
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 27 July 2022
Sec. Terrestrial Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.922149
The continuous application of chemical fertilizers in vegetable cropping has led to deterioration of the soil environment and reduced yield and quality. The objective of this study was to evaluate the effect of combining chemical and bio-organic fertilizers on cauliflower yield, soil biochemical properties, and the bacterial community. Six treatments were established: no fertilizer (CK, control), chemical fertilizers (CF, conventional dosage for this region), balanced fertilization (BF, 30% reduction of chemical fertilizers), and balanced fertilization plus 3,000, 6,000, or 12,000 kg.ha–1 bio-organic fertilizer (Lvneng Ruiqi Biotechnology Co., Ltd., Gansu, China) (BF + OF1, BF + OF2, BF + OF3, respectively). A two-season field experiment with cauliflower was conducted under the different fertilizer treatments in irrigation districts along the Yellow River, Northwest China. The results indicate that the yield, soil organic matter, total potassium content, and enzyme activity under the bio-organic treatments were generally higher than those under the CF treatment. Compared with the CF treatment, the BF treatment increased soil organic matter content, enzyme activity and soil bacterial relative abundance. Moreover, the bacterial alpha-diversity were higher than those of conventional fertilization. The predominant phyla, including Proteobacteria, Actinobacteria, Gemmatimonadetes, and Chloroflexi, were the main contributors to the microbiome shift, as demonstrated by their remarkable enrichment in the soil under BF + OF2 and BF + OF3 treatments. Furthermore, Pearson correlation analyses show significant correlations among the soil organic matter, available P and K, electrical conductivity, and relative abundance of potentially beneficial microbial groups, such as the genera Massilia, Bacillus, Lysobacter, and Nitrosospira. Overall, this study suggests that balanced fertilization and the application of bio-organic fertilizers are essential to ensure soil fertility and long-term sustainable green productivity.
Vegetables are among the foods with the largest per capita consumption globally. Moreover, vegetable production has become one of the primary sources of per capita income growth among rural residents worldwide (Gai et al., 2016). The large rise in chemical fertilizer inputs has resulted in a tremendous boost in vegetable yield in China during the last 20 years (Dawson and Hilton, 2012; Souza et al., 2020). However, chemical fertilizer application also increased production cost and led to the misuse of fertilizers, disruption to the stability of the soil environment, and decreased the quality of vegetables (Wang et al., 2017; Wesenbeeck et al., 2021; Yin et al., 2021). Recently, concerns have been raised about planting pattern productivity and sustainability, which has remained stagnant or even declined. Therefore, coordinating the relationship between chemical fertilizer input and the high-stable yield of vegetables is a problem that urgently needs to be solved in current agricultural vegetable production.
Balanced fertilization and the combined application of organic and chemical fertilizers are beneficial for the sustainable development of vegetable production (Zhao et al., 2016; Wang et al., 2021). Balanced fertilization technology was designed according to the nutrient requirements of specific crops during growth and fruiting. In addition, the effects of climate and environmental factors on crop nutrient absorption were considered (Singh et al., 2015). Balanced fertilization can increase crop yield, create scientific, standardized, and accurate fertilization management, and reduces fertilizer waste (Lin et al., 2012). Bio-organic fertilizer is a compound fertilizer that uses animal and plant residues as raw material, which is produced by adding specific functional microorganisms and harmless decomposed organic substances (Naher et al., 2021). The organic matter content is very high (≥40%), which improves soil quality and enhances the nutrient supply capacity (Aparna et al., 2014; Qiao et al., 2019). Soil organic matter provides all the nutrients needed for crop growth, and prevents nutrient leaching. Soil microorganisms play an indispensable role in terrestrial ecosystems, they are involved in producing and transforming organic matter in the soil, nutrient cycling, and improving the utilization efficiency of nutrients by crops (Paz-Ferreiro et al., 2014; Lin et al., 2016; Paul, 2016). Previous studies have shown that balanced fertilization and the combined application of bio-organic fertilizers can improve crop yield, possibly owing to the alteration of soil properties, enzyme activities, and soil microorganisms. Yu et al. (2019) and Brenzinger et al. (2021) found that balanced fertilization could improve soil fertility by increasing soil cation exchange capacity (ECE), soil organic matter (SOM), and available phosphorus (AP) content as well as transforming soil microorganisms. By combining bio-organic fertilizers with chemical fertilizers, organic matter in the soil can be increased and soil enzymes and microorganisms can be stimulated (Baldi et al., 2016; Jiang et al., 2019).
Cauliflower (Brassica oleracea var. botrytis L.) is one of the main plateau summer vegetables crops in the Yellow River irrigation area of Northwest China. However, there is concern regarding the problem of unbalanced fertilization during cauliflower production. Excessive chemical fertilization leads to low fertilizer use efficiency and yield in cauliflower cropping (Yang et al., 2020). We speculated that balanced fertilization and application of bio-organic fertilizer could improve fertilizer use efficiency and yield of cauliflower by altering the soil biochemical environment. Therefore, we a field experiment was conducted on cauliflower with reduced chemical fertilizers and combination of the later with bio-organic fertilizers. To investigate how fertilizers fertilization affect vegetable cauliflower yields and, soil enzyme activity, and composition and diversity of microbial community by high-throughput sequencing. The results are expected to provide insights to increase fertilizer use efficiency, reduce fertilizer input, and offer a reference for developing effective fertilizer management strategies for vegetable production.
The experimental site was one of main plateau summer vegetables production areas in Lanzhou City, Gansu Province, China (35°87′N, 104°23′E). The region has a temperate semi-arid continental climate with distinct seasons, mean altitude is 1,790 m above sea level, mean annual temperature of 6.6°C, mean annual rainfall of 300–400 mm, mean annual evaporation of 1,343 mm, and a frost-free period of approximately 150 days. The test field has gentle terrain and the fertility level was uniform. The soil is a typical loessal, and the main physicochemical parameters of the 0–20 cm topsoil were pH 7.85, SOM 12.08 g.kg–1, total nitrogen (TN) 1.01 g.kg–1, AP 97.66 mg.kg–1, and available potassium (AK) 138.44 mg.kg–1.
The field experiment was performed in a completely randomized block design with three replicates for each of the following treatments: CK, no fertilizer (control); CF, chemical fertilizers (conventional dosage for this region, N 805.5 kg.ha–1, P2O5 948 kg.ha–1, K2O 420 kg.ha–1); BF, balanced fertilizer (30% reduction of chemical fertilizers); BF + OF1, BF + OF2, and BF + OF3, balanced fertilizer plus 3,000, 6,000, or 12,000 kg.ha–1 bio-organic fertilizer, respectively. The bio-organic fertilizer (Lvneng Ruiqi Biotechnology Co., Ltd., Gansu, China) was N + P2O5 + K2O ≥ 5%; it contained > 40% organic matter, >25% of humic acid, and >20 million effective living bacteria per gram.
The cauliflower cultivation mode was adopted for two seasons per year. For the spring crop, bio-organic fertilizer, phosphorus, potassium, and 30% nitrogen were used as the base fertilizer and applied once before planting and mixed thoroughly. The remaining 70% nitrogen fertilizer was used as topdressing at the rosette stage (50%) and early flowering stage (20%). In the autumn cultivation, the same fertilizer types and methods were used as controls, but reduced by 30%, and the NPK ratio was adjusted. All fertilizers were applied three times at the rosette stage, the early stage of curd formation, and the peak stage of curd formation. The fertilization amounts for each treatment are summarized in Table 1.
Each treatment had three trial plots, each with an area of 31.5 m2 (4.5 × 7 m). When the cauliflower met the harvest standard, the middle section of each plot of 18 m2 (3 × 6 m) was selected; the economic yield (wet weight of curd) within this section was measured and converted into yield per hectares based on the yield per unit area.
Economic benefit (yuan.667 m–2) = Annual total output (yuan.667 m–2) − Total fertilizer input (yuan.667 m–2).
A soil sample was taken at a depth of 0–20 cm after removal of 1 cm deep topsoil during the harvest period of autumn cauliflower. Three parallel samples were collected using a five-spot sampling method for each treatment. We collected soil samples and immediately loaded them into ice boxes and transported them to the laboratory within 1.5 h. The soil from the 15 points in each treatment was thoroughly mixed to remove stones, roots, plastic film debris, and other impurities, and then passed through a sieve of 2 mm in size. We divided each sample into two parts: one part was frozen at −80°C for later extraction of soil DNA. In a laboratory environment, the remaining soil was placed in a ventilated area and dried so that it reached a constant weight, after which its physical and chemical properties and enzyme activities were determined.
The pH of the soil samples was measured using a pH electrode (PHS-3C, Shanghai Yidian Scientific Instrument Co., Ltd., Shanghai, China) at a soil/water ratio of 1:5. The electrical conductivity (EC) of the soil samples was measured using a conductivity meter (DDS-307A, Shanghai Yidian Scientific Instrument Co., Ltd., Shanghai, China) at a soil/water ratio of 1:5. The potassium dichromate volumetric method was used to measure the SOM content. The TN, total phosphorus (TP), and total potassium (TK) contents were measured after the digestion with H2SO4-H2O2 mixture. TN content was analyzed using a Kjeldahl nitrogen analyzer, TP content was measured using the Mo-Sb colorimetric method, and TK content was determined using flame spectrometry. Alkali-hydrolyzed nitrogen (Alk N), AP, and AK contents were analyzed using diffusion, Olsen, and ammonium acetate extraction flame photometry methods, respectively. These methods were modified according to Lu (1999). Soil enzyme (sucrase, urease, and alkaline phosphatase) activity was estimated by colorimetry (Guan et al., 1986). The soil enzyme activities of β-glucosidase and cellulase were measured using a soil β-glucosidase (S-β-GC) activity assay kit (BC0165) and soil cellulase (S-CL) activity assay kit (BC0155, Solarbio Biotech Co., Ltd., Beijing, China), respectively. To guarantee the validity of the experimental data, three independent tests were performed on each sample.
