
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 06 October 2022
Sec. Microbial Symbioses
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.912701
This article is part of the Research Topic Portraying the Phytomicrobiome Studies during Abiotic stresses: Revisiting the past and exploring the future outcomes View all 5 articles
 Kanika Khanna1,2*
Kanika Khanna1,2* Sukhmeen Kaur Kohli1
Sukhmeen Kaur Kohli1 Nandni Sharma3
Nandni Sharma3 Jaspreet Kour1
Jaspreet Kour1 Kamini Devi1
Kamini Devi1 Tamanna Bhardwaj1
Tamanna Bhardwaj1 Shalini Dhiman1
Shalini Dhiman1 Arun Dev Singh1
Arun Dev Singh1 Neerja Sharma1
Neerja Sharma1 Anket Sharma4
Anket Sharma4 Puja Ohri3
Puja Ohri3 Renu Bhardwaj1*
Renu Bhardwaj1* Parvaiz Ahmad5,6*
Parvaiz Ahmad5,6* Pravej Alam7*
Pravej Alam7* Thamer H. Albalawi7
Thamer H. Albalawi7The agricultural sector is a foremost contributing factor in supplying food at the global scale. There are plethora of biotic as well as abiotic stressors that act as major constraints for the agricultural sector in terms of global food demand, quality, and security. Stresses affect rhizosphere and their communities, root growth, plant health, and productivity. They also alter numerous plant physiological and metabolic processes. Moreover, they impact transcriptomic and metabolomic changes, causing alteration in root exudates and affecting microbial communities. Since the evolution of hazardous pesticides and fertilizers, productivity has experienced elevation but at the cost of impeding soil fertility thereby causing environmental pollution. Therefore, it is crucial to develop sustainable and safe means for crop production. The emergence of various pieces of evidence depicting the alterations and abundance of microbes under stressed conditions proved to be beneficial and outstanding for maintaining plant legacy and stimulating their survival. Beneficial microbes offer a great potential for plant growth during stresses in an economical manner. Moreover, they promote plant growth with regulating phytohormones, nutrient acquisition, siderophore synthesis, and induce antioxidant system. Besides, acquired or induced systemic resistance also counteracts biotic stresses. The phytomicrobiome exploration is crucial to determine the growth-promoting traits, colonization, and protection of plants from adversities caused by stresses. Further, the intercommunications among rhizosphere through a direct/indirect manner facilitate growth and form complex network. The phytomicrobiome communications are essential for promoting sustainable agriculture where microbes act as ecological engineers for environment. In this review, we have reviewed our building knowledge about the role of microbes in plant defense and stress-mediated alterations within the phytomicrobiomes. We have depicted the defense biome concept that infers the design of phytomicrobiome communities and their fundamental knowledge about plant-microbe interactions for developing plant probiotics.
A glaring overhaul has been observed in the 21st century in regard to climate change where environmental stresses have caused global threat toward food safety and security. The world population has been projected to be 9 billion (approx.) by 2050, and environmental disturbances cause reduced crop productivities and soil fertility thereby impeding agricultural sustainability (Muller et al., 2017; Chouhan et al., 2021). Consequently, it will likely enhance more than 60% of the food demand, specifically for the cereal crops (wheat, maize, and rice), leguminous crops, pulses (peas, lentils, and soybeans), etc., to feed the massive population (Majeed et al., 2017). The yields of these crops are often challenged with the poor soil and biotic and abiotic stresses (Suzuki et al., 2014; Kumar et al., 2021). Stresses cause a plethora of adverse effects onto plants in terms of morphology; biochemistry; and metabolic, physiological, and molecular reactions. To meet the ongoing demands of food, traditional agricultural practices are exclusively used in which the use of pesticides and fertilizers is most common, but they lead to polluted and contaminated ecosystem. Henceforth, in order to safeguard the agricultural sustainability and boost the crop yields for posterity, we need to adopt novel and environmental friendly methods (Majeed et al., 2017; Chouhan et al., 2021).
Since decades, enormous studies have depicted the sky-scarping diversity of microbiome co-linked to agricultural plants (Lemanceau et al., 2017). Microbiome holobiont refers to huge microflora associated with plants, and plant microbiome involves the microbial genomes in regard to host plants (Brader et al., 2017; Bhatt et al., 2020). Phytomicrobiome is found in various niches in the form of phyllosphere, endosphere, and rhizosphere in order to promote plant growth along with soil fertility (Vorholt, 2012; Etesami and Adl, 2020). Plant microbiome has opened the avenues for the usage of microbes as biofertilizers and biopesticides, and public interest has been considerably enhanced to use this biological resource as alternatives for external inputs toward agriculture. For, this phytomicrobiome interaction enables us to effectively understand the beneficial aspects of microbiome partners to be used as ecological engineers (Glick, 2012). Soil microbiologists have been trying to utilize microbes also called as inoculants for stimulating soil fertility and nutrition. For successful utilization, various trials are first done for regular practice. Phytomicrobiome contains numerous microbes, such as eubacteria, bacteria, fungi, archea, protozoa, virus, etc., that play an imperative role in soil-plant system (Theis et al., 2016).
Phytomicrobiome is the most sustainable and efficient approach that is effective both in terms of productivity and quality (Yadav et al., 2020). Harnessing the biological resources in agriculture improves both productivity as well as social outcomes in a constructive manner. This microbiome technology aids the beneficial plant-linked microbiomes toward enhancing quality and quantity of agriculture with minimal usage of resources along with plummeting environmental stresses (Kour et al., 2020). Microbiomes are basically host-linked microflora that inhabit different tissues onto host surfaces along with inter/intra colonization of host cells. As huge diversity of microbes is linked with plant roots, therefore, they can be used to alter the host metabolism, physiology, and defense systems. This, in turn, substantially induces the growth, yield, vigor, and efficiency of the plants for nutrient acquisition and stress tolerance (Kour et al., 2020). The mutualistic associations among plants and microbes benefit both the partners and microbiomes that help in shaping the host phenotype and act as a shield among host genotypes and environment (Rana et al., 2020). There are abundant of facts that support the plant microbiome and further revolutionize the agriculture sector. To elucidate, the integration of crops with effective management practices for enhancing productivities and the power of eradicating different pests and pathogens through microbes serve to be one of the positive aspect. Most importantly, this minimizes the chemical fertilizer usage and improves crop fitness with minimal interference of chemical stimulants (Kumar et al., 2019).
Rhizosphere, a niche of microbes surrounding plant roots stimulates a plethora of signaling processes of plants along with the exchange of different materials across the plant-microbial interface (Qiao et al., 2017). Among this zone, plant growth-promoting microbes hold the crucial rank in natural ecosystem and in the agriculture sector as it enables to boost the plant growth via nitrogen fixation, mineral acquisition, inhibiting pathogenic organisms and modulating plant defense responses toward stresses (Shameer and Prasad, 2018). There are a plethora of reports that have outlined the direct and indirect mechanisms of microbes in promoting crop yields with enhancing soil nutrition, nutrient uptake, and enhancing overall activities of plants (Qiao et al., 2017; Kumar et al., 2019). Different microflora have been found to improve soil quality, remediate heavy metal-contaminated soil, suppress pathogenic organisms through secreting antagonistic metabolites with enhanced immunity toward pathogens (Berendsen et al., 2018; Shameer and Prasad, 2018). More to the discussion, these beneficial microbes have attained a great reputation in managing various abiotic and biotic stresses. Henceforth, considering their overall beneficial aspects, they have been deployed as alternatives for yield enhancement and used as biofertilizers and inoculants in the fields for elevating the soil fertility and crop production. Keeping in view the prevailing situations, this review has been designed to discuss all the aspects in regard to phytomicrobiome. We have aimed toward exploring each and every facet of microbiomes and its predominant role against stresses. It is quite evident that the agricultural sector is facing the serious challenge in terms of yields and productivities, and, for maintaining its quality and quantity, this has been found to be the most economical and eco-friendly approach. Thus, global urge for a sustainable approach to meet the food security worldwide has led to develop amended and novel sustainable agricultural patterns.
Under natural environmental conditions, plants are closely associated with a plethora of well- orchestrated and complex microbial community in the phytomicrobiome or the plant microbiome. These intimate associations generally involve the rhizosphere region, endosphere region, pollens, and nectar and leaf surfaces of plants, and are collectively termed as phytomicrobiomes. Under altering environments, these associations do not remain static in the phytomicrobiome, however, undergoes abrupt changes with respect to its structural composition as well as the functional activities of its host partners. (Meena et al., 2017; Li et al., 2018). Recent literature has inferred that these continuous oscillations in the microbiomes are not passively controlled by the host plants only, but it is an outcome of millions of years of co-evolution between the microbes and the host partners. This co-evolution likely triggers the plants to initiate active cooperation with these microbes under stressful environmental conditions (Barnawal et al., 2017; Xu et al., 2019; Morcillo and Manzanera, 2021). Plants, in particular, undergo an unusual phenomenon “cry for help” strategy, generally after experiencing a kind of abiotic or biotic stress. Furthermore, they are deployed by a plethora of microbial species through specific signaling molecules to amplify their potential to counter these stresses (Bhat et al., 2020). In general, plant and microbes interactions are critically important for the plant growth, nutrient acquisition, and shielding of multiple stressors (Kosová et al., 2018).
The plant-microbe interface generally includes the key hot spots for the plant microbe interactions, likely to be in the root region and rhizosphere. Plants release specific root exudates to attract a diverse group of microbes under stress conditions, and these exudates account for the 11 to 40% of the photosynthetically prepared organic carbon (Pandey et al., 2017; Ismail et al., 2021). They found that sugar beet roots under pathogen ‘Rhizoctonia saloni’ exposure attract Flavobacterium and Chitinophage inside the endosperm to counteract these fungal pathogens. On the other hand, Ismail et al. (2021) found that leguminous plants intensify flavonoids secretions under nitrogen-limiting conditions to attract diverse groups of nitrogen-fixing microbes. While, the maize crops secrete certain phenolic compounds like benzoxazinoides to counteract the stresses, as a defense mechanism (Singhal et al., 2016; Wang et al., 2016). Biotic and abiotic stressors exhibit extreme losses in growth and productivity of food crops and thereby causing constraints toward global food security. Sivasakthi et al. (2014) reported that there is annual estimated loss in the yield of food crops of approximately USD$220 billion. However, it has also been reported that chemical control methods are not found to be environmentally friendly and economic; thus, biological tools have been recommended over the chemical control agents (Farooq et al., 2009). To achieve this, microbiome engineering is considered to be a sustainable and effective approach to minimize the yield losses and to improve overall productivity. Pandey et al. (2017) found that plant stress-related amino acids 1-aminocyclopropane-1-carboxylate (ACC) is capable of reshaping the soil microbiome composition under salinity stress and triggers the stress responses. Likewise, rhizosphere exhibits a plethora of signaling molecules contributed by plants and microbial partners that possess potential to counteract multiple stresses, therefore needed to be explored.
Rhizosphere is among the primary sites where microbial communities interact with their plant partners. Both the pathogenic and beneficial microbial communities reside in the rhizosphere region. Also their composition varies in response to the variation in the soil composition and its chemical properties (Pérez-de-Luque et al., 2017). Rhizobacterial partners in the rhizosphere trigger multiple responses, i.e., improve the soil nutrients, stabilize soil health, root growth, and further promote the remediation responses (Badri et al., 2009). This alliance and cooperation among the rhizobacteria and roots within the rhizosphere promotes the growth and proliferation of plant root system and improves the nutrient uptake toward the shoots (Traxler and Kolter, 2015; Zhang et al., 2017). However, microbial populations are found to enhance the crop yields and are used as alternatives to the chemical fertilizers in the field.
The rhizobacterial-rhizosphere interactions maintain the levels of root exudates secretions, which are required to attract the microbial populations (el Zahar Haichar et al., 2014; Semchenko et al., 2014). Root exudates act as mediators between the plant-microbe interaction and further impact roots colonization and root growth. These root exudates contain a wide range of organic compounds, i.e., peptides, sugars, vitamins, amino acids, enzymes, and a series of primary and secondary metabolites (Basiliko et al., 2012; Korenblum et al., 2020). Thus, the microbial populations correspond to a variety of root exudates constituents in the rhizosphere region, thus support the growth of beneficial microbial populations and improve the yield and biomass of different crop plants. Whereas, some root exudates inhibit the growth of harmful microbial flora and provide defensive action against them (Berlanas et al., 2019; Raklami et al., 2019). Baysal et al. (2013) assessed through proteomic studies that elimination of soil-borne pathogens occurred by the introduction of Bacillus species in the rhizosphere region. Bona et al. (2019) also carried out his studies to confirm the activities of different microbes in the rhizosphere region by applying the metaproteome approaches. In this approach, they conducted proteomic studies of the rhizosphere of Vitis vinifera and observed that the bacterial species associated with different groups like Pseudomonas, Streptomyces, Bradyrhizobium, Bacillus, and Bulkhorderia genus are found to have high-protein expressions and thus induce multiple regulatory processes in the rhizosphere region.
Strigolactone, a root exudate as well as a novel phytohormone, is found to induce primary root length as well as the root hair elongation in the rhizosphere region. They are released as root exudates in the rhizosphere via a plethora of dicot and monocot plants, where they mediate mutualistic associations between the root system and the arbuscular mycorrhizal fungi (De Cuyper et al., 2015). Also they participate in the symbiosis between the rhizobia and the leguminous crops (Yang et al., 2019). Among these root secretions, organic acids are found to have high-energy compounds for the microbial partners as well as mediate the regulation of different bio-geochemical cyclic secretions in the rhizosphere region (Wu et al., 2013). However, the low carbon compounds in the root exudates intricate the biosynthesis of rhizobacterial-associated phytohormones, whereas the tryptophan exudates act as a precursor for indole-3-acetic acid (IAA) biosynthesis (Li et al., 2018). Also, ACC, an ethylene precursor, is used as carbon and nitrogen sources for the microbial partners (Xiang et al., 2019). On the other hand, leguminous plants provide flavonoids as root exudates, which further mediate the transcriptional regulation of rhizobia Nod factors (NFs). Furthermore, these Nod factors participate in the nodulation initiation as well as the root hair initiation process (Figure 1).
These root-rhizobacterial associations in the rhizosphere region influence the defense-related mechanisms in plants under different environmental stresses. Rhizobacterial partners influence the regulation of stress-responsive genes, i.e., heat shock proteins, ascorbate peroxidase, and S-adenosyl-methionine synthetase (Gontia-Mishra et al., 2016). They also provide resistance against root herbivores and other diseases in the rhizosphere region (Maheshwari et al., 2015). These associations or alliance between the plants and the microbial partners assist the hosts in nutrient acquisition, growth promotion, and scaling up the yields (Rahimi et al., 2020). These intricate associations are of much importance for both plants and soil health (Wang et al., 2018; Anbi et al., 2020; Etesami and Adl, 2020).
Microbes have the potential to effectively undergo colonization of the roots in order to stimulate their growth through direct and indirect methods. The modulation in the root architecture via root-kinked microbes involves the phytohormonal assembly, such as auxins, cytokinins, ethylene, and other signaling molecules that cause lateral root branching and root hair formation. After the microbial proximity toward plant roots, they tend to convert the exudates into plant hormones (Arora et al., 2020; Khan et al., 2020). There occurs the modulation in the root exudation process with the different developmental stages of the plants; thereby, rhizo-microbiome alignment varies accordingly (ALKahtani et al., 2020). They also possess antagonistic properties toward pathogenic organisms through releasing certain enzymes, antibiotics, siderophores, and crucial nutrients for improving the root architecture (Hartman and Tringe, 2019).
Plants face several stresses throughout their life time that hamper their physiological and metabolic activities, therefore need to be managed for their growth and survival. These stresses are directly related to microbes that offer them to understand and alter certain mechanisms to counteract such adversities. Many stress resistance mechanisms are related to microbes that aid modulation in the metabolic network of plants and help in the activation of stress-related metabolites and genes, respectively (de la Fuente Cantó et al., 2020). There is huge popularity of microbes synthesizing stress-related metabolites in current times. In addition, they also alter transcriptional machinery for stress resistance in plants. The upregulation of the ABA-signaling cascade also modulates the expression of WRKY and MYB during stresses (Gamez et al., 2019). A wide range of genetic alterations has been observed in plants in response to microbe-mediated alterations of their metabolic pools (Kousar et al., 2020). A myriad of secondary metabolites, namely, compatible solutes and volatile compounds have been found to be released within the rhizosphere that forms the basis for the microbial-mediated interactions. Different microbes possess traits for stress tolerance, osmoregulation at cellular levels, ionic homeostasis, reducing toxic ions (Kumar and Verma, 2018). Alongside, they also release various volatile compounds or anti-pathogenic metabolites, such as amino acids, sugars, polyols, betaines, etc., in order to tolerate harsh conditions (Kumar and Verma, 2018).
Plant growth is considerably stimulated through the application of plant growth-promoting microbes as well as mycorrhizal fungi. There are different mechanisms (direct or indirect) that enhance the beneficial traits in plants during stresses. Various biochemical and molecular pathways are adopted by microbes for maintaining growth through hormonal regulation and nutrient acquisition, thereby enhancing the plant growth regulators for resistance against adverse conditions (Spence and Bais, 2015). As depicted earlier, microbes release an array of metabolites to reduce the incidence of pathogen attack and also facilitate certain metabolic processes, such as phosphate solubilization, nitrogen fixation, hormonal signaling, exopolysaccharide production, etc., to avoid the undesirable events (Złoch et al., 2016). Along with this, they possess the ability to synthesize crucial enzymes, namely, glucanases, ACC-deaminases, lytic enzymes, and chitinases, to counteract negative situations. Due to these factors, they may act as potent fertilizers as well as pesticides in agricultural practices. Followed by this, this may also improve the overall nutritional quality as well as yield, thereby prove to be a boon to farmers in terms of increasing the financial income and promoting organic and sustainable agriculture.
