
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 25 August 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.956642
This article is part of the Research TopicState of the Art: Biological Control of Diseases of Horticultural CropsView all 6 articles
 Aiting Zhou1
Aiting Zhou1 Fang Wang1,2*
Fang Wang1,2* Jiabi Yin3
Jiabi Yin3 Ruiqi Peng4
Ruiqi Peng4 Jia Deng1
Jia Deng1 Dezhou Shen3
Dezhou Shen3 Jianrong Wu1,4
Jianrong Wu1,4 Xiaoyun Liu2
Xiaoyun Liu2 Huancheng Ma1*
Huancheng Ma1*Anthracnose disease caused by Colletotrichum fructicola is a serious disease that can afflict Camellia oleifera. Biological control is a rapidly growing approach for the management of plant diseases. In this study, we investigated the bio-control efficiency and the defense responses of an endophytic Bacillus tequilensis strain YYC 155, which was isolated from the root nodules of the Crotalaria pallida against anthracnose disease, caused by C. fructicola in C. oleifera. B. tequilensis YYC 155 exhibited significant inhibitory activity against anthracnose disease, caused by C. fructicola in C. oleifera. YYC 155 can secrete extracellular hydrolases, such as chitinase and β-1, 3-glucanase, which produce lipopeptides that are antimicrobial and forms strong biofilms. In addition, in treatment with YYC 155, the cell membranes of C. fructicola were injured and the leakage of cell contents from the mycelia of the pathogen was increased. Spraying 1 × 107 cells mL–1 bacterial suspension of YYC 155 on C. oleifera leaves enhanced the activity of key enzymes in C. oleifera associated with the phenylpropanoid pathway and increased the content of phenolic compounds and flavonoids. Results of our study indicate that B. tequilensis YYC 155 may potentially represent an effective biocontrol agent against anthracnose disease in C. oleifera.
Camellia oleifera is an evergreen shrub in the Camellia family. It is known for its production of tea oil or Camellia oil and is an economically important crop in southern China (Wu et al., 2019). C. oleifera is an important component in many health care products due to the high content of unsaturated fatty acids in its oil and other nutrients (Zhang et al., 2012; Wu et al., 2019). The production of C. camellia, however, is hampered by anthracnose disease, caused by Colletotrichum fructicola, a serious disease that is widespread in Camellia production areas. It can infect various parts of C. oleifera during its entire production cycle, including dropped leaves and fruit. Anthracnose disease has led to a decline in Camellia production and results in huge economic losses every year (Perrett et al., 2003). Traditionally, anthracnose control has mainly been achieved using chemical fungicides. While these agents are effective in controlling anthracnose, the overuse of chemical pesticides may have negative effects on human health and the environment (Kilani-Feki et al., 2016). Therefore, the development of safe and effective biological control methods is attracting increasing public attention.
The use of a variety of different microbial antagonists as biocontrol agents appears to represent a promising alternative for controlling anthracnose (Shiomi et al., 2006). In this regard, endophytic microorganisms (bacteria and fungi) are being extensively studied for their potential as biocontrol agents due to their antagonistic activity against pathogens and their ability to induce resistance in host plants (Shiomi et al., 2006; Mahdi et al., 2014). The bacterial genus Bacillus has been identified as a frequently occurring endophyte in many plant species (Vendan et al., 2010). Cui et al. (2020) reported that Bacillus velezensis 8-4 isolated from potatoes exhibits a strong antagonistic ability against potato scab and increases potato yields. Li et al. (2009) screened 175 endophytic bacteria from C. oleifera to control the anthracnose caused by Colletetrichum gloeosporioides with confront culture method and Bacillus licheniformis YB128 had the best antagonistic effect with 86.1% inhibitory rate. An endophytic bacteria, Bacillus subtilis strain 1-L-29, isolated from Camellia oleifera could efficiently inhibit the C. fructicola growth and colonize the root surfaces of C. oleifera (Xu et al., 2020). It is known that Bacillus species produces a variety of compounds, including antibiotics (Leifertl et al., 1995), lipopeptides (Ahimou et al., 2000), cell wall-degrading enzymes (Leelasuphakul et al., 2006), and volatile organic compounds (VOCs) (Fiddaman and Rossall, 1993), which play a role in the inhibitory activity of Bacillus against many plant pathogens. To further explore the mechanisms of biocontrol endophytes, some researchers have focused on their effects on the physiological metabolism of pathogens. Dai et al. (2021) found that Bacillus pumilus HR10 was capable of injuring the cell membrane of pathogens and increasing the leakage of cell components. Additionally, antagonistic Bacillus species could induce systemic resistance of plants, especially the phenylpropanoid metabolism that plays a very important role in plant disease resistance.
Elicitation of plant defense mechanisms by these Bacillus strains has been demonstrated in root irrigation treatment and root-irrigation with Bacillus strains effectively increased plant defense-related enzyme activities (Chen et al., 2010; Zhang et al., 2022). Whether leaf spraying treatment of biological control agents have the same effects of inducing disease resistance as root irrigation treatment. Therefore, the objective of the present work was to investigate the impacts of foliar spray with YYC 155, which was previously isolated from the root nodule of Crotalaria pallida on the activity of some defense-related enzymes and compounds in the phenylpropanoid pathway of C. oleifera plants. Moreover, we assessed the ability of Bacillus sp. strain YYC 155 (subsequently identified as a strain of B. tequilensis) to inhibit the growth of C. fructicola in vitro, determined the presence of genes in YYC 155 responsible for the synthesis of antimicrobial compounds, and determined the effect of YYC 155 on parameters such as cell membrane integrity and cell leakage in C. fructicola.
Two-year-old shrubs of C. oleifera (“Changlin No. 4”) were obtained from Yingjiang Linli Oil Tea Co., Ltd., in Dehong Prefecture, Yunnan Province of China, and then grown in a greenhouse with the following conditions 25°C and 85% relative humidity.
Pathogen isolation: Leaves of C. oleifera infected exhibiting anthracnose symptoms were collected from Dehong Prefecture, Yunnan Province of China. The leaves were brought to the laboratory and rinsed thoroughly with tap water. Five pieces (3–5 mm) of leaf tissue were taken from the border of lesions of 30 leaves and surface sterilized with 75% ethanol for 10 s and 0.1% mercuric chloride for 3 min. The leaf sections were then rinsed three times with sterile distilled water (SDW), dried on sterile filter paper, and placed on the Potato Dextrose Agar (PDA) medium amended with streptomycin sulfate (300 mg L–1) (w/v) and incubated for 1 week at 25°C. Pathogenicity of the collected C. fructicola isolates was confirmed on C. oleifera plants. The most aggressive isolate of C. fructicola was selected for use in the in vitro and in vivo bioassays.
Eight bacterial strains were isolated from root nodules of Crotalaria pallida in a previous study and maintained in a lysogeny broth (LB) medium at 4°C at the Key Laboratory of the National Forestry and Grassland Administration on Biodiversity Conservation in Southwest China, Southwest Forestry University, Kunming, Yunnan Province of China.
We conducted dual-culture assays to determine the antifungal capacity of the bacterial strains in vitro in Petri plates (diameter, 90 mm). Five microliters (OD600 = 0.1) droplets of each of the eight bacterial strains were then placed on Petri plates. Four droplets were placed around the edge of the Petri dish. An agar-mycelial plug (diameter, 6 mm) obtained from the margin of a C. fructicola culture during active growth was placed upside down in the center of the Petri plate center approximately 30 mm away from the droplets of the bacterial suspension. Three replicates of every bacterial strain were prepared, as well as pathogen-only plates (control). After inoculation, the plates were placed in an incubator at 25°C. The diameter of the colony of C. fructicola was recorded daily beginning at day 4 and ending at day 9 of incubation. Antifungal activity was calculated as the proportion of mycelial growth inhibition relative to the control using the formula, (R1–R2)/R1 × 100%. In the formula, R2 represents pathogen colony diameter in the presence of a bacterial isolate, while R1 is pathogen colony diameter in control plates.
The bacterial strain YYC 155, which had exhibited the best inhibitory activity against C. fructicola in vitro in the previous experiment, was used to assess its inhibitory effect on C. fructicola in detached leaves taken from healthy C. oleifera plants. The detached leaves were washed with sterilized distilled water (SDW), dried on paper towels, and placed abaxial side up in Petri dishes containing two filter papers saturated with SDW (Dorighello et al., 2015). Four artificial wounds were administered at the same position on each leaf using a sterile needle. Each wound was inoculated with 5 μL of a YYC 155 suspension at a concentration of 1 × 107 cells mL–1 (previously cultured for 24 h). Subsequently, 24 h later YYC 155-inoculated leaves were inoculated with mycelial disks (diameter, 5 mm) of actively growing mycelia of C. fructicola. Wounded leaves inoculated with just C. fructicola and leaves inoculated with just SDW served as positive and negative controls, respectively. Three leaves with four wounds each were used for each treatment and the experiment was repeated twice. Lesion diameters on leaves were measured daily beginning on the 3rd day after pathogen inoculation. The inhibition of C. fructicola was determined by comparing the diameter of the lesion.
Genomic DNA was extracted from bacterial strain YYC 155 using an Ezup Column Bacteria Genomic DNA Purification Kit (Sangon Biotech, Shanghai, China), according to the manufacturer’s instructions. We utilized the universal primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) for PCR amplification of the 16S rRNA gene (Fredriksson et al., 2013). The obtained PCR products were purified and sequenced. The consensus 16S rRNA sequence was analyzed at https://www.ezbiocloud.net, to identify phylogenetic neighbors and to calculate the pairwise similarity of the 16S rRNA gene sequence. CLUSTALW in Mega 7.0 was used to conduct a similarity calculation and multiple alignments (Kumar et al., 2016). A 16S rRNA phylogenetic tree was constructed using the neighbor-joining method with 1,000 bootstrap replicates.
