- 1Agriculture and Agri-Food Canada Saskatoon Research and Development Centre, Saskatoon, SK, Canada
- 2Department of Biology, University of Saskatchewan, Saskatoon, SK, Canada
- 3Centre de Recherche et D'innovation sur les Végétaux (CRIV), Faculté des Sciences de L'agriculture et de L'alimentation, Département de Phytologie, Université Laval, Québec, QC, Canada
- 4Institut de Biologie Intégrative et des Systèmes, Université Laval, Québec, QC, Canada
- 5Canadian Food Inspection Agency (CFIA), Sidney Laboratory, Centre for Plant Health, North Saanich, BC, Canada
- 6Department of Veterinary Microbiology, University of Saskatchewan, Saskatoon, SK, Canada
Phytoplasmas are insect-vectored, difficult-to-culture bacterial pathogens that infect a wide variety of crop and non-crop plants, and are associated with diseases that can lead to significant yield losses in agricultural production worldwide. Phytoplasmas are currently grouped in the provisional genus ‘Candidatus Phytoplasma’, which includes 49 ‘Candidatus’ species. Further differentiation of phytoplasmas into ribosomal groups is based on the restriction fragment length polymorphism (RFLP) pattern of the 16S rRNA-encoding operon, with more than 36 ribosomal groups (16Sr) and over 100 subgroups reported. Since disease symptoms on plants are not associated with phytoplasma identity, accurate diagnostics is of critical importance to manage disease associated with these microorganisms. Phytoplasmas are typically detected from plant and insect tissue using PCR-based methods targeting universal taxonomic markers. Although these methods are relatively sensitive, specific and are widely used, they have limitations, since they provide limited resolution of phytoplasma strains, thus necessitating further assessment of biological properties and delaying implementation of mitigation measures. Moreover, the design of PCR primers that can target multiple loci from phytoplasmas that differ at the sequence level can be a significant challenge. To overcome these limitations, a PCR-independent, multilocus sequence typing (MLST) assay to characterize an array of phytoplasmas was developed. Hybridization probe s targeting cpn60, tuf, secA, secY, and nusA genes, as well as 16S and rp operons, were designed and used to enrich DNA extracts from phytoplasma-infected samples for DNA fragments corresponding to these markers prior to Illumina sequencing. This method was tested using different phytoplasmas including ‘Ca. P. asteris’ (16SrI-B), ‘Ca. P. pruni’ (16SrIII-A),‘Ca. P. prunorum’ (16SrX-B), ‘Ca. P. pyri’ (16SrX-C), ‘Ca. P. mali’ (16SrX-A), and ‘Ca. P. solani’ (16SrXII-A). Thousands of reads were obtained for each gene with multiple overlapping fragments, which were assembled to generate full-length (typically >2 kb), high-quality sequences. Phytoplasma groups and subgroups were accurately determined based on 16S ribosomal RNA and cpn60 gene sequences. Hybridization-based MLST facilitates the enrichment of target genes of phytoplasmas and allows the simultaneous determination of sequences corresponding to seven different markers. In this proof-of-concept study, hybridization-based MLST was demonstrated to be an efficient way to generate data regarding ‘Ca. Phytoplasma’ species/strain differentiation.
Introduction
Phytoplasmas are phytopathogenic bacteria that are grouped into the provisional genus ‘Candidatus Phytoplasma’, which includes 49 known species (Bertaccini et al., 2022). These pathogens infect a wide variety of plant species including both crops and weedy species, and can cause agricultural production losses in all production areas of the world. Phytoplasmas are mainly transmitted by hemipteran insects (Weintraub and Beanland, 2006), but they can also be spread through vegetative propagation, grafting, or seeds (Satta et al., 2020; Ranebennur et al., 2022). Phytoplasma-infected plants usually show symptoms such as yellowing, virescence, witches’ broom, phyllody, leaf roll, and generalized decline (Bertaccini, 2022). Symptoms of phytoplasma infection are often overlooked or confused with the response of plants to viral diseases and abiotic stressors; therefore, accurate diagnosis is required for choosing appropriate management strategies. Moreover, like other plant pathogens such as viruses, phytoplasmas are grouped in a variety of groups and subgroups, and genetically distinguishable phytoplasmas can infect the same plant species. Additionally, in many cases, the insect vectors are unknown. Therefore, detecting, monitoring, and controlling diseases associated with these pathogens is very challenging.
Species boundaries for phytoplasmas have been defined by sequence analysis of 16S rRNA-encoding genes, with a sequence identity of the full-length 16S rRNA gene of 98.65% as a cutoff for determining species (Bertaccini et al., 2022). In addition to the ‘Candidatus’ species designations, phytoplasmas have been classified into ribosomal groups and subgroups based on RFLP analysis of a fragment of the 16S rRNA gene (Gundersen and Lee, 1996; Zhao et al., 2009). This classification has resulted in the designation of more than 37 ribosomal groups (16Sr) and over 150 subgroups (Wei and Zhao, 2022). Differentiation of phytoplasma strains based on the sequences of 16S rRNA genes is complicated by the relatively low inter-taxon sequence differences observed, and by the fact that the two copies of the 16S rRNA gene within a single genome are in some cases distinct from one another (16S rRNA-encoding gene heterogeneity) (Liefting et al., 1996; Jomantiene et al., 2002; Davis et al., 2003; Perez-Lopez et al., 2019). Therefore, supplementary sequence information from single-copy, protein-coding gene markers is recommended for resolution of phytoplasma strains. There are 49 recognized ‘Candidatus Phytoplasma’ species, and while whole-genome draft sequence analysis is becoming available for some phytoplasmas (Firrao et al., 2013; Cho et al., 2020), many of them still do not have genome sequences available.
Multilocus sequence typing (MLST) is well recognized as an improvement over single-marker sequencing for differentiation of bacterial strains in general, and particularly for difficult-to-culture bacteria such as phytoplasmas. Accordingly, MLST has been extensively and recently applied to the differentiation of various groups of phytoplasmas. For example, in defining the taxon ‘Ca. P. pruni’, Davis et al. suggested that the strains should best be differentiated by including not only 16S rRNA gene sequences, but also additional sequence information from secY and rp genes (Davis et al., 2013). Moreover, closely related, but distinct, phytoplasmas belonging to ribosomal group 16SrV (“flavescence dorée”) were successfully differentiated using 16S, map, uvrB-degV, and secY sequences (Arnaud et al., 2007). Strains within group 16SrI associated with azalea little leaf disease were discerned using another set of markers, which included 16S, rpsS, rplIV, rpsC, and secY genes (Wei et al., 2011). Phytoplasmas classified within ribosomal groups XI and XIV associated with sugarcane white leaf disease, Napier grass stunt, and Bermuda grass white leaf are difficult to differentiate based only on 16S genes, but a MLST scheme using group-specific primers generating a 1 kb fragment of leuS (leucyl tRNA synthetase), in combination with secA and 16S sequences, provided clarity to their differentiation (Abeysinghe et al., 2016). More recently, MLST has been applied for the differentiation of phytoplasmas within ribosomal group 16SrIV, associated with palm lethal decline. Sequences of the 16S rRNA gene, 16S-23S intergenic spacer region, secA, and groEL (cpn60) demonstrated that three palm lethal decline phytoplasmas could be observed (‘Ca. P. palmae’, ‘Ca. P. aculeata’, and ‘Ca. P. hispanola’), which were distinct from a Tanzanian strain (‘Ca. P. cocotanzaniae’). Furthermore, the sequence identity of multiple genes within ribosomal group IV confirmed that distinct ribosomal RNA gene subgroups are properly considered to be the same species, ‘Ca. P. aculeata’ (Soto et al., 2021). All of these studies confirm and support the utility of determining the sequences of multiple genetic markers for accurate differentiation of phytoplasma strains.
The objective of the current study was to develop and validate a rapid, convenient, and accurate method of determining the sequences of multiple taxonomic markers for diverse phytoplasma strains. A hybridization-based MLST assay using a set of capture probes corresponding to seven taxonomic markers from a variety of phytoplasmas was developed. Hybridization probes were designed targeting the 16S ribosomal RNA-encoding gene as well as six other protein-coding genes. The protein-coding, single-copy genes selected to develop a multilocus panel were cpn60 (also known as groEL or hsp60) (Mitrović et al., 2011; Pérez-López et al., 2016), tuf (Marcone et al., 2000), secA (Hodgetts et al., 2008), secY (Lee et al., 2010), and nusA (Shao et al., 2006). In addition, this MLST sequencing panel includes the ribosomal protein (rp) operon, which consists of several short genes including rplV-rpsC and intergenic regions (Martini et al., 2007).
Moreover, to provide a proof-of-concept that the method can detect and type these sequences accurately from a variety of phytoplasmas, six distinct phytoplasmas were examined: ‘Ca. P. asteris’ (16SrI), ‘Ca. P. pruni’ (16SrIII-A), ‘Ca. P. prunorum’ (16SrX-B), ‘Ca. P. pyri’ (16SrX-C), ‘Ca. P. mali’ (16SrX-A), and ‘Ca. P. solani’ (16SrXII-A). The hybridization-based MLST was used to determine the sequences of these markers from each of these phytoplasmas, and in samples with a wide range of phytoplasma concentrations in infected tissues.
Materials and methods
Infected plant samples
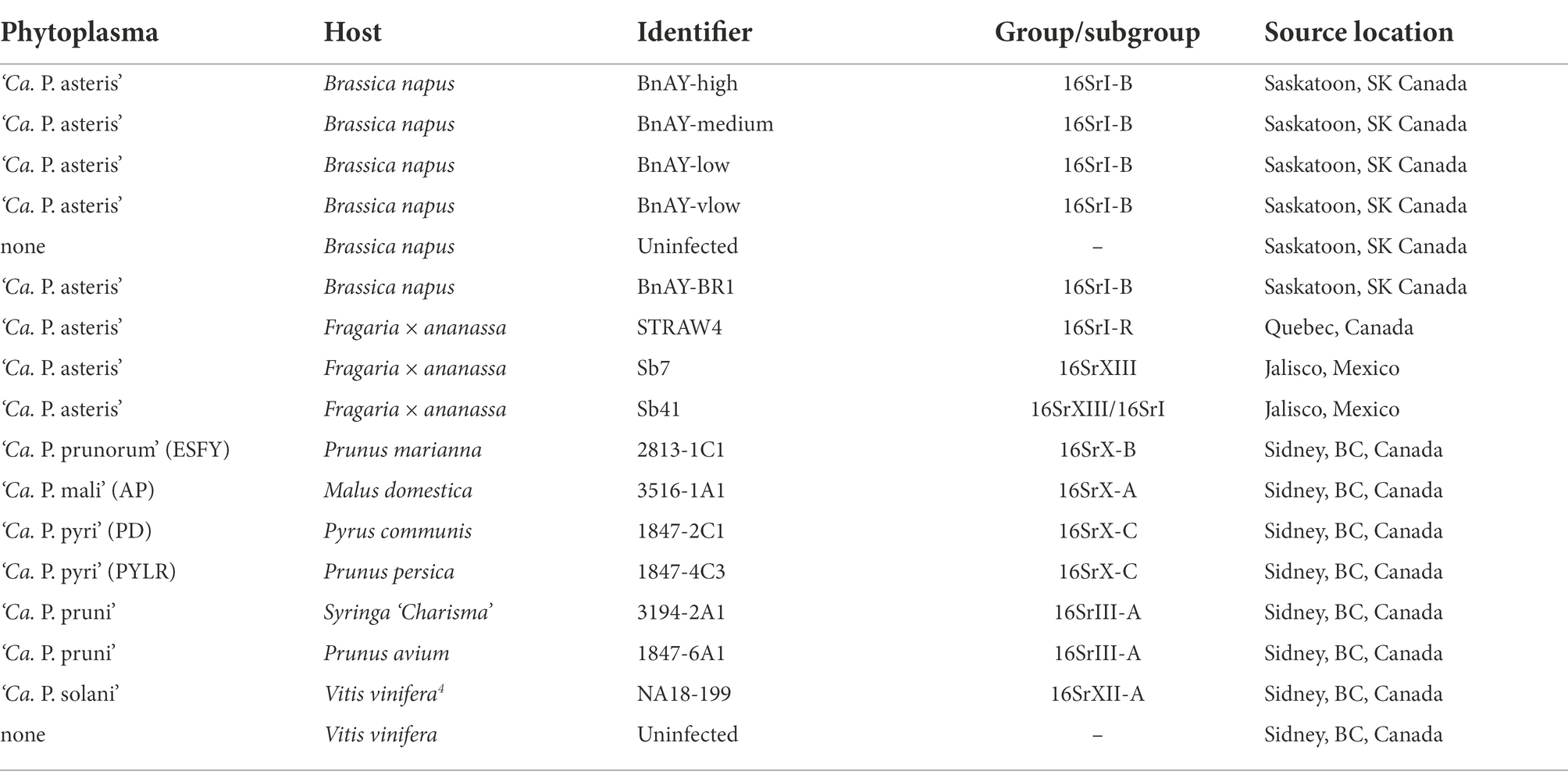
Plant material indicated in Table 1 that was infected with ‘Ca. P. mali’, ‘Ca. P. prunorum’, ‘Ca. P. pyri’, ‘Ca. P. solani’, and ‘Ca. P. pruni’ were maintained in appropriate hosts (Table 1) at the Centre for Plant Health, Canadian Food Inspection Agency, North Saanich, BC. Infected canola was collected at the research farm of Agriculture and Agri-Food Canada in Saskatoon, SK. Strain TW1 was collected in 2018 as previously described (Town et al., 2018), and was used to prepare a dilution series of known phytoplasma concentrations in a background of DNA extracted from healthy B. napus. Uninfected canola DNA was prepared by germinating phytoplasma-free B. napus seeds (Plant Gene Resources of Canada, accession no. CN42942) on filter paper soaked with sterile water, and shoots were collected after 7–10 days of germination in the dark at room temperature. Strain BR1 was collected at the same site as strain TW1, but in 2021. Samples from infected strawberries were collected from fields in Quebec in the summer of 2021 as previously described (Plante et al., 2021). Samples from infected strawberries in Mexican production fields have been described previously (Pérez-López et al., 2017).
