
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 01 August 2022
Sec. Phage Biology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.905343
Viruses that infect bacteria (i.e., phages) are abundant and widespread in the human body, and new anti-infective approaches such as phage therapy are essential for the future of effective medicine. Our understanding of microenvironmental factors such as tissue oxygen availability at the site of phage–bacteria interaction remains limited, and it is unknown whether evolved resistance is sculpted differentially under normoxia vs. hypoxia. We, therefore, analyzed the phage–bacteria interaction landscape via adsorption, one-step, time-kill dynamics, and genetic evolution under both normoxia and hypoxia. This revealed that adsorption of phages to Pseudomonas aeruginosa decreased under 14% environmental oxygen (i.e., hypoxia), but phage time-kill and one-step growth kinetics were not further influenced. Tracking the adaptation of P. aeruginosa to phages uncovered a higher frequency of phage resistance and constrained types of spontaneous mutation under hypoxia. Given the interest in developing phage therapies, developing our understanding of the phage–pathogen interaction under microenvironmental conditions resembling those in the body offers insight into possible strategies to overcome multidrug-resistant (MDR) bacteria.
Multidrug-resistant (MDR) disease-causing bacteria are one of the biggest threats to global public health and security today. The United States Centers for Disease Control and Prevention (CDC) estimate that antibiotic resistance causes over 2.8 million illnesses each year, leading to more than 35,000 deaths as a result, with many more deaths from conditions complicated by resistant infections (CDC, 2019). While we have made significant progress in improving antibiotic stewardship, the discovery and commercialization of novel classes of antibiotics have waned (Sommer et al., 2017). New antimicrobial strategies continue to emerge, which focus on satisfying several criteria for innovation and safety, such as identifying agents with novel mechanisms of action or combinatorial treatments. The main preventive and therapeutic approaches under investigation include nanoparticles, bacteriophages (phages), endolysins, antibodies, vaccines, virulence suppressors, and microbiota modifications (Sommer et al., 2017; Luong et al., 2020b; Burki, 2021).
Phage therapy is widely being reconsidered to treat bacterial infections with virulent (lytic) phages, alone or in combination with traditional antimicrobials, as a promising strategy to combat MDR infections (Pirnay et al., 2018; Luong et al., 2020b; Hatfull et al., 2022). Phages are naturally occurring viruses that infect both gram-positive and gram-negative bacteria. As such, they pose limited adverse health effects in eukaryotic cells in patients. A substantial volume of preclinical data and an increasing body of clinical evidence indicate the immense therapeutic potential across a wide range of infectious diseases. While phages are generally unaffected by mechanisms of antibiotic resistance, there remain several clinical challenges such as their viral nature make it difficult to standardize as a drug, the need to employ multiple phage strains to increase drug spectra, and the lack of understanding the pharmacological properties of phages. Another obstacle in developing effective phage therapies is the evolution of phage resistance. Predicting the future of bacterial phage resistance is difficult, especially for evolutionary processes that are influenced by numerous unknown factors. Still, these are requirements during drug development to assess the risk of resistance arising against a new phage candidate during preclinical development.
Phage susceptibility and possible resistance testing are typically first performed in vitro to predict the in vivo effectiveness of treatment and to help guide the phage choice, dose, and timing of administration. One factor that is often overlooked during in vitro testing is the oxygen level at the site of a bacterial infection within the patient. As a result, the majority of the time, in vitro experiments are performed under ambient (normoxic) conditions, which may not recapitulate disease manifestations and phage activity in the body. Most mammalian tissue cells and organs experience oxygen partial pressures equivalent to 0.5–7% O2 (i.e., physioxia), with the exception of O2 tension ranging from 4 to 14% in the lungs, the liver, and the kidneys (i.e., hypoxia) (Carreau et al., 2011; Gan and Ooi, 2020). Recent studies have suggested that hypoxia can significantly alter the state of a bacterial cell, including differential gene expression, defense activation, metabolic states, receptor expression, and subsequent disease progression (Schaible et al., 2017). Therefore, the phage treatment net effect depends on a number of factors, including infection and replication dynamics of the target bacteria under low oxygen conditions.
To focus on research and development of novel antimicrobials, the WHO published its list of pathogens for which new drugs are in dire need. Within this broad list, ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogens were designated as “priority status” (WHO, 2017). P. aeruginosa is a ubiquitously distributed opportunistic human pathogen commonly associated with severe respiratory infections in patients with compromised immune defenses. Patients with chronic or inherited lung diseases, such as bronchiectasis and cystic fibrosis (CF), are highly susceptible to persistent pulmonary infection, with episodic exacerbations requiring hospitalization and intravenous antibiotics, with a subsequent risk of selection for MDR pathogens (Malhotra et al., 2019). The plasticity and adaptability of the relatively large P. aeruginosa genome (∼6.5 Mbp), conferred by a large repertoire of metabolic and regulatory genes, are the key elements of the pathogen’s ability to chronically persist in the host and evade antimicrobial therapeutics (Moradali et al., 2017). Usually described as a bacterium that favors aerobic growth conditions, P. aeruginosa is a facultative anaerobe well adapted to proliferate in conditions of partial or total oxygen depletion (Worlitzsch et al., 2002; Schaible et al., 2012; Gil-Marques et al., 2018). Therefore, the microenvironment at the site of infection likely plays a vital role in determining the outcome of P. aeruginosa infection.