Relative electrical conductivity was measured as described by Elsherbiny et al. (2021) with slight modification. An agar-mycelial plug (6 mm diameter) taken from the edge of an actively growing culture of C. fructicola was placed in a 50 mL centrifuge tube containing 20 mL of Potato Dextrose Broth (PDB) and incubated for 72 h at 25°C on a rotary shaker set at 160 rpm/min. The resulting mycelia were then washed 3 × with SDW and transferred to a 10 mL centrifuge tube containing 4 mL of a YYC 155 suspension at 1 × 107 cells mL–1. Tubes containing SDW instead of YYC 155 served as a control. The mixtures were incubated at 28°C on a rotary shaker set at 180 rpm/min, and samples were collected at 24 h, 48 h, 72 h, 96 h, and 120 h. The electrical conductivity of the mixtures was determined using an electrical conductivity meter (DDS-307, China), after which the mixtures were heated in a boiling water bath for 15 min. After cooling to room temperature, the final conductivity measurement was taken. Relative conductivity was determined using the formula, relative conductivity (%) = initial conductivity/final conductivity × 100%. Each assay was conducted in triplicate.
The assessment of injury to C. fructicola used the same experimental design used to measure electrical conductivity; however, only the supernatant was used in the assessment. The cultures were incubated at 24 h, 48 h, 72 h, 96 h, and 120 h, and then centrifuged at 5,000 g/min for 2 min at 4°C. The supernatant was used to assess the leakage of extracellular nucleic acids and soluble protein content, while the remaining mycelia were used to measure malondialdehyde (MDA) content (Dai et al., 2021). The values for leakage of extracellular nucleic acids are expressed as the OD260 mL–1, the values for soluble protein content are expressed as milligram per mL of supernatant, and the values for MDA are expressed as nmol per gram of mycelia. Each assessment had three replicates and the experiment was conducted twice.
To test the effect of YYC 155 on fungal mycelia morphology, a scanning electron microscope (SEM) was utilized. Mycelial plugs of C. fructicola with 6-mm-diameter were obtained from the interaction region treated with strain YYC 155 through dual-culture assays according to the above-mentioned method in section “Screening of bacterial strains in vitro” and mycelial plugs of the C. fructicola without strain YYC 155 were as control. The mycelial plugs were fixed with 2.5% glutaraldehyde at 4°C overnight, rinsed with 0.1 M phosphate buffer three times, and dehydrated through a series gradient concentration of ethanol followed by drying in a critical point drainer by liquid CO2. Then the samples were coated with gold and observed using a Hitachi Regulus 8100 Scanning Electron Microscope.
The ability of YYC 155 to form a biofilm was assessed as previously described (Banerjee et al., 2018) with some modifications. A single colony of YYC 155 obtained from a 24 h culture of YYC 155 growing on LB agar plates at 28°C was placed in LB liquid medium and cultured for 24 h at 28°C on a rotary shaker set at 160 rpm/min. The cell density of the 24 h culture was adjusted to 1 × 107 cells mL–1, and 1,500 μL of aliquots were placed into individual wells of a 24-well polystyrene plate that contained 1,000 μL LB medium. Three replicate wells were used to assess biofilm formation at different timepoints. Three wells containing LB medium alone were used as a control. The plates were incubated at 28°C for 12 h, 24 h, 48 h, 72 h, 96 h, and 120 h. At each timepoint, the medium with the pathogen was emptied from the wells and the adherent biofilm was stained with the 0.1% aqueous crystal violet solution for 10 min. The wells were then rinsed 3 × with SDW and the biofilms were dissolved by adding 40% acetic acid solution (2,500 μL) to each well for 5 min. The absorbance of the acetic acid solution was then measured at OD560 to assess the ability of YYC155 to form a biofilm. Three duplicate wells were used for the control and bacteria at each time point, and the assay was conducted twice.
The secretion of extracellular hydrolases by strain YYC 155 was assessed. The bacteria were grown in LB medium for 24 h at 28°C, and a single colony of YYC 155 was then transferred to LB liquid medium and cultured at 28°C on a rotary shaker set at 180 rpm/min. Chitinase and β-1, 3-glucanase activity in the culture medium was evaluated after 12 h, 24 h, 48 h, 72 h, 96 h, and 120 h of incubation. The enzymatic assays were conducted as described by Zhang et al. (2017) with some modifications. Chitinase activity is expressed in units, where one unit is defined as the enzyme activity catalyzing the formation of 1 mg of N-acetyl-D-glucosamine per milliliter of culture medium per hour. B -1, 3-glucanase is also denoted in units, where one unit is defined as the amount of enzyme needed to produce 1 mg of reducing sugar per milliliter of culture suspension. Three replicates were measured in each experiment and the experiment was repeated twice.
Genomic DNA was isolated from B. tequilensis YYC 155 as described in section “Identification of bacterial strain YYC 155.” Lipopeptide biosynthetic genes were then amplified using the genomic DNA as a template. PCR amplification was conducted in 50 μL reaction volume in a thermocycler using the following protocol: initial denaturation at 95°C for 3 min, 32 cycles of 94°C for 30 s, annealing temperature varied depending upon the specific primers, and extension at 72°C for 40 s. The annealing temperature and the primers used in the PCR amplification are listed in Supplementary Table 1.
A total of 20 mL of a bacterial suspension of YYC 155 at the concentration of 1 × 107 cells mL–1, previously cultured at 28°C on a rotary shaker set at 180 rpm/min for 24 h, was evenly sprayed on the leaves of 2-year-old potted shrubs of C. oleifera, with the application of sterile distilled water treatment serving as a control. The effect of the application of s YYC 155 on the level of total phenols, flavonoids, and the activity of phenylalanine ammonia lyase (PAL), polyphenol oxidase (PPO), cinnamic acid-4-hydroxylase (C4H), 4-coumarate: CoA ligase (4CL), and chalcone isomerase (CHI) were assessed in leaves of C. oleifera collected at 1 d (24 h after the application of YYC 155), 5 d, 10 d, 15 d, 20 d, 25 d, and 30 d. The level of total phenols and flavonoids was measured using the method of Wang et al. (2018) in which absorbance was measured at 280 nm or 325 nm. Content is expressed on a fresh weight basis as OD280 g–1 and OD325 g–1, respectively. PAL, C4H, 4CL, and CHI activities were also calculated using the procedure described by Wang et al. (2018). PPO was assayed according to Wang et al. (2021). Results are expressed as units per gram fresh weight of C. oleifera leaves.
A completely randomized design was used in the experiments. The data were statistically analyzed using Duncan’s multiple range test or an independent t-test in SPSS20.0. Statistical significance was set at P < 0.05.
The antifungal activity of eight strains of bacteria isolated from root nodules of C. pallida was assessed in a dual culture plate assay. The eight strains (YYC 22, YYC 38, YYC 72, YYC 79, YYC 135, YYC 150, YYC 155, and YYC 171) exhibited varying levels of inhibition of C. fructicola, relative to the control, after 9 d of co-culture. The percentage of inhibition of the different strains was 18.02%, 33.72%, 43.80%, 33.91%, 45.35%, 21.51%, 56.00%, and 34.88%, respectively (Figure 1). Strain YYC 155 exhibited the greatest level of inhibitory activity against the mycelial growth of C. fructicola and its inhibition level increased with incubation time (Figures 1, 2). Based on these results, strain YYC 155 was used in the subsequent experiments.
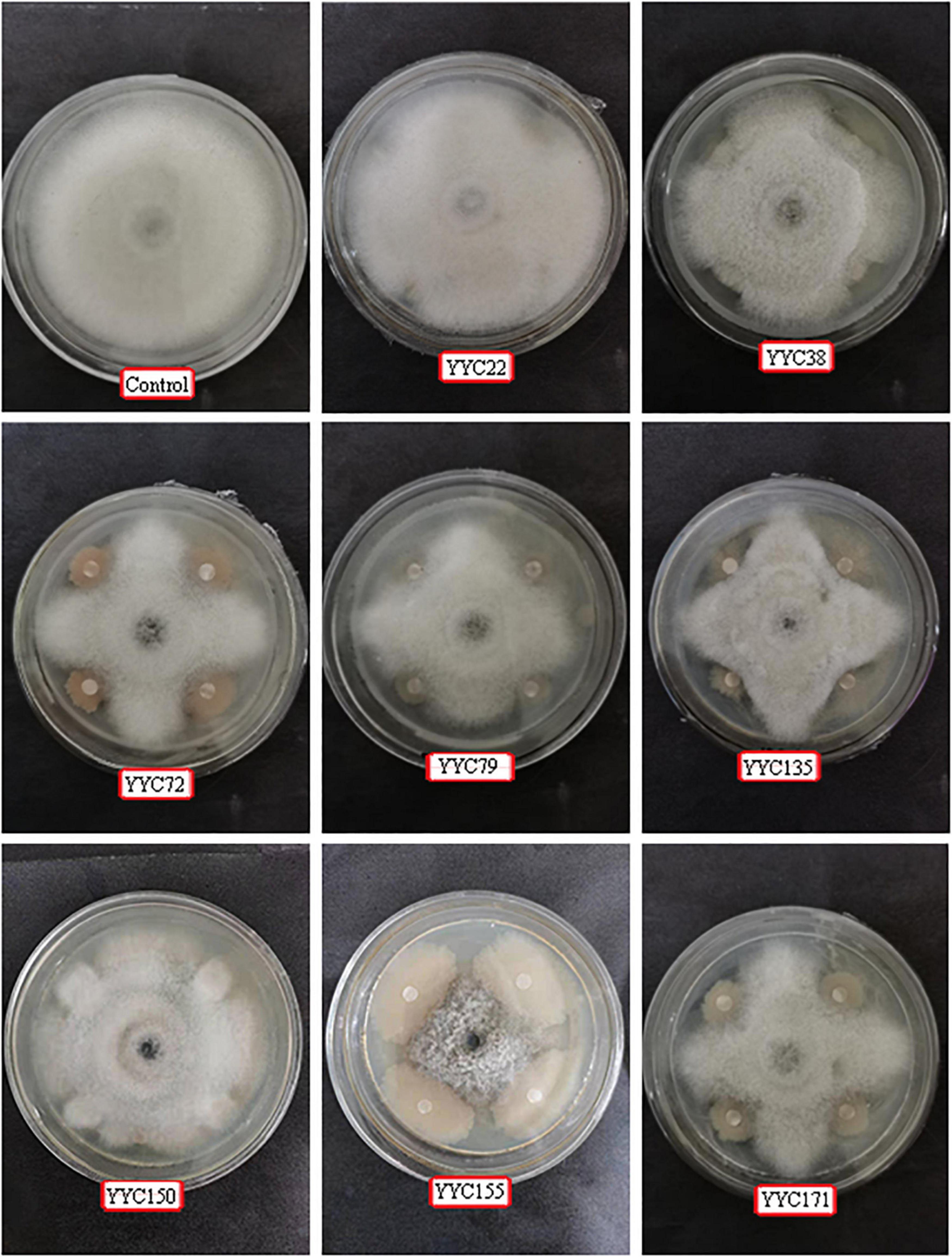
Figure 1. Representative photos illustrating the antagonistic activity of eight bacterial strains isolated from root nodules of C. pallida against C. fructicola in vitro. Three independent replicates were used for different strains.
