
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 11 August 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.960748
This article is part of the Research TopicWomen in Antimicrobials, Resistance and Chemotherapy: 2022View all 6 articles
 Rose M. Collis1,2*
Rose M. Collis1,2* Patrick J. Biggs2,3,4
Patrick J. Biggs2,3,4 Sara A. Burgess2
Sara A. Burgess2 Anne C. Midwinter2
Anne C. Midwinter2 Gale Brightwell1,4
Gale Brightwell1,4 Adrian L. Cookson1,2*
Adrian L. Cookson1,2*
Antimicrobial resistance (AMR) is a global threat to human and animal health, with the misuse and overuse of antimicrobials being suggested as the main driver of resistance. In a global context, New Zealand (NZ) is a relatively low user of antimicrobials in animal production. However, the role antimicrobial usage on pasture-based dairy farms, such as those in NZ, plays in driving the spread of AMR within the dairy farm environment remains equivocal. Culture-based methods were used to determine the prevalence and distribution of extended-spectrum β-lactamase (ESBL)- and AmpC-producing Escherichia coli from farm environmental samples collected over a 15-month period from two NZ dairy farms with contrasting management practices. Whole genome sequencing was utilised to understand the genomic epidemiology and antimicrobial resistance gene repertoire of a subset of third-generation cephalosporin resistant E. coli isolated in this study. There was a low sample level prevalence of ESBL-producing E. coli (faeces 1.7%; farm dairy effluent, 6.7% from Dairy 4 and none from Dairy 1) but AmpC-producing E. coli were more frequently isolated across both farms (faeces 3.3% and 8.3%; farm dairy effluent 38.4%, 6.7% from Dairy 1 and Dairy 4, respectively). ESBL- and AmpC-producing E. coli were isolated from faeces and farm dairy effluent in spring and summer, during months with varying levels of antimicrobial use, but no ESBL- or AmpC-producing E. coli were isolated from bulk tank milk or soil from recently grazed paddocks. Hybrid assemblies using short- and long-read sequence data from a subset of ESBL- and AmpC-producing E. coli enabled the assembly and annotation of nine plasmids from six E. coli, including one plasmid co-harbouring 12 antimicrobial resistance genes. ESBL-producing E. coli were infrequently identified from faeces and farm dairy effluent on the two NZ dairy farms, suggesting they are present at a low prevalence on these farms. Plasmids harbouring several antimicrobial resistance genes were identified, and bacteria carrying such plasmids are a concern for both animal and public health. AMR is a burden for human, animal and environmental health and requires a holistic “One Health” approach to address.
Antimicrobial resistance (AMR) is a complex “One Health” issue which affects human, animal, and environmental health (Queenan et al., 2016). Antimicrobial exposure, particularly the misuse and overuse of antimicrobials, in both human and animal health has been suggested as the main driver of AMR (Laxminarayan et al., 2013; Holmes et al., 2016). However, other factors such as heavy metal (Knapp et al., 2011; Chen et al., 2019; Thomas IV et al., 2020) and biocide use (Romero et al., 2017), and in the dairy farm environment, management practices such as waste milk disposal and feed type, may also influence the development and dissemination of AMR (Collis et al., 2019).
Extended-spectrum β-lactamase- (ESBL) and AmpC-producing Escherichia coli are of concern to human health. ESBL and AmpCs are β-lactamase enzymes which confer resistance to first- and third-generation cephalosporins (3GCs), penicillins, and monobactams. The AmpC β-lactamase enzymes also confer resistance to the second-generation cephalosporins and cephamycins (Rubin and Pitout, 2014). ESBL genes are often encoded on plasmids which can carry multiple antimicrobial resistance genes (ARGs), resulting in a multi-drug resistant phenotype (Ho et al., 2013; McGann et al., 2016). Therefore ESBL-producing Enterobacteriaceae have been classified as critical on the World Health Organisation’s “Priority Pathogens List” (Tacconelli et al., 2018). The most common ESBL variants include the SHV and TEM (excluding the parent type) enzymes and the CTX-M variants (Pitout, 2012). The predominant ESBL variants differ geographically and can exhibit temporal shifts (Livermore et al., 2006; Canton et al., 2008; Bevan et al., 2017), however, CTX-M-14 and CTX-M-15 are the predominant genotypes in most geographic regions (Bevan et al., 2017). In the livestock and animal sectors, the CTX-M-1, CTX-M-14, and CTX-M-15 ESBL types have frequently been detected (Ewers et al., 2012, 2021; Day et al., 2016; Afema et al., 2018; Cormier et al., 2019). In E. coli, an AmpC phenotype can arise from mutations in the promoter region of the chromosomal ampC gene resulting in AmpC hyperproducers (Caroff et al., 1999, 2000; Siu et al., 2003), or from plasmid-mediated AmpC β-lactamase (pAmpC) genes (Pérez-Pérez and Hanson, 2002). Plasmid-mediated resistance is of particular concern as this phenotype can be shared by horizontal gene transfer (HGT) between related bacteria. In addition, amino acid (AA) substitutions in the R2, H-9 and H-10 regions of chromosomal AmpC have been proposed to result in conformational changes within the enzyme, resulting in extended-spectrum AmpC β-lactamases (ESAC) which confer resistance against fourth-generation cephalosporins (4GCs) such as cefepime (Haenni et al., 2014a; Santiago et al., 2018). AA deletions in the H-10 helix have also been reported for plasmid-borne AmpC β-lactamases (CMY-33), which may alter the enzyme’s substrate spectrum (Pires et al., 2015).
Globally, ESBL- and AmpC-producing Enterobacteriaceae have been detected in various agricultural environments including dairy (Gonggrijp et al., 2016; Santman-Berends et al., 2017; Alzayn et al., 2020; Massé et al., 2021), poultry (Daehre et al., 2018; von Tippelskirch et al., 2018; Gazal et al., 2021), swine (von Salviati et al., 2015; Dohmen et al., 2017) and aquaculture (Hamza et al., 2020). The development and transmission of AMR in agricultural environments is complex. Risk factors for ESBL- and AmpC-producing Enterobacteriaceae on dairy farms includes 3GC and 4GC use, increased antimicrobial use in calves (Gonggrijp et al., 2016) and amoxicillin use for AmpC-producing E. coli (Alzayn et al., 2020). ESBL- and AmpC-producing E. coli have also been detected in organic dairy herds with low antimicrobial usage (AMU) (Santman-Berends et al., 2017). A study using mixed effects logistic regression found that AMU only partially explained ESBL/AmpC positive samples on dairy farms in the Netherlands (Hordijk et al., 2019). These findings suggest that multiple factors are involved in the development and transmission of AMR in dairy farm environments.
Few studies have investigated the prevalence of ESBL and AmpC-producing E. coli in pasture-based dairy farm environments such as those found in New Zealand (NZ) where there is relatively low use of antimicrobials in food-producing animals (Hillerton et al., 2017, 2021). One regional cross-sectional study of dairy farms in NZ found a low prevalence of ESBL-producing E. coli (1 of 116; 0.9%) in pooled faecal samples (Burgess et al., 2021a). Similarly, E. coli with pAmpC genes were absent but 7.9% (9 of 114) of faecal samples were positive for AmpC hyperproducers with mutations in the promoter region of the ampC gene (Burgess et al., 2021a). A nationwide cross-sectional study (n = 26 dairy farms) in NZ did not detect any ESBL-producing E. coli (Burgess et al., 2021b) while AmpC-hyperproducing E. coli were isolated from 14% (11 of 78) of pooled faecal enrichments originating from seven farms (Burgess et al., 2021b). Additionally, an NZ study between 2009 and 2010 found no ESBL-producing E. coli (0 of 300) from young calf carcasses (4–10 days old) (Heffernan et al., 2011). Hence, the aims of this study were to utilise culture-based methods to investigate the prevalence of ESBL- and AmpC-producing E. coli from two NZ dairy farm environments over a 15-month period, taking into consideration seasonal variation and farm management practices, and to understand the genomic epidemiology and ARG repertoire of a subset of 3GC resistant E. coli isolated in this study.
The Massey University research farms No. 1 Dairy Farm (referred to hereafter as Dairy 1) and No. 4 Dairy Farm (referred to hereafter as Dairy 4) were recruited for inclusion in this study. The two dairy farms are located in Palmerston North, New Zealand, are <5 km apart and both operate a closed dairy farm system (animals are not introduced into the herd). The two farms are pasture-based, with the use of supplementary feed such as silage (pickled pasture) and baleage (partially dried pasture) when required. Both dairy farms have a spring calving system, use selective dry cow therapy (DCT) and teat sealants are applied to the whole herd. A comparison between farm parameters and management practices is detailed in Supplementary Table 1.
Farm environmental samples were collected concurrently each month from October 2018 to December 2019, spanning a 15-month period and both farms were sampled on the same day. On each sampling occasion, composite soil cores and composite cow faeces from cow pats in a recently grazed paddock, farm dairy effluent (FDE) and bulk tank milk (referred to hereafter as milk) were collected (Supplementary Table 2). Previous studies looking at the prevalence of antimicrobial resistant E. coli in soil from dairy farms have also utilised a sampling strategy which focuses on critical source areas where a large number of dairy cows are concentrated or areas recently impacted by dairy cows, such as housing areas, gateways, recently grazed paddocks and manure fertilised soil (Liu et al., 2019; Gelalcha et al., 2022). The FDE management strategy on Dairy 1 changed during the study period; initially effluent was filtered before being administered to paddocks, but later effluent was disposed of via the municipal waste-water system and as a result, two FDE sample collection points were used (Supplementary Table 2). Samples were transported to the Hopkirk Research Institute (Massey University, Palmerston North, New Zealand) and stored at 4°C until processing (within 8 h of collection). Farm management practice metadata were collected during sampling visits. AMU was reported as individual animal antimicrobial treatments recorded on farm. The total amount administered for each treatment (mg) was calculated according to the concentration of the product (mg/ml), number of doses and volume (ml). AMU was reported as mg/population correction unit (PCU) calculated using the total active ingredient weight (mg)/herd size/average cow size in NZ (453 kg).
The environmental samples were enriched in buffered peptone water (BD Difco,™ Fort Richard Laboratories, Auckland, New Zealand) at 35°C for 18 h (Supplementary Table 2). After incubation, enrichments were mixed by vortex and 1 ml was mixed with glycerol (30% [v/v]) and stored at −80°C.
Frozen enrichments (Dairy 1; n = 101, Dairy 4; n = 103) were plated on (i) MacConkey (MC) agar plates (Fort Richard Laboratories, Auckland, New Zealand) as a positive control to ensure growth of E. coli, (ii) in-house prepared MC agar (BD Difco™) plates supplemented with 1 μg/ml cefotaxime sodium (Sigma-Aldrich, St. Louis, MO, United States), (iii) MC agar (BD Difco™) plates supplemented with 1 μg/ml ceftazidime (Sigma-Aldrich) and (iv) CHROMagar ESBL™ (CHROMagar, Paris, France, Fort Richard Laboratories), as previously described (Burgess et al., 2021a), except Columbia Sheep Blood agar (5% blood) (Fort Richard Laboratories) was used for the purification of isolates.
Presumptive pure E. coli isolates were stored in in-house prepared brain heart infusion broth (Oxoid, Hampshire, United Kingdom) containing glycerol (30% [v/v]) at −80°C. Isolates were identified by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (MS) (Bruker Daltonics, Billerica, CA, United States) using the previously described “on slide formic acid extraction” method (Lévesque et al., 2015). Confirmed E. coli strains isolated from agars (ii – iv) outlined above were subjected to antimicrobial susceptibility testing (ASTs) against six antimicrobials (Supplementary Table 3) using the Kirby-Bauer disc diffusion tests and CLSI guidelines (CLSI, 2019). An AmpC and ESBL positive phenotype was confirmed using either a three-disc (D69C AmpC disc test, Mast Group Ltd., Liverpool, United Kingdom) or double-disc comparison assay (D62C cefotaxime and D64C ceftazidime ESBL disc tests, Mast Group Ltd., Liverpool, United Kingdom), respectively. Isolates which were AmpC positive and had a zone size which could not be differentiated as either positive or negative (≥2–≤5 mm) for either ESBL double-disc comparison assay were further tested using a double-disc assay containing cefepime (D63C cefepime ESBL disc test, Mast Group Ltd., Liverpool, United Kingdom). The AmpC-producing E. coli isolate NZRM4402 and the ESBL-producing Klebsiella pneumoniae isolate NZRM3681 were used as positive controls in the AmpC and ESBL confirmatory disc assays, respectively, and the susceptible E. coli isolate NZRM916 was used as a negative control.
Crude DNA was extracted from pure isolates by adding three or four colonies to 400 μl sterile molecular biology-grade water and heating at 100°C for 10 min in a heating block and storing at −20°C. The supernatant was used for subsequent PCR reactions and the PCR conditions are detailed in Supplementary Table 4. The E. coli phylogroup was determined using the Clermont quadruplex PCR Typing method (Clermont et al., 2000, 2013) with KAPA Hifi HotStart ReadyMix (KAPA BioSystems, Wilmington, United States). E. coli with an AmpC phenotype were tested for pAmpC gene families using a multiplex PCR (Pérez-Pérez and Hanson, 2002) and a PCR targeting the blaCMY gene family (Dierikx et al., 2012) was used on boiled DNA preparations from isolates positive for the CITM primer set, indicative of CMY-positive E. coli. E. coli in which no pAmpC genes were identified were further analysed to identify mutations in the promoter region of the ampC gene (Caroff et al., 1999). PCR products were purified using the QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and the sequencing reaction primers and conditions are detailed in Supplementary Table 4. Capillary separation of sequencing reactions was undertaken by the Massey Genome Service using an ABI3730 DNA analyser (Massey University, Palmerston North, New Zealand). DNA sequence chromatograms were trimmed using Geneious Prime v2019.1.1 (Kearse et al., 2012; Biomatters Geneious Prime, 2022) and the complete PCR products were confirmed against the NCBI database using BLASTN (Altschul et al., 1990). The ESBL blaCTX–M group was confirmed using the CTX-M-1-group PCR (Gonggrijp et al., 2016) for the ESBL-producing E. coli which did not undergo whole genome sequencing (WGS). PCR reactions were performed using HOT FIREPol® Blend Master Mix (Solis BioDyne, Tartu, Estonia) unless stated otherwise.
