
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 June 2022
Sec. Extreme Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.909494
This article is part of the Research Topic Microbial Roles in Caves View all 17 articles
Karst caves are a natural oligotrophic subsurface biosphere widely distributed in southern China. Despite the progress in bacterial and fungal diversity, the knowledge about interactions between bacteria, fungi, and minerals is still limited in caves. Hence, for the first time, we investigated the interaction between bacteria and fungi living on weathered rocks in the Heshang Cave via high-throughput sequencing of 16S rRNA and ITS1 genes, and co-occurrence analysis. The mineral compositions of weathered rocks were analyzed by X-ray diffraction. Bacterial communities were dominated by Actinobacteria (33.68%), followed by Alphaproteobacteria (8.78%), and Planctomycetia (8.73%). In contrast, fungal communities were dominated by Sordariomycetes (21.08%) and Dothideomycetes (14.06%). Mineral substrata, particularly phosphorus-bearing minerals, significantly impacted bacterial (hydroxyapatite) and fungal (fluorapatite) communities as indicated by the redundancy analysis. In comparison with fungi, the development of bacterial communities was more controlled by the environmental selection indicated by the overwhelming contribution of deterministic processes. Co-occurrence network analysis showed that all nodes were positively linked, indicating ubiquitous cooperation within bacterial groups and fungal groups, as well as between bacteria and fungi under oligotrophic conditions in the subsurface biosphere. In total, 19 bacterial ASVs and 34 fungal OTUs were identified as keystone taxa, suggesting the fundamental role of fungi in maintaining the microbial ecosystem on weathered rocks. Ascomycota was most dominant in keystone taxa, accounting for 26.42%, followed by Actinobacteria in bacteria (24.53%). Collectively, our results confirmed the highly diverse bacterial and fungal communities on weathered rocks, and their close cooperation to sustain the subsurface ecosystem. Phosphorus-bearing minerals were of significance in shaping epipetreous bacterial and fungal communities. These observations provide new knowledge about microbial interactions between bacteria, fungi, and minerals in the subterranean biosphere.
Karst caves are thought to be barren with few microorganisms and a weak reflection of the microbiology of surface soils due to oligotrophic conditions (Hess, 1900; Høeg, 1946; Caumartin, 1963; Palmer, 1991). Autotrophic microorganisms in caves usually obtain energy by chemosynthesis using inorganic energy sources (reduced Fe, Mn, and S compounds), organic and inorganic nutrients in host rocks, cave sediments, groundwater, and atmosphere (Jones and Macalady, 2016; Tomczyk-Żak and Zielenkiewicz, 2016; Hershey and Barton, 2018). The occurrence of various metabolic pathways is concurrent in cave microbiota (Kováč, 2018). In consideration of the oligotrophic conditions in caves, the microbial diversity encountered is surprisingly higher than expected as indicated by the application of molecular technology (routinely 106 cells/g; Hershey and Barton, 2018).
Surfaces of various materials have been demonstrated to serve as important niches for microbial communities in diverse ecosystems, such as the surface of the painted murals, heritage buildings, petroglyph panels, and rock surfaces (Ma et al., 2015; Brewer and Fierer, 2018; Zhang et al., 2018; Irit et al., 2019; Ren et al., 2019; He et al., 2021). Rock surfaces are subjected to weathering by physical, chemical, and biological processes, thus offering special niches for microorganisms (Cañveras et al., 2001; Lian et al., 2008). Abundant radiation-resistant bacteria on the surface of limestone outcrops were detected, which were associated with lichens and grew under neutral to alkaline pH conditions (Brewer and Fierer, 2018). Usually, bacterial communities living on outcrop carbonate rocks exposed to sunlight and rainfall were dominated by Actinobacteria, Proteobacteria, Chloroflexi, Bacteroidetes, and Acidobacteria, whereas fungal communities consisted of Eurotiomycetes, Lecanoromycetes, Dothideomycetes, and Leotiomycetes (Choe et al., 2018). Microbial communities inhabiting the surfaces of carbonate rocks in permanently dark caves (Man et al., 2015; Yun et al., 2016; Zhu et al., 2021) are far less studied than those associated with outcrop rocks. Rock surfaces in subsurface caves are directly exposed to air, aerosol, and water vapor (high humidity), as well as a variety of physicochemical and biological dissolution–precipitation and oxidation–reduction reactions, leading with time to weathering and the formation of various secondary minerals (Aloisi et al., 2006; Gallagher et al., 2012). Rock walls act as surface biofilms for exchanges between the bedrock and the atmosphere. Microorganisms can use many minerals as sources of nutrients, carbon, and energy (Tomczyk-Żak et al., 2013). Nevertheless, the current knowledge is ambiguous on microbial communities in relation to mineral substrates and secondary minerals formed during rock weathering in subsurface caves.
Studies have shown that microbial communities in caves are affected by a variety of physical and chemical parameters. Studies in the Heshang Cave, a karst dolomite cave, have shown that pH and TOC are important drivers of the variations in bacterial communities among sediments, overlying soils, dripping water, and weathered rocks (Yun et al., 2016; Cheng et al., 2021). The underlying factors resulting in differences in bacterial and fungal communities living on rocks in caves are poorly understood. The physical properties and chemical compositions of minerals can profoundly impact the composition of microbial communities. Mineral phases in rocks serve as nutrients, substrates, and habitats for microbial communities (Shi L. et al., 2016). Some microbes use conductive mineral particles as conduits of interspecies electron transfer and cooperative catabolism (Kato et al., 2012). Actinobacteria are prevalent in Ca carbonate environments, and alpha-, beta-, and gamma- Proteobacteria are typically associated with complex minerals in cave environments (Barton et al., 2007).
Metabolic activities of microorganisms have important effects on the dissolution and formation of minerals. Moderately halophilic bacteria in liquid media cause the precipitation of calcite, Mg-calcite, and struvite (Arias et al., 2019). Carbonate biomineralization can be enhanced by ureolytic bacteria (Dhami et al., 2013; Arias et al., 2019). Paracoccus versutus XT0.6 isolated from the Xikuangshan antimony mine could dissolve stibnite and oxidize released Sb(III) to Sb(V), subsequently resulting in the formation of secondary Sb(V)-bearing minerals (Loni et al., 2020).
Highly diverse bacterial communities were detected from the weathered rocks in caves with the dominance of Proteobacteria, Actinomycetes, and Firmicutes (Yun et al., 2016; Zhu et al., 2019, 2021; Ma et al., 2021). In contrast, the diversity of fungi is relatively low due to the limited input of organic matter by photosynthesis in caves (Lee et al., 2012; Man et al., 2015). It was estimated that the content of organic matter in caves is three orders of magnitude less than that on the surface (Lavoie et al., 2010). Sunlight penetrates into the cave via the cave entrance to a limited distance and thus divides the cave into three zones: the photic zone with relatively strong light, the twilight zone with weak light, and the totally dark aphotic zone (Yun et al., 2016; Zhao et al., 2018). Due to the different light intensities, physical and chemical conditions vary in the three zones, resulting in different geochemical gradients and biological distribution patterns (Bonacci et al., 2009). In the photic zone, algae and green plants are visible on the rock surface and ground. Light is sufficient to support photosynthesis, thereby supporting CO2 fixation and primary production (Kozlova and Mazina, 2020). The availability of organic matter in the twilight and aphotic zones is extremely limited as it mainly depends on the input of exogenous organic matter via airflow, bat feces, and external organic matter excreted by burrowing animals such as cave rats (Pape and OConnor, 2014; Pfendler et al., 2019). Due to the differences in light, mineral components, and organic matter from different light zones, we hypothesize that (1) microbial communities living on the rocks vary with the zones, and mineral substrates significantly contribute to the variation; (2) bacterial and fungal communities interact intensively to sustain the microbial ecosystem on the rock; and (3) different ecological processes would contribute to the community assembly of bacteria and fungi, respectively.
To test these hypotheses, we collected weathered rock samples from different light zones in the Heshang Cave and subjected them to high-throughput sequencing of bacterial 16S rRNA and fungal ITS1 genes to investigate microbial communities. Variations in microbial communities and the driving forces responsible for these changes would be elucidated via the redundancy analysis. Interactions between bacterial and fungal communities were studied via co-occurrence network analysis to decipher how microbes interact with other microorganisms and sustain the ecosystem. Ecological processes and their contributions to the bacterial and fungal community assembly will be conducted to help understand how the specific microbial communities are established in caves. Our results will provide new insights into the interactions between bacteria and fungi living on carbonate rocks and the ecological processes responsible for microbial communities within subsurface caves.
The Heshang Cave, a dolomite karst cave, is 250 m long, 20–30 m wide, and 15–20 m high. It is located on the steep south bank of the Qingjiang River, northwest Hubei province, central China, with an elevation of 194 m above sea level (Supplementary Figure 1). An intermittent stream develops inside the cave during the rainy season, and dripping water is frequently observed from the ceiling of the cave all year round. The humidity is above 90% till saturated (Hu et al., 2008).
Weathered rock samples from the cave walls were collected along the cave from the entrance to the end on October 25, 2015. In total, nine sites were selected for sampling, with triplicates at each site (Supplementary Figure 1D), and the samples were designated as R1 to R9 accordingly. Green microbial mats were present on the surface of the cave walls close to the cave entrance (Supplementary Figure 2). At each sampling site, weathered rock samples were carefully scraped off with a sterilized scalpel and stored in 50 mL centrifuge tubes. The samples were placed on ice and transported to the Geomicrobiology Laboratory of China University of Geosciences (Wuhan) within 24 h and stored at –80 °C upon arrival until analysis. The wind speed and relative humidity were measured in situ with a handheld weather station (XYZ06E, Dalian Hede Technologies Corporation, China; Supplementary Figures 3A,B). The wind speed and relative humidity at each sampling site were continuously monitored for over 10 min. Values at 3, 6, and 9 min were read, and the mean values were used.
The weathered rock samples were freeze-dried (ALPHA 1-2 LD, Christ, Germany) and fully ground into powder. Mineral phases were analyzed by X-ray powder diffraction (XRD, Shimadzu XRF-1800) at a scanning speed of 0.05 s per step, with a step scan of 0.02° (10° ≤ 2 θ ≤ 90°) via a Cu target Kα radiation source (Ni filter, 40 kV, 40 mA). The phase composition and content of minerals in the samples were qualitatively and semi-quantitatively analyzed by Jade7 and X-powder software.
Genomic DNA was extracted using a FastDNA™ SPIN Kit for soil DNA extraction (MP Biomedicals) in accordance with the instructions. The concentration and quality of extracted DNA were detected by using a micro-nucleic acid protein detector (Nanodrop 2000, Thermo Fisher). The primer set of 520F (5’-AYTGGGYDTAAAGNG-3’) and 802R (5’-TACNVGGGTATCTAATCC-3’) targeting the bacterial 16S rRNA V4 region (Claesson et al., 2009, 2010) and the primer set of ITS5F (5’-GGAAGTAAAAGTCGTAACAAGG-3’) and ITS1R (5’-GCTGCGTTCTTCATCGATGC-3’) targeting the fungal internal transcribed spacer (ITS1) region (Chang et al., 2001) were used for bacterial and fungal sequencing, respectively. All the sequences were conducted using an Illumina MiSeq PE250 platform (Shanghai Personal Biotechnology Co., Shanghai, China). All data are accessible via BioProject IDs of PRJNA337918 and PRJNA821722 for bacteria and fungi, respectively, on the NCBI.
FLASH (v1.2.7)1 was used for quality checking the original readings with Q30 and sequence assembly. Chimeras were removed by a DADA2 plugin in QIIME2 (2019.7, Quantitative Insight into Microbial Ecology). A table of bacterial amplicon sequence variants was constructed from the clean sequences with a 100% similarity, and the representative sequence of each ASV was obtained. Fungal OTUs were identified with a 97% similarity using Vsearch (2.15.1) software. All samples were resampled to the same sequencing depth. Bacterial annotation was carried out against the SILVA database,2 and fungal annotation was based on the UNITE database3 (Nilsson et al., 2019).
