
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 07 July 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.916304
The prevalence of carbapenem-resistant Klebsiella pneumoniae is increasing. Although carbapenemase production is the main resistance mechanism of K. pneumonia to carbapenems, there are still some reports of non-carbapenemase-producing K.pneumoniae showing high-level resistance to carbapenems. In this study, we had also isolated a carbapenemase-negative carbapenem-resistant K. pneumoniae L204 from a patient with an asymptomatic urinary tract infection. Species identification was performed using MALDI-TOF MS, and carbapenemase-encoding genes were detected using both NG-test carba-5 and whole-genome sequencing. Antimicrobial susceptibility testing was performed by the broth microdilution method according to CLSI guidance. The results of antimicrobial susceptibility testing indicated that K. pneumoniae L204 was resistant to meropenem (MIC = 16 mg/L) and imipenem (MIC = 4 mg/L), but susceptible to ceftazidime-avibactam (MIC = 8 mg/L). Through whole-genome sequencing, several resistance genes had been identified, including blaTEM–1B, blaCTX–M–167, blaSHV–1, aac(6’)-1b-cr, qnrS, aadA16, tet(A), fosA, sul1, and mph(A). The efflux pump inhibition testing showed that the efflux pump was not involved in the resistance mechanism to carbapenems. The result of the conjugation experiment indicated that the plasmid with blaCTX–M–167 and blaSHV–1 was transferrable. The sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) demonstrated that K. pneumoniae L204 only contained outer membrane porin OmpK35.
Klebsiella pneumoniae is one of the most common clinical opportunistic pathogens. It can be colonized in the respiratory and intestine tract of the human body (Chen et al., 2014; Hu et al., 2020), causing infection in the respiratory tract, urinary tract, and blood flow system (Amit et al., 2015; Lambelet et al., 2017). Immunosuppressive patients or those who use antibiotics for a long time are easy to be affected (Pantelidou et al., 2015; Xu et al., 2017; Arato et al., 2021). To control the infection caused by carbapenem-resistant K. pneumoniae (CRKP), a lot of new antimicrobial agents have appeared in recent years, bringing hope to patients with CRKP infection (Logan and Weinstein, 2017). Carbapenems have always been the first choice for clinicians to deal with carbapenem-sensitive K. pneumoniae infections due to their wide antibacterial spectrum and high stability (David et al., 2019). However, with the increase of CRKP, medical expenses and the mortality of patients are also increasing year by year (Shields et al., 2017), a systematic study on its resistance mechanism is helpful for clinicians to use antibiotics effectively. Although carbapenemase production is the main resistance mechanism of K. pneumoniae to carbapenems, numerous reports also have described non-carbapenemase-producing K. pneumoniae with high carbapenem levels. In this study, we found a common carbapenemase-negative (including blaKPC, blaNDM, blaIMP, blaVIM, and blaOXA–48) CRKP during routine antimicrobial susceptibility testing and carbapenemase testing (by NG-test Carba-5 kit). To clarify the resistance mechanism of CRKP to carbapenem including the existing of other carbapenemase genes or the new carbapenemase gene, we use various methods to study the mechanism that may mediate carbapenem resistance among this CRKP, and we reported a carbapenemase-negative CRKP clinical isolate bearing blaCTX–M–167 among K. pneumoniae coupled with the inactivation of OmpK36 from a patient. To our best knowledge, this is the first time to identify a blaCTX–M–167-positive K. pneumoniae strain in China.
