
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 28 July 2022
Sec. Microbiological Chemistry and Geomicrobiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.918986
 Zhiqiang Tang1
Zhiqiang Tang1 Liying Zhang1
Liying Zhang1 Na He1
Na He1 Zhiqi Liu2
Zhiqi Liu2 Zuobin Ma1
Zuobin Ma1 Liang Fu1
Liang Fu1 Hui Wang2
Hui Wang2 Changhua Wang1
Changhua Wang1 Guomin Sui2*
Guomin Sui2* Wenjing Zheng1*
Wenjing Zheng1*A comprehensive understanding of rice cultivation techniques and organic amendments affecting soil quality, enzyme activities and bacterial community structure is crucial. We investigated two planting methods (direct seeding and transplanting) of paddy rice (Oryza sativa) and organic amendments with rice straw and biochar on crop yield and soil biological and physicochemical properties. Rhizosphere bacterial communities at the maturity stage of rice growth were characterized through high-throughput 16S rRNA sequencing. Soil biochemical properties and enzyme activity levels were analyzed. Grain yield of paddy rice with transplanting increased 10.6% more than that with direct seeding. The application of rice straw increased grain yield by 7.1 and 8.2%, more than with biochar and the control, respectively. Compared to biochar and the control, the application of rice straw significantly increased sucrase, cellulase, protease, organic carbon, available phosphorus, nitrate, and ammonium. The application of biochar increased microbial biomass nitrogen and carbon, urease, pH, available nitrogen, and available potassium compared to the application of rice straw and the control. Principal coordinate analysis and dissimilarity distances confirmed significant differences among the microbial communities associated with planting methods and organic amendments. Bacteroidetes, Nitrospirae, Firmicutes, and Gemmatimonadetes abundance increased with rice straw relative to biochar and the control. The biochar addition was associated with significant increases in Chloroflexi, Patescibacteria, Proteobacteria, and Actinobacteria abundance. Pearson’s correlation analyzes showed that Chloroflexi, Bacteroidetes and Nitrospirae abundance was positively correlated with grain yield. The relative abundance of these bacteria in soil may be beneficial for improving grain yield. These results suggest that planting methods and organic amendments impact soil biochemical characteristics, enzyme activity levels, and microbial community composition.
Rice production has a long history in China, and results in large volumes of crop residue byproducts (Liu et al., 2008). The two primary approaches to producing paddy rice (Oryza sativa) production are direct seeding and mechanized transplantation. Transplanting is a method to establish rice after growing seedlings in the nursery (Susilawati et al., 2021), and the field needs to go through the process of soaking, raking and settling. Transplanting is labor intensive and financially expensive. Dry direct seeding is a simple approach to rice cultivation in which rice seeds are sown on dry land, after that, a water layer is established following seedling emergence and development to the 3-leaf stage. Direct seeding obviates the need for seedling nurseries, seeding management, or transplanting, thereby improving overall mechanical efficiency and lowering labor costs (Ashraf et al., 2017a,b). Planting methods alter tilling systems in the field, which will affect the root growth environment and may indirectly affect the rhizosphere microbial community structure.
In Northeastern China, a common post-harvest approach to disposing of rice straw entails burning these crop residues after the annual harvest and before planting during the following spring. However, this process leads to the waste of potentially valuable crop resources, induces severe air pollution, and can accelerate the loss of organic matter from the underlying soil (Cao et al., 2008). Thus, more environmental friendly and efficient approaches to utilizing these natural resources are urgently needed. One strategy aimed at remediating this issue is the return of crop straw to the field, which can protect from soil degradation, thereby simultaneously reducing resource waste while maintaining soil fertility in intensive cultivation systems (Sun et al., 2015). Another approach relies on the preparing of rice straw-derived biochar that can then be applied to fields, thereby modulating soil quality characteristics in a manner beneficial to overall soil productivity, nutrient availability, and soil health (Novak et al., 2014). Biochar application can also reportedly improve the pH of acidic soils and reduce nutrient losses through the enhancement of soil water retention (Joseph et al., 2010).
Soil microbes are essential regulators of soil health, crop productivity, and sustainability of cultivated plants (Berendsen et al., 2012). The rhizosphere is the interface between plant roots and soil that is affected by environmental factors, microbial richness and diversity (Phillips et al., 2013). Recent studies have shown that plant roots selectively recruit specific root microbial communities from the soil, which impact crop growth, development, and grain yield (Edwards et al., 2015; Chen et al., 2019). Soil microorganisms and plants live in the same environment, interact and influence each other; many of which help crops obtain nutrients from the soil, promote the maintenance of soil fertility, and make crops grow healthily, and ultimately affect grain yield formation. Therefore, crops usually utilize rhizosphere microorganisms and roots for nutrient-absorbing, growth-promoting, and grain yield-improving (Fierer, 2017; Zhong et al., 2020; Wang et al., 2021). Therefore, understanding the relationship between crops and rhizosphere-related microbial communities is of great agronomic significance for improving agricultural productivity and sustainable development.
The straw application represents a practical approach to improving soil quality through increases in enzymatic activity and regulating soil microbial activity (Bastida et al., 2007). The straw application can influence the composition and diversity of soil microbial communities (Qin et al., 2015; Sun et al., 2016). Biochar has a high organic carbon content, which increases the soil carbon content after application to the soil and affects the community and diversity of microorganisms (Steinbeiss et al., 2009; Cayuela et al., 2013). As biochar is highly porous, it can serve as a favorable habitat in which many microorganisms can thrive and proliferate (Jia et al., 2017).
Soil enzymes are biologically active substances with specific catalytic functions involved in energy flow and the recycling of mineral elements in soil ecosystems. Soil enzyme activity is an essential indicator linking soil microbial community and soil nutrient availability (Nannipieri et al., 2017). Soil enzymes are one of the factors that reflect the activity of soil microorganisms, which can directly and indirectly reflect soil nutrient conditions and serve as an essential indicator for evaluating soil nutrient availability (Cotrufo et al., 2015).
Establishing the linkage between soil bacteria, enzyme activities and soil nutrients has been a long-standing and complex topic in soil ecology. We attempted to tackle this question with specific objectives: (1) to evaluate the impact of planting methods and organic amendments on the grain yield of paddy rice, (2) to determine the response pattern of rhizospere soil microbial taxonomy, microbial composition and soil biochemical properties and enzymes, and (3) to identify differences and similarities in bacteria communities as impacted by planting methods and organic amendments.
This study was conducted at the Liaoning Rice Research Institute (40°57′ N, 122°14′ W, altitude: 41.5 m) in Shenyang, Liaoning Province, China. The field experiment was carried out in 2019. The study site is located in a region with a semi-humid temperate and monsoonal climate, an annual average temperature of 8.3°C, and average yearly precipitation of 545 mm. This experiment was conducted using a split-plot, fully randomized complete block design with three replicates. Experimental treatments included paddy rice seeding methods [direct seeding (DS) and transplanting (TP)] and organic amendments [rice straw (RS) applied at 9750 kg ha–1, biochar (BC) applied at 3450 kg ha–1, and a control (CK) without rice straw or biochar amendment]. Application rate of RS is based on average amount of rice straw produced in the region. The soil at the study site was clay loam and contained 18.6 g kg–1 organic matter, 1.31 g kg–1 total N, 1.24 g kg–1 total P, 24.6 g kg–1 total K, 108.6 mg kg–1 available N, 23.8 mg kg–1 available P, 46.2 mg kg–1 available K, and had a pH (H2O) of 5.3 at the start of the study period. The ‘Liaoxing 21′ rice cultivar was used. This cultivar was selected as it was commonly grown by local farmers in the northern region of Liaoning Province. Rice straw was prepared by chopping dry straw into 20-50 mm pieces, yielding RS containing 35% C and 0.7% N. Biochar was manufactured via the pyrolysis of prepared RS under low-oxygen conditions at 450°C for 1 h (Jin and Fu Agriculture Development Co., Ltd.) (Sui et al., 2016). This approach converted approximately 35% of the RS into small granular (diameter: 2-5 mm) particles. The prepared BC contained 66% C and 0.8% N, and had a pH of 8.7 (1:2.5 H2O). Each plot was 240 m2 in size. RS and BC were applied to appropriate experimental plots in April 2019 before rice planting. These organic amendments were incorporated into the soil by hand with a rake, and all plots were mechanically plowed to a 200-mm depth. Compound fertilizers were applied as a basal fertilizer on the soil surface and mechanically incorporated into the 0–100-mm layer by raking. The soil was additionally amended with compound fertilizers (750 kg ha–1, containing 12% N, 12% P, and 12% K), with additional urea applied at the seedling establishment (270 kg ha–1) and at the mid-tillering stage of rice growth (270 kg ha–1). For the TP, rice seedlings were sown in a nursery bed on April 19, 2019, and were transplanted into paddy fields on May 26, 2019. For the DS, rice seeds were mechanically planted on April 27, 2019 under dry conditions (5.5 million seeds ha–1). All paddies were harvested on October 20, 2019. The same amount of N, P, and K was used for paddy rice with the TP and DS. Water levels, weed growth, insects, and diseases were managed as necessary to minimize yield losses.
On October 2, 2019, five hills of rice were selected from each plot, and soil samples with an area of 200 × 200 mm in a depth of 200 mm were excavated for each hill. Each hill consisted of approximately 15 plants. Briefly, five soil core samples from each plot were pooled together to generate a composite sample. Excessive soil was removed by shaking the roots of the plant until the roots still having soil attached. The roots were separated from the soil by vigorously shaking the root system with sterile forceps. The soil samples separated from the roots were then placed in a 500 mL sterile flask. Following vigorously stirring of this mixture using sterile forceps, the collected rhizosphere soil was separated into two aliquots, with one being frozen at –80°C in a 50 ml tube for subsequent DNA extraction, whereas the other was stored at 4°C to measure enzyme activity levels, microbial biomass, and soil physicochemical properties.
