
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 26 October 2022
Sec. Aquatic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.933413
The potentially deadly and sporadic diarrhea-causing agent, Vibrio cholerae, is present in a great number in the freshwater aquatic environment and can be transmitted to humans by different aquatic organisms. In the perspective of Bangladesh, an anadromous fish species Hilsha (Tenualosa ilisha) can act as a transmission vehicle of V. cholerae from the aquatic to the household kitchen environment. The present study was carried out to investigate the presence of V. cholerae in the aquatic habitat of Bangladesh with a major emphasis on freshly caught Hilsha fish, along with river water and plankton samples from the fish capture site. The study also detected the biofilm formation capability of V. cholerae within Hilsha fish that might help the transmission and persistence of the pathogen in aquatic habitat. Twenty out of 65 freshly caught fish (30.8%) and 1 out of 15 water samples (6.67%) showed the presence of V. cholerae and none of the plankton samples were positive for V. cholerae. The isolated strains were identified as non-O1 and non-O139 serogroups of V. cholerae and contain some major toxin and virulence genes. A few strains showed cellular cytotoxicity on the HeLa cell line. All strains were able to form biofilm on the microtiter plate and the detection of three genes related to biofilm formation (vpsA, vpsL, and vpsR) were also assayed using qPCR. In this study, the in vitro biofilm formation ability of the isolated strains may indicate the long-term persistence of V. cholerae in different parts of Hilsha fish. The abundance of V. cholerae only in freshly caught Hilsha fish and the absence of the pathogen in the surrounding aquatic environment could stipulate the role of Hilsha fish as one of the major transmission routes of V. cholerae from the freshwater aquatic environment of Bangladesh to the household kitchen environment.
Vibrio cholerae is a well-known human pathogen that causes severe watery diarrhea and dehydration and can lead to death if left untreated. Its role in causing and spreading infection has been investigated for over many years. V. cholerae is an indigenous member of the aquatic environment, including both marine, and freshwaters (Schuster et al., 2011). Its transmission from an aquatic environment to a healthy individual is studied widely and some studies proved that aquatic organisms, including planktons, can serve as a reservoir and vector of this pathogen (Colwell et al., 1985). Fishes that use plankton as a food source may also become a reservoir of V. cholerae (Halpern et al., 2008). In Bangladesh, intrusion of saline water from the Bay of Bengal into the river water of the coastal region during the dry season may influence the growth and transmission of V. cholerae (Lipp et al., 2002; Akanda et al., 2011; Hossain et al., 2018).
Cholera epidemics have been seen in many parts of the world, including Bangladesh, with seasonal regularity. Seasonal outbreaks of V. cholerae were found associated with planktonic and zooplanktonic blooms, which were influenced by water, temperature, the hour of sunlight, salinity, rainfall, sea surface height, etc. (Lobitz et al., 2000). In Bangladesh, two bimodal seasonal cholera peaks are observed in the early summer (March–May) and after the monsoon (September–November). Every year hundreds of cholera cases are originated from the southern coastal area of Bangladesh and this area has been considered an endemic region for cholera. It is assumed that aquatic environmental conditions (pH, salinity, plankton bloom, and water temperature) of the Bay of Bengal favor the growth and survival of epidemic strains of V. cholerae (Lipp et al., 2002; Huq et al., 2005; Akanda et al., 2011).
Fishes act as a natural reservoir and vector for V. cholerae (Halpern and Izhaki, 2017), and V. cholerae already has been isolated from marine fishes (Scheelbeek et al., 2009; Senderovich et al., 2010; Kumar and Lalitha, 2013), frozen fish stocks (Acharjee et al., 2012), guts of Zebrafish (Runft et al., 2014), intestines of Tilapia (Traoré et al., 2014), gills of Tilapia (Hounmanou et al., 2016), filets of processed Pangasius fish, and also from the water used to rinse them (Thi et al., 2014). In Japan, non-O1 strains of V. cholerae were identified from diseased Ayu fish, which is one of the important species of Japanese anadromous fish (Kiiyukia et al., 1992). A recent study by Hounmanou et al. (2019) showed that both toxigenic and non-toxigenic strains of V. cholerae are able to colonize in the intestine of Tilapia (Hounmanou et al., 2016) and Zebrafish (Runft et al., 2014) and horizontally transfer to naïve Tilapia fish. A previous study in Bangladesh revealed that Hilsha fish (Tenualosa ilisha), which are an inhabitant of the coastal region, can serve as a vector and reservoir for V. cholerae (Hossain et al., 2018).
The migration pattern of Hilsha (Tenualosa ilisha) is widely studied in the southeast region of Asia. Hilsha migrates from the Bay of Bengal for spawning (Hora, 1938; Pillay, 1958) during the monsoon, grows in freshwater, and returns to the ocean (Ahsan et al., 2014; Bhaumik, 2015; Miah, 2015). In Bangladesh, the spawning migration toward estuaries and rivers starts in July and ends in October (Bhaumik, 2015) and the peak period of breeding is from September to October (Miah, 2015). Due to their unique life cycle, Hilsha fish travel from the Bay of Bengal to freshwater rivers (Dutta and Hazra, 2017), and persistent bacterium within fish may also be transmitted to the freshwater environment.
V. cholerae persists in the aquatic environment of both endemic and non-endemic regions and this persistence is enhanced by the biofilm formation capability on various biotic and abiotic surfaces (Lutz et al., 2013). Biofilm helps the bacteria to stick with one another and also on a solid surface (López et al., 2010). For aquatic bacteria, surface attachment helps to access nutrients at the liquid-surface interface and thus helps the bacteria to survive in the nutrient-limited natural environment (Dawson et al., 1981). Examples of the biotic surfaces in the aquatic environment are ship hulls (Shikuma and Hadfield, 2010), zooplankton (Tamplin et al., 1990; Epstein, 1993; Huq et al., 2005; Turner et al., 2009), microalgae (Hood and Winter, 1997), and floating aggregates (Alam et al., 2006). In aquatic habitats, V. cholerae is capable of attaching to live copepods and their egg sacs (Huq et al., 1983), freshly harvested oyster samples from estuarine (Twedt et al., 1981), internal organs of fishes, for example, digestive tract (Alam et al., 2007; Senderovich et al., 2010), outer scale sample (Du Preez et al., 2010), etc. It has been also suggested that the association of V. cholerae with biotic surfaces increases the survival rate of V. cholerae in the aquatic environment (Huq et al., 1983).
Toxigenic strains of V. cholerae can cause explosive outbreaks when introduced into populations with poor sanitary infrastructure (O’Connor et al., 2011; Barzilay et al., 2013; Jackson et al., 2013). According to WHO, worldwide the annual mortality rate of cholera is 1.4–4.0 million cases with 21,000–143,000 deaths (World Health Organization [WHO], 2020). Bangladesh faces more than 100,000 cases of cholera each year (Ali et al., 2015). Major cholera outbreaks are caused by the O1 and O139 serogroups of V. cholerae containing major virulence factors like cholera toxin (CT) and toxin co-regulated pilus (TCP) (Nair et al., 1988; Pal et al., 1992; Chatterjee et al., 2009; Shin et al., 2011; Awasthi et al., 2013). Other serogroups collectively called non-O1/O139 are also able to cause cholera-like diarrhea, and other intestinal infections in humans (Dziejman et al., 2005; Deshayes et al., 2015). The virulence factors of non-O1/O139 V. cholerae are mostly cholera toxin independent but associated with different types of secretion systems, e.g., type six secretion system (T6SS) (Kostakioti et al., 2005), type III (T3SS) (Alam et al., 2011), heat-stable enterotoxin (NAG-ST), hemolysin (Honda and Finkelstein, 1979; Chatterjee et al., 2009), and cholix exotoxin (Awasthi et al., 2013).
