
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 03 November 2022
Sec. Microbial Symbioses
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.911623
Candida albicans is a symbiotic fungus that commonly colonizes on oral mucosal surfaces and mainly affects immuno-compromised individuals. Polymicrobial interactions between C. albicans and oral microbes influence the cellular and biochemical composition of the biofilm, contributing to change clinically relevant outcomes of biofilm-related oral diseases, such as pathogenesis, virulence, and drug-resistance. Notably, the symbiotic relationships between C. albicans and oral bacteria have been well-documented in dental caries, oral mucositis, endodontic and periodontal diseases, implant-related infections, and oral cancer. C. albicans interacts with co-existing oral bacteria through physical attachment, extracellular signals, and metabolic cross-feeding. This review discusses the bacterial–fungal interactions between C. albicans and different oral bacteria, with a particular focus on the underlying mechanism and its relevance to the development and clinical management of oral diseases.
The oral cavity is one of the main sites of microorganisms colonization on the human body. More than 700 species of microorganisms can be detected in the human oral cavity, including bacteria, fungi, viruses, mycoplasma, rickettsia, and protozoa (Dewhirst et al., 2010). Microbiota that resides in the oral cavity can be symbiotic, competitive, and antagonistic to maintain the balance of microecology, which determines oral health and the development of biofilm-related oral diseases. Oral microbiota can be opportunistic pathogens when the oral microenvironment or personal oral hygiene changes (Wolcott et al., 2013). Recently, cross-kingdom interactions between fungi and oral bacteria have drawn increasing attention. Candida albicans can interact with a variety of oral microbes and their interactions are interdependent and mutually beneficial rather than unidirectional. These polymicrobial interactions have been demonstrated in the pathogenesis of biofilm-related oral diseases, including dental caries, oral candidosis, endodontic diseases, periodontitis, implant-related infections, and oral cancer (Bamford et al., 2009; Shirtliff et al., 2009; Morales and Hogan, 2010; Peleg et al., 2010; Harriott and Noverr, 2011; Diaz et al., 2012; Koo et al., 2018; Lohse et al., 2018).
C. albicans is a symbiotic fungus commonly colonizing on the mucosal surfaces of living bodies. The detection rate of C. albicans in the healthy population is 18.5∼40.9% (Mun et al., 2016; Zomorodian et al., 2016; Babatzia et al., 2020). Individuals with compromised immune systems, such as HIV-positive individuals, newborns, and the elderly, are susceptible to C. albicans infection (Shay et al., 1997; Zomorodian et al., 2016; Diaz et al., 2017; Babatzia et al., 2020; Vila et al., 2020). C. albicans biofilms contain yeast, pseudo-hyphal, and hyphal form cells, surrounded by extracellular matrix (Wang, 2015; Lohse et al., 2018). After adherence to the surface, C. albicans cells proliferate in the form of yeast and begin to form hyphal, elongating and proliferating throughout the biofilm maturation process (Lohse et al., 2018). The yeast-to-hypha transition is widely recognized as a key virulence trait of C. albicans associated with biofilm formation (Garcia-Sanchez et al., 2004). The synergistic effects of C. albicans and commensal bacteria have been well-studied in the context of importance to the microbiological community, which impact on the virulence of polymicrobial biofilms and antibiotic resistance (Morales and Hogan, 2010; Diaz et al., 2014; Allison et al., 2016; Janus et al., 2016; Koo et al., 2018). C. albicans can interact with oral bacteria via physical attachment through fungal cell walls (e.g., surface proteins and extracellular polysaccharides, EPS), extracellular signals, metabolite cross-feeding, and environmental changes (Figure 1; Allison et al., 2016; Koo et al., 2018; Khan et al., 2021). Here, we discusses recent findings on the C. albicans mutualistic interactions with oral commensal bacteria, particularly focusing on the underlying mechanisms and relevance to biofilm-related oral diseases, aiming to provide new insights into prevention and treatment strategies for oral diseases.
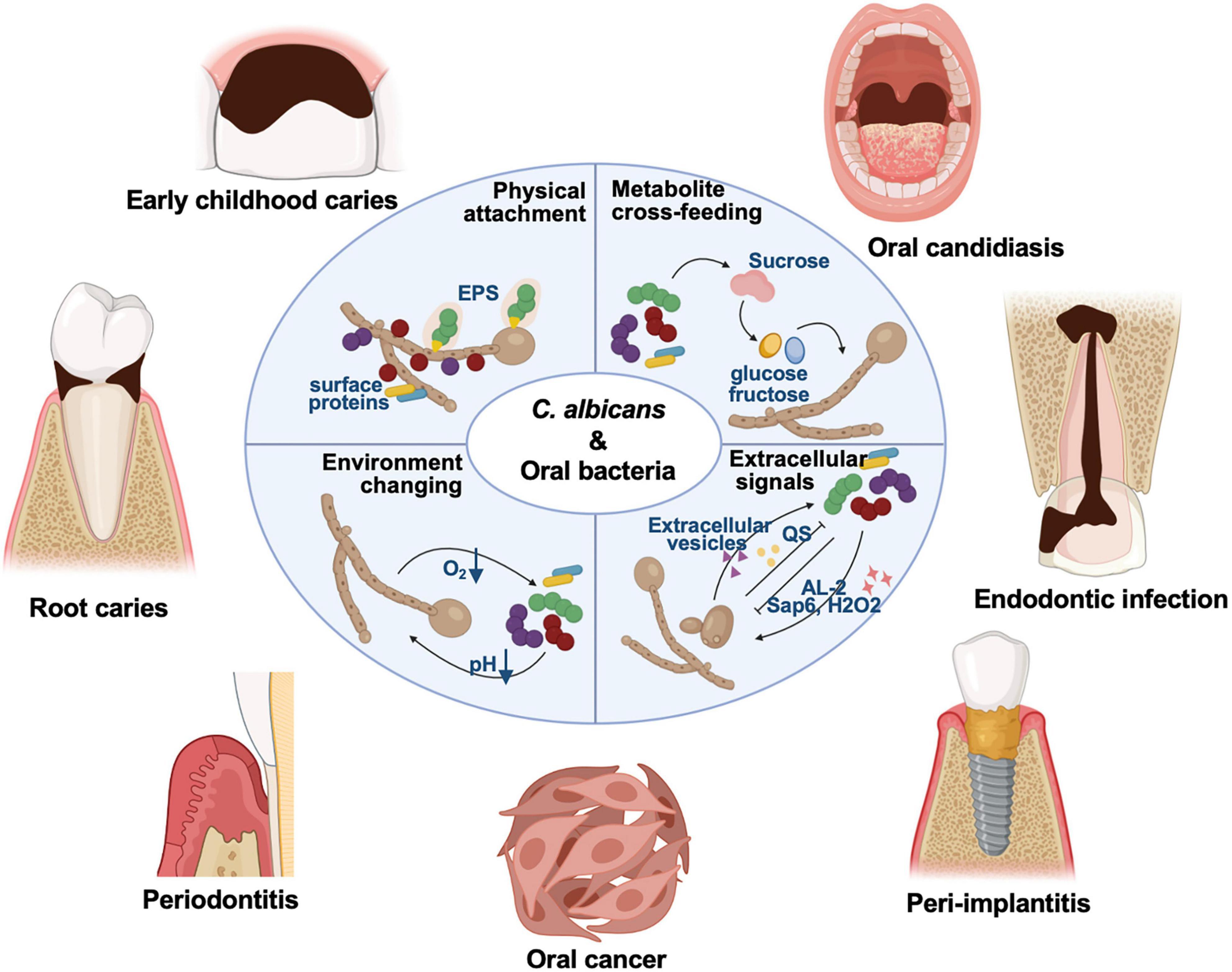
Figure 1. C. albicans can interact with a variety of oral microbes, and these cross-kingdom interactions have been demonstrated involved in the pathogenesis of oral diseases for years, including dental caries, oral candidosis, endodontic disease, periodontitis, and other biofilm-related oral diseases. C. albicans can interact with oral bacteria via physical attachment through fungal cell walls (e.g., surface proteins and EPS), extracellular signals, metabolite cross-feeding, and environment changing.
Cell-cell adhesion is one of the main and common factors mediating C. albicans and oral bacteria interactions, reciprocally aiding the colonization of both fungi and bacteria (Zijnge et al., 2010; Diaz et al., 2012; Falsetta et al., 2014; Xu et al., 2014a; Hwang et al., 2015; Nobbs and Jenkinson, 2015; Koo et al., 2018; Dige and Nyvad, 2019; Kim and Koo, 2020). Fungal cell walls are composed of different polysaccharides, including glucans, mannans, and chitin (Gow et al., 2017). There are a variety of adhesion proteins or receptors existing on the fungal cell wall. C. albicans physically interacts with oral microorganisms through these proteins or receptors to form a well-characterized structure, “cross-kingdom corncob” (Figure 2), such as mitis group streptococci (MGS, e.g., Streptococcus gordonii, Streptococcus sanguinis, and Streptococcus oralis), Porphyromonas gingivalis, and Staphylococcus aureus (Zijnge et al., 2010; Diaz et al., 2012; Peters et al., 2012; Falsetta et al., 2014; Xu et al., 2014a; Hwang et al., 2015; Nobbs and Jenkinson, 2015; Koo et al., 2018; Dige and Nyvad, 2019; Kim and Koo, 2020). Hyphae formation has been confirmed to be the preferred C. albicans morphotype of these microorganisms that adhere to (Brady et al., 2010; Silverman et al., 2010; Peters et al., 2012; Xu et al., 2014a; Bamford et al., 2015; Koo et al., 2018). Visualized by multi-color fluorescence microscopy, numerous clusters of either S. oralis or S. gordonii are observed forming around C. albicans hyphae within a few hours with a sufficient supply of nutrients (Bamford et al., 2009; Diaz et al., 2012), and even in poor nutrient microconditions, S. oralis can also bind to Candida germ tubes (Diaz et al., 2012). C. albicans hyphae-specific cell wall adhesins Als and Hwp1 appear to mediate the binding of this fungus to oral bacteria, such as S. gordonii, S. oralis, P. gingivalis, and S. aureus, and two antigen I/II family members of Streptococcal cell-surface adhesins, SspA and SspB, are illustrated to be the key points of interaction with C. albicans hyphae (Silverman et al., 2010; Peters et al., 2012; Xu et al., 2014a; Bamford et al., 2015; Xu et al., 2017; Koo et al., 2018; Bartnicka et al., 2019). Specifically, S. gordonii SspB protein directly interacts with C. albicans through the N-terminal domain of Als3 on the C. albicans hyphal filament surface (Silverman et al., 2010; Bamford et al., 2015). Meanwhile, C. albicans cell wall mannoproteins and O-mannosylation contribute to the development of inter-kingdom biofilm (Dutton et al., 2014). C. albicans O-mannosylation deficient strain (mnt1Δ mnt2Δ mutant) has defective functionality of adhesins, and its hyphal filaments do not interact with SspB adhesin on the S. gordonii surface or C. albicans Als3/Hwp1 protein, which is different from C. albicans wild-type hyphae (Dutton et al., 2014). Furthermore, the aspartyl proteinase Sap9, which is involved in designing of mono- and dual-species biofilms architecture (Dutton et al., 2016), is absent in this mutant. Increased gene expression of ALS3 and HWP1 has also been observed in C. albicans when co-cultured with P. gingivalis (Bartnicka et al., 2019), and the interaction of these two microorganisms occurs directly through the fungal adhesin Als3 and gingipain RgpA (Bartnicka et al., 2019). Unlike MGS, antigen I/II has been shown to mediate the dual-species biofilm interaction of C. albicans and S. mutans in an Als1/Als3-independent manner, and the underlying mechanism needs further exploration (Yang et al., 2018).
