
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 03 August 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.931180
 Xingxing Li‡
Xingxing Li‡ Weicong Ren‡
Weicong Ren‡ Yihong Li
Yihong Li Yuanyuan Shi
Yuanyuan Shi Hongmin Sun
Hongmin Sun Lifei Wang
Lifei Wang Linzhuan Wu
Linzhuan Wu Yunying Xie
Yunying Xie Yu Du
Yu Du Zhibo Jiang*†
Zhibo Jiang*† Bin Hong*
Bin Hong*Natural products from microorganisms are important sources for drug discovery. With the development of high-throughput sequencing technology and bioinformatics, a large amount of uncharacterized biosynthetic gene clusters (BGCs) in microorganisms have been found, which show the potential for novel natural product production. Nine BGCs containing PKS and/or NRPS in Streptomyces globisporus C-1027 were transcriptionally low/silent under the experimental fermentation conditions, and the products of these clusters are unknown. Thus, we tried to activate these BGCs to explore cryptic products of this strain. We constructed the cluster-situated regulator overexpressing strains which contained regulator gene(s) under the control of the constitutive promoter ermE*p in S. globisporus C-1027. Overexpression of regulators in cluster 26 resulted in significant transcriptional upregulation of biosynthetic genes. With the separation and identification of products from the overexpressing strain OELuxR1R2, three ortho-methyl phenyl alkenoic acids (compounds 1–3) were obtained. Gene disruption showed that compounds 1 and 2 were completely abolished in the mutant GlaEKO, but were hardly affected by deletion of the genes orf3 or echA in cluster 26. The type II PKS biosynthetic pathway of chain-extended cinnamoyl compounds was deduced by bioinformatics analysis. This study showed that overexpression of the two adjacent cluster-situated LuxR regulator(s) is an effective strategy to connect the orphan BGC to its products.
Natural products, especially secondary metabolites from microorganisms, have been widely used in the pharmaceutical industry as antibiotics, immunosuppressants, antifungal, anticancer, and anti-parasitic drugs (Hwang et al., 2014; Qin et al., 2017). Among all microorganisms, actinomycetes, especially Streptomyces, are well-known to be prolific producers of natural products, and about two-thirds of the known natural antibiotics are produced by Streptomyces (Lucas et al., 2013). The traditional programs of natural product discovery are usually based on the guidance of bioactivity screening. However, this strategy leads to the tendency of repeated discovery of known compounds, which requires new strategies to solve this problem (Nielsen et al., 2011; Gaudencio and Pereira, 2015; Patridge et al., 2016). With the rapid development of high-throughput sequencing technology, the massive microbial genomic data have revealed a potential to produce diverse and novel metabolites than previously appreciated (Choi et al., 2015; Mukherjee et al., 2017). Millions of putative biosynthetic gene clusters (BGCs) have been identified, but only a small part of them has been characterized (Rutledge and Challis, 2015). This can be attributed to the fact that most BGCs are silent or poorly expressed in their native hosts under conventional experimental fermentation conditions. On the other hand, a lot of observed natural products have not been linked to their BGCs (Hoskisson and Seipke, 2020). It is still a key challenge to discover novel natural products and link them to their BGCs.
Several approaches have been developed to activate the targeted BGCs and comprehensively reviewed recently (Rutledge and Challis, 2015; Ren et al., 2017; Baral et al., 2018; Covington et al., 2021). Based on genetic manipulation in native hosts, overexpressing putative cluster-situated positive regulators (activators) and deleting negative regulators (repressors) encoded in cryptic clusters are two technically practical and rational approaches. For instance, overexpression of a cluster-situated activator gene chaI led to the production of two novel chattamycins in Streptomyces chattanoogensis (Zhou et al., 2015). Similarly, the constitutive expression of cluster-situated activator hcdR2 triggered the production of herbicidin in Streptomyces mobaraensis (Shi et al., 2019b). In contrast, deletion of alpW, which encodes a TetR-like cluster-situated repressor, led to the identification of a new product kinamycin in Streptomyces ambofaciens (Bunet et al., 2011). The cryptic jadomycin BGC could be activated by disruption of the cluster-situated repressor gene jadR2 in Streptomyces venezuelae (Xu et al., 2010). Instead of genetically manipulating regulators, Wang et al. developed a transcription factor decoy strategy, which interferes with the binding of the regulator to its cognate DNA targets, and successfully activated eight silent BGCs in streptomycetes (Wang et al., 2019).
Streptomyces globisporus C-1027, which was isolated from a soil sample collected in the Qian-jiang area of Hubei Province, China, produces an extremely potent antitumor antibiotic lidamycin (also known as C-1027) (Hu et al., 1988; Shao and Zhen, 2008). The biosynthetic pathway and regulatory mechanism of lidamycin have been elucidated previously (Liu et al., 2002; Wang et al., 2009; Li et al., 2015). The genome sequence of this strain has been reported (Li et al., 2016), and antiSMASH (version 3.0.5) (Weber et al., 2015) analysis indicated that it contained at least 33 secondary metabolite BGCs (Supplementary Table 3), including the potential for the synthesis of compounds such as polyketides (PKs), non-ribosomal peptides (NRPs), ribosomally synthesized and post-translationally modified peptides (RiPPs), and siderophores. Although this strain has been extensively studied, it has not been found to produce secondary metabolites other than lidamycin and its precursors. To explore the ability of this strain to produce different secondary metabolites, especially PKs and NRPs with high druggability, we attempted to overexpress the transcriptional regulatory gene(s) within or near these BGCs to rapidly uncover new secondary metabolites that might be activated, before delving into the specific biosynthetic and regulatory mechanism of these understudied clusters.
Herein, we report the identification, isolation, and structure elucidation of three cinnamoyl fatty acid derivatives with a 2-methylphenyl unit by overexpression of two adjacent LuxR genes in cluster 26 of S. globisporus C-1027 simultaneously. This is the first time to link these chain-extended cinnamoyl fatty acids to their biosynthetic gene cluster.
Genome mining of S. globisporus C-1027 revealed the presence of 10 BGCs of PKs (cluster 8, 31), NRPs (cluster 7, 10, 16), or hybrid polyketide-non-ribosomal peptides (PK-NRPs, cluster 19, 26, 28, 32, 33) (Supplementary Table 3). Analysis of the transcriptome sequencing data indicates that except for the biosynthetic cluster for lidamycin (cluster 33), the other nine clusters were silent or expressed at very low levels [below 300 FPKM (fragments per kilobase of exon per million fragments mapped) value, Supplementary Figure 1]. Thus, we attempted to overexpress the putative cluster-situated regulator(s) within or near these BGCs to activate these clusters to explore uncharacterized metabolites. We constructed the putative cluster-specific regulator overexpression plasmids based on the pICLset vector under the control of the constitutively active ermE* promoter (Hong et al., 2007). Each was integrated into the chromosome of S. globisporus C-1027, and the transcriptional level was detected by reverse transcription-quantitative PCR (RT-qPCR). Notably, the hybrid PKS/NRPS cluster 26 contained four putative regulatory genes (tetR, luxR1, luxR2, and lacI) within or near the cluster (Figure 1A and Supplementary Table 4). Considering that luxR1 and luxR2 were adjacent to each other and were close to the echA (a peptide synthetase, NRPS) gene and the glaE (β-ketoacyl synthase, KS) gene, we first constructed the co-overexpression strain (OELuxR1R2). The transcriptional level of these two regulators and biosynthetic genes in cluster 26 increased significantly (Figure 1B). This result suggested that LuxR1 and/or LuxR2 might be the cluster-situated positive regulators for cluster 26. Then, four recombinant strains harboring individually overexpressed regulator gene (OETetR, OELuxR1, OELuxR2, or OELacI) were constructed. Biosynthetic genes (echA and glaE) showed much higher transcriptional levels in OELuxR1 and OELuxR2 strains than that in OETetR or OELacI strain (Supplementary Figure 2). These data further support the positive regulatory roles for LuxR1 and LuxR2 in the transcription of genes in cluster 26.
