- Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
Pseudostellaria heterophylla is a traditional Chinese herbal medicine, which has been cultivated for hundreds of years. Viral diseases of P. heterophylla occur widely and limit the yield and quality of this medicinal plant. In this study, five leaf samples of P. heterophylla with typical viral symptoms were collected from four main producing regions that are distributed in Fujian, Guizhou, and Anhui Provinces in China and analyzed by next-generation sequencing. Comprehensive bioinformatics analyses revealed that nine viruses in five genera Carlavirus, Potyvirus, Fabavirus, Cucumovirus, and Amalgavirus infected P. heterophylla. Among these viruses, three novel and two known carlaviruses, tentatively designated Pseudostellaria heterophylla carlavirus 1, 2, and 3 (PhCV1, PhCV2, and PhCV3), Jasmine virus C isolate Ph (Ph-JVC) and Stevia carlavirus 1 isolate Ph (Ph-StCV1), respectively, were first identified in P. heterophylla. PhCV1-3 share a similar genomic organization and clear sequence homology with members in the genus Carlavirus and could potentially be classified as new species of this genus. One novel amalgavirus, tentatively designated P. heterophylla amalgavirus 1 (PhAV1), was first identified in P. heterophylla. It had a typical genomic organization of the genus Amalgavirus. In PhAV1, the + 1 programmed ribosomal frameshifting, which is prevalent in most amalgaviruses, was identified and used in the expression of RNA-dependent RNA polymerase (RdRp). Combined with a phylogenetic analysis, PhAV1 could potentially be classified as new species of the genus Amalgavirus. In addition, multiple Broad bean wilt virus 2 (BBWV2) variants, Turnip mosaic virus (TuMV), and Cucumber mosaic virus (CMV), which have been reported in P. heterophylla, were also detected in this study. The distribution of PhCV1-3, Ph-JVC, Ph-StCV1, TuMV, BBWV2, and CMV in four production regions in Fujian, Guizhou, and Anhui Provinces was determined. This study increased our understanding of P. heterophylla virome and provided valuable information for the development of a molecular diagnostic technique and control of viral diseases in P. heterophylla.
Introduction
Pseudostellaria heterophylla (Miq.) Pax is also known as “Tai-zi-shen” or “Hai-er-shen” in China. It is a member of the family Caryophyllaceae and one of the most popular traditional Chinese herbal medicines. It can be used to treat spleen deficiency, anorexia, weakness after illness, and spontaneous perspiration symptoms (Pang et al., 2011; Sheng et al., 2011; Guo et al., 2014; Choi et al., 2017). P. heterophylla is widely distributed in northeastern Asia, including China, Japan, and Korea. In China, P. heterophylla has been cultivated for hundreds of years, and its commercial areas of production are primarily distributed in Guizhou, Fujian, Anhui, Shandong, and Jiangsu Provinces. Most of the P. heterophylla used in the medicinal commodity market is grown in Shibing County (in Guizhou Province), Zherong County (in Fujian Province), and Xuancheng City (in Anhui Province; Kang et al., 2016).
Pseudostellaria heterophylla primarily propagates asexually through root tubers, which facilitates the transmission of viral diseases across generations. The infections then spread to different regions through the transportation of seedlings. Owing to the long-term vegetative propagation of these plants, the incidence of viral diseases increases yearly, which can result in rates of infection as high as 90% or even 100% (Huang and Lin, 2004; Wang et al., 2022). Such severe infections reduce the yield and quality of P. heterophylla. P. heterophylla infected by virus(es) display symptoms, such as foliar mottles, mosaics, ringspots, and leaf malformations, as well as the dwarfing of plants. To date, TuMV (of the genus Potyvirus, family Potyviridae), BBWV2 (of the genus Fabavirus, family Secoviridae), CMV (of the genus Cucumovirus, family Bromoviridae), and Tobacco mosaic virus (TMV, of the genus Tobamovirus, family Virgaviridae) are the four viruses that have been reported to infect P. heterophylla (Song and Pu, 1991).
The genus Carlavirus of the family Betaflexiviridae comprises 61 confirmed species (ICTV).1 Carlaviruses have a monopartite, single-stranded, and positive-sense RNA genome that ranges from 8.3 to 8.7 kb long. Their genomic RNAs encode six open reading frames (ORFs). ORF1 encodes a viral replicase (Rep) and three overlapping ORFs, including ORF2–ORF4, encode the triple gene block (TGB) proteins 1, 2, and 3 that facilitate the movement of viruses. ORF5 encodes the coat protein (CP), and ORF6 encodes a cysteine-rich protein (CRP; Adams et al., 2012a). CRP serves as an RNA silencing suppressor and a determinant of symptoms and/or pathogenicity (Deng et al., 2015; Fujita et al., 2018). Most carlaviruses are transmitted naturally by aphids in a non-persistent manner, while some of them are transmitted by whiteflies (Bemisia tabaci; Adams et al., 2012a). All the carlaviruses are mechanically transmissible (Adams et al., 2012a).
Amalgaviridae is a recently reported family of double-stranded RNA viruses that is an amalgam of the families Partitiviridae and Totiviridae, which encode proteins phylogenetically related to former, but their structure is more closely related to the latter (Martin et al., 2011; Krupovic et al., 2015). It contains two genera, Amalgavirus and Zybavirus. Amalgavirus includes nine viral species that infect plants (ICTV).1 Putative amalgaviruses have also been reported in fungi and bryophytes (Koloniuk et al., 2015; Vendrell-Mir et al., 2021). These amalgaviruses are monopartite and have small dsRNA genomes that are 3.3–3.5 kb long and encode two partially overlapping ORFs. ORF1 encodes a putative CP with unclear function, which is similar to the nucleocapsid proteins of the genera Phlebovirus and Tenuivirus (Krupovic et al., 2015) or the replication factory matrix-like protein (Isogai et al., 2011). In most amalgaviruses, there is a + 1 programmed ribosomal frameshifting (PRF) in ORF1. When it occurs, the codon frame changes from UUU_ CGN (ORF1) to U_UUC_GNN (ORF2; underlining indicates the codon boundary; N, any nucleotide), which causes the fusion of ORF1 and ORF2, and encodes an RdRp (Nibert et al., 2016; Park et al., 2018). Members of this family are transmitted vertically by seeds and are not thought to be capable of efficient extracellular transmission unless mediated by an unknown vector (Martin et al., 2006; Sabanadzovic et al., 2009).
To identify the viral species that infect P. heterophylla and provide a reference for the diagnosis, prevention, and control of viral diseases of P. heterophylla, the virome of P. heterophylla was analyzed by next-generation sequencing (NGS), a rapidly developing technique for viral detection and diagnosis that is suitable for a variety of plants, animals, and fungi (Mu et al., 2018; Jo et al., 2020; Kondo et al., 2020; Zhang et al., 2020). The five libraries used for NGS were prepared with P. heterophylla leaf samples collected from four different geographic regions in Guizhou, Fujian, and Anhui Provinces in China. Three novel and two known carlaviruses, one novel amalgavirus, multiple BBWV2 variants, TuMV, and CMV that infect P. heterophylla were identified.
Materials and methods
Plant samples
Leaf samples were collected from symptomatic P. heterophylla and transported to the laboratory under conditions of low temperature and humidity. The samples were stored at -80°C and used for paired-end RNA-Seq.
RNA extraction, NGS, and data processing
Total RNA was extracted from the leaf tissue using a mirVana™ microRNA (miRNA) Isolation kit (Ambion, Thermo Fisher Scientific, Waltham, MA, United States) and treated with an RNA Clean XP Kit (Beckman Coulter, Brea, CA, United States) and an RNase-Free DNase Set (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s instructions. The quality and quantity of total RNA were measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific) and Agilent2100 (Agilent Technologies, Santa Clara, CA, United States). After the removal of ribosomal RNA using a Ribo-Zero Magnetic Kit (Epicentre, Lucigen, Middleton, WI, United States), the libraries were built using a TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA, United States). Barcoded libraries were paired-end sequenced on an Illumina HiSeq X platform according to the manufacturer’s instructions.2 Sequences of adaptors and low-quality traits were trimmed using the FASTX-Toolkit software,3 and clean reads were assembled de novo using CLC Genomics Workbench 6.0.4 (Qiagen, Valencia, CA, United States) according to the scaffolding contig algorithm. The second assembly was then conducted using CAP3 software.4 The final contigs were compared against the NCBI non-redundant (Nr) database using BLASTX with an E-value < 1e−5.
Recovery of viral genomes
The genome of new P. heterophylla viruses were obtained by overlapping reverse transcription (RT)-PCR and rapid amplification of cDNA ends-PCR (RACE-PCR). The primers used in RT-PCR and RACE-PCR were designed based on the viral contig sequences using Primer Premier 6 (PREMIER Biosoft, Palo Alto, CA, United States; Supplementary Table S1). A RACE-PCR assay was conducted using a SMARTer RACE 5′/3′ Kit (Clontech, Mountain View, CA, United States). The PCR amplicons were purified by a TIANgel Midi Purification Kit (Tiangen, Beijing, China) and cloned into the pMD18 or pUC19 Vector (TaKaRa, Dalian, China). The sequence of each amplicon was determined from both directions of more than five clones by the biotechnology company Tsingke (Beijing, China). The full-length genome of each virus was assembled from all amplicons of the virus using DNAMAN 6.0 (Lynnon Biosoft, Quebec City, Canada).
Sequence analysis and read assembly
Viral genome organizations were analyzed using the ORF finder program.5 The conserved domains of ORFs were analyzed using the Conserved Domain Search Service on the NCBI website.6 The multiple sequence alignment, conserved domains analysis of the RdRps and CRPs of carlaviruses, and prediction of the PRF motif in RdRp of amalgaviruses were performed using CLC Genomics Workbench 21.0.5 (Qiagen). Pairwise comparisons between viruses were performed using MAFFT program7 and displayed by Sequence Demarcation Tool (SDT) software using a color-coded matrix (Muhire et al., 2014).
