
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 September 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.942302
This article is part of the Research Topic Community Series in Plants and Microbial Communities: Diversity, Pathogens and Biological Control, volume II View all 25 articles
 Yuanpeng Fang1†
Yuanpeng Fang1† Qiaoli Du1†
Qiaoli Du1† Qian Yang1†
Qian Yang1† Junmei Jiang2
Junmei Jiang2 Xiaolong Hou1
Xiaolong Hou1 Zaifu Yang1
Zaifu Yang1 Degang Zhao3,4
Degang Zhao3,4 Xiangyang Li2
Xiangyang Li2 Xin Xie1,4*
Xin Xie1,4*The U-box family is one of the main E3 ubiquitin ligase families in plants. The U-box family has been characterized in several species. However, genome-wide gene identification and expression profiling of the U-box family in response to abiotic stress in Sorghum bicolor remain unclear. In this study, we broadly identified 68 U-box genes in the sorghum genome, including 2 CHIP genes, and 1 typical UFD2 (Ub fusion degradation 2) gene. The U-box gene family was divided into eight subclasses based on homology and conserved domain characteristics. Evolutionary analysis identified 14, 66, and 82 U-box collinear gene pairs in sorghum compared with arabidopsis, rice, and maize, respectively, and a unique tandem repeat pair (SbPUB26/SbPUB27) is present in the sorghum genome. Gene Ontology (GO) enrichment analysis showed that U-box proteins were mainly related to ubiquitination and modification, and various stress responses. Comprehensive analysis of promoters, expression profiling, and gene co-regulation networks also revealed that many sorghum U-box genes may be correlated with multiple stress responses. In summary, our results showed that sorghum contains 68 U-box genes, which may be involved in multiple abiotic stress responses. The findings will support future gene functional studies related to ubiquitination in sorghum.
Sorghum (Sorghum bicolor) is used in winemaking, bioenergy, and feed plant industries worldwide, and is the most important winemaking crop in China (Mace et al., 2013). Since the completion of genome sequencing in 2013 (Mace et al., 2013), sorghum breeding based on genome research has expanded rapidly (Vallabhaneni et al., 2010; Zhao et al., 2011; Yang and Wang, 2016; Ding et al., 2019; Yang et al., 2020). However, biological (Colletotrichum graminicola and Mythimna separata infection, etc.) and non-biological stresses (drought, hormonal, salt stress, etc.) affect the quality and quantity of this important crop (Sharma, 1993; Anaso, 2010; Silva et al., 2013, 2021; Dowd et al., 2016; Cuevas et al., 2018; Das et al., 2019; Moharam and Hassan, 2020). Previous studies have shown that epigenetic modification proteins are related to plant stress resistance, providing a potential resource for plant resistance breeding (Callis, 2014; Yang et al., 2018; Miryeganeh and Saze, 2020; Yu et al., 2021; Puyuan et al., 2022).
Epigenetic regulation involves site-specific modification of nucleic acids and proteins under environmental stress, and it occurs widely in all animals, plants, and microorganisms. The epigenetic modification mechanism mainly includes pre-translational modification (DNA modification, non-coding RNA regulation, and chromatin remodeling) and post-translational modification (histone modification and non-histone modification). Post-translational modification (PTM), an important type of epigenetic regulation, mainly includes protein methylation, acetylation, ubiquitination, phosphorylation, etc. (Miryeganeh and Saze, 2020). Methylation and acetylation are generally considered to regulate protein activity, while phosphorylation initiates kinase activity (Yu et al., 2021). By contrast, ubiquitination mainly mediates protein clearance and recycling process by promoting the stability of active proteins and the repair of misfolded proteins to achieve protein equilibrium (Callis, 2014). Reports showed that ubiquitination plays principal roles in the regulation of plant stress resistance, for example, abscisic acid (ABA)-insensitive RING protein (AIRP) in some maize cultivars is more sensitive to ABA than wild varieties, thus achieving higher drought tolerance (Kong et al., 2015; Yang et al., 2018). The ubiquitin ligase TagW2-6a negatively regulates gibberellin (GA) synthesis and signaling (An et al., 2021). E3 ligase MIEL1 negatively regulates jasmonic acid (JA) signaling and thereby reduces cold resistance in apple (Malus domestica) by mediating ubiquitination and degradation of the novel B-box (BBX) protein BBX37 (You and He, 2016). Besides, studies also have shown that ubiquitination is a potential resource for plant disease resistance.
Ubiquitination modifications include conventional ubiquitination modification (with ubiquitin as the substrate) and SUMO-type ubiquitination modification (with the ubiquitin-like SUMO molecule as the substrate) (Zhou and Zeng, 2017). Conventional ubiquitination typically involves defined steps: (1) first, E1 ubiquitin activase (UBA) activates ubiquitin in the presence of ATP, allowing the cysteine residues of UBA to form the thioester-linked intermediate E1-ubiquitin (E1-Ub) (Mandal et al., 2018); (2) subsequently, the E2 ubiquitin-binding enzyme (UBC) interacts with UAB-Ub and transfers activated Ub to an active cysteine residue of UBC to form a thioester-linked UBC-Ub intermediate (Mandal et al., 2018); (3) finally, the ubiquitin ligase (E3) interacts with the target protein and E2-Ub to create an isopeptide bond between the C-terminal glycine residue and the lysine residue of Ub. The establishment of an isopeptide bond between this glycine residue and the lysine residue of the target protein results in the transfer of Ub to the target protein by ubiquitin ligase (E3) (Mandal et al., 2018). The ubiquitination modification system includes a large protein family, with ∼10 E1 ubiquitin activating enzymes, ∼50 E2 ubiquitin conjugating enzymes, and ∼400 E3 ubiquitin ligases in plants (Richard, 2009). In general, the ubiquitination modification pathway includes three important factors; E1 ubiquitin-activating enzyme (UAE), E2 ubiquitin conjugating enzyme (UBC), and E3 ubiquitin ligases, including RING, U-box, HECT (homologous to the E6-AP carboxy terminus), and CRL (cullin-ring ubiquitin ligase) (Richard, 2009). Among them, E3 ubiquitin ligase is the most abundant, and it also influences ubiquitinated substrate diversity (Rennie et al., 2020). The numerous substrate-selective E3 ubiquitin ligases are mainly classified into RING-finger, HECT, and U-box structural domain classes (Mandal et al., 2018). Due to their specific non-spontaneous ubiquitination activity and their ability to participate in the degradation of unfolded or misfolded proteins upon activation by cofactors, U-box-like proteins are important, but they are complex and more difficult to control than other E3 ubiquitin ligases. Therefore, clarifying the unique contributions of different U-box proteins to different stress processes will help us to understand the onset of plant resistance (Li et al., 2017; Kim et al., 2021; Tang et al., 2021).
Most U-box gene family members contain U-box domains, and these main ligases control the conventional ubiquitination modification process that involves attaching ubiquitin to substrates, thereby altering multiple protein activities (Azevedo et al., 2001; Hu et al., 2018; Tong et al., 2021). Specifically, U-box type E3 obtains ubiquitin from E2 through the conserved U-box domain, through salt bridges, ion chelation, and non-covalent hydrogen bonding interactions, and passes it to target proteins. In previous studies, cotton PUBs and banana U-box genes were found to be widely responsive to abiotic stress (Hu et al., 2018; Lu et al., 2020). Examples include OsPUB67 that can enhance tolerance to drought stress by regulating ABA, thereby enhancing the capacity to scavenge reactive oxygen species (ROS) and stomatal closure capacity (Qin et al., 2020). AtPUB11 degrades AtLRR1 (LEUCINE RICH REPEAT PROTEIN 1) and Atkin7 (KINASE 7), and negatively regulates ABA-mediated drought tolerance in Arabidopsis thaliana (Chen X. X. et al., 2020). However, U-box genes have important value for plant resistance, and they have been identified in rice (Oryza sativa), tomato (Solanum lycopersicum), apple (Malus domestica), banana (Musa nana), soybean (Glycine max), Chinese cabbage (Brassica rapa), cotton (Gossypium hirsutum), and cabbage (Brassica oleracea) (Azevedo et al., 2001; Li et al., 2009; Wang et al., 2015, Wang J. et al., 2020; Wang K. L. et al., 2020; Hu et al., 2018, 2019; Sharma and Taganna, 2020). However, comprehensive genome-wide identification of U-box genes in sorghum has not been performed, and expression characteristics have not been analyzed.
In this study, 68 U-box genes were identified in sorghum, and phylogenetic relationships, conserved structures, gene structures, and collinearity were systematically analyzed by bioinformatics. In addition, tissue-specific and stress-related expression patterns were analyzed by RNA sequencing (RNA-seq) under GA, MeJA (methyl jasmonate), ABA, and PEG (polyethylene glycol) 6000 treatments, and the results were validated by real-time quantitative PCR (RT-qPCR). The findings provide a basis for further functional exploration of U-box genes in sorghum resistance breeding, and shed new light on the molecular evolution of the U-box gene family.
Sorghum (BTx623) seeds were obtained from the Key Laboratory of Agricultural Microbiology, College of Agriculture, Guizhou University, Guiyang, China. After surface disinfection, the sorghum BTx623 seeds were soaked in sterilized water for 24 h and then moisturized with gauze for 72 h. The germinated seeds (25°C) were planted in sterilized nutrient soil (PINDSTRUP, Denmark) and cultivated in a greenhouse with a relative humidity of 75% at 25/20°C under a 14 h light/10 h dark cycle. In the three-leaf stage, ABA (200 μM), 20% PEG 6000, GA (100 μM), and MeJA (100 mM) were sprayed onto the whole plants in seedlings separately (Townsley et al., 2013; Chai and Subudhi, 2016; Yimer et al., 2018; Miao et al., 2019; Zeng et al., 2019). Sorghum leaves were collected at 0, 3, 6, 9, and 12 h after treatment. All samples included three biological replicates, with four seedlings treated per replicate. All samples were immediately frozen in liquid nitrogen after sampling and stored at −80°C until RNA extraction (Wu et al., 2019).
Total RNA was extracted using TRIzol reagent (Thermo Fisher, United States), and the cDNA was synthesized using HiScript III RT SuperMix (Vazyme Biotech, Beijing, China) (Wu et al., 2019). Real-time fluorescent quantitative PCR (RT-qPCR) was used to determine gene expression patterns. The reaction system was composed of 4.5 μL of cDNA, 7.5 μL of SYBR mix, 0.3 μL of each primer, and 15 μL of ddH2O. Thermal cycling included 40 cycles at 95°C for 5 min, 95°C for 10 s, and 58°C for 30 s, and one cycle at 95°C for 10 s, 58°C for 60 s, and 95°C for 10 s. Three biological replicates were performed, and SbEIF4a was used as an internal reference gene (Yang et al., 2020; Du et al., 2021; Jin et al., 2021). RT-qPCR data were analyzed using the 2–Δ Δ Ct method (Pfaffl, 2001), and Duncan’s new multiple range test (SPSS software) was used for significance analysis.
To identify members of the sorghum U-box gene family, the basic conserved domain (U-box: PF04564) was searched against EnsemblPlant1 using HMMER 3.6.1 software (E value limited to 0.05, other parameters remained as default values) (Potter et al., 2018), and domain confirmation was carried out using PFAM and SMART (Letunic and Bork, 2018; El-Gebali et al., 2019). Conserved motifs were predicted using MEME2 and 20 motifs were explored (E value limited to 0.05, other parameters remained as default values). Chromosome distribution, conserved motifs, and gene structure visualization were carried out using TBtools 1.0 (Chen C. et al., 2020). Analysis of physicochemical properties was performed by ExPASy3 (Bailey et al., 2009; Artimo et al., 2012; Chen C. et al., 2020). Gene naming uses a combination of homology and sequence naming. The PUB, CHIP, and UFD genes among U-box members were first identified through homology, then genes were named sequentially based on chromosome positioning.