The rationale of the current study was to investigate the transmission of V. cholerae from the Bay of Bengal to the freshwater habitat of Bangladesh via an aquatic reservoir. Previous studies from Bangladesh investigated the environmental transmission of V. cholerae in particular focusing on river water and plankton (Lipp et al., 2003; Ferdous et al., 2018; Rahman et al., 2018; Daboul et al., 2020). However, fish is an integral part of the aquatic environment, which is often not considered an important source of transmission vehicle of V. cholerae. In this study, the prevalence of V. cholerae is analyzed in freshly caught Hilsha fish along with surrounding river water and the plankton samples. As the environmental persistence capability of bacteria largely relies on biofilm formation, the biofilm formation ability of the isolated V. cholerae strains was examined and the virulence profile and cytotoxicity of isolated strains were also investigated. The study was conducted as an extension part of a research project funded by the Danish Government (DANIDA) called “Combating Cholera Caused by Climate Change” (C5).1
The sampling places have been chosen depending on the spawning location of Hilsha fish, which were the estuary points near the bank of Padma, originating from the Ganges river and Meghna river (Ahsan et al., 2014). From June 2016 to April 2017, 65 fish, 15 water, and 9 plankton samples were collected from five different river locations in Bangladesh. Information on sampling locations is listed in Table 1 and Figure 1. Each fish was collected from fishermen in an individual sterile collection bag and transported directly to the laboratory in the Department of Microbiology, the University of Dhaka within 4 h of collection in a cool box maintaining cold condition.

Table 1. Sampling locations and number of collected fish, water, and plankton samples in the presence (%) of the ompW gene.
Four different body parts of fish were taken as samples, they are the outer surface, gill, gut, and rectum, and enriched in alkaline peptone water (APW) overnight. For the river water sample, 100 ml of water was collected in sterile polypropylene bottles from two or three different locations at each sampling site. The water samples were filtered through a 0.22 μm membrane filter and the filter paper was placed in APW for enrichment. In the case of the plankton sample, 100 ml of water was collected from the edge of the river and passed through a 0.22 μm pore size plankton net (Huq et al., 2012). The sample was filtered through a 0.22 μm membrane filter and the filter paper was placed within a stomacher bag with normal saline, and then homogenized in a stomacher (Seward Stomacher 80, Lab UK) and 1 mL of the homogenized sample was enriched in APW.
All overnight enriched samples were further assessed by polymerase chain reaction (PCR) for the presence of the ompW gene specific to V. cholerae (Nandi et al., 2000; Choopun et al., 2002; Huq et al., 2012). A total of 284 samples were taken from fish, water, and plankton. Their Total deoxyribonucleic acid (DNA) was extracted using a previously published protocol (De Medici et al., 2003). Only the ompW-positive samples were cultured in thiosulfate citrate bile salt sucrose (TCBS) agar (Oxoid, UK). The yellow, shiny colonies were picked from the culture plate and further confirmed by the biochemical and molecular tests.
The boiled template method (De Medici et al., 2003) was followed for the extraction of total DNA. In total DNA, the presence of V. cholerae was confirmed by PCR using two primers for the outer membrane protein (ompW) gene (Nandi et al., 2000) (F-CACCAAGAAGGTGACTTTATTGTG and (R- GTTTGTCGAATTAGCTTCACC). PCR reactions were conducted according to the protocol used in by Hossain et al. (2018), and PCR product sizes (304 bp) were estimated using 100 bp DNA size markers (Invitrogen, USA).
For serotyping of isolated strains, the agglutination test was performed on clean glass slides using polyvalent O1 and 139 antisera (Denka Seiken, Japan).
The sequences of oligonucleotide primers were sense (5′-TTTGTTAGGCACGATGATGGAT-3′) and anti-sense (5′-AC CAGACAATATAGTTTGACCCACTAAG-3′), which generate an 84 base pair amplicon within the ctxA gene. 5′ nuclease probe within the ctxA gene (TGTTTCCACCTCAATTAGT TTGAGAAGTGCCC) 5′ labeled with either FAM or TET and a Black Hole Quencher 1 on the 3′ end (Integrated DNA Technologies) (Tag Copenhagen A/S, Denmark) was used. The detection of the ctxA gene was assayed using a protocol previously described by Blackstone et al. (2007). The qPCR thermal cycling was run on ABI StepOne System (Life Technologies, USA) using an initial UNG incubation step at 50°C for 2 min and a polymerase activation step at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15 s and a combined anneal and extension step at 60°C for 1 min. The other parameters of the machine were set as default for analysis.
Both V. cholerae positive total DNA and isolated strains were tested for major toxin and regulatory genes of V. cholerae by performing PCR. The primers used in this study are listed in Supplementary Table 2. The DNA extraction and PCR protocol are described in the previous section.
V. cholerae strains were grown under shaking conditions for 18 h in Trypticase soy broth (TSB; Difco, Detroit, Mich.) supplemented with 0.6% yeast extract (TSB-YE). The culture supernatant was collected by centrifugation and by passing it through a 0.22 μm pore size filter unit (corning incorporated, Germany), which made it a cell-free culture. The cell-free culture filtrate was taken in sterile test tubes that were kept at 4°C until they were used. HeLa cells were grown as monolayers in the Dulbecco’s minimum essential medium (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) supplemented with 10% (vol/vol) horse serum (Gibco Laboratories, Grand Island, N.Y.). Cell lines were maintained in 25-cm2 cell culture flasks (NUNC, Roskilde, Denmark) at 37°C supplemented with 5% CO2. A confluent monolayer of HeLa cells grown for 3–4 days was removed from the cell culture flasks, and 200 μl of the cell suspension (ca. 6.4 × 103 cells) was added to each of the 96-well plates along with 50 μl of the cell-free culture filtrate, and the plates were incubated as described above. Morphological changes in HeLa cells were recorded at 24 h. The un-inoculated culture medium was used as medium control (Sharma et al., 1998).
The isolated strains were tested for their ability to grow at different NaCl concentrations of 0, 3, 5, 6, 8, and 10% in nutrient broth (lab lemco 1 g/L, yeast extract 2 g/L, and peptone 5 g/L) and turbidity (indicates growth) was scored spectrophotometrically at 600 nm wavelength after 24 h.
Antibiotic susceptibility of the V. cholerae strains was conducted by the agar disk diffusion method (Biemer, 1973) using commercial disks (Oxoid, UK). The strains were tested for Tetracycline 30 μg, Sulfamethoxazole-trimethoprim 25 μg, Chloramphenicol 30 μg, Ciprofloxacin 5 μg, Kanamycin 30 μg, Neomycin 30 μg, Oxytetracyclin 30 μg, Gentamicin 10 μg, Ampicillin 10 μg, Ceftriaxone 30 μg, Aztreonam 30 μg, and Nalidixic acid 30 μg, and were used in this study according to the standard guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2013 January) (Patel et al., 2014). The zone standards for Enterobacteriaceae were used when there were no established breakpoint interpretive criteria for V. cholerae. E. coli ATCC 25922 was used as a quality control strain. The experiment was done in duplicate.
For the biofilm formation assay, the isolated bacterial strains along with the strain N-16961 (as a positive control) were assayed both qualitatively and quantitatively for biofilm formation in the microtiter plate (O’Toole, 2011). One replicate for each strain was taken and cell absorbance was measured in 595 nm wavelength. Here, sterile media was taken as the negative control, and OD cut-off value was calculated using the average OD of the negative control + 3 × standard deviation of the negative control. The OD cut-off value was separately calculated for each of the strains and biofilm formation by isolates was calculated and categorized according to the absorbance (Samadi et al., 2019).