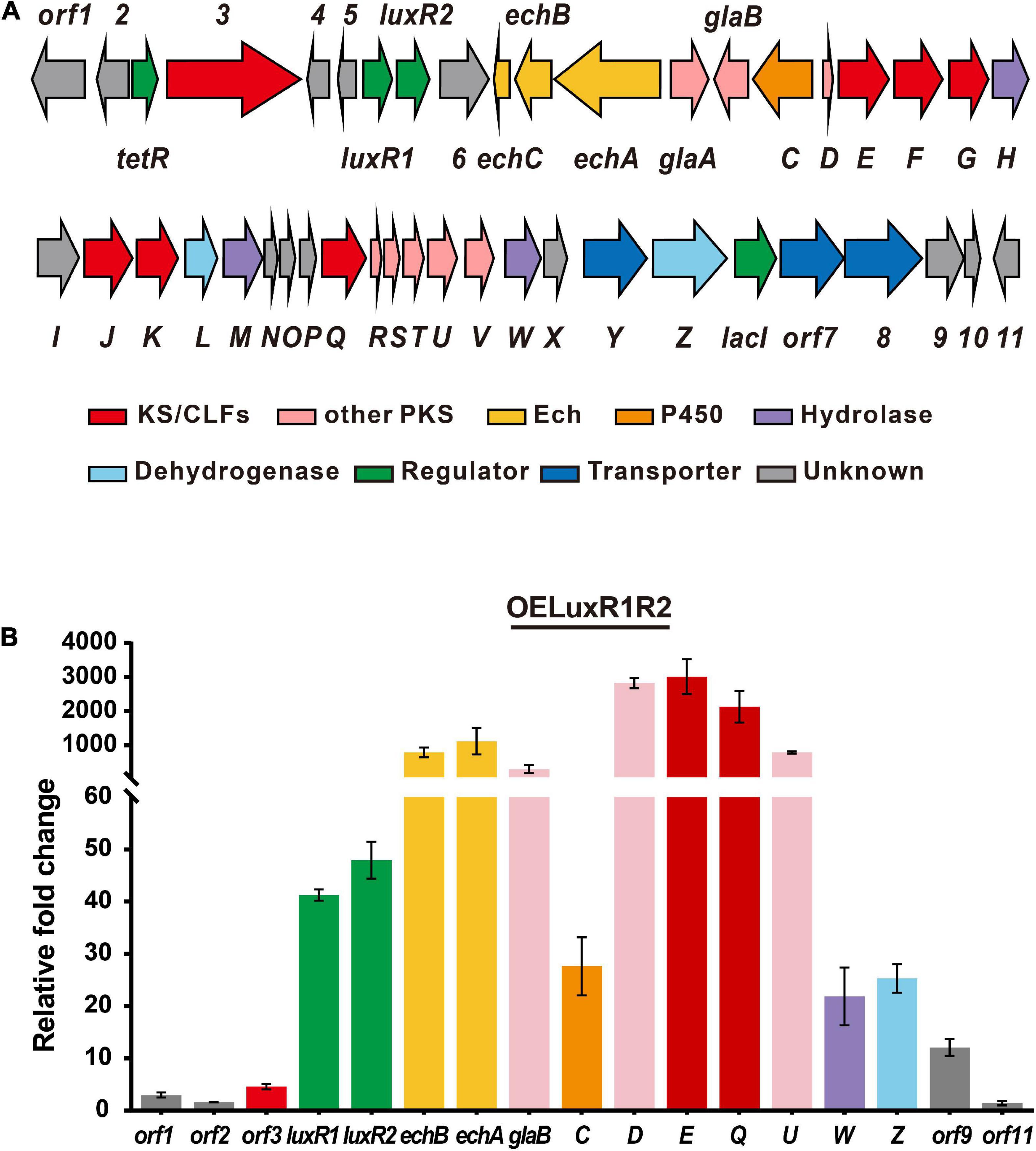
Figure 1. LuxR1 and LuxR2 were the cluster-specific positive regulators for cluster 26. (A) Organization of cluster 26. (B) Transcriptional analysis of cluster 26 genes in the regulator-overexpression strain detected by RT-qPCR analysis. The mycelia of regulator-overexpression strain (OELuxR1R2) were collected for the extraction of total RNAs at 48 h at the beginning of the fermentation in an FMC-1027-1 medium. These samples were subjected to RT-qPCR analysis. The relative mRNA level of the target genes was normalized to the principal sigma factor gene hrdB. The relative expressional level of each sample was represented as the value related to the control strain C-1027/pSET152. Values are presented as means ± SEM (three biological repeats for each strain).
As cluster 26 was increased remarkably at the transcriptional level in the OELuxR1R2 strain, we then compared the metabolic profiles of this overexpressing strain and the control strain C-1027/pSET152. Each of the fermentation broths was extracted with ethyl acetate (EtOAc) and analyzed by thin-layer chromatography (TLC, Figure 2A) or high-performance liquid chromatography (HPLC, Figure 2B). Compared with the control strain, the overexpressed strain OELuxR1R2 produced several new products by TLC analysis (Figure 2A). Moreover, the HPLC results indicated that several peaks with retention times at 28–30 and 39–40 min were significantly increased (Figure 2B). Then, the fermentation of strain OELuxR1R2 was scaled up to 20 L and the separation of target compounds was performed as described in the “Materials and Methods” section. The main components 1 and 2, together with minor congeners 3, were purified.
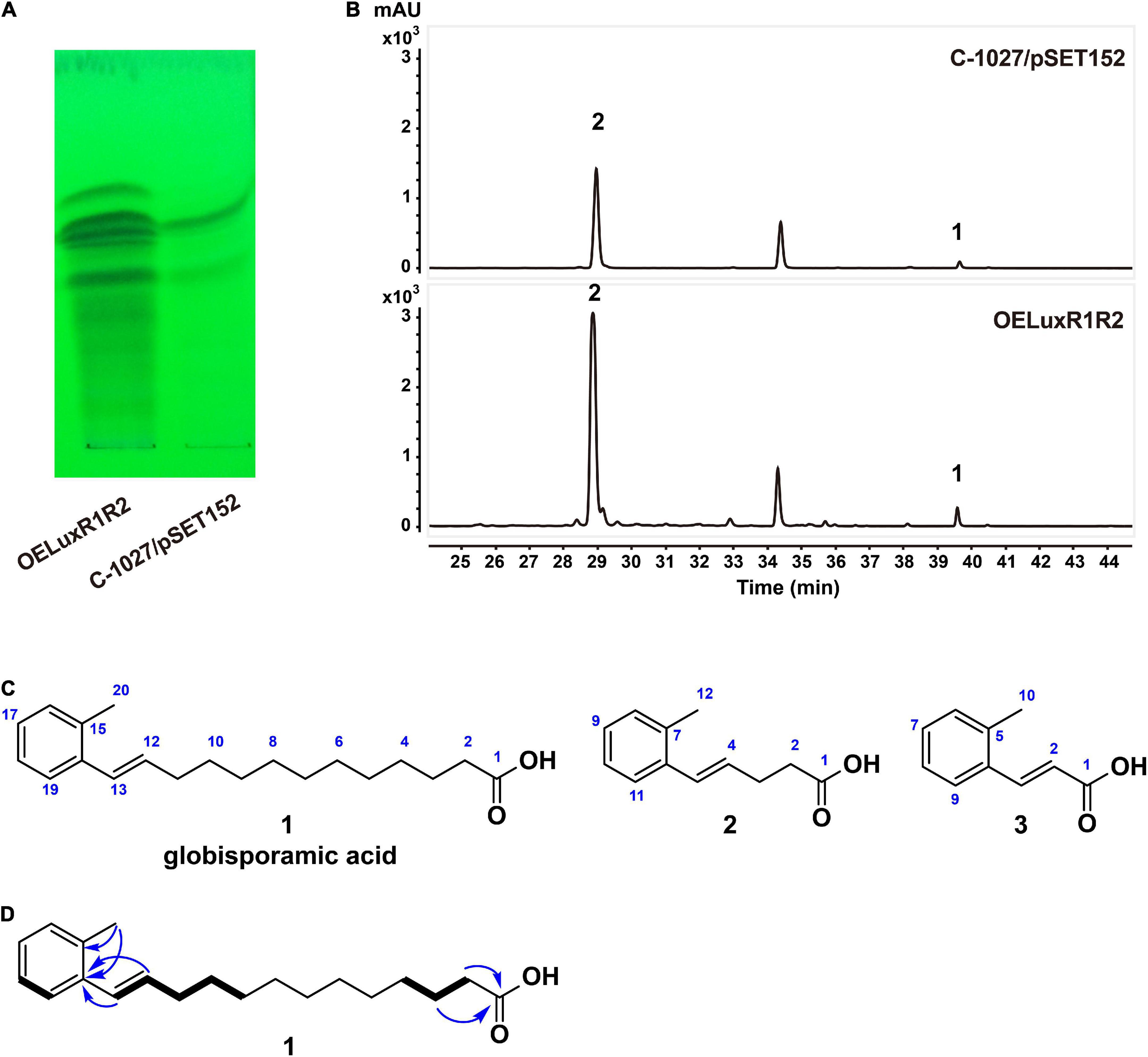
Figure 2. Metabolic profiles and compounds produced by recombinants. Analysis of the metabolic profiles between regulator-overexpression strain OELuxR1R2 and control strain C-1027/pSET152 by TLC (A) and HPLC (B) analysis. (C) Chemical structures of three compounds isolated from OELuxR1R2. (D) The 1H-1H COSY (thick bonds) and key HMBC correlations (arrows) of compound 1.