To explore the evolutionary relationships and subcategories of U-box genes between sorghum and arabidopsis, MEGA 7.0 software (Kumar et al., 2016) was used to construct homologous protein rootless evolutionary trees via the neighbor-joining (NJ) method with 1,000 bootstrap replicates, followed by ITOL.4 Selection of tandem repeat gene pairs in sorghum was carried out with (a) short sequences covering 75% and (b) aligned regions of longer genes with 75% similarity (Gu et al., 2002). Whole-genome replication examination was performed by MCScan X (Wang et al., 2013). OrthoVenn25 was used to query homologous genes in different plants (Xu et al., 2019).
AgriGO6 (Tian et al., 2017) and AIpuFu7 were employed to investigate the main functions and Gene Ontology (GO) annotation features of sorghum U-box genes. To explore the promoter and expression characteristics of sorghum U-box genes, putative promoter sequences were extracted (sequences 2 kb before the 5′ end of the sequence) using TBtools1.0 (Chen C. et al., 2020). Cis-acting elements were predicted by PlantCARE8 and plotted by TBtools1.0 (Lescot et al., 2002; Chen et al., 2020).
To analyze the expression profiling of sorghum U-box genes in different tissues, and under drought stress and osmotic stress, an FPKM (fragments per kilobase of exon per million fragments mapped) expression matrix provided by the Sorghum functional database (Supplementary Table 5) (Tian et al., 2016) and SRA9 (PRJDB1973 and PRJDB1973) was extracted and mapped by TBtools1.0 (Chen C. et al., 2020).
Arabidopsis FPKM data were downloaded from the Expression Atlas website10 for SAMN01041946 and SAMN02440041. Based on the previously obtained expression values in FPKM format for arabidopsis and sorghum U-box genes, Pearson correlation coefficients (PCC) and p-values were calculated to obtain the expression levels of U-box genes by the Pearson method using R software. Correlation heat maps were generated by the corrplot tool in R software (version 0.84). Gene co-regulatory networks were constructed by Cytoscape version 3.7.1 (Shannon et al., 2003) based on the PCCs of U-box gene pairs with a p-value ≤ 0.05 (Zhao et al., 2019).
In this study, a total of 68 U-box genes with complete U-box domains, including a typical ubiquitin fusion degradation 2 (UFD2) gene, two carboxyl terminus of heat shock protein 70-interacting protein (CHIP) genes, and 65 plant U-box (PUB) genes were identified in sorghum genome by using hidden Markov model (HMMER) method (Table 1; Potter et al., 2018).
Sorghum U-box genes were distributed in all chromosomes except chromosome 5, with CHIP genes on chromosomes 7 and 9, and the UFD2 gene on chromosome 1. The number of amino acids per protein ranges from 400 to 1,500, and the relative molecular weight of Sorghum PUB proteins is 40–160 kDa. Sorghum PUB5, PUB17, PUB33, PUB36, PUB46, and PUB49 are among the largest, with relative molecular weights >100 kDa. By contrast, the Sorghum CHIP proteins are relatively small with the protein weight between 30 and 40 kDa. The isoelectric point (pI) ranges from 4.99 to 9.65, from medium strong acid to medium strong base. SbPUB26 has the highest pI and SbPUB3/7 have the lowest pI. UFD2 and CHIP proteins have pI values in the weakly acidic range (Table 1). These indicated that sorghum U-box proteins are divergent in protein characteristics.
Based on the phylogenetic relationships and domain composition of A. thaliana and sorghum U-box genes, they can be divided into eight categories; U-box only, Kinase + U-box, U-box + WD40-1, U-box + WD40-2, U-box + Armadillo 1 (ARM-1), U-box + ARM-2, U-box + Tetratricopeptide repeat (TPR) or CHIP, and U-box + Domain With No Name (DWNN) or UFD2 (Figure 1 and Supplementary Table 1).
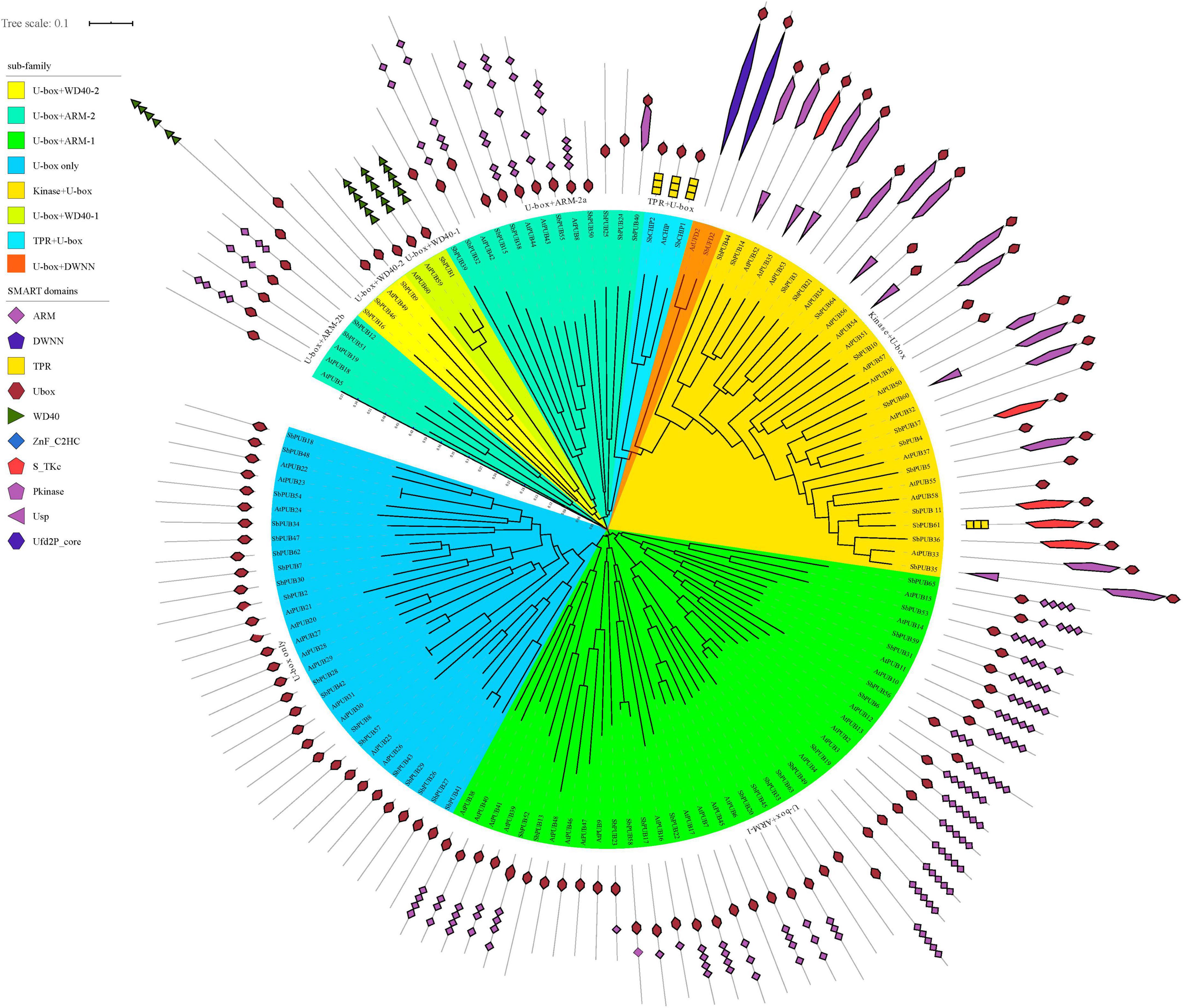
Figure 1. Phylogenetic tree and domain analysis of U-box genes in Sorghum and Arabidopsis. Phylogenetic relationships between U-box genes in Sorghum bicolor and Arabidopsis thaliana were explored. Rootless trees were generated by the neighbor-joining (NJ) method using MEGA7 software. Numbers next to branches indicate 1,000 bootstrap replicates as a percentage. The phylogenetic classification of U-box genes is marked with corresponding colors and different shapes. Colors in the outer ring represent different domains, as shown in the color legend in the upper left corner. U-box, Ub fusion degradation 2 (UFD2) domain; PUB, plant U-box; UFD, Ub fusion degradation; CHIP, carboxyl terminus of heat shock protein 70-interacting protein; At, Arabidopsis thaliana; Sb, Sorghum bicolor.
The sorghum U-box gene family includes an extra CHIP gene and six PUB genes than that in arabidopsis. Most sorghum U-box genes share one ortholog in arabidopsis, as exemplified by AtCHIP/SbCHIP1, SbUFD2/AtUFD2, and SbPUB49/AtPUB9, indicating similar evolutionary pathways (Supplementary Table 2). However, some gene homology branches such as SbPUB16/SbPUB46, SbPUB1/SbPUB12, and AtPUB18/AtPUB19 appear to reflect divergence shortly after the separation in these two species. Within the same category, a large number of structural changes are evident; for example, the Kinase + U-box within the SbPUB61 branch divides into the CHIP branch containing genes with a TPR domain structure, and multiple branches only contain proteins with a U-box domain. Some homologous genes appear to lack intact structural domains, such as U-box + ARM-1 within the SbPUB13 branch that contain only a U-box domain but no ARM structural domain (Figure 1). These indicated that sorghum U-box genes are divergent in structural domains, showing complex evolutionary pathway.
As shown in Figure 1, the domains of CHIP and UFD2 proteins are clearly distinguished, and the motifs of CHIP and UFD2 are also significantly different (Figures 2A,B). CHIP proteins contain at least two motifs (1/2). The UFD2 protein consists of two motifs (1). For PUBs, the distribution of their motifs is not completely conserved. Specifically, U-box + WD40-1 and U-box + WD40-2 branch proteins share the lowest similarity with PUB branch proteins with core motifs 14. Interestingly, the U-box + WD40-2 branch is classified into two categories: a typical branch SbPUB49/AtPUB9, and the sorghum-specific branch SbPUB16/SbPUB46. The U-box + ARM branch is divided to three groups: U-box + ARM1 (motifs 1/2/5/7), U-box + ARM2a (motifs 1/2/3), and U-box + ARM2b (motifs 1/2/3). Finally, the Kinase + U-box and U-box only branches include one core motifs (1) and four core motifs (1/2/4/10), respectively (Figures 2A,B).
