- 1National & Local Joint Engineering Research Center of Northern Horticultural Facilities Design & Application Technology, Shenyang, China
- 2Key Laboratory of Protected Horticulture of Education Ministry and Liaoning Province, College of Horticulture, Shenyang Agriculture University, Shenyang, China
Long-term monoculture cropping and overfertilization degrade soil fertility, which reduces crop growth and promotes the development of soil-borne diseases. However, it remains unclear what the temporal effects of the above factors are on the tomato yield and microbial community structure. Thus, a greenhouse experiment with different amounts of fertilization [2,196 kg ha−1 (control) and 6,588 kg ha−1 (overfertilization) of inorganic fertilizers (NPK)] was carried out with the soils used previously for 1, 2, and 12 years under monoculture of tomato. A 12-year overfertilization decreased soil pH by 1.37 units. Soil electrical conductivity (EC) and concentrations of soil nutrients are enhanced with the increase in tomato cropping duration. Higher content of soil nutrients was found under overfertilization compared to the control in the 12-year soil. Overfertilization decreased the activity of β-1,4-glucosidase (BG) and oxidase compared to the control in the 12-year soil. Bacterial diversity and richness decreased by 6 and 31%, respectively, under overfertilization in 12-year soil compared to the control. The relative abundance of Gemmatimonas and Gp6 in 12-year soil under overfertilization was 17 and 78%, respectively, lower than in control soil. Soil pH and total carbon (TC) were the major factors explaining changes in microbial composition. A 38% decrease in yield was caused by overfertilization in 12-year soil compared to the control. Microbial community composition was the main factor that moderated tomato yield. In addition, fertilization rather than cropping duration had a greater impact on tomato yield. Therefore, our results suggest that long-term overfertilization influenced soil pH, soil TC, and soil microbial community composition to regulate tomato yield.
Introduction
Continuous monoculture cropping is a common cultural pattern in the greenhouse, and lots of research has found that the yield and quality of crops are reduced under this system. This occurs due to (i) soil acidification and secondary salinization (Shi et al., 2009; Han et al., 2014) caused by the reduced leaching with rainwater and application of inorganic fertilizer; (ii) production and accumulation of allelochemicals (fatty acids and phenolic acids) (Bertin et al., 2003), which influence the permeability of root membranes, nutrient uptake, enzyme activities, and microbial community development (Ning et al., 2020; Shi et al., 2021) and, thus, affect the growth of crops (Chung et al., 2000; Wu et al., 2007); and (iii) soil-borne diseases such as Fusarium wilt, which reduce the quality and yield of vegetables (Reeves, 1997; Yu, 2001; Zhao et al., 2019). Besides, selective nutrient accumulation is observed (Magarey, 1999), which, together with consistent fertilization, further promotes nutrient imbalance (Ning et al., 2020). Long-term continuous cropping decreases soil microbial diversity, which simplifies community structure, the number and types of beneficial bacteria (e.g., Mortierella and Pseudaleuria), and keystone taxa (e.g., Acidobacteria Gp1, Ac-idobacteria Gp2, and Acidobacteria 16) abundances, which unbalances the community (Ning et al., 2020; Tan et al., 2021), but not enough information is available about the importance of these impacts on crop growth.
Fertilization can not only increase the content of soil organic matter and stabilize the microbial community and enzyme activity, but it can also improve plants and microorganisms' nutrient use efficiency (Xing and Zhu, 2002; Yang et al., 2018; Puissant et al., 2019). However, excessive inorganic fertilizer input in greenhouse production driven by yield and economic benefit is common, especially for N fertilizer (Guo et al., 2010; Ali et al., 2020). The fertilizer N application rates used for vegetable production in North China ranged from approximately 500 to 1,900 kg N ha−1 (Zhang et al., 1996). N fertilizer application rates reached 4,000 kg N ha−1 in Shandong province (Ju et al., 2007). In tomato cultivation, higher nitrogen use efficiency, biomass, soil organic C and organic N, and lower N leaching were observed in relatively low N application rate (from 143 to 360 kg N ha−1) treatments than in relatively high N application rate (from 480 to 870 kg N ha−1) treatments (Sainju et al., 2001; Liang et al., 2020). Long-term low fertilization (120 kg N ha−1) does not change the pH and the electrical conductivity (EC; Masto et al., 2007; Xu et al., 2012). However, overfertilization (900 kg N ha−1) introduced soil-borne pathogens, such as Fusarium oxysporum and Luteimonas, and suppressed the beneficial microorganisms, including nitrifiers, and Nocardioides, Ilumatobacter, and Gaiella (Zhang X. M. et al., 2017; Zhao et al., 2019). Short-term overfertilization causes beneficial effects due to the provision of sufficient nutrients for crops, while with the increase in cropping duration, the beneficial effects gradually decrease or even disappear (Lazcano et al., 2013; Han et al., 2016). Long-term overfertilization decreased the soil pH (Zhang Y. T. et al., 2017); increased the content of NH (Ma et al., 2021); caused a greater risk of N leaching; enhanced nitrous oxide emissions (Song et al., 2016); reduced the activity of β-glucosidase and phosphatases; affected C, N, and P cycling; and reduced the content of base cations (Chang et al., 2007). Furthermore, long-term fertilization reduced bacterial diversity, richness, and fungal diversity (Wallenstein et al., 2006; Yuan et al., 2020); enhanced fungal richness (Dong et al., 2014); and altered the community structural and functional gene structure of microbes in soil (Edwards et al., 2011; Su et al., 2015). Inorganic fertilization decreased the abundance of microbial taxa (i.e., phyla Proteobacteria, Bacteroidetes, Mortierella, and Pseudaleuria), which are associated with plant growth, organic matter accumulation, and disease suppression (Ma et al., 2018; Ning et al., 2020). However, there is not enough data on how organic fertilization combined with different concentrations of inorganic fertilization could affect soil properties via long-term continuous cropping.
Despite long-term continuous cropping and overfertilization coexist under the greenhouse culture system, there are only a few reports on how these two variables interact and affect the crop yield. Thus, this study used soils from continuous cropping for 1, 2, and 12 years to reveal the effect of usual (2,196 kg ha−1) vs. overfertilization (6,588 kg ha−1) of inorganic fertilizers combined with the same amount of chicken manure on the yield of tomato (Solanum lycopersicum L.), soil chemical properties, and microbial communities. It was hypothesized that (i) control and overfertilization will alter the yield of tomatoes differently under long-term monoculture cropping, and (ii) microbial community activity will differ among control and overfertilization due to alteration of soil properties.
Materials and methods
Soil sampling from the field
Two types of long-term greenhouse fertilized soils, which have been treated for 0, 1, and 11 years of continuous tomato monoculture with rational fertilization (control) and overfertilization, were collected from two adjacent sites in Liaoning Province, China. The fertilization strategies of these two soils were as follows: control—chemical fertilizers [ShiJiaLi Chemical fertilizer company limited (Chengdu, China)] were applied at rates of 286 kg N ha−1, 229 kg P2O5 ha−1, and 190 kg K2O ha−1; and overfertilization—chemical fertilizers were applied at rates of 858 kg N ha−1, 685 kg P2O5 ha−1, and 571 kg K2O ha−1. Chicken manure compost [2.80 g N kg−1, 2.43 g P2O5 kg−1, and 0.13 g K2O kg−1; RuiYuanDe Biotechnology Co., Ltd. (Shenyang, China)] was added to both soils at the rate of 5.04 g kg−1 soil.
The soil is classified as a Hapli-Udic Cambisol (WRB). The initial soil properties of control soil were pH 7.10 (1:2.5, w/v), EC 0.13 ms cm−1 (1:5, w/v), available N 87.7 mg kg−1, available P 18.9 mg kg−1, available K 147.6 mg kg−1, total N 0.15 g kg−1, and total P 1.62 g kg−1. Initial soil properties of overfertilization soil were pH 7.08 (1:2.5, w/v), EC 0.14 ms cm−1 (1:5, w/v), available N 90.5 mg kg−1, available P 23.5 mg kg−1, available K 154 mg kg−1, total N 0.17 g kg−1, and total P 1.81 g kg−1.
Greenhouse experiment
Soils were transported to the greenhouse (123° 24′ E, 41° 31′ N), which was located at Shenyang Agricultural University, Shenyang, Liaoning Province. Soils were sieved through a 2-cm screen to remove debris and stone and then mixed homogeneously with fertilizers according to the treatments. All treatments were arranged in a randomized complete block design, with 3 replicates per treatment.
The experiment was conducted from March 2020 to July 2020, and tomato plants (“Kaide Zunyue”) were planted in each treatment. Each tomato plant had a single branch after pruning, and there were three clusters of fruits on the branch and five fruits per cluster. The air temperature inside the experimental greenhouse ranged from 15 to 35°C. All tomato plants received natural light only. After one growing period of cultivation, 1, 2, and 12 years of soil were obtained.
