- 1State Key Laboratory for Managing Biotic and Chemical Treats to the Quality and Safety of Agro-Products, Institute of Plant Protection and Microbiology, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2The Key Laboratory for Quality Improvement of Agricultural Products of Zhejiang Province, College of Advanced Agricultural Sciences, Zhejiang Agriculture and Forest University, Hangzhou, China
- 3College of Food and Health, Zhejiang Agriculture and Forest University, Hangzhou, China
- 4College of Forestry and Biotechnology, Zhejiang Agriculture and Forest University, Hangzhou, China
Peroxisomes have been proved playing roles in infection of several plant pathogens. Although the contribution of a portion of peroxins in pathogenicity was demonstrated, most of them are undocumented in fungi, especially, Botrytis cinerea. The homologs of Pex8, Pex10, and Pex12 in B. cinerea were functionally characterized in this work using gene disruption strategies. Compared with the wild-type strain (WT), the Δbcpex8, Δbcpex10, and Δbcpex12 mutants exhibited significant reduction in melanin production, fatty acid utilization, and decreased tolerance to high osmotic pressure and reactive oxygen species (ROS). The mycelial growth and conidiation of were significantly inhibited in Δbcpex8, Δbcpex10, and Δbcpex12 strains. The mycelial growth rates of Δbcpex8, Δbcpex10, and Δbcpex12 were reduced by 32, 35, and 34%, respectively, compared with WT and ectopic transformant (ET), and the conidiation was reduced by approximately 89, 27, and 88%, respectively. The conidial germination, germ tube elongation, and the formation of initiate infection structures (IFSs) were also reduced by the deletion of the genes. The pathogenicity was tested on the leaves of tobacco and strawberry, and fruits of tomato. On the leaves of tobacco and strawberry, the Δbcpex8, Δbcpex10, and Δbcpex12 mutants could not induce necrotic lesions, and the lesions on tomato fruits infected with the mutants were significantly reduced than those of the wide type. The results indicated that BcPEX8, BcPEX10, and BcPEX12 are indispensable for the development and pathogenicity of B. cinerea.
Introduction
Botrytis cinerea, a typical phytopathogen with a necrotrophic lifestyle infecting more than 1,000 species worldwide, has induced huge losses in agro products (Qin et al., 2010; Weiberg et al., 2013; Cui et al., 2021). Nearly 40 different strains of B. cinerea have been isolated and identified according to the differences in infection, pathogenicity, and biological traits (Walker et al., 2011). It was nominated for the second-rank order of fungal plant pathogen based on the scientific/economic importance (Dean et al., 2012). The pathogen infection that occurred in the field remains quiescent during the growing season and develops even at 0°C after harvesting (Romanazzi et al., 2016). The pathogen spreads among hosts by aerial mycelial growth and conidia. Effective inhibition of the mycelial growth and conidiation of B. cineara are the key factors to minimize gray mold development.
Peroxisomes are a type of single membrane-bound organelles that are present in almost all kinds of eukaryotes. A variety of crucial metabolic processes are commonly in the peroxisome, such as the fatty acid β-oxidation and the degradation of the reactive oxygen species (ROSs) (Reumann and Bartel, 2016; Zhang et al., 2022). The proteins involved in the peroxisome biogenies were normally assigned as peroxins, the peroxin-encoding genes were named as PEX genes, and up to now, there are 37 PEX proteins described (Kiel et al., 2006; Jansen et al., 2020). The disruption of PEX genes in yeasts impeded the growth on a medium that contain fatty acids as the only carbon source (Maggio and Keller, 2004). In filamentous fungi, peroxisomes also play the roles in some specific biochemical pathways, such as the biosynthesis of β-lactam antibiotics (Weber et al., 2012). Furthermore, Woronin body, a fungal-specific type of peroxisomes is required to plug the septal pores to prevent cytoplasmic leakage upon hyphal damage (Tenney et al., 2000).
A large portion of plant fungal pathogens generate conidia to reproduce and spread. Landing on the host surface, the conidia lose the nutrition resource from hyphae and have to deplete the storage component to complete the infection morphogenesis. For instance, the conidia of rice blast fungus Magnaporthe oryzae form appressorium, a hemispherical swollen infection structure. The appressorium is equipped high-strength cell walls and high concentration of glycerol accumulates in the interior to produce huge osmotic pressure, which pushed the penetration and infection process (Falter and Reumann, 2022). During this progress, the fatty in the conidia of M. oryzae is rapidly depleted to provide material and energy for the infection morphogenesis. In addition, pathogens must overcome ROSs generated in the host cells for successful invasion (Li et al., 2017). As the main organelles for fatty metabolism and ROSs degradation, peroxisomes were shown to be crucial for the pathogenicity of M. oryzae, as well as other fungal plant pathogens (Ramos and Naqvi, 2006; Min et al., 2012). The Δmgpex6 mutant of M. oryzae is incapable of β-oxidation of long-chain fatty acids and lack of appressorial melanin and host penetration (Ramos and Naqvi, 2006). The number of peroxisomes was found upregulated in M. oryzae responding to the oxidative stress generated by the host cells (Chen et al., 2017). FgPEX5 and FgPEX6 genes are critical to virulence and survival of Fusarium graminearum (Min et al., 2012). FgPEX13, FgPEX14, and FgPEX33 play the critical roles in DON biosynthesis and virulence in F. graminearum (Chen et al., 2018). Our previous findings showed that quite a few PEX genes (MoPEX1/5/7/11/13/14/17/19/33) are required for the development and pathogenicity of M. oryzae (Wang et al., 2013, 2015, 2019; Li et al., 2014, 2017).
The conidia of B. cinerea that germinate on plant surfaces form appressorium-like structures to facilitate host penetration (Gourgues et al., 2004). Mycelia of the pathogen are also able to form highly melanized infection cushions to promote the host invasion (Marschall and Tudzynski, 2016; Cao et al., 2016; Liu et al., 2016). In addition, occasionally, host invasion by the pathogen can occur via germ tube apices (Van den Heuvel and Waterreus, 1983; Choquer et al., 2007). These facts reflect the infection processes and mechanism of B. cinerea largely differ to that of M. oryzae. However, to date, whether and how the peroxisome matrix protein import play the roles in the growth, development, and pathogenicity of B. cinerea is still not well studied. In this article, we investigated the roles of three PEX genes (PEX8, PEX10, and PEX12) of B. cinerea in fugal development and pathogenicity. The results certainly bring a value-add to our understanding of the regulation of PEX8 and ring finger complex genes (PEX10 and PEX12) in the growth, development, and pathogenicity of B. cinerea, which is vital to effective disease management.
Materials and methods
Fungal strains, culture conditions, and transformation
Botrytis cinerea wild-type (WT) B05.10 and all transformants (BcPEX8, BcPEX10, and BcPEX12) were cultured on a complete medium (CM) at 22°C for 3–15 days. All fungal transformants were generated by Agrobacterium tumefaciens-mediated transformation (AtMT). CM plates containing 250 μg/ml hygromycin B (Roche, Mannheim, Germany) were used to screen the corresponding transformants. For dry weight measurement, mycelia of the strains were collected by culture in liquid CM with shaking.
Sequence analysis
The homologs of Pex8, Pex10, and Pex12 in B. cinerea were identified by searching the NCBI database.1 The coding sequences of BcPEX8, BcPEX10, and BcPEX12 were determined by amplification (with the primers set bcpex8-innerF1/bcpex8-innerR1, bcpex10-innerF1/bcpex10-innerR1, and bcpex12-innerF1/bcpex12-innerR1) from total RNAs of B. cinerea and sequencing the amplificons. Sequence alignments were performed using the Clustal W method and imported into the software GeneDoc2.0 for type setting and into MEGA version 7.0 to establish the phylogenetic trees.
Gene deletion and mutant verification
Upstream fragments at least 1.5 kb equipped with Bam HI/PstI and downstream fragments with EcoRI/XhoI for BcPEX8, BcPEX10, and BcPEX12 were amplified, respectively, with the primer sets BCPEX8UP-P1/BCPEX8UP-B1, BCPEX10UP-P1/BCPEX10DN-B1, BCPEX8UP-P1/BCPEX8U P-B1, BCPEX8DN-E1/BCPEX8DN-X1, BCPEX10DN-E1/BC PEX10DN-X1, and BCPEX12DN-E1/BCPEX12DN-X1 and the genomic DNA of B. cinerea as a template. The resulting amplicons were then inserted into p1300-KO to generate the gene replacement vector Pko-BcPEX8, Pko-BcPEX10, and Pko-BcPEX12 (Supplementary Table 1).