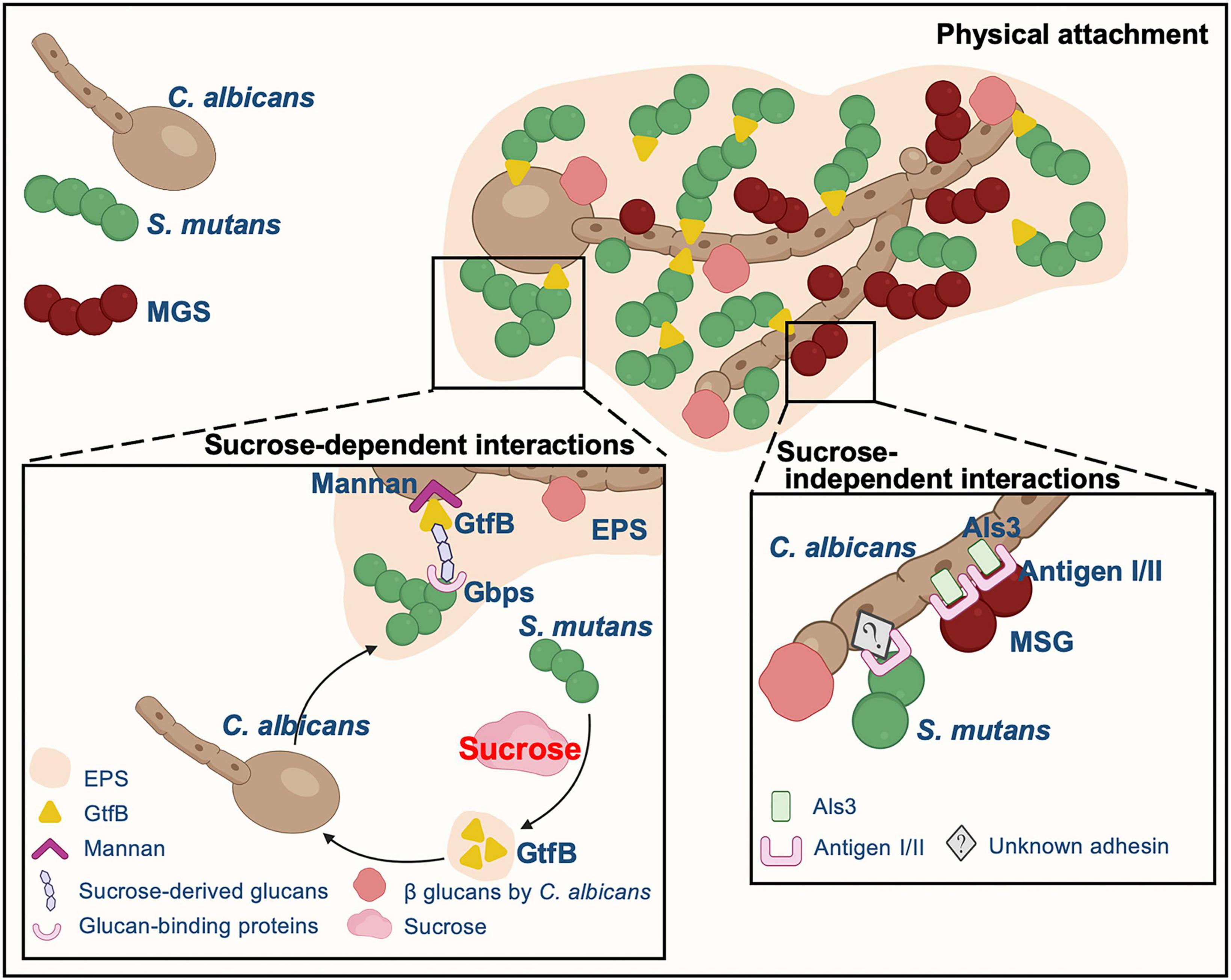
Figure 2. The typical physical attachment mechanisms underlying the interaction between C. albicans and streptococci. C. albicans physically interacts with MGS through proteins/receptors on cell wall surface of both fungi and bacteria whereas EPS is critical to the mutualistic interaction between C. albicans and S. mutans instead of cell-cell physical adhesion.
It is worth noting that although hyphae formation has been validated to be important in C. albicans mutualism effects with MGS (Brady et al., 2010; Silverman et al., 2010; Xu et al., 2014a; Bamford et al., 2015; Koo et al., 2018), hyphae formation may not affect the microorganism composition of more complicated polymicrobial communities (Xu et al., 2016; Montelongo-Jauregui et al., 2019; Du et al., 2021a). Efg1, a key C. albicans hyphae-associated morphological regulator, has been shown to be required for dual-species biofilm coaggregation between C. albicans and streptococci. Lacking hyphal formation, the efg1Δ/Δ strain forms significantly lighter biofilms with S. oralis compared with wild-type strain in vitro. S. oralis cannot upregulate ALS1, ALS3, and HWP1 gene expression of efg1Δ/Δ strain (Xu et al., 2017). Whereas it has been observed that S. oralis is still able to promote efg1Δ/Δ strain colonization in oral mucosa of mice (Xu et al., 2016), and it is especially apparent in synthetic saliva culture conditions that S. gordonii and the C. albicans efg1Δ/Δ and brg1Δ/Δ filamentation-deficient mutants interact in a highly synergistic manner (Montelongo-Jauregui et al., 2019). The occurrence of this phenomenon suggests that hyphae formation is required for Candida interactions with bacteria in vivo, which may be influenced by of oral environment as well as the presence of commensal microbiota.
Interestingly, different from MGS, synergetic collaboration between C. albicans and the main cariogenic bacteria S. mutans is dominated mainly by sucrose-dependent partnership (Figure 2). Sucrose significantly changes the adhesion pattern between S. mutans and C. albicans, increasing the connection between these two microorganisms. Sucrose allows S. mutans to produce EPS (also termed as water insoluble glucans), which is critical to the interaction between C. albicans and S. mutans rather than physical cell-cell adhesion (Bowen and Koo, 2011; Gregoire et al., 2011; Falsetta et al., 2014; Hwang et al., 2015; Hwang et al., 2017; Koo et al., 2018; Kim and Koo, 2020). RNA-Seq data have demonstrated that the presence of C. albicans in biofilms dramatically alters 393 gene expression in S. mutans and most of the upregulated genes are involved in carbohydrate transport and metabolic/catabolic processes (He et al., 2017). In the C. albicans/S. mutans mixed biofilm, S. mutans forms a large amount of microcolonies around fungal cells instead of directly adhering to the cell wall surface of Candida, and the microcolonies are enmeshed in an EPS-rich extracellular matrix (Falsetta et al., 2014; Hwang et al., 2017; Koo et al., 2018; Kim and Koo, 2020). In sucrose-limited environments, C. albicans adheres to the surface of S. gordonii about two times stronger than it adheres to S. mutans. However, in the presence of glucans, the binding force between S. mutans and C. albicans surfaces is dramatically elevated (∼6 folds) (Wan et al., 2021). S. mutans effectively synthesizes EPS from dietary sucrose through glucosyltransferases (Gtfs). All S. mutans Gtfs can adhere to C. albicans cell surfaces; among them, GtfB exhibits the greatest affinity (Bowen and Koo, 2011; Gregoire et al., 2011). The glucans produced abundantly by surface-bound GtfB on both organismal cells and tooth hard tissue concurrently enhance the efficiency and stability of C. albicans in attaching to and colonizing on the teeth. Simultaneously, the glucans on fungal cell walls in turn improve S. mutans-binding sites (Bowen and Koo, 2011; Gregoire et al., 2011; Falsetta et al., 2014; Huffines and Scoffield, 2020; Kim and Koo, 2020). C. albicans has a significantly larger surface area than bacteria with a plentiful source of Gtfs-binding sites for S. mutans colonization (Gregoire et al., 2011). GtfB adhesion to C. albicans is 2.5 folds stronger and 20 folds more stable than adhesion to S. mutans according to atomic force microscopy; however, there is an uneven distribution of GtfB-binding domains on the fungal surface (Hwang et al., 2015). On the other hand, C. albicans can also secrete polysaccharides that mediate mixed biofilm formation (Hall and Gow, 2013; Mitchell et al., 2015; Khoury et al., 2020). β-1,3-glucans synthesized by Candida also contribute to the co-cultured biofilm matrix structure (Falsetta et al., 2014; Khoury et al., 2020). C. albicans cell wall mannan provides sites for GtfB binding and function (Falsetta et al., 2014; Hwang et al., 2017; Kim et al., 2021). GtfB-binding domains have been demonstrated to reside in both the O- and N-terminal structures of mannans in Candida cell walls. C. albicans pmt4Δ/Δ and och1Δ/Δ strains exhibit functional defects in biological synthesis of both N- and O-linked mannans, and GtfB-binding properties are compromised compared to wild-type strain (Hwang et al., 2017). Meanwhile, mannan-degrading endo- and exo-enzymes reduce the GtfB-binding forces to C. albicans by about 15 folds, accompanied with thicker biofilm biomass, under the premise of eliminating the possibility of killing microorganisms (Kim et al., 2021).
A range of extracellular signals appear to facilitate C. albicans synergistic interactions with oral bacteria, including signaling molecules, quorum sensing molecules (QS), and other factors (Table 1; Peleg et al., 2010; Harriott and Noverr, 2011; Xu et al., 2014a; Nobbs and Jenkinson, 2015; Lohse et al., 2018). QS is a microbial cell-to-cell communication process principally dependents on population density. As shown in an in vitro study, early in the exponential growth phase, about 4 h into culturing, S. mutans secretes competence-stimulating peptide (CSP) (Jarosz et al., 2009) and fatty acid signaling molecule trans-2-decenoic (SDSF) (Vilchez et al., 2010), which inhibit germ tube formation of C. albicans, thus favoring fungal proliferation. SDSF activity can be also detected in S. mitis, S. oralis, and S. sanguinis (Vilchez et al., 2010). In the later stage of biofilm maturity, the inhibitory germ tube formation effect on C. albicans disappears, and C. albicans produces a large number of hyphae and becomes more virulent (Jarosz et al., 2009; Vilchez et al., 2010). S. mutans can also upregulate comC of C. albicans, which encodes CSP (He et al., 2017) and the S. gordonii comCDE QS-system, which modulates the dual-species biofilm co-cultured with C. albicans (Jack et al., 2015). Meanwhile, when co-cultured with S. gordonii, C. albicans forms hyphae earlier and more extensively, which is mediated to a certain extent by autoinducer-2, a universal signaling molecule in interactions between bacterial species. S. gordonii effectively suppresses the inhibitory effect of farnesol on C. albicans filamentation in a LuxS-dependent pattern (Bamford et al., 2009). The role of QS molecule farnesol in C. albicans yeast-to-hyphae transition has long been recognized. A non-monotonic response to farnesol concentration is observed in S. mutans growth. In the C. albicans/S. mutans mixed biofilm, low levels of farnesol (∼25 μm) stimulate GtfB expression/activity and increase bacterial growth. In contrast, an abundance of farnesol (> 100 μm) inhibits S. mutans growth (Kim et al., 2017). Moreover, membrane vesicles produced by S. mutans can augment C. albicans biofilm development without significantly affect its planktonic growth (Wu et al., 2020).
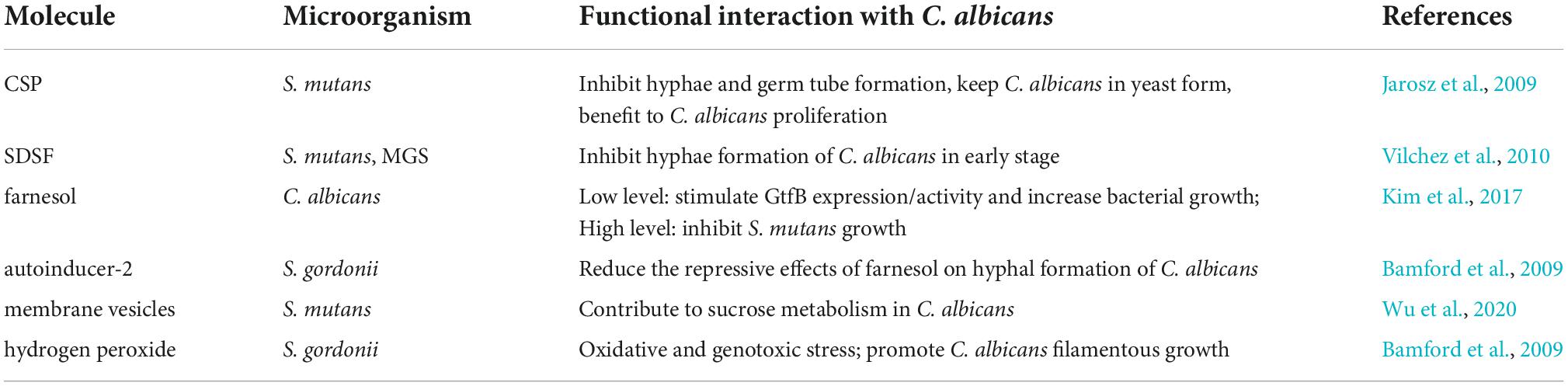
Table 1. Summary of extracellular signals modulating C. albicans interactions between oral bacteria.