Phylogenetic analysis
Each of the new viruses and its closely related viruses retrieved from the NCBI databases were aligned using the ClustalW program. Phylogenetic analyses were performed using the maximum-likelihood method (carlaviruses and amalgaviruses) or the neighbor-joining method (BBWV2) with 1,000 bootstrap replicates in MEGA X (Kumar et al., 2018).
Results
Sample collection and symptom description
Five P. heterophylla leaf samples were collected from the four different main production regions of P. heterophylla in June of 2020 and 2021, including one sample in Shibing County (SB) and one sample in Danzhai (DZ) County in Guizhou Province, two samples in Zherong County (ZR-HB and ZR-GX) in Fujian Province and one sample in Xuancheng city (XC) in Anhui Province, in China (Figure 1A; Supplementary Table S2). The samples displayed typical viral symptoms, such as foliar mosaics, ringspots, mottles, and leaf malformations (Figures 1B–F). Each sample was used to generate a library for paired-end RNA-Seq. For simplicity, the libraries were named based on the geographical regions that the samples were collected from.
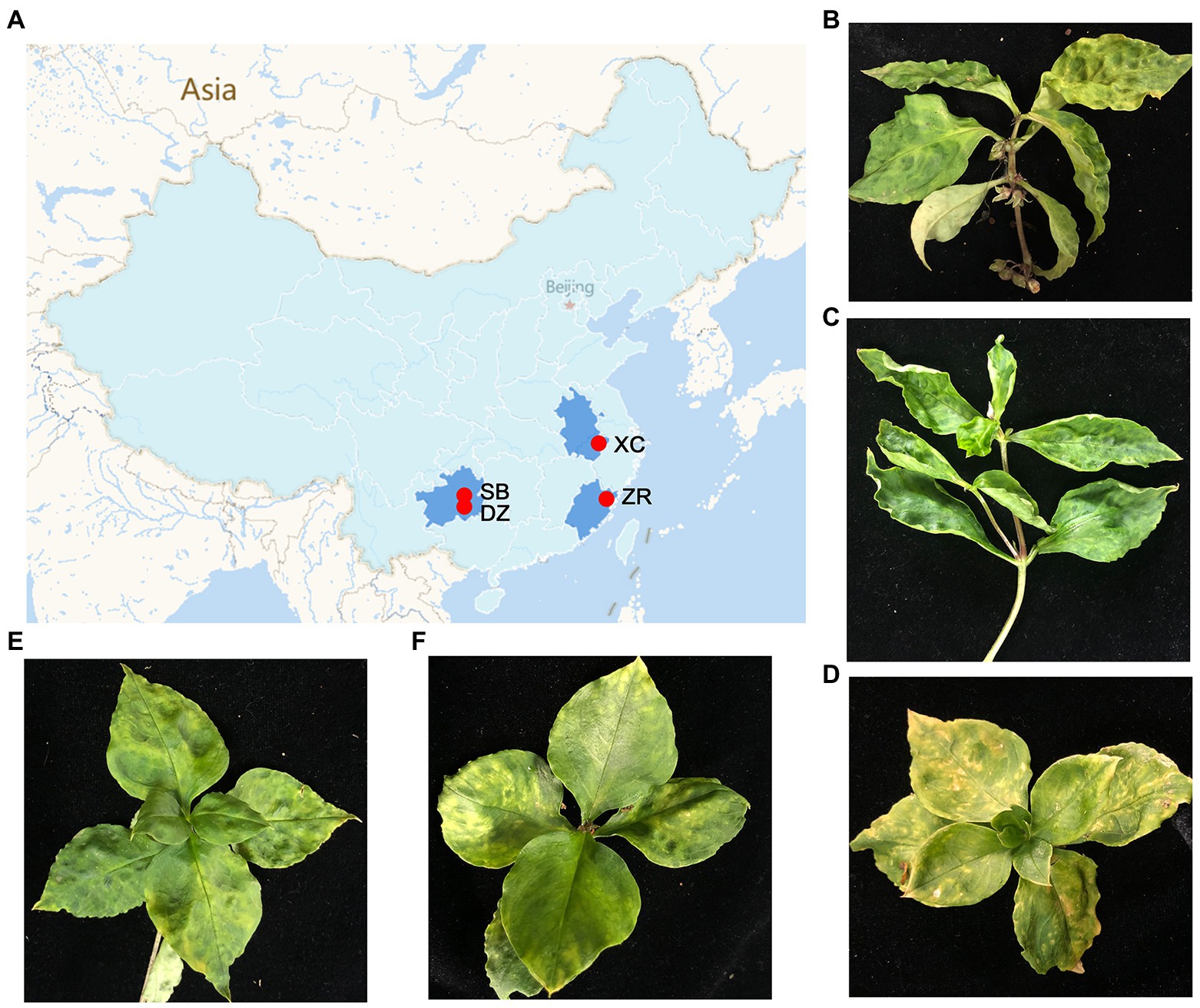
Figure 1. Geographical regions of the Pseudostellaria heterophylla samples collected in China and the leaf symptoms of P. heterophylla viral diseases. (A) A map that displays the four different geographical regions in China in which the P. heterophylla samples were collected. (B–F) Symptoms displayed on P. heterophylla leaves collected from SB, XC, ZR-HB, DZ, and ZR-GX, respectively. DZ and SB represent Danzhai County (DZ) and Shibing County (SB) in Guizhou Province, respectively. ZR-HB and ZR-GX represent Huangbai village (ZR-HB) and Gongxi village (ZR-GX) in Zherong County in Fujian Province, respectively. XC represents Xuancheng city (XC) in Anhui Province.
Transcriptome assembly and virus identification
A total of 59,932,792–142,685,292 trimmed reads were individually generated from five independent libraries (Supplementary Table S3). Assembly of the clean reads generated 95,378 to 277,230 contigs that ranged from 200 to 10,252 nucleotides (nt) in size (Supplementary Table S3). A BLASTX analysis of the contigs revealed that 320 virus-related contigs representing five different viral taxa were obtained (Table 1). They included TuMV, BBWV2, carlaviruses, CMV, and the amalgaviruses. TuMV, BBWV2, and carlaviruses were identified in all five libraries. Approximately 94.06% (301 of 320) of the viral-related contigs, which were assembled from approximately 95.75% (150,587,101 of 157,276,057) viral-related reads, were homologous to the carlaviruses, TuMV or BBWV2 (Figures 2A,B; Table 1). Followed by CMV and the amalgaviruses, CMV was identified in the libraries ZR-HB and XC, and amalgaviruses were identified in the libraries ZR-HB, ZR-GX, DZ, and XC. Approximately 5.94% (19 of 320) of the viral-related contigs, which were assembled from approximately 4.25% (6,688,956 of 157,276,057) viral-related reads, were homologous to CMV or the amalgaviruses (Figure 2A,B; Table 1). TMV, a virus previously reported to infect P. heterophylla (Gao et al., 1993), was not identified in this study. Based on the viral-related reads, carlaviruses were the dominant viruses in the libraries DZ, ZR-HB, and ZR-GX; TuMV was the major virus in library SB, and BBWV2 was the major virus in library XC (Figure 2A; Table 1).
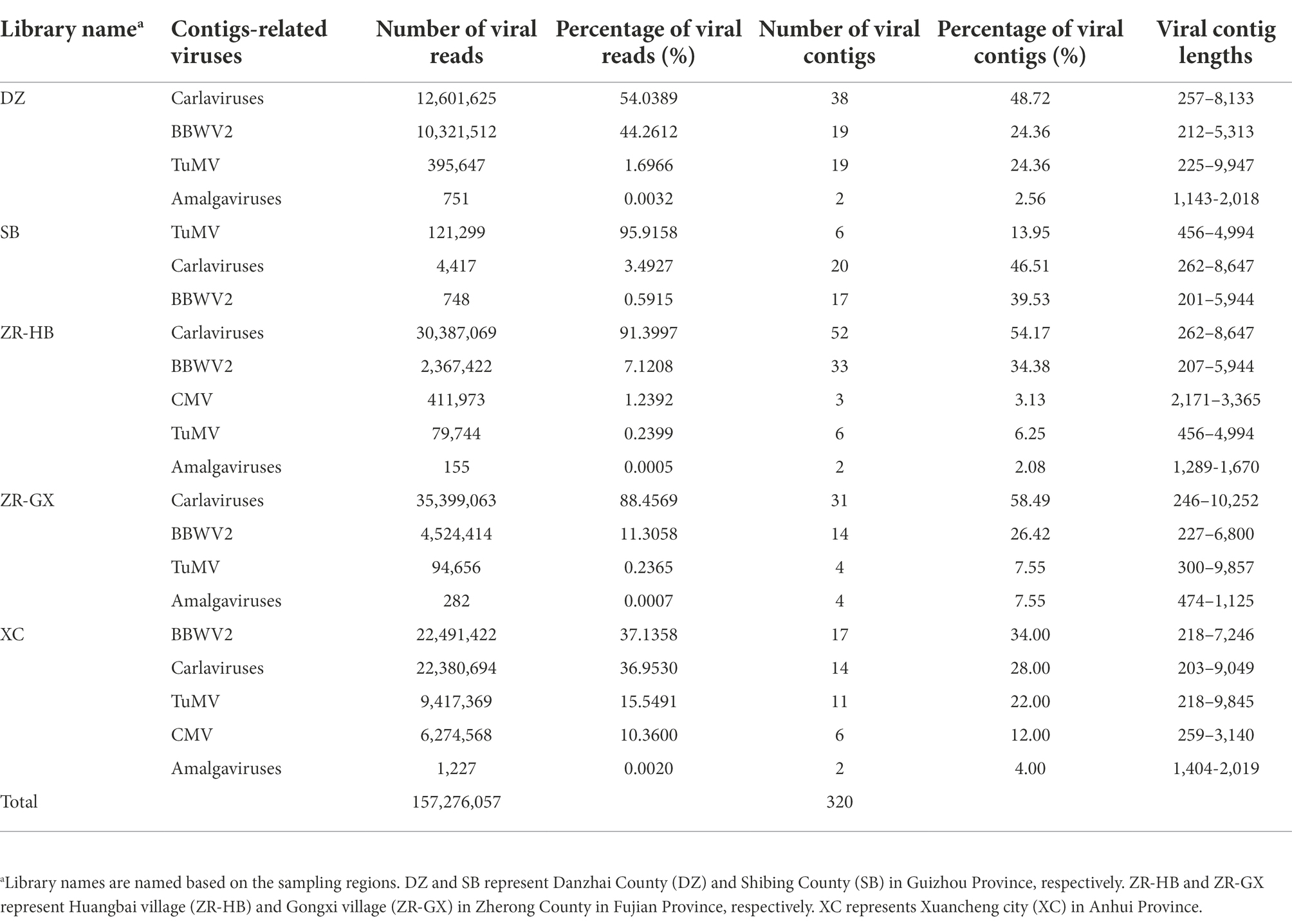
Table 1. Information of the viral reads and contigs for the viruses identified in Pseudostellaria heterophylla in China.