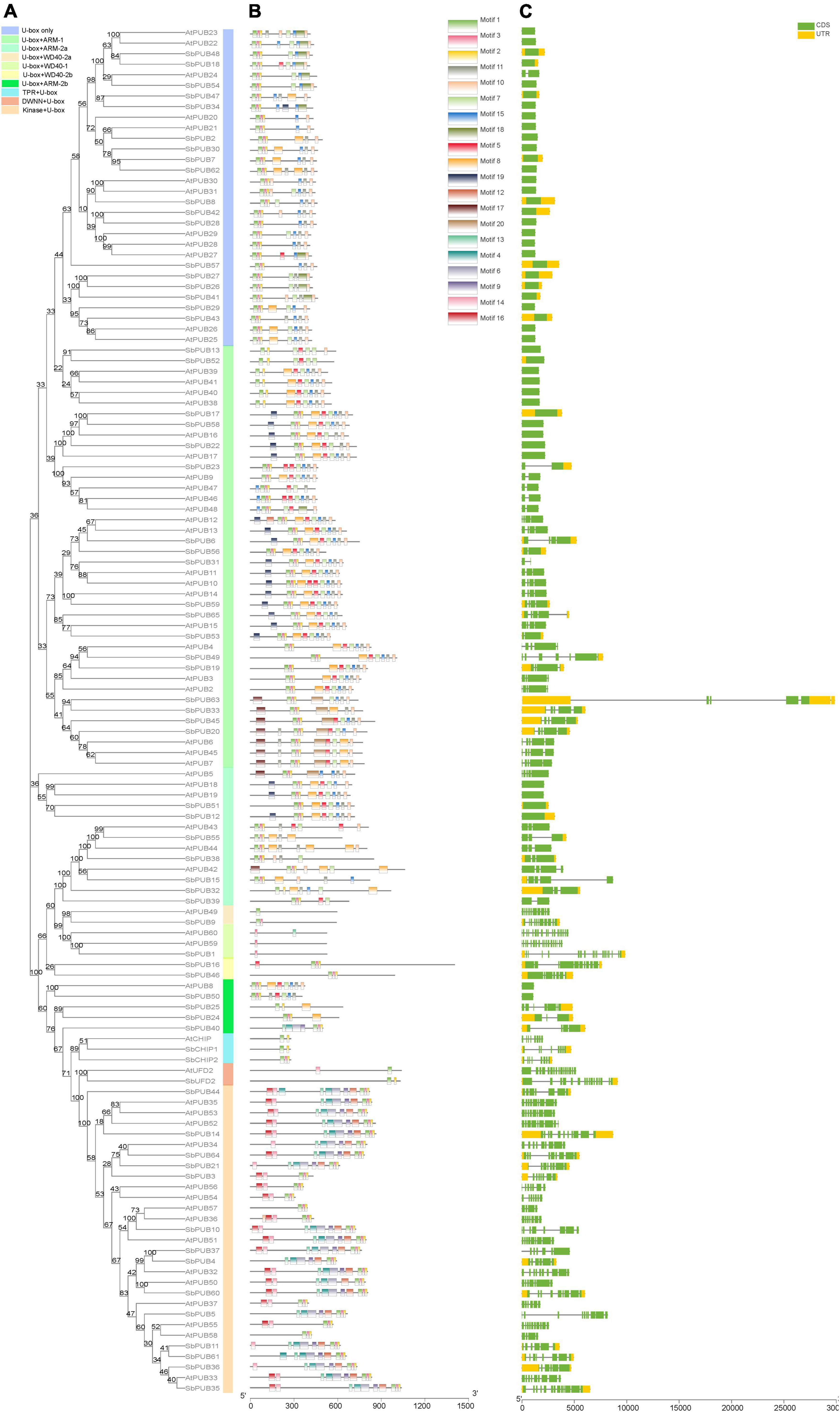
Figure 2. Motif and structural analysis of U-box gene family members in Sorghum and Arabidopsis. (A) Phylogenetic relationships between U-box genes of Sorghum bicolor and Arabidopsis thaliana. Rootless trees were generated by the neighbor-joining (NJ) method in MEGA7 software. Numbers next to branches indicate 1,000 bootstrap replicates as a percentage. Different background colors represent different subfamilies. (B) Motifs in U-box genes. Different colored rectangles represent 30 different motifs, as shown in the color legend in the upper right corner. (C) Structural analysis of U-box genes. Green and yellow indicate coding sequence (CDS) and untranslated region (UTR) features, respectively, and the horizontal line represents gene regions, as shown in the color legend in the upper right corner.
In terms of gene structure, both CHIP and UFD2 genes include a complex gene structure with >6 introns (Figures 1, 2C). Regarding the gene structure of PUB genes, some maintain low complexity, as exemplified by SbPUB6 and SbPUB23 that contain 1–5 introns, while SbPUB1 and SbPUB9 have high complexity (>10 introns). It is worth noting that all gene groups with a single U-box domain have a simple gene structure, mostly with 0 or 1 intron. In the groups containing both U-box and ARM domains, the corresponding gene structure is complex than that of the single U-box groups, and the number of introns varies between 0 and 5. In addition, the groups with multi-domains in the U-box gene (Kinase + U-box, U-box + WD40, U-box + TPR, and U-box + DWNN) have the most complex gene structure (>10 introns; Figures 2A,C).
In order to understand the chromosomal distribution and replication pattern of sorghum U-box genes, gene mapping and tandem duplication were analyzed to explore the diversification of U-box genes. Ten (containing a typical UFD2 gene), 6, 5, 18, 0, 10, 6, and 3 (with 1 CHIP1), 3 (with 1 CHIP2), and 7 U-box genes were found to be distributed on chromosomes 1–10, respectively. Most PUB genes are distributed in the same cluster, but the two CHIP genes are not adjacent to each other, and are relatively distant from the PUB genes (Figure 3 and Table 1). Interestingly, UFD2 genes are located in the low-density distribution region, and CHIP genes are present in the region with high gene density. The CHIP gene is near the start of the chromosome, while UFD2 is in a region near the end of the chromosome. Most PUB genes also form large gene clusters near the end of chromosomes (Figure 3). Additionally, chromosome 4 includes 18 PUB genes, most of which form gene cluster at the end of this chromosome (Figure 3).
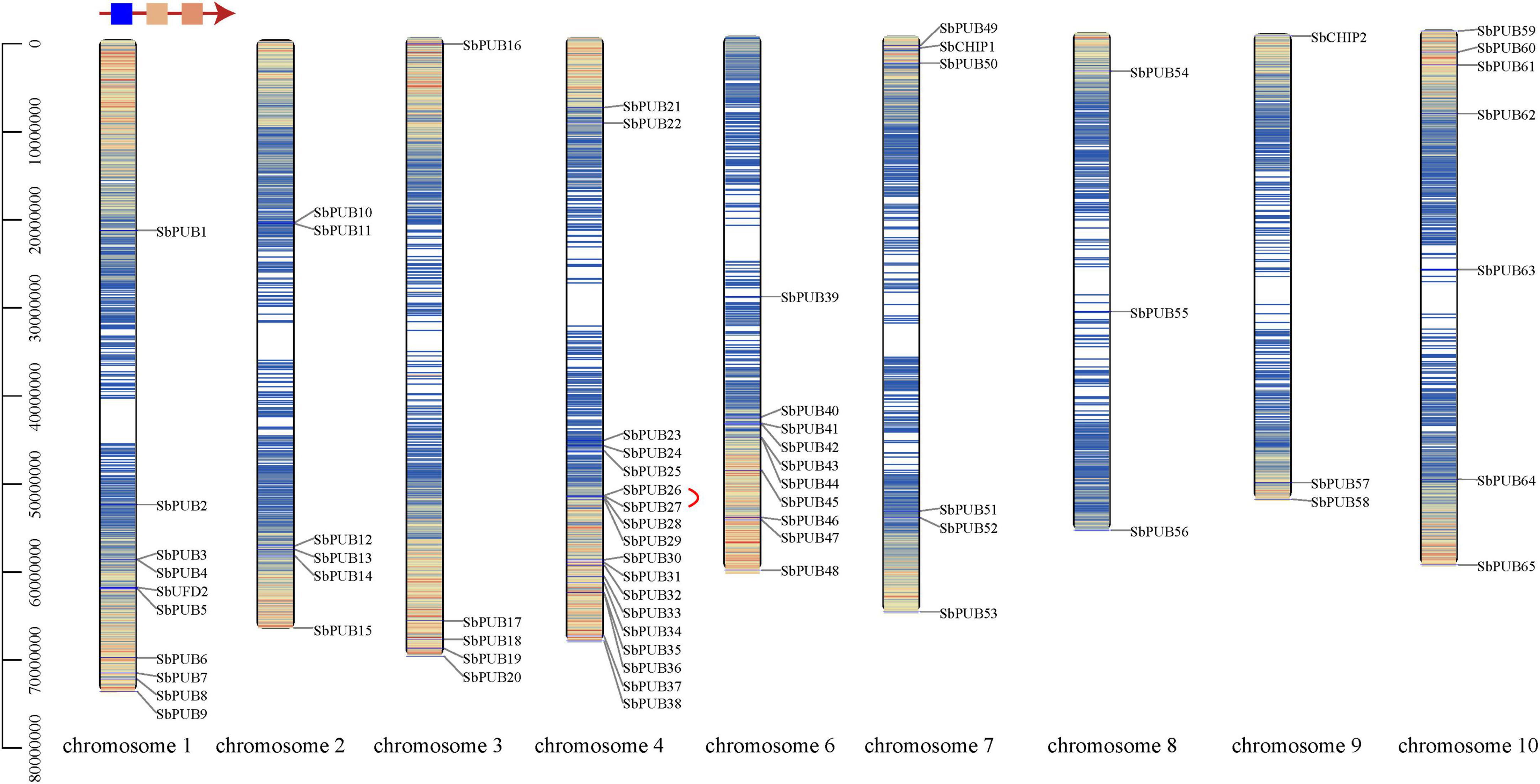
Figure 3. Chromosome location of U-box genes in Sorghum. The different colors of chromosomes represent gene density in this range, and variation is indicated in the upper left corner. Red lines represent tandem repeats.
Based on a cut-off of 75% sequence similarity and coverage (Potter et al., 2018), only one tandem duplicate gene pair was found, the SbPUB26/SbPUB27 gene pair, located in a region of low gene density at the end of chromosome 4 (Figure 3). Gene collinearity results showed that there are 14 collinear relationships between S. bicolor and A. thaliana U-box genes. Among them, chromosome 1 and chromosome 4 contain the largest number of genes, and the highest collinearity (Figure 4A). However, collinearity between rice, maize, and sorghum U-box genes was high, with 66 and 82 showing collinearity, respectively. Among them, in sorghum, chromosomes 4 and 6 include the highest collinearity gene distribution, compared with chromosome 2 in rice and chromosomes 3 and 5 in maize (Figures 4B,C). In particular, neither rice chromosome 11 nor sorghum chromosome 5 showed any collinearity of U-box genes. Furthermore, collinearity was observed for all types of U-box genes (49 sorghum U-box genes), but no collinearity was found for CHIP gene types between S. bicolor and A. thaliana (Figure 4 and Supplementary Table 3).
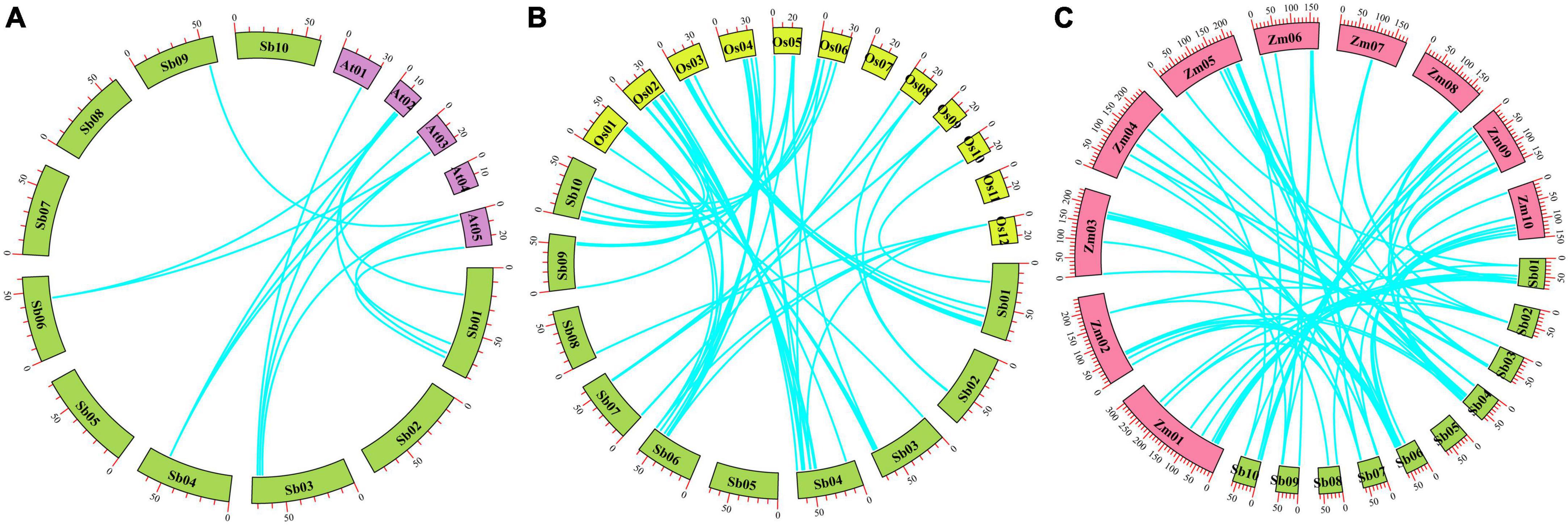
Figure 4. Collinear block identification of sorghum and Arabidopsis, rice, and maize U-box genes. (A) Collinearity relationships between U-box genes in sorghum and Arabidopsis thaliana; (B) collinear relationships between U-box genes in sorghum and rice; (C) collinearity relationships between U-box genes in sorghum and maize.