A subset of ESBL- and AmpC-producing E. coli were selected for WGS according to phenotype, resistance profile, E. coli phylogroup and metadata (including farm, source, and collection date). For samples where multiple isolates were identified, only one isolate per molecular and resistance profile combination was included. Bacterial isolates from glycerol broths stored at −80°C were inoculated on Columbia Sheep Blood agar (5% blood) and incubated for 18 h at 35°C. An individual colony was subsequently sub-cultured onto a fresh Columbia Sheep Blood agar plate (5% blood) and incubated at 35°C for 18 h. An individual colony was inoculated in 4 ml Luria-Bertani Miller broth (Fort Richard Laboratories) and incubated at 35°C for 15 h at 200 rpm. Genomic DNA (for both Illumina and MinION sequencing methods) was extracted using the Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, United States), according to the manufacturer’s instructions for Gram-negative bacteria. Several modifications were included to optimise the protocol for E. coli. Briefly, 2 ml overnight culture was centrifuged at 13,000 × g for 2 min to pellet the cells which were treated with RNAse (100 mg/ml) at 37°C for 1 h. After the Protein Precipitation Solution was added to the cell lysate and incubated on ice for 5 min, the sample was centrifuged at 13,000 × g for 3 min, the supernatant transferred to a clean tube and centrifuged again at 13,000 × g to reduce any residual protein contamination. The DNA was re-hydrated overnight at 4°C in 100 μl 10 mM TrisHCl pH 8.5 (Geneaid Biotech Ltd., New Taipei City, Taiwan).
The DNA concentration was quantified using a Qubit 4.0 fluorometer (Thermo Fisher Scientific Inc., Waltham, MA, United States) and A260/280 and A260/230 ratios determined using the Nanodrop microvolume spectrophotometer (Nanodrop 2000c, Thermo Fisher Scientific Inc.). DNA integrity and size was visualised on a 0.8% [w/v] agarose gel using a high molecular weight Lambda DNA/HindIII ladder (Thermo Fisher Scientific Inc.). The libraries were prepared using the Nextera XT DNA library preparation kit (Illumina, San Diego, CA, United States) and sequencing performed using an Illumina MiSeq v3 with 2 × 300 bp paired-end reads (Massey Genome Service, Massey University, Palmerston North, New Zealand). The Nanopore MinION sequencing was performed at the Molecular Epidemiology and Public Health Laboratory (Massey University, Palmerston North, New Zealand) using a R9.4.1 flow cell (Oxford Nanopore Technologies, United Kingdom). The libraries were prepared using the Rapid Barcoding Sequencing kit (SQK-RBK004; Oxford Nanopore Technologies, United Kingdom) according to the manufacturer’s instructions (“Library Preparation”), with minor modifications for E. coli. Briefly, 600 ng DNA template was used as input and the DNA pellet was resuspended in 10 μl 10 mM TrisHCl/50 mM NaCl by incubating at 50°C for 10 min. The flow cell was primed and loaded according to the manufacturer’s instructions and run for 24–48 h (SQK-RBK004, “Priming and loading the SpotON flow cell,” Oxford Nanopore Technologies, United Kingdom).
The Illumina MiSeq sequencing reads were randomly subsampled down to a 100× genome sequencing depth using Rasusa v0.6.0 (Hall, 2022). The Illumina MiSeq sequencing reads were subsequently processed using the Nullarbor v2.0 (Seemann et al., (n.d.)) pipeline with default parameters. In summary, adapters were removed from raw reads using Trimmomatic v0.39 (Bolger et al., 2014), species identification by k-mer analysis performed using the Kraken v1.1.1 database (Wood and Salzberg, 2014), the genomes were assembled using SKESA v2.4.0 (Souvorov et al., 2018) and annotated with Prokka v1.14.6 (Seemann, n.d.a). The sequence type was determined using mlst v2.19.0 (Seemann, n.d.b) with information downloaded from PubMLST (Jolley and Maiden, 2010), the resistome profile identified with ABRicate v1.0.1 (Seemann, n.d.a) using the Resfinder 4.0 database (Zankari et al., 2012; Bortolaia et al., 2020), and the Centre for Genomic Epidemiology website (Center for Genomic Epidemiology, 2011) was used to detect the virulence genes and serotype in assembled genomes using the VirulenceFinder 2.0.3 database (v2020-05-29) (Joensen et al., 2015) and the SeroTypeFinder 2.0.1 database (v1.0.0), respectively. The presence/absence data from 37 virulence associated genes identified in the whole genome sequences (n = 12) was used to construct a hierarchical cluster tree using Jaccard distances and the tree was further annotated using the Interactive Tree of Life webserver (Letunic and Bork, 2016). The core single nucleotide polymorphism (SNP) variation was assessed using Snippy v4.4.3 (Seemann, 2016) with a ST131 ESBL-producing E. coli JJ1887 as the reference sequence (Genbank accession: CP014316). A maximum-likelihood tree was generated from the core SNP alignment using a general time-reversible model with the Randomised Axelerated Maximum Likelihood (RAxML) Next-Generation tool (Kozlov et al., 2019) and visualised in GrapeTree (Zhou et al., 2018).
For long-read data, the MinION fast5 sequencing read files were basecalled using Guppy v4.2.2. The reads were de-multiplexed using qcat v1.1.0 (Oxford Nanopore Technologies, 2019) and adapters removed with Porechop v0.2.4 (Wick, 2018b) using default settings. Filtlong v0.2.0 (Wick, 2018a) was used to trim the reads with a minimum length of 1 kb. Hybrid assemblies were generated using Unicycler v0.4.9b (Wick et al., 2017) with default settings. Plasmids from hybrid assemblies were re-constructed and typed using MOB-suite v1.4.9.1 (Robertson and Nash, 2018; Robertson et al., 2020) and annotated with Prokka v1.14.6 (Seemann, n.d.a) using a custom database consisting of the best-match “nearest neighbour” plasmids as identified by MOB-suite v1.4.9.1 (Genbank accessions: CP009566, CP015997, CP018107, CP016585, and KF362121). For pMLST, variants within IncI group plasmids from hybrid assemblies and plasmid incompatibility groups for short-read data only were identified (Carattoli et al., 2014). Plasmid core genome variation for selected isolates was assessed using Snippy v4.6.0 (Seemann, 2016) with pDF0049.2e_1 as the reference. Clusters of Orthologous Groups (COG) were identified using eggNOG-mapper v2 (Huerta-Cepas et al., 2017, 2019; Cantalapiedra et al., 2021). The plasmid oriC region was identified using DoriC v10.0 (Luo and Gao, 2019). Statistical tests were performed in Minitab® 19.1.1 (Minitab, 2019) using a one-way analysis of variance (ANOVA) with a 95% confidence interval for sample prevalence comparisons.
Farm samples were obtained from Massey University research farms Dairy 1 and Dairy 4 over a 15-month period from October 2018 to December 2019, consisting of composite faeces (n = 60), milk (n = 13), soil (n = 15), and FDE (n = 13 Dairy 1; n = 15 Dairy 4) from each farm. From the selective agar plates, 52 putative 3GC resistant E. coli (n = 24 Dairy 1; n = 28 Dairy 4) were isolated (Supplementary Table 5).
The susceptibility of E. coli isolated in this study (n = 52) to six clinically relevant antimicrobials, including three β-lactams, was examined (Figure 1 and Supplementary Table 6). All of the E. coli (n = 52) were resistant to cefpodoxime, 46 of 52 (89%) to cefoxitin and 34 of 52 (65%) were resistant or intermediate (15 of 52; 29%) to cefotaxime. Numerous isolates were resistant to streptomycin (41 of 52; 79%) and tetracycline (33 of 52, 64%) although all isolates (n = 52) were susceptible to ciprofloxacin, which is a critically important antimicrobial in human medicine (World Health Organization, 2011). The susceptibility of 20 E. coli to the 4GC cefepime was assessed (Supplementary Table 6) representing at least one E. coli from every sample and all phylotype and resistance profile combinations. Seven isolates from four samples were cefepime resistant and the remaining 13 E. coli were either susceptible or intermediate (Supplementary Table 6). According to phenotypic testing, 33 of 52 E. coli (64%) were multi-drug resistant, but phenotypic testing was only performed using antibiotics representing four classes (β-lactams, tetracyclines, aminoglycosides, and fluoroquinolones).
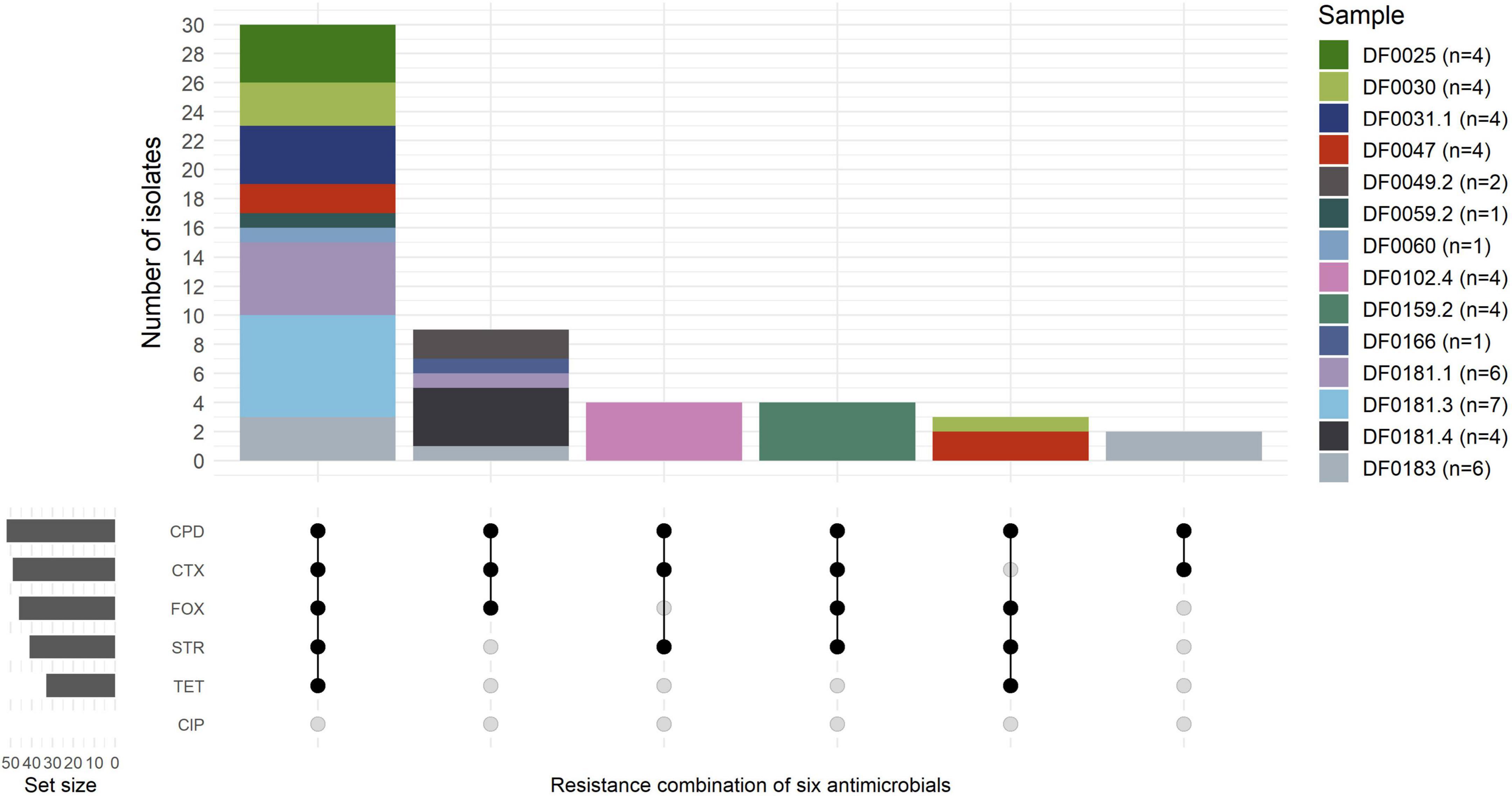
Figure 1. Resistance profiles of E. coli (n = 52) isolated across 14 farm samples. The number of isolates per sample is indicated in the brackets. Isolates with intermediate resistance to cefotaxime (n = 15) were grouped as resistant in the Upset plot. CPD, cefpodoxime (10 μg); FOX, cefoxitin (30 μg); STR, streptomycin (10 μg); CTX, cefotaxime (30 μg); TET, tetracycline (30 μg); CIP, ciprofloxacin (5 μg).
At the isolate level, all putative antimicrobial resistant E. coli (n = 52) were tested for AmpC and ESBL production. In total, 46 of 52 (88.5%) were AmpC-producing E. coli and, 2 of 52 (3.8%) isolates from the same FDE sample were confirmed as ESBL producers (DF0183c and DF0183g), and one (1.9%) isolate (DF0059.2e) was both ESBL and AmpC positive. Four isolates (7.7%) from the same sample (DF0102.4e-h) were both ESBL and AmpC negative according to the phenotypic tests.
Farm samples were investigated for ESBL- and AmpC-producing E. coli (Table 1). At the sample level, one of 60 (1.7%) pooled faecal and one of 15 (6.7%) FDE samples from Dairy 4 were positive for ESBL-producing E. coli, with one and two isolates cultured from these samples, respectively. No ESBL-producing E. coli were isolated from pooled faeces and FDE from Dairy 1 nor from soil from the most recently grazed paddocks or milk samples from either farm. AmpC-producing E. coli were isolated from faeces (2 of 60, 3.3%; 5 of 60, 8.3%) and FDE (5 of 13, 38.5%; 1 of 15, 6.7%) on Dairy 1 and Dairy 4, respectively and none were isolated from soil (0 of 30) or milk (0 of 26) from either farm. The sample level prevalence of AmpC-producing E. coli isolated from Dairy 1 and Dairy 4 (p = 0.526) and between sample types (faeces or FDE; p = 0.408) was not statistically significant. ESBL-producing E. coli were isolated at a low prevalence on Dairy 4 but were not isolated from Dairy 1 during this study period. Dairy 4 had a higher prevalence of AmpC-producing E. coli from faeces (8.3%), whereas Dairy 1 had a higher number isolated from FDE (38.5%).
