
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 23 August 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.940156
This article is part of the Research TopicMicroorganisms and their Metabolites as Biocontrol Agents for Sustainable AgricultureView all 9 articles
 Xiaona Sui1†
Xiaona Sui1† Xiaobin Han2†
Xiaobin Han2† Jianmin Cao1
Jianmin Cao1 Yiqiang Li1
Yiqiang Li1 Yuan Yuan1
Yuan Yuan1 Jianyu Gou2
Jianyu Gou2 Yanfen Zheng1*
Yanfen Zheng1* Chen Meng1*
Chen Meng1* Chengsheng Zhang1*
Chengsheng Zhang1*Tobacco bacterial wilt caused by Ralstonia solanacearum is one of the most devastating diseases. Microbial keystone taxa were proposed as promising targets in plant disease control. In this study, we obtained an antagonistic Bacillus isolate EM-1 from bacterial wilt-suppressive soil, and it was considered rhizosphere-resident bacteria based on high (100%) 16S rRNA gene similarity to sequences derived from high-throughput amplicon sequencing. According to 16S rRNA gene sequencing and MLSA, strain EM-1 was identified as Bacillus velezensis. This strain could inhibit the growth of R. solanacearum, reduce the colonization of R. solanacearum in tobacco roots, and decrease the incidence of bacterial wilt disease. In addition, strain EM-1 also showed a strong inhibitory effect on other phytopathogens, such as Alternaria alternata and Phytophthora nicotianae, indicating a wide antagonistic spectrum. The antimicrobial ability of EM-1 can be attributed to its volatile, lipopeptide and polyketide metabolites. Iturin A (C14, C15, and C16) was the main lipopeptide, and macrolactin A and macrolactin W were the main polyketides in the fermentation broth of EM-1, while heptanone and its derivatives were dominant among the volatile organic compounds. Among them, heptanones and macrolactins, but not iturins, might be the main potential antibacterial substances. Complete genome sequencing was performed, and the biosynthetic gene clusters responsible for iturin A and macrolactin were identified. Moreover, strain EM-1 can also induce plant resistance by increasing the activity of CAT and PPO in tobacco. These results indicated that EM-1 can serve as a biocontrol Bacillus strain for tobacco bacterial wilt control. This study provides a better insight into the strategy of exploring biocontrol agent based on rhizosphere microbiome.
Ralstonia solanacearum (Rs) is a soilborne Gram-negative bacterium that can cause disastrous bacterial wilt (BW) in various plant species, for example, tobacco, tomato, potato, banana, eggplant, and pepper (Cunnac et al., 2004). Bacterial pathogens infect plant roots through wounds or natural openings, produce excessive extracellular polysaccharides (EPS) within the vascular system, and cause wilt, ultimately leading to the death of severely infected plants (Genin, 2010). BW mainly occurs at the maturity stage of tobacco and thus lead to huge economic losses. Due to its genetic diversity and wide geographic distribution, Rs presents a rather complex life cycle and can survive in both soil and water for years (Jiang et al., 2017). Currently, chemical fungicides dominated by antibiotics and inorganic copper-based pesticides are used for bacterial wilt control. However, the use of antibiotics in plant protection is increasingly restricted, and the long-term use of cupric pesticides negatively affects the environment. Therefore, there is an urgent need to find a novel and effective prevention strategy for bacterial wilt control.
Antagonistic microorganisms have been recognized as effective and sustainable for controlling plant diseases. To date, a large number of antagonistic microbes against Rs have been identified, including Bacillus spp. (Gao et al., 2013), Pseudomonas spp. (Maji and Chakrabartty, 2014), Trichoderma spp. (Mohamed et al., 2020), and Streptomyces spp. (Ling et al., 2020). Among them, the genus Bacillus has many advantages, including fast reproduction, strong survival ability, and long storage period. Moreover, Bacillus species can produce a wide range of antibiotic compounds, including cyclic lipopeptides (CLPS) and antifungal proteins, which play an important role in plant pathogen inhibition (Ongena and Jacques, 2008; Kwon and Kim, 2014). Therefore, bacteria of the genus Bacillus are considered as an ideal biocontrol agent and have been widely researched and used.
A large number of Bacillus strains have been screened for controlling tobacco bacterial wilt (Wu et al., 2016; Tahir et al., 2017; Agarwal et al., 2020). However, these strains were isolated mainly based on the interaction between individual microbial strains and plants, resulting in poor field colonization ability and unstable biocontrol effects (Lebeis, 2014). The latest research shows that a few key microorganisms, called “core microbiome,” may play the most important functions in the microbial community (Banerjee et al., 2018); thus, the microbial keystone taxa have been proposed as promising targets in plant disease control (Agler et al., 2016). Qi et al. (2019) reported that healthy soil harbors more key microbes (e.g., Bacillus and Actinobacteria) than bacterial wilt-susceptible soil. Our previous study analyzed the microbial community structure and function of tobacco bacterial wilt-suppressive soil and found that Pseudomonas was enriched in suppressive soil, which displayed strong inhibitory effects on R. solanacearum (Zheng et al., 2021). The current study aims to screen biocontrol Bacillus strains from bacterial wilt-suppressive soil and to evaluate their inhibitory efficacy against tobacco bacterial wilt. The complete genome of the selected strain was analyzed to reveal the possible mechanisms of disease control. The results will provide a better insight into the strategy of exploring biocontrol agent based on rhizosphere microbiome.
R. solanacearum RS10 was isolated from a diseased tobacco plant (Bai et al., 2016). The phytopathogenic fungi used in this study were Alternaria alternata, Phytophthora nicotianae, Botrytis cinerea, Fusarium oxysporum, Cucumber anthracnose, and Rhizoctonia cerealis (Xie et al., 2021; Wang et al., 2022). All strains were preserved in our laboratory and stored in 25% (v/v) glycerol at −80°C.
Seeds of tobacco (Nicotiana tabacum cv Xiaohuangjin 1025) were sown in sterilized soil (volume of soil:vermiculite, 3:1) and grown in a greenhouse with a 16-h light/8-h dark cycle, day and night temperatures of 29 and 28°C, respectively, and a relative humidity of 70–80%.
All the phytopathogenic fungi were cultured on potato dextrose agar (PDA, 6 g potato infusion, 20 g dextrose, 15 g agar, distilled water 1 L) medium, except P. nicotianae, which was cultured on oatmeal agar (OA, oatmeal 30 g, agar 15 g, distilled water 1 L) medium. R. solanacearum RS10 was cultured in nutrient broth (NB, 10 g glucose, 5 g tryptone, 0.5 g yeast extract, 3 g beef extract, distilled water 1 L, pH = 7.0) medium or nutrient agar (NA, NB medium containing an additional 15 g agar per liter) medium. Unless otherwise stated, the isolate EM-1 was cultured on Luria–Bertani (LB, 10 g tryptone, 5 g yeast extract, 10 g sodium chloride, distilled water 1 L) medium.
According to 16S rRNA gene amplicon sequencing data from our previous study (Zheng et al., 2021), an OTU-based analysis was used to analyze the relative abundance of Bacillus. The bacteria were isolated according to Khanh et al. (2020). The biocontrol microbes were selected from a total of 43 Bacillus isolates by using the confrontation culture method (Chen et al., 2020). Specifically, Rs and Bacillus isolates were grown for 18–24 h to OD600 = 0.8–1.0 in NB medium at 28°C in a shaking incubator (180 rpm). Rs (0.2 ml) was included in NA solid medium (20 ml) to make plates, which were used to screen biocontrol Bacillus strains. The suspensions of Bacillus isolates (4 μl) were point-inoculated on the plates containing Rs. Sterile distilled water was used as a control. After incubation at 28°C for 48 h, the bacterial inhibition ability was estimated by measuring the inhibition zone diameter. Based on preliminary experimental results, the isolate EM-1 was selected for further investigation.
The confrontation culture method was used for antifungal activity determination (Kurniawan et al., 2018). An agar block (5 mm in diameter) from the margin of 5-day-old culture of the fungal pathogen was placed in the center of the PDA medium. The strain EM-1 was grown for 18–24 h to OD600 = 1.0 in NB medium at 28°C in a shaking incubator (180 rpm), and 4 μl of EM-1 suspensions were point-inoculated on both sides of the Petri dish. Equal volumes of sterile distilled water served as control. The plates were then incubated at 28°C. The diameter of the fungal colony was measured after the control culture had covered the plate. The percentage of mycelial growth inhibition was calculated using the following equation:
The fumigation effect of EM-1 on Rs was tested according to You et al. (2015). Suspensions (20 μl) of Rs and EM-1 were spread evenly on different NA medium plates, respectively, and two plates (the plate inoculated with Rs was upper) were sealed together with polyethylene (PE) stretch-wrap. After incubation at 28°C for 24 h, the growth of Rs was observed.
Genomic DNA of the strain EM-1 was isolated using the Solarbio DNA Extraction Kit (Solarbio, Beijing, China). 16S rRNA gene of bacterial isolates was amplified by PCR using universal primers 27F (5′ AGAGGTTTGATCCTGGCTCAG 3′) and 1492R (5′ GGTTACCTTGTTACGACTT 3′), and seven housekeeping genes (glpF, ilvD, ptA, purH, pycA, rpoD, and tpiA) were amplified using primers described by Le et al. (2019). PCR amplifications included a 20 μl mixture containing 1 μl of genomic DNA of the strain EM-1 as a template, 0.5 μl of each primer (0.5 μM), 10 μl of polymerase buffer with dNTPs and MgCl2, 0.5 μl of Taq DNA polymerase (TransGen Biotech, China), and distilled water. PCR amplification was performed as follows: one cycle of 5 min at 95°C; followed by 40 cycles of 30 s at 95°C, 30 s at 50°C, and 1.5 min at 72°C; then followed by one cycle of 10 min at 72°C. PCR products were sent to Ruibiotech for Sanger sequencing (Qingdao, China). The 16S rRNA gene sequence was aligned in the EzBioCloud database (https://www.ezbiocloud.net/) (Yoon et al., 2017). The multi-locus sequence analysis (MLSA) (www.pubmlst.org/bsubtilis) was used to analyze housekeeping genes. The nucleotide sequences were concatenated to construct a phylogenetic tree. The phylogenetic tree was constructed using MEGA X software by using the maximum-likelihood method and general time reversible model (Nei and Kumar, 2002; Kumar et al., 2018).
