- 1State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Université Paris-Saclay, INRAE, AgroParisTech, Micalis Institute, Jouy-en-Josas, France
- 3Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
The novel protein MclX (mother cell lysis X) in Bacillus thuringiensis subsp. kurstaki strain HD73 (B. thuringiensis HD73) was characterized in this work. MclX has no known domain and its gene deletion in HD73 resulted in Cry1Ac encapsulation in the mother cell and did not influence Cry1Ac protein production or insecticidal activity. In vitro cell wall hydrolysis experiments showed that MclX cannot hydrolyze the cell wall. In mclX deletion mutants, the expression of cwlC (which encodes a key cell wall hydrolase) was significantly decreased, as shown by the β-galactosidase activity assay. MclX cannot directly bind to the cwlC promoter, based on the electrophoretic mobility shift assay (EMSA). The cwlC was reported to be regulated by σK and GerE. However, the transcriptional activities of sigK and gerE showed no difference between HD73 and the mclX deletion mutant. It is indicated that MclX influenced cwlC expression independently of σK or GerE, through a new pathway to regulate cwlC expression. mclX deletion could be a new approach for insecticidal protein encapsulation in Bacillus thuringiensis.
Introduction
Bacillus thuringiensis is a microbial insecticide used worldwide in agriculture (Jouzani et al., 2017). The most important feature of B. thuringiensis is the formation of spores and the production of insecticidal parasporal crystals. When B. thuringiensis grown to mature, the spore and crystal protein was released from mother cell. The crystal protein is toxic to Lepidoptera, Coleoptera, Diptera and other agricultural pests (Beegle and Yamamoto, 1992; Jouzani et al., 2017). However, the insecticidal activity of crystal protein was frequently reduced by environmental factors such as sunlight exposure (Myasnik et al., 2001; Zogo et al., 2019). Encapsulation of crystal proteins by physicochemical, mechanical, or molecular biological techniques can protect their activity from sunlight exposure (Lereclus et al., 1995; Manasherob et al., 2002; Jallouli et al., 2014; Chen et al., 2018; de Oliveira et al., 2021).
Encapsulation of crystal protein by physicochemical and mechanical techniques has some advantages and disadvantages. Encapsulation by extrusion is simple to use but low productivity, encapsulation by fluidized bed is high productivity but difficult to control, encapsulation by electrospinning is easy to expand production but the media may be toxic (de Oliveira et al., 2021). Crystal protein encapsulation in mother cell by molecular biological techniques was proved to be a good strategy to improve Cry protein stability under ultraviolet (UV) radiation (Sanchis et al., 1999; Zhou et al., 2014). The mother cell wall can protect Cry protein from inactivation by UV radiation. Deletion of sigK led to blocking mother cell lysis, encapsulating crystal proteins within the mother cell wall. This technology not only constructed spore-free strain, which had no pollution to the environment and no competition with wild B. thuringiensis strains, but also improved the UV resistance of crystal proteins in the field condition (Sanchis et al., 1999). Encapsulation Cry1Ba in the mother cell of sigK mutant also increasing UV radiation resistance (Zhou et al., 2014). However, some cry genes are regulated by σK, and cry gene expressions were reduced in the sigK deletion mutant (Adams et al., 1991; Bravo et al., 1996).
Cell wall hydrolases are important in cell wall metabolism, they are classified as glycosidases and peptidases. Glycosidases (glucosaminidase, muramidase, and lytic transglycosylase) and peptidases (amidase, endopeptidase, and carboxypeptidase) cleave different sites of cell wall peptidoglycan (Do et al., 2020a). In B. subtilis, cell wall hydrolases involved in mother cell lysis (CwlB, CwlH, CwlC) are amidases with functional redundancy. Single amidase gene deletion did not affect mother cell lysis, while cwlB cwlH cwlC deletion mutant significant reduced mother cell lysis (Nugroho et al., 1999). In B. thuringiensis, CwlB and CwlC show low sequence identities with B. subtilis hydrolases. Deletion of the cwlB gene can delay lysis of mother cells (Yang et al., 2013). Deletion of the cwlC can block the lysis of mother cells without influence on Cry1Ac production (Chen et al., 2018). As the essential hydrolase in mother cell lysis, CwlC was reported to be regulated by σK and GerE in B. thuringiensis (Chen et al., 2018). No other genes have been reported to be deleted for encapsulation crystal protein except sigK and cwlC.
In this work, we identified a hypothetical gene whose deletion resulted in Cry1Ac encapsulation in the mother cell. The mother cell did not lysis because deletion of this hypothetical gene reduced the cwlC expression. This discovery of the regulatory mechanism of cwlC is independent from the regulatory pathway of σK or GerE. Studies on this new gene may contribute to a deeply understanding of cwlC expression. The deletion of this new gene encapsulated Cry1Ac in the mother cell with no effect on sporulation frequency, Cry1Ac protein production, or insecticidal activity. This could be a potential new approach for Cry protein encapsulation by molecular biology techniques.
Materials and methods
Bacterial strains, growth conditions, and plasmids
Tables showed the strains (Supplementary Table 1), primers (Supplementary Table 2), and plasmids (Supplementary Table 3) used in this work. The B. thuringiensis subsp. kurstaki strain HD73 (GenBank Accession Number: NC_020238.1) was used for transformations (Du and Nickerson, 1996; Liu et al., 2017). All B. thuringiensis strains were cultured in SSM (Schaeffer’s sporulation medium) (Schaeffer et al., 1965) or LB (Luria-Bertani) medium under 30°C, with 100 μg/ml Kan (Kanamycin) or 5 μg/ml Ery (Erythromycin) if needed. The E. coli strain TG1 was used for constructing the vector (Hoffmann et al., 2015). The E. coli ET12567 was used for extracting non-methylated vectors on a large-scale and transforming into B. thuringiensis. All E. coli strains were grown in LB medium at 37°C, with 100 μg/ml Amp (Ampicillin) if needed.
