
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 June 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.908304
This article is part of the Research Topic Secondary Metabolism: An Unlimited Foundation for Synthetic Biology, Volume II View all 6 articles
The accumulation of the compatible solute L-proline by Bacillus subtilis via synthesis is a cornerstone in the cell’s defense against high salinity as the genetic disruption of this biosynthetic process causes osmotic sensitivity. To understand how B. subtilis could potentially cope with high osmolarity surroundings without the functioning of its natural osmostress adaptive L-proline biosynthetic route (ProJ-ProA-ProH), we isolated suppressor strains of proA mutants under high-salinity growth conditions. These osmostress-tolerant strains carried mutations affecting either the AhrC transcriptional regulator or its operator positioned in front of the argCJBD-carAB-argF L-ornithine/L-citrulline/L-arginine biosynthetic operon. Osmostress protection assays, molecular analysis and targeted metabolomics showed that these mutations, in conjunction with regulatory mutations affecting rocR-rocDEF expression, connect and re-purpose three different physiological processes: (i) the biosynthetic pathway for L-arginine, (ii) the RocD-dependent degradation route for L-ornithine, and (iii) the last step in L-proline biosynthesis. Hence, osmostress adaptation without a functional ProJ-ProA-ProH route is made possible through a naturally existing, but inefficient, metabolic shunt that allows to substitute the enzyme activity of ProA by feeding the RocD-formed metabolite γ-glutamate-semialdehyde/Δ1-pyrroline-5-carboxylate into the biosynthetic route for the compatible solute L-proline. Notably, in one class of mutants, not only substantial L-proline pools but also large pools of L-citrulline were accumulated, a rather uncommon compatible solute in microorganisms. Collectively, our data provide an example of the considerable genetic plasticity and metabolic resourcefulness of B. subtilis to cope with everchanging environmental conditions.
Increases in the environmental osmolarity occur frequently in the varied habitats of microorganisms and impose considerable energetic and growth-restricting constrains on bacterial cells (Gunde-Cimerman et al., 2018). The detrimental effects of high osmolarity on cellular physiology are a consequence of the different osmotic potentials of the cell’s crowded cytoplasm and its surroundings, and of the physico-chemical attributes of the semi-permeable cytoplasmic membrane (Wood, 2011; Van den Berg et al., 2017; Bremer and Krämer, 2019). As a result, the bacterial cell faces dehydration at high environmental osmolarity, and concomitantly encounters an undesired increase in molecular crowding and a reduction of turgor to values unsuitable for efficient growth. Many microorganisms counteract the outflow of water through a sustained accumulation of compatible solutes (Brown, 1976). These organic osmolytes are compliant with the biochemistry and physiology of bacterial cells (Bolen and Baskakov, 2001; Ignatova and Gierasch, 2006; Stadmiller et al., 2017), and can thus be amassed to very high intracellular pools in a finely-tuned fashion in a response to the degree of osmotic stress imposed onto the cell. Accordingly, the accumulation of compatible solutes, either through synthesis or import, promotes cellular hydration and growth under osmotically unfavorable conditions (Kempf and Bremer, 1998; Roesser and Müller, 2001; Wood et al., 2001).
L-proline is a prominent member of the compatible solutes and is widely used by both plants and microorganisms as an osmostress protectant and chemical chaperone (Csonka, 1989; Cayley et al., 1992; Chattopadhyay et al., 2004; Ignatova and Gierasch, 2006; Fichman et al., 2014). The Gram-positive soil bacterium Bacillus subtilis is one of the microorganisms that uses the accumulation of L-proline to counteract high osmolarity and high salinity incurred water stress (Whatmore et al., 1990; Von Blohn et al., 1997; Brill et al., 2011a; Hoffmann et al., 2017). The genetic disruption of osmostress-adaptive L-proline biosynthesis causes osmotic sensitivity of B. subtilis (Brill et al., 2011a), thereby attesting to the physiological importance of L-proline accumulation for the cell’s ability to cope with osmotically challenging growth conditions (Hoffmann and Bremer, 2016). L-proline is the only compatible solute that B. subtilis can synthesize de novo (Whatmore et al., 1990; Brill et al., 2011a), as the synthesis of the osmostress protectant glycine betaine by this microorganism requires the prior import of the precursor molecule choline (Boch et al., 1994, 1996). Other members of the genus Bacillus can either rely exclusively on the synthesis of L-glutamate, or produce the compatible solutes ectoine/5-hydroxyectoine to counteract osmotic stress (Kuhlmann and Bremer, 2002; Bursy et al., 2007). Depending on the type of compatible solute synthesized by a given Bacillus species, different degrees of osmostress tolerance are attained. L-proline typically affords a substantial, yet intermediate, level of osmotic stress tolerance (Hoffmann and Bremer, 2016).
Synthesis of L-proline by bacteria typically proceeds from the central metabolite L-glutamate and is mediated by three enzymes: γ-glutamyl kinase (ProB), γ-glutamyl phosphate reductase (ProA), and Δ1-pyrroline-5-carboxylate reductase (ProC), with γ-glutamyl phosphate and γ-glutamate-semialdehyde/Δ1-pyrroline-5-carboxylate as the respective intermediates (Fichman et al., 2014). Bacillus subtilis adheres to this L-proline biosynthetic scheme (Belitsky et al., 2001). However, it possesses several L-proline biosynthetic isoenzymes (Figure 1A). Their activities and the transcription of their structural genes are differently regulated so that L-proline pools adequate for either anabolic or osmostress-protective purposes can be produced (Whatmore et al., 1990; Brill et al., 2011a,b; Forlani et al., 2017; Hoffmann et al., 2017). In order to synthesize the large quantities of L-proline under osmotic stress conditions, B. subtilis and related species need to re-route their metabolism to provide adequate cellular pools for the L-proline biosynthetic precursor L-glutamate (Kohlstedt et al., 2014; Godard et al., 2020). This central metabolite is successively drained as the environmental osmolarity increases and enhanced L-proline synthesis commences (Brill et al., 2011a).
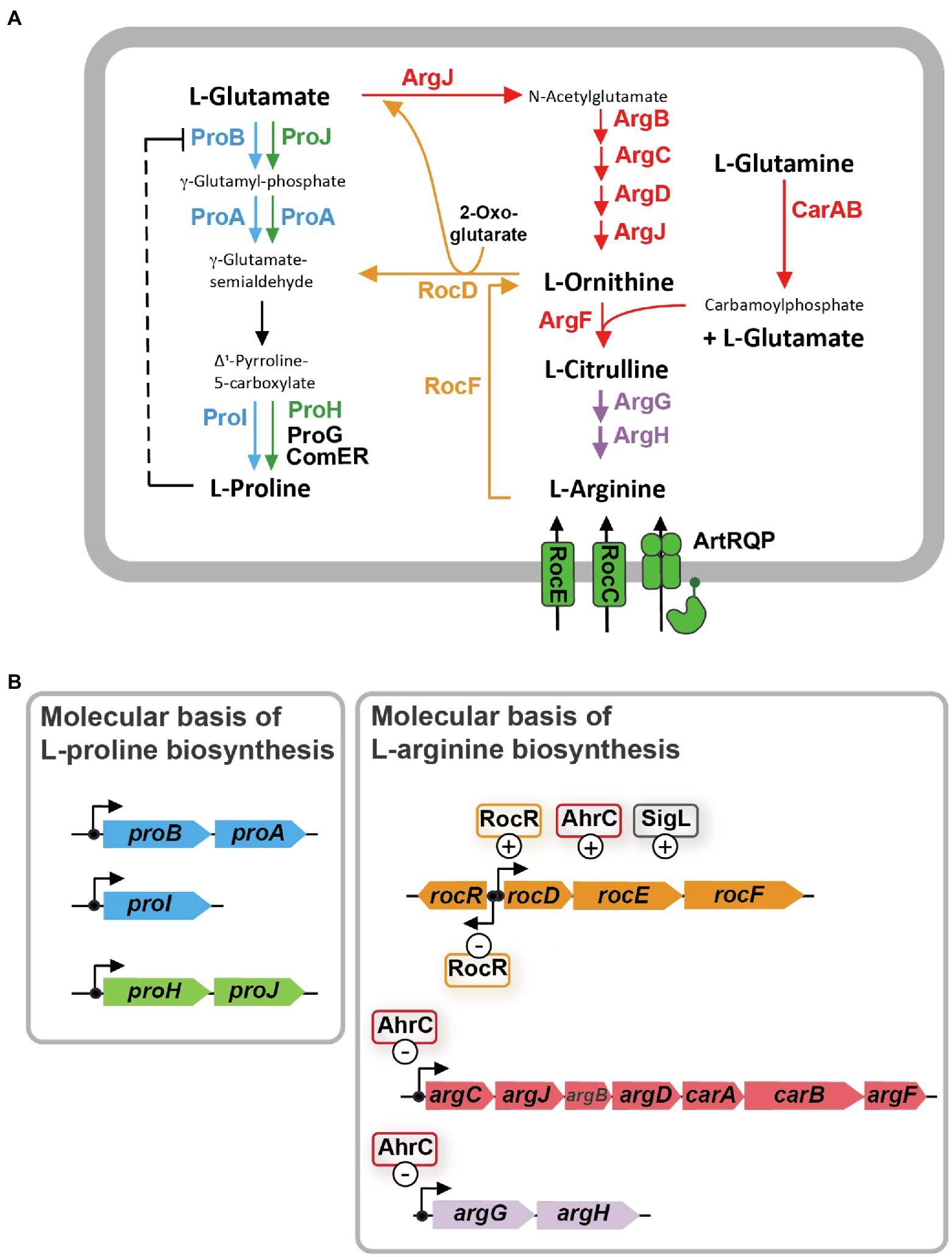
Figure 1. L-arginine biosynthesis and degradation pathways are metabolically interconnected with the L-proline biosynthetic route in Bacillus subtilis. (A) Schematic overview of the precursors, intermediates, and products of L-arginine synthesis and catabolism, and the anabolic and osmostress-responsive L-proline biosynthetic route. The ProB-ProA-ProI enzymes (blue) are used for anabolic L-proline production, while the ProJ-ProA-ProH (green) route is employed for the synthesis of L-proline as an osmostress protectant. γ-glutamate-semialdehyde spontaneously converts to Δ1-pyrroline-5-carboxylate which is the substrate for the Δ1-pyrroline-5-carboxylate reductase; four of these types of enzymes operate in B. subtilis. The precise physiological functions of the ProG and ComER enzymes are not entirely clear. (B) Molecular and regulatory overview of L-proline and L-arginine biosynthetic genes and those for L-arginine degradation. Positive and negative influence of known regulatory proteins on gene expression are highlighted. These data for this figure were compiled from the literature.
Biosynthesis of L-proline is energetically costly (Akashi and Gojobori, 2002). Hence, B. subtilis uses both genetic and biochemical control mechanisms to tie anabolic L-proline production via the ProB-ProA-ProI route to the ongoing protein biosynthetic activities of the cell. At the genetic level, expression of the genes for the proBA operon encoding γ-glutamyl kinase (ProB) and γ-glutamyl phosphate reductase (ProA), and that of the gene (proI) encoding the Δ1-pyrroline-5-carboxylate reductase ProI (Figure 1B) are controlled by a T-box regulatory mechanism. This tRNAPro-responsive riboswitch (Kreuzer and Henkin, 2018) allows only enhanced full-length transcription of the proBA operon and that of the proI gene when the L-proline pool is insufficient to adequately fuel protein biosynthesis (Brill et al., 2011b). At the biochemical level, feed-back inhibition of ProB enzyme activity by L-proline ensures that the flow of the precursor L-glutamate into the L-proline biosynthetic pathway (Figure 1A) is curbed when the L-proline pool is sufficiently high to promote growth (Fujita et al., 2003; Chen et al., 2006; Perez-Arellano et al., 2010). As a result of these combined genetic and biochemical regulatory mechanisms, the steady-state pool of L-proline is kept by non-osmotically stressed B. subtilis cells in a rather narrow range (about 10–20 mM; Whatmore et al., 1990; Brill et al., 2011a; Zaprasis et al., 2013a; Hoffmann et al., 2017).
In contrast, and depending on the severity of the environmentally imposed osmotic stress, B. subtilis amasses several hundred mM of L-proline to increase the osmotic potential of the cytoplasm in order to counteract water outflow (Whatmore and Reed, 1990; Brill et al., 2011a; Zaprasis et al., 2013a; Hoffmann et al., 2017). To provide these large quantities of L-proline, B. subtilis developed an osmostress-responsive L-proline biosynthetic pathway that is freed from the genetic and biochemical constraints imposed onto the anabolic route (Brill et al., 2011a; Hoffmann et al., 2017). The osmostress adaptive L-proline biosynthetic route (ProJ-ProA-ProH) consists of isoenzymes of the first (ProJ) and last (ProH) step of the anabolic L-proline biosynthetic route (ProB/ProI) but shares with it [for unknown reasons (Hoffmann et al., 2017)] the γ-glutamyl phosphate reductase (ProA; Figure 1A). The proHJ operon lacks a T-box, and instead, its transcription is strongly upregulated in response to high osmolarity or salinity (Brill et al., 2011a; Hoffmann et al., 2017). Furthermore, the feed-back control of the enzyme activity of ProJ is probably abolished, or at least strongly reduced, in comparison with the ProB isoenzyme (Fujita et al., 2003; Chen et al., 2006; Perez-Arellano et al., 2010). Despite the T-box control of proBA, enough ProA enzymes are produced in osmotically challenged cells to functionally match the amounts of the ProJ-ProH enzymes needed to produce large amounts of the osmostress protectant L-proline (Hoffmann et al., 2017). In this context, it is noteworthy that Bacillus species other than B. subtilis (e.g., Bacillus licheniformis, Bacillus megaterium) also possess two separate, yet biochemically complete, routes for either anabolic or osmostress protective L-proline production (Schroeter et al., 2013; Godard et al., 2020).
