- 1Department of Medical Laboratory, Henan Provincial Chest Hospital, Zhengzhou, China
- 2School of Pharmaceutical Sciences, Zhengzhou University, Zhengzhou, China
- 3Key Laboratory of Advanced Drug Preparation Technologies, Ministry of Education, Zhengzhou University, Zhengzhou, China
Colistin is considered as an antibiotic of ‘last resort’ for the treatment of lethal infections caused by carbapenem-resistant Enterobacterales (CRE), dissemination of plasmid-borne colistin resistance gene mcr-1, particularly into CRE, resulting in the emergence of strains that approach pan-resistance. A wide variety of plasmid types have been reported for carrying mcr-1. Among which, large IncHI2-type plasmids were multidrug-resistant (MDR) plasmids harbored multiple resistance determinants in addition to mcr-1. Herein, we characterized a novel hybrid IncHI2-like mcr-1-bearing plasmid in an NDM-7-producing ST167 Escherichia coli strain EC15-50 of clinical origin. Antimicrobial susceptibility testing showed E. coli EC15-50 exhibited an extensively drug-resistant (XDR) profile that only susceptible to amikacin and tigecycline. S1-PFGE, Southern hybridization and Whole-genome Sequencing (WGS) analysis identified a 46,161 bp blaNDM-7-harboring IncX3 plasmid pEC50-NDM7 and a 350,179 bp mcr-1-bearing IncHI2/HI2A/N/FII/FIA plasmid pEC15-MCR-50 in E. coli EC15-50. Sequence detail analysis revealed the type IV coupling protein (T4CP) gene was absent on pEC15-MCR-50, explaining that pEC15-MCR-50 was a non-conjugative plasmid. Comparative genetic analysis indicated the hybrid plasmid pEC15-MCR-50 was probably originated from pXGE1mcr-like IncHI2/HI2A/N plasmid and pSJ_94-like IncFII/FIA plasmid, and generated as a result of a replicative transposition process mediated by IS26. Currently, the prevalent mcr-1-carrying IncHI2 plasmids were rarely reported to be fused with other plasmids. The identification of the novel hybrid plasmid pEC15-MCR-50 in this study highlighted the importance of close surveillance for the emergence and dissemination of such fusion MDR plasmids, particularly in NDM-producing Enterobacterales.
Introduction
Carbapenem-resistant Enterobacterales (CRE) strains, which exhibited multi-drug resistant or extensively drug resistant patterns and were associated with high mortality rates, were regarded as a serious threat for public health (Centers for Disease Control and Prevention, 2019). Given that less effective treatment options left for treating infections caused by CRE, colistin that belongs to cationic antimicrobial peptides with neurotoxic effects has been used as a last resort of antibiotic against CRE in clinic. Unfortunately, emergence and global dissemination of plasmid-borne colistin resistance gene mcr-1 that encodes phosphoethanolamine transferase among Enterobacterales, especially in CRE, led to the emergence of potentially untreatable strains.
Since the first mobile colistin resistance (mcr) gene, mcr-1, was identified in Escherichia coli in 2016 from China, more mcr variants (mcr-1 to mcr-10) were reported in different species of Enterobacterales exhibiting MDR phenotypes isolated from animals, food samples and clinical settings origin worldwide, and mcr-1 was the most frequently detected and globally spread variant (Wang et al., 2018, 2020). Various mcr-1-encoding plasmids belonging to IncI2, IncX4, IncP, IncY, IncFII, IncN, IncQ and IncHI2 incompatibility groups have been described as the vector facilitates the dissemination of mcr-1 (Ye et al., 2016; Wang et al., 2018). Among which, IncI2, IncHI2 and IncX4 were the most common plasmid types (Wu et al., 2018; Matamoros et al., 2020). Unlike these two types of small plasmids (IncI2: ~65 kb, IncX4: ~33 kb) that only carried a single resistance gene mcr-1, IncHI2 type plasmids with large size (> 200 kb) always harbored multiple resistance determinants beside mcr-1 (Tang et al., 2020). In addition, hybrid mcr-1-bearing plasmids have also been described, such as pCQ02-121, a transferable IncX3/X4 hybrid plasmid co-harboring mcr-1 and blaNDM-5 (Sun et al., 2016). Recently, the generation of hybrid and cointegrate mcr-1 plasmids mediated by IS26 or ISCR2 were found in the process of conjugation (He et al., 2019; Li et al., 2020), promoting the dissemination of resistance genes and extending the resistance profile and the host spectrum for the fusion plasmid, which poses an enormous threat to public health.
Herein, we characterized a hybrid mcr-1-bearing plasmid containing IncFII, IncFIA, IncHI2, IncHI2A and IncN replicons in an NDM-7-producing extensively drug resistant (XDR) ST167 E. coli clinical isolate. Based on its structural features, we proposed a putative IS26-mediated formation model of this novel IncHI2/HI2A/N/FII/FIA hybrid plasmid.
