- School of Pharmaceutical Sciences, South-Central University for Nationalities, Wuhan, China
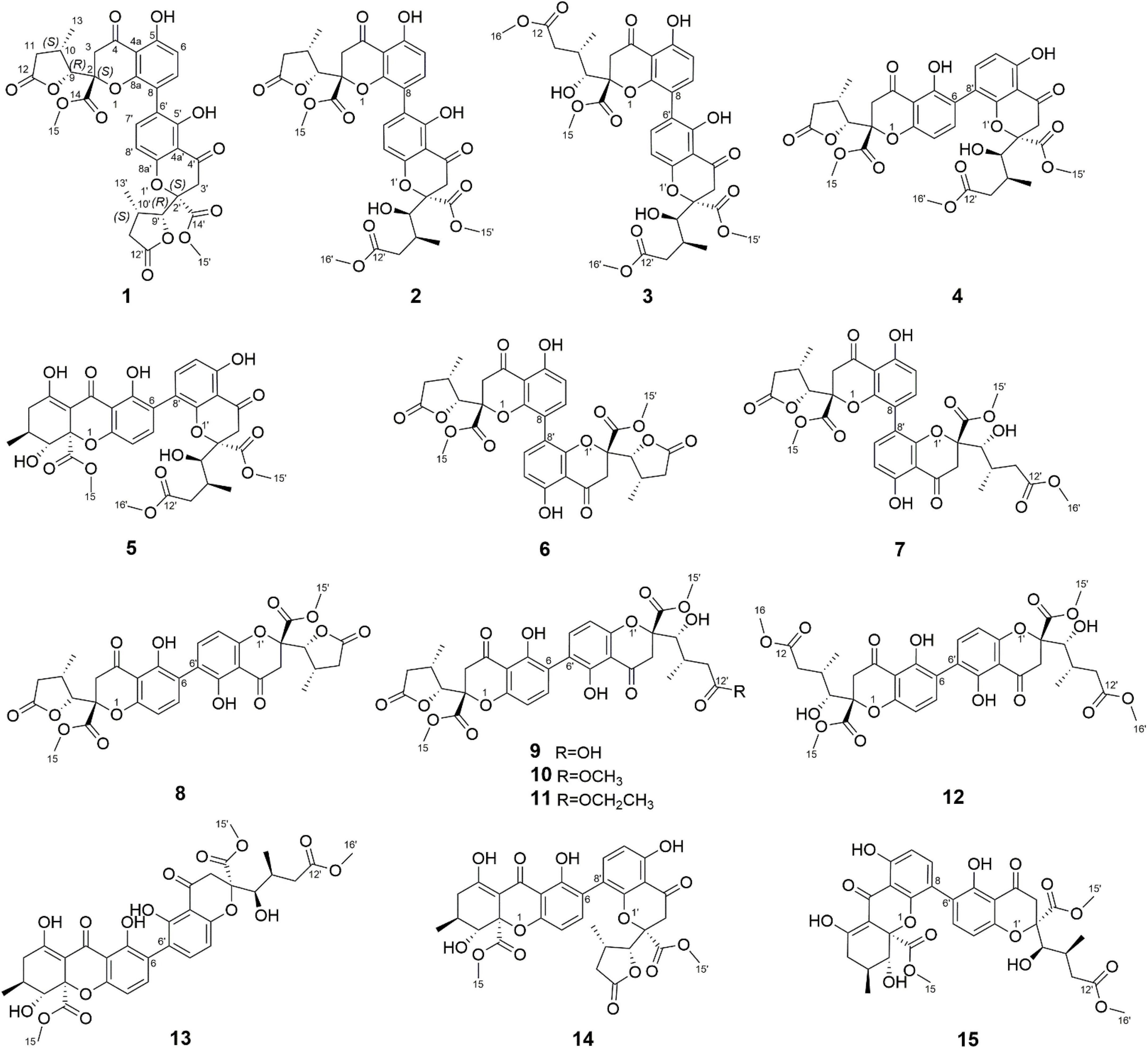
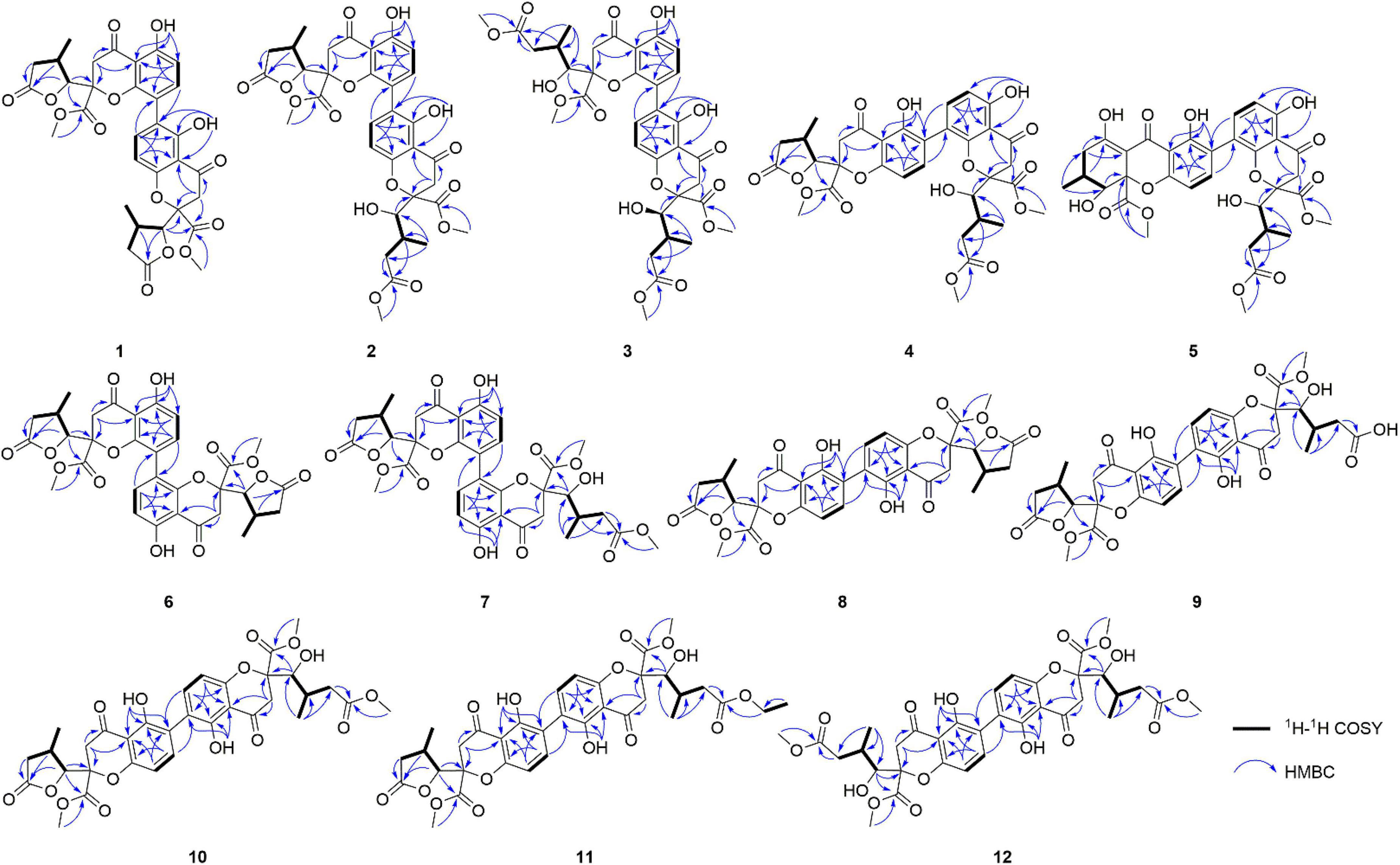
A total of eleven new dimeric chromanones, paecilins F-P (2–12), were isolated from the endophytic fungus Xylaria curta E10, along with four known analogs (1, 13–15). Their structures and absolute configurations were determined by extensive experimental spectroscopic methods, single-crystal X-ray diffraction, and equivalent circulating density (ECD) calculations. In addition, the structure of paecilin A, which was reported to be a symmetric C8-C8′ dimeric pattern, was revised by analysis of the nuclear magnetic resonance (NMR) data, and single-crystal X-ray diffraction. Compound 1 showed antifungal activity against the human pathogenic fungus Candida albicans with a minimum inhibitory concentration of 16 μg/mL, and Compounds 8 and 10 showed antibacterial activity against the gram-negative bacterium Escherichia coli with the same minimum inhibitory concentration of 16 μg/mL.
Introduction
Chromanones are a class of compounds with benzo-γ-pyranone skeletons. They are widely distributed in plants, fungi, and lichens (Duan et al., 2019). Chromanones usually form homodimers or heterodimers with xanthones, the biosynthetic precursors of chromanones, with different linkages, including C6-C6, C6-C8, and C8-C8 (Wu et al., 2015; Cao et al., 2022). To unambiguously determine the absolute or even relative configurations of these dimers remains challenging because of the presence of various chiral centers, sometimes with axial chirality. These compounds have been reported to have good biological activities, such as antitumor (Pinto et al., 2014; El-Elimat et al., 2015; Wu et al., 2015; Arora et al., 2017; Nguyen et al., 2020; Tuong et al., 2020; Lünne et al., 2021; Wang et al., 2021), antifungal (da Silva et al., 2018) and antibacterial (Pontius et al., 2008; Kumla et al., 2017; Wang et al., 2021) activities. Their dimers have attracted much attention from scientists in the fields of chemical synthesis and biosynthesis for many years because of their complex structures and remarkable biological activity (Xiao et al., 2017; Wei and Matsuda, 2020; Wei et al., 2021).
Endophytic fungi are microorganisms that live in healthy plant tissues and do not cause any loss or disease to the host plant (Ribeiro et al., 2021). Endophytic fungi are an important resource of natural bioactive compounds (Li et al., 2008; Liu et al., 2014; Chen et al., 2016; Nalin Rathnayake et al., 2019). For many years, they have attracted much attention because of their ability to produce new bioactive secondary metabolites (Liu et al., 2019). Xylaria curta E10 is an endophytic fungus isolated from Solanum tuberosum. A previous investigation of this fungus led to the isolation of several cytochalasans with novel scaffolds and intriguing biological activity. Curtachalasins A and B are two cytochalasans with 5/6/6/6 tetracyclic skeletons (Wang et al., 2018), curtachalasins C-E have an unprecedented bridged 6/6/6/6 ring system with significant resistance reversal activity against fluconazole-resistant Candida albicans (Wang et al., 2019a), and xylarichalasin A possesses a 6/7/5/6/6/6 fused polycyclic system with remarkable anti-proliferative activity (Wang et al., 2019b).
