
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 05 September 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.927702
This article is part of the Research TopicClimate Impact on Plant Holobiont: Mitigation Strategies and SustainabilityView all 16 articles
An investigation was carried out to understand the mechanism(s) involved in the uptake of sulfur (S) as sulfate in pigeonpea following single inoculation of two sulfur-oxidizing bacteria (SOB), Stenotrophomonas maltophilia and Stenotrophomonas pavanii in the treatments amended with either elemental sulfur (S0) or sulfate (S6). Colonization potential and biofilm formation were analyzed through confocal laser scanning microscope (CLSM) and scanning electron microscope (SEM). Furthermore, the effect of seed inoculation on root architecture, expression of genes involved in sulfur oxidation (sox) in bacterial inoculants, and genes involved in sulfate transport in pigeonpea (PpSULTR) were analyzed to correlate with the higher uptake of S in roots and shoots of pigeonpea. Both the SOB exhibited a good colonization potential and biofilm formation on the roots of pigeonpea. Among the 11 sox genes targeted in rhizosphere of pigeonpea, expression was achieved for seven genes, which showed 2-fold increase in treatments inoculated with S. maltophilia and amended with either S6 or S0. The inoculation of S. maltophilia and amendment of S0 led to increased expression of PpSULTR genes by several folds in roots. The inoculation of SOB had a significant influence on non-enzymatic (osmolytes like proline) and enzymatic (PAL, peroxidase, superoxide dismutase, and catalase) levels. The results revealed a significant increase in sulfur uptake in roots and shoots in treatment inoculated with S. maltophilia and amended with S6. The investigation showed that the SOB-mediated over-expression of PpSULTR genes in roots of pigeonpea and sox genes in the rhizosphere were acting synergistically in facilitating higher uptake and translocation of S in roots and shoots of pigeonpea plants.
Sulfur is an important nutrient for the plant growth and development. Plants take-up sulfur (S) in the form of sulfate (S6), which is +6 oxidation state of sulfur (Takahashi et al., 2011a,b; Giovannetti et al., 2014). In soils, sulfur is chiefly present in bound form as organic compounds (Takahashi et al., 2011a,b; Giovannetti et al., 2014). The plants utilize the oxidized form of S for the biosynthesis of S-rich amino acids such as cysteine, cystine, and methionine, glutathione, secondary metabolites, sulfoflavonoids, S-containing co-enzymes, and prosthetic groups (Giovannetti et al., 2014). In the last two decades, there are the reports of S-deficiency in different soil types across the globe. There are many factors contributing to this decline in S content in the soil. Intensive cropping patterns, low organic matter particularly in tropical soils, and extensive use of chemical fertilizers that are low in sulfur are some of the factors that influence plant growth due to poor availability of sulfur (McGrath et al., 2002; Lewandowska and Sirko, 2008). The conventional solution to this problem is the use of S-based chemical fertilizers. In general, it is recommended to apply elemental sulfur (S0) as compared to sulfate (S+6) for the proper growth and development of plants (Bouranis et al., 2018; Fuentes-Lara et al., 2019). However, there are microorganisms that have the ability to convert S0 to S+6 and are collectively termed sulfur-oxidizing bacteria (SOB) (Frigaard and Dahl, 2009; Zhi-Hui et al., 2010; Wang et al., 2019). Both autotrophic and heterotrophic sulfur-oxidizing bacteria have been isolated from different ecological niches (Rawlings, 2005; Majumder and Palit, 2017; Berben et al., 2019; Wang et al., 2019; Zhang et al., 2019). They are metabolically and nutritionally diverse, which includes autotrophs, heterotrophs and mixotrophs. The autotrophs include Acidithiobacillus ferrooxidans, Acidithiobacillus thiooxidans, Ancylobacter aquaticus, Halothiobacillus kellyi, Mesorhizobium thiogangeticum, Methylobacterium thiocyanatum, Thiobacillus denitrificans, Thiobacillus thioparus, Thiomonas cuprina, Thiomonas intermedia, Thiomonas perometabolis, and Thiomonas thermosulfata (Wood et al., 1998; Bacelar-Nicolau and Johnson, 1999; Sievert et al., 2000; Chen et al., 2004; Kumar et al., 2018, 2022). The heterotrophs include species of Achromobacter, Arthrobacter, Brevibacterium, Dyella thiooxydans, Flavobacterium, Klebsiella, Micrococcus, Mycobacterium, Pandoraea thiooxydans, Paracoccus, Streptomyces, Thiosphaera, and Xanthobacter (Anandham et al., 2009, 2010, 2011; Ghosh and Dam, 2009; Ryan et al., 2009; Sajjad et al., 2016; Chaudhary et al., 2017, 2021; Hou et al., 2018). However, mixotrophs include species of Aeromonas, Alcaligenes, Bacillus, Bordetella, Burkholderia kururiensis subsp. Thiooxydans, Citrobacter, Diaphorobacter, Micrococcus, Pseudomonas, Paenibacillus, Pseudoclavibacter, Rhizobium, and Stenotrophomonas, and they are the key bacterial species playing a key role in nutrient mineralization and promoting plant growth (Anandham et al., 2009; Sultan and Faisal, 2016; Malviya et al., 2022; Sanwani et al., 2022). There are many reports available on the positive influence of inoculation of these SOB on plant growth and yield (Anandham et al., 2007; Berben et al., 2019). In oil seed crops, these bacteria also help in improving the oil recovery and oil quality (Anandham et al., 2007). In legumes, the deficiency of sulfur has been reported to inhibit the process of nodulation and nitrogen fixation (Anandham et al., 2007; Cheng et al., 2017). Rhizosphere engineering of crop plants using SOB as inoculants appears to be a safe alternative to S-containing chemical fertilizers.
There are few reports available on the mechanisms by which SOB exerts its influence on uptake of S in plants. The sulfate taken-up by the plant roots is transported from roots to shoots and to seeds through various sulfate transporters. The sulfate transporters and the genes involved therein have been identified in the model plant Arabidopsis and a few other crop plants (Yoshimoto et al., 2007; Maruyama-Nakashita et al., 2015). In Arabidopsis, about 12 sulfate transporters (SULTR) were identified that vary in their affinity and location (Vidmar et al., 2000; Yoshimoto et al., 2002, 2003, 2007; Maruyama-Nakashita et al., 2015). A number of four groups of sulfur transporters (SULTR1, SULTR2, SULTR3, and SULTR4) have been identified that are involved in translocation of sulfate from soil to roots and in vascular translocation to other parts of the plant (Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002). They are also involved in release of vacuolar sulfate to maintain sustained release and utilization of S-pools in the plant system (Kataoka et al., 2004; Maruyama-Nakashita et al., 2015).
Pigeonpea is the second most important legume grown in India and ranks sixth among the legumes globally (Varshney et al., 2012). In general, pulses are reported to have deficiency of sulfur containing amino acids (Bressani et al., 1986; Singh and Diwakar, 1993; Saxena et al., 2010) and the fulfillment of S requirement in pigeonpea is largely dependent upon the use of chemical fertilizers (Jat and Ahlawat, 2010; Kumar et al., 2012). Consequences of chemical fertilizers use include deterioration soils quality, contamination of the environment, and negative impact on human and animal health. The negative impacts of chemicals have compelled researchers and policymakers to look for alternative strategies. Among them, plant-breeding approach is one of the alternative strategies where plant breeders are targeting this issue through breeding approaches using suitable donor parents. However, the availability of the suitable donor parents and the transfer of desired traits into a suitable commercial cultivar using a backcross/marker-assisted breading program is a great challenge to the pulse breeders. Under these circumstances, the use of microbe-based strategies for S nutrition is an emerging technique/approach, which is environment-friendly and residue-free. The utilization of SOB could be an alternative approach to improve sulfur content in pigeonpea. In our earlier study, we reported strains of Stenotrophomonas maltophilia and S. pavanii isolated from different samples collected from Open Cast Projects of Jharkhand (India), to be efficient for sulfur oxidation and plant growth promotion (Malviya et al., 2022). These strains exhibited multiple plant growth-promoting traits and their inoculation enhanced the activity of reactive oxygen scavenging (ROS) enzymes and uptake of nitrogen, phosphorus, and sulfur in pigeonpea (Malviya et al., 2022). In-depth investigation is required to understand the key mechanisms playing role in the S oxidation in the rhizosphere along with S uptake and translocation in the pigeonpea. In this study, we performed a comprehensive investigation of the pigeonpea SULTR genes family using comparative genomics and phylogenetic analyses. Furthermore, we characterized the biofilm forming S-oxidizing microbial inoculants and attempted to explain the microbe-mediated mechanisms of S-transport in pigeonpea plants using physio-biochemical and molecular approaches. This work presents the analyses of the SULTR genes family, and the results will provide a basis for further investigation on the microbe-mediated modulation of SULTR genes for efficient uptake and translocation of sulfur in other plants.
In total, two sulfur-oxidizing bacterial strains, Stenotrophomonas maltophilia DRC-18-7A (MZ436650) and Stenotrophomonas pavanii DRC-18-7B (MZ436648) previously isolated from coal mines (23°41′42.20″N 85°17′42.99″E), were obtained from Plant–Microbe Interaction and Rhizosphere Biology Lab, ICAR-National Bureau of Agriculturally Important Microorganisms, Kushmaur, Maunath Bhanjan, Uttar Pradesh, India (Malviya et al., 2022). These strains were sub-cultured and maintained on thiosulfate medium (sodium thiosulfate: 5 g, sodium carbonate: 200 mg, ammonium chloride: 100 mg, di-potassium hydrogen phosphate: 100 mg, agar: 20 g, water: 1,000 ml, bromophenol blue: 100 mg, pH 8) (Veerender et al., 2014) at 28°C for 21 days and stored at 4°C.
Pigeonpea seeds (cv. Malviya 13) were obtained from Department of Genetics and Plant Breeding, Banaras Hindu University, Varanasi, India. Seeds were surface sterilized with mercuric chloride (0.1%) for 3 min followed by second sterilization with ethyl alcohol (70%) for 30 s. Thereafter, seeds were washed three times with sterile water and germinated on water agar plates. The germinated seedlings were placed in the Leonard jars filled with 500 g of sterilized river sand. The Leonard jars were inoculated with 1 ml of broth suspension (2 × 108 cells ml–1). In total, two plants were maintained in each Leonard jar. Uninoculated jars were maintained as control. The average mean temperature and relative humidity during the experimentation were 26°C and 80%, respectively.
The Leonard jar experiment was laid out in a completely randomized block design under glasshouse conditions. The experimental set-up consisted of nine different treatments: T1- Stenotrophomonas maltophilia DRC-18-7A + sulfate compound (S6), T2- S. maltophilia DRC-18-7A + elemental S (S0), T3- S. pavanii DRC-18-7B + S6, T4- S. pavanii DRC-18-7B + S0, T5- S. maltophilia DRC-18-7A, T6- S. pavanii DRC-18-7B, T7- S6, T8- S0 and T9- absolute control (No inoculation, -S). Each treatment was replicated 10 times. The amount of sulfur added as S6 or S0 was 54 mg in each Leonard jar. The sulfate was added through nutrient solution, whereas elemental S was mixed with the sterile sand used to fill Leonard jars. The composition of nutrient solution with and without sulfate ions is given in Supplementary Table 1.
The selected strains were inoculated in the thiosulfate broth (Veerender et al., 2014), incubated for 7 days in the incubator shaker at 150 RPM at 28°C. Broth culture of each bacterium (1 ml, 2 × 108 cfu ml–1) was inoculated over seeds in each Leonard jar.
After 15 days of sowing, the plants from three replicates for each treatment were up-rooted gently and washed in running tap water. Confocal laser scanning microscopy was done according to the protocols described by Singh S. et al. (2020). Briefly, clean roots were treated with Syto9 and propidium iodide stains and imaging was performed using 488 and 543 nm channels under confocal scanning laser microscope (Nikon Eclipse Confocal A1, Japan). For scanning electron microscopy, root samples were washed in running tap water, fixed in mixture of formaldehyde (37%) (HiMedia, Mumbai, India) and glutaraldehyde (2.5%) (HiMedia, Mumbai, India) in 1:1 ratio for 24 h at 4°C. Thereafter, the fixed samples were kept into osmium tetroxide solution (HiMedia, Mumbai, India) for 12 h at ambient room temperature (∼27°C). The fixed root samples were dehydrated using gradient of ethyl alcohol, i.e., 30, 50, 70, 90, and 100% (30 min each) and dried under vacuum. After proper drying, the samples were coated with gold (20 nm) and visualized under scanning electron microscope (Hitachi S-3400N, United States) as described by Singh et al. (2021).
After 30 days of sowing, the plants from seven replications of each treatment were uprooted. Roots were washed gently in running tap water and brought to the laboratory. The plant growth parameters such as shoot and root length and fresh and dry biomass of root and shoot were recorded.
To see the effect of seed inoculation on root architecture, roots were washed gently in running tap water and the clean roots were scanned using root scanner (Regent Instrument, Canada). The scanned images were analyzed using image analysis software “WinRhizo Pro 2017” (Client# IN1803202) and different parameters related to root architecture, secondary and tertiary rooting were recorded in the plants inoculated with selected strains and amended with S6 and S0 at 30 days of sowing.
To evaluate the S-oxidizing activity of selected strains in the pigeonpea rhizosphere, expression analyses of genes associated with the S-oxidation were performed. For this, rhizospheric sand samples were collected from seven replicates of each treatment and brought to the laboratory in cool packs. The samples were vortexed to loosen the bacteria from sand particles. Total RNAs were isolated with the MoBio PowerSoil total RNA isolation kit (MO BIO Laboratories, Inc.) following the manufacturer’s protocols. Approximately 1 μg of RNA was used to synthesize cDNA with oligo-dT using cDNA Synthesis Kit (BioRAD, India) following the manufacturer’s instructions and quality as well as concentration of cDNA was determined using Nanodrop 2000c (Thermo Fisher Scientific, United States). For gene expression analysis, a semi-quantitative PCR method was used. The expression of genes related to S-oxidation, that is, soxB, tetH, sdoA, sdoB, tsdA, TQO, and sorAB was analyzed using gene-specific primers (Supplementary Table 2). Gene rpoD was taken as internal control. The final gene product obtained with RT-PCR was separated by electrophoresis in 1.5% agarose gel in TAE buffer (Mini gel electrophoresis unit, Bangalore GeNei, India), and visualization was done with the help of gel documentation system (Bio-Rad, India).
