- 1College of Land and Environment, Shenyang Agricultural University, Shenyang, China
- 2Key Laboratory of Pollution Ecology and Environmental Engineering, Institute of Applied Ecology, Chinese Academy of Sciences, Shenyang, China
- 3College of Bioscience and Biotechnology, Shenyang Agricultural University, Shenyang, China
Plant growth-promoting rhizobacteria (PGPR) can produce hormone-like substances, promote plant nutrient uptake, enhance plant resistance, inhibit the growth of pathogenic bacteria, and induce plant resistance to biotic and abiotic stresses. Bacillus is one of the most studied genera that promote plant root development. Since its discovery in 2009, B. aryabhattai has shown promising properties such as promoting plant growth and improving crop yield. However, the mechanisms of B. aryabhattai promoting plant growth remain to be investigated. In this study, the chromosome of B. aryabhattai strain LAD and five plasmids within the cell were sequenced and annotated. The genome, with a length of 5,194,589 bp and 38.12% GC content, contains 5,288 putative protein-coding genes, 39 rRNA, and 112 tRNA. The length of the five plasmids ranged from 116,519 to 212,484 bp, and a total of 810 putative protein-coding genes, 4 rRNA, and 32 tRNA were predicted in the plasmids. Functional annotation of the predicted genes revealed numerous genes associated with indole-3 acetic acid (IAA) and exopolysaccharides (EPSs) biosynthesis, membrane transport, nitrogen cycle metabolism, signal transduction, cell mobility, stress response, and antibiotic resistance on the genome which benefits the plants. Genes of carbohydrate-active enzymes were detected in both the genome and plasmids suggesting that LAD has the capacity of synthesizing saccharides and utilizing organic materials like root exudates. LAD can utilize different carbon sources of varied carbon chain length, i.e., methanol, acetate, glycerol, glucose, sucrose, and starch for growth and temperature adaptation suggesting a high versatility of LAD for thriving in fluctuating environments. LAD produced the most EPSs with sucrose as sole carbon source, and high concentration of IAA was produced when the maize plant was cultivated with LAD, which may enhance plant growth. LAD significantly stimulated the development of the maize root. The genome-based information and experimental evidence demonstrated that LAD with diverse metabolic capabilities and positive interactions with plants has tremendous potential for adaptation to the dynamic soil environments and promoting plant growth.
Introduction
Plant growth and development is highly dependent on the interactions with other living organisms that habitat the soil ecosystem. These interactions are very complex and critical for maintaining the biodiversity in the below-ground system (Lau and Lennon, 2011; Bever et al., 2013; Shao et al., 2018). Microbes are the most abundant and diverse entities in soil and can directly participate in ecological processes and nutrient cycling. The role of soil microbial community in soil ecosystem functioning and plant production has been widely investigated via isolation of culturable microbes and culture-independent techniques (Liu et al., 2019; Roy et al., 2020). However, the soil environment is extremely heterogeneous imposing great challenges on studies of soil microbial ecology.
Roots harbor a rich abundance of biomass and are the crucial organ of the plant to absorb water and nutrients for the plant. Rhizosphere the narrow region immediately adjacent to the root is the plant root-soil interface and is the hot spot for microbial interactions and cross-kingdom interactions between plants and microbes (Chaparro et al., 2014; Korenblum et al., 2020). Rhizosphere microbes and the plant may form symbiotic relationships in which the root microbiome utilizes root exudates and secretes compounds benefiting plant growth (Berendsen et al., 2012). Rhizosphere microbes may synthesize antibiotics as required by the plant for suppression of soil-borne pathogens and ultimately enhance the plant health (Mendes et al., 2013; Lazcano et al., 2021). Other rhizosphere microbiome-plant mutualistic interactions include microbiome fixing and providing essential elements (e.g., nitrogen) for plant growth (Moreau et al., 2019). Rhizosphere microbiome may also induce systemic root exudation of metabolites and mediate root-root signaling promoting soil conditioning (Korenblum et al., 2020). Meanwhile, many environmental and biotic factors can influence rhizosphere microbiome-plant interactions making it more complicated to study the mechanism of different rhizosphere microbes promoting plant growth.
Rhizospheric microorganisms could produce hormone-like substances, then promote plant growth and nutrient uptake, inhibit the growth of pathogenic bacteria and induce plant resistance to biotic and abiotic stresses (Ahmed and Hasnain, 2014). For example, inoculation with typical beneficial microbial mycorrhizal fungi not only promotes the secretion of organic acids and phosphatases from the roots of symbiotic plants, but also enhances the ability of plant to activate insoluble phosphates in the soil and promotes the uptake of water and minerals, especially phosphate (Berta, 2000; Erik et al., 2000; Philippe, 2001; Maria, 2005). Bacillus amylolyticus strain B3 harbors functional genes capable of directly promoting crop growth, like yhcX and ysnE, key genes for plant growth hormone (IAA) synthesis; and AlsS, AlsD, and AlsR, synthase genes related to the volatile disease-promoting substance 2,3-butanediol. The phytase synthesis gene phy is present intact in the B3 genome, and phytase can catalyze the degradation of phytic acid into inositol, facilitating plant uptake and utilization of nutrients from soil (Idriss et al., 2002; Zhang et al., 2010). The exocrine secretion of Bacillus subtilis BS-2 promotes rice growth, increases chlorophyll content in the crop, slows membrane lipid peroxidation in rice, and promotes indoleacetic acid production in the plant (Ma et al., 2018). Bacterial colonization can trigger plant immune reaction, and the interaction between beneficial bacteria and plant immune system is essential for efficient bacterial colonization, survival, and plant growth promoting hormone production (Tzipilevich et al., 2021).
Bacillus is the one of the most studied genera among the plant growth promoting rhizobacteria and shows tremendous potential in promoting plant growth (Chen et al., 2007; Tahir et al., 2017; Backer et al., 2018). Bacillus aryabhattai are widely distributed in nature but were only discovered in 2009 (Shivaji et al., 2009). More strains of B. aryabhattai were isolated from various environments including plant roots, and evaluations of this rhizosphere bacteria have revealed promising properties of this Bacillus species for promoting plant growth and improving crop yields thus having attracted plenty of attention from researchers (Bhattacharyya et al., 2017; Park et al., 2017; Ghosh et al., 2018). Mehmood et al. (2021) found that B. aryabhattai promoted wheat growth and reduced the effects of salt stress on wheat. However, the mechanism of this rhizobacteria species promoting plant growth remains to be investigated. With the development of sequencing technology, whole genome sequencing of the isolated microbial strains has been widely used to reveal the potential functions of the microbes in enhancing the plant performance (Chu et al., 2020; Chen et al., 2022).
In a recent study, an excellent plant inter-rhizosphere strain, B. aryabhattai LAD, was isolated from maize rhizosphere, and the impact of LAD on rhizosphere microbial structure was also explored in the previous study (Deng et al., 2022). LAD showed promising plant growth promoting properties, including nitrogen fixation and phosphorus solubilization and IAA production. Here, we aimed to reveal the genes related to plant growth-promoting functions of this strain through whole genome sequencing and investigate the genetic differences between this strain and similar plant growth promoting rhizobacterial strains via comparative genomics. The metabolic and phenotypic traits and habitat-specific adaptations of this strain to maize root promotion were also investigated.
Materials and methods
Total DNA extraction and whole genome sequencing
The strain LAD was isolated from maize rhizosphere in our lab, and initial characterization of the newly isolated bacterial strain was performed and the effects of LAD on corn seedlings were also evaluated. In this study, LAD was grown in its optimum growth medium containing 20 g L−1 sucrose, 2 g L−1 beef extract, 0.4 g L−1 KH2PO4, 0.4 g L−1 MgSO4·7H2O, 0.4 g L−1 NaCl, 0.4 g L−1 CaSO4·2H2O, and 2 g L−1 CaCO3 at 180 rpm and 37°C. The bacterial cells were harvested at the late exponential phase (around 22 h) for total DNA extraction with the extraction kit. The extracted DNA was quantified with Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) and sent out to Shanghai Personal Biotechnology Co., Ltd. for next-generation sequencing of the total DNA. Whole genome sequencing of LAD was performed using on PacBio Sequel (Pacific Biosciences of California, Inc., Menlo Park, CA, United States) and Illumina NovaSeq (Illumina, Inc., San Diego, CA, United States) sequencing platforms, respectively.
Whole genome annotation
The obtained sequence reads from PacBio platform were assembled into contigs using HGAP4 WGS-Assembler 8.2 and CANU (Chin et al., 2016; Koren et al., 2017). The Illunima reads were employed for correction of the assembled contigs with Pilon 1.22 to get the final complete genome (Walker et al., 2014). GeneMarkS was used for finding protein-coding genes via GeneMark.hmm program (Besemer et al., 2001). Transfer RNAs (tRNAs) were predicted using tRNAscan-SE, and the prediction of other non-coding RNAs was performed in Rfam (Lowe and Eddy, 1997; Kalvari et al., 2018). Clustered regularly interspaced short palindromic repeats (CRISPRs) were predicted using CRISPRFinder program (Grissa et al., 2007). PHASTER (Phage Search Tool Enhanced Release) was used for detection of prophages on the genome (Arndt et al., 2016).
