
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 July 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.914589
This article is part of the Research Topic Microorganisms and their Metabolites as Biocontrol Agents for Sustainable Agriculture View all 9 articles
Bacterial wilt caused by Ralstonia solanacearum is a fatal disease that affects the production of tomatoes and many other crops worldwide. As an effective strategy to manage bacterial wilt, biological control agents using plant growth-promoting rhizobacteria (PGPR) are being developed. In this study, we screened 2,3-butanediol (BDO)-producing PGPR to control tomato bacterial wilt and investigated the action mechanism of the disease control agent. Of the 943 strains isolated from soil, Klebsiella pneumoniae strain JCK-2201 produced the highest concentration of 2,3-BDO. The culture broth of K. pneumoniae JCK-2201 did not show any direct activity on R. solanacearum in vitro, but a 100-fold dilution effectively controlled tomato bacterial wilt with a control value of 77% in vivo. Fermentation utilizing K. pneumoniae JCK-2201 was optimized to produce 48 g/L of meso-2,3-BDO, which is 50% of the sucrose conversion efficiency. In addition, the control efficacy and mechanism of meso-2,3-BDO produced by JCK-2201 in tomato bacterial wilt were determined by comparative analysis with Bacillus licheniformis DSM13 producing meso-2,3-BDO and B. licheniformis DSM13 ΔalsS that did not produce 2,3-BDO, as the step of converting pyruvate to α-acetolactate was omitted. Tomato seedlings treated with the K. pneumoniae JCK-2201 (500-fold dilution) and B. licheniformis DSM13 (100-fold dilution) culture broth produced meso-2,3-BDO that significantly reduced R. solanacearum-induced disease severity with control values of 55% and 63%, respectively. The formulated meso-2,3-BDO 9% soluble concentrate (SL; 1,000-fold dilution) showed 87% control against tomato bacterial wilt in the field condition. Klebsiella pneumoniae JCK-2201 and B. licheniformis DSM13 treatment induced the expression of plant defense marker genes, such as LePR1, LePR2, LePR5, LePR3, and PI-II, in the salicylic acid and jasmonic acid signaling pathways at 4 days after inoculation. These results show that 2,3-BDO-producing bacteria and 2,3-BDO are potential biological control agents that act through induction of resistance for controlling tomato bacterial wilt.
Ralstonia solanacearum is one of the most important phytopathogenic bacteria that attack many crops and native plants in tropical and subtropical climates worldwide and has a very extensive host range. It enters the roots from the soil and attacks the host’s vascular structures, causing the plant to wilt and perish (Denny, 2007; Sebastià et al., 2021). Tomato bacterial wilt caused by R. solanacearum affects the youngest leaves and causes yield loss through wilting and stunted growth (Ji et al., 2005). Various strategies, such as soil improvement, field sanitation, breeding of resistant varieties, and improvement of cropping systems, have been used to control this disease (Xue et al., 2013; Zheng et al., 2019). However, this disease is very difficult to control owing to the high variability of the pathogen, limited chemical control potential, high viability rate of the pathogen in various environments, and broad host range (Mohammed et al., 2020). Therefore, sustainable, economical, and environment-friendly biological controls have been proposed as alternatives over the past few decades (Van Lenteren, 2001).
Among the biological control agents, plant growth-promoting rhizobacteria (PGPR) have been reported as potential candidates as they promote plant growth, improve quality, and directly or indirectly control diseases (Jetiyanon and Kloepper, 2002). PGPR, such as the genera Pseudomonas, Klebsiella, Enterobacter, Azospirillum, Burkholderia, Bacillus, and Serratia, promote plant growth and control plant diseases (Glick et al., 1998; Shaharoona et al., 2007; Mazumdar et al., 2018). Klebsiella pneumoniae HR1 reduces the incidence of root rot-causing Macrophomina phaseolina in Vigna mungo L. and significantly increases the germination rate, shoot and root length, and shoot and root dry weight in the treated plants (Dey et al., 2019). Klebsiella pneumoniae NG14 has nitrogen-fixing activity and colonizes the root surface and vascular tissue of rice (Liu et al., 2011). Klebsiella jilinsis 2N3 induces resistance to corn leaf blight caused by Setosphaeria turcica, a hemibiotrophic ascomycete fungal pathogen, and promotes maize growth (Zhang et al., 2021). In addition, Bacillus cereus Sneb 560, B. subtilis Sneb 815, Pseudomonas putida Sneb 821, P. fluorescens Sneb 825, and Serratia proteamaculans Sneb 821 isolated from the rhizosphere soil show strong larvicidal and nematicidal activity in vitro and promote higher root and shoot growth compared to the control when tomato seeds were treated in pot experiments (Zhao et al., 2018). Taken together, PGPR isolates can be used as potential biological control agents against various plant diseases in the future.
All plants have defense mechanisms against pathogens. In general, induced resistance acts systemically because it is not limited to the infected plant parts, and this is commonly referred to as systemic acquired resistance (SAR; Van Loon et al., 1998). SAR involves the accumulation of the signal molecule salicylic acid (SA) and pathogenesis-related proteins (PRs) and confers persistent and broad-spectrum resistance. Induced systemic resistance (ISR) relies on pathways regulated by jasmonic acid (JA) and ethylene (ET). Some PRs hydrolyze fungal cell walls with chitinase and β-1,3-glucanases and are used as markers for SA-associated necrosis infection (Yan et al., 2002; Park et al., 2007a). Activation of ISR by beneficial microorganisms in the plant rhizosphere makes plants healthy and is distinct from SAR, which is a response associated with the aerial part (Pieterse et al., 2004).
Induced resistance occurs when the plant’s defense mechanism induced by PGPR, such as Pseudomonas, Klebsiella, and Bacillus, resists infection and does not cause visible damage to the root system. Klebsiella pneumoniae SnebYK treatment promotes soybean growth and induces systemic resistance by expression of PR1, PR2, PR5, and PDF1.2 to inhibit the invasion and development of Heterodera glycines (Liu et al., 2018). Some strains of PGPR also play an important role in plant disease control by enhancing ISR via the production of volatile organic compounds (VOCs), such as acetoin and 2,3-butanediol (BDO; Kirankumar et al., 2008; Lee et al., 2012). 2,3-BDO production in microorganisms was first reported in 1906 from K. pneumoniae (Harden and Walpole, 1906). Moreover, strains of the genera Klebsiella and Enterobacter primarily produce (2S,3S)-BDO (known as dextro-2,3-BDO) and (2R,3S)-BDO (known as meso-2,3-BDO), whereas the genus Bacillus produces (2R,3R)-BDO (known as levo-2,3-BDO) and meso-2,3-BDO (Ji et al., 2011). α-Acetolactate synthase (ALS), acetolactate decarboxylase (ALDC), and acetoin reductase (AR) are involved in the 2,3-BDO production process. These three enzymes catalyze the production of 2,3-BDO in successive metabolic conversion steps: pyruvate to ALS, ALS to acetoin, and acetoin to 2,3-BDO (Song et al., 2019).
Previous reports have demonstrated that activating SAR and ISR is a promising and effective approach to control a wide range of plant diseases (Kong et al., 2018; Liu et al., 2018). Furthermore, because PGPR strains do not normally exhibit direct activity unlike chemical pesticides, they can be controlled without selective pressure and appear to be environment-friendly (Vallad and Goodman, 2004). Klebsiella pneumoniae, a PGPR strain, produces meso-2,3-BDO and induces resistance to the symptoms of wilt and root rot disease and sheath blight of rice (Ji et al., 2014). To the best of our knowledge, previous studies have not investigated the induced resistance effect of K. pneumoniae on tomato bacterial wilt and the control ability of meso-2,3-BDO on tomato bacterial wilt in the field.
During the screening of antagonistic PGPR with high induced resistance activity, we found that K. pneumoniae JCK-2201 strain produces a high amount of meso-2,3-BDO and can effectively suppress the development of tomato bacterial wilt even though it was not active to R. solanacearum in vitro bioassay. In order to elucidate the role of meso-2,3-BDO in the induced resistance activity of K. pneumoniae JCK-2201, we tried to make mutants that do not produce meso-2,3-BDO. However, we failed to make the mutants. Therefore, we tried to make meso-2,3-BDO knockout mutants using Bacillus licheniformis DSM13 which can produce meso-2,3-BDO (Veith et al., 2004). It also did not show any in vitro antibacterial activity against R. solanacearum like K. pneumoniae JCK-2201. Accordingly, the objectives of this study were to (1) optimize 2,3-BDO production by K. pneumoniae JCK-2201, selected through the Voges–Proskauer (VP) test, (2) evaluate the in vivo effects of K. pneumoniae JCK-2201, B. licheniformis DSM13, and B. licheniformis DSM13 ΔalsS strains on tomato bacterial wilt, (3) investigate the effect of meso-2,3-BDO in the tomato field, and (4) elucidate the action mechanism against tomato bacterial wilt by qRT-PCR analysis of defense-related genes.
To isolate acetoin-producing bacteria, we collected soil samples from several sampling sites, viz., Gwang-ju campus of Chonnam National University (35.176°N, 126.906°E), Mt. Mudeung (35.134°N, 126.989°E), Mt. Naejang (35.486°N 126.883°E), and Gok-seong area (35.282°N, 127.292°E), in the South Korea. Each soil sample (1 g) was suspended in 9 ml of sterile distilled water and shaken for 10 min. The solution was then serially diluted to 10−7 in 10-fold increments. A 100-μl aliquot of the diluted soil was plated on tryptic soy agar (TSA; Becton, Dickinson and Co., Sparks, MA, United States) plates. Plates were incubated at 30°C for 1–3 days. A single colony was then isolated and incubated in tryptic soy broth (TSB; Becton, Dickinson and Co., Sparks, MA, United States) at 30°C with constant shaking at 150 rpm for 1–2 days and then stored at −80°C in 30% glycerol until use.
Acetoin-producing bacteria were screened using the VP method that detects acetoin, a precursor of 2,3-BDO (Renna et al., 1993). A single colony isolated from soil samples was inoculated into 500 μl of MR-VP medium (7 g/L peptone, 5 g/L dextrose, 5 g/L dipotassium phosphate) in a 1.7-ml e-tube and cultured for 24 h at 37°C. After 24 h of incubation, the culture broth was used for the VP test to distinguish acetoin-producing cells from the background. Culture broth (100 μl) or acetoin (1 mg/ml; Tokyo Chemical Industry Co., Ltd., Tokyo, Japan) was added to each well, and the VP reaction was induced by sequential addition of 60 μl of 5% 1-naphthol dissolved in ethanol and 20 μl of 40% potassium hydroxide dissolved in distilled water. After mixing for 10 min, the color of the culture broth was noted to analyze the presence of acetoin. Cherry red color indicated a positive result that the culture broth contains acetoin produced by a strain and a yellow–brown color indicated a negative result that the culture broth does not contain acetoin. Each sample was replicated three times.