Detection of the three genes of interest, vpsR, vpsA, and vpsL, was conducted by real-time PCR (Yildiz and Schoolnik, 1999). The sequence of the primers used for real-time PCR is listed in Table 2. The formula of the reaction mixture and cycling conditions were optimized for the detection of vps genes as per supplier instruction. In short, a 25 μl reaction mixture contains 12.5 μl 2X SYBR Green Universal Master Mix (Applied Biosystems, USA, containing AmpliTaq Gold® DNA Polymerase, dNTPs, ROX Passive Reference, Uracil-N glycosylase), 2.5 μl of 100 nM of each primer, and 2.5 μl of DEPC-treated water (Carl Roth, Germany) with 5 μl of the template. The qPCR thermal cycling was run on the ABI StepOne System (Life Technologies, USA) with the following condition: 5 min at 95°C for the activation of Taq polymerase followed by 40 cycles of 30 s at 95°C and a combined annealing and extension step at 60°C for 1 min. DNA amplifications were observed from the fluorescent signal emitted for binding of the SYBR Green dye with double-stranded DNA.
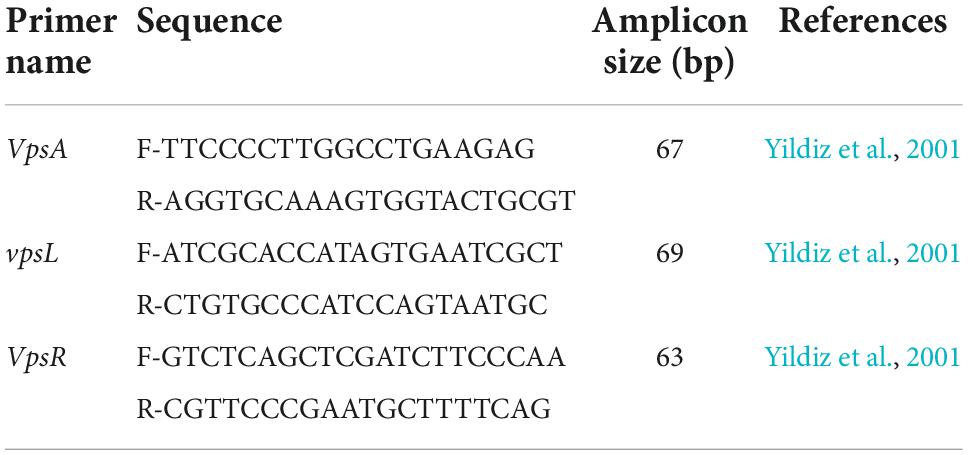
Table 2. Sequences of the primers and amplicon size (bp) in the qPCRs used for the detection of biofilm formation gene vps.
Both exploratory and regression models were used to analyze the data. To explore the influencing factor of different sample types on V. cholerae presence, a logistic regression analysis was performed in the presence of the ompW gene as the dependent variable and the type of samples from different sampling locations as the independent variable. The odds ratio or Exp(B) was considered under a 95% confidence interval.
In an exploratory analysis, different salt tolerance levels were compared with virulence factors. The absorbance of positive control strain N-16961 at different salt concentrations was considered as a cut-off value to categorize isolated 29 V. cholerae strains (absorbance more than cut-off value = 1 and absorbance less than cut off value = 0). Statistical analysis was carried out using SPSS (IBM SPSS Statistic 22) and results were measured as significant for p-value < 0.05.
A total of 284 DNA were extracted from 65 fish, 15 water, and 9 plankton samples. Among them, 25 samples were positive (8.80%) for the presence of the ompW gene in PCR, which is specific for V. cholerae outer membrane protein. Of all samples, 20 fishes (30.8%) and only one (6.7%) water samples were positive for V. cholerae in total DNA, but no V. cholerae was found in the plankton sample. The highest number of V. cholerae-positive Hilsha fish (9 out of 10 fish) was collected from Meghna river, Gosair Upazilla, and Kodalpur than other sampling points. From Tetulia river, Dashmina Upazilla, Bhola, and Meghna river, and Chandpur sampling points, all the samples were found negative for V. cholerae. From four different organs of all collected fish, 11 out of 65 (16.92%) total DNA from outer surface samples were positive for the presence of the V. cholerae-specific ompW gene, which was higher than the other three organs, and only 2 out of 65 (3.08%) total DNA were positive in case of the gut samples (Figure 2). Twenty-nine V. cholerae strains were isolated from the ompW-positive different organ parts of fish samples. No strains were isolated from river water and plankton samples (Figure 2).
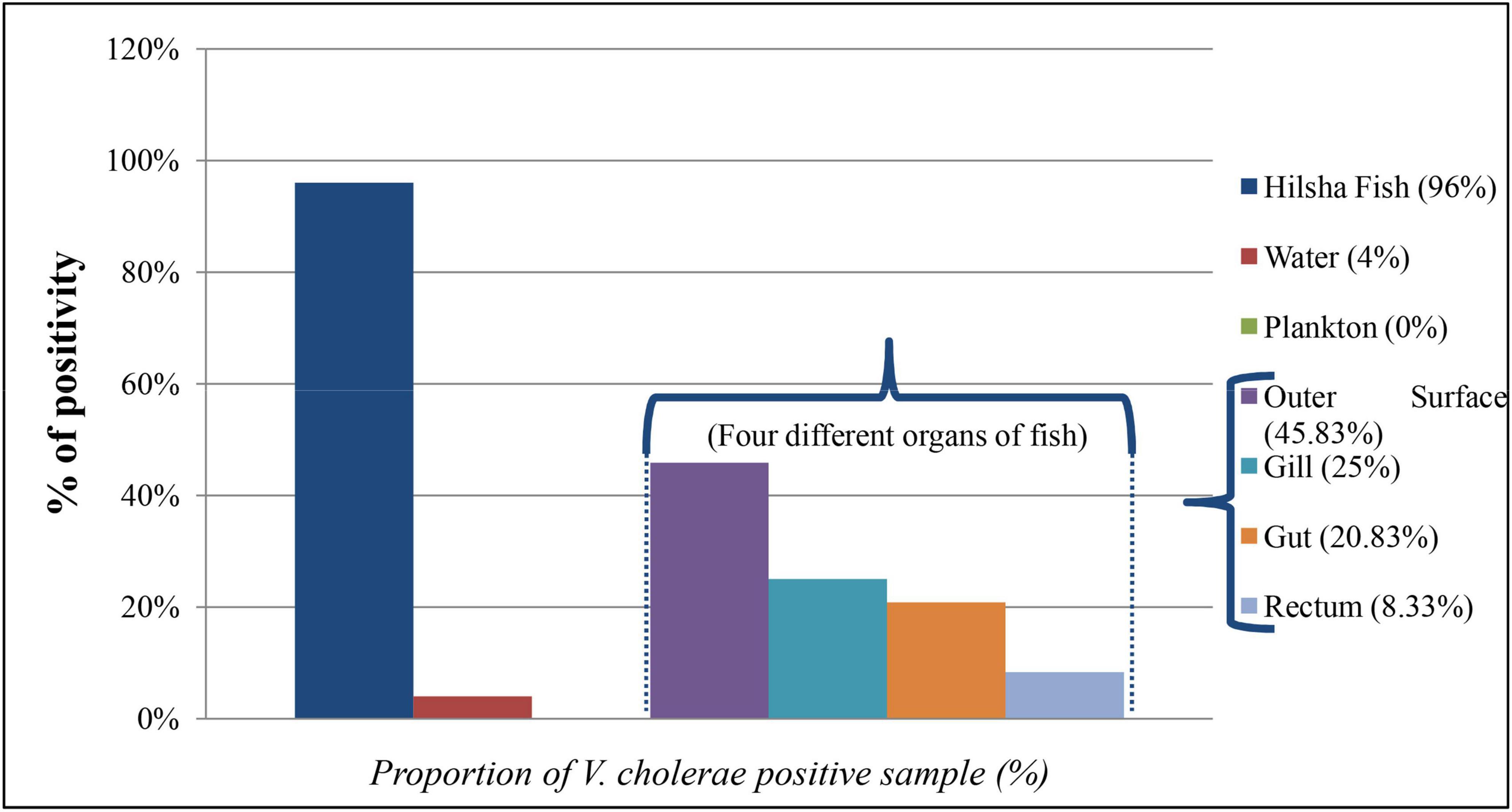
Figure 2. Proportion (%) of V. cholerae-positive direct DNA samples from fish, water, plankton, and different organs of fish (outer surface, gill, rectum, and gut).
