- 1Key Laboratory of Food Quality and Safety, School of Food Science and Biotechnology, Zhejiang Gongshang University, Hangzhou, China
- 2The Second Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, China
This research aimed to investigate the microbial composition and diversity of antimicrobial resistance genes (ARGs) found in Chinese broiler and layer family poultry farms. We focused on the differences in resistance phenotypes and genotypes of multidrug-resistant Enterobacteriaceae (MDRE) isolated from the two farming environments and the existence and transmissibility of colistin resistance gene mcr-1. Metagenomic analysis showed that Firmicutes and Bacteroides were the dominant bacteria in broiler and layer farms. Many aminoglycoside and tetracycline resistance genes were accumulated in these environments, and their absolute abundance was higher in broiler than in layer farms. A total of 526 MDRE were isolated with a similar distribution in both farms. The results of the K-B test showed that the resistance rate to seven antimicrobials including polymyxin B and meropenem in broiler poultry farms was significantly higher than that in layer poultry farms (P ≤ 0.05). PCR screening results revealed that the detection rates of mcr-1, aph(3’)Ia, aadA2, blaoxa–1, blaCTX–M, fosB, qnrD, sul1, tetA, and catA1 in broiler source MDRE were significantly higher than those in layers (P ≤0.05). A chimeric plasmid p20432-mcr which carried the novel integron In1866 was isolated from broiler source MDRE. The high frequency of conjugation (10–1 to 10–3) and a wide range of hosts made p20432-mcr likely to play an essential role in the high detection rate of mcr-1, aph(3’)-Ia, and aadA2 in broiler farms. These findings will help optimize disinfection and improve antimicrobial-resistant bacteria surveillance programs in poultry farms, especially broilers.
Introduction
Antimicrobial resistance has been characterized as a global public health concern, catalyzed by the over-use of antimicrobials used in farming food-producing animals (Winglee et al., 2017; KöCk et al., 2018). Tons of antimicrobials are used annually as treatments and growth promoters in livestock and poultry farms (Udikovic-Kolic et al., 2014; Xiong et al., 2018). It led to the accumulation of antimicrobials in the farming environments, creating the constant selective pressure for antimicrobial resistance to microorganisms (Udikovic-Kolic et al., 2014), and making poultry a reservoir for resistant bacteria and genes (Mceachran et al., 2015). China has the largest layer chicken industry globally and is the second-largest producer of broiler chickens (Gaze et al., 2013; FAO, 2019). In addition to the large-scale poultry farms, family poultry farms are a typical farming pattern with Chinese characteristics. Especially in rural areas, residential buildings and poultry sheds are often housed in the same yard, facilitating poultry management for farmers. More importantly, it reduces the space needed for independent farms and lowers the threshold for poultry farming. Because of the compact space and frequent contact, farmers face a higher risk of antimicrobial resistance in such family farms. These industries cause massive pollution to the environment by animal excretion. Family farming puts people in close contact with breeding environments, making it easier for antimicrobial-resistant bacteria to spread to people and threaten human health (Wei et al., 2013). The prevalence of resistant bacteria often isolated from feces turn farms into the primary sources of spreading resistant bacteria (Rawat et al., 2018). ARGs are mainly derived from the Enterobacteriaceae, which have become increasingly common in human and animal infections (Falagas et al., 2011). Polymyxin is generally considered the last strategy for treating multidrug-resistant Enterobacteriaceae (MDRE) (Wang et al., 2017). Although the application of colistin as a growth-promotor in livestock and poultry production has been banned in China (Shi et al., 2020), the prevalence of colistin resistance genes and increasing rates of MDRE (Buelow et al., 2017) in poultry products remains a significant risk to public health that may jeopardize the treatment of serious bacterial infections (Belluco et al., 2016; Zhai et al., 2020).
Most studies on antimicrobial resistance in poultry production have focused on antimicrobial residues and specific ARGs, and few studies have compared broilers and layers (Zhai et al., 2020). There is a gap in understanding the differences in bacterial community structure, antimicrobial resistome, and mobile genetic elements (MGEs) in layer and broiler chicken environments. It is crucial to assess resistance-transmission risk in family poultry farms carefully.
Previous studies on different poultry (broilers and layers) farms suggest that robust screening of antimicrobials leads to the spreading of antimicrobial resistance genes (ARGs) and entering the human food chain (Su et al., 2015). Therefore, the transmission of resistant genes and their hosts in the farm environment needs to be monitored more frequently. This study aimed to analyze the abundance of ARGs and their potential bacterial hosts in the microbial communities in broilers and layers breeding environments and explore the ARGs’ potential transmission risks in breeding poultry and human, as determined by metagenomics sequencing combining analysis of mcr-1 positive MGEs. These findings will provide a more comprehensive and accurate theoretical basis for risk assessment of antimicrobial resistance in family poultry patterns in China and control further spread of antimicrobial resistance and protect farmers.
Materials and methods
Study area and sample collection
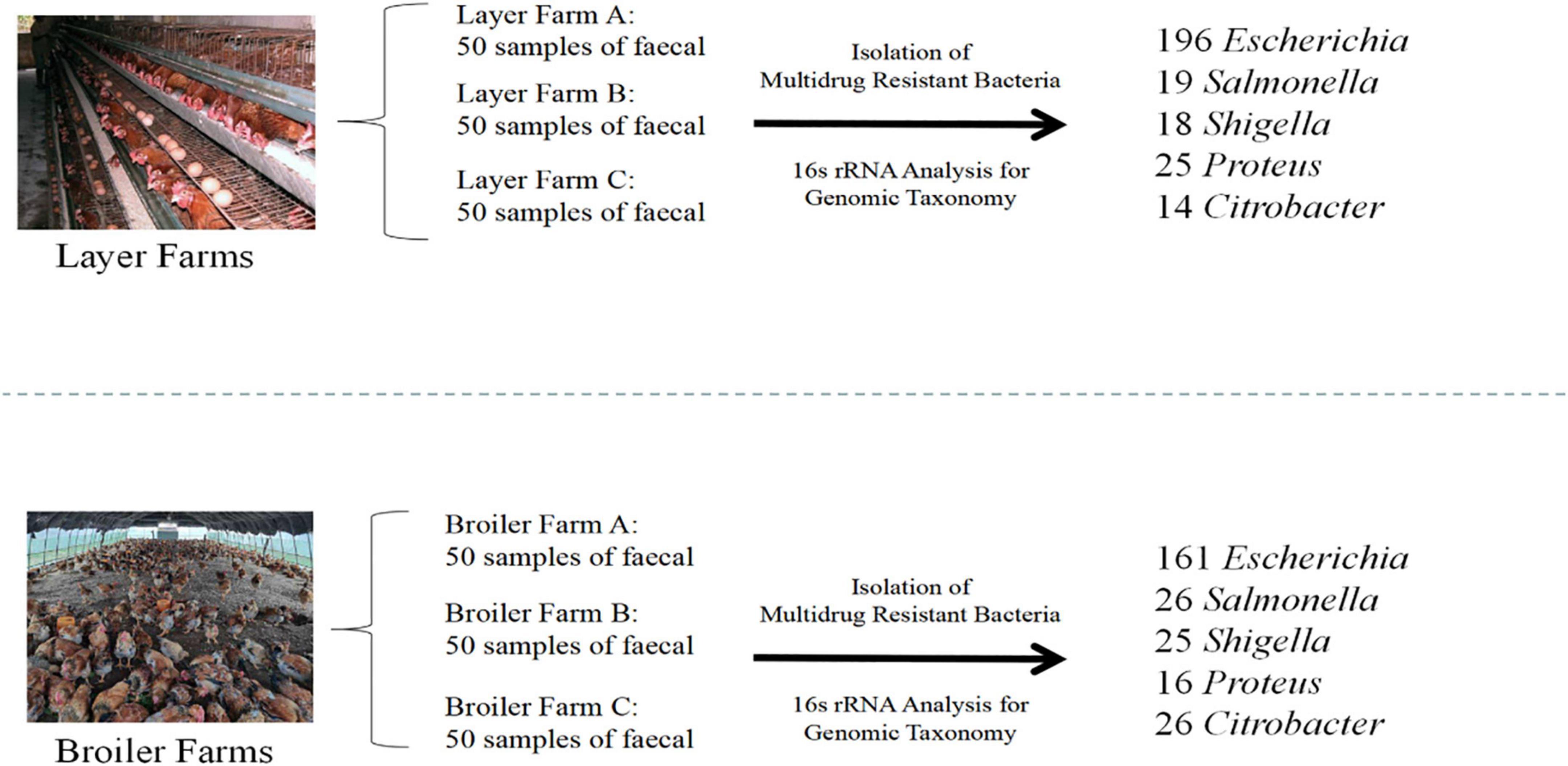
Six broilers and layers poultry farms were selected for a comparative study. All farms were located in Jiaxing, Zhejiang Province. All the six environments were small family farms with chicken houses, which is very common in local areas. Each farm had four to six individual chicken houses and each house contained ∼80 chickens. Twenty fecal samples were randomly collected from each chicken house by swab. A total of 150 fecal samples were collected from broiler farms (BFs), and 150 fecal samples were collected from layer farms (LFs) in September 2021. In addition, soil (n = 3), water (n = 3), and feces (n = 3) samples were collected from broiler farms and mixed into a composite sample. The same sampling scheme was also used for layer farms. Two composite samples were used for metagenomic analysis. All samples were transported to the laboratory in normal saline within 8 h to isolate Enterobacteriaceae, and each sample enriched in 10 mL of Buffered Peptone Water (BPW, Macklin, China) and incubated overnight at 37°C. MacConkey (MAC) agar (Macklin, China) was used to cultivate Enterobacteriaceae strains. A suspected colony of each microorganism was isolated on a slope agar and PCR was used for subsequent identification.
Metagenomic analysis
Metagenomic sequencing was performed using Illumina Hiseq 4000 with the sequencing strategy of Index 250 PE (paired-end sequencing). Raw data is filtered using fastp (Chen et al., 2018) to correct base and quality-filtered. Low-quality reads were filtered to ensure that (1) the reads were aligned to proper adaptors or primers, (2) the reads contained <10% unknown bases, (3) the reads contained >50% high-quality bases (2013). Bowtie2 (Langmead and Salzberg, 2012) was used to remove reads from the host genome or contamination with high similarity. On average, 9.5 GB of clean reads were generated for each sample. ARG-OAP PROTO (Yin et al., 2018), an analysis pipeline that integrates two common resistance gene databases, including ARDB (Liu and Pop, 2009) and CARD (Alcock et al., 2019), identifies assembly sequences based on the hidden Markov model (HMM) was used to characterize the profile and quantify the relative abundance of ARGs.
Antimicrobial susceptibility test
The antimicrobial susceptibility of isolated bacteria was tested with the broth dilution method which is recommended by the Clinical and Laboratory Standards Institute (Institute CLSI, 2020) on Mueller-Hinton (MH) broth and evaluated based on the breakpoint and classified as resistant, intermediate, or susceptible (the breakpoints were shown in Supplementary Table 1). The strains exhibiting resistance to three or more classes of antimicrobials were considered to be multidrug-resistant organisms.