Soil is a complex microbial biome whose community structure, composition, and diversity are mainly influenced by a wild range of physical, chemical, and biological variables of soil. Each gram of soil consisted of around 109–1010 microbial cells with an estimated 104–106 species (Vieira and Nahas, 2005; Roesch et al., 2008). Soil structure formation depends upon the shape and composition of a slit, sand, and clay particle aggregates that further form macro/micropores, giving rise to soil ecosystem (Figure 2; Vadakattu and Germida, 2014). Thus, these mineral particles aggregates can form different microhabitats through migration of smaller bacteria inside them, whereas fungal hyphae migrate within macropores (Kim and Crowley, 2013). Additionally, a molecular investigation on soil clay particles aggregates by Kim and Crowley (2013) was done through the 16S ribosomal gene. They found that the area with more clay particles retains more moisture and possesses high bacterial population. However, more archae bacteria are drawn toward sand particles, which have a lower water retention capability (Figure 2). Between these soil particles, microorganisms form a cluster to secrete polysaccharides responsible for fixing colonies and generating a cementing material between them. They also contribute to building up organic matter in these areas (Pereira et al., 2021). Moreover, the fungal hyphae operate mechanically to connect the soil particles. Apart from the plant–microbe interactions within the rhizospheric zone, different microorganisms interact with one another in these soil aggregates, allowing the evolution of microbial population in this biosphere (Bhat et al., 2020).
Most of the nutrients present inside the soil are not accessible to plants. However, phytomicrobiomes presence inside the soil enhanced the accessibility of these nutrients to plants (Table 1). Many bacterial species like Bacillus, Pseudomonas, etc., and fungal species like Aspergillus and Penicillium lead to the production of organic acids and enzymes that solubilize phosphate, thus making it available to the plants (Sańchez et al., 2017). Additionally, these phytomicrobiomes immobilize phosphates inside the organic matter and serve as nutrient storage with quick release in the dissolved form for the plant (Braos, 2015). Some bacterial genera like Azospirillum, Rhizobium, as well as a few cyanobacteria, fix atmospheric nitrogen and convert this element into an accessible form for plants (Shridhar, 2012). These microorganisms, especially Rhizobium, are used as a bio-fertilizer and have been popular in soybean production, providing a cost-effective and environmentally friendly way to increase nutrient supply inside the soil ecosystem (Shridhar, 2012; Philippot, 2013). Other tropical diazotrophic bacteria, such as Azospirillum, have been investigated in Brazil for their practical application in corn, wheat, sugarcane, and rice, as well as used as co-adjuvants in soya beans, which have shown favorable positive outcomes (Hungria et al., 2010; Fernandes, 2014).
When all the soil components are balanced properly, such as C sources availability, aeration, oxygenation, acidity, alkalinity, and inorganic nutrients like N, P, and S, then, it causes the expansion of the phytomicrobiome variability and population and thus resulting in healthy and fertile soil (Johns, 2017; Wang et al., 2018). Therefore, the presence of phytomicrobiome in the form of archaebacteria, bacteria, and fungi inside the soil system causes the enhancement of soil fertility and soil health. Subsequently, it creates a healthy relationship between the soil-plants system, forming a suitable agricultural management strategy with more sustainability and eco-friendly nature (Pereira et al., 2021).
Abiotic stresses play a major role in dwindling the agricultural productivity and yield. High light intensity, heavy metals, heat, chilling, drought, cold, salinity, nutrient, and ozone are the few abiotic stresses that affect the plants in different aspects (Suzuki et al., 2014). They are responsible for weakening the plant defense mechanism and make it more susceptible for other stresses (Mittler and Blumwald, 2010). Moreover, they impair growth, development, yields, and nutrient uptake in plants during stresses. And these characteristics, such as soil quality, nutritional status, and various physicochemical activities, play a major role for plant growth and development. The adverse effects due to stresses are faced by various agricultural crops globally that decline their overall productivity and yield output. The management strategies adopted against such constraints do not always follow the target-oriented criteria, yet the soil fertility gets affected due to excessive usage of fertilizers as well as pesticides that, no doubt, enhance the yield of the crop but also hinder environment and ecosystem, thereby worsening the living populations. The microbes, however, can be manipulated in agro-farming for best management of abiotic stresses along with stimulating crop yields.
In order to combat these stressful conditions, plant-microbe interactions serve to be one of the most effective mechanism to combat the adverse conditions. Rhizosphere is comprised of numerous microbes that mediate plant growth and are ecologically significant and exist either in free or symbiotic form. Many microbes, endophytic or free-living, stimulate plant growth in a direct or indirect manner (Shameer and Prasad, 2018). The microorganisms present in the rhizosphere help in increasing the plant yield by improving the soil health and root growth (Asati et al., 2016). Several microbial strains, such as Frankia sp., Bacillus sp., Azotobacter sp., Pseudomonas sp., Burkholderia sp., Enterobacter sp., Bradyrhizobium sp., Kocuria sp., Flavobacterium sp., Alcaligenes sp., Serratia sp., Klebsiella sp., Microbacterium sp., Agrobacterium sp., Chromobacter sp., and Rhizobium sp., are known for possessing growth-promoting traits (Sarkar et al., 2018; Shah et al., 2021). The plant-microbe associations are highly complex, and the fundamental step of this interaction is due to the secretions released by roots in the form of organic compounds that act as nutrients within the rhizosphere. Plants play a crucial role for assembling and colonizing rhizospheric entities for their own benefits (Zhalnina et al., 2018). Subsequently, the colonized micro-organisms enhance the overall plant metabolic activities during stresses through different mechanisms, namely, soil restoration and reclamation, synthesis of growth-promoting compounds, inhibiting pathogenic organisms, nutrient acquisition, and phosphate/nitrogen solubilization, respectively (Etesami and Adl, 2020; Kour et al., 2020; Liu et al., 2020). The basic mechanism adopted by microbes includes nitrogen fixation, phosphate solubilization, iron sequestration, and production of plant hormones (Chauhan et al., 2021). This is mainly accomplished by enhancing the metabolic pathways of plants during stresses, thereby reducing the adverse effect of stressors from the plants. Moreover, inducing nitrogen fixation, phosphate solubilization from soil is mediated by microbes at an effective rate in order to overcome any metabolic adversity. The nitrogen and phosphate requirement is fulfilled by the plant under stress conditions as mediated by microbes. Moreover, they also trigger iron sequestration so as to balance their levels in the plants. Alongside, the most important aspect is synthesis of phytohormones by microbes that play an important aspect to prevent from stresses. To illustrate, the salicylic acid and jasmonic acid pathway is induced as a defense strategy in plants to cope with the stresses. Also, indole-3-acetic acid, gibberellins, and abscisic acid are released by the microbes in plants to regulate the normal functioning and metabolic processes. Microbes in the rhizosphere of plants cause induction of 1-aminocyclopropane-1-carboxylic acid (ACC)-deaminase activity and systemic resistance against the pathogens, which help the plants to defend the stressful conditions (Benáková et al., 2017). Several reports depicting the role of microbes during abiotic stresses have been tabulated in Table 2.
When plants are subjected to drought stress, it reduces size of the cell, damages membrane integrity, causes senescence of leaf, enhances the production of reactive oxygen species, thereby reduces the plant productivity (Tiwari and Rana, 2015). Studies conducted by Lata and Prasad (2011) found that drought stress impedes several physiological and molecular alterations within plants. It also alters pigment content and, therefore, damages the photosynthetic apparatus. Ethylene production is also increased in plants. Microbes are able to tolerate drought stress along with plant growth and development. During stress, these microbes form a thick wall, accumulate osmolytes synthesized exopolysaccharides or become dormant to combat the stressed conditions (Porcel et al., 2014). Several microbes playing a critical role under drought stress include Azosprillum brasilense, Pseudomonas aeruginosa, Alcaligenes faecalis, Pseudomonas putida, Bacillus thuringeinsis, etc. (Table 1; Naseem and Bano, 2014; Cohen et al., 2015; Ortiz et al., 2015). There are several direct and indirect mechanisms that are followed by microbes that include production of phytohormones as abscisic acid (ABA), Indole-3-acetoc acid (IAA), cytokinins, formation of bacterial exopolysaccharides, induced systemic tolerance, ACC deaminase production (Porcel et al., 2014), which has been shown in Figure 3. Studies conducted by Goswami et al. (2015) found that IAA under drought stress helps in the differentiation of lateral and adventitious roots, promotes root and shoot growth, and undergoes differentiation of vascular tissues. Similar to IAA, ABA also plays a very important role under drought stress through regulation of root hydraulic conductivity and, also, transcription of genes related to drought (Cohen et al., 2015). Bacteria also control the formation of ethylene by hydrolyzing ACC into ammonia and α-ketobutyrate with the help of bacterial ACC-deaminase (Bal et al., 2013). There are some microbes, such as Pseudomonas aeruginosa, Proteus penneri, and Alcaligenes faecalis that help to combat drought stress through the production of exopolysaccharides within maize plants. They help in increasing relative water content, sugars, and proteins through induction in proline content (Naseem and Bano, 2014). Similarly, combination of two microbes Bacillus thuringiensis and Pseudomonas putida assists in decreasing stomatal conductance and leakage of proline within the roots and shoots of plants (Ortiz et al., 2015).
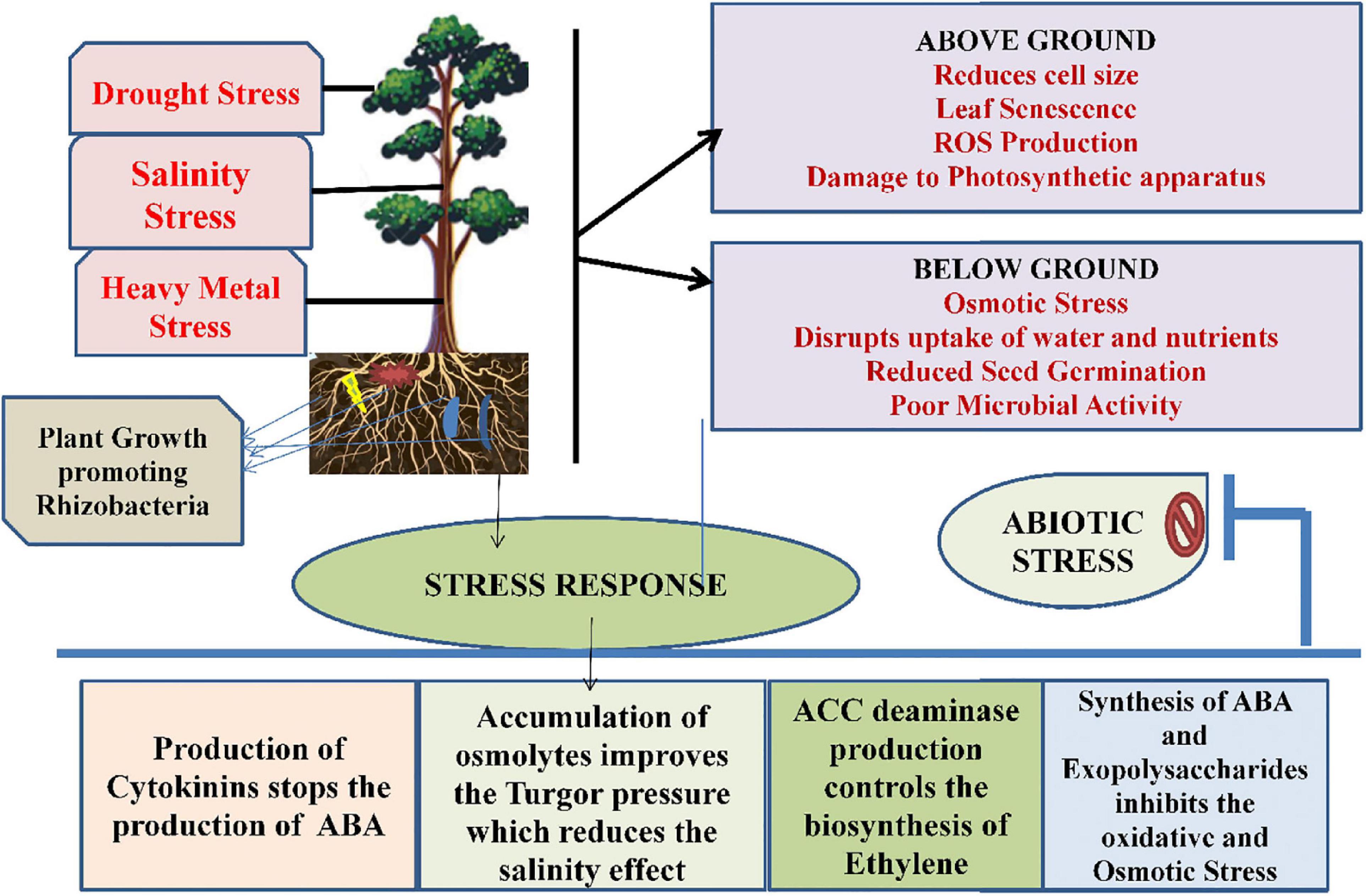
Figure 3. The role of plant growth-promoting rhizobacteria in association with plants under abiotic stress.
Salinity is also considered as an important abiotic stress for modern agriculture. Salinity is responsible for causing low water potential, eventually leading to osmotic stress, and plants are unable to take water and nutrients properly. These salts are present in soil as electrically charged ions (cations as Na+++, Ca2+, Cl–, NO3−, K+) and are generated due to weathering and inadequate rainfall (Shrivastava and Kumar, 2015). Apart from all these consequences, salinity stress also disturbs the ecological and physicochemical balance, affects seed germination, and reduces crop productivity and poor microbial activity as a result of negative effects of ions and osmotic stress (Shrivastava and Kumar, 2015). There are many microbes that are involved in salinity stress tolerance in plants and are tabulated in Table 2. Microbes use both direct and indirect mechanisms for the tolerance in plants. The direct mechanism involves production of phytohormones, nutrient mobilization, nitrogen fixation, siderophore production, and ACC deaminase by rhizobacteria. An enzyme named rhizobitoxine helps in inhibiting the production of ethylene (Hayat et al., 2010; Vardharajula et al., 2011). Reports from Vardharajula et al. (2011) showed that microbes help in salinity stress resistance by the accumulation of osmolytes in the cytoplasm that acts against the osmotic stress regulator, thereby maintaining the turgor pressure of cells and improving the plant growth. Production of exopolysaccharides by microbes also helps in binding onto cations and, therefore, makes them unavailable to plants under stress conditions and facilitates plant growth. The bacterial strains, Bacillus pumilus and Bacillus subtilis, also stimulate defense mechanisms under salinity stress through production of IAA, hydrogen cyanide, ammonia, and phosphate solubilization, respectively (Damodaran et al., 2013).
Advancement in the agricultural practices and industrialization have resulted in heavy metal contamination in soil. Due to increasing heavy metals in soil, the productivity of crops is declining and affecting the growth and development of the plant (Ma et al., 2016). Among all the heavy metals, Pb is widespread and is known to cause root growth inhibition due to impairment of cells near root tips (Zia et al., 2020). Cr toxicity also causes injury to roots and leads to chlorosis of leaves. Similarly, Cd in rhizosphere is also harmful as it causes browning of roots, injuries to shoots and roots, and chlorosis of shoots (Boo et al., 2011; Li et al., 2011). Due to change in climatic conditions, temperature also acts as a stressor for plants in the form of heat and cold conditions. Change in water content, plasma membrane, enzyme functioning, growth of plants, photosynthetic activity, and cell division are some of the toxic effects that are produced by the temperature stress (Alam et al., 2017). Both high- and low- temperature conditions act as stress for plants. In high temperature, the interactions of the roots with microorganisms in the rhizosphere are affected adversely, whereas nitrogen fixation and the nodulation process are adversely affected during low-temperature conditions (Grover et al., 2011). There is a wide variety of microbes that are used for amelioration of heavy metals stresses in plants, such as rhizobacteria and mycorrhizae, and many reports are tabulated in Table 2. The mechanism used by these microbes includes impermeability to metals, the efflux mechanism, metal complexation, EPS sequestration, enzymatic detoxification, and volatilization. Other than these processes, these microbes promote plant growth by producing IAA, ACC deaminase, and reducing ethylene concentration (Glick, 2010). Microbes also cause the bioaccumulation of heavy metals, which also remove heavy metals from soil. Actinobacteria, Proteobacteria, and Firmicutes are reported to remove Mn, Pb, and As from soil (Zhang et al., 2015). The mechanistic approach of PGPRs under heavy metal stress has been shown in Figure 3.
