- Division of Immunobiology and Center for Inflammation and Tolerance, Cincinnati Children’s Hospital Medical Center, University of Cincinnati College of Medicine, Cincinnati, OH, United States
Despite modern therapeutic developments and prophylactic use of antibiotics during birth or in the first few months of life, enteric infections continue to be a major cause of neonatal mortality and morbidity globally. The neonatal period is characterized by initial intestinal colonization with microbiota and concurrent immune system development. It is also a sensitive window during which perturbations to the environment or host can significantly impact colonization by commensal microbes. Extensive research has demonstrated that these early life alterations to the microbiota can lead to enhanced susceptibility to enteric infections and increased systemic dissemination in newborns. Various contributing factors continue to pose challenges in prevention and control of neonatal enteric infections. These include alterations in the gut microbiota composition, impaired immune response, and effects of maternal factors. In addition, there remains limited understanding for how commensal microbes impact host-pathogen interactions in newborns. In this review, we discuss the recent recognition of initial microbiota-epithelial interactions that occur in neonates and can regulate susceptibility to intestinal infection. These studies suggest the development of neonatal prophylactic or therapeutic regimens that include boosting epithelial defense through microbiota-directed interventions.
Introduction
Despite a steady decline in childhood mortality (Sharrow et al., 2022), enteric infections in infants continue to pose a significant health risk (Kotloff et al., 2013; Bagamian et al., 2020). Indeed, infections remain one of the major causes of mortality in both preterm and term infants (Stoll et al., 2011; Isaac et al., 2016; Troeger et al., 2018). Due to the immaturity of their developing immune system and lack of a diverse intestinal microbiota, trillions of microbes residing in the gut lumen that restrain pathogens, neonates are particularly vulnerable to infection (Miller et al., 2018). Newborns are initially dependent on epithelial defense, transfer of maternal immune factors, innate immune cell activation, and intestinal microbial colonization for protection against intestinal pathogens. Studies have emphasized that the development of the intestinal microbiota is critical for the maturation of the immune system, epithelial barrier, and colonization resistance against invading pathogens (Olszak et al., 2012; Cahenzli et al., 2013; El Aidy et al., 2013; An et al., 2014; Gensollen et al., 2016; Yu et al., 2016; Roubaud-Baudron et al., 2019; Singer et al., 2019; Travier et al., 2021). Infants become rapidly colonized during birth with the intestinal microbiota abundance, composition, and diversity continuing to mature throughout neonatal development. Further, the neonatal immune system coevolves with the intestinal microbiota, and this early window of immune cell education and development is critical for healthy immune responses later in life. Antibiotics (ABX) are commonly used to treat or prevent infections in infancy. However, early life (prenatal, perinatal, and postnatal) exposure to ABX can perturb the developing microbiota, induce epithelial barrier dysfunction and may lead to enhanced neonatal susceptibility to enteric pathogens (Schumann et al., 2005; Greenwood et al., 2014; Rogawski et al., 2015; Schulman et al., 2015; Man et al., 2019). In addition, alterations in the intestinal microbiota by ABX promote short- and long-term immunological effects extending well into adulthood. This review examines the critical role and interplay of commensal bacteria and the epithelial barrier that influence enteric infections in neonates.
Neonatal intestinal infection
Newborns are highly susceptible to enteric pathogens, particularly during the first year of life (Liu et al., 2012; Lanata et al., 2013; Darmstadt et al., 2014; Miller et al., 2016). Most common enteric pathogens linked to neonatal morbidity and mortality include Enteropathogenic Escherichia coli (EPEC), Group B streptococci (GBS), Listeria monocytogenes, Salmonella, Rotavirus, and Cryptosporidium parasites (Abba et al., 2009; Stoll et al., 2011; Pedersen et al., 2014; Bergin et al., 2015; Shane et al., 2017; Reju et al., 2022; Yoon et al., 2022). These pathogens can gain entry orally, invade the gastrointestinal (GI) tract, and lead to further blood or systemic infections in neonates.
Enteropathogenic E. coli (EPEC) is responsible for many diarrhea outbreaks in newborns (Nataro and Kaper, 1998; Abba et al., 2009; Hu and Torres, 2015). EPEC infection is characterized by attaching and effacing (A/E) lesions (Cepeda-Molero et al., 2017). While adult C57BL/6 mice are resistant to human EPEC, neonatal mice exhibit age-dependent susceptibility. Pups up to 7 day-old were found to be highly susceptible, with significantly decreased susceptibility observed in 10–13 day-old (Dupont et al., 2016). The neonatal infection depended on EPEC expression of virulent factors IV bundle forming pili (BFP) and type III secretion system (T3SS), both of which are essential for A/E lesion formation and host cell invasion (Cleary et al., 2004; Iizumi et al., 2007; Galan et al., 2014). This unique susceptibility of neonates was attributed to the neonatal gut microbiota as well as epithelial responses to the pathogen that are unique to neonates, including more exaggerated upregulation of TLR-dependent genes compared to adults (Dupont et al., 2016).
Non-typhoidal Salmonella, Salmonella enterica subsp. enterica, also causes enteric diseases. Salmonella is classified as acid-sensitive and can be killed by the acidity in the adult stomach. However, less acidic stomach contents and faster gastric emptying of neonates favor Salmonella survival and small intestinal colonization (Gorden and Small, 1993; Bula-Rudas et al., 2015). Neonatal mice demonstrate a higher susceptibility to infections than adults (Zhang et al., 2014). Salmonella directly invades enterocytes in neonates in a T3SS dependent manner, instead of utilizing microfold cells (M-cell)-mediated uptake of pathogens observed in adults (Zhang et al., 2014). Unlike in adults, infection of C57BL/6 neonatal mice did not require streptomycin pretreatment, suggesting that characteristics of the neonatal microbiota lead to unique susceptibility (Zhang et al., 2014). A more recent study colonized adult germ-free mice with neonatal or adult cecal contents and found that microbiota from adults, but not neonates, prevented Salmonella colonization (Kim et al., 2017).
Group B Streptococcus (GBS) is an intestinal commensal that causes systemic diseases such as septicemia and meningitis in infants, but not in immunocompetent adults (Raabe and Shane, 2019). GBS gains access to the systemic circulation through invasion of the neonatal intestinal epithelium. A recent study demonstrated that immature epithelial barrier function and microbiota composition attributed neonatal susceptibility to GBS infection (Travier et al., 2021). These enteric infection studies in young mice demonstrate strong links between increased pathogen susceptibility with the neonatal microbiota and developing epithelial cell function, discussed in more detail below.
Neonatal gut microbiota
Colonization by commensal microbes at birth is critical for the development of host immunity and defense against pathogens (Negi et al., 2019; Zheng et al., 2020; Fan and Pedersen, 2021). The microbiota composition of neonates differs from adults. In humans, Firmicutes and Bacteroidetes are the two dominant phyla in adults while Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria predominate in term, vaginally delivered neonates (Arboleya et al., 2015; Backhed et al., 2015; Del Chierico et al., 2015; Radjabzadeh et al., 2020). Microbiota colonization begins at birth and is dominated mainly by facultative anaerobes such as Lactobacillus, Enterococcus, and Streptococcus in the initial days of life (Hansen et al., 2015; Robertson et al., 2019). Further, breastfeeding enriches the Bifidobacterium and Bacteroidetes followed by the prevalence of obligate anaerobes such as Clostridia in the gut. The murine commensal Clostridia has been shown to protect against enteric Salmonella infection. This protective effect was attributed to the production of metabolite succinate, though its mechanisms remain to be elucidated (Kim et al., 2017). In addition, Bifidobacterium-produced acetate has been reported to protect against E. coli infection (Fukuda et al., 2011).
The composition of the neonatal microbiota is significantly shaped by the maternal microbiota and diet (Dominguez-Bello et al., 2010; Chu et al., 2016; Asnicar et al., 2017; Shao et al., 2019; Garcia-Mantrana et al., 2020; Maher et al., 2020). There are also reports that maternal microbiota-derived factors and metabolites such as short-chain fatty acids may be sensed or passed during gestation and influence the offspring’s metabolism (Kimura et al., 2020; Pessa-Morikawa et al., 2022). Transmission of microbiota during birth is followed by early development during the first 2–3 years to ultimately more closely resemble adulthood composition (Donnet-Hughes et al., 2010; Rautava et al., 2012). This dynamic bacterial colonization inversely correlates with the occurrence of infections in neonates (Madan et al., 2012; Mai et al., 2013; Matamoros et al., 2013). Additionally, studies have demonstrated that proper microbiota colonization during this early neonatal period may impact long-term health (Torow and Hornef, 2017; Renz et al., 2018). These include murine models showing the importance of early colonization in immune development as described in the following section (Olszak et al., 2012; Al Nabhani et al., 2019). Further, human longitudinal studies have shown that neonatal microbiota compositions associate with clinical manifestations in allergy, neurodevelopment, and metabolic disorders later in childhood (Galazzo et al., 2020; Roze et al., 2020; Jian et al., 2021).