The activity of soil enzymes was measured using the method of Guan et al. (1986). Soil weight for determination of enzyme activity was 5 g. Soil saccharase activity was measured using the 3,5-dinitrosalicylic acid colorimetric method. The incubation was carried out at 37 °C for 24 h. The amount of glucose produced per gram of soil per day was defined as the unit of saccharase activity. Urease activity was determined by sodium phenol colorimetry. The incubation was carried out at 37 °C for 24 h. The production of 1 μg of ammonia nitrogen per gram of soil sample per day was defined as the unit of urease activity. Cellulase activity was determined using carboxymethy cellulose as a substrate and glucose as a product after incubation at 37°C for 72 h. Protease activity was determined by the ninhydrin colorimetric method. The incubation was carried out at 30°C for 24 h. The production of 1 mg of tyrosine per gram of soil per day was defined as the unit of protease activity. A chloroform fumigation-K2SO4 extraction approach was used to measure microbial biomass carbon (MBC) and nitrogen (MBN) (Brookes et al., 1985; Vance et al., 1987). The weight of soil used to determine the microbial biomass was 20 g. Briefly, these samples were fumigated with chloroform steam in a vacuum desiccator for 24 h, and residual chloroform was removed by repeated vacuuming. After adding 0.5 M K2SO4 or NaHCO3, respectively, the mixture was shaken for 30 min and filtered immediately. The filtered extract was used to measure MBC and MBN. The organic carbon and total nitrogen were used to calculate MBC and MBN. The conversion factor for MBC and MBN was 0.45 (Zang et al., 2015).
The pH of soil samples suspended in water at a 1:2.5 soil:water ratio (w/w) was determined with a pH meter. Soil available N (AN) was quantified via a NaOH diffusion approach (Lu, 2000). Two grams of soil samples were weighed and evenly distributed into the outer chamber of the diffusion dish, and 2 ml of the boric acid indicator was added to the inner chamber. 10mol/l sodium hydroxide solution was added to the soil for alkaline hydrolysis and diffusion at 40°C for 24 h. Soil available P (AP) was extracted using NaHCO3, and P concentrations in filtrate samples were measured via a molybdenum blue colorimetric method (Page et al., 1982). Soil available K (AK) was extracted using ammonium acetate at a pH of 7.0 and measured via flame photometry. Soil organic carbon was quantified with a K2Cr2O7-H2SO4 wet oxidation method (Vos et al., 2007).
A Fast DNA SPIN Kit for Soil (Q-BIOgene, CA, United States) was used to isolate total DNA from 0.5 g soil samples based on provided directions, after which the quantity and quality of the isolated DNA were assessed with an automated microplate reader (Synergy HTX, Gene Company Ltd.). The bacterial 16S rRNA V3-V4 region was amplified using the 338F (5′-ACTCCTACGGGAG GCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) primers (Cui et al., 2019). PCR amplification was performed in a total reaction volume of 10 μl containing DNA template (5–50 ng), Vn F (10 μM) 0.3 μl, Vn R (10 μM) 0.3 μl, KOD FX Neo Buffer 5 μl, dNTP (2 mM) 2 μl, KOD FX Neo 0.2 μl, ddH2O up to 10 μl (Yuan et al., 2019). After initial denaturation at 95°C for 5 min, samples underwent 25 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 40 s, with a final incubation at 72°C for 7 min. PCR amplicons were purified with Agencourt AMPure XP Beads (Beckman Coulter, Indianapolis, IN, United States). Finally, these PCR products were sent to Beijing Biomarker Technologies Co., Ltd. for sequencing.
FLASH was used to merge the paired-end read sequencing data produced with an Illumina MiSeq 2500 instrument (Magoč and Salzberg, 2011). The combined tags were compared with the primer sequences, and those tags with more than six mismatches were discarded to filter out low-quality tags, followed by mass filtering (Bolger et al., 2014). High-quality tagged sequences were obtained by eliminating chimeric sequences (Edgar et al., 2011), with the remaining sequences clustered at a 97% similarity level (Edgar, 2013), using an OTU filtering threshold of 0.005% (Schloss et al., 2009), yielding the final effective data (effective tags). The raw FASTQ files obtained in the study for the sequencing libraries have been deposited into NCBI Sequence Read Archive (SRA) under the BioProject accession number PRJNA833904.
The SAS MIXED procedure was used for data analyzes (Littell et al., 2006). A split-plot design was utilized for these analyzes, with seeding methods as the main plot and organic amendments as a subplot. Data were tested for normality (Shapiro-Wilk normality test) and homogeneity of variance, and were transformed as appropriate to ensure that they conformed to a normal distribution. When ANOVAs were performed with significance (P < 0.05) least significant difference (LSD) tests were used to assess the significance of the impact of different treatment conditions on crop yields, enzyme activity levels, microbial biomass, and soil properties. Bioinformatics analyzes were performed using BMK Cloud (www.biocloud.net). Alpha diversity analysis was performed to study microbial community richness and diversity. Principal coordinate analyzes (PCoA) and analysis of similarities based on Jaccard distance dissimilarity matrices were used to visualize soil bacterial communities with the R Vegan package (Oksanen et al., 2018). Relationships between soil enzyme activities, soil physicochemical properties and bacterial communities were visualized via a redundancy analysis (RDA) (The R Development Core Team, 2015). Pearson’s correlation coefficients were used to detect significant relationships among abundant phyla, enzyme activity levels, and soil properties.
Significant differences in grain yield and yield components were observed when comparing planting methods and organic amendments in the present study (Table 1). Specifically, TP was associated with significantly higher grain yields relative to DS (P < 0.05). Grain yield of paddy rice with TP increased 10.6% more than that with DS. RS amendment increased grain yields by 7.1 and 8.2%, more than with biochar and the control, respectively.
The Chao1 and Shannon indexes are shown in Figure 1. Regarding the richness of bacterial communities, the Chao 1 index showed that the bacterial richness with RS was significantly greater than that with BC and CK in DS. There was no difference in Chao 1 index among RS, BC and CK in TP. With TP, the Chao 1 index was higher than with DS. Shannon index in RS and CK treatments was significantly higher than that in the BC treatment under TP. Under DS, the Shannon index was substantially greater with RS than with BC and CK under DS. The Shannon index of TP was significantly higher than that of DS.
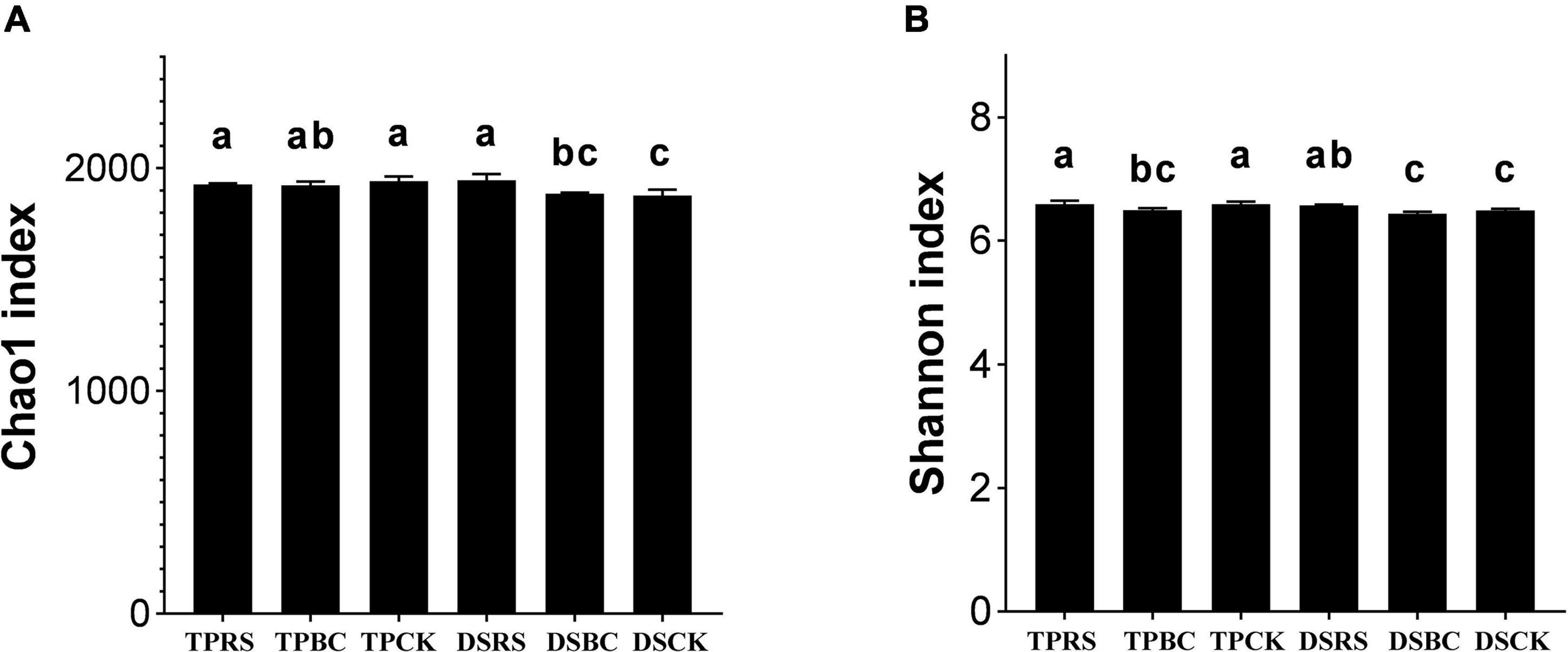
Figure 1. The diversity and richness of bacterial communities in the rhizosphere soil samples subjected to the planting methods and organic amendments characterized by the Chao (A) and Shannon indexes (B) (alpha diversity). Different letters indicate statistical significance at the P < 5% level within planting methods and organic amendments. TP, transplanting; DS, direct seeding; RS, rice straw; BC, biochar; CK, no RS or BC.
