- 1Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Guangdong-Hong Kong Joint Laboratory for RNA Medicine, RNA Biomedical Institute, Medical Research Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China
- 2Department of Obstetrics and Gynecology, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China
- 3Department of Biopharmaceutical Technology, School of Life Sciences, Guangzhou University, Guangzhou, China
- 4Breast Tumor Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China
Pseudouridine (Ψ) is one of the most abundant RNA modifications in cellular RNAs that post-transcriptionally impact many aspects of RNA. However, the metabolic fate of modified RNA nucleotides has long been a question. A pseudouridine kinase (PsuK) and a pseudouridine monophosphate glycosylase (PsuG) in Escherichia coli were first characterized as involved in pseudouridine degradation by catalyzing the phosphorylation of pseudouridine to pseudouridine 5′-phosphate (ΨMP) and further hydrolyzing 5′-ΨMP to produce uracil and ribose 5′-phosphate. Recently, their homolog proteins in eukaryotes were also identified, which were named PUKI and PUMY in Arabidopsis. Here, we solved the crystal structures of apo-EcPsuK and its binary complex with Ψ or N1-methyl-pseudouridine (m1Ψ). The structure of EcPsuK showed a homodimer conformation assembled by its β-thumb region. EcPsuK has an appropriate binding site with a series of hydrophilic and hydrophobic interactions for Ψ. Moreover, our complex structure of EcPsuK-m1Ψ suggested the binding pocket has an appropriate capacity for m1Ψ. We also identified the monovalent ion-binding site and potential ATP-binding site. Our studies improved the understanding of the mechanism of Ψ turnover.
Introduction
Post-transcriptional RNA modifications regulate various RNA species and influence gene expression (Barbieri and Kouzarides, 2020). More than 160 modifications in RNAs have been found until now (Boccaletto et al., 2018). Among them, N6-methylated adenine (m6A) and pseudouridine (Ψ) are the most prevalent naturally occurring modifications (Gilbert et al., 2016; Zaccara et al., 2019); furthermore, Ψ is also considered the first discovered RNA modification (Cohn and Volkin, 1951). As an isoform of uridine, Ψ has been detected in tRNAs, rRNAs, mRNAs, snoRNAs, and snRNAs existing in all three domains of life; therefore, Ψ is sometimes referred to as the fifth RNA nucleoside because of its ubiquitous nature (Zaringhalam and Papavasiliou, 2016; Lin et al., 2021). For instance, Ψs in tRNA molecules account for around 2-5% of all identified tRNA modifications (Lin et al., 2021). Conserved Ψ sites in rRNAs across different species are found to stabilize various local motifs (Sharma and Lafontaine, 2015). H/ACA box snoRNAs that mediate RNA pseudouridylation are known to carry Ψs themselves mostly in the regions involved in base pairing with target sites (Carlile et al., 2014). All snRNAs are predominantly modified with Ψs mainly located in the functionally important regions (Morais et al., 2021). Transcriptome-wide studies have also mapped many Ψ sites in mRNAs in yeast, human, and human pathogens (Carlile et al., 2014; Schwartz et al., 2014; Li et al., 2015; Nakamoto et al., 2017). In general, Ψs are considered to affect the RNA structure, further stabilize the structure of the functionally important areas, and tune ribosome functions for efficient and accurate protein translation (Schwartz et al., 2014). Interestingly, the incorporation of Ψ or its synthetic derivative N1-methyl-pseudouridine (m1Ψ) was validated to escape from degradation by ubiquitous RNases, significantly decreasing the immunogenic nature of mRNA vaccine and improving the antigen production (Kariko et al., 2005, 2008; Andries et al., 2015; Pardi et al., 2018).
RNA modifications are usually dynamically introduced and removed by specific enzymes (Zaccara et al., 2019). The formation of Ψ is catalyzed by the pseudouridine synthases (PUS), which can be further subcategorized into RNA guide-dependent and stand-alone enzyme modes (Ganot et al., 1997; Ni et al., 1997; Rintala-Dempsey and Kothe, 2017). The production of Ψ involves an isomerization process, in which the base reposition of uracil occurred by replacing the carbon–nitrogen glycosidic bond (C1–N1) with a carbon–carbon bond (C1–C5) (Veerareddygari et al., 2016; Motorin and Marchand, 2021), and then a free N1 position is exposed (Deb et al., 2019). The loss of Ψs by mutations in the pseudouridine synthases leads to several diseases, such as growth retardation (Han et al., 2015; Balogh et al., 2020), neuronal dysfunctions, behavior defects (de Brouwer et al., 2018), and Crohn’s disease (Festen et al., 2011).
Compared to the knowledge regarding the biogenesis and functions of RNA modifications, the metabolic fate of non-canonical nucleotides derived from the degradation of modified RNAs has just started. N6-methyl-adenosine monophosphate (N6-mAMP) produced from the metabolic turnover of m6A-containing RNAs was demonstrated to be degraded by an N6-mAMP-specific deaminase named ADAL that hydrolyzes N6-mAMP to inosine monophosphate (IMP), which is an intermediate of either purine nucleotide biosynthesis or catabolism in plants and human cells (Chen et al., 2018; Baccolini and Witte, 2019; Witte and Herde, 2020). Two subsequent structural studies validated the function of ADAL and identified the key residues in the active site that mediate the substrate specificity (Jia and Xie, 2019; Wu et al., 2019). Compared to N6-mAMP turnover, degradation of ΨMP was initially observed in the study with pyrimidine auxotrophic Escherichia coli mutants (Breitman, 1970). Detailed investigations of these mutants led to the discovery of YeiC and YeiN in E. coli, which were also known as pseudouridine kinase (PsuK) and pseudouridine-5′-phosphate glycosidase (PsuG) (Preumont et al., 2008), respectively. The catabolic process for Ψ consists of two steps: PsuK phosphorylates pseudouridine to 5′-ΨMP, and PsuG further hydrolyzes 5′-ΨMP, producing uracil and ribose 5′-phosphate (Figure 1A; Preumont et al., 2008; Chen and Witte, 2020). Recently, the homolog enzymes AtPUKI and AtPUMY in Arabidopsis were also described (Figure 1B; Chen and Witte, 2020). Like IMP, uracil may be reincorporated into uridine monophosphate in the salvage reaction or may enter pyrimidine ring catabolism (Loh et al., 2006; Zrenner et al., 2009). Malfunction of these catabolic enzymes was validated to be toxic and causes delayed seed germination and growth inhibition (Chen and Witte, 2020).
