- 1Jiangxi Key Laboratory of Honeybee Biology and Beekeeping, Jiangxi Agricultural University, Nanchang, China
- 2Honeybee Research Institute, Jiangxi Agricultural University, Nanchang, China
- 3College of Forestry, Jiangxi Agricultural University, Nanchang, China
- 4USDA-ARS Bee Research Laboratory, Beltsville, MD, United States
Standing genetic variation is the predominant source acted on by selection. Organisms with high genetic diversity generally show faster responses toward environmental change. Nosema ceranae is a microsporidian parasite of honey bees, infecting midgut epithelial cells. High genetic diversity has been found in this parasite, but the mechanism for the parasite to maintain this diversity remains unclear. This study involved continuous inoculation of N. ceranae to honey bees. We found that the parasites slowly increased genetic diversity over three continuous inoculations. The number of lost single nucleotide variants (SNVs) was balanced with novel SNVs, which were mainly embedded in coding regions. Classic allele frequency oscillation was found at the regional level along the genome, and the associated genes were enriched in apoptosis regulation and ATP binding. The ratio of synonymous and non-synonymous substitution suggests a purifying selection, and our results provide novel insights into the evolutionary dynamics in microsporidian parasites.
Introduction
Microsporidia comprise a large group of obligate intracellular parasites, which forms an early branch in the fungal kingdom (Capella-Gutiérrez et al., 2012; Choi and Kim, 2017). Microsporidian genomes were generally compact, ranging between 2 and 5 Mbp (Ndikumana et al., 2017), arguably because they rely on host cells for much of their metabolic processes. Interestingly, one exceptionally large genome of 51 Mbp has been reported, and this species maintains transportable elements, which are exceedingly rare in microsporidia generally (Desjardins et al., 2015). Conventional mitochondria are lost during evolution, leaving tiny mitochondrially derived organelles called mitosomes (Burri et al., 2006). Microsporidian infections have been reported to inhibit host apoptosis, suppress host immune responses, and enhance host metabolism (Scanlon et al., 1999; del Aguila et al., 2006; Antunez et al., 2009; Cuomo et al., 2012; Higes et al., 2013; Martín-Hernández et al., 2017). More recently, a parasitic miRNA was found to be a virulence factor for one microsporidian to establish infection in silkworms (Dong et al., 2021).
Nosema ceranae is a microsporidian parasite that infects epithelial cells of honey bee midgut tissue (Higes et al., 2007). N. ceranae was first described in the Asian honey bee Apis cerana, which has established infection in the European honey bee Apis mellifera globally (Fries et al., 1996; Higes et al., 2006; Huang et al., 2007). The genome size of N. ceranae is 8.8 Mbps, with 2,280 protein-coding genes (Huang et al., 2021). The infection starts with the ingestion of N. ceranae spore-contaminated nectar. A parasite proliferation cycle lasts approximately 4 days, defined by life stages from meronts and sporonts to mature spores (Higes et al., 2007). Infected cells eventually burst to release a large number of spores. Parasites can then germinate to infect neighboring healthy epithelial cells or can be expelled into the environment through feces. Honey bees infected by N. ceranae showed reduced life span, enhanced energetic burden, and impaired navigation and memory (Antunez et al., 2009; Mayack and Naug, 2009; Eiri et al., 2015; Gage et al., 2018). In extreme conditions, the infection may lead to colony collapse (Higes et al., 2008; Botías et al., 2013). Using a set of genetic markers, high genetic diversity of N. ceranae has been found. Surprisingly, this genetic diversity can be even higher within a colony than among colonies (Gómez-Moracho et al., 2014, 2015). Additionally, the genetic variation of N. ceranae was minor among geographically distant locations (Pelin et al., 2015). The host shapes the parasite diversity, and the parasite drives the host diversification (Kamiya et al., 2014; Betts et al., 2018; McNew et al., 2021). It remains unclear how N. ceranae could maintain high levels of polymorphism. We hypothesized that mutation and frequency-dependent selection together shape the adaptation of the parasite. To validate the above hypothesis, the single nucleotide variants (SNVs) of N. ceranae were quantified over three successive generations to assess the frequency of novel and lost SNVs, and allele oscillation from parental to offspring spores.
Materials and methods
Parasite isolation, inoculation, and sample collection
To harvest sufficient spores, 300 honey bee foragers were randomly collected from five heavily infected honey bee (A. mellifera) colonies in the experimental apiary of Jiangxi Agricultural University following a regular survey. Midguts of these honey bees were dissected and homogenized to isolate N. ceranae spores via centrifugation (Fries et al., 2013). Then, the spores were purified using Percoll gradient centrifugation and confirmed as pure N. ceranae by species-specific fragment size of a conventional PCR (Chen et al., 2013; Fries et al., 2013). The purified spores were defined as F0_spores. A subset of these F0_spores was used to extract genomic DNA using cetyltrimethylammonium bromide (CTAB) (Chen et al., 2013). The remaining F0_spores were used to inoculate newly emerged honey bees. Sealed honey bee (A. mellifera) brood frames were kept in an incubator (34 ± 1°C, 60% relative humidity). In total, 200 newly emerged honey bee workers were individually fed with 2 μl sucrose solution containing 105 N. ceranae F0 spores.
Additionally, 200 newly emerged honey bee workers were fed with 2 μl sucrose solution as the F1 uninfected group. During the experiment, cohorts were divided into 4 cups containing 50 bees each and maintained on 50% sucrose solution ad libitum in the incubator. To harvest sufficient spores to infect a new batch of honey bees, the midgut was dissected from individual honey bees at 14 days postinfection (dpi), and F1 spores were collected. A subset of F1 spores was again used to extract genomic DNA, and the remaining F1 spores were used to inoculate 100 newly emerged honey bees due to a limited number of spores. In total, 100 newly emerged honey bee workers were fed with 2 μl sucrose solution as the F2 uninfected group. At 14 dpi, the midgut was dissected from individual honey bees and then pooled for spore purification, defined as F2 spores. The experiment was duplicated. Overall, two libraries for F0_spores, two libraries for F1 spores, and two libraries for F2 spores were prepared and sequenced on Illumina Hiseq 2000 platform in Novogene Co.
