
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 20 June 2022
Sec. Extreme Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.918491
This article is part of the Research Topic Extreme Environmental Microbial Products: Structures, Functions, Biosynthesis View all 12 articles
 Jing Guo1,2†
Jing Guo1,2† Zhanling Xie1,2*†
Zhanling Xie1,2*† Hongchen Jiang3,4
Hongchen Jiang3,4 Hongyan Xu1,5
Hongyan Xu1,5 Baolong Liu6
Baolong Liu6 Qing Meng1,2
Qing Meng1,2 Qingqing Peng1,2
Qingqing Peng1,2 Yongpeng Tang7
Yongpeng Tang7 Yingzhu Duan8
Yingzhu Duan8The Qinghai-Tibet Plateau (QTP) is the highest plateau in the world, and its ultraviolet (UV) radiation is much greater than that of other regions in the world. Yellow mushroom (Floccularia luteovirens) is a unique and widely distributed edible fungus on the QTP. However, the molecular mechanism of F. luteovirens’s response to strong UV radiation remains unclear. Herein, we reported the 205 environmental adaptation and information processing genes from genome of F. luteovirens. In addition, we assembled the RNA sequence of UV-affected F. luteovirens at different growth stages. The results showed that in response to strong UV radiation, a total of 11,871 significantly different genes were identified, of which 4,444 genes in the vegetative mycelium (VM) stage were significantly different from the young fruiting bodies (YFB) stage, and only 2,431 genes in the YFB stage were significantly different from fruiting bodies (FB) stage. A total of 225 differentially expressed genes (DEGs) were found to be involved in environmental signal transduction, biochemical reaction preparation and stress response pathway, pigment metabolism pathway, and growth cycle regulation, so as to sense UV radiation, promote repair damage, regulate intracellular homeostasis, and reduce oxidative damage of UV radiation. On the basis of these results, a molecular regulation model was proposed for the response of F. luteovirens to strong UV radiation. These results revealed the molecular mechanism of adaptation of F. luteovirens adapting to strong UV radiation, and provided novel insights into mechanisms of fungi adapting to extreme environmental conditions on the QTP; the production the riboflavin pigment of the endemic fungi (Yellow mushroom) in the QTP was one of the response to extreme environment of the strong UV radiation.
Ultravioet (UV) radiation is a component of solar radiation spectrum, which is the most harmful and mutagenic light band (Chen et al., 2021). UV can damage plant growth, development, biomass accumulation, and metabolism (Li et al., 2019a; Ozel et al., 2021). Plants have developed a series of strategies to cope with UV radiation, including activating light defense system, regulating light response pathway, and producing DNA photoproducts to mitigate DNA sequence changes and mutations caused by UV radiation (Azarafshan et al., 2020; Huarte-Bonnet et al., 2020). In addition, UV radiation will also activate the metabolic pathways of anthocyanin, carotenoid and melanin synthesis and anti-DNA oxidative damage system in plants, resulting in the accumulation of pigments such as anthocyanins, carotenoids, melanin, and riboflavin (Mapari et al., 2005; Gmoser et al., 2017; Qi et al., 2021; Zhu et al., 2021). In contrast for fungi, previous studies suggested that UV radiation would reduce the germination rate of conidia of Magnaporthe oryzae, and limited the population and diffusion of fungi in nature (Braga et al., 2015; Li et al., 2019a).
The Qinghai-Tibet Plateau (QTP) has the highest intensity of UV radiation in the world (Qiao et al., 2016). There are more than 1,500 kinds of fungi growing in the QTP alpine meadow ecosystem, among which yellow mushroom (Floccularia luteovirens) is one of the most flourish and representative macrofungi (Xing et al., 2017; Gan et al., 2020). In the alpine meadow of the QTP, our previous observation found that the wild fruiting bodies of F. luteovirens in high-UV environment were more yellowish brighter and showed more sulfur color than those in low-UV, and Liu et al. extracted riboflavin from yellow mushrooms fruiting bodies (Gan et al., 2022). However, the molecular adaptation mechanism of F. luteovirens in response to strong UV radiation remains unclear during its evolutionary process to extreme environment on QTP. In this work, we studied the gene expression of F. luteovirens at three growth stages [i.e., vegetative mycelium (VM) in lab, young fruiting body (YFB) in natural environment, fruiting body (FB) in natural environment] by using transcriptomics, and we also verified UV stress to cause the accumulation of the riboflavin and chlorophyll pigment in the vegetative mycelium, with the purpose to reveal the molecular adaptation mechanism of F. luteovirens to cope with strong UV radiation in the Qinghai-Tibet Plateau.
In August 2018, three wild fruiting bodies of F. luteovirens were collected to transcriptome sequencing from Haiyan County (100°47′13″E, 37°0′37″N), Qinghai Province, China for transcriptome sequencing (Table 1). The sampling area is 3,220 m above sea level, with annual sun light of 2517.6–2995.3 h and annual average radiation of 5210.2–6568.3 MJ/m2 (Zhao Y. T. et al., 2021). According to the pileus diameter, the stage of F. luteovirens can be classified into young fruiting bodies (YFB, diameter 2.5–4.5 cm) and fruiting bodies (FB, diameter 4.5–6.6 cm), all of which samples were collected for transcriptome sequencing in three replicates. Vegetative mycelium (VM) was isolated from the fruiting bodies of F. luteovirens F18-3 and F. luteovirens DTS10, and were then cultivated in Potato Dextrose agar (PDA) medium (glucose 20.0 g/L, potato extract 200.0 g/L, agar 18.0 g/L) for 30 days at 20°C (at the same temperature of outside) in the dark (Xu et al., 2021). Three replicate plates were prepared for transcriptome sequencing and the determination of pigments. In the end of cultivation, the resulting F. luteovirens fruiting bodies were immediately frozen in liquid nitrogen and stored at −80°C.
The strain of F. luteovirens F18-3 was cultured in PDB medium (glucose 20.0 g/L, potato extract 200.0 g/L) in flask of 500 mL at 200 rpm, at 20°C in liquid for 30 days. Three replicate plates in liquid culture were prepared for the determination of pigments.
The genome of F. luteovirens fruiting body was sequenced with PacBio Sequel and Illumina Hiseq 2500 Systems (Genewiz Biotechnology Co. Ltd., Suzhou, China). Long reads from PacBio could give a high-quality sequencing length while also generating a 2.5-kb library. The genome size and hybrid rate of F. luteovirens was estimated by kmerfreq.1 Genome assembly was performed by using Canu software and was then assessed by using Benchmarking Universal SingleCopy Orthologs program v3.0 (Gan et al., 2020). The genome sequence of F. luteovirens has been deposited at GenBank under accession number of RPFY00000000.1.
Total RNA was extracted from the three samples using TRIzol reagent, according to the manufacturer’s instructions (Sangon Biotech, Shanghai, China). The quality and quantity of total RNA were measured by using electrophoresis in 1% agarose gel. cDNA libraries were prepared using TruSeqTM RNA sample prep kit (Illumina). In brief, cDNA was synthesized from mRNA by reverse transcriptase with the use of random primers. Raw sequence reads were obtained by using an Illumina HiSeq sequencing platform. Each read was trimmed individually by setting quality score below 20 to obtain pure reads. Low quality reads were trimmed out and reads of 25 base pairs were filtered out in order to ensure purity. The sequences obtained in this study were deposited in the NCBI Sequence Read Archive (SRA2) under accession number SRP279887.
Clean data were obtained by removing low-quality reads, adapter sequences, reads with ambiguous bases “N,” and reads of <20 bp. In the absence of a reference genome, contigs, and singletons were generated by de novo assembly using the Trinity software package.3
Differentially expressed genes (DEGs) were identified by comparing gene expression levels using TPM (Transcripts Per Million). TPM is the most commonly used index of gene expression levels, and can reflect the impact of sequencing depth and gene length on read counts (Mortazavi et al., 2008). We used “q Value < 0.05 and | FoldChange| > 2” as the threshold to assess the significance of gene expression differences. BLAST analysis was carried out by searching against the NCBI databases and setting the E-value cut off at 10–3. The upregulated and downregulated unigenes were subject to Venn diagram analysis.
