
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 August 2022
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.966239
This article is part of the Research Topic Community Series in Microbiological Safety and Quality Aspects of Fermented Dairy Products, Volume II View all 8 articles
Time of ripening has a strong impact on shaping the valuable and recognizable characteristics of long-ripened types of cheese such as Parmigiano Reggiano (PR) due to the interrelationship between microbiota and proteolysis that occurs during ripening. The derived peptide profile is linked to cheese quality and represents the canvas for enzymes upon digestion, which could be responsible for the release of potentially bioactive peptides (BPs). In this study, we aimed at investigating the presence of BP in 72 PR cheese samples of different ripening times, from curd to 24 months of ripening, produced in six different dairies, and following their fate after simulated gastrointestinal digestion. A small number of peptide sequences sharing 100% similarity with known antimicrobial, antioxidant, and ACE-inhibitor sequences were found in PR cheeses, while a higher number of potential BPs were found after their simulated gastrointestinal digestion, in different amounts according to ripening time. Taking advantage of the complex organization of the sampling plan, we were able to follow the fate of peptides considered quality drivers during cheese ripening to their release as functional compounds upon digestion.
Fermented dairy products are consumed from the dawn of time, and nowadays are gaining even more attention due to their potential functional features (van der Reijden et al., 2017; Górska-Warsewicz et al., 2019; Castellone et al., 2021). Scientific evidence is accumulating on the role of long-ripened types of cheese in conferring positive health effects to consumers. As an example, cheese consumption has been associated with a significantly reduced risk of blood hypertension, stroke, and coronary heart disease (Petyaev and Bashmakov, 2012), as well as colorectal cancer (Godos et al., 2020). During the manufacturing of long ripened cheese, many processes take place, from milk gathering to the time when the cheese is finally ready. Among all these processes, ripening is regarded as the most important and despite the static appearance of the cheese wheels in the ripening chamber, it is a deeply active period of radical biochemical changes in the matrix. In particular, proteolysis is recognized as one of the most important events during cheese ripening, and the interrelationship between cheese microbiota and aging results in a specific peptide profile (Bottari et al., 2020), which leads to the valuable and recognizable characteristics of long-ripened types of cheese, and where bioactive peptides (BPs) have frequently been found (Coppola et al., 2000; Ardö et al., 2009; López-Expósito et al., 2017; Di Nunzio et al., 2022). BPs are protein fragments produced from parent proteins, involved in carrying out various physiological functions, such as anti-microbial, antioxidant, antihypertensive, and ACE-inhibitory activities, mainly described for BPs identified in various types of cheese (Sforza et al., 2012; Sultan et al., 2018; Martini et al., 2020; Solieri et al., 2020). Recently, some authors have investigated the presence of BPs in Parmigiano Reggiano (PR) cheese (Basiricò et al., 2015; Martini et al., 2020, 2021; Solieri et al., 2020; Tagliazucchi et al., 2020; Di Nunzio et al., 2022). PR is a protected designation of origin (PDO), raw milk, and hard-cooked cheese, with a minimum ripening time of 12 months (Gatti et al., 2014). PR peptide profile and its evolution during ripening have been described in detail (Sforza et al., 2012; Bottari et al., 2020). Long-ripened kinds of cheese are also known for the presence of Non-Proteolytic Amino Acyl Derivatives (NPADs), which are aminoacyl derivates of non-proteolytic origin synthesized in cheese de novo by an enzymatic activity (Bottesini et al., 2014; Zhao et al., 2016; Gazme et al., 2019). These compounds have been reported to exert different bioactivities (Lu et al., 2021). For example, different studies report NPADs being effective in reducing appetite (Yang et al., 2019) and having anti-inflammatory (Xing et al., 2019) and antioxidant effects (Salama et al., 2019), among many others. As they are stable to gastrointestinal digestion, their potential to be absorbed as such and being possibly transported to the body tissues have been suggested (Bottesini et al., 2014). NPADs have been shown to accumulate in PR during ripening, probably due to microbial enzymes (Sforza et al., 2009; Sgarbi et al., 2013).
It is known that gastrointestinal digestion is a key element for determining the biological activities of BPs, which might be degraded or transformed into new sequences released from inactive or less active precursors by pepsin and pancreatic enzymes (Sultan et al., 2018). Lately, the effect of ripening and in vitro digestion on the evolution and fate of BPs in PR were studied (Sultan et al., 2018; Martini et al., 2020). In this work, we have exploited the complex sampling that we have already published in a previous work where we described the peptide profile of 72 PR samples as a function of ripening time and microbial dynamics. Leveraging that unique and strictly controlled production and sampling approach, this work was aimed at evaluating the peptide profile of PR at different ripening times and from different dairies after simulated gastrointestinal digestion, particularly focusing on BPs.
The cheese samples were provided by the “Consorzio del Parmigiano-Reggiano” (Reggio Emilia, Italy) and obtained according to Bottari et al. (2020) from six dairies (called A–F) located in the PR PDO production area. For each dairy, samples were taken from the acidified curd (48 h), after brining (1 month of aging) and after 6, 12, and 24 months. For dairies C, E, and F, samples were also taken at 2, 7, and 9 months. Samples were taken for each dairy at different ripening times from the same original wheel (same production batch) and from different wheels with the same ripening times (Figure 1). For each dairy (A–F), the samples were identified with the letter W followed by a number, indicating the sampled wheel, and a slash followed by a second number, indicating the stage of ripening (e.g., AW1/0 corresponds to the dairy A, wheel 1, 0 months of ripening, i.e., the curd 48 h after cheese making). Cheeses were produced according to the EU PDO Regulation established by Article 11 of Regulation (EU) no. 1151/2012 (Gragnani, 2013). The samples were obtained by coring, thus obtaining a cross-section for each wheel. Each section was completely grated and mixed before analysis to have a representative sample of the entire wheel. Aliquots of the samples were kept at –20°C until digestion and subsequent analysis.
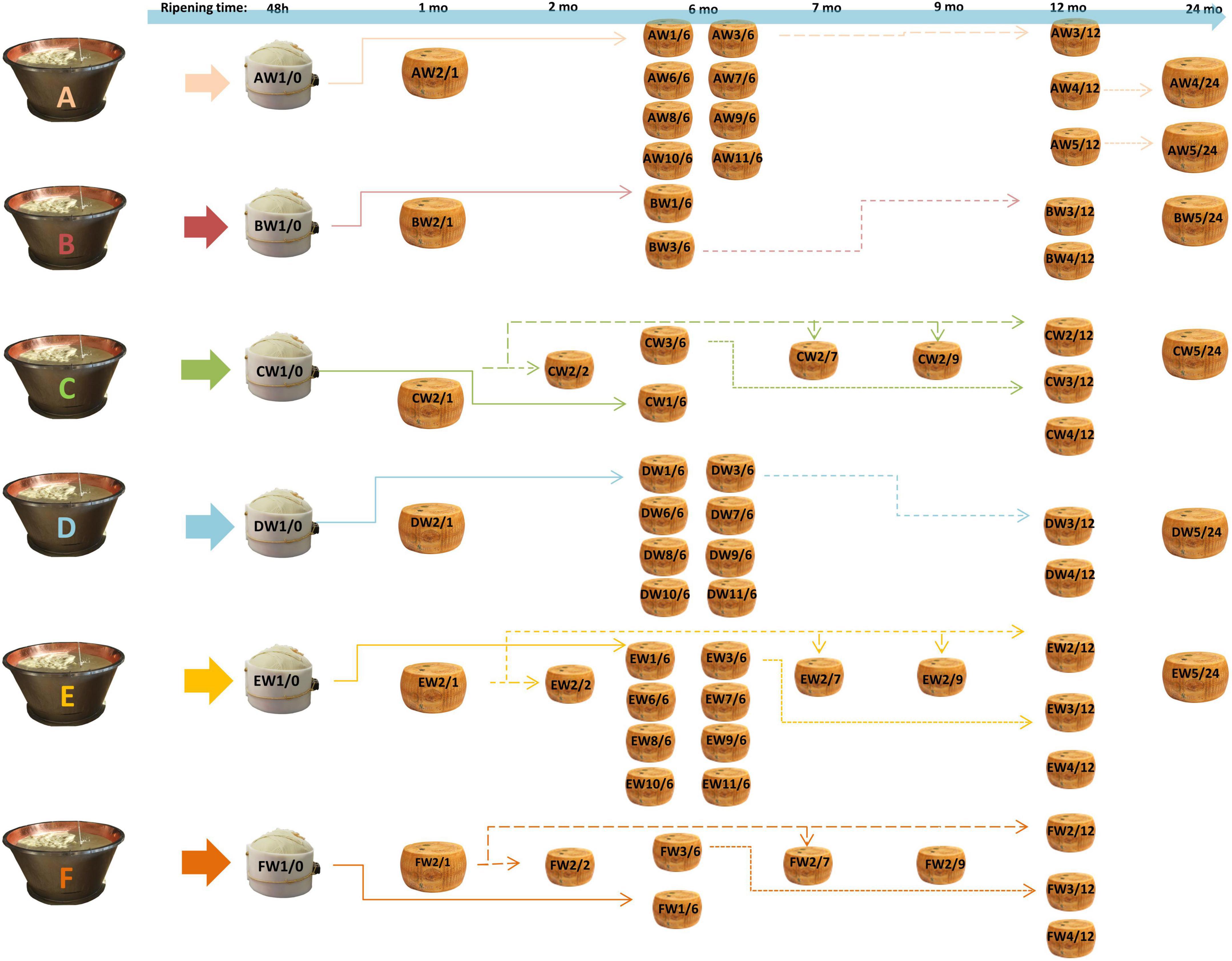
Figure 1. Sampling scheme. For each dairy (A–F), samples were taken from the same cheese-making lot (same wheel, W) at different ripening stages, and from different kinds of cheese-making lots (different wheels) at the same ripening stage. AW1/0 corresponds to dairy A, wheel 1, months of ripening 0, i.e., the curd after 48 h from cheese-making; AW1/6 is the same wheel sampled after 6 months. The same applies to other samples. Samples from the same cheese-making lot are connected by arrows. The time lime of ripening is also indicated at the top of the picture.
The simulated digestion of the samples was performed according to the INFOGEST protocol (Brodkorb et al., 2019) for gastrointestinal digestion in vitro without the addition of gastric lipase. The difficulties in finding the enzyme on the market (despite the high cost), combined with the fact that this study is focused on proteolysis (and not on the lipid fraction) led to this choice. First, the stock solutions were prepared, and then the simulated salivary (SSF), gastric (SGF), and intestinal (SIF) fluids were then prepared by suitably mixing the solutions. The solutions were then brought to 400 ml with demineralized water. In addition to the previous solutions, enzymatic solutions were prepared and suitably dissolved in the corresponding digestive fluid.
