
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 27 September 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.923181
This article is part of the Research Topic Community Series in Plants and Microbial Communities: Diversity, Pathogens and Biological Control, volume II View all 25 articles
 Keqin Peng
Keqin Peng Yintao Pan
Yintao Pan Tingjun Tan
Tingjun Tan Xiangyu Zeng
Xiangyu Zeng Meiling Lin
Meiling Lin Shuang Jiang
Shuang Jiang Zhibo Zhao
Zhibo Zhao Fenghua Tian*
Fenghua Tian* Xiaosheng Zhao*
Xiaosheng Zhao*Sweet cherry is an important fruit crop with high economic and ornamental value in China. However, cherry fruit anthracnose, caused by Colletotrichum species, greatly impacts cherry yield and quality. Here, we surveyed cherry anthracnose in Guizhou, China from 2019–2020. Necrotic sweet cherry fruits were collected from different areas in Guizhou and examined. A total of 116 Colletotrichum strains were isolated from these symptomatic fruits. Based on the morphological characteristics of the isolates and phylogenetic analyses of concatenate internal transcribed spacer (ITS) region and ACT, CHS-1, GAPDH, TUB2, and HIS3 genes, the pathogen responsible for causing sweet cherry anthracnose was identified as Colletotrichum godetiae. Pathogenicity tests were conducted by inoculating healthy sweet cherry fruits with spore suspensions of the fungal pathogen, and Koch’s postulates were confirmed by pathogen re-isolation and identification. The Q-1 isolate showed different sensitivities to 13 fungicides, exhibiting seven different modes of action, and its EC50 values ranged from 0.04 to 91.26 μg ml−1. According to that, the sensitivity of 20 isolates from different samples to ten fungicides with better performance, were measured. The results showed that 6 of the 10 fungicides (difenoconazole, propiconazole, prochloraz-manganese, pyraclostrobin, trifloxystrobin-tebuconazole, and difenoconazole-azoxystrobin) all showed higher sensitive to the 20\u00B0C. godetiae isolates, and no resistance groups appeared. Its EC50 values ranged from 0.013 to 1.563 μg ml−1. In summary, this is the first report demonstrating that C. godetiae causes sweet cherry anthracnose and the results of this study provide insights into how sweet cherry anthracnose could be effectively controlled in China.
Chinese sweet cherry (Cerasus pseudocerasus Lindl.; Rosaceae) is an important native fruit crop with high economic and ornamental value (Chen et al., 2012, 2013b). ‘Manaohong’, approved by Guizhou Provincial Variety Approval Committee in 2011, is an unique local sour cherry variety in Guizhou Province, China. Cherry fruits are rich in vitamins, niacin, phenolic compounds, and minerals (Kim et al., 2005; Zhang et al., 2008). Sweet cherry cultivation is one of the 12 most important agricultural sectors that contribute to the development of characteristic agriculture in Guizhou. With the expansion of ‘Manaohong’ cherry cultivation and the increase in temperature and rainfall, the occurrence of fungal disease of cherry fruits has significantly increased in Guizhou.
Anthracnose is a widespread disease that reduces crop yield and quality, resulting in great economic losses. Colletotrichum, the causal agent of anthracnose, is one of the top 10 fungal genera of economic and scientific importance (Dean et al., 2012). Colletotrichum is the only genus in the Glomerellaceae family (order Glomerellales, class Sordariomycetes) (Wijayawardene et al., 2018, 2020). Species of Colletotrichum are known as pathogens (causing anthracnose and postharvest fruit rot in plants), endophytes (producing a range of secondary metabolites), and saprobes (Bhunjun et al., 2021). Anthracnose, caused by Colletotrichum spp., is an important disease that seriously threatens the production of sweet cherry. Although Colletotrichum spp. are known to infect leaves and young shoots of cherry trees, they most frequently infect cherry fruits at various developmental stages (Børve and Stensvand, 2006a, 2013; Børve et al., 2010). For example, on the sweet cherry fruits infected by Colletotrichum acutatum in Norway, the initial symptoms of anthracnose on young fruit include dark brown spots, which later spread to the whole fruit and block fruit development. On mature fruits, the disease lesions are sunken and dark brown, and sticky piles of orange/yellow spores are formed on the lesion. Damage to the fruit reduces the yield and quality of sweet cherry, causing great economic losses (Børve and Stensvand, 2013).
A clear understanding of Colletotrichum species involved in sweet cherry anthracnose is essential for disease management. The genus Colletotrichum is divided into 14 species complexes, which comprise approximately 189 species (Jayawardena et al., 2021). In previous studies, morphological characterization and molecular characterization were used to accurately identify Colletotrichum species (Cai et al., 2011). Most of the Colletotrichum spp. displayed an unique colony color, mycelial growth rate, and size and shape of conidia and appressoria, which were used as key morphological traits for preliminary identification (Damm et al., 2012). The Colletotrichum spp. were further distinguished based on molecular data, including two intergenic regions, internal transcribed spacer (ITS) region and the intergenic region between apn2 and MAT1-2-1 genes (ApMAT), and partial DNA sequences of five genes, namely actin (ACT), chitin synthase (CHS-1), histone 3 (HIS3), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-tubulin (TUB2) (Damm et al., 2012; Sharma et al., 2013). Based on this method, a growing number of Colletotrichum spp. causing cherry anthracnose has been reported. For example, four Colletotrichum species, C. aenigma based on ITS, DAPDH, ACT, TUB2, and CHS-1 genes (Beijing city; Chethana et al., 2019), C. pseudotheobromicola sp. nov. based on ITS, DAPDH, ACT, TUB2 and CHS-1 genes (Beijing city; Chethana et al., 2019), C. liaoningense based on ITS, DAPDH, ACT, TUB2 and CHS-1 genes (Shandong province; Liu et al., 2021) and C. fructicola based on ITS, DAPDH, ACT, TUB2, and CHS-1 genes (Zhejiang province; Tang et al., 2021), causing leaf spot on cherry, have been reported in China. In Brazil, Colletotrichum theobromicola was determined to cause necrotic and sunken spots on Barbados cherry fruit (Bragança et al., 2014). In southwestern Norway, Colletotrichum acutatum was reported to infect sweet cherry leaves (Børve et al., 2010), and overwinter on the buds (Børve and Stensvand, 2006a; Stensvand et al., 2017) and shoots (Stensvand et al., 2017) of sweet cherry, thus serving as the primary source of inoculum for more infections in the growing season. Among the above six pathogens, C. aenigma, C. pseudotheobromicola, C. fructicola, and C. theobromicola, were classified into the C. gloeosporioides species complex; C. acutatum was assigned to the C. acutatum species complex; and C. liaoningense was determined as a singleton species (Jayawardena et al., 2016).
Although the application of fungicides has adverse effects on the environment, chemical control is still the most effective measure for controlling the anthracnose disease. Recently, a number of studies reported the emergence of fungicide resistant strains of Colletotrichum species. In Hainan province, C. gloeosporioides strains highly resistant to carbendazim were isolated from mango, litchi, and longan (Zhang et al., 2014). Moreover, C. gloeosporioides, which causes grape ripe rot, showed a resistance to thiophanate-methyl and diethofencarb (Chen et al., 2013a). In other studies, fungicides were shown to be highly effective in controlling the anthracnose disease. For example, dithianon was effective in controlling anthracnose in sweet and sour cherry in Norway (Børve and Stensvand, 2006b). Methyl benzimidazole carbamate (MBC) and demethylation inhibitor (DMI) fungicides were used against Colletotrichum recently (Chen et al., 2016). In the USA, quinone outside inhibitors (QoIs) are the most common fungicides used in commercial strawberry fields for controlling anthracnose caused by Colletotrichum spp. (Forcelini et al., 2017). Understanding the sensitivity of Colletotrichum spp. to various fungicides may have significant implications on the effective management of this disease. However, the fungicide sensitivities of Colletotrichum isolates causing sweet cherry anthracnose in China remain unknown.