The PEX8, PEX10, and PEX12 mutants and ET (ectopic transformant) were generated using Agrobacterium tumefaciens-mediated transformation (AtMT) (Rolland et al., 2003). The potential mutants and ET strain were first screened on CM plate with 250 μg/ml hygromycin B. Most primary transformants presumably are heterokaryons; therefore, elimination of the parental nuclei was attempted (Rolland et al., 2003). To this end, three rounds of single spore isolation and subculturing under selective pressure were performed when the transformants produced conidia.
For the verification of mutants and ET transformants, the potential strains that were resistant to hygromycin B were harvested and initially screened by genomic PCR. The Δbcpex8 mutants and ET transformants were checked with the primer sets bcpex8-innerF1/bcpex8-innerR1, bcpex8-outF1/Seq-BP1, bcpex8-outR1/Seq-EX1, and HPH52/HPH34. The Δbcpex10 mutants were checked with the primers sets bcpex10-innerF1/bcpex10-innerR1, bcpex10-outF1/Seq-BP1, bcpex10-outR1/Seq-EX1, and HPH52/HPH34. The Δbcpex10 mutants were checked with the primer sets bcpex12-innerF1/bcpex12-innerR1, bcpex12-outF1/Seq-BP1, bcpex12-outR1/Seq-EX1, and HPH52/HPH34. The potential deletants and ET were further confirmed by quantitative reverse transcription PCR using the primer sets PEX8RTF/PEX8RTR, PEX10RTF/PEX10RTR, and PEX12RTF/PEX12RTR (Supplementary Table 2).
Pathogenicity tests
For pathogenicity assays, the detached leaves from tobacco and strawberry and tomato fruits were inoculated with the hypha stripped from the mycelial plugs (5 mm in diameter, 4-day-old cultures) of the strains and were then darkly incubated in a moistened box at 25°C. The symptoms of inoculated materials were recorded at 3 days.
Phenotypic assay of the strain
Phenotypic assays for mycelial growth, conidiation, sclerotia formation, and conidial germination were performed as previously described (Hou et al., 2020).
For conidiation assay, conidia were harvested from 10-day-old of the tested strains with 2 ml ddH2O. Conidial concentration was estimated by counting the cells on a hemocytometer under a microscope.
For conidial germination and IFSs formation assays, 10 μl of conidia suspension (1 × 105 conidia/ml) in the absence or presence of exogenous fructose (10 mM) was dropped on glass slides or onion epidermis, respectively. Then, the inoculated slides or onion epidermis were incubated in dark in a moistened box at 22°C. The number conidia germination and appressorium formation were counted under a microscope. The number of conidia germination and appressorium were counted under a microscope. At least 100 conidia were counted per replicate in each experiment; three independent experiments with triplicated per experiment were performed. The morphology of conidia and appressorium were stained with Calcofluor white (CFW) (10 μg/ml) and observed under a fluorescence microscope.
For stress adaptation assay, mycelial plugs (5 mm in diameter) taken from the periphery of a 4-day-old colony of each strain were inoculated on CM supplemented with the osmotic stress agents (NaCl and KCl, 1 M each), oxidative stress agent (H2O2, 5 mM and 7.5 mM), cell wall integrity test reagent (Congo red, 200 μg/ml and CFW 50 μg/ml), or sensitivity to fungicides (Fludioxonil,0.05 μg/ml). The inoculated CM plates were incubated at 22°C dark. At 3–4 days post-incubation, the adaptation abilities of the tested strains to the agents were observed and evaluated by their colony diameters. At least three independent experiments were performed, and in each experiment, triplicate colonies for each strain were analyzed.
For the fatty utilization assay, mycelial plugs (5 mm in diameter) taken from the periphery of a 4-day-old colony of each strain were inoculated on minimal medium (MM), which removes the glucose and supplemented with 1% (v/v) Tween80, Olive oil, 50 mM NaAC, 10 mg/ml maltase, or 10 mg/ml sucrose, respectively.
Statistical analysis
All the quantitative data in this study were derived from at least three independent experiments with triplicate treatments. The significance between the control and the other experimental data was assessed using the Student’s t-test. A p < 0.01 was considered as significant difference.
Results
Identification of BcPEX8, BcPEX10, and BcPEX12 gene in Botrytis cinerea
The potential homologous genes of PEX8 (BCIN_16g01260), PEX10 (BCIN_01g02750), and PEX12 (BCIN_12g06240) in B. cinerea were retrieved by searching the databases (see text footnote 1) using the homologs from yeasts and related fungal species and assigned as BcPEX8, BcPEX10, and BcPEX12, respectively. The real coding regions of the three genes were confirmed by amplifying the cDNA fragments and were found to be identical to the hypothetical versions in the genome database. BcPEX8 has a 2070-bp open reading frame (ORF), encoding 689 amino acid residues that show 90.42% identity to Pex8 from Sclerotinia sclerotiorum (XP_001585316.1) and 57% to Pex8 from Colletotrichum tofieldiae (KZL65398.1). Interestingly, a peroxisomal targeting signal 1 (PTS1, Ser-Lys-Leu) was present at the C terminus of Bcpex8p (Figures 1A,D), which is known to be sufficient to target proteins in peroxisome lumen in eukaryotes. Moreover, there was an internal amino acid stretch (amino acids 120–129) that resembled the proposed consensus sequence for a PTS2. The ORF of Bcpex10 is 1,110 bp long, encoding a 369-amino acid peptide with 92.41% identity to Pex10 in Sclerotinia sclerotiorum (XP_001595000.1) and 74.25% to Pex10 in Lachnellula hyalina (XP_031004878.1) (Figures 1B,D). A zinc finger RING-type profile (MATRiX) domain was found at the C terminals of the Bcpex10. The ORF of Bcpex12 is 1,404 bp long, encoding a 467-amino acid peptide with 84.14% identity to Pex12 in Sclerotinia sclerotiorum (XP_001585178.1) and 53.03% to Pex12 in Beauveria bassiana (XP_008600244.1) (Figures 1C,D). SMART analysis revealed that both Bcpex10 and Bcpex12 proteins contain PEX2_PEX12 pfam and RING finger domains (Figures 1E,F). These data suggest that Pex8, Pex10, and Pex12 are the ubiquitous peroxins in eukaryotes.
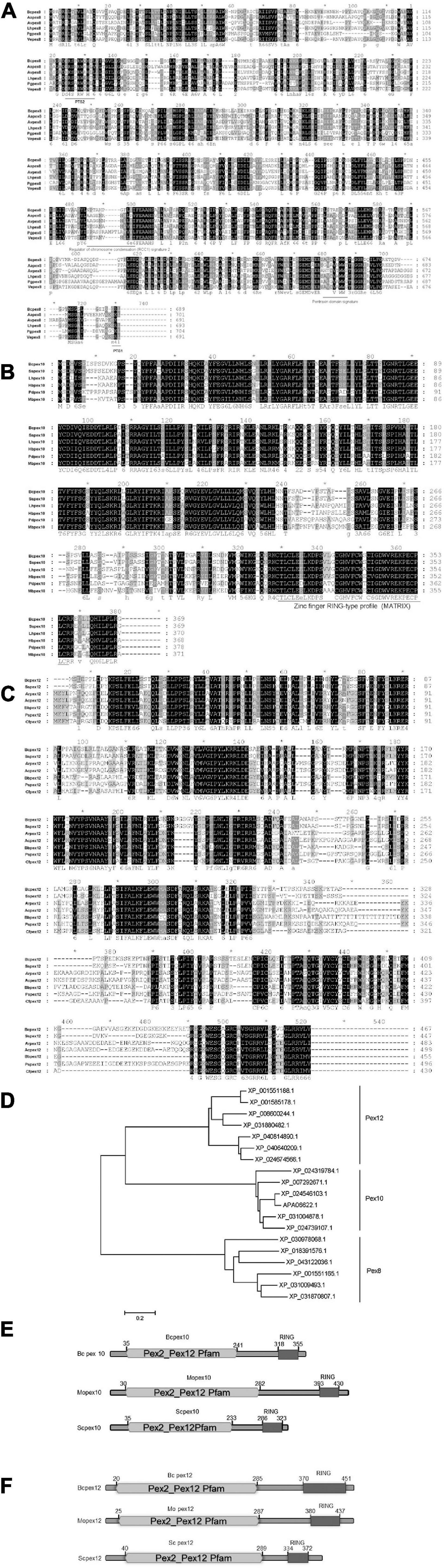
Figure 1. Sequence analysis of Bcpex8, Bcpex10, and Bcpex12 homologs. Protein sequences of Mopex8 (A), Mopex10 (B), and Mopex12 (C) and the homologous proteins from other related fungal species were aligned using Clustal X method. The identical amino acids are highlighted against a black background, the conserved residues on a dark gray background and the similar residues on a light gray background. “*” indicates only the positions each 10 residues. (D) Phylogenetic relationship of Pex8, Pex10, and Pex12 homologs calculated using the neighbor-joining method with the MEGAX program according to the alignment. (E) Domain architectures of PEX10 in B. cinerea, M. oryzae, and S. cerevisiae. (F) Domain architectures of PEX12 in B. cinerea, M. oryzae, and S. cerevisiae.
