- 1Key Laboratory of Tropical Biological Resources of Ministry Education, School of Pharmaceutical Sciences, Hainan University, Haikou, China
- 2Department of Biotechnology, College of Science and Humanities, SRM Institute of Science and Technology, Chennai, Tamil Nadu, India
- 3Department of Biochemistry, Cardiac Metabolic Disease Laboratory, School of Biological Sciences, Madurai Kamaraj University, Madurai, India
- 4Biofouling and Biofilm Processes Section, Water and Steam Chemistry Division, Bhabha Atomic Research Centre Facilities, Kalpakkam, Tamil Nadu, India
Cell-to-cell communication is a fundamental process of bacteria to exert communal behaviors. Sputum samples of patients with cystic fibrosis have often been observed with extensive mycobacterial genetic diversity. The emergence of heterogenic mycobacterial populations is observed due to subtle changes in their morphology, gene expression level, and distributive conjugal transfer (DCT). Since each subgroup of mycobacteria has different hetero-resistance, they are refractory against several antibiotics. Such genetically diverse mycobacteria have to communicate with each other to subvert the host immune system. However, it is still a mystery how such heterogeneous strains exhibit synchronous behaviors for the production of quorum sensing (QS) traits, such as biofilms, siderophores, and virulence proteins. Mycobacteria are characterized by division of labor, where distinct sub-clonal populations contribute to the production of QS traits while exchanging complimentary products at the community level. Thus, active mycobacterial cells ensure the persistence of other heterogenic clonal populations through cooperative behaviors. Additionally, mycobacteria are likely to establish communication with neighboring cells in a contact-independent manner through QS signals. Hence, this review is intended to discuss our current knowledge of mycobacterial communication. Understanding mycobacterial communication could provide a promising opportunity to develop drugs to target key pathways of mycobacteria.
Introduction
Bacteria use a variety of mechanisms to communicate with one another. One of the well-known mechanisms of cell-to-cell communication is QS, in which bacteria secrete, recognize, and respond to chemical messengers with respect to their population density (Mukherjee and Bassler, 2019). However, QS machinery in mycobacteria is still poorly understood. Unlike Gram-positive and Gram-negative bacteria, the cell wall of mycobacteria is rich in lipids, leading to the long-held belief that this rigid lipid layer prevents extracellular ligands/signals from reaching receptor proteins, and thus mycobacteria are considered non-communicating bacteria. However, several experimental pieces of evidence suggest that mycobacteria exhibit QS-related behaviors, such as biofilm, membrane vesicle formation, and horizontal gene transfer (Ojha and Hatfull, 2007; Prados-Rosales et al., 2014; Clark et al., 2018). Cell-to-cell interaction is a prerequisite for the production of QS-related traits. Bacterial interaction may have a significant impact on evolutionary dynamics if interacting bacteria are present at high cell density so that they can perceive each other via direct cell-to-cell contact or QS signaling molecules (Nadell et al., 2016). Mice infected with the active Mycobacterium tuberculosis (Mtb) strain had about 10,000 bacilli per lung after 10 days, whereas 10 million bacilli per lung were observed after 20 days (Valway et al., 1998), suggesting that mycobacteria exist at high density during active tuberculosis. Such high-density mycobacterial cells exist as filamentous or aggregated forms (Chauhan et al., 2006; Julián et al., 2010) and establish cell-to-cell communication through contact-dependent or -independent mechanism to overcome host-mediated stress conditions through the exchange of molecules. The respiratory tract of patients with cystic fibrosis is infected with multiple pathogens, including different genotypes of Mycobacterium abscessus, Pseudomonas aeruginosa, and Staphylococcus aureus (Olivier et al., 2003; Levy et al., 2008; Shaw et al., 2019), implying that mycobacteria might interact with different species of bacteria at the niche. Though mycobacteria majorly infect the lungs, multiple genotypes are disseminated to extrapulmonary organs, such as the liver and the spleen (Lieberman et al., 2016). While infecting other parts of the body, such as skin and soft tissue, mycobacteria might interact with native microbiota to compete for spatial stratification and nutrients. However, there are no supporting studies until now to link mycobacterial interaction with other species of bacteria. The coexistence of different species of mycobacteria along with multiple pathogens in the lungs of the patients arguably functions as a polymicrobial pathogenic community, which could be the reason for recurrent treatment failures, as each species and genotypes will have a different range of antibiotic susceptibility. Hence, this review is intended to discuss the recent evidence on mycobacterial communication and how they use social behaviors to promote their persistence in the host.
Mycobacterial persistence increases with heterogeneity
Owing to the paucity of advancements in molecular techniques in the last few decades, mycobacteria-mediated lung infection was thought to be caused by a single type of genetically conserved strain. However, using recent advancements in molecular techniques, it was evidently proved that multiple strains of mycobacteria enter the lungs through multiple independent transmission, spatially distributed across different sites of the lungs (Cohen et al., 2012; Lieberman et al., 2016). Interestingly, few reports suggest that Mtb-infected patients are also unusually coinfected with non-tuberculosis mycobacteria (NTM) (Huang et al., 2007; Aliyu et al., 2013; Paleiron et al., 2019). NTM causes a broad range of diseases in different parts of the body of immunocompromised patients (Piersimoni and Scarparo, 2009; Misch et al., 2018). The emergence of heterogenic clonal populations among mycobacteria is an inevitable survival strategy that provides fitness to mycobacteria to withstand the hostile host environment. Since factors, such as cell elongation, DNA replication, chromosome segregation, and cell division, are unsynchronized events in mycobacteria, variation in their growth is often observed (Dhar et al., 2016) (Figure 1A). As a result of asymmetrical cell division, mycobacterial cell wall components are unevenly distributed in the dividing daughter cells (Kieser and Rubin, 2014; Hannebelle et al., 2020), resulting in variable responses to antibiotics and host immunity (Dulberger et al., 2020). Empirical evidence revealed that mycobacteria defective to produce LamA have decreased asymmetrical growth and failed to create heterogenic clonal populations (Rego et al., 2017). It was also observed that due to the lack of heterogeneity, cell wall-targeting antibiotics efficiently kill LamA defective mycobacterial cells, implying that cell wall heterogeneity plays an imperative role in fitness. Not surprisingly, the mistranslation of protein also contributes to the emergence of rifampicin resistance in mycobacterial phenotypes under in vitro conditions (Javid et al., 2014).
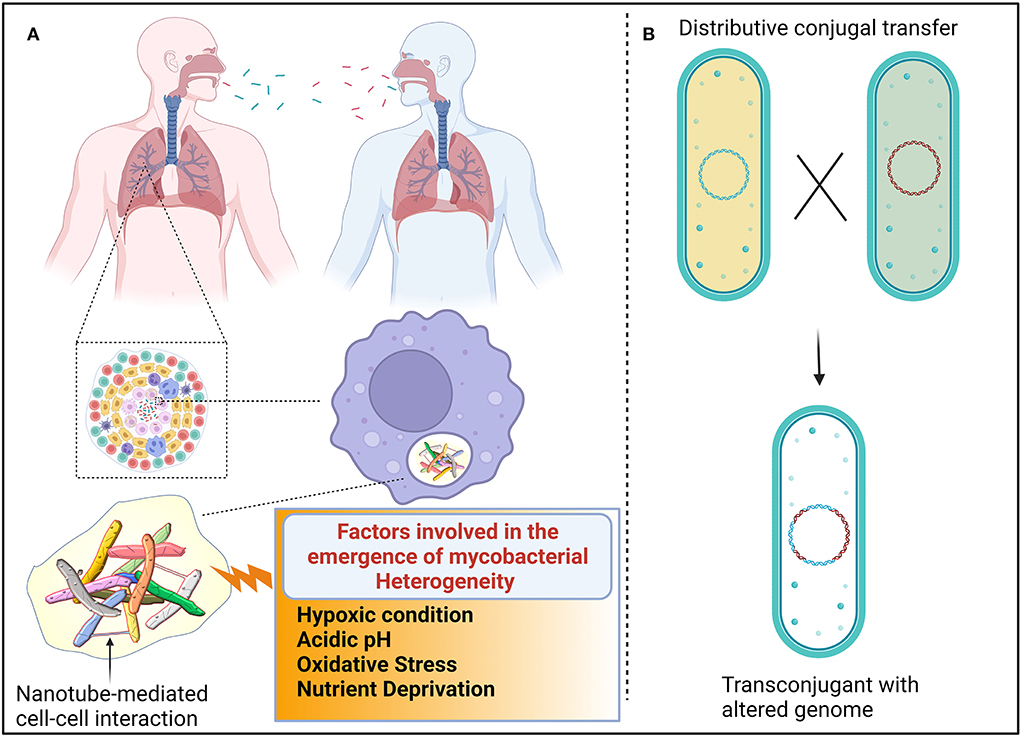
Figure 1. Proposed mechanism for the emergence of heterogenic mycobacteria. (A) Different genotypic mycobacteria emerged due to unsynchronized events, such as cell elongation, DNA replication, chromosome segregation, and cell division. Additionally, host immune cells also induce mycobacterial heterogeneity. The composition of granulomatous tissue can vary greatly between individuals, within an individual, and within a single granuloma. The composition and quantity of nutrients and stress-inducing factors are dynamic because each compartment in granuloma shift over time. Such heterogenic mycobacteria are transmitted from one individual to another. Mycobacteria interact with other cells, possibly via nanotubes, to exchange quorum sensing traits to establish successful colonization. (B) Distributive conjugal transfer (DCT) occurs in mycobacteria wherein multiple segments of DNA from the donor are transferred to the recipient and generate unique genotypes that exert different phenotypic profiles.
It is adequately represented that the heterogeneity of mycobacteria is also governed by the immune status of the host. Upon sensing the bacterial components, immune cells are assembled at the site of infection, resulting in the formation of lesions. Autopsies of lesions from tuberculosis patients revealed that lesions are heterogeneous in nature in terms of cellular composition and structural organization (Irwin et al., 2015; Carow et al., 2019). As a result, antibiotics penetrate easily in certain lesions, while some lesions receive the least concentration of antibiotics, resulting in the emergence of antibiotic-resistant subsets of bacteria (Prideaux et al., 2015). Mice deficient in the production of interferon γ have less heterogeneous mycobacterial populations than wild-type mice, corroborating that host immune cells also influence mycobacterial physiology (Manina et al., 2015). It could be perceived that heterogeneity among the clonal populations enables them to exhibit multiple personalities to tolerate a variety of stress factors which could result in the persistence of substantial cells even in the presence of multiple attacks (Dulberger et al., 2020). However, cooperation among the heterogenic mycobacterial population is essentially required for successful colonization and survival.
