- 1College of Life Science and Technology, Central South University of Forestry and Technology, Changsha, China
- 2College of Arts and Sciences, Governors State University, University Park, IL, United States
- 3Key Laboratory of National Forestry and Grassland Administration on Management of Western Forest Bio-Disaster, College of Forestry, Northwest A&F University, Yangling, China
Photocatalysis, as a novel technique, has been widely used for antibiotic pollution remediation in wastewater. In the processes of degradation and removal of antibiotics, the impact of photocatalysts on microenvironment is very important but remains poorly understood. In the present study, the effect of typical photocatalyst g-C3N4 (Graphitic carbon nitride) on microbial community was investigated in two sediment types (riverbed sediment and pig-farm sediment) polluted by tetracycline (TC) in central southern China. The riverbed sediment and pig farm sediment samples were respectively exposed to g-C3N4 (25, 75, 125 mg⋅kg–1) and TC (60, 120, 180 mg⋅L–1) treatments alone or combination for 30 days, respectively. The bacterial community and antibiotic resistance genes (ARGs) of the treated sediments were analyzed by Illumina sequencing and metagenomic sequencing. Studies had shown that: TC, g-C3N4, and TC/g-C3N4 have significant effects on the changes of microbial communities and components in riverbed sediment, but they do not exist in pig farm sediment. The most alterations of microbial taxa were Acidobacteriota, Actinobacteriota, and Desulfobacterota in riverbed sediment, and Elusimicrobiota in the pig farm sediment under various treatments. Through network analysis, it was found that the distribution of microorganisms in the pig farm sediment is more complex and more stable. The addition of g-C3N4 reduced the absolute abundance of ARGs in the two examined sediments, but not significantly changed their relative abundance of ARGs. The g-C3N4 application was beneficial to the removal of TC residues and to the prevention of the generation and transmission of ARGs in sediments. Our results suggested that g-C3N4 was a suitable photocatalyst with excellent application prospect for the removal of TC residues and the control of ARGs in environment.
Introduction
Antibiotics had been used worldwide for disease treatment in livestock and aquaculture as feed additives in the past decades (Henriksson et al., 2018). Over time, these substances would flow into the surrounding soil in various ways, thus causing serious pollution to the environment and causing great harm to the human body (Zhan et al., 2021). Tetracycline (TC) is one of the most inexpensive classes of antibiotics and it had been widely used as veterinary therapy in livestock and fish farming industry in the world (Gao et al., 2020). Due to its extensive usage and poor bioavailability, relative high concentrations of TC residues have been found in various environments (Mahmoud and Abdel-Mohsein, 2019). The presence of TC residues could inhibit microbial growth and reduce the number of bacteria in sediment (He et al., 2021), cause negative effects on the structure and activity of environmental microbiota in aquaculture (Martinez-Porchas and Vargas-Albores, 2017), and affect soil ecological functions in terrestrial ecosystems (Santas-Miguel et al., 2020). In some cases, TC could induce new antibiotic resistance mutations by promoting microbial communities to defend the attack of antibiotics, developing recombination and repair function (Zhang L. L. et al., 2021).
As a novel remediation way, photocatalysts have been applied for the remediation of a variety of organic pollutants (Guo et al., 2021). By inducing strongly oxidizing reagents they could decompose some pollution substances present in the atmosphere (Nath et al., 2016; Sun et al., 2022a). Owing to numerous active sites, photocatalysts could degrade organic pollutants in air and water (Shi et al., 2022). Many photocatalysts have been applied to remove emerging water pollutants such as drug pollution and shown a remarkable stability in the photocatalytic degradation (Tobajas et al., 2017). Recently, photocatalysts have been applied to remove some pathogenic bacteria in the environment, such as Escherichia coli, Salmonella typhimurium and Staphylococcus aureus (Tang et al., 2019; Zhang C. et al., 2019). Photocatalysis were considered as excellent ecofriendly candidates for water remediation given their outstanding photostability, and minimal ecotoxicological effects on aquatic biota (Tobajas et al., 2017; Serra et al., 2020). Photocatalysts might influence antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARGs) under photocatalytic conditions, so that it might be a worth-trying material for antibiotic resistance control (Guo and Tian, 2019). Among various photocatalysts, graphite carbon nitride (g-C3N4) was a kind of unalloyed polymer with good thermal, chemical and electrochemical stability (Hou et al., 2020). Thus, it has a great potential for applying in the field of catalytic elimination of environmental pollutants (Jun et al., 2015), such as degradation of TC in sewage and sludge samples (Yan et al., 2020; Zhao et al., 2020).
Sediment systems were essential components when assessing aquatic ecosystems functionality (He et al., 2019), in which microorganisms were the most abundant group of organisms in sediment for maintaining sediment biological activity and reducing pollutant levels (Yan et al., 2020). Several studies have been conducted to examine the impact of application of g-C3N4 on removal of TC in environments, but the effect of g-C3N4 usage on microenvironments in TC contaminated sediments was still poorly understand (Gao et al., 2021; Guo et al., 2021). It is particular true in riverbed sediment and pig farm sediment, were less was known about the influence of g-C3N4 on the richness and diversity of ARGs in the TC polluted environments.
This study aimed to examine the influence of g-C3N4 application on the ARGs and the structure of bacterial community in TC contaminated riverbed sediments and pig farm sediments. The specific objectives of this study were: (a): to evaluate the ability of g-C3N4 to remove TC pollutants in the examined sediments, (b): to analyze the effects of TC, g-C3N4 and TC/g-C3N4 treatments on bacterial community structure in the different sediments, (c): to assess the effects of g-C3N4 on ARGs and tetracycline resistance genes (TRGs) in the two sediment types, and (d): to reveal the interrelation and interaction among the TRGs, bacterial species communities and functional metabolism of bacteria in the studied environments. The results from this study would provide theoretical basis and scientific reference for better understanding of photocatalyst application in the degradation of antibiotics in environments.
Materials and methods
Sediment sampling and physicochemical parameters
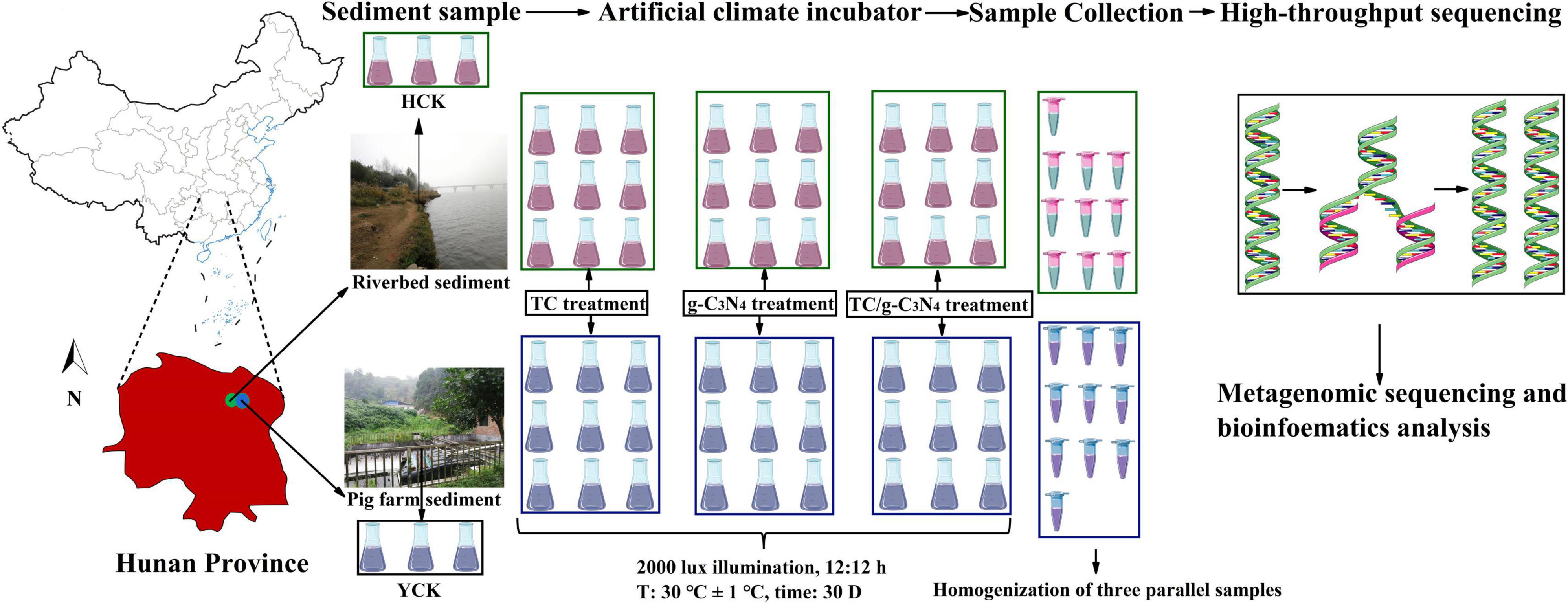
The pig farm sediments in this experiment were collected from several farms located in Liuyang County, Hunan Province of China (113°51′55′′E, 28°28′08′′N) (Figure 1). These selected farms have complete manure treatment facilities, where the treated effluents were released into receiving water-collectors and the deposited manures were applied as fertilizers to the surrounding agricultural lands. About 185.3 ng⋅kg–1 was still detected in the pig farm sediment, which had a serious risk of TC contamination. The riverbed sediments were collected from the Xiangjiang River in Changsha, Hunan Province (112°94′71′′E, 28°14′23′′N), and about 17.46 ng⋅kg–1 TC residue was detected in the river bed sediment, which also had a certain pollution risk. The field sampling work was carried out in July 2020.
The sediment samples were manually taken from the surface sediment (10–20 cm in depth) at five points on a 2 m2 rectangular area, with one point at the center and other four points at the corners. The five samples were pooled to form a mixed sample for a rectangular area. There were five replicated mixed samples (2 kg each) for each of the two sediment types.
Two types of sediment were characterized to determine pH, Total nitrogen (TN), ammonium nitrogen (NH4+-N), nitrate nitrogen (NO3–-N), Total phosphorus (TP) and Total potassium (TK). The pH of the sediment was determined by a composite electrode (pH glass-saturated mercury) in a sediment/water ratio of 1:2.5. TN, NH4+-N, and NO3–-N contents were analyzed using an Elemental analyzer (Vario MACRO cube, Elementar, Germany). TP content was analyzed using the sodium hydroxide fusion-Molybdenum-antimony resistance colorimetric method. TK was determined by flame photometry (Flame photometer 410, sherwood). The physical properties of the sediment were shown in the Supplementary Table 1.
The TC residue in each sample was detected by high performance liquid chromatography (HPLC) according to the method. The standard substances of TC (C22H24N2O8⋅HCl, purity, USP grade, purity 98%) were purchased from Shanghai Aladdin Biochemical Technology (Shanghai, China). TC standard stock solution (1000 mg⋅L–1) was prepared with HPLC grade methanol (Shimadzu Corporation, Kyoto, Japan) and stored at −20°C in the dark. The mobile phase contained 0.1% trifluoroacetic acid and acetonitrile gradient. The flow rate was 1 mL⋅min–1, the equilibrium solution was 10% B solution, and the gradient rose to 30% B solution within 20 min at a flow rate of 1 mL⋅min–1 and a temperature of 35°C. Three samples with the same label were used to measure recoveries of TC in the samples respectively, and it was in the range of 95.53–105.27. Values of 3 and 10 were used as signal-to-noise ratios for measurement limit and quantization limit. The relative standard deviations of TC content in each sample was within 10%. The quantitative standard curve of the measured antibiotics and the confirmation of the measurement method are shown in Supplementary Figure 1 and the Supplementary Table 2.