After the preparation of the solutions containing digestive fluids and enzymes, 2.5 g of grated PR cheese are weighted, and in vitro digestion begins with the oral phase. For the oral phase, we mixed the grated cheese with 1.75 ml of SSF, 250 μL of amylase solution, 12.5 μL of CaCl2, and 487.5 μL of demineralized water. The samples were then homogenized for a few seconds with a vortex and incubated for 2 min at 37°C under constant stirring, to simulate chewing. After the oral phase, the gastric phase was continued by adding 3.75 ml of gastric solution, 800 μL of pepsin solution, 2.5 μL of CaCl2, 0.3475 ml of demineralized water, and sufficient HCl to the bolus to bring the pH to 3. Then, the gastric phase was carried out in incubation for 2 h at 37°C under constant stirring. The last phase is the intestinal phase, in which 5.5 ml of SIF, 2.5 mL of pancreatin, and 1.25 ml of bile have been added to the chyme, along with 20 μL of CaCl2 and enough NaOH to bring the pH to 7. The samples were then incubated at 37°C for an additional 2 h. After incubation, digestion was stopped by bringing the samples to 95°C for 10 min. Samples were then centrifuged at 10,000g for 10 min at 4°C and filtered with 0.45 μm sterile syringe filters to remove all the particulates. Finally, to allow for semi-quantification of peptides, samples were spiked with 1 mM of (L,L)-phenylalanyl-phenylalanine (Phe-Phe) as the internal standard. The samples were then frozen and held at –20°C until ultra-high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry (UHPLC/ESI-MS/MS) analysis. Each sample was extracted and analyzed in triplicate.
UHPLC/ESI-MS/MS analysis was performed on digested PR samples as described in Prandi et al. (2020). Chromatographic separation was achieved using a reversed-phase column (Aeris Peptide 1.7 μm XB-C18, 150 × 2.10 mm, Phenomenex, Torrance, CA) equipped with a Security Guard ULTRA Cartridge (C18-Peptide, ID 2.1 mm; Phenomenex, Torrance, CA, United States) in a UHPLC system (Dionex Ultimate 3000, Thermo Scientific, Waltham, MA, United States). Eluent A was H2O + 0.2% CH3CN + 0.1% HCOOH and eluent B was CH3CN + 0.2% H2O + 0.1% HCOOH. The flow was maintained at 0.2 ml/min and the applied gradient was: 0–7 min, 100% A; 7–50 min, 100% A to 50% A; 50–52.6 min, 50% A; 52.6–53 min, 50% A to 0% A; 53–58.2 min, 0% A; 58.2–59 min, 0% A to 100% A; 59–72 min, 100% A. Total run time: 72 min; column temperature: 35°C; sample temperature: 10°C; injection volume: 2 μL for Full Scan analysis, 4 μL for Product Ion Scan analysis. Detection was achieved using a triple quadrupole TSQ Vantage (Thermo Scientific, Waltham, MA) using the following parameters: positive ion mode, acquisition time: 7–58.2 min (7 min of solvent delay was applied at the beginning of the chromatographic run), acquisition range: 100–1,500 m/z, micro scans: 1, scan time: 0.50, Q1 PW: 0.70, spray voltage: 3,200 V, capillary temperature: 250°C, vaporizer temperature: 250°C, sheath gas flow: 22 units. The samples were first analyzed in Full Scan mode, then in Product Ion Scan mode. The collision energies (CE) were calculated as CE = 3.314 + 0.034 × m/z (MacLean et al., 2011). The peptides were identified as reported in (Boukid et al., 2019). The peptide sequences were assigned based on the obtained tandem mass spectra. In short, the FindPept software1 was used to find the peptide sequences within the target proteins (UniprotKB accessions: P02666, P02662, P02663, P02668, P02754, P00711), whose molecular weight corresponding to the experimental data. Then, the Proteomics Toolkit software2 was used to check the correspondence between the theoretical MS/MS fragmentation and the obtained tandem MS spectra.
After UHPLC/ESI-MS/MS analysis, chromatograms were processed to identify all peptides present in the samples. The peptides from digested samples were semi-quantified against an internal standard (Phe-Phe). After the identification of the peaks and the semi-quantification, the Milk Bioactive Peptide Database (MBPB) was used to identify peptides reported as bioactive in the protein fractions (Nielsen et al., 2017). Peptide sequences with 100% similarity to the recorded BP sequence were considered for the bioactivities. Thirty-four peptide sequences identified in a previous work in the same PR samples before digestion (Bottari et al., 2020), and 105 peptide sequences identified in the present study in digested the PR varieties of cheese were analyzed.
Statistical analyses were carried out using the IBM SPSS Statistics software (version 27.0, Armonk, NY, United States). Bivariate correlation was performed using Pearson’s coefficients, with a two-tailed significance test, and pairwise case exclusion for missing values. Significance was fixed to a p < 0.05. The analysis of variance (ANOVA) followed by Tukey’s HSD test was performed to detect statistical differences (p ≤ 0.05) among peptides in the samples as a function of ripening time. The SIMCA 16.0.1 (Sartorius Stedim Data Analytics, Göttingen, Germany) software was used to create a principal component analysis (PCA) biplot to get a visual interpretation of the analyzed data.
The peptide profile of 72 PR cheeses collected from six different dairies (A–F) throughout the PR area, at different ripening times (0, 1, 2, 6, 7, 9, 12, and 24 months) was analyzed after simulated digestion to detect peptides, and in particular, BP, released after the passage in the first part of the GIT. After in vitro digestion, 105 different peptides and 13 NPADs were detected. Out of 105 identified peptides, 41 were derived from α-S1-casein and 46 from β-casein. Regarding α-S1-casein, the shortest peptides are composed of 2 amino acids and the longest has 14 amino acids, with an average length of 6 amino acids. On the other hand, the peptides derived from β-casein are composed of peptides among which the shortest are dipeptides, and the longest have 16 amino acids, with an average length of 5 amino acids. Eighteen out of 105 of the detected peptides derived from α-S2-casein and whey proteins.
Regarding casein digestibility, our results indicate that β-casein is the PR protein most prone to proteolysis, with coverage close to 68%. No peptides derived from regions f(15–43) and f(154–169) were detected, probably indicating that these two regions of the protein are more resistant to proteolysis. A -S1-casein was also highly digestible, with a coverage of 61%; the regions most resistant to proteolytic digestion are those at the N-term of the protein, f(1–16) and f(42–69). Instead, fewer peptides were found from α-S2-casein and κ-casein. This could be due both to a higher resistance of those fractions to proteolysis compared to α-S1-casein and β-casein, and their lower content in PR. Casein coverage after simulated gastrointestinal digestion is shown in Supplementary Figure 1. Clear cleavage sites could not be identified. In fact, pepsin preferentially cleaves in the P1 or P1’ position at Phe, Tyr, Trp, and Leu, but this specificity is lost at pH ≥ 2. Pancreatin is a mixture of different digestive enzymes produced by the exocrine cells of the pancreas. It is a broad-spectrum protease composed of amylase, trypsin, lipase, ribonuclease, and protease. Hence, the complexity of the proteolytic mixture really broadens the possible spectrum of cleavage sites. The principal component analysis (PCA) was performed to investigate possible correlations among peptides detected after digestion and the ripening time of samples (Figure 2).
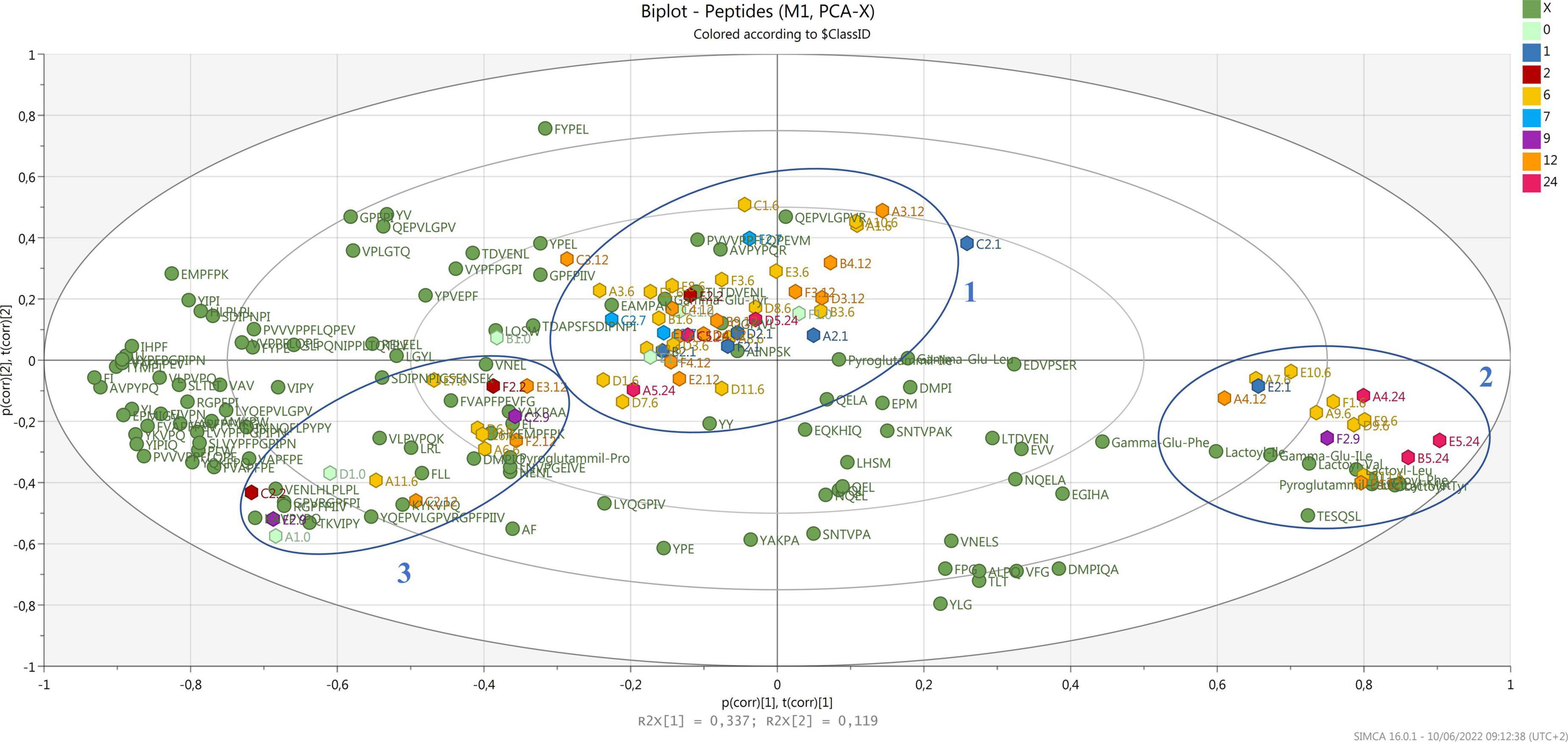
Figure 2. Biplot of samples (hexagons) and all detected peptides (green dots). Samples are colored by ripening time.
