- 1Département de Microbiologie, Infectiologie et Immunologie, Université de Montréal, Montréal, QC, Canada
- 2Département des Sciences Animales, Faculté des Sciences de l’Agriculture et de l’Alimentation, Université Laval, Québec, QC, Canada
- 3Institut de Biologie Intégrative et des Systèmes, Université Laval, Québec, QC, Canada
Vibrio cholerae includes strains responsible for the cholera disease and is a natural inhabitant of aquatic environments. V. cholerae possesses a unique polar flagellum essential for motility, adhesion, and biofilm formation. In a previous study, we showed that motility and biofilm formation are altered in the presence of subinhibitory concentrations of polymyxin B in V. cholerae O1 and O139. In this study, we performed an experimental evolution to identify the genes restoring the motility in the presence of a subinhibitory concentration of polymyxin B. Mutations in five genes have been identified in three variants derived from two different parental strains A1552 and MO10: ihfA that encodes a subunit of the integration host factor (IHF), vacJ (mlaA) and mlaF, two genes belonging to the maintenance of the lipid asymmetry (Mla) pathway, dacB that encodes a penicillin-binding protein (PBP4) and involved in cell wall synthesis, and ccmH that encodes a c-type cytochrome maturation protein. We further demonstrated that the variants derived from MO10 containing mutations in vacJ, mlaF, and dacB secrete more and larger membrane vesicles that titer the polymyxin B, which increases the bacterial survival and is expected to limit its impact on the bacterial envelope and participate in the flagellum’s retention and motility.
Introduction
Vibrio cholerae is a gram-negative bacterium, and the strains belonging to the O1 and O139 serogroups are responsible for the cholera disease, which infects millions of people per year worldwide. More than 200 serotypes have been reported but only the O1 and O139 cause cholera (Harris et al., 2012). The O1 strains are further divided into 2 biotypes: classical and El Tor. The classical biotype was responsible for six pandemics and has been replaced by the El Tor biotype in 1961, which is responsible for the ongoing 7th pandemic (Harris et al., 2012). The O139 serotype, which causes local epidemics, appeared in 1992 and is derived from the O1 El Tor biotype (Faruque et al., 2003). Some strains from non-O1/non-O139 serogroups can cause less serious gastroenteritis (Aydanian et al., 2011).
For most pathogenic bacteria, motility is essential for host colonization and virulence (Josenhans and Suerbaum, 2002). Different types of motility exist: swimming, swarming, twitching, gliding, and sliding, the latest being a passive process that does not involve energy (Kearns, 2010). Swarming, twitching, gliding, and sliding are the surface motility mechanisms whereas swimming allows bacteria to move in liquid suspensions and involves active propulsion by the flagella. Flagella are filamentous apparatus anchored to the bacterial envelope and are divided into three major structures: the basal body, the hook, and the filament (Echazarreta and Klose, 2019). The movement is ensured by the rotation of the flagellum’s filament driven by a motor-like structure. In most bacteria, the motor uses an H + gradient as an energy source (Nakamura and Minamino, 2019). However, in Vibrio, the rotation of the flagellum is driven by a Na + motor (Kojima et al., 1999), which provides a rotation speed up to five times faster than the H + driven flagella of E. coli (Magariyama et al., 1994). V. cholerae possesses a single polar flagellum. Its filament is composed of five homologous subunits: FlaA, FlaB, FlaC, FlaD, and FlaE, and the flagellum is coated by an outer membrane sheath (Fuerst and Perry, 1988; Klose and Mekalanos, 1998). Besides its role in motility, Vibrio’s flagellum is also essential for biofilm formation, colonization, and virulence (Echazarreta and Klose, 2019). In V. cholerae, the expression of the flagellar genes is tightly regulated and is organized in a four-step hierarchical process, resulting in four classes of genes. The FlaA flagellin gene (flaA) belongs to the class II genes and is the only essential flagellin for motility (Klose and Mekalanos, 1998). The other flagellins (flaB, flaC, flaD, and flaE), belong to the class IV genes (Echazarreta and Klose, 2019).
Antimicrobial peptides (AMPs) are molecules of low molecular weight with antimicrobial activities against viruses, fungi, or bacteria (Boparai and Sharma, 2020). They can also modulate the immune response, as a part of innate immunity (Boparai and Sharma, 2020). AMPs are produced by both eukaryotic host cells and bacteria (Bevins and Salzman, 2011; Boparai and Sharma, 2020). Polymyxin B (PmB), an AMP produced by the gram-positive bacteria Bacillus polymyxa, has been revived as a last-resort treatment for multidrug-resistant gram-negative bacteria (Yang et al., 2020). Several mechanisms have been reported to explain the antimicrobial activity of AMPs, the best known being the electrostatic interaction with the bacterial envelope, leading to pore formation and cell death (Daugelavicius et al., 2000; Boparai and Sharma, 2020). There are a growing number of evidence showing that AMPs at subinhibitory concentrations can modulate the resistance and the virulence of gram-negative bacteria (Duperthuy, 2020), including our studies on V. cholerae (Duperthuy et al., 2013; Giacomucci et al., 2019; Mathieu-Denoncourt and Duperthuy, 2021). Because of their co-evolution with their host and with microbial competitors, the Vibrio have evolved mechanisms to resist AMPs that relate to lipopolysaccharides (LPS) modifications, outer membrane remodeling, induction of the envelope stress response, efflux of AMPs, repression of AMPs’ expression by the host cells, and titration by membrane vesicles (MVs) (Destoumieux-Garzón et al., 2014).
Bacteria produce MVs to remodel their membrane and release diverse molecules in the extracellular environment (Bohuszewicz et al., 2016; Toyofuku et al., 2019). In gram-negative bacteria, MVs are produced from blebbing of the bacterial envelope or by explosive cell lysis. Depending on their origin, MVs can contain LPS, peptidoglycan, proteins, lipids, and nucleic acids from extracellular, periplasmic, and cytoplasmic origins (Altindis et al., 2014; Toyofuku et al., 2019). MVs have a role in DNA exchange, virulence, and resistance to phages, plasmas, and antimicrobials (Guerrero-Mandujano et al., 2017). V. cholerae produces MVs through a VacJ/Yrb-dependent process (Roier et al., 2016). This system is implicated in the membrane phospholipids asymmetry and its inactivation increases MVs production (Roier et al., 2016). The MVs of V. cholerae can carry proteins implicated in virulence (Rompikuntal et al., 2015; Zingl Franz et al., 2021), can help bacterial surface modulation during the infection (Zingl et al., 2020), and have a role in AMPs resistance and cross-resistance (Duperthuy et al., 2013; Rompikuntal et al., 2015; Mathieu-Denoncourt et al., 2021).
In our previous study, we showed that the biofilm formation was impaired in the presence of subinhibitory concentrations of PmB, due to a significant reduction in the proportion of flagellated V. cholerae O1 and O139 (Giacomucci et al., 2019). Accordingly, a motility reduction was observed in the presence of PmB and was sometimes associated with a non-homogenous flower-like motility pattern on soft agar. In this study, we investigated the mechanisms explaining the motility restoration of some bacterial cells leading to the flower-like motility pattern in the presence of PmB. To do so, we performed an experimental evolution in the presence of PmB in two strains of V. cholerae, i.e., O1 El Tor strain A1552 and O139 strain MO10. After sequencing, we identified mutations in genes involved in global regulation (ihfA), the outer membrane lipid asymmetry maintenance (vacJ and mlaF), and cell wall synthesis (dacB). We further demonstrated that the increased motility of the MO10 variants containing mutations in vacJ, mlaF and dacB is due to an increased titration of PmB through hypervesiculation and large vesicles production. The mechanism explaining the gain of motility resulting from the mutation in ihfA remains to be explored and is not linked to flagellins’ expression.
Materials and methods
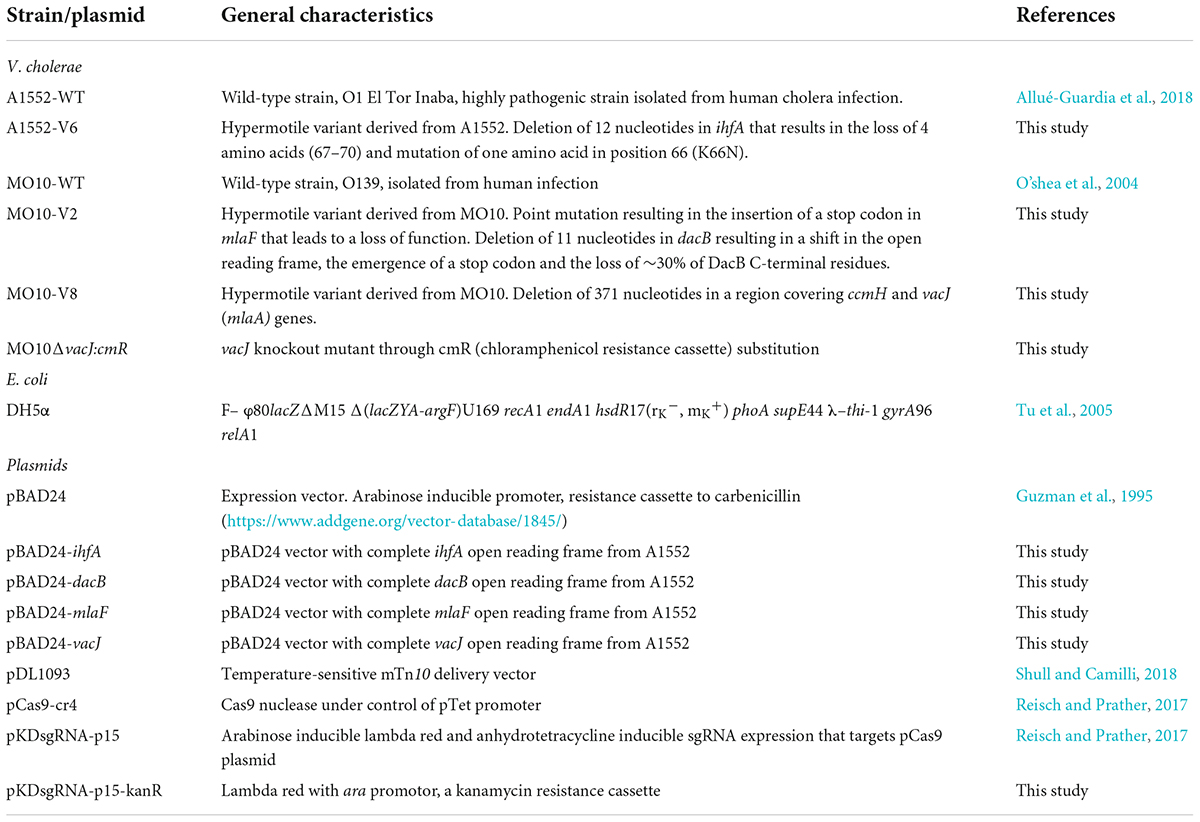
Bacterial strains and growth conditions
Two V. cholerae strains have been used in this study: A1552 (O1, El Tor, Inaba) and MO10 (O139), initially isolated from patients in Peru and India, respectively (O’shea et al., 2004; Allué-Guardia et al., 2018). Bacterial growth was performed in Luria-Bertani (LB) medium supplemented with 25 μg/ml of PmB (PmB sulfate, Millipore-Sigma) (Giacomucci et al., 2019). Growth curves were performed as follows: precultures were started from overnight cultures (1:50) in 10 ml of fresh LB medium at 37°C to an OD600 nm ∼0.2–0.4. They were then diluted to a final OD600nm of 0.001 in LB supplemented with 0.2% L-arabinose when indicated. Growth was followed at 37°C by measuring the OD600 nm each 5 min for 24 h using SpectramaxID3 Molecular Device. Between two readings, plates were constantly and vigorously shaken. All results correspond to, at least, two technical replicates within three biological replicates for each strain and condition.
Complementation with pBAD24 vector
All expression plasmids for complementation were constructed from the multicopy pBAD24 vector (Guzman et al., 1995) and are listed in Table 1. Empty vector was extracted from E. coli DH5α using QIAprep Spin Miniprep Kit (#27106) from Qiagen. The complete open reading frames from the target genes were amplified by polymerase chain reaction (PCR) (Q5® High-Fidelity DNA Polymerase, #M0491S, New England Biolab) with the respective pair of primers containing restriction sites listed in Table 2 and purified with Monarch® PCR & DNA Cleanup Kit (#T1030L, New England Biolab). Vectors and amplicons were double-digested with HindIII-HF and EcoRI-HF (#R3104S and #R3101S, New England Biolab) or with HindIII-HF and XbaI (#R0145S New England Biolab) according to the manufacturer’s guidelines. Digested products were purified from electrophoresis gel (Monarch® DNA Gel Extraction Kit, #T1020L, New England Biolab) and were then ligated with T4 DNA ligase (#M0202S, New England Biolab). Newly ligated plasmids were purified on columns (Monarch® PCR & DNA Cleanup Kit, #T1030L, New England Biolab), then transformed by heat shock, and amplified in E. coli DH5α (Bryant, 1988). The inserts’ sequences were verified using the pBAD24-verif_Fwd and pBAD24-verif_rev primers. Purified plasmids from E. coli were then transformed by electroporation in V. cholerae MO10-WT or A1552-WT strains with 1.25 kV for 5 ms (O’callaghan and Charbit, 1990). Then, respective plasmids were transformed in V. cholerae A1552-V6 or MO10-V2 or –V8 (O’callaghan and Charbit, 1990).