C. albicans can use glucose as a carbon source but possesses insufficient ability to metabolize dietary sucrose. Streptococci may provide a nutritional source for C. albicans and enable its persistence under acidic condition (Bamford et al., 2009; Sztajer et al., 2014; He et al., 2017; Kim et al., 2017). S. mutans can rapidly breakdown sucrose to glucose and fructose, which can be utilized by C. albicans, thus elevating its growth (He et al., 2017; Kim et al., 2017). Sucrose and monosaccharides released by S. mutans from sucrose are depleted in spent medium of S. mutans/C. albicans dual-species biofilms at 10 h, whereas a large amount of carbohydrates remain in S. mutans single-species biofilms (Sztajer et al., 2014). With high salivary glucose levels, Candida colonization increases, insoluble EPS accumulates, and proteins and phospholipase activities increase (Brito et al., 2021). S. mutans and C. albicans dual-species biofilms produce lactate, along with small amounts of formate and fumarate as a result of carbohydrate metabolism, facilitating the growth of S. mutans and C. albicans within cariogenic biofilms, as both these organisms are acid-tolerant (He et al., 2017; Kim et al., 2017). Meanwhile, sucrose supplementation also reduces the inhibitory effect of sugar alcohols (Chan et al., 2020). In low-fermentable carbohydrate environments, C. albicans promotes the growth and biofilm formation of S. gordonii by elevating the enzymatic activities of cell wall-anchored glycoside hydrolases (GHs). GHs mediate the hydrolysis of glycoproteins, which is critical for the growth of S. gordonii in limited nutrient environment, such as saliva. Furthermore, C. albicans gene TEC1 is critical for this cross-kingdom metabolic communication (Zhou et al., 2022).
At the same time, C. albicans consumes oxygen in the local environment to ensure strictly anaerobic conditions within the biofilm that favor the growth of anaerobes (Shirtliff et al., 2009; Fox et al., 2014; Janus et al., 2017; Bartnicka et al., 2019; Du et al., 2021a). Specifically, the presence of C. albicans alters the microbial composition of oral biofilms (Janus et al., 2017; Du et al., 2021a), increasing the abundance of strictly anaerobic Veillonella, Prevotella, Leptotrichia, and Fusobacterium genera under oxygen-rich conditions (Janus et al., 2017). The presence of C. albicans significantly increases Streptococcus in the saliva-derived biofilms in our previous study (Du et al., 2021a). C. albicans can also increase the viability of P. gingivalis biofilm by 20% in a normoxic environment (Bartnicka et al., 2019). Meanwhile, peptidoglycan fragments (Xu et al., 2008; Wang, 2013) and hydrogen peroxide (Nasution et al., 2008; Bamford et al., 2009) produced by catalase-negative bacteria can enhance C. albicans filamentation.
Dental caries, also known as tooth decay, is a representative biofilm-related oral disease and occurs as a result of microbial dysbiosis characterized with the enrichment of acidogenic pathogens and depletion of alkali-generating commensal microbes within the plaque biofilm (Pitts, 2016; Pitts et al., 2017; Bowen et al., 2018). Evidence has supported that C. albicans is closely related to the occurrence of dental caries. A recent meta-analysis reveals that the prevalence of dental caries in individuals carrying Candida is higher than those free of Candida in the oral cavity. Associations between oral Candida carriage and the occurrence of dental caries in children/adolescents and adults have been demonstrated (Eidt et al., 2020). A study of 132 patients with caries and 58 non-caries controls has demonstrated that patients with caries have greater Candida colonization than individuals without caries, and a significant clinical correlation between oral Candida carriage and dental caries incidence has been noted (De-la-Torre et al., 2016). A cross-sectional study on 160 patients demonstrates that caries experience is significantly associated with higher oral Candida carriage (Al-Amad et al., 2021). Consistent with clinical studies, C. albicans and S. mutans co-infection synergistically aggravates the onset of different types of dental caries with severe lesions, including smooth surface, and pit and fissure caries (Falsetta et al., 2014; Koo and Bowen, 2014; Thomas et al., 2016; Khoury et al., 2020; Kim et al., 2020; Du et al., 2021a). C. albicans inoculation significantly enhances the maturity of multi-species biofilm and sustains an acidic environment in the mixed biofilm with oral streptococci (Kim et al., 2020; Du et al., 2021a). More importantly, the growth of cariogenic bacteria S. mutans within the mixed biofilm under cariogenic conditions is dramatically improved by C. albicans (Falsetta et al., 2014; Koo and Bowen, 2014; Thomas et al., 2016; Khoury et al., 2020; Kim et al., 2020; Du et al., 2021a); Oral biofilms with C. albicans also express elevated levels of genes associated with acid production (ldh) and aciduricity (fabM and atpD) of cariogenic bacteria (Falsetta et al., 2014; Du et al., 2021a), whereas genes associated with ammonia production by commensal streptococci (arcA and ureC) are downregulated (Du et al., 2021a). In addition, the presence of C. albicans upregulates genes encoding CiaRH, such as ciaR and ciaH, which are implicated in the biofilm formation, acid-tolerance, and sucrose-dependent adherence of S. mutans (He et al., 2017). C. albicans significantly increases both S. mutans and saliva-derived biofilm demineralization ability on tooth hard tissue in an in vitro artificial caries model, manifested by deeper demineralized lesion and increased mineral loss (Sampaio et al., 2019; Du et al., 2021a). These promotive effects are largely accredited to C. albicans PHR2, and deletion of PHR2 partially restored the microbial ecology of the polymicrobial biofilm, resulting in a biofilm with decreased acidogenicity, compromised demineralizing capability and reduced cariogenicity (Du et al., 2021a). Above all, root caries and early childhood caries (ECC) are most well-documented as associated with C. albicans, both commonly affected on the smooth surface of the teeth.
Root caries develops on the root surface where gingival recede and the root are exposed to the oral environment. C. albicans has been isolated and identified from root caries lesions for decades (Beighton and Lynch, 1993; Shen et al., 2002; Zaremba et al., 2006). Notably, longitudinal studies have revealed positive correlations between root caries increment of older adults and the presence of Candida in saliva (Scheinin et al., 1992; Scheinin et al., 1994). Since the development of root caries involves both hydroxyapatite demineralization and organic materials (such as type I collagen) denaturation and degradation, the increased colonization of C. albicans promoted by S. mutans may play a significant and supplementary role in the disease process. In vitro and animal experiments suggest that C. albicans hyphae can enter dentin tubules and destroy the collagen fibers with proteolytic enzyme, and this collagen hydrolase is most active in acidic environments (Nikawa et al., 1998; Klinke and Klimm, 2002; Klinke et al., 2011). In total, two characteristic colonization patterns are observed in root caries biofilm in vivo. In one pattern, Candida hyphae form a network structure extending the entire biofilm, embedded with coccoid, rod-like, and filamentous bacteria. In the other pattern, Candida and streptococci (usually MGS) form corncob configurations in the surface layers of the biofilm, whereas S. mutans clusters in microcolonies mix with other oral commensal bacteria and does not typically appear close to C. albicans cells (Dige and Nyvad, 2019). Our previous study revealed that C. albicans is detected more frequently at root carious lesions than at sound root surfaces of the same patients as well as non-caries controls. Further quantitative analysis has shown that C. albicans colonization follows the same trend. Meanwhile, root caries lesions possess a larger amount of S. mutans and reduced carriage of commensal organisms (e.g., S. sanguinis). Furthermore, a significant correlation between S. mutans/S. sanguinis ratio and C. albicans carriage is observed in all recruited subjects (Du et al., 2021a). RNA-Seq analysis has demonstrated that C. albicans upregulates the genes associated with C. albicans metabolism, sugar transportation, invasion, stress tolerance, and pH regulation in supragingival plaque of root caries, compared with this fungus in dental plaque of sound root surfaces (Ev et al., 2020). Besides S. mutans, another key root caries pathogen, Actinomyces viscosus, is often found alongside C. albicans in supragingival plaques of root caries patients (Shen et al., 2002). C. albicans co-cultured with A. viscosus exhibits elevated biomass of both microorganisms, and the biofilm is more acidogenic and possesses more microcolonies, which causes more damage to hydroxyapatite, in vitro (Deng et al., 2019).
ECC is one of the aggressive forms of dental caries that occurs in children under six-year age. Oral C. albicans prevalence and carrier rate are both positively correlated with the severity of ECC (Raja et al., 2010; Wu et al., 2015; Xiao et al., 2016; Lozano Moraga et al., 2017; Xiao et al., 2018b; Fakhruddin et al., 2021). Epidemiological studies have found that oral infection rate of C. albicans in children with ECC is higher than that in caries-free children even by different detection methods (24∼100% vs. 10∼100% in saliva, 44∼80% vs. 7∼19% in plaque, 14.7∼44% vs. 6∼7% in swab samples). More importantly, the detection rate of this fungus in carious lesions is up to 60∼100% (Xiao et al., 2018b). Absolute quantitative data show that the colonization amounts of C. albicans in both saliva and dental plaque collected from children with severe ECC are also significantly higher than that in the same samples of caries-free subjects (Zaremba et al., 2006; Thomas et al., 2016; Xiao et al., 2016). Furthermore, higher C. albicans count is detected in dental plaque of ECC children accompanied by increased S. mutans colonization, which is associated with higher prevalence of active caries lesion (Sridhar et al., 2020), severity (DMFT/S) of ECC (Xiao et al., 2016), and caries recurrence (Hajishengallis et al., 2017). For recurrence subjects infected with S. mutans strains harboring collagen-binding proteins (Cbps), higher amounts of Candida and S. mutans are identified in caries dentin compared with those infected with Cbp– strains (Garcia et al., 2021). A recent study using 16s rRNA amplicon sequencing has revealed that oral C. albicans infection is accompanied by characteristic microbial communities comprising bacteria characterized by high acidogenicity and acid tolerance in severe ECC. Dental plaque of severe ECC subjects is harbored by an increased streptococci (particularly S. mutans), Veillonella and Prevotella, certain Lactobacillus/Scardovia species, and a decreased level of Actinomyces (Xiao et al., 2018a). Significant alteration in salivary fungal communities in severe ECC/ECC children is observed compared with the caries-free controls, and the fungal community is distinguished into five types based on the different oral health status, which significantly affect the bacterial profile (Tu et al., 2022).
