- 1Hangzhou Medical College, Hangzhou, China
- 2Ningbo Stomatology Hospital, Ningbo, China
- 3Hwa Mei Hospital, University of Chinese Academy of Sciences, Ningbo, China
- 4Ningbo Institute of Life and Health Industry, University of Chinese Academy of Sciences, Ningbo, China
- 5Key Laboratory of Diagnosis and Treatment of Digestive System Tumors of Zhejiang Province, Ningbo, China
- 6Pilot Gene Technologies (Hangzhou) Co., Ltd., Hangzhou, China
Vibrio vulnificus (V. vulnificus) is one of the most common pathogenic Vibrio species to humans; therefore, the establishment of timely and credible detection methods has become an urgent requirement for V. vulnificus illness surveillance. In this study, an assay combining droplet digital PCR (ddPCR) with propidium monoazide (PMA) treatment was developed for detecting V. vulnificus. The primers/probes targeting the V. vulnificus hemolysin A (vvhA) gene, amplification procedures, and PMA processing conditions involved in the assay were optimized. Then, we analyzed the specificity, sensitivity, and ability to detect live cell DNA while testing the performance of PMA-ddPCR in clinical samples. The optimal concentrations of primers and probes were 1.0 and 0.3 μM, respectively. The annealing temperature achieving the highest accuracy in ddPCR assay was 60°C. With an initial V. vulnificus cell concentration of 108 CFU/mL (colony-forming units per milliliter), the optimal strategy to distinguish live cells from dead cells was to treat samples with 100 μM PMA for 15 min in the dark and expose them to LED light with an output wavelength of 465 nm for 10 min. The specificity of the PMA-ddPCR assay was tested on 27 strains, including seven V. vulnificus strains and 20 other bacterial strains. Only the seven V. vulnificus strains were observed with positive signals in specificity analysis. Comparative experiments on the detection ability of PMA-ddPCR and PMA-qPCR in pure cultures and plasma samples were performed. The limit of detection (LOD) and the limit of quantitation (LOQ) in pure culture solutions of V. vulnificus were 29.33 and 53.64 CFU/mL in PMA-ddPCR, respectively. For artificially clinical sample tests in PMA-ddPCR, V. vulnificus could be detected at concentrations as low as 65.20 CFU/mL. The sensitivity of the PMA-ddPCR assay was 15- to 40-fold more sensitive than the PMA-qPCR in this study. The PMA-ddPCR assay we developed provides a new insight to accurately detect live cells of V. vulnificus in clinical samples, which is of great significance to enhance public health safety and security capability and improve the emergency response level for V. vulnificus infection.
Introduction
Vibrio vulnificus (V. vulnificus), one of the notorious pathogenic Vibrio species in Vibrionaceae, can be found in seawater and aquatic products near the coast in tropical and subtropics (Guan et al., 2021). It could enter the human body through the consumption of raw, under-cooked seafood (like oysters, fish, and shellfish), contact with the open wound on skin, or the bite of insects (Liang et al., 2021). V. vulnificus could cause a wide range of human diseases, such as septicemia (Narendrakumar et al., 2021), wound infections (D’Souza et al., 2018), necrotizing fasciitis (Tsai et al., 2022), meningoencephalitis (He et al., 2019), and acute gastrointestinal reactions (Hannet et al., 2019). Wound infections (mean incubation period of 16 h) and septicemia (mean incubation period of 26 h) caused by V. vulnificus deteriorate very quickly and could be easily misdiagnosed (Baker-Austin and Oliver, 2020), which results in high mortality rates for V. vulnificus infection over 24 h (Yun and Kim, 2018). The accurate diagnosis and debridement of V. vulnificus infections are crucial to significantly reduce the mortality and amputation rate of patients with serious infections, such as necrotizing fasciitis (Yu et al., 2017). There are growing pieces of evidence (Lee et al., 2019; Brehm et al., 2021; Kim and Chun, 2021) that V. vulnificus is posing a more serious threat to human health due to the increasing seawater temperatures in many areas of the world. Additionally, the increase in seafood farming and global seafood trade has led to an increase in exposure risk of V. vulnificus (Liang et al., 2021). Hence, the establishment of rapid and credible detection methods for V. vulnificus has become more important in Vibrio illness surveillance.
The conventional clinical detection of V. vulnificus is based on the microbiological culture method. The complex processes, including incubation, selective plate isolation, and biochemical identification, usually require several days, which not only is time-consuming (Bonny et al., 2022) but also might cause interference to the identification of V. vulnificus due to over-proliferation of the other bacteria in the samples from blood and infected trauma tissue culture fluid. In addition, when V. vulnificus is exposed to adverse growth conditions, such as lower temperature (below 13°C), nutrient deficiency, and disinfectants, it often exists in a viable but non-culturable (VBNC) state (Malayil et al., 2021). However, the traditional pathogenic culture method cannot detect VBNC cells (Zhao et al., 2017), which might result in false-negative results and increase the risk of transmission of V. vulnificus infection. Antibiotics may also affect the results of the pathogenic culture. Some researchers in a clinical study (Lee et al., 2017) analyzed the blood culture results from 23 hospitalized cases of V. vulnificus infection and found that the culture method could not detect V. vulnificus in the blood samples of eight patients who used antibiotics before sample collection. The immunoassay method (Rengpipat et al., 2008; Jadeja et al., 2010) for the detection of V. vulnificus is also questionable due to the difficulty of obtaining antibodies, easy inactivation during storage, and the Vibrio-associated cross-antigens (Lee et al., 2021b), and the high cost of antibody production also leads to difficulties in practical application (Harwood et al., 2004). The traditional pathogenic culture method and immunoassay method are cumbersome, unreliable, and prone to false-negative results, especially in distinguishing homologous strains and VBNC cells. It is necessary to find more accurate detection techniques for V. vulnificus detection (Copin et al., 2021). In recent years, with the development of heat-resistant DNA polymerases (e.g., Taq DNA polymerase) and thermal cycling instruments, molecular biology techniques represented by polymerase chain reaction (PCR) have gradually advanced from the research laboratories. Currently, the emergence of quantitative real-time PCR (qPCR) methods has greatly promoted the application of PCR in the quantitative detection of clinical pathogenic microorganisms (Cao et al., 2019; Guan et al., 2021; Li et al., 2021). The PCR-based methods are also recommended as confirmation methods for the identification and diagnosis of V. vulnificus in the International Organization for Standardization (ISO 21872: 2017) (Hartnell et al., 2019) in addition to the traditional pathogenic culture-based detection methods.
Digital PCR (dPCR), as a representative of the third-generation PCR, is a micro-drop reaction system and assigns templates into 10,000–80,000 independent reaction microchambers, ensuring that the DNA molecules in each microchamber were ≤ 1 (Lei et al., 2021). After PCR amplification, the copy number of the target gene in the initial sample could be calculated directly according to Poisson statistics by collecting the fluorescence signal within each reaction microchamber, which finally achieves the absolute quantification of target DNA without standard curves like qPCR (Wouters et al., 2020). There are two dominant methods of producing microdroplets: the microfluidic chip method represented by the QuantStudio™ 3D chip digital PCR (cdPCR) system and the “oil-in-water” droplet method represented by the QX100/QX200™ droplet digital PCR (ddPCR) system. The ddPCR used in this study could partition the reaction mixture into “oil-in-water” microdroplets and disperse them on the reaction area by droplet generator. The reaction mixture and cycling parameters of ddPCR were no different from those of qPCR assay. However, owing to the microtitration of the ddPCR mixture, the shortcoming of qPCR, such as the influence of inhibitory component in mixture (Lei et al., 2020) and the unstable ability to detect weakly positive samples (Suo et al., 2020), had been overcome. A more stable and consistent amplification efficiency could be obtained on ddPCR.
Since the dead bacterial DNA also remained in the samples for a long time (Zeng et al., 2020), routine PCR assays cannot distinguish the target DNA coming from live or dead bacteria in the actual samples (e.g., contaminated food samples after autoclaving and clinical samples after antibiotic treatment), which results in the overestimation of live cells (Zhao et al., 2020). Recently, this bottleneck could be broken by combining photo-reactive DNA-binding dye, such as propidium monoazide (PMA) and ethidium monoazide (EMA), with nucleic acid molecular diagnostics (Codony et al., 2020). A previous study revealed (Copin et al., 2021) that pre-treatment with different concentration gradients of EMA in seafood and environmental samples had a lethal effect on V. vulnificus strain compared with PMA. PMA could penetrate into the damaged and dead cells and selectively bind to the DNA in these cells, which can effectively inhibit their PCR amplification (Elizaquível et al., 2014). PMA combined with PCR-based correlation assays had been widely used in the research studies for infectious viable bacterial detection, such as Brucella (Zhang et al., 2020), Escherichia coli (Deshmukh et al., 2020), Salmonella (Xie et al., 2020), and SARS-CoV-2 (Hong et al., 2021).
To our knowledge, most researchers have developed qPCR assay for detecting V. vulnificus (Tsai et al., 2019; Guan et al., 2021; Lee et al., 2021a). However, there were few studies on the detection of V. vulnificus by ddPCR, especially based on PMA-ddPCR. In this study, we aimed to establish an accurate and sensitive method based on PMA-ddPCR to detect live cells of V. vulnificus in samples (Figure 1). The specificity, sensitivity, and the ability to detect live V. vulnificus DNA were analyzed, and the performance in clinical samples was evaluated by testing artificially contaminated plasma sample, compared to that of PMA-qPCR.