Nucleotide and protein sequences of sulfate transporters (SULTRs) of Arabidopsis (Arabidopsis thaliana), soybean (Glycin max), field pea (Pisum sativum), rice (Oryza sativa), and wheat (Triticum durum) were retrieved from National Center for Biotechnology Information (NCBI) database (Supplementary Table 3). These sequences were used to search the homologous sequences in pigeonpea genome using nucleotide BLAST (Basic Local Alignment Search Tool), BLASTx (translated nucleotide → protein), tBLASTn (protein → translated nucleotide) program of NCBI. These sequences were analyzed to confirm the presence of the SULTR domain in retrieved pigeonpea homologs SULTRs gene sequences using the SMART program. Furthermore, ExPasy website1 was used to analyze and confirm the primary structure of SULTR proteins and several other parameters such as molecular weight, length, total number of atoms extinction coefficients, isoelectric point, aliphatic index, instability index, grand average of hydropathicity, etc. The phylogenetic tree was constructed based on the alignment of SULTR domains of pigeonpea, Arabidopsis, soybean, field pea, rice, and wheat to elucidate the phylogenetic relationships and classified them into different groups. For this, MEGA X version was used to prepare the phylogenetic tree, and neighbor-joining method was adopted with 1,000 bootstrap replications. Furthermore, primers were designed for qPCR analyses using Primer3 (v. 0.4.0) online software2 (Supplementary Table 4) and validated in silico using primer-BLAST online tools of NCBI3 against pigeonpea transcript sequences (Cajanus cajan taxid:3821).
The quantitative RT-PCR analysis was performed to investigate the expression of genes involved in sulfur uptake and transport in pigeonpea plant under different treatments. After 30 days of sowing, plants from four replications were harvested and divided into roots and shoots. The root and shoot samples were quick-frozen in liquid nitrogen, ground and total RNAs was extracted using RNA isolation kit (Agilent, India) using the manufacturer’s protocols. The cDNA was made as discussed in the previous sections “Expression of genes responsible for S-oxidation in the plant rhizosphere.” The quality and quantification of cDNA was carried out using nanodrop. The housekeeping gene actin was used as an endogenous standard to normalize the quantitative expression data. The expression of PpSULTR genes was analyzed using gene-specific primers designed for the present investigation (Supplementary Table 4). The qRT-PCR was performed using the SYBR Green Master Mix (Thermo Fisher Scientific) on the BioRAD Real Time PCR System (MJ MiniOpticon, BioRAD). The specificity of the amplification was verified by melting-curve analysis. The relative transcript levels were calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001).
A quantitative estimation was done to evaluate the impact of inoculation of SOB on physio-biochemical properties and antioxidant enzymes in the pigeonpea leaves at 30 days of sowing. The total chlorophyll, carotenoids, total soluble sugar, and total protein in the plant leaves were measured (Sadasivam and Manickam, 1996). The accumulation of proline, phenolics, flavonoids, and superoxide dismutase (SOD) in the plant leaves was analyzed according to the procedure described by Thimmaiah (2012). The activities of PAL, peroxidase and catalase were estimated in the plant leaves according to Sadasivam and Manickam (1996).
Histological studies were also carried out to visualize the deposition of superoxide radicals (O2–) in the leaves and program cell death. Plant leaves were sampled randomly from each treatment and used for microscopic localization of superoxide radicals (O2–) using nitroblue tetrazolium (NBT; HiMedia, India) as per the methods described by Rao and Davis (1999), and it was visualized as blue color spots on the leaves. Program cell death (PCD) was examined using Evans Blue staining as described by Baker and Mock (1994).
Sequences of key genes regulating the phenylpropanoid cascade in pigeonpea were retrieved from NCBI. Primers were designed for qPCR analyses and validated in silico (Supplementary Table 5). The nine key genes analyzed were as follows: phenylalanine ammonia-lyase [EC:4.3.1.24], phenylalanine/tyrosine ammonia-lyase [EC:4.3.1.25], 4-coumarate-CoA ligase [EC:6.2.1.12], cinnamoyl-CoA reductase [EC:1.2.1.44], cinnamyl-alcohol dehydrogenase [EC:1.1.1.195], peroxiredoxin 6 [EC:1.11.1.7], Ferulate-5-hydroxylase [EC:1.14.-.-], caffeoyl-CoA O-methyltransferase [EC:2.1.1.104], and coniferyl-aldehyde dehydrogenase [EC:1.2.1.68]. qRT-PCR analyses were performed to estimate the transcript and expression analyses (as mentioned in the previous section: Expression analysis of PpSULTR genes). Actin was taken as internal control.
Phenolics and flavonoids such as gallic acid, ferulic acid, sinapic acid, syringic acid, rutin, and quercetin in the plant leaves were analyzed through HPLC (binary pump model 515, 2414 refractive index (RI), and 2998 photodiode array (PDA) detector; Supelco C-18 column; Waters Pvt. Ltd.). Leaf samples (1 g) were collected from each treatment and cleaned before processing using running tap water. Active principles were extracted using methanol and acetonitrile and individual phenolics and flavonoids were measured as per the methods described by Tiwari et al. (2011).
The total sulfur in plant samples was estimated using barium sulfate turbidimetry method (Garrido, 1964). In principle, during wet digestion of plant samples, sulfur present in the plants tissue is converted into sulfate ions and precipitated as barium sulfate after treatment with barium chloride. Briefly, 1 g of plant sample was taken in a 100-ml Erlenmeyer flask and pre-digested for 8 h using 10 ml of concentrated HNO3. The samples were further digested by addition of 10 and 3 ml of HCIO4 (3 ml) in flasks. The flasks were placed on a hot plate, heated at 100°C for 1 h, and subsequently, the temperature was raised to 200°C. The heating was continued until the contents became colorless and reduced to 3 ml. The flasks were cooled at room temperature. Approximately 1 ml HCl (6N) and 1 ml Gum acacia (0.5%) were added and mixed properly by swirling, and finally, 0.5 g BaCl2.2H2O crystals were added to the flasks. The samples were mixed until BaCl2.2H2O crystals were dissolved completely. The reading was taken at 420 nm using UV-Vis spectrophotometer. The S-content in the plant samples was calculated using the reading of standards.
The data were subjected to the analysis of variance and least significant difference (LSD) at p ≤ 0.05 using SPSS 16.0. Data were compared with Duncan’s multiple range test at p ≤ 0.05. Graphs were prepared using statistical software Origin (Version 9) and Microsoft Office Excel (2010).
In this study, the microbe-mediated mechanisms of S-oxidation and enhanced uptake and translocation of sulfate ions in the pigeonpea at the early stage of crop growth were elucidated.
Confocal laser scanning microscopic and scanning electron microscopic photographs clearly showed that both the strains have the potential to colonize and develop biofilm on pigeonpea roots even under limited S-availability at 15 days of sowing. The colonization pattern/efficiency was different for the two strains on root surface. Confocal microphotograph clearly indicated that strain S. maltophilia DRC-18-7A produced primarily micro-aggregates and later on converted into macro-aggregates on the root surface after 15 days of inoculation (Figure 1A). Microphotograph of S. pavanii DRC-18-7B-treated roots revealed primarily single cells embedded in the root epidermis and rarely formed micro-aggregates (Figure 1B). However, no such evidence of bacterial colonization was observed in untreated control plants (Figure 1C).
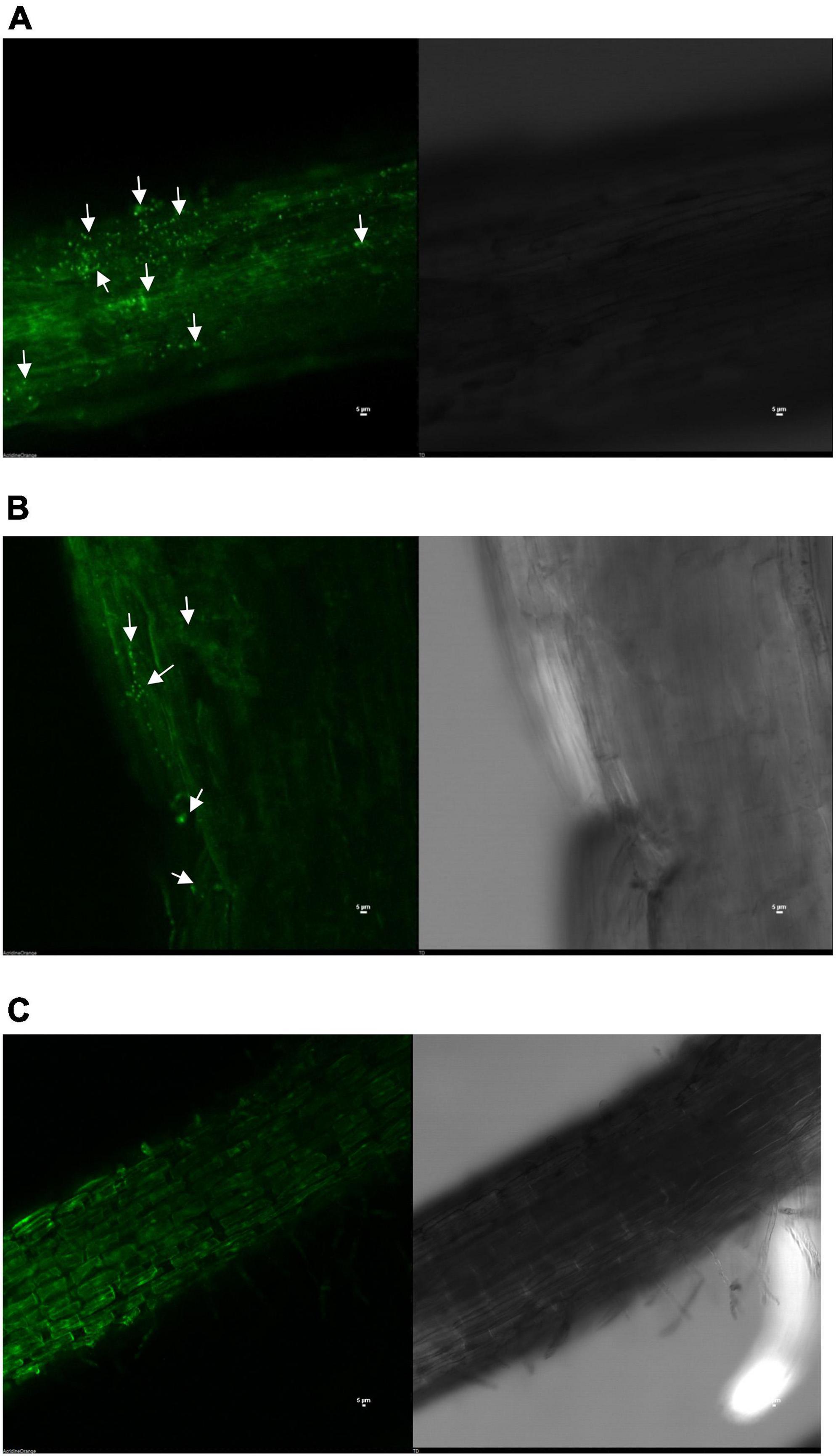
Figure 1. Confocal microphotograph showing root colonization by S. maltophilia DRC-18-7A (A), S. pavanii DRC-18-7B (B) and uninoculated control (C).
Stenotrophomonas maltophilia DRC-18-7A colonized pigeonpea roots at a very high population density which is clearly visible in scanning electron microphotographs where cells were anchored to the root surfaces and to themselves by a network of fibrillar material, exo-polysaccharide produced by them on the root surface (Figure 2A). It is clearly visible in the scanning electron microphotograph that strain S. maltophilia DRC-18-7A produced ample amount of exo-polysaccharide and formed microbiotic crust on the root surface and bacterial cells were embedded/trampled in the crust on the root surface. In general, S. maltophilia DRC-18-7A cover entire root and produced thick biofilm by forming micro-aggregates and macro-aggregates (Figure 2A). From scanning electron microphotograph of S. pavanii DRC-18-7B, it is clear that strain S. pavanii DRC-18-7B is a better root colonizer (Figure 2B). S. pavanii DRC-18-7B population was spread on the entire root by forming micro-aggregates, and sometime, single-single cells are visible. In contrast, it is not producing too much of exo-polysaccharides as compared to S. maltophilia DRC-18-7A (Figure 2B). However, no such evidence of bacterial colonization was observed in untreated control plants (Figure 2C).
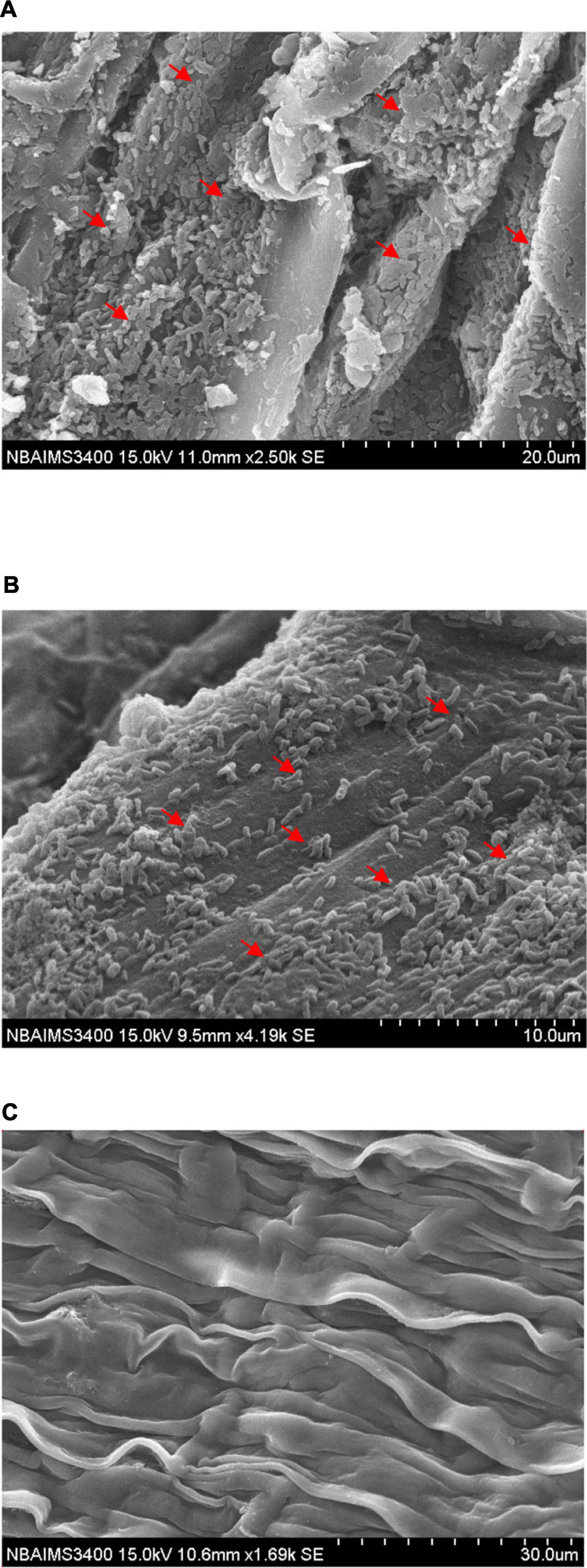
Figure 2. Scanning electron microphotographs showing root colonization by S. maltophilia DRC-18-7A (A), S. pavanii DRC-18-7B (B), and uninoculated control (C).