For metabolic pathway analysis, DEGs were aligned to the KEGG database through the KOBAS program.4 In both analyses, q value < 0.05 and | FoldChange| > 2 were used as thresholds.
Total RNA was extracted from same samples using a Fungal Total RNA Rapid Extraction Kit (Sangon Biotech, Shanghai, China). Then cDNA was synthesized by using an M-MuLV First Strand cDNA Synthesis kit (Sangon Biotech, Shanghai, China). The 18S ribosomal RNA (rRNA) gene was used as an internal control for normalization (Song et al., 2018). Primers were designed by primer 3.0 online5 (Supplementary Table 1). The qRT-PCR reaction was carried out in a LightCycler 96 system (Bio-Rad Laboratories, Hercules, CA, United States) using a SYBR Green kit (2 × SG Fast qPCR Master Mix, Sangon Biotech, Shanghai, China). Amplification conditions used for qPCR were 95°C for 3 min, followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. For each sample, three biological and two technical replicates were done. Fold changes in gene expression were calculated using the 2–ΔΔCt method (Li et al., 2019b).
The cultured mycelia of 30 days in liquid of F18-3 and solid culture of DTS10 were stressed by 5400 mW m–2 UV radiation for 0, 2, 4, 6, 8, 10, 12, 24 h. The mycelia from liquid culture and solid culture were collected and dry at 40°C for 2 h, and then at the weight of 100 mg mycelia were polished with liquid nitrogen for 5 min. For riboflavin measurements, samples were diluted to arrive at the linear range of the spectrophotometer with 0.05 M NaOH. The chlorophyll was determined according to the assay kits of chlorophyll (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The OD444 was immediately measured, the titer of riboflavin is calculated according to the following formula: y = 0.0105x + 0.0089, R2 = 0.9953 (Supplementary Figure 1; Zhang et al., 2021).
The whole genome data of F. luteovirens were reported in previous studies (Gan et al., 2020). By comparing with 357 species-specific gene families from KEGG database, we found 1,648 genes in the whole genome of F. luteovirens. Among them, a total of 205 genes were annotated, which were categorized into three levels: 193 signal transduction genes (94.14%), one membrane transport gene (0.49%) and 11 environmental adaptation genes (5.37%), respectively (Figure 1A).
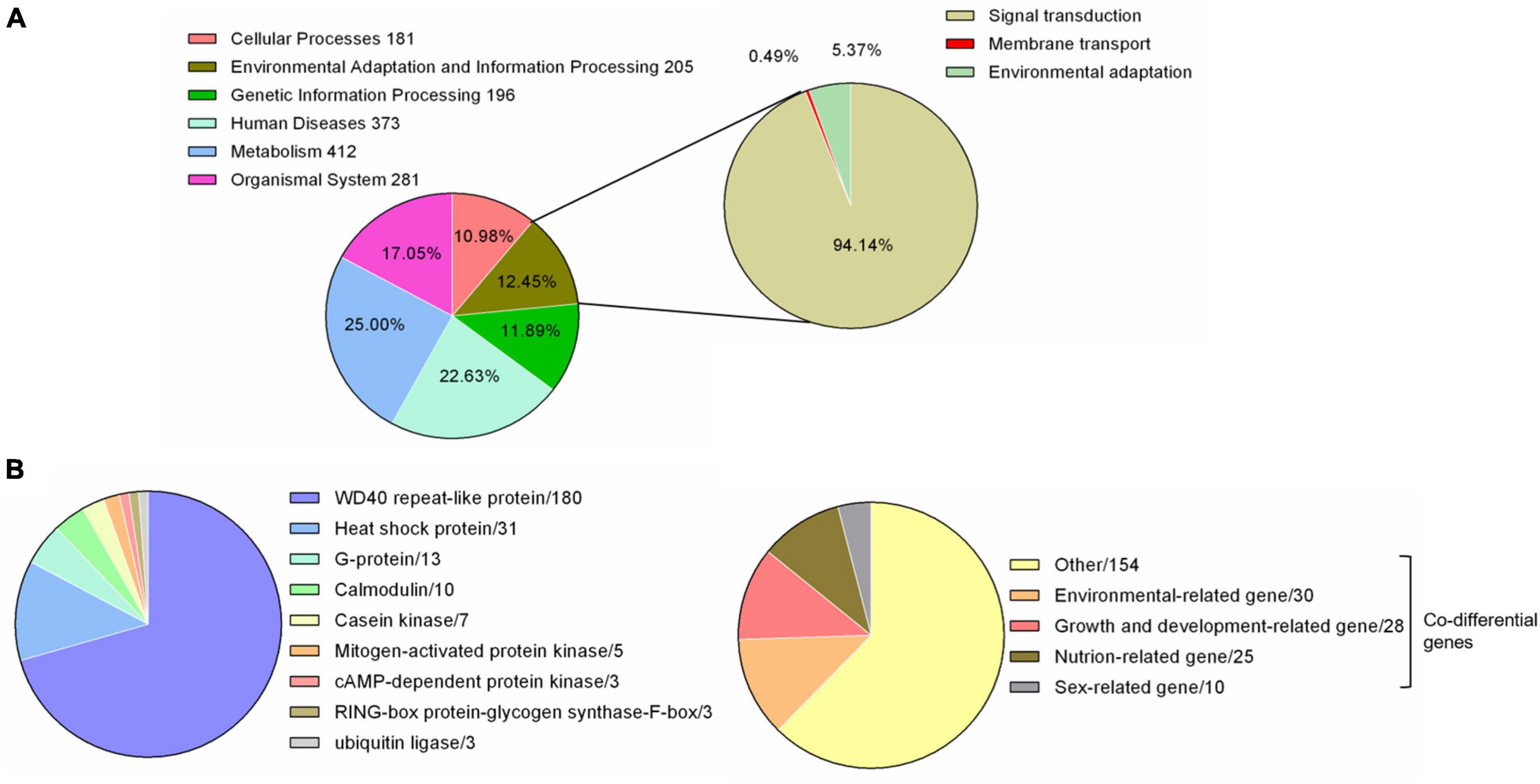
Figure 1. Environmental related genes of F. luteovirens. A total of 205–225 genes involved in responding to environment. (A) Pie chart of the 205 environmental adaptation and information processing genes from genome. (B) Pie chart of the 225 environmental related genes from transcriptome [The 225 genes = DEGs (255) − Co differential genes of environmental-related (30)].
A total of 44,897,648, 53,152,546, and 46,470,082 high-quality sequence reads were obtained from the VM, YFB, and FB samples, respectively (Table 2). These reads were then assembled into 89962 extended transcripts with an average length of 2028.56 bp and a N50 length of 3559 bp (N50 indicates that at least half of the resulting transcripts have a base length of no less than 50 bp). The resulting extended transcripts were used to generate a transcriptome database by using Trinity (see text footnote 3). These extended transcripts consisted of 23823 unigenes, with an average unigene length of 1023.6 bp and a N50 length of 2447 bp (Table 3). The length of transcripts ranged from 200 to ≥2000 bp (Supplementary Figure 2).