Oral candidosis is a common fungal disease of oral mucosa with various subtypes. There are four subtypes of oral candidosis according to the classification of Lehner (1966): pseudomembranous candidosis, acute erythematous candidosis, chronic erythematous candidosis, and chronic hyperplastic candidosis. Oral candidosis is common in “the young, the old and the sick”, also known as “a disease of the diseased”. Overgrowth of C. albicans on the mucosal surface is traditionally regarded as the most common cause of oral candidosis. Recently, it has been increasingly defined as a combination of fungal and bacterial biofilm-induced disease (Dongari-Bagtzoglou et al., 2009; Nett et al., 2010; Johnson et al., 2012; Xu et al., 2014b). Commensal bacteria increase not only the colonization of C. albicans in mucosal niches but also the persistence of C. albicans. The interaction between this fungus and oral bacteria may further modulate the virulence of Candida biofilm. Denture stomatitis is one of the chronic erythematous candidosis. Since C. albicans has a strong affinity for denture materials, Candida-associated stomatitis affects up to 60% of denture wearers (Figueiral et al., 2007; Geerts et al., 2008). Candida species co-exist frequently with S. mutans and Staphylococcus aureus on denture surfaces and oral mucosa of denture users (Baena-Monroy et al., 2005). The amount of S. mutans in saliva of active denture wearers is significantly higher than that of natural oral teeth subjects and fixed denture wearers (Beighton et al., 1990; Tanaka et al., 2009; Valentini et al., 2013). Consistent with clinical findings, in vitro studies exhibit that S. mutans and S. oralis aid C. albicans biofilm formation on hydroxyapatite, polymethyl methacrylate, and soft denture liner disks (Pereira-Cenci et al., 2008). In total, two rodent models of denture stomatitis have identified the biofilm formed on denture surfaces comprising commensal bacteria and C. albicans (Nett et al., 2010; Johnson et al., 2012). Inoculating C. albicans with MGS reveals increased colonization and biofilm efficiency in in vitro human oral mucosae models with salivary flow. Due to low immunity, patients with autosomal-dominant hyper IgE syndrome are predisposed to C. albicans infection, which has shown to maintain severe dysbiosis oral mucosal microbial communities, dominated by C. albicans and particularly increased abundance of S. mutans and S. oralis in patients with active infection (Abusleme et al., 2018). Compared to mono-species, co-cultured C. albicans with either S. oralis or S. sanguinis shows dramatically increased colonization of both Streptococcus and Candida (Diaz et al., 2012).
Besides the enrichment of colonization, interaction with oral bacteria also increases the virulence of C. albicans. Multi-species infections of C. albicans and oral bacteria are characterized by higher proportion of C. albicans hyphae and worse tissue invasion (Nair et al., 2001; Bamford et al., 2009; Diaz et al., 2012; Xu et al., 2014a,b; Bertolini et al., 2015; Cavalcanti et al., 2015; Xu et al., 2016; Xu et al., 2017). As a result of co-infection with S. oralis, C. albicans is better able to invade the mucosa and produces a heightened inflammatory response in comparison with infection by either microorganism alone (Diaz et al., 2012; Xu et al., 2016), reflected by the denser biofilm with longer and higher proportion of C. albicans hyphae extending into the submucosal compartment (Xu et al., 2016; Morse et al., 2019). S. gordonii promotes C. albicans hyphal development, which reaches 60% when S. gordonii is deposited first (Bamford et al., 2009). Furthermore, C. albicans and S. oralis co-infection synergistically increases the level of μ-calpain, a proteolytic enzyme capable of destroying the epithelial E-cadherin (Xu et al., 2016). IL-18 gene expression is upregulated in reconstituted human oral epithelium infected by mixed-species biofilms, along with greater lactate dehydrogenase activity (Cavalcanti et al., 2015). The invasive ability of C. albicans co-infected with either S. oralis or S. gordonii is also tested in an in vivo oral thrush mouse model. Subjects co-infected with Streptococcus and this fungus exhibit significantly worse severity of tongue thrush lesions. Interestingly, quantitative analysis data of C. albicans from the tongue reveals that the Candida burdens are not significantly different with/without S. oralis infection (Diaz et al., 2012). Co-infection with oral Streptococcus and C. albicans leads to a stronger pro-inflammatory response compared with either single microorganism infection. Based on the microarray analysis of the mouse tongue whole genome, dual-species infected animals shows significantly upregulated genes involved in the primary categories of inflammation and neutrophilic response/chemotaxis (Xu et al., 2014b).
Multiple clinical data confirm that C. albicans is the most frequently detected fungus in infected tooth root canals (Baumgartner et al., 2000; Ashraf et al., 2007; Siqueira and Rocas, 2009; Narayanan and Vaishnavi, 2010; Kumar et al., 2015; Sakko et al., 2016; Persoon et al., 2017; Mergoni et al., 2018; Prada et al., 2019; Alberti et al., 2021). Enterococcus faecalis is a well-recognized pathogen of endodontic infection and post-treatment endodontic disease (Siqueira and Rôças, 2014; Delboni et al., 2017; Prada et al., 2019). The co-existence of C. albicans and E. faecalis in the oral cavity is becoming increasingly evident (Dahlen et al., 2012; Abusrewil et al., 2020). E. faecalis is the most common bacteria co-detected with Candida in oral samples (Hermann et al., 1999; Peciuliene et al., 2001). By scanning electron microscopy, it has been demonstrated that E. faecalis adheres to yeast and hyphal cells of C. albicans in infected tooth root canals as well as in dentinal tubules (Siqueira and Rocas, 2009). In an in vitro study, co-culturing C. albicans and E. faecalis yields a thicker and denser biofilm compared with mono-species biofilm, which exhibits higher tolerance to detrimental stresses, including alkalinity starvation, mechanical shear force, and fungicide/bactericide (Gao et al., 2016; Diogo et al., 2017; Du et al., 2021b), such as sodium hypochlorite (NaClO), ethylenediamine tetraacetic acid (EDTA), and chlorhexidine gluconate (CHX) (Diogo et al., 2017; Du et al., 2021b). The same results are also seen for antimicrobial photodynamic therapy (Diogo et al., 2017) and some new root canal medicaments (chitosan, silver nanoparticles, and ozonated olive oil) (Elshinawy et al., 2018). Consistently, significantly increased extent of periapical alveolar bone damage area is observed in an in vivo E. faecalis and C. albicans co-infection rat model, compared with mono-species infection, in conjunction with the increase in proportion of osteoclasts and decrease in osteoblasts (Du et al., 2021b). Moreover, inflammatory cytokines (TNF-a and IL-6) in periapical lesions are also upregulated by E. faecalis/C. albicans co-infection (Du et al., 2021b).
C. albicans is also the most prevalent fungi in the periodontal pockets of periodontitis patients (Reynaud et al., 2001; Urzua et al., 2008; Al Mubarak et al., 2013; Canabarro et al., 2013; De-La-Torre et al., 2018). Clinical study has shown a higher colonization rate of C. albicans in patients with severe chronic periodontitis, especially C. albicans (De-La-Torre et al., 2018). Canabarro et al. (2013) explores that C. albicans is the only fungal species present in all yeast-positive chronic periodontitis cases, and C. albicans subgingival dental plaque colonization is related to the severity of chronic periodontitis. Co-infection with C. albicans and P. gingivalis has also been confirmed to be significantly associated with deep periodontal pockets and bleeding, contributing to active periodontitis (Oka et al., 2022). In vitro studies further demonstrate that C. albicans and P. gingivalis dual-species biofilm exacerbates periodontal disease, with increasing epithelial cells invasion by P. gingivalis. C. albicans may serve as a scaffold to allow P. gingivalis sufficient time for invasion (Tamai et al., 2011). Sequential infection initiated by C. albicans demonstrates a milder inflammation induced by C. albicans and P. gingivalis co-infection. P. gingivalis count is higher over a longer period of time in mice co-infected with C. albicans, suggesting that dual-species infections have a specific chronic nature (Bartnicka et al., 2020).
In terms of implant-related infections, multiple microbial biofilms on implant surfaces are thought to be the primary reason for peri-implant inflammation and peri-implant mucositis (Salvi et al., 2017; Schincaglia et al., 2017). C. albicans can stimulate almost all MGS species to adhere to and form biofilms on titanium surfaces. In an in vitro titanium-mucosal interface model, although co-cultured with Streptococcus does not influence pro-inflammatory cytokine responses, mucosal tissue exhibits worse damage (Souza et al., 2020). In addition, C. albicans/S. gordonii dual-species biofilms exhibit high levels of resistance to combined antifungal–antibacterial therapy (Montelongo-Jauregui et al., 2018).
Oral cancer is one of the most prevalent cancers, with most mouth neoplasms identified as oral squamous cell carcinoma (OSCC). Risk factors for oral cancer include tobacco use, heavy alcohol consumption, and human papillomavirus infection. Recently, dysbiosis in the oral microbiota has been proposed as involved in etiopathogenesis and processes of OSCC. Changes in relative abundance of specific bacteria (e.g., P. gingivalis, Fusobacterium nucleatum, and Streptococcus sp.) and fungi (especially Candida sp.) are associated with OSCC (Vyhnalova et al., 2021). While it is clear that oncological treatments can lead to changes in oral microorganisms (Sami et al., 2020), more specific mechanistic studies are needed to clarify the causality between cancer and alteration in oral microbiota composition. To our current knowledge, there is limited strong evidence on the role of fungi–bacterial interactions on OSCC development (Vyhnalova et al., 2021). Some researchers hold that microbial infections may contribute to the pathogenesis, by increasing pro-inflammatory cytokines due to microbial infection of oral mucosa (Pushalkar et al., 2011; Arzmi et al., 2019b). C. albicans is considered one of the major microbes contributing to oral cancer development (Kazmierczak-Siedlecka et al., 2020). Polymicrobial interactions have been shown to affect biofilm formation of C. albicans, Actinomyces naeslundii, and S. mutans, and biofilm effluents modulate cancer cell phenotype by increasing the adhesion of oral squamous cell carcinoma cells to extracellular matrix and enhance the expression of pro-inflammatory cytokines, particularly IL-6 and IL-8 (Arzmi et al., 2019a). This potentially cancer-promoting effect of oral microbial biofilms occurs at either the early stage of oral carcinogenesis or perhaps as an enhancement during the later stages of tumor progression. Meanwhile, metabolites from polymicrobial biofilm consisting of C. albicans and S. aureus promote the changes in proto-oncogenes and cell cycle gene expression in normal and neoplastic oral epithelial cell lines, such as Bcl-2 and CDKN1A (Amaya Arbelaez et al., 2021).
Cross-kingdom interactions between C. albicans and oral bacteria are not only widely associated with the pathogenesis of oral disease, but will also likely change the treatment strategies for biofilm-related disease. Recent drug susceptibility studies reveal that the co-presence of C. albicans and oral bacteria in biofilms influences the susceptibility of either to antimicrobial agents. Eradicating Candida–bacterial polymicrobial biofilm-induced diseases is challenging (Montelongo-Jauregui et al., 2016; Diogo et al., 2017; Elshinawy et al., 2018; Kim et al., 2018; Chinnici et al., 2019; Ikono et al., 2019; Du et al., 2021b; Gong et al., 2021; Kulshrestha and Gupta, 2022). Montelongo-Jauregui et al. (2016) reports that in both monotherapy and combination therapy with commonly used antifungals and antibacterial antibiotics, C. albicans/S. gordonii mixed-species biofilm becomes more resistant to antimicrobial treatments at all doses regardless of whether they are cultured on conventional media or synthetic saliva. Ampicillin resistant polymicrobial biofilms consisting of S. gordonii and C. albicans appear to be controlled by transcription factors from C. albicans (Sfl2, Tec1) (Chinnici et al., 2019).
Polysaccharides secreted by microorganisms in biofilm may play an important role in antibiotic resistance, which might prevent drug penetration and provide protection for the microorganisms. In situ generated EPS by S. mutants directly binds and sequesters fluconazole, reducing drug uptake and intracellular transport, and the combination of topical bactericidal povidone iodine with fluconazole increase can completely suppress C. albicans carriage and mixed-biofilm formation without increasing bacterial killing activity in vivo (Kim et al., 2018). Meanwhile, C. albicans and its secreted cell wall polysaccharide material, especially β-1,3-glucan cell wall component, significantly enhance the tolerance of S. aureus to drugs. Fluorescence confocal time-lapse microscopy reveals the impairment of drug diffusion through the mixed biofilm matrix. By inhibiting the production of the fungal polysaccharides, a specific antifungal agent indirectly sensitized the bacteria to antimicrobials (Kong et al., 2016).