Materials and methods
Bacterial isolation, antimicrobial susceptibility testing and PCR detection
Escherichia coli EC15-50 was obtained from the sputum culture of a 44-year-old male patient hospitalized in respiratory intensive care unit (RICU) at a tertiary hospital in Zhengzhou city in 2015. This isolate was identified by using the Vitek 2 system (bioMérieux, France) and 16S rDNA gene sequencing. Antimicrobial susceptibility testing was determined by using the broth microdilution method and agar dilution method (for fosfomycin) according to the Clinical and Laboratory Standards Institute guidelines (CLSI, 2020) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST).1 E. coli ATCC25922 was used as the quality-control strain. PCR was used to detect the presence of the colistin resistance gene mcr and carbapenemase-encoding genes, including blaNDM, blaIMP, blaOXA-48-like, blaVIM, and blaKPC, with primers described previously (Qin et al., 2014).
S1-PFGE and Southern blotting
To detect the location of the blaNDM-7 and mcr-1 gene, whole-cell genomic DNA of the E. coli EC15-50 isolate was digested by S1 nuclease (TaKaRa, Dalian, China) in agarose gel plug and then separated by pulsed-field gel electrophoresis (PFGE) under the conditions reported previously (Qin et al., 2014). The location of the blaNDM-7 and mcr-1 was indicated by Southern hybridization using a digoxigenin-labeled blaNDM-7 and mcr-1 gene fragment probe, respectively, using a DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics, Basel, Switzerland) according to the manufacturer’s instructions.
Whole-genome sequencing (WGS) and bioinformatics analysis
Whole-genome DNA of E. coli EC15-50 was extracted with the Wizard genomic DNA purification kit (Promega, Madison, WI, United States) and then sequenced using the Illumina MiSeq platform (San Diego, CA, United States) and the Pacific Biosciences RS II platform (Pacific Biosciences, Menlo Park, CA, United States). Sequencing reads were assembled into scaffolds using GS De Novo Assembler software (version 3.0). The prediction and annotation of ORFs were performed using Glimmer 3.02.2 Antimicrobial resistance genes (ARGs) were identified by using Resfinder (Bortolaia et al., 2020). Plasmid identified in this study was analyzed using PlasmidFinder3 and pMLST4 to investigate the replicons (Carattoli et al., 2014). Sequence alignments for the genetic environments of mcr-1 were performed using BLAST.5
Conjugation experiments
Conjugation assays were performed to assess the transferability of mcr-1 gene according to the method described previously with minor modification (Wang et al., 2019). Briefly, approximately 1 × 108 colony-forming units (CFU)/mL of the donor strain (EC15-50) and the rifampicin-resistant E. coli isolate EC600 as the recipient strain were mixed at a ratio of 1:4 on LB agar and cultured for 12 h. The mixtures were collected and then plated on an LB agar containing rifampicin (64 μg/ml) and colistin (1 μg/ml). All suspected transconjugants were verified by PCR amplification for mcr-1, and antimicrobial susceptibility testing.
Plasmid stability testing
The stability of plasmid pEC15-MCR-50 was studied by passage in antibiotic-free Luria broth (LB) as previously described with minor modification (Sandegren et al., 2012). Briefly, the plasmid-containing E. coli EC15-50 were grown in 1 ml LB broth at 37°C for 12 h. The bacterial cells cultured for 12 h were washed by centrifugation and resuspended in 1 ml LB broth. Transfer 1 μl washed cells into 1 ml LB broth and incubate at 37°C for 12 h. For continuous culture, wash 1 μl cells into 1 ml LB broth once every 12 h, approximately 10 generations of growth per passage, for 10 consecutive days. The 200 generation strains were diluted and spread on LB agar plates. Then, 50 colonies were screened on LB agar plates supplemented with 2 μg/ml colistin, and the stability and integrity of the plasmid pEC15-MCR-50 carrying mcr-1 gene across passages were investigated by PCR mapping technology. PCR primers were summarized in Supplementary Table S1.