Acetoin-producing bacteria were grown in TSA at 37°C for 1–2 days and then cultured in 3 ml of TSB in a 15-ml tube at 37°C with constant shaking at 180 rpm for 12 h. The pre-culture was inoculated at 1% in a basal medium containing 50 g/L glucose, 4 g/L K2HPO4, 1 g/L triammonium citrate, 4 g/L sodium acetate, 10 g/L yeast extract, and 0.8 g/L MgSO4·7H2O and incubated at 37°C with constant shaking at 180 rpm for 24 h. The culture broth was then filtered through a 0.2-μm membrane filter (Advantec Toyo Co., Tokyo, Japan) to obtain axenic culture filtrate.
Ralstonia solanacearum SL341 was cultured in TSB at 30°C with constant shaking at 150 rpm for 2 days, and the concentration of the culture broth was adjusted to 1 × 106 colony-forming units (CFU)/ml. Acetoin-producing bacteria (1 μl) were transferred to each well of a 96-well plate containing R. solanacearum SL341 suspension (99 μl). Samples were tested in two-fold dilutions at concentrations ranging from 10% to 0.078%. Streptomycin sulfate (10 mg/ml) dissolved in distilled water was used as the positive control. Plates were incubated at 30°C for 24 h. The minimum inhibitory concentration (MIC) was defined as the lowest concentration of the culture filtrate that completely inhibited bacterial growth and was observed visually. Three replicates were performed.
Concentrations of acetoin and 2,3-BDO were determined using an HPLC system (Shimadzu Corporation, Kyoto, Japan) equipped with a DGU-20A degassing unit, LC-20AT pump, SIL-20A autosampler, CTO-20A column oven, RID-20A refractive index detector, and CBM-20A communications bus module. All acetoin-producing bacterial culture broths were centrifuged at 10,000 rpm for 5 min at 4°C and then filtered through a 0.45-μm filter to remove the debris before injecting it into the HPLC system. The samples were eluted isocratically on Aminex HPX-87H column (7.8 × 300 mm; Bio-Rad Laboratories, Hercules, CA, United States) and maintained at 60°C with 5 mM H2SO4 in water at a flow rate of 0.6 ml/min.
Standard solutions of acetoin (Tokyo Chemical Industry Co., Japan), levo-2,3-BDO, meso-2,3-BDO, and dextro-2,3-BDO (Sigma-Aldrich, St. Louis, MS, United States) were prepared at a concentration of 1.11, 3.33, 10, 30, and 90 mg/ml, respectively, in water. The solutions were filtered through a 0.45-μm Advantec membrane filter. The calibration curve was plotted by linear regression.
Genomic DNA of JCK-2201 was extracted using the Bacterial genomic DNA purification Kit (ELPIS-Biotech, Daejeon, South Korea) according to the manufacturer’s recommendations. This strain was identified by sequence analysis of the 16S rRNA gene. The 16S rRNA gene was amplified by PCR using the specific primer pair set 9F (5′-GAGTTTGATCCTGGCTCAG-3′)/1512R (5′-ACGGCTACCTTGTTACGACTT-3′) and sequenced (Genotech Co., Daejeon, South Korea; Le et al., 2020). Gene sequences were compared to the GenBank database using BioEdit version 5.0.9.1 and aligned using ClustalW. A phylogenetic tree was constructed using the neighbor-joining method with 1,000 bootstrap replicates in MEGA version 6.06 (Chen et al., 2016).
The strains and plasmids used in this study are described in Supplementary Table 1. Escherichia coli DH5α (Invitrogen, Carlsbad, CA, United States) was used for DNA cloning, whereas E. coli HST04 dam−/dcm− (Takara, Kyoto, Japan) was used for the propagation of plasmids to prevent the host restriction–modification system in B. licheniformis DSM13 before electrotransformation. For genetic analysis, B. licheniformis DSM13 was cultivated in Luria–Bertani (LB) medium at 30°C or 50°C depending on the specific requirement. Kanamycin (Km) was used at final concentrations of 50 μg/ml and 20 μg/ml for E. coli and B. licheniformis, respectively.
pBKN was used for homologous recombination-mediated gene manipulation (Song et al., 2021). For scarless deletion of the alsS gene, homologous regions at ~700 base pairs upstream [primers: lf-F (NheI)/lf-R) and downstream (primers: rt-F/rt-R (AflII)] were subjected to PCR to amplify the homologous fragments H1 and H2, respectively (Supplementary Figure 1). To construct a deletion cassette, fusion PCR was performed using flanking primers with H1 and H2 fragments as the template. The fusion product was then ligated into the NheI/AflII sites of pBKN to construct the recombinant plasmid, pBKN_alsS. pBKN_alsS was used for conventional homologous recombination-mediated gene deletion. To generate a gene knockout mutant, B. licheniformis strain was electrotransformed with the corresponding gene knockout plasmid by the high-osmolarity transformation protocol (Xue et al., 1999). For the sequential 1st/2nd crossover process, the 1st crossover (integration) of the whole plasmid was triggered at a high temperature (50°C) in the cells based on the unstable characteristics of the pE194ts replication origin. PCR was performed to confirm the 1st crossover mutants using genomic-F/screen-R and screen-F/genomic-R primers. Next, several clones were inoculated in LB broth without antibiotics at 50°C. The cells were then sequentially transferred five to eight times in a fresh LB medium to generate a 2nd crossover mutant. Finally, several clones were plated on an LB agar plate without antibiotics. Km-sensitive gene knockout mutants were then screened by replica plating and PCR.
Medium optimization for the production of 2,3-BDO was conducted using the basal medium. Optimum carbon and nitrogen sources were selected and then their optimum contents were determined. Klebsiella pneumoniae JCK-2201 was incubated in LB medium at 37°C with constant shaking at 180 rpm. The pre-culture was inoculated at 1% into 125-ml Erlenmeyer flasks containing 20 ml of each medium and then incubated at 37°C with constant shaking at 180 rpm. The amounts of acetoin and 2,3-BDO in the culture broth were determined by HPLC analysis after 24 h of incubation. Seven carbon sources, viz., glucose, sucrose, starch, maltose, lactose, fructose, and xylose, were tested as alternative carbon sources at 50 g/L. The optimal concentration of the selected carbon source for the production of 2,3-BDO by K. pneumoniae JCK-2201 was tested at 25, 50, 75, 100, 125, and 150 g/L. Similarly, seven nitrogen sources, viz., beef extract, casamino acid, corn steep liquor, NaNO3, NH4NO3, soybean meal, and yeast extract, were tested to replace yeast extract as alternative nitrogen sources at 10 g/L. The optimal concentration of the selected nitrogen source was investigated at 5, 10, 20, and 40 g/L. Finally, after culturing K. pneumoniae JCK-2201 in a medium containing the selected carbon and nitrogen sources, HPLC analysis was performed every 12 h to determine the incubation time.
To perform in vivo test against tomato bacterial wilt, we modified the conditions previously described (Lee et al., 2015). “Seokwang” tomato seeds (Farm Hannong Co., Ltd., Seoul, South Korea) were used for the wilt test. Tomato seeds were sown in plug trays (5 × 8 cm, Bumnong Co., Ltd., Seoul, South Korea) using horticulture nursery soil (Bumnong Co., Ltd., Seoul, South Korea) and then cultivated in a culture room at 25 ± 5°C for 4 weeks under a light/dark cycle of 16 h/8 h. Tomato plants were transplanted into 7.5 cm (ø) plastic pots. Tomato bacterial wilt was evaluated using 4th–5th-leaf stage tomato plants.
Klebsiella pneumoniae JCK-2201 culture broth, containing basal medium, was centrifuged at 10,000 rpm for 10 min to obtain the supernatant was then diluted to 100-, 500-, 1,000-, and 5,000-fold. Optimal K. pneumoniae JCK-2201 culture broth was diluted at 500-fold, and B. licheniformis DSM13 and B. licheniformis DSM13 ΔalsS were prepared by diluting at 100-fold. Meso-2,3-BDO was prepared at concentrations of 1 mM and 0.1 mM. Samples (20 ml) were treated by soil drench 7 and 2 days before inoculation. Sungbocycline (17% oxytetracycline; Sungbo Chemicals Co., Ltd., Seoul, South Korea) was used as the positive control and treated by soil drench 1 day before inoculation at 1,000-fold dilution.
Ralstonia solanacearum SL341 was cultured in TSA at 30°C for 3 days, and the cells were then harvested with water. Ralstonia solanacearum SL341 cell suspension (20 ml; 1 × 108 CFU/ml) was inoculated by soil drench. The inoculated plants were maintained in the dark for 24 h at 30 ± 3°C and then transferred to an incubation room at 30°C, 12 h of daylight per day, and relative humidity of 75% for 10 days. Disease severity was assessed on a scale of 0–5 described previously (Vu et al., 2017): 0 = no wilted leaves, 1 = one or two wilted leaves, 2 = two or three wilted leaves, 3 = three or four wilted leaves, 4 = more than four wilted leaves, and 5 = a dead plant. Each treatment was replicated three times, and each replicate consisted of three tomato plants. The control value was calculated using the Equation 1.
Meso-2,3-BDO-rich [90%] (1.2% levo-2,3-BDO, 93.1% meso-2,3-BDO, 5.7% dextro-2,3-BDO) raw material was produced from GS Caltex Corporation. Meso-2,3-BDO 9% and 0.9% liquid soluble concentrate (SL) formulations were prepared by Yuseong Hwayeon Tech Co., Ltd., Geumsan, South Korea.
The control efficacy of meso-2,3-BDO 9% and 0.9% SL against tomato bacterial wilt was evaluated in a field located in Dunnae-myeon, Hoengseong-gun, Gangwon-do, South Korea. “Seokwang” tomato seeds were sown in a seed tray containing topsoil in a greenhouse and transplanted into a field plot when 6–7 leaves were observed after 5 weeks. The field experiment consisted of 1 m × 7.5-m plots with 0.5-m spacing between the plots, and four replicates of each treatment group were formed using a randomized complete block design. A total of 48 plants were used, with 12 tomato plants per replicate. Bacterial wilt disease was caused naturally because the field had been severely damaged by R. solanacearum in the previous season. Tomatoes were treated with the samples four times at 7-days intervals after transplantation by soil drench.
A positive control, Dazomet (98% dazomet; Farm Hannong Co., Seoul, South Korea), was applied at 300 kg per 1 ha of soil 4 weeks before transplantation of tomato plants. Tap water was used as untreated control. Equation 2 was used to investigate the disease incidence 40 days after the first treatment, and Equation 3 was used to calculate the control value.
The effects of K. pneumoniae JCK-2201, B. licheniformis DSM13, and B. licheniformis DSM13 ΔalsS on the expression of defense-related genes against tomato bacterial wilt were analyzed using 4–5 leaf stage tomato plants. The culture broth of each strain was treated by soil drench 7 and 2 days before inoculation, and water was used for the untreated control group. Two days after the second treatment, 10 ml of R. solanacearum (1 × 108 CFU/ml) was inoculated. Tomato leaves were collected 1, 2, and 4 days after inoculation (DAI). Total RNA of tomato leaves was extracted using the IQeasy™ Plus Plant RNA Extraction Mini Kit (iNtRON Biotechnology, Seong-nam, South Korea) according to the manufacturer’s recommendations. Total RNA was synthesized as cDNA using oligo (dT) primer and SuperScript™ IV reverse transcriptase (Invitrogen Inc., Carlsbad, CA, United States) according to the manufacturer’s instructions. Real-time PCR was performed on a real-time PCR detection system (Bio-Rad CFX 96; Bio-Rad Laboratories, Hercules, CA, United States) using iQ™ SYBR Green Supermix (Bio-Rad Laboratories) according to manufacturer specifications.