In our ongoing research on mining structurally interesting and biologically active constituents from natural resources, a chemical study on the endophytic fungus Xylaria curta E10 was carried out. As a result, eleven new dimeric chromanones named paecilins F-Q (2–12), together with four known compounds, paecilin A (1) (Guo et al., 2007), versixanthone F (13) (Wu et al., 2015), versixanthone A (14) (Wu et al., 2015), and versixanthone E (15) (Wu et al., 2015) (Figure 1) were isolated. Herein, the details of the isolation, structural elucidation, and antimicrobial activities of these compounds are presented.
Materials and methods
Fungal material
The strain required in this study was isolated from fresh healthy potato tissues collected from Dali City, Yunnan Province, China, and identified as Xylaria curta E10 according to the ITS sequence (GenBank Accession No. KJ883611.1, query cover 100%, maximum identity 99%). At present, the strain is stored in the microbial seed bank of the School of Pharmacy, South-Central University for Nationalities. The fungus Xylaria curta E10 was fermented in a solid rice medium (100 g of rice and 100 ml of water, in each 500 ml culture flask, with a total of 15 kg of rice) and was cultured at 24°C for one month.
Fungal fermentation, extraction, and isolation
The fermented materials were soaked in absolute MeOH (20 L × 5). The combined extracts were evaporated under reduced pressure to afford a crude extract, which was further dissolved in water and partitioned against EtOAc (10 L × 5) to yield 130 g of the extract. Five fractions (A-E) were eluted by gradient elution of chloroform/methanol (1:0-0:1, V/V) by normal silica gel column chromatography.
Fraction B (30 g) was eluted by MPLC (MeOH-H2O, 10:90-100:0, V/V) to obtain nine subfractions (B1–B9). Fraction B3 (380 mg) was separated by preparative HPLC using a reversed-phase column (CH3CN-H2O from 35:65 to 50:50 in 20 min, flow rate 4 mL/min) to yield 1 (20.3 mg, tR = 15.6 min). Fraction B4 (6.7 g) was purified over Sephadex LH-20 eluted with MeOH to give six subfractions (B4–1–B4–6). Fraction B4–3 (198 mg) was further purified by preparative HPLC with CH3CN-H2O (from 32:68 to 52:48 in 20 min, flow rate 4 mL/min) to obtain Compounds 2 (30 mg, tR = 16.8 min), 3 (7.8 mg, tR = 17.2 min), 4 (6.6 mg, tR = 28.3 min), and 5 (7.3 mg, tR = 29.6 min). Fraction B4–4 (600 mg) was separated on a silica gel column and eluted with petroleum ether and ethyl acetate (10:1) to yield five subfractions (B4–4–1-B4–4–5). B4–4–3 was purified by prep-HPLC (CH3CN-H2O from 39:61 to 55:45 in 25 min, flow rate 4 mL/min) to give 6 (6.8 mg, tR = 15.6 min), 7 (17.2 mg, tR = 17.9 min) and 8 (20.4 mg, tR = 20.8 min).
Fraction C (43 g) was eluted by MPLC (MeOH-H2O, 10:90-100:0, V/V) to obtain eleven subfractions (C1–C11). Fraction C5 (3.6 g) was separated on a silica gel column and eluted with petroleum ether and ethyl acetate (8:1), to yield eight subfractions (C5–1–C5–8). C5–5 was purified by prep-HPLC (CH3CN-H2O from 37:63 to 56:44 in 20 min, flow rate 4 mL/min) to give 9 (11.8 mg, tR = 18.2 min) and 10 (9.5 mg, tR = 19.7 min). Fraction C6 (12 g) was purified over Sephadex LH-20 eluted with MeOH to give nine subfractions (C6–1–C6–9). Fraction C6–5 (300 mg) was further purified by preparative HPLC with CH3CN-H2O (from 40:60 to 54:46 in 20 min, flow rate 4 mL/min) to obtain Compounds 11 (28 mg, tR = 18.4 min), 12 (6.1 mg, tR = 19 min) and 13 (10.4 mg, tR = 23.6 min). Fraction C7 (380 mg) was separated by preparative HPLC using a reversed-phase column (CH3CN-H2O from 42:58 to 57:43 in 20 min, flow rate 4 mL/min) to yield 14 (16.3 mg, tR = 18.5 min) and 15 (7.5 mg, tR = 21.6 min).
General experimental procedures
Both 1D and 2D spectra were run on Bruker Avance III 600 MHz and Bruker Avance III 500 MHz spectrometers. IR spectra were obtained with a Tenor 27 spectrophotometer using KBr pellets. UV spectra were measured on a UH-5300 spectrometer. CD spectra were recorded by a Chirascan-plus circular dichromatic spectrometer. The optical rotations were measured with a Horiba Sepa-300 polarimeter. According to the solvent signal, the chemical shifts were expressed in ppm. The mass spectra were recorded on a mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Medium-pressure liquid chromatography (MPLC) was applied to the Biotage SP1 system and packed with RP-18 gel. Preparative high-performance liquid chromatography (prep-HPLC) was performed on an Agilent 1260 liquid chromatography system equipped with Zorbax SB-C18 columns (5 μm, 9.4 mm × 150 mm, or 21.2 mm × 150 mm) and a DAD detector. Column chromatography (CC) separations were carried out in silica gel (200–300 mesh, Qingdao Haiyang Chemical Co., Ltd., Qingdao) and Sephadex LH-20 (Sweden’s France Asia Fine Chemical Co., Ltd.). The components were monitored by TLC (GF254, Qingdao Haiyang Chemical Co., Ltd., Qingdao) and the spots were observed by heating the silica gel plate and spraying with vanillin and 10% H2SO4 in ethanol.
Spectroscopic characterization of Compounds 2–12
Paecilin F (2): light yellow crystal (MeOH); [α]25.1D + 2.3 (c.5, MeOH); UV (MeOH) λmax (log ε) 260 (3.28) nm; IR (KBr) νmax 3500, 3000, 2950, 1794, 1736, 1647, 1458, and 1320 cm–1; 1H nuclear magnetic resonance (NMR) (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data, see Table 1; positive ion HRESIMS m/z 693.1814 [M + Na]+ (calcd for C33H34O15Na, 693.1795).
Paecilin G (3): pale yellow gum; [α]21.5D + 66.9 (c.5, MeOH); UV (MeOH) λmax (log ε) 260 (2.70) nm; IR (KBr) νmax 3500, 1734, 1647, 1472, 1437, 1356, 1281, 1233, 1198, 1177, 1070, 1049, and 1011 cm–1; 1H NMR (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data, see Table 1; positive ion HRESIMS m/z 725.2048 [M + Na]+ (calcd for C34H38O16Na, 725.2058).
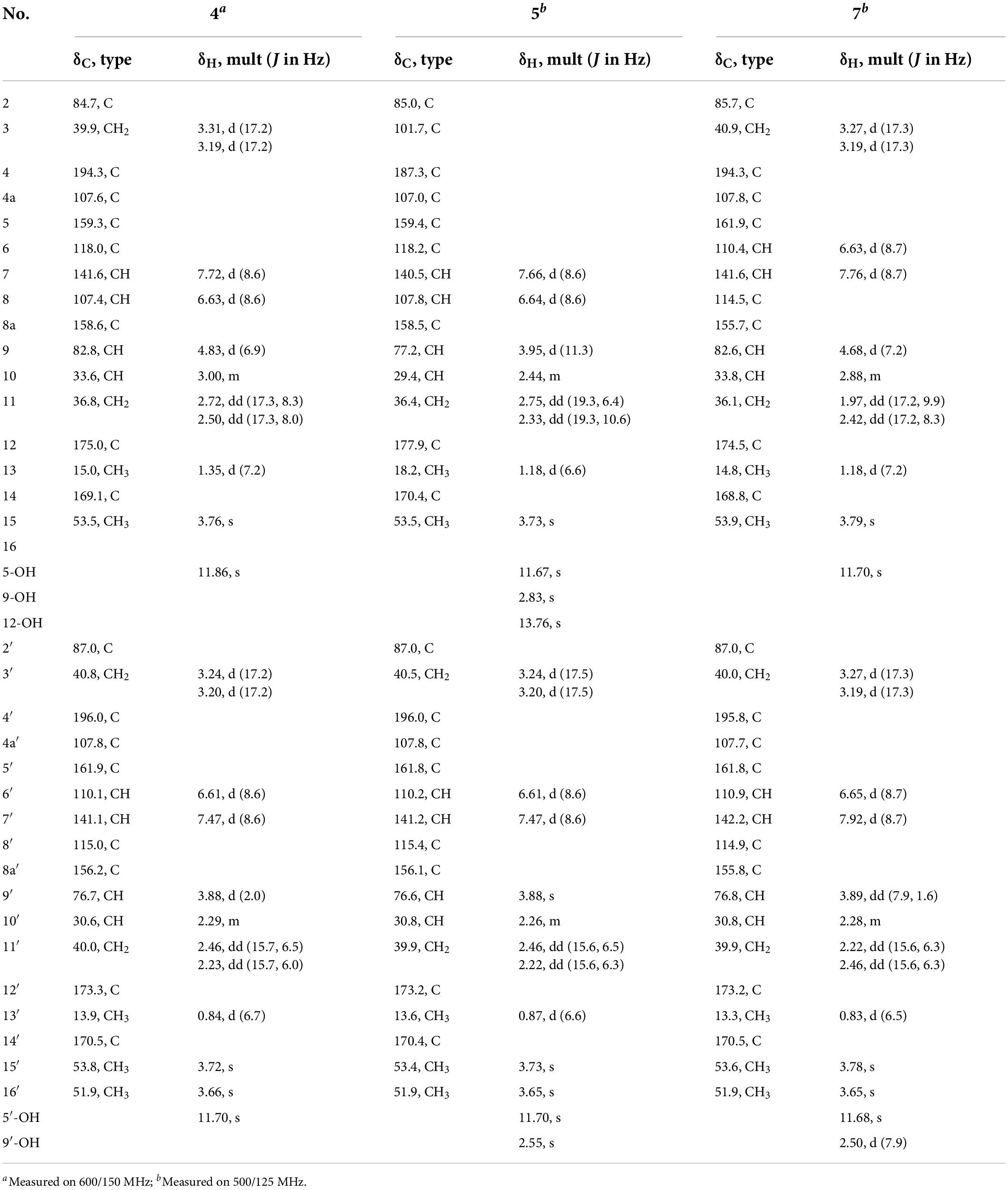
Paecilin H (4): pale yellow gum; [α]21.4D + 69.6 (c.5, MeOH); UV (MeOH) λmax (log ε) 260 (3.20) nm; IR (KBr) νmax 3500, 1792, 1738, 1651, 1472, 1437, 1356, 1281, 1223, 1196, 1177, 1067, 1007, 837, 785, 584, and 536 cm–1; 1H NMR (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data, see Table 2; positive ion HRESIMS m/z 693.1791 [M + Na]+ (calcd for C33H34O15Na 693.1795).