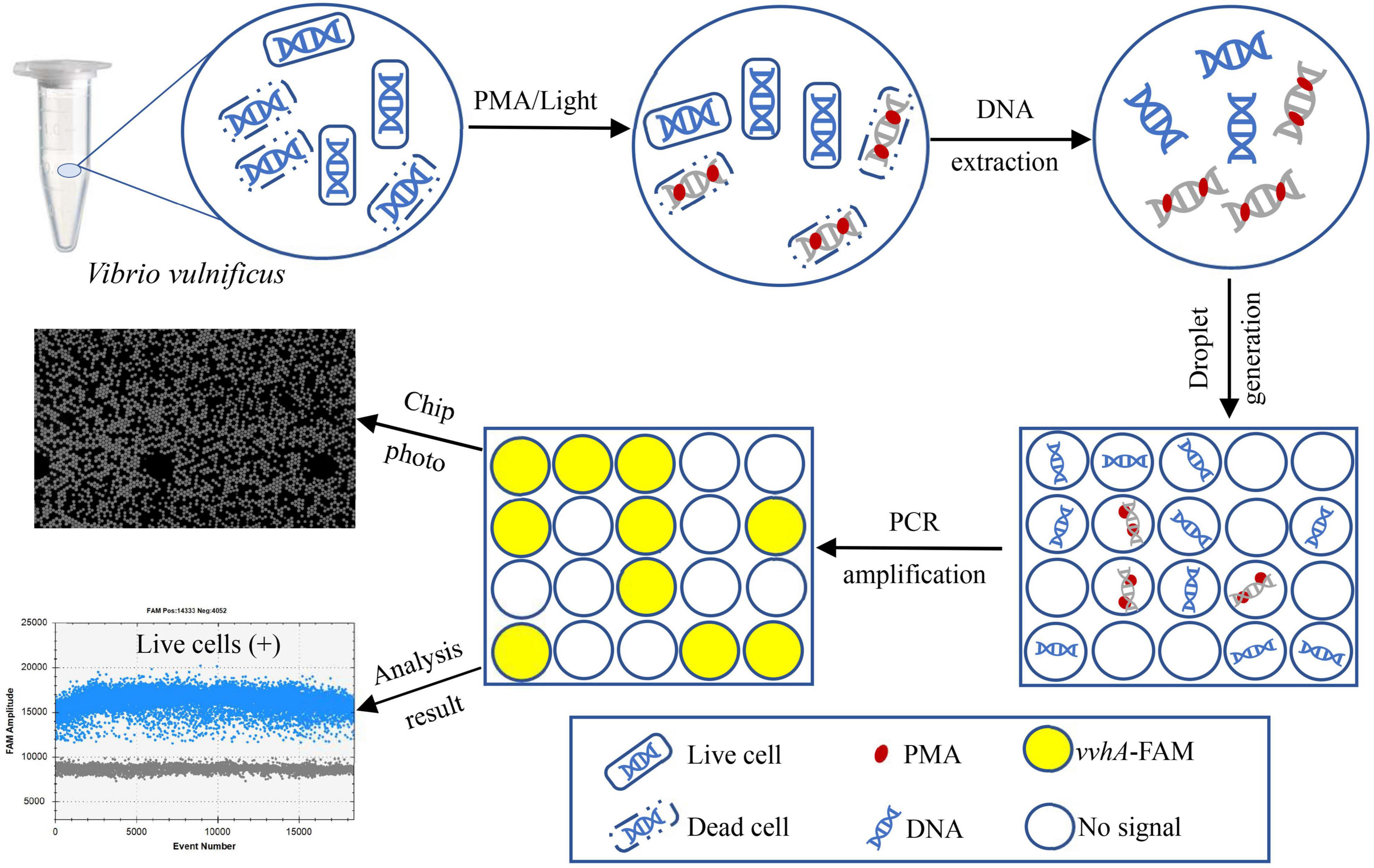
Figure 1. Technical route of PMA-ddPCR for the accurate detection of live cells of V. vulnificus by targeting the vvhA gene.
Materials and Methods
Bacterial Strains and Cultivation
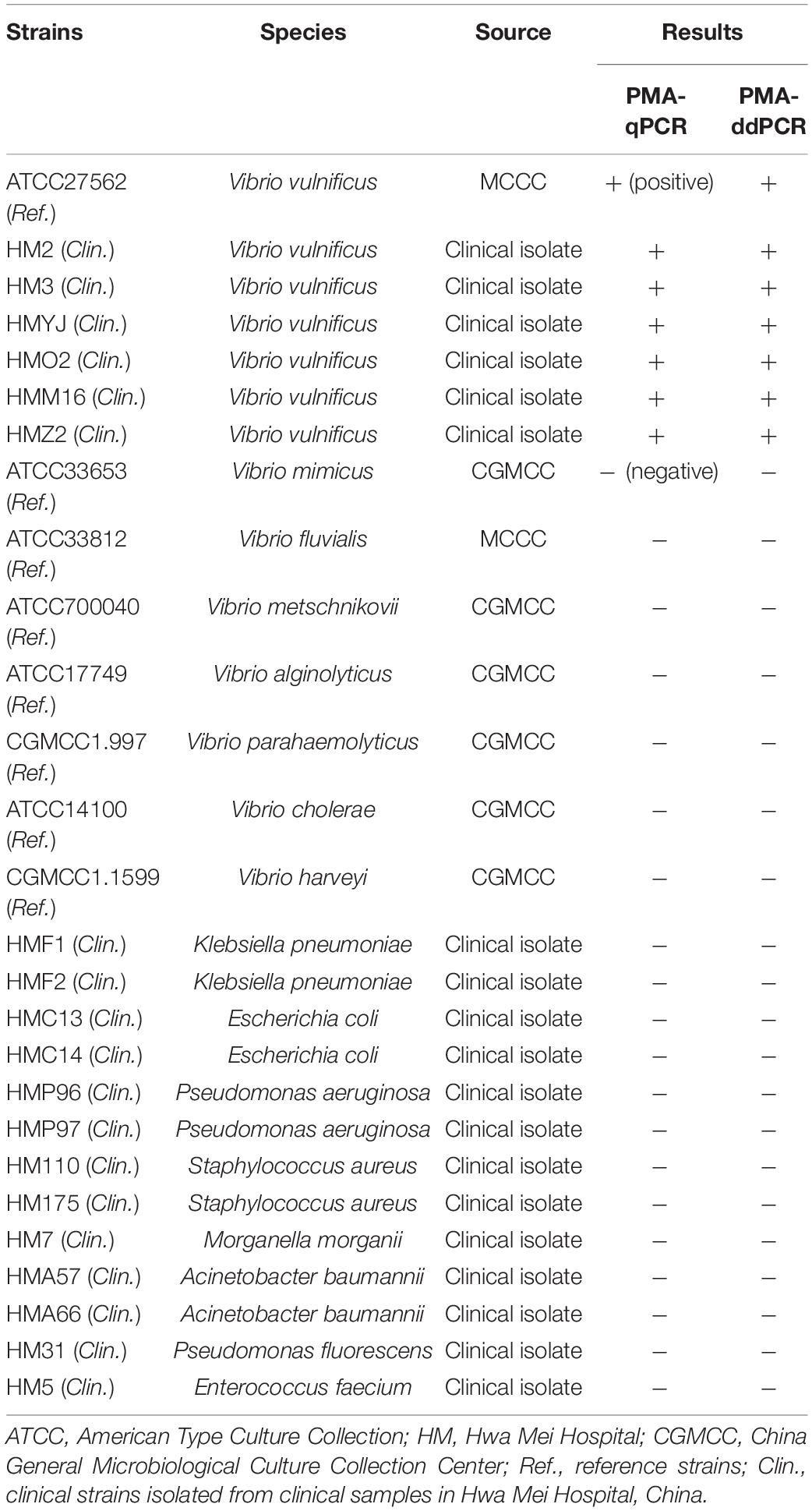
In the PMA-qPCR and PMA-ddPCR assays, optimization and validation assays were performed by using eight reference strains and 19 clinical isolates. Among them, seven strains belonged to V. vulnificus (Table 1). Reference strains (Ref.) were purchased from the Marine Culture Collections of China (MCCC) and the China General Microbiological Culture Collection Center (CGMCC). Clinical strains (Clin.) were isolated from clinical samples in Hwa Mei Hospital, China, and confirmed by an automated microbial identification system. All strains were initially stored in 20% (v/v) glycerol at –80°C. Vibrio species strains were grown on 2216E liquid medium (QDRS BIOTEC, China), and the other 13 bacterial strains were cultured on Luria–Bertani (LB) broth medium. All strains were incubated at 37°C for 48 h under constant shaking (180 rpm) to make live cell suspensions for the subsequent experiments. The V. vulnificus culturable cells were enumerated by dilution plating on 2216E agar medium at 37°C for 48 h.
Preparation of Heat-Killed Cell Suspensions
We divided the counted bacterial solutions in Section “Bacterial Strains and Cultivation” into two equal portions: One was used to prepare live V. vulnificus cell suspensions and the other was incubated at 95°C for 10 min to obtain the heat-killed cell suspensions. The effect of heat killing was assessed by spreading 100 μL heat-killed cell suspension on 2216E agar medium and repeating for three plates. No colony formation after incubation at 37°C for 48 h proved that all V. vulnificus cells were dead.
Genomic DNA Extraction
The genomic DNA was extracted with a MiniBEST Bacterial Genomic DNA Extraction Kit (TaKaRa, Japan) according to the user manual. DNA was eluted in 80 μL nuclease-free water. The concentration and quality of extracted DNA were analyzed using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, United States). All template DNA were stored at −40°C until use.
