- 1School of Ecology and Nature Conservation, Beijing Forestry University, Beijing, China
- 2Key Laboratory of Ecosystem Network Observation and Modeling, Institute of Geographic Sciences and Natural Resources Research, Chinese Academy of Sciences, Beijing, China
The assembly mechanisms and drivers of abundant and rare fungi in dryland montane forest soils remain underexplored. Therefore, in this study, we compared the distribution patterns of abundant and rare fungi and explored the factors determining their assembly processes in a dryland montane forest in China. Stronger distance-decay relationships (DDRs) were found in abundant sub-communities than in rare sub-communities. In addition, abundant fungi exhibited greater presence and wider habitat niche breadth than rare fungi. Both the null model and variation partitioning analysis indicated that dispersal limitation and environmental selection work together to govern both abundant and rare fungal assembly, while dispersal limitation plays a dominant role. Meanwhile, the relative influence of dispersal limitation and environmental selection varied between abundant and rare sub-communities, where dispersal limitation showed greater dominance in abundant fungal assembly. Mantel tests demonstrated that soil pH and phosphorus played critical roles in mediating abundant and rare fungi assembly processes, respectively. Our findings highlight that the distinct biogeographic patterns of abundant and rare fungi are driven by different assembly mechanisms, and the assembly processes of abundant and rare fungi are determined by diverse ecological drivers in dryland montane forest soils.
Introduction
Soil microbes play crucial roles in mediating ecosystem structure and processes, such as nutrient and material cycles (Mooshammer et al., 2014; Delgado-Baquerizo et al., 2016; de Sosa et al., 2018). Elucidating the fundamental assembly process driving microbial richness and composition is vital to predicting the response of ecosystems to global changes (Zhou and Ning, 2017). Indeed, most processes related to community assembly can be classified into two classes: stochastic and deterministic processes (Stegen et al., 2012; Zhou and Ning, 2017; Liu L. et al., 2021). Niche theory postulates that niche processes, including abiotic and biotic selection, determine community assembly (Fargione et al., 2003). In contrast, the neutral theory assumes that all individuals in communities are ecologically equivalent and that communities are regulated by neutral processes, such as dispersal limitation and ecological drift (Chase and Myers, 2011). Multiple ecological processes are generally believed to work together and drive community assembly (Stegen et al., 2013), whereas their relative roles depend on time and space (Stegen et al., 2012; Dini-Andreote et al., 2015).
Soil microbial communities are primarily dominated by a few abundant species, while a mass of other species (“rare biosphere”) have an extremely low abundance (Jia et al., 2018; Egidi et al., 2019). Rare and abundant microbial assembly processes are subjected to divergent controlling mechanisms (Liu et al., 2015; Gao et al., 2020). Owing to the difference in competition capacity and stress tolerance, rare and abundant microbes exhibit quite different biogeographic patterns (Jiao and Lu, 2020b). Hence, comparing the ecological distribution and assembly mechanisms of rare and abundant microbes may be a good way to better infer microbe-driven ecosystem functioning. To date, biogeographical studies on abundant and rare bacteria have been extensively conducted in diverse environments (Jiao et al., 2017; Gao et al., 2020; Hou et al., 2020). Compared with bacteria, fungi have a larger body size (Powell et al., 2015) and can decompose complex molecules from plant litter inaccessible to most bacteria (Boer et al., 2005; Romaní et al., 2006). A previous study has found that soil fungal communities are structured by dispersal limitation, whereas deterministic factors shape bacterial composition in drylands (Wang et al., 2017). Our previous study has demonstrated that abundant and rare bacteria exhibit distinct biogeographic patterns and assembly mechanisms in the dryland montane forest. Therefore, a comparison between abundant and rare fungi is vital to exploring soil microbial assembly processes. However, the difference in distribution patterns and assembly processes between rare and abundant fungi in dryland montane forests has been barely elucidated.
An open question in ecology is whether and how environmental factors regulate the balance among different assembly processes (Tripathi et al., 2018). In fact, the environmental moderators of soil microbial community assembly processes have been widely examined in numerous ecosystems (Delgado-Baquerizo et al., 2020; Liu L. et al., 2021; Ni et al., 2021). Studies have reported that community assembly processes of soil microbes are influenced by different environmental variables, such as aridity, temperature, salinity, soil pH, and nutrients (Zhang et al., 2019; Jiao and Lu, 2020a; Wan et al., 2021), and their relative influence depends on soil microbial taxa, ecosystem types, and inquiry scales. As a particular component of dryland ecosystems, dryland montane forests are mainly distributed in high-elevation regions. Compared with grasslands and deserts, dryland montane forests are characterized by higher nutrient and water availability and lower temperatures (Wang et al., 2021a). Moreover, dryland forests have been reported to be particularly sensitive to climate change and associated increases in water stress (Liu et al., 2013; Poulter et al., 2013). Our previous studies have observed that soil pH and temperature rather than aridity drive the assembly processes of abundant and rare bacteria in dryland montane forests (Wang et al., 2021b). More importantly, studies on agricultural ecosystems and wetlands have demonstrated that divergent environmental factors mediate the assembly processes of abundant and rare fungi (Jiao and Lu, 2020a; Wan et al., 2021). Therefore, exploring the foremost drivers of soil fungal assembly processes in dryland montane forests may provide new evidence for fundamental mechanisms generating and maintaining biodiversity in drylands. However, the relative influence of different environmental factors on abundant and rare fungal assembly processes remains unclear.
Here, we aim to compare the distribution patterns and assembly mechanisms of abundant and rare fungi in dryland montane forest soils and test how different environmental factors jointly drive the assembly processes of abundant and rare fungi. Hence, we collected 24 samples from major distribution regions of dryland montane forests in China and assessed soil fungal communities based on high-throughput sequencing data of ITS. We hypothesized that (1) abundant and rare fungal sub-communities have distinct distribution patterns and assembly mechanisms and (2) divergent environmental factors regulate the assembly processes of abundant and rare fungi.
Materials and methods
Study region and field sampling
According to the distribution range of forest habitat, we selected 24 sites from a mountain forest ecosystem in northern Xinjiang, China, during the peak of the growing season (July–August) in 2016 (Supplementary Figure 1). The study region covers more than 450,000 km2. Its general topography is characterized by two longitudinal mountain systems (i.e., the Tianshan Mountains and Altay Mountains) separated by a basin (the Junggar Basin). The climate is mainly arid or semi-arid, with high variability of precipitation and temperature. At each site, a 20 m × 20 m plot was established from the representative vegetation. After that, 15 soil cores were combined per plot, taken at depths of 0–10 cm, and then mixed into a composite sample. Then all composite samples were sieved through a 2 mm mesh and divided into two portions: one portion was stored in thermally insulated boxes (at 4°C) for determining soil physicochemical properties, and the other was stored at −20°C until DNA extraction.
Soil and climate data
Soil physicochemical properties, including soil pH, total phosphorus (TSP, g/kg), total nitrogen (TSN, g/kg), total organic carbon (TOC, g/kg), soil available nitrogen (AN, mg/kg), moisture content (SM, %), and soil N: P and C: N ratios. SM was measured gravimetrically by drying at 105°C to a constant weight. Soil pH was measured at a soil-to-water ratio of 1:2.5. TOC was measured by the K2Cr2O7 oxidation method. TSN was measured by the Kjeldahl procedure. AN was measured by the alkali diffusion method. TSP was measured by the molybdenum blue method.
