- 1Department of Biology and Biochemistry, University of Bath, Bath, United Kingdom
- 2Department of Pharmacy and Pharmacology, University of Bath, Bath, United Kingdom
New therapeutic options are urgently required for the treatment of Staphylococcus aureus infections. Accordingly, we sought to exploit the vulnerability of S. aureus to naturally occurring polyamines. We have developed and tested the anti-staphylococcal activity of three novel linear polyamines based on spermine and norspermine. Using a panel of genetically distinct and clinically relevant multidrug resistant S. aureus isolates, including the polyamine resistant USA300 strain LAC, compound AHA-1394 showed a greater than 128-fold increase in inhibition against specific S. aureus strains compared to the most active natural polyamine. Furthermore, we show that AHA-1394 has superior biofilm prevention and biofilm dispersal properties compared to natural polyamines while maintaining minimal toxicity toward human HepG2 cells. We examined the potential of S. aureus to gain resistance to AHA-1394 following in vitro serial passage. Whole genome sequencing of two stable resistant mutants identified a gain of function mutation (S337L) in the phosphatidylglycerol lysyltransferase mprF gene. Inactivation of mutant mprF confirmed the importance of this allele to AHA-1394 resistance. Importantly, AHA-1394 resistant mutants showed a marked decrease in relative fitness and increased generation time. Intriguingly, mprF::S337L contributed to altered surface charge only in the USA300 background whereas increased cell wall thickness was observed in both USA300 and SH1000. Lastly, we show that AHA-1394 displays a particular proclivity for antibiotic potentiation, restoring sensitivity of MRSA and VRSA isolates to daptomycin, oxacillin and vancomycin. Together this study shows that polyamine derivatives are impressive drug candidates that warrant further investigation.
Introduction
Staphylococcus aureus has become notorious as a multidrug resistant pathogen. Globally, an estimated 100,000 deaths are attributed to, and almost 1 million deaths associated with resistant S. aureus infections, second only to Escherichia coli in AMR burden (Murray et al., 2022).
Vancomycin (glycopeptide) and daptomycin (lipopeptide) remain the treatments of choice for invasive MRSA infections (Catherine Liu et al., 2011). Vancomycin failure is increasingly being reported in MRSA infections and is associated with a slow increase in vancomycin minimal inhibitory concentrations (MIC), coined “MIC creeping” (Canty et al., 2021). Similarly, daptomycin resistant strains of MRSA have been reported, and have been attributed to single nucleotide polymorphisms (SNPs) housed in cell envelope and two-component system genes (Miller et al., 2016). As such, the current treatment options for MRSA, particularly invasive infections, are limited.
Polyamines are small polycationic molecules that are involved in numerous cellular functions both in eukaryotes and bacteria (Hamana and Matsuzaki, 1992). The uniform distribution of positive charges across a hydrophobic backbone are thought to be essential in how these compounds exert their diverse array of functions (Cohen, 1998). Polyamines interact with highly anionic molecules such as nucleic acids, phospholipids and certain proteins leading to pleiotropic effects in gene expression, membrane stabilization, enzymatic activity, and protein translation (Tabor and Tabor, 1984; Schuber, 1989; Zheliaskova et al., 2000; Sakamoto et al., 2020). Until recently it was believed that polyamines were produced and required by all forms of life. However, certain bacterial species lack de novo polyamine biosynthesis machinery and rely on polyamine import from their host. In these cases, supplementation of growth medium with polyamines such as spermine and spermidine enhance bacterial growth (Joshi et al., 2011). S. aureus is a peculiar example as it lacks polyamine biosynthetic genes and displays hypersensitivity to exogenously added polyamines, where treatment with spermine and spermidine was shown to be bactericidal (Joshi et al., 2011; Thurlow et al., 2013).
The development of new antimicrobials and anti-infectives represents an important step in combatting antimicrobial resistance as outlined by the UN Interagency Coordination Group on AMR (The Ad hoc Interagency Coordination Group (IACG) on Antimicrobial Resistance, 2019). Accordingly, in this study we sought to exploit the existing sensitivity of S. aureus toward polyamines, synthesizing novel linear polyamine compounds (Figure 1) which had superior anti-staphylococcal activity compared to natural compounds. One of these polyamine derivatives, designated AHA-1394, displayed potent bactericidal activity against a range of S. aureus clinical isolates, including hyper virulent community-associated MRSA, vancomycin intermediate S. aureus (VISA) and vancomycin resistant S. aureus (VRSA) strains, whilst displaying mild toxigenic effects to host cells comparably to clinical used antibiotics. Gene inactivation of mprF resulted in enhanced susceptibility whereas mprF gain of function mutations, specifically S337L, conferred resistance. Finally, we report significant synergism of AHA-1394 with oxacillin, vancomycin and daptomycin using a panel of multidrug resistant S. aureus isolates, highlighting the potential of AHA-1394 to resensitize S. aureus to clinically relevant antibiotics.
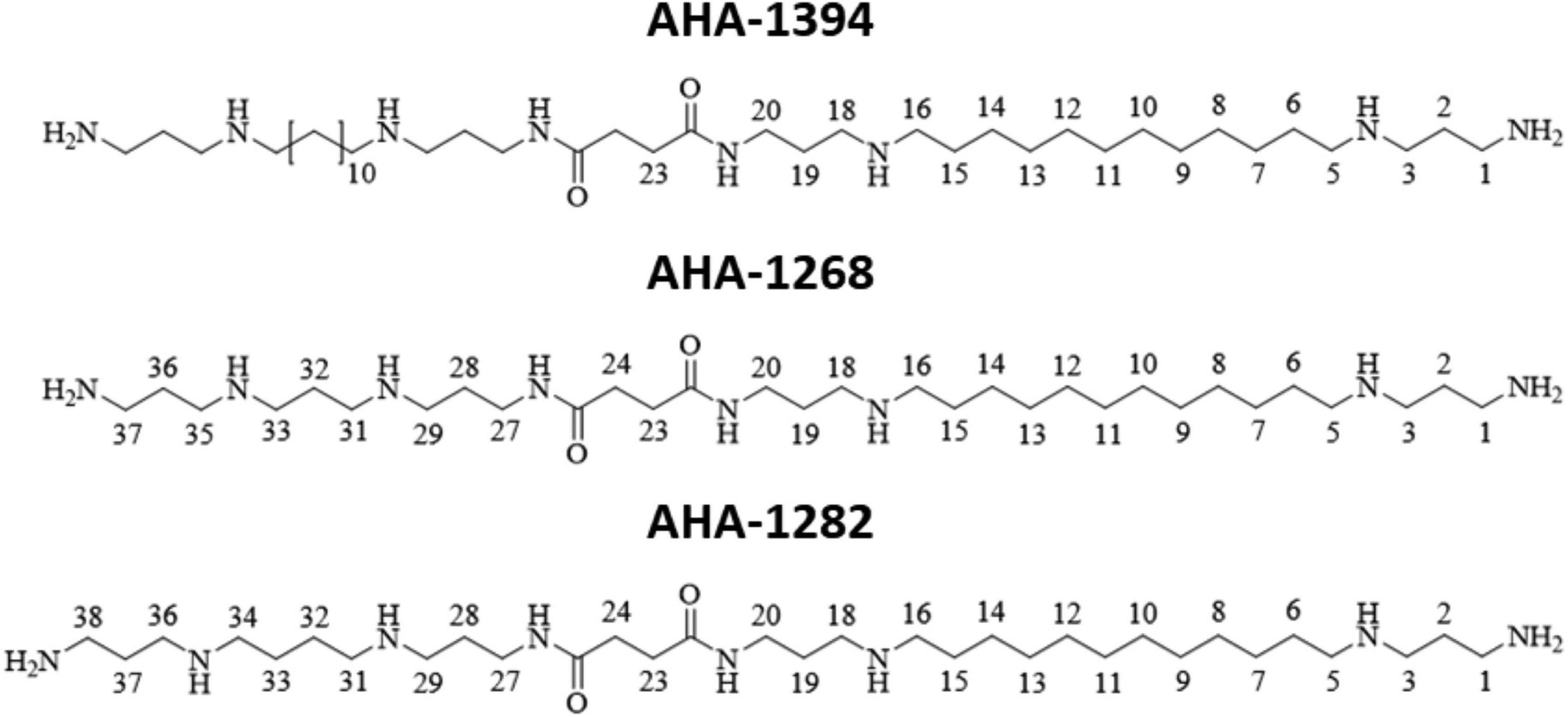
Figure 1. Structures of novel linear polyamines with improved anti-staphylococcal activity used in this study.
Materials and methods
Bacterial strains and culture conditions
S. aureus strains used in this study as listed in Table 1. S. aureus isolates were streaked onto tryptic soy agar (TSA) and incubated for 18 h at 37°C, and single colonies transferred into 3 ml of growth medium in 15 ml polystyrene test tubes. Erythromycin (5 μg/ml) was added to the growth medium for transposon mutants obtained from the Nebraska Transposon Mutant Library (NTML) (Fey et al., 2013) DNA from JE2 mprF::Tn was transduced into SH1000-B and SH1000-128 with Φ11 and colonies containing the inserted transposon were screened on TSA containing erythromycin (10 μg/ml).
Synthesis of polyamine compounds
A two-step procedure was used to form long linear tetraamines (Karigiannis and Papaioannou, 2000) from commercially available 1,12-diaminododecane by reaction with two equivalents of acrylonitrile to undergo two 1,4-Michael addition reactions to obtain the dinitrile in 87% yield (Figure 1). Both the nitrile functional groups were successfully reduced to primary amines using catalytic Raney nickel and sodium hydroxide (co-catalyst) under a hydrogen pressure of 1 bar (Bergeron and Garlich, 1984; Klenke and Gilbert, 2001) to obtain the desired tetraamine in 75 % yield. Trifluoroacetyl is an ideal amine protecting group in the gram-scale protection of polyamines in preparing unsymmetrical polyamine amides (Geall and Blagbrough, 2000). The key intermediate carboxylic acids were then synthesized by the addition of one equivalent of succinic anhydride to a solution of the triBoc-protected long linear polyamines in anhydrous pyridine. All spectral data confirmed that the reactions successfully occurred. The designed target polyamine amides hetero AHA-1268 and AHA-1282, and homo AHA-1394 dimers of these linear polyamine amides incorporating one or two units of 1,12-diaminododecane as 3.12.3 methylene moieties were prepared on a 100–300 mg scale in good yield, and the final poly-TFA salt products were lyophilized and confirmed by mass spectrometry.