DNA extraction and quality control
Midribs were excised from the leaves of infected plants for all samples except TW1 (inflorescence) and healthy canola (shoots from germinated seeds). The tissue (~0.1 g) was cut into ~5 mm pieces, placed into a 2 ml tube with 2 sterile steel beads (3.2 mm, BioSpec Products), and immediately frozen in liquid nitrogen. Frozen tissue was pulverized using a homogenizer (Retsch, model no. MM 400) using 2 pulses of 30 s of shaking at 30 Hz. Powdered samples were then briefly centrifuged, and DNA was extracted using a Qiagen Plant DNA mini kit. DNA was eluted into 100 μl of 10 mM Tris-Cl pH 8.0 (kit elution buffer). DNA concentration was measured using a Qubit Broad Range kit (Invitrogen).
Quantitative PCR (qPCR) was used to determine the level of phytoplasma in each of the DNA extracts from infected tissue samples prior to hybridization and sequencing. Primers and probes were purchased from IDT (Coralville, IA). Their sequences, along with amplification conditions, are provided as supplementary information (Supplementary Table S1). For all samples except ‘Ca. P. pruni’, qPCR used 1x SsoFast Universal Probes Supermix (Bio-Rad), 0.3 μM each primer, and 0.2 μM probe in a final volume of 20 μl. For ‘Ca. P. pruni’, qPCR used 1x SsoFast Universal Probes Supermix, 0.4 μM primer 16SF, 1.2 μM primer 16SR, 0.2 μM probe 16S72, and 1x SsoFast Universal Probes Supermix. qPCR standards were prepared from plasmid DNA containing the cpn60 universal target sequence from the respective phytoplasma (Muirhead et al., 2019a). qPCR standards for ‘Ca. P. pruni’ were prepared using infected cherry DNA calibrated to 200,000 copies/μl, and infected plum DNA at 200 copies/μl. Amplifications used a C1000 thermocycler base with a CFX96 qPCR module and results were analyzed using Bio-Rad CFX Manager v3.1.1517.0823. Statistical correlations between cpn60 input DNA copies measured by qPCR and cpn60 read numbers post-hybridization were calculated using SigmaPlot v14.5.
To prepare a dilution series of ‘Ca. P. asteris’, DNA extracted from infected B. napus inflorescence (strain TW1) was mixed with DNA extracted from uninfected B. napus shoots. Briefly, AY cpn60 copies were measured in the TW1 inflorescence extract using qPCR, then the sample was adjusted to approximately 5×105 cpn60 copies/ul in 10 mM Tris-Cl, pH 8.0. This sample is referred to as BnAY-high. Three serial 10-fold dilutions of BnAY-high were then prepared, with 50 μl of DNA from uninfected canola (Bn-H) added to each dilution and the volume adjusted to 110 μl using 10 mM Tris-Cl, pH 8.0. These “spiked-in” samples are labeled BnAY-medium, BnAY-low, and BnAY-vlow (very low). The DNA concentration and number of AY cpn60 copies/μl were measured in each final pool as described.
PCR amplification and cloning of target gene sequences
The 16S rRNA-encoding gene locus was amplified from DNA extracts using a nested PCR strategy consisting of primers P1 (Deng and Hiruki, 1991) and P7 (Schneider et al., 1995) in a first round, generating a product of >1.8 kb. This PCR product was diluted 1:30, then 2 μl was used as template in a secondary PCR step using primers R16F2n and R16R2 (Gundersen and Lee, 1996), which generated the ~1.2 kb F2nR2 amplicon that is commonly used for phytoplasma detection and typing(Zhao et al., 2009). Both rounds of PCR for 16S used 1x PCR buffer (Invitrogen), 2.5 mM MgCl2, 500 nM dNTPs, and 400 nM each primer using previously described methodologies (Pérez-López et al., 2017). Thermal cycling conditions were 95°C, 10 min (1x), 95°C, 1 min; 55°C, 1 min; 72°C, 1.75 min (35X), 72°C, 10 min (1x). The universal target region from the cpn60 gene (Dumonceaux et al., 2014) was amplified using a primer cocktail as described at cpnclassiphyr.ca (Muirhead et al., 2019a). Amplicons were generated from the rp operon of AY infected samples using nested PCR with primers rpF1 and rpR1 (Lim and Sears, 1992) in the first round, and rp(I)F1A/rp(I)R1A in the second round as described (Lee et al., 2004).
Amplicons were cloned into the vector pGEM-T Easy (Promega, Madison, WI) according to the instructions provided, then plasmids were transformed into chemically competent E. coli TOP10 cells (Life Technologies). 5–6 individual clones were sequenced from each amplicon using a commercial sequencing service (Eurofins Genomics, Toronto, ON).
Hybridization probe design
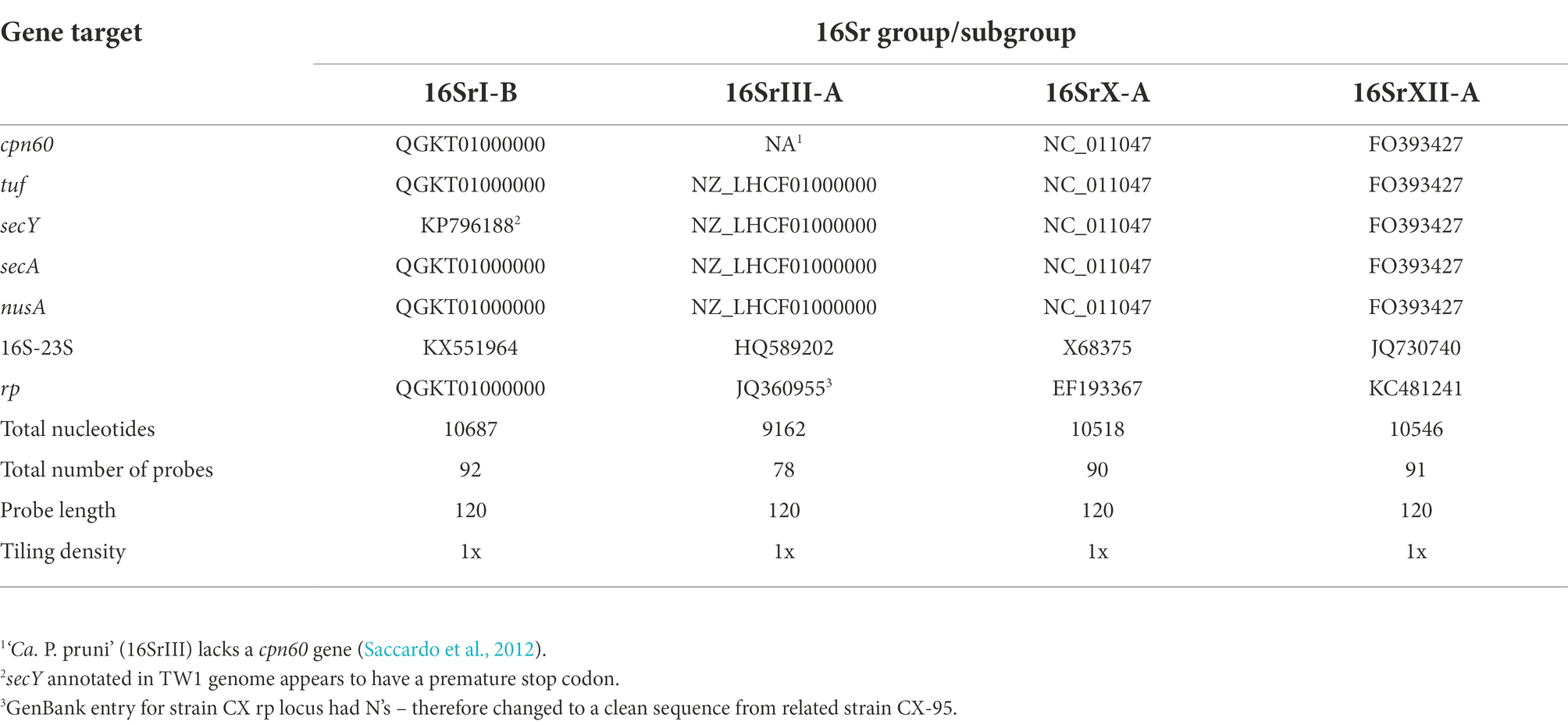
Gene targets for probe design were selected from publicly available sequences at GenBank (Table 2). 27 genes were selected from genome sequences and individual gene sequences, with 7 targets from each of ‘Ca. P. asteris’ (16SrI-B); ‘Ca. P. mali’ (16SrX-A); ‘Ca. P. solani’ (16SrXII-A), and 6 genes from ‘Ca. P. pruni’ (16SrIII-A). A total of 351 probes were designed with a length of 120 nucleotides and 1x tiling density using the IDT X-gen design tool found.1 Probe sequences are provided as Supplementary File S1.
Hybridization and sequencing
Detailed protocols for DNA preparation for hybridization and sequencing have been published elsewhere (Dumonceaux et al., 2017), and these were generally followed. Wherever possible, DNA was diluted to 2.5 ng/ul in a total volume of 100 μl of 10 mM Tris-Cl pH 8.0. If the DNA concentration was below this level, it was used at its extracted concentration, with the volume adjusted to 100 μl for shearing. Shearing proceeded using a Bioruptor 300 (Diagenode # B01020001) in 0.2 ml shearing tubes (Diagenode #C30010015) on a setting of “high” with 30 cycles of 30 s on, 45 s off, and cooling to 4°C. Sheared DNA was concentrated using Amicon YM-30 filter membranes to a volume of 60 μl. DNA concentration was determined using a Qubit Broad Range kit (1 μl). Illumina adaptors and indices were added using a NEBNext Ultra DNA library prep kit for Illumina (NEB, cat no E7370) according to the manufacturer’s instructions. Prior to index addition, size selection was used to isolate fragments of 400–500 bp by using 35 μl of SPRI beads (Cytiva) in the first bead selection and 25 μl in the second bead selection. Index addition proceeded using 8 cycles of PCR under the conditions recommended by the manufacturer, and indexed fragments were purified using a 1:1 volume ratio of SPRI beads (Cytiva) and eluted in a volume of 30 μl. Results were examined using a TapeStation (Agilent) and DNA concentration was determined using a Qubit High Sensitivity (HS) kit.