Recent advancements in metagenomic sequencing have revealed strain-level details of shared bacteria between infants and their mothers, with the mode of delivery affecting microbial exposure in early life (Asnicar et al., 2017; Ferretti et al., 2018; Shao et al., 2019). Vaginally delivered infants are often predominantly colonized by beneficial commensal bacteria such as Bacteroides, Lactobacillus, and Bifidobacterium (Hesla et al., 2014; Chu et al., 2017; Stewart et al., 2018). Interestingly, by 6 months of life, Lactobacillus colonization was found to be the same irrespective of delivery mode in one study (Huurre et al., 2008). In another study, Bacteroides were still found to be high in vaginally born infants (Stinson et al., 2018). Infants delivered via C-section harbor high abundance of opportunistic pathogens such as Enterococcus and Enterobacter (Jakobsson et al., 2014; Shao et al., 2019). They also exhibit delayed colonization with beneficial Bifidobacterium (Reyman et al., 2019).
Breast milk (BM) is often the first diet for newborns and plays a central role in shaping the neonatal microbiota (Turfkruyer and Verhasselt, 2015; Walker and Iyengar, 2015). BM contains high amounts of human milk oligosaccharides (HMOs); thus breastfed infants exhibit increased Bifidobacterium species that are involved in catabolism of HMOs (Backhed et al., 2015; Robertson et al., 2019). Bifidobacterium infantis EVC001 supplementation in term infants positively correlated with abundance of memory Treg, and negatively correlated with Th2/Th17 cytokines in the blood (Henrick et al., 2021). Further, formula-fed newborns possess more Enterobacteriaceae and fewer Bifidobacterium species (Stewart et al., 2018). Secretory IgA (sIgA) in BM also contributes to development of the neonatal microbiota, and neonates lacking sIgA have alterations in commensal communities that persist to adulthood (Rogier et al., 2014). While less characterized, viral communities also colonize the infant gut and are influenced by breastfeeding (Bushman and Liang, 2021). Furthermore, a study shows that breastfed infants harbor more temperate phages of Bifidobacterium or Lactobacillus at 4 months of age, coinciding with higher abundance of these bacteria, in comparison to formula-fed infants. On the other hand, viruses that infect human cells were less abundant in BM-fed infants’ stool, suggestive of the protective roles of BM against viral infection (Liang et al., 2020).
Antibiotics exposure both in utero and early postnatal life alters the gut microbiota composition of both mother and newborn, and therefore can potentially have long-lasting effects (Ohlsson and Shah, 2014; Arboleya et al., 2016; Miller et al., 2018). Exposure to commensal microbes in this early life period is critical in the development of proper immune function. For instance, GF mice harbor elevated numbers of invariant natural killer T (iNKT) cells which predispose them to chemically induced colitis. This abnormal iNKT cell phenotype can only be rescued when GF mice are recolonized on the first day of life but not in adulthood, highlighting the importance of early life immunological imprinting by the microbiota (Olszak et al., 2012). In addition, ABX-inhibition of colonization during the weaning period in mice resulted in decreased RORγt+ regulatory T cells and increased susceptibility to chemically induced colitis (Al Nabhani et al., 2019). Further, another study reported that neonatal mice with transient early life ABX exposure resulted in persistent microbiota alterations and increased susceptibility to enteric bacterial infection as an adult (Roubaud-Baudron et al., 2019). Therefore, these findings suggest that ABX treatment early in life may alter the microbiota in a manner that not only impacts neonatal defense, but may also increase susceptibility later in life. In preterm infants, factors such as gestational age, ABX, reduced BM consumption, environmental microbes of neonatal intensive care unit, and prolonged hospitalization contribute to the colonization with specific microbial strains such as Enterobacter, Enterococcus, Lactobacillus, Photorhabdus, and Tannerella (Ardissone et al., 2014; Collado et al., 2015).
Intestinal epithelial cells
Immune responses in the neonatal intestine are required to be tolerant to newly colonizing commensal bacteria while also protecting against enteric pathogens. Innate immune cells play a central role in protective immunity in the neonatal period, as adaptive immunity is still immature (Kollmann et al., 2012; Lee et al., 2019; Rudd, 2020; Westrom et al., 2020). In addition, intestinal epithelial cells (IECs) are non-hematopoietic cells that serve as the first line of defense and a key barrier to invading pathogens in both mice and humans (Allaire et al., 2018; Eshleman and Alenghat, 2021; Figure 1).
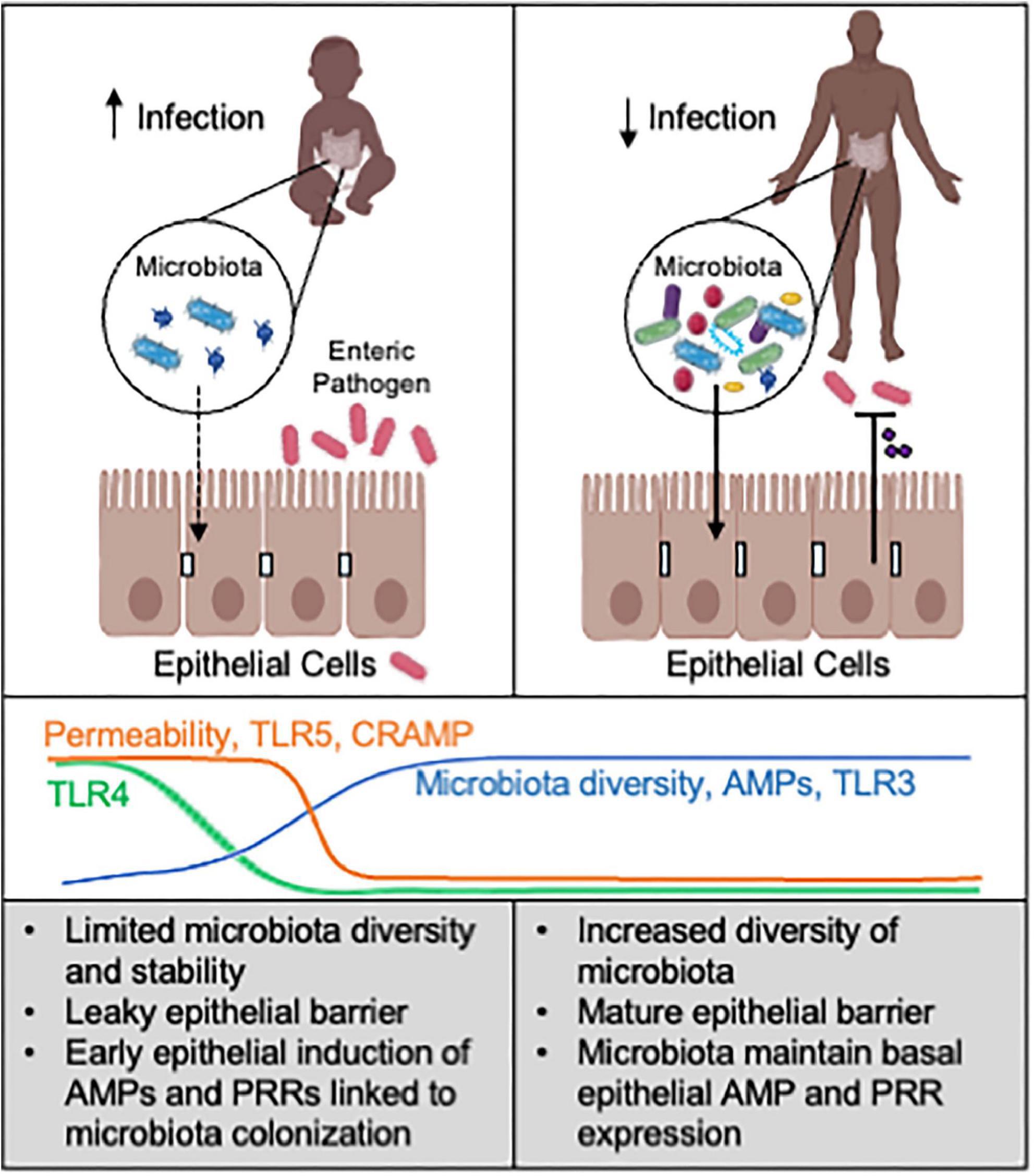
Figure 1. Microbiota-epithelial interactions alter susceptibility to infection. Neonates are highly susceptible to enteric pathogens, relative to older children and adults. In the neonatal intestine, a limited abundance and diversity of commensal microbes correspond with increased epithelial permeability and distinct AMP expression and microbial sensing. This less mature microbiota-epithelial relationship likely increases susceptibility to enteric infection. In contrast, the adult intestine harbors a more complex and stable microbiota that can maintain more effective epithelial defense mechanisms against pathogens. Promoting epithelial defense through microbiota-based interventions may help prevent or combat neonatal enteric infection. AMPs: antimicrobial peptides. PRRs: pattern recognition receptors. CRAMP (cathelin-related antimicrobial peptide) is AMP enriched in neonatal intestine. Line graphs are based on data from animal models. Created with Biorender.com.
