- State Key Laboratory of Pharmaceutical Biotechnology, Nanjing University, Nanjing, China
Laccases are ligninolytic enzymes that play a crucial role in various biological processes of filamentous fungi, including fruiting-body formation and lignin degradation. Lignin degradation is a complex process and its degradation in Schizophyllum commune is greatly affected by the availability of oxygen. Here, a total of six putative laccase genes (ScLAC) were identified from the S. commune 20R-7-F01 genome. These genes, which include three typical Cu-oxidase domains, can be classified into three groups based on phylogenetic analysis. ScLAC showed distinct intron-exon structures and conserved motifs, suggesting the conservation and diversity of ScLAC in gene structures. Additionally, the number and type of cis-acting elements, such as substrate utilization-, stress-, cell division- and transcription activation-related cis-elements, varied between ScLAC genes, suggesting that the transcription of laccase genes in S. commune 20R-7-F01 could be induced by different substrates, stresses, or other factors. The SNP analysis of resequencing data demonstrated that the ScLAC of S. commune inhabiting deep subseafloor sediments were significantly different from those of S. commune inhabiting terrestrial environments. Similarly, the large variation of conserved motifs number and arrangement of laccase between subseafloor and terrestrial strains indicated that ScLAC had a diverse structure. The expression of ScLAC5 and ScLAC6 genes was significantly up-regulated in lignin/lignite medium, suggesting that these two laccase genes might be involved in fungal utilization and degradation of lignite and lignin under anaerobic conditions. These findings might help in understanding the function of laccase in white-rot fungi and could provide a scientific basis for further exploring the relationship between the LAC family and anaerobic degradation of lignin by S. commune.
Introduction
Laccase (benzenediol: oxygen oxidoreductase, EC 1.10.3.2) is a metalloprotein belonging to the group of polyphenol oxidases containing copper atoms in the catalytic site and therefore also called blue multicopper oxidases (Baldrian, 2006). Laccase proteins have three conserved domains (Cu-oxidase, Cu-oxidase_2, and Cu-oxidase_3) that are utilized to identify canonical laccases (Bento et al., 2006; Kudanga et al., 2011). Such an arrangement of copper atoms distributed in three domains is present in most of the bacterial and fungal laccases. The catalytic capacity of laccases is actually non-specific but, in most cases, laccases oxidize a range of aromatic compounds, including phenolic moieties typically found in lignin, aromatic amines, benzenothiols, and hydroxyindols as well as non-aromatic compounds, using molecular oxygen as an electron acceptor (Shiba et al., 2000; Claus, 2004; Rodríguez and Toca Herrera, 2006; Chirivì et al., 2012). Laccases have been implicated in a variety of physiological functions in living organisms due to their non-specific catalytic abilities.
Laccase was initially discovered in the Japanese lacquer tree by Yoshida (1883), and since then, it has been found in all domains of life: higher plants, some insects, a few bacteria, and fungi (Solomon et al., 1996; Alexandre and Zhulin, 2000; Claus, 2003). Basic characteristics and functions of laccases are diverse both within and across biological kingdoms. In plants, laccases participate in the radical-based mechanisms of lignin polymer formation (Berthet et al., 2011; Hu et al., 2018), while in fungi, laccases are hypothesized to play a variety of physiological roles, such as stress defense, melanin synthesis (Hua et al., 2018), fruiting-body formation (Lettera et al., 2010; Zhang et al., 2015), and lignin degradation (Singh and Sharma, 2010; Coconi et al., 2018). Lignin degradation is the most important function of fungal laccase. Laccases can directly depolymerize the lignin macromolecule, either alone or in combination with other enzymes. Laccases catalyze the removal of an electron from natural lignin’s phenolic hydroxyl groups, resulting in free phenoxy radicals, and further oxidizes to quinones. Additionally, laccases decarboxylate phenolic and methoxyphenolic acid structures of lignin and cause their demethylation or demethoxylation (Leonowicz et al., 2001). Laccases also are useful biocatalysts for a wide range of biotechnological applications due to their high non-specific oxidation capacity and the use of readily available molecular oxygen as an electron acceptor (Mayer and Staples, 2002). In addition, laccases have important application values in various industrial processes, including textile refining, dye decolorization, bioremediation, lignocellulose delignification, organic synthesis, and food processing (Bilal et al., 2017; Mtibaà et al., 2018; Zhang et al., 2018).
Schizophyllum commune belongs to the white rot fungi and is one of the most widely distributed mushrooms on Earth. It is an effective wood-degrading basidiomycete that can produce a large number of hydrolases such as xylanase (Gautam et al., 2018), pectinase (Mehmood et al., 2019), cellulase (Kumar B. et al., 2018), endoglucanase, glycoside hydrolase, and oxidoreductase (Tovar-Herrera et al., 2018). Genome sequencing of a terrestrial strain H4-8 revealed two laccase genes and four genes encoding a distant relative of laccase (Ohm et al., 2010). Our previous study indicated that S. commune was the predominant fungal species in deep subseafloor coal-bearing sediments ranging from ∼1.5 to ∼2.0 km below the seafloor (kmbsf), and could grow under both anaerobic and aerobic culture conditions (Liu et al., 2017; Zain Ul Arifeen et al., 2020). Compared with other environmental isolates, strain 20R-7-F01 of S. commune isolated from ∼20-million-year-old coal-bearing sediment at 1,966.3 kmbsf has a stronger ability to adapt to in situ environmental conditions, including carbon (energy) source, temperature, oxygen, and nitrogen source (Zain Ul Arifeen et al., 2020).
Although laccases were identified and classified in various S. commune strains (Kumar et al., 2015, Kumar V. P. et al., 2018; Zhao et al., 2018; Kirtzel et al., 2019), an investigation of the laccase gene family in S. commune at the whole-genome level is yet to be conducted. In this study, we identified all possible laccase-coding genes from the S. commune reference genome (20R-7-F01). We then analyzed the physical and chemical properties, gene structure, amino acid sequence, systematic evolution, and expression patterns of the gene family in media with or without lignin/lignite. The results could facilitate the understanding of the laccase function in white-rot fungi and provide a scientific basis for further exploring the relationship between the LAC family and the anaerobic degradation of lignin by S. commune.
Materials and methods
Strains and culture conditions
The fungal strains were isolated from subseafloor sediment, which was collected by drilling vessel at Site C0020 (41°10.5983′N, 142°12.0328′E) in the Pacific Plate off the Shimokita Peninsula, Japan, during the IODP Expedition 337, at a water depth of 1,180 m (Inagaki et al., 2015; Liu et al., 2017). Briefly, the sediment samples were ground into powder in an anaerobic chamber with a flame-sterilized hammer, placed evenly on three petri dishes containing specific media that simulated to the in situ environmental conditions, and incubated at 30°C for 7∼14 days (Liu et al., 2017). S. commune strains 6R-2-F01, 15R-5-F01, 20R-7-F01, and 24R-3-F01 were obtained from the sediment samples at to 1,496; 1,924; 1,966, and 1,993 mbsf, respectively. Two terrestrial strains CFCC_7252 and CFCC_86625 were purchased from China Forestry Culture Collection Center, which were isolated from Populus wood in Songshan, Beijing and Jurong, Jiangsu of China, respectively. Strain MCCC_3A00233 collected from marine sediment of the Atlantic Ocean was purchased from Third Institute of Oceanography, State Oceanic Administration, People’s Republic of China. and the other five terrestrial strains (225DK, 227DK, MF, Hom2-8, and 207) were obtained from NCBI and JGI database. Details of the habitat and culture conditions of S. commune strains have been described previously (Liu et al., 2022). All the fungal strains were maintained on potato dextrose agar (PDA) at 30°C. For DNA and RNA isolations, the fresh mycelia of S. commune were inoculated into a 250-ml conical flask containing 150 ml PD (200 g/L potato, 20 g/L glucose) and incubated in a shaking chamber at 30°C, 200 rpm for 7 days.