Genomic comparisons
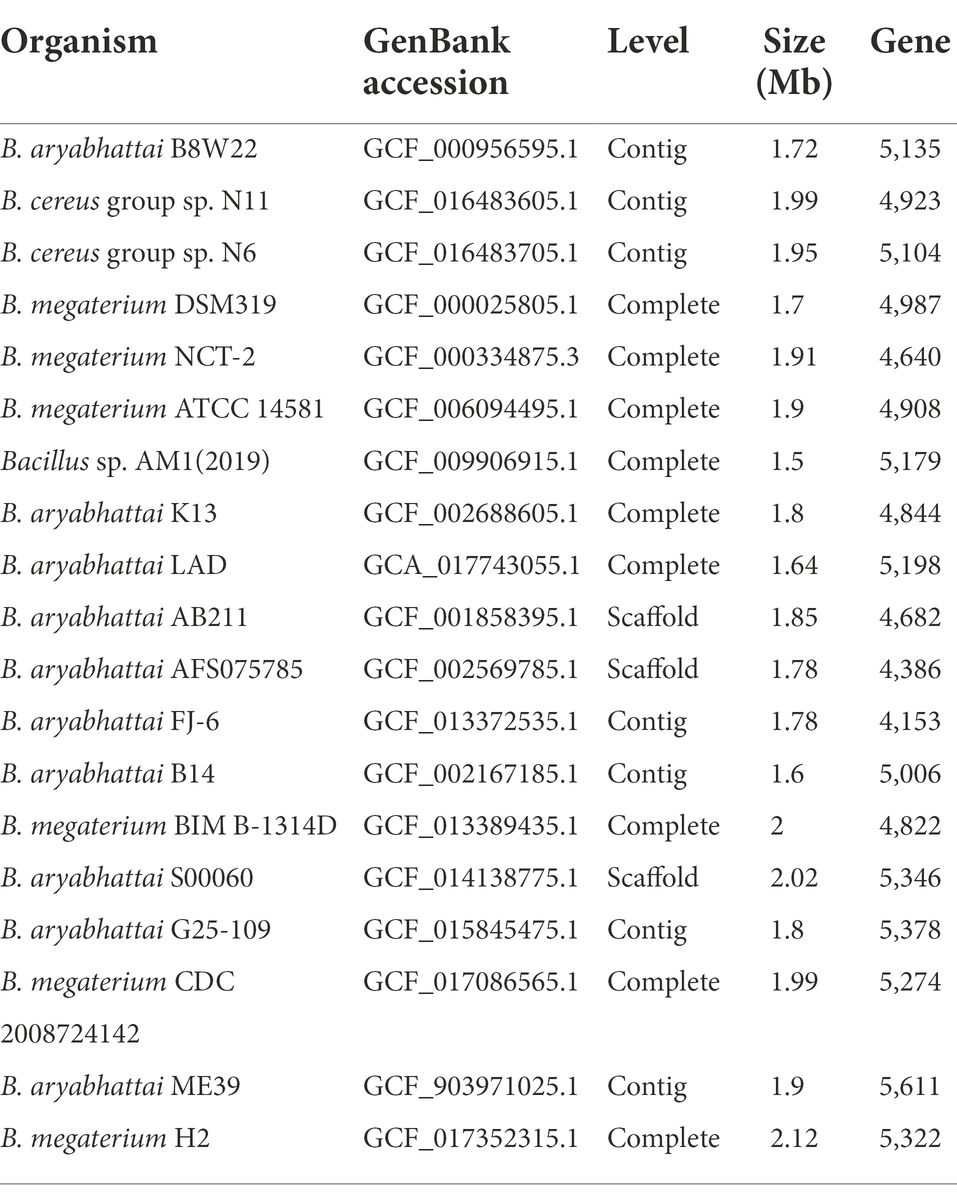
Reference genome sequences of other Bacillus strains were retrieved from NCBI GenBank database for comparative genomic analysis of B. aryabhattai LAD (Table 1). B. aryabhattai is a homotypic synonym of Priestia aryabhattai. Pairwise genome comparisons were performed on JSpeciesWS which evaluates whole genome homologies via BLAST alignments for determination of the average nucleotide identity (ANI) between genome sequences (Richter et al., 2016). The bacterial pan genome analysis (BPGA) pipeline was used to identify the orthologous pan genome profile among the genomes of the B. aryabhattai strains (Chaudhari et al., 2016). Core genome (the set of shared genes by all strains), accessory genome (genes shared by different strains but not by all strains), and unique genes (exclusively belonging to one strain), and pan genome (non-homologous genes) were identified with BPGA. Carbohydrate-active enzymes (CAZy) were characterized based on the CAZy database (Lombard et al., 2014). The whole genomic sequence data of B. aryabhattai LAD was deposited to NCBI GenBank, with the accession number of GCA_017743055.1.
Bacillus aryabhattai LAD growth under different temperatures and carbon sources
The growth of LAD on different carbon sources and at different temperatures was evaluated with Epoch 2 microplate spectrophotometer (BioTek Instruments Inc., Winooski, VT, United States). LAD cells were inoculated in LB medium and grown at 37°C overnight. The LAD culture was used as inoculum (1%) for growth in the optimum growth medium, but the carbon source in the medium was the same concentration (2%, m v−1) of sodium acetate, glycerol, glucose, sucrose, or starch. The growth of LAD on methanol was also examined in the optimum medium but with the carbon source replaced with 0.5% (v v−1) methanol. The growth curves of LAD populations on different carbon sources and at different temperatures (20°C and 37°C) were obtained by culturing the cells in 24-well plates and analyzed at a 1-h interval in the Epoch 2 microplate spectrophotometer. Each treatment included three replicates, and the mean cell density was used to plot the growth curves.
Indole-3-acetic acid production by Bacillus aryabhattai LAD
The maize seeds (Dongdan 1,331) were immersed in 105 CFU ml−1 LAD cell suspensions and cultivated in the artificial climate room for 8 days. As the control treatment, the maize seeds were immersed in sterilized water and cultured for 8 days under the same conditions. Each treatment included three replicates, and t-test was used to reveal the statistical differences between LAD treatment and control. The plant was removed after cultivation, and the liquid culture was concentrated by rotary evaporation and was archived for later use. Liquid chromatography system of Waters ACQUITY UPLC (Milford, MA, USA) with a liquid chromatography column (182.1 mm × 50 mm i.d., 1.7 μm) was used to determine the amount of the produced IAA (Fu et al., 2012). IAA was isolated in a mobile phase consisting of methanol solution (mobile phase A) and 0.1% formic acid aqueous solution (mobile phase B). The gradient elusion was run at a flow rate of 0.2 ml min−1 with initial 20% mobile phase A which was increased to 80% in the next 12 min. The volume fraction of mobile phase A was reduced from 80 to 20% in the time range of 12 to 16 min. The injection volume for all samples was 3 μl and the column temperature was 40°C. The mass spectrometer that affiliated with the UPLC system used an electrospray ion source, with a capillary voltage of 0.8 KV in positive ion mode (ESI+), a taper voltage of 25 V, a dissolvent temperature of 650°C, a dissolvent gas flow rate of 1,000 l h −1, and a conical hole back blow 5 l h −1. The collision voltage was set 30 V at 176 > 103 and 15 V at 176 > 130.
Production of exopolysaccharides
LAD cells were grown in LB medium at 37°C for 24 h before the cells were transferred into the optimum growth medium with different carbon sources, i.e., methanol (0.5, v v−1), sodium acetate (2%, m v−1), glycerol (2%, m v−1), glucose (2%, m v−1), sucrose (2%, m v−1), and starch (2%, m v−1), respectively. The bacterial cells were cultivated at 37°C with the EPSs production being evaluated at 24 and 72 h of the cultivation. For EPSs extraction, 10 ml bacterial culture was centrifuged at 5,000 rpm for 20 min, and the supernatant was transferred to a clean 50 ml centrifuge tube with 2-fold volume of 95% ethanol. The mixture was stirred using a glass rod until clear flocculent precipitation appeared. The mixture was stored at 4°C for 24 h before centrifugation at 10,000 rpm for 15 min. The extracted products, crude EPSs, were air dried at room temperature and weighed for determination of the EPSs production on different carbon sources.
Impacts of LAD on maize root development
LAD was cultivated in selective nutrient broth at 37°C for 72 h (until OD600nm reached 1.4). The LAD cells were harvested by centrifugation and resuspended in sterile water, and the cell suspension was diluted to a final concentration of 105 CFU ml−1 using sterile water. The maize seeds were immersed in the diluted bacterial suspension for 24 h, after which the maize seeds were cultivated in laboratory hydroponics and in the field, respectively. In the control treatment, the maize seeds were soaked in the sterile water for 24 h before being cultivated in laboratory hydroponics and field. The root development of the maize seedlings in hydroponics was measured by a root system analyzer after 14 days, while measurement of the root development of each maize plant grown in field was performed after 60 days (Bucksch et al., 2014). The hydroponic and field cultivation for each treatment included 10 maize plants, respectively. Each treatment was repeated three times. Student’s t-test was used to compare the root development under LAD treatment with that of the control to show the statistical differences.
Results
Functional annotation of Bacillus aryabhattai LAD whole genome
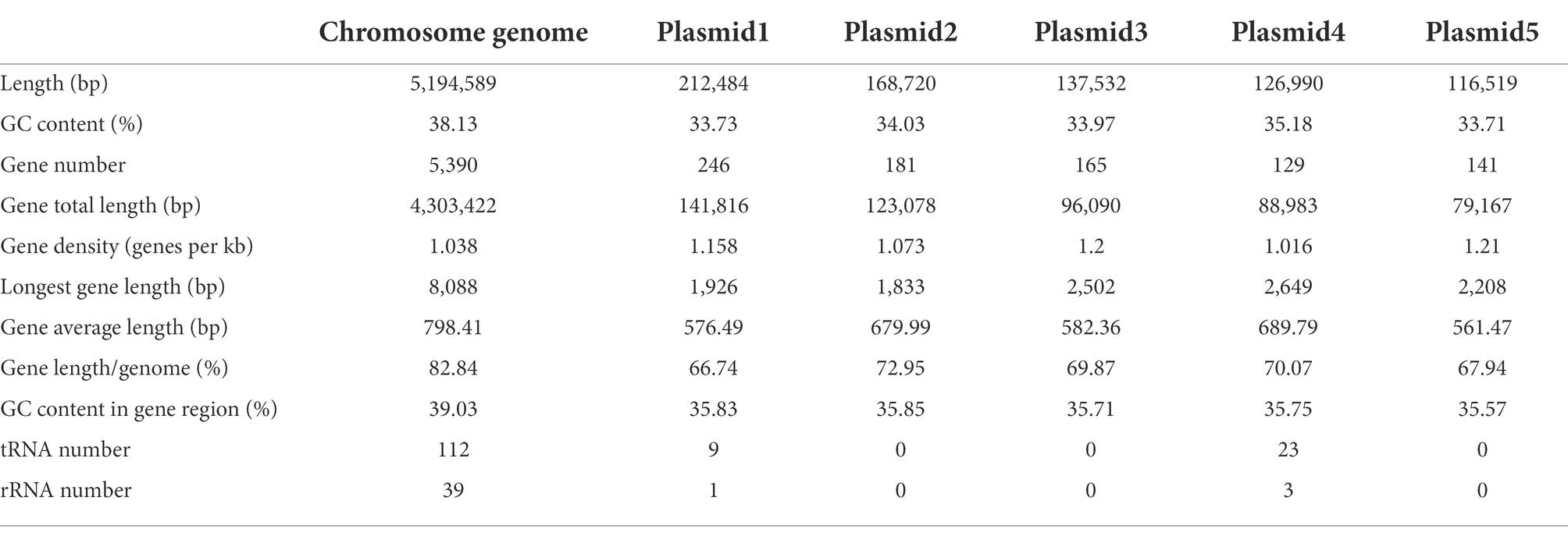
A total of 184,213 sequence reads was obtained in the next-generation sequencing. The de novo genome assembly using HGAP revealed a single circular chromosome genome and five circular plasmids in B. aryabhattai LAD. The newly sequenced genome was composed of 5,194,589 bp with a GC content of 38.12% (Figure 1). The five plasmids contained 212,484, 168,720, 137,532, 126,990, and 116,519 bp, respectively, with GC content ranging from 33.71 to 35.18%. The properties of the chromosome genome and plasmids are shown in Table 2. The genome contained 5,390 predicted genes, and the total length of the predicted genes was 4,303,422 genes accounting for 82.8% of the genome length. The five plasmids contained 246, 181, 165, 129, and 141 genes, respectively, with the coding percentage ranging from 66.7 to 73%. There were 112 tRNA genes and 39 rRNA genes on the chromosome genome. Fifteen genome islands (GIs), 4 CRISPRs, and 2 incomplete prophages were predicted on the genome of B. aryabhattai LAD. There were 13 5S rRNA genes (with an average length of 110 bp), 13 16S rRNA genes (average length of 1,549 bp), and 13 23S rRNA genes (average length of 2,932 bp) on the genome. The genomic annotation also revealed rRNA genes on plasmid1 and plasmid4. Specifically, one 5S rRNA gene of 108 bp was present on plasmid1, and there were one 5S rRNA gene (111 bp), one 16S rRNA gene (1,549 bp), and one 23S rRNA gene (2,932 bp) on plasmid4.