Disruption of BcPEX8, BcPEX10, and BcPEX12
To determine the roles of BcPEX8, BcPEX10, and BcPEX12 in B. cinerea, the gene deletion mutants were generated via homologous recombination. The knockout vector pKO-BcPEX8, pKO-BcPEX10, and pKO-BcPEX12 were introduced into the wild-type strain B05.10 (Figures 2A–C). The hygromycin-resistant transformants of the genes were selected and checked primarily by PCR, respectively. The transformants (with BcPEX10 as the example, and the other two genes are analogous) with the amplicon profile that bcpex10-innerF1/bcpex10-innerR1-negative, bcpex10-outF1/Seq-BP1-positive, bcpex10-outR1/Seq-EX1-positive, and HPH52/HPH34-positive were assigned as the possible deleted mutants, whereas the transformants showing a positive amplicon of bcpex10-innerF1/bcpex10-innerR1 were regarded as ectopic transformants (ET) (Figure 2D). The possible deleted mutants and ectopic transformants were selected randomly and confirmed again using qPCR. The expression of corresponding genes could not be detected in Δbcpex8, Δbcpex10, and Δbcpex12, which indicated that the gene replacement events occurred truly in these mutants (Figure 2E). The confirmed mutants Δbcpex8, Δbcpex10, and Δbcpex12 and one of the confirmed ET strains (as the phenotypes of the ET strains for the three genes are extensively identical, that of BcPEX8 was used as a case in result figures) were used for phenotypic analysis.
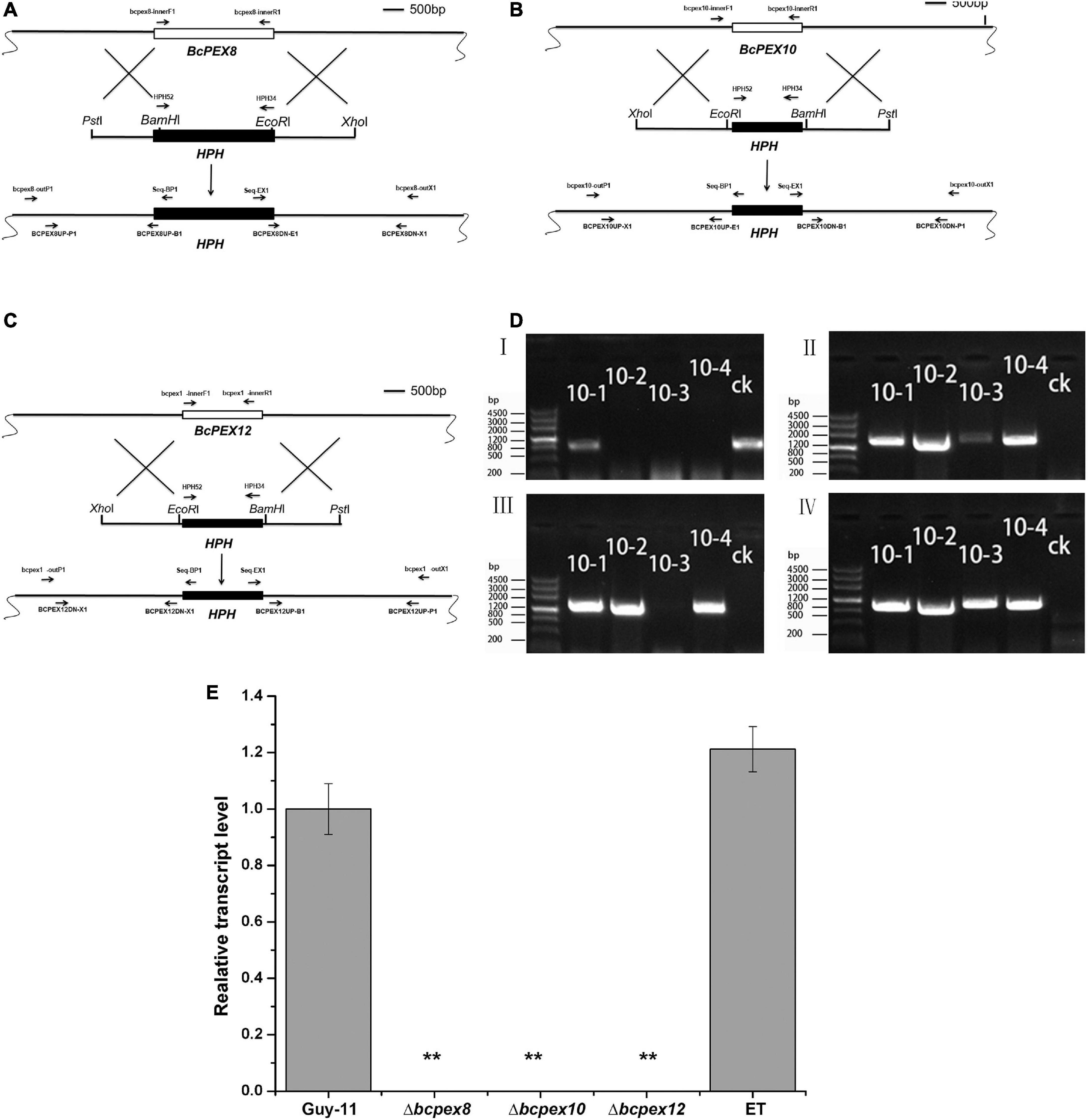
Figure 2. BcPEX8, BcPEX10, and BcPEX12 genes’ deletion. (A) Diagram showing that the 2.07-kb BcPEX8 coding region was replaced by the 1.36-kb HPH cassette. (B) Diagram showing that the 1.10-kb BcPEX10 coding region was replaced by the 1.36-kb HPH cassette. (C) Diagram showing that the 1.40-kb BcPEX12 coding region was replaced by the 1.36-kb HPH cassette. Scale bar, 500 bp. The locations of the primers are indicated with arrows, and primers in the same color were used as a pair. (D) Genomic polymerase chain reaction (PCR) was used to validate the deletion of BcPEX10. The upstream fragment, downstream fragment, BcPEX10, and HPH were amplified with the primer pairs Seq-BP1/bcpex10-outP1, Seq-EX1/bcpex10-outX1, bcpex10-innerF1/bcpex10-innerR1, and HPH52/HPH34, respectively. 10-1, 10-2, 10-3, and 10-4 are the possible deleted BcPEX10 mutants and ectopic transformants and ck is the wild-type strain guy-11. (E) Confirmation of the mutants by transcriptional analysis. BcPEX8/10/12 transcripts were detected in the wild-type (B05.10) and ET strain using quantitative reverse transcription PCR, but completely undetectable in the mutants Δbcpex8, Δbcpex10, and Δbcpex12. Double asterisks indicate significant differences from the transcriptional level of the wild type (p < 0.01).
BcPEX8, BcPEX10, and BcPEX12 are required for vegetative development of Botrytis cinerea
To test whether BcPEX8, BcPEX10, and BcPEX12 play the roles in fungal growth and development, the mycelial growth rates, melanin productions, and sclerotia production of the strains cultured on complete medium (CM) plate were measured. The data indicated that the colony diameter of the Δbcpex8, Δbcpex10, and Δbcpex12 strains was reduced significantly (Figures 3A,B). The mycelial growth of Δbcpex8, Δbcpex10, and Δbcpex12 strains was significantly inhibited, and the growth rates were reduced by 32, 35, and 34%, respectively, compared with wild-type (WT) and ectopic transformant (ET) strains at 3 days (Figure 3B). The mycelial biomass of the mutant strains was significantly reduced at 5 days (Figure 3A). The numbers of sclerotia produced by the mutant strains were more, but smaller in size, than those formed by the WT and ET strains (Figures 3A,C). The melanin productions of Δbcpex8, Δbcpex10, and Δbcpex12 mutants were significantly reduced compared with those of the WT and ET when cultured in liquid CM at 3 days (Figure 3D). These results suggest that BcPEX8, BcPEX10, and BcPEX12 are important for B. cinerea mycelium growth, sclerotium, and melanin productions.