Tobacco seedlings with five leaves were divided into two treatment groups (I and II) with three biological replicates, and each replicate contained at least 20 seedlings: (I) irrigating EM-1 suspensions (OD600 = 0.6) and then inoculating Rs suspensions on the 2nd day; (II) control, irrigating Rs suspensions (OD600 = 0.7). The bacterial suspensions were prepared, as described previously (Section Screening of candidate bio-control Bacillus isolates). The bacterial suspensions of EM-1 and Rs were then centrifuged at 6,000 rpm for 5 min and diluted in water to OD600 of 0.6 and 0.7, respectively. Each inoculation treatment was 10 ml per plant. Plants were cultivated in a greenhouse, as described before (Section Microbial and plant resources) and watered every 3 days to maintain soil moisture. The disease severity of the tobacco plants was scored on the 5th day of Rs inoculation, and tobacco samples were collected for further use. The disease severity investigation and disease index calculation were carried out according to (Zhang et al., 2017).
Total DNA of root samples were extracted using the FastDNA® Spin Kit for Soil (MP Biomedicals, USA). The specific primer Rsol_fliC (F 5′ GAACGCCAACGGTGCGAACT 3′ and R 5′ GGCGGCCTTCAGGGAGGTC 3′) that targets the fliC gene encoding the flagellum subunit was used to quantify Rs densities. Real-time PCR experiments were performed by using the SYBR® Premix Ex TaqTM (Takara Bio Inc, Japan) and analyzed using the 7500 Real-Time PCR System (Applied Biosystems, USA). Standard curves were generated using 10-fold serial dilutions of a plasmid containing a fragment copy of Rs fliC. qPCR amplifications for standard and DNA samples included a 20 μl mixture containing 2 μl of templates, 10 μl of the SYBR Green Premix Ex Taq (2 ×), 0.4 μl of ROX Reference Dye II, 0.4 μl of each primer, and distilled water. PCR amplification was programmed with one cycle of denaturation at 95°C for 30 s and 40 cycles of 95°C for 5 s and 60°C for 34 s. Fluorescence was monitored in each PCR cycle during the annealing and extension phases at 60°C (Zheng et al., 2021).
The crude enzyme extracts of polyphenol oxidase (PPO), superoxide dismutase (SOD), and catalase (CAT) were prepared as follows: Tobacco leaves (0.1 g) were dipped in liquid nitrogen in a pre-cooled mortar, homogenized in 1 ml of ice-cold 50 mM potassium phosphate buffer (pH 6.8), and centrifuged at 12,000 rpm at 4°C for 10 min. The supernatant was collected as the enzyme extract of SOD and CAT. The preparation of crude PPO was the same, except for sodium borate buffer (pH 8.8). The enzyme activity determination was performed according to Ju et al. (2014), Li et al. (2014), and Wang et al. (2019). Each treatment consisted of three replicates.
CFs obtained from the strain EM-1 were prepared, as described by Ezrari et al. (2021). In brief, the strain EM-1 was cultured in LB liquid medium, at 180 rpm for 72 h at 28°C, and then the liquid cultures were centrifuged at 8,000 g for 10 min. The supernatants were collected and filtered using 0.22-μm filter membranes and then dried into powder by freeze drying (the temperature and pressure of the vacuum oven were −80°C and 1.0 Pa, respectively). The inhibition zone method was used to evaluate the antibacterial activity of the supernatants, as described by Zhu and Zhang (2020). The powder was diluted with methanol into 50 mg/ml. Rs was inoculated into NA medium to prepare a plate, as described before. After that, three holes were punched in each of the plate. Then, 60 μl of CFs and methanol as a control were injected into the holes, and the diameter of the inhibition zone around the hole was measured to determine antibacterial activity after incubation at 28°C for 48 h.
To prepare the samples for antagonistic activity experiments, cell-free filtrates (CFs, 50 ml, 100 mg/ml) of the strain EM-1 were passed through an Amberlite XAD-16 column (100 g, 4 × 70 cm), as described by Yuan et al. (2012a) with some modifications. In brief, the active compounds and impurities were bound to the column matrix. To remove the impurities, first, the column was washed with 500 ml of 30% methanol, and then elution of active compounds was performed using 500 ml of 100% methanol. The eluates were dried in a rotary evaporator (<40°C). The secondary metabolites were dissolved in methanol to prepare the solution with a concentration of 50 mg/ml.
For the preparation of polyketides, the CFs were extracted with ethyl acetate (Mondol et al., 2011) after EM-1 was incubated in LB medium at 37°C with shaking at 200 rpm for 16 h. The CFs of EM-1 were mixed with ethyl acetate (EtOAc, 1:1 volume ratio) and extracted two times. The cumulative EtOAc layer was dried in a rotary evaporator (<40°C), and the extract was dissolved in methanol to prepare the solution with a concentration of 30 mg/ml. The iturin A standard (Sigma, USA) was purchased and dissolved in methanol to prepare the solution with concentrations of 10, 5, and 1 mg/ml. The activity of the crude extract against Rs was evaluated by using the well diffusion method with the same conditions previously used.
Crude protein extracts were precipitated using the ammonium sulfate precipitation method with slight modifications (Wu et al., 2020). The strain EM-1 was grown in 200 ml liquid LB medium and incubated at 180 rpm, 28°C for 72 h. Supernatants were collected by centrifugation at 8,000 g, 4°C for 20 min. The soluble proteins in the supernatants were precipitated by slowly adding two volumes of 100% saturated ammonium sulfate solution, and the resulting protein-containing suspensions were then centrifuged at 12,000 rpm for 30 min. The precipitated proteins were suspended in 20 mM Tris–HCl, pH 7.4, and dialyzed against the same buffer overnight to remove residual ammonium sulfate. The complete removal of ammonium sulfate was confirmed by continuous titration of the dialysate with barium chloride solution (0.5 M) until no white precipitation was observed. The resulting protein-containing solutions were dried into powder by freeze drying (the conditions of freeze drying were as described in Section Antibacterial activity of a cell-free culture filtrates (CFs) from strain EM-1). The crude protein extracts were dissolved in methanol to prepare the solution with a concentration of 50 mg/ml. The activity of the crude protein extracts against Rs was evaluated by using the well diffusion method using the same conditions previously used.
LC-MS/MS was carried out by coupling an Agilent 1290 LC System to a LTQ-Orbitrap XL Mass Spectrometer (Thermo Fisher Scientific, USA). Chromatographic separation was performed on a SunFire column (C18, 250 × 2.6 mm, Waters, USA). The mobile phase was 60% A [0.1% (v/v) aqueous acetic acid] and 40% B [acetonitrile] with 0.6 ml/min flow rate, 30°C column temperature, and 10 μl injection volume. The general mass spectrometric parameters were as follows: the spray voltage, capillary voltage, and tube lens voltage were 4.0 kV, 16 V, and 35 V, respectively. The capillary temperature was 300°C with a sheath gas flow rate of 40 L/min and an auxiliary gas flow rate of 10 L/min. External calibration of mass spectra routinely produced a mass accuracy of better than 3 ppm. Full mass spectra were acquired in the positive ionization mode at a resolution of 30,000 with 100–1,500 Da mass range, followed by a data-dependent scan in the collision-induced dissociation (CID) mode. Data acquisition and analysis were performed using Xcalibur software version 4.1 (Thermo Fisher Scientific, USA).
VOCs were extracted by headspace solid-phase microextraction (HS-SPME). Airtight vials were thoroughly cleaned, sterilized, and baked at 120°C for 20 h before use. NA medium was poured into the airtight vials to obtain slants. The suspensions of EM-1 were prepared as described earlier (2.3). A measure of 100 μl of suspensions were transferred to an NA slant in an airtight vial, which was sealed tightly with polytetrafluoroethylene (PTFE) septum, cultured at 28°C for 48 h in dark. The culture medium that was subjected to identical conditions was set as the control. For the sampling of volatile compounds, the solid-phase microextraction (SPME) fiber was inserted into the headspace of the airtight vial containing EM-1 at room temperature for 20 min. When the sampling was completed, the SPME fiber was removed from the sample vial and immediately inserted into the GC injector at 250°C for 5 min with a split ratio of 10:1. The used SPME fiber was conditioned at 250°C for 5 min prior before the next sampling.
GC-MS/MS (an Agilent 7890B GC system coupled to a 7000C GC/MS Triple Quad Mass Detector) (Agilent Technologies, USA) was used to identify VOCs. Chromatographic separation was performed on a 30-m DB-5MS capillary column (Agilent 122-5532UI, USA), the temperature of the injection ports was 250°C, and the flow rate was 1.5 ml/min. The temperature program started in the isocratic mode at 45°C for 2 min, followed by temperature ramping of 5°C/min to a final temperature of 280°C, which was then held constantly for an additional 2 min. The MS operating parameters were as follows: electron energy 70 eV, ion source temperature 280°C, quadrupole temperature 180°C, and m/z scanned area 33–500. The mass spectra data for the VOCs were analyzed using the data in the NIST/EPA/NIH Mass Spectrum Library.
The 2-heptanone standard (98% pure) and 6-methyl-2-heptanone standard (98% pure) were purchased from Aladdin (China), while the 5-methyl-2-heptanone standard (98% pure) was purchased from AndHider (China). These chemical standards were used for the fumigation test with a modified sealed plate method (Tahir et al., 2017). The suspensions of Rs (5 μl) were point-inoculated at the center of NB plates, while the chemical standards (20 μl, 98% pure) were taken on a Petri dish cover, sealed with parafilm, and incubated at 28°C. Sterilized water (20 μl) and EM-1 suspensions were taken on a Petri dish cover as negative control and positive control, respectively. The medium containing Rs was cut down after 24 h and added into the sterilized water and shaken for 5 min. Cell density was measured at OD600 to evaluate T of the VOCs against Rs.