Climatic variables, including mean annual temperature (MAT) and mean annual precipitation (MAP), were extracted from the WorldClim global climate database using the geographic coordinates of each site (resolution: 1 km × 1 km).1 We then obtained annual potential evapotranspiration (PET) from CGIAR-CSI (with a resolution of 1 km × 1 km).2 The aridity index (AI) was estimated as the ratio of MAP to PET (AI = MAP/PET; UNEP, 1992).
Molecular and bioinformatics analysis
Total fungal DNA was extracted from 0.5 g of well-mixed fresh soil samples using E.Z.N.A. soil DNA kits (OMEGA, USA) following the manufacturer’s instructions. The fungal internal transcribed spacer (ITS) region was amplified using universal primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 (5′- TGCGTTCTTCATCGATGC-3′) (Gardes and Bruns, 1993). High-throughput sequencing was performed on an Illumina Miseq PE300 sequencing platform at Beijing Allwegene Tech, Ltd. (Beijing, China).
Fungal sequences > 200 bp with an average quality score > 20 and without ambiguous base calls were quality processed within the QIIME package (Version 2.0). After that, the remaining high-quality sequences were clustered into operational taxonomic units (OTUs) using a 97% similarity threshold within UPARSE. The taxonomy of each ITS gene sequence was analyzed by comparison with sequences within the UNITE database (Version 8.2). OTUs were picked at 97% sequence similarity. Meanwhile, OTUs with reads of less than 20 were discarded to avoid the random influence on the identification of rare taxa (Jiao and Lu, 2020a). To eliminate the influence of sequencing depths on the analyses, sequences were rarefied at 21,042 sequences from each sample. In this study, OTUs with relative abundances above 0.1% of the total sequences were regarded as abundant, while those with relative abundances below 0.01% were defined as rare. Soil fungal raw sequences used in this paper are available in the NCBI Sequence Read Archive under BioProject PRJNA825059.
Statistical analyses
Firstly, 11 environmental variables (MAT and AI for climate; SM, TSN, TOC, SAN, TSP, CN, NP, and pH for soil attributes; Altitude) were used in this study. To reduce strong collinearity between variables, we removed TOC and NP according to Pearson’s > | 0.7| (Supplementary Figure 2). Geographic distance matrices were calculated based on GPS coordinates, and then standardized environmental Euclidean distance matrices were calculated within the “vegan” package (Oksanen et al., 2015). The Bray-Curtis community dissimilarity distance was estimated to reflect the variance in species composition (β-diversity) among soil fungal communities. The slope of ordinary least-square regression between compositional similarity (1-β-diversity) and geographic distance was further used to quantify the distance–decay relationships (DDRs).
Levins’ niche breadth (B) index was employed to elucidate the patterns of stochastic and deterministic processes and their effects on soil fungal communities (Levins, 1968). The B-value of each fungal OTU was calculated following the previous study (Jiao et al., 2020). A higher B-value indicates a wider habitat niche breadth. Community B-values (Bcom) were quantified by abundance-weighted mean B-values from all fungal OTUs occurring within each community (Wu et al., 2018). A fungal community with a higher B-value is expected to be more metabolically flexible (Pandit et al., 2009). Notably, the “niche. width” function of the “spaa” R package was applied to calculate Levins’ niche breadth (B) index.
Abundance-based null model and neutral model analyses were used to infer the influence of ecological processes on soil fungal assembly (Kraft et al., 2011; Myers et al., 2013; Ning et al., 2019). In brief, 999 null local communities were generated by randomly resampling individuals into a local community with probabilities proportional to the regional abundance of the species while maintaining the same species richness and abundance (Ning et al., 2019; Liu W. et al., 2021). Afterward, the standardized effect size (β-deviation) of β-diversity was calculated using the following formula: β-deviation = [β-diversityobs − Mean (β-diversitynull)]/standard deviation (β-diversitynull), where β-diversitynull and β-diversityobs can denote the mean Bray–Curtis dissimilarity of null communities and observed β-diversity, respectively. Stochastic processes dominate community assembly if the β-deviation is statistically indistinguishable from zero; otherwise, the β-deviation remarkably greater than zero indicates a dominant influence on dispersal limitation of heterogeneous selection. Conversely, the domination of homogenizing dispersal or homogeneous selection would be supported if the β-deviation is significantly less than zero (Zhang et al., 2020). The null-model analysis was performed using “tNST” within the NST package (Ning et al., 2019). Additionally, the null-model approach conducted based on phylogenetic β-diversity can better evaluate the relative roles of different assembly processes (Stegen et al., 2013). However, fungal ITS is a variable region and cannot be aligned, so this study did not implement such analyses (Zinger et al., 2019). Meanwhile, the contribution of stochastic processes was further calculated using a neutral model by predicting the association between abundance and frequency of taxonomic occurrence (Sloan et al., 2006). R2 indicates the fit to the neutral model. The neutral model analysis was performed using the “snm” function within the iCAMP package (Ning et al., 2020).
To further identify the relative roles of environmental and dispersal limitation, we partitioned the relative influence of spatial and environmental factors on β-deviations through variation-partitioning analysis (VPA), which was performed using the “vegan” package in R. Multiple regressions on distance matrices (MRMs) were used to select environmental and spatial factors through forwarding selection until P < 0.05. The MRM test was performed using the “ecodist” package in R. The individual influence of spatial factors represents the effect of dispersal limitation, whereas the individual effect of environmental distance indicates the importance of environmental selection (Myers et al., 2013; Zhang et al., 2020). After that, the effect ratio of environmental selection to dispersal limitation (ESDS) was used to elucidate further the relative importance of environmental selection and dispersal limitation. Finally, the Mantel test was conducted to elucidate the influence of different environmental factors on the relative importance of different assembly processes.
Results
General distribution patterns of abundant and rare taxa
After quality filtering and removing chimeric sequences, 505,008 high-quality sequences were clustered into 1,688 OTUs. Across those fungal OTUs, a total of 969 OTUs (57.41%) with 23,647 sequences (4.68%) were identified as rare fungi, while only 172 OTUs (10.19%) with 398,367 sequences (78.88%) were identified as abundant fungi (Supplementary Table 1). Abundant sub-communities were mainly dominated by Ascomycota (54.08%), Basidiomycota (27.17%), and Mortierellomycota (6.82%), whereas rare sub-communities were primarily dominated by Ascomycota (57.30%), Basidiomycota (18.17%), Mortierellomycota (5.03%), Chytridiomycota (3.05%), Rozellomycota (2.04%), and Glomeromycota (1.59%) (Figure 1A).