All the strains (n = 29) with typical biochemical behaviors were subjected to serotyping for “O” antigen using the O1 polyvalent and O139 antisera. The strains did not show a typical serological reaction like that of control strains.
All the total DNA samples (n = 24) have shown negative results for the presence of the ctxA gene.
The major virulence-related genes of V. cholerae like rfbO1, rfbO139, ctxA, ctxB, ace, zot, and tcpI were absent in both total DNA and isolated strains (Figure 3A). The spotting of the tcpA and cep genes was confirmed in 20.0% (5 out of 25) and 32.0% (8 out of 25), respectively, in ompW-positive total DNA samples. Other virulence-related genes were also present in V. cholerae strains, which are represented in Figure 3B. All strains contained vasA, vasK, and HA protease genes. The genes for type 3 secretion system (T3SS), vcsN2, vcsC2, and vopF, were also detected in 20.69%, 24.14%, 24.14% of strains, respectively. There is a pattern in the virulence genes among samples from different sampling points. tcpA and cep genes were present in total DNA extracted from two sampling sites Padma River, Dolarchar Union, Kachikata, and Padma River, Bhedarganj Upazilla. Other major virulence genes like vcsN2, and vcsC2, vopF were present mostly in the V. cholerae strains isolated from Meghna River, Gosair Upazilla, and Kodalpur. Both sxt and chxA were present mostly in the V. cholerae strains isolated from Meghna River, Gosair Upazilla, Kodalpur, and Padma River, Dolarchar Union, Kachikata.
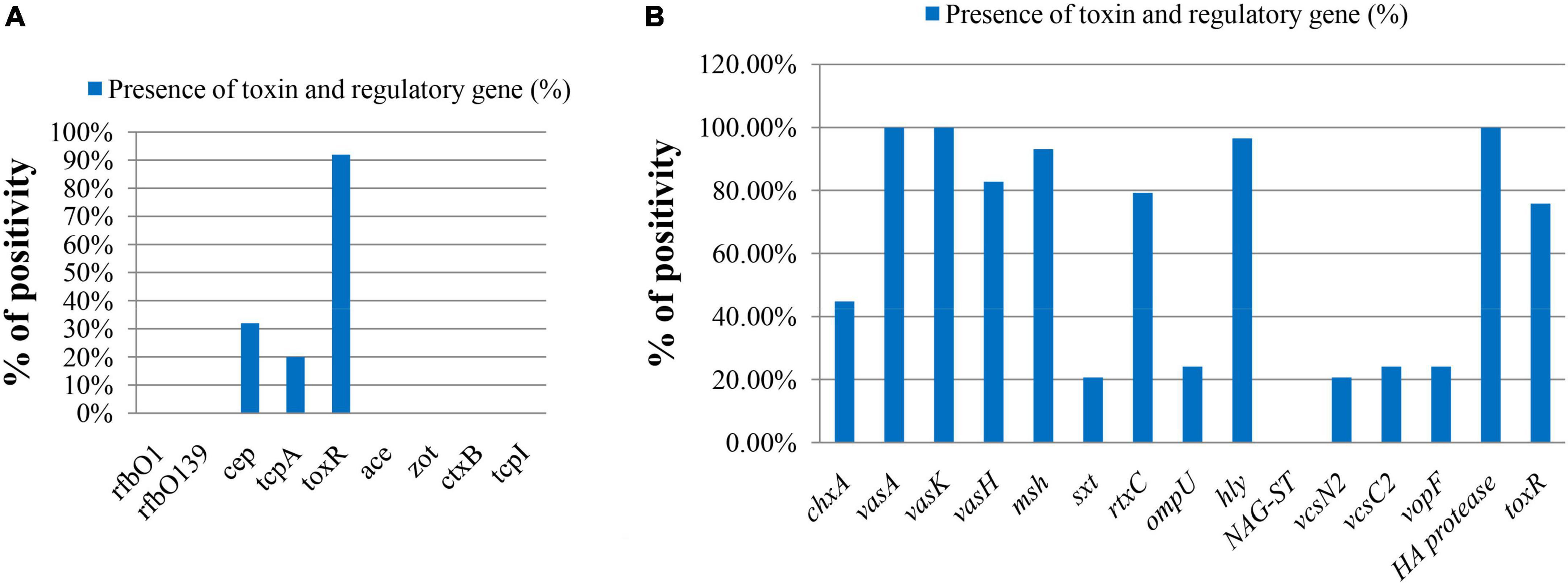
Figure 3. The molecular pattern of ompW-positive sample on the basis of toxin and regulatory genes of V. cholerae. (A) Direct DNA sample (% bar chart); (B) DNA sample of confirmed isolated V. cholerae strains (% bar chart).
In the cellular cytotoxicity test, four strains, EL-1(b), EL-98(b), EL-187(b), and EL-238(b), were selected depending on the maximum presence of toxin and virulence genes. EL-1(b) and EL-187(b) strains showed 95% killing of cells, which is higher than the clinical strain of V. cholerae N-16961 (70–80%).
The average highest cell growth was observed in 3% salt concentration (OD 0.328) and the lowest in 10% salt concentration (O.D. 0.0169). But strain EL-8(b) gave the highest optical density at 5% salt concentration (OD 0.737). The clinical isolate positive control N-16961 showed the highest optical density at 6% salt concentration (OD 0.468) and lowest at 0% salt concentration (OD 0.009) (listed in Supplementary Table 3).
Antibiotics were selected according to their mode of action and frequency of use. The antibiotic response of the strains revealed that all were uniformly susceptible to Ceftriaxone and Aztreonam (100%). There were 9, 4, 3, and 2 strains out of a total of 29 strains that showed resistance to Ampicillin, Sulfamethoxazole-trimethoprim, Nalidixic acid, Neomycin, and Streptomycin, respectively. Five isolates showed intermediate resistance to Streptomycin, three to Ciprofloxacin, Neomycin, and Oxyteracyclin, and four to Neomycin.
The results of the biofilm formation assay in the microtiter plate showed that all strains created a thin purple ring in the microtiter plate wells, which was an indication of the biofilm formation zone. All strains showed higher mean absorbance than the clinical reference strain V. cholerae O1 N16961. The lowest mean absorbance for the V. cholerae strains was 0.19 and the highest mean absorbance for the V. cholerae strains was O1 N16961, whereas the mean absorbance for V. cholerae N1T was 0.16. The mean absorbance of biofilm formation is listed in Supplementary Table 4. Classification of 29 strains for biofilm formation ability on microtiter plate was done according to Samadi et al. (2019). In this study, 41.38% (12 out of 29) of strains were strong, 44.83% (13 out of 29) of strains were moderate, and 13.79% (4 out of 29) of strains were weak biofilm former, and the clinical reference strain V. cholerae O1 N16961 was found as a weak biofilm former in our study.
Measuring the presence of three biofilm-formation-related genes, using real-time PCR, revealed that 25 strains of V. cholerae had CT values lower than 20 in the case of vpsR, which is the main regulatory gene for VPS synthesis. The other two structural genes for biofilm formation vpsA and vpsL were also present among the strains. The number of strains showed CT values lower than 20 for vpsA and vpsL genes, respectively, 19 and 22 out of 29 strains. Only two strains EL-195(a) and EL-195(b) showed the CT value higher than 35 for vpsA and vpsL gene, which indicates the absence of vpsA and vpsL gene in this two strains. But they contained the vpsR gene (CT value: 21.3 and 17.8, respectively) (listed in Supplementary Figure 1) and also formed a purple zone on the microtiter plate.
The logistic regression analysis showed that the odds of V. cholerae presence in water is 0.16 times lower than Hilsha fish. Regarding plankton samples, the odds were undetermined (0.00).