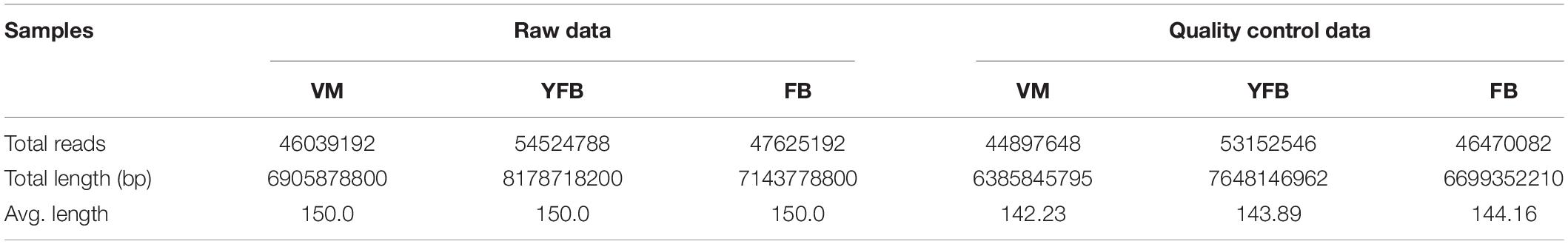
Table 2. Throughput and quality of Illumina sequencing of the Floccularia luteovirens transcriptome.
A total of 11,871 significant differently expressed genes (DEGs) were identified between the VM, YFB, and FB stages, of which 247 DEGs that were mainly involved in the growth and development of F. luteovirens co-existed in all three stages (Supplementary Table 2). A total of 4,444 DEGs were identified between the VM and YFB stages, 4,996 DEGs were identified between the VM and FB stages, and only 2,431 DEGs genes were identified between the YFB stage and FB stages (q ≤ 0.05) (Figure 2). Furthermore, cluster analysis showed the DEGs of YFB and FB belonged to the same branch, but the DEGs of VM formed a separate branch (Figure 2C), suggesting that the expression profiles of the YFB and FB were similar.
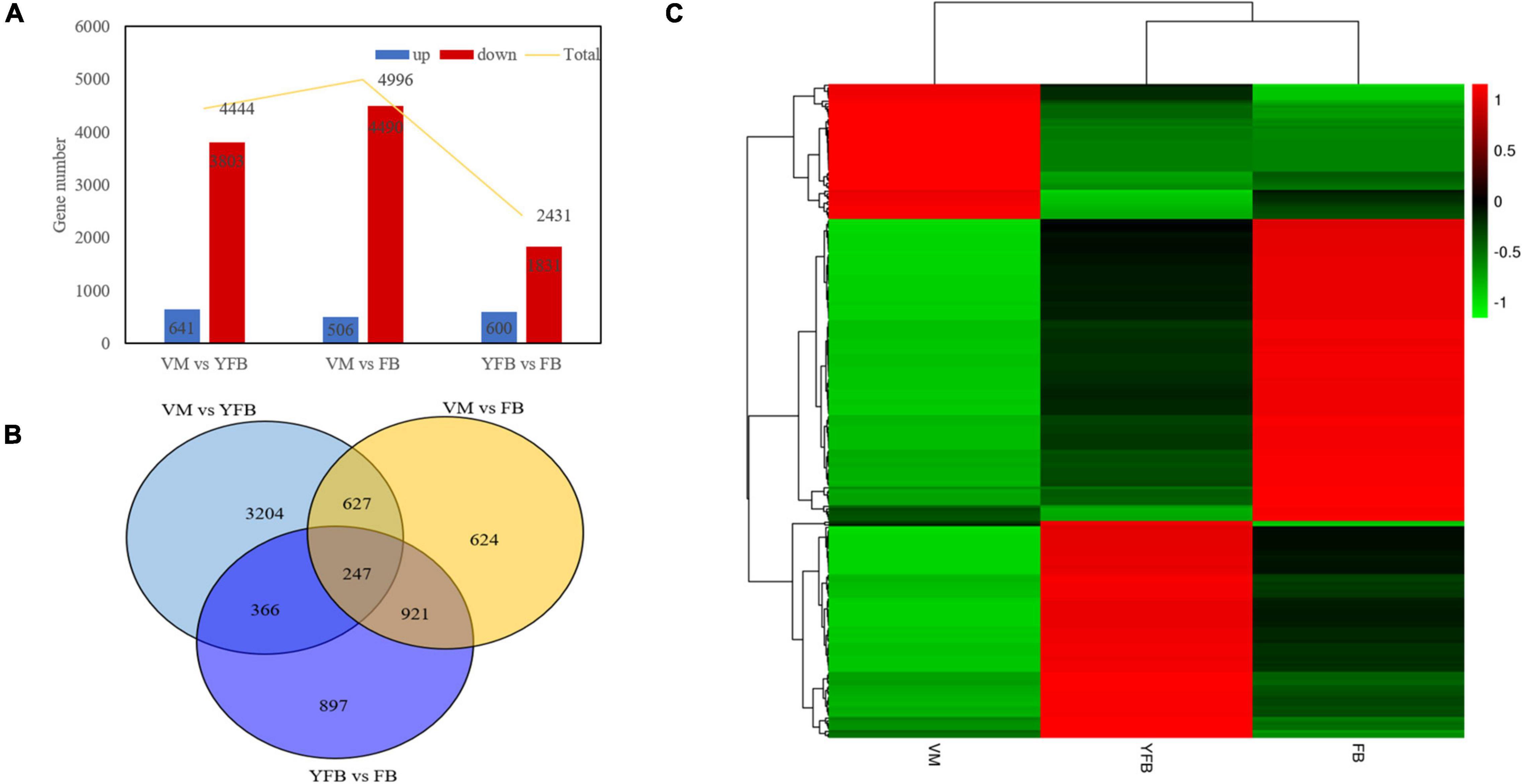
Figure 2. Bar plot, Venn diagram, and heatmap showing co-differentially expressed genes. (A) Bar plot of the differentially expressed genes (DEGs). (B) Venn diagram showing the DEGs exclusive to VM vs. YFB (light blue circle), YFB vs. FB (dark blue circle), and VM vs. FB (yellow circle). (C) Heatmap of co-differentially genes. Red and green colors in the heat-maps represent down- and up- regulated expression levels, respectively.
A total of 225 DEGs were identified to play a regulatory role in adapting to the changes of external environmental factors and UV radiation. The functions of these DEGs included signal transduction, biochemical reaction preparation and effect stress (Figure 1B and Supplementary Table 3). Among these identified DEGs, calmodulin and G proteins play important roles in signal transduction of F. luteovirens. Five calmodulin signaling genes that were up-regulated in the cells of F. luteovirens at the FB stage were identified, and they were calmodulin-binding motif protein (DN6933), calmodulin (DN255, DN3162) and Calcium/calmodulin-dependent protein (DN8891, DN3857). The expression of nine G protein coding genes were significantly up-regulated from the VM stage to the FB stage. Secondly, kinase genes are the key genes regulating the biochemical reaction in F. luteovirens. The expression of five mitogen-activated protein kinase genes were significantly up-regulated in the cells at the VM stage compared with those at the FB stage, the expression of two cAMP-dependent protein kinase genes, three casein kinase genes and cAMP-dependent protein kinase (DN6015) and casein kinase (DN15337) genes were significantly upregulated in the cells at the FB stage, compared with that in the VM stage. In addition, ubiquitin ligase, RING-box protein, glycogen synthase, and F-box protein were involved in the regulation of effector stress. In the present study, genes for ubiquitin ligases, glycogen synthase, and F-box protein were all expressed at a much higher level in the cells at the FB stage than that that of VM state. In addition, heat shock protein and WD40 repeat-like protein play vital role in oxidative stress of F. luteovirens under UV radiation and environmental factors. For instance, a large number of heat shock proteins were up-regulated in the cells at the FB growth stage. Most of the WD40 repeat-like protein genes of F. luteovirens were significantly upregulated in the cells at both YFB and FB stages (Figure 3).