Experimental design, and sampling
Tetracycline and g-C3N4
1 g TC was dissolved in 100 mL sterile water to form a TC solution with a concentration of 10 mg⋅mL–1. Then, the solution was diluted to different TC concentrations (60, 120, 180 mg⋅L–1) with sterile water. Bulk g-C3N4 were obtained by direct polycondensation of melamine according to the procedure (Hou et al., 2020). Typically, a certain amount of melamine was loaded into a porcelain boat with a lid and placed in a tube furnace, then heated up to 550°C for 4 h, after cooled to room temperature naturally, the resultant powder was collected for further use (Liu E. L. et al., 2021; Chen et al., 2022).
Tetracycline was extracted according to the method of Zhang et al. (2015), within 24 h after the sample collection. 2 g of sediment sample was acidified to pH 4.0 by adding HCl, followed by addition of 40 mL of McIlvaine-Na2 EDTA buffer (0.2 M) and acetonitrile (v:v = 1:2) through 60 s vortex for three times, and ultra-sonicated at room temperature (15 min at 300 mA). After centrifugation in air-cooled conditions at 10,000 rpm for 10 min, the supernatant was collected from the mixture. The combined supernatant was then concentrated to about 5 mL and diluted to 100 mL with Milli-Q water. The samples were extracted using Oasis HLB (150 mg, 6 mL) extraction cartridges. The extracts percolated through the adsorbent at a flow rate of approximately 1.0 mL⋅min–1. Afterward, the column was rinsed with 5 mL of ultra-pure water and 5 mL of methanol (5%), dried under vacuum for 10 min, and eluted twice with 5 mL of methanol-ethyl acetate solution (10%, 1:9, v:v). The selected analytes were dried under a gentle nitrogen stream, re-dissolved in 1 mL of mobile phase [acetonitrile: formic-acid (0.1%), 1:9, v:v]. Final extracts were transferred to 2 mL amber vials for HPLC analysis.
Sediment exposure design
Four experimental treatments were conducted for each of the pig farm sediment and the riverbed sediment and they were: (1) Control treatments (CK), (2) TC treatments, (3) g-C3N4 treatments, and (4) TC/g-C3N4 treatments. In CK, 100 g of pig farm sediment or riverbed sediment were added to 100 mL of sterile water and labeled as YCK or HCK. In the TC treatments, 100 g sediment samples were added to 100 mL of TC solutions with different concentrations (60, 120, 180 mg⋅L–1). The treatments for pig farm sediment were named as YTL, YTM, and YTH, and the treatments for riverbed sediment as HTL, HTM, and HTH, respectively. In the g-C3N4 treatments, 100 g sediment samples were mixed with g-C3N4 to different concentration (25, 75, 125 mg⋅kg–1). The treatments for pig farm were labeled as YPL, YPM, and YPH, and the treatments for riverbed sediment as HPL, HPM, and HPH, respectively. In TC/g-C3N4 treatments, 100 g sediment samples were added to 100 mL of solution with various TC/g-C3N4 combination (60 mg⋅L–1/25 mg⋅kg–1, 120 mg⋅L–1/75 mg⋅kg–1, 180 mg⋅L–1/125 mg⋅kg–1). The treatments for pig farm were assigned as YTLPL, YTMPM, YTHPH, and the treatments for riverbed sediment as HTLPL, HTMPM, and HTHPH, respectively. There were a total of 20 treatments (two groups of control + two sediment samples × 9 treatment conditions) were pretreated and each treatment was performed for three replications (Figure 1). All samples were placed in a homeothermic incubator (RGX-350) at 30 ± 1°C under 2000 lux illumination with a light dark period of 12:12 h. After treated for 30 days, 1–2 g sediment sample was taken and stored in a refrigerator at -80°C for the analysis of bacterial community structure, diversity and ARGs.
DNA extraction, library construction, and metagenomic sequencing
DNA extraction and 16S rRNA gene sequencing
Total genomic DNA of each sediment sample was extracted from 2 g of sediment samples using the FastDNA Spin Kit (Omega Bio-tek, Norcross, GA, United States) for pig farm sediment. The DNA extract was checked on 1% agarose gel, and DNA concentration and purity were determined with NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, United States). The hypervariable region V3-V4 of the bacterial 16S rRNA gene was amplified with primer pairs 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) by an ABI GeneAmp 9700 PCR thermocycler (ABI, CA, United States). The details of PCR were fully described in Supplementary materials.
Illumina MiSeq sequencing
Majorbio Bio-Pharm Technology Co., Ltd., (Shanghai, China) pooled purified amplicons in equimolar amounts and paired-end sequenced on an Illumina MiSeq PE300 platform (Illumina, San Diego, United States) using standard protocols.
Processing of sequencing data
Fastp (Version 0.20.0) demultiplexed and quality-filtered the raw 16S rRNA gene sequencing readings before merging them with FLASH (Version 1.2.7) (Magoč and Salzberg, 2011; Chen et al., 2018). Using unique barcodes, sequencing reads were assigned to each sample and shortened by removing the primer sequence and barcode. The original DNA fragments were merged into tags using FLASH (Magoč and Salzberg, 2011). According to QIIME (Version 1.9.1) quality-controlled process, the raw tags was filtered under specific filtering conditions to generate high-quality clean tags (Caporaso et al., 2010).
In order to generate effective tags, the UCHIME algorithm was used to remove the chimeric sequence from the clean tags on the basis of reference to each operational taxonomic unit (OTU). Assign each remaining sequence to an OTU when at least 97% of the threshold identity was obtained using UPARSE software (Version 7.0.1) and chimeric sequence were identified and deleted (Edgar, 2013). According to the 16S rRNA database (Silva v138), each OTU representative sequence was classified by RDP Classifier version 2.2 with a confidence interval of 0.7 (Wang et al., 2007).
Metagenomic sequencing and bioinformatics analysis
Metagenomic shotgun sequencing libraries of the four selected sediment samples were performed by Majorbio, Inc., (Shanghai, China) using Illumina MiSeq PE300 platform. This approach generated 245,754,330 raw reads and 35,398,741,165 total clean bases for the four samples (HCK, YCK, HTHPH, and YTHPH). MEGAHIT was used to clean and assemble the sequence, contigs with length of 300 bp or more selected as the final assembling result, and then contigs were used for further gene prediction and annotation.
Statistical analysis
The statistical analysis of biological information of OTU was performed by using Uparse software. The Bray–Curtis distance was used to generate principal co-ordinates analysis (PCoA) for the visualization of complex multidimensional data. Drawing rarefaction curve and bacterial community composition were determined by R package vegan (Version 3.3.3). Annotation of antibiotic resistance was conducted using Diamond against Comprehensive Antibiotic Resistance Database (CARD, Version 3.0.9) (Buchfink et al., 2015). The correlation coefficient of Spearman rank among the bacteria phylum in the two sediment types was calculated using Network software. Significantly positive correlation (p < 0.05) was used to build network in Cytoscape (Version 3.5.2) in Prefuse Force Directed Layout by betweenness centrality (correlation coefficient > 0.5). The One-way ANOVA analyses were conducted using SPSS (IBM SPSS Statistics 26.0).
Results
Characterizations
As shown in Figure 2A, a strong peak appeared in the diffraction pattern of g-C3N4 at 2θ = 27.8°, corresponding to the (002) planes of g-C3N4, which is the characteristic interlayer stacking peak of g-C3N4. Meanwhile, a weak peak appeared in the diffraction pattern of g-C3N4 at 2θ = 12.8°, corresponding to the (100) planes of g-C3N4. The FT-IR spectra of the g-C3N4 was shown in Figure 2B, sharp absorption peaks at about 800 cm–1 of g-C3N4 was assigned to the stretching vibration modes of the triazine units. The absorption bands at 1251, 1325, and 1437 cm–1 are attributed to the typical stretching vibration modes of the C = N heterocycles. Figure 2C shown the UV-vis diffuse reflectance spectra to characterize the optical properties of the g-C3N4 sample, g-C3N4 nanosheets showed strong absorption both in the UV and in the visible light regions. The morphology and structure of the g-C3N4 samples were investigated using SEM and TEM technology. Figure 2D shown TEM images of g-C3N4 photocatalysts. The g-C3N4 presented a fold Nano thin layer. From the SEM image of Figure 2E, it can be seen that g-C3N4 is made of multiple nanosheets stacked on top of each other, with a typical stacked layer structure. BET analysis of g-C3N4 photocatalyst The N2 adsorption-desorption isotherms and BET specific surface area (SSA) of the g-C3N4 photocatalyst were shown in Figure 2F. g-C3N4 has a mesoporous structure, which facilitates the reduction of mass transfer limitations and collecting light in the photocatalytic process.
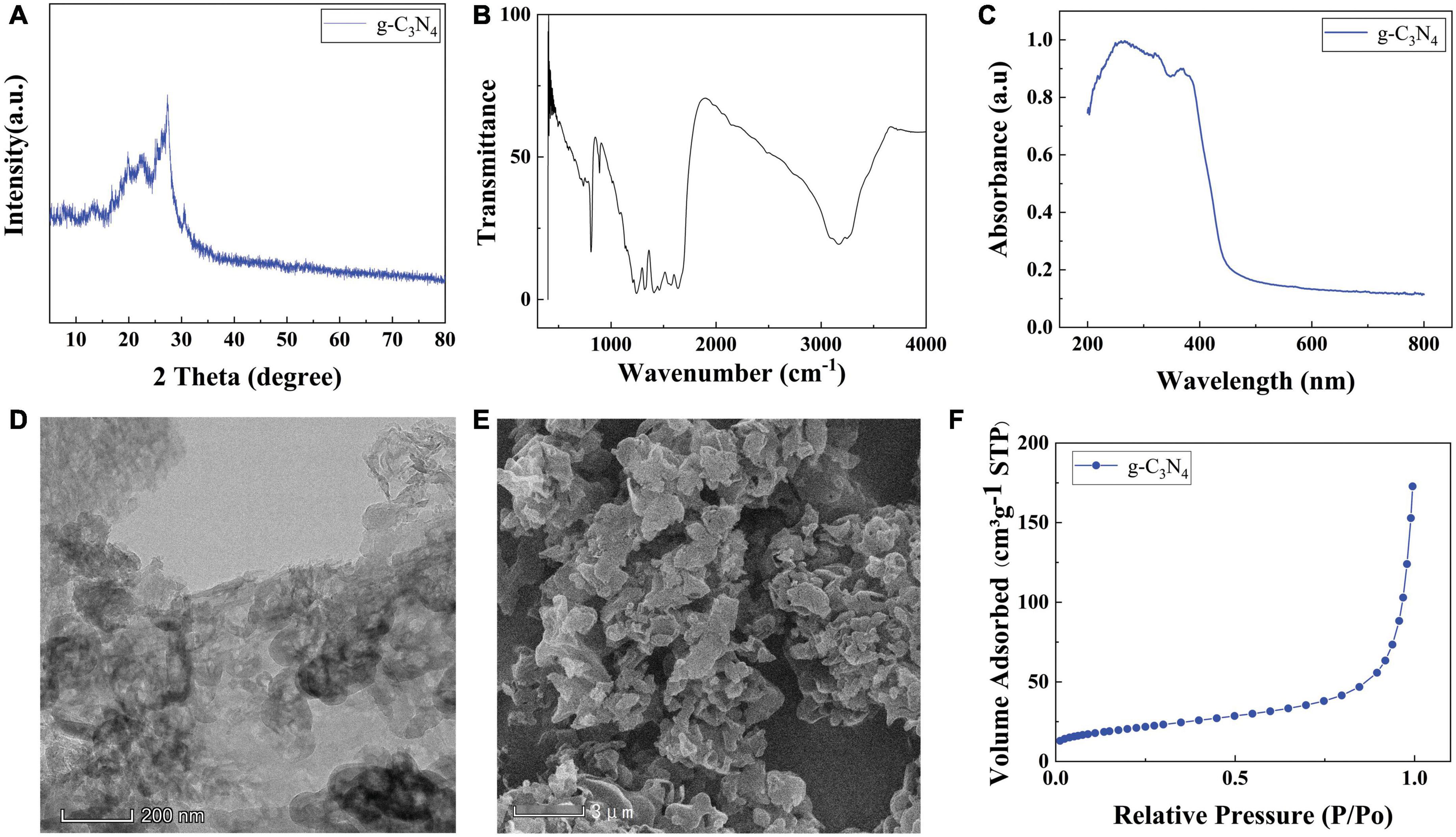
Figure 2. (A) XRD of g-C3N4, FT-IR (B) patterns of the g-C3N4, (C) UV-vis SEM images of (D) g-C3N4, TEM images of (E) g-C3N4, (F) Nitrogen adsorption-desorption isotherms of g-C3N4.