The interactions between C. albicans and oral bacteria play an important role in oral microecology and are closely associated with the occurrence, development, and treatment of biofilm-related oral diseases. Understanding such symbiotic interactions with clinical relevance between microbial species in biofilms will greatly aid in disease prevention and overcoming the limitations of current therapies. Studies focusing on the mutualistic interactions between C. albicans and specific pathogens in vitro are fundamental in simplifying the phenomenon and exploring the underlying mechanisms, making great progress for reveal the underlying mechanisms. Additionally, it is necessary to recognize the fact that Candida–bacterial interaction is not only driven by microbial communication but also relies on the influence of environmental conditions, microbial communities, and host factors. The limitation of most in vitro study not being able to reproduce the oral environment is still a problem that requires a breakthrough. In silico or computational studies performed with genomic and metabolic pathway comparisons may help to capture important interaction mechanisms or molecule in complex interaction networks between C. albicans and oral bacteria. Further in-depth studies using models mimicking health and disease situations and clinical trials are still required to delineate the underlying molecular mechanisms. As the threat of antimicrobial resistance increases, the need for new antimicrobial and antifungal agents is reaching a tipping point. Identifying effective therapeutic techniques for Candida–bacterial polymicrobial biofilm is a new approach, rather than focusing only on the specific pathogen (Hwang, 2022). Although there is a growing awareness of the importance of combatting cross-kingdom biofilms, the majority of treatments rely on broad-spectrum antimicrobial activity that can kill both fungus and bacterium or supplemented with antifungal drugs. New therapeutics targeting the binding mechanism between C. albicans and streptococci are currently being investigated. For instance, Kim et al. (2021) presents that mannan-degrading exo- and endo-enzymes target GtfB–mannan interactions in this cross-kingdom consortium and are highly effective in reducing biofilm biomass without killing microorganisms, as well as alleviating the production of an acidic pH environment conducive to tooth decay. Furthermore, recombinantly expressed human and mouse serum amyloid A1 (rhSAA1) proteins promote cell aggregation and target the C. albicans cell wall adhesin Als3 (Gong et al., 2020). Computational methodologies have become crucial components of many programs used in pharmaceutical production for discovery of new antibacterial targets that may decrease virulence of cariogenic microorganisms present in dental biofilms and quickly predict their spectrum and selectivity (da Silva et al., 2014; Rivera-Quiroga et al., 2020; Alharbi et al., 2022). Indeed, we still have a long way to go in finding effective treatments. It would be meaningful to combine in silico, in vitro, and in vivo studies, examining bacterial–fungal interactions in a high-throughput manner to systemically evaluate both positive and negative effects on proliferation and virulence. Further researches focus on molecular targets and signaling pathways of C. albicans, and oral commensal bacteria interactions and the effects on the virulence of fungi–bacteria cross-kingdom biofilms are necessary.
QD: conceptualization, methodology, software, investigation, and writing—original draft preparation. BR: writing—review and editing. XZ: resources and validation. LZ: supervision, project administration, and writing—review and editing. XX: writing—review and editing, visualization, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
This work was supported by a research grant from Science and Technology Department of Sichuan Province (2021YFQ0064), National Natural Science Foundation of China (81870754), and a research grant from West China Hospital of Stomatology (LCYJ2019-4).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abusleme, L., Diaz, P. I., Freeman, A. F., Greenwell-Wild, T., Brenchley, L., Desai, J. V., et al. (2018). Human defects in STAT3 promote oral mucosal fungal and bacterial dysbiosis. JCI. Insight 3:e122061. doi: 10.1172/jci.insight.122061
Abusrewil, S., Alshanta, O. A., Albashaireh, K., Alqahtani, S., Nile, C. J., Scott, J. A., et al. (2020). Detection, treatment and prevention of endodontic biofilm infections: what’s new in 2020? Crit. Rev. Microbiol. 46, 194–212. doi: 10.1080/1040841X.2020.1739622
Al Mubarak, S., Robert, A. A., Baskaradoss, J. K., Al-Zoman, K., Al Sohail, A., Alsuwyed, A., et al. (2013). The prevalence of oral Candida infections in periodontitis patients with type 2 diabetes mellitus. J. Infect Public Health 6, 296–301. doi: 10.1016/j.jiph.2012.12.007
Al-Amad, S. H., Rahman, B., Khalifa, N., and Awad, M. A. (2021). Oral candidal carriage and its association with dental carious lesions in asymptomatic adults: a cross-sectional study from the UAE. BMC. Oral Health 21:197. doi: 10.1186/s12903-021-01559-3
Alberti, A., Corbella, S., Taschieri, S., Francetti, L., Fakhruddin, K. S., and Samaranayake, L. P. (2021). Fungal species in endodontic infections: A systematic review and meta-analysis. PLoS One 16:e0255003. doi: 10.1371/journal.pone.0255003
Alharbi, A., Alsoliemy, A., Alzahrani, S. O., Alkhamis, K., Almehmadi, S. J., Khalifa, M. E., et al. (2022). Green synthesis approach for new Schiff’s-base complexes; theoretical and spectral based characterization with in-vitro and in-silico screening. J. Mol. Liq. 345:117803. doi: 10.1016/j.molliq.2021.117803
Allison, D. L., Willems, H. M. E., Jayatilake, J., Bruno, V. M., Peters, B. M., and Shirtliff, M. E. (2016). Candida-Bacteria interactions: Their Impact on Human Disease. Microbiol. Spectr. 4, 106–136. doi: 10.1128/microbiolspec.VMBF-0030-2016
Amaya Arbelaez, M. I., de Paula, E. S. A. C. A., Navegante, G., Valente, V., Barbugli, P. A., and Vergani, C. E. (2021). Proto-Oncogenes and cell cycle gene expression in normal and neoplastic oral epithelial cells stimulated with soluble factors from single and dual biofilms of Candida albicans and Staphylococcus aureus. Front. Cell Infect Microbiol. 11:627043. doi: 10.3389/fcimb.2021.627043
Arzmi, M. H., Cirillo, N., Lenzo, J. C., Catmull, D. V., O’Brien-Simpson, N., Reynolds, E. C., et al. (2019a). Monospecies and polymicrobial biofilms differentially regulate the phenotype of genotype-specific oral cancer cells. Carcinogenesis 40, 184–193. doi: 10.1093/carcin/bgy137
Arzmi, M. H., Dashper, S., and McCullough, M. (2019b). Polymicrobial interactions of Candida albicans and its role in oral carcinogenesis. J. Oral Pathol. Med. 48, 546–551. doi: 10.1111/jop.12905
Ashraf, H., Samiee, M., Eslami, G., and Ghodse Hosseini, M. R. (2007). Presence of Candida albicans in root canal system of teeth requiring endodontic retreatment with and without Periapical Lesions. Iran Endod. J 2, 24–28.
Babatzia, A., Papaioannou, W., Stavropoulou, A., Pandis, N., Kanaka-Gantenbein, C., Papagiannoulis, L., et al. (2020). Clinical and microbial oral health status in children and adolescents with type 1 diabetes mellitus. Int. Dent. J. 70, 136–144. doi: 10.1111/idj.12530
Baena-Monroy, T., Moreno-Maldonado, V., Franco-Martinez, F., Aldape-Barrios, B., Quindos, G., and Sanchez-Vargas, L. O. (2005). Candida albicans, Staphylococcus aureus and Streptococcus mutans colonization in patients wearing dental prosthesis. Med. Oral Patol. Oral Cir. Bucal. 10(Suppl 1), E27–E39.
Bamford, C. V., d’Mello, A., Nobbs, A. H., Dutton, L. C., Vickerman, M. M., and Jenkinson, H. F. (2009). Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect. Immun. 77, 3696–3704. doi: 10.1128/IAI.00438-09
Bamford, C. V., Nobbs, A. H., Barbour, M. E., Lamont, R. J., and Jenkinson, H. F. (2015). Functional regions of Candida albicans hyphal cell wall protein Als3 that determine interaction with the oral bacterium Streptococcus gordonii. Microbiology (Reading) 161(Pt 1), 18–29. doi: 10.1099/mic.0.083378-0
Bartnicka, D., Gonzalez-Gonzalez, M., Sykut, J., Koziel, J., Ciaston, I., Adamowicz, K., et al. (2020). Candida albicans shields the periodontal killer Porphyromonas gingivalis from recognition by the host immune system and supports the bacterial infection of gingival tissue. Int. J. Mol. Sci. 21:1984. doi: 10.3390/ijms21061984
Bartnicka, D., Karkowska-Kuleta, J., Zawrotniak, M., Satala, D., Michalik, K., Zielinska, G., et al. (2019). Adhesive protein-mediated cross-talk between Candida albicans and Porphyromonas gingivalis in dual species biofilm protects the anaerobic bacterium in unfavorable oxic environment. Sci. Rep. 9:4376. doi: 10.1038/s41598-019-40771-8
Baumgartner, J. C., Watts, C. M., and Xia, T. (2000). Occurrence of Candida albicans in infections of endodontic origin. J. Endod. 26, 695–698. doi: 10.1097/00004770-200012000-00003
Beighton, D., and Lynch, E. (1993). Relationships between yeasts and primary root-caries lesions. Gerodontology 10, 105–108. doi: 10.1111/j.1741-2358.1993.tb00090.x
Beighton, D., Hellyer, P. H., and Heath, M. R. (1990). Associations between salivary levels of mutans Streptococci, lactobacilli, yeasts and black-pigmented Bacteroides spp. and dental variables in elderly dental patients. Arch. Oral Biol. 35, 173S–175S. doi: 10.1016/0003-9969(90)90151-y
Bertolini, M. M., Xu, H., Sobue, T., Nobile, C. J., Del Bel Cury, A. A., and Dongari-Bagtzoglou, A. (2015). Candida-streptococcal mucosal biofilms display distinct structural and virulence characteristics depending on growth conditions and hyphal morphotypes. Mol. Oral Microbiol. 30, 307–322. doi: 10.1111/omi.12095
Bowen, W. H., and Koo, H. (2011). Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 45, 69–86. doi: 10.1159/000324598
Bowen, W. H., Burne, R. A., Wu, H., and Koo, H. (2018). Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 26, 229–242. doi: 10.1016/j.tim.2017.09.008
Brady, L. J., Maddocks, S. E., Larson, M. R., Forsgren, N., Persson, K., Deivanayagam, C. C., et al. (2010). The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol. Microbiol. 77, 276–286. doi: 10.1111/j.1365-2958.2010.07212.x
Brito, A. C. M., Bezerra, I. M., Borges, M. H. S., Cavalcanti, Y. W., and Almeida, L. F. D. (2021). Effect of different salivary glucose concentrations on dual-species biofilms of Candida albicans and Streptococcus mutans. Biofouling 37, 615–625. doi: 10.1080/08927014.2021.1946519
Canabarro, A., Valle, C., Farias, M. R., Santos, F. B., Lazera, M., and Wanke, B. (2013). Association of subgingival colonization of Candida albicans and other yeasts with severity of chronic periodontitis. J. Periodontal. Res. 48, 428–432. doi: 10.1111/jre.12022
Cavalcanti, Y. W., Morse, D. J., da Silva, W. J., Del-Bel-Cury, A. A., Wei, X., Wilson, M., et al. (2015). Virulence and pathogenicity of Candida albicans is enhanced in biofilms containing oral bacteria. Biofouling 31, 27–38. doi: 10.1080/08927014.2014.996143
Chan, A., Ellepola, K., Truong, T., Balan, P., Koo, H., and Seneviratne, C. J. (2020). Inhibitory effects of xylitol and sorbitol on Streptococcus mutans and Candida albicans biofilms are repressed by the presence of sucrose. Arch. Oral Biol. 119:104886. doi: 10.1016/j.archoralbio.2020.104886
Chinnici, J., Yerke, L., Tsou, C., Busarajan, S., Mancuso, R., Sadhak, N. D., et al. (2019). Candida albicans cell wall integrity transcription factors regulate polymicrobial biofilm formation with Streptococcus gordonii. PeerJ 7:e7870. doi: 10.7717/peerj.7870
da Silva, A. C. B., da Silva, D. R., de Macêdo Ferreira, S. A., Agripino, G. G., Albuquerque, A. R., and do Rêgo, T. G. (2014). In silico approach for the identification of potential targets and specific antimicrobials for Streptococcus mutans. Adv. Biosci. Biotechnol. 5, 373–385.