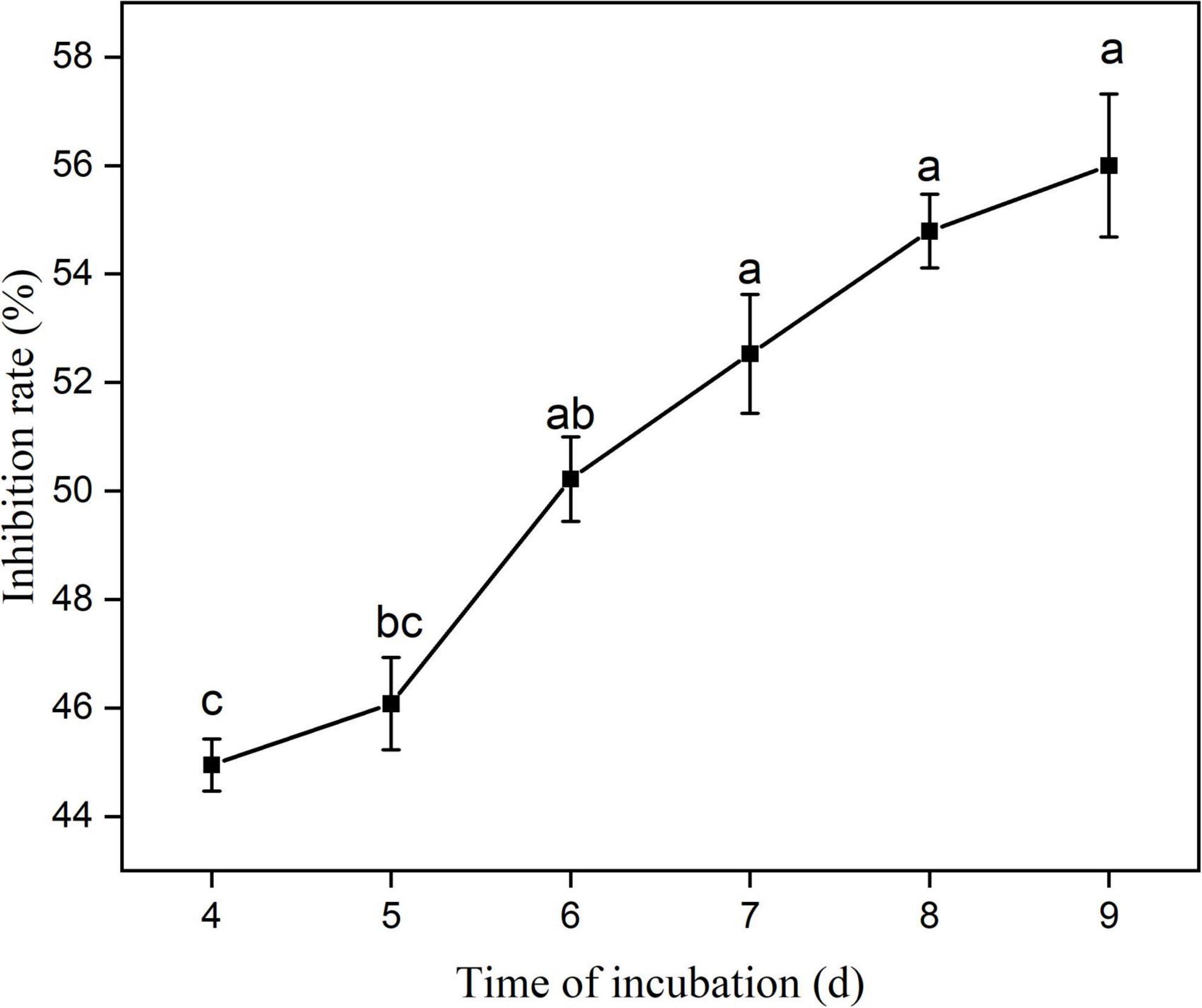
Figure 2. Percentage inhibition of C. fructicola by Bacillus tequilensis strain YYC 155 in vitro during 9 days of co-culture. Three duplicates are used for control and strain YYC 155 at each time point. Data represent the mean ± standard error (se). Different letters indicate a significant difference between the inhibitor rate on different days as determined by Duncan’s multiple range tests (P < 0.05).
Figure 3 demonstrates that wounded, detached leaves of C. oleifera treated with strain YYC 155 exhibited a decreased rate of lesion development. Lesion diameter in C. oleifera leaves treated with YYC 155 was significantly (P < 0.05) smaller than it was in leaves inoculated with C. fructicola alone beginning from the 3rd day after pathogen inoculation (Table 1). No evidence of lesions was observed on leaves treated only with sterile water or only with YYC 155, the latter indicating that strain YYC 155 is not a plant pathogen of C. oleifera.

Figure 3. The effect of Bacillus tequilensis YYC 155 on lesion development in detached leaves of C. oleifera caused by C. fructicola at 5 days after pathogen inoculation. Three independent replicates were used for different treatments and three detached leaves were used for each replicate. (A) Sterile water treatment (control), (B) YYC 155 treatment, (C) C. fructicola treatment, (D) YYC 155 + C. fructicola treatment.
We amplified the 16S rRNA gene sequence of YYC 155 from genomic DNA to determine its taxonomic classification. Then, the nearly complete 16S rRNA gene sequence of 1,452 bp was amplified and sequenced. Nearly, the entire 1,452 bp sequence of the 16S rRNA gene was amplified and sequenced. Sequence analysis indicated that YYC 155 appears to be B. tequilensis having 100% homology to B. tequilensis strains CRRI-HN-4 and SJ33. The phylogenetic tree of the sequenced 16S rRNA also indicated that it is in the same clade as B. tequilensis, further suggesting that YYC 155 is a strain of B. tequilensis (Figure 4). The 16S rRNA sequence of YYC 155 was deposited at DDBJ/ENA/GenBank (accession number, OM131692).