In the present study, we isolated Colletotrichum species associated with sweet cherry anthracnose in Guizhou province, and identified these species based on their morphological traits and multilocus phylogeny. Then, the pathogenicity of Colletotrichum species was determined by the artificial inoculation of sweet cherry fruits. Finally, we determined the sensitivities of the Colletotrichum isolates to different fungicides. The results of these analyses provide important information that could be used to develop an effective strategy for controlling sweet cherry fruit anthracnose.
During 2019–2020, anthracnose disease investigations were conducted on sweet cherry orchards in four regions of China’s Guizhou Province, namely Bijie (26°19′N, 106°46′E), Guiyang (26°19′N, 106°46′E), Liupanshui (26°18′N, 104°51′E) and Qianxinanzhou (25°56′N, 107°18′E). Through the statistics of diseased plant rate, we found that its incidence had reached 10 to 20%, hindering cherry fruit industry development. Therefore, to clarify the cause of the disease, we collected about 40 diseased sweet cherry fruits at different developmental stages from diseased orchards in these four regions.
To isolate the disease-causing pathogen, the infected fruits were washed with tap water, dried on absorbent paper, and then surface-sterilized by rubbing the surface of the lesion three times with a 75% ethanol-soaked cotton ball. The diseased tissue was crushed, immersed in sterilized water, and then subjected to gradient dilution, Low-titer spore suspensions were spread on a potato dextrose agar (PDA) plate (Yuan et al., 2021). It was then placed in the dark at 25°C for 4 days, and the single colonies that grew were picked into new PDA plates. The pure colonies were soaked in 20% (v/v) glycerol at −70°C for long-term storage (Kim et al., 2020). The isolated strains were classified according to the sampling location and strains with the same morphology, and 20 isolates were chosen for further study.
The colony color of each isolate was recorded after culturing on PDA at 25°C for 6 days. Mycelial plugs (5 mm) excised from the margin of 6-day-old colonies were placed at the center of each PDA plate (total five plates), and cultured in darkness at 25°C. The diameter of each colony was measured by cross direction, and its daily growth rate was calculated. The experiment was repeated three times. A small amount of mycelia was scraped from 10-day-old colonies to observe the mycelial appressorium. Hyphal tips were sampled from the agar, transferred to fresh PDA plates, and incubated at 25°C for 10 days to induce the formation of conidia. Appressoria were induced as described previously (Cai et al., 2009). Using a compound light microscope (Zeiss Scope 5 with camera AxioCam 208 color), the shape and size of mycelial appressorium, conidia, and appressorium were recorded and measured.
Genomic DNA was extracted from the mycelia of 6-day-old colonies cultured on PDA plates using the CWBIOTECH Plant Genomic DNA Kit (Changping, Beijing, China). The ITS region of the rDNA gene cluster, a 200-bp intron of the GAPDH gene, and partial sequences of CHS-1, ACT, TUB2, and HIS3 genes were amplified from the genomic DNA using ITS4/ITS5, GDF1/GDR1, CHS-79F/CHS-354R, ACT-512F/ACT-783R, T1/T2, and CYLH3F/CYLH3R primer pairs, respectively (Crous et al., 2004; Damm et al., 2012; López-Moral et al., 2017). The sequences of primers used in this study are listed in Table 1.
PCR was performed on the T100™ Thermal Cycler (Bio-Rad Laboratories Inc., CA, USA) in a 25-μL reaction volume containing 1.6 μl of dNTPs (2.5 mM μL−1 each), 0.2 μl of Taq polymerase (5 U μL−1), 2 μl of polymerase buffer (10× μL−1; Takara, Japan), 1 μl of each primer (25 mM μL−1) and 1 μl of genomic DNA (50 ng μL−1). The following conditions were used for the amplification of all genomic regions (except the ITS region): initial denaturation at 95°C for 5 min, followed by 32 cycles of denaturation at 94°C, annealing at 55°C, and extension at 72°C for 30 s each and a final extension at 72°C for 10 min. The ITS region was amplified under the following conditions: 94°C for 5 min, followed by 30 cycles at 94°C, 52°C and 72°C for 30 s each and lastly 72°C for 10 min. PCR products were sequenced by Sangon Biotech Co., Ltd. (Shanghai, China) using the same PCR primers as those used for PCR amplification.
Sequences of each gene or genomic region generated using forward and reverse primers were assembled with BioEdit v.7.2.5 (Hall, 1999). Then, consensus sequences were then combined with related sequences downloaded from GenBank, and aligned separately using Mafft v7.187 (Katoh and Standley, 2013) or manually when necessary. The nucleotide substitution model for each gene or genomic region was determined based on the Bayesian information criterion (BIC) using jModelTest v2.1.6 (Darriba et al., 2012). Phylogenetic trees based on ITS, ACT, CHS-1, GAPDH, TUB2, and HIS3 datasets as well as a concatenated dataset were constructed using maximum likelihood (ML) and Bayesian inference (BI) analyses at the CIPRES web portal (Miller et al., 2010). The ML analysis was performed using the RAxML-HPC BlackBox tool (Stamatakis, 2014). The Markov Chain Monte Carlo (MCMC) algorithm for BI with two parallel runs of four chains was performed using MrBayes on XSEDE (Ronquist et al., 2012). Trees were sampled every 100 generations, and runs were stopped automatically when the average standard deviation of split frequencies fell below 0.01. A 50% majority rule consensus tree was summarized after discarding the first 25% samples. The resulting trees were visualized in FigTree v1.4.3 (Rambaut, 2016). In this study, 20 isolates were selected for phylogenetic analysis.
To conform the Koch’s postulates, three C. godetiae isolates (Q-1, Q-2, and Q-3) were chosen for pathogenicity testing. Healthy sweet cherry fruits were selected in the field and sprayed with conidial suspensions (1 × 106 conidia ml−1) or sterile distilled water (control). Five replicates of fruit were used for each conidial suspension. Disease development on the inoculated fruit was observed daily, and lesions on fruit were photographed at 5 days post-inoculation. The fungus was re-isolated from infected fruit after the appearance of symptoms. The morphological characteristics of these fungal isolates were compared with those originally used as inoculum.
Referring to the fungicides with high efficiency and low intensity used for the control of other plant anthracnose, 13 kinds of fungicides were selected for preliminary screening of Q-1 isolates (Table 2). Thirteen fungicides, including difenoconazole, propiconazole, prochloraz-manganese, azoxystrobin, pyraclostrobin, chlorothalonil, dithianon, polyantimycin, zhongshengmycin, trifloxystrobin-tebuconazole, difenoconazole-azoxystrobin, bromothalonil, and polysaccharide, were dissolved in sterile water to prepare 10 mg/ml stock solutions. Half maximal effective concentration (EC50) values indicating fungicide sensitivity were determined using mycelial growth assays. The PDA medium plates, containing a series of final concentrations of each fungicide (Table 3), were prepared by adding an appropriate volume of the stock solution (in sterile water). The margin of 6-day-old colonies cultured on PDA was used to generate 5-mm mycelial plugs, which were placed at the center of PDA plates containing different concentrations of fungicides. All plates were incubated at 25°C for 6 days. The growth inhibition rate of mycelia was calculated by the following formula: i = (a1 − a2)/a1 × 100, where i is the growth inhibition rate of mycelia, a1 is the hyphae area of untreated pathogen, and a2 is the hyphae area of treated pathogen (Etebarian et al., 2005). The EC50 (concentration for 50% of maximal effect) values of different plant extracts were calculated using IBM SPSS analytics (SPSS Inc., Chicago, IL, United States) (Mo et al., 2021). Each fungicide treatment and control contained three replicate plates, and the experiment was performed twice.