PCR primers used in this study were synthesized by Genotech (Daejeon, South Korea; Supplementary Table 2). Ubiquitin (UBI) was used as the reference gene (Aimé et al., 2013). Pathogenesis-related protein-1 (LePR1), LePR2, and LePR5 were used as SA signaling pathway-related genes (Jia et al., 2013). Plant proteinase inhibitor-II (PI-II), LePR3, and LeLOX are involved in the JA signaling pathway, and ETR4 is involved in the ET signaling pathway. Catalase (CAT) gene is recognized as a reactive oxygen species (ROS) scavenger-associated gene (Mhamdi and Van Breusegem, 2018).
All statistical analyses were performed using the SPSS statistical analysis software (version 21.0 for Windows; SPSS, IBM Corp., Armonk, NY, United States). Data were expressed as mean ± standard deviation (SD) of replicates, and statistical differences were assessed using one-way ANOVA and determined by Tukey’s HSD test (p < 0.05).
Among the 943 strains isolated from soil, 23 strains showed a strong positive effect in the VP medium. Acetoin, levo-2,3-BDO, dextro-2,3-BDO, and meso-2,3-BDO were confirmed by HPLC analysis using a standard compound. However, levo-2,3-BDO and dextro-2,3-BDO could not be distinguished because samples showed the same retention time in the HPLC chromatogram. Among the 23 strains, K. pneumoniae JCK-2201 produced the most amount of acetoin and meso-2,3-BDO at 8.62 and 9.65 g/L, respectively (Table 1). In the in vitro MIC test against R. solanacearum SL341 causing tomato bacterial wilt, K. pneumoniae JCK-2201 did not show direct antibacterial activity when the culture broth was treated even at a concentration of 10%.
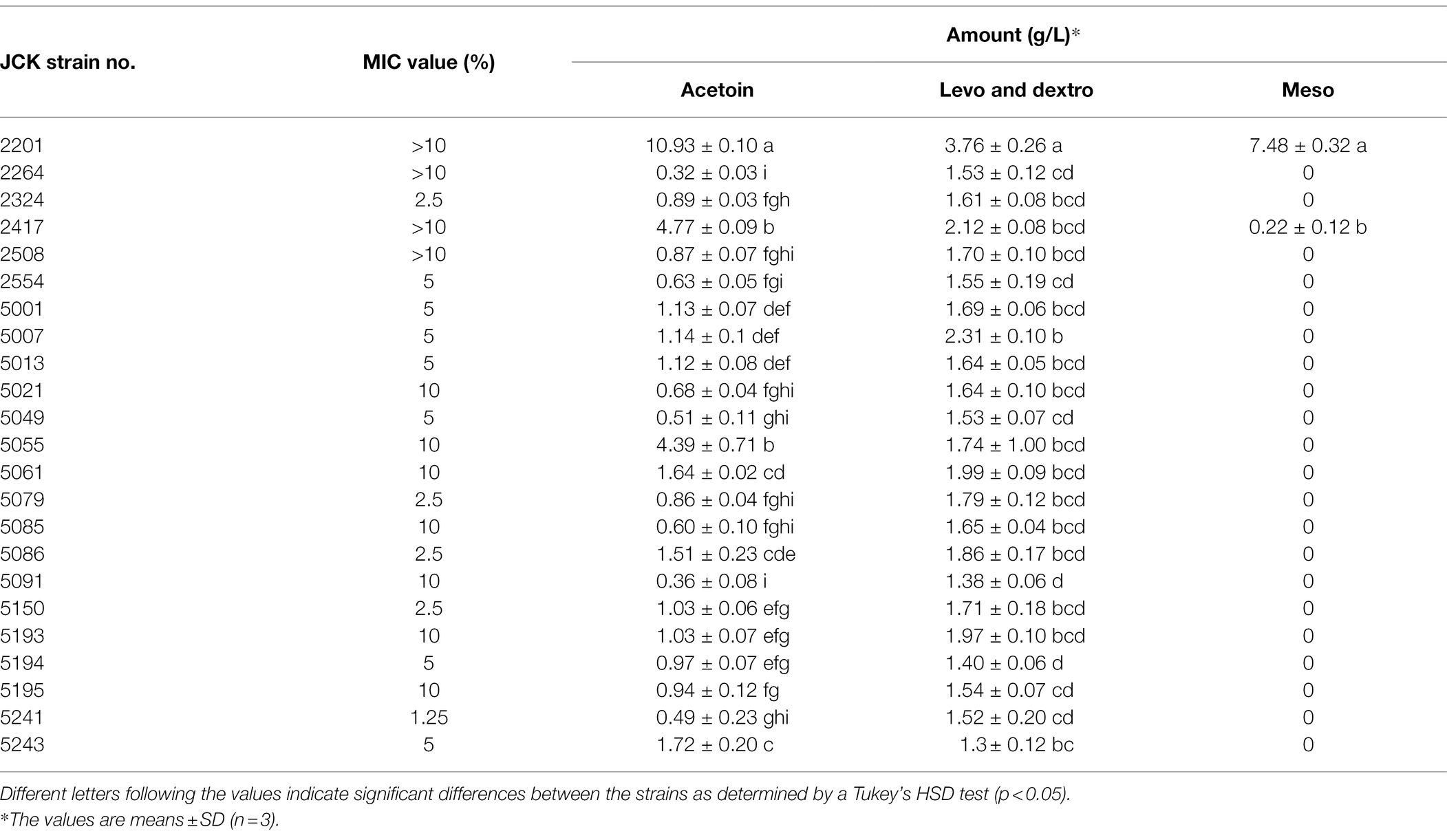
Table 1. Minimum inhibitory concentration (MIC) value against R. solanacearum and 2,3-BDO production amounts by the selected JCK strains.
To investigate the effect of K. pneumoniae JCK-2201 on tomato bacterial wilt, fermentation broth, containing the initial basal medium, of K. pneumoniae JCK-2201 was used (Figure 1). In the basal medium, K. pneumoniae JCK-2201 produced approximately 9 g/L of meso-2,3-BDO. The culture broth was diluted at 100-, 500-, 1,000-, and 5,000-fold, in which the concentrations of 2,3-BDO were 1, 0.2, 0.1, and 0.02 mM, respectively. Among these four dilution rates, 100-fold dilution showed the highest disease control efficacy against tomato bacterial wilt a control value of 77.14%, which was comparable to that of the positive control Sungbocycline.
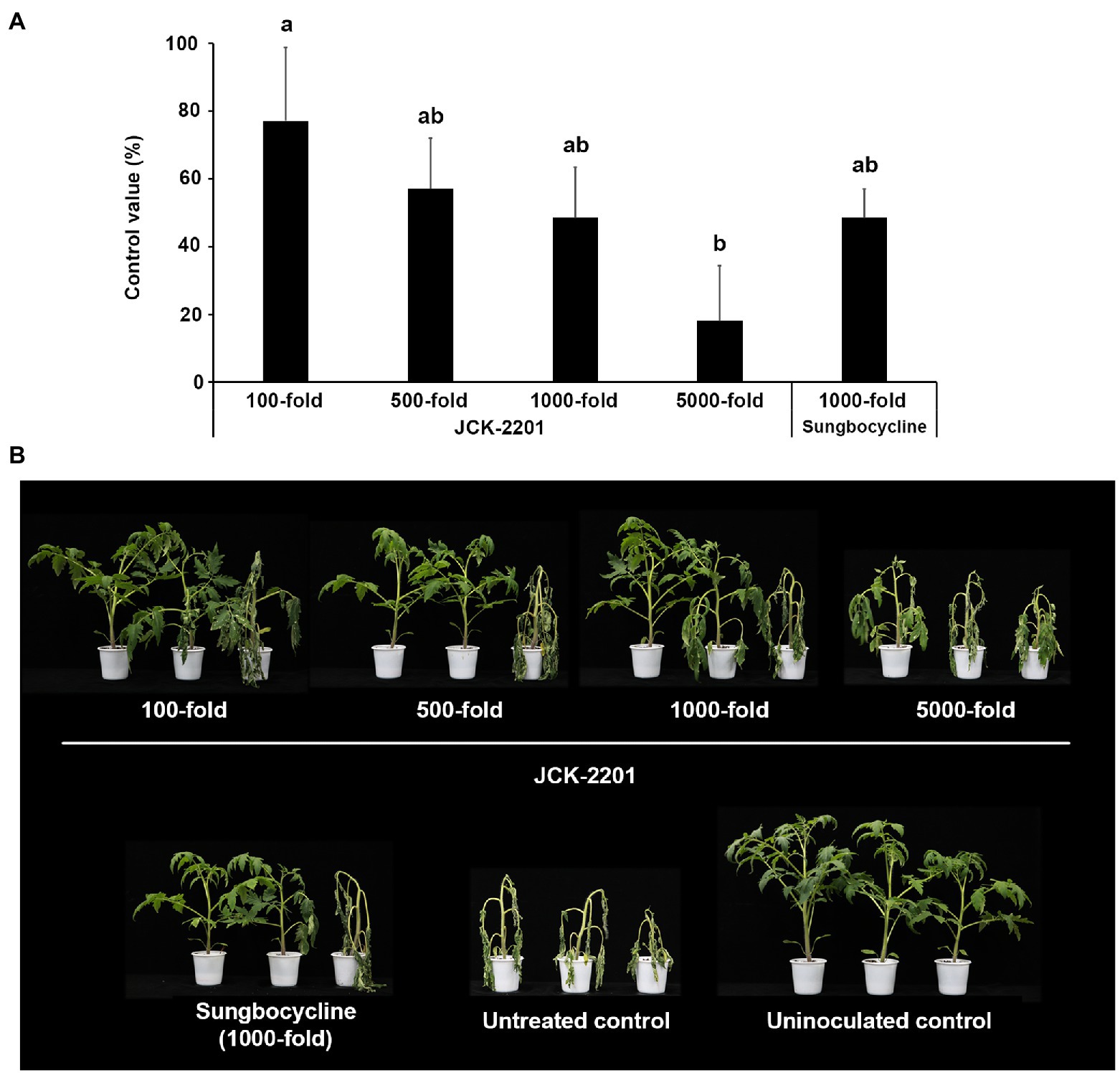
Figure 1. Disease control efficacy of K. pneumoniae JCK-2201 culture supernatant on tomato bacterial wilt caused by R. solanacearum. (A) Control value was investigated 7 days after pathogen inoculation. The data show the means ± SD (n = 3). Lower-case letters indicate statistically significant differences as compared to untreated control plants (p < 0.05) according to Tukey’s HSD test. Error bars indicate the SDs. (B) Representative photographs of tomato seedlings at 7 days after inoculation of R. solanacearum by soil drench. As a positive control, plants were treated with Sungbocycline.
The phylogenetic tree generated using the 16S rRNA gene sequence of JCK-2201 showed the highest homology with K. pneumoniae (Supplementary Figure 2). The JCK-2201 strain was, therefore, identified as K. pneumoniae with accession number OM510396 by phylogenetic analysis. Subsequently, JCK-2201 was named K. pneumoniae JCK-2201. The strain has been deposited in the Korean Agricultural Culture Collection (KACC, Jeonju, South Korea) as KACC 22683.