Genomic DNA was extracted using the SDS method (Lim et al., 2016). The harvested DNA was detected by agarose gel electrophoresis and quantified by a Qubit® 2.0 Fluorometer (Thermo Scientific). Libraries for single-molecule real-time (SMRT) sequencing were constructed with an insert size of 10 kb using the SMRTbell TM Template Kit, version 1.0. A total of 1 μg DNA per sample was used as input material for the DNA sample preparations. Sequencing libraries were generated using a NEBNext® UltraTM DNA Library Prep Kit for Illumina (NEB, USA) following manufacturer's recommendations, and index codes were added to attribute sequences to each sample. These libraries were sequenced on the PacBio Sequel platform (Novogene, China), and PacBio reads were assembled using SMRT Link software v5.0.1. The coding genes, repetitive sequences, and noncoding RNA were predicted using GeneMarkS (http://topaz.gatech.edu/), and the Cluster of Orthologous Groups (COG) database (http://www.ncbi.nlm.nih.gov/COG) was used to annotated the functional information of ORFs by DIAMOND software. The complete genome sequence of the strain EM-1 was accessible from the NCBI with GenBank accession number CP095842.
All statistical analyses were carried out using SPSS 21.0 software. We used the t-test, one-way analysis of variance (ANOVA), and Tukey's multiple comparison test to determine the significant differences (p < 0.05) between treatment groups. All experiments were performed with at least three independent replicates, unless otherwise stated.
The relative abundance of genus Bacillus was significantly higher in disease-suppressive soil and healthy root samples than that in disease-conducive soil and infected roots (Figure 1A), implying Bacillus species might play a critical role in suppressing tobacco bacterial wilt. In this study, we obtained 43 Bacillus strains from healthy tobacco rhizosphere soil and root samples. Among them, four isolates showed antagonistic activity against Rs (Supplementary Table 1). The strain EM-1 displayed the largest bacteriostatic circle and was selected for further evaluation. To determine whether the strain EM-1 represented indigenous species of bacterial wilt-suppressive soil, phylogenetic relationships between strain EM-1 and Bacillus OTUs from high-throughput amplicon data were analyzed. The result indicated that the strain EM-1 clustered together with OTU252 (Figure 1B) and showed 100% 16S rRNA gene similarity to this OTU (Figure 1C), suggesting EM-1 can be considered a resident bacterium in the tobacco rhizosphere.
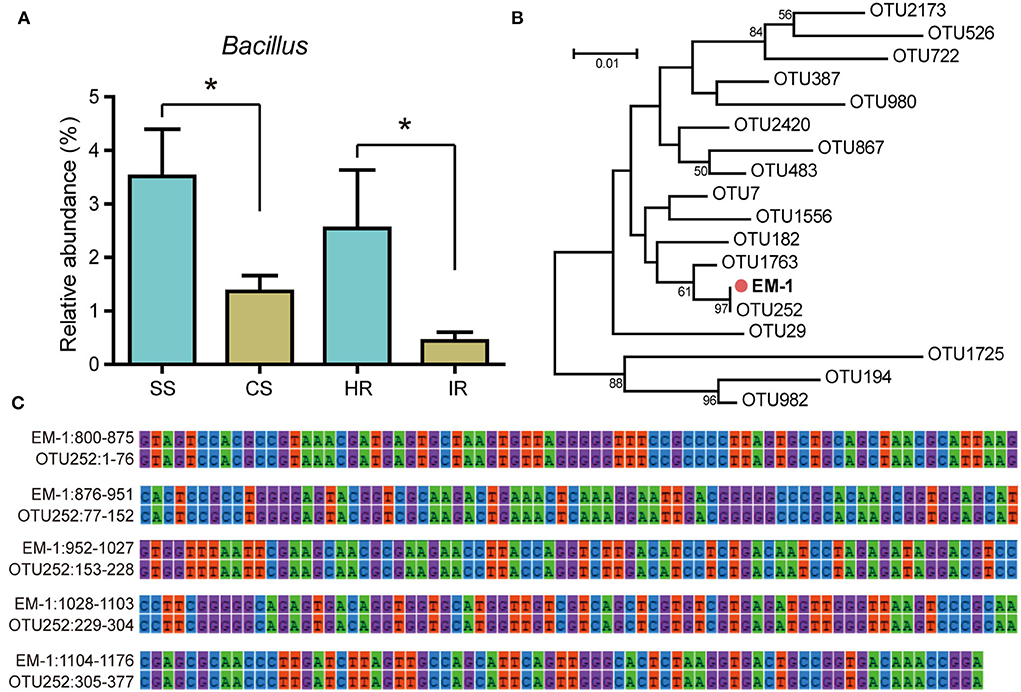
Figure 1. Relative abundance of Bacillus OTUs and their phylogenetic relationships with strain EM-1. (A) Relative abundance of Bacillus OTUs in different samples. SS, suppressive rhizosphere soil; CS, conducive rhizosphere soil; HR, root of healthy tobacco; IR, root of infected tobacco. (B) Neighbor-joining phylogenetic tree constructed with the 16S rRNA gene sequences of all Bacillus OTUs obtained from high-throughput amplicon sequencing and the strain EM-1. (C) 16S rRNA gene alignment between OTU252 and strain EM-1. High-throughput amplicon data used in this figure were derived from our previous study (Zheng et al., 2021).
Bacterial identification to the species level was frequently performed using 16S rRNA gene sequencing. Sequence alignment of 16S rRNA gene (accession number: OK090956) indicated that the strain EM-1 exhibited 99.79% sequence similarity to Bacillus velezensis CR-502T (accession number: AY603658). For the MLSA tree, we used the nucleotide sequences of the seven genus Bacillus-specific MLST genes: glpF, ilvD, ptA, purH, pycA, rpoD, and tpiA. The final sequence length was 2,871 bp. The phylogenetic analyses based on both the 16S rRNA gene and housekeeping gene sequences revealed that the strain EM-1 was closely related to members of B. velezensis species (Figure 2).
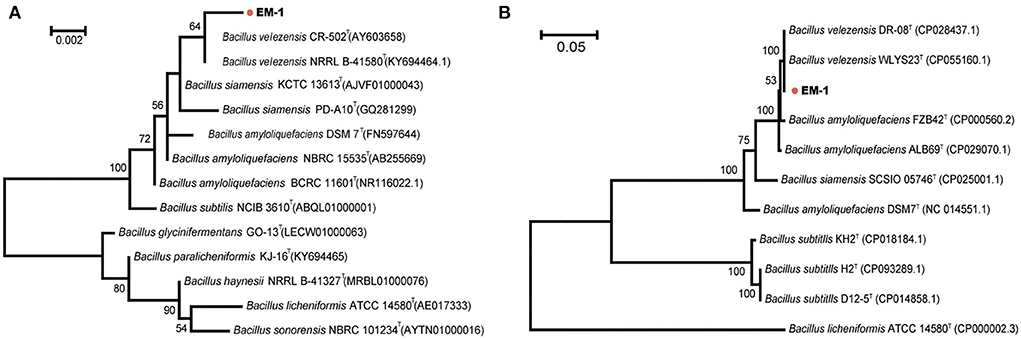
Figure 2. Phylogenetic analysis of strain EM-1. (A) Maximum-likelihood phylogenetic tree constructed with 16S rRNA gene sequences. (B) Maximum-likelihood phylogenetic tree constructed with the concatenated MLST DNA sequences of the seven housekeeping genes.
The growth of Rs was inhibited not only by EM-1 (Figure 3A), but also by VOCs produced by EM-1 (Figure 3B), indicating a strong fumigation effect of EM-1 on Rs. EM-1 also showed a broad-spectrum antagonistic activity against a variety of phytopathogens, including A. alternata, P. nicotianae, B. cinerea, C. anthracnose, F. oxysporum, and R. cerealis (Figure 3C). Among them, the strain EM-1 showed the strongest antagonistic effect on B. cinerea, followed by A. alternata, and the inhibition rates were 89.5 and 79.4%, respectively (Supplementary Figure 1).