AHA-1268 HRMS: Found 585.5538 (m/z), C31H69N8O2 requires 585.5465 (m/z) [M+H]+.
AHA-1282 HRMS: Found 599.5690 (m/z), C32H71N8O2 requires 599.5630 (m/z) [M+H]+.
AHA-1394 HRMS: Found 733.6757 (m/z), C40H86NaN8O2 requires 733.6874 (m/z) [M+Na]+.
Determination of polyamine minimum inhibitory concentration
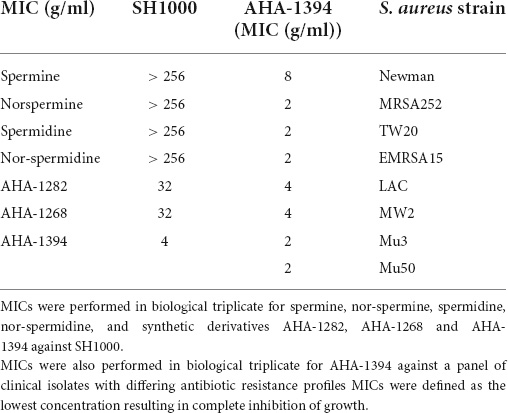
Minimum inhibitory concentration (MIC) of polyamine compounds against S. aureus strains were determined by the microdilution method using Mueller Hinton broth (MHB) (CLSI, 2018). Polyamine compounds were reconstituted in sterile deionized water and diluted in MHB, to a final concentration ranging between 256 and 0.5 μg/ml. The MIC was determined as the lowest concentration that completely inhibited bacterial growth. Experiments were performed using two technical repeats with three biological replicates.
Biofilm prevention and dispersion assays
S. aureus strain SH1000 was used in all biofilm studies. For biofilm prevention, 18 h bacterial cultures were diluted 1:200 in Tryptic soy broth containing 0.5% glucose (TSB-G) and added to polyamines in a 1:1 ratio to a final concentration ranging between 256 and 2 μg/ml in 96-well flat-bottomed microtiter plate, incubated for 24 h at 37°C under static conditions. For biofilm dispersion assays, 18 h bacterial cultures were diluted 1:400 in TSB-G in 96 well plates and incubated at 37°C for 6 h or 24 h, following which non-adhered bacteria were carefully aspirated and biofilms washed three times in sterile PBS. Following washing, 200 μl of individual polyamines were diluted in TSB-G and added to biofilms to a final concentration ranging between 4,096 and 8 μg/ml and incubated for a further 24 h 37°C under static conditions. Biofilm formation was determined using the crystal violet assay as previously described (Lacoma et al., 2019). Experiments were performed with three technical repeats and four biological repeats.
HepG2 liver cell toxicity
HepG2 cells were cultured as previously described (Rajamuthiah et al., 2015). Cells were grown for either 8 or 24 h after which cell viability was assessed using the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. Absorbance (OD595nm) was recorded and % toxicity was measured relative to that of the no-treatment control. Experiments were performed using two technical repeats with three biological replicates.
Multi-passage resistance studies and whole genome sequencing
Multi-passage resistance selection studies using compound AHA-1394 were performed with one MRSA (strain LAC) and one MSSA (strain SH1000) isolate. Serial passage was performed in 15 ml tubes containing 3 ml of MHB with or without varying concentrations of AHA-1394. The tubes were incubated at 37°C for 24 h prior to each serial passage; passages were performed at 24 h intervals by transferring a 30 μl aliquot of culture containing approximately 106 CFU from the tube nearest the MIC which had the same turbidity as antibiotic-free controls. When an MIC for a strain stabilized at 128 μg/ml during four successive passages, serial transfer in the presence of antibiotic was discontinued, and the strains were grown to 10 passages in antibiotic-free medium.
Genome sequencing and bioinformatic analysis was provided by MicrobesNG1. We chose 4 strains to sequence: Two strains that were resistant to AHA-1394 at 128 μg/ml (LAC-128 and SH1000-128) and two control strains (LAC-B and SH1000-B) that were serially passaged in parallel in the absence of AHA-1394. Genomic DNA was isolated according to MicrobesNG protocol and genome sequencing was performed on an Illumina sequencing platform with 2 × 250 bp paired end reads with 30X coverage. Accession numbers can be found here https://www.ncbi.nlm.nih.gov/bioproject/PRJNA859022.
Relative Darwinian fitness
Competitions were established in TSB using 104 CFU/mL of either SH1000-B or LAC-B and corresponding mutant strains. The bacteria were competed at 37°C with shaking (180 rpm) for 24 h. Final cell numbers were enumerated by serial dilutions on TSA plates (total cell count) and TSA plates containing 16 μg/ml AHA-1394 as an indicator for mutant cell count. The fitness of a strain was defined as a measure of the reproductive success of the population, which can be expressed as the natural logarithm of the ratio of the final and initial cell densities of the culture (Lenski et al., 1991), using the following formula:
Where A(0), estimated density of test strain at time 0; M(0), estimated density of marker strain at time 0; A(1), estimated density of test strain at time 1 d; M(1), estimated density of marker strain at time 1 d; ln, natural logarithm (logarithm to the base e).
FITC-poly-L-lysine and cytochrome C binding assays
A FITC-poly-L-lysine (FITC-PLL) binding assay was used to determine the relative surface charge of the bacteria. Overnight S. aureus cultures where diluted to an OD600nm 0.5 in 1 ml of PBS. These suspensions were subsequently incubated with 80 μg/ml FITC-PLL (MW 15,000–30,000; Sigma) for 10 min at room temperature in the dark. Cells were then washed three times in 1 ml of PBS by three rounds of centrifugation (16,000 × g for 1 min). Fluorescence was visualized by using a PHERAstar (BMG Labtech) plate reader (excitation 485 nm: emission 525 nm). The FITC-poly-L-lysine binding assay was compared with another commonly used method to investigate differences in surface charge, a cytochrome C binding assay. For the cytochrome C binding assay, overnight cultures were normalized to an OD600nm of 8. The bacterial suspensions were washed twice in MOPS buffer (20 mM pH 7.0) and finally resuspended in 200 μl of MOPS buffer. Samples were then combined with 50 μl of cytochrome C (equine heart, 2.5 mg/ml in MOPS buffer) and incubated for 10 min at room temperature. Finally, samples were pelleted (16,000 × g for 1 min) and 200 μl of supernatant read for absorbance at Abs530nm using a SUNRISE Tecan microplate reader.
Transmission electron microscopy
S. aureus strains SH1000-B, SH1000-128, SH1000-128 mprF::Tn, LAC-B and LAC-128 were imaged by transmission electron microscopy (TEM) to compare differences in cell wall thickness, using a previously described method (Grigor’eva et al., 2020). Briefly, 1 ml of 18 h overnight cultures grown in TSB were pelleted and resuspended in 500 μl of 4% paraformaldehyde, 2.5% glutaraldehyde mixture. Samples were subsequently incubated at room temperature for 20 min before being pelleted and stored for 24 h at 4°C. Following this, samples were washed to remove fixative, postfixed in a 1% osmium tetroxide solution for 1 h and dehydrated in ethanol acetone. Samples were then embedded in an epon-araldite mixture and left to polymerize at 60°C for 24–48 h to obtain hard blocks. Blocks were sectioned (70 nm) using a Leica UC7 ultramicrotome and imaged using a FEI Tecnai T12 microscope at the Wolfson Bioimaging Facility, University of Bristol. For cell wall thickness analysis, 50 cells were measured for each strain, with each individual cell being measured in 4 equidistant locations before being averaged. Similarly, 50 cells were measured for each strain for the cell area analysis. The diameter was measured across 2 perpendicular planes before being averaged, the radius calculated, yielding cell area values by A = π r2.
Synergistic activity with clinically relevant antibiotics
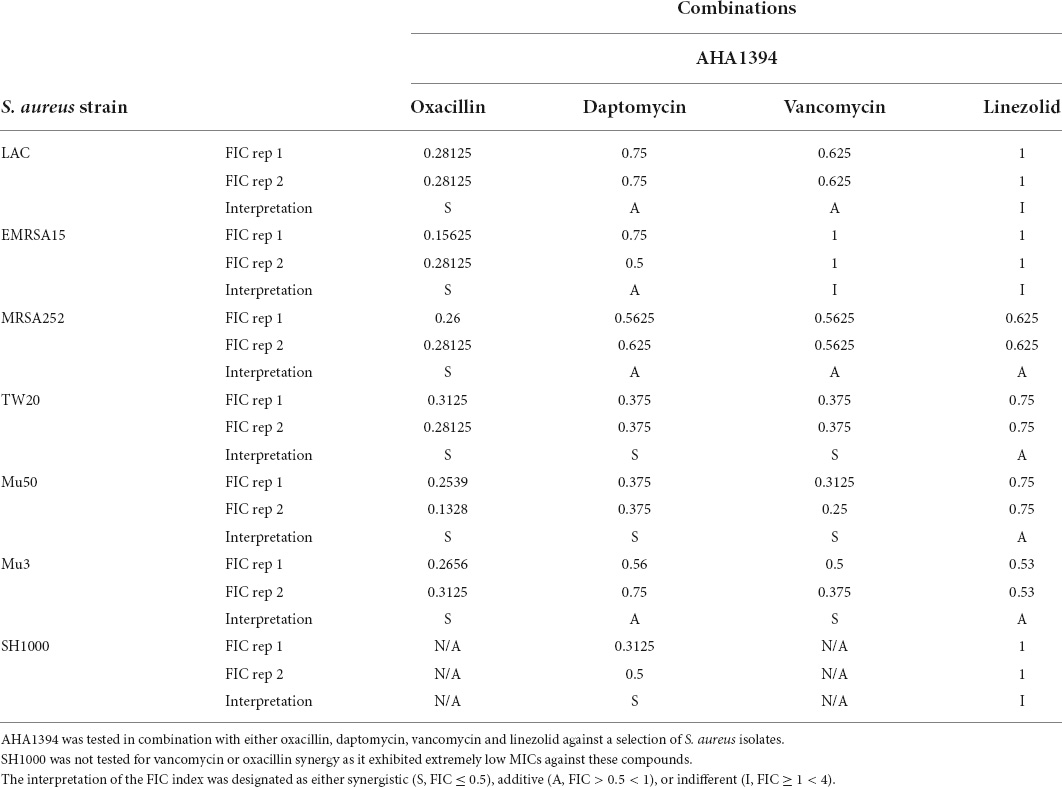
The ability of AHA-1394 to synergize with daptomycin, oxacillin, vancomycin and linezolid was tested using the microbroth dilution method. Overnight S. aureus cultures were diluted 1:200 in MHB and grown to early exponential phase (OD600nm) of 0.5–0.6. The final concentration was 256 μg/ml for oxacillin, 8 μg/ml for daptomycin, 8 μg/ml for linezolid and either 8 μg/ml or 16 μg/ml for vancomycin depending on the strain tested. Following 18 h incubation at 37°C the sum of the fractional inhibitory concentration (ΣFIC) was calculated for each well that corresponded to a minimal inhibitory concentration (MIC) using the equation: ΣFIC = FICA + FICB = (MICA+B combination/MICA) + (MICB+A combination/MICB), where MICA and MICB are the MICs of drugs A and B alone. The FIC index value presented represents the lowest value achieved for each polyamine/drug combination. The interpretation of the FIC index was designated as either synergistic (S, FIC ≤ 0.5), additive (A, FIC >0.5 < 1), or indifferent (I, FIC ≥ 1 < 4).