There is an incomplete understanding of interactions between phages and bacteria under conditions that resemble those present in the body, such as reduced oxygen availability, making the clinical outcome of phage therapy unpredictable. In this study, we explored the impact of “hypoxia” on the interaction between P. aeruginosa and its phages. We demonstrated that exposure to environmental hypoxia did not alter time-kill kinetics of P. aeruginosa phages. Hypoxia, however, promoted a higher frequency of phage resistance and constrained types of spontaneous mutations. Developing our understanding of the phage–pathogen interaction under conditions that resemble those present in humans will provide us with new strategies to improve phage therapy efficacy.
The P. aeruginosa strain K (PAK) was cultured at 37°C overnight in Luria-Bertani (LB) broth, Miller. LB supplemented with 1.5% agar allowed for solid medium growth when required. The myovirus PAK_P1 (Forti et al., 2018) was propagated and purified as previously described (Luong et al., 2020a). Briefly, phages were added to mid-log growing bacteria at a multiplicity of infection (MOI) 0.1 and incubated overnight. Phage lysaes were centrifugred at 8,000 × g and phage supernatants were steriliezed by 0.22 μm dead-end filtraiton. Phage titers are expressed in plaque-forming units (PFUs) per ml and were measured using the serial spot method on LB agar.
Experiments were conducted under different oxygen levels (Figure 1). For hypoxic conditions (14% O2, 5% CO2) and ambient CO2-enriched (21% O2, 5% CO2) conditions, cultures were incubated in a hypoxic incubator chamber (Coy Laboratory Products, Grass Lake, MI, United States) before being transferred to a Clariostar microplate reader (BMG Labtech, Ortenberg, Germany) under the same conditioned atmosphere using compressed N2 and CO2 gases. For normoxic conditions with 21% O2 and 0.04% CO2, cultures were placed in a standard benchtop incubator (Eppendorf, Enfield, CT, United States) before transfer to the microplate reader. All media and buffers were preconditioned for >48 h in the test environment. Microplates were sealed with a BreathEasy™ membrane (Diversified Biotech, Dedham, MA, United States) to allow gas exchange.
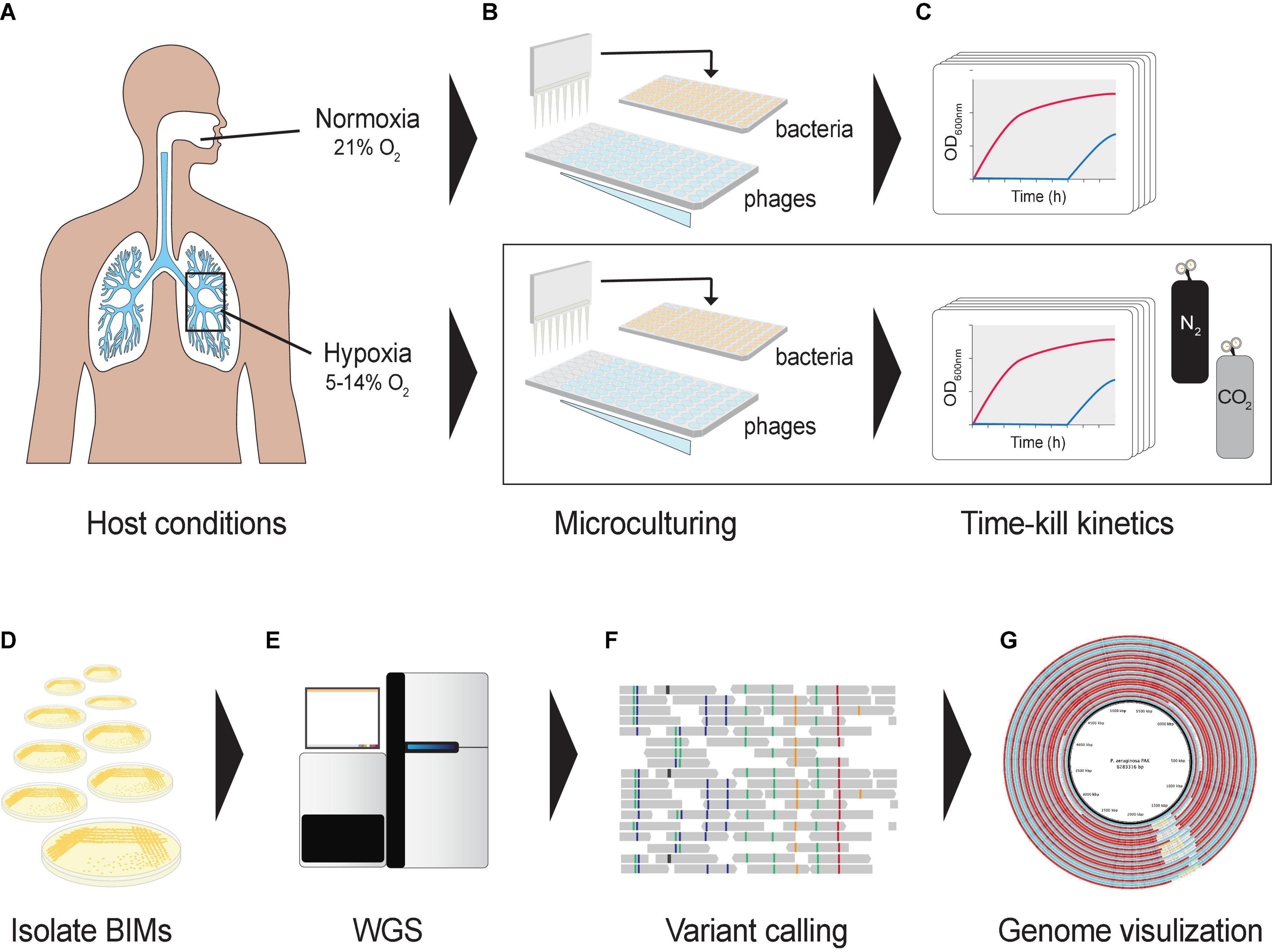
Figure 1. Graphical representation of the experimental design. (A) Oxygen levels in lungs mimicked using a hypoxic growth chamber. (B) A hypoxia culture-based assay was developed in a 96-well microtiter plate format to measure the phage–bacteria dynamics under different oxygen levels. Co-cultures were prepared in a hypoxic chamber fed with CO2 and N2. (C) Cultures were incubated in a microplate reader to assess bacterial optical densities at 600 nm under different oxygen levels. (D) Bacteriophage-insensitive mutants (BIMs) were isolated from cocultures (E) and their DNA extracted for whole-genome sequencing. (F) Detecting and characterizing sequence variation in BIM genomes, and (G) comparing evolution of multiple BIM genomes as a group against the central reference sequence.
