- 1Guangxi Key Laboratory of Agro-Environment and Agro-Product Safety, College of Agriculture, Guangxi University, Nanning, China
- 2Department of Plant Sciences and Plant Pathology, Montana State University, Bozeman, MT, United States
- 3College of Plant Protection, China Agricultural University, Beijing, China
Pseudomonas fluorescens 2P24 is a plant root-associated bacterium that suppresses several soilborne plant diseases due to its production of the antibiotic 2,4-diacetylphloroglucinol (2,4-DAPG). The biosynthesis of 2,4-DAPG is controlled by many regulatory elements, including the global regulator of the Gac/Rsm regulon and the pathway-specific repressor PhlF. In this work, a novel genetic element grxD, which encodes the monothiol glutaredoxin GrxD, was identified and characterized in the production of 2,4-DAPG in P. fluorescens 2P24. Our data showed that the mutation of grxD remarkably decreased 2,4-DAPG production. GrxD lost its ability to alter the production of 2,4-DAPG when the active-site CGFS motif of GrxD was mutated by site-directed mutagenesis. Further studies showed that the RsmA and RsmE proteins were essential for the GrxD-mediated regulation of 2,4-DAPG and exoprotease production. In addition, our data revealed that the deletion of grxD increased the expression of phlF, which negatively regulated the production of 2,4-DAPG. In addition, the grxD mutant was severely impaired in the biocontrol effect against the bacterial wilt of tomato. Overall, our results indicated that the monothiol glutaredoxin GrxD is involved in the production of 2,4-DAPG of P. fluorescens by influencing the Gac/Rsm global signaling pathway and transcriptional regulator PhlF and is essential for the biocontrol properties.
Introduction
Certain strains of Pseudomonas, known as plant growth-promoting rhizobacteria (PGPR), can competitively colonize plant roots, enhance crop growth, and produce antibiotics to control plant diseases (Haas and Défago, 2005). Efficient root colonization by these rhizobacteria is an essential prerequisite for the biocontrol of root pathogens (Benizri et al., 2001). However, microbial colonization by PGPR is a highly complex process and is involved in the interactions among plants, rhizobacteria, and environmental factors. During the early phases of root colonization, some PGPR penetrates the intercellular spaces of host plants, which trigger microbe-associated molecular patterns similar to those of pathogenetic fungi (Pršiæ and Ongena, 2020). This recognition of host cells then activates an oxidative burst (or respiratory burst), leading to the release of reactive oxygen species (ROS) (Segonzac and Zipfel, 2011). Besides the plant-derived ROS, rhizobacteria must cope with the self-made ROS produced as a byproduct of aerobic respiration or environmental cues (Nanda et al., 2010). To counteract the damages caused by ROS, rhizobacteria possess multiple antioxidant mechanisms, such as glutathione (GSH)- and thioredoxin (TRX)-dependent systems, catalases, and superoxide dismutases (Forman et al., 2009; Heck et al., 2010).
The GSH-dependent system, which is also called the glutaredoxin (GRX) system, consists of GRX, GSH, NADPH, and GSH reductase, and controls the cellular redox state of a myriad of proteins together with thioredoxins (Fernandes and Holmgren, 2004). GRXs represent a group of ubiquitous oxidoreductases of the thioredoxin superfamily and are widely conserved in most eukaryotes and prokaryotes. These proteins preferentially reduce the disulfide bonds formed between the cysteine residues of glutathione and its target proteins. GRX family proteins are required for many biological processes, including iron–sulfur cluster (Fe–S) biogenesis, iron homeostasis, symbiotic capacity, cell survival, and production of virulence factors (Meyer et al., 2009; Poor et al., 2014).
Many GRX isoforms have been identified and characterized by different bacteria. On the basis of their structures and catalytic properties, GRXs are divided into two categories: dithiol (C [PG/FY] C motif) and monothiol (CGFS motif) GRXs. The dithiol GRX is discovered in Escherichia coli as an alternative donor to TRXs for ribonucleotide reductase (Holmgren, 1976). Dithiol GRXs Grx1 and Grx3 are small proteins (∼10 kDa) with the CPYC motif as their active site. Grx1 is reported to influence the intracellular disulfides of ribonucleotide reductase Ia and 3′-phophoasenylyl sulfate, whereas Grx3 has only 5% of the catalytic activity of Grx1 for ribonucleotide reductase Ia (Lilling et al., 1999). By contrast, monothiol GRXs have the CGFS motif as their active site and are proposed to function in the Fe–S cluster storage and delivery (Couturier et al., 2015).
Although GRXs have been investigated extensively for many years, knowledge about their functions in the biocontrol ability of PGPR remains limited. Pseudomonas fluorescens 2P24 is a root-associated beneficial bacterium from the rhizosphere of wheat. Strain 2P24 can efficiently colonize wheat roots and exhibits biocontrol activity against phytopathogens, such as Rhizoctonia solani, Ralstonia solanacearum, and Gaeumannomyces graminis var. tritici (Wei et al., 2004a). This biocontrol capacity relies on the production of secondary metabolite 2,4-diacetylphloroglucinol (2,4-DAPG) (Wei et al., 2004b). The biosynthetic cluster for 2,4-DAPG has been identified and characterized in strain 2P24 and other biocontrol pseudomonads (Bangera and Thomashow, 1999; Wu et al., 2010). The 2,4-DAPG biosynthetic gene cluster comprises phlACBD genes. phlA and phlF (encoding a pathway-specific regulator) genes are divergently transcribed (Abbs et al., 2002). The production of 2,4-DAPG by P. fluorescens is a dynamic process regulated by many regulatory elements. The Gac/Rsm signaling cascade is necessary for the production of 2,4-DAPG (Zhang et al., 2020a). This signaling cascade is initiated by the GacS/GacA two-component system. Upon stimulation, the GacS sensor kinase autophosphorylates and then activates the response regulator GacA by phosphotransfer, which directly regulates the expression of the non-coding small RNA (sRNA) genes, i.e., rsmX1, rsmX, rsmY, and rsmZ, in strain 2P24 (Heeb and Haas, 2001; Zhang et al., 2020a). These sRNAs can sequester CsrA family proteins RsmA and RsmE, which can repress the translation of phlA mRNA (Zhang et al., 2020b). Interestingly, GacA negatively controls the transcription of the sRNA gene rgsA, which negatively influences the production of 2,4-DAPG (Zhang et al., 2020a).
In this study, we screen the mutant libraries of P. fluorescens to identify genetic factors that influence the production of 2,4-DAPG. Our data indicate that the mutation of the cytosolic monothiol glutaredoxin-encoding gene grxD remarkably decreases the production of 2,4-DAPG. Further investigations reveal that the protein levels of RsmA/RsmE and PhlF are significantly increased in the grxD mutant. The mutation of phlF, rsmA, or rsmE genes in the grxD mutant background can improve the production of 2,4-DAPG. Furthermore, GrxD regulated the production of 2,4-DAPG and exoprotease by influencing the protein levels of RsmA and RsmE. Overall, this work reveals the essential role of grxD in redox homeostasis, siderophores, and biocontrol ability in P. fluorescens.
Materials and Methods
Strains and Culture Conditions
Bacterial strains and plasmids used in this work are listed in Supplementary Table 1. Pseudomonas fluorescens and E. coli strains were routinely grown in the Lysogeny Broth (LB) medium, King’s B medium (KB) (King et al., 1954), or ABM medium (Chilton et al., 1974) for general genetic procedures. Rhizoctonia solani A01 was routinely cultured in potato dextrose agar (PDA) medium. When required, antibiotics were used at final concentrations as follows: ampicillin (50 μg/ml), kanamycin (50 μg/ml), gentamicin (5 μg/ml), and tetracycline (20 μg/ml).
Tn5 Transposon Mutagenesis
The Tn5-transposon mutagenesis was performed by conjugating a mini-Tn5 suicide plasmid pUT-Km into P. fluorescens 2P24, as previously described (Wu et al., 2010). Approximately 5,000 mutants were screened for attenuating the antifungal activity against R. solani A01. Mutants were grown in 96-well microtiter plates containing KB broth. Overnight cultures (2 μl) were then transferred to the OmniTray plate containing PDA with a suspension of blended fungal hyphae of R. solani A01. Mutants in which the antifungal activity was significantly decreased compared with the parental strain were selected, and the Tn5 insertion site was determined as previously described (Wu et al., 2010).
Construction of Deletion Mutants and Complementary Strains
The grxC, grxD, grxF, and grxG genes were deleted by allelic replacement according to the method of Zhang et al. (2018). In-frame deletion plasmids in grxC, grxD, grxF, and grxG were created by overlapping PCR of the flanking regions of the target genes (primers are listed in Supplementary Table 2). The resulting DNA fragments were then cloned into the suicide vector p2P24Km (Zhang et al., 2018). The constructed plasmids p2P24Km-grxC, p2P24Km-grxD, p2P24Km-grxF, and p2P24Km-grxG were introduced into P. fluorescens 2P24 or its derivatives by the electroporation, and colonies with kanamycin sensitivity and sucrose resistance were selected and confirmed by PCR and verified by Sanger sequencing.
To generate complemented strains, the promoter and open reading frame regions of target genes were cloned into the broad-host range expression plasmid pRK415 (Keen et al., 1988). The resulting plasmids were confirmed by sequencing and introduced into P. fluorescens cells by electroporation. The successful construction was confirmed by sequencing and electroporation into P. fluorescens cells.
Site-Directed Mutagenesis
Site-directed mutagenesis was accomplished using the site-directed mutagenesis Kit (Sangon Biotech, Shanghai, China) with the plasmid p415-grxD to substitute critical residue in the CGFS motif (from CGFS to SGFS) of GrxD. The substitution was confirmed by DNA sequencing.
Antifungal Activity of Pseudomonas fluorescens
Antifungal activity of 2P24 and its derivatives were determined on the PDA agar plate against R. solani A01. A 0.6-cm R. solani A01 plug was inoculated in the center of the PDA agar plate. In total, 5 μl of bacterial cultures grown overnight were then spotted on the plate at a distance of 2.5 cm from the fungal colony. The plates were then incubated at 30°C for 36 h, and the growth inhibition of R. solani was assessed.
Extraction and Detection of 2,4-DAPG and Phloroglucinol
The production of 2,4-DAPG by P. fluorescens 2P24 and its derivatives was measured after being cultured in 30 ml KB medium at 30°C for 24 h. 2,4-DAPG was analyzed in ethyl acetate extract of the culture supernatants using high-performance liquid chromatography (HPLC) as described by Shanahan et al. (1992). A direct colorimetric assay was performed for quantification of the concentration of PG using 0.2% 4-hydroxy-3-methoxycinnamaldehyde solution in ethanol/HCL (v/v 3:1). The optical density of the reaction products was then measured at 550 nm (Kidarsa et al., 2011).