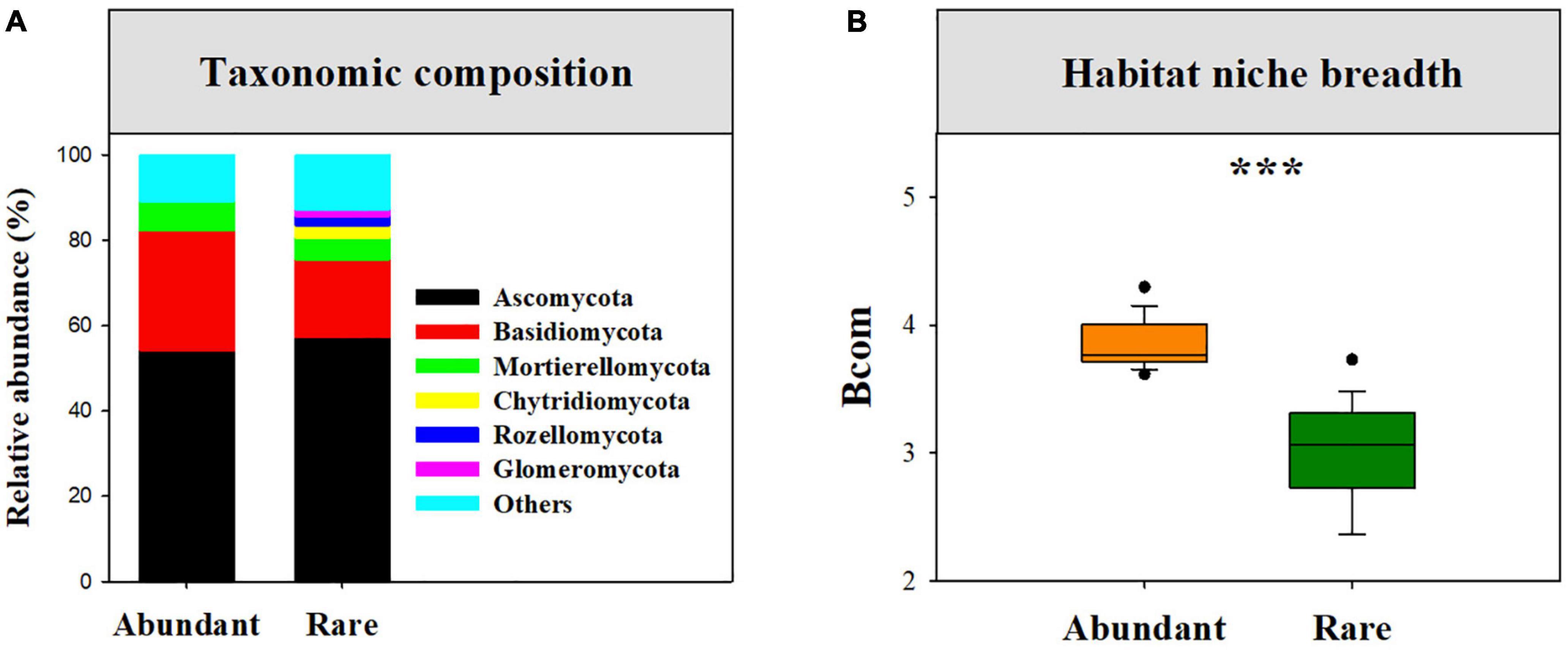
Figure 1. The taxonomic composition of rare and abundant fungal sub-communities (A), and the difference in mean habitat niche breadths (Bcom) between abundant and rare fungal subcommunities (B), and ***P < 0.0001; Wilcoxon rank-sum test.
Our results showed that abundant fungi had a greater presence than rare fungi across soil samples. Specifically, 62.21% of abundant OTUs occurred in > 50% of samples, whereas only 3.20% of the rare OTUS (31 OTUs) were present in > 50% of samples (Figure 2A). Abundance–occupancy relationships indicated that abundant fungi showed weaker positive associations than rare fungi (Figure 2B). Meanwhile, we observed remarkably higher mean Bcom values in abundant sub-communities than in rare sub-communities (Figure 1B). Remarkable DDRs between geographic distance and community similarity were found in both abundant and rare sub-communities (P < 0.001, Figure 3A), and the slope of DDRs was much stronger in abundant sub-communities than in rare sub-communities. Furthermore, abundant and rare sub-communities did not differ significantly in community β-diversity (Figure 3B). Both the species composition of abundant fungi was mainly shaped by spatial factors, followed by SM and TSN (R2 = 0.311 and 0.308, Supplementary Table 2).
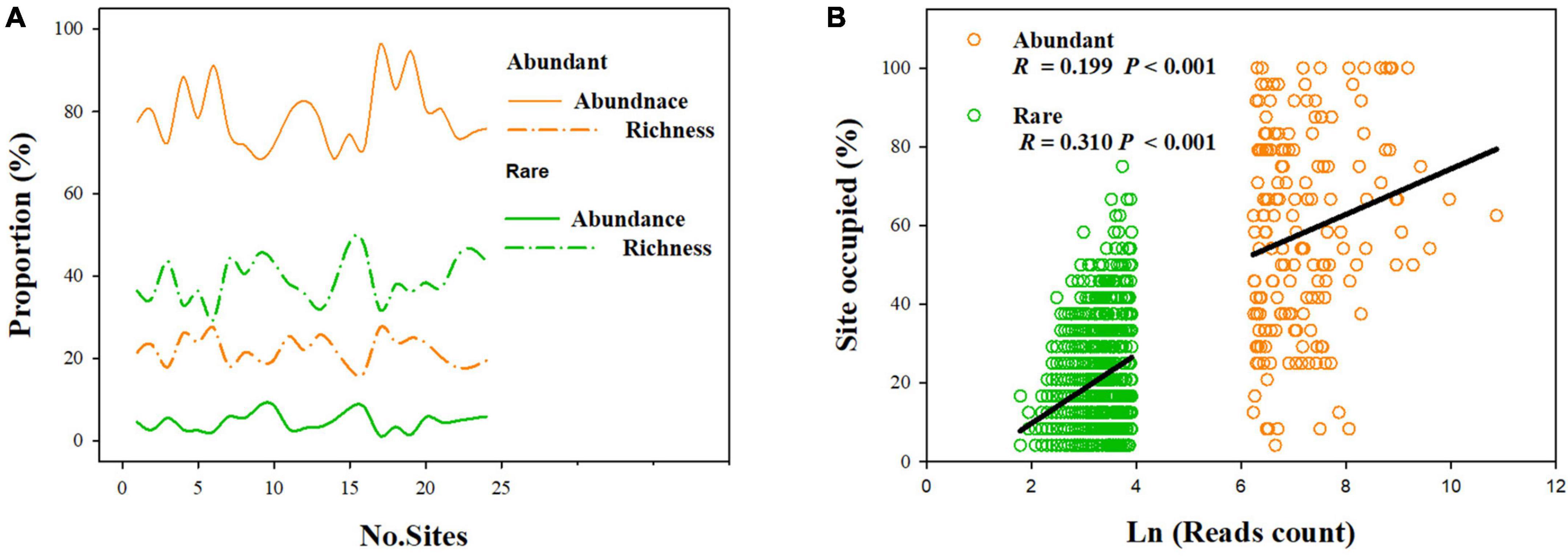
Figure 2. Distributions of rare and abundant fungi in desert soils. (A) The proportion of the OTUs’ richness (“richness”) and relative abundance of abundant and rare fungi compared to the whole fungal community in each sample. (B) Abundance–occupancy relationships for the abundant and rare sub-communities.