Identification of laccase gene family members in Schizophyllum commune 20R-7-F01
The S. commune 20R-7-F01 genome was assembled using SMRT Analysis and deposited in GenBank under the accession number VCHW00000000. Laccase members contain Cu-oxidase, Cu-oxidase_2, and Cu-oxidase_3 (PF00394, PF07731 and PF07732) domains. The three domains were searched in the S. commune 20R-7-F01 genome using HAMMER software (Finn et al., 2011), and protein sequences with three Cu-oxidase domains in the LAC domain were recognized as members of the LAC family. The laccase gene was named using the prefix Sc for S. commune followed by the LAC gene family abbreviation and numbered sequentially according to their position on unitigs.
Physical map of Schizophyllum commune 20R-7-F01 laccase genes and properties of laccase proteins
Using the S. commune 20R-7-F01 genome, the unitig length and the starting position of genes on unitigs were obtained. After statistical analysis, the physical distribution map of their unitigs was visualized using Mapchart 2.32 software (Voorrips, 2002). The theoretical isoelectric point (pI) and molecular weight (MW) of ScLAC proteins were analyzed using the Compute pI/MW tool on the ExPASy server1 (Wilkins et al., 1999). Subcellular locations of the ScLAC members were determined using the online software CELLO2 (Yu et al., 2006). Signal peptides of each laccase were predicted using SignalP algorithm3 (Nielsen et al., 1997). Prediction of transmembrane regions was performed with TMHMM Server4 (Krogh et al., 2001). The glycosylation sites of the ScLAC members were predicted by NetNGlyc 1.05 (Gupta and Brunak, 2002).
Analysis of gene structure and motif composition
The sequence of laccase genes and their coding region were first transformed into FASTA format then matched, and intron/exon structure was determined by comparing the coding sequence of each ScLAC gene with its genomic sequence using the Gene Structure Display Server 2.06 (Hu B. et al., 2015). In addition, the upstream regions (1.5 kb) of the ScLAC gene sequences were extracted and used for the search of cis-elements using YEASTRACT7 (Monteiro et al., 2020). Conserved motifs of laccase proteins were identified statistically using MEME8 (Bailey et al., 2009), and the maximum number of motifs to find was set at 10. Visualization of motif compositions was executed with TBtools V1.09 (Chen et al., 2020).
Sequence alignment and phylogenetic analysis
The identified ScLAC amino acid sequences were aligned separately against each other using ClustalW in MEGA7.0 (Kumar et al., 2016). The conserved regions of ScLAC were used to build the phylogenetic tree. The unrooted phylogenetic tree was created using MEGA7.0 by a neighbor-joining algorithm with bootstrap replication of 100 times. The final phylogenetic tree was visualized and edited in iTOL9 (Letunic and Bork, 2016).
Genome resequencing and variant calling
The genome resequencing and variant detection for S. commune strains were carried out according to our previous methods (Liu et al., 2022). Briefly, the genome DNA of S. commune strains was extracted and fragmented to generate an approximately 300 bp library insert size and sequenced on an Illumina HiSeq 2500 platform at BGI Genomic (Shenzhen, China). The filtered resequencing reads were mapped to the reference genome of S. commune 20R-7-F01 for SNP and variant detection.
Transcriptome analysis
Total RNA was extracted from mycelia of strain 20R-7-F01 that were cultured in bottles containing lignin and lignite medium, and incubated under anaerobic (i.e., LigWO1-3 and CoalWO1-3) and aerobic (i.e., LigO1-3 and CoalO1-3) condition for seven days (30°C), respectively. Each treatment included three replicates. Lignite was collected from coal mine in Xinjiang. It contains N 1.15%, C 68.7%, H 4.123%, S 1.642%, organic component 98.17%, and inorganic component 1.83%, and vitrinite reflectance was 0.49%. Lignin alkali was purchased from Sigma (CAS# 8068-05-1), which contains 5% moisture. After sampling, all mycelia were immediately frozen in a liquid nitrogen tank and delivered to the Personal Biotechnology Company (Shanghai, China) for mRNA extraction, cDNA library construction, and sequencing. After trimming of low-quality reads (Q < 20) and adapter contamination, the clean reads were mapped to the assembled genome of strain 20R-7-F01 using TopHat (Trapnell et al., 2009). Gene prediction was performed using Cufflinks (Roberts et al., 2011). To compare the gene expression level in different libraries, the transcript level of each expressed gene was calculated and normalized to the reads per kilobase of exon model per million mapped reads (RPKM). We used DESeq software for differential analysis of gene expression (Anders and Huber, 2010). Genes with an adjusted p-value ≤0.01 and an absolute value of log2 (expression-fold change) ≥1 were deemed to be differentially expressed (Hu L. et al., 2015). The Pheatmap software package in R language was used to perform bidirectional cluster analysis of differential genes and samples. Distances were calculated using the Euclidean method and clustered by complete linkage.
Results
Laccase gene family of Schizophyllum commune 20R-7-F01
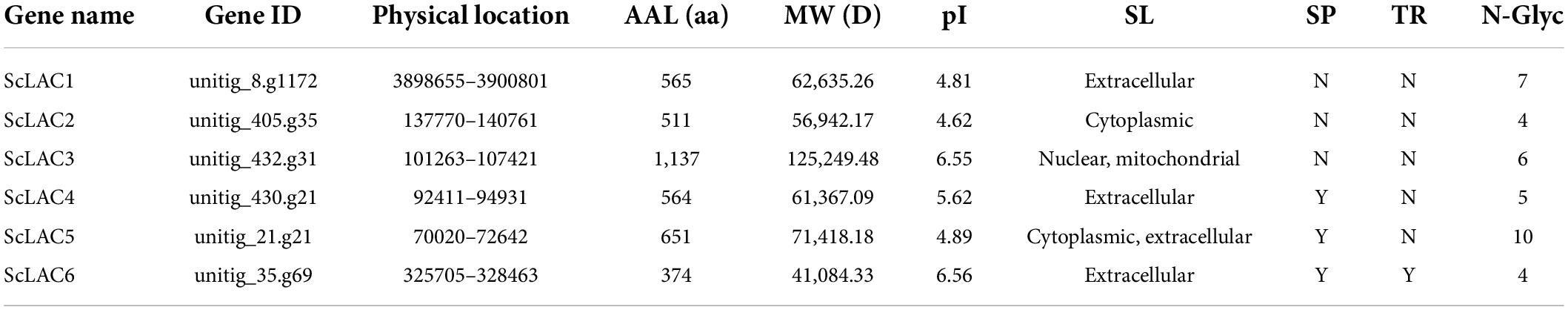
To identify the laccase genes in S. commune 20R-7-F01, we searched the genome with HAMMER software for Cu-oxidase, Cu-oxidase_2, and Cu-oxidase_3 domains (PF00394, PF07731, and PF07732). Six putative laccase genes (ScLAC1 to ScLAC6) were identified (Table 1) and mapped to six of the 162 S. commune 20R-7-F01 unitigs (Figure 1), indicating that the ScLAC gene family did not have the characteristics of tandem replication or clustering.
ScLAC proteins
Basic information on all S. commune 20R-7-F01 laccases, including gene name, physical location, amino acid length, molecular weight, pI value, subcellular localization, signal peptide and transmembrane topology, were presented in Table 1. The length of laccase proteins ranged from 374 aa (ScLAC6) to 1,137 aa (ScLAC3) residues, and the predicted molecular weights were between 41.08 kDa (ScLAC6) and 125.25 kDa (ScLAC3). The predicted pI-values of the laccase proteins were found to be in the range of 4.62 (ScLAC2) to 6.56 (ScLAC6), indicating that they belonged to acidic proteins. The predicted subcellular locations revealed that the six laccase proteins were located in cytoplasm, nucleus, and mitochondria, and were also found extracellularly. ScLAC1, ScLAC4, and ScLAC6 were predicted to be localized only in the extracellular space, whereas ScLAC3 was located either in the nucleus or mitochondria, ScLAC2 was located in the cytoplasm, and ScLAC5 was located in either the cytoplasm or extracellular. ScLAC4–ScLAC6 were probably signal proteins, while ScLAC1–ScLAC3 may not contain any signal regions. ScLAC6 had transmembrane topology, while the other five laccases did not contain transmembrane domains. Additionally, variable N-glycosylation sites were predicted to be present in all ScLAC proteins (Table 1), indicating that ScLAC family exhibited potential post-translational modifications.