Two metabolic pathways including chlorophyll and riboflavin metabolisms, related to pigment synthesis were differentially regulated in F. luteovirens. Several DEGs were enriched in the metabolisms of chlorophyll and riboflavin pathways. The chlorophyll metabolic pathway may affect the accumulation of photosynthetic pigments and the production of photochromes (Supplementary Figure 3). The gene expression of the core enzymes of the chlorophyll metabolic pathway was higher at the FB stage than at the VM stage (Supplementary Table 4). Such core enzyme included the genes of uroporphyrinogen decarboxylase (HemE, 21.24-fold), 5-aminolevulinate synthase (ALAS, 22.29-fold), ferrochelatase (FECH, 21.94-fold), glutamyl-tRNA synthetase (EARS, 21.41-fold), cytochrome c heme-lyase (CYC, 21.40-fold), oxygen-dependent protoporphyrinogen oxidase (HemY, 21.15-fold). In the riboflavin metabolism pathway, the expression levels of tyrosinase (TYR, 27.81-fold) and GTP cyclohydrolase II (ribA, 21.62-fold) genes were much higher than those in the FB stage than in the VM stage (Supplementary Table 5 and Supplementary Figure 4). Another gene for 3,4-dihydroxy 2-butanone 4-phosphate synthase (ribB) and 6,7-dimethyl-8-ribityllumazine synthase (ribH) was differentially expressed only in the FB stage (Figure 4B). The expression of these enzymes may result in the accumulation of riboflavin in F. luteovirens in the FB stage. In general, there were more differential regulation genes and specific genes of pigment synthesis under strong UV than without UV radiation, which was consistent with the fact that the fruiting body of F. luteovirens obtained long-term sunlight radiation and a large amount of UV radiation in the late stage of development.
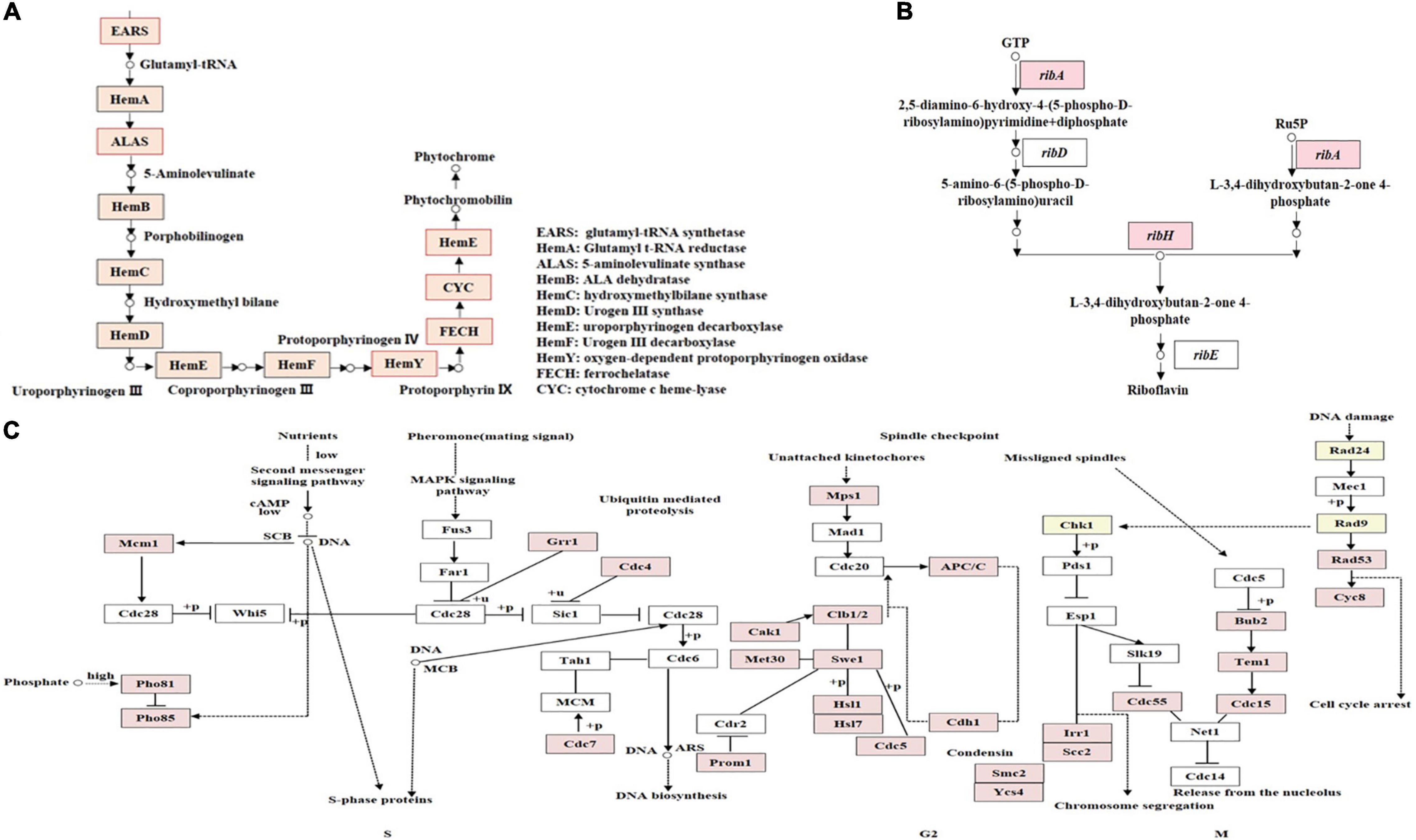
Figure 4. KEGG annotation of DEGs involved in different metabolism pathways. (A) KEGG annotation of proteins encoded by DEGs involved in the phytochrome biosynthesis pathway. The genes whose products are highlighted in red are up-regulated during the FB stages. (B) KEGG annotation of DEGs involved in riboflavin metabolism pathways. Genes highlighted in red are up-regulated during the FB stages. (C) KEGG annotation of DEGs involved in the cell-cycle. DEGs between VM and FB are shown in the cell-cycle pathway at the S, G2, and M phases. Genes whose products are highlighted in red are up-regulated during the FB stages.
In addition, the cell-cycle pathway of F. luteovirens was associated with its cell growth, proliferation, and development. The expression of the genes related to cell-cycle pathway in the cells of F. luteovirens was greatly up-regulated, and such genes included the S phase-related Mcm1 (21.01-fold), Pho81 (22.31-fold), Pho85 (21.58-fold), Grr1 (21.73-fold), Cdc4 (21.42-fold), and Cdc7 (23.88-fold); the G2 phase-related Mps1 (21.70-fold), Cak1 (22.02-fold), and Cdh1 (21.67-fold) (Supplementary Figure 5); and the M phase-related Rad53 (21.61-fold) and Scc2 (21.46-fold), were higher in the FB stage than in the VM stage (Supplementary Table 6). However, the expression of Cdc7 and Cyc8 were significantly down-regulated, which were considered to depress the growth and development of F. luteovirens cells. Moreover, there were 34 DEGs that may be mainly involved in DNA damage repair of F. luteovirens, and they showed higher expression at the FB stage than at the VM stage (Figure 4C).
Twenty-two DEGs from F. luteovirens that were significantly expressed at all the three stages (VM, YFB, and FB) were randomly selected, and their differential expression was confirmed by qRT-PCR (Supplementary Table 7). Among them, 11 environment-related DEGs, eight growth and development-related DEGs, and three sex-related DEGs were investigation, all of which have different expression patterns similar to those in the RNA-Seq (Figure 5), indicating the reliability and accurately of the RNA-Seq data of F. luteovirens.