Plant growth, development, and productivity are majorly affected by biotic stress factors like viruses, bacteria, fungi, arachnids, and nematodes. These pathogens disrupt the normal metabolism of plants and impede agricultural production. Globally, the crop yield is decreased by 21–30%, specifically the cereals crop is greatly affected by plant diseases (Savary et al., 2019). On account of climate change, variation in precipitation and increase in atmospheric temperature has led to the generation of new crop pests and diseases (Naamala and Smith, 2020). Pesticides and chemical fertilizers are not only of high cost, but they also degrade the soil quality and persistence in nature and pollute the environment. Also, due to long-term usage of these chemical fertilizers the pathogens-evolved resistance among them, so phytomicrobiome studies form a great sustainable background for the control of plant pests and diseases. Microbes serve as biocontrol agents for plant protection from biotic stress factors and also improve their nutrient uptake, growth, and health (Chouhan et al., 2021). Antagonistic activity of many rhizospheric microbes removes the infection of soil-borne plant pathogens. Plant growth-promoting rhizobacteria synthesize many organic compounds, phytohormones, siderophores, lytic and antioxidant enzymes, and also elevate the level of the stress-responsive genes in host plants for protection against pathogen attack and thus suppress the onset of the disease (Hashem et al., 2017). They produce antibiotics that inhibit the growth of phytopathogens, e.g., an antifungal activity of 2, 4-diacetyl phloroglucinol (2, 4-DAPG) produced by pigment-forming rhizobacteria that suppress the development of many fungal species (Aspergillus niger, A. flavus, and Sclerotium rolfsii) (Chauhan et al., 2021). Various antibiotics, such as pyrrolnitrin, zwittermicin-A phenazine-1-carboxylic acid, pyoluteorin, kanosamine, and pantocin, are produced by some PGPR strains (Bacillus, Azospirillum, Rhizobium, and Pseudomonas species) that enhance the immunity of plants against invaders and induce their mortality (Kenawy et al., 2019). Bacteriocin, a proteinacious toxin produced by many bacterial species, protects plants against phytopathogens. Pyocins produced by Pseudomonas species that are popular bacteriocins showed antagonistic effect against pathogenic strains of Xanthomonas and Pseudomonas, respectively (Fernandez et al., 2020). A gaseous compound HCN has been recognized as a biocontrol agent against plant diseases, which is produced by many free-living microorganisms (Chromobacterium, Bacillus, Burkholderia, Actinomycetes, Pseudomonas, etc.) (Banerjee et al., 2020). It is also reported that HCN enhanced the availability of phosphate indirectly and thus improved the nutrient uptake of plants (Rijavec and Lapanje, 2016). Different types of hydrolytic enzymes, such as proteases, amylases, lipases, cellulases, chitinases, and β-1, 3-glucanases, are produced by many rhizobacterial strains (Streptomyces sp., Paenibacillus terrae, Bacillus, Paenibacillus, and Brevibacillus), which degrade the cell wall and cause cell destruction of phytopathogens (Thampi and Bhai, 2017; Bhadrecha et al., 2020). Various Volatile Organic Compounds (VOCs) are generated by rhizospheric Pseudomonas sp. (L–Ala–L–Ala–L–Ala, 4-hydroxybenzoic acid, phosphonoacetic acid, and octamethylcyclotetrasiloxane), which inhibit the mycelial growth of harmful fungal strains (Kong et al., 2020). They also trigger plants to induce systemic responses against phytopathogens. It is reported that rhizobacterium Bacillus cereus triggers the expression of Induced Systemic Resistance (ISR), inhibiting the infection of Pseudomonas syringae in Arabidopsis (Niu et al., 2016). It is also evaluated that induced systemic response is enhanced by rhizobacteria Pseudomonas taiwanensis in anthurium infected with bacterium Xanthomonas axonopodis pv. dieffenbachiae, which induces polyphenol oxidase activity and synthesizes phenolic compounds at higher amounts (Dhanya et al., 2020). Several reports depicting the role of microbes during biotic stresses have been tabulated in Table 3.
The plant immune system is activated during biotic stress, which triggers the roots to secrete several exudates, such as organic acids, phytohormones, alexins, etc. These exudates act as signaling components and are enhanced in response to various stresses. The complexes like polyphenols, plant hormones (JA, SA, and Ethylene), AHLs (N-acyl-homoserine lactones) are also involved in plants-microbes associations under stress severity (Bhatt et al., 2020). The AHLs molecules are produced by rhizospheric bacteria, which are then used for communication among themselves and, also, for regulation of genes in the signaling cascade. The root secretions attract various microorganisms that further activate the downstream signaling pathway of stress tolerance. Plants recognize rhizospheric microbes through the perception of MAMPs (microbe-associated molecular patterns) signals by PRRs (pattern-recognition receptors) located on plasma membrane. These high-affinity receptors activate MAPKs (mitogen-activated protein kinases) and generate ROS, followed by the activation of the pattern-triggered immunity of plants with enhanced expression of defense-related genes, respectively (PR genes) (Liu et al., 2020; Figure 4).
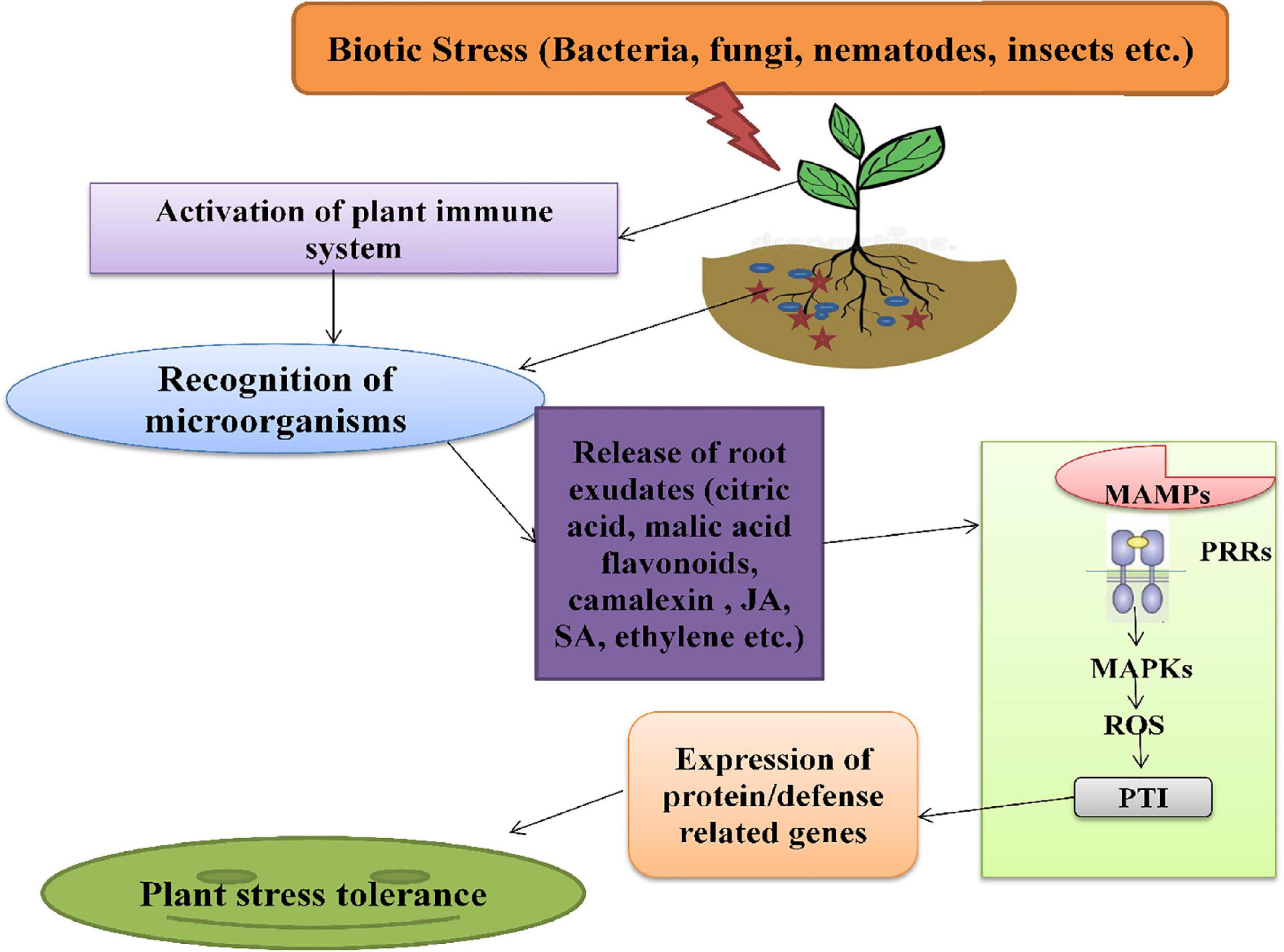
Figure 4. The mechanism of action of phytomicrobiome on plant immune system under biotic stress. MAMPs, microbe-associated molecular patterns; PRRs, pattern-recognition receptor; MAPKs, mitogen-activated protein kinases; ROS, reactive oxygen species; PTI, pattern-triggered immunity.
Until now, the majority of plant microbiome investigations have been found dependent on putative functions obtained from the genomic data, for instance, metagenomics (Liu et al., 2017). However, the recent application of metagenomics in combination with metabolomics and SynComs strategy has demonstrated its efficiency in overcoming the loopholes of plant-microbiome interaction studies (Castrillo et al., 2017; Berendsen et al., 2018; Durán et al., 2018; Kwak et al., 2018; Carrión et al., 2019). Basically, this integrated approach utilizes metagenomics to evaluate the structure and the probable role of plant-consociated microbiological metagenomes, proceeded by the isolation and culturing of fungus and bacteria for SynCom reconstruction (Durán et al., 2018). The SynCom constructed can thus be manipulated by its application to axenic plants (host plant deficient of its usual microbiota) for assessing various factors, for instance, to investigate microbial inter-kingdom interactions and, also, in determining their plant growth-enhancing potential as well as their effects on health and physiology of plants (Berendsen et al., 2018; Durán et al., 2018; Figure 5). Furthermore, by comparing stressed and non-stressed plant microbiomes, stress-induced microbiome alterations can also be observed. Thus, the stress-induced SynComs have been applied to plants in order to determine the biological significance of these modifications (Berendsen et al., 2018; Carrión et al., 2019). Moreover, the biochemical diversity of root exudates can be examined for changes in the presence or quantities of metabolites in order to discover how stress-induced microbiome construction is aided by root exudation. The major metabolic chemicals are isolated and evaluated in vitro for studying their interactions with plant microbial symbionts and pathogens, thus ultimately studying their impact on plant health. However, due to the chemical complexity of varied soil types, root exudation analysis remains an important hurdle in natural settings. A few plant compounds have been discovered in having a positive impact on the rhizosphere microbiota. These compounds, which include flavonoids, peroxidases, oxylipins, coumarins, benzoxazinoids, phenylpropanoids, aromatic chemicals, triterpenes, and mucilage, have all been shown to attract beneficial microorganisms that modulate plant defense system (Lombardi et al., 2018; Schulz-Bohm et al., 2018; Stringlis et al., 2018; Van Deynze et al., 2018; Huang et al., 2019).
Plant-associated microorganisms and metabolites as plant biocontrol agents could be investigated using the methods described above. Furthermore, in recent years, the ability of culturing bacteria for SynComs has substantially improved. As per the reports of Bai et al. (2015), more than 50% of Arabidopsis thaliana-associated bacteria may be isolated and conserved axenically. Isolation can be aided by isolation chips and selective media, such as imipenem-supplemented media, which select for a plant-growth-enhancer bacterium, Stenotrophomonas maltophilia (Bollet et al., 1995; Aleklett et al., 2018). SynComs built with culturable microorganisms provide defined, low-complexity consortia that may be used to investigate plant–microbe interactions in the lab. Due to the considerable intraspecies variability of bacterial genomes and the limited taxonomic precision of amplicon sequencing along with restricted databases, specific bacterial species/strains isolated from metagenomic investigations serve to be a significant problem. Complete-genome metagenomic sequencing with strain-level resolution may be able to reassemble whole bacterial genomes and overcome these limitations, but it comes at a higher cost and necessitates with advanced bioinformatic studies (Ranjan et al., 2016; Kwak et al., 2018; Carrión et al., 2019). Furthermore, when employing omics techniques and the SynComs approach, soil habitat-related characteristics (e.g., heterogeneity and edaphic features) that might significantly influence soil microbial function and composition should be taken into account (Young and Crawford, 2004).
Plant microbiome consists of a colossal range of microorganisms that are associated with various plant parts, be it leaf surfaces, pollens, nectar, and in the rhizosphere (Liu and Brettell, 2019). Plants recruit profitable microbes because of millions of year’s co-evolution. Hence, they offer enhanced approaches to fray both abiotic and biotic stressors and promote plant development by backing nutrient acquisition (Carrión et al., 2019). Root exudates largely influence the composition of microbes (Lombardi et al., 2018). It has been reported in maize that production of benzoxazinoids by roots attracts microbes that assist in strengthening plant defense (Kudjordjie et al., 2019). Likewise, under nitrogen (N)-deficit soils, leguminous plants discharge generous amounts of flavonoids to captivate N-fixing bacteria (Hassan and Mathesius, 2012).
The interactions between plant immune system and microbiome are very complex (Teixeira et al., 2019; Figure 6). Pathogens compete with plant-associated microbes and then interact with plant innate immune system. It includes microbe-associated molecular patterns (MAMPs)-triggered immunity (MTI) and effector-triggered immunity (ETI) (Jones and Dangl, 2006). The nature of interactions between plant-associated commensals and plant immune systems is not definite, but it commands plant-stress interplay. Both pathogens and commensal microbes can trigger MTI and ETI. Their MAMPs are perceived by plant pattern-recognition receptors (PRRs) that reside in the plasma membrane (Teixeira et al., 2019). Nevertheless, commensal microorganisms benefit plants against stress by three means. (i) Lead to production of enzymes that assists in scavenging ROS (Yu et al., 2019) (ii) change MAMP structure that reduces plant immune responses (Teixeira et al., 2019) (iii) possess cell surface molecules that draw out plant defenses, i.e., T3SS and T4SS (Types III and IV secretion systems) (Levy et al., 2018).
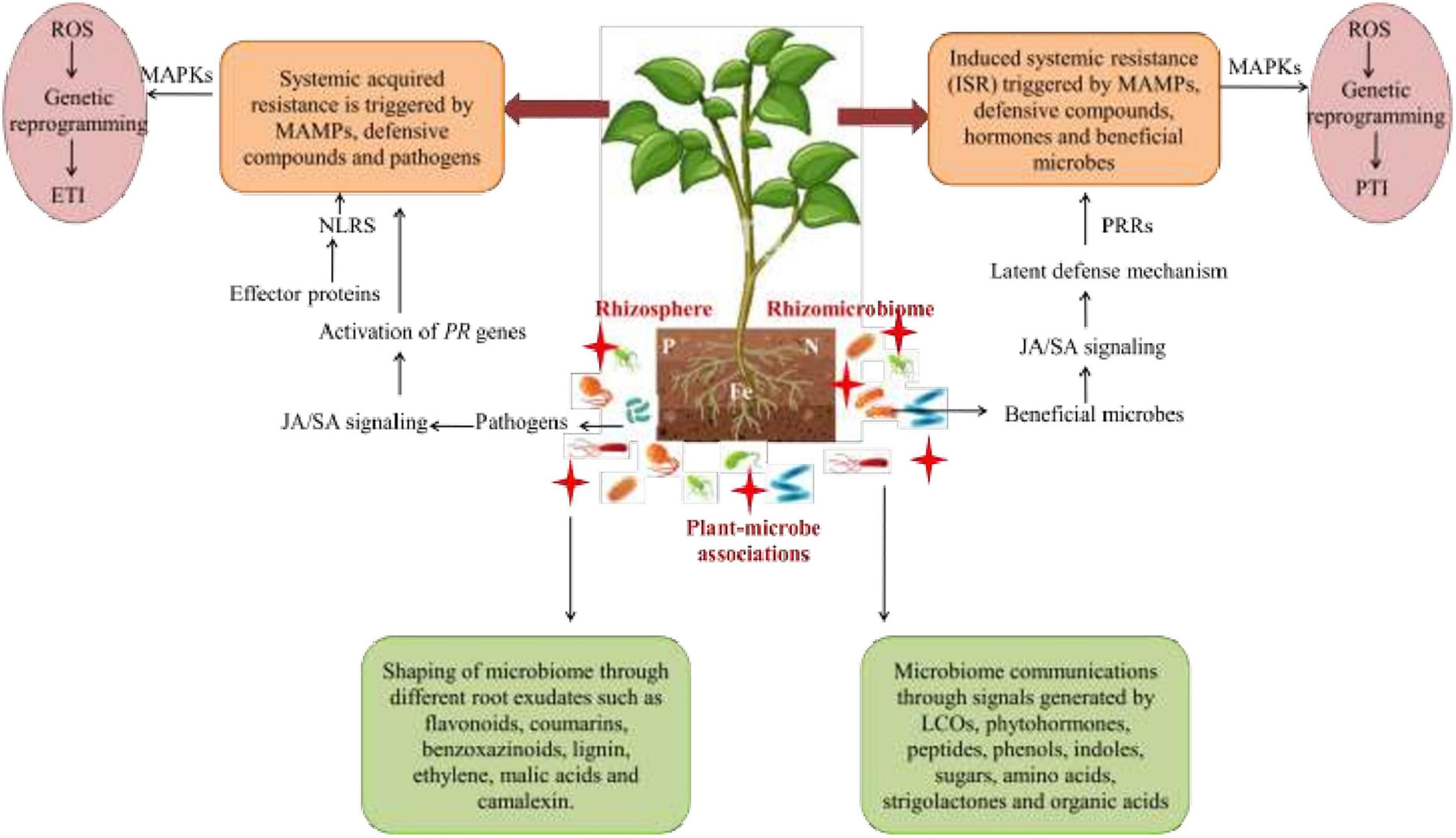
Figure 6. Microbe-mediated signaling mechanisms comprising of systemic acquired resistance (SAR) and induced systemic resistance (ISR) co-linked with plant defense system with the aid of signaling molecules. Beneficial microbes activate ISR that further activates ET, the SA/JA signaling pathway where pathogens stimulate SA levels and trigger SAR. The signaling cascade for immunity against stressors also activates the downstream signaling cascade by which effector proteins are detected by NLRs in order to activate MAPKs that generate ROS through genetic reprogramming for further activation of ETI. Moreover, plants also trigger defense processes to counteract stresses by MAMPs with the aid of PRR receptors in order to activate MAPKs for ROS generation by genetic reprogramming for further activation of PTI. SA, salicylic acid; JA, jasmonic acid; ET, ethylene; PR, pathogenesis; MAMPs, microbe-assisted molecular patterns; NLRs, nucleotide-binding leucine-enriched repeat receptors; MAPKs, mitogen-activated protein kinases; ROS, reactive oxygen species; PRR, pattern-recognition receptors; ETI, effector-triggered immunity; PTI, pattern-triggered immunity.
