- 1Faculty of Biology, University of Bucharest, Bucharest, Romania
- 2Life, Environmental and Earth Sciences Division, Research Institute of the University of Bucharest, Bucharest, Romania
- 3The Romanian Academy, Bucharest, Romania
- 4Academy of Romanian Scientists, Bucharest, Romania
- 5Faculty of Medicine and Biological Sciences, Stefan cel Mare University of Suceava, Suceava, Romania
- 6Suceava Emergency County Hospital, Suceava, Romania
- 7Institute of Biology, Bucharest, Romania
- 8Stefan S. Nicolau Institute of Virology, Bucharest, Romania
- 9Romanian Academy of Scientists, Bucharest, Romania
- 10Bacteriology Branch, Veterinary Sciences Division, Agri-Food and Biosciences Institute, Belfast, United Kingdom
- 11Faculty of Bioengineering of Animal Resources, Banat University of Agricultural Sciences and Veterinary Medicine—King Michael I of Romania, Timisoara, Romania
Antimicrobial and anticancer drug resistance represent two of the main global challenges for the public health, requiring immediate practical solutions. In line with this, we need a better understanding of the origins of drug resistance in prokaryotic and eukaryotic cells and the evolutionary processes leading to the occurrence of adaptive phenotypes in response to the selective pressure of therapeutic agents. The purpose of this paper is to present some of the analogies between the antimicrobial and anticancer drug resistance. Antimicrobial and anticancer drugs share common targets and mechanisms of action as well as similar mechanisms of resistance (e.g., increased drug efflux, drug inactivation, target alteration, persister cells’ selection, protection of bacterial communities/malignant tissue by an extracellular matrix, etc.). Both individual and collective stress responses triggered by the chemotherapeutic agent involving complex intercellular communication processes, as well as with the surrounding microenvironment, will be considered. The common themes in antimicrobial and anticancer drug resistance recommend the utility of bacterial experimental models for unraveling the mechanisms that facilitate the evolution and adaptation of malignant cells to antineoplastic drugs.
Introduction
The emerging and accelerated evolution of drug resistance in microbial and tumor cells share many analogies such as common targets (topoisomerase II) and mechanisms of action; availability of molecules with dual antimicrobial and antineoplastic activity (e.g., antineoplastic antibiotics); similar mechanisms of resistance; common selective drivers for resistance; dual resistance, respectively, intrinsic (failure to obtain an initial response to drugs) and acquired (involving specific and gradual genetic and epigenetic mechanisms); and existence of complex interactions between the gut microbiota and the drugs (Lambert et al., 2011). We will enumerate below some of the most well-known examples of similarities between antimicrobial and anticancer drugs resistance, at single cell and population level, that will be further detailed in dedicated sections of this review.
Single-cell processes involved in antimicrobial and anticancer drugs resistance
One of the best-known examples of multi-drug resistance (MDR) mechanisms shared by bacterial and malignant cells are the efflux pumps (Spratt, 1994; Davies and Davies, 2010). Efflux pump activation is responsible for therapeutic failure in solid and hematological malignancies as well as in many bacterial infections (Sissi and Palumbo, 2003; Bisacchi and Hale, 2016). Another common feature is represented by the adaptation strategies to the stress induced by different microenvironments, which are triggered by the genetic instability of tumor cells and bacterial mutability, respectively. In this regard, stimulation of stress-induced DNA repairing (SOS) genes plays an important role in DNA repair in both prokaryotic and eukaryotic cells exposed to inhibitory agents. Bacterial resistance could occur not only by spontaneous mutations or horizontal gene transfer of resistance genes but also by adaptation through prolonged exposure to sublethal doses (Andersson and Hughes, 2014). It is very likely that the same adaptive mechanism is at work in neoplastic cells.
Collective responses involved in antimicrobial and anticancer drugs resistance
Besides single-cell processes, adaptation at population level has been shown to be involved in resistance to both antimicrobial and antineoplastic agents. The collective stress response triggered by the chemotherapeutic agent involves complex intercellular communication processes, as well as with the surrounding microenvironment (Stewart and Costerton, 2001; Butler et al., 2010). For example, in the absence of nutrients, both microbial and tumor cells adapt to environmental conditions by metabolically shifting to a persistent phenotype in which cells do not grow and divide. In the presence of antibiotics, bacterial cells try to avoid the drugs by flagellar movement to a region of low concentration of the cytocidal agent and by forming a biofilm, which limits access of the drug (Stewart and Costerton, 2001; Butler et al., 2010). The tumor cells avoid the antineoplastic drugs by metastasis or by establishing an altered microenvironment through vascularization (Ahmed et al., 2010; Lambert et al., 2011). The stroma and extracellular matrix (ECM) of neoplastic cells and the microbial biofilm exopolymeric matrix limit the rate of O2 and nutrient diffusion and protect the cellular communities.
Interaction of antimicrobial and anticancer drugs with gut microbiota
The gut microbiome plays an important role in modulating the efficacy and toxicity of antibiotics and antitumoral agents, thus representing an attractive target for improving drug safety and efficacy, by manipulating its composition. However, both antibacterial and anticancer treatments induce perturbations in the normal microbiota (also known as dysbiosis). Dysbiosis is not only associated with diarrhea and fungal infections, it could have a role in neoplastic etiology and cancer risk, influence the efficacy of chemotherapy, radiotherapy, and immunotherapy, and also promote the emergence of antimicrobial and antitumoral drug resistance (Alexander et al., 2017; Lazar et al., 2019; Pinato et al., 2019; Cheng et al., 2020). Antibiotics with broad spectrum have been shown to induce numerical changes in 3% of bacterial species in the gut microbiota and the occurred dysbiosis further alters the therapeutic efficacy of certain antitumor antibiotics and increases their toxicity (Lofmark et al., 2006; Jernberg et al., 2007; Andersson and Hughes, 2014; Francino, 2015). On the other side, antineoplastic chemotherapy indirectly amplifies dysbiosis and has profound effects, mainly on the intestinal epithelium, influencing cancer progression, treatment efficacy, and toxicity.
All these functional analogies and complex interactions can generate common therapeutic strategies. Therefore, the purpose of this paper is to present some of the analogies between the antimicrobial and anticancer drug resistance, starting with the presentation of the common targets and mechanisms of action and of the antineoplastic antibiotics. Both single-cell and collective mechanisms of resistance in bacterial communities and malignant tissues will be presented.
Antimicrobial and anticancer drug resistance: The dimension of the challenge for the public health
Nowadays, cancer and infectious diseases are two of the most problematic and common diseases, exhibiting a great impact on the health status of the general population. According to the World Health Organization (WHO), cancer is one of the leading mortality causes worldwide, with nearly 10 million deaths recorded in 2020.1 The most common neoplasia, also accounting for the highest mortality rates are, in alphabetical order, breast, colorectal, liver, lung, prostate, skin, and stomach cancers (Ferlay et al., 2021). The current therapeutic approaches in cancer include surgery and chemotherapy as first options, followed by radiotherapy, immunotherapy, and targeted therapy (Kanady et al., 1997). Based on experimental tumor induction and propagation models, it is estimated that in neoplasms containing 103–106 cells, the chemotherapeutic treatment can select at least one clone of resistant cells, which can survive and continue to proliferate, leading to tumor recurrence or metastasis formation. The spontaneous mutation rate is directly proportional with the metastatic potential, being 3- to 7-fold higher in case of tumorigenic clones with high metastatic potential (Cifone and Fidler, 1981). Thus, the emergence of resistance to antitumoral drugs currently represents one of the major medical challenges.
Antimicrobial resistance (AMR) is one of the top ten global threats, not only for humans, but also for the environmental health, being considered a typical One Health problem.2 The most threatening bugs, exhibiting MDR, extended-drug and even pan-drug resistance phenotypes are grouped under the acronym ESCAPE (Enterococcus faecium, Staphylococcus aureus, Clostridium difficile, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacteriaceae; Peterson, 2009). The first 18 most fearful resistant microbes are classified by the Center for Disease Control and Prevention (CDC) as either urgent, serious, or concerning threats in the 2019 AR Threats Report (CDC’s Antibiotic Resistance Threats in the United States, 2019).
The antimicrobials crisis, beyond direct consequences related to increased mortality and morbidity rates caused by infectious diseases and huge economic losses, would compromise the success of modern medicine, including invasive diagnosis procedures (e.g., biopsy), surgery, and cancer radio- and chemotherapy. Regardless of the extent of the surgery, the risk of developing healthcare associated infections with resistant strains is very high, causing difficulties in making the most appropriate therapeutic decision. Also, radiotherapy and chemotherapy could induce a temporary immunodeficiency and thus increase the oncologic patient susceptibility to infections, including those produced by MDR bacteria3 (Ariza-Heredia and Chemaly, 2018). Fluoroquinolone and carbapenem resistance, nosocomial outbreaks of Gram-negative sepsis on cancer wards, are often caused by enteric bacteria, with mortality rates of 60%–84% (Papanicolas et al., 2018).
All these facts demonstrate that the continuously growing cancer burden is potentiated by the emergence and spread of AMR.