Removal of tetracycline in different sediments
After stored for 30 days, the removal percentage of TC residues was less than 20% in the blank group with different TC concentrations (Figure 3). The removal proportion of TC residues significantly increased in both of pig farm sediment (YT) and riverbed sediment (HT) when compared to the blank group (p < 0.05). The residue of TC in pig farm sediment was significantly higher (p < 0.05) than that in riverbed sediment after 30 days exposure. Adding g-C3N4 promoted TC removal in the two examined sediment types.
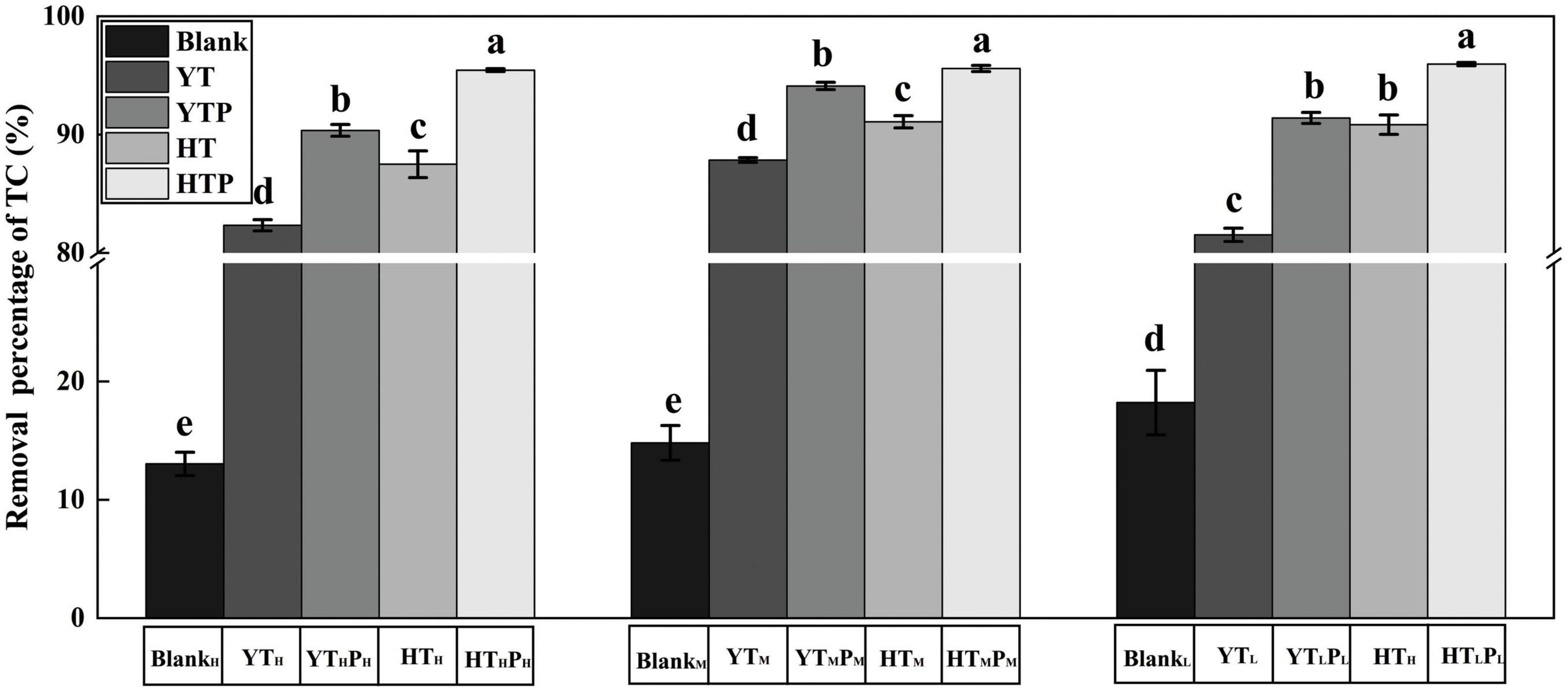
Figure 3. Removal percentage of TC residues from various treated sediments. BlankH, BlankM, and BlankL refer to 180, 120, and 60 mg⋅L– 1 TC solution after stored for 30 days, respectively. HT, HTP refer to the riverbed sediment treated by TC and TC/g-C3N4 for 30 days, respectively. YT, YTP refer to the pig farm sediment treated by TC and TC/g-C3N4 for 30 days, respectively. The error bars represent the SDs (n = 3). Significant differences were determined by Duncan’s test There were significant differences between different letters (a, b, c, d, e) (p < 0.05).
Bacterial diversity in the riverbed sediment and pig farm sediment
A total of 604,406 high-quality readings (average 30,203) were obtained from 20 samples, after eliminating low and short quality reads, replicates, singletons, and chimeras (Supplementary Figure 2). The number of OTU varied for different groups and a total of 8,934 OTUs was detected out in all samples (Supplementary Table 3). There were 1,696 OTUs identified in YCK, and the observed OTUs ranged from 1701 to 2026 in the 9 pig farm sediment samples treated by different concentrations of TC, g-C3N4 and TC/g-C3N4. Moreover, 3,118 OTUs were obtained from HCK, and the range of OTUs was from 3304 to 3850 OTUs in the 9 riverbed sediment samples treated by different level of TC, g-C3N4 and TC/g-C3N4.
The Shannon indices were 5.07, 4.98, and 5.03 for the three kinds of treated pig farm sediments (YT, YP, and YTP), respectively. The mean value was 5.03 ± 0.045, similar to that of the CK sediment (YCK: 5.09). The Simpson index of bacteria in YT, YP, and YTP were not significantly different compared to that in YCK, implying that the addition of TC, g-C3N4 or TC/g-C3N4 had no significant effect on the diversity and richness of microbial community in pig farm sediment. The treated riverbed sediments, HT, HP, and HTP, had a Shannon index of 6.86, 6.85, and 6.85, respectively. The mean value was 6.85 ± 0.005, which was higher than that of the CK sediment (HCK: 6.59). The Simpson index of bacteria was higher in HT, HP, and HTP treatments than in HCK. The results implied that the addition of TC, g-C3N4, TC/g-C3N4 increased the diversity and richness of microbial communities of the riverbed sediments.
Principal co-ordinates analysis was used to investigate the differences of bacterial community between the two sorts of sediment (Figure 4Aa), which explained 90.31 and 2.19% at the first two coordinates. The pig farm sediment treated by TC, g-C3N4 and TC/g-C3N4 clustered nearby axis-2, respectively (p < 0.05), while the riverbed sediment treated by TC, g-C3N4 and TC/g-C3N4 clustered along axis-1. Further PCoA analysis for the pig farm sediment samples revealed that after being treated by TC, g-C3N4 and TC/g-C3N4. The bacterial community in the pig farm sediment samples did not change significantly under the TC, g-C3N4 and TC/g-C3N4 treatments (p > 0.05) (Figure 4Ab), in which the first two axes (PCoA1 and PCoA2) explained 30.30 and 21.16% of the total variance, respectively. However, the addition of TC, g-C3N4 and TC/g-C3N4 lead an obvious separation of the bacterial community in the riverbed sediment samples (p < 0.05) (Figure 4Ac), in which the first two axes (PCoA1 and PCoA2) explained 30.68 and 16.81% of the total variance in the bacterial communities, respectively. These results suggested that the difference of the bacterial community composition between pig farm sediment and riverbed sediment samples depended on the sediment properties rather than the treatment method. Each sediment type had the capacity to form its specific ecosystem with an independent clustering. Additionally, the effect of TC, g-C3N4 and TC/g-C3N4 application on the microbial communities structure was more obvious in riverbed sediment than in pig farm sediment.
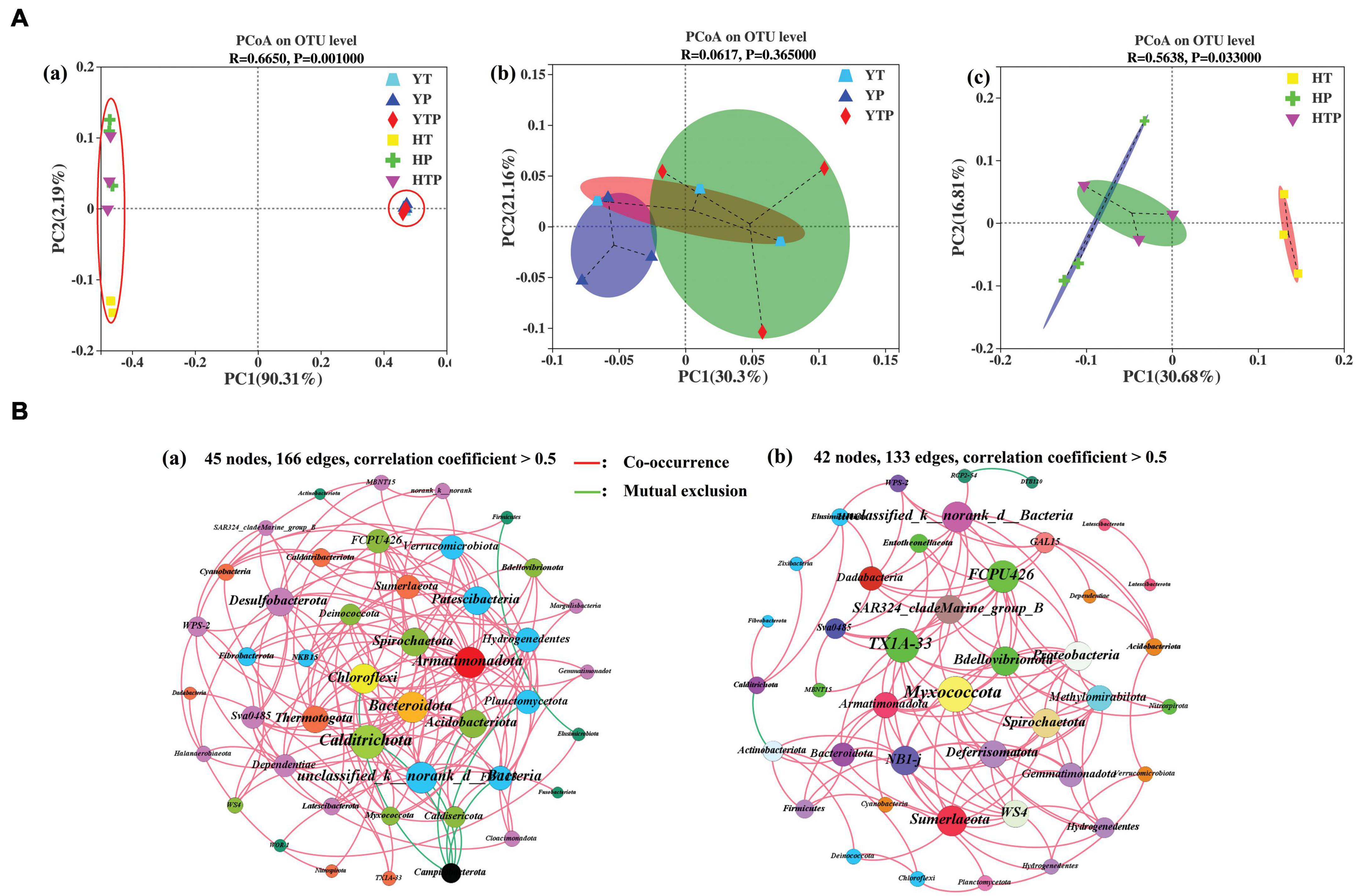
Figure 4. Principal Coordinate Analysis (PCoA) and co-occurrence network analysis illustrates the dissimilarities and stability of microbiome. (A) PCoA illustrate dissimilarities of bacterial communities’ structure, (a) refer to PCoA among two types of sediment system contain of TC, g-C3N4, TC/g-C3N4, respectively, (b) refer to PCoA of the bacterial communities in pig farm sediment system contain of TC, g-C3N4, TC/g-C3N4, respectively, (c) refer to PCoA of the bacterial communities in riverbed sediment system contain of TC, g-C3N4, TC/g-C3N4, respectively. YT, YP, YTP refer to the pig farm sediment treated with different concentrations of TC, g-C3N4 and TC/g-C3N4 for 30 days, respectively; HT, HP, HTP refer to the riverbed sediment treated with different concentrations of TC, g-C3N4 and TC/g-C3N4 for 30 days, respectively. (B) The co-occurrence network analysis of bacterial community at phylum level in pig farm sediment (a) and riverbed sediment (b) (correlation coefficient > 0.5). The nodes of each network are colored according to phylum affiliation (97%) and sized according to degree of connection. The edges connecting the nodes are represented by red lines to indicate co-occurrence interactions or green to indicate mutualistic exclusions.