The microbial communities play an important role to protect plants from insect herbivory and fungal pathogens (Elhady et al., 2018). Other than the normal soils, microbial composition plays an important role in disease suppression as it induces defensive gene expression in plants (Chialva et al., 2018). Many studies clearly validate that there is a connection between root microbiota and plant phenotypes (Zhang et al., 2019). Thus, rice variety having high nitrogen requirements has microbiome of greater nitrogen metabolic capacity than the variety having low nitrogen use (Zhang et al., 2019). Similarly, maize variety adapted to nitrogen-deficit soils has a plethora of diazotrophic bacteria to compensate for the nitrogen (Van Deynze et al., 2018). The microbiota also plays an important role in disease suppression as it is found that tomato variety resistant to Ralstonia solanacearum (causes tomato wilt) has rhizosphere rich in Flavobacterium spp. (Kwak et al., 2018). In Arabidopsis, a phosphate starvation response 1 (PHR1) was found to deal with phosphate stress responses and regulates a set of plant immune system genes, which ultimately lead to changing composition of root microbiome (Castrillo et al., 2017). Hence, ongoing studies are paving ways in understanding the role of microbiome in modulating plant defense, its survival, and growth in the future. Root microbiome has an impact on plant health by turning conductive soils into disease-suppressive soils and contributes to modulating plants’ immune system.
There is a tough competition among microflora populations in soils for available plant-derived nutrients (Raaijmakers et al., 2009). So, in order to infect the host, such soil-borne pathogens are required to maintain an intricately close association with host plants. To do so, they need to grow as saprophytes and in large numbers. Therefore, progress of the pathogen in infectious soils depends on the microbial community. The process of disease elimination in the company of microbial activity is termed as general disease suppression. The process of disease suppression turns highly productive due to organic amendments as it provokes activity of microbes. In order to understand biotic nature of specific disease suppression, the soils are refined (pasteurized) for removal of disease suppressiveness (Mendes et al., 2011).
Soils can maintain their potential of disease suppressiveness for comprehensively long periods even if soils were left bare. Similarly, monoculture of a crop for several years improves soil suppressiveness (Bennett et al., 2012). The soil suppressiveness is developed against multiple crop diseases, such as in damping-off disease of sugar beet caused by Rhizoctonia, wheat diseases caused by Gaeumannomyces graminis var. tritici, Fusarium wilt disease of numerous plant species, and Streptomyces species causing potato scab disease, respectively (Mendes et al., 2011). To confer suppressiveness in conductive soils, microbes are isolated from suppressive soils followed by their inoculation into conductive soils. Raaijmakers et al. (2009) stated the role of microorganisms, such as Proteobacteria and Firmicutes, coupled with fungal members of Ascomycota to confer suppressiveness. The mechanism involved in order to bar soil-borne pathogen is through the production of lytic enzymes and antibiotic compounds (Doornbos et al., 2012).
Many pieces of evidence were reported, stating the role of beneficial soil-borne microorganisms in boosting defensive capacity of aboveground parts of the plants (Zamioudis and Pieterse, 2012). Interestingly, the state of plants for triggered defense response is called induced systemic resistance (ISR) (De Vleesschauwer and Höfte, 2009). Furthermore, Millet et al. (2010) reported that root-associated Rhizobacterium WCS417 elicits production of low molecular weight molecules that subdue flagellin-triggered immune responses (Millet et al., 2010). Credible pieces of evidence state that systemic initiation of the immune signaling cascade aids in delivering resistance against a broad spectrum of pathogens and some insects (Pineda et al., 2010).
Microbial signaling plays an essential role in regulating plant growth, as plants release organic compounds within the rhizosphere in the form of exudates that induce beneficial microbes. These microbes modulate plant behavior by generating inter-organism signaling molecules. The microbes are crucial for signaling, metabolism, and ion and hormonal homeostasis. They release antibiotic-like compounds to inhibit pathogens and induce nutrient acquisition during abiotic/biotic stresses in plants. Moreover, they initiate antibiotic production and elicit defense processes in plants. Signaling is governed by several molecules, such as N-acyl-homoserine lactones and volatile organic compounds, which play critical roles in downstream signaling and modulation of gene activities.
Nevertheless, cell-to-cell communication and signaling in plants take place through pathogen-associated molecular patterns (PAMPs) that enhance immunogenic responses in plants by identifying various chemical cues from microbes, and this immunity is called as PAMP-mediated immunity. In this process, the plants lead to callose deposits within the cell wall and ROS production with the activation of signaling and defense-related genes, such as PR-genes. For example, PR- genes, such as the ethylene-responsive gene (ERF1) and many other jasmonate-responsive genes (LOX2, VSP, PDF, etc.), behave as signaling molecules during stresses (Sharma et al., 2022). Furthermore, series of effector molecules are also released that generate another immune response in the form of effector-triggered immunity. This is an active defense process of plants, also called as gene-for-gene resistance and linked with resistance genes that are activated during stress alleviation. The resistance genes further regulate the expression of PR genes, NO accumulation, SA synthesis, oxidative burst, and programmed cell death, respectively (Wu et al., 2014). Along with this, many molecules are synthesized and transported via plant vascular system in order to behave as a mobile signal for activating a systemic acquired resistance (SAR) mechanism. SAR inhibits many pathogens and leaves prolonged effects through their mobile signals with the ability to activate the transcription of defense-related genes and plant immunity (Enebe and Babalola, 2019). SA has a predominant role in maintaining SAR (Figure 6). During incidence of pathogen infection, ISR is triggered by microbes along with the activation of genes encoding the SA-mediated defense signaling cascade for orchestration of plant defense responses (Ansari, 2018).
The interaction of plants with their proximity occurs through the virtue of some chemical signals that initiate from rhizodeposits, specifically from root exudations and mucilage secretions. And these chemical signals are in the form of primary or secondary metabolites (sugars, organic compounds, proteins, phenols, plant hormones, flavonoids, etc.). Under stresses, plants synthesize chemical cues in excessive amounts, like amino acids and sugars, when released in rhizodeposits and behave as chemo-attractants for beneficial microbes, causing enhancement in the microbial populations within rhizosphere. The mucilage secretions also consist of anti-microbial compounds that prevent the pathogen attack and establish mutual or symbiotic associations among plants and microbes (Basu et al., 2022). The leguminous plants also synthesize flavonoids and isoflavonoids, namely, naringenin and methoxychalcones that act as attractants for rhizospheric communities through regulating nod gene expression. Rhizobia also release lipochitooligosaccharides (LCO) that are mostly recognized by lysine-like receptors, thereby forming a symbiotic pathway to generate the nodulation (Basu and Kumar, 2020). The common symbiotic pathway signals are triggered by specific proteins called as common symbiotic proteins (CSP) that are widely present in nucleoplasm, plasma membrane, and nuclear membrane. Nucleoplasm contains three nucleoporins, cation channels, and core proteins. Others that are localized in cytoplasm are LysM-receptor kinases, leucine-receptor kinases, and NFR1/NFR5 (Basu and Kumar, 2020). The LCO signals generated by rhizobia form a special effect that act as signaling molecules (Clúa et al., 2018). Jasmonic acid also induces the expression levels of genes encoding LCO-biosynthesis, and LCO performs many functions within the plants, such as root nodulation, differentiation, etc. (Smith et al., 2015). In the similar fashion, the isoflavonoids also mediate regulation of nodulation genes (Liu and Murray, 2016). LCO also forms a communication mode during plant-mycorrhizal interactions, for example, strigolactone, a lactone homoserine is involved in plant-mycorrhizal signaling.
Apart from this, microbes also release a plethora of signaling compounds, such as antibiotics and plant hormones. To elucidate, thuricin 17 is a type of bacteriocin released by Bacillus thuringenesis that responds to plant signals and promotes plant growth along with inhibiting pathogenic organisms (Lyu et al., 2020). Moreover, various other compounds, such as lumichrome released by Pseudomonas sp. and Sinorhizobium meliloti and canavanine released by mucilage, are signaling compounds that favor in stress mitigation, root colonization, and symbiosis (Behm et al., 2014). The most probable mechanism by which the signaling molecules get activated is after reception of plant-microbe signaling during stresses, and this is called as positive regulation (Lyu et al., 2020). Contrastingly, the signaling molecules generated without any particular signal are known as negative regulation, and these signals are perceived through external/internal receptors present onto the cell wall. Here, the pattern recognition receptors (PRPs) are able to identify the microbial cell, followed by regulating the function of receptors, and, after activation, they initiate the signaling process to synthesize the crucial molecules (Bhatt et al., 2020).
Plants have developed a unique strategy to confront with stresses by attracting profitable microbes from environment to fight against that stress, which is referred to as Helping hand strategy (Liu and Brettell, 2019). This strategy can pave the way for the survival of future generations as it supports profitable plant-microbe interplay (Yuan et al., 2018). The helping hand strategy provides solution in different stress conditions. To combat pest/pathogen attack, various microbes, such as Chryseobacterium spp., Chitinophaga spp., Flavobacterium spp., and Xanthomonas spp., augment defense mechanisms in rhizosphere. They alleviate plant stress either by suppressing pathogen’s growth (Berendsen et al., 2018) or by activating the plant defense signaling pathway (Chen et al., 2018). There is an increase in plant and microbe symbiosis under phosphate/nitrogen starvation. They accumulate and provide the benefit under stress (Van Deynze et al., 2018). Hence, there exists a co-adaptive strategy between microbes and plants under peculiar stress (Terhorst et al., 2014). Different stressors in plants (metal toxicity, drought, poor nutrition) alter plant root metabolism and its microbial partners. Drought increases Actinobacteria in rhizosphere and/or in endosphere, which aids the plants to retaliate against drought conditions (Xu et al., 2018). Long distance signaling impelled by plant hormones plays a critical role in forming systemic-acquired resistance in plants (Pieterse et al., 2009). Application of hormones affects the microbial congregation in the rhizosphere, and bacterial forms recruited have potential to upgrade the plant defense responses (Lee et al., 2012).
Nevertheless, very less information is available for the impact of root and leaf stresses on phyllosphere microbiomes (Hammerbacher et al., 2019). There are pieces of evidence stating the role of plant volatile emissions in phyllosphere. Although data regarding influence of disease-induced changes on plant microbiome is inadequate but research is still going on for further revelations (Farré-Armengol et al., 2016). Many mechanisms need to be traced that are involved in the ‘Helping hand strategy,’ detecting the metabolites that play a critical role in interacting stressed plants and microbiome. There underlie many mechanisms that increase abundance of these profitable microbes under biotic and abiotic stresses. This phenomenon is termed as a “Defense Biome” mechanism (Figure 7). Under herbivore attack, the leaf and root exudates profile alters, resulting in attracting beneficial bacteria and/or fungi. Similarly, Arabidopsis roots secrete high amounts of citric acid in order to invite favorable bacterium Bacillus subtilis into the rhizosphere (Canarini et al., 2019). Furthermore, these microbes secrete quorum-sensing quenching molecules and antimicrobial compounds to obstruct pathogen growth and virulence (Raaijmakers and Mazzola, 2012). Both bacteria and fungi can have mutualism, where a bacterium supports fungal spore germination and, in turn, fungi provide nutrients and physical support (e.g., biofilms) for bacteria (Hoffman and Arnold, 2010). Likewise, abiotic stress (heat, drought, salinity, and mineral toxicity) led to abundance of plant-associated microbial commensals by (a) altering the root exudates profile and supporting growth of plant-associated microbes (b) changes soil properties, such as pH and nutrients levels (Schimel et al., 2007). Hence, the stress-induced microbial increase (Stenotrophomonas spp., Pseudomonas putida, and Chryseobacterium indologenes) is favorable to plants in enhancing host disease resistance (Neal et al., 2012; Santhanam et al., 2015).
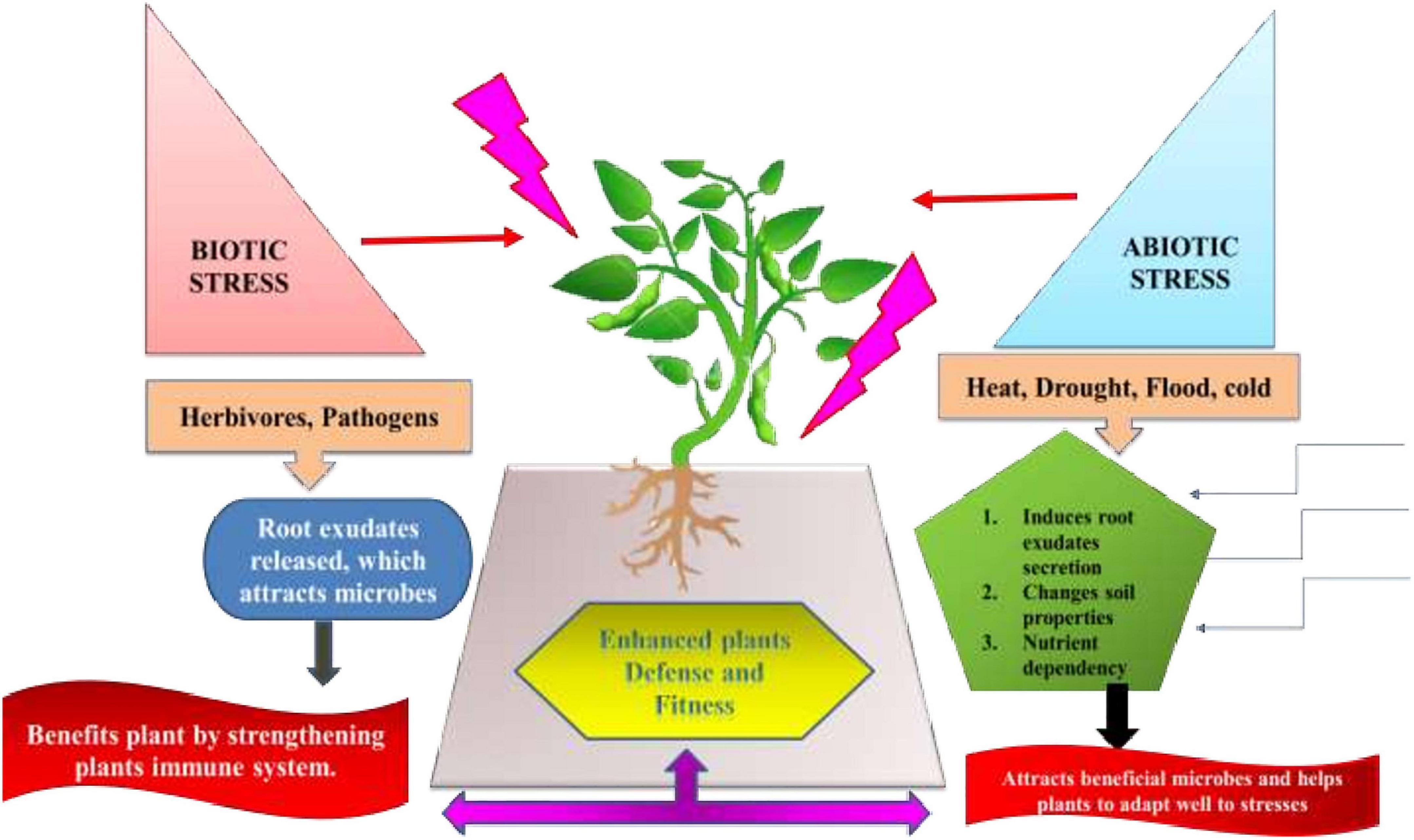
Figure 7. ‘Defense Biome’ mechanisms favoring enrichment of particular microbes under diverse abiotic and biotic stressors by modulating root exudates composition, altering soil properties and nutrients availability.
Artificial microbes are also called as synthetic microbes that form a novel concept for synthetic biology. It is highly useful for remodeling the plant microbiome by modulating its structure as well as function in order to maximize the plant benefits (Arif et al., 2020). There are various steps that are required for preparing efficacious artificial microbes. The initial step includes the determination of microbe and its origin followed by its procurement and cultivation according to its requirements. The next step includes the stimulation of microbial interactions according to their affinities and monitoring the efficiency of the artificially constructed consortium (Kong et al., 2018). With the ability of the plant microbiome to modulate the plant growth and metabolism by synthesizing phytohormones, Tsolakidou et al. (2019) designed two microbes with the ACC-deaminase activity. And inoculation of these microbial strains in Lycopersicum esculentum plants leads to lower the incidence of Fusarium oxysporum infection. Therefore, in conclusion, the microbial strains are best alternatives for stimulating plant growth during biotic and abiotic stresses. They are able to fill the lacunae for using conventional biofertilizers by resolving the issues related to environmental adjustment, host incongruity as well as futile competitive nature along with the native microbes (Hart et al., 2018). Certain microbes, namely, Trichoderma spp., are classified into Microbial Biological Control elements by Plant Protection Products agency (Woo et al., 2014). Furthermore, a plethora of these microbes are also categorized as biopesticides with the potential to enhance the plant growth and development (Lorito and Woo, 2015). Likewise, the arbuscular mycorrhizae, also known as biostimulants, prevent plants from pathogenic attack and other harmful organisms and diseases through activating an induced systemic resistance mechansim (Rouphael et al., 2015). Therefore, such examples elicit the need for registering the novel microbial consortium in order to counteract stresses and able to provide benefit toward plants. This would further boost the effective usage of microbes in the agriculture sector in the form of biopesticides, biofertilizers, biocontrol agents, and biostimulants (Woo and Pepe, 2018).
Phytomicrobiome acts as a stable and effective alternative implication for replenishing the crop yield. It has been studied that microbial products have varied and unreliable effects during trials in different climatic conditions. However, the phytomicrobiome technology has potential, but there was a gap of understanding between the selection of microbes (native or inoculated strains) and its application. Due to the complexity in the mechanism of interactions among soil, plants, and environment, there was difficulty in persisting this phytomicrobiome study because only limited communications have been studied under naive conditions. Many industries showed interest in the use of microbial products that contain specific microbes and carrier-dependent inoculant strains. The implication of emerging technology “Microbiome on a Chip” was effective in studying the multitrophic interplay (microbiome plants) through the combinations of microbial colonies, the response of the host, and several environmental incentives (Stanley and van der Heijden, 2017). Advanced and facile technologies that have prolonged shelf lives would be a gainful strategy in the enhancement of the efficacy of microbial products. The evaluation and activities of the potential or native strains of microbes were helpful in monitoring the harmless microbes in field studies.