Cooperation among heterogenic mycobacteria
In mycobacteria, the exchange of genetic material occurs between donor and recipient cells in a systemic manner in which the type VII secretory system plays a vital role throughout the process. The transfer of chromosomal DNA between mycobacteria generates transconjugants with mosaic genomes of the parental strains. During DCT, novel genes or operons from donor bacteria may be inserted into the genomes of transconjugants, or their essential genes may be replaced with the segment of donor bacteria, thus resulting in the change of biochemical pathways in transconjugants, which could influence the fitness of the transconjugants (Gray and Derbyshire, 2018) (Figure 1B). Due to promiscuous DNA replication, daughter cells might lose essential metabolite/protein-encoding genes, which causes them to depend on neighboring cells for nutrients (Boopathi et al., 2021). Though the reciprocal exchange of essential nutrients has been reported in different bacterial pathogens, such molecular trading between mycobacteria has yet not been defined clearly. Metabolic diversification among the clonal population is one of the key strategies for generating heterogeneous subpopulations to survive even in fluctuating nutrient conditions (Simsek and Kim, 2018). Each heterogeneous subpopulation might be specialized in producing certain essential molecules that are required for survival. Therefore, each subpopulation of mycobacteria must depend on each other for their survival. The sub-population of bacteria becomes metabolically inactive, which enables increased fitness in them. Such inactive bacterial cells get benefit from nearby virulence proteins producing active cells without investing in the production of such virulence proteins (Davis and Isberg, 2019). Division of labor may be the common feature of the mycobacterial community, where one subset of cells may produce virulent effectors to subvert host immune systems, while another subset of the population may produce molecules to acquire essential nutrients to benefit the entire population (Davis, 2020). In response to the microenvironments of heterogeneous granuloma, a subset of residing mycobacteria differentially express granuloma-activated genes (Chan et al., 2002), supporting the above-mentioned hypothesis.
Division of labor
Mycobacteria produce three different siderophores, including mycobactin and the soluble carboxymycobactin and exochelin, to efficiently scavenge iron molecules (Dragset et al., 2019). To reduce the metabolic burden, mycobacteria recycle siderophores rather than synthesizing new siderophore molecules (Jones et al., 2014). They also found that the presence of abundant siderophores in both intracellular and extracellular environments is toxic to mycobacteria defective in siderophore recycling. It could be conceived that siderophore recycling in mycobacteria might facilitate them to share siderophores with their local community (Figure 2).
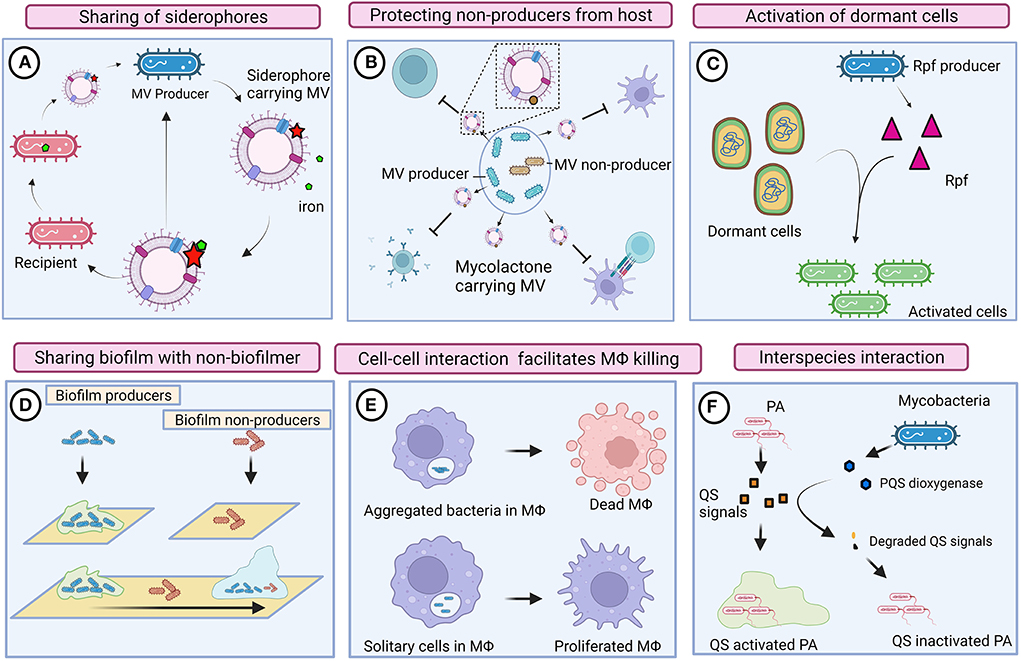
Figure 2. Cooperative and competitive behaviors of mycobacteria. (A) Mycobacteria produces mycobactin-carrying membrane vesicles (MVs) to scavenge iron molecules. Siderophore-bound iron enables the growth of mycobacteria that is defective in mycobactin production (ΔmbtB). (B) Mycobacteria produce mycolactone harboring MV. Mycolactone could protect inactive mycobacterial cells by suppressing the host immune system. (C) Resuscitation-promoting factors (Rpf) could reactivate dormant mycobacteria regardless of producers. (D) Biofilm-producing mycobacteria could also provide shelter to heterogenic populations (defective to produce biofilm). (E) Intrinsic interaction between mycobacteria possibly enables communication and activates macrophage (MΦ) killing pathways. Solitary mycobacterial cells were unable to kill macrophages. (F) Mycobacteria compete with Pseudomonas aeruginosa (PA) by degrading its quorum sensing (QS) signals. Images are drawn using the Biorender tool (https://Biorender.com/).
Essentially, membrane vesicles (MVs) also play an imperative role in inter-bacterial communication. Iron-deprived in vitro condition induces the production of mycobactin carrying MVs in Mtb, and the mycobactin associated with these MVs serves as an iron chelator and supports the growth of bacteria (Prados-Rosales et al., 2014). MVs from Mtb grown in an iron-deficient medium enable the growth of Mtb defective in mycobactin production (ΔmbtB), suggesting that MVs are shared among mycobacteria in the local community (Prados-Rosales et al., 2014). Mycolactone is a toxin required for the development of necrotic cutaneous lesions. Mycolactone harboring MVs have abundantly been found in mycobacterial biofilms (Marsollier et al., 2007), indicating that mycolactone serves as a product of cooperative behavior that also protects other subgroups of bacteria from the host immune system. Another plausible evidence for the cooperative behavior of mycobacteria is resuscitation from a dormant state. Resuscitation-promoting factors (Rpfs) are known to show cell wall hydrolyzing activity and are thus involved in the resuscitation of dormant mycobacterial cells (Nikitushkin et al., 2011). Such cell wall remodeling enzymes are likely to function similar to inter-bacterial signaling enzymes, where Rpfs generate peptidoglycan fragments that act as signaling molecules to activate the resuscitation pathway in dormant mycobacteria (Maitra et al., 2019). Perhaps, resuscitation is not strictly associated with Rpfs producing cells; however, non-producers in the local community also might get benefits by receiving such signals for reactivation.
In response to the relative availability of carbon and nitrogen, mycobacteria either grow as large aggregates or as planktonic cells (DePas et al., 2019). Mycobacteria aggregate in a specific pattern that mimics cords; this phenomenon is known as cord formation. Aggregation, rough morphology, and prolonged persistence of mycobacteria within macrophages have all been found to be strongly correlated (Julián et al., 2010). The rough morphotype of M. abscessus forms clumps through tight cell-to-cell interaction that enables them to kill macrophages efficiently, whereas the smooth morphotype remains as isolated bacilli and fails to kill macrophages (Brambilla et al., 2016), suggesting that mycobacterial cells as a community could efficiently subvert the host immune system through intrinsic interaction. Rough (Rg) Mycobacterium avium infection caused severe pneumonia and enhanced cytotoxicity in mice and macrophages, respectively (Nishimura et al., 2020). When macrophages were infected with Rg, bacilli were aggregated, and the expression of proinflammatory cytokines, such as TNFα and IL-6, increased. However, Rg could not induce the secretion of proinflammatory cytokines or cause cytotoxicity when they were disaggregated (Nishimura et al., 2020), indicating that intrinsic cell–cell interaction is required for exerting high virulence. Surface-exposed glycopeptidolipids (GPLs) are the outermost components of mycobacteria that mask cell wall-associated lipids. Mycobacteria exhibit rough morphology when they lose several surface-exposed GPLs due to the downregulation or mutation of GPL biosynthetic pathways (Torrelles et al., 2002; Gutiérrez et al., 2018). The absence or reduced expression of GPLs exposes cell wall lipids of mycobacteria and thus induces an immune response. The smooth variant of Mycobacterium abscessus is often observed within the phagosome as single bacteria, whereas the Rg variant was found as a group within the phagosome that contains a loose membrane (Roux et al., 2016). Despite the fact that host-induced stress is predominantly prevalent in phagolysosomes, the Rg variant rapidly multiplies and weakens the defense mechanism of macrophages, which results in autophagy and apoptosis (Viljoen et al., 2017), indicating that complex communication between mycobacteria promotes hypervirulence activity. It was also reported that Mtb aggregates might spread via bioaerosols (Dinkele et al., 2021). The presence of Mtb aggregates around the lung cavitary lesions of TB patients denotes that hypervirulent Mtb aggregates could transmit from individual to individual and cause increased inflammation and cell death, thus leading to active disease (Rodel et al., 2021). Infection of cells with heat-killed Mtb aggregates did not show significant cell death, suggesting that live mycobacteria intensively communicate with one another and exchange molecular information to elicit virulence within the host.
Mycobacterial biofilms function as a common shelter for cooperative phenotypes in which multiple genotypes, including mycobacteria defective in biofilm formation, are also assimilated, thereby intercellular communication is established between bacterial cells through cell–cell interaction for the exchange of biomolecules (Nguyen et al., 2010). Thus, biofilms also serve as a common shelter for the local community.
Molecular trading between mycobacteria
Mycobacteria establish communication with neighboring bacteria to exchange multiple fragments of DNA through a unique process of horizontal gene transfer (HGT) that differs from classical oriT-mediated conjugation, which provides evolutionary fitness to the bacteria to adapt to a hostile environment (Gray et al., 2013). Prior to the interaction, bacteria must detect suitable partners. Gene knockout studies revealed that type VII secretion systems (ESX systems) are essentially required for inter-bacterial communication (Gray and Derbyshire, 2018). Based on phylogenetic and comparative genomics analysis, it was found that mycobacteria are evolved and harbor different ESX systems in the order of ESX−4, ESX-1, ESX-3, ESX-2, and ESX-5 (Abdallah et al., 2007). Though mycobacteria harbor five T7SS, each ESX system is deserved for unique functions, which implies that one ESX system will not complement other ESX systems due to their distinct secretion signal and differential regulation pathways (Abdallah et al., 2007). Supportively, HGT between mycobacteria happens through the interaction between ESX-1 and ESX-4 systems (Clark et al., 2018), suggesting that interactions between ESX systems are likely to occur to maintain bacterial physiology. It is vital that mycobacterial interaction does not occur between random cells, whereas mycobacteria which possess a mid locus, encoded by mid gene, confers mating identity. Transconjugants become donors if the donor-conferring mid-locus also transfers to the recipients (Gray et al., 2013).