PCR amplification and sequencing of 16S rRNA
The isolated bacteria were identified by VITEK-2 Compact and 16S rRNA gene identification using the universal 16S primers 27F and 1492R (Lane, 1991). The system amplified by 5 μL PCR was added into 1.2% agarose gel for electrophoresis, and the voltage was set at 120 V for 30 min. PCR amplification consisted of denaturation at 96°C for 5 min followed by denaturation at 95°C for 30 s, annealing at 50°C for 90 s, and polymerization at 72°C for 60 s for a total of 30 cycles, and a final extension at 72°C for 5 min. A Western blot detection system was used to observe whether agarose gel had corresponding bands at 1500 bp after electrophoresis. PCR product of successful amplification was used paired-end sequencing (2 × 150 bp) by Sangon Biotech (Shanghai, China).
PCR amplification and sequencing of antimicrobial resistance genes
The isolated bacteria were screened for nine major classes of antimicrobial resistance genes usually used for human clinical treatment or poultry by PCR using the primers listed in Supplementary Table 2. PCR amplification was carried out as follow: initial denaturation at 95°C for 5 min, denaturation at 94°C for 30 s, annealing at their respective annealing temperature for 30 s, and polymerization at 72°C for 40 s for a total of 30 cycles, and a final extension at 27°C for 10 min. All the PCR products were subjected to Sanger sequencing.
Conjugation experiments
To access the transferability of mcr-1 positive plasmids, conjugal transfer experiments were performed with rifampin-resistant E. coli strain EC600 used as a recipient and each of the mcr-1 positive RB20432 and DB20D28 isolates as a donor. The donor was cultured on Brain Heart Infusion (BHI) agar plates with 2 μg/ml polymyxin, and the recipient E. coli strain EC 600 was cultivated on BHI agar plates with 2.5 mg/mL rifampicin. After incubation at 37 C for 24 h, colonies growing on selective plates were further confirmed by antimicrobial sensitivity experiments and VITEK-2 Compact.
Sequencing and analysis of plasmids
The two conjugative plasmids of the transconjugants RB20432-EC600 and DB20D28-EC600 were sequenced by the Illumina HiSeq platforms. The two plasmid DNA was extracted from the transconjugants RB20432-EC600 and DB20D28-EC600 using the SanPrep Column Plasmid Mini-Preps Kit. After extraction, plasmid sequencing was performed with a pair-end library with an average insert size of 300 bp on a Hiseq sequencer (Illumina). Quality control and removing low-quality data were performed with Trim Momatic 0.36 (Bolger et al., 2014). Gapcloser (Nederbragt, 2014) was used to fill gaps. Cytoscape (Smoot et al., 2011) was used to spline the sequence to obtain the final cyclized plasmids. The plasmid sequences were submitted to Rast (Aziz et al., 2008), Genemarks, Glimmer, and Prodigal library for preliminary gene prediction then submitted to ISFinder, Tn Number Registry, and INTEGRALL (Moura et al., 2009) for MGEs.
Statistical analysis
The data of bacterial and ARGs were processed with Microsoft Excel 2016. The column was generated using Origin 2021. The heat map for the resistance profile of MDRE was visualized via Heatmapper1 (Sasha et al., 2016). Antimicrobial susceptibility and resistance gene detection of MDRE isolates were analyzed by independent sample t-tests and SPSS ver.21, and P ≤ 0.05 was considered statistically significant. Running a gene alignment program in Perl language with Ubuntu 18.04 LTS1 and Inkscape 0.48.1 were used to draw gene organization diagrams.
Results
Characterization of bacterial and antimicrobial resistance gene communities in layer farms and broiler farms
The top ten relative abundance of the microbial communities at the phylum levels are shown in Figure 1. Firmicutes and Bacteroidetes were the top two in all bacteria phyla, with 55.16% and 32.58% in the BF samples, 44.72% and 41.88% in the LF samples. It can be inferred that Firmicutes and Bacteroidetes are the dominant bacteria in broiler and layer breeding environments, which were potential hosts for ARGs. Actinobacteria, Proteobacteria, and Synergistetes were the second dominant phyla.
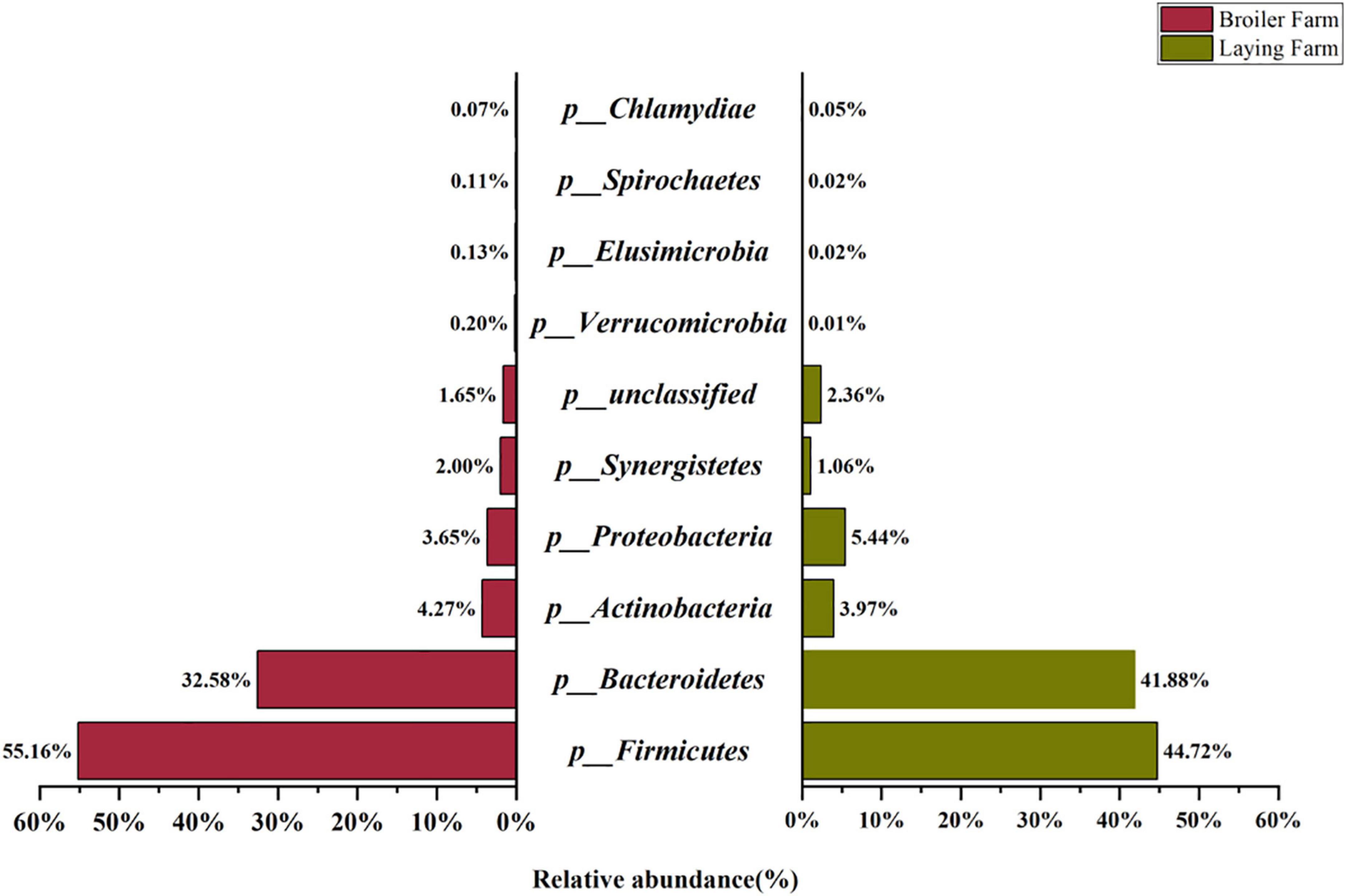
Figure 1. Top ten relative abundance of taxonomic composition at the phylum level in broiler and laying farm samples.
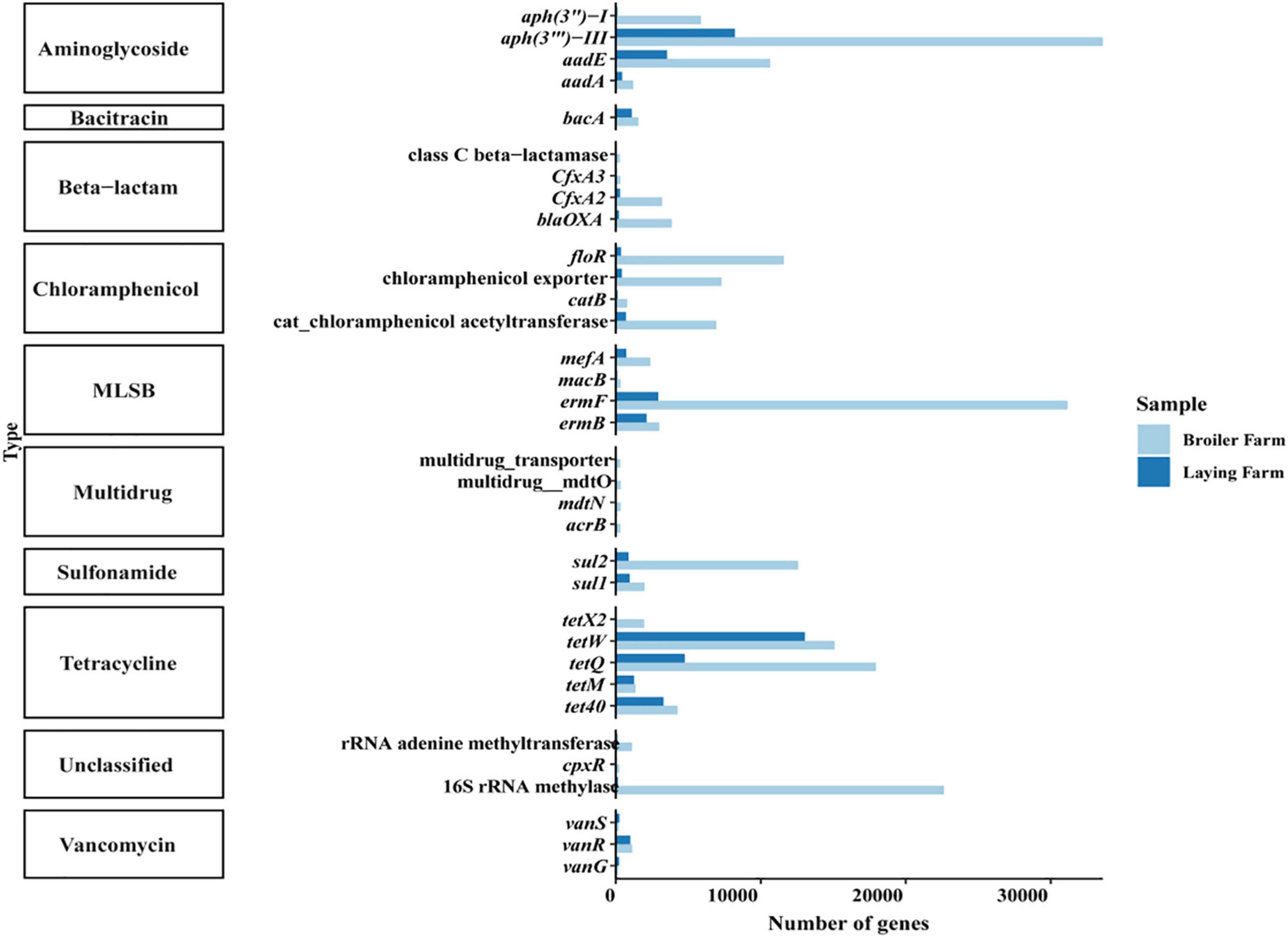
The top ten antimicrobial resistance genes in broiler and laying farms in absolute abundance were aminoglycosides, bacitracin, β-lactam, chloramphenicol, MLSBs (macrolide-lincosamide-streptomycin B), multidrug resistance genes, sulfonamides, tetracyclines, vancomycin and unclassified ARGs (Figure 2). ARGs of all ten classifications were generally detected in broilers, while MDR_transporter and cpxR were not detected in layers. Many aminoglycoside and tetracycline resistance genes were accumulated in broilers and layers, and their absolute abundance in broilers was higher than that in layers. In broiler samples, aph(3″′)-III had the highest absolute abundance, followed by ermF, 16S rRNA methylase, and tetQ. The absolute abundance of tetW in the laying samples was the highest, followed by aph(3″′)-III, tetQ, and aadE. The absolute abundance of vancomycin resistance genes vanG and vanS in the laying samples was higher than in broiler samples.