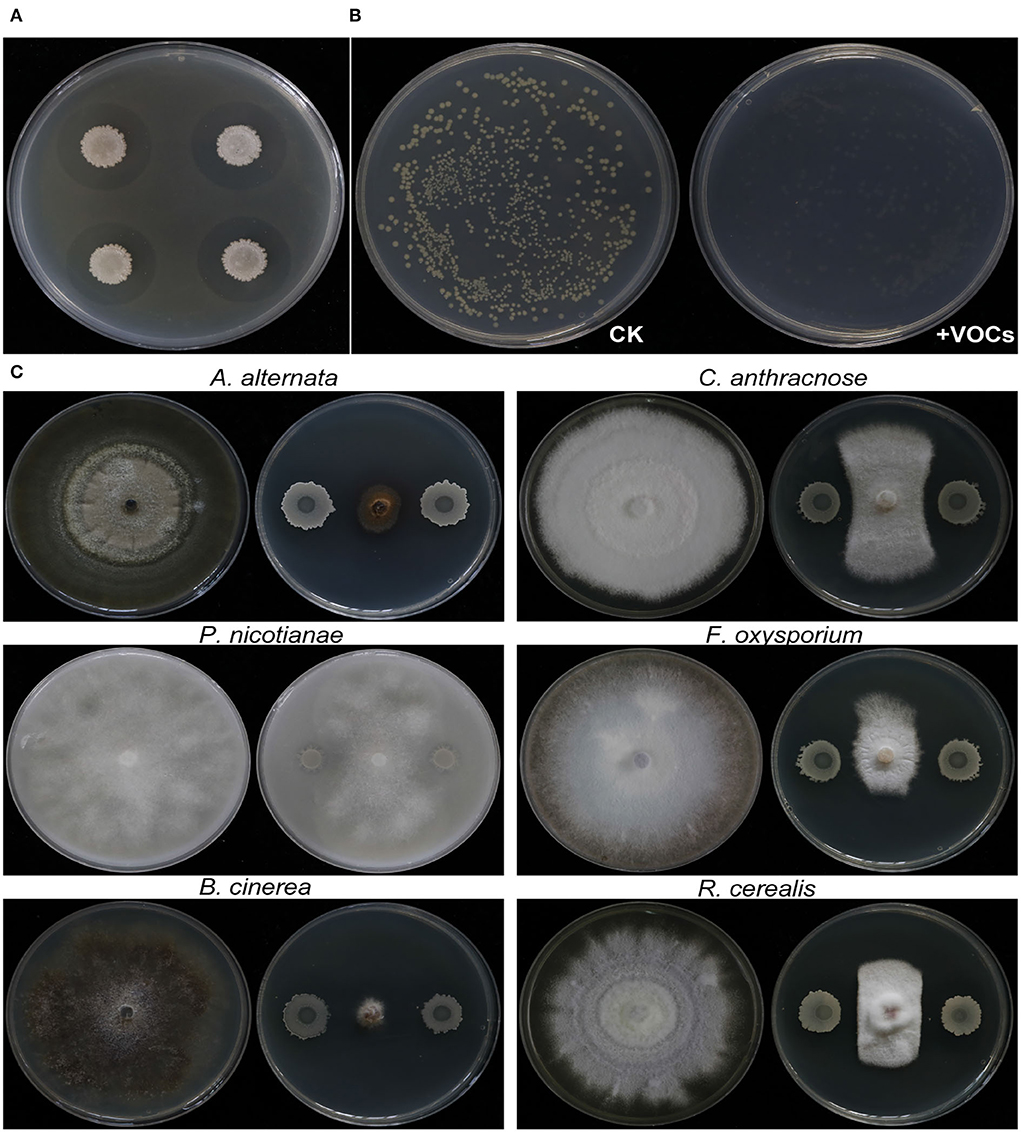
Figure 3. Antagonistic activity of strain EM-1 against R. solanacearum and antifungal activity against different pathogenic fungi. (A) Antagonistic activity of strain EM-1 against R. solanacearum. (B) Effects of VOCs produced by strain EM-1 against R. solanacearum. (C) Antifungal activity of strain EM-1 against different pathogenic fungi.
Based on the qPCR result, the abundance of Rs in roots inoculated with the strain EM-1 was 1.03 × 107 copies/g, while that in the control was 7.8 × 107 copies/g, which was up to 8-fold higher than that in the strain EM-1-treated group (Figure 4A). This result suggested that the strain EM-1 significantly reduces the colonization of Rs in tobacco roots.
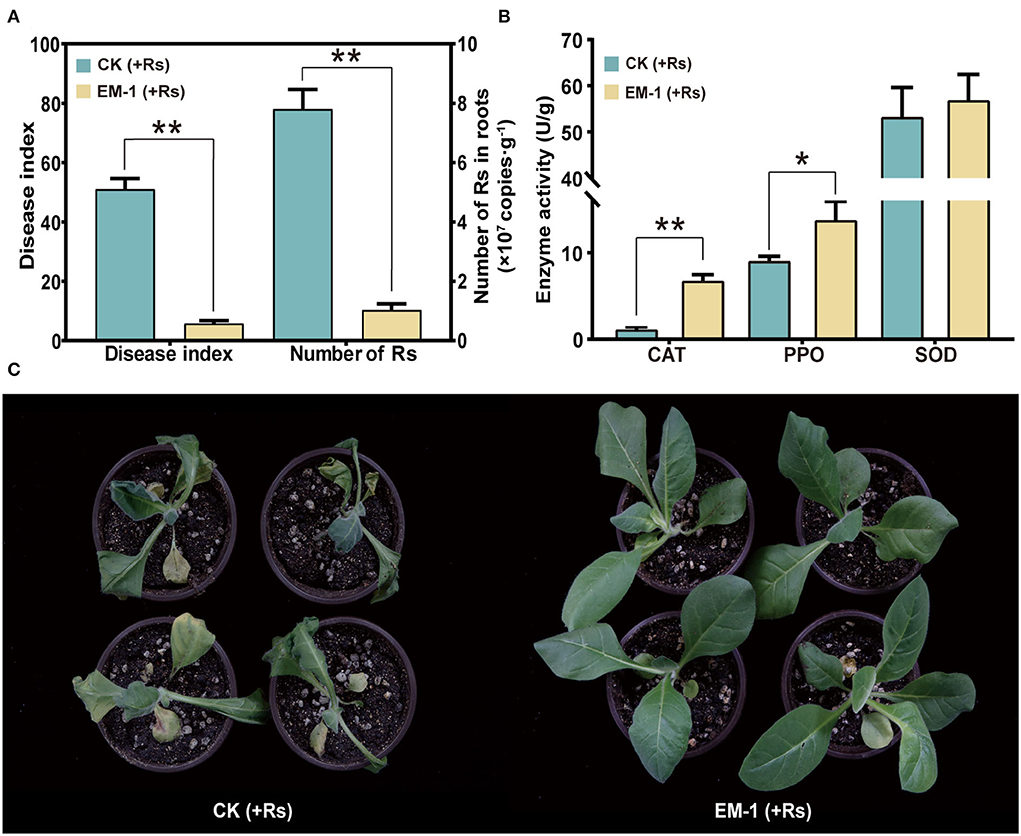
Figure 4. Defense-related enzyme activity and biocontrol effect of strain EM-1 on R. solanacearum. (A) Disease index of strain EM-1 against tobacco bacterial wilt and the absolute abundance of R. solanacearum in root samples. (B) Defense-related enzyme activity in leaves. Asterisks indicate a significant difference between the control and EM-1-treated plants (t-test, *p < 0.05, **p < 0.001). (C) Disease development symptoms in leaves observed at 5 days post-inoculation with R. solanacearum in control and EM-1-treated tobacco.
After 3 days of Rs inoculation, the symptoms of wilt gradually appeared on the control plants. On the 5th day, all the control plants were infected and accompanied by varying degrees of wilting (Figure 4C), reaching a disease index of 51.03. By contrast, a small amount of tobacco plants treated with the strain EM-1 showed disease symptoms, with the disease index of 5.8 (Figure 4A). These results indicated that the strain EM-1 has potential to control tobacco wilt disease.
To analyze the impact of inoculating EM-1 on tobacco resistance to the Rs pathogen, SOD, CAT, and PPO activities in the leaf samples were measured. Compared with the control group, SOD activity had no significant difference, while CAT and PPO activities of the EM-1 treatment were significantly increased (Figure 4B). These results indicate that the application of the strain EM-1 could induce the activity of defense-related enzymes in tobacco seedlings.
A preliminary experiment indicated that the cell-free supernatant of EM-1 can effectively inhibit the growth of Rs. The results showed that the inhibition zone diameter produced by the cell-free supernatant (50 mg/ml) is 2.18 ± 0.14 cm (Figure 5A), which indicated that the biocontrol ability of the strain EM-1 could be partially attributed to the production of extracellular bioactive substances.
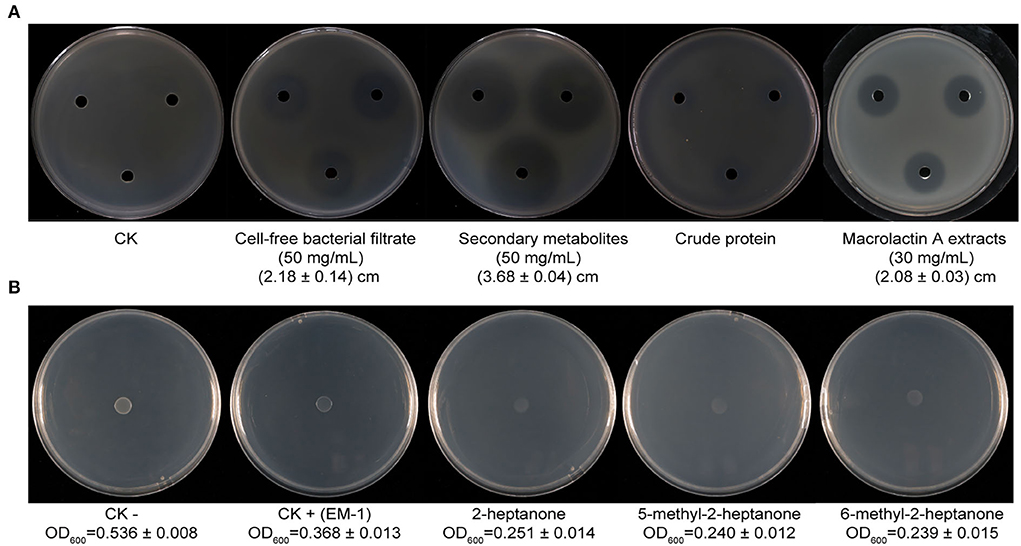
Figure 5. Inhibition of bacterial metabolites of strain EM-1 and VOC standards against R. solanacearum in vitro. (A) Inhibition of bacterial metabolites of strain EM-1 against R. solanacearum. (B) Inhibition of VOC standards against R. solanacearum.
The secondary metabolites and proteins produced by EM-1 were further extracted, and their inhibition ability against Rs was assayed. The results showed that the secondary metabolites (50 mg/ml) have a significant inhibitory effect on Rs with the inhibition zone diameter of 3.68 ± 0.04 cm, which is larger than those produced by the simple cell-free culture filtrates (50 mg/ml, 2.18 ± 0.14 cm). However, there is only weak inhibition zone produced by crude protein extracts (Figure 5A). Thus, it can be concluded that the secondary metabolites may play a major role on the antagonistic ability of EM-1 against Rs.