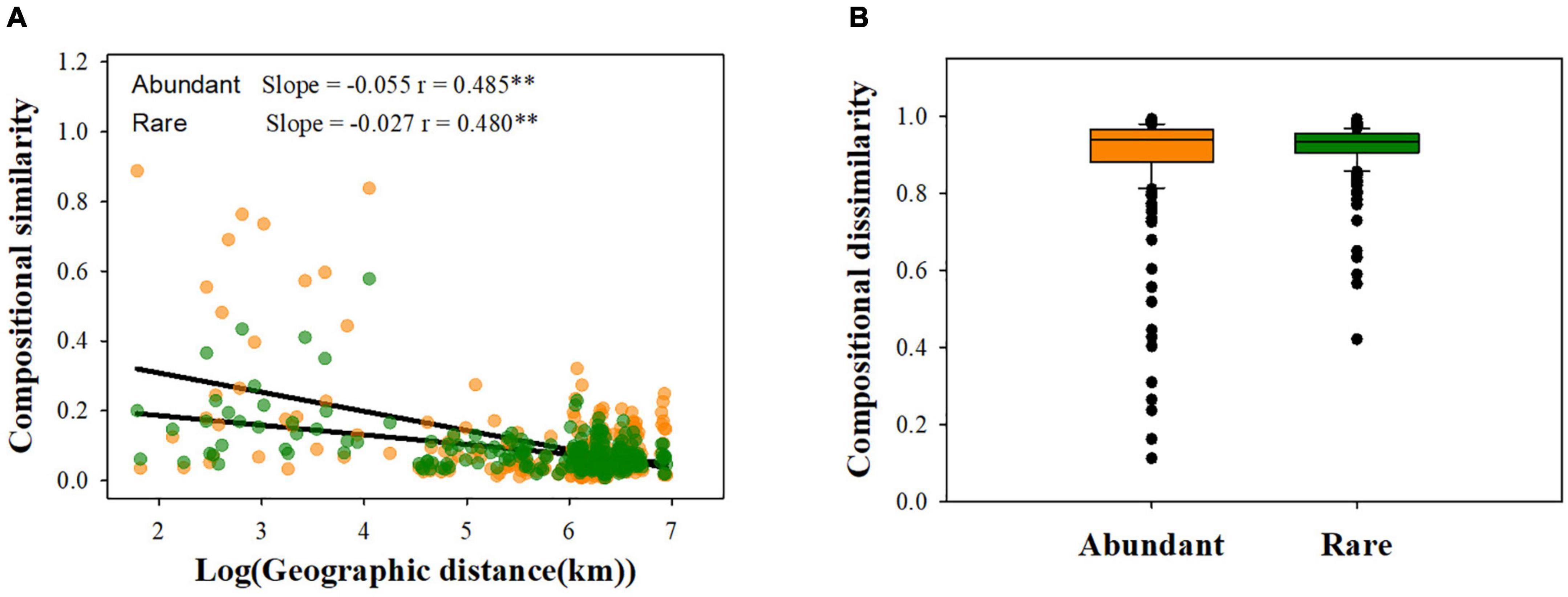
Figure 3. Patterns of rare and abundant fungal β-diversity. (A) Distance- decay curves of community similarity for rare and great fungi; (B) fungal β-diversity between rare and abundant subcommunities. **P < 0.01.
Assembly processes of rare and abundant fungal sub-communities
The neutral community model explained a larger fraction of the variation in the abundant sub-community (R2 = 0.87) than in the rare sub-community (R2 = 0.59; Table 1). The null model analysis showed that β-deviations for both abundant and rare fungal subcommunities were significantly greater than zero (Figure 4A), implying the dominance of dispersal limitation or heterogeneous selection. VPA showed that environment and space explained the total amount of variation in rare fungal β-deviations than in abundant β-deviations (Figure 4B). Environmental and space individually explained 3.40 and 11.2% of the variation in abundant fungal β-deviation, with an ESDR of 0.30. Meanwhile, environmental and space individually explained 8.67 and 21.23% of the variation in rare fungal β-deviation, with an ESDR of 0.408. These results showed that both abundant and rare fungal assembly were mainly regulated by dispersal limitation, while dispersal limitation played a relatively more important role in the abundant fungal assembly.

Table 1. Fit of the neutral model in abundant and rare fungal sub-communities in dryland montane forest soil.

Figure 4. The β-deviations for abundant and rare fungal subcommunities. (A) Difference between abundant and rare fungal β-deviation and 0 values; (B) variation in β-deviations were explained by spatial and environmental variables after forward-model selection. ***P < 0.0001; Wilcoxon rank-sum test.
The Mantel test showed that β-deviation of abundant sub-communities was significantly related to the differences in MAT, soil nutrient, and pH, while that of rare sub-communities was significantly associated with differences in altitude, MAT, AI, soil nutrient, and pH (Table 2). Among these environmental factors, the β-deviation of abundant sub-communities was more influenced by the difference in pH, while that of rare sub-communities was more influenced by the difference in TSP. Increasing differences in soil pH and phosphorus resulted in increased stochasticity for abundant and rare sub-communities, respectively (Figure 5).
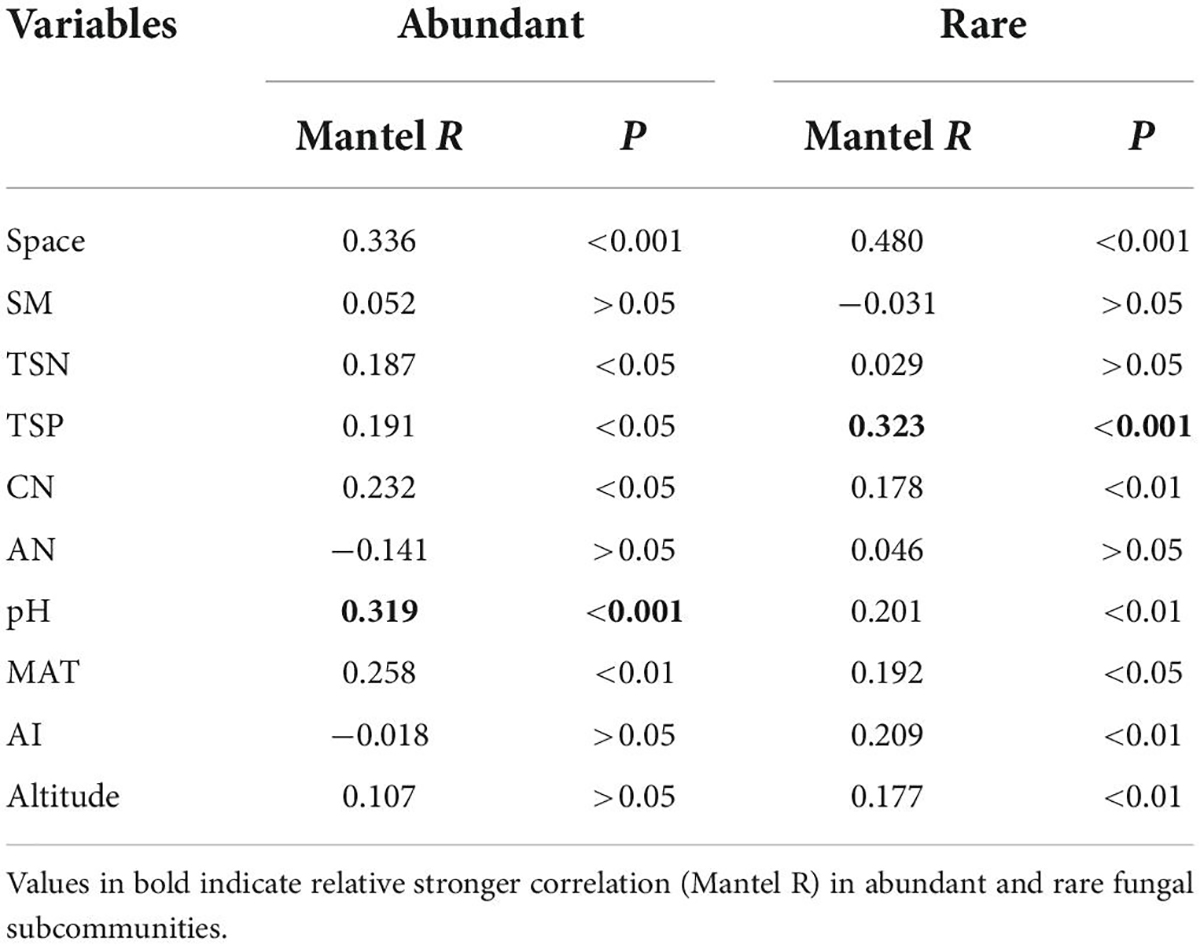
Table 2. Mantel test results showing internal links of β-deviation and environmental and spatial distances.