The epithelium is organized into villi and crypt structures, with stem cells residing at the base of crypts differentiating into absorptive and secretory lineages (Walthall et al., 2005; Allaire et al., 2018). Humans and pigs exhibit mature crypt-villus axes at birth while in mice, crypts become fully functional 2 weeks after birth (Walthall et al., 2005; Harper et al., 2011). Thereafter, stem cells in crypts differentiate into different lineages of IECs such as absorptive enterocytes and secretory goblet cells, Paneth cells and enteroendocrine cells (EECs) (Barker et al., 2007). The proliferation rate of epithelial stem cells in neonates is lower than in adults, which is at least partly due to elevated expression of B lymphocyte-induced maturation protein-1 (Blimp-1), a transcriptional repressor that inhibits adult-like differentiation and regulates structural and biochemical changes of IECs during the suckling-weaning transition in mice (Harper et al., 2011; Muncan et al., 2011).
Barrier function
Intestinal epithelial cells reside directly at the interface of the developing microbiota and underlying immune system (Allaire et al., 2018; Eshleman and Alenghat, 2021). Many intracellular structures such as adherens junctions (AJ), desmosomes and tight junctions (TJ) maintains barrier integrity in both mice and humans. TJ proteins, such as claudins and occludin, seal the intercellular spaces and strictly regulate the macromolecular transport (Van Itallie et al., 2008; Suzuki et al., 2014; Ronaghan et al., 2016). Zonulin, a signaling molecule that triggers phosphorylation of epidermal growth factor receptor (EGFR) and downstream tight junction disassembly, regulates gut permeability (Sturgeon and Fasano, 2016). Fecal and serum zonulin are used as a biomarker for increased intestinal permeability in neonatal studies (Saleem et al., 2017; Kaczmarczyk et al., 2021; Sochaczewska et al., 2022). The development of intestinal barrier function occurs in utero in humans, with IEC tight junction proteins production observed as early as 10 weeks of gestation, followed by secretion of defensins, lysozyme, and mucin that create additional layers of a chemical and mechanical barrier (Polak-Charcon et al., 1980; Mallow et al., 1996; Buisine et al., 1998; Rumbo and Schiffrin, 2005). Epithelial barrier function is affected by oral ABX administration and alterations in microbiota composition (Schumann et al., 2005; Ma et al., 2018; Garcia et al., 2021; Sochaczewska et al., 2022). Reports differ on whether ABX increase or decrease epithelial barrier function, and this may reflect differences in the types of ABX and microbiota composition (Schumann et al., 2005; Soto et al., 2014; Zhou et al., 2020; Chaaban et al., 2022).
Maturation of barrier function continues postnatally in response to factors present in the neonatal intestine. For example, breast milk (BM) components have been described to improve epithelial barrier function in both rodent and human studies (Weaver et al., 1987; Saleem et al., 2017; Chleilat et al., 2020). Lactoferrin, an iron-binding protein, exhibits protective effects in bacterial endotoxin-induced intestinal barrier damage (Hirotani et al., 2008). Further, BM-derived transforming growth factor-beta (TGF-β) inhibits proinflammatory responses in immature human IECs and is associated with the intestinal microbiota composition in neonates (Rautava et al., 2011; Sitarik et al., 2017). BM has been shown to increase the expression of TJ protein occludin in a pediatric enteroid model (Noel et al., 2021). Furthermore, neonatal supplementation of EGF in mice prevented translocation of pathogenic bacteria by inhibiting goblet cell-associated antigen passages (GAPs) that transport luminal antigens across intestinal epithelium (Knoop et al., 2020). Unlike adults, newborn epithelium is characterized by highly endocytic vacuolated enterocytes in the distal immature small intestine that allows passage of BM immunomodulatory components across the intestine (Baba et al., 2002; Arevalo Sureda et al., 2016; Garcia et al., 2021). Further, increased expression of neonatal Fc receptor (FcRn) on epithelial cells enables transport of maternal antibodies that confer passive immunity against pathogens in neonates (Yoshida et al., 2006; Menard et al., 2010; Ben Suleiman et al., 2012).
Antimicrobial peptides
In addition to serving as a physical barrier, IECs also produce antimicrobial peptides (AMPs), mucins, chemokines, and cytokines that prime and regulate innate and adaptive immunity. Additionally, IECs possess various membrane and cytoplasmic patterns-recognition receptors that can detect microbial stimuli (Figure 1). AMPs can inhibit microbial survival or growth, and are among one of the most evolutionarily ancient immune defense mechanisms. A diverse array of AMPs secreted by IECs provide the first line of defense against pathogens. The enzymatic AMPs such as lysozyme and phospholipase A2 (sPLA2) are mainly secreted by Paneth cells, and damage bacterial cell walls through their catalytic activities (Menard et al., 2008; Clevers and Bevins, 2013; Mukherjee and Hooper, 2015; Bel et al., 2017). Other AMPs, such as cathelicidins, C-type lectins of the regenerating islet-derived protein (reg) 3 gamma family, and defensins disrupt microbial cell walls in a non-enzymatic fashion (Ouellette, 2010; Clevers and Bevins, 2013). Defensins produced in crypts possess bactericidal activity and can promote chloride secretion that may facilitate pathogen flushing from the intestine (Baird and O’malley, 1993; Lencer et al., 1997; Ayabe et al., 2000).
Paneth cells, major AMP producers in the small intestinal crypt, develop prenatally at 13 weeks of gestation in humans and postnatally within 2 weeks in mice (Rumbo and Schiffrin, 2005; Menard et al., 2008; Heida et al., 2016). Accordingly, a murine study showed gradual upregulation of Paneth cell specific AMPs, including defensins and lysozyme, by small intestinal IECs in the first 4 weeks of life (Menard et al., 2008). Interestingly, the same study observed neonate-specific expression of cathelin-related antimicrobial peptide (CRAMP) in IECs that likely regulate antibacterial defense and commensal colonization in early life (Menard et al., 2008). In human fetal intestine, Paneth cells and AMPs such as defensin and lysozyme expression have been reported (Heida et al., 2016). Expression of other AMPs whose expression can be induced by microbiota is postulated to occur postnatally (Kai-Larsen et al., 2007). The unique makeup of AMPs in newborns may permit initial commensal establishment (Darnaud et al., 2018; Fulde et al., 2018; Liang et al., 2022) but may be insufficient for defense against early enteric pathogens (Figure 1).
Epithelial sensing of microbes
Intestinal epithelial cells of neonatal mice exhibit variable expression of the pattern recognition receptors (PRRs) that recognize conserved structures on beneficial commensals and harmful pathogens (Stadnyk, 2002). These PRRs activate signaling cascades in IECs that result in induction of AMPs, as well as epithelial cytokine production (Peterson and Artis, 2014). Immediately after birth, epithelial sensing of lipopolysaccharides (LPS) via Toll-like receptor (TLR)4 triggers establishment of LPS tolerance through dampening of TLR4 signaling through repression of interleukin 1 receptor associated kinase 1 (IRAK1) (Lotz et al., 2006). This LPS tolerance in the intestine is crucial for promoting microbiota colonization and inhibiting inflammatory responses as lack of endotoxin resistance leads to bacterial-induced IEC apoptosis and loss of barrier integrity (Lotz et al., 2006; Chassin et al., 2010). Additionally, TLR2 which senses bacterial lipoproteins is expressed in neonatal IECs. TLR2 and TLR4 overexpression was reported in a premature rat necrotizing enterocolitis model and positively correlated with disease severity (Le Mandat Schultz et al., 2007). Further, neonatal IECs exhibit >100-fold higher expression of TLR5, a PRR that recognizes bacterial flagellin, relative to IECs from adults (Fulde et al., 2018). The high expression of TLR5 in neonates may not contribute to protection against pathogens as neonatal TLR5 knockout mice did not exhibit increased susceptibility to Salmonella infection. Instead, TLR5 was essential in the colonization of symbiotic microbiota, partly through upregulation of the AMP Reg3γ (Fulde et al., 2018). In contrast, expression of TLR3, which detects viral dsRNA, was >20-fold lower in neonatal mice compared to adults. Interestingly, epithelial TLR3 expression inversely correlated to rotavirus infection (Pott et al., 2012), suggesting insufficient epithelial TLR3 expression in neonates may underlie their unique susceptibility to this pathogen.
In summary, dynamic epithelial regulation of barrier function, AMP expression, and microbial sensing in the intestine seem to each be critical factors underlying neonatal susceptibility to enteric infection (Figure 1).
Therapeutic approaches and future directions
Given the strong link between neonatal microbiota and enteric infection, microbiota-based therapies may be effective in reducing neonatal morbidity and mortality caused by enteric infection. Probiotics are live microbial supplements that have the potential to impact the host microbiota and suppress pathogenic outgrowth in the intestine (Gerritsen et al., 2011; Quigley, 2019). Probiotics may be used alone or in combination with prebiotics, which include dietary fibers that can promote expansion of beneficial microbial species (Quigley, 2019). Lactobacillus probiotics have shown therapeutic benefits in infants and young children with enteric infection, although these effects have not been observed universally (Szajewska and Mrukowicz, 2001; Van Niel et al., 2002; Szajewska et al., 2007; Freedman et al., 2018; Schnadower et al., 2018). A combination of Lactobacillus plantarum and a prebiotic fructooligosaccharide may provide defense against sepsis in early- and full-term infants based on a large-scale randomized placebo-controlled study (Panigrahi et al., 2017). However, effects of probiotics for sepsis in preterm infants have varied (Rao et al., 2016; Morgan et al., 2020). Although considered rare, probiotic sepsis associated with live strains, particularly in preterm infants with immature barrier functions, could present potential adverse effects (Chiang et al., 2021; Kulkarni et al., 2022). Thus, there is clear need for further investigation into neonatal host-microbiota interactions to develop efficacious and safe microbiota-based therapeutic approaches.