Analysis of co-occurrence network was performed based on the phylum level (Figure 4B). The numbers of nodes and edges were 45 and 166, and 42 and 133 in pig farm sediment and riverbed sediment, respectively. The network of the pig farm sediment was more complex and the stability of the bacterial community structure was higher in pig farm sediment than in riverbed sediment, which was beneficial to resist the changes in the bacterial community caused by external environment. Therefore, TC treatment samples in pig farm sediment groups was clearly separated from either those from g-C3N4 treatment or TC/g-C3N4 treatment (Figure 4Ab).
Microbial community of the pig farm sediment and riverbed sediment
The dominant bacterial phyla were Firmicutes, Proteobacteria, Actinobacteriota, Bacteroidota in the pig farm sediment samples and there was no significant difference in species abundance and genus level among different sample treatments (Figures 5Aa,Ba and Supplementary Table 5). However, bacterial communities were changed significantly within 4 treated groups in the riverbed sediments. The dominant bacteria were Actinobacteriota, Proteobacteria, Acidobacteriota, Chloroflexi in riverbed sediment (Figure 5Ab and Supplementary Table 6), which were different from those in pig farm sediment. As the abundance of Acidobacteriota increased in HP and HTP groups, the addition of g-C3N4 significantly stimulated the growth of Acidobacteriota species, such as RB41 in the riverbed sediment (Figure 5Bb).
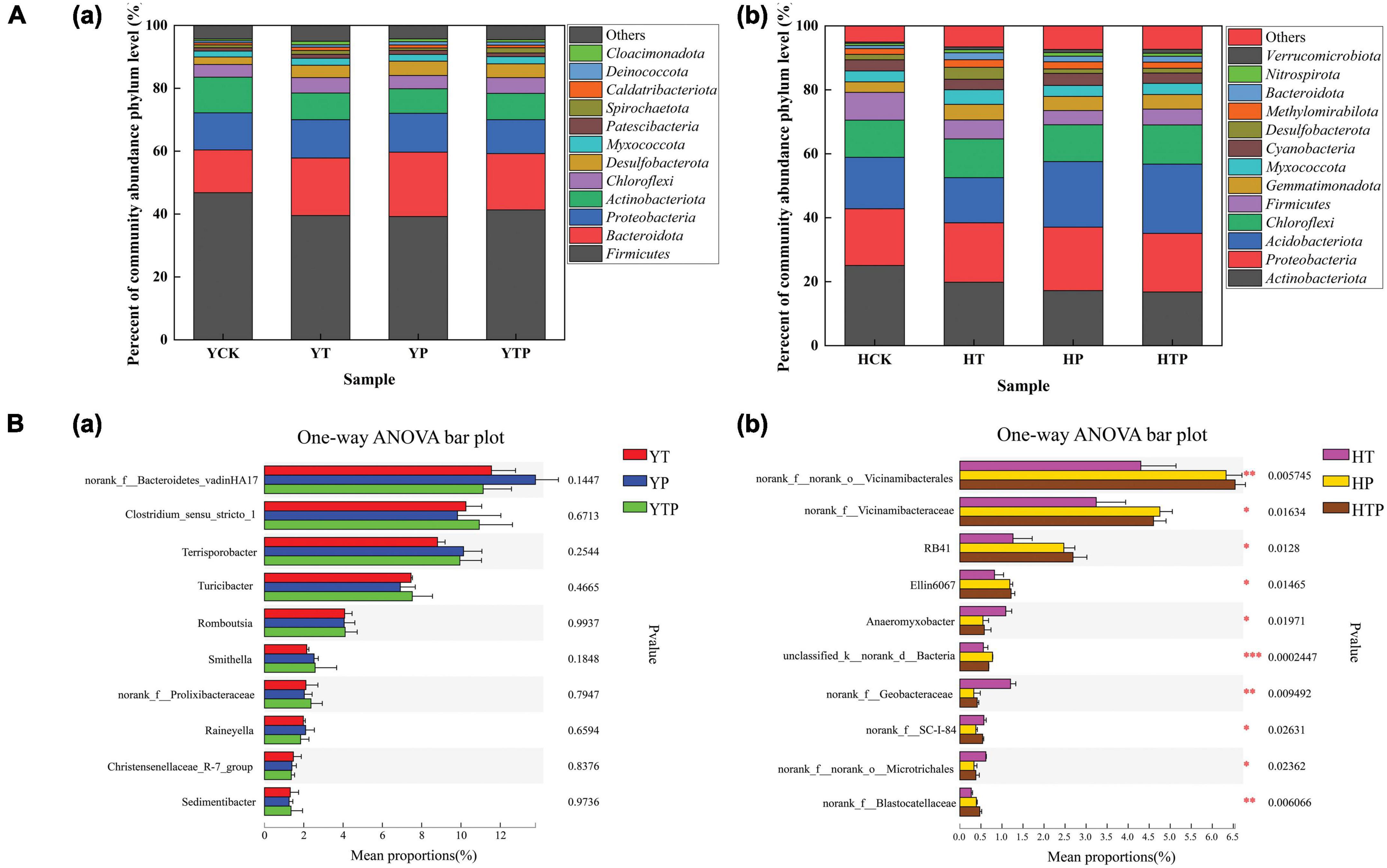
Figure 5. Dynamics of the bacterial community under different treatment. (A) Relative abundance of bacterial taxa at the phylum level in pig farm sediment and riverbed sediment. (B) The One-way ANOVA to compare bacterial community structure changes, (a) Bacterial community structure changes at genus level in pig farm sediment, (b) Bacterial community structure changes at genus level in riverbed sediment. YCK refer to the pig farm sediment for 30 days, HCK refer to the riverbed sediment for 30 days. *p < 0.05, **p < 0.01, ***p < 0.001.
Abundance and diversity of ARGs types and tetracycline resistance genes subtypes
There were 2,415,418,884 high-quality reads after filtering in the four representative metagenomics libraries (YCK, HCK, YTHPH, and HTHPH) (Supplementary Table 7). A total of 836 ARGs subtypes potentially conferring resistance to 20 classes of antibiotics were detected in the two sediment types. Multidrug resistance genes were most abundant (39.19%) with 199 subtypes observed, followed by TRGs, with 60 subtypes that comprised 12.74% of abundance, and MLS resistance genes with 90 subtypes that comprised 11.33% of abundance (Supplementary Figure 3).
The pig farm sediment samples encoded ARGs belonging to 80.48% of antibiotic classes found in these environments while riverbed sediment samples only encoded 19.52% of antibiotic classes (Supplementary Table 8). The ARGs abundance were higher in pig farm sediment than in riverbed sediment, implying a more complex composition of antibiotic resistance mechanisms existed in the pig farm sediment (Figure 6A). The top 50 selected ARGs subtypes accounted for 99.00% of the 20 total ARGs types identified across the four samples, and used to analyze the effect of g-C3N4 on the abundance of ARGs in the samples (Figure 6A and Supplementary Table 8). Four genes [tetA(58), tetB(P), tetT, and tetA(46)] conferring resistance to TC also had higher abundant in the YCK and HCK than in the YTHPH and HTHPH, respectively. There were 45 ARGs subtypes conferring resistance to other antibiotics in the top 50 selected ARGs subtypes, e.g., macB, evgS, novA. The ARGs subtypes were less in g-C3N4 treatment groups (YTHPH, HTHPH) than those in CK groups (YCK, HCK), especially in pig farm sediment (Figure 6A and Supplementary Table 9). The g-C3N4 could reduce the abundance of most ARGs in sediment environment. TC/g-C3N4 treatment not only decreased the abundance of TRGs, but also the occurrence and diversity of non-TC ARGs, such as Multidrug resistance genes and MLS resistance genes.
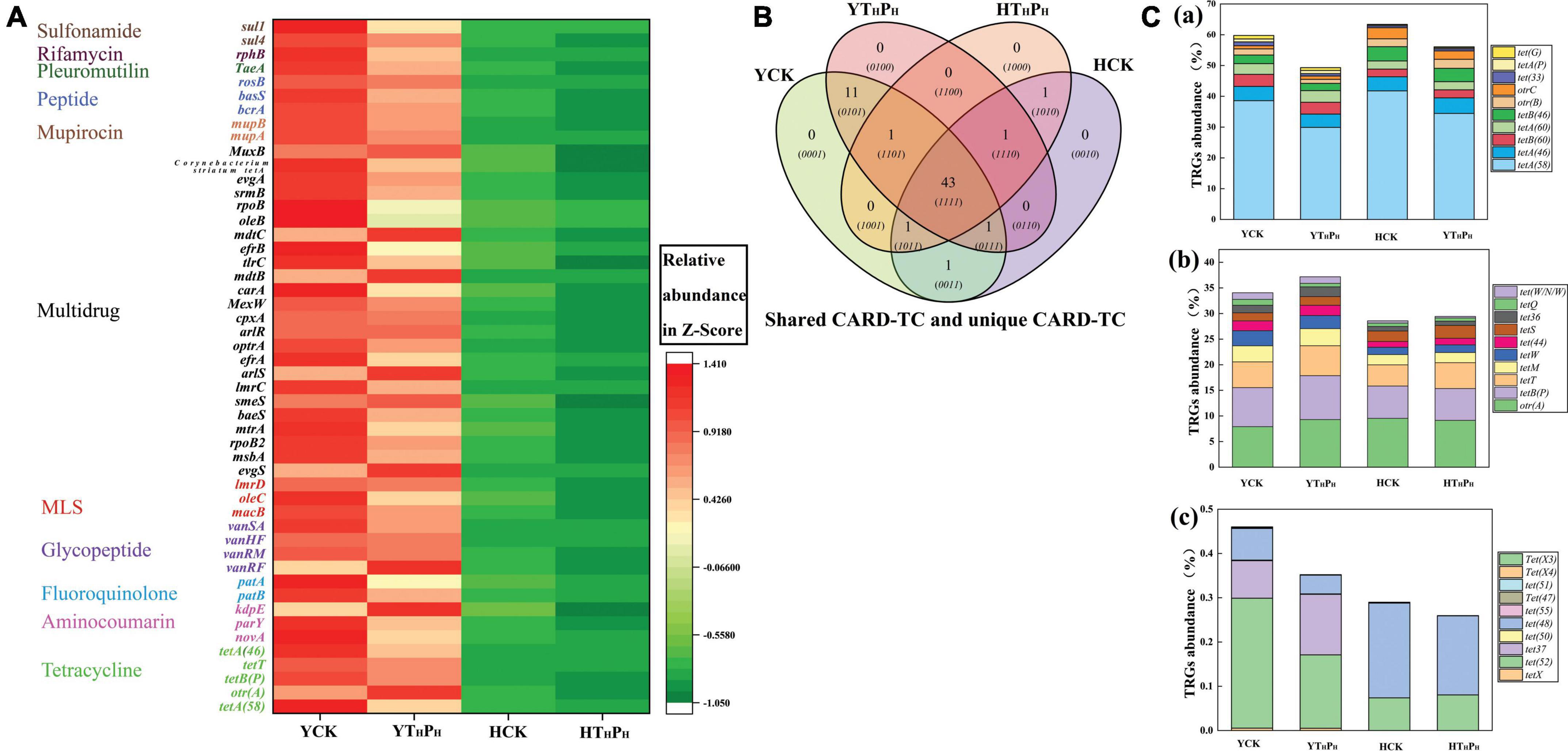
Figure 6. (A) Heatmap showing abundances of 50 background ARGs in 4 sediments. (B) Shared TRGs and unique TRGs. (C) Relative abundances of TRGs among sediment of ecosystems, (a) Efflux pump genes, (b) Ribosomal protection protein genes, (c) TC inactivation genes. YCK means the pig farm sediments alone treated for 30 days, HCK means the riverbed sediments alone treated for 30 days, YTHPH means the pig farm sediment system contain 180 mg⋅L– 1 of TC and 125 mg⋅kg– 1 of g-C3N4 for 30 days, HTHPH means the riverbed sediment system contain 180 mg⋅L– 1 of TC and 125 mg⋅kg– 1 of g-C3N4 for 30 days.