A suitable fungicide was selected based on the preliminary screening of 13 fungicides using the Q-1 isolate. Then, the sensitivity of 20 isolates (5, 5, 2, and 8 isolates from Bijie, Guiyang, Liupanshui, and Qianxinanzhou, respectively) to the suitable fungicide was tested as described above.
A fruit disease emerged in four major ‘Manaohong’ cherry production areas (Bijie, Guiyang, Liupanshui, and Qianxinanzhou) in Guizhou province (Figure 1). Symptomatic sweet cherry fruits displaying lesion collapsed but maintained an intact peel. At the early stage of infection, yellow, caviar-like patches were formed on the fruit surface. With increased disease severity over time, the fruit became rotten, and lesions turned black (Figures 2A–D). Microscopic analysis revealed the presence of brown acervuli (Figure 3A) occasionally branched hyaline conidiophores (Figures 3C–F), black setae (Figure 3B), and aseptate, cylindrical, and slightly curved conidia (Figure 3G) on the surface of diseased fruits, suggesting that the disease was caused by Colletotrichum species. To confirm the identity of the causal agent, a total of 116 Colletotrichum isolates were isolated from diseased sweet cherry fruits using the single-spore separation method. We classified the obtained 116 isolates according to the collection location and colony morphology. We divided these strains into three categories according to the colony morphology, and finally selected 20 strains for the next experiment. In these 20 isolates, 5 (named as B-1 to B-5), 5 (G-1 to G-5), 2 (L-1 to L-2), and 8 (Q-1 to Q-8) Colletotrichum isolates, obtained from Bijie, Guiyang, Liupanshui and Qianxinanzhou, respectively, were chosen for further study.

Figure 2. The symptoms of cherry anthracnose on fruits. (A) Symptoms on fruits in field. (B) Symptoms on fruits at different development stages. (C) Necrotic spot on mature fruit. (D) Margin of lesions on mature fruit.
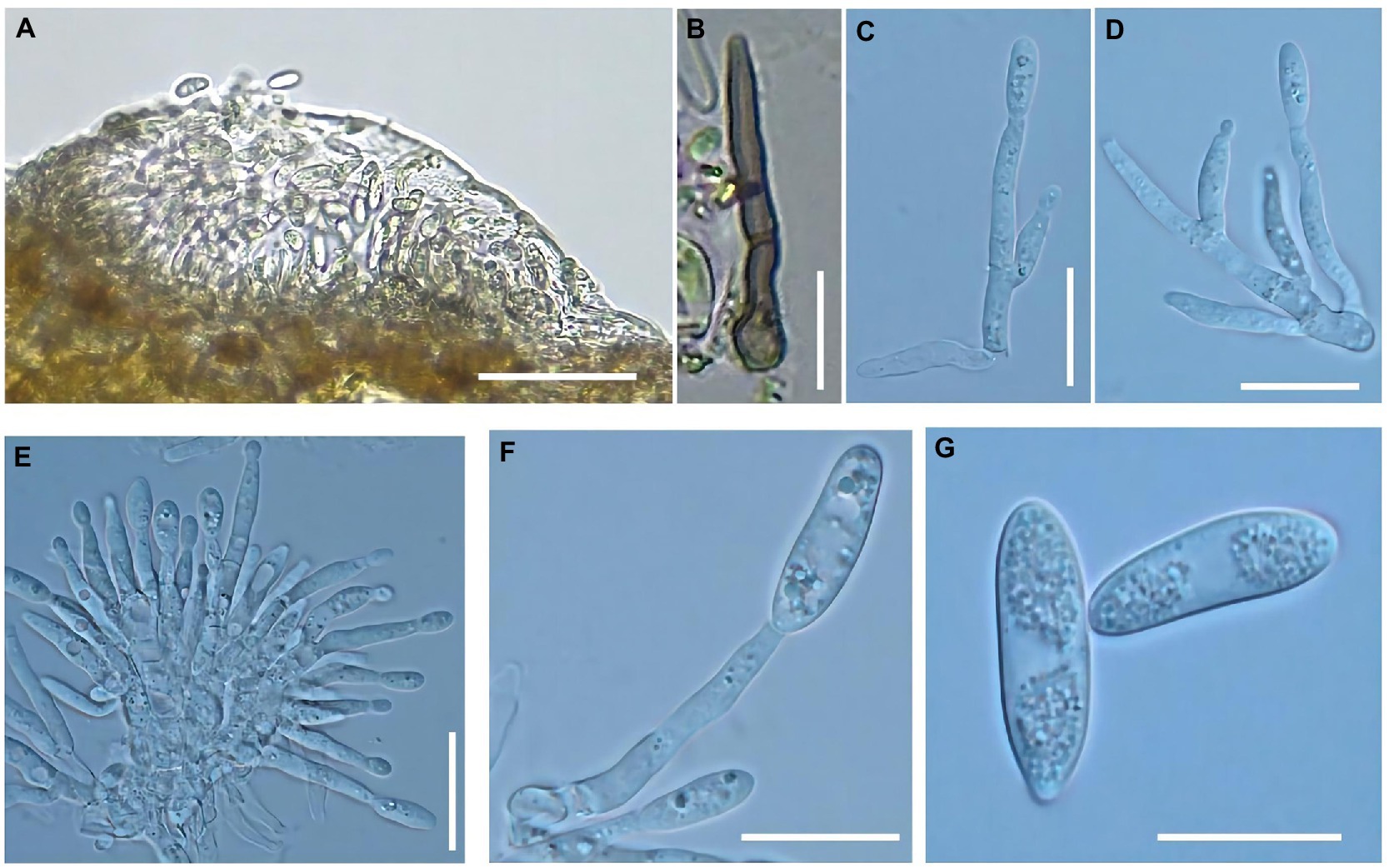
Figure 3. Morphological characteristics of Colletotrichum godetiae were observed on the diseased fruits. (A) Acervuli, bar = 50 μm. (B) Setae, bar = 20 μm. (C–F) Conidiophores, bar = 10 μm. (G) Conidia, bar = 10 μm.
After growth on PDA at 25°C for 6 days, the colonies were light gray at the margin, dark gray at the center and the underside, with dense and cottony aerial mycelium (Figures 4A,B). Its growth rate was 6.6 mm per day, and its growth rate was consistent with each repetition. Setae were dark brown and acicular (Figure 4C). The mycelial appressorium was dark brown or black in color and elliptical or irregular in shape (Figure 4D), and ranged from 6.9–11.1 × 4.2–7.4 μm in size (n = 30). Conidiophores were hyaline, smooth-walled, and crooked with no branches (Figure 4E). Conidia on PDA were transparent with aseptate, cylindrical, and slightly curved (Figure 4F), and ranged in size from 12.5–17.6 × 3.5–5.3 μm (n = 40). The appressoria were dark brown or black and elliptical or irregular (Figures 4G,H), and ranged in size from 7.7–10.8 × 6.2–9.8 μm (n = 30). The color and shape of setae, conidiophores, and conidia formed on PDA plates and on diseased fruit were similar. Overall, the morphological characteristics of the isolates determined in this study were consistent with those of C. godetiae described by Damm et al. (2012).