Numerous tissue-specific elements, environmental response elements, and hormone response elements were identified in sorghum U-box gene promoters. The promoter of SbUFD2 contains two light response, three hormone response, and four environment response elements. The SbCHIP promoter contains drought response elements. Most SbPUB gene promoters are rich in light response elements, followed by hormone response elements such as MeJA, GA, ABA, and drought, and they also contain the environmental response or tissue-specific expression elements. Some gene promoters also include injury response elements, such as SbPUB16 and SbPUB62 (Figure 5).
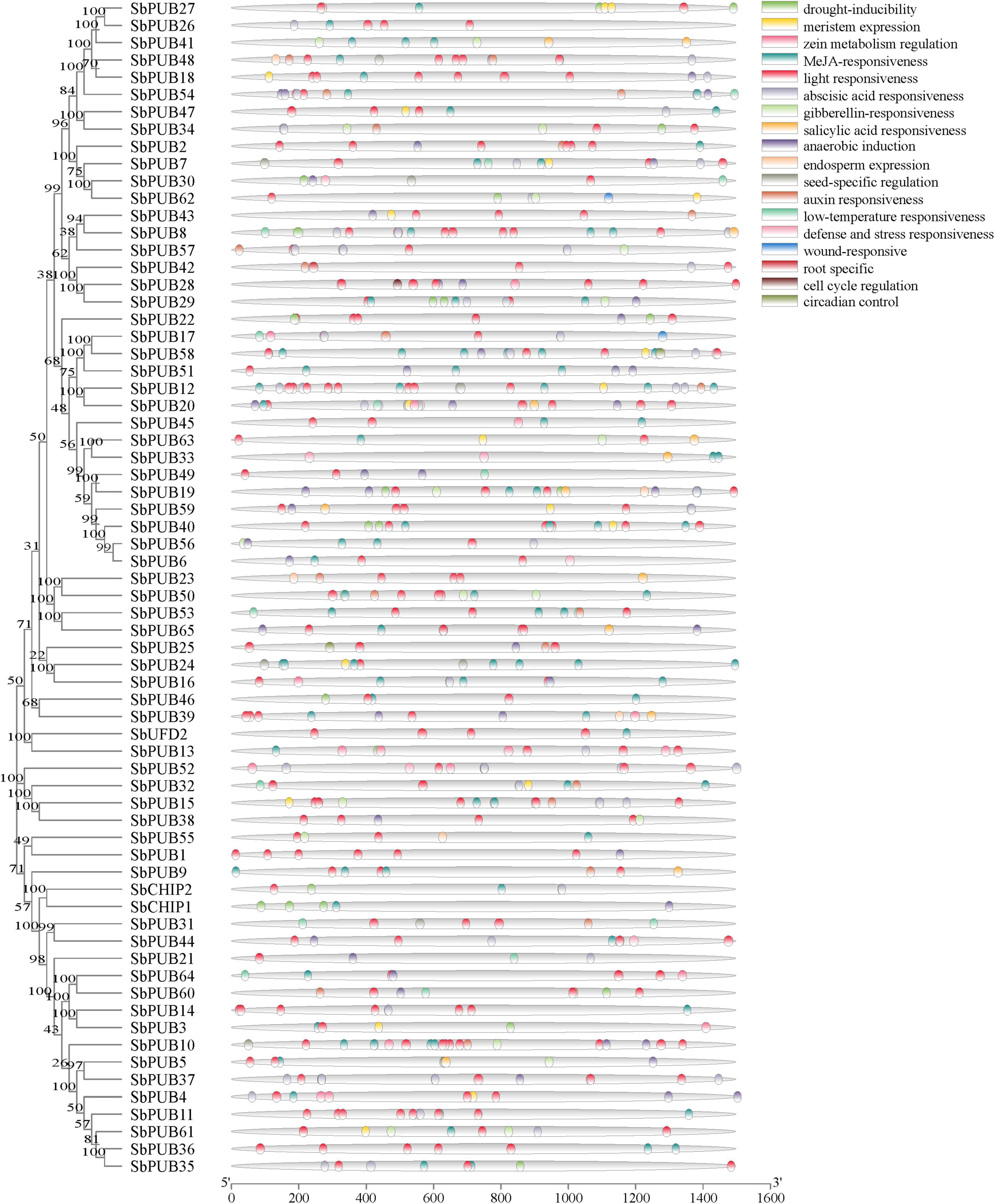
Figure 5. Promoter analysis of sorghum U-box genes. Rootless trees were generated by the neighbor-joining (NJ) method using MEGA7 software. Numbers next to branches indicate 1,000 bootstrap replicates as a percentage. Different promoters are indicated by rectangles of different colors to represent homeopathic elements. Detailed comments are included in the top right panel.
In particular, MeJA or GA response elements occur repeatedly in PUB genes. For example, MeJA response element occurred three times in SbPUB7, SbPUB8, SbPUB9, SbPUB11, SbPUB15, SbPUB18, SbPUB23, SbPUB28, SbPUB29, SbPUB34, SbPUB39, SbPUB41, SbPUB50, SbPUB51, SbPUB53, SbPUB58, and SbPUB61 promoters. Additionally, ABA or drought response elements were also found in SbPUB10, SbPUB11, SbPUB12, SbPUB14, SbPUB19, SbPUB28, SbPUB36, SbPUB52, SbPUB57, SbPUB58, SbPUB65, and SbPUB7 in the sorghum genome when searching the EnsemblPlant database. Collectively, in the promoters of SbPUB11, SbPUB28, SbPUB58, and SbPUB7, three ABA and MeJA response elements were simultaneously detected, indicating that these genes may be involved in responses to stress related to JA and ABA (Figure 5).
In addition, GO enrichment analysis showed that U-box proteins were mainly related to ubiquitination and modification in cells, followed by phosphorylation. Sixty-two genes were enriched in ubiquitination and metabolism (protein metabolic, primary metabolic, macromolecule metabolic, and cellular metabolic processes) and ligase activity, and 13 genes were enriched in phosphorylation. Six genes were enriched in response to stress and stimuli (Figure 6 and Supplementary Table 4).
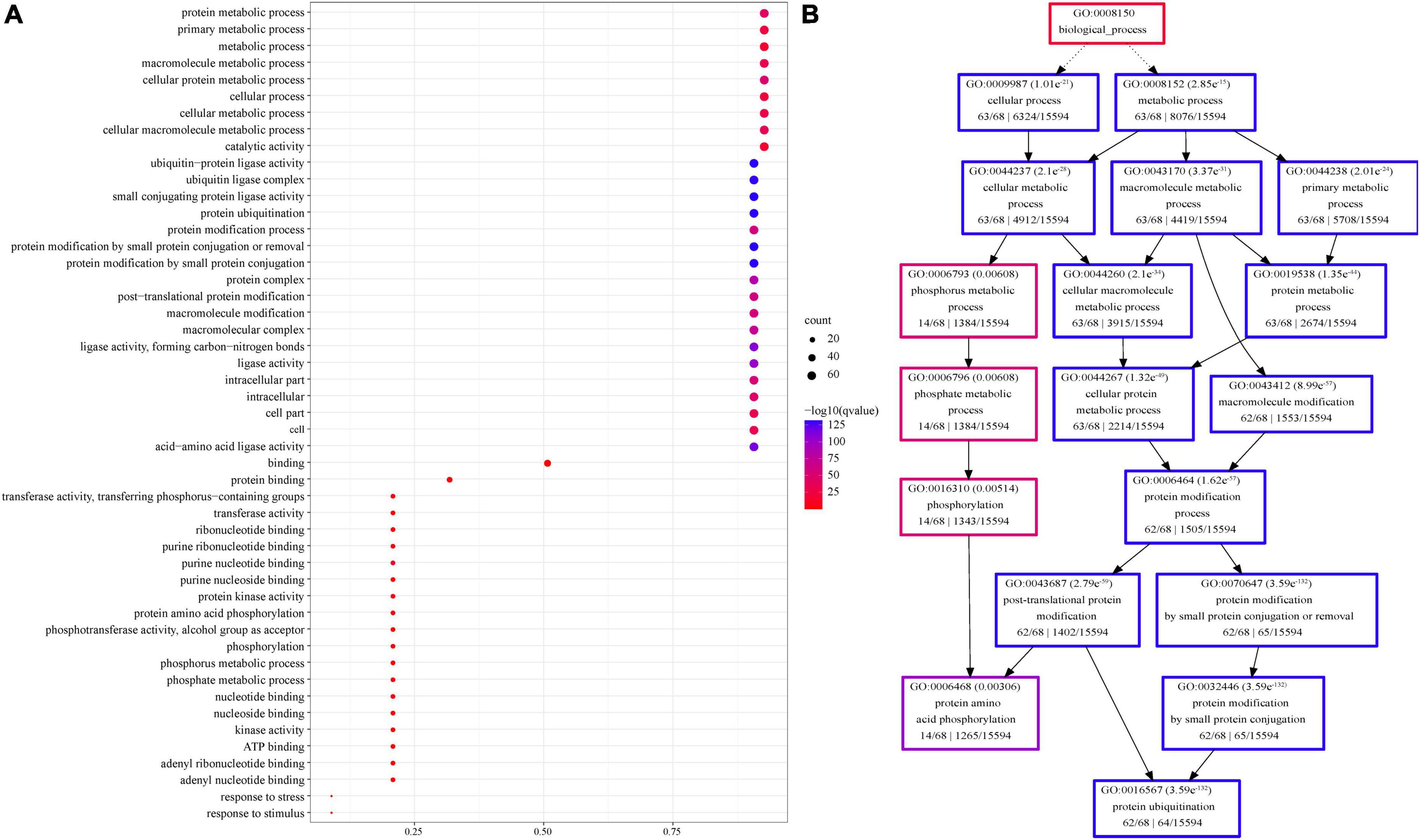
Figure 6. Gene Ontology enrichment of sorghum U-box members. (A) GO enrichment presented as dot bubbles. (B) Corresponding pathway diagram of enriched GO terms. GO enrichment was conducted by Agrigo, and corrected p-values were calculated by R version 3.6.2.
In order to explore the expression characteristics of U-box genes in sorghum, the expression level data were searched against the sorghum database. As shown in Figure 7, expression levels of SbPUBs in drought and osmotic responses were significantly different. In tissue-specific expression profiling, most SbPUB genes displayed tissue and organ expression specificity, especially in roots, meristems, embryos and other tissues associated with vigorous growth. The expression levels of SbPUBs and SbCHIPs were higher in most tissues, while those of UFD2 genes were slightly increased in roots. The expression patterns of PUB genes could be divided into two categories; one group with low expression in the roots, meristems, embryos and other tissues (including the 24 PUB genes PUB1/6/8/9/17/19/22/33/44/49/50/56/61/63/64), and the other group was highly expressed in the roots, meristems, embryos and other tissues (including 41 PUB genes). The first group of PUB genes could be divided into three categories; Stem-specific expression enrichment (PUB8/57/60), root specific expression enrichment (PUB17/20/44/50/55/59), and flower and meristem specific expression enrichment (PUB1/6/9/22/23/33/40/56/63/64). The second group of PUB genes could also be divided into three categories; Root-specific expression enrichment (PUB3/4/11/37/47/48), floral and meristem specific expression enrichment (PUB5/14/45/53/58/65), and no significant expression (another 29 U-box genes; Figure 7A and Supplementary Table 5).