DNA manipulation and transformation
The EasyPure Plasmid MiniPrep Kit (Transgen, Beijing, China) was used to extract vectors from E. coli cells listed in Supplementary Table 3. The PrimeSTAR Max DNA Polymerase (Takara, Beijing, China) was used to perform PCR, using a Mastercycler X50 (Eppendorf). Primers were synthesized in a company (Sangon, China). The EasyPure Quick Gel Extraction Kit (Transgen, Beijing, China) was used to purified DNA fragments, the Seamless Assembly Cloning Kit (Clonesmarter, United States) was used to connect DNA fragment and the linearized vector. Plasmid was introduced into the strain HD73 by electroporation (GenePulser Xcell, Bio-Rad).
Construction of the mother cell lysis X deletion mutant strain HD (ΔmclX)
To delete the mclX gene in the HD73 genome, pMAD (Arnaud et al., 2004) (a temperature-sensitive suicide plasmid) with the mclX mutation box was constructed. The mclX mutation box was amplified as follows. A 725-bp fragment upstream of mclX (mclX fragment A) which contains a 12-bp overlap with the mclX 5′ end, was amplified by PCR, and mclX-1 and mclX-2 were used as primers. A 677-bp fragment downstream of mclX (mclX fragment B) which starts 4-bp away from mclX 3′ end, was amplified by PCR, and mclX-5 and mclX-6 were used as primers. A 1,473-bp fragment of the kan resistance gene was amplified by PCR, and mclX-3 and mclX-4 were used as primers. The overlapping PCR was used in amplifying the mclX mutation box (mclX fragment A, kan fragment, and mclX fragment B), using mclX-1 and mclX-6 as primers, and the resulting fragment of the mclX mutation box was inserted into the pMAD plasmid. Then, the recombinant plasmid was introduced into HD73. The mclX mutation box allelic replacement was performed as reported (Yang et al., 2013). The mclX ORF (12–663) was replaced by kan fragment which detected by PCR with primers mclX-1 and mclX-6.
Construction of the complemented strain
To construct the complemented strain HD (ΔmclX:mclX), the mclX promoter and open reading frame (PmclX-mclX, 998-bp) were amplified by PCR, using HFmclX-F and HFmclX-R as primers. The PmclX-mclX fragment was inserted into the pHT315 plasmid (Arantes and Lereclus, 1991) to generate pHTHFmclX. The plasmid pHTHFmclX was introduced into HD (ΔmclX) to generate complemented strain HD (ΔmclX:mclX).
Construction of a PmclX-lacZ fusion strain
To analyze the transcriptional activity of the mclX promoter, a 335-bp fragment upstream of the mclX ATG start codon was amplified by PCR, using PmclX-F and PmclX-R as primers. The PmclX fragment was inserted into the pHT304-18Z plasmid (Hervé and Lereclus, 1994) carrying the lacZ gene. The recombinant plasmid pHTPmclX was introduced into the HD73 strain, mutant strain HD (ΔsigK) or HD (ΔgerE).
Construction of a mother cell lysis X-His fusion protein
To express the MclX protein in an E. coli strain, an mclX gene fragment without a termination codon was amplified by PCR, using MclX-F and MclX-R as primers. The resulting fragment was inserted into the pET21b plasmid to generate pETmclX with a C-terminal His tag. The plasmid pETmclX was transformed into the BL21 strain for expressing the MclX-His protein.
Expression and purification of mother cell lysis X-His protein
The method of protein expression and purification was previously described (Zhang et al., 2020). A BL21(pETmclX) strain was grown in LB liquid medium until reaching an OD600 of 0.7. Then, 1 mM isopropyl-beta-D-thiogalactoside (Solarbio, Beijing, China) was added to induce MclX-His expression, and bacteria were harvested after overnight growth at 18°C and sonicated (CP750, Cole-Parmer). The supernatant was loaded onto a 2-ml suspension of Ni Sepharose 6 Fast Flow (GE Healthcare, Sweden) with wash buffer (25 mM Tris-HCl, 50 mM imidazole, 0.5 M NaCl, pH 8.0) and elution buffer (25 mM Tris-HCl, 250 mM imidazole, 0.5 M NaCl, pH 8.0). The purified MclX-His protein was analyzed by 10% SDS–PAGE (Mini protein III, Bio-rad).
β-galactosidase assays
The method of the β-galactosidase activity assay was previously described (Miller, 1972). The error bars represent three independent experiment results.
Sporulation frequency analysis
Liquid samples (50-ml) of HD73 and HD (ΔmclX) were harvested at T24 (24 h after the exponential phase ended). Heated samples (under 65°C for 20 min) and unheated samples were diluted in a gradient, coated on the LB agar plates. Then the number of grown colonies in heated and unheated sample plates was counted. The sporulation frequency is the clone number ratio of the heated sample to the unheated sample. The error bars represent three independent experiment results.
Cry1Ac protein production
The method was performed as previous described (Zhang et al., 2018). The wild-type strain HD73 and the strain HD (ΔmclX) were harvested at T24 and lyophilized into powder, same quality of samples was taken and determined by 10% SDS–PAGE (Bio-rad). The results of three independent experiments are consistent.