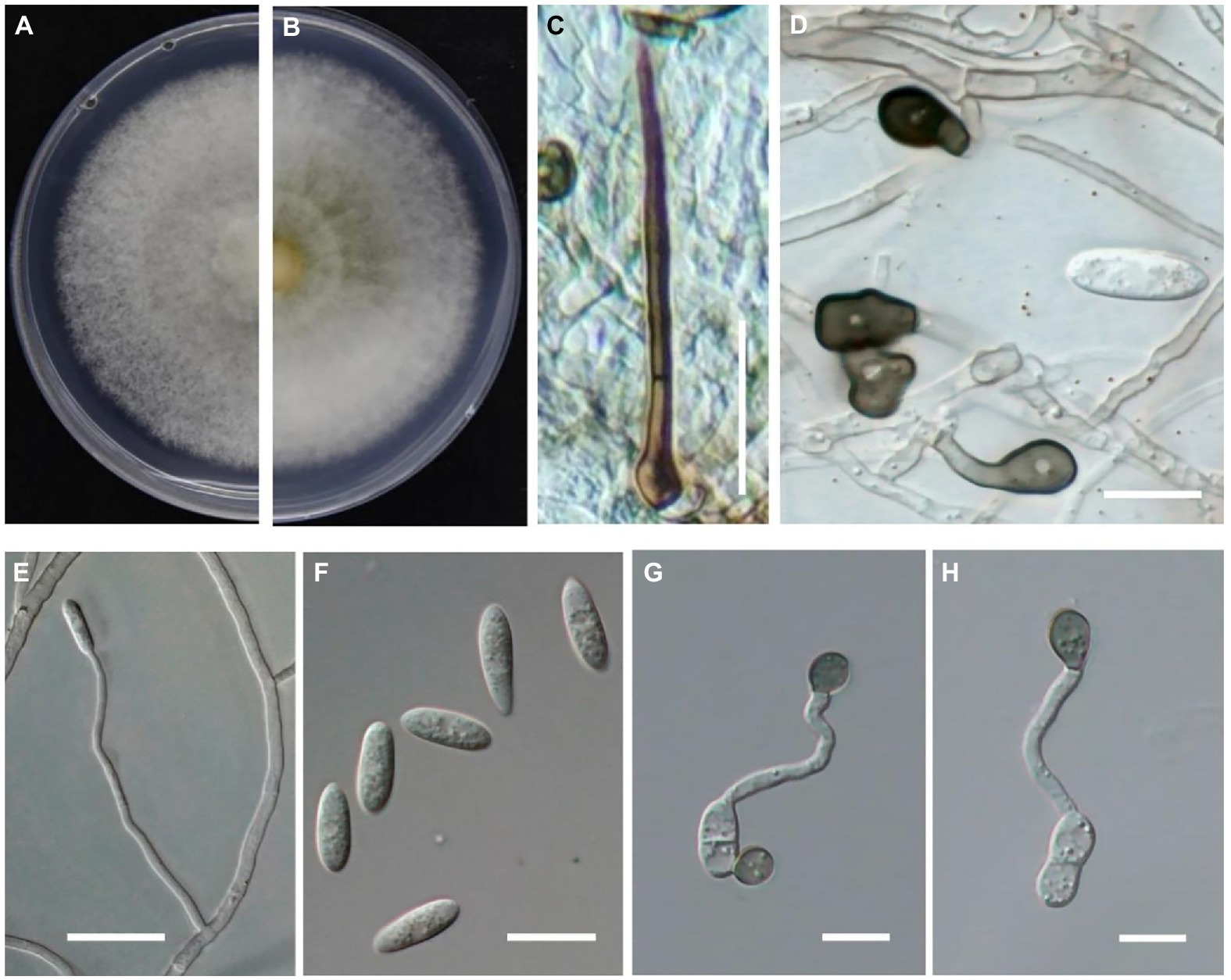
Figure 4. Morphological characteristics of Colletotrichum godetiae cultured on PDA plates. (A,B) Colony morphology on upside (A) and underside (B) of C. godetiae on PDA after 7 days at 25°C. (C) Setae, bar = 20 μm. (D) Mycelial appressorium, bar = 10 μm. (E) Conidiophores, bar = 10 μm. (F) Conidia, bar = 10 μm. (G,H) Appressorium, bar = 10 μm.
Twenty colletotrichum isolates (B-1 to B-5, G-1 to G-5, L-1 to L-2, and Q-1 to Q-8) obtained from the four cities were selected for further analysis. Six DNA fragments (ITS, ACT, CHS-1, GAPDH, TUB2, and HIS3) combined a gene alignment data matrix was used to perform phylogenetic analysis. The sequences of Six PCR fragments of each isolate were deposited in GenBank, and the accession numbers are listed in Table 4. In Figure 5, the results showed that our new collections (20 isolates) were clustered with C. godetiae. Based on the multilocus phylogenetic analyses of five genomic regions and morphological characteristics of colonies, conidia, appressoria, conidiophores, and setae, the isolates were identified as C. godetiae.
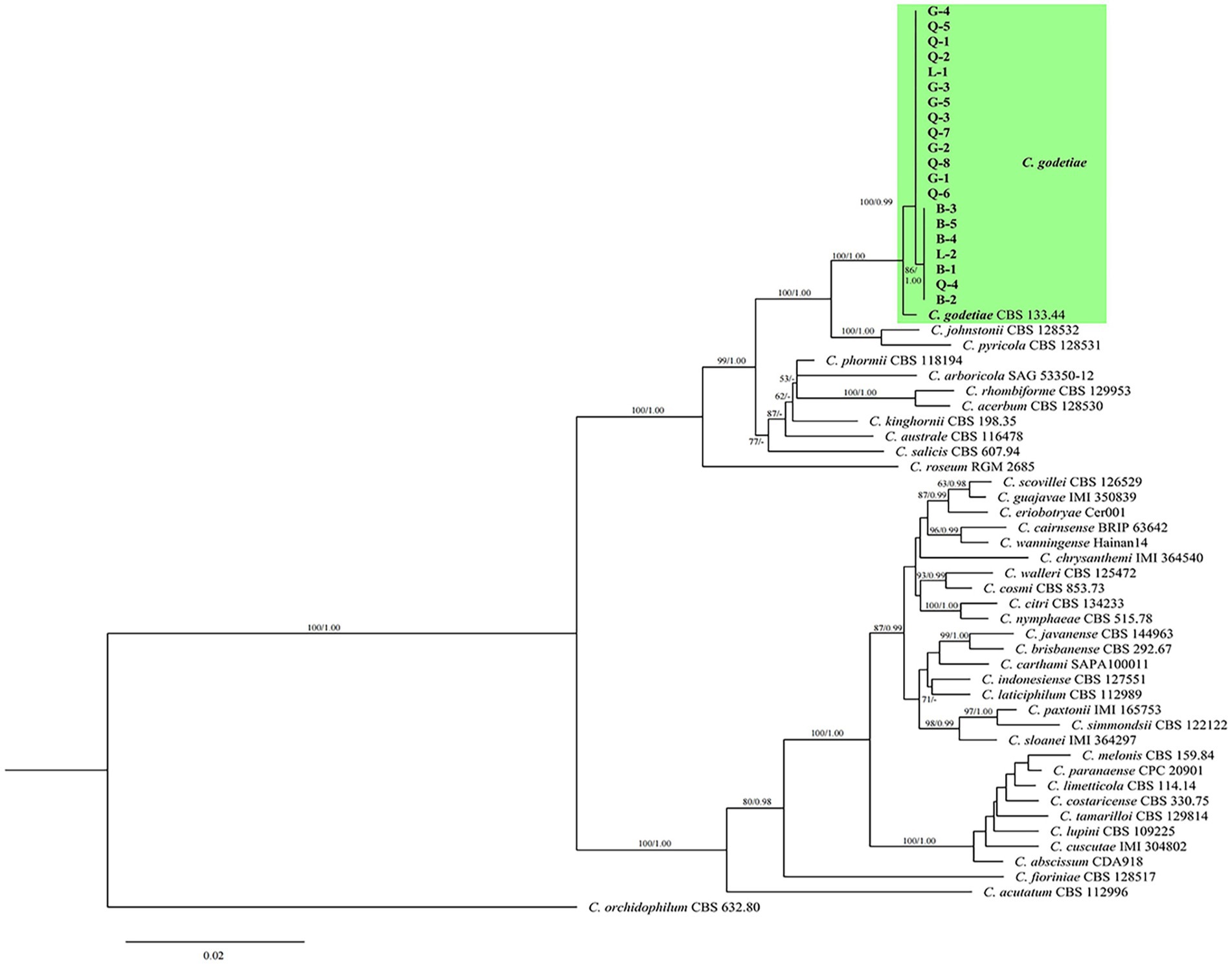
Figure 5. Maximum-likelihood (ML) and Bayesian inference (BI) phylogenetic tree illustrating the relationships with the godetiae species complex and the Colletotrichum strains isolated from diseased cherry fruits in Guizhou. Bootstrap support values for ML greater than 50% and Bayesian posterior probabilities greater than 0.90 are shown next to topological nodes.