Results and discussion
Characteristics of Escherichia coli EC15-50 co-harboring blaNDM-7 and mcr-1
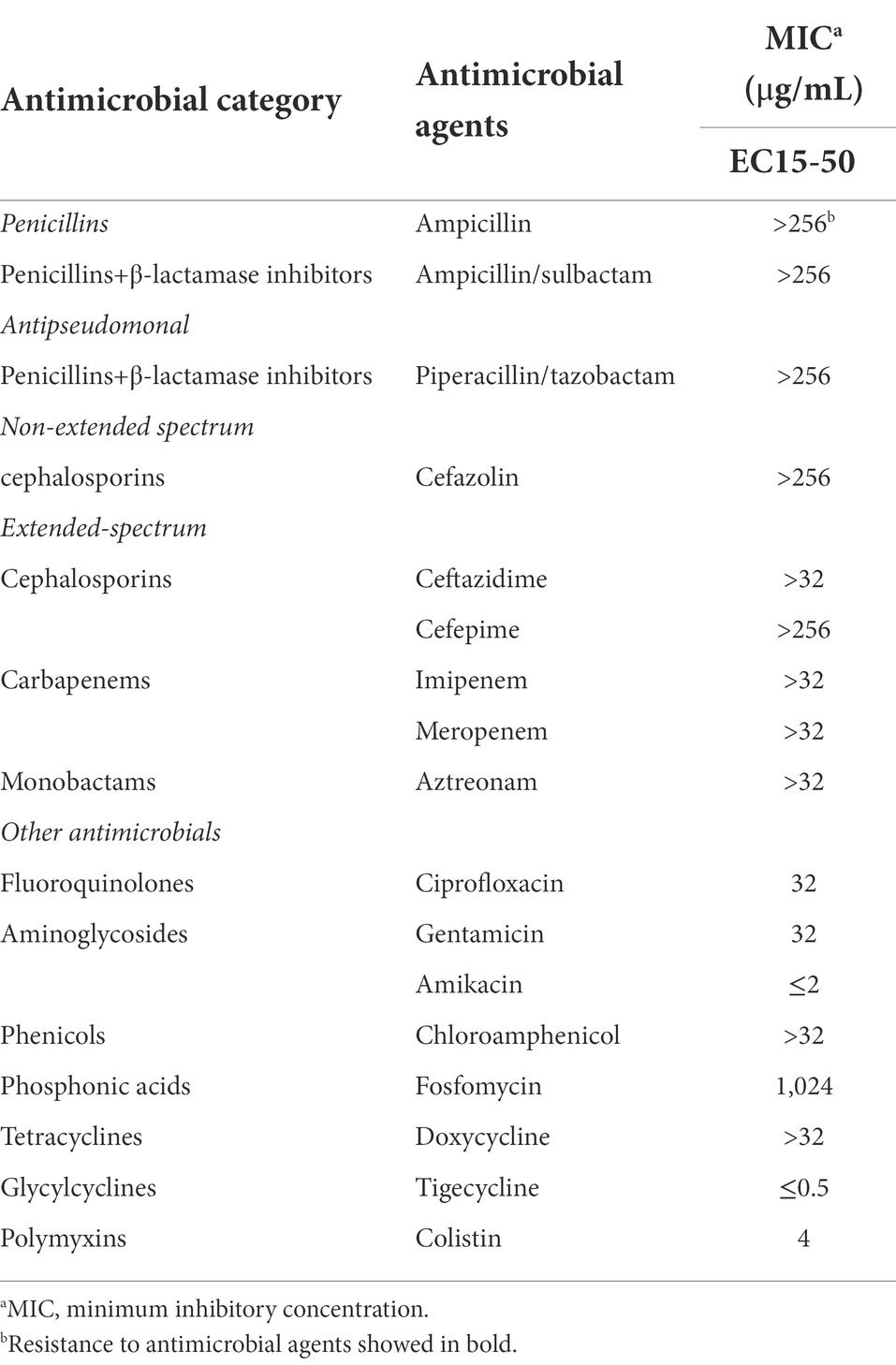
Clinical strain E. coli EC15-50 belonging to ST167 was recovered from the sputum culture of a 44-year-old male patient with acute lymphoblastic leukemia in 2015. In vitro antimicrobial susceptibility testing results showed that strain EC15-50 exhibited extensively drug resistant (XDR) profile for a wide range of antimicrobial agents, resistant to all cephalosporins, carbapenems, β-lactam/β-lactamase inhibitors, fluoroquinolones, gentamicin, chloroamphenicol, fosfomycin and doxycycline as well as colistin, but susceptible to amikacin and tigecycline (Table 1). PCR and sequencing based on carbapenems and colistin resistance profiles revealed the co-occurrence of blaNDM-7 and mcr-1 gene in EC15-50. S1-PFGE and hybridization results further indicated the blaNDM-7 and mcr-1 gene were located on separated plasmids with the sizes of ~45 kb and ~ 350 kb, respectively (Figure 1).
Figure 1. S1-PFGE and Southern hybridization of mcr-1 and blaNDM genes in the Escherichia coli strain EC15-50. The arrows indicate the locations of the plasmids hybridized to the mcr-1 and blaNDM probes. M, reference standard strain H9812 restricted with XbaI; Lane 1, E. coli EC15-50; Lane 2, the plasmid hybridized to the blaNDM-probe; Lane 3, the plasmid hybridized to the mcr-1-probe.