A 16S rDNA amplicon sequencing was carried out by MiSeq at Novogene Biotechnology Co., Ltd. (Beijing, China). Genomic DNA was extracted from the samples using thecetyltrimethylammonium bromide (CTAB) method. The DNA samples were diluted to 1 ng.μL–1 with sterile water. The 16S rRNA genes of distinct regions (16S V3-V4) were amplified using a specific primer [16S V4: 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) −806R (5′-GGACTACHVGGGTWTCTAAT-3′)] with the barcode (Caporaso et al., 2011; Apprill et al., 2015; Parada et al., 2016). All PCRs were carried out with 15 μL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs, Beijing, China), 0.2 μM of forward and reverse primers, and ∼10 ng template DNA. The thermal cycling procedure included initial denaturation at 98°C for 1 min followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and elongation at 72°C for 30 s. The PCR products were then purified. The TruSeq® DNA PCR-free sample preparation kit was used to construct a library. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. After the library had been quantified, it was sequenced using a NovaSeq6000.
According to the barcode sequence and PCR primer sequences, each sample was separated from the offline data. After the barcode and primer sequences were truncated, FLASH (V1.2.7)1 (Magoc and Salzberg, 2011) was used to splice the reads of each sample, and the spliced sequence was counted as the original tags data (raw tags). In order to obtain high-quality tags (clean tags) from spliced raw tags, strict filtering is required (Bokulich et al., 2013). The tag quality control process of Qiime (V1.9.1)2 (Caporaso et al., 2010) was used with the following operations: (a) tag interception: raw tags were truncated at the first low-quality base site when the continuous low-quality value (default quality threshold ≤19) was set to a default length (default length 3); (b) tag length filtering: the tag dataset obtained by interception of tags further filtered out tags with continuous high-quality base lengths of <75% of tag length. Following the above processing, the tags were further processed to remove the chimera sequence3 by comparison with the species annotation database to produce the final effective data (effective tags) (Rognes et al., 2016).
One-way analysis of variance (ANOVA) with Duncan’s honestly significant difference (HSD) test (P < 0.05) was conducted using the SPSS software to analyze the data for each set of physicochemical factors and enzyme activity indices. The Uparse software (Uparse v7.0.1001)4 was used to cluster all effective tags of all the samples (Haas et al., 2011). Based on 97% identity, the sequences were clustered into operational taxonomic units (OTUs) and representative sequences of OTUs were selected. In accordance with the algorithm principle, the sequences with the highest frequency within the OTUs were selected as representative sequences within the OTUs. The species annotation analysis was carried out by using the Mathur method and the SSUrRNA database of SILVA1325 (Wang, 2007; Edgar, 2013). The α-Diversity indices (including goods coverage, PD_whole_tree index, Chao1, Shannon, Simpson, and ACE) were calculated using the Qiime software (version 1.9.1). The Unifrac distance was computed using the Qiime software (version 1.9.1), and the principal component analysis (PCA) diagram was drawn using R (version 2.15.3). PCA for OTUs (97% similarity) can reflect the differences and distances among samples. The higher the similarity between two samples, the closer the distance between them. The similarity of the sample composition under the same conditions can be judged according to the dispersion and aggregation of the samples. Using the corr.test function of the Psych package in R, we calculated the Spearman correlation coefficient between species and environmental factors and tested the significance. To visualize the heatmap, the pheatmap function was used in the “pheatmap” package.
The various fertilizer applications strongly influenced the cauliflower yield (Table 2). The BF + OF2 and BF + OF3 treatments significantly increased cauliflower yield by 7.5 and 5.9% compared to CF, respectively in the spring crop. In autumn crops, the yield difference between BF and CF treatments was not significant. The highest yield was achieved by the BF + OF3 treatment, which was 18.6% higher than that of the CF treatment. The second largest total yield was observed in the BF + OF2 treatment, which increased yield by 11.3% compared with that of the CF treatment. Overall, the treatments with balanced fertilizer plus bio-organic fertilizer produced higher yields than the treatments with only chemical fertilizer (CF) or balanced fertilizer (BF). Compared with CK2, the economic benefits of T2, T3 and T4 treatment increased by 5.8, 6.8, and 6.1%, respectively. The fertilizer cost of T3 treatment increased by 320 yuan.667 m–2 and the economic benefits increased by 190.9 yuan.667 m–2, compared to T2 treatment. The fertilizer cost of T4 treatment increased by 640 yuan.667 m–2 compared with T3 treatment, but the economic benefit decreased. Therefore, the economic benefit of T3 treatment was the best, which was 19777.0 yuan.667 m–2, increased by 1265.5 yuan.667 m–2 compared with CK2 treatment.
Compared with the conventional CF treatment, balanced fertilization combined with biological organic fertilizers increased the soil nutrient content, except for TP, and decreased the pH and EC (Table 3). Compared with that of CF treatment, soil organic matter content in the BF + OF1, BF + OF2, and BF + OF3 treatments increased by 2.0, 2.1, and 3.7%, respectively. The TP content decreased by 10.0 ∼16.9%. The soil TN and TK contents were not significantly different under the different fertilization treatments. The soil AK content in the BF + OF3 treatment was markedly higher than that in the CF treatment, and increased by 20.1%, while the AP contents in the BF, BF + OF1, BF + OF2, and BF + OF3 treatments were significantly lower than that of the CF treatment (decreases of 30.0∼61.3%). The soil pH of the BF + OF2 and BF + OF3 treatments were decreased by 1.1 and 2.5%, respectively, compared with that of the CF treatment. The EC values decreases by 6.4 ∼13.2% in the BF + OF1, BF + OF2, and BF + OF3 treatments compared with the CF treatment.
The soil enzyme activities of the different fertilization treatments are shown in Figure 1. Balanced fertilization and bio-organic fertilizers improved soil enzyme activity. Compared with that of the CF treatment, the sucrase activities of the BF + OF2 and BF + OF3 treatments increased by 17.0 and 20.4%, respectively (Figure 1A). The urease activities of the BF, BF + OF1, BF + OF2, and BF + OF3 treatments increased by 1.3, 1.4, 3.1, and 1.9%, respectively, compared with that of the CF treatment (Figure 1B). The activity of alkaline phosphatase in the CK treatment was the lowest. The alkaline phosphatase activities in the BF + OF1, BF + OF2, and BF + OF3 treatments were higher than that in the CF treatment. Among them, the BF + OF3 treatment reached a significant difference (P < 0.05), increasing the alkaline phosphatase activity by 22.1% (Figure 1C). The order of β-glucosidase activity was BF + OF3 > BF + OF2 > BF + OF1 > CF > BF > CK in six soil treatment. The β-glucosidase activities of BF + OF1, BF + OF2, and BF + OF3 treatment increased by 6.1, 30.2, and 32.0%, respectively (Figure 1D). Compared with that of the CF treatment, the BF, BF + OF1, BF + OF2, and BF + OF3 treatments increased soil cellulase activity by 2.6, 6.1, 9.7, and 13.2%, respectively (Figure 1E).
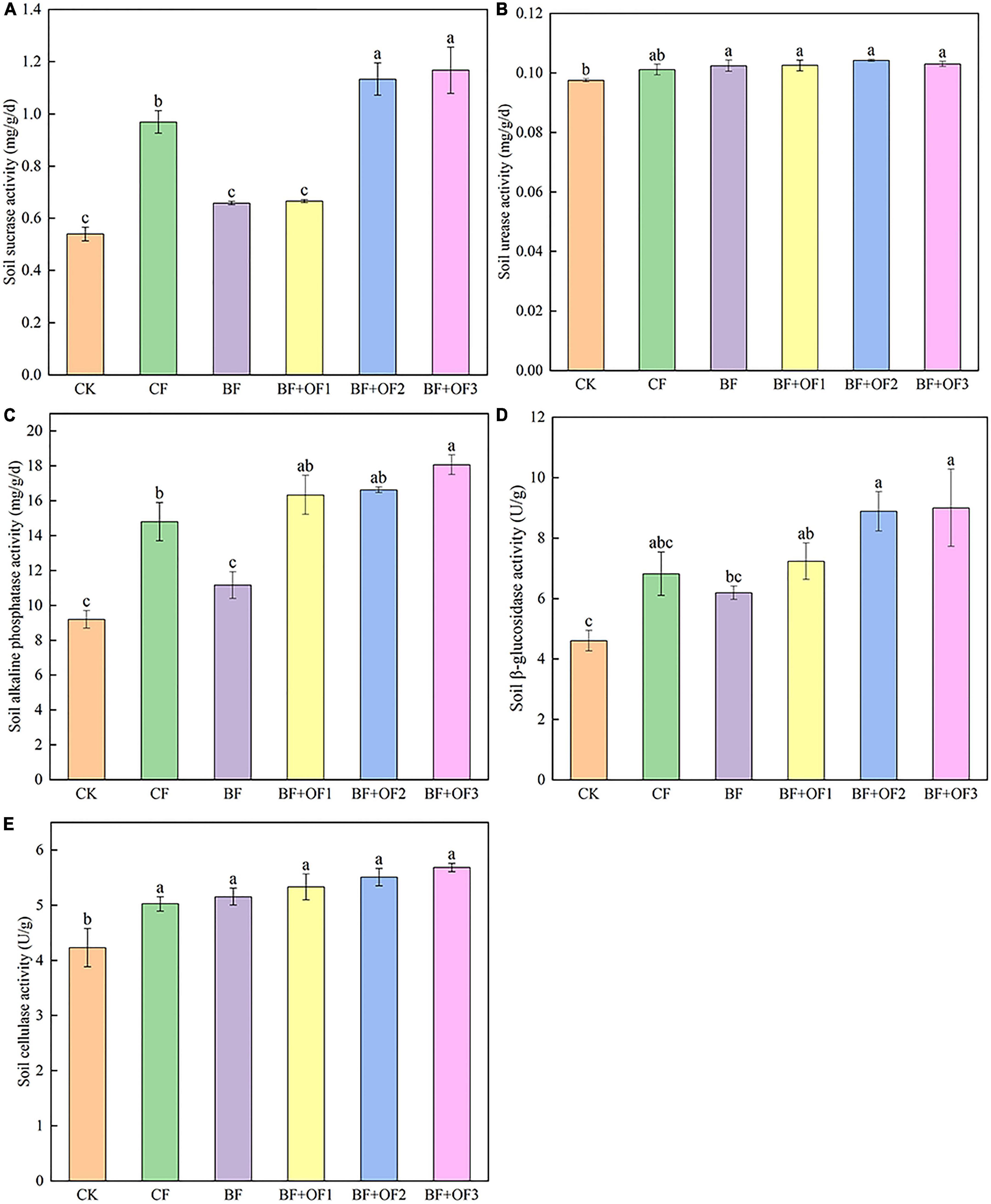
Figure 1. Soil enzyme activities under different fertilization treatments. The sucrase activities (A), urease activities (B), alkaline phosphatase activities (C), β-glucosidase activities (D) and cellulase activity (E). CK, no fertilizer, CF, chemical fertilizers (conventional dosage for this region); BF, balanced fertilizer (70% of total chemical fertilizer); BF + OF1, BF + OF2, and BF + OF3, balanced fertilizer plus 3,000, 6,000, and 12,000 kg⋅ha–1 bio-organic fertilizer, respectively. Each histogram represents the mean ± standard error (SE) of three independent biological experiments (n = 3). Different letters above the bars indicate statistically significant differences by Duncan’s test (p < 0.05).
