- 1State Key Laboratory of Reproductive Regulation and Breeding of Grassland Livestock, School of Life Sciences, Inner Mongolia University, Hohhot, China
- 2College of Life Sciences, Inner Mongolia Agricultural University, Hohhot, China
Trimethylamine N-oxide (TMAO), an important intestinal flora-derived metabolite, plays a role in the development of cardiovascular disease and tumor immunity. Here, we determined the minimum inhibitory concentration (MIC) of antibiotics against Escherichia coli under gradient concentrations of TMAO and performed a bacterial killing analysis. Overall, TMAO (in the range of 10 ~ 100 mM) increased the MIC of quinolones, aminoglycosides, and β-lactams in a concentration-dependent manner, and increased the lethal dose of antibiotics against E. coli. It implies that TMAO is a potential risk for failure of anti-infective therapy, and presents a case for the relationship between intestinal flora-derived metabolites and antibiotic resistance. Further data demonstrated that the inhibition of antibiotic efficacy by TMAO is independent of the downstream metabolic processes of TMAO and the typical bacterial resistance mechanisms (mar motif and efflux pump). Interestingly, TMAO protects E. coli from high-protein denaturant (urea) stress and improves the viability of bacteria following treatment with two disinfectants (ethanol and hydrogen peroxide) that mediate protein denaturation by chemical action or oxidation. Since antibiotics can induce protein inactivation directly or indirectly, our work suggests that disruption of protein homeostasis may be a common pathway for different stress-mediated bacterial growth inhibition/cell death. In addition, we further discuss this possibility, which provides a different perspective to address the global public health problem of antibiotic resistance.
Introduction
Trimethylamine N-oxide (TMAO) is a bioactive molecule produced by gut microbial-derived metabolism (Ufnal et al., 2015; Subramaniam and Fletcher, 2018; Gessner et al., 2020). In humans, diets rich in TMA precursors (e.g., choline and L-carnitine) are the main source of TMAO (Zhang et al., 1999; Wang et al., 2011; Koeth et al., 2013). The intestinal flora converts dietary nutrients to TMA, which is absorbed into the bloodstream through the intestinal mucosa and then converted to TMAO in the liver by flavin-containing monooxygenases (FMOs; Zeisel and Warrier, 2016; Subramaniam and Fletcher, 2018). In recent years, the relevance of TMAO to human diseases has been widely reported. Overall, elevated TMAO concentrations increase the risk of diseases such as diabetes (Tai et al., 2015), heart failure (Trøseid et al., 2015), and atherosclerosis (Dalla Via et al., 2019). TMAO has also been associated with mortality and hospitalization rates for cardiac and renal diseases such as atrial fibrillation (Tang et al., 2014), acute myocardial infarction (Suzuki et al., 2017), and chronic kidney disease (Tang et al., 2015). In addition, TMAO has been reported to be strongly correlated with the development of inflammatory bowel disease (IBD) and Alzheimer’s disease (AD; Wilson et al., 2015, 2016; Xu and Wang, 2016), and even with the outcome of immunotherapy in triple-negative breast cancer (Wang et al., 2022). However, the effect of TMAO on antibiotic efficacy is still unknown.
Recently, an interesting study has found that a high-fat diet leads to dysbiosis of intestinal flora and depletion of the microbial metabolite indole-3-acetic acid (IAA), leading to reduced antibiotic efficacy against bacterial infections (Liu et al., 2021). The study by Oliver et al. found that a diet high in fiber and low in animal protein established an association with antibiotic resistance by shaping the human gut microbiota, but the role played by the corresponding gut metabolites triggered by a specific diet needs further elucidation (Oliver et al., 2022). The intestinal microbiota carries a large number of antibiotic resistance genes (Ruppé et al., 2019; Forster et al., 2022), and those metabolites that reduce antibiotic susceptibility may exacerbate the emergence and spread of resistant bacteria. The inhibition of antibiotic efficacy by intestinal microbiota metabolites may drive the enrichment and evolution of antibiotic-resistant bacteria (Becattini et al., 2016). If so, bacterial resistance will become even more problematic. Because the influence of diet on intestinal metabolites is extensive (Koh et al., 2016; Sonnenburg and Bäckhed, 2016; Cani et al., 2019), the dietary structure of different individuals is complex. This makes it particularly important to explore the effect of intestinal flora metabolites on antibiotic efficacy.
This work explored the effect of a gut microbial-associated metabolite, TMAO, on antibiotic efficacy in vitro. Interestingly, TMAO reduced the susceptibility of Escherichia coli to a variety of antibiotics. Further data suggest that TMAO does not rely on activation of the TMAO sense-regulatory system and the classical multidrug efflux system to mediate the inhibition of antibiotic efficacy. In addition, TMAO enhanced the survival of E. coli under lethal urea (protein denaturant) stress and also protected E. coli from killing by two disinfectants (ethanol and H2O2) that cause protein damage. In conclusion, our study suggests that TMAO has the potential to improve bacterial survival under anti-biotics and other lethal stresses through its protective effect against protein denaturation/damage.