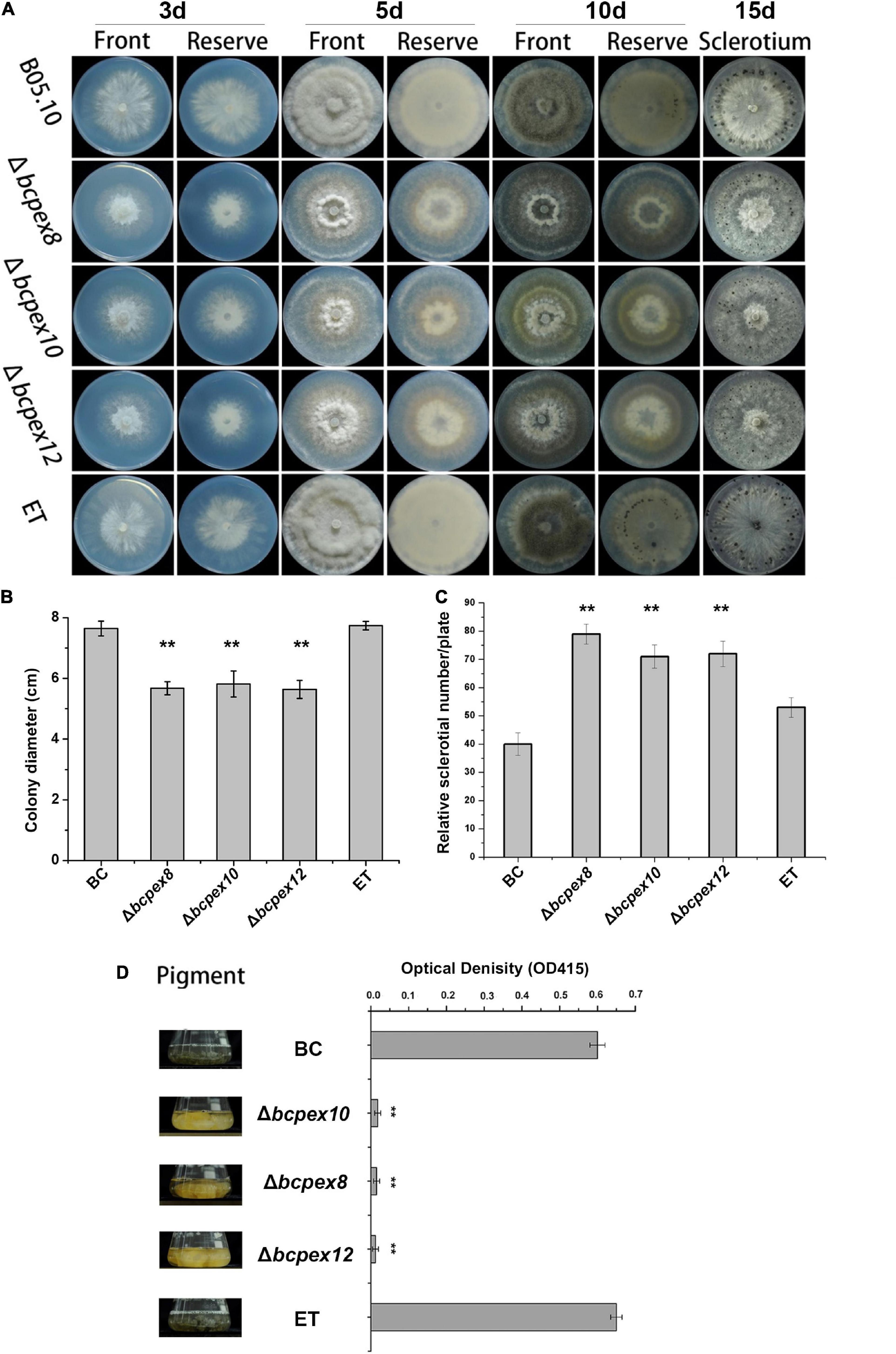
Figure 3. Defects in lipid utilization in Δbcpex8, Δbcpex10, and Δbcpex12. (A) The mycelial growth of the Δbcpex8, Δbcpex10, Δbcpex12 mutants and WT and ET strains cultured on CM plate were measured at 3, 5, 10, and 15 days. (B) The colony diameters of the strains cultured on CM plate were measured at 3 days. (C) The sclerotium production of the strains cultured on CM plates was investigated at 15 days. (D) The melanin production of the strains was investigated by measuring the optical density of the sample at 415 nm (**p < 0.01).
BcPEX8, BcPEX10, and BcPEX12 are involved in asexual and IFSs development
Compared to the WT and ET strains, the conidiation of Δbcpex8, Δbcpex10, and Δbcpex12 was reduced by approximately 89, 27, and 88%, respectively (Figures 4A,C). The conidiophore of the mutants exhibited normal morphology, but the conidial germination of the mutants was significantly inhibited (Figure 4D). Using a fluorescence microscope, shorter germ tubes were observed in the mutants compared with WT and ET strains when incubated at 4, 6, 8, and 10 h (Figure 4B). These results show that BcPEX8, BcPEX10, and BcPEX12 play the important roles in conidiation and germination of the fungus.

Figure 4. The asexual development of Δbcpex8, Δbcpex10, Δbcpex12 mutants and WT and ET strains. (A) Conidiation ofΔbcpex8,Δbcpex10, Δbcpex12 mutants and WT and ET strains. (B) The conidial germination rate ofΔbcpex8,Δbcpex10, Δbcpex12 mutants and WT and ET strains. (C) Observation of conidiophore cluster of Δbcpex8, Δbcpex10, and Δbcpex12 mutants and WT and ET strains. Bar = 200 μm. (D) Germ tube elongation of Δbcpex8, Δbcpex10, and Δbcpex12 mutants and WT and ET strains. The spores were stained with CFW. Bar = 20 μm.
IFSs, including appressoria and infection cushions (ICs), play the crucial roles in host penetration in many phytopathogenic fungi. To test the effect of BcPEX8, BcPEX10, and BcPEX12 deletion on IFSs development and melanization, we inoculated conidial suspension of WT, Δbcpex8, Δbcpex10, and Δbcpex12 mutants and ET strains on glass slides and onion epidermis to induce the formation of appressoria and ICs. All the strains formed appressoria or appressoria-like structures on glass slides at 10 h without fructose or 8 h with 10 mM fructose. The WT, Δbcpex10, and ET strains formed numerous ICs at 24 h post-inoculation/incubation (hpi) (Figure 5A). However, appressoria and ICs formed by the Δbcpex8, Δbcpex10, and Δbcpex12 mutants were significantly reduced. The appressoria number of Δbcpex8, Δbcpex10, and Δbcpex12 mutants was reduced by 77.2, 58, and 74%, respectively, compared with WT and ET strains at 10 hpi (Figures 5B,C). The ICs number of Δbcpex8, Δbcpex10, and Δbcpex12 mutants was reduced by 100, 80.8, and 100%, respectively, compared with WT and ET strains at 24 hpi (Figures 5D,E). These findings demonstrated that disruption of BcPEX8, BcPEX10, and BcPEX12 dramatically reduced appressoria and ICs formation in the mutant strains.
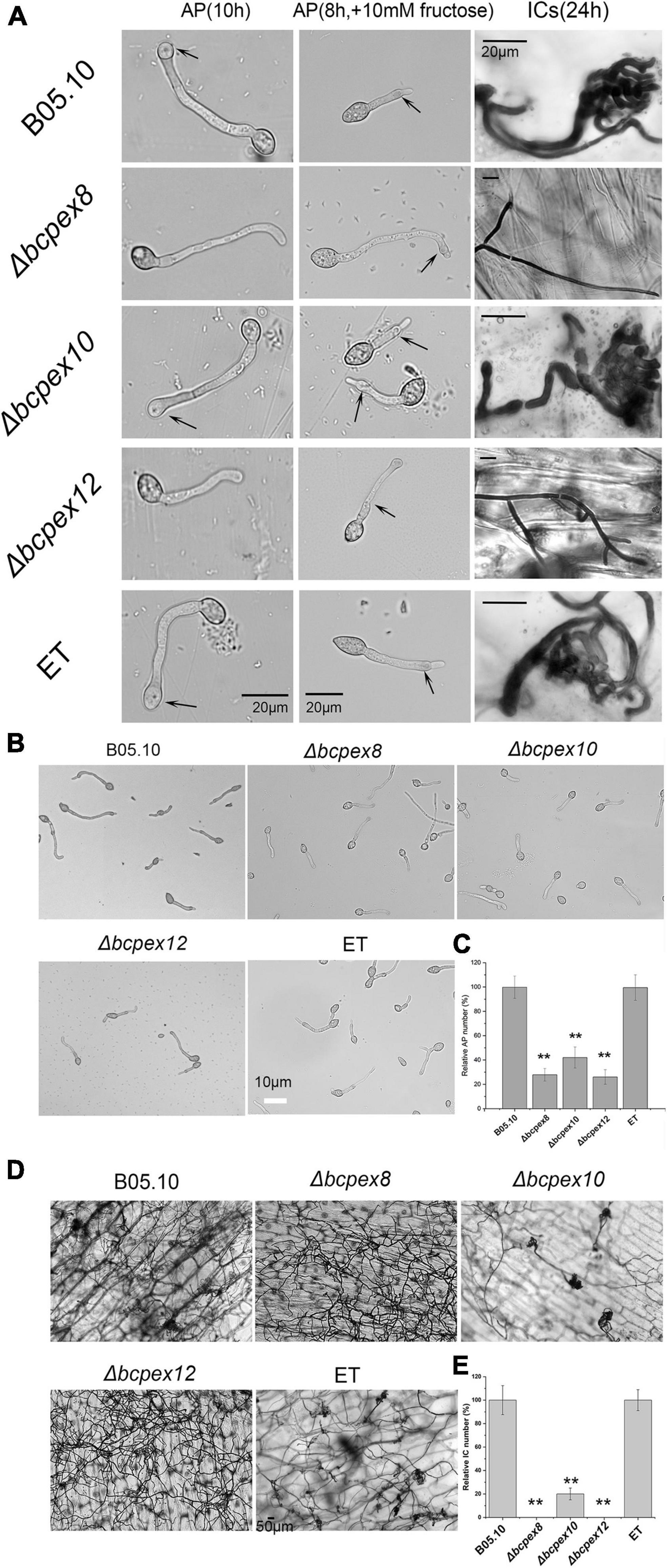
Figure 5. BcPEX8, BcPEX10, and BcPEX12 are essential for the formation of IFSs. (A) Loss of BcPEX8 and BcPEX12 in of B. cinerea impairs appressorial or appressorium-like structure formation in the absence (leaf panel) or presence (middle panel) of 10 mM fructose at 10 and 8 hpi, respectively, and abolishes infection cushion formation (right panel) on onion epidermis at 24 hpi. (B) Conidia of the Δbcpex8,Δbcpex10, and Δbcpex12 mutants and WT and ET strains were performed appressorium formation assay with 10 mM fructose at 8 hpi. (C,E) Quantification of the relative number of formed appressoria at 10 h post-inoculation with 10 mM fructose and infection cushions at 30 hpi (h) by the indicated strains. (D) The indicated strains were performed infection cushion formation assay using conidia suspensions (**p < 0.01).