Paecilin I (5): pale yellow gum; [α]21.4D + 147.3 (c.5, MeOH); UV (MeOH) λmax (log ε) 265 (2.39) nm; IR (KBr) νmax 3500, 1734, 1653, 1616, 1458, 1356, 1273, 1233, 1067, 1049, 1016, and 820 cm–1; 1H NMR (600 MHz, CDCl3) and 13C NMR (150 MHz, CDCl3) data, see Table 2; positive ion HRESIMS m/z 693.1810 [M + Na]+ (calcd for C33H34O15Na, 693.1795).
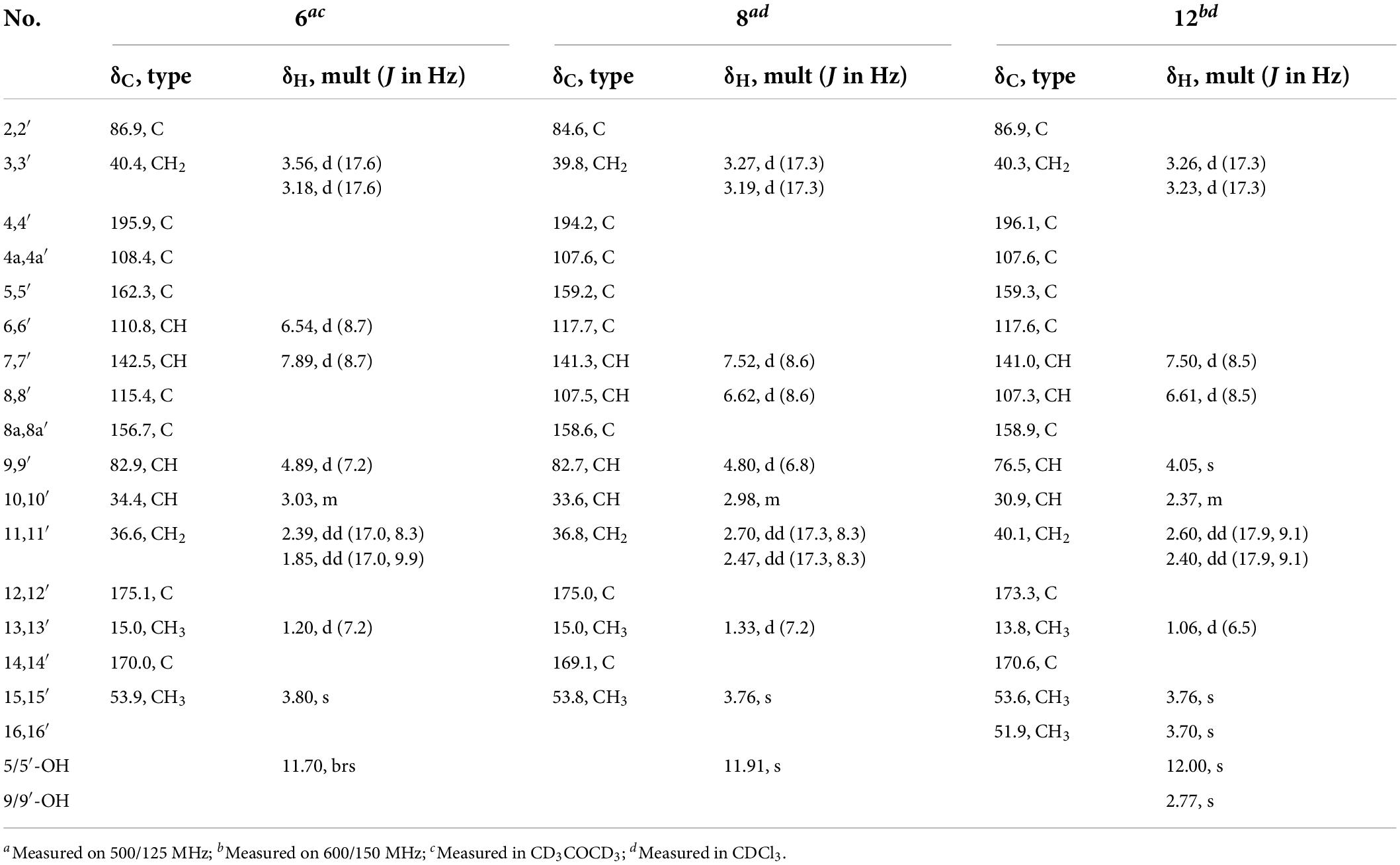
Paecilin J (6): brown crystal (MeOH); [α]22.5D + 36.9 (c.5, MeOH); UV (MeOH) λmax (log ε) 260 (1.40) nm; IR (KBr) νmax 3500, 2922, 2851, 1790, 1742, 1647, 1466, 1383, 1346, 1234, 1202, 1179, 1049, and 1024 cm–1; 1H NMR (500 MHz, CD3COCD3) and 13C NMR (125 MHz, CD3COCD3) data, see Table 3; positive ion HRESIMS m/z 661.1526 [M + Na]+ (calcd for C32H30O14Na 661.1533).
Paecilin K (7): pale yellow gum; [α]21.3D + 46 (c.5, MeOH); UV (MeOH) λmax (log ε) 260 (1.76) nm; IR (KBr) νmax 3500, 1794, 1740, 1647, 1468, 1348, 1283, 1236, 1051, 1020, 837, 783, 758, 737, 648, and 550 cm–1; 1H NMR (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data, see Table 2; positive ion HRESIMS m/z 693.1785 [M + Na]+ (calcd for C33H34O15Na, 693.1795).
Paecilin L (8): pale yellow gum; [α]21.0D + 52.9 (c.5, MeOH); UV (MeOH) λmax (log ε) 265 (1.85) nm; IR (KBr) νmax 3500, 2920, 1792, 1740, 1645, 1626, 1435, 1362, 1215, 1177, 1150, 1061, 1024, 804, and 586 cm–1; 1H NMR (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data, see Table 3; positive ion HRESIMS m/z 661.1538 [M + Na]+ (calcd for C32H30O14Na, 661.1533).
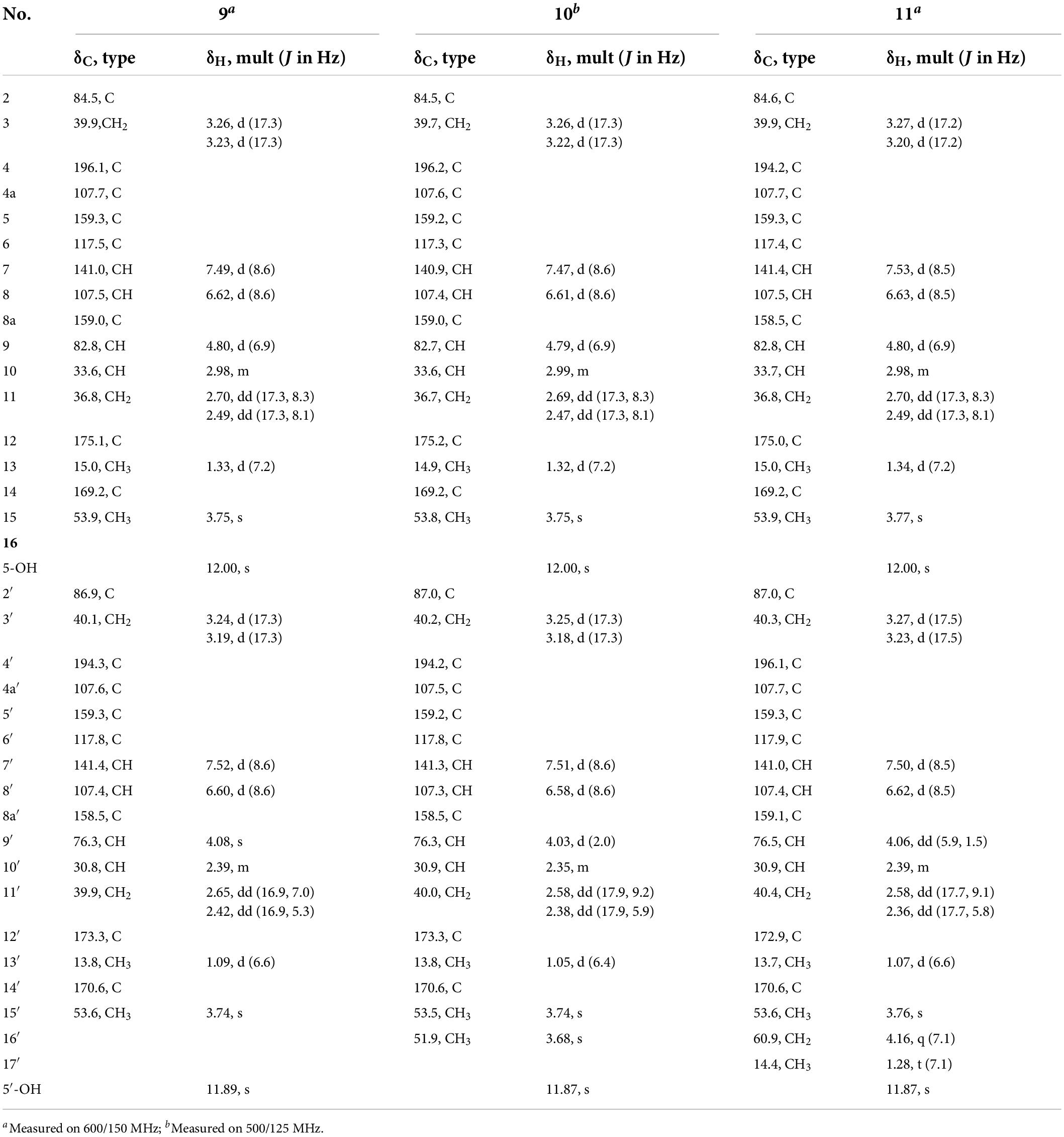
Paecilin M (9): pale yellow gum; [α]20.8D – 96.9 (c.5, MeOH); UV (MeOH) λmax (log ε) 265 (1.66) nm; IR (KBr) νmax 3500, 2924, 1788, 1736, 1626, 1578, 1437, 1362, 1288, 1271, 1250, 1217, 1119, 1065, 839, 816, 785, and 586 cm–1; 1H NMR (600 MHz, CDCl3) and 13C NMR (150 MHz, CDCl3) data, see Table 4; positive ion HRESIMS m/z 679.1627 [M + Na]+ (calcd for C32H32O15Na, 679.1639).