In the Chi-square test, the test of independence implied there is no significant association exists between most of the virulence genes and salt tolerance (as the p > 0.05). However, few exceptions were also observed here, for example, the msh gene showed significant relation with both 5% (p = 0.011) and 10% (p = 0.008) salt tolerance, whereas rtxC showed a significant relationship with 0% salt tolerance (p = 0.033) and ompU showed with 3% salt tolerance (p = 0.002) (listed in Table 3).
In our study, we investigated the prevalence of V. cholerae on aquatic surfaces to define the important transmission source of V. cholerae in a freshwater environment. In Bangladesh, the freshwater habitat is a possible main source of V. cholerae (Faruque et al., 1998; Ferdous et al., 2018) and studies showed that fishes are one of the intermediate reservoirs of V. cholerae in various aquatic ecosystems (Halpern and Izhaki, 2017; Hossain et al., 2018). A recent study on Tilapia fish was done in a laboratory to prove that fish act as a reservoir host of V. cholerae O1 El Tor strain and showed the survival and transmission of the pathogen horizontally to other fish (Hounmanou et al., 2019). And another recent study showed the population dynamics and virulence profile of V. cholerae strains isolated from freshly caught and local market Hilsha fish of Bangladesh and tried to find out the missing link between environmental and household contamination in Bangladesh (Hossain et al., 2018). The current study is an extension work of Hossain et al. (2018), and here, we have focused on the aquatic environmental transmission of V. cholerae with particular emphasis on Hilsha fish. We have detected V. cholerae in freshly collected fish along with surrounding river water and plankton samples. Our aim was to validate Hilsha fish as V. cholerae transmission vehicle in the riverine environment of Bangladesh. Hence, the abundance of V. cholerae within different parts of Hilsha has been investigated and the biofilm formation ability of the isolated strains was also studied.
In our study, we found V. cholerae attached to the outer surface, gill, gut, and rectum of freshly caught Hilsha fish (Figure 2). Also, 6.7% of the collected river water samples showed the presence of V. cholerae in PCR. People who live near the river area can be infected with V. cholerae while using river water for cleaning cloths, other utensils, and bathing. Previous reports revealed the incidence of cholera cases between 2016 and 2017 near Narayonganj, Matlab, and Mathbaria areas coinciding with the duration of our sampling period and close to our sampling points (Khan et al., 2020; Sack et al., 2021). The molecular profiling of both extracted total DNA and isolated strain DNA revealed that major virulence related genes like tcpA and cep are present in extracted total DNA, collected from different parts of Hilsha fish but they are not present in isolated V. cholerae strains. This is due to the tendency of V. cholerae to remain in VBNC (Viable But Non-Culturable State) or dormant state in the environment (Huq et al., 1990; Alam et al., 2007; Du Preez et al., 2010; Bhuyan et al., 2016).
In our study, all 29 strains isolated from fish samples were non-O1/non-O139 and negative for ctx and zot genes, which are the major virulence factors for O1 and O139 V. cholerae. However, the isolated strains contain other virulent genes like vasA (100%), vasK (100%), vasH (82.76%), vcsN2 (20.69%), vcsC2, and vopF (24.14%), and chxA (44.83%) that are associated with cholera or cholera-like diarrhea by ctx-independent mechanism (Park et al., 2004; Dziejman et al., 2005; Kostakioti et al., 2005; Alam et al., 2011; Shin et al., 2011; Awasthi et al., 2013). In V. cholerae, genes comprising the virulence-associated (VAS) clusters, namely, vasH, vasK, vasF, and vasA, have been suggested to be of structural importance and critical for the Type Six Secretion System (T6SS) machinery (Kostakioti et al., 2005). Type Three Secretion System (T3SS) contains vcsN2, vcsC2, and vopF, and vcsN2 is an ATPase and vcsC2 is an inner-membrane protein. The domain structure and conservation of vopF suggested that it is a putative effector for the V. cholerae T3SS and likely to interact with the host actin cytoskeleton (Shin et al., 2011). Cholix toxin (ChxA) is a recently discovered exotoxin in V. cholerae that has been characterized as a third member of the eukaryotic elongation factor 2-specific ADP-ribosyltransferase toxins, and in a recent study, it is observed that chxA is an important virulence factor among non-O1/non-O139 V. cholerae strains (Awasthi et al., 2013).
Four selected strains showed a cytotoxic effect on HeLa cells though they lacked ctxA or tcp genes. They contained other toxin genes along with that hemolysin toxin (encoded by the hlyA gene) and previous evidence showed that hemolysin toxin contains cytolytic activity when demonstrated on Vero and other mammalian cells in culture (Honda and Finkelstein, 1979; Chatterjee et al., 2009). Average highest cell growth was observed in 3% salt concentration (OD 0.328) (Supplementary Table 3), which is much closer to seawater salinity (3.5%) (United States Geological Survey [USGS], 2020) and much higher than freshwater salinity (less than 0.1%) (United States Geological Survey [USGS], 2020).
V. cholerae biofilm could play a critical role in pathogenesis and disease transmission as they are more resistant to a stressful condition in the host and requires a lower infectious dose (Kostakioti et al., 2013; Silva and Benitez, 2016). It has been reported that surface colonization and biofilm formation enhance the persistence of V. cholerae in the environment and biofilm-encased cells are protected from predation and starving, as VPS synthesis allows the exploitation of attached surface nutrients (Matz et al., 2005; Lutz et al., 2013). Data also showed that persister cells of V. cholerae could survive in nutrient-poor “filter sterilized” lake water (FSLW) for about 700 days, these persister cells had a cell-to-cell aggregation and biofilm formation ability (Jubair et al., 2014). From the analysis of mean absorbance data of biofilm assay, it has been found that all of the strains showed positive in vitro biofilm formation in a microtiter plate. The presence of three major genes in all isolates behind the regulation (vpsR) and formation (vpsA and vpsL) of biofilm was quite remarkable in our study.
In conclusion, tracking the possible route of V. cholerae dissemination to fresh water is crucial in a densely populated country like Bangladesh. The absence of V. cholerae in surrounding river water and plankton samples reduces the chance of alternative sources of contamination in Hilsha fish with V. cholerae. This may indicate the traveling of the pathogen with fish to the freshwater aquatic environment from the Bay of Bengal. A limitation of the present study is the lack of the molecular typing data of the isolated V. cholerae strains that would strengthen the data to prove the possible transmission route of the reported strains. Further molecular typing experiments, i.e., multilocus sequence typing, next generation sequencing, PFGE, etc. using the V. cholerae strains from different sampling points would identify the clonal relationship within the environmental and clinical strains. This study also provides data about producing a high degree of in vitro biofilm formation (VPS synthesis) by isolated V. cholerae strains from freshly caught Hilsha fish. These findings imply that biofilm plays a part in the persistence of V. cholerae during transmission with Hilsha, but further studies are needed to elucidate the proper mechanism and function of in vivo biofilm formation and their role in the persistence of V. cholerae in the environment. The public health impact of the toxigenic V. cholerae strains from Hilsha fish is a matter of concern. People from rural and low-income urban populations with poor access to safe water and sanitation are particularly at risk of V. cholerae infection, as proper kitchen hygiene during the cleaning and processing of fish is not regularly maintained. So, proper kitchen hygiene is mandatory during the handling and preparation of Hilsha fish to avoid illness of consumers. The health safety and security of the fishermen during the catching and transportation of the fish to the market should be considered with priority. National policymakers and stakeholders should formulate policies regarding the safe capture, transportation, and storage of fish to ensure the health and safety of the people who earn their livelihood from the fish sector.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by the Faculty of Biological Science, University of Dhaka.
SD designed the research methodology, carried out formal analysis and laboratory work, and prepared the original draft. ZH conceptualized the research method, helped in data curation, and participated in editing the original draft. AB was the principal supervisor of the research and contributed to the revision of the draft, and final approval of the article. PJ participated in a critical reviewing of the original draft. HA helped in the revision and editing of the original draft. All authors have read and agreed to the published version of the manuscript.