It is well known that the biosynthesis of L-proline and L-arginine are interconnected in many bacteria, including B. subtilis (Figure 1A; Belitsky, 2002; Fischer and Debarbouille, 2002; Csonka and Leisinger, 2007). As a result, suppressor mutations located in genes for L-arginine metabolism can bypass particular genetic blocks in L-proline biosynthesis (Itikawa et al., 1968; Kuo and Stocker, 1969; Berg and Rossi, 1974; Rossi et al., 1977). L-proline and L-arginine are both produced from the central metabolite L-glutamate, and γ-glutamate-semialdehyde/Δ1-pyrroline-5-carboxylate is an common intermediate in their respective biosynthetic routes (Baumberg and Harwood, 1979; Belitsky, 2002; Fischer and Debarbouille, 2002; Fichman et al., 2014; Figure 1A).
The γ-glutamate-semialdehyde/Δ1-pyrroline-5-carboxylate-dependent metabolic shunt between the L-arginine and L-proline biosynthetic routes operates very inefficiently in B. subtilis wild-type cells. This is evidenced by the fact that a proA mutant forms only tiny colonies on L-proline-free minimal medium agar plates with glucose as the carbon and ammonium as the nitrogen source (Zaprasis et al., 2013b). However, spontaneously occurring suppressor strains with increased growth performance can readily be isolated. In these suppressors, the genetic block in the ProA-catalyzed step is bypassed through a transcriptional up-regulation of the L-arginine catabolic rocDEF operon (Figure 1A; Zaprasis et al., 2013b). This allows the rocD-encoded ornithine aminotransferase (Figure 1B) to convert increased amounts of L-ornithine, an intermediate in L-arginine biosynthesis, to produce increased amounts of γ-glutamate-semialdehyde/Δ1-pyrroline-5-carboxylate, metabolites that are also the product(s) of the ProA enzyme (Figure 1A; Belitsky et al., 2001; Fichman et al., 2014). Hence, ProA enzyme activity can be bypassed as Δ1-pyrroline-5-carboxylate can be converted into L-proline by Δ1-pyrroline-5-carboxylate reductase, a type of enzyme present in B. subtilis in four different forms (ProI, ProH, ProG, ComER; Figure 1A; Belitsky et al., 2001; Brill et al., 2011a; Forlani et al., 2017; Hoffmann et al., 2017). Accordingly, only a proA rocD double-mutant of B. subtilis exhibits a tight L-proline auxotrophic growth phenotype (Zaprasis et al., 2013b).
While the previously reported suppressor strains of a proA defect in B. subtilis allow the production of L-proline pools sufficiently large to restore growth in a L-proline free minimal medium, none of them was able to attain osmostress protective cellular levels of L-proline (Zaprasis et al., 2013b). Building on the idea that microorganisms can almost always find a way to circumvent metabolic constraints through mutational changes (Barrick and Lenski, 2013), we have now investigated if proA bypass suppressors can be found that would restore osmostress tolerance to B. subtilis. Indeed, we found such suppressor strains, and in each of these mutants the same physiological process, L-arginine biosynthesis, was targeted, albeit as the consequence of different genetic events. Our analysis of the metabolome of some of these osmostress tolerant suppressors suggests that not only L-proline but also L-citrulline might function as a compatible solute for B. subtilis under special circumstances.
All antibiotics used in this study and the chromogenic substrate (para-nitrophenyl-α-glycopyranoside; α-PNPG) for activity assays of the TreA enzyme, a phospho-α-(1,1)-glucosidase, were obtained from Sigma-Aldrich (Steinheim, Germany). Bacillus subtilis strains were maintained at room temperature on LB-agar plates. Liquid cultures of B. subtilis strains were grown in shake flasks (20 ml culture volume in 100-ml Erlenmeyer flasks) set in a rotating water bath (220 rpm at 37° C) until they reached an OD578 indicated in the individual experiments. Throughout our study, we used Spizizen’s minimal medium (SMM) with glucose (0.5% w/v) as the carbon source, ammonium as the nitrogen source, and a solution of trace elements (Harwood and Archibald, 1990). Because the B. subtilis wild-type strain JH642 (Smith et al., 2014) and its derivatives (Table 1; Supplementary Table S1) are auxotrophic for L-phenylalanine and L-tryptophan, these two amino acids were added to the growth medium (L-Phe at 18 mg L−1 and L-Trp at 20 mg L−1). When high-salinity growth medium was used, pre-cultures were grown in SMM without additional NaCl to early-exponential growth phase (OD578 of about 1.5) and were then used to inoculate main cultures, to an OD578 of about 0.1, containing increased NaCl concentrations as indicated in the individual experiments.
To verify integration of PargC-treA (catR) reporter fusion constructs via a double recombination event into the chromosomal amyE gene as a single copy (Harwood and Archibald, 1990), corresponding B. subtilis strains were grown on LB-agar plates containing 1% starch. The plates were then flooded with Gram’s iodine stain for 1 min and scored for zones of starch degradation around colonies to identify those transformants that were no longer able to degrade starch (the AmyE phenotype; Harwood and Archibald, 1990). To select chloramphenicol resistant (catR) B. subtilis strains after transformation with linearized plasmid DNA encoding various PargC-treA transcriptional reporter constructs, LB agar plates containing 5 μg of this antibiotic were used. PargC-treA reporter fusion derivatives of plasmid pPINK1 (Supplementary Table S2) were selected in the Escherichia coli strain TOP10 (Invitrogen, Darmstadt, Germany), by plating competent cells on LB agar plates containing 100 μg ml−1 of ampicillin.
All recombinant DNA work used established procedures. To construct transcriptional fusions between the argC regulatory region to the promoter-less treA reporter gene (Schöck et al., 1996), a 427 bp DNA fragment of the wild-type sequence and of six AhrC-operator mutants (Figure 2) was amplified by PCR using the DNA primers DS27_5′-ATTGGGCCCGAGTGGATTGATGATGATGA-3′ and DS28_5′-ATTGGATCCTCGTATTCATATCAATCGGGC-3′. The amplified fragment carrying the argC promoter and its AhrC responsive regulatory sequences was inserted in front of the promoter-less treA+ gene carried by the low-copy number plasmid pPINK1 that had been cut with the restriction enzymes BamHI and SmaI. The resulting PargC-treA reporter plasmids are listed in Supplementary Table S2. The DNA of each of these plasmids was linearized by cutting with the restriction enzymes XhoI and PstI and was then used to transform the B. subtilis strain GNB37 [(treA::ery)2 argC+)] (Nau-Wagner et al., 2012). Each of the PargC-treA constructs is followed by a chloramphenicol resistance gene (catR), and the entire cassette is flanked by 5′ and 3′ segments of the amyE amylase gene. This allows the selection of recombinant strains via their chloramphenicol resistance and the identification of chromosomal amyE::PargC-treA-catR::amyE insertions by scoring the AmyE− phenotype on starch plates. To combine the various PargC-treA reporter fusions with an ahrC gene disruption mutation, we used DNA of the linearized reporter plasmids to transform strain DRB47 (ΔahrC::ery treA::kan) by selecting chloramphenicol-resistant colonies and scoring their EmyE−phenotype on starch plates. The resulting strains are listed in Supplementary Table S1. To combine the wild-type PargC-treA transcriptional reporter fusion with various ahrC suppressor alleles (Table 2), we used DNA of linearized plasmid pDST40 (argC-treA) to transform corresponding suppressor strains, again selecting chloramphenicol-resistant colonies. The resulting strains are listed in Supplementary Table S1.
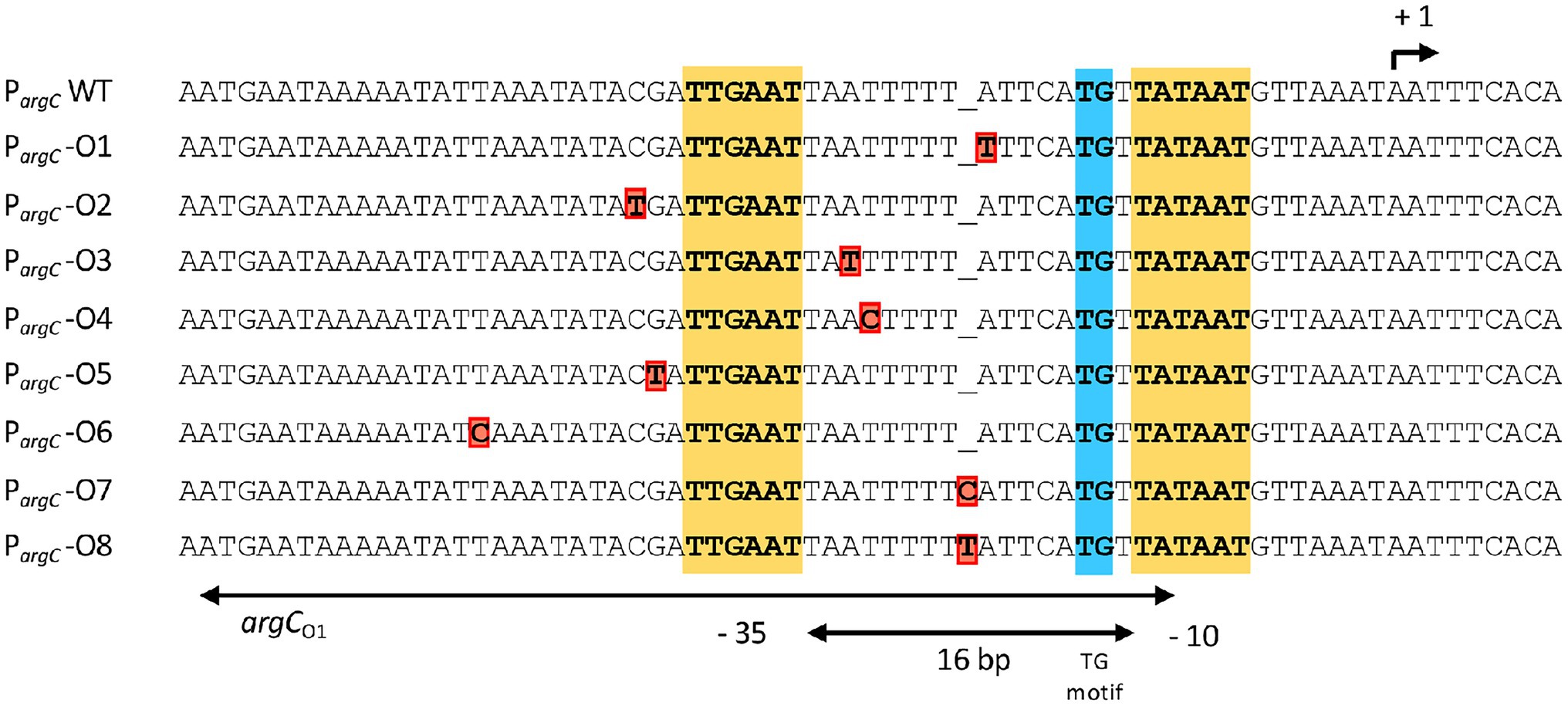
Figure 2. DNA sequence of the argC promoter region and that of the overlapping main operator for the AhrC regulatory protein. AhrC operator mutations derived from the salt-stress resistance suppressor screen are indicated in red. The −35 and of the extended −10 regions (with a TG motif) of the argC promoter are highlighted. The argC promoter possesses a sub-optimal spacing of 16 bp in comparison with typical SigA-type promoters of B. subtilis (Helmann, 1995). The transcriptional start site for the argCJBD-carAB-argF operon is indicated by a bent arrow. The AhrC operator(s) and its interactions with the AhrC regulatory protein, which is a homohexamer, have been defined through molecular and structural analysis (Garnett et al., 2007a, 2008). Three ArgC operators are present to transcriptionally control argCJBD-carAB-argF expression. One of these is present in the coding region of the argC gene, while the two other operators are positioned such that one of them overlaps the −10 and −35 regions of the SigA-type promoter. The other operator is positioned further upstream, with a 11 bp DNA sequence separating them. Binding of AhrC to the operators overlapping with and juxta positioned to the argC promoter leads to the bending of the DNA (Garnett et al., 2008). The entire regulatory region has been termed argCO1 while the ArgC operator present in argC is referred to as argC02 (Czaplewski et al., 1992).
Promoter activities of PargC-treA transcriptional reporter fusions inserted into the B. subtilis genome at the amyE gene were measured as described before (Schöck et al., 1996; Brill et al., 2011a; Zaprasis et al., 2013b). treA encodes a salt-tolerant phospho-α-(1,1)-glucosidase whose enzyme activity can be quantitated using the chromogenic synthetic substrate α-PNPG (Schöck et al., 1996). Bacillus subtilis strains carrying PargC-treA transcriptional fusions all harbored a gene disruption of the native treA gene (Supplementary Table S1), so that the measured TreA enzyme reporter activity reflects exclusively that of the reporter gene construct. To measure the promoter activities of the various PargC-treA transcriptional fusions, strains were grown in SMM at 37° C (20 ml culture volume in 100-ml Erlenmeyer flasks) in the absence or presence of 20 mM L-arginine (Gardan et al., 1997) until they reached an OD578 of 1.5. 1.8-ml samples were withdrawn from the culture and assayed for TreA enzyme activity as described (Brill et al., 2011a; Zaprasis et al., 2013b).