Bioinformatic and statistics
The quality of DNA sequencing reads was processed with Fastp and validated with FastQC (Chen et al., 2018). DNA sequencing reads were aligned to the N. ceranae genome (Ncer 3.0, GCA_004919615.1) using BWA with default parameters (Li and Durbin, 2009; Huang et al., 2021). The SNVs were identified and annotated using the Picard-GATK-SNPEFF pipeline (Van der Auwera et al., 2013). The SNVs with a quality score of less than 50 and locus alignment coverage of less than 10 were removed. Enrichment GO terms were analyzed using TopGO, R (Alexa and Rahnenfuhrer, 2021). The SNVs among F0, F1, and F2 spores were analyzed using chi-squared tests implemented using R (R Core Team, 2013). The lost and novel SNVs among the two generations were analyzed using paired t-test, implemented using R (R Core Team, 2013). The ratio of πnon–synonymous/πsynonymous (πa/πs) was calculated using SNPGenie (Nelson et al., 2015). The genome diversity π, Watterson’s θ, and corrected Tajima’s D were calculated using Popoolation (Kofler et al., 2011a). The fixation index FST was calculated using Popoolation2 (Kofler et al., 2011b). The recombination rate was calculated based on the allele frequency using ReLERNN (Adrion et al., 2020). DNA sequencing reads have been deposited in NCBI under BioProject PRJNA784016.
Results
The number of novel and lost single nucleotide variants were balanced between parental and offspring spores
Nosema ceranae spores were not found in the uninfected groups. For the spores purified from infected honey bees, 104 ± 7 million reads (mean ± SD) were aligned to the N. ceranae genome (150 bp paired reads, over 1,000 genome coverage), which accounted for 93.2% of all sequenced reads. In F0 spores, 97,279 and 98,314 SNVs were identified in two replicates with 96.8% overlap, reflecting approximately 11 SNVs per Kbp (Figure 1). Additionally, over 95% SNVs were heterozygous. Within coding regions, 7,352 non-synonymous and 8,695 synonymous SNVs were identified in F0 spores. From F0 to F1 spores, 103 synonymous and 200 non-synonymous SNVs were lost. Comparatively, 314 synonymous and 350 non-synonymous novel SNVs were gained in F1 spores. From F1 to F2 spores, 302 synonymous and 314 non-synonymous SNVs were lost. Meanwhile, 982 synonymous and 1,228 non-synonymous novel