Common features in antimicrobial and antineoplastic agents’ structures, targets, and mechanisms of action
The currently available antibiotics act through: (a) inhibition of cell wall synthesis (beta-lactams); (b) alteration of cell surface structures (cell wall, outer membrane, cytoplasmic membrane; polypeptides, daptomycin), (c) inhibition of protein synthesis (aminoglycosides, tetracyclines, macrolides, lincosamides, streptogramins, oxazolidinones, chloramphenicol, etc.) (d) inhibition of DNA synthesis and transcription (quinolones, rifamycins); and inhibition of essential metabolites (trimethoprim-sulfamethoxazole; Mihaescu et al., 2008, 2009). The main mechanisms of action of the antitumoral drugs are (a) alteration of DNA structure by intercalation or ROS release, (b) inhibition of enzymes involved in replication and transcription, (c) inhibition of essential metabolites, and (d) inhibition of cellular growth by preventing the activation of certain proteins (Figure 1).
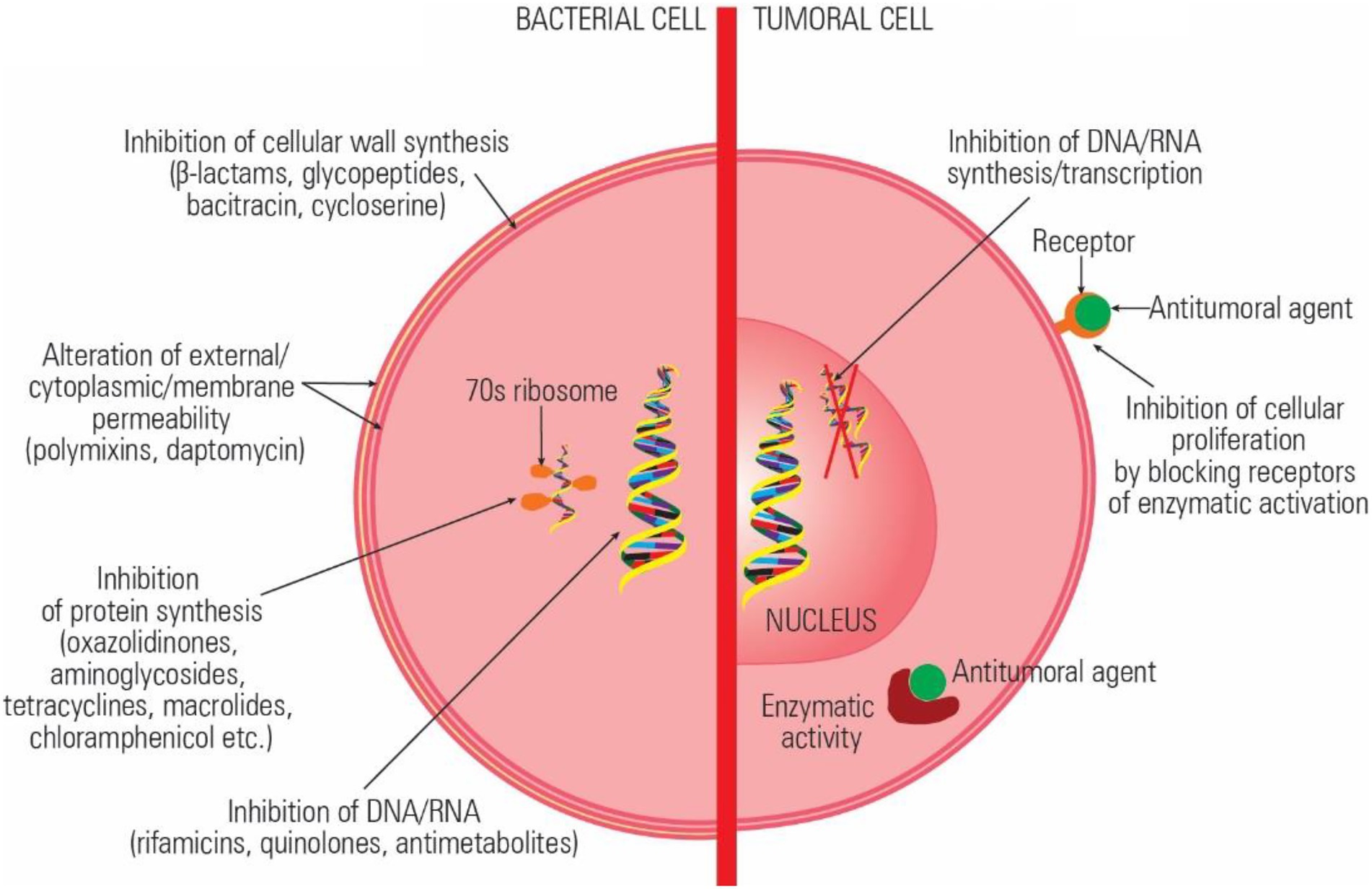
Figure 1. Comparative representation of the antibiotics and antitumoral agents’ mechanisms of action.
Some of the current antibiotics were reported to have the same target and similar mechanisms of action exhibiting both antimicrobial and antitumor activity (Gao et al., 2020). Antineoplastic antibiotics with major therapeutic impact are anthracyclines, peptides, and quinolones, which interact with topoisomerases both in prokaryotic and eukaryotic cells (Pommier et al., 2010). Anthracyclines react with topoisomerase DNA complex, stabilize it, and induce cell apoptosis (D’Arpa and Liu, 1989). Quinolones and aminocoumarin antibiotics target type II topoisomerases (gyrase) and topoisomerase IV, converting these enzymes into physiological poisons, both in tumoral and bacterial cells (Barrett, 1996; Salerno et al., 2010). Moreover, besides their direct antitumoral effects, quinolones proved to have immunomodulators activity and to stimulate the antineoplastic immune response (Dalhoff and Shalit, 2003).
These aspects suggest that the identification of inhibitory compounds with multiple targets (i.e., topoisomerases I and II and DNA repairing enzymes) could represent a promising lead to efficiently fight bacterial infections and cancer and to avoid emergence of bacterial and tumor cells resistance (Skok et al., 2020). Even a low level of inhibition for multiple bacterial and tumor cell functions would have a high cumulative effect and limit the risk of resistance development (Avner et al., 2012).
Common features in antimicrobial and antineoplastic agents’ resistance
The accelerated evolution of resistance of neoplastic cells to chemotherapeutic agents and of bacterial cells to antibiotics is the result of common mechanisms acting at individual (Figure 2) and population levels (Lambert et al., 2011). The current research is focused on the identification of these mechanisms in order to be able to develop efficient drugs to overcome them (Nikolaou et al., 2018).
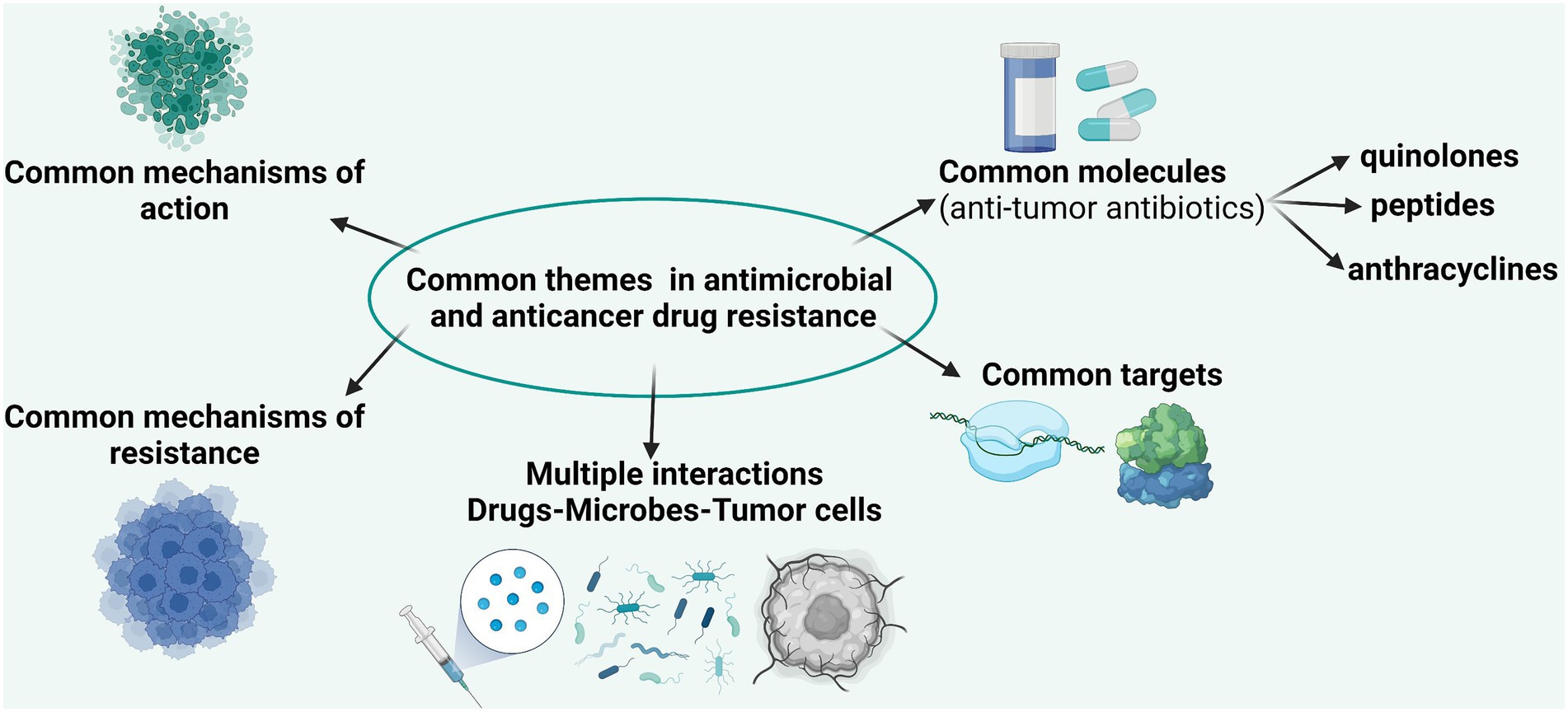
Figure 2. Comparative representation of the main targets, mechanisms of action, and mechanisms of resistance in bacterial and tumoral cells.