The common number of OTUs in the six treatments are 2,948, and the number of unique OTUs in each treatment was 158 (CK), 131 (CF), 175 (BF), 175 (BF + OF1), 127 (BF + OF2), and 189 (BF + OF3) (Figure 2). Generally, the number of unique OTUs in the chemical fertilizer reduction combined with bio-organic fertilizers increased compared with that of the CF treatment. The number of unique OTUs in BF and BF + OF1 increased by 44%, while those in BF + OF3 increased by 58% compared with that of the CF treatment.
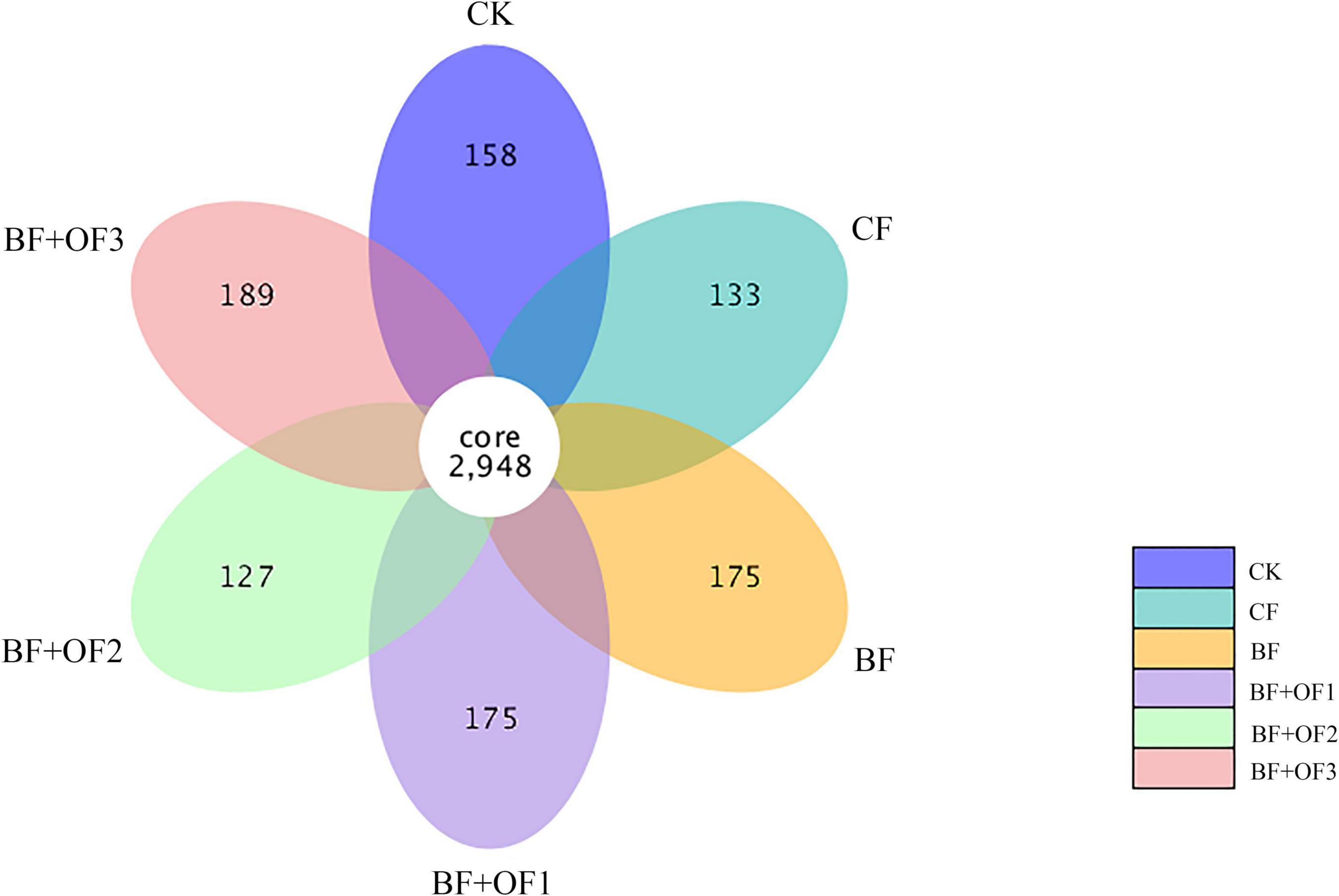
Figure 2. Petal plot of soils with different fertilization treatments based on operational taxonomic unit (OTU) distribution. CK, no fertilizer, CF, chemical fertilizers (conventional dosage for this region); BF, balanced fertilizer (70% of total chemical fertilizer); BF + OF1, BF + OF2, and BF + OF3, balanced fertilizer plus 3,000, 6,000, 12,000 kg.ha–1 bio-organic fertilizer, respectively.
Table 4 shows that the sequencing data can effectively express almost 99% of the bacteria in soil samples and cover almost all possible species. Fertilizer reduction based on Balanced Fertilization did not reduce the diversity of soil microorganisms, and there was no significant difference in Chao1 and ACE indexes among fertilization treatments. Compared with the CF treatment, the bacterial Shannon indices of BF, BF + OF1, and BF + OF3 treatments increased to different degrees. The Shannon index of the BF + OF3 was the highest. The greater the Simpson index value, the lower the community diversity. In general, the Simpson index difference between treatments was not significant. The Simpson index of The BF and BF + OF1 treatment are slightly lower than of CF treatment. The results show that chemical fertilizer reduction combined with bio-organic fertilizers could improve the diversity of soil bacteria to a certain extent.
The results show that the dominant phyla were Proteobacteria, Actinobacteria, Acidobacteria, Gemmatimonadetes, Bacteroidetes, and Chloroflexi, with average relative abundances of 45.9, 13.9, 12.7, 7.8, 5.7, and 5.2%, respectively (Figure 3). In addition, some bacterial phyla with low relative abundance were detected, including Cyanobacteria, Verrucomicrobia, Firmicutes, and Rokubacteria, with relative abundances of 0.8∼4.7%.
