- 1National Risk Assessment Laboratory for Antimicrobial Resistance of Animal Original Bacteria, South China Agricultural University, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, South China Agricultural University, Guangzhou, China
- 3National Reference Laboratory of Veterinary Drug Residues, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
- 4Guangdong Provincial Key Laboratory of Microbial Safety and Health, State Key Laboratory of Applied Microbiology Southern China, Institute of Microbiology, Guangdong Academy of Sciences, Guangzhou, China
- 5Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China
We conducted a molecular surveillance study for carbapenem-resistant Enterobacteriaceae (CRE) colonization in food-producing animals in China that included primarily swine and poultry for three consecutive years. A total of 2,771 samples from food-producing animals and their surrounding environments were collected from different regions in China from 2015 to 2017. Enrichment cultures supplemented with meropenem were used to isolate carbapenem non-susceptible isolates and these were subsequently identified by MALDI-TOF MS. Resistance phenotypes and genotypes were confirmed using antimicrobial susceptibility testing and molecular biological techniques. Genomic characteristics of the carbapenemase-producing isolates were investigated using whole genome sequencing (WGS) and bioinformatic analysis. In total, 88 NDM-positive Enterobacteriaceae were identified from 2,771 samples and 96.6% were Escherichia coli. The New Delhi metallo-β-lactamase (NDM)-positive E. coli displayed a diversity of sequence types (ST), and ST48 and ST165 were the most prevalent. Three variants of blaNDM (blaNDM-1, blaNDM-4, and blaNDM-5) were detected and WGS indicated that blaNDM-5 predominated and was carried primarily on IncX3 plasmids. All these isolates were also multiply-drug resistant. These results revealed that food-producing animals in China are an important reservoir for NDM-positive E. coli and pose a potential threat to public health.
Introduction
Antimicrobial resistance (AMR) is rising to high levels in both human and veterinary health. New threats arising from multidrug-resistant bacteria and the very limited number of antibiotics coming to the market and bring us perilously to the end of antibiotic era where common treatable infections can once again be fatal. The abuse of human antibiotics in food-producing animals as prophylactic agents or growth promoters has become an important contributing factor to increasing AMR (Van et al., 2020). Food-producing animals could serve as reservoir for antibiotic-resistant bacteria that can be transmitted to humans through direct contact or the food chain (Founou et al., 2016; Wang et al., 2017). Therefore, the prevalence of AMR will continue to rise worldwide and become one of the most serious threats to human and animal health.
Among the major multidrug-resistant bacteria, the emergence and dissemination of carbapenem-resistant Enterobacteriaceae (CRE) is identified as one of the most serious threats to human and animal health worldwide (Mariana Huband et al., 2017; Zhang et al., 2017, 2018; Bonomo et al., 2018). Notably, the recent increase in CRE was primarily because of the acquisition of carbapenem hydrolyzing enzymes such as the New Delhi metallo-β-lactamase (NDM; Kumarasamy et al., 2010; Munoz-Price et al., 2013). Since the first report of NDM-1 in a Klebsiella pneumoniae isolated from a patient in 2009, NDM-positive Enterobacteriaceae have been rapidly spreading in hospitals as well as in food-producing animals around the world and especially in China (Yong et al., 2009; Logan and Weinstein, 2017). To date, NDM-positive Enterobacteriaceae have been found on swine, geese farms, cow and chicken in China (Kong et al., 2017a; Köck et al., 2018). The spread of NDM-positive Escherichia coli has been documented among chickens, swallows, farmers, dogs and even flies in a chicken farm (Berrazeg et al., 2014; Logan and Weinstein, 2017; Li et al., 2019).
However, the dissemination of NDM-positive E. coli among food-producing animals in China is currently poorly understood. A large-scale survey of the NDM-positive Enterobacteriaceae covering a large region of China has yet to be performed. In this study, we examined the long-term and large-scale prevalence and molecular epidemiological features of NDM-positive E. coli from livestock animals from northern to southern China for three consecutive years, 2015–2017.
Materials and Methods
Sample Collection and Bacterial Isolation
A total of 2,771 samples, including 526 samples in 2015, 812 samples in 2016, and 1,433 samples in 2017 were collected from food-producing animals (fecal samples) on poultry and swine farms and their surrounding environments (Supplementary Table S1). Sixteen provinces that play important roles in the swine and poultry industry from north to south China were randomly selected for sampling. According to the north–south demarcation zone, eight provinces that include Jiangsu, Henan, Heilongjiang, Hebei, Jilin, Ningxia, Jiangsu, Inner Mongolia are situated in the north China while the remainder were located in south China (Figure 1).