Single-cell resistance mechanisms
After surgical resection of the primary tumor, the next step in the therapeutic approach is chemotherapy. This can be given before and/or after the primary tumor is resected to inhibit the proliferation of malignant cells and induce their apoptosis, but its disadvantage is the selection of resistant clones. According to Goldie–Coldman, the clones of tumor resistant cells survive because of spontaneous mutations, which occur in 1/1,000,000 cells; thus, the probability of the emergence of tumor resistance is very high (considering that 1 g of tumor tissue contains 109 cells) (Goldie and Coldman, 1979, 1983, 1984). In bacteria, resistance is acquired either by spontaneous mutations or by horizontal gene transfer mediated by phage transduction, transformation with external DNA, and resistance plasmid transfer during conjugation.
Malignant cells could develop resistance through one/some of the following biochemical mechanisms: (a) inhibition of drug distribution through circulation or blood–brain barrier, (b) inhibition of intracellular accumulation, (c) inhibition of interaction between the chemotherapeutic agent and target molecules (i.e., proteins and nucleic acids), (d) transformation of the active agent or of its specific target, (e) enzymatic or chemical drug inactivation, and (f) removal of the chemotherapeutic agent from the cells via efflux pumps.
In bacteria, the biochemical mechanisms of resistance include (a) decreased porin permeability, (b) active efflux of the antibiotic, (c) target alteration, and (d) chemical modification or inactivation of antibiotics (Patel et al., 2021). Thus, one of the most important resistance mechanisms present in both bacteria and tumor cells is represented by the activation/hyperactivation of efflux pumps. In neoplastic cells, the overexpression of P-glycoprotein occurred through the amplification of the encoding gene is leading to MDR to hydrophobic compounds: (a) anthracyclines (doxorubicin, daunorubicin, epirubicin, and idarubicin); (b) aminoacridines (AMSA); (c) taxans (taxol, etc.), vinca alkaloids, and actinomycin D; and (d) mitomycin C and topoisomerase I inhibitors (Patel et al., 2021) In bacteria, there are at least four families of secondary transporters conferring MDR and assuring toxic compounds elimination (Mihaescu et al., 2008).
Bacterial biofilms versus tumoral tissue – The power of the collective
The ability to resist a chemotherapeutic treatment or to survive in stressful environments is the result of the ability of both bacterial and tumor cells to cooperate and elaborate a collective adaptive response (Figure 3).
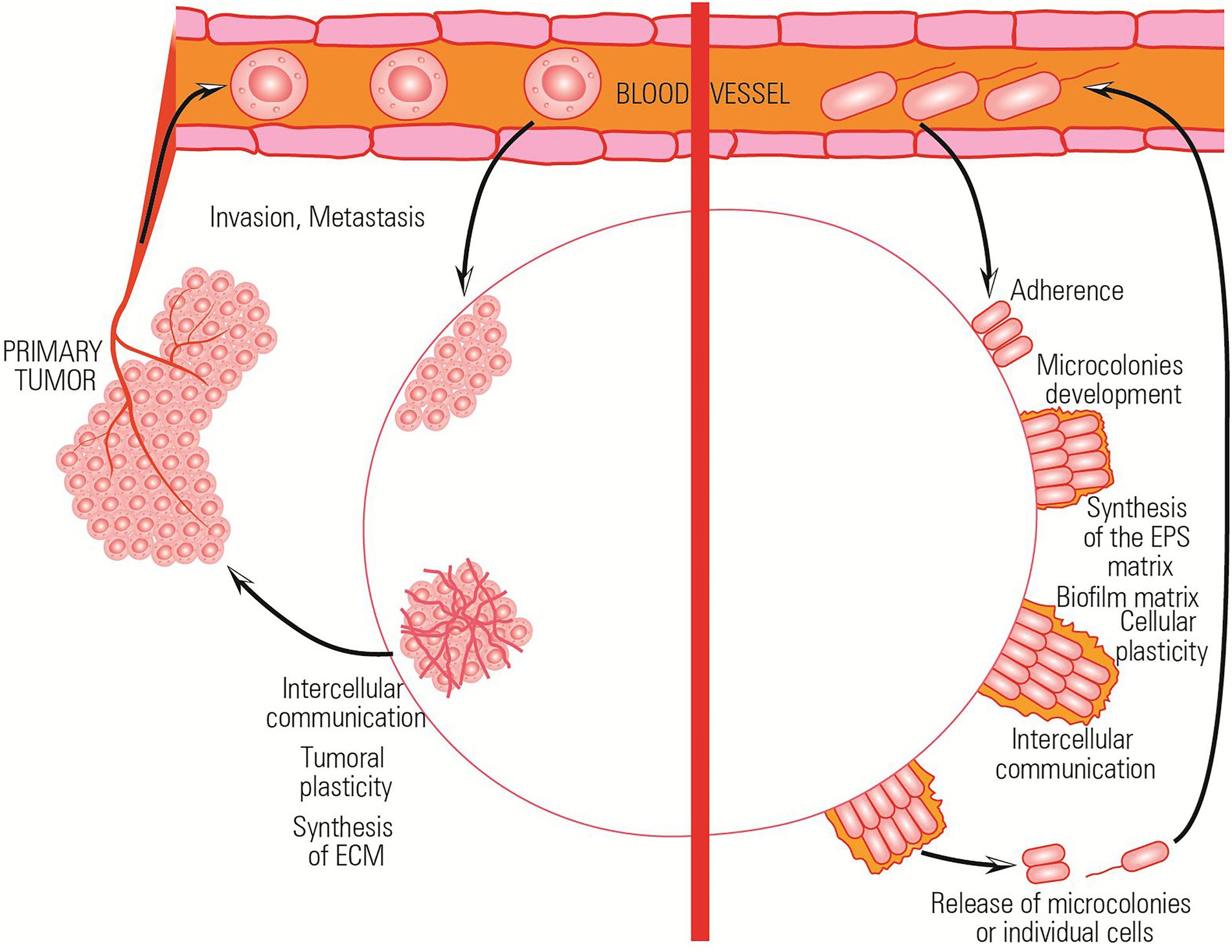
Figure 3. Comparative representation of the collective adaptative mechanisms exhibited by bacterial and tumor cells in stressful environments.
Both bacterial and tumor cells adapt to different microenvironmental conditions by changing their phenotype and activating the stress response processes. For example, in the absence of nutrients, both bacterial and tumor cells adapt to environmental conditions by metabolically shifting to a persister phenotype in which cells do not grow and divide.
Also, both bacterial and tumor cells transiently increase their mutation rates (e.g., hypermutator bacterial cells) and can reduce DNA replication fidelity to foster diversity. This process is known as adaptive mutability and leads to accelerated evolution and adaptation. As a result, biofilm and tumor cells could develop MDR much more rapidly. However, the hypermutability state is maintained only in a small population fraction reducing the risk of detrimental or fatal mutations in the rest of the clonal population (Taddei et al., 1997; Macia et al., 2005).
Both bacterial and tumor cells adopt a running behavior (mediated by flagellar motility in case of bacteria and by metastasis process in case of tumoral cells) in the presence of stressor agents.
The bacterial communities/tumoral tissues isolate in citadels which are hard to penetrate by the antibiotics/antitumoral agents. Inside these isolated communities, a collective stress response, involving complex intercellular and with the surrounding microenvironment communication, is achieved (Lambert et al., 2011). All these functional analogies and a better understanding of the mechanisms and implications of individual and collective adaptive evolution are of relevance to generate common therapeutic strategies (Table 1).