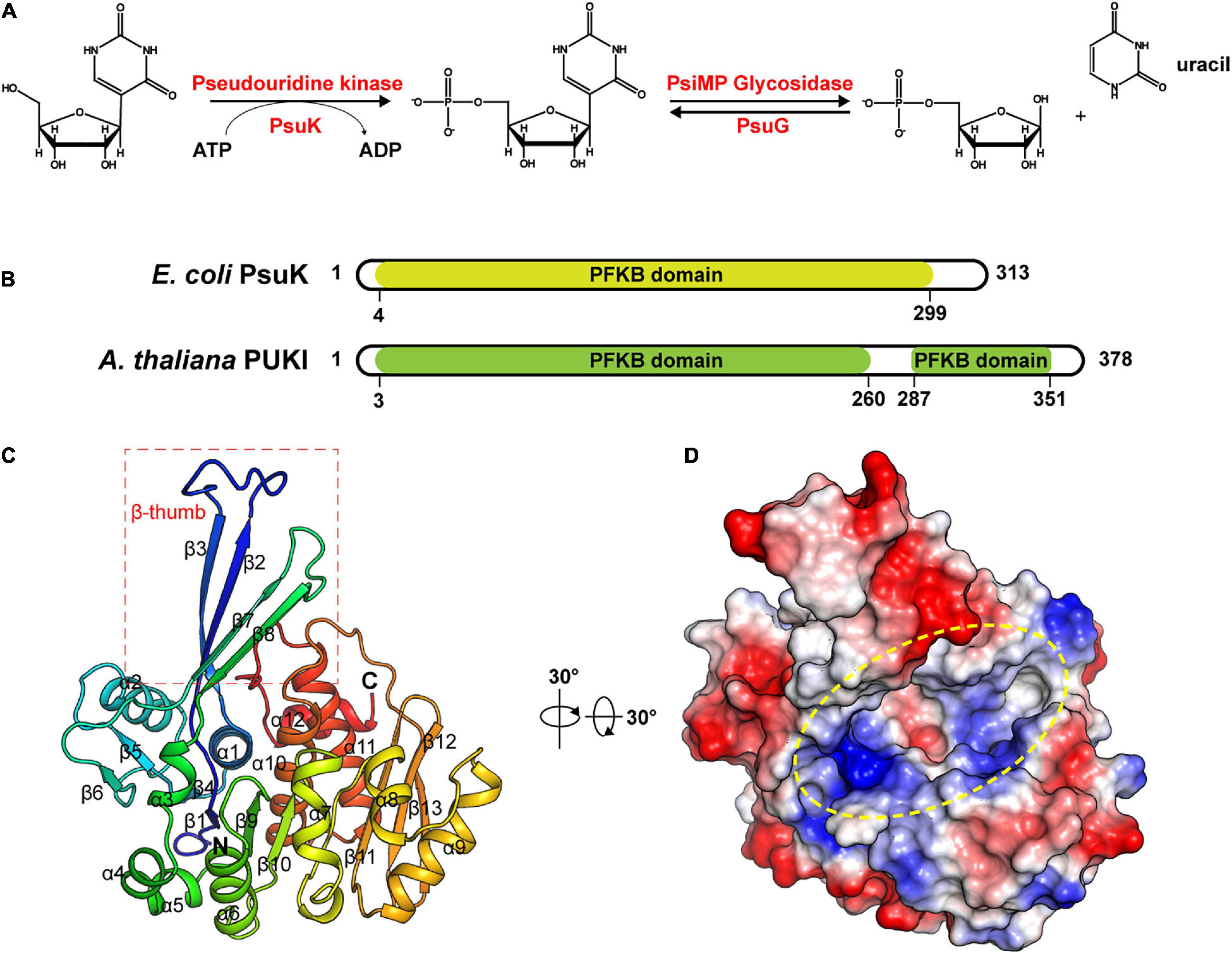
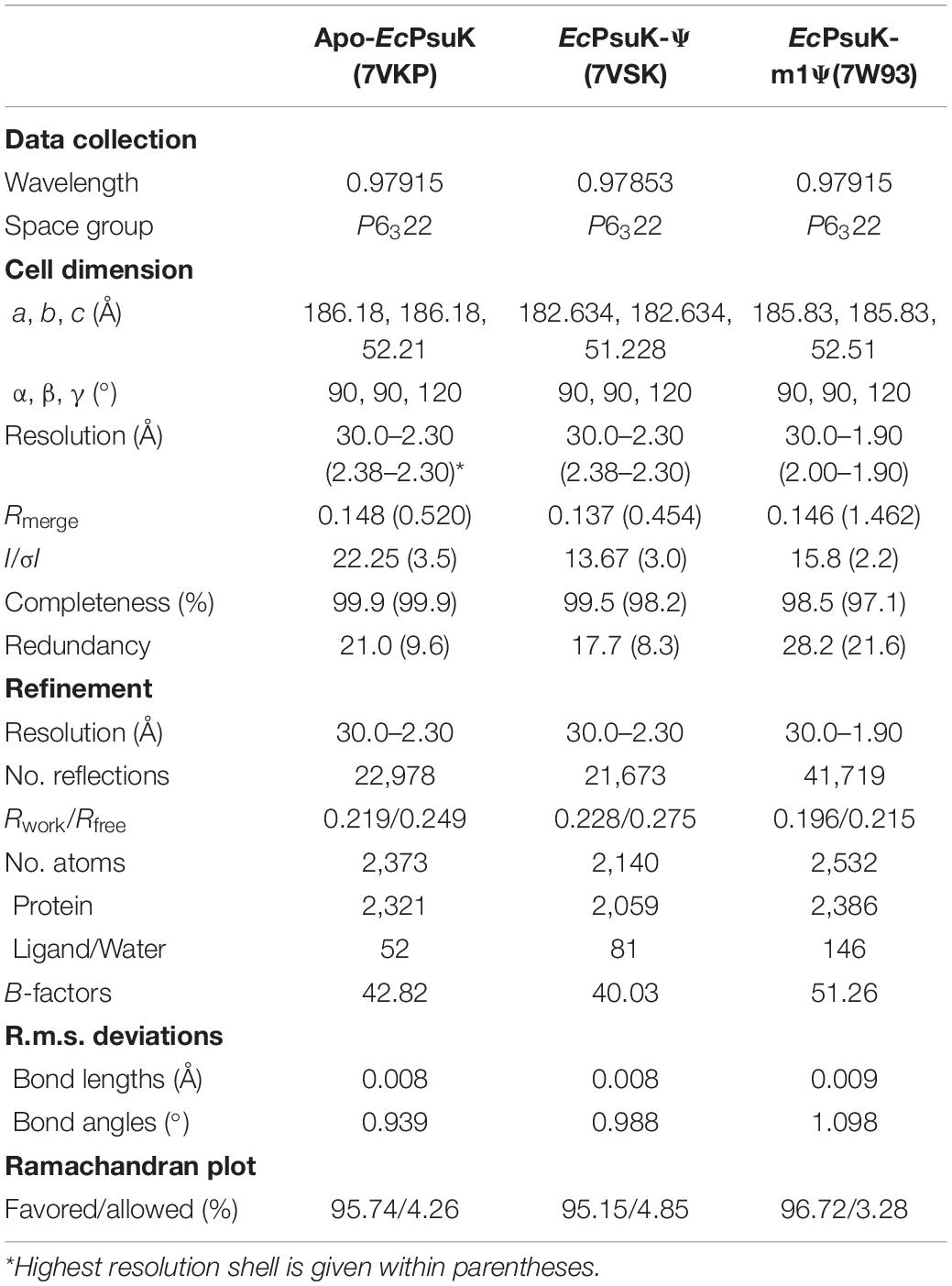
Figure 1. Structure of apo-EcPsuK. (A) Reaction scheme of pseudouridine metabolism. (B) Domain organization of Arabidopsis PUKI and E. coli PsuK. (C) Overall structure of EcPsuK is colored in the rainbow. β-Thumb region is indicated by a red box. (D) Surface representation of apo-EcPsuK, and the potential substrate-binding pocket is indicated by a yellow oval.
Intriguingly, PsuK and PsuG are present in many organisms from bacteria to eukaryotes (Preumont et al., 2008; Chen and Witte, 2020), whereas in metazoa, amoebozoa, and fungi, homologs of PsuG and PsuK reside on a single polypeptide chain, representing eukaryotic ΨMP glycosylase physically linked to pseudouridine kinase. By contrast, mammals generally lack these enzymes, except for platypus (Ornithorhynchus anatinus) (Chen and Witte, 2020). Previous biochemical and structural observations for AtPUKI showed its high specificity toward pseudouridine (Chen and Witte, 2020; Kim et al., 2021) and illustrated how AtPUKI discriminates pseudouridine from other structurally similar pyrimidine nucleosides or derivates. Nevertheless, although EcPsuK and its homolog protein AtPUKI both belong to the phosphofructokinase B (PfkB) family of carbohydrate kinases, they share a low sequence identity of about 21% (Park and Gupta, 2008; Chen and Witte, 2020). The specific catalytic mechanism for EcPsuK to Ψ has just begun to be uncovered; a residue Ser30 was suggested to play a key role in promoting the catalytic reaction by inducing the conformational change in this specific kinase (Kim et al., 2022).
In this study, we determined crystal structures of E. coli PsuK in apo-form and its binary complex with Ψ or m1Ψ. Our results provide a structural rationale for the high preference of EcPsuK for the non-canonical nucleoside pseudouridine and N1-methyl-pseudouridine. Furthermore, the undiscovered side effect of Ψ-containing RNAs appears strikingly advantageous for the development of generations of mRNA-based vaccines. Our studies put forward a hypothesis for the nucleoside-modified mRNA vaccine degradation pathway.
Results
Structure of Apo-EcPsuK
To unveil the catalytic mechanism of EcPsuK for the pseudouridine substrate, we first obtained the crystal structure of apo-EcPsuK; the crystal of apo-EcPsuK was determined at 2.3Å which belonged to the space group P6322. Detailed diffraction statistics can be found in Table 1. There is one EcPsuK molecule in the asymmetric unit, which can be modeled from Arg2 to Asn308 and folded into a conformation employing the β-α unit as a basic structural motif composed of a central α/β region, and a β-stranded region protruding from the N-terminal (Figure 1C). In detail, the interlaced β-α units in the α/β domain prompt the formation of a central β-sheet, which contains eight β-strands that are positioned in the order of β6-β5(β4)-β1-β9-β10-β11-β12-β13 with a parallel orientation, except for β12; seven α-helices (α3-α9) were further arranged on one side of the central β-sheet and the remaining α-helices (α1, α2, α10–α12) on the other side. Furthermore, two β-strands β2 and β3 between the β1–α1, β7, and β8 between the β6–α3 extend from the central α/β structure region. These two consecutive β-strands are corporately constituting the antiparallel β-sheet in an edge-to-edge orientation (hereafter named β-thumb region) (Figure 1C). These structural conformations of EcPsuK present a groove alongside the β-thumb region above the central β-sheet that is full of charged residues (Figure 1D).
Dimeric Structure of EcPsuK
The calculated molecular weight of monomeric EcPsuK (aa. 1–313) is 33.6 kDa, but the purified protein had a molecular weight of ∼67 kDa determined by size exclusion chromatography (SEC), indicating that it is a dimer in solution (Supplementary Figure 1A). Consistently, our structure also showed that EcPsuK was a dimer when we analyzed the adjacent asymmetric unit (Figure 2A). Therefore, although there is only one EcPsuK molecule in the asymmetric unit, it is a homodimer. Dimerization of EcPsuK is mainly assembled by two regions including the β-thumb region and α2 on the edge of the central β-sheet (Figures 2A,B). The dimer interface adopts a face-to-face mode that looks like a butterfly with the four β-strands in the β-thumb region almost perpendicular to these elements from the adjacent asymmetric unit, which created a cross β-barrel-like fold (Figure 2A; Sigrell et al., 1998).
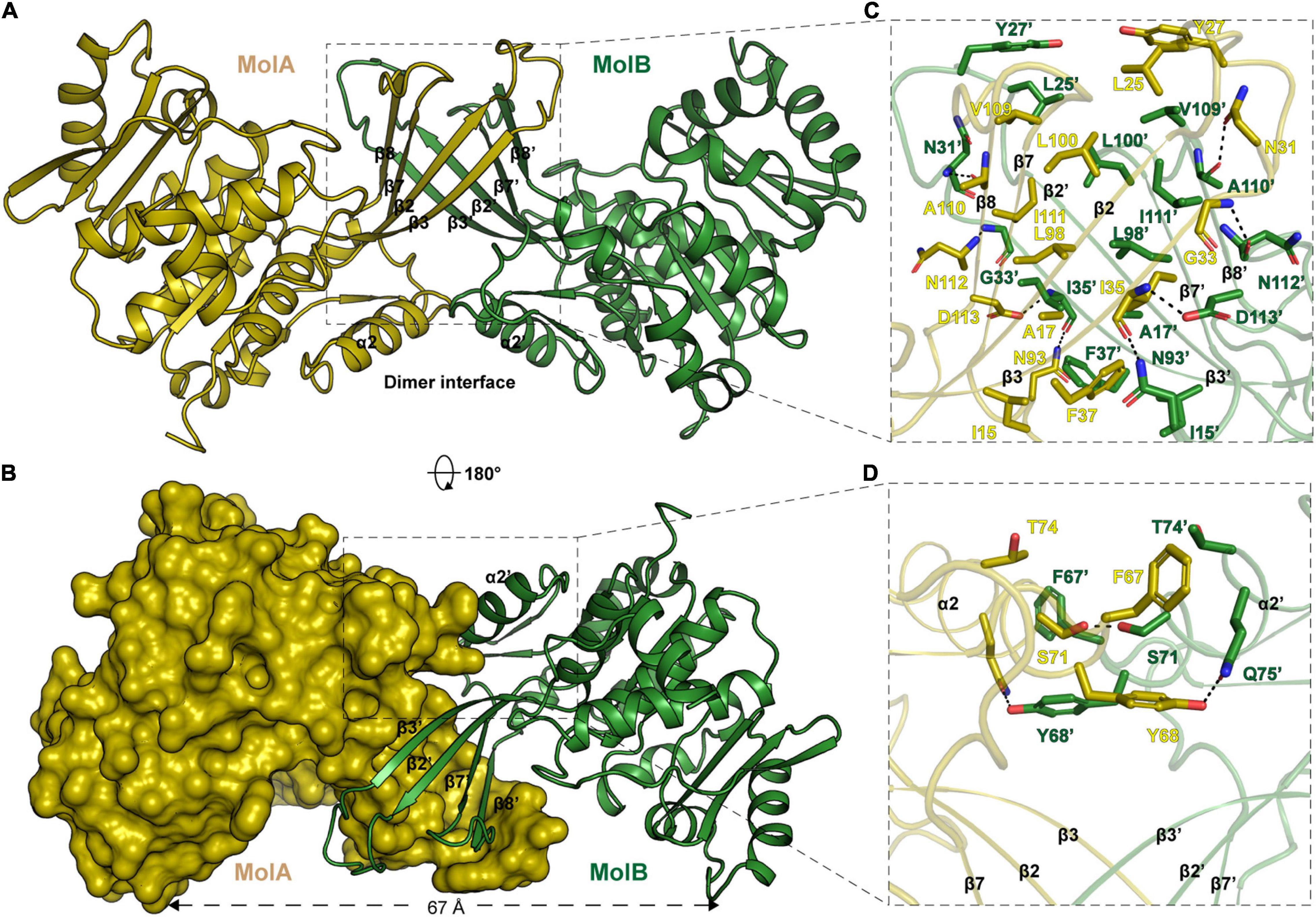
Figure 2. Structure of EcPsuK homodimer. (A) Dimeric structure of EcPsuK is shown as the cartoon; the two protomers are colored yellow and green. The dimer interface is indicated by a black box. (B) Surface representation of the dimer interface. (C) Detailed interactions for dimerization within the β-thumb region. The hydrogen bonds are shown as black dashed lines. (D) Detailed interactions between α2 and α2′. The hydrogen bonds are shown as black dashed lines.