A carbapenem-resistant K. pneumoniae L204 clinical strain was isolated from a urine sample of a patient with a urinary tract infection in Huashan Hospital, Fudan University in Shanghai, China. Species identification was performed using MALDI-TOF MS (bioMeìrieux, France). Escherichia coli ATCC25922 and Pseudomonas aeruginosa ATCC 27853 were used as controls for Antimicrobial Susceptibility Testing. K. pneumoniae ATCC 35657 that contains intact OmpK35 and OmpK36 used as a control for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
The minimal inhibitory concentration (MIC) was determined by the broth microdilution method recommended by the Clinical and Laboratory Standards Institute (CLSI), and the results were interpreted according to CLSI M100-31th breakpoints for all agents except tigecycline, polymyxin, and cefepime–tazobactam (Yang et al., 2020; Clinical and Laboratory Standards Institute, 2021). Cefepime–tazobactam referred to the standards of cefepime in CLSI. Tigecycline MICs were interpreted using US FDA MIC breakpoints for Enterobacterales and polymyxin MICs were interpreted using the European Committee for Antimicrobial Susceptibility Testing (EUCAST) MIC breakpoints for Enterobacterales. Efflux pump inhibition testing (Sanchez-Carbonel et al., 2021) using Carbonyl cyanide 3-chlorophenylhydrazone (CCCP, 25 mg/L) as an inhibitor was conducted to determine whether the efflux pump takes a role in the resistance of K. pneumoniae L204 to imipenem and meropenem.
A conjugation experiment was performed to explore the transferability of the plasmid. Briefly, the blaCTX–M–167 and blaSHV–1 -positive isolate K. pneumoniae L204 was used as the donor, while the E. coli J53 (azide resistant) was used as the recipient strain. Conjugants were selected on Mueller–Hinton (MH) agar supplemented with ampicillin (50 mg/L) and azide (100 mg/L). The presence of the blaCTX–M–167 and blaSHV–1 gene and other resistance genes in conjugants was confirmed by antimicrobial susceptibility, PCR, and DNA sequencing.
The cultured bacteria growing in 3ml LB broth was subjected to washing, crushed by ultrasonic, and then the membrane porin was obtained after centrifugation. Phosphate buffered saline (PBS) was used to resuspend the membrane protein. Extracted samples were separated using SDS-PAGE at a constant voltage of 130 V for about 50 min in both a 12% (w/v) polyacrylamide gel and a gradient gel (ExpressPlus™ PAGE Gel, GenScript, Piscataway, NJ, United States) as previous reported (Kurien and Scofield, 2012). The bands were detected using Coomassie brilliant blue R-250 staining.
Five common carbapenemase genes (KPC, NDM, IMP, VIM, and OXA-48) were initially detected by PCR amplification and DNA sequencing (Gülmez et al., 2008; Poirel et al., 2011; Weiß et al., 2017). To fully understand the resistance genes among K. pneumoniae L204, in particular the carbapenemase, ESBL, and AmpC, we performed whole-genome sequencing analysis for this isolate. The genomic DNA of K. pneumoniae L204 was extracted by using a commercial Qiagen Midi kit (Qiagen, Hilden, Germany). The sequencing libraries were prepared using the Nextera XT DNA Library preparation protocol (Illumina, San Diego, CA, United States) and then the genomic DNA was subjected to Illumina (Illumina, San Diego, CA, United States) short-read sequencing (150 bp paired-end reads). Reads were trimmed with a sickle (GitHub) and were de novo assembled using SPAdes 3.12.0. Antimicrobial resistance genes analysis was performed using ResFinder 4.11 and further verified using NCBI BLAST and the annotation process was done using RAST version 2.0.2
Klebsiella pneumoniae L204 was isolated from a urine sample of a 55-year-old male patient with hydrocephalus who underwent a ventriculoperitoneal shunt. CT showed a small amount of inflammation in the lungs and a small amount of pleural effusion on both sides. A isolate of extended-spectrum β-lactamase-negative K. pneumoniae was initially isolated from the sputum, and cefoperazone-sulbactam (3g q8h) was given anti-infective treatment. Thereafter, the infection comes under control. On the 7th postoperative day, K. pneumoniae L204 was isolated from the urine and the anti-infection regimen was switched to meropenem (2g q8h) combined with amikacin (0.8g qd). The culture of the urine sample was negative three days later, and the patient was discharged 21 days after admission and was subsequently transferred to a rehabilitation hospital for further rehabilitation.