A total of 60 TRGs subtypes potentially conferring resistance to TC were detected out in the four sediment samples, most belonging to antibiotic efflux (34 TRGs subtypes), antibiotic inactivation (13 TRGs subtypes), and antibiotic target protection (13 TRGs subtypes). The shared and unique TRGs subtypes were examined by Venn diagrams for sediment samples (Figure 6B). 43 TRGs subtypes were shared by 4 sediments, accounting for 71.7% of TRGs abundance across all samples. Additionally, 11 TRGs subtypes were shared by 2 pig farm sediment samples (YCK, YTHPH), however, only 1 TRGs subtypes was shared by 2 riverbed sediments samples (HCK, HTHPH).
Much of the TRGs abundance was contributed by the gene components of efflux pump complexes, such as tetA(58), tetA(46), tetB(60), and tetA(60) (Figure 6C). Interestingly, tetA(58) had higher abundance than tetA(46), tetB(60), and tetA(60) by an order of magnitude. Compared to YCK and HCK, the percentage of tetA(58) decreased respectively from 37.59 and 41.77% to 29.91 and 34.44% in YTHPH and HTHPH, suggesting that g-C3N4 could decrease the abundance of tetA(58) in the environment, but there was no significant effect on other low abundance TRGs.
Networking plots were constructed to figure out the TRGs that were significantly correlated (correlation coefficient > 0.5) with dominant bacteria (p < 0.05) (Figure 7A). The tetA(58), tetA(46), tetB(60), and tetB(46) were the dominant genes in the samples and were positively related with Proteobacteria, Actinobacteria, Euryarchaeota, Acidobacteria, Firmicutes, Planctomycetes, Chloroflexi, unclassified_d__Bacteria. The genes of And tet(C), tet(H), tet(T), tet(S), tet(H), tetA(60), and tet(A) were negatively related with Acidobacteria. Thus, the elimination of TRGs by g-C3N4 in different sediments might attributed to the influence of g-C3N4 on their bacteria community structure. g-C3N4 reduced the relative abundance of bacteria with TRGs, such as Actinobacteria (21.96–12.07%), Proteobacteria (29.54–27.82%), Euryarchaeota (18.94–13.43%), and Firmicutes (8.06–5.71%) in the pig farm sediment. Similarly, g-C3N4 also reduced the relative abundance of bacteria with TRGs, such as Acidobacteria (13.05–10.58%) in the riverbed sediment (Figure 7B).
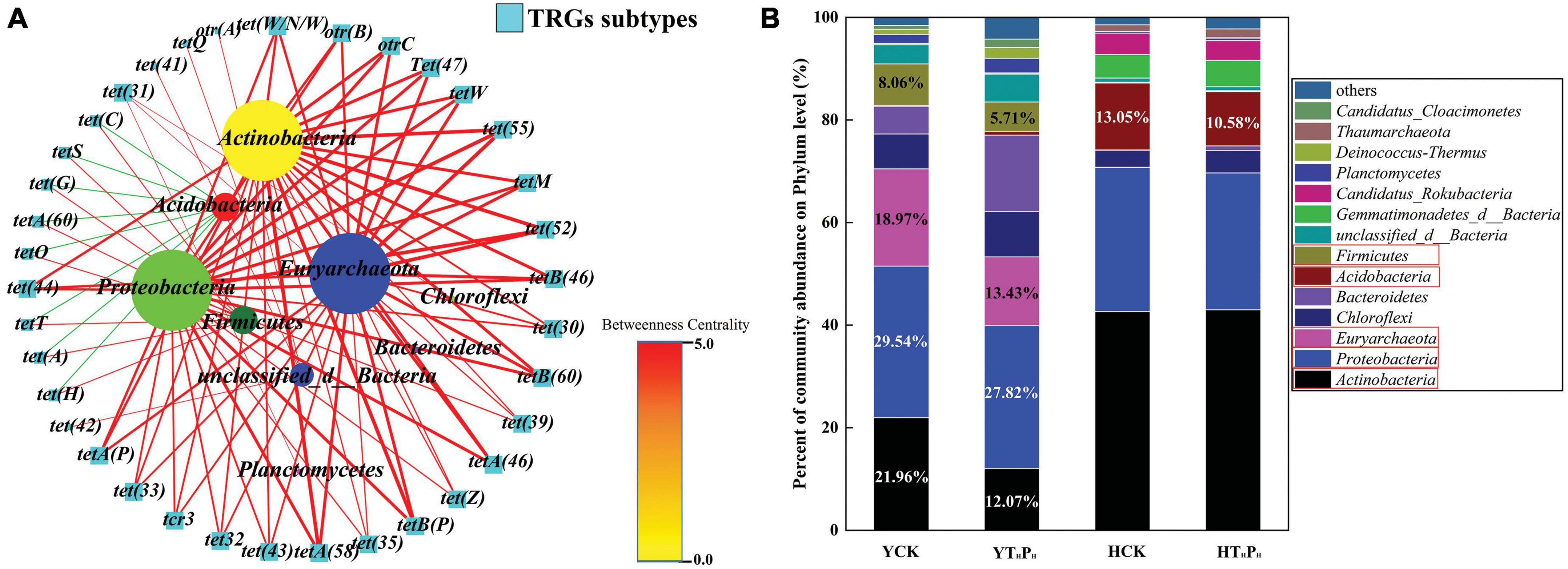
Figure 7. Network analyses among the TRGs, samples, and bacterial community across: YCK, HCK, YTHPH, and HTHPH. (A) TRGs-bacterial community network analyses. Red and green edges represent the positive and negative correlations, respectively. The network graphs were displayed with Cytoscape 3.5.2 in Prefuse Force Directed Layout by Betweenness centrality. (B) Distribution histograms of the bacteria with TRGs at the phylum levels. YCK means the pig farm sediments alone treated for 30 days, HCK means the riverbed sediments alone treated for 30 days, YTHPH means the pig farm sediment system contain 180 mg⋅L– 1 of TC and 125 mg⋅kg– 1 of g-C3N4 for 30 days, HTHPH means the riverbed sediment system contain 180 mg⋅L– 1 of TC and 125 mg⋅kg– 1 of g-C3N4 for 30 days.
Bacterial metabolic functional analysis
To obtain a better comprehension of the impact of g-C3N4 on the functional trait of TRGs during the removal of TC in sediment, the function level metabolism of eggNOG (evolutionary genealogy of genes: Non-supervised Orthologous Groups) category of the four sediment samples were performed and were depicted in Figure 8A and Supplementary Table 10. Obviously, the number of total predicted functional genes was higher in pig farm sediment than that in riverbed sediment (about 4.7 times higher). Overall, the predominant functional assignment across the whole dataset were mainly related to “Translation, ribosomal structure and biogenesis,” “Defense mechanism,” “Amino acid transport and metabolism,” “Inorganic ion transport and metabolism,” and “Carbohydrate transport and metabolism.” In addition, the abundance of the TRGs related to “Defense mechanism” were 30.37 and 29.89% in YCK and YTHPH, respectively, which were higher than those in HCK (24.41%) and HTHPH (22.93%). The genes related to “Amino acid transport and metabolism” were less prevalent in pig farm sediments than that in riverbed sediments. Relative abundance of genes related to “Carbohydrate transport and metabolism” and “Translation, ribosomal structure and biogenesis” were more prevalent in pig farm sediments than those in riverbed sediments. Additionally, for the above four functional genes, g-C3N4 treatment had very slight effect on the abundance of the other three functional genes except increasing the abundance of the functional genes related to “Translation, ribosomal structure and biogenesis.”
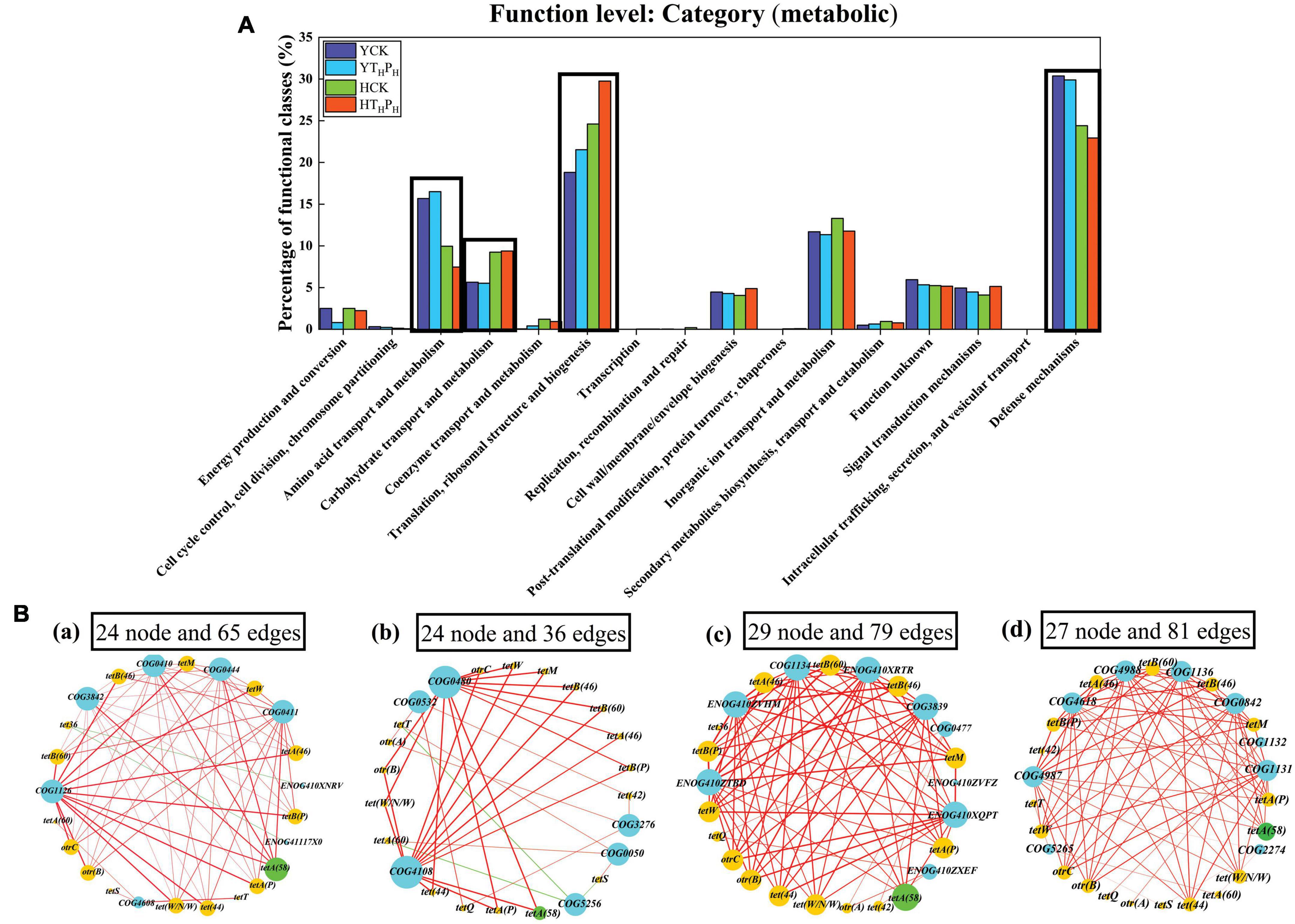
Figure 8. Bacterial metabolic functional analysis. (A) Metabolic functional traits and categories of the four sediment samples according to the eggNOG database. (B) Correlation network analysis of TRGs and the predominant metabolic functional, (a) Correlation network analysis of TRGs and “Amino acid transport and metabolism,” (b) Correlation network analysis of TRGs and “Carbohydrate transport and metabolism,” (c) Correlation network analysis of TRGs and “Translation, ribosomal structure and biogenesis,” (d) Correlation network analysis of TRGs and “Defense mechanism.” The network graphs were displayed with Cytoscape 3.5.2 in Prefuse Force Directed Layout by betweenness centrality (correlation coefficient > 0.5).