Phenotypic Assays
The swimming and swarming motilities of P. fluorescens 2P24 were tested on an LB plate as described previously (Ha et al., 2014). Overnight bacterial cultures were inoculated and adjusted to the same bacterial density. In total, 5 μl aliquots were then inoculated onto the center of the swimming and swarming plates, respectively. The diameter of swim zones was measured after 24 h at 30°C.
The production of siderophore was determined by chrome azurol S (CAS) assay (Schwyn and Neilands, 1987). In total, 5 μl of overnight bacterial cultures were spotted on the center of the CAS plate. The production of siderophore was determined by measuring the diameter of an orange zone after incubation at 30°C for 24 h.
To visualize the production of exoprotease production of strain 2P24 and its derivatives, 5 μL drops of overnight bacterial cultures were inoculated onto a 2.5% skimmed milk agar plate. A visible halo appeared on the plate was measured after 24 h of incubation at 30°C.
For quantification of iron concentration, overnight bacterial cultures were collected and resuspended with 1 ml H2O and 0.5 ml fresh iron-releasing buffer containing 0.142 M KMnO4 and 0.6 M HCl. The mixtures were incubated at 60°C for 2 h and then added with 100 μl iron-reaction buffer containing 13.1 mM neocuproine, 6.5 mM ferrozine, 2 M ascorbic acid, and 5 M ammonium acetate at 25°C for 0.5 h. The subsequent cultures were centrifuged, and the absorbance was determined at 562 nm. The iron concentration was calculated using a FeCl3 standard curve.
To assess survival in response to oxidative stress, P. fluorescens 2P24 and its derivatives were grown overnight in LB broth, and cells were collected and re-suspended in distilled H2O to obtain an OD at 600 nm of 1.0. Suspended cells were treated with 0.6 mM H2O2 and 5 mM CuOOH and incubated at 30°C for 1 h. Cell cultivability was assessed by plating serial dilutions of the cultures on ABM plates.
The P. fluorescens 2P24 and its derivatives were grown overnight in the LB broth and were adjusted to an optical density at 600 nm of 1.0. In total, 20 μl of the bacterial cultures were then inoculated into 20 ml ABM medium in Erlenmeyer flasks at 30°C with shaking at 180 rpm. The growth of bacteria was indicated by optical density at 600 nm.
Western Blot Analyses
The P. fluorescens 2P24 and its derivatives were grown in LB at 30°C for 18 or 36 h and a 1 ml sample was taken. Cells were then suspended in phosphate-buffered saline (PBS) buffer and lysed by sonication. Samples containing 30 μg of total protein were separated by a 12% (vol/vol) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blot analysis using an anti-Flag antibody (1:2,000) (Sangon) and anti-RNA polymerase monoclonal antibody (1:2,000) (Neoclone), respectively. The resulting blots were developed using the chemiluminescence detection kit (Thermo Fisher, Waltham, MA, United States).
β-Galactosidase Activity Assay
Pseudomonas fluorescens cells were grown in BL broth at 30°C with shaking at 200 rpm. Cells were collected at different points of the growth curve. β-Galactosidase activities were then determined using the Miller method (Miller, 1972).
Plant Disease Suppression Assay
For the plant assays, the surface sterilized tomato seeds were cultured on the Murashige and Skoog (MS) medium with vitamins in 1-L sterile vials. After germination, tomato seedlings were maintained under sterile conditions up to the four-leaf stage. Plants were then uprooted carefully, and the root tips of the plants were cut off. Injured roots were immersed in the culture suspension of R. solanacearum GX02 (concentration 1 × 107 CFU per milliliter). One hour after infection, tomato seedlings were planted in separate flasks containing sterile soil. Soil inoculation was performed by mixing with sterile water or P. fluorescens strains at concentrations of 1 × 107 CFU per gram of soil, respectively. At 7 or 14 days after inoculation, plant survival was determined. The disease index scale from 0 to 4 was determined as described previously (Liang et al., 2020): 0 – no wilt symptoms; 1 – wilt symptoms on 1–25% of the leaves; 2 – wilt symptoms on 26–50% of the leaves; 3 –wilt symptoms on 51–75% of the leaves; and 4 – wilt symptoms on more than 76% of the leaves.
Statistical Analysis
The signification of differences was assessed with Student’s t-test. Asterisks or different letters indicated p-values (p < 0.05), and results were presented as the mean ± standard deviation. Each experiment was performed three times with similar results.
Results
Identification of the grxD Gene in Pseudomonas fluorescens 2P24
A Tn5 transposon mutant library was constructed, and ca. 5,000 mutants were screened for loss of inhibition against the fungal pathogen R. solani to identify novel regulators of the production of 2,4-DAPG in P. fluorescens 2P24. Six mutants were found to have a reduced ability to repress the mycelial growth of R. solani (Supplementary Table 3). One of the mutants, 2P24M-39, fully abolished its ability to inhibit the mycelial growth of R. solani. DNA sequence analysis showed that the Tn5 transposon was inserted into the C0J56_22945 gene, encoding the monothiol glutaredoxin GrxD (accession AUM71418.1).
An in silico analysis of the 2P24 genome identified three paralogs of GrxD, namely, GrxC (locus C0J56_02105, accession AUM67817.1), GrxF (locus C0J56_23915, accession AUM71586.1), and GrxG (locus C0J56_21615, accession AUM72833.1). Phylogenetic analysis revealed that these four proteins belonged to two GRX classes (data not shown). GrxD (113 aa, 12.12 kDa) is a class I GRX with the CGFS motif that is typical of monothiol GRXs, whereas GrxC (84 aa, 9.34 kDa), GrxF (118 aa, 13.4 kDa), and GrxG (123 aa, 14.16 kDa) belong to class II GRX, which contain the dithiol C(PG/FY)C active site.
Effect of GRXs on the Production of 2,4-Diacetylphloroglucinol in Pseudomonas fluorescens
The grxD, grxC, grxF, and grxG mutants were constructed to investigate the role of different GRXs on the production of 2,4-DAPG of P. fluorescens. HPLC indicated that the production of 2,4-DAPG in the grxD mutant was significantly impaired compared with that in strain 2P24 (Figure 1A). In addition, the production of PG, the intermediate of 2,4-DAPG, was severely decreased in the grxD mutant (Figure 1B). The complementation of grxD by introducing the complementary plasmid p415-grxD restored the 2,4-DAPG and PG production to wild-type levels (Figures 1A,B). In addition, the expression of the phlA′-′lacZ translational fusion on the plasmid p6013-phlA in the grxD mutant was significantly decreased compared with that in strain 2P24 (Figure 1C). GrxD contained the characterized monothiol glutaredoxin CGFS motif. To further determine whether this motif was required for GrxD-mediated 2,4-DAPG production in P. fluorescens, we generated a grxDC29S variant by using site-directed mutagenesis, which converted the cysteine residue C29 into serine. The grxD mutant expressing GrxDC29S was unable to restore 2,4-DAPG and PG production to those in strain 2P24, suggesting that the CGFS motif was necessary for the function of GrxD (Figures 1A,B). Furthermore, the grxD mutant failed to inhibit the growth of R. solani, whereas the grxD mutant expressing GrxD but not GrxDC29S restored this inhibition to wild-type levels (Figure 1D). Interestingly, no differences in 2,4-DAPG production were observed among grxC, grxF, grxG, and the wild-type strain (Supplementary Figure 1). Considering that the grx genes belonging to the same class were functionally redundant, we then constructed the grxC grxF double mutant and the grxC grxF grxG triple mutant. Similarly, the grxC grxF double mutant and the grxC grxF grxG triple mutant produced similar levels of 2,4-DAPG as the wild-type strain 2P24 (Supplementary Figure 1). Furthermore, the quantification of acetyl-CoA (the substrate for PG biosynthesis) by using the acetyl-CoA assay kit showed no significant difference in the concentration of acetyl-CoA in strain 2P24 and its grxD mutant (Supplementary Figure 2). Overall, these results indicated that only grxD was specifically involved in the production of 2,4-DAPG in P. fluorescens.
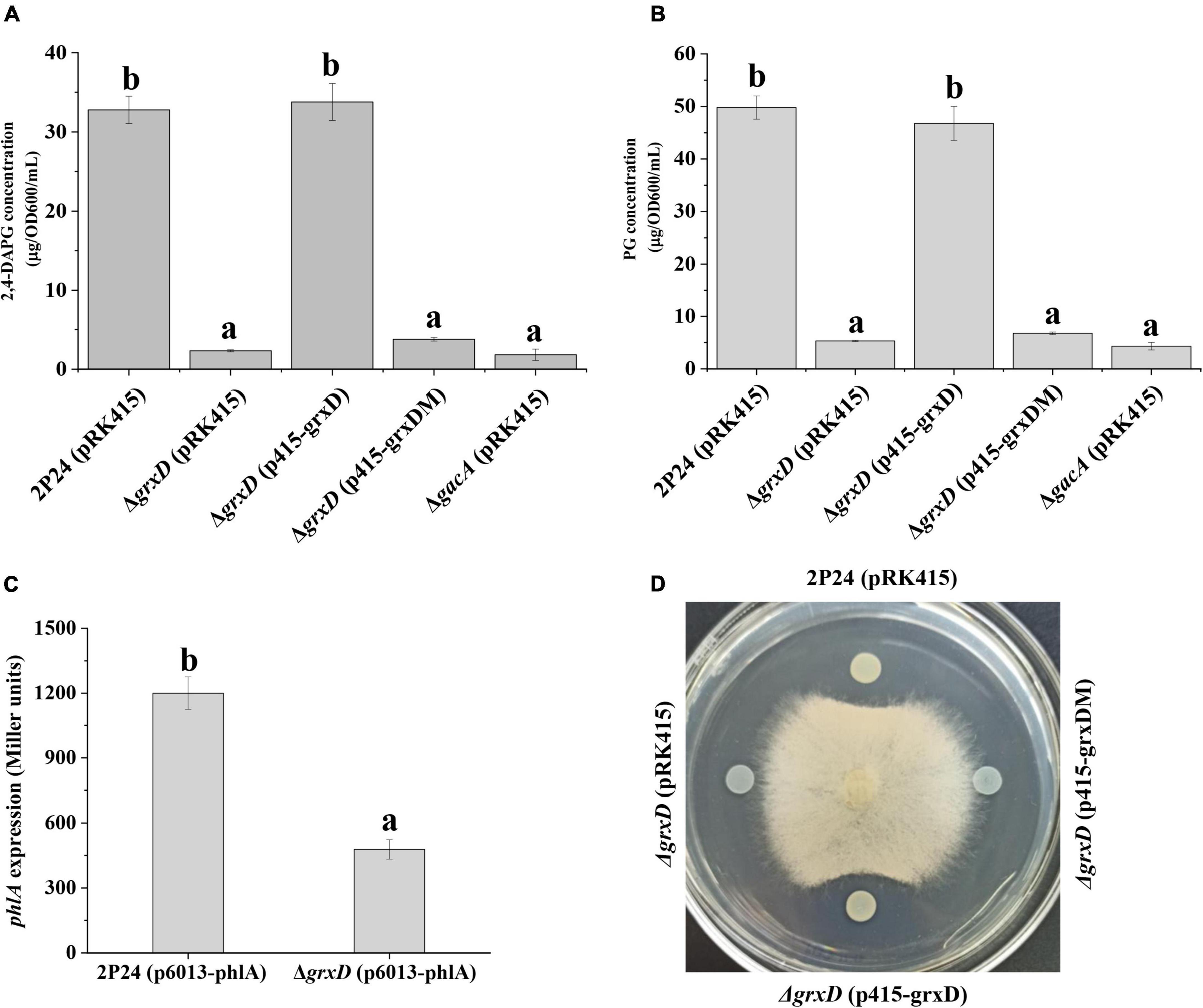
Figure 1. The effect of grxD gene on 2,4-diacetylphloroglucinol (2,4-DAPG) biosynthesis (A), phloroglucinol (PG) production (B), phlA expression (C), and antifungal activity (D) of Pseudomonas fluorescens 2P24. (A) High-performance liquid chromatography (HPLC) analysis of the concentration of 2,4-DAPG by strains 2P24 (pRK415), the grxD mutant (pRK415), the grxD mutant (p415-grxD), and the grxD mutant (p415-grxDM). (B) PG quantification from strains 2P24 (pRK415), the grxD mutant (pRK415), the grxD mutant (p415-grxD), and the grxD mutant (p415-grxDM) was measured by a colorimetric method. (C) The expression of phlA was positively regulated by GrxD. The β-galactosidase activity of the translational fusion phlA′-′lacZ was measured in strain 2P24 and its grxD mutant. (D) Inhibition of Rhizoctonia solani mycelial growth by 2P24 (pRK415), the grxD mutant (pRK415), the grxD mutant (p415-grxD), and the grxD mutant (p415-grxDM). All experiments were performed in triplicate, and letter represents significant differences between samples where p < 0.05.