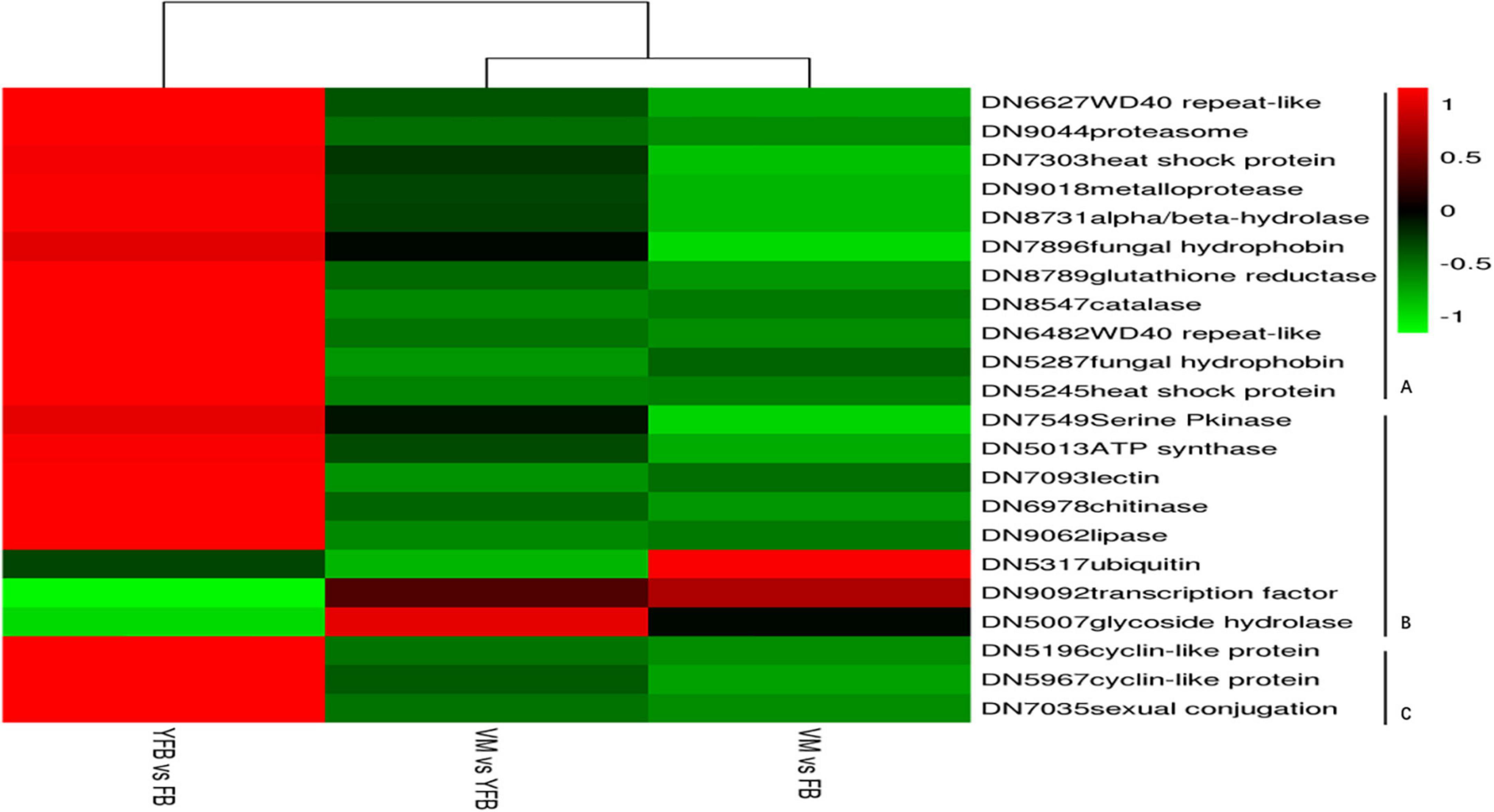
Figure 5. qRT-PCR validation of DEGs. The 18S gene was selected as the internal control in this study. The expression patterns determined by qRT-PCR were consistent with those obtained by RNA-Seq, thus confirming the reliability of our RNA-Seq data. (A) Eleven environment-related DEGs were analyzed by qRT-PCR. (B) Eight growth and development-related DEGs were analyzed by qRT-PCR. (C) Three sex-related DEGs were analyzed by qRT-PCR.
Under UV irradiation for 8–10 h, the riboflavin concentration of mycelium reached the concentration of 42.35–47.21 ug/mL (Figure 6). Meanwhile, the chlorophyll content reached the 1.56–2.10 mg/g after UV stress for 4–6 h. Both DTS 10 and F18-3 of F. luteovirens reached the highest concentration after simultaneous exposure to UV radiation of 5400 mW m–2, although they were cultured and exposed in different states. The highest of riboflavin concentration occurred at UV radiation of 8–10 h, which was consistent with the daytime illumination time of F. luteovirens.
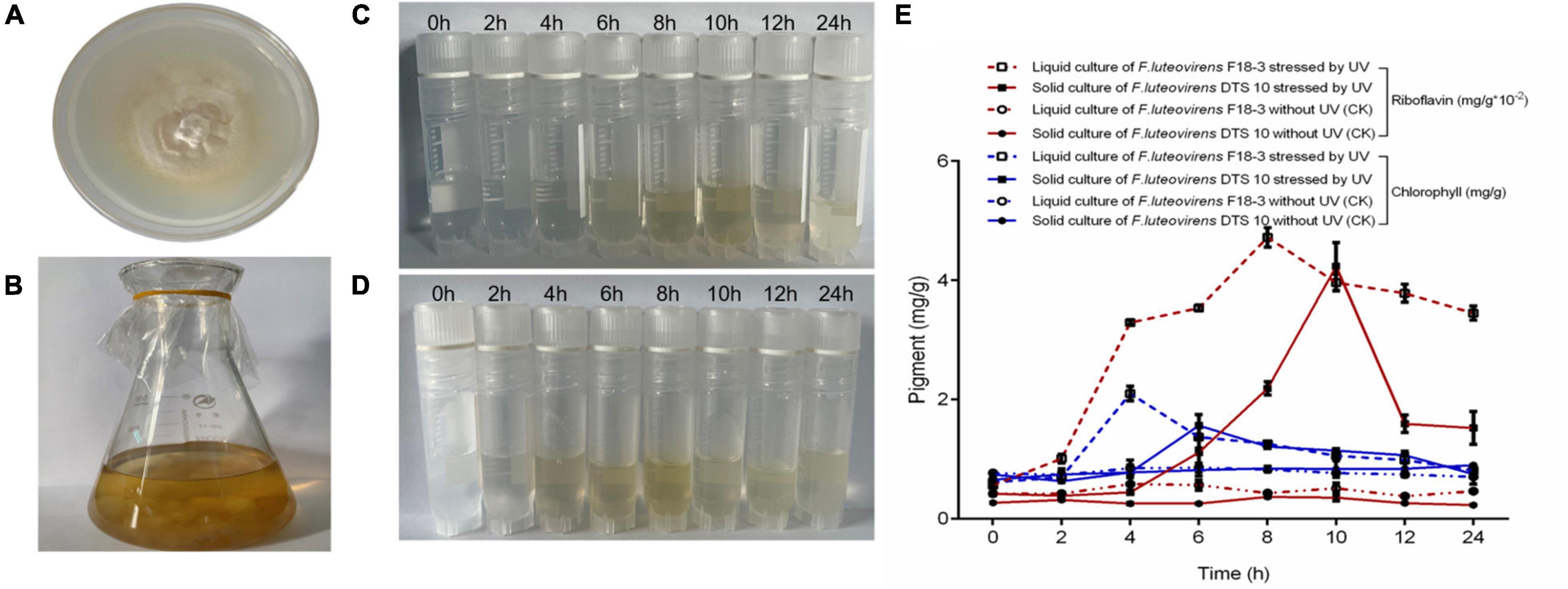
Figure 6. Riboflavin and chlorophyll induced by UV of F. luteovirens in mycelium. (A) Plate culture of F. luteovirens F18-3. (B) Liquid culture of F. luteovirens DTS10. (C) Abstracted riboflavin of F. luteovirens F18-3 in mycelium stressed by UV in solid culture. (D) Abstracted riboflavin of F. luteovirens DTS10 in mycelium stressed by UV in liquid culture. (E) The production of riboflavin and chlorophyll stressed by UV of F. luteovirens in mycelium. The red line represents riboflavin; the blue line represents chlorophyll.