Soil sampling
In each treatment, soil samples were collected in July 2020 via a five-point sampling method, and each treatment was repeated three times. The roots and rocks were removed, and soil was passed through a 2-mm sieve. One part of the samples was air-dried to measure basal soil properties, one part was frozen in liquid N2 immediately and saved at −80°C for future DNA extraction, and the other fresh soil samples were kept in ice boxes to be transported to the lab and stored at 4°C for enzymes and NH/NO-N analyses.
Yield and basal soil properties analysis
Tomatoes were harvested as fresh vegetables. Soil pH (soil:distilled water, 1:2.5) was determined using a Thunder Magnetic SJ-3F pH Meter (INESA, Shanghai, China). The soil EC (soil:distilled water, 1:5) value was determined using a Thunder Magnetic DDS-307 EC Meter (INESA, Shanghai, China). Soil NH-N and NO-N were extracted by 2 M KCl (5:1 v/w) and analyzed using the SAN++ Continuous Flow Analyzer (Skalar, Netherlands; Blakemore et al., 1987). Soil total C (TC) and total N (TN) contents were measured using an elemental analyzer (Elementar III, Germany). Total phosphorus (TP) was determined by the molybdenum-blue method (Nobile et al., 2020). Total potassium (TK) was measured using flame photometry after soil digestion with NaOH (Lu, 2000). Soil available phosphorus (AP) was determined in sodium bicarbonate extraction following colorimetric measurement (Olsen and Sommers, 1982). Available potassium (AK) was determined in the ammonium acetate extraction following the flame ionization photometry (Lu, 2000).
Enzyme assays
Activities of β-1,4-glucosidase (βG, substrate: 4-MUB-β-D-glucopyranoside), leucine aminopeptidase (LAP, substrate: L-Leucine-7-AMC), and peroxidase and polyphenol oxidase (PER and PPO, substrate: L-DOPA) were measured using fluorogenic methods with 4-methylumbelliferyl (MUB), 7-amino-4-methylcoumarin (AMC), and L-dihydroxyphenylalanine (DOPA), respectively. Enzyme assays were performed following the protocol established by German et al. (2011). For each sample, soil slurry was prepared by adding 125 ml of 50 mM sodium acetate buffer (pH 5.0) to 1.5 g of fresh soil, then homogenizing for 1 min. The supernatant was continuously stirred using a magnetic stir plate, and 200 μl aliquots were dispensed into 96-well microplates and further used for the analyses. For hydrolytic enzymes (i.e., βG and LAP), the final concentrations of the substrates were 224.7 and 189.6 μM, and for oxidative enzymes (i.e., PER and PPO), it was 68.4 μM. The hydrolytic enzymes were incubated for 2.5 h, and oxidative enzymes were incubated for 24 h in the dark at 25°C. The quantity of fluorescence (hydrolytic enzymes) was read at 360 nm excitation and 460 nm emission; absorbance (oxidative enzymes) was read at 450 nm using a Microplate Reader (BioTek, Synergy2, United States).
16S, ITS rRNA Gene Amplification, and Sequencing
Soil DNA was extracted using the E.Z.N.A.® Soil DNA Kit (Omega Bio-Tek, Norcross, GA, United States) according to the manufacturer's instructions. DNA concentration and purity were determined using a spectrophotometer, and DNA integrity was checked on a 1% agarose gel. These primers target the V3-V4 regions of bacterial 16S rRNA genes and ITS2 regions of fungal ITS rRNA genes (Ghannoum et al., 2010; Mori et al., 2013). The primers F338/R806 (F338, 5′-ACTCCTACGGGAGGCAGCAG-3′; R806, 5′-GGACTACHVGGGTWTCTAAT-3′) and ITS1/ITS2 (ITS1, 5′-CTTGGTCATTTAGAGGAAGTAA-3′; ITS2, 5′-GCTGCGTTCTTCATCGATGC-3′) were selected. The PCR amplification of the 16S rRNA gene was performed as follows: initial denaturation at 95°C for 3 min, then 27 cycles of denaturing at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 45 s, and a single extension at 72°C for 10 min. The PCR amplification of the ITS rRNA gene was performed as follows: initial denaturation at 95°C for 3 min, then 35 cycles of denaturing at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 45 s, and a single extension at 72°C for 10 min. The PCR products were purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) according to the manufacturer's instructions and quantified by the real-time quantitative PCR. Purified amplicons were pooled in equimolar and paired-end sequenced on an Illumina MiSeq PE300 platform (Illumina, San Diego, United States) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: PRJNA839183).
Sequencing reads were demultiplexed, quality-filtered using fastp version 0.20.0 (Chen et al., 2018), and merged by FLASH version 1.2.7 (Magoč and Salzberg, 2011). Poor-quality sequences shorter than 50 bp and with an average quality score of less than 20 were discarded, and reads containing ambiguous characters were also discarded. Operational taxonomic units (OTUs) with a 97% similarity cutoff (Stackebrandt and Goebel, 1994; Edgar, 2013) were clustered using UPARSE version 7.1, and chimeric sequences were identified and removed. The taxonomy of each OTU representative sequence was analyzed using RDP Classifier version 2.2 (Wang et al., 2007) against the 16S rRNA database (Rdp v11.5) and the ITS rRNA database (Unite v8.0), using a confidence threshold of 0.7.
Statistical analyses
One-way ANOVA, mixed-effects ANOVA, and independent sample T-test were performed to reveal the significant differences in the crop yields and soil properties in the control and overfertilization treatment groups, respectively. Residuals of all ANOVA were checked for normality and homogeneity, and if assumptions were met, the Tukey's test was done and p < 0.05. When the interaction of fertilization and cropping duration is significant on soil properties, an independent sample T-test was used for cultivation durations in each fertilization treatment, and one-way ANOVA was used for cultivation durations in each fertilization treatment. When the interaction of fertilization and cropping duration is not significant on soil properties, one-way ANOVA was used for all treatments. All ANOVA analyses were conducted in SPSS 19.0 (IBM Corp., New York, United States). All experimental data are expressed as an average of three replicates with standard deviations.
Bacterial alpha-diversity (Shannon index and Chao1 index) was calculated with 10 times subsampling using Mothur software (version 1.30.2). Non-metric multidimensional scaling (NMDS) analyses based on Bray-Curtis dissimilarity matrices were performed to describe the structure of microbial community under treatments. Relationships between soil properties and microbial community composition were revealed by redundancy analysis (RDA). Variance inflation factors (VIFs) were used to detect multicollinearity, and factors with VIFs below 10 were retained. A multiple stepwise linear regression was used in SPSS 19.0 to select the most important contributors to explain the microbial communities. Residuals from the regression model were checked for normality as well.
Structural equation modeling (SEM) has been used to examine the direct or indirect effects of cropping duration, soil properties, and microbial community characteristics on crop yields. SEM analysis was performed using AMOS 24.0 (AMOS IBM, United States). The fitness of the model was evaluated via a non-significant chi-square test (p > 0.05), low χ2 (<7), high goodness-of-fit index (GFI > 0.95), and low root square mean errors of approximation (RMSEA < 0.08; Byrne and Erlbaums, 2009).
Results
Tomato yields and soil chemical properties
Tomato yield significantly enhanced from 1- to 2-year-old soils in both control and overfertilization. However, overfertilization decreased tomato yield by 39% from 2 to 12 years. Under overfertilization, the yield decreased by 38% in the 12-year soil, compared to the control (Figure 1).
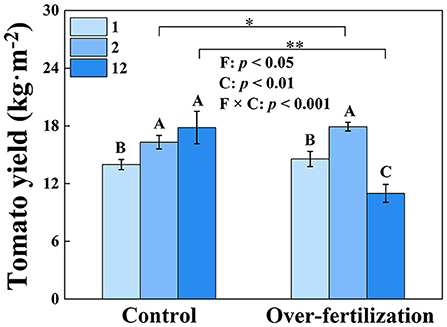
Figure 1. Effects of fertilization on tomato yield. Values are means ± SD (n = 3). * and ** indicate significant differences between fertilization under the same cropping duration at p < 0.05 and p < 0.01, respectively. Different letters (A, B, and C) above the bars indicate significant (p < 0.05) differences between cropping durations under the same fertilization treatments. F, fertilization; C, cropping duration.