Paecilin N (10): pale yellow gum; [α]25.1D – 62.7 (c.5, MeOH); UV (MeOH) λmax (log ε) 260 (1.88) nm; IR (KBr) νmax 3500, 2960, 1800, 1736, 1624, 1540, 1437, 1320, 1063, and 800 cm–1; 1H NMR (500 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3) data, see Table 4; positive ion HRESIMS m/z 671.2001 [M + H]+ (calcd for C33H35O15, 671.1976).
Paecilin O (11): pale yellow gum; [α]25.1D – 46.7 (c.5, MeOH); UV (MeOH) λmax (log ε) 260 (1.37) nm; IR (KBr) νmax 3500, 2970, 1792, 1734, 1626, 1520, 1480, and 1204 cm–1; 1H NMR (600 MHz, CDCl3) and 13C NMR (150 MHz, CDCl3) data, see Table 4; positive ion HRESIMS m/z 707.1934 [M + Na]+ (calcd for C34H36O15Na, 707.1952).
Paecilin P (12): pale yellow gum; [α]21.4D – 59.6 (c.5, MeOH); UV (MeOH) λmax (log ε) 265 (3.25) nm; IR (KBr) νmax 3500, 1736, 1647, 1626, 1437, 1364, 1204, 1119, 1065, 1011, and 584 cm–1; 1H NMR (600 MHz, CDCl3) and 13C NMR (150 MHz, CDCl3) data, see Table 3; positive ion HRESIMS m/z 725.2049 [M + Na]+ (calcd for C34H38O16Na, 725.2058).
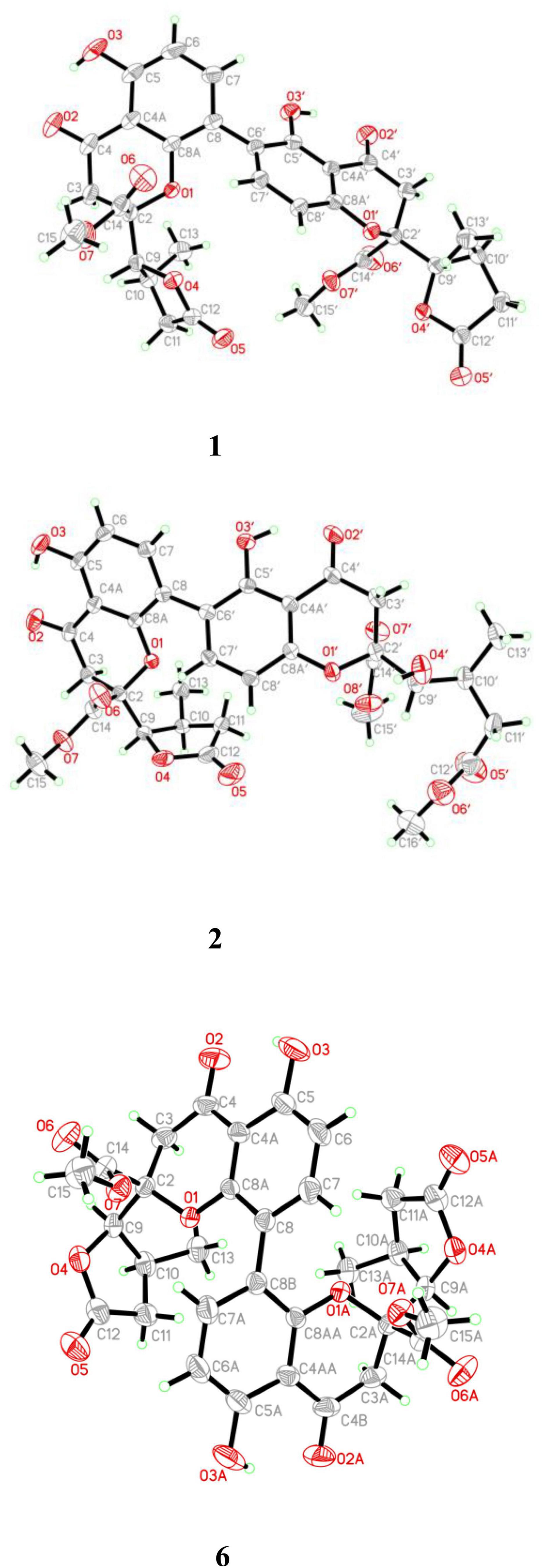
X-ray crystallographic analysis of Compounds 1, 2 and 6
X-ray crystallographic analysis of paecilin A (1)
C32H30O14, M = 638.56, a = 13.492(2) Å, b = 8.2053(15) Å, c = 14.726(3) Å, α = 90°, β = 114.945(5)°, γ = 90°, V = 1488.7(5) Å3, T = 294(2) K, space group P1211, Z = 2, μ(Cu Kα) = 1.54178 mm–1. The final anisotropic full-matrix least-squares refinement on F2 with 424 variables converged at R1 = 2.99% for the observed data and wR2 = 8.38% for all data. The goodness-of-fit was 1.061. The absolute configuration was determined by the Flack parameter = 0.03(5). CCDC: 2155089. Available online: https://www.ccdc.cam.ac.uk (accessed on 28 February 2022).
X-ray crystallographic analysis of paecilin F (2)
C33H34O15, M = 670.6, a = 14.3975(11) Å, b = 14.4598(11) Å, c = 15.2705(12) Å, α = 90°, β = 90°, γ = 90°, V = 3179.1(4) Å3, T = 295(2). K, space group P212121, Z = 4, μ(Cu Kα) = 1.54178 mm–1. The final anisotropic full-matrix least-squares refinement on F2 with 448 variables converged at R1 = 3.63% for the observed data and wR2 = 10.01% for all data. The goodness-of-fit was 1.058. The absolute configuration was determined by the Flack parameter = 0 (3). CCDC: 2155090. Available online: https://www.ccdc.cam.ac.uk (accessed on 28 February 2022).
X-ray crystallographic analysis of paecilin J (6)
C32H30O14, M = 638.56, a = 12.0791(14) Å, b = 13.3887(17) Å, c = 18.223(2) Å, α = 90°, β = 126.144(2)°, γ = 90°, V = 2947.1(6) Å3, T = 299(2) K, space group C2221, Z = 4, μ(Cu Kα) = 1.54178 mm–1. The final anisotropic full-matrix least-squares refinement on F2 with 212 variables converged at R1 = 3.67% for the observed data and wR2 = 11.05% for all data. The goodness-of-fit was 1.18. The absolute configuration was determined by the Flack parameter = 0.04(8). CCDC: 2155091. Available online: https://www.ccdc.cam.ac.uk (accessed on 28 February 2022).
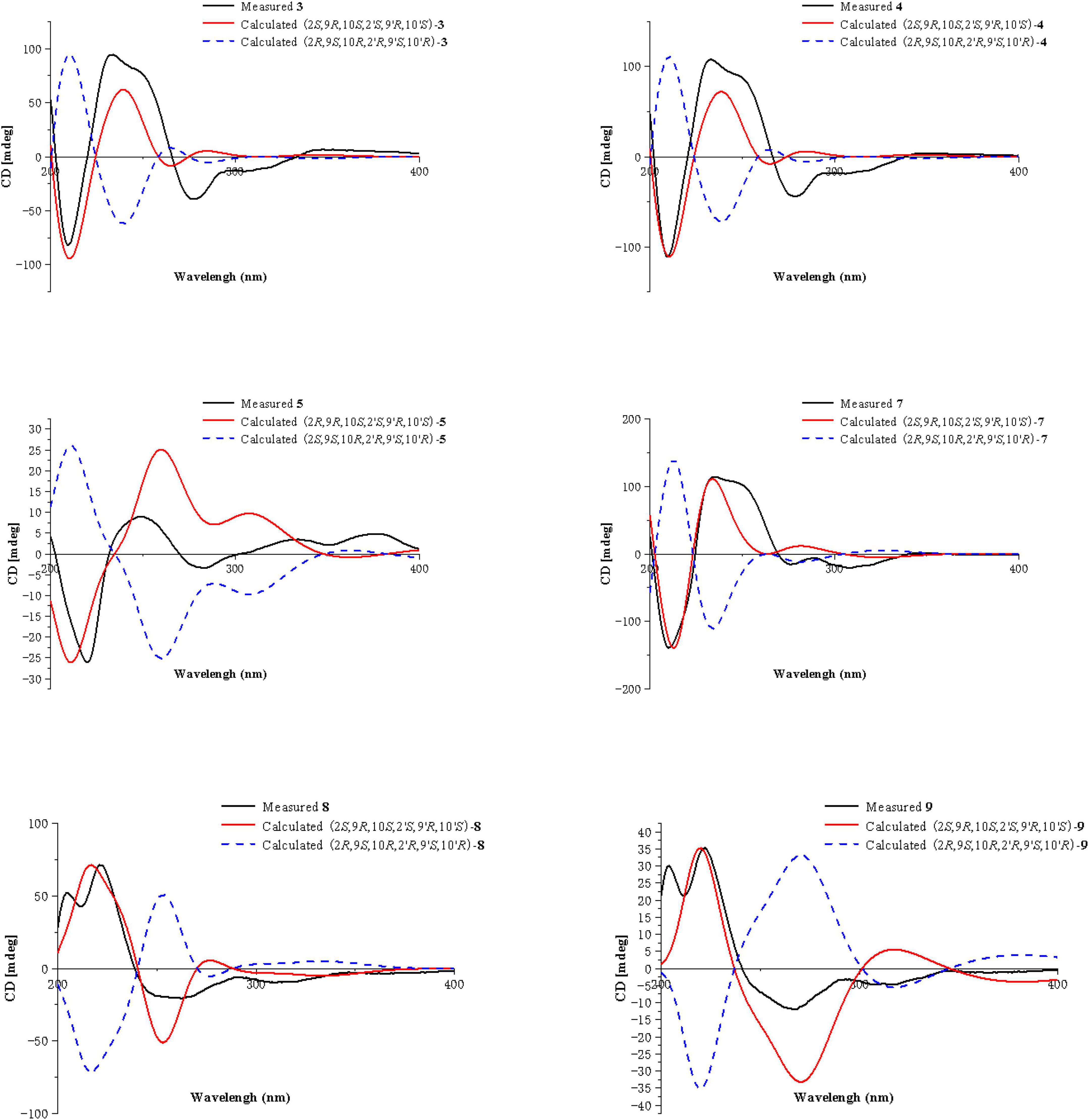
Equivalent circulating density calculations
Equivalent circulating density (ECD) calculations were carried out by the Gaussian 16 software package (Frisch et al., 2013). The conformation of the system was analyzed by Spartan 14 and calculated by the MMFF94s molecular mechanics’ force field. The cutoff energy was 10 kcal/mol. The conformational optimization and frequency were calculated at the B3LYP/def2svp theoretical level by using the IEFPCM solvent model (MeOH). The optimized conformations with proportions greater than 2% were selected for ECD calculations. Compounds 3, 4, 8, 10, 11, and 12 used the IEFPCM solvent model (MeOH) to calculate ECD (TDDFT) at the wB97xd/def2svp theoretical level. Compounds 5 and 9 used the IEFPCM solvent model (MeOH) to calculate ECD (TDDFT) at the B3LYP/DGD2VP theoretical level. Compound 7 used the IEFPCM solvent model (MeOH) to calculate ECD (TDDFT) at the wB97xd/TZVP theoretical level. The ECD curves were simulated in SpecDis v1.71 using a Gaussian function (Bruhn et al., 2013). The calculated ECD data of all conformers were Boltzmann averaged by Gibbs free energies.