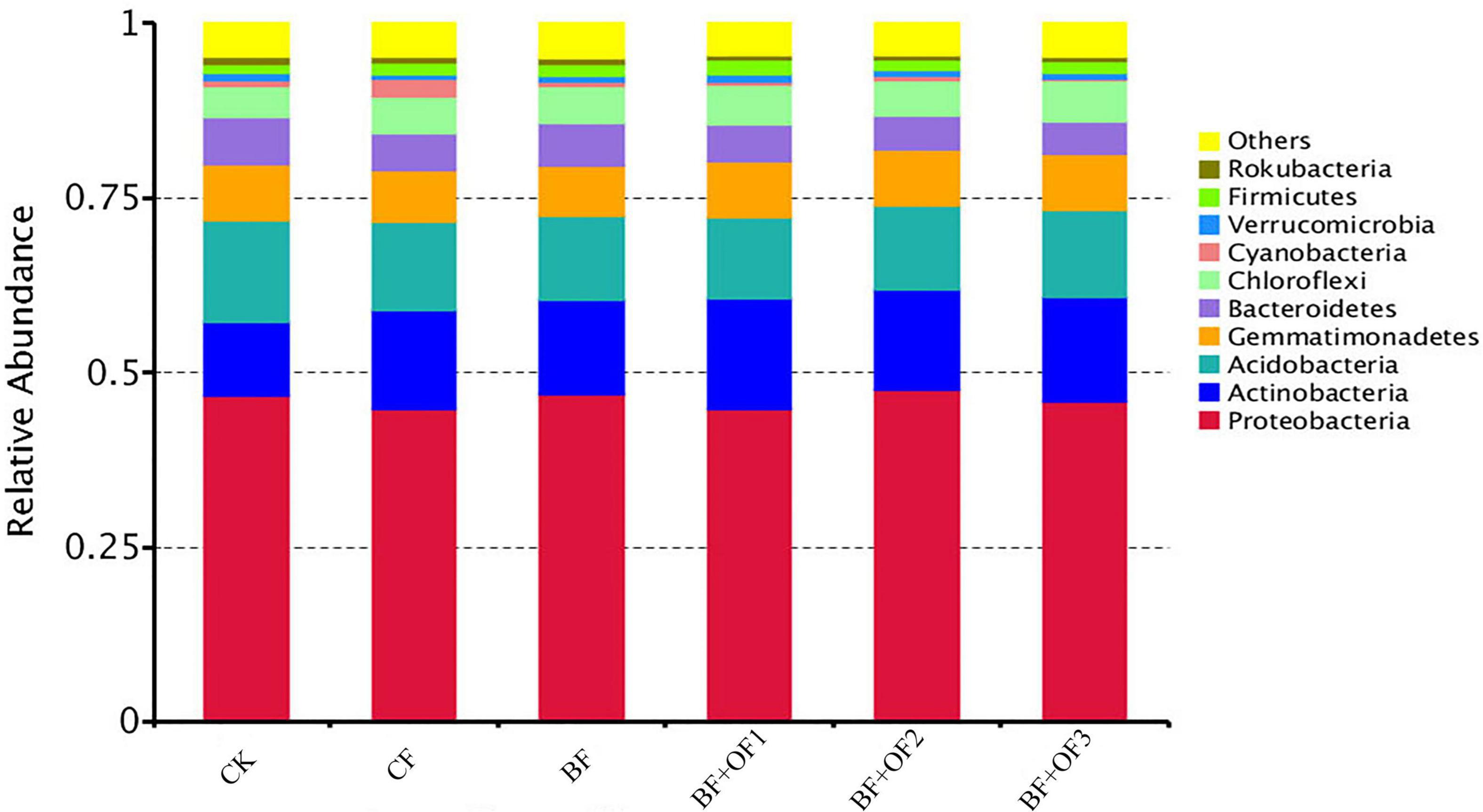
Figure 3. Relative abundances of species at phylum level under different fertilization treatments. CK, no fertilizer, CF, chemical fertilizers (conventional dosage for this region); BF, balanced fertilizer (70% of total chemical fertilizer); BF + OF1, BF + OF2, and BF + OF3, balanced fertilizer plus 3,000, 6,000, 12,000 kg.ha–1 bio-organic fertilizer, respectively.
The effects of different fertilization treatments on the relative abundances of the six dominant bacterial phyla are shown in Figure 4. No significant differences were observed in the relative abundance of Proteobacteria among the different treatments. The relative abundance of Actinobacteria in BF decreased by 3.4% compared with that of CF, while the abundances in BF + OF1, BF + OF2, and BF + OF3 increased by 11.7, 2.0, and 6.2%, respectively. The relative abundances of Gemmatimonadetes in the BF + OF1, BF + OF2 and BF + OF3 treatments was 9.9, 10.9, and 10.3% higher than that in the CF treatment, respectively. The BF + OF3 treatment significantly improved the relative abundance of Chloroflexi compared with that in the CF treatment.
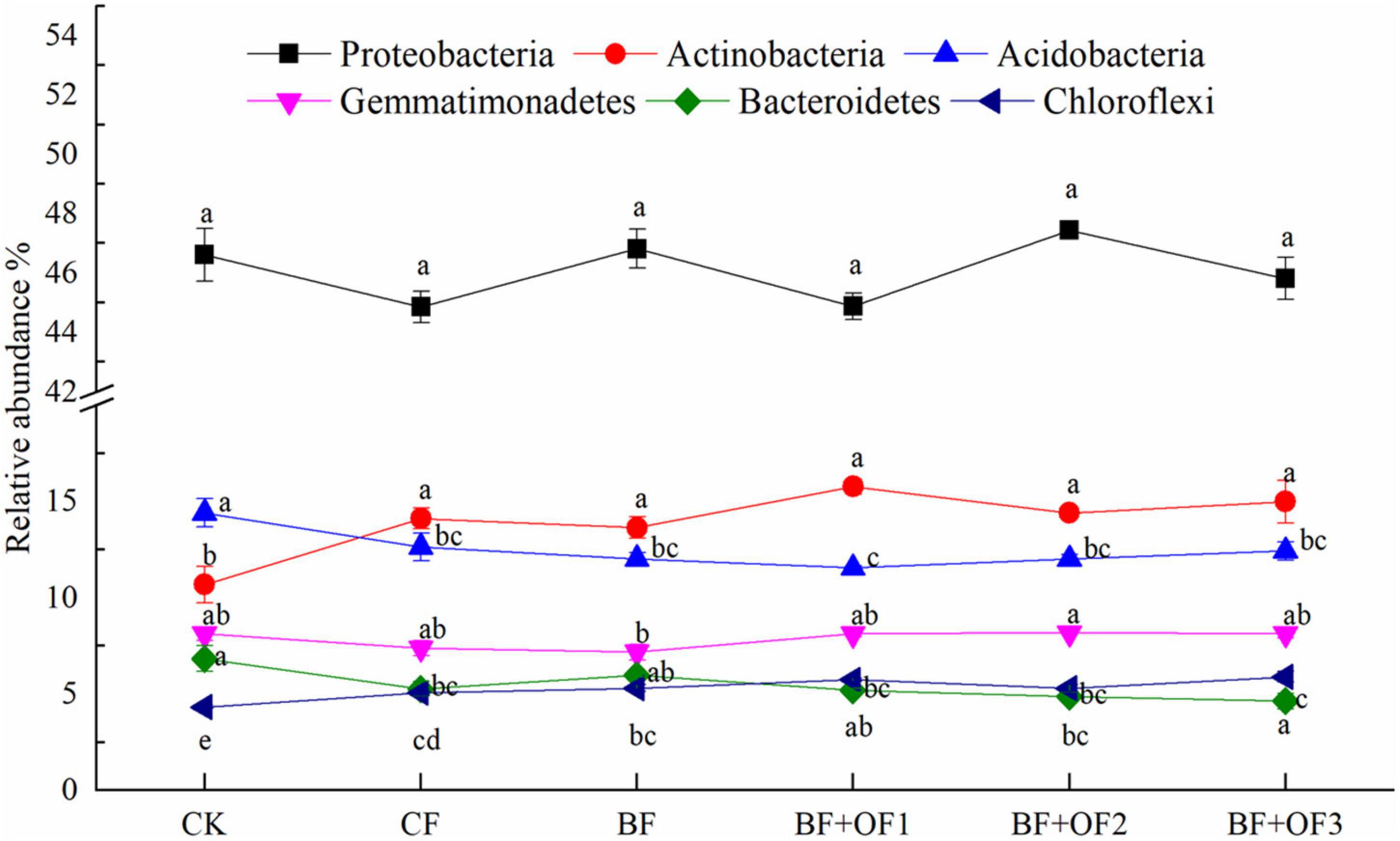
Figure 4. Relative abundances of six dominant bacteria at the phylum level under different fertilization treatments. CK, no fertilizer; CF, chemical fertilizers (conventional dosage for this region); BF, balanced fertilizer (70% of total chemical fertilizer); BF + OF1, BF + OF2, and BF + OF3, balanced fertilizer plus 3,000, 6,000, and 12,000 kg.ha–1 bio-organic fertilizer, respectively. Different letters above the bars indicate statistically significant differences by Duncan’s test (p < 0.05).
The differences and similarities of species communities were verified by the color change gradients and similarities of the heat maps (Figure 5). Heat maps of the top 35 genera of soil bacteria were analyzed. Significant changes in bacterial community structure was observed in all six treatments. In the CF treatment, the relative abundances of unidentified_Nostocales and unidentified_Cyanobacteria were the highest. The application of bio-organic fertilizer enriched the soil with Bacillus, llumatobacter, Kouria, and Nocardiodes in BF + OF1 and Massilia, Dongia, Woeseia, and Altereythrobacter in BF + OF2, compared with the CF treatment. The relative abundances of Pseudoxanthomonas, Gemmatimonas, and Nitrospirae increased significantly in the BF + OF3 treatment compared with in the CF treatment. Moreover, the soil of the BF + OF series exhibited a more even bacterial composition than the CF soil, as shown by the color intensity.