Primer and Probe Design
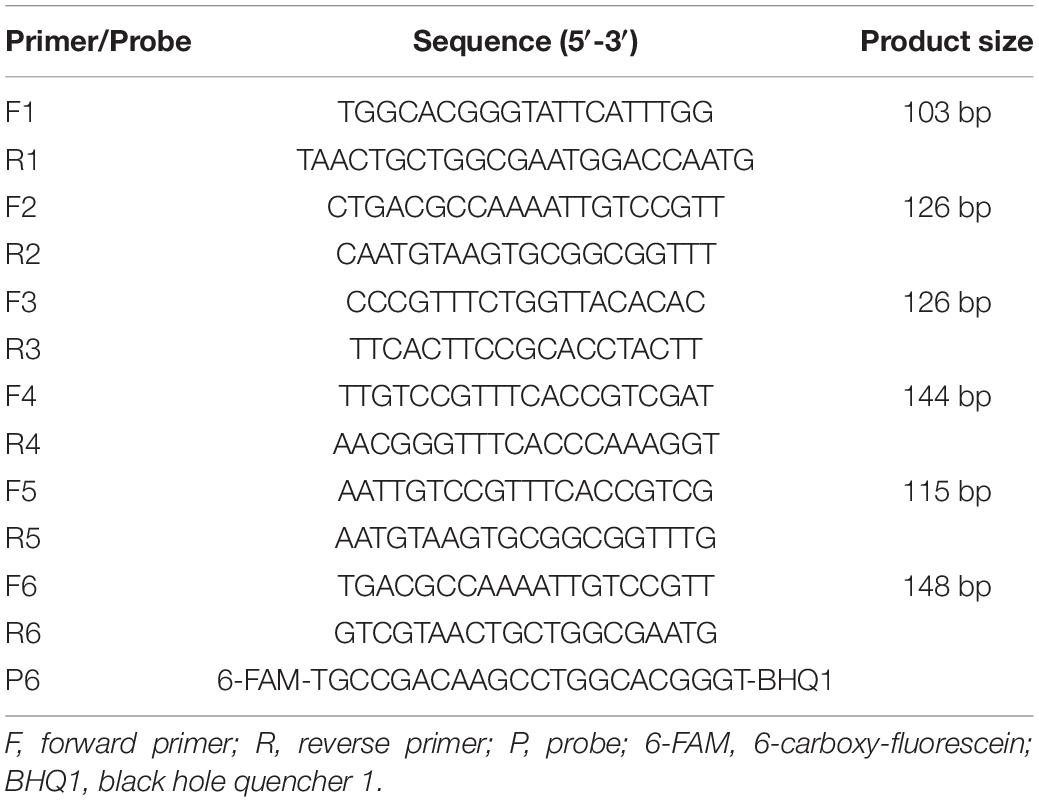
Multiple sequence alignments were performed on the vvhA gene sequence based on the published sequences from GenBank (Accession nos.: KC821520.1, MW132717.1, AF376032.1, KU680790.1, KP224256.1, KF255393.1, and CP0112740.1), and the similarity regions among those sequences were entered into the National Center for Biotechnology Information (NCBI) Primer-BLAST (Basic Local Alignment Search Tool) for primer design. At the same time, for probe design, the internal hybridization oligo-parameters in Primer-BLAST was chosen and performed with the following criteria: (1) The probe size was set as minimum at 18–30 bp; (2) the melting temperature (Tm) was set between 50 and 70°C; (3) and the GC content was set as 40–60%. Other parameters were set as default. The probe had a 6-FAM (6-carboxy-fluorescein) at the 5′ end and a BHQ1 (black hole quencher 1) at the 3′ end. All primers and probe used in this study are listed in Table 2 and synthesized at Sangon Biotech, China.
Optimization of the Primer Pairs and Probe
The performance of primers and probes is key to evaluating the quality of nucleic acid amplification. Not all primer pairs screened by Primer-BLAST could produce stable positive signals. We used qPCR assay to screen the optimal primer pairs and designed the corresponding probe for subsequent PMA-qPCR and PMA-ddPCR assays.
Quantitative Real-Time PCR
The reaction was carried out in a total volume of 20 μL, containing 10 μL of a PerfectStart II Probe qPCR SuperMix (Trans, China), 1 μL of primer (10 μM), 1 μL of probe (10 μM), 2 μL of template DNA, and 5 μL of nuclease-free water. The cycling parameters were performed as follows: 95°C for 30 s (pre-denaturation), followed by 40 cycles of 95°C for 5 s (denature), 65°C for 30 s (annealing and extension), and then cooling to 50°C for 30 s. The qPCR assay was performed on the LightCycler 480 thermocycler (Roche, United States).
To increase the detection accuracy and effectiveness, the concentration of primer and probe and the annealing temperature were optimized. Totally, five concentrations of the primer (0.1, 0.2, 0.3, 0.4, and 0.5 μM) were tested together with four concentrations of the probe (0.3, 0.4, 0.5, and 0.6 μM), and then five different annealing temperatures (55, 57, 60, 63, and 65°C) were tested. The optimal qPCR protocol was judged by the threshold cycle (Ct) and the fluorescence intensity enhancement (ΔRn) of the amplification curves under different amplification conditions. The qPCR parameter optimization was performed with two independent replicates.
Droplet Digital PCR
The ddPCR was carried out in a total volume of 14 μL, containing 7 μL of the PerfectStart II Probe qPCR SuperMix (Trans, China), 1.4 μL of each primer (10 μM), 0.42 μL of probe (10 μM), 1.4 μL of template DNA, and 2.38 μL of nuclease-free water. The reaction mixtures were injected into the spiking well on chip. Pipettes were used to drain the air bubbles in spiking wells, and then the spiking wells were covered with silica caps. Subsequently, the chip was put into the ddPCR instrument. Approximate 30000 droplets were produced and assigned on a digital chip using a droplet generator (Pilot Gene, China). Then, an independent PCR was conducted to end point using the Thermo Cycler PCR (Pilot Gene, China) on chip, and the cycling parameters were performed as qPCR in 2.6. Following PCR amplification, the chip was transferred to a chip scanner (Pilot Gene, China) for the reading and analysis of fluorescence amplitudes in each droplet.
To increase the ddPCR detection accuracy and effectiveness, the concentration of primers/probe and the annealing temperature were optimized. Totally, three concentrations of the primer (0.6, 0.8, and 1.0 μM) were tested together with three concentrations of the probe (0.2, 0.3, and 0.4 μM), and then three annealing temperatures (58, 60, and 62°C) were tested. The optimal ddPCR protocol was judged by the degree of separation between all positive and negative spots in the amplification images and the amplitude of the positive cluster. The ddPCR parameter optimization was performed using three independent replicates.
PMA Treatments
PMA (Biotium, United States) was dissolved in 20% dimethyl sulfoxide (Solarbio, China) to generate a stock concentration of 10 mM and stored at −40°C in the dark. Optimization of the PMA treatment was conducted at the room temperature of 25°C. To find the minimum PMA concentration that can completely inhibit the amplification of DNA from dead cells without affecting that from live cells of V. vulnificus, a series of concentrations (0, 10, 20, 40, 60, 100, 140, and 180 μM) of PMA were added to live cell suspensions and heat-killed cell suspensions, following an incubation period of 15 min in the dark. Then, the samples were exposed for 20 min using a 200 W blue LED lighting array with an output wavelength of 465 nm. Under the optimal PMA concentration, different exposure times (0, 1, 3, 5, 10, 15, 20, 25, and 30 min) were tested on heat-killed cell suspensions to find the optimal exposure time. PMA parameter optimization was performed with three independent replicates.
Clinical Sample Detection
To validate the diagnostic performance of PMA-ddPCR in clinical samples compared to that of PMA-qPCR, a series of plasma samples artificially spiked with different concentrations of V. vulnificus were used to simulate the clinical samples infected by V. vulnificus. The spiked samples were treated with PMA and extracted genomic DNA with a MiniBEST Bacterial Genomic DNA Extraction Kit (TaKaRa, Japan). Clinical sample detection assays were performed using three independent replicates.
Statistical Analysis
The LOD is the target level that would be detected in 95% of cases (Zhu et al., 2021). In this study, probit regression analysis was used to fit the limit of detection (LOD) of PMA-qPCR and PMA-ddPCR. The positive rates obtained by testing the V. vulnificus samples of different concentrations were transformed into probability values using probability function. The analysis of variance (ANOVA) was applied to test whether the effect of the primer/probe concentration (section “Optimization of qPCR Amplification Conditions”) or the PMA concentration (section “Screening the Optimal Concentration and Light-Exposed Time of PMA Treatment”) based on the Ct value was significant or not. The results of the positive and negative samples from the corresponding assays were used to perform receiver operating characteristic (ROC) curve analysis. ROC curve analysis was used to calculate the maximum value of the Youden index and cut-off value. The cut-off value and the calibration curve in sensitivity analysis were used to determine the limit of quantitation (LOQ) of PMA-qPCR and PMA-ddPCR (Nutz et al., 2011). The probit regression analysis and ANOVA were performed using IBM SPSS Statistics 23 software. ROC curve analysis and calibration curve in sensitivity analysis were performed by GraphPad Prism 8.0 software. All figures were drawn by using GraphPad Prism 8.0 software.
Results
Screening of Specific Primers and Probes
Totally, six pairs of potential primers were designed by Primer-BLAST and are listed in Table 2. Comparing the Ct value and ΔRn in amplification curves (Supplementary Table 1), the primer pair (F6/R6) showed the best performance. Therefore, F6/R6 and the corresponding probe (P6) were selected for subsequent qPCR and ddPCR assays.
Optimization of qPCR Amplification Conditions
To optimize the conditions of qPCR amplification, a total of 20 primer/probe concentration combinations in Supplementary Table 2 were compared. The Ct values obtained from these combinations were subjected to two-way ANOVA. The results indicated that there was no statistically significant interaction for “Primer*Probe” (P > 0.05). There was also no statistically significant difference among probe concentrations (P > 0.05), but there were statistically significant differences among primer concentrations (P < 0.05). For further screening the optimal primer concentration, the Tukey’s post hoc test was performed, and the estimated marginal mean of Ct value was calculated. The results showed that “Primer = 0.5 μM” was the optimal group. In this group, there was the highest ΔRn when the concentration of probe was 0.5 μM. So, the combination (primer = 0.5 μM and probe = 0.5 μM) was selected as the optimal primer/probe concentration combination (Supplementary Table 2). Thereafter, five different annealing temperatures were tested (Supplementary Table 3). In the subsequent qPCR assays, 65°C, at which the highest detection accuracy can be achieved, was chosen as the optimal annealing temperature.