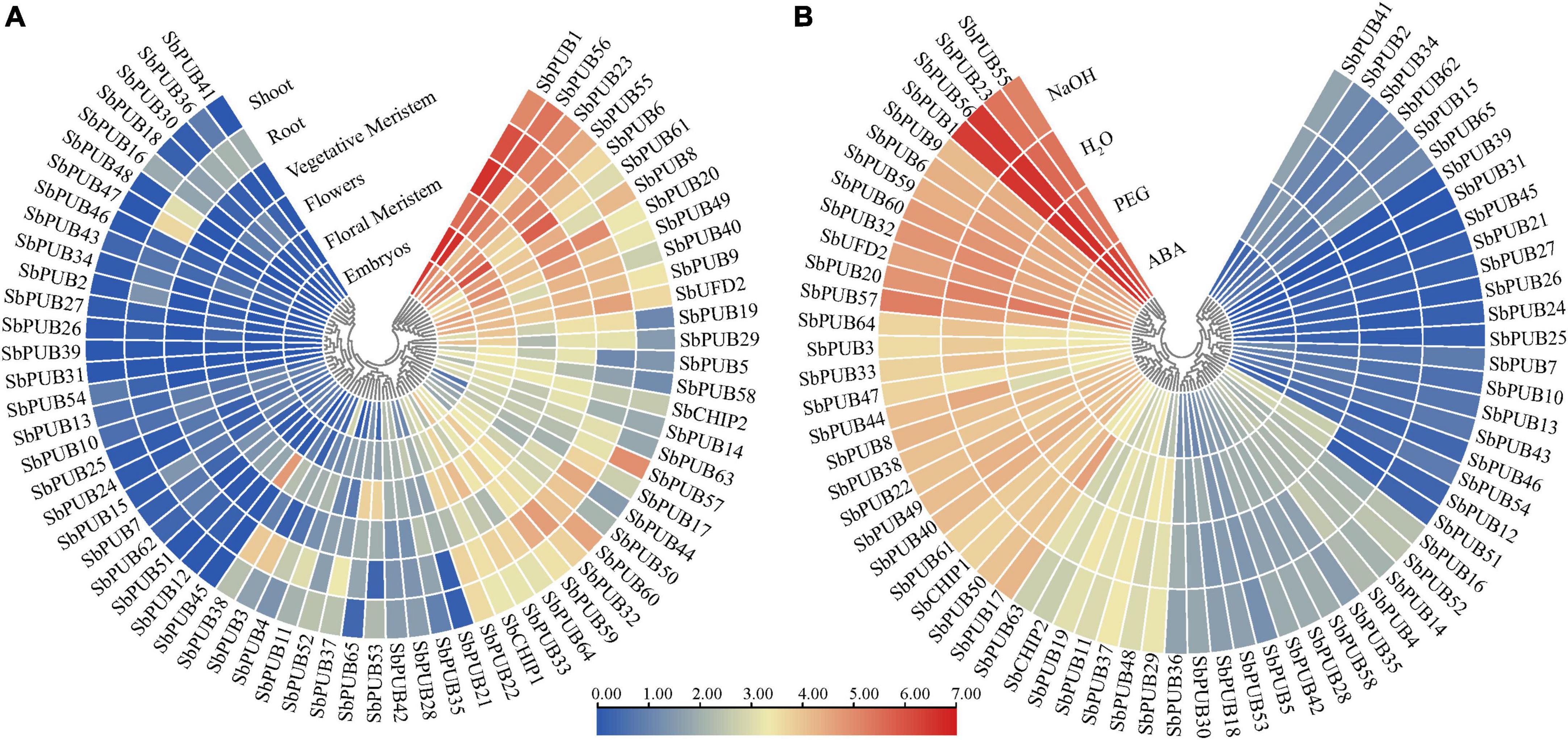
Figure 7. Heatmap of tissue-specific expression and abiotic stress expression of sorghum U-box genes. (A) Expression of U-box genes in different tissues. (B) Expression of U-box genes under the influence of NaOH, ABA, and PEG. The color bar represents the log2 expression level of each gene (FPKM, fragments per kilobase of exon per million fragments mapped). Color bar annotation is included at the top of the image. The heatmap is colored according to expression values, with blue, yellow, and red representing at low, medium, and high transcription abundance, respectively.
Regarding stress-related expression, SbPUB genes exhibited different expression characteristics when treated with PEG, ABA, and NaOH, distributing into three main types. Type I in which expression was inhibited (ABA response inhibition = PUB2/15/21/27/29/30/34/36/41/44/46/48/50/54/57/60/64/65, PEG response inhibition = PUB15/41/44/65, NaOH response inhibition = PUB65). Type II in which expression was promoted (ABA response upregulation = PUB7/10/12/23/51, PEG response upregulation = PUB2/12/13/17/18/21/24/25/26/34/35/39/46/54/62, NaOH response upregulation = PUB12/51/62), and type III with no significant expression (other U-box genes; Figure 7B and Supplementary Table 5).
Based on the FPKM method (Tian et al., 2016), the PCC of U-box gene expression levels in S. bicolor and A. thaliana was calculated, and a co-regulatory network was constructed (Zhao et al., 2019). The results showed that ∼50% U-box genes were positively correlated. Additionally, the same gene type was mostly correlated, similar to those of A. thaliana (Figure 8A, Supplementary Figure 1, and Supplementary Table 6).
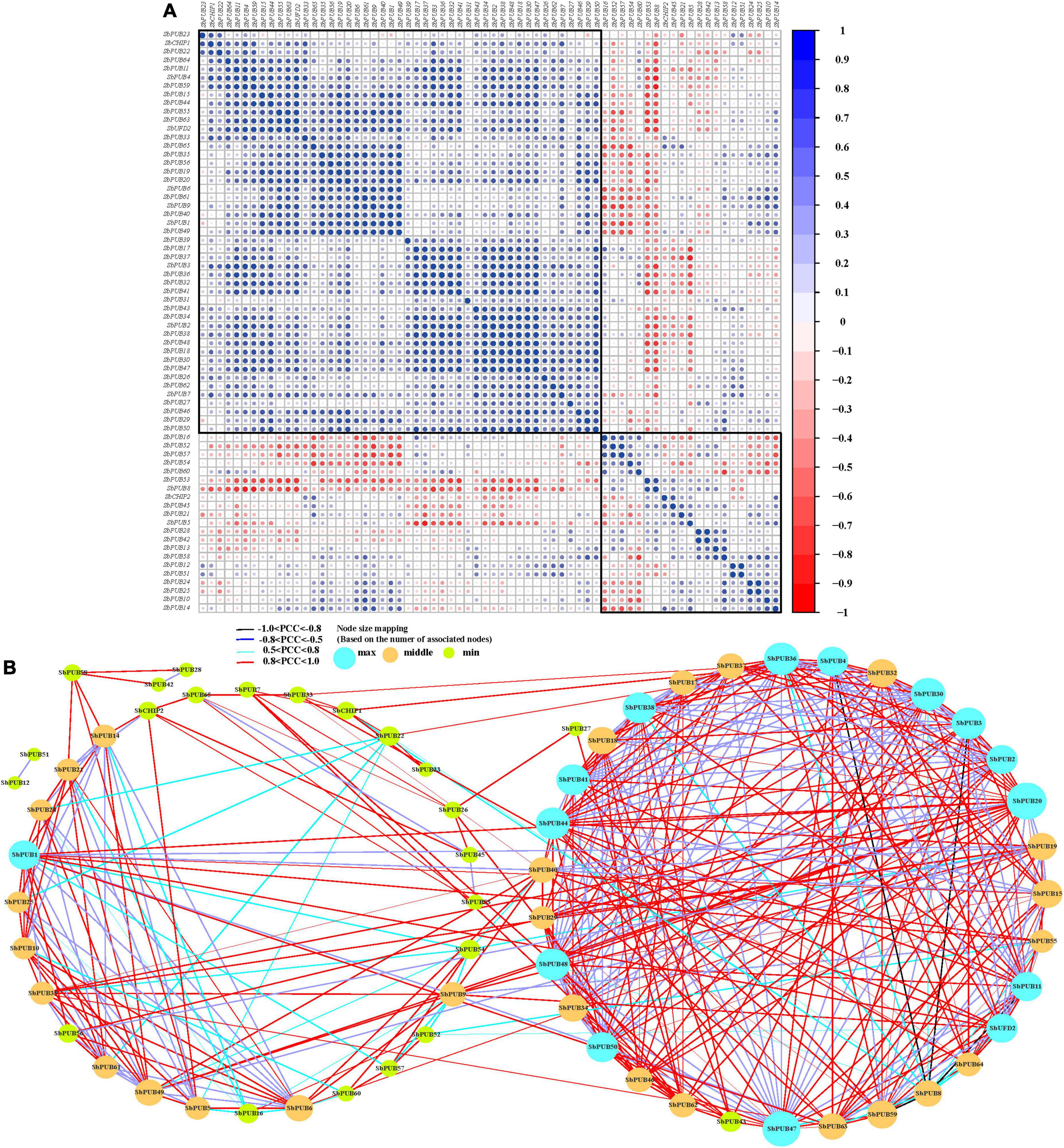
Figure 8. Correlation and co-regulation network of sorghum U-box genes. (A) Correlation analysis of U-box genes conducted by Pearson correlation coefficients (PCCs). Correlations are represented by the size and color of the circles. Heatmap labels indicate PCC values. (B) Co-regulatory network of sorghum U-box genes generated based on gene pairs with significant PCCs (p-value ≤ 0.05). The strength of correlations between gene pairs is marked at the bottom using different colored lines.
All significant PCCs (p-value ≤ 1e−5 and PCC >0.5) of U-box genes were extracted and Cytoscape was used to construct a co-regulation network. The co-regulation network of S. bicolor consists of 65 nodes and 406 edges (not including SbPUB13/SbPUB31/SbPUB39; Figure 8B). The results showed that the U-box genes in S. bicolor and A. thaliana were mainly positively correlated (1 > PCC > 0). Specifically, there were 123 strong positive correlations (0.8 < PCC < 1) and four strong negative correlations (−1 < PCC ≤ 0.8) in sorghum: SbPUB3/SbPUB8, SbPUB4/SbPUB8, SbPUB11/SbPUB8, and SbPUB57/SbPUB8 (Figure 8). Meanwhile, there were 110 strong positive associations but no negative associations in A. thaliana (Supplementary Figure 1). In addition, there were weak negative correlations (−0.8 < PCC < 0.5) between S. bicolor and A. thaliana U-box genes (25.37 and 20.86%, respectively), and a dominant weak positive correlation (0.5 < PCC < 0.8) accounting for 45.4 and 43.35%, respectively (Supplementary Table 6).
To explore the key roles of the U-box gene family in the resistance process in sorghum, RT-qPCR was employed to assess the impact of plant in stress conditions. Figure 5 shows that drought response elements and ABA response elements were enriched in some genes, indicating that they may be stress tolerance genes (for those with >3 ABA response elements and drought response elements). Meanwhile, Figure 6 shows that many sorghum U-box genes exhibited obvious responses to ABA and PEG. Overall, these sets of results suggest that these genes may have important function for sorghum drought stress tolerance.
Seven U-box genes (SbPUB7/17/18/21/25/26/62) were characterized by high responses (>1.5-fold variation in expression) and abundant elements (>3 ABA and drought elements). Therefore, we quantitatively assessed these potential drought-regulated U-box genes. The results showed that the expression levels of these PUB genes were also significantly different according to RT-qPCR determination. Specifically, SbPUB7 was significantly downregulated after ABA treatment. Expression of SbPUB17 was significantly upregulated after ABA and PEG treatment, and the highest expression value was reached at 6 h. For SbPUB18, expression was significantly downregulated (6 h) after ABA treatment, but rapidly upregulated under PEG treatment. Similarly, SbPUB21 was significantly downregulated by ABA treatment, but significantly upregulated by PEG treatment. After ABA treatment, expression of SbPUB25 was first significantly upregulated then significantly downregulated to below the initial level, while expression of SbPUB25 was significantly upregulated following PEG treatment at 6 h. Expression of SbPUB26 was significantly downregulated after ABA treatment, and the expression level was significantly decreased with the extension of ABA treatment time, whereas expression of SbPUB26 was significantly upregulated under PEG treatment. SbPUB62 was significantly downregulated following ABA treatment at 6 and 12 h, and following PEG treatment at 3 h (Figure 9). These results indicate that there were differences in the response patterns of U-box genes to drought stress in sorghum, and there were significant differences in the response patterns of U-box genes to physiological drought induced by different conditions in sorghum. Most sorghum U-box genes were sensitive to ABA-induced physiological drought, but they were also linked to resistance to PEG-induced physiological drought. Interestingly, SbPUB17 was upregulated under drought stress, while SbPUB62 was downregulated.