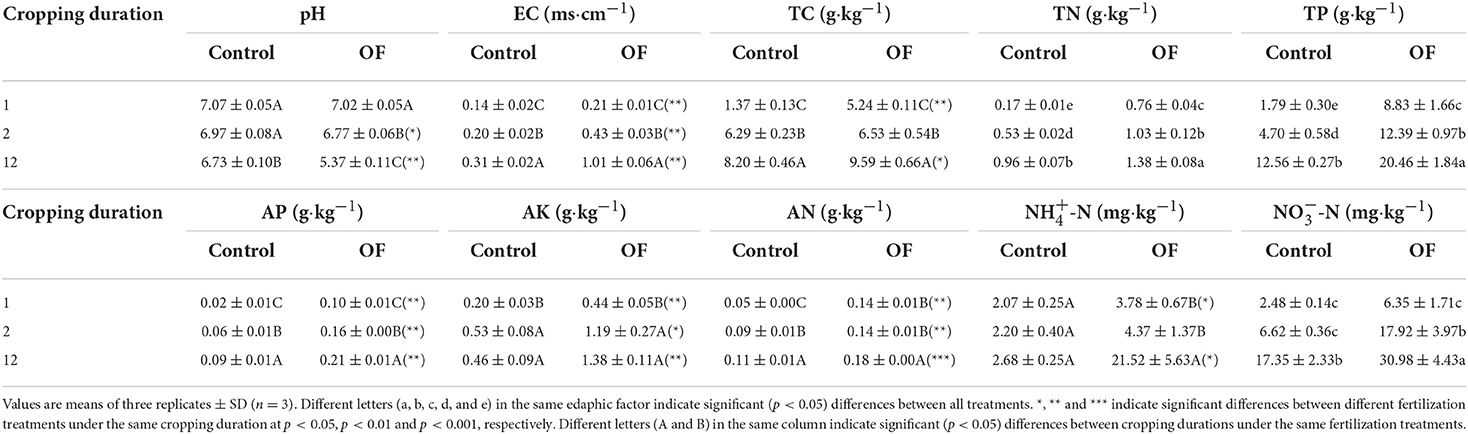
Soil pH was significantly reduced by 0.34 and 1.65 units from 1 to 12 years in control and overfertilization, respectively. Overfertilization showed a greater decrease in soil pH (by 4.85 times) than the control in 12-year soil (Table 2). Soil EC and concentrations of soil TC, TN, TP, AN, AP, AK, NH4+-N, and NO-N enhanced with the increase of tomato cropping duration. Higher content of soil TC, TN, TP, AN, AP, AK, NH4+-N, and NO-N were found under overfertilization compared to the control in the 12-year soil (Tables 1, 2).
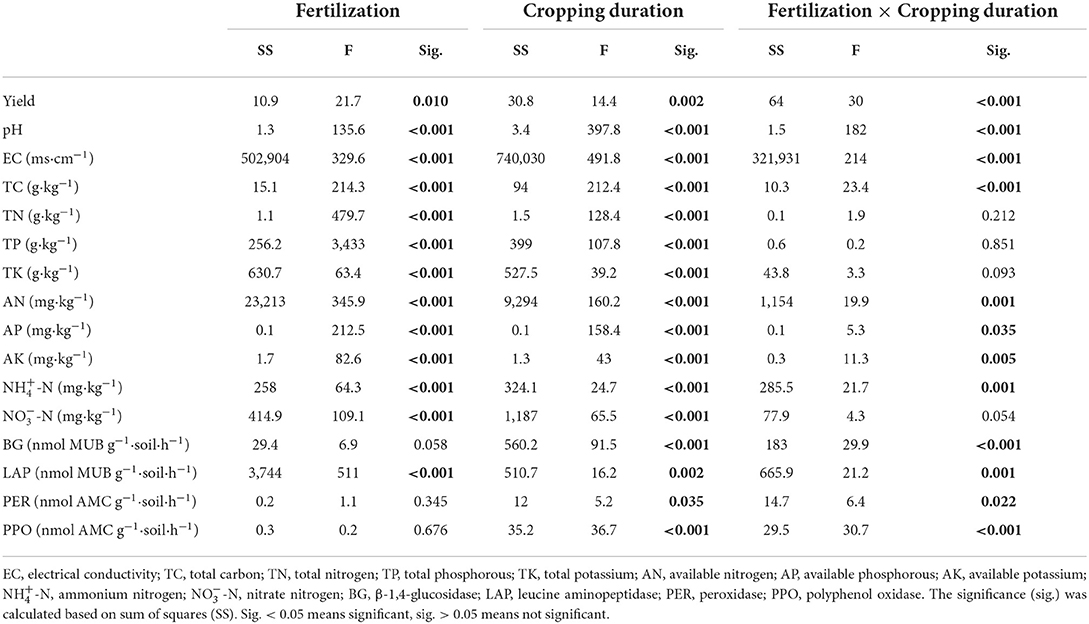
Table 1. Mixed effect ANOVA for the effects of fertilization, crops, and their interaction on soil properties.
The activities of BG, LAP, PER, and PPO increased with the cropping duration under control, while the activity of PER stayed constant among different continuous cropping years under overfertilization. Overfertilization caused an increase in the activities of BG, LAP, and PPO from 1- to 2-year soils and a decrease in the activities of LAP and PPO from 2- to 12-year soils (Table 1 and Figure 2).
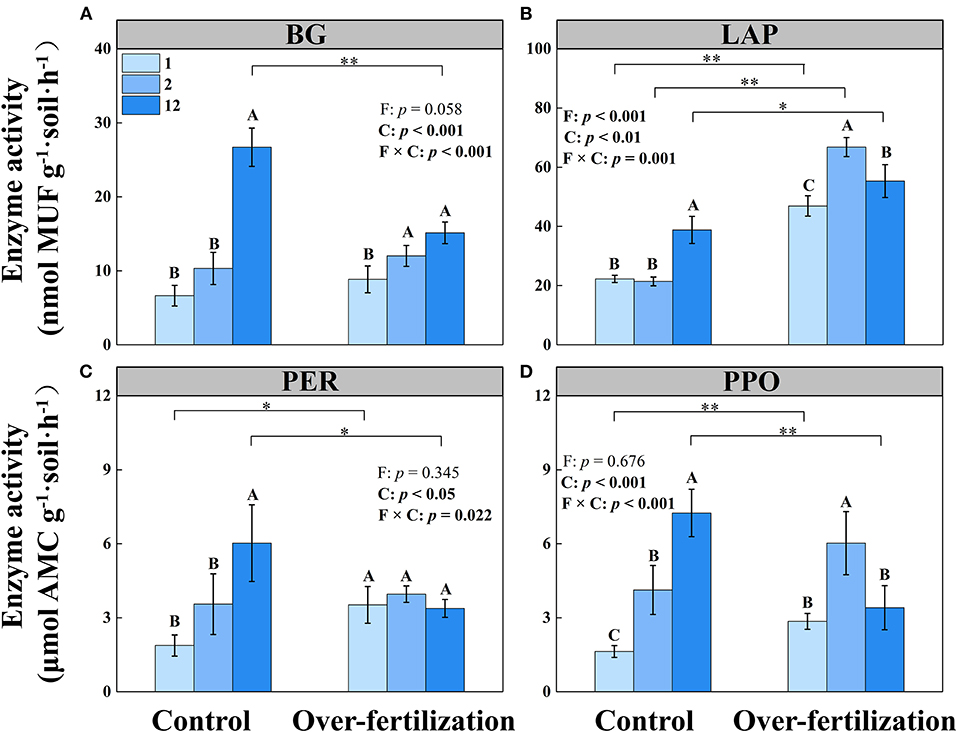
Figure 2. Effects of fertilization on soil enzyme activity under tomato cropping. (A) The activity of β-1,4-glucosidase (BG). (B) The activity of leucine aminopeptidase (LAP). (C) The activity of peroxidase (PER). (D) The activity of polyphenol oxidase (PPO). Values are means ± SD (n = 3). * and ** indicate significant differences between fertilization under the same cropping duration at p < 0.05 and p < 0.01, respectively. Different letters (A, B, and C) above the bars indicate significant (p < 0.05) differences between cropping durations under the same fertilization treatments. F, fertilization; C, cropping duration.
Soil microbial alpha-diversity
A higher tomato cropping duration resulted in a decrease in the microbial diversity and richness of bacteria under control. Overfertilization enhanced the microbial diversity and richness of bacteria in 1- to 2-year soils and then decreased them in 2- to 12-year soils. Compared with control, overfertilization significantly decreased the bacterial diversity and richness under the same cropping duration.
The fungal diversity and richness decreased with the increase in tomato cropping duration for the control. Overfertilization resulted in an increase in fungal diversity from 1- to 2-year soils, followed by a decrease (Figure 3). Lower fungi diversity and richness were observed in the overfertilization than in the control (1- and 2-year soils).