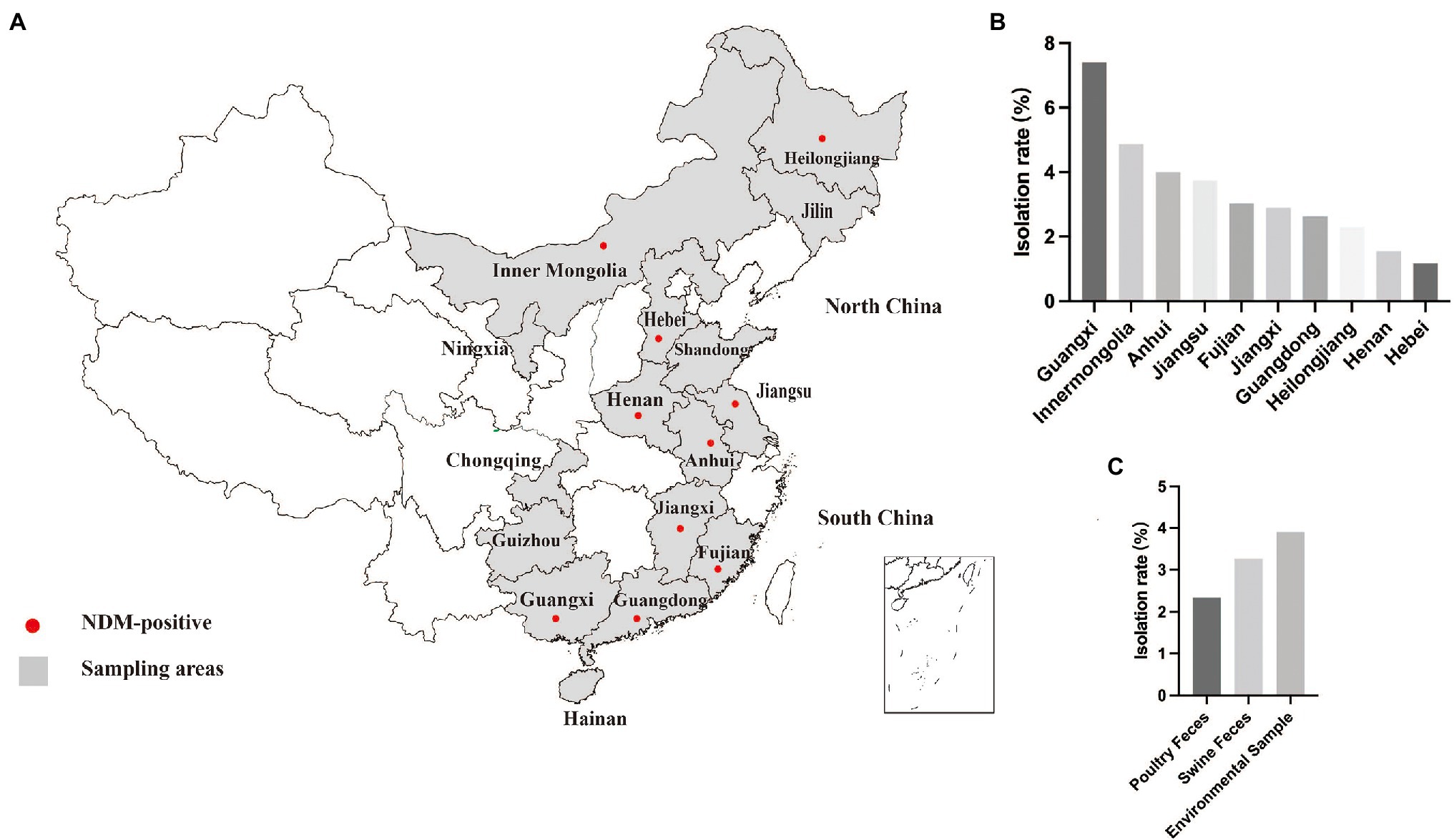
Figure 1. Sampling areas and detection rate. (A) Sampling areas for poultry and swine farms in China examined in this study. (B) Detection rate of blaNDM-positive isolates in different provinces. (C) Detection rate of blaNDM-positive isolates in different type of samples.
Environmental samples (sewage and soil) were incubated overnight in Luria Bertani broth without antibiotics. Environmental cultures and diluted feces samples were directly plated on MacConkey agar with 1 mg/l meropenem and incubated at 37°C. Carbapenem non-susceptible isolates were identified using MALDI-TOF MS (Shimadzu Biotech) and 16S rDNA sequencing (Seng et al., 2009; Walsh and Duffy, 2013) Carbapenemase production was examined for all isolates using the Carbapenemase Nordmann-Poirel test (Nordmann et al., 2012b). All the carbapenemase-producing isolates were screened using polymerase chain reaction (PCR) for the presence of blaNDM, blaOXA-48-like, blaVIM, blaKPC, and blaIMP (Poirel et al., 2011). In addition, the entire blaNDM amplicons were sequenced (Sanger) and the 813 bp ORF of blaNDM was typed using BLAST.1
Antimicrobial Susceptibility Testing
The minimal inhibitory concentrations (MIC) of 14 antibiotics (amikacin, gentamicin, meropenem, imipenem, ertapenem, cefoxitin, cefotaxime, ceftazidime, aztreonam, cefoxitin, tetracycline, fosfomycin, trimethoprim/sulfamethoxazole, and florfenicol) for the isolates were determined by agar dilution and interpreted according to the clinical and laboratory standard institute (CLSI) documents M100-S28 and VET01-S2. The breakpoints of tigecycline and colistin for Enterobacteriaceae were interpreted according to the EUCAST criteria (Version 6.0). E. coli American Type Culture Collection (ATCC) 25922 served as a quality control strain for the testing.
Pulsed-Field Gel Electrophoresis (PFGE) Typing, Whole-Genome Sequencing and Conjugation Assay
The genetic relatedness of NDM-positive isolates was investigated using XbaI-PFGE for E. coli. PFGE patterns were analyzed with the Dice coefficient and the unweighted pair group with arithmetic mean clustering method using BioNumerics (Applied Maths, Ghent, Belgium), resulting in PFGE patterns with > 90% similarity between clusters. Based on different resistance phenotypes and genetic relationships, 76 strains were selected for Whole-Genome Sequencing (WGS). Genomic DNA was extracted from isolates using the Genomic DNA Purification Kit (Tiangen, Beijing, China). WGS was performed with the Illumina HiSeq 2,500 system (Novogene Guangzhou, China). Gene annotation and prediction were performed using RAST2 and BLAST. MLST sequence types (ST), plasmid incompatibility (Inc) groups and antibiotic resistance genes (ARGs) were analyzed using software from IS finder3 and the Center for Genomic Epidemiology.4
Streptomycin-resistant E. coli strain C600 was used as the recipient to determine the transferability of the resistance genes. The conjugation assays were performed using the filter mating method and the proportion of donor to recipient bacteria stands 1:1. Transconjugants were selected using MacConkey agar plates containing both 2,000 mg/l streptomycin and 1.0 mg/l meropenem. Selected transconjugants were confirmed by PCR.