The antimicrobial susceptibility profiles of K.pneumoniae L204 to antimicrobial agents are presented in Table 1. The isolate was resistant to aztreonam (MIC ≥ 128 mg/L), ceftolozane-tazobactam (MIC ≥ 128 mg/L), meropenem (MIC = 16 mg/L), imipenem (MIC = 4 mg/L), levofloxacin (MIC ≥ 16mg/L), ciprofloxacin (MIC > 8mg/L), and tigecycline (MIC = 8 mg/L), but susceptible to amikacin (MIC = 2 mg/L), polymyxin B (MIC = 1 mg/L), and ceftazidime-avibactam (MIC = 8 mg/L). After adding CCCP, the MIC of meropenem and imipenem hasn’t changed indicating that the efflux pump may not play a role in the resistance mechanism of K. pneumoniae L204 to β-lactams such as ceftazidime, cefepime, piperacillin-tazobactam, aztreonam, meropenem, and imipenem.
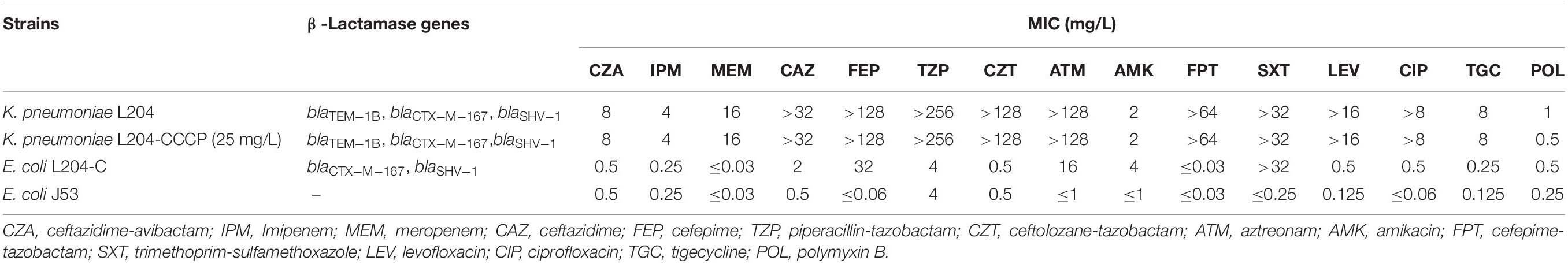
Table 1. Susceptibility of K. pneumoniae L204 clinical isolate, conjugant, and recipient to antimicrobial agents.
The blaCTX–M–167 and blaSHV–1-carrying plasmid was successfully transferred from K. pneumoniae L204 to E. coli J53, PCR-based sequencing demonstrated the presence of blaCTX–M–167 and blaSHV–1 in the conjugant E. coli L204-C. Compared with the recipient E. coli J53, the MICs of ceftazidime, cefepime, and aztreonam increased four times (from 0.5 to 2 mg/L), ≥512 times (from ≤0.06to 32 mg/L), and ≥16 times (from ≤1to 16 mg/L), respectively; while meropenem, imipenem, ceftazidime-avibactam, piperacillin-tazobactam, and cefepime-tazobactam make no difference for MIC between recipient and conjugant.
The results of whole genome sequencing indicated that none of carbapenemase gene was detected in K. pneumoniae L204. However, a lot of resistance genes had been identified according to the whole genome sequencing, including the β-lactamase genes blaTEM–1B, blaCTX–M–167, and blaSHV–1, the fluoroquinolone resistance gene qnrS1, and the aminoglycoside resistance genes aac(6’)-1b-cr and aadA16, the tetracycline resistance gene tet(A), the fosfomycin resistance gene fosA, the sulfonamide resistance gene sul1, and the macrolide resistance gene mph(A). Compared with blaCTX–M–15, blaCTX–M–167 has two amino acid substitutions: G at position 241 and 242 are replaced by C,D, respectively. Compared with blaCTX–M–1, except that the GG at 241-242 was replaced by CD, there was also a single base replacement, but it did not cause changes at the amino acid level. The genetic platform analysis revealed that the blaCTX–M–67 gene was flanked by ISEcp1 and Tn2-like transposase downstream and upstream, respectively. The full genetic environment surrounding blaCTX–M–67 is: Tn2-like transposase–blaTEM–1B–blaCTX–M–67–ISEcp1 (Figure 1). K. pneumoniae L204 has a complete ompk35, while Ompk36 has many mutations at many sites such as P.n218H and P.n217H were also reflected in whole gene sequencing. The result of multilocus sequence typing of K. pneumonia L204 indicated that the strain belonged to ST893. The results of SDS-PAGE of outer membrane protein showed that membrane porins Ompk35 (40 kDa) was complete while Ompk36 (37 kDa) in K. pneumonia L204 was not detected (Figure 2).