In detail, the β-thumb region of each monomer of the EcPsuK homodimer rigidly contacts with each other through many hydrophobic interactions, creating a hydrophobic core (Figure 2C). The residues involved in the contacts contain Ile15, Ala17, Leu25, Tyr27, Asn31, Gly33, Ile35, Phe37, Leu98, Leu100, Val109, Ala110, and Ile111 with a face-to-face mode to their counterparts from another molecule. Hydrophilic interactions are also observed in this β-barrel-like region, the side chain of Asn31 forms a H-bond with the main chain of Ala110′, and the main chain of Ile35 is synchronously hydrogen-bonded to the side chain of Asp113′ and Asn93′ (Figure 2C). In the α2-α2′ region, two aromatic residues Phe67 and Tyr68 in the center account for the hydrophobic interactions together with Thr74; meanwhile, the hydroxyl group of Tyr68 interacts with the side chain of Gln75′ via a hydrogen bond, and the side chain of serine interacts with its counterparts of the other molecule (Figure 2D). Taken together, these interactions anchored the EcPsuK homodimer.
The Binary Complex of EcPsuK-Ψ
To further illustrate the substrate binding properties of EcPsuK with pseudouridine, we determined the crystal structure of EcPsuK complexed with pseudouridine through co-crystallization, and further soaking of the crystals with both pseudouridine and ADP. We successfully obtained the complex structure, which also belongs to the space group P6322, and diffracted it to 2.30 Å (Table 1). However, we can only achieve electron density for pseudouridine, and the ADP cannot be modeled (Figure 3A); thus, we named the structure of the binary complex EcPsuK-Ψ. The superposition of both the monomeric and dimeric structures of the EcPsuK-Ψ complex with apo-EcPsuK revealed minor conformational differences (r.m.s.d about 0.295 Å) (Supplementary Figure 1B). However, a loop region (Ile244 to Gly253) and the C-terminal (Met285 to Asn308) are lost in this binary structure, which is discussed later.
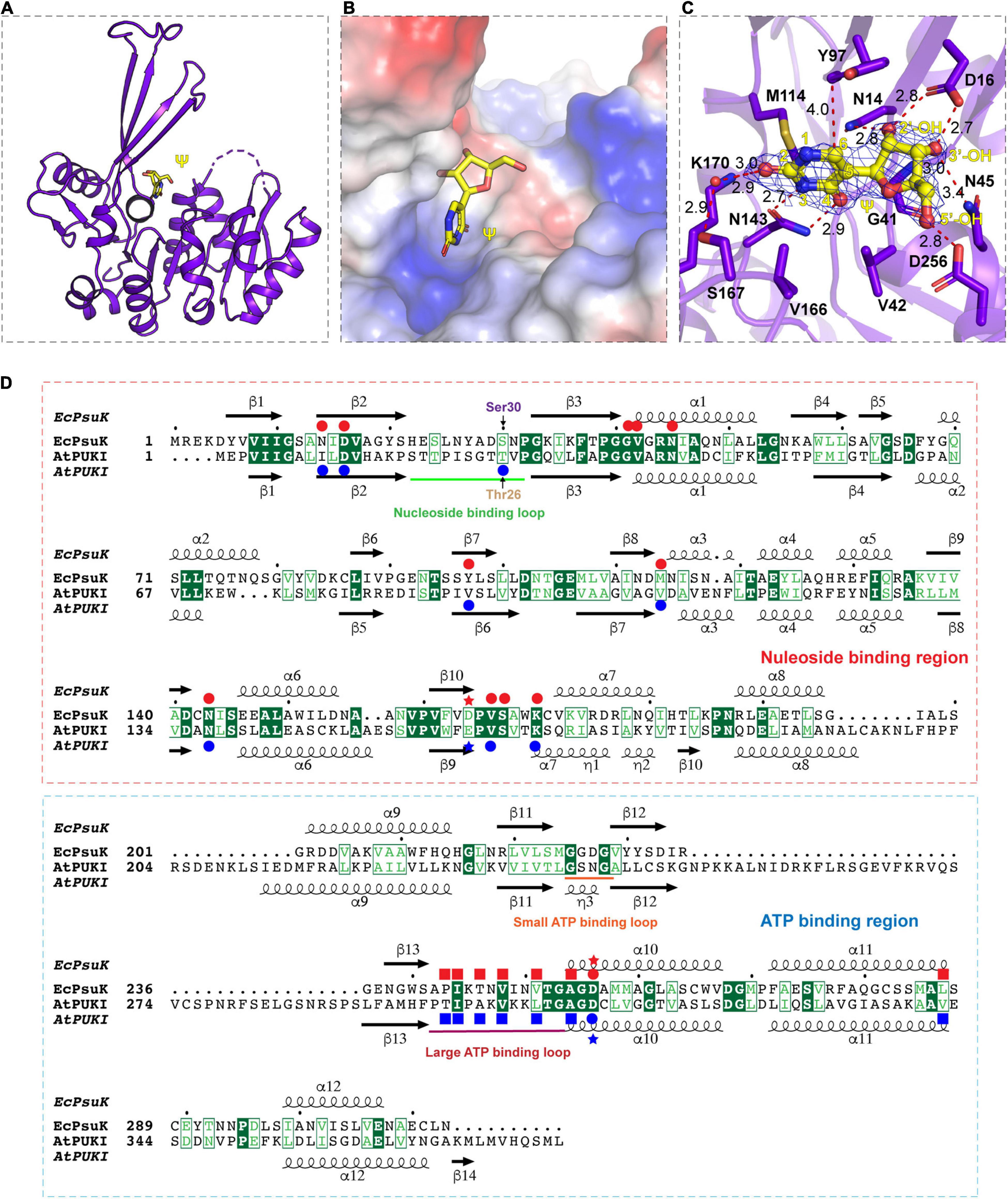
Figure 3. Complex structure of EcPsuK with Ψ. (A) Overall structure of EcPsuK in complex with Ψ. EcPsuK is shown as cartoon and colored in purple, and Ψ is shown as the stick and colored in yellow. (B) Substrate-binding surface underneath the β-thumb region and above the α/β fold region. (C) Detailed interactions between EcPsuK and Ψ. EcPsuK is shown as cartoon and colored in purple, the residues in contact with Ψ are shown as sticks, and Ψ is shown as stick colored in yellow. The hydrogen bonds are shown as red dashed lines. The red sphere represents the water molecule. The 2| Fo| –| Fc| σ-weighted map is contoured at 1.5σ. (D) Structure-based sequence alignment between AtPUKI and EcPsuK. The residues involved in substrate binding are indicated by red (EcPsuK) or blue (AtPUKI) circles, respectively. The residues involved in ATP binding are indicated by blue squares for AtPUKI, and the red squares indicated the potential residues in EcPsuK involved in ATP binding. The nucleoside-binding region and the ATP-binding region are indicated by red and blue rectangles, respectively. The red and blue stars indicate the residues that may involve in catalytic reaction.
In the EcPsuK-Ψ complex, pseudouridine is well accommodated at the cleft alongside the β-thumb domain, with the nucleobase inserted into the pocket and the 5′-OH of the ribose group points to the orientation of the unoccupied region in the cleft (Figure 3B). The base of pseudouridine is well recognized by a number of hydrophobic and hydrophilic interactions with good densities (Figures 3C,D and Supplementary Figure 1C). For instance, the O2 position hydrogen binds to the side chain of K170 and the side chain of Ser167 mediated by a water molecule, respectively. The N3 and O4 positions are bound to the side chain of Asn143. The base is also well clamped by the side chain of Met114, Val166, and Tyr97, which forms a hydrophobic environment with the nucleobase of pseudouridine. The ribose group is also well anchored by hydrogen contacts from Asn14, Asp16, and Asn45 (Figures 3C,D). The 2′-OH forms hydrogen binds with the side chain NH2 group of Asn14 and the side chain of Asp16; similarly, the 3′-OH of pseudouridine is hydrogen-bonded to the side chain Asp16, main chain amino group of Gly41, and the side chain of Asn45. The 5′-OH position forms a hydrogen bond with the side chain of Asp256, which is considered one of the residues involved in catalyzing the reaction (Figure 3D, indicated by asterisks) (Kang et al., 2019; Kim et al., 2021). Furthermore, Gly41 and Val42 hydrophobically interact with the ribose ring to stabilize the conformation of pseudouridine. It is interesting to find that the residues involved in the recognition of the pseudouridine are mainly located in the N-terminal part of the EcPsuK end to α7, except for the Asp256 (Figure 3D), and the following region is a loop between α7 and α8 with good density; moreover, these residues are very conserved in AtPUKI (Figure 3D; Kim et al., 2021). Taken together, the structure of the EcPsuK-Ψ binary complex suggested that pseudouridine is specifically recognized by EcPsuK mainly by the N-terminal half.
Structure Comparisons of EcPsuK With AtPUKI
To further dissect the catalytic mechanism of EcPsuK to the substrate, we compare the structures of apo-EcPsuK and EcPsuK-Ψ with the AtPUKI-Ψ-ADP complex (PDB code:7C1Y) (Kim et al., 2021). Although AtPUKI has an extra 65 residues length longer than EcPsuK (Figure 1B), which has 313 residues, the superposition of monomeric apo-EcPsuK to AtPUKI monomer showed quite a minor difference with r.m.s.d about 1.263 Å (Supplementary Figure 2A). There are two insertions in AtPUKI when compared with EcPsuK; they are the loop region between α8 and α9 and a long-disordered region between β12 and β13 (Figure 3D). However, according to the complex structure of AtPUKI-Ψ-ADP, the homodimeric structure of AtPUKI adopts a transition mode to catalyze the reaction and leads to the differences in each protomer of AtPUKI (Kim et al., 2021). Consistent with these observations, the superposition is quite different between dimeric EcPsuK and AtPUKI, when one molecule is superimposed well, and the other one has a high deviation (Supplementary Figure 2B). We then superimposed the structure of apo-EcPsuK with another protomer of AtPUKI, and the results showed that the r.m.s.d is 1.988 Å with the β-thumb region of AtPUKI closer to the central α/β region, which is also observed when we compared the complex structure of EcPsuK-Ψ with the same protomer of AtPUKI (Figure 4A and Supplementary Figures 2C,D). This set of AtPUKI can represent the dimeric structure status of the apo-state because the overall conformation of all the solved AtPUKI structures has the same dimeric structure, and these two molecules represent the different catalytic-associated states (Supplementary Figure 2E) (Kim et al., 2021).