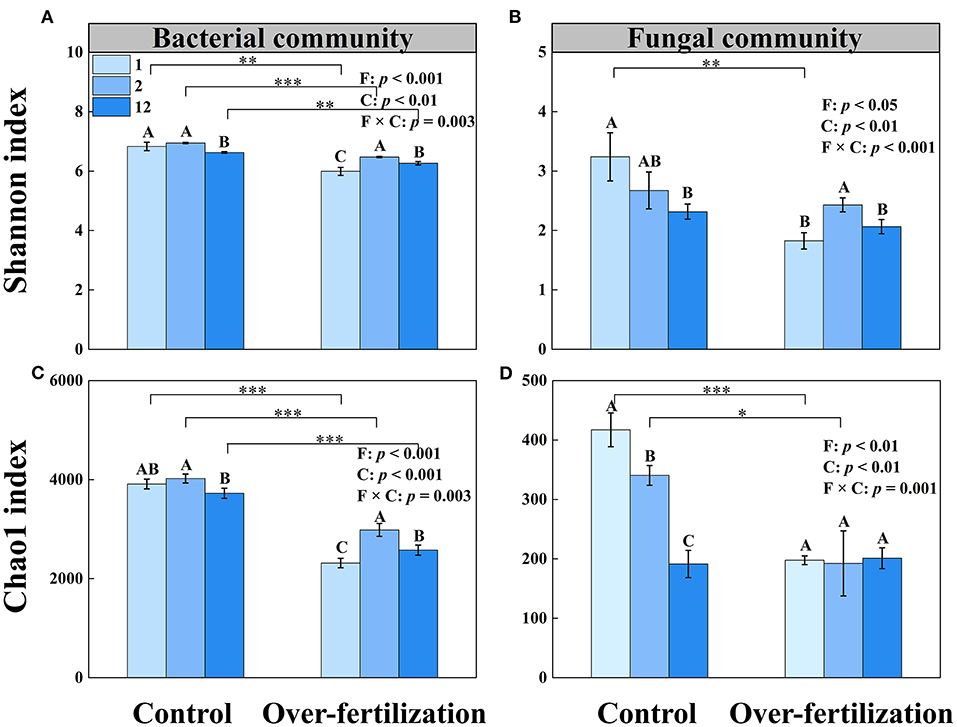
Figure 3. Effects of fertilization on soil microbial α-diversity under tomato cropping. (A) The variations of bacterial Shannon index. (B) The variations of fungal Shannon index. (C) The variations of bacterial Chao1 index. (D) The variations of fungal Chao1 index. Values are means ± SD (n = 3). *, **, and *** indicate significant differences between fertilization under the same cropping duration at p < 0.05, p < 0.01, and p < 0.001, respectively. Different letters (A, B, and C) above the bars indicate significant (p < 0.05) differences between cropping durations under the same fertilization treatments. F, fertilization; C, cropping duration.
Soil microbial communities
For the bacterial community structure, a distinct separation was observed between control and overfertilization by NMDS axis1 (ANOSIM, p = 0.001). Fungal community structures in 1- and 2-year soils were separated between the control and overfertilization by NMDS axis1, and a distinct separation between various cropping duration was only observed under overfertilization by NMDS axis1 (ANOSIM, p = 0.002; Figures 4A,B).
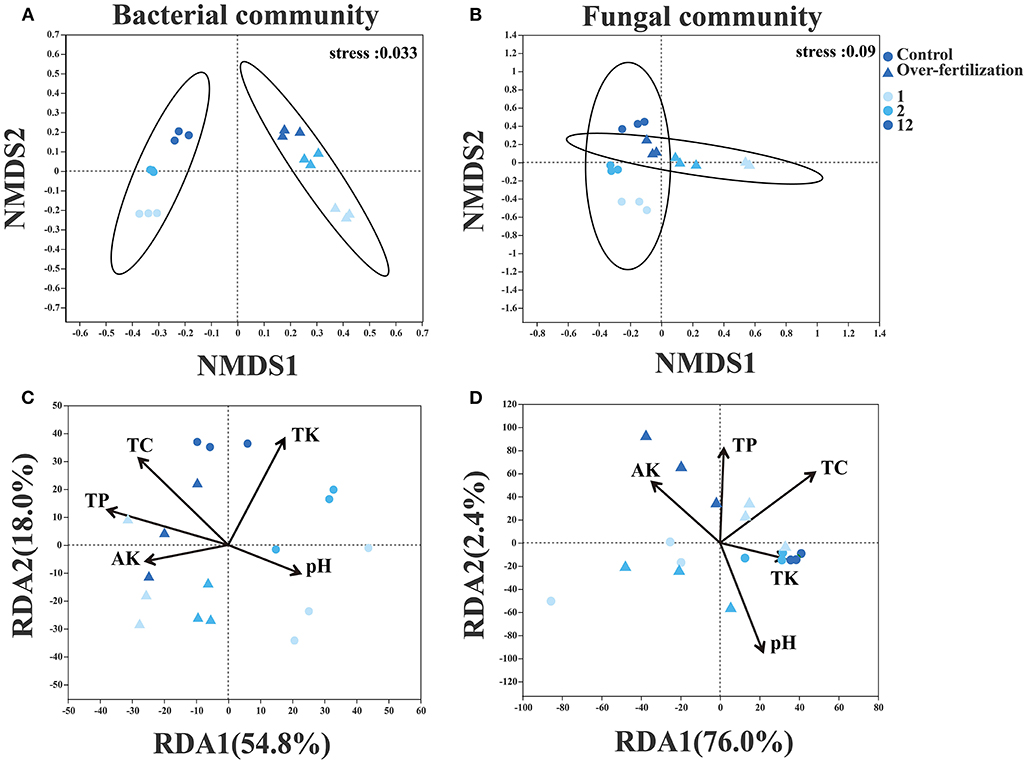
Figure 4. Effects of fertilization on microbial community composition and structure under tomato cropping. (A) The variations of bacterial NMDS under different treatments. (B) The variations of fungal NMDS under different treatments. (C) The variations of bacterial RDA under different treatments. (D) The variations of fungal RDA under different treatments. Variance inflation factors (VIFs) were used to detect multicollinearity, and factors with VIFs under 10 were retained. TC, total carbon; TP, total phosphorous; TK, total potassium; AK, available potassium.
Edaphic factors (i.e., pH, TC, TK, TP, and AK) explained 72.8% and 78.4% of the variation by the first two constrained axes of RDA in the community structure of bacteria and fungi, respectively. Soil TC, TK, and TP were positively correlated with the variations in the bacterial community. Soil pH, TP, and TC were positively correlated with the variations in the fungal community (Figures 4C,D).
Soil-specific microbial taxa enriched
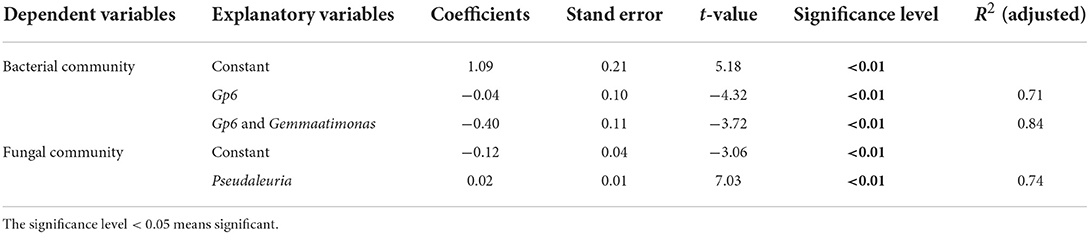
The multiple stepwise linear regression analysis showed that the bacterial community was influenced by Gp6 and Gemmatimonas as they explained up to 83.9% (p < 0.01) of the variability. The soil fungal community was primarily explained by Pseudaleuria as it explained up to 74.0% (p < 0.01) of the variation (Table 3).
The relative abundance of Gp6 was constant for the soils having various cultivation ages in both control and overfertilization. The relative abundance of Gp6 was lower under overfertilization compared to the control (Figure 5A). The relative abundance of Gemmatimonas was the same in control, while overfertilization enhanced it with the increase in tomato cropping duration. The relative abundance of Gemmatimonas was lower under overfertilization compared to the control (Figure 5B). Relative abundance of Pseudaleuria was reduced under overfertilization, while control enhanced it with the increase in tomato cropping duration. Overfertilization led to a higher relative abundance of Pseudaleuria than control in 1- and 2-year soils (Figure 5C), but no difference was observed in 12-year soil (Figure 5C).
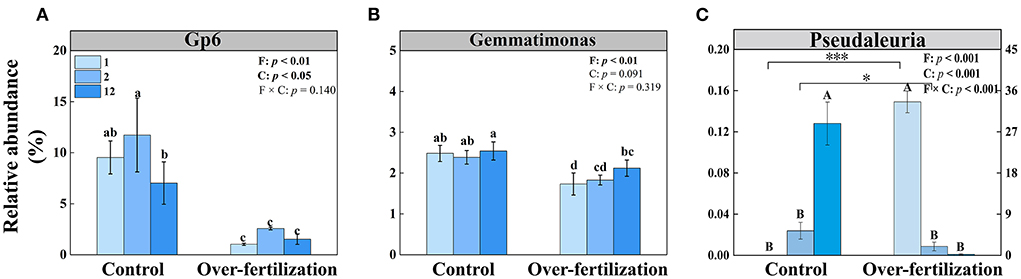
Figure 5. Effects of fertilization on the relative abundances of bacteria and fungi under tomato cropping. (A) The relative abundance of Gp6. (B) The relative abundance of Gemmatimonas. (C) The relative abundance of Pseudaleuria. Values are means ± SD (n = 3). Different letters (a, b, c, d, and e) above the bars indicate significant (p < 0.05) differences between all treatments. * and *** indicate significant differences between different fertilization treatments under the same cropping duration at p < 0.05 and p < 0.001, respectively. Different letters (A and B) above the bars indicate significant (p < 0.05) differences between cropping durations under the same fertilization treatments. F, fertilization; C, cropping duration.