Inoculation with S. maltophilia DRC-18-7A and S. pavanii DRC-18-7B significantly enhanced the plant growth attributes in pigeonpea both in the presence of S6 and S0. In general, all the growth parameters recorded (root and shoot length, root and shoot fresh weight, root and shoot dry weight) were significantly higher in treatment inoculated with S. maltophilia and amended with SO42– compound (Table 1).
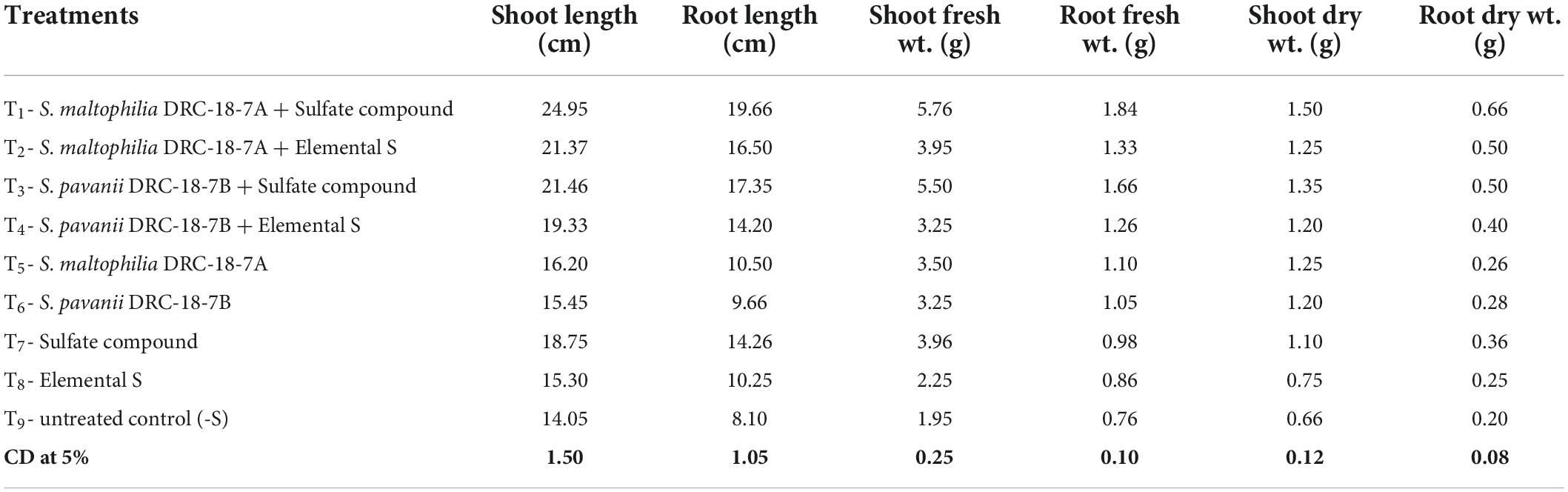
Table 1. Effects of inoculation on plant growth attributes in pigeonpea at 30 days of sowing under glasshouse conditions.
The inoculation of SOB along with sulfate compound significantly enhanced the root parameters as analyzed through root scanner as compared to all other treatments (Table 2). Among the two strains, inoculation of S. maltophilia DRC-18-7A significantly influenced root architecture in the presence of both sulfate and elemental sulfur. Treatments with only inoculation of SOB or only amendment of SO42– or S0 could not significantly influence the root parameters compared to absolute control (no inoculation and no S amendment) (Table 2).
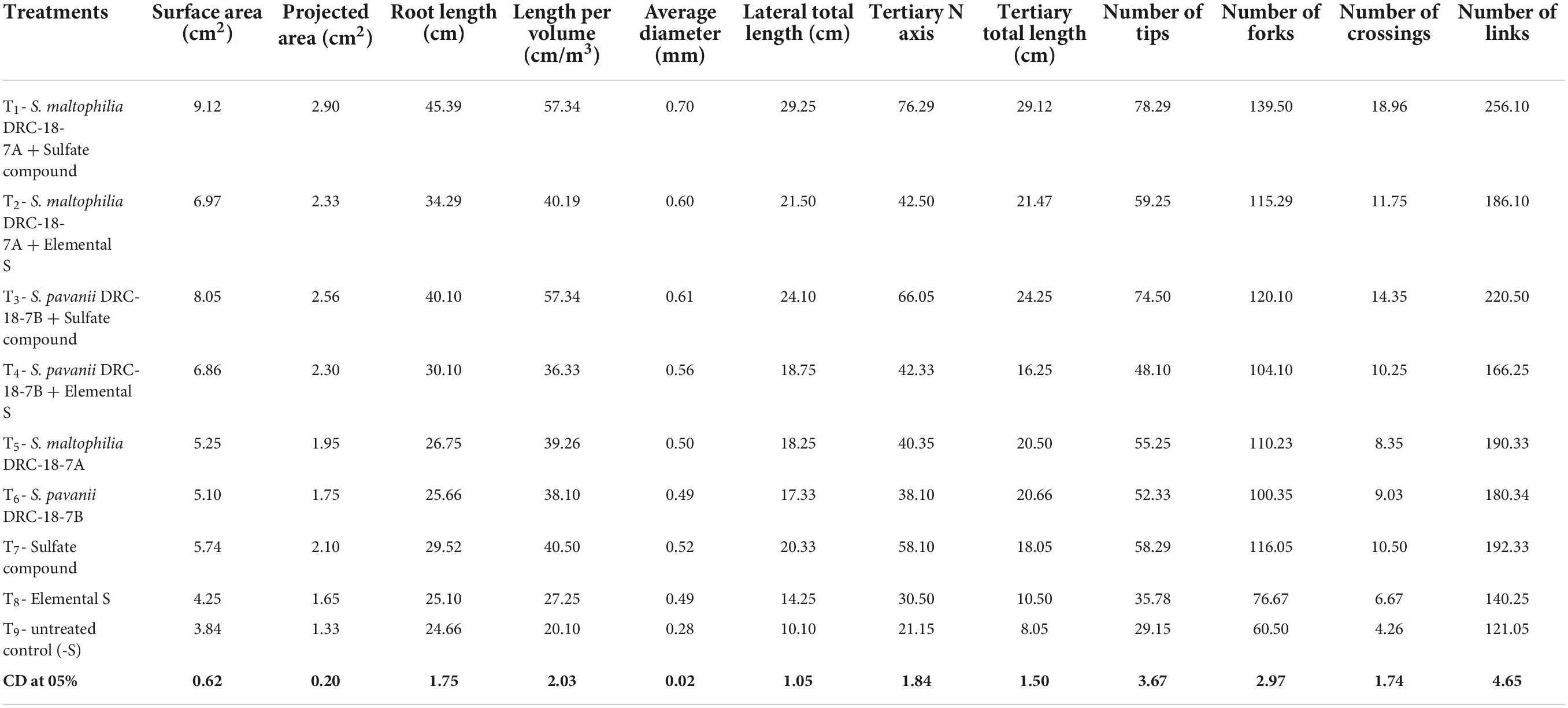
Table 2. Effects of inoculation on root architect and root development in pigeonpea leaves at 30 days of sowing under glasshouse conditions.
Among the 11 sox genes targeted in rhizosphere of pigeonpea, expression was achieved for 7 genes (soxB, tetH, sdoA, sdoB, TQO, sorAB, and tsdA), which showed 2-fold increase in treatments inoculated with S. maltophilia and amended with either S6 or S0. Similar tends were not observed in respective treatments inoculated with S. pavanii (Figure 3). The results revealed significantly higher transcript accumulation for genes sdoB, TQO, sorAB, and tsdA in the rhizosphere of plants inoculated with S. maltophilia (T-1). In general, expression and transcript accumulation of genes tsdA, soxB, and tetH were significantly lower across the treatments as compared to other genes.
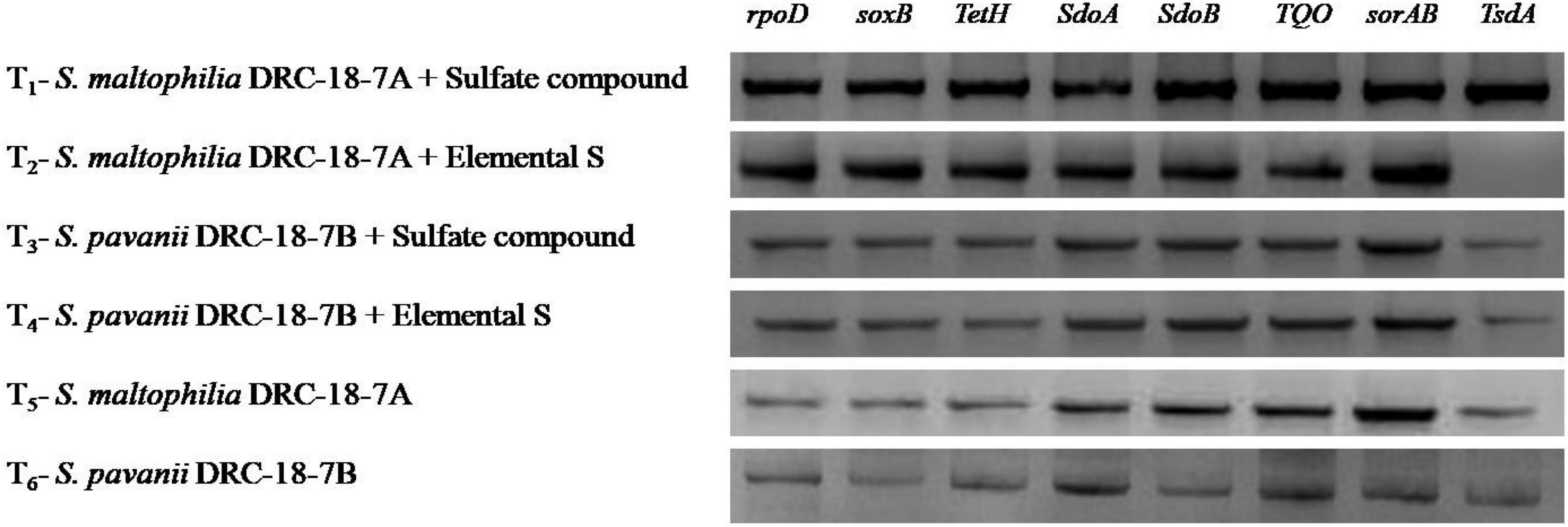
Figure 3. Effects of different treatments on expression of genes responsible for S-oxidation in the plant rhizosphere, treatments were as follows: T1-Stenotrophomonas maltophilia DRC-18-7A + Sulfate compound, T2-S. maltophilia DRC-18-7A + Elemental S, T3-S. pavanii DRC-18-7B + Sulfate compound, T4-S. pavanii DRC-18-7B + Elemental S, T5-S. maltophilia DRC-18-7A, T6-S. pavanii DRC-18-7B.
For identification of SULTR genes in pigeonpea, 10 AtSULTRs, 4 GmSULTRs, 9 PsSULTRs, 1 OsSULTRs, and 3 TdSULTRs were used as query sequences for BLASTn searches of the pigeonpea database (Cajanus cajan, taxid:3821) in NCBI with default parameters and redundant sequences were discarded manually. As a result, 11 SULTR genes, i.e., PpSULTR1.1, PpSULTR1.2, PpSULTR1.3, PpSULTR2.1, PpSULTR2.2, PpSULTR3.1, PpSULTR3.3, PpSULTR3.3-like, PpSULTR3.4, PpSULTR3.5, PpSULTR4.1, and PpSULTR4.2 were identified in the pigeonpea genome. These putative SULTR genes are located on different chromosomes. Their proteins contain STAS domain and the C-terminal region, which are critical for sulfate transporter activity and stability. To gain insights into the biological function of these genes and close relatives, a phylogenetic tree was constructed based on the full-length amino acid sequence alignment of SULTRs including 26 putative pigeonpea SULTR sequences, 10 AtSULTRs, 4 GmSULTRs, 9 PsSULTRs, 1 OsSULTRs, and 3 TdSULTRs (Figure 4). Based on phylogeny, the PpSULTRs are closely related to soybean SULTRs (GmSULTRs) and classified into four groups based on phylogenetic analyses. These pigeonpea SULTR genes were named corresponding to the homologous genes from other species.