This research was funded by the project entitled “Combating Cholera Caused by Climate Change” (C5) (grant no. 12-040KU) from the Danish International Development Agency (DANIDA) and the University Grant Commission of Bangladesh (UGC), Dhaka University.
We thank M. Niamul Naser, Professor and Chairman, Dept. of Zoology, the University of Dhaka for helping us with sample collection, and Masud Parvez for his continuous assistance during our laboratory work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.933413/full#supplementary-material
Acharjee, M., Fatema, K., Jahan, F., Siddique, S. J., Uddin, M. A., and Noor, R. (2012). Prevalence of Vibrio cholerae in different food samples in the city of Dhaka, Bangladesh. Int. Food Res. J. 20, 1017–1022.
Ahsan, D. A., Naser, M. N., Bhaumik, U., Hazra, S., and Battacharya, S. B. (2014). Migration, spawning patterns and conservation of hilsa shad in Bangladesh and India. New Delhi: Academic Foundation. doi: 10.1007/s13398-014-0173-7.2
Akanda, A. S., Jutla, A. S., Alam, M., de Magny, G. C., Siddique, A. K., Sack, R. B., et al. (2011). Hydroclimatic influences on seasonal and spatial cholera transmission cycles: Implications for public health intervention in the Bengal Delta. Water Resour. Res. 47:W00H07. doi: 10.1029/2010WR009914
Alam, A., Miller, K. A., Chaand, M., Butler, J. S., and Dziejman, M. (2011). Identification of Vibrio cholerae type III secretion system effector proteins. Infect. Immun. 79, 1728–1740. doi: 10.1128/IAI.01194-10
Alam, M., Sultana, M., Nair, G. B., Sack, R. B., Sack, D. A., Siddique, A. K., et al. (2006). Toxigenic Vibrio cholerae in the aquatic environment of Mathbaria, Bangladesh. Appl. Environ. Microbiol. 72, 2849–2855. doi: 10.1128/AEM.72.4.2849-2855.2006
Alam, M., Sultana, M., Nair, G. B., Siddique, A. K., Hasan, N. A., Sack, R. B., et al. (2007). Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proc. Natl. Acad. Sci. U.S.A. 104, 17801–17806. doi: 10.1073/pnas.0705599104
Ali, M., Nelson, A. R., Lopez, A. L., and Sack, D. A. (2015). Updated global burden of cholera in endemic countries. PLoS Negl. Trop. Dis. 9:e0003832. doi: 10.1371/journal.pntd.0003832
Awasthi, S. P., Asakura, M., Chowdhury, N., Neogi, S. B., Hinenoya, A., Golbar, H. M., et al. (2013). Novel cholix toxin variants, ADP-ribosylating toxins in Vibrio cholerae non-O1/non-O139 strains, and their pathogenicity. Infect. Immun. 81, 531–541. doi: 10.1128/IAI.00982-12
Barzilay, E. J., Schaad, N., Magloire, R., Mung, K. S., Boncy, J., Dahourou, G. A., et al. (2013). Cholera surveillance during the Haiti epidemic – the first 2 years. N. Engl. J. Med. 368, 599–609. doi: 10.1056/NEJMoa1204927
Bhaumik, U. (2015). Migration of hilsa shad in the Indo – Pacific region – a review. Int. J. Curr. Res. Acad. Rev. 3, 139–155.
Bhuyan, S. K., Vairale, M. G., Arya, N., Yadav, P., Veer, V., Singh, L., et al. (2016). Molecular epidemiology of Vibrio cholerae associated with flood in Brahamputra river valley, Assam, India. Infect. Genet. Evol. 40, 352–356. doi: 10.1016/j.meegid.2015.11.029
Biemer, J. J. (1973). Antimicrobial susceptibility testing by the kirby-bauer disc diffusion method. Ann. Clin. Lab. Sci. 3, 135–140.
Blackstone, G. M., Nordstrom, J. L., Bowen, M. D., Meyer, R. F., Imbro, P., and DePaola, A. (2007). Use of a real time PCR assay for detection of the ctxA gene of Vibrio cholerae in an environmental survey of Mobile Bay. J. Microbiol. Methods 68, 254–259. doi: 10.1016/j.mimet.2006.08.006
Chatterjee, S., Nordstrom, J. L., Bowen, M. D., Meyer, R. F., Imbro, P., and DePaola, A. (2009). Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J. Clin. Microbiol. 47, 1087–1095. doi: 10.1128/JCM.02026-08
Choopun, N. N., Louis, V., Huq, A., and Colwell, R. R. (2002). Simple procedure for rapid identification of Vibrio cholerae from the aquatic environment. Appl. Environ. Microbiol. 68, 995–998. doi: 10.1128/aem.68.2.995-998.2002
CLSI (2013). Performance standards for antimicrobial susceptibility testing; Twenty-third informational supplement. CLSI document M100-S23. Wayne, PA: Clinical and Laboratory Standards Institute.
Colwell, R. R., Brayton, P. R., Grimes, D. J., Roszak, D. B., Huq, S. A., and Palmer, L. M. (1985). Viable but non-culturable Vibrio cholerae and related pathogens in the environment: Implications for release of genetically engineered microorganisms. Bio Technol. 3, 817–820. doi: 10.1038/nbt0985-817
Daboul, J., Weghorst, L., DeAngelis, C., Plecha, S. C., Saul-McBeth, J., and Matson, J. S. (2020). Characterization of Vibrio cholerae isolates from freshwater sources in northwest Ohio. PLoS One 15:e0238438. doi: 10.1371/journal.pone.0238438
Dawson, M. P., Humphrey, B. A., and Marshall, K. C. (1981). Adhesion: A tactic in the survival strategy of a marine vibrio during starvation. Curr. Microbiol. 6, 195–199. doi: 10.1007/BF01566971
De Medici, D., Croci, L., Delibato, E., Di Pasquale, S., Filetici, E., and Toti, L. (2003). Evaluation of DNA extraction methods for use in combination with SYBR green I real-time PCR to detect Salmonella enterica serotype enteritidis in poultry. Appl. Environ. Microbiol. 69, 3456–3461. doi: 10.1128/aem.69.6.3456-3461.2003
Deshayes, S., Daurel, C., Cattoir, V., Parienti, J. J., Quilici, M. L., and de La Blanchardière, A. (2015). Non-O1, non-O139 Vibrio cholerae bacteraemia: Case report and literature review. Springerplus 4:575. doi: 10.1186/s40064-015-1346-3
Du Preez, M., van der Merwe, M. R., Cumbana, A., and le Roux, W. (2010). A survey of Vibrio cholerae O1 and O139 in estuarine waters and sediments of Beira, Mozambique. Water SA 36, 615–620. doi: 10.4314/wsa.v36i5.61995
Dutta, S., and Hazra, S. (2017). From biology to management: A critical review of Hilsa Shad (Tenualosa ilisha). Indian J. Geo Mar. Sci. 46, 1503–1510.
Dziejman, M., Serruto, D., Tam, V. C., Sturtevant, D., Diraphat, P., Faruque, S. M., et al. (2005). Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc. Natl. Acad. Sci. U.S.A. 102, 3465–3470. doi: 10.1073/pnas.0409918102
Epstein, P. R. (1993). Algal blooms in the spread and persistence of cholera. Biosystems 31, 209–221. doi: 10.1016/0303-2647(93)90050-M
Faruque, S. M., Albert, M. J., and Mekalanos, J. J. (1998). Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62, 1301–1314. doi: 10.1128/MMBR.62.4.1301-1314.1998
Ferdous, J., Sultana, R., Rashid, R. B., Tasnimuzzaman, M., Nordland, A., Begum, A., et al. (2018). A comparative analysis of Vibrio cholerae contamination in point-of-drinking and source water in a low-income urban community, Bangladesh. Front. Microbiol. 9:489. doi: 10.3389/fmicb.2018.00489
Halpern, M., and Izhaki, I. (2017). Fish as hosts of Vibrio cholerae. Front. Microbiol. 8:282. doi: 10.3389/fmicb.2017.00282
Halpern, M., Senderovich, Y., and Izhaki, I. (2008). Waterfowl—the missing link in epidemic and pandemic cholera dissemination? PLoS Pathog. 4:e1000173. doi: 10.1371/journal.ppat.1000173
Honda, T., and Finkelstein, R. A. (1979). Purification and characterization of a hemolysin produced by Vibrio cholerae biotype El Tor: Another toxic substance produced by cholera vibrios. Infect. Immun. 26, 1020–1027. doi: 10.1128/IAI.26.3.1020-1027.1979
Hood, M. A., and Winter, P. A. (1997). Attachment of Vibrio cholerae under various environmental conditions and to selected substrates. FEMS Microbiol. Ecol. 22, 215–223. doi: 10.1111/j.1574-6941.1997.tb00373.x
Hora, S. L. (1938). A preliminary note on the spawning grounds and bionomics of the so-called Indian Shad, Hilsa Ilisha (Ham.) in the river Ganges. Rec. Indian Mus. 40, 147–158.