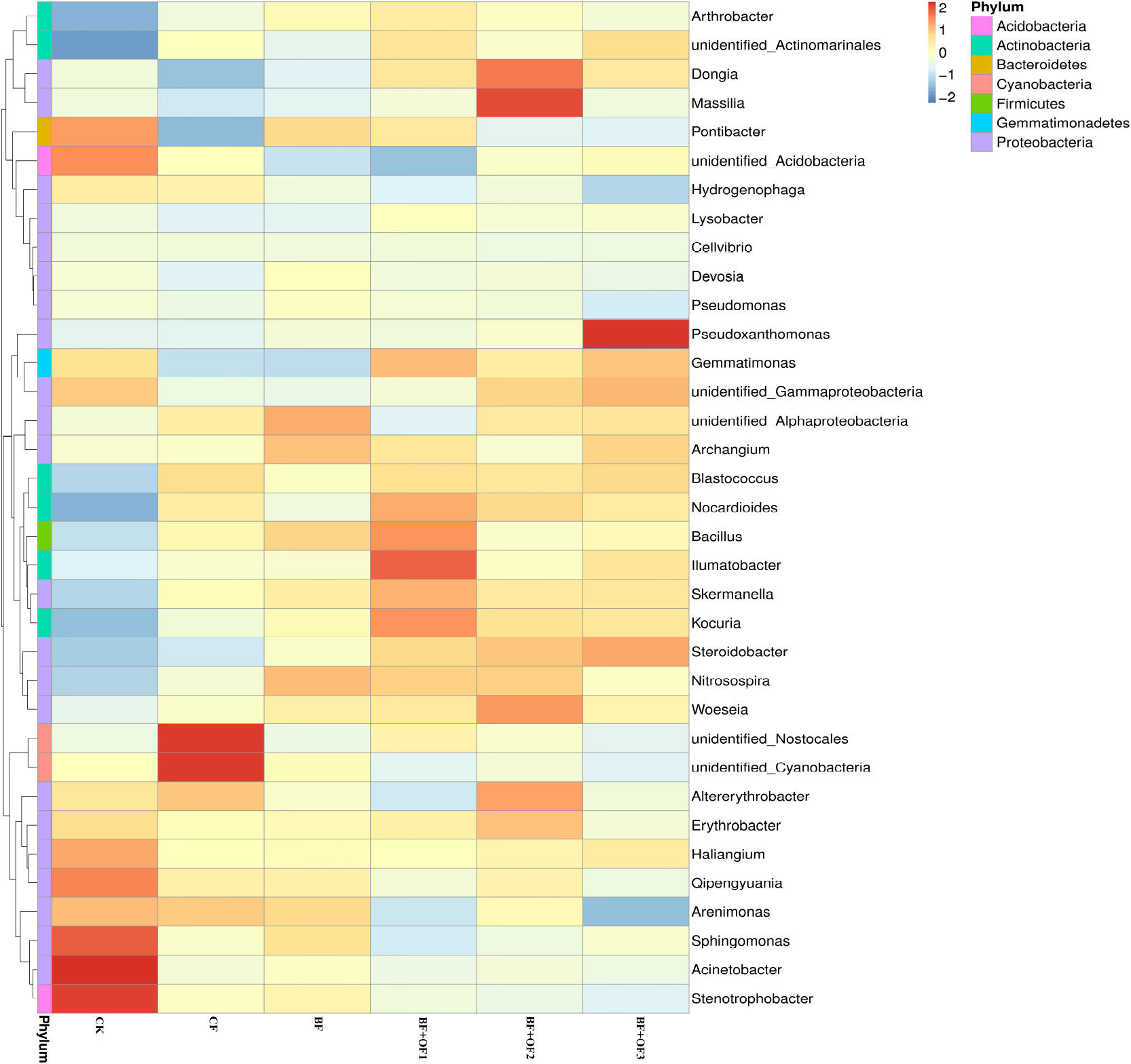
Figure 5. Heat map analysis of bacterial community in soil of different fertilization treatments. CK, no fertilizer, CF, chemical fertilizers (conventional dosage for this region); BF, balanced fertilizer (70% of total chemical fertilizer); BF + OF1, BF + OF2, and BF + OF3, balanced fertilizer plus 3,000, 6,000, and 12,000 kg.ha–1 bio-organic fertilizer, respectively. (Colors from blue to red represent that the relative abundance of the community from low to high).
Figure 6 shows that, principal components 1 and 2 explain 9.3 and 7.7% of the variance of variables, respectively, and the cumulative variance contribution rate is 17.0%. The samples were divided into two groups (Figure 6). The treatments with no fertilizer and only chemical fertilizer (CK, CF, and BF) clustered more closely together.
Figure 6. Principal component analysis (PCA) of soil under different fertilization treatments based on the operational taxonomic unit (OTU) level. CK, no fertilizer, CF: chemical fertilizers (conventional dosage for this region); BF, balanced fertilizer (70% of total chemical fertilizer); BF + OF1, BF + OF2, and BF + OF3, balanced fertilizer plus 3,000, 6,000, and 12,000 kg.ha–1 bio-organic fertilizer, respectively.
In contrast, BF + OF1, BF + OF2, and BF + OF3, in which chemical fertilizer reduction was based on balanced fertilization combined with biological organic fertilizers, formed one group. There was an obvious difference between the two groups, showing a good dispersion pattern.
Correlation analysis of different bacterial genera and soil physicochemical properties showed that SOM was positively correlated with the abundances of Blastococcus, Kocuria, Nocardioides, Steroidobacter, and Skermanella (Figure 7). Soil alkaline nitrogen, AK, and AP contents were positively correlated with the abundances of Kocuria, Nocardioides, Skermanella, Nitrosospira, Blastococcus, Lysobacter, and Bacillus. The abundances of these bacteria were significantly higher in the chemical fertilizer reduction combined with biological organic fertilization treatments (BF + OF1, BF + OF2, and BF + OF3) than in the conventional chemical fertilization (CF) treatment. In contrast, the abundances of Stenotrophobacter, Haliangium, Acinetobacter, Cellvibrio, and Sphingomonas was negatively correlated with TN, TP, SOM, and PH.
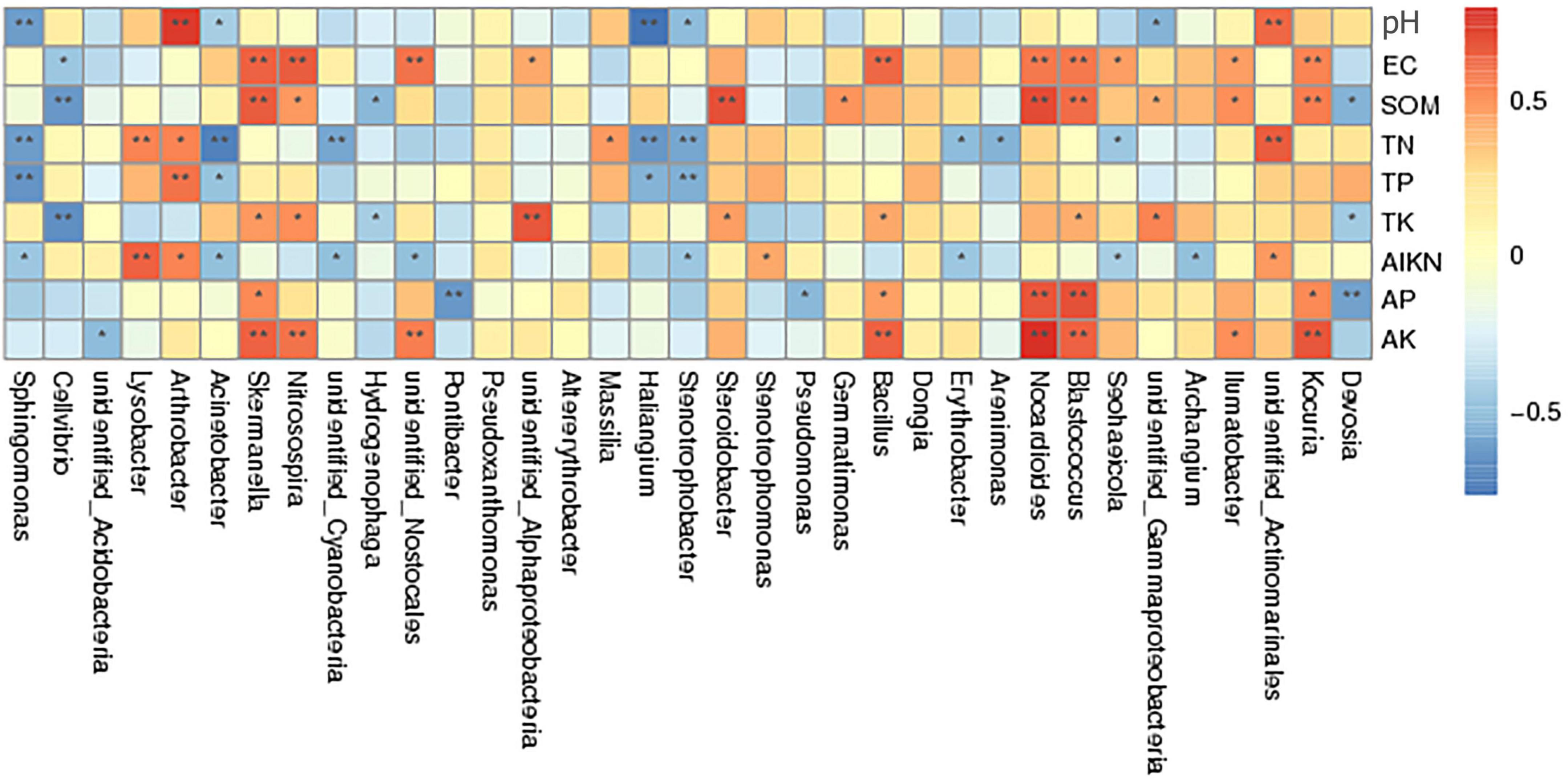
Figure 7. Spearman’s correlation analysis of soil bacterial community and soil physicochemical properties under different treatments. CK, no fertilizer; CF, chemical fertilizers (conventional dosage for this region); BF, balanced fertilizer (70% of total chemical fertilizer); BF + OF1, BF + OF2, and BF + OF3, balanced fertilizer plus 3,000, 6,000, and 12,000 kg.ha–1 bio-organic fertilizer, respectively. **Correlation is significant at the 0.01 level; *correlation is significant at the 0.05 level.
It is generally considered a practical and reasonable practice to apply both chemical and organic fertilizers to maintain the sustainable development of agricultural ecosystems (Gentile et al., 2008). In our study, we found that reducing chemical fertilizers in conjunction with bio-organic fertilizer use could improve SOM, nutrient availability, enzyme activity, bacterial community diversity and richness, and crop yield.