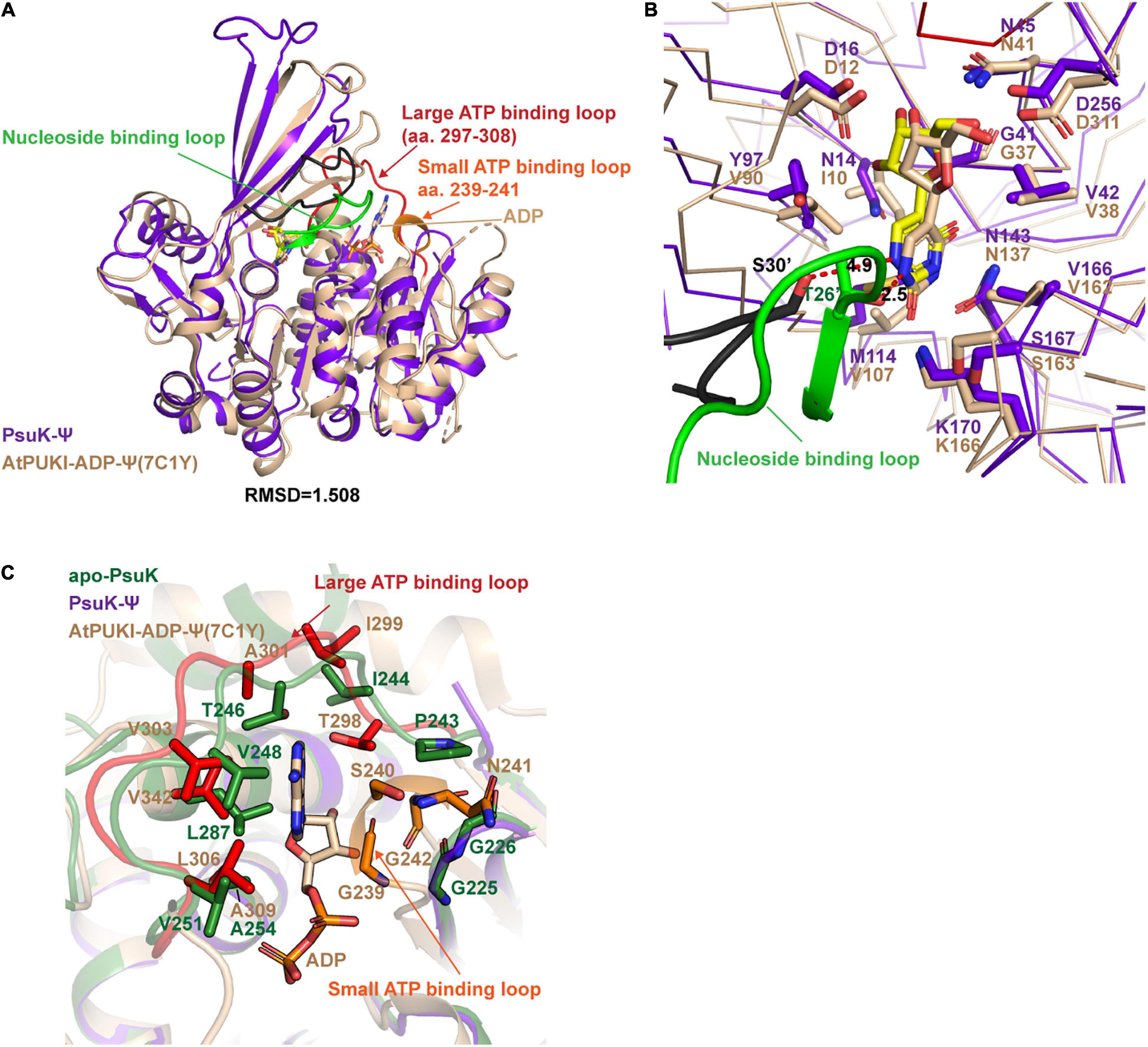
Figure 4. Structure comparisons between EcPsuK and AtPUKI. (A) Structure superposition of the EcPsuK-Ψ complex with the AtPUKI-ADP-Ψ complex (PDB code: 7C1Y). The nucleoside-binding loop, ATP-binding loop, and ADP in AtPUKI are all indicated by arrows. (B) Comparisons of the substrate-binding pocket between EcPsuK and AtPUKI. The nucleoside-binding loop containing T26′ is colored green. The residues involved in substrate recognition are shown as stick. The black-colored loop is the corresponding nucleoside-binding loop in EcPsuK, and the residue S30’ is colored in black and shown as stick. (C) Detailed comparisons of the ATP-binding site between EcPsuK and AtPUKI. The residues used for binding ADP in AtPUKI, and the corresponding residues in EcPsuK are all shown as stick.
We further compared the structure of EcPsuK-Ψ with the active state of the AtPUKI-Ψ-ADP complex in detail, which showed an r.m.s.d. about 1.508 Å (Figure 4A). The substrate cleft in the AtPUKI-Ψ-ADP complex contains a Ψ and an ADP molecule. A comparison of the pseudouridine recognition revealed that almost all the residues involved in specific interactions of the pseudouridine are well conserved with EcPsuK, although with some substitution such as Val90 in AtPUKI but a Tyr97 in EcPsuK; Ile10 and Val107 in AtPUKI are corresponding to Asn14 and Met114 of EcPsuK, respectively; these substitutions will not impact the substrate binding properties (Figures 3D, 4B). Importantly, a key residue Thr26’ located in the so-called nucleoside-binding loop in the dimeric AtPUKI plays a key role in recognizing the pseudouridine because the side chain hydroxyl group of Thr26’ can form a 2.5 Å hydrogen bond with the N1 position of pseudouridine (Kim et al., 2021), whereas in EcPsuK, it is a Ser30’ with 4.9 Å to the N1 position (Figure 4B). Previous studies showed that the T26S mutant of AtPUKI has similar Km and kcat values for the pseudouridine, like that of the wild-type AtPUKI (Kim et al., 2021). These analyses suggested that the Ser30’ in EcPsuK has the potential to bind the N1 position of pseudouridine and further improve the specificity of the substrate.
The ADP is bound by two so-called ATP-binding loops including the large ATP-binding loop and the small ATP-binding loop in AtPUKI (Kim et al., 2021) (Figures 3D, 4C). However, in our binary complex structure of EcPsuK-Ψ, the large ATP-binding loops have been lost and without any electron density of ADP (Figure 3A). The two regions, the loop (Ile244 to Gly253) and the C-terminal (Met285 to Asn308) of EcPsuK, are corresponding to the region wherein AtPUKI was observed to bind to the monovalent ion. Although we failed to obtain the complex structure of EcPsuK with ADP, we compared the ATP-binding pocket of EcPsuK to AtPUKI with the apo-EcPsuK structure. Combined with sequence alignment, the ATP-binding region is also conserved within the large ATP-binding loop full of hydrophobic residues to stack contact with the nucleobase, and the sequence of the small ATP-binding loop showed a conserved “GXXG” motif (Figures 3D, 4C). Whereas the small ATP-binding loop in AtPUKI has a much more rigid conformation for the nucleobase of ATP accommodation than that in EcPsuK, these differences may result from the existence of ADP, which pushes the “GXXG” motif closer to the nucleobase in AtPUKI (Figure 4C). Compared to the pseudouridine-binding site, the residues involved in ATP binding are all located in the C-terminal part of EcPsuK; thus, the α/β domain may also be divided into two parts in EcPsuK and AtPUKI, which can be defined as the nucleotide-binding region and ATP-binding region (Figure 3D).
The Complex Structure of EcPsuK With N1-Methyl-Pseudouridine (m1Ψ)
During the crystallization process of the EcPsuK-Ψ complex, we used the gel filtration buffer containing 10mM Tris pH 8.0 and 100mM NaCl to purify the EcPsuK protein. The disorder of the loop region (Ile244 to Gly253) and the C-terminal (Met285 to Asn308) may be attributed to the weak interactions of sodium with EcPsuK for anchoring these regions in the presence of ADP. Therefore, we changed the gel filtration buffer to 10mM Tris pH 8.0 and 100mM KCl, which contains the substituted monovalent ion potassium to purify the EcPsuK protein. Furthermore, compared to the Ser26’ in AtPUKI structures that are directly involved in the binding to the N1 position of pseudouridine (Kim et al., 2021), Ser30’ in EcPsuK is much far from the N1 position of pseudouridine observed in our EcPsuK-Ψ complex (Figure 4B). Despite it may impact the transition status for catalytic reaction, we aimed to understand if EcPsuK has the binding ability to N1-substituted pseudouridine, one such example is m1Ψ, in which the hydrogen of the N1 position of pseudouridine is substituted by a methyl group. Previous studies tested many nucleotide analogs of Ψ such as 5-methyl uridine (Chen and Witte, 2020; Kim et al., 2021); however, there is still no evidence of the binding ability of EcPsuK to Ψ derivates.
To test this hypothesis, we then co-crystallized EcPsuK with m1Ψ in the presence of potassium and ADP. We then obtained the complex structure of EcPsuK with m1Ψ with higher resolution at 1.90 Å (Table 1); however, the density of ADP still could not be found in this complex structure. Compared to the EcPsuK-Ψ complex with an r.m.s.d only 0.285, in this EcPsuK-m1Ψ structure, the lost regions can be well modeled (Figure 5A). The density of K+ can be achieved due to the high resolution and bound by the main chain of surrounding residues including Asn250, Thr252, Ala286, Cys289, and Tyr291 (Figure 5B). We further analyzed the binding environment of m1Ψ with good densities (Supplementary Figure 2F), and the comparisons between Ψ and m1Ψ in the substrate-binding pocket revealed that the interactions between EcPsuK and m1Ψ are a little more stringent than those in EcPsuK-Ψ (Figures 3C, 5C). Without major conformational changes, Tyr97 donates more hydrophobic interactions with the N1-methyl group in m1Ψ, and the pairing of Lys170 and Asn143 with m1Ψ is more intensive than that with Ψ (Figure 5C, Supplementary Figures 1C,2E). Meanwhile, the side chain hydroxyl group showed a 3.9 Å distance from the N1-methyl group (Figure 5C). These observations suggested that EcPsuK has the binding capacity for the substrate of N1-substituted pseudouridine.