The microbial diversity index was calculated by a diversity plugin of QIIME2. The one-way analysis of variance (ANOVA) and Pearson correlation analysis were carried out using SPSS (v10.0). The heatmap analysis was performed using the OmicShare tools, a free online platform for data analysis.4 The vegan package of R software was used for NMDS analysis and output of rarefaction curves (Dixon, 2003), and the box diagram and stacked bar chart were visualized by the ggplot2 package of R software (Wickham et al., 2016). The Venn diagram was drawn using the plotrix package (Lemon et al., 2021). The redundancy analysis (RDA) of bacterial communities and physicochemical parameters was conducted by Canoco5 software. Indicator groups in the different light zones were obtained online with the interactive web pages5 by using the least discriminant analysis effect size (LEfSe) method, in which the linear discriminant analysis (LDA) threshold was set to 3.0. To reduce the network complexity, bacterial ASVs and fungal OTUs detected in at least three samples were selected. The relative abundance of retained bacterial ASVs accounted for > 36.6%, and the numbers of ASVs accounted for 6.6% of the total. The relative abundance of retained fungal OTUs accounted for > 94.4%, and the numbers of OTUs accounted for 15.2% of the total. The Gephi software (v.0.9.2) was used to visualize the co-occurrence network of bacteria and fungi. The ecological roles of each node were determined based on within-module connectivity (Zi) and among-module connectivity (Pi; Guimera and Amaral, 2005). Highly associative “hubs” in the microbiome can be identified from the network and are considered keystone taxa (Banerjee et al., 2018). Node topologies were classified as module hubs (highly connected nodes within modules, Zi ≥ 2.5 and Pi ≤ 0.62), network hubs (highly connected nodes within the entire network, Zi ≥ 2.5 and Pi > 0.62), connectors (nodes that connect modules, Zi < 2.5 and Pi ≥ 0.62), and peripherals (nodes connected in modules with few outside connections, Zi < 2.5 and Pi < 0.62; Deng et al., 2012; Shi S. et al., 2016). The assessment of ecological processes was performed with a mature workflow from the published literature (Stegen et al., 2012, 2013). The nearest taxon index (NTI), mean nearest taxon distance (MNTD), weighted β-mean nearest taxon distance (βMNTD), and weighted β-nearest taxon index (βNTI) were utilized to measure the phylogenic turnover using the theory of null model analyses across samples using the picante package (Kembel et al., 2010). Ecological processes were divided into deterministic processes with a |βNTI| > 2 and stochastic processes with a |βNTI| < 2. If βNTI values are above 2, variable selection is the crucial assembly process in the microbial community; otherwise, homogeneous selection dominates (βNTI < –2). To further specify the stochastic processes, the Raup–Crick matrix (RCbray) based on the Bray–Curtis matrix of microbial community was calculated using the vegan package. Dispersal limitation acting with the drift, undominated processes, and homogenizing dispersal were, respectively, dominant in bacterial communities under the conditions of RCbray values > 0.95, | RCbray| < 0.95, and RCbray < –0.95 (Stegen et al., 2012, 2013).
Minerals of the weathered rocks in the photic zone were dolomite, calcite, Mg-calcite, quartz, and amorphous materials (Supplementary Table 1). More minerals were detected in samples collected in the twilight zone, with the dominance of dolomite and quartz and a minor proportion of amorphous materials (Supplementary Table 1). Phosphate minerals (hydroxyapatite and fluorapatite), gypsum, and illite were also observed in twilight zone samples. Samples in the aphotic zone harbored eight minerals with Mg-calcite (18.6%) and phosphate minerals as the most abundant.
The average wind speed decreased from the cave entrance inward to the cave throughout the year. It was relatively stable (< 0.3 m/s) in the aphotic zone (Supplementary Figure 3A and Supplementary Table 2). The average relative humidity increased from inward to the cave and stabilized in the aphotic zone (>80%; Supplementary Figure 3B and Supplementary Table 3).
A total of 6,885 bacterial ASVs and 5,320 fungal OTUs were obtained in this study, and 511 bacterial ASVs and 1,269 fungal OTUs were found in samples from photic, twilight, and aphotic zones as indicated by the Venn diagram (Supplementary Figure 4). The rarefaction curves of bacterial communities were leveled off across all sampling sites, whereas those of fungal communities at the R5 sampling site were rather steeper (Supplementary Figure 5).
No significant differences were observed in the alpha diversity of bacterial communities among different zones. The Shannon indices varied between 7.56 and 8.12, and ACE values ranged between 4.18 and 10.60 (Figures 1A,B and Supplementary Table 4). The alpha diversity of fungi showed significant differences in the Shannon indices but not in ACE values between the three zones (Figures 1C,D and Supplementary Table 4).
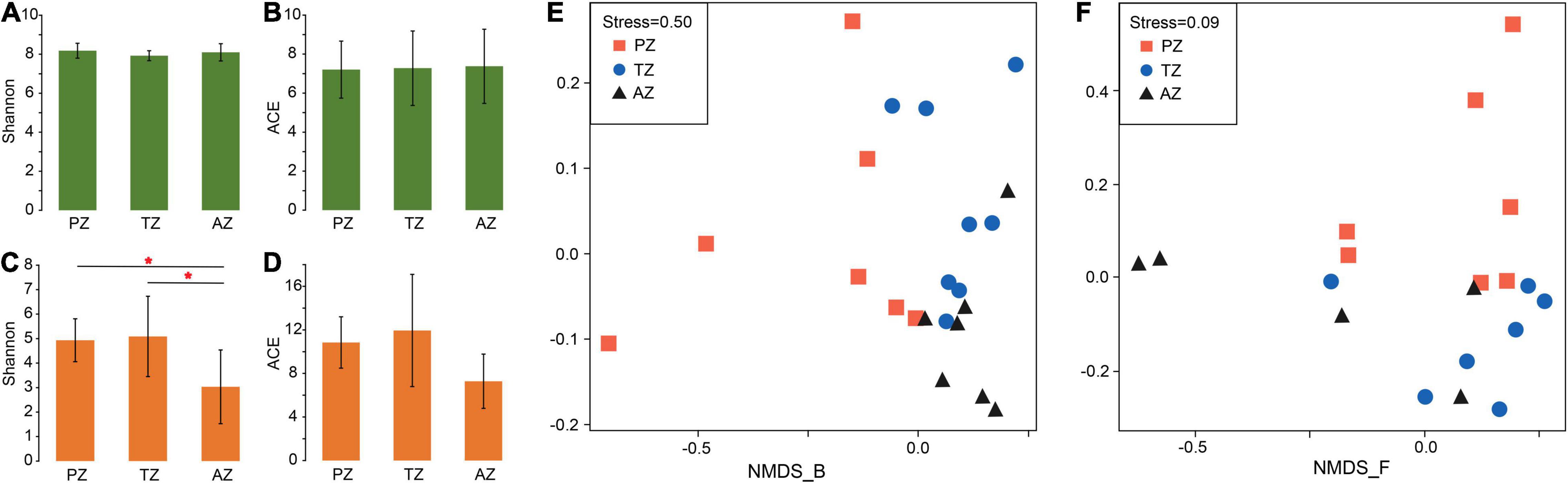
Figure 1. Alpha-diversity indices and non-metric multidimensional scaling (NMDS) analysis of bacterial (A,B,E) and fungal (C,D,F) communities of weathered rocks in the Heshang Cave, Hubei province, China. PZ: photic zone; TZ: twilight zone; AZ: aphotic zone. The mark with * indicates significant differences between the two groups (P < 0.05).
The NMDS analysis showed poor interpretation of bacterial communities between the three light zones (stress > 0.2; Figure 1E), but clearly separated the fungal communities between the photic zone and the aphotic zone, while those in the twilight zone were scattered around (Figure 1F).
Taxonomically, 26 phyla and 55 classes of bacteria were detected with the dominance of Actinobacteria (relative abundance 33.68%). The most abundant class in all samples was Actinobacteria, followed by Alphaproteobacteria and Planctomycetia. The class of Cyanobacteria (4.31%) was only detected in the photic zone, which matched well with the presence of light (Figure 2A). At the genus level, Rubrobacter (4.61%), Gemmatimonas (2.25%), and Solirubrobacter (1.61%) ranked the top three in relative abundance in the photic zone. Rubrobacter (2.87%), Gaiella (1.92%), and Thermoleophilum (1.49%) were the three most abundant genera in the twilight zone, whereas Gaiella (3.00%), Rubrobacter (2.50%), and Gemmatimonas (2.41%) were the top three genera in the aphotic zone (Figure 2B). LEfSe identified Cyanobacteria (4.31%), Gemmatimonas (2.25%), Nitrolancea (0.94%), Brevundimonas (0.63%), and Zavarzinella (0.58%) as indicator groups in the photic zones with an LDA score of 3 (Figure 2E). Indicator groups in the twilight zone included Aquisphaera (0.83%), Gimesia (0.1%), and Streptomyces (0.58%), whereas Conexibacter (1.90%), Actinomadura (0.32%), Aciditerrimonas (0.35%), Planctopirus (0.16%), and Chthonomonas (0.19%) were indicator groups of the aphotic zone (Figure 2E).
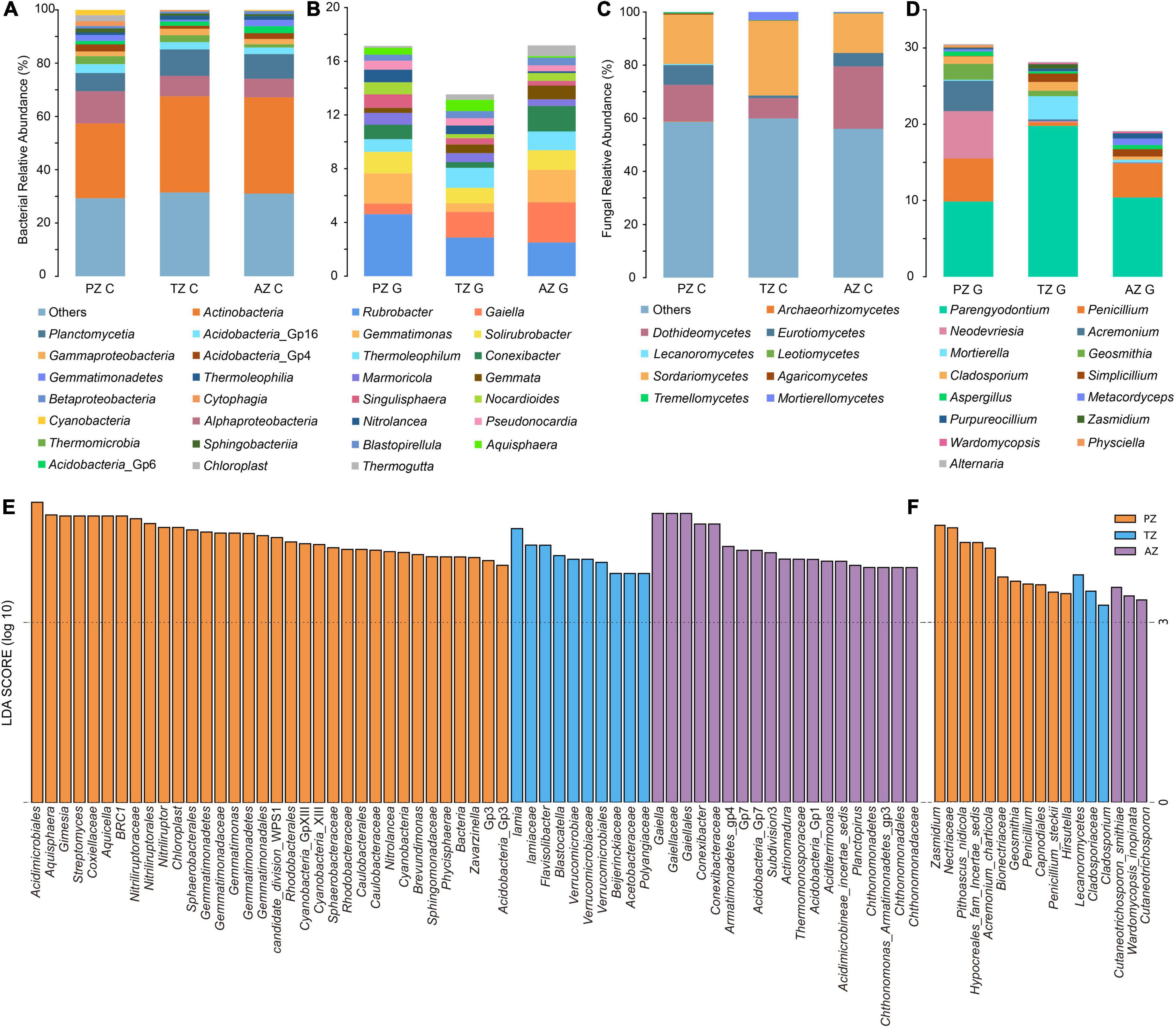
Figure 2. Bacterial (A,B) and fungal community (C,D) compositions of the top 15 classes (A,C) and genera (B,D) of weathered rocks in the Heshang Cave. Linear discriminant analysis (LDA) effect size taxonomic histogram with LDA = 4 of bacterial (E) and LDA = 3 of fungal (F) communities of the weathered rocks in the Heshang Cave, Hubei province, China. PZ: photic zone, TZ: twilight zone, AZ: aphotic zone. C and G after the sampling zone abbreviation indicate classes and genera of microbial taxonomy in the corresponding sampling location.