Hybridization was performed using an xGen hybridization and wash kit (IDT) and the recommended protocols (IDT xGen hybridization capture of DNA libraries manual version 4, 2019). Up to 500 ng of indexed DNA was added to each hybridization reaction along with xGen universal blockers (TS mix, IDT). All samples were hybridized with the same set of capture probes containing sequences from all seven markers from all of the targeted phytoplasmas (Table 1). Hybridizations proceeded at 65°C for 16 h. After washing, libraries were purified using a 1:1 volume ratio of SPRI beads (Cytiva) and amplified for 14 cycles using the x-Gen library amplification primer (IDT) and KAPA HiFi HotStart ReadyMix (Roche) under cycling conditions recommended by the manufacturer. After another SPRI purification, DNA concentration was determined using a Qubit HS kit. If the DNA yield was insufficient (under approx. 1 ng/ul), a second amplification reaction was performed using 1 μl of the first amplification reaction with 19 μl water and the same primer; products were then re-purified using SPRI beads. Samples were pooled to 4 nM and sequenced using an Illumina nano kit v2 (500 cycles) using a MiSeq instrument according to the manufacturer’s recommendations.
Data analysis
Illumina reads were processed with trimmomatic (v0.39) to remove adapters and low quality bases (<Q15) and R1/R2 reads were merged using flash2 (v2.2.00). Merged reads were mapped to the sequences of the genes used for hybridization probe design using bowtie2 --local (v2.4.4). The reference sequences used for mapping are included as Supplementary File S2. Reads mapped to each target sequence were extracted using samtools (v1.9), and the corresponding fastq files were then assembled as individual bins using transabyss (v2.0.1). Assemblies were filtered to include only contigs >500 nucleotides.
To remove off-target reads that mapped to undesired 16S sequences (16S genes from other bacteria, along with host chloroplast and mitochondrial genes), assembled contigs that were not phytoplasma 16S sequences were identified using BLAST. These contigs were then added as reference sequences for a second round of mapping for 16S-mapped reads only using bowtie2 --local to remove these non-target sequences from the assembly dataset. This was done only in cases where the initial assembly using the mapped 16S genes generated short or only nonspecific contigs. The code used for mapping and assembly is provided as Supplementary File S3.
Phylogenetic analysis
Sequences assembled from the post-hybridization sequencing and mapping data were oriented manually by BLAST to determine sequence orientation (coding, or reverse-complement), then reverse-complementing as necessary. Oriented sequences were aligned using clustalw, then trimmed manually to the length of the shortest sequence in the alignment. Phylogenetic relationships among the sequences were inferred by using the Neighbor Joining algorithm (Saitou and Nei, 1987) in MEGA X (Kumar et al., 2018). Trees were bootstrapped using 1,000 replications, and consensus trees were reported with percentages of trees in which the associated taxa clustered together indicated next to each branch. The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al., 2004).
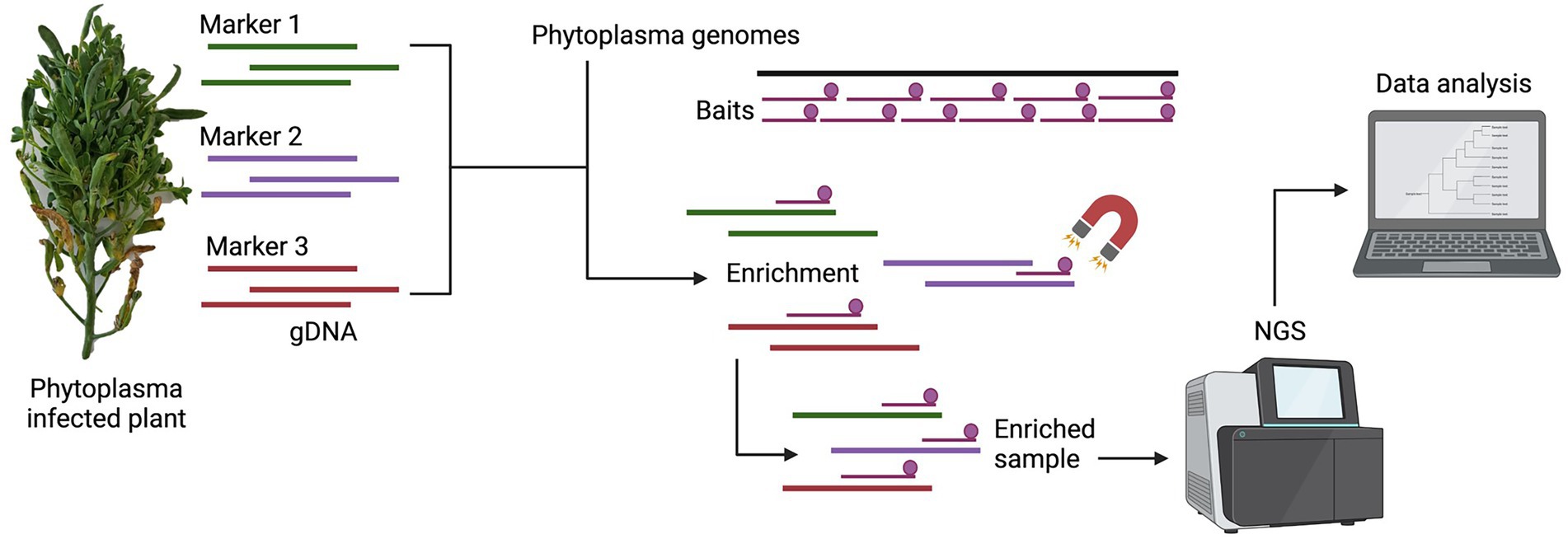
A schematic representation of the methodology used in this study is presented in Figure 1.
Results
DNA extraction and qPCR
DNA yield from infected leaf midribs was somewhat variable, ranging from under 1 ng/ul (‘Ca. P. solani’, Sb41) to approximately 15 ng/ul in lilac infected with ‘Ca. P. pruni’ (Table 3). The mean DNA concentration for all samples was 8.74 ng/ul. Similarly, samples displayed a wide range of phytoplasma levels in the infected plant tissue; Cq values were quite high in some samples (25–27 for ESFY, AP, and PD) but lower in other samples approximately 20 in the ‘Ca. P. pruni’ and ‘Ca. P. solani’-infected samples (Table 3). The sample from the canola plant infected with ‘Ca. P. asteris’ strain BR1 had the lowest Cq (14) of the samples examined, reflecting the highest phytoplasma concentration. Samples of canola DNA that were purposefully constructed to contain various levels of phytoplasma DNA had Cq values ranging from 17.62 (BnAY-TW1) (‘Ca. P. asteris’-high) to 27.12 (BnAY-TW1-vlow). All samples were successfully sheared and indexed, regardless of the DNA input amounts.
Hybridization and Illumina sequencing
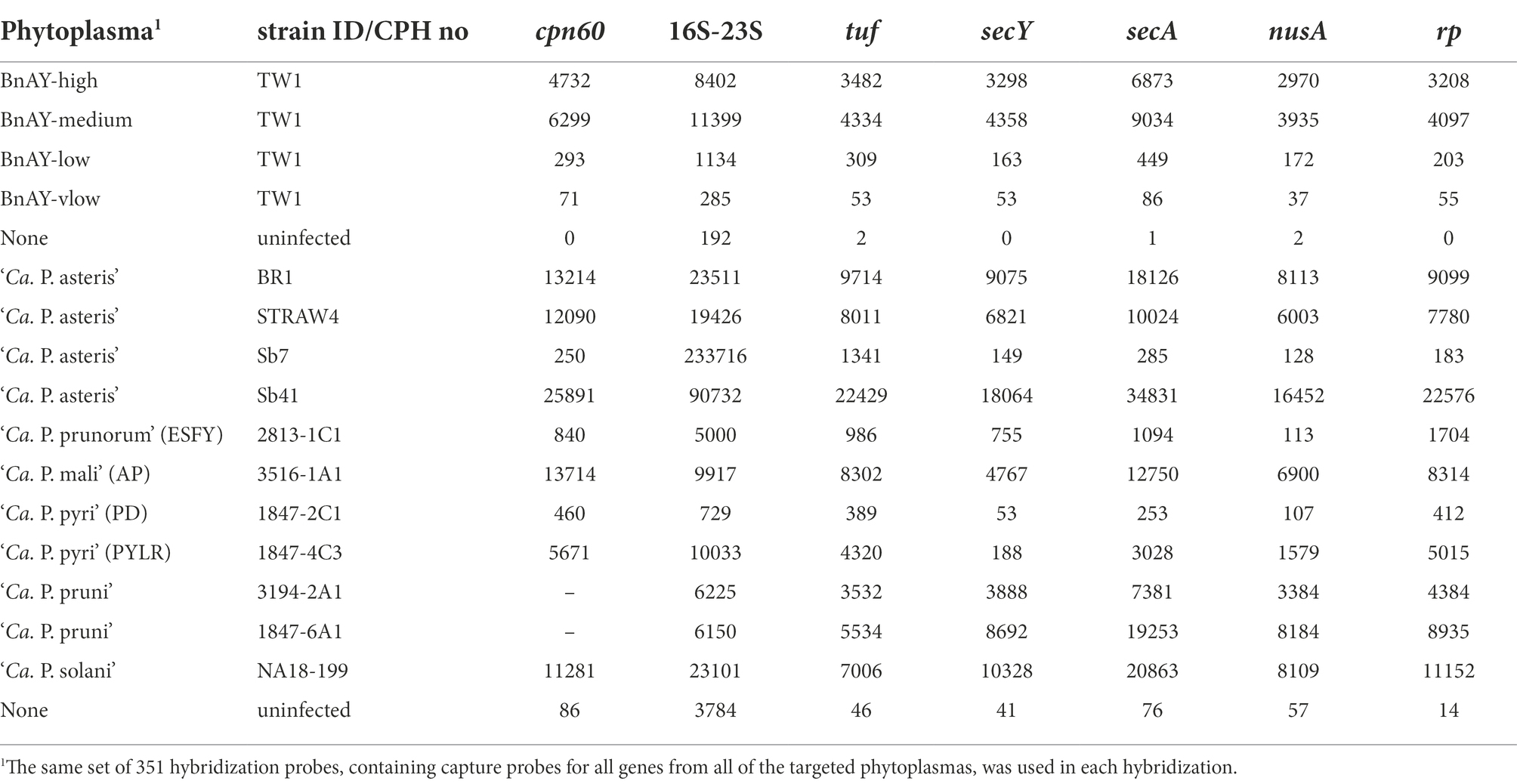
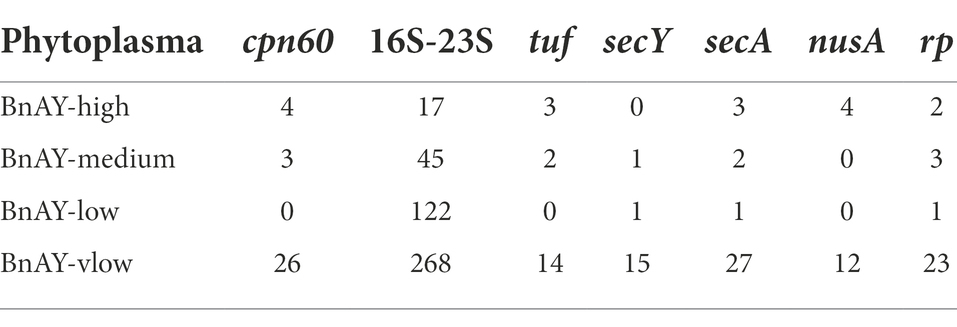
Post-hybridization read numbers mapping to each gene are shown in Table 4. Depending on the amount of phytoplasma DNA in the sample, the configuration of the sequencing run, and other technical variables, hundreds to thousands of reads typically mapped to each gene. Fewer reads were observed in the lower concentration samples such as ESFY and PD, with the latter showing one of the lowest proportions of reads mapping at ~3% (Table 4). In most cases, the 16S ribosomal gene had the highest number of mapped reads. Samples from uninfected canola and grape showed lower numbers of mapped reads compared to the corresponding infected plants, especially for the protein-coding genes (Table 4). Without hybridization, samples from the canola phytoplasma mixtures produced very few reads mapping to the target genes (Table 5).