Gene structure, motif compositions, and phylogeny of ScLAC
To reveal the structural diversity of S. commune 20R-7-F01 laccase genes, we constructed the exon/intron organization and searched for conservative motifs based on the phylogenetic tree of all S. commune 20R-7-F01 laccase alignments (Figure 2). Phylogenomic analysis showed that the S. commune 20R-7-F01 laccase gene family was clustered into three branches, of which ScLAC1 and ScLAC2 were one clade, ScLAC4 and ScLAC6 were another clade, and ScLAC5 and ScLAC3 were the last clade (Figure 2). In addition, to evaluate the number of laccase genes in the genome of S. commune 20R-7-F01, the total number of laccase genes was determined in other Agaricales. The total number of laccase genes varied significantly among species, ranging from 4 in Hebeloma cylindrosporum and Postia placenta to 55 in Dendrothele bispora (Supplementary Figure 1). The amount of laccase in Schizophyllaceae was relatively small compared to other species. In addition, the total number of laccases and protein-coding genes were normalized by genome assembly (in Mb) to avoid potentially misleading comparisons due to differences in genome size and total number of genes among the investigated species. No positive correlation was found between genome size or total number of predicted genes and the number of laccase genes in the corresponding genome (Supplementary Figure 1). For instance, D. bispora showed the highest number of laccases (55), but Moniliophthora perniciosa had the highest proportion of laccases per total number of genes (0.22%).
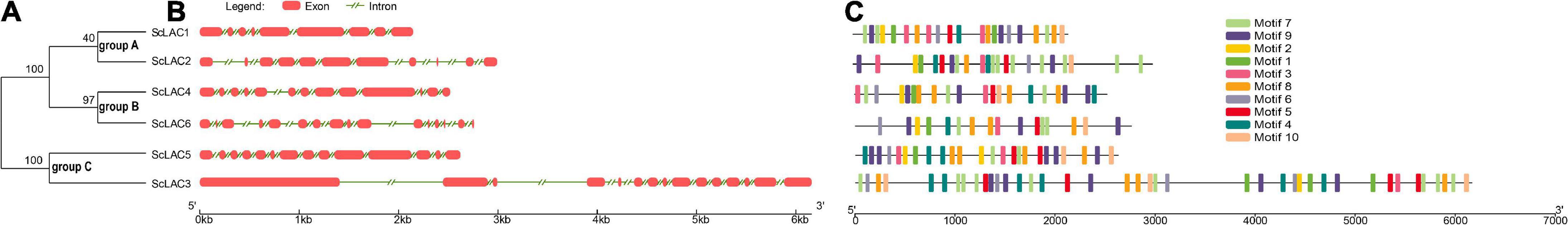
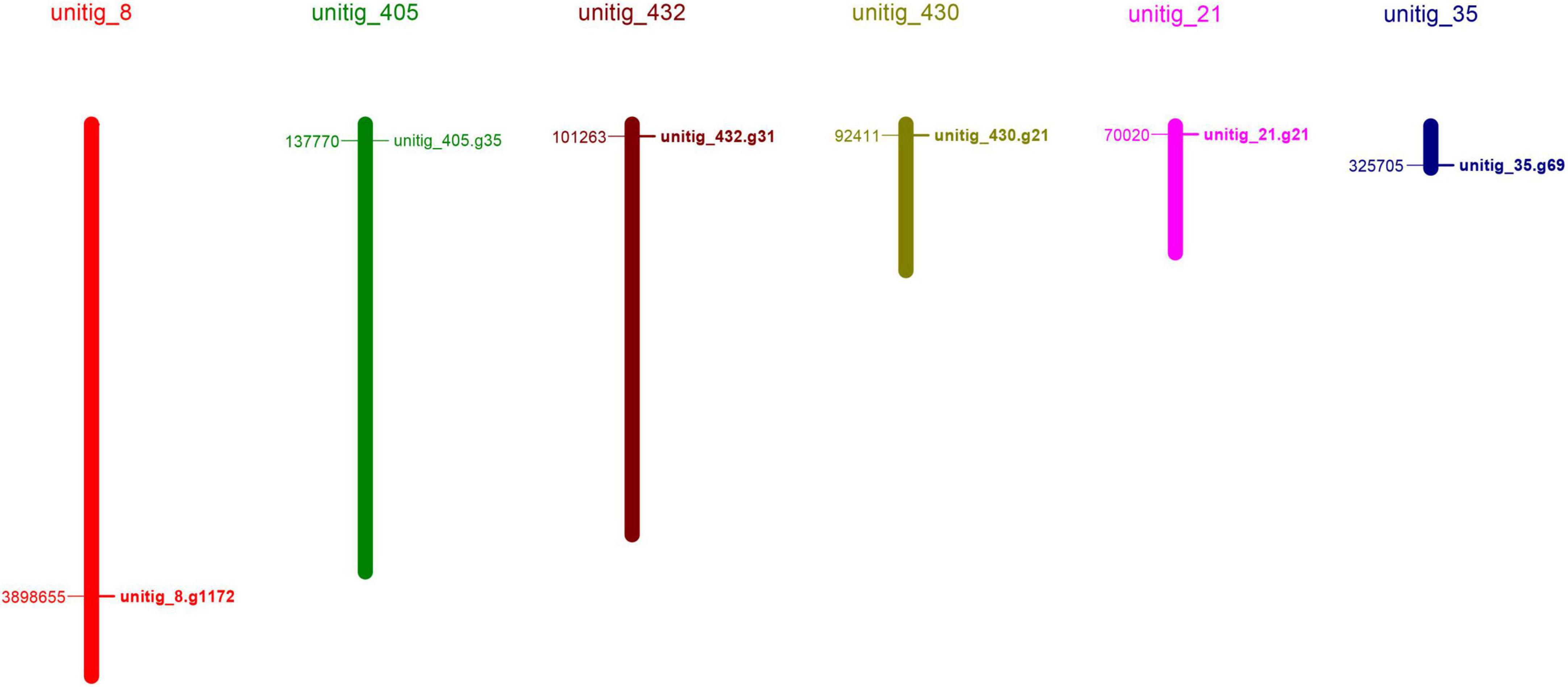
Figure 2. Phylogenetic relationships, gene structure, and motif compositions of the Schizophyllum commune 20R-7-F01 laccase gene family. (A) A neighbor-joining tree of six ScLAC protein sequences constructed using MEGA v7.0. (B) The structure of the six ScLAC genes. Red squares correspond to exons and shrinked green lines indicate introns. (C) Schematic motif composition of six ScLAC genes. The colored boxes represent the different motifs, indicated in the top right-hand corner. The scales at the bottom of the image indicate the estimated exon/intron and motif length in kb.
The number of introns of ScLAC family members varied from 8 to 15. Surprisingly, nearly all of the closest genes on the phylogenetic tree showed remarkably different gene structures. For instance, the introns and exons of ScLAC5 were most closely arranged, whereas its nearby paralogous gene ScLAC3 had the longest intron, although their evolutionary relationship reached a 100% bootstrap value. Additionally, ScLAC6 had the most introns; its coding sequences were divided into 15 parts by introns. In short, ScLAC3, ScLAC5, and ScLAC6 were more complicated than the other laccase genes with respect to their structure.
To further reveal the conserved motifs of the ScLAC proteins, we analyzed six ScLAC proteins and identified 10 motifs using the MEME program (Figure 2). As expected, the motif compositions of peer groups had different structures and organizations, which indicated the possibility of functional divergence among those proteins. Although 10 motifs were found in every ScLAC protein, there were some differences in the number of occurrences. For instance, motif-4 was repeated three and six times in ScLAC5 and ScLAC3, respectively. This difference in motif number across ScLAC proteins indicated that different ScLAC proteins may have different functions.