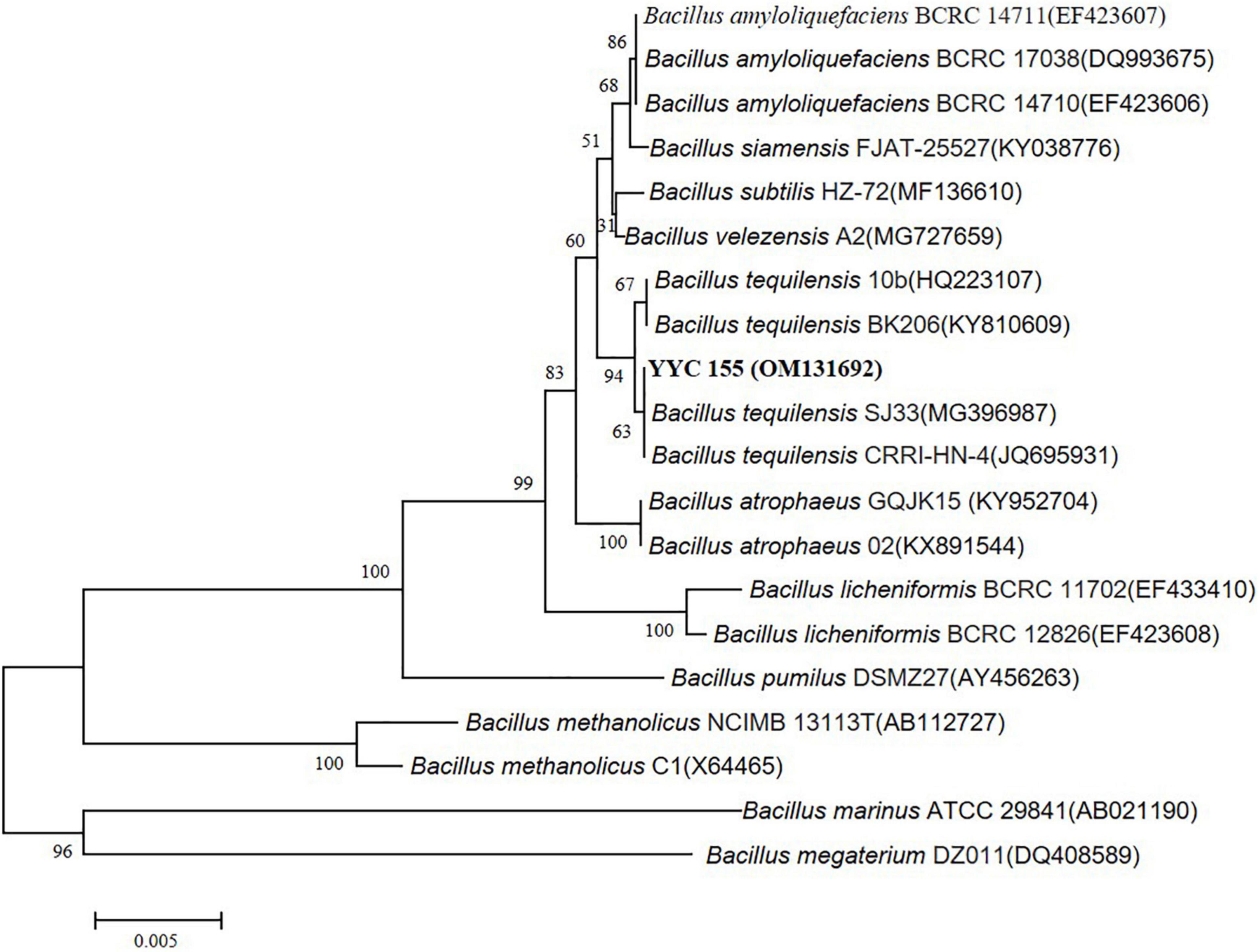
Figure 4. Phylogenetic tree of YYC 155 based on the 16S rRNA gene sequence. The tree was constructed using the neighbor-joining method with 1,000 bootstrap replicates. Bootstraps greater than 50% are indicated. Bars = 0.005 nucleotide substitutions per site.
The permeability of the cell membrane of C. fructicola was assessed as the relative electrical conductivity of mycelia (Figure 5A). The electrical conductivity of C. fructicola mycelia co-incubated with YYC 155 increased significantly over the entire course of co-incubation relative to the control treatment.
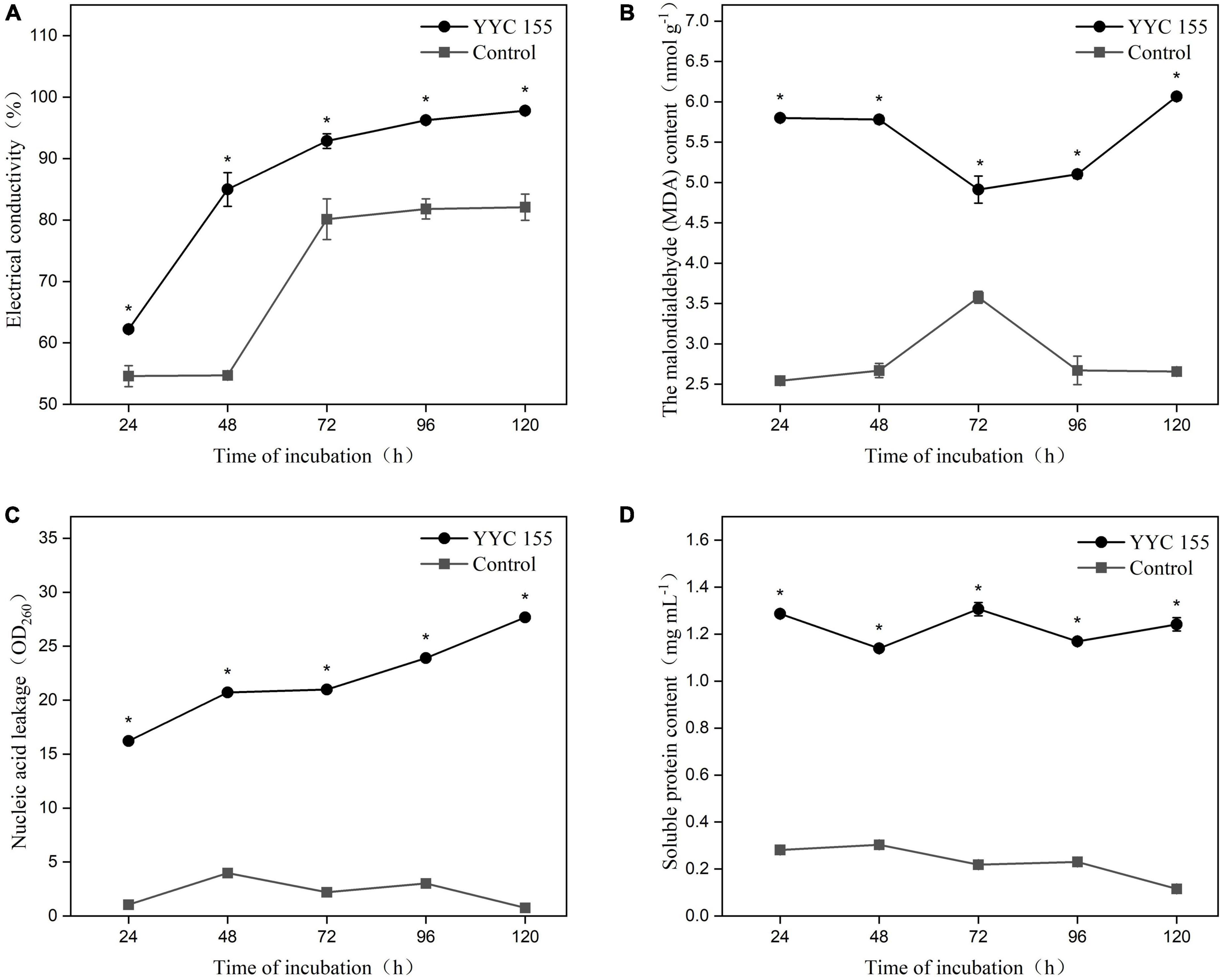
Figure 5. Effect of strain YYC 155 on cell membrane permeability and the content of cellular compounds in C. fructicola. (A) Electrical conductivity; (B) the malondialdehyde (MDA) content; (C) nucleic acid leakage; (D) soluble protein content. Three duplicates are used for the control and YYC 155 treatment against C. fructicola at each time point and were measured three times for each replicate. Data represent the mean ± standard error (se). Asterisks indicate a significant difference between the control and YYC 155 treatment groups at the designated time point as determined by the t-test (P < 0.05).
Lipid peroxidation in C. fructicola mycelia was assessed by measuring malondialdehyde (MDA), a marker of oxidative stress injury. The MDA level in C. fructicola mycelia co-cultured with YYC 155 was significantly higher (P < 0.05), relative to the control mycelia. The highest level of MDA (6.07 ± 0.05 nmol g–1) in C. fructicola co-cultured with YYC 155 was observed after 120 h of co-incubation (Figure 5B). These results indicate that exposure of C. fructicola to YYC 155 induces injury to the cell membranes of mycelia of C. fructicola and as a result increases the permeability of the cell membrane, as evidenced by the increased level of electrolyte leakage.
The effect of YYC 155 on the leakage of nucleic acids and protein from the mycelia of C. fructicola is also an indicator of cell membrane damage. The level of leaked nucleic acids and soluble proteins present in the growth medium was significantly higher (P < 0.05) in the YYC 155 treatment group than in the control during the entire time of co-incubation (Figures 5C,D).
Scanning electron microscope analysis showed that C. fructicola hyphae changed significantly under the action of strain YYC 155 compared with the control (Figure 6). Hyphae were round and full in the control, with complete morphology and structure, uniform thickness, and smooth surface, whereas the hyphae of C. fructicola affected by strain YYC 155 were seriously damaged, with surface shrinkage and leakage of contents.

Figure 6. The morphological changes of C. fructicola hyphae under SEM. (A) Untreated hyphae of C. fructicola (control). (B) Hyphae of C. fructicola treated with strain YYC 155.
B. tequilensis YYC 155 exhibited the ability to form biofilms on good surfaces of polystyrene plates after 12 h of incubation at 28°C (Figure 7). The data suggest that the bacteria undergo a period of initial adhesion, film-formation, and then a steady dissociation at 12–24 h, 24–72 h, and 72–120 h, respectively. Notably, biofilm formation was significantly higher (P < 0.05) at 24–72 h than at the other time points. These results indicate that YYC 155 cultured for 24–72 h has a strong ability to form a biofilm, which contributes to its successful colonization of plant surfaces, both external and internal.