Motility assays
Motility assays were performed as previously described (Giacomucci et al., 2019). Briefly, bacteria were spotted on LB plates containing 0.3% agar (soft agar) and incubated for 24–48 h at 37°C. The motility was evaluated by measuring the diameter of bacterial growth. PmB was added to the plate at a concentration of 25 μg/ml when indicated. The selection of variants was performed on technical triplicates. Experiments using variants were performed in biological triplicates. Statistical analyses were performed using a one-way analysis of variance (ANOVA) test (p < 0.05).
Sequencing, genome assembly, and analysis
The DNA extraction of the bacterial strains was performed using Promega Wizard Genomic DNA Purification Kit according to the manufacturer’s guidelines. The wild-type strains MO10 and A1552 were sequenced by PacBio RS II and Illumina MiSeq at the Génome Québec Innovation Center (Centre Hospitalier Universitaire Sainte-Justine, Montréal, QC, Canada). The PacBio sequencing reads were assembled with Flye 2.8.1 (Kolmogorov et al., 2019). The sequences corresponding to chromosomes I and II were circularized with various tools from the EMBOSS version 6.6.0 suite (Rice et al., 2000). Illumina reads were subsequently used to polish assemblies with BWA version 0.7.17-r1188 (Li, 2013), SAMtools version 1.12 (Li et al., 2009), and Pilon version 1.23 (Walker et al., 2014).
The DNA of the variants sequenced by Illumina MiSeq at the Génome Québec Innovation Center (McGill University, Montréal, QC, Canada). The reads were filtered using FASTP version 0.20.1 (Chen et al., 2018). The mutations were identified and validated in relation to the genomes of the parental strains, beforehand annotated with Prokka version 1.14.5 (Seemann, 2014), with both Snippy version 4.6.0 Seemann (2015) and Breseq version 0.35.2 (Barrick and Lenski, 2009; Barrick et al., 2009).
Sequences homologies, alignments, and features analysis
Escherichia coli K12 MG1655 and Vibrio cholerae IhfA protein sequences’ identity and similarity were calculated with protein–protein BLAST (Altschul et al., 1997, 2005). Alignment of two or multiple sequences was respectively performed using semi-global alignment “glocal” (Brudno et al., 2003) and MUSCLE (Edgar, 2004) from Snapgene®. Figure 4B was edited from Easyfig graphical sequence alignment (Sullivan et al., 2011), and panels Figures 4A,C,D were edited from Snapgene® alignments. The signal peptide was identified with protcompB (V9.0) (Protcompb) and SignalP 5.0 (Nielsen et al., 2019). Putative –35 and –10 promoter boxes were identified with BPROM service from Softberry (Solovyev and Salamov, 2011; Supplementary Figures S1–S3). Vibrio cholerae genes and protein sequence homologies were compared to Escherichia coli K12 MG1655 (NC_000913.3).
Membrane vesicles isolation, protein content analysis, and lipopolysaccharides quantification
Membrane vesicles were isolated as previously described (Duperthuy et al., 2013). Briefly, V. cholerae wild-type strain MO10 and its hypermotile variants MO10-V2 and MO10-V8 were grown in 125 ml LB broth in 250-ml glass Erlenmeyer flasks for 16 h at 37°C with agitation. Bacterial cells were removed completely by centrifugation and filtration of the supernatant using acetate cellulose 0.2-μm filters (Millipore). MVs were recovered from 25 ml cell-free supernatant by ultracentrifugation at 100,000 × g for 3 h at 4°C in a 70-Ti rotor (Beckman Instruments) and suspended in 250 μl of phosphate-buffered saline (PBS).
The vesicle lipopolysaccharides (LPSs) were then quantified using Pierce Chromogenic Endotoxin Quant Kit according to the manufacturer’s instructions, and the protein content of the vesicles was quantified using a Bradford assay (Bio-Rad 500-0006) and a bovine serum albumin (BSA) standard curve. Data were obtained from at least 3 independent experiments done in technical duplicates. Statistical analysis was done using Student’s t-tests (p < 0.05).
Survival in the presence of a lethal concentration of PmB
Midlog cultures of MO10-WT, MO10-V2, or MO10-V8 were incubated for 30 min at 37°C with 200 μg/ml of PmB. Afterward, 10 μl of 10-fold dilutions of the mixture was spotted on LB Agar (LBA), incubated for 18 h at 37°C, and numerated. For the MVs protection assays, 90 μl of a midlog culture of MO10 was incubated for 1 h at 37°C with 200 μg/ml of PmB, and 5 μl of the MVs preparations isolated from MO10-WT, MO10-V2, or MO10-V8. A final concentration representing 5 times the concentration of vesicles produced by an overnight culture has been used, and the MV concentrations have been normalized to the MO10-WT MV concentration ([norm]) according to the LPS dosage, or not. Similarly, 10-fold dilutions were spotted on LBA and numbered. Data are presented as relative survival percentage (CFU/ml of each condition with PmB divided by CFU/ml of the condition without PmB) of at least three independent experiments done in technical duplicates. Statistical analysis was performed using a one-way ANOVA test.
Electron microscopy
The bacteria were visualized by transmitted electron microscopy (TEM Hitachi H-7100) as previously described (Giacomucci et al., 2019). Bacterial cultures were obtained by dilution 1:50 of overnight culture in a fresh LB medium, followed by incubation at 37°C until the culture reached an OD600nm of 0.08. Bacteria in the exponential growth phase were then diluted in a fresh LB medium containing 25 μg/ml of PmB to an OD600nm of ∼ 0.5. A volume of 50 μl of fresh bacterial culture was spread on formvar and carbon coated 200 Mesh hexagonal slim bar grids. Mounting was washed with 50 mM cacodylate acid saccharose 3% and labeled using 1% phosphotungstic acid for 2 s. Bacteria were visualized at 75 kV and pictures were obtained using a digital camera AMT XR-111 (Advanced Microscopy Techniques).
Sequestration of the PmB by the membrane vesicles
To assess the sequestration of PmB by the MVs, 30 ml of the filtered supernatants from overnight cultures of MO10, MO10-V2, or MO10-V8 was incubated at 37°C for 1 h with or without 10 or 25 μg/ml of PmB (Sigma-Aldrich). Samples (1.5 ml) were taken directly after the incubation with PmB (T). Vesicles (V) were then isolated by ultracentrifugation. Samples (1.5 ml) from the vesicle-free supernatants (S) were taken, and then, the vesicles were suspended in 200 μl of PBS.
Proteins from total (T) and vesicle-free supernatants (S) were precipitated with 150 μl trichloro-acetic acid (100% w/v) for 16 h at 4°C. Samples were then centrifuged at 20,000 × g for 20 min at 4°C, and proteins were suspended in 20 μl of Laemmli buffer. For the vesicle fraction, 10 μl of the MVs preparation was suspended in 20 μl of Laemmli buffer. As a control, 5 μl of PmB at 20 mg/ml (Sigma-Aldrich) was suspended in 120 μl of Laemmli buffer. All the samples were boiled for 10 min at 100°C and 7 μl of each sample migrated on 13% sodium dodecyl sulfate (SDS) gels. Precision plus dual color protein ladder from Bio-Rad was used as a molecular weight scale. Gels were stained with Coomassie blue and silver nitrate 0.2%.
Quantitative PCR analysis of the flagellins genes’ expression
RNA extraction, cDNA construction, and qPCR analysis were performed as described before (Giacomucci et al., 2019; Mathieu-Denoncourt and Duperthuy, 2021). V. cholerae strains A1552, MO10, and their variants were grown in LB to an OD600nm of 0.4 at 37°C with agitation. The bacterial cells from 10 ml of culture were recovered by centrifugation at 5,500 × g and suspended in 1 ml TRIzol reagent (Invitrogen). Total RNAs were extracted according to the manufacturer’s instructions, and 500 ng of RNA was retrotranscribed to cDNA using QuantiNova Reverse Transcription Kit (Qiagen). RNA and cDNA were quantified with nanodrop and visualized on 2% agarose gel.
Flagellins’ expression (flaA and flaE) was assessed by qPCR using primers as previously described (Giacomucci et al., 2019) and normalized with the expression of recA as the housekeeping gene. The relative expression of flaA and flaE of A1552-V6 and MO10-V2 and –V8 was calculated compared to their expression in A1552-WT and MO10-WT, respectively. Flagellins genes were amplified using PerfeCTa SYBR Green FastMix Low ROX in QantStudio 3 real-time PCR system (Thermo Fisher) with a protocol consisting of 1 × 30 s at 95°C; 40 × 95°C for 5 s, 58°C for 17 s, and 70°C for 12 s.
MO10ΔvacJ::cmR construction
A MO10 mutant lacking vacJ has been constructed through CmR substitution using the lambda red recombination system, as previously described (Yamamoto et al., 2009). Briefly, up and downstream regions of the target gene of approximatively 1,000 pb were amplified by PCR using primers listed in Table 2. The chloramphenicol resistance cassette (cmR) from pCas9 CR4 (Reisch and Prather, 2017), with its promotor and terminator, was also amplified by PCR. The cmR and the flanking regions of the target genes were bound together by overlap PCR and purified using Monarch DNA Gel Extraction Kit (NEB). A kanamycin resistance cassette (kanR) from pDL1093 (Shull and Camilli, 2018) was amplified using primers listed in Table 2, adding XhoI et XbaI restriction sites in 3′ and 5′. The purified amplicon and pKDsgRNA-p15 (Reisch and Prather, 2017) were digested using XhoI and XbaI (NEB), purified, and ligated. The resulting plasmid was amplified in E. coli and then inserted in MO10-WT by electroporation. The cmR construction was then introduced in MO10 containing pKDsgRNA-p15-kanR and grown at 30°C with 0.2% arabinose and 50 μg/ml of kanamycin, by electroporation at 1.25 kV, 5 ms, in 1 mm electroporation cuvettes. Mutants were selected on LB agar plates supplemented with 2 μg/ml of chloramphenicol at 37°C and verified by sequencing.
Results
Experimental evolution procedure
In our previous study (Giacomucci et al., 2019), we observed a heterogenous pattern on soft agar with large protuberances in the presence of PmB (Figure 1). No protuberances were observed in the control without PmB. We hypothesized that they correspond to a mutation in the bacterial chromosome that compensates for the loss of motility in the presence of PmB. Therefore, we performed an experimental evolution to select for adaptative variants that gained motility in the presence of PmB (Figure 2). To do so, we plated the wild-type strains on soft agar supplemented with a subinhibitory concentration of PmB that causes no pore formation in the envelope of both strains used in this study (Giacomucci et al., 2019). After 48 h, we sampled bacteria at the edge of the protuberances and streaked them on LB medium to obtain isolated colonies. Two wild-type mother strains were used: V. cholerae O1 El Tor A1552 (A1552-WT) and O139 MO10 (MO10-WT). Eight samplings have been performed for each strain tested, and one colony representative of each sample has been selected and conserved for a total of 16 variants: A1552-V1 to V8 and MO10-V1 to V8. Then, we confirmed the motility phenotype of the variants in the presence of PmB (25 μg/ml) on soft agar. After 48 h of incubation, three variants of each strain were selected based on their increased motility in the presence of PmB (Figure 3). The variants selected are A1552-V1, A1552-V2, and A1552-V6 and MO10-V2, MO10-V7, and MO10-V8 from the A1552 and MO10 parental strains, respectively. Most of the remaining variants were less motile than the ones that were selected (n = 9), and one did not show any increase in motility at the confirmation step. As expected, the WT strains in the presence of PmB demonstrated reduced motility with a diameter of ∼2.5 cm. Without PmB, the motility pattern of the variants was comparable with the control, displaying a more circular pattern (data not shown) and a motility diameter of ∼4.3 cm on average (Giacomucci et al., 2019; Figure 3). Altogether, these results suggest that the variants have compensated for the loss of motility observed in the presence of PmB.