Cis-regulatory elements predicted in the ScLAC promoters
To obtain further insights into the possible regulatory patterns of ScLAC, we analyzed the cis-acting elements of the 1.5 kb regulatory sequence upstream of the six ScLAC gene sequences using the Yeastract database. The promoter regions of ScLAC1–ScLAC6 included various functional cis-acting elements (Figure 3 and Supplementary Table 1) associated with substrate utilization, stress, cell division, and transcription activation. Among them, ScLAC6 had the most cis-elements, including 16 stress-related, 15 substrate utilization-related, nine cell division-related, and eight amino acid transcription-related cis-elements. In addition, these laccases also contained specific cis-elements; for example, ScLAC2 contained one specific cis-element, named Nrg2p, which mediated glucose repression and negatively regulated filamentous growth, while ScLAC3 contained four specific cis-elements, which negatively regulated nitrogen catabolic gene expression and were involved in induction of CLN3 transcription in response to glucose (Supplementary Figure 2 and Supplementary Table 1). The differences in the number and types of cis-acting elements in S. commune 20R-7-F01 suggested that the transcription of laccase genes may be regulated by substrates, stresses, or other factors.
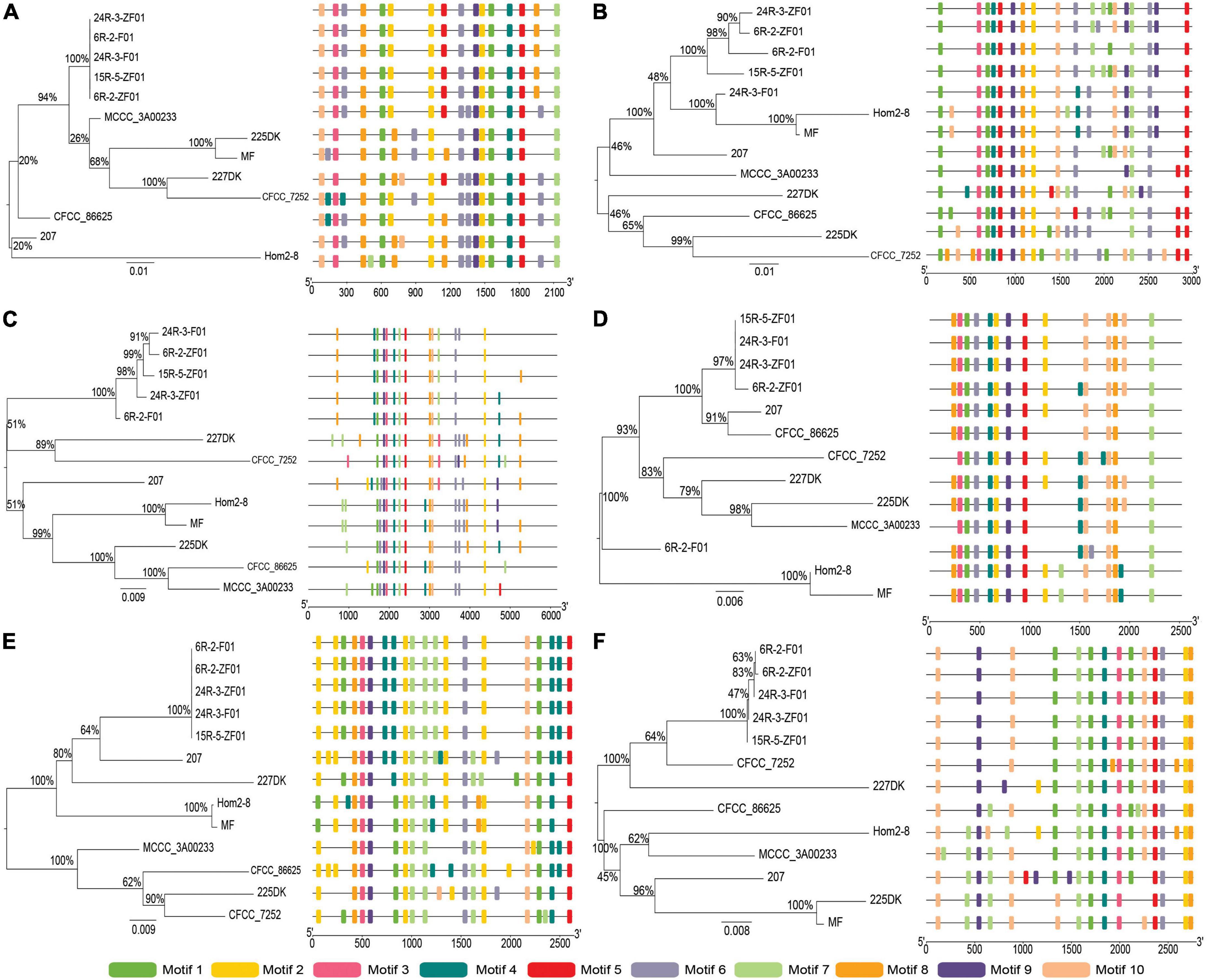
Figure 3. Phylogenetic and motif compositions of the Schizophyllum commune population laccase gene family. (A–F) Represent neighbor-joining tree of ScLAC1-ScLAC6 protein sequences in S. commune population constructed using MEGA v7.0, and schematic motif composition of ScLAC1-ScLAC6 genes in S. commune population. The colored boxes represent the different motifs, indicated in the top right-hand corner. The scales at the bottom of the image indicate the motif length in kb.
Laccase differences between subseafloor and terrestrial environments
To understand the evolutionary relationships between subseafloor and terrestrial laccase genes, five subseafloor and eight terrestrial strains were re-sequenced. Phylogenetic analysis based on SNP mutation sites showed that the ScLAC of S. commune strains inhabiting deep subseafloor sediments differed significantly from that of S. commune strains inhabiting terrestrial environments (Figure 3). The average number of SNP mutation sites of the five ScLAC in the terrestrial strains was greater than those in subseafloor strains, with the exception of ScLAC3, although it contained the most SNP mutation sites (Supplementary Tables 2, 3). In addition, the number and arrangement of conserved motifs in laccases between subseafloor and terrestrial strains also showed various differences, suggesting that ScLAC possessed a diverse structure. Among them, the conserved domains of ScLAC2–ScLAC4 were different in both subseafloor and terrestrial strains, indicating that the evolution of these three laccase genes was not only related to habitat but also related to strains (Figure 3).
Transcriptome analysis of six putative laccase in lignite/lignin degradation
RNA-seq analysis of strain 20R-7-F01 cultured in lignin/lignite-containing medium under aerobic and anaerobic conditions at 30°C for 7 days showed that the six laccase genes could be classified into three groups (I, II, and III) (Figure 4). The relative expression levels of ScLAC1 and ScLAC4 were lower under anaerobic conditions than under aerobic conditions. In contrast, the laccase genes of group II (ScLAC2 and ScLAC3) and group III (ScLAC5 and ScLAC6) tended to be induced by anaerobic conditions (Figure 4). Additionally, compared with aerobic condition, the expression of ScLAC5 and ScLAC6 genes was upregulated by 2.48- and 2.10-fold in the anaerobic conditions (Supplementary Table 4), suggesting that these two laccase genes may be involved in anaerobic utilization and degradation of lignite and lignin by fungi.