In the biplot, all the detected peptides and the NPADs (green dots) are reported together with the samples (hexagons) which are differently colored by months of ripening. Samples tend to create a big cluster (cluster 1) in the center of the biplot, where PR from all the dairies of origin and ripening times are present. Most samples (9 out of 16) of 12 months, the minimum ripening time for a PR cheese to be sold, are included in this cluster. A second cluster (cluster 2), characterized by the majority of detected NPAD, can be observed at the bottom right-hand side, where 7 samples of 6 months are present together with 3 samples of 24 months, 2 of 12 months, and 1 of 9 months. Finally, 1 more cluster (cluster 3) can be observed at the bottom left-hand side, grouping samples of different ripening times, and produced in different dairies. These data suggest that after digestion, samples share a common core of a few peptides, while on the other hand, most peptides are widely distributed along the two components, and they do not characterize a specific ripening time. This is not surprising as during ripening the profile of cheese changes a lot because of microbial dynamics (Bottari et al., 2020), with peptides of different sizes and sequences characterizing each time point. Thus, the peptides that after digestion are common to several ripening stages may have been released by both proteins and already hydrolyzed peptides with similar specific cleavage sites (Bottari et al., 2017). Differently from what was previously observed (Bottari et al., 2020), after digestion, samples do not cluster strongly according to the ripening time but are more heterogeneously distributed along the PCOs, as for the 12-month-old PR that is included in all the three clusters. It is known that the most relevant proteolytic transformations in PR cheese occur in the first 12 months (Sforza et al., 2009; Bottari et al., 2020) with a huge peptide evolution, thus peptides at this stage can present many different cleavage sites for digestive enzymes. On the contrary, the majority of NPADs are very strongly related to each other as they have been shown to be perfectly stable to gastrointestinal digestion (Bottesini et al., 2014). These compounds accumulate during ripening, reaching their maximum amount at the end of cheese aging (Sforza et al., 2009). In fact, they were associated with the longer ripened PR varieties of cheese in a former study (Bottari et al., 2020), where all the PR samples from 7 to 24 months clustered with the identified NPAD. However, in this study, where the same samples were analyzed after simulated digestion, only some of the longer ripened cheese samples resulted strongly associated with the majority of the identified NPADs, particularly lactoyl-amino acids. This seems to suggest that the presence of different NPADs is peculiar to different types of cheese.
Interestingly, we observed that samples of 12–24 months of ripening present in cluster 2 have been produced during winter, while those present in cluster 1 were also from summer production (Supplementary Table 1). Seasonal variability in PR production has been previously investigated, both for milk and natural whey starter, although still not exhaustively explained due to the many factors that can affect it (Gatti et al., 2014; Franceschi et al., 2019). In particular, the microbial composition of raw milk has been recently correlated to season (Montel et al., 2014; Garroni et al., 2020). Garroni et al. (2020) showed that lactic acid bacteria were variable from summer to winter, but with a higher abundance of Lacticaseibacillus species in winter samples. Considering that this genus, which is coming from raw milk, is dominant in PR cheese up to 20 months of ripening (Neviani et al., 2009) and it has been reported to be responsible for the accumulation of NPADs during PR ripening (Bottari et al., 2020), the correlation found in the present study among PR samples produced in winter and the NPADs in cluster 2 could be due to a higher abundance of Lacticaseibacillus species. On the other hand, no specific correlation has been found between ripening and peptides in clusters 1 and 3 despite for those peptides located in the center of cluster 1, namely AINPSK [α-S2-CN f(27-32)], TDVENL [β-CN f(128-133)], γ-Glu-Tyr and QGPIVL [α-S2-CN f(101-106)]. These peptides were in fact more strongly correlated with 12- and 24-months ripened PR that were produced during summer.
As the majority of identified peptides are derived from α-S1-casein and β-casein, two separate PCA were run on data, to evaluate the distribution of the samples and peptides coming from the two native proteins according to ripening time.
Most samples were grouped according to component 2, correlating with few peptides released after digestion that were derived from α-S1-casein (Figure 3), with no relation to ripening time and area of production. Considering that the same samples clustered according to ripening time when analyzed before digestion (Bottari et al., 2020), the result obtained confirms that in vitro digestion greatly influences the peptide profile of PR cheese (Martini et al., 2021). It must be noticed that several samples are grouped together on the right side of the plot, opposite to most peptides derived from α-S1-casein. Most of these peptides contain a proline that is known to increase the resistance to gastro-pancreatic protease action (Tagliazucchi et al., 2016).
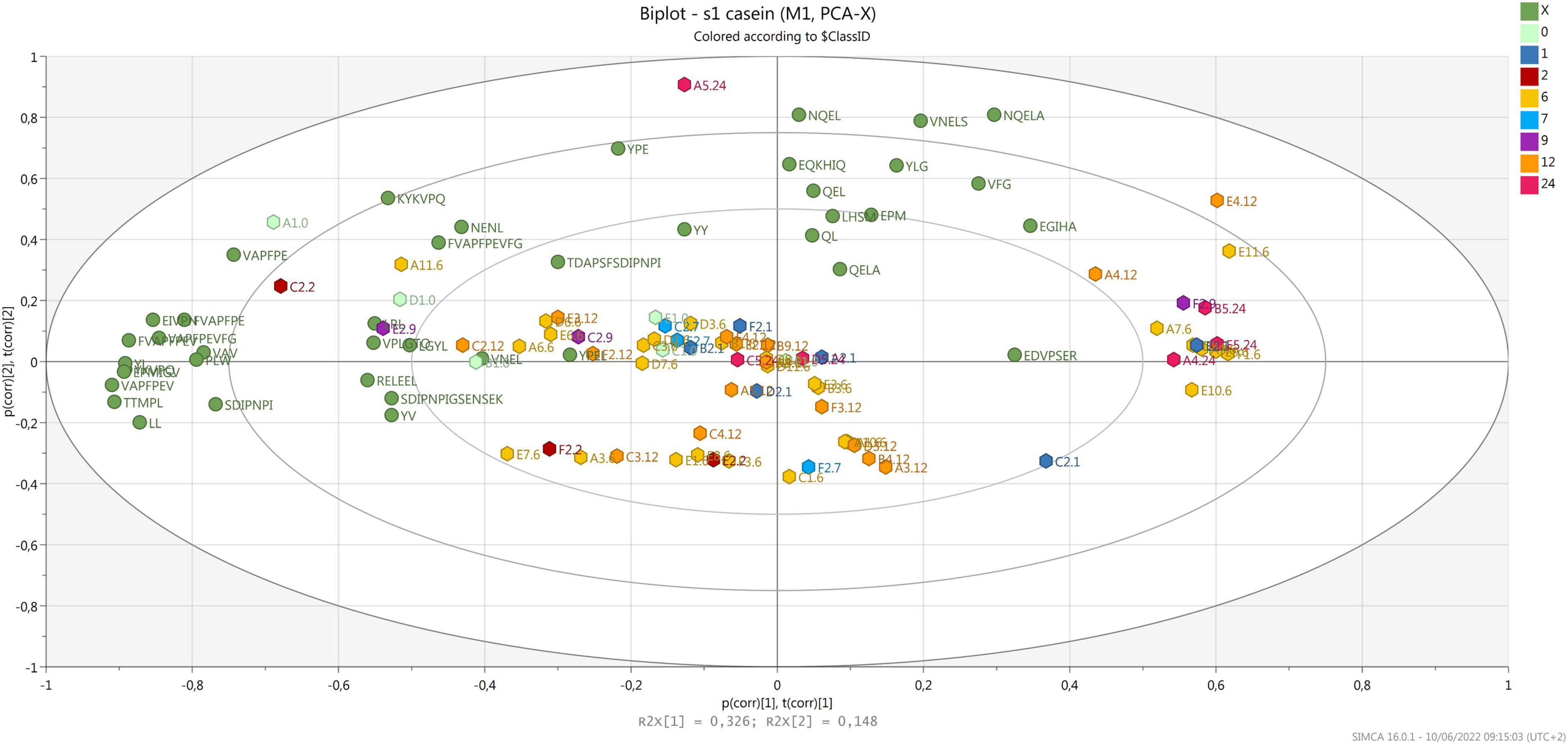
Figure 3. Biplot of samples (hexagons) and peptides derived from α-S1-casein (green dots). Samples are colored by ripening time.
Figure 4 shows the biplot with the variance explained for the peptides released after digestion, which are derived from β-casein. In this case, the samples are grouped in three different clusters. Few peptides, characterize one central cluster, where most samples are, meaning that those peptides are released from all cheese samples after digestion, independently from the ripening time and the dairy of origin. Interestingly, most of the 12-month ripened cheese and 2 of the 24-month ripened samples were grouped together in this cluster. A second cluster, characterized by several peptides, is present on the bottom left side of the biplot, where samples from 2 to 12 months of ripening group. Three samples derived from the same cheesemaking (dairy C, same wheel sampled at 2, 9, and 12 months of ripening) are present in this cluster, suggesting that the peptide profile originating from the first biochemical events and evolving during ripening, show a common trend after digestion. Finally, 12 samples mainly from 6 months of ripening group in a third cluster on the right-hand side of the plot, opposite to most peptides derived from β-casein. Also, in this case, these peptides contain a PXP sequence or a proline residue near the carboxylic end, which increases the resistance to gastro-pancreatic proteases action (Tagliazucchi et al., 2016; Martini et al., 2020).