As shown in Figure 8B, the majority of the TRGs and bacterial metabolism function presented clearly strong correlations (correlation coefficient > 0.5), the interactions between TRGs and bacterial metabolic functions were complex, 29 nodes and 79 edges were observed from the interaction of TRGs with “Translation, ribosomal structure and biogenesis,” and 27 nodes and 81 edges were observed from the interaction between TRGs and “Defense mechanism.” Among these correlations, the correlation between tetA(58) and some of bacterial metabolism function had the most remarkable edges and the highest betweenness centrality. There were 5, 2, 6, and 6 edges of significant and strong positive correlations (p < 0.05) were identified between tetA(58) and “Amino acid transport and metabolism,” “Carbohydrate transport and metabolism,” “Translation, ribosomal structure and biogenesis,” and “Defense mechanisms,” respectively. Compared with other TRGs, tetA(58) had a higher degree centrality, closeness centrality, betweenness centrality and abundance in sediment environment, indicating it was very important in the structure of networks (Supplementary Figure 4). Overall, tetA(58) had high relative abundance in the sediment, and strongly related with most bacterial communities and metabolism functions. Therefore, tetA(58) had great contribution to the shifts in bacterial metabolism functions.
Discussion
The main active substances in the g-C3N4 photocatalytic process might be h+, and •O2– (Yang and Bian, 2020; Zheng et al., 2021). Photogenerated holes (h+) and superoxide radical (•O2–) are the main active species for TC oxidative degradation, but hydroxyl free radicals are not the main oxidizing substances, and the influence order of active species is h+ > •O2– > •OH (Liu et al., 2022). The removal efficiency of TC residues was lower in pig farm sediment than that in riverbed sediment (Figure 3), reflecting the TC removal capacity of sediment itself and the photocatalytic degradation capacity of g-C3N4. The reasons were mainly resulted from (a): The pig farm sediment had more complex nutrients than riverbed sediment, such as proteins, sugars and polyunsaturated fatty acids, which could provide more carbon sources for microorganisms. Perrin et al. (2020) pointed out that when multiple carbon sources were available, microorganisms metabolize them sequentially and some substances would be used preferentially. As a carbon source with poor bioavailability, TC would not be easily metabolized by microorganisms, especially in pig farm sediment where there were some other carbon sources that could be metabolized preferentially. (b) The removal efficiency of TC residues by g-C3N4 could be influenced by sediment environment conditions and characteristics of transmission light. The low transmittance of pig farm sediment might be due to the decrease of g-C3N4 light absorption efficiency due to the increased turbidity of pig farm sediment (Zhong et al., 2020; Sun et al., 2022b). Consequently, less light energy was available for g-C3N4 to degrade TC in pig farm sediments (Shi et al., 2021). Moreover, g-C3N4 often possesses small specific surface area and serious charge recombination, which limits its application in sediments (Liu et al., 2022; Shi et al., 2022).
Microorganisms was the most abundant group of organisms on earth and played an important role in maintaining soil biological activity and removing contaminants from the environment (Yan et al., 2020). The diversity and richness of microbial in sediment were an important indicator of environmental pollution. The bacterial diversity and richness were usually higher in most of uncontaminated sites of riverbed sediment than in contaminated sediment (Wang et al., 2015). The microbiota had higher richness and diversity in soil irrigated by river water than in soil irrigated by wastewater (He et al., 2019). In our study, the bacterial diversity and abundance in riverbed sediment were higher than those in pig farm sediment (Supplementary Tables 3, 4). These differences might mainly be attributed to (a): Bacterial growth was strongly dependent on nutrient conditions. The complex components in the pig farm sediment were not only used as nutrients for some bacteria but also used to induce toxic effects on other bacteria for inhibiting their growth and production (Cerro-Galvez et al., 2019). (b) the amounts of nutrients were different between the two sediment types, especially for organic matter and NH4+-N, (Supplementary Table 1), which might be the main factors in affecting bacteria diversity (Huang et al., 2016; Zhai et al., 2021). (c) TC concentration might be another key factor influencing the microbial community compositions of the two types of sediment samples (Gao et al., 2020). Moreover, the presence of TC altered the microbial communities and diversity, and microbial abundance also was associated with its ability to acquire ARGs (Zhang M. Q. et al., 2019). In the environment with excess nutrients, bacterial communities were able to remain a stable status for a long time despite of the perturbations of external conditions (Kearns et al., 2016). Islam and Gilbride (2019) reported that the bacterial community of swine sediment were diverse and remained stable during the TC treatment experiment. In the current study, we found that the addition of TC, g-C3N4 or TC/g-C3N4 had more obvious effect on bacterial communities’ structure in riverbed sediment than in pig farm sediment. Since the pig farm sediment possessed much more nutrients, it could effectively resist the impact of external perturbation on bacterial diversity and, thus, the diversity changes were not obvious in this sediment type. In contrast, the nutrients were inadequate in riverbed sediments and the richness of microbial community increased in response to resist the perturbations of external conditions. The microbial community might reach to a new stability in the environment (Bai et al., 2019). Therefore, TC, g-C3N4 or TC/g-C3N4 significantly increased the richness and diversity of the bacterial community in riverbed sediment type (Tang et al., 2021).
The network stability was normally strongly correlated with the complexity of molecular ecological networks (Dubey et al., 2021; Yuan et al., 2021). If the bacterial community network was simplified, the stability of the bacterial community might be reduced (Dubey et al., 2021). Previous study revealed that the accumulation of nutrients enhanced positive interaction between bacterial communities but reduced the stability of sediment ecosystem (Zhang L. L. et al., 2021). However, our research showed that pig farm sediment had abundant nutrition substances and had a significant complex bacterial network (Figure 4B). Therefore, TC, g-C3N4 and TC/g-C3N4 application had less effects on the bacterial community structure in pig farm sediment.
Studies had found that Firmicutes and Bacteroidetes were the dominant bacteria in pig manure (Luo et al., 2020). Xie et al. (2021) studied the microbial composition of riverbed sediment at different geographical locations and found that Proteobacteria was the most dominant bacterial flora, orderly followed by Actinobacteria and Firmicutes (Figure 5). The finding was consistent with our results for dominant microorganisms in both sediments. The relative abundances of Acidobacteriota in the g-C3N4 treatment was higher than the original communities. Acidobacteriota might be a significant component of the natural environment and played significant ecological roles in key carbon, nitrogen, and sulfur biogeochemical circuits (Kalam et al., 2020). Our result confirmed that g-C3N4 would be beneficial to the metabolism of certain microorganisms in riverbed sediment. Recently, photocatalysis had been found to exhibit remarkable antibacterial activity against some of bacterial under light and dark conditions (Balapure et al., 2020). Our study illustrated that for top ten dominant genera, only two genera, Anaeromyxobacter and norank_f__Geobacteraceae were in low abundance and have been significantly inhibited by g-C3N4 in riverbed sediment (Figure 5Bb). This suggests that g-C3N4 might produce weak toxicity in some low-abundance species, and had no obvious adverse effects on high-abundance species. In a word, when g-C3N4 degraded TC, the bacterial community structure in the two sediment types was basically stable, indicating that g-C3N4 did not obvious adverse impact on the bacterial community.
ARGs might have higher abundance in the presence of antibiotics (Zhang et al., 2020; Liu C. et al., 2021). Our research showed that ARGs such as Multidrug, TC and MLS resistance genes were much higher in pig farm sediment than in riverbed sediment (Supplementary Figure 8). Numerous studies showed that the increase of TC concentration in the environment contributed to the generation and transmission of TRGs (Gao et al., 2018; Zhang M. Q. et al., 2019). Hence, the higher prevalence and a more diverse repertory of virulence and ARGs in TC polluted samples (Jia et al., 2018). g-C3N4 treatment not only decreased the abundance of TRGs, but also reduced the occurrence and diversity of non-TC ARGs (Figure 6). Under the illumination of visible light, g-C3N4 could be involved in the oxidation/reduction of various organic substances, which significantly decreased the TC concentration as well as the level of ARGs and TRGs. Three phyla (Actinobacteria, Proteobacteria, and Euryarchaeota) showed strong positive correlations with different TRGs (Figure 7A), suggesting that these phyla should be the dominant hosts for these TRGs. g-C3N4 could reduce the relative abundance of TRGs by decreasing the relative abundance of potential hosts of TRGs (Figure 7B). These results were well consistent with the findings obtained by other researchers (Wang et al., 2019; Tang et al., 2021). Luo et al. (2021b) reported that photocatalyst could effectively remove TC in sediment through photocatalysis which might weaken the transport of TC from inside microbial cells, and result in the decrement of antibiotic efflux pump genes [tetA(58)] in sediment. Their results were further confirmed by our study, since g-C3N4 could reduce the abundance of tetA(58) in sediment. Thereby, like other photocatalysts (Yu et al., 2020; Gonzalez-Gaya et al., 2021), g-C3N4 could lower the abundance of bacteria with TRGs in the environment and reduced the risk of TRGs transmission in the environment.
Our results demonstrated the pig farm sediment contained more TRGs related to “Amino acid transport and metabolism” and “Defense mechanism” than riverbed sediment. Previous studies showed that amino “Amino acid transport and metabolism” function was correlated with TRGs (Wang et al., 2019; Zhang T. T. et al., 2021). Therefore, this metabolic function was extremely vital for bacteria to counteract the detrimental influence of antibiotic like TC (Zhang T. T. et al., 2021). The complex bacterial network structure and rich nutrients in the pig farm sediment made it have a strong “Defense mechanism.” These results implied the potential effect of TRGs on the metabolic functions of the specific communities in different sediment (Shen et al., 2020). Meanwhile, g-C3N4 treatment also affected the metabolic function of the sediment bacterial community (Hou et al., 2020). It is because g-C3N4 could damage bacterial cell membranes by destroying the structures of nucleic acids, proteins and other macromolecules embedded in the membranes, thus affecting function of bacterial metabolic (Zhang M. Q. et al., 2019; Zhang T. T. et al., 2021), especially in “Translation, ribosomal structure and biogenesis” (Figure 8A).