To identify the suppressor mutations in the evolved B. subtilis proA suppressor strains (Figures 3B,C), genomic DNAs were subjected to DNA sequencing, which was kindly performed by the Göttingen Genomics Laboratory (Göttingen, Germany) on Illumina instruments. The reads were mapped onto the reference genome sequence of the B. subtilis strain JH642 (GenBank accession number CM000489.1; Smith et al., 2014) as previously described (Widderich et al., 2016) using the Geneious software package (Kearse et al., 2012). Single nucleotide polymorphisms were considered as significant when the total coverage depth exceeded 25 reads with a frequency variance of >90%. The genome sequence of the following salt-tolerant suppressor strains was determined: DRB16, DRB17, DRB20, DRB28, and DRB40 (Table 1; Supplementary Table S1).
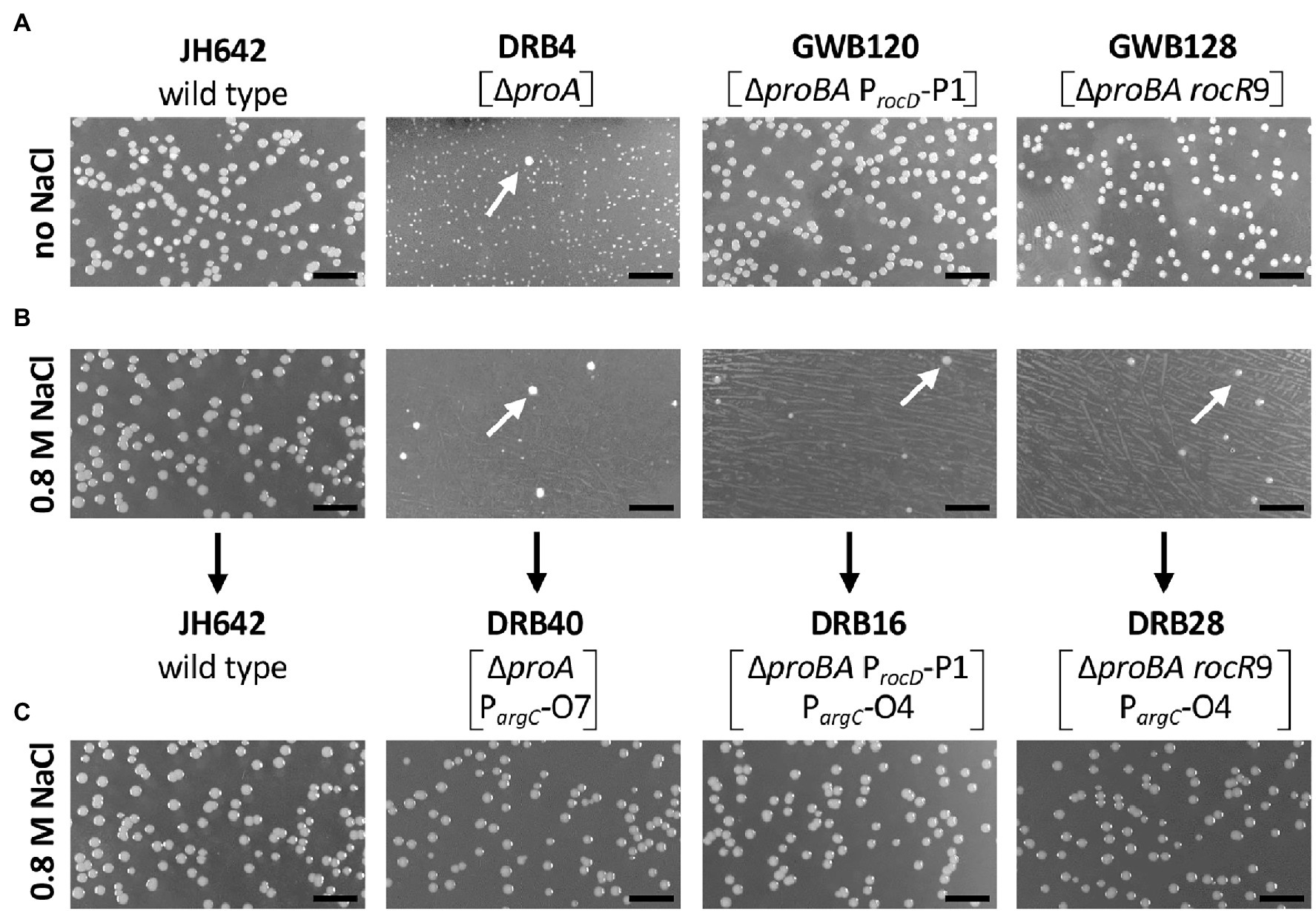
Figure 3. Selection for osmostress resistant suppressor mutants bypassing a defect in the proA-encoded γ-glutamyl phosphate reductase. Equal dilutions of the indicated strains were plated onto either (A) SMM agar plates lacking L-proline or (B) SMM agar plates lacking L-proline but that contained 0.8 M NaCl. The plates were incubated for 72 h at 37° C. (C) Spontaneously arising faster growing suppressor colonies (indicated by the white arrows) were picked from the high-salinity agar plates, purified by streaking single colonies on the same medium, and dilution of liquid cultures were replated onto the high-salinity L-proline-free agar plates. The length bar corresponds to 5 mm. Bacillus subtilis JH642 is the wild-type strain (Smith et al., 2014) from which the first generation suppressor strains DRB4, GWB120 and GWB128 were derived (Zaprasis et al., 2013b). Strain DRB4 and its derivatives contain a deletion of the proA gene but proB is intact. In strains GWB120 and GWB128, the entire proBA operon is deleted but ProB enzyme activity is provided in these strains via the amino acid sequence related ProJ L-glutamate kinase; hence these strains are not L-proline auxotrophs (Belitsky et al., 2001; Brill et al., 2011a; Zaprasis et al., 2013b). Strains GWB120 and GWB128, carry mutations either in the regulatory region of the rocDEF operon, or in the gene for the RocR activator protein; both mutations increase rocDEF expression (Zaprasis et al., 2013b).
In addition to suppressor mutants whose entire genome was re-sequenced, we obtained the DNA sequence of the ahrC gene and of the argC regulatory region of the argCJBD-carAB-argF operon in 22 suppressor stains by amplifying the corresponding DNA segment via PCR (ahrC: 715 bp; argC: 598 bp) and subsequent DNA sequence analysis. The DNA sequence of the DNA primers used to amplify the ahrC gene were: DS23_5’-TGCGCGTTGTAGAAGAAGCA-3′ and DS24_5’-GCCCGCGTTCAAAAGAAACC-3′. Those used for the amplification of the argC regulatory region were DS25_5’-CCATTATGCTCGGGGGCTTT-3′ and DS26_5’-AACCGTAATTCCCGCGTCTG-3′. DNA sequencing was carried out by Eurofins MWG Operon (Ebersberg, Germany).
To detect and quantify metabolites of the L-proline and L-arginine biosynthetic pathways, we used targeted metabolomics. For these experiments, two independently prepared cultures of the B. subtilis strain JH642 and its mutant derivatives were grown in SMM or SMM containing 1.2 M NaCl. Cells were cultured in shake flasks (20 ml culture volume in 100-ml Erlenmyer flasks) to early-exponential growth phase (OD578 of about 1.5). 1 ml culture aliquots were withdrawn from the cultures and the cells were rapidly vacuum-filtered onto a 0.45 μm pore size filter (HVLP02500, Merck Millipore). The filters were immediately transferred into a acetonitrile/methanol/water (40:40:20) extraction solution at −20°C. The filters were incubated in the extraction solution for 30 min. Subsequently, the metabolite extracts were centrifuged for 15 min at 13,000 rpm at −9° C and the supernatant was stored at −80° C until analysis.
Metabolite extracts were mixed with a 13C-labeled internal standard in a 1:1 ratio. LC-MS/MS analysis was performed with an Agilent 6495 triple quadrupole mass spectrometer (Agilent Technologies) as described previously (Guder et al., 2017). An Agilent 1290 Infinity II UHPLC system (Agilent Technologies) was used for liquid chromatography. The temperature of the column oven was 30°C, and the injection volume was 3 μl. LC solvents in channel A were either water with 10 mM ammonium formate and 0.1% formic acid (v/v; for acidic conditions), or water with 10 mM ammonium carbonate and 0.2% ammonium hydroxide (for basic conditions). LC solvents in channel B were either acetonitrile with 0.1% formic acid (v/v; for acidic conditions) or acetonitrile without additive (for basic conditions). LC columns were an Acquity BEH Amide (30 × 2.1 mm, 1.7 μm) for acidic conditions, and an iHILIC-Fusion(P; 50 × 2.1 mm, 5 μm) for basic conditions. The gradient for basic and acidic conditions was: 0 min 90% B; 1.3 min 40% B; 1.5 min 40% B; 1.7 min 90% B; 2 min 90% B. The ratio of 12C and 13C peak heights was used to quantify metabolites and absolute concentrations were determined by calibrating the 13C standard with authentic standards. Intracellular concentrations of metabolites were calculated using an intracellular volume of 0.65 μl per 1 ml B. subtilis culture grown to an OD578nm = 1. The B. subtilis cell volume was estimated from previously reported values for the internal and total water spaces that were determined by measuring the distribution of membrane-permeable 3H2O and membrane-impermeable inulin-[14C]carboxylic acid (Bakker and Mangerich, 1981; Holtmann et al., 2003; Hoffmann et al., 2013). The cellular extracts of two biological replicates were assessed and each of them was measured in at least two technical parallels.
Statistical assessment of values obtained for assays of the TreA reporter enzyme (Figures 4B–D) or individual compounds detected and quantitated in the course of targeted metabolomics (Figure 5; Supplementary Table S4), was carried out with the “unpaired t test” analysis tool as implemented in the Prism 9 software suite (GraphPad Software, San Diego, CA, United States; see Supplementary Tables S3, S5).1
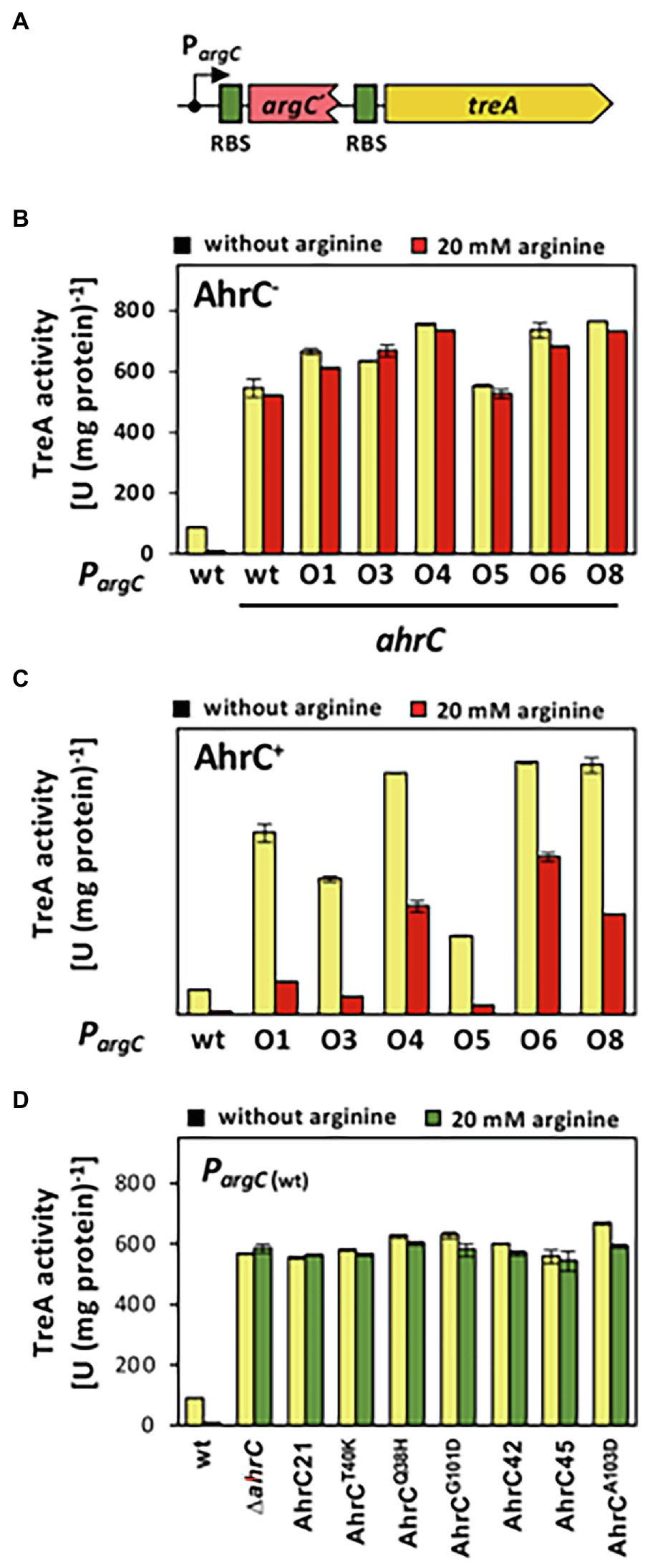
Figure 4. Reporter gene assays assessing the influence of argC operator or ahrC regulator mutations on argC promoter activity. (A) Scheme of a PargC-treA transcriptional reporter fusion. The treA gene encodes a salt-tolerant a phospho-α-(1,1)-glucosidase (Schöck et al., 1996). In the reporter fusion construct, treA lacks its own promoter and its transcription is thus dependent on the promoter activity mediating the expression of the argCJBD-carAB-argF operon. treA possesses its own ribosome binding site (RBS; green box). (B, C) The PargC-treA transcriptional fusion was stably integrated via a double recombination event as a single copy into the chromosomal amyE gene of an ahrC mutant (B), or into the chromosome of the ahrC+ FIGURE 4 | B. subtilis wild-type strain JH642 (C). Mutant derivatives of the PargC-treA reporter fusion strains carrying various mutations in the AhrC operator (O1 to O8) in the argC-treA construct were similarly constructed. (D) The wild-type PargC-treA transcriptional fusion was inserted either into the chromosome of the wild-type strain JH642, or into derivatives of this strain carrying various ahrC mutant alleles. For details on the types of argC operator mutants see Figure 2 and for the description the ahrC mutant alleles see Table 3. All strains carry a disruption of the chromosomal treA gene so that the measured TreA enzyme activities reflect solely that encoded by the PargC-treA reporter fusion. The shown data represent experiments from two biological replicates and each sample was assayed twice. SDs for TreA reporter enzyme activity are indicated by bars and the statistical significands of the reported values are listed in Supplementary Table S3.