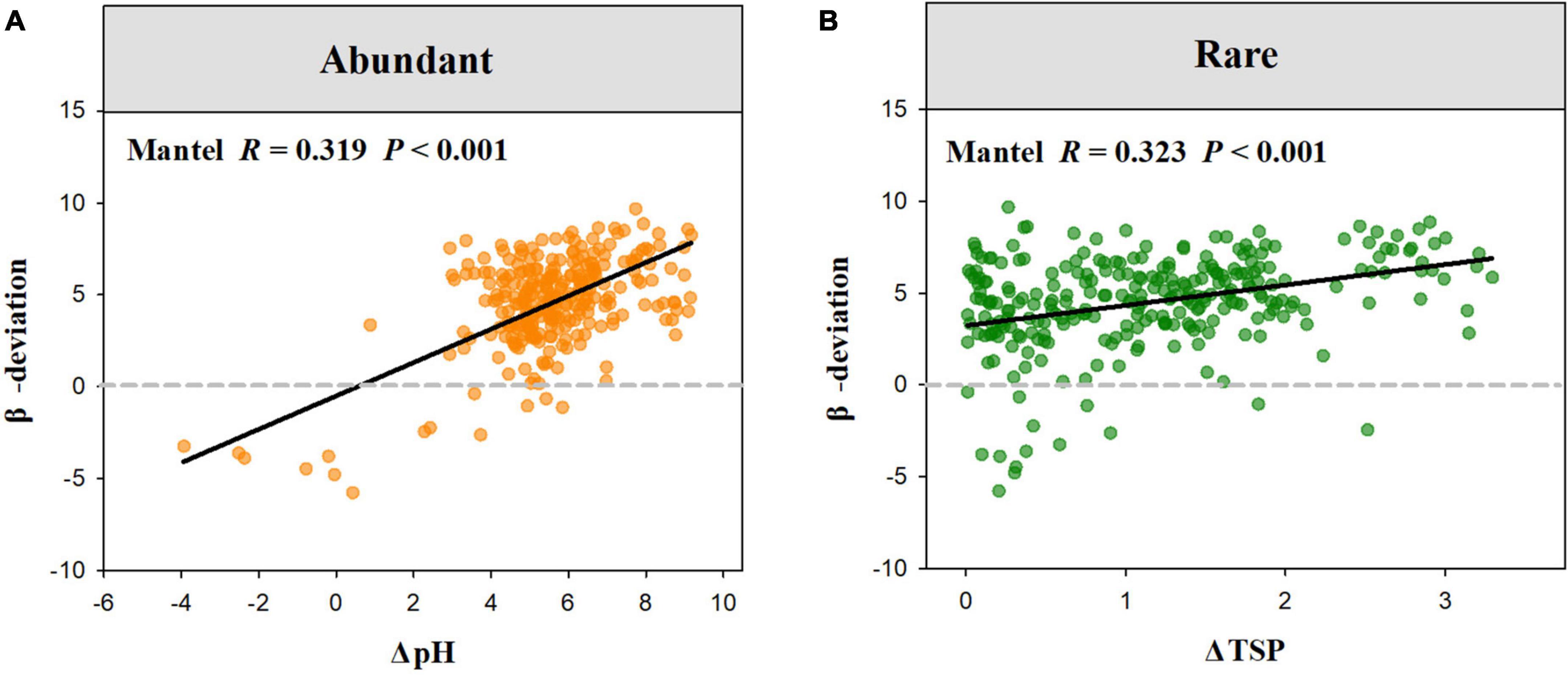
Figure 5. The relationships between abundant (A) and rare (B) fungal β-deviation and difference in pH and TSP.
Discussion
Differential distribution and environment preference of abundant and rare fungi
Understanding species distribution patterns and ecological preferences is critical for predicting how species respond to ongoing environmental changes (Maharjan et al., 2021). Consistent with the findings reported for the whole fungal community (Li et al., 2021), robust DDRs were found for abundant and rare fungi. But the steeper distance-decay slope of abundant fungi indicated that the turnover rate of abundant fungi was considerably faster than that of rare fungi (Figure 3A). The divergence in distribution patterns of abundant and rare fungi may be attributed to differences in dispersal potential and tolerance capability. We also found narrower habitat niche breadth and less ubiquity for rare fungi than abundant fungi (Figure 1B and Figure 2), indicating that rare taxa have lower tolerance and adaptability to harsh environments than abundant taxa (Delgado-Baquerizo et al., 2018). This phenomenon may reflect that rare taxa are ill-suited to most desert habitats (Brown, 1984) and therefore are limited by habitat specificity (Barberán et al., 2014; Jousset et al., 2017). Taken together, these findings reveal differential distribution patterns of rare and abundant fungi in dryland montane forests.
Dominant role of dispersal limitation in abundant and rare fungal assembly
Disentangling the relative contributions of deterministic and stochastic processes to microbial soil assembly can help better infer microbially driven ecosystem processes and functions (Nemergut et al., 2013). In this study, the neutral model analysis indicated that abundant sub-communities were more affected by neutral processes (Table 1). Both the null model and VPA analysis further demonstrated that dispersal limitation and environmental selection work together to govern both soil abundant and rare fungal assembly, whereas dispersal limitation showed a dominating effect on both abundant and rare fungal assembly (Figure 4). Meanwhile, our results also revealed that dispersal limitation has a greater relative contribution in abundant fungal assembly than in the rare, which supports prior reports that abundant sub-communities are more limited by dispersion than rare sub-communities (Wu et al., 2017; Jiao and Lu, 2020a). Most abundant species are more prone to dispersal limitation because more individuals can potentially be involved in a dispersal event (Liu et al., 2015). Moreover, it is noteworthy that a large proportion of the variation in fungal β-deviations remained unexplained by selected environmental and spatial factors (Supplementary Table 2). This result was consistent with the findings of previous studies, which may reflect the influence of other unidentified biotic factors (i.e., plant litter or plant traits; Yang et al., 2019; Guo et al., 2020; Wang et al., 2022). Together, these results implied that dispersal limitation played a greater role than environmental selection in shaping the community assembly of abundant and rare fungi.