Deletion of grxD Enhances the RsmA/RsmE Protein Levels
In P. fluorescens 2P24, the Gac/Rsm signaling cascade plays a key role in controlling the production of 2,4-DAPG (Zhang et al., 2020b). Considering that the grxD mutant showed a 2,4-DAPG production phenotype similar to the gacA mutant (Figure 1), we speculated that the Gac/Rsm signaling cascade was involved in the positive role of 2,4-DAPG production by GrxD. To test this hypothesis, we initially assessed whether GrxD negatively regulated the expression of rsmA and rsmE in P. fluorescens. Translation fusion assays showed that the deletion of grxD significantly enhanced the rsmA′-′lacZ expression, whereas the rsmE′-′lacZ expression was slightly decreased in the grxD mutant compared with those in strain 2P24 (Figure 2A). To further gain insight into the effect of grxD on RsmA and RsmE, the protein levels of RsmA and RsmE were examined in the wild-type 2P24 and the grxD mutant. At 18 h after inoculation, the grxD mutant produced significantly enhanced levels of RsmE and similar levels of RsmA compared with the wild-type strain 2P24 (Figures 2B,C). However, at 36 h after inoculation, the levels of RsmA and RsmE were significantly increased in the grxD mutant (Figures 2B,C). Later, the effect of grxD on the transcription of rsmZ, rsmY, rsmX, rsmX1, and rgsA was then investigated. As shown in Supplementary Figure 2, the grxD mutant had unchanged expression levels of rgsA, rsmX1, rsmY, and rsmZ, but significantly increased expression of rsmX compared with the 2P24 strain (Supplementary Figure 3A). Our previous data showed that the lack of rsmX in 2P24 could not influence 2,4-DAPG production (Zhang et al., 2020a). Interestingly, the production of 2,4-DAPG in the grxD rsmX double mutant was significantly decreased compared with the grxD mutant (Supplementary Figure 3B), suggesting that the mutation of grxD could improve the expression of rsmX, which competitively binds to the RsmA/RsmE proteins and thus influence the function of the RsmA/RsmE proteins. Therefore, these results indicated that the RsmA/RsmE proteins played an important role in the GrxD-mediated 2,4-DAPG production in strain 2P24.
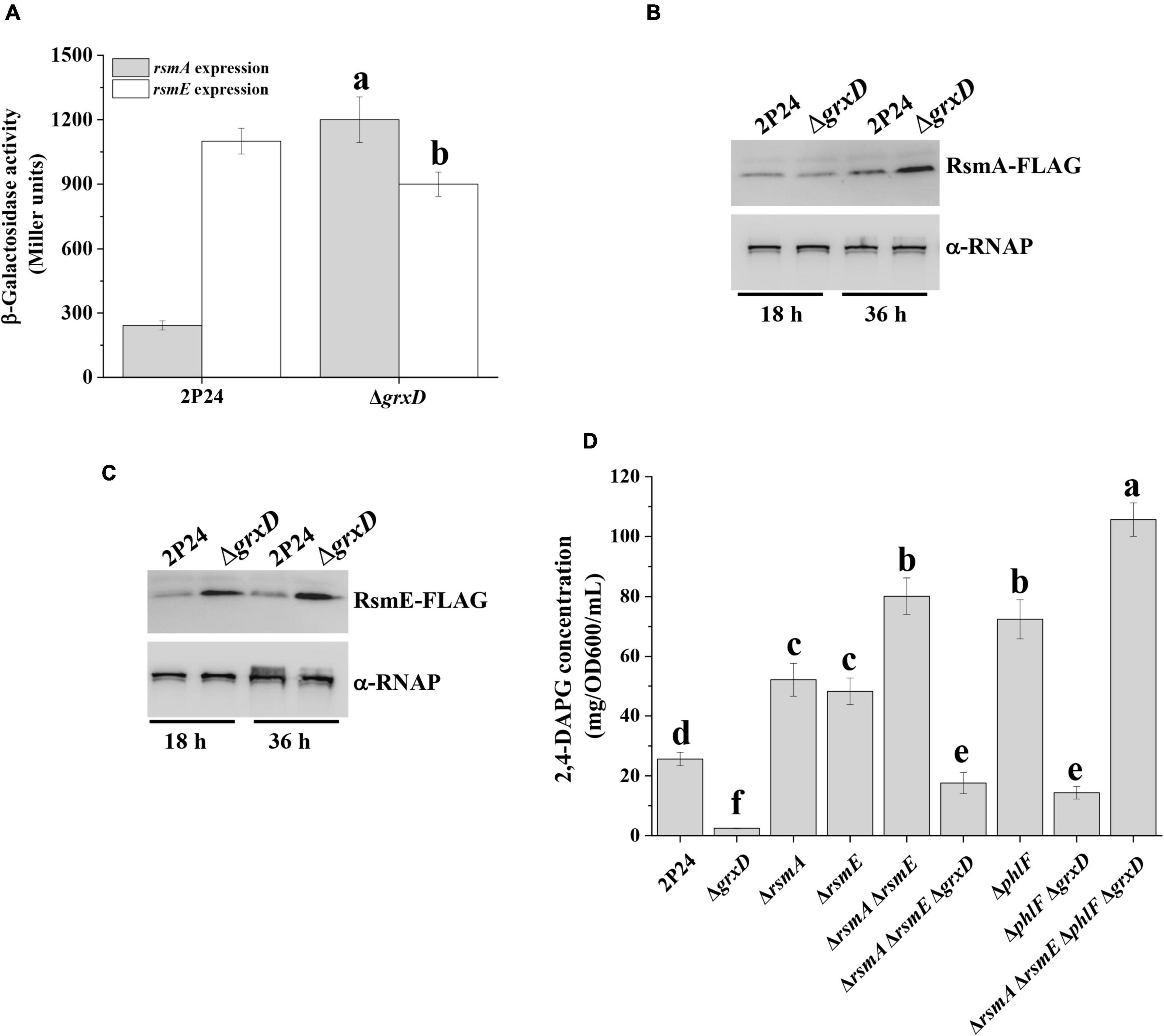
Figure 2. GrxD influenced the levels of RsmA and RsmE proteins. (A) The β-galactosidase activity of rsmA′-′lacZ and rsmE′-′lacZ translational fusions was determined in strain 2P24 and its grxD mutant at 36 h after inoculation. Analysis of RsmA-FLAG (B) or RsmE-FLAG (C) levels in total crude lysates prepared from strain 2P24 and its grxD mutant. An antibody directed against RNA polymerase β subunit (α-RNAP) was used as a loading control in this and later assays. Quantification of the production of 2,4-DAPG (D) from strain 2P24 and its derivatives. All experiments were performed in triplicate, and different letters indicate a significant difference at the 0.05 level.
The overexpression of RsmA and RsmE resulted in decreased 2,4-DAPG production (Zhang et al., 2020b). Thus, we investigated the effect of RsmA and RsmE on GrxD-mediated 2,4-DAPG production. Results indicated that the rsmA and the rsmE mutants produced a significantly higher amount of 2,4-DAPG than strain 2P24 (Figure 2D). The deletion of rsmA and rsmE genes in the grxD mutant background partially restored the production of 2,4-DAPG to the wild-type levels (Figure 2D). In addition, the production of PG and 2,4-DAPG in the grxD mutant was unchanged at 18 h after inoculation and significantly decreased at 36 h after inoculation compared with that in the wild-type strain (Supplementary Figure 4). Our previous study showed that the production of exoprotease is negatively regulated by RsmA/E proteins (Wei et al., 2004a). To further examine whether GrxD controlled the production of exoprotease in strain 2P24, we compared the production of exoprotease in the wild-type strain and its derivatives. The deletion of the grxD gene in the wild-type background significantly influenced the production of exoprotease (Supplementary Figure 5A). By contrast, the production of exoprotease in the rsmA rsmE double mutant was significantly increased compared with that in the wild type. As expected, the lack of grxD in the rsmA rsmE double mutant could not influence exoprotease production, suggesting that GrxD affected the production of exoprotease through RsmA/E proteins (Supplementary Figure 5A). A similar regulation was observed when the lacZ-fused translational reporter of aprA (exoprotease biosynthetic gene) was determined in 2P24 and its derivatives (Supplementary Figure 5B). Our results indicated that GrxD repressed the levels of RsmA and RsmE, which contributed, at least partially, to the positive regulatory effect of GrxD on the 2,4-DAPG production in strain 2P24.