According to antimicrobial treatment data, between October 2018 and December 2019 the estimated AMU on Dairy 1 and Dairy 4 was 17.09 mg/PCU and 5.36 mg/PCU, respectively (Supplementary Tables 7,8). Months with higher AMU are consistent with spring calving in NZ. The New Zealand Veterinary Association (NZVA) has classified antimicrobials as green, yellow or red tier according to the World Health Organisation classes (Anonymous, 2018). Of the total AMU used on Dairy 1 and Dairy 4, the majority of antimicrobials were green tier (91.1 and 30.5%), followed by yellow tier (6.2 and 67.9%) while red tier antimicrobials were infrequently used (2.75 and 1.6%), respectively.
As shown in Figure 4, AmpC-producing E. coli were isolated in spring and summer and after months of both high and low AMU. On Dairy 1, the highest AMU occurred in October 2018 (12.3%) and August to November 2019 (8.7–18.6%); whereas on Dairy 4 the highest AMU predominantly occurred in September 2019 (56.7%) (Figure 4 and Supplementary Tables 7–9).
At the isolate level, the predominant E. coli phylogroups were B1 (18 of 52, 35%) and C (17 of 52, 33%), followed by E (6 of 52, 12%), D (5 of 52, 10%), A (4 of 52, 8%), and F (2 of 52, 4%). The blaCTX–M–15 gene was identified in the ESBL-producing E. coli (DF0183c and DF0183g). Isolate DF0059.2e was confirmed as both AmpC and ESBL positive using phenotypic and genotypic testing and co-harboured the blaCMY–2 and blaCTX–M–1 genes. The plasmid-mediated gene blaCMY–2 was detected in 28 of 46 isolates (61%). Plasmid-mediated AmpC strains were predominantly isolated from Dairy 4 (27 of 28, 96%). For the remaining 18 AmpC-producing E. coli, mutations were identified in the promoter region of the ampC gene. All AmpC hyperproducers (n = 18) were isolated from Dairy 1. The 18 isolates were all phylogroup C and had mutations in the promoter region of the ampC gene at positions -42 (C → T), -18 (G → A), -1 (C → T) and +58 (C → T), excluding DF0025c in which position −42 could not be determined [positions relative to the E. coli K12 transcriptional start base (+1) (Olsson et al., 1982)]. No pAmpC or ESBL genes were detected in 3GC resistant four E. coli which were AmpC and ESBL negative in the phenotypic testing (DF0102.4e-h). Therefore, the mechanism for resistance to 3GCs was unassigned for these isolates (designated as ‘unknown’ in subsequent figures).
To determine whether the cefepime resistant phenotype of the five isolates which were not ESBLs was a result of extended-spectrum AmpC beta-lactamase production, the deduced AA sequence of AmpC was compared between isolates DF0102.4g (chromosomal AmpC), DF0159.2g (plasmid-mediated AmpC), previously identified ESAC producers and a negative control. AA substitutions were identified in the omega loop at positions 191 (K191Q) for both isolates and at positions 201 (N201T) and 209 (S209P) for DF0102.4g and DF0159.2g, respectively; DF0159.2g also had an AA substitution in the R2 loop region, at position 300 (I300V) (Table 2). Neither isolate had any substitutions in the H-9 or H-10 helices.
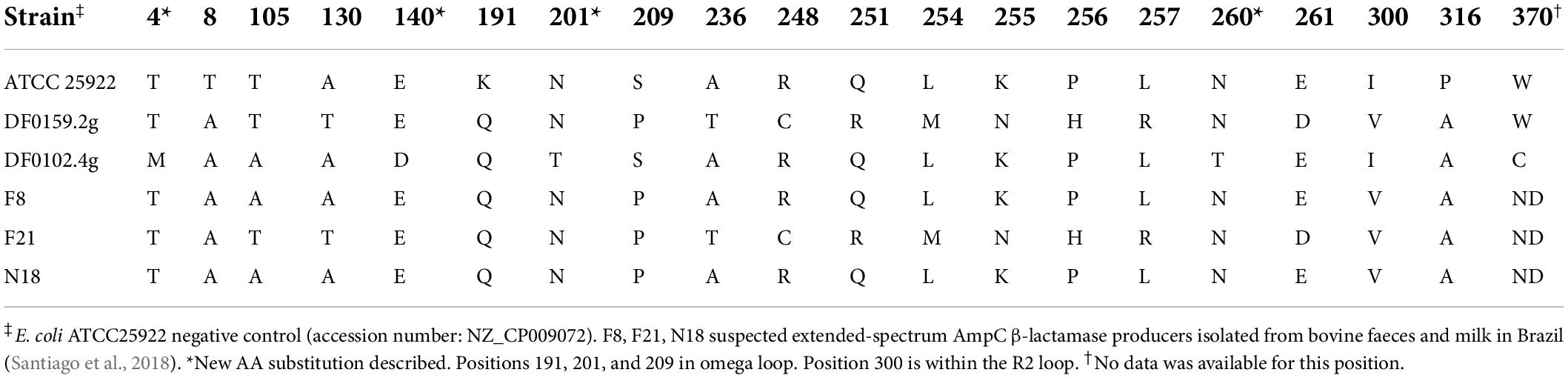
Table 2. Amino acid substitution profiles of suspected extended-spectrum AmpC β-lactamase producing E. coli from this study.
A subset of 3GC resistant E. coli were analysed using WGS to understand the genomic epidemiology and transmission dynamics of these bacteria on farm (Table 3). E. coli isolates with a plasmid-mediated AmpC-producing phenotype (n = 4), ESBL-producing (n = 1), ESBL/AmpC-producing (n = 1) and an unassigned mechanism for resistance to 3GCs (n = 1) were selected for Oxford Nanopore Technologies (ONT) MinION long-read sequencing, with a focus on generating complete genomes and examining plasmids in detail. The population structure of the AmpC- and ESBL-producing E. coli which were sequenced in this project (n = 12) was diverse and the isolates belonged to eight sequence types including ST56 (3 of 12, 25%), ST57 (2 of 12, 17%), ST88 (2 of 12, 17%) and singletons for ST442, ST973, ST2541, ST4553, and ST5135 (1 of 12, 8%) (Table 3). All isolates had a genome size ranging from 4,839,855 to 5,431,661 bp and a GC content between 51.7 and 52.0%.
Core genome SNP analysis separated the strains into eight clusters, each representing a different ST (Figure 2). The separation of clusters according to ST was reflected in the large number of SNPs identified in the core genome of these strains (102,841 SNPs), accounting for approximately 2.0% of the E. coli genome. However, within two clusters genetically similar isolates representing two STs were identified. Three blaCMY–2-positive E. coli were isolated in December 2019 on Dairy 4 from two faeces and one FDE sample, all belonged to ST56 and were genetically similar (58 – 65 SNPs; isolates DF0181.1c, DF0181.3c, and DF0183e). Similarly, two AmpC hyperproducers, were isolated in December 2018 and January 2019 from Dairy 1 and differed by 82 SNPs (DF0031.1c and DF0047c). In each case, the E. coli were isolated from both faeces (ST56 n = 2; ST88 n = 1) and FDE (ST56 n = 1; ST88 n = 1).
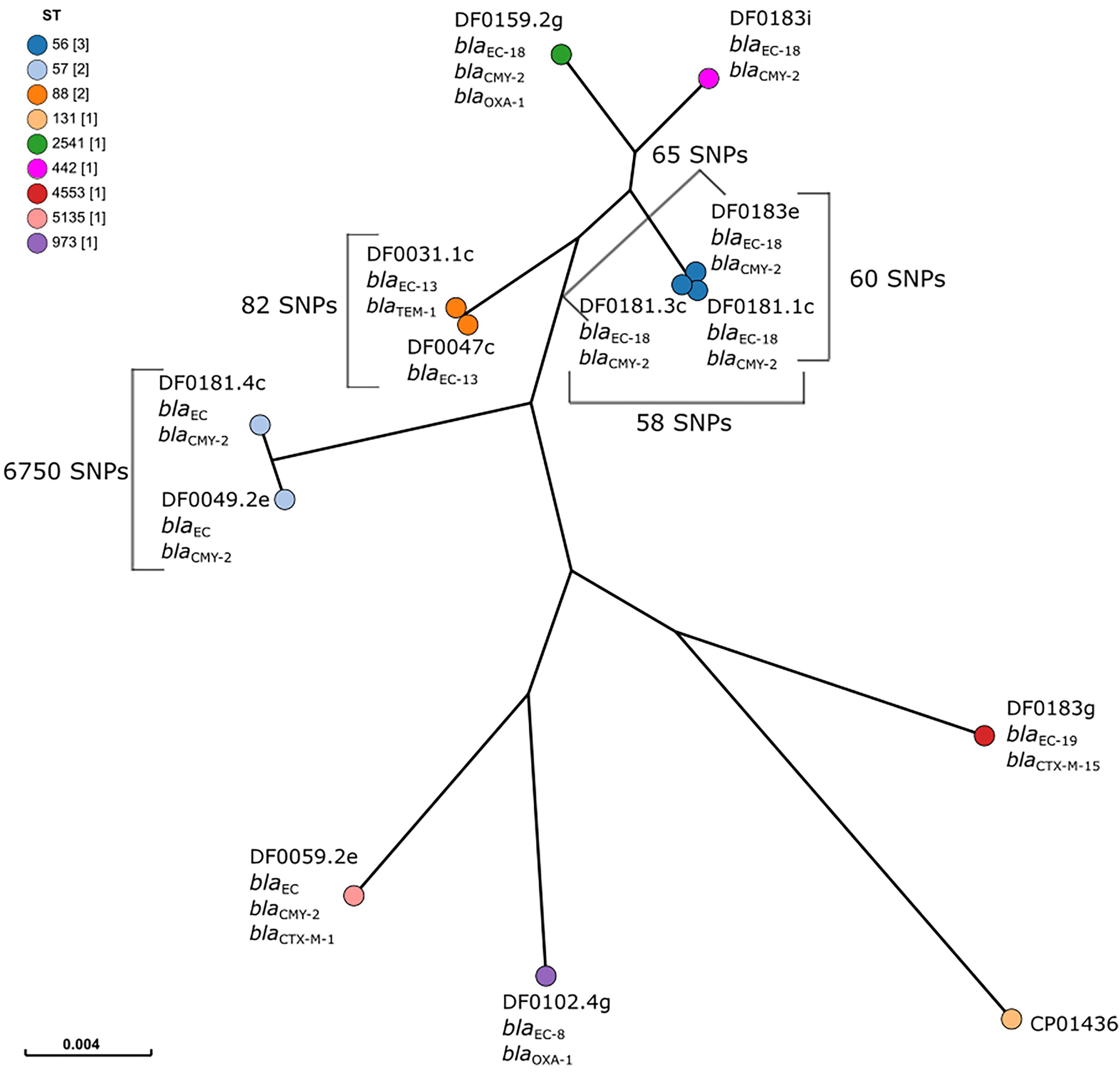
Figure 2. Maximum-likelihood tree of core genome single nucleotide polymorphism (SNP) analysis of ESBL and AmpC-producing E. coli (n = 12). E. coli CP014316 (ST131, blaCTX–M–15 positive) was used as the reference and nodes are coloured by sequence type, as indicated in the figure legend. The scale bar indicates the proportion of the core genome alignment over which core SNPs have been calculated. The number of SNPs between isolates in the same cluster is indicated on the figure.
Analysis of sequence data of 12 E. coli confirmed the presence of the β-lactam resistance genes blaCMY–2 (8 of 12, 66.7%), blaCTX–M–1 and blaCTX–M–15 (1 of 12 each, 8.3%) and blaOXA–1 (2 of 12, 16.7%) (Table 4). The genotypic results were in agreement with the confirmed ESBL and AmpC phenotypes. All isolates carried the ampC gene (a synonym for the blaEC gene (Mammeri et al., 2006) and gene-specific variation observed was broadly associated with the different E. coli phylogroups.
Other ARGs including mph(A), catA1, dfrA5, dfrA17, sul1, and sul2 were identified during analysis of the assembled genomes but representative phenotypes were not examined. Initial PCR methods failed to identify any ESBL and AmpC genetic determinants associated with four DF0102.4e-h isolates resistant to 3GC and 4GCs (cefotaxime, cefpodoxime, and cefepime). Further analysis of DF0102.4g using WGS also failed to determine the genetic basis for the AMR phenotype. However, this isolate carried the blaOXA–1 gene which encodes a narrow-spectrum Class D β-lactamase and had several AA substitutions in the chromosomally encoded AmpC, although no substitutions were identified in the R2 loop of H-9 or H-10 helices. According to the phenotypic confirmation tests, this isolate was AmpC negative and this phenotype was confirmed by in silico analysis of the promoter region of the ampC gene which failed to identify any mutations in the promoter elements, although mutations within the ampC gene (Caroff et al., 1999) were identified at positions +70 (C → T) and +81 (A → G).
The E. coli sequenced in this study carried a range of virulence factors, which were mainly involved in adhesion, protection/serum resistance, iron uptake and toxins, hemolysins, proteases or autotransporters (Figure 3). E. coli strains of the same ST clustered together, based on the presence/absence of virulence factors. The number of virulence factors each E. coli harboured varied, with strains DF0049.2e and DF0183i carrying the fewest virulence factors (3 of 139) and DF0031.1c and DF0047c carrying the most (28 of 139).