Deletion of BcPEX8, BcPEX10, and BcPEX12 alters stress adaptation as well as cell wall integrity
To determine whether BcPEX8, BcPEX10, and BcPEX12 mediate fungal adaption to pathogenesis-associated stress, the WT, Δbcpex8, Δbcpex10, Δbcpex12, and ET strains were incubated on CM plates supplemented with 5 mM or 7.5 mM H2O2, 200 μg/ml Congo red (CR), 50 μg/ml CFW, 1 M NaCl/KCl, or 0.05 μg/ml fludioxonil, and colony diameters of the strains were measured at 4 days post-incubation (dpi). The growth of the Δbcpex8, Δbcpex10, and Δbcpex12 was significantly inhibited compared with the wild-type strain cultured on these media (Figure 6). The results indicated that BcPEX8, BcPEX10, and BcPEX12 are required for the cell wall integrity, sensitivity to fungicides, and adaptation of the pathogen to oxygen stress and osmotic stress.
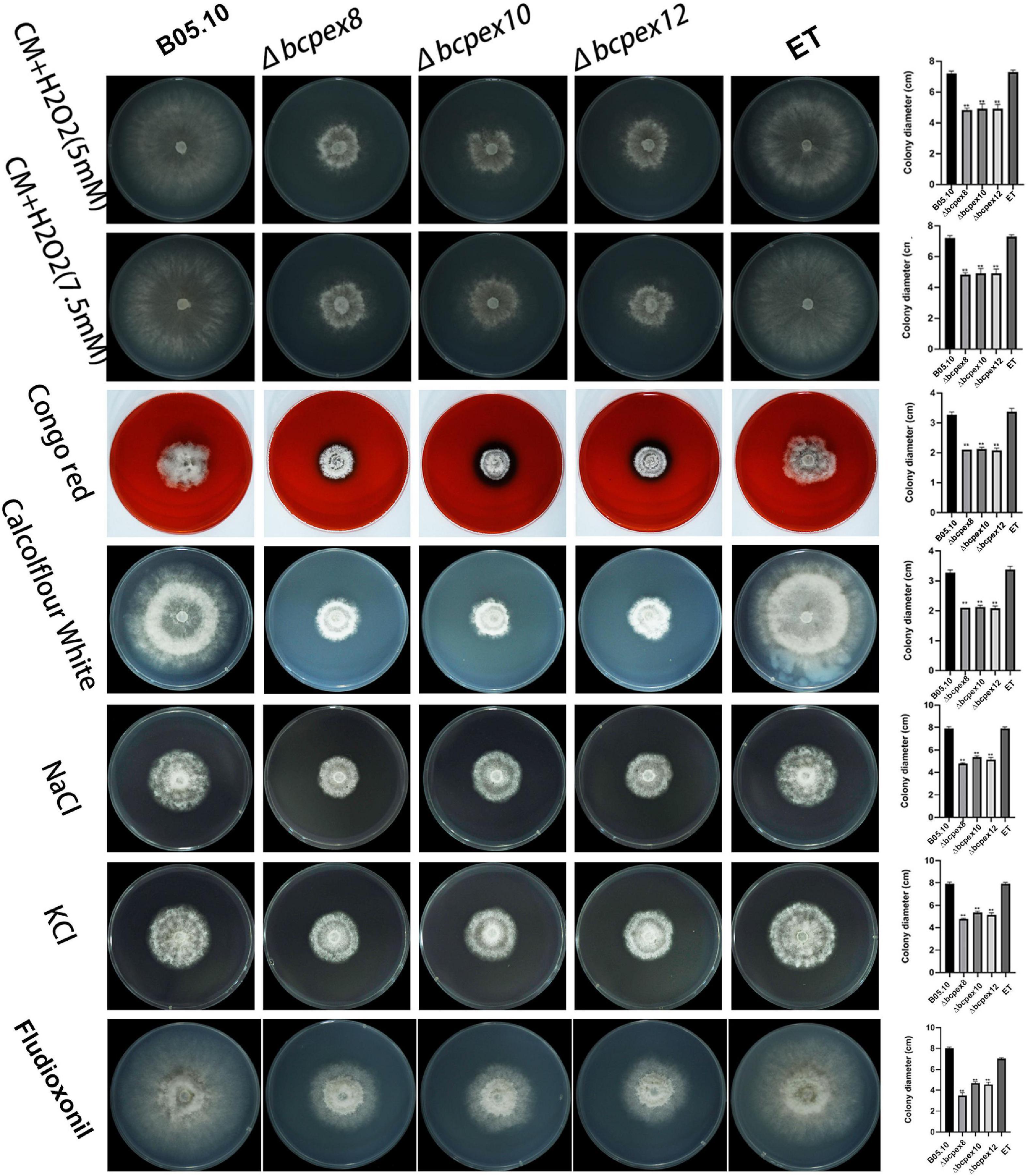
Figure 6. Tolerance test of Δbcpex8, Δbcpex10, and Δbcpex12 mutants and WT and ET strains to osmotic and oxidative stress, cell wall interference agent, and fludioxonil. The strains were cultured on CM plate supplemented with 5 or 7.5 mM H2O2, 200 μg/ml Congo red, 50 μg/mL Calcofluor white, 1 M sodium chloride or 1 M potassium chloride, or 0.05 μg/ml fludioxonil and were observed at 4 dpi (**p < 0.01).
BcPEX8, BcPEX10, and BcPEX12 are involved in lipid metabolism
To assess the effects of BcPEX8, BcPEX10, and BcPEX12 deletion on peroxisomal lipid metabolism, we investigated the lipid utilization capacity of the mutants. The development of Δbcpex8, Δbcpex10, and Δbcpex12 mutants cultured on the minimum medium (MM) complemented with Tween80, olive oil, or NaAC was significantly inhibited compared with the WT and ET strains, indicating the disruption of lipid metabolism occurred in the mutants. Moreover, Δbcpex8, Δbcpex10, and Δbcpex12 were lower efficient in the utilization of olive oil. When cultured on the MM complemented with Tween80, all the mutants failed to utilize the fatty acid (Figure 7). The results suggest that the deletion of BcPEX8, BcPEX10, and BcPEX12 leads to the defects in lipid metabolism.