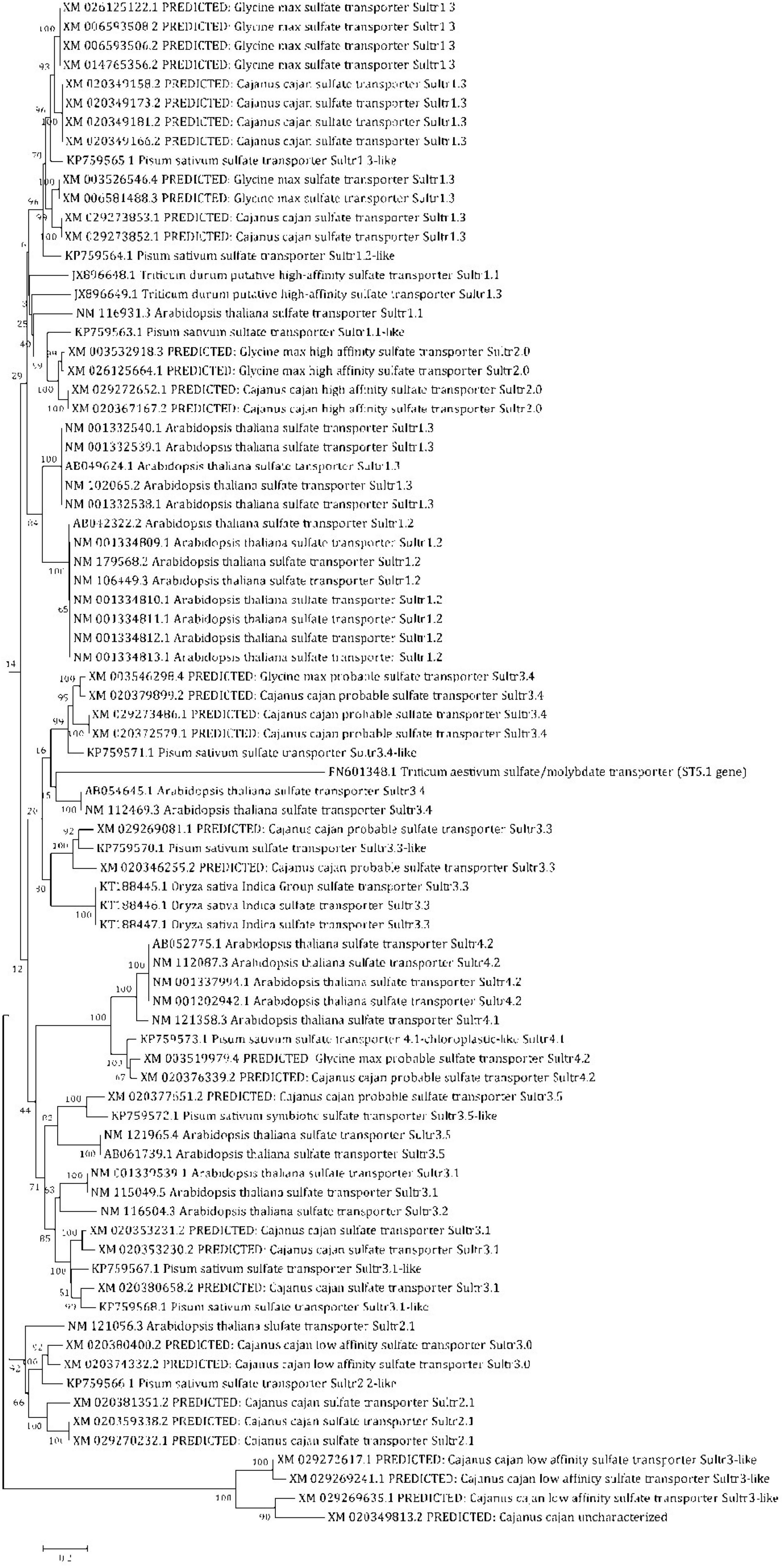
Figure 4. Phylogenetic tree showing the relationships of SULTR domains of pigeonpea, with other crop plants, Arabidopsis, soybean, field pea, rice, and wheat and classified into different groups.
To validate the reliability of the expression profile, in silico PCR amplification as well as validation was done using genomic sequences of pigeonpea as query sequence. Based on the in silico amplification, a set of primers were selected for real-time gene expression analysis in the pigeonpea grown under different treatments.
Transcript profiling of the PpSULTR genes was done in the pigeonpea plants inoculated with S. maltophilia DRC-18-7A and S. pavanii DRC-18-7B. It was found that sulfur sources and microbial inoculation significantly influenced the expression profile of PpSULTR genes in pigeonpea which was also evident from sulfur content in pigeonpea root and shoot. Furthermore, the expression profiles of PpSULTRs also varied in root and shoot of the same plant. Significantly higher expression (upregulation) of all the 11 PpSULTR genes was recorded in the roots and shoots of pigeonpea inoculated with S. maltophilia DRC-18-7A and amended with elemental sulfur (Figure 5A). Likewise, the expression of these genes in the roots and shoots of plants from treatment inoculated with S. pavanii and amended with S0 was higher and the fold increase closely followed treatment with S. maltophilia + S0. In general, the expression levels of PpSULTR genes in the roots were significantly higher (3–5-folds) as compared to the shoots. Interestingly, it was found that the expression level (fold change) of PpSULTRs was slightly higher in the negative control (-S) as compared to positive control (+S) (Figure 5B).
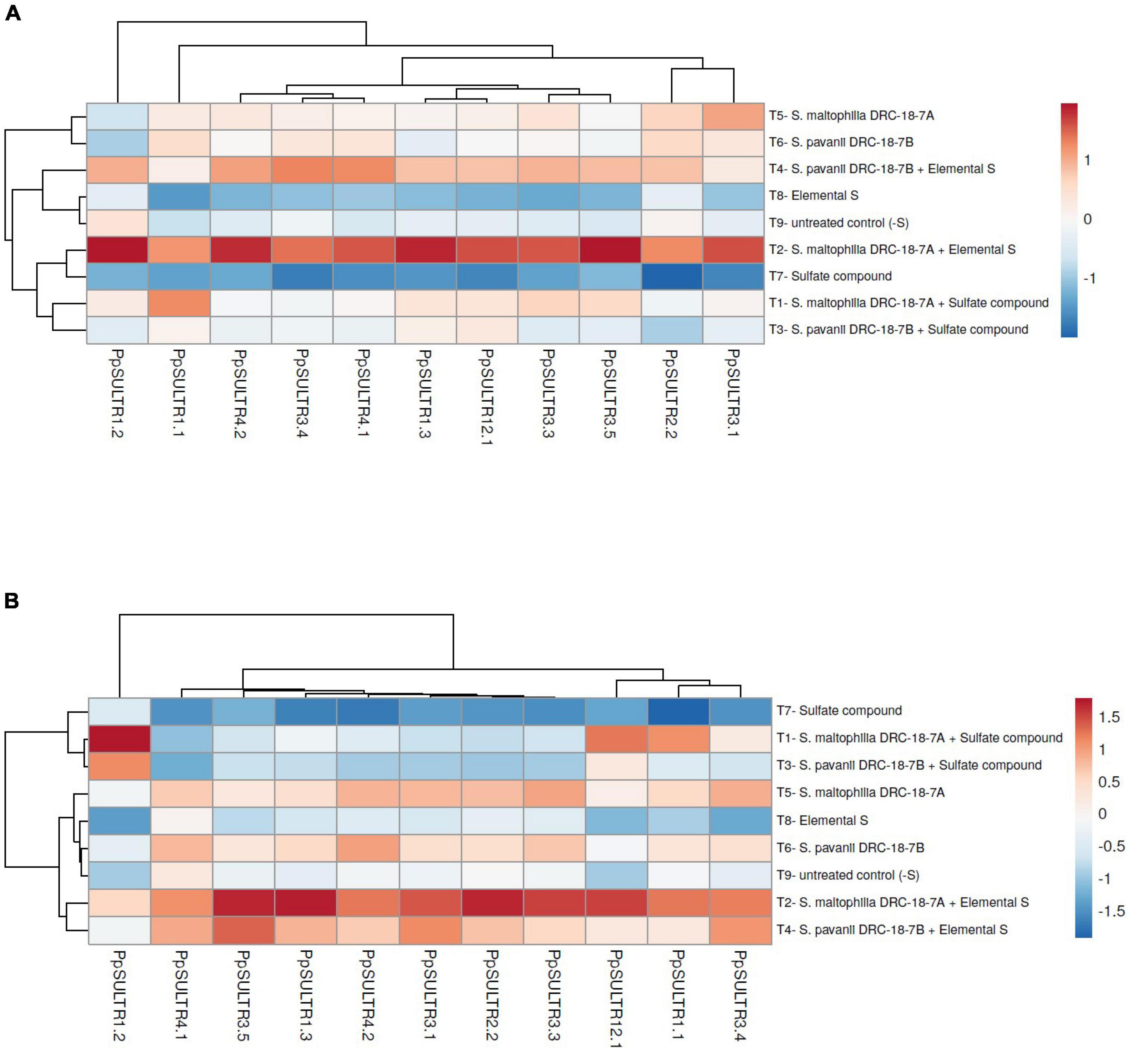
Figure 5. Heatmap showing the effects of microbial inoculation on expression of SULTR genes in pigeonpea (A) root and (B) shoot at 30 days of sowing T1-Stenotrophomonas maltophilia DRC-18-7A + Sulfate compound, T2-S. maltophilia DRC-18-7A + Elemental S, T3-S. pavanii DRC-18-7B + Sulfate compound, T4-S. pavanii DRC-18-7B + Elemental S, T5-S. maltophilia DRC-18-7A, T6-S. pavanii DRC-18-7B, T7-Sulfate compound, T8-Elemental S, and T9-untreated control (-S).
The inoculation of the selected strains, S. maltophilia DRC-18-7A and S. pavanii DRC-18-7B, modulated the physio-biochemical pathways and accumulation of antioxidants in the pigeonpea plants. The quantitative analysis revealed that the accumulation of total chlorophyll, carotenoids, soluble sugars, and protein content was significantly enhanced in the treatment inoculated with S. maltophilia and supplemented with sulfate compound (Figure 6). Inoculation of SOB alone could not influence the accumulation and was significantly lower than treatment amended with S6 compound. In contrast, the accumulation of proline, flavanoids, total phenolics, and activities of antioxidant enzymes (PAL, POx, APx, catalase, and SOD) were significantly enhanced in the treatment inoculated with either of the SOB and amended with elemental sulfur. The presence of S6 in the treatments with or without inoculation led to significantly lower accumulation of proline and flavonoids (Figure 7).
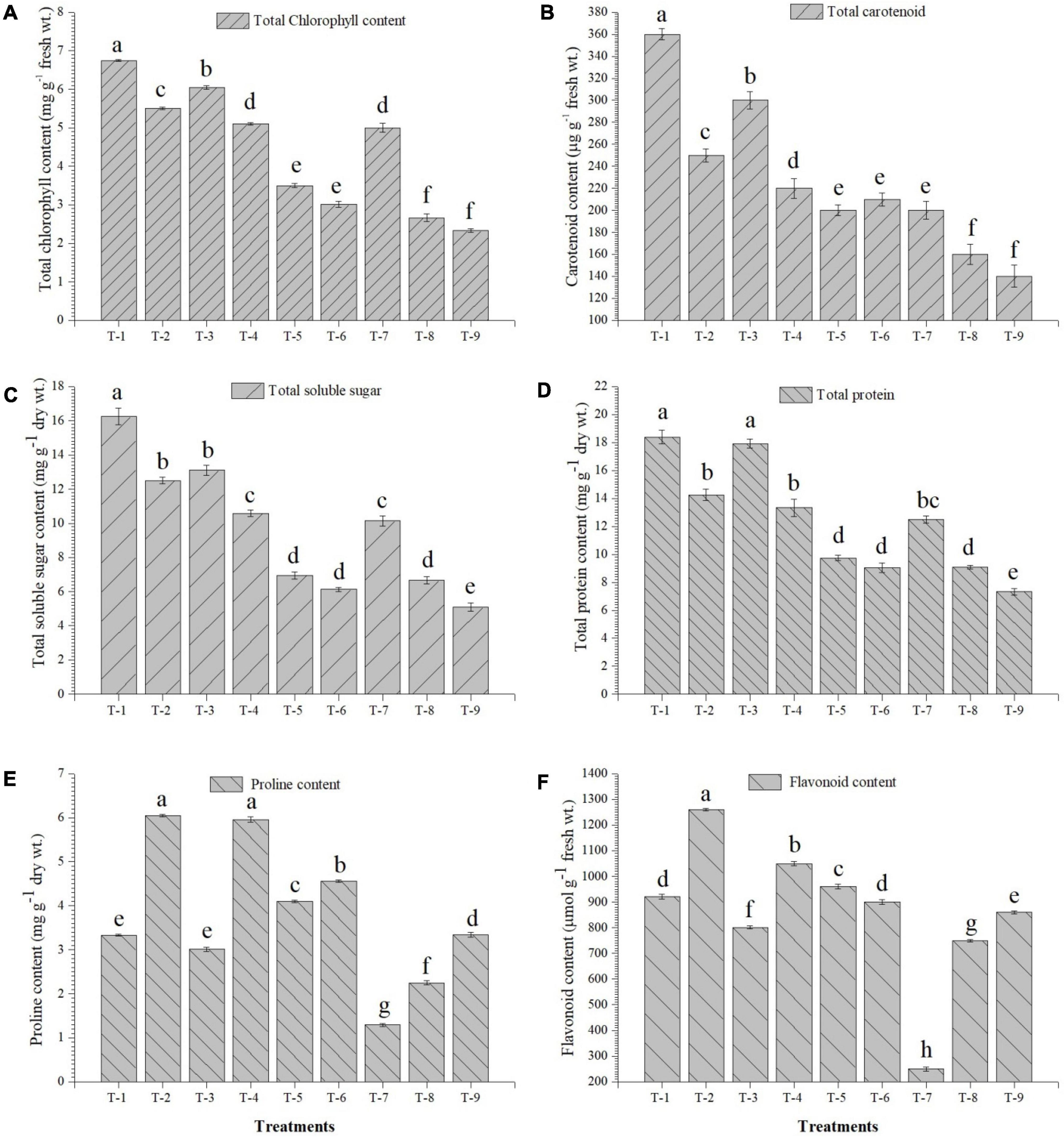
Figure 6. Effects of seed treatment on accumulation of (A) total chlorophyll, (B) total carotenoids, (C) total soluble sugar, (D) total protein (E) proline, and (F) flavonoids in the pigeonpea leaves at 30 days of sowing Treatments were as follows: T1-Stenotrophomonas maltophilia DRC-18-7A + Sulfate compound, T2-S. maltophilia DRC-18-7A + Elemental S, T3-S. pavanii DRC-18-7B + Sulfate compound, T4-S. pavanii DRC-18-7B + Elemental S, T5-S. maltophilia DRC-18-7A, T6-S. pavanii DRC-18-7B, T7-Sulfate compound, T8-Elemental S and T9-untreated control (-S). Data are mean (n = 10) and vertical bar represents standard deviation. Data with different letters show significant difference in column data in randomized block design test at p < 0.05 under Duncan’s multiple range test.