Antimicrobial activity assays
The Microbroth dilution drug sensitivity test was used for MIC determination of the tested compounds. The bacterial liquid growth was observed on a clean workbench when the concentrations of the compounds to be measured were 8, 16, 32, 64, 128, 256, and 512 μg/mL. The concentrations of Escherichia coli ATCC 25922 and Salmonella enteritidis ATCC 25923 after secondary activation were adjusted to 1.0 × 108 CFU/mL using MH medium (Qingdao High-tech Industrial Park Haibo Biotechnology Co., Ltd.). The concentration of C. albicans ATCC 10231 was adjusted to 1.0 × 105 CFU/mL using a PDB medium (Qingdao High-tech Industrial Park Haibo Biotechnology Co., Ltd.). The 96-well plate was rationally planned, and the experimental groups were set as 100 μL compound and 100 μL bacterial solution, with two multiple Wells. The first group of negative control medium was 200 μL MH or PDB medium, which was evaluated in a single well. In the second group, 100 μL DMSO and 100 MH or PDB medium were used as negative controls. The positive control was a 200 μL bacterial solution with two duplicate wells. Amphenicol, penicillin, and fluconazole were used as positive controls for E. coli, S. enteritidis, and C. albicans, respectively. The 96-well plate was cultured in a high-throughput growth curve analyzer for 24 h, and the results were observed. The positive control well showed turbidity, while the negative control was clear. The quality control was within the specified range, and the MIC values of compounds were the lowest concentrations without bacterial growth observed by the naked eye.
Results and discussion
Structure elucidations
Compound 1 was obtained as light-yellow crystals, whose molecular formula was determined to be C32H30O14 by HRESIMS at m/z 661.1532 [M + Na]+ (calcd for C32H30O14Na+, 661.1533), with 18 degrees of unsaturation. In the 1H NMR spectrum (Table 1), two hydroxy protons at δH 11.58 (1H, s, 5-OH) and 11.82 (1H, s, 5′-OH), two pairs of adjacent aromatic protons at δH 7.58 (1H, d, J = 8.6 Hz, H-7′), 6.61 (1H, d, J = 8.6 Hz, H-8′), 7.45 (1H, d, J = 8.6 Hz, H-7) and 6.62 (1H, d, J = 8.6 Hz, H-6), two methoxy protons at δH 3.79 (3H, s, H-15) and 3.73 (3H, s, H-15′), two methyl doublets at δH 1.29 (3H, d, J = 7.2 Hz, H-13) and 1.22 (3H, d, J = 7.1 Hz, H-13′), two oxymethine protons at δH [4.86 (1H, d, J = 6.6 Hz, H-9) and 4.65 (1H, d, J = 7.5 Hz, H-9′) were clearly shown. The 13C NMR and DEPT spectra (Table 1) of 1 showed oxymethine carbons, including four methyl carbons (two oxygenated carbons), four methylene carbons, eight methine carbons (four sp2 carbons), and 16 quaternary carbons (two ketone carbonyl carbons, four ester carbonyl carbons, and eight sp2 carbons). The 1H NMR and 13C NMR spectral data (Table 1) of Compound 1 were nearly the same as those of paecilin A (Guo et al., 2007). Paecilin A is a symmetric dimer and should have one set of NMR data; however, there are two sets of data in its nuclear magnetic attribution table in the reference (Guo et al., 2007), so the structure of paecilin A was incorrectly assigned by erroneously linking C-8 with C-8′. Actually, C-8 was linked with C-6′, which was confirmed by the HMBC correlations from 5′-OH (δH 11.82) to δC C-6′ (δC 117.7)/C-4a’ (δC 107.4), from H-7 (δH 7.45) to C-6′ (δC 117.7), and from H-7′ (δH 7.58) to C-8 (δC 115.3) (Figure 2). The cis configurations of H-9/H-10 and H-9′/H-10′ in the γ-butyrolactone moiety were supported by the coupling constants (3JH–9,H–10 = 6.6 Hz, 3JH–9’,H–10’ = 7.5 Hz) (Wu et al., 2015). In addition, the relative configuration of Compound 1 was deduced as 2S*,9R*,10S*,2′S*,9′R*,10′S* by comparing the chemical shifts of H-2′/H-3′/H-4′ and the coupling constants of the four differential isomers in the reference (Tietze et al., 2014). Finally, the structure of 1 was confirmed by single-crystal X-ray diffraction analysis [Flack parameter = 0.03(5), CCDC: 2155089; Figure 3]. The absolute configuration of 1 was determined to be 2S,9R,10S,2′S,9′R,10′S. Therefore, the structure of paecilin A was revised.
Compound 2 was obtained as light-yellow crystals, whose molecular formula was determined to be C33H34O15 by HRESIMS analysis at m/z 693.1814 [M + Na]+ (calcd for C33H34O15Na+, 693.1795), with 17 degrees of unsaturation. Analysis of the 1H and 13C NMR data (Table 1) suggested that the structure of 2 was similar to that of 1, and the only observed difference was that 2 has an additional methoxy group compared to that of 1. Combined with the unsaturation degree, the γ-butyrolactone ring of one monomer part was opened. This change was confirmed by the key HMBC correlations (Figure 2) from H3-16′ (δH 3.70) to C-12′ (δC 173.3). By comparing the chemical shifts and coupling constants of H-9′ of Compound 2 (δH 4.08, s) with H-5′ in versixanthones E and F (δH 4.06, d, J = 1.8 Hz) (Wu et al., 2015), the relative configuration of Compound 2 was thus determined to be 2S*,9R*,10S*,2′S*,9′R*,10′S*. Finally, the absolute configuration of Compound 2 was determined to be 2S,9R,10S,2′S,9′R,10′S by single-crystal X-ray diffraction [Flack parameter = 0.00(3), CCDC: 2155090; Figure 3]. In conclusion, Compound 2 was named paecilin F.
Compound 3 was obtained as a pale-yellow gum, whose molecular formula was determined to be C34H38O16 by HRESIMS analysis at m/z 725.2048 [M + Na]+ (calcd for C34H38O16Na+, 725.2058), with 16 degrees of unsaturation. The 1H and 13C NMR spectroscopic data of 3 (Table 1) showed that 3 was a congener of 2. The only difference was that Compound 3 has an additional methoxy group compared to 2. By considering the degrees of unsaturation, it was suggested that the γ-butyrolactone groups of the two monomer moieties were opened. These changes were confirmed by the key HMBC correlations (Figure 2) from H3-16 (δH 3.66) to C-12 (δC 173.3) and H3-16′ (δH 3.70) to C-12′ (δC 173.3). Referring to the chemical shifts and coupling constants of 2, the relative configuration of Compound 3 was deduced as 2S*,9R*,10S*,2′S*,9′R*,10′S*. Finally, the absolute configuration of Compound 3 was determined to be 2S,9R,10S,2′S,9′R,10′S by ECD calculations (Figure 4). In summary, the structure of 3 was established and named paecilin G.
Compound 4 was obtained as a pale-yellow gum. Its molecular formula of C33H34O15 was determined based on the HRESIMS data (found at m/z 693.1791 [M + Na]+, calcd for C33H34O15Na+ 693.1795), corresponding to 17 degrees of unsaturation. Compound 4 had the same molecular formula and highly similar 1D NMR data (Table 2) to those of 2. The only difference was that the two monomers of Compound 4 were connected by C-6 and C-8′. The 1H-1H COSY cross peaks (Figure 2) of H-7 (δH 7.72)/H-8 (δH 6.63), H-6′ (δH 6.61)/H-7′ (δH 7.47), and the key HMBC correlations from H-7′ (δH 7.47) to C-6 (δC 118), from H-7 (δH 7.72) to C-8′ (δC 115), from 5-OH (δH 11.86) to C-4a (δC 107.6)/C-5 (δC 159.3)/C-6 (δC 118), and from 5′-OH (δH 11.7) to C-4a’ (δC 107.8)/C-5′ (δC 161.9)/C-6′ (δC 110.1) confirmed the above inference. The relative configuration of Compound 4 was deduced by comparison with the chemical shifts and coupling constants of 2. Finally, the absolute configuration of Compound 4 was determined to be 2S,9R,10S,2′S,9′R,10′S by ECD calculations (Figure 4). In summary, the structure of 4 was established and named paecilin H.