GrxD Regulates 2,4-Diacetylphloroglucinol Production Through the Transcriptional Regulator PhlF
To further investigate how GrxD influences 2,4-DAPG production, we attempted to identify potential target genes regulated by GrxD. Previous work demonstrated that the expression of phlACBD was negatively regulated by the pathway-specific repressor PhlF (Abbs et al., 2002; Zhou et al., 2010). We hypothesized that GrxD negatively influenced phlF and then positively affected the production of 2,4-DAPG in P. fluorescens. To test this hypothesis, we assessed the expression of phlF in the wild-type strain and the grxD mutant. The deletion of grxD significantly increased phlF expression, and the complementation of the grxD mutant restored phlF expression to wild-type levels (Figure 3A). We then monitored the levels of the PhlF protein in strain 2P24 and its grxD mutant by Western blotting. In agreement with the phlF transcriptional levels, the Western blot assay showed that levels of the PhlF protein in the grxD mutant were increased compared with that of 2P24 (Figure 3B). These data supported our hypothesis and suggested that GrxD negatively controlled the expression of phlF.
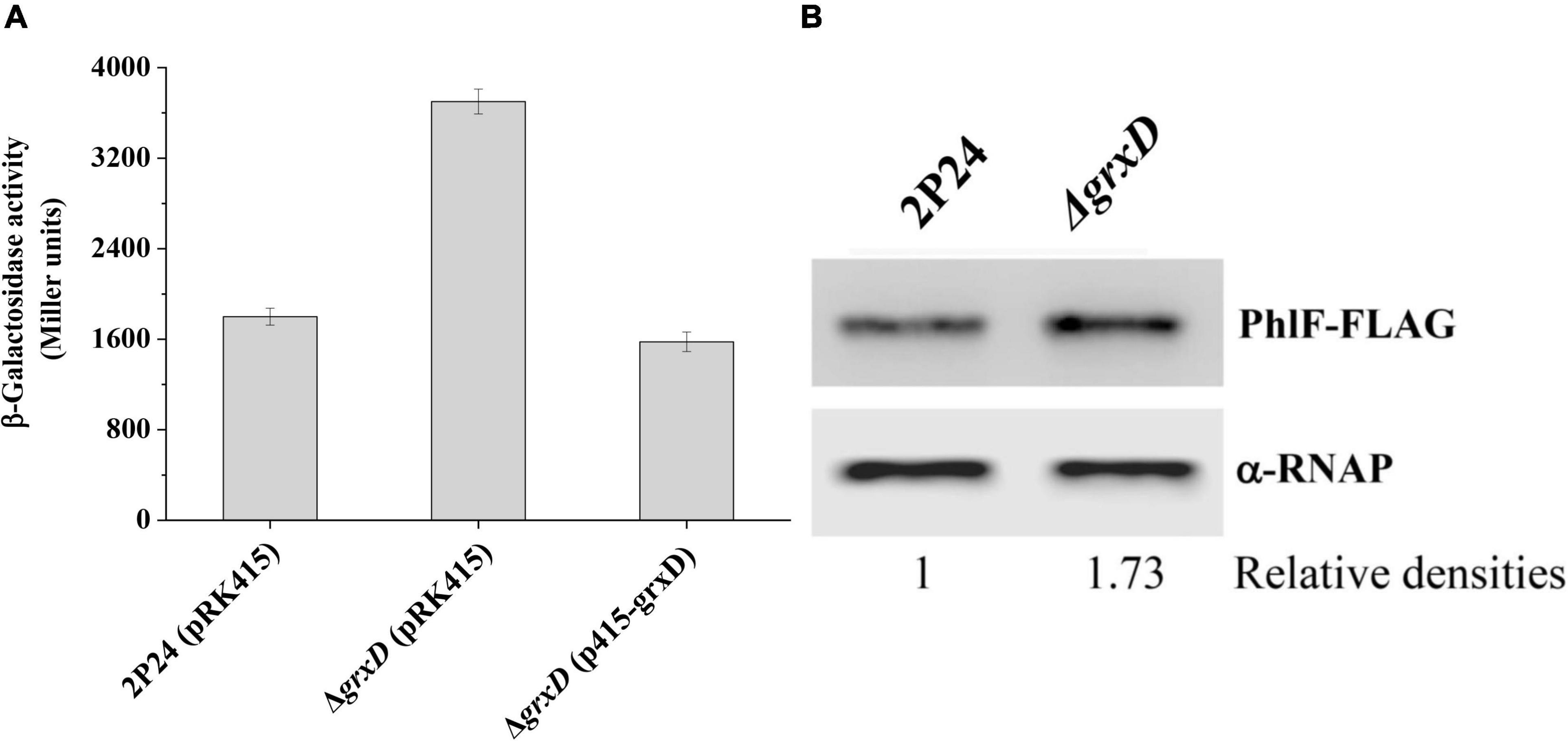
Figure 3. GrxD influenced phlF in Pseudomonas fluorescens. (A) The β-galactosidase activity of phlF-lacZ transcriptional fusion was determined in strains 2P24 (pRK415), the grxD mutant (pRK415), and the grxD mutant (p415-grxD). (B) Western blot analysis was performed to detect the level of PhlF-FLAG. The relative intensities of PhlF-FLAG bands were marked below the blot. All experiments were performed in triplicate, and different letters indicate a significant difference at the 0.05 level.
Later, we constructed the grxD phlF double mutant and compared its 2,4-DAPG production to that of the wild-type strain 2P24 and its derivatives. Our data indicated that the production of 2,4-DAPG in the grxD phlF double mutant was higher compared with that in the grxD mutant but was significantly lower compared with the wild-type strain (Figure 2D). Furthermore, the rsmA rsmE phlF grxD quadruple mutant was constructed, and HPLC showed that the production of 2,4-DAPG in the rsmA rsmE phlF grxD quadruple mutant was significantly increased compared with that in the phlF mutant or the rsmA rsmE double mutant (Figure 2D). These results suggested that the transcriptional regulator PhlF and RsmA/E proteins were involved in the GrxD-mediated regulation of 2,4-DAPG.
GrxD Influences the Growth of Pseudomonas fluorescens
Given the imbalance of redox state in grx mutants and the involvement of GRXs in regulating bacterial growth (Cao et al., 2020), the growth of all grx mutants was measured in an ABM medium. Our data indicated that the grxD mutant showed a significantly lower than the wild-type strain in the ABM minimal medium (Figure 4). This growth defect could be restored to wild-type levels by complementation with p415-grxD but not p415-grxDM (Figure 4). By contrast, no difference could be observed among the grxC, grxF, and grxG mutants, and strain 2P24 in the ABM medium. Furthermore, the grxC grxF double mutant and the grxC grxF grxG triple mutant were indistinguishable from the wild-type strain (Supplementary Figure 6). Thus, the monothiol glutaredoxin GrxD was important for the normal growth of P. fluorescens.
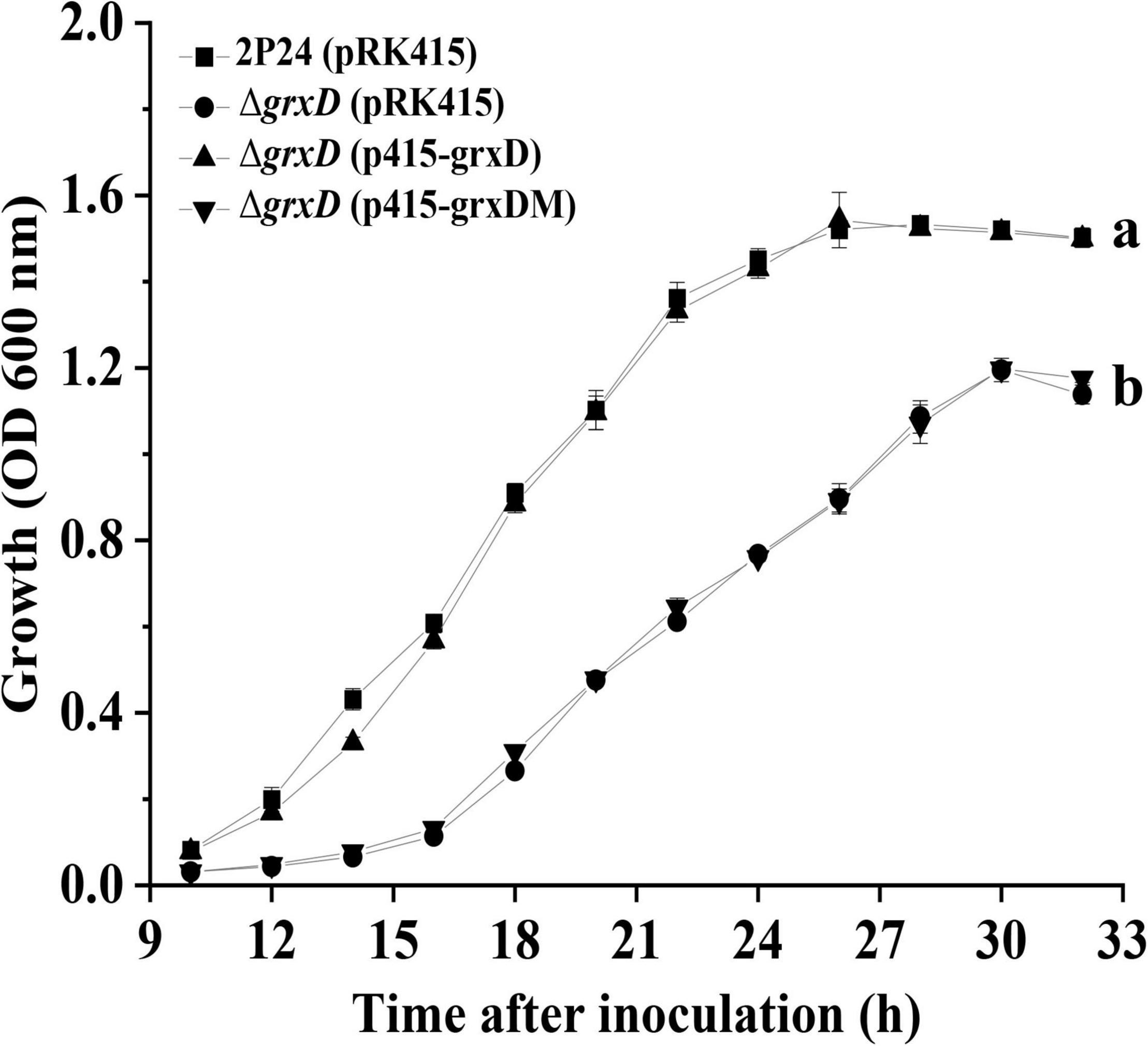
Figure 4. The effect of grxD gene on the growth of Pseudomonas fluorescens. Strain 2P24 and its derivatives were cultured in ABM medium and the absorbance at 600 nm were measured at different points of the growth curve. All experiments were performed in triplicate, and different letters indicate a significant difference at the 0.05 level.
grxD Mutant Is Sensitive Under Oxidative Stress Conditions
The survival of strain 2P24 and its derivatives was examined via killing assays. As shown in Figure 5, the grxD mutant showed significantly increased susceptibility to relatively high H2O2 (0.6 mM) or CuOOH (5 mM). The complementation of the grxD mutant by expression of grxD in trans restored H2O2 or CuOOH resistance to that of the wild-type strain, whereas the grxD mutant expressing GrxDC29S exhibited the same phenotype of the grxD mutant (Figure 5A). In addition, the grxC, grxF, or grxG mutant was as sensitive as the wild-type strain to H2O2 or CuOOH (Figure 5B), suggesting that the monothiol GRX GrxD played an important role in the resistance of P. fluorescens against oxidative stress.