The chromatograms of LC-MS analyses are shown in Supplementary Figure 2, and the prominent masses from the LC-MS spectra are listed in Table 1. The spectra of compounds revealed that compound A with a molecular mass (m/z) of 1,043.56 Da [M + H]+ might be iturin A (C14; Supplementary Figure 2B), compounds C–E with an m/z of 1,057.57 Da [M + H]+ might be iturin A (C15; Supplementary Figures 2D–F), and compound F with an m/z of 1,071.59 Da[M + H]+ might be iturin A (C16; Supplementary Figure 2G). The three substances with m/z values of 1,043.56, 1,057.57, and 1,071.59 Da exhibited a 14-Da (–CH2) difference in molecular weights, the same difference previously reported for iturin A (Yuan et al., 2012b). Compounds C–E with the same molecular mass might be isomers of iturin A (C15); two other compounds B and G with molecular masses of 687.3 Da and 425.23 Da [M + Na]+ were found to be macrolactin W (Supplementary Figure 2C) and macrolactin A (Supplementary Figure 2H), respectively. The polyketides were extracted and identified as macrolactin A by the UV absorbance spectrum and LC-MS/MS (Supplementary Figure 3). The macrolactin A extracts (30 mg/ml) showed an inhibitory effect on Rs with an inhibition zone diameter of 2.08 ± 0.03 cm (Figure 5A). In addition, the iturin A standard did not show an inhibitory effect on Rs at the tested concentration (10, 5, and 1 mg/ml) (Supplementary Figure 4), indicating that macrolactins might be the main active substances against Rs.
A total of 23 different substances were detected as VOCs of EM-1 by GC-MS analysis (Table 2). Among them, 2-heptanone (27.94%), 6-methyl-2-heptanone (12.66%), and 5-methyl-2-heptanone (10.82%) were the main components, representing approximately 51% of total VOCs. The standard chemicals of the three components were purchased and tested for their fumigation effect on Rs. The VOC standards exhibited significant antibacterial activity against Rs, where the cell densities of Rs exposed to 2-heptanone, 6-methyl-2-heptanone, or 5-methyl-2-heptanone were OD600 = 0.251 ± 0.014, OD600 = 0.239 ± 0.015, and OD600 = 0.240 ± 0.012, respectively, while cell densities of the negative and positive controls were OD600 = 0.536 ± 0.008 and OD600 = 0.368 ± 0.013, respectively (Supplementary Figure 5).
To investigate the genetic basis of the antimicrobial phenotypes of the strain EM-1, we further analyzed its complete genome (Figure 6). The genome of B. velezensis EM-1 consisted of a circular chromosome of 3,929,796 bp in size, with an average GC content of 46.5%. Further analysis predicted that the chromosome contains 4,026 coding DNA sequences (CDSs), 86 tRNAs, 27 rRNAs, and nine sRNAs (Supplementary Table 2).
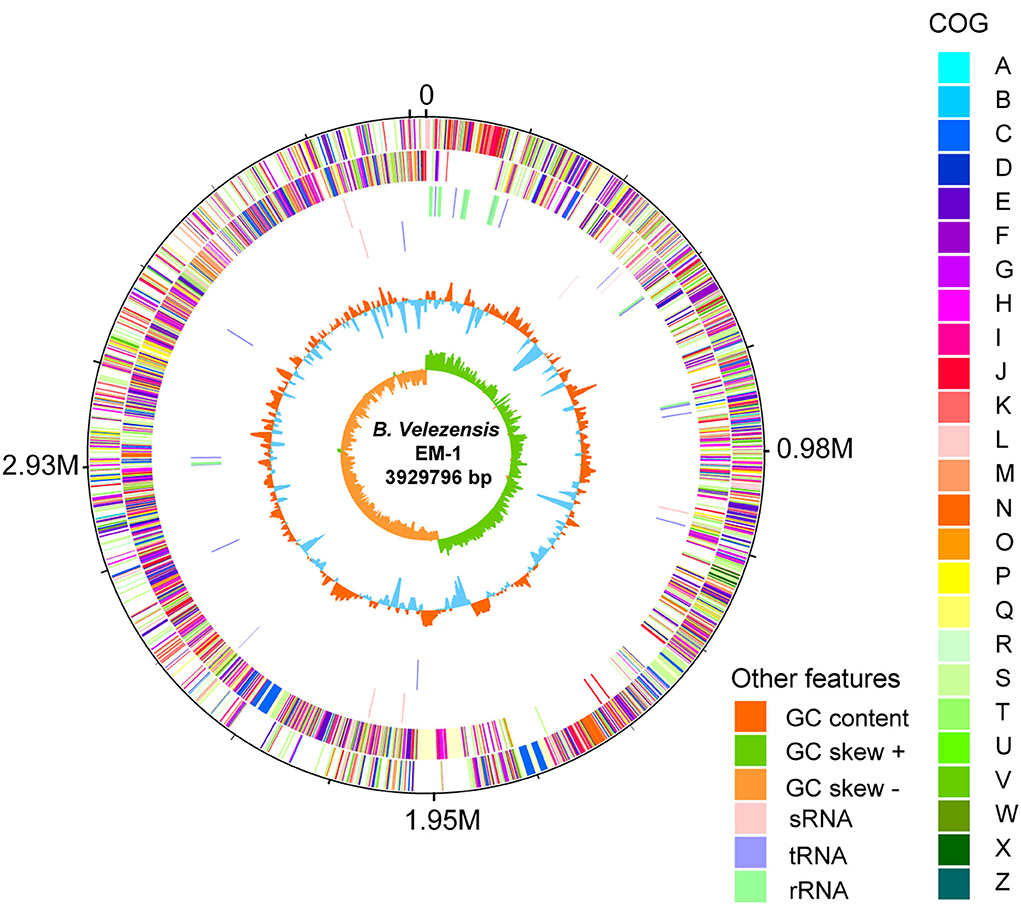
Figure 6. Circular representation of B. velezensis EM-1 genome. The outer scale is in mega bases (M). Circle 1 (from outside to inside): the marker of genome size. Circle 2 and 3: CDS with positive and negative chains; different colors represent different functional classifications. Circle 4 and 5: sRNA, tRNA, and rRNA with positive and negative chains. Circle 6: GC content; higher values are redder, and lower are bluer. Circle 7: the GC skew value; the algorithm is (G – C)/(G + C). Generally, when GC skew > 0, the histogram is outward and expressed in green; when GC skew < 0, the histogram is inward and expressed in orange.
Based on COG analysis (Tatusov et al., 2003), 2,900 proteins were classified into 25 different functional categories. Among them, 42.97% of proteins were associated with metabolism including “energy production and conversion (5.21%),” “amino acid transport and metabolism (9.08%),” “nucleotide transport and metabolism (2.61%),” “carbohydrate transport and metabolism (7.97%),” “coenzyme transport and metabolism (5.48%),” “lipid transport and metabolism (4.10%),” “inorganic ion transport and metabolism (5.21%),” and “secondary metabolites biosynthesis, transport, and catabolism (3.30%)” (Supplementary Figure 6).
Bacillus species has potential to produce secondary metabolites with a wide structural variability that exhibit strong antibacterial and antifungal activity (Sansinenea and Ortiz, 2011). Approximately 18.43% of B. velezensis EM-1 genome encodes for genes involved in the production of secondary metabolites with antimicrobial properties. Overall, 12 secondary metabolite biosynthesis gene clusters were identified (Table 3), including surfactin, butirosin, macrolactin, bacillaene, fengycin, difficidin, bacillibactin, bacilysin, and some unknown substances. Cluster 5 displayed high similarity to the macrolactin biosynthetic gene cluster. Macrolactin has been correlated with the gene organization of the mln operon, which comprises nine genes mlnA–I located in cluster 5 (Figure 7A). Interestingly, the biosynthetic cluster of iturin A was not found using antiSMASH. Based on in-depth analysis of the prediction results, cluster 7 had two core operons: one operon showed high sequence similarity to the fengycin operon of Bacillus amyloliquefaciens FZB42, and the other operon showed high sequence similarity to the iturin A operon of B. subtilis RB14 (Supplementary Table 3). Iturin A has been correlated with the gene organization of the itu operon, which comprises four genes ituA–D located in cluster 7 (Figure 7B).