Pathogenicity tests were performed to satisfy Koch’s postulates by artificially inoculating sweet cherry fruits with spore suspensions of three isolates (Q-1, Q-2, and Q-3) of C. godetiae. After 5 days, all C. godetiae inoculated fruits exhibited necrotic lesions with yellowish colonies (Figures 6B–D), similar to the symptoms initially observed on naturally infected fruits. Fruits inoculated with distilled water were asymptomatic (Figure 6A). The morphological characteristics of the fungal pathogen re-isolated from the inoculated fruits were identical to those of the C. godetiae strains originally obtained from sweet cherry fruits. Therefore, C. godetiae was confirmed as the causal agent of anthracnose on sweet cherry fruits.
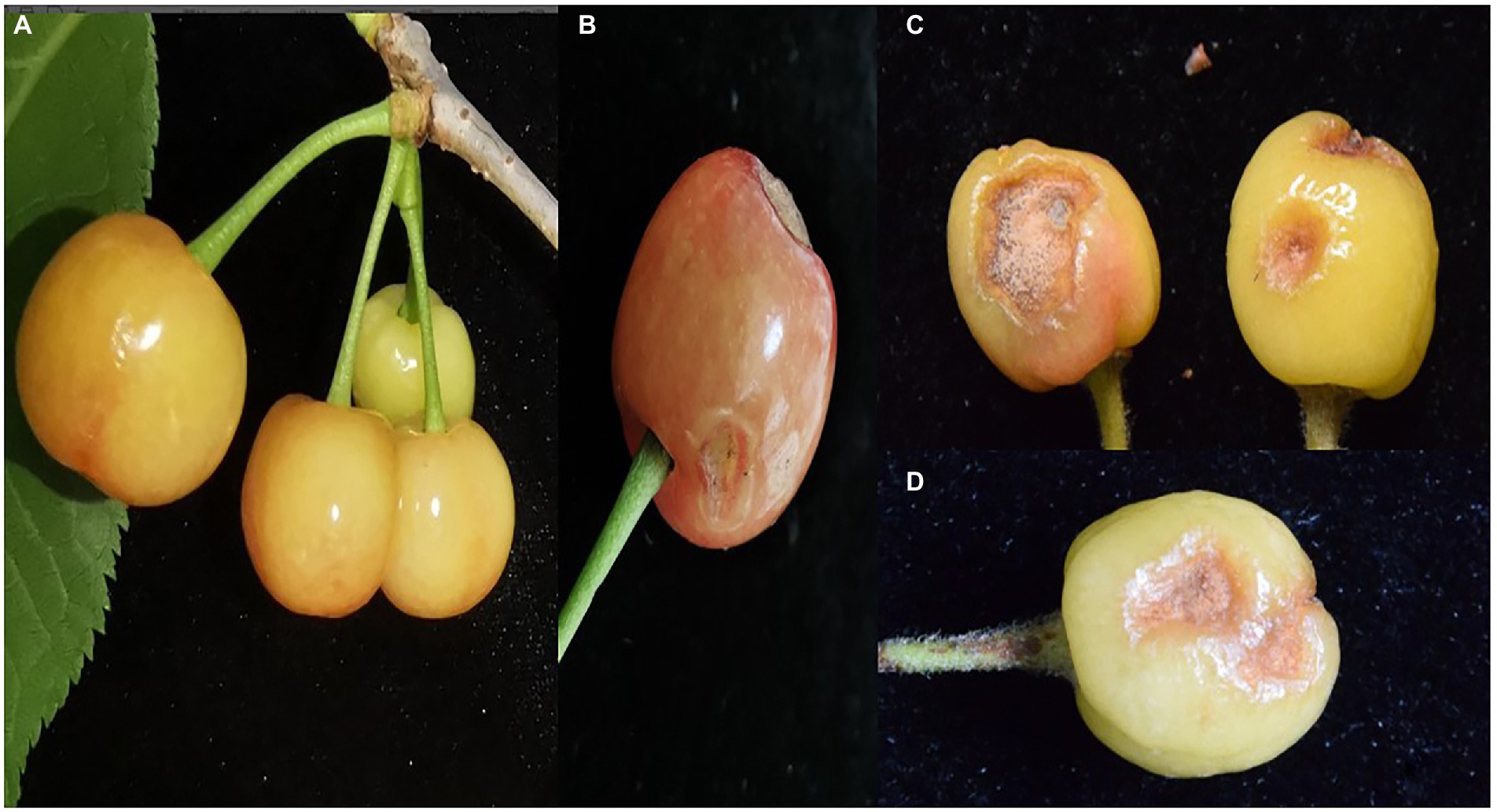
Figure 6. Pathogenicity test of Colletotrichum godetiae isolates obtained from Qianxinanzhou. (A) CK was treated with sterilized distilled water. (B–D) Lesions on cherry fruits were inoculated with Q-1, Q-2 and Q-3 isolates, respectively.
To identify fungicides effective in controlling the anthracnose disease, 13 fungicides were divided into seven classes (DMIs, QoIs, glycolysis inhibitors, antibiotics, compounds of DMI and QoI, bromothalonil, and polysaccharides) were used in this study. First, the Q-1 isolate of C. godetiae was selected to perform the sensitivity assay. The 13 fungicides all had different degrees of inhibitory effect on the growth of C. godetiae, and the inhibitory effect gradually increased with the increase of chemical concentration. The results in Table 3 show that among the different control agents, prochloraz-manganese have the best inhibitory effects, EC50 were 0.04 μg ml−1, respectively, of which the inhibitory effect of trifloxystrobin-tebuconazole and difenoconazole-azoxystrobin followed, with EC50 values of 0.08 and 0.10 μg ml−1, and the worst inhibitory effect was chlorothalonil, whose EC50 was 91.26 μg ml−1.
We selected 10 fungicides with better inhibitory effect among 13 fungicides and carried out extensive inhibition tests, and found that six fungicides in these 10 fungicides showed high sensitivity to the 20 isolates (B-1 to B-5, G-1 to G-5, L-1 to L-2, Q-1 to Q-8) of C. godetiae, and no drug-resistant groups appeared. The six fungicides are DMIS fungicide (difenoconazole, propiconazole, and prochloraz-manganese), QOIS fungicide (pyraclostrobin), and DMI and QoI fungicide compounds (trifloxystrobin-tebuconazole and difenoconazole-azoxystrobin), and their EC50 values ranged from 0.013 to 1.563 μg ml−1 (Table 5). These results suggest that these five fungicides could be used for controlling C. godetiae.
Colletotrichum species cause anthracnose disease of the leaf, young shoot, and especially fruit of sweet cherry trees, resulting in great economic losses (Dean et al., 2012). Recently, sweet Cherry fruit anthracnose has occurred in a large area in Guizhou province of China, and the disease has become a major factor restricting the development of this industry. However, the cause of the disease is still unclear, and there is no targeted prevention and treatment method. To determine the cause of this problem, 116 isolates of Colletotrichum species were isolated from four Guizhou cities with high anthracnose incidence. Based on morphological characteristics (colony color, mycelial growth rate, and shape and size of the mycelial appressorium, conidia, and appressorium) and molecular data (sequences of ITS, ACT, CHS-1, GAPDH, TUB2, and HIS3), the causal agent was identified as C. godetiae (belonging to the acutatum species complex) (Damm et al., 2012). This is also the first report showing that C. godetiae (belonging to the acutatum species complex) is responsible for causing sweet cherry anthracnose in China.