Results
Trimethylamine N-Oxide Increases the MIC of Antibiotics in a Concentration-Dependent Manner
To investigate the correlation between TMAO and antibiotic efficacy in vitro, we added 5 ~ 100 mM TMAO to LB medium to determine the effect of different concentrations of TMAO on the minimum inhibitory concentration (MIC) of antibiotic. In the absence of TMAO, the MIC of ciprofloxacin against the wild-type strain (BW25113) was about 0.02 mg/l; under the condition of 10 mM TMAO, the MIC of ciprofloxacin was mostly 0.03 mg/l; however, when the concentration of TMAO was increased to 100 mM, the MIC of ciprofloxacin measured in six independent experiments was as high as 0.16 ~ 0.32 mg/l (Figure 1A). Similarly, TMAO also increased the MIC of moxifloxacin, another quinolone antibiotic, in a concentration-dependent manner (Figure 1B). Next, we measured the MIC of two aminoglycosides and two β-lactams under the same TMAO concentration gradient. The results showed that 10 mM TMAO more significantly increased the MIC of gentamicin and kanamycin against wild-type strain (Figures 1C,D), and the MIC promoted with increasing of TMAO concentrations. Differently, the effect of TMAO on the MIC of β-lactams required higher concentrations than that for the other two types of antibiotics. The MIC of meropenem increased to 0.06 mg/l in three out of six MIC determinations under 20 mM TMAO, compared to 0.03 mg/l (2 times)/0.04 mg/l (4 times) in the absence of TMAO. And in 40 mM TMAO conditions, the MIC of meropenem increased to 0.06 ~ 0.08 mg/l (Figure 1E). The MIC of ampicillin only increased from 4 to 6 mg/l (1 time)/8 mg/l (5 times) with 60 mM TMAO (Figure 1F). The above results suggest that TMAO increased the MIC of the three types of antibiotics against E. coli in a concentration-dependent manner in the range of 10 ~ 100 mM. Furthermore, by measuring the growth curve, we ruled out the possibility that TMAO affects bacterial growth and thus alters the susceptibility of E. coli to antibiotics (Supplementary Figure 1).
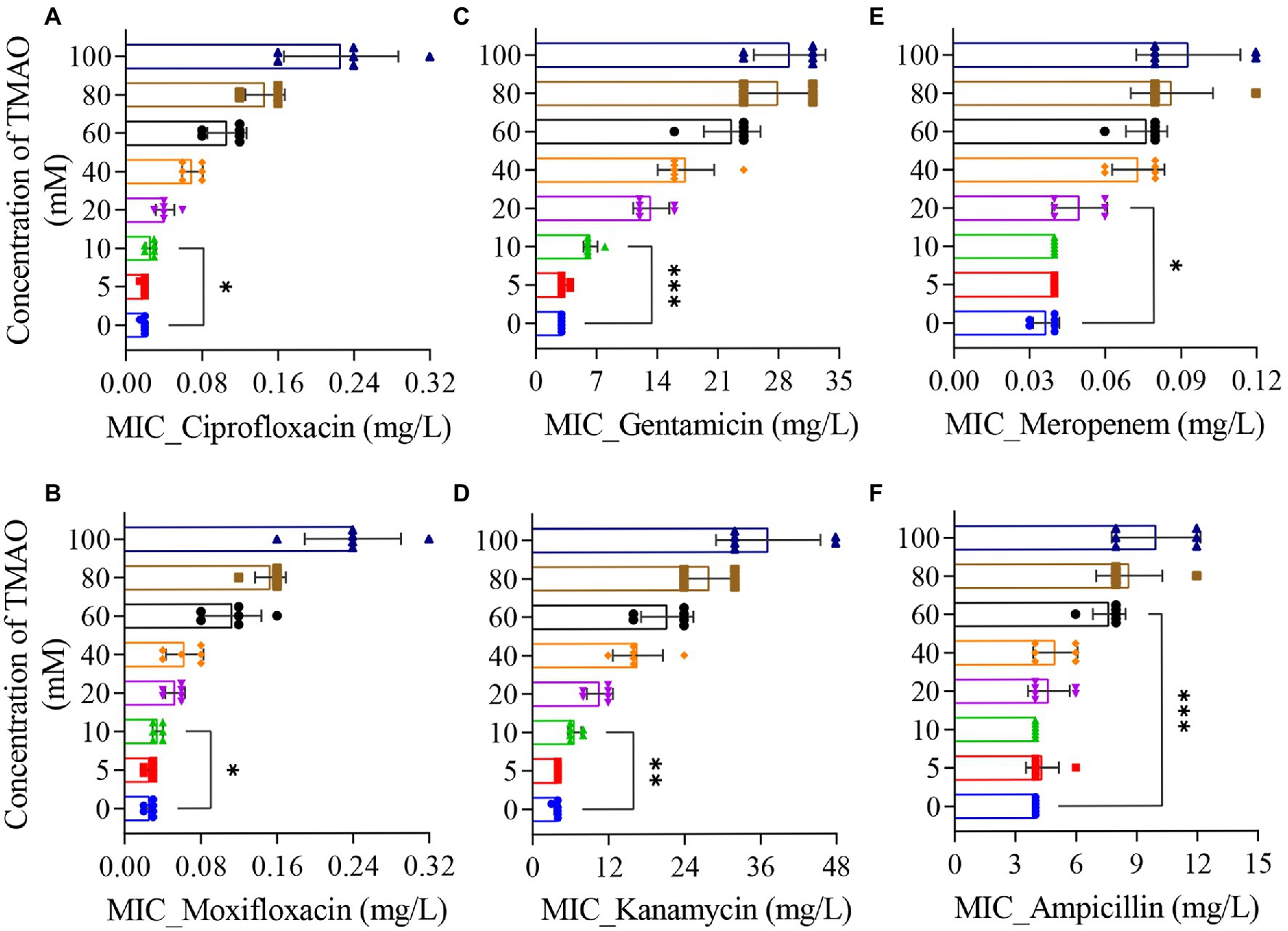
Figure 1. Effect of Trimethylamine N-oxide (TMAO) on minimum inhibitory concentration (MIC) of antibiotics. (A–F) MIC was determined by the 2-fold broth dilution method. Cultures incubated to OD600 = 0.2 were diluted to ~105 cells/ml, mixed with various amounts of drug, and incubated at 37°C for 12 h. As needed, different concentrations of TMAO were added to the LB medium. Six individual experiments were performed. Each data plotted value represents mean ± SD. Significance determined by paired t-test. *p < 0.05; **p < 0.01; ***p < 0.001.