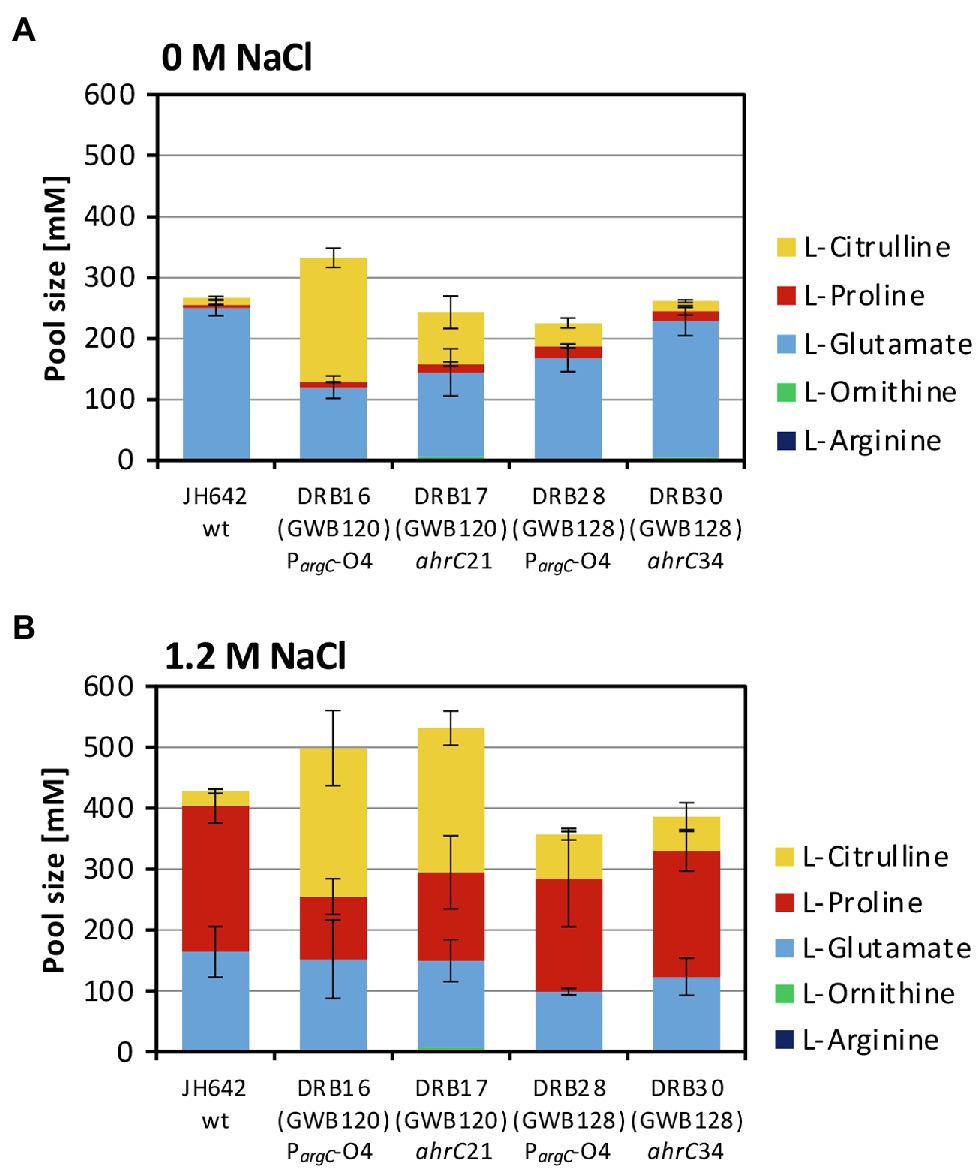
Figure 5. Targeted metabolome analysis of the B. subtilis wild-type strain and selected osmostress tolerant suppressor derivatives. Metabolites from the B. subtilis wild-type (WT) strain JH642 and of four osmostress tolerant suppressor derivatives were analyzed by focusing on L-proline and on key metabolites of the L-arginine synthesis and degradation intermediates. The corresponding metabolites were assessed and quantitated form cells that had been grown in SMM to early exponential growth phase (OD578 of about 1.5) containing either no (A) additional NaCl or (B) 1.2 M NaCl. The values given are averages and SDs of two biological replicates and each of the cellular extracts was measured in at least two technical parallels’. The precise values of the measured metabolites are documented in Supplementary Table S4. SDs for the measurements of the indicated metabolites are indicated by bars and the statistical significands of the reported values are listed in Supplementary Table S5.
Faster growing Pro+ suppressor strains arising spontaneously in a proA mutant fall genetically into two classes (Zaprasis et al., 2013b). In one class [represented by strain GWB120 (ΔproBA ProcD-P1); Figure 3A], point mutations activate a cryptic SigA-type promoter present in front of the rocDEF operon (Zaprasis et al., 2013b), a gene cluster whose transcription is otherwise dependent on the alternative Sig-54-type sigma factor SigL and the NtrC/NifA-type activator protein RocR (Debarbouille et al., 1991; Calogero et al., 1994; Gardan et al., 1995, 1997). In the second class of suppressor mutants (represented by strain GWB128 (ΔproBA rocR-9; Figure 3A), single amino acid substitutions in RocR result in partial effector-independent variants of this activator protein (Zaprasis et al., 2013b). Hence, enhanced rocDEF expression occurs even when the RocR effector molecules L-arginine, L-ornithine, or L-citrulline are not added to the growth media (Calogero et al., 1994; Gardan et al., 1995, 1997; Ali et al., 2003).
While the above described suppressor mutants allow wild-type level growth in an L-proline-free minimal medium (SMM; Figure 3A), none of them was able to produce L-proline pools sufficiently large to confer osmostress resistance (Zaprasis et al., 2013b). This is seen when the first generation of the suppressor strains are plated on SMM agar plates containing 0.8 M NaCl, conditions under which the wild-type B. subtilis strain JH642 can readily grow, while the suppressor strains cannot (Figure 3B). However, we observed that faster growing colonies spontaneously arose from the background lawn of strains DRB4 (ΔproA), GWB120 (ΔproBA ProcD-P1) and GWB128 (ΔproBA rocR-9; Table 1) when these strains were plated on a L-proline-free minimal medium with increased salinity (0.8 M NaCl; Figure 3B). Isolation and re-plating of the suppressor strains on high salinity SMM agar plates containing 0.8 M NaCl yielded colonies with a growth behavior and visual appearance resembling that of the B. subtilis wild-type strain JH642 (Figure 3C). Collectively, these observations indicated that the second generation of suppressor strains had gained the ability to withstand high salinity-incurred cellular stress, despite the fact that its natural biosynthetic route (ProJ-ProA-ProH; Brill et al., 2011a) for the osmostress protectant L-proline, the only compatible solute that B. subtilis can synthesize de novo (Whatmore et al., 1990; Brill et al., 2011a), was not intact due to the disruption of the proA gene (Figure 1A).
We chose four independently isolated suppressor strains with increased salt tolerance that were derived from strain DRB4 (ΔproA), 12 suppressor colonies derived from strain GWB120 (ΔproBA ProcD-P1) and 11 suppressor strains derived from strain GWB128 (ΔproBA rocR-9) for further analysis. The genome sequence of the B. subtilis laboratory strain JH642, a derivative of strain 168, is known (Smith et al., 2014). We therefore resorted to whole genome re-sequencing of the JH642-derived suppressor strains to reveal possible leads for the type(s) of mutations giving rise to increased salt stress resistance. To this end, we sequenced the genomes of five randomly picked strains from our collection of 27 suppressor isolates. Four strains carried single base-pair substitutions in the high-affinity operator sequence for the AhrC repressor protein overlapping the −10 and −35 promoter sequence of the argCJBD-carAB-argF arginine biosynthetic operon of B. subtilis (Smith et al., 1989; Czaplewski et al., 1992; Miller et al., 1997; Garnett et al., 2008). One strain carried a mutation in the ahrC regulatory gene (Dennis et al., 2002; Garnett et al., 2007a,b, 2008; Tables 2, 3). Mutations targeting the AhrC operator sequence (Parg-O1, Parg-O4, Parg-O7; Figure 2) were found among the suppressors derived from each of the three B. subtilis strains used for the original genetic selection scheme. The same point mutation (Parg-O4; Figure 2) was even present in suppressors derived either from strain GWB120 or from strain GWB128 (Table 3). As expected, the chromosome of the five suppressor strains chosen for genome re-sequencing also contained the mutations that were originally present in the parental strains. No other mutations outside of the AhrC operator or of the ahrC gene were found.
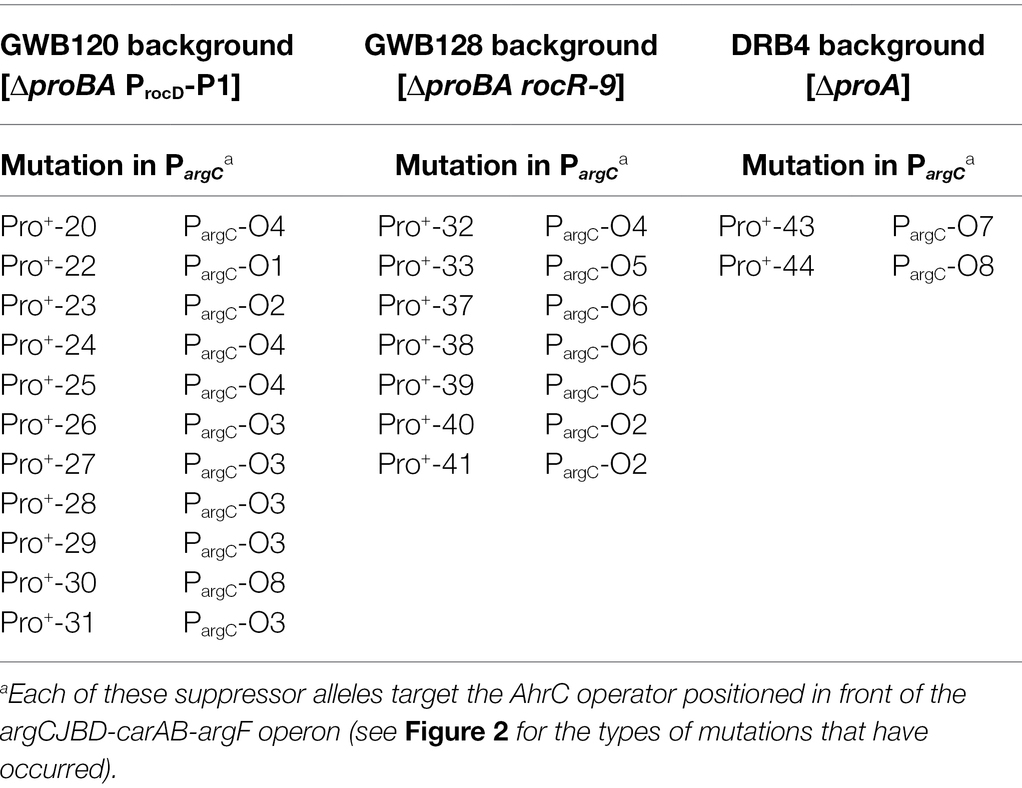
Table 3. Suppressor mutations targeting the AhrC operator of the argCJBD-carAB-argF L-ornithine/L-citrulline/L-arginine biosynthetic operon.
While the number of suppressor strains subjected to whole genome re-sequencing is restricted, our data provided a consistent picture about the molecular underpinnings for the salt-stress-resistant phenotype of the suppressor strains. In each of them, the same genetic process was targeted; namely the genetic control of the biosynthetic route for L-arginine and for its main intermediates, the non-proteinogenic amino acids L-ornithine and L-citrulline (Figure 1A). To corroborate this conclusion further, we amplified by PCR the regulatory region of the argCJBD-carAB-argF operon and of the ahrC regulatory gene from the remaining 22 strains of our suppressor collection. In each case, we found a point mutation in either of these DNA regions. Seventeen strains carried mutations in the AhrC operator sequence positioned in front of the argCJBD-carAB-argF gene cluster (Miller et al., 1997; Garnett et al., 2008; Table 3). This increased the total number of different mutations in the AhrC operator sequence up to eight (Figure 2). Additional six mutations occurred in the ahrC gene. Among the seven ahrC variants that we recovered in our genetic suppressor screen, four single amino acid substitution mutations were found, and three mutations led to truncated AhrC proteins caused by frame-shift mutations (Table 2).
The B. subtilis AhrC regulatory protein serves as a repressor for the L-arginine biosynthetic gene clusters argCJBD-carAB-argF and argGH (Czaplewski et al., 1992; Figures 1A,B). However, it also functions as a transcriptional activator for the L-arginine catabolic operons rocABC and rocDEF (Gardan et al., 1995; Klingel et al., 1995; Figure 1B). The B. subtilis AhrC protein comprises 149 amino acids and the crystal structure of this hexameric protein (Figure 6) is known (Dennis et al., 2002). The monomer adopts a two-domain structural organization comprising an N-terminal DNA-reading head that contains a winged helix-turn-helix DNA binding motif, while the C-terminal domain mediates oligomerization into a hexamer and binding of the corepressor molecule L-arginine (Figure 6; Garnett et al., 2007a,b).