The enhancement in the production of goods was achieved by modulating the beneficial microflora of the soil at the blooming phase of seeds. This technology introduced harmless microbial strains and protected the plant biome from other competitive strains that further enhanced the survival and colonization of inoculated microbial strains. The chain of multiple harmless strains of microbes has more compatibility as compared to single-strain designed experiments because the networking and the grouping of beneficial strains with the existing microflora of soil favor the host. There were various systematic approaches to isolating microbial strains that are associated with phyllo- and rhizosphere (Bai et al., 2015; Mitter et al., 2017; Wallenstein, 2017).
Some reports depicted the emerging solutions to harness the ability of beneficial strains of microbes through in situ engineering of the microbiome for the profit in the agriculture and food sector. In this technology, the crops probiotics formulation was composed of the engineered microbiome that further manipulated the microflora (soil and plants) and its activity during its application (Mueller and Sachs, 2015; Sheth et al., 2016). Engineered strains affected the physiology of crops and microbiome by secreting the chemicals that effectively stimulated the activities of beneficial microbial strains, thereby providing resistance to crops against stresses. Based upon the phenotypic and genetic makeup of the microbiome, it was selected for the modulation in plants and soil native microbiome. Another proposed approach, domestication, was used for the elimination of traits facilitated by the microbiome through spraying of fertilizers, insecticides, and growth hormones for increasing crop productivity (Pérez-Jaramillo et al., 2016). The application of eco-friendly biofertilizers that contained beneficial microbial strains was an emerging solution for various crops and soil productivity. The genetic basis for interaction between plants and microbiome was understood by breeding microbe-enhanced designed plants during breeding programs, which, in turn, maintained the harmless strains of microbes.
Plants secreted specific chemicals (exudates) that engineered their own microbiome and enhanced the interactions with other beneficial microbiomes. It was reported that the miRNA was associated with maintaining the structural property of the microbiome of the rhizosphere. Therefore, these interactions are defined as a key to allocating the miRNA and engineering the beneficial microbiome from the target to beneficiary soil for desirable products. Different experiments were conducted with engineered soil or plant-enhanced microbes to manage the ecological engineering of the microbiome of plants and soil in an artificial selection system (Archana et al., 2020). This artificial ecosystem selection of microbial strains helped in improving the fitness of plants, resulting in evolving microbiome. In situ engineering of the genome was an effective and specific technology for the manipulation of the microbiome at a certain level. Another solution for enhancing disease and stress resistance in plants includes the combination of both the plant hormones and improved microbiomes, which activates the defense responses of the microbiome. Moreover, the interaction of microbiome allocation with diverse species of plants can be proved by discovering functional overlapped core microflora of the different plant species (Sheth et al., 2016).
Food security and sustainable agri-products will be a considerable challenge in the coming years for the growing population. The stressed conditions reduce the productivity and availability of food for the growing population that creates rigidity in sustainable farming. Therefore, for enhancing the sustainability and productivity of soil, the phytomicrobiome is being used as an imperative biological toolkit to help and improve agricultural soil flora and fertility. It is a forgoing resource that counteracts various stresses by enhancing the synthesis of phytohormones, enzymes, nutrients, etc. This toolkit has a solution of boosting plant resistance against stresses by supplementing beneficial microbial stress-tolerant strains in soil or plants. The interface of the plant-soil microbe will promote productivity and growth by enhancing the production of hormones, siderophores, and the antioxidative defense system. Another technique, i.e., the application of plant growth-promoting rhizobacteria will also support the development of internal and external resistance or tolerance in plants under stress conditions. The plant growth-promoting bacteria may be used as priming agents, in the form of biopesticides and biofertilizers or as a direct supplement. It is revealed that the rhizo-microflora play an important role in plant growth and health, and the plant itself manages the composition of microbiomes. This mechanism of selection and regulation of microbial strains by plants is mostly favored for their maturation that appears as a natural-picking pressure whenever plants need microbial help.
The microbes act as ecological engineers to cope with environmental stresses. Still, stupendous research will be needed in the field of sustainable agriculture approaches by knowing the durability and survival, commercial design, inconsistency, and host relevance in extensive conditions to meet full utilization of these approaches and provide food security demands in the future. Also, there are other challenges in interpreting plant or soil physiological data and multi-omics with appropriate validation via lab and field experiments. This approach of understanding plant immunity, physiology, genetics, and biome under stress conditions needs to be explored more for improving productivity and ecological sustainability. Moreover, the biological explanation of the mechanism of microbiome shifts induced by stress would also allow the secretion of chemicals in biomes against stress is still unclear. The interplay between plants and microbes is a complex mechanism that needs a thorough investigation as it is still in its initial stage of experiments. Scientific policy organizers and associates need to make a toolkit to identify the plant growth-promoting microbes that are stress tolerant, and, also, the effective strains should be verified to overcome the harmful impact of the stressed environment. Proper research and study will achieve the process of selection and engineering of phytomicrobiome to make them stress resistant, resulting in enhanced production and yield of crops. Plant-microbe interactions and biotic stress-induced alterations have been mostly shown in plants, but the mechanism associated with metabolic pathways of microbes is still yet to be explored. Research should be focused on the characterization and effect of the microbiome on the gene expression in plants, resulting in altering the plant functioning both at proteomics and genomics levels. The CRISPR-Cas9-based forward genetic screen is a comprehensive approach to explain the interactions of plant and microbiome that will be beneficial in the future. To expand the knowledge of plant-soil-microbes interaction, information at the genomic level and engineering of beneficial microbial factories should be explored.
KK, RB, SK, ASh, PAh, PAl, and TA designed the outline and revised and finalized the draft. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abo-Elyousr, K. A., Khalil Bagy, H. M., Hashem, M., Alamri, S. A., and Mostafa, Y. S. (2019). Biological control of the tomato wilt caused by Clavibacter michiganensis subsp. michiganensis using formulated plant growth-promoting bacteria. Egypt. J. Biol. Pest Cont. 29:54. doi: 10.1186/s41938-019-0152-6
Alam, M. A., Seetharam, K., Zaidi, P. H., Dinesh, A., Vinayan, M. T., and Nath, U. K. (2017). Dissecting heat stress tolerance in tropical maize (Zea mays L.). Field Crops Res. 204, 110–119. doi: 10.1016/j.fcr.2017.01.006
Aleklett, K., Kiers, E. T., Ohlsson, P., Shimizu, T. S., Caldas, V. E., and Hammer, E. C. (2018). Build your own soil: exploring microfluidics to create microbial habitat structures. ISME J. 12, 312–319. doi: 10.1038/ismej.2017.184
ALKahtani, M. D., Fouda, A., Attia, K. A., Al-Otaibi, F., Eid, A. M., Ewais, E. E. D., et al. (2020). Isolation and characterization of plant growth promoting endophytic bacteria from desert plants and their application as bioinoculants for sustainable agriculture. Agronomy 10:1325. doi: 10.3390/agronomy10091325
An, Y., Wang, X., Ji, S., Han, J., Wang, Y., Ma, L., et al. (2021). A novel Trichodermaasperellum isolate promoting the growth, detoxification, and antifungal abilities of Arabidopsis thaliana seedlings under Fusarium oxysporum toxin stress. Plant Pathol. J. 103, 1207–1220. doi: 10.1007/s42161-021-00871-9
Anbi, A. A., Mirshekari, B., Eivazi, A., Yarnia, M., and Behrouzyar, E. K. (2020). PGPRs affected photosynthetic capacity and nutrient uptake in different Salvia species. J. Plant Nutr. 43, 108–121. doi: 10.1080/01904167.2019.1659342
Ansari, M. I. (2018). Plant microbiome and its functional mechanism in response to environmental stress. Int. J. Green Pharm. (IJGP) 12, S81–S92.
Archana, T., Rajendran, L., Manoranjitham, S. K., Santhana Krishnan, V. P., Paramasivan, M., and Karthikeyan, G. (2020). Culture-dependent analysis of seed bacterial endophyte, Pseudomonas spp. EGN 1 against the stem rot disease (Sclerotium rolfsii Sacc.) in groundnut. Egypt. J. Biol. Pest Cont. 30, 1–13.
Araú,jo, R. C., Ribeiro, M. S., Rodrigues, F. A., Silva, B. S., Dória, J., and Pasqual, M. (2022). Association of growth-promoting bacteria and hydroponic system aiming at reducing the time of production of banana seedlings. Arch. Agronomy Soil Sci. 1–14. doi: 10.1080/03650340.2022.2078965
Arif, I., Batool, M., and Schenk, P. M. (2020). Plant microbiome engineering: expected benefits for improved crop growth and resilience. Trends Biotechnol. 38, 1385–1396. doi: 10.1016/j.tibtech.2020.04.015
Arora, N. K., Fatima, T., Mishra, J., Mishra, I., Verma, S., Verma, R., et al. (2020). Halo-tolerant plant growth promoting rhizobacteria for improving productivity and remediation of saline soils. J. Adv. Res. 26, 69–82. doi: 10.1016/j.jare.2020.07.003
Asati, A., Pichhode, M., and Nikhil, K. (2016). Effect of heavy metals on plants: an overview. Int. J. Appl. Innov. Eng. Manag. 5, 56–66.
Badri, D. V., Weir, T. L., Van der Lelie, D., and Vivanco, J. M. (2009). Rhizosphere chemical dialogues: plant–microbe interactions. Curr. Opi. Biotechnol. 20, 642–650. doi: 10.1016/j.copbio.2009.09.014
Bai, Y., Müller, D. B., Srinivas, G., Garrido-Oter, R., Potthoff, E., Rott, M., et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369. doi: 10.1038/nature16192
Bal, H. B., Nayak, L., Das, S., and Adhya, T. K. (2013). Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant Soil 366, 93–105. doi: 10.1007/s11104-012-1402-5
Banerjee, S., Singh, S., Pandey, S., Bhandari, M. S., Pandey, A., and Giri, K. (2020). Biocontrol potential of Pseudomonas azotoformans, Serratiam arcescens and Trichoderma virens against Fusarium wilt of Dalbergiasissoo. For. Pathol. 50:e12581. doi: 10.1111/efp.12581
Barnawal, D., Bharti, N., Pandey, S. S., Pandey, A., Chanotiya, C. S., and Kalra, A. (2017). Plant growth-promoting rhizobacteria enhance wheat salt and drought stress tolerance by altering endogenous phytohormone levels and TaCTR1/TaDREB2 expression. Physiol. Plant. 161, 502–514. doi: 10.1111/ppl.12614
Basiliko, N., Stewart, H., Roulet, N. T., and Moore, T. R. (2012). Do root exudates enhance peat decomposition? Geomicrobiol. J. 29, 374–378. doi: 10.1080/01490451.2011.568272
Basu, S., and Kumar, G. (2020). “Stress signalling in the phytomicrobiome: breadth and potential,” in Phyto-microbiome in Stress Regulation, eds M. Kumar, V. Kumar, and R. Prasad (Singapore: Springer), 245–268. doi: 10.1007/978-981-15-2576-6_12
Basu, S., Priyadarshini, P., Prasad, R., and Kumar, G. (2022). “Effects of microbial signaling in plant growth and development,” in Beneficial Microorganisms in Agriculture, eds R. Prasad, and S-H. Zhang (Singapore: Springer), 329–348. doi: 10.1007/978-981-19-0733-3_14
Baysal, Ö, Lai, D., Xu, H. H., Siragusa, M., Çalışkan, M., Carimi, F., et al. (2013). A proteomic approach provides new insights into the control of soil-borne plant pathogens by Bacillus species. PLoS One 8:e53182. doi: 10.1371/journal.pone.0053182
Behm, J. E., Geurts, R., and Kiers, E. T. (2014). Parasponia: a novel system for studying mutualism stability. Trends Plant Sci. 19, 757–763. doi: 10.1016/j.tplants.2014.08.007
Benáková, M., Ahmadi, H., Dučaiová, Z., Tylová, E., Clemens, S., and Tůuma, J. (2017). Effects of Cd and Zn on physiological and anatomical properties of hydroponically grown Brassica napus plants. Environ. Sci. Pollut. Res. 24, 20705–20716. doi: 10.1007/s11356-017-9697-7
Bennett, A. J., Bending, G. D., Chandler, D., Hilton, S., and Mills, P. (2012). Meeting the demand for crop production: the challenge of yield decline in crops grown in short rotations. Biol. Rev. 87, 52–71. doi: 10.1111/j.1469-185X.2011.00184.x
Berendsen, R. L., Vismans, G., Yu, K., Song, Y., de Jonge, R., Burgman, W. P., et al. (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 12, 1496–1507. doi: 10.1038/s41396-018-0093-1
Berlanas, C., Berbegal, M., Elena, G., Laidani, M., Cibriain, J. F., Sagües, A., et al. (2019). The fungal and bacterial rhizosphere microbiome associated with grapevine rootstock genotypes in mature and young vineyards. Front. Microbiol. 10:1142. doi: 10.3389/fmicb.2019.01142
Berni, R., Hausman, J. F., Villas-Boas, S., and Guerriero, G. (2022). Impact of Pseudomonas sp. SVB-B33 on stress-and cell wall-related genes in roots and leaves of hemp under salinity. Horticulturae 8:336. doi: 10.3390/horticulturae8040336
Bhadrecha, P., Bala, M., Khasa, Y. P., Arshi, A., Singh, J., and Kumar, M. (2020). Hippophaerhamnoides L. rhizobacteria exhibit diversified cellulase and pectinase activities. Physiol. Mol. Biol. Plants 26, 1075–1085. doi: 10.1007/s12298-020-00778-2
Bhat, M. A., Kumar, V., Bhat, M. A., Wani, I. A., Dar, F. L., Farooq, I., et al. (2020). Mechanistic insights of the interaction of plant growth-promoting rhizobacteria (PGPR) with plant roots toward enhancing plant productivity by alleviating salinity stress. Front. Microbiol. 11:1952. doi: 10.3389/fmicb.2020.01952
Bhatt, P., Verma, A., Verma, S., Anwar, M., Prasher, P., Mudila, H., et al. (2020). Understanding phytomicrobiome: a potential reservoir for better crop management. Sustainability 12:5446. doi: 10.3390/su12135446
Bilal, L., Asaf, S., Hamayun, M., Gul, H., Iqbal, A., Ullah, I., et al. (2018). Plant growth promoting endophytic fungi Asprgillus fumigatus TS1 and Fusarium proliferatum BRL1 produce gibberellins and regulates plant endogenous hormones. Symbiosis 76, 117–127. doi: 10.1007/s13199-018-0545-4
Bisht, S., Singh, S., Singh, M., and Sharma, J. G. (2022). Augmentative role of Piriformospora indica fungus and plant growth promoting bacteria in mitigating salinity stress in Trigonella foenum-graecum. J. Appl. Biol. Biotechnol. 10, 8–4.