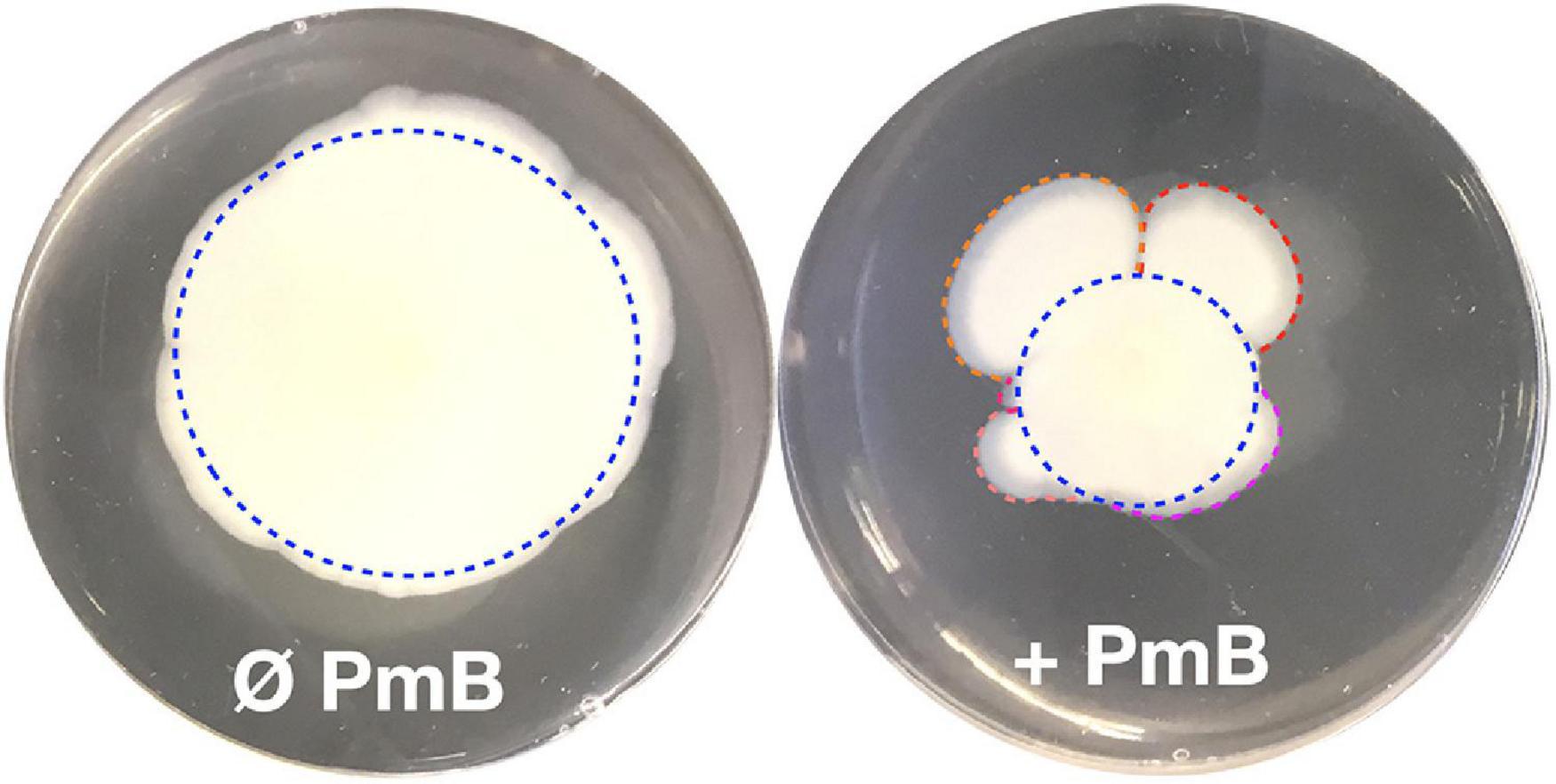
Figure 1. A flower-like pattern often appears in V. cholerae motility tests in the presence of polymyxin B. Motility assay on soft agar in the absence (Ø) or in the presence (+) of PmB. In the presence of PmB, the motility pattern of V. cholerae is not symmetrical as we would expect (blue dotted circles) but shows protrusions (red, orange, violet, salmon, and pink curved lines). In absence of PmB, the motility pattern is much less heterogenous than in the presence of PmB.
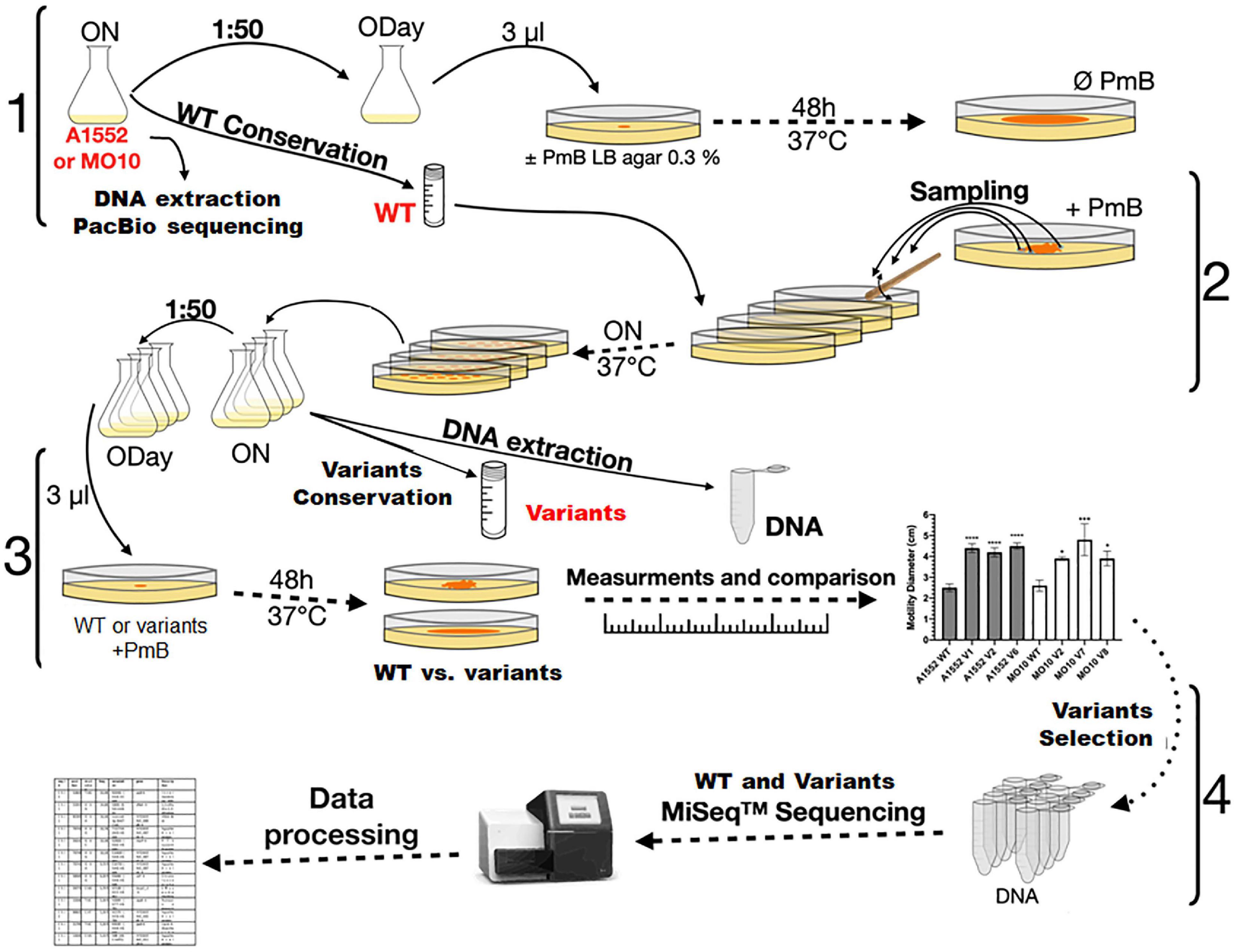
Figure 2. Experimental evolution design. V. cholerae A1552 and MO10 strains were grown overnight in LB broth. Genomic DNA from both strains was extracted and sequenced by Pac Bio. After dilution, the bacteria were grown to an optical density at 600 nm (OD600 nm) of 0.3 (ODay). Aliquots were spotted in technical triplicates on soft LB-agar supplemented or not with PmB. After incubation, samples of the different strains were taken from the edge of different protuberances of the flower-like patterns and streaked on LB agar. A colony from each of the different WT strains and their putative variants were grown overnight in LB broth and conserved at –80°C. The motility of the WT strains and variants was compared on soft agar plates supplemented with PmB. Motility diameters of the variants were measured and compared to those of their respective WT strains. The genomic DNA of selected hypermotile variants was extracted and sequenced by MiSeq.
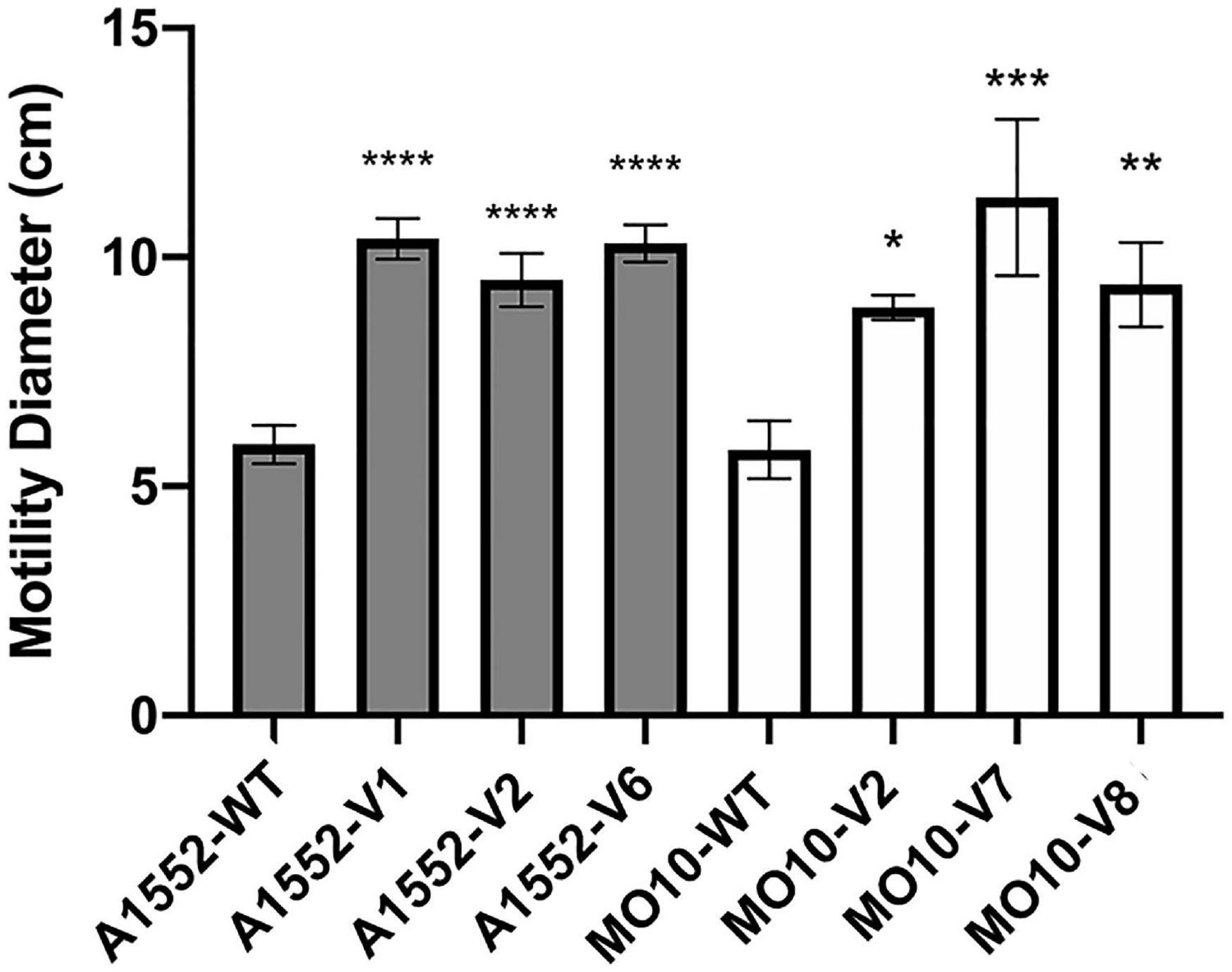
Figure 3. Variants of A1552 and MO10 present increased motility in the presence of PmB. Motility assay on soft agar supplemented with PmB after 48 h. Data are represented as mean ± SD of at least 3 independent experiments. Statistical significance between the variants’ motility diameter and their respective WT stains was tested by Dunnett’s one-way ANOVA. (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
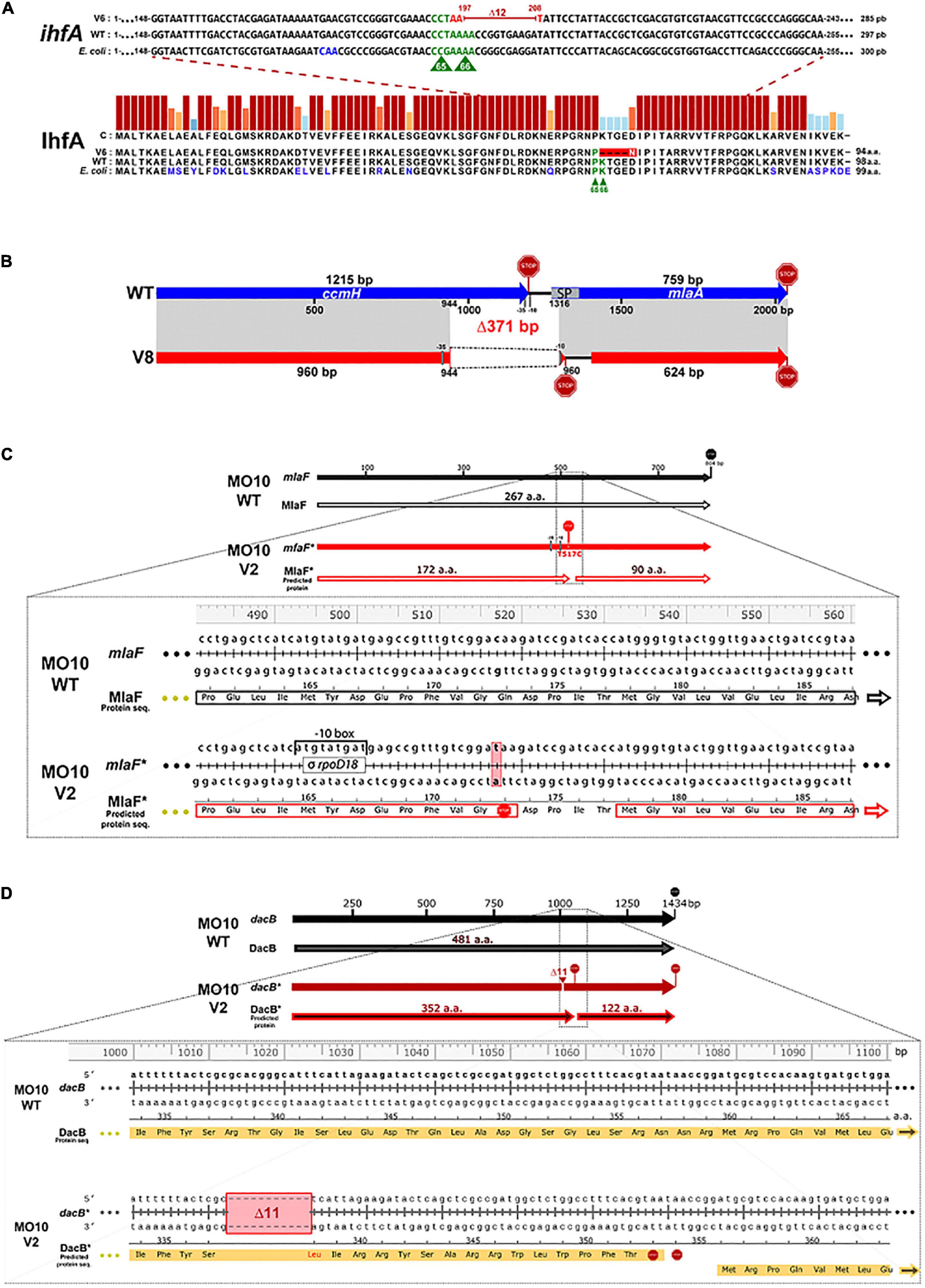
Figure 4. Identification of the mutations in the variant strains and their repercussions on the final product. (A) The deletion of 12 nucleotides in V. cholerae A1552-V6 suggests the loss of essential amino acid for IhfA function. Gene sequence (ihfA) and protein sequence (IhfA) alignments of V. cholerae A1552-V6 (V6), A1552-WT (WT), and E. coli MG1655 (E. coli). The IhfA protein sequences of V. cholerae A1552-WT and E. coli MG1655 share 94% similarity and 81% identity. P65 and K66 amino acids (in green), essential for IhfA function in E. coli, are also present in V. cholerae A1552-WT strain. However, in A1552-V6, the deletion of 12 nucleotides (197–208, in red) in the ihfA sequence leads to the substitution of a lysin residue in position 66 by an asparagine residue (in white) and the loss of 4 amino acids. (B) The deletion of 371 bp between ccmH-1 and vacJ locus in MO10-V8 probably causes a loss of function of CcmH and VacJ. Schematic representation of ccmH-1 (VchoM_01282) and vacJ (mlaA, VchoM_01281) locus alignment (in gray) between V. cholerae MO10-WT (blue) and MO10-V8 (red). The 371-bp deletion in MO10-V8 leads to the loss of ∼20% of ccmH 3’ end and a frameshift leading to the emergence of a new stop codon in the vacJ’s 5’-ORF region. The vacJ gene also lost important features, including its -35 and -10 promoter regions and the first 30 coding nucleotides, including the signal peptide (SP) sequence (nucleotides 1–51, codons 1–17). The vacJ predicted ORF in MO10-V8 starts 123 nucleotides (41 codons) forward and new putative –35 and –10 promoter regions for vacJ have been detected (see Supplementary Figure S1). (C) A nonsense mutation in mlaF results in the loss of 26% of its original product. Alignments of mlaF gene sequence and MlaF protein translation sequence from MO10-WT and MO10-V2 (*). The substitution of the thymine-517 to cytosine in MO10-V2 resulted in the insertion of a premature stop codon (codon 173). MO10-V2 MlaF* has lost 95 amino acids (a. a.) corresponding to the ∼36% C-terminal end. A new ORF of 90 codons is created in the same reading frame, and a new promoter is predicted (refer to Supplementary Figure S2). (D) Deletion of 11 nucleotides in dacB in MO10-V2. Alignments of dacB gene sequence and DacB predicted amino acid sequence from MO10-WT and MO10-V2 (*). MO10-V2 had lost 11 nucleotides (1,013–1,023) resulting in a frame shift, causing the modification of 16 amino acids, the emergence of two early stop codons (STOP signs), and a new ORF starting at codon 360 of the WT sequence (refer to Supplementary Figure S3).
Identification of the mutations