Dahlen, G., Blomqvist, S., Almstahl, A., and Carlen, A. (2012). Virulence factors and antibiotic susceptibility in enterococci isolated from oral mucosal and deep infections. J. Oral Microbiol. 4:10855. doi: 10.3402/jom.v4i0.10855
De-la-Torre, J., Marichalar-Mendia, X., Varona-Barquin, A., Marcos-Arias, C., Eraso, E., Aguirre-Urizar, J. M., et al. (2016). Caries and Candida colonisation in adult patients in Basque Country (Spain). Mycoses 59, 234–240. doi: 10.1111/myc.12453
De-La-Torre, J., Quindos, G., Marcos-Arias, C., Marichalar-Mendia, X., Gainza, M. L., Eraso, E., et al. (2018). Oral Candida colonization in patients with chronic periodontitis. Is there any relationship? Rev. Iberoam Micol. 35, 134–139. doi: 10.1016/j.riam.2018.03.005
Delboni, M. G., Gomes, B. P., Francisco, P. A., Teixeira, F. B., and Drake, D. (2017). Diversity of Enterococcus faecalis genotypes from multiple oral sites associated with endodontic failure using repetitive sequence-based polymerase chain reaction and arbitrarily primed polymerase chain reaction. J. Endod. 43, 377–382. doi: 10.1016/j.joen.2016.10.042
Deng, L., Li, W., He, Y., Wu, J., Ren, B., and Zou, L. (2019). Cross-kingdom interaction of Candida albicans and Actinomyces viscosus elevated cariogenic virulence. Arch. Oral Biol. 100, 106–112. doi: 10.1016/j.archoralbio.2019.02.008
Dewhirst, F. E., Chen, T., Izard, J., Paster, B. J., Tanner, A. C., Yu, W. H., et al. (2010). The human oral microbiome. J. Bacteriol. 192, 5002–5017. doi: 10.1128/JB.00542-10
Diaz, P. I., Hong, B. Y., Dupuy, A. K., and Strausbaugh, L. D. (2017). Mining the oral mycobiome: Methods, components, and meaning. Virulence 8, 313–323. doi: 10.1080/21505594.2016.1252015
Diaz, P. I., Strausbaugh, L. D., and Dongari-Bagtzoglou, A. (2014). Fungal-bacterial interactions and their relevance to oral health: linking the clinic and the bench. Front. Cell Infect Microbiol. 4:101. doi: 10.3389/fcimb.2014.00101
Diaz, P. I., Xie, Z., Sobue, T., Thompson, A., Biyikoglu, B., Ricker, A., et al. (2012). Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun. 80, 620–632. doi: 10.1128/IAI.05896-11
Dige, I., and Nyvad, B. (2019). Candida species in intact in vivo biofilm from carious lesions. Arch. Oral Biol. 101, 142–146. doi: 10.1016/j.archoralbio.2019.03.017
Diogo, P., Fernandes, C., Caramelo, F., Mota, M., Miranda, I. M., Faustino, M. A. F., et al. (2017). Antimicrobial photodynamic therapy against endodontic Enterococcus faecalis and Candida albicans mono and mixed biofilms in the presence of photosensitizers: A comparative study with classical endodontic irrigants. Front. Microbiol. 8:498. doi: 10.3389/fmicb.2017.00498
Dongari-Bagtzoglou, A., Kashleva, H., Dwivedi, P., Diaz, P., and Vasilakos, J. (2009). Characterization of mucosal Candida albicans biofilms. PLoS One 4:e7967. doi: 10.1371/journal.pone.0007967
Du, Q., Ren, B., He, J., Peng, X., Guo, Q., Zheng, L., et al. (2021a). Candida albicans promotes tooth decay by inducing oral microbial dysbiosis. ISME J. 15, 894–908. doi: 10.1038/s41396-020-00823-8
Du, Q., Yuan, S., Zhao, S., Fu, D., Chen, Y., Zhou, Y., et al. (2021b). Coexistence of Candida albicans and Enterococcus faecalis increases biofilm virulence and periapical lesions in rats. Biofouling 37, 964–974. doi: 10.1080/08927014.2021.1993836
Dutton, L. C., Jenkinson, H. F., Lamont, R. J., and Nobbs, A. H. (2016). Role of Candida albicans secreted aspartyl protease Sap9 in interkingdom biofilm formation. Pathog. Dis. 74:ftw005. doi: 10.1093/femspd/ftw005
Dutton, L. C., Nobbs, A. H., Jepson, K., Jepson, M. A., Vickerman, M. M., Aqeel Alawfi, S., et al. (2014). O-mannosylation in Candida albicans enables development of interkingdom biofilm communities. mBio 5:e00911. doi: 10.1128/mBio.00911-14
Eidt, G., Waltermann, E. D. M., Hilgert, J. B., and Arthur, R. A. (2020). Candida and dental caries in children, adolescents and adults: A systematic review and meta-analysis. Arch. Oral Biol. 119:104876. doi: 10.1016/j.archoralbio.2020.104876
Elshinawy, M. I., Al-Madboly, L. A., Ghoneim, W. M., and El-Deeb, N. M. (2018). Synergistic effect of newly introduced root canal medicaments; Ozonated olive oil and chitosan nanoparticles, against persistent endodontic pathogens. Front. Microbiol. 9:1371. doi: 10.3389/fmicb.2018.01371
Ev, L. D., Dame-Teixeira, N., Do, T., Maltz, M., and Parolo, C. C. F. (2020). The role of Candida albicans in root caries biofilms: an RNA-seq analysis. J. Appl. Oral Sci. 28:e20190578. doi: 10.1590/1678-7757-2019-0578
Fakhruddin, K. S., Perera Samaranayake, L., Egusa, H., Ngo, H. C., and Pesee, S. (2021). Profuse diversity and acidogenicity of the candida-biome of deep carious lesions of Severe Early Childhood Caries (S-ECC). J. Oral Microbiol. 13, 1964277. doi: 10.1080/20002297.2021.1964277
Falsetta, M. L., Klein, M. I., Colonne, P. M., Scott-Anne, K., Gregoire, S., Pai, C. H., et al. (2014). Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 82, 1968–1981. doi: 10.1128/IAI.00087-14
Figueiral, M. H., Azul, A., Pinto, E., Fonseca, P. A., Branco, F. M., and Scully, C. (2007). Denture-related stomatitis: identification of aetiological and predisposing factors - a large cohort. J. Oral Rehabil. 34, 448–455. doi: 10.1111/j.1365-2842.2007.01709.x
Fox, E. P., Cowley, E. S., Nobile, C. J., Hartooni, N., Newman, D. K., and Johnson, A. D. (2014). Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr. Biol. 24, 2411–2416. doi: 10.1016/j.cub.2014.08.057
Gao, Y., Jiang, X., Lin, D., Chen, Y., and Tong, Z. (2016). The starvation resistance and biofilm formation of Enterococcus faecalis in Coexistence with Candida albicans, Streptococcus gordonii, Actinomyces viscosus, or Lactobacillus acidophilus. J. Endod. 42, 1233–1238. doi: 10.1016/j.joen.2016.05.002
Garcia, B. A., Acosta, N. C., Tomar, S. L., Roesch, L. F. W., Lemos, J. A., Mugayar, L. R. F., et al. (2021). Association of Candida albicans and Cbp(+) Streptococcus mutans with early childhood caries recurrence. Sci. Rep. 11, 10802. doi: 10.1038/s41598-021-90198-3
Garcia-Sanchez, S., Aubert, S., Iraqui, I., Janbon, G., Ghigo, J. M., and d’Enfert, C. (2004). Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot. Cell 3, 536–545. doi: 10.1128/EC.3.2.536-545.2004
Geerts, G. A., Stuhlinger, M. E., and Basson, N. J. (2008). Effect of an antifungal denture liner on the saliva yeast count in patients with denture stomatitis: a pilot study. J. Oral Rehabil. 35, 664–669. doi: 10.1111/j.1365-2842.2007.01805.x
Gong, J., Bing, J., Guan, G., Nobile, C. J., and Huang, G. (2020). The Als3 Cell Wall Adhesin Plays a Critical Role in Human Serum Amyloid A1-Induced Cell Death and Aggregation in Candida albicans. Antimicrob. Agents Chemother. 64, e24–e20. doi: 10.1128/AAC.00024-20
Gong, J., Hu, D., He, J., Zou, L., Chen, Z., and Li, M. (2021). Effect of longzhang gargle on dual-species biofilm of Candida albicans and Streptococcus mutans. Biomed. Res. Int. 2021:6654793. doi: 10.1155/2021/6654793
Gow, N. A. R., Latge, J. P., and Munro, C. A. (2017). The fungal cell wall: structure, biosynthesis, and function. Microbiol. Spectr. 5. doi: 10.1128/microbiolspec.FUNK-0035-2016
Gregoire, S., Xiao, J., Silva, B. B., Gonzalez, I., Agidi, P. S., Klein, M. I., et al. (2011). Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle on hydroxyapatite surfaces. Appl. Environ. Microbiol. 77, 6357–6367. doi: 10.1128/AEM.05203-11
Hajishengallis, E., Parsaei, Y., Klein, M. I., and Koo, H. (2017). Advances in the microbial etiology and pathogenesis of early childhood caries. Mol. Oral Microbiol. 32, 24–34. doi: 10.1111/omi.12152
Hall, R. A., and Gow, N. A. (2013). Mannosylation in Candida albicans: role in cell wall function and immune recognition. Mol. Microbiol. 90, 1147–1161. doi: 10.1111/mmi.12426
Harriott, M. M., and Noverr, M. C. (2011). Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 19, 557–563. doi: 10.1016/j.tim.2011.07.004
He, J., Kim, D., Zhou, X., Ahn, S. J., Burne, R. A., Richards, V. P., et al. (2017). RNA-Seq reveals enhanced sugar metabolism in Streptococcus mutans Co-cultured with Candida albicans within Mixed-Species Biofilms. Front. Microbiol. 8:1036. doi: 10.3389/fmicb.2017.01036
Hermann, C., Hermann, J., Munzel, U., and Ruchel, R. (1999). Bacterial flora accompanying Candida yeasts in clinical specimens. Mycoses 42, 619–627. doi: 10.1046/j.1439-0507.1999.00519.x
Huffines, J. T., and Scoffield, J. A. (2020). Disruption of Streptococcus mutans and Candida albicans synergy by a commensal streptococcus. Sci. Rep. 10:19661. doi: 10.1038/s41598-020-76744-5
Hwang, G. (2022). In it together: Candida-bacterial oral biofilms and therapeutic strategies. Environ. Microbiol. Rep. 14, 183–196. doi: 10.1111/1758-2229.13053
Hwang, G., Liu, Y., Kim, D., Li, Y., Krysan, D. J., and Koo, H. (2017). Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 13:e1006407. doi: 10.1371/journal.ppat.1006407
Hwang, G., Marsh, G., Gao, L., Waugh, R., and Koo, H. (2015). Binding force dynamics of Streptococcus mutans-glucosyltransferase B to Candida albicans. J. Dent Res. 94, 1310–1317. doi: 10.1177/0022034515592859
Ikono, R., Vibriani, A., Wibowo, I., Saputro, K. E., Muliawan, W., Bachtiar, B. M., et al. (2019). Nanochitosan antimicrobial activity against Streptococcus mutans and Candida albicans dual-species biofilms. BMC Res. Notes 12:383. doi: 10.1186/s13104-019-4422-x
Jack, A. A., Daniels, D. E., Jepson, M. A., Vickerman, M. M., Lamont, R. J., Jenkinson, H. F., et al. (2015). Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology (Reading) 161(Pt 2), 411–421. doi: 10.1099/mic.0.000010
Janus, M. M., Crielaard, W., Volgenant, C. M., van der Veen, M. H., Brandt, B. W., and Krom, B. P. (2017). Candida albicans alters the bacterial microbiome of early in vitro oral biofilms. J. Oral Microbiol. 9:1270613. doi: 10.1080/20002297.2016.1270613
Janus, M. M., Willems, H. M., and Krom, B. P. (2016). Candida albicans in Multispecies Oral Communities; A Keystone Commensal? Adv. Exp. Med. Biol. 931, 13–20. doi: 10.1007/5584_2016_5
Jarosz, L. M., Deng, D. M., van der Mei, H. C., Crielaard, W., and Krom, B. P. (2009). Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot. Cell 8, 1658–1664. doi: 10.1128/EC.00070-09
Johnson, C. C., Yu, A., Lee, H., Fidel, P. L. Jr., and Noverr, M. C. (2012). Development of a contemporary animal model of Candida albicans-associated denture stomatitis using a novel intraoral denture system. Infect Immun. 80, 1736–1743. doi: 10.1128/IAI.00019-12
Kazmierczak-Siedlecka, K., Dvorak, A., Folwarski, M., Daca, A., Przewlocka, K., and Makarewicz, W. (2020). Fungal gut microbiota dysbiosis and its role in colorectal, oral, and pancreatic carcinogenesis. Cancers (Basel) 12:1326. doi: 10.3390/cancers12051326
Khan, F., Bamunuarachchi, N. I., Pham, D. T. N., Tabassum, N., Khan, M. S. A., and Kim, Y. M. (2021). Mixed biofilms of pathogenic Candida-bacteria: regulation mechanisms and treatment strategies. Crit. Rev. Microbiol. 47, 699–727. doi: 10.1080/1040841X.2021.1921696
Khoury, Z. H., Vila, T., Puthran, T. R., Sultan, A. S., Montelongo-Jauregui, D., Melo, M. A. S., et al. (2020). The Role of Candida albicans secreted polysaccharides in augmenting Streptococcus mutans adherence and mixed biofilm formation: In vitro and in vivo Studies. Front. Microbiol. 11:307. doi: 10.3389/fmicb.2020.00307
Kim, D., and Koo, H. (2020). Spatial design of polymicrobial oral biofilm in its native disease state. J. Dent Res. 99, 597–603. doi: 10.1177/0022034520909313
Kim, D., Liu, Y., Benhamou, R. I., Sanchez, H., Simon-Soro, A., Li, Y., et al. (2018). Bacterial-derived exopolysaccharides enhance antifungal drug tolerance in a cross-kingdom oral biofilm. ISME J. 12, 1427–1442. doi: 10.1038/s41396-018-0113-1
Kim, D., Sengupta, A., Niepa, T. H., Lee, B. H., Weljie, A., Freitas-Blanco, V. S., et al. (2017). Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci. Rep. 7:41332. doi: 10.1038/srep41332
Kim, H. E., Dhall, A., Liu, Y., Bawazir, M., Koo, H., and Hwang, G. (2021). Intervening in symbiotic cross-kingdom biofilm interactions: a binding mechanism-based nonmicrobicidal approach. mBio 12, e00651–00621. doi: 10.1128/mBio.00651-21
Kim, H. E., Liu, Y., Dhall, A., Bawazir, M., Koo, H., and Hwang, G. (2020). Synergism of Streptococcus mutans and Candida albicans reinforces biofilm maturation and acidogenicity in saliva: An in vitro study. Front. Cell Infect Microbiol. 10:623980. doi: 10.3389/fcimb.2020.623980
Klinke, T., and Klimm, W. (2002). Induction of caries-like lesions by Candida albicans in an artificial mouth. Caries Res. 36, 195–196.