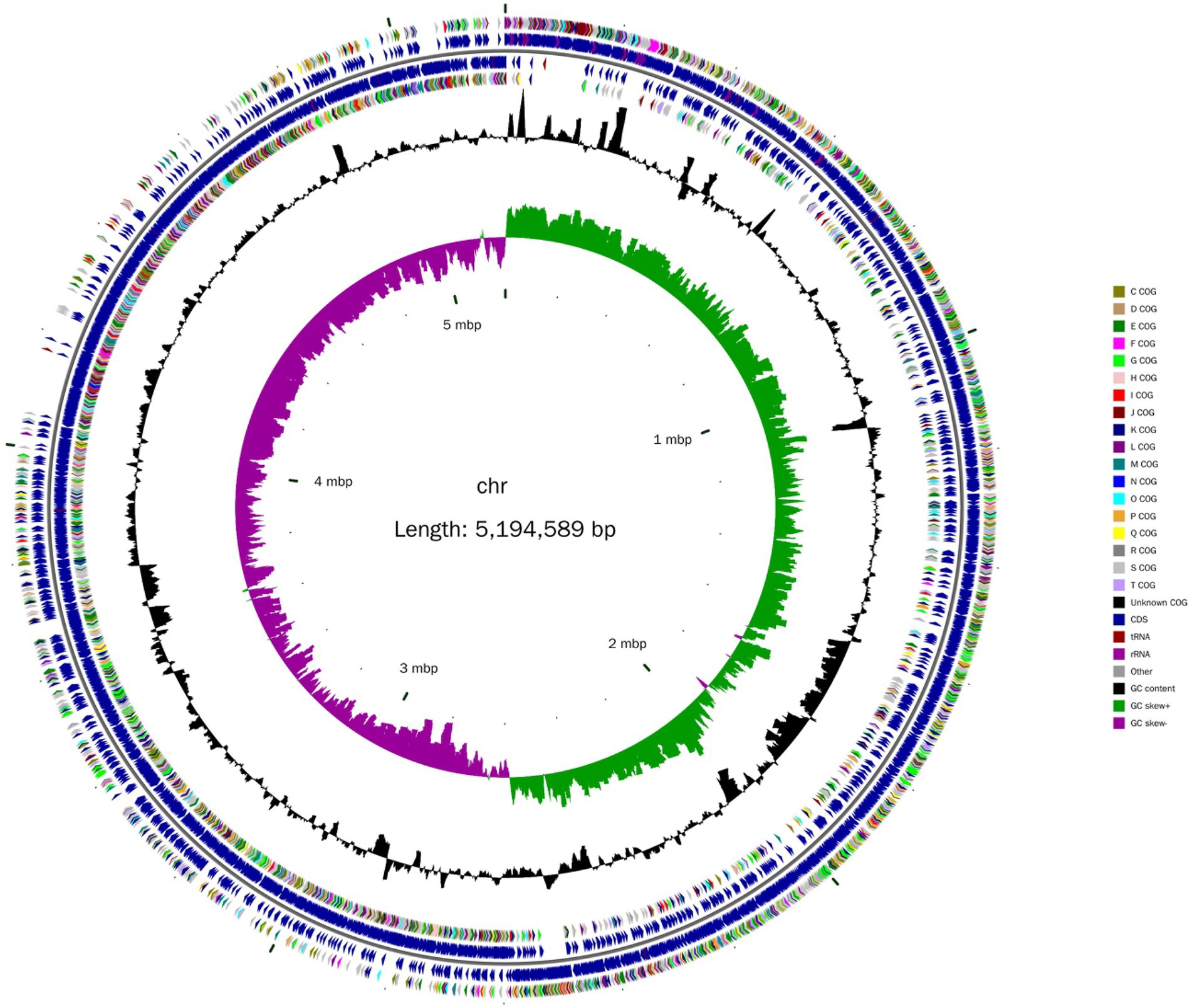
Figure 1. Circular chromosome diagram of B. aryabhattai LAD. From the inner circle to the outer circle, the information on each of the seven circles shows genome position, GC skew, GC content, coding sequence on the positive and strands (classified by Cluster of Orthologous Groups of protein), and the position of the coding sequence, tRNA and rRNA on the genome.
There were 148 CAZymes, including 35 glycosyltransferases (GTs), 1 polysaccharide lyases (PLs), 34 carbohydrate esterases (CEs), 12 auxiliary activities (AAs), 17 carbohydrate binding modules (CBMs), and 49 glycoside hydrolases (GHs) identified in the B. aryabhattai LAD chromosome genome via the CAZy annotation pipeline (Figure 2A). CAZymes were also present in two plasmids, with 1 GTs, 2 CEs, 2AAs, and 2GHs identified on plasmid 1, and 1 GTs and 1 GHs identified on plasmid 2. The eggNOG (evolutionary genealogy of genes: Non-supervised Orthologous Groups) annotation revealed an abundance of genes for basic cellular functions including amino acid transport and metabolism (381 genes, accounting for 7.07% of the total gene abundance), carbohydrate transport and metabolism (305, 5.7%), inorganic ion transport and metabolism (268, 5%), energy production and conversion (249, 4.6%), cell wall/membrane/envelope biogenesis (202, 3.7%), signal transduction mechanisms (188, 3.5%), and replication, recombination and repair (166, 3.1%; Figure 2B). The Gene Ontology (GO) classification revealed that ion binding, oxidoreductase activity, and DNA binding were very active molecular functions, and cellular nitrogen compound metabolic process, biosynthetic process, small molecule metabolic process, and transport were major biological processes (Figure 2B). The analysis against the Comprehensive Antibiotic Resistance Database (CARD) identified 28 antibiotic resistance genes and 4 antibiotic biosynthesis genes on the genome and 3 more antibiotic resistance genes on the plasmids.
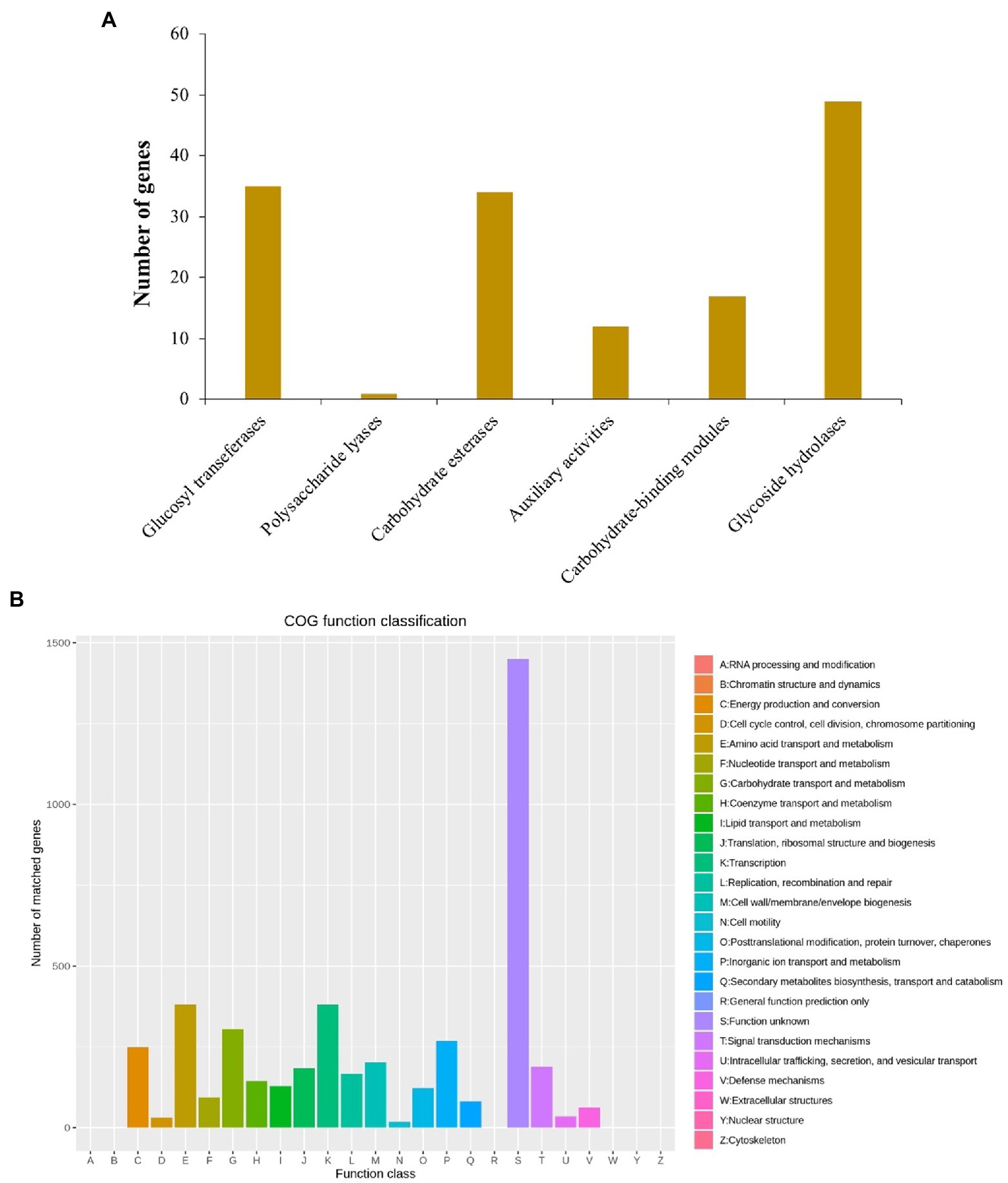
Figure 2. (A) Annotation of carbohydrate active enzymes (CAZymes) genes on B. aryabhattai LAD genome. The hmmscan program was used for estimation of the CAZy genes (alignment length > 80 amino acids, E-value <1e-5; alignment length < 80 amino acids, E-value <1e-3). (B) eggNOG annotation. The protein-coding gene sequences were compared to COG database using DIAMOND blastp (cutoff at 1e-6).