Once recipient cells perceive the signal from donor cells, a series of molecular events happen in recipients for the acquisition of molecules. For instance, through unknown signals, once recipient cells recognize donor cells, transmembrane anti-SigM is inactivated, which in turn releases extracytoplasmic transcription factor SigM that activates the ESX-4 system, resulting in the modulation of gene expression that facilitates HGT (Clark et al., 2018). ESX-1 also plays an imperative role in inter-bacterial communication. For instance, donor cells defective in ESX-1 become hyper conjugative (Flint et al., 2004), whereas ESX-1 defective recipient cells do not successfully receive DNA from donors (Coros et al., 2008). Upon interaction with neighboring donor cells, ESX-4-mediated esxUT transcripts are upregulated in the recipient cells, whereas such induction of esx4 transcripts is not observed in the monoculture of either donor or recipient, which gives the notion that ESX-4 is also critically involved in inter-bacterial interaction (Clark et al., 2018). RNA sequencing analysis of coculture revealed that the expression of many other genes was also modulated apart from the genes required for HGT, suggesting that many molecular events occur between donor and recipient cells upon interaction (Gray et al., 2016). Since SigM and ESX-4-mediated signaling system is conserved throughout mycobacteria, such an interaction network might be an active process in different species of mycobacteria. It could be concluded that esxUT of recipient cells could be used as a biomarker for intercellular communication (Clark et al., 2018). Molecular trading between bacteria could dramatically alter the mycobacterial molecular pool by directly acquiring foreign molecules, which in turn can influence the colonized niche and infection outcome in the host. Though structural components of ESX-1 similarly exist across different mycobacterial species, their secretory repertoire may be varied between species, even that could be species-specific, which gives the notion that ESX-1 defines the cell surface morphology of mycobacterial cells for cell–cell recognition (Gray and Derbyshire, 2018). Thus, molecular trading in terms of HGT has been well-established in mycobacteria. However, identification of other exchanging molecules could provide deeper insights into mycobacterial physiology.
Biofilm formation
Mycobacteria can survive in the host for a longer period of time, possibly within the biofilm. Biofilm is a typical organized structure, which harbors a heterogeneous bacterial population that interacts cooperatively to acquire and share resources (Chakraborty and Kumar, 2019). Mycobacteria produce such biofilm in response to environmental cues, such as osmolarity, nutrient availability, pH, oxygen, and CO2 concentration (Xiang et al., 2014). Biofilm formation can be viewed as a representation of coordinated behavior, which occurs in both tuberculous and non-tuberculous mycobacteria, including Mycobacterium tuberculosis, Mycobacterium smegmatis, Mycobacterium marinum, Mycobacterium fortuitum, Mycobacterium chelonae, Mycobacterium ulcerans, M. abscessus, M. avium, and Mycobacterium bovis, affirming that cell–cell communication occurs in mycobacteria (Sharma et al., 2014). Biofilms of non-tuberculous mycobacteria have been reported in different environmental surfaces, including medical devices, dental cavities, membrane filters, and drinking water systems (Schulze-Röbbecke et al., 1995; Falkinham et al., 2015).
Cell–cell interaction is a prerequisite for biofilm formation, where multiple factors critically govern such an interaction. Bacteria effectively communicate with each other if they are present in an aggregated form in the niche. Supportively, rough morphotypic M. abscessus makes biofilms that confer resistance against antibiotics (Clary et al., 2018). Lsr2 is a non-specific DNA binding protein, essentially required in donor cells but not in recipient cells for a successful DNA transfer process (Nguyen et al., 2010). Mycobacteria defective in Lsr2 production exhibit altered colony morphology and mutant in biofilm formation (Chen et al., 2006; Arora et al., 2008). Additionally, Mtb Lsr2 regulates the genes involved in adaptation during hypoxia conditions (Bartek et al., 2014), implying that the Lsr2 is a key regulator essentially required for exhibiting multicellular behaviors.
Electron microscopic studies of the lung cavity from the patients infected with M. abscessus revealed that mycobacteria exist within the matrix embedded biofilm, suggesting that mycobacteria can efficiently form biofilms under in vivo conditions (Qvist et al., 2015; Fennelly et al., 2016). Acid-fast staining of NTM patients' sputum revealed the presence of biofilm-like aggregates (Qvist et al., 2015). It is possible that unsterilized lesions promote the formation of in vivo biofilms, contributing to the progression of chronic infection. The lung cavity of the patient harbors a high proportion of mycobacteria along with different species of bacteria (Fennelly et al., 2016), indicating that different strains of Mtb might exist as a community in mixed biofilms in vivo. Aggregated mycobacterial cells at the bottom of the liquid media are not able to exchange DNA from donor to recipient, suggesting that biofilm not only serves as a platform to facilitate DNA transfer by enabling cell–cell physical contact but also provides other essential conditions for DNA transfer (Nguyen et al., 2010). Furthermore, molecular evidence is required to elaborate on the mechanism. The emergence of drug-tolerant mycobacteria in vivo is possibly due to biofilm formation (Chakraborty and Kumar, 2019). Thus, drug tolerance is a hallmark of the biofilm-resident mycobacterial population. The existence of mycobacteria as a community within the biofilm could be the fact for increased drug tolerance.
Mycobacteria critically govern the mycolic acid production through FAS I and FAS II complexes, where FAS II is involved in the elongation of the FAS I product. FAS II complex consists of six enzymes, including MabA, InhA, KasA, KasB, HadAB, and HadBC, for completing the elongation cycle of fatty acids (Stokas et al., 2020). In addition to FAS I and II, GroEL1 chaperonin plays a significant role in the mycolic acid synthesis and biofilm formation. M. smegmatis defective in the groEL1 gene does not affect the planktonic growth but restricts the formation of mature biofilm and physically interacts with KasA (Ojha et al., 2005). Since the overexpression of GroEL1 protein is positively associated with mature biofilm formation, this protein could be viewed as the hallmark of biofilm formation. Mtb genome consists of 11 serine/threonine kinases (STPKs) that are involved in regulating drug resistance, metabolism, and pathogenicity (Baer et al., 2014). Since STPK phosphorylation regulates the condensation activity of KasA and KasB, changes in the expression of mycobacterial kinase in response to stress conditions may have an impact on the production of mycolic acid (Molle et al., 2006). Furthermore, the role of STPKs in QS traits (e.g., biofilm formation) was demonstrated, stating that overexpression of PknF in M. smegmatis resulted in irregular cell structure, as well as defects in sliding motility and biofilm formation (Gopalaswamy et al., 2008). They also discovered that overexpression of PknF alters the composition of glycopeptidolipids, resulting in aggregation. Interestingly, PknF of M. smegmatis (PknFMsm) does not phosphorylate GroEL1 of M. smegmatis (GroEL1Msm), but PknFMsm phosphorylates GroEL1 of Mtb (GroEL1Mtb) in vitro, suggesting that STPKs might involve in the crosstalk between different mycobacterial species (Canova et al., 2009). Mycobacteria lacking functional HadC are less aggregated, exhibit a distinctive mycolic acid profile, have a unique morphology, are deficient in sliding motility and biofilm formation, and are also detergent-resistant (Jamet et al., 2015). HadC mutants are defective in cording formation and exhibit less virulence in the tissue of Mtb-infected mice (Slama et al., 2016). Furthermore, they found that HadC mutants are rifampicin sensitive, implying that mutation or deletion of hadC results in virulence loss. Mycobacteria transform from a replication state to a non-replicating persistence state by altering their metabolism and behavior in response to host-induced stress. HadAB and HadBC are downregulated during the stationary phase as a result of STPK-mediated phosphorylation, which suggests that mycobacteria stop producing mycolic acid in a non-replicating condition (Slama et al., 2011). Since cording and biofilm formation are strongly associated with cell-to-cell communication/interaction, HadBC could be a potential target for drug development.
Within the granulomatous lesions, mycobacteria encounter several environmental stresses, including hypoxia, altered pH, nutrient starvation, reactive oxygen and nitrogen species, cell membrane disturbing factors, toxic lipid components, and other cues. To adapt to such a harsh environment, mycobacteria utilize 11 two-component regulatory systems (TCSs) (Bretl et al., 2011). Mycobacteria utilize such TCSs for regulating physiological and metabolic processes, including cell division, stress response, and virulence. The role of TCSs in mycobacterial physiology is extensively reviewed elsewhere (Bretl et al., 2011). This review focuses specifically on the role of TCSs in QS mechanisms and cell-to-cell interaction.
In response to the hypoxic condition in the anaerobic non-replicating persistence (NRP) stage 2 model, it was found that Mtb increases the expression of fabG, kasB, and fbpA (Gopinath et al., 2015). FabG and Kas are involved in the biofilm formation of mycobacteria (Ojha et al., 2005; Hegde, 2020). Proteins like SigK and DevR are found to be unregulated in both microaerophilic and hypoxic conditions (NRP stage 1 and 2). DevR is one of the components of DosS/DosR that regulate approximately 50 genes during dormancy (Peddireddy et al., 2017). The deletion of the dosS/dosR regulon from Mtb resulted in less biofilm formation than the wild type (Flores-Valdez et al., 2020), suggesting that DoS/DosR components are crucially involved in the biofilm formation.