Higher Doses of Antibiotics Are Required to Kill Escherichia coli Cells in the Presence of TMAO
Then, we examined the effect of TMAO on antibiotic sterilization in E. coli. Since ampicillin showed a significant increase in MIC under the 60 mM TMAO condition (Figure 1F), the concentration was used for subsequent experiments, and one of each of the three types of antibiotics tested was chosen as a representative. In the absence of TMAO, treatment with 0.08 mg/l (4 × MIC) ciprofloxacin (Figure 2A) or 6 mg/l (2 × MIC) gentamicin (Figure 2B) for 2 h reduced the survival of log-phase wild-type E. coli by approximately three orders of magnitude; treatment with 0.32 mg/l (8 × MIC) meropenem (Figure 2C) for 6 h reduced the survival of wild-type E. coli by about four orders of magnitude. In contrast, under 60 mM TMAO condition, neither ciprofloxacin, gentamicin, nor meropenem at equivalent absolute concentrations exhibited lethality against the wild-type strain because of the elevated MIC (Figures 2A–C, comparison of the second column with the first column). Ciprofloxacin and gentamicin demonstrated better bactericidal efficacy when sterilized with a standardized MIC (Figures 2A,B, fourth column compared to the first column). With 60 mM TMAO, meropenem required a higher multiple of MIC (10 × MIC) to achieve the bactericidal level of 8 × MIC meropenem in the absence of TMAO (Figure 2C, fourth column compared to the first column). Overall, the presence of TMAO requires higher concentrations of antibiotics for growth inhibition or bacterial killing.
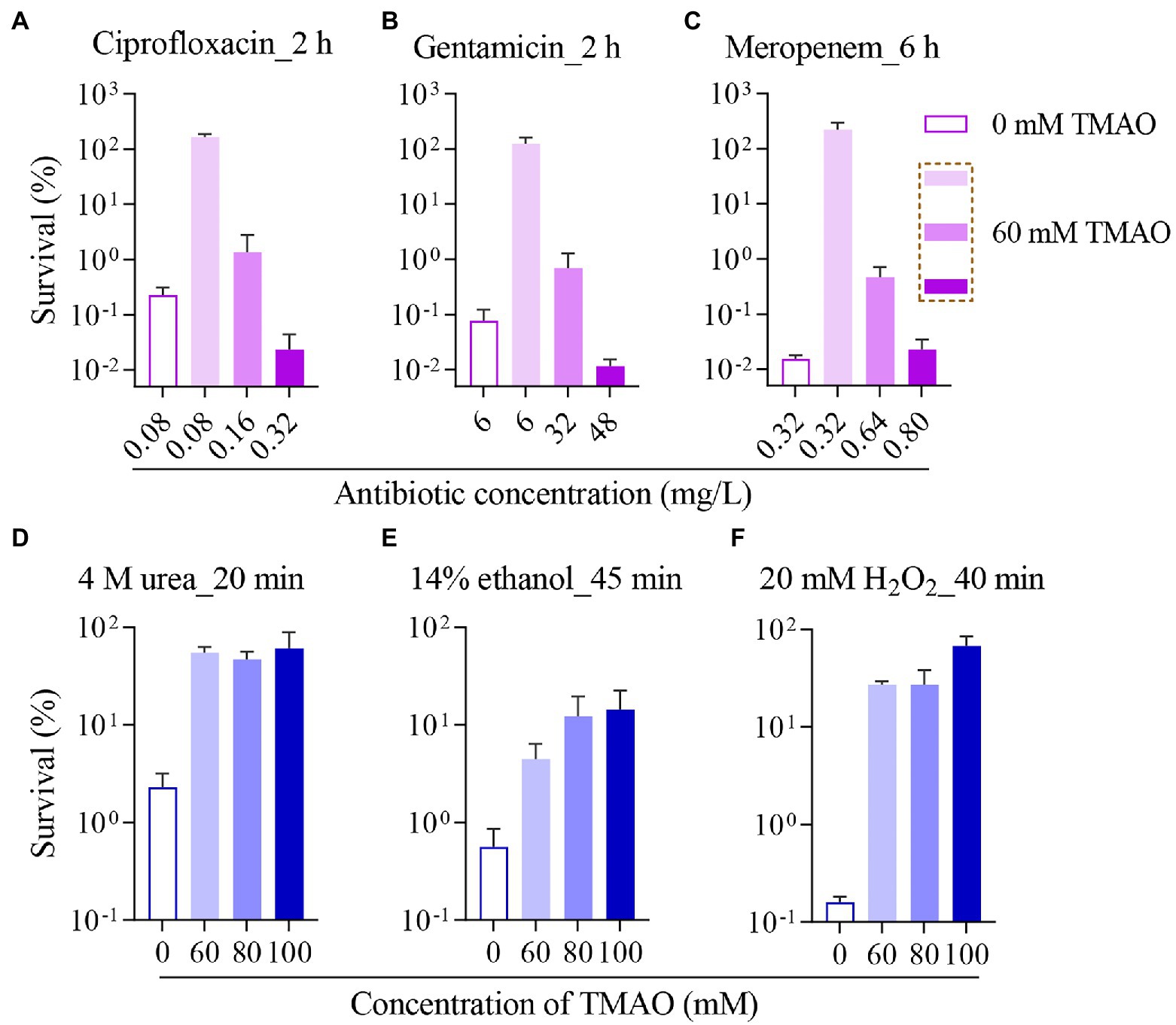
Figure 2. Trimethylamine N-oxide protects Escherichia coli from killing by antibiotics or other lethal stressors. Survival of wild-type strain incubated with 0 or 60 mM TMAO, after ciprofloxacin treatment for 2 h (A), gentamicin treatment for 2 h (B), or meropenem treatment for 6 h (C) was measured. And survival of wild-type strain incubated with different concentrations of TMAO, after treatment with 4 M urea for 20 min (D), 14% ethanol treatment for 45 min (E), or after 20 mM H2O2 treatment for 40 min (F) was measured. TMAO was added to the corresponding bacterial culture for co-incubation 30 min before antibiotic addition. Experiments were performed independently for three times. Each data plotted value represents mean ± SD.