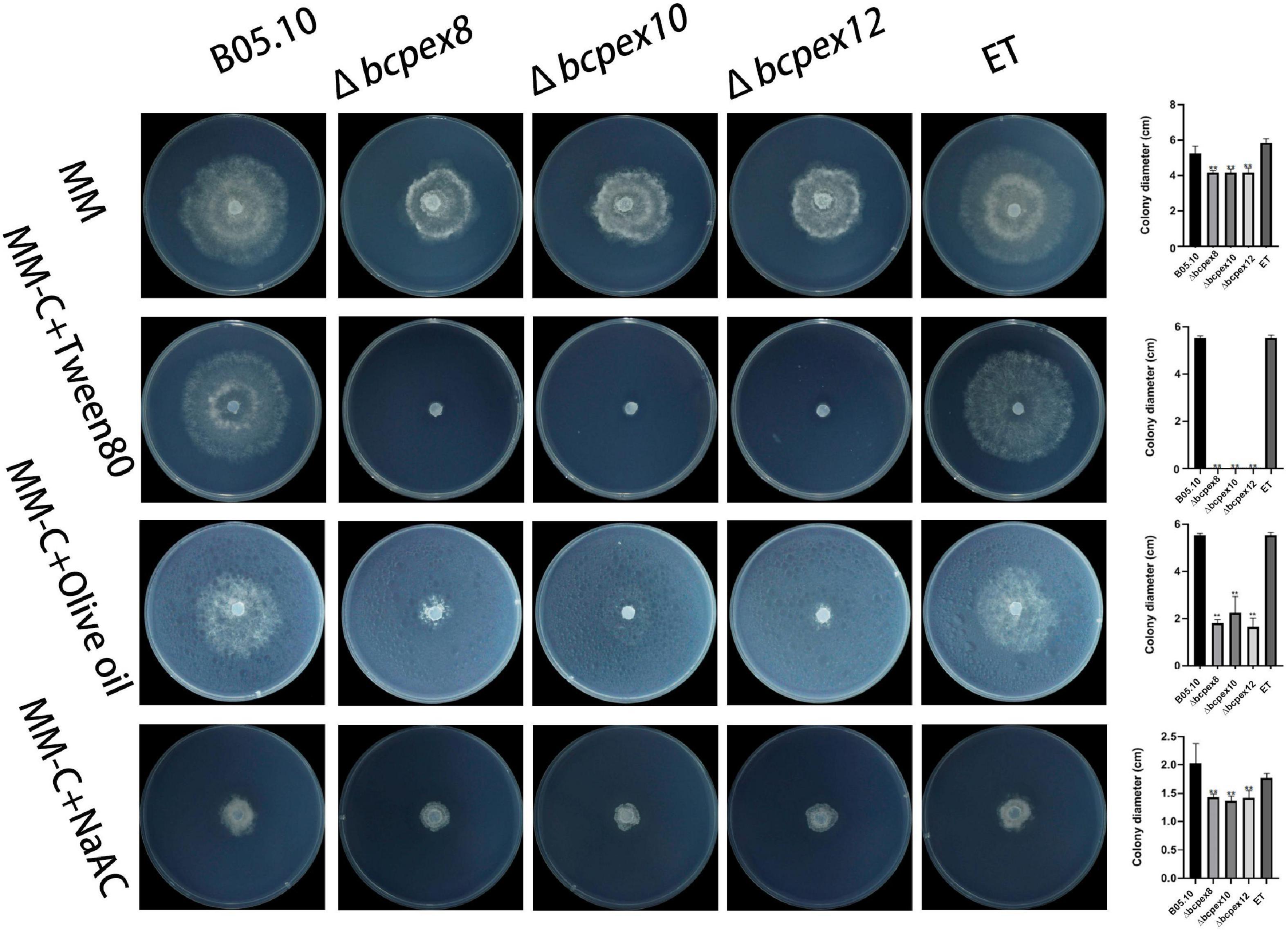
Figure 7. Lipid utilization assays of Δbcpex8, Δbcpex10, and Δbcpex12 mutants and WT and ET strains. The strains were cultured on MM plate supplemented with 1% tween 80, 1% olive oil, 50 mM sodium acetate, 10 g/L maltase, or 10 g/L sucrose and were observed at 3 dpi (**p < 0.01).
BcPEX8, BcPEX10, and BcPEX12 are the virulence determinants of Botrytis cinerea
To investigate whether BcPEX8, BcPEX10, and BcPEX12 are associated with virulence, we performed pathogenicity assay with mycelia plus of the WT, Δbcpex8, Δbcpex10, Δbcpex12, and ET strains on different plant hosts. Our findings indicated that Δbcpex8, Δbcpex10, and Δbcpex12 strains were non-pathogenic on tobacco and strawberry leaves, whereas the WT and ET strains induced severe rot symptoms on the host leaves. The Δbcpex8, Δbcpex10, and Δbcpex12 mutants could cause lesions on wounded tomato fruit in a wound-inoculation approach, but the lesions caused by the mutant strains were significantly smaller compared with those caused by the WT and ET strains (Figure 8). These findings suggest that BcPEX8, BcPEX10, and BcPEX12 are the virulence determinants in B. cinerea and are required for the pathogen invasive growth upon penetration into host cells.
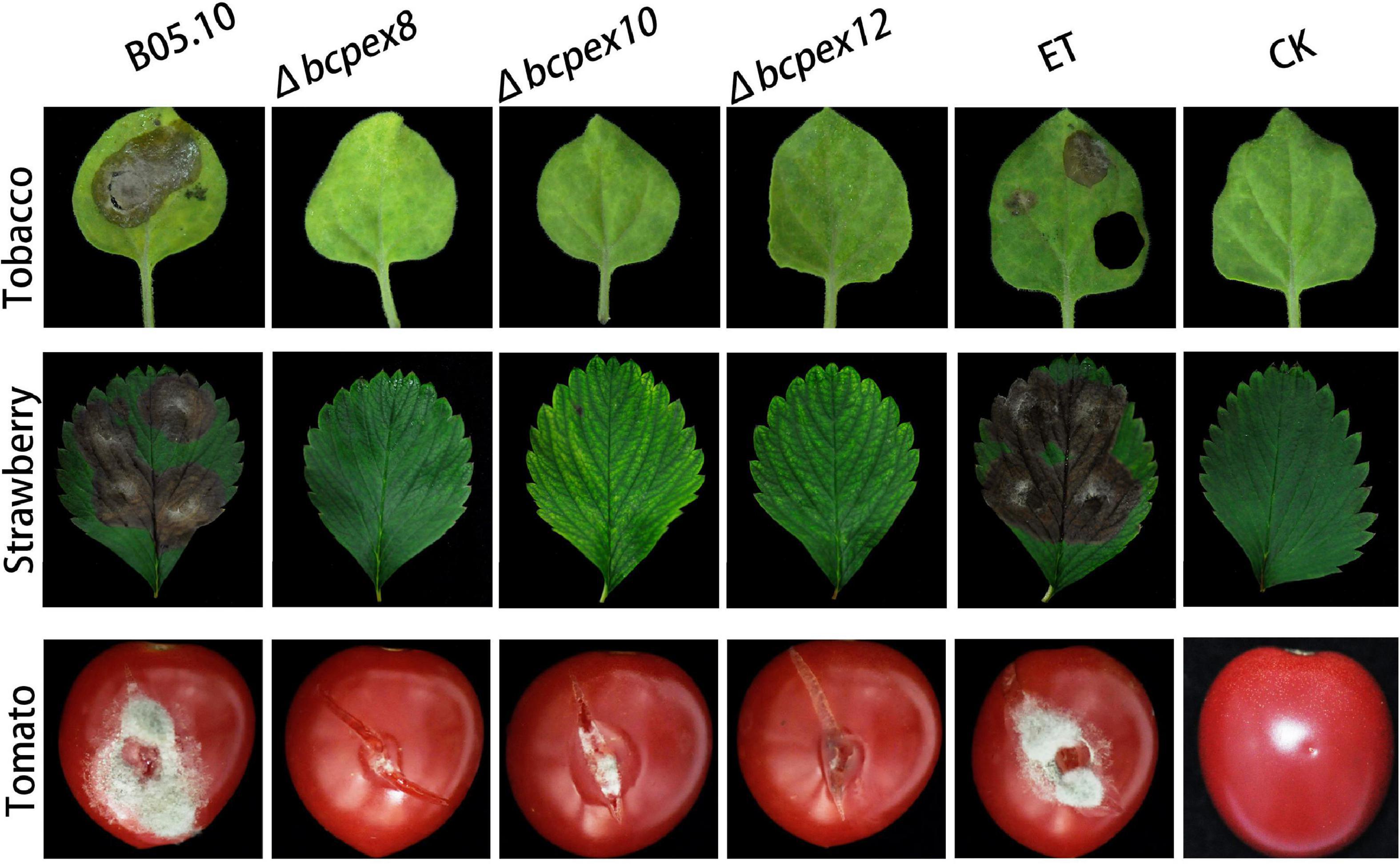
Figure 8. Pathogenicity of the bcpex8/10/12 deletion mutants on tobacco leaves, strawberry leaves, and tomato fruits.