E. coli has been regarded as the predominant species in Enterobacterales for carrying blaNDM, as well as mcr-1 gene, and co-occurrence of blaNDM and mcr was most frequently detected in E. coli strains (Han et al., 2020). Among these blaNDM and mcr positive strains, blaNDM-5 and mcr-1 were the most common reported variants combination. However, the blaNDM-7 and mcr-1 gene co-harbored E. coli have not been described. Our study represented the first report of clinical E. coli strain co-producing NDM-7 and MCR-1 that belongs to sequence type ST167. Notably, E. coli ST167 is classified as an internationally disseminated clonal lineage associated with the global spread of blaCTX-M-15 and blaNDM genes. It has been considered as high-risk clone and is emerging associated with β-lactamases mainly in clinical samples origin throughout the world (Sánchez-Benito et al., 2017; Nukui et al., 2019; Xu et al., 2019). Dissemination of mcr-1 within the successful epidemic clone may facilitate further spread of colistin resistance in β-lactamases-producing E. coli isolates.
Identification of a novel hybrid mcr-1-bearing plasmid pEC15- MCR-50
Sequence analysis showed that the 46,161 bp blaNDM-7-harboring plasmid pEC50-NDM7 (KX470735), which was nearly identical to a reported plasmid pKpN01-NDM7 (CP012990) with a single nucleotide change from A to G at 31,701 bp, was a typical IncX3 plasmid carrying only one resistance gene blaNDM-7. However, the mcr-1-bearing plasmid pEC15-MCR-50 (MG656414), with a length of 350,179 bp and an average GC content of 48.0%, was a hybrid plasmid containing multiple replicons including IncFII, IncFIA, IncHI2, IncHI2A and IncN. ResFinder analysis identified a number of antimicrobial resistance genes in pEC15-MCR-50, including aminoglycosides resistance genes aac(3)-IV, aph(3′)-Ia, aph(4)-Ia, aadA1 and aadA2b, macrolides resistance gene mph(A), β-lactamases resistance gene blaCTX-M-14, phenicols resistance genes cmlA1 and floR, tetracyclines resistance gene tet(M), fosfomycin resistance gene fosA3, sulfonamides resistance genes sul2 and sul3 in addition to colistin resistance gene mcr-1 (Figure 2A). Interestingly, these resistance genes were distributed in three distinct regions of pEC15-MCR-50, a 3,673 bp mcr-1-containing cassette ISApl1-mcr-1-pap2 and a 6,867 bp mph(A)-harboring segment organized as IS15DI-repB(N)-IS5075-IS26-mph(A) were located in the backbone. However, the remaining resistance genes which were dispersed between multiple insertion sequences were located in the variable region. Additionally, genes responsible for plasmid maintenance including parB, sopA (plasmid partition) and ccdAB (toxin-antitoxin system) and conjugation and transfer (tra-relative genes) were also observed in the backbone.
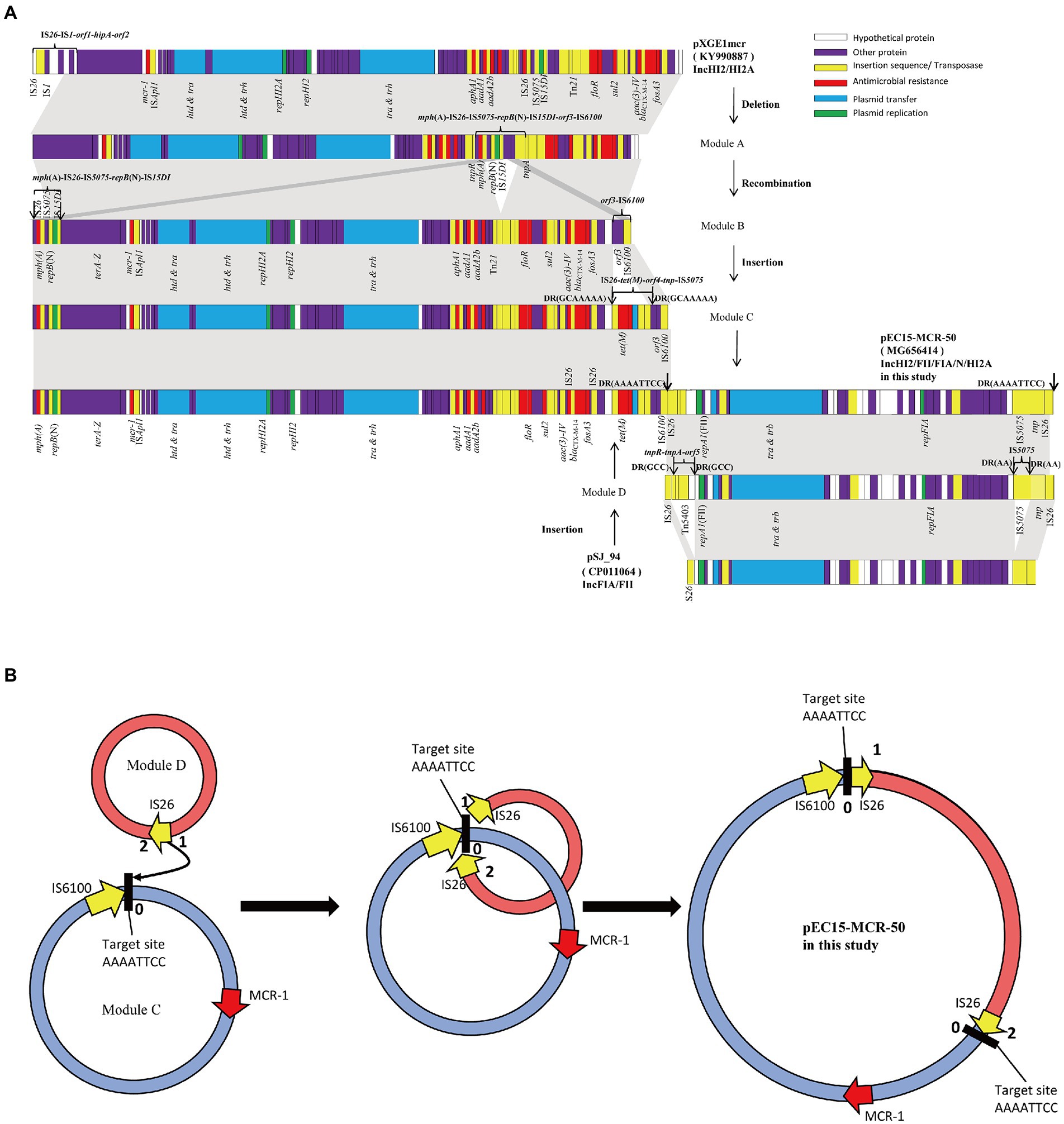