Optimization of ddPCR Amplification Conditions
The amplification efficiencies of different primers and probe concentration combinations were compared in ddPCR assay. The results in Figure 2A reveal that the clusters in the combination (F/R = 1 μM, P = 0.3 μM) were divided clearly (largest separation of the two red lines) with highest fluorescence amplitudes. Hence, this combination was chosen as the optimal primer/probe concentration combination. Similarly, as shown in Figure 2B, the annealing temperature of 60°C was considered to be the optimum, with the largest discrimination in fluorescence intensity between blue and gray dots.
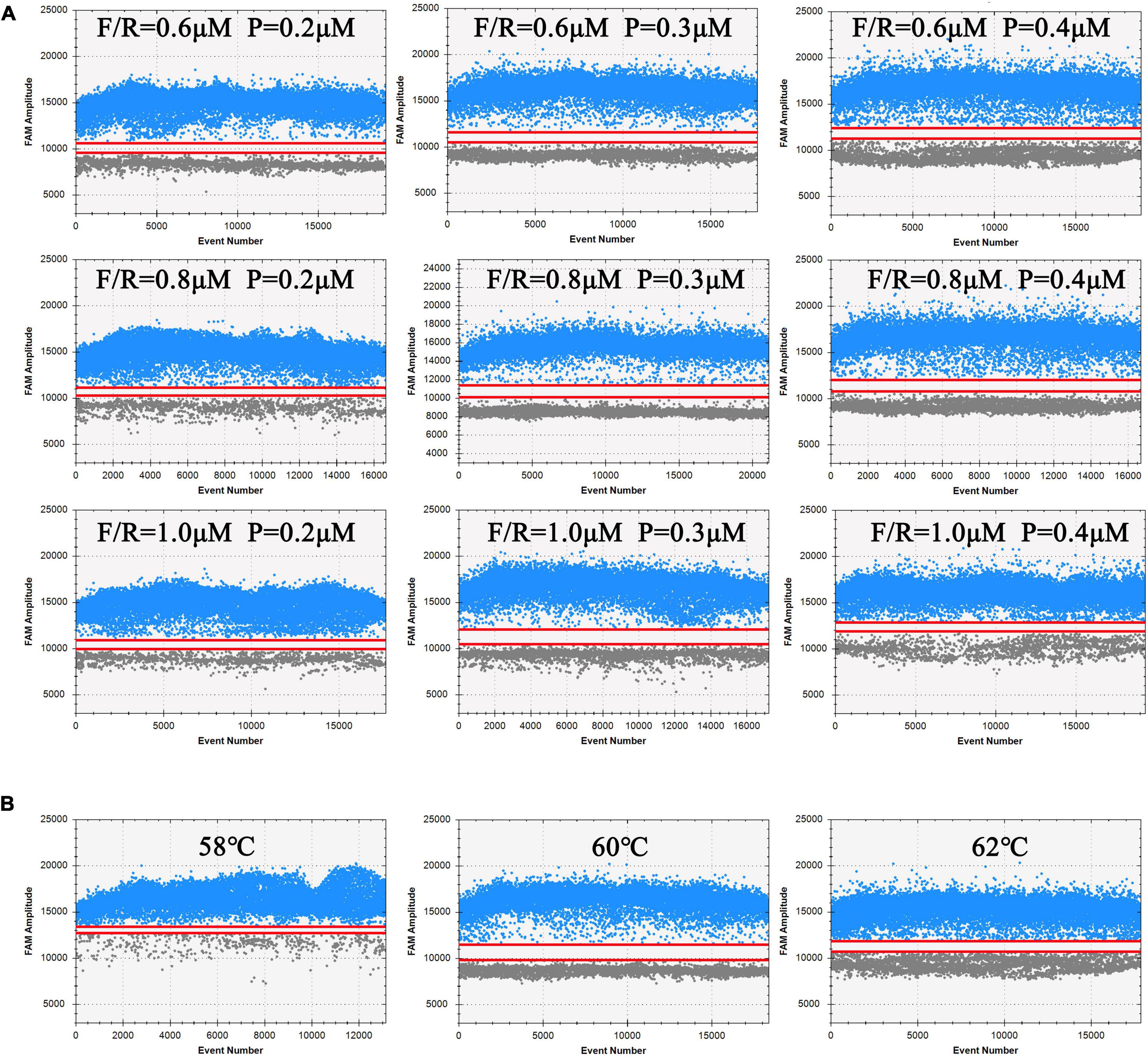
Figure 2. Analysis results of ddPCR under different processing conditions. Blue dots represent positive droplets, and gray dots represent negative droplets. The figures show the most available results of three independent replicates. (A) Optimization of primers and probe concentration combinations on ddPCR. (B) Optimization of annealing temperature on ddPCR.
Screening the Optimal Concentration and Light-Exposed Time of PMA Treatment
The different concentrations and light-exposed time of PMA treatment had different effects on the cells (Wu et al., 2015). Considering the unknown initial concentration of bacteria in actual samples, we chose a concentration (2.65 × 108 CFU/mL) for the following experiments. As shown in Figure 3A, for the heat-killed cell group, as the concentration increased, the Ct value gradually raised and leveled off at 100 μM. For live cell group, there was no statistical significance (one-way ANOVA, P > 0.05) in the Ct value under different concentrations of PMA treatment, which showed that the different concentrations (0–180 μM) of PMA treatment had no inhibitory effect on amplification of DNA from live cells of V. vulnificus. The results comparing the different light-exposed times during PMA treatments are shown in Figure 3B. The Ct value is highest at 10 min, which improved that this light-exposed time had the strongest inhibitory effect on the dead V. vulnificus DNA amplification. These results indicated that the optimal strategy for this study was to treat the samples with PMA at a final concentration of 100 μM and expose them for 10 min afterward, which could completely inhibit the DNA amplification from the dead cells of V. vulnificus.
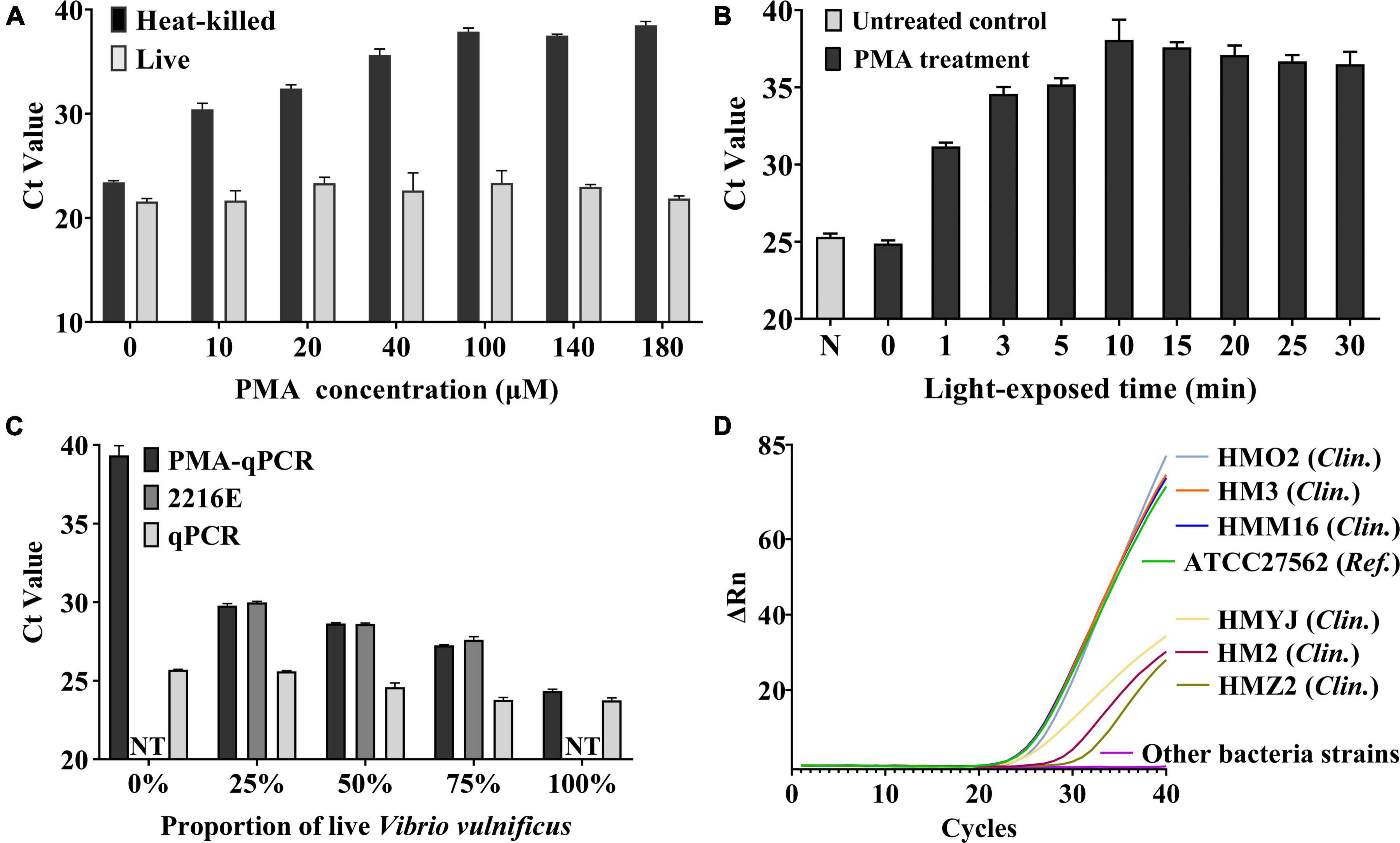
Figure 3. (A) Optimization of final PMA concentration. The different concentrations (10–180 μM) of PMA were treated on live V. vulnificus cell suspensions and heat-killed cell suspensions. Error bars represent the standard deviation based on three replicates. (B) Optimization of light-exposed time. The different light-exposed times were treated on heat-killed cell suspensions. N: PMA untreated control. Error bars represent the standard deviation based on three replicates. (C) Verification of the ability to detect live V. vulnificus cells by PMA-qPCR. The different concentrations (0–100%) of live cells were mixed with heat-killed cells and treated with PMA (PMA-qPCR group) or not (qPCR group). While the 2216E group was the blank control which was the corresponding concentration of live cells mixed with 2216E liquid medium. NT, no test. When the proportion of live V. vulnificus was 0% and 100%, the samples were pure dead bacterial solution or live bacterial solution, and it is not necessary to mix with 2216E medium. Error bars represent the standard deviation based on three replicates. (D) Specificity test of PMA-qPCR. Only the above seven V. vulnificus strains were observed positive signals. Clin., clinical strain of V. vulnificus isolated from clinical samples; Ref., reference strain of V. vulnificus.