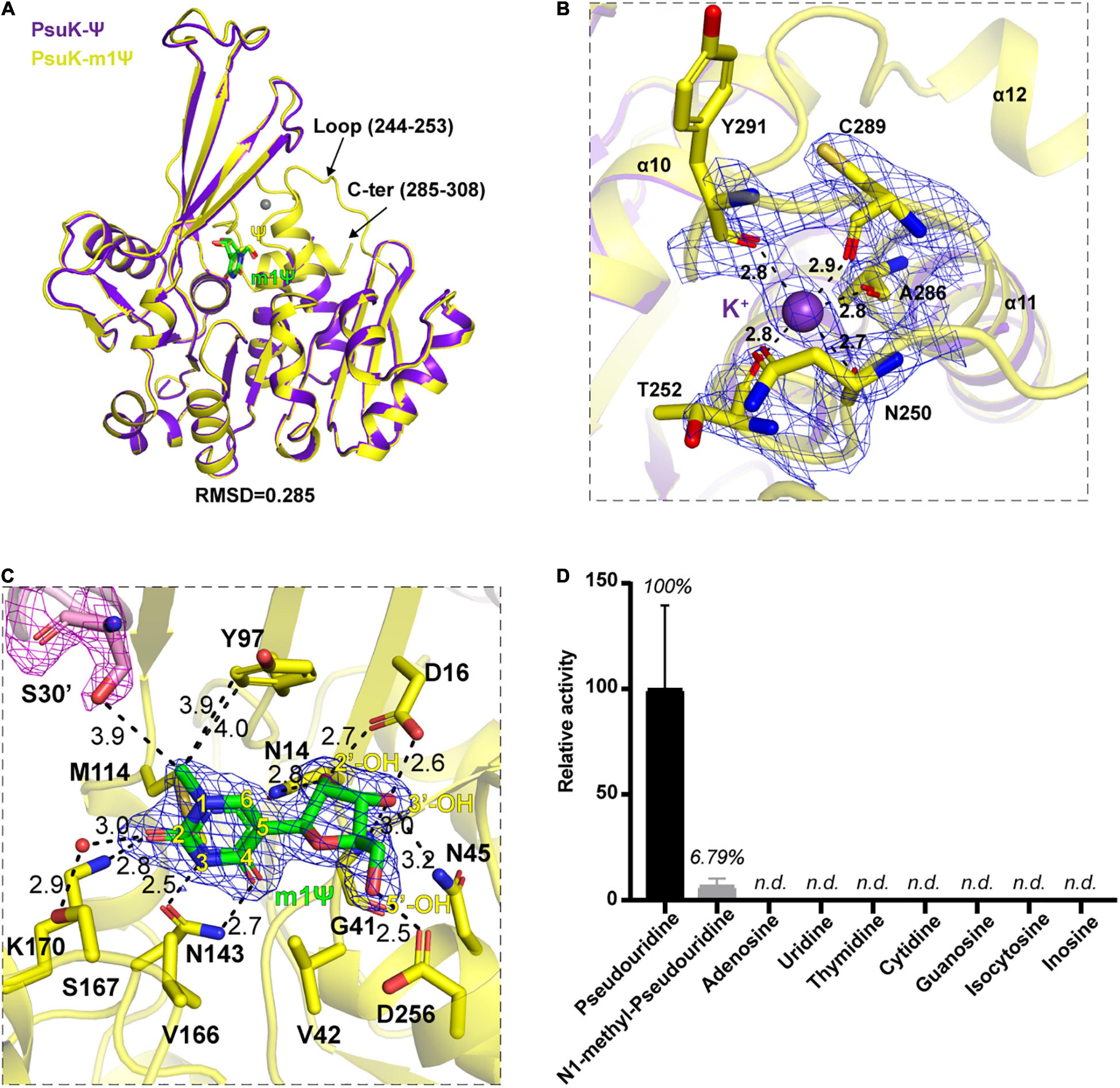
Figure 5. Structure complex of EcPsuK with m1Ψ. (A) Structure superposition of EcPsuK-m1Ψ and EcPsuK-Ψ. m1Ψ is colored in green, and Ψ is colored in yellow. The lost regions in the EcPsuK-Ψ complex are indicated by black arrows. (B) Binding site of a monovalent ion. The 2| Fo| –| Fc| σ-weighted map is contoured at 1.5σ. K+ is colored purple shown as a sphere. (C) Detailed interactions between EcPsuK and m1Ψ. EcPsuK is shown as cartoon and colored in yellow, the residues in contact with Ψ are shown as sticks, and m1Ψ is shown as stick colored in green. The hydrogen bonds are shown as black dashed lines. The red sphere represents the water molecule. The 2| Fo| –| Fc| σ-weighted map is contoured at 1.5σ. (D) Substrate specificity of EcPsuK for pseudouridine and other nucleoside analogs. n.d., not detected.
We then compared the catalytic activity of EcPsuK to Ψ, m1Ψ, and many other nucleoside analogs by a direct activity assay method (Andersson and Mowbray, 2002). The assay results showed that EcPsuK kept a weak activity of about 6.79% toward the m1Ψ substrate relative to the Ψ substrate; nevertheless, EcPsuK revealed no catalytic activity to the other nucleoside analogs including adenosine, uridine, thymidine, cytidine, guanosine, isocytosine, and inosine (Figure 5D). Taken together, these results suggested that although m1Ψ can be bound by EcPsuK just as our structure showed, it is not the most suitable substrate for EcPsuK.
Structural Homologs of EcPsuK
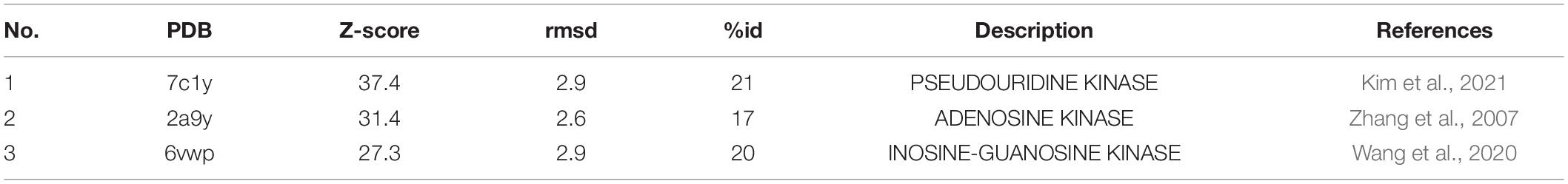
EcPsuK is a member of the PfkB family (Park and Gupta, 2008); accordingly, structural features of monomeric and dimeric EcPsuK are also highly homologous to those of this protein family. A structure similarity search using the program DALI (Holm, 2020) indicated that the EcPsuK monomer exhibits high structural homology with the PfkB family kinases catalyzing the phosphorylation of ribose. Based on the DALI search results, we selected the published structures with nucleoside kinase activity to further analyze the similarities and differences among the nucleoside kinase of the PfkB family (Table 2). They are two known function nucleoside kinases containing the adenosine kinase from Toxoplasma gondii (TgAK, PDB code: 2A9Y) and inosine–guanosine kinase from E. coli K12 (Gsk, PDB: 6VWP) (Figures 6A–C). DALI results revealed that the EcPsuK and TgAK share only 17% sequence identity, but with the structural homology of about 4.194 Å, and the EcPsuK and EcGsk share about 20% sequence identity with 5.393 Å (Figures 6B,C and Supplementary Figure 3). The TgAK presents as a monomeric conformation that contains a nucleotide-binding pocket essentially at an equivalent location to where it is found in EcPsuK and AtPUKI (Figure 4A). Compared with EcPsuK, the pocket of TgAK is larger to allow the accommodation of a nucleoside with a purine base (Figures 6D,E). In the complex structure of TgAK with N6,N6-dimethyladenosine (DMA), the nucleobase-binding part of the pocket lacks hydrophilic residues that could mediate specific hydrogen bonds to the adenine moiety (Figure 6E). By contrast, guanosine–inosine kinase (Gsk) protein can recognize guanine with high specificity, in which the nucleobase is specifically bound via many hydrophilic interactions (Figure 6F).
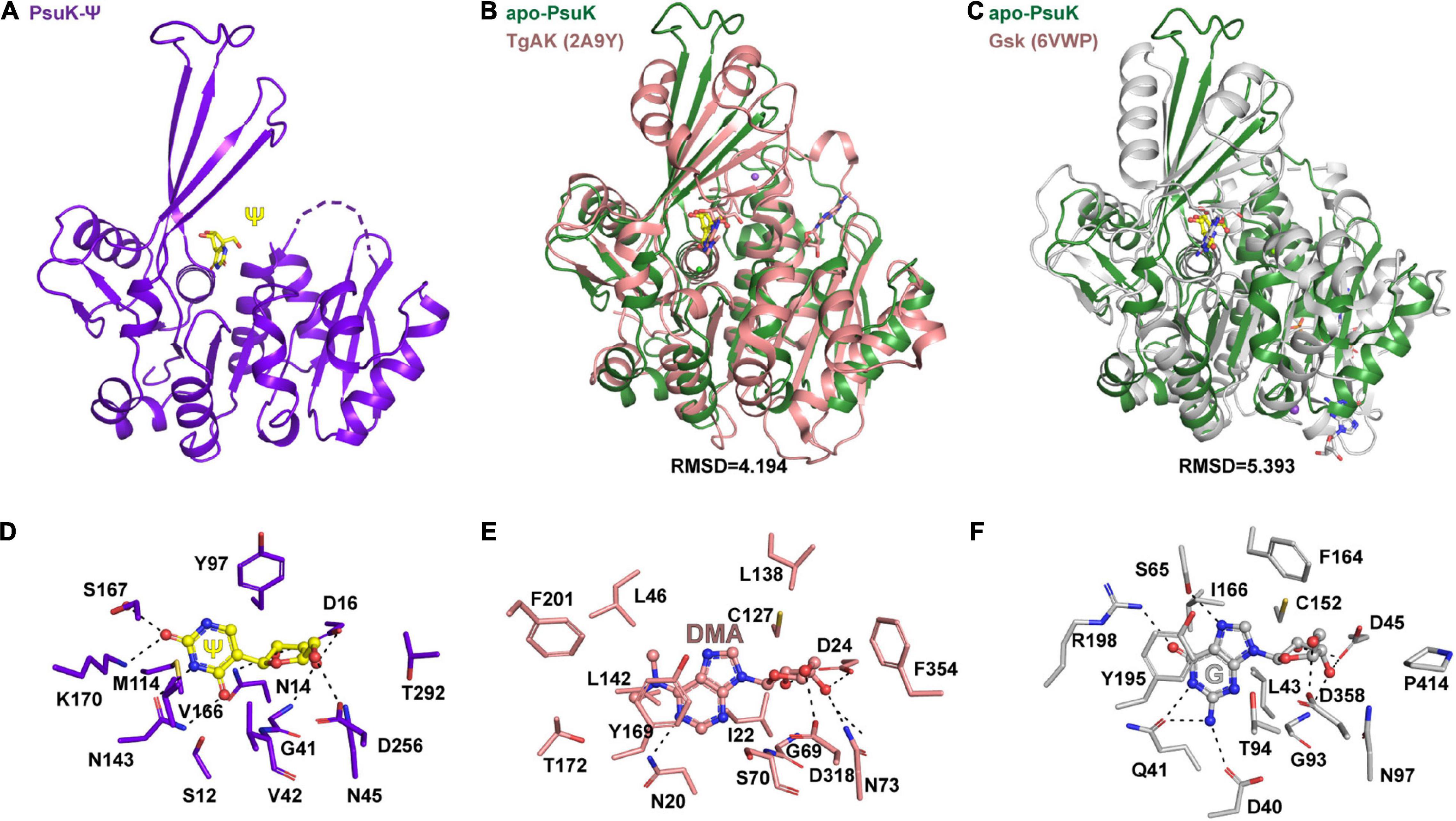
Figure 6. Comparisons of substrate binding properties of EcPsuK with TgAK and Gsk. (A) The overall structure of EcPsuK in complex with Ψ. (B) Structure superposition of EcPsuK-Ψ with TgAK-DMA complex (PDB code: 2A9Y). The latter complex structure is colored in pink. (C) Structure superposition of EcPsuK-Ψ with Gsk-guanosine complex (PDB code: 6VWP). The latter complex structure is colored in white. (D) Detailed interactions between PusK with Ψ. (E) Detailed interactions between TgAK with DMA (N6, N6-dimethyladenosine). (F) Detailed interactions between Gsk with guanosine.