UV radiation affected the biochemical process and physiological characteristics of organisms. In this study, 205–225 DEGs potentially involved in coping with external strong UV radiation stress were obtained from F. luteovirens living on the Qinghai-Tibet Plateau. The genome environmental adaptation and information processing genes of these DEGs were basically consistent with the transcriptome genes of their environment-related genes. When F. luteovirens cells received signals of UV and external stimuli, G protein and calmodulin were activated as signal molecules. G protein interacted directly with downstream enzymes or ion channels and was involved in regulating cell-specific changes caused by UV and external stimuli (Smrcka and Fisher, 2019; Wang et al., 2019; Patel et al., 2020). Calmodulin, as a ubiquitous sensor protein and calcium receptor in eukaryotic cells, was involved in regulating the response of organisms to various environmental stresses and transmitting stimulation signals (Kiselev et al., 2021; Li et al., 2021). Our results showed that the significantly upregulated genes of DN255 and DN90411 were considered as candidate genes that played a crucial role in coping with UV stress and signal transduction in response to external factors. This finding was consistent with the expression trend of signal transduction genes of CaM and CML in Vitis amurensis under abiotic stress (Kiselev et al., 2021).
In addition, phosphorylation pathway is a key biological process in signal transduction, and can regulate the expression of kinase genes. When the kinase genes sense stimulation signals, cAMP-dependent protein kinase was the core enzyme that regulated the protein phosphorylation pathway (Wu Z. et al., 2019), and MAPK transduced signals through the phosphorylation pathway of downstream targets (Li C. et al., 2018). DEGs of DN7039 and DN88351 were found to be involved in the regulation of UV and external stress response of F. luteovirens and showed differential expression patterns. Furthermore, casein kinase is another gene essential for cell proliferation. It is an evolutionarily conserved Ser/Thr protein kinase and is also pivotal in signal transduction (Li Y. et al., 2018). Nagatoshi et al. (2018) reported that casein kinase is a pleiotropic enzyme involved in a variety of developmental and stress response processes. Our results suggested that casein kinase may be independently involved in signal transduction of F. luteovirens on the Qinghai-Tibet Plateau.
The cells of F. luteovirens often undergo post-translational modifications of proteins they sense and transmit internal or external UV stimulation signals (Han et al., 2019; Jiao and Duan, 2019; Yu et al., 2021). In F. luteovirens, RING-box protein, ubiquitin ligase, F-box protein, and glycogen synthase were involved in the post-translational modification of proteins under UV and external stimulation: on the one hand, RING-box proteins and ubiquitin ligases were identified as key regulators of basic cellular processes and participated in plant growth and development (Abd-Hamid et al., 2020); On the other hand, F-box protein and glycogen synthase depended on the regulation of protein stability under abiotic stress, and participated in the regulation of antioxidant system after translational modification (Rameneni et al., 2018; Jiao and Duan, 2019). Thus it is reasonable to speculate that RING-box protein and ubiquitin ligase may mediate the ubiquitination of key factors in the process of UV and external stress, and then regulate the adaptability of F. luteovirens to strong UV radiation and external factors.
Heat shock protein and WD40-repeat like protein were important molecular regulators involved in controlling cellular processes in response to stresses (Guz et al., 2021; Kim et al., 2021). The former helped to maintain the folding of damaged and newly formed proteins under stress conditions, and also responded to environmental stress by regulating the expression dynamics of numerous genes involved in maintaining intracellular homeostasis (Guz et al., 2021; Kim et al., 2021); while the latter mainly regulated signal transduction, transcriptional regulation, and immune response (Jain and Pandey, 2018). Our results suggested that under UV the stimulation and external factors, F. luteovirens cells produced denatured peptides and new proteins, so the genes of DN13641 and DN7704 were significantly upregulated.
Chlorophyll metabolism includes chlorophyll biosynthesis, chlorophyll cycle, and chlorophyll degradation. The produced chlorophyll can regulate the host’s defense response and survival under different extreme environmental conditions (Wang et al., 2020). In this study, 20 candidate genes related to porphyrin and chlorophyll metabolic pathway were analyzed. It is found that the core genes of HemE, ALAS, FECH, EARS, CYC, HemY were involved in the synthesis of phytochrome and cytochrome in F. luteovirens under strong UV radiation. Previous studies showed that ALAS can promote plant growth under abiotic stress and participate in the regulation of plant photosynthesis, nutrient absorption, antioxidant properties and osmotic balance (Wu Y. et al., 2019). Therefore, ALAS may mediate the color change of F. luteovirens to adapt to the strong UV radiation on the Qinghai-Tibet Plateau. The role of ALAS in F. luteovirens may be consistent with that of HY5 in regulating the color change of Brassica campestris L. leaf under cold stress (Yuan et al., 2021).
In addition to chlorophyll, riboflavin also participated in the response to UV radiation and the other external environment stimuli in the Qinghai-Tibet Plateau. The coding genes of FXN and BLVRA, the main derivatives of riboflavin, were other genes that mainly regulate plant growth and color rendering (Bai et al., 2019; Rouault, 2019; Britti et al., 2021). We found that the coding genes of FXN and BLVRA were involved in controlling the growth and division of F. luteovirens under strong UV radiation, and regulated its yellow color at the later growth stage of F. luteovirens (Figure 4A). FMN and FAD, the main riboflavin derivatives, were cofactors of enzymes that mediated many redox reactions in cells, and riboflavin operon ribA gene encoded GTP, which was the first step of riboflavin biosynthesis (Cisternas et al., 2017; Liu et al., 2020; Zhao G. et al., 2021). Accordingly, we found that F. luteovirens may have the capability of de novo riboflavin biosynthesis, and the ribA and ribH genes were directly involved in riboflavin synthesis. Their differential expression patterns may be related to the regulation of redox reaction and electron respiratory chain transmission under UV radiation. This is similar to the role of the anthocyanins of Lycium ruthenicum on Qinghai-Tibet Plateau (Qi et al., 2021). These results suggested that the metabolic pathway of fungal pigment synthesis was closely related to the response to UV radiation.
Furthermore, fungi have evolved complex signal transduction pathways to repair direct DNA damage. DNA damage checkpoints transmit signals of damaged DNA to effector molecules, thereby regulating cell cycle pathways to repair DNA oxidative damages (Zhou et al., 2016; Gkouskou et al., 2020). In the cell cycle pathway of F. luteovirens, the key genes Rad53, Pho85, Pho81, and Cdh1 were significantly upregulated in the FB stage under strong UV radiation. The Rad53 gene was responsible for the transduction of UV stimulated signals and was significantly upregulated under strong UV radiation to regulate DNA damage repair, replication, bifurcation stability, cell cycle progression, and transcription (Gkouskou et al., 2020). Pho81 and Pho85 gene were significantly upregulated in F. luteovirens, indicating that their encoded enzymes may play a key role in maintaining phosphate homeostasis under UV stimulation; Pho85 controlled the cell cycle by regulating phosphate metabolism when it binds to the Pho80 family of cyclins, while the CDK inhibitor Pho81 regulated the cell cycle by inhibiting thePho80-Pho85 complex (Zhou et al., 2016; Zheng et al., 2020). The Cdh1 gene encoded cell division cycle 20-like protein 1, which was highly upregulated in the FB stage under strong UV radiation. This result suggested that Cdh1 may be a key regulatory gene in the cell cycle of the fruiting body of F. luteovirens from the S phase to G2 phase, which may be consistent with the role of Chl1p as G1/S phase checkpoint and its role in the damage checkpoint pathway in budding yeast (Katheeja et al., 2021).