To evaluate the lytic activity of phage PAK_P1 under normoxic and hypoxic conditions, 2 × 106 CFU of strain PAK was exposed to phages at MOIs 0.1, 1, and 10, and bacterial growth was monitored (OD600) at 37°C with an atmosphere controlled microplate reader. Media and buffers were preconditioned and microtiter plates were prepared in a hypoxic chamber. The area under the curve (AUC) was calculated Σ(Y1 + Y2)/2*(T2−T1). A total of five replicate microplates were performed and inoculated with different CFUs for each condition.
The phage adsorption rate was determined as previously described (Kropinski, 2009) under the aforementioned atmospheric conditions (Figure 1). Briefly, 1 ml of pre-warmed phage suspension was added to 9.0 ml of bacterial culture at 2 × 108 CFU ml–1 at an MOI of 0.001 and incubated at 37°C with shaking at 120 rpm. About 50 μl of samples was diluted in 950 μl of fresh LB and 50 μl of chloroform (10% v/v) and placed on ice for 10 min before titering. The adsorption constant (k) was calculated (k = −slope/N), where N is the density of bacteria. A total of nine replicates were performed on different CFUs.
The one-step growth curves were determined by monitoring the dynamic changes in the number of phage particles during a lytic cycle (Hyman and Abedon, 2009). Briefly, phage suspension was mixed with 5 × 108 CFU mid-exponential phase host cells to obtain MOI 0.1. After 6 min, the mixture was diluted 1,000 times in an LB broth to prevent infection by the new phage progeny and further incubated at 37°C. The results are the mean of three replicates ± standard deviation.
Microtiter wells from time-kill curves were streaked onto LB agar to isolate bacteriophage-insensitive mutants (BIMs). Bacterial genomic DNA was extracted by NucleoSpin™ Microbial DNA Isolation Kit (Macherey-Nagel; Bethlehem, PA, United States), and sequencing libraries were prepared by MiGS (Pittsburg, PA, United States) using the Illumina Nextera Kit and then sequenced on the Illumina NextSeq 550 platform (San Diego, CA, United States). Raw 2 × 150 bp reads were trimmed by FASTP v.0.20.11 and de novo assembled using SPAdes v.3.15.1 (Prjibelski et al., 2020), and reference guided assembled using MeDuSa v1.6 (Bosi et al., 2015) and the PAK reference genome (NC_002516.2). Annotation of a specific function of open reading frames (ORFs) was conducted using rapid annotations of subsystem technology (RAST v2.0) (Overbeek et al., 2014). Mutational analyses relative to ancestral PAK isolates grown under atmospheric conditions in the absence of phages were conducted using breseq v.0.35.5 (Deatherage and Barrick, 2014) and visualized using Circa software (OMG genomics).
Statistical analyses were performed using RStudio (Boston, United States) and visualized using the Jupyter Notebook. Tests used include the analysis of variance (ANOVA) and when required Dunnett’s and Tukey’s post hoc tests and Student’s t-test. A p-value of <0.05 was considered significant.
Our understanding of how the course of opportunistic bacterial infection is influenced by the microenvironment is limited. We first determined P. aeruginosa growth rate (μmax) and growth potential (AUC) under hypoxia (14% O2/5% CO2) and under normoxia (21% O2/0.5% CO2) in the absence of phages. We observed P. aeruginosa grown under both atmospheric conditions. As expected, however, decreased oxygen availability by 7% caused a 19% reduction in μmax (normoxia 0.091 OD600 nm⋅h–1 vs. hypoxia 0.074 OD600 nm⋅h–1) between 2 and 5 h (Figure 2A). Overtime, the reduced growth rate decreased the maximum OD600 nm by 11% (OD600 nm 0.958 ± 0.057 vs. 0.85 ± 0.02). Together, hypoxia caused a significant growth potential reduction of P. aeruginosa over 18 h at 37°C (AUC 10.38 ± 0.18 vs. 11.9 ± 0.58, p < 0.001) of 12.9% (Figure 2B). Nonetheless, changes in metabolism likely allow the pathogen to grow even under unfavorable conditions.