Detection of multidrug-resistant Enterobacteriaceae in layer farms and broiler farms
Five hundred twenty-six MDR Enterobacteriaceae were recovered from 300 fresh fecal samples, 272 in BFs and 254 in LFs. According to the general prevalence of MDRE layers and broilers, E. coli, whose percent reached 67.8% (357/526), was the dominant bacteria. The rest were Salmonella (8.6%, 45/526), Shigella (8.2%, 43/526), Proteus (7.8%, 41/526), and Citrobacter (7.6%, 40/526) (Figure 3).
Differences in antimicrobial-resistant phenotypes of MDRE in broiler farms and layer farms
MDRE isolated from 6 farms showed different resistance rates to 25 antimicrobials (Figure 4). Amoxicillin had the highest resistance rate among the 25 antimicrobials, 89.33% in BFs and 94.3% in LFs. In addition, MDRE in BFs and LFs showed more than 80% resistance rates to five antimicrobials: Amoxicillin, sulfamethoxazole, trimethoprim-sulfamethoxazole, and tetracycline. Comparison between the two farms showed that resistance rates of polymyxin B, meropenem, amikacin, gentamicin, ciprofloxacin, sulfamethoxazole, and chloramphenicol were significantly higher in BFs than in LFs (P ≤ 0.05). Polymyxin B, banned in poultry production in recent years, has a high resistance rate in BFs, indicating that its resistance gene has been widely spread in the broiler environment.
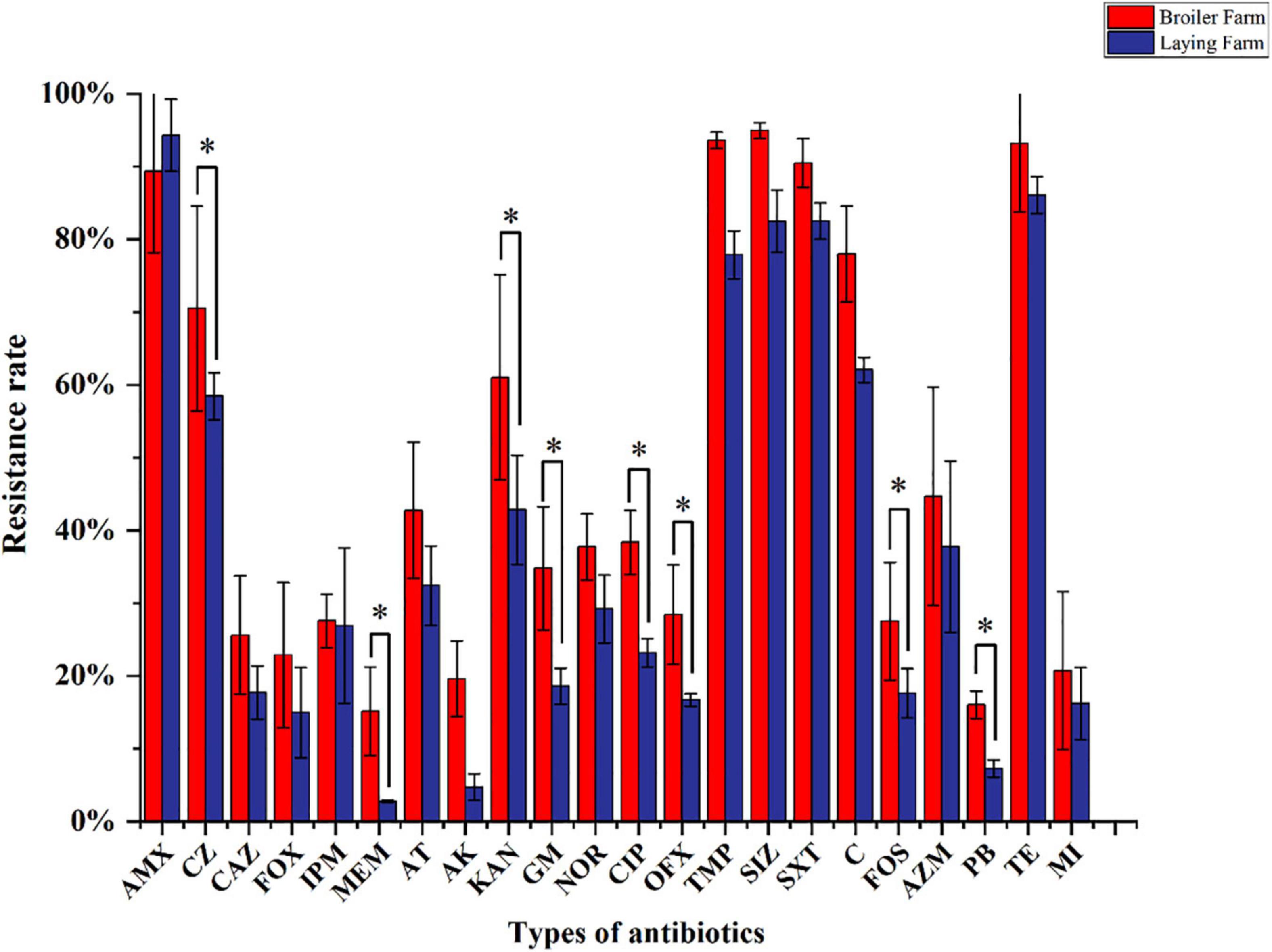
Figure 4. Resistance rates of MDRE isolated from broilers and layers to 25 antimicrobials. AMX, Amoxicillin; CZ, Cefazolin; CAZ, Ceftazidime; FOX, Cefoxitin; IPM, Imipenem; MEM, Meropenem; AT, Aztreonam; AK, Amikacin; KAN, Kanamycin; GM, Gentamicin; CIP, Ciprofloxacin; NOR, Norfloxacin; OFX, Ofloxacin; TMP, Trimethoprim; SIZ, Sulfamethoxazole; SXT, Trimethoprim-Sulfamethoxazole; C, Chloramphenicol; FOS, Fosfomycin; AZM, Azithromycin; PB, Polymyxin B; TE, Tetracycline; MI, Minocycline.
Antimicrobial resistance of Enterobacteriaceae in broilers was more severe than in layers
To reflect the multidrug-resistance profile of MDRE in the culture environment more directly, they were divided into three resistance levels: mild antimicrobial resistance (4R∼9R, the number represents the variety of antimicrobials, and “R” represents “Resistance”), moderate antimicrobial resistance (10R∼16R), and severe antimicrobial resistance (17R∼22R) according to the variety of antimicrobials which MDRE was resistance to Figure 5. The proportion of moderate and severe resistance MDRE in BFs was higher than LFs, indicating that the MDR profile of Enterobacteriaceae in BFs was more severe than LFs. Most of the MDRE in both LFs and BFs were at the level of moderate antimicrobial resistance, based on which the resistance spectrum of BFs and LFs was deduced. The MDRE in BFs were mainly resistant to ciprofloxacin, cefazolin, kanamycin, minocycline, polymyxin B, and trimethoprim (Figure 6A). However, the heatmaps showed that the MDRE in LFs were most resistant to ofloxacin, fosfomycin, aztreonam, ceftazidime, polymyxin B, and tetracycline (Figure 6B). It should be noted that most 11R and 12R MDRE which have a large base in LFs and BFs were resistant to polymyxin B. Severe antimicrobial resistance MDRE especially 21R and 22R in BFs, was resistant to polymyxin B, meropenem, and imipenem, which should be monitored more frequently. In addition, tetracycline, macrolides, sulfonamides in folic acid metabolic pathway inhibitors, and amoxicillin in β-lactam were resistant; these antimicrobials should be avoided in poultry.