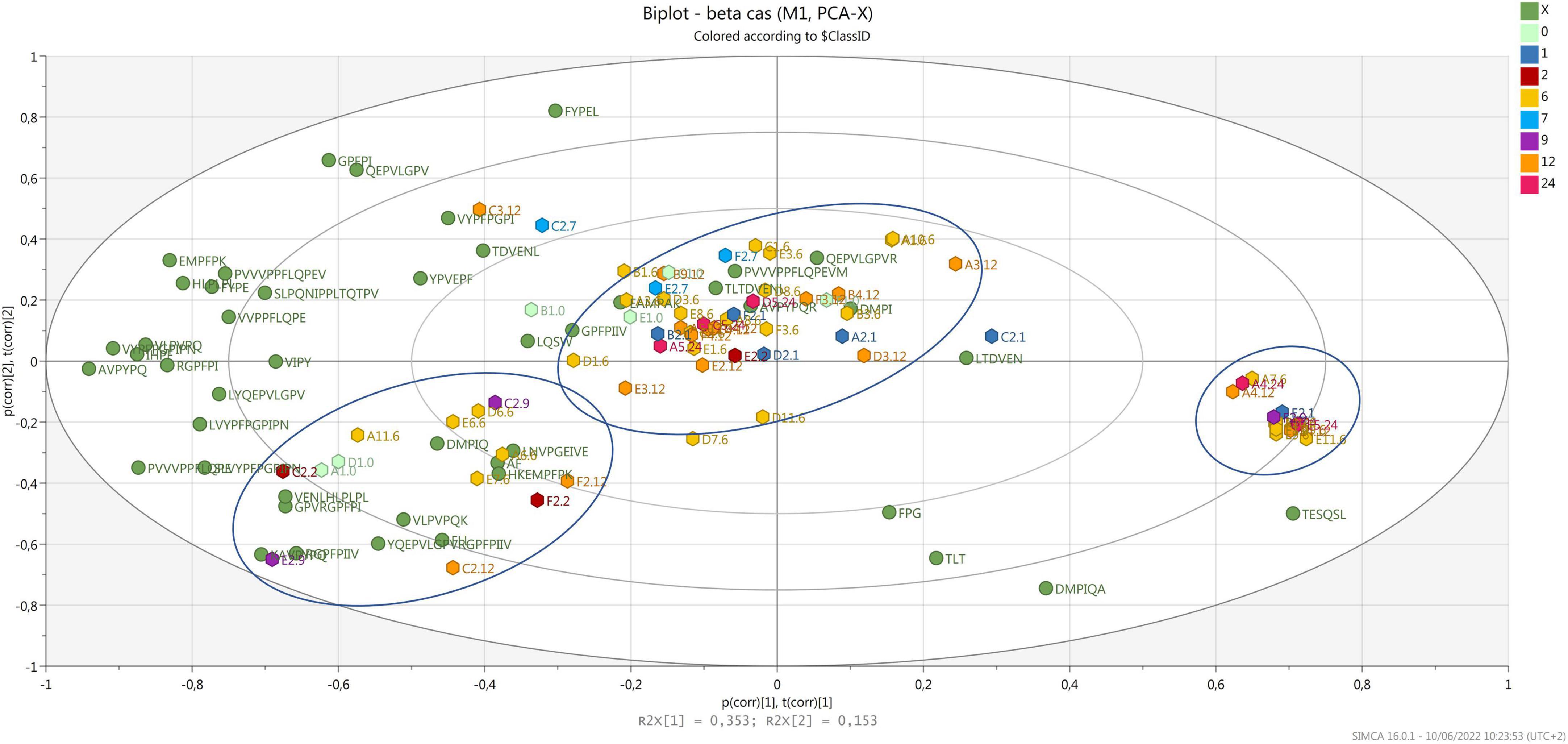
Figure 4. Biplot of samples (hexagons) and peptides d from β-casein (green dots). Samples are colored by ripening time.
The presence of potential BPs was investigated for both undigested (Table 1) and digested cheese samples (Table 2). Analyzing the 34 peptide sequences previously identified in the same PR varieties of cheese before digestion, four potential BPs, and three NPADs were found. One possible BP was derived from α-S1-casein while the other three originated from β-casein. As shown in Table 1, the functionality of BPs present in the PR varieties of cheese of different ripening times, can be multiple, with a single peptide expressing many bioactivities. In fact, 2 of these peptides are reported to exert more than one bioactivity, specifically, the peptide RPKHPIKHQGLPQEVLNENLLRF [α-S1-CN f(1-23)] that was derived from α-S1-casein shows immunomodulatory and antimicrobial activities. However, this potentially bioactive sequence was found only in the peptide profile of PR curd samples. This is not surprising as the length of the peptide makes it easily susceptible to proteolysis in the further ripening stages. On the other hand, the peptides YQEPVLGPVRGPFPIIV [β-CN f(193-209)], DKIHPF [β-CN f(47-52)], and RELEEL [β-CN f(1-6)], respectively, reported as immunomodulator/antithrombin/antimicrobial/ACE-inhibitor, ACE-inhibitor, and antioxidant, were found in almost all the considered ripening stages, albeit in a different amount, with the latter particularly present in 6 months ripened cheeses (data not shown).
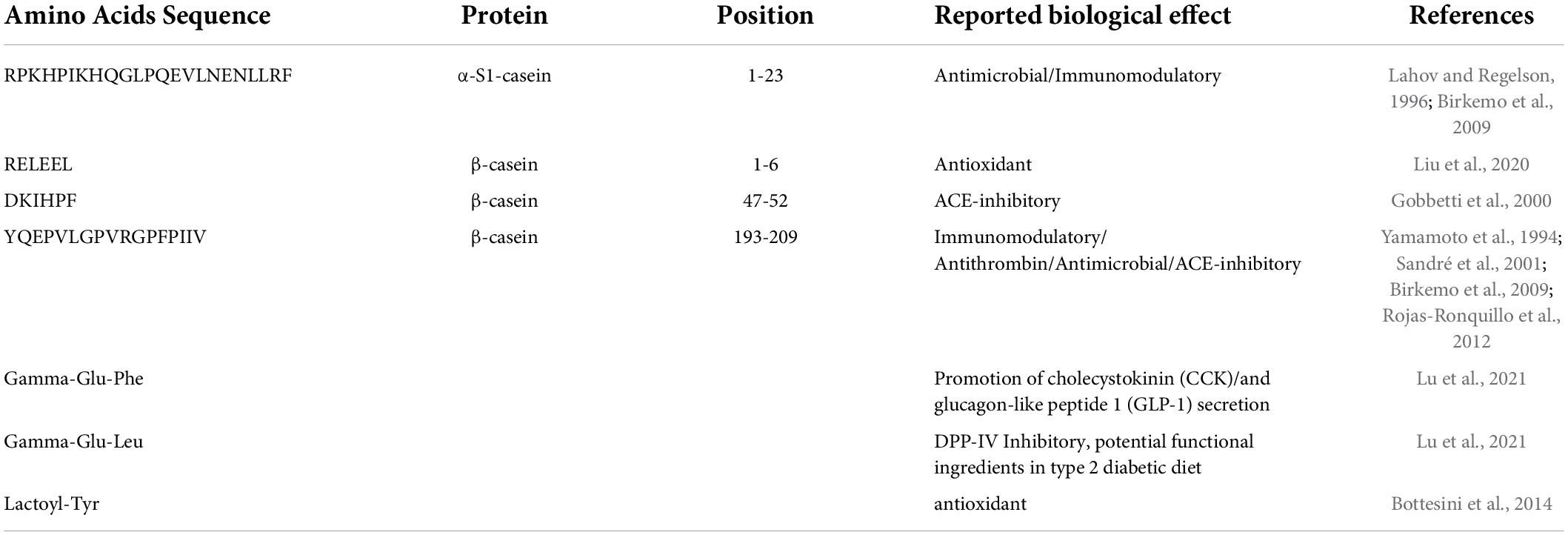
Table 1. List of reported bioactive peptides present in undigested Parmigiano Reggiano (PR) cheese samples.
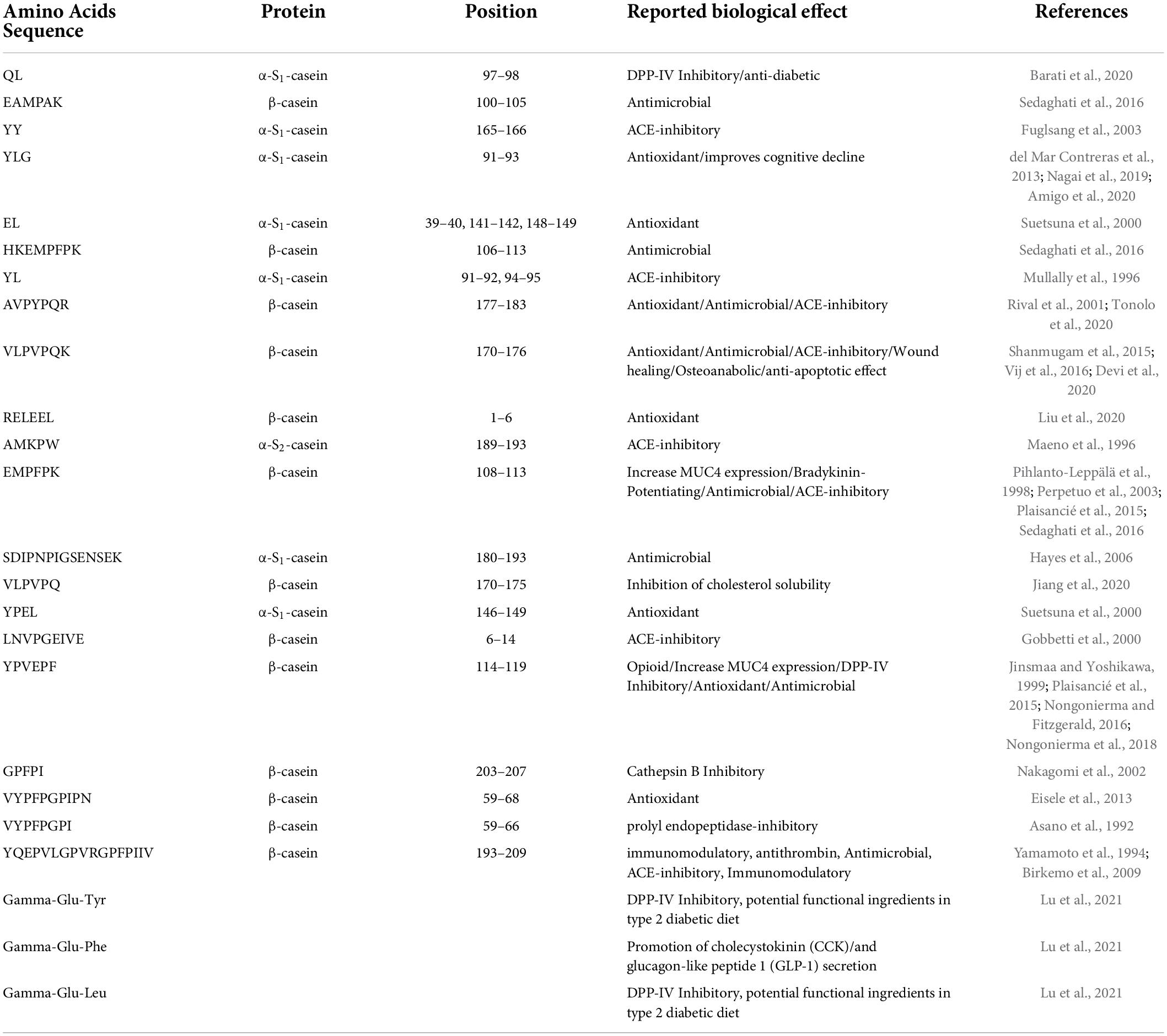
Table 2. List of the best-known bioactive peptides detected in PR cheese samples after in vitro simulated digestion.