To determine the genes involved in the motility restoration of the variants in the presence of PmB, we performed comparative genomic analysis. To do so, we first sequenced the genome of the WT strains using PacBio technology. The variants were sequenced using a MiSeq technology and compared to the genome of their respective WT strain. The results of the comparative genomic analysis are presented in Supplementary Table S1 for A1552-V1, A1552-V2, A1552-V6, MO10-V2, and MO10-V8. The sequencing of MO10-V7 failed, and thus, we decided to pursue our investigation of the 5 variants with successful sequencing. Since the sequencing analysis was performed on DNA extracted from isolated variants, the only mutations that were considered are those with frequencies of 100%, which means that all the reads mapping this region identified the indicated mutations. The mutations were then confirmed by Sanger sequencing (Supplementary Materials and Methods).
The only mutation detected in the A1552 variants occurs in A1552-V6 and consists of a deletion of 12 nucleotides in ihfA (VC1222) (Figure 4A). This mutation results in the loss of 4 amino acids (67-70) and a mutation of one amino acid in position 66 (K66N). A comparison of the ihfA sequence in A1552 with its homolog in E. coli demonstrates that the sequence is highly conserved (94% similarity and 81% identity). Interestingly, the amino acids responsible for the IHF function in E. coli have been identified (P65 and K66) and are conserved in the wild-type sequence of A1552. Conversely, K66 is deleted in A1552-V6 (Figure 4A). Therefore, it is possible that the protein remains transcribed but with an attenuated functionality.
In MO10-V8, we noticed a deletion of 371 nucleotides in a region covering ccmH (VC2050) and vacJ (mlaA, VC2048) genes (Figure 4B). Regarding vacJ, which encodes a lipoprotein of the Mla (maintenance of lipid asymmetry) pathway, the first 17 codons were deleted, which results in the loss of the signal peptide for the secretion through the outer membrane as determined using SignalP 5.0 software (Almagro Armenteros et al., 2019). Therefore, it is expected that the mutation in vacJ induces a loss of function. Regarding ccmH, which encodes a c-type cytochrome maturation protein (Thony-Meyer, 1997; Braun and Thony-Meyer, 2005), the mutation results in the modification of four amino acids and the deletion of the 85 subsequent amino- acids in the C-terminal, which represents more than 20% of the sequence and the periplasmic domain of the protein.
In MO10-V2, we observed a point mutation resulting in the insertion of a stop codon in the mlaF (VC2520) sequence, which encodes an ATPase of the Mla pathway (Figure 4C). In addition, we observed in this variant an 11-nucleotide deletion in dacB (VC0632) in positions 1,013–1,023 (Figure 4D). This deletion results in a shift in the open reading frame and the emergence of a stop codon. In addition, DacB protein has lost ∼30% of its C-terminal residues. Therefore, it is very likely that dacB mutation in MO10-V2 results in a loss of function.
To confirm that the mutations identified through the experimental evolution are responsible for the gain of motility, we complemented the variants with a WT copy of the genes of interest using the pBAD24 vector. The results demonstrate that the growth default of A1552-V8 is restored upon complementation (Supplementary Figure S4). Besides, the motility is partially restored after complementation for all the strains tested (Figure 5).
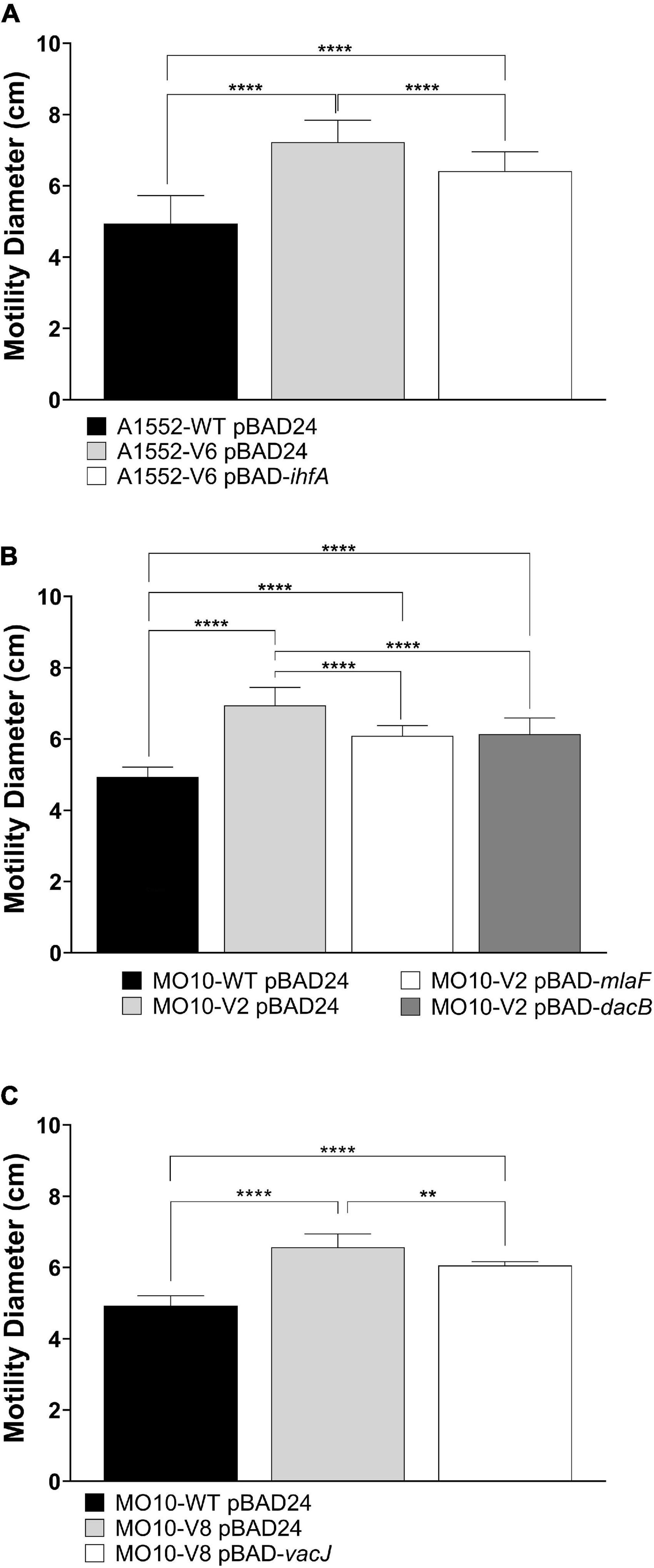
Figure 5. Effect of ihfA, vacJ, mlaF, or dacB overexpression on WT and variant strains’ motility on soft agar in the presence of polymyxin B. (A) A1552-WT and A1552-V6 carrying pBAD24 or pBAD24-ihfA, (B) MO10-WT and MO10-V2 carrying pBAD24, pBAD24-mlaF or pBAD24-dacB, and (C) MO10-WT and MO10-V8 carrying pBAD24 or pBAD24-vacJ. Data are represented as mean ± SD of at least three independent experiments (**p < 0.01; ****p < 0.0001).
Electron microscopy observations of the variants
To determine the impact of the mutations on the bacterial morphology, we observed the variants by electron microscopy. No major structural differences were observed between A1552-WT and A1552-V6 cells (Figure 6A). By looking closer into the flagellum’s structure, we observed that they both have an enveloped flagellum. However, while the A1552-WT filament is centered in the outer membrane envelope, the positioning of the A1552-V6 filament appears disorganized (Figure 6A). Regarding MO10-V2, we observed no difference in the flagellum and the cell structure while the flagellum of MO10-V8 appears thinner (Figure 6B). We also observed that MO10-V2 produces very large vesicles in comparison with MO10-WT (Figure 6B). In the presence of PmB, both MO10-V2 and MO10-V8 are entirely coated with large MVs, which is not the case with the MO10-WT (Figure 6B). In all variants, we can observe that the flagellum remains attached in the presence of PmB (Figures 6A,B) and not for the A1552-WT and MO10-WT strains, which is in line with the motility results.