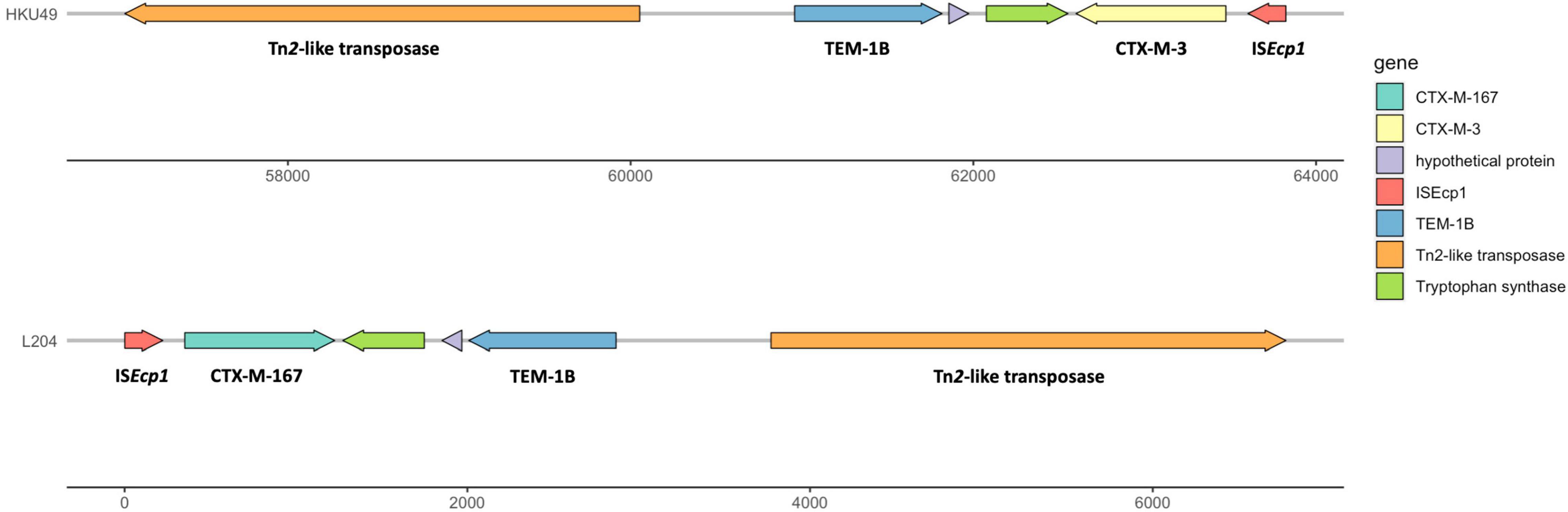
Figure 1. Multidrug resistance region (MRR) of blaCTX–M–167 in K. pneumoniaeL204 and blaCTX–M–3 in HKU49. Resistance genes are indicated by yellow symbols. Transposon-related genes and insertion sequences are indicated by green symbols. Light gray shading indicated homologous regions (>99% DNA identity).