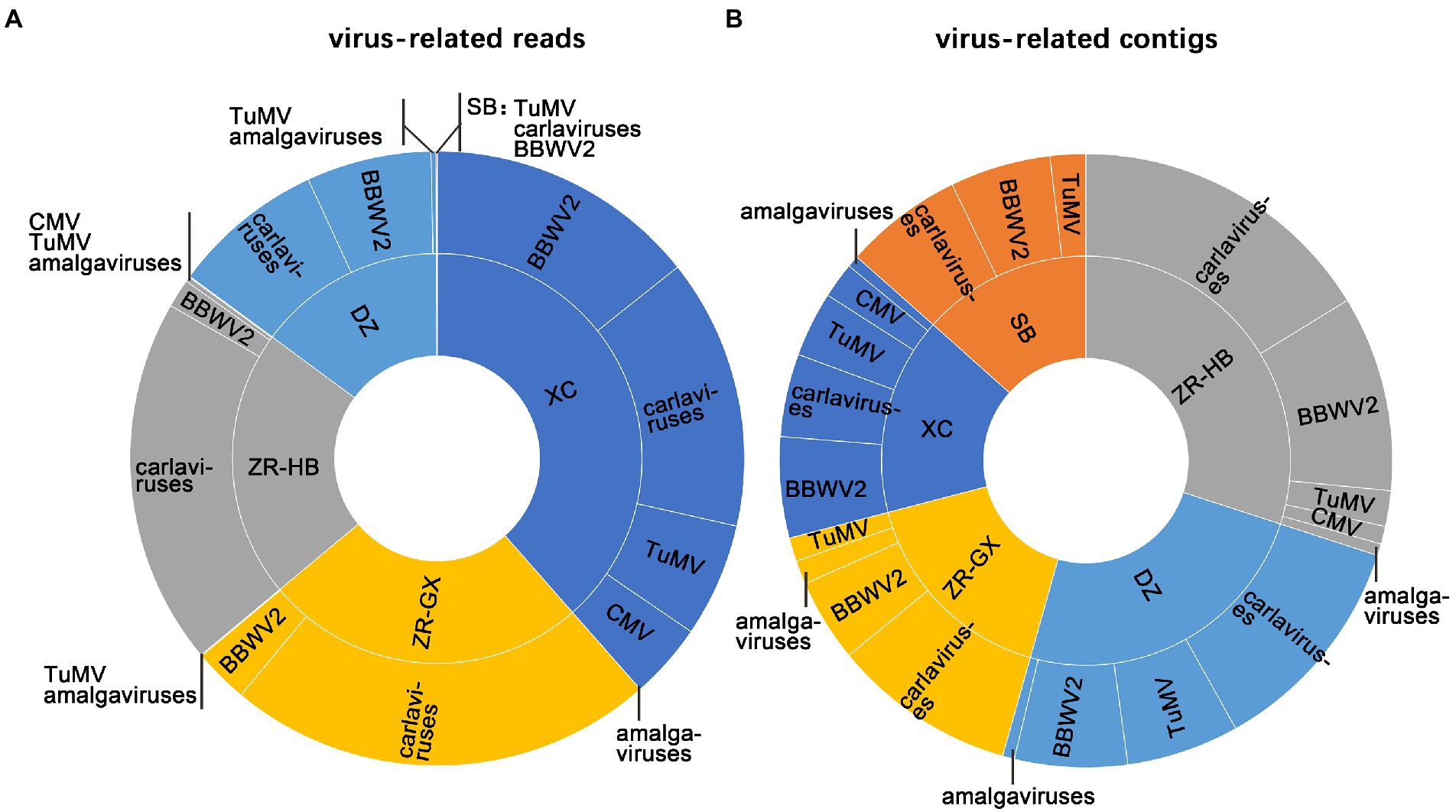
Figure 2. The identification of viruses in each sample. The proportion of identified viruses from all five libraries based on viral-associated reads (A), and viral-associated contigs (B). SB, DZ, ZR-GX, ZR-HB, and XC were library names designated based on geographical regions.
Turnip mosaic virus, CMV-II, and multiple BBWV2 variants were identified in Pseudostellaria heterophylla
The TuMV- and CMV-related contigs shared more than 96% nt sequence identity with TuMV and CMV in subgroup II (CMV-II). These results confirmed the presence of both viruses in P. heterophylla.
Six BBWV2 isolates were identified in P. heterophylla. One BBWV2 isolate was identified in libraries DZ and ZR-HB and designated BBWV2-DZ and -ZR-HB, respectively. Two BBWV2 isolates were identified in libraries XC and ZR-GX and designated BBWV2-XC1, -XC2, −ZR-GX1 and -ZR-GX2, respectively. Pairwise comparisons showed that the polyprotein encoded by RNA1 shared 73.41 to 98.15% nt sequence identities and 82.71 to 99.09% amino acid (aa) sequence identities with each other (Tables 2; Supplementary Table S4). The polyprotein encoded by RNA2 shared 70.12 to 98.18% nt sequence identities and 78.97 to 98.78% aa sequence identities with each other (Tables 2; Supplementary Table S4). Among them, RNA1 of ZR-GX1 and XC1 had lower nt identities (< 75.66%) compared with the other reported BBWV2 isolates. RNA2 of ZR-GX1 and XC2 had low nt similarities (< 74.19%) with the isolates reported (Table 2). The high proportion of nucleotide sequence changes did not result in significant changes in the aa sequences of the CP and Pro-Pol region, two indicators of the definition of new species (Sanfaçon et al., 2012). The Pro-Pol regions of ZR-GX1 and XC1 shared the highest aa sequence identities (89.81 and 87.04%, respectively) with that of the LNSY isolate (GenBank accession No. MN786954; Supplementary Table S4). The CP of ZR-GX1 and XC2 shared the highest aa sequence identities (85.31 and 85.81%, respectively) with that of the Yunnan (MW271032) and PC (MW939477) isolate, respectively (Supplementary Table S4). Since these sequence identity values are higher than the species demarcation threshold (80% aa sequence identity in Pro-Pol region or 75% aa sequence identity in CP) for the genus Fabavirus (Sanfaçon et al., 2012), these BBWV2 isolates cannot be defined as a new species in the genus Fabavirus. However, these results confirmed the variability of BBWV2 isolates in P. heterophylla.
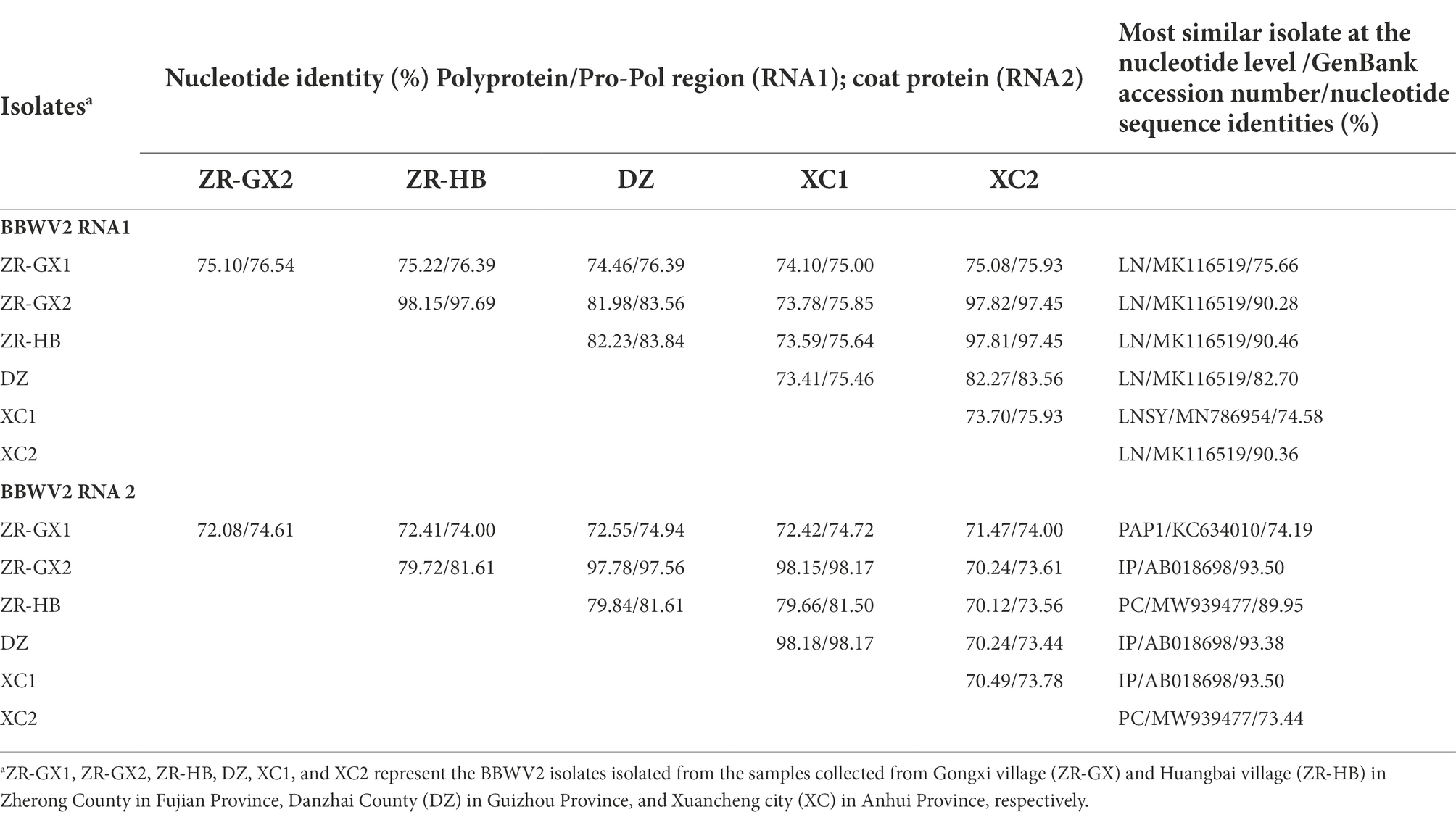
Table 2. Pairwise nucleotide sequence identities of the BBWV2 isolates identified in Pseudostellaria heterophylla.