TorSTR Signaling Pathway and Typical Resistance Mechanisms Are Not Involved in the TMAO Effect on Antibiotic Sensitivity
TorSTR signaling pathway senses TMAO and activates downstream pathways in E. coli (Baraquet et al., 2006). TorZ (TMAO reductase), and the torCAD operon under positive regulation of TorR can reduce TMAO to TMA (Gon et al., 2000, 2001). To investigate the mechanism by which TMAO inhibits antibiotic susceptibility of E. coli, we determined the MIC of ciprofloxacin on mutant strains related to the TorSTR signaling pathway. The results showed that the MIC of ciprofloxacin against ∆torT, ∆torR, ∆torC, ∆torZ, and wild-type strains with or without 60 mM TMAO was essentially the same (Supplementary Figure 2). In addition, neither the addition of 60 mM nor 100 mM TMA changed the MIC of the above six antibiotics against the wild-type strain (Supplementary Table 1). Another TMAO-binding protein (Pietrzyk-Brzezinska and Cociurovscaia, 2022), RcdA in E. coli, is suggested to be involved in regulating many stress response genes (Shimada et al., 2012). The rcdA mutation also considerably increased the MIC of ciprofloxacin against E. coli (Supplementary Figure 2). This suggests that the activation of intracellular TMAO-related downstream pathways does not serve as a condition to reduce the susceptibility of E. coli to antibiotics.
The typical mechanisms that cause larger MIC for antibiotics include: (1) the mar (multiple antibiotic resistance) motif, in which MarA can alter the expression of multiple genes to confer bacterial resistance (Cohen et al., 1993; Barbosa and Levy, 2000; Duval and Lister, 2013) and (2) a more direct way of causing bacterial resistance is the efflux pump, and eight efflux pump complexes have been reported in E. coli, and they all contain TolC proteins (Koronakis et al., 2000; Koronakis, 2003). The results showed that deletion of the marA gene did not alter the MIC of ciprofloxacin or meropenem, and the increase in MIC of ciprofloxacin or meropenem against the ∆marA strain in the presence of 60 mM TMAO was not significantly different from that against the wild-type cells (Supplementary Figures 3A,B left). Deletion of the tolC gene reduced the MIC of ciprofloxacin from 0.02 to 0.0025 mg/l and that of meropenem from 0.04 to 0.0075 mg/l against E. coli, but TMAO significantly increased the MIC of ciprofloxacin or meropenem against the ∆tolC strain (Supplementary Figures 3A,B right).
Trimethylamine N-Oxide Reduces the Lethality of Urea, Ethanol, and H2O2 Against Escherichia coli
After excluding the role of the above factors in the inhibition of antibiotic efficacy by TMAO, we focused on the biological activity of TMAO. It is known that TMAO is present in high concentrations in some aquatic organisms and counteracts the denaturing effects of urea and salt (Lin and Timasheff, 1994; Seibel and Walsh, 2002). Under laboratory conditions, TMAO has likewise been shown to protect protein stability under urea stress (Zou et al., 2002), and TMAO may stabilize proteins by acting as a heterogeneous surfactant for folded proteins (Liao et al., 2017). Four molar urea is commonly used as a high-intensity protein denaturant, and our experiments revealed that treated with 4 M urea for 20 min, different concentrations of TMAO (60 ~ 100 mM) showed 1 ~ 2 orders of magnitude of protection against the wild-type strain (Figure 2D), and treated with 4 M urea for 40 min, 60 mM TMAO also demonstrated significant protection, and the degree of protection by TMAO is positively correlated with the concentration of TMAO (Supplementary Figure 4A).
Further, we selected a common disinfectant, ethanol, to verify the role of the property of TMAO to stabilize proteins in protecting bacterial survival. Ethanol denatures proteins by breaking the hydrogen bonds originally present in the protein (Thomas and Dill, 1993). The data showed that 14% ethanol decreased the survival of the wild-type strain by two orders of magnitude or more than three orders of magnitude for 45 or 60 min treatment, respectively (Figure 2E; Supplementary Figure 4B), for both 45 and 60 min treatment, 60 ~ 100 mM TMAO showed protective effects. H2O2, a reactive oxide, denatures proteins through covalent modification of specific amino acid side chains, resulting in oxidative damage (Dahl et al., 2015). Multiple antibiotics have the effect of inducing protein aggregation (Cardoso et al., 2010; Tran et al., 2011; Wu et al., 2015). If TMAO can improve the survival of E. coli under H2O2 stress, it may also suggest that protection against protein denaturation could be a potential mechanism for TMAO to inhibit the efficacy of different antibiotics. Surprisingly, as shown in Figure 2F; Supplementary Figure 4C, 60 mM TMAO significantly increased the survival of E. coli under 20 mM H2O2 stress, and the protection of TMAO to bacteria against H2O2 was more concentration-dependent for 60 min of H2O2 treatment. Since TMAO is a weak oxidant, this suggests that the contribution of TMAO to bacterial survival under H2O2 is not directly by antioxidant effect.
Discussion and Future Directions
In this work, we report the inhibitory effect of TMAO on the efficacy of various antibiotics and disinfectants. The concentration of TMAO showed a positive correlation with the MIC of quinolones, aminoglycosides, and β-lactams (Figure 1), and the lethal dose of different antibiotics against E. coli increased accordingly when TMAO was included (Figures 2A–C). Unexpectedly, the effect of TMAO on antibiotic MIC did not depend on downstream metabolic regulatory pathways (Supplementary Figure 2), mar motif, and efflux pump (Supplementary Figure 3). Similar to the direct protection of E. coli under urea stress (Figure 2D), TMAO also improved the survival of E. coli after treatment with two disinfectants, ethanol and hydrogen peroxide (Figures 2E,F).