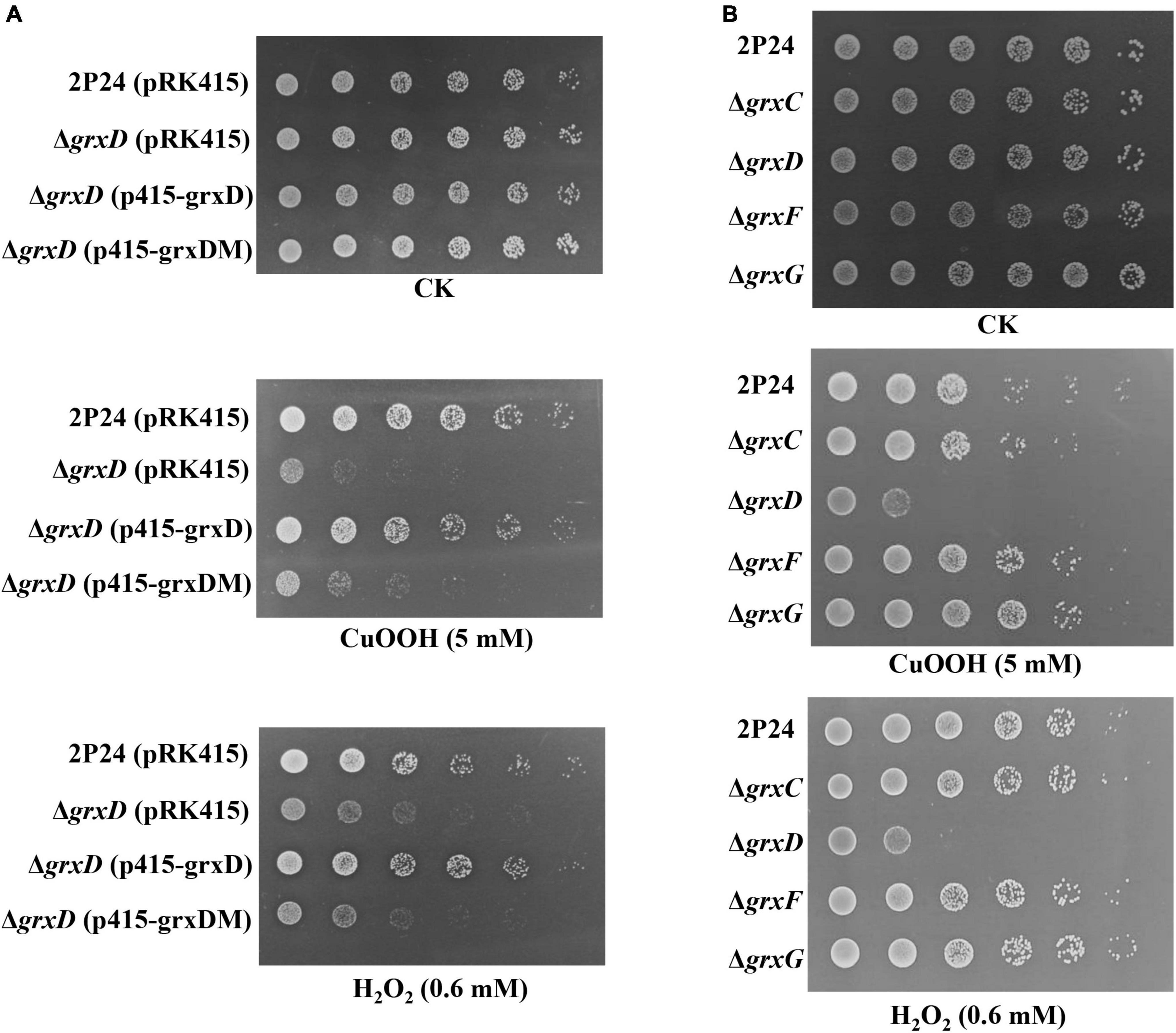
Figure 5. Effect of the grx mutants on tolerance of Pseudomonas fluorescens to peroxides. Pseudomonas fluorescens 2P24 (pRK415), the grxD mutant (pRK415), the grxD mutant (p415-grxD), and the grxD mutant (p415-grxDM) (A), or 2P24, the grxC mutant, the grxD mutant, the grxF mutant, and the grxG mutant (B) were grown in Lysogeny Broth (LB) broth, serially diluted and spotted onto ABM plates or ABM plates containing 5 mM CuOOH or 0.6 mM H2O2. Plates were incubated at 28°C for 2 days.
Mutation of the grxD Gene Decreases the Production of Siderophores in Pseudomonas fluorescens
To determine whether GrxD influenced iron availability in the environment, we compared the production of siderophores in the wild-type strain and its grx mutants. The siderophore activity was analyzed using the CAS assay. The grxD mutant showed a significantly decreased amount of siderophores, which was restored in the complemented strain (Figure 6). By contrast, the amount of siderophores produced by the grxC, grxF, grxG mutants, the grxC grxF double mutant, or the grxC grxF grxG triple mutant was indistinguishable from that produced by the wild-type strain (Figure 6). These results demonstrated the important role of GrxD in the production of siderophores of P. fluorescens.
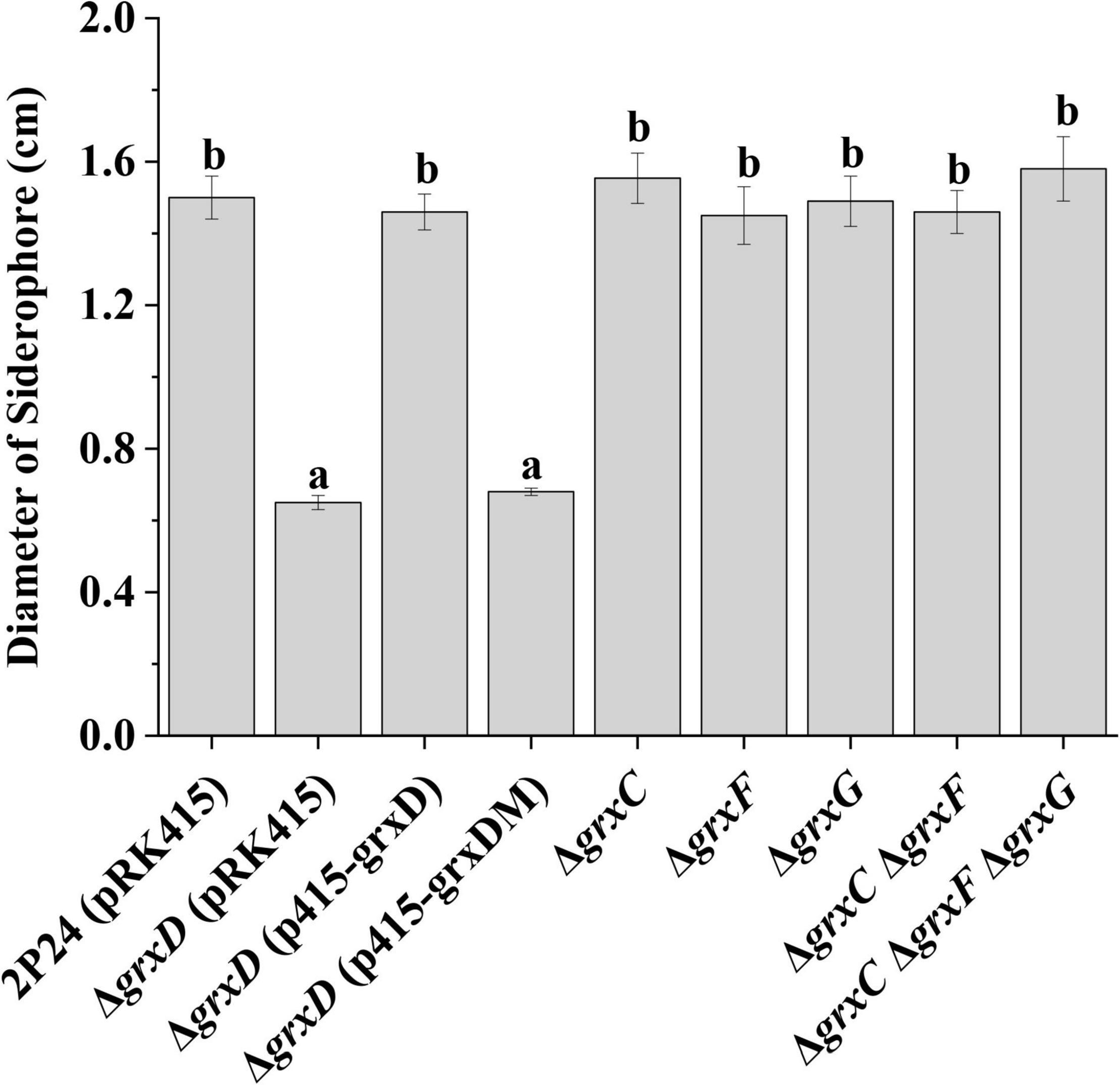
Figure 6. Effects of grx mutants on siderophore production of Pseudomonas fluorescens. Siderophore production of strain 2P24 and its derivatives was indicated by the diameter of the orange circle on the chrome azurol S (CAS) plate. The experiment was performed in triplicate, and different letters indicate a significant difference at the 0.05 level.
Effects of grx Mutants on the Concentration of Iron
To assess the effect of grx genes on the accumulation of iron in P. fluorescens cells, we then measured the intracellular Fe content in 2P24 and its derivatives. The Fe content in the grxC, grxF, and grxG mutants was significantly lower than that in the 2P24 strain (Figure 7). By contrast, the deletion of the monothiol grxD gene resulted in a dramatic accumulation of total Fe content in the cells of P. fluorescens (Figure 7). These results suggested that the monothiol GrxD played an important role in the Fe homeostasis of P. fluorescens.