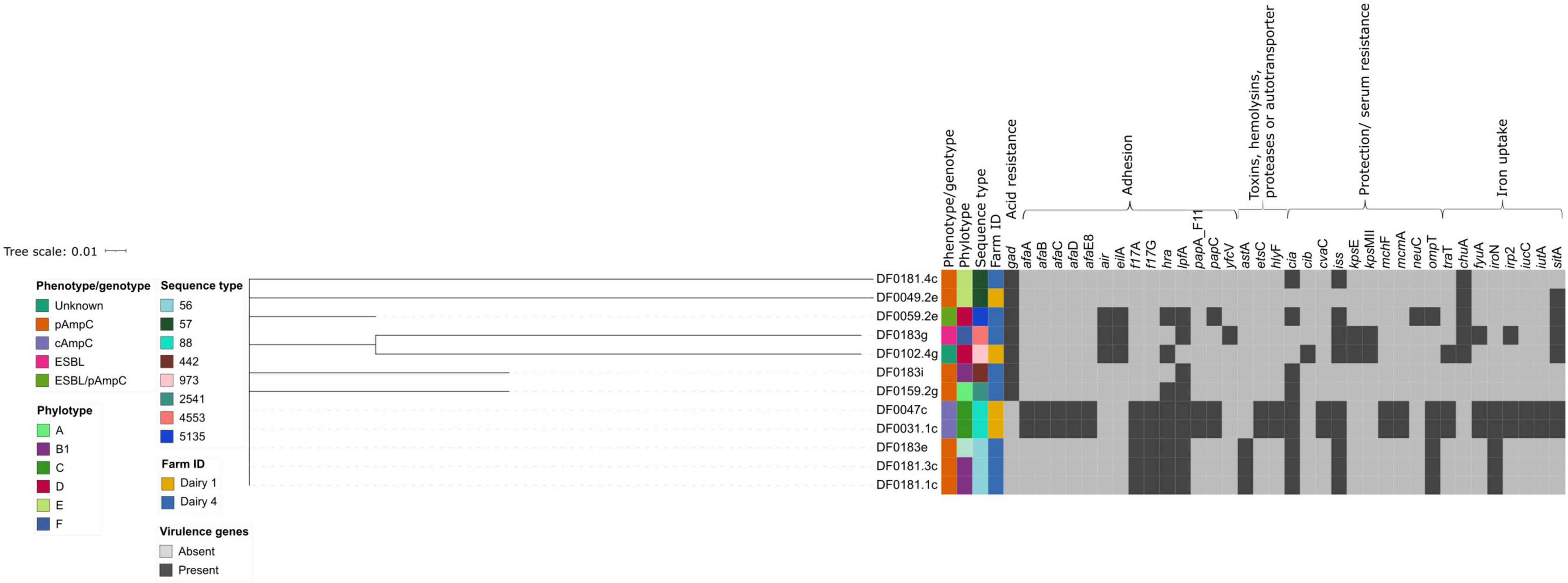
Figure 3. Hierarchical cluster tree constructed using Jaccard distances for the presence or absence data from 37 virulence genes identified using Virulence Finder. The tree was edited using the Interactive Tree of Life webserver. Isolate metadata is included for ESBL or AmpC phenotype, farm and sequence type, as indicated by the colour keys. The virulence genes are grouped and annotated by general function.
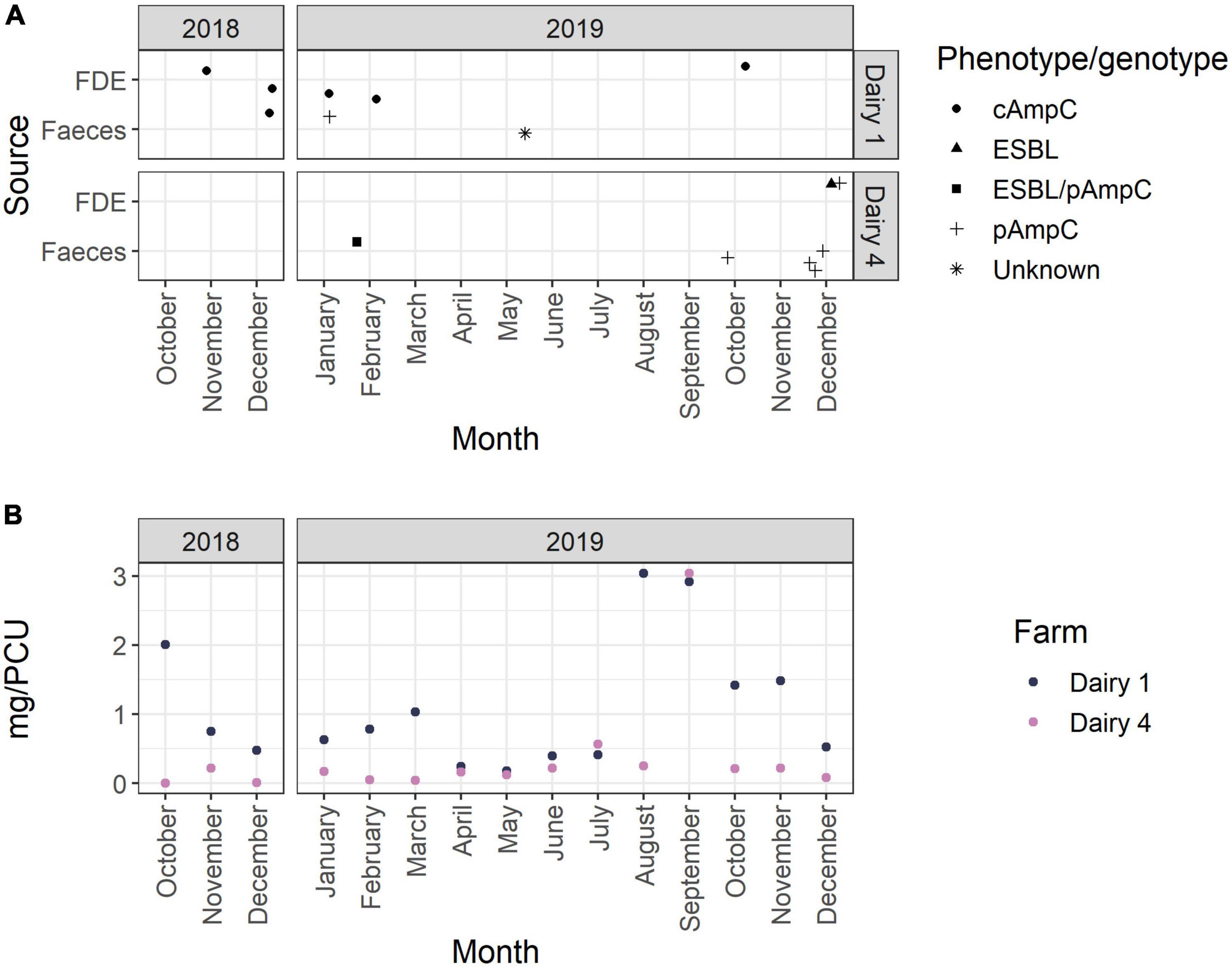
Figure 4. (A) Collection dates of samples positive for ESBL- and/or AmpC-producing E. coli over the 15-month study period. The phenotype is represented by shape as indicated in the figure legend. (B) Antimicrobial use per month (mg active ingredient per population correction unit) on Dairy 1 and Dairy 4 during the 15-month study period.
Five E. coli strains were putatively classified as avian pathogenic E. coli (APEC), a subgroup of extraintestinal pathogenic E. coli, due to the presence of the following virulence factors: salmochelin siderophore receptor (iroN), outer membrane protease (ompT) and increased serum survival (iss) (in DF0181.1c, DF0181.3c, and DF0183e) as well as ferric aerobactin receptor (iutA) and avian hemolysin (hlyF) which were also detected in strains DF0031.1c and DF0047c. These virulence factors have been associated with APEC and are often encoded on plasmids (Johnson et al., 2008). No E. coli harboured the Shiga toxin genes (stx1 or stx2) or the locus of enterocyte effacement pathogenicity island, indicative of Shiga-toxin producing E. coli (STEC) or enteropathogenic E. coli (EPEC), respectively (Robins-Browne et al., 2016). Three strains carried the air gene for the enteroaggregative immunoglobulin repeat protein Air which is commonly found in enteroaggregative E. coli (EAEC), however, other virulence genes such as EAST-1 and pic associated with this pathotype were not detected (Johnson et al., 2008). Five strains harboured enterotoxigenic E. coli (ETEC) associated fimbriae F17 (f17A and f17G) encoding genes, although no other ETEC-related toxin or fimbriae genes were detected in these strains.
A large proportion of the AmpC- (8 of 10, 80%) and ESBL-encoding (1 of 2, 50%) genes identified from E. coli isolated within this study are plasmid-associated. Four isolates sequenced using MinION long-read methods carried one IncI1 plasmid harbouring the blaCMY–2 gene, one isolate carried an IncI1 plasmid encoding the ARGs blaOXA–1, sul1 and aadA1 and an IncI1/IncFII plasmid which did not carry any ARGs. One isolate carried three plasmids, an IncI1 plasmid encoding the blaCMY–2 gene, an IncY plasmid and an IncFIA/IncQ1 which harboured 12 ARGs including blaCTX–M–1 (Table 5). No plasmids were identified from the assembled sequence data of DF0183g, instead the blaCTX–M–15 gene was chromosomally encoded. All plasmids contained a relaxase and mate-pair formation marker sequences and were therefore classified as conjugative except for plasmid pDF0059.2e_2 which belonged to the IncY group and was non-mobilisable.
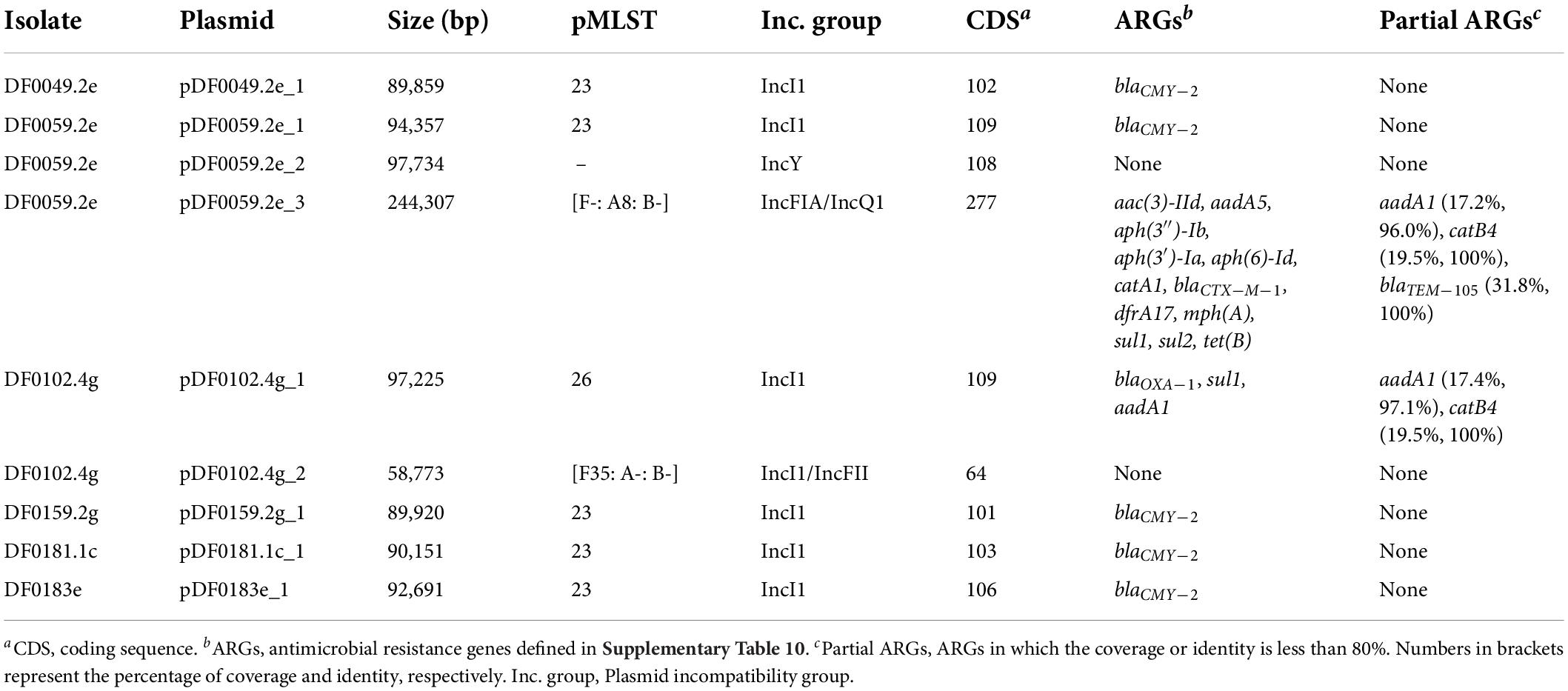
Table 5. Characteristics of plasmids from E. coli sequenced with short-read Illumina and long-read MinION methods.
DF0059.2e carried an IncFIA/IncQ1 plasmid (244,307 bp) of unknown pMLST for which the nearest plasmid neighbour was the E. coli strain T23 multi-drug resistant plasmid pEQ1 (Genbank accession: KF362121). This plasmid carried multiple ARGs (n = 12), potentially conferring resistance to aminoglycoside, β-lactam, phenicol, trimethoprim, macrolide, sulfonamide and tetracycline antibiotic classes (Table 5). Physical linkage of ARGs and the location of mobile genetic elements on pDF0059.2e_3 is shown in Supplementary Figure 1. In addition, a partial copy of the blaTEM–105 (279 bp; 31.82% coverage), the catB4 (106 bp; 19.49% coverage), and the aadA1 (166 bp; 17.18% coverage) genes were detected. However, the partial copy of the catB4 and aadA1 genes could not be confirmed by visual inspection of the annotated plasmid. The genetic region surrounding the partial blaTEM–105 gene and the upstream IS26 transposase was extracted (2,438 bp) from the pDF0059.2e_3 plasmid and aligned to the complete blaTEM–105 reference gene sequence (Genbank accession number: NG_050150, 966 bp). Pairwise alignment showed that 274 of 279 bp of the partial blaTEM–105 gene was an identical match to the distal end of the reference gene sequence, with the IS26 transposase disrupting the upstream region of the blaTEM–105 gene (Supplementary Figure 2). We can hypothesise that plasmid pDF0059.2e_3 once harboured a complete copy of the blaTEM–105 gene and a recombination event involving IS26 transposase disrupted this gene sequence.