The qPCR assays targeting the cpn60 gene (Supplementary Table S1) provided input concentrations of phytoplasma. Examining the relationship between the initial phytoplasma levels in each sample and the number of reads mapping to cpn60 generated revealed a significant correlation (Pearson r2 = 0.696, p < 0.05; Spearman’s ρ = 0.770, p < 0.05), suggesting that the number of reads mapping was related to the starting concentration of phytoplasma in the sample, at least within the same Illumina run. Such a determination could not be made for the other protein-coding genes due to the lack of gene-targeted qPCR assays, but this correlation was not evident in the 16S ribosomal gene datasets (Table 4). The actual number of reads mapping to each target was variable across samples and was quite high in some samples, even those that had relatively low concentration of phytoplasma such as STRAW4 (Table 4).
A large number of reads typically mapped to the 16S ribosomal gene target in all samples, including uninfected tissue. In fact, the large number of reads mapping to the 16S ribosomal gene in each sample were found to include many reads that did not correspond to the target gene, including host chloroplast and mitochondrial genes, 16S ribosomal genes from non-phytoplasma bacteria, and other nonspecific DNA sequences. Examination of the nucleotide identities of the post-hybridization reads that mapped to the 16S ribosomal gene compared to the probe sequences using BLAST revealed that the lower spike levels contained a substantial proportion of reads with <95% nucleotide identity to any of the probes (Supplementary Figure S1A). These non-target reads interfered in some samples with the assembly process, to the extent that the ‘Ca. P. asteris’ samples spiked at low and very low levels assembled only nonspecific host chloroplast sequences (Supplementary Table S2). Other samples, such as PYLR, assembled only 16S contigs that were too short for classification using the iPhyClassifier (Supplementary Table S2). Incorporating a second mapping step that included these nonspecific assembled sequences as mapping targets provided a much higher proportion of reads (average read length 274 bp) with >98% identity to a probe sequence (Supplementary Figure S1B), and greatly improved most of the 16S assemblies (Supplementary Table S2). This was most pronounced with the ‘Ca. P. asteris’ mixtures, with the medium and low titer samples yielding 16S assemblies that were > 2 kb after this step, and the very low sample yielding a 925 nucleotide 16S assembly compared to no detectable 16S sequence after the initial mapping (Supplementary Table S2). This two-step mapping strategy for 16S rRNA sequences was not required for other samples. In particular, the 16SrIII samples generated 16S assemblies >2.4 kb by mapping only to the target 16S gene sequence, so in this case a two-step mapping strategy was not necessary (Supplementary Table S2). In contrast, the initially mapped reads from the spiked canola samples corresponding to a protein-coding gene (cpn60) had >98% sequence identity to a probe sequence at all spike levels, and therefore represented high-quality sequence reads that were used in the assemblies (Supplementary Figure S1C).
Assembly of the mapped reads for each gene generated sequences that ranged from 599 nucleotides (PYLR secY) to 3,225 nucleotides (‘Ca. P. pruni’ 6A1 secA) (Table 6). Most of the sequences were at least 1,500 nucleotides, with an average contig length of 1927 nucleotides and a median of 1867 nucleotides. The secA assemblies were the longest (average of 2,812 nucleotides), while the nusA assemblies tended to be shorter (average of 1,438 nucleotides). This is consistent with the target gene lengths (secA is the longest target gene at ~2,500 nucleotides, and nusA the shortest at ~1,075 nucleotides).
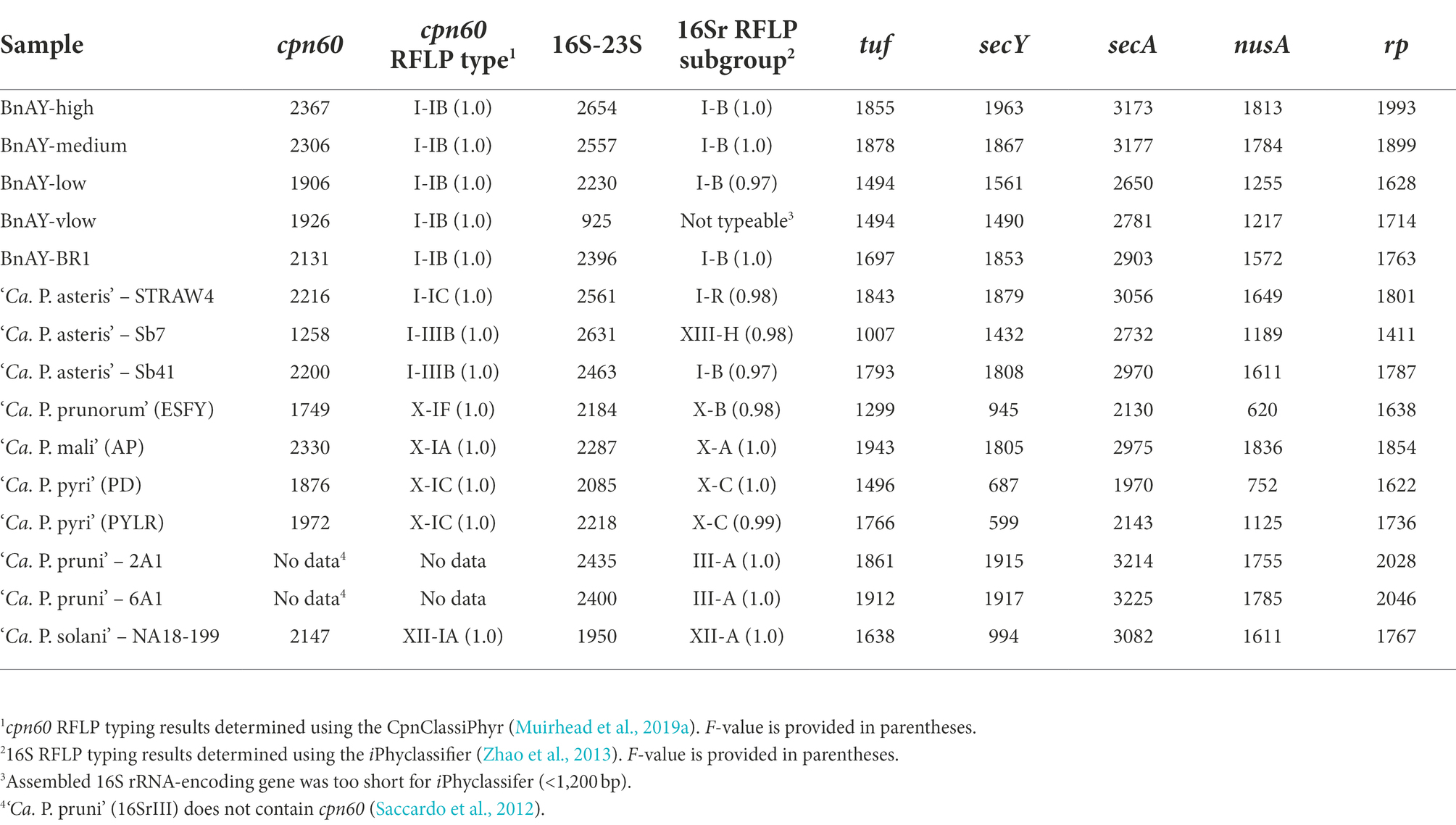
Table 6. Longest assembled contig lengths (in base pairs) and RFLP-based typing results (16S and cpn60 only) for each gene in each sample.
Sequence typing and phylogenetic analysis
16S ribosomal RNA gene and cpn60 sequence types
All samples yielded 16S ribosomal RNA gene and cpn60 sequences that typed correctly using the corresponding RFLP-based classifiers (Table 6). In all cases except Sb7 (Strawberry – Mexico) and STRAW4 (Strawberry-Quebec), the cpn60 and 16S sequence types were in agreement. For Sb7, the cpn60 sequence generated typed as cpn60 I-IIIB: maize bushy stunt (MBS), while the 16S sequence corresponded to the group 16SrXIII-H. For STRAW4, the cpn60 typed as I-IC while the 16S sequence typed as 16SrI-R (Table 6).
Phylogenetic analysis of target gene assemblies
16S rRNA gene sequences
16S rRNA-encoding gene sequences generated from the samples that were infected with group 16SrX phytoplasmas (AP, ESFY, PYLR, PD) clustered with their respective reference sequences (Figure 2A), in most cases with nearly zero branch length, indicating sequence identity. Although the hybridization probes were designed using the 16S rRNA-encoding gene sequence of 16SrX-A (strain AT; Table 1), hybridization of other subgroups 16SrX-C (PYLR/PD) and 16SrX-B (ESFY) was also successful. Similarly, the 16S ribosomal RNA gene sequences generated from samples infected with 16SrIII clustered with their respective reference sequences with near sequence identity (Figure 2A). The grape sample infected with 16SrXII was slightly different from the strain used for hybridization probe design, and clustered with a “stolbur” phytoplasma strain within the same subgroup 16SrXII-A.