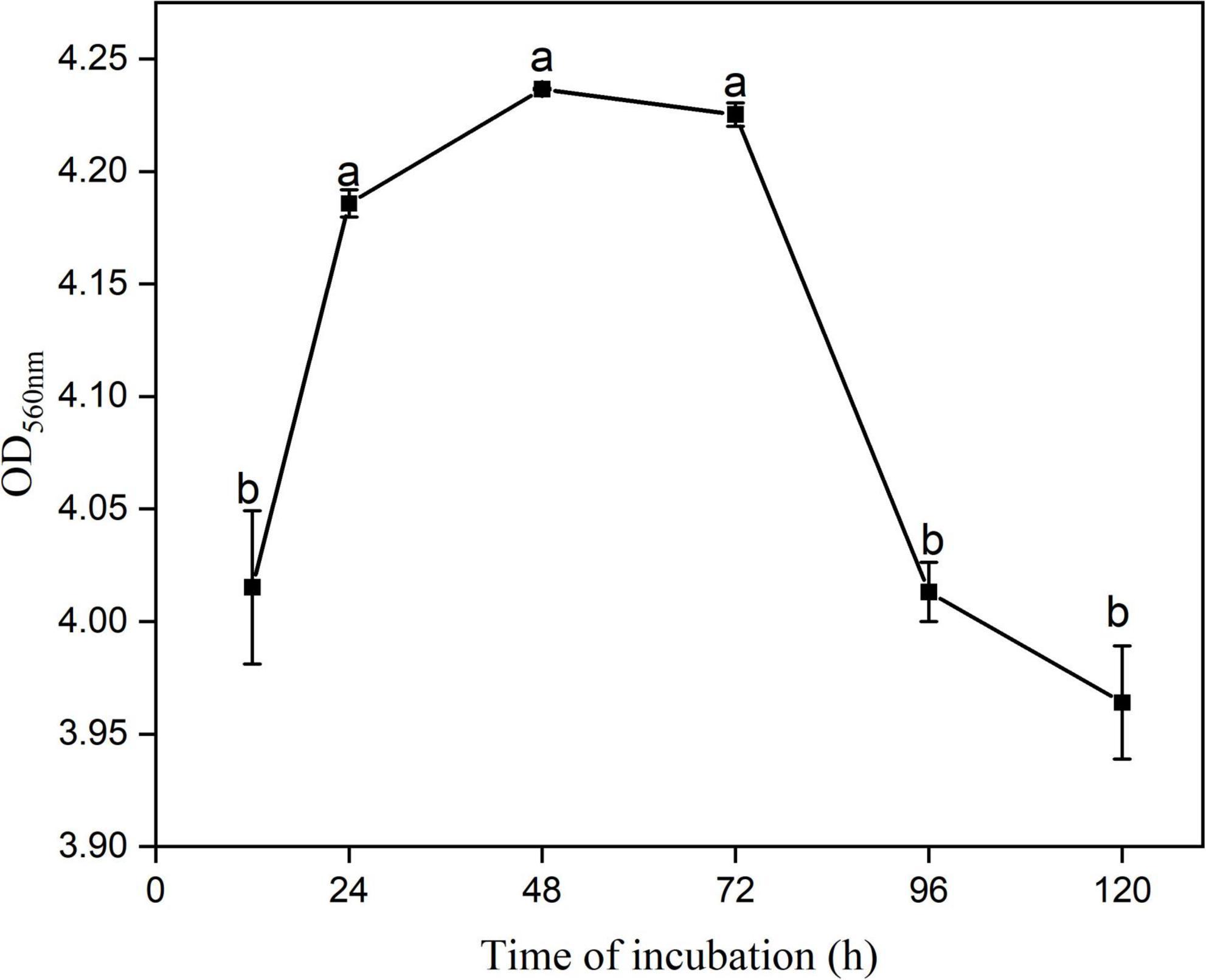
Figure 7. The ability of B. tequilensis YYC 155 to form a biofilm at 28°C. Three duplicate wells are used for YYC 155 at each time point. Data represent the mean ± standard error (se). Different letters represent significant differences in biofilm-forming capacity at the designated timepoint as determined by Duncan’s multiple range test (P < 0.05).
Figure 8A indicates that strain YYC 155 can secrete defense-related chitinase extracellularly. Chitinase enzyme activity reached its highest level at 96 h (1.13 ± 0.0029 U mL–1), then decreased sharply. Figure 8B indicates that strain YYC 155 can secrete β-1, 3-glucanase enzyme extracellularly, and that there was a linear increase in activity over the course of incubation. YYC 155 maintained a high level of extracellular β-1, 3-glucanase activity (6.81 ± 0.0016 U mL–1) up to 120 h of incubation.
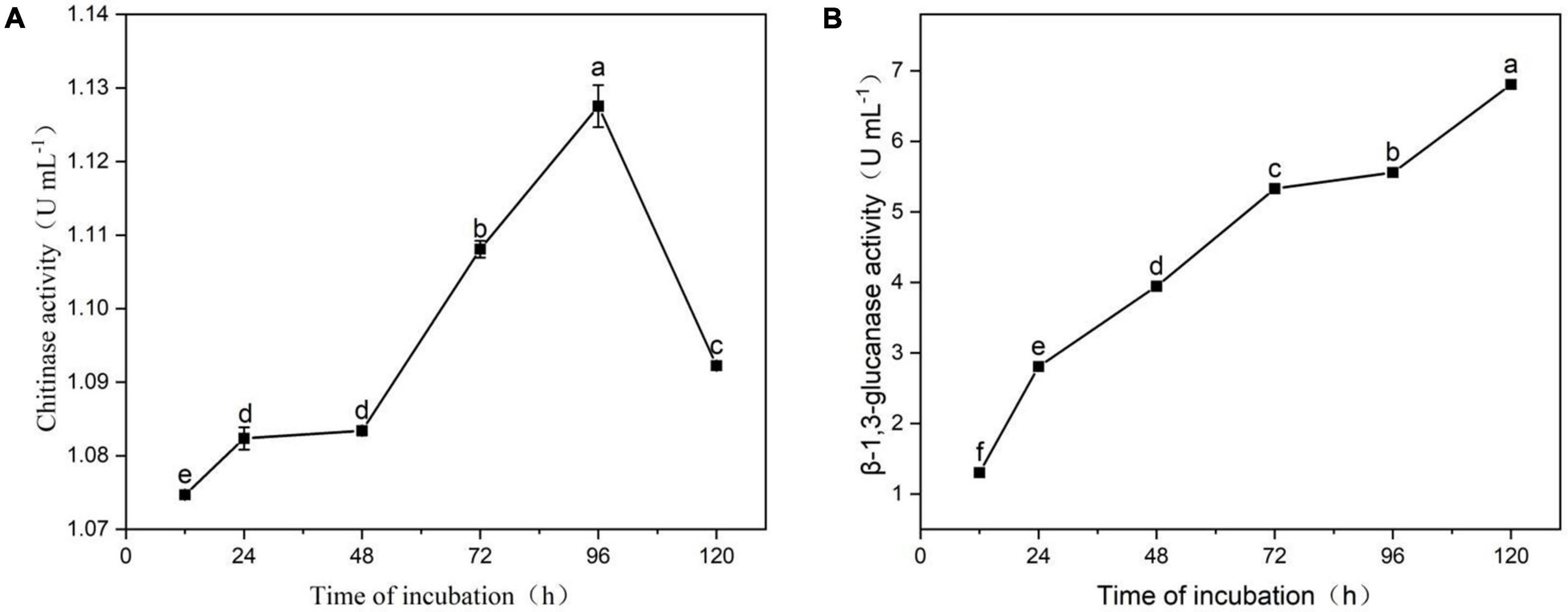
Figure 8. Extracellular chitinase (A) and β-1, 3-glucanase (B) activity by strain YYC 155. Three duplicates were measured for YYC 155 at each time point and were measured three times for each replicate. Data represent the mean ± standard error (se). Different letters represent a significant difference in enzyme activity between the different timepoints as determined by Duncan’s multiple range test (P < 0.05).
PCR products for the lipopeptide synthesis genes bacA (500 bp), ppsD (350 bp), and srfAA (200 bp), which encode bacylisin, plipastatin, and surfacing, respectively, were obtained from strain YYC 155 (Figure 9). PCR products for baC, baE, bmyB, and ituB were not obtained.
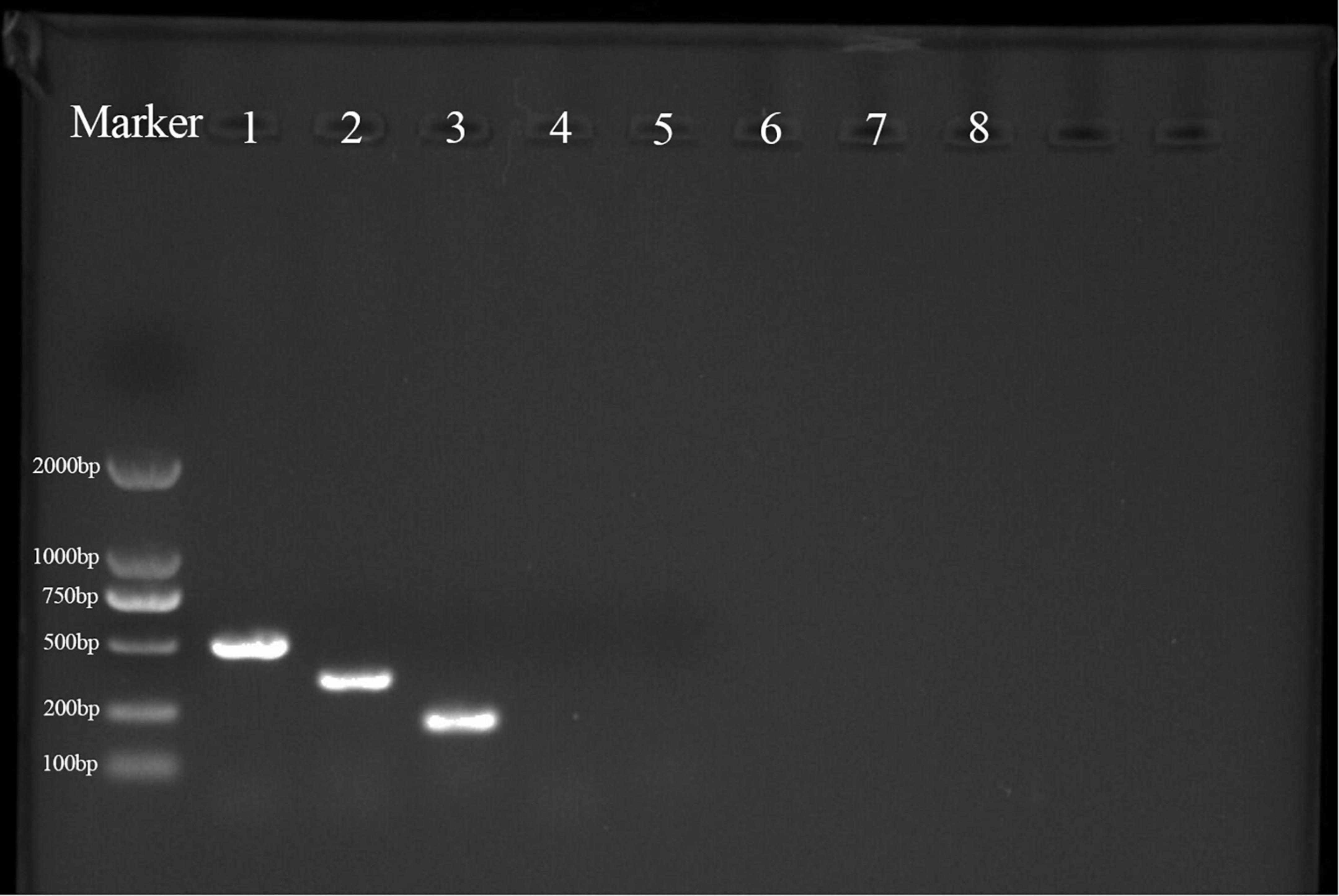
Figure 9. PCR products were obtained from the amplification of lipopeptide biosynthetic genes in strain YYC 155. M is a size marker; 1–8 are PCR products for genes encoding bacA, ppsD, srfAA, fenD, baC, baE, bmyB, and ituB, respectively. Only primers for the first three genes produced a PCR product.