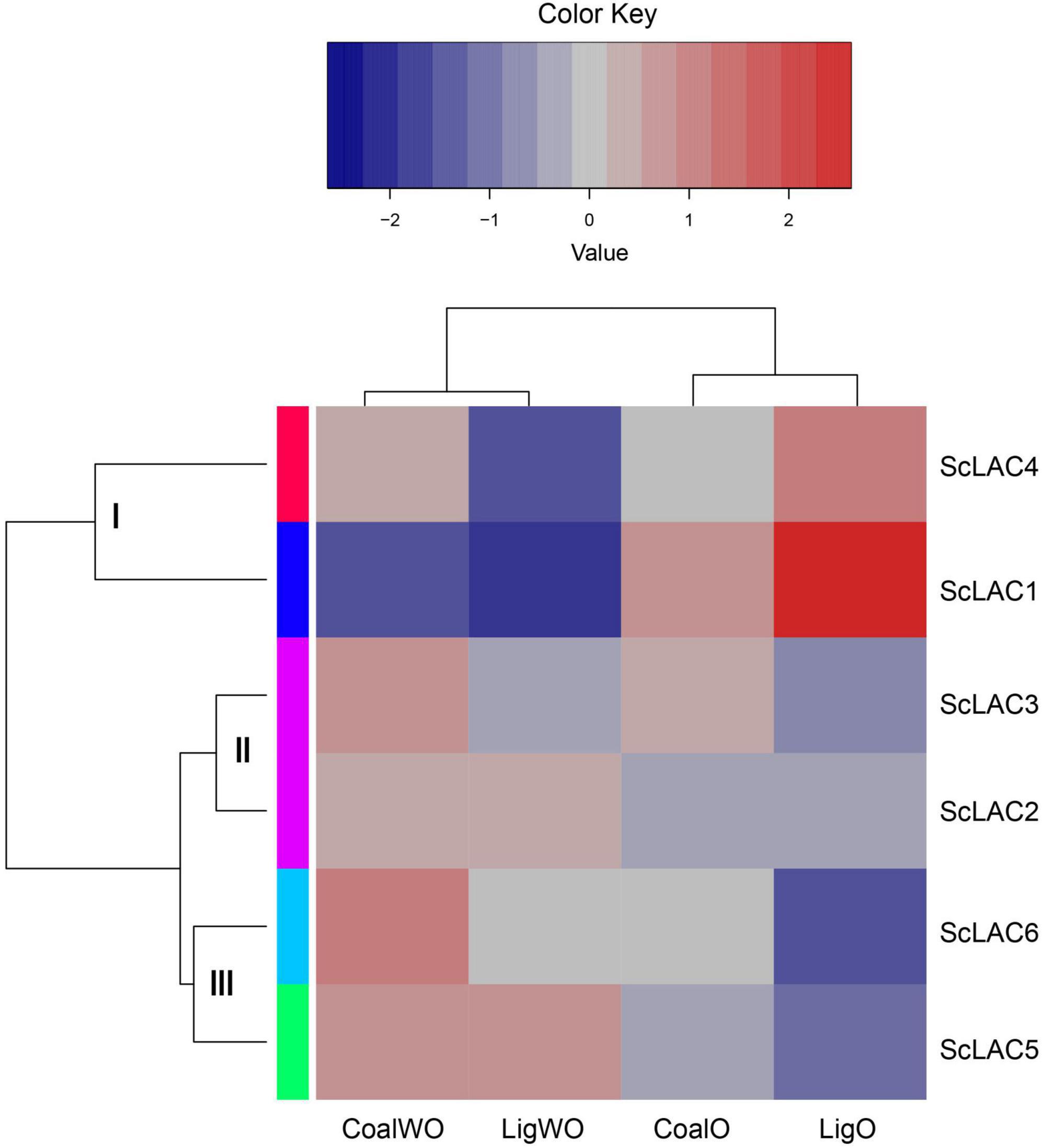
Figure 4. Heatmap of the expression profiles of laccase genes from Schizophyllum commune 20R-7-F01 in various carbon sources with or without oxygen. The heatmaps with hierarchical clustering were visualized using the software heatmap2 and the values were log2-transformed with normalization. The blue and red elements indicate low and high relative expression levels, respectively.
Discussion
Laccases as ligninolytic enzymes play important roles in various biological processes of fungi, including lignin degradation and fruiting-body formation, are typically encoded by gene families (Vasina et al., 2015). Through genome-sequencing analysis, we identified six laccase genes in subseafloor S. commune 20R-7-F01, all of which contained three conserved Cu-oxidase domains. However, significant differences were found among these six genes; for instance, very few amino acid sequence similarities were found, and their exon-intron structures were diversified. This suggests that the laccase genes of subseafloor S. commune have vast evolutional and functional diversity.
The subcellular localization of proteins is invaluable for understanding their functions and interactions with other proteins (Peng and Gao, 2014). Based on subcellular localization analysis, we found that the ScLAC were located in extracellular, cytoplasmic and nuclear, and mitochondrial. The wide distribution of ScLAC in S. commune indicated that these enzymes might have distinct roles in response to various environments (Yang et al., 2021).
The amino acid sequence of fungal laccase generally contains a signal peptide sequence at the N-terminus to guide transmembrane transfer (Jiao et al., 2018). However, some fungal strains have no signal peptide sequence in the laccase gene. For instance, LeLAC3 of Lentinula edodes contained a signal peptide sequence in strain D703PP-9, but was absent in strain W1-26 (Sakamoto et al., 2015; Yan et al., 2019). The deficiency of signal peptide sequence was also reported in Flammulina velutipes and Setosphaeria turcica (Wang et al., 2015; Liu et al., 2019), as well as in plants (Xu et al., 2019). Our study also proved that ScLAC4–ScLAC6 in subseafloor S. commune 20R-7-F01 possessed the signal peptide, while ScLAC1–ScLAC3 did not. These data suggest that the laccase genes differ not only between species but also within species.
In fungi, laccase genes differentiate into many paralogous genes and perform various functions throughout the fungal life cycle (Kumar et al., 2003). They are usually clustered in the form of scaffolds; for instance, the 12 laccase genes in Pleurotus ostreatus and 13 laccase genes in L. edodes were mapped into six and seven scaffolds, respectively (Sakamoto et al., 2015; Jiao et al., 2018). Here, we identified six laccase genes scattered in six unitigs, which were thought to be only specific to S. commune 20R-7-F01 genome; similarly, six laccase genes were distributed on five scaffolds of S. commune strain H4-8 genome. Therefore, we inferred that this difference might be related to species and strain differences.
Gene structure and protein motif analyses can provide a theoretical basis for the function and classification of laccase family genes (Wang et al., 2019). In general, genes in the same group and subgroup should have a similar conserved domain and motif distribution with closely related members in the phylogenetic tree, revealing the functional similarity between proteins in the same subgroup proteins (Yang et al., 2021). Inconsistent with the results of previous studies, we found that the most closely related members of laccase genes in the phylogenetic tree of S. commune had highly diverse motif compositions, and that the conserved motifs of laccases between subseafloor and terrestrial strains were highly diverse. The number and length of introns and exons in ScLAC genes were significantly different. In general, groups B and C had more exons and introns than group A (Figure 2). A small number of introns in a gene usually are the result of genetic evolution, which can rapidly regulate genes in response to stress (Stival Sena et al., 2014). Introns are another source of sequence variation (Jacob and Smith, 2017; Naro and Sette, 2017), and intron retention may increase the diversity of proteins and the complexity of genes expression (Zhang et al., 2004).
Cis-elements play significant roles in the regulatory process to respond to multiple abiotic stresses (Feng et al., 2016). The various cis-elements found in the promoter regions of ScLAC genes were classified into four major groups: substrate utilization-related, stress-related, cell division-related, and amino acid transcription-related cis-elements. These cis-elements may be recognized by some transcription factors and were thus involved in the regulation and expression of ScLAC genes. The presence of multiple cis-elements suggests that ScLAC genes could be involved in fungal response to multiple stresses. Laccases are thought to play an important role in fruiting-body formation (Lettera et al., 2010; Zhang et al., 2015) and our recent investigation found that the biosynthesis of amino acids also helps in the formation of fruiting bodies (Zain Ul Arifeen et al., 2021). Thus, the activation of amino acid transcription-related cis-elements in ScLAC genes could explain the possible role of ScLAC genes in fruiting-body formation.
Lignocellulose degradation by S. commune is an important but complex process, which needs to be thoroughly understood. S. commune utilizes more than 150 genera of woody plants and can also colonize softwood and grass silage (Ohm et al., 2010). As a model mushroom, S. commune H4-8 has complete genome sequence and annotation, and possesses the most extensive polysaccharide decomposition mechanism. The genome of strain H4-8 is rich in the glycoside hydrolase family (hemicellulose and pectin degradation) and polysaccharide lyase family (pectin degradation), which enables it to degrade all plant cell wall components, including lignin (Ohm et al., 2010; Sornlake et al., 2017). Fungi are known to possess a variety of lignin degrading enzymes including lignin peroxidase, manganese peroxidase, dye decolorizing peroxidase, multifunctional peroxidase, and laccase (Floudas et al., 2012). Among these enzymes, laccases are the primary tool lignin degradation in most basidiomycetes (white-rot fungi) and litter-decomposing saprotrophic fungi (Janusz et al., 2020). Laccase catalyzes the one-electron oxidation of substituted phenols, aniline, and aromatic thiols to corresponding free radicals, and reduces molecular oxygen to water (Qi et al., 2015). The broad substrate specificities of laccases, coupled with their use of molecular oxygen as the final electron acceptor rather than the hydrogen peroxide used by ligninolytic peroxidases, makes these enzymes suitable for lignin degradation. However, laccase can only directly degrade phenolic compounds with low-redox-potential, but cannot oxidize the most recalcitrant aromatic hydrocarbons. Nevertheless, some low-molecular-weight compounds produced by fungal degradation of lignin can act as redox mediators to promote the oxidation of refractory substrates (e.g., the non-phenolic lignin moiety) by laccases (Eggert et al., 1996; Camarero et al., 2005).