SNVs were gained. The dynamic of non-synonymous SNVs was significantly stronger than synonymous SNVs from F0 to F2 spores (paired t-test, P < 0.01). Among the novel non-synonymous SNVs in F1 spores, 198 SNVs were maintained in F2 spores located within 54 genes. Using the genome as the background, the 54 genes were significantly enriched in the metabolism pathway (Fisher’s exact test, P < 0.001). After normalizing the contig length, the contig SMUP01000049.1 (13.1 novel SNVs per 10 kbp), SMUP01000091.1 (8.6 novel SNVs per 10 kbp), SMUP01000014.1 (8.0 novel SNVs per 10 kbp), SMUP01000071.1 (6.7 novel SNVs per 10 kbp), and SMUP01000003.1 (6.2 novel SNVs per 10 kbp) showed a significantly higher tendency to generate novel SNVs compared with the genome (one sample t-test, adjusted P < 0.05) (Figure 2A). There were 269 genes embedded in the above contigs, which were enriched in the cell cycle (GO: 0010971, adjusted P < 0.05) and ATP binding (GO: 0005524, adjusted P < 0.001).
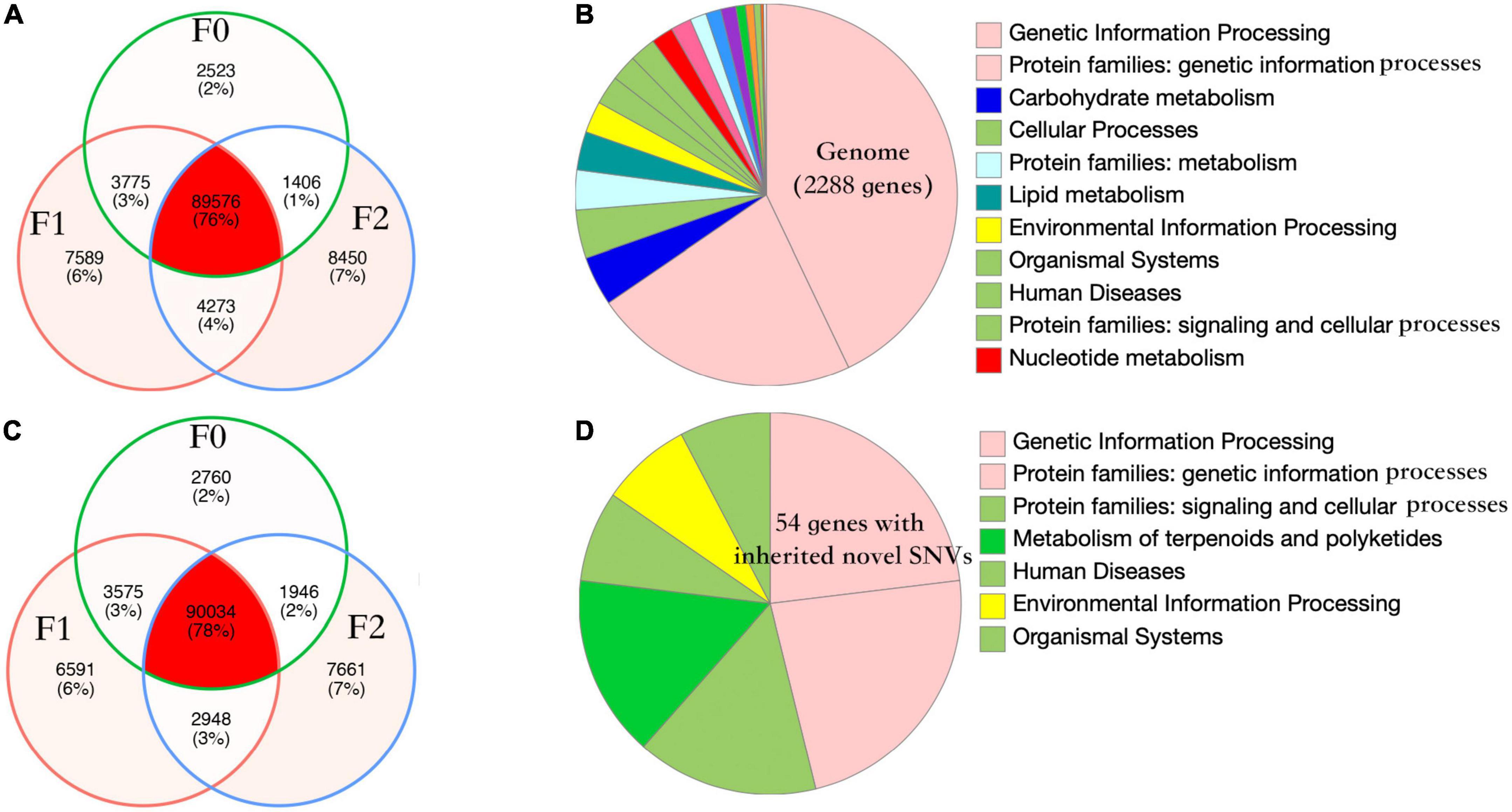
Figure 1. Number of single nucleotide variants (SNVs) and the gene function of Nosema ceranae. Venn diagram of parasite SNVs isolated from the three generations in two replicates (A,C). The number of lost SNVs was balanced with the number of novel SNVs, which could be the mechanism for the parasite to maintain high polymorphism. The pattern of SNVs was highly congruent between the two replicates (Pearson’s chi-squared test, P > 0.05). The function of protein-coding genes in the parasite genome (B,D) and the function of 54 genes with novel non-synonymous SNVs. The genes were enriched in metabolism compared with the genome (Fisher’s exact test, P < 0.001).
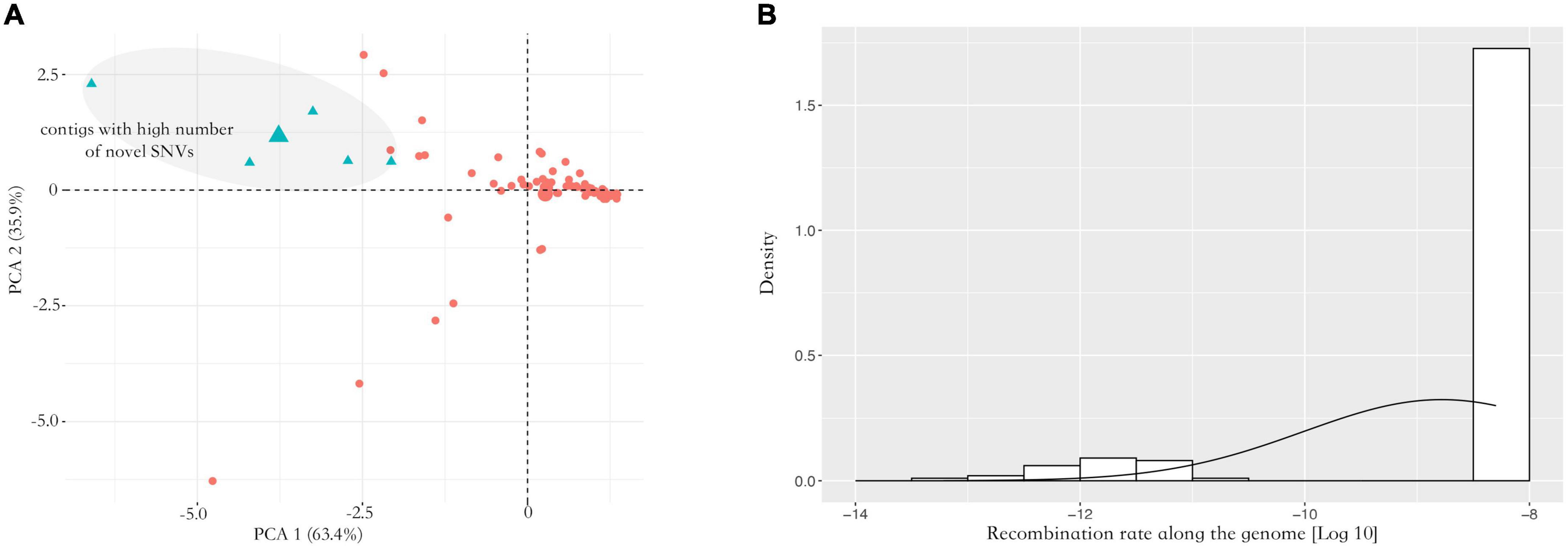
Figure 2. Analysis of allele frequency. (A) PCA analysis of the novel single nucleotide variants (SNVs) along the genome. The contigs showed a significant difference in generating novel alleles. The contigs highlighted with blue showed a higher tendency to generate novel SNVs across two replicates (one sample t-test, adjusted P < 0.05). (B) Inferred recombination rate along the genome. Overall, the inferred recombination rate was small along the genome with 5.14e-9 crossover per bp. The X-axis indicates the recombination rate after the log10 transformation. The Y-axis indicates density.