Results
Bacterial Isolation and Detection of Carbapenemase Genes
In this study, 654 carbapenem non-susceptible bacteria (≥ 1 mg/l) were recovered from 2,771 fecal and environmental samples that included sewage and soil samples from 16 provinces in China from 2015 to 2017. We identified the presence of the blaNDM gene in 88 isolates. In 2015, the prevalence of NDM-positive Enterobacteriaceae detected in samples was 1.52% (8/526) and the positive samples were primarily from Jiangsu and Guangdong. The prevalence for 2016 was almost identical (1.48%, 12/812). Interestingly, the prevalence increased dramatically to 4.82% (69/1433) in 2017 (Supplementary Table S1). Among all the provinces, blaNDM was the most prevalent in Guangxi. Notably, samples collected from six provinces, including Jilin, Shandong, Ningxia, Chongqing, Guizhou and Hainan did not detected NDM-positive strain (Figure 1). Species identification and WGS analysis indicated that the 88 blaNDM-carrying isolates belonged to three Enterobacteriaceae species, including E. coli (n = 82), K. pneumoniae (n = 5) and Citrobacter freundii (n = 1).
Antimicrobial Susceptibility Testing and Conjugation Assays
Our collection of 88 NDM-positive isolates were found to be resistant to imipenem (MIC 4 to > 64 mg/l), meropenem (MIC 4 to > 64 mg/l) and ertapenem (MIC 16 to > 64 mg/l) as well as concurrently resistant to the other β-lactams cefotaxime, cefoxitin and ceftazidime. Moreover, most were also resistant to trimethoprim/sulfanilamide (96.6%; n = 85; MIC 40 to > 320 mg/l), tetracycline (94.3%; n = 83; MIC 16 to > 256 mg/l), florfenicol (80.6%; n = 71; MIC 16 to > 256 mg/l), ciprofloxacin (72.7%; n = 60; MIC 4 to > 256 mg/l), and gentamicin (59.1%; n = 52; MIC 16 to > 256 mg/l). However, we found relatively a lower prevalence for resistance to aztreonam (45.5%), amikacin (34.1%), Fosfomycin (29.5%), colistin (28.4%) and tigecycline (11.4%). These results showed that NDM-positive isolates displayed reduced susceptibilities to β-lactam antibiotics, but remained susceptible to colistin and tigecycline. Conjugation experiments using these isolates resulted in the transfer of blaNDM-carrying plasmids from 76 isolates (86.5%) to E. coli C600str recipient (Supplementary Table S2).
Phylogenetic Analysis of NDM-Positive Escherichia coli
PFGE was successfully performed on these 77 blaNDM-carrying E. coli and resulted in 74 PFGE patterns. There were four groups of isolates that shared similar PFGE patterns including FS21-4 and FSCK25-1, HB14-1 and HB14-3, JZ33 and JZ41-2; most isolates of each PFGE group were from the same area, while HN68-2 and FCG47-1 were from different provinces and years but shared > 90% similarity in PFGE patterns. The remaining 69 isolates belonged to different phylogenetic groups (Supplementary Figure S1). Based on PFGE profiles, a total of 72 isolates were selected for WGS and this analysis demonstrated that these E. coli isolates had 38 distinct STs. The most prevalent STs were ST48 (10/72; 13.9%), ST165 (6/72; 8.3%) and ST405 (4/72; 5.6%).
Resistance Profiles
Whole genome sequencing analysis revealed that NDM-positive E. coli isolates possessed > 30 distinct ARGs conferring resistance to nearly all currently available antibiotics. Among the other 24 ARG types, 11 ARGs were common with a prevalence > 50% and these included blaTEM (n = 43), aph (n = 64), ant (n = 36), aadA (n = 49), aac (n = 40), cmlA (n = 37), dfrA (n = 68), sul (n = 65), tet (n = 65), mdf (n = 72), and flor (n = 53). These genes conferred resistance to seven classes of antibiotics; β-lactams, sulphonamides, quinolones, tetracyclines, chloramphenicol, aminoglycosides and macrolides. Notably, gene mcr-1 mediating resistance to the last-resort antibiotic colistin (Poirel et al., 2017; Li et al., 2020) was detected in 19 NDM-positive E. coli isolates. Additional analysis of plasmid sequences indicated that 72 NDM-positive E. coli isolates in this study carried 14 types of Inc. plasmids. IncX3 predominated and IncFIB (65.3%, 47/72) and IncFII (34.7%, 25/72) were also highly represented (Figure 2).

Figure 2. MLST, Resistance Phenotype, ARG and plasmid replicons of 72 NDM-positive Escherichia coli isolates from food-producing animals in China. The heatmap was generated after aligning the contigs of sequenced genomes of each strain to MLST, Resfinder and PlasmidFinder. ARGs and plasmid Inc. types are indicated by yellow and blue squares, respectively. AMK, amikacin; GEN, gentamicin; MEM, meropenem; IMP, imipenem; ERT, ertapenem; CTX, cefotaxime; CAZ, ceftazidime; FOX, cefoxitin; ATM, aztreonam; FOS, fosfomycin; CS, colistin; TET, tetracycline; TGC, tigecycline; CIP, ciprofloxacin; SXT, sulfamethoxazole-trimethoprim. FFC, florfenicol.