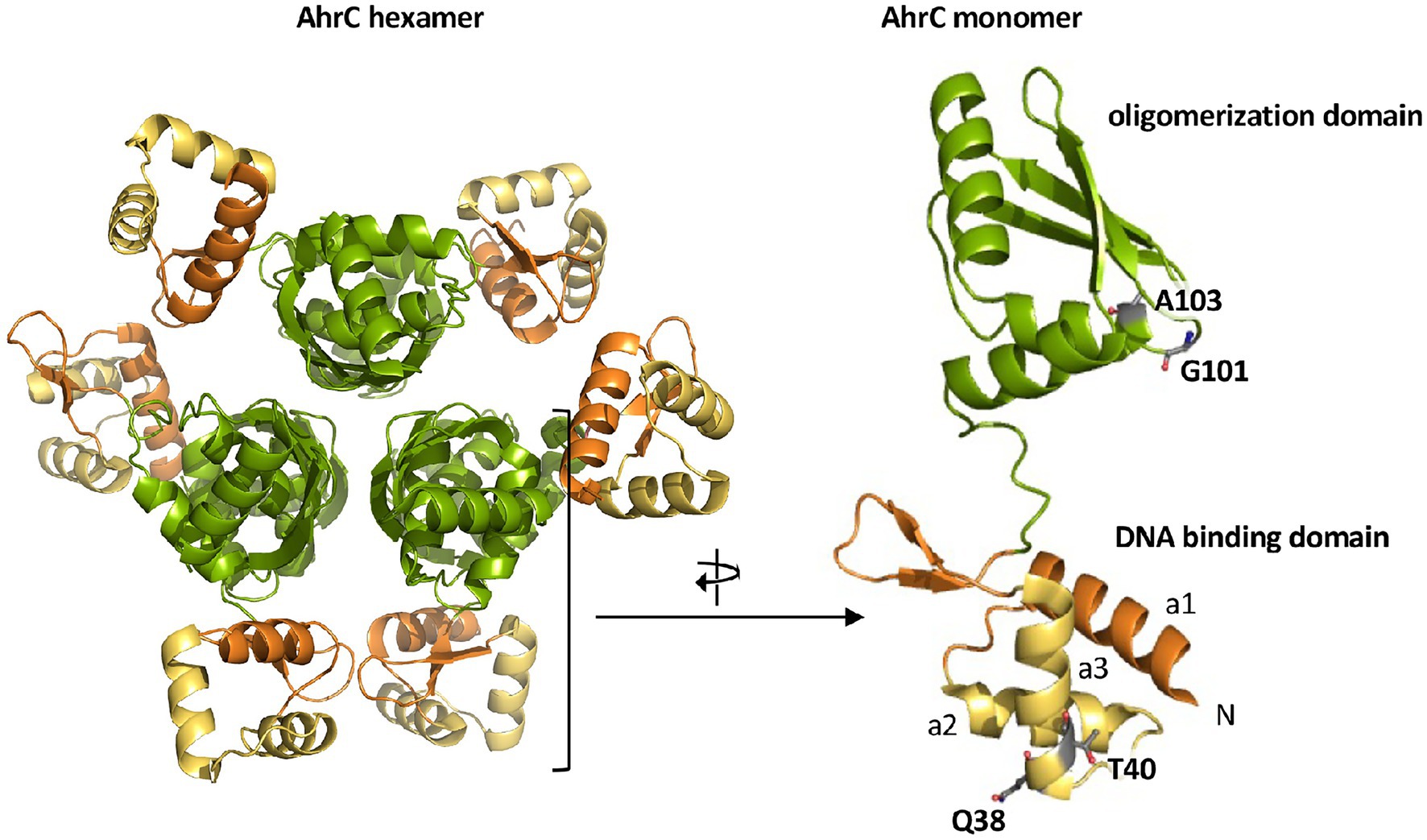
Figure 6. Crystal structure of the AhrC regulatory protein. The B. subtilis AhrC protein is a homo-hexamer and its monomer possesses two functionally distinct domains that are connected by a flexible linker region (Dennis et al., 2002). The N-terminal domain is involved in DNA-binding and contains a winged helix-turn-helix DNA-binding motif (yellow); the C-terminal part represents the effector (L-arginine) and multimerization domain (green; Garnett et al., 2007a,b). The representation of the AhrC hexamer and monomer was rendered using PyMol (Delano, 2002) and by using structural information deposited in the RCSB Protein Data Bank (pdb code: 1F9N). The positions of four single amino acid substitutions in AhrC leading to an osmostress-tolerant growth phenotype (Table 3) are indicated.
Four of the ahrC suppressor alleles that we recovered encode AhrC variants containing single amino acid substitutions (Table 2). Two of these (Q38/A and T40/K) are present in the winged helix-turn-helix-DNA binding motif of AhrC, while the second pair (G101/D and A103/D) are located in the oligomerization and L-arginine effector-binding domain (Figure 6; Czaplewski et al., 1992; Garnett et al., 2007b, 2008).
So far, we have assumed that the argC operator mutations (Figure 2) will lead to enhanced expression of the argCJBD-carAB-argF arginine biosynthetic operon (Smith et al., 1989; Czaplewski et al., 1992; Garnett et al., 2008). To show this directly, we constructed a set of transcriptional reporter fusions by linking the regulatory region of the argCJBD-carAB-argF gene cluster to a promoter-less treA reporter gene, which encodes a salt-tolerant phospho-α-(1,1)-glucosidase (Schöck et al., 1996). We stably inserted these PargC-treA transcriptional reporter gene constructs (Figure 4A) in single copy into the chromosomal non-essential amyE gene of B. subtilis through a double-homologous recombination event. We then measured TreA reporter enzyme activity in cells of these strains grown in SMM with glucose as the carbon source either in the absence or the presence of 20 mM L-arginine (Gardan et al., 1997). L-arginine serves as an AhrC-dependent co-repressor for argCJBD-carAB-argF transcription (Garnett et al., 2007b). The presence of increased L-arginine pools signals the B. subtilis cell that only low transcriptional levels of the corresponding biosynthetic operon are required to sustain adequate protein biosynthesis and hence growth (Czaplewski et al., 1992; Garnett et al., 2008). Several L-arginine import systems are present in B. subtilis (Figure 1A) so that changes in the transcription of the argCJBD-carAB-argF operon can be triggered by adding L-arginine to the growth medium.
When the wild-type PargC-treA transcriptional reporter construct was inserted into a B. subtilis strain proficient in AhrC, the reporter fusion was expressed at moderate levels, and the presence of L-arginine in the growth medium largely repressed promoter activity (Figure 4A). Conversely, transcription of the same fusion was about 50-fold de-repressed when AhrC was absent. Furthermore, the transcriptional reporter fusion was no longer responsive to the presence of L-arginine in the growth medium (Figures 4B,C). This transcriptional profile of the argC-treA reporter fusion reflects the pattern reported in previous studies on the transcriptional control of the argCJBD-carAB-argF operon (Smith et al., 1989; Czaplewski et al., 1992; Garnett et al., 2008).
The transcriptional profile of the six studied PargC-treA reporter fusions carrying argC operator mutations (Figure 2) was strikingly different from that of the wild-type strain. In the absence of AhrC, all strains expressed the reporter fusion constitutively at a level resembling that of the wild-type reporter fusion (Figure 4B). However, when AhrC was present, all reporter fusions carrying mutant argC operators were expressed at a much higher level, between 54-fold and 60-fold, than the wild-type fusion in the absence of L-arginine (Figure 4C), indicating that the binding of the AhrC regulatory protein to the mutant operators was probably reduced. However, AhrC-responsiveness of the argC operator variants was not completely lost as the promoter activity was still reduced by the presence of L-arginine in the medium, albeit to various degrees (Figure 4C). Collectively, the data obtained with the PargC-treA transcriptional reporter fusion show that the argCJBD-carAB-argF operon is expressed at higher levels in each of the suppressor strains carrying the operator mutations compared with the wild-type strain that possesses an intact ahrC gene. This is true for both the absence and for the presence of AhrC effector molecule L-arginine in the growth medium (Figures 4B,C).
To study the influence of the seven recovered suppressor mutations located in the ahrC regulatory gene (Table 2) on the expression of the argCJBD-carAB-argF operon, we introduced a wild-type PargC-treA reporter fusion into both a ΔahrC strain and into each of the seven suppressor strains possessing gene variants of ahrC. In each of the strains carrying the ahrC suppressor alleles, the PargC-treA transcriptional reporter fusion was expressed at high levels in an L-arginine non-responsive manner (Figure 4D). Consequently, all AhrC variants obtained in our suppressor screen (Table 2) are non-functional, and hence, the defects in this regulatory protein should influence L-arginine metabolism of B. subtilis.
After clarification of the molecular underpinnings of the types of mutations present in the collection of 27 suppressor strains isolated on SMM plates with 0.8 M NaCl (Tables 2 and 3), we chose six representatives for further physiological studies. In these strains, the original SigA promoter-up mutation for the rocDEF operon, or the original partial effector independent mutation in rocR (Zaprasis et al., 2013b) were combined with argC operator variants or mutations in ahrC (see Table 1 for the relevant genotypes of the strains; Tables 2 and 3). These six strains were grown in SMM containing 1.2 M NaCl, a severe osmotic challenge for B. subtilis (Boch et al., 1994). Their growth pattern was compared to that of the B. subtilis wild-type strain JH642, strain JSB8 (ΔproHJ) carrying a defect in the osmostress adaptive L-proline biosynthesis route (Brill et al., 2011a), and that of the three parent strains (DRB4 (ΔproA), GWB120 (ΔproBA ProcD-P1) and GWB128 (ΔproBA rocR-9; Table 1) used for the suppressor selection (Figures 3B,C). Under the imposed high salinity conditions through the presence of 1.2 M NaCl in the medium, the strain with a defective osmostress adaptive L-proline biosynthesis system (strain JSB8; ΔproHJ; Brill et al., 2011a), and the parent strains used for the selection of osmostress resistant suppressors barely grew. However, growth was noticeably improved for each of the six spontaneous suppressor strains newly isolated in this study (Figures 7A–C).
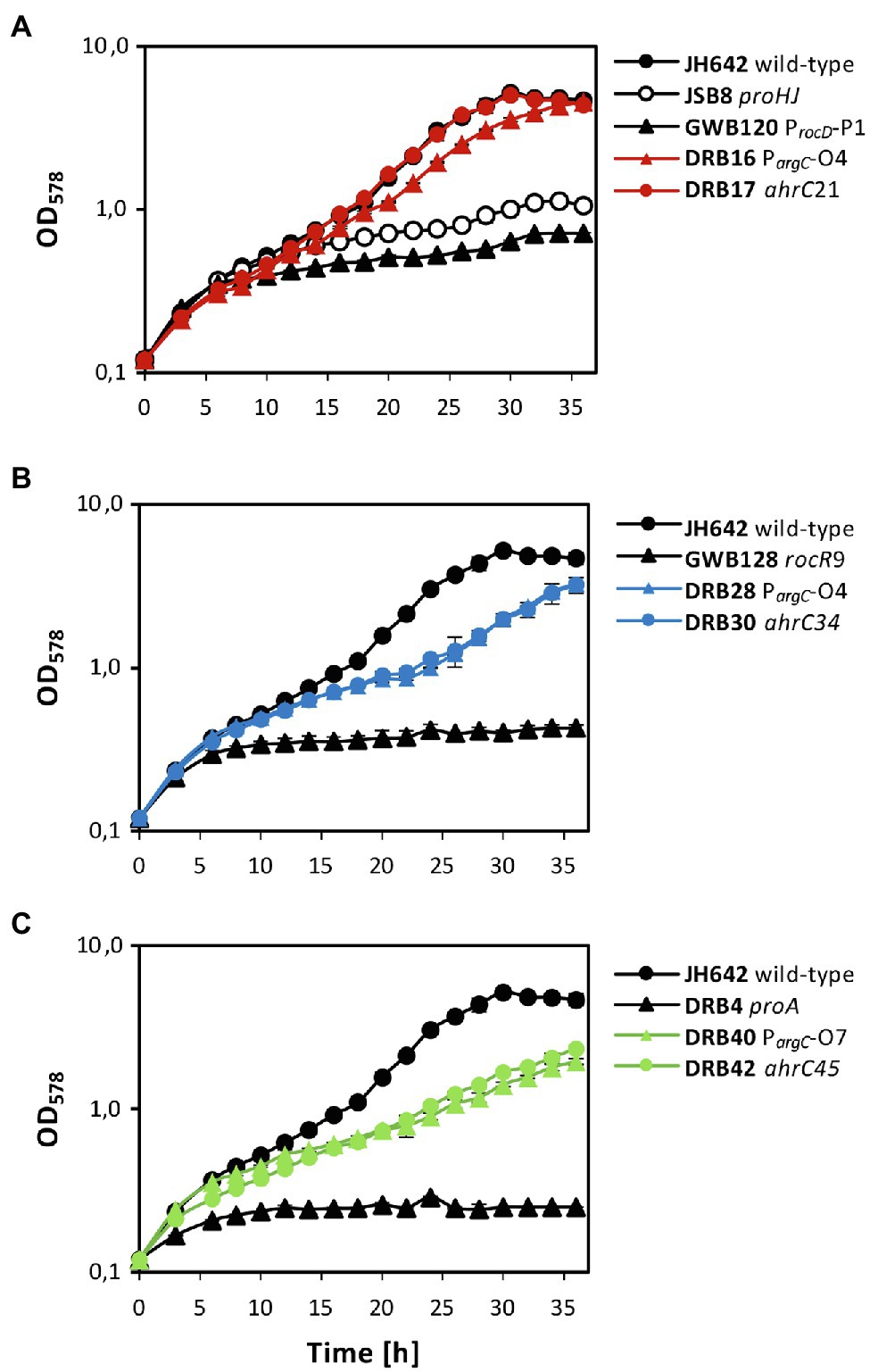
Figure 7. Osmostress-resistant growth phenotype of various proA suppressor mutants. The cells were grown in shake-flasks at 37° C in SMM containing 1.2 M NaCl. For comparison of the various suppressor strains, the growth pattern of the B. subtilis JH642 wild-type strain and that of a strain (JSB8) defective in the osmostress-adaptive L-proline biosynthetic route (ΔproHJ; Brill et al., 2011a) is shown. To aid clarity of the presentation, the same growth data documented for the wild-type strain JH642 in (A), were re-used in panels (B,C). In each of the panels, growth of the starting strains used for the suppressor selection (GBW120, GWB128, DRB4) and two of their suppressor derivatives is shown. The shown growth curves reflect a representative growth experiment performed with two biological replicates. These experiments were repeated in an independently conducted experiment, which yielded the same results.