Klinke, T., Guggenheim, B., Klimm, W., and Thurnheer, T. (2011). Dental caries in rats associated with Candida albicans. Caries Res. 45, 100–106. doi: 10.1159/000324809
Kong, E. F., Tsui, C., Kucharikova, S., Andes, D., Van Dijck, P., and Jabra-Rizk, M. A. (2016). Commensal protection of Staphylococcus aureus against Antimicrobials by Candida albicans biofilm matrix. mBio 7, e1365–e1316. doi: 10.1128/mBio.01365-16
Koo, H., and Bowen, W. H. (2014). Candida albicans and Streptococcus mutans: a potential synergistic alliance to cause virulent tooth decay in children. Fut. Microbiol. 9, 1295–1297. doi: 10.2217/fmb.14.92
Koo, H., Andes, D. R., and Krysan, D. J. (2018). Candida-streptococcal interactions in biofilm-associated oral diseases. PLoS Pathog. 14:e1007342. doi: 10.1371/journal.ppat.1007342
Kulshrestha, A., and Gupta, P. (2022). Polymicrobial interaction in biofilm: mechanistic insights. Pathog. Dis. 80:ftac010. doi: 10.1093/femspd/ftac010
Kumar, J., Sharma, R., Sharma, M., Prabhavathi, V., Paul, J., and Chowdary, C. D. (2015). Presence of Candida albicans in root canals of teeth with apical periodontitis and evaluation of their possible role in failure of endodontic treatment. J. Int. Oral Health 7, 42–45.
Lehner, T. (1966). “Classification and clinico-pathological features of Candida infections in the mouth,” in Symposium on Candida Infection, eds. H. I. Winner and R. Hurley (Edinburgh and London: Churchill Livingstone).
Lohse, M. B., Gulati, M., Johnson, A. D., and Nobile, C. J. (2018). Development and regulation of single- and multi-species Candida albicans biofilms. Nat. Rev. Microbiol. 16, 19–31. doi: 10.1038/nrmicro.2017.107
Lozano Moraga, C. P., Rodriguez Martinez, G. A., Lefimil Puente, C. A., Morales Bozo, I. C., and Urzua Orellana, B. R. (2017). Prevalence of Candida albicans and carriage of Candida non-albicans in the saliva of preschool children, according to their caries status. Acta Odontol. Scand. 75, 30–35. doi: 10.1080/00016357.2016.1244560
Mergoni, G., Percudani, D., Lodi, G., Bertani, P., and Manfredi, M. (2018). Prevalence of Candida species in endodontic infections: Systematic review and meta-analysis. J. Endod. 44, 1616–1625. doi: 10.1016/j.joen.2018.07.016
Mitchell, K. F., Zarnowski, R., Sanchez, H., Edward, J. A., Reinicke, E. L., Nett, J. E., et al. (2015). Community participation in biofilm matrix assembly and function. Proc. Natl. Acad. Sci. U. S. A. 112, 4092–4097. doi: 10.1073/pnas.1421437112
Montelongo-Jauregui, D., Saville, S. P., and Lopez-Ribot, J. L. (2019). Contributions of Candida albicans dimorphism, adhesive interactions, and extracellular matrix to the formation of dual-species biofilms with Streptococcus gordonii. mBio 10, e01179–01119. doi: 10.1128/mBio.01179-19
Montelongo-Jauregui, D., Srinivasan, A., Ramasubramanian, A. K., and Lopez-Ribot, J. L. (2016). An in vitro model for oral mixed biofilms of Candida albicans and Streptococcus gordonii in Synthetic Saliva. Front. Microbiol. 7:686. doi: 10.3389/fmicb.2016.00686
Montelongo-Jauregui, D., Srinivasan, A., Ramasubramanian, A. K., and Lopez-Ribot, J. L. (2018). An in vitro model for Candida albicans(-)Streptococcus gordonii biofilms on titanium surfaces. J. Fungi (Basel) 4:66. doi: 10.3390/jof4020066
Morales, D. K., and Hogan, D. A. (2010). Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 6:e1000886. doi: 10.1371/journal.ppat.1000886
Morse, D. J., Wilson, M. J., Wei, X., Bradshaw, D. J., Lewis, M. A. O., and Williams, D. W. (2019). Modulation of Candida albicans virulence in in vitro biofilms by oral bacteria. Lett. Appl. Microbiol. 68, 337–343. doi: 10.1111/lam.13145
Mun, M., Yap, T., Alnuaimi, A. D., Adams, G. G., and McCullough, M. J. (2016). Oral candidal carriage in asymptomatic patients. Aust. Dent J. 61, 190–195. doi: 10.1111/adj.12335
Nair, R. G., Anil, S., and Samaranayake, L. P. (2001). The effect of oral bacteria on Candida albicans germ-tube formation. APMIS 109, 147–154. doi: 10.1034/j.1600-0463.2001.d01-116.x
Narayanan, L. L., and Vaishnavi, C. (2010). Endodontic microbiology. J. Conserv. Dent 13, 233–239. doi: 10.4103/0972-0707.73386
Nasution, O., Srinivasa, K., Kim, M., Kim, Y. J., Kim, W., Jeong, W., et al. (2008). Hydrogen peroxide induces hyphal differentiation in Candida albicans. Eukaryot Cell 7, 2008–2011. doi: 10.1128/EC.00105-08
Nett, J. E., Marchillo, K., Spiegel, C. A., and Andes, D. R. (2010). Development and validation of an in vivo Candida albicans biofilm denture model. Infect Immun. 78, 3650–3659. doi: 10.1128/IAI.00480-10
Nikawa, H., Hamada, T., and Yamamoto, T. (1998). Denture plaque–past and recent concerns. J. Dent 26, 299–304. doi: 10.1016/s0300-5712(97)00026-2
Nobbs, A. H., and Jenkinson, H. F. (2015). Interkingdom networking within the oral microbiome. Microbes Infect. 17, 484–492. doi: 10.1016/j.micinf.2015.03.008
Oka, I., Shigeishi, H., and Ohta, K. (2022). Co-Infection of Oral Candida albicans and Porphyromonas gingivalis is associated with active periodontitis in middle-aged and older japanese people. Medicina (Kaunas) 58:723. doi: 10.3390/medicina58060723
Peciuliene, V., Reynaud, A. H., Balciuniene, I., and Haapasalo, M. (2001). Isolation of yeasts and enteric bacteria in root-filled teeth with chronic apical periodontitis. Int. Endod. J. 34, 429–434. doi: 10.1046/j.1365-2591.2001.00411.x
Peleg, A. Y., Hogan, D. A., and Mylonakis, E. (2010). Medically important bacterial-fungal interactions. Nat. Rev. Microbiol. 8, 340–349. doi: 10.1038/nrmicro2313
Pereira-Cenci, T., Deng, D. M., Kraneveld, E. A., Manders, E. M., Del Bel Cury, A. A., Ten Cate, J. M., et al. (2008). The effect of Streptococcus mutans and Candida glabrata on Candida albicans biofilms formed on different surfaces. Arch. Oral Biol. 53, 755–764. doi: 10.1016/j.archoralbio.2008.02.015
Persoon, I. F., Crielaard, W., and Ozok, A. R. (2017). Prevalence and nature of fungi in root canal infections: a systematic review and meta-analysis. Int. Endod. J. 50, 1055–1066. doi: 10.1111/iej.12730
Peters, B. M., Ovchinnikova, E. S., Krom, B. P., Schlecht, L. M., Zhou, H., Hoyer, L. L., et al. (2012). Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology (Reading) 158(Pt 12), 2975–2986. doi: 10.1099/mic.0.062109-0
Pitts, N. (2016). Understanding Dental Caries–from Pathogenesis to Prevention and Therapy,” in Understanding Dental Caries. Berlin: Springer, 3–9.