Genetic Environments of blaNDM
A sequence comparison analysis revealed that all 64 IncX3 plasmids were similar to a blaNDM-5-carrying IncX3 plasmid pNDM-MGR194 (Acc. No. NC_022740) from a K. pneumoniae isolate. These 64 plasmids shared an identical and conserved IncX3 backbone, and their inserted blaNDM sites were all near the umuD gene. Differences between these plasmids were primarily contained within the blaNDM-5 region between the parA and IS3000 genes. The diversity within this region was used to separate these 64 plasmids into four groups (Types A–D). Type A plasmids (n = 47) shared the highest similarity with the reference plasmid. Types B (n = 5), C (n = 11), and D (n = 1) omitted IS5, ΔISAba125 and ΔISAba125-IS3000-ΔTn2, respectively (Figure 3A). In the Type B group, blaNDM was located in the structure IS26-tat-trpF-bleMBL-blaNDM-ISAba125-IS3000-ΔTn2. The structure was inserted into the plasmid pEC14-35 (Acc. No. NC_019083) and therefore formed the genetic environments of blaNDM for Type B. Type A most likely was formed by IS5 insertion into the region between blaNDM and ISAba125. Types C and D emerged after losing ISAba125 and ISAba125-IS3000-ΔTn2 most likely due to homologous recombination (Figure 3B).
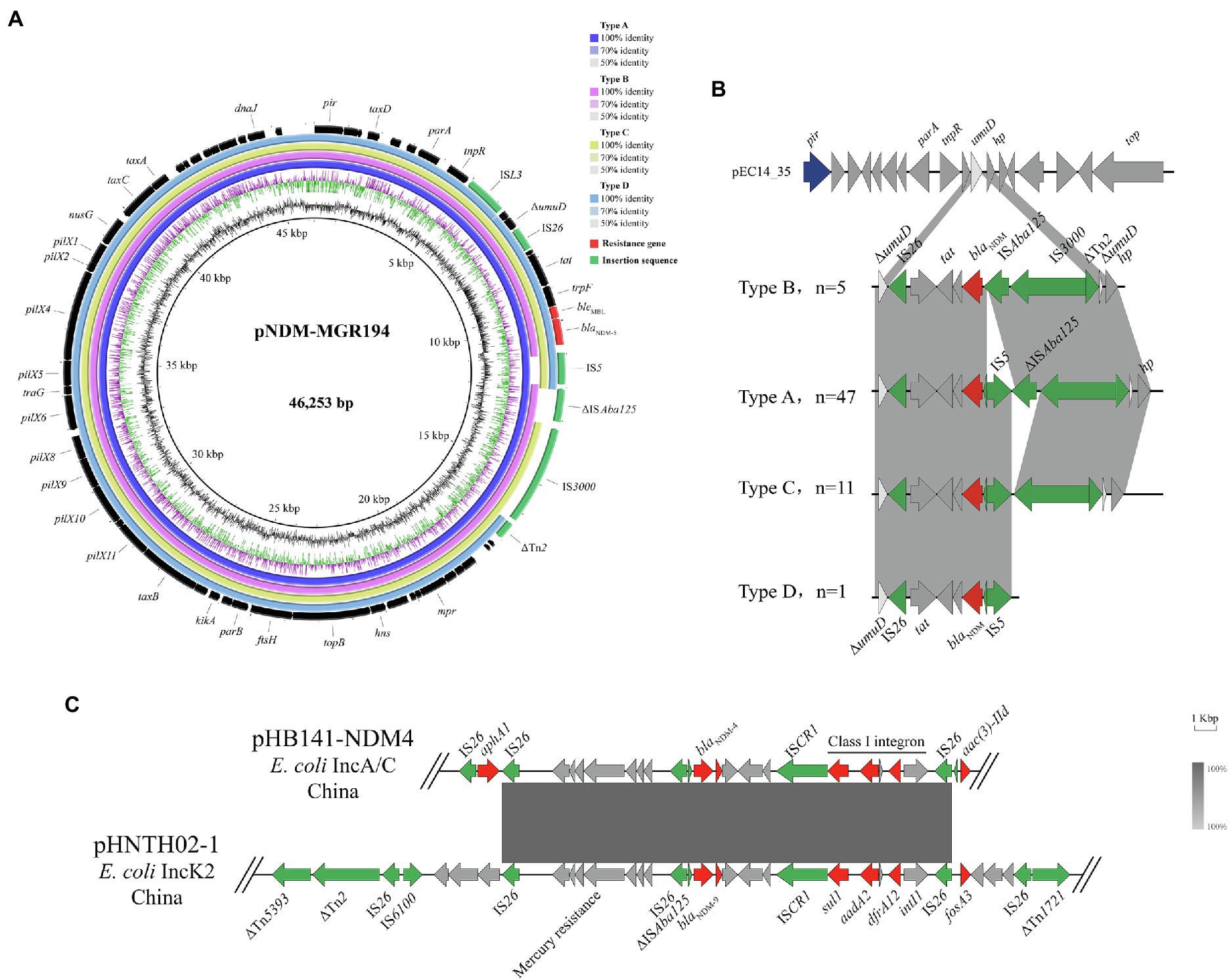
Figure 3. Genetic environments for blaNDM. (A) Comparison of blaNDM-5-carrying IncX3 plasmids with reference sequence pNDM-MGR194. (B) Four different genetic environments on the IncX3 plasmid harboring blaNDM. (C) Linear sequence comparison of pHB141-NDM4 and pHNTH02-1.