Both suppressor derivatives of strain GBW120 (ΔproBA ProcD-P1) were able to cope effectively with the high salinity growth conditions. The strain carrying a defective ahrC allele grew like the wild-type strain with growth rates of 0.125 h−1 (wild-type strain JH642) and 0.124 h−1 (suppressor strain DRB17), respectively. Likewise, the strain possessing the arg-O4 operator mutation (strain DRB16) was almost as osmotolerant as the wild-type strain JH642 and exhibited a growth rate of 0.110 h−1 (Figure 7A). Growth was also significantly improved in the two strains derived from strain GWB128 (ΔproBA rocR-9) that carried either an additional argC operator mutation (strain DRB28), or a mutation disrupting the function of the ahrC regulatory gene (strain DRB30). Yet, the growth rates of these two suppressor strains (DRB28: 0.084 h−1; DRB30: 0.081 h−1) were reduced in comparison to that exhibited by the wild-type strain JH642 (0.125 h−1; Figure 7B). As expected from their phenotype on minimal agar plates containing 0.8 M salt used originally for the suppressor section (Figures 3A,B), the DRB4 (ΔproA) derived strains DRB40 (carrying a argC operator mutation) and DRB42 (carrying a ahrC gene disruption mutation) also showed notably improved growth in comparison with their parent strain. However, the improvement of growth at a salinity of 1.2 M NaCl in liquid cultures was only moderate in comparison with that of the wild-type strain JH642 (Figure 7C). Hence, the suppressor strains in which the RocD-mediated flow of γ-glutamate-semialdehyde/Δ1-pyrroline-5-carboxylate into the L-proline biosynthetic route (Figure 1A) was enhanced through pre-existing mutations increasing rocDEF expression (Zaprasis et al., 2013b) fared best under osmotically very challenging growth conditions (Boch et al., 1994; Figure 7).
We examined the metabolic changes that resulted from the suppressor mutations in the ahrC gene or in the argC operator. For this analysis we focused on the L-proline and L-arginine biosynthetic routes from their common precursor L-glutamate (Figure 1A). We analyzed the metabolites of suppressor strain DRB16, DRB17, DRB28 and DRB30 (Table 1), as these four suppressor strains showed a considerable improvement of growth at high salinity in comparison with their corresponding parent strains (Figures 7A,B).
The size of the steady-state pools of the precursor L-glutamate, key intermediates in L-arginine biosynthesis (L-ornithine and L-citrulline), and of the products L-arginine and L-proline in the absence and presence of salt stress are summarized in Figures 5A,B and in Supplementary Table S3. Consistent with previous studies (Whatmore et al., 1990; Brill et al., 2011a), the pool size of the compatible solute L-proline was about 48-fold increased (from 5 to 239 mM), while that of the L-proline biosynthetic precursor L-glutamate was concomitantly noticeably drained by about 35% (from 249 to 162 mM) when the B. subtilis wild-type cells were subjected to a severe and persistent salt stress (Figures 5A,B; Supplementary Table S3). The pools of the intermediates in L-arginine synthesis, L-ornithine and L-citrulline (Figure 1A) were only marginally affected in the wild-type strain JH642 by an increase in salinity. With pool sizes about 2 mM (L-ornithine) and 24 mM (L-citrulline), they were rather moderate in comparison with those of L-glutamate (126 mM) and L-proline (239 mM) pools found in the salt-stressed cells (Figures 5A,B; Supplementary Table S4).
The metabolic profile of the four studied suppressor strains differed from that of the salt-stressed B. subtilis wild-type strain JH642. Like the wild-type strain, each of these strains contained a substantial L-proline pool, but the most noticeable difference was an increase in the L-citrulline content of the cells. The L-citrulline pool (24 mM) was increased only by about twofold to threefold in the suppressor strains (DRB28 and DRB30) derived from strain GWB128 (ΔproBA rocR-9; Figure 3), but it was enhanced by about tenfold in those strains (DRB16 and DRB17) derived from strain GWB120 (ΔproBA ProcD-P1; Figures 5A,B; Supplementary Table S4). Each of the suppressor strains also contained substantial pools of L-glutamate, while the cellular content of L-ornithine or L-arginine were very low and comparable to that of the wild-type strain JH642 grown under persistent high salinity conditions (Figures 5A,B; Supplementary Table S4). γ-glutamate-semialdehyde, the reaction product formed either by the ProA or RocD enzymes (Figure 1A), was not detected in our analysis of the metabolome, probably because it is unstable. Large cellular pools of this metabolite are also not expected due to the high chemical reactivity and ensuing toxicity of this aldehyde.
Bacterial genome evolution is highly dynamic and the ability of microorganisms to finely-tune their metabolism through mutations to circumvent cellular or environmental constraints is one of the reasons for their ecological success (Barrick and Lenski, 2013). Here, we describe a series of spontaneous osmostress-tolerant suppressor strains of B. subtilis that overcame a proA-dependent genetic block of the osmostress-adaptive biosynthetic route (ProJ-ProA-ProH) for the compatible solute L-proline (Figures 1A, 3, 7). L-proline is the only compatible solute that B. subtilis can synthesize de novo (Whatmore et al., 1990), and disruption of its osmostress adaptive synthesis route causes osmotic sensitivity (Brill et al., 2011a; Hoffmann and Bremer, 2016). The suppressor strains isolated in this study achieve osmostress protection through a combination of point mutations in two different regulatory sequences, either for the argCJBD-carAB-argF or for the rocDEF operon, and in two different regulatory genes, either in rocR or in ahrC. Pairs of mutations thus upregulate the transcription of the genes for L-arginine biosynthesis and the degradation of L-ornithine, an intermediate in L-arginine synthesis, to synthesize enhanced amounts of γ-glutamate-semialdehyde/Δ1-pyrroline-5-carboxylate. These RocD-produced metabolites are also the products of the γ-glutamyl phosphate reductase ProA (Belitsky et al., 2001), thereby providing a metabolic by-pass for the loss of the ProA enzyme (Figure 1A).
To overcome the salt-sensitive growth phenotype of a B. subtilis proA mutant (Figure 7), the opening of the RocD-dependent metabolic shunt between L-ornithine catabolism and the conversion of the generated γ-glutamate-semialdehyde/Δ1-pyrroline-5-carboxylate by Δ1-pyrroline-5-carboxylate reductases is a pre-requisite to efficiently produce L-proline (Figures 5A,B; Supplementary Table S4). The suppressor strains thus make use of a naturally existing, but weakly effective (Zaprasis et al., 2013b), by-pass route interconnecting L-arginine and L-proline synthesis in B. subtilis (Figure 1A). The combined regulatory mutations in either of the argCJBD-carAB-argF/rocDEF operons and in either of the rocR/ahrC activator/repressor genes make this metabolic by-pass route more efficient, thereby resulting in the enhanced production of L-proline (Figures 5A,B; Supplementary Table S4). As B. subtilis repurposes this route physiologically to attain osmostress tolerance (Figures 7A,B), one can view the corresponding metabolic shunt as a form of underground metabolism (Rosenberg and Commichau, 2019). However, in contrast to a traditional view on underground metabolism (Copley, 2020; Cotton et al., 2020; Tawfik, 2020), no enzyme activities had to be evolved to produce substantial amounts of the osmostress protectant L-proline (Figures 5A,B; Supplementary Table S4) via the L-arginine—L-ornithine—γ-glutamate-semialdehyde/Δ1-pyrroline-5-carboxylate metabolic shunt.
In studies addressing the cellular adjustment of B. subtilis to potassium limitation (0.5 mM K+ present in the growth medium), suppressor mutants similar to those reported here were recovered (Gundlach et al., 2017a). Two mutations occurred in the AhrC operator of the argCJBD-carAB-argF operon (one of which is actually identical to suppressor PargC-O8; Figure 2), and one mutation was found that causes an amino acid substitution (Q22/R) in the AhrC regulatory protein (Gundlach et al., 2017a). The authors of this study concluded that increased pools of the positively charged amino acids L-ornithine, L-citrulline, and L-arginine resulting from these mutations might functionally substitute, at least to some extent, for the crucial role played by potassium for the general physiology of microbial cells (Danchin and Nikel, 2019; Korolev, 2021).
The size of potassium pools plays an important role both during the initial and the sustained cellular adjustment of B. subtilis to high osmolarity surroundings (Whatmore et al., 1990; Whatmore and Reed, 1990; Holtmann et al., 2003; Hoffmann and Bremer, 2016). However, we consider it unlikely that the physiological consequences of the suppressors that we isolated in the ahrC operator of the argCJBD-carAB-argF operon and in the ahrC gene are somehow related to potassium limitations of osmotically stressed cells. The minimal medium used in our study contains 205 mM potassium (Harwood and Archibald, 1990). Furthermore, the isolated suppressor strains are derived from the B. subtilis laboratory strain JH642 (Smith et al., 2014), which is proficient in both the high- and the low-affinity potassium import systems KtrAB and KtrCD, respectively (Holtmann et al., 2003; Gundlach et al., 2017b).
The four suppressor strains that we studied in greater detail, both with respect to their growth under high-salinity conditions (Figures 7A,B) and their metabolome, contained substantial amounts of L-proline, as observed for the B. subtilis wild-type strain (Figures 5A,B; Supplementary Table S4), despite the fact that their natural osmostress-responsive L-proline biosynthetic route is not intact (Figure 1A). The amassing of L-proline could have been guessed from the way the suppressor screen was set-up (Brill et al., 2011a; Zaprasis et al., 2013b). However, the suppressor mutants also contained, unexpectantly, substantial amounts of L-citrulline (Figures 5A,B; Supplementary Table S4). We note, that similar to the suppressors that we have isolated, those found by Gundlach et al. possessed only moderately increased pools of L-ornithine and L-arginine, while the pool of L-citrulline was substantial increased (Gundlach et al., 2017a).
The suppressor strains derived from strain GWB120 and those derived from strain GWB128 contained different amounts of L-citrulline (Figures 5A,B; Supplementary Table S4). This difference might stem from the fact that the mutation in the AhrC operator of the argCJBD-carAB-argF operon will only affect the expression of this particular gene cluster. In contrast, mutations in the ahrC regulator gene will de-repress the transcription of the argCJBD-carAB-argF and argGH operons as AhrC serves as a repressor for both gene clusters (Figure 1B; Czaplewski et al., 1992). However, mutations in ahrC will simultaneously also affect the level of rocDEF transcription because AhrC functions as an activator for this operon (Gardan et al., 1995; Klingel et al., 1995). Furthermore, the RocR-9 variant (L250/H) used in some of our strains introduces another level of complexity into this regulatory network as L-proline can serve as an effector of the mutant RocR-9 protein, a feature apparently not shared by the wild-type RocR protein (Zaprasis et al., 2013b). Hence, intricacies in the regulatory mechanisms and the ensuing substantial changes in the cellular pool size of potential effector molecules for RocR and AhrC probably provide the backdrop for the different compatible solute pools produced in the various suppressor strains. While the molecular events underpinning the different pools sizes of L-proline and L-citrulline under osmotically challenging conditions are not entirely resolved, our data collectively suggest that the substantial L-citrulline pools present in some of our suppressor strains (Figures 5A,B; Supplementary Table S4) should contribute to the balancing of turgor in B. subtilis (Whatmore et al., 1990; Whatmore and Reed, 1990).
There seems to be a difference in the way L-citrulline contributes to osmostress tolerance of B. subtilis when it is generated intracellularly through a metabolic process as observed in our suppressor strains (Figures 5A,B; Supplementary Table S4), and when it is added to the growth medium of osmotically stressed cells (Zaprasis et al., 2015). In the latter case, L-citrulline is imported and is enzymatically converted to L-proline and no osmostress-protective L-citrulline pools can seemingly be formed as the osmostress-relieving attributes of externally added L-citrulline depend entirely on an intact proHJ operon (Zaprasis et al., 2015). Although not explicitly tested, there were apparently no citrulline pools sufficiently high to confer osmostress tolerance in the absence of the osmostress adaptative L-proline biosynthetic route. Hence, the apparent physiological difference in the use of externally provided and internally synthesized citrulline as an osmostress protectant for B. subtilis warrants further study.