Regarding the analysis of potential BP after digestion, 21 potential BPs and 3 NPADs were detected. Out of these 21 potential BPs, 13 were derived from β-casein, 7 from α-S1-casein, and 1 from α-S2-casein (Table 2). Some of them, specifically 7 out of 20, seem to exert more than one functionality. All the NPADs show more than 1 functionality. Among the range of bioactivities reported for bioactive peptides, one of the most studied features is the ability to inhibit angiotensin-converting enzyme (ACE), preventing a sharp rise in blood pressure, and limiting the risk of heart failure and stroke. Eight BPs {YY [α-S1-CN f(165-166)], YL, AVPYPQR [β-CN f(177-183)], VLPVPQK [β-CN f(170-176)], AMKPW [α-S2-CN f(189-193)], EMPFPK [β-CN f(108-113)], LNVPGEIVE [β-CN f(6-14)], and YQEPVLGPVRGPFPIIV [β-CN f(193-209)]} known for their ACE inhibitory effect (Minervini et al., 2003; Daliri et al., 2018) were detected in the analyzed cheese samples after digestion. One of the BPs {EMPFPK [β-CN f(108-113)]} detected possesses a potential bradykinin-enhancing effect, which in combination with the ACE-inhibitory effect helps to maintain a regular blood pressure level (Castellone et al., 2021). One peptide (QL) is reported to exert antidiabetic effects (Barati et al., 2020). Other known bioactivities attributed to BPs found in the samples include opioid effect and delayed cognitive decline along with inhibition of prolyl-endopeptidase and cathepsin B. These latter effects, combined, could be useful in delaying the progression of tumors (Lee and Lee, 2000; Perez et al., 2020) and fighting the onset and development of Parkinson’s disease (Kalinina et al., 2019). A potential beneficial effect of cheese consumption on cardiovascular health related to the presence of potential BP, or their release after digestion, had already been hypothesized by other authors (Petyaev and Bashmakov, 2012; Hjerpsted and Tholstrup, 2016; Sultan et al., 2018) and for PR (Basiricò et al., 2015; Martini et al., 2020). However, further studies to establish whether the absorption of these peptides in the body is sufficient to develop the bioactive effect are needed (Hjerpsted and Tholstrup, 2016; Santiago-López et al., 2018).
In our experiments, only four potentially BPs and three NPADs were detected in undigested cheese samples, while among the 105 different peptides revealed by the analysis of digested cheese samples, 21 peptides and 3 NPADs showed at least one bioactivity as reported by the Milk Bioactive Peptides Database (MBDP). This is in agreement with the literature that reports a higher number of BP released after digestion than undigested food (Egger and Ménard, 2017; Giromini et al., 2019; Fernández-Tomé and Hernández-Ledesma, 2020; Di Nunzio et al., 2022). Digestion is a key step to freeing BPs from their cryptic form and increasing the probability of having a positive effect exerted by the food components. Only one BP sequence, namely YQEPVLGPVRGPFPIIV [β-CN f(193-209)], was found both before and after digestion meaning that neither digestive enzymes nor gastric acids can hydrolyze it. Indeed, this multifunctional peptide is known to be resistant to digestion, due to a particular conformation (Rojas-Ronquillo et al., 2012). Other BP found in undigested PR cheese does not resist simulated digestion, although some bioactive fragments found in digested cheese may result from larger bioactive sequences found in undigested PR cheese (Liu and Pischetsrieder, 2017; Martini et al., 2020). For example, the antioxidant peptide “EL” in digested PR cheese may be derived from “RELEEL” [β-CN f(1-6)], a peptide with the same reported bioactivity, present in undigested cheese.
Table 3 shows the semi-quantitative data of peptides known as bioactive released during the digestion of differently ripened PR cheese samples. BPs show different trends: some of them display a constant increasing or decreasing trend according to ripening time, while others increase until reaching a plateau, then start to decrease with aging time. It is also fundamental to consider the effect of digestion in releasing encrypted peptides that otherwise are locked in bigger and not active sequences (Udenigwe et al., 2013).
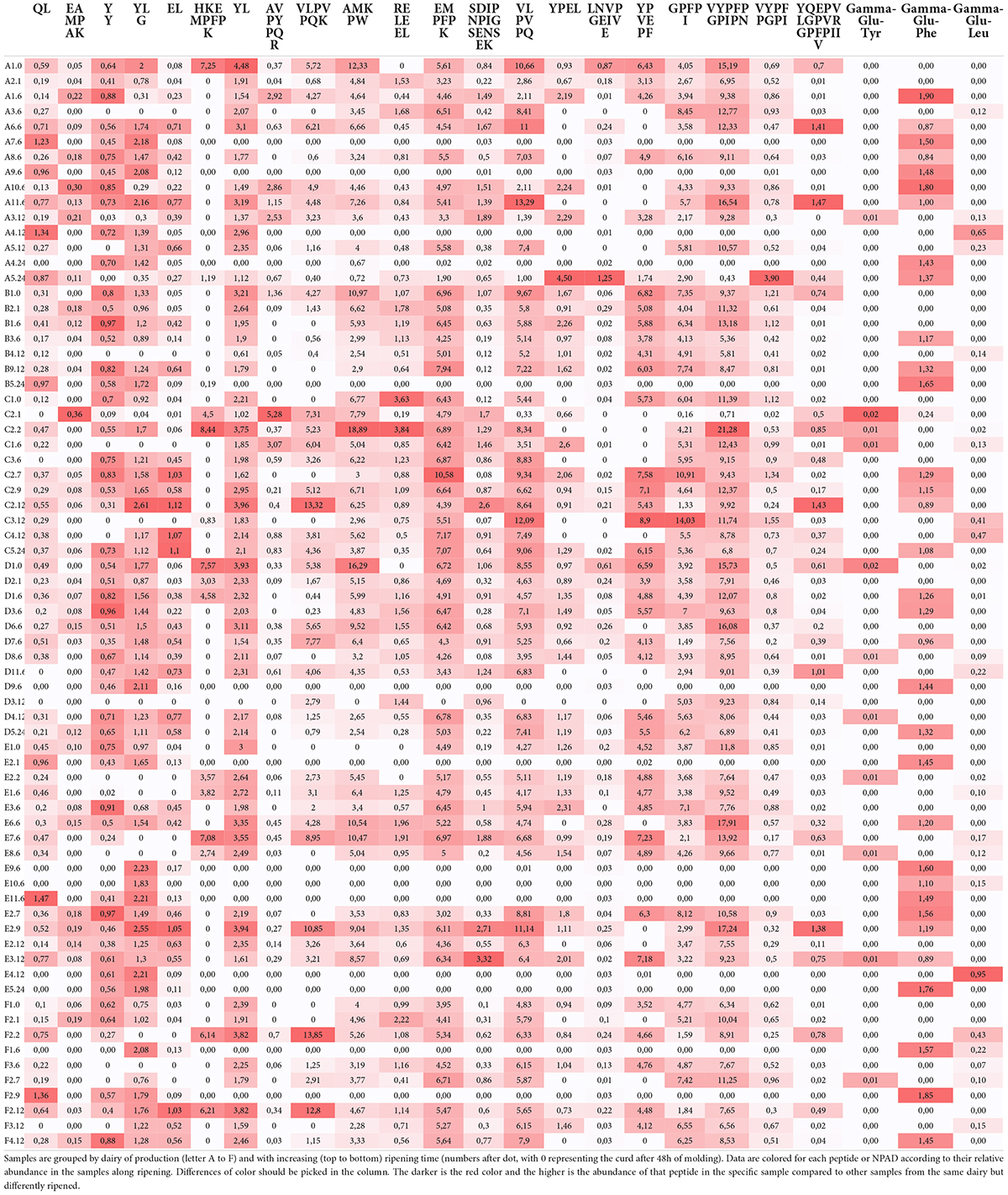
Table 3. Relative quantification of potential bioactive peptides (BPs) in PR cheese with different ripening times after in vitro simulated human digestion.
To obtain a statistical value for these observations, semi-quantification data were analyzed by Pearson’s correlation and ANOVA. The results are reported in Table 4. Statistical analysis shows that peptide “EL” and the NPAD Gamma-glu-phe follow an increasing trend in digested samples according to ripening time. On the contrary, “HKEMPFPK” [β-CN f(105-113)], “YL,” “AMKPW” [α-S2-CN f(189-193)], and “VYPFPGPIPN” [β-CN f(59-68)] follow a decreasing trend in longer ripened kinds of cheese after digestion. In fact, within their sequences, there are different cleavage sites of digestive enzymes (e.g., K, Y, M, and W). The ANOVA analysis confirms that the presence of BPs was statistically different among the samples for 9 out of 20 detected potentially as BPs.
Since PDO regulation allows the PR cheese to be sold only after a minimum of 12 months of ripening, differences in BP presence were then searched between 12- and 24-months ripened cheeses. Results showed higher amounts (p-value of 0.03) of potential BPs in 24 months ripened samples compared to 12 months ripened ones. This is in agreement with other authors (Martini et al., 2021) and can be explained by the higher proteolytic activity of lactic acid bacteria exerted in the longer ripened cheeses (Bottari et al., 2020).
Potential bioactivities of digested cheese samples resulting from BP profiles showed small variations based on ripening time (Figure 5). According to the graph, the main bioactivities possibly present at each ripening time are antimicrobial, ACE-inhibitor, and antioxidant. These three bioactivities make up almost 60% of the total bioactivities reported for each single time point. These results agree with Solieri et al. (2020), who reported ACE inhibitory and antimicrobial activities as the most present in PR samples. During digestion, peptides evolve, and their relative quantity increases or decreases, giving rise to changes in the associated bioactivities as well. However, it is not always possible to obtain the maximum beneficial effect from the potential BP present in the PR cheese, since, as indicated by the EU PDO regulation, the PR cheese can only be sold after 12 months of ripening while some bioactivities have the highest peak before reaching the minimum ripening time to be marketed (Figure 5).