More importantly, our results indicated that environmental selection had a stronger influence on a rare fungal assembly than the abundant. It is widely believed that abundant species occupy diverse niches and have higher resource competitiveness and greater tolerance and adaptability to environmental changes than rare species (Kraft and Ackerly, 2010; Delgado-Baquerizo et al., 2018). Hence, a rare fungal assembly is more easily influenced by environmental selection than an abundant one. Additionally, our results are also inconsistent with previous reports that environmental selection dominates in rare fungal sub-communities in agricultural and apple orchard soil (Jiao and Lu, 2020a; Zheng et al., 2021), probably due to the difference in environmental regime and geography among studies (Chase, 2010; Zhou et al., 2014). Taken together, our findings reveal dominant roles for stochastic processes in abundant and rare fungal assembly.
Soil pH and phosphorus drove the variation in the assembly process of abundant and rare fungi in dryland montane forests
Uncovering drivers mediating the balance between deterministic and stochastic processes in soil microbial communities is vital to gaining an advanced mechanistic understanding of microbial ecology (Feng et al., 2018; Tripathi et al., 2018). Previous studies have reported that community assembly processes of soil microbes are regulated by a wide range of environmental factors, such as soil pH, salinity, nutrients, and temperature (Shen et al., 2019; Zhang et al., 2019; Jiao and Lu, 2020a,b; Ni et al., 2021). In this study, we found both the difference in soil pH, nutrients, MAT, and AI were significantly related to the variation in the balance between different assembly processes of abundant and rare fungi. However, the assembly process of abundant and rare fungi was more affected by soil pH and phosphorus (STP). Increasing differences in soil pH and STP resulted in increased stochasticity for abundant and rare sub-communities, respectively (Figure 5).
Soil pH and nutrients are the key determinants of ecosystem structure and processes at multiple scales (Förstner, 1994; Xu et al., 2018; Neina, 2019; Qin et al., 2020). We further found that the relative frequency of abundant fungal β-deviation in high-pH sites (pH 6.7–7.5) was higher than in low-pH sites (pH 4.3–6.3) (Figure 6), which indicated that the relative importance of dispersal limitation on abundant fungi was higher in neutral soil than weakly acid soils. Neutral soils were suitable for most soil microbes due to their weakened environmental stress and selection strength in them (Tripathi et al., 2018), which may induce the increased role of dispersal limitation in high-pH sites (neutral soil). Furthermore, we observed that the relative frequency of rare fungal β-deviation in high-TSP sites (TSP 0.71–1.04) was higher than in low-TSP sites (TSP 0.36–0.70) (Figure 6), which demonstrated that dispersal limitation on rare fungi was more important in high-TSP sites. The increased role of dispersal limitation in high-STP sites may be owing to higher nutrient availability that could enhance the ability of rare fungi to disperse, which is inconsistent with the resource supply–stochasticity relationships (Dini-Andreote et al., 2015). Together, these findings revealed that the relative influence of environmental selection and dispersal limitation on abundant and rare fungi in dryland montane forests was driven by variations in pH and STP, respectively.

Figure 6. β-deviations distributions of abundant and rare fungi in different soil pH and STP gradients, respectively. These two categories (i.e., high-pH vs. low-pH) were divided by standardizing the number of samples in each category, which is more reasonable when comparing distinct categories (Jiao et al., 2021).
Conclusion
This study compared abundant and rare fungi distribution patterns and assembly mechanisms in dryland montane forests along wide environmental gradients. Abundant and rare fungal community similarities showed different relationships with geographic distance. Abundant fungi exhibited greater presence and wider habitat niche breadth than rare fungi. Dispersal limitations of stochastic processes dominated abundant and rare fungal sub-communities, whereas they exerted relatively greater effects on abundant fungal sub-communities. Soil pH and phosphorus played critical roles in mediating the assembly processes of abundant and rare fungi, respectively. Our study highlights the distinct distribution patterns and assembly mechanisms of abundant and rare fungal sub-communities and reveals that the assembly processes of abundant and rare fungi are determined by diverse ecological drivers in dryland montane forest soils.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA825059.
Author contributions
JW and JL designed the study. JW and YW performed the field investigation and collected the data. JW and MQ developed the methods. JW, YW, MQ, and JL wrote the manuscript.
All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Third Xinjiang Scientific Expedition Program (Grant no. 2021xjkk0600) and the National Natural Science Foundation of China (item identification nos. 32001186 and 31971538, respectively).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.929772/full#supplementary-material
Footnotes
References
Barberán, A., Ramirez, K. S., Leff, J. W., Bradford, M. A., Wall, D. H., and Fierer, N. (2014). Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria. Ecol. Lett. 17, 794–802. doi: 10.1111/ele.12282
Boer, W. D., Folman, L. B., Summerbell, R. C., and Boddy, L. (2005). Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29, 795–811. doi: 10.1016/j.femsre.2004.11.005
Brown, J. H. (1984). On the relationship between abundance and distribution of species. Am. Nat. 124, 255–279. doi: 10.1086/284267
Chase, J. M. (2010). Stochastic community assembly causes higher biodiversity in more productive environments. Science 328, 1388–1391. doi: 10.1126/science.1187820
Chase, J. M., and Myers, J. A. (2011). Disentangling the importance of ecological niches from stochastic processes across scales. Philos. Trans. R. Soc. B Biol. Sci. 366, 2351–2363. doi: 10.1098/rstb.2011.0063
de Sosa, L. L., Glanville, H. C., Marshall, M. R., Schnepf, A., Cooper, D. M., Hill, P. W., et al. (2018). Stoichiometric constraints on the microbial processing of carbon with soil depth along a riparian hillslope. Biol. Fertil. Soils 54, 949–963. doi: 10.1007/s00374-018-1317-2
Delgado-Baquerizo, M., Doulcier, G., Eldridge, D. J., Stouffer, D. B., Maestre, F. T., Wang, J., et al. (2020). Increases in aridity lead to drastic shifts in the assembly of dryland complex microbial networks. Land Degrad. Dev. 31, 346–355. doi: 10.1002/ldr.3453
Delgado-Baquerizo, M., Maestre, F. T., Reich, P. B., Jeffries, T. C., Gaitan, J. J., Encinar, D., et al. (2016). Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 7:10541. doi: 10.1038/ncomms10541
Delgado-Baquerizo, M., Oliverio, A. M., Brewer, T. E., Benavent-González, A., Eldridge, D. J., Bardgett, R. D., et al. (2018). A global atlas of the dominant bacteria found in soil. Science 359, 320–325. doi: 10.1126/science.aap9516
Dini-Andreote, F., Stegen, J. C., Van Elsas, J. D., and Salles, J. F. (2015). Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. U.S.A. 112, E1326–E1332. doi: 10.1073/pnas.1414261112
Egidi, E., Delgado-Baquerizo, M., Plett, J. M., Wang, J., Eldridge, D. J., Bardgett, R. D., et al. (2019). A few Ascomycota taxa dominate soil fungal communities worldwide. Nat. Commun. 10:2369. doi: 10.1038/s41467-019-10373-z
Fargione, J., Brown, C. S., and Tilman, D. (2003). Community assembly and invasion: An experimental test of neutral versus niche processes. Proc. Natl. Acad. Sci. U.S.A. 100, 8916–8920. doi: 10.1073/pnas.1033107100
Feng, Y., Chen, R., Stegen, J. C., Guo, Z., Zhang, J., Li, Z., et al. (2018). Two key features influencing community assembly processes at regional scale: Initial state and degree of change in environmental conditions. Mol. Ecol. 27, 5238–5251. doi: 10.1111/mec.14914
Förstner, U. (1994). “Land contamination by metals: Global scope and magnitude of problem,” in Metal speciation and contamination of soil, Vol. 1, eds H. E. Allen, C. P. Huang, G. W. Bailey, and A. R. Bowers (Boca Raton, FL: CRC Press), 1–33.