Genome recombination and allele frequency oscillation from parental to offspring spores
A total of 87,129 SNVs were inherited from F0 to F2 spores in both replicates. Over 95% of loci were heterozygous with an average alternative allele frequency of 0.38. Contigs with lower recombination rates were rarely found. Most contigs were homogeneous with 5.14e-9 cross-over per bp (Figure 2B). To quantify the allele oscillation, the allele frequency variation was calculated between parents and offspring, denoted as coefficient S. When offspring inherits the same allele frequency from their parents, the coefficient S equals 0. In our data, the coefficient S significantly deviated from F0 to F1, as well as from F1 to F2 in both replicates (one sample t-test, adjusted P < 0.001) (Figure 3). By comparing with the genome, the genes associated with the selected loci were significantly enriched in negative regulation of apoptosis (GO: 0043066, adjusted P < 0.05) and ATP binding (GO: 0005524, adjusted P < 0.001).
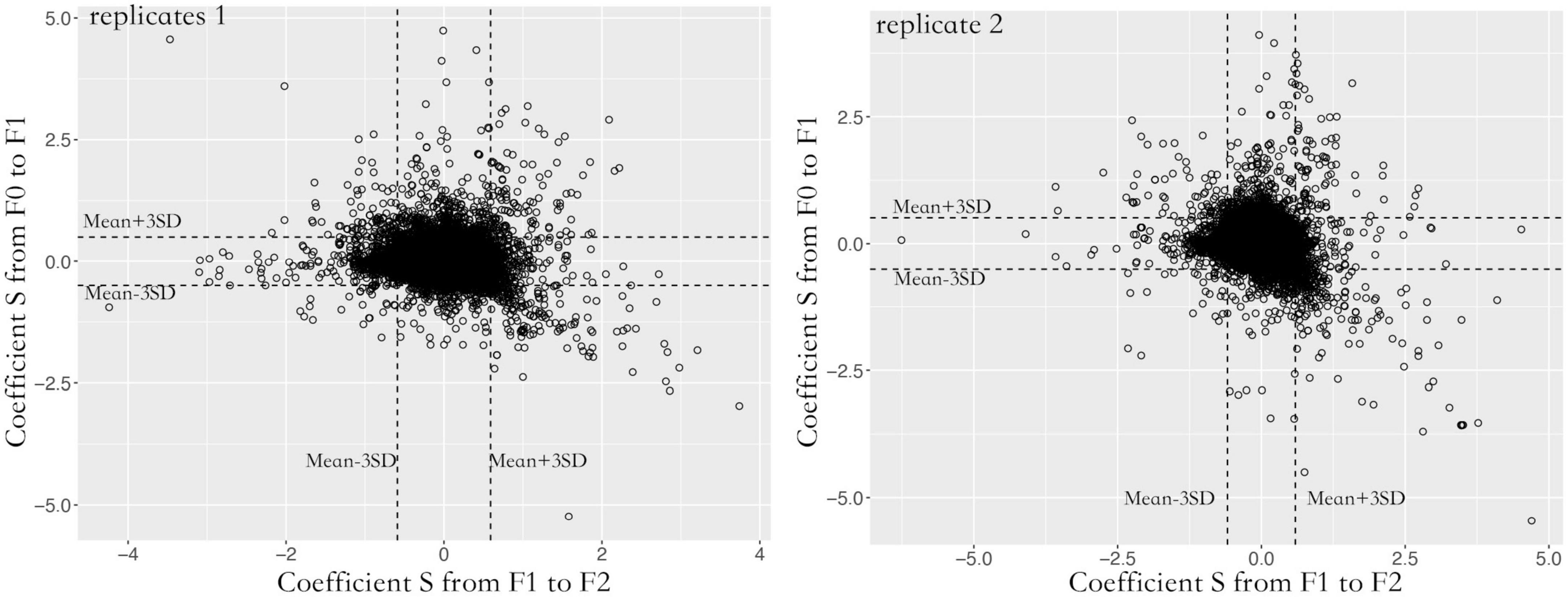
Figure 3. Allele frequency oscillation over three generations in two replicates. For each generation, the coefficient S significantly deviated from random, suggesting the existence of oscillation. To reduce the false positive calling, only the loci that fell within 99.9% confidence were considered under oscillation. The X-axis indicates allele frequency variation (coefficient S) from F1 to F2 spores. The Y-axis indicates allele frequency variation (coefficient S) from F0 to F1 spores.
Genetic differentiation accumulated over generations
Genome diversity (π) slowly increased from F0 (0.0084 ± 0.0001) and F1 (0.0087 ± 0.0001) to F2 (0.0088 ± 0.0001). Tajima’s D was positive, and the ratio of πa/πs was less than 1 for the parasites in all three generations. The fixation index FST was 0.0015 ± 0.0001 between F0 and F1 spores, which increased by twofold (0.0042 ± 0.0001) between F1 and F2 spores. After two parasite generations, the index FST increased by fourfold from F0 to F2 spores (Table 1).