Dig of promoting growth genes
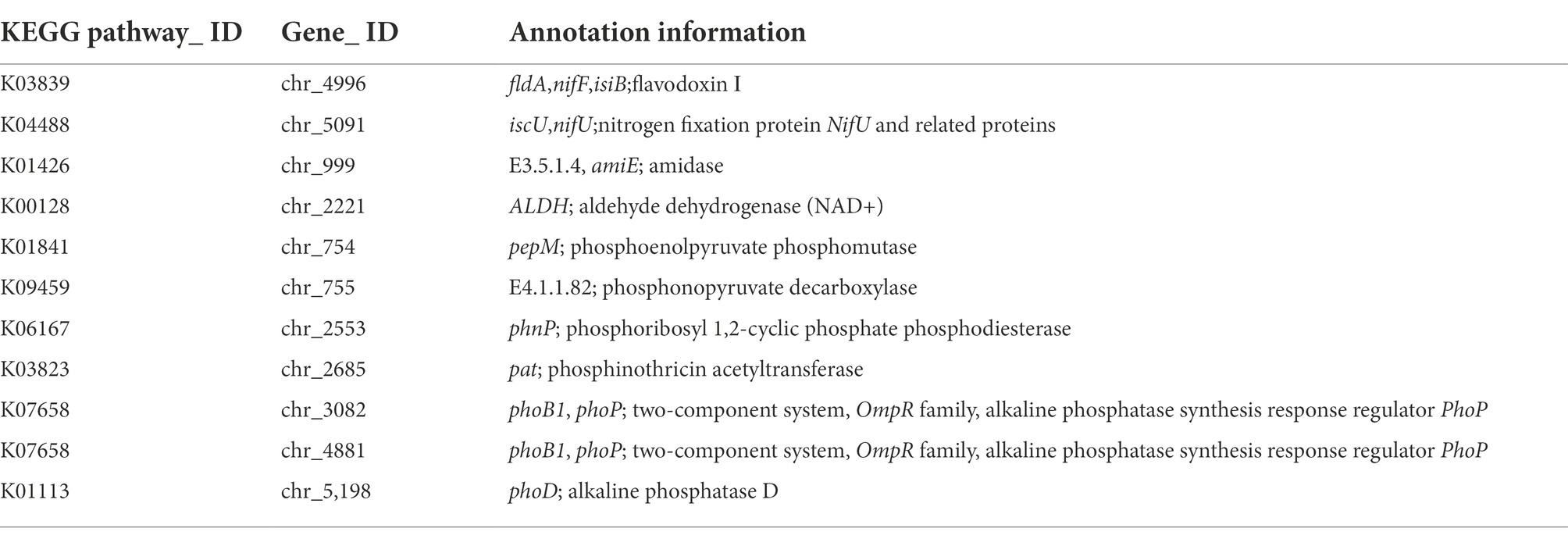
In the whole genome of LAD strain, there were many genes related to the promotion of plant growth, some of which are shown in the Table 3. Ten related genes were found to be involved in the Nitrogen metabolism pathway in LAD strains, and the NifF gene was found at chr_4496 and NifU gene at chr_5091. In the KEGG pathway of Tryptophan metabolism, 25 genes related to IAA synthesis were identified, including amidase (amiE), which hydrolyzes indole-3-acetamide to indoleacetic acid, and aldehyde dehydrogenase (ALDH), which hydrolyzes indole-3-acetaldehyde to indoleacetic acid. We identified 20 genes related to biofilm formation. Four genes encoding related proteins in LAD strains mapped to the KEGG pathway of phosphonate and phosphinate metabolism, including phosphoenolpyruvate phosphomutase (pepM), phosphonopyruvate decarboxylase, phosphoribosyl 1,2-cyclic phosphate phosphodiesterase (phnP) and phosphinothricin acetyltransferase (pat, bar), and a total of 53 phosphatase genes were identified in the LAD genome, mapping to 34 KEGG metabolic pathways. The extracellular polysaccharide production by LAD strains is closely related to carbohydrate metabolism, a total of 315 carbohydrate related genes were found for carbohydrate metabolism. The two-component regulatory system is a major mechanism for biotransduction and response to external environmental changes, which can sense environmental changes, regulate internal gene expression and play an important role in the survival and adaptation of microorganisms to different environments, and help microorganisms preserve their competitive advantage in the environment. A total of 98 KEGG pathways of two-component system were identified in LAD strains, including phosphate limitation, oxygen limitation, and temperature limitation system. A total of 106 genes related to the ABC transporters metabolic pathway were identified in LAD strains, including mineral and organic ion transporters, phosphate and amino acid transporters, peptide and nickel transporters, monoasccharide transporters, oligosaccharide, polyol, lipid transporters, metallic cation and iron-siderop hore and vitamin B12 transporters.
Comparative genomics analyses
The ANI between the whole genome sequences was measured by pairwise genome comparisons using JSpeciesWS Online Service to evaluate the homologies of the whole genomes and determine if the strains belong to the same species. The matrix of nucleotide identities between the whole genomes of the Bacillus strains was shown in Table 4. The ANI value of B. aryabhattai LAD based on BLAST against other Bacillus strains ranged from 67.7 to 95.6%, and the aligned percentage of the genome sequence ranged from 24.2 to 82.4%. The ANI values of B. aryabhattai LAD compared with other B. aryabhattai strains and B. megaterium strains were higher than 95%, which was the threshold identity for species boundaries.
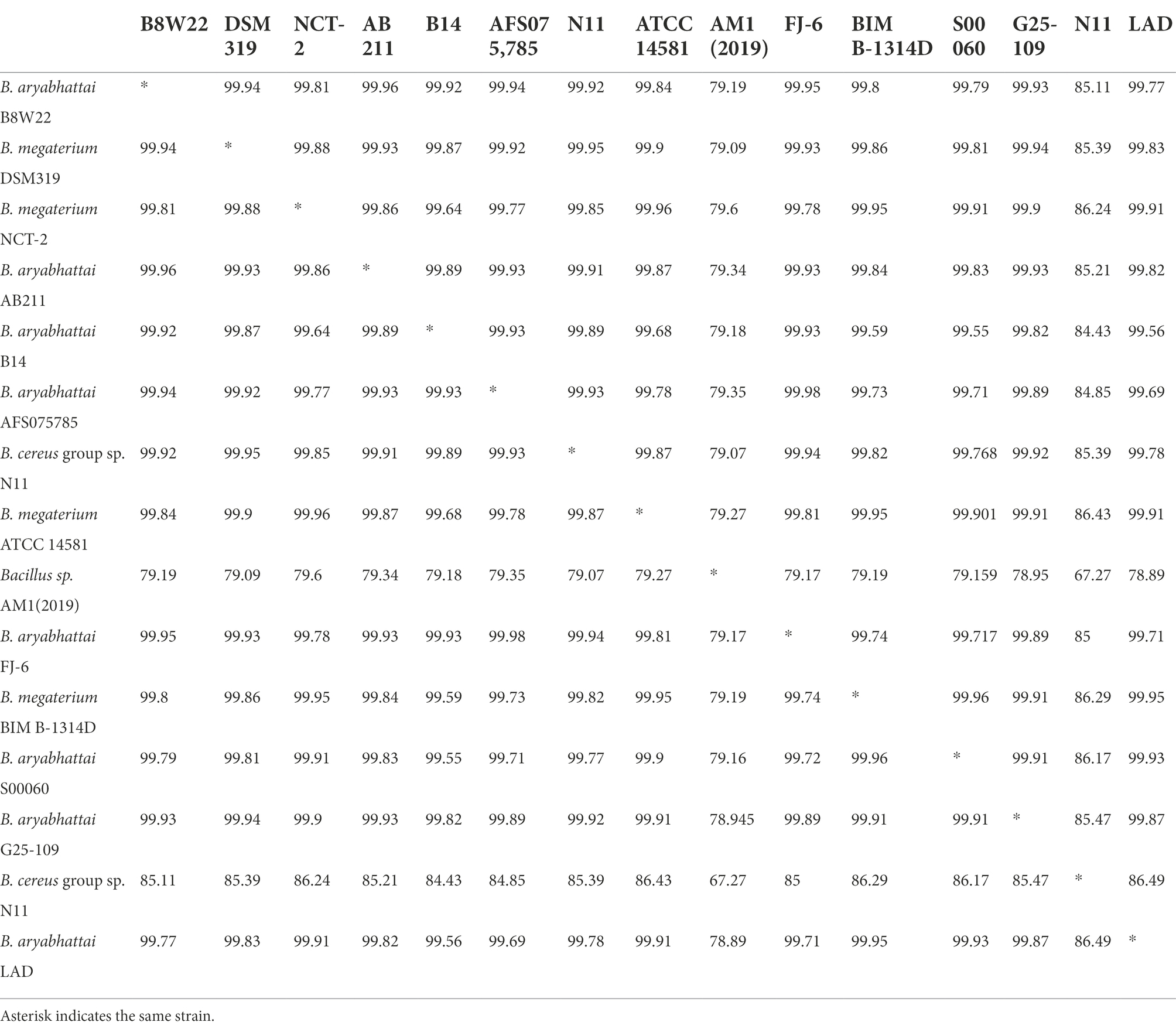
Table 4. Average nucleotide identities (ANI) analysis for pairwise genome comparison between B. aryabhattai LAD and other Bacillus strains.
PGAP pipeline was used to determine the pan-genome for B. aryabhattai LAD and 18 other Bacillus strains. The 19 compared Bacillus strains had a total pan-genome consisted of 94,898 putative protein-coding genes, and 1,111 of them (accounting for 1.17% the pan-genome) were core conserved genes across the genomes of the 19 strains (Figure 3). The number of strain-specific genes for each train ranged from 0 to 288, and Priestia aryabhattai FJ-16 had 288 strain-specific genes which was the highest among the 20 strains. The isolate in this study, B. aryabhattai LAD, had 1 strain-specific gene.