Statistical analysis
Paired two-tailed Student’s t-test or one-way analysis of variance (ANOVA) with a Dunnett’s multiple comparisons test, with single pooled variance (GraphPad Prism v9.0), were used to analyze the observed differences between experimental results. A P-value < 0.05 was considered statistically significant.
Results
AHA-1394 displays antimicrobial activity against an extended panel of MSSA, MRSA, VISA and VRSA S. aureus strains
In this study we synthesized three novel linear synthetic polyamines designated AHA-1282, AHA-1268 and AHA-1394 with an increased total net charge compared to their monomeric counterparts. To test the activity of these new compounds, the MIC was compared to spermine, nor-spermine, spermidine and nor-spermidine against SH1000, an MSSA laboratory strain (Table 2 and Supplementary Figure 1). SH1000 was able to grow in the presence of all four of the naturally occurring polyamines up to concentrations of 256 μg/ml. Both AHA-1282 and AHA-1268 displayed an MIC of 32 μg/ml, a greater than eightfold increase in activity whereas AHA-1394 displayed the most potent anti-staphylococcal activity with an MIC of 2 μg/ml, a greater than 64-fold increase in activity. MIC of AHA-1394 was further tested using a cohort of clinically relevant, multi-drug resistant and genetically diverse strains summarized in Table 1, with MIC values ranging from 2 to 8 μg/ml (Table 2 and Supplementary Figure 2). The minimum bactericidal concentration of AHA1394 against both MSSA (SH1000) and MRSA (LAC) was determined as 16 μg/ml (Supplementary Figure 3).
AHA-1394 is a potent biofilm inhibitor
We investigated whether the synthetic polyamines displayed enhanced biofilm inhibitory activity against the strong biofilm-forming isolate, SH1000 (Figure 2 and Supplementary Figure 4). Of the natural polyamines, only spermine displayed limited biofilm reduction (20%) at 256 μg/ml. Comparably, AHA-1268 and AHA-1282 reduced biofilm formation by greater than 50% at 64 μg/ml, whereas AHA-1394 displayed complete biofilm inhibition at a concentration of 16 μg/ml. We were also interested in whether these compounds could penetrate and contribute to biofilm dispersal. SH1000 biofilm was allowed to form for 6 h or 24 h followed by the addition of the relevant polyamines and a further incubation for 24 h to allow for biofilm dispersal (Figure 2). Spermine and nor-spermine displayed limited biofilm reduction (20%) for both 6 h and 24 h biofilms at 256 μg/ml and 2,048 μg/ml respectively. AHA-1268 and AHA-1282 caused a 50% reduction in 6 h biofilm formation at 256 μg/ml with AHA-1394 displaying 50% biofilm reduction at 128 μg/ml. AHA-1394 also performed best at biofilm dispersal of 24 h biofilms, where a 50% reduction was observed following treatment with 1,024 μg/ml.
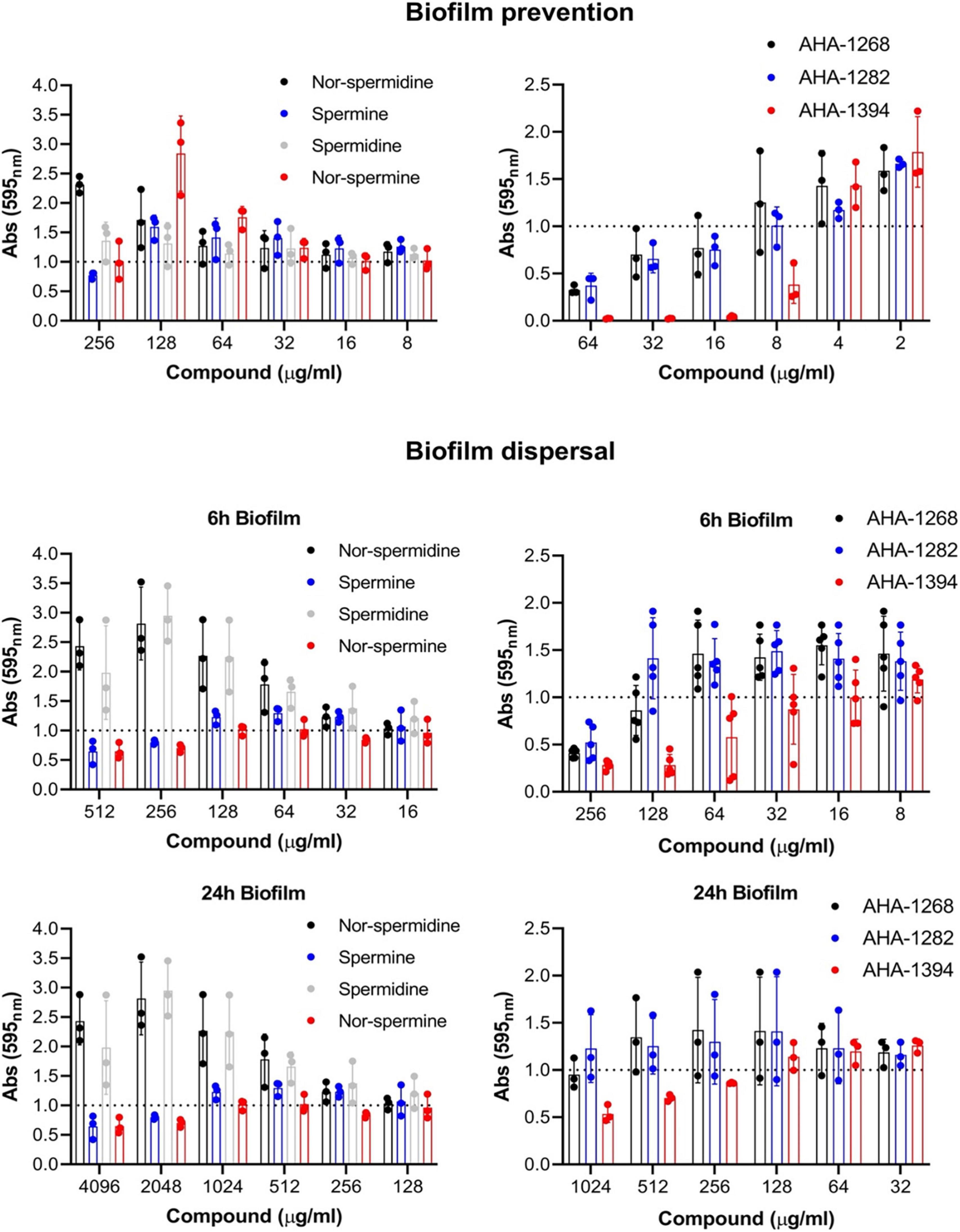
Figure 2. AHA-1394 performs better than natural polyamines in biofilm inhibition. The biofilm prevention and dispersal activity of natural polyamines were compared to AHA-1268, AHA-1282 and AHA-1394. For biofilm prevention polyamines were added at a final concentration ranging 256–2 μg/ml, whereas for biofilm dispersal polyamines were added at a final concentration ranging 4,096–8 μg/ml. Absorbance 595nm values of the polyamines below the normalized threshold of 1.0 showed biofilm preventative and dispersal activity. The dots represent biological replicates with the bars representing the mean and error bars the standard deviation.
AHA-1394 displays minimal toxicity against human cells
The cytotoxicity of the most active synthetic polyamines, AHA-1394, toward the human hepatocellular carcinoma cell line (HepG2) was evaluated at both 8 h and 24 h incubation (Figure 3). The IC50 of AHA-1394 against HepG2 cells was 144.2 μg/ml and 50.95 μg/ml following 8 h and 24 h incubation respectively. The MIC50 (defined as the median concentration for which 50% of the isolates were inhibited (Schwarz et al., 2010) for AHA-1394 from the 10 S. aureus strains tested was 2 μg/ml giving a selectivity index (defined as IC50/ MIC50 (Ong et al., 2013)) of 25.48, providing a robust therapeutic window, indicating that AHA-1394 is selective for bacteria over human cells.