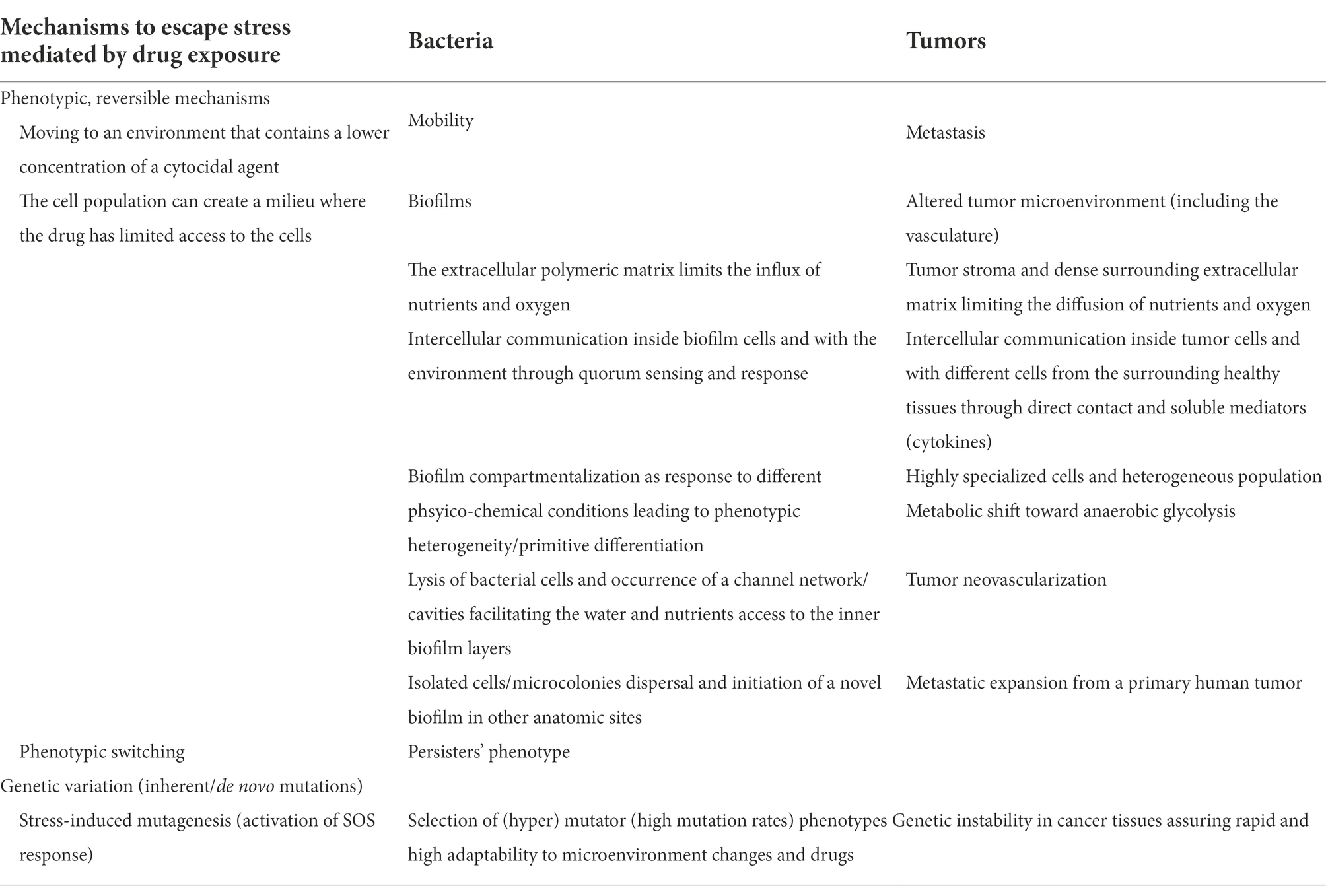
Table 1. Summarization of comparative mechanisms adopted by bacterial and tumor cells seeking survival under stress (Lambert et al., 2011).
Gut microbiota–antibiotics–antitumoral agents trialogue
Given the similarities regarding the antibiotics and some antitumoral agents’ structure, targets, and resistance mechanisms, concerns have been raised regarding their bidirectional interactions with the host microbiota. The human microbiota is considered both an essential organ, with important roles in nutrition, pathogenesis, and immunity, and the second genome, represented by bacteria, archaea, fungi, and viruses (Neu, 2015).
The normal gut microbiota has a density of 1011–1012 cells/ml content and contains mainly two anaerobic phylotypes: Bacteroidetes (48%) and Firmicutes (51%) together with Proteobacteria, Fusobacteria, Spirochaetes, etc. (Mandal et al., 2016). The intestinal microbiome plays an important role in integration of the dietary signals with immune system reactivity and maintenance of the intestinal homeostasis (Thaiss et al., 2016), assuring (i) the mechanical and physiological barrier against colonization with pathogenic agents and overgrowth of existing microorganisms and (ii) anti-tumoral effects, mucosa protection from the side effects and potentiation of antitumoral treatments (Cheng et al., 2020). It is well-known that many factors, from ethnicity, diet, and other lifestyle factors and long-term or frequent use of common antibiotics influence the microbiota composition and can induce malignant transformation (Chelariu et al., 2016, 2017; Sommer et al., 2017; Lazar et al., 2018, 2019; Pircalabioru et al., 2019; Mihai et al., 2021). There are studies stating that the elimination of the infective process with amoxicillin and clarithromycin in early stage of neoplasia has beneficial effects (Van Nuffel et al., 2015). On the contrary, an analysis of 125,441 cancer patients and 490,510 subjects in the control group proved that the antibiotic treatment increased the cancer risk. It has been shown that the incidence of cancer in people treated on long-term with antibiotics increased by 18%, especially for lung, kidney, pancreatic, lymphoma, and myeloma cancers (Friedman et al., 2009). In case of colorectal cancer (CRC), the highest risk was associated with anti-anaerobic antibiotics (vancomycin and penicillins) and the lowest, with tetracyclines (Bufill, 1990; Kilkkinen et al., 2008; Friedman et al., 2009; Dik et al., 2016; Cao et al., 2018).
Neoplasia-associated changes in microbiota are frequently seen, particularly in tissues/systems in contact with gut or respiratory tract microbiota (Wotherspoon et al., 1991; Bayerdörffer et al., 1995; Isaacson and Du, 2004; Yamamoto and Schiestl, 2014; Mima et al., 2017; Mao et al., 2018). The decrease of mucopolysaccharide layer thickness exposes epithelium to direct contact with microbiota and to the development of invasive biofilms (Costerton et al., 1999). It has been shown that the adenomatous polyps, CRC, and healthy epithelium at distance from benign or malignant tumors are covered with biofilms formed by tumorigenic invasive polymicrobial associations incorporated in a polysaccharide matrix (Bufill, 1990; Swidsinski et al., 2005; Dejea et al., 2014, 2018; Tomkovich et al., 2019). In the colon epithelium covered with invasive biofilms, the density of E-cadherin adherence molecules decreases, and the level of IL-6, a marker of increased permeability and inflammatory reaction, increases. By increasing the permeability of the epithelium, the excess lipopolysaccharides (LPS) from the outer membrane of the Gram-negative bacterial cell pass into the blood and induce endotoxemia, inflammation in visceral adipose tissue, disruption of glucose metabolism, insulin resistance, and obesity, playing a role in the development of metabolic syndrome, type 2 diabetes, inflammatory bowel disease, autoimmunity, and carcinogenesis (Halmos and Suba, 2016). The risk of such pathologies is increased by prolonged antibiotic exposure (Hernandez, 2016).
Dysbiosis associated with an increase in pathogenic bacteria such as Fusobacterium nucleatum stimulates the growth of different intestinal tumor types, including CRC, by inhibiting the activity of NK cells.
During dysbiosis, the pro-inflammatory Th17 and anti-inflammatory Treg lymphocytes ratio changes in the favor of the last ones, probably due to changes in the concentration and ratios of short-chain fatty acids (SCFAs). It has been shown that butyrate and propionate, but not acetate, stimulate extrathymic production of foxp3 anti-inflammatory Treg lymphocytes (Arpaia et al., 2013). Importantly, SCFA can act as histone deacetylases inhibitors to induce hyperacetylation of histones modulating gene expression leading to apoptosis and growth arrest (Schilderink et al., 2013).
Modulation of gut microbiota by antitumoral agents and influence of antibiotics
Antitumoral treatments influence the eubiosis by different mechanisms such as: (a) alteration of the quantitative and qualitative panel of molecules produced by the epithelium; (b) induction of changes in gut microbiota, followed by increase of the epithelial barrier permeability, facilitating bacterial translocation into the internal environment in cancer patients that are already immunosuppressed due to cytostatic treatment or malignancy, hence increasing the risk of sepsis (Sullivan et al., 2001; Samet et al., 2013; Papanicolas et al., 2018); (c) alteration of immunological homeostasis due to antibiotic treatment, increasing immunosuppression and susceptibility to infectious diseases and neoplasia (Willing et al., 2011; Alexander et al., 2017); (d) decrease in the efficacy of some anticancer agents and an increase in their toxicity; and (e) triggering of oxidative stress (ROI release) and genotoxicity since bacterial DNA double-strand breaking under antibiotic/cytostatic agents action activate the mutation-inducing SOS repair system leading to antibiotic resistance, while other antibiotics favor the spread of resistant strains (Podolsky, 2002; Cho, 2008).