Basically, laccase use molecular oxygen as the final electron acceptor, and its activity is driven by the concentration of available oxygen (Qi et al., 2015). However, it has been proved that laccase can also oxidize catechol, o-aminophenol, p-aminophenol, o-phenylenediamine, and p-phenylenediamine under anaerobic conditions, with activities of 0.978, 0.707, 0.437, 3.603, and 1.039 mg μmol–1 min–1, respectively (Xie et al., 1999). Shleev et al. (2005) observed direct electron transfer (DET) between the gold electrode and the laccase of Trametes hirsuta under anaerobic conditions. Our previous study also proved that laccase may be involved in the anaerobic degradation of phenanthrene by S. commune 20R-7-F01 (Zain Ul Arifeen et al., 2022). Based on the transcriptome analysis of S. commune 20R-7-F01 during lignin/lignite degradation, we found for the first time that the expression of ScLAC1 was significantly downregulated under anaerobic conditions, while the expression of ScLAC5 and ScLAC6 was significantly up-regulated, compared with that under aerobic conditions (Figure 4 and Supplementary Table 4). These data suggested that ScLAC5 and ScLAC6 may play an important role in the utilization of lignite/lignin and other carbon sources by fungi in anaerobic environment. However, the anaerobic catalytic mechanism of laccase and its effect on fungi to obtain nutrition and energy in the anaerobic subseafloor environments need to be further studied.
Conclusion
A total of six putative laccase genes (ScLAC) with three typical Cu-oxidase domains were identified in S. commune 20R-7-F01 genome. The physical locations of these genes were mapped in six of 162 unitigs of S. commune 20R-7-F01. The theoretical pI of deduced ScLAC proteins widely ranged from 4.62 to 6.56. The MW of the ScLAC proteins ranged from 41.08 to 125.25 kDa and the length varied between 374 and 1,137 amino acids. Based on phylogenetic analysis, the six ScLAC genes were classified into three groups with distinct intron-exon structures and conserved motif. All of the ScLAC had cis-elements related to substrate utilization, stress, cell division, and activates transcription of amino acid in the promoter regions, while the number and type of cis-elements had difference between each other. The phylogenetic tree of resequencing data shows that there are many differences in the number and arrangement of conserved motifs between the ScLAC gene of S. commune strains inhabiting deep subseafloor sediments and the ScLAC gene of strains inhabiting terrestrial environments. The expressions of ScLAC5 and ScLAC6 genes were significantly upregulation under anaerobic conditions, implying that these two laccase genes might be involved in anaerobic utilization and degradation of lignite and lignin by fungi.
In summary, we identified all possible laccase-coding genes from the S. commune reference genome (20R-7-F01) and analyzed the physical and chemical properties, gene structure, amino acid sequence, and systematic evolution, also studied the expression patterns of the gene family under anaerobic and aerobic by growing in lignin/lignite medium. Our data and analysis could facilitate the understanding of the laccase function of white-rot fungi and provide a scientific basis for further exploring the relationship between the ScLAC genes family and the anaerobic degradation of lignin by S. commune.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA858196; https://www.ncbi.nlm.nih.gov/, PRJNA738972.
Author contributions
XL performed data analysis and wrote the first draft of the manuscript. MZ helped with data analysis and edited the manuscript. YX cultivated strains of S. commune and edited the manuscript. CL conceived the study and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (nos. 91951121, 41973073, and 41773083).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.923451/full#supplementary-material
Supplementary Figure 1 | Comparative analysis of the total number of laccases genes and their distribution across the different subclades in the genome of Agaricales. Percentage of laccases in total number of genes was calculated as following: (total number of laccases/total number of predicted genes in the genome) ×100. Genome size and number of predicted genes were retrieved from NCBI or JGI and refer to the current version of the assembled genome of each species.
Supplementary Figure 2 | Venn analysis of predicted cis-elements in the promoter regions of laccase genes from Schizophyllum commune 20R-7-F01.
Footnotes
- ^ http://web.expasy.org/
- ^ http://cello.life.nctu.edu.tw/
- ^ https://services.healthtech.dtu.dk/service.php?SignalP-5.0
- ^ https://services.healthtech.dtu.dk/service.php?TMHMM-2.0
- ^ https://services.healthtech.dtu.dk/service.php?NetNGlyc-1.0
- ^ http://gsds.gao-lab.org/index.php
- ^ http://www.yeastract.com/index.php
- ^ http://meme-suite.org/tools/meme
- ^ http://itol.embl.de/
References
Alexandre, G., and Zhulin, I. B. (2000). Laccases are widespread in bacteria. Trends Biotechnol. 18, 41–42. doi: 10.1016/S0167-7799(99)01406-7
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106.
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Baldrian, P. (2006). Fungal laccases–occurrence and properties. FEMS Microbiol. Rev. 30, 215–242. doi: 10.1111/j.1574-4976.2005.00010.x
Bento, I., Carrondo, M. A., and Lindley, P. F. (2006). Reduction of dioxygen by enzymes containing copper. J. Biol. Inorg. Chem. 11, 539–547. doi: 10.1007/s00775-006-0114-9
Berthet, S., Demont-Caulet, N., Pollet, B., Bidzinski, P., Cézard, L., Le Bris, P., et al. (2011). Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23, 1124–1137. doi: 10.1105/tpc.110.082792
Bilal, M., Asgher, M., Parra-Saldivar, R., Hu, H., Wang, W., Zhang, X., et al. (2017). Immobilized ligninolytic enzymes: An innovative and environmental responsive technology to tackle dye-based industrial pollutants-A review. Sci. Total Environ. 576, 646–659. doi: 10.1016/j.scitotenv.2016.10.137
Camarero, S., Ibarra, D., Martínez, M. J., and Martínez, A. T. (2005). Lignin-derived compounds as efficient laccase mediators for decolorization of different types of recalcitrant dyes. Appl. Environ. Microbiol. 71, 1775–1784. doi: 10.1128/AEM.71.4.1775-1784.2005
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chirivì, C., Fontana, G., Monti, D., Ottolina, G., Riva, S., and Danieli, B. (2012). The quest for new mild and selective modifications of natural structures: Laccase-catalysed oxidation of ergot alkaloids leads to unexpected stereoselective C-4 hydroxylation. Chemistry 18, 10355–10361. doi: 10.1002/chem.201201076
Claus, H. (2003). Laccases and their occurrence in prokaryotes. Arch. Microbiol. 179, 145–150. doi: 10.1007/s00203-002-0510-7
Claus, H. (2004). Laccases: Structure, reactions, distribution. Micron 35, 93–96. doi: 10.1016/j.micron.2003.10.029
Coconi, L. N., Fernández, F., Loske, A. M., and Gómez-Lim, M. A. (2018). Enhanced delignification of lignocellulosic biomass by recombinant fungus Phanerochaete chrysosporium overexpressing laccases and peroxidases. J. Mol. Microbiol. Biotechnol. 28, 1–13.