The medium was optimized for 2,3-BDO production by K. pneumoniae JCK-2201. Among the various carbon and nitrogen sources, sucrose and casamino acid were selected as the carbon and nitrogen source, respectively. When 50 g/L of sucrose was used as the carbon source, K. pneumoniae JCK-2201 produced acetoin and meso-2,3-BDO at 12.00 and 11.62 g/L, respectively (Figure 2A), and when sucrose was incorporated at 100 g/L, the concentrations of acetoin and meso-2,3-BDO increased to 4.72 and 36.46 g/L, respectively (Figure 2B). When 10 g of casamino acid was used as the nitrogen source, the production of meso-2,3-BDO increased to 42.25 g/L (Figure 2C); when casamino acid was incorporated at 20 g/L, the production of acetoin and meso-2,3-BDO increased to 4.62 and 47.57 g/L, respectively (Figure 2D). In the optimum medium, the production of meso-2,3-BDO reached 47.57 g/L 24 h after inoculation (Figure 2E), after which it decreased. Based on these results, we decided to incubate K. pneumoniae JCK-2201 in a medium containing sucrose (100 g) and casamino acid (20 g) at 37°C with constant shaking at 180 rpm for 24 h.
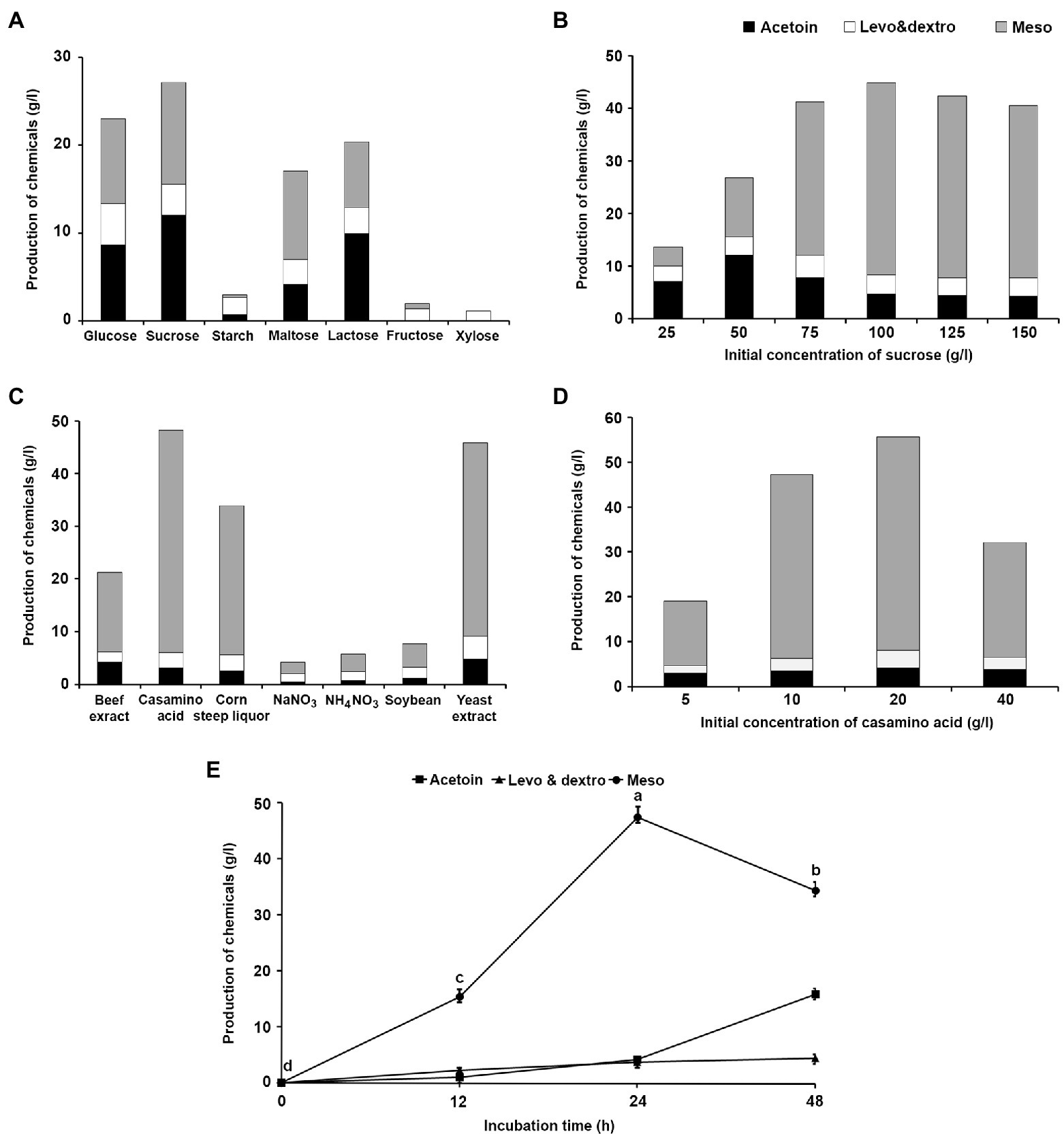
Figure 2. Effect of different carbon and nitrogen sources on the production of acetoin and 2,3-BDO by K. pneumoniae JCK-2201. The data show the means ± SD (n = 3). Black bar: acetoin, white bar: levo-2,3-BDO and dextro-2,3-BDO, gray bar: meso-2,3-BDO. (A) Carbon sources. (B) Sucrose concentration. (C) Nitrogen sources. (D) Casamino acid concentration. (E) Comparison of production according to incubation time when cultured in optimum medium containing 100 g/L sucrose and 20 g/L casamino acid. Lower-case letters indicate statistically significant differences as compared to meso-2,3-BDO production by incubation time (p < 0.05) according to Tukey’s HSD test.
In the optimum medium, K. pneumoniae JCK-2201 produced approximately 48 g/L meso-2,3-BDO. Bacillus licheniformis DSM13 and B. licheniformis DSM13 △alsS produced approximately 9 and 0 g/L of meso-2,3-BDO, respectively (Supplementary Figure 3). Treatment with 500-fold diluent of K. pneumoniae JCK-2201 culture broth and 100-fold diluent of B. licheniformis DSM13 culture broth containing approximately 1 mM meso-2,3-BDO significantly reduced the disease severity by 55.26% and 63.16% compared with the untreated control, respectively (Figure 3). The 100-fold diluent of the culture broth of B. licheniformis DSM13 ΔalsS, which does not produce meso-2,3-BDO, showed weaker disease control efficacy than the culture broths of K. pneumoniae JCK-2201 and B. licheniformis DSM13.
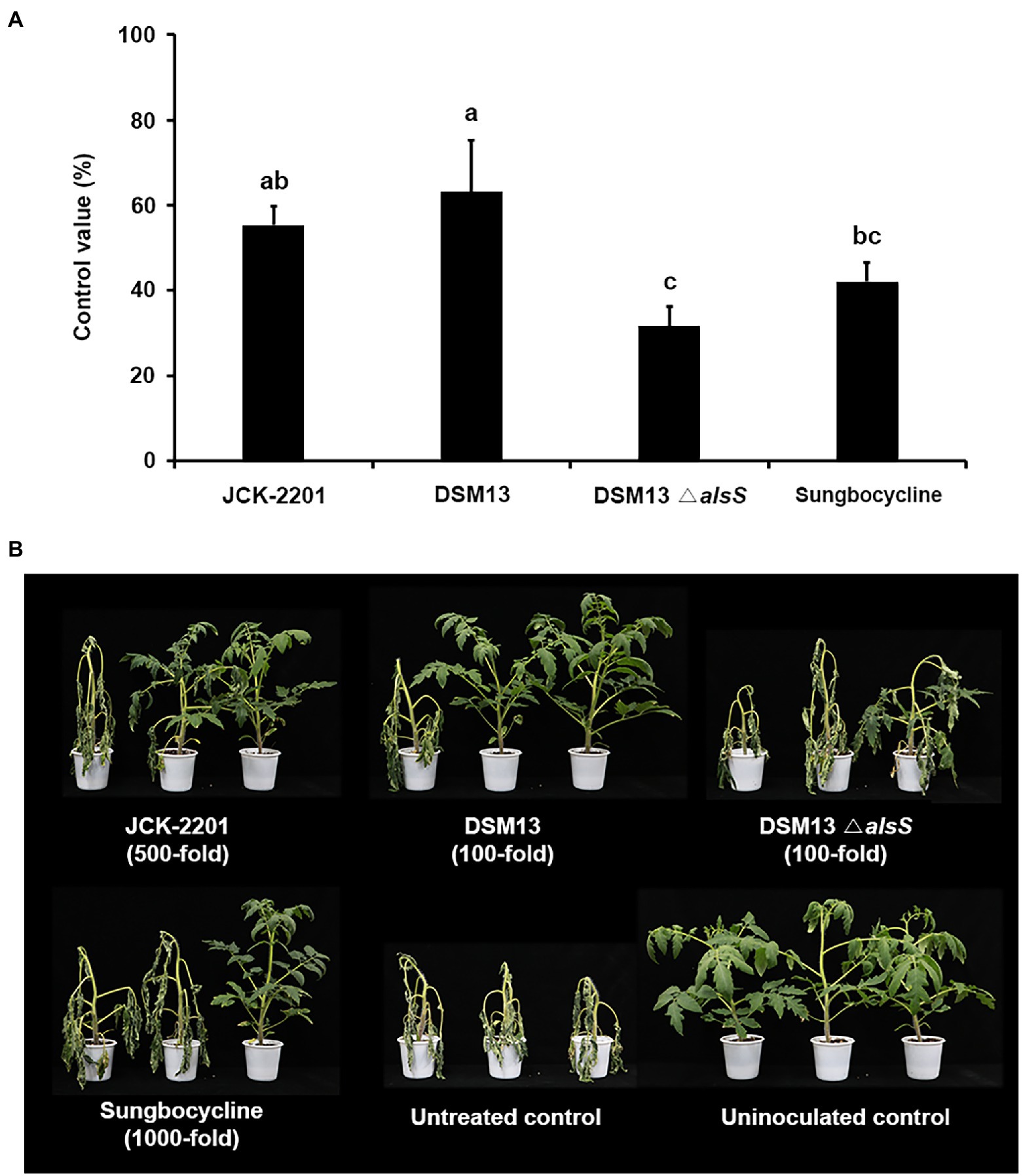
Figure 3. Disease control efficacy of culture supernatant of K. pneumoniae JCK-2201, B. licheniformis DSM13, and B. licheniformis DSM13ΔalsS on tomato bacterial wilt caused by R. solanacearum. (A) Control value was investigated 7 days after pathogen inoculation. The data show the means ± SD (n = 3). Lower-case letters indicate statistically significant differences as compared to untreated control plants (p < 0.05) according to Tukey’s HSD test. Error bars indicate the SDs. (B) Representative photographs of tomato seedlings at 7 days after inoculation of R. solanacearum by soil drench. As a positive control, plants were treated with Sungbocycline.