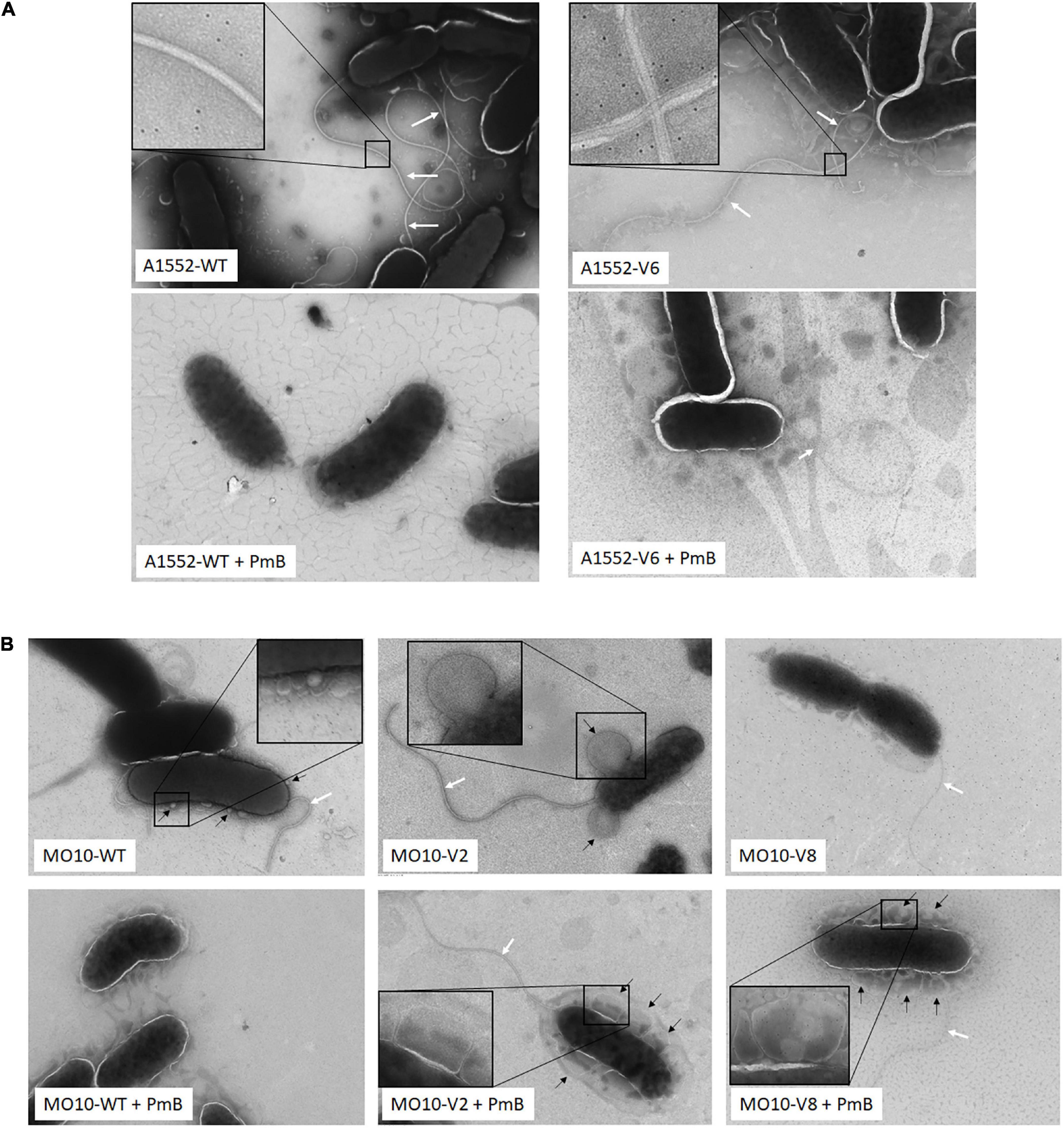
Figure 6. Electron microscopy images. (A) A1552-WT and A1552-V6 and (B) MO10-WT, MO10-V2, and MO10-V6. Bacteria were grown to an exponential phase in the absence and the presence of PmB (+ PmB). White arrows indicate the bacterial flagellum and black arrows indicate membrane vesicles.
Determination of the role of the membrane vesicles in PmB titration and resistance
Because we observed the presence of large vesicles in MO10-V2 and MO10-V8 variants, we analyzed the vesiculation of these strains. To do so, the MVs of MO10-V2, MO10-V8, and MO10-WT were isolated by ultracentrifugation of the cell-free supernatant. The amount of MVs produced was estimated by quantification of the LPS and the protein content. MO10-V2 and MO10-V8 vesicle fractions are richer in LPS than the vesicle fraction of the MO10-WT, with MO10-V2 containing the largest amount (Figure 7B), suggesting an increase in the quantity of vesicles released. Moreover, a nitrate coloration of an SDS gel (Figure 7C) shows larger bands corresponding to the LPS size (gray arrows) in the vesicles of MO10-V2 and –V8 than in the MVs of MO10-WT. Similarly, the protein concentration of the vesicle fraction, estimated by the Bradford assay, showed an increase in MO10-V2 and -V8 in comparison with MO10-WT MVs (Figure 7A). MO10-V2 and -V8 complementation restored the protein concentrations in the vesicles to the MO10-WT level (data not shown). Similar results were obtained using a MO10ΔvacJ:cmR mutant in terms of MVs formation and PmB sequestration. The protein concentration increased by 1.68-folds, and we observed larger LPS bands on a sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel upon vacJ mutation (Supplementary Figures S5A,B). Besides, the PmB sequestration assay showed that the MVs from MO10ΔvacJ:cmR sequestered more PmB than the MVs from MO10 (Supplementary Figure S5B).
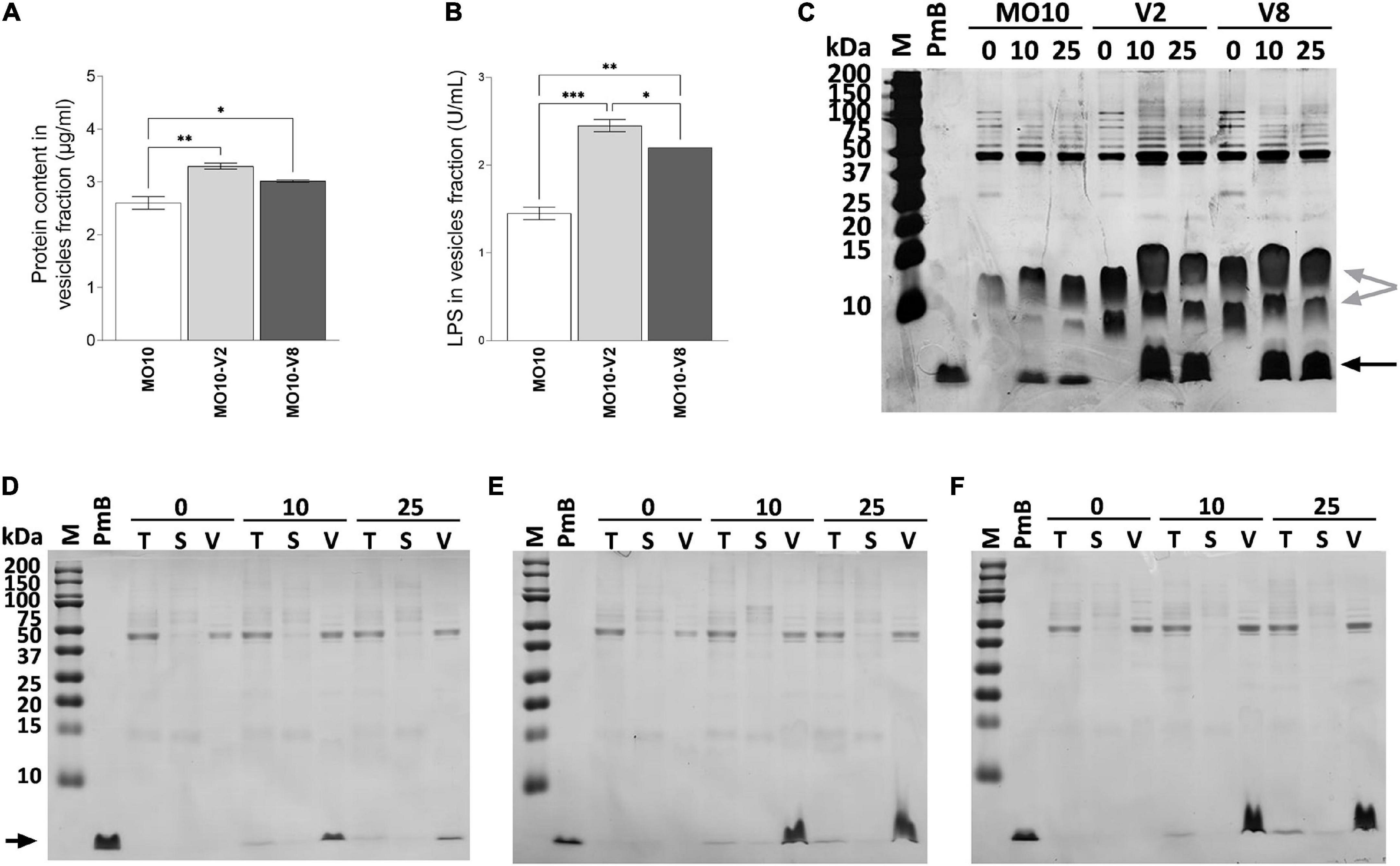
Figure 7. MO10-V2 and MO10-V8 variants produce more membrane vesicles and titer more polymyxin B than MO10-WT. The membrane vesicles were isolated by ultracentrifugation. The protein content (A) and LPS (B) of the vesicles isolated from MO10-WT (MO10) and its hypermotile variants, MO10-V2 and MO10-V8 cultures were quantified. Data represent mean values ± standard error of the mean from at least three independent experiments. (*p < 0.05; **p < 0.005; ***p < 0.0001). The polymyxin B (PmB) titration capacity of the vesicles from MO10-WT (D), -V2 (E), and -V8 (F) was assessed by incubating the cell-free culture supernatant with 0, 10, or 25 μg/ml of PmB (0, 10, and 25, respectively). Samples from total culture supernatant (T), vesicles (V), and vesicles-free supernatant (S) were analyzed. Proteins were visualized on SDS-PAGE after Coomassie blue staining. PmB alone was used as control of the PmB size (black arrow). (C) The vesicle fraction from MO10-WT, -V2, and -V8 incubated with or without 10 or 25 μg/ml of PmB migrated on an SDS-PAGE gel. A silver nitrate coloration of the gel shows LPS (gray arrows) and PmB (black arrow). Gel pictures are representative of at least three independent experiments.
To further investigate the role of the MVs in the resistance of the MO10 variants to AMPs, we analyzed their ability to sequester PmB. To do so, we incubated the cell-free supernatant of MO10-V2, MO10-V8, and MO10-WT with 0, 10, and 25 μg/ml of PmB. After 1 h of interaction, we collected a sample of the total supernatant (T) and performed ultracentrifugation to separate the MVs (V) from the vesicle-free supernatant (S). The samples from the total supernatant (T), the vesicle fraction (V), and the vesicle-free supernatant (S) migrated on an SDS-PAGE electrophoresis gel. The PmB alone was used as control of the PmB size. The band corresponding to PmB size is not found in samples incubated without PmB (Figures 7D–F). We observed a band corresponding to the size of the PmB in the total supernatant (T) that is darker than in the vesicles-free supernatant (S). In addition, an intense band is observed in the vesicles fraction (V) (Figures 7D–F), suggesting that most of the PmB is associated with the MVs. In addition, we observed that the MVs isolated from an equal volume of MO10-V2 and MO10-V8 cell-free supernatants can sequester more PmB than the ones from MO10-WT, as shown by a larger band of PmB size (Figures 7D–F). To determine the ability of the MVs to protect the bacteria against PmB, we performed a protection assay using MVs. To do so, MO10-WT bacteria were incubated with the MVs from MO10-WT, –V2, and –V8, or without MVs, and a lethal concentration of PmB (200 μg/ml). We observed that the MVs from MO10-V2 and –V8 increased the survival of MO10-WT (Figure 8A), which is in line with the sequestration assay results. Conversely, the MVs from MO10-WT did not protect the bacteria from PmB. After concentration normalization of the MVs from MO10-WT, –V2, and –V8 according to the LPS dosage, the protection conferred by the MVs from MO10-V2 and –V8 was similar to that conferred by the vesicles of MO10-WT (Figure 8A). It indicates that the higher protection and sequestration of PmB by MO10-V2 and –V8 are probably due to the number of MVs produced rather than their composition. Altogether, our results suggest that the increased production of MVs by the MO10 variants allows more efficient sequestration of the PmB by the MVs, resulting in bacterial protection toward PmB.