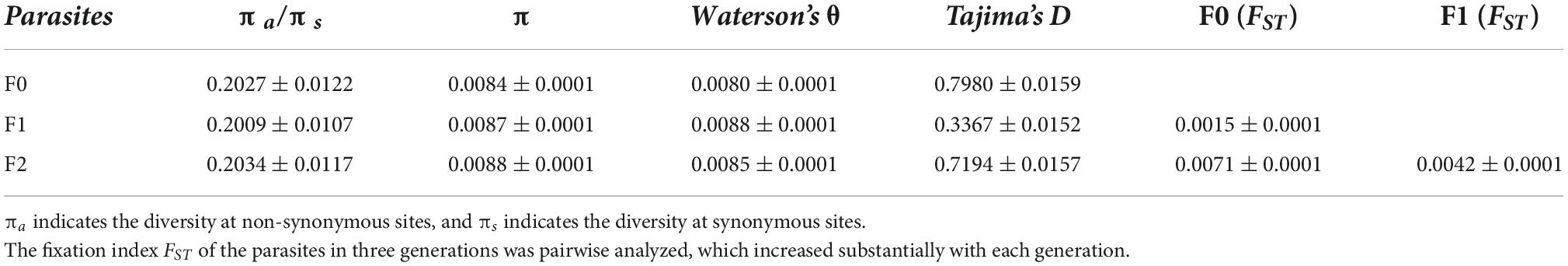
Table 1. Population genetic statistics for the three generations of Nosema ceranae spores (mean ± SE).
Discussion
Host-parasite coevolution is a process of reciprocal allelic oscillation between antagonist species and is often characterized by the Red Queen Hypothesis, which is a central theorem in coevolutionary biology (Liow et al., 2011; Brockhurst et al., 2014; Rabajante et al., 2016; Martínez-López et al., 2021). High-standing genetic variation allows fast responses to environmental change (Lai et al., 2019). N. ceranae is a native parasite of Eastern honey bees (A. cerana), which has successfully established widespread infections in Western honey bees (A. mellifera). The parasite showed high virulence in the novel host, and infected honey bees showed a shortened life span (Eiri et al., 2015), suppressed apoptosis (Antunez et al., 2009; Kurze et al., 2015; Martín-Hernández et al., 2017), and arguably are prone to colony collapses (Higes et al., 2008; Botías et al., 2013). Using a set of genetic markers, the diversity of this parasite was found higher within a honey bee colony than among colonies (Gómez-Moracho et al., 2014, 2015). In our data, the number of identified SNVs was congruent with a previous study (Ke et al., 2021). The numbers of inherited, lost, and novel SNVs were highly congruent between the two replicates, suggesting that selection is not random at the colony level. Additionally, we found minor variance between the replicates, suggesting that genetic diversity is higher within a colony than among colonies, as indicated previously (Gómez-Moracho et al., 2014; Pelin et al., 2015). Previously, the recombination event was reported in N. ceranae (Pelin et al., 2015). In our study, the calculated recombination rate was orders of magnitude smaller than those of other fungal pathogens (Heinzelmann et al., 2020; Wyka et al., 2022). Inhibiting apoptosis and ATP acquisition were essential to the success of N. ceranae infection. When apoptosis was facilitated, infected honey bees were tolerant of the infection (Kurze et al., 2015). Suppressing the expression of ATP transporters also reduced N. ceranae proliferation (Paldi et al., 2010). In our data, the alleles associated with apoptosis and ATP binding were under constant oscillation, which supports the antagonistic allele oscillation under the Red Queen hypothesis (Papkou et al., 2019). Anthropocene could also shape the genome selection of the parasite (Ptaszyńska et al., 2021), as well as the foraging behavior of honey bees. For example, parasites can quickly disperse through commonly visited flowers, which may decrease colony level variance (Graystock et al., 2015). The transporting pollination activities may further enhance the parasite dispersal at a larger scale (Proesmans et al., 2021). Furthermore, plant diversity can strongly impact the honey bee diet quality, which may shape the susceptibility to pathogens (Ptaszyńska et al., 2021). Genetic drift and selection decrease genetic variation, while mutation, gene flow, and horizontal gene transfer increase genetic variation (Schulte et al., 2013; Ellegren and Galtier, 2016). In our data, a strong fraction of SNVs was lost during the bottleneck caused by a single inoculation. Meanwhile, mutations led to ample novel SNVs. As the parasite inoculation was employed in lab conditions, the impacts of gene flow were minimal. This suggests that a balance between lost and novel SNVs maintains the high genetic diversity in N. ceranae. In our data, the accumulating π suggests a slow accumulation of genetic diversity over generations. Genome diversity π was slightly higher than Watterson’s θ, leading to a small positive Tajima’s D, which suggests a small number of low-frequency alleles (Fu and Li, 1993; Stajich and Hahn, 2005). The ratio of πa/πs was less than 1 in all three parasite generations, which suggests that the parasite genome is going through purifying selection as found in historical samples (Huang et al., 2016). The fixation index increased fourfold from F0 to F2 spores, suggesting a genome diversification (Jackson et al., 2015). The parasite can transmit among bee species, which might have posed additional selective pressure on the parasite to adapt to multiple hosts. In yeast, static environments favor small numbers of fit genotypes, dramatically decreasing genetic diversity. In contrast, fluctuating environments enrich genotypes and partially contribute to the maintenance of genetic diversity through balancing selection (Abdul-Rahman et al., 2021). Additionally, parasites show elevated diversity after infecting diverse host populations compared with monocultures (Ekroth et al., 2021), suggesting that host genotype shapes parasite diversity. To adapt to a highly polyandrous honey bees, Nosema parasites may be under the pressure to maintain high polymorphism. In Denmark, honey bees tolerant toward N. ceranae infection showed positive selection (Huang et al., 2014).
N. ceranae is a notorious parasite that weakens honey bee health. In this study, we partially explained the mechanism underlying this parasite’s observed high genetic diversity. We need to point out that multiple proliferation cycles may have occurred for each sampling generation because the infection period was three times more than a typical parasite life cycle. We found that a balance between the novel and lost SNVs drives the maintenance of genetic diversity in this species. Additionally, the allele frequencies of genes essential for parasite infection showed oscillation, supporting the Red Queen hypothesis. Our study enhances the understanding of the genetic structure, facilitated by the global spread of this parasite while providing a blueprint for parasite control.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA784016.