However, meso-2,3-BDO suppressed the development of tomato bacterial wilt in a dose-dependent manner, with control values of 54.29% and 31.43% at 1 and 0.1 mM, respectively (Figure 4). The control value of 1 mM meso-2,3-BDO was almost similar to that of the 500-fold diluent of K. pneumoniae JCK-2201 culture broth and the 100-fold diluent of B. licheniformis DSM13 culture broth.
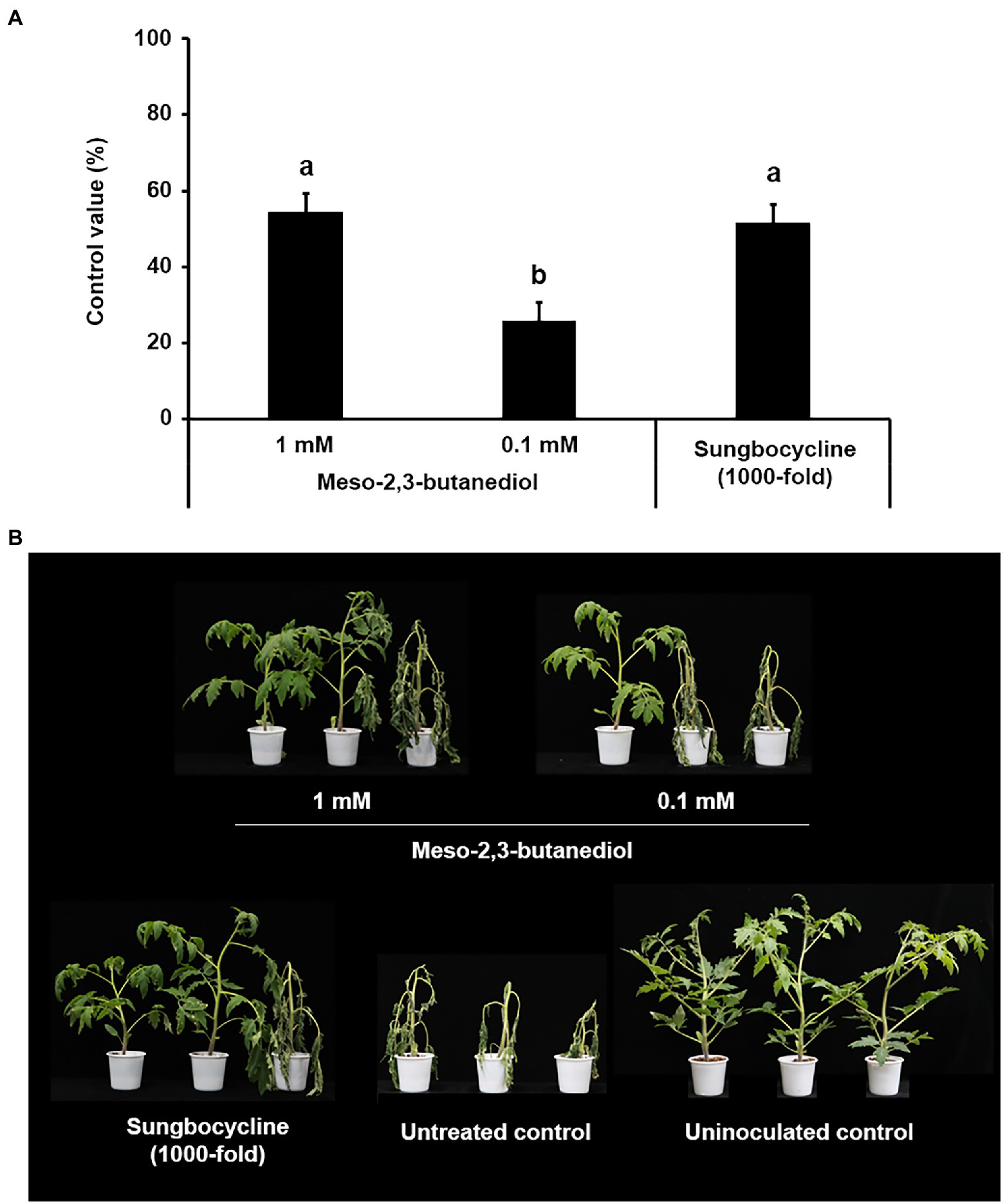
Figure 4. Disease control efficacy of meso-2,3-BDO on tomato bacterial wilt caused by R. solanacearum. (A) Control value was investigated 7 days after pathogen inoculation. The data show the means ± SD (n = 3). Lower-case letters indicate statistically significant differences as compared to untreated control plants (p < 0.05) according to Tukey’s HSD test. Error bars indicate the SDs. (B) Representative photographs of tomato seedlings at 7 days after inoculation of R. solanacearum by soil drench. As a positive control, plants were treated with Sungbocycline.
The disease control efficacy of two SL formulations of meso-2,3-BDO (9% SL and 0.9% SL) was evaluated under field conditions. When samples were applied at 1,000-fold dilution by soil drench, the control values were 87% for meso-2,3-BDO (9% SL) and 59% for meso-2,3-BDO (0.9% SL; Figure 5). These results were similar to those of Dazomet, the positive control.
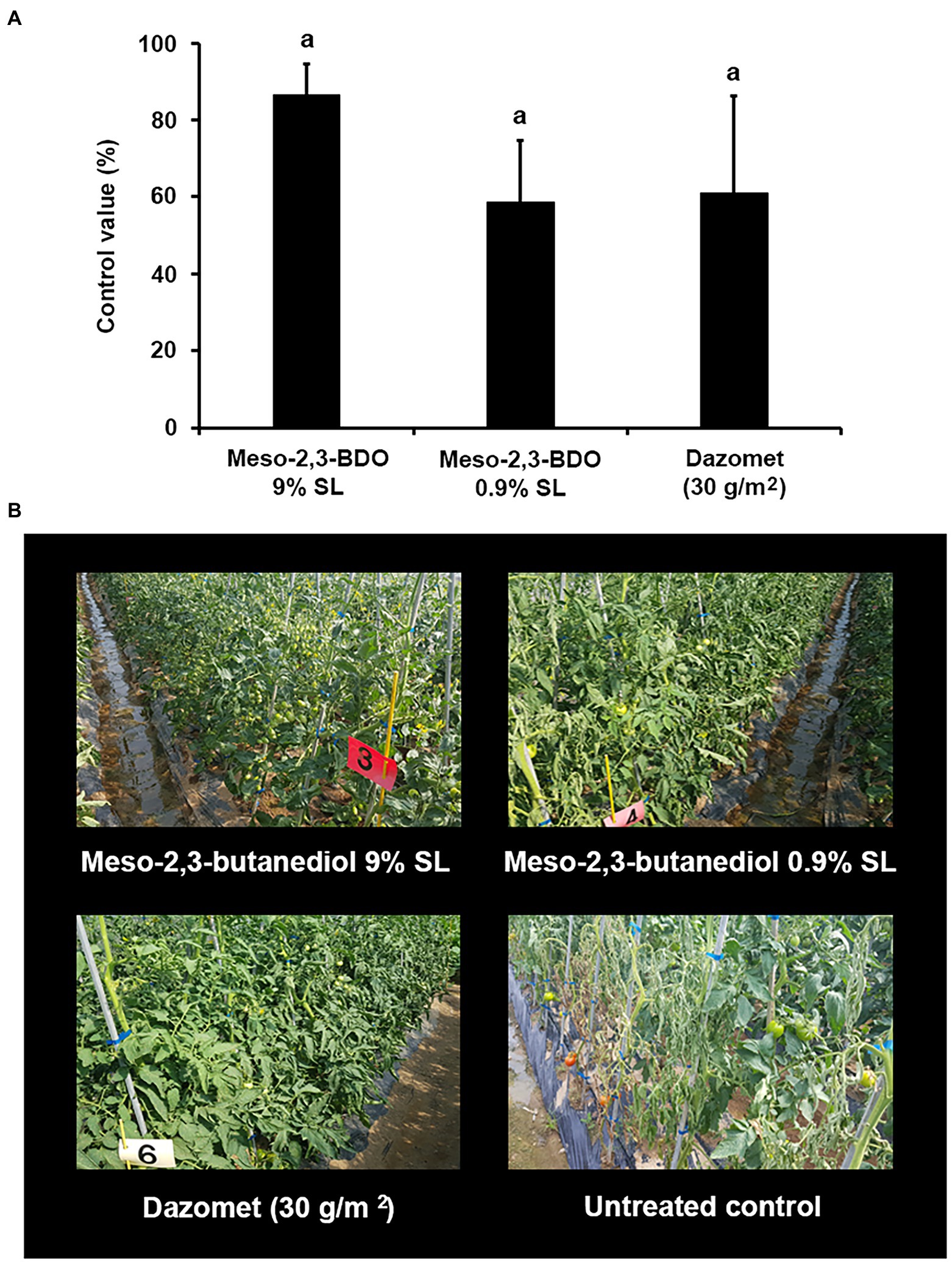
Figure 5. Disease control efficacy of meso-2,3-BDO on tomato bacterial wilt caused by R. solanacearum under the field conditions. 2,3-BDO treatment was applied by preparing meso-2,3-BDO in 9% soluble concentrate (SL) and 0.9% SL. After transplantation, the tomatoes were irrigated four times at 7-day intervals at 200 ml of the 1,000-fold diluent of meso-2,3-BDO 9% SL or meso-2,3-BDO 0.9% SL per plant by soil drench. As a positive control, plants were treated with Dazomet at 300 kg per 1 ha. (A) Control value was investigated 40 days after pathogen inoculation. The data show the means ± SD (n = 4). Lower-case letters indicate statistically significant differences as compared to untreated control plants (p < 0.05) according to Tukey’s HSD test. Error bars indicate the SDs. (B) Representative photographs of tomato plants after naturally occurring tomato bacterial wilt.
To investigate the effect of K. pneumoniae JCK-2201 and B. licheniformis DSM13 producing 2,3-BDO on the induction of plant defense-related genes, tomato leaves were assessed by qRT-PCR. The expression levels of SA-related pathway genes LePR1, LePR2, and LePR5 in the K. pneumoniae JCK-2201 (500-fold dilution)-treated leaves were significantly upregulated by 15.14-, 1.65-, and 19.08-fold, respectively, compared to the control group at 4 DAI (Figures 6A–C). The expression levels of LePR1, LePR2, and LePR5 in the B. licheniformis DSM13 (100-fold dilution) treatment group also increased significantly by 15.96-, 1.27-, and 32.58-fold, respectively (Figures 6A–C). Among the genes involved in the JA-related pathway, LePR3 and PI-II, except for LeLOX, were upregulated significantly in the tomato plants at 1 DAI and gradually increased until 4 DAI (Figures 6D–F). In the K. pneumoniae JCK-2201-treated group, LePR3 and PI-II expression levels were 1.45- and 9.85-fold higher than those in the untreated group and 1.09- and 12.45-fold higher in the B. licheniformis DSM13 group at 4 DAI (Figures 6D,F). Conversely, CAT and ETR4 were not significantly expressed after inoculation (Figures 6G,H).