Interspecies interaction
Mycobacteria were thought to be present within granuloma, but they have also been found as extracellular microcolonies outside of granulomatous lesions (Lenaerts et al., 2007). Such extracellular bacteria are recognized as persisters that function as a source for reactivation. Patients with cystic fibrosis are infected with multiple pathogens, where interaction is likely to occur between extracellular bacteria for space and nutrients. Pseudomonas aeruginosa and Aspergillus spp. were found in great abundance in the sputum samples of CF patients who had been reported to have NTM infections (Levy et al., 2008). Another study found a positive correlation between NTM infection and Staphylococcus aureus, but an inverted correlation between NTM and P. aeruginosa abundance (Olivier et al., 2003), suggesting the possibility of interspecies communication. During the active phase of tuberculosis, millions of tubercle bacilli are released, which might interact with other bacterial species to subvert the host immune system. Accordingly, they also found that mycobacteria can modulate the pathogenesis of coexisting P. aeruginosa and S. aureus. For instance, Mycobacteria could influence the QS mechanisms of Pseudomonas sp. by degrading their QS signals through PQS dioxygenase and phosphotriesterase-like lactonases (Afriat et al., 2006; Chow et al., 2009; Birmes et al., 2019; Wullich et al., 2019). Likewise, Mycobacterium fortuitum modulates the biofilm formation of P. aeruginosa by producing pyocyanin demethylase (PodA) that degrades phenazines, thereby inhibiting the pyocyanin-dependent release of extracellular DNA (Costa et al., 2015, 2017). Thus, mycobacteria execute multiple pathways to influence the QS mechanisms of P. aeruginosa. Though interspecies interactions are well-defined in other bacterial species (Boopathi et al., 2017a), such interactions are not well-studied in mycobacteria. Moreover, the interaction between slow-growing mycobacteria and fast-growing P. aeruginosa is still highly debatable. Further in vivo studies are required to support these findings.
Cell–cell communication
Since HGT occurs in a contact-dependent manner in both fast and slow-growing mycobacteria, it could be conceived that such interaction might prevail across the genus (Gray and Derbyshire, 2018). The constant interaction between mycobacterial cells either on a solid medium or within the biofilm facilitates HGT, whereas HGT does not occur between mobilizing cells (Nguyen et al., 2010). Supportively, HGT is unsuccessful if donor and recipient cells are physically separated using a transwell plate (Gray et al., 2016), denoting that cell–cell contact is essentially required for the exchange of genetic material and for mycobacterial evolution. A previous study has shown that the exchange of genetic material between mycobacteria mechanistically does not happen due to transduction or cell fusion or transformation, suggesting that some specialized mechanisms exist for trading biomolecules between mycobacterial cells (Parsons et al., 1998). However, intercellular communication between mycobacterial species through specialized physical structures is an unexplored phenomenon to date. Since hydrophobic lipid-rich cell wall hinders access to DNA for transformation, it is postulated that HGT occurs in mycobacteria through the specialized structure (Gray and Derbyshire, 2018).
Interestingly, electron microscopy studies suggest that different species of mycobacteria, including Mycobacterium paratuberculosis, M. avium, Mycobacterium intracellulare, M. bovis, M. tuberculosis, M. marinum, Mycobacterium phlei, and Mycobacterium xenopi, produce hollow extracellular filaments (Draper and Rees, 1970; Merkal et al., 1973; Draper, 1974; Dahl, 2005; Alteri et al., 2007). Furthermore, it was observed that aged mycobacterial cultures produce more extracellular filaments (Dahl, 2005). These structures were referred to by varied nomenclatures as pili, fibrils, or simply extracellular filaments. Despite in-depth insights into the mycobacterial genome and available biochemical and structural data, the functional relevance of these mycobacterial filaments remains elusive. Hence, an in-depth understanding of mycobacterial filaments could provide insights into dimensions, chemical composition, and functionality. Recent molecular evidence reaches certain milestones in deciphering such filaments. For instance, Mtb produces EspC that multimerizes and self-assembles into an extended filament-like structure that is required for the export of EsxA (Ates and Brosch, 2017; Lou et al., 2017). Since ESX-4 tightly regulates cell–cell communication, it could be hypothesized that Esx-4 secreted molecules may function as building blocks to form the tubular structure for the trading of molecules between mycobacteria (Gray and Derbyshire, 2018). Nevertheless, to date, there are no reports on specialized physical structures for establishing communication between mycobacterial strains. Owing to the thick hydrophobic mycomembrane, the mycobacterial cell wall was thought to be highly impermeable, giving the notion that specialized secreting machinery might exist for the transport of molecules (Abdallah et al., 2007).
Nanotube-mediated communication
Several studies reported that different types of bacteria, including Bacillus subtilis, Bacillus megaterium, S. aureus, and E. coli, produce nanotubes that form a network between cells for exchanging cytoplasmic molecules, such as amino acids, toxins, proteins, and non-conjugative plasmids, in an intercellular manner (Dubey and Ben-Yehuda, 2011; Pande et al., 2015; Stempler et al., 2017), giving the notion that the above discussed extracellular filaments of mycobacteria are likely to be conduits for transporting cytoplasmic molecules between cells. When neighboring cells are abundantly present in the niche, bacteria establish communication through a network of nanotubes; however, bacteria produce elongated nanotubes in the absence of neighbors (Dubey et al., 2016). In nutrient-deprived conditions, nanotube formation is induced among bacteria to facilitate the movement of nutrients to the intended bacteria (Pande et al., 2015). Supportively, the interaction between auxotroph and donor cells (amino acid producer) via nanotubes results in the overproduction of amino acids in donor cells, which is required for auxotroph, thereby deferring feedback inhibition in donor cells (Shitut et al., 2019). Therefore, nanotubes offer selective fitness advantages to bacteria to adapt to the changing environment (D'Souza and Kost, 2016). According to these findings, nanotube formation in bacteria is most likely essential for overcoming nutrient deficiency. Recently, channel-forming membrane integrated proteins (FliO, FlioP, FliQ, FliR, FlhB, and Flh), called CORE, have been found to be essential for nanotube synthesis (Bhattacharya et al., 2019). In silico analysis revealed that such CORE complex is widely distributed in different bacterial phyla, including actinobacteria. Since mycobacteria come under the phylum actinobacteria, it could be an indication that mycobacteria are likely to possess such machinery for the synthesis of nanotubes.
Bacteria utilize the cell wall remodeling enzyme LytC and its activator LytB for the extrusion of nanotubes from donor cells and penetration into the recipient cells (Baidya et al., 2020). Mycobacteria are known to harbor several cell wall remodeling enzymes that might be involved in establishing the mycobacterial nanotube network. For instance, M. smegmatis has a membrane fission protein called iniA that participates in membrane remodeling activities (Wang et al., 2019). Upon physical interaction between mycobacteria, SigM directly regulates the expression of several genes that include ESX-4 apparatus, cell wall hydrolase, and nuclease (Clark et al., 2018), supporting that SigM and ESX-4 pathway might be involved in mycobacterial nanotube extrusion and penetration through the activation of cell wall hydrolase. Upregulation of peptidoglycan amidase (Rv0024) in M. smegmatis induces cell–cell interaction that leads to increased biofilm formation, which in turn increases the antibiotic resistance pattern (Padhi et al., 2016), suggesting that cell–cell interaction exists within the mycobacterial biofilm, probably through a nanotube network.
Contact-dependent interaction not only occurs between mycobacterial cells but also between host cells. For instance, Mtb utilizes the ESX-1 system to tear the host cell membrane in a contact-dependent manner (Conrad et al., 2017), which facilitates successful colonization in the host. However, they failed to show tube-like structures that connect bacteria and host cells. However, in a recent study, the mechanism by which host-invaded E.coli acquire nutrients from the host cells through nanotube machinery has been demonstrated (Pal et al., 2019). Perceiving messages via MVs could be one method of mycobacterial communication. Microscopic analysis revealed that B. subtilis is likely to produce MVs from nanotubes through a pinching-off mechanism (Dubey et al., 2016).
Contact-independent interaction
Although there is sufficient indirect evidence indicating the existence of a possible QS mechanism, no direct evidence has been reported to date in mycobacteria. For instance, a planktonic culture of M. avium produces a higher biofilm after the addition of supernatant from biofilm culture, which implies that some QS signaling molecules are present in the supernatant (Carter et al., 2003). C-di-GMP is a secondary messenger that plays an important role in mycobacteria for their survival and adaptation. The C-di-GMP level is determined by diguanylate cyclase (DGC) and phosphodiesterase (PDE-A) (Gupta et al., 2010). As a master regulator, C-di-GMP inter-connected with many regulatory pathways, including QS (Srivastava and Waters, 2012), phosphorylation networks (Lori et al., 2015), and other signal-mediated pathways (An et al., 2013; Gupta et al., 2015). With reference to the protein domain structure, Rv1354c of Mtb is likely to be the crucial component in the C-di-GMP signaling pathway, which may be involved in sensing external signals (Cui et al., 2009). Deletion of C-di-GMP genomic copy in M. smegmatis affects its prolonged survival under nutrient-starved conditions, whereas the overexpression of MSDGC-1 (an ortholog of Rv1354c) modulates the morphotypes and growth under in vitro conditions (Bharati et al., 2012). When mice are immunized with a BCG strain (ΔBCG1416c) deficient in Delta C-di-GMP diguanylate cyclase, specific CD4+ T and CD8+ T cells are increased, and host protection is conferred by reducing the burden of Mtb. This finding suggests that C-di-GMP is necessary for mycobacterial survival within the host (Segura-Cerda et al., 2018). Recently, crosstalk between C-di-GMP and two-component regulatory systems is elegantly demonstrated (Hu et al., 2019). They found that when C-di-GMP interacts with DevR, the expression of the devR operon increases, allowing mycobacteria to survive under oxidative stress. Thus, the QS mechanism is associated with the DosS/DosR system.
In response to stress or starvation conditions, bacteria produce alarmone (p)ppGpp that regulates several processes, including biofilm, virulence, survival, and persistence (Potrykus and Cashel, 2008). Rel is an alarmone synthetase/hydrolase enzyme that produces (p)ppGpp by hydrolyzing C-di-GMP or transferring pyrophosphate from ATP to GDP (Prusa et al., 2018). When mice are infected with H37RvΔrelMtb, sustained chronic infection is impaired, suggesting that RelMtb strongly modulates the expression of components involved in the latent infection (Dahl et al., 2003). Deletion of Rel in M. smegmatis resulted in altered cell wall components, decreased long-term survival, and increased vulnerability to hypoxia and nutrient starvation (Dahl et al., 2005). In order to succeed in host-generated stresses, mycobacteria exert several stringent responses that are associated with virulence gene expression, drug tolerance, persistence, and latency. Mtb deficient in RelMtb replicates uncontrollably during nutrient deprivation, just like wild-type bacteria, and is unable to enter a dormant state. Mtb lacking RelMtb is more sensitive to isoniazid treatment during nutrient starvation and in the lungs of infected mice (Dutta et al., 2019). Therefore, (p)ppGpp homeostasis is required during chronic infection in animals to regulate metabolism and virulence in the face of host-mediated stress.