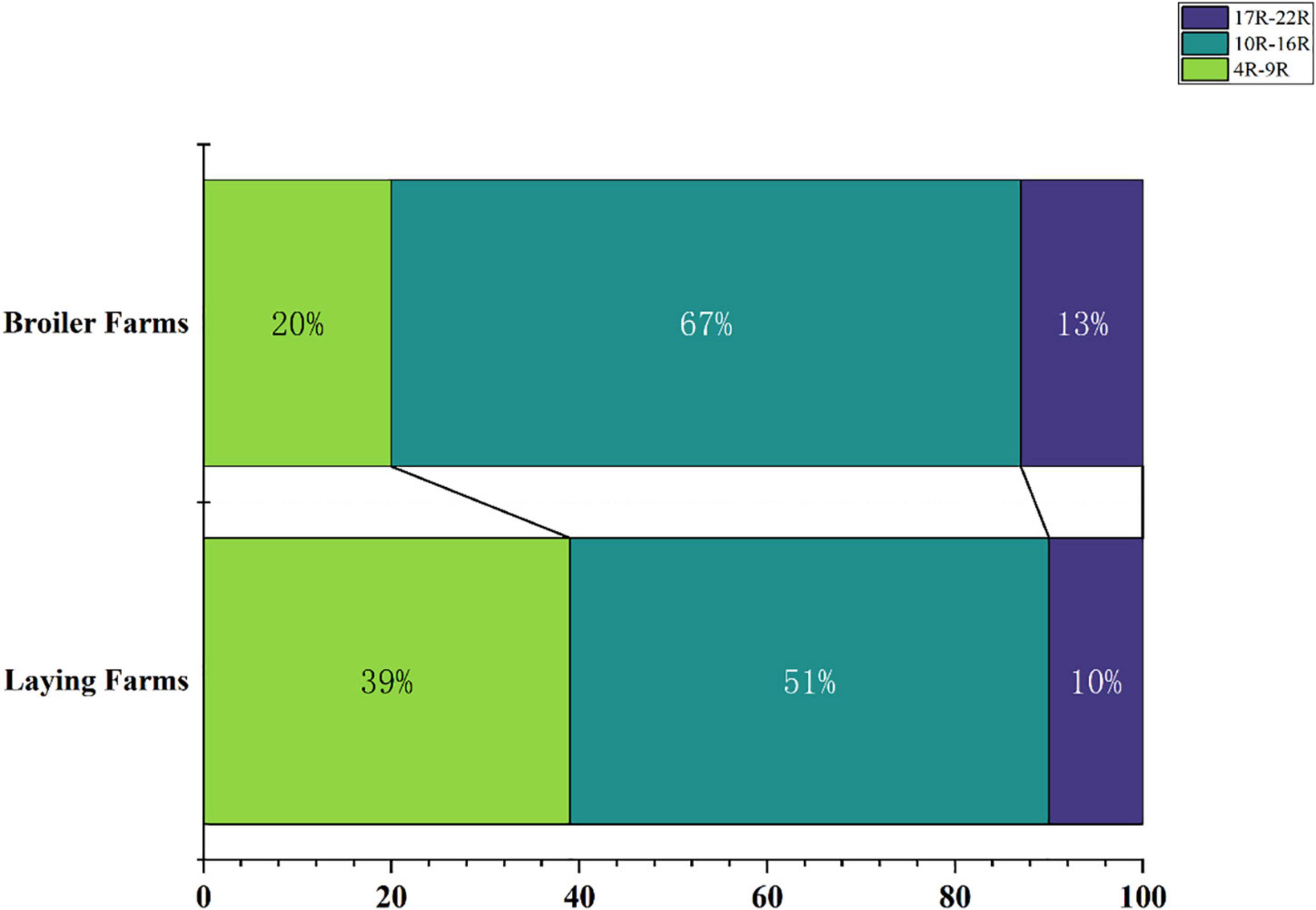
Figure 5. Proportion of resistant bacteria in different drug resistance situations in broiler farms and laying farms.
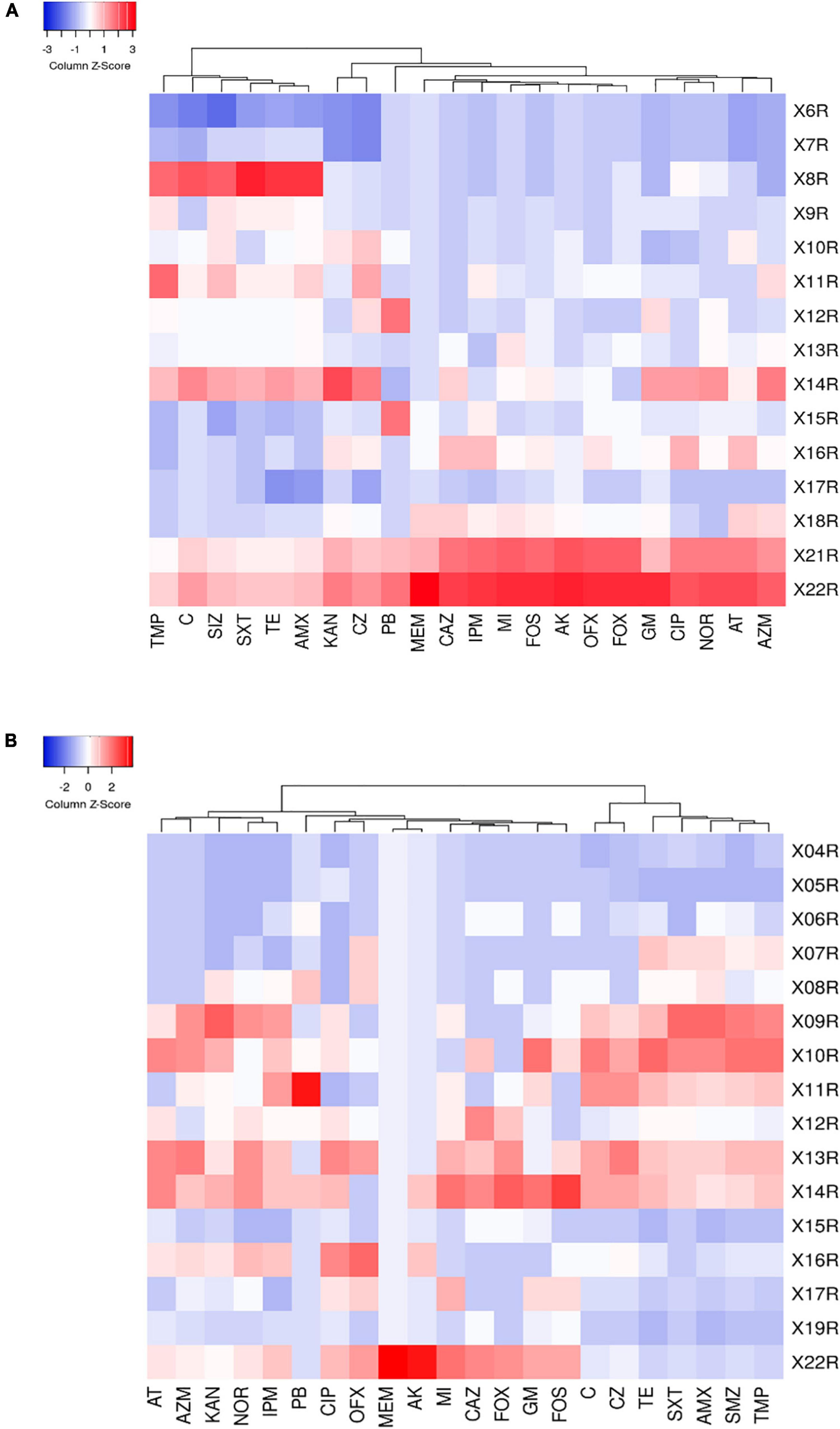
Figure 6. (A) Heat map of multidrug-resistance of resistant strains in broiler farms. (B) Heat map of multidrug-resistance of resistant strains in laying farms. The right column shows the number of antimicrobials which were resisted by MDRE.
Differences in antimicrobial resistance genes detection between broiler and layer farms
To investigate the prevalence of ARGs in the poultry environment, resistance genes of nine categories of antimicrobials were detected by PCR analysis. The detection rates of ARGs, blaTEM (β-lactamases), tetA (tetracycline), sul1 (sulfonamides), floR (chloramphenicol), fosA3 (fosfomycin), mph(A) (macrolides), mcr-1 (polymyxins), qnrS (quinolones) and aac(6′)-Ib- cr (aminoglycoside), were significantly higher than the other ARGs in the same category (P ≤ 0.05). The detection rates of each gene are detailed in Supplementary Data 1 and Supplementary Figure 1. In addition, we focused on antimicrobial resistance genes that are banned in poultry production and used differently in layers and broilers for subsequent analysis. Some of these antimicrobials, such as carbapenems and polymyxin, are only available for clinical use but not for use in animals. To avoid antimicrobial residues in eggs, some antimicrobials are banned during the laying period, such as tetracycline, quinolones (danofloxacin, pefloxacin, norfloxacin, ciprofloxacin), macrolides (kitasamycin), β-lactam (amoxicillin, ampicillin), sulfonamides (sulfachlorpyridazine sodium), polymyxin (polymyxin B).
Three kinds of tetracycline resistance genes were detected, and the detection rates of tetA were all above 60% in LFs and BFs. The detection rate of tetA in BFs was significantly higher than in LFs (P ≤ 0.05). Although tetD was detected in both LFs and BFs, there was no significant (P > 0.05) difference (Figure 7A). MDRE containing quinolone resistance genes qnrA, qnrB, qnrD, qnrS were detected in LFs and BFs. There is a significant difference in the detection rates of qnrD between BFs and LFs (P ≤ 0.05), and the detection rates of qnrA, qnrB, and qnrD were all higher in BFs. In contrast, the detection rate of qnrS was higher in LFs than in BFs (Figure 7B). Among the macrolide resistance genes, the detection rate of mph(A) was the highest in broiler and layer farms, both exceeding 60% (Figure 7C). The detection rates from high to low were mph(A), mph(E), mph(B), mph(D), ermA, respectively, and other macrolide resistance genes were not detected. The detection rate of mph(B) in BFs was significantly higher (P ≤ 0.05). Only three β-lactam resistance genes were identified: blaTEM, blaOXA–1, and blaCTX–M. Except that blaTEM had high detection rates in both broiler and layer farms, the detection rates of blaOXA–1 and blaCTX–M in broiler farms were significantly higher than those in LFs (P ≤ 0.05) (Figure 7D). Among the sulfonamides resistance genes, only sul1 and folp1 were amplified in MDRE. The total detection rate of sul1 was higher than folp1 in BFs and LFs (Figure 7E). Moreover, the detection rate of sul1 in BFs was significantly higher than that in LFs (P ≤ 0.01). The detection rate of polymyxin resistance gene in broiler and layer farms was not high, and mcr-1 was detected in both farms and was significantly higher in BFs than in LFs (P ≤ 0.01) (Figure 7F). However, mcr-2 was only detected in BFs, and the detection rate was 0.32%.