In addition, the genetic environments of a blaNDM-4-bearing IncA/C plasmid (pHB141-NDM4) demonstrated that the ARGs sul1, aadA2 and dfrA12 were packaged in Class I integrons located within the structure of ISAba125-blaNDM-4-bleMBL-trpF-tat-ΔcutA. There were also a series of genes mediating Hg resistance downstream of blaNDM-4. Two IS26 elements flanking the aphA1 gene were also downstream of blaNDM-4. A sequence comparison analysis revealed that all the 64 IncX3 plasmids were similar to a blaNDM-9-carrying IncK2 plasmid pHNTH02-1 from E. coli in China (Acc. No. NZ_MG196294). The reference sequence featured a composite transferable structure of IS26-ΔISAba125-blaNDM-bleMBL-trpF-tat-ΔcutA-ISCR1-sul1-aad2-gcuF-dfrA12-intI1-IS26, with two inverted repeat sequences ΔTn1721 and ΔTn5393 (Figure 3C).
Discussion
The presence of NDM-positive Enterobacteriaceae in livestock animals is of concern because this may facilitate expansion of the gene pool from which pathogenic bacteria can acquire ARGs and consumers may subsequently be exposed via the food chain (Woodford et al., 2014; Mollenkopf et al., 2017). For this reason, we conducted a long-term and large-scale characterization of the prevalence and genetic relationships between NDM-positive bacteria from livestock animals in China from 2015 to 2017. Since its first report in British, blaNDM-5 has been detected in clinical isolates from Europe, Asia, South America, the United States of America and Australia (Nakano et al., 2014; Wailan et al., 2015; Baek et al., 2019; López-Hernández et al., 2020; Ramadan et al., 2020; Costa et al., 2021). Notably, the prevalence of blaNDM-5 isolates from food-producing animals has been sporadic in aboard (Ghatak et al., 2013), although there are more relevant reports in China (Yang et al., 2016; Kong et al., 2017a; Yao et al., 2020; Zhao et al., 2021; Lv et al., 2022). In the current study, our results revealed a wide contamination of NDM-positive Enterobacteriaceae in ten different provinces. In particular, the prevalence of blaNDM-carrying Enterobacteriaceae in swine and poultry in Guangxi province (10.3%, 32/312) was much higher than for 2017 and suggested that food-producing animals in Guangxi province have been severely contaminated by carbapenem-resistant Enterobacteriaceae. Fortunately, three northern provinces Jilin, Ningxia and Shandong, and three southern provinces Chongqing, Guizhou and Hainan had not been contaminated by NDM-positive Enterobacteriaceae over our 3-year study period.
While carbapenem use is prohibited in swine and poultry production chains, different Enterobacteriaceae species and various blaNDM variants have been found on livestock farms (Zhai et al., 2020; Wang et al., 2021). In the current study, we identified 88 NDM-positive isolates belonging to three Enterobacteriaceae species including E. coli, K. pneumoniae and C. freundii. For the NDM-positive E. coli isolates, the predominant variant of blaNDM was blaNDM-5. There are to date seven blaNDM variants (blaNDM-1, blaNDM-4, blaNDM-5, blaNDM-7, blaNDM-9, blaNDM-17, and blaNDM-20) that have been detected in food-producing animals in China and blaNDM-5 was the most prevalent and was found in poultry, swine, geese and ducks (Liu et al., 2017, 2018; Pruthvishree et al., 2017; Cen et al., 2021; Wang et al., 2021). Furthermore, our PFGE analysis illustrated that the genetic background of NDM-positive E. coli varied considerably across the country although there were a small number of closely related strains in the same regions. Therefore, clonal spread was not the primary mode of blaNDM gene transmission and was consistent with the results of previous studies (Mantilla-Calderon et al., 2016; Zhang et al., 2018). Our WGS results also demonstrated that these E. coli isolates had 38 distinct STs and could be distinguished by their geographical distribution. For instance, ST48, ST165 and ST405 were more prevalent than the others. ST48 E. coli isolates have been linked to blaNDM spread in livestock farms and in retail meat in China (Liu et al., 2019; Zhang et al., 2019b).
IncX3 plasmids have been identified as the primary vectors for the horizontal transfer of blaNDM-5 on farms (Ho et al., 2018; Williams et al., 2018) and our study further confirms this. IncX3 plasmids normally encode a type IV secretion system (pilX1-11) that allows the exchange of genetic material within bacteria. IncX3 plasmids are also highly conjugatable and stable and exert no fitness costs on their bacterial hosts (Zhai et al., 2020). This characteristic may explain why NDM-positive bacteria still have been found in these settings even though carbapenem use is prohibited on livestock farms. In addition to the IncX3 plasmids, IncFIB and IncFII were also highly represented. IncFIB plasmids have been frequently detected in E. coli and has already been described as an ESBL gene carrier (Zhang et al., 2019a). IncFII plasmids are important in ARGs spread and are also the primary mcr-1 vector for E. coli (Xavier et al., 2016; Sugawara et al., 2019). Therefore, the three types of plasmids discussed above may serve as an important vehicles for the spread of ARGs among animals and humans.
Conclusion
In this study we identified 88 NDM-positive isolates from animal farms and their surrounding environments in China. Notably, this is a large-scale survey of CRE covering a large region of China over three consecutive years. Phylogenetic analysis indicated considerable diversity for this population of CRE isolates taken from livestock farms. WGS analysis further determined that blaNDM-5 coexisted with other ARGs and also demonstrated a great diversity in the plasmid population; the latter provides important epidemiological information for the global dissemination of blaNDM-5. These results indicated that continuous monitoring of NDM-positive E. coli in livestock farms and their surrounding environments is required to ensure public health safety.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: “NCBI BioProject—PRJNA826982.”