Some of the most frequently reported alterations of gut microbiota composition and diversity induced by chemotherapy are (i) increased abundance of pathogenic bacteria (e.g., enterococci, staphylococci, Escherichia coli, other Enterobacteriaceae, Pseudomonas, clostridia, and Gram-negative anaerobes); (ii) augmented bacterial translocation to mesenteric lymph nodes and spleen, accompanied by the occurrence of Th1 and Th17 immune responses (Panebianco et al., 2018); (iii) decreased gut bacterial total numbers and biodiversity; (iv) decrease of beneficial bacteria, such as Lactobacillus spp., Bacteroides spp., Faecalibacterium prausnitzii, Bifidobacterium spp., and Firmicutes (Panebianco et al., 2018). It has been shown that oncologic patients harbor a marked reduction in intestinal bacterial diversity, with >30% of fecal samples dominated by a single bacterial genus, increasing the risk of bacterial translocation and explaining the fact that bloodstream infections are causing death in ~10% of cancer patients (Liang et al., 2019). In 50% of patients, bacteremia was preceded by intestinal domination with a corresponding organism for 7 days, the enterococcal domination of the gut microbiota being associated with a 9-fold increase in the risk of vancomycin-resistant enterococcus (VRE) bacteremia, while proteobacterial domination with a 5-fold increase in the risk of Gram-negative bacteremia (Liang et al., 2019).
The majority of antitumoral agents have been shown to modulate the gut microbiota. Anthracyclines have a bacteriostatic effect on the microbiota. Irinotecan is toxic to commensal microbiota, causing an increase in the number of potentially pathogenic species (Enterobacteriaceae, Clostridium, and Fusobacterium nucleatum). Cyclophosphamide, an alkylating compound, is located at the intersection of immunotherapy and chemotherapy. Its anti-neoplastic efficacy is based on stimulating the anti-tumor immune response, which is mainly pro-inflammatory. As a consequence, it also alters the permeability of the small intestine and induces translocation of Gram-negative and Gram-positive species into the secondary lymphoid organs, where bacteria stimulate the Th1 and Th2 memory lymphocytes. Germ-free mice and conventional mice treated with anti-Gram-positive antibiotics have minimal Th17 lymphocyte response, and tumors do not respond to cyclophosphamide (Viaud et al., 2013).
Besides their direct effect on gut eubiosis, cystostatic derivatives are eliminated by biliary secretion in the gut and could, on their turn, stimulate or inhibit the microbiota (Alexander et al., 2017).
Many studies demonstrate that antibiotic treatment could influence the efficiency of cytostatic treatments. For example, cyclophosphamide efficiency was diminished by vancomycin and colistin, while Lactobacillus murinus, Lactobacillus johnsonii, Barnesiella intestinihominis, and Enterococcus hirae increased its antitumor activity or restored the response to cyclophosphamide in tumor-bearing antibiotic-treated mice (White et al., 2008; Viaud et al., 2013; Li et al., 2021). The cytotoxicity of CB 1954 was strongly enhanced by E. coli nitroreductase activity (Chen et al., 2004). Therefore, gut microbiota modulation could maximize the response to anticancer treatments. For example, starting from the capacity of intravenously injected spores of strictly anaerobic Clostridium species to germinate in the hypoxic regions present in solid tumors, a genetically engineered, non-toxinogenic strain of C. novyi-NT has been used to carry the anticancer drug and deliver it inside the tumor (Staedtke et al., 2016; Feng et al., 2021). Not only chemotherapy, but also radiotherapy has been shown to induce persistent selective killing of commensal anaerobes as well as expansion of potentially pathogenic enterococci and Enterobacteriaceae (Garajova et al., 2021).
Influence of gut microbiota on antitumoral treatment efficacy
It has been shown that gut microbiota influences the efficacy of conventional chemotherapy, immunotherapy, radiotherapy and surgery, the drug toxicity, and utltimately, the oncologic patient’ prognosis. Gut microbiota influences the efficacy of chemotherapy and immunotherapy due to its ability to metabolize drugs and xenobiotics, and to modulate host inflammation and immune responses.
The healthy microbiota enhances the efficacy of the platinum-based agent oxaliplatin, by inducing ROI release from myeloid cells, thereby enhancing inflammatory cytokine production and tumor regression (Goldszmid et al., 2015). This effect was decreased in germ-free / antibiotic-treated mice, while a better response of mice to cisplatin was obtained when combined with Lactobacillus (Iida et al., 2013). On the contrary, Fusobacterium nucleatum has been shown to increase resistance platinum-based agents through increasing authophagy (Yu et al., 2017).
Doxorubicin has been shown to be inactivated by microbial deglycosylation performed by strains of Streptomyces and Raoultella (Westman et al., 2012; Blaustein et al., 2021).
The efficacy of 5-Fluorouracil has been reduced by the presence of mycoplasmas or F. nucleatum, via upregulation of an inhibitor apoptotic protein (IAP). It has been shown that F. nucleatum increases tumor-associated neutrophils, dendritic cells, and pro-cancer M2 macrophages and prevents the cytotoxicity of T and NK cells (Zhang et al., 2019). When present inside the tumor microenvironment, Mycoplasma hyorhinis can induce a dramatic antitumor activity decrease (20–150-fold) of 5-Fluorouracil by degradation to its less active derivatives (Bronckaers et al., 2008; Garajova et al., 2021).
Gemcitabine can be metabolized by different microbes, such as Mycoplasma, F. nucleatum, and E. coli which express nucleoside analog-catabolizing enzymes to its inactive metabolite 2′,2′-difluoro-2′-deoxyuridine (dFdU) and resistance could be neutralized by some antibiotics such as ciprofloxacin, which eliminate these species from gut microbiota (Geller et al., 2017; Garajova et al., 2021).
Gut microbiota dysbiosis has been also shown to interfere with the efficacy of anticancer immunotherapy (Garajova et al., 2021), inducing resistance to antibodies against programmed cell death 1 (PD-1; Routy et al., 2018). Moreover, the efficacy of Ipilimumab, a monoclonal antibody that neutralizes the cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), is dependent on the presence of Bacteroides fragilis polysaccharide that induces a splenic Th1 cell memory response (Beck et al., 2006; Vetizou et al., 2015). Besides increasing its efficiency, many members of gut microbiota such as Bacteroidaceae, Rikenellaceae, and Barnesiellaceae protect from Ipilimumab adverse events (Dubin et al., 2016). The favorable clinical outcome during anti-PD1 therapy was linked to the presence of Akkermansia muciniphila, as well as to high diversity and abundance of Clostridiales/Ruminococcaceae/Faecalibacterium, Bifidobacterium longum, Collinsella aerofaciens, and Enterococcus faecium. Conversely, the poor responders harbored a low diversity and high abundance of Bacteroidales (Robert et al., 2011; Sivan et al., 2015; Gopalakrishnan et al., 2018; Mao et al., 2021; Wong et al., 2021; Sevcikova et al., 2022).
Chemotherapy and emergence of AMR
Chemotherapy could trigger the emergence of de novo antimicrobial resistance, one of the mechanisms being the increase of mutation rate by activating the bacterial SOS response to DNA damage (Poulin-Laprade et al., 2015). Mitomycin C has been shown to be a potent inducer of the bacterial SOS response, being used to select resistant E. coli clones with resistance mediated by mdfA efflux pump overexpression (Wei et al., 2001). It has been shown that activation of the SOS response by mitomycin C increases the transcription of genes necessary to horizontal transfer the SXT element that encodes resistance to multiple antibiotics in Vibrio cholerae (Beaber et al., 2004). Methotrexate, a dihydrofolate reductase inhibitor, acts by blocking dihydrofolic acid synthesis similarly to trimethoprim antibiotic and thus, co-selects bacterial cells carrying the trimethoprim resistance gene on the same plasmid (Guethmundsdottir et al., 2021). Non-lethal doses of cisplatin resulted in a 3-7-fold increase in mutation frequency, leading to resistance to rifampicin and ciprofloxacin and the administration of antioxidants (ascorbic acid) decreased genotoxicity by 41% and bacterial mutation rates by 65% (Lofmark et al., 2006; Chistyakov et al., 2018). Some chemotherapeutic agents have been shown to increase the horizontal transfer of bacterial resistance genes by phage transduction, transformation with external DNA, or plasmid transfer by conjugation (Wei et al., 2001).
Conclusion
The evolutionary strategies used by bacteria and tumors to individually and collectively adapt to continuously changing and stressful environments share many similarities, opening new avenues in the study of drug resistance within cancer tissues, using more simple and reproducible bacterial models, as well as in the development of novel antitumor agents and microbiome-based therapeutic interventions that may be able to correct dysbiosis and thus to maximize the treatment efficiency and prevent selection of drug resistance. The gut microbiota is influenced by and influences the treatment efficacy and drug toxicity. Chemotherapy is likely to produce de novo antimicrobial resistance in gut microbiota by inducing dysbiosis, increasing the horizontal gene transfer and the mutation rates consequently to the bacterial SOS system activation. On the other side, the disruption of commensal gut microbiota and alteration of host physiology might influence both the efficacy of the antitumoral treatments and their toxicity. Therefore, a better knowledge of the complex interactions among gut microbiota, antibiotics, and anticancer drugs will enable us to develop novel anticancer treatment strategies and subsequently improve the cancer patients’ outcome, minimizing the risk of antibiotic-resistant bacteria carriage and of associated infections.