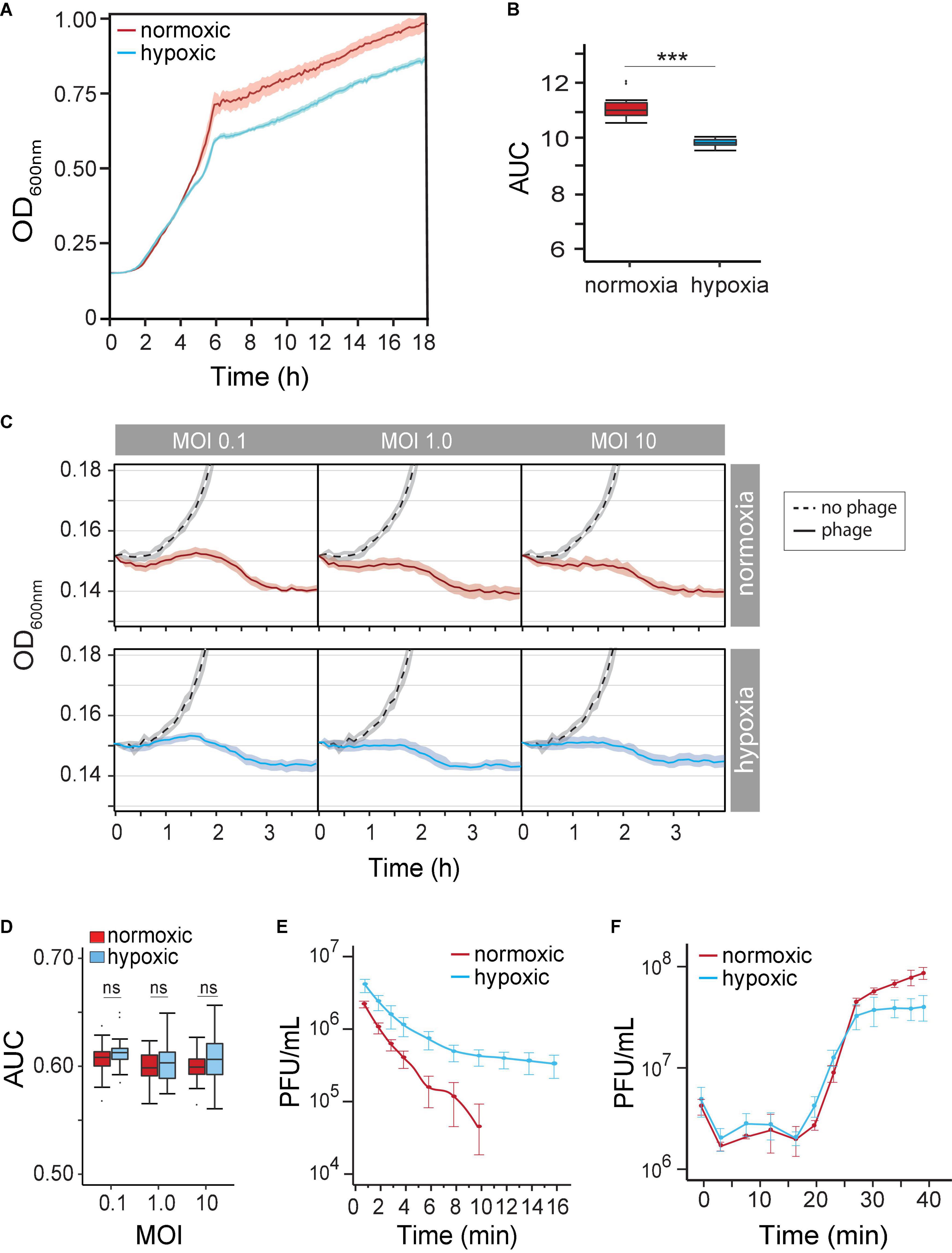
Figure 2. (A) Pseudomonas aeruginosa strain PAK population growth curves (based on OD at 600 nm) under normoxia versus hypoxia (14% O2/5% CO2) in the LB medium. (B) The difference in the P. aeruginosa strain K (PAK) population growth potential was measured as the area under the curve (AUC) over 18 h under normoxia (norm) and hypoxia (hypo). p < 0.001 (C) time-kill curves of no phage (dash line), phage under normoxia (red lines), and phage under hypoxia (blue lines) at different multiplicities of infection (MOIs). Microcultures were inoculated with 2 × 106 CFU of bacteria. (D) AUC of time-kill kinetics over 4 h at different phage MOIs under normoxia (red) and hypoxia (blue). (E) The adsorption rate of phage PAK_P1 at MOI 0.001 under normoxia (red; n = 9) and hypoxia (blue; n = 9). (F) One-step growth curves of phage PAK_PI under normoxia (red circles) and hypoxia (blue circles). Phages were mixed with 5 × 108 CFU bacteria at MOI 0.1. ***p < 0.001.
Because the physiological growth state of host bacterial cells may also influence the life cycle of phages, we next observed the phage’s lytic activity over time under hypoxia. Time-kill curves that monitor bacterial growth (and death) over a range of phage concentrations (i.e., MOIs) can evaluate the effect of hypoxia on phage lysis. However, the myovirus PAK_P1 time-kill kinetics of hypoxia-induced slower growth of P. aeruginosa was seemingly unchanged to that induced under normoxia (Figure 2C). In addition, bacterial growth potential (AUC) over 4 h was not significantly different between normoxia and hypoxia conditions (p = 0.17) after phage exposure (Figure 2D).
There are three stages of a lytic phage’s life cycle corresponding to the processes of adsorption, replication, and lysis. The rates and timing of these processes can thus be characterized as the phage’s life-history traits. Although the phage time-kill kinetics appeared not to be influenced by hypoxia, the phage–host interaction is a mass action, which assumes an equal influence of host cell density and phage adsorption rate (Shao and Wang, 2008). We sought to determine whether the altered state of the host under hypoxia influenced phage virion adsorption onto the surface of a susceptible host cell. Figure 2E shows a 66% decrease in phage PAK_P1 adsorption rate due to host cells grown under hypoxia (0.2288 vs. 0.0771 Log10PFU/ml min–1, p < 0.001). Because the influence of host density cell was controlled, this phenomenon might indicate a distinct effect on the bacterial outer membrane that delays phage binding under different oxygen levels.