Five strains carried IncI1 plasmids (also called IncIα) that were pMLST 23, had a similar number of coding sequences (101–109), encoded the blaCMY–2 gene and ranged in size from 89,859 to 94,357 bp. Using the MOB-suite database (consisting of 17,779 complete plasmids) the most similar reference to the IncI1 plasmids identified using Mash distances, was the Salmonella enterica subspecies enterica serovar Newport plasmid pCVM22462 (Genbank accession number: CP009566.1). Core genome SNP analysis of these five plasmids, using pDF0049.2e_1 as the reference, suggested they were genetically similar. There were no SNP differences between the plasmid-core genome of DF0181.1c and DF0183e, which were isolated from composite faeces and FDE respectively on Dairy 4 in December 2019 (Supplementary Table 5). The core chromosomal genome of these isolates was also genetically similar, differing by only 65 SNPs. E. coli DF0181.3c (only short-read sequencing data available) was isolated from the same sampling month as DF0181.1c and DF0183e and had an identical serotype and resistance profile. IncI1 plasmid incompatibility factors were detected in the draft genome for DF0181.3c, and therefore this isolate may carry a similar plasmid as the aforementioned strains, however, this plasmid could not be reconstructed and compared in detail in the absence of long-read sequencing data. Plasmid DF0049.2e_1 was annotated as a representative of the IncI1 plasmids showing the location of the blaCMY–2 gene, mobile genetic elements and the tra genes involved in the conjugal transfer system (Supplementary Figure 3). All plasmids showed variation near the shufflon protein, a genetic region involved in conjugation of IncI1 plasmids (Brouwer et al., 2019).
Two chromosomal mediated AmpC-producing E. coli (DF0031.1c and DF0047c) underwent Illumina short-read sequencing only and additional ARGs which potentially confer resistance to aminoglycoside, trimethoprim, sulfonamide and tetracycline antibiotics (trimethoprim and sulfonamide resistance was not phenotypically confirmed) as well as plasmid incompatibility factors were identified. These findings suggest that these isolates harbour additional ARGs, some of which are likely encoded on plasmids due to the co-location of ARGs [aph(3′′)-Ib, aph(6)-Id, sul2] and plasmid incompatibility factors (IncQ1) on the same contig and therefore could potentially spread via HGT.
Using VirulenceFinder, the five IncI1 plasmids harboured virulence genes encoding colicins, namely cia (n = 4, blaCMY–2 positive plasmids) and cib (n = 1, pDF0102.4g_1). Plasmid pDF0102.4g_2 harboured the traT gene, which encodes an outer membrane protein involved in complement resistance, although the traT gene is also a part of the transfer operon in conjugative plasmids and is involved in surface exclusion between identical or closely related plasmids by preventing stable mating aggregates (Sukupolvi and O’Connor, 1990). Plasmids pDF0059.2e_2 and pDF0059.2e_3 did not carry any known virulence genes.
Overall, the sample level prevalence of ESBL-producing E. coli from faeces and FDE was low on Dairy 4 and they were not detected on Dairy 1 (Table 1). These results are consistent with a previous regional-based cross-sectional study of NZ dairy farms, which found a low prevalence of ESBL-producing E. coli in pooled faecal samples and no E. coli with pAmpC genes were identified (Burgess et al., 2021a). Similarly, a nationwide cross-sectional study on dairy farms did not detect any ESBL- or plasmid-mediated AmpC-producing E. coli (Burgess et al., 2021b). Overseas studies have found a higher prevalence of AmpC- and/or ESBL-producing E. coli from dairy farms, with herd level prevalence estimates ranging from 13% on organic dairy farms in the Netherlands (Santman-Berends et al., 2017) and 5.2–86.7% on conventional farms (Ohnishi et al., 2013; Schmid et al., 2013; Dahms et al., 2015; Gonggrijp et al., 2016; Heuvelink et al., 2019; Schubert et al., 2021).
The sample level prevalence of AmpC-producing E. coli in this study was also relatively low (Table 1). AmpC-producing E. coli were isolated after months of both high and low AMU, highlighting that additional factors other than total AMU may also play a role in the development and transmission of antimicrobial resistant bacteria in the dairy farm environment. E. coli with pAmpC genes were infrequently identified on Dairy 1 (1 of 18; 5.6%) and all AmpC-producing E. coli isolated from Dairy 4 were plasmid-mediated (27 of 27; 100%; blaCMY–2). This suggests that although Dairy 1 had a higher sample level prevalence of AmpC-producing E. coli, particularly in FDE, these putative AmpC hyperproducers predominantly spread by vertical transmission. The differences in sample prevalence from FDE may be due to different effluent management strategies between farms, with Dairy 4 storing FDE in a large pond prior to spraying onto paddocks which likely results in the dilution of bacteria. Univariable analysis of data obtained from British beef farms found that spreading of farm manure was significantly associated with an animal testing positive for AmpC-producing E. coli (Velasova et al., 2019). A higher number of 3GC resistant E. coli were isolated from Dairy 4 in December 2019, which may have been due to several farm management factors including increased AMU during calving and mating. However, there was no increase in 3GC and 4GC use on Dairy 4 during this time period, which has been associated with ESBL/AmpC positive herd status (Gonggrijp et al., 2016). Climatic conditions such as increased temperatures in December, may also play a role. A recent study suggested that lower monthly ambient temperatures were associated with a lower rate of identifying blaCTX–M positive E. coli samples (Schubert et al., 2021). However, the small number of samples (n = 2) positive for ESBL-producing E. coli in this study does not allow for any associations between seasonality and ESBL positive samples to be observed. December 2019 was at the end of the study sampling period and no further sampling was undertaken to assess whether this increased trend of 3GC resistant E. coli continued in the following months, but no comparable increase was observed during the same 2018 months. However, these observations and climatic factors may assist with future sampling strategies for investigating the prevalence of 3GC E. coli on pasture-based dairy farms.
Of the AmpC-producing E. coli isolated during this study, 18 of 46 (39.1%) were AmpC hyperproducers. Variable proportions of AmpC hyperproducers have been detected in previous studies, with a small proportion of extended-spectrum cephalosporin resistant E. coli isolated from livestock in the Netherlands being AmpC hyperproducers (217 of 2,034; 9.4%) (Ceccarelli et al., 2019) compared to a higher proportion (46.2% of cefotaxime-resistant E. coli) that were identified across 53 farms in the United Kingdom (Alzayn et al., 2020; Schubert et al., 2021). Higher use of amoxicillin/clavulanate and sampling of faecal samples from the environment of young calves has been associated with an increased risk of identifying putative ampC hyperproducers on dairy farms (Alzayn et al., 2020) and in vitro studies have shown an association between amoxicillin use and AmpC-producing E. coli arising from mutations in the promoter region of the ampC gene (Händel et al., 2014; Stohr et al., 2020). Amoxicillin and clavulanic acid are classified as yellow tier antimicrobials by the NZVA, indicating their use should be restricted in NZ veterinary practices (Anonymous, 2018). Amoxicillin and clavulanic acid were not used on Dairy 1 and Dairy 4 during the study period. Cephalosporin use (including 3GCs) was not identified as a risk factor for putative ampC hyperproducing E. coli (Alzayn et al., 2020) and 3GC use on Dairy 1 and Dairy 4 was very low during the 15-month study period (<1% total mg/PCU). The lack of known risk factors for AmpC hyperproducers on Dairy 1 highlights the complexity of factors involved in the development of AMR and suggests additional studies are required to identify risk factors for AmpC hyperproducers, particularly in pasture-based dairy farms.
Infections caused by ESBL/AmpC positive Enterobacterales pose significant treatment option challenges for clinicians, with cefepime (a 4GC) or carbapenems being suggested as the main treatment options (Meini et al., 2019), both of which are critically important antimicrobials for human medicine (Collignon et al., 2016). One E. coli isolated from faeces in this study (1.9%, 1 of 52) was both AmpC and ESBL positive, carrying the blaCMY–2 and blaCTX–M–1 genes on two distinct plasmids (Table 4). A low proportion of E. coli isolates displayed an ESBL/AmpC phenotype in a study of dairy and beef cattle and sheep farms in Spain (5.2%, 7 of 135 isolates) (Tello et al., 2020). A cross-sectional study of dairy farms in Canada (n = 102) also found a low proportion of ESBL/AmpC positive E. coli (2%) in comparison to AmpC (51%) and ESBL (46%) phenotypes (Massé et al., 2021). These findings suggest that E. coli displaying both an ESBL and AmpC phenotype are infrequently isolated from dairy farm environments.
ESBL- and AmpC-producing E. coli were not detected in soil from recently grazed paddocks or bulk tank milk samples on either farm. Despite a small sample size, the lack of detection from these matrices over a 15-month period indicates that they may be potentially uncommon sources of ESBL- and AmpC-producing E. coli in the NZ dairy farm environment, whereas faeces and FDE are more probable sources. At the sample level, blaCTX–M or blaCMY–2 positive E. coli were infrequently detected from soil samples across 17 commercial beef farms in the United States (3.89%; 3 of 77) (Lee et al., 2020) and the prevalence of ESBL- and AmpC-producing Enterobacteriaceae from bulk tank milk was more varied, ranging from 0 to 9.5% (Geser et al., 2012; Skockova et al., 2015; Sudarwanto et al., 2015; Odenthal et al., 2016). The lack of detection of ESBL-producing E. coli in bulk tank milk in NZ is not unexpected due to the stringent hygiene and food safety standards for dairy farming and milk storage in NZ (Fonterra Co-operative Group Limited, 2018).
It is important to consider variations in study design (sample size and animal age/health status), sample matrices and culture selection methods when comparing between prevalence studies. For example, this study did not use a pre-enrichment step for ESBL- and AmpC-producing E. coli prior to plating on selective agar, which is a technique used in some prevalence studies (Hordijk et al., 2013; Gonggrijp et al., 2016; Hutchinson et al., 2017). In addition, selecting one isolate per sample may not be sufficient to reflect the bacterial heterogeneity associated with an ecological niche (Venturini et al., 2022), therefore, analysing multiple isolates per sample is beneficial in prevalence studies. The age of the study population is also a crucial factor to consider. For example, a study of 101 dairy farms in Canada detected ESBL- or AmpC-producing E. coli at least once during the study in 85% of farms, although the majority were isolates from calves (Massé et al., 2021). A longitudinal study of a United Kingdom dairy farm in which CTX-M-15 ESBL E. coli had previously been identified from a septic neonatal calf, found a higher proportion of E. coli positive for the blaCTX–M gene in milking cows (30.3%) compared with non-milking cows (3.0%) (Watson et al., 2012), although this study did not look for other ESBL enzyme types and other management factors (e.g., AMU) differed between cattle groups.
Variation in farming systems between countries is also important to consider when comparing studies. Intensive farming systems, particularly indoor housing, have been associated with a higher prevalence of mastitis (Lacy-Hulbert et al., 2002), which can lead to higher AMU and subsequently increased levels of AMR. It has been proposed that the NZ pasture-based farming system, in conjunction with low AMU in food-producing animals, may contribute to lower levels of AMR (Collis et al., 2019). The low sample level prevalence of ESBL- and AmpC-producing E. coli from Dairy 1 and Dairy 4 supported this hypothesis. Factors linked to less intensive farming practices have been associated with fewer samples positive for cefotaxime resistant E. coli samples from both beef and dairy cattle (Hille et al., 2017). A cross-sectional study of grazing beef cattle farms (n = 17) in the United States found that larger farming operations (>500 cattle) were associated with a 58% higher likelihood of detecting cefotaxime resistant bacteria from faecal samples (Markland et al., 2019).
Both farms in this present study operated a closed herd system (no introduction of off-farm animals), which may reduce the risk of introductions of antimicrobial resistant bacteria into the herd from outside cattle sources. For example, on British beef farms, buying bulls or fattening cattle have been identified as risk factors for blaCTX–M positive Enterobacteriaceae or AmpC-producing E. coli, respectively (Velasova et al., 2019). However, the impact that introductions of antimicrobial resistant bacteria has compared to AMR selection in agricultural environments is unknown.
AMU on farms is influenced by a number of management factors including average age of the herd, disease outbreaks, hygiene practices, the use of teat sealants as well as the farmers perception toward antimicrobial stewardship. Interestingly, Dairy 1 had a higher total AMU during the study period, yet no ESBL-producing E. coli were detected on Dairy 1 (Supplementary Tables 8, 9). However, a higher proportion of the total AMU on Dairy 1 was classified as green tier antimicrobials by the NZVA with Dairy 4 using more of yellow tier antimicrobials. The variation, predominantly green on Dairy 1 and yellow tier classes on Dairy 4, is likely associated with the predominant illnesses treated per farm. Other factors may have an impact on the AMU on farms, such as the average age of the cows, since the incidence of clinical mastitis is higher in older cows (Petrovski et al., 2009). The estimated base rate for use of antimicrobials in food-producing animals in NZ in 2018 was 10.21 mg/PCU (Hillerton et al., 2021). The representative total AMU during the study period on Dairy 1 and Dairy 4 was higher (17.09 mg/PCU) and lower (5.36 mg/PCU) than this estimate, respectively. The AMU on Dairy 1 and Dairy 4 was also within the range reported in a cross-sectional study of 26 dairy farms across NZ (4.39 – 20.92 mg/PCU) (Burgess et al., 2021b), albeit at the higher and lower end of the spectrum, respectively. The total AMU was estimated using sales data in the aforementioned study, whereas individual antimicrobial treatments were used for calculating the total AMU in this study, which makes comparisons difficult. The use of 3GCs (<1% total mg/PCU), which have been identified as a risk factor for ESBL-producing Enterobacteriaceae (Gonggrijp et al., 2016), and use of NZVA red tier classified antimicrobials.
The ESBL- and AmpC-producing E. coli which were sequenced in this study belonged to a diverse range of STs and serotypes. Similar findings have been reported for blaCMY–2 positive E. coli from human clinical cases, livestock and food matrices in which a diverse range of STs were reported (Pietsch et al., 2018). In contrast, E. coli ST131, which are frequently multi-drug resistant and harbour blaCTX–M genes, are widely disseminated in humans globally (Banerjee and Johnson, 2014; Jafari et al., 2020). The two sequenced AmpC hyperproducers belonged to ST88 (phylogroup C), which is consistent with previous findings that AmpC hyperproducers predominantly belong to this sequence type (Haenni et al., 2014b; Alzayn et al., 2020). In contrast, three AmpC hyperproducing E. coli previously isolated from dairy cattle in NZ belonged to ST1148 (n = 2) and ST298 (n = 1) (Burgess et al., 2021a). ESBL-producing E. coli were isolated from two samples (faeces and FDE) in this study. Isolate DF0183g, belonging to ST4553, harboured a chromosomally encoded blaCTX–M–15. ST4553 E. coli positive for the blaCTX–M–15 gene have also been detected in dog faeces (n = 1) (Toombs-Ruane et al., 2021) and storm water (n = 3) in NZ (Burgess et al., 2021c). E. coli DF0059.2e belongs to ST5135 and to the best of our knowledge, this is the first E. coli ST5135 blaCTX–M–1 to be reported. Few E. coli ST5135 have been reported in Enterobase Zhou et al., 2020, and those identified were isolated from human, livestock, canine and poultry samples (accessed 23rd May 2022).