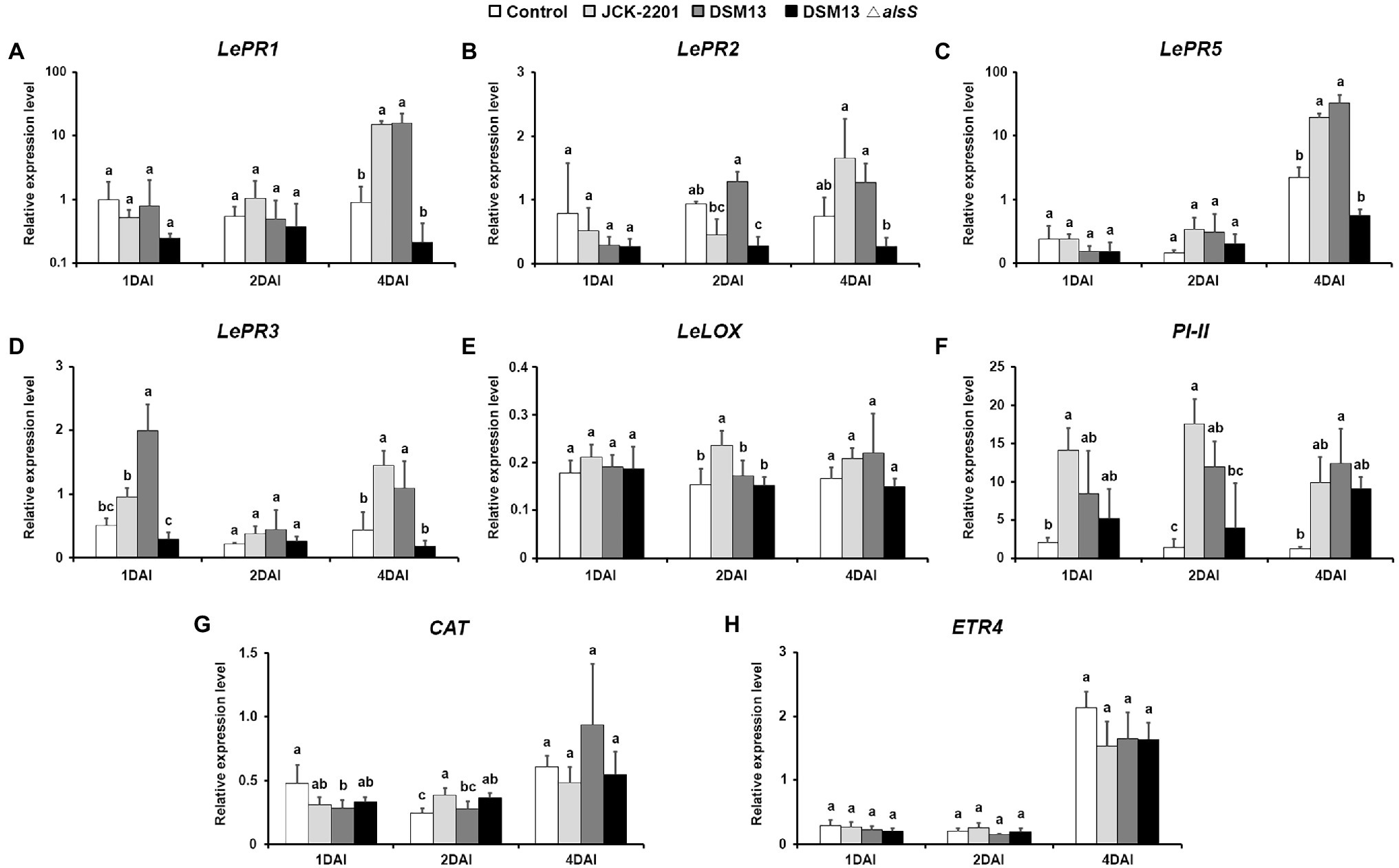
Figure 6. Induction of tomato defense-related gene expression by culture supernatant of K. pneumoniae JCK-2201, B. licheniformis DSM13, and B. licheniformis DSM13ΔalsS at 1, 2, 4 days after inoculation. Expression of defense genes of the SA signaling pathway-related genes (A) LePR1, (B) LePR2, and (C) LePR5, JA signaling pathway-related genes (D) LePR3, (E) LeLOX, (F) PI-II, ROS-related gene (G) CAT, and ET signaling pathway-related gene (H) ETR4 in tomato plants were quantified. The housekeeping gene LeUBI was used for normalization. The data show the means ± SD (n = 3). Lower-case letters indicate statistically significant differences as compared to untreated control plants (p < 0.05) according to Tukey’s HSD test. Error bars indicate the SDs.
Acetoin is a flavor compound present in oils, dairy products, and some fruits. Moreover, it is a physiological metabolite produced by many microorganisms and a precursor to 2,3-BDO (Xu et al., 2011). In this study, we selected acetoin-producing bacteria through the VP test and confirmed the production of acetoin and 2,3-BDO by HPLC analysis. The selected strain K. pneumoniae JCK-2201 produced acetoin and meso-2,3-BDO but showed no in vitro antibacterial activity against R. solanacearum. Some PGPR strains control plant diseases by the induced resistance mechanism without directly affecting the pathogens (Vallad and Goodman, 2004). Park et al. (2007b) reported that Bacillus vallismortis EXTN-1 does not exhibit direct in vitro antibiosis against R. solanacearum but effectively inhibits tomato bacterial wilt by inducing systemic resistance in vivo and significantly hampering the movement of R. solanacearum. Therefore, we predicted that K. pneumoniae JCK-2201 potentially induced resistance against plant diseases. To this end, we performed an in vivo experiment on tomato bacterial wilt and demonstrated that a 100-fold dilution of K. pneumoniae JCK-2201 effectively inhibited tomato bacterial wilt to a control value of 77%.
2,3-BDO is produced by several microorganisms, including K. pneumoniae, B. licheniformis, and Paenibacillus polymyxa (Nilegaonkar et al., 1996; Shrivastav et al., 2013). Generally, K. pneumoniae produces meso-2,3-BDO with a small amount of dextro-2,3-BDO, and B. licheniformis usually produces a mixture of meso-2,3-BDO and levo-2,3-BDO (Chen et al., 2014; Qiu et al., 2016). 2,3-BDO is used as a promising chemical in a wide range of applications owing to its low toxicity and higher yield in microbial systems (Conveerti and Perego, 2002). Klebsiella has the advantage of easy and rapid proliferation in a simple medium. It has also been reported that 1,3-propanediol production is reduced under microaerobic and low pH conditions and sugar is primarily directed for the production of 2,3-BDO (Biebl et al., 1998). Therefore, we conducted an experiment to select the optimal medium to increase the production of 2,3-BDO. When K. pneumoniae JCK-2201 was cultured in flasks for 24 h using 100 g/L sucrose and 20 g/L casamino acid, meso-2,3-BDO was produced at the maximum concentration of approximately 48 g/L. Klebsiella pneumoniae SDM produces a maximum of 36.7 g/L 2,3-BDO in flasks containing optimal medium when incubated at 37°C with constant shaking at 200 rpm for 38 h (Ma et al., 2009). After 10 h of batch fermentation using glucose as the carbon source, the maximum production of 2,3-BDO by K. pneumoniae DSM 2026 and K. oxytoca ATCC 43863 were 17.6 and 10.9 g/L, respectively (Cho et al., 2012). Klebsiella sp. XMR21 produces the highest amount of 2,3-BDO (14.5 g/L) after 48 h when sucrose was used as a substrate among the four carbon sources, viz., glycerol, xylose, glucose, and sucrose (60 g/L; Xin et al., 2015). Thus, K. pneumoniae JCK-2201 produced higher amounts of 2,3-BDO compared to that reported in previous studies. Sucrose as the carbon source can provide a sustainable supply at a lower production cost and is more environment-friendly than glucose (Ji et al., 2011; Zeng and Sabra, 2011).
However, it could not be confirmed whether 2,3-BDO is responsible for the disease control efficacy of K. pneumoniae JCK-2201. Therefore, we prepared a mutant strain in which alsS was deleted using the B. licheniformis DSM13 strain, which produces meso-2,3-BDO, to investigate the role of 2,3-BDO. The fermentation broth diluents of K. pneumoniae JCK-2201 and B. licheniformis DSM13 showed comparable and higher disease control efficacy against tomato bacterial wilt compared to the positive control of Sungbocycline. The two samples also had significantly higher disease control efficacy than the B. licheniformis DSM13 ΔalsS strain. Kang et al. (2021) reported that deletion of alsS from 2,3-BDO-producing Saccharomyces cerevisiae CBMMP1P2 blocked the 2,3-BDO production pathway. Indeed, the B. licheniformis DSM13 ΔalsS strain, which is the knocked out α-acetolactate synthase B. licheniformis DSM13 strain, produced neither acetoin nor 2,3-BDO and showed very low disease control efficacy against tomato bacterial wilt. Volatile metabolites, such as acetoin and 2,3-BDO, induce plant defense mechanisms that help plants overcome bacterial diseases and promote plant growth (Ryu et al., 2004; Magno-Pérez-Bryan et al., 2015). These reports confirm that the disease control efficacy of K. pneumoniae JCK-2201 and B. licheniformis DSM13 was due to induced resistance by 2,3-BDO. Because K. pneumoniae JCK-2201 and B. licheniformis DSM13 produced small amounts of levo-2,3-BDO and dextro-2,3-BDO rather than meso-2,3-BDO, we expect that the effects of K. pneumoniae JCK-2201 and B. licheniformis DSM13 on tomato bacterial wilt is primarily related to meso-2,3-BDO. Meso-2,3-BDO production at 500-fold dilution of K. pneumoniae JCK-2201 and 100-fold dilution of B. licheniformis DSM13 was approximately 1 mM. In this study, when meso-2,3-BDO was applied at 1 and 0.1 mM, its control values against tomato bacterial wilt were 54 and 31%, respectively. Levo-2,3-BDO treatment reduces diseased leaf area of Agrostis stolonifera by 20%–40% against Microdochium nivale, Rhizoctonia solani, or Sclerotinia homoeocarpa (Cortes-Barco et al., 2010). Meso-2,3-BDO treatment also significantly reduced the incidence of naturally occurring cucumber mosaic virus and tobacco mosaic virus compared to untreated controls in a field experiment (Kong et al., 2018). Although the effect of 2,3-BDO on various plant diseases has been reported, its effect on tomato bacterial wilt has not been investigated yet under field conditions. Therefore, we evaluated the disease control efficacy of meso-2,3-BDO against tomato bacterial wilt under field conditions. Meso-2,3-BDO (1 mM) effectively inhibited the development of tomato bacterial wilt at a control value of 87%, with a higher efficacy than dazomet (used as the positive control). This effect of 2,3-BDO treatment is not direct and may be due to the induced resistance effect by the expression of plant defense marker genes in the SA, JA, and ET signaling pathways (Kong et al., 2018). Therefore, we investigated the action mechanism of these genes.