However, we found that the timing of phage PAK_P1 replication and host lysis were similar. One-step growth curves allow us to define the timing of phage-induced host cell lysis (i.e., latent period), which is under the control of the phage gene products (e.g., holin). Phage PAK_P1 had a similar latent period of ∼14 min and a growth plateau at 30 min post-exposure (Figure 2F). Although one-step growth curves, which incorporate adsorption rate but avoid the effects of reinfections by phage progeny produced during replication, suggest that hypoxia did not affect phage generation time and the burst size, it was consistent with time-kill curves (Figure 2C).
Next, we reasoned that the evolution of BIMs is perhaps inevitable in vitro. However, the altered state of host cells due to reduced O2 availability may also influence evolution phage resistance. We observed that the long-term phage time-kill kinetics were similar under both normoxia and hypoxia, with extended periods of controlled bacterial growth. There was, however, about a 1-h delay of bacterial regrowth that crossed the LOD under hypoxia (11.8 ± 0.61 vs. 10.56 ± 0.35 h, p = 0.002) (Figure 3A vs. Figure 3B). Furthermore, phage PAK_P1 appeared to have greater overall bacterial population control under hypoxia. The maximum rate of BIM growth (μmax) (i.e., bacterial regrowth) was reduced by 11% compared to BIM growth under normoxia, which also resulted in a lower OD600max (Figure 3A vs. Figure 3B). In addition, we show in Figure 3C that phage treatment promoted a further 3% decrease in bacterial growth potential (AUC) compared to the exposure under ambient conditions after 18 h, normalized to bacterial growth in the absence of phages under the respective growth condition.
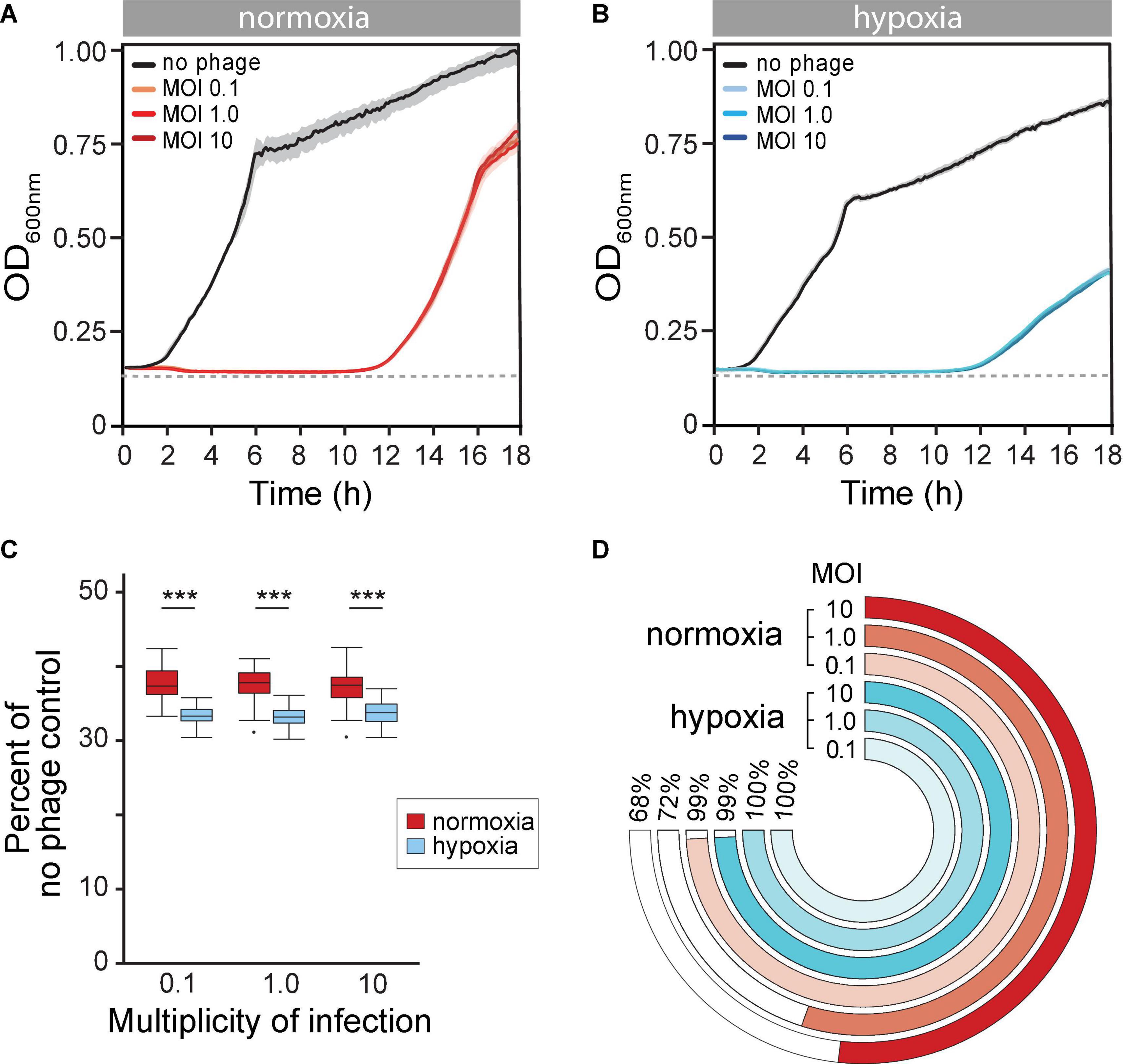
Figure 3. (A,B) Mean time-kill curves of phage PAK_P1 at different a multiplicity of infections (MOIs) under (A) normoxia and (B) hypoxia conditions. Microcultures under both atmospheric conditions inoculated with 2 × 106 CFU and treated with phages at MOIs 0.1, 1.0, or 10 (n ≅ 42 per MOI per condition) showed gradual reductions to the limit of detection (dotted line) of bacterial density until bacteriophage-insensitive mutant (BIMs) outgrowth. (C) The phage time-kill area under the curve (AUC) for 18 h under normoxia (red) and hypoxia (blue) conditions normalized to no treatment control were significantly different (p < 0.001). (D) Percent of turbid microcultures 18 h after phage treatment at different MOIs. Treatment at MOI 0.1, 1.0, and 10 prevented bacterial growth under normoxia in 4 of 465, 209 of 741, and 119 of 372 microcultures, respectively. In contrast, bacteria growth was observed after 18 h under hypoxia in all 465 microcultures at each treatment MOI.