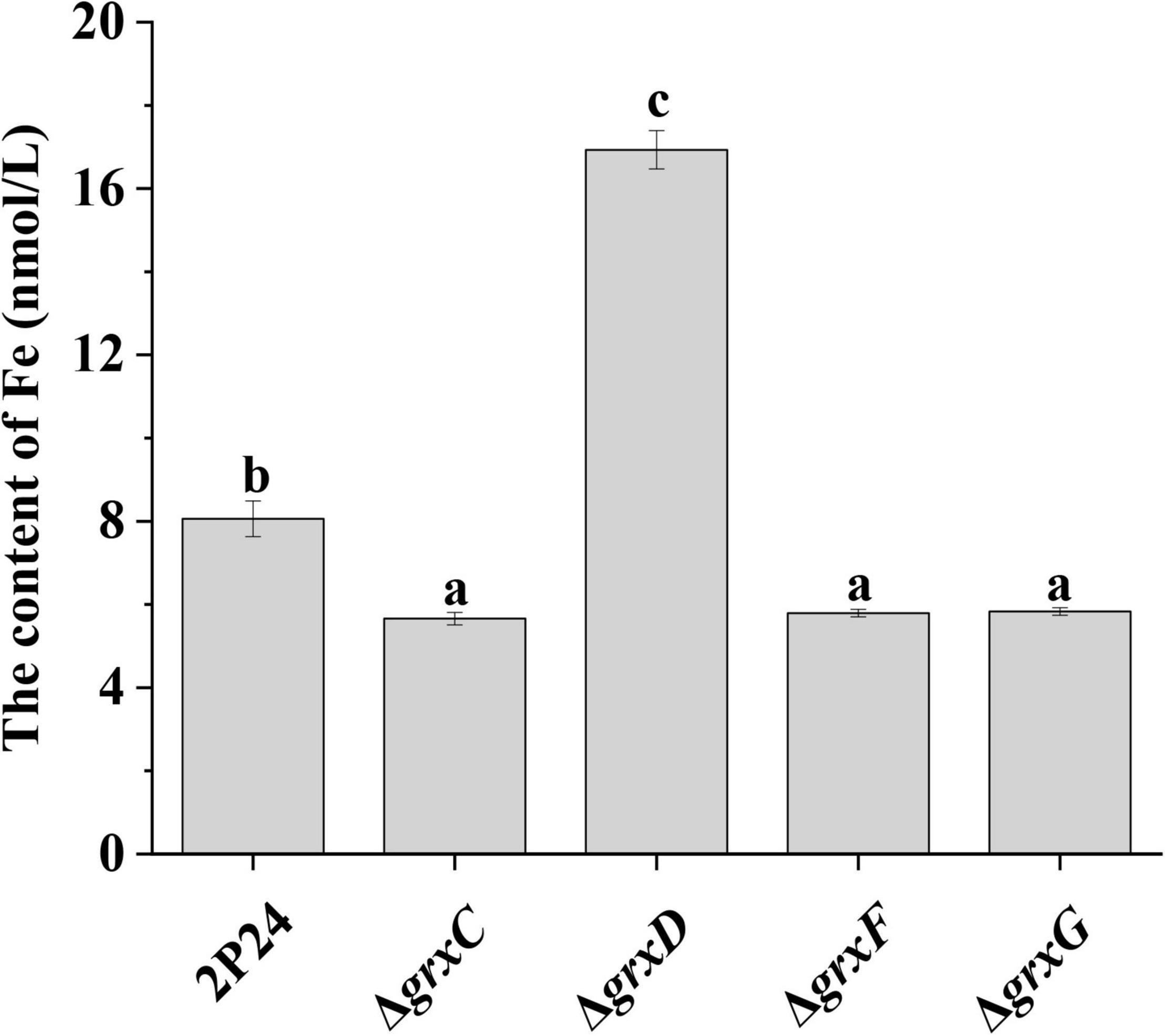
Figure 7. Deletion of grx genes influenced the iron content of Pseudomonas fluorescens. The Fe content was measured in 3 × 109 cells of strain 2P24 and its derivatives using the ferrozine assay. The experiment was performed in triplicate, and different letters indicate a significant difference at the 0.05 level.
Effect of the grx Mutation on the Motility of Pseudomonas fluorescens
Motility is important for the establishment of the bacteria in the rhizosphere and bacterial performance as PGPR (Barret et al., 2011). The effect of the grx mutants on motility was then measured. Compared with the wild-type strain, the loss of the GrxD function affected the swarming or swimming motility (Figure 8). This observed phenotype in the grxD mutant could be complemented by introducing a copy of wild-type grxD. Furthermore, the characterized active site (CGFS) motif of GrxD played a vital role in bacterial motility (Figure 8). However, no such difference in swarming or swimming motility could be observed between the wild-type strain and the grxC, grxF, and grxG mutants, the grxC grxF double mutant, or the grxC grxF grxG triple mutant (data not shown). These results indicated that GrxD positively affected motility in P. fluorescens.
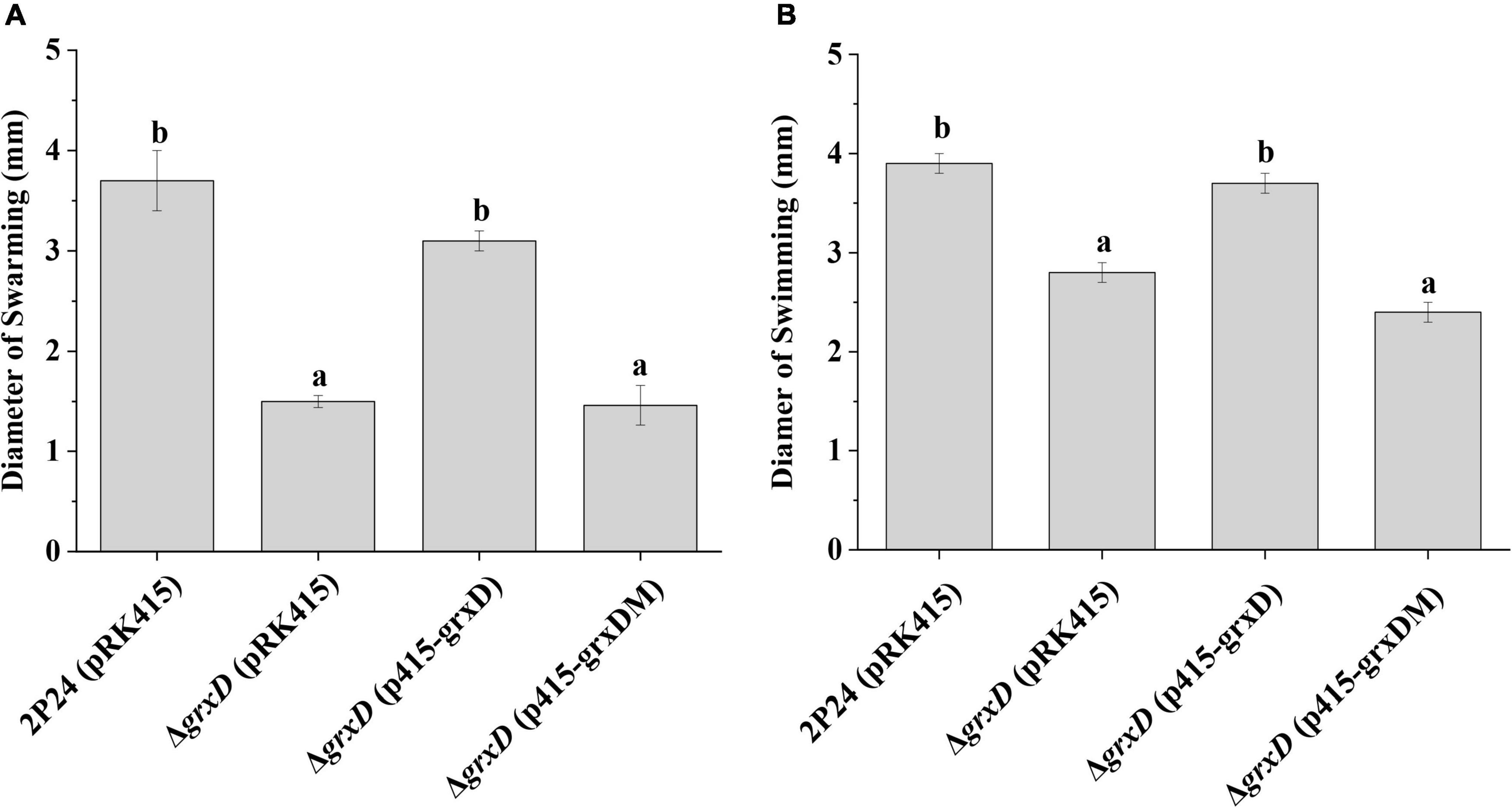
Figure 8. Effect of the grxD gene on the motility of Pseudomonas fluorescens. (A) Swarming agar plate assay. The swarming abilities of strain 2P24 and its derivatives were determined by the diameter of the zone of motility. (B) Swimming agar plate assay. The swimming abilities of strain 2P24 and its derivatives were determined by the diameter of the zone of motility. The experiments were performed in triplicate, and different letters indicate a significant difference at the 0.05 level.
GrxD Is Required for the Biocontrol Capacity of Pseudomonas fluorescens
GrxD positively affects the production of 2,4-DAPG, which is essential for protecting tomatoes against R. solanacearum. To assess the relative contribution of grx genes to the biocontrol capacity of P. fluorescens, we then investigated the effect of strain 2P24 and its derivatives in controlling the bacterial wilt of tomatoes. The grxD mutant was significantly impaired in its capacity to protect tomatoes from root diseases (Table 1). The biocontrol activity of the grxD mutant could be restored to the wild-type level by complementing the grxD mutant with p415-grxD, but not p451-grxDM. The deletion of grxC, grxF, or grxG in 2P24 did not result in a significant alteration in biocontrol efficacy (Table 1). These results indicated that GrxD played an important role in the biocontrol capacity of P. fluorescens.