Eggert, C., Temp, U., Dean, J. F., and Eriksson, K. E. (1996). A fungal metabolite mediates degradation of non-phenolic lignin structures and synthetic lignin by laccase. FEBS Lett. 391, 144–148. doi: 10.1016/0014-5793(96)00719-3
Feng, K., Yu, J., Cheng, Y., Ruan, M., Wang, R., Ye, Q., et al. (2016). The SOD gene family in tomato: Identification, phylogenetic relationships, and expression patterns. Front. Plant Sci. 7:1279. doi: 10.3389/fpls.2016.01279
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Floudas, D., Binder, M., Riley, R., Barry, K., Blanchette, R. A., Henrissat, B., et al. (2012). The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336, 1715–1719. doi: 10.1126/science.1221748
Gautam, A., Kumar, A., Bharti, A. K., and Dutt, D. (2018). Rice straw fermentation by Schizophyllum commune ARC-11 to produce high level of xylanase for its application in pre-bleaching. J. Genet. Eng. Biotechnol. 16, 693–701. doi: 10.1016/j.jgeb.2018.02.006
Gupta, R., and Brunak, S. (2002). Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 7, 310–322.
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, L., Li, H., Chen, L., Lou, Y., Amombo, E., and Fu, J. (2015). RNA-seq for gene identification and transcript profiling in relation to root growth of bermudagrass (Cynodon dactylon) under salinity stress. BMC Genomics 16:575. doi: 10.1186/s12864-015-1799-3
Hu, Q., Min, L., Yang, X., Jin, S., Zhang, L., Li, Y., et al. (2018). Laccase Ghlac1 modulates broad-spectrum biotic stress tolerance via manipulating phenylpropanoid pathway and jasmonic acid synthesis. Plant Physiol. 176, 1808–1823. doi: 10.1104/pp.17.01628
Hua, S., Zhang, B., Fu, Y., Qi, B., Li, Y., Tian, F., et al. (2018). Enzymatic gene expression by Pleurotus tuoliensis (Bailinggu): Differential regulation under low temperature induction conditions. World J. Microbiol. Biotechnol. 34:160.
Inagaki, F., Hinrichs, K. U., Kubo, Y., Bowles, M. W., Heuer, V. B., Hong, W. L., et al. (2015). Exploring deep microbial life in coal-bearing sediment down to ~2.5 km below the ocean floor. Science 349, 420–424. doi: 10.1126/science.aaa6882
Jacob, A. G., and Smith, C. W. J. (2017). Intron retention as a component of regulated gene expression programs. Hum. Genet. 136, 1043-1057. doi: 10.1007/s00439-017-1791-x
Janusz, G., Pawlik, A., Świderska-Burek, U., Polak, J., Sulej, J., Jarosz-Wilkołazka, A., et al. (2020). Laccase properties, physiological functions, and evolution. Int. J. Mol. Sci. 21:966.
Jiao, X., Li, G., Wang, Y., Nie, F., Cheng, X., Abdullah, M., et al. (2018). Systematic analysis of the Pleurotus ostreatus laccase Gene (PoLac) family and functional characterization of PoLac2 involved in the degradation of cotton-straw lignin. Molecules 23:880.
Kirtzel, J., Scherwietes, E. L., Merten, D., Krause, K., and Kothe, E. (2019). Metal release and sequestration from black slate mediated by a laccase of Schizophyllum commune. Environ. Sci. Pollut. Res. Int. 26, 5–13. doi: 10.1007/s11356-018-2568-z
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kudanga, T., Nyanhongo, G. S., Guebitz, G. M., and Burton, S. (2011). Potential applications of laccase-mediated coupling and grafting reactions: A review. Enzyme Microb. Technol. 48, 195–208. doi: 10.1016/j.enzmictec.2010.11.007
Kumar, B., Bhardwaj, N., Alam, A., Agrawal, K., Prasad, H., and Verma, P. (2018). Production, purification and characterization of an acid/alkali and thermo tolerant cellulase from Schizophyllum commune NAIMCC-F-03379 and its application in hydrolysis of lignocellulosic wastes. AMB Express 8:173.
Kumar, S. V., Phale, P. S., Durani, S., and Wangikar, P. P. (2003). Combined sequence and structure analysis of the fungal laccase family. Biotechnol. Bioeng. 83, 386–394. doi: 10.1002/bit.10681
Kumar, S. V., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kumar, V. P., Naik, C., and Sridhar, M. (2015). Production, purification and characterization of novel laccase produced by Schizophyllum commune NI-07 with potential for delignification of crop residues. Appl. Biochem. Microbiol. 51, 432–441. doi: 10.1134/S0003683815040080
Kumar, V. P., Naik, C., and Sridhar, M. (2018). Morphological and phylogenetic identification of a hyper laccase producing strain of Schizophyllum commune NI-07 exhibiting delignification potential. Indian J. Biotechnol. 17, 302–315.
Leonowicz, A., Cho, N. S., Luterek, J., Wilkolazka, A., Wojtas-Wasilewska, M., Matuszewska, A., et al. (2001). Fungal laccase: Properties and activity on lignin. J. Basic Microbiol. 41, 185–227. doi: 10.1002/1521-4028(200107)41:3/4<185::AID-JOBM185>3.0.CO;2-T
Lettera, V., Piscitelli, A., Leo, G., Birolo, L., Pezzella, C., and Sannia, G. (2010). Identification of a new member of Pleurotus ostreatus laccase family from mature fruiting body. Fungal Biol. 114, 724–730. doi: 10.1016/j.funbio.2010.06.004
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Liu, C. H., Huang, X., Xie, T. N., Duan, N., Xue, Y. R., Zhao, T. X., et al. (2017). Exploration of cultivable fungal communities in deep coal-bearing sediments from ∼1.3 to 2.5 km below the ocean floor. Environ. Microbiol. 19, 803–818. doi: 10.1111/1462-2920.13653
Liu, N., Cao, Z. Y., Cao, K. K., Ma, S. X., Gong, X. D., Jia, H., et al. (2019). Identification of laccase-like multicopper oxidases from the pathogenic fungus Setosphaeria turcica and their expression pattern during growth and infection. Eur. J. Plant Pathol. 153, 1149–1163. doi: 10.1007/s10658-018-01632-8
Liu, X., Huang, X., Chu, C., Xu, H., Wang, L., Xue, Y. R., et al. (2022). Genome, genetic evolution, and environmental adaptation mechanisms of Schizophyllum commune in deep subseafloor coal-bearing sediments. iScience 25:104417.
Mayer, A. M., and Staples, R. C. (2002). Laccase: New functions for an old enzyme. Phytochemistry 60, 551–565. doi: 10.1016/S0031-9422(02)00171-1
Mehmood, T., Saman, T., Irfan, M., Anwar, F., Ikram, M. S., and Tabassam, Q. (2019). Pectinase production from Schizophyllum commune through central composite design using citrus waste and its immobilization for industrial exploitation. Waste Biomass Valori. 10, 2527–2536. doi: 10.1007/s12649-018-0279-9
Monteiro, P. T., Oliveira, J., Pais, P., Antunes, M., Palma, M., Cavalheiro, M., et al. (2020). YEASTRACT+: A portal for cross-species comparative genomics of transcription regulation in yeasts. Nucleic Acids Res. 48, D642–D649. doi: 10.1093/nar/gkz859
Mtibaà, R., Barriuso, J., de Eugenio, L., Aranda, E., Belbahri, L., Nasri, M., et al. (2018). Purification and characterization of a fungal laccase from the ascomycete Thielavia sp. and its role in the decolorization of a recalcitrant dye. Int. J. Biol. Macromol. 120, 1744–1751. doi: 10.1016/j.ijbiomac.2018.09.175
Naro, C., and Sette, C. (2017). Timely-regulated intron retention as device to fine-tune protein expression. Cell Cycle 16, 1321–1322. doi: 10.1080/15384101.2017.1337983
Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. doi: 10.1093/protein/10.1.1
Ohm, R. A., de Jong, J. F., Lugones, L. G., Aerts, A., Kothe, E., Stajich, J. E., et al. (2010). Genome sequence of the model mushroom Schizophyllum commune. Nat. Biotechnol. 28, 957–963. doi: 10.1038/nbt.1643
Peng, C., and Gao, F. (2014). Protein localization analysis of essential genes in prokaryotes. Sci. Rep. 4:6001.