Compound 1 was obtained as a white amorphous powder and determined to have a molecular formula of C20H30O2 from its HRESIMS analysis (m/z 301.2174 [M-H]–, 0.33 ppm; Supplementary Figure 4), requiring six double-bond equivalents (DBE). Analysis of the 1H, 13C NMR, and DEPT spectra of compound 1 (Supplementary Table 5 and Supplementary Figures 5–7) returned a carbonyl (δC 174.9), a disubstituted benzene [δC 137.9, 135.6, 128.7, 127.7, 127.0, 126.3 and δH 7.41 (d, J = 7.5 Hz, 1H), 7.02 (m, 3H)], an alkene [δC 133.1, δH 6.12 (dt, J = 13.5, 5.5, 1H) and δC 131.0, δH 6.62 (dt, J = 13.5, 1.5, 1H)], a methyl [δC 20.0, δH 2.29 (s, 3H)], and 10 methylene groups [δC 34.4, 34.1, 30.4, 30.4, 30.3, 30.3, 30.2, 30.0, 30.0, 25.8 and δH 2.28 (t, J = 7.0 Hz, 2H), 2.25 (ddd, J = 13.5, 5.5, 1.5, 2H), 1.59 (m, 2H), 1.50 (m, 2H), 1.29–1.39 (m, 12H)]. A free carboxylic acid was inferred by the downfield chemical shift of the carbonyl group in compound 1 and no ester methyl or methylene signals in 1H and 13C NMR spectra. Two hydrocarbon fragments, -CH = CH-CH2-CH2-CH2- (F1) and -CH2-CH2-CH2- (F2), were indicated by the 1H-1H COSY correlations of H-13/H-12/H-11/H-10/H-9 and H-2/H-3/H-4, respectively (Figure 2D and Supplementary Figure 9). The above proposed disubstituted benzene in compound 1 was proved to be decorated by the methyl group and the alkene moiety of fragment F1, respectively, in ortho position according to the key HMBC correlations from the methyl protons H3-20 to both the sp2 quaternary carbons C-14 and C-15, and from both the alkene protons H-12 and H-13 to C-14 (Figure 2D and Supplementary Figure 10), as well as by comparison with the chemical shifts of its analog (Venugopal et al., 2002). Additionally, the free carboxylic acid was confirmed to be linked to fragment F2 according to the HMBC correlations from H-2 and H-3 to C-1 (Figure 2D and Supplementary Figure 10). Based on the above interpretation, only four methylene groups were still unassigned. Although these four methylene groups cannot be distinguished from each other due to their severely overlapped signals, it is reasonable to deduce that they are parts of the fatty acid chain and located between fragments F1 and F2 based on their nearly identical chemical shifts and by comparison with those of reported fatty acids (Gunstone et al., 1977). Therefore, compound 1 was identified as E-13-(2-methyl phenyl)-12-tridecenoic acid (named globisporamic acid).
Compounds 2 and 3 were identified by NMR data analysis as E-5-(2-methylphenyl)-4-pentenoic acid (2) and E-3-(2-methylphenyl)-acrylic acid (3) (Supplementary Table 5 and Supplementary Figures 11–22). Compounds 1 to 3 are all ortho-methyl phenyl alkenoic acids. Among them, compound 2 was previously found in a terrestrial Streptomycete GW 10/2517 (Venugopal et al., 2002) and a marine-derived Streptomyces isolate B8112 (Shaaban et al., 2011). o-Methylcinnamide (U-77863), the amide form of compound 3, was reported in Streptomyces griseoluteus (Harper and Welch, 1992). During our preparation of the manuscript, the compound with the same structure as globisporamic acid was isolated from Streptomyces sp. QHA10 (Zhao et al., 2021). However, no related biosynthetic mechanisms have been reported for these compounds.
In particular, C-2 substituted phenyl alkenoic group known as cinnamoyl lipid moieties have been reported as part of the structure of a small family of cyclodepsipeptides, such as skyllamycin (Pohle et al., 2011), cinnapeptin (Zhang and Seyedsayamdost, 2020), atratumycin (Sun et al., 2019; Figure 3A), WS9326A (Hayashi et al., 1992), kitacinnamycin (Shi et al., 2019a), pepticinnamin E (Santa Maria et al., 2019), atrovimycin (Liu et al., 2019), and so on. It is noteworthy that the unusual ortho-methyl phenyl alkenoic group has only been found in atratumycin (Sun et al., 2019) and cinnapeptin (Zhang and Seyedsayamdost, 2020; Figure 3A). Among the BGCs of these compounds, the biosynthesis of cinnamoyl lipid moieties is presumed to be accomplished by type II PKS.
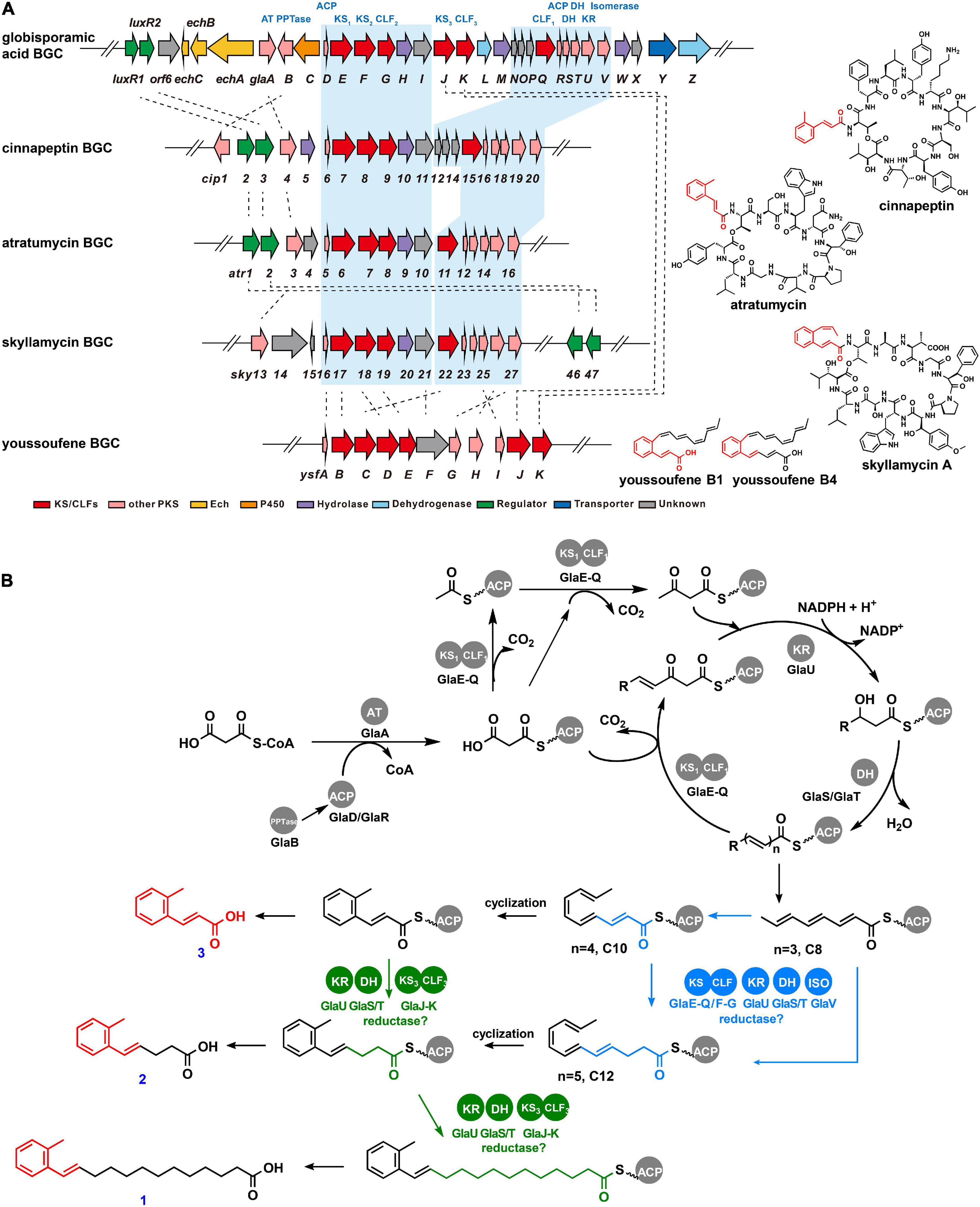
Figure 3. Gene cluster mining and proposed biosynthetic pathway of globisporamic acid. (A) Genetic organizations of the BGCs of globisporamic acid and cinnamoyl moiety-containing compounds. The blue-shaded region represented the amino acid sequence alignment of PKS genes with their homologs. The chemical structure of the natural products from the corresponding BGCs was shown on the right. (B) Proposed biosynthetic process of globisporamic acid. The pathway putatively catalyzed by KS1-CLF1 (GlaE-GlaQ) or KS2-CLF2 (GlaF-GlaG) was highlighted in blue, while that catalyzed by KS3-CLF3 (GlaJ-GlaK) was highlighted in green.