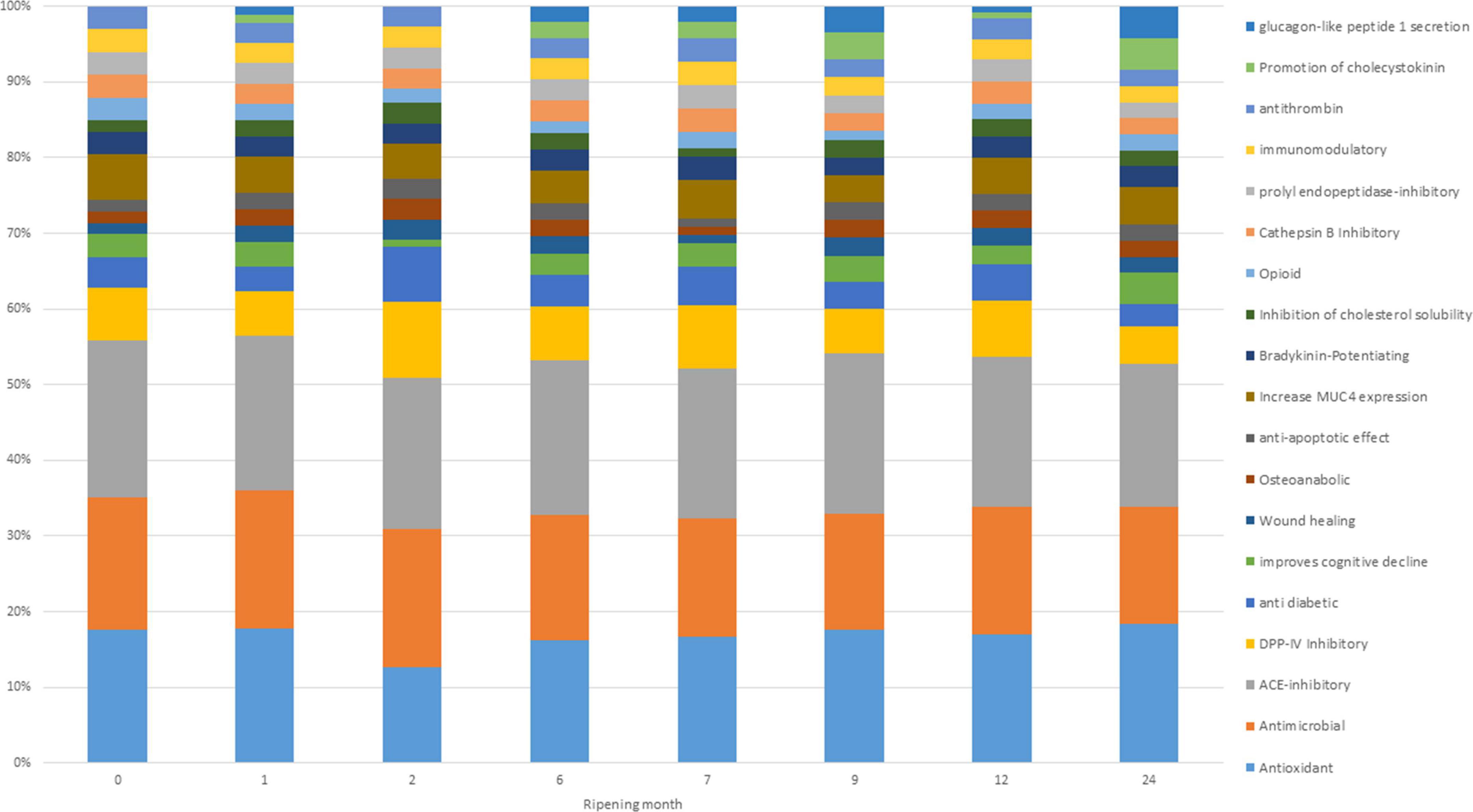
Figure 5. Frequency of identification (in percentage) of bioactivities exerted by detected bioactive peptides (BPs) and non-proteolytic amino acyl derivatives (NPADs) at different ripening times.
The link between dairy intake and health benefits is not entirely clear. Many studies in the scientific literature try to evaluate the production of BP and its absorption in the human body. The big drawback to these experiments is the difficulty in studying the effects in vivo. In fact, many of these works concern in vitro, in silico, or animal approaches (Fu et al., 2016; Cao et al., 2017; Li et al., 2018).
The regular introduction of dairy products into the diet is suggested by many nutritional guidelines for maintaining a healthy diet. The indications also suggest introducing dairy products, especially long-ripened ones, more than once a week. The consumption of long-ripened cheeses has long been debated due to possible negative effects, such as a high content of salt and saturated fats, well known to be linked to an increased risk of heart failure and cardiovascular disease, together with other compounds with possible positive effects. Despite this, diets that involve high consumption of cheese, such as the Mediterranean and French diets, have been shown to reduce the risk of developing non-communicable diseases such as heart attacks and angina pectoris (Petyaev and Bashmakov, 2012; Higurashi et al., 2016; Lallès, 2016), suggesting that somehow the positive outcomes of a moderate but constant introduction to the diet of long-ripened cheeses may outweigh the risk posed by the high introduction of salt, saturated fat, and cholesterol (Petyaev and Bashmakov, 2012; Hirahatake et al., 2020; Feeney et al., 2021).
The study of the presence of BP in cheese must take into account not only the production, release, and therefore the presence of peptides in the cheese, but also the mechanisms of release and absorption by the intestinal wall cells (Lorenzo et al., 2018; Sultan et al., 2018). Since many studies in the literature reporting the positive effect of BP are conducted in vitro, BP should be tested for its effect in vivo.
The complexity of enzymatic activities that occur during the production of long-ripened cheese has been widely described. This also applies to PR cheese, which despite its restricted geographical area of production is known almost all over the world. Further steps have recently been taken in the literature regarding the understanding of what happens to this treasure of nutrients and possibly bioactive compounds generated after digestion. With this study, conducted on a large number of PR cheese samples with different ripening times and produced by different dairies, we were able to expand what was previously known about the effect of gastrointestinal digestion on the peptide profile of cheese during ripening. The presence of potential BP in samples before and after digestion was also investigated. Samples treated with simulated digestion were characterized by peptide profiles that shared few common peptides, regardless of the ripening time and dairy farm of production. On the other hand, most peptides were released during the digestion of specific samples. The peptide profile is similar after digestion for most PR varieties of cheese at the required minimum ripening time of 12 months. However, differences could be observed among samples of both 12 and 24 months of ripening, possibly linked to the season of production of the cheese. Longer ripened PR varieties of cheese produced during winter showed a stronger relation with NPADs. As the production of these compounds was previously reported to be associated with specific microbial species, such as Lacticacaseibacillus spp., further studies are needed to investigate this aspect. As for the potential BP revealed in the samples, they increased with digestion, particularly for long-ripened PR cheeses. Any differences related to the production of dairy could be considered to guide and predict proteolysis from the milk to cheese ready for consumption.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
VC and BP: analysis. VC, BP, EB, and BB: conceptualization, statistical analysis, and writing. MG and TT: critical review, writing, and editing. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.966239/full#supplementary-material
Amigo, L., Martinez-Maqueda, D., and Hernandez-Ledessma, B. (2020). In silico and in vitro analysis of multifunctionality. Foods 9:991.
Ardö, Y., Pripp, A. H., and Lillevang, S. K. (2009). Impact of heat-treated Lactobacillus helveticus on bioactive peptides in low-fat, semi-hard cheese. Aust. J. Dairy Technol. 64, 58–62.
Asano, M., Nio, N., and Ariyoshi, Y. (1992). Inhibition of prolyl endopeptidase by synthetic β-casein peptides and their derivatives with a C-terminal prolinol or prolinal. Biosci. Biotechnol. Biochem. 56, 976–977.
Barati, M., Javanmardi, F., Jabbari, M., Mokari-Yamchi, A., Farahmand, F., Eş, I., et al. (2020). An in silico model to predict and estimate digestion-resistant and bioactive peptide content of dairy products: A primarily study of a time-saving and affordable method for practical research purposes. Lwt 130:109616. doi: 10.1016/j.lwt.2020.109616
Basiricò, L., Catalani, E., Morera, P., Cattaneo, S., Stuknytë, M., Bernabucci, U., et al. (2015). Release of angiotensin converting enzyme-inhibitor peptides during in vitro gastrointestinal digestion of Parmigiano Reggiano PDO cheese and their absorption through an in vitro model of intestinal epithelium. J. Dairy Sci. 98, 7595–7601. doi: 10.3168/jds.2015-9801
Birkemo, G. A., O’Sullivan, O., Ross, R. P., and Hill, C. (2009). Antimicrobial activity of two peptides casecidin 15 and 17, found naturally in bovine colostrum. J. Appl. Microbiol. 106, 233–240. doi: 10.1111/j.1365-2672.2008.03996.x
Bottari, B., Levante, A., Bancalari, E., Sforza, S., Bottesini, C., Prandi, B., et al. (2020). The interrelationship between microbiota and peptides during ripening as a driver for Parmigiano Reggiano cheese quality. Front. Microbiol. 11:581658. doi: 10.3389/fmicb.2020.581658
Bottari, B., Quartieri, A., Prandi, B., Raimondi, S., Leonardi, A., Rossi, M., et al. (2017). Characterization of the peptide fraction from digested Parmigiano Reggiano cheese and its effect on growth of lactobacilli and bifidobacteria. Int. J. Food Microbiol. 255, 32–41. doi: 10.1016/j.ijfoodmicro.2017.05.015
Bottesini, C., Tedeschi, T., Dossena, A., and Sforza, S. (2014). Enzymatic production and degradation of cheese-derived non-proteolytic aminoacyl derivatives. Amino Acids 46, 441–447. doi: 10.1007/s00726-013-1637-3
Boukid, F., Prandi, B., Faccini, A., and Sforza, S. (2019). A complete mass spectrometry (MS)-based peptidomic description of gluten peptides generated during in vitro gastrointestinal digestion of durum wheat: Implication for celiac disease. J. Am. Soc. Mass Spectrom. 30, 1481–1490. doi: 10.1007/s13361-019-02212-8
Brodkorb, A., Egger, L., Alminger, M., Alvito, P., Assunção, R., Ballance, S., et al. (2019). INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 14, 991–1014. doi: 10.1038/s41596-018-0119-1
Cao, Y., Miao, J., Liu, G., Luo, Z., Xia, Z., Liu, F., et al. (2017). Bioactive peptides isolated from casein phosphopeptides enhance calcium and magnesium uptake in Caco-2 cell monolayers. J. Agric. Food Chem. 65, 2307–2314. doi: 10.1021/acs.jafc.6b05711
Castellone, V., Bancalari, E., Rubert, J., Gatti, M., Neviani, E., and Bottari, B. (2021). Eating fermented: Health benefits of LAB-fermented foods. Food 10, 2639. doi: 10.3390/foods10112639
Coppola, R., Nanni, M., Iorizzo, M., Sorrentino, A., Sorrentino, E., Chiavari, C., et al. (2000). Microbiological characteristics of Parmigiano Reggiano cheese during the cheese making and the first months of the ripening. Lait 80, 479–490. doi: 10.1051/lait:2000139
Daliri, E. B. M., Lee, B. H., Park, B. J., Kim, S. H., and Oh, D. H. (2018). Antihypertensive peptides from whey proteins fermented by lactic acid bacteria. Food Sci. Biotechnol. 27, 1781–1789. doi: 10.1007/s10068-018-0423-0
del Mar Contreras, M., Sanchez, D., Sevilla, M. Á., Recio, I., and Amigo, L. (2013). Resistance of casein-derived bioactive peptides to simulated gastrointestinal digestion. Int. Dairy J. 32, 71–78. doi: 10.1016/j.idairyj.2013.05.008
Devi, S., Pal, G. K., Kapila, R., and Kapila, S. (2020). C-terminal sequence deletion effect on antioxidative characteristics of VLPVPQK bioactive peptide from buffalo milk casein. Lwt 119:108816. doi: 10.1016/j.lwt.2019.108816
Di Nunzio, M., Loffi, C., Chiarello, E., Dellafiora, L., Picone, G., Antonelli, G., et al. (2022). Impact of a shorter brine soaking time on nutrient bioaccessibility and peptide formation in 30-months-ripened Parmigiano Reggiano cheese. Molecules 27:664. doi: 10.3390/molecules27030664
Egger, L., and Ménard, O. (2017). Update on bioactive peptides after milk and cheese digestion. Curr. Opin. Food Sci. 14, 116–121. doi: 10.1016/j.cofs.2017.03.003