Hossain, Z. Z., Farhana, I., Tulsiani, S. M., Begum, A., and Jensen, P. K. M. (2018). Transmission and toxigenic potential of Vibrio cholerae in Hilsha fish (Tenualosa ilisha) for human consumption in Bangladesh. Front. Microbiol. 9:222. doi: 10.3389/fmicb.2018.00222
Hounmanou, Y. M. G., Mdegela, R. H., Dougnon, T. V., Madsen, H., Withey, J. H., Olsen, J. E., et al. (2019). Tilapia (Oreochromis niloticus) as a putative reservoir host for survival and transmission of Vibrio cholerae O1 biotype El Tor in the aquatic environment. Front. Microbiol. 10:1215. doi: 10.3389/fmicb.2019.01215
Hounmanou, Y. M., Mdegela, R. H., Dougnon, T. V., Mhongole, O. J., Mayila, E. S., Malakalinga, J., et al. (2016). Toxigenic Vibrio cholerae O1 in vegetables and fish raised in wastewater irrigated fields and stabilization ponds during a non-cholera outbreak period in Morogoro, Tanzania: An environmental health study. BMC Res. Notes 9:466. doi: 10.1186/s13104-016-2283-0
Huq, A., Colwell, R. R., Rahman, R., Ali, A., Chowdhury, M. A., Parveen, S., et al. (1990). Detection of Vibrio cholerae O1 in the aquatic environment by fluorescent-monoclonal antibody and culture methods. Appl. Environ. Microbiol. 56, 2370–2373. doi: 10.1128/AEM.56.8.2370-2373.1990
Huq, A., Grim, C., Colwell, R. R., and Nair, G. B. (2012). Detection, isolation, and identification of Vibrio cholerae from the environment. Curr. Protoc. Microbiol. Chapter 6:Unit6A.5. doi: 10.1002/9780471729259.mc06a05s26
Huq, A., Sack, R. B., Nizam, A., Longini, I. M., Nair, G. B., Ali, A., et al. (2005). Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl. Environ. Microbiol. 71, 4645–4654. doi: 10.1128/AEM.71.8.4645-4654.2005
Huq, A., Small, E. B., West, P. A., Huq, M. I., Rahman, R., and Colwell, R. R. (1983). Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 45, 275–283. doi: 10.1128/aem.45.1.275-283.1983
Jackson, B. R., Talkington, D. F., Pruckler, J. M., Fouché, M. D. B., Lafosse, E., Nygren, B., et al. (2013). Seroepidemiologic survey of epidemic cholera in Haiti to assess spectrum of illness and risk factors for severe disease. Am. J. Trop. Med. Hyg. 89, 654–664. doi: 10.4269/ajtmh.13-0208
Jubair, M., Atanasova, K. R., Rahman, M., Klose, K. E., Yasmin, M., Yilmaz, O., et al. (2014). Vibrio cholerae persisted in microcosm for 700 days inhibits motility but promotes biofilm formation in nutrient-poor lake water microcosms. PLoS One 9:e92883. doi: 10.1371/journal.pone.0092883
Khan, A. I., Rashid, M. M., Islam, M. T., Afrad, M. H., Salimuzzaman, M., Hegde, S. T., et al. (2020). Epidemiology of cholera in Bangladesh: Findings from nationwide hospital-based surveillance, 2014-2018. Clin. Infect. Dis. 71, 1635–1642. doi: 10.1093/cid/ciz1075
Kiiyukia, C., Nakajima, A., Nakai, T., Muroga, K., Kawakami, H., Hashimoto, H., et al. (1992). Vibrio cholerae non-O1 isolated from ayu fish (Plecoglossus altivelis) in Japan. Appl. Environ. Microbiol. 58, 3078–3082. doi: 10.1128/aem.58.9.3078-3082.1992
Kostakioti, M., Hadjifrangiskou, M., and Hultgren, S. J. (2013). Bacterial biofilms: Development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 3:a010306. doi: 10.1101/cshperspect.a010306
Kostakioti, M., Newman, C. L., Thanassi, D. G., and Stathopoulos, C. (2005). Mechanisms of protein export across the bacterial outer membrane. J. Bacteriol. 187, 4306–4314. doi: 10.1128/JB.187.13.4306-4314.2005
Kumar, R., and Lalitha, K. V. (2013). Prevalence and molecular characterization of Vibrio cholerae O1, non-O1 and non-O139 in tropical seafood in Cochin, India. Foodborne Pathog. Dis. 10, 278–283. doi: 10.1089/fpd.2012.1310
Lipp, E. K., Huq, A., and Colwell, R. R. (2002). Effects of global climate on infectious disease: The cholera model. Clin. Microbiol. Rev. 15, 757–770. doi: 10.1128/cmr.15.4.757-770.2002
Lipp, E. K., Rivera, I. N., Gil, A. I., Espeland, E. M., Choopun, N., Louis, V. R., et al. (2003). Direct detection of Vibrio cholerae and ctxA in Peruvian coastal water and plankton by PCR. Appl. Environ. Microbiol. 69, 3676–3680. doi: 10.1128/AEM.69.6.3676-3680.2003
Lobitz, B., Beck, L., Huq, A., Wood, B., Fuchs, G., Faruque, A. S., et al. (2000). Climate and infectious disease: Use of remote sensing for detection of Vibrio cholerae by indirect measurement. Proc. Natl. Acad. Sci. U.S.A. 97, 1438–1443. doi: 10.1073/pnas.97.4.1438
López, D., Vlamakis, H., and Kolter, R. (2010). Biofilms. Cold Spring Harb. Perspect. Biol. 2:a000398. doi: 10.1101/cshperspect.a000398
Lutz, C., Erken, M., Noorian, P., Sun, S., and McDougald, D. (2013). Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front. Microbiol. 4:375. doi: 10.3389/fmicb.2013.00375
Matz, C., McDougald, D., Moreno, A. M., Yung, P. Y., Yildiz, F. H., and Kjelleberg, S. (2005). Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl Acad. Sci. U.S.A. 102, 16819–16824. doi: 10.1073/pnas.0505350102
Miah, M. S. (2015). Climatic and anthropogenic factors changing spawning pattern and production zone of Hilsa fishery in the Bay of Bengal. Weather Clim. Extremes 7, 109–115. doi: 10.1016/j.wace.2015.01.001
Nair, G. B., Oku, Y., Takeda, Y., Ghosh, A., Ghosh, R. K., Chattopadhyay, S., et al. (1988). Toxin profiles of Vibrio cholerae non-O1 from environmental sources in Calcutta, India. Appl. Environ. Microbiol. 54, 3180–3182. doi: 10.1128/aem.54.12.3180-3182.1988
Nandi, B., Nandy, R. K., Mukhopadhyay, S., Nair, G. B., Shimada, T., Ghose, A. C., et al. (2000). Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J. Clin. Microbiol. 38, 4145–4151. doi: 10.1128/JCM.38.11.4145-4151.2000
O’Connor, K. A., Cartwright, E., Loharikar, A., Routh, J., Gaines, J., Fouché, M. D., et al. (2011). Risk factors early in the 2010 cholera epidemic, Haiti. Emerg. Infect. Dis. 17, 2136–2138. doi: 10.3201/eid1711.110810
O’Toole, G. A. (2011). Microtiter dish biofilm formation assay. J. Vis. Exp. JoVE 30:2437. doi: 10.3791/2437
Pal, A., Ramamurthy, T., Bhadra, R. K., Takeda, T., Shimada, T., Takeda, Y., et al. (1992). Reassessment of the prevalence of heat-stable enterotoxin (NAG-ST) among environmental Vibrio cholerae non-O1 strains isolated from Calcutta, India, by using a NAG-ST DNA probe. Appl. Environ. Microbiol. 58, 2485–2489. doi: 10.1128/AEM.58.8.2485-2489.1992
Park, K.-S., Ono, T., Rokuda, M., Jang, M. H., Okada, K., Iida, T., et al. (2004). Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72, 6659–6665. doi: 10.1128/IAI.72.11.6659-6665.2004
Patel, J., Cockerill, F. III., Alder, J., Bradford, P., Eliopoulos, G., and Hardy, D. (2014). Performance standards for antimicrobial susceptibility testing; twenty-fourth information supplement. Wayne, IL: SettiClinical and Laboratory Standards Institute.