Figure 2. Schematic diagrams depicting the linear structures of the plasmid pEC15- MCR-50 and the proposed mechanism of plasmid fusion. (A) Schematic diagrams depicting the linear structure of the plasmid pEC15- MCR-50. White, purple, yellow, red, blue and green arrows indicate hypothetical protein, other genes, insertion sequence or transposase, resistance genes, plasmid transfer, and plasmid replication, respectively; The shaded area indicates 99–100% identity. (B) Proposed model for the formation of the fusion plasmid pEC15-MCR-50 mediated by IS26. Red arrow, mcr-1 gene; Yellow arrow, IS26; Black bold string labelled with the number 0 represent the 8 bp target site (AAAATTCC).
Plasmid pEC15-MCR-50 failed to transfer from the donor strain E. coli EC15-50 to the recipient isolate E. coli EC600 in the conjugation assay. Sequence analysis by oriTfinder6 identified the oriT region, relaxase gene and T4SS gene cluster on plasmid pEC15-MCR-50, but the type IV coupling protein (T4CP) gene was absent. Given that the four components are necessary for conjugation in self-transmissible plasmids (Smillie et al., 2010), incomplete MOB module might be responsible for the non-conjugative profile of pEC15-MCR-50.
Proposed formation mechanism of the plasmid pEC15-MCR-50
Based on the detailed sequence analysis of pEC15-MCR-50, we found it was a cointegrate plasmid with chimeric characteristics consisting of a ~243-kb pXGE1mcr (KY990877) -derived module (Module C) and a ~107-kb pSJ_94 (CP011064) -derived module (Module D; Figure 2A). Comparative genomic analysis between the hypothetical ancestor plasmid pXGE1mcr and the Module C sequence indicated that formation of this module was due to a series of genetic events. It is proposed that the deletion of a 12,747 bp of IS26-IS1-orf1-hipA-orf2 in pXGE1mcr formed Module A. Subsequently, the genetic rearrangement of a 9,379 bp fragment organized as IS6100-orf3-IS15DI-repB(N)-IS5075-IS26-mph(A) immediately upstream of the tnpA gene and downstream of the tnpR gene, causing the insertion of macrolide resistance determinant containing part IS15DI-repB(N)-IS5075-IS26-mph(A) in the backbone region and the integration of the remnant part of the 2,512 bp fragment between the tnpA gene and the IS15DI, resulting in the formation of Module B. Finally, insertion of the IS26-tet(M)-orf4-tnp-IS5075 element, conferring resistance to tetracyclines, as previously described in pHS13-1-IncHI2 (Liu et al., 2020a), upstream of the orf3 gene in Module B formed Module C. On the other hand, Module D was proposed formation from an IncFIA/FII plasmid pSJ_94 (CP011064) by insertion of Tn5403 (tnpR-tnpA-orf5) downstream of repA1 flanked by a 3 bp direct repeat (GCC) and a 1,005 bp of IS5075 upstream of the tnp gene at the end of the sequence flanked by a 2 bp direct repeat (AA), respectively.