Discussion
Peroxisomes are a class of monolayer organelles ubiquitously present in almost all eukaryotes, mainly involved in the β-oxidation of fatty acids and the detoxification of ROSs (Chen et al., 2018). Previous studies have shown that peroxisome and peroxisomal formation-related genes (PEX genes) are involved in fungal growth and development, sporulation, invasion, and parasitizing in several plant pathogenic fungi (Kimura et al., 2001; Min et al., 2012; Li et al., 2017; Wang et al., 2019). However, the roles of peroxisome and PEX genes in the growth, development, and pathogenicity of B. cinerea have not been well investigated. In this study, Pex8-, Pex10-, and Pex12-deficient mutants of B. cinerea showed similar phenotypic changes. The loss of BcPEX8, BcPEX10, or BcPEX12 led to a plethora of defects in polar growth, melanin and sclerotium production, lipid metabolism, conidiation, and stress adaptation of the fungus.
Pex8p is an intraperoxisomal peripheral membrane protein, and it links the docking and RING finger complexes (PEX2/10/12) in fungi (Agne et al., 2003; Jansen et al., 2020). Less is known about PEX8, but has been implicated in cargo release from the PTS1 receptor PEX5 (Ma et al., 2013). In this study, we found that the Δbcpex8 mutant exhibited the defects in lipid utilization and cell wall integrity, caused less lesions on the host leaves and fruits inoculated, impaired growth on agar plate, and reduced conidiation and melanin production, indicating that BcPEX8, as a peroxisome biogenesis factor, is involved in regulating development and pathogenicity in B. cinerea. It is well known that fatty acid β-oxidation is exclusively located in peroxisomes, and lipid metabolism is crucial for conidial germination and the development of infection structures in filamentous plant-pathogenic fungi (Falter and Reumann, 2022). The previous study reported that Δpex8 mutant of Saccharomyces cerevisiae failed to grow on the medium containing oleic acid (Geraghty et al., 1999), indicating that Pex8 play the roles in fatty acid utilization S. cerevisiae. The results in this study confirmed the association between BcPEX8 and lipid metabolism in B. cinerea. Plant-pathogenic fungi use mycotoxins to alter plant metabolism for their advantage in invasion (Pusztahelyi et al., 2015). Peroxisomes are essential for mycotoxin biosynthesis in several fungal species (Falter and Reumann, 2022). Although the role of PEX8 gene in toxin biosynthesis has not been well elucidated, a recent study reported that FvPEX8 was a key component in F. verticillioides docking module affected peroxisome function and fumonisin biosynthesis (Yu et al., 2021). It will be an interesting and our undergoing topic whether the involvement of BcPEX8 in pathogenicity is associated with mycotoxin biosynthesis in B. cinerea.
Pex10p and Pex12p are RING finger peroxins that have ubiquitin (E3) ligase activity and are involved in PEX5 recycling in filamentous plant-pathogenic fungi (Falter and Reumann, 2022). To date, there are only a few reports regarding phenotypic analysis of PEX10 and PEX12, and their roles in phytopathogenic fungi remain to be clearly established. Previous studies demonstrated that in the Δpex10 and Δpex12 mutants of F. graminearum, the conidia and hyphae were prone to be broken, the cell walls were more sensitive to the cell wall-perturbing agents, and the mycelial growth, conidiation, and lipid metabolism are impaired, compared with the wild type. F. graminearum Δpex10 and Δpex12 mutants also showed downregulation of selected TRI genes and reduced deoxynivalenol production compared with the wild type (Zhang et al., 2019; Wang et al., 2020). However, less is known about the roles of PEX10 and PEX12 in fungal development and pathogenicity in B. cinerea. Our results reveal that both BcPEX10 and BcPEX12 are involved in hyphal growth, asexual reproduction, fatty acid utilization, maintenance of cell wall integrity, and pathogenicity in B. cinerea. Comparison of the findings with the documented studies confirmed that PEX8, PEX10, and PEX12 play the roles in the development and pathogenicity in both B. cinerea and F. graminearum. We demonstrated previously that MoPEX1, MoPEX11A, MoPEX13, MoPEX14, MoPEX14/17, and MoPEX19 were required for development and pathogenicity in M. oryzae (Li et al., 2014, 2017; Wang et al., 2015, 2019). The collected evidence provides further support for the hypothesis that peroxisomes play numerous essential roles in fungal development and pathogenicity, and PEX gene has now emerged as an important virulence factor bearing the possibility to block multiple biogenesis pathways of different virulence factors simultaneously.
In the ongoing attempts to develop novel control agents against plant fungal pathogens, some peroxisomal enzymes essential for virulence are regarded as elegant targets, which implied that the peroxisome is a potential organelle used as a target. For lowering the risk of rapid evolution of fungicidal resistance in fungi, multiple gene targets were deemed to be preferred to single gene targets. Thus, more PEX genes in plant-pathogenic fungi are required to be further characterized. In addition, recent studies have shown that RNAs can move from a host to the interacting pathogen to inhibit infection (Cai et al., 2019). The exogenous small interfering RNAs against the PEX genes of plant-pathogenic fungi also have the potentiality as environmentally friendly RNA fungicides for crop protection.
In summary, the Pex8, Pex10, and Pex12 play the crucial roles in the development and pathogenicity in plant pathogenic fungus, B. cinerea.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
J-YW: conceptualization. LL, ZZ, and JG: data curation. M-XY, Z-QL, and X-MZ: formal analysis. LL: funding acquisition. J-YW and G-CS: investigation. M-XY, Z-QL, ZZ, and J-YW: methodology. Y-LW and G-CS: project administration. LL, X-MZ, and ZZ: software. J-YW, G-CS, and F-CL: supervision. Z-QL and LL: validation. All authors contributed to the article and approved the submitted version.
Funding
The authors gratefully acknowledged the financial support provided by the Zhejiang Key Research and Development Program (grant nos. 2019C02010 and 2021C02010), the National Natural Science Foundation of China (grant no. 31900126), Zhejiang Provincial Natural Science Foundation (grant no. LZ20C140001), Fundamental Research Funds for the Provincial Universities of Zhejiang (grant no. 2020YQ002), Bio-Health Inner-Cooperation Plan in Zhejiang Academy of Agricultural Sciences, and State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products (grant no. 2021DG700024-KF202110).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.962500/full#supplementary-material
Footnotes
References
Agne, B., Meindl, N. M., Niederhoff, K., Einwächter, H., Rehling, P., Sickmann, A., et al. (2003). Pex8p: An intraperioxisomal organizer of the peroxisomal import machinery. Mol. Cell 11, 635–646.
Cai, Q., He, B., Weiberg, A., Buck, A. H., and Jin, H. (2019). Small RNAs and extracellular vesicles: New mechanisms of cross-species communication and innovative tools for disease control. PLoS Pathog. 15:e1008090. doi: 10.1371/journal.ppat.1008090
Cao, H., Huang, P., Zhang, L., Shi, Y., Sun, D., Yan, Y., et al. (2016). Characterization of 47 Cys2-His2 zinc finger proteins required for the development and pathogenicity of the rice blast fungus Magnaporthe oryzae. New Phytol. 211, 1035–1051. doi: 10.1111/nph.13948
Chen, X. L., Shen, M., Yang, J., Xing, Y., Chen, D., Li, Z., et al. (2017). Peroxisomal fission is induced during appressorium formation and is required for full virulence of the rice blast fungus. Mol. Plant Pathol. 18, 222–237. doi: 10.1111/mpp.12395
Chen, Y., Zheng, S., Ju, Z., Zhang, C., Tang, G., Wang, J., et al. (2018). Contribution of peroxisomal docking machinery to mycotoxin biosynthesis, pathogenicity and pexophagy in the plant pathogenic fungus Fusarium graminearum. Environ. Microbiol. 20, 3224–3245. doi: 10.1111/1462-2920.14291
Choquer, M., Fournier, E., Kunz, C., Levis, C., Pradier, J. M., Simon, A., et al. (2007). Botrytis cinerea virulence factors: New insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 277, 1–10. doi: 10.1111/j.1574-6968.2007.00930.x
Cui, X., Ma, D., Liu, X., Zhang, Z., Li, B., Xu, Y., et al. (2021). Magnolol inhibits gray mold on postharvest fruit by inducing autophagic activity of Botrytis cinerea. Postharvest Biol. Technol. 180:111596.
Dean, R., Vankan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430.
Falter, C., and Reumann, S. (2022). The essential role of fungal peroxisomes in plant infection. Mol. Plant Pathol. 23, 781–794.
Geraghty, M. T., Bassett, D., Morrell, J. C., Gatto, G. J. Jr., Bai, J., Geisbrecht, B. V., et al. (1999). Detecting patterns of protein distribution and gene expression in silico. Proc. Natl. Acad. Sci.U.S.A. 96, 2937–2942.