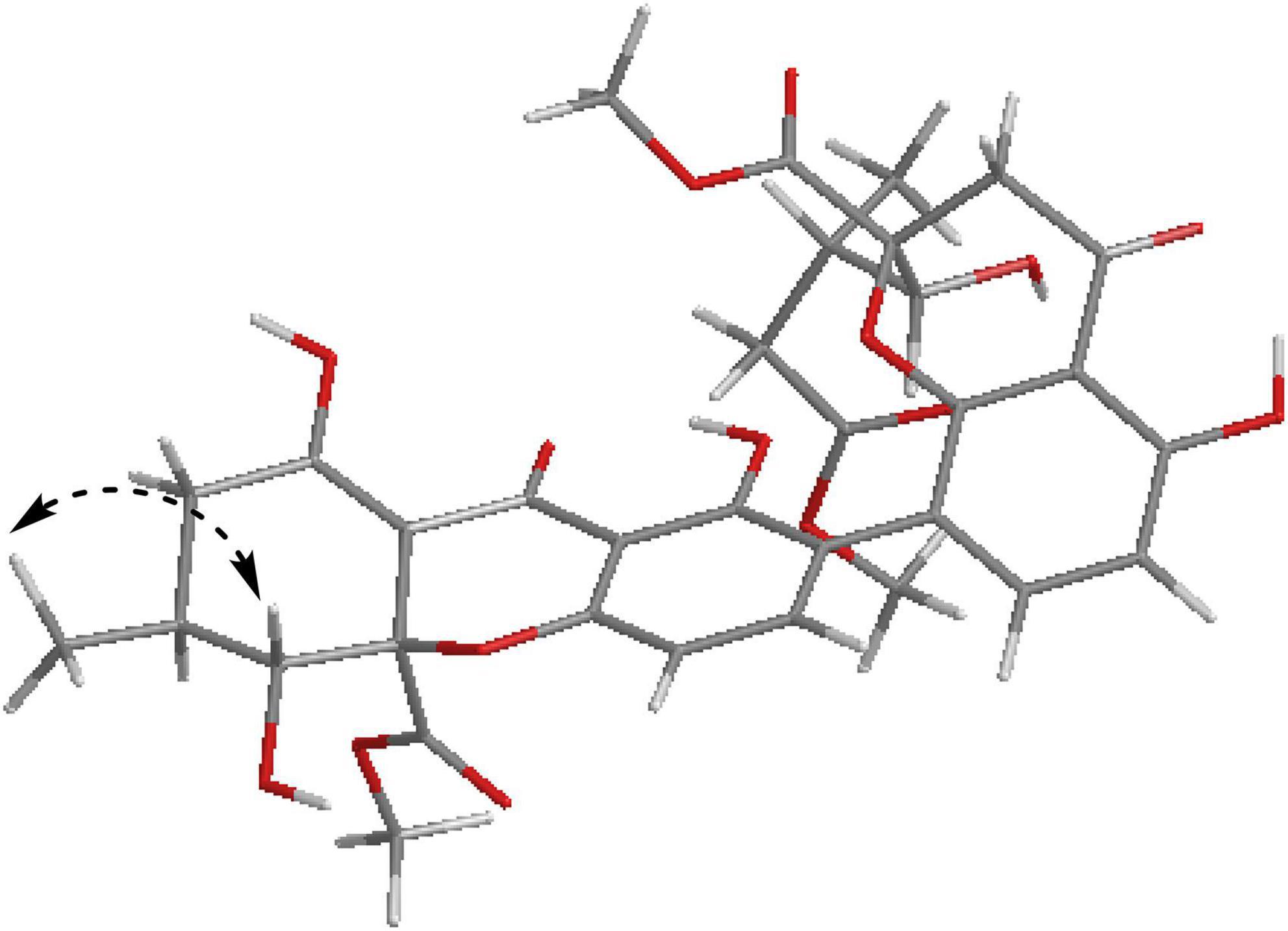
Compound 5 was obtained as a pale-yellow gum, and it possessed the molecular formula of C33H34O15 as indicated by the HRESIMS analysis at m/z 693.1810 [M + Na]+ (calcd for C33H34O15Na+, 693.1795), with 17 degrees of unsaturation. The 1H and 13C NMR spectroscopic data of Compound 5 (Table 2) were highly similar to those of versixanthone B (Wu et al., 2015). The mass spectrometry data showed that Compound 5 had one more methoxy group than versixanthone B, which suggested that Compound 5 was the γ-butyrolactone ring-opening analog of versixanthone B. This conclusion was confirmed by the correlation between H-11′ (δH 2.46, 2.22) and H-16′ (δH 3.65 to C-12′) (δC 173.2) in the HMBC spectrum (Figure 2). Therefore, the planar structure of 5 was determined. The relative configuration of the tetrahydroxanthone monomer was readily established to be the same as that of versixanthone A by the coupling constant (3JH–9,H–10 = 11.3 Hz), the NOESY correlation between H-9 and H3-13 (Figure 5), and the chemical shifts. By considering the biosynthesis of this family of congeners, together with the chemical shifts, and coupling constants of H-5′, H-6′, and H-7′ in versixanthone E (Wu et al., 2015), it was deduced that the relative configuration of Compound 5 was 2R*,9R*,10S*,2′S*,9′R*,10′S*. Finally, the absolute configuration was deduced as 2R,9R,10S,2′S,9′R,10′S by ECD calculations (Figure 4). Therefore, Compound 5 could be fully assigned and named paecilin I.
Compound 6 was obtained as a brown crystal, whose molecular formula was determined to be C32H30O14 by HRESIMS analysis at m/z 661.1526 [M + Na]+ (calcd for C32H30O14Na+, 661.1533), with 18 degrees of unsaturation. According to the 1H NMR and 13C NMR data (Table 3) and the mass spectra, it could be inferred that Compound 6 was also a dimeric compound that was structurally similar to Compound 1. There was only one set of 1H and 13C NMR spectroscopic data for 6, indicating that the structure of 6 was symmetric. Therefore, Compound 6 had either a C8-C8′ or a C6-C6′ linkage pattern. Due to the lack of the key HMBC correlations of the signals 5-OH and 5′-OH, the connection position of its monomer remained to be determined. Fortunately, single-crystals of 6 were obtained, which finally enabled us to determine the absolute configuration as 2S,9R,10S,2′S,9′R,10′S [Flack parameter = 0.04(8), CCDC: 2155091; Figure 3]. In summary, the structure of 6 was established and named paecilin J.
Compound 7 was obtained as a pale-yellow gum, whose molecular formula was determined to be C33H34O15 by HRESIMS analysis at m/z 693.1785 [M + Na]+ (calcd for C33H34O15Na+, 693.1795), with 17 degrees of unsaturation. By comparing the 1H and 13C NMR spectra of Compound 7 (Table 2) with those of 2 (Table 1), it was found that 7 was a structural congener of 2. The only difference between the two compounds was that Compound 7 was linked via C8-C8′ while 2 was linked by C8-C6′. This change was proved by the key HMBC correlations (Figure 2) from H-7′ (δH 7.92) to C-8 (δC 114.5), from H-7 (δH 7.76) to C-8′ (δC 114.9), from 5-OH (δH 11.70) to C-4a (δC 107.8)/C-5 (δC 161.9)/C-6 (δC 110.4), and from 5′-OH (δH 11.68) to C-4a′ (δC 107.7)/C-5′ (δC 161.8)/C-6′ (δC 110.9). By comparison of the coupling constants of Compounds 7 and 2, it was deduced that the relative configuration of Compound 7 was 2S*,9R*,10S*,2′S*,9′R*,10′S*. Finally, the absolute configuration of Compound 7 was determined to be 2S,9R,10S,2′S,9′R,10′S by ECD calculations (Figure 4). Therefore, Compound 7 was elucidated and named paecilin K.
Compound 8 was obtained as a pale-yellow gum, whose molecular formula was determined to be C32H30O14 by the HRESIMS analysis at m/z 661.1538 [M + Na]+ (calcd for C32H30O14Na+, 661.1533), with 18 degrees of unsaturation. The 1D NMR spectrum of Compound 8 (Table 3) showed high similarity to that of Compound 6 (Table 3). The only difference was that the two monomers of Compound 6 were C8-C8′ linked, while the two monomers of Compound 8 were C6-C6′ linked. This conclusion was established by the key HMBC correlations (Figure 2) from H-7/H-7′ (δH 7.52) to C-6/C-6′ (δC 117.7), from 5-OH/5′-OH (δH 11.91) to C-4a/C-4a’ (δC 107.6), C-5/C-5′ (δC 159.2), and C-6/C-6′ (δC 117.7). By biosynthetic considerations, and comparison with the chemical shifts and coupling constants with those of 6, the relative configuration of Compound 8 was deduced as 2S*,9R*,10S*,2′S*,9′R*,10′R*. The ECD calculation for 8 was performed, and the results of 8 matched well with the experimental ECD curve (Figure 4). Therefore, the absolute configuration of 8 was 2S,9R,10S,2′S,9′R,10′S, and it was named paecilin L.
Compound 9 was obtained as a pale-yellow gum, whose molecular formula was determined to be C32H32O15 by HRESIMS analysis at m/z 679.1627 [M + Na]+ (calcd for C32H32O15Na+, 679.1639), with 17 degrees of unsaturation. Comparing the 1H and 13C NMR data of Compounds 8 (Table 3) and 9 (Table 4), it was found that these two compounds are structural analogs. The mass spectrum showed that Compound 9 was 18 Da more than Compound 8, corresponding to a molecule of H2O. Combined with the degrees of unsaturation, it was speculated that one of the γ-butyrolactone rings was opened. In the HMBC spectrum (Figure 2), H-9′ (δH 4.80) of Compound 8 was correlated with C-12′ (δC 175.0), while for Compound 9, this correlation was absent. This evidence helped to prove the above conclusion. The relative configurations in the two monomeric units of 9 were proposed to be the same as 8, as indicated by the similar chemical shifts and coupling constants and by consideration of the biogenetic origin. Finally, the absolute configuration of Compound 9 was determined to be 2S,9R,10S,2′S,9′R,10′S by ECD calculations (Figure 2). In summary, Compound 9 was named paecilin M.