Conserved Substrate-Binding Site in the Eukaryotic PsuG-PsuK Fusion Proteins
In some eukaryotic organisms, the putative enzymes responsible for the catabolism of pseudouridine are physically fused to a polypeptide (Chen and Witte, 2020). We then performed a multiple sequence alignment with AtPUKI, AtPUMY, EcPsuK, EcPsuG, and their potential homolog proteins in yeast (Schizosaccharomyces pombe), zebrafish (Danio rerio), fly (Drosophila melanogaster), and nematode (Caenorhabditis elegans). However, all these enzymes we selected have not been defined and were without a formal name; thus, we named these bifunctional enzymes pseudouridine kinase glycosidase (PsuKG) following the nomenclature of PsuK and PsuG in E. coli. The N-terminal of the PsuKG proteins represents PsuG, which is very conserved in these species mentioned earlier (Supplementary Figure 4). In the C-terminal half, about 350 residues are well-conserved putative pseudouridine kinases (Supplementary Figure 4). We performed a structure-based sequence analysis, whose structures are derived from the solved structure in E. coli, plants, and the structures predicted by Alphafold2 (Supplementary Figures 5A–H) (Jumper et al., 2021). Based on our observations, the residues that participate in the recognition of pseudouridine are all well conserved, except in flies, in which the key residue Lys170 pairing with the nucleobase of pseudouridine in E. coli is substituted with isoleucine (Supplementary Figures 4, 5E). The residue for N1 position recognition is either a serine or threonine in all the protein sequences we showed, indicating the transition reaction scheme for pseudouridine may be necessary and conserved during evolution (Supplementary Figure 4). For the ATP-binding region, the small ATP-binding loop is GXXG in yeast and zebrafish, but a serine in flies and alanine in the nematode (Supplementary Figures 4, 5B,D,F,H). The large ATP-binding loop is much more conserved and is full of aromatic residues. These comparisons indicated the potential catabolic function of the PsuKG proteins for pseudouridine. Whether or not these proteins metabolize pseudouridine and also use ATP as the phosphate group donor needs further investigation.
Discussion
Pseudouridine is a type of widespread naturally occurred modification existing in almost every type of RNA. It was demonstrated that pseudouridine modification in RNAs can regulate many aspects of RNA fate such as RNA stability, translation efficiency, and base-pairing properties due to its additional N1 position compared with uridine. Although the catalytic installation mechanism has been well studied, the metabolism of modified nucleotides including pseudouridine has just started to be uncovered. Previous studies revealed that N6-mAMP was catalyzed by a specific deaminase ADAL and produced inosine, which can be further utilized by the purine salvage pathway (Chen et al., 2018; Jia and Xie, 2019; Wu et al., 2019). The enzyme that can recognize N6-methyl-adenosine was also determined not long ago (Jiang et al., 2021).
Recently, the metabolic pathway of pseudouridine in plants has been uncovered; two enzymes named AtPUKI and AtPUMY can sequentially phosphorylate the Ψ to ΨMP and hydrolyze 5′-ΨMP to produce uracil and ribose 5′-phosphate (Chen and Witte, 2020). This study in plants confirmed the initial finding in E. coli, in which the study showed that EcPsuK and EcPsuG have a similar function in metabolizing pseudouridine as AtPUKI and AtPUMY (Preumont et al., 2008). Here, we determined the structure of E. coli PsuK in a complex with Ψ or m1Ψ, and our studies revealed that the overall structure of EcPsuK adopts a homodimer conformation with a face-to-face mode through the interaction of the β-thumb region and α2 with their counterparts (Figure 2A), and the monomeric EcPsuK-Ψ showed high structural similarity with the inactive status of AtPUKI (Supplementary Figure 2A). Ψ was captured by the α/β region mainly located in the N-terminal part of EcPsuK alongside the β-thumb region. We identified the key residues in recognizing the pseudouridine substrate, and these residues are well conserved in EcPsuK homolog proteins from different species (Supplementary Figure 4). We also obtained the complex structure of EcPsuK with m1Ψ; comparisons of the binding properties between EcPsuK-m1Ψ and EcPsuK-Ψ suggested that EcPsuK has binding capacity for an additional N1-methyl substituent group with weak catalytic activity.
Although we solved these complex structures, there are still many questions to be clarified. (1) We attempted to capture ATP or ADP, but no density could be observed in all the diffraction data we collected. The ATP-binding loop between EcPsuK and AtPUKI has some differences, especially in the small ATP-binding loop. Whether the binding of ATP can induce the conformational change in the small ATP-binding loop is still a question. (2) We have observed some extra density around the residues including Asp164, Glu190, Asn187, and Asp256; however, due to the lack of a phosphate group, we cannot model the bivalent metal ions because of the existence of some water molecules. Given that there is magnesium chloride in the crystallization conditions, we believed that the extra densities around these residues contain two Mg2+ ions; furthermore, all the crystals grown in the conditions lacking Mg2+ showed weak diffraction quality. These observations were consistent with the results shown in previous studies that the active site contains two Mg2+ in AtPUKI (Kim et al., 2021). (3) We have attempted to change the crystallization conditions to rule out the crystal packing effect, but in all conditions, the crystals of apo-EcPsuK or its complex belong to the P6322 space group. Therefore, m1Ψ in our crystal structures seems more suitable in the substrate-binding pocket excluding the impact of Ser30’ from another molecule in the homodimer when in the inactive form. A similar study was published online when we prepared our manuscript (Kim et al., 2022); the transition status was also observed in that study. In their EcPUKI-Ψ complex structure, there are eight molecules that can be considered four homodimers (Supplementary Figure 6A). Intriguingly, only one molecule of each homodimer contains the Ψ substrate; comparisons of these homodimer structures suggested a dynamic sensing mechanism with the active protomers bound to the substrate assistant by the bent β-thumb regions, and the other protomers without substrate were much more flexible (Supplementary Figure 6B). We analyzed the dimerization status in these homodimer structures in detail and found that the distance of Ser30’ with the N1-position presents a variable distance of about 4.6 Å in the Mol12 homodimer, 5.1 Å in the Mol34 homodimer, 3.3 Å and 3.9 Å in Mol56 and Mol78 homodimer structures, respectively (Supplementary Figures 6C–F). By contrast, our structure of either apo-EcPsuK or EcPsuK-(m1)Ψ complex showed more rigid homodimer conformation due to the crystallographic symmetry than the multiple structure status of EcPUKI (Supplementary Figures 6G–J). Furthermore, EcPUKI was suggested to bind the uridine and cytidine with both of the two protomers containing substrates in their binding pockets; however, these structures together with the structures of apo-EcPUKI and EcPUKI-S30A-Ψ showed the inactive form belong to the space group P3. Structure comparisons of our EcPsuK-m1Ψ with these EcPUKI structures revealed minor RMSD, these results validated that our EcPsuK structures are in the inactive form (Supplementary Figures 6K–N), and this form leads to the omissive identification of the important function of Ser30’ in inducing the conformational change for catalysis. Therefore, although our studies demonstrated that the m1Ψ substrate could be bound comfortably in our EcPsuK-m1Ψ complex in the inactive status, the methyl group of m1Ψ will impact the phosphorylation effectiveness. It is worth noting that all the dimers of AtPUKI presented a transition status even in the unliganded AtPUKI structure (Supplementary Figure 2E). (4) Last but not least, previous structure studies suggested that the PsuG in E. coli was a trimer (Huang et al., 2012), and EcPsuK is a homodimer; the real tertiary and quaternary structure arrangement of the fusion protein PsuKG is still unknown.
mRNA vaccines have been demonstrated as a highly effective technique to cope with the COVID-19 pandemic; this successful experience promotes mRNA-based technologies as a promising method in cell therapies (Baden et al., 2021; Thomas et al., 2021). However, two major issues including the widespread degradation of exogenous RNA by ubiquitous RNases and the immunogenic nature of exogenous RNA need to be overcome while making use of the mRNA-based technologies. The incorporation of naturally occurring RNA modifications is an effective method to avoid these undesirable results (Kariko et al., 2005; Lockhart et al., 2019). Among these RNA modifications, fully replacing uridines with Ψ has been demonstrated to be a robust method that can enhance the stability of the parent mRNA and lead to strongly increased protein expression compared to the unmodified mRNAs (Kariko et al., 2008, 2011). Furthermore, incorporation of the synthetic derivative N1-methyl-pseudouridine can further improve translation efficiency and evade innate immune response (Andries et al., 2015; Svitkin et al., 2017). The hypermodified m1Ψ is formed via further methylation of Ψ. At present, at least five types of Ψ hypermodification, including Ψm and m1Ψ, are found in all domains of life (Spenkuch et al., 2014). For m1Ψ, a specific RNA methyltransferase Nep1 was demonstrated to responsible for the N1-specific Ψ methylation in the small ribosomal subunit RNA (Leulliot et al., 2008; Taylor et al., 2008; Wurm et al., 2010; Meyer et al., 2011). Mja_1640 from M. jannaschii was also validated to catalyze the N1-methylation of position Ψ54 located in the T-arm of tRNAs in vitro with the proper sequence specificity (Wurm et al., 2012). Mutation in human Nep1 results in a fatal developmental disorder known as Bowen-Conradi syndrome (Armistead et al., 2009). However, the specific demethylase of m1Ψ is still unknown, and the metabolic fate of these modified nucleotides does not attract much notice. PsuK and PsuG are present in organisms from bacteria to eukaryotes (Preumont et al., 2008; Chen and Witte, 2020), although in metazoa, amoebozoa, and fungi, these two proteins are fused to a single polypeptide chain. To our knowledge, there is still no homolog proteins of PsuK found in mammals, and how the pseudouridine-modified nucleosides of mRNA vaccine are degraded is still a question.