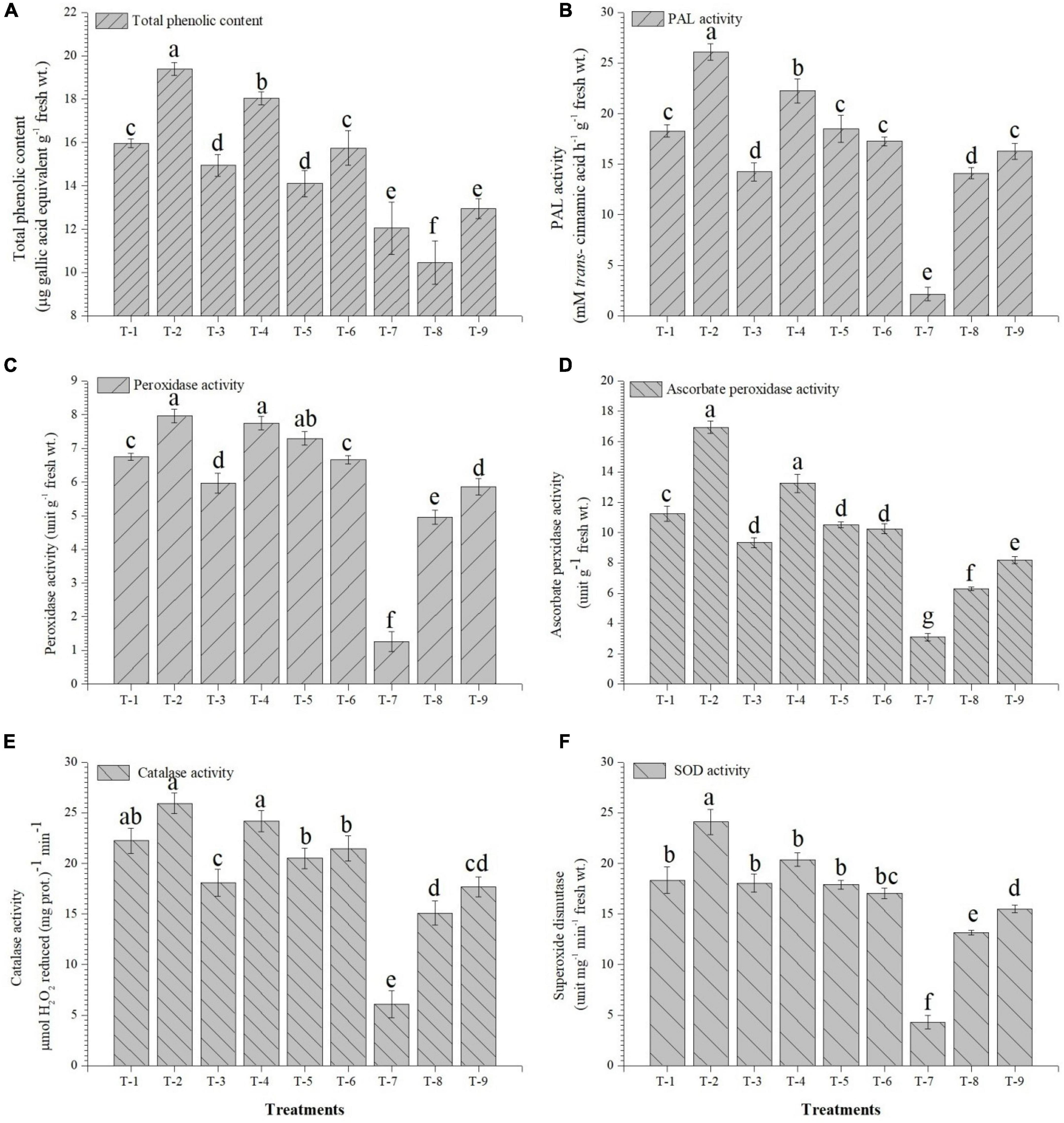
Figure 7. Effects of seed treatment on activities and accumulation of antioxidant biomolecules and enzymes (A) total phenolic, (B) PAL, (C) peroxidase, (D) ascorbate peroxidase (E) catalase, and (F) superoxide dismutase in the pigeonpea leaves at 30 days of sowing treatments were: T1-Stenotrophomonas maltophilia DRC-18-7A + Sulfate compound, T2-S. maltophilia DRC-18-7A + Elemental S, T3-S. pavanii DRC-18-7B + Sulfate compound, T4-S. pavanii DRC-18-7B + Elemental S, T5-S. maltophilia DRC-18-7A, T6-S. pavanii DRC-18-7B, T7-Sulfate compound, T8-Elemental S and T9-untreated control (-S). Data are mean (n = 10) and vertical bar represents standard deviation. Data with different letters show significant difference in column data in randomized block design test at p < 0.05 under Duncan’s multiple range test.
Attenuation of superoxide levels was observed as a blue formazan, which is the outcome of NBT dye and superoxide interactions (Figure 8A). Stereoscopic visualization clearly showed dense localization of superoxide radicals in the petioles near veins and midrib of leaves of the untreated control plant (negative control) followed by plants grown with elemental S. Least accumulation of superoxide radical was observed in the plants inoculated with either of strains and supplemented with sulfate compounds compared to all other treatments (Figure 8A). Similarly, program cell death was observed as greenish polymerization product of Evans Blue stain. The bacterial inoculation and supplementation of sulfate compound in the nutrient solution significantly reduced greenish discoloration compared to other treatments. Similar to superoxide radicals, maximum program cell death was observed in the untreated control plants (Figure 8B).
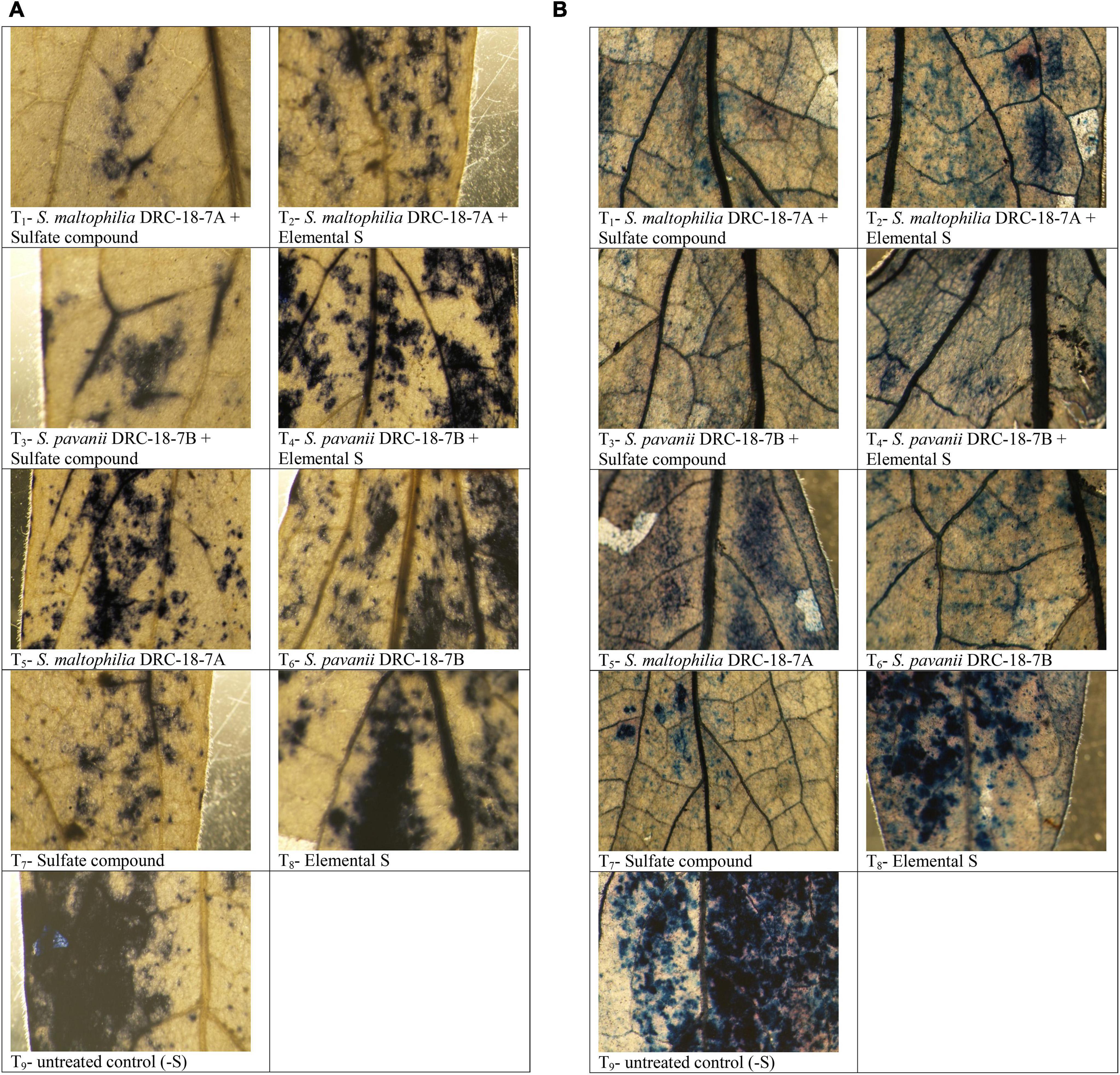
Figure 8. Microscopic detection of superoxide radical by NBT staining (A) and program cell death (B) in leaves of pigeonpea after treatment with T1-Stenotrophomonas maltophilia DRC-18-7A + Sulfate compound, T2-S maltophilia DRC-18-7A + Elemental S, T3-S. pavanii DRC-18-7B + Sulfate compound, T4-S. pavanii DRC-18-7B + Elemental S, T5-S. maltophilia DRC-18-7A, T6-S. pavanii DRC-18-7B, T7-Sulfate compound, T8-Elemental S and T9-untreated control (-S) at 30 days of sowing.
The up-/downregulation of nine key genes (phenylalanine ammonia-lyase [EC:4.3.1.24], phenylalanine/tyrosine ammonia-lyase [EC:4.3.1.25], 4-coumarate-CoA ligase [EC:6.2.1.12], cinnamoyl-CoA reductase [EC:1.2.1.44], cinnamyl-alcohol dehydrogenase [EC:1.1.1.195], peroxiredoxin 6 [EC:1.11.1.7], ferulate-5-hydroxylase [EC:1.14.-.-], caffeoyl-CoA O-methyltransferase [EC:2.1.1.104], and coniferyl-aldehyde dehydrogenase [EC:1.2.1.68]) involved in phenylpropanoid pathway was investigated. The results revealed that these genes were upregulated in treatment inoculated with S. maltophilia DRC-18-7A and supplemented with elemental S in the leaves of pigeonpea. The highest expression of 4-coumarate-CoA ligase [EC: 6.2.1.12] was recorded in the leaves of pigeonpea plants across the treatments taken into consideration followed by phenylalanine ammonia-lyase [EC:4.3.1.24] and phenylalanine/tyrosine ammonia-lyase [EC:4.3.1.25]. However, in the inoculated plants, expression level was significantly higher in comparison with untreated positive and negative control plants (Figure 8). In contrast, comparatively less expression was recorded in the plants harvested from treatments amended with S6 compounds (Figure 9) as compared to plants grown with elemental S. It revealed that plants grown in the presence of S0 experienced stress and tend to over-express antioxidant genes.
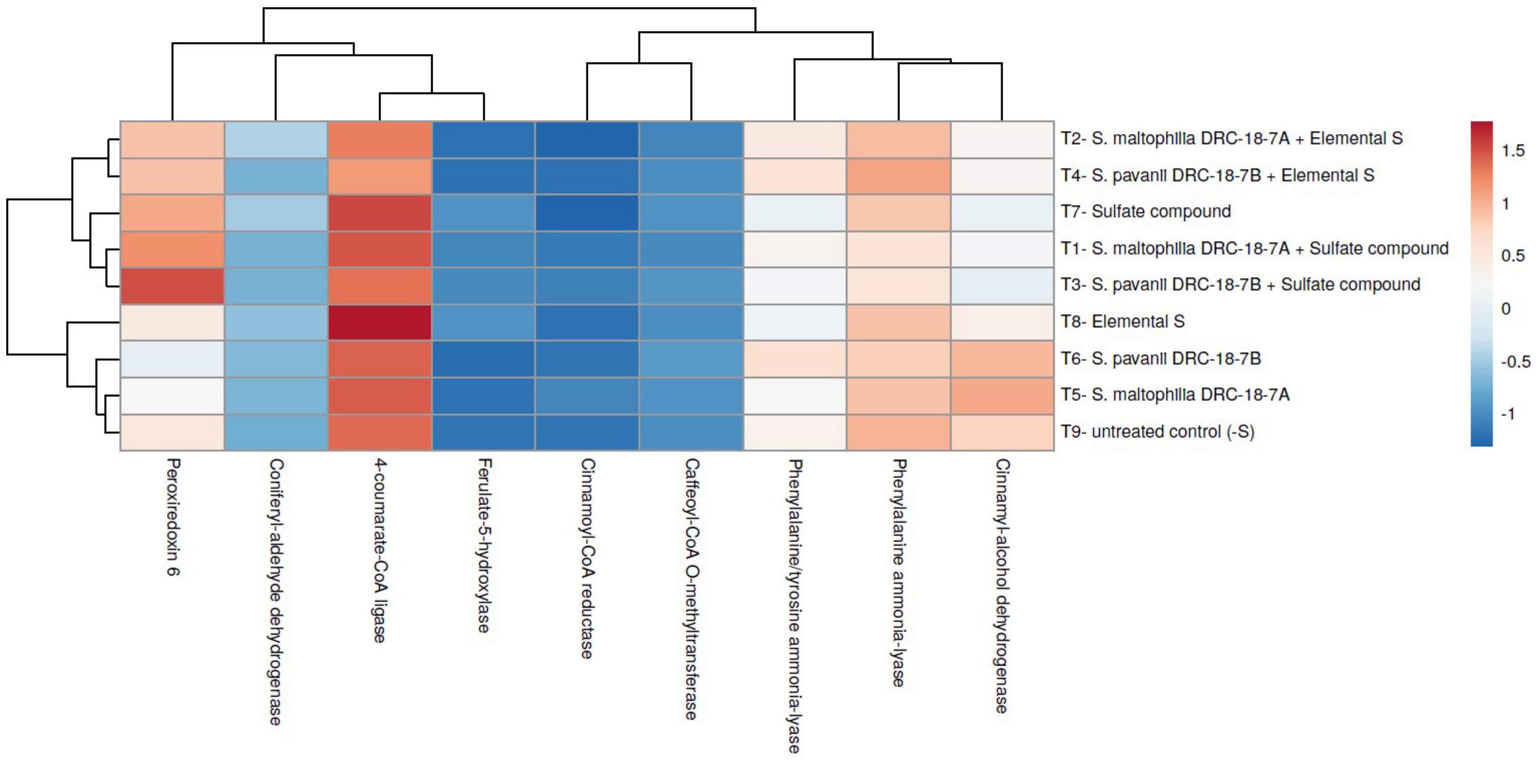
Figure 9. Heatmap showing the effects of microbial inoculation on expression profile of key genes of phenylpropanoid pathways in leaves of pigeonpea at 30 days of sowing T1- Stenotrophomonas maltophilia DRC-18-7A + Sulfate compound, T2- S. maltophilia DRC-18-7A + Elemental S, T3- S. pavanii DRC-18-7B + Sulfate compound, T4- S. pavanii DRC-18-7B + Elemental S, T5- S. maltophilia DRC-18-7A, T6- S. pavanii DRC-18-7B, T7- Sulfate compound, T8- Elemental S and T9- untreated control (-S).