Totally, 6 phyla and 21 classes of fungi were detected in all samples. Sordariomycetes was most abundant in photic (18.54%) and twilight zones (28.08%), followed by Dothideomycetes (13.74%) and Eurotiomycetes (7.43%) in the photic zone, Dothideomycetes (7.64%) and Mortierellomycetes (3.06%) in the twilight zone. The top three classes in the aphotic zone were Dothideomycetes (23.49%), Sordariomycetes (14.84%), and Eurotiomycetes (5.05%; Figure 2C). Among the top 15 genera, Parengyodontium ranked first in relative abundance among all samples. In the photic zone, Neodevriesia (6.23%) and Penicillium (5.64%) were the second and third abundant genera, whereas those in the twilight zone were Mortierella (3.06%), Cladosporium (1.15%), and in the aphotic zone, Penicillium (4.46%) and Simplicillium (0.97%) ranked second and third most abundant genera. To be noted, unidentified fungal genera in the aphotic zone accounted for a relatively high proportion (80.9%; Figure 2D). Lecanoromycetes (0.37%), Hypocreales fam incertae sedis (4.14%), Bionectriaceae (2.11%), and Cladosporiaceae (0.99%) were indicator groups in the photic zone, and those in the twilight zone were mainly Nectriaceae (0.15%) and Zasmidium (0.59%). The main indicator groups in the aphotic zone were Cutaneotrichosporon (0.05%) and Wardomycopsis inopinata (0.21%; Figure 2F).
RDA was conducted to investigate the relationship between mineral phases and microbial communities living on the weathered rock. All minerals such as quartz, carbonate minerals (including dolomite, calcite, Mg-calcite, and aragonite), gypsum, illite, phosphate minerals (including white apatite, hydroxyapatite, and fluorapatite), and amorphous materials were included to elucidate their impact on bacterial and fungal communities. The results showed that hydroxyapatite and fluorapatite significantly affected bacterial and fungal communities with an explanation of 22.9% (P = 0.018) and 31.2% (P = 0.006), respectively (Figures 3A,B).
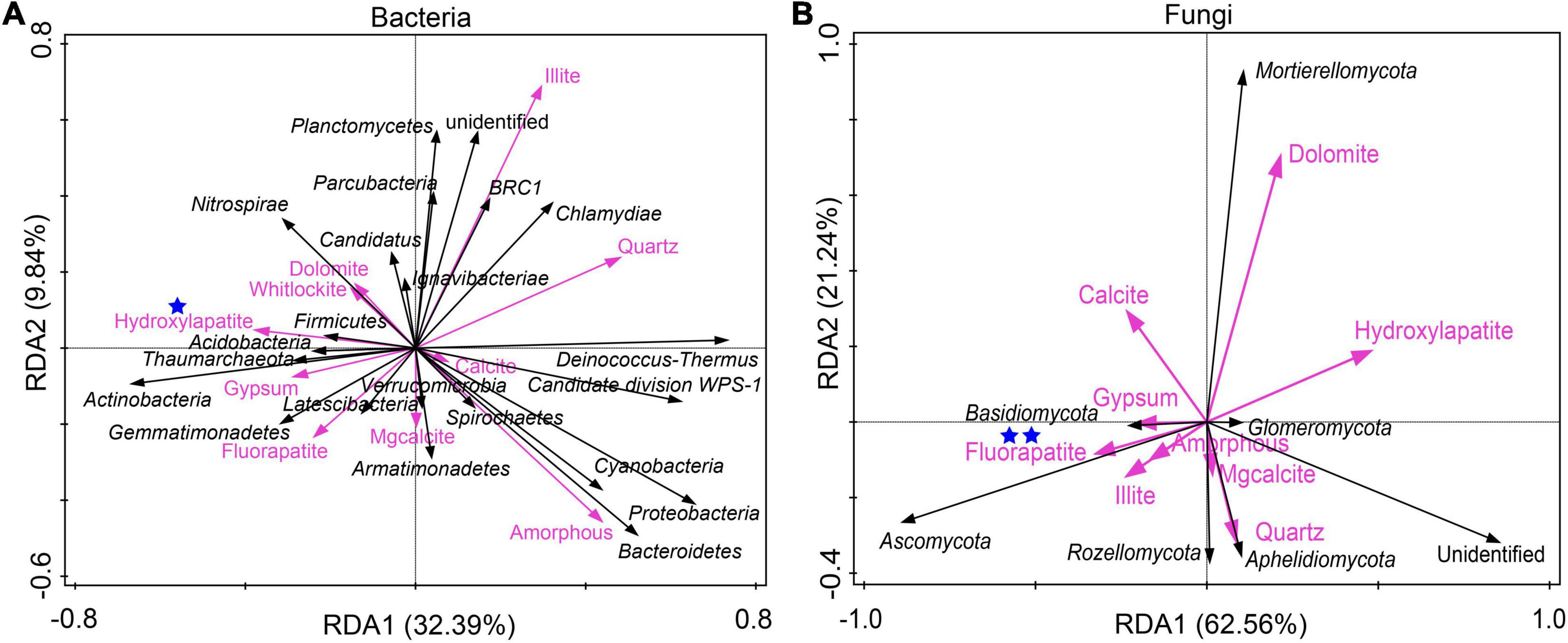
Figure 3. Redundancy analysis of mineral compositions and bacterial communities (A) and fungal communities (B) of the weathered rocks in the Heshang Cave, China. Mineral phases significantly impacting microbial communities were marked with one (with P < 0.05) or two blue stars (P < 0.01), respectively.
The median of the βNTI matrix was below –2, indicating that the deterministic process controlled the assembly of bacterial communities (Figure 4A). Bacterial community assembly was overwhelmingly dominated by the homogeneous selection, with a contribution from 71.43 to 92.86% (Figure 4C). By contrast, the median of the βNTI matrix of fungal communities was between –2 and 2, indicating the dominance of the stochastic process in fungal community assembly (Figure 4B). Thus, ecological processes responsible for fungal community assembly were more complex than those for bacterial communities. Variable selection contributed 42.86 and 47.62% in the photic and twilight zones, respectively, but decreased to 10% in the aphotic zone, which was dominated by homogeneous selection (40%; Figure 4C). The contribution of dispersal limitation decreased inward to the cave from 23.81% in the photic zone, 19.05% in the twilight zone, to no contribution in the aphotic zone (Figure 4C).
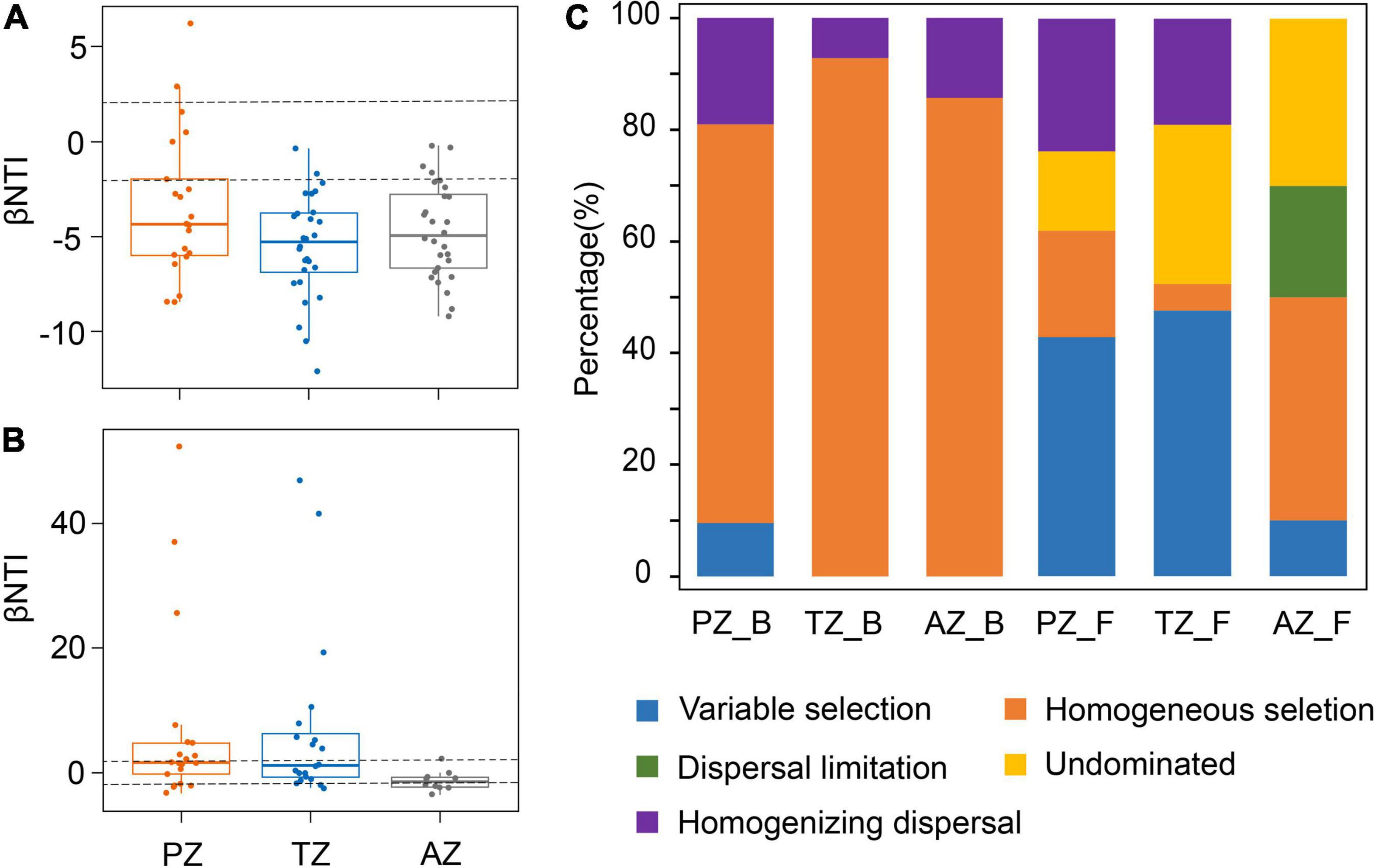
Figure 4. Phylogenetic and null model analyses show the assembly processes of bacterial (A) and fungal communities (B) in the photic zone (PZ), twilight zone (TZ), and aphotic zone (AZ) tested by βNTI values. The contribution of each ecological process to microbial community assembly in different zones in the Heshang Cave (C). PZ_B, TZ_B, and AZ_B indicate the ecological processes responsible for bacterial community assembly in the photic zone, twilight zone, and aphotic zone, respectively, whereas those PZ_F, TZ_F, and AZ_F denote fungal community assembly.
In total, 924 nodes and 7,425 links were observed in the co-occurrence network of bacteria and fungi with good modularity (0.788). Among the links, 800 connected fungal and bacterial nodes, 2,776 connected bacterial nodes, and 3,849 connected fungal nodes, respectively (Figure 5A). All nodes were positively linked. The average path length, diameter, and clustering coefficient were 5.634, 14, and 0.618, respectively.