Materials and Methods
Protein Expression and Purification
Plasmids encoding E. coli PsuK (Uniprot ID: A0A140N873) were PCR amplified from E. coli BL21 (DE3) genome. The PCR product was double-digested with restriction endonuclease BamHI and XhoI and then ligated into a modified pET-28a plasmid carrying the Ulp1 cleavage site. Recombinant plasmids were confirmed by DNA sequencing and transformed into Escherichia coli BL21 (DE3) to produce target proteins with N-terminal His6-sumo fusions. E. coli cells were cultured in the LB medium at 37°C with 50 mg/L kanamycin until the OD600 reached 0.6–0.8, then the bacteria were induced with 0.2 mM isopropyl-β-d-thiogalactoside (IPTG) at 18°C for 16 h. Bacteria were collected by centrifugation; resuspended in buffer containing 20 mM Tris–HCl pH 8.0, 500 mM NaCl, 20 mM imidazole pH 8.0; and lysed by high pressure. Cell extracts were centrifuged at 18,000 rpm for 1 h at 4°C. Supernatants were purified with Ni-NTA (GE), the target protein was washed with lysis buffer, and then eluted with a buffer containing 20 mM Tris–HCl, pH 8.0, 500 mM NaCl, and 500 mM imidazole. Ulp1 protease was added to remove the N-terminal tag and fusion protein of the recombinant protein and dialyzed with lysis buffer for 3 h. The mixture was applied to another Ni-NTA resin to remove the protease and uncleaved proteins. Eluted proteins were concentrated by centrifugal ultrafiltration, loaded onto a pre-equilibrated HiLoad 16/60 Superdex 200-pg column, and eluted at a flow rate of 1 ml/min with the buffer containing 10 mM Tris–HCl pH8.0, 100 mM NaCl or 10 mM Tris–HCl pH8.0, 100 mM KCl. Peak fractions were analyzed by SDS-PAGE (15%, w/v) and stained with Coomassie Brilliant Blue R-250. Purified fractions were pooled together and concentrated by centrifugal ultrafiltration. The protein was concentrated at 10 mg/ml for crystallization trials determined by A280.
Crystallization and Data Collection
Apo-EcPsuK was crystallized using the hanging drop vapor diffusion method by mixing 1 μl of protein and 1 μL of reservoir solution at 18°C. The crystal suitable for X-ray diffraction was grown in a reservoir solution consisting of 0.02 M magnesium chloride hexahydrate, 0.1 M HEPES pH 7.5, 22% w/v poly (acrylic acid sodium salt) 5,100 (Hampton Research). For the EcPsuK-Ψ complex, the crystals suitable for data collection were first co-crystallized of EcPsuK with ADP and Ψ in the reservoir solution containing 20% (w/v) polyacrylic acid 5,100, 0.1 M HEPES/sodium hydroxide pH 7.0, 0.02 M magnesium chloride, and were further soaked with cryoprotectant containing the solution supplied with 25% glycerol, 1mM Ψ and 4mM ADP. For the EcPsuK-m1Ψ complex, the crystals suitable for data collection were first co-crystallized of EcPsuK with ADP and m1Ψ in the reservoir solution containing 20% (w/v) polyacrylic acid 5,100, 0.1M HEPES/sodium hydroxide pH 7.0, and 0.02M magnesium chloride and were further soaked with a cryoprotectant containing the solution supplied with 25% glycerol, 1mM m1Ψ, and 4mM ADP.
Structure Determination and Refinement
For the apo-EcPsuK structure, the diffraction data set was processed and scaled using HKL3000 or imosflm (Minor et al., 2006; Battye et al., 2011). The phase was determined by molecular replacement using the program PHASER with the structure of AtPUKI (PDB code: 7C1Y) as the search model (McCoy et al., 2007). Cycles of refinement and model building were carried out using REFMAC5 and COOT, respectively (Emsley and Cowtan, 2004; Murshudov et al., 2011). For the EcPsuK-Ψ and EcPsuK-m1Ψ complex, the phase was determined by molecular replacement using the PHASER program with apo-EcPsuK as the search model. The details of data collection and processing are presented in Table 1. All structure figures were prepared with PyMOL.
Activity Assay
A direct assay was utilized to measure EcPsuK activity to the nucleoside substrates (Andersson and Mowbray, 2002), as reported previously for the assays of AtPUKI and EcPUKI (Kim et al., 2021, 2022). In this direct assay, the reaction mixture contained 40 mM Tris–HCl pH 7.5, 20 mM MgCl2, 50 mM KCl, 0.003% phenol red, 4 mM ATP, and 200 nM wild-type EcPsuK. The mixture was incubated at 25°C for 2 min, then the absorbance was monitored using the Thermo Evolution 201 at 430 nm. To trigger the enzyme reaction, 1.25 mM pseudouridine or other nucleosides were added to this mixture, and the absorbance at 430 nm was monitored again after 30 s. The substrates used for testing the activity contain pseudouridine, N1-methyl-pseudouridine, adenosine, uridine, thymidine, cytidine, guanosine, isocytosine, and inosine. All assays were conducted in triplicate.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
BW conceived the project, solved the structures and interpreted the experimental data. XL and KL expressed, purified, and grew crystals of the EcPsuK. BW, YW, WG, CM, and XL collected X-ray diffraction data. BW and XL wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31900435 to BW) and the Guangdong Science and Technology Department (2020B1212060018 and 2020B1212030004 to BW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the staff from BL17B/BL18U1/BL19U1 beamline of the National Facility for Protein Science in Shanghai (NFPS) at Shanghai Synchrotron Radiation Facility, for assistance during data collection.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.926099/full#supplementary-material
References
Andersson, C. E., and Mowbray, S. L. (2002). Activation of ribokinase by monovalent cations. J. Mol. Biol. 315, 409–419. doi: 10.1006/jmbi.2001.5248
Andries, O., Mc Cafferty, S., De Smedt, S. C., Weiss, R., Sanders, N. N., and Kitada, T. (2015). N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 217, 337–344. doi: 10.1016/j.jconrel.2015.08.051
Armistead, J., Khatkar, S., Meyer, B., Mark, B. L., Patel, N., Coghlan, G., et al. (2009). Mutation of a gene essential for ribosome biogenesis, EMG1, causes Bowen-Conradi syndrome. Am. J. Hum. Genet. 84, 728–739. doi: 10.1016/j.ajhg.2009.04.017
Baccolini, C., and Witte, C. P. (2019). AMP and GMP catabolism in Arabidopsis converge on xanthosine, which is degraded by a nucleoside hydrolase heterocomplex. Plant Cell 31, 734–751. doi: 10.1105/tpc.18.00899
Baden, L. R., El Sahly, H. M., Essink, B., Follmann, D., Neuzil, K. M., August, A., et al. (2021). Phase 3 trial of mRNA-1273 during the delta-variant surge. N. Engl. J. Med. 385, 2485–2487. doi: 10.1056/NEJMc2115597
Balogh, E., Chandler, J. C., Varga, M., Tahoun, M., Menyhard, D. K., Schay, G., et al. (2020). Pseudouridylation defect due to DKC1 and NOP10 mutations causes nephrotic syndrome with cataracts, hearing impairment, and enterocolitis. Proc. Natl. Acad. Sci. U S A. 117, 15137–15147. doi: 10.1073/pnas.2002328117
Barbieri, I., and Kouzarides, T. (2020). Role of RNA modifications in cancer. Nat. Rev. Cancer 20, 303–322. doi: 10.1038/s41568-020-0253-252
Battye, T. G., Kontogiannis, L., Johnson, O., Powell, H. R., and Leslie, A. G. (2011). iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D. Biol. Crystallogr. 67(Pt 4), 271–281. doi: 10.1107/S0907444910048675
Boccaletto, P., Machnicka, M. A., Purta, E., Piatkowski, P., Baginski, B., Wirecki, T. K., et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307. doi: 10.1093/nar/gkx1030
Breitman, T. R. (1970). Pseudouridulate synthetase of Escherichia coli: correlation of its activity with utilization of pseudouridine for growth. J. Bacteriol. 103, 263–264. doi: 10.1128/jb.103.1.263-264.1970
Carlile, T. M., Rojas-Duran, M. F., Zinshteyn, B., Shin, H., Bartoli, K. M., and Gilbert, W. V. (2014). Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146. doi: 10.1038/nature13802
Chen, M., and Witte, C. P. (2020). A kinase and a glycosylase catabolize pseudouridine in the peroxisome to prevent toxic pseudouridine monophosphate accumulation. Plant Cell 32, 722–739. doi: 10.1105/tpc.19.00639
Chen, M., Urs, M. J., Sanchez-Gonzalez, I., Olayioye, M. A., Herde, M., and Witte, C. P. (2018). m(6)A RNA degradation products are catabolized by an evolutionarily conserved N(6)-Methyl-AMP deaminase in plant and mammalian cells. Plant Cell 30, 1511–1522. doi: 10.1105/tpc.18.00236
Cohn, W. E., and Volkin, E. (1951). Nucleoside-5′-phosphates from ribonucleic acid. Nature 167, 483–484. doi: 10.1038/167483a0
de Brouwer, A. P. M., Abou Jamra, R., Kortel, N., Soyris, C., Polla, D. L., Safra, M., et al. (2018). Variants in PUS7 cause intellectual disability with speech delay, microcephaly, short stature, and aggressive behavior. Am. J. Hum. Genet. 103, 1045–1052. doi: 10.1016/j.ajhg.2018.10.026
Deb, I., Popenda, L., Sarzynska, J., Malgowska, M., Lahiri, A., Gdaniec, Z., et al. (2019). Computational and NMR studies of RNA duplexes with an internal pseudouridine-adenosine base pair. Sci. Rep. 9:16278. doi: 10.1038/s41598-019-52637-52630
Emsley, P., and Cowtan, K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132. doi: 10.1107/S0907444904019158
Festen, E. A., Goyette, P., Green, T., Boucher, G., Beauchamp, C., Trynka, G., et al. (2011). A meta-analysis of genome-wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for Crohn’s disease and celiac disease. PLoS Genet. 7:e1001283. doi: 10.1371/journal.pgen.1001283
Ganot, P., Bortolin, M. L., and Kiss, T. (1997). Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89, 799–809. doi: 10.1016/s0092-8674(00)80263-80269
Gilbert, W. V., Bell, T. A., and Schaening, C. (2016). Messenger RNA modifications: form, distribution, and function. Science 352, 1408–1412. doi: 10.1126/science.aad8711