Tetracycline efflux pump genes [tetA(58)], which had the largest proportion of TRGs, was found in the riverbed sediment and pig farm sediment samples (Figures 6, 7). Moreover, antibiotic efflux pump genes [tetA(58)] had stable and positive connectivity with metabolic function of bacteria (Luo et al., 2021b). Classifying TRGs by aligning them to the eggNOG, we found that tetA(58) had significant and strong correlations with bacterial metabolism functions, especially with “Translation, ribosomal structure and biogenesis” and “Defense mechanisms,” indicating that tetA(58) was the dominant TRGs of functional metabolism in bacterial communities. The relative abundance of tetA(58) in g-C3N4 treatment was lower than in the CK samples, suggesting the bacterial cell membranes destruction induced by g-C3N4 increased the permeability of bacterial cells and caused the TRGs being released and eliminated (Luo et al., 2021a). Thus, g-C3N4 affected the metabolic function of bacteria and played a positive role in the removal of TRGs.
Conclusion
This study investigated the responses of TRGs and microbial community in two sediment types against exposure of TC, g-C3N4 and TC/g-C3N4. g-C3N4 could effectively remove TC residues in pig farm sediment and riverbed sediment, with a high removal efficiency of TC in riverbed sediment type. The microbial community was shifted under the TC, g-C3N4 and TC/g-C3N4 exposure in riverbed sediment type, especially Acidobacteriota, Actinobacteriota, and Desulfobacterota. On the contrary, no significant change was found in the microbial community in pig farm sediment type under the TC, g-C3N4 and TC/g-C3N4 exposure. It was because there was a complex bacterial network in the pig farm sediment environment. In general, g-C3N4 treatment had little effect on the structure of the bacterial community in the two sediments, and the community structure was basically stable, especially in the pig farm sediment. Meanwhile, g-C3N4 treatment not only decreased the abundance of TRGs, but also reduced occurrence and diversity of non-TC ARGs, due to the inhibitory effects of g-C3N4 on the growth of related resistant bacteria. Interestingly, TRGs were significantly associated with metabolic functions of bacteria. Therefore, g-C3N4 could affect bacterial related metabolic functions by affecting TRGs and contributed to the removal of TRGs. Our results suggested that g-C3N4 was an environmentally friendly photocatalyst and had no obvious adverse effect on the bacterial community and the abundance of TRGs in the sediments. This study could provide some new insights for g-C3N4 for removal of TC contaminants from sediment.
Data availability statement
All data generated or analysed during this study are included in this published article and its supplementary information files. The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA760830.
Author contributions
XH designed the sampling and experiments and drafted the manuscript. XC, YT, and ZX performed the experiment and manuscript preparation. YeZ, YWa, and YuZ performed the data analyses. YWu and GW helped perform the analysis with constructive discussions and revisions of text passages. All authors commented on an early draft of the manuscript.
Funding
This work was supported in Key Research and Development Project of Hunan Province (2021NK20180), Research Foundation of Education Bureau of Hunan Province (19A509), Key Research and Development Project of Guangxi Province (AB21220026), Natural Science Foundation of Changsha, China (kq2202280), and National Science Foundation of China (31971462).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.964401/full#supplementary-material
References
Bai, Y., Xu, R., Wang, Q. P., Zhang, Y. R., and Yang, Z. H. (2019). Sludge anaerobic digestion with high concentrations of tetracyclines and sulfonamides: dynamics of microbial communities and change of antibiotic resistance genes. Bioresour. Technol. 276, 51–59. doi: 10.1016/j.biortech.2018.12.066
Balapure, A., Nikhariya, Y., Boppudi, N. S. S., Ganesan, R., and Dutta, J. R. (2020). Highly dispersed nanocomposite of AgBr in g-C3N4 matrix exhibiting efficient antibacterial effect on drought-resistant pseudomonas putida under dark and light conditions. ACS Appl. Mater. Inter. 12, 21481–21493. doi: 10.1021/acsami.0c05158
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cerro-Galvez, E., Sala, M. M., Marrase, C., Gasol, J. M., Dachs, J., and Vila-Costa, M. (2019). Modulation of microbial growth and enzymatic activities in the marine environment due to exposure to organic contaminants of emerging concern and hydrocarbons. Sci. Total. Environ. 678, 486–478. doi: 10.1016/j.scitotenv.2019.04.361
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chen, Z. H., Guo, F., Sun, H. R., Shi, Y. X., and Shi, W. L. (2022). Well-designed three-dimensional hierarchical hollow tubular g-C3N4/ZnIn2S4 nanosheets heterostructure for achieving efficient visible-light photocatalytic hydrogen evolution. J. Colloid Interf. Sci. 607, 1391–1401. doi: 10.1016/J.JCIS.2021.09.095
Dubey, M., Hadadi, N., Pelet, S., Carraro, N., Johnson, D. R., and van der Meer, J. R. (2021). Environmental connectivity controls diversity in soil microbial communities. Commun. Biol. 4:492.
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/NMETH.2604
Gao, B., Dou, M. M., Wang, J., Li, S. M., Wang, D. Y., Ci, L., et al. (2021). Efficient persulfate activation by carbon defects g-C3N4 containing electron traps for the removal of antibiotics, resistant bacteria and genes. Chem. Eng. J. 426, 131677–131677. doi: 10.1016/j.cej.2021.131677
Gao, F. Z., He, L. Y., He, L. X., Zou, H. Y., Zhang, M., Wu, D. L., et al. (2020). Untreated swine wastes changed antibiotic resistance and microbial community in the soils and impacted abundances of antibiotic resistance genes in the vegetables. Sci. Total. Environ. 741:140482. doi: 10.1016/j.scitotenv.2020.140482
Gao, P., Xu, W. L., Ruan, X. H., Qian, Y. J., Xue, G., and Jia, H. Z. (2018). Long-term impact of a tetracycline concentration gradient on the bacterial resistance in anaerobic-aerobic sequential bioreactors. Chemosphere 205, 308–316. doi: 10.1016/j.chemosphere.2018.04.135
Gonzalez-Gaya, B., Garcia-Bueno, N., Buelow, E., Marin, A., and Rico, A. (2021). Effects of aquaculture waste feeds and antibiotics on marine benthic ecosystems in the Mediterranean Sea. Sci. Total Environ. 806:151190. doi: 10.1016/j.scitotenv.2021.151190
Guo, F., Chen, Z. H., Huang, X. L., Gao, L. W., Cheng, X. F., Shi, W. L., et al. (2021). Cu3P nanoparticles decorated hollow tubular carbon nitride as a superior photocatalyst for photodegradation of tetracycline under visible light. Sep. Purif. Technol. 275:119223. doi: 10.1016/j.seppur.2021.119223
Guo, M. T., and Tian, X. B. (2019). Impacts on antibiotic-resistant bacteria and their horizontal gene transfer by graphene-based TiO2 &Ag composite photocatalysts under solar irradiation. J. Hazard. Mater. 380:120877. doi: 10.1016/j.jhazmat.2019.120877
He, G., Jiang, X. L., Yao, L. G., Liu, G. H., Yang, Y. Y., Jiang, Y., et al. (2021). Effects of tetracycline on nitrogen and carbon cycling rates and microbial abundance in sediments with and without biochar amendment. Chemosphere 270:129509. doi: 10.1016/j.chemosphere.2020.129509
He, L. Y., He, L. K., Liu, Y. S., Zhang, M., Zhao, J. L., Zhang, Q. Q., et al. (2019). Microbial diversity and antibiotic resistome in swine farm environments. Sci. Total. Environ. 685, 197–207. doi: 10.1016/j.scitotenv.2019.05.369
Henriksson, P. J. G., Rico, A., Troell, M., Klinger, D., Buschmann, A. H., Saksida, S., et al. (2018). Unpacking factors influencing antimicrobial use in global aquaculture and their implication for management: a review from a systems perspective. Sustain. Sci. 13, 1105–1120. doi: 10.1007/s11625-017-0511-8
Hou, F., Liu, J. N., Zhang, Y. H., Zhao, C., Xiao, X. D., Zou, J. L., et al. (2020). Synthesis of metallic copper modified g-C3N4 by molecular self-assembly structure and its combined catalytic performance with activated sludge. J. Hazard. Mater. 388:121754. doi: 10.1016/j.jhazmat.2019.121754
Huang, H. N., Chen, Y. G., Zheng, X., Su, Y. L., Wan, R., and Yang, S. Y. (2016). Distribution of tetracycline resistance genes in anaerobic treatment of waste sludge: the role of pH in regulating tetracycline resistant bacteria and horizontal gene transfer. Bioresour. Technol. 218, 1284–1289. doi: 10.1016/j.biortech.2016.07.097
Islam, G. M., and Gilbride, K. A. (2019). The effect of tetracycline on the structure of the bacterial community in a wastewater treatment system and its effects on nitrogen removal. J. Hazard. Mater. 371, 130–137. doi: 10.1016/j.jhazmat.2019.02.032
Jia, Y. Y., Khanal, S. K., Shu, H. Y., Zhang, H. Q., Chen, G. H., and Lu, H. (2018). Ciprofloxacin degradation in anaerobic sulfate-reducing bacteria (SRB) sludge system: mechanism and pathways. Water. Res. 136, 64–74. doi: 10.1016/j.watres.2018.02.057
Jun, P., Xi, C., Wu, Y., Wu, H., and Tan, P. (2015). “Fabrication of g-C3N4 based organic-inorganic nanocomposite and application in the field of catalytic elimination of environmental pollutants,” in Proceedings of the fourth international conference on new materials, Chongqing.