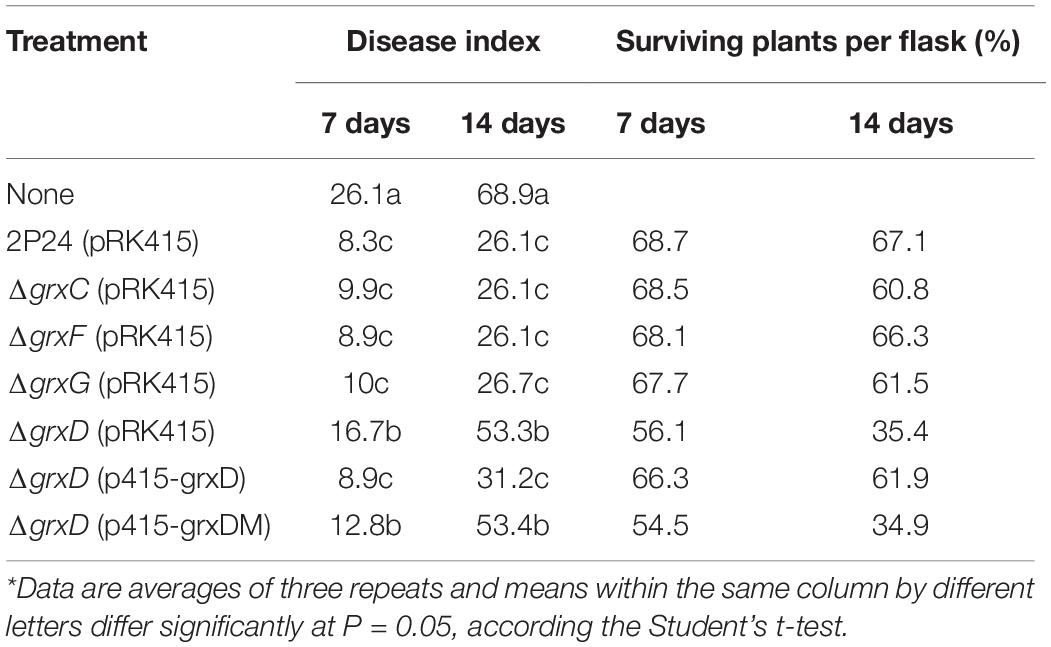
Table 1. Contribution of grx genes in strain 2P24 to the suppression of tomato bacterial wilt caused by Ralstonia solanacearum*.
Discussion
Glutaredoxin is an antioxidant protein that plays an important role in various physiological processes, such as stress response, petal development, Fe–S cluster assembly, signaling and pathogen response in bacteria, fungi, and mammals (Couturier et al., 2015). The results of this study have advanced our knowledge in the functions of glutaredoxin and have shown that monothiol GRX GrxD plays roles in many important biocontrol-related traits, such as the production of the antibiotic 2,4-DAPG, siderophores, motility, and biocontrol efficacy of the plant beneficial bacterium P. fluorescens 2P24.
Previous work showed that 2,4-DAPG-producing pseudomonads have evolved sophisticated mechanisms to fine-tune the concentration of 2,4-DAPG in the cells. Among these mechanisms is the Gac/Rsm signal cascade. This cascade involves the GacS/GacA two-component system, which induces the expression of sRNAs, and the RNA-binding proteins RsmA and RsmE (Lapouge et al., 2008). RsmA and RsmE are predicted to bind to the ribosome-binding site of phlACBD mRNA directly, thereby repressing its translation and influencing the production of 2,4-DAPG (Zhang et al., 2020b). To date, some genetic elements that regulate the expression of sRNAs have been described in detail. In P. aeruginosa, the expression levels of rsmZ are negatively regulated by the phosphotransfer protein HptB and H-NS family member MvaT (Brencic et al., 2009). In addition, IHF, PsrA, and AlgR are involved in inducing the expression of rsmZ of P. protegens CHA0 (Humair et al., 2010). Compared with the well-studied regulation of these sRNAs, few regulators are reported to have regulatory effects on the expression of their cognate small regulatory proteins RsmA or RsmE. Our previous work showed that c-di-GMP positively regulates the expression levels of rsmA and rsmE in strain 2P24 (Liang et al., 2020). Our findings here showed that GrxD controls the expression of RsmA and RsmE (Figure 2). However, the regulatory effects of GrxD on rsmA and rsmE are unique. Unlike c-di-GMP, GrxD could influence the levels of RsmA protein at the stationary phase (36 h), whereas RsmE was significantly altered during the exponential (18 h) and stationary (36 h) phases (Figure 2). Regarding the complex effect of RsmA and RsmE on 2,4-DAPG, this work suggested that the effect of 2,4-DAPG by GrxD is achieved through fine-tuning the protein levels of RsmA and RsmE in the cells.
Besides the RsmA and RsmE proteins, our data revealed that GrxD positively influenced the production of 2,4-DAPG by repressing the expression of phlF (Figure 3). PhlF is a transcriptional regulator for the 2,4-DAPG biosynthetic operon. Sequence analysis reveals that PhlF is a cytoplasmic protein with four cysteine residues (data not shown). Considering that the GRX can influence the disulfide bonds formed between cysteine residues of target proteins and glutathione, we hypothesize that GrxD directly interacts with PhlF. However, the bacterial two-hybrid assay shows no interaction between GrxD and PhlF in vivo (data not shown). Previous studies indicated that GrxD could directly interact with the iron-responsive transcriptional regulators Aft1 and Aft2 in Saccharomyces cerevisiae (Ojeda et al., 2006). Aft1 induces the expression of RNA-binding protein CTH2, which degrades the transcripts of iron-requiring enzymes to conserve iron in the cell. Since the mutation of grxD could induce the accumulation of iron in P. fluorescens 2P24. Homologs of CTH2, such as C0J56_01730, C0J56_04495, and C0J56_06345, can be found in strain 2P24. It is possible that the iron-responsive transcriptional factors could be activated in the grxD mutant to induce these RNA-binding proteins, which could inhibit the translation of RsmA/E proteins. Furthermore, the mutation of grxD could affect the enzyme activity of PhlA, PhlC, PhlB, or PhlD. Thus, the mechanism behind GrxD in the production of 2,4-DPAG remains to be elucidated.
The characterization of grx mutants indicates that monothiol GRX is involved in influencing cellular redox state, and this finding is in agreement with phenotypes previously described in Azorhizobium caulinodans and Rhizobium leguminosarum bv. viciae (Cao et al., 2020; Zou et al., 2021). In P. fluorescens, the mutation of grxD is hypersensitive to H2O2 and CuOOH, whereas the grxC, grxF, grxG mutants, and the grxC grxF grxG triple mutant display no such effect (Figure 5). Monothiol GRX is essential for the Fe–S cluster biosynthesis, and our data also indicate that GrxD regulates siderophore production and is involved in the iron homeostasis of P. fluorescens (Figures 6, 7). Overall, our data have characterized the essential role of GrxD of P. fluorescens in oxidative stress.
In plant assays carried out under greenhouse conditions, the mutation of grxD severely impairs the biocontrol activity against tomato bacterial wilt caused by R. solanacearum (Table 1). By contrast, the grxC, grxF, and grxG mutants provide a similar effect in protecting tomatoes from tomato bacterial wilt as the wild-type strain. Our previous data suggested that 2,4-DAPG plays a vital role in the biocontrol activity of P. fluorescens (Table 1). Given the effect of GrxD on many biocontrol traits, such as motility, bacterial resistance against peroxides, and the production of siderophore, defects in biocontrol capacity can be a combined result. Therefore, our work indicates that GrxD plays a vital role in the regulation of the biocontrol ability and is necessary for the ecological fitness of P. fluorescens in a constantly changing environment (Figure 9).
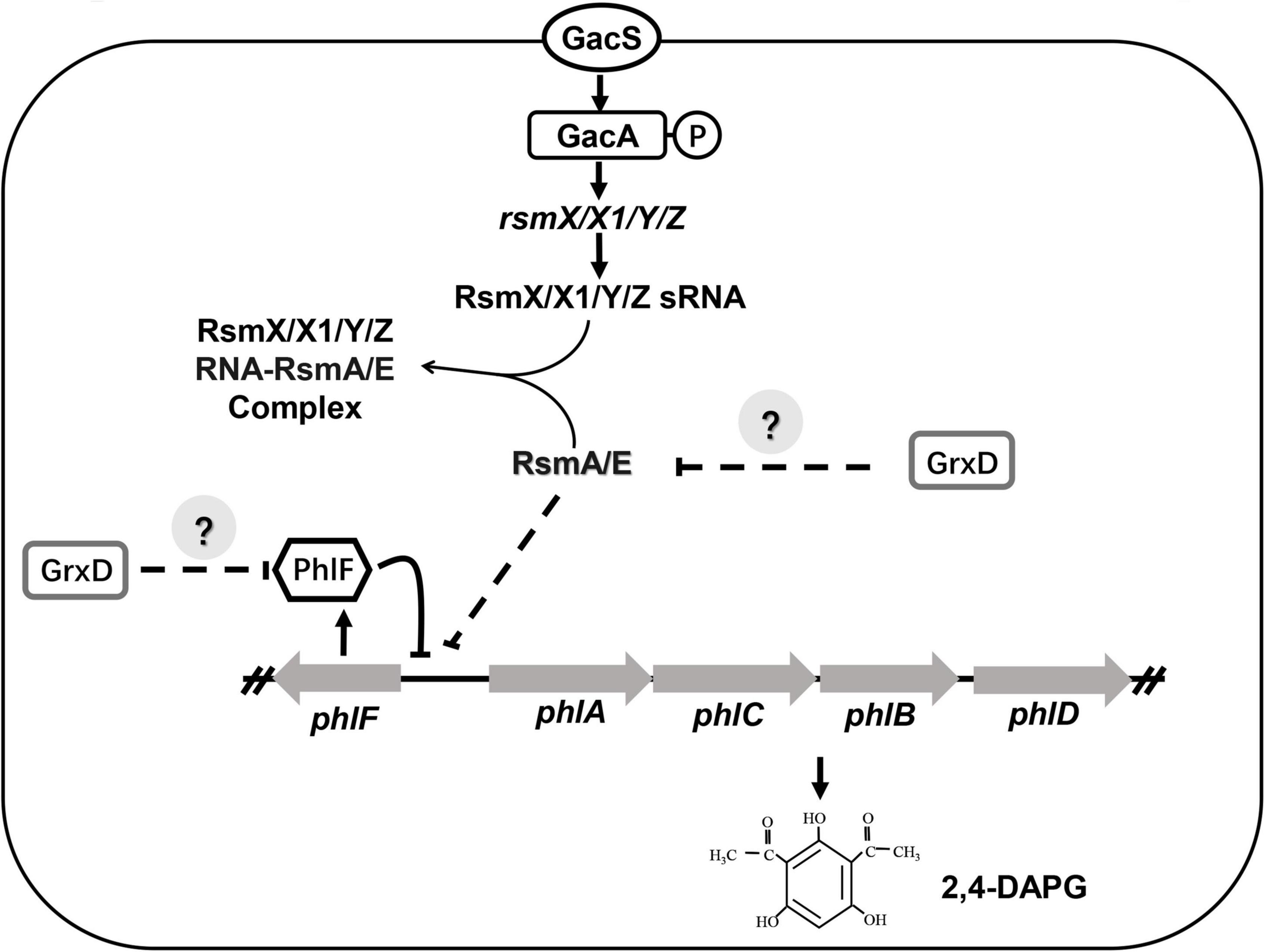
Figure 9. Model of GrxD affects the production of 2,4-DAPG in Pseudomonas fluorescens 3P24. The production of 2,4-diacetylphloroglucinol (2,4-DAPG) was previously reported to be regulated by the Gac/Rsm signaling pathway and the transcriptional factor PhlF. In this study, the levels of RsmA/E and PhlF proteins were modulated by GrxD. In addition, GrxD could interact directly with an unknown transcriptional regulator, which may influence the expression of rsmA/E and phlF.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
XW conceived and designed the study. QD, QY, BZ, and XW performed the assays. QD, QY, and BZ collected the data. L-QZ and XW analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Guangxi (2021GXNSFDA220006) and the National Natural Science Foundation of China (31760533).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.920793/full#supplementary-material
References
Abbs, A., Morrissey, J. P., Marquez, P. C., Sheehan, M. M., Delany, I. R., and O’Gara, F. (2002). Characterization of interactions between the transcriptional repressor PhlF and its binding site at the phlA promoter in Pseudomonas fluorescens F113. J. Bacteriol. 184, 3008–3016. doi: 10.1128/JB.184.11.3008-3016.2002
Bangera, M. G., and Thomashow, L. S. (1999). Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphlorogluconil form Pseudomonas fluorescens Q2-87. J. Bacteriol. 181, 3155–3163. doi: 10.1128/JB.181.10.3155-3163.1999
Barret, M., Morrissey, J. P., and O’Gara, F. (2011). Functional genomics analysis of the plant growth-promoting rhizobacterial traits involved in rhizosphere competence. Biol. Fertil. Soils 47, 729–743.