Han, L., Kon, Y., and Phizicky, E. M. (2015). Functional importance of Psi38 and Psi39 in distinct tRNAs, amplified for tRNAGln(UUG) by unexpected temperature sensitivity of the s2U modification in yeast. RNA 21, 188–201. doi: 10.1261/rna.048173.114
Holm, L. (2020). Using dali for protein structure comparison. Methods Mol. Biol. 2112, 29–42. doi: 10.1007/978-1-0716-0270-6_3
Huang, S., Mahanta, N., Begley, T. P., and Ealick, S. E. (2012). Pseudouridine monophosphate glycosidase: a new glycosidase mechanism. Biochemistry 51, 9245–9255. doi: 10.1021/bi3006829
Jia, Q., and Xie, W. (2019). Alternative conformation induced by substrate binding for Arabidopsis thalianaN6-methyl-AMP deaminase. Nucleic Acids Res. 47, 3233–3243. doi: 10.1093/nar/gkz070
Jiang, Z., Wang, C., Wu, Z., Chen, K., Yang, W., Deng, H., et al. (2021). Enzymatic deamination of the epigenetic nucleoside N6-methyladenosine regulates gene expression. Nucleic Acids Res. 49, 12048–12068. doi: 10.1093/nar/gkab1124
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-3812
Kang, P. A., Oh, J., Lee, H., Witte, C. P., and Rhee, S. (2019). Crystal structure and mutational analyses of ribokinase from Arabidopsis thaliana. J. Struct. Biol. 206, 110–118. doi: 10.1016/j.jsb.2019.02.007
Kariko, K., Buckstein, M., Ni, H., and Weissman, D. (2005). Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175. doi: 10.1016/j.immuni.2005.06.008
Kariko, K., Muramatsu, H., Ludwig, J., and Weissman, D. (2011). Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 39:e142. doi: 10.1093/nar/gkr695
Kariko, K., Muramatsu, H., Welsh, F. A., Ludwig, J., Kato, H., Akira, S., et al. (2008). Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16, 1833–1840. doi: 10.1038/mt.2008.200
Kim, S. H., Kim, M., Park, D., Byun, S., and Rhee, S. (2022). Substrate-binding loop interactions with pseudouridine trigger conformational changes that promote catalytic efficiency of pseudouridine kinase PUKI. J. Biol. Chem. 298:101869. doi: 10.1016/j.jbc.2022.101869
Kim, S. H., Witte, C. P., and Rhee, S. (2021). Structural basis for the substrate specificity and catalytic features of pseudouridine kinase from Arabidopsis thaliana. Nucleic Acids Res. 49, 491–503. doi: 10.1093/nar/gkaa1144
Leulliot, N., Bohnsack, M. T., Graille, M., Tollervey, D., and Van Tilbeurgh, H. (2008). The yeast ribosome synthesis factor Emg1 is a novel member of the superfamily of alpha/beta knot fold methyltransferases. Nucleic Acids Res. 36, 629–639. doi: 10.1093/nar/gkm1074
Li, X., Zhu, P., Ma, S., Song, J., Bai, J., Sun, F., et al. (2015). Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 11, 592–597. doi: 10.1038/nchembio.1836
Lin, T. Y., Mehta, R., and Glatt, S. (2021). Pseudouridines in RNAs: switching atoms means shifting paradigms. FEBS Lett. 595, 2310–2322. doi: 10.1002/1873-3468.14188
Lockhart, J., Canfield, J., Mong, E. F., VanWye, J., and Totary-Jain, H. (2019). Nucleotide modification alters MicroRNA-Dependent silencing of MicroRNA switches. Mol. Ther. Nucleic Acids 14, 339–350. doi: 10.1016/j.omtn.2018.12.007
Loh, K. D., Gyaneshwar, P., Markenscoff Papadimitriou, E., Fong, R., Kim, K. S., Parales, R., et al. (2006). A previously undescribed pathway for pyrimidine catabolism. Proc. Natl. Acad. Sci. U S A. 103, 5114–5119. doi: 10.1073/pnas.0600521103
McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C., and Read, R. J. (2007). Phaser crystallographic software. J. Appl. Crystallogr. 40(Pt 4), 658–674. doi: 10.1107/S0021889807021206
Meyer, B., Wurm, J. P., Kotter, P., Leisegang, M. S., Schilling, V., Buchhaupt, M., et al. (2011). The Bowen-Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of Psi1191 in yeast 18S rRNA. Nucleic Acids Res. 39, 1526–1537. doi: 10.1093/nar/gkq931
Minor, W., Cymborowski, M., Otwinowski, Z., and Chruszcz, M. (2006). HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D. Biol. Crystallogr. 62(Pt 8), 859–866. doi: 10.1107/S0907444906019949
Morais, P., Adachi, H., and Yu, Y. T. (2021). Spliceosomal snRNA epitranscriptomics. Front. Genet. 12:652129. doi: 10.3389/fgene.2021.652129
Motorin, Y., and Marchand, V. (2021). Analysis of RNA modifications by second- and third-generation deep sequencing: 2020 update. Genes (Basel) 12:278. doi: 10.3390/genes12020278
Murshudov, G. N., Skubak, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., et al. (2011). REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D. Biol. Crystallogr. 67(Pt 4), 355–367. doi: 10.1107/S0907444911001314
Nakamoto, M. A., Lovejoy, A. F., Cygan, A. M., and Boothroyd, J. C. (2017). mRNA pseudouridylation affects RNA metabolism in the parasite Toxoplasma gondii. RNA 23, 1834–1849. doi: 10.1261/rna.062794.117
Ni, J., Tien, A. L., and Fournier, M. J. (1997). Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89, 565–573. doi: 10.1016/s0092-8674(00)80238-x
Pardi, N., Hogan, M. J., Porter, F. W., and Weissman, D. (2018). mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279. doi: 10.1038/nrd.2017.243
Park, J., and Gupta, R. S. (2008). Adenosine kinase and ribokinase–the RK family of proteins. Cell Mol. Life. Sci. 65, 2875–2896. doi: 10.1007/s00018-008-8123-8121
Preumont, A., Snoussi, K., Stroobant, V., Collet, J. F., and Van Schaftingen, E. (2008). Molecular identification of pseudouridine-metabolizing enzymes. J. Biol. Chem. 283, 25238–25246. doi: 10.1074/jbc.M804122200
Rintala-Dempsey, A. C., and Kothe, U. (2017). Eukaryotic stand-alone pseudouridine synthases - RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 14, 1185–1196. doi: 10.1080/15476286.2016.1276150
Schwartz, S., Bernstein, D. A., Mumbach, M. R., Jovanovic, M., Herbst, R. H., Leon-Ricardo, B. X., et al. (2014). Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162. doi: 10.1016/j.cell.2014.08.028
Sharma, S., and Lafontaine, D. L. J. (2015). ‘View from a bridge’: a new perspective on eukaryotic rRNA Base Modification. Trends Biochem. Sci. 40, 560–575. doi: 10.1016/j.tibs.2015.07.008
Sigrell, J. A., Cameron, A. D., Jones, T. A., and Mowbray, S. L. (1998). Structure of Escherichia coli ribokinase in complex with ribose and dinucleotide determined to 1.8 a resolution: insights into a new family of kinase structures. Structure 6, 183–193. doi: 10.1016/s0969-2126(98)00020-3
Spenkuch, F., Motorin, Y., and Helm, M. (2014). Pseudouridine: still mysterious, but never a fake (uridine)! RNA Biol. 11, 1540–1554. doi: 10.4161/15476286.2014.992278
Svitkin, Y. V., Cheng, Y. M., Chakraborty, T., Presnyak, V., John, M., and Sonenberg, N. (2017). N1-methyl-pseudouridine in mRNA enhances translation through eIF2alpha-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 45, 6023–6036. doi: 10.1093/nar/gkx135
Taylor, A. B., Meyer, B., Leal, B. Z., Kotter, P., Schirf, V., Demeler, B., et al. (2008). The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site. Nucleic Acids Res. 36, 1542–1554. doi: 10.1093/nar/gkm1172
Thomas, S. J., Moreira, E. D. Jr., Kitchin, N., Absalon, J., Gurtman, A., et al. (2021). Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine through 6 months. N. Engl. J. Med. 385, 1761–1773. doi: 10.1056/NEJMoa2110345
Veerareddygari, G. R., Singh, S. K., and Mueller, E. G. (2016). The pseudouridine synthases proceed through a glycal intermediate. J. Am. Chem. Soc. 138, 7852–7855. doi: 10.1021/jacs.6b04491
Wang, B., Grant, R. A., and Laub, M. T. (2020). ppGpp coordinates nucleotide and amino-acid synthesis in E. coli during starvation. Mol. Cell 80, 29–42.e10. doi: 10.1016/j.molcel.2020.08.005
Witte, C. P., and Herde, M. (2020). Nucleotide metabolism in plants. Plant Physiol. 182, 63–78. doi: 10.1104/pp.19.00955
Wu, B., Zhang, D., Nie, H., Shen, S., Li, Y., and Li, S. (2019). Structure of Arabidopsis thaliana N(6)-methyl-AMP deaminase ADAL with bound GMP and IMP and implications for N(6)-methyl-AMP recognition and processing. RNA Biol. 16, 1504–1512. doi: 10.1080/15476286.2019.1642712
Wurm, J. P., Griese, M., Bahr, U., Held, M., Heckel, A., Karas, M., et al. (2012). Identification of the enzyme responsible for N1-methylation of pseudouridine 54 in archaeal tRNAs. RNA 18, 412–420. doi: 10.1261/rna.028498.111
Wurm, J. P., Meyer, B., Bahr, U., Held, M., Frolow, O., Kotter, P., et al. (2010). The ribosome assembly factor Nep1 responsible for Bowen-Conradi syndrome is a pseudouridine-N1-specific methyltransferase. Nucleic Acids Res. 38, 2387–2398. doi: 10.1093/nar/gkp1189
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624. doi: 10.1038/s41580-019-0168-165
Zaringhalam, M., and Papavasiliou, F. N. (2016). Pseudouridylation meets next-generation sequencing. Methods 107, 63–72. doi: 10.1016/j.ymeth.2016.03.001
Zhang, Y., El Kouni, M. H., and Ealick, S. E. (2007). Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase. Acta Crystallogr. D. Biol. Crystallogr. 63(Pt 2), 126–134. doi: 10.1107/S0907444906043654
Keywords: crystal structrue, pseudouridine (Ψ), kinase, nucleoside, N1-methyl-pseudouridine
Citation: Li X, Li K, Guo W, Wen Y, Meng C and Wu B (2022) Structure Characterization of Escherichia coli Pseudouridine Kinase PsuK. Front. Microbiol. 13:926099. doi: 10.3389/fmicb.2022.926099
Received: 22 April 2022; Accepted: 31 May 2022;
Published: 17 June 2022.
Edited by:
Yong-Gui Gao, Nanyang Technological University, SingaporeReviewed by:
Jack A. Dunkle, University of Alabama, United StatesXinfu Yan, Nanyang Technological University, Singapore
Copyright © 2022 Li, Li, Guo, Wen, Meng and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baixing Wu, d3VieDI4QG1haWwuc3lzdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Xiaojia Li
Xiaojia Li Kangjie Li
Kangjie Li Wenting Guo1
Wenting Guo1 Baixing Wu
Baixing Wu