L-citrulline is rarely used in bacteria as a compatible solute (Da Costa et al., 1998; He et al., 2017; Chun et al., 2019) and has not been previously detected as a newly synthesized osmostress protectant in Bacilli (Kuhlmann and Bremer, 2002; Bursy et al., 2007). However, it possesses, like L-proline, chemical chaperone activity (Held et al., 2010; Choudhary et al., 2015; Held and Sadowski, 2016). This raises the question why nature has not chosen L-citrulline as a frequently used compatible solute by microorganisms (Da Costa et al., 1998; Kempf and Bremer, 1998). In contrast to L-proline that possesses a net-neutral charge at physiological pH, L-citrulline is positively charged. Its high-level accumulation might thus disturb the cells attempt to avoid a long-lasting high ionic strength cytoplasm under high osmolarity growth conditions (Wood, 2011; Van den Berg et al., 2017; Bremer and Krämer, 2019). In addition, the solubility of L-citrulline (200 g L−1; =1.14 M) at 20° C in water is much lower than that of L-proline (1,500 g L−1; =13 M), a key physico-chemical determinant for most compatible solutes used by microorganisms (Da Costa et al., 1998; Held et al., 2010; Held and Sadowski, 2016). Nevertheless, we note in this context that enhanced synthesis of L-citrulline occurs under osmotic and drought-stress in various plants where this non-proteogenic amino acid might not only serve as a protectant against water stress but also functions as a hydroxyl radical scavenger (Kawasaki et al., 2000; Akashi et al., 2001; Joshi and Fernie, 2017; Song et al., 2020). Such functions have also been suggested for newly synthesized L-citrulline in the moderate halophilic lactic acid bacterium Tetragenococcus halophilus exposed on a sustained basis to high-salinity environments (He et al., 2017; Chun et al., 2019).
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
EB conceived and supervised this study. DS conducted the physiological and genetic experiments. HL and TH performed metabolomic analysis. FC conducted the analysis of the re-sequenced genomes of various suppressor strains. TH designed all figures. EB, TH, and FC wrote the manuscript. All authors contributed to the article and approved the submitted version.
Financial support for this work was provided to EB by the Deutsche Forschungsgemeinschaft (DFG) through the Collaborative Research Council CRC 987 (SFB 987), the LOEWE Program of the State of Hessen via the SNMIKRO Research Center (University of Marburg; to EB and HL), the Max Planck Society (to HL), and by the DFG (to FC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors greatly appreciate the work of Anja Pöhlein and Rolf Daniel from the Göttingen Genomics Laboratory (Göttingen, Germany) for kindly performing genome resequencing of some of the B. subtilis suppressor mutant strains analyzed in this study. The authors are very grateful to Jutta Gade for expert technical assistance during part of this study and thank Vickie Koogle for her kind help in the language editing of our manuscript. The authors thank Felix Dempwolff (SYNMIKRO; University of Marburg) for critical reading of the manuscript. FC thanks Jörg Stülke (University of Göttingen) for financial support and for providing laboratory space. EB greatly appreciates the kind hospitality and support of his colleagues Anke Becker and Gert Bange (both at SYNMIKRO; University of Marburg).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.908304/full#supplementary-material
Akashi, H., and Gojobori, T. (2002). Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 99, 3695–3700. doi: 10.1073/pnas.062526999
Akashi, K., Miyake, C., and Yokota, A. (2001). Citrulline, a novel compatible solute in drought-tolerant wild watermelon leaves, is an efficient hydroxyl radical scavenger. FEBS Lett. 508, 438–442. doi: 10.1016/S0014-5793(01)03123-4
Ali, N. O., Jeusset, J., Larquet, E., Le Cam, E., Belitsky, B., Sonenshein, A. L., et al. (2003). Specificity of the interaction of RocR with the rocG-rocA intergenic region in Bacillus subtilis. Microbiology 149, 739–750. doi: 10.1099/mic.0.26013-0
Bakker, E. P., and Mangerich, W. E. (1981). Interconversion of components of the bacterial proton motive force by electrogenic potassium transport. J. Bacteriol. 147, 820–826. doi: 10.1128/jb.147.3.820-826.1981
Barrick, J. E., and Lenski, R. E. (2013). Genome dynamics during experimental evolution. Nat. Rev. Genet. 14, 827–839. doi: 10.1038/nrg3564
Baumberg, S., and Harwood, C. R. (1979). Carbon and nitrogen repression of arginine catabolic enzymes in Bacillus subtilis. J. Bacteriol. 137, 189–196. doi: 10.1128/jb.137.1.189-196.1979
Belitsky, B.R. (2002). “Biosynthesis of amino acids of the glutamate and aspartate families, alanine, and polyamines,” in Bacillus subtilis and its Closes Relatives, eds. A. L. Sonenshein, J. A. Hoch, and R. Losick. (Washington, D.C.: ASM Press), 203–231.
Belitsky, B. R., Brill, J., Bremer, E., and Sonenshein, A. L. (2001). Multiple genes for the last step of proline biosynthesis in Bacillus subtilis. J. Bacteriol. 183, 4389–4392. doi: 10.1128/JB.183.14.4389-4392.2001
Berg, C. M., and Rossi, J. J. (1974). Proline excretion and indirect suppression in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 118, 928–934. doi: 10.1128/jb.118.3.928-934.1974
Boch, J., Kempf, B., and Bremer, E. (1994). Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J. Bacteriol. 176, 5364–5371. doi: 10.1128/jb.176.17.5364-5371.1994
Boch, J., Kempf, B., Schmid, R., and Bremer, E. (1996). Synthesis of the osmoprotectant glycine betaine in Bacillus subtilis: characterization of the gbsAB genes. J. Bacteriol. 178, 5121–5129. doi: 10.1128/jb.178.17.5121-5129.1996
Bolen, D. W., and Baskakov, I. V. (2001). The osmophobic effect: natural selection of a thermodynamic force in protein folding. J. Mol. Biol. 310, 955–963. doi: 10.1006/jmbi.2001.4819
Bremer, E., and Krämer, R. (2019). Responses of microorganisms to osmotic stress. Annu. Rev. Microbiol. 73, 313–334. doi: 10.1146/annurev-micro-020518-115504
Brill, J., Hoffmann, T., Bleisteiner, M., and Bremer, E. (2011a). Osmotically controlled synthesis of the compatible solute proline is critical for cellular defense of Bacillus subtilis against high osmolarity. J. Bacteriol. 193, 5335–5346. doi: 10.1128/JB.05490-11
Brill, J., Hoffmann, T., Putzer, H., and Bremer, E. (2011b). T-box-mediated control of the anabolic proline biosynthetic genes of Bacillus subtilis. Microbiology 157, 977–987. doi: 10.1099/mic.0.047357-0
Brown, A. D. (1976). Microbial water stress. Bacteriol. Rev. 40, 803–846. doi: 10.1128/br.40.4.803-846.1976
Bursy, J., Pierik, A. J., Pica, N., and Bremer, E. (2007). Osmotically induced synthesis of the compatible solute hydroxyectoine is mediated by an evolutionarily conserved ectoine hydroxylase. J. Biol. Chem. 282, 31147–31155. doi: 10.1074/jbc.M704023200
Calogero, S., Gardan, R., Glaser, P., Schweizer, J., Rapoport, G., and Debarbouille, M. (1994). RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J. Bacteriol. 176, 1234–1241. doi: 10.1128/jb.176.5.1234-1241.1994
Cayley, S., Lewis, B. A., and Record, M. T. Jr. (1992). Origins of the osmoprotective properties of betaine and proline in Escherichia coli K-12. J. Bacteriol. 174, 1586–1595. doi: 10.1128/jb.174.5.1586-1595.1992
Chattopadhyay, M. K., Kern, R., Mistou, M. Y., Dandekar, A. M., Uratsu, S. L., and Richarme, G. (2004). The chemical chaperone proline relieves the thermosensitivity of a dnaK deletion mutant at 42 degrees C. J. Bacteriol. 186, 8149–8152. doi: 10.1128/JB.186.23.8149-8152.2004
Chen, M., Cao, J., Zheng, C., and Liu, Q. (2006). Directed evolution of an artificial bifunctional enzyme, gamma-glutamyl kinase/gamma-glutamyl phosphate reductase, for improved osmotic tolerance of Escherichia coli transformants. FEMS Microbiol. Lett. 263, 41–47. doi: 10.1111/j.1574-6968.2006.00397.x
Choudhary, S., Kishore, N., and Hosur, R. V. (2015). Inhibition of insulin fibrillation by osmolytes: mechanistic insights. Sci. Rep. 5:17599. doi: 10.1038/srep17599
Chun, B. H., Han, D. M., Kim, K. H., Jeong, S. E., Park, D., and Jeon, C. O. (2019). Genomic and metabolic features of Tetragenococcus halophilus as revealed by pan-genome and transcriptome analyses. Food Microbiol. 83, 36–47. doi: 10.1016/j.fm.2019.04.009
Copley, S. D. (2020). Evolution of new enzymes by gene duplication and divergence. FEBS J. 287, 1262–1283. doi: 10.1111/febs.15299
Cotton, C. A., Bernhardsgrutter, I., He, H., Burgener, S., Schulz, L., Paczia, N., et al. (2020). Underground isoleucine biosynthesis pathways in E. coli. elife 9:e54207. doi: 10.7554/eLife.54207
Csonka, L. N. (1989). Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53, 121–147. doi: 10.1128/mr.53.1.121-147.1989
Csonka, L. N., and Leisinger, T. (2007). “Chapter 34.6.1.4, Biosynthesis of proline” in EcoSal-Escherichia coli and Salmonella: Cellular and Molecular Biology. eds. A. Böck, R. Curtis, J. B. K. Iii, P. D. Karp, F. C. Neidhardt, and T. Nystrom, et al. (Washington, D.C: ASM Press).
Czaplewski, L. G., North, A. K., Smith, M. C., Baumberg, S., and Stockley, P. G. (1992). Purification and initial characterization of AhrC: the regulator of arginine metabolism genes in Bacillus subtilis. Mol. Microbiol. 6, 267–275. doi: 10.1111/j.1365-2958.1992.tb02008.x
Da Costa, M. S., Santos, H., and Galinski, E. A. (1998). An overview of the role and diversity of compatible solutes in Bacteria and Archaea. Adv. Biochem. Eng. Biotechnol. 61, 117–153. doi: 10.1007/BFb0102291
Danchin, A., and Nikel, P. I. (2019). Why nature chose potassium. J. Mol. Evol. 87, 271–288. doi: 10.1007/s00239-019-09915-2
Debarbouille, M., Martin-Verstraete, I., Kunst, F., and Rapoport, G. (1991). The Bacillus subtilis sigL gene encodes an equivalent of sigma 54 from gram-negative bacteria. Proc. Natl. Acad. Sci. U. S. A. 88, 9092–9096. doi: 10.1073/pnas.88.20.9092
Dennis, C. C., Glykos, N. M., Parsons, M. R., and Phillips, S. E. (2002). The structure of AhrC, the arginine repressor/activator protein from Bacillus subtilis. Acta Crystallogr. D Biol. Crystallogr. 58, 421–430. doi: 10.1107/S0907444901021692
Fichman, Y., Gerdes, S. Y., Kovacs, H., Szabados, L., Zilberstein, A., and Csonka, L. N. (2014). Evolution of proline biosynthesis: enzymology, bioinformatics, genetics, and transcriptional regulation. Biol. Rev. Camb. Philos. Soc. 90, 1065–1099. doi: 10.1111/brv.12146
Fischer, S. H., and Debarbouille, M. (2002). “Nitrogen Source Utilization and Its Regulation” in Bacillus subtilis and Its Closes Relatives. eds. A. L. Sonenshein, J. A. Hoch, and R. Losick (Washington, D.C.: ASM Press), 181–191.
Forlani, G., Nocek, B., Chakravarthy, S., and Joachimiak, A. (2017). Functional characterization of four putative delta(1)-pyrroline-5-carboxylate reductases from Bacillus subtilis. Front. Microbiol. 8:1442. doi: 10.3389/fmicb.2017.01442
Fujita, T., Maggio, A., Garcia-Rios, M., Stauffacher, C., Bressan, R. A., and Csonka, L. N. (2003). Identification of regions of the tomato gamma-glutamyl kinase that are involved in allosteric regulation by proline. J. Biol. Chem. 278, 14203–14210. doi: 10.1074/jbc.M212177200
Gardan, R., Rapoport, G., and Debarbouille, M. (1995). Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. J. Mol. Biol. 249, 843–856. doi: 10.1006/jmbi.1995.0342
Gardan, R., Rapoport, G., and Debarbouille, M. (1997). Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis. Mol. Microbiol. 24, 825–837. doi: 10.1046/j.1365-2958.1997.3881754.x
Garnett, J. A., Baumberg, S., Stockley, P. G., and Phillips, S. E. (2007a). A high-resolution structure of the DNA-binding domain of AhrC, the arginine repressor/activator protein from Bacillus subtilis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 914–917. doi: 10.1107/S1744309107048166
Garnett, J. A., Baumberg, S., Stockley, P. G., and Phillips, S. E. (2007b). Structure of the C-terminal effector-binding domain of AhrC bound to its corepressor L-arginine. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 918–921. doi: 10.1107/S1744309107049391
Garnett, J. A., Marincs, F., Baumberg, S., Stockley, P. G., and Phillips, S. E. (2008). Structure and function of the arginine repressor-operator complex from Bacillus subtilis. J. Mol. Biol. 379, 284–298. doi: 10.1016/j.jmb.2008.03.007
Godard, T., Zuhlke, D., Richter, G., Wall, M., Rohde, M., Riedel, K., et al. (2020). Metabolic rearrangements causing elevated proline and polyhydroxybutyrate accumulation during the ssmotic adaptation response of Bacillus megaterium. Front. Bioeng. Biotechnol. 8:47. doi: 10.3389/fbioe.2020.00047
Guder, J. C., Schramm, T., Sander, T., and Link, H. (2017). Time-optimized isotope ratio LC-MS/MS for high-throughput quantification of primary metabolites. Anal. Chem. 89, 1624–1631. doi: 10.1021/acs.analchem.6b03731
Gunde-Cimerman, N., Plemenitas, A., and Oren, A. (2018). Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 42, 353–375. doi: 10.1093/femsre/fuy009
Gundlach, J., Herzberg, C., Hertel, D., Thurmer, A., Daniel, R., Link, H., et al. (2017a). Adaptation of Bacillus subtilis to life at extreme potassium limitation. mBio 8, e00861–e00816. doi: 10.1128/mBio.00861-17
Gundlach, J., Herzberg, C., Kaever, V., Gunka, K., Hoffmann, T., Weiss, M., et al. (2017b). Control of potassium homeostasis is an essential function of the second messenger cyclic di-AMP in Bacillus subtilis. Sci. Signal. 10:eaal 3011. doi: 10.1126/scisignal.aal3011
Harwood, C. R., and Archibald, A. R. (1990). “Growth, maintenance and general techniques,” in Molecular Biological Methods for Bacillus. eds. C. R. Harwood and S. M. Cutting (Chichester: John Wiley & Sons), 1–26.