Bollet, C., Davin-Regli, A., and De Micco, P. (1995). A simple method for selective isolation of Stenotrophomonas maltophilia from environmental samples. Appl. Environ. Microbiol. 61, 1653–1654. doi: 10.1128/aem.61.4.1653-1654.1995
Bona, E., Massa, N., Novello, G., Boatti, L., Cesaro, P., Todeschini, V., et al. (2019). Metaproteomic characterization of the Vitis vinifera rhizosphere. FEMS Microbiol. Ecol. 95:fiy204. doi: 10.1093/femsec/fiy204
Boo, H. O., Heo, B. G., Gorinstein, S., and Chon, S. U. (2011). Positive effects of temperature and growth conditions on enzymatic and antioxidant status in lettuce plants. Plant Sci. 181, 479–484. doi: 10.1016/j.plantsci.2011.07.013
Brader, G., Compant, S., Vescio, K., Mitter, B., Trognitz, F., Ma, L. J., et al. (2017). Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 55, 61–83. doi: 10.1146/annurev-phyto-080516-035641
Braos, L. B. (2015). Organic phosphorus fractions in soil fertilized with cattle manure. Rev. Bras. Ci^enc. Solo 39, 140–150. doi: 10.1590/01000683rbcs20150137
Bruno, L. B., Karthik, C., Ma, Y., Kadirvelu, K., Freitas, H., and Rajkumar, M. (2020). Amelioration of chromium and heat stresses in Sorghum bicolor by Cr6+ reducing-thermotolerant plant growth promoting bacteria. Chemosphere 244:125521. doi: 10.1016/j.chemosphere.2019.125521
Canarini, A., Kaiser, C., Merchant, A., Richter, A., and Wanek, W. (2019). Root exudation of primary metabolites: mechanisms and their roles in plant responses to environmental stimuli. Front. Plant Sci. 10:157. doi: 10.3389/fpls.2019.00157
Carlucci, A., Raimondo, M. L., Colucci, D., and Lops, F. (2022). Streptomyces albidoflavus strain CARA17 as a biocontrol agent against fungal soil-borne pathogens of fennel plants. Plants 11:1420. doi: 10.3390/plants11111420
Carrión, V. J., Perez-Jaramillo, J., Cordovez, V., Tracanna, V., De Hollander, M., Ruiz-Buck, D., et al. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 366, 606–612. doi: 10.1126/science.aaw9285
Castrillo, G., Teixeira, P. J. P. L., Paredes, S. H., Law, T. F., de Lorenzo, L., Feltcher, M. E., et al. (2017). Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518. doi: 10.1038/nature21417
Castro-Restrepo, D., Dominguez, M. I., Gaviria-Gutiérrez, B., Osorio, E., and Sierra, K. (2022). Biotization of endophytes Trichoderma asperellum and Bacillus subtilis in mentha spicata microplants to promote growth, pathogen tolerance and specialized plant metabolites. Plants 11:1474. doi: 10.3390/plants11111474
Chandarana, K. A., Pramanik, R. S., and Amaresan, N. (2022). Interaction between ciliate and plant growth promoting bacteria influences the root structure of rice plants, soil PLFAs and respiration properties. Rhizosphere 21:100466. doi: 10.1016/j.rhisph.2021.100466
Chao, W., and Zhuang, W. Y. (2019). Evaluating effective Trichoderma isolates for biocontrol of Rhizoctonia solani causing root rot of Vigna unguiculata. J. Integr. Agric. 18, 2072–2079. doi: 10.1016/S2095-3119(19)62593-1
Chattopadhyay, P., Banerjee, G., and Handique, P. J. (2022). Use of an abscisic acid-producing Bradyrhizobium japonicum isolate as biocontrol agent against bacterial wilt disease caused by Ralstonia solanacearum. J. Plant Diseases Protect. 129, 869–879. doi: 10.1007/s41348-022-00604-9
Chauhan, A., Saini, R., and Sharma, J. C. (2021). Plant growth promoting rhizobacteria and their biological properties for soil enrichment and growth promotion. J. Plant Nutr. 45, 273–299. doi: 10.1080/01904167.2021.1952221
Chen, Y., Wang, J., Yang, N., Wen, Z., Sun, X., Chai, Y., et al. (2018). Wheat microbiome bacteria can reduce virulence of a plant pathogenic fungus by altering histone acetylation. Nat. Commun. 9:3429. doi: 10.1038/s41467-018-05683-7
Chi, Y., You, Y., Wang, J., Chen, X., Chu, S., Wang, R., et al. (2022). Two plant growth-promoting bacterial Bacillus strains possess different mechanisms in affecting cadmium uptake and detoxification of Solanum nigrum L. Chemosphere 305:135488. doi: 10.1016/j.chemosphere.2022.135488
Chialva, M., Salvioli, di Fossalunga, A., Daghino, S., Ghignone, S., Bagnaresi, P., et al. (2018). Native soils with their microbiotas elicit a state of alert in tomato plants. New Phytol. 220, 1296–1308. doi: 10.1111/nph.15014
Chouhan, G. K., Verma, J. P., Jaiswal, D. K., Mukherjee, A., Singh, S., de Araujo Pereira, A. P., et al. (2021). Phytomicrobiome for promoting sustainable agriculture and food security: opportunities, challenges, and solutions. Microbiol. Res. 248:126763. doi: 10.1016/j.micres.2021.126763
Clúa, J., Roda, C., Zanetti, M. E., and Blanco, F. A. (2018). Compatibility between legumes and rhizobia for the establishment of a successful nitrogen-fixing symbiosis. Genes 9:125. doi: 10.3390/genes9030125
Cohen, A. C., Bottini, R., Pontin, M., Berli, F. J., Moreno, D., Boccanlandro, H., et al. (2015). Azospirillum brasilense ameliorates the response of Arabidopsis thaliana to drought mainly via enhancement of ABA levels. Physiol. Plant. 153, 79–90. doi: 10.1111/ppl.12221
Damodaran, T., Sharma, D. K., Mishra, V. K., Jha, S. K., Kannan, R., Sah, V., et al. (2013). Isolation of salt tolerant endophytic and rhizospheric bacteria by natural selection and screening for promising plant growth promoting rhizobacteria [PGPR] and growth vigour in Tomato under sodic soil. African J. Microbiol. Res. 7, 5082–5089.
Daroodi, Z., Taheri, P., and Tarighi, S. (2021). Direct antagonistic activity and tomato resistance induction of the endophytic fungus Acrophia lophorajodhpurensis against Rhizoctonia solani. Biol. Control 160:104696. doi: 10.1016/j.biocontrol.2021.104696
De Cuyper, C., Fromentin, J., Yocgo, R. E., De Keyser, A., Guillotin, B., Kunert, K., et al. (2015). From lateral root density to nodule number, the strigolactone analogue GR24 shapes the root architecture of Medicago truncatula. J. Exp. Bot. 66, 137–146. doi: 10.1093/jxb/eru404
de la Fuente Cantó, C., Simonin, M., King, E., Moulin, L., Bennett, M. J., Castrillo, G., et al. (2020). An extended root phenotype: the rhizosphere, its formation and impacts on plant fitness. Plant J. 103, 951–964. doi: 10.1111/tpj.14781
De Vleesschauwer, D., and Höfte, M. (2009). Rhizobacteria-induced systemic resistance. Adv. Bot. Res. 51, 223–281. doi: 10.1016/S0065-2296(09)51006-3
Desoky, E. S. M., Merwad, A. R. M., Semida, W. M., Ibrahim, S. A., El-Saadony, M. T., and Rady, M. M. (2020). Heavy metals-resistant bacteria (HM-RB): potential bioremediators of heavy metals-stressed Spinacia oleracea plant. Ecotoxicol. Environ. Saf. 198:110685. doi: 10.1016/j.ecoenv.2020.110685
Dhanya, S., Sherin, V., Divya, K., Sreekumar, J., and Jisha, M. S. (2020). Pseudomonas taiwanensis (MTCC11631) mediated induction of systemic resistance in Anthurium andreanum L. against blight disease and visualisation of defence related secondary metabolites using confocal laser scanning microscopy. Biocatal. Agric. Biotechnol. 24:101561. doi: 10.1016/j.bcab.2020.101561
Diaconu, M., Pavel, L. V., Hlihor, R. M., Rosca, M., Fertu, D. I., Lenz, M., et al. (2020). Characterization of heavy metal toxicity in some plants and microorganisms—a preliminary approach for environmental bioremediation. New Biotechnol. 56, 130–139. doi: 10.1016/j.nbt.2020.01.003
Djebaili, R., Pellegrini, M., Ercole, C., Farda, B., Kitouni, M., and Del Gallo, M. (2021). Biocontrol of soil-borne pathogens of Solanum lycopersicum L. and Daucus carota L. by plant growth-promoting actinomycetes: in vitro and in planta antagonistic activity. Pathogens 10:1305. doi: 10.3390/pathogens10101305
Doornbos, R. F., van Loon, L. C., and Bakker, P. A. (2012). Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. a review. Agro Sustainable Dev. 32, 227–243. doi: 10.1007/s13593-011-0028-y
Durán, P., Thiergart, T., Garrido-Oter, R., Agler, M., Kemen, E., Schulze-Lefert, P., et al. (2018). Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175, 973–983. doi: 10.1016/j.cell.2018.10.020
el Zahar Haichar, F., Santaella, C., Heulin, T., and Achouak, W. (2014). Root exudates mediated interactions belowground. Soil Biol. Biochem. 77, 69–80. doi: 10.1016/j.soilbio.2014.06.017
Elhady, A., Adss, S., Hallmann, J., and Heuer, H. (2018). Rhizosphere microbiomes modulated by pre-crops assisted plants in defense against plant-parasitic nematodes. Front. Microbiol. 9:1133. doi: 10.3389/fmicb.2018.01133
Elshahawy, I. E., and El-Mohamedy, R. S. (2019). Biological control of Pythium damping-off and root-rot diseases of tomato using Trichoderma isolates employed alone or in combination. Plant Pathol. J. 101, 597–608. doi: 10.1007/s42161-019-00248-z
Enebe, M. C., and Babalola, O. O. (2019). The impact of microbes in the orchestration of plants’ resistance to biotic stress: a disease management approach. Appl. Microbio. Biotechnol. 103, 9–25. doi: 10.1007/s00253-018-9433-3
Etesami, H., and Adl, S. M. (2020). “Plant growth-promoting rhizobacteria (PGPR) and their action mechanisms in availability of nutrients to plants,” in Phyto-Microbiome Stress Regulation, (Singapore: Springer Nature). doi: 10.1007/978-981-15-2576-6_9
Farooq, M., Wahid, A., Kobayashi, N., Fujita, D. B. S. M. A., and Basra, S. M. A. (2009). Sustainable Agriculture. Plant Drought Stress: Effects, Mechanisms and Management. Dordrecht: Springer, 153–188. doi: 10.1007/978-90-481-2666-8_12
Farré-Armengol, G., Filella, I., Llusia, J., and Peñuelas, J. (2016). Bidirectional interaction between phyllospheric microbiotas and plant volatile emissions. Trends Plant Sci. 21, 854–860. doi: 10.1016/j.tplants.2016.06.005
Fernandes, V. H. (2014). Cultivo de Azospirillum amazonense BR11145 para produção de inoculantes promotores de crescimento vegetal. Rev. Ci^enc. Exatas 33, 87–97.
Fernandez, M., Godino, A., Principe, A., Ramírez, V. L., Quesada, J. M., Rigo, V., et al. (2020). Characterization of the bacteriocins and the PrtR regulator in a plant-associated Pseudomonas strain. J. Biotechnol. 307, 182–192. doi: 10.1016/j.jbiotec.2019.11.003
Figueredo, M. S., Tonelli, M. L., Ibáñez, F., Morla, F., Cerioni, G., del Carmen Tordable, M., and Fabra, A. (2017). Induced systemic resistance and symbiotic performance of peanut plants challenged with fungal pathogens and co-inoculated with the biocontrol agent Bacillus sp. CHEP5 and Bradyrhizobium sp. SEMIA6144. Microbiol. Res. 197, 65–73.
Forouzi, A., Ghasemnezhad, A., and Nasrabad, R. G. (2020). Phytochemical response of Stevia plant to growth promoting microorganisms under salinity stress. South Afr. J. Botany 134, 109–118. doi: 10.1016/j.sajb.2020.04.001
Gamez, R., Cardinale, M., Montes, M., Ramirez, S., Schnell, S., and Rodriguez, F. (2019). Screening, plant growth promotion and root colonization pattern of two rhizobacteria (Pseudomonas fluorescens Ps006 and Bacillus amyloliquefaciens Bs006) on banana cv. Williams (Musa acuminata Colla). Microbiol Res. 220, 12–20. doi: 10.1016/j.micres.2018.11.006
Glick, B. R. (2010). Using soil bacteria to facilitate phytoremediation. Biotechnol. Adv. 28, 367–374. doi: 10.1016/j.biotechadv.2010.02.001
Glick, B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012:963401. doi: 10.6064/2012/963401
Gontia-Mishra, I., Sapre, S., Sharma, A., and Tiwari, S. (2016). Amelioration of drought tolerance in wheat by the interaction of plant growth-promoting rhizobacteria. Plant Biol. 18, 992–1000. doi: 10.1111/plb.12505
Goswami, D., Thakker, J. N., and Dhandhukia, P. C. (2015). Simultaneous detection and quantification of indole-3-acetic acid (IAA) and indole-3-butyric acid (IBA) produced by rhizobacteria from l-tryptophan (Trp) using HPTLC. J. Microbiol. Methods 110, 7–14. doi: 10.1016/j.mimet.2015.01.001
Grover, M., Ali, S. Z., Sandhya, V., Rasul, A., and Venkateswarlu, B. (2011). Role of microorganisms in adaptation of agriculture crops to abiotic stresses. World J. Microbiol. Biotechnol. 27, 1231–1240. doi: 10.1007/s11274-010-0572-7
Guarino, F., Miranda, A., Castiglione, S., and Cicatelli, A. (2020). Arsenic phytovolatilization and epigenetic modifications in Arundodonax L. assisted by a PGPR consortium. Chemosphere 251:126310. doi: 10.1016/j.chemosphere.2020.126310
Gupta, S., and Pandey, S. (2019). ACC deaminase producing bacteria with multifarious plant growth promoting traits alleviates salinity stress in French bean (Phaseolus vulgaris) plants. Front. Microbiol. 10:1506. doi: 10.3389/fmicb.2019.01506
Hammerbacher, A., Coutinho, T. A., and Gershenzon, J. (2019). Roles of plant volatiles in defence against microbial pathogens and microbial exploitation of volatiles. Plant Cell Environ. 42, 2827–2843. doi: 10.1111/pce.13602
Hart, M. M., Antunes, P. M., Chaudhary, V. B., and Abbott, L. K. (2018). Fungal inoculants in the field: is the reward greater than the risk? Funct. Ecol. 32, 126–135. doi: 10.1111/1365-2435.12976
Hartman, K., and Tringe, S. G. (2019). Interactions between plants and soil shaping the root microbiome under abiotic stress. Biochemical J. 476, 2705–2724. doi: 10.1042/BCJ20180615
Hashem, A., Abd_Allah, E. F., Alqarawi, A. A., Radhakrishnan, R., and Kumar, A. (2017). Plant defense approach of Bacillus subtilis (BERA 71) against Macrophomina phaseolina (Tassi) Goid in mung bean. J. Plant Interact. 12, 390–401. doi: 10.1080/17429145.2017.1373871
Hassan, S., and Mathesius, U. (2012). The role of flavonoids in root–rhizosphere signalling: opportunities and challenges for improving plant–microbe interactions. J. Exp. Bot. 63, 3429–3444. doi: 10.1093/jxb/err430
Hayat, R., Ali, S., Amara, U., Khalid, R., and Ahmed, I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review. Annals Microbiol. 60, 579–598. doi: 10.1007/s13213-010-0117-1
Helaly, A. A., Mady, E., Salem, E. A., and Randhir, T. O. (2022). Stimulatory effects of growth-promoting bacteria on growth, nutritional composition, and yield of kale plants. J. Plant Nutrition. 1–13. doi: 10.1080/01904167.2022.2046084
Hoffman, M. T., and Arnold, A. E. (2010). Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl. Environ. Microbiol. 76, 4063–4075. doi: 10.1128/AEM.02928-09
Hristozkova, M., Geneva, M., Stancheva, I., Boychinova, M., and Djonova, E. (2016). Contribution of arbuscular mycorrhizal fungi in attenuation of heavy metal impact on Calendula officinalis development. Appl. Soil Ecol. 101, 57–63. doi: 10.1016/j.apsoil.2016.01.008
Huang, A. C., Jiang, T., Liu, Y. X., Bai, Y. C., Reed, J., Qu, B., et al. (2019). A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364:eaau6389. doi: 10.1126/science.aau6389
Hungria, M., Campo, R. J., Souza, E. M., and Pedrosa, F. O. (2010). Inoculation with selected strains of Azospirillum brasilense and A. lipoferum improves yields of maize and wheat in Brazil. Plant Soil 331, 413–425. doi: 10.1007/s11104-009-0262-0
Hussain, M. B., Zahir, Z. A., Asghar, H. N., and Asgher, M. (2014). Can catalase and exopolysaccharides producing rhizobia ameliorate drought stress in wheat? Int. J. Agricul. Biol. 16, 3–13.
Ismail, M. A., Amin, M. A., Eid, A. M., Hassan, S. E. D., Mahgoub, H. A., Lashin, I., et al. (2021). Comparative study between exogenously applied plant growth hormones versus metabolites of microbial endophytes as plant growth-promoting for Phaseolus vulgaris L. Cells 10:1059. doi: 10.3390/cells10051059
Jabborova, D., Enakiev, Y., Sulaymanov, K., Kadirova, D., Ali, A., and Annapurna, K. (2021). Plant growth promoting bacteria Bacillus subtilis promote growth and physiological parameters of Zingiber officinale roscoe. Plant Sci. Today 8, 66–71. doi: 10.14719/pst.2021.8.1.997
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kenawy, A., Dailin, D. J., Abo-Zaid, G. A., Malek, R. A., Ambehabati, K. K., Zakaria, K. H. N., et al. (2019). “Biosynthesis of antibiotics by PGPR and their roles in biocontrol of plant diseases,” in Plant Growth Promoting Rhizobacteria for Sustainable Stress Management, ed. R. Z. Sayyed (Singapore: Springer), 1–35. doi: 10.1007/978-981-13-6986-5_1
Khan, M. A., Asaf, S., Khan, A. L., Adhikari, A., Jan, R., Ali, S., et al. (2020). Plant growth-promoting endophytic bacteria augment growth and salinity tolerance in rice plants. Plant Biol. 22, 850–862. doi: 10.1111/plb.13124
Khan, M. A., Hamayun, M., Asaf, S., Khan, M., Yun, B. W., Kang, S. M., et al. (2021). Rhizospheric Bacillus spp. rescues plant growth under salinity stress via regulating gene expression, endogenous hormones, and antioxidant system of Oryza sativa L. Front. Plant Sci. 12:665590. doi: 10.3389/fpls.2021.665590
Kim, J. S., and Crowley, D. E. (2013). Size fractionation and microbial community structure of soil aggregates. J. Agric. Chem. Environ. 2, 75–80. doi: 10.4236/jacen.2013.24011
Kong, W. L., Li, P. S., Wu, X. Q., Wu, T. Y., and Sun, X. R. (2020). Forest tree associated bacterial diffusible and volatile organic compounds against various phytopathogenic fungi. Microorganisms 8:590. doi: 10.3390/microorganisms8040590
Kong, Z., Hart, M., and Liu, H. (2018). Paving the way from the lab to the field: using synthetic microbial consortia to produce high-quality crops. Front. Plant Sci. 9:1467. doi: 10.3389/fpls.2018.01467
Korenblum, E., Dong, Y., Szymanski, J., Panda, S., Jozwiak, A., Massalha, H., et al. (2020). Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. U S A. 117, 3874–3883. doi: 10.1073/pnas.1912130117
Kosová, K., Vítámvás, P., Urban, M. O., Prášil, I. T., and Renaut, J. (2018). Plant abiotic stress proteomics: The major factors determining alterations in cellular proteome. Front. Plant Sci. 9:122. doi: 10.3389/fpls.2018.00122
Kour, D., Rana, K. L., Yadav, A. N., Sheikh, I., Kumar, V., Dhaliwal, H. S., et al. (2020). Amelioration of drought stress in Foxtail millet (Setaria italica L.) by P-solubilizing drought-tolerant microbes with multifarious plant growth promoting attributes. Environ. Sustainability 3, 23–34. doi: 10.1007/s42398-020-00094-1
Kousar, B., Bano, A., and Khan, N. (2020). PGPR modulation of secondary metabolites in tomato infested with Spodoptera litura. Agronomy 10:778. doi: 10.3390/agronomy10060778
Kudjordjie, E. N., Sapkota, R., Steffensen, S. K., Fomsgaard, I. S., and Nicolaisen, M. (2019). Maize synthesized benzoxazinoids affect the host associated microbiome. Microbiome 7:59. doi: 10.1186/s40168-019-0677-7
Kumar, A., and Verma, J. P. (2018). Does plant—microbe interaction confer stress tolerance in plants: a review? Microbiol. Res. 207, 41–52. doi: 10.1016/j.micres.2017.11.004
Kumar, A., Singh, S., Mukherjee, A., Rastogi, R. P., and Verma, J. P. (2021). Salt-tolerant plant growth-promoting Bacillus pumilus strain JPVS11 to enhance plant growth attributes of rice and improve soil health under salinity stress. Microbiol. Res. 242:126616. doi: 10.1016/j.micres.2020.126616
Kumar, M., Kour, D., Yadav, A. N., Saxena, R., Rai, P. K., Jyoti, A., et al. (2019). Biodiversity of methylotrophic microbial communities and their potential role in mitigation of abiotic stresses in plants. Biologia 74, 287–308. doi: 10.2478/s11756-019-00190-6
Kwak, M. J., Kong, H. G., Choi, K., Kwon, S. K., Song, J. Y., Lee, J., et al. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1100–1109. doi: 10.1038/nbt.4232
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Lava, A., and Babaeizad, V. (2021). Effects of Trichoderma and endophytic fungus Piriformospora indica on cucumber physiology and Fusarium wilt disease of Cucumber. Annals Romanian Soc. Cell Biol. 25, 20742–20755.