It has been reported that the VOCs from the PGPR strain P. polymyxa E681 induce ISR via SA and/or JA signaling pathways against P. syringae by priming the defense genes PR1, ChiB, and VSP2 (Lee et al., 2012). In our experiment, LePR1, LePR2, and LePR5 were induced by both K. pneumoniae JCK-2201 and B. licheniformis DSM13 treatments than by B. licheniformis DSM13 ΔalsS and untreated control at 4 DAI. The results suggest that meso-2,3-BDO produced by K. pneumoniae JCK-2201 and B. licheniformis DSM13 activated the SA signaling pathway in tomato after pathogen infection. Similarly, 1 mM 2,3-BDO treatment induced resistance against R. solanacearum at 3 DAI, as evidenced by the expression of SA signaling-related PR genes, such as CaPR2, CaSAR8.2, and CaPAL (Yi et al., 2016). Moreover, our results showed that K. pneumoniae JCK-2201 and B. licheniformis DSM13 treatment induced the expression of JA pathway-related genes, such as LePR3 and PI-II, than that by B. licheniformis DSM13 ΔalsS and untreated control at 1 DAI. However, LeLOX expression was not statistically significant. These results were consistent with the report by Park et al. (2018) that 2,3-BDO produced by P. polymyxa DSM365 increases the expression levels of PR protein-encoding genes, such as PR1, PR3, PR4-b, PR5, and PR6, in tobacco at 48 h after Phytophthora parasitica var. nicotianae inoculation. Meso-2,3-BDO 9% SL treatment upregulated the expression of the SA-, JA-, and ET-related pathway genes AsNPR1, AsPR4, AsLOX, AsAOS1, and AsERF as well as ROS-related genes AsDHAR, AsCAT, AsMDHAR, AsSOD, AsGr, and AsPOD, resulting in reduced incidence of dollar spot disease in creeping bentgrass (Duraisamy et al., 2022). However, the ROS-related gene CAT and ET pathway-related gene ETR4 showed relatively low expression levels or were not statistically significantly upregulated by meso-2,3-BDO in this study. These differences may be due to the different plants used. Consequently, we demonstrated that meso-2,3-BDO produced by K. pneumoniae JCK-2201 and B. licheniformis DSM13 increased the gene expression of SA- and JA-related pathways after R. solanacearum pathogen inoculation, resulting in the induction of SAR and ISR and activating defense-related systems against pathogens.
These findings indicate that the upregulated expression of PR genes and plant defense-related genes are responsible for the reduced incidence of R. solanacearum. Plant pathogens are divided into biotrophs and necrotrophs according to their life cycle, and hemi-biotrophs act as biotrophs and necrotrophs according to conditions or life cycle stages (Glazebrook, 2005). In general, plant defense responses include SA-dependent defenses acting on biotrophs and JA- and ET-dependent responses acting on necrotrophs; resistance is induced depending on the attacking pathogen (McDowell and Dangl, 2000). However, one recent study confirmed the resistance to Plasmodiophora brassicae in Arabidopsis by ARGAH2 and THI2.1 gene expression, suggesting that JA-mediated defense contributes to resistance to some biotrophic or antibiotic pathogens (Thaler et al., 2004; Lemarie et al., 2015; Ding et al., 2021). Klebsiella pneumoniae JCK-2201 and B. licheniformis DSM13 induced resistance to R. solanacearum by the involvement of both SA and JA signaling pathways and was demonstrated to be related to meso-2,3-BDO produced by K. pneumoniae JCK-2201 and B. licheniformis DSM13. Klebsiella pneumoniae JCK-2201 and meso-2,3-BDO are potential biopesticides for the control of tomato bacterial wilt, and further studies are required on the development of optimum formulation, toxicity test, and evaluation of their disease control efficacy under various field conditions.
In the present study, the PGPR strain K. pneumoniae JCK-2201 effectively suppressed the development of tomato bacterial wilt, and its disease control efficacy was demonstrated to be primarily because of meso-2,3-BDO production in the fermentation broth. This is the first study on the disease control efficacy of meso-2,3-BDO on tomato bacterial wilt under field conditions. The fermentation broth of K. pneumoniae JCK-2201 enhanced the expression of plant defense marker genes in both SA and JA signaling pathways. These results show that 2,3-BDO-producing bacteria and 2,3-BDO are potential biopesticides for controlling tomato bacterial wilt.
The data presented in the study are deposited in the GeneBank repository, accession number OM510396.
BK and J-CK designed this study. BK isolated and identified strain K. pneumoniae JCK-2201, determined the optimal medium conditions for 2,3-BDO production, and performed in vitro and in vivo antibacterial bioassays and field experiments. CS and HS provided 2,3-BDO and produced the B. licheniformis DSM13 mutant. BK and AP performed qRT-PCR analyses. BK, AP, and J-CK wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
CS and HS were employed by the GS Caltex Corporation of South Korea.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.914589/full#supplementary-material
Aimé, S., Alabouvette, C., Steinberg, C., and Olivain, C. (2013). The endophytic strain Fusarium oxysporum Fo47: a good candidate for priming the defense responses in tomato roots. Mol. Plant-Microbe Interact. 26, 918–926. doi: 10.1094/MPMI-12-12-0290-R
Biebl, H., Zeng, A. P., Menzel, K., and Deckwer, W. D. (1998). Fermentation ofo glycerol to 1,3-propanediol and 2,3-butanediol by Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 50, 24–29. doi: 10.1007/s002530051251
Chen, M., Li, Y., Li, S., Tang, L., Zheng, J., and An, Q. (2016). Genomic identification of nitrogen-fixing Klebsiella variicola, K. pneumoniae and K. quasipneumoniae. J. Basic Microbiol. 56, 78–84. doi: 10.1002/jobm.201500415
Chen, C., Wei, D., Shi, J., Wang, M., and Hao, J. (2014). Mechanism of 2, 3-butanediol stereoisomer formation in Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 98, 4603–4613. doi: 10.1007/s00253-014-5526-9
Cho, J. H., Rathnasingh, C., Song, H., Chung, B. W., Lee, H. J., and Seung, D. (2012). Fermentation and evaluation of Klebsiella pneumoniae and K. oxytoca on the production of 2,3-butanediol. Bioprocess Biosyst. Eng. 35, 1081–1088. doi: 10.1007/s00449-012-0691-7
Conveerti, A., and Perego, P. (2002). Use of carbon and energy balances in the study of the anaerobic metabolism of Enterobacter aerogenes at variable starting glucose concentrations. Appl. Microbiol. Biotechnol. 59, 303–309. doi: 10.1007/s00253-002-1009-5
Cortes-Barco, A. M., Hsiang, T., and Goodwin, P. H. (2010). Induced systemic resistance against three foliar diseases of Agrostis stolonifera by (2R, 3R)-butanediol or an isoparaffin mixture. Ann. Appl. Biol. 157, 179–189. doi: 10.1111/j.1744-7348.2010.00417.x|
Denny, T. (2007). “Plant pathogeic Ralstonia species,” in Plant-Associated Bacteria. ed. S. S. Gnanamanickam (Dordrecht: Spirnger).
Dey, S., Dutta, P., and Majumdar, S. (2019). Biological control of Macrophomina phaseolina in Vigna mungo L. by Endophytic Klebsiella pneumoniae HR1. Jordan J. Biol. Sci. 12, 219–227.
Ding, S., Shao, X., Li, J., Ahanmmed, G. J., Yao, Y., Ding, J., et al. (2021). Nitrogen forms and metabolism affect plant defence to foliar and root pathogens in tomato. Plant Cell Environ. 44, 1596–1610. doi: 10.1111/pce.14019|
Duraisamy, K., Ha, A., Kim, J., Park, A. R., Kim, B., Song, C. W., et al. (2022). Enhancement of disease control efficacy of chemical fungicides combined with plant resistance inducer 2,3-butanediol against turfgrass fungal diseases. Plant Pathol. J. 38, 182–193. doi: 10.5423/PPJ.OA.02.2022.0022
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Glick, B. R., Penrose, D. M., and Li, J. (1998). A model for the lowering of plant ethylene concentrations by plant growth promoting bacteria. J. Theor. Biol. 190, 63–68. doi: 10.1006/jtbi.1997.0532
Harden, A., and Walpole, G. S. (1906). 2, 3-Butylene glycol fermentation by Aerobacter aerogenes. In Proc. Royal. Soc. 77, 399–405.
Jetiyanon, K., and Kloepper, J. W. (2002). Mixtures of plant growth promoting rhizobacteria for induction of systemic resistance against multiple plant diseases. J. Biol. Control. 24, 285–291. doi: 10.1016/S1049-9644(02)00022-1
Ji, S. H., Gururani, M. A., and Chun, S. (2014). Isolation and characterization of plant growth promoting endophytic diazotrophic bacteria from Korean rice cultivars. Microbiol. Res. 169, 83–98. doi: 10.1016/j.micres.2013.06.003
Ji, X.-J., Huang, H., and Ouyang, P.-K. (2011). Microbial 2,3-butnaediol production: a state-of-the-art review. Biotechnol. Adv. 29, 351–364. doi: 10.1016/j.biotechadv.2011.01.007
Ji, P., Momol, M. T., Olson, S. M., Pradhanang, P. M., and Jones, J. B. (2005). Evaluation of thymol as biofumigant for control of bacterial wilt of tomato under field conditions. Plant Dis. 89, 497–500. doi: 10.1094/PD-89-0497
Jia, C., Zhang, L., Liu, L., Wang, J., Li, C., and Wang, Q. (2013). Multiple phytohormone signalling pathways modulate susceptibility of tomato plants to Alternaria alternata f. sp. lycopersici. J. Exp. Bot. 64, 637–650. doi: 10.1093/jxb/ers360
Kang, N. K., Lee, J. W., Ort, D. R., and Jin, Y. S. (2021). L-malic acid production from xylose by engineered Saccharomyces cerevisiae. Biotechnol. J. 17:e2000431. doi: 10.1002/biot.202000431
Kirankumar, R., Jagadeesh, K. S., Krishnaraj, P. U., and Patil, M. S. (2008). Enhanced growth promotion of tomato and nutrient uptake by plant growth promoting rhizobacterial isolates in presence of tobacco mosaic virus pathogen. Karnataka J. Agric. Sci. 21, 309–311.