The accumulation of phenolics (gallic, ferrulic, sinapic, and syringic acids) and flavonoids (rutin and quercetin) was differentially influenced by inoculation of SOB and supplementation of two different sources of sulfur (S6– or S0). In treatments inoculated with either S. maltophilia or S. pavanii and amended with S0, the levels of all analyzed phenolics and flavonoids were significantly higher than all other treatments. Addition of S6 compound to SOB inoculated treatments did not significantly influence the synthesis of phenolics acids and flavonoids (Table 3).
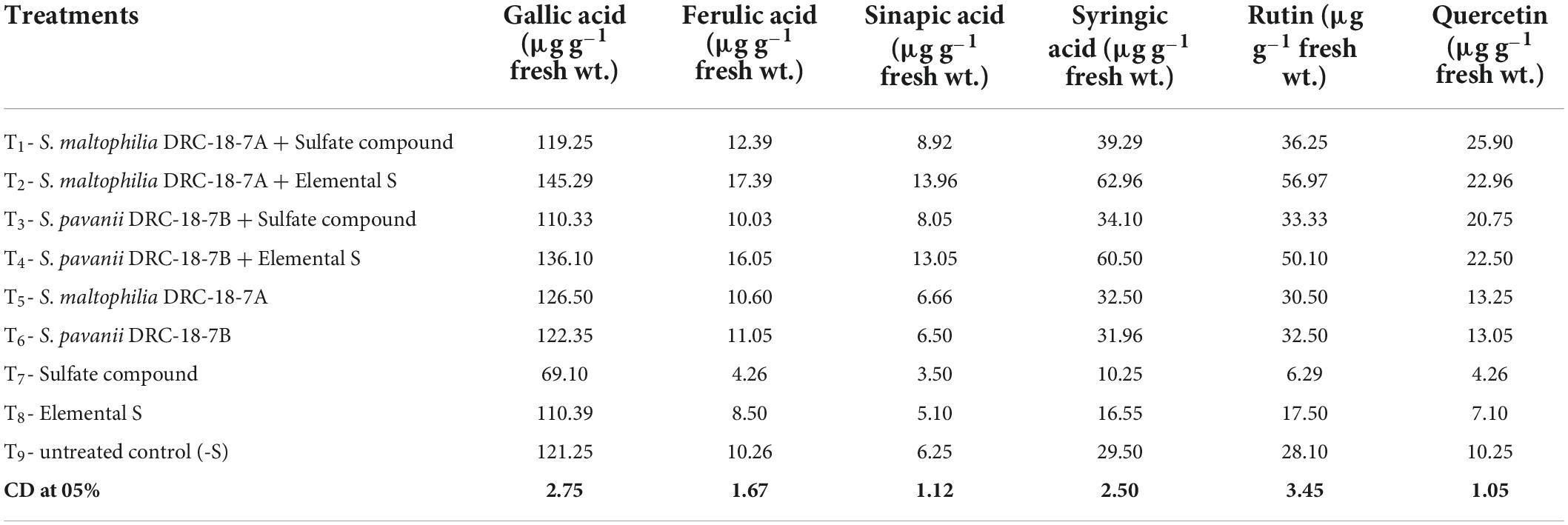
Table 3. Effects of inoculation on individual phenolic and flavonoids content in pigeonpea leaves at 30 days of sowing under glasshouse conditions.
The uptake of sulfur in the roots and shoots was significantly influenced by inoculation of SOB and supplementation of sulfur in different forms (Table 4). In general, for all the treatments, the S content was higher in roots as compared to shoots. Among the treatments, maximum S content was recorded due to the inoculation of S. maltophilia and supplementation of S6 compound. The S-content was 48 and 42% higher in roots and shoots, respectively, as compared to treatment where only S6 compound was supplemented. Similar trend was observed in treatment inoculated with S. pavanii. The S-content in roots and shoots of plants from treatments inoculated with SOB and supplemented with elemental S was significantly lower as compared to other treatments (Table 4).
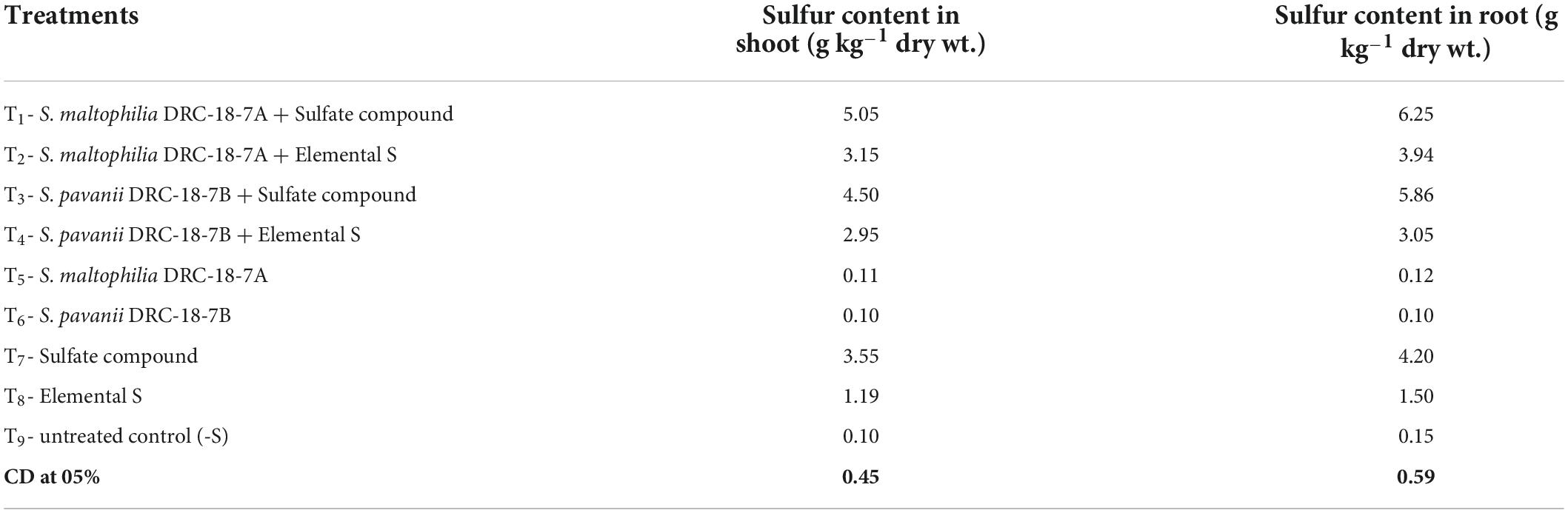
Table 4. Effects of inoculation on sulfur content in pigeonpea at 30 days of sowing under glasshouse conditions.
Besides the importance of three major nutrients, i.e., nitrogen, phosphorus, and potassium, the focus of research work has been shifted to investigate the key role of other macro- and micro-nutrients in major crop plants including pulses. The intensive agriculture has led to the deficiency of these nutrients such as S, Bo, Zn, and Fe. In the last two decades, the losses in crop yield due to the deficiency of these nutrients are now reported often from different parts of the world. Sulfur nutrition is important as it influences different metabolic pathways as a structural component of many secondary metabolites, vitamins, amino acids, and enzymes. Among the crop plants, legumes are strikingly affected by deficiency of S in soil (Chandler et al., 1984; Scherer, 2001). Besides influencing the plant growth, the process of nitrogen fixation and nodulation is hampered due to sulfur deficiency in soil (Watkinson, 1989; Scherer, 2001; Stamford et al., 2002; Cheng et al., 2017). Inoculation of SOB has been reported to enhance the growth and yield of different crop plants such as groundnut by 11% (Anandham et al., 2007), mustard by 6.6% (Chaudhary et al., 2017), onion by 45-50% (Awad et al., 2011), and mustard by 14.50–30.60% (Abhijit et al., 2014). We earlier reported the isolation of SOB from mud, coal, and drainage waters collected from open cost coal mines in India. Strains of Stenotrophomonas maltophilia and S. pavanii were identified to be most efficient for promotion of plant growth and sulfur nutrition in pigeonpea (Malviya et al., 2022). The detailed study was required to study the mechanisms by which S is transported from soil to roots and to shoots need to be deciphered.
Root colonization is an important attribute for any of the inoculant strains and provides clue for a commensal association between the two partners mediated through root exudates (Bais et al., 2006). S. maltophilia and S. pavanii were found to be good colonizers and formed biofilm on the root system. It has been reported that during the plant–microbe interaction, the expression of several genes of both plant and bacterial origin is modulated (Beauregard et al., 2013; de Souza et al., 2015). The bacterial genes associated with exo-polysaccharide production and biofilm formation are triggered by the root exudates during compatible interaction (Rudrappa et al., 2008; Meneses et al., 2011; Lopes et al., 2021). The formation of aggregates (micro-colonies) particularly by S. maltophilia on the roots indicates the copious production of EPS in the rhizosphere. It is worthwhile to mention that both S. maltophilia and S. pavanii form dry colonies on the growth medium. It has also been reported that the efficiency of bacteria in stimulating growth occurs in a density-dependent manner (Rudrappa et al., 2008). The stage at which the threshold level of microbial density is achieved, the biofilms work as a single unit to coordinate the release of molecules that helps in the promotion of plant growth through different mechanisms (McNear, 2013). A good colonization potential by both the SOB also gives an indication about rhizosphere competence as reported earlier (de Souza et al., 2015). This was further confirmed by the enhanced expression of sox genes in the treatments inoculated with SOB and S6 compound. Under sterile conditions, the enhanced expression of genes involved in S oxidation is directly related to population build-up of SOB. When S. maltophilia was inoculated along with S6 compound, the population build-up and root colonization were enhanced and the same was manifested in the higher expression of genes responsible for S-oxidation in the soil. Similar observations were made by Berben et al. (2019) and Zhang et al. (2019). The variations in the expression levels of sox genes in treatments where two different sources of S were amended (S6 or S0) irrespective of the inoculant strain indicate that the population build-up of SOB was higher in the presence of readily available source of sulfur (S6) as compared to elemental sulfur (S0).
The root system architecture (RSA) was also analyzed in both inoculated and uninoculated treatments. It is believed that the RSA is controlled by different biological and edaphic conditions (McNear, 2013). In this study, RSA was greatly influenced by inoculation of S. maltophilia and amendment of S6 compound. However, the same strain in presence of S0 could not influence the RSA to that extent. There are many reports regarding the modification of root architecture and anatomy in response to agriculturally important microorganisms so as to enhance the uptake of nutrients by the plants from the soil (Ortiz-Castro et al., 2008; Tian et al., 2014; Singh et al., 2017). It is well-known that larger root volume, root hair density, and increased number of lateral roots not only provide a better stand to the plant but also enhance the uptake and translocation of different nutrients from the soil to the plant (Singh et al., 2021).
Besides the root system architecture, the effect of inoculation of SOB and amendment of two different sources of sulfur (S6 and S0) was also studied at the enzymatic, non-enzymatic, and gene expression levels. The presence of unavailable form of sulfur (S0) is perceived by the plant as nutritional stress. In turn, the plant responds by regulating the antioxidative reaction and accumulation of polyphenolics in plant (Singh S. et al., 2020). Stress conditions, in general, accelerate the production of reactive oxygen species (ROS) in the plant system (Meng et al., 2016). To overcome the burst of ROS, the plants have developed both non-enzymatic (organic osmolyte like glycine betaine, proline, glutathione, etc.) and enzymatic (catalase, superoxide dismutase, ascorbate peroxidase, glutathione reductase, etc.) components (Nawaz and Wang, 2020; Singh D. P. et al., 2020). In this study, there was a significant increase in the accumulation of proline, flavonoids, total phenolics, and activities of antioxidant enzymes in the treatment amended with elemental sulfur and inoculated with SOB. The inoculation of SOB induced the synthesis of both enzymatic and non-enzymatic component which in turn provided protection to the plant from ROS. Similar results have been reported in different studies related to alleviation of abiotic stress by microbial inoculation (Singh et al., 2015, 2021). It is worthwhile to mention that microbial inoculants need to be developed that provide protection in the presence of elemental sulfur. Moreover, in different studies, use of S0 is recommended over that of sulfate, since it not only improves plant growth and nutrition but also increases systemic tolerance to different abiotic stresses (Degryse et al., 2016; Fuentes-Lara et al., 2019).
Sulfate transporters (SULTRs) are the key gene family responsible for the S-uptake and translocation in the higher plants. These are encoded by a large gene family, comprising of 12 genes in Arabidopsis thaliana, 10 in wheat (Triticum spp.), 12 in rice (Oryza sativa), 16 in Populus (Populus stremula × P. alba), and 28 in soybean (Glycine max). However, the literature is silent about the pigeonpea SULTRs and their role in S-nutrition. In-depth research is a pre-requisite to establish the relative contribution of the pigeonpea sulfate transporter genes to overall sulfate transport in plants. It is also necessary to explore whether all SULTRs are involved in sulfate acquisition, translocation, and remobilization of sulfur in the plant system. In this study, we performed a comprehensive investigation of the pigeonpea SULTRs gene family using comparative genomic and phylogenetic analyses. For this 10 AtSULTRs, 4 GmSULTRs, 9 PsSULTRs, 1 OsSULTRs, and 3 TdSULTRs were used as query sequences for BLASTn searches of the pigeonpea database (Cajanus cajan, taxid:3821) in NCBI with default parameters, and redundant sequences were discarded manually. Furthermore, qPCR analysis was done in the presence and absence of S-oxidizing bacteria in pigeonpea. This is the first report on the microbe-mediated induction of PpSULTR genes in pigeonpea and their role in S-uptake and translocation. A 7.56- to 27.33-fold changes in the expression of PpSULTRs were recorded at early crop growth stage (30 days after sowing), which is further confirmed by the enhanced sulfur content in the roots and shoots of pigeonpea. The expression of SULTRs in the plants supplemented with elemental S was significantly higher as compared to plants supplemented with S6 compounds at 30 days after sowing. Interestingly, the expression of SULTRs in the plants was significantly increased in the presence of potential SOB in the rhizosphere, suggesting their versatility in controlling SULTRs transcription.