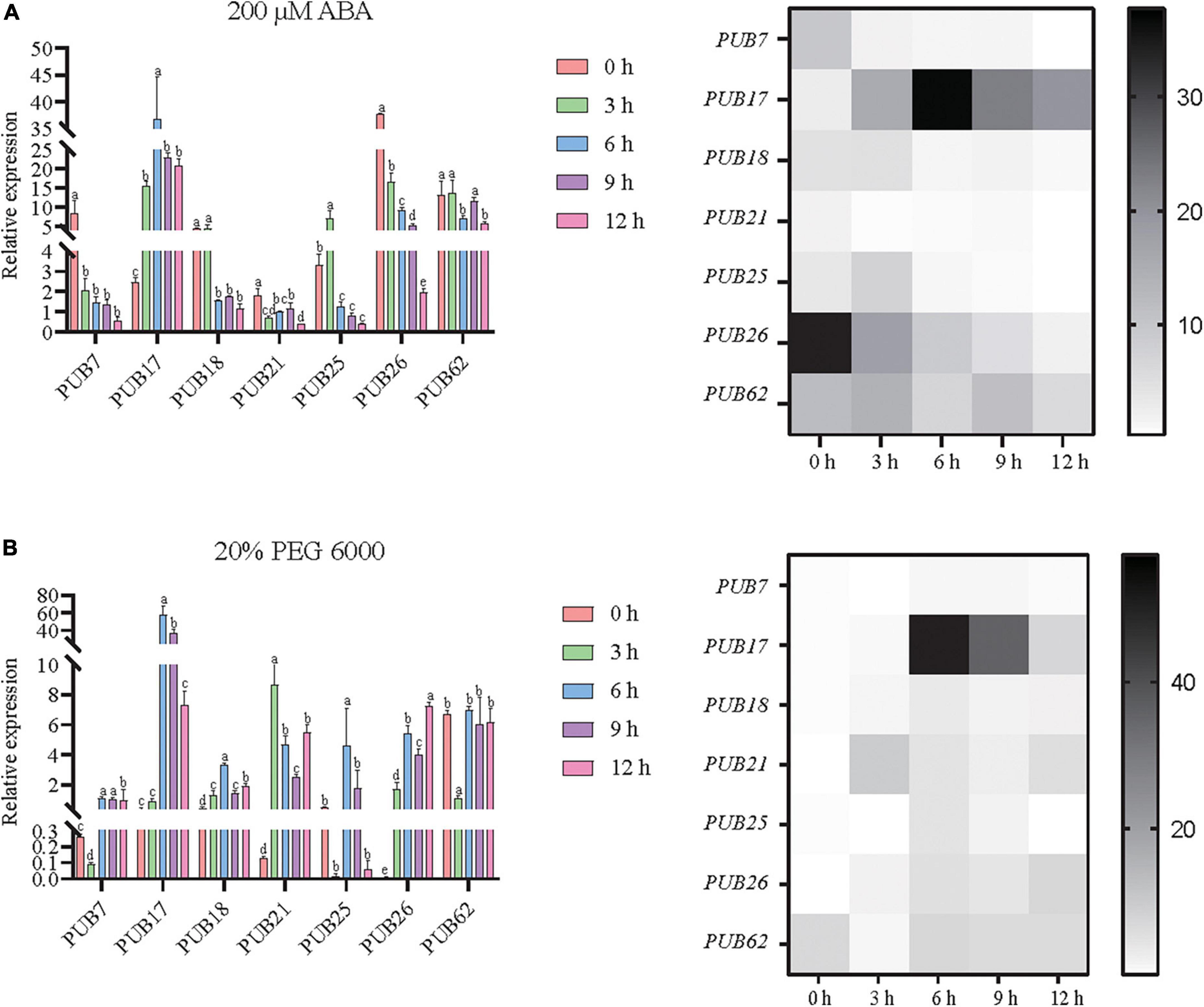
Figure 9. Potential responses of seven sorghum U-box genes to drought. All seven selected sorghum PUB genes (SbPUB7, SbPUB17, SbPUB18, SbPUB21, SbPUB25, SbPUB26, and SbPUB62) showed obvious expression differences under different conditions in heatmaps, and contain numerous drought elements. (A) Expression patterns of these genes in response to ABA-induced physiological drought. (B) Expression patterns of these genes in response to physiological drought induced by PEG 6000. Different lowercase letters indicate a significant difference determined by the Duncans new multiple range test (P-value < 0.05).
As shown in Figure 5, the promoter regions of these PUB genes were rich in hormone response elements, especially those of JA and GA. In order to explore the response characteristics of sorghum PUBs under JA and GA, we further studied the previously selected seven genes by RT-qPCR. The results showed that all seven genes were upregulated in response to MeJA and GA3 treatment over a period (Figure 10).
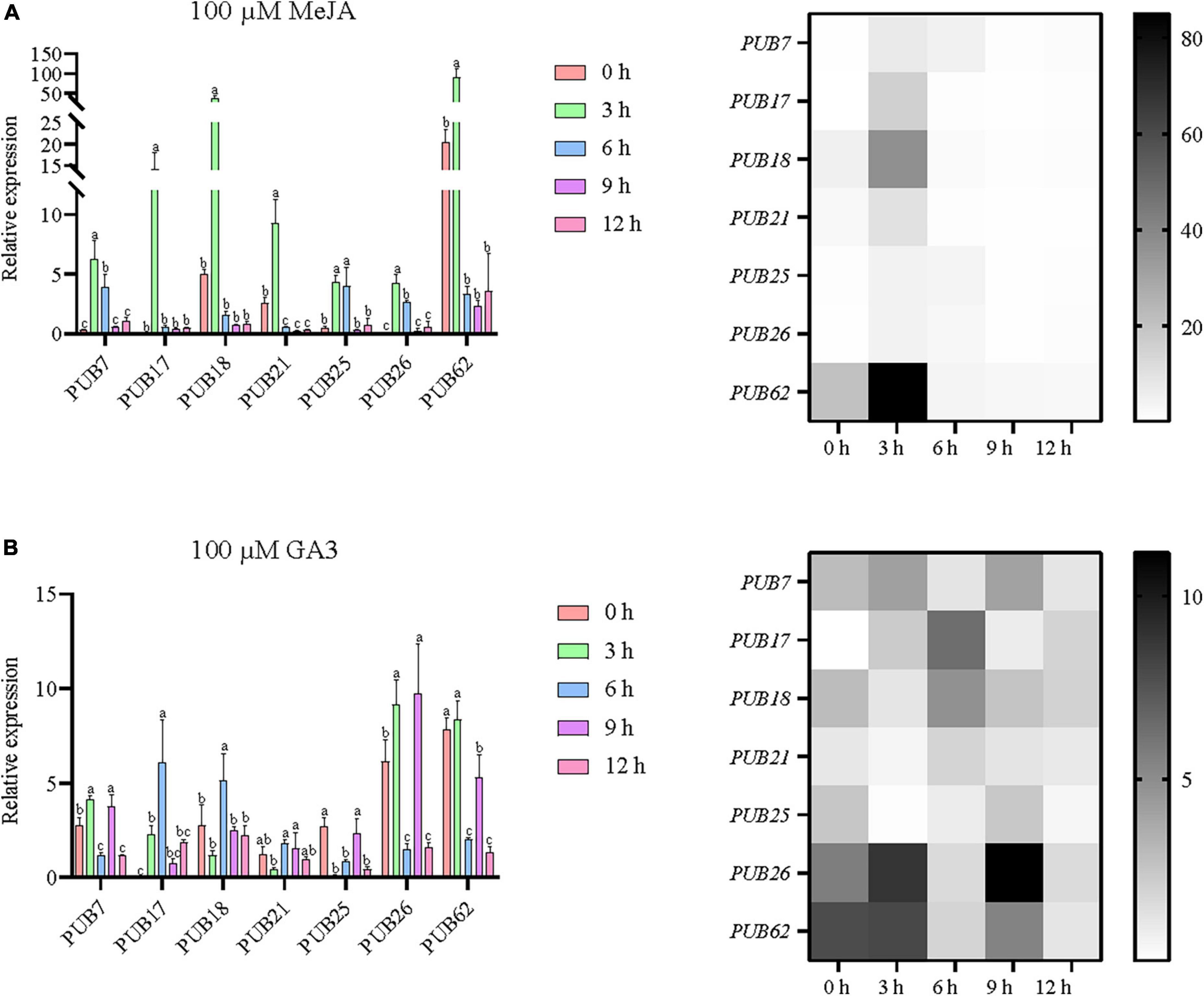
Figure 10. Potential responses of the seven sorghum U-box genes to methyl jasmonate (A) and gibberellin (B). All seven selected sorghum PUB genes (SbPUB7, SbPUB17, SbPUB18, SbPUB21, SbPUB25, SbPUB26, and SbPUB62) show obvious expression differences under different conditions in heatmaps, and contain numerous JA/GA3 response elements. Different lowercase letters indicate a significant difference determined by the Duncans new multiple range test (P-value < 0.05).
Under MeJA treatment, expression of all seven sorghum PUBs was upregulated rapidly, and the expression level reached a peak at 3 h. Under longer treatment, expression of these genes dropped to initial levels or below. Specifically, SbPUB7, SbPUB25, and SbPUB26 were significantly upregulated after 3 and 6 h of treatment then fell to the initial level after 9 h of treatment. Both SbPUB17 and SbPUB18 genes were significantly upregulated only after 3 h compared with initial levels, and expression then dropped to the initial level after 6 h of treatment. Both SbPUB21 and SbPUB62 genes were significantly upregulated after 3 h of treatment compared with the initial level, but dropped to below the initial level after 6 h of treatment (Figure 10A).
Under GA3 treatment, the expression patterns of the seven sorghum PUBs were significantly altered. In general, with the extension of treatment time, expression levels were first upregulated then downregulated. Specifically, expression levels of SbPUB7, SbPUB25, SbPUB26, and SbPUB62 were significantly reduced at 6 and 12 h. The highest expression levels for SbPUB17, SbPUB18, and SbPUB21 were obtained after continuous treatment for 6 h. In particular, the initial expression level of SbPUB17 was extremely low, while the expression level was higher after GA3 treatment (Figure 10B).
Ubiquitylation, one of the most important modification types in plants, mediates the degradation of plant proteins and the process of protein repair. Most members of the U-box gene family are ligases, mediating the ubiquitination modification of protein, which enables ubiquitin to be combined with substrates to manipulate various protein activities (Azevedo et al., 2001; Monaghan et al., 2009; Wang et al., 2015; You and He, 2016; Qin et al., 2020). It has been reported that the expression of U-box genes can affect their functions both in plant development and stress conditions (Hu et al., 2018, 2019; Lu et al., 2020; Sharma and Taganna, 2020; Wang J. et al., 2020; Wang N. et al., 2020).
In the present study, we identified 68 U-box genes (including 1 UFD2 and 2 CHIP genes) in S. bicolor. Additionally, CHIPs, UFD2, and U-box + WD40 branches were found to differ from other U-box genes in their characteristic sequence regions, at both protein and gene structure levels. U-box only and U-box + ARM branches have a relatively simple structure, consistent with the conclusions of previous studies. Most of the U-box gene families constitute about 50-80 members, such as rice, tomato, citrus, and Arabidopsis. Some plants have nearly 100 U-box genes (bananas = 91, cabbages = 101, and soybeans = 127). There are 93–208 U-box genes in cotton and 99 in cabbage, most within 6–8 in terms of sub-branch classification, such as apple, banana, cotton, and cabbage. Some contain more than 10, such as Chinese cabbage (Azevedo et al., 2001; Li et al., 2017; Hu et al., 2018; Chen X. X. et al., 2020; Lu et al., 2020; Qin et al., 2020; Rennie et al., 2020; Tang et al., 2021).