Cell wall preparation and hydrolysis
For cell wall preparation and hydrolysis, method was previously described (Yang et al., 2013; Chen et al., 2018). CwlC added to cell wall was used as a positive control. The cell wall itself was used as a negative control, and the weight of MclX was the same as that of CwlC. The results of three independent experiments are consistent.
Bioassay of insecticidal activity
A bioassay was carried out as previous described (Chen et al., 2018). Each test was conducted with three independent cultures replicated.
Western blot analysis
Cell lysates (the equivalent of 2 ml at the time point) were suspended in 600 μl of Tris buffer (pH 8.0) and analyzed by 10% SDS–PAGE (Bio-rad). Anti-CwlC was generated by a company (Beijing Protein Innovation Inc., China), and the second antibody (HRP-conjugated goat anti-mouse IgG) was bought (CWBiotech, Beijing, China). The results of three independent experiments are consistent.
Electrophoretic mobility shift assays
EMSA (Electrophoretic mobility shift assay) was carried out as previous described (Shen et al., 2021). The cwlC promoter was labeled with 6-carboxyfluorescein (FAM) (Supplementary Table 3). The results of three independent experiments are consistent.
Results
Deletion of the hypothetical gene HD73_RS12920 can encapsulate Cry1Ac in mother cell
To search for key genes involved in mother cell lysis, T7 transcriptome data (Tn stands for n hours after the exponential phase ended) of the strain HD73 was analyzed (Peng et al., 2015). T7 stands for the late sporulation stage, at which the exosporium genes of the spore coat begin to be transcribed (Peng et al., 2016). Transcription of cwlC had not started at T7, which means that mother cell lysis also had not started. Highly expressed hypothetical genes were screened (Supplementary Table 4) and genetic deletion mutants were constructed.
Fortunately, deletion of a hypothetical gene showed a phenotype in which the mother cell did not lyse. This hypothetical gene is HD73_RS12920 (which encodes a protein in NCBI with the reference sequence WP_000101499.1). HD73_RS12920 is 663-bp in length and has no known domains. According to the deletion mutant phenotype, which displayed no mother cell lysis, HD73_RS12920 was named mclX (mother cell lysis X).
In the mclX deletion mutant HD (ΔmclX) (method described in “Materials and Methods”), the mother cell did not lyse, while the mother cells of wild-type strain HD73 lysed at 1, 3, and 5 days after the bacterial growth reached the end of the exponential phase (Figure 1A). There was no difference in vegetative growth between the wild-type strain HD73 and HD (ΔmclX) (Supplementary Figure 1). To further demonstrate the role of MclX in mother cell lysis, we constructed a genetically complemented strain HD (ΔmclX:mclX), as described in section “Materials and methods.” In the mother cell lysis stage, most wild-type and HD (ΔmclX:mclX) mother cells have lysed (Supplementary Figure 2). The results showed that MclX played an important role in mother cell lysis.
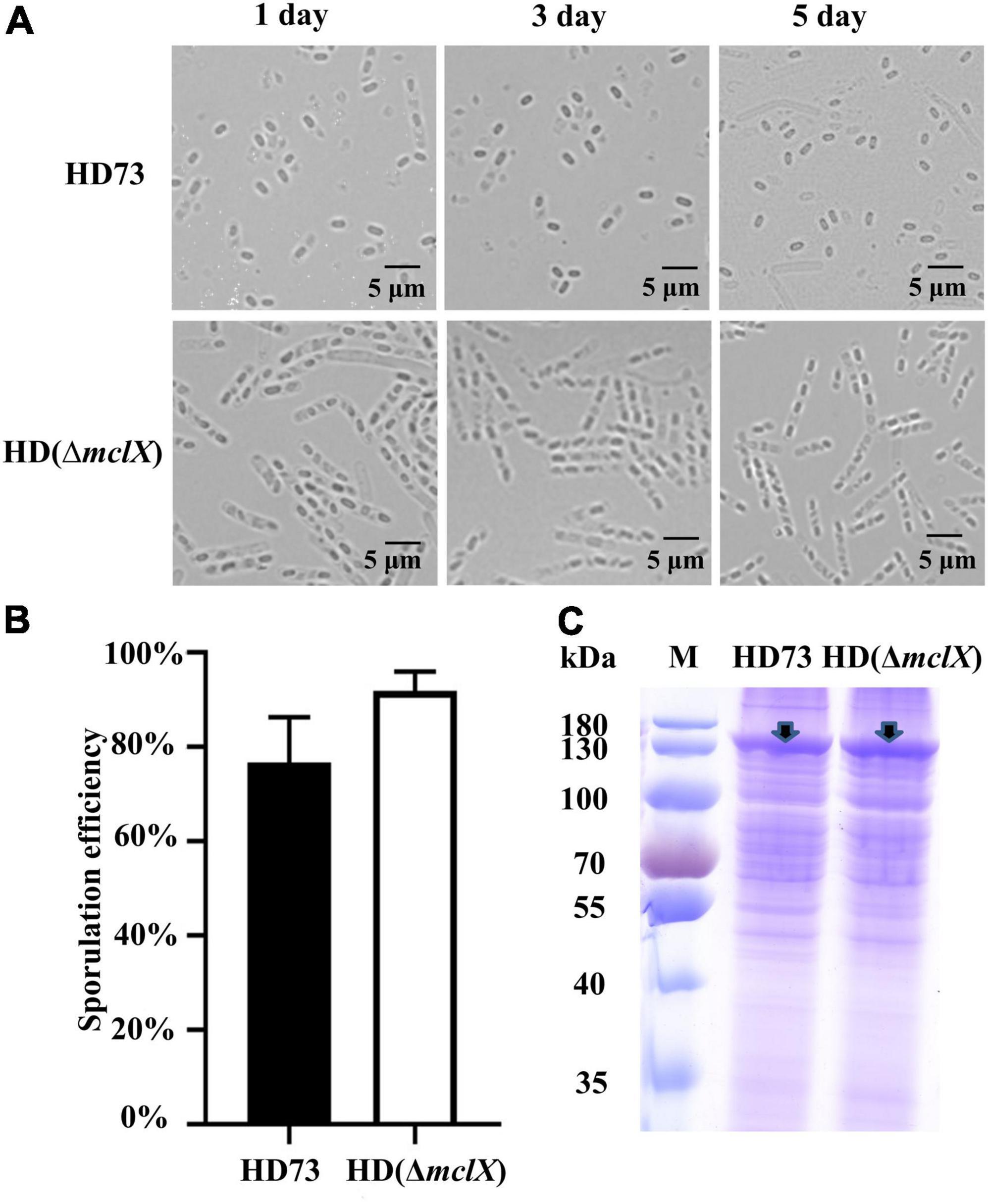
Figure 1. Phenotype analysis of the HD (ΔmclX) strain. (A) Phenotype of the wild-type strain HD73 and HD (ΔmclX) at 1, 3, and 5 days after the bacterial growth reached the end of the exponential phase. Scale bars, 5 μm; (B) sporulation frequency of the wild-type strain HD73 and HD (ΔmclX). Error bars show the standard error of the mean; (C) Cry1Ac protein production in the wild-type strain HD73 and HD (ΔmclX). Cry1Ac production bands are marked by arrows. Lane M, protein marker (26616).