C. godetiae was originally isolated from the seeds of Clarkia (syn. Godetia) (Damm et al., 2012). Currently, C. godetiae has a number of hosts worldwide, including plant species belonging to the Adoxaceae, Anacardiaceae, Berberidaceae, Fabaceae, Juglandaceae, Myrtaceae, Oleaceae, Onagraceae, Podocarpaceae, Rosaceae, Rhamnaceae, Rutaceae, Solanaceae, and Vitaceae families, resulting in leaf spots, fruit rot, die back and stem end rot (Afanador-Kafuri et al., 2014; Baroncelli et al., 2014, 2015, 2017; Mosca et al., 2014; Munda, 2014; Talhinhas et al., 2015; Wang et al., 2017, 2019, 2020; Zhang et al., 2020; Shi et al., 2021). Recently, a study on walnut anthracnose identified C. fioriniae and C. godetiae in nuts and buds in the same orchard (Da Lio et al., 2018), suggesting that the pathogen population responsible for walnut anthracnose was complex. Therefore, distinguishing among the different Colletotrichum species is particularly important for accurate pathogen identification. In this study, we obtained 116 Colletotrichum isolates from 40 diseased sweet cherry fruits collected from four regions in Guizhou. The morphological characteristics of these isolates, including colony color, the shape, and size of mycelial appressorium, conidia, and appressorium, were similar to those of C. godetiae (Damm et al., 2012). To fulfill Koch’s postulates, spore suspensions of three isolates of C. godetiae (Q-1, Q-2, and Q-3) were sprayed on sweet cherry fruits in this study. The inoculation results showed that all isolates could infect sweet cherry fruits, causing yellow and sunken lesions, which was consistent with the naturally infected sweet cherry fruit samples. The morphological and molecular data of the re-isolated isolates confirmed that C. godetiae is associated with sweet cherry anthracnose in Guizhou province.
No crop completely immune to the various isolates of Colletotrichum species has been reported to date (Dean et al., 2012). As a result, chemical control is still considered as the most effective and important management strategy for controlling the anthracnose disease. At present, QoIs and DMIs are the major groups of fungicides used to control anthracnose in agricultural crops worldwide (Li et al., 2005; Ji et al., 2014; Hu et al., 2015; Forcelini et al., 2016; Yokosawa et al., 2017; Baggio et al., 2018; Wang et al., 2019; Kongtragoul et al., 2020; Shi et al., 2020). However, several reports have described a few QoI (azoxystrobin) resistant isolates belonging to the C. gloeosporioides species complex, such as C. gloeosporioides (Inada et al., 2008; Kim et al., 2016), C. siamense (Hu et al., 2015; Zhang et al., 2020) and C. fructicola (Yokosawa et al., 2017; Zhang et al., 2020), indicating that these fungicides may not be effective in controlling the anthracnose disease in some areas. Therefore, to identify suitable fungicides for controlling sweet cherry anthracnose-causing C. godetiae, 13 fungicides with different modes of action were evaluated in this study. Our results showed that the EC50 values of antibiotics (polyantimycin and zhongshengmycin), glycolysis inhibitors (chlorothalonil and dithianon), bromothalonil, and polysaccharide fungicides effective against the Q-1 isolate ranged from 4.07 to 91.26 μg ml−1, while those of DMIs (prochloraz-manganese, difenoconazole, and propiconazole), QoIs (azoxystrobin and pyraclostrobin), compounds of DMI and QoI (trifloxystrobin-tebuconazole and difenoconazole-azoxystrobin) ranged from 0.04 to 1.18 μg ml−1. These results indicated that the Q-1 isolate is quite sensitive to DMI, QoI and compound of DMI and QoI fungicides. At the same time, we selected 10 fungicides with better inhibitory effects for extensive inhibition tests on C. godetiae isolates, and found that these 20\u00B0C. godetiae isolates (B-1 to B-5, G-1 to G-5, L-1 to L-2, Q-1 to Q-8) showed strong sensitivity to 5 fungicides (difenoconazole, propiconazole, prochloraz-manganese, pyraclostrobin, trifloxystrobin-tebuconazole, and difenoconazole-azoxystrobin), and their EC50 values ranged from is 0.013 to 1.563 μg ml−1, suggesting that these fungicides would be ideal for controlling sweet cherry anthracnose in Guizhou, China. However, the effect of 6 fungicides on C. godetiae remains unknown under field conditions and needs further study.
In this study, we showed for the first time that C. godetiae causes anthracnose in sweet cherry in Guizhou province, China. C. godetiae was identified as the causal agent of sweet cherry anthracnose based on morphological characterization, phylogenetic analyses, and pathogenicity assays. Additionally, fungicide sensitivity assays showed that the isolates were highly sensitive to difenoconazole, propiconazole, prochloraz-manganese, pyraclostrobin, trifloxystrobin-tebuconazole, and difenoconazole-azoxystrobin. Overall, this study provides crucial information for the effective control of sweet cherry anthracnose in China.
The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/nuccore. The accession numbers of the sequences deposited in GenBank are: ITS: OK336098-OK336117; ACT: ON241033-ON241052; CHS-1: ON241053-ON241072; GAPDH: ON241073-ON241092; HIS3: ON241093-ON241112; TUB2: ON241113-ON241132.
KP and YP conducted the experiments. TT and XZe analyzed the data. ML and SJ prepared the figures and tables. FT and ZZ designed the project and supervised the experiments. XZh drafted the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the Guizhou Provincial Science and Technology Project, grant number Support of QKH [2021] General 199 and the National Natural Science Foundation of China (NSFC: 32000013).
We would like to thank all authors listed for their substantial, direct, and intellectual contributions to the work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Afanador-Kafuri, L., González, A., Gañán, L., Mejía, J. F., Cardona, N., and Alvarez, E. (2014). Characterization of the Colletotrichum species causing anthracnose in Andean blackberry in Colombia. Plant Dis. 98, 1503–1513. doi: 10.1094/pdis-07-13-0752-RE
Baggio, J. S., Wang, N. Y., Peres, N. A., and Amorim, L. (2018). Baseline sensitivity of Colletotrichum acutatum isolates from Brazilian strawberry fields to azoxystrobin, difenoconazole, and thiophanate-methyl. Trop Plant Pathol. 43, 533–542. doi: 10.1007/s40858-018-0232-2
Baroncelli, R., Sarrocco, S., Zapparata, A., Tavarini, S., Angelini, L. G., and Vannacci, G. (2015). Characterization and epidemiology of Colletotrichum acutatum sensu lato (C. chrysanthemi) causing C. arthamus tinctorius anthracnose. Plant Pathol. 64, 375–384. doi: 10.1111/ppa.12268
Baroncelli, R., Sreenivasaprasad, S., Lane, C. R., Thon, M. R., and Sukno, S. A. (2014). First report of Colletotrichum acutatum sensu lato (Colletotrichum godetiae) causing anthracnose on grapevine (Vitis vinifera) in the United Kingdom. New Disease Reports 29, 26. doi: 10.5197/j.2044-0588.2014.029.026
Baroncelli, R., Talhinhas, P., Pensec, F., Sukno, S. A., Le Floch, G., and Thon, M. R. (2017). The Colletotrichum acutatum species complex as a model system to study evolution and host specialization in plant pathogens. Front. Microbiol. 8, 2001. doi: 10.3389/fmicb.2017.02001
Bhunjun, C. S., Phukhamsakda, C., Jayawardena, R. S., Jeewon, R., Promputtha, I., and Hyde, K. D. (2021). Investigating species boundaries in Colletotrichum. Fungal Divers. 107, 107–127.