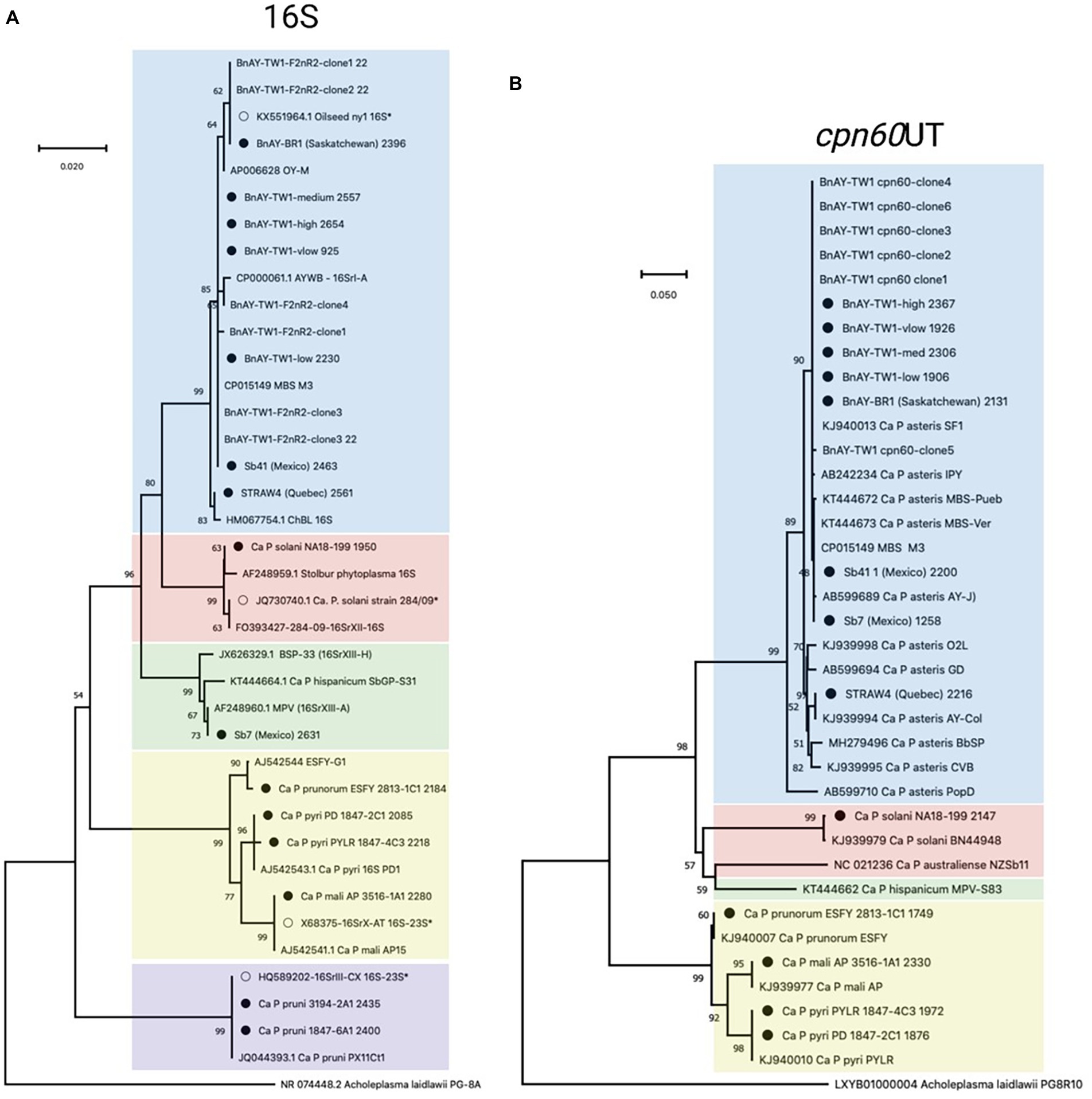
Figure 2. Phylogenetic analysis of 16S (A) and cpn60 UT (B) sequences generated in this study. 16S sequences corresponded to the F2nR2 fragment (~1.2 kb), and cpn60 sequences were trimmed to the universal target length (~550 bp) using the CpnClassiPhyR (Muirhead et al., 2019a). Phylogenetic analysis was performed using the Neighbor Joining algorithm using 1000 replicates, as described in Methods. Sequences corresponding to the samples analyzed in this study (Table 1) are indicated with a filled circle, while samples with an open circle represent the sequences used for hybridization probe design (Table 2). Samples are color-coded according to the 16Sr groups represented within the samples and the probes described in this study – 16SrI (blue); 16SrXII (pink); 16XrXIII (green); 16SrX (yellow); 16SrIII (purple; 16S only).
The samples that were infected with various 16SrI subgroups generated 16S rRNA gene sequences that were generally consistent with the expected groupings. The AY-infected strawberry from Quebec (STRAW4) yielded a sequence that clustered with a strain of 16SrI-R (Figure 2A), consistent with the RFLP-based typing results (Table 6). The AY-infected canola samples from Saskatchewan provided 16S gene sequences that mostly clustered with strains from subgroup 16SrI-B, as expected from the RFLP typing results. This included strain BnAY-BR1, which was collected in 2021 from the same field as strain BnAY-TW1 (collected in 2017). Examination of F2nR2 clone sequences generated from strain BnAY-TW1 revealed that 5/6 clones clustered with 16SrI-B, while one (clone 4) was more closely related to 16SrI-A. The clones that clustered within 16SrI-B were therefore differentiated into two types, consistent with the RFLP typing results for these clones. The sequences that were assembled from the hybridization, however, all appeared to cluster with the 16SrI-B sequences. These sequences were slightly distinct from the sequence used for the design of hybridization probes (Figure 2A).
Strawberry samples Sb41 and Sb7, which were collected in Jalisco, Mexico (Pérez-López et al., 2017), provided 16S gene sequences that clustered with 16SrI-B (Sb41) and 16SrXIII-A (Sb7) (Figure 2A). Despite the previously reported double infection of sample Sb41 with 16SrXIII and 16SrI (Pérez-López et al., 2017), no evidence of 16S rRNA gene sequences from 16SrXIII was found in sample Sb41.
cpn60
The length of the cpn60 assemblies (Table 6) allowed the use of more than the ~550 bp universal target region (Muirhead et al., 2019b) for phylogenetic analysis. The sample infected with 16SrX-A generated a cpn60 sequence that was identical to that of the strain that was used for hybridization probe design (Supplementary Figure S2). There is no full-length cpn60 sequence available for ESFY (16SrX-B), but the PYLR/PD (16SrX-C) cpn60 sequences were identical to a strain from the GenBank database (Supplementary Figure S2), indicating that the assemblies were correct for the 16SrX-C samples. In addition, the cpn60 sequence from the 16SrXII-infected sample clustered with the corresponding sequence used for hybridization probe design (Supplementary Figure S2).
Sequences generated from infected canola plants in Saskatchewan, including BnAY-TW1 and BnAY-BR1, clustered with the sequence used to generate the hybridization probes with zero branch length (Supplementary Figure S2). However, the sequences from infected strawberries clustered independently with no reference sequence (STRAW4, Quebec), or with subgroup 16SrI-B detected in samples Sb7 and Sb41 from Mexico.
Trimming the sequences to the universal target using the CpnClassiPhyr (Muirhead et al., 2019a) permitted the use of an expanded set of reference sequences, along with RFLP analysis. This trimming provided results that were consistent with the longer sequences (Figure 3A), and further showed that the 16SrX samples corresponding to ESFY, PYLR, and PD clustered with their respective reference sequences, and that the sample from infected strawberries in Quebec (STRAW4) clustered with the cpn60 UT I-IC sample (AY-Col). Moreover, all six cpn60 UT amplicon clones generated from strain BnAY-TW1 were identical to the assembled sequence and to the reference sequence used to design the hybridization probes (Figure 2B).
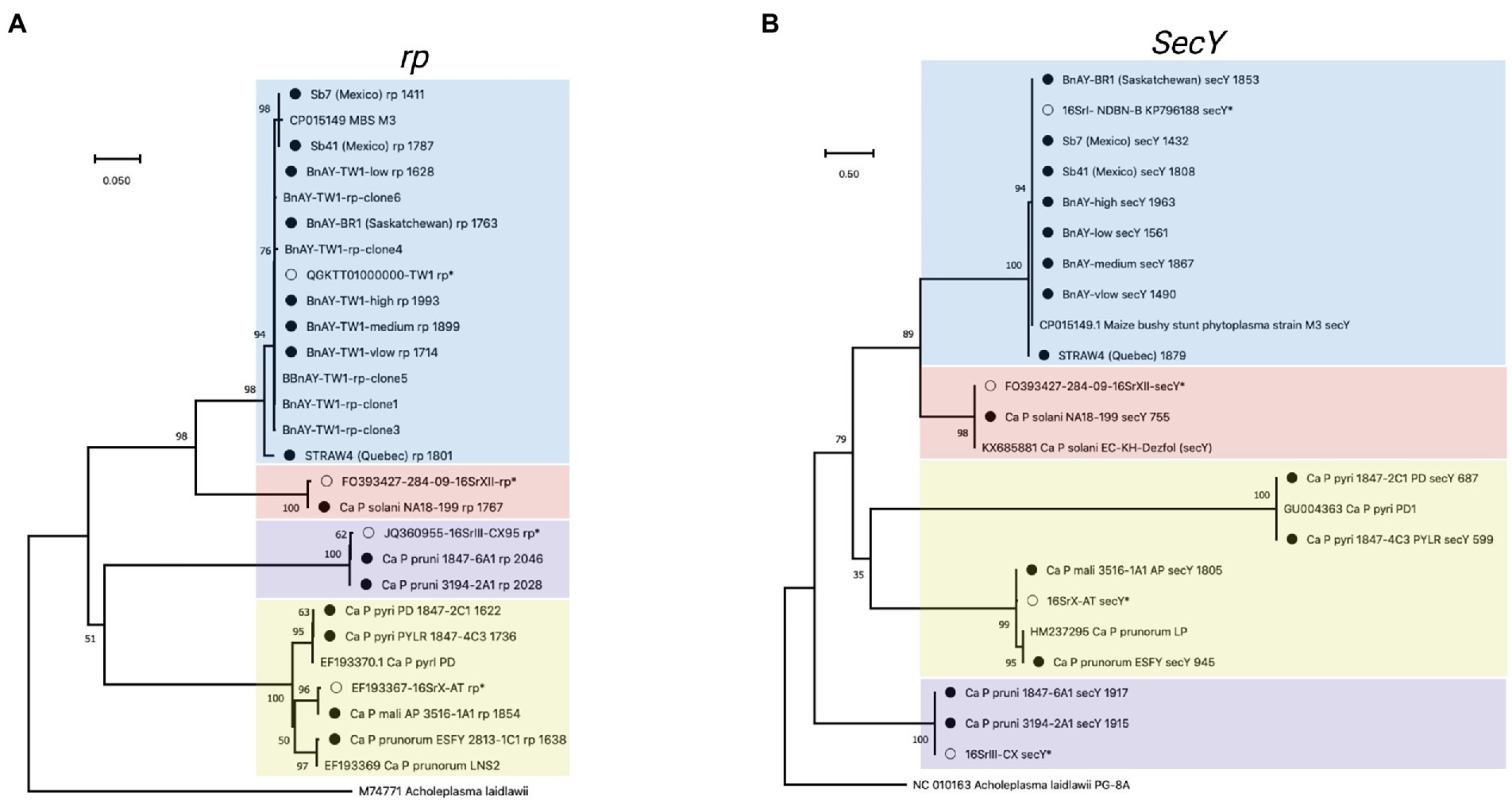
Figure 3. Phylogenetic analysis of rp (A), and (B) secY sequences generated in this study. Sequences were trimmed manually to the length of the shortest sequence analyzed (see Methods). Samples are indicated as described for Figure 2, and the length of the original assembly prior to trimming (Table 6) is indicated for each sample (filled circles).
rp
Results of phylogenetic analysis using the rp sequences assembled from hybridized infected samples are shown in Figure 3A. A similar theme was observed; sequences obtained from infected ‘Ca. P. mali’ clustered with the sequence used to generate the probes, while the sequences from the other 16SrX subgroups represented by strains ESFY, PYLR, and PD clustered separately, and were identical to their respective reference sequences. The samples infected with 16SrIII and 16SrXII also generated sequences that were identical to the sequence used for hybridization probe design. The AY infected canola samples were all virtually identical to one another and to the probe sequence, along with 5 PCR-amplified rp clones from BnAY-TW1. The infected strawberry from Quebec generated a distinct sequence, while the sequences from infected strawberries in Mexico clustered with the aster yellows rp sequences.
secY
Despite the fact that the secY reads generated assemblies that were, on average across all strains analyzed, longer (average length 1,514 bp – Table 6) than those of the shorter target gene nusA (average length 1,438 bp – Table 6), the alignment and trimming of the secY sequences to a common fragment resulted in the shortest sequence comparison among the genes analyzed; phylogenetic analysis was based on a fragment of only ~530 nucleotides. This was mostly due to the shorter assemblies that were generated for ‘Ca. P. prunorum’ (ESFY) and ‘Ca. P. pyri’ (PD and PYLR) – see Table 6. The secY sequences assembled from infected samples showed that this gene is highly discriminatory for 16SrX strains (Figure 3B). The >1,800 nucleotide sequence that was assembled from the ‘Ca. P. mali’-infected sample, as expected, was almost identical to the sequence used for probe design, while the ESFY (16SrX-B) sequence was identical to its reference sequence and distinct from the ‘Ca. P. mali’ (16SrX-A) sequence. However, the secY sequences that were assembled from ‘Ca. P. pyri’ (16SrX-C) displayed a long branch length compared to the other 16SrX samples, consistent with their lower nucleotide sequence identity (~90%). As with other genes, the secY sequences generated from 16SrIII samples were identical to the target sequence, and the sequence assembled from the ‘Ca. P. solani’-infected sample was nearly identical to its target sequence. secY sequences were less discriminatory with the 16SrI samples, as all sequences except STRAW4 were identical, including the closely related samples generated from the infected strawberries in Mexico that were also identical to MBS secY (Figure 3B).
secA
The longest target gene in the MLST scheme was secA, which was approximately 2,500 nucleotides. Phylogenetic analysis was therefore based on a relatively long fragment of approximately 1,970 nucleotides, corresponding to the length of the shortest sequence generated (‘Ca. P. pyri’ PD). The sequence generated for ‘Ca. P. mali’ was identical to the sequence used for group 16SrX probe design, while the sequences obtained from the other 16SrX-infected samples clustered separately (Figure 4A). While there was no reference sequence available for ESFY, the PYLR and PD sequences were identical to a reference sequence from ‘Ca. P. pyri’ (isolate PD1; Figure 4A). Similarly, the 16SrXII- and 16SrIII-infected samples generated secA sequences that were identical to the sequence used for probe design (Figure 4). The secA sequences of the infected strawberries from Mexico clustered with MBS phytoplasma (16SrI-B) secA sequence, although the sequence differences were not as great as those observed with other markers. The infected strawberry sample from Quebec (STRAW4) provided a secA sequence that clustered separately from the other AY-infected samples. Finally, the sequences assembled from infected canola in Saskatchewan were all identical to the sequence used for probe design, corresponding to the BnAY-TW1 secA sequence (Figure 4A).