There were no differences between the wild-type strain HD73 and HD (ΔmclX) in sporulation frequency or Cry1Ac protein production at T24 (Figures 1B,C). The second instar larva of Plutella xylostella was used to determine the insecticidal activity of the wild-type strain HD73 and HD (ΔmclX). P. xylostella larvae were fed prepared cabbages treated with a mixture of spores and crystals of the wild-type strain or the HD (ΔmclX) strain. The LC50 of the wild-type strain HD73 was 8.89 μg/ml and the LC50 of the HD (ΔmclX) strain was 10.2 μg/ml, displaying no significant differences (Table 1). Therefore, the deletion of the mclX encapsulated Cry1Ac in HD73 mother cells, without effect on sporulation frequency, Cry1Ac protein production, or insecticidal activity.
Transcriptional regulation of the mother cell lysis X gene
MclX has no known domain and a molecular size of 25.6 kDa. Secondary structure alignment (PSIPRED)1 revealed the presence of several α-helixes and β-strands in MclX (Supplementary Figure 3). In the B. thuringiensis HD73 genome, mclX has an upstream gene, RS12915, which encodes a YozE family protein, and a downstream gene, RS12925, which encodes a hypothetical protein (Figure 2A). In the B. cereus group, MclX is very conserved (Supplementary Figure 4). Streptococcus pneumoniae, Bacillus subtilis, and Rhodococcus qingshengii, have a MclX homologous protein with 100, 62, and 65% similarity, respectively.
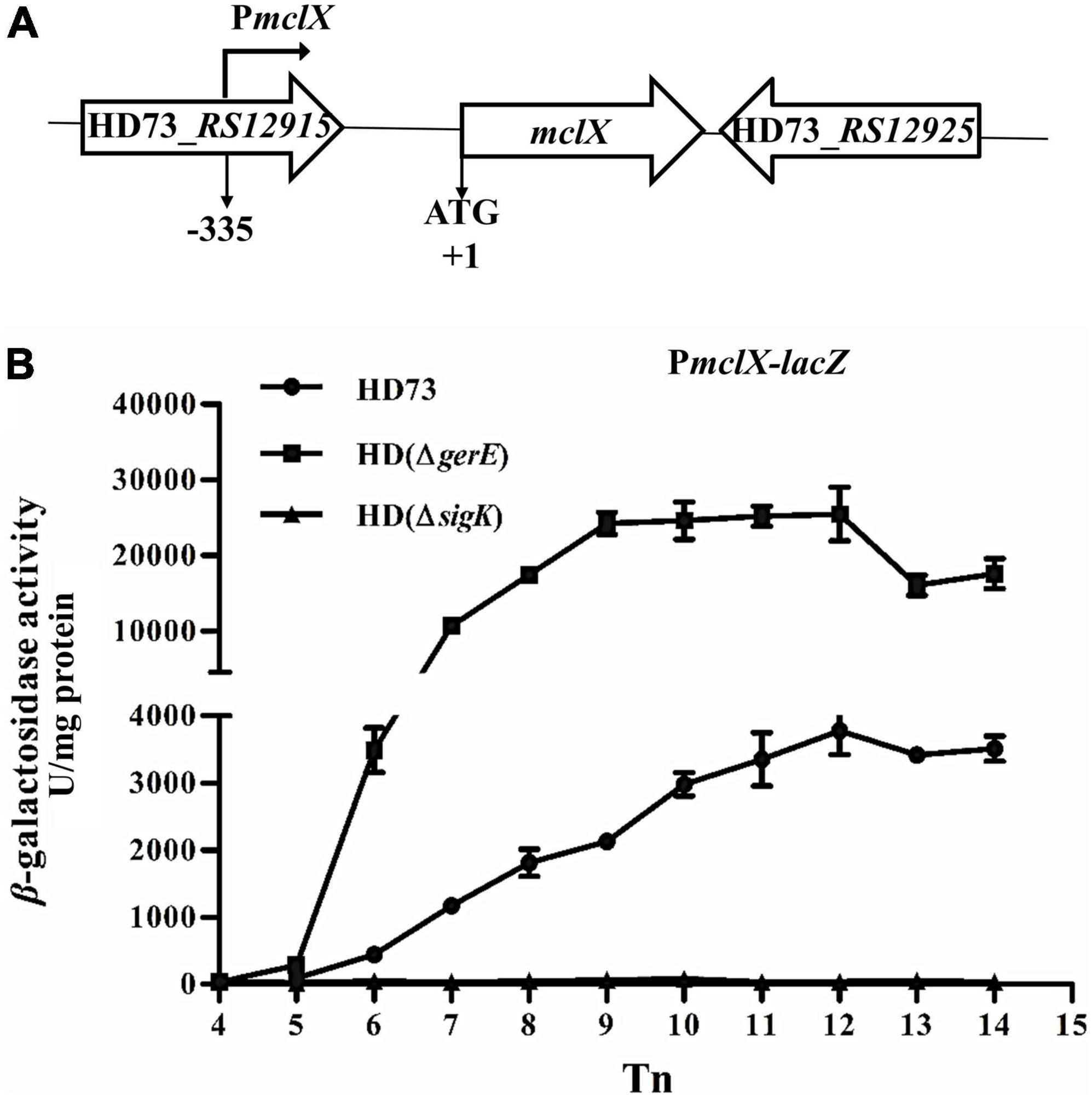
Figure 2. mclX transcription activity. (A) Gene map of the RS12915-RS12925; (B) mclX transcription activities in the wild-type strain HD73, HD (ΔgerE), and HD (ΔsigK) (HD73-PmclX, circles; HDΔgerE-PmclX, squares; HDΔsigK-PmclX, triangles). Tn means n hours after the exponential phase ended. Error bars show standard error of the mean.