Verification of the Ability to Detect Live Cells of Vibrio vulnificus
We used the qPCR assay to assess the ability to detect live cells of V. vulnificus under the optimal condition of PMA treatment. The different concentrations of live cells were mixed with heat-killed cells and treated with PMA (PMA-qPCR group) or not (qPCR group). The corresponding concentration of live cells mixed with 2216E liquid medium was used as the blank control (2216E group). As shown in Figure 3C, the qPCR group showed the results of DNA amplification from total cells (dead and live cells). PMA-qPCR and 2216E groups reflected the status of DNA amplification only from live cells. As the live cells of V. vulnificus cells increased in samples, the Ct values of the two groups decreased accordingly, and were close to each other. The aforementioned results indicated that the condition of PMA treatment used in this study was capable of detecting live cells of V. vulnificus.
Specificity of PMA-qPCR and PMA-ddPCR
To evaluate the specificity of PMA-qPCR and PMA-ddPCR, the genomic DNA isolated from V. vulnificus and non-V. vulnificus strains (Table 1) were tested, respectively. The positive signals were only observed in the V. vulnificus reference strain and six clinical strains, while other strains (non-V. vulnificus) showed negative signals (Figure 3D).
Sensitivity and Regression Analysis of PMA-qPCR and PMA-ddPCR
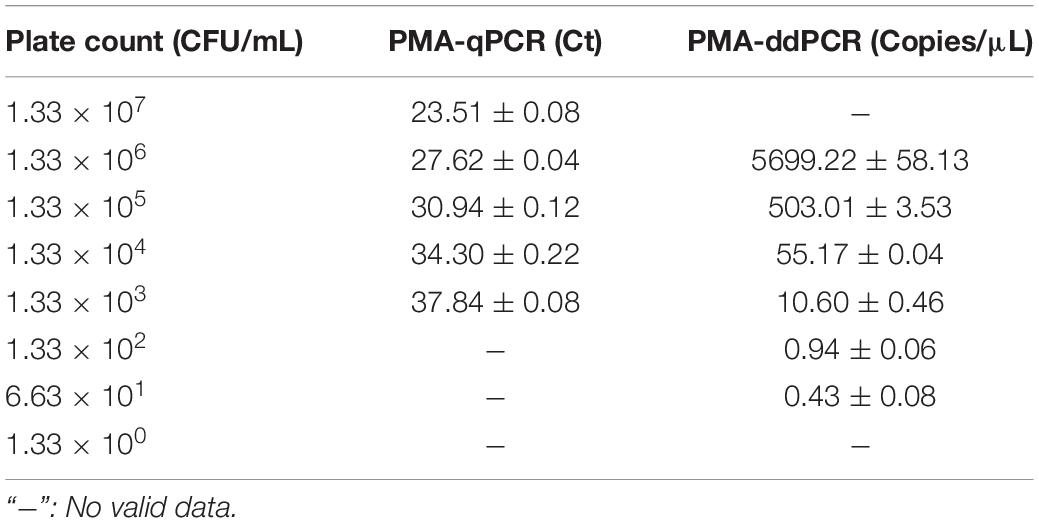
For sensitivity, a series of dilution of V. vulnificus solutions approximately ranging from 1.33 × 101 to 1.33 × 107 CFU/mL were, respectively, detected by PMA-qPCR and PMA-ddPCR (Table 3). The amplification curves of qPCR and ddPCR amplification images with different concentrations of V. vulnificus are shown in Figures 4A,C, and the constructed standard curve is shown in Figures 4B,D. The LOD was determined by the fitting of a probit regression analysis. The results showed the LODs of PMA-qPCR and PMA-ddPCR in detecting V. vulnificus from the pure culture were, respectively, 1.14 × 103 and 29.33 CFU/mL (Supplementary Figure 1). The LOQ was defined as the amount of DNA corresponding to the Ct value or the ddPCR measurement value (copies/μL) at which the sum of specificity and sensitivity of the assay was maximized (optimal cut-off point in ROC curve) and calculated using the regression equation, the Youden index, and the cut-off value. The results showed the LOQ was 1.29 × 103 CFU/mL (PMA-qPCR) and 53.64 CFU/mL (PMA-ddPCR) in pure culture bacterial solutions.
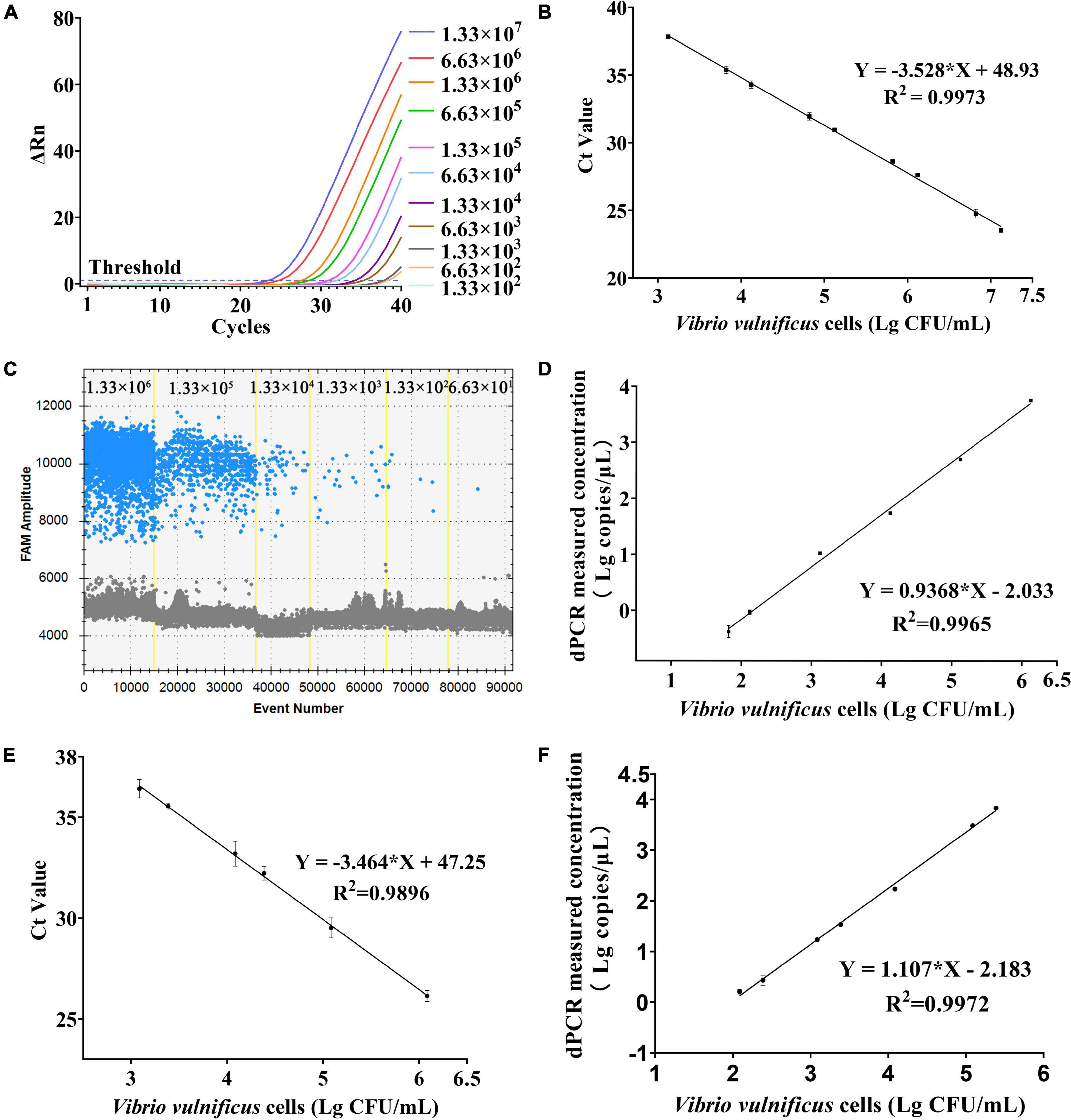
Figure 4. Sensitivity and regression analysis of PMA-qPCR and PMA-ddPCR. (A) Amplification curves of different concentrations of V. vulnificus from pure culture on qPCR. (B) Linear regression analysis based on qPCR in pure culture solutions. Error bars represent the standard deviation based on three replicates. (C) Picture of FAM channel from different concentrations of V. vulnificus in pure culture on ddPCR. (D) Linear regression analysis based on ddPCR in pure culture solutions. Error bars represent the standard deviation based on three replicates. (E) Linear regression analysis based on qPCR in plasma samples. Error bars represent the standard deviation based on three replicates. (F) Linear regression analysis based on ddPCR in plasma samples. Error bars represent the standard deviation based on three replicates.