Eisele, T., Stressler, T., Kranz, B., and Fischer, L. (2013). Bioactive peptides generated in an enzyme membrane reactor using Bacillus lentus alkaline peptidase. Eur. Food Res. Technol. 236, 483–490. doi: 10.1007/s00217-012-1894-5
Feeney, E. L., Lamichhane, P., and Sheehan, J. J. (2021). The cheese matrix: Understanding the impact of cheese structure on aspects of cardiovascular health – a food science and a human nutrition perspective. Int. J. Dairy Technol. 74, 656–670. doi: 10.1111/1471-0307.12755
Fernández-Tomé, S., and Hernández-Ledesma, B. (2020). Gastrointestinal digestion of food proteins under the effects of released bioactive peptides on digestive health. Mol. Nutr. Food Res. 64:2000401. doi: 10.1002/mnfr.202000401
Franceschi, P., Malacarne, M., Formaggioni, P., Cipolat-Gotet, C., Stocco, G., and Summer, A. (2019). Effect of season and factory on cheese-making efficiency in Parmigiano Reggiano manufacture. Foods 8:315. doi: 10.3390/foods8080315
Fu, Y., Wu, W., Zhu, M., and Xiao, Z. (2016). In silico assessment of the potential of patatin as a precursor of bioactive peptides. J. Food Biochem. 40, 366–370. doi: 10.1111/jfbc.12213
Fuglsang, A., Nilsson, D., and Nyborg, N. C. B. (2003). Characterization of new milk-derived inhibitors of angiotensin converting enzyme in vitro and in vivo. J. Enzyme Inhib. Med. Chem. 18, 407–412. doi: 10.1080/1475636031000138723
Garroni, E., Doulgeraki, A. I., Pavli, F., Spiteri, D., and Valdramidis, V. P. (2020). Characterization of indigenous lactic acid bacteria in cow milk of the Maltese Islands: A geographical and seasonal assessment. Microorganisms 8:812. doi: 10.3390/microorganisms8060812
Gatti, M., Bottari, B., Lazzi, C., Neviani, E., and Mucchetti, G. (2014). Invited review: Microbial evolution in raw-milk, long-ripened cheeses produced using undefined natural whey starters. J. Dairy Sci. 97, 573–591. doi: 10.3168/jds.2013-7187
Gazme, B., Boachie, R. T., Tsopmo, A., and Udenigwe, C. C. (2019). Occurrence, properties and biological significance of pyroglutamyl peptides derived from different food sources. Food Sci. Hum. Wellness 8, 268–274. doi: 10.1016/j.fshw.2019.05.002
Giromini, C., Cheli, F., Rebucci, R., and Baldi, A. (2019). Invited review: Dairy proteins and bioactive peptides: Modeling digestion and the intestinal barrier. J. Dairy Sci. 102, 929–942. doi: 10.3168/jds.2018-15163
Gobbetti, M., Ferranti, P., Smacchi, E., Goffredi, F., and Addeo, F. (2000). Production of angiotensin-I-converting-enzyme-inhibitory peptides in fermented milks started by Lactobacillus delbrueckii subsp. bulgaricus SS1 and Lactococcus lactis subsp. cremoris FT4. Appl. Environ. Microbiol. 66, 3898–3904. doi: 10.1128/AEM.66.9.3898-3904.2000
Godos, J., Tieri, M., Ghelfi, F., Titta, L., Marventano, S., Lafranconi, A., et al. (2020). Dairy foods and health: An umbrella review of observational studies. Int. J. Food Sci. Nutr. 71, 138–151. doi: 10.1080/09637486.2019.1625035
Górska-Warsewicz, H., Rejman, K., Laskowski, W., and Czeczotko, M. (2019). Milk and dairy products and their nutritional contribution to the average polish diet. Nutrients 11:1771. doi: 10.3390/nu11081771
Gragnani, M. (2013). The EU regulation 1151/2012 on quality schemes for agricultural products and foodstuffs. Eur. Food Feed Law Rev. 8, 376–385.
Hayes, M., Ross, R. P., Fitzgerald, G. F., Hill, C., and Stanton, C. (2006). Casein-derived antimicrobial peptides generated by Lactobacillus acidophilus DPC6026. Appl. Environ. Microbiol. 72, 2260–2264. doi: 10.1128/AEM.72.3.2260-2264.2006
Higurashi, S., Ogawa, A., Nara, T. Y., Kato, K., and Kadooka, Y. (2016). Cheese consumption prevents fat accumulation in the liver and improves serum lipid parameters in rats fed a high-fat diet. Dairy Sci. Technol. 96, 539–549. doi: 10.1007/s13594-016-0288-z
Hirahatake, K. M., Astrup, A., Hill, J. O., Slavin, J. L., Allison, D. B., and Maki, K. C. (2020). Potential cardiometabolic health benefits of full-fat dairy: The evidence base. Adv. Nutr. 11, 533–547. doi: 10.1093/advances/nmz132
Hjerpsted, J., and Tholstrup, T. (2016). Cheese and cardiovascular disease risk: A review of the evidence and discussion of possible mechanisms. Crit. Rev. Food Sci. Nutr. 56, 1389–1403. doi: 10.1080/10408398.2013.769197
Jiang, X., Pan, D., Zhang, T., Liu, C., Zhang, J., Su, M., et al. (2020). Novel milk casein–derived peptides decrease cholesterol micellar solubility and cholesterol intestinal absorption in Caco-2 cells. J. Dairy Sci. 103, 3924–3936. doi: 10.3168/jds.2019-17586
Jinsmaa, Y., and Yoshikawa, M. (1999). Enzymatic release of neocasomorphin and β-casomorphin from bovine β-casein. Peptides 20, 957–962. doi: 10.1016/S0196-9781(99)00088-1
Kalinina, A. P., Kapitsa, I. G., Ivanova, E. A., and Voronina, T. A. (2019). Antiparkinsonian action of the prolyl endopeptidase inhibitor in mice. Moscow Univ. Biol. Sci. Bull. 74, 69–74. doi: 10.3103/S0096392519020044
Lahov, E., and Regelson, W. (1996). Antibacterial and immunostimulating casein-derived substances from milk: Casecidin, isracidin peptides. Food Chem. Toxicol. 34, 131–145. doi: 10.1016/0278-6915(95)00097-6
Lallès, J. P. (2016). Dairy products and the French paradox: Could alkaline phosphatases play a role? Med. Hypotheses 92, 7–11. doi: 10.1016/j.mehy.2016.04.033
Lee, H. S., and Lee, K. J. (2000). Cathepsin B inhibitory peptides derived from β-casein. Peptides 21, 807–809. doi: 10.1016/S0196-9781(00)00212-6
Li, S., Liu, L., He, G., and Wu, J. (2018). Molecular targets and mechanisms of bioactive peptides against metabolic syndromes. Food Funct. 9, 42–52. doi: 10.1039/c7fo01323j
Liu, Y., Huang, F., Xu, J., Yang, P., Qin, Y., Cao, M., et al. (2020). Anti-hypertensive angiotensin II receptor blockers associated to mitigation of disease severity in elderly COVID-19 patients. medRxiv [Preprint]. doi: 10.1101/2020.03.20.20039586
Liu, Y., and Pischetsrieder, M. (2017). Identification and relative quantification of bioactive peptides sequentially released during simulated gastrointestinal digestion of commercial kefir. J. Agric. Food Chem. 65, 1865–1873. doi: 10.1021/acs.jafc.6b05385
López-Expósito, I., Miralles, B., Amigo, L., and Hernandez-Ledesma, B. (2017). “Health effects of cheese components with a focus on bioactive peptides,” in Fermented foods in health and disease prevention, 1st Edn, eds J. Frias, C. Martinez-Villaluenga, and E. Peñas (London: Academic Press), 239–265.
Lorenzo, J. M., Munekata, P. E. S., Gómez, B., Barba, F. J., Mora, L., Pérez-Santaescolástica, C., et al. (2018). Bioactive peptides as natural antioxidants in food products – a review. Trends Food Sci. Technol. 79, 136–147. doi: 10.1016/j.tifs.2018.07.003
Lu, Y., Wang, J., Soladoye, O. P., Aluko, R. E., Fu, Y., and Zhang, Y. (2021). Preparation, receptors, bioactivity and bioavailability of γ-glutamyl peptides: A comprehensive review. Trends Food Sci. Technol. 113, 301–314. doi: 10.1016/j.tifs.2021.04.051
MacLean, B., Tomazela, D. M., Abbatiello, S. E., Zhang, S., Whiteaker, J. R., Paulovich, A. G., et al. (2011). Effect of collision energy optimization on the measurement of peptides by selected reaction monitoring (SRM) mass spectrometry. Anal. Chem. 82, 10116–10124. doi: 10.1021/ac102179j
Maeno, M., Yamamoto, N., and Takano, T. (1996). Identification of an antihypertensive peptide from casein hydrolysate produced by a proteinase from L. helveticus CP790. J. Dairy Sci. 79, 1316–1321. doi: 10.3168/jds.S0022-0302(96)76487-1
Martini, S., Conte, A., and Tagliazucchi, D. (2020). Effect of ripening and in vitro digestion on the evolution and fate of bioactive peptides in Parmigiano-Reggiano cheese. Int. Dairy J. 105:104668. doi: 10.1016/j.idairyj.2020.104668
Martini, S., Solieri, L., Cattivelli, A., Pizzamiglio, V., and Tagliazucchi, D. (2021). An integrated peptidomics and in silico approach to identify novel anti-diabetic peptides in Parmigiano-Reggiano cheese. Biology 10:563. doi: 10.3390/biology10060563
Minervini, F., Algaron, F., Rizzello, C. G., Fox, P. F., Monnet, V., and Gobbetti, M. (2003). Angiotensin I-converting-enzyme-inhibitory and antibacterial peptides from Lactobacillus helveticus PR4 proteinase-hydrolyzed caseins of milk from six species. Appl. Environ. Microbiol. 69, 5297–5305. doi: 10.1128/AEM.69.9.5297-5305.2003
Montel, M. C., Buchin, S., Mallet, A., Delbes-Paus, C., Vuitton, D. A., Desmasures, N., et al. (2014). Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 177, 136–154. doi: 10.1016/j.ijfoodmicro.2014.02.019
Mullally, M. M., Meisel, H., and Fitzgerald, R. J. (1996). Synthetic peptides corresponding to α-lactalbumin and β-lactoglobulin sequences with angiotensin-i-converting enzyme inhibitory activity. Biol. Chem. Hoppe Seyler 377, 259–260. doi: 10.1515/bchm3.1996.377.4.259
Nagai, A., Mizushige, T., Matsumura, S., Inoue, K., and Ohinata, K. (2019). Orally administered milk-derived tripeptide improved cognitive decline in mice fed a high-fat diet. FASEB J. 33, 14095–14102. doi: 10.1096/fj.201900621R