Author contributions
MCh, GM, NC, and RF original draft preparation. EI, LB, and CB reviewing and editing. GP figures and manuscript submission. MCo figure preparation. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by CNFIS-FDI-2022-0675, UEFISCDI - PN-III-P4-PCE2021-1797, PN-III-P1-1.1-36PD-2019-0499, Grant number 224/2021 and the Ministry of Research, Innovation, and Digitalization through Program 1—Development of the national R&D system, Subprogram 1.2—Institutional performance—Financing projects for excellence in RDI, Contract no. 41 PFE/30.12.2021. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1. ^https://www.who.int/news-room/fact-sheets/detail/cancer
2. ^https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance
3. ^https://www.reactgroup.org/news-and-views/news-and-opinions/year-2020/new-react-policy-brief-successful-cancer-treatment-relies-on-effective-antibiotics/
References
Ahmed, N., Abubaker, K., Findlay, J., and Quinn, M. (2010). Epithelial mesenchymal transition and cancer stem cell-like phenotypes facilitate chemoresistance in recurrent ovarian cancer. Curr. Cancer Drug Targets 10, 268–278. doi: 10.2174/156800910791190175
Alexander, J. L., Wilson, I. D., Teare, J., Marchesi, J. R., Nicholson, J. K., and Kinross, J. M. (2017). Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 14, 356–365. doi: 10.1038/nrgastro.2017.20
Andersson, D. I., and Hughes, D. (2014). Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 12, 465–478. doi: 10.1038/nrmicro3270
Ariza-Heredia, E. J., and Chemaly, R. F. (2018). Update on infection control practices in cancer hospitals. CA Cancer J. Clin. 68, 340–355. doi: 10.3322/caac.21462
Arpaia, N., Campbell, C., Fan, X., Dikiy, S., van der Veeken, J., deRoos, P., et al. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. doi: 10.1038/nature12726
Avner, B. S., Fialho, A. M., and Chakrabarty, A. M. (2012). Overcoming drug resistance in multi-drug resistant cancers and microorganisms: a conceptual framework. Bioengineered. 3, 262–270. doi: 10.4161/bioe.21130
Barrett, J. F. (1996). Section reviews; anti-infectives: section review anti-infectives: quinolone antibacterials and derivatives as antineoplastic agents. Expert Opin. Investig. Drugs 5, 1021–1031. doi: 10.1517/13543784.5.8.1021
Bayerdörffer, E., Rudolph, B. D., Neubauer, A., Thiede, C., Lehn, N., Eidt, S., et al. (1995). Regression of primary gastric lymphoma of mucosa-associated lymphoid tissue type after cure of Helicobacter pylori infection. Lancet 345, 1591–1594. doi: 10.1016/S0140-6736(95)90113-2
Beaber, J. W., Hochhut, B., and Waldor, M. K. (2004). SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427, 72–74. doi: 10.1038/nature02241
Beck, K. E., Blansfield, J. A., Tran, K. Q., Feldman, A. L., Hughes, M. S., Royal, R. E., et al. (2006). Enterocolitis in patients with cancer after antibody blockade of cytotoxic T-lymphocyte-associated antigen 4. J. Clin. Oncol. 24, 2283–2289. doi: 10.1200/JCO.2005.04.5716
Bisacchi, G. S., and Hale, M. R. (2016). A “Double-Edged” scaffold: antitumor power within the antibacterial quinolone. Curr. Med. Chem. 23, 520–577. doi: 10.2174/0929867323666151223095839
Blaustein, R. A., Seed, P. C., and Hartmann, E. M. (2021). Biotransformation of doxorubicin promotes resilience in simplified intestinal microbial communities. mSphere. 6:e0006821. doi: 10.1128/mSphere.00068-21
Bronckaers, A., Balzarini, J., and Liekens, S. (2008). The cytostatic activity of pyrimidine nucleosides is strongly modulated by Mycoplasma hyorhinis infection: implications for cancer therapy. Biochem. Pharmacol. 76, 188–197. doi: 10.1016/j.bcp.2008.04.019
Bufill, J. A. (1990). Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumor location. Ann. Intern. Med. 113, 779–788. doi: 10.7326/0003-4819-113-10-779
Butler, M. T., Wang, Q., and Harshey, R. M. (2010). Cell density and mobility protect swarming bacteria against antibiotics. Proc. Natl. Acad. Sci. U. S. A. 107, 3776–3781. doi: 10.1073/pnas.0910934107
Cao, Y., Wu, K., Mehta, R., Drew, D. A., Song, M., Lochhead, P., et al. (2018). Long-term use of antibiotics and risk of colorectal adenoma. Gut 67, 672–678. doi: 10.1136/gutjnl-2016-313413
CDC’s Antibiotic Resistance Threats in the United States (2019). Available at: https://www.cdc.gov/drugresistance/biggest-threats.html (Accesed May 31, 2022).
Chelariu, M., Curutiu, C., Ditu, L., Ionescu, D., Ionescu, B., Chifiriuc, M., et al. (2016). Composition of cultivable intestinal microbiota in patients suffering from obesity and type II diabetes. Biointerface Res. Appl. Chem. 6, 1580–1584.
Chelariu, M., Grosu, M., Gheorghe, I., Gradisteanu, G., Picu, A., Petcu, L., et al. (2017). Host metabolic syndrome can disrupt the intestinal microbiota and promote the acquisition of resistance and virulence genes in Enterobacteriaceae stains. Rom Biotech. Lett. 22, 12643–12650.
Chen, M. J., Green, N. K., Reynolds, G. M., Flavell, J. R., Mautner, V., Kerr, D. J., et al. (2004). Enhanced efficacy of Escherichia coli nitroreductase/CB1954 prodrug activation gene therapy using an E1B-55K-deleted oncolytic adenovirus vector. Gene Ther. 11, 1126–1136. doi: 10.1038/sj.gt.3302271
Cheng, W. Y., Wu, C.-Y., and Yu, J. (2020). The role of gut microbiota in cancer treatment: friend or foe? Gut 69, 1867–1876. doi: 10.1136/gutjnl-2020-321153
Chistyakov, V. A., Prazdnova, E. V., Mazanko, M. S., Churilov, M. N., and Chmyhalo, V. K. (2018). Increase in bacterial resistance to antibiotics after cancer therapy with platinum-based drugs. Mol. Biol. 52, 232–236. doi: 10.1134/S0026893317050077
Cho, J. H. (2008). The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 8, 458–466. doi: 10.1038/nri2340
Cifone, M. A., and Fidler, I. J. (1981). Increasing metastatic potential is associated with increasing genetic instability of clones isolated from murine neoplasms. Proc. Natl. Acad. Sci. U. S. A. 78, 6949–6952. doi: 10.1073/pnas.78.11.6949
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Dalhoff, A., and Shalit, I. (2003). Immunomodulatory effects of quinolones. Lancet Infect. Dis. 3, 359–371. doi: 10.1016/S1473-3099(03)00658-3
D'Arpa, P., and Liu, L. F. (1989). Topoisomerase-targeting antitumor drugs. Biochim. Biophys. Acta 989, 163–177.
Davies, J., and Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433. doi: 10.1128/MMBR.00016-10
Dejea, C. M., Fathi, P., Craig, J. M., Boleij, A., Taddese, R., Geis, A. L., et al. (2018). Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597. doi: 10.1126/science.aah3648
Dejea, C. M., Wick, E. C., Hechenbleikner, E. M., White, J. R., Mark Welch, J. L., Rossetti, B. J., et al. (2014). Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. U. S. A. 111, 18321–18326. doi: 10.1073/pnas.1406199111
Dik, V. K., van Oijen, M. G., Smeets, H. M., and Siersema, P. D. (2016). Frequent use of antibiotics is associated with colorectal cancer risk: results of a nested case-control study. Dig. Dis. Sci. 61, 255–264. doi: 10.1007/s10620-015-3828-0
Dubin, K., Callahan, M. K., Ren, B., Khanin, R., Viale, A., Ling, L., et al. (2016). Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 7:10391. doi: 10.1038/ncomms10391
Feng, X., He, P., Zeng, C., Li, Y. H., Das, S. K., Li, B., et al. (2021). Novel insights into the role of Clostridium novyi-NT related combination bacteriolytic therapy in solid tumors. Oncol. Lett. 21:110. doi: 10.3892/ol.2020.12371
Ferlay, J., Colombet, M., Soerjomataram, I., Parkin, D. M., Pineros, M., Znaor, A., et al. (2021). Cancer statistics for the year 2020: an overview. Int. J. Cancer 149, 778–789. doi: 10.1002/ijc.33588
Francino, M. P. (2015). Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front. Microbiol. 6:1543. doi: 10.3389/fmicb.2015.01543
Friedman, G. D., Udaltsova, N., Chan, J., Quesenberry, C. P. Jr., and Habel, L. A. (2009). Screening pharmaceuticals for possible carcinogenic effects: initial positive results for drugs not previously screened. Cancer Causes Control: CCC. 20, 1821–1835. doi: 10.1007/s10552-009-9375-2
Gao, Y., Shang, Q., Li, W., Guo, W., Stojadinovic, A., Mannion, C., et al. (2020). Antibiotics for cancer treatment: a double-edged sword. J. Cancer 11, 5135–5149. doi: 10.7150/jca.47470
Garajova, I., Balsano, R., Wang, H., Leonardi, F., Giovannetti, E., Deng, D., et al. (2021). The role of the microbiome in drug resistance in gastrointestinal cancers. Expert. Rev. Anticancer. Ther. 21, 165–176. doi: 10.1080/14737140.2021.1844007
Geller, L. T., Barzily-Rokni, M., Danino, T., Jonas, O. H., Shental, N., Nejman, D., et al. (2017). Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160. doi: 10.1126/science.aah5043
Goldie, J. H., and Coldman, A. J. (1979). A mathematic model for relating the drug sensitivity of tumors to their spontaneous mutation rate. Cancer Treat. Rep. 63, 1727–1733.