Cluster 26 was predicted to contain more than 40 genes by antiSMASH (version 3.0.5, Supplementary Table 4), which are encoding NRPS, type II-PKS, isomerase, dehydrogenase, hydrolase, cytochrome P450 enzyme, transporters, and regulators. The genes on the left boundary (orf1-orf2) and right boundary (orf11) might not be involved in the biosynthesis of globisporamic acid because their transcriptional levels were not as increased as that of core genes (Figure 1B).
Since orf3 and glaE were predicted as ketosynthases (KS) and the transcriptional levels of echA (echABC are homologous to biosynthetic genes for echosides) and glaE were both increased in OELuxR1R2 strain (Figure 1B), we constructed gene inactivation mutants to determine whether they were responsible for the biosynthesis of globisporamic acid. In the gene disrupted strains S. globisporus orf3KO, echAKO, and glaEKO, parts of their coding region were replaced by a thiostrepton-resistant gene (tsr) via gene recombination strategy and the double crossover mutants were verified by PCR (Supplementary Figures 23–25). In the corresponding mutant, the relative transcriptional level of the knockout gene was undetectable (Figure 4A). This further confirmed that the target gene was disrupted successfully. We next detected the products of the wild-type strain (C-1027) and the aforementioned three mutants by HPLC-MS analysis. It was revealed that the production of compounds 1 and 2 was completely abolished in the glaEKO strain, but was hardly affected by the deletion of orf3 or echA gene (Figure 4B and Supplementary Figure 26). It was suggested that glaE, but not orf3 or echA, was essential for the biosynthesis of compounds 1 and 2, and these two compounds were indeed the natural products encoded by cluster 26.
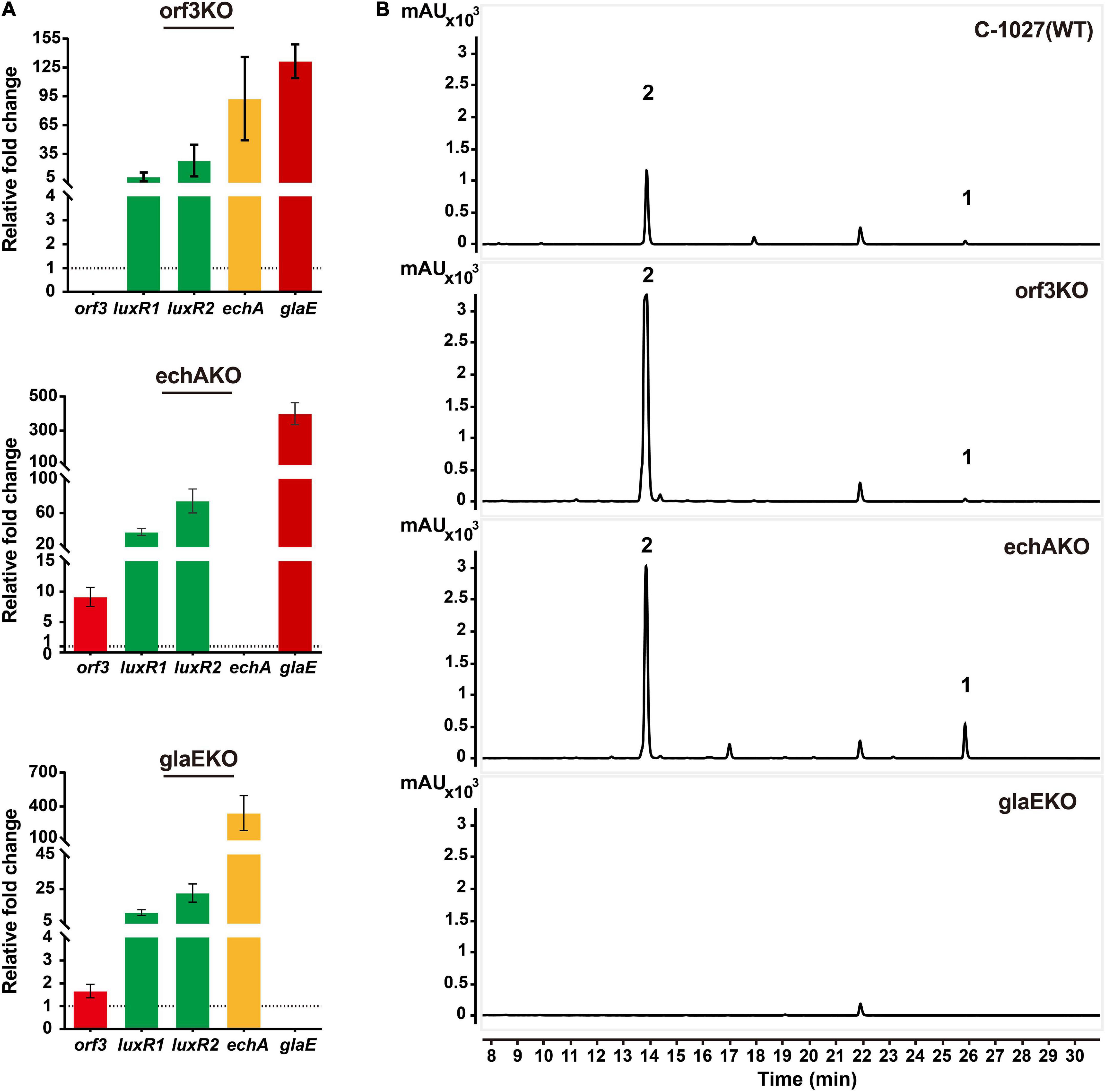
Figure 4. Compounds 1 and 2 were the cluster-specific products of cluster 26. (A) Transcriptional analysis of knockout mutants detected by RT-qPCR analysis. The relative mRNA level of the target genes was normalized to the principal sigma factor gene hrdB. The relative expressional level of each sample was represented as the value related to the wild-type strain S. globisporus C-1027. Values are presented as means ± SEM (two biological repeats for orf3KO mutant and echAKO mutant and three biological repeats for glaEKO mutant). (B) Analysis of the metabolic profiles of knockout mutants by HPLC-MS. The production of compounds 1 and 2 was abolished when the glaE gene was inactivated in the S. globisporus C-1027.
Interestingly, the transcriptional levels of the regulatory genes LuxR1 and LuxR2 were increased in all three gene knockout mutants (orf3KO, echAKO, and glaEKO), and the underlying mechanism is worth further investigation. The transcriptional level of glaE and the yield of compounds 1 and 2 were also significantly increased in orf3KO and echAKO strains (Figure 4). The results were consistent with the improved yield of compounds 1 and 2 in the OELuxR1R2 strain (Figure 2), and further verified that LuxR1 and LuxR2 were the positive regulators for the biosynthesis of compounds 1 and 2.
Genome mining revealed that GlaA-W (SGL6302-SGL6324) in cluster 26 had high sequence identities (from 35% to 77%) to type II PKSs implicated in cinnamoyl lipid moiety construction during skyllamycin (Pohle et al., 2011), cinnapeptin (Zhang and Seyedsayamdost, 2020), atratumycin (Sun et al., 2019), and youssoufenes (Deng et al., 2021) biosynthesis (Figure 3A and Supplementary Table 4). Unlike the canonical type II PKS, the PKS system involved in cinnamoyl lipid biosynthesis was phylogenetically different (Supplementary Figure 27; Shi et al., 2019a; Deng et al., 2021), which generally contained two or three pairs of ketosynthase/chain length factor (KS/CLF) complexes and harboring an isomerase-dependent polyunsaturated chain elongation process (Deng et al., 2021). In particular, there were three pairs of KS-CLF (GlaE-GlaQ, GlaF-GlaG, and GlaJ-GlaK) complexes in the gla cluster, similar to that of youssoufene’s BGC (ysf). The three ysf KS-CLFs were deciphered to modularly assemble different parts of the youssoufene skeleton. YsfB-YsfC initiated chain elongation, and then YsfD-YsfE yield the short-chain youssoufenes (C18), followed by YsfJ-YsfK to generate the long-chain youssoufenes (C20, C22). Of note, GlaJ-GlaK exhibits homology to YsfJ-YsfK (Supplementary Figure 27), which recognizes ACP-linked benzene ring containing intermediates instead of acyclic polyene intermediates in youssoufene’s biosynthesis. It is tempting to speculate that GlaJ-GlaK may have similar catalytic characteristics. GlaV showed 47% identity to the glutathione-dependent cis–trans isomerase YsfG of youssoufene’s cluster, suggesting that it may possess the ability to transform configuration for cyclization and participate in the chain elongation process.