The observed presence of an 8 bp direct repeat (AAAATTCC) bracketed the Module D segment indicated cointegrate formation is much more likely to be mediated by IS26 element. Close inspection of the overall structure of pEC15-MCR-50, we proposed a possible model of cointegrate formation indicated by Figure 2B. In this model, IS26 in Module D attacked the target site 0 (AAAATTCC) in mcr-1-harbored Module C and then replicative cointegrate formation occurred. Module D was incorporated into the downstream of IS6100 in Module C creating the cointegrate pEC15-MCR-50 and resulting in an 8 bp target site duplications (TSD) and acquisition of an additional copy of IS26. Eventually, two direct repeats of the TSDs were present surrounding Module D.
IS26 was regarded as a vital mobile element contributed to the dissemination and rearrangement of resistance determinants, as well as plasmid reorganization (Harmer and Hall, 2015; He et al., 2015; Wei et al., 2022). A previous study demonstrated that IS26 was involved in recombination of an IncN1-F33: A-: B- plasmid and an mcr-1-carrying phage-like plasmid during conjugation process, facilitating the dissemination of mcr-1 (He et al., 2019). Additionally, IS26 was also reported to play an important role in generation of blaNDM and blaKPC-bearing MDR cointegrate plasmids. A 59 kb cointegrate plasmid pT18 (CP017086) from Proteus mirabilis was likely to be generated by IS26-mediated cointegrate formation between the IncN-type plasmid pT211 (containing blaKPC-2) and the IncFII-33 plasmid (blaTEM-1B, blaCTX-M-65, rmtB and fosA3; Hua et al., 2020), and a 122 kb plasmid pL53T in the transconjugant of E. coli L53 was formed due to IS26-mediated fusion of pL53-4 (CP034737), a typical blaNDM-5-bearing IncX3 plasmid, and pL53-3 (CP034736), an IncFII-type plasmid harboring fosA, blaCTX-M-55 and blaTEM-1, during the conjugation process (Liu et al., 2020b). IS26-mediated plasmid recombination can not only lead to the emergence of large-size MDR plasmids but also accelerate the evolution of virulence plasmids. The entire Salmonella Enteritidis-specific virulence plasmid pSEN (HG970000) was observed to fuse with another IncHI2 MDR plasmid (KX518743) mediated by IS26 to generate a cointegrate virulence–resistance plasmid pSE380T (KY401053; Wong et al., 2017). Similar to pSE380T, the 244 kb plasmid pCR-HvKP3TC-2_Vir-p3 (OM001475) was also found to be a fusion plasmid generated by IS26-mediated recombination of virulence plasmid pVir-CR-HvKP3 (MW598234) and plasmid p3-CR-HvKP3 (MW598238; Yang et al., 2022).
In addition to IS26, ISCR2 has also been reported to reorganize the mcr-1-bearing large MDR plasmid and was responsible for mcr-1-bearing large fused MDR plasmids (Li et al., 2020). Compared with previous reported IS26-mediated N1-F33: A-: B- type mcr-1-bearing hybrid plasmid (MK419512), the HI2/HI2A/N/FII/FIA type plasmid pEC15-MCR-50 which possibly generated as a result of a replicative transposition process mediated by IS26, was identified as an MDR plasmid, carrying more resistance determinants. Although this plasmid could not be transferred by conjugation, it remained stable in strain in antimicrobial agent-free environment and could be served as a reservoir of multiple resistance genes. Based on the important role of IS26 in mediating plasmid reorganization, further attention should be paid to focusing on this element in hybrid plasmids to trace the evolution of MDR or virulence plasmids.
The stability of the plasmid pEC15-MCR-50
The serial passages and PCR mapping results showed that the plasmid pEC15-MCR-50 from E. coli EC15-50 could be retained at 100% over 200 generations of passage, revealing that the large fusion plasmid pEC15-MCR-50 remained stable in antimicrobial agent-free environment, and the resistance genes in this novel hybrid plasmid can be stably inherited.