In this study, we could speculate that in the process of U-box gene cluster expansion, the original cluster contained only U-box genes with a few introns, from which substructures became embedded and shuffled, and mutations led to WD-40, ARM, TPR, and DWNN structures gradually appearing. Furthermore, U-box genes form large gene clusters at specific ends of chromosomes, and this cluster arrangement is also reflected in other plants (Qin et al., 2020; Wang N. et al., 2020). These gene clusters are likely to have occurred in recent years, because CHIP and some PUB genes are not only distributed at the beginning of chromosomes, and the beginning of chromosomes contain no clusters. We speculate that these genes differentiated early, and the beginning of chromosomes was not conducive to the enrichment of U-box genes. In addition, we only found one tandem repeat gene pair, namely SbPUB26/SbPUB27, which indicates that U-box genes only underwent one tandem repeat recently, hence we speculate that differentiation was minimal.
Collinearity analysis demonstrated that 9 arabidopsis U-box genes, 46 rice U-box genes, 59 maize U-box genes and 10, 46, and 45 sorghum U-box genes formed 14, 66, and 82 collinearity gene pairs, respectively. In addition, analysis of homologous U-box genes suggested that a large number of genes were derived from the differentiation of monocotyledons, such as SbCHIP2. We speculated that numerous U-box genes may exist in early sorghum plants, principally on chromosomes 1 and 4. During the course of evolution, a few U-box genes underwent various repetitions (e.g., fragment repetition, transposition, and tandem duplication) (Tan et al., 2016; Jiang et al., 2019; Vicient and Casacuberta, 2020; Wang J. et al., 2020; Wang N. et al., 2020; Xiao et al., 2020), resulting in the characteristic gene distribution (i.e., genes are widely distributed except on chromosome 5).
Furthermore, numerous tissue-specific elements were found in the U-box promoter regions. Analysis of expression characteristics revealed that U-box genes displayed obvious tissue specificity, with highest expression levels in roots, flowers, meristem, and stems. Similarly, obvious tissue expression was evident for U-box genes in bananas, with high expression levels in roots (Hu et al., 2018). In soybean, selective tissue expression was also observed in the roots of young tissues and flowers (Wang N. et al., 2020). Similar expression patterns were also found in cotton, cabbage, and other plants (Wang et al., 2015; Song et al., 2017; Hu et al., 2018; Sharma and Taganna, 2020). In the sorghum U-box promoter regions, we identified numerous abiotic response elements, especially those related to drought, ABA, JA, and GA responses, and this was confirmed by RT-qPCR (Figures 5, 7, 9, 10). Previous studies have shown that many U-box genes mediate drought tolerance in plants through multiple pathways. For example, OsPUB67, AtPUB11, AtPUB18, AtPUB19, AtPUB22, and AtPUB23 are involved in drought tolerance in an ABA-dependent manner (Chen X. X. et al., 2020; Qin et al., 2020). Drought-responsive genes were significantly upregulated in GmPUB6-overexpressing plants, indicating that this gene mediates osmotic stress and ABA signaling pathways to enhance drought tolerance in plants (Wang N. et al., 2020). In response to physiological drought, the SbU-box genes displayed different expression pattern under PEG and ABA treatments, and the responses could be mainly divided into three types: type I (suppressed expression), type II (upregulated expression), and type III (no significant change in expression). In our study, seven genes (PUB7/17/18/21/25/26/62) differentially expressed at different time points following abiotic stress were examined. Most U-box genes in sorghum are physiological drought-sensitive genes induced by ABA, but some are physiological drought resistance genes induced by PEG. Interestingly, SbPUB17 was upregulated following physiological drought caused by two different reagents, while SbPUB62 was downregulated following physiological drought caused by two different reagents. Previous studies have shown that many U-box genes can respond to JA and GA hormones, thereby promoting plant stress resistance. For example, AtPUB10 and StPUB17 are induced by JA, and they promote the stability of the JA hormone pathway, whereas StPHOR1 is induced by GA, and this mediates the GA hormone pathway (Amador et al., 2001). In our study, under MeJA treatment, expression of the seven PUB genes in sorghum was rapidly upregulated, and expression peaked at 3 h. Over a longer period, expression of these genes dropped to initial levels or lower. Under GA3 treatment, the expression patterns of the seven sorghum PUB genes were changed significantly. In general, with the extension of treatment time, expression levels were typically upregulated initially then downregulated. In particular, the initial expression level of SbPUB17 was very low, and this increased following GA3 treatment.
The co-regulatory network and correlations between plant U-box genes were similar to those of most plant gene families. Among the same groups, the co-regulatory network and expression correlations were relatively high and mainly positive (Lv et al., 2017; Zhao et al., 2019; Zhang et al., 2020). However, we only carried out genome-wide identification and structural analysis of sorghum U-box genes, hence the detailed involvement of U-box genes in plant development remains unclear. Previous studies have shown that U-box genes may be associated with cold stress (Hu et al., 2018; Lu et al., 2020). Additionally, functional differentiation and functional responses have been linked to sorghum U-box genes, but the details remain to be verified.
In this study, we identified and characterized 68 sorghum U-box genes, which are distributed across all chromosomes except chromosome 5, and divided into eight subclasses. The gene co-regulation network and expression correlations were relatively high and mainly positive. RNA-seq and RT-qPCR experiments showed that the sorghum U-box genes could be tissue-specific, and are involved in responses to various stresses. The results provide a list of potential genes involved in the sorghum ubiquitination system, and expand our understanding of the evolution of the sorghum genome.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
YF, JJ, and QD conducted the experiment. YF wrote the manuscript. ZY and XL directed the experiments and participated in the revision of the manuscript. YF, XH, QY, DZ, XL, and XX completed the revision of the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by the National Natural Science Foundation of China (32060614), the Guizhou Provincial Science and Technology Project ([2022]091), the China Postdoctoral Science Foundation (2022MD713740), and Department of Education of Guizhou Province (QianJiaoHe YJSKYJJ[2021]056).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.942302/full#supplementary-material
PUB, plant U-box protein; CHIP, heat shock protein 70-interacting protein; UFD2, Ub fusion degradation 2; PEG, polyethylene glycol; ABA, abscisic acid; GA, gibberellin; MeJA, methyl jasmonate; HMM, hidden Markov model; PI, isoelectric point; FPKM, fragments per kilobase of exon per million fragments mapped; PCC, Pearson correlation coefficient; NJ, neighbor-joining; RT-qPCR, real-time quantitative PCR; PTM, post-translational modification; UAE, ubiquitin-activating enzyme; UBC, ubiquitin-conjugating enzyme; HECT, homologous to the E6-AP carboxy terminus; CRL, cullin-ring ubiquitin ligase; GO, gene ontology; BBX, novel B-box protein; AIRP, abscisic acid (ABA)-insensitive RING protein; LRR1, leucine rich repeat protein 1; kin7, Kinase 7.
Amador, V., Monte, E., Garcia-Martinez, J. L., and Prat, S. (2001). Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106, 343–354. doi: 10.1016/S0092-8674(01)00445-7
An, J. P., Wang, X. F., Zhang, X. W., You, C. X., and Hao, Y. J. (2021). Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 229, 2707–2729. doi: 10.1111/nph.17050
Anaso, C. E. (2010). Efficacy of neem pesticides on whorl larva, stem-borer and panicle insect pests of sorghum in Nigeria. Arch. Phytopathol. Plant Prot. 43, 856–862. doi: 10.1080/03235400801972335
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., de Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Azevedo, C., Santosrosa, M. J., and Shirasu, K. (2001). The U-box protein family in plants. Trends Plant Sci. 6, 354–358. doi: 10.1016/S1360-1385(01)01960-4
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Callis, J. (2014). The ubiquitination machinery of the ubiquitin system. Arabid. Book 12:e0174. doi: 10.1199/tab.0174
Chai, C., and Subudhi, P. K. (2016). Comprehensive analysis and expression profiling of the OsLAX and OsABCB auxin transporter gene families in rice (Oryza sativa) under phytohormone stimuli and abiotic stresses. Front. Plant Sci. 7:593. doi: 10.3389/fpls.2016.00593
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools - an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, X. X., Wang, T. T., Rehman, A. U., Wang, Y., Qi, J. S., Li, Z., et al. (2020). Arabidopsis U-box E3 ubiquitin ligase PUB11 negatively regulates drought tolerance by degrading the receptor-like protein kinases LRR1 and KIN7. J. Integr. Plant Biol. 63, 494–509. doi: 10.1111/jipb.13058
Cuevas, H. E., Prom, L. K., and Rosavalentin, G. (2018). Population structure of the NPGS Senegalese sorghum collection and its evaluation to identify new disease resistant genes. PLoS One 13:e0191877. doi: 10.1371/journal.pone.0191877
Das, I. K., Kumar, S., Kannababu, N., and Tonapi, V. A. (2019). Pokkah boeng resistance in popular rabi sorghum cultivars and effects of the disease on leaf chlorophyll. Ind. Phytopathol. 72, 421–426. doi: 10.1007/s42360-019-00146-5
Ding, S., Cai, Z., Du, H., and Wang, H. (2019). Genome-wide analysis of TCP family genes in Zea mays L. identified a role for ZmTCP42 in drought tolerance. Int. J. Mol. Sci. 20:2762. doi: 10.3390/ijms20112762
Dowd, P. F., Funnellharris, D. L., and Sattler, S. E. (2016). Field damage of sorghum (Sorghum bicolor) with reduced lignin levels by naturally occurring insect pests and pathogens. J. Pest Sci. 89, 885–895. doi: 10.1007/s10340-015-0728-1
Du, Q. L., Qu, Z. G., Wang, L., Jiang, J. M., Fu, X. D., Fang, Y. P., et al. (2021). Histone deacetylase SbHDT701 in Sorghum bicolor reveals functions in response to stress factors by enhancing acetylation. Pest. Biochem. Physiol. 178:104908. doi: 10.1016/J.PESTBP.2021.104908
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Gu, Z. L., Cavalcanti, A., Chen, F. C., Bouman, P., and Li, W. H. (2002). Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 19, 256–262. doi: 10.1093/oxfordjournals.molbev.a004079
Hu, D., Xie, Q., Liu, Q., Zuo, T., Zhang, H., Zhang, Y., et al. (2019). Genome-wide distribution, expression and function analysis of the U-box gene family in Brassica oleracea L. Genes 10:1000. doi: 10.3390/genes10121000
Hu, H., Dong, C., Sun, D., Hu, Y., and Xie, J. (2018). Genome-wide identification and analysis of U-box E3 ubiquitin-protein ligase gene family in banana. Int. J. Mol. Sci. 19:3874. doi: 10.3390/ijms19123874
Jiang, S., Wang, X., Shi, C., and Luo, J. (2019). Genome-Wide identification and analysis of high-copy-number LTR retrotransposons in Asian pears. Genes 10:156. doi: 10.3390/genes10020156
Jin, X. Y., Long, Y. F., Xiong, S. Q., Yang, Z., Chen, W., Hawar, A., et al. (2021). SbNAC2 enhances abiotic stress tolerance by upregulating ROS scavenging activities and inducing stress-response genes in sorghum. Environ. Exp. Bot. 192:104664. doi: 10.1016/J.ENVEXPBOT.2021.104664
Kim, E. J., Lee, S. H., Park, C. H., Kim, S. H., Hsu, C. C., Xu, S., et al. (2021). Corrigendum to: plant U-Box 40 mediates degradation of the brassinosteroid-responsive transcription factor BZR1 in Arabidopsis roots. Plant Cell 33:2900. doi: 10.1093/plcell/koab127
Kong, L. Y., Cheng, J. K., Zhu, Y. J., Ding, Y. L., Meng, J. J., Chen, Z. Z., et al. (2015). Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 6:8630. doi: 10.1038/ncomms9630
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., and Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Li, L., Bai, H., Cao, Y., Wang, X., and Liu, G. (2009). Expression analysis of rice U-box proteins at different developmental stages. Prog. Biochem. Biophys. 36, 1208–1214. doi: 10.3724/Sp.J.1206.2009.00075
Li, Q. Y., Li, L. Q., Liu, Y., Lv, Q., Zhang, H., Zhu, J., et al. (2017). Influence of TaGW2-6A on seed development in wheat by negatively regulating gibberellin synthesis. Plant Sci. 263, 226–235. doi: 10.1016/j.plantsci.2017.07.019
Lu, X., Shu, N., Wang, D., Wang, J., Chen, X., Zhang, B., et al. (2020). Genome-wide identification and expression analysis of PUB genes in cotton. BMC Genomics 21:213. doi: 10.1186/s12864-020-6638-5
Lv, G. Y., Guo, X. G., Xie, L. P., Xie, C. G., Zhang, X. H., Yang, Y., et al. (2017). Molecular characterization, gene evolution, and expression analysis of the fructose-1, 6-bisphosphate aldolase (FBA) gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 8:1030. doi: 10.3389/fpls.2017.01030
Mace, E. S., Tai, S., Gilding, E. K., Li, Y., Prentis, P. J., Bian, L., et al. (2013). Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat. Commun. 4:2320. doi: 10.1038/ncomms3320
Mandal, A., Sharma, N., Muthamilarasan, M., and Prasad, M. (2018). Ubiquitination: a tool for plant adaptation to changing environments. Nucleus 61, 253–260. doi: 10.1007/s13237-018-0255-6
Miao, J., Yang, Z., Zhang, D., Wang, Y., Xu, M., Zhou, L., et al. (2019). Mutation of RGG2, which encodes a type B heterotrimeric G protein gamma subunit, increases grain size and yield production in rice. Plant Biotechnol. J. 17, 650–664. doi: 10.1111/pbi.13005
Miryeganeh, M., and Saze, H. (2020). Epigenetic inheritance and plant evolution. Popul. Ecol. 62, 17–27. doi: 10.1002/1438-390x.12018
Moharam, M. H. A., and Hassan, M. A. A. (2020). Defense response of seedling plumule and mesocotyl of sorghum to infection by Sporisorium sorghi, causing covered kernel smut in relation to disease resistance classes. Austral. Plant Pathol. 49, 1–14. doi: 10.1007/s13313-019-00670-y
Monaghan, J., Xu, F., Gao, M., Zhao, Q., Palma, K., Long, C., et al. (2009). Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog. 5:e1000526. doi: 10.1371/journal.ppat.1000526
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Puyuan, Y., Jianghui, C., Genzeng, R., Yukun, G., Shilin, W., and Jinhua, C. (2022). Identification, classification and expression characterization of PHD-FINGER gene family in Sorghum. Mountain Agricult. Biol. J. 33, 10–20.