Goldie, J. H., and Coldman, A. J. (1983). Quantitative model for multiple levels of drug resistance in clinical tumors. Cancer Treat. Rep. 67, 923–931.
Goldie, J. H., and Coldman, A. J. (1984). The genetic origin of drug resistance in neoplasms: implications for systemic therapy. Cancer Res. 44, 3643–3653.
Goldszmid, R. S., Dzutsev, A., Viaud, S., Zitvogel, L., Restifo, N. P., and Trinchieri, G. (2015). Microbiota modulation of myeloid cells in cancer therapy. Cancer Immunol. Res. 3, 103–109. doi: 10.1158/2326-6066.CIR-14-0225
Gopalakrishnan, V., Spencer, C. N., Nezi, L., Reuben, A., Andrews, M. C., Karpinets, T. V., et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103. doi: 10.1126/science.aan4236
Guethmundsdottir, J. S., Fredheim, E. G. A., Koumans, C. I. M., Hegstad, J., Tang, P. C., Andersson, D. I., et al. (2021). The chemotherapeutic drug methotrexate selects for antibiotic resistance. EBioMedicine 74:103742. doi: 10.1016/j.ebiom.2021.103742
Halmos, T., and Suba, I. (2016). Physiological patterns of intestinal microbiota. The role of dysbacteriosis in obesity, insulin resistance, diabetes and metabolic syndrome. Orv. Hetil. 157, 13–22. doi: 10.1556/650.2015.30296
Hernandez, D. L. (2016). Letter to the editor: use of antibiotics, gut microbiota, and risk of type 2 diabetes: epigenetics regulation. J. Clin. Endocrinol. Metab. 101, L62–L63. doi: 10.1210/jc.2016-1151
Iida, N., Dzutsev, A., Stewart, C. A., Smith, L., Bouladoux, N., Weingarten, R. A., et al. (2013). Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342, 967–970. doi: 10.1126/science.1240527
Isaacson, P. G., and Du, M. Q. (2004). MALT lymphoma: from morphology to molecules. Nat. Rev. Cancer 4, 644–653. doi: 10.1038/nrc1409
Jernberg, C., Lofmark, S., Edlund, C., and Jansson, J. K. (2007). Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 1, 56–66. doi: 10.1038/ismej.2007.3
Kanady, K. E., Shipley, W. U., Zietman, A. L., Kaufman, D. S., Althausen, A. F., and Heney, N. M. (1997). Treatment strategies using transurethral surgery, chemotherapy, and radiation therapy with selection that safely allows bladder conservation for invasive bladder cancer. Semin. Surg. Oncol. 13, 359–364. doi: 10.1002/(SICI)1098-2388(199709/10)13:5<359::AID-SSU10>3.0.CO;2-I
Kilkkinen, A., Rissanen, H., Klaukka, T., Pukkala, E., Heliovaara, M., Huovinen, P., et al. (2008). Antibiotic use predicts an increased risk of cancer. Int. J. Cancer 123, 2152–2155. doi: 10.1002/ijc.23622
Lambert, G., Estevez-Salmeron, L., Oh, S., Liao, D., Emerson, B. M., Tlsty, T. D., et al. (2011). An analogy between the evolution of drug resistance in bacterial communities and malignant tissues. Nat. Rev. Cancer 11, 375–382. doi: 10.1038/nrc3039
Lazar, V., Ditu, L. M., Pircalabioru, G. G., Gheorghe, I., Curutiu, C., Holban, A. M., et al. (2018). Aspects of gut microbiota and immune system interactions in infectious diseases, immunopathology, and cancer. Front. Immunol. 9:1830. doi: 10.3389/fimmu.2018.01830
Lazar, V., Ditu, L. M., Pircalabioru, G. G., Picu, A., Petcu, L., Cucu, N., et al. (2019). Gut microbiota, host organism, and diet trialogue in diabetes and obesity. Front. Nutr. 6:21. doi: 10.3389/fnut.2019.00021
Li, W., Deng, X., and Chen, T. (2021). Exploring the modulatory effects of gut microbiota in anti-cancer therapy. Front. Oncol. 11:644454. doi: 10.3389/fonc.2021.644454
Liang, W., Yang, Y., Wang, H., Wang, H., Yu, X., Lu, Y., et al. (2019). Gut microbiota shifts in patients with gastric cancer in perioperative period. Medicine 98:e16626. doi: 10.1097/MD.0000000000016626
Lofmark, S., Jernberg, C., Jansson, J. K., and Edlund, C. (2006). Clindamycin-induced enrichment and long-term persistence of resistant Bacteroides spp. and resistance genes. J. Antimicrob. Chemother. 58, 1160–1167. doi: 10.1093/jac/dkl420
Macia, M. D., Blanquer, D., Togores, B., Sauleda, J., Perez, J. L., and Oliver, A. (2005). Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob. Agents Chemother. 49, 3382–3386. doi: 10.1128/AAC.49.8.3382-3386.2005
Mandal, S. M., Pati, B. R., Chakraborty, R., and Franco, O. L. (2016). New insights into the bioactivity of peptides from probiotics. Front. Biosci. 8, 450–459. doi: 10.2741/e779
Mao, Q., Jiang, F., Yin, R., Wang, J., Xia, W., Dong, G., et al. (2018). Interplay between the lung microbiome and lung cancer. Cancer Lett. 415, 40–48. doi: 10.1016/j.canlet.2017.11.036
Mao, J., Wang, D., Long, J., Yang, X., Lin, J., Song, Y., et al. (2021). Gut microbiome is associated with the clinical response to anti-PD-1 based immunotherapy in hepatobiliary cancers. J. Immunother. Cancer 9:e003334. doi: 10.1136/jitc-2021-003334
Mihaescu, G., Chifiriuc, M., and Ditu, L. (2008). Antibiotice şi substanţe chimioterapeutice antimicrobiene (Antibiotics and Antimicrobial Chemotherapeutic Agents). Romanian Academy Publishing House Bucharest. ISBN 978-973-27-1573-4.
Mihaescu, G., Chifiriuc, C., and Ditu, L. M. (2009). Imunobiologie (imunobiology). Bucharest, Romania: University of Bucharest Publishing House. ISBN 978-973-737-734-0.