Lee, B., Lee, S., and Ryu, C. M. (2012). Foliar aphid feeding recruits rhizosphere bacteria and primes plant immunity against pathogenic and non-pathogenic bacteria in pepper. Annals Bot. 110, 281–290. doi: 10.1093/aob/mcs055
Lemanceau, P., Blouin, M., Muller, D., and Moënne-Loccoz, Y. (2017). Let the core microbiota be functional. Trends Plant Sci. 22, 583–595. doi: 10.1016/j.tplants.2017.04.008
Levy, A., Gonzalez, I. S., Mittelviefhaus, M., Clingenpeel, S., Paredes, S. H., Miao, J., et al. (2018). Genomic features of bacterial adaptation to plants. Nat. Genet. 50, 138–150. doi: 10.1038/s41588-017-0012-9
Li, G., Zhao, H., Liu, Z., Wang, H., Xu, B., and Guo, X. (2018). The wisdom of honeybee defenses against environmental stresses. Front. Microbiol. 9:722. doi: 10.3389/fmicb.2018.00722
Li, S., Fu, Q., Chen, L., Huang, W., and Yu, D. (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233, 1237–1252. doi: 10.1007/s00425-011-1375-2
Liu, C. W., and Murray, J. D. (2016). The role of flavonoids in nodulation host-range specificity: an update. Plants 5:33. doi: 10.3390/plants5030033
Liu, H., and Brettell, L. E. (2019). Plant defense by VOC-induced microbial priming. Trends Plant Sci. 24, 187–189. doi: 10.1016/j.tplants.2019.01.008
Liu, H., Brettell, L. E., Qiu, Z., and Singh, B. K. (2020). Microbiome-mediated stress resistance in plants. Trends Plant Sci. 25, 733–743. doi: 10.1016/j.tplants.2020.03.014
Liu, H., Carvalhais, L. C., Crawford, M., Singh, E., Dennis, P. G., Pieterse, C. M., et al. (2017). Inner plant values: diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 8:2552. doi: 10.3389/fmicb.2017.02552
Liu, K., McInroy, J. A., Hu, C. H., and Kloepper, J. W. (2018). Mixtures of plant-growth-promoting rhizobacteria enhance biological control of multiple plant diseases and plant-growth promotion in the presence of pathogens. Plant Dis. 102, 67–72. doi: 10.1094/PDIS-04-17-0478-RE
Lombardi, N., Vitale, S., Turrà, D., Reverberi, M., Fanelli, C., Vinale, F., et al. (2018). Root exudates of stressed plants stimulate and attract Trichoderma soil fungi. Mol. Plant Microbe Interact. 31, 982–994. doi: 10.1094/MPMI-12-17-0310-R
Lorito, M., and Woo, S. L. (2015). “Trichoderma: a multi-purpose tool for integrated pest management,” in Principles of Plant-microbe Interactions, ed. B. Lugtenberg (Cham: Springer), 345–353. doi: 10.1007/978-3-319-08575-3_36
Lyu, D., Backer, R., Subramanian, S., and Smith, D. L. (2020). Phytomicrobiome coordination signals hold potential for climate change-resilient agriculture. Front. Plant Sci. 11:634. doi: 10.3389/fpls.2020.00634
Ma, Y. H., Zhang, X. F., Song, J., and Wu, E. (2016). Bistability and steady-state spin squeezing in diamond nanostructures controlled by a nanomechanical resonator. Annals Phys. 369, 36–44. doi: 10.1016/j.aop.2016.03.001
Madriz-Ordeñana, K., Pazarlar, S., Jørgensen, H. J. L., Nielsen, T. K., Zhang, Y., Nielsen, K. L., et al. (2022). The Bacillus cereus strain EC9 primes the plant immune system for superior biocontrol of Fusarium oxysporum. Plants 11:687. doi: 10.3390/plants11050687
Maheshwari, D. K., Dheeman, S., and Agarwal, M. (2015). “Phytohormone-producing PGPR for sustainable agriculture,” in Bacterial Metabolites in Sustainable Agroecosystem, ed. D. K. Maheshwari (Cham: Springer), 159–182. doi: 10.1007/978-3-319-24654-3_7
Mahgoub, H. A., Fouda, A., Eid, A. M., Ewais, E. E. D., and Hassan, S. E. D. (2021). Biotechnological application of plant growth-promoting endophytic bacteria isolated from halophytic plants to ameliorate salinity tolerance of Vicia faba L. Plant Biotechnol. Rep. 15, 819–843. doi: 10.1007/s11816-021-00716-y
Majeed, A., Muhammad, Z., Islam, S., Ullah, Z., and Ullah, R. (2017). Cyanobacterial application as bio-fertilizers in rice fields: role in growth promotion and crop productivity. PSM Microbiol. 2, 47–50.
Mani, D., Kumar, C., and Patel, N. K. (2016). Integrated micro-biochemical approach for phytoremediation of cadmium and lead contaminated soils using Gladiolus grandiflorus L cut flower. Ecotoxicol. Environ. Saf. 124, 435–446. doi: 10.1016/j.ecoenv.2015.11.016
Meena, K. K., Sorty, A. M., Bitla, U. M., Choudhary, K., Gupta, P., Pareek, A., et al. (2017). Abiotic stress responses and microbe-mediated mitigation in plants: the omics strategies. Front. Plant Sci. 8:172. doi: 10.3389/fpls.2017.00172
Mendes, R., Kruijt, M., De Bruijn, I., Dekkers, E., van der Voort, M., Schneider, J. H., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Miguel-Ferrer, L., Romero-Arenas, O., Andrade-Hoyos, P., Sánchez-Morales, P., Rivera-Tapia, J. A., and Fernández-Pavía, S. P. (2021). Antifungal activity of Trichoderma harzianum and T. koningiopsis against Fusariumsolani in seed germination and vigor of Miahua tecochili seedlings. Rev. Mex. Fitopatol 39, 228–247. doi: 10.18781/R.MEX.FIT.2101-5
Millet, Y. A., Danna, C. H., Clay, N. K., Songnuan, W., Simon, M. D., Werck-Reichhart, D., et al. (2010). Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22, 973–990. doi: 10.1105/tpc.109.069658
Mitter, B., Pfaffenbichler, N., Flavell, R., Compant, S., Antonielli, L., Petric, A., et al. (2017). A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front. Microbiol. 8:11. doi: 10.3389/fmicb.2017.00011
Mittler, R., and Blumwald, E. (2010). Genetic engineering for modern agriculture: challenges and perspectives. Annu. Rev. Plant Biol. 61, 443–462. doi: 10.1146/annurev-arplant-042809-112116
Morcillo, R. J., and Manzanera, M. (2021). The effects of plant-associated bacterial exopolysaccharides on plant abiotic stress tolerance. Metabolites 11:337. doi: 10.3390/metabo11060337
Moreira, H., Pereira, S. I., Vega, A., Castro, P. M., and Marques, A. P. (2020). Synergistic effects of arbuscular mycorrhizal fungi and plant growth-promoting bacteria benefit maize growth under increasing soil salinity. J. Environ. Manag. 257:109982. doi: 10.1016/j.jenvman.2019.109982
Mueller, U. G., and Sachs, J. L. (2015). Engineering microbiomes to improve plant and animal health. Trends Microbiol. 23, 606–617. doi: 10.1016/j.tim.2015.07.009
Muller, A., Schader, C., El-Hage Scialabba, N., Brüggemann, J., Isensee, A., Erb, K. H., et al. (2017). Strategies for feeding the world more sustainably with organic agriculture. Nat. Commun. 8:1290. doi: 10.1038/s41467-017-01410-w
Naamala, J., and Smith, D. L. (2020). Relevance of plant growth promoting microorganisms and their derived compounds, in the face of climate change. Agronomy 10:1179. doi: 10.3390/agronomy10081179
Naseem, H., and Bano, A. (2014). Role of plant growth-promoting rhizobacteria and their exopolysaccharide in drought tolerance of maize. J. Plant Interac. 9, 689–701. doi: 10.1080/17429145.2014.902125
Nawrocka, J., Malolepsza, U., Szymczak, K., and Szczech, M. (2018). Involvement of metabolic components, volatile compounds, PR proteins, and mechanical strengthening in multilayer protection of cucumber plants against Rhizoctonia solani activated by Trichoderma atroviride TRS25. Protoplasma, 255, 359–373.
Neal, A. L., Ahmad, S., Gordon-Weeks, R., and Ton, J. (2012). Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS One 7:e35498. doi: 10.1371/journal.pone.0035498
Nisa, S., Shoukat, M., Bibi, Y., Al Ayoubi, S., Shah, W., Masood, S., et al. (2022). Therapeutic prospects of endophytic Bacillus species from Berberis lycium against oxidative stress and microbial pathogens. Saudi J. Biol. Sci. 29, 287–295. doi: 10.1016/j.sjbs.2021.08.099
Niu, D., Wang, X., Wang, Y., Song, X., Wang, J., Guo, J., et al. (2016). Bacillus cereus AR156 activates PAMP-triggered immunity and induces a systemic acquired resistance through a NPR1-and SA-dependent signaling pathway. Biochem. Biophys. Res. Commun. 469, 120–125. doi: 10.1016/j.bbrc.2015.11.081
Ortiz, N., Armada, E., Duque, E., Roldán, A., and Azcón, R. (2015). Contribution of arbuscular mycorrhizal fungi and/or bacteria to enhancing plant drought tolerance under natural soil conditions: effectiveness of autochthonous or allochthonous strains. J. Plant Physiol. 174, 87–96. doi: 10.1016/j.jplph.2014.08.019
Özer, N., Coşkuntuna, A., and Şabudak, T. (2021). Trichoderma harzianum-induced defense in sunflower (Helianthus annuus L.) against Plasmopara halstedii with changes in metabolite profiling of roots. Biocontrol. Sci. Technol. 31, 1403–1418. doi: 10.1080/09583157.2021.1963417
Pandey, P., Irulappan, V., Bagavathiannan, M. V., and Senthil-Kumar, M. (2017). Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front. Plant Sci. 8:537. doi: 10.3389/fpls.2017.00537
Passari, A. K., Upadhyaya, K., Singh, G., Abdel-Azeem, A. M., Thankappan, S., Uthandi, S., et al. (2019). Enhancement of disease resistance, growth potential, and photosynthesis in tomato (Solanum lycopersicum) by inoculation with an endophytic actinobacterium, Streptomyces thermocarboxydus strain BPSAC147. PLoS One 14:e0219014. doi: 10.1371/journal.pone.0219014
Pellegrini, M., Spera, D. M., Ercole, C., and Del Gallo, M. (2021). Allium cepa L. inoculation with a consortium of plant growth-promoting bacteria: effects on plants, soil, and the autochthonous microbial community. Microorganisms 9:639. doi: 10.3390/microorganisms9030639
Pereira, A. P., de Souza, A. J., de Chaves, M. G., Fracetto, G. G., Garcia, K. G., Paulo Filho, F. M., et al. (2021). “Mechanisms of the phytomicrobiome for enhancing soil fertility and health,” in New and Future Developments in Microbial Biotechnology and Bioengineering, eds. J. P. Verma, C. A. Macdonald, V. K. Gupta, and A. R. Podile (Amsterdam: Elsevier), 1–14. doi: 10.1016/B978-0-444-64325-4.00001-8
Pérez-de-Luque, A., Tille, S., Johnson, I., Pascual-Pardo, D., Ton, J., and Cameron, D. D. (2017). The interactive effects of arbuscular mycorrhiza and plant growth-promoting rhizobacteria synergistically enhance host plant defences against pathogens. Sci. Rep. 7:16409. doi: 10.1038/s41598-017-16697-4
Pérez-Jaramillo, J. E., Mendes, R., and Raaijmakers, J. M. (2016). Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 90, 635–644. doi: 10.1007/s11103-015-0337-7
Philippot, L. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Pieterse, C. M., Leon-Reyes, A., Van der Ent, S., and Van Wees, S. C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. doi: 10.1038/nchembio.164
Pineda, A., Zheng, S. J., van Loon, J. J., Pieterse, C. M., and Dicke, M. (2010). Helping plants to deal with insects: the role of beneficial soil-borne microbes. Trends Plant Sci. 15, 507–514. doi: 10.1016/j.tplants.2010.05.007
Porcel, R., Zamarreño, ÁM., García-Mina, J. M., and Aroca, R. (2014). Involvement of plant endogenous ABA in Bacillus megaterium PGPR activity in tomato plants. BMC Plant Biol. 14:36. doi: 10.1186/1471-2229-14-36
Qiao, Q., Wang, F., Zhang, J., Chen, Y., Zhang, C., Liu, G., et al. (2017). The variation in the rhizosphere microbiome of cotton with soil type, genotype and developmental stage. Sci. Rep. 7:3940. doi: 10.1038/s41598-017-04213-7
Raaijmakers, J. M., and Mazzola, M. (2012). Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Ann. Rev. Phytopathol. 50, 403–424. doi: 10.1146/annurev-phyto-081211-172908
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., and Moënne-Loccoz, Y. (2009). The rhizosphere: a playground and battlefield for soil borne pathogens and beneficial microorganisms. Plant Soil 321, 341–361. doi: 10.1007/s11104-008-9568-6
Rahimi, S., Talebi, M., Baninasab, B., Gholami, M., Zarei, M., and Shariatmadari, H. (2020). The role of plant growth-promoting rhizobacteria (PGPR) in improving iron acquisition by altering physiological and molecular responses in quince seedlings. Plant Physiol. Biochem. 155, 406–415. doi: 10.1016/j.plaphy.2020.07.045
Raklami, A., Bechtaoui, N., Tahiri, A. I., Anli, M., Meddich, A., and Oufdou, K. (2019). Use of rhizobacteria and mycorrhizae consortium in the open field as a strategy for improving crop nutrition, productivity and soil fertility. Front. Microbiol. 10:1106. doi: 10.3389/fmicb.2019.01106
Rana, K. L., Kour, D., Yadav, N., and Yadav, A. N. (2020). “Endophytic microbes in nanotechnology: current development, and potential biotechnology applications,” in Microbial Endophytes, ed. S. Elias (Sawston: Woodhead Publishing), 231–262. doi: 10.1016/B978-0-12-818734-0.00010-3
Ranjan, R., Rani, A., Metwally, A., McGee, H. S., and Perkins, D. L. (2016). Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 469, 967–977. doi: 10.1016/j.bbrc.2015.12.083
Rijavec, T., and Lapanje, A. (2016). Hydrogen cyanide in the rhizosphere: not suppressing plant pathogens, but rather regulating availability of phosphate. Front. Microbiol. 7:1785. doi: 10.3389/fmicb.2016.01785
Roesch, L. F. W., Camargo, F. A., Bento, F. M., and Triplett, E. W. (2008). Biodiversity of diazotrophic bacteria within the soil, root and stem of field-grown maize. Plant Soil 302, 91–104. doi: 10.1007/s11104-007-9458-3
Rojas, E. C., Sapkota, R., Jensen, B., Jørgensen, H. J., Henriksson, T., Jørgensen, L. N., et al. (2020). Fusarium head blight modifies fungal endophytic communities during infection of wheat spikes. Microb. Ecol. 79, 397–408. doi: 10.1007/s00248-019-01426-3
Rouphael, Y., Franken, P., Schneider, C., Schwarz, D., Giovannetti, M., Agnolucci, M., et al. (2015). Arbuscular mycorrhizal fungi act as biostimulants in horticultural crops. Sci. Horticul. 196, 91–108. doi: 10.1016/j.scienta.2015.09.002
Sańchez, O. J., Ospina, D. A., and Montoya, S. (2017). Compost supplementation with nutrients and microorganisms in composting process. Waste Manag. 69, 136–153. doi: 10.1016/j.wasman.2017.08.012
Santhanam, R., Luu, V. T., Weinhold, A., Goldberg, J., Oh, Y., and Baldwin, I. T. (2015). Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc. Natl. Acad. Sci. U S A. 112, E5013–E5020. doi: 10.1073/pnas.1505765112
Sarkar, A., Ghosh, P. K., Pramanik, K., Mitra, S., Soren, T., Pandey, S., et al. (2018). A halotolerant Enterobacter sp. displaying ACC deaminase activity promotes rice seedling growth under salt stress. Res. Microbiol. 169, 20–32. doi: 10.1016/j.resmic.2017.08.005
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., and Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Schimel, J., Balser, T. C., and Wallenstein, M. (2007). Microbial stress-response physiology and its implications for ecosystem function. Ecology 88, 1386–1394. doi: 10.1890/06-0219
Schulz-Bohm, K., Gerards, S., Hundscheid, M., Melenhorst, J., de Boer, W., and Garbeva, P. (2018). Calling from distance: attraction of soil bacteria by plant root volatiles. ISME J. 12, 1252–1262. doi: 10.1038/s41396-017-0035-3
Semchenko, M., Saar, S., and Lepik, A. (2014). Plant root exudates mediate neighbour recognition and trigger complex behavioural changes. New Phytol. 204, 631–637. doi: 10.1111/nph.12930
Seneviratne, M., Gunaratne, S., Bandara, T., Weerasundara, L., Rajakaruna, N., Seneviratne, G., et al. (2016). Plant growth promotion by Bradyrhizobium japonicum under heavy metal stress. South African J. Bot. 105, 19–24. doi: 10.1016/j.sajb.2016.02.206
Shah, A., Tyagi, S., Saratale, G. D., Guzik, U., Hu, A., Sreevathsa, R., et al. (2021). A comprehensive review on the influence of light on signaling cross-talk and molecular communication against phyto-microbiome interactions. Crit. Rev. Biotechnol. 41, 370–393. doi: 10.1080/07388551.2020.1869686
Shameer, S., and Prasad, T. N. V. K. V. (2018). Plant growth promoting rhizobacteria for sustainable agricultural practices with special reference to biotic and abiotic stresses. Plant Growth Regul. 84, 603–615. doi: 10.1007/s10725-017-0365-1
Sharma, A., Raina, M., Kumar, D., Singh, A., Chugh, S., Jain, S., et al. (2022). Harnessing phytomicrobiome signals for phytopathogenic stress management. J. Biosci. 47, 1–21. doi: 10.1007/s12038-021-00240-9
Sheth, R. U., Cabral, V., Chen, S. P., and Wang, H. H. (2016). Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 32, 189–200. doi: 10.1016/j.tig.2016.01.005
Shrivastava, P., and Kumar, R. (2015). Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 22, 123–131. doi: 10.1016/j.sjbs.2014.12.001
Shultana, R., Kee Zuan, A. T., Yusop, M. R., Saud, H. M., and El-Shehawi, A. M. (2021). Bacillus tequilensis strain ‘UPMRB9’improves biochemical attributes and nutrient accumulation in different rice varieties under salinity stress. PLoS One 16:e0260869. doi: 10.1371/journal.pone.0260869
Singh, A., Kumari, R., Yadav, A. N., Mishra, S., Sachan, A., and Sachan, S. G. (2020). “Tiny microbes, big yields: microorganisms for enhancing food crop production for sustainable development,” in New and Future Developments in Microbial Biotechnology and Bioengineering, eds. A. A Rastegari, A. N Yadav, and N. Yadav (Amsterdam: Elsevier), 1–15. doi: 10.1016/B978-0-12-820526-6.00001-4
Singh, D. P., Singh, V., Gupta, V. K., Shukla, R., Prabha, R., Sarma, B. K., et al. (2020). Microbial inoculation in rice regulates antioxidative reactions and defense related genes to mitigate drought stress. Sci. Rep. 10:4818. doi: 10.1038/s41598-020-61140-w
Singh, M., and Tiwari, N. (2021). Microbial amelioration of salinity stress in HD 2967 wheat cultivar by up-regulating antioxidant defense. Commun. Int. Biol. 14, 136–150. doi: 10.1080/19420889.2021.1937839
Singhal, P., Jan, A. T., Azam, M., and Haq, Q. M. R. (2016). Plant abiotic stress: a prospective strategy of exploiting promoters as alternative to overcome the escalating burden. Front. Life Sci. 9, 52–63. doi: 10.1080/21553769.2015.1077478
Sivasakthi, S., Usharani, G., and Saranraj, P. (2014). Biocontrol potentiality of plant growth promoting bacteria (PGPR)-Pseudomonas fluorescens and Bacillus subtilis: A review. Afr. J. Agri. Res. 9, 1265–1277.