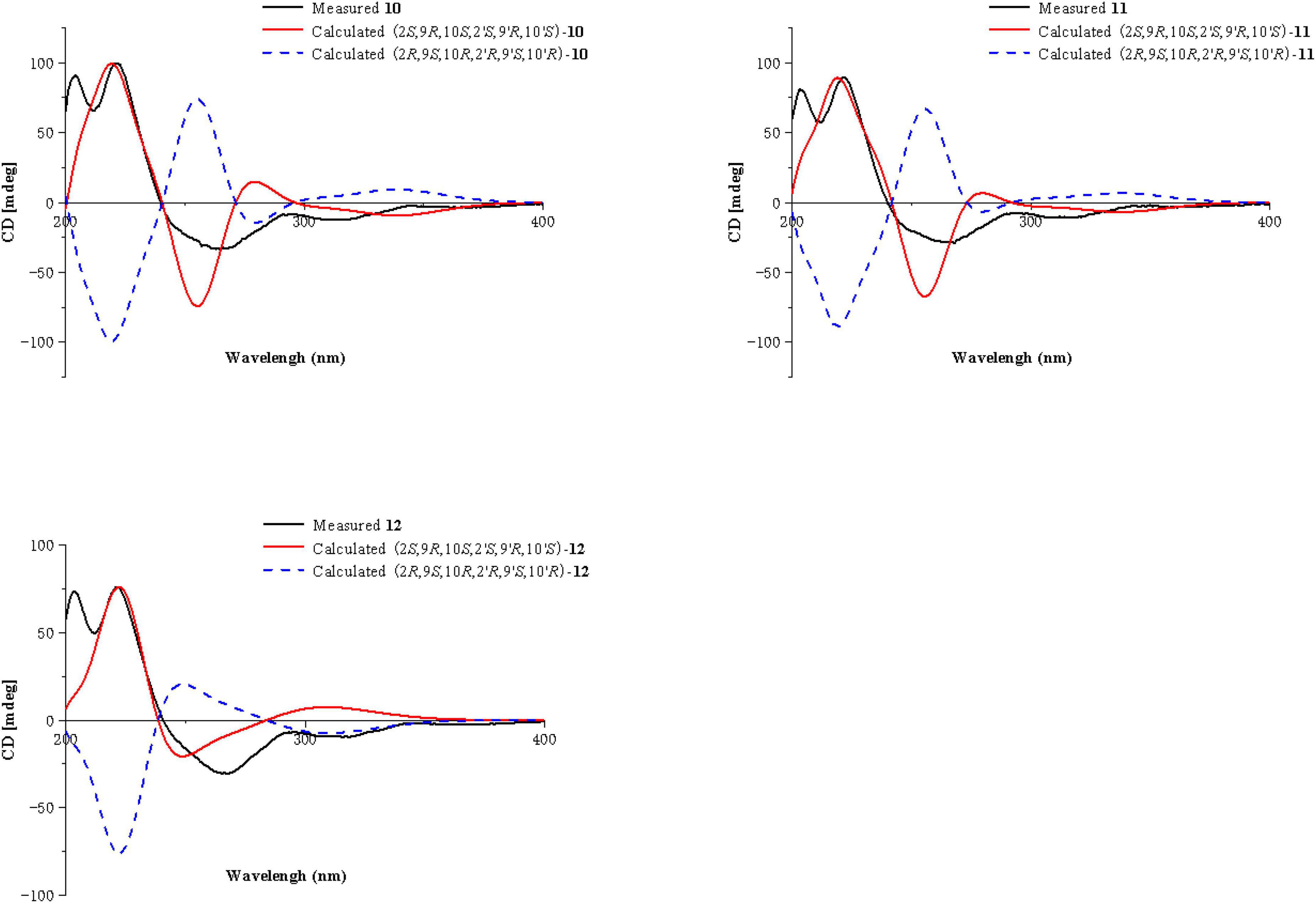
Compounds 10 and 11 were obtained as pale-yellow gum. The molecular formula of 10 was determined to be C33H34O15 by HRESIMS at m/z 671.2001 [M + H]+ (calcd for C33H35O15+, 671.1976), with 17 degrees of unsaturation. The molecular formula of 11 was determined to be C34H36O15 by HRESIMS at m/z 707.1934 [M + Na]+ (calcd for C34H36O15Na+, 707.1952), also with 17 degrees of unsaturation. By comparing the 1H and 13C NMR spectroscopic data of Compounds 9-11 (Table 4), it was found that the data of these three compounds were highly similar, with the only difference being that Compound 10 had one additional methoxy group compared to 9, while Compound 11 had one additional ethoxy group compared to 9. The key HMBC correlations (Figure 2) from H-10′ (δH 2.35)/H-11′ (δH 2.58, 2.38)/H-16′ (δH 3.68) to C-12′ (δC 173.3) indicated that the carboxylic acid group at C-12′ was methyl-esterified in Compound 10. The key HMBC correlations (Figure 2) from H-10′ (δH 2.39)/H-11′ (δH 2.58, 2.36)/H-16′ (δH 4.16) C-12′ (δC 172.9) and from H-17′ (δH 1.28) to C-16′ (δC 60.9), in combination with the key 1H-1H COSY correlations (Figure 2) between H-16′ (δH 4.16) and H-17′ (δH 1.28) indicated that the carboxylic acid group at C-12′ was ethyl-esterified in Compound 11. Combining the chemical shifts and coupling constants of Compound 9, it was deduced that the relative configurations of Compounds 10 and 11 were 2S*,9R*,10S*,2′S*,9′R*,10′S*. Finally, by ECD calculations (Figure 6), the absolute configurations of Compounds 10 and 11 were determined to be 2S,9R,10S,2′S,9′R,10′S. Therefore, Compounds 10 and 11 were named paecilin N and paecilin O, respectively.
Compound 12 was obtained as a pale-yellow gum, and it possessed the molecular formula of C34H38O16, as indicated by the HRESIMS analysis at m/z 725.2049 [M + Na]+ (calcd for C34H38O16Na, 725.2058). By comparing the 1D spectroscopic data of Compound 12 (Table 3) with those of Compound 8 (Table 3), it was suggested that Compound 12 was highly similar to Compound 8. The only difference was that Compound 12 has two additional methoxyl groups compared to Compound 8. Considering the reduction of two degrees of unsaturation, it was assumed that both γ-butyrolactone groups of 12 were opened. This conclusion was established by the key HMBC correlations (Figure 2) from H-10/H-10′ (δH 2.37), H-11/H-11′ (δH 2.60, 2.40), and H-16/H-16′ (δH 3.70) to C-12/C-12′ (δC 173.3). By comparison with the chemical shifts and coupling constants of the ring-opening monomer of Compound 10, it was deduced that the relative configuration of Compound 12 was 2S*,9R*,10S*,2′S*,9′R*,10′S*. Finally, the absolute configuration of Compound 12 was determined to be 2S,9R,10S,2′S,9′R,10′S by ECD calculations (Figure 6). In summary, Compound 12 was named paecilin P.
The other known analogs isolated in this study were identified as versixanthone F (13) (Wu et al., 2015), versixanthone A (14) (Wu et al., 2015), and versixanthone E (15) (Wu et al., 2015) by comparison of their NMR data with those reported in the literature.
Antimicrobial activities
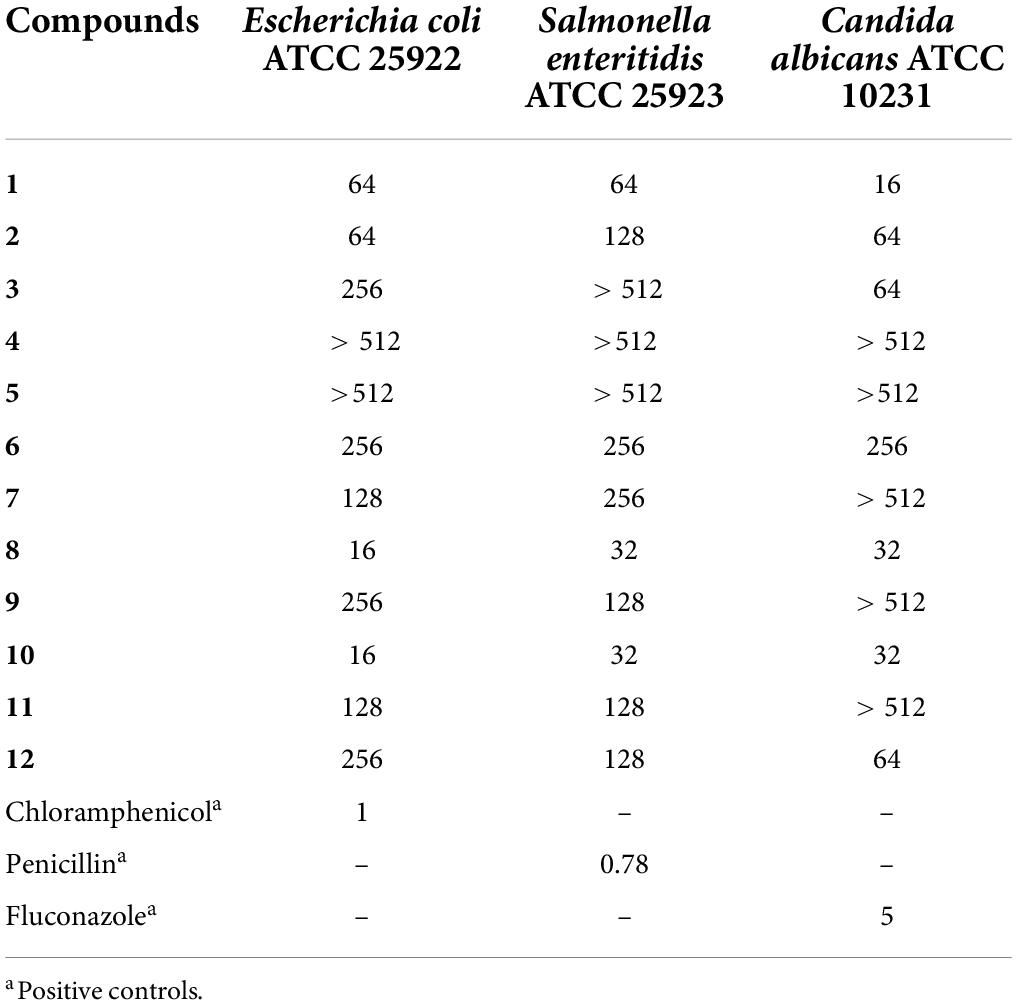
Compound 1 and new compounds (2–12) were evaluated for their antimicrobial activities against the bacteria E. coli and S. enteritidis and the fungus C. albicans. As a result, Compound 1 showed significant inhibitory activity against C. albicans, with an MIC of 16 μg/mL, while Compounds 8 and 10 showed inhibitory activity against the gram-negative bacterium E. coli, with the same MIC value of 16 μg/mL (Table 5).
Conclusion
In conclusion, a total of 15 compounds, including 11 new compounds, were identified from the endophytic fungus Xylaria curta E10. The absolute configurations of Compounds 1, 2, and 6 were determined by single-crystal X-ray diffraction analysis. The absolute configurations of 3, 4, 5, 7, 8, 9, 10, 11, and 12 were determined by ECD calculations and referred to the NMR data with their structural analogs. In the antimicrobial activity assays, Compound 1 showed antifungal activity against C. albicans, and Compounds 8 and 10 showed antibacterial activity against Escherichia coli.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
H-LA isolated and provided the fungus. Z-HL and X-XL designed experiments. P-PW was responsible for compound isolation, elucidated the structures and wrote the manuscript. P-PW, B-BS, KY, X-YP, X-JM, and DX performed chemical calculations. XL and Z-HL were responsible for the evaluation of the immunosuppressive activity. All authors read and approved the final manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (31870513 and 32000011) and the Fundamental Research Funds for the Central University, South-Central MinZu University (CZD21003).