Changes in soil physicochemical properties were closely linked to application of organic fertilizers. The SOM, AN, AP, and AK in the soil increased after using the bio-organic fertilizer. Similarly, Baldi et al. (2016) reported that SOM and TN contents of orchard soils increased with organic fertilizer application. Zhao et al. (2016) found that the combined application of organic and inorganic fertilizers could improve the availability of soil nutrients and increase crop economic yields to a certain extent. Furthermore, Organic fertilizer can improve saline soil by improving soil aggregate structure, soil bulk density, soil permeability and alkalization (Li-ping et al., 2015). The soil pH and EC values of the BF + OF2 and BF + OF3 treatments were significantly lower than those of CF treatment, which is similar to the findings of Jiang et al. (2019). This indicated that adding appropriate amount of bio-organic fertilizer can maintain the stability of soil pH and prevent the partial acid or alkali of soil from affecting the growth and development of crops. According to Chang et al. (2007) there may be a reason why a high organic matter content can diminish the harmful effects of soluble salts on plant growth. Soil enzymes play an essential role in regulating soil nutrient turnover and are derived primarily from soil microorganisms (Yang et al., 2008). These enzymes are involved in the transformation of soil C, N, and P, such as sucrase, urease, alkaline phosphatase, and β-glucosidase. Using bio-organic fertilizer can improve soil enzyme activity and promote nutrient cycling, a finding that is in agreement with that published elsewhere (Zhao et al., 2016).
Adopting bio-organic fertilizers effectively improves soil environmental conditions and the soil microbiome (Tao et al., 2015). Therefore, soil microflora play a central role in decomposing loaded organic matter and cycling nutrients, particularly the most abundant group of bacteria, which are essential for ecological services (Zhang et al., 2012). Ding et al. (2016) reported that soil microbial diversity increased by combining organic and inorganic fertilizers, while the Shannon, Chao1, and Simpson indices of inorganic fertilizers with manure (MNPK) treatment significantly exceeded those of nitrogen, phosphorus and potassium inorganic fertilizer (NPK). We found that chemical fertilizer reduction combined with bio-organic fertilizers significantly increased soil bacterial diversity and richness compared to with conventional fertilization.
Different fertilization methods affect microbial community composition and structure. According to PCA analysis (Figure 5), the bacterial community composition were distinguished by different fertilization regimes. As confirmed by previous studies (Gu et al., 2019; Jiang et al., 2019; Brenzinger et al., 2021), the soil bacterial community structure changed after organic fertilizer application. In this study, the bio-organic fertilizer combination treatments increased the relative abundances of beneficial bacteria, such as Proteobacteria, Actinobacteria, Gemmatimonadetes, and Chloroflexi. Proteobacteria are thought to play an essential role in soil carbon, nitrogen and sulfur cycling (Rampelotto et al., 2013), and also have a strong resistance to heavily contaminated soils, especially under heavy metal stress (Burkhardt et al., 2011; Li et al., 2016). Actinobacteria play a critical role in the decomposition of organic matter and are highly resistant to UV radiation, heat, and desiccation (Essoussi et al., 2012; Hamedi et al., 2013). Gemmatimonadetes can promote the accumulation of P in the soil and root growth (Zhang et al., 2003). Chloroflexi can generate energy through photosynthesis and promote carbon conversion and utilization under conditions of low nutrient availability (Fullerton and Moyer, 2016). As with most fertilization experiments, these results are consistent. For example, the application of bio-organic fertilizer increased the abundances of Actinobacteria, Bacteroidetes, and Chloroflexi, and these bacteria suppress bacterial wilt in tomatoes (Zheng et al., 2020). With the addition of organic fertilizer, the microbial community composition was altered, leading to an increase in Proteobacteria, Bacteroidetes, and Gemmatimonadetes (Zhang J. et al., 2019). Higher relative abundances of Massilia, Dongia, Pseudoxanthomonas, Gemmatimonas, and Nitrospirae was observed at the genus level in the BF + OF1, BF + OF2, and BF + OF3 treatments compared with the CF treatment (Figure 4). The genus Massilia has been frequently reported as a rhizosphere and endorhizal colonizer. Furthermore, Massilia was reported to promote plant growth by producing auxin and siderophore, and by antagonizing the growth of Phytophthora infestans in vitro (Ofek et al., 2012). The genus Dongia is frequently reported as a beneficial microbe that suppresses the growth of soil-borne pathogens (Han et al., 2018). At present, the severity of soil and air pollution are increased by the extensive use of fertilizers and pesticides (Jianmin et al., 2007). Bioremediation is the most powerful and influential method of decontaminating polluted ecosystems. As confirmed by previous studies, the genus Pseudoxanthomonas functions in phenanthrene degradation and pesticide residue degradation (Patel et al., 2012; Talwar and Ninnekar, 2015). Wu et al. (2016) reported that Gemmatimonas was beneficial for bacteria wilt control. Nitrospirae can promote soil nutrient cycles and effectively reduce the salinity and alkalinity of soil to improve soil fertility (Zhang Y. et al., 2019; Xu et al., 2021). The increase of beneficial microorganisms in soil can promote plant growth and increase yield (Srinivasarao and Manjunath, 2017; Brenzinger et al., 2021). Moreover, Pseudomonas fluorescens can produce plant hormones and iron carriers (iron chelators) that promote growth, and also help to dissolve minerals in soil (Doni et al., 2019). In summary, chemical fertilizer reduction combined with bio-organic fertilizers enhances the structure and function of soil microorganism communities. As a result, beneficial bacteria are more abundant, which contributes to soil nutrient and yield improvement.
The soil environment plays a critical role in influencing the structure of microbial communities, as evidenced by numerous studies. For example, Jin et al. (2021) observed that AP, TN, SOM, EC, and pH were the primary drivers of microbial community structure, which is consistent with the results of this study. Redundancy analysis (RDA) revealed that organic matter (OM) and available phosphorus (AP) were the main factors driving the bacterial and fungal community structure (Ji et al., 2021). Zheng et al. (2022) results suggest a strong link between microbial community traits and SOC accumulation rate. Chemical fertilizer reduction combined with organic fertilizer and straw can effectively improve soil fertility and nutrient availability, altered the community structure of both bacteria and fungi (Shen et al., 2018; Tripathi et al., 2018; Wang et al., 2019). The redundancy analysis (RDA) results showed that the structure of the bacterial community was significantly correlated with SOM and TN, and the fungal community structure was strongly correlated with AP, SOM, and TN (Wu et al., 2021). Soil pH is one of the main factors affecting soil chemical, physical and biological properties (Buckman and Brady, 2002). The pH value in soil can be decreased due to inappropriate fertilization patterns through long-term fertilization experiments (Cai et al., 2015). Previous studies have proved that soil pH changes are closely related to soil microbial changes. The composition of soil bacterial community is significantly affected by soil pH (Shen et al., 2018; Tripathi et al., 2018). Xiong et al. (2012) believe that pH is the most decisive factor determining the structure of soil microbiomes. Soil pH is more important than nutrients in shaping bacterial communities in agricultural soils, including their ecological functions and biogeographic distribution (Wang et al., 2019). This may be because the fertilizer regime is one of the most influential determinants of soil environmental change and further affects the composition of the soil microbial community. Organic fertilization is the most efficient approach for increasing SOM content and improving soil fertility (Lin et al., 2019). Qiao et al. (2019) found that combined organic and bio-organic fertilizers provide more available nutrients, which is advantageous to crop growth and development. Bio-organic fertilizer application significantly affected the bacterial community in the cauliflower cropping soil. The relative abundances of Blastococcus, Kocuria, Nocardioides, Nitrospirae, and Skermanella bacteria were positively correlated with SOM content. The Blastococcus genus can degrade organic material through proteolysis (Duan et al., 2019). These results showed that the addition of microbial fertilizer could increase the content of organic matter and the amounts of beneficial bacteria in soil, then promote the absorption and utilization of crop nutrients. Kocuria abundance was significantly correlated with soil EC, which may be due to its higher salt tolerance during growth (Takarada et al., 2008). After the application of bio-organic fertilizer, the relative abundance of Bacillus was positively related to AK, owing to the ability of Bacillus to dissolve K (Tahir et al., 2018; Pramanik et al., 2019).
Chemical fertilizer reduction combined with appropriate bio-organic fertilizers (6,000 kg.ha–1 and 12,000 kg.ha–1) can increase cauliflower yield by improving soil fertility, promoting soil enzyme activity, altering microbial community composition, and stimulating potentially beneficial microbial functions. The fertilization pattern is adequate for the sustainable development of cauliflower production and soil environment protection in the northwest plateau region of China.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
XX and JLi designed the study and wrote the manuscript. XX, JLi, CG, HY, and ZF performed the work. JLi and LJ analyzed the data. JY, JLy, and GZ revised the manuscript and provided resources. All authors contributed to the article and approved the submitted version.
This research was supported by the Top Leading Talent Plan of Gansu Province (GSBJLJ-2021-14), the National Natural Science Foundation of China (No. 32160703), China Agriculture Research System (CARS-23-C-07), Research Program Sponsored by State Key Laboratory of Arid-land Crop Science, Gansu Agricultural University (GSCS-2020-12), and General Project of Scientific Research in Colleges and Universities in Gansu Province (2021B-141).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the graduated students from Gansu Agri-cultural University, for their help in managing the field experiment and collecting field data. We also wish to thank the referees for their constructive comments, which significantly improved the manuscript.
Aparna, K., Pasha, M. A., Rao, D. L. N., and Krishnaraj, P. U. (2014). Organic amendments as ecosystem engineers: microbial, biochemical and genomic evidence of soil health improvement in a tropical arid zone field site. Ecol. Eng. 71, 268–277. doi: 10.1016/j.ecoleng.2014.07.016
Apprill, A., Mcnally, S., Parsons, R., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137.