This study provides the key evidence on molecular mechanism underlying microbial-induced expression of SULTRs in pigeonpea roots and shoots in the presence of the two possible enhancers, S. maltophilia DRC-18-7A and S. pavanii DRC-18-7B at an early stage of crop growth. Figure 10 depicts the possible interactions contributing to S-uptake in the plants. It is suggested that S. maltophilia DRC-18-7A and S. pavanii DRC-18-7B-dependent transcriptional induction and post-transcriptional regulation allow fine-tuning of the SULTRs transcript levels in roots and shoots of pigeonpea.
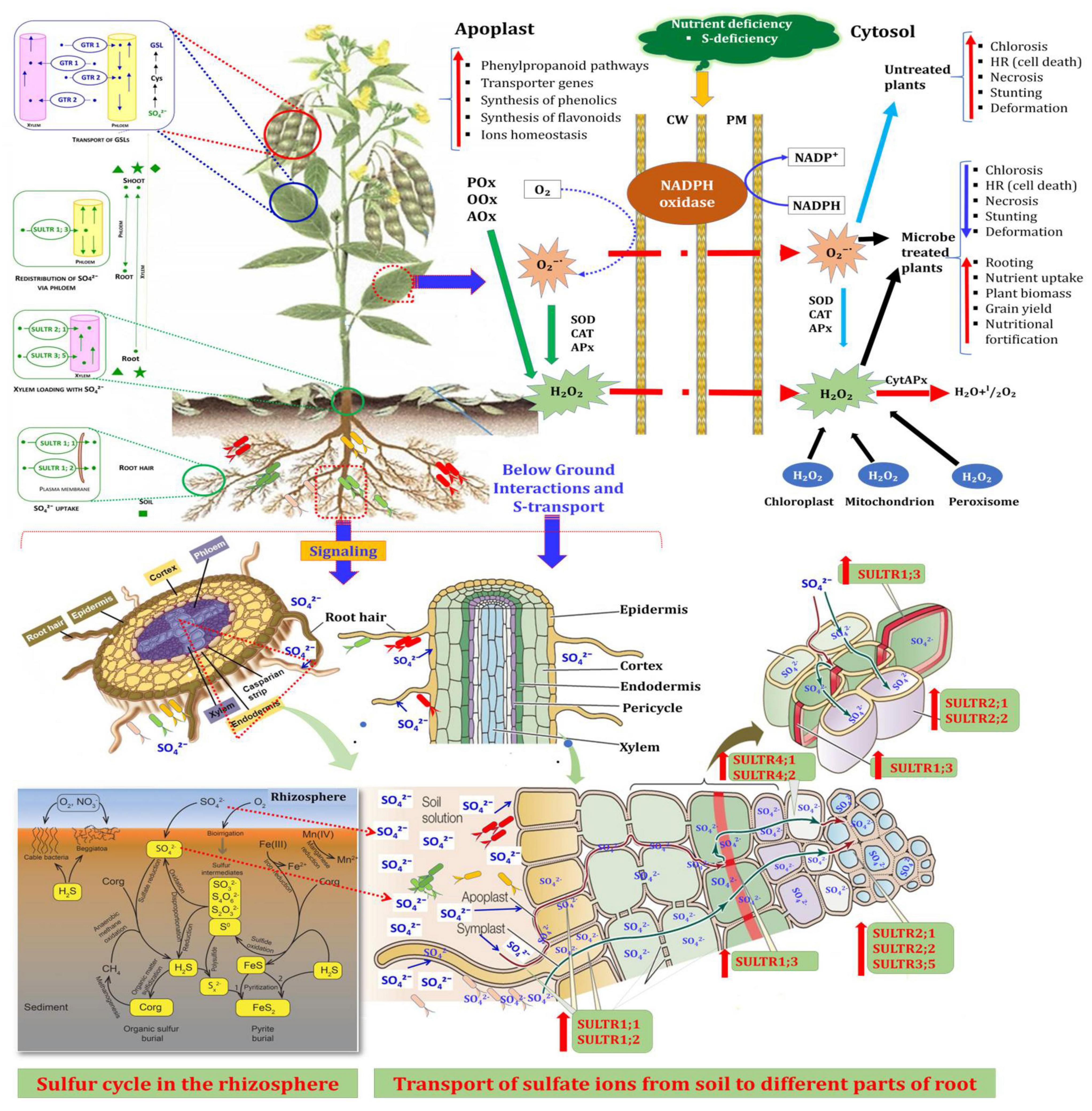
Figure 10. A comprehensive overview of plant-microbe interactions contributing to S-uptake in plants The oxidation of sulfur to sulfate by SOB in rhizosphere, its uptake by roots and its transport to shoots through involvement of S-transporter genes. The interaction results changes in the activity of radical scavenging enzymes and led to increase in growth and yield of pigeonpea.
The microorganisms and plant take up sulfur in the form of sulfate (S6). The elemental sulfur (S0) applied/present in the soil undergoes change in the oxidation state from S0 to S6 due to the action of specific group of bacteria collectively termed as sulfur-oxidizing bacteria. Inoculation of two potential SOB (S. maltophilia and S. pavanii) to pigeonpea led to the modifications in the root architecture that supports efficient uptake of nutrients. The enhanced activity of sulfur oxidation genes in inoculated treatments and PpSULTR genes in plants contributed to the enhanced uptake of sulfur in roots and shoots of pigeonpea. The increase in non-enzymatic and enzymatic components to counter ROS due to inoculation also contributed to the enhanced growth of pigeonpea. SOB with additional plant growth-promoting attributes could be recommended as potential inoculants for pigeonpea for commercial production after extensive field evaluation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
DM, AS, US, and AV conceived and designed the experiments. DM, SS, and US performed the experiments. DM, AS, and US analyzed the data. DM and US wrote the manuscript. AS and AV have edited and given final touch to the manuscript. All authors have reviewed the manuscript and have given approval to the final version.
This research was supported by the Indian Council of Agricultural Research, New Delhi (India).
We sincerely thank Renu, ICAR-NBAIM, for technical assistance for HPLC analyses. We sincerely acknowledge Hillol Chakdar and Prasanna Choudhary ICAR-NBAIM for providing instrumentation support to carry out the root scanning. We gratefully acknowledge Pramod K. Sahu and Manish Roy, ICAR-NBAIM, for providing technical support to carry out confocal and scanning electron microscopy, respectively. We gratefully acknowledge the Indian Council of Agricultural Research, Ministry of Agriculture and Farmers Welfare, Government of India, for providing financial support for the study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MK declared a shared parent affiliation with the author AV to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.927702/full#supplementary-material
Abhijit, D., Chandra, K. S., and Joseph, M. (2014). Evaluation of the efficacy of different sulphur amendments and sulphur oxidizing bacteria in relation to its transformation in soil and yield of mustard (Brassica juncea). Res. Crops 15, 578–584. doi: 10.5958/2348-7542.2014.01380.1
Anandham, R., Indira Gandhi, P., Kwon, S. W., Sa, T. M., Kim, Y. K., and Jee, H. J. (2009). Mixotrophic metabolism in Burkholderia kururiensis subsp. thiooxydans subsp. nov., a facultative chemolithoautotrophic thiosulfate oxidizing bacterium isolated from rhizosphere soil and proposal for classification of the type strain of Burkholderia kururiensis as Burkholderia kururiensis subsp. kururiensis subsp. nov. Arch. Microbiol. 191, 885–894. doi: 10.1007/s00203-009-0517-4
Anandham, R., Indiragandhi, P., Kwon, S. W., Sa, T. M., Jeon, C. O., Kim, Y. K., et al. (2010). Pandoraea thiooxydans sp. nov., a facultatively chemolithotrophic, thiosulfate-oxidizing bacterium isolated from rhizosphere soils of sesame (Sesamum indicum L.). Int. J. Syst. Evol. Microbiol. 60, 21–26. doi: 10.1099/ijs.0.012823-0
Anandham, R., Kwon, S. W., Gandhi, P. I., Kim, S. J., Weon, H. Y., Kim, Y. S., et al. (2011). Dyella thiooxydans sp. nov., a facultatively chemolithotrophic, thiosulfate-oxidizing bacterium isolated from rhizosphere soil of sunflower (Helianthus annuus L.). Int. J. Syst. Evol. Microbiol. 61, 392–398. doi: 10.1099/ijs.0.022012-0
Anandham, R., Sridar, R., Nalayini, P., Poonguzhali, S., and Madhaiyan, M. (2007). Potential for plant growth promotion in groundnut (Arachis hypogaea L.) cv. ALR-2 by co-inoculation of sulfur-oxidizing bacteria and Rhizobium. Microbiol. Res. 162, 139–153. doi: 10.1016/j.micres.2006.02.005
Awad, N. M., El-Kader, A., Attia, M. K. A. A., and Alva, A. K. (2011). Effects of nitrogen fertilization and soil inoculation of sulfur-oxidizing or nitrogen-fixing bacteria on onion plant growth and yield. Int. J. Agron. 2011:316856. doi: 10.1155/2011/316856
Bacelar-Nicolau, P., and Johnson, D. B. (1999). Leaching of pyrite by acidophilic heterotrophic iron-oxidizing bacteria in pure and mixed cultures. Appl. Environ. Microbiol. 65, 585–590. doi: 10.1128/AEM.65.2.585-590.1999
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Ann. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Baker, J., and Mock, C. N. M. (1994). An improved method for monitoring cell death in cell suspension and leaf disc assays using evans blue. Plant Cell Tissue Organ. Cult. 39, 7–12. doi: 10.1007/BF00037585
Beauregard, P. B., Chai, Y., Vlamakis, H., Losick, R., and Kolter, R. (2013). Bacillus subtilis biofilm induction by plant polysaccharides. PNAS 110, E1621–E1630. doi: 10.1073/pnas.1218984110
Berben, T., Overmars, L., Sorokin, D. Y., and Muyzer, G. (2019). Diversity and distribution of sulfur oxidation-related genes in Thioalkalivibrio, a genus of chemolithoautotrophic and haloalkaliphilic sulfur-oxidizing bacteria. Front. Microbiol. 10:160. doi: 10.3389/fmicb.2019.00160
Bouranis, D. L., Chorianopoulou, S. N., Margetis, M., Saridis, G. I., and Sigalas, P. P. (2018). Effect of elemental sulfur as fertilizer ingredient on the mobilization of iron from the iron pools of a calcareous soil cultivated with durum wheat and the crop’s iron and sulfur nutrition. Agriculture 8:20. doi: 10.3390/agriculture8020020
Bressani, R., Gómez-Brenes, R. A., and Elías, L. G. (1986). Calidad nutricional de la proteína del gandul, tierno y maduro, y su valor suplementario a los cereales [Nutritional quality of pigeonpea protein, immature/e and ripe, and its supplementary value for cereals]. Arch. Latinoam. Nutr. 36, 108–116.
Chandler, P. M., Spencer, D., Randall, P. J., and Higgins, T. J. (1984). Influence of sulfur nutrition on developmental patterns of some major pea seed proteins and their mRNAs. Plant Physiol. 75, 651–657. doi: 10.1104/pp.75.3.651
Chaudhary, S., Dhanker, R., and Tanvi Goyal, S. (2017). Characterization and optimization of culture condition for sulphur oxidizing bacteria after isolation from thizospheric mustard soil, decomposing sites and pit house. Int. J. Biol. Biomol. Agril. Food Biotechnol. Eng. 11, 379–383.
Chaudhary, T., Gera, R., and Shukla, P. (2021). Deciphering the potential of rhizobium pusense MB-17a, a plant growth-promoting root endophyte, and functional annotation of the genes involved in the metabolic pathway. Front. Bioeng. Biotechnol. 8:617034. doi: 10.3389/fbioe.2020.617034
Chen, X. G., Geng, A. L., Yan, R., Gould, W. D., Ng, Y. L., and Liang, D. T. (2004). Isolation and characterization of sulfur-oxidizing Thiomonas sp. and its potential application in biological deodorization. Lett. Appl. Microbiol. 39, 495–503. doi: 10.1111/j.1472-765X.2004.01615.x
Cheng, G., Karunakaran, R., East, A. K., Munoz-Azcarate, O., and Poole, P. S. (2017). Glutathione affects the transport activity of Rhizobium leguminosarum 3841 and is essential for efficient nodulation. FEMS Microbiol. Lett. 364:fnx045. doi: 10.1093/femsle/fnx045
de Souza, R., Ambrosini, A., and Passaglia, L. M. P. (2015). Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 38, 401–419. doi: 10.1590/S1415-475738420150053
Degryse, F., Ajiboye, B., Baird, R., Da Silva, R. C., and McLaughlin, M. J. (2016). Availability of fertilizer sulfate and elemental sulfur to canola in two consecutive crops. Plant Soil 398, 313–325. doi: 10.1007/s11104-015-2667-2
Frigaard, N. U., and Dahl, C. (2009). Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 54, 103–200. doi: 10.1016/S0065-2911(08)00002-7
Fuentes-Lara, L. O., Medrano-Macías, J., Pérez-Labrada, F., Rivas-Martínez, E. N., García-Enciso, E. L., González-Morales, S., et al. (2019). From elemental sulfur to hydrogen sulfide in agricultural soils and plants. Molecules 24:2282. doi: 10.3390/molecules24122282
Garrido, M. L. (1964). Determination of sulfur in plant material. Analyst 89, 61–66. doi: 10.1039/an9648900061
Ghosh, W., and Dam, B. (2009). Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol. Lett. 33, 999–1043. doi: 10.1111/j.1574-6976.2009.00187.x
Giovannetti, M., Tolosano, M., Volpe, V., Kopriva, S., and Bonfante, P. (2014). Identification and functional characterization of a sulfate transporter induced by both sulfur starvation and mycorrhiza formation in Lotus japonicus. New Phytol. 204, 609–619. doi: 10.1111/nph.12949
Hou, N., Xia, Y., Wang, X., Liu, H., Liu, H., and Xun, L. (2018). H2S biotreatment with sulfide-oxidizing heterotrophic bacteria. Biodegradation 29, 511–524. doi: 10.1007/s10532-018-9849-6
Jat, R. A., and Ahlawat, I. P. S. (2010). Effect of organic manure and sulphur fertilization in pigeonpea (Cajanus cajan) + groundnut (Arachis hypogaea) intercropping system. Indian J. Agron. 55, 276–281.