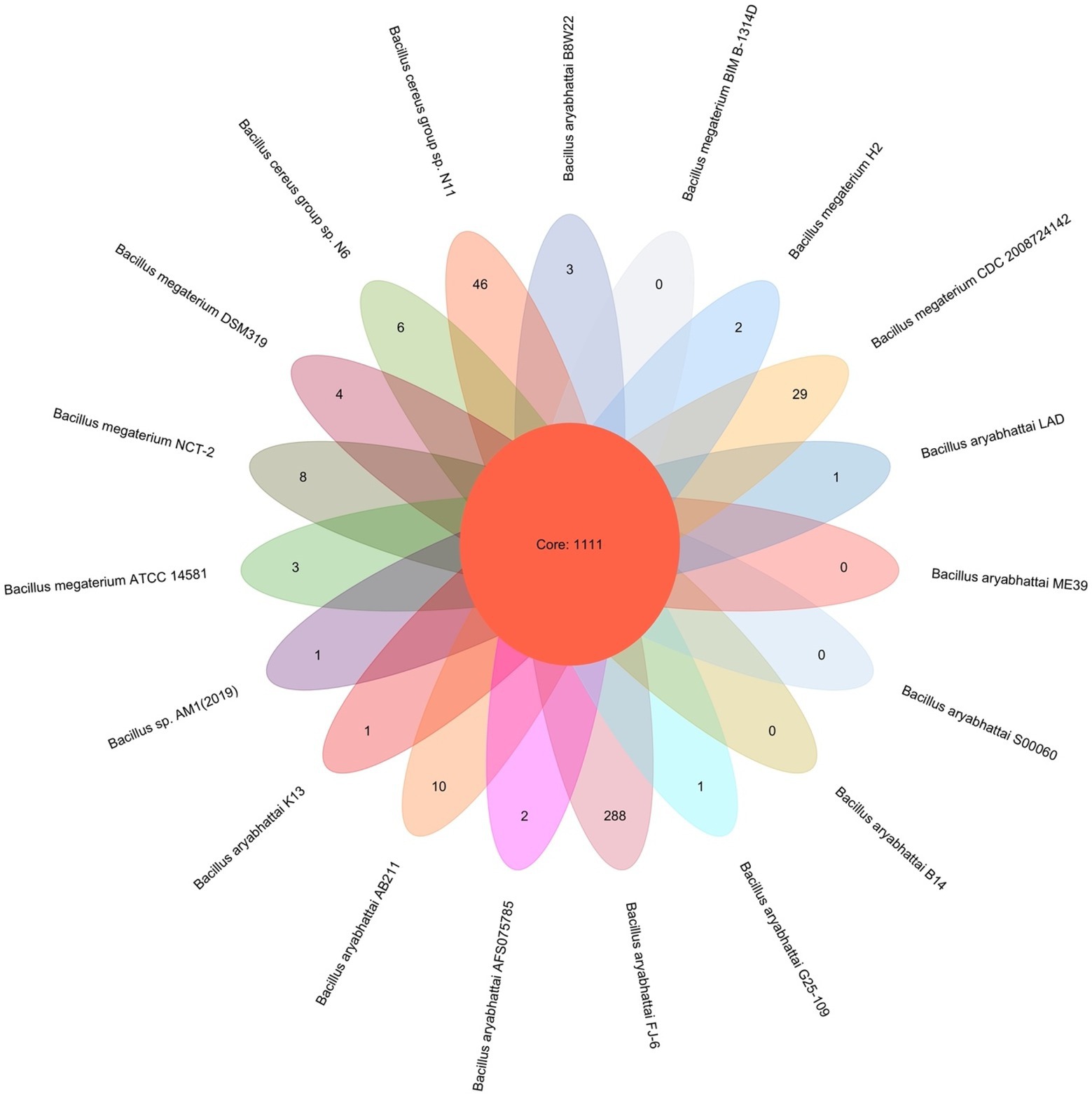
Figure 3. Pan-genome analysis of 19 Bacillus strains. The number in the overlapping center represents the number of orthologous coding sequences (core genome) shared by all analyzed strains. The number of coding sequences specific to each strain was shown in the non-overlapping portion of the oval.
Highly reliable pan-genome analysis can be obtained by mathematical extrapolation with more than five genomes (Vernikos et al., 2015). The analysis via BPGA showed that the pan-genome of the 19 Bacillus genomes had a parameter γ of 0.4 in the reduced power-fit curve equation [f(x) = 4175n0.4] suggesting that the pan-genome was open (Figure 4). The exponential curve equation of core genome [f1(x) = 4228.01e-0.07] had a steep slope and reached a minimum of 1,111 gene families.
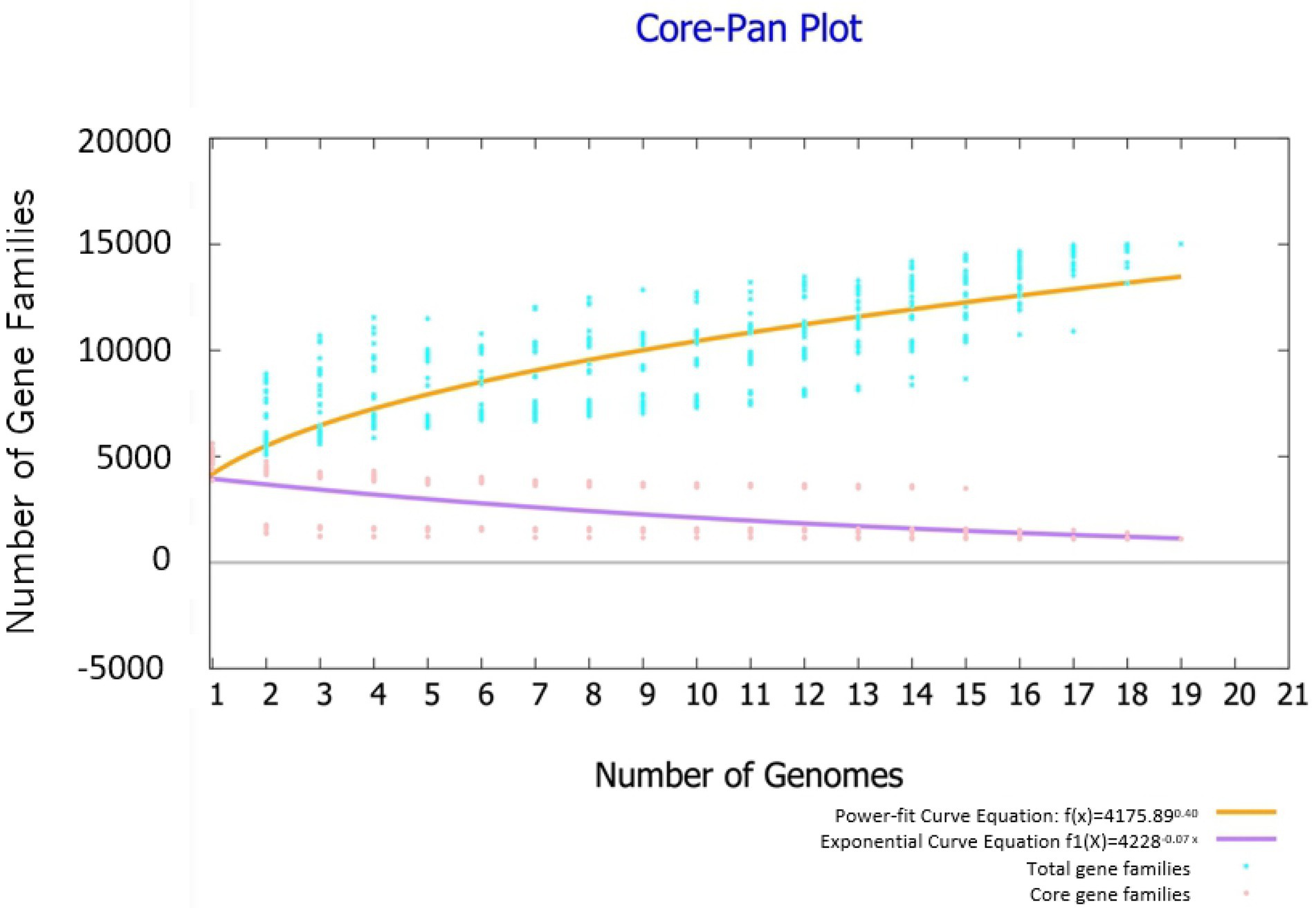
Figure 4. Pan genome and core genome profile plot of 19 Bacillus strains. The analysis was performed through the Bacterial Pan Genome Analysis (BPGA) software package.
Bacillusaryabhattai LAD growth and production of IAA and EPSs
The growth of B. aryabhattai LAD on different carbon sources was evaluated with Epoch 2 microplate spectrophotometer at 20°C and 37°C. The results showed that LAD can utilize all the six carbon sources, including acetate, glycerol, glucose, sucrose, starch, and methanol, under appropriate temperatures (Figure 5). LAD had the best growth on sucrose at 37°C, and the bacterial density reached the highest OD600 value (as high as 1.9). Glucose and glycerol supported good LAD growth both at 20°C and 37°C, with the OD600 value of glucose reaching 1.4 at 20°C and 1.5 at 37°C, and the OD600 value of glycerol reaching 1.1 at 20°C and 1.8 at 37°C. Interestingly, LAD can efficiently utilize methanol as carbon source for growth with the ability to reach a high bacterial density (an OD600 value of 0.4 at 20°C and 0.2 at 37°C) which demonstrates that B. aryabhattai LAD belongs to methylotrophic bacteria. Acetate also supported the growth of LAD with the bacterial cell density reaching an OD600 of 0.4 at 37°C. LAD was able to utilize starch, but the bacterial growth rate and final cell density were both lower than using other carbon sources.
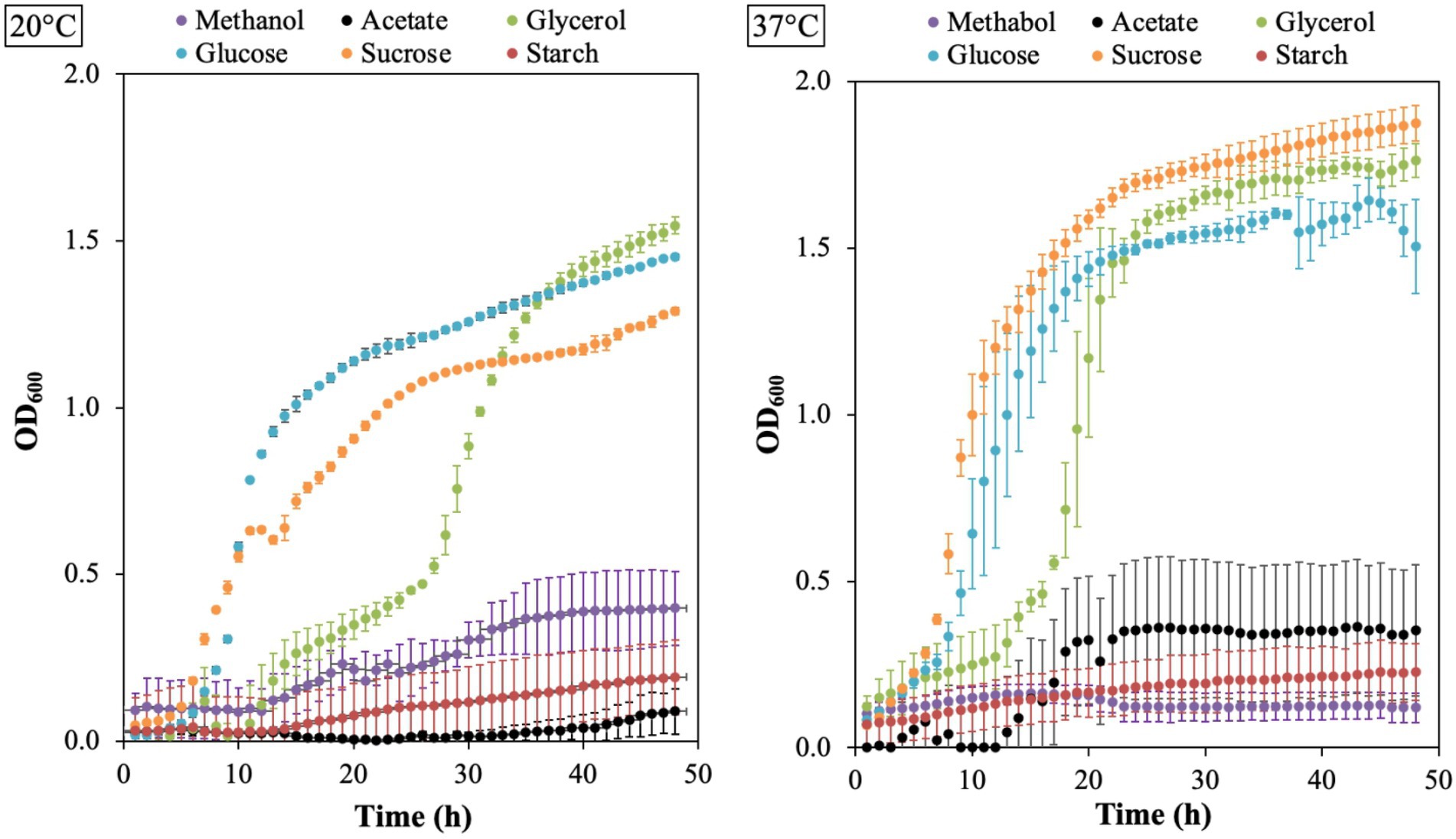
Figure 5. The growth plots for B. aryabhattai LAD. LAD cells were cultivated in the optimum growth medium with different carbon sources, i.e., methanol (0.5, v v−1), sodium acetate (2%), glycerol (2%), glucose (2%), sucrose (2%), and starch (2%), respectively. The temperature for LAD growth included 20°C and 37°C. Each data point represents the mean value of three replicates.