Ethics Statement
The Institutional Review Board of South China Agricultural University (SCAU-IRB) approved the samples and bacteria protocols. All animal faeces were sampled under authorization from Animal Research Committees of South China Agricultural University (SCAU-IACUC).
Author Contributions
YY and Y-HL conceived of this study. R-SY, XK, YZ, and JL performed the experiments and collected the data. XK, R-SY, YZ, and Z-YQ analyzed and interpreted the data. XK drafted the manuscript. YY revised the manuscript. X-PL and JS coordinated the whole project. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31730097), Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (2019BT02N054), the Program for Innovative Research Team in the Ministry of Education of China (IRT_17R39), the Innovation Team Project of Guangdong University (2019KCXTD001) and the 111 Project (D20008). R-SY was funded by GDAS’ Project of Science and Technology Development (2020GDASYL-20200103031).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.912260/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | PFGE analysis of 77 blaNDM-positive Escherichia coli isolates from food-producing animals. Triangles represent that the corresponding isolates sharing similar PFGE patterns.
Footnotes
References
Baek, J. Y., Cho, S. Y., Kim, S. H., Kang, C. I., Peck, K. R., Song, J. H., et al. (2019). Plasmid analysis of Escherichia coli isolates from South Korea co-producing NDM-5 and OXA-181 carbapenemases. Plasmid 104:102417. doi: 10.1016/J.PLASMID.2019.102417
Berrazeg, M., Diene, S. M., Medjahed, L., Parola, P., Drissi, M., Raoult, D., et al. (2014). New Delhi metallo-beta-lactamase around the world: An ereview using google maps. Eur. Secur. 19:20809. doi: 10.2807/1560-7917.ES2014.19.20.20809
Bonomo, R. A., Burd, E. M., Conly, J., Limbago, B. M., Poirel, L., Segre, J. A., et al. (2018). Carbapenemase-producing organisms: a global scourge. Clin. Infect. Dis. 66, 1290–1297. doi: 10.1093/cid/cix893
Cen, D. J., Sun, R. Y., Mai, J. L., Jiang, Y. W., Wang, D., Guo, W. Y., et al. (2021). Occurrence and transmission of blaNDM-carrying Enterobacteriaceae from geese and the surrounding environment on a commercial goose farm. Appl. Environ. Microbiol. 87, 1–13. doi: 10.1128/AEM.00087-21
Costa, A., Figueroa-Espinosa, R., Gaudenzi, F., Lincopan, N., Fuga, B., Ghiglione, B., et al. (2021). Co-occurrence of NDM-5 and RmtB in a clinical isolate of Escherichia coli belonging to CC354 in Latin America. Front. Cell. Infect. Microbiol. 11:654852. doi: 10.3389/FCIMB.2021.654852/PDF
Founou, L. L., Founou, R. C., and Essack, S. Y. (2016). Antibiotic resistance in the food chain: a developing country-perspective. Front. Microbiol. 7:1881. doi: 10.3389/fmicb.2016.01881
Ghatak, S., Singha, A., Sen, A., Guha, C., Ahuja, A., Bhattacharjee, U., et al. (2013). Detection of New Delhi metallo-beta-lactamase and extended-spectrum beta-lactamase genes in Escherichia coli isolated from mastitic milk samples. Transbound. Emerg. Dis. 60, 385–389. doi: 10.1111/TBED.12119
Ho, P. L., Wang, Y., Liu, M. C. J., Lai, E. L. Y., Law, P. Y. T., Cao, H., et al. (2018). IncX3 epidemic plasmid carrying blaNDM-5 In Escherichia coli from swine in multiple geographic areas in China. Antimicrob. Agents Chemother. 62, e02295–17. doi: 10.1128/AAC.02295-17
Köck, R., Daniels-Haardt, I., Becker, K., Mellmann, A., Friedrich, A. W., Mevius, D., et al. (2018). Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin. Microbiol. Infect. 24, 1241–1250. doi: 10.1016/j.cmi.2018.04.004
Kong, L. H., Lei, C. W., Ma, S. Z., Jiang, W., Liu, B. H., Wang, Y. X., et al. (2017a). Various sequence types of Escherichia coli isolates coharboring blaNDM-5 and mcr-1 genes from a commercial swine farm in China. Antimicrob. Agents Chemother. 61, e02167–16. doi: 10.1128/AAC.02167-16
Kumarasamy, K. K., Toleman, M. A., Walsh, T. R., Bagaria, J., Butt, F., Balakrishnan, R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. doi: 10.1016/S1473-3099(10)70143-2
Li, J., Bi, Z., Ma, S., Chen, B., Cai, C., He, J., et al. (2019). Inter-host transmission of carbapenemase-producing Escherichia coli among humans and backyard animals. Environ. Health Perspect. 127:107009. doi: 10.1289/EHP5251
Li, B., Yin, F., Zhao, X., Guo, Y., Wang, W., Wang, P., et al. (2020). Colistin resistance gene mcr-1 mediates cell permeability and resistance to hydrophobic antibiotics. Front. Microbiol. 10, 1–7. doi: 10.3389/fmicb.2019.03015
Liu, Z., Li, J., Wang, X., Liu, D., Ke, Y., Wang, Y., et al. (2018). Novel variant of New Delhi metallo-β-lactamase, NDM-20, in Escherichia coli. Front. Microbiol. 9:497. doi: 10.3389/fmicb.2018.00248