Performance of PMA-qPCR and PMA-dPCR to Detect Vibrio vulnificus in Spiked Plasma Samples
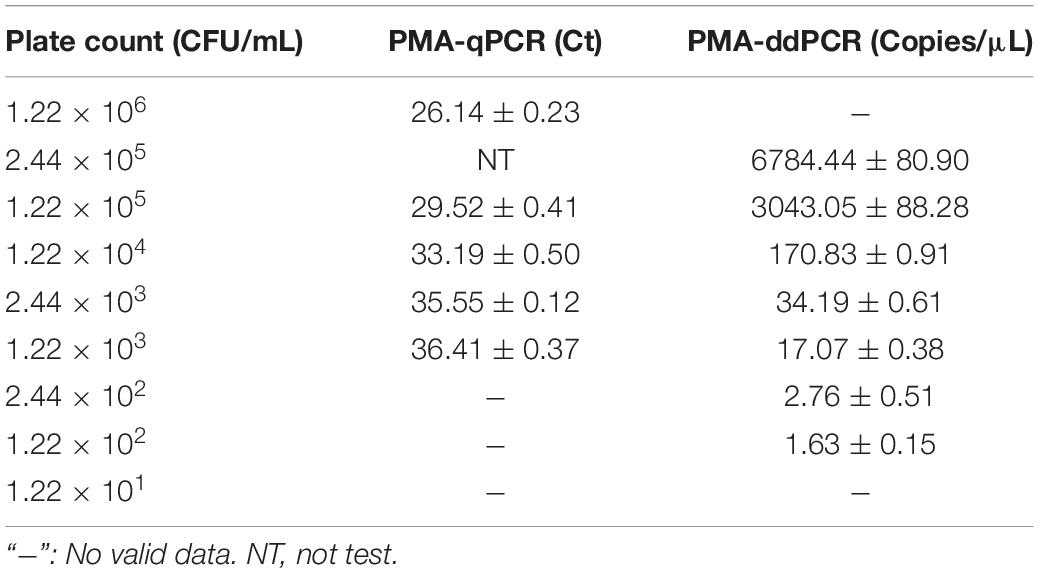
The performance of PMA-ddPCR to detect V. vulnificus in spiked plasma samples was compared to that of PMA-qPCR (Table 4). The results are shown in Figure 4E (PMA-qPCR) and Figure 4F (PMA-ddPCR), and the LODs were 1.00 × 103 CFU/mL (PMA-qPCR) and 65.20 CFU/mL (PMA-ddPCR) (Supplementary Figure 2). The results were consistent with those of pure culture bacterial solution, and it confirmed that PMA-ddPCR was applicable for the determination of V. vulnificus in clinical samples.
Discussion
Using the traditional pathogenic culture to detect V. vulnificus required pre-enrichment, isolation and purification, biochemical identification, and serological identification, which would take 3–7 days. The traditional culture method is prone to give false-negative results, especially in distinguishing Vibrio homologous strains and VBNC cells. Meanwhile, the atypical clinical signs of V. vulnificus infection with increasingly unclear factors of exposure risk have caused difficulties in its early diagnosis and treatment (Liang et al., 2021). With the development of molecular diagnosis technology, qPCR based on nucleic acid fragment detection has provided a rapid, efficient, and specific way to detect V. vulnificus (Abioye et al., 2021; Lee et al., 2021a; Álvarez-Contreras et al., 2021; Bonny et al., 2022). However, there is no official authority that can provide V. vulnificus DNA calibrations with clear quantitative values. Moreover, some factors, such as the quality of primers and probes, the inhibitory components in the samples, the laboratory environment, and the experimental skills of the operator, could directly affect the quality of the standard curve in qPCR assay, which limit the accuracy of qPCR in the practical detection. Digital PCR, as a representative of the third-generation PCR assay, does not need to rely on standard curves and Ct values for quantification, which could improve the accuracy of detection compared with qPCR (Lei et al., 2021). In this study, we developed a ddPCR method to detect V. vulnificus and evaluated the performance of this method in plasma samples, which would provide valuable insights for accurate detection of V. vulnificus in future.
Pathogens only pose a human health risk if viable (Wang et al., 2021). The genomic DNA of the pathogen remains in the body for a long time after the patient has been treated with antibiotics (Kobayashi et al., 2010), which made it difficult to effectively distinguish the DNAs from dead cells and those from live cells by traditional PCR assay. The DNAs from dead cells could lead to an overestimation of live cell counts and produce a false-positive result, which is not conducive to clinical monitoring of V. vulnificus infection. Since the first study on using PMA to differentiate live cells from dead cells was reported in 2006 (Nocker et al., 2006), PMA has become one of the most widely used dyes (Codony et al., 2020) and been applied to many fields of microbiology (Deshmukh et al., 2020; Cechova et al., 2021; Hong et al., 2021). PMA could not bind to the DNA of live cells with the intact bacterial plasma membrane structure. Therefore, the PMA treatment with PCR (PMA-PCR) is considered a successful approach to detect live cells (Lazou et al., 2021), reducing the rate of misdiagnosis in the detection of pathogen infection.
The suitable conditions of PMA treatment are key to conduct PMA-PCR as overtreatment could damage live cells and lead to wrong results (Young et al., 2017). The concentration of PMA and light-exposed time during PMA treatment are two important factors that should be optimized according to the target strain, initial concentration, exposure lamps, etc. In this study, the optimal strategy to distinguish the live cells from the dead cells of V. vulnificus was to treat the samples with 100 μM PMA for 15 min in the dark and expose them to 465 nm blue LED lighting array for 10 min. We mixed live cells with dead cells of V. vulnificus in different proportions (0–100%) and detected the Ct value by qPCR. The results verified the ability to detect live cells of V. vulnificus by PMA-qPCR and also revealed that the PMA concentration (100 μM) we chose had no effect on live cells. So, we believed that the combination of PMA treatment and detection of DNA by PCR (qPCR or ddPCR) would provide a reference for the evaluation of the therapeutic effect on clinical V. vulnificus infections.
It is necessary to choose proper target gene for successful detection of V. vulnificus in samples. An exocrine cytotoxin hemolysin A encoded by the single-copy V. vulnificus hemolysin A gene (vvhA) is an essential virulence factor causing cell infection and tissue damage by forming small pores on the cell membrane surface of target cells (Guan et al., 2021). Therefore, the vvhA gene has been considered the important virulence marker of V. vulnificus (Yuan et al., 2020). The vvhA gene is present in the majority of V. vulnificus strains that are closely related to human diseases (Wong et al., 2005; Baker-Austin and Oliver, 2020; Lin et al., 2021) and is often used as a marker for the detection of V. vulnificus (Wei et al., 2014; Gyraite et al., 2019; Bullington et al., 2022). Thus, in this study, we chose the vvhA gene as the target gene and tested its performance of PMA-qPCR and PMA-ddPCR on pure cultures and plasma samples. The results showed only the positive signals were observed in those strains of V. vulnificus, and there was no cross-reactions with DNA from no-V. vulnificus strains. This clearly validated that the vvhA gene is suitable for V. vulnificus detection. As well known, the standard for V. vulnificus detection is a culture-based method. So, based on the results of our experiments (Supplementary Table 4), we evaluated the agreement between PMA-ddPCR developed in this study and traditional culture-based method by using the kappa test. The results from the kappa test showed that there is a good agreement between the results of the PMA-ddPCR method and culture-based method (kappa = 0.800). Considering the sensitivity, the LOD of the PMA-ddPCR in this study was 29.33 CFU/mL, which performed better than those methods in previous studies, such as the LOD of V. vulnificus, was 1 × 103 CFU/mL with qPCR (Panicker and Bej, 2005), or 1.2 × 102–1 × 103 CFU/mL with recombinase polymerase amplification (RPA) (Yang et al., 2020; Zhu et al., 2021), or 2 × 103 CFU/mL with loop-mediated isothermal amplification (LAMP) (Zhang et al., 2018). In accurate detection in artificially spiked plasma samples, the performance of the PMA-ddPCR was also superior to that of PMA-qPCR (Supplementary Figure 2), which means that PMA-ddPCR is more capable in handling clinical samples. What Regrettably, the samples with higher concentrations of V. vulnificus cells (greater than 106 CFU/mL) needed to be diluted before detection because of the limitation of “oil-in-water” microdroplet number on PMA-ddPCR. Otherwise, the microdroplets would be oversaturated (target DNA molecules in each microchamber ≥ 1), leading to no valid results (Table 3). So, the microdroplet number led to a lower upper limit of the detection for ddPCR than that of qPCR. In brief, in our study, the PMA-ddPCR is suitable for the detection of V. vulnificus at the range from 1.33 × 106 to 6.63 × 101 CFU/mL in samples. It is more sensitive and accurate in the detection of target DNA at low copy numbers.
It is worth noting that the measured concentration of V. vulnificus by PMA-ddPCR was 4–7 times higher than that using by the conventional plate counting method in the theoretical situation in this study (Table 3). Which method is more accurate? It may be PMA-ddPCR. There may be two reasons as follows for this question. First, as a technology of absolute quantification, ddPCR did not rely on the calibration curve. Meanwhile, the divided droplets in ddPCR could minimize the interference of inhibitory components during the amplification of target DNA, enhancing the accuracy of ddPCR results to a large extent. Second, some inherent disadvantages of the agar plate culture, such as the overlapping of multiple colonies during growth, the inability to cultivate V. vulnificus in the VBNC state, or the decline of bacterial growth activity often led to the lower counts of V. vulnificus by using the conventional plate counting method.
Conclusion
In summary, an assay combining ddPCR with PMA (PMA-ddPCR) to accurately detect the live cells of V. vulnificus based on vvhA gene was successfully established and evaluated in artificially contaminated plasma samples. The PMA-ddPCR assay developed in our study provides a reliable tool for the clinical diagnosis and the evaluation of the therapeutic effect in V. vulnificus infection, which is very important in Vibrio illness surveillance.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
LH: methodology, data curation, software, and writing—original draft preparation. YF: data curation and software. SZ: investigation. ZP: investigation and data curation. JX: software. PZ: conceptualization writing – reviewing and editing and supervision. JG: supervision. All authors contributed to the final version of the manuscript and approved the final manuscript.