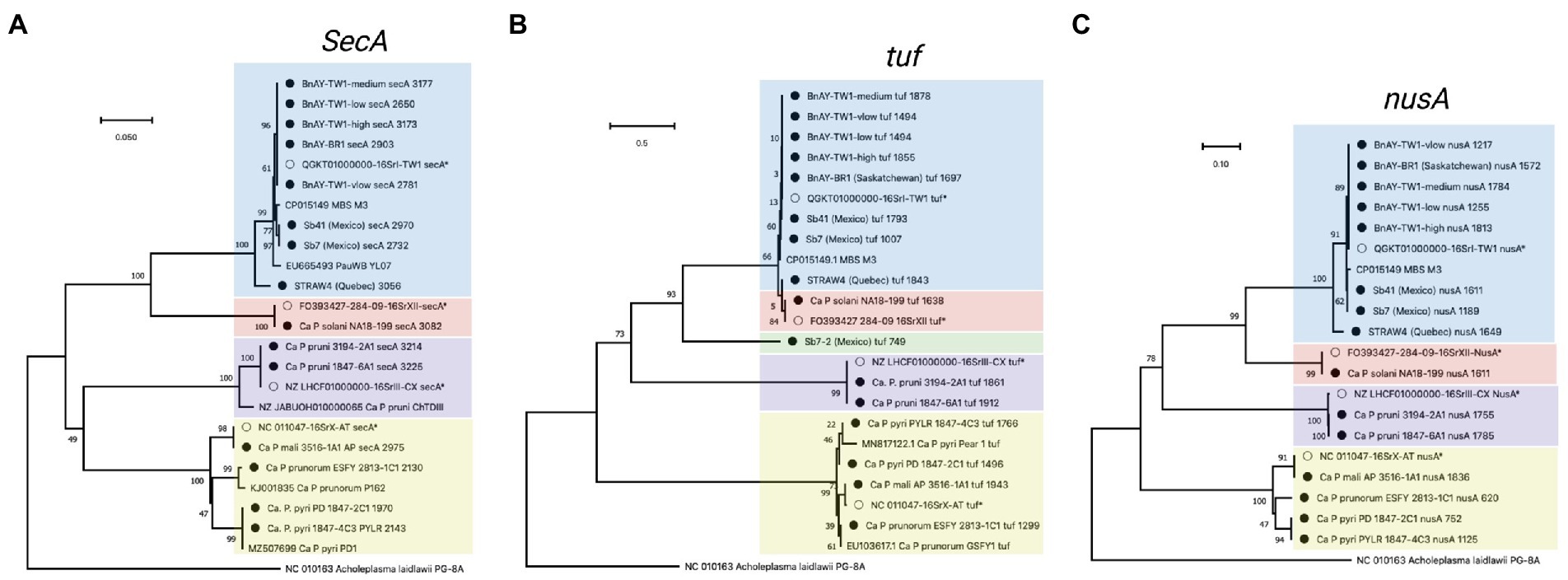
Figure 4. Phylogenetic analysis of secA (A), tuf (B), and nusA (C) sequences generated in this study. Sequences were trimmed manually to the length of the shortest sequence analyzed (see Methods). Samples are indicated as described for Figure 2, and the length of the original assembly prior to trimming (Table 6) is indicated for each sample (filled circles).
Tuf
The tuf gene sequences also provided results that were consistent with expectations. The 16SrX samples yielded sequences that clustered together more tightly than the secY sequences, but the different subgroups were readily differentiated (Figure 4B). Again, the ‘Ca. P. mali’ sequence was nearly identical to the hybridization target sequence, while the other 16SrX infected samples yielded sequences that were somewhat distinct. The sample infected with ‘Ca. P. prunorum’ (ESFY) provided a sequence that was nearly identical to a reference sequence from ESFY. The ‘Ca. P. pyri’ samples were identical to one another, and clustered with a short reference sequence for this taxon with a sequence identity of >95%. The 16SrIII and 16SrXII-infected samples also generated sequences that were identical to the respective hybridization target sequence. Unlike secY, tuf gene sequences were able to differentiate the MBS phytoplasma sequences (Sb7 and Sb41) from the canola samples that were also infected with 16SrI-B, all of which yielded identical sequences. The infected strawberry sample from Quebec (STRAW4) provided a distinct tuf sequence from the other 16SrI infected samples. The infected strawberry sample from Mexico, Sb7, generated a second, shorter tuf sequence (749 nucleotides) that was unlike any previously reported tuf sequence, but shared ~88% sequence identity with 16SrXII-B and 16SrXII-A 284/09 strain.
nusA
The nusA assemblies tended to be the shortest across all samples analyzed, consistent with the length of the nusA gene (1,074 nucleotides, the shortest gene analyzed). These sequences were trimmed to 620 bp, which was the length of the shortest nusA assembly (from ESFY). Like secY and tuf, nusA sequences from 16SrX phytoplasmas placed ‘Ca. P. pyri’ (PYLR/PD) in a clade with ‘Ca. P. prunorum’ (ESFY), with ‘Ca. P. mali’ (AP) forming a distinct but related group (Figure 4C). No nusA reference sequences are available for ESFY and PYLR/PD. Sequences assembled from 16SrIII and 16SrXII were identical to the hybridization target sequences, as observed with other genes. The nusA sequences could differentiate weakly the AY sequences assembled from the infected Mexican strawberries, and all nusA sequences assembled from the infected canola were identical to one another and to the target sequence (Figure 4C). STRAW4 clustered separately but within the AY group, as with the other genes (Figure 4C).
Discussion
The detection, identification, and classification of phytoplasma strains has typically relied upon PCR amplification and sequencing of the 16S rRNA-encoding gene (Zhao et al., 2013). Due to the well-recognized limitations of this approach for various phytoplasma groups, a wide variety of MLST schemes has been described, nearly all of which use PCR amplification and sequencing of specific protein-coding marker genes to improve strain differentiation. An alternative approach to PCR-based MLST was developed and applied to a variety of phytoplasma ribosomal groups, which produces the sequences of seven molecular markers simultaneously and with high accuracy. An important feature of this hybridization-based MLST approach is that it is independent of the design of “universal” primers targeting a subset of phytoplasmas, which overcomes a limitation of some MLST schemes that have been described. In this MLST scheme, hybridization-based gene enrichment was demonstrated to be advantageous. For example, the number of sequencing reads for the BnAY-high sample that mapped to all genes before hybridization was 33, compared to 32,965 reads post-hybridization. These numbers of reads resulted in the assembly of rather long sequences for most of the gene targets (global average of 1929 nucleotides) that are commonly supported by at least hundreds or thousands of reads, providing confidence in the results that are obtained. Reliable reads of this length would be very difficult to obtain using Sanger sequencing from the ends of a clone or amplicon. The assembly of longer protein-coding sequences containing both coding and flanking non-coding regions greatly improves phylogenetic resolution (Gardner et al., 2020). Unlike PCR-based methods, hybridization and assembly does not provide reads with ends that are defined by primer binding locations; therefore, the phylogenetic analysis used fragments that were trimmed manually to the length of the shortest assembly. The ends of the assembled sequence are roughly defined by the sequences of the hybridization probes and are affected by the number of reads mapping to that gene in a given sample. This, in turn, is related to the amount of phytoplasma in the analyzed sample.
Hybridization probe-based detection, differentiation, and classification of difficult-to-culture phytopathogenic bacteria has been previously investigated. For example, in a pioneering study of phytoplasmas, dot and Southern hybridization were used to differentiate phytoplasma strains (Lee et al., 1990, 1992). More recently, enrichment approaches for determining whole-genome sequences of phytoplasmas have been described, using an antibody-based protocol that depletes a sample of eukaryotic DNA (Nijo et al., 2021). An alternative protocol using hybridization probes, similar to that described here, was used to enrich samples infected with the phloem-limited citrus pathogen, ‘Candidatus Liberibacter asiaticus’ for target bacterial DNA. This facilitated the assembly of the complete genome of the pathogen, including from samples with low levels (Cq ~30) of the target DNA (Cai et al., 2019).
The results presented here demonstrate that, similar to the observations of (Cai et al., 2019), phytoplasma-infected samples with a low concentration can still generate very long, phylogenetically informative assemblies. Ultimately, a standard fragment length for MLST analysis could be implemented bioinformatically using a tool such as cutadapt, which can trim sequences between two specified, degenerate sequences- this tool is used to trim cpn60 sequences to the universal target by the CpnClassiPhyr (Muirhead et al., 2019a). In almost all cases, for 16S ribosomal and cpn60 sequences, the hybridization-based MLST approach provided sequences that were of sufficient length and quality for typing using the relevant RFLP-based classifiers, providing additional data that was useful for accurate classification of the samples.
While the hybridization-based approach obviates the need for the design of PCR primers, clearly the phytoplasmas that are targeted are limited to those that possess sequences that are closely related to those of the hybridization probes. The capture of phytoplasma-derived DNA fragments that do not quite match the sequences of the hybridization probes (“off-target” hybridization) is desirable because it can increase the number of distinct phytoplasmas that can be profiled (Ranwez et al., 2011). Some amount of off-target hybridization was observed, consistent with other hybridization-based approaches for determining molecular marker sequences (Gasc and Peyret, 2018). For example, the ribosomal group X-infected samples examined in this work included 16SrX-A, which exactly matches the hybridization target sequence, along with other group X subgroups (16SrX-C and 16SrX-B), which did not exactly match the hybridization probe sequences. In most cases, the lack of a perfect match to the capture probes did not prevent the appearance of the off-target reads in the assembly dataset, since relatively long, robust assemblies were observed for even low concentration samples such as PD. However, secY generated off-target assemblies of lesser quality, which is presumably related to the fact that these sequences are more distinct among 16SrX strains. Off-target hybridization was also observed in samples that were infected by phytoplasmas from 16Sr groups that were not represented at all in the hybridization panel, such as Sb7 and Sb41. For example, we observed evidence of a novel tuf sequence most likely corresponding to that of 16SrXIII (‘Ca. P. hispanicum’), which was found in the Sb7 tuf assembly and had no match to anything known at the GenBank database. In a previous study (Pérez-López et al., 2017), sample Sb41 showed evidence of double infection with an on-target (16SrI) and off-target (16SrXIII) phytoplasma. Sample Sb41 had low levels of 16SrXIII and slightly higher levels of 16SrI (Table 3), but only assemblies s corresponding to 16SrI (which typed as closely related to MBS phytoplasma, consistent with their geographic origin in Mexico) were observed. Sample Sb7, however, had much higher levels of 16SrXIII, and was demonstrated to be additionally infected with 16SrI (Table 3), although the levels of the latter were not measured. Nevertheless, assemblies generated from Sb7 mostly corresponded to 16SrI, except for the 16S rRNA gene, which was from 16SrXIII (Table 6). It appears therefore that off-target hybridization does occur, but inefficiently in the case of sequences from phytoplasmas from distinct ribosomal groups (as opposed to subgroups). The protein-coding genes from 16SrXIII also lacked reference genes to use for mapping, which may have resulted in these reads not being represented in the assembly dataset even if they had hybridized to the probes.