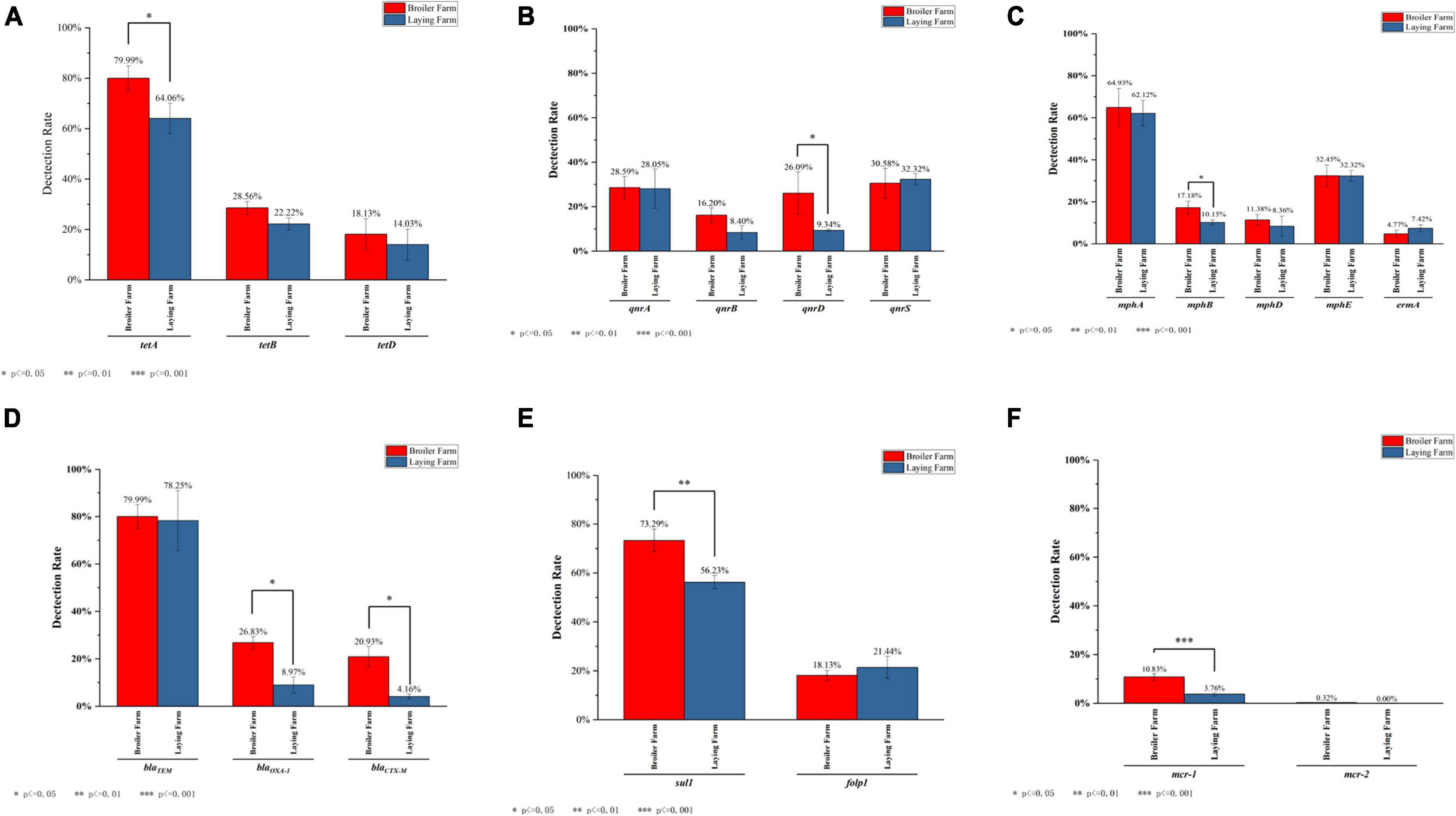
Figure 7. The detection rate of ARGs which are banned in layers or both. (A) The detection rate of tetracyclines resistance genes. (B) The detection rate of quinolones resistance genes. (C) The detection rate of macrolides resistance genes. (D) The detection rate of β-lactam resistance genes. (E) The detection rate of sulfonamide resistance genes. (F) The detection rate of polymyxins resistance genes.
The mcr-1 positive plasmids p20432-mcr and p2021-mcr were conjugative
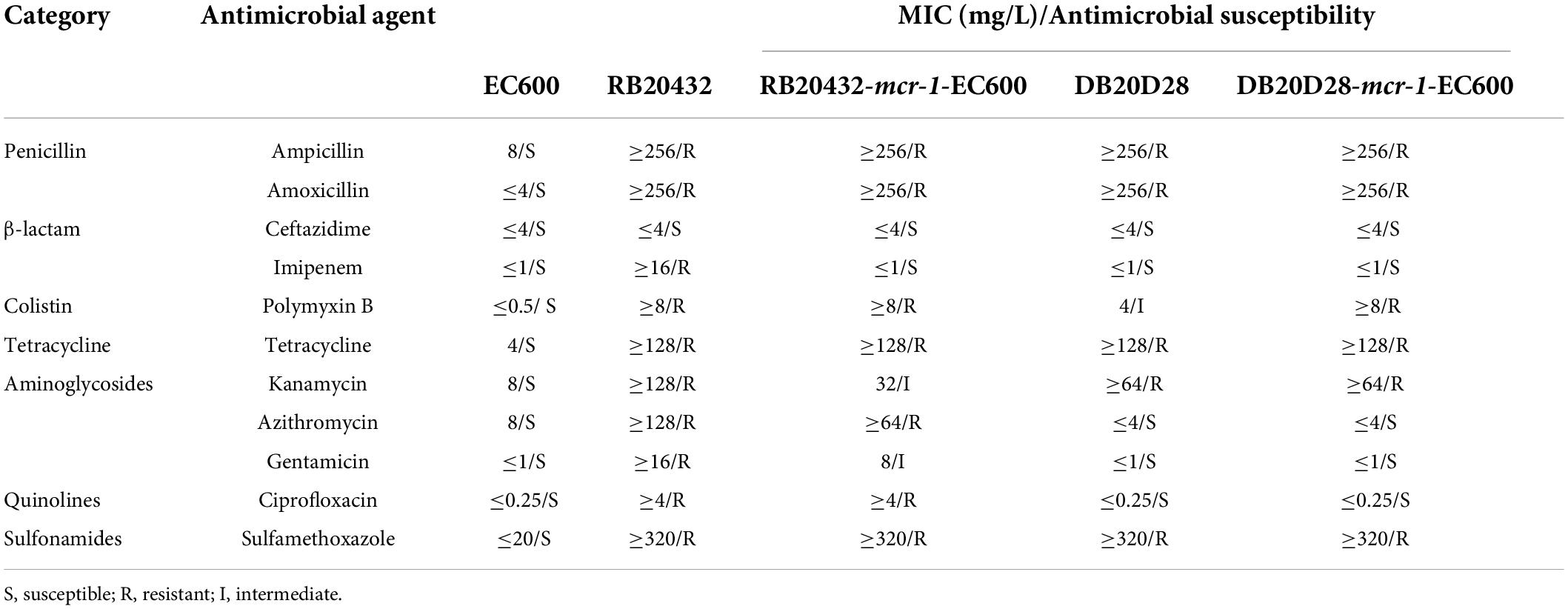
The transconjugants isolated by mating RB20432 and DB20D28 strains with EC600 were resistant to rifampicin and polymyxin antimicrobials, possibly indicating plasmids transfer. Experiments showed that polymyxin resistance could be successfully transferred from RB20432 and DB20DB to EC600 at 10–1 to 10–3 (p20432-mcr) and 10–2 to 10–3 (p2021-mcr) cell per recipient cell by conjugation. As shown in the table (Table 1), the minimum inhibitory concentrations for polymyxin B for the transconjugant increased 16-fold compared with the untransformed control. In addition, the minimum inhibitory concentration for ampicillin (32-fold), amoxicillin (32-fold), tetracycline (32-fold), gentamicin (eightfold, only RB20432), and sulfamethoxazole (16-fold) were increased to different degrees, suggesting that resistance genes of these antimicrobials also existed in p20432-mcr and p2021-mcr and were transferred through conjugation.
Characterization of multidrug-resistant plasmids p20432-mcr and p2021-mcr
Two mcr-1 positive multidrug-resistant plasmids p20432-mcr and p2021-mcr were found in the polymyxin-resistant MDRE, isolated from layers and broilers fecal samples, respectively. As shown in Figure 8, the total length of plasmid 20432-mcr was 228.7 kb, containing 275 ORFs. Furthermore, it had a complex region with two replicons (IncHI2 and IncN), and the backbone was interrupted by accessory modules. The content of GC was 47.3%, and the plasmid contained four accessory modules which were MDR-1 region, MDR-2 region, In1866-related region, and metal resistant region. The single replicon IncN plasmid R46 (GenBank: AY046276.1) was chosen as the reference plasmid with similar repA, intI1, and resistance genes.
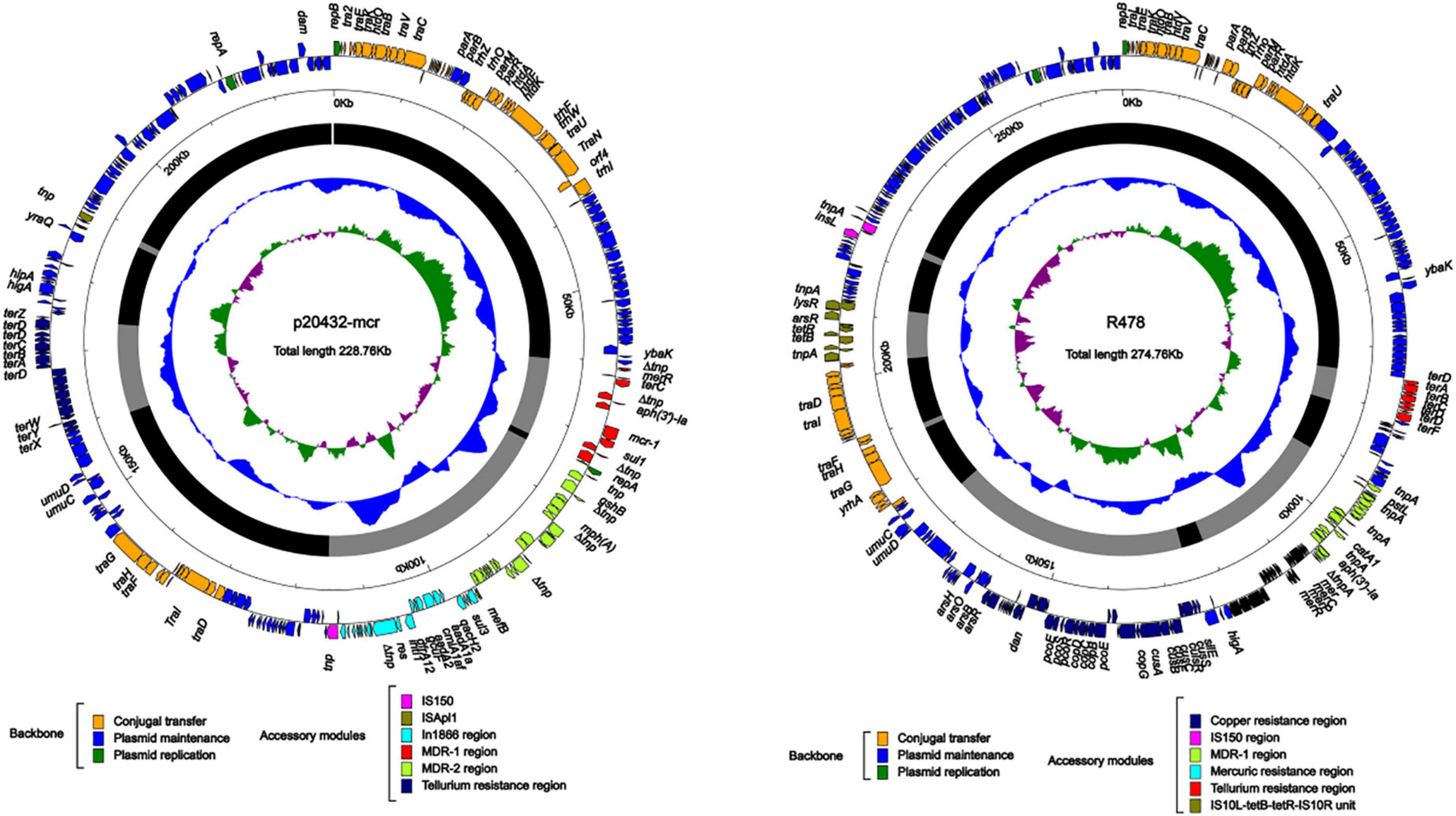
Figure 8. The complete sequence circular diagram of p20432-mcr and R478. The innermost ring represents (G–C)/(G + C); the blue ring indicates GC content, and the outward depression indicates that the GC content is higher than the mean value. Black area in the outer circle represents the backbone area and gray area represents the accessory modules. The outermost circle shows the distribution of genes represented by colored arrows in the plasmid.