Protein Inactivation Is a Potentially Common Pathway for Different Stress-Mediated Bacterial Cell Death
What is the mechanism by which TMAO helps E. coli cope with different stresses? Understanding this question will help in the development of antimicrobial strategies and guide the development of novel antimicrobial agents or adjuvants. Quinolones act on DNA replication, aminoglycosides target ribosomal subunits, β-lactams inhibit cell wall synthesis, urea and ethanol mediate protein denaturation, and hydrogen peroxide can cause oxidative damage. Our data suggest that TMAO can reduce the stress of E. coli by these different stressors. One of the possible explanations is that TMAO has an unknown wide range of biological activities to modulate stress processes under different stresses, like the universal molecule metformin (Lv and Guo, 2020). The reports of TMAO in humans involved in different diseases such as cardiovascular or tumor (Janeiro et al., 2018; Yang et al., 2019; Gatarek and Kaluzna-Czaplinska, 2021) may support this opinion.
However, we prefer an alternative explanation, which is the direction of our coming studies, that TMAO may help bacteria to cope with different external stresses in a pervasive way, such as by stabilizing proteins (Krywka et al., 2008; Ganguly et al., 2020). Many conditions including temperature, osmotic pressure, ionic strength, and pH, can denature proteins (Anfinsen and Scheraga, 1975), or oxidative stress occurring from hydrogen peroxide exposure can damage proteins through covalent modification of specific amino acid side chains (Dahl et al., 2015). In terms of antibiotics, sublethal concentrations of kanamycin induce protein aggregation by reducing ribosome fidelity and causing protein misfolding (Tamás et al., 2018; Schramm et al., 2019), and these mistranslated proteins can further exacerbate aggregation by promoting the production of reactive oxygen species and lead to oxidation-sensitive protein damage (Ling et al., 2012). More importantly, several antibiotics can induce protein aggregation (Cardoso et al., 2010; Tran et al., 2011; Wu et al., 2015), and Kohanski et al. (2007) found that ROS is a common pathway for the lethality of different antibiotics and that antibiotic-induced oxidative damage is part of the lethal effects of antibiotics (Dwyer et al., 2014). Proteins are responsible for accomplishing most cellular functions and protein activity is closely related to cell death. Acute external stresses can interfere with protein quality control mechanisms, leading to extensive protein denaturation and aggregation, which further leads to the loss of protein function, impairment of critical cellular functions required for growth and survival, and ultimately cell death (Mogk et al., 2018). For example, in the recently discovered new mode of cell death, cuproptosis, lipid acylated protein aggregation, and loss of iron–sulfur cluster proteins can trigger proteotoxic stress and ultimately cell death (Tsvetkov et al., 2022). These studies on protein homeostasis and cell death strengthen our confidence in the second explanation mentioned above and suggest that problem of antibiotic resistance caused by TMAO can be alleviated by finding adjuvants that promote protein denaturation. Alternatively, other possible mechanisms in TMAO-mediated antibiotic resistance events cannot be excluded. In addition, the discovery of common pathways that can respond to different lethal stresses contributes to a better understanding of the interaction of antibiotic resistance with adverse environments, where the emergence of resistance is not only limited to the drug itself.
Dietary-Mediated Changes in Intestinal Flora Metabolites May Influence the Effectiveness of Clinical Anti-infective Therapy
Different diets can alter the composition of the intestinal flora and the levels of microbiome-dependent intestinal metabolites (Sonnenburg et al., 2016; Frame et al., 2020; Mayneris-Perxachs et al., 2022). In particular, decreased levels of the tryptophan metabolite IAA in the intestine of mice fed a high-fat diet could antagonize the treatment of Methicillin-resistant Staphylococcus aureus (MRSA) or E. coli with several antibiotics (Liu et al., 2021). TMAO, as an intestine-derived metabolite, is formed by specific intestinal microorganisms that metabolize dietary choline and betaine into precursor TMA, which is oxidized to TMAO in the liver (Wang et al., 2011). A diverse diet rich in carnitine and choline from red meat, eggs, and shellfish (Li et al., 2018), betaine present in plants (Zeisel et al., 2003), may influence TMAO levels. Rath et al. (2021) showed that microbial flora and diet were associated with high levels of trimethylamine N-oxide in the plasma of senior individuals.