The neonatal period represents a unique window of opportunity for guiding improved microbiota-based strategies that could have a greater impact on infection prevention. Thus, further taxonomic, and functional characterization of neonatal microbiota in relation to maturity, diet, and age would aid development of such therapeutic and prophylactic approaches. Preventing the disruption of initial microbial colonization while strengthening neonatal intestinal immunity can impart protection against enteric pathogens. Additionally, current studies linking effects of breastfeeding and microbiota warrant continued exploration. Given the inefficiency of neonatal adaptive immunity, microbiota-targeted therapies, including during the prenatal period, may allow induction of epithelial defenses that can boost early innate defense. Further investigation into microbiota-based strategies to enhance basal epithelial antimicrobial and barrier responses may guide improved strategies for reducing neonatal morbidity and mortality to enteric infection.
Author contributions
SN, SH-H, and TA wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This review was supported by the National Institutes of Health (DK114123 and DK116868). TA holds a Kenneth Rainin Foundation Award and Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Acknowledgments
We thank members of the Alenghat lab for discussions and reading this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abba, K., Sinfield, R., Hart, C. A., and Garner, P. (2009). Pathogens associated with persistent diarrhoea in children in low and middle income countries: Systematic review. BMC Infect. Dis. 9:88. doi: 10.1186/1471-2334-9-88
Al Nabhani, Z., Dulauroy, S., Marques, R., Cousu, C., Al Bounny, S., Dejardin, F., et al. (2019). A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity 50, 1276–1288.e5. doi: 10.1016/j.immuni.2019.02.014
Allaire, J. M., Crowley, S. M., Law, H. T., Chang, S. Y., Ko, H. J., and Vallance, B. A. (2018). The intestinal epithelium: Central coordinator of mucosal immunity. Trends Immunol. 39, 677–696. doi: 10.1016/j.it.2018.04.002
An, D., Oh, S. F., Olszak, T., Neves, J. F., Avci, F. Y., Erturk-Hasdemir, D., et al. (2014). Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 156, 123–133. doi: 10.1016/j.cell.2013.11.042
Arboleya, S., Sanchez, B., Milani, C., Duranti, S., Solis, G., Fernandez, N., et al. (2015). Intestinal microbiota development in preterm neonates and effect of perinatal antibiotics. J. Pediatr. 166, 538–544. doi: 10.1016/j.jpeds.2014.09.041
Arboleya, S., Sanchez, B., Solis, G., Fernandez, N., Suarez, M., Hernandez-Barranco, A. M., et al. (2016). Impact of prematurity and perinatal antibiotics on the developing intestinal microbiota: A functional inference study. Int. J. Mol. Sci. 17:649. doi: 10.3390/ijms17050649
Ardissone, A. N., De La Cruz, D. M., Davis-Richardson, A. G., Rechcigl, K. T., Li, N., Drew, J. C., et al. (2014). Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS One 9:e90784. doi: 10.1371/journal.pone.0090784
Arevalo Sureda, E., Westrom, B., Pierzynowski, S. G., and Prykhodko, O. (2016). Maturation of the intestinal epithelial barrier in neonatal rats coincides with decreased FcRn expression, replacement of vacuolated enterocytes and changed Blimp-1 expression. PLoS One 11:e0164775. doi: 10.1371/journal.pone.0164775
Asnicar, F., Manara, S., Zolfo, M., Truong, D. T., Scholz, M., Armanini, F., et al. (2017). Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems 2:e00164-16. doi: 10.1128/mSystems.00164-16
Ayabe, T., Satchell, D. P., Wilson, C. L., Parks, W. C., Selsted, M. E., and Ouellette, A. J. (2000). Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 1, 113–118. doi: 10.1038/77783
Baba, R., Fujita, M., Tein, C. E., and Miyoshi, M. (2002). Endocytosis by absorptive cells in the middle segment of the suckling rat small intestine. Anat. Sci. Int. 77, 117–123. doi: 10.1046/j.0022-7722.2002.00017.x
Backhed, F., Roswall, J., Peng, Y., Feng, Q., Jia, H., Kovatcheva-Datchary, P., et al. (2015). dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703. doi: 10.1016/j.chom.2015.04.004
Bagamian, K. H., Anderson, J. D. T., Muhib, F., Cumming, O., Laytner, L. A., Wierzba, T. F., et al. (2020). Heterogeneity in enterotoxigenic Escherichia coli and shigella infections in children under 5 years of age from 11 African countries: A subnational approach quantifying risk, mortality, morbidity, and stunting. Lancet Glob. Health 8, e101–e112. doi: 10.1016/S2214-109X(19)30456-5
Baird, A. W., and O’malley, K. E. (1993). Epithelial ion transport - possible contribution to parasite expulsion. Parasitol. Today 9, 141–143. doi: 10.1016/0169-4758(93)90180-N
Barker, N., Van Es, J. H., Kuipers, J., Kujala, P., Van Den Born, M., Cozijnsen, M., et al. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. doi: 10.1038/nature06196
Bel, S., Pendse, M., Wang, Y., Li, Y., Ruhn, K. A., Hassell, B., et al. (2017). Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 357, 1047–1052. doi: 10.1126/science.aal4677
Ben Suleiman, Y., Yoshida, M., Nishiumi, S., Tanaka, H., Mimura, T., Nobutani, K., et al. (2012). Neonatal Fc receptor for IgG (FcRn) expressed in the gastric epithelium regulates bacterial infection in mice. Mucosal Immunol. 5, 87–98. doi: 10.1038/mi.2011.53
Bergin, S. P., Thaden, J. T., Ericson, J. E., Cross, H., Messina, J., Clark, R. H., et al. (2015). Neonatal Escherichia coli bloodstream infections: Clinical outcomes and impact of initial antibiotic therapy. Pediatr. Infect. Dis. J. 34, 933–936. doi: 10.1097/INF.0000000000000769
Buisine, M. P., Devisme, L., Savidge, T. C., Gespach, C., Gosselin, B., Porchet, N., et al. (1998). Mucin gene expression in human embryonic and fetal intestine. Gut 43, 519–524. doi: 10.1136/gut.43.4.519
Bula-Rudas, F. J., Rathore, M. H., and Maraqa, N. F. (2015). Salmonella infections in childhood. Adv. Pediatr. 62, 29–58. doi: 10.1016/j.yapd.2015.04.005
Bushman, F., and Liang, G. (2021). Assembly of the virome in newborn human infants. Curr. Opin. Virol. 48, 17–22. doi: 10.1016/j.coviro.2021.03.004
Cahenzli, J., Koller, Y., Wyss, M., Geuking, M. B., and Mccoy, K. D. (2013). Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe 14, 559–570. doi: 10.1016/j.chom.2013.10.004
Cepeda-Molero, M., Berger, C. N., Walsham, A. D. S., Ellis, S. J., Wemyss-Holden, S., Schuller, S., et al. (2017). Attaching and effacing (A/E) lesion formation by enteropathogenic E. coli on human intestinal mucosa is dependent on non-LEE effectors. PLoS Pathog. 13:e1006706. doi: 10.1371/journal.ppat.1006706
Chaaban, H., Patel, M. M., Burge, K., Eckert, J. V., Lupu, C., Keshari, R. S., et al. (2022). Early antibiotic exposure alters intestinal development and increases susceptibility to necrotizing enterocolitis: A mechanistic study. Microorganisms 10:519. doi: 10.3390/microorganisms10030519
Chassin, C., Kocur, M., Pott, J., Duerr, C. U., Gutle, D., Lotz, M., et al. (2010). miR-146a mediates protective innate immune tolerance in the neonate intestine. Cell Host Microbe 8, 358–368. doi: 10.1016/j.chom.2010.09.005
Chiang, M. C., Chen, C. L., Feng, Y., Chen, C. C., Lien, R., and Chiu, C. H. (2021). Lactobacillus rhamnosus sepsis associated with probiotic therapy in an extremely preterm infant: Pathogenesis and a review for clinicians. J. Microbiol. Immunol. Infect. 54, 575–580. doi: 10.1016/j.jmii.2020.03.029
Chleilat, F., Klancic, T., Ma, K., Schick, A., Nettleton, J. E., and Reimer, R. A. (2020). Human milk oligosaccharide supplementation affects intestinal barrier function and microbial composition in the gastrointestinal tract of young sprague dawley rats. Nutrients 12:1532. doi: 10.3390/nu12051532
Chu, D. M., Antony, K. M., Ma, J., Prince, A. L., Showalter, L., Moller, M., et al. (2016). The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 8:77. doi: 10.1186/s13073-016-0330-z
Chu, D. M., Ma, J., Prince, A. L., Antony, K. M., Seferovic, M. D., and Aagaard, K. M. (2017). Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 23, 314–326. doi: 10.1038/nm.4272
Cleary, J., Lai, L. C., Shaw, R. K., Straatman-Iwanowska, A., Donnenberg, M. S., Frankel, G., et al. (2004). Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: Role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiology 150, 527–538. doi: 10.1099/mic.0.26740-0
Clevers, H. C., and Bevins, C. L. (2013). Paneth cells: Maestros of the small intestinal crypts. Annu. Rev. Physiol. 75, 289–311. doi: 10.1146/annurev-physiol-030212-183744
Collado, M. C., Cernada, M., Neu, J., Perez-Martinez, G., Gormaz, M., and Vento, M. (2015). Factors influencing gastrointestinal tract and microbiota immune interaction in preterm infants. Pediatr. Res. 77, 726–731. doi: 10.1038/pr.2015.54