Therefore, a biosynthetic pathway for compounds 1 and 2 can be proposed based on bioinformatic analysis of the gla cluster (Figure 3B). To begin with, a phosphopantetheinyl transferase (PPTase, GlaB) post-translationally transfers the pantetheine prosthetic group to convert apo-acyl carrier protein (ACP, GlaD/GlaR) into its activated form, holo-ACP. Initially, malonyl-CoA, as the basic building block, is loaded onto the holo-ACP via acyltransferase (AT, GlaA) to obtain the malonyl-ACP. The acetyl-ACP is generated by decarboxylation of malonyl-ACP and is condensed with malonyl-ACP by the catalysis of KS1-CLF1 (GlaE-GlaQ) to form the acetoacetyl-ACP. The chain elongation process carries out a repeating cycle of condensation, reduction, and dehydration, which were catalyzed by KS1-CLF1 (GlaE-GlaQ), ketoreductase (KR, GlaU), and dehydratase (DH, GlaS/GlaT), respectively, to generate a polyene with three carbon–carbon double bonds (n = 3, C8). Then, the polyene precursor was transformed from trans- to cis-configuration by isomerase (GlaV), bringing the carbon atoms within the required distance for cyclization. As it remains unclear whether the putative different length polyene precursor for compounds 1, 2, and 3 (C20, C12, and C10) was generated after the benzene ring closure, we consequently speculated two pathways (Figure 3B), and the difference is that the chain elongation of polyene precursors was catalyzed by KS1-CLF1 (GlaE-GlaQ) and KS2-CLF2 (GlaF-GlaG) (blue pathway), or catalyzed by KS3-CLF3 (GlaJ-GlaK) (green pathway), as KS3-CLF3 was proposed to participate in the chain elongation of benzene ring containing intermediates.
Like other cinnamoyl moiety-containing clusters, the enoyl-ACP reductase (ER), involved in further reduction of the double bond to a saturated intermediate, is absent in the gla cluster. In addition, the functions of some modified enzymes, including cytochrome P450 enzyme, dehydrogenase, and hydrolase, need to be further investigated. Moreover, the homologs of LuxR1 and LuxR2 also appeared in skyllamycin (Pohle et al., 2011), cinnapeptin (Zhang and Seyedsayamdost, 2020), and atratumycin (Sun et al., 2019) BGCs (Supplementary Table 4 and Supplementary Figure 28). Given that LuxR1 and LuxR2 have been identified as positive regulators in cluster 26, we hypothesized that they might be conservative cluster-situated regulators for cinnamoyl moiety-containing natural products biosynthesis.
The cinnamoyl moiety containing depsipeptides exhibited a broad range of biological activities, for example, atratumycin and atrovimycin exerted specific activity against Mycobacteria tuberculosis (Liu et al., 2019; Sun et al., 2019). The benzoic polyene acid youssoufene A1 was moderately active against multidrug-resistant Enterococcus faecalis (Li et al., 2020). However, both compounds 1 and 2 failed to display antibacterial activities at the concentration of 1,024 μg/mL against Pseudomonas aeruginosa, Mycobacterium smegmatis, Staphylococcus aureus, Escherichia coli, E. coliΔtolC mutant, and Bacillus subtilis. Also, no antiviral activities were observed at the concentration of 500 μg/mL of compound 1 or 2 against coxsackievirus B3, herpes simplex virus type 1, and influenza A virus (H3N2). Zhao et al. (2021) showed compound 1 had potential activity to inhibit nitric oxide (NO) production in lipopolysaccharide (LPS)-induced mouse macrophages. Their biological activity is worthy of further exploration.
The genome of Streptomyces has the potential to encode a variety of natural metabolites. One of the challenges in natural product discovery is that the majority of BGCs of the natural product have not been characterized, partially because they are either transcriptionally silent or expressed at a very low level under standard laboratory conditions. BGCs in streptomycete often contain at least one ultimate positive transcriptional regulator for the activation of the cluster, and sometimes contain negative regulators as well. Not surprisingly, increasing positive or decreasing negative cluster-situated regulatory gene expression levels were the effective method for improving secondary metabolite titers (Rutledge and Challis, 2015). Since the genetic manipulation of overexpression regulatory genes in Streptomyces is far more convenient than gene knockout, we tried to overexpress the predicted regulatory genes situated in the clusters to rapidly discover novel secondary metabolites without deciphering their positive or negative regulatory mechanisms. Applying this strategy to nine cryptic clusters of PKS, NRPS, or PKS/NRPS (usually encoding compounds with high druggability) in S. globisporus C-1027, we significantly upregulated the transcription of cluster 26, and isolated and identified its products, including globisporamic acid and two congeners compounds 2 and 3. We also linked the structures of the compounds to their biosynthetic gene cluster.
LuxR-family proteins have often been found as a class of cluster-situated transcriptional regulators that are widespread in actinomycetes’ secondary metabolism BGCs. Examples include HcdR2 (from herbicidin BGC in S. mobaraensis) (Shi et al., 2019b), GdmRI and GdmRII (from geldanamycin BGC in S. hygroscopicus) (He et al., 2008), PikD (from pikromycin in S. venezuelae) (Wilson et al., 2001), PimM (from pimaricin in S. natalensis) (Anton et al., 2007), PteF (from filipin in S. avermitilis) (Vicente et al., 2014), Tcp29 (Tei16*, from teicoplanin in Actinoplanes teichomyceticus) (Horbal et al., 2014), and so on. Herein, LuxR1 (SGL6295) and LuxR2 (SGL6296), two LuxR family regulators encoded within the gla cluster of S. globisporus C-1027, are demonstrated to positively regulate the biosynthesis of compounds 1 and 2, especially when overexpressed simultaneously. Genome mining suggested that the highly conserved homologs of LuxR1 and LuxR2 were also present in the BGCs for some known cinnamoyl moiety-containing natural products, such as skyllamycin (Pohle et al., 2011), cinnapeptin (Zhang and Seyedsayamdost, 2020), and atratumycin (Sun et al., 2019; Figure 3A and Supplementary Table 4). Noteworthy, these LuxRs in the BGCs of cinnamoyl moiety-containing natural products, especially two adjacent LuxRs in the same cluster, tended to group together in the phylogenetic tree (Supplementary Figure 28). Thus, we speculated that overexpression of the adjacent LuxR-family regulators may be used to explore the orphan BGCs of novel cinnamoyl moiety-containing natural products discovered by genome mining.
In this study, three cinnamoyl fatty acid analogs were isolated and structurally elucidated from the recombinant strain OELuxR1R2. They all contained an ortho-methylphenyl unit with different lengths of the alkenoic acid side chain. Among them, compound 1, globisporamic acid, contains a unique long-chain composed of 13 carbons, while compounds 2 and 3 contain a shorter side chain. Compound 3 is o-methyl cinnamic acid with an acrylic side chain. The biosynthesis of these congeners was demonstrated by gene knockout to be catalyzed by type II PKS, and we speculate that the chain length is determined by three pairs of KS-CLF complexes as in the biosynthesis of youssoufene. The KS-CLFs encoded in gla cluster along with reductase and dehydratase were proposed to catalyze the formation of different length polyene precursors, and the detailed mechanism needs to be further investigated.
Different from several reported cinnamoyl lipid-containing compounds [such as skyllamycin (Pohle et al., 2011), cinnapeptin (Zhang and Seyedsayamdost, 2020), and atratumycin (Sun et al., 2019)], which contain cyclic depsipeptide assembled by NRPS, the structure of the products of cluster 26 in S. globisporus C-1027 was much simpler. The only NRPS in cluster 26 was EchA (SGL6301). BLASTP alignment showed that SGL6301, SGL6299, and SGL6300 had high similarity with EchA, EchB, and EchC, which are involved in the biosynthesis of echosides, a class of para-terphenyl natural products isolated from Streptomyces sp. LZ35 (Zhu et al., 2014). Through the adenylation (A) domain sequence analysis (Rottig et al., 2011), 10 active-site residues, which are responsible for amino acid substrate specificity, were identified as VAEFVGAAGK and the substrate recognized by SGL6301 should be the same as that of EchA for echosides. The T and TE domains also possessed the established signature active motifs (LGGTSL and GYSFG, respectively) (Schwarzer et al., 2003; Zhu et al., 2014). These suggested that the product of the ech cassette in cluster 26 should be echoside’s congener. Since ech cassette and PKS core genes in cluster 26 were adjacent to each other and the transcriptional levels were both upregulated in the regulator overexpression strain OELuxR1R2, it was speculated that they might synergistically synthesize an NRPS-like product containing cinnamoyl moiety. However, neither echoside’s congener nor possible hybrid NRPS-PKS product has been found under our current fermentation and isolation conditions. This is worthy of further exploration.
In summary, we activated the gla cluster by simultaneous overexpression of two adjacent cluster-situated positive LuxR regulator(s) in S. globisporus C-1027. Three ortho-methyl phenyl alkenoic acids were isolated and identified from the recombinant strain OELuxR1R2. Its biosynthetic pathway was proposed according to in vivo gene inactivation and the bioinformatic analysis of the type II PKS assembly line. This strategy can be easily and rapidly applied to the exploration of much more of such orphan gene clusters present in actinomycete genomes.