Another indirect evidence also has speculated the presence of the QS system in mycobacteria. It showed that a putative transcriptional regulator's (whiB3) expression relied on bacterial density, reflecting the possible existence of the QS mechanism (Banaiee et al., 2006). Mycobacteria overcome host-mediated stress through the use of Whib3, which reprogrammes metabolic activity in response to oxidative, reductive, and nitrosative stresses. Through comparative genomics study, numerous proteins in Mtb are found to be associated with QS mechanisms (Hegde, 2020). The LuxR transcriptional regulator is a component of the QS circuit, which controls the expression of several genes, including those encoding bioluminescence, virulence factors, and biofilm formation. Sequencing analysis revealed that seven LuxR family proteins, such as Rv0386, Rv0195, Rv0491, Rv0890c, Rv0894, Rv2488c, and Rv3133c, are found in the genome of Mtb (Chen and Xie, 2011). A QS signal, such as autoinducer-2 (AI-2), induces oxidative stress which in turn induces biofilm formation in M. avium (Geier et al., 2008), suggesting that AI-2 could be recognized by mycobacteria which allow downstream QS events. In silico analysis has revealed that Mtb possesses a gene that has 59% homology and 41% identity with LuxS, which is responsible for AI-2 production (Surette et al., 1999). Like AI-2, diffusible signal factor (DSF) activity was detected in different mycobacterial species, including M. avium, M. chelonae, M. smegmatis, M. intracellulare, and Mycobacterium kansasii (Wang et al., 2004). Thus, mycobacteria presumably execute contact-dependent and contact-independent interactions concurrently.
Conclusion and future directions
In light of the findings of several reports, it can be concluded that mycobacteria also execute inter-bacterial communication. Unambiguously, mycobacteria also detect and respond to the neighboring cells that could be either their clone mates or other bacterial species. Despite the existence of a heterogeneous population, mycobacteria exhibit multicellular behavior under in vivo conditions, in which each subset of cells divides their work themselves (a division of labor) rather than producing all essential molecules to reduce their metabolic burden. The fact is that cell-to-cell communication is an integral part of the evolution of mycobacteria and the increase of rich diversity. While considering the importance of inter-bacterial communication for the exchange of genetic material, many fascinating questions are also raised. Whether mycobacteria exchange any other cytoplasmic molecules, such as proteins, RNAs, toxins, nutrients, and plasmids, with the neighboring cells? The existence of multiple subsets of the mycobacterial population at the site of infection, intriguing to speculate that one subset of cells confers resistance to certain antibiotics, while other subsets of cells produce some other antibiotic resistant-conferring proteins. The frequent incidence of failures in antibiotic therapy might be due to the exchange of those resistance-conferring molecules among the population. However, the coexistence of both subtypes benefits the entire population during antibiotic treatment. The exchange of antibiotic resistance-conferring proteins/mRNAs between different genotypes of B.subtilis has been well-documented (Dubey and Ben-Yehuda, 2011). The response of mycobacteria to AI-2 suggests the presence of common QS mechanisms across the mycobacterial species that might control several vital processes in mycobacteria.
Since virulence proteins are crucial during infection, it is imperative to find the factors that control/regulate the expression of these proteins. Hence, understanding the regulatory pathways could give insights into the molecular nature of mycobacterial infection (i.e., tuberculosis and leprosy). Key regulatory proteins of QS mechanisms could be used as a target to control pathogenesis (Boopathi et al., 2022b). As mentioned before, key proteins that are conserved across mycobacteria may be involved in QS mechanisms of various mycobacterial species, which could be targeted as a common target to control various mycobacterial diseases. Analogs of QS signals could be used as competitive inhibitors to attenuate the QS mechanisms (Boopathi et al., 2017b, 2022a), thereby mycobacterial virulence could be inhibited during pathogenesis. Moreover, identifying drugs that specifically target key regulatory proteins of QS mechanisms could be the potential approach for drug development.
Author contributions
SB designed the overall idea and wrote the manuscript. RM and BH assisted in the artwork. SR, MV, A-QJ, and JA performed the review of the literature, reviewed, and corrected the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (82160664).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdallah, A. M., Gey van Pittius, N. C., DiGiuseppe Champion, P. A., Cox, J., Luirink, J., Vandenbroucke-Grauls, C. M., et al. (2007). Type VII secretion—mycobacteria show the way. Nat. Rev. Microbiol. 5, 883–891. doi: 10.1038/nrmicro1773
Afriat, L., Roodveldt, C., Manco, G., and Tawfik, D. S. (2006). The latent promiscuity of newly identified microbial lactonases is linked to a recently diverged phosphotriesterase. Biochemistry 45, 13677–13686. doi: 10.1021/bi061268r
Aliyu, G., El-Kamary, S. S., Abimiku, A., Brown, C., Tracy, K., Hungerford, L., et al. (2013). Prevalence of non-tuberculous mycobacterial infections among tuberculosis suspects in Nigeria. PLoS ONE 8, e63170. doi: 10.1371/journal.pone.0063170
Alteri, C. J., Xicohténcatl-Cortes, J., Hess, S., Caballero-Olín, G., Girón, J. A., and Friedman, R. (2007). Mycobacterium tuberculosis produces pili during human infection. Proc. Natl. Acad. Sci. USA. 104, 5145–5150. doi: 10.1073/pnas.0602304104
An, S. Q., Chin, K. H., Febrer, M., McCarthy, Y., Yang, J. G., Liu, C. L., et al. (2013). A cyclic GMP-dependent signalling pathway regulates bacterial phytopathogenesis. EMBO J. 32, 2430–2438. doi: 10.1038/emboj.2013.165
Arora, K., Whiteford, D. C., Lau-Bonilla, D., Davitt, C. M., and Dahl, J. L. (2008). Inactivation of lsr2 results in a hypermotile phenotype in Mycobacterium smegmatis. J. Bacteriol. 190, 4291–4300. doi: 10.1128/JB.00023-08
Ates, L. S., and Brosch, R. (2017). Discovery of the type VII ESX-1 secretion needle? Mol. Microbiol. 103, 7–12. doi: 10.1111/mmi.13579
Baer, C. E., Iavarone, A. T., Alber, T., and Sassetti, C. M. (2014). Biochemical and spatial coincidence in the provisional ser/thr protein kinase interaction network of mycobacterium tuberculosis. Biol. Chem. 289, 20422–20433. doi: 10.1074/jbc.M114.559054
Baidya, A. K., Rosenshine, I., and Ben-Yehuda, S. (2020). Donor-delivered cell wall hydrolases facilitate nanotube penetration into recipient bacteria. Nat. Commun. 11, 1–13. doi: 10.1038/s41467-020-15605-1
Banaiee, N., Jacobs, W. Jr, and Ernst, J. (2006). Regulation of Mycobacterium tuberculosis whiB3 in the mouse lung and macrophages. Infect. Immun. 74, 6449–6457. doi: 10.1128/IAI.00190-06
Bartek, I., Woolhiser, L., Baughn, A., Basaraba, R., Jacobs, W. Jr., Lenaerts, A., et al. (2014). Mycobacterium tuberculosis Lsr2 is a global transcriptional regulator required for adaptation to changing oxygen levels and virulence. MBio 5, e01106–01114. doi: 10.1128/mBio.01106-14
Bharati, B. K., Sharma, I. M., Kasetty, S., Kumar, M., Mukherjee, R., and Chatterji, D. (2012). A full-length bifunctional protein involved in c-di-GMP turnover is required for long-term survival under nutrient starvation in Mycobacterium smegmatis. Microbiology 158, 1415–1427. doi: 10.1099/mic.0.053892-0
Bhattacharya, S., Baidya, A. K., Pal, R. R., Mamou, G., Gatt, Y. E., Margalit, H., et al. (2019). A ubiquitous platform for bacterial nanotube biogenesis. Cell Rep. 27, 334–342. e310. doi: 10.1016/j.celrep.2019.02.055
Birmes, F. S., Säring, R., Hauke, M. C., Ritzmann, N. H., Drees, S. L., Daniel, J., et al. (2019). Interference with Pseudomonas aeruginosa quorum sensing and virulence by the mycobacterial Pseudomonas quinolone signal dioxygenase AqdC in combination with the N-acylhomoserine lactone lactonase QsdA. Infect. Immun. 87, e00278–e00219. doi: 10.1128/IAI.00278-19
Boopathi, S., Liu, D., and Jia, A.-Q. (2021). Molecular trafficking between bacteria determines the shape of gut microbial community. Gut Microbes 13, 1959841. doi: 10.1080/19490976.2021.1959841
Boopathi, S., Selvakumar, G., and Sivakumar, N. (2017a). Quorum quenching potentials of probiotic Enterococcus durans LAB38 against methicillin resistant Staphylococcus aureus. Asian J. Pharmaceut. Clin. Res. 10, 445–550. doi: 10.22159/ajpcr.2017.v10i4.17039
Boopathi, S., Vashisth, R., Manoharan, P., Kandasamy, R., and Sivakumar, N. (2017b). Stigmatellin Y–An anti-biofilm compound from Bacillus subtilis BR4 possibly interferes in PQS–PqsR mediated quorum sensing system in Pseudomonas aeruginosa. Bioorganic Med. Chem. Lett. 27, 2113–2118. doi: 10.1016/j.bmcl.2017.03.074