Liu, Z., Wang, Y., Walsh, T. R., Liu, D., Shen, Z., Zhang, R., et al. (2017). Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a sequence type 48 Escherichia coli strain. Antimicrob. Agents Chemother. 61. doi: 10.1128/AAC.02233-16
Liu, Z., Xiao, X., Li, Y., Liu, Y., Li, R., and Wang, Z. (2019). Emergence of IncX3 plasmid-harboring blaNDM–5 dominated by Escherichia coli ST48 in a goose farm in Jiangsu. China. Front. Microbiol. 10:2002. doi: 10.3389/fmicb.2019.02002
Logan, L. K., and Weinstein, R. A. (2017). The epidemiology of Carbapenem-resistant enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis. 215, S28–S36. doi: 10.1093/infdis/jiw282
López-Hernández, I., García Barrionuevo, A., Díaz de Alba, P., Clavijo, E., and Pascual, A. (2020). Characterization of NDM-1- and CMH-3-producing Enterobacter cloacae complex ST932 in a patient transferred from Ukraine to Spain. Enfermedades Infecc. y Microbiol. Clin. (English ed.) 38, 327–330. doi: 10.1016/J.EIMC.2019.10.007
Lv, L. C., Lu, Y. Y., Gao, X., He, W. Y., Gao, M. Y., Mo, K. B., et al. (2022). Characterization of NDM-5-producing Enterobacteriaceae isolates from retail grass carp (Ctenopharyngodon idella) and evidence of Bla NDM-5-bearing IncHI2 plasmid transfer between ducks and fish. Zool. Res. 43, 255–264. doi: 10.24272/J.ISSN.2095-8137.2021.426
Mantilla-Calderon, D., Jumat, M. R., Wang, T., Ganesan, P., Al-Jassim, N., and Hong, P. Y. (2016). Isolation and characterization of NDM-positive Escherichia coli from municipal wastewater in Jeddah, Saudi Arabia. Antimicrob. Agents Chemother. 60, 5223–5231. doi: 10.1128/AAC.00236-16
Mariana Huband, M. D., Mendes, R. E., and Flamm, R. K. F. (2017). Meropenem-vaborbactam tested against contemporary gram-negative isolates collected worldwide during 2014, including carbapenem-resistant, KPC-producing, multidrug-resistant, and extensively drug-resistant Enterobacteriaceae. Am. Soc. Microbiol. 61, 1–12. doi: 10.1128/AAC.00567-17
Mollenkopf, D. F., Stull, J. W., Mathys, D. A., Bowman, A. S., Feicht, S. M., Grooters, S. V., et al. (2017). Carbapenemase-producing Enterobacteriaceae recovered from the environment of a swine farrow-to-finish operation in the United States. Antimicrob. Agents Chemother. 61, e01298–16. doi: 10.1128/AAC.01298-16
Munoz-Price, L. S., Poirel, L., Bonomo, R. A., Schwaber, M. J., Daikos, G. L., Cormican, M., et al. (2013). Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13, 785–796. doi: 10.1016/S1473-3099(13)70190-7
Nakano, R., Nakano, A., Hikosaka, K., Kawakami, S., Matsunaga, N., Asahara, M., et al. (2014). First report of metallo-β-lactamase NDM-5-producing Escherichia coli in Japan. Antimicrob. Agents Chemother. 58, 7611–7612. doi: 10.1128/AAC.04265-14
Nordmann, P., Boulanger, A. E., and Poirel, L. (2012a). NDM-4 metallo-β-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob. Agents Chemother. 56, 2184–2186. doi: 10.1128/AAC.05961-11
Nordmann, P., Poirel, L., and Dortet, L. (2012b). Rapid detection of carbapenemase-producing enterobacteriaceae. Emerg. Infect. Dis. 18, 1503–1507. doi: 10.3201/eid1809.120355
Poirel, L., Jayol, A., Nordmann, P., Aurélie, J., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes Laurent Poirela,b,c, Aurélie Jayola,b,c and Patrice Nordmann. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Pruthvishree, B. S., Vinodh Kumar, O. R., Sinha, D. K., Malik, Y. P. S., Dubal, Z. B., Desingu, P. A., et al. (2017). Spatial molecular epidemiology of carbapenem-resistant and New Delhi metallo beta-lactamase (blaNDM)-producing Escherichia coli in the piglets of organized farms in India. J. Appl. Microbiol. 122, 1537–1546. doi: 10.1111/jam.13455
Ramadan, H., Gupta, S. K., Sharma, P., Ahmed, M., Hiott, L. M., Barrett, J. B., et al. (2020). Circulation of emerging NDM-5-producing Escherichia coli among humans and dogs in Egypt. Zoonoses Public Health 67, 324–329. doi: 10.1111/ZPH.12676
Seng, P., Drancourt, M., Gouriet, F., Scola, B.La, Fournier, P. E., Rolain, J. M., et al. (2009). Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49, 543–551. doi:doi: 10.1086/600885
Sugawara, Y., Akeda, Y., Hagiya, H., Sakamoto, N., Takeuchi, D., Shanmugakani, R. K., et al. (2019). Spreading patterns of NDM-producing Enterobacteriaceae in clinical and environmental settings in Yangon. Myanmar. Antimicrob. Agents Chemother. 63, e01924–18. doi: 10.1128/AAC.01924-18
Van, T. T. H., Yidana, Z., Smooker, P. M., and Coloe, P. J. (2020). Antibiotic use in food animals worldwide, with a focus on Africa: pluses and minuses. J. Glob. Antimicrob. Resist. 20, 170–177. doi: 10.1016/j.jgar.2019.07.031