Extended-spectrum β-lactamase(ESBLs) are plasmid-mediated β-lactamases produced by gram-negative bacteria that are capable of conferring resistance to the penicillins, cephalosporins, and aztreonam by hydrolysis of these antibiotics, but usually have no activity against cephamycins or carbapenems (Paterson and Bonomo, 2005). According to the China Antimicrobial Surveillance Network, the detection rates of ESBL-producing strains in E. coli, K. pneumoniae, and Proteus mirabilis were 52.4, 41.5, and 38.2%, respectively, and P. mirabilis showed an increasing trend year by year.3 According to the Ambler classification, ESBLs are mostly from the class A TEM, SHV, and CTX-M families (Denkel et al., 2020; Verschuuren et al., 2021). CTX-M type ESBLs have surpassed TEM type and SHV type, and become the most widespread extended-spectrum β-lactamase in Gram-negative bacilli. Carrying CTX-M ESBLs with high hydrolysis activity to cephalosporin antibiotics is one of the main resistance mechanisms of K. pneumonia to antimicrobial agents, and their characteristics are carried by plasmids, which is also conducive to the generation, dissemination, and prevalence of new CTX-M ESBLs (Zhao and Hu, 2013). Although Enterobacterales from Asia, Europe, and America generally carry CTX-M ESBLs, and CTX-M-1 and CTX-M-9 are the main types, in different regions, the detection rate of the CTX-M is different (Daoud et al., 2015).
In this study, a multi-drug resistant K. pneumoniae L204 clinical strain with three β-Lactamase genes including blaCTX–M–167, blaTEM–1B, and blaSHV–1, only showed sensitivity to ceftazidime-avibactam, amikacin, and polymyxin B was isolated from a urine sample from a 55-year-old male patient with hydrocephalus who underwent a ventriculoperitoneal shunt. The result of the conjugation experiment showed that the plasmid containing blaCTX–M–167 and blaSHV–1 was successfully transferred from the donor to the recipient, and made the conjugant resistant to cefepime, and aztreonam while still susceptible to ceftazidime, meropenem, and imipenem.
To further explore the mechanism of resistance to carbapenems in K. pneumonia L204, we analyzed the efflux pump and outer membrane porins. The efflux pump inhibition testing indicated that the efflux pump does not play a role in the resistance of K. pneumoniae L204 to carbapenems. Combined with the previous study, Ompk36 loss may be one of the factors contributing to the resistance among ESBL-producing K. pneumoniae, and OmpK36 may play an essential role in the resistance or reduced susceptibility of K. pneumoniae to carbapenems coupled with ESBL and/or AmpC (Doménech-Sánchez et al., 2003; Tsai et al., 2011). In our study, The results of SDS-PAGE showed that OmpK36 was not detected in K. pneumonia L204, and whole-genome sequencing showed that it has many mutations at many sites such as P.n218H and P.n217H. we speculate that it was inactivated after genetic mutation and the loss of outer membrane porins may take the lead for the resistance of K. pneumoniae L204 to carbapenems, or other temporarily unknown mechanisms play an important role.
OmpK35 and OmpK36 belong to the Porin_1 (PF00267) group of bacterial outer membrane proteins. Both OmpK35 and OmpK36 form trimers composed of 16-stranded β-barrels integrated into the outer membrane, and the crystal structures of two of these proteins showed polar residues lining the internal porespolar molecules of less than 600 Da in size would permeate the channels formed by OmpK35 and OmpK36 (Dutzler et al., 1999; Acosta-Gutiérrez et al., 2018). Current knowledge on the regulation of porin gene expression in K. pneumoniae is limited, a study of Klebsiella aerogenes shows that overexpression of the small RNAs MicF and MicC can suppress the expression of omp35 and omp36, respectively (Hao et al., 2018).
Based on PLACNETw, the blaCTX–M–167 genes were located in contigs with homology to the plasmid that belonged to IncFII (K) type. The plasmid location of blaCTX–M genes has already been reported, namely, compared with the chromosomal location of the resistance genes that can benefit the stable propagation of resistance, the plasmid location of the resistance genes can make the horizontal transmission easier (Zhang et al., 2017, 2022; Yang et al., 2021). BLAST comparison disclosed that the blaCTX–M–167 gene-environment of K. pneumoniae L204 shared >99% similarity with blaCTX–M–3-carrying plasmid pHKU49_CIP (GenBank accession number: MN543570) with 99% nucleotide identity and 100% query coverage, which was isolated in a K.pneumonia strain HKU49 from Hong Kong, China. The genetic platform analysis revealed that the blaCTX–M gene was flanked by Tn2-like transposase and ISEcp1 downstream and upstream in both genetic environments, respectively. ISEcp1, belonging to the IS1380 family, is one of the most important insertion sequences associated with blaCTX–M genes (Zhao and Hu, 2013; Leão et al., 2021). Tn3 transposon, which is conjugative, was also detected. These mobile genetic elements found in the plasmids may help promote excision and reintegration or transfer to other plasmids by horizontal propagation (Naseer and Sundsfjord, 2011; Cantón et al., 2012).