Conclusion
Polymyxin is regarded as the last line of defense in the clinical treatment of CRE infections. Emergence of mcr-1 in NDM-producing Enterobacterales can seriously limit clinical treatment options. Our study identified and characterized an mcr-1-bearing IncHI2/HI2A/N/FII/FIA hybrid plasmid in NDM-7-producing XDR ST167-type E. coli of clinical origin. Detail analysis showed that IS26 might be responsible for this mcr-1-bearing large fused plasmid. The formation of fusion plasmid was likely to maintain the stability of mcr-1 gene, which may facilitate bacteria to adapt to the antibiotic stress. Close surveillance is urgently needed to monitor the emergence and dissemination of E. coli co-producing NDM and MCR-1, as well as such fusion MDR plasmids.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI – KX470735, MG656414.
Author contributions
SQ, XS, and YL designed the study. SX, WW, JC, and YH performed the experiments. TZ, JC, ZX, and XZ analyzed the bioinformatics data. SQ and JC wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grant U2004125 from the National Natural Science Foundation of China (SQ), the Key R&D and Promotion Project of Henan Province (No. 212102310246, 2018010038) of China.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.950087/full#supplementary-material
Footnotes
2. ^http://cbcb.umd.edu/software/glimmer/
3. ^https://cge.cbs.dtu.dk/services/PlasmidFinder/
4. ^https://cge.cbs.dtu.dk/services/pMLST/
References
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/aac.02412-14
Centers for Disease Control and Prevention. (2019). Antibiotic resistance threats in the United States. Available at https://stacks.cdc.gov/view/cdc/82532
CLSI. (2020). Performance Standards for Antimicrobial Susceptibility Testing; 30th Informational Supplement. CLSI Document M100. Wayne, PA: CLSI.
Han, H., Liu, W., Cui, X., Cheng, X., and Jiang, X. (2020). Co-existence of mcr-1 and blaNDM-5 in an Escherichia coli strain isolated from the pharmaceutical industry, WWTP. Infect. Drug Resist. 13, 851–854. doi: 10.2147/idr.S245047
Harmer, C. J., and Hall, R. M. (2015). IS26-mediated precise excision of the IS26-aphA1a translocatable unit. mBio 6, e01866–e01815. doi: 10.1128/mBio.01866-15
He, S., Hickman, A. B., Varani, A. M., Siguier, P., Chandler, M., Dekker, J. P., et al. (2015). Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. MBio 6:e00762. doi: 10.1128/mBio.00762-15
He, D., Zhu, Y., Li, R., Pan, Y., Liu, J., Yuan, L., et al. (2019). Emergence of a hybrid plasmid derived from IncN1-F33: A-: B- and mcr-1-bearing plasmids mediated by IS26. J. Antimicrob. Chemother. 74, 3184–3189. doi: 10.1093/jac/dkz327
Hua, X., Zhang, L., Moran, R. A., Xu, Q., Sun, L., van Schaik, W., et al. (2020). Cointegration as a mechanism for the evolution of a KPC-producing multidrug resistance plasmid in Proteus mirabilis. Emerg. Microbes Infect. 9, 1206–1218. doi: 10.1080/22221751.2020.1773322
Li, R., Lu, X., Peng, K., Liu, Y., Xiao, X., and Wang, Z. (2020). Reorganization of mcr-1-bearing large MDR plasmids resolved by nanopore sequencing. J. Antimicrob. Chemother. 75, 1645–1647. doi: 10.1093/jac/dkaa046
Liu, Y.-y., Sun, Y.-w., Sun, H.-r., Luo, X.-w., He, D.-d., Wu, H., et al. (2020a). Characterization of the IncHI2 plasmid pTW4 harboring tet(M) from an isolate of Escherichia coli ST162. J. Antibiot. 73, 876–880. doi: 10.1038/s41429-020-0337-y
Liu, Z., Xiao, X., Liu, Y., Li, R., and Wang, Z. (2020b). Recombination of NDM-5-producing plasmids mediated by IS26 among Escherichia coli. Int. J. Antimicrob. Agents 55:105815. doi: 10.1016/j.ijantimicag.2019.09.019
Matamoros, S., van Hattem, J. M., Arcilla, M. S., Willemse, N., Melles, D. C., Penders, J., et al. (2020). Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 7, 15364. doi: 10.1038/s41598-020-58953-0
Nukui, Y., Ayibieke, A., Taniguchi, M., Aiso, Y., Shibuya, Y., Sonobe, K., et al. (2019). Whole-genome analysis of EC129, an NDM-5-, CTX-M-14-, OXA-10- and MCR-1-co-producing Escherichia coli ST167 strain isolated from Japan. J. Glob. Antimicrob. Resist. 18, 148–150. doi: 10.1016/j.jgar.2019.07.001