Jaccard distance values were used to conduct a principal coordinate analysis at the OTU level comparing the composition of the rhizosphere microbial community as a function of planting methods and organic amendments (Figure 2). Apparent differences in microbial communities were evident among these different treatment conditions, with significant differences between the microbial communities associated with TP and DS. Moreover, within each of TP and DS, the microbial communities were distinctly separated from one another.
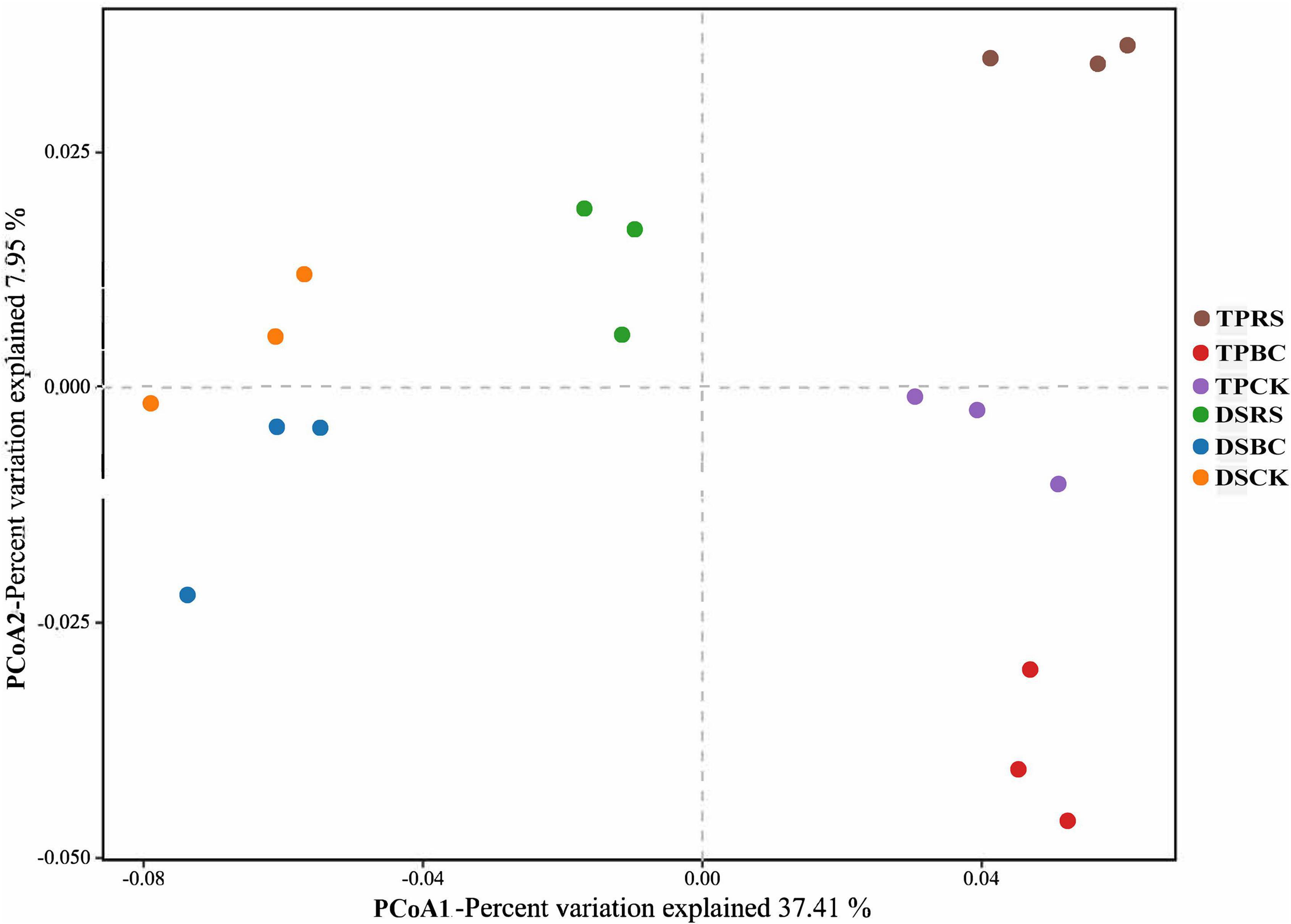
Figure 2. Principle coordinate analysis (PCoA) of the bacterial communities in rhizosphere soil samples. PCoA distances were based on the Jaccard distance algorithm at the OTU level. TP, transplanting; DS, direct seeding; RS, rice straw; BC, biochar; CK, no RS or BC.
Dissimilarity distances among these planting methods and organic amendments were further computed (Figure 3), revealing different distances in the rhizosphere soil bacterial community structures associated with TP. With DS differences in the rhizosphere bacterial communities exhibited more similar distance values with BC and CK, whereas they were distinct from the OTUs associated with the RS amendment.
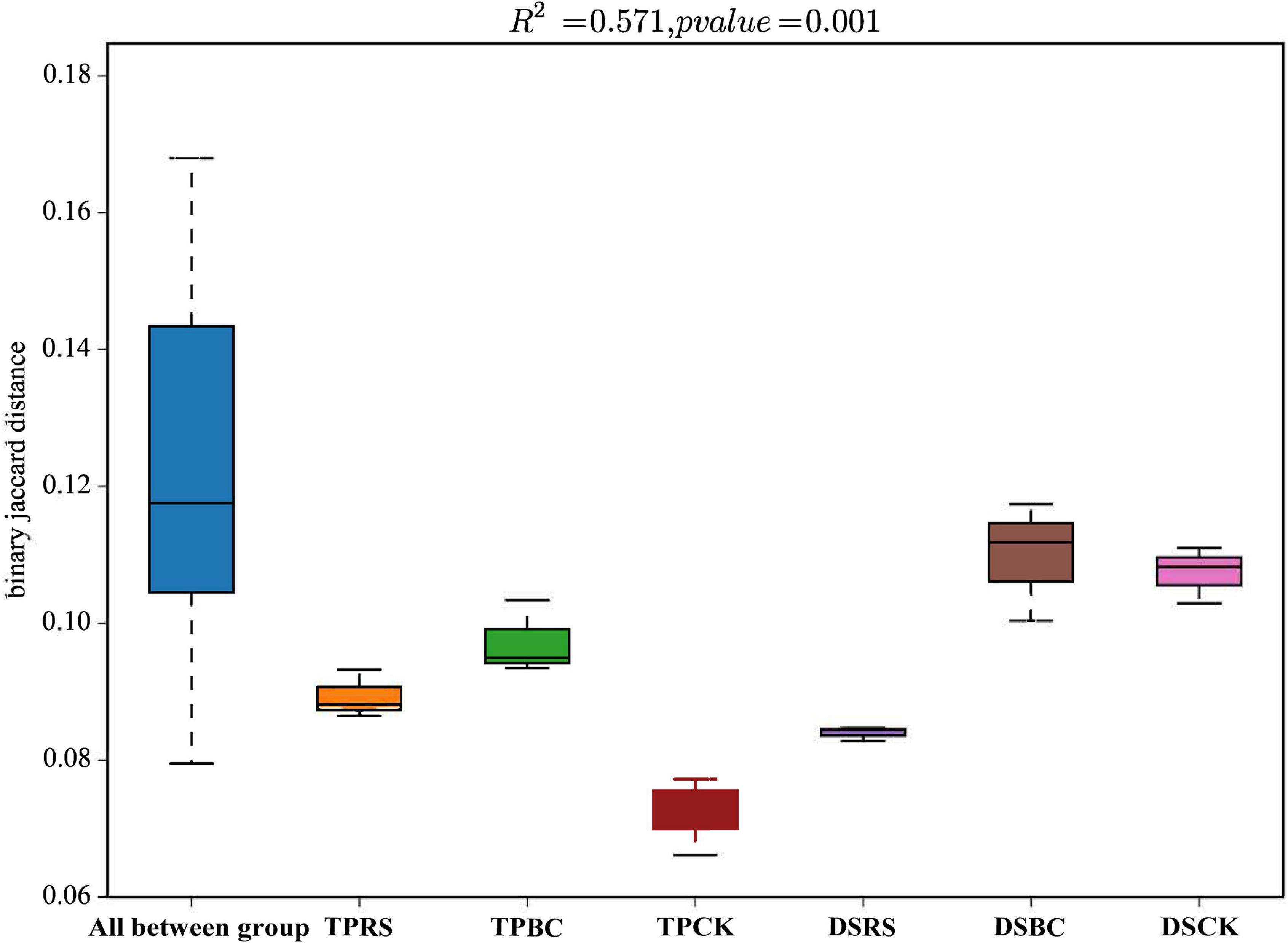
Figure 3. Dissimilarity distances showing the differences in bacterial communities in rhizosphere soil. Dissimilarity distances were based on the Jaccard distance algorithm at the OTU level. TP, transplanting; DS, direct seeding; RS, rice straw, BC, biochar; CK, no RS or BC.
High-throughput sequencing was used to explore the composition of the rhizosphere soil bacterial communities associated with planting methods and organic amendments (Figure 4). Diverse phylum level distributions were determined for each treatment. Among the 29 phyla (Figure 4), 193 orders, 299 families and 480 genus identified in the post-processed dataset, we found that across all investigated depths, the abundant phyla (relative abundance>1%) were Proteobacteria (28.6%), Acidobacteria (23.9%), Chloroflexi (21.5%), Actinobacteria (7.3%), Gammaproteobacteria (6.2%), Verrucomicrobia (2.4%), Nitrospirae (2.0%), Patescibacteria (2.0%) and Bacteroidetes (1.8%), reflecting abundance patterns in planting methods and organic amendments. Their relative abundances, as well as those of all orders and genus exceeding 1% relative abundance, are shown in Supplementary Figures 1–3.
