1 with broad antimicrobial spectrum. J. Mol. Microbiol. Biotechnol. 24, 59–66. doi: 10.1159/000356838
Villadsen, N. L., Jacobsen, K. M., Keiding, U. B., Weibel, E. T., Christiansen, B., Vosegaard, T., et al. (2017). Synthesis of ent-BE-43547A(1) reveals a potent hypoxia-selective anticancer agent and uncovers the biosynthetic origin of the APD-CLD natural products. Nat. Chem. 9, 264–272. doi: 10.1038/nchem.2657
Wang, B., Yuan, J., Jian, Z., Zhang, M., Li, R., Ruan, Y., et al. (2013). Effects of novel bioorganic fertilizer produced by Bacillus amyloliquefaciens W19 on antagonism of Fusarium wilt of banana. Biol. Fertil. Soils 49, 435–446. doi: 10.1007/s00374-012-0739-5
Wang, P., Chen, H., Qian, G., and Liu, F. (2017). LetR is a TetR family transcription factor from Lysobacter controlling antifungal antibiotic biosynthesis. Appl. Microbiol. Biotechnol. 101, 3273–3282. doi: 10.1007/s00253-017-8117-8
Wang, Y., Zhao, Y., Zhang, J., Zhao, Y., Shen, Y., Su, Z., et al. (2014). Transcriptomic analysis reveals new regulatory roles of Clp signaling in secondary metabolite biosynthesis and surface motility in Lysobacter enzymogenes OH11. Appl. Microbiol. Biotechnol. 98, 9009–9020. doi: 10.1007/s00253-014-6072-1
Xia, J., Chen, J., Chen, Y., Qian, G., and Liu, F. (2018). Type IV pilus biogenesis genes and their roles in biofilm formation in the biological control agent Lysobacter enzymogenes OH11. Appl. Microbiol. Biotechnol. 102, 833–846. doi: 10.1007/s00253-017-8619-4
Xie, Y., Wright, S., Shen, Y., and Du, L. (2012). Bioactive natural products from Lysobacter. Nat. Prod. Rep. 29, 1277–1287. doi: 10.1039/c2np20064c
Xu, S., Zhao, Y., Peng, Y., Shi, Y., Xie, X., Chai, A., et al. (2022). Comparative genomics assisted functional characterization of Rahnella aceris ZF458 as a novel plant growth promoting Rhizobacterium. Front. Microbiol. 13:850084. doi: 10.3389/fmicb.2022.850084
Yu, F., Zaleta-Rivera, K., Zhu, X., Huffman, J., Millet, J. C., Harris, S. D., et al. (2007). Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob. Agents Chemother. 51, 64–72. doi: 10.1128/aac.00931-06
Yuan, L., Li, L., Zheng, F., Shi, Y., Xie, X., Chai, A., et al. (2020). The complete genome sequence of Rahnella aquatilis ZF7 reveals potential beneficial properties and stress tolerance capabilities. Arch. Microbiol. 202, 483–499. doi: 10.1007/s00203-019-01758-1
Yuen, G. Y., Broderick, K. C., Jochum, C. C., Chen, C. J., and Caswell-Chen, E. P. (2018). Control of cyst nematodes by Lysobacter enzymogenes strain C3 and the role of the antibiotic HSAF in the biological control activity. Biol. Control 117, 158–163. doi: 10.1016/j.biocontrol.2017.11.007
Yuen, G. Y., Steadman, J. R., Lindgren, D. T., Schaff, D., and Jochum, C. (2001). Bean rust biological control using bacterial agents 1 1 University of Nebraska, Agricultural Research Division Journal Number 12922. Crop Prot. 20, 395–402. doi: 10.1016/S0261-2194(00)00154-X
Zhang, W., Li, Y., Qian, G., Wang, Y., Chen, H., Li, Y. Z., et al. (2011). Identification and characterization of the anti-methicillin-resistant Staphylococcus aureus WAP-8294A2 biosynthetic gene cluster from Lysobacter enzymogenes OH11. Antimicrob. Agents Chemother. 55, 5581–5589. doi: 10.1128/aac.05370-11
Zhang, Y., Fan, Q., and Loria, R. (2016). A re-evaluation of the taxonomy of phytopathogenic genera Dickeya and Pectobacterium using whole-genome sequencing data. Syst. Appl. Microbiol. 39, 252–259. doi: 10.1016/j.syapm.2016.04.001
Zhang, Z., Chen, G., and Tang, Y. (2018). Towards selenium recovery: Biocathode induced selenate reduction to extracellular elemental selenium nanoparticles. Chem. Eng. J. 315, 1095–1103. doi: 10.1016/j.cej.2018.06.172
Zhang, Z., and Yuen, G. Y. (1999). Biological control of bipolaris sorokiniana on tall fescue by Stenotrophomonas maltophilia strain C3. Phytopathology 89, 817–822. doi: 10.1094/phyto.1999.89.9.817
Zhang, Z., and Yuen, G. Y. (2000). The role of chitinase production by Stenotrophomonas maltophilia Strain C3 in biological control of bipolaris sorokiniana. Phytopathology 90, 384–389. doi: 10.1094/phyto.2000.90.4.384
Zhang, Z., Yuen, G. Y., Sarath, G., and Penheiter, A. R. (2001). Chitinases from the Plant Disease Biocontrol Agent, Stenotrophomonas maltophilia C3. Phytopathology 91, 204–211. doi: 10.1094/phyto.2001.91.2.204
Zhao, Y., Qian, G., Ye, Y., Wright, S., Chen, H., Shen, Y., et al. (2016). Heterocyclic aromatic N-oxidation in the biosynthesis of phenazine antibiotics from Lysobacter antibioticus. Org. Lett. 18, 2495–2498. doi: 10.1021/acs.orglett.6b01089
Zhou, X., Qian, G., Chen, Y., Du, L., Liu, F., and Yuen, G. Y. (2015). PilG is involved in the regulation of twitching motility and antifungal antibiotic biosynthesis in the biological control agent Lysobacter enzymogenes. Phytopathology 105, 1318–1324. doi: 10.1094/PHYTO-12-14-0361-R
Keywords: Lysobacter enzymogenes, comparative genomic analysis, secondary metabolites, antagonistic activity, bacterial secretory system, biological control
Citation: Xu S, Zhang Z, Xie X, Shi Y, Chai A, Fan T, Li B and Li L (2022) Comparative genomics provides insights into the potential biocontrol mechanism of two Lysobacter enzymogenes strains with distinct antagonistic activities. Front. Microbiol. 13:966986. doi: 10.3389/fmicb.2022.966986
Received: 12 June 2022; Accepted: 27 July 2022;
Published: 11 August 2022.
Edited by:
Ernesto Perez-Rueda, Universidad Nacional Autónoma de México, MexicoReviewed by:
Long Lin, Nanjing Agricultural University, ChinaZhijun Wang, Shanghai Jiao Tong University, China
Copyright © 2022 Xu, Zhang, Xie, Shi, Chai, Fan, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Li, bGlsZWkwMUBjYWFzLmNu; Baoju Li, bGliYW9qdWl2ZkAxNjMuY29t
 Shuai Xu
Shuai Xu Ziyu Zhang
Ziyu Zhang Yanxia Shi
Yanxia Shi Baoju Li
Baoju Li Lei Li
Lei Li