Conversely, the capture of non-phytoplasma DNA targets (nonspecific hybridization) is potentially less desirable and can interfere with the assembly of the correct DNA sequences. The number of reads mapping to the different taxonomic marker genes was quite variable across samples and in some cases was quite high, even for samples with relatively low phytoplasma levels (Table 4). This is explained at least in part by variability in read numbers generated in each Illumina run, and variation in the number of samples that were simultaneously processed on a single flowcell (which affects the number of reads allocated to each sample). There may also be technical variability in hybridization stringency and washing efficacy between repeats, which affects the number of reads that are observed in each mapped dataset. The actual number of reads mapping to a given target gene is less important than the quality and length of the assemblies that are produced from these reads. For protein-coding genes such as cpn60, virtually all of the mapped reads had very high sequence identity to a probe sequence (Supplementary Figure S1C), representing high-quality sequences that enabled the assembly of long target sequences that were supported by tens to thousands of reads. This contrasts with PCR-based MLST, which typically has two slightly overlapping Sanger reads to support the relatively short contigs that are produced.
In the case of the 16S rRNA gene, many reads were observed in the initial mapping datasets for all phytoplasma-infected samples that corresponded to non-phytoplasma bacteria, along with host chloroplast and mitochondrial genes. In some cases, these reads interfered with the assembly of the correct taxonomic markers, and resulted in samples from uninfected plants showing post-hybridization reads (uninfected grape and canola). The most likely explanation for the preponderance of nonspecific reads in the 16S datasets is that the 16S gene is insufficiently distinct between different bacterial taxa to provide selectivity at both the hybridization and mapping steps. In the case of 16S rRNA genes, many of these “bleed-through” reads corresponded to 16S genes from non-phytoplasma bacteria, as well as 16S-like genes from host chloroplast and mitochondrial genomes. For 16S rRNA target genes only, a second mapping step was required that incorporated these reads; once so cleansed of the nonspecific reads, the assemblies improved considerably. In the case of the small number of protein-coding gene reads that appeared in the uninfected datasets, some amount of nonspecific hybridization and/or mapping can be expected, but these reads did not assemble into the target genes and so may be considered background noise. Therefore, this method is not suited to differentiating phytoplasma positive from negative samples, but that is not its intended use. In most cases, the MLST method will be applied to known positive samples. In addition, the non-hybridized samples showed higher read numbers mapping to all markers, including 16S rRNA, in the samples with lower amounts of phytoplasma (Table 5). While the explanation for this is not obvious, it is worth noting that these relatively small number of reads did not permit the assembly of the complete taxonomic markers in these non-hybridized samples, which demonstrates that necessity of the hybridization step.
This MLST scheme is effective for ribosomal groups 16SrI, 16SrIII, 16SrX, 16SrXII, and various subgroups within each. In addition, this relatively small panel of 351 probes could easily be expanded to design probes to include other phytoplasma groups and subgroups. This process would be straightforward, as it would use already available sequences and software for probe design and follows the hybridization procedures, protocols, and manufacturer guidelines that are well-tested and proven. Another advantage of this method is its potential to analyze a higher number of samples at the same time. For example, by including multiple samples (up to 12) in each hybridization and utilizing a plate format for simultaneous processing of 32 samples, up to 32 × 12 = 384 samples could be processed simultaneously, making high throughput MLST for phytoplasma strains a possibility. Inclusion of other distinct phytoplasmas in the MLST panel is limited only by the availability of reference sequences, as demonstrated by ‘Ca. P. hispanicum’ (16SrXIII), which would be impossible to represent due to the lack of reference sequences for several genes included in the MLST scheme.
A challenge faced by all molecular methods for differentiating phytoplasmas, including PCR-based methods, is dealing with samples containing either two distinct strains representing a mixed infection, or heterogeneous phytoplasma strains that feature two distinct but closely related phytoplasma 16S rRNA genes. For example, BnAY-TW1 from canola (Town et al., 2018) was found, by PCR and cloning, to contain 2 distinct 16S rRNA-encoding genes, which typed as 16SrI-A and 16SrI-B. Clone sequences generated from the more divergent genes cpn60 and rp were identical to one another, consistent with the presence of a single strain that contains two 16S rRNA genes that type distinctly using RFLP analysis. The genome sequence of strain TW1 was identified as a possible chimeric artifact resulting from the combination of a long-read genome containing 16SrI-A scaffolds, polished with Illumina reads from a 16SrI-B phytoplasma strain (Cho et al., 2020). The hybridization results presented here are inconsistent with this hypothesis, since all the assembled protein-coding genes, and the clone sequences generated, clustered with 16SrI-B sequences. Moreover, the sequences that were assembled were identical or very nearly identical in all cases to the genes identified within the reported TW-1 genome sequence that were used to design the hybridization probes. In addition, the AY-infected strawberry sample from Quebec, STRAW4, provided distinct typing results using the assembled cpn60 and 16S rRNA genes (Table 6). This sample has also been shown through cloning to contain two distinct 16S genes, which type as 16SrI-R and 16SrI-S along with a single cpn60 sequence type (Brochu, AS et al., manuscript submitted). In cases of strains with heterogeneous 16S rRNA genes, the hybridization-based method described here would presumably produce a composite sequence from the two distinct loci, much like direct sequencing of PCR-generated amplicon would. Therefore, caution must be used in applying this method to strains that are known to feature 16S rRNA gene heterogeneity, and this should be investigated in each case using PCR amplification, cloning, and sequencing.
In conclusion, this hybridization-based MLST scheme is a method for phytoplasma characterization and provides a proof-of-concept for molecular characterization of other bacterial pathogens that are difficult to culture, despite the limitations listed above. All single-locus classification systems will suffer from a lack of taxonomic resolution due to the limited amount of sequencing information that can be generated from a single marker. This MLST scheme is based on gene targets of proven utility and generates high-quality sequences corresponding to seven different molecular markers. High resolution molecular marker sequences can be determined in this way for phytoplasmas within host plant tissue, even those with low concentrations of this pathogen. Given how straightforward the probe design process is and its high throughput potential, this hybridization-based MLST scheme can be a very efficient molecular tool that provides resolution of closely related phytoplasmas.
The use of DNA sequencing for classification and typing of phytoplasmas will continue to be essential for understanding and monitoring the detrimental effects of phytoplasma infections on crop production. Implementation of a novel, universal, standardized approach for MLST could benefit these efforts and will result in an increased understanding of the spread and effects of these organisms on crop plants worldwide.
Data availability statement
The datasets generated for this study can be found in GenBank under BioProject accession number 642 PRJNA837572. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA837572.
Author contributions
HB, TD, DS, and MG were responsible for experimental design. TD, KP-B, DS, and MG performed the experiments. TD and EL analyzed the data. TW and HB provided intellectual input. EL and TD prepared the figures and Supplementary material. TD and KP-B wrote the initial draft of the manuscript. HB, DS, MG, EL, TW, TD, and HB edited the manuscript. TD, TW, and HB acquired the funding, and supervised the project. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by Canadian Food Inspection Agency grant # SID-P-1802, “Evaluation of Next generation Sequencing (NGS) for the detection and identification of Phytoplasmas.” Graduate stipends for KP-B were provided through two projects funded by Western Grains Research Foundation (Project 1 and 2) and SaskCanola (Project 2), to TW and TD, “An early warning system to predict aster yellows outbreaks in Western Canada: origin and arrival of migrant leafhoppers (AAFC AGR-14988; WGRF # AGR1817), and “Continuing to watch the winds: the origin and arrival of migrant aster leafhoppers and diamondback moths” (AAFC AGR-17913; SaskCanola Ref: CARP ADF2020.409; WGRF Ref: AGR2105.
Acknowledgments
We thank Jennifer Town for advice and assistance on the data analysis code, and Christine Hammond for DNA sequencing support and commentary on the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.959562/full#supplementary-material
Footnotes
References
Abeysinghe, S., Abeysinghe, P. D., Kanatiwela-De Silva, C., Udagama, P., Warawichanee, K., Aljafar, N., et al. (2016). Refinement of the taxonomic structure of 16SrXI and 16SrXIV phytoplasmas of gramineous plants using multilocus sequence typing. Plant Dis. 100, 2001–2010. doi: 10.1094/PDIS-02-16-0244-RE
Arnaud, G., Malembic-Maher, S., Salar, P., Bonnet, P., Maixner, M., Marcone, C., et al. (2007). Multilocus sequence typing confirms the close genetic interrelatedness of three distinct flavescence doree phytoplasma strain clusters and group 16SrV phytoplasmas infecting grapevine and alder in Europe. Appl. Environ. Microbiol. 73, 4001–4010. doi: 10.1128/AEM.02323-06
Bertaccini, A. (2022). Plants and Phytoplasmas: when bacteria modify plants. Plan. Theory 11:11. doi: 10.3390/plants11111425
Bertaccini, A., Arocha-Rosete, Y., Contaldo, N., Duduk, B., Fiore, N., Montano, H. G., et al. (2022). Revision of the ‘Candidatus Phytoplasma’ species description guidelines. Int. J. Syst. Evol. Microbiol. 72:72. doi: 10.1099/ijsem.0.005353
Cai, W., Nunziata, S., Rascoe, J., and Stulberg, M. J. (2019). SureSelect targeted enrichment, a new cost effective method for the whole genome sequencing of Candidatus Liberibacter asiaticus. Sci. Rep. 9:18962. doi: 10.1038/s41598-019-55144-4
Cho, S.-T., Kung, H.-J., Huang, W., Hogenhout, S. A., and Kuo, C.-H. (2020). Species boundaries and molecular markers for the classification of 16SrI phytoplasmas inferred by genome analysis. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01531
Davis, R. E., Jomantiene, R., Kalvelyte, A., and Dally, E. L. (2003). Differential amplification of sequence heterogeneous ribosomal RNA genes and classification of the 'Fragaria multicipita' phytoplasma. Microbiol. Res. 158, 229–236. doi: 10.1078/0944-5013-00201
Davis, R. E., Zhao, Y., Dally, E. L., Lee, I. M., Jomantiene, R., and Douglas, S. M. (2013). 'Candidatus Phytoplasma pruni', a novel taxon associated with X-disease of stone fruits, Prunus spp.: multilocus characterization based on 16S rRNA, secY, and ribosomal protein genes. Int. J. Syst. Evol. Microbiol. 63, 766–776. doi: 10.1099/ijs.0.041202-0
Deng, S., and Hiruki, C. (1991). Amplification of 16S rRNA genes from culturable and nonculturable Mollicutes. J. Microbiol. Meth. 14, 53–61. doi: 10.1016/0167-7012(91)90007-D
Dumonceaux, T. J., Green, M., Hammond, C., Perez, E., and Olivier, C. (2014). Molecular diagnostic tools for detection and differentiation of phytoplasmas based on chaperonin-60 reveal differences in host plant infection patterns. PLoS One 9:e116039. doi: 10.1371/journal.pone.0116039
Dumonceaux, T. J., Links, M. G., Town, J. R., Hill, J. E., and Hemmingsen, S. M. (2017). Targeted capture of cpn60 gene fragments for PCR-independent microbial community profiling. Nat. Protoc. Exch. doi: 10.1038/protex.2017.100
Firrao, G., Martini, M., Ermacora, P., Loi, N., Torelli, E., Foissac, X., et al. (2013). Genome wide sequence analysis grants unbiased definition of species boundaries in 'Candidatus Phytoplasma'. Syst. Appl. Microbiol. 36, 539–548. doi: 10.1016/j.syapm.2013.07.003
Gardner, E. M., Johnson, M. G., Pereira, J. T., Puad, A. S. A., Arifiani, D., Sahromi, W., et al. (2020). Paralogs and off-target sequences improve phylogenetic resolution in a densely sampled study of the breadfruit genus (Artocarpus, Moraceae). Syst. Biol. 70, 558–575. doi: 10.1093/sysbio/syaa073
Gasc, C., and Peyret, P. (2018). Hybridization capture reveals microbial diversity missed using current profiling methods. Microbiome 6:61. doi: 10.1186/s40168-018-0442-3
Gundersen, D. E., and Lee, I. M. (1996). Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol. Mediterr. 35, 144–151.