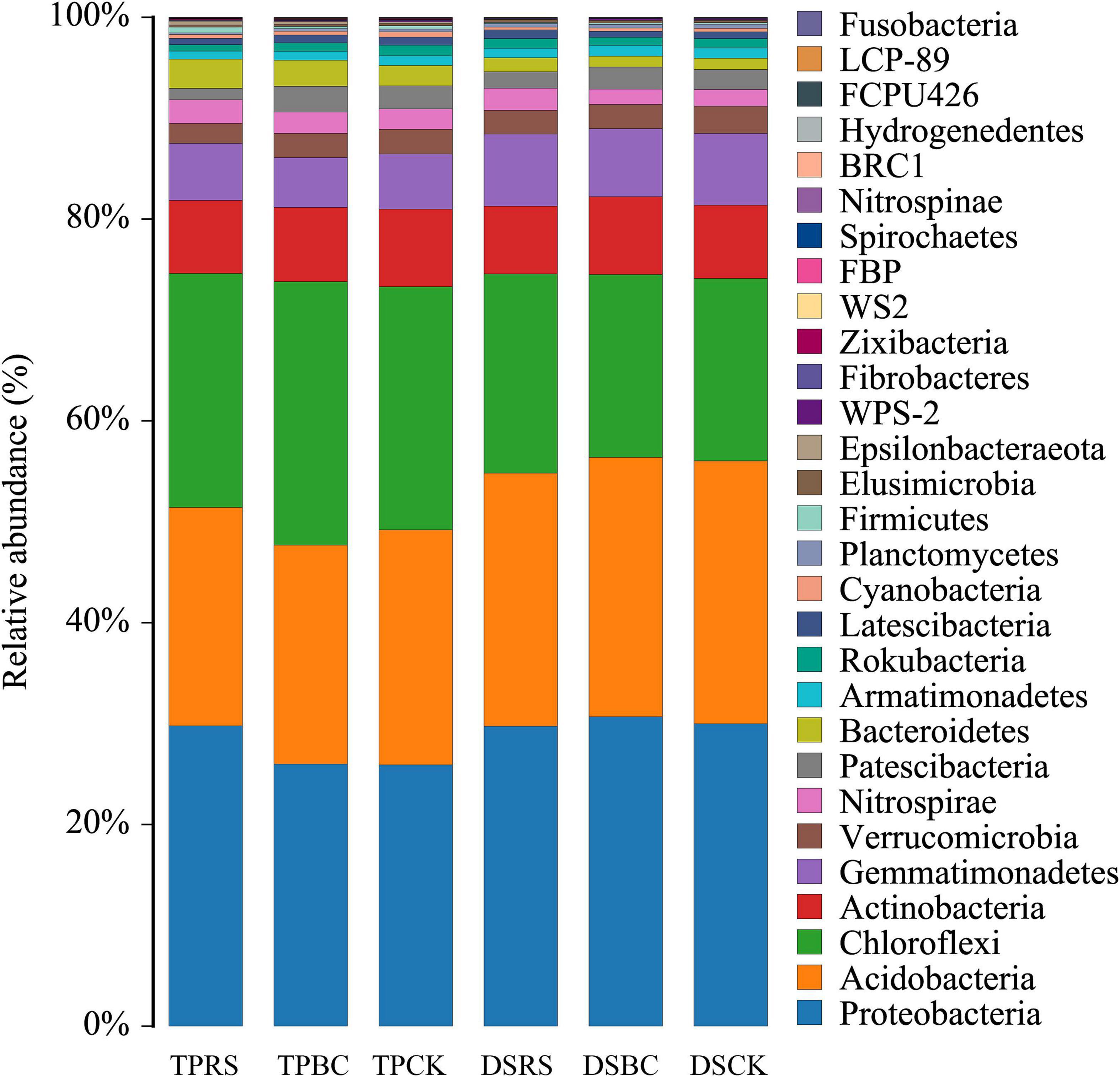
Figure 4. Phylum level bacterial community composition following the rhizosphere treatments. Relative abundance of different bacterial phyla in each treatments. TP, transplanting; DS, direct seeding; RS, rice straw; BC, biochar; CK, no RS or BC.
Bacteroidetes, Nitrospirae, and Firmicutes abundance were increased under the RS for TP relative to other treatment conditions, while Gemmatimonadetes abundance was significantly increased under the RS for DS. The BC addition was associated with significant increases in Chloroflexi and Patescibacteria abundance for TP and with significant increases in Proteobacteria and Actinobacteria abundance for DS. Acidobacteria, Actinobacteria, Patescibacteria, Gemmatimonadetes, and Verrucomicrobia were less abundant in the soil collected from TP relative to DS, while Chloroflexi, Bacteroidetes, Nitrospirae, and Firmicutes were more abundant in the soil collected from TP.
Soil sucrase activity (SA), urease activity (UA), cellulose activity (CA), protease activity (PA), MBN, and MBC differed significantly among treatment conditions (Table 2). These selected enzymes were used to evaluate the level of nutrients and the decomposition of organic matter. Specifically, MBN, MBC, and CA decreased significantly for TP relative to DS, whereas SA and PA exhibited the opposite trend. RS amendment was associated with an increase in SA, CA, and PA relative to BC and CK amendment conditions, while MBN, MBC, and UA increased significantly under conditions of BC amendment relative to RS and CK amendment. RS application improved rhizosphere soil SA and PA more effectively for TP related to DS. Under conditions of DS, BC amendment was associated with higher MBN, MBC, and UA relative to RS and CK treatment conditions, while RS amendment improved rhizosphere soil CA relative to BC and CK treatment conditions.
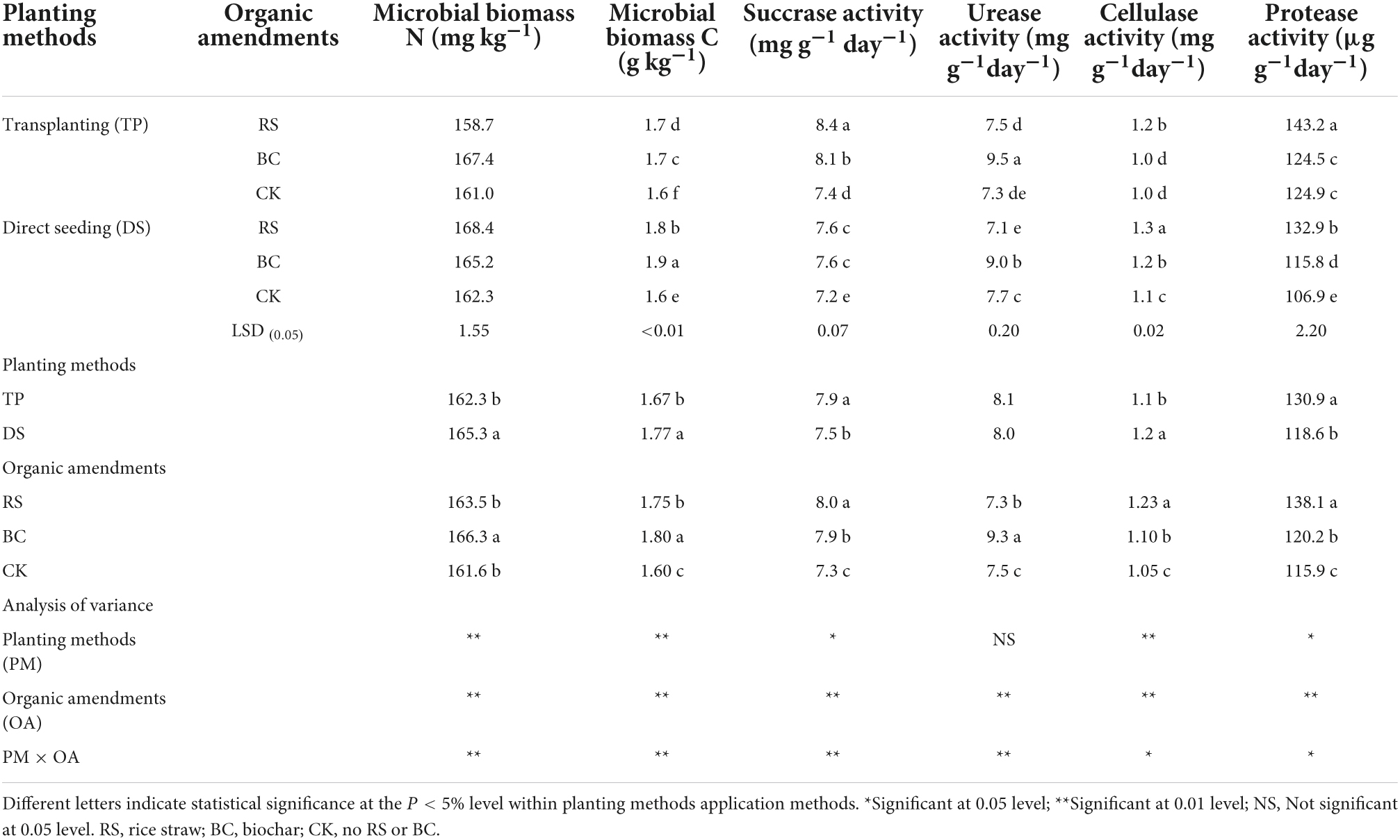
Table 2. Soil enzyme activity and microbial biomass under different treatments of planting methods and organic amendments.
There were significant differences in soil pH, organic C (OC), available N (AN), available P (AP), available K (AK), NO3–-N (NON), and NH4+-N (NHN) between the different seeding methods and among organic amendments (P < 0.05) (Table 3). Specifically, OC and NHN were significantly higher for TP relative to DS, while the opposite trend was observed for AP and AK. RS amendment was associated with increases in OC, AP, NON, and NHN contents relative to BC and CK amendment conditions. BC application led to increases in pH, AN, and AK relative to RS and CK. RS application was associated with increased rhizosphere soil NHN and OC for TP and with increased rhizosphere soil AP for DS, while BC application improved rhizosphere soil AK contents.