A phylogenetic analysis based on the amino acid sequences of the Pro-Pol region of the polymerase showed that the BBWV2 isolates DZ, ZR-HB, ZR-GX2, and XC2 clustered closely with the isolates LN and LNSY, while the isolates ZR-GX1 and XC1 did not cluster (Figure 3).

Figure 3. Phylogenetic relationships of BBWV2 isolates from Pseudostellaria heterophylla (ZR-GX1-, ZR-GX2-, ZR-HB-, DZ-, XC1-, and XC2-BBWV2) and the nine isolates that were the most similar to those of BBWV2 based on the amino acid sequences of the Pro-Pol region. The phylogenetic trees were constructed using the neighbor-joining method in MEGA X software with a bootstrap of 1,000 replicates. The black dots represent isolates identified in this study. ZR-GX1-, ZR-GX2-, ZR-HB-, DZ-, XC1- and XC2-BBWV2 represent the isolates isolated from the samples collected from Gongxi village (ZR-GX) and Huangbai village (ZR-HB) in Zherong County in Fujian Province, Danzhai County (DZ) in Guizhou Province, and Xuancheng city (XC) in Anhui Province, respectively.
Three novel and two known carlaviruses were identified in Pseudostellaria heterophylla
Carlavirus-related contigs were detected in all five libraries, and carlavirus-related reads accounted for the high proportions (91.40, 88.46, and 54.04%, respectively) of the viral-related reads in the ZR-HB, ZR-GX, and DZ libraries (Table 1). Three novel carlaviruses, PhCV1, PhCV2, and PhCV3, and two known carlaviruses, Ph-JVC and Ph-StCV1, were identified in this study. The genomic sequences of PhCV1, PhCV2, and PhCV3 were 8,744, 8,533, and 8,497 bp long, respectively, excluding the poly(A) tail at the 3′ end. Their genome organizations are typical of carlaviruses and contain six open reading frames (ORFs) that encode an Rep, three TGB proteins, a CP, and a CRP (Figure 4A; Adams et al., 2012a).
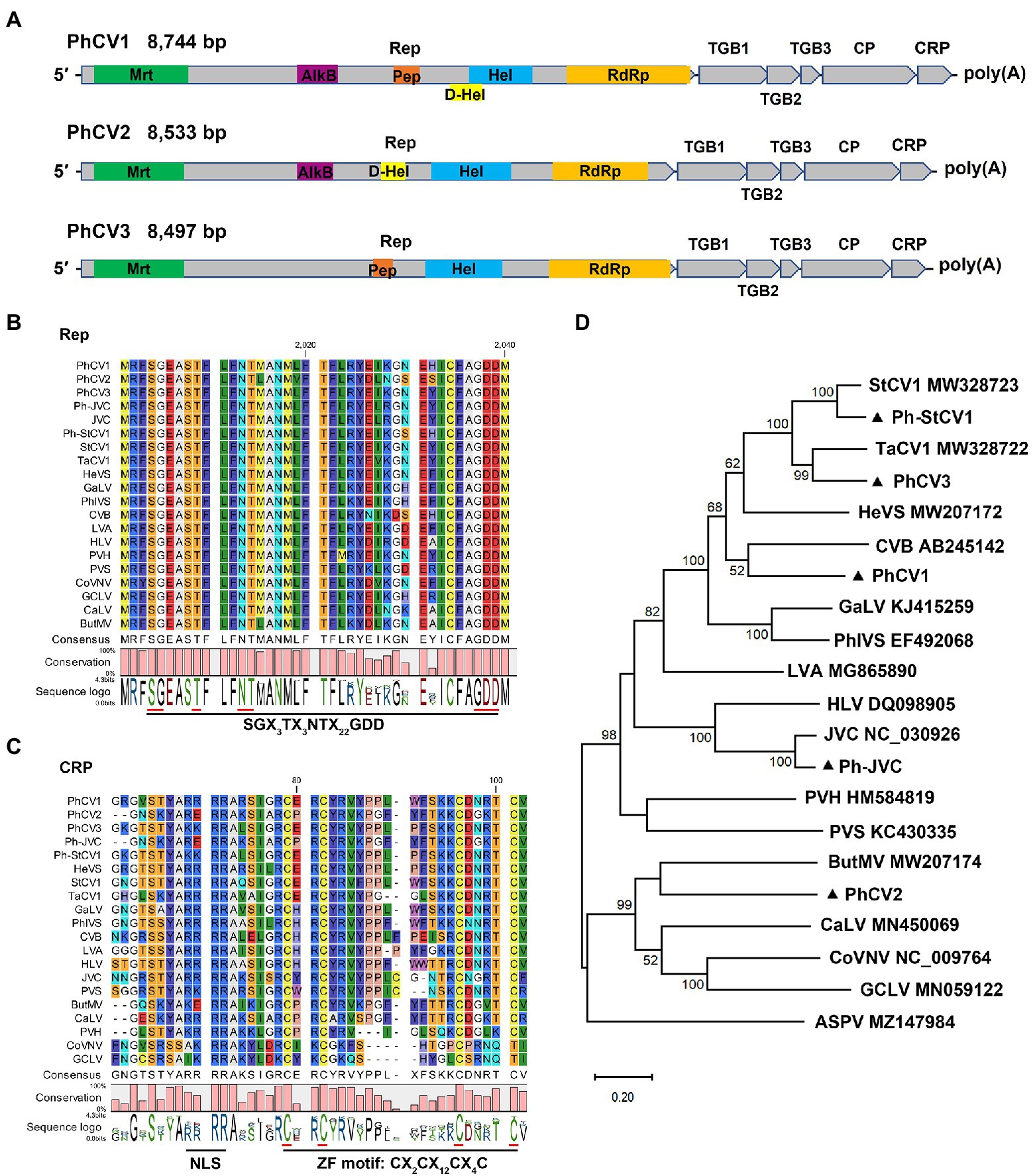
Figure 4. Characterization of three novel carlaviruses, pseudostellaria heterophylla carlavirus 1 (PhCV1), pseudostellaria heterophylla carlavirus 2 (PhCV2), and pseudostellaria heterophylla carlavirus 3 (PhCV3) isolated from Pseudostellaria heterophylla in China. (A) Genomic organization of PhCV1, PhCV2, and PhCV3. Rep, viral replicase; TGB, triple gene block; CP, coat protein; CRP, cysteine-rich protein; Mrt, methyltransferase; AlkB, alkylated DNA repair protein; Hel, viral helicase; D-Hel, DEAD-like helicase; Pep, Carlavirus endopeptidase; RdRp, RNA-dependent RNA polymerase. (B) Amino acid alignment between the conserved RdRp motifs of PhCV1, PhCV2, PhCV3, Ph-JVC, and Ph-StCV1 and 15 selected carlaviruses. (C) Amino acid alignment between NLS and ZF in the CRP of PhCV1, PhCV2, PhCV3, Ph-JVC, Ph-StCV1, and 15 selected carlaviruses. NLS, nuclear localization signal. ZF, zinc finger-like motif. (D) Phylogenetic analysis based on the amino acid sequences of the Rep of PhCV1, PhCV2, PhCV3, Ph-JVC, Ph-StCV1, and the 15 most similar carlaviruses, performed by the maximum-likelihood method in MEGA X software. Bootstrap values (1,000 replicates) are shown below the branches. The black triangles represent carlaviruses identified in this study.
Six, five, and four domains were identified in the Rep of PhCV1, PhCV2, and PhCV3, respectively (Figure 4A). Viral methyltransferase (Mtr; pfam01660), viral helicase (Hel; cl26263), and RdRp (cl03049) domains were identified in the Rep of these three viruses (Figure 4A). Alkylated DNA repair protein (AlkB; cl21496) and DEAD-like helicase (D-Hel; cl28899) domains were identified in the Rep of PhCV1 and PhCV2 but not PhCV3 (Figure 4A). The carlavirus endopeptidase (Pep; cl05111) domain was identified in the Rep of PhCV1 and PhCV3 but not PhCV2 (Figure 4A). A conserved motif, SGX3TX3NTX22GDD (X, any aa residue), was identified near the C-terminus of the RdRp domain of these three carlaviruses (Figure 4B). Zinc finger-like motifs (ZFs), CX2CX12CX4C, were identified in the CRPs of PhCV1, PhCV2, and PhCV3 (Figure 4C). Nuclear localization signals (NLSs), including RRRR or KKRR, were identified at aa 51–54 in the CRPs of PhCV1 and PhCV3 but not in the CRP of PhCV2 (Figure 4C).