In summary, this is the first report of K. pneumoniae carrying blaCTX–M–167 globally, indicating a new evolutionary starting point for CTX-M ESBLs. Our study suggested that the monitoring and prevention of novel ESBLs should be strengthened to prevent the spread of multidrug-resistant gram-negative bacilli in healthcare facilities.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The study protocol was approved by the Institutional Review Board of Huashan Hospital, Fudan University (Number: 2018-408).
FH and MG designed the study. SL, SS, LD, RH, DY, and YG collected clinical samples and performed the experiments. SL, SS, FH, and MG analyzed the data. SS and SL wrote the manuscript. FH and MG reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
This work was supported by the National Key Research and Development Program of China (2021YFC2701800 and 2021YFC2701803); National Natural Science Foundation of China (81872311 and 81861138052); Shanghai Public Health System Construction Three-Year Action Plan (2020–2022) Key Disciplines (GWV-10.1-XK04) and the China Antimicrobial Surveillance Network (2020QD049). The funders had no role in study design, data collection, analysis, decision to publish, or manuscript preparation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acosta-Gutiérrez, S., Ferrara, L., Pathania, M., Masi, M., Wang, J., and Bodrenko, I. (2018). Getting Drugs into Gram-Negative Bacteria: rational Rules for Permeation through General Porins. ACS Infect. Dis. 4, 1487–1498. doi: 10.1021/acsinfecdis.8b00108
Amit, S., Mishali, H., Kotlovsky, T., Schwaber, M. J., and Carmeli, Y. (2015). Bloodstream infections among carriers of carbapenem-resistant Klebsiella pneumoniae: etiology, incidence and predictors. ClinMicrobiol. Infect. 21, 30–34. doi: 10.1016/j.cmi.2014.08.001
Arato, V., Raso, M. M., Gasperini, G., BerlandaScorza, F., and Micoli, F. (2021). Prophylaxis and Treatment against Klebsiella pneumoniae: current Insights on This Emerging Anti-Microbial Resistant Global Threat. Int. J. Mol. Sci. 22:4042. doi: 10.3390/ijms22084042
Cantón, R., González-Alba, J. M., and Galán, J. C.. (2012). CTX-M Enzymes: origin and Diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Chen, L., Mathema, B., Chavda, K. D., DeLeo, F. R., Bonomo, R. A., and Kreiswirth, B. N. (2014). Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 22, 686–696. doi: 10.1016/j.tim.2014.09.003
Clinical and Laboratory Standards Institute. (2021). Performance Standards for Antimicrobial Susceptibility testing, 31th ed. Clsi supplement m100. Wayne, PA: Clinical and Laboratory Standards Institute.
Daoud, Z., Salem Sokhn, E., Masri, K., Matar, G. M., and Doron, S. (2015). Escherichia coli Isolated from Urinary Tract Infections of Lebanese Patients between 2005 and 2012: epidemiology and Profiles of Resistance. Front. Med. 2:26. doi: 10.3389/fmed.2015.00026
David, S., Reuter, S., Harris, S. R., Glasner, C., Feltwell, T., and Argimon, S. (2019). Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 4, 1919–1929. doi: 10.1038/s41564-019-0492-8
Denkel, L. A., Maechler, F., Schwab, F., Kola, A., Weber, A., Gastmeier, P., et al. (2020). Infections caused by extended-spectrum β-lactamase-producing Enterobacterales after rectal colonization with ESBL-producing Escherichia coli or Klebsiella pneumoniae. Clin. Microbiol. Infect. 26, 1046– 1051.