Mihai, M. M., Ion, A., Giurcaneanu, C., Nitipir, C., Popa, A. M., Chifiriuc, M. C., et al. (2021). The impact of long-term antibiotic therapy of cutaneous adverse reactions to EGFR inhibitors in colorectal cancer patients. J. Clin. Med. 10:3219. doi: 10.3390/jcm10153219
Mima, K., Nakagawa, S., Sawayama, H., Ishimoto, T., Imai, K., Iwatsuki, M., et al. (2017). The microbiome and hepatobiliary-pancreatic cancers. Cancer Lett. 402, 9–15. doi: 10.1016/j.canlet.2017.05.001
Neu, J. (2015). Developmental aspects of maternal-fetal, and infant gut microbiota and implications for long-term health. Matern. Health Neonatol. Perinatol. 1:6. doi: 10.1186/s40748-015-0007-4
Nikolaou, M., Pavlopoulou, A., Georgakilas, A. G., and Kyrodimos, E. (2018). The challenge of drug resistance in cancer treatment: a current overview. Clin. Exp. Metastasis 35, 309–318. doi: 10.1007/s10585-018-9903-0
Panebianco, C., Andriulli, A., and Pazienza, V. (2018). Pharmacomicrobiomics: exploiting the drug-microbiota interactions in anticancer therapies. Microbiome. 6:92. doi: 10.1186/s40168-018-0483-7
Papanicolas, L. E., Gordon, D. L., Wesselingh, S. L., and Rogers, G. B. (2018). Not just antibiotics: is cancer chemotherapy driving antimicrobial resistance? Trends Microbiol. 26, 393–400. doi: 10.1016/j.tim.2017.10.009
Patel, H., Wu, Z. X., Chen, Y., Bo, L., and Chen, Z. S. (2021). Drug resistance: from bacteria to cancer. Mol. Biomed. 2:27. doi: 10.1186/s43556-021-00041-4
Peterson, L. R. (2009). Bad bugs, no drugs: no ESCAPE revisited. Clin. Infect. Dis. 49, 992–993. doi: 10.1086/605539
Pinato, D. J., Gramenitskaya, D., Altmann, D. M., Boyton, R. J., Mullish, B. H., Marchesi, J. R., et al. (2019). Antibiotic therapy and outcome from immune-checkpoint inhibitors. J. Immunother. Cancer 7:287. doi: 10.1186/s40425-019-0775-x
Pircalabioru, G. G., Stoica, R. A., Petcu, L., Picu, A., Suceveanu, A. P., Salmen, T., et al. (2019). Microbiota signatures in type-2 diabetic patients with chronic kidney disease – a pilot study. J Mind Med Sci 6, 130–136. doi: 10.22543/7674.61.P130136
Podolsky, D. K. (2002). Inflammatory bowel disease. N. Engl. J. Med. 347, 417–429. doi: 10.1056/NEJMra020831
Pommier, Y., Leo, E., Zhang, H., and Marchand, C. (2010). DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17, 421–433. doi: 10.1016/j.chembiol.2010.04.012
Poulin-Laprade, D., Matteau, D., Jacques, P. E., Rodrigue, S., and Burrus, V. (2015). Transfer activation of SXT/R391 integrative and conjugative elements: unraveling the SetCD regulon. Nucleic Acids Res. 43, 2045–2056. doi: 10.1093/nar/gkv071
Robert, C., Thomas, L., Bondarenko, I., O'Day, S., Weber, J., Garbe, C., et al. (2011). Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N. Engl. J. Med. 364, 2517–2526. doi: 10.1056/NEJMoa1104621
Routy, B., Le Chatelier, E., Derosa, L., Duong, C. P. M., Alou, M. T., Daillere, R., et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. doi: 10.1126/science.aan3706
Salerno, S., Da Settimo, F., Taliani, S., Simorini, F., La Motta, C., Fornaciari, G., et al. (2010). Recent advances in the development of dual topoisomerase I and II inhibitors as anticancer drugs. Curr. Med. Chem. 17, 4270–4290. doi: 10.2174/092986710793361252
Samet, A., Sledzinska, A., Krawczyk, B., Hellmann, A., Nowicki, S., Kur, J., et al. (2013). Leukemia and risk of recurrent Escherichia coli bacteremia: genotyping implicates E. coli translocation from the colon to the bloodstream. Eur. J Clin. Microbiol. Infect. Dis. 32, 1393–1400. doi: 10.1007/s10096-013-1886-9
Schilderink, R., Verseijden, C., and de Jonge, W. J. (2013). Dietary inhibitors of histone deacetylases in intestinal immunity and homeostasis. Front. Immunol. 4:226. doi: 10.3389/fimmu.2013.00226
Sevcikova, A., Izoldova, N., Stevurkova, V., Kasperova, B., Chovanec, M., Ciernikova, S., et al. (2022). The impact of the microbiome on resistance to cancer treatment with chemotherapeutic agents and immunotherapy. Int. J. Mol. Sci. 23:488. doi: 10.3390/ijms23010488
Sissi, C., and Palumbo, M. (2003). The quinolone family: from antibacterial to anticancer agents. Curr. Med. Chem. Anticancer Agents 3, 439–450. doi: 10.2174/1568011033482279
Sivan, A., Corrales, L., Hubert, N., Williams, J. B., Aquino-Michaels, K., Earley, Z. M., et al. (2015). Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089. doi: 10.1126/science.aac4255
Skok, Z., Zidar, N., Kikelj, D., and Ilas, J. (2020). Dual inhibitors of human DNA topoisomerase II and other cancer-related targets. J. Med. Chem. 63, 884–904. doi: 10.1021/acs.jmedchem.9b00726
Sommer, F., Anderson, J. M., Bharti, R., Raes, J., and Rosenstiel, P. (2017). The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 15, 630–638. doi: 10.1038/nrmicro.2017.58
Spratt, B. G. (1994). Resistance to antibiotics mediated by target alterations. Science 264, 388–393. doi: 10.1126/science.8153626
Staedtke, V., Roberts, N. J., Bai, R. Y., and Zhou, S. (2016). Clostridium novyi-NT in cancer therapy. Genes Dis. 3, 144–152. doi: 10.1016/j.gendis.2016.01.003
Stewart, P. S., and Costerton, J. W. (2001). Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138. doi: 10.1016/S0140-6736(01)05321-1
Sullivan, A., Edlund, C., and Nord, C. E. (2001). Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 1, 101–114. doi: 10.1016/S1473-3099(01)00066-4
Swidsinski, A., Weber, J., Loening-Baucke, V., Hale, L. P., and Lochs, H. (2005). Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43, 3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005
Taddei, F., Radman, M., Maynard-Smith, J., Toupance, B., Gouyon, P. H., and Godelle, B. (1997). Role of mutator alleles in adaptive evolution. Nature 387, 700–702. doi: 10.1038/42696
Thaiss, C. A., Zmora, N., Levy, M., and Elinav, E. (2016). The microbiome and innate immunity. Nature 535, 65–74. doi: 10.1038/nature18847
Tomkovich, S., Dejea, C. M., Winglee, K., Drewes, J. L., Chung, L., Housseau, F., et al. (2019). Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J. Clin. Invest. 129, 1699–1712. doi: 10.1172/JCI124196
Van Nuffel, A. M., Sukhatme, V., Pantziarka, P., Meheus, L., Sukhatme, V. P., and Bouche, G. (2015). Repurposing Drugs in Oncology (ReDO)-clarithromycin as an anti-cancer agent. Ecancermedicalscience. 9, 513. doi: 10.3332/ecancer.2015.513
Vetizou, M., Pitt, J. M., Daillere, R., Lepage, P., Waldschmitt, N., Flament, C., et al. (2015). Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084. doi: 10.1126/science.aad1329
Viaud, S., Saccheri, F., Mignot, G., Yamazaki, T., Daillere, R., Hannani, D., et al. (2013). The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342, 971–976. doi: 10.1126/science.1240537
Wei, Y., Vollmer, A. C., and LaRossa, R. A. (2001). In vivo titration of mitomycin C action by four Escherichia coli genomic regions on multicopy plasmids. J. Bacteriol. 183, 2259–2264. doi: 10.1128/JB.183.7.2259-2264.2001
Westman, E. L., Canova, M. J., Radhi, I. J., Koteva, K., Kireeva, I., Waglechner, N., et al. (2012). Bacterial inactivation of the anticancer drug doxorubicin. Chem. Biol. 19, 1255–1264. doi: 10.1016/j.chembiol.2012.08.011
White, C. L., Menghistu, T., Twigger, K. R., Searle, P. F., Bhide, S. A., Vile, R. G., et al. (2008). Escherichia coli nitroreductase plus CB1954 enhances the effect of radiotherapy in vitro and in vivo. Gene Ther. 15, 424–433. doi: 10.1038/sj.gt.3303081
Willing, B. P., Russell, S. L., and Finlay, B. B. (2011). Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat. Rev. Microbiol. 9, 233–243. doi: 10.1038/nrmicro2536
Wong, M. K., Barbulescu, P., Coburn, B., and Reguera-Nunez, E. (2021). Therapeutic interventions and mechanisms associated with gut microbiota-mediated modulation of immune checkpoint inhibitor responses. Microbes Infect. 23:104804. doi: 10.1016/j.micinf.2021.104804
Wotherspoon, A. C., Ortiz-Hidalgo, C., Falzon, M. R., and Isaacson, P. G. (1991). Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet 338, 1175–1176. doi: 10.1016/0140-6736(91)92035-Z
Yamamoto, M. L., and Schiestl, R. H. (2014). Intestinal microbiome and lymphoma development. Cancer J. 20, 190–194. doi: 10.1097/PPO.0000000000000047
Yu, T., Guo, F., Yu, Y., Sun, T., Ma, D., Han, J., et al. (2017). Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563.e16. doi: 10.1016/j.cell.2017.07.008
Keywords: microbial biofilms, persister cells, efflux pumps, stress response, gut microbiota, mutator phenotype
Citation: Chifiriuc MC, Filip R, Constantin M, Pircalabioru GG, Bleotu C, Burlibasa L, Ionica E, Corcionivoschi N and Mihaescu G (2022) Common themes in antimicrobial and anticancer drug resistance. Front. Microbiol. 13:960693. doi: 10.3389/fmicb.2022.960693
Edited by:
Nagendran Tharmalingam, Rhode Island Hospital, United StatesReviewed by:
Bindu Subhadra, Long Island University, United StatesCharuta Agashe, Icahn School of Medicine at Mount Sinai, United States
Jagpreet Singh Nanda, Cedars Sinai Medical Center, United States
Copyright © 2022 Chifiriuc, Filip, Constantin, Pircalabioru, Bleotu, Burlibasa, Ionica, Corcionivoschi and Mihaescu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gratiela Gradisteanu Pircalabioru, Z3JhdGllbGEuZ3JhZGlzdGVhbnVAaWN1Yi51bmlidWMucm8=; Coralia Bleotu, Y29yYWxpYS5ibGVvdHVAdmlyb2xvZ3kucm8=; Y2JsZW90dUB5YWhvby5jb20=
†These authors have contributed equally to this work
 Mariana Carmen Chifiriuc
Mariana Carmen Chifiriuc Roxana Filip5,6†
Roxana Filip5,6† Gratiela Gradisteanu Pircalabioru
Gratiela Gradisteanu Pircalabioru Coralia Bleotu
Coralia Bleotu Liliana Burlibasa
Liliana Burlibasa Elena Ionica
Elena Ionica Nicolae Corcionivoschi
Nicolae Corcionivoschi