A BLASTN analysis with the nucleotide sequence of genome revealed that PhCV1, PhCV2, and PhCV3 were the most similar to StCV1 (MW328723), Potato virus S (PVS, KC430335), and Tagetes carlavirus 1 (TaCV1, MW328722) with nt identities of 69.95% (55% query coverage), 69.75% (19% query coverage), and 71.15% (87% query coverage), respectively. Pairwise comparisons between PhCV1, PhCV2, PhCV3, Ph-JVC, Ph-StCV1, and 15 of the most similar carlaviruses were performed based on the nt and aa sequences of Rep and CP using MAFFT program and SDT software. The Reps of PhCV1, PhCV2, and PhCV3 shared 53.50–61.30%, 52.80–54.90%, and 53.70–66.90% nt sequence identities and 38.50–56.40%, 36.70–41.90%, and 39.20–72.20% aa sequence identities with Reps in the other aligned carlaviruses (Figures 5A,C; Supplementary Tables S5, S6). The CPs of PhCV1, PhCV2, and PhCV3 shared 47.80–66.40%, 46.10–58.30%, and 50.60–73.50% nt sequence identities and 25.70–67.10%, 24.20–50.50%, and 35.60–85.70% aa sequence identities with CPs in the other aligned carlaviruses (Figures 5B, D; Supplementary Tables S7, S8). The threshold for species demarcation of the genus Carlavirus is < 72% nt sequence identity and < 80% aa sequence identity between their CP or Rep. Since the CP and Rep of PhCV1 and PhCV2, Rep of PhCV3 meet the threshold, they can be recognized as new species of the genus Carlavirus.

Figure 5. The pairwise identities plot of Reps and CPs in the genus of Carlavirus based on amino acid (aa) sequence (A) (B) and nucleotide (nt) sequence (C) (D) aligned by MAFFT and displayed by SDT software. Rep, viral replicase; CP, coat protein.
Phylogenetic trees were constructed based on the aa sequences of the Reps and CPs of PhCV1-3, Ph-JVC, and Ph-StCV1, and 15 of the most similar carlaviruses were reported. Apple stem pitting virus (ASPV; MZ147984) was used as an outgroup. The phylogenetic tree based on Rep revealed that PhCV1 clusters closely with Chrysanthemum virus B (CVB); PhCV2 clusters closely with Butterbur mosaic virus (ButMV), and PhCV3 clusters closely with TaCV1 (Figure 4D). A similar result was obtained in the phylogenetic tree based on CP sequences (Supplementary Figure S1). These results demonstrate that PhCV1, PhCV2, and PhCV3 meet the criteria to be new species of the genus Carlavirus.
A novel double-strand RNA virus was identified in Pseudostellaria heterophylla
Amalgavirus-related contigs were identified in all five libraries except for SB. The genome of the novel amalgavirus, PhAV1, was 3,430 bp long and contained two partially overlapping ORFs in its positive strand (Figure 6A). ORF1 (nt 171–1,325) followed a short 5’-UTR of 170 nt and was predicted to encode a CP of 384 aa residues, with an estimated molecular weight of 43.3 kDa. A + 1 PRF motif sequence 999UUU_CGN1,004 (underline indicates the codon boundary; N, any nucleotide) was found in the ORF1 of PhAV1 (Figure 6B). When the + 1 PRF event occurs, the codon frame changes from 999UUU_CGU1,004 to 999U_UUC_GUC1,005, causing the fusion of ORF1 + 2 (nt 171–998, 1,000–3,336), which is predicted to encode RdRp with 1,054 aa residues, with an estimated molecular weight of 120.4 kDa. A BLASTP analysis showed that the CP of PhAV1 had the highest degree of identity (30.73% aa sequence) with that of Phalaenopsis equestris amalgavirus 1 (PeAV1; NC_040590), and RdRp of PhAV1 had the most identity (51.01% aa sequence) with that of Cucumis melo amalgavirus 1 (CmAV1; MH479774; Zhan et al., 2019). A phylogenetic analysis based on the aa sequences of the RdRp indicated that PhAV1 is in a clade with approved and putative amalgaviruses and in a different clade with Zygosaccharomyces bailii virus Z (ZbV-Z), the only species of the genus Zybavirus that infects fungi (Figure 6C). Within the amalgaviruses clade, PhAV1 clusters closely with a branch that includes three putative amalgaviruses, PeAV1 (Nibert et al., 2016), Cucumis melon amalgavirus 1 (CmAV1; Zhan et al., 2019), and rubber dandelion latent virus 1 (RdLV1; unpublished results; Figure 6C). These results demonstrate that PhAV1 could be a new species of the genus Amalgavirus.
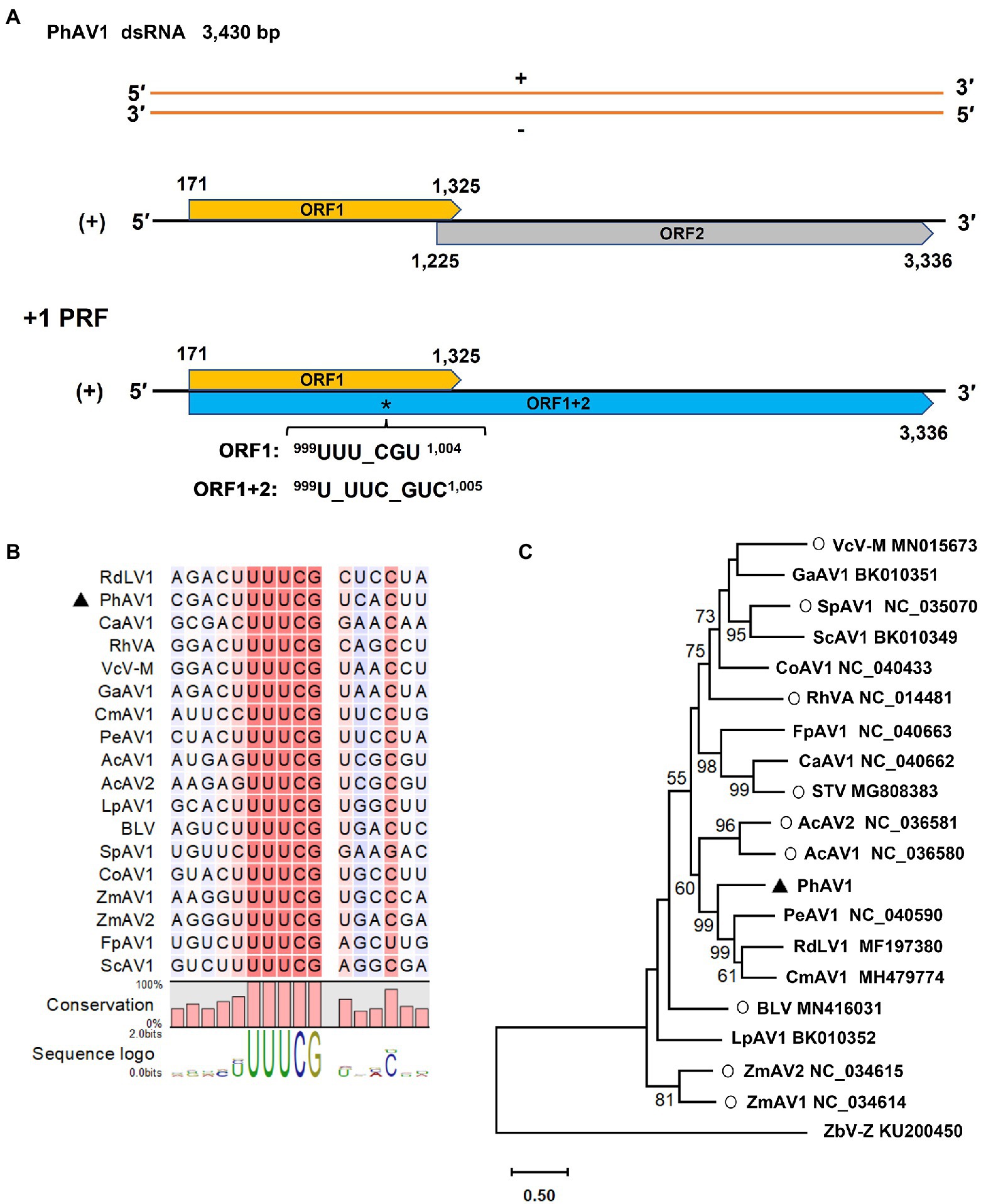
Figure 6. Characterization of a novel amalgavirus, pseudostellaria heterophylla amalgavirus 1 (PhAV1), isolated from Pseudostellaria heterophylla in China. Genomic organization of PhAV1 (A), and the putative + 1 programmed ribosomal frameshifting (PRF) motif of PhAV1 (B). (C) A phylogenetic analysis based on the amino acid sequences of the RNA-dependent RNA polymerase (RdRp) of PhAV1, nine confirmed (marked with a circle) and nine putative species in the genus Amalgavirus, performed by the maximum-likelihood method in MEGA X. The black triangle represents the novel amalgavirus identified in this study.
The distribution of PhCV1-3, Ph-JVC, Ph-StCV1, TuMV, BBWV2, and CMV in Pseudostellaria heterophylla
To identify the incidence of these nine viruses in different P. heterophylla growing regions, 7, 16, 8, 12, and 12, P. heterophylla leaf samples were collected from ZR-GX, ZR-HB, SB, DZ, and XC, respectively. Primers that specifically detect PhCV1, PhCV2, PhCV3, Ph-JVC, Ph-StCV1, TuMV, CMV, and RNA1 of BBWV2 were designed and are shown in Supplementary Table S1. The results showed that the detection rate of BBWV2 was the highest (78.18%), followed by TuMV (56.36%), PhCV1 (41.82%), and Ph-StCV1 (40.00%; Table 3). All eight viruses were detected in ZR-GX and ZR-HB in Fujian Province. BBWV2 (100%), TuMV (95.65%), and PhCV1 (65.22%) were present at higher levels (Figure 7; Supplementary Figure S2; Table 3). Except for PhCV2, all the viruses were detected in XC in Anhui Province with BBWV2 (83.33%), CMV (83.33%), and PhCV3 (83.33%) present at higher levels (Figure 7; Supplementary Figure S2; Table 3). Except for PhCV1 and PhCV3, all the viruses were detected in SB or DZ in Guizhou Province with BBWV2 (50.0%), PhCV2 (20.0%), and Ph-StCV1 (20.0%) present at higher levels (Figure 7; Supplementary Figure S2; Table 3). Due to the low content of PhAV1 (Table 1) and the long-term frozen storage of the cDNA samples, this virus was not detected in this experiment.