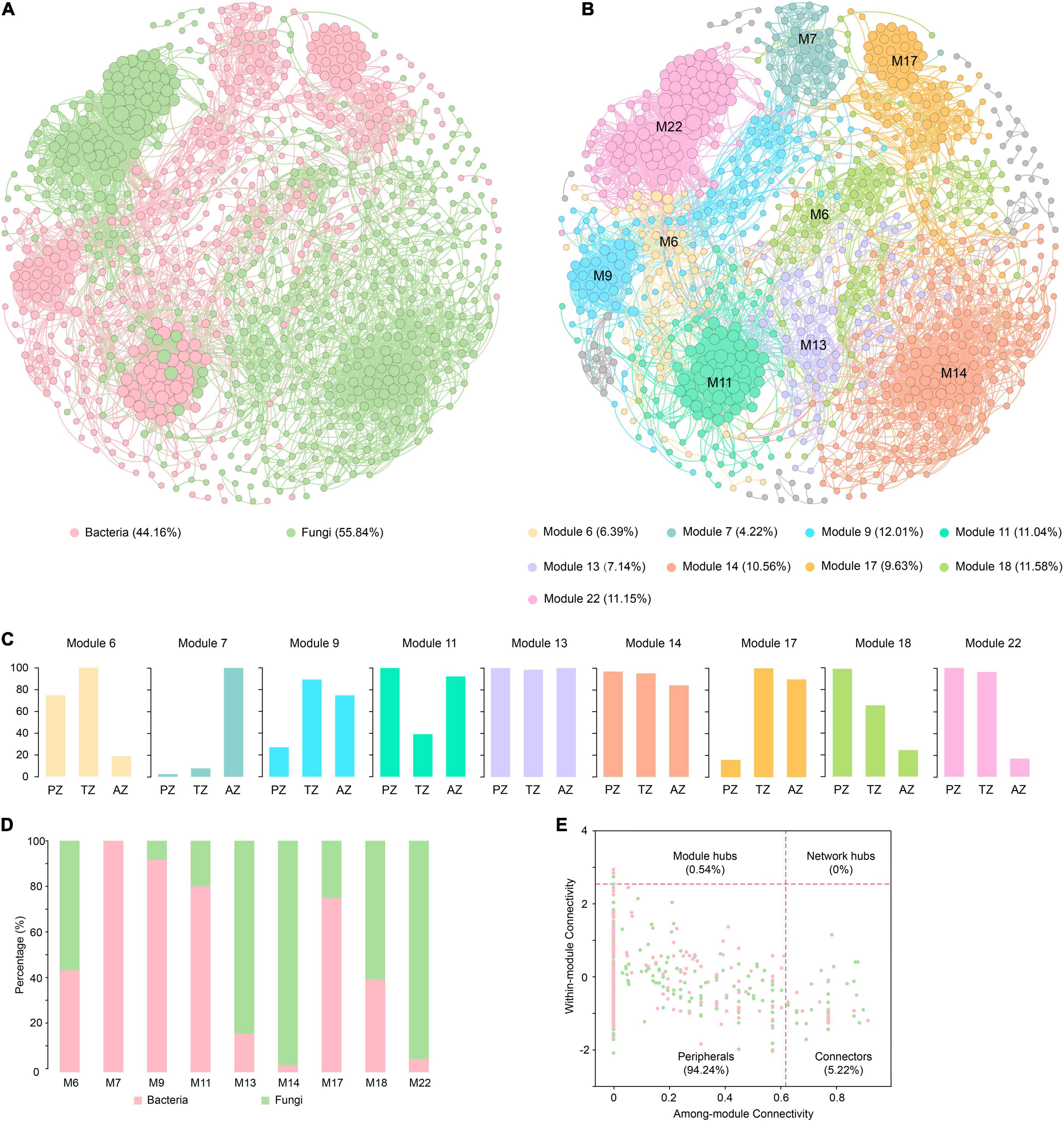
Figure 5. Co-occurrence networks of bacterial and fungal communities of weathered rocks in the Heshang Cave colored by bacteria and fungi (A) and by modules (B). The proportion of ASVs/OTUs based on sampling zonation in each module (C). The proportion of bacterial ASVs and fungal OTUs in each module (D). The within-module connectivity (Zi)–among-module connectivity (Pi) plot shows distribution of ASVs/OTUs according to their module-based topological roles in the co-occurrence networks of bacterial and fungal communities (E). Each dot represents an ASV in the dataset of bacteria (pink) or an OTU of fungi (green). PZ: photic zone, TZ: twilight zone, AZ: aphotic zone.
Totally 28 modules were observed in the fungal–bacterial interaction network, and nine of the modules had a node relative abundance of over 4%, which were subject to further analysis (Figure 5B). Module 14 had the most nodes, accounting for 20.56% of the total nodes in the network. Nodes in modules 6, 18, and 22 mainly originated from PZ and TZ, whereas those in module 7 were mainly from AZ (Figure 5C). Nodes in modules 13 and 14 were evenly contributed by the three zones. Fungi dominated in modules 6, 13, 14, 18, and 22 (Figure 5D), and nodes in these modules were mainly affiliated with Ascomycetes. By contrast, bacteria dominated in modules 7, 9, 11, and 17 (Figure 5D), with the dominance of Actinobacteria. Actinobacteria and Ascomycota accounted for 19.59 and 16.99% of the total nodes, respectively.
Microbial keystone taxa were highly connected taxa that individually or in a guild exerted a considerable influence on the microbiome structure and functioning (Banerjee et al., 2018). Overall, 53 keystone taxa were identified (Figure 5E), five of them were module hubs (0.54% of all nodes), and the rest were connectors (5.19% of the total nodes). The module hubs and connectors accounted for 35.85, and 64.15% of all keystone taxa, respectively. Among 53 keystone taxa, 34 were affiliated with fungi and 19 were from bacteria. Ascomycota was the most dominant in keystone taxa, accounting for 26.42%, of which the most dominant class is Sordariomycetes, accounting for 15.09%, followed by Actinobacteria (24.53%). Module 13 harbored most keystone taxa, accounting for 22.64% of the total keystone taxa, followed by those in modules 6 (18.87%), 18 (13.21%), and 14 (13.21%).
Proteobacteria and Actinomycetes were previously reported to dominate bacterial communities living on the weathered rocks (Tomczyk-Żak and Zielenkiewicz, 2016), which matched well with our observation of the dominance of Actinomycetes (37.66%) and Proteobacteria (12.68%) in the Heshang Cave. Actinomycetes can interact with a variety of mineral phases (Abdulla, 2009; Cuezva et al., 2012) and metals (Manimaran and Kannabiran, 2017) and are capable of degrading various organic compounds (Ningthoujam et al., 2012), which enables them to survive in extreme environments like caves. Proteobacteria (12.68%) ranked the second most abundant linage in the Heshang Cave, indicating highly diverse metabolic capabilities and important roles in the global carbon (Huügler et al., 2005; Kang et al., 2019; Schada von Borzyskowski et al., 2019), nitrogen (Shapleigh, 2011; De Mandal et al., 2017; Delmont et al., 2017), and sulfur cycles (Campbell et al., 2006; Marshall and Morris, 2013). The relative abundance of Planctomycetes (8.92%) in the Heshang Cave was relatively higher than that previously reported for other caves (<5%; Ortiz et al., 2013; Ma et al., 2015; Wu et al., 2015; Gulecal-Pektas, 2016), suggesting the uniqueness of bacterial communities in our study. The dominance of Ascomycetes (40%) in the Heshang Cave was consistent with previous reports on fungal communities in many other caves (Ogórek et al., 2013; Vaughan et al., 2015). Some of the fungal top 15 fungal genera observed in our study were also common in other caves, such as Acremonium, Aspergillus, Cladosporium, and Penicillium (Popović et al., 2015; Zhang et al., 2017; Zhang and Cai, 2019). Parengyodontium (13.66%) was the most abundant genus, which was frequently observed in other caves (Leplat et al., 2017, 2019). Parengyodontium album had often been the most abundant detected species when salt efflorescence was present (Ponizovskaya et al., 2019; Trovão et al., 2019). The high abundance of Penicillium (3.42%) may be related to bat activity (Karkun et al., 2012). The species Penicillium janczewskii is characterized by habitation in dry substrates depleted in organic matter, which is partially consistent with its presence in the cave (Mazina et al., 2020). Neodevriesia (2.37%) was reported as a dominant group on mural paintings in North Thai temples (Suphaphimol et al., 2022).
Alpha diversities and bacterial community compositions on the rocks did not show significant differences as indicated by Shannon and ACE indices and NMDS analysis in the sampling zones (the photic zone, twilight zone, and aphotic zone), suggesting a fundamental role of the rock substrate (carbonate especially), rather than sunlight in the shaping of bacterial communities. In contrast, the alpha diversity of fungal communities in the aphotic zone was significantly lower than that in the other two zones, which may be related to the isolation of the aphotic zone from light and plants. Fungal communities are known to be tightly associated with plant communities (Tedersoo et al., 2014; Purahong et al., 2018; Adamczyk et al., 2019), and almost 20% of total fungal species are symbiotic with lichens (Kirk et al., 2001; Honegger, 2009). Our results also found high fungal diversity at the entrance of the Heshang Cave, where lichens were observed on the rock surface. The decrease in fungal diversity inward to the cave may result from the impediment in the transfer of fungal spores due to the slow wind speed inside the cave (Supplementary Figure 3A). In fact, our observation of the significant positive correlations between the relative abundances of fungal classes such as Wallemiomycetes, Agaricomycetes, Tremellomycetes, Lecanoromycetes, Exobasidiomycetes, and the wind speed (Supplementary Figure 3C) also supported the decrease in the fungal diversity inward to the cave.
Despite small differences in bacterial communities, indicator groups were distinct in three light zones. The indicator groups in the photic zone were Cyanobacteria, Gemmatimonas, Nitrolancea, Brevundimonas, and Zavarzinella, which are closely related to light-dependent metabolism. Many cyanobacterial species can survive on soil and other terrestrial habitats and play important roles in the carbon and nitrogen cycles (Wegener et al., 2010; Forchhammer and Selim, 2020). Some species in Gemmatimonas such as Gemmatimonas aurantiaca can accumulate polyphosphate (Zhang et al., 2003), and other species like Gemmatimonas phototrophica are able to use light as an energy source (Zeng et al., 2015). Nitrolancea spp. are obligately aerobic, which are chemolithoautotrophic nitrite-oxidizers (Sorokin et al., 2014). Flavisolibacter are aerobic chemoheterotrophs in the phylum Bacteroidetes (Lee J. J. et al., 2016). The main indicator groups in the twilight zone were Aquisphaera, Gimesia, and Streptomyces. Aquisphaera and Gimesia belong to the phylum Planctomycetes. Aquisphaera forms large visible cell aggregates during growth, which may affect the survival of other microorganisms (Bondoso et al., 2011). Isolates of Gimesia chilikensis are salt- and alkali-tolerant (Kumar et al., 2020). Many Streptomyces species produce multiple antibiotics or other secondary metabolites, which act synergistically or contingently against biological competition (Challis and Hopwood, 2003). Conexibacter, Actinomadura, Aciditerrimonas, Planctopirus, and Chthonomonas were the indicator groups in the aphotic zone. They can survive in extreme environments, which is consistent with the lack of organic matter in the depths of the cave. Conexibacter, Actinomadura, and Aciditerrimonas all belong to class Actinobacteria and have been frequently isolated from extreme environments. For example, Conexibacter stalactiti was isolated from samples of stalactites collected from a lava cave (Lee, 2017), and Actinomadura namibiensis was isolated from desert sandy soil in Namibia (Wink et al., 2003). Chthonomonas calidirosea was thermophilic and isolated from geothermal soil (Lee K. C. et al., 2016; Compton et al., 2020). Microbes in the aphotic zone can also be involved in iron reduction. For example, Aciditerrimonas ferrireducens is capable of reducing ferric iron aerobically or anaerobically (Itoh et al., 2011; Hu et al., 2014).
The indicator groups of fungi also varied in the zones. In the photic zone, Lecanoromycetes, Hypocreales, Bionectriaceae, and Cladosporiaceae were identified as indicator groups. Lecanoromycetes is the largest class of lichenized fungi and includes most of the lichen-forming fungal species (> 13,500 species; Miadlikowska et al., 2006; Hofstetter et al., 2007). The majority of lichenized fungi in Lecanoromycetes are obligate mutualists, which can obtain organic nutrients from photosynthetic symbiotes and N compounds fixed by cyanobacteria (Gueidan et al., 2015; Jüriado et al., 2017). The Bionectriaceae is a family in the order Hypocreales, which tends to grow on plant materials (Khodosovtsev et al., 2012), and some species are related to algae (Khodosovtsev et al., 2012), bryophytes (Döbbeler et al., 2015), or other fungi. Cladosporiaceae is also associated with plants (Alghuthaymi, 2017) and is dominant in highly saline environments (Yang and Sun, 2020). The indicator groups in the twilight zone were Nectriaceae and Zasmidium. Numerous members in the family Nectriaceae (Hypocreales) are plant and human pathogens, and several species belonging to this family are used extensively in industrial and commercial applications for biodegradation and as biocontrol agents (Lombard et al., 2014, 2015). Zasmidium cellare has been reported in the Mammoth Cave, Kentucky (Barr, 1968), and is a strain known for living in dark and ethanol-rich environments (Cailhol et al., 2020). The indicator groups in the aphotic zone included Cutaneotrichosporon and Wardomycopsis inopinata. Cutaneotrichosporon oleaginosus is an oleaginous yeast with fast growth and also grows as filamentous fungus in the soil and leaf litter (Bracharz et al., 2017). Information on the function of Wardomycopsis inopinata in the environment is not available.
RDA indicated that bacterial communities were significantly affected by hydroxyapatite, mainly due to phosphate-solubilizing bacteria Streptomyces and Pseudomonas. An isolate of Pseudomonas fluorescens from the Heshang Cave has been demonstrated to solubilize fluorapatite, making phosphate bioavailable for microorganisms (Zhou et al., 2017). Fungal communities were positively impacted by fluorapatite, attributed to phosphorus-solubilizing soilborne fungi Aspergillus niger and Penicillium spp. (Reyes et al., 2007; Farhat et al., 2009; Li et al., 2016). The mechanism of phosphorus-solubilizing microorganisms is generally attributed to organic acid metabolites produced by bacteria and fungi (Kavanagh, 2005; Jones and Oburger, 2011). Phosphate is a common minor element in many silicate, sulfide, and carbonate minerals. Under phosphate-limited conditions in soil environments, microorganisms colonizing minerals may acquire bioavailable phosphate as it solubilizes from the mineral phase (Rogers and Bennett, 2004). Our observation of the significant impact of phosphate-bearing minerals on bacterial and fungal communities on the weathered rocks substantiates the development of microbial communities nutritionally dependent on the mineral-phase substrate.