Børve, J., Djønne, R. T., and Stensvand, A. (2010). Colletotrichum acutatum occurs asymptomatically on sweet cherry leaves. Eur J of Plant Pathol. 127, 325–332. doi: 10.1007/s10658-010-9597-x
Børve, J., and Stensvand, A. (2006a). Colletotrichum acutatum overwinters on sweet cherry buds. Plant Dis. 90, 1452–1456. doi: 10.1094/pd-90-1452
Børve, J., and Stensvand, A. (2006b). Timing of fungicide applications against anthracnose in sweet and sour cherry production in Norway. Crop Prot. 25, 781–787. doi: 10.1016/j.cropro.2005.10.012
Børve, J., and Stensvand, A. (2013). Colletotrichum acutatum can establish on sweet and sour cherry trees throughout the growing season. Eur Hortic Sci. 78, 258–266.
Bragança, C., Junior, A. N., Rogério, F., and Massola Jr, N. (2014). First report of anthracnose caused by Colletotrichum theobromicola on Barbados cherry (Malpighia emarginata) in Brazil. Plant Dis. 98, 1272. doi: 10.1094/pdis-01-14-0099-PDN
Cai, L., Giraud, T., Zhang, N., Begerow, D., Cai, G., and Shivas, R. G. (2011). The evolution of species concepts and species recognition criteria in plant pathogenic fungi. Fungal Divers. 50, 121–133. doi: 10.1007/s13225-011-0127-8
Cai, L., Hyde, K., Taylor, P., Weir, B., Waller, J., Abang, M., et al. (2009). A polyphasic approach for studying colletotrichum. Fungal Divers. 39, 183–204.
Carbone, I., and Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91, 553–556. doi: 10.2307/3761358
Chen, S., Luo, C., Hu, M., and Schnabel, G. (2016). Sensitivity of Colletotrichum species, including C. fioriniae and C. nymphaeae, from peach to demethylation inhibitor fungicides. Plant Dis. 100, 2434–2441. doi: 10.1094/pdis-04-16-0574-re
Chen, D., Shi, H., Wu, H., Xu, Z., and Zhang, C. (2013a). Resistance of Colletotrichum gloeosporioides causing grape ripe rot to thiophanate-methyl and tebuconazole in Zhejiang. J. Fruit Sci. 30, 665–668.
Chen, T., Wang, X. R., Luo, H., Wang, C. T., and Luo, M. M. (2012). Chloroplast DNA trnQ-rps16 variation and genetic structure of nine wild Chinese cherry (Cerasus pseudocerasus Lindl.) populations. Hereditas (Beijing) 34, 1475–1483. doi: 10.3724/sp.j.1005.2012.01475
Chen, T., Wang, X. R., Tang, H. R., Chen, Q., Huang, X. J., and Chen, J. (2013b). Genetic diversity and population structure of Chinese cherry revealed by chloroplast DNA trnQ-rps16 intergenic spacers variation. Genet. Resour. Crop Evol. 60, 1859–1871. doi: 10.1007/s10722-013-9960-9
Chethana, K., Jayawardene, R., Zhang, W., Zhou, Y., Liu, M., Hyde, K., et al. (2019). Molecular characterization and pathogenicity of fungal taxa associated with cherry leaf spot disease. Mycosphere 10, 490–530. doi: 10.5943/mycosphere/10/1/8
Crous, P. W., Groenewald, J. Z., Risede, J., and Hyweljones, N. (2004). Calonectria species and their cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Stud. Mycol. 157, 127–135. doi: 10.1023/b:myco.0000012225.79969.29
Da Lio, D., Cobo-Díaz, J. F., Masson, C., Chalopin, M., Kebe, D., Giraud, M., et al. (2018). Combined metabarcoding and multi-locus approach for genetic characterization of Colletotrichum species associated with common walnut (Juglans regia) anthracnose in France. Sci. Rep. 8, 1–17. doi: 10.1038/s41598-018-29027-z
Damm, U., Cannon, P., Woudenberg, J., and Crous, P. (2012). The Colletotrichum acutatum species complex. Stud. Mycol. 73, 37–113. doi: 10.3114/sim0010
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772. doi: 10.1038/nmeth.2109
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2012.00822.x
Etebarian, H. R., Sholberg, P. L., Eastwell, K. C., and Sayler, R. J. (2005). Biological control of apple blue mold with Pseudomonas fluorescens. Can. J. Microbiol. 51, 591–598. doi: 10.1139/w05-039
Forcelini, B. B., Rebello, C. S., Wang, N. Y., and Peres, N. (2017). Fitness, competitive ability and mutation stability of isolates of Colletotrichum acutatum from strawberry resistant to qoi fungicides. Phytopathology 108, 462–468. doi: 10.1094/phyto-09-17-0296-R
Forcelini, B. B., Seijo, T. E., Amiri, A., and Peres, N. A. (2016). Resistance in strawberry isolates of Colletotrichum acutatum from Florida to quinone-outside inhibitor fungicides. Plant Dis. 100, 2050–2056. doi: 10.1094/pdis-01-16-0118-re
Hall, T. A. (1999). Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt. Nuclc Acids Symposium Series 734, 95–98. doi: 10.1021/bk-1999-0734.ch008
Hu, M. J., Grabke, A., Dowling, M. E., Holstein, H. J., and Schnabel, G. (2015). Resistance in Colletotrichum siamense from peach and blueberry to thiophanate-methyl and azoxystrobin. Plant Dis. 99, 806–814. doi: 10.1094/pdsi-10-14-1077-RE
Inada, M., Ishii, H., Chung, W. H., Yamada, T., Yamaguchi, J. I., and Furuta, A. (2008). Occurrence of strobilurin-resistant strains of Colletotrichum gloeosporioides (Glomerella cingulata), the causal fungus of strawberry anthracnose. Jpn. J. Phytopathol. 74, 114–117. doi: 10.3186/jjphytopath.74.114
Jayawardena, R. S., Bhunjun, C. S., Hyde, K. D., Gentekaki, E., and Itthayakorn, P. (2021). Colletotrichum: lifestyles, biology, morpho-species, species complexes and accepted species. Mycosphere 12, 519–669. doi: 10.5943/mycosphere/12/1/7
Jayawardena, R., Hyde, K., Damm, U., Cai, L., Liu, M., Li, X., et al. (2016). Notes on currently accepted species of Colletotrichum. Mycosphere 7, 1192–1260. doi: 10.5943/mycosphere/si/2c/9
Ji, M., Wu, X., Yao, K., Chen, H., Yang, J., Wang, L., et al. (2014). Identification of strawberry anthracnose pathogens and screening of germicides. Agric. Sci. Technol. 15, 94.
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, C. H., Hassan, O., and Chang, T. (2020). Diversity, pathogenicity, and fungicide sensitivity of Colletotrichum species associated with apple anthracnose in South Korea. Plant Dis. 104, 2866–2874. doi: 10.1094/PDIS-01-20-0050-RE
Kim, D. O., Heo, H. J., Kim, Y. J., Yang, H. S., and Lee, C. Y. (2005). Sweet and sour cherry phenolics and their protective effects on neuronal cells. J. Agr. Food Chem. 53, 9921–9927. doi: 10.1021/jf0518599
Kim, S., Min, J., Back, D., Kim, H., Lee, S., and Kim, K. (2016). Assessment of QoI resistance in Colletotrichum spp. isolated from boxthorn and apple in Korea. Phytopathology 48, 6–71. doi: 10.1016/j.cimid.2016.07.001
Kongtragoul, P., Imamoto, K., and Ishii, H. (2020). Resistance to quinone-outside inhibitor (QoI) fungicides in Colletotrichum species isolated from anthracnose disease occurring in Thailand. Curr. Appl. Sci. Technol. 48, 6–13. doi: 10.1016/j.cimid.2016.07.001
Li, H. X., Liu, Z. Y., Wang, J. X., and Zhou, M. G. (2005). Baseline sensitivity of Colletotrichum gloeosporioides and C. capsici from capsium to azoxystrobin. Acta Phytopathol. Sin. 35, 73–77.