Author contributions
XW conducted the experiment. QH designed the experiment. XW, JZ, JE, and QH organized the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by the initiation package of Jiangxi Agricultural University (050014/923230722) and the National Natural Science Foundation of China #32260862.
Acknowledgments
We appreciate the technical support from Zhihao Wu, Bing Liu, and Lizheng Zhang.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdul-Rahman, F., Tranchina, D., and Gresham, D. (2021). Fluctuating environments maintain genetic diversity through neutral fitness effects and balancing selection. Mol. Biol. Evol. 38, 4362–4375. doi: 10.1093/molbev/msab173
Adrion, J. R., Galloway, J. G., and Kern, A. D. (2020). Predicting the landscape of recombination using deep learning. Mol. Biol. Evol. 37, 1790–1808. doi: 10.1093/molbev/msaa038
Alexa, A., and Rahnenfuhrer, J. (2021). TopGO: Enrichment Analysis for Gene Ontology. Brighton, MA: Galaxy Integrated.
Antunez, K., Martin-Hernandez, R., Prieto, L., Meana, A., Zunino, P., and Higes, M. (2009). Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia). Environ. Microbiol. 11, 2284–2290. doi: 10.1111/j.1462-2920.2009.01953.x
Betts, A., Gray, C., Zelek, M., MacLean, R. C., and King, K. C. (2018). High parasite diversity accelerates host adaptation and diversification. Science 360, 907–911. doi: 10.1126/science.aam9974
Botías, C., Martín-Hernández, R., Barrios, L., Meana, A., and Higes, M. (2013). Nosema spp. infection and its negative effects on honey bees (Apis mellifera iberiensis) at the colony level. Vet. Res. 44:25. doi: 10.1186/1297-9716-44-25
Brockhurst, M. A., Chapman, T., King, K. C., Mank, J. E., Paterson, S., and Hurst, G. D. D. (2014). Running with the Red Queen: the role of biotic conflicts in evolution. Proc. Biol. Sci. 281:20141382. doi: 10.1098/rspb.2014.1382
Burri, L., Williams, B. A. P., Bursac, D., Lithgow, T., and Keeling, P. J. (2006). Microsporidian mitosomes retain elements of the general mitochondrial targeting system. Proc. Natl. Acad. Sci. U.S.A. 103, 15916–15920. doi: 10.1073/pnas.0604109103
Capella-Gutiérrez, S., Marcet-Houben, M., and Gabaldón, T. (2012). Phylogenomics supports microsporidia as the earliest diverging clade of sequenced fungi. BMC Biol. 10:47. doi: 10.1186/1741-7007-10-47
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chen, Y. P., Pettis, J. S., Zhao, Y., Liu, X., Tallon, L. J., Sadzewicz, L. D., et al. (2013). Genome sequencing and comparative genomics of honey bee microsporidia, Nosema apis reveal novel insights into host-parasite interactions. BMC Genomics 14:451. doi: 10.1186/1471-2164-14-451
Choi, J., and Kim, S.-H. (2017). A genome tree of life for the fungi kingdom. Proc. Natl. Acad. Sci. U.S.A. 114, 9391–9396. doi: 10.1073/pnas.1711939114
Cuomo, C. A., Desjardins, C. A., Bakowski, M. A., Goldberg, J., Ma, A. T., Becnel, J. J., et al. (2012). Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res. 22, 2478–2488. doi: 10.1101/gr.142802.112
del Aguila, C., Izquierdo, F., Granja, A. G., Hurtado, C., Fenoy, S., Fresno, M., et al. (2006). Encephalitozoon microsporidia modulates p53-mediated apoptosis in infected cells. Int. J. Parasitol. 36, 869–876. doi: 10.1016/j.ijpara.2006.04.002
Desjardins, C. A., Sanscrainte, N. D., Goldberg, J. M., Heiman, D., Young, S., Zeng, Q., et al. (2015). Contrasting host-pathogen interactions and genome evolution in two generalist and specialist microsporidian pathogens of mosquitoes. Nat. Commun. 6:7121. doi: 10.1038/ncomms8121
Dong, Z., Zheng, N., Hu, C., Deng, B., Fang, W., Wu, Q., et al. (2021). Nosema bombycis microRNA-like RNA 8 (Nb-milR8) increases fungal pathogenicity by modulating BmPEX16 gene expression in its host, Bombyx mori. Microbiol. Spectr. 9:e0104821. doi: 10.1128/Spectrum.01048-21
Eiri, D. M., Suwannapong, G., Endler, M., and Nieh, J. C. (2015). Nosema ceranae can infect honey bee larvae and reduces subsequent adult longevity. PLoS One 10:e0126330. doi: 10.1371/journal.pone.0126330
Ekroth, A. K. E., Gerth, M., Stevens, E. J., Ford, S. A., and King, K. C. (2021). Host genotype and genetic diversity shape the evolution of a novel bacterial infection. ISME J. 15, 2146–2157. doi: 10.1038/s41396-021-00911-3
Ellegren, H., and Galtier, N. (2016). Determinants of genetic diversity. Nat. Rev. Genet. 17, 422–433. doi: 10.1038/nrg.2016.58
Fries, I., Chauzat, M.-P., Chen, Y.-P., Doublet, V., Genersch, E., Gisder, S., et al. (2013). Standard methods for Nosema research. J. Apic. Res. 52, 1–28. doi: 10.3896/IBRA.1.52.1.14
Fries, I., Feng, F., da Silva, A., Slemenda, S. B., and Pieniazek, N. J. (1996). Nosema ceranae n. sp. (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eur. J. Protistol. 32, 356–365. doi: 10.1016/S0932-4739(96)80059-9