Darmstadt, G. L., Kinney, M. V., Chopra, M., Cousens, S., Kak, L., Paul, V. K., et al. (2014). Who has been caring for the baby? Lancet 384, 174–188. doi: 10.1016/S0140-6736(14)60458-X
Darnaud, M., Dos Santos, A., Gonzalez, P., Augui, S., Lacoste, C., Desterke, C., et al. (2018). Enteric delivery of regenerating family member 3 alpha alters the intestinal microbiota and controls inflammation in mice with colitis. Gastroenterology 154, 1009–1023.e14. doi: 10.1053/j.gastro.2017.11.003
Del Chierico, F., Vernocchi, P., Petrucca, A., Paci, P., Fuentes, S., Pratico, G., et al. (2015). Phylogenetic and metabolic tracking of gut microbiota during perinatal development. PLoS One 10:e0137347. doi: 10.1371/journal.pone.0137347
Dominguez-Bello, M. G., Costello, E. K., Contreras, M., Magris, M., Hidalgo, G., Fierer, N., et al. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U.S.A. 107, 11971–11975. doi: 10.1073/pnas.1002601107
Donnet-Hughes, A., Perez, P. F., Dore, J., Leclerc, M., Levenez, F., Benyacoub, J., et al. (2010). Potential role of the intestinal microbiota of the mother in neonatal immune education. Proc. Nutr. Soc. 69, 407–415. doi: 10.1017/S0029665110001898
Dupont, A., Sommer, F., Zhang, K., Repnik, U., Basic, M., Bleich, A., et al. (2016). Age-dependent susceptibility to enteropathogenic Escherichia coli (EPEC) infection in mice. PLoS Pathog. 12:e1005616. doi: 10.1371/journal.ppat.1005616
El Aidy, S., Hooiveld, G., Tremaroli, V., Backhed, F., and Kleerebezem, M. (2013). The gut microbiota and mucosal homeostasis: Colonized at birth or at adulthood, does it matter? Gut Microbes 4, 118–124. doi: 10.4161/gmic.23362
Eshleman, E. M., and Alenghat, T. (2021). Epithelial sensing of microbiota-derived signals. Genes Immun. 22, 237–246. doi: 10.1038/s41435-021-00124-w
Fan, Y., and Pedersen, O. (2021). Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 19, 55–71. doi: 10.1038/s41579-020-0433-9
Ferretti, P., Pasolli, E., Tett, A., Asnicar, F., Gorfer, V., Fedi, S., et al. (2018). Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24, 133–145.e5. doi: 10.1016/j.chom.2018.06.005
Freedman, S. B., Williamson-Urquhart, S., Farion, K. J., Gouin, S., Willan, A. R., Poonai, N., et al. (2018). Multicenter trial of a combination probiotic for children with gastroenteritis. N. Engl. J. Med. 379, 2015–2026. doi: 10.1056/NEJMoa1802597
Fukuda, S., Toh, H., Hase, K., Oshima, K., Nakanishi, Y., Yoshimura, K., et al. (2011). Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547. doi: 10.1038/nature09646
Fulde, M., Sommer, F., Chassaing, B., Van Vorst, K., Dupont, A., Hensel, M., et al. (2018). Neonatal selection by Toll-like receptor 5 influences long-term gut microbiota composition. Nature 560, 489–493. doi: 10.1038/s41586-018-0395-5
Galan, J. E., Lara-Tejero, M., Marlovits, T. C., and Wagner, S. (2014). Bacterial type III secretion systems: Specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 68, 415–438. doi: 10.1146/annurev-micro-092412-155725
Galazzo, G., Van Best, N., Bervoets, L., Dapaah, I. O., Savelkoul, P. H., Hornef, M. W., et al. (2020). Development of the microbiota and associations with birth mode, diet, and atopic disorders in a longitudinal analysis of stool samples, collected from infancy through early childhood. Gastroenterology 158, 1584–1596. doi: 10.1053/j.gastro.2020.01.024
Garcia, T. M., Van Roest, M., Vermeulen, J. L. M., Meisner, S., Smit, W. L., Silva, J., et al. (2021). Early life antibiotics influence in vivo and in vitro mouse intestinal epithelium maturation and functioning. Cell Mol. Gastroenterol. Hepatol. 12, 943–981. doi: 10.1016/j.jcmgh.2021.05.019
Garcia-Mantrana, I., Selma-Royo, M., Gonzalez, S., Parra-Llorca, A., Martinez-Costa, C., and Collado, M. C. (2020). Distinct maternal microbiota clusters are associated with diet during pregnancy: Impact on neonatal microbiota and infant growth during the first 18 months of life. Gut Microbes 11, 962–978. doi: 10.1080/19490976.2020.1730294
Gensollen, T., Iyer, S. S., Kasper, D. L., and Blumberg, R. S. (2016). How colonization by microbiota in early life shapes the immune system. Science 352, 539–544. doi: 10.1126/science.aad9378
Gerritsen, J., Smidt, H., Rijkers, G. T., and De Vos, W. M. (2011). Intestinal microbiota in human health and disease: The impact of probiotics. Genes Nutr. 6, 209–240. doi: 10.1007/s12263-011-0229-7
Gorden, J., and Small, P. L. (1993). Acid resistance in enteric bacteria. Infect. Immun. 61, 364–367. doi: 10.1128/iai.61.1.364-367.1993
Greenwood, C., Morrow, A. L., Lagomarcino, A. J., Altaye, M., Taft, D. H., Yu, Z., et al. (2014). Early empiric antibiotic use in preterm infants is associated with lower bacterial diversity and higher relative abundance of Enterobacter. J. Pediatr. 165, 23–29. doi: 10.1016/j.jpeds.2014.01.010
Hansen, R., Scott, K. P., Khan, S., Martin, J. C., Berry, S. H., Stevenson, M., et al. (2015). First-pass meconium samples from healthy term vaginally-delivered neonates: An analysis of the microbiota. PLoS One 10:e0133320. doi: 10.1371/journal.pone.0133320
Harper, J., Mould, A., Andrews, R. M., Bikoff, E. K., and Robertson, E. J. (2011). The transcriptional repressor Blimp1/Prdm1 regulates postnatal reprogramming of intestinal enterocytes. Proc. Natl. Acad. Sci. U.S.A. 108, 10585–10590. doi: 10.1073/pnas.1105852108
Heida, F. H., Beyduz, G., Bulthuis, M. L., Kooi, E. M., Bos, A. F., Timmer, A., et al. (2016). Paneth cells in the developing gut: When do they arise and when are they immune competent? Pediatr. Res. 80, 306–310. doi: 10.1038/pr.2016.67
Henrick, B. M., Rodriguez, L., Lakshmikanth, T., Pou, C., Henckel, E., Arzoomand, A., et al. (2021). Bifidobacteria-mediated immune system imprinting early in life. Cell 184, 3884–3898.e11. doi: 10.1016/j.cell.2021.05.030
Hesla, H. M., Stenius, F., Jaderlund, L., Nelson, R., Engstrand, L., Alm, J., et al. (2014). Impact of lifestyle on the gut microbiota of healthy infants and their mothers-the ALADDIN birth cohort. FEMS Microbiol. Ecol. 90, 791–801. doi: 10.1111/1574-6941.12434
Hirotani, Y., Ikeda, K., Kato, R., Myotoku, M., Umeda, T., Ijiri, Y., et al. (2008). Protective effects of lactoferrin against intestinal mucosal damage induced by lipopolysaccharide in human intestinal Caco-2 cells. Yakugaku Zasshi 128, 1363–1368. doi: 10.1248/yakushi.128.1363
Hu, J., and Torres, A. G. (2015). Enteropathogenic Escherichia coli: Foe or innocent bystander? Clin. Microbiol. Infect. 21, 729–734. doi: 10.1016/j.cmi.2015.01.015
Huurre, A., Kalliomaki, M., Rautava, S., Rinne, M., Salminen, S., and Isolauri, E. (2008). Mode of delivery - effects on gut microbiota and humoral immunity. Neonatology 93, 236–240. doi: 10.1159/000111102
Iizumi, Y., Sagara, H., Kabe, Y., Azuma, M., Kume, K., Ogawa, M., et al. (2007). The enteropathogenic E. coli effector EspB facilitates microvillus effacing and antiphagocytosis by inhibiting myosin function. Cell Host Microbe 2, 383–392. doi: 10.1016/j.chom.2007.09.012
Isaac, B. M., Masonbrink, A., Kennedy, J., Greene, S. K., Hennessy, R. R., Rosen, J. B., et al. (2016). Reportable bacterial infections among New York City-Born Infants, 2001-2009. J. Pediatr. 174, 218–225e4. doi: 10.1016/j.jpeds.2016.03.033
Jakobsson, H. E., Abrahamsson, T. R., Jenmalm, M. C., Harris, K., Quince, C., Jernberg, C., et al. (2014). Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 63, 559–566. doi: 10.1136/gutjnl-2012-303249
Jian, C., Carpen, N., Helve, O., De Vos, W. M., Korpela, K., and Salonen, A. (2021). Early-life gut microbiota and its connection to metabolic health in children: Perspective on ecological drivers and need for quantitative approach. Ebiomedicine 69:103475. doi: 10.1016/j.ebiom.2021.103475
Kaczmarczyk, M., Lober, U., Adamek, K., Wegrzyn, D., Skonieczna-Zydecka, K., Malinowski, D., et al. (2021). The gut microbiota is associated with the small intestinal paracellular permeability and the development of the immune system in healthy children during the first two years of life. J. Transl. Med. 19:177. doi: 10.1186/s12967-021-02839-w
Kai-Larsen, Y., Bergsson, G., Gudmundsson, G. H., Printz, G., Jornvall, H., Marchini, G., et al. (2007). Antimicrobial components of the neonatal gut affected upon colonization. Pediatr. Res. 61, 530–536. doi: 10.1203/pdr.0b013e318045be83
Kim, Y. G., Sakamoto, K., Seo, S. U., Pickard, J. M., Gillilland, M. G. III, Pudlo, N. A., et al. (2017). Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science 356, 315–319. doi: 10.1126/science.aag2029
Kimura, I., Miyamoto, J., Ohue-Kitano, R., Watanabe, K., Yamada, T., Onuki, M., et al. (2020). Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice. Science 367:eaaw8429. doi: 10.1126/science.aaw8429