Streptomyces globisporus C-1027 and its derivatives were grown at 28°C on S5 agar (Wang et al., 2009) for sporulation and in an FMC-1027-1 medium (Wang et al., 2009) for fermentation. Mannitol soya flour (MS) agar (Kieser et al., 2000) was used for conjugation. Trypticase Soy Broth (TSB, BD Difco, Sparks, MD, United States) was used for the isolation of genomic DNA. E. coli DH5α (Sambrook, 2001) was used as a host for cloning experiments. E. coli ET12567/pUZ8002 (Kieser et al., 2000) was used for conjugal transfer. E. coli strains were grown in liquid or solid Luria-Bertani (LB) medium at 37°C. When required, strains were incubated with apramycin (Am, 50 μg/mL), ampicillin (Amp, 100 μg/mL), kanamycin (Km, 50 μg/mL), thiostrepton (Th, 30 μg/mL), and chloramphenicol (Cm, 25 μg/mL). The bacterial strains and plasmids used in this study are listed in Supplementary Table 1.
Strains were collected for the extraction of total RNAs at 48 h at the beginning of the fermentation in an FMC-1027-1 medium (Li et al., 2015). Total RNAs were isolated using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States) and chloroform followed by a PureLink RNA Mini Kit (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. Samples were treated with DNase I to eliminate the genomic DNA. The quantity and quality of RNA samples were assessed as described previously (Uguru et al., 2005).
cDNA library construction and Illumina sequencing were performed by Beijing Genomics Institute (Shenzhen, China). The sequencing reads were aligned to the S. globisporus C-1027 reference genome. The distribution of the transcriptome reads was shown in the Integrated Genome Browser (Nicol et al., 2009). The gene expression quantifications were estimated using FPKM (fragments per kilobase of exon per million fragments mapped) values by the RSEM tool (Li and Dewey, 2011).
The vector pL646 (Hong et al., 2007), a pSET152 (Bierman et al., 1992)-derived expression plasmid with the strong constitutive promoter ermE*p, was used for gene overexpression. The coding region of regulators was amplified from S. globisporus C-1027 genomic DNA by PCR with specific primers (Supplementary Table 2). The amplified DNA fragments were cloned into NdeI - BamHI or NdeI - XbaI sites of pL646 to obtain the overexpression of plasmids. The recombinant vectors and control plasmid pSET152 (Bierman et al., 1992) were introduced into S. globisporus C-1027 by intergeneric conjugation from E. coli ET12567/pUZ8002 according to the established protocol (Li et al., 2015).
Streptomyces globisporus C-1027 or its derivative strains were collected at 48 h at the beginning of the fermentation in an FMC-1027-1 medium (Li et al., 2015). Total RNAs were isolated as described above. The first-strand synthesis of cDNA was performed with the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, Beijing, China) using random primers according to the manufacturer’s instructions. Primers were designed to amplify fragments of about 100 to 180 bp from the target genes (Supplementary Table 2). Quantitative PCR (qPCR) reaction was performed as described previously (Wang et al., 2009). Two qPCR replicates were performed for each cDNA, and the relative cDNA level of target genes was normalized to the level of hrdB according to Pfaffl’s method (Pfaffl, 2001).
Streptomyces globisporus C-1027 or its derivative strains were cultured in 50 mL TSB medium at 28°C for 2 days, and then 10% culture was transferred into 100 mL FMC-1027-1 medium for continuous fermentation at 28°C for 5 days. Each fermented supernatant was extracted three times with an equal volume of ethyl acetate (EtOAc). The organic phase was combined and vacuum-dried and then dissolved with 2 mL acetone or methanol. The extract was analyzed by TLC, HPLC, or HPLC-MS.
In the silica gel TLC, EtOAc extract was separated using a mixture of solvent system with chloroform:methanol:acetic acid 100:20:1, v/v/v. The dark bands were observed under ultraviolet light at a λ of 254 nm.
High-performance liquid chromatography was performed on an Agilent 1100 HPLC using a Waters XBridge C18 column (4.6 mm × 150 mm, 3.5 μm) in a two-phase system, with phase A for methanol and phase B for 0.1% formic acid–water solution. The optimized program included column elution with a linear gradient of A/B = 10/90–100/0, v/v, over 40 min; flow rate = 0.8 mL/min, λ = 254 nm.
High-performance liquid chromatography mass spectrometry analysis was performed on an Agilent 1100 HPLC coupled to Agilent 6410 Triple Quadrupole mass spectrometer equipped with an electrospray ionization source. Chromatographic separation was performed using a Waters XBridge C18 column (4.6 mm × 150 mm, 3.5 μm) in a two-phase system, with phase A for ACN and phase B for 0.1% formic acid–water solution. The optimized program included column elution with a linear gradient of A/B = 15/85 – 100/0, v/v, over 30 min; flow rate = 1.0 mL/min, λ = 254 nm. MS spectral data were collected in the negative-ion mode, with a mass range of m/z 100 to 1,000. The capillary voltage was set to 3,500 V and fragmentor voltage was 120 V. The desolvation temperature was 350°C, with a desolvation gas flow of 11 L/min and a nebulizer pressure of 50 psi.
Fermentation supernatant (20 L) was extracted two times with an equal volume of EtOAc. The combined organic phases were evaporated under reduced pressure and mixed with 1.1 g silica gel and then separated by silica column chromatography using chloroform:methanol [100:0, v/v, (0.2 g, Fr-1); 50:1, v/v, (0.1 g, Fr-2); 10:1, v/v, (0.12 g, Fr-3); 5:1, v/v, (0.15 g, Fr-4); and 1:1, v/v, (0.12 g, Fr-5)] to obtain the five fractions. TLC analysis was carried out using chloroform:methanol:acetic acid (100:20:1, v/v/v), and the target compound was found to be distributed in Fr-2 and Fr-3 fractions. Fr-2 and Fr-3 were merged and separated by preparation TLC [developing solvent as chloroform:methanol:acetic acid (100:10:1, v/v/v)]. Two darker bands on the TLC were collected, and the silicon powder was eluted with chloroform. The eluted product was evaporated and further purified by semi-preparative HPLC using different concentrations of ACN–water solution to obtain compounds 1–2: compound 1 (35 mg, 95% ACN–water solution); compound 2 (20 mg, 80% ACN–water solution). Fr-4 (0.15 g) was a pale-yellow oily substance, and a color band (chloroform:methanol:acetic acid 50:10:1, v/v/v) was separated by preparation TLC. After semi-preparation and HPLC purification, compound 3 (20.5 mg) was obtained.
Genes glaE (SGL6306), echA (SGL6301), and orf3 (SGL6292) were inactivated by substituting each of the core regions with a thiostrepton resistance gene via homologous recombination (Supplementary Figures 23–25). The primers (Supplementary Table 2) for glaE disruption introduced restriction sites into the arms (HindIII and XbaI in the upstream arm, and XbaI and BamHI in the downstream arm). The amplified flanking DNA fragments were ligated into the pUC119 vector to give pUC-LR. Then, the thiostrepton resistance gene was amplified from pIJ680 and was cloned into the XbaI sites of the pUC-LR vector to yield pUC-LtR. The disruption box of glaE was ligated into plasmid pKC1139 to give pKC-LtR. Following the transformation into E. coli ET12567/pUZ8002, disruption plasmid pKC-LtR was conjugated into S. globisporus C-1027. Recombinant strains were selected for thiostrepton resistance, and correct knockout recombinants were confirmed by PCR (Supplementary Figure 25). With the same strategy, knockout mutants for echA and orf3 genes were selected on MS agar containing thiostrepton resistance and confirmed by PCR (Supplementary Figures 23, 24).
The in vitro antibacterial activity of compounds against Pseudomonas aeruginosa 11, Mycobacterium smegmatis CPCC 240556, Staphylococcus aureus CPCC 100051, Escherichia coli O11, E. coliΔtolC mutant, and Bacillus subtilis CMCC(B) 63501 were determined through the broth microdilution method. The bacteria (5 × 105 per well) were grown in Mueller–Hinton broth (Oxoid) in a 96-well plate at 37°C. Compounds were tested at a concentration range of two-fold dilution from 1,024 μg/mL to 8 μg/mL. Streptomycin was tested at a concentration range of two-fold dilution from 64 to 0.5 μg/mL and used as a positive control. DMSO (2%) was used as a negative control. The minimum inhibitory concentration (MIC) was defined as the lowest concentration of compound that prevents turbidity after 16 h incubation at 37°C. The experiment was performed in duplicate.
The antiviral activity of compounds against coxsackievirus B3, herpes simplex virus type 1, and influenza A virus (H3N2) were performed as described previously (Ji et al., 2010; Ma et al., 2013). The 50% cytotoxic concentration (TC50) and the 50% inhibitory concentration (IC50) of the test samples were calculated using the Reed–Muench method (Pizzi, 1950).