Although phages exhibited greater control of P. aeruginosa growth under hypoxia, we observed that nearly all phage-treated microcultures were turbid (>OD600 0.3) after 18 h under hypoxia (Figure 3D, blue rings). This suggested that the hypoxia promoted a rising rate of phage resistance. In contrast, phage treatments at MOIs 0.1, 1.0, and 10 under normoxia were able to prevent regrowth in 1 (4 of 465), 28 (209 of 741), and 32% (119 of 372) of microcultures, respectively (Figure 3D, red rings). This implied that evolutionary dynamics were different under different atmospheric conditions, with high rates of phage resistance evolving quickly under hypoxia.
To look at genetic differences between evolutions of phage resistance under hypoxia compared to normoxia, we isolated 44 single BIM CFUs from 44 randomly selected individual time-kill cultures (i.e., wells) for whole-genome sequencing. We observed several types of genetic variants (or mutations) in the genome of each BIM isolated under either hypoxia or normoxia, including non-synonymous single-nucleotide polymorphisms (nSNPs), insertion–deletions (Indels), and large deletions (LD) (Figures 4A,B and Supplementary Table 1). The mean number of total mutations in each BIM was not significantly different between normoxia and hypoxia (1.30 ± 0.54 and 1.63 ± 0.62, respectively; p = 0.06). However, an evolution of resistance under hypoxia resulted in increased nSNPs (21%) and indels (14.9%), as well as a decrease in LDs (36%) compared to normoxia counterparts (Figure 4C). Thus, phage resistance evolution under normoxia resulted in a significant increase in large chromosomal deletions (85–438 kbp), which generally excised upward of 400 genes (Figures 4A,B and Supplementary Table 1). Interestingly, large chromosomal deletions of hundreds of genes were also observed in the evolution and diversification of P. aeruginosa associated with chronic infections in patients with CF (Hocquet et al., 2016).
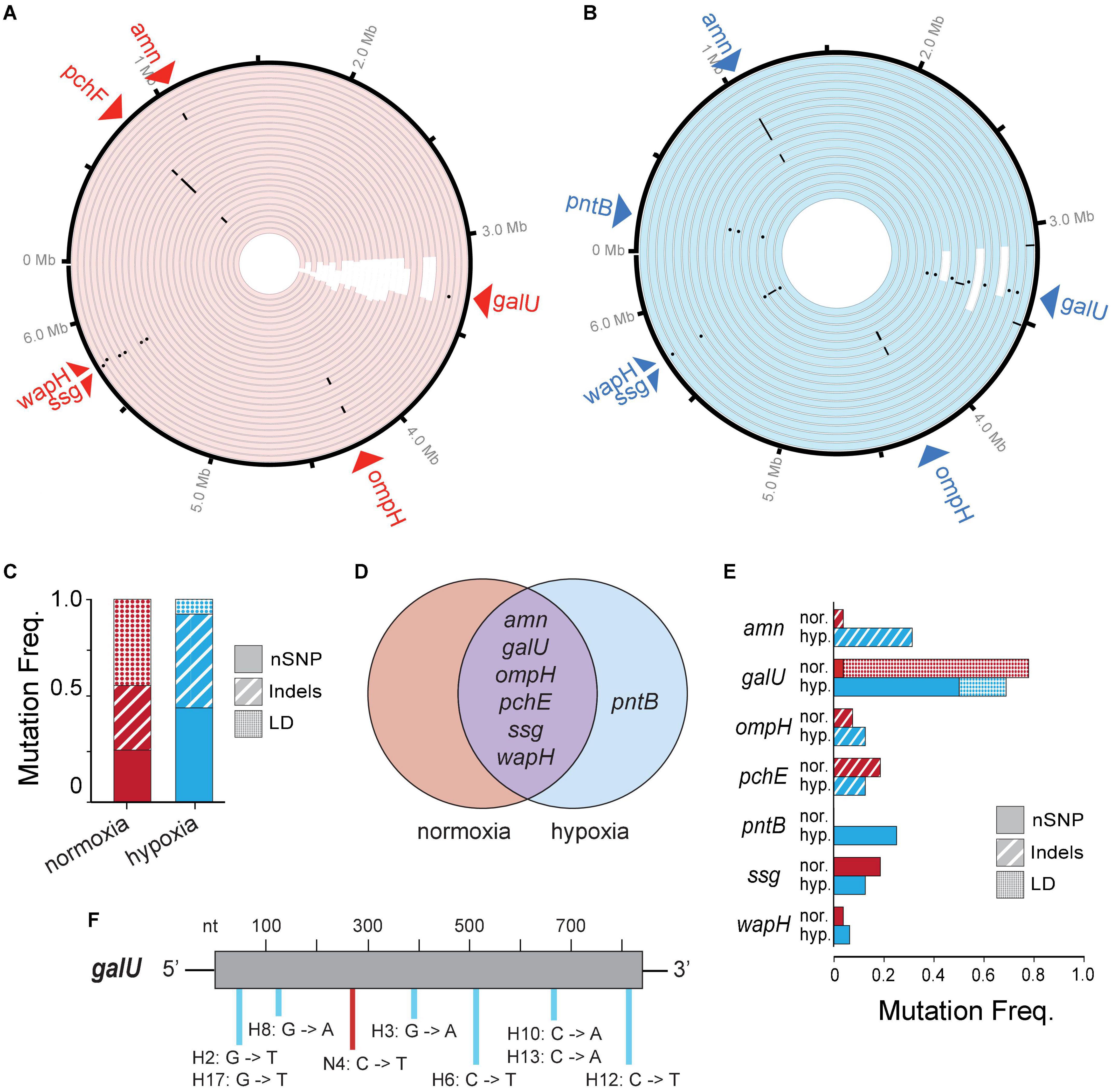
Figure 4. (A,B) A Circa plot showing spontaneous mutations in randomly isolated bacteriophage-insensitive mutants (BIMs) evolved under (A) normoxia (n = 27) and (B) hypoxia (n = 17) relative to no phage exposure. Both conditions exhibited mutations as non-synonymous single-nucleotide polymorphisms (nSNPs) (dots), insertion–deletions (dash), and large deletions (LD) (white rectangles). (C) Proportion of different mutation types identified in BIMs and (D) a Venn diagram showing the shared and unique genes with nSNPs. (E) Frequencies of BIMs shared mutated gene and frequency of gene mutation type. (F) Spectra of galU (UTP–glucose–1–phosphate uridylyltransferase) point mutations.