Knoop, K. A., Coughlin, P. E., Floyd, A. N., Ndao, I. M., Hall-Moore, C., Shaikh, N., et al. (2020). Maternal activation of the EGFR prevents translocation of gut-residing pathogenic Escherichia coli in a model of late-onset neonatal sepsis. Proc. Natl. Acad. Sci. U.S.A. 117, 7941–7949. doi: 10.1073/pnas.1912022117
Kollmann, T. R., Levy, O., Montgomery, R. R., and Goriely, S. (2012). Innate immune function by Toll-like receptors: Distinct responses in newborns and the elderly. Immunity 37, 771–783. doi: 10.1016/j.immuni.2012.10.014
Kotloff, K. L., Nataro, J. P., Blackwelder, W. C., Nasrin, D., Farag, T. H., Panchalingam, S., et al. (2013). Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 382, 209–222. doi: 10.1016/S0140-6736(13)60844-2
Kulkarni, T., Majarikar, S., Deshmukh, M., Ananthan, A., Balasubramanian, H., Keil, A., et al. (2022). Probiotic sepsis in preterm neonates-a systematic review. Eur. J. Pediatr. 181, 2249–2262. doi: 10.1007/s00431-022-04452-5
Lanata, C. F., Fischer-Walker, C. L., Olascoaga, A. C., Torres, C. X., Aryee, M. J., and Black, R. E. (2013). Global causes of diarrheal disease mortality in children <5 years of age: A systematic review. PLoS One 8:e72788. doi: 10.1371/journal.pone.0072788
Le Mandat Schultz, A., Bonnard, A., Barreau, F., Aigrain, Y., Pierre-Louis, C., Berrebi, D., et al. (2007). Expression of TLR-2, TLR-4, NOD2 and pNF-kappaB in a neonatal rat model of necrotizing enterocolitis. PLoS One 2:e1102. doi: 10.1371/journal.pone.0001102
Lee, A. H., Shannon, C. P., Amenyogbe, N., Bennike, T. B., Diray-Arce, J., Idoko, O. T., et al. (2019). Dynamic molecular changes during the first week of human life follow a robust developmental trajectory. Nat. Commun. 10:1092. doi: 10.1038/s41467-019-08794-x
Lencer, W. I., Cheung, G., Strohmeier, G. R., Currie, M. G., Ouellette, A. J., Selsted, M. E., et al. (1997). Induction of epithelial chloride secretion by channel-forming cryptdins 2 and 3. Proc. Natl. Acad. Sci. U.S.A. 94, 8585–8589. doi: 10.1073/pnas.94.16.8585
Liang, G., Zhao, C., Zhang, H., Mattei, L., Sherrill-Mix, S., Bittinger, K., et al. (2020). The stepwise assembly of the neonatal virome is modulated by breastfeeding. Nature 581, 470–474. doi: 10.1038/s41586-020-2192-1
Liang, W., Enee, E., Andre-Vallee, C., Falcone, M., Sun, J., and Diana, J. (2022). Intestinal cathelicidin antimicrobial peptide shapes a protective neonatal gut microbiota against pancreatic autoimmunity. Gastroenterology 162, 1288–1302.e16. doi: 10.1053/j.gastro.2021.12.272
Liu, L., Johnson, H. L., Cousens, S., Perin, J., Scott, S., Lawn, J. E., et al. (2012). Global, regional, and national causes of child mortality: An updated systematic analysis for 2010 with time trends since 2000. Lancet 379, 2151–2161. doi: 10.1016/S0140-6736(12)60560-1
Lotz, M., Gutle, D., Walther, S., Menard, S., Bogdan, C., and Hornef, M. W. (2006). Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J. Exp. Med. 203, 973–984. doi: 10.1084/jem.20050625
Ma, B., Mccomb, E., Gajer, P., Yang, H., Humphrys, M., Okogbule-Wonodi, A. C., et al. (2018). Microbial biomarkers of intestinal barrier maturation in preterm infants. Front. Microbiol. 9:2755. doi: 10.3389/fmicb.2018.02755
Madan, J. C., Salari, R. C., Saxena, D., Davidson, L., O’toole, G. A., Moore, J. H., et al. (2012). Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch. Dis. Child. Fetal Neonatal Ed. 97, F456–F462. doi: 10.1136/fetalneonatal-2011-301373
Maher, S. E., O’brien, E. C., Moore, R. L., Byrne, D. F., Geraghty, A. A., Saldova, R., et al. (2020). The association between the maternal diet and the maternal and infant gut microbiome: A systematic review. Br. J. Nutr. 1–29. Online ahead of print. doi: 10.1017/S0007114520000847
Mai, V., Torrazza, R. M., Ukhanova, M., Wang, X., Sun, Y., Li, N., et al. (2013). Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS One 8, e52876. doi: 10.1371/journal.pone.0052876
Mallow, E. B., Harris, A., Salzman, N., Russell, J. P., Deberardinis, R. J., Ruchelli, E., et al. (1996). Human enteric defensins. gene structure and developmental expression. J. Biol. Chem. 271, 4038–4045. doi: 10.1074/jbc.271.8.4038
Man, W. H., Clerc, M., De Steenhuijsen Piters, W. A. A., Van Houten, M. A., Chu, M., Kool, J., et al. (2019). Loss of microbial topography between oral and nasopharyngeal microbiota and development of respiratory infections early in life. Am. J. Respir. Crit. Care Med. 200, 760–770. doi: 10.1164/rccm.201810-1993OC
Matamoros, S., Gras-Leguen, C., Le Vacon, F., Potel, G., and De La Cochetiere, M. F. (2013). Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 21, 167–173. doi: 10.1016/j.tim.2012.12.001
Menard, S., Cerf-Bensussan, N., and Heyman, M. (2010). Multiple facets of intestinal permeability and epithelial handling of dietary antigens. Mucosal Immunol 3, 247–259. doi: 10.1038/mi.2010.5
Menard, S., Forster, V., Lotz, M., Gutle, D., Duerr, C. U., Gallo, R. L., et al. (2008). Developmental switch of intestinal antimicrobial peptide expression. J. Exp. Med. 205, 183–193. doi: 10.1084/jem.20071022
Miller, J. E., Hammond, G. C., Strunk, T., Moore, H. C., Leonard, H., Carter, K. W., et al. (2016). Association of gestational age and growth measures at birth with infection-related admissions to hospital throughout childhood: A population-based, data-linkage study from Western Australia. Lancet Infect. Dis. 16, 952–961. doi: 10.1016/S1473-3099(16)00150-X
Miller, J. E., Wu, C., Pedersen, L. H., De Klerk, N., Olsen, J., and Burgner, D. P. (2018). Maternal antibiotic exposure during pregnancy and hospitalization with infection in offspring: A population-based cohort study. Int. J. Epidemiol. 47, 561–571. doi: 10.1093/ije/dyx272
Morgan, R. L., Preidis, G. A., Kashyap, P. C., Weizman, A. V., Sadeghirad, B., Mcmaster Probiotic, P., et al. (2020). Probiotics reduce mortality and morbidity in preterm, low-birth-weight infants: A systematic review and network meta-analysis of randomized trials. Gastroenterology 159, 467–480. doi: 10.1053/j.gastro.2020.05.096
Mukherjee, S., and Hooper, L. V. (2015). Antimicrobial defense of the intestine. Immunity 42, 28–39. doi: 10.1016/j.immuni.2014.12.028
Muncan, V., Heijmans, J., Krasinski, S. D., Buller, N. V., Wildenberg, M. E., Meisner, S., et al. (2011). Blimp1 regulates the transition of neonatal to adult intestinal epithelium. Nat. Commun. 2:452. doi: 10.1038/ncomms1463
Nataro, J. P., and Kaper, J. B. (1998). Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11, 142–201. doi: 10.1128/CMR.11.1.142
Negi, S., Das, D. K., Pahari, S., Nadeem, S., and Agrewala, J. N. (2019). Potential role of gut microbiota in induction and regulation of innate immune memory. Front. Immunol. 10:2441. doi: 10.3389/fimmu.2019.02441
Noel, G., In, J. G., Lemme-Dumit, J. M., Devine, L. R., Cole, R. N., Guerrerio, A. L., et al. (2021). Human breast milk enhances intestinal mucosal barrier function and innate immunity in a healthy pediatric human enteroid model. Front. Cell Dev. Biol. 9:685171. doi: 10.3389/fcell.2021.685171
Ohlsson, A., and Shah, V. S. (2014). Intrapartum antibiotics for known maternal group B streptococcal colonization. Cochrane Database Syst. Rev. 6:CD007467. doi: 10.1002/14651858.CD007467.pub4
Olszak, T., An, D., Zeissig, S., Vera, M. P., Richter, J., Franke, A., et al. (2012). Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493. doi: 10.1126/science.1219328
Ouellette, A. J. (2010). Paneth cells and innate mucosal immunity. Curr. Opin. Gastroenterol. 26, 547–553. doi: 10.1097/MOG.0b013e32833dccde
Panigrahi, P., Parida, S., Nanda, N. C., Satpathy, R., Pradhan, L., Chandel, D. S., et al. (2017). A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 548, 407–412. doi: 10.1038/nature23480
Pedersen, S. H., Wilkinson, A. L., Andreasen, A., Warhurst, D. C., Kinung’hi, S. M., Urassa, M., et al. (2014). Cryptosporidium prevalence and risk factors among mothers and infants 0 to 6 months in rural and semi-rural Northwest Tanzania: A prospective cohort study. PLoS Negl. Trop. Dis. 8:e3072. doi: 10.1371/journal.pntd.0003072
Pessa-Morikawa, T., Husso, A., Karkkainen, O., Koistinen, V., Hanhineva, K., Iivanainen, A., et al. (2022). Maternal microbiota-derived metabolic profile in fetal murine intestine, brain and placenta. BMC Microbiol. 22:46. doi: 10.1186/s12866-022-02457-6
Peterson, L. W., and Artis, D. (2014). Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14, 141–153. doi: 10.1038/nri3608
Polak-Charcon, S., Shoham, J., and Ben-Shaul, Y. (1980). Tight junctions in epithelial cells of human fetal hindgut, normal colon, and colon adenocarcinoma. J. Natl. Cancer Inst. 65, 53–62.