Secondary metabolite biosynthesis gene clusters were predicted by the online resource antiSMASH1. PCR and qPCR primers were designed with the online resource primer3web2. BLASTP was used for genome mining of potential cinnamoyl moiety-containing natural products’ clusters. Amino acid sequence similarity of protein homologous was retrieved using Blastp tools. The phylogenetic tree was calculated using the MEGA-X program based on the Neighbor-joining method (Kumar et al., 2018).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number can be found below: The RNA-sequencing data is publicly available at NCBI’s repository under BioProject ID: RJNA854280, (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA854280), and SRA ID: SRR19905466. The HPLC-MS raw data have been deposited in the EMBL-EBI MetaboLights database (https://www.ebi.ac.uk/metabolights/) with the identifier MTBLS5152. The HPLC-MS data will be released to public on 21-July according to the database’s policies.
XL, WR, and ZJ performed most of the experiments, analyzed the primary data, and wrote the draft manuscript. YL, YS, HS, LFW, LZW, YX, and YD assisted the microbiologic and chemical work in this study. BH supervised the whole research work and revised the manuscript extensively. All authors have read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (81872780, 81703398, 81803410, and 82104046), the CAMS Innovation Fund for Medical Sciences (2021-I2M-1-055), and the Beijing Natural Science Foundation (7214286).
We are grateful to Yuhuan Li and Rongmei Gao for the antiviral activity assay.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.931180/full#supplementary-material
Anton, N., Santos-Aberturas, J., Mendes, M. V., Guerra, S. M., Martin, J. F., and Aparicio, J. F. (2007). PimM, a PAS domain positive regulator of pimaricin biosynthesis in Streptomyces natalensis. Microbiology 153, 3174–3183. doi: 10.1099/mic.0.2007/009126-0
Baral, B., Akhgari, A., and Metsa-Ketela, M. (2018). Activation of microbial secondary metabolic pathways: avenues and challenges. Synth. Syst. Biotechnol. 3, 163–178. doi: 10.1016/j.synbio.2018.09.001
Bierman, M., Logan, R., O’Brien, K., Seno, E. T., Rao, R. N., and Schoner, B. E. (1992). Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49. doi: 10.1016/0378-1119(92)90627-2
Bunet, R., Song, L., Mendes, M. V., Corre, C., Hotel, L., Rouhier, N., et al. (2011). Characterization and manipulation of the pathway-specific late regulator AlpW reveals Streptomyces ambofaciens as a new producer of Kinamycins. J. Bacteriol. 193, 1142–1153. doi: 10.1128/JB.01269-10
Choi, S.-S., Kim, H.-J., Lee, H.-S., Kim, P., and Kim, E.-S. (2015). Genome mining of rare actinomycetes and cryptic pathway awakening. Process Biochem. 50, 1184–1193. doi: 10.1007/s12010-020-03361-3
Covington, B. C., Xu, F., and Seyedsayamdost, M. R. (2021). A natural product chemist’s guide to unlocking silent biosynthetic gene clusters. Annu. Rev. Biochem. 90, 763–788. doi: 10.1146/annurev-biochem-081420-102432
Deng, Z., Liu, J., Li, T., Li, H., Liu, Z., Dong, Y., et al. (2021). An unusual type II polyketide synthase system involved in cinnamoyl lipid biosynthesis. Angew Chem. Int. Ed. Engl. 60, 153–158. doi: 10.1002/anie.202007777
Gaudencio, S. P., and Pereira, F. (2015). Dereplication: racing to speed up the natural products discovery process. Nat. Prod. Rep. 32, 779–810. doi: 10.1039/c4np00134f
Gunstone, F. D., Polard, M. R., Scrimgeour, C. M., and Vedanayagam, H. S. (1977). Fatty acids. Part 50. 13C nuclear magnetic resonance studies of olefinic fatty acids and esters. Chem. Phys. Lipids 18, 115–129. doi: 10.1016/0009-3084(77)90031-7
Harper, D. E., and Welch, D. R. (1992). Isolation, purification, synthesis, and antiinvasive/antimetastatic activity of U-77863 and U-77864 from Streptomyces griseoluteus, strain WS6724. J. Antibiot. 45, 1827–1836. doi: 10.7164/antibiotics.45.1827
Hayashi, K., Hashimoto, M., Shigematsu, N., Nishikawa, M., Ezaki, M., Yamashita, M., et al. (1992). WS9326A, a novel tachykinin antagonist isolated from Streptomyces violaceusniger no. 9326. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J. Antibiot. 45, 1055–1063. doi: 10.7164/antibiotics.45.1055
He, W., Lei, J., Liu, Y., and Wang, Y. (2008). The LuxR family members GdmRI and GdmRII are positive regulators of geldanamycin biosynthesis in Streptomyces hygroscopicus 17997. Arch. Microbiol. 189, 501–510. doi: 10.1007/s00203-007-0346-2
Hong, B., Phornphisutthimas, S., Tilley, E., Baumberg, S., and McDowall, K. J. (2007). Streptomycin production by Streptomyces griseus can be modulated by a mechanism not associated with change in the adpA component of the A-factor cascade. Biotechnol. Lett. 29, 57–64. doi: 10.1007/s10529-006-9216-2
Horbal, L., Kobylyanskyy, A., Truman, A. W., Zaburranyi, N., Ostash, B., Luzhetskyy, A., et al. (2014). The pathway-specific regulatory genes, tei15* and tei16*, are the master switches of teicoplanin production in Actinoplanes teichomyceticus. Appl. Microbiol. Biotechnol. 98, 9295–9309. doi: 10.1007/s00253-014-5969-z
Hoskisson, P. A., and Seipke, R. F. (2020). Cryptic or Silent? The known unknowns, unknown knowns, and unknown unknowns of secondary metabolism. mBio 11:e02642-20. doi: 10.1128/mBio.02642-20
Hu, J. L., Xue, Y. C., Xie, M. Y., Zhang, R., Otani, T., Minami, Y., et al. (1988). A new macromolecular antitumor antibiotic, C-1027. I. Discovery, taxonomy of producing organism, fermentation and biological activity. J. Antibiot. 41, 1575–1579. doi: 10.7164/antibiotics.41.1575
Hwang, K. S., Kim, H. U., Charusanti, P., Palsson, B. O., and Lee, S. Y. (2014). Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol. Adv. 32, 255–268. doi: 10.1016/j.biotechadv.2013.10.008
Ji, X. Y., Zhong, Z. J., Xue, S. T., Meng, S., He, W. Y., Gao, R. M., et al. (2010). Synthesis and antiviral activities of synthetic glutarimide derivatives. Chem. Pharm. Bull. 58, 1436–1441. doi: 10.1248/cpb.58.1436
Kieser, T. B. M., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical Streptomyces Genetics. Norwich: John Innes Foundation.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, H., Liu, J., Deng, Z., Li, T., Liu, Z., Che, Q., et al. (2020). Correction to genetic manipulation of an aminotransferase family gene dtlA activates youssoufenes in marine-derived Streptomyces youssoufiensis. Org. Lett. 22:7773. doi: 10.1021/acs.orglett.0c02844
Li, X., Lei, X., Zhang, C., Jiang, Z., Shi, Y., Wang, S., et al. (2016). Complete genome sequence of Streptomyces globisporus C-1027, the producer of an enediyne antibiotic lidamycin. J. Biotechnol. 222, 9–10. doi: 10.1016/j.jbiotec.2016.02.004
Li, X. X., Yu, T. F., He, Q., McDowall, K. J., Jiang, B. Y., Jiang, Z. B., et al. (2015). Binding of a biosynthetic intermediate to AtrA modulates the production of lidamycin by Streptomyces globisporus. Mol. Microbiol. 96, 1257–1271. doi: 10.1111/mmi.13004
Liu, Q., Liu, Z., Sun, C., Shao, M., Ma, J., Wei, X., et al. (2019). Discovery and Biosynthesis of Atrovimycin, an Antitubercular and Antifungal Cyclodepsipeptide Featuring Vicinal-dihydroxylated Cinnamic Acyl Chain. Org. Lett. 21, 2634–2638. doi: 10.1021/acs.orglett.9b00618
Liu, W., Christenson, S. D., Standage, S., and Shen, B. (2002). Biosynthesis of the enediyne antitumor antibiotic C-1027. Science 297, 1170–1173. doi: 10.1126/science.1072110
Lucas, X., Senger, C., Erxleben, A., Gruning, B. A., Doring, K., Mosch, J., et al. (2013). StreptomeDB: a resource for natural compounds isolated from Streptomyces species. Nucleic Acids Res. 41, D1130–D1136. doi: 10.1093/nar/gks1253
Ma, S. G., Gao, R. M., Li, Y. H., Jiang, J. D., Gong, N. B., Li, L., et al. (2013). Antiviral spirooliganones A and B with unprecedented skeletons from the roots of Illicium oligandrum. Org. Lett. 15, 4450–4453. doi: 10.1021/ol401992s
Mukherjee, S., Stamatis, D., Bertsch, J., Ovchinnikova, G., Verezemska, O., Isbandi, M., et al. (2017). Genomes OnLine Database (GOLD) v.6: data updates and feature enhancements. Nucleic Acids Res. 45, D446–D456. doi: 10.1093/nar/gkw992
Nicol, J. W., Helt, G. A., Blanchard, S. G. Jr., Raja, A., and Loraine, A. E. (2009). The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25, 2730–2731. doi: 10.1093/bioinformatics/btp472
Nielsen, K. F., Mansson, M., Rank, C., Frisvad, J. C., and Larsen, T. O. (2011). Dereplication of microbial natural products by LC-DAD-TOFMS. J. Nat. Prod. 74, 2338–2348. doi: 10.1021/np200254t
Patridge, E., Gareiss, P., Kinch, M. S., and Hoyer, D. (2016). An analysis of FDA-approved drugs: natural products and their derivatives. Drug Discov. Today 21, 204–207. doi: 10.1016/j.drudis.2015.01.009
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pizzi, M. (1950). Sampling variation of the fifty percent end-point, determined by the Reed-Muench (Behrens) method. Hum. Biol. 22, 151–190.