Gao, G.-F., Peng, D., Tripathi, B. M., Zhang, Y., and Chu, H. (2020). Distinct community assembly processes of abundant and rare soil bacteria in coastal wetlands along an inundation gradient. mSystems 5:e01150-20. doi: 10.1128/mSystems.01150-20
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Guo, J., Ling, N., Chen, Z., Xu, C., Li, L., Liu, L., et al. (2020). Soil fungal assemblage complexity is dependent on soil fertility and dominated by deterministic processes. New Phytol. 226, 232–243. doi: 10.1111/nph.16345
Hou, J., Wu, L., Liu, W., Ge, Y., Mu, T., Zhou, T., et al. (2020). Biogeography and diversity patterns of abundant and rare bacterial communities in rice paddy soils across China. Sci. Total Environ. 730:139116. doi: 10.1016/j.scitotenv.2020.139116
Jia, X., Dini-Andreote, F., and Salles, J. F. (2018). Community assembly processes of the microbial rare biosphere. Trends Microbiol. 26, 738–747. doi: 10.1016/j.tim.2018.02.011
Jiao, S., Chen, W., and Wei, G. (2017). Biogeography and ecological diversity patterns of rare and abundant bacteria in oil-contaminated soils. Mol. Ecol. 26, 5305–5317. doi: 10.1111/mec.14218
Jiao, S., Chen, W., and Wei, G. (2021). Linking phylogenetic niche conservatism to soil archaeal biogeography, community assembly and species coexistence. Glob. Ecol. Biogeogr. 30, 1488–1501. doi: 10.1111/geb.13313
Jiao, S., and Lu, Y. (2020b). Soil pH and temperature regulate assembly processes of abundant and rare bacterial communities in agricultural ecosystems. Environ. Microbiol. 22, 1052–1065. doi: 10.1111/1462-2920.14815
Jiao, S., and Lu, Y. (2020a). Abundant fungi adapt to broader environmental gradients than rare fungi in agricultural fields. Glob. Change Biol. 26, 4506–4520. doi: 10.1111/gcb.15130
Jiao, S., Yang, Y., Xu, Y., Zhang, J., and Lu, Y. (2020). Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J. 14, 202–216. doi: 10.1038/s41396-019-0522-9
Jousset, A., Bienhold, C., Chatzinotas, A., Gallien, L., Gobet, A., Kurm, V., et al. (2017). Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 11, 853–862. doi: 10.1038/ismej.2016.174
Kraft, N. J., and Ackerly, D. D. (2010). Functional trait and phylogenetic tests of community assembly across spatial scales in an Amazonian forest. Ecol. Monogr. 80, 401–422. doi: 10.1890/09-1672.1
Kraft, N. J., Comita, L. S., Chase, J. M., Sanders, N. J., Swenson, N. G., Crist, T. O., et al. (2011). Disentangling the drivers of β diversity along latitudinal and elevational gradients. Science 333, 1755–1758. doi: 10.1126/science.1208584
Levins, R. (1968). Evolution in changing environments: Some theoretical explorations. Princeton, NJ: Princeton University Press. doi: 10.1515/9780691209418
Li, S., Li, Y., Zheng, X., Zhang, J., Zhang, H., Bai, N., et al. (2021). Stochastic processes drive bacterial and fungal community assembly in sustainable intensive agricultural soils of Shanghai, China. Sci. Total Environ. 778:146021. doi: 10.1016/j.scitotenv.2021.146021
Liu, H., Park Williams, A., Allen, C. D., Guo, D., Wu, X., Anenkhonov, O. A., et al. (2013). Rapid warming accelerates tree growth decline in semi-arid forests of Inner Asia. Glob. Change Biol. 19, 2500–2510. doi: 10.1111/gcb.12217
Liu, L., Yang, J., Yu, Z., and Wilkinson, D. M. (2015). The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J. 9, 2068–2077. doi: 10.1038/ismej.2015.29
Liu, L., Zhu, K., Krause, S. M., Li, S., Wang, X., Zhang, Z., et al. (2021). Changes in assembly processes of soil microbial communities during secondary succession in two subtropical forests. Soil Biol. Biochem. 154:108144. doi: 10.1016/j.soilbio.2021.108144
Liu, W., Graham, E. B., Dong, Y., Zhong, L., Zhang, J., Qiu, C., et al. (2021). Balanced stochastic versus deterministic assembly processes benefit diverse yet uneven ecosystem functions in representative agroecosystems. Environ. Microbiol. 23, 391–404. doi: 10.1111/1462-2920.15326
Maharjan, S. K., Sterck, F. J., Dhakal, B. P., Makri, M., and Poorter, L. (2021). Functional traits shape tree species distribution in the Himalayas. J. Ecol. 109, 3818–3834. doi: 10.1111/1365-2745.13759
Mooshammer, M., Wanek, W., Hämmerle, I., Fuchslueger, L., Hofhansl, F., Knoltsch, A., et al. (2014). Adjustment of microbial nitrogen use efficiency to carbon: Nitrogen imbalances regulates soil nitrogen cycling. Nat. Commun. 5:3694. doi: 10.1038/ncomms4694
Myers, J. A., Chase, J. M., Jiménez, I., Jørgensen, P. M., Araujo-Murakami, A., Paniagua-Zambrana, N., et al. (2013). Beta-diversity in temperate and tropical forests reflects dissimilar mechanisms of community assembly. Ecol. Lett. 16, 151–157. doi: 10.1111/ele.12021
Neina, D. (2019). The role of soil pH in plant nutrition and soil remediation. Appl. Environ. Soil Sci. 2019:5794869. doi: 10.1155/2019/5794869
Nemergut, D. R., Schmidt, S. K., Fukami, T., O’Neill, S. P., Bilinski, T. M., Stanish, L. F., et al. (2013). Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356. doi: 10.1128/MMBR.00051-12
Ni, Y., Yang, T., Ma, Y., Zhang, K., Soltis, P. S., Soltis, D. E., et al. (2021). Soil pH determines the bacterial distribution and assembly processes in natural mountain forests of eastern China. Glob. Ecol. Biogeogr. 30, 2164–2177. doi: 10.1111/geb.13373
Ning, D., Deng, Y., Tiedje, J. M., and Zhou, J. (2019). A general framework for quantitatively assessing ecological stochasticity. Proc. Natl. Acad. Sci. U.S.A. 116, 16892–16898. doi: 10.1073/pnas.1904623116
Ning, D., Yuan, M., Wu, L., Zhang, Y., Guo, X., Zhou, X., et al. (2020). A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat. Commun. 11:4717. doi: 10.1038/s41467-020-18560-z
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P., O’Hara, R., et al. (2015). Vegan: Communityecologypackage. R package ver, 2.3–1. Available online at: https://cran.rproject.org
Pandit, S. N., Kolasa, J., and Cottenie, K. (2009). Contrasts between habitat generalists and specialists: An empirical extension to the basic metacommunity framework. Ecology 90, 2253–2262. doi: 10.1890/08-0851.1
Poulter, B., Pederson, N., Liu, H., Zhu, Z., D’Arrigo, R., Ciais, P., et al. (2013). Recent trends in Inner Asian forest dynamics to temperature and precipitation indicate high sensitivity to climate change. Agric. For. Meteorol. 178, 31–45. doi: 10.1016/j.agrformet.2012.12.006