Floccularia luteovirens can adapt to the changes of external environmental factors, especially it can respond to UV radiation in three ways by regulating key genes and obtain the ability to deal with UV radiation (as shown in the conceptual scheme Figure 7). Firstly, at the nuclear level, cell cycle was regulated to repair DNA oxidative damage of ultraviolet radiation to cells. Secondly, at the cytoplasmic level, differentially expressed genes of environmental signal transduction, biochemical reaction preparation and stress response involved in maintaining intracellular homeostasis, and the metabolic pathways of chlorophyll and riboflavin were activated to involved in the pigment synthesis of ultraviolet radiation to cells. In addition, after metabolizing gene clusters at the fruiting body and mycelium levels, the pigment of yellowish brightness and sulfur color emerged in the cells of yellow mushroom, which contribute to the characteristic yellow color of ultraviolet radiation to cells. This regulation model can reveal the molecular mechanism of F. luteovirens to deal with the strong UV radiation, which is of great significance for us to understand the environmental adaptation mechanism of fungi on the Qinghai-Tibet Plateau.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI–RPFY00000000.1, SRP279887.
JG: conceptualization, methodology, and writing-original draft. ZX: supervision, validation, and writing-review and editing. HJ: writing-review and editing. HX: investigation. BL: supervision. QM: visualization. All authors contributed to the article and approved the submitted version.
This study was supported by funding from the Natural Science Planning Project of Qinghai Province (grant no. 2021-HZ-802).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.918491/full#supplementary-material
Abd-Hamid, N. A., Ahmad-Fauzi, M. I., Zainal, Z., and Ismail, I. (2020). Diverse and dynamic roles of F-box proteins in plant biology. Planta 251:68.
Azarafshan, M., Peyvandi, M., Abbaspour, H., Noormohammadi, Z., and Majd, A. (2020). The effects of UV-B radiation on genetic and biochemical changes of Pelargonium graveolens L’Her. Physiol. Mol. Biol. Plants 26, 605–616. doi: 10.1007/s12298-020-00758-6
Bai, D. P., Lin, X. Y., Wu, Y., Zhou, S. Y., Huang, Z. B., Huang, Y. F., et al. (2019). Isolation of blue-green eggshell pigmentation-related genes from Putian duck through RNA-seq. BMC Genomics. 20:66. doi: 10.1186/s12864-019-5436-4
Braga, G. U., Rangel, D. E., Fernandes, E. K., Flint, S. D., and Roberts, D. W. (2015). Molecular and physiological effects of environmental UV radiation on fungal conidia. Curr. Genet. 61, 405–425. doi: 10.1007/s00294-015-0483-0
Britti, E., Delaspre, F., Tamarit, J., and Ros, J. (2021). Calpain-inhibitors protect frataxin-deficient dorsal root Ganglia Neurons from loss of mitochondrial Na (+)/Ca (2+) exchanger, NCLX, and apoptosis. Neurochem. Res. 46, 108–119. doi: 10.1007/s11064-020-03020-3
Chen, H., Huang, K., Liu, S., and Lu, S. (2021). Effects of ultraviolet (UV) radiation on outdoor- and indoor-cultured Prorocentrum lima, a toxic benthic dinoflagellate. J. Ocean Univ. China 20, 619–628.
Cisternas, I. S., Torres, A., Flores, A. F., and Angulo, V. A. G. (2017). Differential regulation of riboflavin supply genes in Vibrio cholerae. Gut Pathog. 9:10. doi: 10.1186/s13099-017-0159-z
Gan, X. L., Bao, X. M., Liu, B. L., Li, Y., Cao, D., Zhang, H., et al. (2022). Chemical constituents and molecular mechanisms of the yellow phenotype of yellow mushroom (Floccularia luteovirens). J. Fungi 8:314. doi: 10.3390/jof8030314
Gan, X., Cao, D., Zhang, Z., Cheng, S., Wei, L., Li, S., et al. (2020). Draft genome assembly of Floccularia luteovirens, an edible and symbiotic mushroom on Qinghai-Tibet Plateau. G3 (Bethesda) 10, 1167–1173. doi: 10.1534/g3.120.401037
Gkouskou, K., Fragiadakis, G. S., Voutsina, A., and Alexandraki, D. (2020). Distinct associations of the Saccharomyces cerevisiae Rad9 protein link Mac1-regulated transcription to DNA repair. Curr. Genet. 66, 531–548. doi: 10.1007/s00294-019-01047-w
Gmoser, R., Ferreira, J., Lennartsson, P., and Taherzadeh, M. (2017). Filamentous ascomycetes fungi as a source of natural pigments. Fungal Biol. Biotechnol. 4:4. doi: 10.1186/s40694-017-0033-2
Guz, N., Dageri, A., Altincicek, B., and Aksoy, S. (2021). Molecular characterization and expression patterns of heat shock proteins in Spodoptera littoralis, heat shock or immune response? Cell Stress Chaperon. 26, 29–40.
Han, P. L., Dong, Y. H., Gu, K. D., Yu, J. Q., Hu, D. G., and Hao, Y. J. (2019). The apple U-box E3 ubiquitin ligase MdPUB29 contributes to activate plant immune response to the fungal pathogen Botryosphaeria dothidea. Planta 249, 1177–1188.
Huarte-Bonnet, C., Pereira-Junior, R. A., Paixão, F. R. A., Braga, G. U. L., Roberts, D. W., Luz, C., et al. (2020). Metarhizium robertsii and M. acridum conidia produced on riboflavin-supplemented medium have increased UV-A tolerance and upregulated photoprotection and photoreactivation genes. BioControl 65, 211–222.
Jain, B. P., and Pandey, S. (2018). WD40 repeat proteins: signalling scaffold with diverse functions. Protein J. 37, 391–406. doi: 10.1007/s10930-018-9785-7
Jiao, C., and Duan, Y. (2019). The role of glycogen synthase kinase-3 in Gibberellic acid-induced chilling tolerance and defense response in postharvest peach fruit. Food Bioprocess Tech. 12, 1733–1740.
Katheeja, M. N., Das, S. P., and Laha, S. (2021). The budding yeast protein Chl1p is required for delaying progression through G1/S phase after DNA damage. Cell Div. 16:4. doi: 10.1186/s13008-021-00072-x
Kim, T., Samraj, S., Jimenez, J., Gomez, C., Liu, T., and Begcy, K. (2021). Genome-wide identification of heat shock factors and heat shock proteins in response to UV and high intensity light stress in lettuce. BMC Plant Biol. 21:185. doi: 10.1186/s12870-021-02959-x
Kiselev, K. V., Aleynova, O. A., Ogneva, Z. V., Suprun, A. R., Ananev, A. A., Nityagovsky, N. N., et al. (2021). The effect of stress hormones, UV-C, and stilbene precursors on calmodulin (CaM) and calmodulin-like gene (CML) expression in Vitis amurensis Rupr. Plant Cell Tiss Org. 146, 59–68.
Li, C., Kong, F., Sun, P., Bi, G., Li, N., Mao, Y., et al. (2018). Genome-wide identification and expression pattern analysis under abiotic stress of mitogen-activated protein kinase genes in Pyropia yezoensis. J. Appl. Phycol. 30, 2561–2572.
Li, H., Gao, D., and Yan, X. (2021). Identification and expression analysis of a calmodulin gene in Phycocalida chauhanii (Rhodophyceae, Bangiales). J. Appl. Phycol. 33, 1915–1923.
Li, X., Huang, L., He, Y., Xie, C., Zhan, F., Zu, Y., et al. (2019a). Effects of enhanced UV-B radiation on the interaction between rice and Magnaporthe oryzae in Yuanyang terrace. Photochem. Photobiol. 18, 2965–2976. doi: 10.1039/c8pp00556g
Li, X., Wang, F., Liu, Q., Li, Q., Qian, Z., Zhang, X., et al. (2019b). Developmental transcriptomics of Chinese cordyceps reveals gene regulatory network and expression profiles of sexual development-related genes. BMC Genomics 20:337. doi: 10.1186/s12864-019-5708-z
Li, Y., Min, L., Zhang, L., Hu, Q., Wu, Y., Li, J., et al. (2018). Promoters of Arabidopsis casein kinase I-like 2 and 7 confer specific high-temperature response in anther. Plant Mol. Biol. 98, 33–49.