Boopathi, S., Vashisth, R., Mohanty, A. K., Jia, A.-Q., Sivakumar, N., Alharbi, N. S., et al. (2022a). Investigation of interspecies crosstalk between probiotic Bacillus subtilis BR4 and Pseudomonas aeruginosa using metabolomics analysis. Microb. Pathog. 166, 105542. doi: 10.1016/j.micpath.2022.105542
Boopathi, S., Vashisth, R., Mohanty, A. K., Jia, A. Q., Sivakumar, N., Arockiaraj, J. J. J., et al. (2022b). Bacillus subtilis BR4 derived stigmatellin Y interferes Pqs-PqsR mediated quorum sensing system of Pseudomonas aeruginosa. J. Basic Microbiol. 62, 801–814. doi: 10.1002/jobm.202200017
Brambilla, C., Llorens-Fons, M., Julian, E., Noguera-Ortega, E., Tomas-Martinez, C., Perez-Trujillo, M., et al. (2016). Mycobacteria clumping increase their capacity to damage macrophages. Front. Microbiol. 7, 1562. doi: 10.3389/fmicb.2016.01562
Bretl, D. J., Demetriadou, C., and Zahrt, T. C. (2011). Adaptation to environmental stimuli within the host: two-component signal transduction systems of Mycobacterium tuberculosis. Microbiol. Mol. Biol. Rev. 75, 566–582. doi: 10.1128/MMBR.05004-11
Canova, M. J., Kremer, L., and Molle, V. (2009). The Mycobacterium tuberculosis GroEL1 chaperone is a substrate of Ser/Thr protein kinases. J. Bacteriol. 191, 2876–2883. doi: 10.1128/JB.01569-08
Carow, B., Hauling, T., Qian, X., Kramnik, I., Nilsson, M., and Rottenberg, M. E. (2019). Spatial and temporal localization of immune transcripts defines hallmarks and diversity in the tuberculosis granuloma. Nat. Commun. 10, 1823. doi: 10.1038/s41467-019-09816-4
Carter, G., Wu, M., Drummond, D. C., and Bermudez, L. E. (2003). Characterization of biofilm formation by clinical isolates of Mycobacterium avium. J. Med. Microbiol. 52, 747–752. doi: 10.1099/jmm.0.05224-0
Chakraborty, P., and Kumar, A. (2019). The extracellular matrix of mycobacterial biofilms: could we shorten the treatment of mycobacterial infections? Microb. Cell. 6, 105–122. doi: 10.15698/mic2019.02.667
Chan, K., Knaak, T., Satkamp, L., Humbert, O., Falkow, S., and Ramakrishnan, L. (2002). Complex pattern of Mycobacterium marinum gene expression during long-term granulomatous infection. Proc. Natl. Acad. Sci. USA. 99, 3920–3925. doi: 10.1073/pnas.002024599
Chauhan, A., Madiraju, M. V., Fol, M., Lofton, H., Maloney, E., Reynolds, R., et al. (2006). Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J. Bacteriol. 188, 1856–1865. doi: 10.1128/JB.188.5.1856-1865.2006
Chen, J., and Xie, J. (2011). Role and regulation of bacterial LuxR-like regulators. J. Cell. Biochem. 112, 2694–2702. doi: 10.1002/jcb.23219
Chen, J. M., German, G. J., Alexander, D. C., Ren, H., Tan, T., and Liu, J. (2006). Roles of Lsr2 in colony morphology and biofilm formation of Mycobacterium smegmatis. J. Bacteriol. 188, 633–641. doi: 10.1128/JB.188.2.633-641.2006
Chow, J. Y., Wu, L., and Yew, W. S. (2009). Directed evolution of a quorum-quenching lactonase from Mycobacterium avium subsp. paratuberculosis K-10 in the amidohydrolase superfamily. Biochemistry 48, 4344–4353. doi: 10.1021/bi9004045
Clark, R. R., Judd, J., Lasek-Nesselquist, E., Montgomery, S. A., Hoffmann, J. G., Derbyshire, K. M., et al. (2018). Direct cell-cell contact activates SigM to express the ESX-4 secretion system in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. USA. 115, E6595–E6603. doi: 10.1073/pnas.1804227115
Clary, G., Sasindran, S. J., Nesbitt, N., Mason, L., Cole, S., Azad, A., et al. (2018). Mycobacterium abscessus smooth and rough morphotypes form antimicrobial-tolerant biofilm phenotypes but are killed by acetic acid. Antimicrob. Agents Chemother. 62, e01782–e01717. doi: 10.1128/AAC.01782-17
Cohen, T., van Helden, P. D., Wilson, D., Colijn, C., McLaughlin, M. M., Abubakar, I., et al. (2012). Mixed-strain mycobacterium tuberculosis infections and the implications for tuberculosis treatment and control. Clin. Microbiol. Rev. 25, 708–719. doi: 10.1128/CMR.00021-12
Conrad, W. H., Osman, M. M., Shanahan, J. K., Chu, F., Takaki, K. K., Cameron, J., et al. (2017). Mycobacterial ESX-1 secretion system mediates host cell lysis through bacterium contact-dependent gross membrane disruptions. Proc. Natl. Acad. Sci. USA. 114, 1371–1376. doi: 10.1073/pnas.1620133114
Coros, A., Callahan, B., Battaglioli, E., and Derbyshire, K. M. (2008). The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol. Microbiol. 69, 794–808. doi: 10.1111/j.1365-2958.2008.06299.x
Costa, K. C., Bergkessel, M., Saunders, S., Korlach, J., and Newman, D. K. (2015). Enzymatic degradation of phenazines can generate energy and protect sensitive organisms from toxicity. MBio 6, e01520–e01515. doi: 10.1128/mBio.01520-15
Costa, K. C., Glasser, N. R., Conway, S. J., and Newman, D. K. (2017). Pyocyanin degradation by a tautomerizing demethylase inhibits Pseudomonas aeruginosa biofilms. Science 355, 170–173. doi: 10.1126/science.aag3180
Cui, T., Zhang, L., Wang, X., and He, Z.-G. (2009). Uncovering new signaling proteins and potential drug targets through the interactome analysis of Mycobacterium tuberculosis. BMC Gen. 10, 1–10. doi: 10.1186/1471-2164-10-118
Dahl, J. L. (2005). Scanning electron microscopy analysis of aged Mycobacterium tuberculosis cells. Can. J. Microbiol. 51, 277–281. doi: 10.1139/w05-001
Dahl, J. L., Arora, K., Boshoff, H. I., Whiteford, D. C., Pacheco, S. A., Walsh, O. J., et al. (2005). The relA homolog of Mycobacterium smegmatis affects cell appearance, viability, and gene expression. J. Bacteriol. 187, 2439–2447. doi: 10.1128/JB.187.7.2439-2447.2005
Dahl, J. L., Kraus, C. N., Boshoff, H. I., Doan, B., Foley, K., Avarbock, D., et al. (2003). The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc. Natl. Acad. Sci. USA. 100, 10026–10031. doi: 10.1073/pnas.1631248100
Davis, K. M. (2020). For the greater (bacterial) good: heterogeneous expression of energetically costly virulence factors. Infect. Immun. 88, 1–13. doi: 10.1128/IAI.00911-19
Davis, K. M., and Isberg, R. R. (2019). One for all, but not all for one: social behavior during bacterial diseases. Trends Microbiol. 27, 64–74. doi: 10.1016/j.tim.2018.09.001
DePas, W. H., Bergkessel, M., and Newman, D. K. (2019). Aggregation of nontuberculous mycobacteria is regulated by carbon-nitrogen balance. MBio 10, e01715–01719. doi: 10.1128/mBio.01715-19
Dhar, N., McKinney, J., and Manina, G. (2016). Phenotypic heterogeneity in Mycobacterium tuberculosis. Microbiol. Spectr. 671–697. doi: 10.1128/microbiolspec.TBTB2-0021-2016
Dinkele, R., Gessner, S., McKerry, A., Leonard, B., Seldon, R., Koch, A. S., et al. (2021). Capture and visualization of live Mycobacterium tuberculosis bacilli from tuberculosis patient bioaerosols. PLoS Pathog. 17, e1009262. doi: 10.1371/journal.ppat.1009262
Dragset, M. S., Ioerger, T. R., Zhang, Y. J., Maerk, M., Ginbot, Z., Sacchettini, J. C., et al. (2019). Genome-wide phenotypic profiling identifies and categorizes genes required for mycobacterial low iron fitness. Sci. Rep. 9, 11394. doi: 10.1038/s41598-019-47905-y
Draper, P. (1974). The mycoside capsule of Mycobacterium avium 357. Microbiology 83, 431–433. doi: 10.1099/00221287-83-2-431
Draper, P., and Rees, R. (1970). Electron-transparent zone of mycobacteria may be a defence mechanism. Nature 228, 860–861. doi: 10.1038/228860a0
D'Souza, G., and Kost, C. (2016). Experimental evolution of metabolic dependency in bacteria. PLoS Genet. 12, e1006364. doi: 10.1371/journal.pgen.1006364
Dubey, G. P., and Ben-Yehuda, S. (2011). Intercellular nanotubes mediate bacterial communication. Cell 144, 590–600. doi: 10.1016/j.cell.2011.01.015
Dubey, G. P., Mohan, G. B. M., Dubrovsky, A., Amen, T., Tsipshtein, S., Rouvinski, A., et al. (2016). Architecture and characteristics of bacterial nanotubes. Dev. Cell 36, 453–461. doi: 10.1016/j.devcel.2016.01.013
Dulberger, C. L., Rubin, E. J., and Boutte, C. C. (2020). The mycobacterial cell envelope - a moving target. Nat. Rev. Microbiol. 18, 47–59. doi: 10.1038/s41579-019-0273-7
Dutta, N. K., Klinkenberg, L. G., Vazquez, M.-J., Segura-Carro, D., Colmenarejo, G., Ramon, F., et al. (2019). Inhibiting the stringent response blocks Mycobacterium tuberculosis entry into quiescence and reduces persistence. Sci Adv. 5, eaav2104. doi: 10.1126/sciadv.aav2104
Falkinham, J. O. 3rd, Hilborn, E. D., Arduino, M. J., Pruden, A., and Edwards, M. A. (2015). Epidemiology and ecology of opportunistic premise plumbing pathogens: legionella pneumophila, Mycobacterium avium, and Pseudomonas aeruginosa. Environ. Health Perspect. 123, 749–758. doi: 10.1289/ehp.1408692
Fennelly, K. P., Ojano-Dirain, C., Yang, Q., Liu, L., Lu, L., Progulske-Fox, A., et al. (2016). Biofilm formation by mycobacterium abscessus in a lung cavity. Am. J. Respir. Crit. Care Med. 193, 692–693. doi: 10.1164/rccm.201508-1586IM
Flint, J. L., Kowalski, J. C., Karnati, P. K., and Derbyshire, K. M. (2004). The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. USA. 101, 12598–12603. doi: 10.1073/pnas.0404892101
Flores-Valdez, M. A., Aceves-Sánchez, M., Peterson, E. J., Baliga, N., Bravo-Madrigal, J., la Cruz-Villegas, D., et al. (2020). Transcriptional portrait of M. bovis BCG during biofilm production shows genes differentially expressed during intercellular aggregation and substrate attachment. Sci Rep. 10, 1–15. doi: 10.1038/s41598-020-69152-2