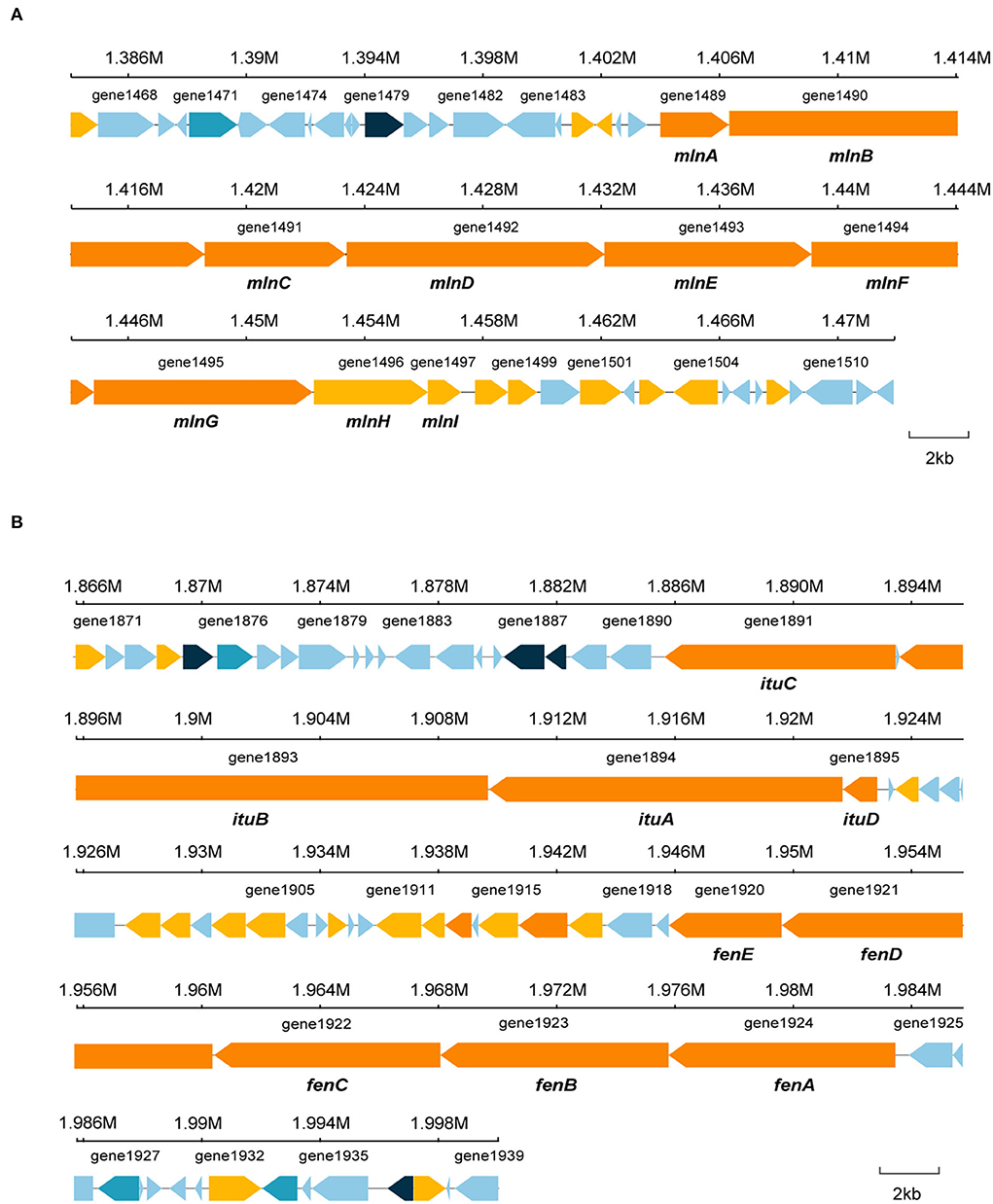
Figure 7. Specific gene structures in cluster 5 and cluster 7. (A) Secondary metabolite gene in cluster 5. (B) Secondary metabolite gene in cluster 7.
The microbe–microbe and microbe–root interactions are very complex, which result in different microbial fitness, population dynamics, and functional capacities in the rhizosphere (Banerjee et al., 2018). However, the majority of microbes used as biocontrol agents have been obtained by conventional methods such as isolation, identification, and evaluation. Recently, the core microbiome has been suggested to play a crucial role in an ecosystem; thus, the keystone species can be a promising way to explore effective biocontrol microbes. In our previous study, three Pseudomonas strains were identified as the keystone species in suppressive soil and/or healthy plants (Zheng et al., 2021) and showed multiple functions in disease control (Shang et al., 2021). Although the field control effect needs to be further confirmed, these results have indicated that the strategy based on keystone species exploring is effective in finding suitable biocontrol agents. More microbiome-based bioagents will be applied in future. In this study, B. velezensis EM-1 associated with suppressive rhizosphere soil microbiome was screened.
Recently, there is increasing research and application of B. velezensis in agriculture. Due to its pronounced ability for disease prevention and plant growth promotion, this species has been widely reported for plant disease biocontrol, including potato late blight (Yan et al., 2021), cucumber Fusarium wilt (Luo et al., 2019), lotus rot (Wang et al., 2020), tomato bacterial wilt (Agarwal et al., 2020), rice blast (Jing et al., 2020), and other plant diseases. However, there are few reports on the use of B. velezensis to control tobacco bacterial wilt. The current study presents a new B. velezensis strain EM-1 with potential for tobacco bacterial wilt biocontrol. EM-1 inoculation can significantly reduce the root colonization of Rs and decrease the disease severity of bacterial wilt. Moreover, EM-1 also exhibited inhibitory activity against other fungal and oomycete pathogens. The broad-spectrum antagonistic activity can be beneficial for its biocontrol ability since many pathogens often coexist in the same niche (Chávez-Ramírez et al., 2020).
Currently, the biocontrol mechanism of B. velezensis reported mainly include plant growth promotion (PGP), induced system resistance, and antibiotic production (Azabou et al., 2020; Guo et al., 2020; Xu et al., 2021); one antagonist will often exhibit more than one biocontrol mechanism. For instance, strain B. velezensis C2 can effectively control Verticillium wilt disease not only by antifungal activity but also associating its PGP traits such as siderophore and indole-3-acetic acid production and inorganic phosphate solubilization (Dhouib et al., 2019). As reported previously, the strain EM-1 also showed multiple biocontrol abilities, including significant inhibitory activity on Rs, fumigation effect, lipopeptides, and polyketide production and induced resistance of tobacco. These traits can contribute to disease controlling ability directly, indirectly, or synergistically. Here, we pay more attention on the inhibitory substances produced by EM-1.
Previous reports indicated that antimicrobial substances produced by B. velezensis mainly consisted of proteins (Guo et al., 2020), lipopeptides, and polyketide antibiotics, which are synthesized by non-ribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs) (Rabbee et al., 2019; Liu et al., 2020). Our results indicated that secondary metabolites were the main inhibitory substances secreted by EM-1, which showed a significant inhibition effect on Rs even at a concentration of 50 mg/ml. They were further identified as iturin A (C14, C15, and C16), macrolactin A, and macrolactin W. To investigate the biosynthetic pathway of iturin A and macrolactin, the whole-genome sequencing of the strain EM-1 was conducted in our study. A total of 12 biosynthetic gene clusters (BGCs) for secondary metabolite synthesis were predicted by antiSMASH. It revealed that cluster 5 in the strain EM-1 encoded a polyketide synthase (PKS), and nine core genes in this cluster were proposed to be responsible for macrolactin biosynthesis. Cluster 7 encoded a hybrid modular polyketide synthase–non-ribosomal peptide synthetase (PKS-NRPS), and four core genes in this cluster were proposed to be responsible for iturin A biosynthesis. Although cluster 7 was predicted to produce “fengycin,” in-depth mining of the prediction results showed that cluster 7 has two core operons, where one has been implicated in regulating fengycin synthesis (Koumoutsi et al., 2004) and the other related to synthesis of iturin A (Tsuge et al., 2001). As outlined previously, the iturin A standard had no inhibitory effect on Rs growth, which was consistent with the finding that iturin A was often related to antifungal activity (Arrebola et al., 2010). It has been reported that macrolactin family compounds are effective bacteriostatic antibiotics that inhibit a number of Gram-positive bacterial pathogens (Yuan et al., 2014). The aforementioned results of this study have also demonstrated that macrolactin A extracts could inhibit the growth of Rs. Polyketides, but not iturins, might be the main active substances in CFs of EM-1 against Rs.
Several B. velezensis strains have been found to have a fumigation effect on phytopathogens (Lim et al., 2017; Myo et al., 2019; Zhang et al., 2021). The potent active compounds include benzaldehyde, diacetyl, pyrazine (2,5-dimethyl), benzothiazole, phenol (4-chloro-3-methyl), and 2,4-dimethyl-6-tert-butylphenol (Gao et al., 2017; Calvo et al., 2020; Li et al., 2020). However, these reports were mainly concerned with the antifungal activity of VOCs produced by B. velezensis. The components against bacterial pathogens are not elucidated yet. The current study identified three compounds (2-heptanone, 6-methyl-2-heptanone, and 5-methyl-2-heptanone) from VOCs of EM-1, which showed a significant fumigation effect on Rs. Among them, 6-methyl-2-heptanone was also detected in the VOCs of B. velezensis C16 and exhibited antifungal activity against Alternaria solani (Zhang et al., 2021). To the best of our knowledge, this is the first report of the three volatile compounds having an inhibitory effect on Rs. A comprehensive evaluation of these compounds is definitely worth exploring.
In summary, the strain B. velezensis EM-1 associated with suppressive rhizosphere soil microbes showed high potential for tobacco bacterial wilt control. Antibiotic production, fumigation effect, and induced resistance of tobacco appeared to be involved in its biocontrol mechanisms. Polyketides and VOCs might be the main active substances against Rs, and 2-heptanone, 6-methyl-2-heptanone, and 5-methyl-2-heptanone were identified as promising volatile compounds. Overall, these results indicated that EM-1 can act as a biocontrol strain for tobacco bacterial wilt control. Future work should identify the effective polyketides and evaluate the biocontrol effect of EM-1 under field conditions.
The data presented in the study are deposited in the https://www.ncbi.nlm.nih.gov/genbank repository, accession number CP095842.
CZ, CM, and YZ provided the experimental ideas and the design of this study. XS performed the experiment and wrote the manuscript with assistance of XH and JC. YL and YY helped analyze the data and revise the manuscript. JG helped revise the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by Key Science and Technology Projects of China National Tobacco Corporation (110201902003), Science and Technology Project of Guizhou Tobacco Corporation (201809), National Natural Science Foundation of China (32101289), and the Agricultural Science and Technology Project of Southwest Guizhou [110202101057(LS-17)].
Authors XH and JG were employed by Guizhou Tobacco Company, Zunyi.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.940156/full#supplementary-material
Supplementary Figure 1. Percentage inhibition of strain EM-1 against different pathogenic fungi. Percentage inhibition, relative to the control, of six plant pathogenic fungi by strain EM-1. All experiments were independently performed in triplicate. Data represent the mean ± standard deviation of three replicates in each treatment group. Different letters indicate a significant difference (p < 0.05; Tukey's test) between the treatment and control.
Supplementary Figure 2. Secondary active metabolite compounds detected by LC-MS/MS. (A) LC chromatogram of the seven detected antimicrobial compounds. (B) Mass spectra of iturin A (C14). (C) Mass spectra of macrolactin W. (D) Mass spectra of iturin A (C15). (E) Mass spectra of iturin A (C15). (F) Mass spectra of iturin A (C15). (G) Mass spectra of iturin A (C16). (H) Mass spectra of macrolactin A. The mass spectra were all detected in the positive ion mode.