The promoter region of the mclX gene (335-bp upstream of the mclX ATG start codon) was fused to the lacZ gene, and transformed into HD (ΔsigK), HD (ΔgerE), and wild-type HD73. In HD73, the transcription of PmclX initiated at T6 and reached a peak at T12 (Figure 2B, circles). In HD (ΔsigK), the transcriptional activity of PmclX was completely abolished (Figure 2B, triangles). In HD (ΔgerE), PmclX transcriptional activity was increased compared to that in wild-type strain HD73 (Figure 2B, squares). All above results demonstrate that mclX was highly expressed in the late sporulation stage, controlled by σK, and negatively regulated by GerE.
Mother cell lysis X cannot hydrolyze the cell wall in vitro
HD (ΔmclX) displayed no mother cell lysis. MclX has no known domains, including DNA binding domains or any functional domains. Regarding its role in mother cell lysis, MclX may be an essential cell wall hydrolase or it may affect the function of the cell wall hydrolase CwlC.
To test whether MclX could be a cell wall hydrolase, the MclX protein hydrolyzed cell wall was analyzed in vitro. The MclX-His protein was expressed and analyzed by SDS–PAGE, its molecular weight was approximately 30 kDa (Figure 3A). The MclX-His protein and the wide-type strain HD73 cell wall were incubated together. The optical density at 540 nm (OD540) showed cell wall turbidity. CwlC is a known hydrolase that can reduce the cell wall turbidity (Figure 3B, squares). However, there was no reduction in the OD540 values of MclX protein-containing cell wall (Figure 3B), suggesting that MclX cannot hydrolyze the cell wall in vitro.
Figure 3. Characterization of the MclX protein. (A) MclX protein expressed in the recombinant strain E. coli BL21 (pETMclX) by SDS–PAGE analysis. M, protein marker, 26614; (B) purified MclX protein was unable to hydrolyze the wild-type strain HD73 cell wall. The strain HD73 cell wall was mixed with MclX protein (triangles). CwlC protein (known cell wall hydrolase) plus cell wall was used as a positive control (squares), and the cell wall itself was used as a negative control (circles).
Mother cell lysis X gene deletion decreases cwlC expression
Previous research indicated that cwlC is controlled by σK and positively regulated by GerE (Chen et al., 2018). Since HD (ΔmclX) exhibited a similar phenotype to HD (ΔcwlC), whether the deletion of the mclX affected the transcriptional activity or protein expression of CwlC was analyzed.
The transcriptional activity of cwlC was investigated in HD (ΔmclX). β-galactosidase activity assays indicated that cwlC expression significantly decreased in HD (ΔmclX), compared to that in wild-type strain (Figure 4A). Whether MclX could directly bind to the cwlC promoter was tested by EMSA. MclX was found to be unable to directly bind to the cwlC promoter (Figure 4B). It is speculated that mclX deletion might affect cwlC transcription levels by affecting transcriptional activities or protein functions of σK and GerE. To investigate this, the transcriptional activities of sigK and gerE in HD (ΔmclX) and the wild-type strain were analyzed, showing no differences (Figures 4C,D). The protein functions of σK and GerE in HD (ΔmclX) were detected. bxpB, which was found to be regulated by σK and GerE (Peng et al., 2016), showed no differences in transcription between the wild-type strain and HD (ΔmclX) (Supplementary Figure 5). Thus, MclX indirectly affected cwlC transcriptional activity, not by affecting σK or GerE.
Figure 4. MclX indirectly affects the transcription of cwlC. (A) cwlC promoter activities in the wild-type strain HD73 and HD (ΔmclX) (HD73-PcwlC, circles; HDΔmclX-PcwlC, squares); (B) the cwlC promoter was unable to interact with the MclX protein based on EMSA. Lane 1, 1 ng FAM-labeled PcwlC probe; lanes 2–5, 1 ng labeled probe with an increasing mass of purified MclX protein; (C) sigK promoter activities in the wild-type strain HD73 and HD (ΔmclX) (HD73-PsigK, circles; HDΔmclX-PsigK, squares); (D) gerE promoter activities in the wild-type strain HD73 and HD (ΔmclX) (HD73-PgerE, circles; HDΔmclX-PgerE, squares). Error bars show standard error of the mean.