Relationship between soil properties and crop yields
The SEM model explained 86% of the variation in yield (Figure 6A). Cropping duration had direct positive effects on soil TC and the fungal community but had direct negative effects on soil pH, bacterial diversity, and community structure. These pathways led to a negative total effect of cropping duration on tomato yield (effect size = −0.23, Supplementary Table 1-1). Fertilization amount had direct positive effects on soil TC and bacterial community but had direct negative effects on soil pH, bacterial diversity, and the fungal community. Fertilization amount had a negative total effect on tomato yield (effect size = −0.33, Supplementary Table 1-1). Standardized total effects showed that the change in tomato yield was mainly driven by the bacterial community, followed by soil pH, fungal community, soil TC, bacterial diversity, fertilization amount, fungal diversity, and cropping year (Figure 6B).
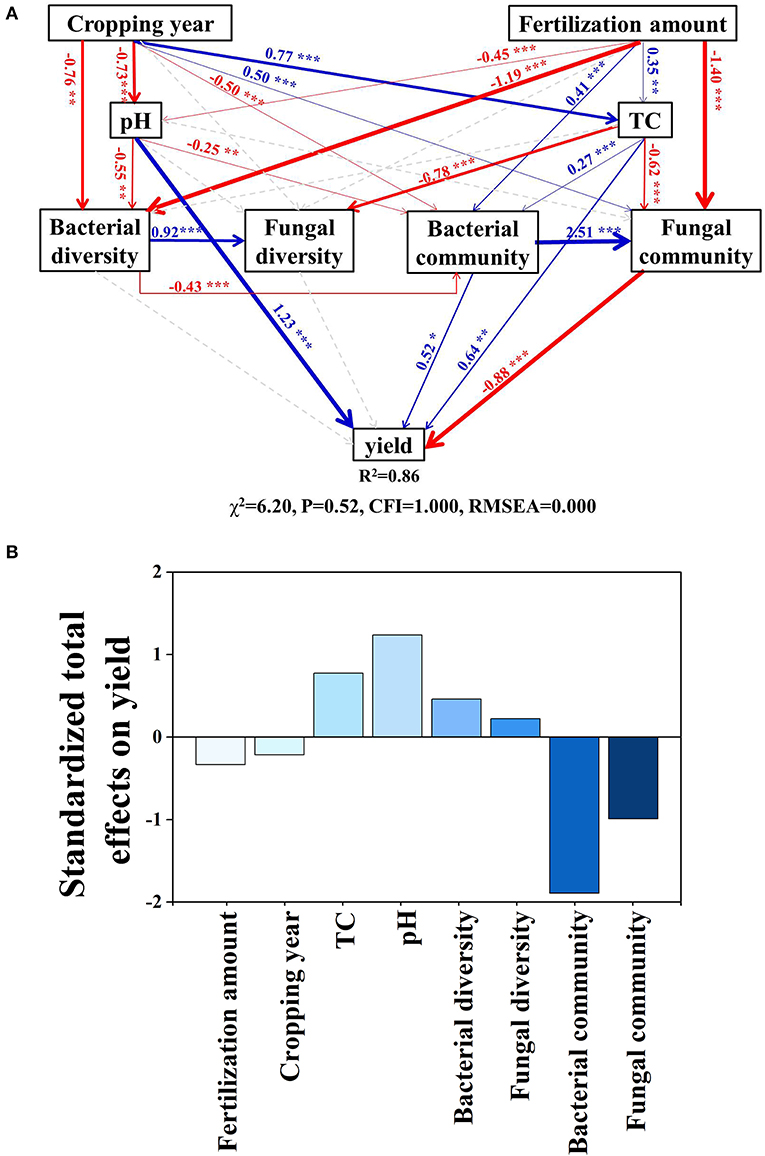
Figure 6. Effects of multiple factors on crop yield. (A) Structural equation modeling based on the effects of cropping durations, fertilization, soil properties (pH and TC), bacterial diversity and community composition, fungal diversity and community composition on crop yield. (B) Standardized total effects. Red arrows indicate significant negative and blue positive relationships. Dashed arrows represent non-significant relationships. Arrow width indicates the strength of the relationships. Numbers at arrows are standardized path coefficients. R2 denotes the proportion of variance explained by the model. *p < 0.05; **p < 0.01; ***p < 0.001. TC, total carbon.
Discussion
Overfertilization deteriorated the tomato yield
Fertilization generated a greater effect on tomato yield than cropping duration (Figure 1). A decrease in tomato yield was observed for overfertilization in 12-year soil. Overfertilization can alter soil properties and influence tomato shoot and root growth (Omay et al., 1997; Sainju et al., 2000). When the N application rate was increased from 375 kg N ha−1 to 1,123 kg ha−1, the nitrogen use efficiency was significantly reduced (Liu et al., 2004). Over NO3–N accumulation resulting from long-term fertilizer input (Table 1) increased N leaching and N2O, and NH3 emissions, which may cause water and air pollution, resulting in high N loss risk and causing a negative effect on the growth of tomatoes (Min et al., 2011; Wang and Xing, 2016; Yao et al., 2019). Furthermore, long-term overfertilization can drive soil acidification (Table 1), which degrades soil and environmental quality (Guo et al., 2010). Soil acidification promoted soil nitrification (Shang et al., 2014) and inhibited root growth (Wang et al., 2017). Long-term tomato monoculture with intensive inorganic fertilizer addition enhanced soil salinity (shown as a high EC value; Zhao et al., 2019), which reduced the availability of plant water uptake by decreasing the osmotic potential of the soil solution, causing disruption of nutritional balance, which eventually reduced tomato yield (Kahlaoui et al., 2011). In addition, overfertilization suppressed the production of BG, PER, and PPO (Figure 2) because enzyme activity is predominantly controlled by temperature and pH, which affect enzyme kinetics through changes in substrate binding and stability (Puissant et al., 2019). Thus, overfertilization has a negative effect on tomato yield under long-term continuous cropping.
The response of microbial communities to overfertilization under long-term tomato cropping
Overuse of inorganic fertilization reduced the microbial diversity and richness and changed the microbial community composition (Figure 3). The bacterial community and the fungal community were influenced by variations of Gp6, Gemmatimonas, and Pseudaleuria, respectively (Table 3). The relatively high abundances of Gp4, Gp6, and Gp16, which belong to Acidobacteria, reflected the neutral pH of the soil (Jones et al., 2009; Zheng et al., 2019). The relative abundance of Gp6 tends to be more enriched in mildly acidic soil, which is found in control. Overfertilization decreased soil pH and reduced the relative abundance of Gp6 because N enrichment inhibited the growth of oligotrophic bacteria (Dai et al., 2018). Control induced a high relative abundance of Gemmatimonas, which contributes to the degradation of cellulose (Wang et al., 2017). Members of the Gemmatimonas genus are known to contribute to soil organic carbon sequestration (Guo et al., 2016), and it can be against plant pathogens and improvement of soil nutrients (Liu et al., 2020). Furthermore, Gemmatimonas is related to the metabolism and transformation of nitrogen, which showed strong positive associations with total nitrogen and reduced the potent greenhouse gas N2O under both aerobic and anaerobic conditions (Li et al., 2017; Park et al., 2017). Some bacterial strains belonging to the genus Gemmatimonas could perform bacteriochlorophyll-based chlorophototrophy, which possesses an expanded gene repository for coping with oxidative stresses (Zeng et al., 2021). In addition, a high abundance of Gemmatimonas was found in the healthy wheat plant rhizosphere, which indicated that Gemmatimonas can help suppress diseases and promote plant growth (Yin et al., 2013; Li et al., 2017). A combination of organic and inorganic fertilizers is beneficial for the relative abundance of certain favorable fungal taxa such as Pseudaleuria, which inhibits crop pathogens (Ding et al., 2017; Xiang et al., 2020). Pseudaleuria was the most abundant in healthy soils, which was involved in the interactions with plant roots to suppressive soil diseases (Xu et al., 2012). Relative abundance of Pseudaleuria increased under overfertilization in the short term but reduced markedly in the long term, while control showed the contrast effects. Thus, soil beneficial microorganisms decrease when the greenhouse ecosystem is exposed to consistent overfertilization.