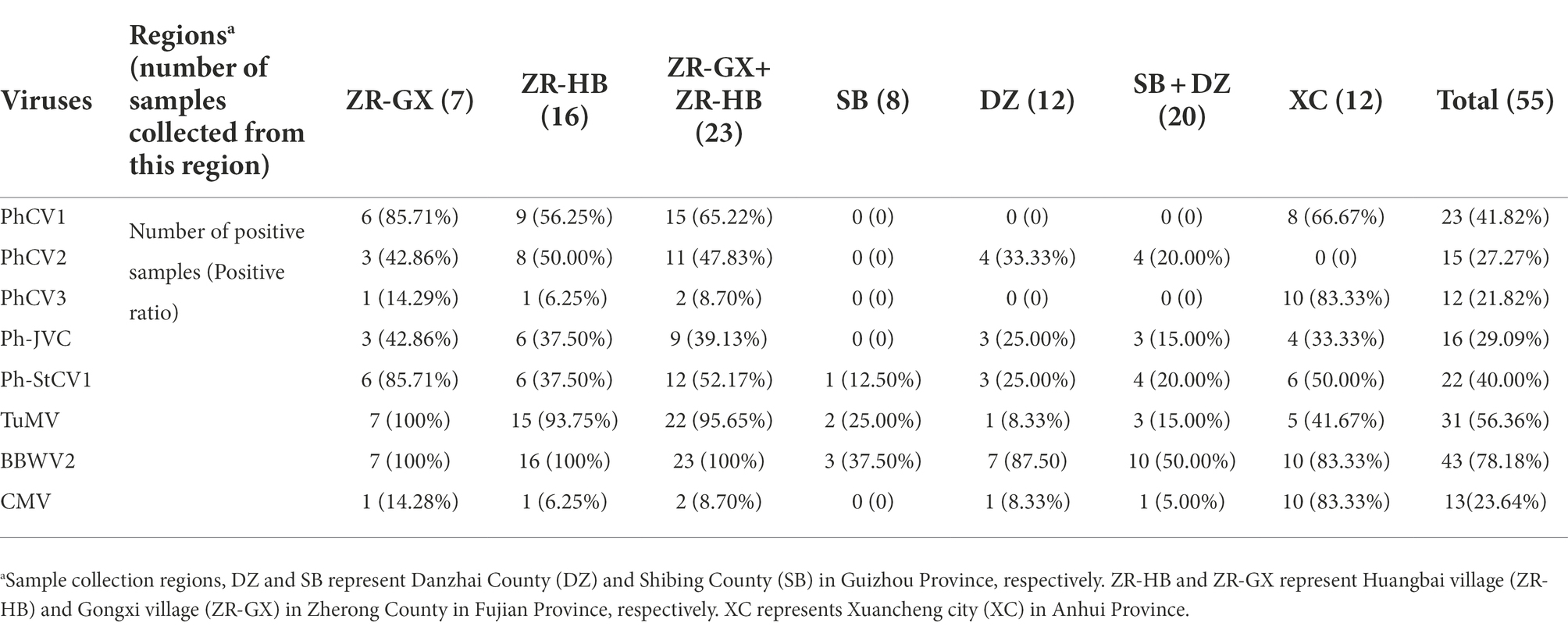
Table 3. RT-PCR analysis of the incidence of PhCV1, PhCV2, PhCV3, Ph-JVC, Ph-StCV1, TuMV, BBWV2, and CMV in Pseudostellaria heterophylla collected from five independent locations, including ZR-GX, ZR-HB, SB, DZ, and XC.
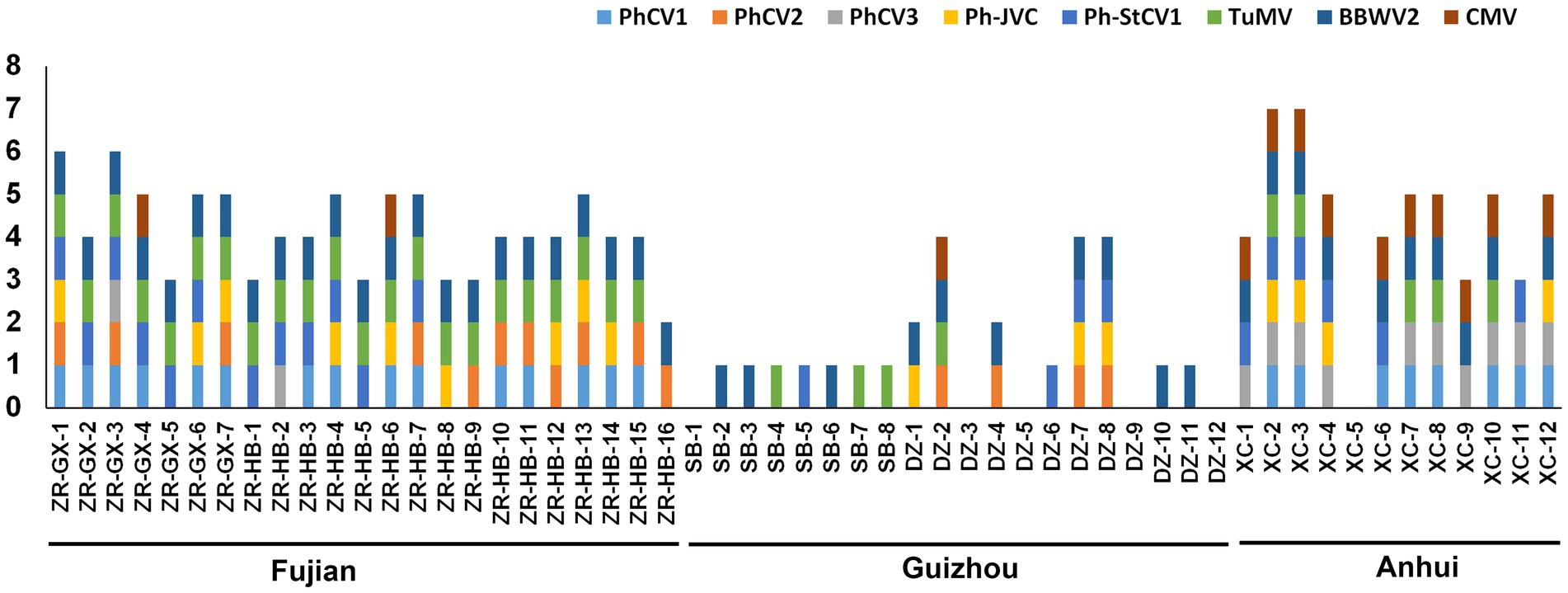
Figure 7. Distribution of PhCV1, PhCV2, PhCV3, Ph-JVC, Ph-StCV1, TuMV, BBWV2, and CMV in Pseudostellaria heterophylla collected from five independent locations, including ZR-GX, ZR-HB, SB, DZ, and XC.
Discussion
Pseudostellaria heterophylla is one of the most popular traditional Chinese herbal medicines and has been cultivated for hundreds of years. Historically, viral infections have always resulted in important diseases that affect the production of P. heterophylla. In 1991, four viruses, including TuMV, BBWV2, CMV, and TMV, were reported to infect P. heterophylla (Song and Pu, 1991). To our knowledge, no other viruses have been reported to infect P. heterophylla in recent decades. Metagenomics based on NGS can be used to discover the biodiversity of plant viruses in nature (Roossinck et al., 2015). Therefore, we used NGS to detect viruses in P. heterophylla. Five samples collected from the different primary growing regions of P. heterophylla were analyzed, and the results revealed the presence of TuMV, CMV, BBWV2, five carlaviruses, including Ph-JVC, Ph-StCV1, PhCV1, PhCV2, and PhCV3, and one amalgavirus, PhAV1, in P. heterophylla. TuMV, CMV, and BBWV2 are three viruses known to infect P. heterophylla, and they have very wide host ranges (Song and Pu, 1991; Bujarski et al., 2012; Sanfaçon et al., 2012; Adams et al., 2012b). JVC and StCV1 are two known unclassified carlaviruses that were isolated from Arabian jasmine (Jasminum sambac) and stevia (Stevia rebaudiana), respectively. Based on informatic analyses of the genomic features and phylogeny, PhCV1, PhCV2, and PhCV3 were proposed to be new members of the genus Carlavirus, and PhAV1 was proposed to be a new member of the genus Amalgavirus. These data indicate that a rich diversity of viruses infect P. heterophylla.
The incidence of these nine viruses in different P. heterophylla growing regions was detected by RT-PCR (Supplementary Figure S2). The occurrence of P. heterophylla virus disease was very serious in Fujian, Anhui and Guizhou Provinces with incidences of 100, 91.67 and 75.0%, respectively. Of the 23 samples collected in Fujian Province, 22 were simultaneously infected with TuMV, BBWV2, and one or more carlavirus(es). Of the 12 samples collected in Anhui Province, 11 were simultaneously infected with CMV, BBWV2, and one or more carlavirus(es). A total of 20 samples were collected in Guizhou Province. One virus was detected in 10 samples, two in two samples, four in three samples, and no viruses were detected in five samples. This showed that severe viral infections in P. heterophylla were present in all three provinces, particularly in Fujian and Anhui Provinces. BBWV2 was the most common virus in the three provinces with an incidence of 100% in Fujian Province, 83.33% in Anhui Province, and 50.0% in Guizhou Province (Table 3). In addition, TuMV (95.65%) and PhCV1 (65.22%) were also common in Fujian Province, and CMV (83.33%) and PhCV3 (83.33%) were common in Anhui Province. The difference and similarity of pathogens of P. heterophylla virus disease in different planting regions may be related to regional differences and/or the long-distance transportation of seedlings.