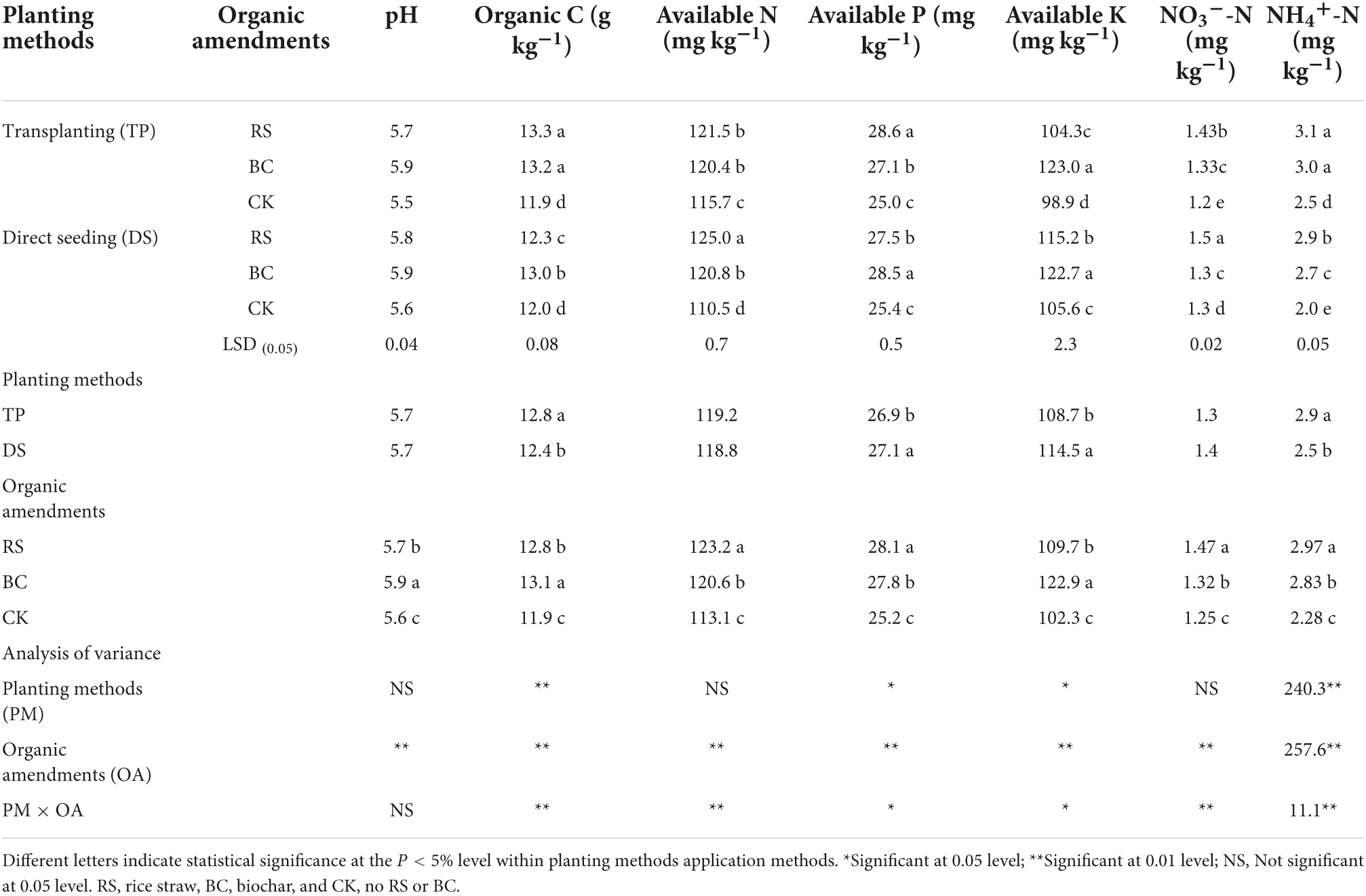
Table 3. Soil physicochemical characteristics under different treatments of planting methods and organic amendments.
RDA ordination plots were further used to explore relationships among soil bacterial communities, soil physicochemical properties, and soil enzyme activity levels. Both planting methods and organic amendments were associated with distinct impacts on soil bacterial communities under these tested conditions (Figure 5). While rhizosphere soil bacteria associated with the CK and BC amendments were similar to one another under both planting methods, they were clearly separated from those bacterial communities associated with the RS. Separation was also evident between bacterial communities associated with DS and TP. SA, PA, AN, AP, SOC, NON, and NHN were significantly correlated with soil bacterial community composition under conditions of RS amendment, while CA and NON were significantly correlated with soil bacterial communities when RS was applied under DS (Figure 5).
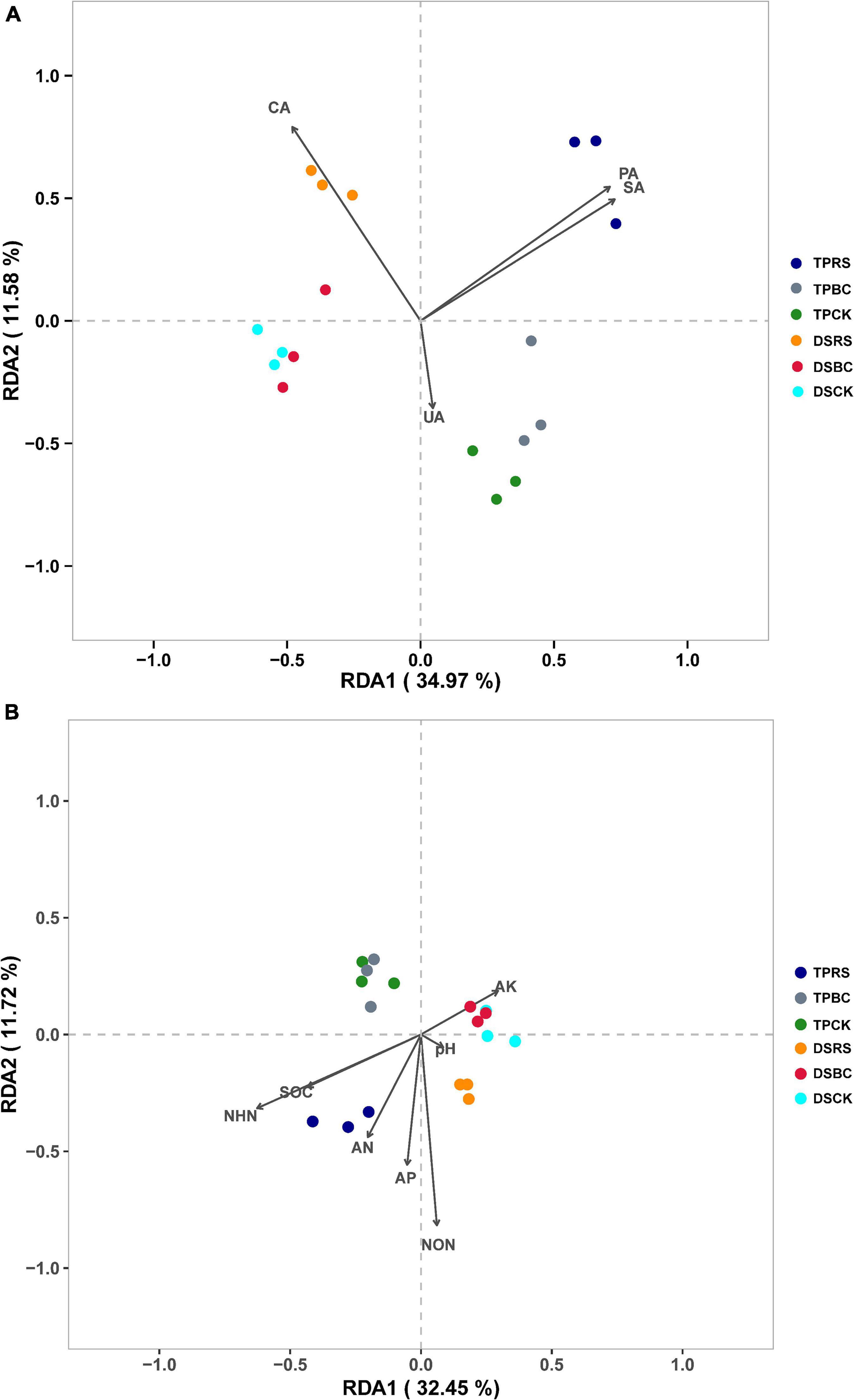
Figure 5. (A,B) Redundancy analysis of rhizosphere soil bacterial genera and soil enzyme activity and physicochemical characteristics.
Pearson’s correlation analyzes were used to examine relationships among rhizosphere bacterial phyla abundance, soil properties, and soil enzyme activity levels (Tables 4–6). Bacteroidetes abundance was positively correlated with SA, PA, SOC, NHN, full grains per panicle and yield. Chloroflexi abundance was negatively correlated with CA and NON, but positively correlated with soil SA, NHN, full grains per panicle and yield. Gemmatimonadetes abundance was positively correlated with CA and number of panicles, but negatively correlated with full grains per panicle and yield. Nitrospirae abundance was positively correlated with SA, PA, NHN and yield. Patescibacteria abundance was negatively correlated with CA, PA and NON, but positively correlated with soil UA. Proteobacteria abundance was positively correlated with CA, number of panicles.and 1000 seed weight. Verrucomicrobia abundance was negatively correlated with UA, PA, SOC, AN, AP, NHN and yield. The Supplementary Tables 1, 2 showed that SA was positively correlated with full grains per panicle and yield, but negatively correlated with number of panicles. Protease activity and NHN were positively correlated with full grains per panicle and yield. Cellulose activity was positively correlated with number of panicles and 1000 seed weight. The Supplementary Table 3 showed that SA was positively correlated with OC, AN, AP, NON and NHN, and UA was positively correlated with pH, OC and AK. Cellulose activity was positively correlated with pH, AN, AP and NON. Protease activity was positively correlated with AN, AP, NON and NHN.