Pohle, S., Appelt, C., Roux, M., Fiedler, H. P., and Sussmuth, R. D. (2011). Biosynthetic gene cluster of the non-ribosomally synthesized cyclodepsipeptide skyllamycin: deciphering unprecedented ways of unusual hydroxylation reactions. J. Am. Chem. Soc. 133, 6194–6205. doi: 10.1021/ja108971p
Qin, Z., Munnoch, J. T., Devine, R., Holmes, N. A., Seipke, R. F., Wilkinson, K. A., et al. (2017). Formicamycins, antibacterial polyketides produced by Streptomyces formicae isolated from African Tetraponera plant-ants. Chem. Sci. 8, 3218–3227. doi: 10.1039/c6sc04265a
Ren, H., Wang, B., and Zhao, H. (2017). Breaking the silence: new strategies for discovering novel natural products. Curr. Opin. Biotechnol. 48, 21–27. doi: 10.1016/j.copbio.2017.02.008
Rottig, M., Medema, M. H., Blin, K., Weber, T., Rausch, C., and Kohlbacher, O. (2011). NRPSpredictor2–a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 39, W362–W367. doi: 10.1093/nar/gkr323
Rutledge, P. J., and Challis, G. L. (2015). Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 13, 509–523. doi: 10.1038/nrmicro3496
Sambrook, J. R. D. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Santa Maria, K. C., Chan, A. N., O’Neill, E. M., and Li, B. (2019). targeted rediscovery and biosynthesis of the farnesyl-transferase inhibitor pepticinnamin E. Chembiochem 20, 1387–1393. doi: 10.1002/cbic.201900025
Schwarzer, D., Finking, R., and Marahiel, M. A. (2003). Nonribosomal peptides: from genes to products. Nat. Prod. Rep. 20, 275–287. doi: 10.1039/b111145k
Shaaban, K. A., Helmke, E., Kelter, G., Fiebig, H. H., and Laatsch, H. (2011). Glucopiericidin C: a cytotoxic piericidin glucoside antibiotic produced by a marine Streptomyces isolate. J. Antibiot. 64, 205–209. doi: 10.1038/ja.2010.125
Shao, R. G., and Zhen, Y. S. (2008). Enediyne anticancer antibiotic lidamycin: chemistry, biology and pharmacology. Anticancer Agents Med. Chem. 8, 123–131. doi: 10.2174/187152008783497055
Shi, Y., Gu, R., Li, Y., Wang, X., Ren, W., Li, X., et al. (2019b). Exploring novel herbicidin analogues by transcriptional regulator overexpression and MS/MS molecular networking. Microb. Cell Fact. 18:175. doi: 10.1186/s12934-019-1225-7
Shi, J., Liu, C. L., Zhang, B., Guo, W. J., Zhu, J., Chang, C. Y., et al. (2019a). Genome mining and biosynthesis of kitacinnamycins as a STING activator. Chem. Sci. 10, 4839–4846. doi: 10.1039/c9sc00815b
Sun, C., Yang, Z., Zhang, C., Liu, Z., He, J., Liu, Q., et al. (2019). Genome Mining of Streptomyces atratus SCSIO ZH16: Discovery of Atratumycin and Identification of Its Biosynthetic Gene Cluster. Org. Lett. 21, 1453–1457. doi: 10.1021/acs.orglett.9b00208
Uguru, G. C., Stephens, K. E., Stead, J. A., Towle, J. E., Baumberg, S., and McDowall, K. J. (2005). Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor. Mol. Microbiol. 58, 131–150. doi: 10.1111/j.1365-2958.2005.04817.x
Venugopal, J. R. V., Mukku, R. P., Maskey, P. M., Iris, G.-W., and Laatsch, H. (2002). 5-(2-Methylphenyl)-4-pentenoic Acid from a terrestrial Streptomycete. Z. Naturforsch. 57, 335–337.
Vicente, C. M., Santos-Aberturas, J., Payero, T. D., Barreales, E. G., de Pedro, A., and Aparicio, J. F. (2014). PAS-LuxR transcriptional control of filipin biosynthesis in S. avermitilis. Appl. Microbiol. Biotechnol. 98, 9311–9324. doi: 10.1007/s00253-014-5998-7
Wang, B., Guo, F., Dong, S. H., and Zhao, H. (2019). Activation of silent biosynthetic gene clusters using transcription factor decoys. Nat. Chem. Biol. 15, 111–114. doi: 10.1038/s41589-018-0187-0
Wang, L., Hu, Y., Zhang, Y., Wang, S., Cui, Z., Bao, Y., et al. (2009). Role of sgcR3 in positive regulation of enediyne antibiotic C-1027 production of Streptomyces globisporus C-1027. BMC Microbiol. 9:14. doi: 10.1186/1471-2180-9-14
Weber, T., Blin, K., Duddela, S., Krug, D., Kim, H. U., Bruccoleri, R., et al. (2015). antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243. doi: 10.1093/nar/gkv437
Wilson, D. J., Xue, Y., Reynolds, K. A., and Sherman, D. H. (2001). Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J. Bacteriol. 183, 3468–3475. doi: 10.1128/JB.183.11.3468-3475.2001
Xu, G., Wang, J., Wang, L., Tian, X., Yang, H., Fan, K., et al. (2010). “Pseudo” gamma-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J. Biol. Chem. 285, 27440–27448. doi: 10.1074/jbc.M110.143081
Zhang, C., and Seyedsayamdost, M. R. (2020). Discovery of a Cryptic Depsipeptide from Streptomyces ghanaensis via MALDI-MS-Guided High-Throughput Elicitor Screening. Angew Chem. Int. Ed. Engl. 59, 23005–23009. doi: 10.1002/anie.202009611
Zhao, H. W., Li, J. Q., and Ding, W. J. (2021). Chemical constituents from salt lake-derived Streptomyces sp. QHA10. J. Asian. Nat. Prod. Res. 23, 26–32. doi: 10.1080/10286020.2019.1700230
Zhou, Z., Xu, Q., Bu, Q., Guo, Y., Liu, S., Liu, Y., et al. (2015). Genome mining-directed activation of a silent angucycline biosynthetic gene cluster in Streptomyces chattanoogensis. Chembiochem 16, 496–502. doi: 10.1002/cbic.201402577
Zhu, J., Chen, W., Li, Y. Y., Deng, J. J., Zhu, D. Y., Duan, J., et al. (2014). Identification and catalytic characterization of a nonribosomal peptide synthetase-like (NRPS-like) enzyme involved in the biosynthesis of echosides from Streptomyces sp. LZ35. Gene 546, 352–358. doi: 10.1016/j.gene.2014.05.053
Keywords: LuxR-family regulator, ortho-methyl phenyl alkenoic acids, cinnamoyl moiety biosynthetic gene cluster, Streptomyces globisporus C-1027, genome mining
Citation: Li X, Ren W, Li Y, Shi Y, Sun H, Wang L, Wu L, Xie Y, Du Y, Jiang Z and Hong B (2022) Production of chain-extended cinnamoyl compounds by overexpressing two adjacent cluster-situated LuxR regulators in Streptomyces globisporus C-1027. Front. Microbiol. 13:931180. doi: 10.3389/fmicb.2022.931180
Received: 28 April 2022; Accepted: 04 July 2022;
Published: 03 August 2022.
Edited by:
Paul Dyson, Swansea University Medical School, United KingdomReviewed by:
Christophe Corre, University of Warwick, United KingdomCopyright © 2022 Li, Ren, Li, Shi, Sun, Wang, Wu, Xie, Du, Jiang and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhibo Jiang, ajgzNjgxMUAxNjMuY29t; Bin Hong, YmluaG9uZzY5QGhvdG1haWwuY29t, aG9uZ2JpbkBpbWIucHVtYy5lZHUuY24=
†Present address: Zhibo Jiang, Department of Pharmaceutical Engineering, School of Chemistry and Chemical Engineering, Key Laboratory of Chemical Engineering and Technology of State Ethnic Affairs Commission, North Minzu University, Yinchuan, China
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.