The IncI2-type plasmid p2021-mcr was extracted from E. coli strain DB20D28, with a length of 60.2 kb, the content was 42.3%, and a total of 78 ORFs (Figure 9A). Its structure was relatively simple as the whole plasmid consisted of a backbone region and a single mcr-1-pap2. mcr-1 was the only ARG in p2021-mcr. The conjugation transfer region accounted for a large proportion of the plasmid, including flagellum formation gene pilP and conjugation transfer regulation gene virB9. Among them, the Shufflon DNA-specific recombinase secreted by the rci gene in this structure is an essential member of the recombinant system of the IncI plasmid, which obtained exogenous DNA through specific recombination. While there were no MGEs such as IS, Tn, and integron in p2021-mcr, mcr-1 was the only ARG. IncI2 plasmids have a high overall structural similarity, while most of the difference is caused by gene-specific recombination upstream and downstream of rci (Gyohda et al., 2002). Linear comparison between p2021-mcr and reference plasmid pAH62-1 (Genbank: CP055260.1) (Figure 9B) showed that mcr-1-pap2 structure also existed upstream of rci and the main difference was that partial sequence inversion occurred in the downstream of this gene due to recombination (Figure 9C).
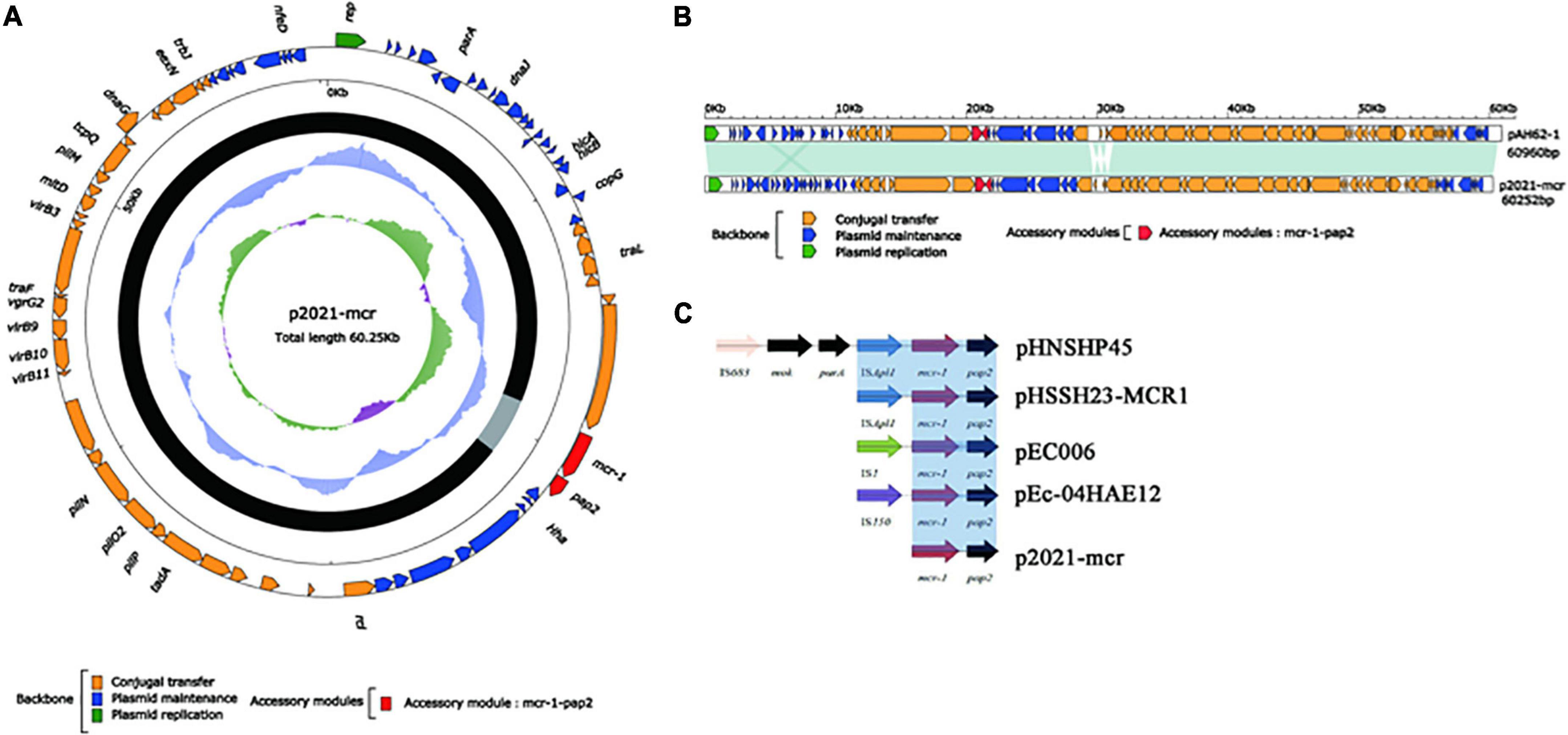
Figure 9. The complete sequence circular diagram of p2021-mcr. (A) The innermost ring represents (G-C)/(G + C); the blue ring indicates GC content, and the outward depression indicates that the GC content is higher than the mean value. Black area in the outer circle represents the backbone area and gray area represents the accessory modules. The outermost circle shows the distribution of genes represented by colored arrows in the plasmid. (B) Linear comparison of p2021-mcr and pAH62-1. Shadows indicate nucleotide homology is greater than 95%. (C) The diversity of mcr-1 upstream environment of rci in Inc2 plasmid.
Analysis of multidrug-resistant region in p20432-mcr
The MDR-1 region of p20432-mcr is composed of MGEs and ARGs in order: △IS2-merR-terC-△IS2-aph(3′)-Ia-IS186B-mcr-1-PAP2-1-sul-△Tn2. mcr-1 was followed by a PAP2 family of proteins, and mcr-1-pap2 has been reported in many plasmids, most of which were related to ISApl1. However, in p20432-mcr, ISApl1 has been replaced by IS186B. IS186B-mcr-1-PAP2 has no similar sequence in Genebank, but IS186B can transpose mcr-1 into chromosomes (Saisree et al., 2001), promoting the vertical transmission of polymyxin resistance genes (Figure 10A). The multidrug -resistance region MDR-2 of the following mobile element in sequence IS4321, IS6100, IS26, TnAsl. IS26-mph(A)-mrx-mphR(A)-IS6100 is a classic antimicrobial resistance locus that contains a macrolide resistance gene mph(A) and a regulator mphR(A). A new type 1 integron was found and the number In1866 was assigned by INTEGRALL database. The ARGs in gene cassette of In1866 contained dfrA12 (Trimethoprim), aadA2 (aminoglycoside), cmlAlaf (chloramphenicol), aadA1 (aminoglycoside) and the 3 prime-CS region contained sul3 (sulfanilamide) (Figure 10B). It is worth noting that the chloramphenicol resistance gene cmlA1af is a new variant of cmlA1, which was first discovered in the gene cassette. The main difference is that the final two amino acids of the product encoded by cmlA1af changed from arginine—valine to serine—methionine.
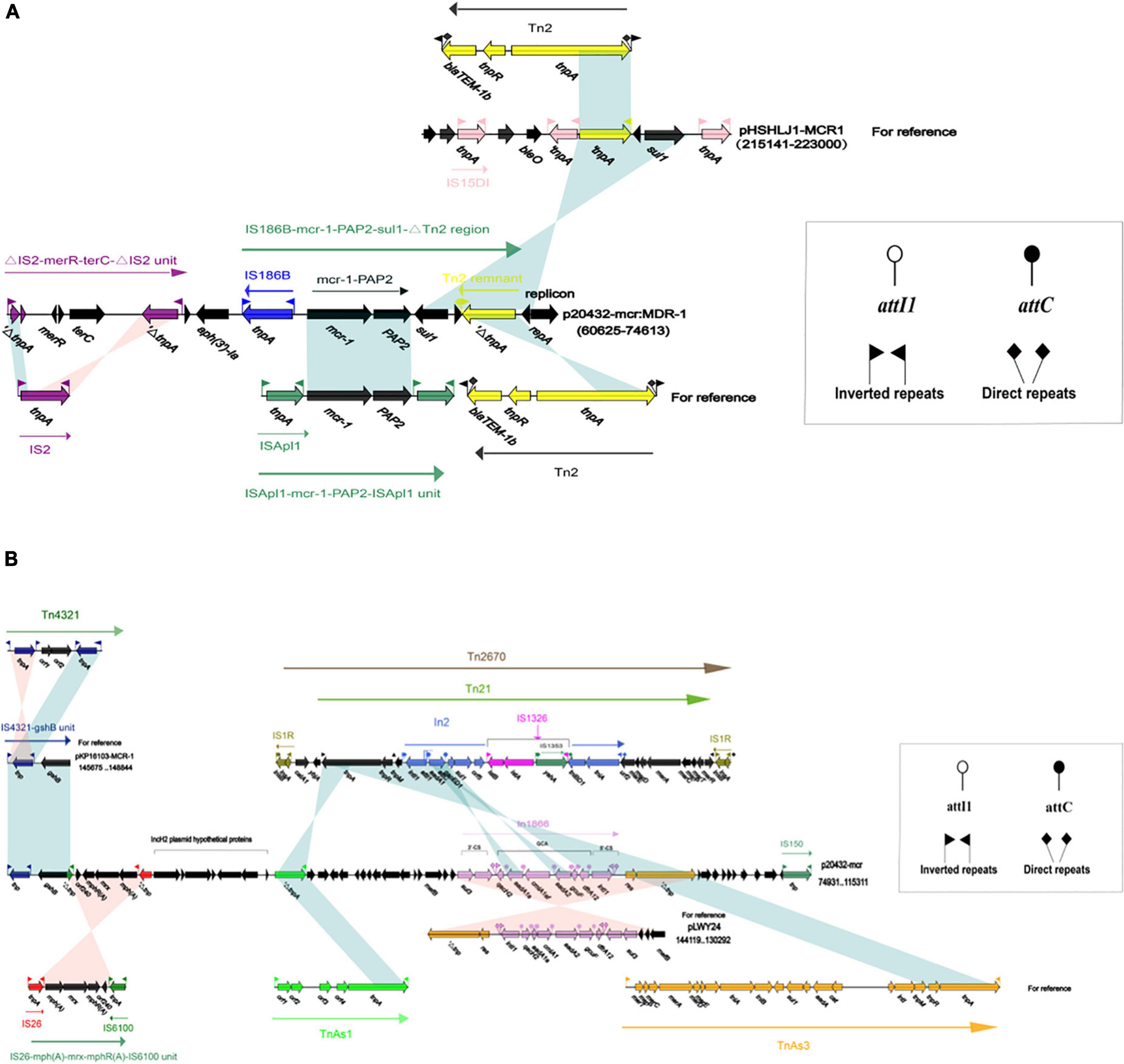
Figure 10. The organization and alignment of MDR-1 region of p20432-mcr. (A) Different colors indicate different moving elements, and arrows indicate genes. Shading indicates that the homology of the two parts of nucleic acids is greater than 95%, and the number in parentheses indicates the location of the fragment in the plasmid. (B) The organization and alignment of MDR-2 region and In1866-related region of p20432-mcr. Different colors indicate different moving elements, and arrows indicate genes. Shading indicates that the homology of the two parts of nucleic acids is greater than 95%, and the number in parentheses indicates the organization and alignment of MDR-2 region and In1866-related region of p20432-mcr.