Kong, H. G., Shin, T. S., Kim, T. H., and Ryu, C.-M. (2018). Stereoisomers of the bacterial volatile compound 2,3-butanediol differently elicit systemic defense responses of pepper against multiple viruses in the field. Front. Plant Sci. 9:90. doi: 10.3389/fpls.2018.00090
Le, K. D., Kim, J., Yu, N. H., Kim, B., Lee, C. W., and Kim, J.-C. (2020). Biological control of tomato bacterial wilt, kimchi cabbage soft rot, and red pepper bacterial leaf spot using Paenibacillus elgii JCK-5075. Front. Plant Sci. 11:775. doi: 10.3389/fpls.2020.00775
Lee, B., Frarag, M. A., Park, H. B., Kloepper, J. W., Lee, S. H., and Ryu, C.-M. (2012). Induced resistance by a long-chain bacterial volatile: elicitation of plant systemic defense by a C13 volatile produced by Paenibacillus polymyxa. PLoS One 7, 1–11. doi: 10.1371/journal.pone.0048744
Lee, J. H., Jang, K. S., Choi, Y. H., Kim, J.-C., and Choi, G. J. (2015). Development of an efficient screening system for resistance of tomato cultivars to Ralstonia solanacearum. Res. Plant Dis. 21, 290–296. doi: 10.5423/RPD.2015.21.4.290
Lemarie, S., Robert-Seilaniantz, A., Lariagon, C., Lemoine, J., Marnet, N., Jubault, M., et al. (2015). Both the jasmonic acid and the salicylic acid pathways contribute to resistance to the biotrophic clubroot agent Plasmodiophora brassicae in Arabidopsis. Plant Cell Physiol. 56, 2158–2168. doi: 10.1093/pcp/pcv127
Liu, D., Chen, L., Zhu, X., Wang, Y., Xuan, Y., Liu, X., et al. (2018). Klebsiella pneumoniae SnebYK mediates resistance against Heterodera glycines and promotes soybean growth. Front. Microbiol. 9:1134. doi: 10.3389/fmicb.2018.01134
Liu, Y., Wang, H., Sun, X., Yang, H., Wang, Y., and Song, W. (2011). Study on mechanisms of colonization of nitrogen-fixing PGPB, Klebsiella pneumoniae NG14 on the root surface of rice and the formatiion of biofilm. Curr. Microbiol. 62, 1113–1122. doi: 10.1007/s00284-010-9835-7
Ma, C., Wang, A., Qin, J., Li, L., Ai, X., Jiang, T., et al. (2009). Enhanced 2,3-butanediol production by Klebsiella pneumoniae SDM. Appl. Microbiol. Biotechnol. 82, 49–57. doi: 10.1007/s00253-008-1732-7
Magno-Pérez-Bryan, M. C., Martínez-García, P. M., Hierrezuelo, J., Rodríguez-Palenzuela, P., Arrebola, E., Ramos, C., et al. (2015). Comparative genomics within the Bacillus genus reveal the sigularities of two robust Bacillus amyloliquefaciens biocontrol strains. Mol. Plant-Microbe Interact. 28, 1102–1116. doi: 10.1094/MPMI-02-15-0023-R
Mazumdar, D., Saha, S. P., and Ghosh, S. (2018). Klebsiella pneumoniae rs26 as a potent PGPR isolated from chikpea (Cicer arietinum) rhizosphere. J. Pharm. Innov. 7, 56–62.
McDowell, J. M., and Dangl, J. L. (2000). Signal transduction in the plant immune response. Trends Biochem. Sci. 25, 79–82. doi: 10.1016/S0968-0004(99)01532-7
Mhamdi, A., and Van Breusegem, F. (2018). Reactive oxygen species in plant development. Development 145:dev164376. doi: 10.1242/dev.164376
Mohammed, A. F., Oloyede, A. R., and Odeseye, A. O. (2020). Biological control of bacterial wilt of tomato caused by Ralstonia solanacearum using Pseudomonas species isolated from the rhizosphere of tomato plants. Arch. Phytopathol. Plant Protect. 53, 1–16. doi: 10.1080/03235408.2020.1715756
Nilegaonkar, S. S., Bhosale, S. B., Dandage, C. N., and Kapadi, A. H. (1996). Potential of Bacillus licheniformis for the production of 2, 3-butanediol. J. Ferment. Bioeng. 82, 408–410. doi: 10.1016/0922-338X(96)89161-6
Park, S.-W., Kaimoyo, E., Kiumar, D., Mosher, S., and Klessig, D. F. (2007a). Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318, 113–116. doi: 10.1126/science.1147113
Park, K., Paul, D., Kim, Y. K., Nam, K. W., Lee, Y. K., Choi, H. W., et al. (2007b). Induced systemic resistance by Bacillus vllismortis EXTN-1 suppressed bacterial wilt in tomato caused by Ralstonia solanacearum. Plant Pathol. J. 23, 22–25. doi: 10.5423/PPJ.2007.23.1.022
Park, K. Y., Seo, S. Y., Oh, B.-R., Seo, J.-W., and Kim, Y. J. (2018). 2,3-Butanediol induces systemic acquired resistance in the plant immune response. J. Plant Biol. 61, 424–434. doi: 10.1007/s12374-018-0421-z
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C., and Bakker, P. A. H. M. (2004). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Qiu, Y., Zhang, J., Li, L., Wen, Z., Nomura, C. T., Wu, S., et al. (2016). Engineering Bacillus licheniformis for the production of meso-2, 3-butanediol. Biotechnol. Biofuels 9, 1–13. doi: 10.1186/s13068-016-0522-1
Renna, M. C., Najimudin, N., Winik, L. R., and Zahler, S. A. (1993). Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J. Bacteriol. 175, 3863–3875. doi: 10.1128/jb.175.12.3863-3875.1993
Ryu, C.-M., Farag, M. A., Hu, C. H., Reddy, M. S., Kloepper, J. W., and Paré, P. W. (2004). Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 134, 1017–1026. doi: 10.1104/pp.103.026583
Sebastià, P., De Pedro Jové, R., Daubech, B., Kashyap, A., Coll, N. S., and Valls, M. (2021). The bacterial wilt reservoir host Solanum dulcamara shows resistance to Ralstonia solanacearum infection. Front. Plant Sci. 12:755708. doi: 10.3389/fpls.2021.755708
Shaharoona, B., Jamro, G. M., Zahir, Z. A., Arshad, M., and Memon, K. S. (2007). Effectiveness of various Pseudomonas spp. and Burkholderia caryophylli containing ACC-deaminase for imporving growth and yeild of wheat (Triticum aestivum L.). J. Microbiol. Biotechnol. 17, 1300–1307.
Shrivastav, A., Lee, J., Kim, H.-Y., and Kim, Y.-R. (2013). Recent insights in the removal of Klebsiella pathogenicity factors for the industrial production of 2,3-butanediol. J. Microbiol. Biotechnol. 23, 885–896. doi: 10.4014/jmb.1302.02066
Song, C. W., Park, J. M., Chung, S. C., Lee, S. Y., and Song, H. (2019). Micirobial production of 2,3-butanediol for industrial applications. J. Ind. Microbiol. Biotechnol. 46, 1583–1601. doi: 10.1007/s10295-019-02231-0
Song, C. W., Rathnasingh, C., Park, J. M., Kwon, M., and Song, H. (2021). CRISPR-Cas9 mediated engineering of Bacillus licheniformis for industrial production of (2R,3S)-butanediol. Biotechnol. Prog. 37:e3072. doi: 10.1002/btpr.3072
Thaler, J. S., Owen, B., and Higgins, V. J. (2004). The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifecycles. Plant Physiol. 135, 530–538. doi: 10.1104/pp.104.041566
Vallad, G. E., and Goodman, R. M. (2004). Systemic acquired resistance and induced systemic resistance in conbentional agriculture. Crop Sci. 44, 1920–1934. doi: 10.2135/cropsci2004.1920|
Van Lenteren, J. C. (2001). “Harvesting safely from biodiversity: natural enemies as sustainable and environmentally friendly solution for pest control,” in Balancing Nature: Assessing the Impact of Importing Non-native Biological Control Agents (an International Perspective). eds. J. A. Lockwood, F. G. Howarth, and M. F. Purcell (Lanham, MD: Entomological Society of America), 15–30.
Van Loon, L. C., Bakker, P. A. H. M., and Pieterse, C. M. J. (1998). Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36, 453–483. doi: 10.1146/annurev.phyto.36.1.453
Veith, B., Herzberg, C., Steckel, S., Feesche, J., Maurer, K. H., Ehrenreich, P., et al. (2004). The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. Microb. Physiol. 7, 204–211. doi: 10.1159/000079829
Vu, T. T., Kim, H., Tran, V. K., Vu, H. D., Hoang, T. X., Han, J. W., et al. (2017). Antibacterial activity of tannins isolated from Sapium baccatum extract and use for control of tomato bacterial wilt. PLoS One 12:e0181499. doi: 10.1371/journal.pone.0181499
Xin, F., Basu, A., Weng, M. C., Yang, K.-L., and He, J. (2015). Production of 2,3-butanediol from sucrose by a Klebsiella species. Bioenergy Res. 9, 15–22. doi: 10.1007/s12155-015-9653-7
Xu, H., Jia, S., and Liu, J. (2011). Development of a mutant strain of Bacillus subtilis showing enhanced production of acetoin. Afr. J. Biotechnol. 10, 779–788. doi: 10.5897/AJB10.1455
Xue, Q. Y., Ding, G. C., Li, S. M., Yang, Y., Lan, C. Z., Guo, J. H., et al. (2013). Rhizocompetence and antagonistic activity towards genetically diverse Ralstonia solanacearum strains-an improved strategy for selecting biocontrol agents. Appl. Microbiol. Biotechnol. 97, 1361–1371. doi: 10.1007/s00253-012-4021-4
Xue, G. P., Johnson, J. S., and Dalrymple, B. P. (1999). High osmolarity improves the electro-transformation efficiency of the gram-positive bacteria Bacillus subtils and Bacillus licheniformis. J. Microbiol. Methods 34, 183–191. doi: 10.1016/s0167-7012(98)00087-6
Yan, Z., Reddy, M. S., Yyu, J. A., Mclnroy, M., Wilson, M., and Kloepper, J. W. (2002). Induced systemic protection against tomato late blight by plant growth-promoting rhizobacteria. Phytopathology 92, 1329–1333. doi: 10.1094/PHYTO.2002.92.12.1329
Yi, H.-S., Ahn, Y.-R., Song, G. C., Ghim, S.-Y., Lee, S., Lee, G., et al. (2016). Impact of a bacterial volatile 2,3-butanediol on Bacillus subtilis rhizosphere robustness. Front. Microbiol. 7:993. doi: 10.3389/fmicb.2016.00993
Zeng, A. P., and Sabra, W. (2011). Microbial production of diols as platform chemicals: recent progresses. Curr. Opin. Biotechnol. 22, 749–757. doi: 10.1016/j.copbio.2011.05.005
Zhang, M., Zhang, C., Zhang, S., Yu, H., Pan, H., and Zhang, H. (2021). Klebsiella jilinsis 2N3 promotes maize growth and induces resistance to northern corn leaf blight. Biol. Control 156:104554. doi: 10.1016/j.biocontrol.2021.104554
Zhao, D., Zhao, H., Zhao, D., Zhu, X., Wang, Y., Duan, Y., et al. (2018). Isolation and identification of bacteria from rhizosphere soil and their effect on plant growth promotion and root-knot nematode disease. Biol. Control 119, 12–19. doi: 10.1016/j.biocontrol.2018.01.004
Keywords: Klebsiella pneumoniae, tomato bacterial wilt, 2,3-butanediol, induced resistance, qRT-PCR
Citation: Kim B, Park AR, Song CW, Song H and Kim J-C (2022) Biological Control Efficacy and Action Mechanism of Klebsiella pneumoniae JCK-2201 Producing Meso-2,3-Butanediol Against Tomato Bacterial Wilt. Front. Microbiol. 13:914589. doi: 10.3389/fmicb.2022.914589
Received: 07 April 2022; Accepted: 15 June 2022;
Published: 14 July 2022.
Edited by:
Ileana Vigentini, University of Milan, ItalyReviewed by:
Becky Nancy Aloo, University of Eldoret, KenyaCopyright © 2022 Kim, Park, Song, Song and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-Cheol Kim, a2ppbmNAam51LmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.