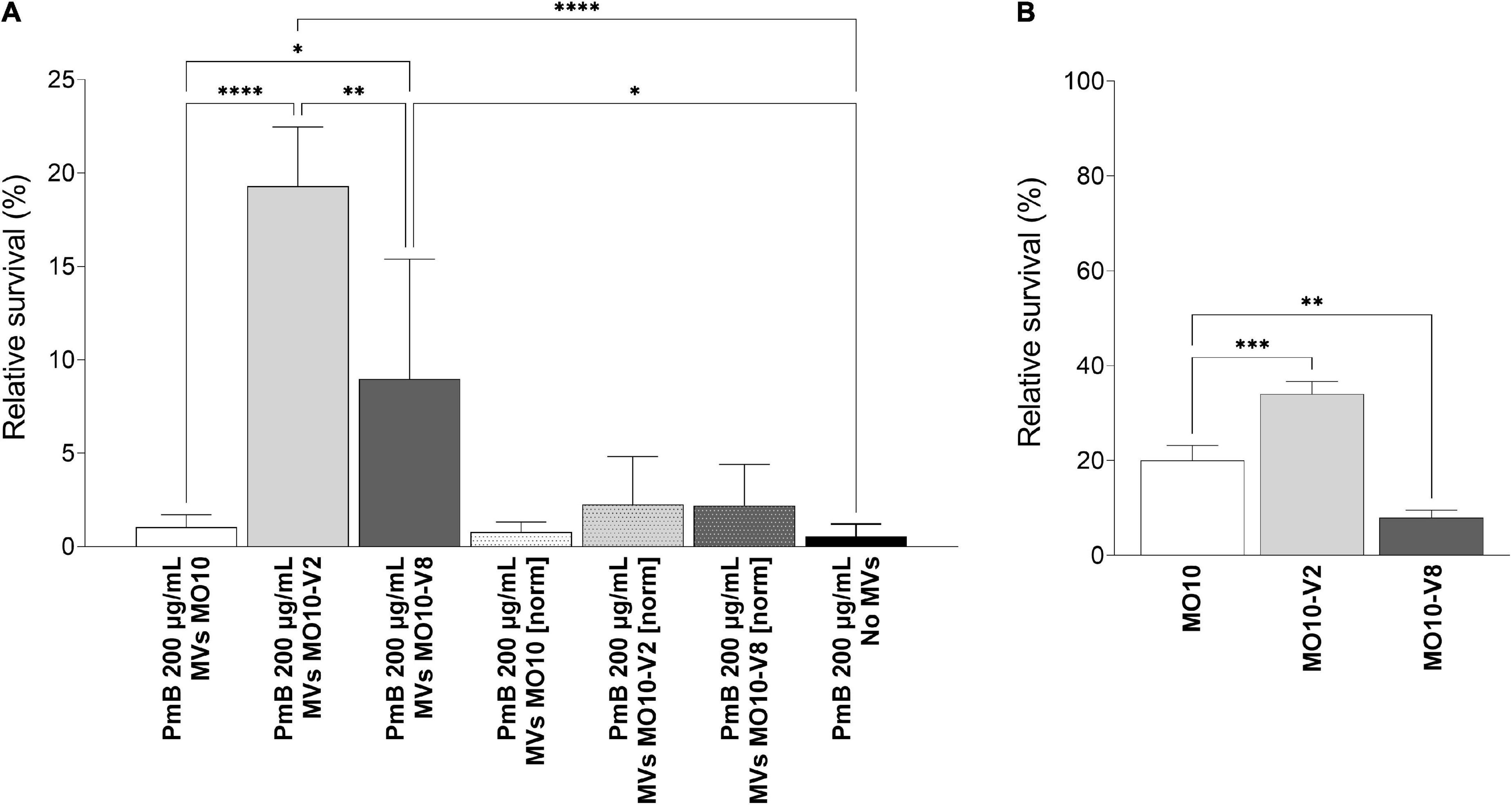
Figure 8. Relative survival to lethal concentration of polymyxin B. (A) Relative survival of MO10-WT in the presence of 200 μg/mL of PmB for 1 h, with or without MVs from MO10-WT, -V2, or -V8, normalized (MVs MO10-WT [norm], MVs MO10-V2 [norm], and MVs MO10-V8 [norm]) to the same concentration or not (MVs MO10-WT, MVs MO10-V2, and MVs MO10-V8). (B) Relative survival of MO10-WT, -V2, or -V8 after 30 min in the presence of a lethal concentration of PmB. Data are presented as mean ± standard error from at least 3 independent experiments. The % of survival was calculated in comparison with the condition without PmB (p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
We further investigated the resistance of MO10-WT and its variants to PmB by first determining the minimum inhibitory concentration (MIC) and observed no difference between the MO10-WT, –V2, and –V8 strains (data not shown). In parallel, we tested their ability to survive a lethal concentration of PmB. Our results show that MO10-V2 survival rate after 1 h is higher than the survival rate of MO10-WT (Figure 8B), indicating that MO10-V2 is more resistant to PmB than its parental strain. Conversely, the MO10-V8 appeared to be more sensitive to the PmB than MO10-WT (Figure 8B).
Analysis of the flagellins genes’ expression
A1552-V6 presents a mutation on the predicted binding site of IhfA. IHF is an important regulator of virulence, colonization, and motility in V. cholerae. Thus, we wondered whether this mutation has an impact on the expression of the flagellar genes. To do so, we determined the relative expression of flaA and flaE, the first genes of both flagellin operons, in A1552-V6. We observed no difference in flaA and flaE expression in A1552-V6 in comparison with the parental strain A1552-WT (Supplementary Figure S6A), suggesting that the impact of the mutation in ihfA on the increased motility in A1552-V6 is not due to a modulation in the flagella-related genes. We also determined the expression of flaA and flaE in MO10-WT and its MO10-V2 and –V8 variants (Supplementary Figure S6B). Our results demonstrate a significant reduction of the flagellins’ expression in both variants.
Discussion
To identify the genes that are important for motility in the presence of a sub-inhibitory concentration of PmB, we performed a short experimental evolution protocol. Experimental evolution has been widely used to validate evolutionary theories and to identify genes involved in responses to various stresses in bacteria (Mcdonald, 2019). In the latter case, de novo mutations driven by stresses led to the identification of genes that are important for survival in the presence of oxidative stress and antibiotics, or during a temperature shift (Hoffmann and Hercus, 2000; Justin et al., 2019). In this study, we used a targeted approach to identify the variants from single clones, which might have obscure other mutations. We identified mutations in genes potentially important for motility in the presence of PmB in two different strains of V. cholerae, i.e., A1552 (O1 El Tor) and MO10 (O139). These mutations in vacJ-ccmH mlaF, dacB, and ihfA have been identified in three variants, two in MO10, and one in A1552.
In our previous study, we demonstrated that the loss of motility in the presence of PmB was due to a high proportion of aflagellated bacteria (Giacomucci et al., 2019). Here, we show that there is no increase in the expression of the flagellins in the variants of both strains that could explain the gain of motility. However, if no difference is observed in A1552-V6, we observed a downregulation of both flagellin genes in the M10-V2 and –V8 variants in comparison with MO10-WT. This downregulation has no impact on their motility in the absence of PmB. V. cholerae has one single polar flagellum coated by an outer membrane sheath. In MO10-V8, the flagellum appears thicker in the electron microscopy pictures, whereas the flagellum of MO10-V2 and MO10-WT appeared to be alike. It has been demonstrated in V. cholerae that the structural integrity of the flagellum is linked to the composition of the outer membrane (Bari et al., 2012). The MO10 variants have a mutation in genes involved in the envelope biogenesis and maintenance. Thus, it is possible that the mutations affect the outer membrane composition and thereby the regulation of the flagellins’ expression.
We previously showed that the absence of the flagellum in the presence of PmB was not resulting in the repression of the flagellins genes, but likely to a default in the flagellum’s assembly or a flagellum miss-anchoring. In any case, the absence of the flagellum is related to envelope perturbations due to PmB, while no pores in the inner membrane were observed (Giacomucci et al., 2019). In this study, we identified 2 genes belonging to the maintenance of lipid asymmetry (Mla) pathway, i.e., vacJ (mlaA) and mlaF. VacJ is homologous to MlaA, an alpha-helical outer membrane lipoprotein, while MlaF is an ATPAse associated with the inner membrane, in complex with MlaD and MlaE. MlaC and MlaB are periplasmic and cytoplasmic proteins, respectively (Du Toit, 2019). It has been previously described in E. coli that the Mla pathway is essential for preserving the outer membrane lipid asymmetry by preventing phospholipids’ accumulation in the outer leaflet through an anterograde phospholipid transport (Malinverni and Silhavy, 2009; Du Toit, 2019) and is involved in the virulence toward silkworm (Nasu et al., 2022). The Mla pathway is important for antimicrobial tolerance in several gram-negative bacteria, including Burkholderia and Pseudomonas (Munguia et al., 2017; Bernier et al., 2018). In V. cholerae, it has been demonstrated that the Mla pathway is involved in MVs biogenesis and is important for serum resistance (Roier et al., 2016). The authors proposed that the hypervesiculation mediated by defects in the Mla pathway is a conserved bacterial mechanism to prevent phospholipids accumulation in the outer membrane (Roier et al., 2016). In this study, we demonstrate that the Mla pathway involved in hypervesiculation leads to the production of very large vesicles that can coat the bacteria in the presence of subinhibitory concentrations of PmB.
The deletion identified in MO10-V8 also affects the sequence of the gene adjacent to vacJ, i.e., ccmH, which encodes for a protein involved in a c-type cytochrome maturation. The cytochromes are important in bacterial metabolism as they function as electron transfer proteins. C-type cytochromes are characterized by the covalent liaison of the polypeptide with a heme (Kranz et al., 2009). In MO10-V8, a deletion of 371 nucleotides is observed, which represents ∼20% of the protein. Therefore, it is likely that CcmH has lost its function in this variant. In V. cholerae, there are 14 proteins containing a CXXCH motif and a signal peptide in N-terminal for the association with the membrane (Braun and Thony-Meyer, 2005). Among them, 7 have been characterized for their function in respiration in aerobic and anaerobic conditions, and one is a peroxidase probably involved in oxidative stress response (Braun and Thony-Meyer, 2005). CcmH is a part of the 6 other c-type cytochrome maturation proteins that are clustered together (ccmA-I) on V. cholerae’s chromosomes and are not required for growth under aerobic conditions in rich media (Braun and Thony-Meyer, 2005). To our knowledge, there is no evidence that ccmH is important for motility or membrane integrity, and its role in bacterial metabolism is probably very limited in our experimental conditions. Therefore, the implication of the deletion that covers both ccmH and vacJ sequences in bacterial motility recovery in the presence of PmB most probably relies on the inactivation of vacJ.
In another variant of the O139 strain MO10, a mutation has been observed in dacB, which encodes PBP4, a low molecular weight cell wall-synthesizing enzymes/penicillin-binding proteins (PBPs) involved in the peptidoglycan biosynthesis pathway (Typas et al., 2011). In Vibrio parahaemolyticus, dacB has a role in the formation of abnormal cell shape during the transition to a viable, but not culturable, state (Hung et al., 2013). In E. coli, low molecular weight PBPs also have a role in cell shape maintenance (Nelson and Young, 2001). The dacB mutant cells are wider and display localized transparent bulges at the poles (Yang et al., 2018). The authors also demonstrated that an accumulation of soluble peptidoglycan occurs in a dacB knockout mutant. Additionally, it has been demonstrated that peptidoglycan remodeling, especially the alteration in the peptidoglycan-outer membrane cross-linking, regulates the formation of MVs (Schwechheimer et al., 2015). This alteration involves the endopeptidase activities of PBP4 and another protein, Spr, under the regulation by NlpI. Altogether, the roles of dacB in cell shape maintenance, peptidoglycan biosynthesis, and vesicles formation pinpoint its importance in the cell wall remodeling that might be important for the stability of the flagellum in V. cholerae.
The role of vesiculation during V. cholerae infection in mammals has been investigated, and the results demonstrated that the Mla pathway is silenced at the early stages of infection to facilitate the outer membrane remodeling through hypervesiculation (Zingl et al., 2020). This remodeling confers resistance to AMPs and bile salts (Zingl et al., 2020). We previously demonstrated that the MVs of Vibrio can sequester the AMPs interacting with the outer membrane and the LPS, resulting in a reduction in the residual antimicrobial quantity interacting with the bacterial envelope (Duperthuy et al., 2013; Vanhove et al., 2015). In this study, we show that the MVs of both MO10 variants, MO10-V2 and MO10-V8, can sequester more PmB than the MVs of the parental strain. We further demonstrated that the MVs from MO10-V2 and –V8 can better protect the bacteria than their WT counterparts. This higher protection is most likely linked to the higher quantity of MVs produced, as protection assays using a similar quantity of MVs showed no differences between the MVs from the variants and the WT stain. Thus, the protection by the variants’ MVs is due to the hypervesiculation, as determined by LPS and protein quantification, and leads to the sequestration of a larger quantity of PmB. Besides, electron microscopy observations revealed that the MVs secreted by the MO10 variants are larger in size than those of the parental strain. Therefore, it is likely that the increased motility observed in the MO10 variants results from a reduction of the impact of the PmB on the bacterial cell membrane through MV sequestration, leading to increased stability of the flagellum.
While determining the ability of MO10-V2 and –V8 to resist PmB, we observed that more MO10-V2 survived to a lethal concentration of PmB, even though its MIC is similar to that of the WT strain. While performing MIC assays, bacteria are harvested at the early exponential growth phase and are heavily diluted (OD600 nm = 0.0001). Thus, few MVs have been produced at this stage of growth and they have been diluted. Conversely, the survival assay was performed on bacteria at an OD600 nm of 1 (midlog phase), without washing or diluting. Consequently, the MVs produced by the bacteria remained in the media during the assay. Thus, the increased survival of MO10-V2 to a lethal concentration of PmB is probably due to the sequestration of PmB by its MVs, which are more abundant than in MO10-WT cultures. Conversely, less MO10-V8 than MO10-WT appeared to survive while facing a lethal concentration of PmB, while its MIC is also similar to that of the MO10-WT strain. It has been previously reported that the MIC of PmB is not affected upon vacJ mutation in V. cholerae (Roier et al., 2016). It is, however, possible that the envelope integrity is slightly disturbed, not enough to observe a difference in MIC, but sufficient to observe a deficient survival to lethal concentrations of PmB. This result also implies that the quantity of MVs produced by MO10-V8 at the midlog phase is not sufficient to protect the bacteria from PmB, even though our protection results using MO10-V8 MVs from an overnight culture can protect MO10-WT. A thorough analysis of the dynamic of MV production in both MO10-V2 and –V8 would certainly increase our understanding of the differences between MO10-V8, –V2, and –WT in terms of survival to lethal concentrations of PmB and MV production.
Altogether, the results of our previous study (Giacomucci et al., 2019) and the current study show that, in the presence of PmB, most of the V. cholerae cells are aflagellated and consequently non-motile (Figure 9). However, some cells can acquire mutations in important vesiculation regulatory pathways, leading to hypervesiculation through large MVs production and PmB titration. Consequently, the flagellum remains attached to the bacterial cell and motility is maintained (Figure 9). Thus, membrane remodeling and hypervesiculation are involved in V. cholerae adaptation to subinhibitory concentrations of PmB. However, since our study is based on a targeted approach by single variant isolation, it is difficult to determine to what extent the proposed mechanism is common at the population level. A global approach using parental and evolved populations would answer this question.