The level of total phenols exhibited a similar pattern in C. oleifera leaves in both the YYC 155 treatment group and the control over the entire period of assessment (Figure 10A). C. oleifera treated with B. tequilensis YYC 155, however, had a higher level of total phenols, relative to the control, which increased significantly over the measured time period and peaked at the 15th day, and then subsequently decreased from day 15–25. The YYC 155 treatment group had a significantly higher (P < 0.05) level of total phenols, relative to the control, on days 10–20. The content of flavonoids in the YYC 155 treatment group was also higher than it was in the control group (Figure 10B). Flavonoid content exhibited an increasing pattern in both the control and the YYC 155 treatment group; however, the increase was very dramatic in the latter group, reaching its highest level at 20 days after treatment, after which, the levels exhibited a continuous decrease. Significantly higher levels (P < 0.05) of flavonoids were observed in the YYC 155 treatment group, relative to the control, at all timepoints except days 1 and 25. As indicated in Figure 10C, PAL activity in the YYC 155 treatment group exhibited an “M” pattern, peaking on day 15. PAL activity in the YYC 155 treatment group was significantly higher (P < 0.05), relative to the control throughout the assessment period, except at the initial stage. C4H activity in the two treatment groups exhibited a similar pattern of variation, steadily increasing up to day 25 and then decreasing from day 25 to day 30. A significantly higher (P < 0.05) level of C4H activity was exhibited in the YYC 155 treatment group, relative to the control, except on day 5 and day 10 (Figure 10D). In correspondence with Figure 10E, 4CL activity in the YYC 155 treatment group was mostly significantly higher (P < 0.05), relative to the control, reaching a maximum level of activity at day 15. CHI activity in the YYC 155 treatment group had a significantly higher (P < 0.05) level of activity, relative to the control, during the first 20 days, reaching a maximum of activity, after which CHI activity decreased (Figure 10F). Significant differences were not observed, however, on day 15. Lastly, the YYC 155 treatment group exhibited enhanced PPO activity, relative to the control, beginning on day 1 (Figure 10G) and was significantly higher (P < 0.05), relative to the control during the entire period of assessment (30 days).
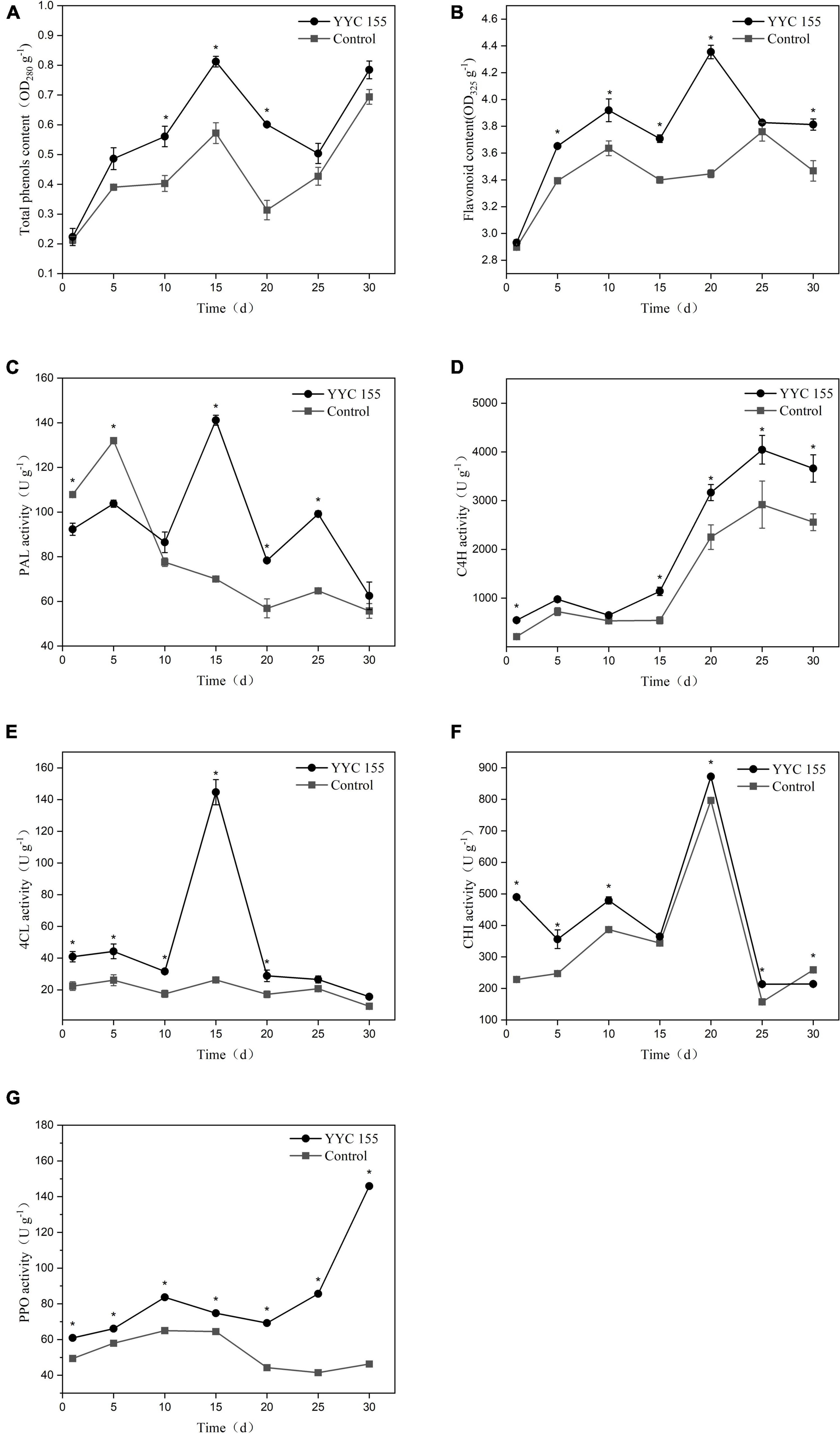
Figure 10. Effect of B. tequilensis YYC 155 on the level of total phenols (A) and flavonoids (B), and the activity of PAL (C), C4H (D), 4CL (E), CHI (F), and PPO (G) in C. oleifera. Three duplicates are used for the control and YYC 155 treatment C. oleifera leaves at each time point and were measured three times for each replicate. Data represent the mean ± standard error (se). Asterisks represent a significant difference between the two treatment groups at the designated timepoint as determined by the t-test (P < 0.05).
In recent years, the use of plant-associated endophytes for biological control has received a great deal of attention and ongoing research efforts (Hong and Park, 2016). In this study, strain YYC 155, an endophytic bacterium isolated from root nodules of Crotalaria pallida, was identified as B. tequilensis by 16S rRNA gene sequencing. B. tequilensis YYC 155 grew faster than other isolated strains, and effectively inhibit the mycelial growth of C. fructicola in vitro, and also inhibited the development of anthracnose lesions caused by C. fructicola on detached leaves of C. oleifera, an economically important shrub that serves as a source of seed oil (Figures 1, 3). The inhibitory activity of strain YYC 155 may potentially be attributed to its ability to successfully compete for nutrients and space with pathogens (Liu et al., 2013).
Biological agents also have direct or indirect impacts on the cell membrane of fungi, affecting both membrane permeability and structure, resulting in increased leakage of critical cellular components, such as nucleic acids and proteins (Elsherbiny et al., 2021). Relative conductivity, MDA content, as well as nucleic acid and soluble protein leakage are often used to determine the degree of cell membrane damage (Dai et al., 2021; Elsherbiny et al., 2021). We demonstrated that exposure to B. tequilensis YYC 155 significantly increased relative conductivity and MDA levels in the anthracnose pathogen, C. fructicola, thus increasing membrane permeability, and providing evidence of membrane structural damage (Figure 5). Alterations in membrane structure increase membrane permeability and result in the release of proteins and nucleic acids outside the cell. We observed significant differences in membrane permeability and the leakage of cellular contents in the mycelia of C. fructicola exposed to B. tequilensis YYC 155, relative to the control, throughout the entire period of co-incubation, indicating that the cell membrane of C. fructicola was damaged by YYC 155. YYC 155 could affect the cell integrity and physiological metabolism of the pathogen, thereby affecting the growth of pathogens to achieve the purpose of inhibiting pathogens.