Funding
This study was funded by the Zhejiang Provincial Public Welfare Technology Program of China (LGF22H190003 and LGC19H190001), Zhejiang Science and Technology Plan Project (2022C01007), and Zhejiang Ningbo Clinical Research Center for Digestive System Tumors (Grant No. 2019A21003).
Conflict of Interest
JX is employed by Pilot Gene Technologies (Hangzhou) Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank LetPub (www.letpub.com) for providing linguistic assistance during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.927285/full#supplementary-material
References
Abioye, O. E., Osunla, A. C., and Okoh, A. I. (2021). Molecular detection and distribution of six medically important Vibrio spp. in selected freshwater and brackish water resources in eastern cape province. South Africa. Front. Microbiol. 12:617703. doi: 10.3389/fmicb.2021.617703
Álvarez-Contreras, A. K., Quiñones-Ramírez, E. I., and Vázquez-Salinas, C. (2021). Prevalence, detection of virulence genes and antimicrobial susceptibility of pathogen Vibrio species isolated from different types of seafood samples at “La Nueva Viga” market in Mexico City. Antonie. Van. Leeuwenhoek. 114, 1417–1429. doi: 10.1007/s10482-021-01591-x
Baker-Austin, C., and Oliver, J. D. (2020). Vibrio vulnificus. Trends Microbiol. 28, 81–82. doi: 10.1016/j.tim.2019.08.006
Bonny, S. Q., Hossain, M. A. M., Uddin, S. M. K., Pulingam, T., Sagadevan, S., and Johan, M. R. (2022). Current trends in polymerase chain reaction based detection of three major human pathogenic vibrios. Crit. Rev. Food Sci. Nutr. 62, 1317–1335. doi: 10.1080/10408398.2020.1841728
Brehm, T. T., Berneking, L., Sena Martins, M., Dupke, S., Jacob, D., Drechsel, O., et al. (2021). Heatwave-associated Vibrio infections in Germany, 2018 and 2019. Euro. Surveill. 26:2002041. doi: 10.2807/1560-7917.Es.2021.26.41.2002041
Bullington, J. A., Golder, A. R., Steward, G. F., McManus, M. A., Neuheimer, A. B., Glazer, B. T., et al. (2022). Refining real-time predictions of Vibrio vulnificus concentrations in a tropical urban estuary by incorporating dissolved organic matter dynamics. Sci Total Environ. 829:154075. doi: 10.1016/j.scitotenv.2022.154075
Cao, X., Zhao, L., Zhang, J., Chen, X., Shi, L., Fang, X., et al. (2019). Detection of viable but nonculturable Vibrio parahaemolyticus in shrimp samples using improved real-time PCR and real-time LAMP methods. Food Control 103, 145–152. doi: 10.1016/j.foodcont.2019.04.003
Cechova, M., Beinhauerova, M., Babak, V., Slana, I., and Kralik, P. (2021). A novel approach to the viability determination of Mycobacterium avium subsp. paratuberculosis using platinum compounds in combination with quantitative PCR. Front. Microbiol. 12:748337. doi: 10.3389/fmicb.2021.748337
Codony, F., Dinh-Thanh, M., and Agustí, G. (2020). Key factors for removing bias in viability PCR-Based methods: a review. Curr. Microbiol. 77, 682–687. doi: 10.1007/s00284-019-01829-y
Copin, S., Mougin, J., Raguenet, V., Robert-Pillot, A., Midelet, G., Grard, T., et al. (2021). Ethidium and propidium monoazide: comparison of potential toxicity on Vibrio sp. viability. Lett. Appl. Microbiol. 72, 245–250. doi: 10.1111/lam.13412
Deshmukh, R., Bhand, S., and Roy, U. (2020). A novel method for rapid and sensitive detection of viable Escherichia coli cells using UV-induced PMA-coupled quantitative PCR. Braz. J. Microbiol. 51, 773–778. doi: 10.1007/s42770-019-00161-8
D’Souza, C., Kumar, B. K., Kapinakadu, S., Shetty, R., Karunasagar, I., and Karunasagar, I. (2018). PCR-based evidence showing the presence of Vibrio vulnificus in wound infection cases in Mangaluru. India. Int. J. Infect. Dis. 68, 74–76. doi: 10.1016/j.ijid.2018.01.018
Elizaquível, P., Aznar, R., and Sánchez, G. (2014). Recent developments in the use of viability dyes and quantitative PCR in the food microbiology field. J. Appl. Microbiol. 116, 1–13. doi: 10.1111/jam.12365
Guan, H., Xue, P., Zhou, H., Sha, D., Wang, D., Gao, H., et al. (2021). A multiplex PCR assay for the detection of five human pathogenic Vibrio species and Plesiomonas. Mol. Cell Probes 55:101689. doi: 10.1016/j.mcp.2020.101689
Gyraite, G., Katarzyte, M., and Schernewski, G. (2019). First findings of potentially human pathogenic bacteria Vibrio in the south-eastern Baltic Sea coastal and transitional bathing waters. Mar. Pollut. Bull. 149:110546. doi: 10.1016/j.marpolbul.2019.110546
Hannet, I., Engsbro, A. L., Pareja, J., Schneider, U. V., Lisby, J. G., Pružinec-Popović, B., et al. (2019). Multicenter evaluation of the new QIAstat gastrointestinal panel for the rapid syndromic testing of acute gastroenteritis. Eur. J. Clin. Microbiol. Infect. Dis. 38, 2103–2112. doi: 10.1007/s10096-019-03646-4
Hartnell, R. E., Stockley, L., Keay, W., Rosec, J. P., Hervio-Heath, D., Van den Berg, H., et al. (2019). A pan-European ring trial to validate an international standard for detection of Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus in seafoods. Int. J. Food Microbiol. 288, 58–65. doi: 10.1016/j.ijfoodmicro.2018.02.008
Harwood, V. J., Gandhi, J. P., and Wright, A. C. (2004). Methods for isolation and confirmation of Vibrio vulnificus from oysters and environmental sources: a review. J. Microbiol. Methods 59, 301–316. doi: 10.1016/j.mimet.2004.08.001
He, R., Zheng, W., Long, J., Huang, Y., Liu, C., Wang, Q., et al. (2019). Vibrio vulnificus meningoencephalitis in a patient with thalassemia and a splenectomy. J. Neurovirol. 25, 127–132. doi: 10.1007/s13365-018-0675-8
Hong, W., Xiong, J., Nyaruaba, R., Li, J., Muturi, E., Liu, H., et al. (2021). Rapid determination of infectious SARS-CoV-2 in PCR-positive samples by SDS-PMA assisted RT-qPCR. Sci. Total Environ. 797:149085. doi: 10.1016/j.scitotenv.2021.149085
Jadeja, R., Janes, M. E., and Simonson, J. G. (2010). Immunomagnetic separation of Vibrio vulnificus with antiflagellar monoclonal antibody. J. Food Prot. 73, 1288–1293. doi: 10.4315/0362-028x-73.7.1288
Kim, J., and Chun, B. C. (2021). Effect of seawater temperature increase on the occurrence of coastal Vibrio vulnificus cases: korean national surveillance data from 2003 to 2016. Int. J. Environ. Res. Public Health 18:4439. doi: 10.3390/ijerph18094439
Kobayashi, H., Oethinger, M., Tuohy, M. J., Hall, G. S., and Bauer, T. W. (2010). Distinction between intact and antibiotic-inactivated bacteria by real-time PCR after treatment with propidium monoazide. J. Orthop. Res. 28, 1245–1251. doi: 10.1002/jor.21108
Lazou, T. P., Gelasakis, A. I., Chaintoutis, S. C., Iossifidou, E. G., and Dovas, C. I. (2021). Method-Dependent implications in foodborne pathogen quantification: the case of campylobacter coli survival on meat as comparatively assessed by colony count and viability PCR. Front. Microbiol. 12:604933. doi: 10.3389/fmicb.2021.604933
Lee, J. H., Park, S. K., Khan, F., Jo, D. M., Lee, D. H., Kang, M. G., et al. (2021a). Simultaneous isolation and enumeration of virulent Vibrio cholerae and Vibrio vulnificus using an advanced MPN-PCR method. Arch. Microbiol. 204:5. doi: 10.1007/s00203-021-02613-y
Lee, J. S., Ahn, J. J., Kim, S. J., Yu, S. Y., Koh, E. J., Kim, S. H., et al. (2021b). POCT detection of 14 respiratory viruses using multiplex RT-PCR. Biochip J. Online ahead of print. doi: 10.1007/s13206-021-00037-w
Lee, J. Y., Kim, S. W., Kim, D. M., Yun, N. R., Kim, C. M., and Lee, S. H. (2017). Clinical usefulness of real-time polymerase chain reaction for the diagnosis of Vibrio vulnificus infection using skin and soft tissues. Am. J. Trop. Med. Hyg. 97, 443–446. doi: 10.4269/ajtmh.16-0512
Lee, S. H., Lee, H. J., Myung, G. E., Choi, E. J., Kim, I. A., Jeong, Y. I., et al. (2019). Distribution of pathogenic Vibrio species in the coastal seawater of South Korea (2017-2018). Osong. Public Health Res. Perspect. 10, 337–342. doi: 10.24171/j.phrp.2019.10.6.03
Lei, S., Chen, S., and Zhong, Q. (2021). Digital PCR for accurate quantification of pathogens: principles, applications, challenges and future prospects. Int. J. Biol. Macromol. 184, 750–759. doi: 10.1016/j.ijbiomac.2021.06.132
Lei, S., Gu, X., Zhong, Q., Duan, L., and Zhou, A. (2020). Absolute quantification of Vibrio parahaemolyticus by multiplex droplet digital PCR for simultaneous detection of tlh, tdh and ureR based on single intact cell. Food Control 114:107207. doi: 10.1016/j.foodcont.2020.107207