Qin, Q., Wang, Y. X., Huang, L. Y., Du, F. P., Zhao, X. Q., Li, Z. K., et al. (2020). A U-box E3 ubiquitin ligase OsPUB67 is positively involved in drought tolerance in rice. Plant Mol. Biol. 102, 89–107. doi: 10.1007/s11103-019-00933-8
Rennie, M. L., Chaugule, V. K., and Walden, H. (2020). Modes of allosteric regulation of the ubiquitination machinery. Curr. Opin. Struct. Biol. 62, 189–196. doi: 10.1016/j.sbi.2020.02.003
Richard, D. V. (2009). The ubiquitin–26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10, 385–397. doi: 10.1038/nrm2688
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sharma, B., and Taganna, J. (2020). Genome-wide analysis of the U-box E3 ubiquitin ligase enzyme gene family in tomato. Sci. Rep. 10:9581. doi: 10.1038/s41598-020-66553-1
Sharma, H. C. (1993). Host-plant resistance to insects in sorghum and its role in integrated pest management. Crop Prot. 12, 11–34. doi: 10.1016/0261-2194(93)90015-b
Silva, A. O., Aliyeva-Schnorr, L., Wirsel, S. G. R., and Deising, H. B. (2021). Fungal pathogenesis-related cell wall biogenesis, with emphasis on the maize anthracnose fungus Colletotrichum graminicola. Plants 11:849. doi: 10.3390/plants11070849
Silva, R. B., Cruz, I., Zanuncio, J. C., Figueiredo, M., Canevari, G. D. C., Pereira, A. G., et al. (2013). Biological aspects of Eriopis connexa (Germar) (Coleoptera: Coccinellidae) fed on different insect pests of maize (Zea mays L.) and sorghum [Sorghum bicolor L. (Moench.)]. Brazil. J. Biol. 73, 419–424. doi: 10.1590/S1519-69842013000200025
Song, J., Mo, X., Yang, H., Yue, L., Song, J., and Mo, B. (2017). The U-box family genes in Medicago truncatula: key elements in response to salt, cold, and drought stresses. PLoS One 12:e0182402. doi: 10.1371/journal.pone.0182402
Tan, S., Cardoso-Moreira, M., Shi, W., Zhang, D., Huang, J., Mao, Y., et al. (2016). LTR-mediated retroposition as a mechanism of RNA-based duplication in metazoans. Genome Res. 26, 1663–1675. doi: 10.1101/gr.204925.116
Tang, X., Ghimire, S., Liu, W., Fu, X., Zhang, H., Sun, F., et al. (2021). Genome-wide identification of U-box genes and protein ubiquitination under PEG-induced drought stress in potato. Physiol. Plant 174, doi: 10.1111/ppl.13475
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). AgriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129. doi: 10.1093/nar/gkx382
Tian, T., You, Q., Zhang, L., Yi, X., Yan, H., Xu, W., et al. (2016). SorghumFDB: sorghum functional genomics database with multidimensional network analysis. Database 2016:baw099. doi: 10.1093/database/baw099
Tong, S., Chen, N., Wang, D., Ai, F., Liu, B., Ren, L., et al. (2021). The U-box E3 ubiquitin ligase PalPUB79 positively regulates ABA-dependent drought tolerance via ubiquitination of PalWRKY77 in Populus. Plant Biotechnol. J. 19, 2561–2575. doi: 10.1111/pbi.13681
Townsley, B. T., Sinha, N. R., and Kang, J. (2013). KNOX1 genes regulate lignin deposition and composition in monocots and dicots. Front. Plant Sci. 4:121. doi: 10.3389/fpls.2013.00121
Vallabhaneni, R., Bradbury, L. M. T., and Wurtzel, E. T. (2010). The carotenoid dioxygenase gene family in maize, sorghum, and rice. Arch. Biochem. Biophys. 504, 104–111. doi: 10.1016/j.abb.2010.07.019
Vicient, C. M., and Casacuberta, J. M. (2020). Additional ORFs in plant LTR-retrotransposons. Front. Plant Sci. 11:555. doi: 10.3389/fpls.2020.00555
Wang, C., Duan, W., Riquicho, A. R., Jing, Z., Liu, T., Hou, X., et al. (2015). Genome-wide survey and expression analysis of the PUB family in Chinese cabbage (Brassica rapa ssp. pekinesis). Mol. Genet. Genomics 290, 2241–2260. doi: 10.1007/s00438-015-1075-x
Wang, J., Ling, L., Cai, H., and Guo, C. (2020). Gene-wide identification and expression analysis of the PMEI family genes in soybean (Glycine max). 3 Biotech 10:335. doi: 10.1007/s13205-020-02328-9
Wang, N., Liu, Y. D., Cai, Y. Y., Tang, J. J., Li, Y., and Gai, J. Y. (2020). The soybean U-box gene GmPUB6 regulates drought tolerance in Arabidopsis thaliana. Plant Physiol. Biochem. 155, 284–296. doi: 10.1016/j.plaphy.2020.07.016
Wang, K. L., Yang, Q. Y., Boen, L. H., Lin, H. T., Shi, Y., Dhanasekaran, S., et al. (2020). Genome-wide investigation and analysis of U-box Ubiquitin-Protein ligase gene family in apple: expression profiles during Penicillium expansum infection process. Physiol. Mol. Plant Pathol. 111:101487. doi: 10.1016/j.pmpp.2020.101487
Wang, Y., Li, J., and Paterson, A. H. (2013). MCScanX-transposed: detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 29, 1458–1460. doi: 10.1093/bioinformatics/btt150
Wu, R., Shi, Y., Zhang, Q., Zheng, W., Chen, S., Du, L., et al. (2019). Genome-wide identification and characterization of the UBP gene family in moso bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 20:4309. doi: 10.3390/ijms20174309
Xiao, H., Wang, C., Khan, N., Chen, M., Fu, W., Guan, L., et al. (2020). Genome-wide identification of the class III POD gene family and their expression profiling in grapevine (Vitis vinifera L). BMC Genomics 21:444. doi: 10.1186/s12864-020-06828-z
Xu, L., Dong, Z., Fang, L., Luo, Y., Wei, Z., Guo, H., et al. (2019). OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 47, W52–W58. doi: 10.1093/nar/gkz333
Yang, L., Wu, L. T., Chang, W., Li, Z., Miao, M. J., Li, Y. J., et al. (2018). Overexpression of the maize E3 ubiquitin ligase gene ZmAIRP4 enhances drought stress tolerance in Arabidopsis. Plant Physiol. Biochem. 123, 34–42. doi: 10.1016/j.plaphy.2017.11.017
Yang, X., and Wang, J. (2016). Genome-wide analysis of NBS-LRR genes in sorghum genome revealed several events contributing to NBS-LRR gene evolution in grass species. Evol. Bioinf. 12, 9–21. doi: 10.4137/EBO.S36433
Yang, Z., Chi, X. Y., Guo, F. F., Jin, X. Y., Luo, H. L., Hawar, A., et al. (2020). SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, in sorghum. J. Plant Physiol. 246–247. doi: 10.1016/j.jplph.2020.153142
Yimer, H. Z., Nahar, K., Kyndt, T., Haeck, A., Van Meulebroek, L., Vanhaecke, L., et al. (2018). Gibberellin antagonizes jasmonate-induced defense against Meloidogyne graminicola in rice. New Phytol. 218, 646–660. doi: 10.1111/nph.15046
You, Q., and He, Z. (2016). Research advances on ubiquitin-26S proteasome-mediated plant immunity. Plant Physiol. J. 52, 771–781. doi: 10.13592/j.cnki.ppj.2016.1002
Yu, F., Li, M., He, D., and Yang, P. (2021). Advances on post-translational modifications involved in seed germination. Front. Plant Sci. 12:642979. doi: 10.3389/fpls.2021.642979
Zeng, W., Peng, Y., Zhao, X., Wu, B., Chen, F., Ren, B., et al. (2019). Comparative proteomics analysis of the seedling root response of drought-sensitive and drought-tolerant maize varieties to drought stress. Int. J. Mol. Sci. 20:31181633. doi: 10.3390/ijms20112793
Zhang, X., Li, X., Zhao, R., Zhou, Y., and Jiao, Y. (2020). Evolutionary strategies drive a balance of the interacting gene products for the CBL and CIPK gene families. New Phytol. 226, 1506–1516. doi: 10.1111/nph.16445
Zhao, W., Liu, H., Zhang, L., Hu, Z., Liu, J., Hua, W., et al. (2019). Genome-wide identification and characterization of FBA gene family in polyploid crop Brassica napus. Int. J. Mol. Sci. 20:5749. doi: 10.3390/ijms20225749
Zhao, Y., Li, X., Chen, W., Peng, X., Cheng, X., Zhu, S., et al. (2011). Whole-genome survey and characterization of MADS-box gene family in maize and sorghum. Plant Cell Tiss. Organ. Cult. 105, 159–173. doi: 10.1007/s11240-010-9848-8
Keywords: U-box, expression pattern, abiotic stress, co-regulatory networks, Sorghum bicolor
Citation: Fang Y, Du Q, Yang Q, Jiang J, Hou X, Yang Z, Zhao D, Li X and Xie X (2022) Identification, characterization, and expression profiling of the putative U-box E3 ubiquitin ligase gene family in Sorghum bicolor. Front. Microbiol. 13:942302. doi: 10.3389/fmicb.2022.942302
Received: 12 May 2022; Accepted: 15 August 2022;
Published: 15 September 2022.
Edited by:
Jesús Navas-Castillo, La Mayora Experimental Station (CSIC), SpainReviewed by:
Mohamed Fokar, Texas Tech University, United StatesCopyright © 2022 Fang, Du, Yang, Jiang, Hou, Yang, Zhao, Li and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Xie, aXBweGlleGluQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.