The ESAC production was examined for DF0102.4g and any ESBL negative E. coli isolated from CHROMagar™ ESBL plates (DF0159.2g). SNPs previously identified in ESAC producers (Santiago et al., 2018) were detected in the areas coding for the omega loop region for both DF0102.4g and DF0159.2g (K191Q and S209P). These SNPs result in AA substitutions with different biochemical characteristics, which may result in modifications in the omega loop (Santiago et al., 2018). However, these AA substitutions have also been described in E. coli which were susceptible to cefepime, and therefore unlikely to be ESAC producers (Burgess et al., 2021b). Isolate DF0102.4g had a further substitution in the omega loop (N201T), but this resulted in substitution of an AA with the same charge which is likely to have a minimal impact on the secondary, tertiary and quaternary structure of the enzyme. DF0159.2g contained a SNP leading to an AA substitution (I300V) in the R2 loop region, which is part of the catalytic site of AmpC and substitutions in this region may result in conformational changes and flexibility in the hydrolysis spectrum of AmpC (Santiago et al., 2018). Due to the combination of AA substitutions in DF0159.2g in both the omega and R2 loops, we hypothesise that this isolate is potentially an ESAC producer. ESAC production could not be inferred for isolate DF0102.4g due to the absence of AA substitutions in the R2 loop region (Table 2), however, this isolate had AA substitutions (E140D, N201T, N260T, and W370C), which to the best of our knowledge, have not previously been reported (Haenni et al., 2014a; Santiago et al., 2018; Burgess et al., 2021b) and the role these may have on the AmpC β-lactamase spectrum of activity are unknown.
E. coli DF0102.4g underwent whole genome sequencing using both Illumina and MinION sequencing methods and carried a plasmid which harboured the blaOXA–1, sul1, and aadA1 genes as well as partial copies of the aadA1 and catB4 gene (Table 4). The blaOXA–1 gene encodes a narrow-spectrum β-lactamase which traditionally confers resistance to aminopenicillins, carboxypenicillins and ureidopenicllins (Torres et al., 2015). However, overexpression of this gene has been shown to confer reduced susceptibility to 4GCs such as cefepime and susceptibility to cefotaxime and ceftazidime when coupled with porin loss (OmpC and/or OmpF) (Beceiro et al., 2011). Three AmpC-producing E. coli, with mutations in the promoter region of the ampC gene, isolated from United Kingdom dairy farms also carried the blaOXA–1 gene and showed reduced susceptibility to cefepime (Alzayn et al., 2020). Possible resistance mechanisms of E. coli DF0102.4g to 3GC and 4GC warranting further investigation include the overexpression of the blaOXA–1 gene coupled with porin loss assessed using expression studies and proteomics, respectively or modification and mutations in other genes such as the mar genes.
Plasmids play a major role in the dissemination of AMR in Enterobacteriaceae (Rozwandowicz et al., 2018). The detection of ARGs on plasmids is of particular concern due to the potential for dissemination within bacterial populations, particularly to pathogenic bacteria. ESBL genes are often encoded on plasmids that carry several ARGs, resulting in a multi-drug resistant phenotype (Ho et al., 2013; McGann et al., 2016). Isolate DF0059.2e (ST5135) harboured three plasmids, two of which encoded ARGs including pDF0059.2e_2 which carried the blaCMY–2 gene. pDF0059.2e_1 belonged to the IncY group, which are prophages that autonomously replicate and have been reported to be co-associated with other plasmid types including IncF and/or IncI (Rozwandowicz et al., 2018), as was seen in this isolate. Although no ARGs were encoded on plasmid pDF0059.2e_1, IncY plasmids have been reported to carry resistance genes including blaCTX–M–15 in K. pneumoniae (Ribeiro et al., 2016) and mcr-1 in an E. coli isolated from a pig farm in China (Chunping et al., 2021). The third plasmid, pDF0059.2e_3, harboured 12 ARGs including blaCTX–M–1 which were all physically linked. Detection of mobile genetic elements surrounding the ARGs suggests that this plasmid has undergone recombination events, potentially contributing to the development of a multi-drug resistant plasmid in this isolate.
Plasmid associated β-lactamase gene types have been shown to have a highly homologous genetic environment, regardless of source (Lee et al., 2020). In this study, five strains harboured highly similar IncI1 plasmids which encoded the blaCMY–2 gene. IncI plasmids are conjugative, have a low copy number and a narrow host range (Rozwandowicz et al., 2018). A high proportion of ESBL and AmpC genetic determinants have also been found on IncI1 plasmids in other studies (Tello et al., 2020; Ewers et al., 2021), with IncI1 plasmids being frequently identified from extended-spectrum cephalosporin resistant E. coli isolated from healthy livestock in the Netherlands over a 10 years period (Ceccarelli et al., 2019). However, the plasmid and ESBL/AmpC gene combinations in E. coli isolated from dairy cattle and veal calves was more variable compared to the plasmid and gene combinations identified in E. coli isolated from other livestock sectors (Ceccarelli et al., 2019). A diverse range of STs and phylogenetic groups were detected among 3GC resistant Escherichia spp. isolated from healthy cattle, pigs and chickens. This suggested that the clonal spread of single lineages is unlikely to have occurred (Ewers et al., 2021). Together, these data and this study highlight the importance of horizontal plasmid transfer in the dissemination of 3GC resistance among Enterobacteriaceae.
This study assessed the sample level prevalence of ESBL- and AmpC-producing E. coli on two NZ dairy farms over a 15-month period. No ESBL or AmpC-producing E. coli were isolated from bulk tank milk or soil from recently grazed paddocks in this study, suggesting they are less likely reservoirs in the NZ dairy farm environment compared with faeces and FDE. ESBL-producing E. coli were infrequently identified during the study, suggesting they are present at a low prevalence in these two dairy farms. Plasmid-mediated AmpC-producing E. coli were isolated at a low prevalence in faeces and FDE but were isolated at a higher frequency on the larger farm (Dairy 4) compared with Dairy 1. In contrast, AmpC-producing E. coli with mutations in the promoter region of the chromosomal ampC gene were detected at a higher prevalence, particularly in FDE but only on Dairy 1. The detection of AmpC-producing E. coli with mutations in the promoter region of the ampC gene is less concerning in regard to the dissemination of AMR within bacterial populations. These findings highlight the necessity to confirm the genotype of an AmpC phenotype in E. coli.
Both ESBL- and AmpC-producing E. coli were isolated at various time points throughout the 15-month study period and their detection was not associated with periods of elevated AMU on NZ dairy farms. Dairy 1 had higher AMU (mg/PCU) than Dairy 4 during this study period, however, both farms used a low proportion of 3GC and 4GCs which are known risk factors for ESBL-producing Enterobacteriaceae. In addition, the farms used low levels of antimicrobials classed as red tier by the NZVA and these results support the hypothesis that low AMU on NZ dairy farms influences the prevalence of AMR in the dairy farm environment. Additional studies should focus on high-risk animals such as young calves and a larger number of dairy farms throughout NZ, including farms with known risk factors or management practices which may influence AMR such as high use of 3GC and 4GCs, an open herd system, feeding waste milk to calves and the use of blanket dry cow therapy.
The BioProject PRJNA844174 and BioSample accession numbers have been included in Supplementary Table 5.
RC, PB, SB, AM, GB, and AC involved in the project conceptualisation. RC, AC, and GB acquired project funding. RC, PB, SB, AM, and AC conducted investigation, formal analysis, and methodology development. AC and GB involved in the project administration. RC, PB, SB, AM, GB, and AC wrote the manuscript. All authors contributed to the manuscript revision, read, and approved the submitted version.
This work was funded by the AgResearch Strategic Science Investment funding (Food Integrity programme), the New Zealand Food Safety Science and Research Centre and the School of Veterinary Science, Massey University Postgraduate research fund. RC was the recipient of a New Zealand Food Safety Science and Research Centre Ph.D. scholarship.
We gratefully acknowledge Hamish Doohan, Joshua Mitchell, Jolanda Amoore, and Fiona Sharland from the Massey University Research Farms for their assistance with the sample collection and the on-farm components of the research; Xiaoxiao Lin (Massey University Genome Service) for his sequencing advice; Ahmed Fayaz (Massey University) for the database development and maintenance, and Scott McDougall (Anexa FVC) for his study design advice. This publication is based on a research chapter from RC’s Ph.D. thesis (Collis, 2022).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.960748/full#supplementary-material
Afema, J. A., Ahmed, S., Besser, T. E., Jones, L. P., Sischo, W. M., and Davis, M. A. (2018). Molecular epidemiology of dairy cattle-associated Escherichia coli carrying bla(CTX–M) genes in Washington state. Appl. Environ. Microbiol. 84:e02430-17. doi: 10.1128/AEM.02430-17
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/s0022-2836(05)80360-2
Alzayn, M., Findlay, J., Schubert, H., Mounsey, O., Gould, V. C., Heesom, K. J., et al. (2020). Characterization of AmpC-hyperproducing Escherichia coli from humans and dairy farms collected in parallel in the same geographical region. J. Antimicrob. Chemother. 75, 2471–2479. doi: 10.1093/jac/dkaa207
Anonymous. (2018). Antibiotic Judicious Use Guidelines For The New Zealand Veterinary Profession: Dairy. Wellington: Anonymous.
Banerjee, R., and Johnson, J. R. (2014). A new clone sweeps clean: The enigmatic emergence of Escherichia coli sequence type 131. Antimicrob. Agents Chemother. 58, 4997–5004. doi: 10.1128/AAC.02824-14
Beceiro, A., Maharjan, S., Gaulton, T., Doumith, M., Soares, N. C., Dhanji, H., et al. (2011). False extended-spectrum β-lactamase phenotype in clinical isolates of Escherichia coli associated with increased expression of OXA-1 or TEM-1 penicillinases and loss of porins. J. Antimicrob. Chemother. 66, 2006–2010. doi: 10.1093/jac/dkr265
Bevan, E. R., Jones, A. M., and Hawkey, P. M. (2017). Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Biomatters Geneious Prime (2022). Geneious prime. Available Online at: https://www.geneious.com/
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Brouwer, M. S. M., Jurburg, S. D., Harders, F., Kant, A., Mevius, D. J., Roberts, A. P., et al. (2019). The shufflon of IncI1 plasmids is rearranged constantly during different growth conditions. Plasmid 102, 51–55. doi: 10.1016/j.plasmid.2019.03.003
Burgess, S. A., Aplin, J., Biggs, P. J., Breckell, G., Benschop, J., Fayaz, A., et al. (2021a). Characterisation of AmpC and extended-spectrum beta-lactamase producing E. coli from New Zealand dairy farms. Int. Dairy J. 117:104998. doi: 10.1016/j.idairyj.2021.104998
Burgess, S. A., Cookson, A. L., Brousse, L., Ortolani, E., Benschop, J., Akhter, R., et al. (2021b). The epidemiology of AmpC-producing Escherichia coli isolated from dairy cattle faeces on pasture-fed farms. J. Med. Microbiol. 70:001447. doi: 10.1099/jmm.0.001447
Burgess, S. A., Francois, M., Midwinter, A. C., Biggs, P. J., and Rasko, D. (2021c). Draft genome sequences of seven extended-spectrum β-lactamase-producing Escherichia coli strains isolated from New Zealand waterways. Microbiol. Resour. Announc. 10:e01445–20. doi: 10.1128/MRA.01445-20
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 9, 5825–5829. doi: 10.1093/molbev/msab293
Canton, R., Novais, A., Valverde, A., Machado, E., Peixe, L., Baquero, F., et al. (2008). Prevalence and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 14, 144–153. doi: 10.1111/j.1469-0691.2007.01850.x
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Larsen, M. V., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/aac.02412-14
Caroff, N., Espaze, E., Bérard, I., Richet, H., and Reynaud, A. (1999). Mutations in the ampC promoter of Escherichia coli isolates resistant to oxyiminocephalosporins without extended spectrum beta-lactamase production. FEMS Microbiol. Lett. 173, 459–465. doi: 10.1111/j.1574-6968.1999.tb13539.x
Caroff, N., Espaze, E., Gautreau, D., Richet, H., and Reynaud, A. (2000). Analysis of the effects of –42 and –32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing AmpC. J. Antimicrob. Chemother. 45, 783–788. doi: 10.1093/jac/45.6.783
Ceccarelli, D., Kant, A., van Essen-Zandbergen, A., Dierikx, C., Hordijk, J., Wit, B., et al. (2019). Diversity of plasmids and genes encoding resistance to extended spectrum cephalosporins in commensal Escherichia coli from Dutch livestock in 2007–2017. Front. Microbiol. 10:76. doi: 10.3389/fmicb.2019.00076
Center for Genomic Epidemiology (2011). Overview of Services. Available Online at: http://www.genomicepidemiology.org/ (accessed May 23, 2022).
Chen, J., Li, J., Zhang, H., Shi, W., and Liu, Y. (2019). Bacterial heavy-metal and antibiotic resistance genes in a copper tailing dam area in Northern China. Front. Microbiol. 10:1916. doi: 10.3389/fmicb.2019.01916
Chunping, Z., Yuqing, F., Fei, L., Hui, J., Zhina, Q., Meng, L., et al. (2021). A phage-like IncY plasmid carrying the mcr-1 gene in Escherichia coli from a pig farm in China. Antimicrob. Agents Chemother. 61:e02035–16. doi: 10.1128/AAC.02035-16
Clermont, O., Bonacorsi, S., and Bingen, E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000
Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013). The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
Collignon, P. C., Conly, J. M., Andremont, A., McEwen, S. A., Aidara-Kane, A., and Group, W. H. O. A. (2016). World Health Organization ranking of antimicrobials according to their importance in human medicine: A critical step for developing risk management strategies to control antimicrobial resistance from food animal production. Clin. Infect. Dis. 63, 1087–1093. doi: 10.1093/cid/ciw475
Collis, R. M. (2022). Metagenomic Analysis And Culture-Based Methods To Examine The Prevalence And Distribution Of Antimicrobial Resistance On Two New Zealand Dairy Farms. Palmerston North: Massey University.