To examine the IAA-producing capacity of LAD, maize seedlings were cultivated with LAD cell suspensions (105 CFU ml−1) and with the same volume of sterile water, respectively, for 8 days. The concentrations of IAA in the bacterial suspensions and water were measured with UPLC. The concentration of IAA in the LAD suspension was 0.191 ± 0.014 μg ml−1, which was significantly higher than that in the hydroponic system without LAD (0.036 ± 0.0096 μg ml−1; t-test, p < 0.001; Figure 6). The results showed that LAD can produce IAA or promote IAA secretion from maize roots. The test of EPSs in the LAD cultures showed that stable production of EPSs by LAD was only detected in medium with sucrose as the sole carbon source. The EPSs production in sucrose supplemented medium was 3.18 ± 0.0198 g L−1 at 24 h and 0.63 ± 0.0041 g L−1 at 72 h.
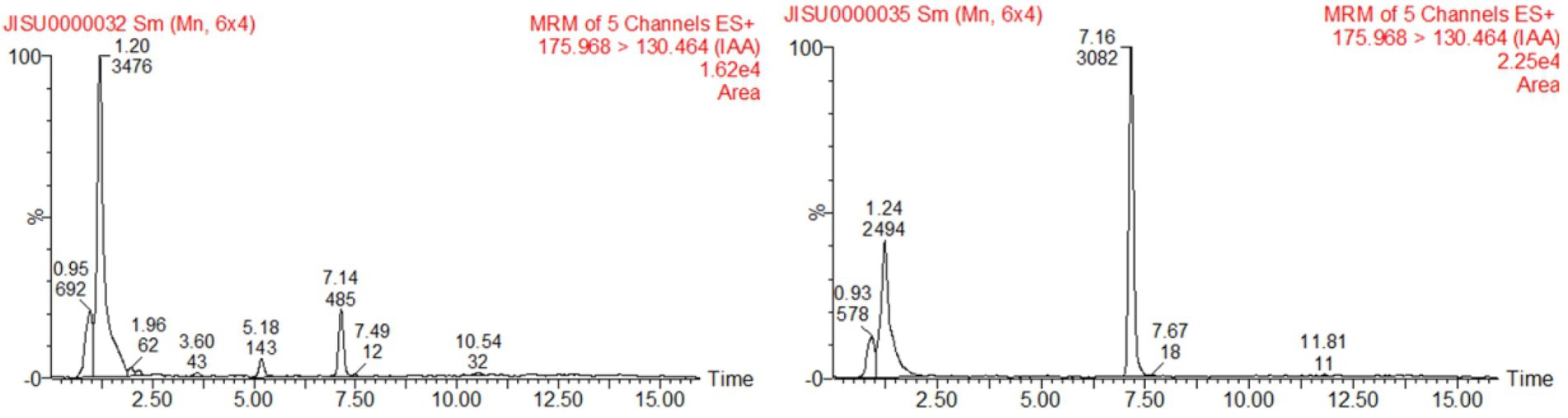
Figure 6. Detection of IAA production by UPLC. The left figure shows the sample of the control, and the right shows the sample of B. aryabhattai LAD culture cultivated with maize seedlings.
Impacts on maize root development
To investigate how LAD affects the maize root development, the maize seeds were treated with 105 CFU ml−1 LAD cell suspensions and grown in laboratory and filed soils, respectively. The roots were collected and analyzed with root analyzer. The laboratory cultivation results showed that the total root length, total root surface area and total root volume of the LAD treatment group is 97.0, 90.1, and 75.6% higher than that of the control, respectively (t-test, p < 0.001; Figure 7). Like the laboratory cultivation, the results of field experiments revealed an increase of 47.6, 43.1, and 42.9% in the total root length, surface area, and volume in the LAD treatment compared with the control (t-test, p < 0.001). The longest root in the LAD treatment was 197% longer than the longest root in the control. The experiments demonstrated that LAD treatment significantly increased the maize root length, surface area, and volume (p < 0.001).
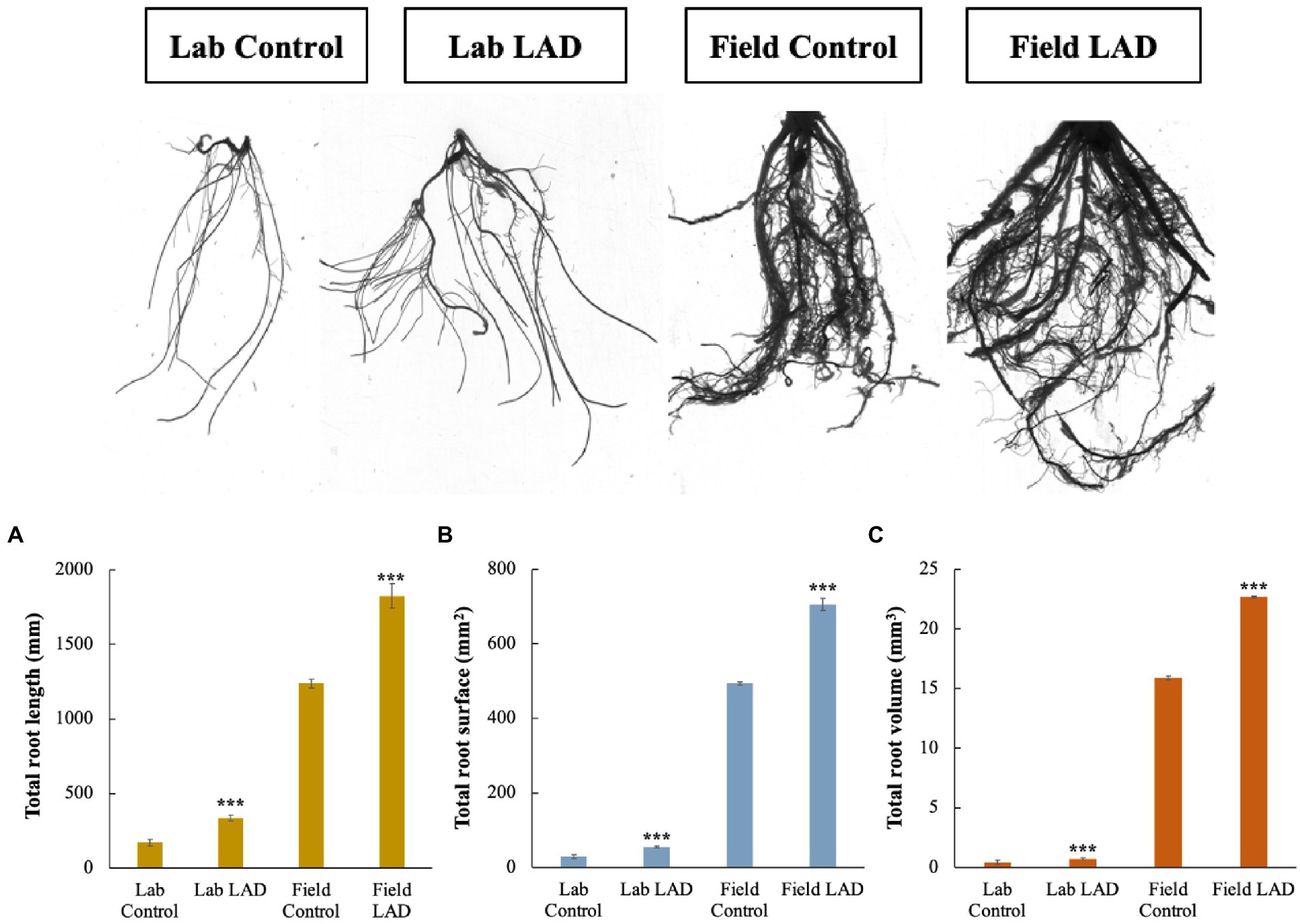
Figure 7. Maize root development in laboratory and field soils impacted by B. aryabhattai LAD. (A) The impacts of LAD on the maize root length. (B) The impacts of LAD on the maize root surface area. (C) The impacts of LAD on the maize root volume. Lab control: the root of the maize seedlings treated with water and cultivated in laboratory soil for 14 d; Lab LAD: the root of the maize seedlings treated with LAD and cultivated in laboratory soil for 14 d; Field control: the root of the maize seedlings treated with water and cultivated in laboratory soil for 60 d; Field LAD: the root of the maize seedlings treated with LAD and cultivated in laboratory soil for 60 d. Each treatment had 10 maize plants and was repeated three times. Student’s t-test was used to compare the root development under LAD treatment with that of the control. Statistical differences were shown with asterisks (i.e., *** representing p < 0.001).