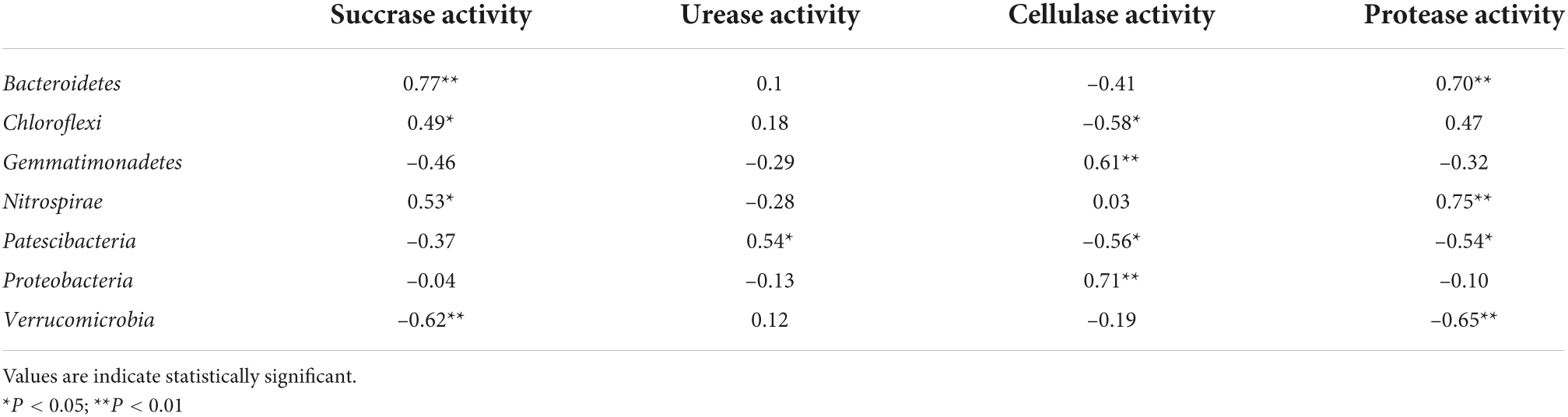
Table 4. Pearson’s correlation coefficients between abundant phyla, microbial biomass and enzyme activities.
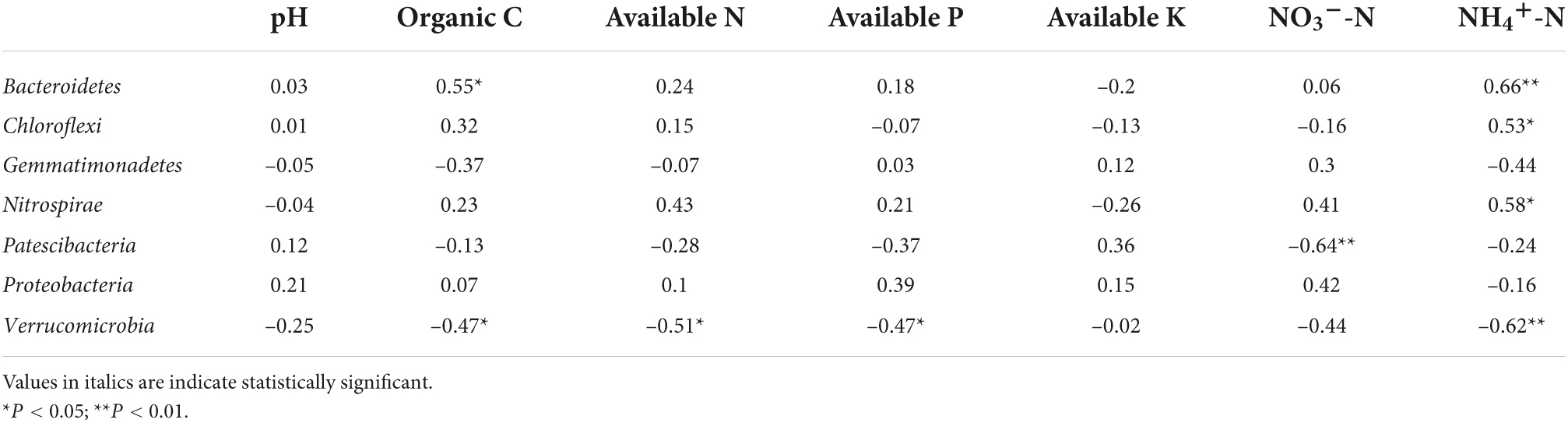
Table 5. Pearson’s correlation coefficients between abundant phyla and soil physicochemical properties.
DS and TP are the two primary approaches to paddy rice planting. Incorporation of both RS and BC has shown to be a practical approach to enhancing soil quality and productivity (Zhao et al., 2014a; Wang et al., 2015). Crop straw return in the present study was associated with increases in grain yields (Table 1), in line with prior evidence that RS, combined with the surface application of chemical fertilizers, can significantly enhance crop yields (Zhao et al., 2017; Kumari et al., 2018). However, these results are not universal, with some researchers reporting an absence of grain yield increase in the context of crop straw return and fertilizer application (Sun et al., 2018). In this study, RS significantly increased grain yields concerning the number of panicles and the number of full grains per panicle. In prior reports, TP was shown to be necessary to achieve reliably high crop yields with respect to an increase in the spikelet number per panicle, consistent with an increase in the number of filled grains per panicle relative to DS (Kamiji et al., 2011; Xing et al., 2017). These results are consistent with our findings. The observed increase in grain yields for the TP relative to DS was primarily attributable to the rise in the number of filled grains per panicle. With TP, the sowing date was earlier and the duration before heading was longer than that with DS, which were conducive to improve dry matter of crops, resulting in a larger panicle and improving grain yield (Chen et al., 2009). Compared to DS, SA, PA and NHN improved with TP (Tables 2,3), and SA, PA and NHN were positively correlated with full grains per panicle (Supplementary Tables 1, 2). Transplanting had more filled grains per panicle which may be an essential factor for improving grain yield.
The positive impact of RS and BC amendments in the present study is mainly likely attributable in large part to improvements in soil physicochemical properties. For instance, RS amendment was associated with increases in OC, AP, NON, and NHN contents and BC application increased pH, AN, and AK (Ding et al., 2016; Blanco-Canqui, 2017; Chen et al., 2017). Indeed, improvements in several soil properties were observed following RS or BC amendment relative to CK, in line with prior reports (Chen et al., 2017; Andrey et al., 2020). Specifically, RS application was associated with significant increases in paddy soil OC, AP, NON and NHN, potentially owing to the enhanced decomposition of RS by enzymes and microbes within the soil, leading to enhanced straw nutrient release into the soil. Biochar was also improved soil pH, AN and AK contents. Prior reports suggest that BC application can increase the pH of acidic soils and reduce nutrient loss through the enhancement of water retention (Joseph et al., 2010), consistent with our results.
The activity of soil enzymes can offer valuable insight regarding nutrient cycling (Nannipieri et al., 2003; Aon and Colaneri, 2011), as these activity levels regulate rates of nutrient cycling and organic matter decomposition in the soil (Nannipieri et al., 2012). In this study, both planting methods and organic amendments were associated with altered rhizosphere soil enzyme activity and microbial biomass (Table 2). Significant improvements in soil SA and PA were observed for TP, which may be owing to the high content of NHN, that was positively correlated with soil SA and PA. In contrast, DS was associated with improved rhizosphere soil MBN, MBC and CA, because of the higher AK content in DS (Supplementary Table 3). The RS application improved the rhizosphere soil AN, AP, NON, and NHN. The AN, AP, NON and NHN were positively correlated with SA, CA and PA (Supplementary Table 3), The application of RS increased SA, CA, and PA. Because of the addition of readily degraded substrate RS contributed to increases in sucrase, cellulase, and protease activity, in line with prior reports (Sardans et al., 2008; Wu et al., 2013; Wei et al., 2015; Zhu et al., 2017; Bao et al., 2020). These soil enzymes and associated microbes break RS down into cellulose and hemicelluloses that are then further degraded to yield carbohydrates, starches, polysaccharides, and simple sugars (Hansen et al., 2016). Soil enzymes and microbes associated with the RS-amended soil are more efficient in carbohydrate substrate utilization. In contrast, the porous structure of BC is associated with enhanced N and C fixing in the soil, enabling certain beneficial species of microbes to thrive (Hagemann et al., 2017). This is likely the primary explanation for the observed increases in UA and soil MBC and MBN in BC-amended soil relative to those observed in CK or RS amendment conditions.
Sustainable and productive cropping systems necessitate the maintenance of pronounced microbial diversity and richness (Zhao et al., 2014b). A loss of such diversity often occurs in the context of prolonged agricultural cultivation, while RS and BC amendments can positively impact both bacterial diversity and abundance (Sun et al., 2016; Chen et al., 2017), consistent with the present results.
The abundance of Chloroflexi, Bacteroidetes, and Nitrospirae increased in the RS-amended TP (Figure 4). In TP, RS amendment improved soil NHN, SA and PA, whereas the Chloroflexi, Bacteroidetes and Nitrospirae abundance were positively correlated with SA, PA, NHN, and grain yield (Tables 4–6). The increased relative abundance of these microbial in the soil is beneficial for improving grain yield. Gemmatimonadetes abundance was significantly increased in the RS-amended DS. Gemmatimonadetes, highly abundant phyla associated with RS amendment, have been shown to reduce cellulose metabolic byproducts, thus indirectly facilitating the degradation of cellulose (Takaichi et al., 2010). Gemmatimonadetes abundance was positively correlated with CA, but negatively correlated with full grains per panicle and yield. This may be the reason why the grain yield of DS was lower than TP.
The RDA ordination plots indicated that both seeding methods and organic amendments had an impact on the structure and rhizosphere soil bacterial communities (Figure 5). Zhao et al. (2017) found that straw addition was associated with pronounced separation from control soil bacterial communities, with the composition of these communities significantly correlated with soil physicochemical properties. In the present study, the soil bacterial communities associated with the CK and BC amendment conditions were similar and clearly separated from those associated with RS amendment for both DS and TP. These data suggest that RS can markedly influence rhizosphere soil bacterial communities. Under conditions of RS amendment, SA, CA, PA, AN, AP, NON, and NHN were significantly correlated with bacterial communities, owing to the participation of many microbes and soil enzymes in the context of straw decomposition and nutrient consumption, in line with prior reports (Zhao et al., 2017; Chen et al., 2020). Under conditions of BC amendment, UA and pH were significantly correlated with bacterial communities (Figure 5). This may be attributable to the porous structure of BC, resulting in soil nutrient immobilization, or to its alkalinity. RDA ordination plots indicated that all three organic amendments were separated from one another under both DS and TP. For DS, the original plow layer was destroyed and the water layer could not be maintained throughout the entirety of the rice-growing period. The consequent alternating wetting and drying of the soil may have contributed to changes in rhizosphere soil microorganisms.