Linkage of soil microbial community composition and tomato yield in response to fertilization
Based on SEM model analyses, the decrease in tomato yield caused by overfertilization was mainly due to the direct effect of pH and the indirect effects of bacterial community composition (Figure 6 and Supplementary Table 1). Continuous cropping and fertilization not only directly but also indirectly affected soil microorganisms through soil pH (Ning et al., 2020), salt (Zhou et al., 2016), nitrogen content (Liu et al., 2021), and availability of organic compounds (Ning et al., 2021). The pH played a more important role in the microbial diversity, while TC played a more important role in the microbial community structure (Supplementary Table 1), especially the bacterial one because fungi have wider enzymatic capabilities and a higher capacity for the decomposition of plant polymers than bacteria do, which means that fungi provide bacteria with resources that the bacteria are not able to acquire on their own (Romani et al., 2006; Sun et al., 2018).
The microbial community played an important role in tomato yield (Figure 6). There are several reasons for this: (i) continuous high fertilizer loading increased N availability (Table 2), which reduces the belowground allocation of recent photosynthate C by plants (Högberg et al., 2010), and as a consequence, the microbial biomass C and basal respiration rates (Ramirez et al., 2012), and the abundance of community members decline, such as ectomycorrhizal fungi (Choma et al., 2017); (ii) fertilization intensities can alter microbial community structure (Figures 3, 4), and Olpidium (a potential phytopathogenic), which caused the negative effect on tomato plants (Usero et al., 2021), was positively affected by inorganic fertilizer; (iii) the relative abundance of Actinobacteria was decreased under long-term overfertilization, which inhibited the direct antagonism between Actinobacteria and antibiotics produced by fungi pathogens (Zhao et al., 2019). The number and types of beneficial microorganisms decreased, and harmful microorganisms enhanced in the soil unbalanced the microbial community, which can cause a significant decrease in soil quality and crop growth (Li et al., 2019; Zhou et al., 2021). Thus, the decrease in tomato yield under overfertilization in our study has a close relationship with the altering of soil pH, soil TC, and microbial community composition.
Conclusion
Long-term overfertilization enhanced soil EC, increased nutrient enrichment, and decreased soil pH. In contrast, the control had a lower soil EC and soil nutrient concentrations and had a higher soil pH than overfertilization. Overfertilization significantly decreased bacterial diversity and richness than control in 12-year soils. The relative abundance of Gp6 and Gemmatimonas was significantly decreased in overfertilization compared to the control, which destroyed the balance of the microbial community. Furthermore, overfertilization decreased the tomato yield by 38% in 12 years compared to the control. The results confirmed that overfertilization has a negative effect on tomato yield under long-term continuous cropping. Collectively, long-term overfertilization intensified soil acidification and altered microbial community composition, thus decreasing tomato yield. Therefore, appropriate reductions in the use of inorganic fertilizers in greenhouse cultivation systems are critical to soil sustainability.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
TL, HF, ZS, and QiaS designed the experiment. QiaS, XS, ZW, and QinS conducted the experiment and collected data for preliminary analysis. QiaS and HF further analyzed the data and prepared the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 31902093) and the National Key R&D Program of China (Grant No. 2019YFD1001900).
Acknowledgments
The authors thank the staff for collecting samples who are not listed as coauthors.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.952021/full#supplementary-material
Abbreviations
AK, Available potassium; AMC, 7-amino-4-methylcoumarin; AN, Available nitrogen; AP, Available phosphorous; BG, β-1, 4-glucosidase; C, Cropping duration; DOPA, L-dihydroxyphenylalanine; EC, Electrical conductivity; F, Fertilization; LAP, Leucine aminopeptidase; MUB, 4-methylumbelliferyl; NH-N, Ammonium nitrogen; NMDS, Non-metric multidimensional scaling; NO-N, Nitrate nitrogen; OF, Overfertilization; PER, Peroxidase; PPO, Polyphenol oxidase; RDA, Redundancy analysis; SEM, Structural equation modeling; SRA, Sequence read archive; TC, Total carbon; TK, Total potassium; TN, Total nitrogen; TP, Total phosphorous; VIFs, Variance inflation factors.
References
Ali, A., Ghani, M. I., Ding, H. Y., Iqbal, M., Cheng, Z. H., and Cai, Z. C. (2020). Garlic substrate induces cucumber growth development and decreases Fusarium Wilt through regulation of soil microbial community structure and diversity in replanted disturbed soil. Int. J. Mol. Sci. 21, 6008–6030. doi: 10.3390/ijms21176008
Bertin, C., Yang, X., and Weston, L. A. (2003). The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 256, 67–83. doi: 10.1023/A:1026290508166
Blakemore, L. C., Searle, P. L., and Daly, B. K. (1987). Methods for Chemical Analysis of Soils. New Zealand Soil Bureau Scientific Report.
Byrne, B. M., and Erlbaums, L. (2009). Structural Equation Modeling with AMOS: Basic Concepts, Applications, and Programming. New York, NY: Routledge.
Chang, E. H., Chung, R. S., and Tsai, Y. H. (2007). Effect of different application rates of organic fertilizer on soil enzyme activity and microbial population (soil biology). Soil Sci. Plant Nutr. 53, 132–140. doi: 10.1111/j.1747-0765.2007.00122.x
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, 884–890. doi: 10.1093/bioinformatics/bty560
Choma, M., Rappe-George, M. O., Bárta, J., Capek, P., Gärdenäs, A.I., and Šantručková, H. (2017). Recovery of the ectomycorrhizal community after termination of long-term nitrogen fertilisation of a boreal Norway spruce forest. Fungal Ecol. 29, 116–122. doi: 10.1016/j.funeco.2016.10.002
Chung, I. M., Seigler, D., Miller, D. A., and Kyung, S. H. (2000). Autotoxic compounds from fresh alfalfa leaf extracts: identification and biological activity. J. Chem. Ecol. 26, 315–327. doi: 10.1023/A:1005466200919
Dai, Z. M., Su, W. Q., Chen, H. H., Barberan, A., Zhao, H. C., Yu, M. J., et al. (2018). Long-term nitrogen fertilization decreases bacterial diversity and favors the growth of Actinobacteria and Proteobacteria in agro-ecosystems across the globe. Global Change Biol. 24, 1–10. doi: 10.1111/gcb.14163
Ding, J., Jiang, X., Guan, D., Zhao, B., Ma, M., Zhou, B., et al. (2017). Influence of inorganic fertilizer and organic manure application on fungal communities in a long-term field experiment of Chinese Mollisols. Appl. Soil Ecol. 111, 114–122. doi: 10.1016/j.apsoil.2016.12.003
Dong, W. Y., Zhang, X. Y., Dai, X. Q., Fu, X. L., Yang, F. T., Liu, X. Y., et al. (2014). Changes in soil microbial community composition in response to fertilization of paddy soils in subtropical China. Appl. Soil Ecol. 84, 140–147. doi: 10.1016/j.apsoil.2014.06.007
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edwards, I. P., Zak, D. R., Kellner, H., Eisenlord, S. D., and Pregitzer, K. S. (2011). Simulated atmospheric N deposition alters fungal community composition and suppresses ligninolytic gene expression in a northern hardwood forest. PLoS ONE 6, e20421. doi: 10.1371/journal.pone.0020421
German, D. P., Weintraub, M. N., Grandy, A. S., Lauber, C. L., Rinkes, Z. L., and Allison, S. D. (2011). Optimization of hydrolytic and oxidative enzyme methods for ecosystem studies. Soil Biol. Biochem. 43, 1387–1397. doi: 10.1016/j.soilbio.2011.03.017
Ghannoum, M. A., Jurevic, R. J., Mukherjee, P. K., Cui, F., Sikaroodi, M., Naqvi, A., et al. (2010). Characterization of the oral fungal microbiome (mcobiome) in healthy individuals. PLoS Pathog. 6, e1000713. doi: 10.1371/journal.ppat.1000713