Doménech-Sánchez, A., Martínez-Martínez, L., Hernández-Allés, S., del Carmen Conejo, M., Pascual, A., and Tomás, J. M. (2003). Role of Klebsiella pneumoniae OmpK35 porin in antimicrobial resistance. Antimicrob. Agents Chemother. 47, 3332–3335. doi: 10.1128/AAC.47.10.3332-3335.2003
Dutzler, R., Rummel, G., Albertí, S., Hernández-Allés, S., Phale, P., Rosenbusch, J., et al. (1999). Crystal structure and functional characterization of OmpK36, the osmoporin of Klebsiella pneumoniae. Structure 7, 425–434. doi: 10.1016/s0969-2126(99)80055-0
Gülmez, D., Woodford, N., Palepou, M. F., Mushtaq, S., Metan, G., Yakupogullari, Y., et al. (2008). Carbapenem-resistant Escherichia coli and Klebsiella pneumoniae isolates from Turkey with OXA-48-like carbapenemases and outer membrane protein loss. Int. J. Antimicrob. Agents 31, 523–526. doi: 10.1016/j.ijantimicag.2008.01.017
Hao, M., Ye, M., Shen, Z., Hu, F., Yang, Y., Wu, S., et al. (2018). Porin Deficiency in Carbapenem-Resistant Enterobacter aerogenes Strains. Microb. Drug Resist. 24, 1277–1283. doi: 10.1089/mdr.2017.0379
Hu, Y., Liu, C., Shen, Z., Zhou, H., Cao, J., Chen, S., et al. (2020). Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008–2018. Emerg. Microbes Infect. 9, 1771–1779.
Kurien, B. T., and Scofield, R. H. (2012). Extraction of proteins from gels: a brief review. Methods Mol. Biol. 869, 403–405. doi: 10.1007/978-1-61779-821-4_33
Lambelet, P., Tascini, C., Fortunato, S., Stefanelli, A., Simonetti, F., Vettori, C., et al. (2017). Oral gentamicin therapy for Carbapenem-resistant Klebsiella pneumoniae gut colonization in hematologic patients: a single center experience. New Microbiol. 40, 161–164.
Leão, C., Clemente, L., Moura, L., Seyfarth, A. M., Hansen, I. M., Hendriksen, R. S., et al. (2021). Emergence and Clonal Spread of CTX-M-65-Producing Escherichia coli From Retail Meat in Portugal. Front. Microbiol. 12:653595. doi: 10.3389/fmicb.2021.653595
Logan, L. K., and Weinstein, R. A. (2017). The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: the Impact and Evolution of a Global Menace. J. Infect. Dis. 215, S28–S36. doi: 10.1093/infdis/jiw282
Naseer, U., and Sundsfjord, A. (2011). The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb. Drug Resist. 17, 83–97. doi: 10.1089/mdr.2010.0132
Pantelidou, I. M., Galani, I., Georgitsi, M., Daikos, G. L., and Giamarellos-Bourboulis, E. J. (2015). Interactions of Klebsiella pneumoniae with the innate immune system vary in relation to clone and resistance phenotype. Antimicrob. Agents Chemother. 59, 7036–7043. doi: 10.1128/AAC.01405-15
Paterson, D. L., and Bonomo, R. A. (2005). Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18, 657–686.