Fu, Y. X., and Li, W. H. (1993). Statistical tests of neutrality of mutations. Genetics 133, 693–709. doi: 10.1093/genetics/133.3.693
Gage, S. L., Kramer, C., Calle, S., Carroll, M., Heien, M., and DeGrandi-Hoffman, G. (2018). Nosema ceranae parasitism impacts olfactory learning and memory and neurochemistry in honey bees (Apis mellifera). J. Exp. Biol. 221, jeb161489. doi: 10.1242/jeb.161489
Gómez-Moracho, T., Bartolomé, C., Bello, X., Martín-Hernández, R., Higes, M., and Maside, X. (2015). Recent worldwide expansion of Nosema ceranae (Microsporidia) in Apis mellifera populations inferred from multilocus patterns of genetic variation. Infect. Genet. Evol. 31, 87–94. doi: 10.1016/j.meegid.2015.01.002
Gómez-Moracho, T., Maside, X., Martin-Hernandez, R., Higes, M., and Bartolome, C. (2014). High levels of genetic diversity in Nosema ceranae within Apis mellifera colonies. Parasitology 141, 475–481. doi: 10.1017/S0031182013001790
Graystock, P., Goulson, D., and Hughes, W. O. H. (2015). Parasites in bloom: flowers aid dispersal and transmission of pollinator parasites within and between bee species. Proc. Biol. Sci. 282:20151371. doi: 10.1098/rspb.2015.1371
Heinzelmann, R., Rigling, D., Sipos, G., Münsterkötter, M., and Croll, D. (2020). Chromosomal assembly and analyses of genome-wide recombination rates in the forest pathogenic fungus Armillaria ostoyae. Heredity 124, 699–713. doi: 10.1038/s41437-020-0306-z
Higes, M., Garcia-Palencia, P., Martin-Hernandez, R., and Meana, A. (2007). Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia). J. Invertebr. Pathol. 94, 211–217. doi: 10.1016/j.jip.2006.11.001
Higes, M., Juarranz, A., Dias-Almeida, J., Lucena, S., Botias, C., Meana, A., et al. (2013). Apoptosis in the pathogenesis of Nosema ceranae (Microsporidia: Nosematidae) in honey bees (Apis mellifera). Environ. Microbiol. Rep. 5, 530–536. doi: 10.1111/1758-2229.12059
Higes, M., Martin, R., and Meana, A. (2006). Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J. Invertebr. Pathol. 92, 93–95. doi: 10.1016/j.jip.2006.02.005
Higes, M., Martín-Hernández, R., Botías, C., Bailón, E. G., González-Porto, A. V., Barrios, L., et al. (2008). How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 10, 2659–2669. doi: 10.1111/j.1462-2920.2008.01687.x
Huang, Q., Chen, Y. P., Wang, R. W., Cheng, S., and Evans, J. D. (2016). Host-parasite interactions and purifying selection in a microsporidian parasite of honey bees. PLoS One 11:e0147549. doi: 10.1371/journal.pone.0147549
Huang, Q., Lattorff, H. M. G., Kryger, P., Le Conte, Y., and Moritz, R. F. A. (2014). A selective sweep in a microsporidian parasite Nosema-tolerant honeybee population, Apis mellifera. Anim. Genet. 45, 267–273. doi: 10.1111/age.12114
Huang, Q., Wu, Z. H., Li, W. F., Guo, R., Xu, J. S., Dang, X. Q., et al. (2021). Genome and evolutionary analysis of Nosema ceranae: a microsporidian parasite of honey bees. Front. Microbiol. 12:645353. doi: 10.3389/fmicb.2021.645353
Huang, W.-F., Jiang, J.-H., Chen, Y.-W., and Wang, C.-H. (2007). A Nosema ceranae isolate from the honeybee Apis mellifera. Apidologie 38, 30–37. doi: 10.1051/apido:2006054
Jackson, B. C., Campos, J. L., and Zeng, K. (2015). The effects of purifying selection on patterns of genetic differentiation between Drosophila melanogaster populations. Heredity 114, 163–174. doi: 10.1038/hdy.2014.80
Kamiya, T., O’Dwyer, K., Nakagawa, S., and Poulin, R. (2014). Host diversity drives parasite diversity: meta-analytical insights into patterns and causal mechanisms. Ecography 37, 689–697. doi: 10.1111/j.1600-0587.2013.00571.x
Ke, L., Yan, W. Y., Zhang, L. Z., Zeng, Z. J., Evans, J. D., and Huang, Q. (2021). Honey bee habitat sharing enhances gene flow of the parasite Nosema ceranae. Microb. Ecol. 83, 1105–1111. doi: 10.1007/s00248-021-01827-3
Kofler, R., Orozco-terWengel, P., De Maio, N., Pandey, R. V., Nolte, V., Futschik, A., et al. (2011a). PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One 6:e15925. doi: 10.1371/journal.pone.0015925
Kofler, R., Pandey, R. V., and Schlötterer, C. (2011b). PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27, 3435–3436. doi: 10.1093/bioinformatics/btr589
Kurze, C., Le Conte, Y., Dussaubat, C., Erler, S., Kryger, P., Lewkowski, O., et al. (2015). Nosema tolerant honeybees (Apis mellifera) escape parasitic manipulation of apoptosis. PLoS One 10:e0140174. doi: 10.1371/journal.pone.0140174
Lai, Y.-T., Yeung, C. K. L., Omland, K. E., Pang, E.-L., Hao, Y., Liao, B.-Y., et al. (2019). Standing genetic variation as the predominant source for adaptation of a songbird. Proc. Natl. Acad. Sci. U.S.A. 116, 2152–2157. doi: 10.1073/pnas.1813597116
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Liow, L. H., Van Valen, L., and Stenseth, N. C. (2011). Red Queen: from populations to taxa and communities. Trends Ecol. Evol. 26, 349–358. doi: 10.1016/j.tree.2011.03.016