In vitro, our results suggest that TMAO reduces the susceptibility of E. coli to a variety of antibiotics in the range of 10 ~ 100 mM. After setting up the test concerning the concentration range of TMAO in normal human plasma, TMAO did not demonstrate an inhibitory effect on antibiotic efficacy (Supplementary Table 2). It might be a matter of concentration effects. Our results imply that TMAO may be a detrimental factor in anti-infective therapy. Considering the practical differences that exist between laboratory conditions and clinical situations, further clinical data are needed on what concentration of TMAO in humans interferes with antibiotic efficacy. Whether microbial flora and diet may influence anti-infective treatment efficacy by modulating TMAO levels in clinical situations also needs to be elucidated in large cohort studies that take into account individual differences and other factors. Moreover, our study and that of Liu et al. (2021) and Oliver et al. (2022) suggest that more possible intestinal metabolites should be included when testing for effects on antibiotic susceptibility and that promising protocols to counteract the inhibition of antibiotic efficacy by metabolites should be sought to inform clinical practice. If a regulated pathway of “diet-flora-metabolites-antibiotic resistance” is widespread, the control of bacterial resistance will require greater engineering.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
JQ, YL, and YW performed experiments and analyzed data. JQ, YL, and Morigen conceived the experiments and wrote/revised the paper. Morigen provided experimental materials and supervised the work. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (NSFC grant no. 32060016 to Morigen), Inner Mongolia Key Laboratory for Molecular Regulation of the Cell (no. 2021PT0002 to Morigen), and Research Program of science and technology at Universities of Inner Mongolia Autonomous Region (no. NJZZ20036 to YL).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to National BioResource Project (NIG, Japan): E. coli for providing strains.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.956673/full#supplementary-material
References
Anfinsen, C. B., and Scheraga, H. A. (1975). Experimental and theoretical aspects of protein folding. Adv. Protein Chem. 29, 205–300. doi: 10.1016/s0065-3233(08)60413-1
Baraquet, C., Théraulaz, L., Guiral, M., Lafitte, D., Méjean, V., and Jourlin-Castelli, C. (2006). TorT, a member of a new periplasmic binding protein family, triggers induction of the Tor respiratory system upon trimethylamine N-oxide electron-acceptor binding in Escherichia coli. J. Biol. Chem. 281, 38189–38199. doi: 10.1074/jbc.M604321200
Barbosa, T. M., and Levy, S. B. (2000). Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182, 3467–3474. doi: 10.1128/jb.182.12.3467-3474.2000
Becattini, S., Taur, Y., and Pamer, E. G. (2016). Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol. Med. 22, 458–478. doi: 10.1016/j.molmed.2016.04.003
Cani, P. D., Van Hul, M., Lefort, C., Depommier, C., Rastelli, M., and Everard, A. (2019). Microbial regulation of organismal energy homeostasis. Nat. Metab. 1, 34–46. doi: 10.1038/s42255-018-0017-4
Cardoso, K., Gandra, R. F., Wisniewski, E. S., Osaku, C. A., Kadowaki, M. K., Felipach-Neto, V., et al. (2010). DnaK and GroEL are induced in response to antibiotic and heat shock in Acinetobacter baumannii. J. Med. Microbiol. 59, 1061–1068. doi: 10.1099/jmm.0.020339-0
Cohen, S. P., Hächler, H., and Levy, S. B. (1993). Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175, 1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993
Dahl, J. U., Gray, M. J., and Jakob, U. (2015). Protein quality control under oxidative stress conditions. J. Mol. Biol. 427, 1549–1563. doi: 10.1016/j.jmb.2015.02.014
Dalla Via, A., Gargari, G., Taverniti, V., Rondini, G., Velardi, I., Gambaro, V., et al. (2019). Urinary TMAO levels are associated with the taxonomic composition of the gut microbiota and with the choline TMA-lyase gene (cutC) harbored by Enterobacteriaceae. Nutrients 12:62. doi: 10.3390/nu12010062
Duval, V., and Lister, I. M. (2013). MarA, SoxS and Rob of Escherichia coli – global regulators of multidrug resistance, virulence and stress response. Int. J. Biotechnol. Wellness Ind. 2, 101–124. doi: 10.6000/1927-3037.2013.02.03.2
Dwyer, D. J., Belenky, P. A., Yang, J. H., MacDonald, I. C., Martell, J. D., Takahashi, N., et al. (2014). Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. U. S. A. 111, E2100–E2109. doi: 10.1073/pnas.1401876111
Forster, S. C., Liu, J., Kumar, N., Gulliver, E. L., Gould, J. A., Escobar-Zepeda, A., et al. (2022). Strain-level characterization of broad host range mobile genetic elements transferring antibiotic resistance from the human microbiome. Nat. Commun. 13:1445. doi: 10.1038/s41467-022-29096-9
Frame, L. A., Costa, E., and Jackson, S. A. (2020). Current explorations of nutrition and the gut microbiome: a comprehensive evaluation of the review literature. Nutr. Rev. 78, 798–812. doi: 10.1093/nutrit/nuz106
Ganguly, P., Polák, J., van der Vegt, N. F. A., Heyda, J., and Shea, J. E. (2020). Protein stability in TMAO and mixed urea-TMAO solutions. J. Phys. Chem. B 124, 6181–6197. doi: 10.1021/acs.jpcb.0c04357
Gatarek, P., and Kaluzna-Czaplinska, J. (2021). Trimethylamine N-oxide (TMAO) in human health. EXCLI J. 20, 301–319. doi: 10.17179/excli2020-3239