Both RS and BC additions affected rhizosphere soil nutrient concentrations, enzymatic activity, microbial diversity, and abundance. Copitrophic bacteria thrive when resource levels are abundant, whereas oligotrophs exhibit greater relative abundance when resource levels are limited (Eilers et al., 2010). The results of the present study revealed a positive correlation between Proteobacteria abundance and CA, while Gemmatimonadetes abundance was positively correlated with CA. In addition, Nitrospirae abundance was positively correlated with SA, PA and NHN, whereas Bacteroidetes abundance was positively correlated with SA, PA, OC and NHN. These data align well with prior reports (Ramirez et al., 2012), suggesting that RS and BC can increase local nutrient contents, thereby preferentially promoting the growth of bacteria best suited to nutrient-rich environments, including Proteobacteria and Nitrospirae. These data thus support the ability of organic amendments to promote the growth of copiotrophic bacteria, which has the potential to enhance the productivity and sustainability of soil under intensive agricultural systems.
Our study proved that both planting methods and organic amendments altered the abundance and diversity of bacterial communities in the rhizosphere soil. RS application could enhance yields of paddy rice under DS and TP. Moreover, both RS and BC applications were sufficient to improve soil nutrient availability and enzymatic activity under DS and TP. RS application thus significantly influenced soil microbial community structure, promoted the preferential growth of copiotrophic bacteria, and increased grain yield. This study marks a crucial step toward a more in-depth knowledge of the mechanisms by which planting methods and organic amendments influence the composition and variety of rhizosphere soil microbial communities, soil enzyme activity levels and soil physicochemical attributes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: SUB11374070, PRJNA833904.
ZT contributed significantly to the field experiment, data collection and analysis, and wrote the original manuscript. LZ, NH, ZM, LF, HW, and CW performed the field experiment. WZ and GS helped to perform the analysis with constructive discussions and contributed to the conception of the study. All authors contributed to the article and approved the submitted version.
We gratefully acknowledge the earmarked fund for China Agriculture Research System (CARS-01-55); China-Russia Cooperation Rice Breeding Laboratory (2019LHSYS04); Shenyang Scientific and technological Innovation Talents Support Program (RC210489); and Key Project of Institute of Atmospheric Environment, CMA, Shenyang (2021SYIAEZD1) support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.918986/full#supplementary-material
Andrey, V. G., Tatiana, M. M., and Saglara, S. (2020). The mechanisms of biochar interactions with microorganisms in soil. Environ. Geochem. Health 42, 2495–2518. doi: 10.1007/s10653-019-00412-5
Aon, M. A., and Colaneri, A. C. (2011). II. Temporal and spatial evolution of enzymatic activities and physico-chemical properties in an agricultural soil. Appl. Soil Ecol. 18, 255–270. doi: 10.1016/S0929-1393(01)00161-5
Ashraf, U., Mo, Z., and Tang, X. (2017a). “Direct seeded vs transplanted rice in Asia: climatic effects, relative performance and constraints-a comparative outlook,” in Recent Advances and Innovation in Rice Research, eds D. K. Verma, P. P. Srivastav, and B. Nadaf (Oakville, ON: Tylor and Francis, Apple Academic Press).
Ashraf, U., Hussain, S., Sher, A., Abrar, M., Khan, I., and Anjum, S. A. (2017b). “Planting geometry and herbicides for weed control in rice: implications and challenges,” in Grasses as Food and Feed, ed. Z. Tadele (London: InTech - open science- open minds).
Bao, Y., Feng, Y. J., Stegen, J., Wu, M., and Lin, X. (2020). Straw chemistry links the assembly of bacterial communities to decomposition in paddy soils. Soil Biol. Biochem. 148:107866. doi: 10.1016/j.soilbio.2020.107866
Bastida, F., Kandeler, E., Hernández, T., and García, C. (2007). Long-term effect of municipal solid waste amendment on microbial abundance and humus-associated enzyme activities under semiarid conditions. Microb. Ecol. 55, 651–661. doi: 10.1007/s00248-007-9308-0
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Blanco-Canqui, H. (2017). Biochar and soil physical properties. Soil Sci. Soc. Am. J. 81:687. doi: 10.2136/sssaj2017.01.0017
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brookes, P. C., Landman, A., Pruden, G., and Jenkinson, D. S. (1985). Chloroform fumigation and the release of soil nitrogen: a rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 17, 837–842. doi: 10.1016/0038-0717(85)90144-0
Cao, G., Zhang, X., Wang, Y., and Zheng, F. (2008). Estimation of emissions from field burning of crop straw in China. Chinese Sci. Bull. 53, 784–790. doi: 10.1007/s11434-008-0145-4
Cayuela, M. L., Sánchez-Monedero, M. A., Roig, A., Hanley, K., Enders, A., and Lehmann, J. (2013). Biochar and denitrifcation in soils: when, how much and why does biochar reduce N2O emissions? Sci. Rep. 3:1732. doi: 10.1038/srep01732
Chen, S., Cai, S.-G., Chen, X., and Zhang, G. P. (2009). Genotypic differences in growth and physiological responses to transplanting and direct seeding cultivation in rice. Rice Sci. 16, 143–150. doi: 10.1016/S1672-6308(08)60071-2
Chen, S., Waghmode, T. R., Sun, R., Kuramae, E. E., and Liu, B. (2019). Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 7:136. doi: 10.1186/s40168-019-0750-2
Chen, Y. L., Xin, L., Liu, J. T., Yuan, M. Z., Liu, S. T., Jiang, W., et al. (2017). Changes in bacterial community of soil induced by long-term straw returning. Sci. Agricola 74, 349–356. doi: 10.1590/1678-992x-2016-0025
Chen, Y. Y., Fan, P. F., Lin, L., Tian, H., and Pan, S. G. (2020). Straw incorporation coupled with deep placement of nitrogen fertilizer improved grain yield and nitrogen use efficiency in direct-seeded rice. J. Soil Sci. Plant Nutrition 20, 2338–2347. doi: 10.1007/s42729-020-00301-2
Cotrufo, M. F., Soong, J. L., Horton, A. J., Campbell, E. E., Haddix, M., Wall, D. H., et al. (2015). Formation of soil organic matter via biochemical and physical pathways of litter mass loss. Nat. Geo. sci. 7, 1–13. doi: 10.1038/ngeo2520
Cui, Y. X., Bing, H. J., Fang, L. C., Wu, Y. H., Yu, J. L., Shen, M. J., et al. (2019). Diversity patterns of the rhizosphere and bulk soil microbial communities along an altitudinal gradient in an alpine ecosystem of the eastern Tibetan Plateau. Geoderma 338, 118–127. doi: 10.1016/j.geoderma.2018.11.047
Ding, Y., Liu, Y., Liu, S., Li, Z., Tan, X., Huang, X., et al. (2016). Biochar to improve soil fertility. a review. Agronomy Sustainable Dev. 36:36. doi: 10.1007/s13593-016-0372-z
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., and Sundaresan, V. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U S A. 112, E911–E920. doi: 10.1073/pnas.1414592112
Eilers, K. G., Lauber, C. L., Knight, R., and Fierer, N. (2010). Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol. Biochem. 42, 896–903. doi: 10.1016/j.soilbio.2010.02.003
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Guan, S. Y., Zhang, D., and Zhang, Z. (1986). Soil Enzyme and its Research Methods. Beijing: China Agriculture Press.
Hagemann, N., Joseph, S., Schmidt, H. P., Kammann, C. I., Harter, J., Borch, T., et al. (2017). Organic coating on biochar explains its nutrient retention and stimulation of soil fertility. Na. Commun. 8:1089. doi: 10.1038/s41467-017-01123-0
Hansen, V., Muller-Stover, D., Munkholm, L. J., Peltre, C., Hauggaard, N. H., and Jensen, L. S. (2016). The effect of straw and wood gasification biochar on carbon sequestration, selected soil fertility indicators and functional groups in soil: an incubation study. Geoderma 269, 99–107. doi: 10.1016/j.geoderma.2016.01.033
Jia, R., Qu, Z., You, P., and Qu, D. (2017). Efect of biochar on photosynthetic microorganism growth and iron cycling in paddy soil under diferent phosphate levels. Sci. Total Environ. 612, 223–230. doi: 10.1016/j.scitotenv.2017.08.126
Joseph, S. D., Camps-Arbestain, M., Lin, Y., Munroe, P., Chia, C. H., Hook, J., et al. (2010). An investigation into the reactions of biochar in soil. Soil Res. 48, 501–515. doi: 10.1071/SR10009
Kamiji, Y., Yoshida, H., Palta, J. A., Sakuratani, T., and Shiraiwa, T. (2011). N applications that increase plant N during panicle development are highly effective in increasing spikelet number in rice. Field Crops Res. 122, 242–247. doi: 10.1016/j.fcr.2011.03.016
Kumari, K., Prasad, J., Solanki, I. S., and Chaudhary, R. (2018). Long-term effect of crop residues incorporation on yield and soil physical properties under rice-wheat cropping system in calcareous soil. J. Soil Sci. Plant Nutrition 18, 27–40. doi: 10.4067/S0718-95162018005000103
Littell, R. C., Milliken, G. A., Stroup, W. W., Wolfinger, R. D., and Schabenberger, O. (2006). SAS for Mixed Models, 2nd Edn. Cary, NC: SAS Institute Inc.