Pott, J., Stockinger, S., Torow, N., Smoczek, A., Lindner, C., Mcinerney, G., et al. (2012). Age-dependent TLR3 expression of the intestinal epithelium contributes to rotavirus susceptibility. PLoS Pathog. 8:e1002670. doi: 10.1371/journal.ppat.1002670
Quigley, E. M. M. (2019). Prebiotics and probiotics in digestive health. Clin. Gastroenterol. Hepatol. 17, 333–344. doi: 10.1016/j.cgh.2018.09.028
Raabe, V. N., and Shane, A. L. (2019). Group B Streptococcus (Streptococcus agalactiae). Microbiol. Spectr. 7, 1–13. doi: 10.1128/microbiolspec.GPP3-0007-2018
Radjabzadeh, D., Boer, C. G., Beth, S. A., Van Der Wal, P., Kiefte-De Jong, J. C., Jansen, M. A. E., et al. (2020). Diversity, compositional and functional differences between gut microbiota of children and adults. Sci. Rep. 10:1040. doi: 10.1038/s41598-020-57734-z
Rao, S. C., Athalye-Jape, G. K., Deshpande, G. C., Simmer, K. N., and Patole, S. K. (2016). Probiotic supplementation and late-onset sepsis in preterm infants: A meta-analysis. Pediatrics 137:e20153684. doi: 10.1542/peds.2015-3684
Rautava, S., Luoto, R., Salminen, S., and Isolauri, E. (2012). Microbial contact during pregnancy, intestinal colonization and human disease. Nat. Rev. Gastroenterol. Hepatol. 9, 565–576. doi: 10.1038/nrgastro.2012.144
Rautava, S., Nanthakumar, N. N., Dubert-Ferrandon, A., Lu, L., Rautava, J., and Walker, W. A. (2011). Breast milk-transforming growth factor-beta(2) specifically attenuates IL-1beta-induced inflammatory responses in the immature human intestine via an SMAD6- and ERK-dependent mechanism. Neonatology 99, 192–201. doi: 10.1159/000314109
Reju, S., Srikanth, P., Selvarajan, S., Thomas, R. K., Barani, R., Amboiram, P., et al. (2022). A shift in circulating rotaviral genotypes among hospitalized neonates. Sci. Rep. 12:2842. doi: 10.1038/s41598-022-06506-y
Renz, H., Adkins, B. D., Bartfeld, S., Blumberg, R. S., Farber, D. L., Garssen, J., et al. (2018). The neonatal window of opportunity-early priming for life. J. Allergy Clin. Immunol. 141, 1212–1214. doi: 10.1016/j.jaci.2017.11.019
Reyman, M., Van Houten, M. A., Van Baarle, D., Bosch, A., Man, W. H., Chu, M., et al. (2019). Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat. Commun. 10:4997. doi: 10.1038/s41467-019-13373-1
Robertson, R. C., Manges, A. R., Finlay, B. B., and Prendergast, A. J. (2019). The human microbiome and child growth - first 1000 days and beyond. Trends Microbiol. 27, 131–147. doi: 10.1016/j.tim.2018.09.008
Rogawski, E. T., Westreich, D., Becker-Dreps, S., Adair, L. S., Sandler, R. S., Sarkar, R., et al. (2015). The effect of early life antibiotic exposures on diarrheal rates among young children in Vellore, India. Pediatr. Infect. Dis. J. 34, 583–588. doi: 10.1097/INF.0000000000000679
Rogier, E. W., Frantz, A. L., Bruno, M. E., Wedlund, L., Cohen, D. A., Stromberg, A. J., et al. (2014). Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc. Natl. Acad. Sci. U.S.A. 111, 3074–3079. doi: 10.1073/pnas.1315792111
Ronaghan, N. J., Shang, J., Iablokov, V., Zaheer, R., Colarusso, P., Dion, S., et al. (2016). The serine protease-mediated increase in intestinal epithelial barrier function is dependent on occludin and requires an intact tight junction. Am. J. Physiol. Gastrointest. Liver Physiol. 311, G466–G479. doi: 10.1152/ajpgi.00441.2015
Roubaud-Baudron, C., Ruiz, V. E., Swan, A. M. Jr., Vallance, B. A., Ozkul, C., Pei, Z., et al. (2019). Long-term effects of early-life antibiotic exposure on resistance to subsequent bacterial infection. mBio 10:e02820-19. doi: 10.1128/mBio.02820-19
Roze, J. C., Ancel, P. Y., Marchand-Martin, L., Rousseau, C., Montassier, E., Monot, C., et al. (2020). Assessment of neonatal intensive care unit practices and preterm newborn gut microbiota and 2-year neurodevelopmental outcomes. JAMA Netw. Open 3:e2018119. doi: 10.1001/jamanetworkopen.2020.18119
Rudd, B. D. (2020). Neonatal T cells: A reinterpretation. Annu. Rev. Immunol. 38, 229–247. doi: 10.1146/annurev-immunol-091319-083608
Rumbo, M., and Schiffrin, E. J. (2005). Ontogeny of intestinal epithelium immune functions: Developmental and environmental regulation. Cell Mol. Life Sci. 62, 1288–1296. doi: 10.1007/s00018-005-5033-3
Saleem, B., Okogbule-Wonodi, A. C., Fasano, A., Magder, L. S., Ravel, J., Kapoor, S., et al. (2017). Intestinal barrier maturation in very low birthweight infants: Relationship to feeding and antibiotic exposure. J Pediatr 183, 31–36e1. doi: 10.1016/j.jpeds.2017.01.013
Schnadower, D., Tarr, P. I., Casper, T. C., Gorelick, M. H., Dean, J. M., O’connell, K. J., et al. (2018). Lactobacillus rhamnosus GG versus placebo for acute gastroenteritis in children. N. Engl. J. Med. 379, 2002–2014. doi: 10.1056/NEJMoa1802598
Schulman, J., Dimand, R. J., Lee, H. C., Duenas, G. V., Bennett, M. V., and Gould, J. B. (2015). Neonatal intensive care unit antibiotic use. Pediatrics 135, 826–833. doi: 10.1542/peds.2014-3409
Schumann, A., Nutten, S., Donnicola, D., Comelli, E. M., Mansourian, R., Cherbut, C., et al. (2005). Neonatal antibiotic treatment alters gastrointestinal tract developmental gene expression and intestinal barrier transcriptome. Physiol. Genomics 23, 235–245. doi: 10.1152/physiolgenomics.00057.2005
Shane, A. L., Sanchez, P. J., and Stoll, B. J. (2017). Neonatal sepsis. Lancet 390, 1770–1780. doi: 10.1016/S0140-6736(17)31002-4
Shao, Y., Forster, S. C., Tsaliki, E., Vervier, K., Strang, A., Simpson, N., et al. (2019). Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 574, 117–121. doi: 10.1038/s41586-019-1560-1
Sharrow, D., Hug, L., You, D., Alkema, L., Black, R., Cousens, S., et al. (2022). Global, regional, and national trends in under-5 mortality between 1990 and 2019 with scenario-based projections until 2030: A systematic analysis by the UN Inter-agency Group for Child Mortality Estimation. Lancet Glob. Health 10, e195–e206. doi: 10.1016/S2214-109X(21)00515-5
Singer, J. R., Blosser, E. G., Zindl, C. L., Silberger, D. J., Conlan, S., Laufer, V. A., et al. (2019). Preventing dysbiosis of the neonatal mouse intestinal microbiome protects against late-onset sepsis. Nat. Med. 25, 1772–1782. doi: 10.1038/s41591-019-0640-y
Sitarik, A. R., Bobbitt, K. R., Havstad, S. L., Fujimura, K. E., Levin, A. M., Zoratti, E. M., et al. (2017). Breast milk transforming growth factor beta is associated with neonatal gut microbial composition. J. Pediatr. Gastroenterol. Nutr. 65, e60–e67. doi: 10.1097/MPG.0000000000001585