Guo, J. H., Liu, X. J., Zhang, Y., Shen, J. L., Han, W. X., Zhang, W. F., et al. (2010). Significant acidification in major chinese croplands. Science 327, 1008–1010. doi: 10.1126/science.1182570
Guo, L. Z., Zheng, S. X., Cao, C. G., and Li, C. F. (2016). Tillage practices and straw-returning methods affect topsoil bacterial community and organic c under a rice-wheat cropping system in central China. Sci. Rep. 6, 33155–33165. doi: 10.1038/srep33155
Han, J., Luo, Y., Yang, L., Liu, X., Wu, L., and Xu, J. (2014). Acidification and salinization of soils with different initial pH under greenhouse vegetable cultivation. J. Soil Sediment. 14, 1683–1692. doi: 10.1007/s11368-014-0922-4
Han, J., Shi, J., Zeng, L., Xu, J., and Wu, L. (2016). Impacts of continuous excessive fertilization on soil potential nitrification activity and nitrifying microbial community dynamics in greenhouse system. J. Soil Sediment. 17, 471–480. doi: 10.1007/s11368-016-1525-z
Högberg, M. N., Briones, M. J. I., Keel, S. G., Metcalfe, D. B., Campbell, C., Midwood, A. J., et al. (2010). Quantification of effects of season and nitrogen supply on tree below-ground carbon transfer to ectomycorrhizal fungi and other soil organisms in a boreal pine forest. N. Phytol. 187, 485–493. doi: 10.2307/40792394
Jones, R. T., Robeson, M. S., Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 3, 442–453. doi: 10.1038/ismej.2008.127
Ju, X. T., Kou, C. L., Christie, P., Dou, Z. X., and Zhang, F. S. (2007). Changes in the soil environment from excessive application of fertilizers and manures to two contrasting intensive cropping systems on the north China plain. Environ. Pollut. 145, 497–506. doi: 10.1016/j.envpol.2006.04.017
Kahlaoui, B., Hachicha, M., Rejeb, S., Misle, E., Rouaissi, M., Rejeb, M. N., et al. (2011). Effect of saline water on tomato under subsurface drip irrigation: yield and fruit quality. J. Appl. Sci. R. 11, 69–86. doi: 10.4067/S0718-95162011000100007
Lazcano, C., Gómez-Brandón, M., Revilla, P., and Dominguez, J. (2013). Short-term effects of organic and inorganic fertilizers on soil microbial community structure and function. Biol. Fert. Soils 49, 723–733. doi: 10.1007/s00374-012-0761-7
Li, F., Chen, L., Zhang, J., Yin, J., and Huang, S. (2017). Bacterial community structure after long-term organic and inorganic fertilization reveals important associations between soil nutrients and specific taxa involved in nutrient transformations. Front. Microbiol. 8, 187. doi: 10.3389/fmicb.2017.00187
Li, X. G., Jousset, A., de Boer, W., Carrión, V. J., Zhang, T. L., Wang, X. X., et al. (2019). Legacy of land use history determines reprogramming of plant physiology by soil microbiome. ISME J. 13, 738–751. doi: 10.1038/S41396-018-0300-0
Liang, H., Chen, Q., Liang, B., Hu, K., Hansen, J. W., Thornton, P. K., et al. (2020). Modeling the effects of long-term reduced n application on soil n losses and yield in a greenhouse tomato production system. Agric. Syst. 185, 1–11. doi: 10.1016/j.agsy.2020.102951
Liu, H. B., Li, Z. H., and Zhang, W. L. (2004). Study on n use efficiency of chinese cabbage and nitrate leaching under open field cultivation. Plant Nutr. Ferti. Sci. 10, 286–291. doi: 10.1300/J064v24n01_09
Liu, S., Wang, Z. Y., Niu, J. F., Dang, K. K., Zhang, S. K., Wang, S. Q., et al. (2021). Changes in physicochemical properties, enzymatic activities, and the microbial community of soil significantly influence the continuous cropping of Panax quinquefolius L. (American ginseng). Plant Soil 463, 427–446. doi: 10.1007/s11104-021-04911-2
Liu, Z. X., Liu, J. J., Yu, Z. H., Yao, Q., Li, Y. S., Liang, A. Z., et al. (2020). Long-term continuous cropping of soybean is comparable to crop rotation in mediating microbial abundance, diversity and community composition. Soil Till. Res. 197, 104503–104515. doi: 10.1016/j.still.2019.104503
Lu, R. K. (2000). Methods for Soil Agro-Chemistry Analysis. China Agricultural Science and Technology Press, Beijing.
Ma, L. F., Yang, X. D., Shi, Y. Z., Yi, X. Y., Ji, L. F., Cheng, Y., et al. (2021). Response of tea yield, quality and soil bacterial characteristics to long-term nitrogen fertilization in an eleven-year field experiment. Appl. Soil Ecol. 166, 103976–103987. doi: 10.1016/j.apsoil.2021.103976
Ma, M. C., Zhou, J., Ongena, M., Liu, W. Z., Wei, D., Zhao, B. S., et al. (2018). Effect of long-term fertilization strategies on bacterial community composition in a 35-year field experiment of chinese mollisols. AMB Express. 8, 20–31. doi: 10.1186/s13568-018-0549-8
Magarey, R. C. (1999). Reduced productivity in long term monoculture: where are we placed? Australas. Plant Path. 28, 11–20. doi: 10.1071/AP99002
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Masto, R. E., Chhonkar, P. K., Singh, D., and Patra, A. K. (2007). Soil quality response to long-term nutrient and crop management on a semi-arid inceptisol. Agric. Ecosyst. Environ. 118, 130–142. doi: 10.1016/j.agee.2006.05.008
Min, J., Zhao, X., Shi, W. M., Xing, G. X., and Zhao, Z. L. (2011). Nitrogen balance and loss in a greenhouse vegetable system in southeastern China. Pedosphere 21, 464–472. doi: 10.1016/S1002-0160(11)60148-3
Mori, H., Maruyama, F., Kato, H., Toyoda, A., Dozono, A., Ohtsubo, Y., et al. (2013). Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res. 21, 217–227. doi: 10.1093/dnares/dst052
Ning, Q., Chen, L., Jia, Z. J., Zhang, C. Z., Ma, D. H., Li, F., et al. (2020). Multiple long-term observations reveal a strategy for soil pH-dependent fertilization and fungal communities in support of agricultural production. Agric. Ecosyst. Environ. 293, 1–10. doi: 10.1016/j.agee.2020.106837
Ning, Q. S., Hattenschwiler, S., Lü, X. T., Kardol, P., Zhang, Y. H., Wei, C. Z., et al. (2021). Carbon limitation overrides acidification in mediating soil microbial activity to nitrogen enrichment in a temperate grassland. Global Change Biol. 8, 1–13. doi: 10.1111/gcb.15819
Nobile, C. M., Bravin, M. N., Becquer, T., and Paillat, J. M. (2020). Phosphorus sorption and availability in an andosol after a decade of organic or mineral fertilizer applications: importance of pH and organic carbon modifications in soil as compared to phosphorus accumulation. Chemosphere 239, 124709–124719. doi: 10.1016/j.chemosphere.2019.124709
Omay, A. B., Rice, C. W., Maddux, L. D., and Gordon, W. B. (1997). Changes in soil microbial and chemical properties under long-term crop rotation and fertilization. Soil Sci. Soc. Am. J. 61, 1672–1678. doi: 10.2136/sssaj1997.03615995006100060019x
Park, D., Kim, H., and Yoon, S. (2017). Nitrous oxide reduction by an obligate aerobic bacterium, Gemmatimonas aurantiaca strain T-27. Appl. Environ. Microbiol. 83, 502–517. doi: 10.1128/AEM.00502-17
Puissant, J., Jones, B., Goodall, T., Mang, D., Blaud, A., Gweon, H. S., et al. (2019). The pH optimum of soil exoenzymes adapt to long term changes in soil pH. Soil Biol. Biochem. 138, 107601–107610. doi: 10.1016/j.soilbio.2019.107601
Ramirez, K. S., Craine, J. M., and Fierer, N. (2012). Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob. Change Biol. 18, 1918–1927. doi: 10.1111/j.1365-2486.2012.02639.x
Reeves, D. W. (1997). The role of soil organic matter in maintaining soil quality in continuous cropping systems. Soil Till. Res. 43, 131–167. doi: 10.1016/S0167-1987(97)00038-X
Romani, A. M., Fischer, H., Mille-Lindblom, C., and Tranvik, L. J. (2006). Interactions of bacteria and fungi on decomposing litter: differential extracellular enzyme activities. Ecology 87, 2559–2569. doi: 10.1890/0012-9658(2006)87[2559:IOBAFO[2.0.CO;2
Sainju, U. M., Singh, B. P., Rahman, S., and Reddy, V. R. (2000). Tillage, cover cropping and nitrogen fertilization influence tomato yield and nitrogen uptake. Hortscience 35, 217–221. doi: 10.1016/S0304-4238(99)00110-7
Sainju, U. M., Singh, B. P., and Whitehead, W. F. (2001). Comparison of the effects of cover crops and nitrogen fertilization on tomato yield, root growth, and soil properties. Sci. Hortic. 91, 201–214. doi: 10.1016/S0304-4238(01)00264-3