Ecological processes responsible for the development of microbial communities were also investigated in the Heshang Cave. The assembly of bacterial communities is mainly a deterministic process, with the dominance of homogeneous selection across different sampling zones, indicating a spatial homogeneity in environmental conditions and the convergence in bacterial communities (Dini-Andreote et al., 2015). This conclusion is also consistent with the result of the NMDS (Figure 1E). In contrast, different ecological processes contributed to fungal assembly in sampling zones. Variable selection dominated the assembly of fungal communities in both the photic and twilight zones, indicating the strong environmental selection of fungal communities partially due to sunlight. Many fungal groups were associated with algae, cyanobacteria, and plant debris, all of which were highly dependent on sunlight. Homogeneous selection only dominated the fungal assembly in the aphotic zone, indicating that a homogeneous environmental condition resulted in the convergence in fungal communities. The occurrence of dispersal limitation in the aphotic zone indicates a weakening selection, which may have resulted from slow wind speed and high relative humidity toward the end of the cave entrance (Supplementary Tables 3,4). Slow airflow is known to decrease airborne transmission of fungal spores (Quintero et al., 2010), and fungal survival increases with relative humidity (Oliveira et al., 2005; Hameed et al., 2012). The relative abundances of fungal classes Wallemiomycetes, Agaricomycetes, Tremellomycetes, Lecanoromycetes, and Exobasidiomycetes showed significant positive correlations with the wind speed (Supplementary Figure 3C), and similar phenomena in air have also been reported (Begerow and Kemler, 2018; Els et al., 2019).
The co-occurrence network of bacteria and fungi showed good modularity and positive links between nodes, indicating that the bacterial and fungal communities formed a close organization through cooperation among different species. Their interactions increased the complexity of the community structure and the stability of the ecosystem (Chen et al., 2018).
In total, 53 keystone taxa were identified, which usually interact with many other groups, thus promoting the exchange and flow of energy, information, metabolites, and nutrients among different species and maintaining the balance of microbial communities (Berry and Widder, 2014; Ma et al., 2016). There are many unknown functions of the keystone taxa in this study, but some clues can be garnered from the previous investigations. The fungal keystone taxa mainly belonged to classes of Sordariomycetes, Dothideomycetes, and Eurotiomycetes. Members of Sordariomycetes are ubiquitous in natural environments (Zhang and Wang, 2015). They participate in the decomposition of organic matter and nutrient cycling as saprotrophs in almost all ecosystems, and they include endophytes and pathogens of plants (Arnold and Lutzoni, 2007), arthropods (Samuels and Blackwell, 2001), and mammals (O’Donnell et al., 2010), and even as mycoparasites attacking other fungi (Broberg et al., 2021). Sordariomycetes also contain species known to produce secondary metabolites such as potent mycotoxins (Zhang and Wang, 2015). Dothideomycetes is the largest class of Ascomycetes, which includes numerous rock-inhabiting fungi, adapted well to nutrient-poor and dry habitats on rock surfaces (Ruibal et al., 2009; Haridas et al., 2020). Eurotiomycetes can degrade a wide variety of organic substrates and can tolerate high salinity, complete darkness, and oligotrophic conditions (Greiner et al., 2014; Zhang and Wang, 2015), which favor their survival in caves.
Nocardioidaceae, Planctomycetaceae, and Rubrobacteraceae were keystone taxa of bacterial classes in the network. Nocardioidaceae uses a wide range of carbon and nitrogen sources, including unusual organic compounds and toxic environmental pollutants, and has versatile metabolic pathways (Evtushenko et al., 2015). Like many other actinomycetes, Nocardioidaceae can survive under extreme conditions such as desiccation, low and high temperatures, UV damage, and toxic compounds (Kirby et al., 2006; Brodie et al., 2007; Griffin, 2007; Polymenakou et al., 2008). Members of the Nocardioidaceae are symbiotic with plants and may exist as reciprocal endophytes. They can also occur in association with lichens (Li et al., 2007) and fungi (Lauer et al., 2007). Marmoricola is a genus of Nocardioidaceae, which are found in different environments, such as marble, beach sediments, and agricultural soil (Lee, 2007; Freed et al., 2019). Some species are alkali-tolerant or display alkaliphilic properties (Evtushenko, 2015). The family Planctomycetaceae is distributed in both soil and water (Wiegand et al., 2018) and shows strong tolerance to seawater, acid peat bog, hot springs, and low temperature (Kulichevskaya et al., 2007, 2008; Fuerst and Sagulenko, 2011). It can use a special mechanism to absorb and digest complex polysaccharides, which may help it compete for nutrients in natural habitats (Boedeker et al., 2017; Schubert et al., 2020). The family Rubrobacteraceae is usually represented by its thermophilic and radiation-resistant types (Albuquerque et al., 2014). The genus Rubrobacter of Rubrobacteraceae is involved in chlorophyll biosynthesis (Gupta and Khadka, 2016). These keystone bacteria and fungi impact microbial community composition and symbiotic or parasitic relationships on rock walls. Some of their secondary metabolites have antibiotic or toxin properties that can promote or inhibit effector species and modulate interactions in microbial communities.
The co-occurrence network analysis provides us with a new way to find potential symbiotic or parasitic associations between bacteria and fungi. Bacteria can be endosymbiotic with fungi (termed endohyphal bacteria), which are phylogenetically and ecologically widespread. Endohyphal bacteria recently have been documented in diverse Ascomycota, including members of multiple classes (Pezizomycetes, Eurotiomycetes, Dothideomycetes, and Sordariomycetes; Hoffman and Arnold, 2010; Shaffer et al., 2016, 2017). Our network analysis found that many unannotated groups within Eurotiomycetes, Dothideomycetes, and Sordariomycetes serve as keystone taxa in the network, which are also tightly connected with bacteria. For example, Betaproteobacteria has a connection with unidentified fungi, and related studies have demonstrated that some genera of Betaproteobacteria can parasitize as endosymbionts in fungal hyphae (Hoffman and Arnold, 2010). Xanthomonadaceae has been found to subsist in fungal hyphae (Hoffman and Arnold, 2010), as also notices for unidentified fungi in our network. The observation of the connection between Caldiline and unidentified fungi in our study is consistent with a previous report of a negative correlation between Caldiline and an unclassified fungus of Pezizomycotina (Fouts et al., 2012). Gammaproteobacteria in the network are linked to many OTUs in Dothideomycetes, Sordariomycetes, and unidentified fungi. Arendt et al. (2016) demonstrated that some species of Gammaproteobacteria can be successfully introduced into Dothideomycetes and Sordariomycetes, thus substantiating their symbiotic relationships.
It should be emphasized that co-occurrence networks visualize the correlations between microbial groups, including real ecological interactions (such as reciprocity), and also include non-random processes (such as niche overlap). They do not, therefore, necessarily reflect direct interactions between the groups. Future experiments will assess whether the identified key species directly affect other members of the microbiome or indirectly affect the performance and fitness of the host, thus affecting other community members.
We studied the ecological processes responsible for the community assembly, endeavoring to provide new insights into microbial interactions and establishment of microbial communities under extreme conditions in the subsurface cave biosphere.
1. Microbial indicator groups varied with different light zones, indicating functional changes in response to the light variation in karst caves. Phosphate minerals such as hydroxyapatite and fluorapatite significantly influenced bacterial and fungal communities living on weathered rock surfaces.
2. The co-occurrence network analysis showed that bacteria and fungi on rock surfaces were mainly positively linked, indicating extensive, close cooperations among microbial groups across different domains connected under stark environmental conditions to maintain the microbial ecosystem. The co-occurrence network may also provide us with a new robust way to decipher symbiosis and parasitism between bacterial and fungal groups.
3. Homogeneous selection dominated bacterial community assembly throughout the cave, resulting in the convergence of bacterial communities. Variable selection contributed most to the fungal community assembly in the photic and twilight zones, whereas homogeneous selection with dispersal limitation contributed more to fungal assembly in the dark zone.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/337918, http://www.ncbi.nlm.nih.gov/bioproject/821722.
YW, JZ, and XL completed the sample collection. JZ finished DNA extraction. YW and XC completed data analysis, visualization, and original draft preparation. HW designed this experiment, supervised data analysis, acquired funding, and wrote and edited this manuscript. OT participated in data discussion and manuscript drafting. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (91951208 and 41130207).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.909494/full#supplementary-material
Abdulla, H. (2009). Bioweathering and biotransformation of granitic rock minerals by actinomycetes. Microb. Ecol. 58, 753–761. doi: 10.1007/s00248-009-9549-1
Adamczyk, M., Hagedorn, F., Wipf, S., Donhauser, J., Vittoz, P., Rixen, C., et al. (2019). The soil microbiome of Gloria Mountain summits in the Swiss Alps. Front. Microbiol. 10:1080. doi: 10.3389/fmicb.2019.01080
Albuquerque, L., Johnson, M. M., Schumann, P., Rainey, F. A., and da Costa, M. S. (2014). Description of two new thermophilic species of the genus Rubrobacter, Rubrobacter calidifluminis sp. nov. and Rubrobacter naiadicus sp. nov., and emended description of the genus Rubrobacter and the species Rubrobacter bracarensis. Syst. Appl. Microbiol. 37, 235–243. doi: 10.1016/j.syapm.2014.03.001
Alghuthaymi, M. A. (2017). Nanotools for molecular identification two novels Cladosporium cladosporioides species (Cladosporiaceae) collected from tomato phyloplane. J. Yeast Fungal Res. 8, 11–18. doi: 10.5897/JYFR2017.0178
Aloisi, G., Gloter, A., Kruger, M., Wallmann, K., Guyot, F., and Zuddas, P. (2006). Nucleation of calcium carbonate on bacterial nanoglobules. Geology 34, 1017–1020. doi: 10.1130/G22986A.1
Arendt, K. R., Hockett, K. L., Araldi-Brondolo, S. J., Baltrus, D. A., and Arnold, A. E. (2016). Isolation of endohyphal bacteria from foliar ascomycota and in vitro establishment of their symbiotic associations. Appl. Environ. Microbiol. 82, 2943–2949. doi: 10.1128/AEM.00452-16
Arias, D., Cisternas, L. A., Miranda, C., and Rivas, M. (2019). Bioprospecting of ureolytic bacteria from Laguna Salada for biomineralization applications. Front. Bioeng. Biotechnol. 6:209. doi: 10.3389/fbioe.2018.00209
Arnold, A. E., and Lutzoni, F. (2007). Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology 88, 541–549. doi: 10.1890/05-1459
Banerjee, S., Schlaeppi, K., and van der Heijden, M. G. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576. doi: 10.1038/s41579-018-0024-1
Barr, T. C. Jr. (1968). Ecological studies in the Mammoth Cave system of Kentucky. Int. J. Speleol. 3, 147–204. doi: 10.5038/1827-806x.3.1.10
Barton, H. A., Taylor, N. M., Kreate, M. P., Springer, A. C., Oehrle, S. A., and Bertog, J. L. (2007). The impact of host rock geochemistry on bacterial community structure in oligotrophic cave environments. Int. J. Speleol. 36, 93–104. doi: 10.5038/1827-806x.36.2.5
Begerow, D., and Kemler, M. (2018). Phylogeny, biogeography and host specificity of smut fungi. Biosyst. Ecol. Ser. 34, 311–329.
Berry, D., and Widder, S. (2014). Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 5:219. doi: 10.3389/fmicb.2014.00219
Boedeker, C., Schüler, M., Reintjes, G., Jeske, O., van Teeseling, M. C., Jogler, M., et al. (2017). Determining the bacterial cell biology of Planctomycetes. Nat. Commun. 8:14853. doi: 10.1038/ncomms14853
Bonacci, O., Pipan, T., and Culver, D. C. (2009). A framework for karst ecohydrology. Environ. Geol. 56, 891–900. doi: 10.1007/s00254-008-1189-0
Bondoso, J., Albuquerque, L., Nobre, M. F., Lobo-da-Cunha, A., da Costa, M. S., and Lage, O. M. (2011). Aquisphaera giovannonii gen. nov., sp. nov., a planctomycete isolated from a freshwater aquarium. Int. J. Syst. Evol. Microbiol. 61, 2844–2850. doi: 10.1099/ijs.0.027474-0
Bracharz, F., Beukhout, T., Mehlmer, N., and Brück, T. (2017). Opportunities and challenges in the development of Cutaneotrichosporon oleaginosus ATCC 20509 as a new cell factory for custom tailored microbial oils. Microb. Cell Fact. 16:178. doi: 10.1186/s12934-017-0791-9
Brewer, T. E., and Fierer, N. (2018). Tales from the tomb: the microbial ecology of exposed rock surfaces. Environ. Microbiol. 20, 958–970. doi: 10.1111/1462-2920.14024
Broberg, M., Dubey, M., Iqbal, M., Gudmundssson, M., Ihrmark, K., Schroers, H. J., et al. (2021). Comparative genomics highlights the importance of drug efflux transporters during evolution of mycoparasitism in Clonostachys subgenus Bionectria (Fungi, Ascomycota, Hypocreales). Evol. Appl. 14, 476–497. doi: 10.1111/eva.13134
Brodie, E. L., DeSantis, T. Z., Parker, J. P. M., Zubietta, I. X., Piceno, Y. M., and Andersen, G. L. (2007). Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 104, 299–304. doi: 10.1073/pnas.0608255104
Cailhol, D., Ciadamidaro, L., Dupuy, D., Allegra, S., Girardot, F., and Pfendler, S. (2020). Fungal and bacterial outbreak in the wine vinification area in the Saint-Marcel show cave. Sci. Total Environ. 733:138756. doi: 10.1016/j.scitotenv.2020.138756
Campbell, B. J., Engel, A. S., Porter, M. L., and Takai, K. (2006). The versatile ε-proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4, 458–468. doi: 10.1038/nrmicro1414
Cañveras, C., Sanchez-Moral, S., Sloer, V., and Saiz-Jimenez, C. (2001). Microorganisms and microbially induced fabrics in cave walls. Geomicrobiol. J. 18, 223–240. doi: 10.1080/01490450152467769
Caumartin, V. (1963). Review of the microbiology of underground environments. Bull. Natl. Speleol. Soc. 25, 1–14.