Supplementary Figure 3. LC-MS/MS spectrums of macrolactin A. (A) LC chromatogram of macrolactin A. (B) UV absorption spectra of macrolactin A. (C) MS/MS spectrums of macrolactin A.
Supplementary Figure 4. The inhibitory effect of iturin A standard on Rs.
Supplementary Figure 5. The cell density of the Rs after VOCs treatments.
Supplementary Figure 6. COG functional classification in B. velezensis EM-1 coding sequences.
Supplementary Table 1. Details of all Bacillus strains isolated from tobacco rhizosphere soil and their inhibition effect against R. solanacearum.
Supplementary Table 2. General genome feature of B. velezensis EM-1.
Supplementary Table 3. Comparison of core biosynthetic genes of B. velezensis EM-1 with known gene clusters in cluster 7.
Agarwal, H., Dowarah, B., Baruah, P. M., Bordoloi, K. S., Krishnatreya, D. B., and Agarwala, N. (2020). Endophytes from Gnetum gnemon L. can protect seedlings against the infection of phytopathogenic bacterium Ralstonia solanacearum as well as promote plant growth in tomato. Microbiol. Res. 238, 126503. doi: 10.1016/j.micres.2020.126503
Agler, M. T., Ruhe, J., Kroll, S., Morhenn, C., Kim, S. T., Weigel, D., et al. (2016). Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14, e1002352. doi: 10.1371/journal.pbio.1002352
Arrebola, E., Jacobs, R., and Korsten, L. (2010). Iturin A is the principal inhibitor in the biocontrol activity of Bacillus amyloliquefaciens PPCB004 against postharvest fungal pathogens. J. Appl. Microbiol. 108, 386–395. doi: 10.1111/j.1365-2672.2009.04438.x
Azabou, M. C., Gharbi, Y., Medhioub, I., Ennouri, K., Barham, H., Tounsi, S., et al. (2020). The endophytic strain Bacillus velezensis OEE1: an efficient biocontrol agent against Verticillium wilt of olive and a potential plant growth promoting bacteria. Biol. Control. 142, 104168. doi: 10.1016/j.biocontrol.2019.104168
Bai, W., Kong, F., Lin, Y., and Zhang, C. (2016). Extract of Syringa oblata: a new biocontrol agent against tobacco bacterial wilt caused by Ralstonia solanacearum. Pestic. Biochem. Physiol. 134, 79–83. doi: 10.1016/j.pestbp.2016.04.002
Banerjee, S., Schlaeppi, K., and Heijden, M. G. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576. doi: 10.1038/s41579-018-0024-1
Calvo, H., Mendiara, I., Arias, E., Gracia, A. P., Blanco, D., and Venturini, M. E. (2020). Antifungal activity of the volatile organic compounds produced by Bacillus velezensis strains against postharvest fungal pathogens. Postharvest Biol. Technol. 166, 111208. doi: 10.1016/j.postharvbio.2020.111208
Chávez-Ramírez, B., Kerber-Díaz, J. C., Acoltzi-Conde, M. C., Ibarra, J. A., Vásquez-Murrieta, M. S., and Santos, E. D. L. (2020). Inhibition of Rhizoctonia solani RhCh-14 and Pythium ultimum PyFr-14 by Paenibacillus polymyxa NMA1017 and Burkholderia cenocepacia CACua-24: a proposal for biocontrol of phytopathogenic fungi. Microbiol. Res. 230, 126347. doi: 10.1016/j.micres.2019.126347
Chen, M., Wang, J., Liu, B., Zhu, Y., Xiao, R., Yang, W., et al. (2020). Biocontrol of tomato bacterial wilt by the new strain Bacillus velezensis FJAT-46737 and its lipopeptides. BMC Microbiol. 20, 160. doi: 10.1186/s12866-020-01851-2
Cunnac, S., Occhialini, A., Barberis, P., Boucher, C., and Genin, S. (2004). Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53, 115–128. doi: 10.1111/j.1365-2958.2004.04118.x
Dhouib, H., Zouari, I., Abdallah, D. B., Belbahri, L., Taktak, W., Triki, M. A., et al. (2019). Potential of a novel endophytic Bacillus velezensis in tomato growth promotion and protection against Verticillium wilt disease. Biol. Control. 139, 104092. doi: 10.1016/j.biocontrol.2019.104092
Ezrari, S., Mhidra, O., Radouane, N., Tahiri, A., Polizzi, G., Lazraq, A., et al. (2021). Potential role of rhizobacteria isolated from citrus rhizosphere for biological control of citrus dry root rot. Plants 10, 872. doi: 10.3390/plants10050872
Gao, S., Wu, H., Wang, W., Yang, Y., Xie, S., Xie, Y., et al. (2013). Efficient colonization and harpins mediated enhancement in growth and biocontrol of wilt disease in tomato by Bacillus subtilis. Lett. Appl. Microbiol. 57, 526–533. doi: 10.1111/lam.12144
Gao, Z., Zhang, B., Liu, H., Han, J., and Zhang, Y. (2017). Identification of endophytic Bacillus velezensis ZSY-1 strain and antifungal activity of its volatile compounds against Alternaria solani and Botrytis cinerea. Biol. Control. 105, 27–39. doi: 10.1016/j.biocontrol.2016.11.007
Genin, S. (2010). Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytol. 187, 920–928. doi: 10.1111/j.1469-8137.2010.03397.x
Guo, D., Yuan, C., Luo, Y., Chen, Y., Lu, M., Chen, G., et al. (2020). Biocontrol of tobacco black shank disease (Phytophthora nicotianae) by Bacillus velezensis Ba168. Pestic. Biochem. Physiol. 165, 104523. doi: 10.1016/j.pestbp.2020.01.004
Jiang, G., Wei, Z., Xu, J., Chen, H., Zhang, Y., She, X., et al. (2017). Bacterial wilt in China: history, current status, and future perspectives. Front. Plant Sci. 8, 1549. doi: 10.3389/fpls.2017.01549
Jing, R., Li, N., Wang, W., and Liu, Y. (2020). An endophytic strain JK of genus Bacillus isolated from the seeds of super hybrid rice (Oryza sativa L., Shenliangyou 5814) has antagonistic activity against rice blast pathogen. Microb. Pathog. 147, 104422. doi: 10.1016/j.micpath.2020.104422
Ju, R., Zhao, Y., Li, J., Jiang, H., Liu, P., Yang, T., et al. (2014). Identification and evaluation of a potential biocontrol agent, Bacillus subtilis, against Fusarium sp. in apple seedlings. Ann. Microbiol. 64, 377–383. doi: 10.1007/s13213-013-0672-3
Khanh, T. L. V., Tan, L. N., Thi, M. L., Thi, M. P., and Hai, T. L. (2020). Selecting Bacillus spp. antagonist of fungal phytopathogen phytophthora infestans causing tomato late blight. Annu. Res. Rev. Biol. 35, 32–40. doi: 10.9734/arrb/2020/v35i1230308
Koumoutsi, A., Chen, X-H., Henne, A., Liesegang, H., Hitzeroth, G., Franke, P., et al. (2004). Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 186, 1084–1096. doi: 10.1128/JB.186.4.1084-1096.2004
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kurniawan, E., Panphon, S., and Leelakriangsak, M. (2018). Potential of marine chitinolytic Bacillus isolates as biocontrol agents of phytopathogenic fungi. IOP Conf. Ser. Earth Environ. Sci. 217, 012044. doi: 10.1088/1755-1315/217/1/012044
Kwon, J. W., and Kim, S. D. (2014). Characterization of an antibiotic produced by Bacillus subtilis JW-1 that suppresses Ralstonia solanacearum. J. Microbiol. Biotechnol. 24, 13–18. doi: 10.4014/jmb.1308.08060
Le, X. T., Pham, D. T., Pham, T. A., Tran, T. T., Khuat, T. H., Le, H. Q., et al. (2019). Exploration of genetic diversity of Bacillus spp. from industrial shrimp ponds in Vietnam by multi-locus sequence typing. Fish. Aquat. Sci. 22, 17. doi: 10.1186/s41240-019-0132-5
Lebeis, S. L. (2014). The potential for give and take in plant-microbiome relationships. Front. Plant Sci. 5, 287. doi: 10.3389/fpls.2014.00287
Li, P., Luo, H., Meng, J., Sun, W., Wang, X., Lu, S., et al. (2014). Effects of oligosaccharides from endophytic Fusarium oxysporum Dzf17 on activities of defense-related enzymes in Dioscorea zingiberensis suspension cell and seedling cultures. Electron. J. Biotechnol. 17, 156–161. doi: 10.1016/j.ejbt.2014.04.012
Li, X., Wang, X., Shi, X., Wang, B., Li, M., Wang, Q., et al. (2020). Antifungal effect of volatile organic compounds from Bacillus velezensis CT32 against Verticillium dahliae and Fusarium oxysporum. Processes 8, 1674. doi: 10.3390/pr8121674
Lim, H. J., Lee, E. H., Yoon, Y., Chua, B., and Son, A. (2016). Portable lysis apparatus for rapid single-step DNA extraction of Bacillus subtilis. J. Appl. Microbiol. 120, 379–387. doi: 10.1111/jam.13011
Lim, S. M., Yoon, M. Y., Choi, G. J., Choi, Y. H., Jang, K. S., Shin, T. S., et al. (2017). Diffusible and volatile antifungal compounds produced by an antagonistic Bacillus velezensis G341 against various phytopathogenic fungi. Plant Pathol. J. 33, 488–489. doi: 10.5423/PPJ.OA.04.2017.0073
Ling, L., Han, X., Li, X., Zhang, X., Wang, H., Zhang, L., et al. (2020). A Streptomyces sp. NEAU-HV9: isolation, identification, and potential as a biocontrol agent against Ralstonia solanacearum of tomato plants. Microorganisms 8, 351. doi: 10.3390/microorganisms8030351
Liu, Y., Teng, K., Wang, T., Dong, E., Zhang, M., Tao, Y., et al. (2020). Antimicrobial Bacillus velezensis HC6: production of three kinds of lipopeptides and biocontrol potential in maize. J. Appl. Microbiol. 128, 242–254. doi: 10.1111/jam.14459
Luo, W., Liu, L., Qi, G., Yang, F., Shi, X., and Zhao, X. (2019). Embedding Bacillus velezensis NH-1 in microcapsules for biocontrol of cucumber Fusarium wilt. Appl. Environ. Microbiol. 85, e03128–e03118. doi: 10.1128/AEM.03128-18
Maji, S., and Chakrabartty, P. K. (2014). Biocontrol of bacterial wilt of tomato caused by Ralstonia solanacearum by isolates of plant growth promoting rhizobacteria. Aust. J. Crop Sci. 8, 208–214.