Qin, S., Fu, Y., Zhang, Q., Qi, H., Wen, J. G., Xu, H., et al. (2014). High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province, China. Antimicrob. Agents Chemother. 58, 4275–4282. doi: 10.1128/aac.02813-13
Sánchez-Benito, R., Iglesias, M. R., Quijada, N. M., Campos, M. J., Ugarte-Ruiz, M., Hernández, M., et al. (2017). Escherichia coli ST167 carrying plasmid mobilisable mcr-1 and blaCTX-M-15 resistance determinants isolated from a human respiratory infection. Int. J. Antimicrob. Agents 50, 285–286. doi: 10.1016/j.ijantimicag.2017.05.005
Sandegren, L., Linkevicius, M., Lytsy, B., Melhus, Å., and Andersson, D. I. (2012). Transfer of an Escherichia coli ST131 multiresistance cassette has created a Klebsiella pneumoniae-specific plasmid associated with a major nosocomial outbreak. J. Antimicrob. Chemother. 67, 74–83. doi: 10.1093/jac/dkr405
Smillie, C., Garcillán-Barcia, M. P., Francia, M. V., Rocha, E. P., and de la Cruz, F. (2010). Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452. doi: 10.1128/mmbr.00020-10
Sun, J., Yang, R. S., Zhang, Q., Feng, Y., Fang, L. X., Xia, J., et al. (2016). Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat. Microbiol. 1, 16176. doi: 10.1038/nmicrobiol.2016.176
Tang, B., Chang, J., Zhang, L., Liu, L., Xia, X., Hassan, B. H., et al. (2020). Carriage of distinct mcr-1-harboring plasmids by unusual serotypes of salmonella. Adv. Biosyst. 4:e1900219. doi: 10.1002/adbi.201900219
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., and Zong, Z. (2020). Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, R., van Dorp, L., Shaw, L. P., Bradley, P., Wang, Q., Wang, X., et al. (2018). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9, 1179. doi: 10.1038/s41467-018-03205-z
Wang, X., Wang, Y., Zhou, Y., Wang, Z., Wang, Y., Zhang, S., et al. (2019). Emergence of Colistin resistance gene mcr-8 and its variant in Raoultella ornithinolytica. Front. Microbiol. 10, 228. doi: 10.3389/fmicb.2019.00228
Wei, D. W., Wong, N. K., Song, Y., Zhang, G., Wang, C., Li, J., et al. (2022). IS26 veers genomic plasticity and genetic rearrangement toward carbapenem hyperresistance under sublethal antibiotics. MBio 13:e0334021. doi: 10.1128/mbio.03340-21
Wong, M. H., Chan, E. W., and Chen, S. (2017). IS26-mediated formation of a virulence and resistance plasmid in Salmonella Enteritidis. J. Antimicrob. Chemother. 72, 2750–2754. doi: 10.1093/jac/dkx238
Wu, R., Yi, L. X., Yu, L. F., Wang, J., Liu, Y., Chen, X., et al. (2018). Fitness advantage of mcr-1-bearing Inc I2 and IncX4 plasmids in vitro. Front. Microbiol. 9:331. doi: 10.3389/fmicb.2018.00331
Xu, L., Wang, P., Cheng, J., Qin, S., and Xie, W. (2019). Characterization of a novel bla NDM-5-harboring IncFII plasmid and an mcr-1-bearing IncI2 plasmid in a single Escherichia coli ST167 clinical isolate. Infect. Drug Resist. 12, 511–519. doi: 10.2147/idr.S192998
Yang, X., Xie, M., Xu, Q., Ye, L., Yang, C., Dong, N., et al. (2022). Transmission of pLVPK-like virulence plasmid in Klebsiella pneumoniae mediated by an Incl 1 conjugative helper plasmid. iScience 25:104428. doi: 10.1016/j.isci.2022.104428
Keywords: Escherichia coli, ST167, mcr-1, cointegrate plasmid, Is26
Citation: Xia S, Wang W, Cheng J, Zhang T, Xia Z, Zhao X, Han Y, Li Y, Shi X and Qin S (2022) Emergence of a novel hybrid mcr-1-bearing plasmid in an NDM-7-producing ST167 Escherichia coli strain of clinical origin. Front. Microbiol. 13:950087. doi: 10.3389/fmicb.2022.950087
Edited by:
Alberto Antonelli, University of Florence, ItalyReviewed by:
Hui Li, Beijing Center for Disease Prevention and Control, ChinaIlaria Baccani, University of Florence, Italy
Copyright © 2022 Xia, Wang, Cheng, Zhang, Xia, Zhao, Han, Li, Shi and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yonghong Li, bHloMjI0QDE2My5jb20=; Xiufang Shi, eGZzaGlAenp1LmVkdS5jbg==; Shangshang Qin, cWluc2hhbmdzaGFuZ0AxMjYuY29t
†These authors have contributed equally to this work
 Shuang Xia1†
Shuang Xia1† Jing Cheng
Jing Cheng Xiaoyu Zhao
Xiaoyu Zhao Shangshang Qin
Shangshang Qin