Hodgetts, J., Boonham, N., Mumford, R., Harrison, N., and Dickinson, M. (2008). Phytoplasma phylogenetics based on analysis of sec a and 23S rRNA gene sequences for improved resolution of candidate species of 'Candidatus Phytoplasma'. Int. J. Syst. Evol. Microbiol. 58, 1826–1837. doi: 10.1099/ijs.0.65668-0
Jomantiene, R., Davis, R. E., Valiunas, D., and Alminaite, A. (2002). New group 16SrIII phytoplasma lineages in Lithuania exhibit rRNA interoperon sequence heterogeneity. Eur. J. Plant Pathol. 108, 507–517. doi: 10.1023/A:1019982418063
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Molec. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lee, I. M., Bottner-Parker, K. D., Zhao, Y., Davis, R. E., and Harrison, N. A. (2010). Phylogenetic analysis and delineation of phytoplasmas based on secY gene sequences. Int. J. Syst. Evol. Microbiol. 60, 2887–2897. doi: 10.1099/ijs.0.019695-0
Lee, I. M., Davis, R. E., Chen, T. A., Chiykowski, L. N., Fletcher, J., Hiruki, C., et al. (1992). A genotype-based system for identification and classification of mycoplasmalike organisms (MLOs) in the aster yellows MLO strain cluster. Phytopathology 82, 977–986. doi: 10.1094/Phyto-82-977
Lee, I. M., Davis, R. E., and Dewitt, N. D. (1990). Nonradioactive screening method for isolation of disease-specific probes to diagnose plant diseases caused by mycoplasmalike organisms. Appl. Environ. Microbiol. 56, 1471–1475. doi: 10.1128/aem.56.5.1471-1475.1990
Lee, I. M., Gundersen-Rindal, D. E., Davis, R. E., Bottner, K. D., Marcone, C., and Seemüller, E. (2004). ‘Candidatus Phytoplasma asteris’, a novel phytoplasma taxon associated with aster yellows and related diseases. Int. J. Syst. Evol. Microbiol. 54, 1037–1048. doi: 10.1099/ijs.0.02843-0
Liefting, L. W., Andersen, M. T., Beever, R. E., Gardner, R. C., and Forster, R. L. (1996). Sequence heterogeneity in the two 16S rRNA genes of Phormium yellow leaf phytoplasma. Appl. Environ. Microbiol. 62, 3133–3139. doi: 10.1128/aem.62.9.3133-3139.1996
Lim, P. O., and Sears, B. B. (1992). Evolutionary relationships of a plant-pathogenic mycoplasmalike organism and Acholeplasma laidlawii deduced from two ribosomal protein gene sequences. J. Bacteriol. 174, 2606–2611. doi: 10.1128/jb.174.8.2606-2611.1992
Marcone, C., Lee, I. M., Davis, R. E., Ragozzino, A., and Seemuller, E. (2000). Classification of aster yellows-group phytoplasmas based on combined analyses of rRNA and tuf gene sequences. Int. J. Syst. Evol. Microbiol. 50, 1703–1713. doi: 10.1099/00207713-50-5-1703
Martini, M., Lee, I. M., Bottner, K. D., Zhao, Y., Botti, S., Bertaccini, A., et al. (2007). Ribosomal protein gene-based phylogeny for finer differentiation and classification of phytoplasmas. Int. J. Syst. Evol. Microbiol. 57, 2037–2051. doi: 10.1099/ijs.0.65013-0
Mitrović, J., Kakizawa, S., Duduk, B., Oshima, K., Namba, S., and Bertaccini, A. (2011). The gro EL gene as an additional marker for finer differentiation of 'Candidatus Phytoplasma asteris'-related strains. Ann. Appl. Biol. 159, 41–48. doi: 10.1111/j.1744-7348.2011.00472.x
Muirhead, K., Pérez-López, E., Bahder, B. W., Hill, J. E., and Dumonceaux, T. (2019a). The Cpn Classi PhyR is a resource for cpn60 universal target-based classification of phytoplasmas. Plant Dis. 103, 2494–2497. doi: 10.1094/PDIS-03-19-0454-A
Muirhead, K., Pérez-López, E., Bahder, B. W., Hill, J. E., and Dumonceaux, T. J. (2019b). “The Cpn Classi PhyR facilitates phytoplasma classification and taxonomy using cpn60 universal target sequences” in Sustainable Management of Phytoplasma Diseases in Crops Grown in the Tropical Belt: Biology and Detection. eds. C. Y. Olivier, T. J. Dumonceaux, and E. Pérez-López (Cham: Springer International Publishing), 1–27.
Nijo, T., Iwabuchi, N., Tokuda, R., Suzuki, T., Matsumoto, O., Miyazaki, A., et al. (2021). Enrichment of phytoplasma genome DNA through a methyl-CpG binding domain-mediated method for efficient genome sequencing. J. Gen. Plant Pathol. 87, 154–163. doi: 10.1007/s10327-021-00993-z
Pérez-López, E., Olivier, C. Y., Luna-Rodríguez, M., and Dumonceaux, T. J. (2016). Phytoplasma classification and phylogeny based on in silico and in vitro RFLP analysis of cpn60 universal target sequences. Int. J. Syst. Evol. Microbiol. 66, 5600–5613. doi: 10.1099/ijsem.0.001501
Pérez-López, E., Rodríguez-Martínez, D., Olivier, C. Y., Luna-Rodríguez, M., and Dumonceaux, T. J. (2017). Molecular diagnostic assays based on cpn60 UT sequences reveal the geographic distribution of subgroup 16SrXIII-(a/I) I phytoplasma in Mexico. Sci. Rep. 7:950. doi: 10.1038/s41598-017-00895-1
Perez-Lopez, E., Vincent, C., Moreau, D., Hammond, C., Town, J., and Dumonceaux, T. J. (2019). A novel ‘Candidatus Phytoplasma asteris’ subgroup 16SrI-(E/AI) AI associated with blueberry stunt disease in eastern Canada. Int. J. Syst. Evol. Microbiol. 69, 322–332. doi: 10.1099/ijsem.0.003100
Plante, N., Brochu, A.-S., Goulet, C., Thibault, P., Fournier, V., and Pérez-López, E. (2021). First evidence of the occurrence of a putative new subgroup of ‘Candidatus Phytoplasma asteris’ (16SrI) associated with strawberry green petal disease in Quebec, Canada. New Dis. Rep. 44:e12038. doi: 10.1002/ndr2.12038
Ranebennur, H., Rawat, K., Rao, A., Kumari, P., Chalam, V. C., Meshram, N., et al. (2022). Transmission efficiency of a ‘Candidatus Phytoplasma australasia’ (16SrII-D) related strain associated with sesame phyllody by dodder, grafting and leafhoppers. Eur. J. Plant Pathol. doi: 10.1007/s10658-022-02550-6
Ranwez, V., Harispe, S., Delsuc, F., and Douzery, E. J. P. (2011). MACSE: multiple alignment of coding SEquences accounting for frameshifts and stop codons. PLoS One 6:e22594. doi: 10.1371/journal.pone.0022594
Saccardo, F., Martini, M., Palmano, S., Ermacora, P., Scortichini, M., Loi, N., et al. (2012). Genome drafts of four phytoplasma strains of the ribosomal group 16SrIII. Microbiology 158, 2805–2814. doi: 10.1099/mic.0.061432-0
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Satta, E., Carminati, G., and Bertaccini, A. (2020). Phytoplasma presence in carrot seedlings. Aust. Plant. Dis. Notes. 15:11. doi: 10.1007/s13314-020-0377-y
Schneider, B., Seemuller, E., Smart, C. D., and Kirkpatrick, B. C. (1995). “Phylogenetic classification of plant pathogenic mycoplasmalike organisms or phytoplasmas” in Molecular and Diagnostic Procedures in Mycoplasmology. eds. R. Razin and J. G. Tully (San Diego: Academic Press), 369–380.
Shao, J. Y., Jomantiene, R., Dally, E. L., Zhao, Y., Lee, I.-M., Nuss, D. L., et al. (2006). Phylogeny and characterization of phytoplasmal nus a and use of the nus a gene in detection of group 16SrI strains. J. Plant Pathol. 88, 193–201.
Soto, N., Helmick, E. E., Harrison, N. A., and Bahder, B. W. (2021). Genetic variability of palm lethal decline phytoplasmas in the Caribbean basin and Florida, U.S.a., based on a multilocus analysis. Phytopathology 111, 2203–2212. doi: 10.1094/PHYTO-04-21-0130-R
Tamura, K., Nei, M., and Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101, 11030–11035. doi: 10.1073/pnas.0404206101
Town, J. R., Wist, T., Perez-Lopez, E., Olivier, C. Y., and Dumonceaux, T. J. (2018). Genome sequence of a plant-pathogenic bacterium, “Candidatus Phytoplasma asteris” strain TW1. Microbiol. Resour. Announc. 7:e01109-18. doi: 10.1128/MRA.01109-18
Wei, W., Cai, H., Jiang, Y., Lee, I.-M., Davis, R. E., Ding, Y., et al. (2011). A new phytoplasma associated with little leaf disease in azalea: multilocus sequence characterization reveals a distinct lineage within the aster yellows phytoplasma group. Ann. Appl. Biol. 158, 318–330. doi: 10.1111/j.1744-7348.2011.00468.x
Wei, W., and Zhao, Y. (2022). Phytoplasma taxonomy: nomenclature, classification, and identification. Biology (Basel) 11:11. doi: 10.3390/biology11081119
Weintraub, P. G., and Beanland, L. (2006). Insect vectors of phytoplasmas. Annu. Rev. Entomol. 51, 91–111. doi: 10.1146/annurev.ento.51.110104.151039
Zhao, Y., Wei, W., Lee, I.-M., Shao, J., Suo, X., and Davis, R. E. (2009). Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the peach X-disease phytoplasma group (16SrIII). Int. J. Syst. Evol. Microbiol. 59, 2582–2593. doi: 10.1099/ijs.0.010249-0
Keywords: ‘Candidatus Phytoplasma’, phytoplasma taxonomy, hybridization probes, aster yellows, apple proliferation, pear decline, bois noir, X-disease
Citation: Pusz-Bochenska K, Perez-Lopez E, Wist TJ, Bennypaul H, Sanderson D, Green M and Dumonceaux TJ (2022) Multilocus sequence typing of diverse phytoplasmas using hybridization probe-based sequence capture provides high resolution strain differentiation. Front. Microbiol. 13:959562. doi: 10.3389/fmicb.2022.959562
Edited by:
Florence Tardy, Agence Nationale de Sécurité Sanitaire de l’Alimentation, de l’Environnement et du Travail (ANSES), FranceReviewed by:
Jun-Yi Yang, National Chung Hsing University, TaiwanXavier Foissac, Institut National de Recherche pour L’agriculture, L’alimentation et L’environnement (INRAE), France
Copyright © 2022 Pusz-Bochenska, Perez-Lopez, Wist, Bennypaul, Sanderson, Green and Dumonceaux. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tim J. Dumonceaux, dGltLmR1bW9uY2VhdXhAYWdyLmdjLmNh
 Karolina Pusz-Bochenska
Karolina Pusz-Bochenska Edel Perez-Lopez
Edel Perez-Lopez Tyler J. Wist
Tyler J. Wist Harvinder Bennypaul
Harvinder Bennypaul Daniel Sanderson5
Daniel Sanderson5 Tim J. Dumonceaux
Tim J. Dumonceaux