Common spontaneous mutations after the evolution of phage resistance under either atmosphere include mutations in the amn, galU, ompH, ssg, and wapH genes (Figure 4D). Of these, galU, ssg, and wapH are each involved in lipopolysaccharide (LPS) biosynthesis, ompH encodes the TolC efflux pump, amn is involved in cellular respiration, and pchE encodes a siderophore. Because phages can use various cell surface structures to infect hosts, and bacteria often lose or modify these structures to gain resistance to phages (Labrie et al., 2010), phage PAK_P1 appears to bind LPS as its receptor. Under hypoxia, phage pressure was selected for the increased frequency of amn indels and galU nSNPs (Figure 4E). The latter is likely the result of opposing pressure against LD due to hypoxia. Point mutations occurred randomly throughout the galU gene (Figure 4F). This suggests that, while phage receptor mutations occur independent of the oxygen level tested, the variant type and frequency are correlated with the growth state of the bacterial host.
Several BIMs evolved under normoxia and displayed an easily observed reddish-brown pigment called pyomelanin (Figure 5A). Pyomelanin was produced in 17.9% of all phage-resistant microcultures at an MOI of 0.1 (n = 465), 36% at an MOI of 1 (n = 745), and 52.7% at an MOI of 10 (n = 372) under normoxia (Figure 5B). Melanogenic BIM genomes were identified to have LD (85–438 kbp), which attributed to the loss of both hmgA and the neighboring galU, among several other genes. Previous studies have shown that hmgA synthesizes homogentisate 1,2-dioxygenase, which is an essential component of the tyrosine catabolism pathway that converts homogentisic acid to 4-maleylacetoacetate (Hocquet et al., 2016). Mutation of hmgA causes an accumulation of homogentisic acid, which is secreted as pyomelanin. A total of three BIMs evolved under hypoxia (Figure 4B and Supplementary Table 1) with LDs that deleted hmgA, did not exhibit pyomelanin pigmentation (Figure 5A), even after further 24-h incubation under normoxia (data not shown).