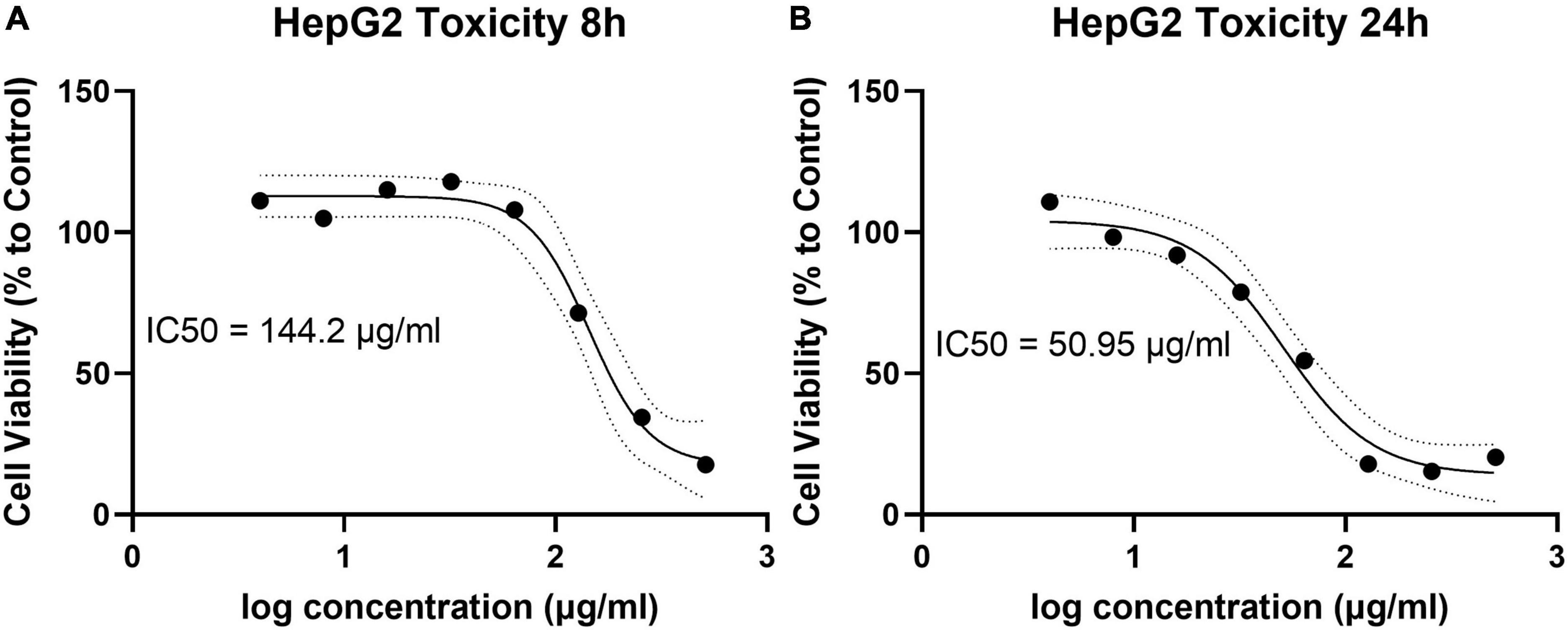
Figure 3. AHA-1394 HepG2 cytotoxicity and IC50. Log AHA-1394 concentration was plotted against the percentage of live HepG2 cells at (A) 8 h and (B) 24 h following normalization of HepG2 cells incubated without AHA-1394. The IC50 was calculated according to standard curve interpretation and defined as the concentration responsible for 50% HepG2 cell death.
AHA-1394 is active against previously described polyamine resistant strains
S. aureus polyamine resistance involves either the acquisition of the speG N-1 acetyltransferase or spontaneous mutation resulting in small colony variant formation (SCV) (Joshi et al., 2011). To test whether these two polyamine resistance mechanisms could be associated with a loss of susceptibility to AHA-1394, MICs were performed against JE2 WT and isogenic speG transposon mutant, and Newman WT and menD SCV associated mutant. Inactivation of speG had no effect on the susceptibility toward AHA-1394 with both JE2 and JE2 speG::Tn having an MIC of 8 μg/ml, suggesting the inability of SpeG to acetylate this novel polyamine. Deletion of menD in the Newman background was associated with a slight increase of MIC from 8 μg/ml to 16 μg/ml.
Fitness cost associated with AHA1394 resistance
Two S. aureus strains (LAC and SH1000) were serially passaged in increasing concentrations of AHA-1394 up to 128 μg/ml, in parallel to broth controls (Figure 4A). This resulted in the generation of two stable, resistant strains denoted LAC-128 and SH1000-128 after 11 and 12 passages respectively. Spontaneously acquired resistance mechanisms are often associated with characteristic growth defects and fitness costs, typified by SCVs (Proctor et al., 2006). Therefore, the relative growth kinetics of LAC-128 and SH1000-128 were compared to their serially passaged broth controls, denoted LAC-B and SH1000-B (Figures 4B,C). LAC-128 and SH1000-128 displayed increased generation times (doubling time = ln2/k) of 3.4 min and 13.2 min respectively, compared to control strains when grown in TSB, confirming an inherent growth defect, suggesting AHA-1394 resistance is energetically costly. Accordingly, the relative Darwinian fitness of LAC-128 and SH1000-128 were calculated following 18 h growth. Both SH1000-128 and LAC-128 showed a marked decrease in relative fitness, with mean values of 0.692 (SD = 0.008) and 0.696 (SD = 0.035) respectively.
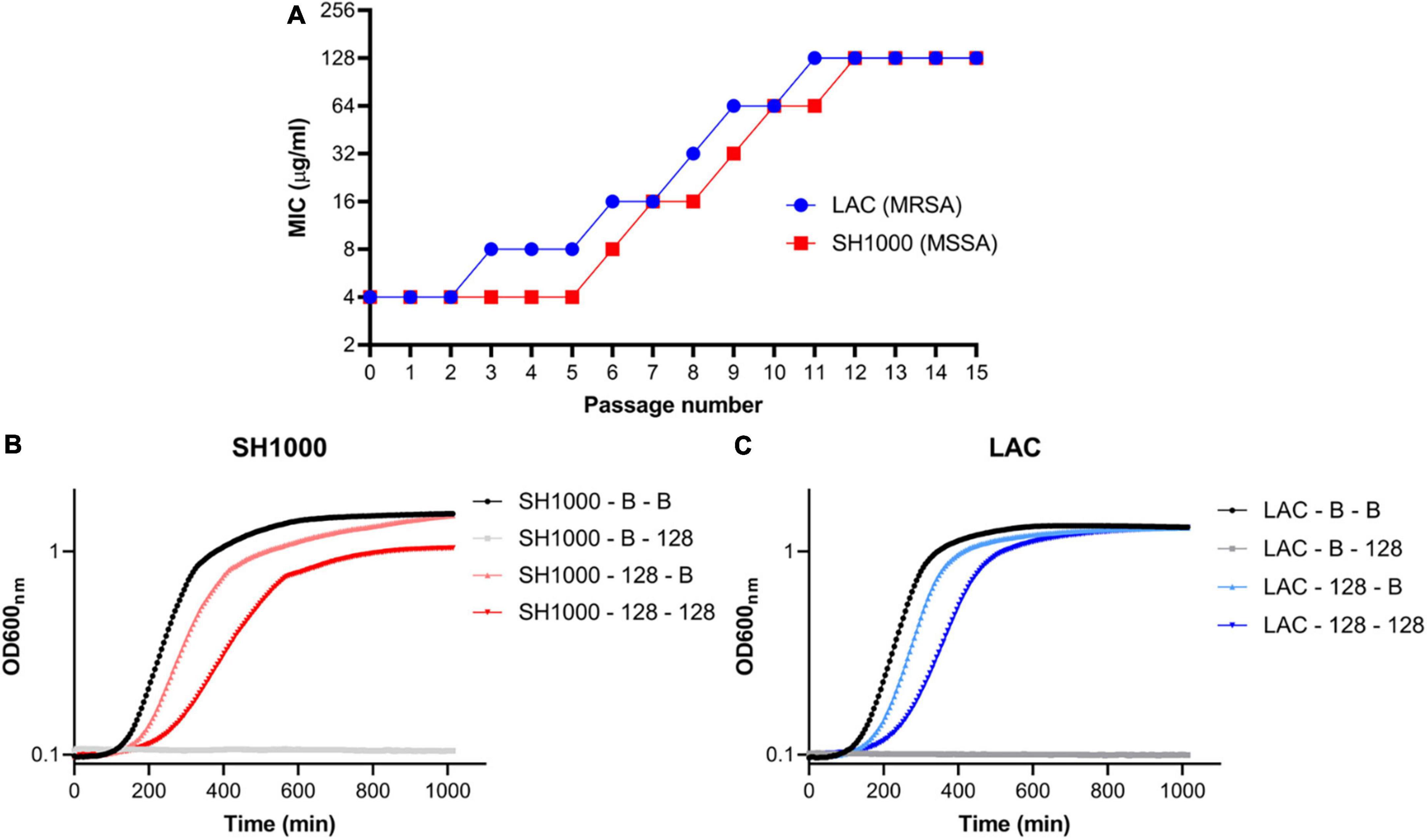
Figure 4. Stable AHA-1394 resistant mutants emerge following AHA-1394 serial passage. (A) Increase of MIC following passage every 24 h of LAC (blue) and SH1000 (red). Bacterial growth curves of (B) SH1000-B and SH1000-128 and (C) LAC-B and LAC-128 grown in MHB (SH1000-B-B; SH1000-128-B; LAC-B-B; LAC-128-B) or MHB containing AHA-1394 (128 μg/ml) (SH1000-B-128; SH1000-128-128; LAC-B-128; LAC-128-128). OD600nm was plotted against time following 18 h incubation. The dots represent the mean of three biological replicates.
Resistance to AHA-1394 is associated with gain of function mutations in mprF
To investigate the mechanism of AHA-1394 resistance, LAC-128 and SH1000-128, alongside respective controls (LAC-B and SH1000-B) were genome sequenced. Subsequent pairwise single nucleotide polymorphism (SNP) comparison between resistant and sensitive controls identified non-synonymous mutations within resistant mutants (22 for SH1000-128 and 15 for LAC-128) (Figure 5A). Both LAC-128 and SH1000-128 had mutations in three genes: mprF, rrfA and SAUPAN00861000 (Figure 5B). The exact rrfA and SAUPAN00861000 mutation differed between LAC-128 and SH1000-128, however, both contained the same S337L mprF mutation, indicating a gain of function mprF mutation may be responsible for AHA-1394 resistance. To confirm, we performed AHA-1394 MIC experiments using wild-type JE2 and isogenic JE2 mprF::Tn. JE2 displayed an MIC of 8 μg/ml, whereas JE2 mprF::Tn was more susceptible to AHA-1394 with an MIC of 4 μg/ml. We next phage transduced the bursa aurealis transposon using Φ11 from JE2 mprF::Tn into two recipient strains: SH1000-B and SH1000-128. AHA-1394 MICs were repeated on SH1000-B, SH1000-B mprF::Tn, SH1000-128 and SH1000-128 mprF::Tn. Both SH1000-B mprF::Tn and SH1000-128 mprF::Tn were more susceptible to AHA-1394 killing with MICs of 2 μg/ml and 8 μg/ml compared with 4 μg/ml and > 128 μg/ml for SH1000-B and SH1000-128 respectively. Importantly, deletion of mprF restored the susceptibility of SH1000-128 to one doubling above SH1000-B, indicating the importance of mprF mutation S337L in AHA-1394 resistance.