Sochaczewska, D., Zietek, M., Dolegowska, B., Kordek, A., and Szczuko, M. (2022). Implications of indirect biomarkers of intestinal permeability in the stools of newborns and infants with perinatal risk factors for intestinal colonization disorders and infant feeding patterns. Nutrients 14:2224. doi: 10.3390/nu14112224
Soto, A., Martin, V., Jimenez, E., Mader, I., Rodriguez, J. M., and Fernandez, L. (2014). Lactobacilli and bifidobacteria in human breast milk: Influence of antibiotherapy and other host and clinical factors. J. Pediatr. Gastroenterol. Nutr. 59, 78–88. doi: 10.1097/MPG.0000000000000347
Stadnyk, A. W. (2002). Intestinal epithelial cells as a source of inflammatory cytokines and chemokines. Can. J. Gastroenterol. 16, 241–246. doi: 10.1155/2002/941087
Stewart, C. J., Ajami, N. J., O’brien, J. L., Hutchinson, D. S., Smith, D. P., Wong, M. C., et al. (2018). Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562, 583–588. doi: 10.1038/s41586-018-0617-x
Stinson, L. F., Payne, M. S., and Keelan, J. A. (2018). A critical review of the bacterial baptism hypothesis and the impact of cesarean delivery on the infant microbiome. Front. Med. 5:135. doi: 10.3389/fmed.2018.00135
Stoll, B. J., Hansen, N. I., Sanchez, P. J., Faix, R. G., Poindexter, B. B., Van Meurs, K. P., et al. (2011). Early onset neonatal sepsis: The burden of group B Streptococcal and E. coli disease continues. Pediatrics 127, 817–826.
Sturgeon, C., and Fasano, A. (2016). Zonulin, a regulator of epithelial and endothelial barrier functions, and its involvement in chronic inflammatory diseases. Tissue Barriers 4:e1251384. doi: 10.1080/21688370.2016.1251384
Suzuki, H., Nishizawa, T., Tani, K., Yamazaki, Y., Tamura, A., Ishitani, R., et al. (2014). Crystal structure of a claudin provides insight into the architecture of tight junctions. Science 344, 304–307. doi: 10.1126/science.1248571
Szajewska, H., and Mrukowicz, J. Z. (2001). Probiotics in the treatment and prevention of acute infectious diarrhea in infants and children: A systematic review of published randomized, double-blind, placebo-controlled trials. J. Pediatr. Gastroenterol. Nutr. 33(Suppl. 2), S17–S25. doi: 10.1097/00005176-200110002-00004
Szajewska, H., Skorka, A., Ruszczynski, M., and Gieruszczak-Bialek, D. (2007). Meta-analysis: Lactobacillus GG for treating acute diarrhoea in children. Aliment. Pharmacol. Ther. 25, 871–881. doi: 10.1111/j.1365-2036.2007.03282.x
Torow, N., and Hornef, M. W. (2017). The neonatal window of opportunity: Setting the stage for life-long host-microbial interaction and immune homeostasis. J. Immunol. 198, 557–563. doi: 10.4049/jimmunol.1601253
Travier, L., Alonso, M., Andronico, A., Hafner, L., Disson, O., Lledo, P. M., et al. (2021). Neonatal susceptibility to meningitis results from the immaturity of epithelial barriers and gut microbiota. Cell Rep. 35:109319. doi: 10.1016/j.celrep.2021.109319
Troeger, C., Blacker, B. F., Khalil, I. A., Rao, P. C., Cao, S., Zimsen, S. R., et al. (2018). Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 18, 1211–1228. doi: 10.1016/S1473-3099(18)30362-1
Turfkruyer, M., and Verhasselt, V. (2015). Breast milk and its impact on maturation of the neonatal immune system. Curr. Opin. Infect. Dis. 28, 199–206. doi: 10.1097/QCO.0000000000000165
Van Itallie, C. M., Holmes, J., Bridges, A., Gookin, J. L., Coccaro, M. R., Proctor, W., et al. (2008). The density of small tight junction pores varies among cell types and is increased by expression of claudin-2. J. Cell Sci. 121, 298–305. doi: 10.1242/jcs.021485
Van Niel, C. W., Feudtner, C., Garrison, M. M., and Christakis, D. A. (2002). Lactobacillus therapy for acute infectious diarrhea in children: A meta-analysis. Pediatrics 109, 678–684. doi: 10.1542/peds.109.4.678
Walker, W. A., and Iyengar, R. S. (2015). Breast milk, microbiota, and intestinal immune homeostasis. Pediatr. Res. 77, 220–228. doi: 10.1038/pr.2014.160
Walthall, K., Cappon, G. D., Hurtt, M. E., and Zoetis, T. (2005). Postnatal development of the gastrointestinal system: A species comparison. Birth Defects Res. B Dev. Reprod. Toxicol. 74, 132–156. doi: 10.1002/bdrb.20040
Weaver, L. T., Laker, M. F., Nelson, R., and Lucas, A. (1987). Milk feeding and changes in intestinal permeability and morphology in the newborn. J. Pediatr. Gastroenterol. Nutr. 6, 351–358. doi: 10.1097/00005176-198705000-00008
Westrom, B., Arevalo Sureda, E., Pierzynowska, K., Pierzynowski, S. G., and Perez-Cano, F. J. (2020). The immature gut barrier and its importance in establishing immunity in newborn mammals. Front. Immunol. 11:1153. doi: 10.3389/fimmu.2020.01153
Yoon, H. S., Lim, J., Sohn, Y. H., and Kim, S. Y. (2022). Incidence, clinical characteristics, and genotype distribution of rotavirus in a neonatal intensive care unit 5 years after introducing rotavirus vaccine. Front. Pediatr. 10:850839. doi: 10.3389/fped.2022.850839
Yoshida, M., Kobayashi, K., Kuo, T. T., Bry, L., Glickman, J. N., Claypool, S. M., et al. (2006). Neonatal Fc receptor for IgG regulates mucosal immune responses to luminal bacteria. J. Clin. Invest. 116, 2142–2151. doi: 10.1172/JCI27821
Yu, Y., Lu, L., Sun, J., Petrof, E. O., and Claud, E. C. (2016). Preterm infant gut microbiota affects intestinal epithelial development in a humanized microbiome gnotobiotic mouse model. Am. J. Physiol. Gastrointest. Liver Physiol. 311, G521–G532. doi: 10.1152/ajpgi.00022.2016
Zhang, K., Dupont, A., Torow, N., Gohde, F., Leschner, S., Lienenklaus, S., et al. (2014). Age-dependent enterocyte invasion and microcolony formation by Salmonella. PLoS Pathog. 10:e1004385. doi: 10.1371/journal.ppat.1004385
Zheng, D., Liwinski, T., and Elinav, E. (2020). Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492–506. doi: 10.1038/s41422-020-0332-7
Keywords: neonate, intestine, epithelial, microbiota, infection
Citation: Negi S, Hashimoto-Hill S and Alenghat T (2022) Neonatal microbiota-epithelial interactions that impact infection. Front. Microbiol. 13:955051. doi: 10.3389/fmicb.2022.955051
Received: 28 May 2022; Accepted: 31 July 2022;
Published: 25 August 2022.
Edited by:
Axel Cloeckaert, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceReviewed by:
Kathryn A. Knoop, Mayo Clinic, United StatesAdora Okogbule-Wonodi, Howard University, United States
Erick Saul Sánchez Salguero, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, Mexico
Copyright © 2022 Negi, Hashimoto-Hill and Alenghat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Theresa Alenghat, dGhlcmVzYS5hbGVuZ2hhdEBjY2htYy5vcmc=; Seika Hashimoto-Hill, U2Vpa2EuSGFzaGltb3RvLUhpbGxAY2NobWMub3Jn
†These authors have contributed equally to this work
 Shikha Negi
Shikha Negi Seika Hashimoto-Hill
Seika Hashimoto-Hill Theresa Alenghat
Theresa Alenghat