Challis, G. L., and Hopwood, D. A. (2003). Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. U. S.A. 100(Suppl. 2), 14555–14561. doi: 10.1073/pnas.1934677100
Chang, H. C., Leaw, S. N., Huang, A. H., Wu, T. L., and Chang, T. C. (2001). Rapid identification of yeasts in positive blood cultures by a multiplex PCR method. J. Clin. Microbiol. 39, 3466–3471. doi: 10.1128/jcm.39.10.3466-3471.2001
Chen, S., Qi, G., Luo, T., Zhang, H., Jiang, Q., Wang, R., et al. (2018). Continuous-cropping tobacco caused variance of chemical properties and structure of bacterial network in soils. Land. Degrad. Dev. 29, 4106–4120. doi: 10.1002/ldr.3167
Cheng, X., Yun, Y., Wang, H., Ma, L., Tian, W., Man, B., et al. (2021). Contrasting bacterial communities and their assembly processes in karst soils under different land use. Sci. Total Environ. 751:142263. doi: 10.1016/j.scitotenv.2020.142263
Choe, Y. H., Kim, M., Woo, J., Lee, M. J., Lee, J. I., Lee, E. J., et al. (2018). Comparing rock-inhabiting microbial communities in different rock types from a high arctic polar desert. FEMS Microbiol. Ecol. 94:fiy070. doi: 10.1093/femsec/fiy070
Claesson, M. J., O’Sullivan, O., Wang, Q., Nikkilä, J., Marchesi, J. R., Smidt, H., et al. (2009). Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669. doi: 10.1371/journal.pone.0006669
Claesson, M. J., Wang, Q., O’Sullivan, O., Greene-Diniz, R., Cole, J. R., Ross, R. P., et al. (2010). Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38:e200. doi: 10.1093/nar/gkq873
Compton, B. J., Lagutin, K., Dyer, B. S., Ryan, J., MacKenzie, A., Stott, M. B., et al. (2020). Isolation and synthesis of glycophospholipids from the extremophile Chthonomonas calidirosea. Asian J. Org. Chem. 9, 1802–1814. doi: 10.1002/ajoc.202000357
Cuezva, S., Fernandez-Cortes, A., Porca, E., Pašić, L., Jurado, V., Hernandez-Marine, M., et al. (2012). The biogeochemical role of Actinobacteria in Altamira Cave, Spain. FEMS Microbiol. Ecol. 81, 281–290. doi: 10.1111/j.1574-6941.2012.01391.x
De Mandal, S., Chatterjee, R., and Kumar, N. S. (2017). Dominant bacterial phyla in caves and their predicted functional roles in C and N cycle. BMC Microbiol. 17:90. doi: 10.1186/s12866-017-1002-x
Delmont, T. O., Quince, C., Shaiber, A., Esen, ÖC., Lee, S. T., Lücker, S., et al. (2017). Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in the surface ocean. bioRxiv Preprint. doi: 10.1101/129791
Deng, Y., Jiang, Y. H., Yang, Y., He, Z., Luo, F., and Zhou, J. (2012). Molecular ecological network analyses. BMC Bioinform. 13:13. doi: 10.1186/1471-2105-13-113
Dhami, N. K., Reddy, M. S., and Mukherjee, A. (2013). Biomineralization of calcium carbonates and their engineered applications: a review. Front. Microbiol. 4:314. doi: 10.3389/fmicb.2013.00314
Dini-Andreote, F., Stegen, J. C., Van Elsas, J. D., and Salles, J. F. (2015). Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. U. S. A. 112, E1326–E1332. doi: 10.1073/pnas.1414261112
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Döbbeler, P., Davison, P. G., and Buck, W. R. (2015). Two new hypocrealean ascomycetes on bryophytes from North America. Nova Hedwigia 100, 383–390. doi: 10.1127/nova_hedwigia/2015/0244
Els, N., Larose, C., Baumann-Stanzer, K., Tignat-Perrier, R., Keuschnig, C., Vogel, T. M., et al. (2019). Microbial composition in seasonal time series of free tropospheric air and precipitation reveals community separation. Aerobiologia 35, 671–701. doi: 10.1007/s10453-019-09606-x
Evtushenko, L. I. (2015). “Marmoricola,” in Bergey’s Manual of Systematics of Archaea and Bacteria, ed. W. B. Whitman (Hoboken, NJ: John Wiley & Sons), 1–27. doi: 10.1002/9781118960608.gbm00158
Evtushenko, L. I., Krausova, V. I., and Yoon, J. H. (2015). “Nocardioidaceae,” in Bergey’s Manual of Systematics of Archaea and Bacteria, ed. W. B. Whitman (Hoboken: John Wiley & Sons), 1–81. doi: 10.1002/9781118960608.fbm00042
Farhat, M. B., Farhat, A., Bejar, W., Kammoun, R., Bouchaala, K., Fourati, A., et al. (2009). Characterization of the mineral phosphate solubilizing activity of Serratia marcescens CTM 50650 isolated from the phosphate mine of Gafsa. Arch. Microbiol. 191, 815–824. doi: 10.1007/s00203-009-0513-8
Forchhammer, K., and Selim, K. A. (2020). Carbon/nitrogen homeostasis control in cyanobacteria. FEMS Microbiol. Rev. 44, 33–53. doi: 10.1093/femsre/fuz025
Fouts, D. E., Szpakowski, S., Purushe, J., Torralba, M., Waterman, R. C., MacNeil, M. D., et al. (2012). Next generation sequencing to define prokaryotic and fungal diversity in the bovine rumen. PLoS One 7:e48289. doi: 10.1371/journal.pone.0048289
Freed, S. Jr., Ramaley, R. F., and Kyndt, J. A. (2019). Whole-genome sequence of the novel Rubrobacter taiwanensis strain Yellowstone, isolated from Yellowstone National Park. Microbiol. Resour. Announce. 8:e00287-e19. doi: 10.1128/mra.00287-19
Fuerst, J. A., and Sagulenko, E. (2011). Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 9, 403–413. doi: 10.1038/nrmicro2578
Gallagher, K. L., Kading, T. J., Braissant, O., Dupraz, C., and Visscher, P. T. (2012). Inside the alkalinity engine: the role of electron donors in the organomineralization potential of sulfate-reducing bacteria. Geobiology 10, 518–530. doi: 10.1111/j.1472-4669.2012.00342.x
Greiner, K., Peršoh, D., Weig, A., and Rambold, G. (2014). Phialosimplex salinarum, a new species of Eurotiomycetes from a hypersaline habitat. IMA Fungus 5, 161–172. doi: 10.5598/imafungus.2014.05.02.01
Griffin, D. W. (2007). Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin. Microbiol. Rev. 20, 459–477. doi: 10.1128/CMR.00039-06
Gueidan, C., Hill, D. J., Miadlikowska, J., and Lutzoni, F. (2015). “Pezizomycotina: Lecanoromycetes,” in Systematics and Evolution, eds D. J. McLaughlin and J. W. Spatafora (Heidelberg: Springer), 89–120.
Guimera, R., and Amaral, L. A. N. (2005). Cartography of complex networks: modules and universal roles. J. Stat. Mech. Theory Exp. 2005, P02001-1–P02001-13. doi: 10.1088/1742-5468/2005/02/p02001
Gulecal-Pektas, Y. (2016). Bacterial diversity and composition in Oylat Cave (Turkey) with combined Sanger/pyrosequencing approach. Pol. J. Microbiol. 65, 69–75. doi: 10.5604/17331331.1197277
Gupta, R. S., and Khadka, B. (2016). Evidence for the presence of key chlorophyll-biosynthesis-related proteins in the genus Rubrobacter (Phylum Actinobacteria) and its implications for the evolution and origin of photosynthesis. Photosyn. Res. 127, 201–218. doi: 10.1007/s11120-015-0177-y
Hameed, A. A., Khoder, M., Ibrahim, Y., Saeed, Y., Osman, M., and Ghanem, S. (2012). Study on some factors affecting survivability of airborne fungi. Sci. Total Environ. 414, 696–700. doi: 10.1016/j.scitotenv.2011.10.042
Haridas, S., Albert, R., Binder, M., Bloem, J., LaButti, K., Salamov, A., et al. (2020). 101 Dothideomycetes genomes: a test case for predicting lifestyles and emergence of pathogens. Stud. Mycol. 96, 141–153. doi: 10.1016/j.simyco.2020.01.003
He, D., Wu, F., Ma, W., Zhang, Y., Gu, J. D., Duan, Y., et al. (2021). Insights into the bacterial and fungal communities and microbiome that causes a microbe outbreak on ancient wall paintings in the Maijishan Grottoes. Int. Biodeterior. Biodegrad. 163:105250. doi: 10.1016/j.ibiod.2021.105250
Hershey, O. S., and Barton, H. A. (2018). “The microbial diversity of caves,” in Cave Ecology, eds O. T. Moldovan, L’. Kováč, and S. Halse (Heidelberg: Springer), 69–90.
Hess, W. H. (1900). The origin of nitrates in cavern earths. J. Geol. 8, 129–134. doi: 10.1086/620781
Høeg, O. (1946). Cyanophyceae and bacteria in calcareous sediments in the interior of limestone caves in Nord-Rana, Norway. Nytt Mag. Naturvidensk. 85, 99–104.
Hoffman, M. T., and Arnold, A. E. (2010). Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl. Environ. Microbiol. 76, 4063–4075. doi: 10.1128/aem.02928-09
Hofstetter, V., Miadlikowska, J., Kauff, F., and Lutzoni, F. (2007). Phylogenetic comparison of protein-coding versus ribosomal RNA-coding sequence data: a case study of the Lecanoromycetes (Ascomycota). Mol. Phylogenet. Evol. 44, 412–426. doi: 10.1016/j.ympev.2006.10.016
Honegger, R. (2009). “Lichen-forming fungi and their photobionts,” in Plant Relationships, 2nd Edn, ed. H. B. Deising (Heidelberg: Springer), 307–333.
Hu, C., Henderson, G. M., Huang, J., Chen, Z., and Johnson, K. R. (2008). Report of a three-year monitoring programme at Heshang Cave, Central China. Int. J. Speleol. 37, 143–151. doi: 10.5038/1827-806x.37.3.1
Hu, L., Cao, L., and Zhang, R. (2014). Bacterial and fungal taxon changes in soil microbial community composition induced by short-term biochar amendment in red oxidized loam soil. World J. Microbiol. Biotechnol. 30, 1085–1092. doi: 10.1007/s11274-013-1528-5
Huügler, M., Wirsen, C. O., Fuchs, G., Taylor, C. D., and Sievert, S. M. (2005). Evidence for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the ε subdivision of Proteobacteria. J. Bacteriol. 187, 3020–3027. doi: 10.1128/jb.187.9.3020-3027.2005
Irit, N., Hana, B., Yifat, B., Esti, K. W., and Ariel, K. (2019). Insights into bacterial communities associated with petroglyph sites from the Negev Desert, Israel. J. Arid Environ. 166, 79–82. doi: 10.1016/j.jaridenv.2019.04.010
Itoh, T., Yamanoi, K., Kudo, T., Ohkuma, M., and Takashina, T. (2011). Aciditerrimonas ferrireducens gen. nov., sp. nov., an iron-reducing thermoacidophilic actinobacterium isolated from a solfataric field. Int. J. Syst. Evol. Microbiol. 61, 1281–1285. doi: 10.1099/ijs.0.023044-0
Jones, D. L., and Oburger, E. (2011). “Solubilization of phosphorus by soil microorganisms,” in Phosphorus in Action, eds E. K. Bünemann, A. Oberson, and E. Frossard (Heidelberg: Springer), 169–198.