Pitts, N. B., Zero, D. T., Marsh, P. D., Ekstrand, K., Weintraub, J. A., Ramos-Gomez, F., et al. (2017). Dental caries. Nat. Rev. Dis. Primers 3:17030. doi: 10.1038/nrdp.2017.30
Prada, I., Mico-Munoz, P., Giner-Lluesma, T., Mico-Martinez, P., Collado-Castellano, N., and Manzano-Saiz, A. (2019). Influence of microbiology on endodontic failure. Literature review. Med. Oral Patol. Oral Cir. Bucal. 24, e364–e372. doi: 10.4317/medoral.22907
Pushalkar, S., Mane, S. P., Ji, X., Li, Y., Evans, C., Crasta, O. R., et al. (2011). Microbial diversity in saliva of oral squamous cell carcinoma. FEMS Immunol. Med. Microbiol. 61, 269–277. doi: 10.1111/j.1574-695X.2010.00773.x
Raja, M., Hannan, A., and Ali, K. (2010). Association of oral candidal carriage with dental caries in children. Caries Res. 44, 272–276. doi: 10.1159/000314675
Reynaud, A. H., Nygaard-Ostby, B., Boygard, G. K., Eribe, E. R., Olsen, I., and Gjermo, P. (2001). Yeasts in periodontal pockets. J. Clin. Periodontol. 28, 860–864. doi: 10.1034/j.1600-051x.2001.028009860.x
Rivera-Quiroga, R. E., Cardona, N., Padilla, L., Rivera, W., Rocha-Roa, C., Diaz De Rienzo, M. A., et al. (2020). In silico selection and in vitro evaluation of new molecules that inhibit the adhesion of Streptococcus mutants through Antigen I/II. Int. J. Mol. Sci. 22:377. doi: 10.3390/ijms22010377
Sakko, M., Tjaderhane, L., and Rautemaa-Richardson, R. (2016). Microbiology of root canal infections. Prim. Dent J. 5, 84–89. doi: 10.1308/205016816819304231
Salvi, G. E., Cosgarea, R., and Sculean, A. (2017). Prevalence and mechanisms of peri-implant Diseases. J. Dent Res. 96, 31–37. doi: 10.1177/0022034516667484
Sami, A., Elimairi, I., Stanton, C., Ross, R. P., and Ryan, C. A. (2020). The role of the microbiome in oral squamous cell carcinoma with insight into the microbiome-treatment axis. Int. J. Mol. Sci. 21:8061. doi: 10.3390/ijms21218061
Sampaio, A. A., Souza, S. E., Ricomini-Filho, A. P., Del Bel Cury, A. A., Cavalcanti, Y. W., and Cury, J. A. (2019). Candida albicans increases dentine demineralization provoked by Streptococcus mutans Biofilm. Caries Res. 53, 322–331. doi: 10.1159/000494033
Scheinin, A., Pienihakkinen, K., Tiekso, J., and Holmberg, S. (1992). Multifactorial modeling for root caries prediction. Commun. Dent Oral Epidemiol. 20, 35–37. doi: 10.1111/j.1600-0528.1992.tb00670.x
Scheinin, A., Pienihakkinen, K., Tiekso, J., Holmberg, S., Fukuda, M., and Suzuki, A. (1994). Multifactorial modeling for root caries prediction: 3-year follow-up results. Commun. Dent Oral Epidemiol. 22, 126–129. doi: 10.1111/j.1600-0528.1994.tb01587.x
Schincaglia, G. P., Hong, B. Y., Rosania, A., Barasz, J., Thompson, A., Sobue, T., et al. (2017). Clinical, immune, and microbiome traits of gingivitis and peri-implant mucositis. J. Dent Res. 96, 47–55. doi: 10.1177/0022034516668847
Shay, K., Truhlar, M. R., and Renner, R. P. (1997). Oropharyngeal candidosis in the older patient. J. Am. Geriatr. Soc. 45, 863–870. doi: 10.1111/j.1532-5415.1997.tb01517.x
Shen, S., Samaranayake, L. P., Yip, H. K., and Dyson, J. E. (2002). Bacterial and yeast flora of root surface caries in elderly, ethnic Chinese. Oral Dis. 8, 207–217. doi: 10.1034/j.1601-0825.2002.01796.x
Shirtliff, M. E., Peters, B. M., and Jabra-Rizk, M. A. (2009). Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol. Lett. 299, 1–8. doi: 10.1111/j.1574-6968.2009.01668.x
Silverman, R. J., Nobbs, A. H., Vickerman, M. M., Barbour, M. E., and Jenkinson, H. F. (2010). Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun. 78, 4644–4652. doi: 10.1128/IAI.00685-10
Siqueira, J. F. Jr., and Rocas, I. N. (2009). Diversity of endodontic microbiota revisited. J. Dent Res. 88, 969–981. doi: 10.1177/0022034509346549
Siqueira, J. F., and Rôças, I. N. (2014). Present status and future directions in endodontic microbiology. Endod. Topics 30, 3–22. doi: 10.1111/etp.12060
Souza, J. G. S., Bertolini, M., Thompson, A., Barao, V. A. R., and Dongari-Bagtzoglou, A. (2020). Biofilm interactions of Candida albicans and mitis group Streptococci in a titanium-mucosal interface model. Appl. Environ. Microbiol. 86, e2950–e2919. doi: 10.1128/AEM.02950-19
Sridhar, S., Suprabha, B. S., Shenoy, R., Suman, E., and Rao, A. (2020). Association of Streptococcus Mutans, Candida albicans and oral health practices with activity status of caries lesions among 5-year-old children with early childhood caries. Oral Health Prev. Dent. 18, 911–919. doi: 10.3290/j.ohpd.a45411
Sztajer, H., Szafranski, S. P., Tomasch, J., Reck, M., Nimtz, M., Rohde, M., et al. (2014). Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 8, 2256–2271. doi: 10.1038/ismej.2014.73
Tamai, R., Sugamata, M., and Kiyoura, Y. (2011). Candida albicans enhances invasion of human gingival epithelial cells and gingival fibroblasts by Porphyromonas gingivalis. Microb. Pathog. 51, 250–254. doi: 10.1016/j.micpath.2011.06.009
Tanaka, J., Tanaka, M., and Kawazoe, T. (2009). Longitudinal research on the oral environment of elderly wearing fixed or removable prostheses. J. Prosthodont Res. 53, 83–88. doi: 10.1016/j.jpor.2008.10.003
Thomas, A., Mhambrey, S., Chokshi, K., Chokshi, A., Jana, S., Thakur, S., et al. (2016). Association of oral Candida albicans with severe early childhood caries - a pilot study. J. Clin. Diagn. Res. 10, ZC109–ZC112. doi: 10.7860/JCDR/2016/19387.8357
Tu, Y., Zhou, Z., Shu, C., Zhou, Y., and Zhou, X. (2022). The crosstalk between saliva bacteria and fungi in early childhood caries. Front. Cell Infect Microbiol. 12:845738. doi: 10.3389/fcimb.2022.845738
Urzua, B., Hermosilla, G., Gamonal, J., Morales-Bozo, I., Canals, M., Barahona, S., et al. (2008). Yeast diversity in the oral microbiota of subjects with periodontitis: Candida albicans and Candida dubliniensis colonize the periodontal pockets. Med. Mycol. 46, 783–793. doi: 10.1080/13693780802060899
Valentini, F., Luz, M. S., Boscato, N., and Pereira-Cenci, T. (2013). Biofilm formation on denture liners in a randomised controlled in situ trial. J. Dent 41, 420–427. doi: 10.1016/j.jdent.2013.02.012
Vila, T., Sultan, A. S., Montelongo-Jauregui, D., and Jabra-Rizk, M. A. (2020). Oral candidiasis: A Disease of Opportunity. J. Fungi (Basel) 6:15. doi: 10.3390/jof6010015
Vilchez, R., Lemme, A., Ballhausen, B., Thiel, V., Schulz, S., Jansen, R., et al. (2010). Streptococcus mutans inhibits Candida albicans hyphal formation by the fatty acid signaling molecule trans-2-decenoic acid (SDSF). Chembiochem 11, 1552–1562. doi: 10.1002/cbic.201000086
Vyhnalova, T., Danek, Z., Gachova, D., and Linhartova, P. B. (2021). The role of the oral microbiota in the etiopathogenesis of oral squamous cell carcinoma. Microorganisms 9:1549. doi: 10.3390/microorganisms9081549
Wan, S. X., Tian, J., Liu, Y., Dhall, A., Koo, H., and Hwang, G. (2021). Cross-Kingdom cell-to-cell interactions in cariogenic biofilm initiation. J. Dent Res. 100, 74–81. doi: 10.1177/0022034520950286
Wang, Y. (2013). Fungal adenylyl cyclase acts as a signal sensor and integrator and plays a central role in interaction with bacteria. PLoS Pathog. 9:e1003612. doi: 10.1371/journal.ppat.1003612
Wang, Y. (2015). Looking into Candida albicans infection, host response, and antifungal strategies. Virulence 6, 307–308. doi: 10.1080/21505594.2014.1000752
Wolcott, R., Costerton, J. W., Raoult, D., and Cutler, S. J. (2013). The polymicrobial nature of biofilm infection. Clin. Microbiol. Infect 19, 107–112. doi: 10.1111/j.1469-0691.2012.04001.x
Wu, N., Lin, J., Wu, L., and Zhao, J. (2015). Distribution of Candida albicans in the oral cavity of children aged 3-5 years of Uygur and Han nationality and their genotype in caries-active groups. Genet. Mol. Res. 14, 748–757. doi: 10.4238/2015.January.30.18
Wu, R., Tao, Y., Cao, Y., Zhou, Y., and Lin, H. (2020). Streptococcus mutans membrane vesicles harboring glucosyltransferases augment Candida albicans biofilm development. Front. Microbiol. 11:581184. doi: 10.3389/fmicb.2020.581184
Xiao, J., Grier, A., Faustoferri, R. C., Alzoubi, S., Gill, A. L., Feng, C., et al. (2018a). Association between Oral Candida and Bacteriome in Children with Severe ECC. J. Dent Res. 97, 1468–1476. doi: 10.1177/0022034518790941
Xiao, J., Huang, X., Alkhers, N., Alzamil, H., Alzoubi, S., Wu, T. T., et al. (2018b). Candida albicans and early childhood caries: A systematic review and meta-analysis. Caries Res. 52, 102–112. doi: 10.1159/000481833
Xiao, J., Moon, Y., Li, L., Rustchenko, E., Wakabayashi, H., Zhao, X., et al. (2016). Candida albicans carriage in children with severe early childhood caries (S-ECC) and maternal relatedness. PLoS One 11:e0164242. doi: 10.1371/journal.pone.0164242
Xu, H., Jenkinson, H. F., and Dongari-Bagtzoglou, A. (2014a). Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol. Oral Microbiol. 29, 99–116. doi: 10.1111/omi.12049
Xu, H., Sobue, T., Bertolini, M., Thompson, A., and Dongari-Bagtzoglou, A. (2016). Streptococcus oralis and Candida albicans Synergistically Activate mu-Calpain to Degrade E-cadherin From Oral Epithelial Junctions. J. Infect Dis. 214, 925–934. doi: 10.1093/infdis/jiw201
Xu, H., Sobue, T., Bertolini, M., Thompson, A., Vickerman, M., Nobile, C. J., et al. (2017). S. oralis activates the Efg1 filamentation pathway in C. albicans to promote cross-kingdom interactions and mucosal biofilms. Virulence 8, 1602–1617. doi: 10.1080/21505594.2017.1326438
Xu, H., Sobue, T., Thompson, A., Xie, Z., Poon, K., Ricker, A., et al. (2014b). Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. 16, 214–231. doi: 10.1111/cmi.12216
Xu, X. L., Lee, R. T., Fang, H. M., Wang, Y. M., Li, R., Zou, H., et al. (2008). Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe. 4, 28–39. doi: 10.1016/j.chom.2008.05.014
Yang, C., Scoffield, J., Wu, R., Deivanayagam, C., Zou, J., and Wu, H. (2018). Antigen I/II mediates interactions between Streptococcus mutans and Candida albicans. Mol. Oral Microbiol. 33, 283–291. doi: 10.1111/omi.12223
Zaremba, M. L., Stokowska, W., Klimiuk, A., Daniluk, T., Rozkiewicz, D., Cylwik-Rokicka, D., et al. (2006). Microorganisms in root carious lesions in adults. Adv. Med. Sci. 51(Suppl 1), 237–240.
Zhou, Z., Ren, B., Li, J., Zhou, X., Xu, X., and Zhou, Y. (2022). The role of glycoside hydrolases in S. gordonii and C. albicans Interactions. Appl. Environ. Microbiol. 88:e0011622. doi: 10.1128/aem.00116-22
Zijnge, V., van Leeuwen, M. B., Degener, J. E., Abbas, F., Thurnheer, T., Gmur, R., et al. (2010). Oral biofilm architecture on natural teeth. PLoS One 5:e9321. doi: 10.1371/journal.pone.0009321
Keywords: Candida albicans, bacteria, co-infection, biofilm, oral diseases
Citation: Du Q, Ren B, Zhou X, Zhang L and Xu X (2022) Cross-kingdom interaction between Candida albicans and oral bacteria. Front. Microbiol. 13:911623. doi: 10.3389/fmicb.2022.911623
Received: 02 April 2022; Accepted: 28 September 2022;
Published: 03 November 2022.
Edited by:
Cristiane Duque, São Paulo State University, BrazilReviewed by:
Rodrigo Alex Arthur, Universidade Federal do Rio Grande do Sul, BrazilCopyright © 2022 Du, Ren, Zhou, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling Zhang, emhhbmdsaW5nMTk4MzA4MjRAMTYzLmNvbQ==; Xin Xu, eGluLnh1QHNjdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.