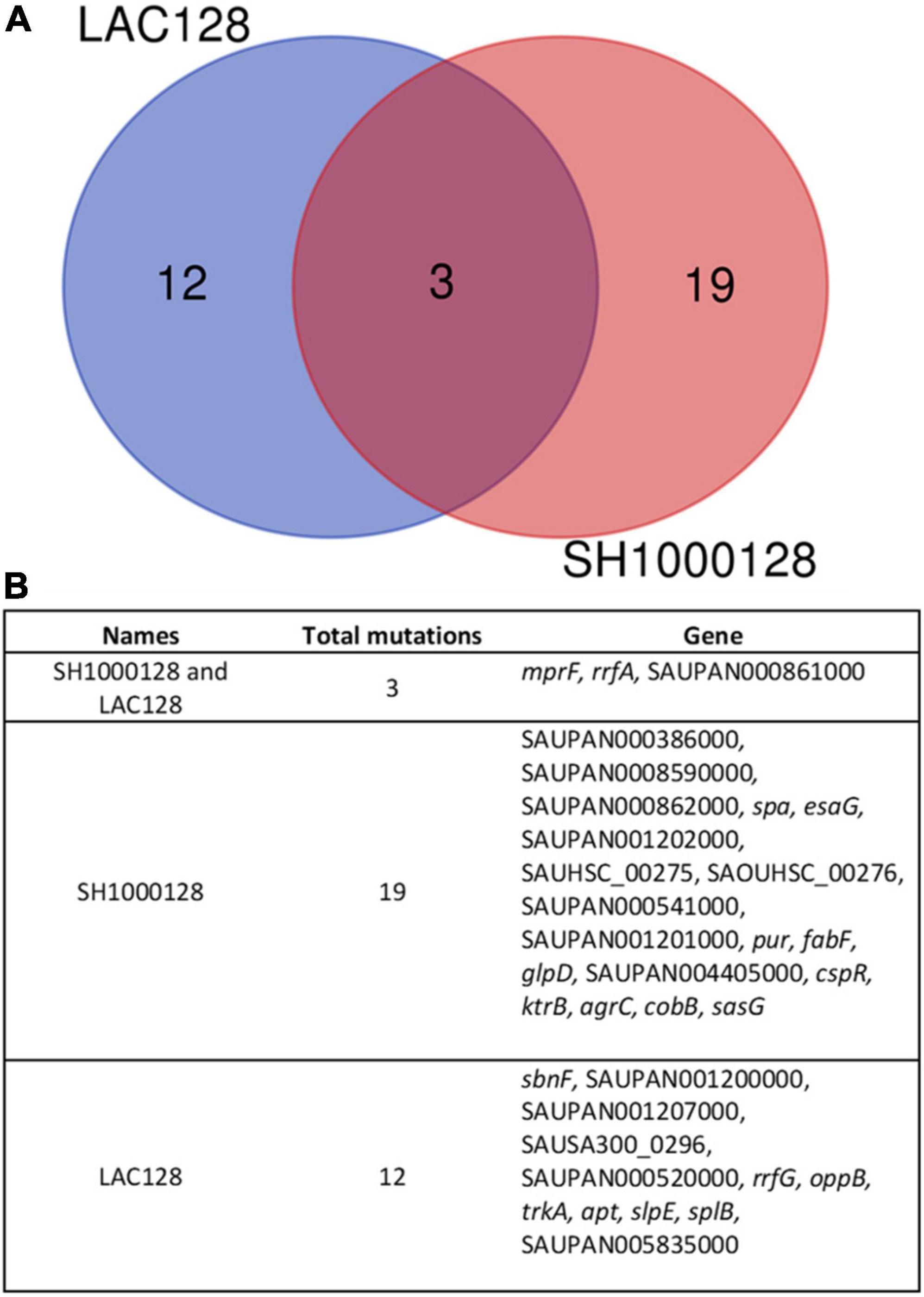
Figure 5. Comparison of mutations indicate a role for MprF in AHA-1394 resistance. (A) Venn diagram showing mutations unique to LAC-128 (blue), unique to SH1000-128 (red) and mutation shared by both mutants (mauve) when compared to the parental strains LAC-B and SH1000-B. (B) Descriptions of mutations present and shared in LAC-128 and SH1000-128. Pangenome locus tags are shown (“SAUPAN”). Where a pangenome locus tag is missing the relevant USA300 “SAUSA300” or NCTC8325 “SAOUHSC” locus tag is used.
mprF S337L mutation is associated with altered surface charge in strain LAC
Mutations in mprF that confer elevated daptomycin MIC are classically associated with increased surface charge due to elevated levels of lysyl-phosphatidylglycerol in the bacterial membrane (Jones et al., 2008). To examine whether the S337L mutation observed in AHA1394 resistant strains resulted in an increase in membrane charge, both an FITC-poly-l-lysine and cytochrome C binding assay were employed. In agreement with the literature, a significant increase in FITC-poly-l-lysine and cytochrome C binding was detected for the SH1000-B mprF::Tn control strain confirming the suitability of both assays (Figure 6) (Ernst et al., 2018). Interestingly, there was no notable difference in the FITC-poly-l-lysine binding ability of either LAC-128 or SH1000-128 compared to LAC-B and SH1000-B (Figure 6A), whereas a significant difference in bound cytochrome C was observed only between the LAC-B and LAC-128 and not in the SH1000 background. This suggests the S337L mutation present in LAC-128 may alter the levels of lysyl-phosphatidylglycerol in the membrane. We also sought to test whether the S337L mutation contributed to daptomycin non-susceptibility (DNS) in these two backgrounds by performing daptomycin MICs. The growth of SH1000-B and LAC-B were inhibited at 1 μg/ml, while SH1000-128 and LAC-128 could survive at higher daptomycin concentrations of > 8 μg/ml and 4 μg/ml, respectively. Deletion of mprF in the SH1000-B background reduced the MIC to 0.25 μg/ml while deletion of mprF in the SH1000-128 also increased daptomycin susceptibility to 2 μg/ml confirming the involvement of S337L in conferring daptomycin resistance.
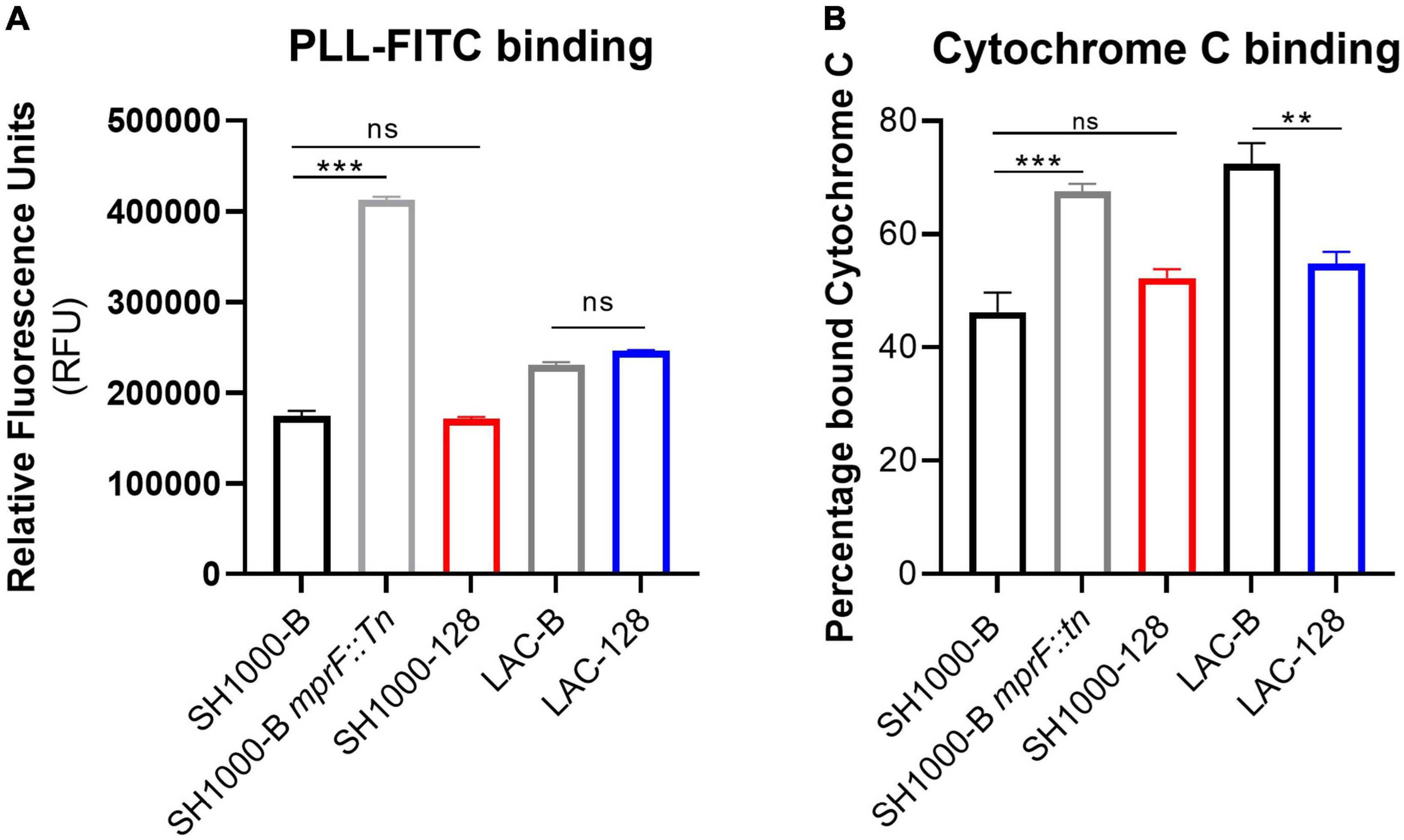
Figure 6. S337L is associated with altered cell surface charge in strain LAC. An indication of cell surface charge was estimated through the binding ability of (A) PLL-FITC, shown by relative fluorescence units (RFU) normalized to a PBS blank and (B) cytochrome C binding depicted as percentage of bound cytochrome C normalized to a MOPS blank. The bars represent the mean of three biological replicates and the error bars the standard deviation. A one-way ANOVA was performed on SH1000-B, SH1000-B mprF::Tn, and SH1000-128, whereas a paired Student’s t-test was performed between LAC-B and LAC-128. “ns” denotes not significant, significance was determined as the following p values: **0.01; ***0.001.
mprF S337L mutation is associated with increased cell wall thickness
As SH1000-128 did not display reduced FITC-poly-l-lysine or cytochrome C binding we wanted to investigate other factors that may underline AHA-1394 resistance. Previous work has indicated that certain mprF mutations are linked to the over expression of the two-component regulatory system, vraRS, conferring an increase in cell wall thickness resulting in decreased susceptibility to daptomycin (Mehta et al., 2012). Accordingly, we investigated cell wall thickness and cell area in the sensitive, resistant and SH1000-128 mprF::Tn mutant strains using transmission electron microscopy (TEM) (Figure 7A). We observed that both SH1000-128 and LAC-128 had significantly thicker cell walls with mean cell wall thickness values of 36.94 nm (SD = 6.23 nm) and 39.24 nm (SD = 6.12 nm), respectively, compared to SH1000-B and LAC-B which displayed cell wall thickness values of 31.67 nm (SD = 2.84 nm) and 28.98 nm (SD = 2.82 nm), respectively (Figure 7B). Interestingly, we found that the increase in cell wall thickness exhibited by the SH1000-128 polyamine resistant strain was ablated when mprF was deleted. This provides insight into the potential role of mprF in regulating cell wall homeostasis.