Moreover, CwlC protein production was investigated in HD (ΔmclX). In HD (ΔmclX) cell lysate, CwlC protein was barely visible in the anti-CwlC immunoblot, whereas significant amounts of CwlC protein were produced in the wild-type strain cell lysate (Figure 5), with the same bacterial volumes taken at the same time point and using the same loading volumes.
Figure 5. CwlC was determined by Western blot in wild-type HD73 and HD (ΔmclX) lysates. The same volumes were taken at the time points, and the same loading volumes were analyzed. CwlC-His served as positive control.
Altogether, CwlC expression was dramatically decreased and barely detectable in HD (ΔmclX).
Discussion
In this work, the novel gene mclX was characterized, whose deletion resulted in Cry1Ac encapsulation in the mother cell of the HD73 strain (Figure 1A). MclX was unable to hydrolyze the cell wall in vitro (Figure 3B), and it is a newly identified mother cell lysis-associated protein which is not an amidase. MclX can be found not only in the B. cereus group but also in some S. pneumoniae strains (Supplementary Figure 4). S. pneumoniae is a Gram-positive human pathogen leading to global health problems (Engholm et al., 2017). Its MclX homologous protein shares 100% similarity with that of HD73, and the role of MclX in S. pneumoniae cell lysis maybe very interesting and warrants further studies.
cwlC encodes a key cell wall hydrolase, and its deletion resulted in Cry1Ac encapsulation in the HD73 strain. Only a few reports about the regulation of cwlC, while the regulation mechanisms of some other hydrolases have been reported (Do et al., 2020a). Transcription of the hydrolases lytE and cwlO is regulated by the WalKR signal transduction pathway in B. subtilis (Dobihal et al., 2019). The endopeptidase EagA is regulated through bacterial second messenger molecule c-di-GMP signaling in Erwinia amylovora (Kharadi and Sundin, 2020). Protein interactions with hydrolases can activate (Uehara et al., 2009, 2014; Yang et al., 2011; Domínguez-Cuevas et al., 2013; Meisner et al., 2013; Do et al., 2020b) or inhibit hydrolase activities (Clarke et al., 2010). The mother cell of the HD (ΔmclX) strain did not lyse because the cwlC expression decreased (Figure 4A). The expression of mclX was detected earlier than that of cwlC (Figures 2B, 4A), although MclX cannot directly bind the cwlC promoter (Figure 4B). The way in which MclX affects the transcriptional activity of cwlC remains unknown. MclX may influence cwlC transcription by affecting other factors, and not by affecting σK or GerE (Figures 4C,D). The function of MclX requires further research, to gain a better understanding of the regulatory mechanism of cwlC.
The HD (ΔmclX) strain displayed no mother cell lysis and did not have an altered sporulation frequency, Cry1Ac protein production, or insecticidal activity (Figure 1 and Table 1). mclX deletion could be a new biotechnological approach in the encapsulation of Cry proteins resistant to adverse environmental factors. The deletion of cwlC or sigK in the Bacillus thuringiensis var. israelensis (Bti) strain did not block mother cell lysis (Xu et al., 2020; Huang et al., 2022), because other cell wall hydrolases, like CwlE, are involved in mother cell lysis in Bti (Huang et al., 2022). However, the deletion of the mclX homolog gene in Bti also did not block mother cell lysis (Supplementary Figure 6). In comparison to the deletion of cwlC in Bti, the deletion of the mclX homolog gene in Bti led to more effective delays in mother cell lysis (Supplementary Figure 6). Thus, insecticidal protein encapsulation in mother cells requires targeted strategies in different B. thuringiensis strains.
Conclusion
Deletion of mclX provides a novel approach for encapsulation of Cry protein in B. thuringiensis. MclX has no known domains and it is a key protein in the mother cell lysis of B. thuringiensis strain HD73. It is a discovery of blocking mother cell lysis which not cause by cell wall hydrolases. This strategy could be utilized to genetically modify the B. thuringiensis products for enhancing UV resistance.