Baldi, E., Marcolini, G., Quartieri, M., Sorrenti, G., Muzzi, E., and Toselli, M. (2016). Organic fertilization in nectarine (Prunus persica var. nucipersica) orchard combines nutrient management and pollution impact. Nutr. Cycl. Agroecosyst. 105, 39–50. doi: 10.1007/s10705-016-9772-3
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Brenzinger, K., Costa, O., Ho, A., Koorneef, G., Robroek, B., Molenaar, D., et al. (2021). Steering microbiomes by organic amendments towards climate-smart agricultural soils. Biol. Fertil. Soils 57, 1053–1074.
Burkhardt, E. M., Bischoff, S., Akob, D. M., Buchel, G., and Kusel, K. (2011). Heavy metal tolerance of Fe(III)-reducing microbial communities in contaminated creek bank soils. Appl. Environ. Microbiol. 77, 3132–3136. doi: 10.1128/AEM.02085-10
Cai, Z., Wang, B., Xu, M., Zhang, H., He, X., Zhang, L., et al. (2015). Intensified soil acidification from chemical N fertilization and prevention by manure in an 18-year field experiment in the red soil of southern China. J. Soil Sediments 15, 260–270.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336.
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chang, E.-H., Chung, R.-S., and Tsai, Y.-H. (2007). Effect of different application rates of organic fertilizer on soil enzyme activity and microbial population. Soil Sci. Plant Nutr. 53, 132–140. doi: 10.1111/j.1747-0765.2007.00122.x
Dawson, C. J., and Hilton, J. (2012). Fertiliser availability in a resource-limited world: production and recycling of nitrogen and phosphorus. Food Policy 36, S14–S22.
Ding, J., Jiang, X., Ma, M., Zhou, B., Guan, D., Zhao, B., et al. (2016). Effect of 35 years inorganic fertilizer and manure amendment on structure of bacterial and archaeal communities in black soil of northeast China. Appl. Soil Ecol. 105, 187–195. doi: 10.1016/j.apsoil.2016.04.010
Doni, F., Mispan, M. S., Suhaimi, N., Ishak, N., and Uphoff, N. (2019). Roles of microbes in supporting sustainable rice production using the system of rice intensification. Appl. Microbiol. Biotechnol. 103, 5131–5142. doi: 10.1007/s00253-019-09879-9
Duan, Y., Awasthi, S. K., Liu, T., Verma, S., Wang, Q., Chen, H., et al. (2019). Positive impact of biochar alone and combined with bacterial consortium amendment on improvement of bacterial community during cow manure composting. Bioresour. Technol. 280, 79–87. doi: 10.1016/j.biortech.2019.02.026
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10:996. doi: 10.1038/nmeth.2604
Essoussi, I., Boujmil, R., Nouioui, I., Abbassi-Ghozzi, I., Hamza, A., Boudabous, A., et al. (2012). Genetic Diversity and Esterase-Profiling of Actinobacteria Isolated from Sahara Desert Stones and Monuments. Geomicrobiol. J. 29, 23–28. doi: 10.1080/01490451.2010.521367
Fullerton, H., and Moyer, C. L. (2016). Comparative Single-Cell Genomics of Chloroflexi from the Okinawa Trough Deep-Subsurface Biosphere. Appl. Environ. Microbiol. 82, 3000–3008. doi: 10.1128/AEM.00624-16
Gai, X., Liu, H., Zhai, L., Tan, G., Liu, J., Ren, T., et al. (2016). Vegetable yields and soil biochemical properties as influenced by fertilization in Southern China. Appl. Soil Ecol. 107, 170–181. doi: 10.1016/j.apsoil.2016.06.001
Gentile, R., Vanlauwe, B., Chivenge, P., and Six, J. (2008). Interactive effects from combining fertilizer and organic residue inputs on nitrogen transformations. Soil Biol. Biochem. 40, 2375–2384. doi: 10.1016/j.soilbio.2008.05.018
Gu, S., Hu, Q., Cheng, Y., Bai, L., Liu, Z., Xiao, W., et al. (2019). Application of organic fertilizer improves microbial community diversity and alters microbial network structure in tea (Camellia sinensis) plantation soils. Soil Tillage Res. 195:104356. doi: 10.1016/j.still.2019.104356
Guan, S. Y., Zhang, D., and Zhang, Z. (1986). Soil Enzyme and its Research Methods. Beijing: Agriculteral Press.
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hamedi, J., Mohammadipanah, F., and Ventosa, A. (2013). Systematic and biotechnological aspects of halophilic and halotolerant actinomycetes. Extremophiles 17, 1–13. doi: 10.1007/s00792-012-0493-5
Han, L., Wang, Z., Li, N., Wang, Y., Feng, J., and Zhang, X. (2018). Bacillus amyloliquefaciens B1408 suppresses Fusarium wilt in cucumber by regulating the rhizosphere microbial community. Appl. Soil Ecol. 136, 55–66.
Ji, L., Si, H., He, J., Fan, L., and Li, L. (2021). The shifts of maize soil microbial community and networks are related to soil properties under different organic fertilizers. Rhizosphere 19:100388.
Jiang, S. Q., Yu, Y. N., Gao, R. W., Wang, H., Zhang, J., Li, R., et al. (2019). High-throughput absolute quantification sequencing reveals the effect of different fertilizer applications on bacterial community in a tomato cultivated coastal saline soil. Sci. Total Environ. 687, 601–609. doi: 10.1016/j.scitotenv.2019.06.105
Jianmin, M. A., Yi-Fan, L. I., Harner, T., and Cao, Z. (2007). Assessing the Relationship between Extensive Use of Organochlorine Pesticides and Cooling Trend during the Mid-20th Century in the Southeastern United States. Environ. Sci. Technol. 41, 7209–7214. doi: 10.1021/es0617744
Jin, L., Yu, J., Jin, N., Xie, J., and Jian, L. (2021). Effects of different vegetable rotations on the rhizosphere bacterial community and tomato growth in a continuous tomato cropping substrate. PLoS One 16:e0257432. doi: 10.1371/journal.pone.0257432
Li, X., Zhang, W., Liu, T., Chen, L., Chen, P., and Li, F. (2016). Changes in the composition and diversity of microbial communities during anaerobic nitrate reduction and Fe(II) oxidation at circumneutral pH in paddy soil. Soil Biol. Biochem. 94, 70–79. doi: 10.1016/j.soilbio.2015.11.013
Lin, X., Feng, Y., Zhang, H., Chen, R., Wang, J., Zhang, J., et al. (2012). Long-Term Balanced Fertilization Decreases Arbuscular Mycorrhizal Fungal Diversity in an Arable Soil in North China Revealed by 454 Pyrosequencing. Environ. Sci. Technol. 46, 5764–5771. doi: 10.1021/es3001695
Lin, Y., Ye, G., Kuzyakov, Y., Liu, D., Fan, J., and Ding, W. (2019). Long-term manure application increases soil organic matter and aggregation, and alters microbial community structure and keystone taxa. Soil Biol. Biochem. 134, 187–196. doi: 10.1016/j.soilbio.2019.03.030
Lin, Z., Zhen, Z., Wu, Z., Yang, J., Zhong, L., Hu, H., et al. (2016). The impact on the soil microbial community and enzyme activity of two earthworm species during the bioremediation of pentachlorophenol-contaminated soils. J. Hazard Mater. 301, 35–45. doi: 10.1016/j.jhazmat.2015.08.034
Li-ping, L., Xiao-hua, L., Hong-bo, S., Zhao-Pu, L., Ya, T., Quan-suo, Z., et al. (2015). Ameliorants improve saline–alkaline soils on a large scale in northern Jiangsu Province. China. Ecol. Eng. 81, 328–334. doi: 10.1016/j.ecoleng.2015.04.032
Lu, K. (1999). Analytical Methods of Soil and Agricultural Chemistry. Beijing: China Agricultural Science and Technology Press.
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Naher, U. A., Biswas, J. C., Maniruzzaman, M., Khan, F. H., and Kabir, M. S. (2021). Bio-Organic Fertilizer: a Green Technology to Reduce Synthetic N and P Fertilizer for Rice Production. Front. Plant Sci. 12:602052. doi: 10.3389/fpls.2021.602052
Ofek, M., Hadar, Y., and Minz, D. (2012). Ecology of root colonizing Massilia (Oxalobacteraceae). PLoS One 7:e40117. doi: 10.1371/journal.pone.0040117
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Patel, V., Cheturvedula, S., and Madamwar, D. (2012). Phenanthrene degradation by Pseudoxanthomonas sp. DMVP2 isolated from hydrocarbon contaminated sediment of Amlakhadi canal, Gujarat, India. J. Hazard. Mater. 20, 43–51. doi: 10.1016/j.jhazmat.2011.11.002
Paul, E. A. (2016). The nature and dynamics of soil organic matter: plant inputs, microbial transformations, and organic matter stabilization. Soil Biol. Biochem. 98, 109–126. doi: 10.1016/j.soilbio.2016.04.001
Paz-Ferreiro, J., Fu, S., Mendez, A., and Gasco, G. (2014). Interactive effects of biochar and the earthworm Pontoscolex corethrurus on plant productivity and soil enzyme activities. J. Soils Sediments 14, 483–494.