Shang, Q. Y., Ling, N., Feng, X. M., Yang, X. X., Wu, P. P., Zou, J. W., et al. (2014). Soil fertility and its significance to crop productivity and sustainability in typical agroecosystem: a summary of long-term fertilizer experiments in China. Plant Soil 381, 13–23. doi: 10.1007/s11104-014-2089-6
Shi, G. Y., Sun, H. Q., Calderón-Urrea, A., Li, M. Q., Yang, H. Y., Wang, W. Z., et al. (2021). Bacterial communities as indicators for soil health under continuous cropping system. Land Degrad. Dev. 32, 2393–2408. doi: 10.1002/ldr.3919
Shi, W. M., Yao, J., and Yan, F. (2009). Vegetable cultivation under greenhouse conditions leads to rapid accumulation of nutrients, acidification and salinity of soils and groundwater contamination in South-Eastern China. Nutr. Cycl. Agroecosyst. 83, 73–84. doi: 10.1007/s10705-008-9201-3
Song, H., Chen, Z., Cao, W. C., Huang, T., Wang, J. G., and Dong, Z. R. (2016). Changing roles of ammonia-oxidizing bacteria and archaea in a continuously acidifying soil caused by over-fertilization with nitrogen. Environ. Sci. Pollut. R. 23, 11964–11974. doi: 10.1007/s11356-016-6396-8
Stackebrandt, E., and Goebel, B. M. (1994). Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44, 846–849. doi: 10.1099/00207713-44-4-846
Su, J. Q., Ding, L. J., Xue, K., Yao, H. Y., Quensen, J., Bai, S. J., et al. (2015). Long-term balanced fertilization increases the soil microbial functional diversity in a phosphorus-limited paddy soil. Mol. Ecol. 24, 136–150. doi: 10.1111/mec.13010
Sun, R., Li, W., Dong, W., Tian, Y., Hu, C., and Liu, B. (2018). Tillage changes vertical distribution of soil bacterial and fungal communities. Front. Microbiol. 9, 699. doi: 10.3389/fmicb.2018.00699
Tan, G., Liu, Y. J., Peng, S. G., Yin, H. Q., Meng, D. L., Tao, J. M., et al. (2021). Soil potentials to resist continuous cropping obstacle: three field cases. Environ. Res. 200, 111319–111329. doi: 10.1016/j.envres.2021.111319
Usero, F. M., Armas, C., Morillo, J. A., Gallardo, M., and Pugnaire, F. I. (2021). Effects of soil microbial communities associated to different soil fertilization practices on tomato growth in intensive greenhouse agriculture. Appl. Soil Ecol. 162, 103896–103907. doi: 10.1016/j.apsoil.2021.103896
Wallenstein, M. D., McNulty, S., Fernandez, I. J., Boggs, J., and Schlesinger, W. H. (2006). Nitrogen fertilization decreases forest soil fungal and bacterial biomass in three long-term experiments. Forest Ecol. Manag. 222, 459–468. doi: 10.1016/j.foreco.2005.11.002
Wang, H., Nie, Y., Butterrly, C. R., Wang, L., Chen, Q., Tian, W., et al. (2017). Fertilization alters microbial community composition and functional patterns by changing the chemical nature of soil organic carbon: a field study in a Halosol. Geoderma 292, 17–24. doi: 10.1016/j.geoderma.01.006
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, X., and Xing, Y. (2016). Effects of irrigation and nitrogen fertilizer input levels on soil NO3–N content and vertical distribution in greenhouse tomato (Lycopersicum esculentum Mill.). Scientifica 2016, 370–377. doi: 10.1155/2016/5710915
Wu, H. W., Pratley, J., Lemerle, D., An, M., and Liu, D. L. (2007). Autotoxicity of wheat (Triticum aestivum L.) as determined by laboratory bioassays. Plant Soil 296, 85–93. doi: 10.1007/s11104-007-9292-7
Xiang, X., Liu, J., Zhang, J., Li, D., Xu, C., and Kuzyakov, Y. (2020). Divergence in fungal abundance and community structure between soils under long-term mineral and organic fertilization. Soil Till. Res. 196, 104491–104498. doi: 10.1016/j.still.2019.104491
Xing, G. X., and Zhu, Z. L. (2002). Regional nitrogen budgets for China and its major watersheds. Biogeochemistry 57, 405–427. doi: 10.1023/A:1016508323200
Xu, Y. G., Yu, W. T., Ma, Q., and Hua, Z. (2012). Responses of bacterial and archaeal ammonia oxidisers of an acidic luvisols soil to different nitrogen fertilization rates after 9 years. Biol. Fert. Soils 48, 827–837. doi: 10.1007/s00374-012-0677-2
Yang, X. D., Ni, K., Shi, Y. Z., Yi, X. Y., Zhang, Q. F., Fang, L., et al. (2018). Effects of long-term nitrogen application on soil acidification and solution chemistry of a tea plantation in China. Agric. Ecosyst. Environ. 252, 74–82. doi: 10.1016/j.agee.2017.10.004
Yao, Z. S., Yan, G. X., Wang, R., Zheng, X. H., Liu, C. Y., and Butterbach-Bahl, K. (2019). Drip irrigation or reduced N-fertilizer rate can mitigate the high annual N2O + NO fluxes from Chinese intensive greenhouse vegetable systems. Atmos. Environ. 212, 183–193. doi: 10.1016/j.atmosenv.2019.05.056
Yin, C. T., Hulbert, S. H., Schroeder, K. L., Mavrodi, O., Mavrodi, D., Dhingra, A., et al. (2013). Role of bacterial communities in the natural suppression of Rhizoctonia solani bare patch disease of wheat (Triticum aestivum L.). Appl. Environ. Microbiol. 79, 7428–7438. doi: 10.1128/aem.01610-13
Yu, J. Q. (2001). Autotoxic potential of cucurbit crops. J. Crop Prod. 4, 335–348. doi: 10.1300/J144v04n02_15
Yuan, X. B., Niu, D. C., Weber-Grullon, L., and Fu, H. (2020). Nitrogen deposition enhances plant-microbe interactions in a semiarid Grassland: the role of soil physicochemical properties. Geoderma 373, 114446–114458. doi: 10.1016/j.geoderma.2020.114446
Zeng, Y., Nupur, W.u N., Madsen, A. M., and Koblíek, M. (2021). Gemmatimonas groenlandica sp. nov. is an aerobic anoxygenic phototroph in the phylum gemmatimonadetes. Front. Microbiol. 11, 606612. doi: 10.3389/fmicb.2020.606612
Zhang, W. L., Tian, Z. X., Zhang, N., and Li, X. Q. (1996). Nitrate pollution of groundwater in northern China. Agric. Ecosyst. Environ. 59, 223–231. doi: 10.1016/0167-8809(96)01052-3
Zhang, X. M., Zhang, Q., Liang, B., and Li, J. L. (2017). Changes in the abundance and structure of bacterial communities in the greenhouse tomato cultivation system under long-term fertilization treatments. Appl. Soil Ecol. 121, 82–89. doi: 10.1016/j.apsoil.2017.08.016
Zhang, Y. T., Shen, H., He, X. H., Thomas, B. W., Lupwayi, N. Z., Hao, X. Y., et al. (2017). Fertilization shapes bacterial community structure by alteration of soil pH. Front. Microbiol. 8, 1325–1336. doi: 10.3389/fmicb.2017.01325
Zhao, F. Y., Zhang, Y. Y., Dong, W. G., Zhang, Y. Q., Zhang, G. X., Sun, Z. P., et al. (2019). Vermicompost can suppress fusarium oxysporum F. sp. lycopersici via generation of beneficial bacteria in a long-term tomato monoculture soil. Plant Soil 440, 491–505. doi: 10.1007/s11104-019-04104-y
Zheng, Q., Hu, Y. T., Zhang, S. S., Noll, L., Bockle, T., Dietrich, M., et al. (2019). Soil multifunctionality is affected by the soil environment and by microbial community composition and diversity. Soil Biol. Biochem. 136, 107521–107534. doi: 10.1016/j.soilbio.2019.107521
Zhou, J., Jiang, X., Zhou, B. K., Zhao, B. S., Ma, M. C., Guan, D. W., et al. (2016). Thirty four years of nitrogen fertilization decreases fungal diversity and alters fungal community composition in black soil in northeast China. Soil Biol. Biochem. 95, 135–143. doi: 10.1016/j.soilbio.2015.12.012
Zhou, R. R., Wang, Y., Tian, M. M., Jahan, M. S., Shu, S., Sun, J., et al. (2021). Mixing of biochar, vinegar and mushroom residues regulates soil microbial community and increases cucumber yield under continuous cropping regime. Appl. Soil Ecol. 161, 103883–103895. doi: 10.1016/j.apsoil.2021.103883
Keywords: long-term continuous cropping, overfertilization, soil acidification, microbial community, structural equation model
Citation: Song Q, Fu H, Shi Q, Shan X, Wang Z, Sun Z and Li T (2022) Overfertilization reduces tomato yield under long-term continuous cropping system via regulation of soil microbial community composition. Front. Microbiol. 13:952021. doi: 10.3389/fmicb.2022.952021
Received: 24 May 2022; Accepted: 11 July 2022;
Published: 04 August 2022.
Edited by:
Xiaogang Li, Nanjing Forestry University, ChinaReviewed by:
Xingang Zhou, Northeast Agricultural University, ChinaDeyan Liu, Institute of Soil Science (CAS), China
Copyright © 2022 Song, Fu, Shi, Shan, Wang, Sun and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongdan Fu, ZnVob25nZGFuQHN5YXUuZWR1LmNu; Tianlai Li, dGlhbmxhaWxpQDEyNi5jb20=
 Qiaobo Song
Qiaobo Song Hongdan Fu
Hongdan Fu Qingwen Shi
Qingwen Shi Xuan Shan1,2
Xuan Shan1,2