Li, Z., Guan, H., Wang, W., Gao, H., Feng, W., Li, J., et al. (2021). Development of a rapid and fully automated multiplex real-time PCR assay for identification and differentiation of Vibrio cholerae and Vibrio parahaemolyticus on the BD MAX platform. Front. Cell Infect. Microbiol. 11:639473. doi: 10.3389/fcimb.2021.639473
Liang, J. H., Liang, W. H., Deng, Y. Q., Fu, Z. G., Deng, J. L., and Chen, Y. H. (2021). Vibrio vulnificus infection attributed to bee sting: a case report. Emerg. Microbes. Infect. 10, 1890–1895. doi: 10.1080/22221751.2021.1977589
Lin, I. C., Hussain, B., Hsu, B. M., Chen, J. S., Hsu, Y. L., Chiu, Y. C., et al. (2021). Prevalence, genetic diversity, antimicrobial resistance, and toxigenic profile of Vibrio vulnificus isolated from aquatic environments in Taiwan. Antibiotics (Basel) 10:505. doi: 10.3390/antibiotics10050505
Malayil, L., Chattopadhyay, S., Mongodin, E. F., and Sapkota, A. R. (2021). Coupled DNA-labeling and sequencing approach enables the detection of viable-but-non-culturable Vibrio spp. in irrigation water sources in the Chesapeake Bay watershed. Environ. Microbiome 16:13. doi: 10.1186/s40793-021-00382-1
Narendrakumar, L., Gopinathan, A., Sreekrishnan, T. P., Asokan, A., Kumar, A., Kumar, G., et al. (2021). The bane of coastal marine environment: a fatal case of Vibrio vulnificus associated cellulitis and septicaemia. Indian J. Med. Microbiol. 39, 386–388. doi: 10.1016/j.ijmmb.2021.05.016
Nocker, A., Cheung, C. Y., and Camper, A. K. (2006). Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67, 310–320. doi: 10.1016/j.mimet.2006.04.015
Nutz, S., Döll, K., and Karlovsky, P. (2011). Determination of the LOQ in real-time PCR by receiver operating characteristic curve analysis: application to qPCR assays for Fusarium verticillioides and F. proliferatum. Anal. Bioanal. Chem. 401, 717–726. doi: 10.1007/s00216-011-5089-x
Panicker, G., and Bej, A. K. (2005). Real-time PCR detection of Vibrio vulnificus in oysters: comparison of oligonucleotide primers and probes targeting vvhA. Appl. Environ. Microbiol. 71, 5702–5709. doi: 10.1128/aem.71.10.5702-5709.2005
Rengpipat, S., Pusiririt, S., and Rukpratanporn, S. (2008). Differentiating between isolates of Vibrio vulnificus with monoclonal antibodies. J. Microbiol. Methods 75, 398–404. doi: 10.1016/j.mimet.2008.07.015
Suo, T., Liu, X., Feng, J., Guo, M., Hu, W., Guo, D., et al. (2020). ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes. Infect. 9, 1259–1268. doi: 10.1080/22221751.2020.1772678
Tsai, Y. H., Chen, P. H., Yu, P. A., Chen, C. L., Kuo, L. T., and Huang, K. C. (2019). A multiplex PCR assay for detection of Vibrio vulnificus, Aeromonas hydrophila, methicillin-resistant Staphylococcus aureus, Streptococcus pyogenes, and Streptococcus agalactiae from the isolates of patients with necrotizing fasciitis. Int. J. Infect. Dis. 81, 73–80. doi: 10.1016/j.ijid.2019.01.037
Tsai, Y. H., Huang, T. Y., Kuo, L. T., Chuang, P. Y., Hsiao, C. T., and Huang, K. C. (2022). Comparison of surgical outcomes and predictors in patients with monomicrobial necrotizing fasciitis and sepsis caused by Vibrio vulnificus, Aeromonas hydrophila, and Aeromonas sobria. Surg. Infect. (Larchmt) 23, 288–297. doi: 10.1089/sur.2021.337
Wang, Y., Yan, Y., Thompson, K. N., Bae, S., Accorsi, E. K., Zhang, Y., et al. (2021). Whole microbial community viability is not quantitatively reflected by propidium monoazide sequencing approach. Microbiome 9:17. doi: 10.1186/s40168-020-00961-3
Wei, S., Zhao, H., Xian, Y., Hussain, M. A., and Wu, X. (2014). Multiplex PCR assays for the detection of Vibrio alginolyticus, Vibrio parahaemolyticus, Vibrio vulnificus, and Vibrio cholerae with an internal amplification control. Diagn. Microbiol. Infect. Dis. 79, 115–118. doi: 10.1016/j.diagmicrobio.2014.03.012
Wong, H. C., Liu, S. H., and Chen, M. Y. (2005). Virulence and stress susceptibility of clinical and environmental strains of Vibrio vulnificus isolated from samples from Taiwan and the United States. J. Food Prot. 68, 2533–2540. doi: 10.4315/0362-028x-68.12.2533
Wouters, Y., Dalloyaux, D., Christenhusz, A., Roelofs, H. M. J., Wertheim, H. F., Bleeker-Rovers, C. P., et al. (2020). Droplet digital polymerase chain reaction for rapid broad-spectrum detection of bloodstream infections. Microb. Biotechnol. 13, 657–668. doi: 10.1111/1751-7915.13491
Wu, B., Liang, W., and Kan, B. (2015). Enumeration of viable non-culturable Vibrio cholerae using propidium monoazide combined with quantitative PCR. J. Microbiol. Methods 115, 147–152. doi: 10.1016/j.mimet.2015.05.016
Xie, G., Yu, S., Li, W., Mu, D., Aguilar, Z. P., and Xu, H. (2020). Simultaneous detection of Salmonella spp., Pseudomonas aeruginosa, Bacillus cereus, and Escherichia coli O157:H7 in environmental water using PMA combined with mPCR. J. Microbiol. 58, 668–674. doi: 10.1007/s12275-020-0084-6
Yang, X., Zhang, X., Wang, Y., Shen, H., Jiang, G., Dong, J., et al. (2020). A real-time recombinase polymerase amplification method for rapid detection of vibrio Vulnificus in seafood. Front. Microbiol. 11:586981. doi: 10.3389/fmicb.2020.586981
Young, G. R., Smith, D. L., Embleton, N. D., Berrington, J. E., Schwalbe, E. C., Cummings, S. P., et al. (2017). Reducing viability bias in analysis of gut microbiota in preterm infants at risk of NEC and sepsis. Front. Cell Infect. Microbiol. 7:237. doi: 10.3389/fcimb.2017.00237
Yu, W., Shen, X., Pan, H., Xiao, T., Shen, P., and Xiao, Y. (2017). Clinical features and treatment of patients with Vibrio vulnificus infection. Int. J. Infect. Dis. 59, 1–6. doi: 10.1016/j.ijid.2017.03.017
Yuan, Y., Feng, Z., and Wang, J. (2020). Vibrio vulnificus hemolysin: biological activity, regulation of vvha expression, and role in pathogenesis. Front. Immunol. 11:599439. doi: 10.3389/fimmu.2020.599439
Yun, N. R., and Kim, D. M. (2018). Vibrio vulnificus infection: a persistent threat to public health. Korean J. Intern. Med. 33, 1070–1078. doi: 10.3904/kjim.2018.159
Zeng, D., Chen, S., Jiang, L., Ren, J., Ling, N., Su, J., et al. (2020). A polymerase chain reaction based lateral flow test strip with propidium monoazide for detection of viable Vibrio parahaemolyticus in codfish. Microchem. J. 159:105418. doi: 10.1016/j.microc.2020.105418
Zhang, L., Wang, M., Cong, D., Ding, S., Cong, R., Yue, J., et al. (2018). Rapid, specific and sensitive detection of Vibrio vulnificus by loop-mediated isothermal amplification targeted to vvhA gene. Acta. Oceanologica Sinica 37, 83–88. doi: 10.1007/s13131-018-1182-8
Zhang, S. J., Wang, L. L., Lu, S. Y., Hu, P., Li, Y. S., Zhang, Y., et al. (2020). A novel, rapid, and simple PMA-qPCR method for detection and counting of viable brucella organisms. J. Vet. Res. 64, 253–261. doi: 10.2478/jvetres-2020-0033
Zhao, L., Lv, X., Cao, X., Zhang, J., Gu, X., Zeng, H., et al. (2020). Improved quantitative detection of VBNC Vibrio parahaemolyticus using immunomagnetic separation and PMAxx-qPCR. Food Control 110:106962. doi: 10.1016/j.foodcont.2019.106962
Zhao, X., Zhong, J., Wei, C., Lin, C. W., and Ding, T. (2017). Current perspectives on viable but non-culturable state in foodborne pathogens. Front. Microbiol. 8:580. doi: 10.3389/fmicb.2017.00580
Keywords: Vibrio vulnificus, droplet digital PCR, propidium monoazide, vvhA gene, clinical, accurate detection
Citation: Hu L, Fu Y, Zhang S, Pan Z, Xia J, Zhu P and Guo J (2022) An Assay Combining Droplet Digital PCR With Propidium Monoazide Treatment for the Accurate Detection of Live Cells of Vibrio vulnificus in Plasma Samples. Front. Microbiol. 13:927285. doi: 10.3389/fmicb.2022.927285
Received: 24 April 2022; Accepted: 24 June 2022;
Published: 15 July 2022.
Edited by:
Jens Andre Hammerl, Bundesinstitut für Risikobewertung, GermanyReviewed by:
Ballamoole Krishna Kumar, Nitte University, IndiaMarília Miotto, Federal University of Santa Catarina, Brazil
Copyright © 2022 Hu, Fu, Zhang, Pan, Xia, Zhu and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Zhu, emh1cGVuZ0B1Y2FzLmFjLmNu; Jing Guo, Z3VvamluZ0BzZHUuZWR1LmNu
 Ling Hu
Ling Hu Yidong Fu3,4,5
Yidong Fu3,4,5 Shun Zhang
Shun Zhang Jing Guo
Jing Guo