Martínez-López, V., Ruiz, C., Muñoz, I., Ornosa, C., Higes, M., Martín-Hernández, R., et al. (2021). Detection of microsporidia in pollinator communities of a mediterranean biodiversity hotspot for wild bees. Microb. Ecol. 84, 638–642. doi: 10.1007/s00248-021-01854-0
Martín-Hernández, R., Higes, M., Sagastume, S., Juarranz, Á, Dias-Almeida, J., Budge, G. E., et al. (2017). Microsporidia infection impacts the host cell’s cycle and reduces host cell apoptosis. PLoS One 12:e0170183. doi: 10.1371/journal.pone.0170183
Mayack, C., and Naug, D. (2009). Energetic stress in the honeybee Apis mellifera from Nosema ceranae infection. J. Invertebr. Pathol. 100, 185–188. doi: 10.1016/j.jip.2008.12.001
McNew, S. M., Barrow, L. N., Williamson, J. L., Galen, S. C., Skeen, H. R., DuBay, S. G., et al. (2021). Contrasting drivers of diversity in hosts and parasites across the tropical Andes. Proc. Natl. Acad. Sci. U.S.A. 118:e2010714118. doi: 10.1073/pnas.2010714118
Ndikumana, S., Pelin, A., Williot, A., Sanders, J. L., Kent, M., and Corradi, N. (2017). Genome analysis of Pseudoloma neurophilia: a microsporidian Parasite of Zebrafish (Danio rerio). J. Eukaryot. Microbiol. 64, 18–30. doi: 10.1111/jeu.12331
Nelson, C. W., Moncla, L. H., and Hughes, A. L. (2015). SNPGenie: estimating evolutionary parameters to detect natural selection using pooled next-generation sequencing data. Bioinformatics 31, 3709–3711. doi: 10.1093/bioinformatics/btv449
Paldi, N., Glick, E., Oliva, M., Zilberberg, Y., Aubin, L., Pettis, J., et al. (2010). Effective gene silencing in a microsporidian parasite associated with honeybee (Apis mellifera) colony declines. Appl. Environ. Microbiol. 76, 5960–5964. doi: 10.1128/AEM.01067-10
Papkou, A., Guzella, T., Yang, W., Koepper, S., Pees, B., Schalkowski, R., et al. (2019). The genomic basis of Red Queen dynamics during rapid reciprocal host–pathogen coevolution. Proc. Natl. Acad. Sci. U.S.A. 116, 923–928. doi: 10.1073/pnas.1810402116
Pelin, A., Selman, M., Aris-Brosou, S., Farinelli, L., and Corradi, N. (2015). Genome analyses suggest the presence of polyploidy and recent human-driven expansions in eight global populations of the honeybee pathogen Nosema ceranae. Environ. Microbiol. 17, 4443–4458. doi: 10.1111/1462-2920.12883
Proesmans, W., Albrecht, M., Gajda, A., Neumann, P., Paxton, R. J., Pioz, M., et al. (2021). Pathways for novel epidemiology: plant-pollinator-pathogen networks and global change. Trends Ecol. Evol. 36, 623–636. doi: 10.1016/j.tree.2021.03.006
Ptaszyńska, A. A., Latoch, P., Hurd, P. J., Polaszek, A., Michalska-Madej, J., Grochowalski, Ł, et al. (2021). Amplicon sequencing of variable 16S rRNA from Bacteria and ITS2 regions from fungi and plants, reveals honeybee susceptibility to diseases results from their forage availability under anthropogenic landscapes. Pathogens 10:381. doi: 10.3390/pathogens10030381
R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rabajante, J. F., Tubay, J. M., Ito, H., Uehara, T., Kakishima, S., Morita, S., et al. (2016). Host-parasite Red Queen dynamics with phase-locked rare genotypes. Sci. Adv. 2:e1501548. doi: 10.1126/sciadv.1501548
Scanlon, M., Leitch, G. J., Shaw, A. P., Moura, H., and Visvesvara, G. S. (1999). Susceptibility to apoptosis is reduced in the Microsporidia-infected host cell. J. Eukaryot. Microbiol. 46, 34–35.
Schulte, R. D., Makus, C., and Schulenburg, H. (2013). Host-parasite coevolution favours parasite genetic diversity and horizontal gene transfer. J. Evol. Biol. 26, 1836–1840. doi: 10.1111/jeb.12174
Stajich, J. E., and Hahn, M. W. (2005). Disentangling the effects of demography and selection in human history. Mol. Biol. Evol. 22, 63–73. doi: 10.1093/molbev/msh252
Van der Auwera, G. A., Carneiro, M. O., Hartl, C., Poplin, R., Del Angel, G., Levy-Moonshine, A., et al. (2013). From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinforma. 43, 1–33. doi: 10.1002/0471250953.bi1110s43
Keywords: microsporidian, SNV, selection, honey bee, mutation
Citation: Wei X, Zheng J, Evans JD and Huang Q (2022) Transgenerational genomic analyses reveal allelic oscillation and purifying selection in a gut parasite Nosema ceranae. Front. Microbiol. 13:927892. doi: 10.3389/fmicb.2022.927892
Received: 25 April 2022; Accepted: 11 October 2022;
Published: 01 November 2022.
Edited by:
Andrew Paul Jackson, University of Liverpool, United KingdomReviewed by:
Christopher Mayack, Sabancı University, TurkeyRui Guo, Fujian Agriculture and Forestry University, China
Copyright © 2022 Wei, Zheng, Evans and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Huang, cWlhbmctaHVhbmdAbGl2ZS5jb20=
 Xiuxiu Wei1,2
Xiuxiu Wei1,2 Jay D. Evans
Jay D. Evans Qiang Huang
Qiang Huang