Mohamed, B. F., Sallam, N. M., Alamri, S. A., Abo-Elyousr, K. A., Mostafa, Y. S., and Hashem, M. (2020). Approving the biocontrol method of potato wilt caused by Ralstonia solanacearum (Smith) using Enterobacter cloacae PS14 and Trichoderma asperellum T34. Egypt. J. Biol. Pest Co. 30, 61. doi: 10.1186/s41938-020-00262-9
Mondol, M., Kim, J. H., Lee, H. S., Lee, Y. J., and Shin, H. J. (2011). Macrolactin W, a new antibacterial macrolide from a marine Bacillus sp. Bioorg. Med. Chem. Lett. 21, 3832–3835. doi: 10.1016/j.bmcl.2010.12.050
Myo, E. M., Liu, B., Ma, J., Shi, L., Jiang, M., Zhang, K., et al. (2019). Evaluation of Bacillus velezensis NKG-2 for bio-control activities against fungal diseases and potential plant growth promotion. Biol. Control. 134, 23–31. doi: 10.1016/j.biocontrol.2019.03.017
Nei, M., and Kumar, S. (2002). Molecular evolution and phylogenetics. Mol. Phylogenet. Evol. 25, 569–570. doi: 10.1016/S1055-7903(02)00245-2
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Qi, G., Ma, G., Chen, S., Lin, C., and Zhao, X. (2019). Microbial network and soil properties are changed in bacterial wilt-susceptible soil. Appl. Environ. Microbiol. 85, e00162–e00119. doi: 10.1128/AEM.00162-19
Rabbee, M., Ali, M., Choi, J., Hwang, B., Jeong, S., and Baek, K. (2019). Bacillus velezensis: a valuable member of bioactive molecules within plant microbiomes. Molecules 24, 1046. doi: 10.3390/molecules24061046
Romero-Tabarez, M., Jansen, R., Sylla, M., Lünsdorf, H., Häubler, S., Santosa, D., et al. (2006). 7-O-malonyl macrolactin A, a new macrolactin antibiotic from Bacillus subtilis active against methicillin-resistant Staphylococcus aureus, vancomycin-resistant enterococci, and a small-colony variant of Burkholderia cepacia. Antimicrob. Agents Chemother. 50, 1701–1709. doi: 10.1128/AAC.50.5.1701-1709.2006
Sansinenea, E., and Ortiz, A. (2011). Secondary metabolites of soil Bacillus spp. Biotechnol. Lett. 33, 1523–1538. doi: 10.1007/s10529-011-0617-5
Shang, X-,c., Cai, X., Zhou, Y., Han, X., Zhang, C-S., Ilyas, N., et al. (2021). Pseudomonas inoculation stimulates endophytic Azospira population and induces systemic resistance to bacterial wilt. Front. Plant Sci. 12, 738611. doi: 10.3389/fpls.2021.738611
Tahir, H. A. S., Gu, Q., Wu, H., Niu, Y., Huo, R., and Gao, X. (2017). Bacillus volatiles adversely affect the physiology and ultra-structure of Ralstonia solanacearum and induce systemic resistance in tobacco against bacterial wilt. Sci. Rep. 7, 40481. doi: 10.1038/srep40481
Tatusov, R. L., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Kiryutin, B., Koonin, E. V., et al. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinf. 4, 1–41. doi: 10.1186/1471-2105-4-41
Tsuge, K., Akiyama, T., and Shoda, M. (2001). Cloning, sequencing, and characterization of the iturin A operon. J. Bacteriol. 181, 6265–6273. doi: 10.1128/JB.183.21.6265-6273.2001
Wang, G., Meng, J., Tian, T., Xiao, X., Zhang, B., and Xiao, Y. (2020). Endophytic Bacillus velezensis strain B-36 is a potential biocontrol agent against lotus rot caused by Fusarium oxysporum. J. Appl. Microbiol. 128, 1153–1162. doi: 10.1111/jam.14542
Wang, L., Dou, G., Guo, H., Zhang, Q., Qin, X., Yu, W., et al. (2019). Volatile organic compounds of Hanseniaspora uvarum increase strawberry fruit flavor and defense during cold storage. Food Sci. Nutr. 7, 2625–2635. doi: 10.1002/fsn3.1116
Wang, X., Han, X., Wang, S., Wang, Y., Wang, P., Zhao, Z., et al. (2022). Extraction of honokiol from Artemisia argyi and in vitro and in vivo investigation of its antifungal activity. Nat. Prod. Res. doi: 10.1080/14786419.2022.2071887
Wu, B., Wang, X., Yang, L., Yang, H., Zeng, H., Qiu, Y., et al. (2016). Effects of Bacillus amyloliquefaciens ZM9 on bacterial wilt and rhizosphere microbial communities of tobacco. Appl. Soil Ecol. 103, 1–12. doi: 10.1016/j.apsoil.2016.03.002
Wu, J., Abbas, H. M. K., Li, J., Yuan, Y., Liu, Y., Wang, G., et al. (2020). Cell membrane-interrupting antimicrobial peptides from Isatis indigotica fortune isolated by a Bacillus subtilis expression system. Biomolecules 10, 30. doi: 10.3390/biom10010030
Xie, Z., Li, M., Wang, D., Wang, F., Shen, H., Sun, G., et al. (2021). Biocontrol efficacy of Bacillus siamensis LZ88 against brown spot disease of tobacco caused by Alternaria alternata. Biol. Control. 154, 104508. doi: 10.1016/j.biocontrol.2020.104508
Xu, Y., Wang, L., Liang, W., and Liu, M. (2021). Biocontrol potential of endophytic Bacillus velezensis strain QSE-21 against postharvest grey mould of fruit. Biol. Control. 161, 104711. doi: 10.1016/j.biocontrol.2021.104711
Yan, H., Qiu, Y., Yang, S., Wang, Y., Wang, K., Jiang, L., et al. (2021). Antagonistic activity of Bacillus velezensis SDTB038 against Phytophthora infestans in potato. Plant Dis. 105, 1738–1747. doi: 10.1094/PDIS-08-20-1666-RE
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
You, C., Zhang, C., Feng, C., Wang, J., and Kong, F. (2015). Myroides odoratimimus, a biocontrol agent from the rhizosphere of tobacco with potential to control Alternaria alternata. Biocontrol 60, 555–564. doi: 10.1007/s10526-015-9654-7
Yuan, J., Li, B., Zhang, N., Waseem, R., Shen, Q., and Huang, Q. (2012a). Production of bacillomycin- and macrolactin-type antibiotics by Bacillus amyloliquefaciens NJN-6 for suppressing soilborne plant pathogens. J. Agric. Food Chem. 60, 2976–2981. doi: 10.1021/jf204868z
Yuan, J., Raza, W., Huang, Q., and Shen, Q. (2012b). The ultrasound-assisted extraction and identification of antifungal substances from B. amyloliquefaciens strain NJN-6 suppressing Fusarium oxysporum. J. Basic Microbiol. 52, 1–10. doi: 10.1002/jobm.201100560
Yuan, J., Zhang, F., Wu, Y., Zhang, J., Raza, W., Shen, Q., et al. (2014). Recovery of several cell pellet-associated antibiotics produced by Bacillus amyloliquefaciens NJN-6. Lett. Appl. Microbiol. 59, 169–176. doi: 10.1111/lam.12260
Zhang, C., Gao, J., Han, T., Tian, X., and Wang, F. (2017). Integrated control of tobacco black shank by combined use of riboflavin and Bacillus subtilis strain Tpb55. BioControl 62, 835–845. doi: 10.1007/s10526-017-9849-1
Zhang, D., Yu, S., Zhao, D., Zhang, J., Pan, Y., Yang, Y., et al. (2021). Inhibitory effects of non-volatiles lipopeptides and volatiles ketones metabolites secreted by Bacillus velezensis C16 against Alternaria solani. Biol. Control. 152, 104421. doi: 10.1016/j.biocontrol.2020.104421
Zheng, Y., Han, X., Zhao, D., Wei, K., Yuan, Y., Li, Y., et al. (2021). Exploring biocontrol agents from microbial keystone taxa associated to suppressive soil: a new attempt for a biocontrol strategy. Front. Plant Sci. 12, 655673. doi: 10.3389/fpls.2021.655673
Keywords: Ralstonia solanacearum, biocontrol agent, volatile compounds, lipopeptides, polyketides, induced resistance
Citation: Sui X, Han X, Cao J, Li Y, Yuan Y, Gou J, Zheng Y, Meng C and Zhang C (2022) Biocontrol potential of Bacillus velezensis EM-1 associated with suppressive rhizosphere soil microbes against tobacco bacterial wilt. Front. Microbiol. 13:940156. doi: 10.3389/fmicb.2022.940156
Received: 10 May 2022; Accepted: 29 July 2022;
Published: 23 August 2022.
Edited by:
Gustavo Cordero-Bueso, University of Cádiz, SpainReviewed by:
Lin Chen, Chinese Academy of Forestry, ChinaCopyright © 2022 Sui, Han, Cao, Li, Yuan, Gou, Zheng, Meng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanfen Zheng, emhlbmd5YW5mZW5AY2Fhcy5jbg==; Chen Meng, bWVuZ2NoZW4wMUBjYWFzLmNu; Chengsheng Zhang, emhhbmdjaGVuZ3NoZW5nQGNhYXMuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.