Acknowledgments
We thank the Analytical & Measuring Centre, South-Central University for Nationalities for the spectra measurements.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.922444/full#supplementary-material
References
Arora, D., Chashoo, G., Singamaneni, V., Sharma, N., Gupta, P., and Jaglan, S. (2017). Bacillus amyloliquefaciens induces production of a novel blennolide K in coculture of Setophoma terrestris. J. Appl. Microbiol. 124, 730–739. doi: 10.1111/jam.13683
Bruhn, T., Schaumloffel, A., Hemberger, Y., and Bringmann, G. (2013). SpecDis: Quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 25, 243–249. doi: 10.1002/chir.22138
Cao, H. Y., Cheng, Y., Sun, S. F., Li, Y., and Liu, Y. B. (2022). Anti-inflammatory dimeric tetrahydroxanthones from an endophytic Muyocopron laterale. J. Nat. Prod. 85, 148–161. doi: 10.1021/acs.jnatprod.1c00878
Chen, L., Zhang, Q. Y., Jia, M., Ming, Q. L., Yue, W., Rahman, K., et al. (2016). Endophytic fungi with antitumor activities: Their occurrence and anticancer compounds. Crit. Rev. Microbiol. 42, 454–473. doi: 10.3109/1040841X.2014.959892
da Silva, P. H., de Souza, M. P., Bianco, E. A., da Silva, S. R., Soares, L. N., Costa, E. V., et al. (2018). Antifungal polyketides and other compounds from Amazonian endophytic Talaromyces fungi. J. Braz. Chem. Soc. 29, 622–630. doi: 10.21577/0103-5053.20170176
Duan, Y. D., Jiang, Y. Y., Guo, F. X., Chen, L. X., Xu, L. L., Zhang, W., et al. (2019). The antitumor activity of naturally occurring chromones: A review. Fitoterapia 135, 114–129. doi: 10.1016/j.fitote.2019.04.012
El-Elimat, T., Figueroa, M., Raja, H. A., Graf, T. N., Swanson, S. M., and Falkinham, J. O. III, et al. (2015). Biosynthetically distinct cytotoxic polyketides from Setophoma terrestris. Eur. J. Org. Chem. 2015, 109–121. doi: 10.1002/ejoc.201402984
Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., et al. (2013). Gaussian 09, revision E. 01. Wallingford, CT: Gaussian, Inc.
Guo, Z. Y., She, Z. G., Shao, C. L., Wen, L., Liu, F., Zheng, Z. H., et al. (2007). 1H and 13C NMR signal assignments of paecilin A and B, two new chromone derivatives from mangrove endophytic fungus Paecilomyces sp (tree 1-7). Magn. Reson. Chem. 45, 777–780. doi: 10.1002/mrc.2035
Kumla, D., Shine Aung, T., Buttachon, S., Dethoup, T., Gales, L., Pereira, J. A., et al. (2017). A new dihydrochromone dimer and other secondary metabolites from cultures of the marine sponge-associated fungi Neosartorya fennelliae KUFA 0811 and Neosartorya tsunodae KUFC 9213. Mar. Drugs. 15:375. doi: 10.3390/md15120375
Li, E., Tian, R., Liu, S. C., Chen, X. L., Guo, L. D., and Che, Y. S. (2008). Pestalotheols A-D, bioactive metabolites from the plant endophytic fungus Pestalotiopsis theae. J. Nat. Prod. 71, 664–668. doi: 10.1021/np700744t
Liu, S., Zhao, Y. P., Heering, C., Janiak, C., Müller, W. E. G., Akoneì, S. H., et al. (2019). Sesquiterpenoids from the endophytic fungus Rhinocladiella similis. J. Nat. Prod. 82, 1055–1062. doi: 10.1021/acs.jnatprod.8b00938
Liu, Y., Wray, V., Abdel-Aziz, M. S., Wang, C. Y., Lai, D., and Proksch, P. (2014). Trimeric anthracenes from the endophytic fungus Stemphylium globuliferum. J. Nat. Prod. 77, 1734–1738. doi: 10.1021/np500113r
Lünne, F., Koöhler, J., Stroh, C., Müller, L., Daniliuc, C. G., Mück-Lichtenfeld, C., et al. (2021). Insights into ergochromes of the plant pathogen Claviceps purpurea. J. Nat. Prod. 84, 2630–2643. doi: 10.1021/acs.jnatprod.1c00264
Nalin Rathnayake, G. R., Savitri Kumar, N., Jayasinghe, L., Araya, H., and Fujimoto, Y. (2019). Secondary metabolites produced by an endophytic fungus Pestalotiopsis microspora. Nat. Prod. Bioprospect. 9, 411–417. doi: 10.1007/s13659-019-00225-0
Nguyen, V. K., Genta-Jouve, G., Duong, T. H., Beniddir, M. A., Gallard, J. F., Ferron, S., et al. (2020). Eumitrins C-E: Structurally diverse xanthone dimers from the vietnamese lichen Usnea baileyi. Fitoterapia 141:104449. doi: 10.1016/j.fitote.2019.104449
Pinto, M. M., Castanheiro, R. A., and Kijjoa, A. (2014). Xanthones from marine-derived microorganisms: Isolation, structure elucidation and biological activities. Encycl. Anal. Chem. 27, 1–21. doi: 10.1002/9780470027318.a9927
Pontius, A., Krick, A., Kehraus, S., Foegen, S. E., Müller, M., Klimo, K., et al. (2008). Noduliprevenone: A novel heterodimeric chromanone with cancer chemopreventive potential. Chem. Eur. J. 14, 9860–9863. doi: 10.1002/chem.200801574
Ribeiro, B. A., da Mata, T. B., Canuto, G. A. B., and Silva, E. O. (2021). Chemical diversity of secondary metabolites produced by Brazilian endophytic fungi. Curr. Microbiol. 78, 33–54. doi: 10.1007/s00284-020-02264-0
Tietze, L. F., Ma, L., Jackenkroll, S., Reiner, J. R., Hierold, J., Gnanaprakasam, B., et al. (2014). The paecilin puzzle enantioselective synthesis of the proposed structures of paecilin A and B. Heterocycles 88, 1101–1119. doi: 10.3987/COM-13-S(S)68
Tuong, T. L., Do, L. T., Aree, T., Wonganan, P., and Chavasiri, W. (2020). Tetrahydroxanthone–chromanone heterodimers from lichen Usnea aciculifera and their cytotoxic activity against human cancer cell lines. Fitoterapia 147:104732. doi: 10.1016/j.fitote.2020.104732
Wang, W. X., Lei, X. X., Ai, H. L., Bai, X., Li, J., He, J., et al. (2019a). Cytochalasans from the endophytic fungus Xylaria cf. curta with resistance reversal activity against fluconazole-resistant Candida albicans. Org. Lett. 21, 1108–1111. doi: 10.1021/acs.orglett.9b00015
Wang, W. X., Lei, X. X., Yang, Y. L., Li, Z. H., Ai, H. L., Li, J., et al. (2019b). Xylarichalasin A, a halogenated hexacyclic ytochalasin from the fungus Xylaria cf. curta. Org. Lett. 21, 6957–6960. doi: 10.1021/acs.orglett.9b02552
Wang, W. X., Li, Z. H., Feng, T., Li, J., Sun, H., Huang, R., et al. (2018). Curtachalasins A and B, two cytochalasans with a tetracyclic skeleton from the endophytic fungus Xylaria curta E10. Org. Lett. 20, 7758–7761. doi: 10.1021/acs.orglett.8b03110
Wang, Z. J., Jiang, Y., Xin, X. J., and An, F. L. (2021). Bioactive indole alkaloids from insect derived endophytic Aspergillus lentulus. Fitoterapia 153:104973. doi: 10.1016/j.fitote.2021.104973
Wei, X., and Matsuda, Y. (2020). Unraveling the fungal strategy for tetrahydroxanthone biosynthesis and diversification. Org. Lett. 22, 1919–1923. doi: 10.1021/acs.orglett.0c00285
Wei, X. X., Chen, X. X., Chen, L., Yan, D. X., Wang, W. G., and Matsuda, Y. (2021). Heterologous biosynthesis of tetrahydroxanthone dimers: Determination of key factors for selective or divergent synthesis. J. Nat. Prod. 84, 1544–1549. doi: 10.1021/acs.jnatprod.1c00022
Wu, G. W., Yu, G. H., Kurtán, T., Mándi, A., Peng, J. X., Mo, X. M., et al. (2015). Versixanthones A–F, cytotoxic xanthone–chromanone dimers from the marine-derived fungus Aspergillus versicolor HDN1009. J. Nat. Prod. 78, 2691–2698. doi: 10.1021/acs.jnatprod.5b00636
Keywords: dimeric chromanones, endophytic fungus, Xylaria curta E10, antimicrobial activities, structural identification
Citation: Wei P-P, Ai H-L, Shi B-B, Ye K, Lv X, Pan X-Y, Ma X-J, Xiao D, Li Z-H and Lei X-X (2022) Paecilins F–P, new dimeric chromanones isolated from the endophytic fungus Xylaria curta E10, and structural revision of paecilin A. Front. Microbiol. 13:922444. doi: 10.3389/fmicb.2022.922444
Received: 01 June 2022; Accepted: 15 August 2022;
Published: 02 September 2022.
Edited by:
Giuseppantonio Maisetta, University of Pisa, ItalyReviewed by:
Guiyang Xia, Beijing University of Chinese Medicine, ChinaDa-Le Guo, Chengdu University of Traditional Chinese Medicine, China
Satish Sreedharamurthy, University of Mysore, India
Zhangshuang Deng, China Three Gorges University, China
Copyright © 2022 Wei, Ai, Shi, Ye, Lv, Pan, Ma, Xiao, Li and Lei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zheng-Hui Li, bGl6aGVuZ2h1aUBtYWlsLnNjdWVjLmVkdS5jbg==; Xin-Xiang Lei, eHhsZWlAbWFpbC5zY3VlYy5lZHUuY24=
†These authors have contributed equally to this work
 Pan-Pan Wei†
Pan-Pan Wei† Hong-Lian Ai
Hong-Lian Ai Zheng-Hui Li
Zheng-Hui Li Xin-Xiang Lei
Xin-Xiang Lei