Gourgues, M., Brunet-Simon, A., Lebrun, M. H., and Levis, C. (2004). The tetraspanin BcPls1 is required for appressorium-mediated penetration of Botrytis cinerea into host plant leaves. Mol. Microbiol. 51, 619–629. doi: 10.1046/j.1365-2958.2003.03866.x
Hou, J., Feng, H. Q., Chang, H. W., Liu, Y., Li, G. H., Yang, S., et al. (2020). The H3K4 demethylase Jar1 orchestrates ROS production and expression of pathogenesis-related genes to facilitate Botrytis cinerea virulence. New Phytol. 225, 930–947. doi: 10.1111/nph.16200
Jansen, R., Molina, C. S., Noort, M., Devos, D. P., and van der Klei, I. J. (2020). Comparative genomics of peroxisome biogenesis proteins: Making sense of the PEX mess. Front. Cell Develop. Biol. 9:654163. doi: 10.3389/fcell.2021.654163
Kiel, J. A. K. W., Veenhuis, M., and van der Klei, I. J. (2006). PEX genes in fungal genomes: Common, rare or redundant. Traffic 7, 1291–1303.
Kimura, A., Takano, Y., Furusawa, I., and Okuno, T. (2001). Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell 13, 1945–1957. doi: 10.1105/tpc.010084
Li, L., Wang, J. Y., Zhang, Z., Wang, Y., Liu, M., Jiang, H., et al. (2014). MoPex19, which is essential for maintenance of peroxisomal structure and woronin bodies, is required for metabolism and development in the rice blast fungus. PLoS One 9:e85252. doi: 10.1371/journal.pone.0085252
Li, L., Wang, J. Y., Chen, H. L., Chai, R., Zhang, Z., Mao, X., et al. (2017). Pex14/17, a filamentous fungus-specific peroxin, is required for the import of peroxisomal matrix proteins and full virulence of Magnaporthe oryzae. Mol. Plant Pathol. 18, 1238–1252. doi: 10.1111/mpp.12487
Liu, J., Lemonds, T. R., Marden, J. H., and Popadiæ, A. (2016). A pathway analysis of melanin patterning in a hemimetabolous insect. Genetics 203, 403–413. doi: 10.1534/genetics.115.186684
Ma, C., Hagstrom, D., Polley, S. G., and Subramani, S. (2013). Redox regulated cargo binding and release by the peroxisomal targeting signal receptor. Pex5. J. Biol. Chem. 288, 27220–27231. doi: 10.1074/jbc.m113.492694
Maggio, L. A., and Keller, N. P. (2004). Mitochondrial β−oxidation in Aspergillus nidulans. Mol. Microbiol. 54, 1173–1185.
Marschall, R., and Tudzynski, P. (2016). Reactive oxygen species in development and infection processes. Semin. Cell Dev. Biol. 57, 138–146.
Min, K., Son, H., Lee, J., Choi, G. J., Kim, J. C., and Lee, Y. W. (2012). Peroxisome function is required for virulence and survival of Fusarium graminearum. Mol. Plant Microbe. Interact. 25, 1617–1627. doi: 10.1094/MPMI-06-12-0149-R
Pusztahelyi, T., Holb, I., and Pócsi, I. (2015). Secondary metabolites in fungus–plant interactions. Front. Plant Sci. 6:573. doi: 10.3389/fpls.2015.00573
Qin, G., Zhong, Y., Chen, Q., Hua, D., and Tian, S. (2010). Inhibitory effet of boron against Botrytis cinerea on table grapes and its possible mechanisms of active. Int. J. Food Microbiol. 138, 145–150. doi: 10.1016/j.ijfoodmicro.2009.12.018
Ramos, M., and Naqvi, N. I. (2006). Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Mol. Microbiol. 61, 61–75. doi: 10.1111/j.1365-2958.2006.05194.x
Reumann, S., and Bartel, B. (2016). Plant peroxisomes: Recent discoveries in functional complexity, organelle homeostasis, and morphological dynamics. Curr. Opin. Plant Biol. 34, 17–26. doi: 10.1016/j.pbi.2016.07.008
Romanazzi, G., Smilanick, J. L., Feliziani, E., and Droby, S. (2016). Integrated management of postharvest gray mold on fruit crops. Postharvest Biol. Technol. 113, 69–76.
Rolland, S., Jobic, C., Fèvre, M., and Bruel, C. (2003). Agrobacterium-mediated transformation of Botrytis cinerea, simple purification of monokaryotic transformants and rapid conidia-based identification of the transfer-DNA host genomic DNA flanking sequences. Curr. Genet. 44, 164–171. doi: 10.1007/s00294-003-0438-8
Tenney, K., Hunt, I., Sweigard, J., Pounder, J. I., McClain, C., Bowman, E. J., et al. (2000). Hex-1, a gene unique to filamentous fungi, encodes the major protein of the woronin body and functions as a plug for septal pores. Fung. Genet. Biol. 31, 205–217. doi: 10.1006/fgbi.2000.1230
Van den Heuvel, J., and Waterreus, L. P. (1983). Conidial concentration as an important factor determining the type of prepenetration structures formed by Botrytis cinerea on leaves of French bean (Phaseolus vulgaris). Plant Pathol. 32, 263–272.
Walker, A. S., Gautier, A. L., Confais, J., Martinho, D., Viaud, M., Le P Cheur, P., et al. (2011). Botrytis pseudocinerea, a new cryptie species causing gray mold in French vineyards in sympatry with Botrytis cinerea. Phytopathology 101, 1433–1445. doi: 10.1094/PHYTO-04-11-0104
Wang, J. Y., Li, L., Zhang, Z., Qiu, H., Li, D., Li, D., et al. (2015). One of Three Pex11 Family Members Is Required for Peroxisomal Proliferation and Full Virulence of the Rice Blast Fungus Magnaporthe oryzae. PLoS One 10:e0134249. doi: 10.1371/journal.pone.0134249
Wang, J. Y., Li, L., Chai, R. Y., Qiu, H. P., Zhang, Z., Wang, Y. L., et al. (2019). Pex13 and Pex14, the key components of the peroxisomal docking complex, are required for peroxisome formation, host infection and pathogenicity-related morphogenesis in Magnaporthe oryzae. Virulence 10, 292–314. doi: 10.1080/21505594.2019.1598172
Wang, L., Zhang, L., Liu, C., Sun, S., Liu, A., Liang, Y., et al. (2020). The roles of FgPEX2 and FgPEX12 in virulence and lipid metabolism in Fusarium graminearum. Fung. Genet. Biol. 135:103288. doi: 10.1016/j.fgb.2019.103288
Wang, J. Y., Zhang, Z., Wang, Y. L., Li, L., Chai, R., Chai, R., et al. (2013). PTS1 Peroxisomal Import Pathway Plays Shared and Distinct Roles to PTS2 Pathway in Development and Pathogenicity of Magnaporthe oryzae. PLoS One 8:e55554. doi: 10.1371/journal.pone.0055554
Weber, S. S., Bovenberg, R. A., and Driessen, A. J. (2012). Biosynthetic concepts for the production of β-lactam antibiotics in Penicillium chrysogenum. Biotechnol. J. 7, 225–236. doi: 10.1002/biot.201100065
Weiberg, A., Wang, M., Lin, F. M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Yu, W., Lin, M., Peng, M., Yan, H., Wang, J., Zhou, J., et al. (2021). Fusarium verticillioides FvPex8 Is a key component of the peroxisomal docking/translocation module that serves important roles in fumonisin biosynthesis but Not in virulence. Mol. Plant Microbe. Interact. 34, 803–814. doi: 10.1094/MPMI-10-20-0273-R
Zhang, L., Liu, C., Wang, L., Sun, S., Liu, A., Liang, Y., et al. (2019). FgPEX1 and FgPEX10 are required for the maintenance of Woronin bodies and full virulence of Fusarium graminearum. Curr. Genet. 65, 1383–1396. doi: 10.1007/s00294-019-00994-8
Keywords: Botrytis cinerea, peroxisome, peroxins, fatty acid metabolism, pathogenicity
Citation: Li L, Yu M-x, Guo J, Hao Z-n, Zhang Z, Lu Z-q, Wang J-y, Zhu X-m, Wang Y-l, Chen J, Sun G-C and Lin F-c (2022) The peroxins BcPex8, BcPex10, and BcPex12 are required for the development and pathogenicity of Botrytis cinerea. Front. Microbiol. 13:962500. doi: 10.3389/fmicb.2022.962500
Received: 06 June 2022; Accepted: 02 August 2022;
Published: 06 September 2022.
Edited by:
Synan F. AbuQamar, United Arab Emirates University, United Arab EmiratesCopyright © 2022 Li, Yu, Guo, Hao, Zhang, Lu, Wang, Zhu, Wang, Chen, Sun and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiao-yu Wang, d2FuZ2ppYW95dTc4QHNpbmEuY29t; Guo-Chang Sun, c3VuZ2MwMUBzaW5hLmNvbQ==
†These authors have contributed equally to this work
 Ling Li
Ling Li Meng-xue Yu1†
Meng-xue Yu1† Jiao-yu Wang
Jiao-yu Wang Xue-ming Zhu
Xue-ming Zhu Fu-cheng Lin
Fu-cheng Lin