He, G., Wu, C., Huang, J., and Zhou, R. (2017). Metabolic response of Tetragenococcus halophilus under salt stress. Biotechnol. Bioprocess Eng. 22, 366–375. doi: 10.1007/s12257-017-0015-5
Held, C., Neuhaus, T., and Sadowski, G. (2010). Compatible solutes: thermodynamic properties and biological impact of ectoines and prolines. Biophys. Chem. 152, 28–39. doi: 10.1016/j.bpc.2010.07.003
Held, C., and Sadowski, G. (2016). Compatible solutes: thermodynamic properties relevant for effective protection against osmotic stress. Biophys. Chem. 152, 28–39. doi: 10.1016/j.bpc.2010.07.003
Helmann, J. D. (1995). Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23, 2351–2360. doi: 10.1093/nar/23.13.2351
Hoffmann, T., Bleisteiner, M., Sappa, P. K., Steil, L., Mäder, U., Völker, U., et al. (2017). Synthesis of the compatible solute proline by Bacillus subtilis: point mutations rendering the osmotically controlled proHJ promoter hyperactive. Environ. Microbiol. 19, 3700–3720. doi: 10.1111/1462-2920.13870
Hoffmann, T., and Bremer, E. (2016). “Management of osmotic stress by Bacillus subtilis: genetics and physiology,” in Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria ed. F. J. De Bruijn. (Wiley-Blackwell Publishers), 657–676.
Hoffmann, T., Wensing, A., Brosius, M., Steil, L., Völker, U., and Bremer, E. (2013). Osmotic control of opuA expression in Bacillus subtilis and its modulation in response to intracellular glycine betaine and proline pools. J. Bacteriol. 195, 510–522. doi: 10.1128/JB.01505-12
Holtmann, G., Bakker, E. P., Uozumi, N., and Bremer, E. (2003). KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J. Bacteriol. 185, 1289–1298. doi: 10.1128/JB.185.4.1289-1298.2003
Ignatova, Z., and Gierasch, L. M. (2006). Inhibition of protein aggregation in vitro and in vivo by a natural osmoprotectant. Proc. Natl. Acad. Sci. U. S. A. 103, 13357–13361. doi: 10.1073/pnas.0603772103
Itikawa, H., Baumberg, S., and Vogel, H. J. (1968). Enzymic basis for a genetic suppression: accumulation and deacylation of N-acetylglutamic gamma-semialdehyde in enterobacterial mutants. Biochim. Biophys. Acta 159, 547–550. doi: 10.1016/0005-2744(68)90142-3
Joshi, V., and Fernie, A. R. (2017). Citrulline metabolism in plants. Amino Acids 49, 1543–1559. doi: 10.1007/s00726-017-2468-4
Kawasaki, S., Miyake, C., Kohchi, T., Fujii, S., Uchida, M., and Yokota, A. (2000). Responses of wild watermelon to drought stress: accumulation of an ArgE homologue and citrulline in leaves during water deficits. Plant Cell Physiol. 41, 864–873. doi: 10.1093/pcp/pcd005
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kempf, B., and Bremer, E. (1998). Uptake and synthesis of compatible solutes as microbial stress responses to high osmolality environments. Arch. Microbiol. 170, 319–330. doi: 10.1007/s002030050649
Klingel, U., Miller, C. M., North, A. K., Stockley, P. G., and Baumberg, S. (1995). A binding site for activation by the Bacillus subtilis AhrC protein, a repressor/activator of arginine metabolism. Mol. Gen. Genet. 248, 329–340. doi: 10.1007/BF02191600
Kohlstedt, M., Sappa, P. K., Meyer, H., Maass, S., Zaprasis, A., Hoffmann, T., et al. (2014). Adaptation of Bacillus subtilis carbon core metabolism to simultaneous nutrient limitation and osmotic challenge: a multi-omics perspective. Environ. Microbiol. 16, 1898–1917. doi: 10.1111/1462-2920.12438
Korolev, N. (2021). How potassium came to be the dominant biological cation: of metabolism, chemiosmosis, and cation selectivity since the beginnings of life. BioEssays 43:e2000108. doi: 10.1002/bies.202000108
Kreuzer, K. D., and Henkin, T. M. (2018). The T-box riboswitch: tRNA as an effector to modulate gene regulation. Microbiol. Spectr. 6:RWR-0028-2018. doi: 10.1128/9781683670247.ch6
Kuhlmann, A. U., and Bremer, E. (2002). Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl. Environ. Microbiol. 68, 772–783. doi: 10.1128/AEM.68.2.772-783.2002
Kuo, T. T., and Stocker, B. A. (1969). Suppression of proline requirement of proA and proAB deletion mutants in Salmonella typhimurium by mutation to arginine requirement. J. Bacteriol. 98, 593–598. doi: 10.1128/jb.98.2.593-598.1969
Miller, C. M., Baumberg, S., and Stockley, P. G. (1997). Operator interactions by the Bacillus subtilis arginine repressor/activator, AhrC: novel positioning and DNA-mediated assembly of a transcriptional activator at catabolic sites. Mol. Microbiol. 26, 37–48. doi: 10.1046/j.1365-2958.1997.5441907.x
Nau-Wagner, G., Opper, D., Rolbetzki, A., Boch, J., Kempf, B., Hoffmann, T., et al. (2012). Genetic control of osmoadaptive glycine betaine synthesis in Bacillus subtilis through the choline-sensing and glycine betaine-responsive GbsR repressor. J. Bacteriol. 194, 2703–2714. doi: 10.1128/JB.06642-11
Perez-Arellano, I., Carmona-Alvarez, F., Gallego, J., and Cervera, J. (2010). Molecular mechanisms modulating glutamate kinase activity. Identification of the proline feedback inhibitor binding site. J. Mol. Biol. 404, 890–901. doi: 10.1016/j.jmb.2010.10.019
Roesser, M., and Müller, V. (2001). Osmoadaptation in bacteria and archaea: common principles and differences. Env. Microbiol. 3, 743–754. doi: 10.1046/j.1462-2920.2001.00252.x
Rosenberg, J., and Commichau, F. M. (2019). Harnessing underground metabolism for pathway development. Trends Biotechnol. 37, 29–37. doi: 10.1016/j.tibtech.2018.08.001
Rossi, J. J., Vender, J., and Berg, C. M. (1977). Proline excretion in Escherichia coli: a comparison of an argD+ strain and a proline-excreting argD− derivative. Biochem. Genet. 15, 287–296. doi: 10.1007/BF00484460
Schöck, F., Gotsche, S., and Dahl, M. K. (1996). Vectors using the phospho-alpha-(1,1)-glucosidase-encoding gene treA of Bacillus subtilis as a reporter. Gene 170, 77–80. doi: 10.1016/0378-1119(95)00860-8
Schroeter, R., Hoffmann, T., Voigt, B., Meyer, H., Bleisteiner, M., Muntel, J., et al. (2013). Stress responses of the industrial workhorse Bacillus licheniformis to osmotic challenges. PLoS One 8:e80956. doi: 10.1371/journal.pone.0080956
Smith, M. C., Czaplewski, L., North, A. K., Baumberg, S., and Stockley, P. G. (1989). Sequences required for regulation of arginine biosynthesis promoters are conserved between Bacillus subtilis and Escherichia coli. Genome Announc. 3, 23–28. doi: 10.1111/j.1365-2958.1989.tb00099.x
Smith, J. L., Goldberg, J. M., and Grossman, A. D. (2014). Complete genome sequences of Bacillus subtilis subsp. subtilis laboratory strains JH642 (AG174) and AG1839. Genome Announc. 2, e00663–e00614.
Song, Q., Joshi, M., Dipiazza, J., and Joshi, V. (2020). Functional relevance of citrulline in the vegetative tissues of watermelon during abiotic stresses. Front. Plant Sci. 11:512. doi: 10.3389/fpls.2020.00512
Stadmiller, S. S., Gorensek-Benitez, A. H., Guseman, A. J., and Pielak, G. J. (2017). Osmotic shock induced protein destabilization in living cells and its reversal by glycine betaine. J. Mol. Biol. 429, 1155–1161. doi: 10.1016/j.jmb.2017.03.001
Tawfik, D. S. (2020). Enzyme promiscuity and evolution in light of cellular metabolism. FEBS J. 287, 1260–1261. doi: 10.1111/febs.15296
van den Berg, J., Boersma, A. J., and Poolman, B. (2017). Microorganisms maintain crowding homeostasis. Nat. Rev. Microbiol. 15, 309–318. doi: 10.1038/nrmicro.2017.17
von Blohn, C., Kempf, B., Kappes, R. M., and Bremer, E. (1997). Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol. Microbiol. 25, 175–187. doi: 10.1046/j.1365-2958.1997.4441809.x
Whatmore, A. M., Chudek, J. A., and Reed, R. H. (1990). The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol. 136, 2527–2535. doi: 10.1099/00221287-136-12-2527
Whatmore, A. M., and Reed, R. H. (1990). Determination of turgor pressure in Bacillus subtilis: a possible role for K+ in turgor regulation. J. Gen. Microbiol. 136, 2521–2526. doi: 10.1099/00221287-136-12-2521
Widderich, N., Rodrigues, C. D., Commichau, F. M., Fischer, K. E., Ramirez-Guadiana, F. H., Rudner, D. Z., et al. (2016). Salt-sensitivity of sigma(H) and Spo0A prevents sporulation of Bacillus subtilis at high osmolarity avoiding death during cellular differentiation. Mol. Microbiol. 100, 108–124. doi: 10.1111/mmi.13304
Wood, J. M. (2011). Bacterial osmoregulation: a paradigm for the study of cellular homeostasis. Annu. Rev. Microbiol. 65, 215–238. doi: 10.1146/annurev-micro-090110-102815
Wood, J. M., Bremer, E., Csonka, L. N., Krämer, R., Poolman, B., Van Der Heide, T., et al. (2001). Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Compara. Biochem. Physiol. Part A, Mol. Integra. Physiol. 130, 437–460. doi: 10.1016/S1095-6433(01)00442-1
Zaprasis, A., Bleisteiner, M., Kerres, A., Hoffmann, T., and Bremer, E. (2015). Uptake of amino acids and their metabolic conversion into the compatible solute proline confers osmoprotection to Bacillus subtilis. Appl. Environ. Microbiol. 81, 250–259. doi: 10.1128/AEM.02797-14
Zaprasis, A., Brill, J., Thüring, M., Wünsche, G., Heun, M., Barzantny, H., et al. (2013a). Osmoprotection of Bacillus subtilis through import and proteolysis of proline-containing peptides. Appl. Environ. Microbiol. 79, 576–587. doi: 10.1128/AEM.01934-12
Zaprasis, A., Hoffmann, T., Wünsche, G., Florez, L. A., Stülke, J., and Bremer, E. (2013b). Mutational activation of the RocR activator and of a cryptic rocDEF promoter bypass loss of the initial steps of proline biosynthesis in Bacillus subtilis. Environ. Microbiol. 16, 701–717. doi: 10.1111/1462-2920.12193
Keywords: osmotic stress, suppressor mutations, metabolomics, gene regulation, compatible solutes, L-proline
Citation: Stecker D, Hoffmann T, Link H, Commichau FM and Bremer E (2022) L-Proline Synthesis Mutants of Bacillus subtilis Overcome Osmotic Sensitivity by Genetically Adapting L-Arginine Metabolism. Front. Microbiol. 13:908304. doi: 10.3389/fmicb.2022.908304
Received: 30 March 2022; Accepted: 17 May 2022;
Published: 16 June 2022.
Edited by:
Anne Galinier, Centre National de la Recherche Scientifique, FranceReviewed by:
Meijuan Xu, Jiangnan University, ChinaCopyright © 2022 Stecker, Hoffmann, Link, Commichau and Bremer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erhard Bremer, YnJlbWVyQHN0YWZmLnVuaS1tYXJidXJnLmRl
†ORCID: Tamara Hoffmann, orcid.org/0000-0002-5218-8506
Hannes Link, orcid.org/0000-0002-6677-555x
Fabian M. Commichau, orcid.org/0000-0003-0323-7479
Erhard Bremer, orcid.org/0000-0002-2225-7005
‡Present addresses: Hannes Link, University Tübingen Cluster of Excellence CMFI – Bacterial Metabolomics, Tübingen, Germany
Fabian M. Commichau, Department of Molecular Microbiology, Institute for Biology, University of Hohenheim, Stuttgart, Germany
§These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.