Pramanik, P., Goswami, A. J., Ghosh, S., and Kalita, C. (2019). An indigenous strain of potassium-solubilizing bacteria Bacillus pseudomycoides enhanced potassium uptake in tea plants by increasing potassium availability in the mica waste-treated soil of North-east India. J. Appl. Microbiol. 126, 215–222. doi: 10.1111/jam.14130
Qiao, C., Penton, C. R., Xiong, W., Liu, C., Wang, R., Liu, Z., et al. (2019). Reshaping the rhizosphere microbiome by bio-organic amendment to enhance crop yield in a maize-cabbage rotation system. Appl. Soil Ecol. 142, 136–146. doi: 10.1016/j.apsoil.2019.04.014
Rampelotto, P. H., de Siqueira Ferreira, A., Barboza, A. D., and Roesch, L. F. (2013). Changes in diversity, abundance, and structure of soil bacterial communities in Brazilian Savanna under different land use systems. Microb. Ecol. 66, 593–607. doi: 10.1007/s00248-013-0235-y
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahe, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Shen, C., Shi, Y., Fan, K., Jin-Sheng, H., Adams, J. M., Yuan, G., et al. (2018). Soil pH dominates elevational diversity pattern for bacteria in high elevation alkaline soils on the Tibetan Plateau. FEMS Microbiol. Ecol. 95:fiz003. doi: 10.1093/femsec/fiz003
Singh, S. R., Kundu, D. K., Tripathi, M. K., Dey, P., Saha, A. R., Kumar, M., et al. (2015). Impact of balanced fertilization on nutrient acquisition, fibre yield of jute and soil quality in New Gangetic alluvial soils of India. Appl. Soil Ecol. 92, 24–34. doi: 10.1016/j.apsoil.2015.03.007
Souza, C. Jr., Ribeiro, V., Mendes, M., and Soares, P. H. (2020). Enhanced-Efficiency Phosphorous Fertilizer Impacts on Corn and Common Bean Crops and Soil Phosphorus Diffusion. J. Agric. Sci. 12:15.
Srinivasarao, C., and Manjunath, M. (2017). “Potential of Beneficial Bacteria as Eco-friendly Options for Chemical-Free Alternative Agriculture,” in Plant-Microbe Interactions in Agro-Ecological Perspectives, eds D. Singh, H. Singh, and R. Prabha (Singapore: Springer), 473–493.
Tahir, M., Khalid, U., Ijaz, M., Shah, G. M., Naeem, M. A., Shahid, M., et al. (2018). Combined application of bio-organic phosphate and phosphorus solubilizing bacteria (Bacillus strain MWT 14) improve the performance of bread wheat with low fertilizer input under an arid climate. Braz. J. Microbiol. 49, 15–24. doi: 10.1016/j.bjm.2017.11.005
Takarada, H., Sekine, M., Kosugi, H., Matsuo, Y., Fujisawa, T., Omata, S., et al. (2008). Complete genome sequence of the soil actinomycete Kocuria rhizophila. J. Bacteriol. 190, 4139–4146. doi: 10.1128/JB.01853-07
Talwar, M. P., and Ninnekar, H. Z. (2015). Biodegradation of pesticide profenofos by the free and immobilized cells of Pseudoxanthomonas suwonensis strain HNM. J. Basic Microbiol. 55, 1094–1103. doi: 10.1002/jobm.201400978
Tao, R., Liang, Y., Wakelin, S. A., and Chu, G. (2015). Supplementing chemical fertilizer with an organic component increases soil biological function and quality. Appl. Soil Ecol. 96, 42–51. doi: 10.1016/j.apsoil.2015.07.009
Tripathi, B. M., Stegen, J. C., Kim, M., Dong, K., Adams, J. M., and Lee, Y. K. (2018). Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 12, 1072–1083.
Wang, C. Y., Zhou, X., Guo, D., Zhao, J. H., and Zhao, L. P. (2019). Soil pH is the primary factor driving the distribution and function of microorganisms in farmland soils in northeastern China. Ann. Microbiol. 69, 1461–1473.
Wang, H., Hu, R., Chen, X., Zhong, X., Zheng, Z., Huang, N., et al. (2017). Reduction in nitrogen fertilizer use results in increased rice yields and improved environmental protection. Int. J. Agric. Sustain. 15, 681–692. doi: 10.1016/j.scitotenv.2018.08.392
Wang, J. L., Liu, K. L., Zhao, X. Q., Zhang, H. Q., Li, D., Li, J. J., et al. (2021). Balanced fertilization over four decades has sustained soil microbial communities and improved soil fertility and rice productivity in red paddy soil. Sci. Total Environ. 793:148664. doi: 10.1016/j.scitotenv.2021.148664
Wang, Q. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5264–5267. doi: 10.1128/AEM.00062-07
Wesenbeeck, C., Keyzer, M. A., Veen, W., and Qiu, H. (2021). Can China’s overuse of fertilizer be reduced without threatening food security and farm incomes? Agric. Syst. 190:103093.
Wu, B., Wang, X., Yang, L., Yang, H., Zeng, H., Qiu, Y., et al. (2016). Effects of Bacillus amyloliquefaciens ZM9 on bacterial wilt and rhizosphere microbial communities of tobacco. Appl. Soil Ecol. 103, 1–12. doi: 10.1016/j.apsoil.2016.03.002
Wu, X., Zhang, T., Zhao, J., Wang, L., Yang, D., Li, G., et al. (2021). Variation of soil bacterial and fungal communities from fluvo-aquic soil under chemical fertilizer reduction combined with organic materials in north China plain. J. Soil Sci. Plant Nutr. 21, 349–363.
Xiong, J., Liu, Y., Lin, X., Zhang, H., Zeng, J., Hou, J., et al. (2012). Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau. Environ. Microbiol. 14, 2457–2466. doi: 10.1111/j.1462-2920.2012.02799.x
Xu, H., Yu, M., and Cheng, X. (2021). Abundant fungal and rare bacterial taxa jointly reveal soil nutrient cycling and multifunctionality in uneven-aged mixed plantations. Ecol. Indic. 129:107932.
Yang, L., Li, T., Li, F., Lemcoff, J. H., and Cohen, S. (2008). Fertilization regulates soil enzymatic activity and fertility dynamics in a cucumber field. Entia Hortic. 116, 21–26.
Yang, P., Lyu, J., Sohail, H., Yu, J., and Li, J. (2020). Partial substitution of mineral fertilizer with biofertilizer enhances cauliflower nutritional quality, yield, and soil characteristics. Crop Sci. 60, 934–944.
Yin, J., Yuan, L., and Huang, J. (2021). New Functions of Ceriporia lacerata HG2011: mobilization of Soil Nitrogen and Phosphorus and Enhancement of Yield and Quality of Ketchup-Processing Tomato. J. Agric. Food Chem. 69, 4056–4063. doi: 10.1021/acs.jafc.0c06783
Yu, Y., Wu, M., Petropoulos, E., Zhang, J., Nie, J., Liao, Y., et al. (2019). Responses of paddy soil bacterial community assembly to different long-term fertilizations in southeast China. Sci. Total Environ. 656, 625–633. doi: 10.1016/j.scitotenv.2018.11.359
Zhang, H., Sekiguchi, Y., Hanada, S., Hugenholtz, P., Kim, H., Kamagata, Y., et al. (2003). Gemmatimonas aurantiaca gen. nov., sp. nov., a gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int. J. Syst. Evol. Microbiol. 53, 1155–1163. doi: 10.1099/ijs.0.02520-0
Zhang, J., Bei, S., Li, B., Zhang, J., Christie, P., and Li, X. (2019). Organic fertilizer, but not heavy liming, enhances banana biomass, increases soil organic carbon and modifies soil microbiota. Appl. Soil Ecol. 136, 67–79. doi: 10.1016/j.apsoil.2018.12.017
Zhang, Y., Li, Q., Chen, Y., Dai, Q., and Hu, J. (2019). Dynamic Change in Enzyme Activity and Bacterial Community with long-term rice Cultivation in Mudflats. Curr. Microbiol. 76, 361–369. doi: 10.1007/s00284-019-01636-5
Zhang, Q. C., Shamsi, I. H., Xu, D. T., Wang, G. H., Lin, X. Y., Jilani, G., et al. (2012). Chemical fertilizer and organic manure inputs in soil exhibit a vice versa pattern of microbial community structure. Appl. Soil Ecol. 57, 1–8.
Zhao, J., Ni, T., Li, J., Lu, Q., Fang, Z., Huang, Q., et al. (2016). Effects of organic–inorganic compound fertilizer with reduced chemical fertilizer application on crop yields, soil biological activity and bacterial community structure in a rice–wheat cropping system. Appl. Soil Ecol. 99, 1–12. doi: 10.1016/j.apsoil.2015.11.006
Zheng, F., Wu, X., Zhang, M., Liu, X., Song, X., Lu, J., et al. (2022). Linking soil microbial community traits and organic carbon accumulation rate under long-term conservation tillage practices. Soil Tillage Res. 220:105360.
Zheng, X., Zhu, Y., Wang, Z., Zhang, H., Chen, M., Chen, Y., et al. (2020). Effects of a novel bio-organic fertilizer on the composition of rhizobacterial communities and bacterial wilt outbreak in a continuously mono-cropped tomato field. Appl. Soil Ecol. 156:103717. doi: 10.1016/j.apsoil.2020.103717
Keywords: yield, soil property, bacterial community, bio-organic fertilizer, cauliflower
Citation: Xiao X, Li J, Lyu J, Feng Z, Zhang G, Yang H, Gao C, Jin L and Yu J (2022) Chemical fertilizer reduction combined with bio-organic fertilizers increases cauliflower yield via regulation of soil biochemical properties and bacterial communities in Northwest China. Front. Microbiol. 13:922149. doi: 10.3389/fmicb.2022.922149
Received: 17 April 2022; Accepted: 08 July 2022;
Published: 27 July 2022.
Edited by:
Chee Kong Yap, Putra Malaysia University, MalaysiaReviewed by:
Touqeer Abbas, University of Minnesota Twin Cities, United StatesCopyright © 2022 Xiao, Li, Lyu, Feng, Zhang, Yang, Gao, Jin and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jihua Yu, eXVqaWh1YUBnc2F1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.