Powell, J. R., Karunaratne, S., Campbell, C. D., Yao, H., Robinson, L., and Singh, B. K. (2015). Deterministic processes vary during community assembly for ecologically dissimilar taxa. Nat. Commun. 6:8444. doi: 10.1038/ncomms9444
Qin, Z., Zhang, H., Feng, G., Christie, P., Zhang, J., Li, X., et al. (2020). Soil phosphorus availability modifies the relationship between AM fungal diversity and mycorrhizal benefits to maize in agricultural soil. Soil Biol. Biochem. 144:107790. doi: 10.1016/j.soilbio.2020.107790
Romaní, A. M., Fischer, H., Mille-Lindblom, C., and Tranvik, L. J. (2006). Interactions of bacteria and fungi on decomposing litter: Differential extracellular enzyme activities. Ecology 87, 2559–2569. doi: 10.1890/0012-9658(2006)87[2559:IOBAFO]2.0.CO;2
Shen, C., Shi, Y., Fan, K., He, J.-S., Adams, J. M., Ge, Y., et al. (2019). Soil pH dominates elevational diversity pattern for bacteria in high elevation alkaline soils on the Tibetan Plateau. FEMS Microbiol. Ecol. 95:fiz003. doi: 10.1093/femsec/fiz003
Sloan, W. T., Lunn, M., Woodcock, S., Head, I. M., Nee, S., and Curtis, T. P. (2006). Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ. Microbiol. 8, 732–740. doi: 10.1111/j.1462-2920.2005.00956.x
Stegen, J. C., Lin, X., Fredrickson, J. K., Chen, X., Kennedy, D. W., Murray, C. J., et al. (2013). Quantifying community assembly processes and identifying features that impose them. ISME J. 7, 2069–2079. doi: 10.1038/ismej.2013.93
Stegen, J. C., Lin, X., Konopka, A. E., and Fredrickson, J. K. (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6, 1653–1664. doi: 10.1038/ismej.2012.22
Tripathi, B. M., Stegen, J. C., Kim, M., Dong, K., Adams, J. M., and Lee, Y. K. (2018). Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 12, 1072–1083. doi: 10.1038/s41396-018-0082-4
Wan, W., Gadd, G. M., Yang, Y., Yuan, W., Gu, J., Ye, L., et al. (2021). Environmental adaptation is m for abundant rather than rare microorganisms in wetland soils from the Qinghai-Tibet Plateau. Mol. Ecol. 30, 2390–2403. doi: 10.1111/mec.15882
Wang, J., He, N., Wang, Y., Li, J., and Li, M. (2021a). Divergent drivers determine soil bacterial β-diversity of forest and grassland ecosystems in Northwest China. Glob. Ecol. Conserv. 28:e01622. doi: 10.1016/j.gecco.2021.e01622
Wang, J., Li, M., and Li, J. (2021b). Soil pH and moisture govern the assembly processes of abundant and rare bacterial communities in a dryland montane forest. Environ. Microbiol. Rep. 13, 862–870. doi: 10.1111/1758-2229.13002
Wang, J., Zhang, T., Li, L., Li, J., Feng, Y., and Lu, Q. (2017). The patterns and drivers of bacterial and fungal β-diversity in a typical dryland ecosystem of northwest China. Front. Microbiol. 8:2126. doi: 10.3389/fmicb.2017.02126
Wang, Y., Wang, J., Qu, M., and Li, J. (2022). Root attributes dominate the community assembly of soil fungal functional guilds across arid inland river basin. Front. Microbiol. 13:938574. doi: 10.3389/fmicb.2022.938574
Wu, W., Logares, R., Huang, B., and Hsieh, C. H. (2017). Abundant and rare picoeukaryotic sub-communities present contrasting patterns in the epipelagic waters of marginal seas in the northwestern Pacific Ocean. Environ. Microbiol. 19, 287–300. doi: 10.1111/1462-2920.13606
Wu, W., Lu, H.-P., Sastri, A., Yeh, Y.-C., Gong, G.-C., Chou, W.-C., et al. (2018). Contrasting the relative importance of species sorting and dispersal limitation in shaping marine bacterial versus protist communities. ISME J. 12, 485–494. doi: 10.1038/ismej.2017.183
Xu, J., Liu, S., Song, S., Guo, H., Tang, J., Yong, J. W., et al. (2018). Arbuscular mycorrhizal fungi influence decomposition and the associated soil microbial community under different soil phosphorus availability. Soil Biol. Biochem. 120, 181–190. doi: 10.1016/j.soilbio.2018.02.010
Yang, T., Tedersoo, L., Soltis, P. S., Soltis, D. E., Gilbert, J. A., Sun, M., et al. (2019). Phylogenetic imprint of woody plants on the soil mycobiome in natural mountain forests of eastern China. ISME J. 13, 686–697. doi: 10.1038/s41396-018-0303-x
Zhang, K., Shi, Y., Cui, X., Yue, P., Li, K., Liu, X., et al. (2019). Salinity is a key determinant for soil microbial communities in a desert ecosystem. mSystems 4:e00225-18. doi: 10.1128/mSystems.00225-18
Zhang, X., Liu, S., Wang, J., Huang, Y., Freedman, Z., Fu, S., et al. (2020). Local community assembly mechanisms shape soil bacterial β diversity patterns along a latitudinal gradient. Nat. Commun. 11:5428. doi: 10.1038/s41467-020-19228-4
Zheng, W., Zhao, Z., Lv, F., Wang, R., Wang, Z., Zhao, Z., et al. (2021). Assembly of abundant and rare bacterial and fungal sub-communities in different soil aggregate sizes in an apple orchard treated with cover crop and fertilizer. Soil Biol. Biochem. 156:108222. doi: 10.1016/j.soilbio.2021.108222
Zhou, J., Deng, Y., Zhang, P., Xue, K., Liang, Y., Van Nostrand, J. D., et al. (2014). Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc. Natl. Acad. Sci. U.S.A. 111, E836–E845. doi: 10.1073/pnas.1324044111
Zhou, J., and Ning, D. (2017). Stochastic community assembly: Does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 81:e00002-17. doi: 10.1128/MMBR.00002-17
Keywords: dryland montane forest, biogeographic patterns, habitat niche, stochastic processes, abundant and rare fungi
Citation: Wang J, Wang Y, Qu M and Li J (2022) Dispersal limitation dominates the community assembly of abundant and rare fungi in dryland montane forests. Front. Microbiol. 13:929772. doi: 10.3389/fmicb.2022.929772
Received: 27 April 2022; Accepted: 29 August 2022;
Published: 27 September 2022.
Edited by:
Angel Valverde, Spanish National Research Council (CSIC), SpainReviewed by:
Shuo Jiao, Northwest A&F University, ChinaWenjie Wan, Wuhan Botanical Garden (CAS), China
Wenjing Zhang, Xiamen University, China
Yu Shi, Henan University, China
Copyright © 2022 Wang, Wang, Qu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingwen Li, bGlqaW5nd2VuaHlAYmpmdS5lZHUuY24=
†These authors share first authorship
 Jianming Wang
Jianming Wang Yin Wang1†
Yin Wang1† Jingwen Li
Jingwen Li