Benizri, E., Baudoin, E., and Guckert, A. (2001). Root colonization by inoculated plant growth-promoting rhizobacteria. Biocontrol. Sci. Techn. 5, 557–574.
Brencic, A., McFarland, K. A., McManus, H. R., Castang, S., Mogno, I., Dove, S. L., et al. (2009). The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73, 434–445. doi: 10.1111/j.1365-2958.2009.06782.x
Cao, Y., Jiang, G., Li, M., Fang, X., Zhu, D., Qiu, W., et al. (2020). Glutaredoxins play an important role in the redox homeostasis and symbiotic capacity of Azorhizobium caulinodans OR571. Mol. Plant-Microbe. Interact. 33, 1381–1393. doi: 10.1094/MPMI-04-20-0098-R
Chilton, M. D., Currier, T. C., Farrand, S. K., Bendich, A. J., Gordon, M. P., and Nester, E. W. (1974). Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc. Natl. Acad. Sci. U. S. A. 71, 3672–3676. doi: 10.1073/pnas.71.9.3672
Couturier, J., Przybyla-Toscano, J., Roret, T., Didierjean, C., and Rouhier, N. (2015). The roles of glutaredoxins ligating Fe-S clusters: sensing, transfer or repair functions? Biocinica. Biophysica. Acta 1853, 1513–1527.
Fernandes, A. P., and Holmgren, A. (2004). Glutaredoxins: glutathione-dependent redox enzymes with functions for beyond a simple thioredoxin backup system. Antioxid. Redox Signal. 6, 63–74. doi: 10.1089/152308604771978354
Forman, H. J., Zhang, H., and Rinna, A. (2009). Glutathione: overview of its protective roles, measurement, and biosynthesis. Mol. Aspects. Med. 30, 1–12.
Ha, D. G., Richman, M. E., and O’Toole, G. A. (2014). Deletion mutant library for investigation of functional outputs of cyclic diguanylate metabolism in Pseudomonas aeruginosa PA14. Appl. Environ. Microbiol. 80, 3384–3393. doi: 10.1128/AEM.00299-14
Haas, D., and Défago, D. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319.
Heck, D. E., Shakajian, M., Kim, H. D., Laskin, J. D., and Vetrano, A. M. (2010). Mechanisms of oxidant generation by catalase. Ann. N. Y. Acad. Sci. 1203, 120–125.
Heeb, S., and Haas, D. (2001). Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe. Interact. 14, 1351–1363. doi: 10.1094/MPMI.2001.14.12.1351
Holmgren, A. (1976). Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc. Natl. Acad. Sci. U. S. A. 73, 2275–2279. doi: 10.1073/pnas.73.7.2275
Humair, B., Wackwitz, B., and Haas, D. (2010). GacA-controlled activation of promoters for small RNA genes in Pseudomonas fluorescens. Appl. Environ. Microbiol. 76, 1497–1506. doi: 10.1128/AEM.02014-09
Keen, N. T., Tamaki, S., Kobayashi, D., and Trollinger, D. (1988). Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70, 191–197.
Kidarsa, T. A., Goebel, N. C., Zabriskie, T. M., and Loper, J. E. (2011). Phloroglucinol mediates cross-talk between the pyoluteorin and 2,4-diacetylphloroglucinol biosynthetic pathways in Pseudomonas fluorescens Pf-5. Mol. Microbiol. 81, 395–414. doi: 10.1111/j.1365-2958.2011.07697.x
King, E. O., Ward, M. K., and Raney, D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab Clin. Med. 44, 301–307.
Lapouge, K., Schubert, M., Allain, F. H. T., and Haas, D. (2008). Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behavior. Mol. Microbiol. 67, 241–253. doi: 10.1111/j.1365-2958.2007.06042.x
Liang, F., Zhang, B., Yang, Q., Zhang, Y., Zheng, D., Zhang, L. Q., et al. (2020). Cyclic-di-GMP regulates the quorum-sensing system and biocontrol activity of Pseudomonas fluorescens 2P24 through the RsmA and RsmE proteins. Appl. Environ. Microbiol. 86:e2016–e2020. doi: 10.1128/AEM.02016-20
Lilling, C., Prior, A., Schwenn, J. D., Åslund, F., Ritz, D., Vlamis-Gardikas, A., et al. (1999). New thioredoxins and glutaredoxins as electron donors of 3’-phosphoadenylylsulfate reductase. J. Biol. Chem 274, 7695–7698. doi: 10.1074/jbc.274.12.7695
Meyer, Y., Buchanan, B. B., Vignols, F., and Reichheld, J. P. (2009). Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu. Rev. Genet. 9, 335–367. doi: 10.1146/annurev-genet-102108-134201
Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, N. Y: Cold Spring Harbor Laboratory Press.
Nanda, A. K., Andrio, E., Marino, D., Pauly, N., and Dunand, C. (2010). Reactive oxygen species during plant-microorganism early interactions. J. Integr. Plant Biol. 52, 195–204. doi: 10.1111/j.1744-7909.2010.00933.x
Ojeda, L., Keller, G., Muhlenhoff, U., Rutherford, J. C., and Winge, D. R. (2006). Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J. Biol. Chem. 281, 17661–17669. doi: 10.1074/jbc.M602165200
Poor, C. B., Wegner, S. V., Li, H., Dlouhy, A. C., Schuermann, J. P., Snishvili, R., et al. (2014). Molecular mechanism and structure of the Saccharomyces cerevisiae iron regulator Aft2. Proc. Natl. Acad. Sci. U. S. A. 111, 4043–4048.
Pršiæ, J., and Ongena, M. (2020). Elicitors of plant immunity triggered by beneficial bacteria. Front. Plant Sci. 11:594530. doi: 10.3389/fpls.2020.594530
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56.
Segonzac, C., and Zipfel, C. (2011). Activation of plant pattern-recognition receptors by bacteria. Curr. Opin. Microbiol. 14, 54–61.
Shanahan, P., O’Sullivan, D. J., Simpson, P., Glennon, J. D., and O’Gata, F. (1992). Isolation of 2,4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl. Environ. Microbiol. 58, 353–358. doi: 10.1128/aem.58.1.353-358.1992
Wei, H. L., Wang, Y., Zhang, L. Q., and Tang, W. H. (2004a). Identification and characterization of biocontrol bacterial strain 2P24 and CPF-10. Acta Phytopathol. Sin. 34, 80–85.
Wei, H. L., Zhou, H. Y., Zhang, L. Q., Wang, Y., and Tang, W. H. (2004b). Experimental evidence on the functional agent of 2,4-diacetylphloroglucinol in biocontrol activity of Pseudomonas fluorescens 2P24. Acta Microbiol. Sin. 44, 663–666.
Wu, X., Duan, H., Tian, T., Yao, N., Zhou, H., and Zhang, L. Q. (2010). Effect of the hfq gene on 2,4-diacetylphloroglucinol production and the PcoI/PcoR quorum-sensing system in Pseudomonas fluorescens 2P24. FEMS Microbiol. Lett. 309, 16–24. doi: 10.1111/j.1574-6968.2010.02009.x
Zhang, Y., Zhang, B., Wu, X. G., and Zhang, L. Q. (2020a). Characterization the role of GacA-dependent small RNAs and RsmA family proteins on 2,4-diacetylphloroglucinol production in Pseudomonas fluorescens 2P24. Microbiol. Res. 233:126391. doi: 10.1016/j.micres.2019.126391
Zhang, Y., Zhang, B., Wu, H., Wu, X., Yan, Q., and Zhang, L. Q. (2020b). Pleiotropic effects of RsmA and RsmE proteins in Pseudomonas fluorescens 2P24. BMC Microiol. 20:191. doi: 10.1186/s12866-020-01880-x
Zhang, Y., Zhang, Y., Zhang, B., Wu, X., and Zhang, L. Q. (2018). Effect of carbon sources on production of 2,4-diacetylphloroglucinol in Pseudomonas fluorescens 2P24. Acta Microbiol. Sin. 58, 1202–1212.
Zhou, Y. P., Wu, X. G., Zhou, H. Y., He, Y. Q., and Zhang, L. Q. (2010). Effect of gene phlF on 2,4-diacetylphloroglucinol production in Pseudomonas fluorescens 2P24. Acta Phytopathol. Sin. 40, 144–150.
Keywords: Pseudomonas fluorescens, GrxD, RsmA/RsmE, PhlF, 2,4-DAPG
Citation: Dong Q, Yan Q, Zhang B, Zhang L-q and Wu X (2022) Effect of the Monothiol Glutaredoxin GrxD on 2,4-Diacetylphloroglucinol Biosynthesis and Biocontrol Activity of Pseudomonas fluorescens 2P24. Front. Microbiol. 13:920793. doi: 10.3389/fmicb.2022.920793
Received: 15 April 2022; Accepted: 17 June 2022;
Published: 08 July 2022.
Edited by:
Haike Antelmann, Freie Universität Berlin, GermanyReviewed by:
Claudio Valverde, National University of Quilmes, ArgentinaEunhye Goo, Seoul National University, South Korea
Copyright © 2022 Dong, Yan, Zhang, Zhang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaogang Wu, NDE0NjMwODUwNUBvdXRsb29rLmNvbQ==
 Qiuling Dong
Qiuling Dong Qing Yan
Qing Yan Bo Zhang
Bo Zhang Li-qun Zhang
Li-qun Zhang Xiaogang Wu
Xiaogang Wu