Discussion
Plant-microbiome interactions are critical for plant health and growth. Rhizosphere microbes reside near or on the plant root and have direct contact and intense interactions with the plant tissue, therefore the root microbiome can utilize root exudates and secrete compounds that remarkably influence plant growth (Haney et al., 2015; Pascale et al., 2020). The interactions between plant and rhizosphere microbes are extremely complex, and the mechanism, ecological significance, and potential for application remain to be clarified. The data in this study shed light on the mechanism of B. aryabhattai LAD promoting plant growth based on both genomic analysis and experimental evidence. Whole genome sequencing of B. aryabhattai LAD showed that the rhizobacteria had diverse pathways for carbohydrate metabolism and mechanisms for environmental adaptations. The growth experiments of B. aryabhattai LAD demonstrated that LAD cells can utilize different types of carbons sources ranging from one-carbon methanol to polymeric carbohydrate starch, and the LAD cells can also grow well in a wide range of temperatures (20°C to 37°C). These properties are important for LAD to adapt to the fluctuating conditions in soil environments and further forming possible symbiosis with plant hosts, as plant root secretion of different carbon chain length may serve as nutrient supply for the root-associated bacteria while the bacterial activities may facilitate plant performance (Blom et al., 2011; Cheng et al., 2019; Lucke et al., 2020).
IAA, one of the most common and physiologically active plant hormones in nature, can induce cell elongation and division and is important for plant development and growth. Bacterial production of IAA as plant growth promoting strategy has been demonstrated in previous studies (Mohite, 2013; Wagi and Ahmed, 2019). For example, the IAA production by B. cereus So3II and B. subtilis Mt3b was shown by, and the growth conditions of the two Bacillus strains could be optimized to enhance optimum bacterial growth and production of IAA which ultimately may be used for plant growth stimulation. B. aryabhattai LAD in the present study produced high yield of IAA when cultivated with maize plant suggesting the symbiosis of the rhizobacteria with maize plant. Besides IAA production, B. aryabhattai LAD can also produce EPSs which is another plant growth-promoting trait (Wagi and Ahmed, 2019). A recent study by showed that EPS-producing bacteria stimulated the salt tolerance of plant because the bacterial produced EPSs can bind cations significantly decreasing the Na+ content in the environment (Upadhyay et al., 2011). Interestingly, the EPSs production decreased from 3.18 g L−1 at 24 h to 0.63 g L−1 at 72 h, suggesting that the produced EPSs might be utilized by LAD cells when available nutrients are depleted.
Microbial metabolisms associated with one-carbon compound conversion were prevalent in rhizosphere, and the methanol metabolism by B. aryabhattai LAD may contribute to the rhizosphere nutrient cycling (Knief et al., 2012; Li et al., 2019). The genome annotation revealed that B. aryabhattai LAD genome contains genes of alcohol dehydrogenases, and the alcohol dehydrogenases enzymes can facilitate the conversion of alcohols to aldehydes with production of NADH. Aldehyde dehydrogenase genes were also detected on B. aryabhattai LAD genome, and the products of the genes catalyze the oxidation of the aldehydes generated from methanol. The other possible pathway for methanol metabolism in B. aryabhattai LAD was through alcohol oxidase (AOX), catalyzing the oxidation of methanol to formaldehyde with production of hydrogen peroxide, and catalase, breaking down hydrogen peroxide, as genes for AOX and peroxidase were detected on B. aryabhattai LAD genome. Plant roots secrete a variety of carbon-containing compounds which significantly influence rhizobacterial activities and community structure (O’Neal et al., 2020; Okutani et al., 2020; Liang et al., 2021). The root exudates of varied molecular weight and complexity may serve as organic carbon sources for microbial growth, and the ability for rhizobacteria utilizing root exudates is critical for both the bacterial populations and the host plant (Mark et al., 2005; Fethel et al., 2014; Olanrewaju et al., 2019). The CAZymes annotation of B. aryabhattai LAD genome revealed an abundance of genes associated with the assimilation, modification, and decomposition of carbohydrates.
The whole genome of B. aryabhattai LAD was obtained via next-generation sequencing, and the comparative analysis showed that B. aryabhattai LAD had high similarities to other B. aryabhattai strains and strains in B. megaterium. For genomic characterization of B. aryabhattai LAD, the pan and core genome analysis were performed to cluster the genes in all the examined genomes. The eggNOG annotation and GO annotation revealed large groups of genes associated with carbohydrate transport and metabolism, oxidoreductase activity, cellular nitrogen compound metabolic process, and biosynthetic process, all of which are important for the symbiotic relationship between bacterial populations and plants and were also found in other plant growth promoting bacterial strains (Taghavi et al., 2010; Han et al., 2011; Bhattacharyya et al., 2017). The CARD analysis identified 31 antibiotic resistance genes and 4 antibiotic biosynthesis genes on the genome and plasmids of B. aryabhattai LAD, suggesting that B. aryabhattai LAD can be resistant to various antibiotics and have potential for synthesizing antibiotics. Antibiotics production were also reported in other Bacillus strains which has a major role for rhizobacteria to suppress plant diseases. For example, the whole genome sequencing of B. amyloliquefaciens FZB42 showed a high potential for production of polyketides bacillaene and difficidin, and over 8.5% of the genome is committed antibiotics and siderophores biosynthesis (Chen et al., 2007). Kinsella et al. (2008) also showed a high yield of antibiotics (i.e., surfactin and iturin) by Bacillus subtilis on cucumber roots (Kinsella et al., 2008).
Conclusion
Plant roots harbor a diverse microbial community, and the complicated interactions between plant and rhizosphere microbes have major ecological importance and critical implications for agricultural practices. The whole genome sequencing of methylotrophic B. aryabhattai LAD revealed many signature genes that are functionally associated with plant growth promotion. Genome analyses revealed the ability of B. aryabhattai LAD to adapt to the environments with antibiotics, oxidative, cold temperature, and heavy metal stresses. Our experiments confirmed that B. aryabhattai LAD has versatile metabolism and can utilize a wide range of carbon sources. B. aryabhattai LAD is also capable of synthesizing IAA and EPSs that are beneficial for plant growth and bacterial colonization. The genomics analysis and experimental studies collectively demonstrated the plant growth promoting capacities of B. aryabhattai LAD making it an exceptional bacterial strain for application in agricultural practices. The impacts of B. aryabhattai LAD on maize root development was directly evaluated, and the results demonstrated the root development stimulation and plant growth promoting capacities of the Bacillus strain. Future efforts are needed for studying the molecular mechanisms of B. aryabhattai LAD in promoting plant growth and evaluating the ecological impacts of bioaugmentation with LAD on soil microbial community.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
Study protocols of plant materials comply with the IUCN Policy Statement on Research Involving Species at Risk of Extinction and the Convention on the Trade in Endangered Species of Wild Fauna and Flora.
Author contributions
NZh and BL conceived and designed the study. CD conducted the experiments. CD, XW, NZe, and XL performed data analysis and prepared the figures. XL wrote the main manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2017YFD0200807), the Liaoning Province Rural Science and Technology Special Action Project (2022-09), and Shenyang Science and Technology Project (20-207-3-35).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmed, A., and Hasnain, S. (2014). Auxins as one of the factors of plant growth improvement by plant growth promoting rhizobacteria. Pol. J. Microbiol. 63, 261–266. doi: 10.33073/pjm-2014-035
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9:1473. doi: 10.3389/fpls.2018.01473
Berendsen, R. L., Pieterse, C. M., and Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berta, B. (2000). Putative sites for nutrient uptake in arbuscular mycorrhizal fungi. Plant Soil 226, 263–274. doi: 10.1023/A:1026456818903
Besemer, J., Lomsadze, A., and Borodovsky, M. (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618. doi: 10.1093/nar/29.12.2607
Bever, J. D., Broadhurst, L. M., and Thrall, P. H. (2013). Microbial phylotype composition and diversity predicts plant productivity and plant–soil feedbacks. Ecol. Lett. 16, 167–174. doi: 10.1111/ele.12024
Bhattacharyya, C., Bakshi, U., Mallick, I., Mukherji, S., Bera, B., and Ghosh, A. (2017). Genome-guided insights into the plant growth promotion capabilities of the physiologically versatile bacillus aryabhattai strain AB211. Front. Microbiol. 8:411. doi: 10.3389/fmicb.2017.00411
Blom, D., Fabbri, C., Connor, E. C., Schiestl, F. P., Klauser, D. R., Boller, T., et al. (2011). Production of plant growth modulating volatiles is widespread among rhizosphere bacteria and strongly depends on culture conditions. Environ. Microbiol. 13, 3047–3058. doi: 10.1111/j.1462-2920.2011.02582.x
Bucksch, A., Burridge, J., York, L. M., Das, A., Nord, E., Weitz, J. S., et al. (2014). Image-based high-throughput field phenotyping of crop roots. Plant Physiol. 166, 470–486. doi: 10.1104/pp.114.243519
Chaparro, J. M., Badri, D. V., and Vivanco, J. M. (2014). Rhizosphere microbiome assemblage is affected by plant development. ISME J. 8, 790–803. doi: 10.1038/ismej.2013.196
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6:24373. doi: 10.1038/srep24373
Chen, X. H., Koumoutsi, A., Scholz, R., Eisenreich, A., Schneider, K., Heinemeyer, I., et al. (2007). Comparative analysis of the complete genome sequence of the plant growth–promoting bacterium bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 25, 1007–1014. doi: 10.1038/nbt1325
Chen, Y. F., Yang, H., Shen, Z. Z., and Ye, J. R. (2022). Whole-genome sequencing and potassium-solubilizing mechanism of bacillus aryabhattai SK1-7. Front. Microbiol. 12:722379. doi: 10.3389/FMICB.2021.722379