Like other perennial herbs, P. heterophylla can germinate the following year through its seeds and root tubers. To preserve unique characters and the high yield of P. heterophylla, root tubers are mostly used for propagation and cultivation. Vegetative propagation using root tubers facilitates the transmission of viruses and viral-plant symbiosis and symbiogenesis (Roossinck, 2008). Viruses can also be transmitted from one plant to another through vectors, such as aphids, which often results in mixed infections. Most of the extant viruses, except for the amalgavirus, related to those identified from P. heterophylla in this study have specific vectors for their dispersal. TuMV and CMV can be transmitted by more than 80 species of aphids, including Myzus persicae and Aphis gossypii which are two efficient and important vectors (Shattuck, 1992; Wang et al., 1998; Palukaitis and García-Arenal, 2003); most fabaviruses and carlaviruses have also been demonstrated to be transmitted by M. persicae (Machado Caballero et al., 2009; Belliure et al., 2011; Sanfaçon et al., 2012; Adams et al., 2012a; Wang et al., 2019). As a result, the viral diseases of P. heterophylla are serious, and multiple virus infection is widespread. Amalgaviruses are not thought to be capable of efficient extracellular transmission; they are primarily transmitted vertically through seeds to maintain their levels and spread among host plants (Martin et al., 2011; Fukuhara et al., 2020). However, this did not affect the widespread existence of amalgaviruses in P. heterophylla. Long-term vegetative propagation could be responsible for the widespread existence of amalgaviruses in P. heterophylla.
Three new (PhCV1, PhCV2, and PhCV3) and two known (Ph-JVC and Ph-StCV1) carlaviruses were identified in P. heterophylla for the first time. Carlavirus-related reads accounted for high proportions (91.37, 88.45, and 54.03% respectively) in the ZR-HB, ZR-GX, and DZ libraries (Figure 2; Table 1), suggesting that carlaviruses are major viruses that infect P. heterophylla. Leaf malformation and whole plant stunting are the types of symptoms caused by carlaviruses (Fujita et al., 2018). The symptoms caused by carlaviruses in P. heterophylla could not be confirmed because such viruses usually combine with TuMV or BBWV2 to infect P. heterophylla, and they were identified in all five libraries. Previous studies have shown that the CRPs encoded by carlaviruses play critical roles in determining symptoms and pathogenicity (Lukhovitskaya et al., 2013; Deng et al., 2015). The N-terminal region of CRPs, which contains conserved NLS and ZF motifs, determined the types of symptoms exhibited (Fujita et al., 2018). The CRPs of PhCV1, PhCV3, and Ph-StCV1 all contain NLS and ZF motifs, while the CRPs of PhCV2 and Ph-JVC contain the ZF motif but not the NLS one (Figure 4C). The association between symptoms and these conserved motifs merits further research.
Most of the putative amalgaviruses that have been recently identified were discovered by the analyses of a transcriptome dataset. Similarly, we identified PhAV1 in P. heterophylla by exploring its transcriptome. BLASTP and phylogenetic analyses based on the aa sequences of PhAV1 RdRp showed that PhAV1 clusters in the amalgaviruses clade. It displayed <51.01% aa sequence identity with the other amalgaviruses, indicating that PhAV1 could be a new species of the genus Amalgavirus. The +1 PRF is used in the expression of many genes of viruses, such as amalgaviruses, fijiviruses, and the influenza A virus (Firth and Atkins, 2009; Firth et al., 2012; Nibert et al., 2016). The shift site (999UUUCG1,003) of +1 PRF was identified in the ORF1 of PhAV1. This could result in the direction (+1) of the frameshifting that is involved in the expression of RdRp.
“BBWV2 exhibits a high genetic variation with sequence variants that are continuously distributed in a wide sequence space compared with most plant viruses. This sequence distribution (genetic structure) is not associated with geographic locations, suggesting long-distance gene flow, and it has been primarily shaped by negative selection and recombination (Ferrer et al., 2011).” Six isolates of BBWV2 were identified in P. heterophylla, and the RNA1 of isolates ZR-GX1 and XC1, RNA2 of isolates ZR-GX1 and XC2 shared a low degree of nt similarity (RNA1 < 75.66% and RNA2 < 74.19%) with other isolates in P. heterophylla and other isolates in the NCBI database (Table 2). Although these BBWV2 isolates did not meet the criteria for definition as new species, they confirmed the high genetic variation of BBWV2. Alternatively, they provide new genomic references for the molecular detection of BBWV2 in P. heterophylla.
In conclusion, this study is the first report of the complete nucleotide sequences of new viruses, including PhCV1, PhCV2, PhCV3, and PhAV1, that infect P. heterophylla. It is also the first report of the infection of P. heterophylla by JVC, StCV1, and multiple BBWV2 variants. The findings of this study provide useful information for the development of a molecular diagnostic technique to establish a more effective P. heterophylla viral disease control strategy.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, ON241319, ON241320, ON241321, ON241322, ON241323, ON241324, ON241325, ON241326, ON241327, ON241328, ON241329, ON241330, ON241331, ON241332, ON241333, ON241334, ON241335, ON241336.
Author contributions
RW and WD contributed to conception and design of the study, collected the samples and conducted the experiments. RW, YL, and SL performed the statistical analysis. RW, KG, YL, and SL discussed the results, drafted and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Key project at central government level: The ability establishment of sustainable use for valuable Chinese medicine resources (2060302), CAMS Innovation Fund for Medical Sciences (CIFMS) (2021-I2M-1-032), and the National Natural Science Foundation of China (81873095).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.955089/full#supplementary-material
SUPPLEMENTARY TABLE S5 | Pairwise nucleotide identities of the Rep of PhCV1, PhCV2, PhCV3, Ph-JVC, Ph-StCV1, and the 15 most similar carlaviruses.
SUPPLEMENTARY TABLE S6 | Pairwise amino acid identities of the Rep of PhCV1, PhCV2, PhCV3, Ph-JVC, Ph-StCV1, and the 15 most similar carlaviruses.
SUPPLEMENTARY TABLE S7 | Pairwise nucleotide identities of the CP of PhCV1, PhCV2, PhCV3, Ph-JVC, Ph-StCV1, and the 15 most similar carlaviruses.
SUPPLEMENTARY TABLE S8 | Pairwise amino acid identities of the CP of PhCV1, PhCV2, PhCV3, Ph-JVC, Ph-StCV1, and the 15 most similar carlaviruses.
Footnotes
1. ^https://talk.ictvonline.org/taxonomy/
3. ^http://hannonlab.cshl.edu/fastx_toolkit/index.html
4. ^http://doua.prabi.fr/software/cap3
5. ^https://www.ncbi.nlm.nih.gov/orffinder
References
Adams, M. J., Candresse, T., Hammond, J., Kreuze, J. F., Martelli, G. P., Namba, S., et al. (2012a). “Family Betaflexiviridae,” in Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. eds. A. M. Q. King, M. J. Adams, E. B. Carstens, and E. J. Lefkowitz (London: Elsevier Academic Press), 920–941.
Adams, M. J., Zerbini, F. M., French, R., Rabenstein, F., Stenger, D. C., and Valkonen, J. P. T. (2012b). “Family Potyviridae,” in Virus Taxonomy: Ninth Report Of The International Committee On Taxonomy Of Viruses. eds. A. M. Q. King, M. J. Adams, E. B. Carstens, and E. J. Lefkowitz (London: Elsevier Academic Press), 1069–1089.
Belliure, B., Amorós-Jiménez, R., Fereres, A., and Marcos-García, M. Á. (2011). Antipredator behaviour of Myzus persicae affects transmission efficiency of broad bean wilt virus 1. Virus Res. 159, 206–214. doi: 10.1016/j.virusres.2011.04.019
Bujarski, J., Figlerowicz, M., Gallitelli, D., Roossinck, M. J., and Scott, S. W. (2012). “Family Bromoviridae,” in Virus Taxonomy: Ninth Report Of The International Committee On Taxonomy Of Viruses. eds. A. M. Q. King, M. J. Adams, E. B. Carstens, and E. J. Lefkowitz (London: Elsevier Academic Press), 965–976.
Choi, Y. Y., Kim, M. H., Ahn, K. S., Um, J. Y., Lee, S. G., and Yang, W. M. (2017). Immunomodulatory effects of Pseudostellaria heterophylla (Miquel) Pax on regulation of Th1/Th2 levels in mice with atopic dermatitis. Mol. Med. Rep. 15, 649–656. doi: 10.3892/mmr.2016.6093
Deng, X. G., Peng, X. J., Zhu, F., Chen, Y. J., Zhu, T., Qin, S. B., et al. (2015). A critical domain of sweet potato chlorotic fleck virus nucleotide binding protein (NaBp) for RNA silencing suppression, nuclear localization and viral pathogenesis. Mol. Plant Pathol. 16, 365–375. doi: 10.1111/mpp.12186
Ferrer, R. M., Ferriol, I., Moreno, P., Guerri, J., and Rubio, L. (2011). Genetic variation and evolutionary analysis of broad bean wilt virus 2. Arch. Virol. 156, 1445–1450. doi: 10.1007/s00705-011-0990-3
Firth, A. E., and Atkins, J. F. (2009). Analysis of the coding potential of the partially overlapping 3’ ORF in segment 5 of the plant fijiviruses. Virol. J. 6:32. doi: 10.1186/1743-422X-6-32
Firth, A. E., Jagger, B. W., Wise, H. M., Nelson, C. C., Parsawar, K., Wills, N. M., et al. (2012). Ribosomal frameshifting used in influenza a virus expression occurs within the sequence UCC_UUU_CGU and is in the +1 direction. Open Biol. 2:120109. doi: 10.1098/rsob.120109
Fujita, N., Komatsu, K., Ayukawa, Y., Matsuo, Y., Hashimoto, M., Netsu, O., et al. (2018). N-terminal region of cysteine-rich protein (CRP) in Carlaviruses is involved in the determination of symptom types. Mol. Plant Pathol. 19, 180–190. doi: 10.1111/mpp.12513
Fukuhara, T., Tabara, M., Koiwa, H., and Takahashi, H. (2020). Effect of asymptomatic infection with southern tomato virus on tomato plants. Arch. Virol. 165, 11–20. doi: 10.1007/s00705-019-04436-1
Gao, W., Zhang, J. S., Zhang, J. H., and Zhang, L. R. (1993). Detection and control of Taizishen mosaic virus. Virol. Sin. 8, 390–393.