Geier, H., Mostowy, S., Cangelosi, G. A., Behr, M. A., and Ford, T. E. (2008). Autoinducer-2 triggers the oxidative stress response in Mycobacterium avium, leading to biofilm formation. Appl. Environ. Microbiol. 74, 1798–1804. doi: 10.1128/AEM.02066-07
Gopalaswamy, R., Narayanan, S., Jacobs Jr, W. R., and Av-Gay, Y. (2008). Mycobacterium smegmatis biofilm formation and sliding motility are affected by the serine/threonine protein kinase PknF. FEMS Microbiol. Lett. 278, 121–127. doi: 10.1111/j.1574-6968.2007.00989.x
Gopinath, V., Raghunandanan, S., Gomez, R. L., Jose, L., Surendran, A., Ramachandran, R., et al. (2015). Profiling the proteome of Mycobacterium tuberculosis during dormancy and reactivation. Mol. Cell. Proteomics. 14, 2160–2176. doi: 10.1074/mcp.M115.051151
Gray, T. A., Clark, R. R., Boucher, N., Lapierre, P., Smith, C., and Derbyshire, K. (2016). Intercellular communication and conjugation are mediated by ESX secretion systems in mycobacteria. Science 354, 347–350. doi: 10.1126/science.aag0828
Gray, T. A., and Derbyshire, K. M. (2018). Blending genomes: distributive conjugal transfer in mycobacteria, a sexier form of HGT. Mol. Microbiol. 108, 601–613. doi: 10.1111/mmi.13971
Gray, T. A., Krywy, J. A., Harold, J., Palumbo, M. J., and Derbyshire, K. M. (2013). Distributive conjugal transfer in mycobacteria generates progeny with meiotic-like genome-wide mosaicism, allowing mapping of a mating identity locus. PLoS Biol. 11, e1001602. doi: 10.1371/journal.pbio.1001602
Gupta, K., Kumar, P., and Chatterji, D. (2010). Identification, activity and disulfide connectivity of C-di-GMP regulating proteins in Mycobacterium tuberculosis. PLoS ONE. 5, e15072. doi: 10.1371/journal.pone.0015072
Gupta, K. R., Kasetty, S., and Chatterji, D. (2015). Novel functions of (p) ppGpp and cyclic di-GMP in mycobacterial physiology revealed by phenotype microarray analysis of wild-type and isogenic strains of Mycobacterium smegmatis. Appl. Environ. Microbiol. 81, 2571–2578. doi: 10.1128/AEM.03999-14
Gutiérrez, A. V., Viljoen, A., Ghigo, E., Herrmann, J.-L., and Kremer, L. (2018). Glycopeptidolipids, a double-edged sword of the Mycobacterium abscessus complex. Front. Microbiol. 9, 1145. doi: 10.3389/fmicb.2018.01145
Hannebelle, M. T. M., Ven, J. X. Y., Toniolo, C., Eskandarian, H. A., Vuaridel-Thurre, G., McKinney, J. D., et al. (2020). A biphasic growth model for cell pole elongation in mycobacteria. Nat. Commun. 11, 452. doi: 10.1038/s41467-019-14088-z
Hegde, S. R. (2020). Computational identification of the proteins associated with quorum sensing and biofilm formation in Mycobacterium tuberculosis. Front. Microbiol. 10, 3011. doi: 10.3389/fmicb.2019.03011
Hu, Q., Zhang, J., Chen, Y., Hu, L., Li, W., and He, Z.-G. (2019). Cyclic di-GMP co-activates the two-component transcriptional regulator DevR in Mycobacterium smegmatis in response to oxidative stress. J. Biol. Chem. 294, 12729–12742. doi: 10.1074/jbc.RA119.008252
Huang, H. C., Yu, W. L., Shieh, C. C., Cheng, K. C., and Cheng, H. H. (2007). Unusual mixed infection of thoracic empyema caused by Mycobacteria tuberculosis, nontuberculosis mycobacteria and Nocardia asteroides in a woman with systemic lupus erythematosus. J. Infect. 54, e25–28. doi: 10.1016/j.jinf.2006.03.024
Irwin, S. M., Driver, E., Lyon, E., Schrupp, C., Ryan, G., Gonzalez-Juarrero, M., et al. (2015). Presence of multiple lesion types with vastly different microenvironments in C3HeB/FeJ mice following aerosol infection with Mycobacterium tuberculosis. Dis. Model. Mech. 8, 591–602. doi: 10.1242/dmm.019570
Jamet, S., Slama, N., Domingues, J., Laval, F., Texier, P., Eynard, N., et al. (2015). The non-essential mycolic acid biosynthesis genes hadA and hadC contribute to the physiology and fitness of Mycobacterium smegmatis. PLoS ONE. 10, e0145883. doi: 10.1371/journal.pone.0145883
Javid, B., Sorrentino, F., Toosky, M., Zheng, W., Pinkham, J. T., Jain, N., et al. (2014). Mycobacterial mistranslation is necessary and sufficient for rifampicin phenotypic resistance. Proc. Natl. Acad. Sci. USA. 111, 1132–1137. doi: 10.1073/pnas.1317580111
Jones, C. M., Wells, R. M., Madduri, A. V., Renfrow, M. B., Ratledge, C., Moody, D. B., et al. (2014). Self-poisoning of Mycobacterium tuberculosis by interrupting siderophore recycling. Proc. Natl. Acad. Sci. USA. 111, 1945–1950. doi: 10.1073/pnas.1311402111
Julián, E., Roldán, M., Sánchez-Chardi, A., Astola, O., Agust, G., and Luquin, M. (2010). Microscopic cords, a virulence-related characteristic of Mycobacterium tuberculosis, are also present in nonpathogenic mycobacteria. J. Bacteriol. 192, 1751–1760. doi: 10.1128/JB.01485-09
Kieser, K. J., and Rubin, E. J. (2014). How sisters grow apart: mycobacterial growth and division. Nat. Rev. Microbiol. 12, 550–562. doi: 10.1038/nrmicro3299
Lenaerts, A. J., Hoff, D., Aly, S., Ehlers, S., Andries, K., Cantarero, L., et al. (2007). Location of persisting mycobacteria in a Guinea pig model of tuberculosis revealed by r207910. Antimicrob. Agents Chemother. 51, 3338–3345. doi: 10.1128/AAC.00276-07
Levy, I., Grisaru-Soen, G., Lerner-Geva, L., Kerem, E., Blau, H., Bentur, L., et al. (2008). Multicenter cross-sectional study of nontuberculous mycobacterial infections among cystic fibrosis patients, Israel. Emerging Infect. Dis. 14, 378. doi: 10.3201/eid1403.061405
Lieberman, T. D., Wilson, D., Misra, R., Xiong, L. L., Moodley, P., Cohen, T., et al. (2016). Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated Mycobacterium tuberculosis. Nat. Med. 22, 1470–1474. doi: 10.1038/nm.4205
Lori, C., Ozaki, S., Steiner, S., Böhm, R., Abel, S., Dubey, B. N., et al. (2015). Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature. 523, 236–239. doi: 10.1038/nature14473
Lou, Y., Rybniker, J., Sala, C., and Cole, S. T. (2017). EspC forms a filamentous structure in the cell envelope of Mycobacterium tuberculosis and impacts ESX-1 secretion. Mol. Microbiol. 103, 26–38. doi: 10.1111/mmi.13575
Maitra, A., Munshi, T., Healy, J., Martin, L. T., Vollmer, W., Keep, N. H., et al. (2019). Cell wall peptidoglycan in Mycobacterium tuberculosis: an achilles' heel for the TB-causing pathogen. FEMS Microbiol. Rev. 43, 548–575. doi: 10.1093/femsre/fuz016
Manina, G., Dhar, N., and McKinney, J. D. (2015). Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe 17, 32–46. doi: 10.1016/j.chom.2014.11.016
Marsollier, L., Brodin, P., Jackson, M., Kordulakova, J., Tafelmeyer, P., Carbonnelle, E., et al. (2007). Impact of Mycobacterium ulcerans biofilm on transmissibility to ecological niches and Buruli ulcer pathogenesis. PLoS Pathog. 3, e62. doi: 10.1371/journal.ppat.0030062
Merkal, R., Rhoades, K., Gallagher, J., and Ritchie, A. (1973). Scanning electron microscopy of mycobacteria. Am. Rev. Respir. Dis. 108, 381–387.
Misch, E. A., Saddler, C., and Davis, J. M. (2018). Skin and soft tissue infections due to nontuberculous mycobacteria. Curr. Infect. Dis. Rep. 20, 6. doi: 10.1007/s11908-018-0611-3
Molle, V., Brown, A. K., Besra, G. S., Cozzone, A. J., and Kremer, L. (2006). The condensing activities of the Mycobacterium tuberculosis type II fatty acid synthase are differentially regulated by phosphorylation. J. Biol. Chem. 281, 30094–30103. doi: 10.1074/jbc.M601691200
Mukherjee, S., and Bassler, B. L. (2019). Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382. doi: 10.1038/s41579-019-0186-5
Nadell, C. D., Drescher, K., and Foster, K. R. (2016). Spatial structure, cooperation and competition in biofilms. Nat. Rev. Microbiol. 14, 589–600. doi: 10.1038/nrmicro.2016.84
Nguyen, K. T., Piastro, K., Gray, T. A., and Derbyshire, K. M. (2010). Mycobacterial biofilms facilitate horizontal DNA transfer between strains of Mycobacterium smegmatis. J. Bacteriol. 192, 5134–5142. doi: 10.1128/JB.00650-10
Nikitushkin, V. D., Demina, G. R., and Kaprelyants, A. S. (2011). Effect of secreted Rpf protein on intracellular contacts in Micrococcus luteus and Mycobacterium smegmatis cultures. Microbiology 80, 143–149. doi: 10.1134/S0026261711020123
Nishimura, T., Shimoda, M., Tamizu, E., Uno, S., Uwamino, Y., Kashimura, S., et al. (2020). The rough colony morphotype of Mycobacterium avium exhibits high virulence in human macrophages and mice. J. Med. Microbiol. 69, 1020–1033. doi: 10.1099/jmm.0.001224
Ojha, A., Anand, M., Bhatt, A., Kremer, L., Jacobs, W. R. Jr, and Hatfull, G. F. (2005). GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell. 123, 861–873. doi: 10.1016/j.cell.2005.09.012
Ojha, A., and Hatfull, G. F. (2007). The role of iron in Mycobacterium smegmatis biofilm formation: the exochelin siderophore is essential in limiting iron conditions for biofilm formation but not for planktonic growth. Mol. Microbiol. 66, 468–483. doi: 10.1111/j.1365-2958.2007.05935.x
Olivier, K. N., Weber, D. J., Wallace, R. J. Jr., Faiz, A. R., Lee, J. H., Zhang, Y., et al. (2003). Nontuberculous mycobacteria. I: multicenter prevalence study in cystic fibrosis. Am. J. Respir. Crit. Care Med. 167, 828–834. doi: 10.1164/rccm.200207-678OC