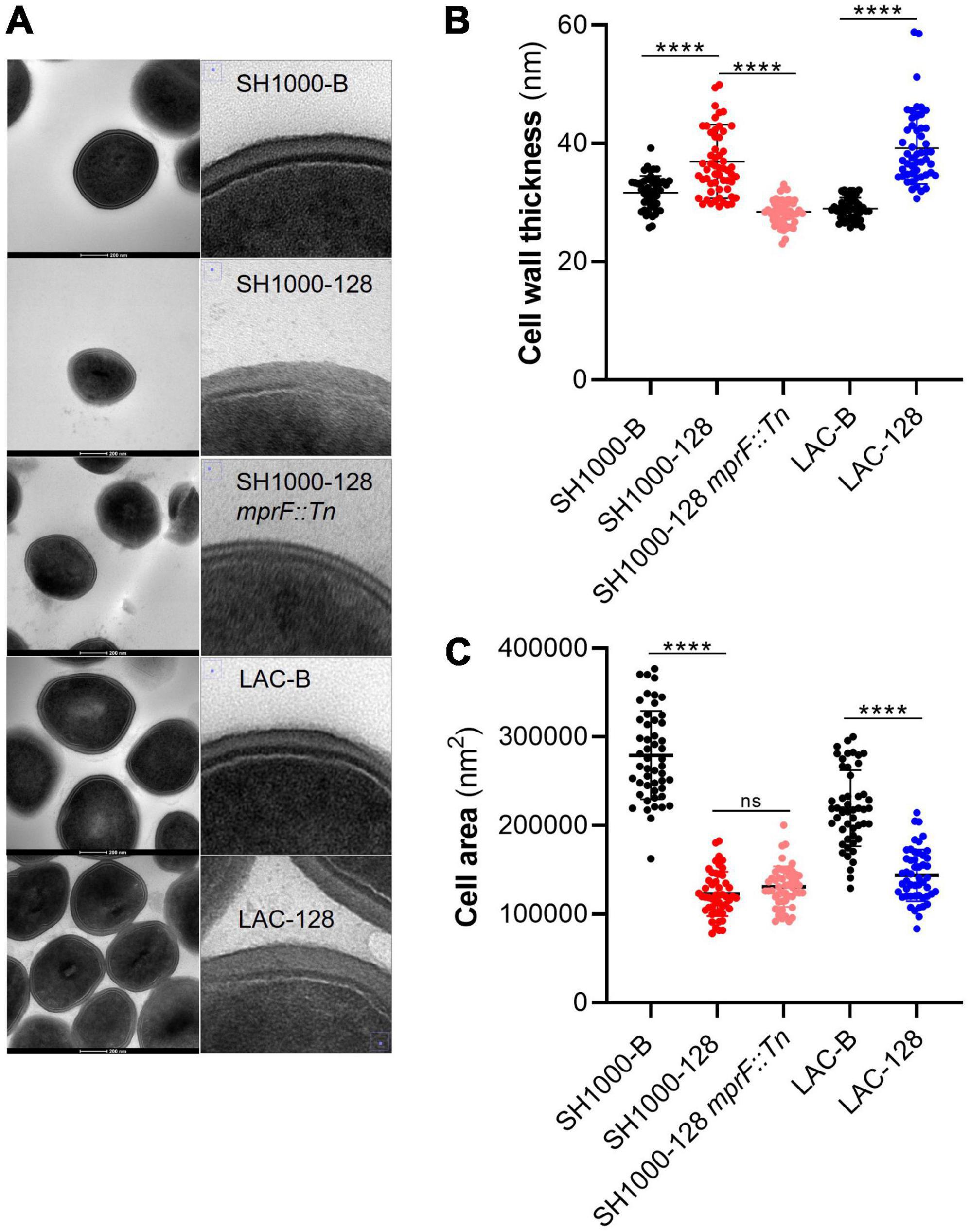
Figure 7. AHA-1394 resistance is associated with increased cell wall thickness. (A) TEM images of each strain at 300,000x magnification are shown with a 200 nm scale bar. For each strain zoomed in images of just the cell wall are shown on the right. (B) The cell wall thickness in nm of 50 individual cells for each of the strains SH1000-B, SH1000-128, SH1000-128 mprF::Tn, LAC-B and LAC-128. The horizontal line represents the mean and the error bars the standard deviation. (C) The cell area in nm2 of 50 individual cells for the same panel of strains. The horizontal line represents the mean and the error bars the standard deviation. A one-way ANOVA was performed on SH1000-B, SH1000-128, SH1000-128 mprF::Tn, whereas a paired Student’s t-test was performed between LAC-B and LAC-128. “ns” denotes not significant, significance was determined as the following p values: ****0.0001.
When performing the cell wall thickness analysis, we noticed that both SH1000-128 and LAC-128 appeared to be smaller. Subsequent analysis of cell area (Figure 7C) indeed showed that both resistant strains were significantly smaller. Importantly, although deletion of mprF in SH1000-128 restored cell wall thickness to near WT-levels, the same effect was not seen for cell area, perhaps indicating that other mutations could be causing this phenotype. Importantly, increased cell wall thickness and altered cell area seems to reaffirm why we observed a decrease in growth rate and relative Darwinian fitness in the AHA-1394 resistant strains.
AHA-1394 synergizes with clinically relevant anti-staphylococcal antibiotics
Previous studies have indicated that natural polyamines synergize with and increase the susceptibility to certain antibiotics (Kwon and Lu, 2007; Liu et al., 2018). Accordingly, synergy measurement by checkerboard MIC analysis were performed using AHA-1394 partnered with the clinically important anti-staphylococcal antibiotics; oxacillin, daptomycin, vancomycin, and linezolid against a selection of clinical isolates (Table 3 and Supplementary Figure 4). We show an impressive ability of AHA-1394 to synergize with oxacillin against all six strains tested as indicated by FIC index. The oxacillin MIC was reduced from > 256 μg/ml to 16 μg/ml (LAC), 16 μg/ml (EMRSA15), 4 μg/ml (MRSA252), 16 μg/ml (TW20), 8 μg/ml (Mu3), and 8 μg/ml (Mu50) with the addition of 0.5–1 μg/ml of AHA-1394. Furthermore, AHA-1394 was able to potentiate the activity of vancomycin against strains TW20, Mu3 and Mu50, restoring vancomycin susceptibility for the VISA strains Mu3 and Mu50 from an initial MIC of 4 μg/ml and 8 μg/ml to 1 μg/ml and 0.5 μg/ml with the addition of 0.5–1 μg/ml of AHA-1394. Strain dependent synergy was also observed for daptomycin partnered with AHA-1394, restoring daptomycin susceptibility toward TW20, Mu50 and SH1000, decreasing the MIC from 4 μg/ml, 8 μg/ml and 8 μg/ml to 0.5 μg/ml,1 μg/ml and 2 μg/ml, respectively, with the addition of 1 μg/ml of AHA-1394. Finally, AHA-1394 was tested for its synergy with the oxazolidinone linezolid, however, it did not improve the bactericidal activity of this antibiotic.
We next tested the activity of antibiotic combinations against the AHA-1394 resistant mutants LAC-128 and SH1000-128 using AHA-1394/oxacillin or AHA-1394/daptomycin against LAC-128 or SH1000-128. AHA-1394 showed impressive synergy with oxacillin against LAC-128 with an FIC of 0.19-0.25. However, AHA-1394 could not improve the killing activity of daptomycin against SH1000-128. Interestingly, we found that the S337L mprF mutation was associated with a “see-saw” effect against oxacillin, whereby increased AHA-1394 MICs were associated with a concomitant decrease in oxacillin MIC (Jiang et al., 2022). For LAC-128 and SH1000-128 the oxacillin MIC was 32 μg/ml and 0.125 μg/ml compared with 256 μg/ml and 2 μg/ml for LAC-B and SH1000-B respectively.
Discussion
We have shown that polyamines can be used as scaffolds for the design of novel antimicrobial compounds effective against S. aureus. We hypothesize that increasing the positive charge along the linear polyamines leads to an increase in their electrostatic attraction to the S. aureus lipid bilayer culminating in enhanced bactericidal activity and biofilm prevention as observed with compound AHA-1394 against a panel of multidrug resistant S. aureus.
S. aureus resistance to polyamines has been described and linked to the acquisition of the arginine catabolic mobile element (ACME), housing the N-1 spermidine acetyltransferase SpeG (Joshi et al., 2011). It is unclear how SpeG-mediated acetylation of polyamines prevents toxicity to S. aureus, however, it is proposed the enzyme acts intracellularly, modifying polyamines that are rapidly imported preventing their modification into toxic intermediates (Kramer et al., 2008). Acquisition of ACME distinguishes the highly virulent USA300 community-associated (CA)-MRSA lineage from other CA- MRSA lineages and has been proposed to have contributed to the hypervirulence, hyper-transmissibility, and increased propensity to cause skin and soft tissue infections (SSTIs) of USA300 (Diep et al., 2006; Thurlow et al., 2013). Importantly, AHA1394 was effective against ACME-positive strain (LAC) as well as a panel of genetically diverse, virulent, and clinically relevant isolates suggesting that SpeG is unable to acetylate AHA-1394. Polyamine resistance can also arise through mutations of the menadione biosynthesis machinery and generation of SCV. Accordingly, we tested the susceptibility of a previously constructed menD SCV mutant and observed an increase in MIC from 8 to 16 μg/ml. Considering SCVs are typically associated with a fourfold increase in MIC against β-lactams and a 4–8-fold increase in MIC against aminoglycosides (R. A. Proctor et al., 1995; Proctor and von Humboldt, 1998; Proctor et al., 2006), our data indicates the therapeutic potential of AHA-1394 against difficult to eradicate S. aureus SCVs.