Guo, R., Wei, W., Wang, Y., Li, K., Cui, X., Zhu, C., et al. (2014). Protective effects of radix Pseudostellariae extract against retinal laser injury. Cell. Physiol. Biochem. 33, 1643–1653. doi: 10.1159/000362947
Huang, Y., and Lin, C. (2004). Occurrence and control of Tizhi jinseng virus diseases in Mindong. J. Ningde Teachers College (Nat. Sci.) 16, 65–68.
Isogai, M., Nakamura, T., Ishii, K., Watanabe, M., Yamagishi, N., and Yoshikawa, N. (2011). Histochemical detection of blueberry latent virus in highbush blueberry plant. J. Gen. Plant Pathol. 77, 304–306. doi: 10.1007/s10327-011-0323-0
Jo, Y., Kim, S. M., Choi, H., Yang, J. W., Lee, B. C., and Cho, W. K. (2020). Sweet potato viromes in eight different geographical regions in Korea and two different cultivars. Sci. Rep. 10:2588. doi: 10.1038/s41598-020-59518-x
Kang, C. Z., Zhou, T., Guo, L. P., Huang, L. Q., Zhu, S. D., and Xiao, C. H. (2016). Ecological suitability and regionalization of Pseudostellaria heterophylla (Miq.) Pax ex Pax et Hoffm. In China. Acta Ecol. Sin. 36, 2934–2944. doi: 10.5846/stxb201409231880
Koloniuk, I., Hrabáková, L., and Petrzik, K. (2015). Molecular characterization of a novel amalgavirus from the entomopathogenic fungus Beauveria bassiana. Arch. Virol. 160, 1585–1588. doi: 10.1007/s00705-015-2416-0
Kondo, H., Fujita, M., Hisano, H., Hyodo, K., Andika, I. B., and Suzuki, N. (2020). Virome analysis of aphid populations that infest the barley field: the discovery of two novel groups of Nege/Kita-like viruses and other novel RNA viruses. Front. Microbiol. 11:509. doi: 10.3389/fmicb.2020.00509
Krupovic, M., Dolja, V. V., and Koonin, E. V. (2015). Plant viruses of the Amalgaviridae family evolved via recombination between viruses with double-stranded and negative-strand RNA genomes. Biol. Direct 10:12. doi: 10.1186/s13062-015-0047-8
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lukhovitskaya, N. I., Solovieva, A. D., Boddeti, S. K., Thaduri, S., Solovyev, A. G., and Savenkov, E. I. (2013). An RNA virus-encoded zinc-finger protein acts as a plant transcription factor and induces a regulator of cell size and proliferation in two tobacco species. Plant Cell 25, 960–973. doi: 10.1105/tpc.112.106476
Machado Caballero, J. E., Lockhart, B. E., Mason, S. L., and Daughtrey, M. (2009). Identification and properties of a carlavirus causing chlorotic mottle of florists’ hydrangea (H. macrophylla) in the United States. Plant Dis. 93, 891–895. doi: 10.1094/PDIS-93-9-0891
Martin, R. R., Tzanetakis, I. E., Sweeney, M., and Wegener, L. (2006). A virus associated with blueberry fruit drop disease. Acta Hortic. 715, 497–502. doi: 10.17660/ActaHortic.2006.715.76
Martin, R. R., Zhou, J., and Tzanetakis, I. E. (2011). Blueberry latent virus: an amalgam of the Partitiviridae and Totiviridae. Virus Res. 155, 175–180. doi: 10.1016/j.virusres.2010.09.020
Mu, F., Xie, J., Cheng, S., You, M. P., Barbetti, M. J., Jia, J., et al. (2018). Virome characterization of a collection of S. sclerotiorum from Australia. Front. Microbiol. 8:2540. doi: 10.3389/fmicb.2017.02540
Muhire, B. M., Varsani, A., and Martin, D. P. (2014). SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9:e108277. doi: 10.1371/journal.pone.0108277
Nibert, M. L., Pyle, J. D., and Firth, A. E. (2016). A + 1 ribosomal frameshifting motif prevalent among plant amalgaviruses. Virology 498, 201–208. doi: 10.1016/j.virol.2016.07.002
Palukaitis, P., and García-Arenal, F. (2003). Cucumoviruses. Adv. Virus Res. 62, 241–323. doi: 10.1016/s0065-3527(03)62005-1
Pang, W., Lin, S., Dai, Q., Zhang, H., and Hu, J. (2011). Antitussive activity of Pseudostellaria heterophylla (Miq) Pax extracts and improvement in lung function via adjustment of multi-cytokine levels. Molecules 16, 3360–3370. doi: 10.3390/molecules16043360
Park, D., Goh, C. J., Kim, H., and Hahn, Y. (2018). Identification of two novel amalgaviruses in the common eelgrass (Zostera marina) and in silico analysis of the amalgavirus +1 programmed ribosomal frameshifting sites. Plant Pathol. J. 34, 150–156. doi: 10.5423/PPJ.NT.11.2017.0243
Roossinck, M. J., Martin, D. P., and Roumagnac, P. (2015). Plant virus metagenomics: advances in virus discovery. Phytopathology 105, 716–727. doi: 10.1094/PHYTO-12-14-0356-RVW
Sabanadzovic, S., Valverde, R. J., Martin, R., and Tzanetakis, I. (2009). Southern tomato virus: the link between the families Totiviridae and Partitiviridae. Virus Res. 140, 130–137. doi: 10.1016/j.virusres.2008.11.018
Sanfaçon, H., Iwanami, T., Karasev, A. V., van der Vlugt, R., Wellink, J., Wetzel, T., et al. (2012). “Family Secoviridae” in Virus Taxonomy: Ninth Report Of The International Committee On Taxonomy Of Viruses. eds. A. M. Q. King, M. J. Adams, E. B. Carstens, and E. J. Lefkowitz (London: Elsevier Academic Press), 881–899.
Shattuck, V. I. (1992). The biology, epidemiology, and control of turnip mosaic virus. Horticult. Rev. 14, 199–238. doi: 10.1002/9780470650523.ch4
Sheng, R., Xu, X., Tang, Q., Bian, D., Li, Y., Qian, C., et al. (2011). Polysaccharide of radix pseudostellariae improves chronic fatigue syndrome induced by poly I: C in mice. Evid. Based Complement. Alternat. Med. 2011:840516. doi: 10.1093/ecam/nep208
Song, R., and Pu, Z. (1991). Studies of Taizishen (Pseudostellaria heterophylla) virus diseases. Acta Agric. Shanghai 7, 80–85.
Vendrell-Mir, P., Perroud, P. F., Haas, F. B., Meyberg, R., Charlot, F., Rensing, S. A., et al. (2021). A vertically transmitted amalgavirus is present in certain accessions of the bryophyte Physcomitrium patens. Plant J. 108, 1786–1797. doi: 10.1111/tpj.15545
Wang, R., Li, Y., Wei, R., Chen, B., Wang, T., and Ding, W. (2022). Investigation on the occurrence of virus diseases in the main producing areas of Pseudostellaria heterophylla in China. Plant Prot. 48, 204–210. doi: 10.16688/j.zwbh.2020584
Wang, R. Y., Powell, G., Hardie, J., and Pirone, T. P. (1998). Role of the helper component in vector-specific transmission of potyviruses. J. Gen. Virol. 79, 1519–1524. doi: 10.1099/0022-1317-79-6-1519
Wang, R. L., Zhu-Salzman, K., Elzaki, M. E. A., Huang, Q. Q., Chen, S., Ma, Z. H., et al. (2019). Mikania micrantha wilt virus alters insect vector's host preference to enhance its own spread. Viruses 11:336. doi: 10.3390/v11040336
Zhan, B., Cao, M., Wang, K., Wang, X., and Zhou, X. (2019). Detection and characterization of Cucumis melo crypticvVirus, Cucumis melo amalgavirus 1, and melon necrotic spot virus in Cucumis melo. Viruses 11:81. doi: 10.3390/v11010081
Keywords: Pseudostellaria heterophylla, virome, next-generation sequencing, novel viruses, Carlavirus, Amalgavirus
Citation: Li Y, Liu S, Guo K, Ding W and Wang R (2022) Virome of Pseudostellaria heterophylla: Identification and characterization of three novel carlaviruses and one novel amalgavirus associated with viral diseases of Pseudostellaria heterophylla. Front. Microbiol. 13:955089. doi: 10.3389/fmicb.2022.955089
Edited by:
Giuseppe Parrella, Institute for Sustainable Plant Protection of the National Research Council (IPSP-CNR), ItalyReviewed by:
Eui-Joon Kil, Andong National University, South KoreaTomofumi Mochizuki, Osaka Metropolitan University, Japan
Fan Li, Yunnan Agricultural University, China
Copyright © 2022 Li, Liu, Guo, Ding and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rong Wang, cndhbmdAaW1wbGFkLmFjLmNu, d2FuZ3JvbmcxMDE5QDE2My5jb20=
 Yong Li
Yong Li Rong Wang
Rong Wang