Several studies have demonstrated that Bacillus species can produce multiple types of antimicrobial compounds that inhibit the growth of pathogens and negatively affect their metabolism (Stein, 2005; Fan et al., 2017; Guardado-Valdivia et al., 2018). Antimicrobial substances produced by Bacillus spp. are mainly divided into two categories, ribosome-synthesized peptides, such as bacteriocin, and small antimicrobial peptides enzymatically synthesized in non-ribosomal pathways, which primarily comprise cyclic lipopeptides (CLPs) (Romero et al., 2007). Lipopeptides are an important Bacillius metabolite, and comprise three major families, iturins, fengycins, and surfactins (Liu et al., 2020). In this study, genes involved in the synthesis of lipopeptides were detected in YYC 155 by PCR amplification, including bacA, ppsD, and srfAA, which function in the synthesis of bacylisin, plipastatin, and surfactin, respectively (Figure 9). Lipopeptides, such as surfactin and other CLPs, produced by antagonistic microorganisms, not only have antimicrobial properties, but have also been reported to act as elicitors that induce disease resistance responses in plants (Ongena et al., 2007; Gond et al., 2015). B. tequilensis has also been reported to inhibit diverse fungal pathogens through the activity of antifungal enzymes (Defilippi et al., 2018). Therefore, their production by bacterial biocontrol agents should be investigated as a mechanism of action. Antifungal enzymes, such as chitinase and β-1, 3-glucanase, are known to inhibit the growth of many pathogens and have the ability to disrupt and degrade fungal cell walls (Aktuganov et al., 2008; Zhang et al., 2017). We demonstrated that B. tequilensis YYC 155 strain secreted chitinase and β-1, 3-glucanase and that these enzymes had a high level of activity (Figure 8), which consequently destroyed the cell wall of pathogenic fungi C. fructicola, as observed under SEM analysis (Figure 6).
In addition to antimicrobial compounds and antifungal enzymes, other features of strain YYC 155 are also likely contributed to its biocontrol activity. Therefore, we also examined biofilm formation. Biofilms are defined as a group of organized bacteria surrounded by a self-produced matrix of extracellular polymeric substances, that allow the colonies to adhere to the surface of both living and inanimate objects (Mah et al., 2003; Dai et al., 2021). Bacteria can adhere to plant tissues, hyphae, and spores, and form a biofilm. The biofilm allows them to compete against fungal pathogens, interfering with their ability to acquire nutrients from the host and blocking their ability to occupy space or receive developmental signals from plant host tissues (Liu et al., 2013). Our assay confirmed that B. tequilensis YYC 155 exhibited a strong ability to form a biofilm in vitro when cultured in LB liquid (Figure 7). The ability to form biofilms also increases their ability to colonize plants and provide protection against infection by pathogens (Haggag and Timmusk, 2008). When biocontrol agents pre-colonize host plants, antifungal substances are produced and are present at a level that protects plants to varying degrees from pathogen infection (Hang et al., 2005; Chen et al., 2020).
Previous studies have demonstrated that antagonistic microorganisms can both inhibit fungal pathogens and produce elicitors that activate defense responses in plants and enhance disease resistance (Chen et al., 2020). In this regard, the phenylpropanoid pathway represents a major pathway for the synthesis of plant secondary metabolites that are associated with plant disease resistance (Xu et al., 2019). PAL, C4H, 4CL, and CHI are important enzymes in the phenylpropanoid pathway. Plants produce numerous pathogen compounds via the phenylpropanoid pathway, such as flavonoids and phenolic substances (Chen et al., 2006). PPO is responsible for oxidizing phenolic substances while producing toxic quinines that restrict and induce mortality in fungal pathogens (Wang et al., 2021). The results of this study indicated that the activity of these defense-related enzymes in the phenylpropanoid pathway was maintained at a significantly higher level in C. oleifera after the plant host was sprayed with B. tequilensis YYC 155 on leaves. This induction of defense-related enzymes may have contributed to the reduced lesion development observed in YYC 155 treated leaves. Our results suggest that the enhancement of defense-related enzymatic activity in C. oleifera by foliar spray with B. tequilensis YYC 155 potentially contributes to increased disease resistance to anthracnose in C. oleifera. Foliar spray with B. tequilensis YYC 155 also elevated the level of both flavonoids and total phenols in C. oleifera, relative to the controls, over the entire course of the assessment period (30 days). Such results showed that the endophytic B. tequilensis strain YYC 155 might colonize the plant phylloplane and enter the plant leaves through the leaf stomata, thus improving plant-induced disease resistance. However, further studies are needed to investigate the interaction of endophytic B. tequilensis strain YYC 155 with the plant leaf surface.
Collectively, the results of this study demonstrated that B. tequilensis YYC 155 inhibited anthracnose development in leaves of C. oleifera by inhibiting the growth of C. fructicola. The mechanism of control by B. tequilensis YYC 155 was associated with a direct antifungal effect on C. fructicola and the induction of enhanced resistance in C. oleifera. It appears that B. tequilensis YYC 155 targets the cell membrane of C. fructicola, increasing membrane permeability and inducing the leakage of cellular compounds, which together inhibit the growth of C. fructicola. Foliar spray with B. tequilensis YYC 155 also enhanced defense response in C. oleifera by enhancing the activity of defense-related enzymes that function in the phenylpropanoid pathway, and also promoted the accumulation of flavonoids and phenols. Thus, B. tequilensis YYC 155 may potentially represent an effective biocontrol agent against the anthracnose pathogen, C. fructicola, in the oil seed shrub, C. oleifera.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
AZ: formal analysis, data curation, visualization, validation, and writing—original draft. FW: conceptualization, supervision, data curation, and writing—review and editing. JY, DS, and JW: methodology and resources. RP: data curation and visualization. JD: methodology and data curation. XL: methodology and formal analysis. HM: project administration, funding acquisition, and writing—review and editing. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (Nos. 32160395, 31860208, and 31560207), the National Key Research and Development Program of China (No. 2019YFD100200X), the Talent Introducing Initial Scientific Research Fund of Southwest Forestry University (No. 111923).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.956642/full#supplementary-material
Ahimou, F., Jacques, P., and Deleu, M. (2000). Surfactin and iturin A effects on Bacillus subtilis surface hydrophobicity. Enzyme Microb. Technol. 27, 749–754. doi: 10.1016/S0141-0229(00)00295-7
Aktuganov, G., Melentjev, A., Galimzianova, N., Khalikova, E., Korpela, T., and Susi, P. (2008). Wide-range antifungal antagonism of Paenibacillus ehimensis IB-X-b and its dependence on chitinase and β-1,3-glucanase production. Can. J. Microbiol. 54, 577–587. doi: 10.1139/W08-043
Banerjee, S., Das, S., Rai, C., Bhattacharyya, S., and Roy, A. (2018). Evaluation of detection methods of biofilm formation by Bacillus Cereus and Staphylococcus Aureus isolates from foods. Res. J. Life Sci. Chem. Sci. 4, 147–155. doi: 10.26479/2018.0403.12
Chen, F., Wang, M., Zheng, Y., Luo, J., Yang, X., and Wang, X. (2010). Quantitative changes of plant defense enzymes and phytohormone in biocontrol of cucumber Fusarium wilt by Bacillus subtilis B579. World J. Microbiol. Biotechnol. 26, 675–684. doi: 10.1007/s11274-009-0222-0
Chen, J. Y., Wen, P. F., Kong, W. F., Pan, Q. H., Wan, S. B., and Huang, W. D. (2006). Changes and subcellular localizations of the enzymes involved in phenylpropanoid metabolism during grape berry development. J. Plant Physiol. 163, 115–127. doi: 10.1016/j.jplph.2005.07.006
Chen, K., Tian, Z., He, H., Long, C., and Jiang, F. (2020). Bacillus species as potential biocontrol agents against citrus diseases. Biol. Control 151:104419. doi: 10.1016/j.biocontrol.2020.104419
Cui, L., Yang, C., Wei, L., Li, T., and Chen, X. (2020). Isolation and identification of an endophytic bacteria Bacillus velezensis 8-4 exhibiting biocontrol activity against potato scab. Biol. Control 141:104156. doi: 10.1016/j.biocontrol.2019.104156