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Sanchez-Carbonel, A., Mondragón, B., López-Chegne, N., Peña-Tuesta, I., Huayan-Dávila, G., and Blitchtein, D. (2021). The effect of the efflux pump inhibitor Carbonyl Cyanide m-Chlorophenylhydrazone (CCCP) on the susceptibility to imipenem and cefepime in clinical strains of Acinetobacter baumannii. PLoS One 16:e0259915. doi: 10.1371/journal.pone.0259915
Shields, R. K., Nguyen, M. H., Chen, L., Press, E. G., Potoski, B. A., and Marini, R. V. (2017). Ceftazidime-Avibactam Is Superior to Other Treatment Regimens against Carbapenem-Resistant Klebsiella pneumoniae Bacteremia. Antimicrob. Agents Chemother. 61, e883–e817. doi: 10.1128/AAC.00883-17
Tsai, Y. K., Fung, C. P., Lin, J. C., Chen, J. H., Chang, F. Y., Chen, T. L., et al. (2011). Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 55, 1485–1493. doi: 10.1128/AAC.01275-10
Verschuuren, T. D., van Hout, D., Arredondo-Alonso, S., Fluit, A. C., Reuland, E. A., and Top, J. (2021). Comparative genomics of ESBL-producing Escherichia coli (ESBL-Ec) reveals a similar distribution of the 10 most prevalent ESBL-Ec clones and ESBL genes among human community faecal and extra-intestinal infection isolates in the Netherlands (2014-17). J. Antimicrob. Chemother. 76, 901–908. doi: 10.1093/jac/dkaa534
Weiß, D., Engelmann, I., Braun, S. D., Monecke, S., and Ehricht, R. (2017). A multiplex real-time PCR for the direct, fast, economic and simultaneous detection of the carbapenemase genes blaKPC, blaNDM, blaVIM, and blaOXA-48. J. Microbiol. Methods 142, 20–26. doi: 10.1016/j.mimet.2017.08.017
Xu, L., Sun, X., and Ma, X. (2017). Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann. Clin. Microbiol. Antimicrob. 16:18. doi: 10.1186/s12941-017-0191-3
Yang, Y. M., Osawa, K., Kitagawa, K., Hosoya, S., Onishi, R., and Ishii, A. (2021). Differential effects of chromosome and plasmid blaCTX-M-15 genes on antibiotic susceptibilities in extended-spectrum beta-lactamase-producing Escherichia coli isolates from patients with urinary tract infection. Int. J. Urol. 28, 623–628. doi: 10.1111/iju.14498
Yang, Y., Guo, Y., Yin, D., Zheng, Y., Wu, S., Zhu, D., et al. (2020). In Vitro Activity of Cefepime-Zidebactam, Ceftazidime-Avibactam, and Other Comparators against Clinical Isolates of Enterobacterales, Pseudomonas aeruginosa, and Acinetobacter baumannii: results from China Antimicrobial Surveillance Network (CHINET) in 2018. Antimicrob. Agents Chemother. 65, e1726–e1720. doi: 10.1128/AAC.01726-20
Zhang, D., Yin, Z., Zhao, Y., Feng, J., Jiang, X., Zhan, Z., et al. (2017). p1220-CTXM, a pKP048-related IncFII(K) plasmid carrying blaCTX-M-14 and qnrB4. Futur. Microbiol. 12, 1035–1043. doi: 10.2217/fmb-2017-0026
Zhang, Y., Peng, S., Xu, J., Li, Y., Pu, L., Han, X., et al. (2022). Genetic context diversity of plasmid-borne blaCTX-M-55 in Escherichia coli isolated from waterfowl. J. Glob. Antimicrob. Resist. 28, 185–194. doi: 10.1016/j.jgar.2022.01.015
Keywords: Klebsiella pneumoniae, blaCTX–M–167, blaSHV–1, blaTEM–1B, carbapenem
Citation: Li S, Shen S, Ding L, Han R, Guo Y, Yin D, Guan M and Hu F (2022) First Report of blaCTX–M–167, blaSHV–1, and blaTEM–1B Carrying Klebsiella pneumonia Showing High-Level Resistance to Carbapenems. Front. Microbiol. 13:916304. doi: 10.3389/fmicb.2022.916304
Received: 09 April 2022; Accepted: 02 June 2022;
Published: 07 July 2022.
Edited by:
Alba Edith Vega, National University of San Luis, ArgentinaReviewed by:
Bela Kocsis, Semmelweis University, HungaryCopyright © 2022 Li, Shen, Ding, Han, Guo, Yin, Guan and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Guan, Z3Vhbm1pbmc4OEB5YWhvby5jb20=; Fupin Hu, aHVmdXBpbkBmdWRhbi5lZHUuY24=
†These authors have contributed equally to this work and share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.