Smith, D. L., Praslickova, D., and Ilangumaran, G. (2015). Inter-organismal signaling and management of the phytomicrobiome. Front. Plant Sci. 6:722. doi: 10.3389/fpls.2015.00722
Spence, C., and Bais, H. (2015). Role of plant growth regulators as chemical signals in plant–microbe interactions: a double edged sword. Curr. Opi. Plant Biol. 27, 52–58. doi: 10.1016/j.pbi.2015.05.028
Stanley, C. E., and van der Heijden, M. G. (2017). Microbiome-on-a-chip: new frontiers in plant–microbiota research. Trends Microbiol. 25, 610–613. doi: 10.1016/j.tim.2017.05.001
Staudinger, C., Mehmeti-Tershani, V., Gil-Quintana, E., Gonzalez, E. M., Hofhansl, F., Bachmann, G., et al. (2016). Evidence for a rhizobia-induced drought stress response strategy in Medicago truncatula. J. Proteom. 136, 202–213. doi: 10.1016/j.jprot.2016.01.006
Stringlis, I. A., Yu, K., Feussner, K., De Jonge, R., Van Bentum, S., Van Verk, M. C., et al. (2018). MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. U S A. 115, E5213–E5222. doi: 10.1073/pnas.1722335115
Suzuki, N., Rivero, R. M., Shulaev, V., Blumwald, E., and Mittler, R. (2014). Abiotic and biotic stress combinations. New Phytol. 203, 32–43. doi: 10.1111/nph.12797
Takishita, Y., Charron, J. B., and Smith, D. L. (2018). Biocontrol rhizobacterium Pseudomonas sp. 23S induces systemic resistance in tomato (Solanum lycopersicum L.) against bacterial canker Clavibacter michiganensis subsp. michiganensis. Front. Microbiol. 9:2119. doi: 10.3389/fmicb.2018.02119
Teixeira, P. J. P., Colaianni, N. R., Fitzpatrick, C. R., and Dangl, J. L. (2019). Beyond pathogens: microbiota interactions with the plant immune system. Curr. Opi. Microbiol. 49, 7–17. doi: 10.1016/j.mib.2019.08.003
Terhorst, C. P., Lennon, J. T., and Lau, J. A. (2014). The relative importance of rapid evolution for plant-microbe interactions depends on ecological context. Proc. Royal Society B: Biol. Sci. 281:20140028. doi: 10.1098/rspb.2014.0028
Thampi, A., and Bhai, R. S. (2017). Rhizospherea ctinobacteria for combating Phytophthora capsici and Sclerotium rolfsii, the major soil borne pathogens of black pepper (Piper nigrum L.). Biol. Control 109, 1–13. doi: 10.1016/j.biocontrol.2017.03.006
Theis, K. R., Dheilly, N. M., Klassen, J. L., Brucker, R. M., Baines, J. F., Bosch, T. C., et al. (2016). Getting the hologenome concept right: an eco-evolutionary framework for hosts and their microbiomes. Msystems 1:e00028-16. doi: 10.1128/mSystems.00028-16
Tiepo, A. N., Constantino, L. V., Madeira, T. B., Gonçalves, L. S. A., Pimenta, J. A., Bianchini, E., et al. (2020). Plant growth-promoting bacteria improve leaf antioxidant metabolism of drought-stressed neotropical trees. Planta 251:83. doi: 10.1007/s00425-020-03373-7
Tiwari, R., and Rana, C. S. (2015). Phytomedicine for the diabetes: a traditional approach. Ann. Phytomed. 4, 108–110.
Tiwari, S., Lata, C., Chauhan, P. S., and Nautiyal, C. S. (2016). Pseudomonas putida attunes morphophysiological, biochemical and molecular responses in Cicerarietinum L. during drought stress and recovery. Plant Physiol. Biochem. 99, 108–117. doi: 10.1016/j.plaphy.2015.11.001
Traxler, M. F., and Kolter, R. (2015). Natural products in soil microbe interactions and evolution. Nat. Prod. Rep. 32, 956–970. doi: 10.1039/C5NP00013K
Tsolakidou, M. D., Stringlis, I. A., Fanega-Sleziak, N., Papageorgiou, S., Tsalakou, A., and Pantelides, I. S. (2019). Rhizosphere-enriched microbes as a pool to design synthetic communities for reproducible beneficial outputs. FEMS Microbiol. Ecol. 95:fiz138. doi: 10.1093/femsec/fiz138
Vadakattu, G., and Germida, J. J. (2014). Soil aggregation: influence on microbial biomass and implications for biological processes. Res. J. Soil Biol. 80, A3–A9. doi: 10.1890/13-0616.1
Van Deynze, A., Zamora, P., Delaux, P. M., Heitmann, C., Jayaraman, D., Rajasekar, S., et al. (2018). Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLoS Biol. 16:e2006352. doi: 10.1371/journal.pbio.2006352
Vardharajula, S., Zulfikar Ali, S., Grover, M., Reddy, G., and Bandi, V. (2011). Drought-tolerant plant growth promoting Bacillus spp.: effect on growth, osmolytes, and antioxidant status of maize under drought stress. J. Plant Interact. 6, 1–14. doi: 10.1080/17429145.2010.535178
Verma, S. K., Kingsley, K., Bergen, M., English, C., Elmore, M., Kharwar, R. N., et al. (2018). Bacterial endophytes from rice cut grass (Leersia oryzoides L.) increase growth, promote root gravitropic response, stimulate root hair formation, and protect rice seedlings from disease. Plant Soil 422, 223–238. doi: 10.1007/s11104-017-3339-1
Vieira, F. C. S., and Nahas, E. (2005). Comparison of microbial numbers in soils by using various culture media and temperatures. Microbiol. Res. 160, 197–202. doi: 10.1016/j.micres.2005.01.004
Vimal, S. R., Patel, V. K., and Singh, J. S. (2019). Plant growth promoting Curtobacterium albidum strain SRV4: an agriculturally important microbe to alleviate salinity stress in paddy plants. Ecol. Ind. 105, 553–562. doi: 10.1016/j.ecolind.2018.05.014
Vorholt, J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840. doi: 10.1038/nrmicro2910
Wallenstein, M. D. (2017). Managing and manipulating the rhizosphere microbiome for plant health: a systems approach. Rhizosphere 3, 230–232. doi: 10.1016/j.rhisph.2017.04.004
Wang, C., Liu, D., and Bai, E. (2018). Decreasing soil microbial diversity is associated with decreasing microbial biomass under nitrogen addition. Soil Biol. Biochem. 120, 120–133. doi: 10.1016/j.soilbio.2018.02.003
Wang, Q., Dodd, I. C., Belimov, A. A., and Jiang, F. (2016). Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase growth and photosynthesis of pea plants under salt stress by limiting Na+ accumulation. Funct. Plant Biol. 43, 161–172. doi: 10.1071/FP15200
Woo, S. L., and Pepe, O. (2018). Microbial consortia: promising probiotics as plant biostimulants for sustainable agriculture. Front. Plant Sci. 9:1801. doi: 10.3389/fpls.2018.01801
Woo, S. L., Ruocco, M., Vinale, F., Nigro, M., Marra, R., Lombardi, N., et al. (2014). Trichoderma-based products and their widespread use in agriculture. Open Mycol. J. 8, 71–126. doi: 10.2174/1874437001408010071
Wu, D., Zhao, M., Shen, S., Fu, Y., Sasaki, T., Yamamoto, Y., et al. (2013). Al-induced secretion of organic acid, gene expression and root elongation in soybean roots. Acta Physiol. Plant. 35, 223–232. doi: 10.1007/s11738-012-1067-y
Wu, L., Chen, H., Curtis, C., and Fu, Z. Q. (2014). Go in for the kill: how plants deploy effector-triggered immunity to combat pathogens. Virulence 5, 710–721. doi: 10.4161/viru.29755
Xiang, G., Ma, W., Gao, S., Jin, Z., Yue, Q., and Yao, Y. (2019). Transcriptomic and phosphoproteomic profiling and metabolite analyses reveal the mechanism of NaHCO3-induced organic acid secretion in grapevine roots. BMC Plant Biol. 19:383. doi: 10.1186/s12870-019-1990-9
Xu, K., Lee, Y. S., Li, J., and Li, C. (2019). Resistance mechanisms and reprogramming of microorganisms for efficient biorefinery under multiple environmental stresses. Synthetic Systems Biotechnol. 4, 92–98. doi: 10.1016/j.synbio.2019.02.003
Xu, L., Naylor, D., Dong, Z., Simmons, T., Pierroz, G., Hixson, K. K., et al. (2018). Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl. Acad. Sci. U S A. 115, E4284–E4293. doi: 10.1073/pnas.1807275115
Yadav, A. N., Rastegari, A. A., Yadav, N., and Kour, D. (2020). Advances in Plant Microbiome and Sustainable Agriculture. Singapore: Springer. doi: 10.1007/978-981-15-3208-5
Yadav, V. K., Jha, R. K., Kaushik, P., Altalayan, F. H., Al Balawi, T., and Alam, P. (2021). Traversing arbuscular mycorrhizal fungi and Pseudomonas fluorescens for carrot production under salinity. Saudi J. Biol. Sci. 28, 4217–4223. doi: 10.1016/j.sjbs.2021.06.025
Yang, F., Zhang, J., Zhang, H., Ji, G., Zeng, L., Li, Y., et al. (2020). Bacterial blight induced shifts in endophytic microbiome of rice leaves and the enrichment of specific bacterial strains with pathogen antagonism. Front. Plant Sci. 11:963. doi: 10.3389/fpls.2020.00963
Yang, J. L., Fan, W., and Zheng, S. J. (2019). Mechanisms and regulation of aluminum-induced secretion of organic acid anions from plant roots. J. Zhejiang University-SCIENCE B 20, 513–527. doi: 10.1631/jzus.B1900188
Young, I. M., and Crawford, J. W. (2004). Interactions and self-organization in the soil-microbe complex. Science 304, 1634–1637. doi: 10.1126/science.1097394
Yu, K., Liu, Y., Tichelaar, R., Savant, N., Lagendijk, E., van Kuijk, S. J., et al. (2019). Rhizosphere-associated Pseudomonas suppress local root immune responses by gluconic acid-mediated lowering of environmental pH. Curr. Biol. 29, 3913–3920. doi: 10.1016/j.cub.2019.09.015
Yuan, J., Zhao, J., Wen, T., Zhao, M., Li, R., Goossens, P., et al. (2018). Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome 6, 1–12. doi: 10.1186/s40168-018-0537-x
Zamioudis, C., and Pieterse, C. M. (2012). Modulation of host immunity by beneficial microbes. Mol. Plant-Microbe Interac. 25, 139–150. doi: 10.1094/MPMI-06-11-0179
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhang, J., Liu, Y. X., Zhang, N., Hu, B., Jin, T., Xu, H., et al. (2019). NRT1. 1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 37, 676–684. doi: 10.1038/s41587-019-0104-4
Zhang, J., Wang, L. H., Yang, J. C., Liu, H., and Dai, J. L. (2015). Health risk to residents and stimulation to inherent bacteria of various heavy metals in soil. Sci. Totl. Environ. 508, 29–36. doi: 10.1016/j.scitotenv.2014.11.064
Zhang, R., Vivanco, J. M., and Shen, Q. (2017). The unseen rhizosphere root–soil–microbe interactions for crop production. Curr. Opi. Microbiol. 37, 8–14. doi: 10.1016/j.mib.2017.03.008
Zhang, Y., and Zhuang, W. Y. (2020). Trichoderma brevicrassum strain TC967 with capacities of diminishing cucumber disease caused by Rhizoctonia solani and promoting plant growth. Biol. Control 142:104151. doi: 10.1016/j.biocontrol.2019.104151
Zia, R., Nawaz, M. S., Siddique, M. J., Hakim, S., and Imran, A. (2020). Plant survival under drought stress: implications, adaptive responses, and integrated rhizosphere management strategy for stress mitigation. Microbiol. Res. 242:126626. doi: 10.1016/j.micres.2020.126626
Złoch, M., Thiem, D., Gadzała-Kopciuch, R., and Hrynkiewicz, K. (2016). Synthesis of siderophores by plant-associated metallotolerant bacteria under exposure to Cd2+. Chemosphere 156, 312–325. doi: 10.1016/j.chemosphere.2016.04.130
Keywords: phytomicrobiome, rhizosphere, stresses, metabolomics, metagenomics, defense mechanisms, plant probiotics
Citation: Khanna K, Kohli SK, Sharma N, Kour J, Devi K, Bhardwaj T, Dhiman S, Singh AD, Sharma N, Sharma A, Ohri P, Bhardwaj R, Ahmad P, Alam P and Albalawi TH (2022) Phytomicrobiome communications: Novel implications for stress resistance in plants. Front. Microbiol. 13:912701. doi: 10.3389/fmicb.2022.912701
Received: 04 April 2022; Accepted: 15 July 2022;
Published: 06 October 2022.
Edited by:
Mahaveer P. Sharma, ICAR Indian Institute of Soybean Research, IndiaReviewed by:
Riyazali Zafarali Sayyed, P.S.G.V.P.M’s Arts, Science and Commerce College, IndiaCopyright © 2022 Khanna, Kohli, Sharma, Kour, Devi, Bhardwaj, Dhiman, Singh, Sharma, Sharma, Ohri, Bhardwaj, Ahmad, Alam and Albalawi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kanika Khanna, a2FuaWthLjI3NTkwQGdtYWlsLmNvbQ==; Renu Bhardwaj, cmVudWJoYXJkd2FqODJAZ21haWwuY29t; Parvaiz Ahmad, cGFydmFpemJvdEB5YWhvby5jb20=; Pravej Alam, YWxhbXByZXpAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.