Collis, R. M., Burgess, S. A., Biggs, P. J., Midwinter, A. C., French, N. P., Toombs-Ruane, L., et al. (2019). Extended-spectrum beta-lactamase-producing Enterobacteriaceae in dairy farm environments: A New Zealand perspective. Foodborne Pathog. Dis. 16, 5–22. doi: 10.1089/fpd.2018.2524
Cormier, A., Zhang, P. L. C., Chalmers, G., Weese, J. S., Deckert, A., Mulvey, M., et al. (2019). Diversity of CTX-M-positive Escherichia coli recovered from animals in Canada. Vet. Microbiol. 231, 71–75. doi: 10.1016/j.vetmic.2019.02.031
CLSI (2019). Performance standards for antimicrobial susceptibility testing. 29 ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute.
Daehre, K., Projahn, M., Semmler, T., Roesler, U., and Friese, A. (2018). Extended-spectrum beta-lactamase-/AmpC beta-lactamase-producing Enterobacteriaceae in broiler farms: Transmission dynamics at farm level. Microb. Drug Resist. 24, 511–518. doi: 10.1089/mdr.2017.0150
Dahms, C., Hubner, N. O., Kossow, A., Mellmann, A., Dittmann, K., and Kramer, A. (2015). Occurrence of ESBL-producing Escherichia coli in livestock and farm workers in Mecklenburg-Western Pomerania, Germany. PLoS One 10:e0143326. doi: 10.1371/journal.pone.0143326
Day, M. J., Rodríguez, I., van Essen-Zandbergen, A., Dierikx, C., Kadlec, K., Schink, A.-K., et al. (2016). Diversity of STs, plasmids and ESBL genes among Escherichia coli from humans, animals and food in Germany, the Netherlands and the UK. J. Antimicrob. Chemother. 71, 1178–1182. doi: 10.1093/jac/dkv485
Dierikx, C. M., van Duijkeren, E., Schoormans, A. H. W., van Essen-Zandbergen, A., Veldman, K., Kant, A., et al. (2012). Occurrence and characteristics of extended-spectrum-β-lactamase- and AmpC-producing clinical isolates derived from companion animals and horses. J. Antimicrob. Chemother. 67, 1368–1374. doi: 10.1093/jac/dks049
Dohmen, W., Van Gompel, L., Schmitt, H., Liakopoulos, A., Heres, L., Urlings, B. A., et al. (2017). ESBL carriage in pig slaughterhouse workers is associated with occupational exposure. Epidemiol. Infect. 145, 2003–2010. doi: 10.1017/s0950268817000784
Ewers, C., Bethe, A., Semmler, T., Guenther, S., and Wieler, L. H. (2012). Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 18, 646–655. doi: 10.1111/j.1469-0691.2012.03850.x
Ewers, C., de Jong, A., Prenger-Berninghoff, E., El Garch, F., Leidner, U., Tiwari, S. K., et al. (2021). Genomic diversity and virulence potential of ESBL- and AmpC-β-Lactamase-producing Escherichia coli strains from healthy food animals across Europe. Front. Microbiol. 12:626774. doi: 10.3389/fmicb.2021.626774
Fonterra Co-Operative Group Limited (2018). “Fonterra farmers’ handbook 2018/2019”. Palmerston North: Fonterra.
Gazal, L. E. S., Medeiros, L. P., Dibo, M., Nishio, E. K., Koga, V. L., Gonçalves, B. C., et al. (2021). Detection of ESBL/AmpC-producing and fosfomycin-resistant Escherichia coli from different sources in poultry production in Southern Brazil. Front. Microbiol. 11:604544. doi: 10.3389/fmicb.2020.604544
Gelalcha, B. D., Ensermu, D. B., Agga, G. E., Vancuren, M., Gillespie, B. E., D’Souza, D. H., et al. (2022). Prevalence of antimicrobial resistant and extended-spectrum beta-Lactamase-producing Escherichia coli in dairy cattle farms in East Tennessee. Foodborne Pathog. Dis. 19, 408–416. doi: 10.1089/fpd.2021.0101
Geser, N., Stephan, R., and Hachler, H. (2012). Occurrence and characteristics of extended-spectrum beta-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet. Res. 8:21. doi: 10.1186/1746-6148-8-21
Gonggrijp, M. A., Santman-Berends, I. M. G. A., Heuvelink, A. E., Buter, G. J., van Schaik, G., Hage, J. J., et al. (2016). Prevalence and risk factors for extended-spectrum beta-lactamase- and AmpC-producing Escherichia coli in dairy farms. J. Dairy Sci. 99, 9001–9013. doi: 10.3168/jds.2016-11134
Haenni, M., Châtre, P., and Madec, J.-Y. (2014a). Emergence of Escherichia coli producing extended-spectrum AmpC β-lactamases (ESAC) in animals. Front. Microbiol. 5:53. doi: 10.3389/fmicb.2014.00053
Haenni, M., Chatre, P., Metayer, V., Bour, M., Signol, E., Madec, J. Y., et al. (2014b). Comparative prevalence and characterization of ESBL-producing Enterobacteriaceae in dominant versus subdominant enteric flora in veal calves at slaughterhouse, France. Vet. Microbiol. 171, 321–327. doi: 10.1016/j.vetmic.2014.02.023
Hall, M. B. (2022). Rasusa: Randomly subsample sequencing reads to a specified coverage. J. Open Source Softw. 7:3941. doi: 10.21105/joss.03941
Hamza, D., Dorgham, S., Ismael, E., El-Moez, S. I. A., Elhariri, M., Elhelw, R., et al. (2020). Emergence of β-lactamase- and carbapenemase- producing Enterobacteriaceae at integrated fish farms. Antimicrob. Resist. Infect. 9:67. doi: 10.1186/s13756-020-00736-3
Händel, N., Schuurmans, J. M., Feng, Y., Brul, S., and ter Kuile, B. H. (2014). Interaction between mutations and regulation of gene expression during development of de novo antibiotic resistance. Antimicrob. Agents Chemother. 58, 4371–4379. doi: 10.1128/AAC.02892-14
Heffernan, H., Wong, T. L., Lindsay, J., Bowen, B., and Woodhouse, R. (2011). A Baseline Survey Of Antimicrobial Resistance In Bacteria From Selected New Zealand Foods, 2009-2010. Christchurch: Institute of Environmental Science and Research.
Heuvelink, A. E., Gonggrijp, M. A., Buter, R. G. J., ter Bogt-Kappert, C. C., van Schaik, G., Velthuis, A. G. J., et al. (2019). Prevalence of extended-spectrum and AmpC beta-lactamase-producing Escherichia coli in Dutch dairy herds. Vet. Microbiol. 232, 58–64. doi: 10.1016/j.vetmic.2019.04.005
Hille, K., Ruddat, I., Schmid, A., Hering, J., Hartmann, M., von Münchhausen, C., et al. (2017). Cefotaxime-resistant E. coli in dairy and beef cattle farms- Joint analyses of two cross-sectional investigations in Germany. Prev. Vet. Med. 142, 39–45. doi: 10.1016/j.prevetmed.2017.05.003
Hillerton, J. E., Bryan, M. A., Beattie, B. H., Scott, D., Millar, A., and French, N. P. (2021). Use of antimicrobials for food animals in New Zealand: Updated estimates to identify a baseline to measure targeted reductions. N. Z. Vet. J. 69, 180–185. doi: 10.1080/00480169.2021.1890648
Hillerton, J. E., Irvine, C. R., Bryan, M. A., Scott, D., and Merchant, S. C. (2017). Use of antimicrobials for animals in New Zealand, and in comparison with other countries. N. Z. Vet. J. 65, 71–77. doi: 10.1080/00480169.2016.1171736
Ho, P. L., Chan, J., Lo, W. U., Law, P. Y., Li, Z., Lai, E. L., et al. (2013). Dissemination of plasmid-mediated fosfomycin resistance fosA3 among multidrug-resistant Escherichia coli from livestock and other animals. J. Appl. Microbiol. 114, 695–702. doi: 10.1111/jam.12099
Holmes, A. H., Moore, L. S. P., Sundsfjord, A., Steinbakk, M., Regmi, S., Karkey, A., et al. (2016). Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387, 176–187. doi: 10.1016/s0140-6736(15)00473-0
Hordijk, J., Fischer, E. A. J., van Werven, T., Sietsma, S., Van Gompel, L., Timmerman, A. J., et al. (2019). Dynamics of faecal shedding of ESBL- or AmpC-producing Escherichia coli on dairy farms. J. Antimicrob. Chemother. 74, 1531–1538. doi: 10.1093/jac/dkz035
Hordijk, J., Wagenaar, J. A., Kant, A., van Essen-Zandbergen, A., Dierikx, C., Veldman, K., et al. (2013). Cross-sectional study on prevalence and molecular characteristics of plasmid mediated ESBL/AmpC-producing Escherichia coli isolated from veal calves at slaughter. PLoS One 8:e65681. doi: 10.1371/journal.pone.0065681
Huerta-Cepas, J., Forslund, K., Coelho, L. P., Szklarczyk, D., Jensen, L. J., von Mering, C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucl. Acids Res. 47:D309–D314. doi: 10.1093/nar/gky1085
Hutchinson, H., Finney, S., Munoz-Vargas, L., Feicht, S., Masterson, M., and Habing, G. (2017). Prevalence and transmission of antimicrobial resistance in a vertically integrated veal calf production system. Foodborne Pathog. Dis. 14, 711–718. doi: 10.1089/fpd.2017.2310
Jafari, A., Falahatkar, S., Delpasand, K., Sabati, H., and Sedigh Ebrahim-Saraie, H. (2020). Emergence of Escherichia coli ST131 causing urinary tract infection in Western Asia: A systematic review and meta-analysis. Microb. Drug Resist. 26, 1357–1364. doi: 10.1089/mdr.2019.0312
Joensen, K. G., Tetzschner, A. M. M., Iguchi, A., Aarestrup, F. M., and Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/jcm.00008-15
Johnson, T. J., Wannemuehler, Y., Doetkott, C., Johnson, S. J., Rosenberger, S. C., and Nolan, L. K. (2008). Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J. Clin. Microbiol. 46, 3987–3996. doi: 10.1128/JCM.00816-08
Jolley, K. A., and Maiden, M. C. J. (2010). BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 11:595. doi: 10.1186/1471-2105-11-595
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Knapp, C. W., McCluskey, S. M., Singh, B. K., Campbell, C. D., Hudson, G., and Graham, D. W. (2011). Antibiotic resistance gene abundances correlate with metal and geochemical conditions in archived Scottish soils. PLoS One 6:e27300. doi: 10.1371/journal.pone.0027300
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B., and Stamatakis, A. (2019). RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455. doi: 10.1093/bioinformatics/btz305
Lacy-Hulbert, S., Kolver, E., Williamson, J., and Napper, A. (2002). “Incidence of mastitis among cows of different genotypes in differing nutritional environments,” in Proceedings of New Zealand Society of Animal Production, (Wellington: New Zealand Society of Animal Production), 24–29. doi: 10.3168/jds.2020-18782
Laxminarayan, R., Duse, A., Wattal, C., Zaidi, A. K. M., Wertheim, H. F. L., Sumpradit, N., et al. (2013). Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 13, 1057–1098. doi: 10.1016/s1473-3099(13)70318-9
Lee, S., Teng, L., DiLorenzo, N., Weppelmann, T. A., and Jeong, K. C. (2020). Prevalence and molecular characteristics of extended-spectrum and AmpC β-Lactamase producing Escherichia coli in grazing beef cattle. Front. Microbiol. 10:3076. doi: 10.3389/fmicb.2019.03076
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucl. Acids Res. 44, 242–245. doi: 10.1093/nar/gkw290
Lévesque, S., Dufresne, P. J., Soualhine, H., Domingo, M. C., Bekal, S., Lefebvre, B., et al. (2015). A side by side comparison of Bruker Biotyper and VITEK MS: Utility of MALDI-TOF MS technology for microorganism identification in a public health reference laboratory. PLoS One 10:e0144878. doi: 10.1371/journal.pone.0144878
Liu, J., Zhao, Z., Avillan, J. J., Call, D. R., Davis, M., Sischo, W. M., et al. (2019). Dairy farm soil presents distinct microbiota and varied prevalence of antibiotic resistance across housing areas. Environ. Pollut. 254:113058. doi: 10.1016/j.envpol.2019.113058
Livermore, D. M., Canton, R., Gniadkowski, M., Nordmann, P., Rossolini, G. M., Arlet, G., et al. (2006). CTX-M: Changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59, 165–174. doi: 10.1093/jac/dkl483
Luo, H., and Gao, F. (2019). DoriC 10.0: An updated database of replication origins in prokaryotic genomes including chromosomes and plasmids. Nucl. Acids Res. 47:D74–D77. doi: 10.1093/nar/gky1014
Mammeri, H., Poirel, L., Fortineau, N., and Nordmann, P. (2006). Naturally occurring extended-spectrum cephalosporinases in Escherichia coli. Antimicrob. Agents Chemother. 50, 2573–2576. doi: 10.1128/AAC.01633-05
Markland, S., Weppelmann, T. A., Ma, Z., Lee, S., Mir, R. A., Teng, L., et al. (2019). High prevalence of cefotaxime resistant bacteria in grazing beef cattle: A cross sectional study. Front. Microbiol. 10:176. doi: 10.3389/fmicb.2019.00176
Massé, J., Lardé, H., Fairbrother, J. M., Roy, J. P., Francoz, D., Dufour, S., et al. (2021). Prevalence of antimicrobial resistance and characteristics of Escherichia coli isolates from fecal and manure pit samples on dairy farms in the province of Québec, Canada. Front. Vet. Sci. 8:654125. doi: 10.3389/fvets.2021.654125
McGann, P., Snesrud, E., Maybank, R., Corey, B., Ong, A. C., Clifford, R., et al. (2016). Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: First report of mcr-1 in the United States. Antimicrob. Agents Chemother. 60, 4420–4421. doi: 10.1128/aac.01103-16
Meini, S., Tascini, C., Cei, M., Sozio, E., and Rossolini, G. M. (2019). AmpC β-lactamase-producing Enterobacterales: What a clinician should know. Infection 47, 363–375. doi: 10.1007/s15010-019-01291-9
Minitab, I. (2019). Minitab 19 Statistical Software. Available Online at: https://www.minitab.com/en-us/ (accessed May 25, 2022).