Liu, H., Jiang, G. M., Zhuang, H. Y., and Wang, K. J. (2008). Distribution, utilization structure and potential of biomass resources in rural China: with special references of crop residues. Renew Sust. Energ. Rev. 12, 1402–1418. doi: 10.1016/j.rser.2007.01.011
Lu, R. K. (2000). Soil Agricultural Chemical Analysis Method. Beijing: China Agricultural Science and Technology Press.
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Nannipieri, P., Ascher, J., Ceccherini, M., Landi, L., Pietramellara, G., and Renella, G. (2003). Microbial diversity and soil functions. Eur. J. Soil Sci. 54, 655–670. doi: 10.1046/j.1351-0754.2003.0556.x
Nannipieri, P., Giagnoni, L., Renella, G., Puglisi, E., Ceccanti, B., Masciandaro, G., et al. (2012). Soil enzymology: classical and molecular approaches. Biol. Fertil. Soils 48, 743–762. doi: 10.1007/s00374-012-0723-0
Nannipieri, P., Trasar-Cepeda, C., and Dick, R. P. (2017). Soil enzyme activity: a brief history and biochemistry as a basis for appropriate interpretations and meta-analysis. Biol. Fertil. Soils 54, 11–19. doi: 10.1007/s00374-017-1245-6
Novak, J. M., Cantrell, K. B., Watts, D. W., Busscher, W. J., and Johnson, M. G. (2014). Designing relevant biochars as soil amendments using lignocellulosic-based and manure-based feed stocks. Soils Sediments 14, 330–343. doi: 10.1007/s11368-013-0680-8
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R. O’Hara, R. B., et al. (2018). Vegan: Community Ecology Package. Ordination Methods, Diversity Analysis and Other Functions for Community and Vegetation Ecologists. Version 2.5-1. Available online at: http://CRAN.R-project.org/package=vegan (accessed March 15, 2021).
Page, A. L., Miller, R. H., and Keeney, D. R. (ed.). (1982). “Methods of soil analysis,” in Chemical and Microbiological Properties (Madison, WI: American Society of Agronomy), 1035–1049.
Phillips, R. P., Brzostek, E., and Midgley, M. G. (2013). The mycorrhizal-associated nutrient economy: a new framework for predicting carbon–nutrient couplings in temperate forests. New Phytol. 199, 41–51. doi: 10.1111/nph.12221
Qin, S., Jiao, K., Lyu, D., Shi, L., and Liu, L. (2015). Effects of maize residue and cellulose-decomposing bacteria inocula on soil microbial community, functional diversity, organic fractions, and growth of Malus hupehensis Rehd. Arch. Agron. Soil Sci. 61, 173–184. doi: 10.1080/03650340.2014.928927
Ramirez, K. S., Craine, J. M., and Fierer, N. (2012). Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob. Chang Biol. 18, 1918–1927. doi: 10.1111/j.1365-2486.2012.02639.x
Sardans, J., Peñuelas, J., and Estiarte, M. (2008). Changes in soil enzymes related to C and N cycle and in soil C and N content under prolonged warming and drought in a Mediterranean shrubland. Appl. Soil Ecol. 39, 223–235. doi: 10.1016/j.apsoil.2007.12.011
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Steinbeiss, S., Gleixner, G., and Antonietti, M. (2009). Effect of biochar amendment on soil carbon balance and soil microbial activity. Soil Biol. Biochem. 41, 1301–1310. doi: 10.1016/j.soilbio.2009.03.016
Sui, Y. H., Gao, J. P., Liu, C. H., Zhang, W. Z., Lan, Y., Li, S. H., et al. (2016). Interactive effects of straw-derived biochar and N fertilization on soil C storage and rice productivity in rice paddies of Northeast China. Sci. Total Environ. 544, 203–210. doi: 10.1016/j.scitotenv.2015.11.079
Sun, L., Wu, Z., Ma, Y., Liu, Y., and Xiong, Z. (2018). Ammonia volatilization and atmospheric N deposition following straw and urea application from a rice-wheat rotation in Southern China. Atmos Environ. 181, 97–105. doi: 10.1016/j.atmosenv.2018.02.050
Sun, L., Xun, W., Huang, T., Zhang, G., Gao, J., Ran, W., et al. (2016). Alteration of the soil bacterial community during parent material maturation driven by different fertilization treatments. Soil Biol. Biochem. 96, 207–215. doi: 10.1016/j.soilbio.2016.02.011
Sun, R., Zhang, X. X., Guo, X., Wang, D., and Chu, H. (2015). Bacterial diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biol. Biochem. 88, 9–18. doi: 10.1016/j.soilbio.2015.05.007
Susilawati, H. L., Kartikawati, R., and Setyanto, P. (2021). Response of planting methods to rice productivity and greenhouse gas emissions. IOP Conf. Series: Earth Environ. Sci. 648:012121. doi: 10.1088/1755-1315/648/1/012121
Takaichi, S., Maoka, M., Takasaki, K., and Hanada, S. (2010). Carotenoids of Gemmatimonas aurantiaca (Gemmatimonadetes): identifcation of a novel carotenoiddeoxyoscillol 2-rhamnoside, and proposed biosynthetic pathway of oscillol 2, 29-dirhamnoside. Microbiology 156, 757–763. doi: 10.1099/mic.0.034249-0
The R Development Core Team (2015). R: A Language and Environment For Statistical Computing. Vienna: R Foundation for Statistical Computing.
Vance, E. D., Brookes, P. C., and Jenkinson, D. S. (1987). An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 19, 703–707. doi: 10.1016/0038-0717(87)90052-6
Vos, B. D., Lettens, S., Muys, B., and Deckers, J. A. (2007). Walkley–Black analysis of forest soil organic carbon: recovery, limitations and uncertainty. Soil Use Manag. 23, 221–229. doi: 10.1111/j.1475-2743.2007.00084.x
Wang, J., Wang, X., Xu, M., Feng, G., Zhang, W., and Lu, C. A. (2015). Crop yield and soil organic matter after long-term straw return to soil in China. Nutr. Cycling Agroecosyst. 102, 371–381. doi: 10.1007/s10705-015-9710-9
Wang, P., Kong, X., Chen, H., Liu, H., Li, X., Zhang, Z., et al. (2021). Exploration of intrinsic microbial community modulators in the rice endosphere indicates a key role of distinct bacterial taxa across different cultivars. Front. Microbiol. 12:629852. doi: 10.3389/fmicb.2021.629852
Wei, T., Zhang, P., Wang, K., Ding, R. X., Yang, B. P., Nie, J. F., et al. (2015). Effects of wheat straw incorporation on the availability of soil nutrients and enzyme activities in semiarid areas. PLoS One 10:e0120994. doi: 10.1371/journal.pone.0120994
Wu, F. P., Jia, Z. K., Wang, S. G., Chang, S. X., and Startsev, A. (2013). Contrasting effects of wheat straw and its biochar on greenhouse gas emissions and enzyme activities in a Chernozemic soil. Biol. Fertil. Soils 49, 555–565. doi: 10.1007/s00374-012-0745-7
Xing, Z. P., Ya-Jie, H. U., Qian, H. J., Cao, W. W., Guo, B. W., Wei, H. Y., et al. (2017). Comparison of yield traits in rice among three mechanized planting methods in a rice-wheat rotation system. J. Int. Agriculture 16, 1451–1466. doi: 10.1016/S2095-3119(16)61562-9
Yuan, H. Z., Zhu, Z. K., Wei, M. M., Liu, S. L., Peng, P. Q., Anna, G., et al. (2019). Straw and biochar strongly affect functional diversity of microbial metabolism in paddy soils. J. Int. Agriculture 18, 1474–1485. doi: 10.1016/S2095-3119(18)62102-1
Zang, Y. F., Hao, M. D., Zhang, L. Q., and Zhang, H. Q. (2015). Effects of wheat cultivation and fertilization on soil microbial biomass carbon, soil microbial biomass nitrogen and soil basal respiration in 26 years. Acta Ecol. Sinica 35, 1445–1451.
Zhao, J., Ni, T., Huang, X., Huang, Q., Ran, W., et al. (2017). Influence of straw incorporation with and without straw decomposer on soil bacterial community structure and function in a rice-wheat cropping system. Appl. Microbiol. Biotechnol. 101, 4761–4773. doi: 10.1007/s00253-017-8170-3
Zhao, X., Wang, J., Wang, S., and Xing, G. (2014a). Successive straw biochar application as a strategy to sequester carbon and improve fertility: a pot experiment with two rice/wheat rotations in paddy soil. Plant Soil. 378, 279–294. doi: 10.1007/s11104-014-2025-9
Zhao, J., Zhang, R., Xue, C., Xun, W., Sun, L., Xu, Y., et al. (2014b). Pyrosequencing reveals contrasting soil bacterial diversity and community structure of two main winter wheat cropping systems in China. Microb. Ecol. 67, 443–453. doi: 10.1007/s00248-013-0322-0
Zhong, Y. Q. W., Hu, J. H., Xia, Q. M., Zhang, S. L., Li, X., Pan, X. Y., et al. (2020). Soil microbial mechanisms promoting ultrahigh rice yield. Soil Biol. Biochem. 143:107741. doi: 10.1016/j.soilbio.2020.107741
Keywords: paddy rice, biochar, high-throughput 16S rRNA sequencing, bacterial communities, enzyme activities
Citation: Tang Z, Zhang L, He N, Liu Z, Ma Z, Fu L, Wang H, Wang C, Sui G and Zheng W (2022) Influence of planting methods and organic amendments on rice yield and bacterial communities in the rhizosphere soil. Front. Microbiol. 13:918986. doi: 10.3389/fmicb.2022.918986
Received: 13 April 2022; Accepted: 01 July 2022;
Published: 28 July 2022.
Edited by:
Xi-En Long, Nantong University, ChinaReviewed by:
Wenhao Yang, Fujian Agriculture and Forestry University, ChinaCopyright © 2022 Tang, Zhang, He, Liu, Ma, Fu, Wang, Wang, Sui and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guomin Sui, Z3VvbWluNjY2QDEyNi5jb20=; Wenjing Zheng, endqMjdAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.