Pillay, T. V. R. (1958). Biology of the hilsa, hilsa Ilisha (Hamilton) of the river Hooghly. Indian J. Fish 5, 201–257.
Rahman, Z., Rahman, M. A., Rashid, M. U., Monira, S., Johura, F. T., Mustafiz, M., et al. (2018). Vibrio cholerae transmits through water among the household contacts of cholera patients in cholera endemic coastal villages of Bangladesh, 2015-2016 (CHoBI7 Trial). Front. Public Health 6:238. doi: 10.3389/fpubh.2018.00238
Runft, D. L., Mitchell, K. C., Abuaita, B. H., Allen, J. P., Bajer, S., Ginsburg, K., et al. (2014). Zebrafish as a natural host model for Vibrio cholerae colonization and transmission. Appl. Environ. Microbiol. 80, 1710–1717. doi: 10.1128/AEM.03580-13
Sack, D. A., Debes, A. K., Ateudjieu, J., Bwire, G., Ali, M., Ngwa, M. C., et al. (2021). Contrasting epidemiology of cholera in Bangladesh and Africa. J. Infect. Dis. 224(Suppl. 7) S701–S709. doi: 10.1093/infdis/jiab440
Samadi, R., Ghalavand, Z., Mirnejad, R., Nikmanesh, B., and Eslami, G. (2019). Antimicrobial resistance and molecular characteristics of methicillin-resistant Staphylococcus aureus isolates from children patients in Iran. Infect. Drug Resist. 12, 3849–3857. doi: 10.2147/IDR.S229394
Scheelbeek, P., Treglown, S., Reid, T., and Maes, P. (2009). Household fish preparation hygiene and cholera transmission in Monrovia, Liberia. J. Infect. Dev. Ctries. 3, 727–731. doi: 10.3855/jidc.615
Schuster, B. M., Tyzik, A. L., Donner, R. A., Striplin, M. J., Almagro-Moreno, S., Jones, S. H., et al. (2011). Ecology and genetic structure of a northern temperate Vibrio cholerae population related to toxigenic isolates. Appl. Environ. Microbiol. 77, 7568–7575. doi: 10.1128/AEM.00378-11
Senderovich, Y., Izhaki, I., and Halpern, M. (2010). Fish as reservoirs and vectors of Vibrio cholerae. PLoS One 5:e8607. doi: 10.1371/journal.pone.0008607
Sharma, C., Thungapathra, M., Ghosh, A., Mukhopadhyay, A. K., Basu, A., Mitra, R., et al. (1998). Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J. Clin. Microbiol. 36, 756–763. doi: 10.1128/JCM.36.3.756-763.1998
Shikuma, N. J., and Hadfield, M. G. (2010). Marine biofilms on submerged surfaces are a reservoir for Escherichia coli and Vibrio cholerae. Biofouling 26, 39–46. doi: 10.1080/08927010903282814
Shin, O. S., Tam, V. C., Suzuki, M., Ritchie, J. M., Bronson, R. T., Waldor, M. K., et al. (2011). Type III secretion is essential for the rapidly fatal diarrheal disease caused by non-O1, non-O139 Vibrio cholerae. mBio 2:e00106-11. doi: 10.1128/mBio.00106-11
Silva, A. J., and Benitez, J. A. (2016). Vibrio cholerae biofilms and cholera pathogenesis. PLoS Negl. Trop. Dis. 10:e0004330. doi: 10.1371/journal.pntd.0004330
Tamplin, M. L., Gauzens, A. L., Huq, A., Sack, D. A., and Colwell, R. R. (1990). Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl. Environ. Microbiol. 56, 1977–1980. doi: 10.1128/AEM.56.6.1977-1980.1990
Thi, T. A. N., Jacxsens, L., Noseda, B., Samapundo, S., Ly Nguyen, B., Heyndrickx, M., et al. (2014). Evaluation of the microbiological safety and quality of Vietnamese Pangasius Hypophthalmus during processing by a microbial assessment scheme in combination with a self-assessment questionnaire. Fish. Sci. 80, 1117–1128. doi: 10.1007/s12562-014-0786-y
Traoré, O., Martikainen, O., Siitonen, A., Traoré, A. S., Barro, N., and Haukka, K. (2014). Occurrence of Vibrio cholerae in fish and water from a reservoir and a neighboring channel in Ouagadougou, Burkina Faso. J. Infect. Dev. Ctries. 8, 1334–1338. doi: 10.3855/jidc.3946
Turner, J. W., Good, B., Cole, D., and Lipp, E. K. (2009). Plankton composition and environmental factors contribute to Vibrio seasonality. ISME J. 3, 1082–1092. doi: 10.1038/ismej.2009.50
Twedt, R. M., Madden, J. M., Hunt, J. M., Francis, D. W., Peeler, J. T., Duran, A. P., et al. (1981). Characterization of Vibrio cholerae isolated from oysters. Appl. Environ. Microbiol. 41, 1475–1478. doi: 10.1128/aem.41.6.1475-1478.1981
United States Geological Survey [USGS] (2020). Saline water and salinity. Available online at: https://www.usgs.gov/special-topic/water-science-school/science/saline-water-and-salinity?qt-science_center_objects=0#qt-science_center_objects (accessed June 24, 2020).
World Health Organization [WHO] (2020). Cholera. Available online at: https://www.who.int/news-room/fact-sheets/detail/cholera (accessed January 17, 2019).
Yildiz, F. H., and Schoolnik, G. K. (1999). Vibrio cholerae O1 El Tor: Identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 96, 4028–4033. doi: 10.1073/pnas.96.7.4028
Yildiz, F. H., Dolganov, N. A., and Schoolnik, G. K. (2001). VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183, 1716–1726. doi: 10.1128/JB.183.5.1716-1726.2001
Keywords: cholera, aquatic environment, Hilsha fish, biofilm, persistence, cellular cytotoxicity, biofilm formation assay
Citation: Dey SS, Hossain ZZ, Akhter H, Jensen PKM and Begum A (2022) Abundance and biofilm formation capability of Vibrio cholerae in aquatic environment with an emphasis on Hilsha fish (Tenualosa ilisha). Front. Microbiol. 13:933413. doi: 10.3389/fmicb.2022.933413
Received: 30 April 2022; Accepted: 30 September 2022;
Published: 26 October 2022.
Edited by:
Joshua A. Steele, Southern California Coastal Water Research Project, United StatesReviewed by:
Mohammad Tarequl Islam, University of Alberta, CanadaCopyright © 2022 Dey, Hossain, Akhter, Jensen and Begum. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anowara Begum, YW5vd2FyYUBkdS5hYy5iZA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.