Jones, D. S., and Macalady, J. L. (2016). “The snotty and the stringy: energy for subsurface life in caves,” in Their World: A Diversity of Microbial Environments, ed. C. J. Hurst (Heidelberg: Springer), 203–224.
Jüriado, I., Kaasalainen, U., and Rikkinen, J. (2017). Specialist taxa restricted to threatened habitats contribute significantly to the regional diversity of Peltigera (Lecanoromycetes, Ascomycota) in Estonia. Fungal Ecol. 30, 76–87. doi: 10.1016/j.funeco.2017.08.004
Kang, C. S., Dunfield, P. F., and Semrau, J. D. (2019). The origin of aerobic methanotrophy within the Proteobacteria. FEMS Microbiol. Lett. 366:fnz096. doi: 10.1093/femsle/fnz096
Karkun, A., Tiwari, K., and Jadhav, S. (2012). Fungal diversity of Mandeepkhol cave in Chhattisgarh. India Adv. Biores 3, 119–123.
Kato, S., Hashimoto, K., and Watanabe, K. (2012). Microbial interspecies electron transfer via electric currents through conductive minerals. Proc. Natl. Acad. Sci. U. S. A. 109, 10042–10046. doi: 10.1073/pnas.1117592109
Kavanagh, K. (2005). “Fungal fermentation systems and products,” in Fungi: Biology and Applications, ed. K. Kavanagh (Hoboken, NJ: John Wiley & Sons), 125–146.
Kembel, S. W., Cowan, P. D., Helmus, M. R., Cornwell, W. K., Morlon, H., Ackerly, D. D., et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. doi: 10.1093/bioinformatics/btq166
Khodosovtsev, A., Vondrák, J., Naumovich, A., Kocourková, J., Vondráková, O., and Motiejnait, J. (2012). Three new Pronectria species in terricolous and saxicolous microlichen communities (Bionectriaceae, Ascomycota). Nova Hedwigia 95, 211–220. doi: 10.1127/0029-5035/2012/0026
Kirby, B. M., Le Roes, M., and Meyers, P. R. (2006). Kribbella karoonensis sp. nov. and Kribbella swartbergensis sp. nov., isolated from soil from the Western Cape, South Africa. Int. J. Syst. Evol. Microbiol. 56, 1097–1101. doi: 10.1099/ijs.0.63951-0
Kirk, P. M., Cannon, P. F., David, J., and Stalpers, J. A. (2001). Ainsworth and Bisby’s Dictionary of the Fungi. Wallingford: CABI publishing.
Kováč, L’. (2018). “Caves as oligotrophic ecosystems,” in Cave Ecology, eds O. T. Moldovan, L’. Kováč, and S. Halse (Cham: Springer), 297–307.
Kozlova, E. V., and Mazina, S. E. (2020). Biodiversity of Fungi in the photic and aphotic zones of Montenegro caves. Aerobiologia 36, 589–604. doi: 10.1007/s10453-020-09654-8
Kulichevskaya, I. S., Ivanova, A. O., Baulina, O. I., Bodelier, P. L., Damste, J. S. S., and Dedysh, S. N. (2008). Singulisphaera acidiphila gen. nov., sp. nov., a non-filamentous, Isosphaera-like planctomycete from acidic northern wetlands. Int. J. Syst. Evol. Microbiol. 58, 1186–1193. doi: 10.1099/ijs.0.65593-0
Kulichevskaya, I. S., Ivanova, A. O., Belova, S. E., Baulina, O. I., Bodelier, P. L., Rijpstra, W. I. C., et al. (2007). Schlesneria paludicola gen. nov., sp. nov., the first acidophilic member of the order Planctomycetales, from Sphagnum-dominated boreal wetlands. Int. J. Syst. Evol. Microbiol. 57, 2680–2687. doi: 10.1099/ijs.0.65157-0
Kumar, D., Gaurav, K., Sreya, P., Shabbir, A., Uppada, J., and Ch, S. (2020). Gimesia chilikensis sp. nov., a haloalkali-tolerant planctomycete isolated from Chilika lagoon and emended description of the genus Gimesia. Int. J. Syst. Evol. Microbiol. 70, 3647–3655. doi: 10.1099/ijsem.0.004211
Lauer, A., Simon, M. A., Banning, J. L., André, E., Duncan, K., and Harris, R. N. (2007). Common cutaneous bacteria from the eastern red-backed salamander can inhibit pathogenic fungi. Copeia 2007, 630–640.
Lavoie, K. H., Northup, D. E., and Barton, H. A. (2010). “Microbe-mineral interactions: cave geomicrobiology,” in Geomicrobiology, eds S. K. Jain, A. A. Khan, and M. K. Rai (New York, NY: CRC Press), 1–46.
Lee, J. J., Kang, M. S., Kim, G. S., Lee, C. S., Lim, S., Lee, J., et al. (2016). Flavisolibacter tropicus sp. nov., isolated from tropical soil. Int. J. Syst. Evol. Microbiol. 66, 3413–3419. doi: 10.1099/ijsem.0.001207
Lee, K. C., Stott, M. B., Dunfield, P. F., Huttenhower, C., McDonald, I. R., and Morgan, X. C. (2016). The Chthonomonas calidirosea genome is highly conserved across geographic locations and distinct chemical and microbial environments in New Zealand’s Taupō Volcanic Zone. Appl. Environ. Microbiol. 82, 3572–3581. doi: 10.1128/aem.00139-16
Lee, N. M., Meisinger, D. B., Aubrecht, R., Kovacik, L., Saiz-Jimenez, C., Baskar, S., et al. (2012). “Caves and karst environments,” in Life at Extremes: Environments, Organisms and Strategies for Survival, ed. E. M. Bell (Oxfordshire: CAB International), 320–344.
Lee, S. D. (2007). Marmoricola aequoreus sp. nov., a novel actinobacterium isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 57, 1391–1395. doi: 10.1099/ijs.0.64696-0
Lee, S. D. (2017). Conexibacter stalactiti sp. nov., isolated from stalactites in a lava cave and emended description of the genus Conexibacter. Int. J. Syst. Evol. Microbiol. 67, 3214–3218. doi: 10.1099/ijsem.0.002083
Lemon, J., Bolker, B., Oom, S., Klein, E., Rowlingson, B., Wickham, H., et al. (2021). Package ‘plotrix’. Vienna: R Development Core Team.
Leplat, J., Francois, A., and Bousta, F. (2017). White fungal covering on the wall paintings of the Saint-Savin-sur-Gartempe Abbey church crypt: a case study. Int. Biodeterior. Biodegrad. 122, 29–37. doi: 10.1016/j.ibiod.2017.04.007
Leplat, J., François, A., Touron, S., Galant, P., and Bousta, F. (2019). Aerobiological behavior of Paleolithic decorated caves: a comparative study of five caves in the Gard department (France). Aerobiologia 35, 105–124. doi: 10.1007/s10453-018-9546-2
Li, B., Xie, C. H., and Yokota, A. (2007). Nocardioides exalbidus sp. nov., a novel actinomycete isolated from lichen in Izu-Oshima Island, Japan. Actinomycetologica 21, 22–26. doi: 10.3209/saj.saj210103
Li, Z., Bai, T., Dai, L., Wang, F., Tao, J., Meng, S., et al. (2016). A study of organic acid production in contrasts between two phosphate solubilizing fungi: Penicillium oxalicum and Aspergillus niger. Sci. Rep. 6:25313. doi: 10.1038/srep25313
Lian, B., Chen, Y., Zhu, L., and Yang, R. (2008). Progress in the study of the weathering of carbonate rock by microbes. Earth Sci. Front. 15, 90–99.
Lombard, L., Van Der Merwe, A., Groenewald, J. Z., and Crous, P. W. (2014). Lineages in Nectriaceae: re-evaluating the generic status of Ilyonectria and allied genera. Phytopathol. Mediterr. 53, 515–532. doi: 10.14601/Phytopathol_Mediterr-14976
Lombard, L., Van der Merwe, N., Groenewald, J., and Crous, P. W. (2015). Generic concepts in Nectriaceae. Stud. Mycol. 80, 189–245. doi: 10.1016/j.simyco.2014.12.002
Loni, P. C., Wu, M., Wang, W., Wang, H., Ma, L., Liu, C., et al. (2020). Mechanism of microbial dissolution and oxidation of antimony in stibnite under ambient conditions. J. Hazard. Mater. 385:121561. doi: 10.1016/j.jhazmat.2019.121561
Ma, B., Wang, H., Dsouza, M., Lou, J., He, Y., Dai, Z., et al. (2016). Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J. 10, 1891–1901. doi: 10.1038/ismej.2015.261
Ma, L., Huang, X., Wang, H., Yun, Y., Cheng, X., Liu, D., et al. (2021). Microbial interactions drive distinct taxonomic and potential metabolic responses to habitats in karst cave ecosystem. Microbiol. Spectr. 9:e011521. doi: 10.1128/spectrum.01152-21.
Ma, Y., Zhang, H., Du, Y., Tian, T., Xiang, T., Liu, X., et al. (2015). The community distribution of bacteria and fungi on ancient wall paintings of the Mogao Grottoes. Sci. Rep. 5:7752. doi: 10.1038/srep07752
Man, B., Wang, H., Xiang, X., Wang, R., Yun, Y., and Gong, L. (2015). Phylogenetic diversity of culturable fungi in the Heshang Cave, central China. Front. Microbiol. 6:1158. doi: 10.3389/fmicb.2015.01158
Manimaran, M., and Kannabiran, K. (2017). Actinomycetes-mediated biogenic synthesis of metal and metal oxide nanoparticles: progress and challenges. Lett. Appl. Microbiol. 64, 401–408. doi: 10.1111/lam.12730
Marshall, K. T., and Morris, R. M. (2013). Isolation of an aerobic sulfur oxidizer from the SUP05/Arctic96BD-19 clade. ISME J. 7, 452–455. doi: 10.1038/ismej.2012.78
Mazina, S., Kozlova, E., Popkova, A., Kochetkov, S., Mannapova, R., and Yakushev, A. (2020). A first assessment of the microbiota of Taurida Cave. Ecol. Montenegrina 37, 1–10. doi: 10.37828/em.2020.37.1
Miadlikowska, J., Kauff, F., Hofstetter, V., Fraker, E., Grube, M., Hafellner, J., et al. (2006). New insights into classification and evolution of the Lecanoromycetes (Pezizomycotina, Ascomycota) from phylogenetic analyses of three ribosomal RNA-and two protein-coding genes. Mycologia 98, 1088–1103. doi: 10.1080/15572536.2006.11832636
Nilsson, R. H., Larsson, K. H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2019). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264. doi: 10.1093/nar/gky1022
Ningthoujam, D. S., Sanasam, S., and Mutum, A. (2012). Characterization of p-nitrophenol degrading actinomycetes from Hundung limestone deposits in Manipur, India. Afr. J. Biotechnol. 11, 10210–10220. doi: 10.5897/ajb11.4123
O’Donnell, K., Sutton, D. A., Rinaldi, M. G., Sarver, B. A., Balajee, S. A., Schroers, H. J., et al. (2010). Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J. Clin. Microbiol. 48, 3708–3718. doi: 10.1128/jcm.00989-10
Ogórek, R., Lejman, A., and Matkowski, K. (2013). Fungi isolated from Niedźwiedzia cave in Kletno (lower Silesia, Poland). Int. J. Speleol. 42, 161–166. doi: 10.5038/1827-806x.42.2.9
Oliveira, M., Ribeiro, H., and Abreu, I. (2005). Annual variation of fungal spores in atmosphere of Porto: 2003. Ann. Agric. Environ. Med. 12, 309–315.