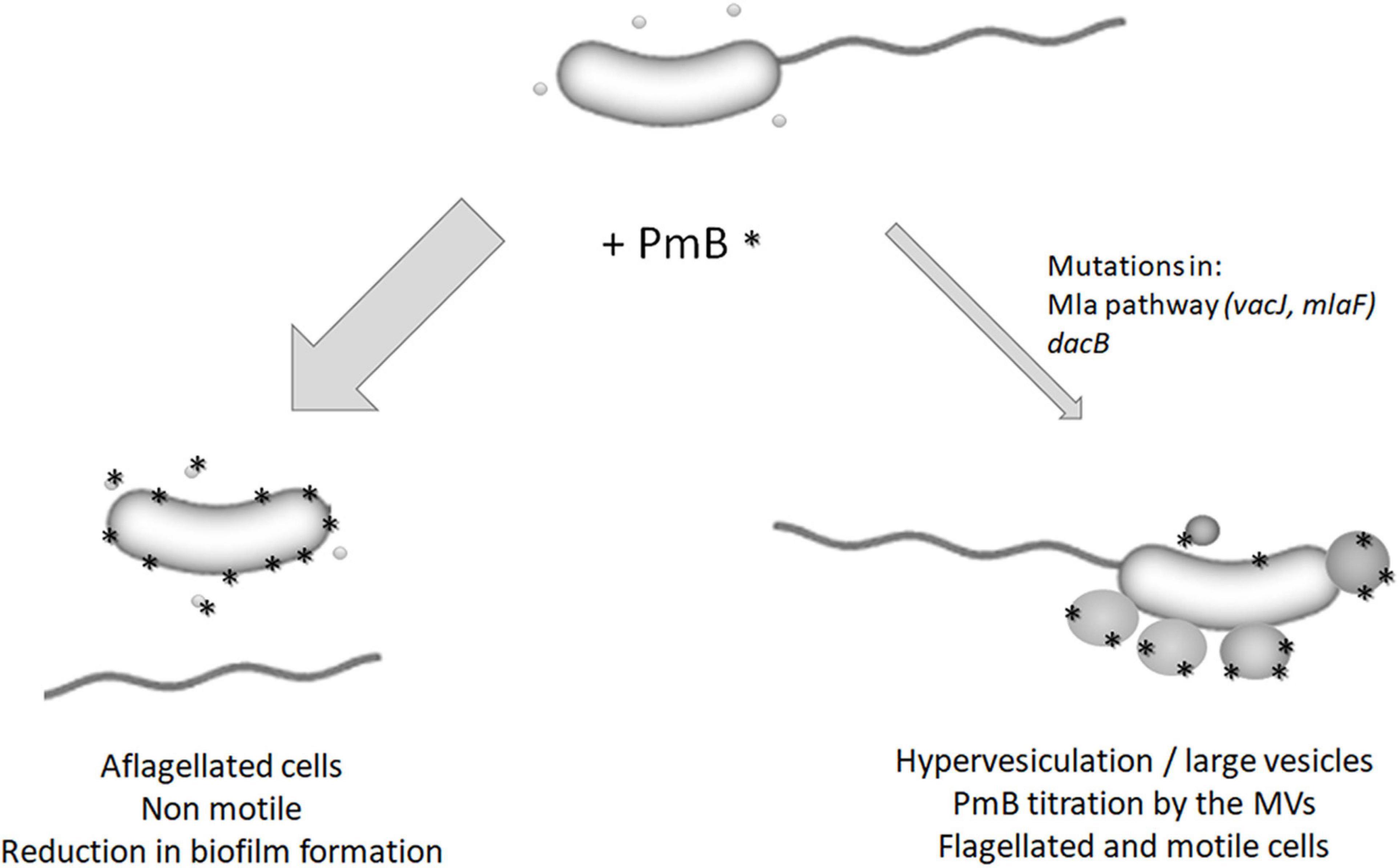
Figure 9. Model for the impact of polymyxin B on V. cholerae MO10. In the presence of subinhibitory and non-pore-forming concentrations of PmB, most of the bacterial cells are aflagellated, which results in non-motile cells and a reduction in biofilm formation (Giacomucci et al., 2019). However, some cells can adapt to the presence of PmB by mutating genes involved in the regulation of the vesiculation process (mlaF, vacJ, and dacB). It results in hypervesiculation and the formation of large MVs that can titer the PmB, thereby reducing its impact on the bacterial membrane and flagellum’s assembly and anchoring. These cells are flagellated and motile.
The only mutation identified in the O1 El Tor strain A1552 is a deletion of 12 nucleotides in ihfA. ihfA encodes IhfA, one of the IHF subunits, together with IhfB. IHF is a histone-like DNA-binding protein that regulates many functions in bacterial cells, including transcription, replication, and virulence (Freundlich et al., 1992). In V. cholerae, IHF is essential for conjugation and virulence genes’ expression (Mcleod et al., 2006). The deletion of 12 nucleotides likely results in the attenuation of IHF function since one of the two essential amino acids for DNA binding has been deleted. It has been demonstrated in a V. cholerae O1 El tor that IHF binds to rpoN and flrA, two major regulators of flagellar genes’ expression (Wang et al., 2012). The binding of IHF to rpoN and flrA promoter regions leads to a limitation of H-NS binding to these regions (Wang et al., 2012). The role of H-NS as a negative regulator of rpoN and flrA has previously been described (Prouty et al., 2001). Thus, an IHF inactivation would lead to the binding of H-NS and eventually to the repression of the flagellum-related genes. However, we noticed that the IHF-binding site in the promoter region of rpoN is absent in A1552. We previously demonstrated that the expression of the flagellin subunits is not reduced in the presence of PmB (Giacomucci et al., 2019), and in this study, we observed no difference in flagellins’ expression in A1552-V6. Since IHF-binding sites have been identified all over the bacterial genome (Prieto et al., 2012; Reverchon et al., 2021), it is reasonable to hypothesize that the mechanisms behind the gain in motility in the presence of PmB involving ihfA mutation are not related to the flagellum itself. As stated above, the loss of motility in the presence of PmB is likely due to a structural defect in the flagellum or the cell envelope. It has been recently demonstrated that a deletion of ihfA in Dickeya, a bacterium belonging to the Enterobacteriaceae, results in strong transcriptomic modifications, reflecting a “survival mode” associated with cell envelope modifications (peptidoglycan, porins, lipopolysaccharide, and exopolysaccharides) (Reverchon et al., 2021). In addition, the expression of major porins of E. coli is controlled by IHF (Huang et al., 1990; Ramani et al., 1992). In Salmonella enterica, an ihfA mutation induces the expression of genes involved in peptidoglycan and lipopolysaccharides biosynthesis (Mangan et al., 2006). Therefore, it is possible that a mutation in ihfA influences the expression of genes that strengthen the bacterial envelope, leading to the retention of the flagellum in the presence of PmB.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
SG, AM-D, and MD: conceptualization. SG, AM-D, and AV: data curation and formal analysis. SG, AM-D, AV, and HJ: investigation. SG and AM-D: methodology. AV: software. SG, AM-D, and MD: validation and visualization. AM-D, SG, AV, and MD: writing – original draft. SG, AM-D, AV, HJ, and MD: writing, reviewing, and editing. MD: resources, supervision, funding acquisition, and project administration. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC http://www.nserc-crsng.gc.ca/index_eng.asp) Discovery grant (grant number: RGPIN-2017-05322; MD). SG received salary from the RAQ (Ressources Aquatiques Québec), an inter-institutional group supported financially by the Fonds de recherche du Québec – Nature et technologies (FRQNT) (Programme regroupements stratégiques). AM-D received financial support from the NSERC scholarship program (BESC D3 – 558624 – 2021). HJ received financial support from the faculty of medicine of Université de Montréal (Bourse de mérite).
Acknowledgments
The sequences of the bacterial strains were performed by Genome Québec Center of expertise and services. Electron microscopy acquisitions were performed by Arnaldo Nakamura from the Plateforme de Microscopie Electronique de l’Institut Armand-Frappier, INRS. We would also like to thank Julien Pauzé-Foixet for the technical support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.932165/full#supplementary-material
References
Allué-Guardia, A., Echazarreta, M., Koenig, S. S. K., Klose, K. E., and Eppinger, M. (2018). Closed Genome Sequence of Vibrio cholerae O1 El Tor Inaba Strain A1552. Genome Announc. 6, e00098–18. doi: 10.1128/genomeA.00098-18
Almagro Armenteros, J. J., Tsirigos, K. D., Sonderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Altindis, E., Fu, Y., and Mekalanos, J. J. (2014). Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proc. Natl. Acad. Sci. U.S.A. 111, E1548–E1556. doi: 10.1073/pnas.1403683111
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Altschul, S. F., Wootton, J. C., Gertz, E. M., Agarwala, R., Morgulis, A., Schaffer, A. A., et al. (2005). Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272, 5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x
Aydanian, A., Tang, L., Morris, J. G., Johnson, J. A., and Stine, O. C. (2011). Genetic diversity of O-antigen biosynthesis regions in Vibrio cholerae. Appl. Environ. Microbiol. 77, 2247–2253. doi: 10.1128/AEM.01663-10
Bari, W., Lee, K.-M., and Yoon, S. S. (2012). Structural and functional importance of outer membrane proteins in Vibrio cholerae flagellum. J. Microbiol. 50, 631–637. doi: 10.1007/s12275-012-2116-3
Barrick, J. E., and Lenski, R. E. (2009). Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harb. Symp. Quant. Biol. 74, 119–129. doi: 10.1101/sqb.2009.74.018
Barrick, J. E., Yu, D. S., Yoon, S. H., Jeong, H., Oh, T. K., Schneider, D., et al. (2009). Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461, 1243–1247. doi: 10.1038/nature08480
Bernier, S. P., Son, S., and Surette, M. G. (2018). The Mla Pathway Plays an Essential Role in the Intrinsic Resistance of Burkholderia cepacia Complex Species to Antimicrobials and Host Innate Components. J. Bacteriol. 200, e00156–18. doi: 10.1128/JB.00156-18
Bevins, C. L., and Salzman, N. H. (2011). Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356–368. doi: 10.1038/nrmicro2546
Bohuszewicz, O., Liu, J., and Low, H. H. (2016). Membrane remodelling in bacteria. J. Struct. Biol. 196, 3–14. doi: 10.1016/j.jsb.2016.05.010
Boparai, J. K., and Sharma, P. K. (2020). Mini Review on Antimicrobial Peptides Sources, Mechanism and Recent Applications. Protein Pept. Lett. 27, 4–16. doi: 10.2174/18755305MTAwENDE80
Braun, M., and Thony-Meyer, L. (2005). Cytochrome c maturation and the physiological role of c-type cytochromes in Vibrio cholerae. J. Bacteriol. 187, 5996–6004. doi: 10.1128/JB.187.17.5996-6004.2005
Brudno, M., Malde, S., Poliakov, A., Do, C. B., Couronne, O., Dubchak, I., et al. (2003). Glocal alignment: Finding rearrangements during alignment. Bioinformatics 19, i54–i62. doi: 10.1093/bioinformatics/btg1005
Bryant, F. R. (1988). Construction of a Recombinase-Deficient Mutant Reca Protein That Retains Single-Stranded DNA-Dependent Atpase Activity. J. Biol. Chem. 263, 8716–8723. doi: 10.1016/S0021-9258(18)68364-4
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Daugelavicius, R., Bakiene, E., and Bamford, D. H. (2000). Stages of polymyxin B interaction with the Escherichia coli cell envelope. Antimicrob. Agents Chemother. 44, 2969–2978. doi: 10.1128/AAC.44.11.2969-2978.2000
Destoumieux-Garzón, D., Duperthuy, M., Vanhove, A. S., Schmitt, P., and Wai, S. N. (2014). Resistance to Antimicrobial Peptides in Vibrios. Antibiotics 3, 540–563. doi: 10.3390/antibiotics3040540
Du Toit, A. (2019). Phospholipid export from the inside out. Nat. Rev. Microbiol. 17:528. doi: 10.1038/s41579-019-0239-9
Duperthuy, M. (2020). Antimicrobial Peptides: Virulence and Resistance Modulation in Gram-Negative Bacteria. Microorganisms 8:280. doi: 10.3390/microorganisms8020280
Duperthuy, M., Sjöström, A. E., Sabharwal, D., Damghani, F., Uhlin, B. E., and Wai, S. N. (2013). Role of the Vibrio cholerae matrix protein Bap1 in cross-resistance to antimicrobial peptides. PLoS Pathog. 9:e1003620. doi: 10.1371/journal.ppat.1003620
Echazarreta, M. A., and Klose, K. E. (2019). Vibrio Flagellar Synthesis. Front. Cell. Infect. Microbiol. 9:131. doi: 10.3389/fcimb.2019.00131
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Faruque, S. M., Sack, D. A., Sack, R. B., Colwell, R. R., Takeda, Y., and Nair, G. B. (2003). Emergence and evolution of Vibrio cholerae O139. Proc. Natl. Acad. Sci. U.S.A. 100, 1304–1309. doi: 10.1073/pnas.0337468100
Freundlich, M., Ramani, N., Mathew, E., Sirko, A., and Tsui, P. (1992). The role of integration host factor in gene expression in Escherichia coli. Mol. Microbiol. 6, 2557–2563. doi: 10.1111/j.1365-2958.1992.tb01432.x
Fuerst, J. A., and Perry, J. W. (1988). Demonstration of lipopolysaccharide on sheathed flagella of Vibrio cholerae O:1 by protein A-gold immunoelectron microscopy. J. Bacteriol. 170, 1488–1494. doi: 10.1128/jb.170.4.1488-1494.1988
Giacomucci, S., Cros, C. D., Perron, X., Mathieu-Denoncourt, A., and Duperthuy, M. (2019). Flagella-dependent inhibition of biofilm formation by sub-inhibitory concentration of polymyxin B in Vibrio cholerae. PLoS One 14:e0221431. doi: 10.1371/journal.pone.0221431
Guerrero-Mandujano, A., Hernández-Cortez, C., Ibarra, J. A., and Castro-Escarpulli, G. (2017). The outer membrane vesicles: Secretion system type zero. Traffic 18, 425–432. doi: 10.1111/tra.12488
Guzman, L. M., Belin, D., Carson, M. J., and Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995
Harris, J. B., Larocque, R. C., Qadri, F., Ryan, E. T., and Calderwood, S. B. (2012). Cholera. Lancet 379, 2466–2476. doi: 10.1016/S0140-6736(12)60436-X
Hoffmann, A. A., and Hercus, M. J. (2000). Environmental Stress as an Evolutionary Force. BioScience 50, 217–226 doi: 10.1641/0006-3568(2000)050[0217:ESAAEF]2.3.CO;2
Huang, L., Tsui, P., and Freundlich, M. (1990). Integration host factor is a negative effector of in vivo and in vitro expression of ompC in Escherichia coli. J. Bacteriol. 172, 5293–5298. doi: 10.1128/jb.172.9.5293-5298.1990
Hung, W. C., Jane, W. N., and Wong, H. C. (2013). Association of a D-alanyl-D-alanine carboxypeptidase gene with the formation of aberrantly shaped cells during the induction of viable but nonculturable Vibrio parahaemolyticus. Appl. Environ. Microbiol. 79, 7305–7312. doi: 10.1128/AEM.01723-13
Josenhans, C., and Suerbaum, S. (2002). The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291, 605–614. doi: 10.1078/1438-4221-00173
Justin, T., Connor, A. O., Joon Ho, P., S, V., Patrick, V. P., Laurance, Y., et al. (2019). Experimental evolution reveals the genetic basis and systems biology of superoxide stress tolerance. bioRxiv [Preprint].