Kalam, S., Basu, A., Ahmad, I., Sayyed, R. Z., ElEnshasy, H. A., Dailin, D. J., et al. (2020). Recent understanding of soil Acidobacteria and their ecological significance: a critical review. Front. Microbiol. 11:580024. doi: 10.3389/fmicb.2020.580024
Kearns, P. J., Angell, J. H., Howard, E. M., Deegan, L. A., Stanley, R. H. R., and Bowen, J. L. (2016). Nutrient enrichment induces dormancy and decreases diversity of active bacteria in salt marsh sediments. Nat. Commun. 7:12881. doi: 10.1038/ncomms12881
Liu, C., Li, X. H., Zheng, S. N., Kai, Z., Jin, T., Shi, R. G., et al. (2021). Effects of wastewater treatment and manure application on the dissemination of antimicrobial resistance around swine feedlots. J. Clean. Prod. 280:123794. doi: 10.1016/j.jclepro.2020.123794
Liu, E. L., Lin, X., Hong, Y. Z., Yang, L., Luo, B. F., Shi, W. L., et al. (2021). Rational copolymerization strategy engineered C self-doped g-C3N4 for efficient and robust solar photocatalytic H2 evolution. Renew. Energ. 178, 757–765. doi: 10.1016/j.renene.2021.06.066
Liu, H., Huo, W. C., Zhang, T. C., Ouyang, L., and Yuan, S. J. (2022). Photocatalytic removal of tetracycline by a Z-scheme heterojunction ofbismuth oxyiodide/exfoliated g-C3N4: performance, mechanism, and degradation pathway. Mater. Today Chem. 23:100729. doi: 10.1038/nm.3639
Luo, J. Y., Cheng, X. S., Su, Y. L., Zhang, L., Du, W., Bao, X. C., et al. (2021b). Metagenomic assembly deciphered the type-dependent effects of surfactants on the fates of antibiotics resistance genes during sludge fermentation and the underlying mechanisms. Sci. Total Environ. 807:150822. doi: 10.1016/j.scitotenv.2021.150822
Luo, J. Y., Huang, W. X., Zhang, Q., Wu, Y., Fang, F., Cao, J. S., et al. (2021a). Distinct effects of hypochlorite types on the reduction of antibiotic resistance genes during waste activated sludge fermentation: insights of bacterial community, cellular activity, and genetic expression. J. Hazard. Mater. 403:124010. doi: 10.1016/j.jhazmat.2020.124010
Luo, L., Zhang, C. J., Zhang, Z., Peng, J., Han, Y. Q., Han, Y. Q., et al. (2020). Differences in tetracycline antibiotic resistance genes and microbial community structure during aerobic composting and anaerobic digestion. Front. Microbiol. 11:583995. doi: 10.3389/fmicb.2020.583995
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mahmoud, M. A. M., and Abdel-Mohsein, H. S. (2019). Hysterical tetracycline in intensive poultry farms accountable for substantial gene resistance, health and ecological risk in Egypt-manure and fish. Environ. Pollut. 255:113039. doi: 10.1016/j.envpol.2019.113039
Martinez-Porchas, M., and Vargas-Albores, F. (2017). Microbial metagenomics in aquaculture: a potential tool for a deeper insight into the activity. Rev. Aquacult. 9, 42–56. doi: 10.1111/raq.12102
Nath, R. K., Zain, M., and Jamil, M. (2016). An environment-friendly solution for indoor air purification by using renewable photocatalysts in concrete: a review. Renew. Sust. Energ. Rev. 62, 1184–1194. doi: 10.1016/j.rser.2016.05.018
Perrin, E., Ghini, V., Giovannini, M., Di Patti, F., Cardazzo, B., Carraro, L., et al. (2020). Diauxie and co-utilization of carbon sources can coexist during bacterial growth in nutritionally complex environments. Nat. Commun. 11:3135. doi: 10.1038/s41467-020-16872-8
Santas-Miguel, V., Fernandez-Sanjurjo, M. J., Nunez-Delgado, A., Alvarez-Rodriguez, E., Diaz-Ravina, M., Arias-Estevez, M., et al. (2020). Use of biomass ash to reduce toxicity affecting soil bacterial community growth due to tetracycline antibiotics. J. Environ. Manage. 269:110838. doi: 10.1016/j.jenvman.2020.110838
Serra, A., Zhang, Y., Sepulveda, B., Gomez, E., Nogues, J., Michler, J., et al. (2020). Highly reduced ecotoxicity of ZnO-based micro/nanostructures on aquatic biota: influence of architecture, chemical composition, fixation, and photocatalytic efficiency. Water Res. 169, 115210.1–115210.12. doi: 10.1016/j.watres.2019.115210
Shen, J., Liu, Z. Y., Yu, H. N., Ye, J. S., Long, Y., Zhou, P. L., et al. (2020). Systematic stress adaptation of Bacillus subtilis to tetracycline exposure. Ecotoxi. Environ. Safe. 188:109910. doi: 10.1016/j.ecoenv.2019.109910
Shi, Y. Y., Li, L. L., Sun, H. R., Zheng, X., Yi, C., Shi, W. L., et al. (2022). Engineering ultrathin oxygen-doped g-C3N4 nanosheet for boosted photoredox catalytic activity based on a facile thermal gas-shocking exfoliation effect. Sep. Purif. Technol. 292:121038. doi: 10.1016/j.seppur.2022.121038
Shi, Z. Y., Rao, L., Wang, P. F., Wang, Y. X., and Zhang, L. X. (2021). Characteristics of transmission light in tetracycline hydrochloride polluted wastewater and the response of g-C3N4 under different transmission spectral range during the photodegradation process. Chemosphere 263:128196. doi: 10.1016/j.chemosphere.2020.128196
Sun, H. R., Shi, Y. X., Shi, W. L., and Guo, F. (2022a). High-crystalline/amorphous g-C3N4 S-scheme homojunction for boosted photocatalytic H2 production in water/simulated seawater: interfacial charge transfer and mechanism insight. Appl. Surf. Sci. 593:153281. doi: 10.1016/J.APSUSC.2022.153281
Sun, H. R., Wang, L. J., Guo, F., Shi, Y. X., Li, L. L., Xu, Z., et al. (2022b). Fe-doped g-C3N4 derived from biowaste material with Fe-N bonds for enhanced synergistic effect between photocatalysis and Fenton degradation activity in a broad pH range. J. Alloy. Compd. 900:163410. doi: 10.1016/J.JALLCOM.2021.163410
Tang, C. Y., Liu, C., Han, Y., Guo, Q. Q., Ouyang, W., Feng, H. J., et al. (2019). Nontoxic carbon quantum dots/g-C3N4 for efficient photocatalytic inactivation of staphylococcus aureus under visible light. Adv. Healthc. Mater. 8:1801534. doi: 10.1002/adhm.201801534
Tang, H. F., Shang, Q., Tang, Y. H., Liu, H. L., Zhang, D. Y., Du, Y., et al. (2021). Filter-membrane treatment of flowing antibiotic-containing wastewater through peroxydisulfate-coupled photocatalysis to reduce resistance gene and microbial inhibition during biological treatment. Water Res. 207:117819. doi: 10.1016/j.watres.2021.117819
Tobajas, M., Belver, C., and Rodriguez, J. J. (2017). Degradation of emerging pollutants in water under solar irradiation using novel TiO2-ZnO/clay nanoarchitectures. Chem. Eng. J. 309, 596–606. doi: 10.1016/j.cej.2016.10.002
Wang, J. B., Li, X., Zhou, Z. W., and Fan, X. Y. (2019). Bacterial communities, metabolic functions and resistance genes to antibiotics and metals in two saline seafood wastewater treatment systems. Bioresource. Technol. 287:121460. doi: 10.1016/j.biortech.2019.121460
Wang, L., Shen, J., and Calvin, K. L. C. (2015). City profile: suzhou-a Chinese city under transformation. Cities 44, 60–72. doi: 10.1016/j.cities.2014.12.005
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb 73, 5261–5267. doi: 10.1128/AEM.00062-07
Xie, N., Zhong, L. P., Ouyang, L., Xu, W., Zeng, Q., Zeng, Q. H., et al. (2021). Community composition and function of bacteria in activated sludge of municipal wastewater treatment plants. Water 13:852. doi: 10.3390/w13060852
Yan, Y. C., Zhou, X. Q., Yu, P., Li, Z. F., and Zheng, T. L. (2020). Characteristics, mechanisms and bacteria behavior of photocatalysis with a solid Z-scheme Ag/AgBr/g-C3N4 nanosheet in water disinfection. Appl. Catal. A Gen. 590:117282. doi: 10.1016/j.apcata.2019.117282
Yang, Y. J., and Bian, Z. Y. (2020). Oxygen doping through oxidation causes the main active substance in g-C3N4 photocatalysis to change from holes to singlet oxygen. Sci. Total. Environ. 753:141908. doi: 10.1016/j.scitotenv.2020.141908
Yu, P., Zhou, X. Q., Li, Z. F., and Yan, Y. C. (2020). Inactivation and change of tetracycline-resistant escherichia coli in secondary effluent by visible light-driven photocatalytic process using Ag/AgBr/g-C3N4. Sci. Total Environ. 705:135639. doi: 10.1016/j.scitotenv.2019.135639
Yuan, M. M., Guo, X., Wu, L. W., Zhang, Y., Xiao, N. J., Ning, D. L., et al. (2021). Climate warming enhances microbial network complexity and stability. Nat. Clim. Change 11, 343–348. doi: 10.1038/s41558-021-00989-9
Zhai, Y. J., Chen, L. H., Liu, G., Song, L., ArenasLago, D., Kong, L. C., et al. (2021). Compositional and functional responses of bacterial community to titanium dioxide nanoparticles varied with soil heterogeneity and exposure duration. Sci. Total Environ. 773:144895. doi: 10.1016/j.scitotenv.2020.144895
Zhan, L., Xia, Z. W., Xu, Z. M., and Xie, B. (2021). Study on the remediation of tetracycline antibiotics and roxarsone contaminated soil. Environ. Pollut. 271:116312. doi: 10.1016/j.envpol.2020.116312
Zhang, C., Li, Y., Shuai, D. M., Shen, Y., Xiong, W., and Wang, L. Q. (2019). Graphitic carbon nitride (g-C3N4)-based photocatalysts for water disinfection and microbial control: a review. Chemosphere 214, 462–479. doi: 10.1016/j.chemosphere.2018.09.137
Zhang, H., Du, M., Jiang, H., Zhang, D., Lin, L., and Ye, H. (2015). Occurrence, seasonal variation and removal efficiency of antibiotics and their metabolites in wastewater treatment plants, Jiulongjiang River Basin, South China. Environ. Sci. Proc. Imp. 17, 225–234. doi: 10.1039/c4em00457d
Zhang, L. L., Zhang, C., Lian, K. T., and Liu, C. X. (2021). Effects of chronic exposure of antibiotics on microbial community structure and functions in hyporheic zone sediments. J. Hazard. Mater. 416:126141. doi: 10.1016/j.jhazmat.2021.126141
Zhang, M. Q., Yuan, L., Li, Z. H., Zhang, H. C., and Sheng, G. P. (2019). Tetracycline exposure shifted microbial communities and enriched antibiotic resistance genes in the aerobic granular sludge. Environ. Int. 130:104902. doi: 10.1016/j.envint.2019.06.012
Zhang, T. T., Zhu, X., Guo, J. H., Gu, A. Z., Li, D., and Chen, J. M. (2021). Toxicity Assessment of Nano-ZnO Exposure on the Human Intestinal Microbiome, Metabolic Functions, and Resistome Using an In Vitro Colon Simulator. Environ. Sci. Technol. 55, 6884–6896. doi: 10.1021/acs.est.1c00573
Zhang, X., Wang, J., Dong, X. X., and Lv, Y. K. (2020). Functionalized metal-organic frameworks for photocatalytic degradation of organic pollutants in environment. Chemosphere 242, 125144.1–125144.15. doi: 10.1016/j.chemosphere.2019.125144
Zhao, R., Yu, K., Zhang, J., Zhang, G., Huang, J. Y., Zhang, G. J., et al. (2020). Deciphering the mobility and bacterial hosts of antibiotic resistance genes under antibiotic selection pressure by metagenomic assembly and binning approaches. Water Res. 186:116318. doi: 10.1016/j.watres.2020.116318
Zheng, X., Shi, Y. X., Li, L. L., Sun, H. R., Amin, M. D. S., Gao, F., et al. (2021). Fabrication of 2D/2D Z-scheme highly crystalline carbon nitride/δ-Bi2O3 heterojunction photocatalyst with enhanced photocatalytic degradation of tetracycline. J. Alloy Compd. 895:162772. doi: 10.1016/J.JALLCOM.2021.162667
Keywords: tetracycline, g-C3N4, bacterial community, metagenomic analysis, TRGs, functional analysis
Citation: Hu X, Chen X, Tang Y, Xu Z, Zeng Y, Wang Y, Zhao Y, Wu Y and Wang G (2022) Effects of g-C3N4 on bacterial community and tetracycline resistance genes in two typical sediments in tetracycline pollution remediation. Front. Microbiol. 13:964401. doi: 10.3389/fmicb.2022.964401
Received: 08 June 2022; Accepted: 17 August 2022;
Published: 16 September 2022.
Edited by:
Alba Edith Vega, National University of San Luis, ArgentinaReviewed by:
Chen Cai, University of Science and Technology of China, ChinaFeng Guo, Jiangsu University of Science and Technology, China
Copyright © 2022 Hu, Chen, Tang, Xu, Zeng, Wang, Zhao, Wu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaohui Wu, d3loNzUyMTAwQDE2My5jb20=; Guangjun Wang, Y3N1ZnR3Z2pAMTYzLmNvbQ==
 Xuemei Hu1
Xuemei Hu1 Zhenggang Xu
Zhenggang Xu Yonghong Wang
Yonghong Wang Yaohui Wu
Yaohui Wu