Gessner, A., di Giuseppe, R., Koch, M., Fromm, M. F., Lieb, W., and Maas, R. (2020). Trimethylamine-N-oxide (TMAO) determined by LC-MS/MS: distribution and correlates in the population-based PopGen cohort. Clin. Chem. Lab. Med. 58, 733–740. doi: 10.1515/cclm-2019-1146
Gon, S., Giudici-Orticoni, M. T., Méjean, V., and Iobbi-Nivol, C. (2001). Electron transfer and binding of the c-type cytochrome TorC to the trimethylamine N-oxide reductase in Escherichia coli. J. Biol. Chem. 276, 11545–11551. doi: 10.1074/jbc.M008875200
Gon, S., Patte, J. C., Méjean, V., and Iobbi-Nivol, C. (2000). The torYZ (yecK bisZ) operon encodes a third respiratory trimethylamine N-oxide reductase in Escherichia coli. J. Bacteriol. 182, 5779–5786. doi: 10.1128/jb.182.20.5779-5786.2000
Janeiro, M. H., Ramírez, M. J., Milagro, F. I., Martínez, J. A., and Solas, M. (2018). Implication of trimethylamine N-oxide (TMAO) in disease: potential biomarker or new therapeutic target. Nutrients 10:1398. doi: 10.3390/nu10101398
Koeth, R. A., Wang, Z., Levison, B. S., Buffa, J. A., Org, E., Sheehy, B. T., et al. (2013). Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585. doi: 10.1038/nm.3145
Koh, A., De Vadder, F., Kovatcheva-Datchary, P., and Bäckhed, F. (2016). From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345. doi: 10.1016/j.cell.2016.05.041
Kohanski, M. A., Dwyer, D. J., Hayete, B., Lawrence, C. A., and Collins, J. J. (2007). A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810. doi: 10.1016/j.cell.2007.06.049
Koronakis, V. (2003). TolC--the bacterial exit duct for proteins and drugs. FEBS Lett. 555, 66–71. doi: 10.1016/s0014-5793(03)01125-6
Koronakis, V., Sharff, A., Koronakis, E., Luisi, B., and Hughes, C. (2000). Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405, 914–919. doi: 10.1038/35016007
Krywka, C., Sternemann, C., Paulus, M., Tolan, M., Royer, C., and Winter, R. (2008). Effect of osmolytes on pressure-induced unfolding of proteins: a high-pressure SAXS study. ChemPhysChem 9, 2809–2815. doi: 10.1002/cphc.200800522
Li, X. S., Wang, Z., Cajka, T., Buffa, J. A., Nemet, I., Hurd, A. G., et al. (2018). Untargeted metabolomics identifies trimethyllysine, a TMAO-producing nutrient precursor, as a predictor of incident cardiovascular disease risk. JCI. Insight 3:e99096. doi: 10.1172/jci.insight.99096
Liao, Y. T., Manson, A. C., DeLyser, M. R., Noid, W. G., and Cremer, P. S. (2017). Trimethylamine N-oxide stabilizes proteins via a distinct mechanism compared with betaine and glycine. Proc. Natl. Acad. Sci. U. S. A. 114, 2479–2484. doi: 10.1073/pnas.1614609114
Lin, T. Y., and Timasheff, S. N. (1994). Why do some organisms use a urea-methylamine mixture as osmolyte? Thermodynamic compensation of urea and trimethylamine N-oxide interactions with protein. Biochemistry 33, 12695–12701. doi: 10.1021/bi00208a021
Ling, J., Cho, C., Guo, L. T., Aerni, H. R., Rinehart, J., and Söll, D. (2012). Protein aggregation caused by aminoglycoside action is prevented by a hydrogen peroxide scavenger. Mol. Cell 48, 713–722. doi: 10.1016/j.molcel.2012.10.001
Liu, Y., Yang, K., Jia, Y., Shi, J., Tong, Z., Fang, D., et al. (2021). Gut microbiome alterations in high-fat-diet-fed mice are associated with antibiotic tolerance. Nat. Microbiol. 6, 874–884. doi: 10.1038/s41564-021-00912-0
Lv, Z., and Guo, Y. (2020). Metformin and its benefits for various diseases. Front. Endocrinol. 11:191. doi: 10.3389/fendo.2020.00191
Mayneris-Perxachs, J., Castells-Nobau, A., Arnoriaga-Rodríguez, M., Martin, M., de la Vega-Correa, L., Zapata, C., et al. (2022). Microbiota alterations in proline metabolism impact depression. Cell Metab. 34, 681–701. doi: 10.1016/j.cmet.2022.04.001
Mogk, A., Bukau, B., and Kampinga, H. H. (2018). Cellular handling of protein aggregates by disaggregation machines. Mol. Cell 69, 214–226. doi: 10.1016/j.molcel.2018.01.004
Oliver, A., Xue, Z., Villanueva, Y. T., Durbin-Johnson, B., Alkan, Z., Taft, D. H., et al. (2022). Association of diet and antimicrobial resistance in healthy U. S. adults. mBio 13:e0010122. doi: 10.1128/mbio.00101-22
Pietrzyk-Brzezinska, A. J., and Cociurovscaia, A. (2022). Structures of the TetR-like transcription regulator RcdA alone and in complexes with ligands. Proteins 90, 33–44. doi: 10.1002/prot.26183
Rath, S., Rox, K., Kleine Bardenhorst, S., Schminke, U., Dörr, M., Mayerle, J., et al. (2021). Higher trimethylamine-N-oxide plasma levels with increasing age are mediated by diet and trimethylamine-forming bacteria. mSystems 6:e0094521. doi: 10.1128/mSystems.00945-21
Ruppé, E., Ghozlane, A., Tap, J., Pons, N., Alvarez, A. S., Maziers, N., et al. (2019). Prediction of the intestinal resistome by a three-dimensional structure-based method. Nat. Microbiol. 4, 112–123. doi: 10.1038/s41564-018-0292-6
Schramm, F. D., Schroeder, K., Alvelid, J., Testa, I., and Jonas, K. (2019). Growth-driven displacement of protein aggregates along the cell length ensures partitioning to both daughter cells in Caulobacter crescentus. Mol. Microbiol. 111, 1430–1448. doi: 10.1111/mmi.14228
Seibel, B. A., and Walsh, P. J. (2002). Trimethylamine oxide accumulation in marine animals: relationship to acylglycerol storage. J. Exp. Biol. 205, 297–306. doi: 10.1242/jeb.205.3.297
Shimada, T., Katayama, Y., Kawakita, S., Ogasawara, H., Nakano, M., Yamamoto, K., et al. (2012). A novel regulator RcdA of the csgD gene encoding the master regulator of biofilm formation in Escherichia coli. Microbiology 1, 381–394. doi: 10.1002/mbo3.42
Sonnenburg, J. L., and Bäckhed, F. (2016). Diet-microbiota interactions as moderators of human metabolism. Nature 535, 56–64. doi: 10.1038/nature18846