An important aspect of any antibacterial candidate is the selectivity of the drug to bacteria over human cells. Accordingly, we tested the toxicity of AHA-1394 using the highly sensitive liver-derived HepG2 cell line. We determined the IC50 of AHA-1394 at 24 h to be 50.95 μg/ml, which is comparable to many current antibiotics approved for use in the clinic; azithromycin (Abdel-Hamid et al., 2017) and linezolid (Sun et al., 2019) commonly used to treat S. aureus SSTIs have HepG2 IC50s of 42.8 μg/ml, 10.1 μg/ml, respectively.
Resistant mutants were selected using a broth multi-passage experiment. Repeated exposure for 11 (LAC) and 12 (SH1000) days resulted in a 32-fold increase in MIC. This is comparable to clinical used anti-staphylococcal antibiotics; ciprofloxacin, mupirocin and fusidic acid MICs increased 256-, 64-, and 256-fold, respectively, following 25 passages (D’Lima et al., 2012). Genome sequencing of resistant mutants LAC-128 and SH1000-128 identified a common mprF S337L mutation. MprF is a bifunctional enzyme; the synthase domain functions as a phosphatidylglycerol lysyltransferase, while the flippase domain mediates the transfer of newly synthesized lysyl-phosphatidylglycerol from the inner to the outer leaflet of the bacterial membrane (Ernst et al., 2009). The S337L mutation present in both LAC-128 and SH1000-128 is located within the overlapping region of the synthase and flippase domains (Jiang et al., 2022). We show that S337L confers increased resistance to AHA-1394, contributes to DNS and resistance is abolished upon deletion of mprF::S337L.
Gain of function mutations in MprF are suspected to cause either increased lysyl-phosphatidylglycerol synthase or flippase activity which contributes to greater levels of lysyl-phosphatidylglycerol in the outer leaflet of the membrane, serving to repel positively charge antimicrobials (Ernst and Peschel, 2011; Bayer et al., 2013). As both daptomycin and AHA-1394 are cationic we reasoned that increased positive charge as a result of S337L mutation conferred resistance. Using a FITC-poly-l-lysine binding assay, we found no difference in surface charge for the AHA-1394 resistant mutants. However, analysis of surface charge via cytochrome C binding analysis revealed a significant difference in strain LAC-128 compared to LAC-B, where the resistant mutant displayed reduced cytochrome C binding and thus increased positive charge. MprF mutations that cause DNS but that are not associated with a detectable difference in lysyl-phosphatidylglycerol abundance have been reported (Mehta et al., 2012; Ernst et al., 2018; Jiang et al., 2022). Mehta et al. described isogenic daptomycin sensitive and resistant strains which displayed no difference in the levels of cytochrome c binding, despite containing MprF mutations (Mehta et al., 2012). These MprF mutations, of which S337L was included, did contribute to increased vraRS expression, and corresponded to elevated vancomycin MICs. Increased vraRS expression in daptomycin resistant strains contributed to increased cell wall thickness, preventing access of daptomycin to the membrane. Accordingly, we investigated the cell wall thickness of AHA-1394 susceptible (SH1000-B; LAC-B) and resistant (SH1000-128l LAC-128) strains using transmission electron microscopy. Here we observed that AHA-1394 resistant strains had significantly thicker cell walls which may prevent AHA-1394 entry and access to the membrane, resulting in resistance. Investigation is currently underway to decipher the molecular mechanisms underpinning the altered cell wall dynamics associated with AHA-1394 resistance.
Alongside drug discovery, optimizing the current panel of antimicrobials and identifying synergistic combinations can aid in treatment of AMR infections and limit the development of resistance. Polyamines can increase the susceptibility of S. aureus to kanamycin (Wang et al., 2016) and some β-lactams (Kwon and Lu, 2007). Molecular dissection revealed that spermine directly interacted with PBP2 reducing transglycosylase activity and rendering it more sensitive to oxacillin (Yao and Lu, 2012). Here we report that AHA-1394 restores sensitivity of MRSA isolates to oxacillin and VISA and VRSA strains to vancomycin. In addition, synergy with daptomycin has been observed in a limited number of genetic backgrounds. The most impressive synergistic activity was observed in combination with oxacillin, whereby oxacillin/AHA-1394 was also active against the AHA-1394 resistant mutant LAC-128. The exact mechanism(s) underlying AHA-1394 synergism with antibiotics is not fully understood and research is ongoing to examine this further. The combinatorial effect of a possible interaction of AHA-1394 with PBP2 resulting in the observed β-lactam synergy, and the association of AHA-1394 resistance with increased oxacillin susceptibility suggests that AHA-1394 partnered with a β-lactam could be a viable treatment option for S. aureus disease.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI; SAMN29765906, SAMN29765907, SAMN29765908, SAMN29765909.
Author contributions
IB and ML: conceptualization, resources, and project administration.. ED, AA, TW, SL, TJW, and ML: methodology, validation, formal analysis, and investigation. ED and ML: writing first draft of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
ML would like to acknowledge funding from the GW4 Generator Award (GW4-GF2-015). We would like to thank the Government of the Kingdom of Saudi Arabia for fully funding this studentship to AA.
Acknowledgments
We would like to thank Matthew Case of the Material and Chemical Characterization Facility (MC2) at the University of Bath and Lorna Hodgson of the Wolfson Bioimaging Facility, University of Bristol for their technical support and assistance in this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.948343/full#supplementary-material
Footnotes
References
Abdel-Hamid, N. I., El-Azab, M. F., and Moustafa, Y. M. (2017). Macrolide antibiotics differentially influence human hepg2 cytotoxicity and modulate intrinsic/extrinsic apoptotic pathways in rat hepatocellular carcinoma model. Naunyn Schmiedeberg’s Arch. Pharmacol. 390, 379–395. doi: 10.1007/s00210-016-1337-0
Altwiley, D., Brignoli, T., Edwards, A., Recker, M., Lee, J. C., and Massey, R. C. (2021). A functional menadione biosynthesis pathway is required for capsule production by staphylococcus aureus. Microbiology 167:11. doi: 10.1099/mic.0.001108
Baba, T., Takeuchi, F., Kuroda, M., Yuzawa, H., Aoki, K. I., Oguchi, A., et al. (2002). Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359, 1819–1827. doi: 10.1016/S0140-6736(02)08713-5
Bayer, A. S., Schneider, T., and Sahl, H. G. (2013). Mechanisms of daptomycin resistance in staphylococcus aureus?: Role of the cell membrane and cell wall. Ann. N.Y. Acad. Sci. 1277, 139–158. doi: 10.1111/j.1749-6632.2012.06819.x
Bergeron, R. J., and Garlich, J. R. (1984). Amines and polyamines from nitriles. Synthesis 1984, 782–784. doi: 10.1055/s-1984-30973
Canty, E., Carnahan, B., Curley, T., Anususinha, E., Hamdy, R. F., and Ericson, J. E. (2021). Reduced vancomycin susceptibility, MRSA and treatment failure in pediatric staphylococcus aureus bloodstream infections. Pediat. Infect. Dis. J. 40, 429–433. doi: 10.1097/INF.0000000000002992
CLSI (2018). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: CLSI standard M07, 11th Edn. Wayne, PA: Clinical and Laboratory Standards Insitute.
Diep, B. A., Gill, S. R., Chang, R. F., Phan, T. H., Chen, J. H., Davidson, M. G., et al. (2006). Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant staphylococcus aureus. Lancet 367, 731–739. doi: 10.1016/S0140-6736(06)68231-7
D’Lima, L., Friedman, L., Wang, L., Xu, P., Anderson, M., and Debabov, D. (2012). No decrease in susceptibility to NVC-422 in multiple-passage studies with methicillin-resistant staphylococcus aureus, s. aureus, Pseudomonas Aeruginosa, and Escherichia Coli. Antim. Agents Chemother. 56, 2753–2755. doi: 10.1128/AAC.05985-11
Ernst, C. M., and Peschel, A. (2011). Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids. Mol. Microbiol. 80, 290–299. doi: 10.1111/j.1365-2958.2011.07576.x
Ernst, C. M., Slavetinsky, C. J., Kuhn, S., Hauser, J. N., Nega, M., Mishra, N. N., et al. (2018). Gain-of-function mutations in the phospholipid flippase MprF confer specific daptomycin resistance. MBio 9:6. doi: 10.1128/mBio.01659-18
Ernst, C. M., Staubitz, P., Mishra, N. N., Yang, S. J., Hornig, G., Kalbacher, H., et al. (2009). The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 5:e1000660. doi: 10.1371/journal.ppat.1000660
Fey, P. D., Endres, J. L., Yajjala, V. K., Widhelm, T. J., Boissy, R. J., Bose, J. L., et al. (2013). A genetic resource for rapid and comprehensive phenotype screening of nonessential staphylococcus aureus genes. MBio 4:12. doi: 10.1128/mBio.00537-12
Geall, A. J., and Blagbrough, I. S. (2000). Homologation of polyamines in the rapid synthesis of lipospermine conjugates and related lipoplexes. Tetrahedron 56, 2449–2460. doi: 10.1016/S0040-4020(99)01082-0
Grigor’eva, A., Bardasheva, A., Tupitsyna, A., Amirkhanov, N., Tikunova, N., Pyshnyi, D., et al. (2020). Changes in the ultrastructure of staphylococcus aureus treated with cationic peptides and chlorhexidine. Microorganisms 8:1991. doi: 10.3390/microorganisms8121991
Hamana, K., and Matsuzaki, S. (1992). Polyamines as a chemotaxonomic marker in bacterial systematics. Crit. Rev. Microbiol. 18, 261–283. doi: 10.3109/10408419209113518
Hawiger, J., Niewiarowski, S., Gurewich, V., and Thomas, D. P. (1970). Measurement of fibrinogen and fibrin degradation products in serum by staphylococcal clumping test. J. Laborat. Clin. Med. 75, 93–108.