Kataoka, T., Hayashi, N., Yamaya, T., and Takahashi, H. (2004). Root-to-shoot transport of sulfate in arabidopsis evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol. 136, 4198–4204. doi: 10.1104/pp.104.045625
Kumar, S., Tomar, J., Kishore, G. R., Kumar, A., and Singh, S. (2012). Effect of phosphorus and sulphur on growth and yield of pigeon pea (Cajanus cajan). Adv. Res. J. Crop Improv. 3, 50–52.
Kumar, U., Kaviraj, M., Panneerselvam, P., and Nayak, A. K. (2022). Conversion of mangroves into rice cultivation alters functional soil microbial community in subhumid tropical paddy soil. Front. Environ. Sci. 10:858028. doi: 10.3389/fenvs.2022.858028
Kumar, U., Panneerselvam, P., Gupta, V. V., Manjunath, M., Priyadarshinee, P., Sahoo, A., et al. (2018). “Diversity of sulfur-oxidizing and sulfur-reducing microbes in diverse ecosystems,” in Advances in Soil Microbiology: Recent Trends and Future Prospects, eds T. K. Adhya, B. Lal, B. Mohapatra, D. Paul, and S. Das (Singapore: Springer), 65–89. doi: 10.1007/978-981-10-6178-3_4
Lewandowska, M., and Sirko, A. (2008). Recent advances in understanding plant response to sulfur-deficiency stress. Acta Biochim. Pol. 55, 457–471. doi: 10.18388/abp.2008_3051
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lopes, M. J. S., Dias-Filho, M. B., and Gurgel, E. S. C. (2021). Successful plant growth-promoting microbes: inoculation methods and abiotic factors. Front. Sustain. Food Syst. 5:606454. doi: 10.3389/fsufs.2021.606454
Majumder, P., and Palit, D. (2017). Isolation, identification and characterization of bacteria of coal mine soil at Sonepur bazari of Raniganj coalfield, West Bengal. Int. J. Appl. Environ. Sci. 12, 1131–1140.
Malviya, D., Varma, A., Singh, U. B., Singh, S., Singh, H. V., and Saxena, A. K. (2022). Sulfur-oxidizing bacteria from coal mine enhance sulfur nutrition in pigeon pea (Cajanus cajan L.). Front. Environ. Sci. 10:932402. doi: 10.3389/fenvs.2022.932402
Maruyama-Nakashita, A., Watanabe-Takahashi, A., Inoue, E., Yamaya, T., Saito, K., and Takahashi, H. (2015). Sulfur-responsive elements in the 39-nontranscribed intergenic region are essential for the induction of SULFATE TRANSPORTER 2;1 gene expression in Arabidopsis roots under sulfur deficiency. Plant Cell 27, 1279–1296. doi: 10.1105/tpc.114.134908
McGrath, S. P., Zhao, F. J., and Blake-Kalff, M. (2002). History and outlook for sulfur fertilizers in Europe. Proc. Fertil. Soc. 497, 1–22.
McNear, D. H. Jr. (2013). The rhizosphere - roots, soil and everything in between. Nat. Educ. Knowl. 4:1.
Meneses, C. H., Rouws, L. F., Simões-Araújo, J. L., Vidal, M. S., and Baldani, J. I. (2011). Exopolysaccharide production is required for biofilm formation and plant colonization by the nitrogen-fixing endophyte Gluconacetobacter diazotrophicus. Mol. Plant Microbe Interact. 24, 1448–1458. doi: 10.1094/MPMI-05-11-0127
Meng, C., Zeleznik, O. A., Thallinger, G. G., Kuster, B., Gholami, A. M., and Culhane, A. C. (2016). Dimension reduction techniques for the integrative analysis of multi-omics data. Brief. Bioinform. 17, 628–641. doi: 10.1093/bib/bbv108
Nawaz, M., and Wang, Z. (2020). Abscisic acid and glycine betaine mediated tolerance mechanisms under drought stress and recovery in Axonopus compressus: a new insight. Sci. Rep. 10, 1–10. doi: 10.1038/s41598-020-63447-0
Ortiz-Castro, R., Martinez-Trujillo, M., and Lopez-Bucio, J. (2008). N-acyl-L-homoserine lactones: a class of bacterial quorum-sensing signals alter post-embryonic root development in Arabidopsis thaliana. Plant Cell Environ. 31, 1497–1509. doi: 10.1111/j.1365-3040.2008.01863.x
Rao, M. V., and Davis, K. R. (1999). Ozone-induced cell death occurs via two distinct mechanisms in Arabidopsis: the role of salicylic acid. Plant J. 17, 603–614. doi: 10.1046/j.1365-313X.1999.00400.x
Rawlings, D. E. (2005). Characteristics and adaptability of iron-and sulfur-oxidizing microorganisms used for the recovery of metals from minerals and their concentrates. Microb. Cell Fact. 4, 1–15. doi: 10.1186/1475-2859-4-13
Rudrappa, T., Biedrzycki, M. L., and Bais, H. P. (2008). Causes and consequences of plant-associated biofilms. FEMS Microbiol. Ecol. 64, 153–166. doi: 10.1111/j.1574-6941.2008.00465.x
Ryan, R. P., Monchy, S., Cardinale, M., Taghavi, S., Crossman, L., Avison, M. B., et al. (2009). The Versatility and Adaptation of Bacteria from the Genus Stenotrophomonas. Nat. Rev. Microbiol. 7, 514–525. doi: 10.1038/nrmicro2163
Sajjad, W., Bhatti, T. M., Hasan, F., Khan, S., Badshah, M., Naseem, A. A., et al. (2016). Characterization of Sulfur-Oxidizing Bacteria Isolated from Acid Mine Drainage and Black Shale Samples. Pak. J. Bot. 48, 1253–1262.
Sanwani, E., Jeremy, E., Chaerun, S. K., Mufakhir, F. R., and Astuti, W. (2022). Use of mixotrophic bacteria as flocculating agents to separate iron from red mud (Alumina Refinery Residue). J. Sustain. Metall. 8, 443–457. doi: 10.1007/s40831-021-00479-4
Saxena, K. B., Kumar, R. V., and Sultana, R. (2010). Quality nutrition through pigeonpea-a review. Health 2, 1335–1344. doi: 10.4236/health.2010.211199
Scherer, H. W. (2001). Sulfur in crop production-invited paper. Eur. J. Agron. 14, 81–111. doi: 10.1016/S1161-0301(00)00082-4
Shibagaki, N., Rose, A., McDermott, J. P., Fujiwara, T., Hayashi, H., Yoneyama, T., et al. (2002). Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J. 29, 475–486. doi: 10.1046/j.0960-7412.2001.01232.x
Sievert, S. M., Heidorn, T., and Kuever, J. (2000). Halothiobacillus kellyi sp. nov., a mesophilic, obligately chemolithoautotrophic, sulfur-oxidizing bacterium isolated from a shallow-water hydrothermal vent in the Aegean Sea, and emended description of the genus Halothiobacillus. Int. J. Syst. Evol. Microbiol. 50, 1229–1237. doi: 10.1099/00207713-50-3-1229
Singh, D., Rajawat, M. V. S., Kaushik, R., Prasanna, R., and Saxena, A. K. (2017). Beneficial role of endophytes in biofortification of Zn in wheat genotypes varying in nutrient use efficiency grown in soils sufficient and deficient in Zn. Plant Soil 416, 107–116. doi: 10.1007/s11104-017-3189-x
Singh, D. P., Singh, V., Gupta, V. K., Shukla, R., Prabha, R., Sarma, B. K., et al. (2020). Microbial inoculation in rice regulates antioxidative reactions and defense related genes to mitigate drought stress. Sci. Rep. 10, 1–17. doi: 10.1038/s41598-020-61140-w
Singh, F., and Diwakar, B. (1993). Nutritive Value and Uses of Pigeonpea and Groundnut. Patancheru: International Crops Research Institute.
Singh, R. P., Jha, P., and Jha, P. N. (2015). The plant-growth-promoting bacterium Klebsiella sp. SBP-8 confers induced systemic tolerance in wheat (Triticum aestivum) under salt stress. J. Plant Physiol. 184, 57–67. doi: 10.1016/j.jplph.2015.07.002
Singh, S., Singh, U. B., Malviya, D., Paul, S., Sahu, P. K., Trivedi, M., et al. (2020). Seed biopriming with microbial inoculant triggers local and systemic defense responses against Rhizoctonia solani causing banded leaf and sheath blight in maize (Zea mays L.). Int. J. Environ. Res. Public Health 17:1396. doi: 10.3390/ijerph17041396
Singh, S., Singh, U. B., Trivedi, M., Malviya, D., Sahu, P. K., Roy, M., et al. (2021). Restructuring the cellular responses: connecting microbial intervention with ecological fitness and adaptiveness to the maize (Zea mays L.) grown in saline–sodic soil. Front. Microbiol. 11:568325. doi: 10.3389/fmicb.2020.568325
Stamford, N. P., Silva, A. J. N., Freitas, A. D. S., and Araujo Filho, J. T. (2002). Effect of sulfur inoculated with Thiobacillus on soil salinity and growth of tropical tree legumes. Bioresour. Technol. 81, 53–59. doi: 10.1016/S0960-8524(01)00099-2
Sultan, S., and Faisal, M. (2016). Isolation and characterization of iron and sulfur oxidizing bacteria from coal mines. J. Environ. Earth Sci. 6, 153–157.
Takahashi, H., Buchner, P., Yoshimoto, N., Hawkesford, M. J., and Shiu, S. H. (2011a). Evolutionary relationships and functional diversity of plant sulfate transporters. Front. Plant Sci. 2:119. doi: 10.3389/fpls.2011.00119
Takahashi, H., Kopriva, S., Giordano, M., Saito, K., and Hell, R. (2011b). Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Ann. Rev. Plant Biol. 62, 157–184. doi: 10.1146/annurev-arplant-042110-103921
Takahashi, H., Watanabe-Takahashi, A., Smith, F. W., Blake-Kalff, M., Hawkesford, M. J., and Saito, K. (2000). The roles of three functional sulfate transporters involved in uptake and translocation of sulfate in Arabidopsis thaliana. Plant J. 23, 171–182. doi: 10.1046/j.1365-313x.2000.00768.x
Thimmaiah, S. R. (2012). Standard Methods of Biochemical Analysis. New Delhi: Kalyani Publishers, 421–426.
Tian, H., De Smet, I., and Ding, Z. (2014). Shaping a root system: regulating lateral versus primary root growth. Trends Plant Sci. 19, 426–431. doi: 10.1016/j.tplants.2014.01.007
Tiwari, S., Singh, P., Tiwari, R., Meena, K. K., Yandigeri, M., Singh, D. P., et al. (2011). Salt-tolerant rhizobacteria-mediated induced tolerance in wheat (Triticum aestivum) and chemical diversity in rhizosphere enhance plant growth. Biol. Fertil. Soils 47, 907–916. doi: 10.1007/s00374-011-0598-5
Varshney, R. K., Chen, W., Li, Y., Bharti, A. K., Saxena, R. K., Schlueter, J. A., et al. (2012). Draft genome sequence of pigeonpea (Cajanus cajan L.), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 30, 83–92. doi: 10.1038/nbt.2022
Veerender, K., Sridar, R., and Sivaji, M. (2014). Isolation and characterization of Sulphur oxidizing bacteria from different ecosystems. Prog. Res. 9, 104–107.
Vidmar, J. J., Tagmount, A., Cathala, N., Touraine, B., and Davidian, J. E. (2000). Cloning and characterization of a root specific high-affinity sulfate transporter from Arabidopsis thaliana. FEBS Lett. 475, 65–69. doi: 10.1016/S0014-5793(00)01615-X
Wang, R., Lin, J. Q., Liu, X. M., Pang, X., Zhang, C. J., Yang, C. L., et al. (2019). Sulfur oxidation in the acidophilic autotrophic Acidithiobacillus spp. Front. Microbiol. 9:3290. doi: 10.3389/fmicb.2018.03290
Watkinson, J. H. (1989). Measurement of the oxidation rate of elemental sulfur in soil. Aust. J. Soil Sci. 27, 365–375. doi: 10.1071/SR9890365
Wood, A. P., Kelly, D. P., McDonald, I. R., Jordan, S. L., Morgan, T. D., Khan, S., et al. (1998). A novel pink-pigmented facultative methylotroph, Methylobacterium thiocyanatum sp. nov., capable of growth on thiocyanate or cyanate as sole nitrogen sources. Arch. Microbiol. 169, 148–158. doi: 10.1007/s002030050554
Yoshimoto, N., Inoue, E., Saito, K., Yamaya, T., and Takahashi, H. (2003). Phloem-localizing sulfate transporter, Sultr1;3, mediates redistribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol. 131, 1511–1517. doi: 10.1104/pp.014712
Yoshimoto, N., Inoue, E., Watanabe-Takahashi, A., Saito, K., and Takahashi, H. (2007). Posttranscriptional regulation of high-affinity sulfate transporters in Arabidopsis by sulfur nutrition. Plant Physiol. 145, 378–388. doi: 10.1104/pp.107.105742
Yoshimoto, N., Takahashi, H., Smith, F. W., Yamaya, T., and Saito, K. (2002). Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J. 29, 465–473. doi: 10.1046/j.0960-7412.2001.01231.x
Zhang, R., Neu, T. R., Li, Q., Blanchard, V., Zhang, Y., Schippers, A., et al. (2019). Insight into interactions of thermoacidophilic archaea with elemental sulfur: biofilm dynamics and EPS analysis. Front. Microbiol. 10:896. doi: 10.3389/fmicb.2019.00896
Keywords: sulfur-oxidizing bacteria, pigeon pea (Cajanus cajan), sulfate transporter, PpSULTR, root colonization, root architecture, ROS
Citation: Malviya D, Varma A, Singh UB, Singh S and Saxena AK (2022) Unraveling the mechanism of sulfur nutrition in pigeonpea inoculated with sulfur-oxidizing bacteria. Front. Microbiol. 13:927702. doi: 10.3389/fmicb.2022.927702
Received: 24 April 2022; Accepted: 11 August 2022;
Published: 05 September 2022.
Edited by:
Milko Alberto Jorquera, University of La Frontera, ChileReviewed by:
Divjot Kour, Eternal University, IndiaCopyright © 2022 Malviya, Varma, Singh, Singh and Saxena. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anil K. Saxena, c2F4ZW5hNDYxQHlhaG9vLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.