Wailan, A. M., Paterson, D. L., Kennedy, K., Ingram, P. R., Bursle, E., and Sidjabat, H. E. (2015). Genomic characteristics of NDM-producing Enterobacteriaceae isolates in Australia and their blaNDM genetic contexts. Antimicrob. Agents Chemother. 60, 136–141. doi: 10.1128/AAC.01243-15
Walsh, F., and Duffy, B. (2013). The culturable soil antibiotic Resistome: a community of multi-drug resistant bacteria. PLoS One 8:e65567. doi: 10.1371/journal.pone.0065567
Wang, Y., Zhang, R., Li, J., Wu, Z., Yin, W., Schwarz, S., et al. (2017). Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2:16260. doi: 10.1038/nmicrobiol.2016.260
Wang, M. G., Zhang, R. M., Wang, L. L., Sun, R. Y., Bai, S. C., Han, L., et al. (2021). Molecular epidemiology of carbapenemase-producing Escherichia coli from duck farms in south-east coastal China. J. Antimicrob. Chemother. 76, 322–329. doi: 10.1093/jac/dkaa433
Williams, S. H., Che, X., Paulick, A., Guo, C., Lee, B., Muller, D., et al. (2018). New York City house mice (Mus musculus) as potential reservoirs for pathogenic bacteria and antimicrobial resistance determinants. mBio 9, e00624–18. doi: 10.1128/mBio.00624-18
Woodford, N., Wareham, D. W., Guerra, B., and Teale, C. (2014). Carbapenemase-producing Enterobacteriaceae and non-Enterobacteriaceae from animals and the environment: an emerging public health risk of our own making? J. Antimicrob. Chemother. 69, 287–291. doi: 10.1093/jac/dkt392
Xavier, B. B., Lammens, C., Butaye, P., Goossens, H., and Malhotra-Kumar, S. (2016). Complete sequence of an IncFII plasmid harbouring the colistin resistance gene mcr-1 isolated from Belgian pig farms. J. Antimicrob. Chemother. 71, 2342–2344. doi: 10.1093/jac/dkw191
Yang, R. S., Feng, Y., Lv, X. Y., Duan, J. H., Chen, J., Fang, L. X., et al. (2016). Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a single Muscovy duck (Cairina moschata). Antimicrob. Agents Chemother. 60, 6899–6902. doi: 10.1128/AAC.01365-16
Yao, H., Li, A., Yu, R., Schwarz, S., Dong, H., and Du, X. D. (2020). Multiple copies of Bla NDM-5 located on conjugative Megaplasmids from porcine Escherichia coli sequence type 218 isolates. Antimicrob. Agents Chemother. 64, e02134–19. doi: 10.1128/AAC.02134-19
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, Bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Zhai, R., Fu, B., Shi, X., Sun, C., Liu, Z., Wang, S., et al. (2020). Contaminated in-house environment contributes to the persistence and transmission of NDM-producing bacteria in a Chinese poultry farm. Environ. Int. 139:105715. doi: 10.1016/j.envint.2020.105715
Zhang, R., Liu, L., Zhou, H., Chan, E. W., Li, J., Fang, Y., et al. (2017). Nationwide surveillance of clinical Carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine 19, 98–106. doi: 10.1016/j.ebiom.2017.04.032
Zhang, Q., Lv, L., Huang, X., Huang, Y., Zhuang, Z., Lu, J., et al. (2019b). Rapid increase in carbapenemase-producing Enterobacteriaceae in retail meat driven by the spread of the blaNDM-5-carrying IncX3 plasmid in China from 2016 to 2018. Antimicrob. Agents Chemother. 63, e00573–19. doi: 10.1128/AAC.00573-19
Zhang, P., Wang, J., Wang, X., Bai, X., Ma, J., Dang, R., et al. (2019a). Characterization of five escherichia coli isolates co-expressing ESBL and mcr-1 resistance mechanisms from different origins in China. Front. Microbiol. 10, 1–9. doi: 10.3389/fmicb.2019.01994
Zhang, Y., Wang, Q., Yin, Y., Chen, H., Jin, L., Gu, B., et al. (2018). Epidemiology of carbapenem-resistant enterobacteriaceae infections: report from the China CRE network. Antimicrob. Agents Chemother. 62, 1–11. doi: 10.1128/AAC.01882-17
Keywords: NDM, food-producing animals, epidemiology, antimicrobial resistance, the environment
Citation: Kuang X, Zhang Y, Liu J, Yang R-S, Qiu Z-Y, Sun J, Liao X-P, Liu Y-H and Yu Y (2022) Molecular Epidemiology of New Delhi Metallo-β-Lactamase-Producing Escherichia coli in Food-Producing Animals in China. Front. Microbiol. 13:912260. doi: 10.3389/fmicb.2022.912260
Edited by:
Shaolin Wang, China Agricultural University, ChinaReviewed by:
Guyue Cheng, Huazhong Agricultural University, ChinaYonghong Xiao, Zhejiang University, China
Copyright © 2022 Kuang, Zhang, Liu, Yang, Qiu, Sun, Liao, Liu and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Yu, eXlzY2F1QDEyNi5jb20=
†These authors have contributed equally to this work
 Xu Kuang
Xu Kuang Yan Zhang
Yan Zhang Juan Liu1,2,3
Juan Liu1,2,3 Jian Sun
Jian Sun Xiao-Ping Liao
Xiao-Ping Liao Ya-Hong Liu
Ya-Hong Liu Yang Yu
Yang Yu