Data Availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
JW and QY conducted the experiments, analyzed the data, and produced the data displays. FS designed the research. QY wrote the manuscript. LS, DL, and QP edited the manuscript. RZ, SH, and QP proposed amendments. FS and QP funded the research. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation (31530095 and 31772243).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.951830/full#supplementary-material
Footnotes
References
Adams, L. F., Brown, K. L., and Whiteley, H. R. (1991). Molecular cloning and characterization of two genes encoding sigma factors that direct transcription from a Bacillus thuringiensis crystal protein gene promoter. J. Bacteriol. 173, 3846–3854. doi: 10.1128/jb.173.12.3846-3854.1991
Arantes, O., and Lereclus, D. (1991). Construction of cloning vectors for Bacillus thuringiensis. Gene 108, 115–119. doi: 10.1016/0378-1119(91)90495-w
Arnaud, M., Chastanet, A., and Debarbouille, M. (2004). New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70, 6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004
Beegle, C. C., and Yamamoto, T. (1992). Invitation paper (C.P. Alexander Fund): history of Bacillus thuringiensis Berliner research and development. Can. Entomol. 124, 587–616. doi: 10.4039/Ent124587-4
Bravo, A., Agaisse, H., Salamitou, S., and Lereclus, D. (1996). Analysis of cryIAa expression in sigE and sigK mutants of Bacillus thuringiensis. Mol. Gen. Genet. 250, 734–741. doi: 10.1007/BF02172985
Chen, X., Gao, T., Peng, Q., Zhang, J., Chai, Y., and Song, F. (2018). Novel cell wall hydrolase CwlC from Bacillus thuringiensis is essential for mother cell lysis. Appl. Environ. Microb. 84:e02640-17. doi: 10.1128/AEM.02640-17
Clarke, C. A., Scheurwater, E. M., and Clarke, A. J. (2010). The vertebrate lysozyme inhibitor Ivy functions to inhibit the activity of lytic transglycosylase. J. Biol. Chem. 285, 14843–14847. doi: 10.1074/jbc.C110.120931
de Oliveira, J. L., Fraceto, L. F., Bravo, A., and Polanczyk, R. A. (2021). Encapsulation strategies for Bacillus thuringiensis: from now to the future. J. Agric. Food Chem. 69, 4564–4577. doi: 10.1021/acs.jafc.0c07118
Do, T., Page, J. E., and Walker, S. (2020a). Uncovering the activities, biological roles, and regulation of bacterial cell wall hydrolases and tailoring enzymes. J. Biol. Chem. 295, 3347–3361. doi: 10.1074/jbc.REV119.010155
Do, T., Schaefer, K., Santiago, A. G., Coe, K. A., and Walker, S. (2020b). Staphylococcus aureus cell growth and division are regulated by an amidase that trims peptides from uncrosslinked peptidoglycan. Nat. Microbiol. 5, 291–303. doi: 10.1038/s41564-019-0632-1
Dobihal, G. S., Brunet, Y. R., Flores-Kim, J., and Rudner, D. Z. (2019). Homeostatic control of cell wall hydrolysis by the WalRK two-component signaling pathway in Bacillus subtilis. eLife 8:e52088. doi: 10.7554/eLife.52088
Domínguez-Cuevas, P., Porcelli, I., Daniel, R. A., and Errington, J. (2013). Differentiated roles for MreB-actin isologues and autolytic enzymes in Bacillus subtilis morphogenesis. Mol. Microbiol. 89, 1084–1098. doi: 10.1111/mmi.12335
Du, C., and Nickerson, K. W. (1996). Bacillus thuringiensis HD73 spores have surface localized Cry1Ac toxin: physiological and pathogenic consequences. Appl. Environ. Microb. 62, 3722–3726. doi: 10.1128/aem.62.10.3722-3726.1996
Engholm, D. H., Kilian, M., Goodsell, D. S., Andersen, E. S., and Kjærgaard, R. S. (2017). A visual review of the human pathogen Streptococcus pneumoniae. FEMS Microbiol. Rev. 41, 854–879. doi: 10.1093/femsre/fux037
Hervé, A., and Lereclus, D. (1994). Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol. Microbiol. 13, 97–107. doi: 10.1111/j.1365-2958.1994.tb00405.x
Hoffmann, F., Schmidt, M., and Rinas, U. (2015). Simple technique for simultaneous on-line estimation of biomass and acetate from base consumption and conductivity measurements in high-cell density cultures of Escherichia coli. Biotechnol. Bioeng. 70, 358–361.
Huang, L., Xu, L., Han, G., Crickmore, N., Song, F., and Xu, J. (2022). Characterization of CwlC, an autolysin, and its role in mother cell lysis of Bacillus thuringiensis subsp. israelensis. Lett. Appl. Microbiol. 74, 92–102. doi: 10.1111/lam.13590
Jallouli, W., Sellami, S., Sellami, M., and Tounsi, S. (2014). Efficacy of olive mill wastewater for protecting Bacillus thuringiensis formulation from UV radiations. Acta Trop. 140, 19–25. doi: 10.1016/j.actatropica.2014.07.016
Jouzani, G. S., Valijanian, E., and Sharafi, R. (2017). Bacillus thuringiensis: a successful insecticide with new environmental features and tidings. Appl. Microbiol. Biotechnol. 101, 2691–2711. doi: 10.1007/s00253-017-8175-y
Kharadi, R. R., and Sundin, G. W. (2020). Cyclic-di-GMP regulates autoaggregation through the putative peptidoglycan hydrolase, EagA, and regulates transcription of the znuABC zinc uptake gene cluster in Erwinia amylovora. Front. Microbiol. 11:605265. doi: 10.3389/fmicb.2020.605265
Lereclus, D., Agaisse, H., Gominet, M., and Chaufaux, J. (1995). Overproduction of encapsulated insecticidal crystal proteins in a Bacillus thuringiensis spo0A mutant. Biotechnology 13, 67–71. doi: 10.1038/nbt0195-67
Liu, G., Lai, S., Shu, C., Wang, P., and Song, F. (2017). Complete genome sequence of Bacillus thuringiensis subsp. kurstaki strain HD73. Genome Announc. 1:e0008013. doi: 10.1128/genomeA.00080-13