Dai, Y., Wu, X. Q., Wang, Y. H., and Zhu, M. L. (2021). Biocontrol potential of Bacillus pumilus HR10 against Sphaeropsis shoot blight disease of pine. Biol. Control 152:104458. doi: 10.1016/j.biocontrol.2020.104458
Defilippi, S., Groulx, E., Megalla, M., Mohamed, R., and Avis, T. J. (2018). Fungal competitors affect production of antimicrobial lipopeptides in Bacillus subtilis strain B9 – 5. J. Chem. Ecol. 44, 374–383. doi: 10.1007/s10886-018-0938-0
Dorighello, D. V., Bettiol, W., Maia, N. B., and de Campos Leite, R. M. V. B. (2015). Controlling Asian soybean rust (Phakopsora pachyrhizi) with Bacillus spp. and coffee oil. Crop Prot. 67, 59–65. doi: 10.1016/j.cropro.2014.09.017
Elsherbiny, E. A., Taher, M. A., Abd El-Aziz, M. H., and Mohamed, S. Y. (2021). Action mechanisms and biocontrol of Purpureocillium lilacinum against green mould caused by Penicillium digitatum in orange fruit. J. Appl. Microbiol. 131, 1378–1390. doi: 10.1111/jam.15016
Fan, H., Zhang, Z., Li, Y., Zhang, X., Duan, Y., and Wang, Q. (2017). Biocontrol of bacterial fruit blotch by Bacillus subtilis 9407 via surfactin-mediated antibacterial activity and colonization. Front. Microbiol. 8:1973. doi: 10.3389/fmicb.2017.01973
Fiddaman, P. J., and Rossall, S. (1993). The production of antifungal volatiles by Bacillus subtilis. J. Appl. Bacteriol. 74, 119–126. doi: 10.1111/j.1365-2672.1993.tb03004.x
Fredriksson, N. J., Hermansson, M., and Wilén, B. (2013). The choice of PCR primers has great impact on assessments of bacterial community diversity and dynamics in a wastewater treatment plant. PLoS One 8:e76431. doi: 10.1371/journal.pone.0076431
Gond, S. K., Bergen, M. S., Torres, M. S., and White, J. F. (2015). Endophytic Bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiol. Res. 172, 79–87. doi: 10.1016/j.micres.2014.11.004
Guardado-Valdivia, L., Tovar-Pérez, E., Chacón-López, A., López-García, U., Gutiérrez-Martínez, P., Stoll, A., et al. (2018). Identification and characterization of a new Bacillus atrophaeus strain B5 as biocontrol agent of postharvest anthracnose disease in soursop (Annona muricata) and avocado (Persea americana). Microbiol. Res. 210, 26–32. doi: 10.1016/j.micres.2018.01.007
Haggag, W. M., and Timmusk, S. (2008). Colonization of peanut roots by biofilm-forming Paenibacillus polymyxa initiates biocontrol against crown rot disease. J. Appl. Microbiol. 104, 961–969. doi: 10.1111/j.1365-2672.2007.03611.x
Hang, N. T. T., Oh, S. O., Kim, G. H., Hur, J. S., and Koh, Y. J. (2005). Bacillus subtilis s1-0210 as a biocontrol agent against Botrytis cinerea in strawberries. Plant Pathol. J. 21, 59–63. doi: 10.5423/PPJ.2005.21.1.059
Hong, C. E., and Park, J. M. (2016). Endophytic bacteria as biocontrol agents against plant pathogens: Current state-of-the-art. Plant Biotechnol. Rep. 10, 353–357. doi: 10.1007/s11816-016-0423-6
Kilani-Feki, O., Ben Khedher, S., Dammak, M., Kamoun, A., Jabnoun-Khiareddine, H., Daami-Remadi, M., et al. (2016). Improvement of antifungal metabolites production by Bacillus subtilis V26 for biocontrol of tomato postharvest disease. Biol. Control 95, 73–82. doi: 10.1016/j.biocontrol.2016.01.005
Kumar, S., Stecher, G., Tamura, K., and Medicine, E. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Leelasuphakul, W., Sivanunsakul, P., and Phongpaichit, S. (2006). Purification, characterization and synergistic activity of β-1, 3-glucanase and antibiotic extract from an antagonistic Bacillus subtilis NSRS 89-24 against rice blast and sheath blight. Enzyme Microb. Technol. 38, 990–997. doi: 10.1016/j.enzmictec.2005.08.030
Leifertl, C., Li, H., Chidbureel, S., Hampson, S., Workman, S., Sigee, D., et al. (1995). Antibiotic production and biocontrol activity by Bacillus subtilis CL27 and Bacillus pumilus CL45. J. Appl. Bacteriol. 78, 97–108. doi: 10.1111/j.1365-2672.1995.tb02829.x
Li, H., Zhou, G., and Liu, J. (2009). “Isolation and identification of endophytic bacteria antagonistic to Camellia oleifera leaf blight base on informatics,” in Proc. 2009 2nd International Conference Biomedical Engineering Informatics, (Tianjin), doi: 10.1109/BMEI.2009.5304770
Liu, J., Sui, Y., Wisniewski, M., Droby, S., and Liu, Y. (2013). Utilization of antagonistic yeasts to manage postharvest fungal diseases of fruit. Int. J. Food Microbiol. 167, 153–160. doi: 10.1016/j.ijfoodmicro.2013.09.004
Liu, Y., Teng, K., Wang, T., Dong, E., Zhang, M., Tao, Y., et al. (2020). Antimicrobial Bacillus velezensis HC6: Production of three kinds of lipopeptides and biocontrol potential in maize. J. Appl. Microbiol. 128, 242–254. doi: 10.1111/jam.14459
Mah, T. F., Pitts, B., Pellock, B., Walker, G. C., Stewart, P. S., and O’Toole, G. A. (2003). A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426, 306–310. doi: 10.1038/nature02122
Mahdi, T., Mohamed, I., and Yagi, S. (2014). Endophytic fungal communities associated with ethno-medicinal plants from Sudan and their antimicrobial and antioxidant prospective. J. For. Prod. Ind. 3, 248–256.
Ongena, M., Jourdan, E., Adam, A., Paquot, M., Brans, A., Joris, B., et al. (2007). Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 9, 1084–1090. doi: 10.1111/j.1462-2920.2006.01202.x
Perrett, C. M., Evans, A. V., and Russell-Jones, R. (2003). Tea tree oil dermatitis associated with linear IgA disease. Clin. Exp. Dermatol. 28, 167–170. doi: 10.1046/j.1365-2230.2003.01229.x
Romero, D., De Vicente, A., Rakotoaly, R. H., Dufour, S. E., Veening, J. W., Arrebola, E., et al. (2007). The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca. Mol. Plant Microbe Interact. 20, 430–440. doi: 10.1094/MPMI-20-4-0430
Shiomi, H. F., Silva, H. S. A., de Melo, I. S., Nunes, F. V., and Bettiol, W. (2006). Bioprospecting endophytic bacteria for biological control of coffee leaf rust. Sci. Agric. 63, 32–39.
Stein, T. (2005). Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 56, 845–857. doi: 10.1111/j.1365-2958.2005.04587.x
Vendan, R. T., Yu, Y. J., Lee, S. H., and Rhee, Y. H. (2010). Diversity of endophytic bacteria in ginseng and their potential for plant growth promotion. J. Microbiol. 48, 559–565. doi: 10.1007/s12275-010-0082-1
Wang, F., Xiao, J., Zhang, Y., Li, R., Liu, L., and Deng, J. (2021). Biocontrol ability and action mechanism of Bacillus halotolerans against Botrytis cinerea causing grey mould in postharvest strawberry fruit. Postharvest Biol. Technol. 174:111456. doi: 10.1016/j.postharvbio.2020.111456
Wang, X., Shi, J., and Wang, R. (2018). Effect of Burkholderia contaminans on postharvest diseases and induced resistance of strawberry fruits. Plant Pathol. J. 34, 403–411. doi: 10.5423/PPJ.OA.02.2018.0031
Wu, B., Ruan, C., Han, P., Ruan, D., Xiong, C. W., Ding, J., et al. (2019). Comparative transcriptomic analysis of high- and low-oil Camellia oleifera reveals a coordinated mechanism for the regulation of upstream and downstream multigenes for high oleic acid accumulation. 3 Biotech 9:257. doi: 10.1007/s13205-019-1792-7
Xu, J. X., Li, Z. Y., Lv, X., Yan, H., Zhou, G. Y., Cao, L. X., et al. (2020). Isolation and characterization of Bacillus subtilis strain 1-L-29, an endophytic bacteria from Camellia oleifera with antimicrobial activity and efficient plant-root colonization. PLoS One 15:e0232096. doi: 10.1371/journal.pone.0232096
Xu, J., Zhang, Z., Li, X., Wei, J., and Wu, B. (2019). Effect of nitrous oxide against Botrytis cinerea and phenylpropanoid pathway metabolism in table grapes. Sci. Hortic. 254, 99–105. doi: 10.1016/j.scienta.2019.04.061
Zhang, J., Xie, J., Zhou, Y., Deng, L., Yao, S., and Zeng, K. (2017). Inhibitory effect of Pichia membranaefaciens and Kloeckera apiculata against Monilinia fructicola and their biocontrol ability of brown rot in postharvest plum. Biol. Control 114, 51–58. doi: 10.1016/j.biocontrol.2017.07.013
Zhang, X., Kong, W., Wu, X., and Ye, J. (2022). Bacillus velezensis JK-XZ8 prevents and controls crown gall disease on Prunus subhirtella by colonizing and inducing resistance. J. For. Res. 33, 1019–1031. doi: 10.1007/s11676-021-01393-x
Keywords: Camellia oleifera, anthracnose, Colletotrichum fructicola, biological control, endophytic bacteria
Citation: Zhou A, Wang F, Yin J, Peng R, Deng J, Shen D, Wu J, Liu X and Ma H (2022) Antifungal action and induction of resistance by Bacillus sp. strain YYC 155 against Colletotrichum fructicola for control of anthracnose disease in Camellia oleifera. Front. Microbiol. 13:956642. doi: 10.3389/fmicb.2022.956642
Received: 30 May 2022; Accepted: 03 August 2022;
Published: 25 August 2022.
Edited by:
Jia Liu, Chongqing University of Arts and Sciences, ChinaCopyright © 2022 Zhou, Wang, Yin, Peng, Deng, Shen, Wu, Liu and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Wang, ZmFuZ2VyZnJpZW5kMjAyMUAxNjMuY29t; Huancheng Ma, bWhjQHN3ZnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.