Odenthal, S., Akineden, O., and Usleber, E. (2016). Extended-spectrum beta-lactamase producing Enterobacteriaceae in bulk tank milk from German dairy farms. Int. J. Food Microbiol. 238, 72–78. doi: 10.1016/j.ijfoodmicro.2016.08.036
Ohnishi, M., Okatani, A. T., Esaki, H., Harada, K., Sawada, T., Murakami, M., et al. (2013). Herd prevalence of Enterobacteriaceae producing CTX-M-type and CMY-2-lactamases among Japanese dairy farms. J. Appl. Microbiol. 115, 282–289. doi: 10.1111/jam.12211
Olsson, O., Bergström, S., and Normark, S. (1982). Identification of a novel ampC beta-lactamase promoter in a clinical isolate of Escherichia coli. EMBO J. 1, 1411–1416.
Oxford Nanopore Technologies, L. (2019). Qcat, Github. Available Online at: https://github.com/nanoporetech/qcat (accessed May 2, 2022).
Pérez-Pérez, F. J., and Hanson, N. D. (2002). Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40, 2153–2162. doi: 10.1128/jcm.40.6.2153-2162.2002
Petrovski, K. R., Heuer, C., Parkinson, T. J., and Williamson, N. B. (2009). The incidence and aetiology of clinical bovine mastitis on 14 farms in Northland, New Zealand. N. Z. Vet. J. 57, 109–115. doi: 10.1080/00480169.2009.36887
Pietsch, M., Irrgang, A., Roschanski, N., Brenner Michael, G., Hamprecht, A., Rieber, H., et al. (2018). Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genom. 19:601. doi: 10.1186/s12864-018-4976-3
Pires, J., Taracila, M., Bethel, C. R., Doi, Y., Kasraian, S., Tinguely, R., et al. (2015). In vivo evolution of CMY-2 to CMY-33 beta-lactamase in Escherichia coli sequence type 131: Characterization of an acquired extended-spectrum AmpC conferring resistance to cefepime. Antimicrob. Agents Chemother. 59, 7483–7488. doi: 10.1128/AAC.01804-15
Pitout, J. D. D. (2012). Extraintestinal pathogenic Escherichia coli: An update on antimicrobial resistance, laboratory diagnosis and treatment. Expert Rev. Anti Infect. Ther. 10, 1165–1176. doi: 10.1586/eri.12.110
Queenan, K., Häsler, B., and Rushton, J. (2016). A One Health approach to antimicrobial resistance surveillance: Is there a business case for it? Int. J. Antimicrob. Agents 48, 422–427. doi: 10.1016/j.ijantimicag.2016.06.014
Ribeiro, T. G., Novais, Â, Peixe, L., and Machado, E. (2016). Atypical epidemiology of CTX-M-15 among Enterobacteriaceae from a high diversity of non-clinical niches in Angola. J. Antimicrob. Chemother. 71, 1169–1173. doi: 10.1093/jac/dkv489
Robertson, J., and Nash, J. H. E. (2018). MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 4:e000206. doi: 10.1099/mgen.0.000206
Robertson, J., Bessonov, K., Schonfeld, J., and Nash, J. H. E. (2020). Universal whole-sequence-based plasmid typing and its utility to prediction of host range and epidemiological surveillance. Microb. Genom. 6:mgen000435. doi: 10.1099/mgen.0.000435
Robins-Browne, R. M., Holt, K. E., Ingle, D. J., Hocking, D. M., Yang, J., and Tauschek, M. (2016). Are Escherichia coli pathotypes still relevant in the era of whole-genome sequencing? Front. Cell. Infect. Microbiol. 6:141. doi: 10.3389/tcimb.2016.00141
Romero, J. L., Burgos, M. J. G., Perez-Pulido, R., Galvez, A., and Lucas, R. (2017). Resistance to antibiotics, biocides, preservatives and metals in bacteria isolated from seafoods: Co-Selection of strains resistant or tolerant to different classes of compounds. Front. Microbiol. 8:1650. doi: 10.3389/fmicb.2017.01650
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
Rubin, J. E., and Pitout, J. D. D. (2014). Extended-spectrum beta-lactamase, carbapenemase and AmpC producing Enterobacteriaceae in companion animals. Vet. Microbiol. 170, 10–18. doi: 10.1016/j.vetmic.2014.01.017
Santiago, G. S., Coelho, I. S., Bronzato, G. F., Moreira, A. B., Gonçalves, D., Alencar, T. A., et al. (2018). Short communication: Extended-spectrum AmpC–producing Escherichia coli from milk and feces in dairy farms in Brazil. J. Dairy Sci. 101, 7808–7811. doi: 10.3168/jds.2017-13658
Santman-Berends, I., Gonggrijp, M. A., Hage, J. J., Heuvelink, A. E., Velthuis, A., Lam, T., et al. (2017). Prevalence and risk factors for extended-spectrum beta-lactamase or AmpC-producing Escherichia coil in organic dairy herds in the Netherlands. J. Dairy Sci. 100, 562–571. doi: 10.3168/jds.2016-11839
Schmid, A., Hormansdorfer, S., Messelhausser, U., Kasbohrer, A., Sauter-Louis, C., and Mansfeld, R. (2013). Prevalence of extended-spectrum beta-lactamase-producing Escherichia coli on Bavarian dairy and beef cattle farms. Appl. Environ. Microbiol. 79, 3027–3032. doi: 10.1128/aem.00204-13
Schubert, H., Morley, K., Puddy, E. F., Arbon, R., Findlay, J., Mounsey, O., et al. (2021). Reduced antibacterial drug resistance and blaCTX–M β-Lactamase gene carriage in cattle-associated Escherichia coli at low temperatures, at sites dominated by older animals, and on pastureland: Implications for surveillance. Appl. Environ. Microbiol. 87:e01468–20. doi: 10.1128/AEM.01468-20
Seemann, T. (n.d.a). Abricate, Github. Available Online at: https://github.com/tseemann/abricate
Seemann, T. (n.d.b). Mlst, Github. Available Online at: https://github.com/tseemann/mlst
Seemann, T. (2014). Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Seemann, T. (2016). Snippy. Available Online at: https://github.com/tseemann/snippy/blob/master/README.md (accessed March 30, 2022).
Seemann, T., Goncalves, D. S. A., Bulach, D. M., Schultz, M. B., Kwong, J. C., and Howden, B. P. (n.d.). Nullarbor Github. Available Online at: https://github.com/tseemann/nullarbor
Siu, L. K., Lu, P.-L., Chen, J.-Y., Lin, F. M., and Chang, S.-C. (2003). High-level expression of AmpC beta-lactamase due to insertion of nucleotides between -10 and -35 promoter sequences in Escherichia coli clinical isolates: Cases not responsive to extended-spectrum-cephalosporin treatment. Antimicrob. Agents Chemother. 47, 2138–2144. doi: 10.1128/AAC.47.7.2138-2144.2003
Skockova, A., Bogdanovicova, K., Kolackova, I., and Karpiskova, R. (2015). Antimicrobial-resistant and extended-spectrum beta-lactamase–producing Escherichia coli in raw cow’s milk. J. Food Prot. 78, 72–77. doi: 10.4315/0362-028x.jfp-14-250
Souvorov, A., Agarwala, R., and Lipman, D. J. (2018). SKESA: Strategic k-mer extension for scrupulous assemblies. Genome Biol. 19, 153–153. doi: 10.1186/s13059-018-1540-z
Stohr, J. J. J. M., Kluytmans-van den Bergh, M. F. Q., Verhulst, C. J. M. M., Rossen, J. W. A., and Kluytmans, J. A. J. W. (2020). Development of amoxicillin resistance in Escherichia coli after exposure to remnants of a non-related phagemid-containing E. coli: An exploratory study. Antimicrob. Resist. Infect. Control 9:48. doi: 10.1186/s13756-020-00708-7
Sudarwanto, M., Akineden, O., Odenthal, S., Gross, M., and Usleber, W. (2015). Extended spectrum beta-lactamase (ESBL)-producing Klebsiella pneumoniae in bulk tank milk from dairy farms in Indonesia. Foodborne Pathog. Dis. 12, 585–590. doi: 10.1089/fpd.2014.1895
Sukupolvi, S., and O’Connor, C. D. (1990). TraT lipoprotein, a plasmid-specified mediator of interactions between gram-negative bacteria and their environment. Microbiol. Rev. 54, 331–341. doi: 10.1128/mr.54.4.331-341.1990
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/s1473-3099(17)30753-3
Tello, M., Ocejo, M., Oporto, B., and Hurtado, A. (2020). Prevalence of cefotaxime-resistant Escherichia coli isolates from healthy cattle and sheep in Northern Spain: Phenotypic and genome-based characterization of antimicrobial susceptibility. Appl. Environ. Microbiol. 86:e00742–20. doi: 10.1128/aem.00742-20
Thomas IV, J. C., Oladeinde, A., Kieran, T. J., Finger, J. W. Jr., Bayona-Vásquez, N. J., Cartee, J. C., et al. (2020). Co-occurrence of antibiotic, biocide, and heavy metal resistance genes in bacteria from metal and radionuclide contaminated soils at the Savannah River site. Microb. Biotechnol. 13, 1179–1200. doi: 10.1111/1751-7915.13578
Toombs-Ruane, L. J., Benschop, J., French, N. P., Biggs, P. J., Midwinter, A. C., Marshall, J. C., et al. (2021). Carriage of extended-spectrum-beta-lactamase- and AmpC beta-lactamase-producing Escherichia coli strains from humans and pets in the same households. Appl. Environ. Microbiol. 86:e01613–e01620. doi: 10.1128/AEM.01613-20
Torres, E., López-Cerero, L., Rodríguez-Martínez, J. M., and Pascual, Á (2015). Reduced susceptibility to cefepime in clinical isolates of Enterobacteriaceae producing OXA-1 beta-lactamase. Microb. Drug Resist. 22, 141–146. doi: 10.1089/mdr.2015.0122
Velasova, M., Smith, R. P., Lemma, F., Horton, R. A., Duggett, N. A., Evans, J., et al. (2019). Detection of extended-spectrum beta-lactam, AmpC and carbapenem resistance in Enterobacteriaceae in beef cattle in Great Britain in 2015. J. Appl. Microbiol. 126, 1081–1095. doi: 10.1111/jam.14211
Venturini, C., Bowring, B., Partridge, S. R., Zakour, N. L. B., Fajardo-Lubian, A., Ayala, A. L., et al. (2022). Co-Occurrence of multidrug resistant Klebsiella pneumoniae pathogenic clones of human relevance in an equine pneumonia case. Microbiol. Spectr. 10:e0215821. doi: 10.1128/spectrum.02158-21
von Salviati, C., Laube, H., Guerra, B., Roesler, U., and Friese, A. (2015). Emission of ESBL/AmpC-producing Escherichia coli from pig fattening farms to surrounding areas. Vet. Microbiol. 175, 77–84. doi: 10.1016/j.vetmic.2014.10.010
von Tippelskirch, P., Golz, G., Projahn, M., Daehre, K., Friese, A., Roesler, U., et al. (2018). Prevalence and quantitative analysis of ESBL and AmpC beta-lactamase producing Enterobacteriaceae in broiler chicken during slaughter in Germany. Int. J. Food Microbiol. 281, 82–89. doi: 10.1016/j.ijfoodmicro.2018.05.022
Watson, E., Jeckel, S., Snow, L., Stubbs, R., Teale, C., Wearing, H., et al. (2012). Epidemiology of extended spectrum beta-lactamase E. coli (CTX-M-15) on a commercial dairy farm. Vet. Microbiol. 154, 339–346. doi: 10.1016/j.vetmic.2011.07.020
Wick, R. (2018b). Porechop, Github. Available Online at: https://github.com/rrwick/Porechop (accessed April 4, 2022).
Wick, R. (2018a). Filtlong, Github. Available Online at: https://github.com/rrwick/Filtlong (accessed April 4, 2022).
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wood, D. E., and Salzberg, S. L. (2014). Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15:R46. doi: 10.1186/gb-2014-15-3-r46
World Health Organization (2011). Critically Important Antimicrobials For Human Medicine. Geneva: World Health Organization.
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zhou, Z., Alikhan, N. F., and Mohamed, K. The Agama Study Group Achtman, M. (2020), “The enterobase user s guide, with case studies on salmonella transmissions, Yersinia pestis phylogeny and escherichia core genomic diversity”. Genome Res. 30: 138–152. doi: 10.1101/gr.251678.119
Keywords: Escherichia coli, antimicrobial resistance, AmpC, ESBL, dairy, third-generation cephalosporin resistance, genomics
Citation: Collis RM, Biggs PJ, Burgess SA, Midwinter AC, Brightwell G and Cookson AL (2022) Prevalence and distribution of extended-spectrum β-lactamase and AmpC-producing Escherichia coli in two New Zealand dairy farm environments. Front. Microbiol. 13:960748. doi: 10.3389/fmicb.2022.960748
Received: 03 June 2022; Accepted: 25 July 2022;
Published: 11 August 2022.
Edited by:
Cindy Shuan Ju Teh, University of Malaya, MalaysiaReviewed by:
Marisa Haenni, Agence Nationale de Sécurité Sanitaire de l’Alimentation, de l’Environnement et du Travail (ANSES), FranceCopyright © 2022 Collis, Biggs, Burgess, Midwinter, Brightwell and Cookson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adrian L. Cookson, QWRyaWFuLkNvb2tzb25AYWdyZXNlYXJjaC5jby5ueg==; Rose M. Collis, Um9zZS5Db2xsaXNAYWdyZXNlYXJjaC5jby5ueg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.