Discussion
China has the highest use of antimicrobials in husbandry each year and there was a long period that antimicrobials (e.g., C, TET, AMP, TMP) were used to treat bacterial infections or as antimicrobial growth promoters (Niu et al., 2020). As a result, selection for resistant bacteria has promoted gene mutation, recombination, and horizontal gene transfer in poultry farming environments, leading to the prevalence of multidrug-resistant bacteria. China is the largest producer and consumer of eggs globally and the second only to the United States in chicken production (Wang et al., 2015). The enormous and still increasing demands for poultry products in China have meant plenty of room to expand the production of layer and broiler poultry farms (Van Boeckel et al., 2015). Furthermore, the family poultry pattern will still be the primary choice for rural farmers in China. Identifying the phenotype and genotype of MDRE isolated from BFs and LFs indicated that different treatment methods and culture cycles could cause antimicrobial resistance in the environment and different ARGs. Several studies have confirmed that animals and the food chain play critical roles in the emergence and spreading of antimicrobial-resistant bacteria and ARGs (Luo et al., 2020; Tartor et al., 2021). Based on the “One Health” concept proposed by WHO, a coordinated, multisectoral approach is needed to address this complex problem of antimicrobial resistance in the environment, human health and animal husbandry (McEwen and Collignon, 2018). However, the close spatial distribution of residential and farm areas in the family poultry pattern makes the environment, human health and animal husbandry greatly influenced by each other. Thus, surveillance and research are essential to identify critical resistance points and prevent them. As the surveillance of the antimicrobial is used in both humans and non-humans, it is also necessary to implement differentiated and targeted surveillance strategies in broiler and layer poultry farms (Sahoo et al., 2010). Understanding of the differences in antimicrobial resistance profiles between broilers and layers and how they spread can help make effective antimicrobial resistance strategies in BFs and LFs.
This study demonstrated a high prevalence of MDR Enterobacteriaceae and high resistance-gene diversity in poultry farms. By identifying the resistance phenotype of MDRE, the results showed that tetracycline, sulfonamides, amoxicillin, and macrolides antimicrobials all had high resistance rates in both types of farms. The comparison of MDRE in BFs and LFs showed that the risk of MDRE isolated from BFs was higher than those from LFs. It is mainly reflected in two aspects: (1) multidrug-resistance rates were higher in BFs samples; (2) in terms of meropenem, amoxicillin, gentamicin, ciprofloxacin, trimethoprim-sulfamethoxazole, enrofloxacin, ofloxacin, and polymyxin B, the resistance rates of MDRE isolated from BFs were higher than LFs. These results may be caused by: (1) short breeding period and weak resistance of broilers; (2) broilers have a higher range of activities in comparison with the caged laying chickens, and therefore they were more susceptible to MDR infection; (3) some antimicrobials are forbidden in laying chicken breeding or during the laying period, but not as strict in broilers. The resistance rates of amoxicillin, trimethoprim, tetracycline, and sulfamethoxazole used in this study have reached more than 80%, while the resistance rates of polymyxin B and meropenem were lower than 20%. Previous studies detected polymyxin resistance in 40% of bacterial isolates from poultry samples, much higher than what was detected in our study (Lay et al., 2021). However, significant differences between broiler and layer samples (P ≤ 0.05) suggest different transmission characteristics of mcr genes in the two poultry farms. In addition, most of the polymyxin-resistant broiler sources MDRE were resistant to imipenem, gentamicin, ceftazidime, meropenem, cefoxitin, and norfloxacin. Moreover, relatively similar patterns were reported for E. coli against antimicrobials in Croatia and Egypt (Singh et al., 2018; Tartor et al., 2021). The polymyxin-resistant layer sources of MDRE were mainly resistant to imipenem, gentamicin, cefazolin, chloramphenicol, amoxicillin and tetracycline, which was inconsistent with other studies. Resistance of ESBL-producing Enterobacteriaceae to penicillins, cephalosporins and monobactams may force the use of last-resort antimicrobials as carbapenems and colistin (Elmonir et al., 2021). According to PCR results, blaTEM was the predominant β-lactamase gene in both BFs (79.99%) and LFs (78.25%), but blaSHV was not detected. In contrast, all K. pneumoniae isolates from broilers and their environment carried blaSHV–2 in Germany (Daehre et al., 2018). However, in Indonesia, blaTEM (100%) was detected in all K. pneumoniae isolates of chicken origin and only 9.1% of the isolates carried blaSHV showing a similar pattern to the results of this study (Hayati et al., 2019).
Since the plasmid carrying mcr-1 was first reported in China in 2015, colistin resistance has spread rapidly worldwide, and the resulting development of Enterobacteriaceae from extensive drug resistance to pan-drug resistance is inevitable and will ultimately become global (Kumarasamy et al., 2010; Liu et al., 2016). A study based on the prevalence of mcr-1 in the human intestine showed that a decline in colistin-resistant bacteria isolated from both animals and humans decreased significantly after the implementation of the colistin prohibition policy in China in 2017 (Wang et al., 2020; Pyw et al., 2020). It is agreed that the mcr-1 is one of the few clear examples of animal origin resistance traits that can hit the entire human health system (Poirel and Nordmann, 2016). In this study, two mcr-1 positive plasmids were found to have high in vitro transfer rates between E. coli, which are key human pathogens in broilers and layers. In particular, p20432-mcr, found in broiler MDRE, carries a novel integron In1866 and multiple drug resistance genes. The emergence of In1866 confirms that selection pressure is an important condition for the generation of new mobile components (Che et al., 2022). However, it remains to be studied how MGEs evolve through high-frequency HGT in a particular pattern of the family farms. The plasmid p20432-mcr isolated from broiler source E. coli RB20432 belongs to a complex chimeric plasmid, which likely contains both IncHI2 type and IncN type replicons. In addition, the structure of multiple replicons is conducive to increasing the host range of this plasmid, which is likely to make mcr-1 spread more widely (Pilla and Tang, 2018). In a previous study, E. coli isolate possessed multiple resistance genes to β-lactams, aminoglycosides, tetracyclines, sulfonamides, and macrolides, which might be attributed to the presence IncFII, IncHI2, IncHI2A, and IncI1α replicon types (Hayati et al., 2019). In addition to polymyxin resistance genes, ARGs of chloramphenicol, aminoglycoside, and metal resistance genes carried by p20432-mcr make the plasmid transfer under a variety of selective pressures and the family poultry pattern is characterized by a greatly increased risk of transmission to humans. This problem will be more directly reflected in the health examination of the farmers in family farms. So, strategies for preventing the spread of antimicrobials resistance in such farming patterns need to be further improved.
Conclusion
We conducted a comprehensive analysis of MDRE from typical broiler and laying farms in Zhejiang Province, China. The results showed that antimicrobials prohibition had a positive effect on reducing resistance genomes in the breeding environments. The detection rate of some ARGs banned in layer farms that were not suppressed in broilers was significantly increased in broiler source MDRE (P ≤ 0.05). However, our study showed that although certain antimicrobials such as polymyxin have been banned in poultry farming, the polymyxin resistance gene mcr-1 persists in the environment for a long time due to spreading mobile elements like plasmids. The multidrug-resistant plasmid p20432-mcr analysis showed that the multi-replicon structure enabled it to transmit mcr-1 and other ARGs to different hosts, resulting in a higher detection rate for mcr-1, aph(3′)-Ia and aadA2 in broiler farms. Therefore, it is necessary to implement and improve cleaning and disinfection procedures in family broiler farms and detect resistant bacteria from broilers and the environment to prevent further transmission to humans through the poultry production chain. It is also necessary to assess the antimicrobial resistance of bacteria from farmers on family farms.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA798875.
Author contributions
SZ analyzed the data and wrote the manuscript. DX and XZ analyzed the data and prepared figures. DZ contributed in review and editing. PW collected samples and helped with software. DQ conceived the study and revised the original manuscript. All authors edited the manuscript and approved the final draft.
Funding
This work was supported in part by the National Natural Science Foundation of China (No. 32172188).
Acknowledgments
We thank the reviewers for their insightful suggestions and comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.912652/full#supplementary-material
Footnotes
References
Alcock, B. P., Raphenya, A. R., Lau, T., Tsang, K. K., and Mc Arthur, A. G. (2019). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48:D517–D525. doi: 10.1093/nar/gkz935
Aziz, R. K., Bartels, D., Best, A. A., Dejongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 9:75. doi: 10.1186/1471-2164-9-75
Belluco, S., Barco, L., Roccato, A., and Ricci, A. (2016). Escherichia coli and Enterobacteriaceae counts on poultry carcasses along the slaughterline: a systematic review and meta-analysis. Food Control 60, 269–280. doi: 10.1016/j.foodcont.2015.07.033
Bolger, A. M., Marc, L., and Bjoern, U. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Buelow, E., González, T. B., Fuentes, S., De, S., Lahti, L., Bayjanov, J. R., et al. (2017). Comparative gut microbiota and resistome profiling of intensive care patients receiving selective digestive tract decontamination and healthy subjects. Microbiome 5:88. doi: 10.1186/s40168-017-0309-z
Che, Y., Xu, X., Yang, Y., Bøinda, K., Hanage, W., Yang, C., et al. (2022). High-resolution genomic surveillance elucidates a multilayered hierarchical transfer of resistance between WWTP- and human/animal-associated bacteria. Microbiome 10:16. doi: 10.1186/s40168-021-01192-w
Chen, S., Zhou, Y., Chen, Y., and Jia, G. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560
Daehre, K., Projahn, M., Friese, A., Semmler, T., Guenther, S., and Roesler, U. H. (2018). ESBL-Producing Klebsiella pneumoniae in the Broiler Production Chain and the First Description of ST3128. Front. Microbiol. 9:2302. doi: 10.3389/fmicb.2018.02302
Elmonir, W., Abd El-Aziz, N. K., Tartor, Y. H., Moustafa, S. M., Abo Remela, E. M., Eissa, R., et al. (2021). Emergence of Colistin and Carbapenem Resistance in Extended-Spectrum β-Lactamase Producing Klebsiella pneumoniae Isolated from Chickens and Humans in Egypt. Biology 10:373. doi: 10.3390/biology10050373
Falagas, M. E., Karageorgopoulos, D. E., and Nordmann, P. (2011). Therapeutic options for infections with Enterobacteriaceae producing carbapenem-hydrolyzing enzymes. Future Microbiol. 6, 653–666.