Liu, S., Hu, W., Wang, Z., and Chen, T. (2020). Production of riboflavin and related cofactors by biotechnological processes. Microb. Cell Fact. 19:31. doi: 10.1186/s12934-020-01302-7
Mapari, S., Nielsen, K., Larsen, T., Frisvad, J., Meyer, A., and Thrane, U. (2005). Exploring fungal biodiversity for the production of water-soluble pigments as potential natural food colorants. Curr. Opin. Biotechnol. 16, 231–238. doi: 10.1016/j.copbio.2005.03.004
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nagatoshi, Y., Fujita, M., and Fujita, Y. (2018). Casein kinase 2 alpha and beta subunits inversely modulate ABA signal output in Arabidopsis protoplasts. Planta 248, 571–578. doi: 10.1007/s00425-018-2919-5
Ozel, H. B., Abo Aisha, A. E. S., Cetin, M., Sevik, H., and Zeren Cetin, I. (2021). The effects of increased exposure time to UV-B radiation on germination and seedling development of Anatolian black pine seeds. Environ. Monit. Assess. 193:388. doi: 10.1007/s10661-021-09178-9
Patel, J. S., Selvaraj, V., Gunupuru, L. R., Kharwar, R. N., and Sarma, B. K. (2020). Plant G-protein signaling cascade and host defense. 3 Biotech. 10:219. doi: 10.1007/s13205-020-02201-9
Qi, Y., Wei, H., Gu, W., Shi, W., Jiang, L., Deng, L., et al. (2021). Transcriptome profiling provides insights into the fruit color development of wild Lycium ruthenicum Murr. from Qinghai-Tibet Plateau. Protoplasma 258, 33–43. doi: 10.1007/s00709-020-01542-9
Qiao, Q., Wang, Q., Han, X., Guan, Y. L., Sun, H., Zhong, Y., et al. (2016). Transcriptome sequencing of Crucihimalaya himalaica Brassicaceae reveals how Arabidopsis close relative adapt to the Qinghai-Tibet Plateau. Sci. Rep. 6:21729. doi: 10.1038/srep21729
Rameneni, J. J., Dhandapani, V., Paul, P., Devaraj, S. P., Choi, S. R., Yi, S. Y., et al. (2018). F-Box genes in Brassica rapa: genome-wide identification, structural characterization, expressional validation, and comparative analysis. Plant Mol. Biol. Rep. 36, 500–517.
Rouault, T. A. (2019). The indispensable role of mammalian iron sulfur proteins in function and regulation of multiple diverse metabolic pathways. Biometals 32, 343–353. doi: 10.1007/s10534-019-00191-7
Smrcka, A. V., and Fisher, I. (2019). G-protein betagamma subunits as multi-functional scaffolds and transducers in G-protein-coupled receptor signaling. Cell Mol. Life Sci. 76, 4447–4459. doi: 10.1007/s00018-019-03275-2
Song, H. Y., Kim, D. H., and Kim, J. M. (2018). Comparative transcriptome analysis of dikaryotic mycelia and mature fruiting bodies in the edible mushroom Lentinula edodes. Sci. Rep. 8:8983. doi: 10.1038/s41598-018-27318-z
Wang, M., Chen, L., Liang, Z., He, X., Liu, W., Jiang, B., et al. (2020). Metabolome and transcriptome analyses reveal chlorophyll and anthocyanin metabolism pathway associated with cucumber fruit skin color. BMC Plant Biol. 20:386. doi: 10.1186/s12870-020-02597-9
Wang, Y., Wang, Y., and Deng, D. (2019). Multifaceted plant G protein: interaction network, agronomic potential, and beyond. Planta 249, 1259–1266. doi: 10.1007/s00425-019-03112-7
Wu, Y., Liao, W., Dawuda, M. M., Hu, L., and Yu, J. (2019). 5-Aminolevulinic acid (ALA) biosynthetic and metabolic pathways and its role in higher plants: a review. Plant Growth Regul. 87, 357–374.
Wu, Z., Jin, Y., Chen, B., Gugger, M. K., Wilkinson-Johnson, C. L., Tiambeng, T. N., et al. (2019). Comprehensive characterization of the recombinant catalytic subunit of cAMP-Dependent protein Kinase by Top-down mass spectrometry. J. Am. Soc. Mass Spectrom. 30, 2561–2570. doi: 10.1007/s13361-019-02341-0
Xing, R., Gao, Q. B., Zhang, F. Q., Fu, P. C., Wang, J. L., Yan, H. Y., et al. (2017). Genetic variation and phylogenetic relationships of the ectomycorrhizal Floccularia luteovirens on the Qinghai-Tibet Plateau. J. Microbiol. 55, 600–606. doi: 10.1007/s12275-017-7101-4
Xu, H. Y., Xie, Z. L., Jiang, H. C., Guo, J., Meng, Q., Zhao, Y., et al. (2021). Transcriptome analysis and expression profiling of molecular responsed to Cd toxicity in morchella spongiola. Mycobiology 49, 421–433. doi: 10.1080/12298093.2021.1937882
Yu, Y., Yang, S., Bian, L., Yu, K., Meng, X., Zhang, G., et al. (2021). Identification of C3H2C3-type RING E3 ubiquitin ligase in grapevine and characterization of drought resistance function of VyRCHC114. BMC Plant Biol. 21:422. doi: 10.1186/s12870-021-03162-8
Yuan, L., Zhang, L., Wu, Y., Zheng, Y., Nie, L., Zhang, S., et al. (2021). Comparative transcriptome analysis reveals that chlorophyll metabolism contributes to leaf color changes in wucai (Brassica campestris L.) in response to cold. BMC Plant Biol. 21:438. doi: 10.1186/s12870-021-03218-9
Zhang, M. X., Zhao, X. C., Chen, X., Li, M. Y., and Wang, X. D. (2021). Enhancement of riboflavin production in Bacillus subtilis via in vitro and in vivo metabolic engineering of pentose phosphate pathway. Biotechnol. Lett. 43, 2209–2216. doi: 10.1007/s10529-021-03190-2
Zhao, G., Dong, F., Lao, X., and Zheng, H. (2021). Strategies to increase the production of biosynthetic riboflavin. Mol. Biotechnol. 63, 909–918. doi: 10.1007/s12033-021-00318-7
Zhao, Y. T., Pu, Y. F., Lin, H. L., and Tang, R. (2021). Examining soil erosion responses to grassland conversation policy in three-river headwaters, China. Sustainability 13, 1–14.
Zheng, Q., Guan, G., Cao, C., Li, Q., and Huang, G. (2020). The PHO pathway regulates white-opaque switching and sexual mating in the human fungal pathogen Candida albicans. Curr Genet. 66, 1155–1162. doi: 10.1007/s00294-020-01100-z
Zhou, Y., Yuikawa, N., Nakatsuka, H., Maekawa, H., Harashima, S., Nakanishi, Y., et al. (2016). Core regulatory components of the PHO pathway are conserved in the methylotrophic yeast Hansenula polymorpha. Curr. Genet. 62, 595–605. doi: 10.1007/s00294-016-0565-7
Keywords: Floccularia luteovirens, strong UV radiation, molecular regulation model, riboflavin, chlorophyll, Qinghai-Tibet Plateau
Citation: Guo J, Xie Z, Jiang H, Xu H, Liu B, Meng Q, Peng Q, Tang Y and Duan Y (2022) The Molecular Mechanism of Yellow Mushroom (Floccularia luteovirens) Response to Strong Ultraviolet Radiation on the Qinghai-Tibet Plateau. Front. Microbiol. 13:918491. doi: 10.3389/fmicb.2022.918491
Received: 12 April 2022; Accepted: 17 May 2022;
Published: 20 June 2022.
Edited by:
Xian-Wen Yang, Ministry of Natural Resources, ChinaReviewed by:
Li Taotao, South China Botanical Garden (CAS), ChinaCopyright © 2022 Guo, Xie, Jiang, Xu, Liu, Meng, Peng, Tang and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhanling Xie, eGllemhhbmxpbmcyMDEyQDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.