Nakagomi, K., Fujimura, A., Maeda, H., Sadakane, Y., Fujii, N., Akizawa, T., et al. (2002). Isolation of novel peptides, cabin–1, –2, –3, and –4, that inhibit cathepsin B from a thermolysin digest of human plasma. Biol. Pharm. Bull. 25, 564–568. doi: 10.1248/bpb.25.564
Neviani, E., De Dea Lindner, J., Bernini, V., and Gatti, M. (2009). Recovery and differentiation of long ripened cheese microflora through a new cheese-based cultural medium. Food Microbiol. 26, 240–245. doi: 10.1016/j.fm.2009.01.004
Nielsen, S. D., Beverly, R. L., Qu, Y., and Dallas, D. C. (2017). Milk bioactive peptide database: A comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 232, 673–682. doi: 10.1016/j.foodchem.2017.04.056
Nongonierma, A. B., and Fitzgerald, R. J. (2016). Structure activity relationship modelling of milk protein-derived peptides with dipeptidyl peptidase IV (DPP-IV) inhibitory activity. Peptides 79, 1–7. doi: 10.1016/j.peptides.2016.03.005
Nongonierma, A. B., Paolella, S., Mudgil, P., Maqsood, S., and FitzGerald, R. J. (2018). Identification of novel dipeptidyl peptidase IV (DPP-IV) inhibitory peptides in camel milk protein hydrolysates. Food Chem. 244, 340–348. doi: 10.1016/j.foodchem.2017.10.033
Perez, R. E., Calhoun, S., Shim, D., Levenson, V. V., Duan, L., and Maki, C. G. (2020). Prolyl endopeptidase inhibitor Y-29794 blocks the IRS1-AKT-mTORC1 pathway and inhibits survival and in vivo tumor growth of triple-negative breast cancer. Cancer Biol. Ther. 21, 1033–1040. doi: 10.1080/15384047.2020.1824989
Perpetuo, E. A., Juliano, L., and Lebrun, I. (2003). Biochemical and pharmacological aspects of two bradykinin-potentiating peptides obtained from tryptic hydrolysis of casein. J. Protein Chem. 22, 601–606. doi: 10.1023/B:JOPC.0000008724.98339.ff
Petyaev, I. M., and Bashmakov, Y. K. (2012). Could cheese be the missing piece in the French paradox puzzle? Med. Hypotheses 79, 746–749. doi: 10.1016/j.mehy.2012.08.018
Pihlanto-Leppälä, A., Rokka, T., and Korhonen, H. (1998). Angiotensin I converting enzyme inhibitory peptides derived from bovine milk proteins. Int. Dairy J. 8, 325–331. doi: 10.1016/S0958-6946(98)00048-X
Plaisancié, P., Boutrou, R., Estienne, M., Henry, G., Jardin, J., Paquet, A., et al. (2015). β-Casein(94-123)-derived peptides differently modulate production of mucins in intestinal goblet cells. J. Dairy Res. 82, 36–46. doi: 10.1017/S0022029914000533
Prandi, B., Lambertini, F., Varani, M., Faccini, A., Suman, M., Leporati, A., et al. (2020). Assessment of enzymatic improvers in flours using LC-MS/MS detection of marker tryptic peptides. J. Am. Soc. Mass Spectrom. 31, 240–248. doi: 10.1021/jasms.9b00109
Rival, S. G., Boeriu, C. G., and Wichers, H. J. (2001). Caseins and casein hydrolysates. 2. Antioxidative properties and relevance to lipoxygenase inhibition. J. Agric. Food Chem. 49, 295–302. doi: 10.1021/jf0003911
Rojas-Ronquillo, R., Cruz-Guerrero, A., Flores-Nájera, A., Rodríguez-Serrano, G., Gómez-Ruiz, L., Reyes-Grajeda, J. P., et al. (2012). Antithrombotic and angiotensin-converting enzyme inhibitory properties of peptides released from bovine casein by Lactobacillus casei Shirota. Int. Dairy J. 26, 147–154. doi: 10.1016/j.idairyj.2012.05.002
Salama, S. A., Arab, H. H., Hassan, M. H., Al Robaian, M. M., and Maghrabi, I. A. (2019). Cadmium-induced hepatocellular injury: Modulatory effects of γ-glutamyl cysteine on the biomarkers of inflammation, DNA damage, and apoptotic cell death. J. Trace Elem. Med. Biol. 52, 74–82. doi: 10.1016/j.jtemb.2018.12.003
Sandré, C., Gleizes, A., Forestier, F., Gorges-Kergot, R., Chilmonczyk, S., Leìonil, J., et al. (2001). A peptide derived from bovine β-casein modulates functional properties of bone marrow-derived macrophages from germfree and human flora-associated mice. J. Nutr. 131, 2936–2942. doi: 10.1093/jn/131.11.2936
Santiago-López, L., Aguilar-Toalá, J. E., Hernández-Mendoza, A., Vallejo-Cordoba, B., Liceaga, A. M., and González-Córdova, A. F. (2018). Invited review: Bioactive compounds produced during cheese ripening and health effects associated with aged cheese consumption. J. Dairy Sci. 101, 3742–3757. doi: 10.3168/jds.2017-13465
Sedaghati, M., Ezzatpanah, H., Mashhadi Akbar Boojar, M., Tajabadi Ebrahimi, M., and Kobarfard, F. (2016). Isolation and identification of some antibacterial peptides in the plasmin-digest of β-casein. LWT 68, 217–225. doi: 10.1016/j.lwt.2015.12.019
Sforza, S., Cavatorta, V., Galaverna, G., Dossena, A., and Marchelli, R. (2009). Accumulation of non-proteolytic aminoacyl derivatives in Parmigiano-Reggiano cheese during ripening. Int. Dairy J. 19, 582–587. doi: 10.1016/j.idairyj.2009.04.009
Sforza, S., Cavatorta, V., Lambertini, F., Galaverna, G., Dossena, A., and Marchelli, R. (2012). Cheese peptidomics: A detailed study on the evolution of the oligopeptide fraction in Parmigiano-Reggiano cheese from curd to 24 months of aging. J. Dairy Sci. 95, 3514–3526. doi: 10.3168/jds.2011-5046
Sgarbi, E., Lazzi, C., Iacopino, L., Bottesini, C., Lambertini, F., Sforza, S., et al. (2013). Microbial origin of non proteolytic aminoacyl derivatives in long ripened cheeses. Food Microbiol. 35, 116–120. doi: 10.1016/j.fm.2013.02.013
Shanmugam, V. P., Kapila, S., Sonfack, T. K., and Kapila, R. (2015). Antioxidative peptide derived from enzymatic digestion of buffalo casein. Int. Dairy J. 42, 1–5. doi: 10.1016/j.idairyj.2014.11.001
Solieri, L., Baldaccini, A., Martini, S., Bianchi, A., Pizzamiglio, V., and Tagliazucchi, D. (2020). Peptide profiling and biological activities of 12- month ripened Parmigiano Reggiano cheese. Biology 9:170. doi: 10.3390/biology9070170
Suetsuna, K., Ukeda, H., and Ochi, H. (2000). Isolation and characterization of free radical scavenging activities peptides derived from casein. J. Nutr. Biochem. 11, 128–131. doi: 10.1016/S0955-2863(99)00083-2
Sultan, S., Huma, N., Butt, M. S., Aleem, M., and Abbas, M. (2018). Therapeutic potential of dairy bioactive peptides: A contemporary perspective. Crit. Rev. Food Sci. Nutr. 58, 105–115. doi: 10.1080/10408398.2015.1136590
Tagliazucchi, D., Baldaccini, A., Martini, S., Bianchi, A., Pizzamiglio, V., and Solieri, L. (2020). Cultivable non-starter lactobacilli from ripened Parmigiano Reggiano cheeses with different salt content and their potential to release anti-hypertensive peptides. Int. J. Food Microbiol. 330:108688. doi: 10.1016/j.ijfoodmicro.2020.108688
Tagliazucchi, D., Helal, A., Verzelloni, E., Bellesia, A., and Conte, A. (2016). Composition and properties of peptides that survive standardised in vitro gastro-pancreatic digestion of bovine milk. Int. Dairy J. 61, 196–204. doi: 10.1016/j.idairyj.2016.06.002
Tonolo, F., Folda, A., Cesaro, L., Scalcon, V., Marin, O., Ferro, S., et al. (2020). Milk-derived bioactive peptides exhibit antioxidant activity through the Keap1-Nrf2 signaling pathway. J. Funct. Foods 64:103696. doi: 10.1016/j.jff.2019.103696
Udenigwe, C. C., Gong, M., and Wu, S. (2013). In silico analysis of the large and small subunits of cereal RuBisCO as precursors of cryptic bioactive peptides. Process Biochem. 48, 1794–1799. doi: 10.1016/j.procbio.2013.08.013
van der Reijden, O. L., Zimmermann, M. B., and Galetti, V. (2017). Iodine in dairy milk: Sources, concentrations and importance to human health. Best Pract. Res. Clin. Endocrinol. Metab. 31, 385–395. doi: 10.1016/j.beem.2017.10.004
Vij, R., Reddi, S., Kapila, S., and Kapila, R. (2016). Transepithelial transport of milk derived bioactive peptide VLPVPQK. Food Chem. 190, 681–688. doi: 10.1016/j.foodchem.2015.05.121
Xing, L., Zhang, H., Majumder, K., Zhang, W., and Mine, Y. (2019). γ-Glutamylvaline prevents low-grade chronic inflammation via activation of a calcium-sensing receptor pathway in 3T3-L1Mouse adipocytes. J. Agric. Food Chem. 67, 8361–8369. doi: 10.1021/acs.jafc.9b02334
Yamamoto, N., Akino, A., and Takano, T. (1994). Antihypertensive effect of the peptides derived from casein by an extracellular proteinase from Lactobacillus helveticus CP790. J. Dairy Sci. 77, 917–922. doi: 10.3168/jds.S0022-0302(94)77026-0
Yang, J., Sun-Waterhouse, D., Zhou, W., Cui, C., and Wang, W. (2019). Glutaminase-catalyzed γ-glutamylation to produce CCK secretion-stimulatory γ-[Glu]n-Trp peptides superior to tryptophan. J. Funct. Foods 60:103418. doi: 10.1016/j.jff.2019.103418
Keywords: long-ripened cheese, Parmigiano Reggiano, bioactive peptides, digestion, bioactivities
Citation: Castellone V, Prandi B, Bancalari E, Tedeschi T, Gatti M and Bottari B (2022) Peptide profile of Parmigiano Reggiano cheese after simulated gastrointestinal digestion: From quality drivers to functional compounds. Front. Microbiol. 13:966239. doi: 10.3389/fmicb.2022.966239
Received: 10 June 2022; Accepted: 31 July 2022;
Published: 23 August 2022.
Edited by:
Uelinton Manoel Pinto, University of São Paulo, BrazilReviewed by:
Débora Parra Baptista, State University of Campinas, BrazilCopyright © 2022 Castellone, Prandi, Bancalari, Tedeschi, Gatti and Bottari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Benedetta Bottari, YmVuZWRldHRhLmJvdHRhcmlAdW5pcHIuaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.