Ortiz, M., Neilson, J. W., Nelson, W. M., Legatzki, A., Byrne, A., Yu, Y., et al. (2013). Profiling bacterial diversity and taxonomic composition on speleothem surfaces in Kartchner Caverns, AZ. Microb. Ecol. 65, 371–383. doi: 10.1007/s00248-012-0143-6
Pape, R., and OConnor, B. (2014). Diversity and ecology of the macro-invertebrate fauna (Nemata and Arthropoda) of Kartchner Caverns, Cochise County, Arizona, United States of America. Check List 10:761. doi: 10.15560/10.4.761
Pfendler, S., Karimi, B., Alaoui-Sosse, L., Bousta, F., Alaoui-Sossé, B., Abdel-Daim, M. M., et al. (2019). Assessment of fungi proliferation and diversity in cultural heritage: reactions to UV-C treatment. Sci. Total Environ. 647, 905–913. doi: 10.1016/j.scitotenv.2018.08.089
Polymenakou, P. N., Mandalakis, M., Stephanou, E. G., and Tselepides, A. (2008). Particle size distribution of airborne microorganisms and pathogens during an intense African dust event in the eastern Mediterranean. Environ. Health Perspect. 116, 292–296. doi: 10.1289/ehp.10684
Ponizovskaya, V. B., Rebrikova, N. L., Kachalkin, A. V., Antropova, A. B., Bilanenko, E. N., and Mokeeva, V. L. (2019). Micromycetes as colonizers of mineral building materials in historic monuments and museums. Fungal Biol. 123, 290–306. doi: 10.1016/j.funbio.2019.01.002
Popović, S., Subakov Simić, G., Stupar, M., Unković, N., Predojević, D., Jovanović, J., et al. (2015). Cyanobacteria, algae and microfungi present in biofilm from Božana Cave (Serbia). Int. J. Speleol. 44:4. doi: 10.5038/1827-806x.44.2.4
Purahong, W., Wubet, T., Krüger, D., and Buscot, F. (2018). Molecular evidence strongly supports deadwood-inhabiting fungi exhibiting unexpected tree species preferences in temperate forests. ISME J. 12, 289–295. doi: 10.1038/ismej.2017.177
Quintero, E., Rivera-Mariani, F., and Bolaños-Rosero, B. (2010). Analysis of environmental factors and their effects on fungal spores in the atmosphere of a tropical urban area (San Juan, Puerto Rico). Aerobiologia 26, 113–124. doi: 10.1007/s10453-009-9148-0
Ren, G., Yan, Y., Nie, Y., Lu, A., Wu, X., Li, Y., et al. (2019). Natural extracellular electron transfer between semiconducting minerals and electroactive bacterial communities occurred on the rock varnish. Front. Microbiol. 10:293. doi: 10.3389/fmicb.2019.00293
Reyes, I., Valery, A., and Valduz, Z. (2007). “Phosphate-solubilizing microorganisms isolated from rhizospheric and bulk soils of colonizer plants at an abandoned rock phosphate mine,” in First International Meeting on Microbial Phosphate Solubilization, eds E. Velázquez and C. Rodríguez-Barrueco (Dordrecht: Springer), 69–75.
Rogers, J. R., and Bennett, P. C. (2004). Mineral stimulation of subsurface microorganisms: release of limiting nutrients from silicates. Chem. Geol. 203, 91–108. doi: 10.1016/j.chemgeo.2003.09.001
Ruibal, C., Gueidan, C., Selbmann, L., Gorbushina, A., Crous, P., Groenewald, J., et al. (2009). Phylogeny of rock-inhabiting fungi related to Dothideomycetes. Stud. Mycol. 64, 123–133. doi: 10.3114/sim.2009.64.06
Samuels, G. J., and Blackwell, M. (2001). “Pyrenomycetes—fungi with perithecia,” in Systematics and Evolution, eds D. J. McLaughlin, E. G. McLaughlin, and P. A. Lemke (Heidelberg: Springer), 221–255.
Schada von Borzyskowski, L. S., Severi, F., Krüger, K., Hermann, L., Gilardet, A., Sippel, F., et al. (2019). Marine Proteobacteria metabolize glycolate via the β-hydroxyaspartate cycle. Nature 575, 500–504. doi: 10.1038/s41586-019-1748-4
Schubert, T., Kallscheuer, N., Wiegand, S., Boedeker, C., Peeters, S. H., Jogler, M., et al. (2020). Calycomorphotria hydatis gen. nov., sp. nov., a novel species in the family Planctomycetaceae with conspicuous subcellular structures. Antonie van Leeuwenhoek 113, 1877–1887. doi: 10.1007/s10482-020-01419-0
Shaffer, J. P., Sarmiento, C., Zalamea, P. C., Gallery, R. E., Davis, A. S., Baltrus, D. A., et al. (2016). Diversity, specificity, and phylogenetic relationships of endohyphal bacteria in fungi that inhabit tropical seeds and leaves. Front. Ecol. Evol. 4:116. doi: 10.3389/fevo.2016.00116
Shaffer, J. P., U’ren, J. M., Gallery, R. E., Baltrus, D. A., and Arnold, A. E. (2017). An endohyphal bacterium (Chitinophaga, Bacteroidetes) alters carbon source use by Fusarium keratoplasticum (F. solani species complex, Nectriaceae). Front. Microbiol. 8:350. doi: 10.3389/fmicb.2017.00350
Shapleigh, J. P. (2011). Oxygen control of nitrogen oxide respiration, focusing on α-proteobacteria. Biochem. Soc. Trans. 39, 179–183. doi: 10.1042/bst0390179
Shi, L., Dong, H., Reguera, G., Beyenal, H., Lu, A., Liu, J., et al. (2016). Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 14, 651–662. doi: 10.1038/nrmicro.2016.93
Shi, S., Nuccio, E. E., Shi, Z. J., He, Z., Zhou, J., and Firestone, M. K. (2016). The interconnected rhizosphere: high network complexity dominates rhizosphere assemblages. Ecol. Lett. 19, 926–936. doi: 10.1111/ele.12630
Sorokin, D. Y., Vejmelkova, D., Lücker, S., Streshinskaya, G. M., Rijpstra, W. I. C., Damste, J. S. S., et al. (2014). Nitrolancea hollandica gen. nov., sp. nov., a chemolithoautotrophic nitrite-oxidizing bacterium isolated from a bioreactor belonging to the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 64, 1859–1865. doi: 10.1099/ijs.0.062232-0
Stegen, J. C., Lin, X., Fredrickson, J. K., Chen, X., Kennedy, D. W., Murray, C. J., et al. (2013). Quantifying community assembly processes and identifying features that impose them. ISME J. 7, 2069–2079. doi: 10.1038/ismej.2013.93
Stegen, J. C., Lin, X., Konopka, A. E., and Fredrickson, J. K. (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6, 1653–1664. doi: 10.1038/ismej.2012.22
Suphaphimol, N., Suwannarach, N., Purahong, W., Jaikang, C., Pengpat, K., Semakul, N., et al. (2022). Identification of microorganisms dwelling on the 19th century Lanna mural paintings from Northern Thailand using culture-dependent and-independent approaches. Biology 11:228. doi: 10.3390/biology11020228
Tedersoo, L., Bahram, M., Põlme, S., Kõljalg, U., Yorou, N. S., Wijesundera, R., et al. (2014). Global diversity and geography of soil fungi. Science 346:1256688. doi: 10.1126/science.1256688
Tomczyk-Żak, K., Kaczanowski, S., Drewniak, Ł., Dmoch, Ł., Sklodowska, A., and Zielenkiewicz, U. (2013). Bacteria diversity and arsenic mobilization in rock biofilm from an ancient gold and arsenic mine. Sci. Total Environ. 461, 330–340. doi: 10.1016/j.scitotenv.2013.04.087
Tomczyk-Żak, K., and Zielenkiewicz, U. (2016). Microbial diversity in caves. Geomicrobiol. J. 33, 20–38. doi: 10.1080/01490451.2014.1003341
Trovão, J., Portugal, A., Soares, F., Paiva, D. S., Mesquita, N., Coelho, C., et al. (2019). Fungal diversity and distribution across distinct biodeterioration phenomena in limestone walls of the old cathedral of Coimbra, UNESCO World Heritage Site. Int. Biodeterior. Biodegrad. 142, 91–102. doi: 10.1016/j.ibiod.2019.05.008
Vaughan, M. J., Nelson, W., Soderlund, C., Maier, R. M., and Pryor, B. M. (2015). Assessing fungal community structure from mineral surfaces in Kartchner Caverns using multiplexed 454 pyrosequencing. Microb. Ecol. 70, 175–187. doi: 10.1007/s00248-014-0560-9
Wegener, K. M., Singh, A. K., Jacobs, J. M., Elvitigala, T., Welsh, E. A., Keren, N., et al. (2010). Global proteomics reveal an atypical strategy for carbon/nitrogen assimilation by a cyanobacterium under diverse environmental perturbations. Mol. Cell. Proteomics 9, 2678–2689. doi: 10.1074/mcp.m110.000109
Wickham, H., Chang, W., and Wickham, M. H. (2016). Package ‘ggplot2’. Create Elegant Data Visualisations Using the Grammar of Graphics. Available online at: https://cran.r-project.org/web/packages/ggplot2/index.html [accessed on May 3, 2022].
Wiegand, S., Jogler, M., and Jogler, C. (2018). On the maverick Planctomycetes. FEMS Microbiol. Rev. 42, 739–760. doi: 10.1093/femsre/fuy029
Wink, J., Kroppenstedt, R. M., Seibert, G., and Stackebrandt, E. (2003). Actinomadura namibiensis sp. nov. Int. J. Syst. Evol. Microbiol. 53, 721–724. doi: 10.1099/ijs.0.02286-0
Wu, Y., Tan, L., Liu, W., Wang, B., Wang, J., Cai, Y., et al. (2015). Profiling bacterial diversity in a limestone cave of the western Loess Plateau of China. Front. Microbiol. 6:244. doi: 10.3389/fmicb.2015.00244
Yang, C., and Sun, J. (2020). Soil salinity drives the distribution patterns and ecological functions of fungi in saline-alkali land in the Yellow River Delta, China. Front. Microbiol. 11:594284. doi: 10.3389/fmicb.2020.594284
Yun, Y., Wang, H., Man, B., Xiang, X., Zhou, J., Qiu, X., et al. (2016). The relationship between pH and bacterial communities in a single karst ecosystem and its implication for soil acidification. Front. Microbiol. 7:1955. doi: 10.3389/fmicb.2016.01955
Zeng, Y., Selyanin, V., Lukeš, M., Dean, J., Kaftan, D., Feng, F., et al. (2015). Characterization of the microaerophilic, bacteriochlorophyll a-containing bacterium Gemmatimonas phototrophica sp. nov., and emended descriptions of the genus Gemmatimonas and Gemmatimonas aurantiaca. Int. J. Syst. Evol. Microbiol. 65, 2410–2419. doi: 10.1099/ijs.0.000272
Zhang, H., Sekiguchi, Y., Hanada, S., Hugenholtz, P., Kim, H., Kamagata, Y., et al. (2003). Gemmatimonas aurantiaca gen. nov., sp. nov., a Gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int. J. Syst. Evol. Microbiol. 53, 1155–1163. doi: 10.1099/ijs.0.02520-0
Zhang, N., and Wang, Z. (2015). “3 Pezizomycotina: Sordariomycetes and Leotiomycetes,” in Systematics and Evolution, eds D. McLaughlin and J. Spatafora (Heidelberg: Springer), 57–88.
Zhang, X., Ge, Q., Zhu, Z., Deng, Y., and Gu, J. D. (2018). Microbiological community of the Royal Palace in Angkor Thom and Beng Mealea of Cambodia by Illumina sequencing based on 16S rRNA gene. Int. Biodeterior. Biodegrad. 134, 127–135. doi: 10.1016/j.ibiod.2018.06.018
Zhang, Z., Liu, F., Zhou, X., Liu, X., Liu, S., and Cai, L. (2017). Culturable mycobiota from karst caves in China, with descriptions of 20 new species. Persoonia 39, 1–31. doi: 10.3767/persoonia.2017.39.01
Zhang, Z. F., and Cai, L. (2019). Substrate and spatial variables are major determinants of fungal community in karst caves in Southwest China. J. Biogeogr. 46, 1504–1518. doi: 10.1111/jbi.13594
Zhao, R., Wang, H., Cheng, X., Yun, Y., and Qiu, X. (2018). Upland soil cluster γ dominates the methanotroph communities in the karst Heshang Cave. FEMS Microbiol. Ecol. 94:fiy192. doi: 10.1093/femsec/fiy192
Zhou, J., Wang, H., Cravotta, I. I. I. C. A., Dong, Q., and Xiang, X. (2017). Dissolution of fluorapatite by Pseudomonas fluorescens P35 resulting in fluorine release. Geomicrobiol. J. 34, 421–433. doi: 10.1080/01490451.2016.1204376
Zhu, H. Z., Zhang, Z. F., Zhou, N., Jiang, C. Y., Wang, B. J., Cai, L., et al. (2019). Diversity, distribution and co-occurrence patterns of bacterial communities in a karst cave system. Front. Microbiol. 10:1726. doi: 10.3389/fmicb.2019.01726
Keywords: subsurface biosphere, karst caves, phosphate-bearing minerals, microbial interaction, community assembly, co-occurrence network
Citation: Wang Y, Cheng X, Wang H, Zhou J, Liu X and Tuovinen OH (2022) The Characterization of Microbiome and Interactions on Weathered Rocks in a Subsurface Karst Cave, Central China. Front. Microbiol. 13:909494. doi: 10.3389/fmicb.2022.909494
Received: 31 March 2022; Accepted: 19 May 2022;
Published: 29 June 2022.
Edited by:
Cesareo Saiz-Jimenez, Institute of Natural Resources and Agrobiology of Seville (CSIC), SpainReviewed by:
Ariel Kushmaro, Ben-Gurion University of the Negev, IsraelCopyright © 2022 Wang, Cheng, Wang, Zhou, Liu and Tuovinen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongmei Wang, d2FuZ2htZWkwNEAxNjMuY29t; aG13YW5nQGN1Zy5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.