Gaze, W. H., Krone, S. M., Larsson, D., Li, X. Z., and Zhu, Y. G. (2013). Influence of Humans on Evolution and Mobilization of Environmental Antibiotic Resistome. Emerg. Infect. Dis. 19:e120871. doi: 10.3201/eid1907.120871
Gyohda, A., Furuya, N., Kogure, N., and Komano, T. (2002). Sequence-specific and Non-specific Binding of the Rci Protein to the Asymmetric Recombination Sites of the R64 Shufflon. J. Mol. Biol. 318, 975–983. doi: 10.1016/S0022-2836(02)00195-X
Hayati, M., Indrawati, A., Mayasari, N. L. P. I., Istiyaningsih, I., and Atikah, N. (2019). Molecular detection of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates of chicken origin from East Java. Indonesia. Vet. World 12, 578–583. doi: 10.14202/vetworld.2019.578-583
Institute CLSI (2020). Performance Standards for Antimicrobial Susceptibility Testing—Thirty Edition: M100”. Wayne, PA: CLSI.
KöCk, R., Daniels-Haardt, I., Becker, K., Mellmann, A., Friedrich, A. W., Mevius, D., et al. (2018). Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin. Microbiol. Infect. 24, 1241–1250. doi: 10.1016/j.cmi.2018.04.004
Kumarasamy, K. K., Toleman, M. A., Walsh, T. R., Bagaria, J., and Woodford, N. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 578–579. doi: 10.1016/S1473-3099(10)70143-2
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (New York, NY: John Wiley and Sons).
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lay, K. K., Jeamsripong, S., Sunn, K. P., Angkititrakul, S., Prathan, R., Srisanga, S., et al. (2021). Colistin Resistance and ESBL Production in Salmonella and Escherichia coli From Pigs and Pork in the Thailand, Cambodia, Lao PDR, and Myanmar Border Area. Antibiotics 10:657. doi: 10.3390/ANTIBIOTICS10060657
Liu, B., and Pop, M. (2009). ARDB–Antibiotic Resistance Genes Database. Nucleic Acids Res. 37:D443–D447. doi: 10.1093/nar/gkn656
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Luo, Q., Wang, Y., and Xiao, Y. (2020). Prevalence and Transmission of Mobilized Colistin Resistance (Mcr) Gene in Bacteria Common to Animals and Humans. Biosaf. Heal 2, 71–78. doi: 10.1016/j.bsheal.2020.05.001
Mceachran, A. D., Blackwell, B. R., Hanson, J. D., Wooten, K. J., and Smith, P. N. (2015). Antibiotics, Bacteria, and Antibiotic Resistance Genes: Aerial Transport from Cattle Feed Yards via Particulate Matter. Environ. Health Perspect. 123, 337–343. doi: 10.1289/ehp.1408555
McEwen, S. A., and Collignon, P. J. (2018). Antimicrobial Resistance: a One Health Perspective. Microbiol. Spectr. 6. doi: 10.1128/microbiolspec.ARBA-0009-2017
Moura, A., Soares, M., Pereira, C., Leitão, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Nederbragt, A. J. (2014). On the middle ground between open source and commercial software - the case of the Newbler program. Genome Biol. 15:113. doi: 10.1186/gb4173
Niu, J.-L., Peng, J.-J., Ming, Y.-Y., Ma, Q.-C., Liu, W.-C., and Ma, Y. (2020). Identification of drug resistance genes and drug resistance analysis of Salmonella in the duck farm environment of Zhanjiang, China. Environ. Sci. Pollut. Res. Int. 27, 24999–25008. doi: 10.1007/s11356-020-09007-5
Pilla, G., and Tang, C. M. (2018). Going around in circles: virulence plasmids in enteric pathogens. Nat. Rev. Microbiol. 16, 484–495. doi: 10.1038/s41579-018-0031-2
Poirel, L., and Nordmann, P. (2016). Emerging Plasmid-Encoded ColistinResistance: The Animal World as the Culprit? J. Antimicrob. Chemother. 71, 2326–2327. doi: 10.1093/jac/dkw074
Pyw, A., Cxm, A., Prz, C., Pyc, B., Ys, D., Pfhe, F., et al. (2020). Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect. Dis. 20, 1161–1171. doi: 10.1016/S1473-3099(20)30149-3
Rawat, N., Maansi, Kumar, D., and Upadhyay, A. K. (2018). Virulence typing and antibiotic susceptibility profiling of thermophilic Campylobacters isolated from poultry, animal, and human species. Vet. World 11, 1698–1705. doi: 10.14202/vetworld.2018.1698-1705
Sahoo, K. C., Tamhankar, A. J., Johansson, E., and Lundborg, C. S. (2010). Antibiotic use, resistance development and environmental factors: a qualitative study among healthcare professionals in Orissa. India. BMC Public Health 10:629. doi: 10.1186/1471-2458-10-629
Saisree, L., Reddy, M., and Gowrishankar, J. (2001). IS186 Insertion at a Hot Spot in the lon Promoter as a Basis for Lon Protease Deficiency of Escherichia coli B: Identification of a Consensus Target Sequence for IS186 Transposition. J. Bacteriol. 183, 6943–6946. doi: 10.1128/JB.183.23.6943-6946.2001
Sasha, B., David, A., Ana, M., Liang, Y., Grant, J. R., Adam, M., et al. (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44:W147–W153. doi: 10.1093/nar/gkw419
Shi, X., Li, Y., Yang, Y., Shen, Z., and Wang, S. (2020). High prevalence and persistence of carbapenem and colistin resistance in livestock farm environments in China. J. Hazard. Mater. 406:124298. doi: 10.1016/j.jhazmat.2020.124298
Singh, A., Chhabra, D., Sikrodia, R., Shukla, S., Sharda, R., Audarya, S. (2018). Isolation of E. coli from bovine mastitis and their antibiotic sensitivity pattern. Int. J. Curr. Microbiol. Appl. Sci. 7, 11–18. doi: 10.20546/ijcmas.2018.710.002
Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P. L., Ideker, T., et al. (2011). Cytoscape 2.8: new features for data integration and networ-k visualization. Bioinformatics 27, 431–432. doi: 10.1093/bioinfor-matics/btq675
Su, J. Q., Wei, B., Ou-Yang, W. Y., Huang, F. Y., Zhao, Y., Xu, H. J., et al. (2015). Antibiotic Resistome and Its Association with Bacterial Communities during Sewage Sludge Composting. Environ. Sci. Technol. 49, 7356–7363. doi: 10.1021/acs.est.5b01012
Tartor, Y. H., Gharieb, R. M. A., Abd El-Aziz, N. K., El Damaty, H. M., Enany, S., Khalifa, E., et al. (2021). Virulence Determinants and Plasmid-Mediated Colistin Resistance mcr Genes in Gram-Negative Bacteria Isolated From Bovine Milk. Front. Cell. Infect. Microbiol. 11:761417. doi: 10.3389/fcimb.2021.761417
Udikovic-Kolic, N., Wichmann, F., Broderick, N. A., and Handelsman, J. (2014). Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc. Natl. Acad. Sci. U.S.A. 111, 15202–15207. doi: 10.1073/pnas.1409836111
Van Boeckel, T. P., Brower, C., Gilbert, M., Grenfell, B. T., Levin, S. A., Robinson, T. P., et al. (2015). Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. U.S.A. 112, 5649–5654. doi: 10.1073/pnas.1503141112
Wang, Y., Lv, Y., Cai, J., Schwarz, S., Cui, L., Hu, Z., et al. (2015). A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 70, 2182–2190. doi: 10.1093/jac/dkv116
Wang, Y., Xu, C., Zhang, R., Chen, Y., Shen, Y., Hu, F., et al. (2020). Dynamics of mcr-1 prevalence and mcr-1 -positive Escherichia coli after the cessation of colistin use as a feed additive for animals in China: a prospective cross-sectional and whole genome sequencing-based molecular epidemiological study. Lancet Microbe 1:e34–e43. doi: 10.1016/s2666-5247(20)30005-7
Wang, Y., Zhang, R., Li, J., Wu, Z., Yin, W., Schwarz, S., et al. (2017). Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2:16260. doi: 10.1038/nmicrobiol.2016.260
Wei, S., Gutek, A., Lilburn, M., and Yu, Z. (2013). Abundance of pathogens in the gut and litter of broiler chickens as affected by bacitracin and litter management. Vet. Microbiol. 166, 595–601. doi: 10.1016/j.vetmic.2013.06.00
Winglee, K., Howard, A. G., Sha, W., Gharaibeh, R. Z., Liu, J., Jin, D., et al. (2017). Recent urbanization in China is correlated with a Westernized microbiome encoding increased virulence and antibiotic resistance genes. Microbiome 5:121. doi: 10.1186/s40168-017-0338-7
Xiong, W., Wang, Y., Sun, Y., Ma, L., Zeng, Q., Jiang, X., et al. (2018). Antibiotic-mediated changes in the fecal microbiome of broiler chickens define the incidence of antibiotic resistance genes. Microbiome 6:34. doi: 10.1186/s40168-018-0419-2
Yin, X., Jiang, X. T., Chai, B., Li, L., Yang, Y., Cole, J. R., et al. (2018). ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics 34, 2263–2270. doi: 10.1093/bioinformatics/bty053
Keywords: metagenomic, ARGs, MGEs, multidrug-resistant Enterobacteriaceae, poultry farms, mcr-1
Citation: Zong S, Xu D, Zheng X, Zaeim D, Wang P, Han J and Qu D (2022) The diversity in antimicrobial resistance of MDR Enterobacteriaceae among Chinese broiler and laying farms and two mcr-1 positive plasmids revealed their resistance-transmission risk. Front. Microbiol. 13:912652. doi: 10.3389/fmicb.2022.912652
Received: 04 April 2022; Accepted: 20 June 2022;
Published: 04 August 2022.
Edited by:
Marcelo Tolmasky, California State University, Fullerton, United StatesReviewed by:
German Matias Traglia, Universidad de la República, UruguayAhmed Mohamed Ammar, Zagazig University, Egypt
Copyright © 2022 Zong, Xu, Zheng, Zaeim, Wang, Han and Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daofeng Qu, MTI1ODAwMTYwQHFxLmNvbQ==
 Shuaizhou Zong1
Shuaizhou Zong1 Daofeng Qu
Daofeng Qu