Kearns, D. B. (2010). A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8, 634–644. doi: 10.1038/nrmicro2405
Klose, K. E., and Mekalanos, J. J. (1998). Differential regulation of multiple flagellins in Vibrio cholerae. J. Bacteriol. 180, 303–316. doi: 10.1128/JB.180.2.303-316.1998
Kojima, S., Yamamoto, K., Kawagishi, I., and Homma, M. (1999). The polar flagellar motor of Vibrio cholerae is driven by an Na+ motive force. J. Bacteriol. 181, 1927–1930. doi: 10.1128/JB.181.6.1927-1930.1999
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
Kranz, R. G., Richard-Fogal, C., Taylor, J. S., and Frawley, E. R. (2009). Cytochrome c biogenesis: Mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol. Mol. Biol. Rev. 73, 510–528. doi: 10.1128/MMBR.00001-09
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint]. doi: 10.48550/arXiv.1303.3997
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Magariyama, Y., Sugiyama, S., Muramoto, K., Maekawa, Y., Kawagishi, I., Imae, Y., et al. (1994). Very fast flagellar rotation. Nature 371:752. doi: 10.1038/371752b0
Malinverni, J. C., and Silhavy, T. J. (2009). An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc. Natl. Acad. Sci. U.S.A. 106, 8009–8014. doi: 10.1073/pnas.0903229106
Mangan, M. W., Lucchini, S., Danino, V., Croinin, T. O., Hinton, J. C., and Dorman, C. J. (2006). The integration host factor (IHF) integrates stationary-phase and virulence gene expression in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59, 1831–1847. doi: 10.1111/j.1365-2958.2006.05062.x
Mathieu-Denoncourt, A., and Duperthuy, M. (2021). A secretome analysis reveals a role of subinhibitory concentrations of polymyxin B in the survival of Vibrio cholerae mediated by the type VI secretion system. Environ. Microbiol. 24, 1133–1149 doi: 10.1111/1462-2920.15762
Mathieu-Denoncourt, A., Giacomucci, S., and Duperthuy, M. (2021). The Secretome of Vibrio Cholerae. London: IntechOpen. doi: 10.5772/intechopen.96803
Mcdonald, M. J. (2019). Microbial Experimental Evolution - a proving ground for evolutionary theory and a tool for discovery. EMBO Rep. 20:e46992. doi: 10.15252/embr.201846992
Mcleod, S. M., Burrus, V., and Waldor, M. K. (2006). Requirement for Vibrio cholerae integration host factor in conjugative DNA transfer. J. Bacteriol. 188, 5704–5711. doi: 10.1128/JB.00564-06
Munguia, J., Larock, D. L., Tsunemoto, H., Olson, J., Cornax, I., Pogliano, J., et al. (2017). The Mla pathway is critical for Pseudomonas aeruginosa resistance to outer membrane permeabilization and host innate immune clearance. J. Mol. Med. 95, 1127–1136. doi: 10.1007/s00109-017-1579-4
Nakamura, S., and Minamino, T. (2019). Flagella-Driven Motility of Bacteria. Biomolecules 9:279 doi: 10.3390/biom9070279
Nasu, H., Shirakawa, R., Furuta, K., and Kaito, C. (2022). Knockout of mlaA increases Escherichia coli virulence in a silkworm infection model. PLoS One 17:e0270166. doi: 10.1371/journal.pone.0270166
Nelson, D. E., and Young, K. D. (2001). Contributions of PBP 5 and DD-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J. Bacteriol. 183, 3055–3064. doi: 10.1128/JB.183.10.3055-3064.2001
Nielsen, H., Tsirigos, K. D., Brunak, S., and Von Heijne, G. (2019). A Brief History of Protein Sorting Prediction. Protein J. 38, 200–216. doi: 10.1007/s10930-019-09838-3
O’callaghan, D., and Charbit, A. (1990). High efficiency transformation of Salmonella typhimurium and Salmonella typhi by electroporation. Mol. Gen. Genet. 223, 156–158. doi: 10.1007/BF00315809
O’shea, Y. A., Reen, F. J., Quirke, A. M., and Boyd, E. F. (2004). Evolutionary genetic analysis of the emergence of epidemic Vibrio cholerae isolates on the basis of comparative nucleotide sequence analysis and multilocus virulence gene profiles. J. Clin. Microbiol. 42, 4657–4671. doi: 10.1128/JCM.42.10.4657-4671.2004
Prieto, A. I., Kahramanoglou, C., Ali, R. M., Fraser, G. M., Seshasayee, A. S., and Luscombe, N. M. (2012). Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 40, 3524–3537. doi: 10.1093/nar/gkr1236
Protcompb. Prediction sub-Cellular Protein Localization. Available online at: http://www.softberry.com/berry.phtml?topic=pcompb&group=programs&subgroup=proloc (accessed 20 april 2021).
Prouty, M. G., Correa, N. E., and Klose, K. E. (2001). The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39, 1595–1609. doi: 10.1046/j.1365-2958.2001.02348.x
Ramani, N., Huang, L., and Freundlich, M. (1992). In vitro interactions of integration host factor with the ompF promoter-regulatory region of Escherichia coli. Mol. Gen. Genet. 231, 248–255. doi: 10.1007/BF00279798
Reisch, C. R., and Prather, K. L. J. (2017). Scarless Cas9 Assisted Recombineering (no-SCAR) in Escherichia coli, an Easy-to-Use System for Genome Editing. Curr. Protoc. Mol. Biol. 117, 1–31 doi: 10.1002/cpmb.29
Reverchon, S., Meyer, S., Forquet, R., Hommais, F., Muskhelishvili, G., and Nasser, W. (2021). The nucleoid-associated protein IHF acts as a ‘transcriptional domainin’ protein coordinating the bacterial virulence traits with global transcription. Nucleic Acids Res. 49, 776–790. doi: 10.1093/nar/gkaa1227
Rice, P., Longden, I., and Bleasby, A. (2000). EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277.
Roier, S., Zingl, F. G., Cakar, F., Durakovic, S., Kohl, P., Eichmann, T. O., et al. (2016). A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun. 7:10515. doi: 10.1038/ncomms10515
Rompikuntal, P. K., Vdovikova, S., Duperthuy, M., Johnson, T. L., Åhlund, M., Lundmark, R., et al. (2015). Outer Membrane Vesicle-Mediated Export of Processed PrtV Protease from Vibrio cholerae. PLoS One 10:e0134098. doi: 10.1371/journal.pone.0134098
Schwechheimer, C., Rodriguez, D. L., and Kuehn, M. J. (2015). NlpI-mediated modulation of outer membrane vesicle production through peptidoglycan dynamics in Escherichia coli. Microbiologyopen 4, 375–389. doi: 10.1002/mbo3.244
Seemann, T. (2014). Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shull, L. M., and Camilli, A. (2018). Transposon Sequencing of Vibrio cholerae in the Infant Rabbit Model of Cholera. Methods Mol. Biol. 1839, 103–116. doi: 10.1007/978-1-4939-8685-9_10
Seemann, T. (2015). Snippy: Fast Bacterial Variant Calling From NGS Reads. Available online at: https://github.com/tseemann/snippy/ (accessed August 11, 2020).
Solovyev, V., and Salamov, A. (2011). Automatic Annotation of Microbial Genomes and Metagenomic Sequences. Metagenomics and its Applications in Agriculture, Biomedicine, and Environmental Studies. Hauppauge, NY: Nova Science Publisher’s.
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: A genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Thony-Meyer, L. (1997). Biogenesis of respiratory cytochromes in bacteria. Microbiol. Mol. Biol. Rev. 61, 337–376. doi: 10.1128/mmbr.61.3.337-376.1997
Toyofuku, M., Nomura, N., and Eberl, L. (2019). Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 17, 13–24. doi: 10.1038/s41579-018-0112-2
Tu, Z. M., He, G. Y., Li, K. X. X., Chen, M. J. J., Chang, J. L., Chen, L., et al. (2005). An improved system for competent cell preparation and high efficiency plasmid transformation using different Escherichia coli strains. Electron. J. Biotechnol. 8, 113–120. doi: 10.2225/vol8-issue1-fulltext-8
Typas, A., Banzhaf, M., Gross, C. A., and Vollmer, W. (2011). From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136. doi: 10.1038/nrmicro2677
Vanhove, A. S., Duperthuy, M., Charrière, G. M., Le Roux, F., Goudenège, D., Gourbal, B., et al. (2015). Outer membrane vesicles are vehicles for the delivery of Vibrio tasmaniensis virulence factors to oyster immune cells. Environ. Microbiol. 17, 1152–1165. doi: 10.1111/1462-2920.12535
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wang, H., Ayala, J. C., Benitez, J. A., and Silva, A. J. (2012). Interaction of the histone-like nucleoid structuring protein and the general stress response regulator RpoS at Vibrio cholerae promoters that regulate motility and hemagglutinin/protease expression. J. Bacteriol. 194, 1205–1215. doi: 10.1128/JB.05900-11
Yamamoto, S., Izumiya, H., Morita, M., Arakawa, E., and Watanabe, H. (2009). Application of λ Red recombination system to Vibrio cholerae genetics: Simple methods for inactivation and modification of chromosomal genes. Gene 438, 57–64. doi: 10.1016/j.gene.2009.02.015
Yang, H., Lu, X., Hu, J., Chen, Y., Shen, W., and Liu, L. (2018). Boosting Secretion of Extracellular Protein by Escherichia coli via Cell Wall Perturbation. Appl. Environ. Microbiol. 84, e01382–18. doi: 10.1128/AEM.01382-18
Yang, Q., Pogue, J. M., Li, Z., Nation, R. L., Kaye, K. S., and Li, J. (2020). Agents of Last Resort: An Update on Polymyxin Resistance. Infect. Dis. Clin. North Am. 34, 723–750. doi: 10.1016/j.idc.2020.08.003
Zingl Franz, G., Thapa Himadri, B., Scharf, M., Kohl, P., Anna, M., Schild, S., et al. (2021). Outer Membrane Vesicles of Vibrio cholerae Protect and Deliver Active Cholera Toxin to Host Cells via Porin-Dependent Uptake. mBio 12, e534–e521. doi: 10.1128/mBio.00534-21
Keywords: Vibrio cholera, flagella, motility, membrane vesicles, experimental evolution, polymyxin, subinhibitory concentration, resistance
Citation: Giacomucci S, Mathieu-Denoncourt A, Vincent AT, Jannadi H and Duperthuy M (2022) Experimental evolution of Vibrio cholerae identifies hypervesiculation as a way to increase motility in the presence of polymyxin B. Front. Microbiol. 13:932165. doi: 10.3389/fmicb.2022.932165
Received: 29 April 2022; Accepted: 27 July 2022;
Published: 22 August 2022.
Edited by:
Etienne Giraud, Institut National de la Recherche Agronomique de Toulouse, FranceReviewed by:
Ansel Hsiao, University of California, Riverside, United StatesFranz Zingl, Harvard Medical School, United States
Copyright © 2022 Giacomucci, Mathieu-Denoncourt, Vincent, Jannadi and Duperthuy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marylise Duperthuy, bWFyeWxpc2UuZHVwZXJ0aHV5QHVtb250cmVhbC5jYQ==
†These authors have contributed equally to this work and share first authorship
 Sean Giacomucci
Sean Giacomucci Annabelle Mathieu-Denoncourt
Annabelle Mathieu-Denoncourt Antony T. Vincent
Antony T. Vincent Hanen Jannadi1
Hanen Jannadi1 Marylise Duperthuy
Marylise Duperthuy