Qi, Y. B., Wang, X. L., Shi, T., Liu, S., Xu, Z. H., Li, X., et al. (2015). Multicomponent kinetic analysis and theoretical studies on the phenolic intermediates in the oxidation of eugenol and isoeugenol catalyzed by laccase. Phys. Chem. Chem. Phys. 17, 29597–29607. doi: 10.1039/C5CP03475B
Roberts, A., Pimentel, H., Trapnell, C., and Pachter, L. (2011). Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27, 2325–2329. doi: 10.1093/bioinformatics/btr355
Rodríguez, C. S., and Toca Herrera, J. L. (2006). Industrial and biotechnological applications of laccases: A review. Biotechnol. Adv. 24, 500–513. doi: 10.1016/j.biotechadv.2006.04.003
Sakamoto, Y., Nakade, K., Yoshida, K., Natsume, S., Miyazaki, K., Sato, S., et al. (2015). Grouping of multicopper oxidases in Lentinula edodes by sequence similarities and expression patterns. AMB Express 5:63.
Shiba, T., Xiao, L., Miyakoshi, T., and Chen, C. L. (2000). Oxidation of isoeugenol and coniferyl alcohol catalyzed by laccases isolated from Rhus vernicifera Stokes and Pycnoporus coccineus. J. Mol. Catal. B Enzym. 10, 605–615. doi: 10.1016/S1381-1177(00)00184-3
Shleev, S., Christenson, A., Serezhenkov, V., Burbaev, D., Yaropolov, A., Gorton, L., et al. (2005). Electrochemical redox transformations of T1 and T2 copper sites in native Trametes hirsuta laccase at gold electrode. Biochem. J. 385, 745–754. doi: 10.1042/BJ20041015
Singh, A. D., and Sharma, R. K. (2010). Ligninolytic fungal laccases and their biotechnological applications. Appl. Biochem. Biotechnol. 160, 1760–1788. doi: 10.1007/s12010-009-8676-y
Solomon, E. I., Sundaram, U. M., and Machonkin, T. E. (1996). Multicopper oxidases and oxygenases. Chem. Rev. 96, 2563–2606. doi: 10.1021/cr950046o
Sornlake, W., Rattanaphanjak, P., Champreda, V., Eurwilaichitr, L., Kittisenachai, S., Roytrakul, S., et al. (2017). Characterization of cellulolytic enzyme system of Schizophyllum commune mutant and evaluation of its efficiency on biomass hydrolysis. Biosci. Biotechnol. Biochem. 81, 1289–1299. doi: 10.1080/09168451.2017.1320937
Stival Sena, J., Gigue‘re, I., Boyle, B., Rigault, P., Birol, I., Zuccolo, A., et al. (2014). Evolution of gene structure in the conifer Picea glauca: A comparative analysis of the impact of intron size. BMC Plant Biol. 14:95. doi: 10.1186/1471-2229-14-95
Tovar-Herrera, O. E., Martha-Paz, A. M., Pérez, L. Y., Aranda, E., Tacoronte-Morales, J. E., Pedroso-Cabrera, M. T., et al. (2018). Schizophyllum commune: An unexploited source for lignocellulose degrading enzymes. Microbiologyopen 7:e00637.
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Vasina, D. V., Mustafaev, O. N., Moiseenko, K. V., Sadovskaya, N. S., Glazunova, O. A., Tyurin, À, et al. (2015). The Trametes hirsuta 072 laccase multigene family: Genes identification and transcriptional analysis under copper ions induction. Biochimie 116, 154–164. doi: 10.1016/j.biochi.2015.07.015
Voorrips, R. E. (2002). MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, Q., Li, G., Zheng, K., Zhu, X., Ma, J., Wang, D., et al. (2019). The Soybean laccase gene family: Evolution and possible roles in plant defense and stem strength selection. Genes 10:701.
Wang, W., Liu, F., Jiang, Y., Wu, G., Guo, L., Chen, R., et al. (2015). The multigene family of fungal laccases and their expression in the white rot basidiomycete Flammulina velutipes. Gene 563, 142–149. doi: 10.1016/j.gene.2015.03.020
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552.
Xie, X. Y., Yan, C. N., Wu, D. Q., and Qu, S. S. (1999). Reactivity properties of laccase with various types of substrates. J. Cent. China Norm. Univ. 2, 246–249.
Xu, X., Zhou, Y., Wang, B., Ding, L., Wang, Y., Luo, L., et al. (2019). Genome-wide identification and characterization of laccase gene family in Citrus sinensis. Genes 689, 114–123.
Yan, L., Xu, R., Bian, Y., Li, H., and Zhou, Y. (2019). Expression profile of laccase gene family in white-rot basidiomycete Lentinula edodes under different environmental stresses. Genes 10:1045.
Yang, Y., Sossah, F. L., Li, Z., Hyde, K. D., Li, D., Xiao, S., et al. (2021). Genome-wide identification and analysis of chitinase GH18 gene family in Mycogone perniciosa. Front. Microbiol. 11:596719. doi: 10.3389/fmicb.2020.596719
Yoshida, H. (1883). Chemistry of lacquer (urushi). J. Chem. Soc. 43, 472–486. doi: 10.1039/CT8834300472
Yu, C. S., Chen, Y. C., Lu, C. H., and Hwang, J. K. (2006). Prediction of protein subcellular localization. Proteins 64, 643–651. doi: 10.1002/prot.21018
Zain Ul Arifeen, M., Chu, C., Yang, X. Y., Liu, J. Z., Huang, X., Ma, Y. N., et al. (2021). The anaerobic survival mechanism of Schizophyllum commune 20R-7-F01, isolated from deep sediment 2 km below the seafloor. Environ. Microbiol. 23, 1174–1185. doi: 10.1111/1462-2920.15332
Zain Ul Arifeen, M., Ma, Y. N., Wu, T. S., Chu, C., Liu, X., Jiang, J. P., et al. (2022). Anaerobic biodegradation of polycyclic aromatic hydrocarbons (PAHs) by fungi isolated from anaerobic coal-associated sediments at 2.5 km below the seafloor. Chemosphere 9:135062.
Zain Ul Arifeen, M., Yang, X. Y., Li, F. F., Xue, Y. R., Gong, P. X., and Liu, C. H. (2020). Growth behaviors of deep subseafloor Schizophyllum commune in response to various environmental conditions. Acta Microbiol. Sin. 60, 1882–1892.
Zhang, G. W., Song, H. D., and Chen, Z. (2004). Molecular mechanism of mRNA alternative splicing. J. Genet. Genomics 31, 102–107.
Zhang, J., Chen, H., Chen, M., Ren, A., Huang, J., Wang, H., et al. (2015). Cloning and functional analysis of a laccase gene during fruiting body formation in Hypsizygus marmoreus. Microbiol. Res. 179, 54–63. doi: 10.1016/j.micres.2015.06.005
Zhang, Y., Lv, Z., Zhou, J., Xin, F., Ma, J., Wu, H., et al. (2018). Application of eukaryotic and prokaryotic laccases in biosensor and biofuel cells: Recent advances and electrochemical aspects. Appl. Microbiol. Biotechnol. 102, 10409–10423. doi: 10.1007/s00253-018-9421-7
Keywords: Schizophyllum commune, subseafloor, laccase, lignin degradation, fungi, phylogeny
Citation: Liu X, Zain ul Arifeen M, Xue Y and Liu C (2022) Genome-wide characterization of laccase gene family in Schizophyllum commune 20R-7-F01, isolated from deep sediment 2 km below the seafloor. Front. Microbiol. 13:923451. doi: 10.3389/fmicb.2022.923451
Received: 19 April 2022; Accepted: 19 July 2022;
Published: 08 August 2022.
Edited by:
Zheng Zhang, Shandong University, ChinaReviewed by:
Hanif Ullah, COMSATS Institute of Information Technology, PakistanRegina Sharmila Dass, Pondicherry University, India
Copyright © 2022 Liu, Zain ul Arifeen, Xue and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changhong Liu, Y2hsaXVAbmp1LmVkdS5jbg==
 Xuan Liu
Xuan Liu Muhammad Zain ul Arifeen
Muhammad Zain ul Arifeen Yarong Xue
Yarong Xue Changhong Liu
Changhong Liu