Sonnenburg, E. D., Smits, S. A., Tikhonov, M., Higginbottom, S. K., Wingreen, N. S., and Sonnenburg, J. L. (2016). Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215. doi: 10.1038/nature16504
Subramaniam, S., and Fletcher, C. (2018). Trimethylamine N-oxide: breathe new life. Br. J. Pharmacol. 175, 1344–1353. doi: 10.1111/bph.13959
Suzuki, T., Heaney, L. M., Jones, D. J., and Ng, L. L. (2017). Trimethylamine N-oxide and risk stratification after acute myocardial infarction. Clin. Chem. 63, 420–428. doi: 10.1373/clinchem.2016.264853
Tai, N., Wong, F. S., and Wen, L. (2015). The role of gut microbiota in the development of type 1, type 2 diabetes mellitus and obesity. Rev. Endocr. Metab. Disord. 16, 55–65. doi: 10.1007/s11154-015-9309-0
Tamás, M. J., Fauvet, B., Christen, P., and Goloubinoff, P. (2018). Misfolding and aggregation of nascent proteins: a novel mode of toxic cadmium action in vivo. Curr. Genet. 64, 177–181. doi: 10.1007/s00294-017-0748-x
Tang, W. H., Wang, Z., Fan, Y., Levison, B., Hazen, J. E., Donahue, L. M., et al. (2014). Prognostic value of elevated levels of intestinal microbe-generated metabolite trimethylamine-N-oxide in patients with heart failure: refining the gut hypothesis. J. Am. Coll. Cardiol. 64, 1908–1914. doi: 10.1016/j.jacc.2014.02.617
Tang, W. H., Wang, Z., Kennedy, D. J., Wu, Y., Buffa, J. A., Agatisa-Boyle, B., et al. (2015). Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 116, 448–455. doi: 10.1161/circresaha.116.305360
Thomas, P. D., and Dill, K. A. (1993). Local and nonlocal interactions in globular proteins and mechanisms of alcohol denaturation. Protein Sci. 2, 2050–2065. doi: 10.1002/pro.5560021206
Tran, T. D., Kwon, H. Y., Kim, E. H., Kim, K. W., Briles, D. E., Pyo, S., et al. (2011). Decrease in penicillin susceptibility due to heat shock protein ClpL in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 55, 2714–2728. doi: 10.1128/aac.01383-10
Trøseid, M., Ueland, T., Hov, J. R., Svardal, A., Gregersen, I., Dahl, C. P., et al. (2015). Microbiota-dependent metabolite trimethylamine-N-oxide is associated with disease severity and survival of patients with chronic heart failure. J. Intern. Med. 277, 717–726. doi: 10.1111/joim.12328
Tsvetkov, P., Coy, S., Petrova, B., Dreishpoon, M., Verma, A., Abdusamad, M., et al. (2022). Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 375, 1254–1261. doi: 10.1126/science.abf0529
Ufnal, M., Zadlo, A., and Ostaszewski, R. (2015). TMAO: a small molecule of great expectations. Nutrition 31, 1317–1323. doi: 10.1016/j.nut.2015.05.006
Wang, Z., Klipfell, E., Bennett, B. J., Koeth, R., Levison, B. S., Dugar, B., et al. (2011). Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63. doi: 10.1038/nature09922
Wang, H., Rong, X., Zhao, G., Zhou, Y., Xiao, Y., Ma, D., et al. (2022). The microbial metabolite trimethylamine N-oxide promotes antitumor immunity in triple-negative breast cancer. Cell Metab. 34, e588, 581–594. doi: 10.1016/j.cmet.2022.02.010
Wilson, A., McLean, C., and Kim, R. B. (2016). Trimethylamine-N-oxide: a link between the gut microbiome, bile acid metabolism, and atherosclerosis. Curr. Opin. Lipidol. 27, 148–154. doi: 10.1097/mol.0000000000000274
Wilson, A., Teft, W. A., Morse, B. L., Choi, Y. H., Woolsey, S., DeGorter, M. K., et al. (2015). Trimethylamine-N-oxide: a novel biomarker for the identification of inflammatory bowel disease. Dig. Dis. Sci. 60, 3620–3630. doi: 10.1007/s10620-015-3797-3
Wu, X., Held, K., Zheng, C., Staudinger, B. J., Chavez, J. D., Weisbrod, C. R., et al. (2015). Dynamic proteome response of Pseudomonas aeruginosa to tobramycin antibiotic treatment. Mol. Cell. Proteomics 14, 2126–2137. doi: 10.1074/mcp.M115.050161
Xu, R., and Wang, Q. (2016). Towards understanding brain-gut-microbiome connections in Alzheimer’s disease. BMC Syst. Biol. 10:63. doi: 10.1186/s12918-016-0307-y
Yang, S., Li, X., Yang, F., Zhao, R., Pan, X., Liang, J., et al. (2019). Gut microbiota-dependent marker TMAO in promoting cardiovascular disease: inflammation mechanism, clinical prognostic, and potential as a therapeutic target. Front. Pharmacol. 10:1360. doi: 10.3389/fphar.2019.01360
Zeisel, S. H., Mar, M. H., Howe, J. C., and Holden, J. M. (2003). Concentrations of choline-containing compounds and betaine in common foods. J. Nutr. 133, 1302–1307. doi: 10.1093/jn/133.5.1302
Zeisel, S. H., and Warrier, M. (2016). Trimethylamine N-oxide, the microbiome, and heart and kidney disease. Annu. Rev. Nutr. 37, 157–181. doi: 10.1146/annurev-nutr-071816-064732
Zhang, A. Q., Mitchell, S. C., and Smith, R. L. (1999). Dietary precursors of trimethylamine in man: a pilot study. Food Chem. Toxicol. 37, 515–520. doi: 10.1016/s0278-6915(99)00028-9
Keywords: trimethylamine N-oxide, antibiotics, urea stress, disinfectants, protein denaturation, cell death, intestinal flora metabolite, Escherichia coli
Citation: Qiao J, Liang Y, Wang Y and Morigen (2022) Trimethylamine N-Oxide Reduces the Susceptibility of Escherichia coli to Multiple Antibiotics. Front. Microbiol. 13:956673. doi: 10.3389/fmicb.2022.956673
Edited by:
Benno H. Ter Kuile, University of Amsterdam, NetherlandsReviewed by:
Jed F. Fisher, University of Notre Dame, United StatesCopyright © 2022 Qiao, Liang, Wang and Morigen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Morigen, bW9yaWdlbm1AaG90bWFpbC5jb20=
†These authors have contributed equally to this work
 Jiaxin Qiao
Jiaxin Qiao Yan Liang2†
Yan Liang2† Morigen
Morigen