Liu, Y., An, F., Zhang, Y., Fu, C., and Su, Y. (2021). First report of anthracnose on Jerusalem Cherry caused by Colletotrichum liaoningense in Shandong, China. Plant Dis. 105:2248. doi: 10.1094/pdis-01-21-0124-PDN
López-Moral, A., Raya-Ortega, M. C., Agustí-Brisach, C., Roca, L. F., Lovera, M., Luque, F., et al. (2017). Morphological, pathogenic, and molecular characterization of Colletotrichum acutatum isolates causing almond anthracnose in Spain. Plant Dis. 101, 2034–2045. doi: 10.1094/pdis-03-17-0318-re
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees.” in 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, 1–8.
Mo, F., Hu, X., Ding, Y., Li, R., and Li, M. (2021). Naturally produced magnolol can significantly damage the plasma membrane of Rhizoctonia solani. Pestic. Biochem. Physiol. 178:104942. doi: 10.1016/j.pestbp.2021.104942
Mosca, S., Li Destri Nicosia, M. G., Cacciola, S. O., and Schena, L. (2014). Molecular analysis of Colletotrichum species in the carposphere and phyllosphere of olive. PloS One 9:e114031. doi: 10.1371/journal.pone.0114031
Munda, A. (2014). First report of Colletotrichum fioriniae and C. godetiae causing apple bitter rot in Slovenia. Plant Dis. 98, 1282. doi: 10.1094/pdis-04-14-0419-PDN
O’Donnell, K., and Cigelnik, E. (1997). Two divergent intragenomic rdna its2 types within a monophyletic lineage of the fungus fusarium are nonorthologous. Mol. Phylogenet. Evol. 7, 103–116. doi: 10.1006/mpev.1996.0376
Rambaut, A. (2016). FigTree version v1.4.3 [online]. Available at: https://github.com/rambaut/figtree/releases/tag/v1.4.3 (Accessed October 04, 2016).
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sharma, G., Kumar, N., Weir, B. S., Hyde, K. D., and Shenoy, B. D. (2013). The ApMat marker can resolve Colletotrichum species: a case study with Mangifera indica. Fungal Divers. 61, 117–138. doi: 10.1007/s13225-013-0247-4
Shi, N., Ruan, H., Gan, L., Dai, Y., Yang, X., Du, Y., et al. (2020). Evaluating the sensitivities and efficacies of fungicides with different modes of action against Phomopsis asparagi. Plant Dis. 104, 448–454. doi: 10.1094/pdis-05-19-1040-RE
Shi, N. N., Ruan, H. C., Jie, Y. L., Chen, F. R., and Du, Y. X. (2021). Characterization, fungicide sensitivity and efficacy of Colletotrichum spp. from chili in Fujian. China. Crop Prot. 143:105572. doi: 10.1016/j.cropro.2021.105572
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post - analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stensvand, A., Børve, J., and Talgø, V. (2017). Overwintering diseased plant parts and newly infected flowers and fruit as sources of inoculum for Colletotrichum acutatum in sour cherry. Plant Dis. 101, 1207–1213. doi: 10.1094/pdis-11-16-1599-re
Talhinhas, P., Gonçalves, E., Sreenivasaprasad, S., and Oliveira, H. (2015). Virulence diversity of anthracnose pathogens (Colletotrichum acutatum and C. gloeosporioides species complexes) on eight olive cultivars commonly grown in Portugal. Eur. J. Plant. Pathol 142, 73–83. doi: 10.1007/s10658-014-0590-7
Tang, Z., Lou, J., He, L., Wang, Q., Chen, L., Zhong, X., et al. (2021). First report of Colletotrichum fructicola causing anthracnose on cherry (Prunus avium) in China. Plant Dis. 106, 317. doi: 10.1094/pdis-03-21-0544-pdn
Templeton, M. D., Rikkerink, E. H. A., Solon, S. L., and Crowhurst, R. (1992). Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase-encoding gene and cDNA from the plant pathogenic fungus glomerella cingulata. Gene. 122, 225–230. doi: 10.1016/0378-1119(92)90055-t
Wang, Q. H., Fan, K., Li, D. W., Han, C. M., Qu, Y. Y., Qi, Y. K., et al. (2020). Identification, virulence and fungicide sensitivity of Colletotrichum gloeosporioides s. s. Responsible for walnut anthracnose disease in China. Plant Dis. 104, 1358–1368. doi: 10.1094/pdis-12-19-2569-re
Wang, Q. H., Fan, K., Li, D. W., Niu, S. G., Hou, L. Q., and Wu, X. Q. (2017). Walnut anthracnose caused by Colletotrichum siamense in China. Australas. Plant Path. 46, 585–595. doi: 10.1007/s13313-017-0525-9
Wang, X. H., Wang, R., Fa, L., Zhang, Y. A., Wang, H. X., Liu, X., et al. (2019). Pathogen dentification of anthracnose on juglans regia in shaanxi province. J. Northeast For. Univ. 47, 113–119. doi: 10.13759/j.cnki.dlxb.2019.11.022
White, T. J., Bruns, T., Lee, S., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics”, in PCR protocols: a guide to methods and applications. 315–322.
Wijayawardene, N., Hyde, K., Al-Ani, L., Tedersoo, L., Haelewaters, D., and Rajeshkumar, K. (2020). Outline of fungi and fungus-like taxa. Mycosphere 11, 1060–1456. doi: 10.5943/mycosphere/11/1/8
Wijayawardene, N. N., Hyde, K. D., Lumbsch, H. T., Liu, J. K., Maharachchikumbura, S. S., Ekanayaka, A. H., et al. (2018). Outline of ascomycota: 2017. Fungal Divers. 88, 167–263. doi: 10.1007/s13225-018-0394-8
Yokosawa, S., Eguchi, N., Kondo, K. I., and Sato, T. (2017). Phylogenetic relationship and fungicide sensitivity of members of the Colletotrichum gloeosporioides species complex from apple. J. Gen. Plant Pathol. 83, 291–298. doi: 10.1007/s10327-017-0732-9
Yuan, X., Peng, K., Li, C., Zhao, Z., Zeng, X., Tian, F., et al. (2021). Complete genomic characterization and identification of Saccharomycopsis phalluae sp. nov., a novel pathogen causes yellow rot disease on phallus rubrovolvatus. J Fungi. 7, 707. doi: 10.3390/jof7090707
Zhang, X. Y., Li, X., and Gao, Z. Y. (2014). Carbendazim resistance of Colletotrichum gloeosporioides on tropical and subtropical fruits. Chinese J. Tropical Agr. 34, 71–74.
Zhang, L., Song, L., Xu, X., Zou, X., Duan, K., and Gao, Q. (2020). Characterization and fungicide sensitivity of Colletotrichum species causing strawberry anthracnose in eastern China. Plant Dis. 104, 1960–1968. doi: 10.1094/pdis-10-19-2241-re
Keywords: Cerasus pseudocerasus, plant disease, Colletotrichum, multi-gene, sweet cherry fruit anthracnose
Citation: Peng K, Pan Y, Tan T, Zeng X, Lin M, Jiang S, Zhao Z, Tian F and Zhao X (2022) Characterization and fungicide sensitivity of Colletotrichum godetiae causing sweet cherry fruit anthracnose in Guizhou, China. Front. Microbiol. 13:923181. doi: 10.3389/fmicb.2022.923181
Received: 19 April 2022; Accepted: 12 August 2022;
Published: 27 September 2022.
Edited by:
Amin Uddin Mridha, University of Chittagong, BangladeshReviewed by:
Latiffah Zakaria, Universiti Sains Malaysia, MalaysiaCopyright © 2022 Peng, Pan, Tan, Zeng, Lin, Jiang, Zhao, Tian and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fenghua Tian, Zmh0aWFuQGd6dS5lZHUuY24=; Xiaosheng Zhao, enhzNzE4QDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.