Cheng, Y. T., Zhang, L., and He, S. Y. (2019). Plant-microbe interactions facing environmental challenge. Cell Host Microbe 26, 183–192. doi: 10.1016/j.chom.2019.07.009
Chin, C. S., Peluso, P., Sedlazeck, F. J., Nattestad, M., Concepcion, G. T., Clum, A., et al. (2016). Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054. doi: 10.1038/nmeth.4035
Chu, T. N., Bui, L. V., and Hoang, M. T. (2020). Pseudomonas PS01 isolated from maize rhizosphere alters root system architecture and promotes plant growth. Microorganisms 8:471. doi: 10.3390/microorganisms8040471
Deng, C., Zhang, N., Liang, X., Huang, T., and Li, B. (2022). Bacillus aryabhattai LAD impacts rhizosphere bacterial community structure and promotes maize plant growth. J. Sci. Food Agric. 102, 6650–6657. doi: 10.1002/jsfa.12032
Erik, J. J., Sabine, R., and Iver, J. (2000). Arbuscular mycorrhizal phosphate transport under monoxenic conditions using radio-labelled inorganic and organic phosphate. Biotechnol. Lett. 22, 1705–1708. doi: 10.1023/A:1005684031296
Fethel, Z. H., Santaella, C., Heulin, T., and Achouak, W. (2014). Root exudates mediated interactions belowground. Soil Biol. Biochem. 77, 69–80. doi: 10.1016/j.soilbio.2014.06.017
Fu, J. H., Chu, J. F., Sun, X. H., Wang, J. D., and Yan, C. Y. (2012). Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal. Sci. 28, 1081–1087. doi: 10.2116/analsci.28.1081
Ghosh, P. K., Maiti, T. K., Pramanik, K., Ghosh, S. K., Mitra, S., and De, T. K. (2018). The role of arsenic resistant bacillus aryabhattai MCC3374 in promotion of rice seedlings growth and alleviation of arsenic phytotoxicity. Chemosphere 211, 407–419. doi: 10.1016/j.chemosphere.2018.07.148
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57. doi: 10.1093/nar/gkm360
Han, J. I., Choi, H. K., Lee, S. W., Orwin, P. M., Kim, J., LaRoe, S. L., et al. (2011). Complete genome sequence of the metabolically versatile plant growth-promoting endophyte Variovorax paradoxus S110. J. Bacteriol. Res. 193, 1183–1190. doi: 10.1128/JB.00925-10
Haney, C. H., Samuel, B. S., Bush, J., and Ausubel, F. M. (2015). Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat. Plants 1, 1–9. doi: 10.1038/NPLANTS.2015.51
Idriss, E. E., Makarewicz, O., Farouk, A., Rosner, K., Greiner, R., Bochow, H., et al. (2002). Extracellular phytase activity of Bacillus amyloliquefaciens FZB45 contributes to its plant-growth-promoting effect. Microbiology 148, 2097–2109. doi: 10.1099/00221287-148-7-2097
Kalvari, I., Argasinska, J., Quinones-Olvera, N., Nawrocki, E. P., Rivas, E., Eddy, S. R., et al. (2018). Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 46, D335–D342. doi: 10.1093/nar/gkx1038
Kinsella, K., Schulthess, C. P., Morris, T. F., and Stuart, J. D. (2008). Rapid quantification of Bacillus subtilis antibiotics in the rhizosphere. Soil Biol. Biochem. 41, 374–379. doi: 10.1016/j.soilbio.2008.11.019
Knief, C., Delmotte, N., Chaffron, S., Stark, M., Innerebner, G., Wassmann, R., et al. (2012). Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 6, 1378–1390. doi: 10.1038/ismej.2011.192
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Korenblum, E., Dong, Y., Szymanski, J., Panda, S., Jozwiak, A., Massalha, H., et al. (2020). Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. U. S. A. 117, 3874–3883. doi: 10.1073/pnas.1912130117
Lau, J. A., and Lennon, J. T. (2011). Evolutionary ecology of plant–microbe interactions: soil microbial structure alters selection on plant traits. New Phytol. 192, 215–224. doi: 10.1111/j.1469-8137.2011.03790.x
Lazcano, C., Boyd, E., Holmes, G., Hewavitharana, S., Pasulka, A., and Ivors, K. (2021). The rhizosphere microbiome plays a role in the resistance to soil-borne pathogens and nutrient uptake of strawberry cultivars under field conditions. Sci. Rep. 11:3188. doi: 10.1038/s41598-021-82768-2
Li, Z., Yao, Q., Guo, X., Crits-Christoph, A., Mayes, M. A., Lebeis, S. L., et al. (2019). Genome-resolved proteomic stable isotope probing of soil microbial communities using 13CO2 and 13C-methanol. Front. Microbiol. 10:2706. doi: 10.3389/fmicb.2019.02706
Liang, X., Wang, Y., Zhang, Y., Zhuang, J., and Radosevich, M. (2021). Viral abundance, community structure and correlation with bacterial community in soils of different cover plants. Appl. Soil Ecol. 168:104138. doi: 10.1016/J.APSOIL.2021.104138
Liu, J., Cui, X., Liu, Z., Guo, Z., Yu, Z., Yao, Q., et al. (2019). The diversity and geographic distribution of cultivable bacillus-like bacteria across black soils of Northeast China. Front. Microbiol. 10:1424. doi: 10.3389/fmicb.2019.01424
Lombard, V., Golaconda, R. H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Lucke, M., Correa, M. G., and Levy, A. (2020). The role of secretion systems, effectors, and secondary metabolites of beneficial rhizobacteria in interactions with plants and microbes. Front. Plant Sci. 11:589416. doi: 10.3389/fpls.2020.589416
Ma, J., Li, Y., Hu, D., Peng, J. L., Ja, N., Zhang, C. M., et al. (2018). Progress on mechanism and applications of bacillus as a biocontrol microbe. Chin. J. Bio. Con. 34, 639–648. doi: 10.16409/j.cnki.2095-039x.2018.04.019
Maria, J. H. (2005). Signaling in the arbuscular mycorrhizal symbiosis. Annu. Rev. Microbiol. 59, 19–42. doi: 10.1146/annurev.micro.58.030603.123749
Mark, G. L., Dow, J. M., Kiely, P. D., Higgins, H., Haynes, J., Baysse, C., et al. (2005). Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc. Natl. Acad. Sci. U. S. A. 102, 17454–17459. doi: 10.1073/pnas.0506407102
Mehmood, S., Khan, A. A., Shi, F., Tahir, M., Sultan, T., Munis, M. F. H., et al. (2021). Alleviation of salt stress in wheat seedlings via multifunctional bacillus aryabhattai PM34: an in-vitro study. Sustainability 13, 8030–8030. doi: 10.3390/SU13148030
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Mohite, B. (2013). Isolation and characterization of indole acetic acid (IAA) producing bacteria from rhizospheric soil and its effect on plant growth. J. Soil Sci. Plant Nutr. 13, 638–649. doi: 10.4067/S0718-95162013005000051
Moreau, D., Bardgett, R. D., Finlay, R. D., Jones, D. L., and Philippot, L. (2019). A plant perspective on nitrogen cycling in the rhizosphere. Funct. Ecol. 33, 540–552. doi: 10.1111/1365-2435.13303
O’Neal, L., Vo, L., and Alexandre, G. (2020). Specific root exudate compounds sensed by dedicated chemoreceptors shape Azospirillum brasilense chemotaxis in the rhizosphere. Appl. Environ. Microbiol. 86:e01026. doi: 10.1128/AEM.01026-20
Okutani, F., Hamamoto, S., Aoki, Y., Nakayasu, M., Nihei, N., Nishimura, T., et al. (2020). Rhizosphere modelling reveals spatiotemporal distribution of daidzein shaping soybean rhizosphere bacterial community. Plant Cell Environ. 43, 1036–1046. doi: 10.1111/pce.13708
Olanrewaju, O. S., Ayangbenro, A. S., Glick, B. R., and Babalola, O. O. (2019). Plant health: feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 103, 1155–1166. doi: 10.1007/s00253-018-9556-6
Park, Y. G., Mun, B. G., Kang, S. M., Hussain, A., Shahzad, R., Seo, C. W., et al. (2017). Bacillus aryabhattai SRB02 tolerates oxidative and nitrosative stress and promotes the growth of soybean by modulating the production of phytohormones. PLoS One 12:e0173203. doi: 10.1371/journal.pone.0173203
Pascale, A., Proietti, S., Pantelides, I. S., and Stringlis, I. A. (2020). Modulation of the root microbiome by plant molecules: the basis for targeted disease suppression and plant growth promotion. Front. Plant Sci. 10:1741. doi: 10.3389/fpls.2019.01741
Philippe, H. (2001). Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review. Plant Soil 237, 173–195. doi: 10.1023/A:1013351617532
Richter, M., Rosselló-Móra, R., Oliver, G. F., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Roy, K., Ghosh, D., DeBruyn, J. M., Dasgupta, T., Wommack, K. E., Liang, X., et al. (2020). Temporal dynamics of soil virus and bacterial populations in agricultural and early plant successional soils. Front. Microbiol. 11:1494. doi: 10.3389/fmicb.2020.01494
Shao, Y., Liu, T., Eisenhauer, N., Zhang, W., Wang, X., Xiong, Y., et al. (2018). Plants mitigate detrimental nitrogen deposition effects on soil biodiversity. Soil Biol. Biochem. 127, 178–186. doi: 10.1016/j.soilbio.2018.09.022
Shivaji, S., Chaturvedi, P., Begum, Z., Pindi, P. K., Manorama, R., Padmanaban, D. A., et al. (2009). Janibacter hoylei sp. nov., bacillus isronensis sp. nov. and bacillus aryabhattai sp. nov., isolated from cryotubes used for collecting air from the upper atmosphere. Int. J. Syst. Evol. Microbiol. 59, 2977–2986. doi: 10.1099/ijs.0.002527-0
Taghavi, S., Lelie, D., Hoffman, A., Zhang, Y. B., Walla, M. D., Vangronsveld, J., et al. (2010). Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PLoS Genet. 6:e1000943. doi: 10.1371/journal.pgen.1000943
Tahir, H. A., Gu, Q., Wu, H., Raza, W., Hanif, A., Wu, L., et al. (2017). Plant growth promotion by volatile organic compounds produced by Bacillus subtilis SYST2. Front. Microbiol. 8:171. doi: 10.3389/fmicb.2017.00171
Tzipilevich, E., Russ, D., Dangl, J. L., and Benfey, P. N. (2021). Plant immune system activation is necessary for efficient root colonization by auxin-secreting beneficial bacteria. Cell Host Microbe 29, 1507–1520.e4. doi: 10.1016/j.chom.2021.09.005
Upadhyay, S. K., Singh, J. S., and Singh, D. P. (2011). Exopolysaccharide-producing plant growth-promoting rhizobacteria under salinity condition. Pedosphere 21, 214–222. doi: 10.1016/S1002-0160(11)60120-3
Vernikos, G., Medini, D., Riley, D. R., and Tettelin, H. (2015). Ten years of pan-genome analyses. Curr. Opin. Microbiol. 23, 148–154. doi: 10.1016/j.mib.2014.11.016
Wagi, S., and Ahmed, A. (2019). Bacillus spp.: potent microfactories of bacterial IAA. PeerJ 7:e7258. doi: 10.7717/peerj.7258
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Keywords: carbohydrate-active enzymes, sequencing, rhizosphere, growth promoting, indole acetic acid
Citation: Deng C, Liang X, Zhang N, Li B, Wang X and Zeng N (2022) Molecular mechanisms of plant growth promotion for methylotrophic Bacillus aryabhattai LAD. Front. Microbiol. 13:917382. doi: 10.3389/fmicb.2022.917382
Edited by:
Víctor Manuel Ruíz-Valdiviezo, Tecnológico Nacional de México/Instituto Tecnológico de Tuxtla Gutiérrez, MexicoReviewed by:
Jesús Muñoz-Rojas, Meritorious Autonomous University of Puebla, MexicoPradeep Kumar Rai, Indian Farmers Fertilizer Cooperative Limited, India
Copyright © 2022 Deng, Liang, Zhang, Li, Wang and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingxue Li, bGliaW5neHVlQHN5YXUuZWR1LmNu; Ning Zhang, emhhbmduaW5nQHN5YXUuZWR1LmNu
†These authors have contributed equally to this work
 Chao Deng
Chao Deng Xiaolong Liang
Xiaolong Liang Ning Zhang3*
Ning Zhang3* Bingxue Li
Bingxue Li