Manasherob, R., Ben-Dov, E., Wu, X., and Zaritsky, B. A. (2002). Protection from UV-B damage of mosquito larvicidal toxins from Bacillus thuringiensis subsp. israelensis expressed in Anabaena PCC 7120. Curr. Microbiol. 45, 217–220. doi: 10.1007/s00284-001-0106-5
Meisner, J., Llopis, P. M., Sham, L. T., Garner, E., Bernhardt, T. G., and Rudner, D. Z. (2013). FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol. Microbiol. 89, 1069–1083. doi: 10.1111/mmi.12330
Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Myasnik, M., Manasherob, R., Ben-Dov, E., Zaritsky, A., and Barak, M. Z. (2001). Comparative sensitivity to UV-B radiation of two Bacillus thuringiensis subspecies and other Bacillus sp. Curr. Microbiol. 43, 140–143. doi: 10.1007/s002840010276
Nugroho, F. A., Yamamoto, H., Kobayashi, Y., and Sekiguchi, J. (1999). Characterization of a new Sigma-K-dependent peptidoglycan hydrolase gene that plays a role in Bacillus subtilis mother cell lysis. J. Bacteriol. 181, 6230–6237. doi: 10.1128/JB.181.20.6230-6237.1999
Peng, Q., Kao, G., Qu, N., Zhang, J., Li, J., and Song, F. (2016). The regulation of exosporium-related genes in Bacillus thuringiensis. Sci. Rep. 6:19005. doi: 10.1038/srep19005
Peng, Q., Wang, G., Liu, G., Zhang, J., and Song, F. (2015). Identification of metabolism pathways directly regulated by sigma (54) factor in Bacillus thuringiensis. Front. Microbiol. 6:407. doi: 10.3389/fmicb.2015.00407
Sanchis, V., Gohar, M., Chaufaux, J., Arantes, O., and Lereclus, D. (1999). Development and field performance of a broad-spectrum nonviable asporogenic recombinant strain of Bacillus thuringiensis with greater potency and UV resistance. Appl. Environ. Microb. 65, 4032–4039. doi: 10.1128/AEM.65.9.4032-4039.1999
Schaeffer, P., Millet, J., and Aubert, J. P. (1965). Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. U.S.A. 54, 704–711. doi: 10.1073/pnas.54.3.704
Shen, X., Yu, Q., Liu, H., Wang, J., Zhang, R., Peng, Q., et al. (2021). Transition phase regulator AbrB positively regulates the sip1Ab1 gene expression in Bacillus thuringiensis. Microbiol. Spectr. 9:e0007521. doi: 10.1128/Spectrum.00075-21
Uehara, T., Dinh, T., and Bernhardt, T. G. (2009). LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J. Bacteriol. 191:5094. doi: 10.1128/JB.00505-09
Uehara, T., Parzych, K. R., Dinh, T., and Bernhardt, T. G. (2014). Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 29, 1412–1422. doi: 10.1038/emboj.2010.36
Xu, L., Han, G., Fan, X., Lv, J., Zhang, X., Peng, Q., et al. (2020). Characteristics of the sigK deletion mutant from Bacillus thuringiensis var. israelensis strain Bt-59. Curr. Microbiol. 77, 3422–3429. doi: 10.1007/s00284-020-02150-9
Yang, D. C., Peters, N. T., Parzych, K. R., Uehara, T., Markovski, M., and Bemhardt, T. G. (2011). An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc. Natl. Acad. Sci. U.S.A. 108, E1052–E1060. doi: 10.1073/pnas.1107780108
Yang, J., Peng, Q., Chen, Z., Deng, C., and Song, F. (2013). Transcriptional regulation and characteristics of a novel N-acetylmuramoyl-L-alanine amidase gene involved in Bacillus thuringiensis mother cell lysis. J. Bacteriol. 195, 2887–2897. doi: 10.1128/JB.00112-13
Zhang, R., Slamti, L., Tong, L., Verplaetse, E., Ma, L., Lemy, C., et al. (2020). The stationary phase regulator CpcR activates cry gene expression in non-sporulating cells of Bacillus thuringiensis. Mol. Microbiol. 113, 740–754. doi: 10.1111/mmi.14439
Zhang, X., Gao, T., Peng, Q., Song, L., Zhang, J., Chai, Y., et al. (2018). A strong promoter of a non-cry gene directs expression of the cry1Ac gene in Bacillus thuringiensis. Appl. Microbiol. Biot. 102, 3687–3699. doi: 10.1007/s00253-018-8836-5
Zhou, C., Zheng, Q., Peng, Q., Du, L., Shu, C., Zhang, J., et al. (2014). Screening of cry-type promoters with strong activity and application in Cry protein encapsulation in a sigK mutant. Appl. Microbiol. Biotechnol. 98, 7901–7909. doi: 10.1007/s00253-014-5874-5
Zogo, B., Tchiekoi, B. N., Koffi, A. A., Dahounto, A., Ahoua Alou, L. P., Dabiré, R. K., et al. (2019). Impact of sunlight exposure on the residual efficacy of biolarvicides Bacillus thuringiensis israelensis and Bacillus sphaericus against the main malaria vector, Anopheles gambiae. Malar. J. 18:55. doi: 10.1186/s12936-019-2687-0
Keywords: Bacillus thuringiensis, mother cell lysis, cell wall lytic enzyme C, Cry1Ac encapsulation, transcriptional regulation
Citation: Wang J, Yu Q, Peng Q, Slamti L, Zhang R, Hou S, Lereclus D and Song F (2022) Deletion of the novel gene mother cell lysis X results in Cry1Ac encapsulation in the Bacillus thuringiensis HD73. Front. Microbiol. 13:951830. doi: 10.3389/fmicb.2022.951830
Received: 24 May 2022; Accepted: 14 July 2022;
Published: 09 August 2022.
Edited by:
Samina Mehnaz, Forman Christian College, PakistanReviewed by:
Deeba Noreen Baig, Forman Christian College, PakistanSalma Mukhtar, Connecticut Agricultural Experiment Station, United States
Copyright © 2022 Wang, Yu, Peng, Slamti, Zhang, Hou, Lereclus and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuping Song, ZnBzb25nQGlwcGNhYXMuY24=
†These authors have contributed equally to this work and share first authorship
 Jiaojiao Wang1†
Jiaojiao Wang1† Qi Peng
Qi Peng Fuping Song
Fuping Song