Hiramatsu, K., Aritaka, N., Hanaki, H., Kawasaki, S., Hosoda, Y., Hori, S., et al. (1997). Dissemination in japanese hospitals of strains of staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350, 1670–1673. doi: 10.1016/S0140-6736(97)07324-8
Holden, M. T. G., Feil, E. J., Lindsay, J. A., Peacock, S. J., Day, N. P. J., Enright, M. C., et al. (2004). Complete genomes of two clinical staphylococcus aureus strains: Evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U.S.A. 101, 9786–9791. doi: 10.1073/pnas.0402521101
Holden, M. T. G., Lindsay, J. A., Corton, C., Quail, M. A., Cockfield, J. D., Pathak, S., et al. (2010). Genome sequence of a recently emerged, highly transmissible, multi-antibiotic- and antiseptic-resistant variant of methicillin-resistant staphylococcus aureus, sequence type 239 (TW). J. Bacteriol. 192, 888–892. doi: 10.1128/JB.01255-09
Horsburgh, M. J., Aish, J. L. I, White, J., Shaw, L., Lithgow, J. K., and Foster, S. J. (2002). σ B modulates virulence determinant expression and stress resistance: Characterization of a functional RsbU strain derived from staphylococcus aureus 8325-4. J. Bacteriol. 184, 5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002
Jiang, S., Zhuang, H., Zhu, F., Wei, X., Zhang, J., Sun, L., et al. (2022). The role of MprF mutations in seesaw effect of daptomycin-resistant methicillin-resistant staphylococcus aureus isolates. Antim. Agents Chemother. 66:21. doi: 10.1128/AAC.01295-21
Jones, T., Yeaman, M. R., Sakoulas, G., Yang, S. J., Proctor, R. A., Sahl, H. G., et al. (2008). Failures in clinical treatment of staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antim. Agents Chemother. 52, 269–278. doi: 10.1128/AAC.00719-07
Joshi, G. S., Spontak, J. S., Klapper, D. G., and Richardson, A. R. (2011). Arginine catabolic mobile element encoded SpeG abrogates the unique hypersensitivity of staphylococcus aureus to exogenous polyamines. Mol. Microbiol. 82, 9–20. doi: 10.1111/j.1365-2958.2011.07809.x
Karigiannis, G., and Papaioannou, D. (2000). Structure, biological activity and synthesis of polyamine analogues and conjugates. Eur. J. Organic Chem. 2000, 1841–1863. doi: 10.1002/(SICI)1099-0690(200005)2000:10<1841::AID-EJOC1841<3.0.CO;2-9
Klenke, B., and Gilbert, I. H. (2001). Nitrile reduction in the presence of boc-protected amino groups by catalytic hydrogenation over palladium-activated raney-nickel. J. Organic Chem. 66, 2480–2483. doi: 10.1021/jo005637f
Kramer, D. L., Diegelman, P., Jell, J., Vujcic, S., Merali, S., and Porter, C. W. (2008). Polyamine acetylation modulates polyamine metabolic flux, a prelude to broader metabolic consequences. J. Biol. Chem. 283, 4241–4251. doi: 10.1074/jbc.M706806200
Kwon, D. H., and Lu, C. D. (2007). Polyamine effects on antibiotic susceptibility in bacteria. Antim. Agents Chemother. 51, 2070–2077. doi: 10.1128/AAC.01472-06
Lacoma, A., Edwards, A. M., Young, B. C., Domínguez, J., Prat, C., and Laabei, M. (2019). Cigarette smoke exposure redirects staphylococcus aureus to a virulence profile associated with persistent infection. Sci. Rep. 9:10798. doi: 10.1038/s41598-019-47258-6
Lenski, R. E., Rose, M. R., Simpson, S. C., and Tadler, S. C. (1991). Long-term experimental evolution in Escherichia coli. I. adaptation and divergence during 2,000 generations. Am. Nat. 138, 1315–1341. doi: 10.1086/285289
Liu, C., Bayer, A., Cosgrove, S. E., Daum, R. S., Fridkin, S. K., Gorwitz, R. J., et al. (2011). Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin. Infect. Dis. 52, e18–e55. doi: 10.1093/cid/ciq146
Liu, C., Shen, H., Wang, S., Cao, X., Xu, H., Xia, Y., et al. (2018). Spermine increases bactericidal activity of silver-nanoparticles against clinical methicillin-resistant Staphylococcus aureus. Chin. Chem. Lett. 29, 1824–1828. doi: 10.1016/j.cclet.2018.10.025
Mehta, S., Cuirolo, A. X., Plata, K. B., Riosa, S., Silverman, J. A., Rubio, A., et al. (2012). VraSR two-component regulatory system contributes to MprF -mediated decreased susceptibility to daptomycin in in vivo -selected clinical strains of methicillin-resistant Staphylococcus aureus. Antim. Agents Chemother. 56, 92–102. doi: 10.1128/AAC.00432-10
Miller, W. R., Bayer, A. S., and Arias, C. A. (2016). Mechanism of action and resistance to daptomycin in Staphylococcus aureus and enterococci. Cold Spring Harbor Perspect. Med. 6:a026997. doi: 10.1101/cshperspect.a026997
Murray, C., Ikuta, K. S., Sharara, F., Swetschinski, L., Aguilar, G. R., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Ong, Z. Y., Gao, S. J., and Yang, Y. Y. (2013). Short synthetic β -sheet forming peptide Amphiphiles as broad spectrum antimicrobials with antibiofilm and endotoxin neutralizing capabilities. Adv. Funct. Mater. 23, 3682–3692. doi: 10.1002/adfm.201202850
Proctor, R. A., van Langevelde, P., Kristjansson, M., Maslow, J. N., and Arbeit, R. D. (1995). Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 20, 95–102. doi: 10.1093/clinids/20.1.95
Proctor, R. A., von Eiff, C., Kahl, B. C., Becker, K., McNamara, P., Herrmann, M., et al. (2006). Small colony variants: A pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4, 295–305. doi: 10.1038/nrmicro1384
Proctor, R. A., and von Humboldt, A. (1998). Bacterial energetics and Antimicrobial resistance. Drug Resist. Updates 1, 227–235. doi: 10.1016/S1368-7646(98)80003-4
Rajamuthiah, R., Fuchs, B. B., Conery, A. L., Kim, W., Jayamani, E., Kwon, B., et al. (2015). Repurposing salicylanilide anthelmintic drugs to combat drug resistant Staphylococcus aureus. PLoS One 10:e0124595. doi: 10.1371/journal.pone.0124595
Richardson, J. F., and Reith, S. (1993). Characterization of a strain of methicillin-resistant Staphylococcus aureus (EMRSA-15) by conventional and molecular methods. J. Hospit. Infect. 25, 45–52. doi: 10.1016/0195-6701(93)90007-M
Sakamoto, A., Terui, Y., Uemura, T., Igarashi, K., and Kashiwagi, K. (2020). Polyamines regulate gene expression by stimulating translation of histone acetyltransferase MRNAs. J. Biol. Chem. 295, 8736–8745. doi: 10.1074/jbc.RA120.013833
Schuber, F. (1989). Influence of polyamines on membrane functions. Biochem. J. 260, 1–10. doi: 10.1042/bj2600001
Schwarz, S., Silley, P., Simjee, S., Woodford, N., van Duijkeren, E., Johnson, A. P., et al. (2010). Editorial: Assessing the antimicrobial susceptibility of bacteria obtained from animals. J. Antimicrob. Chemother. 65, 601–604. doi: 10.1093/jac/dkq037
Sun, A. W., Bulterys, P. L., Bartberger, M. D., Jorth, P. A., O’Boyle, B. M., Virgil, S. C., et al. (2019). Incorporation of a chiral gem-disubstituted nitrogen heterocycle yields an oxazolidinone antibiotic with reduced mitochondrial toxicity. Bio. Med. Chem. Lett. 29, 2686–2689. doi: 10.1016/j.bmcl.2019.07.024
Tabor, C. W., and Tabor, H. (1984). POLYAMINES. Ann. Rev. Biochem. 53, 749–790. doi: 10.1146/annurev.bi.53.070184.003533
The Ad hoc Interagency Coordination Group (IACG) on Antimicrobial Resistance (2019). No time to wait: Securing the future from drug-resistant infections. WHO. Available online at: https://www.who.int/publications/i/item/no-time-to-wait-securing-the-future-from-drug-resistant-infections
Thurlow, L. R., Joshi, G. S., Clark, J. R., Spontak, J. S., Neely, C. J., Maile, R., et al. (2013). Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13, 100–107. doi: 10.1016/j.chom.2012.11.012
Wang, B., Pachaiyappan, B., Gruber, J. D., Schmidt, M. G., Zhang, Y. M., and Woster, P. M. (2016). Antibacterial diamines targeting bacterial membranes. J. Med. Chem. 59, 3140–3151. doi: 10.1021/acs.jmedchem.5b01912
Yao, X., and Lu, C. D. (2012). A PBP 2 mutant devoid of the transpeptidase domain abolishes spermine–β-lactam synergy in Staphylococcus aureus Mu50. Antim. Agents Chemother. 56, 83–91. doi: 10.1128/AAC.05415-11
Keywords: polyamines, Staphylococcus aureus, antibacterial activity, antibiotic synergy, antimicrobial resistance (AMR)
Citation: Douglas EJA, Alkhzem AH, Wonfor T, Li S, Woodman TJ, Blagbrough IS and Laabei M (2022) Antibacterial activity of novel linear polyamines against Staphylococcus aureus. Front. Microbiol. 13:948343. doi: 10.3389/fmicb.2022.948343
Received: 19 May 2022; Accepted: 04 August 2022;
Published: 22 August 2022.
Edited by:
Kunyan Zhang, University of Calgary, CanadaReviewed by:
Lili Zhang, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaMuriel Masi, Aix-Marseille Université, France
Copyright © 2022 Douglas, Alkhzem, Wonfor, Li, Woodman, Blagbrough and Laabei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maisem Laabei, bWw0MThAYmF0aC5hYy51aw==
 Edward J. A. Douglas
Edward J. A. Douglas Abdulaziz H. Alkhzem
Abdulaziz H. Alkhzem Toska Wonfor
Toska Wonfor Shuxian Li1
Shuxian Li1 Ian S. Blagbrough
Ian S. Blagbrough Maisem Laabei
Maisem Laabei