Padhi, A., Naik, S. K., Sengupta, S., Ganguli, G., and Sonawane, A. (2016). Expression of Mycobacterium tuberculosis NLPC/p60 family protein Rv0024 induce biofilm formation and resistance against cell wall acting anti-tuberculosis drugs in Mycobacterium smegmatis. Microbes Infect. 18, 224–236. doi: 10.1016/j.micinf.2015.11.007
Pal, R. R., Baidya, A. K., Mamou, G., Bhattacharya, S., Socol, Y., Kobi, S., et al. (2019). Pathogenic E. coli extracts nutrients from infected host cells utilizing injectisome components. Cell 177, 683–696.e618. doi: 10.1016/j.cell.2019.02.022
Paleiron, N., Soler, C., Hassan, M. O., Andriamanantena, D., Vong, R., Pourcel, C., et al. (2019). First description of Mycobacterium tuberculosis and M. canettii concomitant infection: report of two cases. Int. J. Tuberc. Lung. Dis. 23, 232–235. doi: 10.5588/ijtld.18.0261
Pande, S., Shitut, S., Freund, L., Westermann, M., Bertels, F., Colesie, C., et al. (2015). Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat. Commun. 6, 1–13. doi: 10.1038/ncomms7238
Parsons, L. M., Jankowski, C. S., and Derbyshire, K. M. (1998). Conjugal transfer of chromosomal DNA in Mycobacterium smegmatis. Mol. Microbiol. 28, 571–582. doi: 10.1046/j.1365-2958.1998.00818.x
Peddireddy, V., Doddam, S. N., and Ahmed, N. (2017). Mycobacterial dormancy systems and host responses in tuberculosis. Front. Immunol. 8, 84. doi: 10.3389/fimmu.2017.00084
Piersimoni, C., and Scarparo, C. (2009). Extrapulmonary infections associated with nontuberculous mycobacteria in immunocompetent persons. Emerg. Infect. Dis. 15, 1351–1358; quiz 1544. doi: 10.3201/eid1509.081259
Potrykus, K., and Cashel, M. (2008). (p) ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51. doi: 10.1146/annurev.micro.62.081307.162903
Prados-Rosales, R., Weinrick, B. C., Pique, D. G., Jacobs, W. R. Jr., Casadevall, A., and Rodriguez, G. M. (2014). Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J. Bacteriol. 196, 1250–1256. doi: 10.1128/JB.01090-13
Prideaux, B., Via, L. E., Zimmerman, M. D., Eum, S., Sarathy, J., O'Brien, P., et al. (2015). The association between sterilizing activity and drug distribution into tuberculosis lesions. Nat. Med. 21, 1223–1227. doi: 10.1038/nm.3937
Prusa, J., Zhu, D. X., Stallings, C. L., and disease (2018). The stringent response and Mycobacterium tuberculosis pathogenesis. Pathog Dis. 76, fty054. doi: 10.1093/femspd/fty054
Qvist, T., Eickhardt, S., Kragh, K. N., Andersen, C. B., Iversen, M., Høiby, N., et al. (2015). Chronic pulmonary disease with Mycobacterium abscessus complex is a biofilm infection. Eur. Respir. J. 46, 1823–1826. doi: 10.1183/13993003.01102-2015
Rego, E. H., Audette, R. E., and Rubin, E. J. (2017). Deletion of a mycobacterial divisome factor collapses single-cell phenotypic heterogeneity. Nature 546, 153–157. doi: 10.1038/nature22361
Rodel, H. E., Ferreira, I. A., Ziegler, C. G., Ganga, Y., Bernstein, M., Hwa, S.-H., et al. (2021). Aggregated Mycobacterium tuberculosis enhances the inflammatory response. Front. Microbiol. 12, 1–13. doi: 10.3389/fmicb.2021.757134
Roux, A.-L., Viljoen, A., Bah, A., Simeone, R., Bernut, A., Laencina, L., et al. (2016). The distinct fate of smooth and rough Mycobacterium abscessus variants inside macrophages. Open Biol. 6, 160185. doi: 10.1098/rsob.160185
Schulze-Röbbecke, R., Feldmann, C., Fischeder, R., Janning, B., Exner, M., and Wahl, G. (1995). Dental units: an environmental study of sources of potentially pathogenic mycobacteria. Tuberc. Lung Dis. 76, 318–323. doi: 10.1016/S0962-8479(05)80030-9
Segura-Cerda, C. A., de Jesús Aceves-Sánchez, M., Marquina-Castillo, B., Mata-Espinoza, D., Barrios-Payán, J., Vega-Domínguez, P. J., et al. (2018). Immune response elicited by two rBCG strains devoid of genes involved in c-di-GMP metabolism affect protection versus challenge with M. tuberculosis strains of different virulence. Vaccine 36, 2069–2078. doi: 10.1016/j.vaccine.2018.03.014
Sharma, I. M., Petchiappan, A., and Chatterji, D. (2014). Quorum sensing and biofilm formation in mycobacteria: role of c-di-GMP and methods to study this second messenger. IUBMB Life 66, 823–834. doi: 10.1002/iub.1339
Shaw, L. P., Doyle, R. M., Kavaliunaite, E., Spencer, H., Balloux, F., Dixon, G., et al. (2019). Children with cystic fibrosis are infected with multiple subpopulations of mycobacterium abscessus with different antimicrobial resistance profiles. Clin. Infect. Dis. 69, 1678–1686. doi: 10.1093/cid/ciz069
Shitut, S., Ahsendorf, T., Pande, S., Egbert, M., and Kost, C. (2019). Nanotube-mediated cross-feeding couples the metabolism of interacting bacterial cells. Environ. Microbiol. 21, 1306–1320. doi: 10.1111/1462-2920.14539
Simsek, E., and Kim, M. (2018). The emergence of metabolic heterogeneity and diverse growth responses in isogenic bacterial cells. ISME J. 12, 1199–1209. doi: 10.1038/s41396-017-0036-2
Slama, N., Jamet, S., Frigui, W., Pawlik, A., Bottai, D., Laval, F., et al. (2016). The changes in mycolic acid structures caused by hadC mutation have a dramatic effect on the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 99, 794–807. doi: 10.1111/mmi.13266
Slama, N., Leiba, J., Eynard, N., Daffé, M., Kremer, L., Quémard, A., et al. (2011). Negative regulation by Ser/Thr phosphorylation of HadAB and HadBC dehydratases from Mycobacterium tuberculosis type II fatty acid synthase system. Biochem. Biophys. Res. Commun. 412, 401–406. doi: 10.1016/j.bbrc.2011.07.051
Srivastava, D., and Waters, C. M. (2012). A tangled web: regulatory connections between quorum sensing and cyclic di-GMP. J. Bacteriol. 194, 4485–4493. doi: 10.1128/JB.00379-12
Stempler, O., Baidya, A. K., Bhattacharya, S., Malli Mohan, G. B., Tzipilevich, E., Sinai, L., et al. (2017). Interspecies nutrient extraction and toxin delivery between bacteria. Nat. Commun. 8, 1–9. doi: 10.1038/s41467-017-00344-7
Stokas, H., Rhodes, H. L., and Purdy, G. E. (2020). Modulation of the M. tuberculosis cell envelope between replicating and non-replicating persistent bacteria. Tuberculosis 125, 102007. doi: 10.1016/j.tube.2020.102007
Surette, M. G., Miller, M. B., and Bassler, B. L. (1999). Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA. 96, 1639–1644. doi: 10.1073/pnas.96.4.1639
Torrelles, J., Ellis, D., Osborne, T., Hoefer, A., Orme, I., Chatterjee, D., et al. (2002). Characterization of virulence, colony morphotype and the glycopeptidolipid of Mycobacterium avium strain 104. Tuberculosis 82, 293–300. doi: 10.1054/tube.2002.0373
Valway, S. E., Sanchez, M. P. C., Shinnick, T. F., Orme, I., Agerton, T., Hoy, D., et al. (1998). An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N. Engl. J. Med. 338, 633–639. doi: 10.1056/NEJM199803053381001
Viljoen, A., Herrmann, J.-L., Onajole, O. K., Stec, J., Kozikowski, A. P., and Kremer, L. (2017). Controlling extra-and intramacrophagic Mycobacterium abscessus by targeting mycolic acid transport. Front. Cell. Infect. Microbiol. 7, 388. doi: 10.3389/fcimb.2017.00388
Wang, L. H., He, Y., Gao, Y., Wu, J. E., Dong, Y. H., He, C., et al. (2004). A bacterial cell–cell communication signal with cross-kingdom structural analogues. Mol. Microbiol. 51, 903–912. doi: 10.1046/j.1365-2958.2003.03883.x
Wang, M., Guo, X., Yang, X., Zhang, B., Ren, J., Liu, A., et al. (2019). Mycobacterial dynamin-like protein IniA mediates membrane fission. Nat. Commun. 10, 1–13. doi: 10.1038/s41467-019-11860-z
Wullich, S. C., Kobus, S., Wienhold, M., Hennecke, U., Smits, S. H. J., and Fetzner, S. (2019). Structural basis for recognition and ring-cleavage of the Pseudomonas quinolone signal (PQS) by AqdC, a mycobacterial dioxygenase of the alpha/beta-hydrolase fold family. J. Struct. Biol. 207, 287–294. doi: 10.1016/j.jsb.2019.06.006
Keywords: quorum sensing, nanotubes, mycobacterial communication, T7SS, biofilm
Citation: Boopathi S, Ramasamy S, Haridevamuthu B, Murugan R, Veerabadhran M, Jia A-Q and Arockiaraj J (2022) Intercellular communication and social behaviors in mycobacteria. Front. Microbiol. 13:943278. doi: 10.3389/fmicb.2022.943278
Received: 25 May 2022; Accepted: 22 August 2022;
Published: 13 September 2022.
Edited by:
Soumita Das, University of California, San Diego, United StatesReviewed by:
Blanca Estela Garcia-Perez, Instituto Politécnico Nacional (IPN), MexicoAndaleeb Sajid, National Cancer Institute (NIH), United States
Copyright © 2022 Boopathi, Ramasamy, Haridevamuthu, Murugan, Veerabadhran, Jia and Arockiaraj. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ai-Qun Jia, YWppYUBoYWluYW51LmVkdS5jbg==; Jesu Arockiaraj, amVzdWFyYWpAaG90bWFpbC5jb20=; amVzdWFyb2FAc3JtaXN0LmVkdS5pbg==
†ORCID: Seenivasan Boopathi orcid.org/0000-0003-3537-9340
B. Haridevamuthu orcid.org/0000-0003-4598-5993
Raghul Murugan orcid.org/0000-0002-0864-4818
Ai-Qun Jia orcid.org/0000-0002-8089-6200
Jesu Arockiaraj orcid.org/0000-0002-0240-7141
 Seenivasan Boopathi
Seenivasan Boopathi Subbiah Ramasamy
Subbiah Ramasamy B. Haridevamuthu
B. Haridevamuthu Raghul Murugan
Raghul Murugan Maruthanayagam Veerabadhran
Maruthanayagam Veerabadhran Ai-Qun Jia
Ai-Qun Jia Jesu Arockiaraj
Jesu Arockiaraj