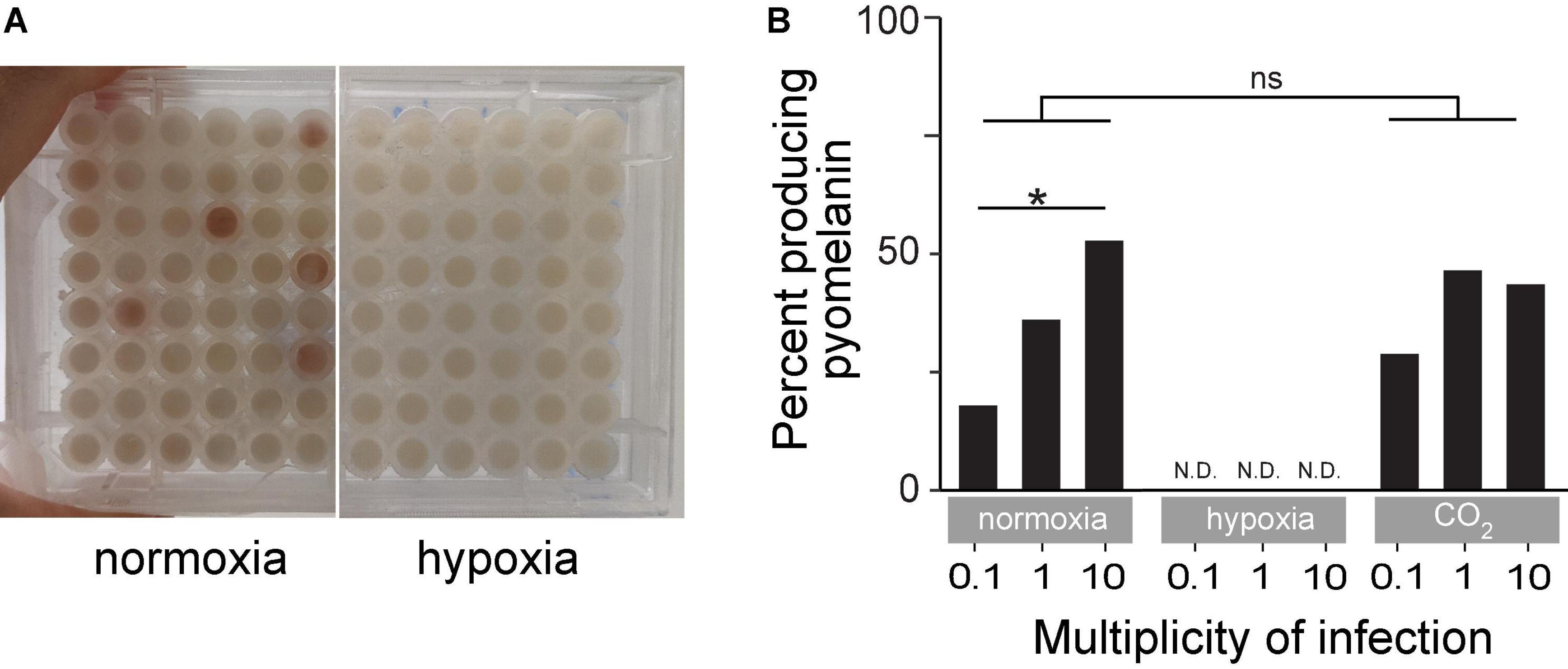
Figure 5. (A) An example of time-kill assay microplates after 18 h of incubation under normoxia or hypoxia. The reddish-brown pigmentation is pyomelanin. (B) Percent of turbid microcultures that also exhibited reddish-brown pigmentation under normoxia, hypoxia, or ambient CO2-enriched (21% O2/5% CO2) conditions. *p < 0.05, ns: not significant.
Developing an understanding of the mechanisms underpinning the phage–bacteria interactions in both the broader context of parasite-host and predator-prey dynamics is key to improving the success of the phage therapy. In this study, we observed that the P. aeruginosa phage PAK_P1 lytic time-kill kinetics were largely indistinguishable under low-oxygen conditions compared to standard laboratory conditions. Our observations were consistent with our previous report (Mizuno et al., 2020) showing phages efficiently lysed Citrobacter rodentium under hypoxic conditions. We noted that reduced oxygen levels slowed phage adsorption to LPSs, suggesting that P. aeruginosa’s response to lower O2 was to alter its outer membrane. However, the slower growth rate of host cells under hypoxic conditions did not influence the phage PAK_P1 growth parameters (latent and burst size). Tracking the evolutionary dynamics within a bacterial population, as it adapted to phage selective pressures under low oxygen environments, we found that hypoxia promoted the evolution of phage resistance in nearly all phage–bacteria interactions compared to standard laboratory conditions. Among the mutants, we demonstrated that conserved galU mutation enabled the pathogen to adapt and persist, suggesting a reproducibility of evolution independent of the oxygen level. We also observed that hypoxic conditions switched the dominant genetic mutation type associated with resistance.
Phage replication is shaped by the cellular microenvironment; however, our current understanding of the influence underlying hypoxia-induced phage–bacteria dynamics is limited (reviewed in refs Hodges et al., 2021; Shareefdeen and Hynes, 2021). The local oxygen level can vary widely depending on the metabolic demand and blood supply of the tissue or organ (Carreau et al., 2011). While most in vitro studies on phage replication utilize cells cultured at atmospheric oxygen levels (∼21% O2), the vast majority of human tissues have oxygen levels much lower than this (referred to as hypoxia). Normal lungs have a partial pressure of oxygen (pO2) of between 42.8 and 110 mmHg (5.6–14% O2) and 5–9% CO2 (Le et al., 2006; Donovan et al., 2010). P. aeruginosa is preferentially an aerobic pathogen (Schaible et al., 2017), although we believe that P. aeruginosa could survive and grow under hypoxic stress to high cell densities, albeit at a reduced growth potential than under normoxic conditions. We observed that there was no significant decline in phage-induced lysis under mild hypoxia. However, mild hypoxia at 14% O2 may not recapitulate bacterial growth and phage activity inside the body. Mammalian tissue cells and organs experience oxygen partial pressures below 14% O2 (e.g., 0.5–14% O2) (Carreau et al., 2011; Gan and Ooi, 2020). Further limitations in oxygen could further alter the state of a bacterial cell, including decreasing gene expression, metabolism, and receptor expression. In our previous study, we indicated that lower oxygen hypoxia by 5% did not decrease phage lysis of C. rodentium in time-kill analysis at similar MOIs (Mizuno et al., 2020). We, however, found that a more extreme case of hypoxia caused a marked decrease in phage lytic activity at low MOIs (e.g., 0.01–0.001), which were not tested in the current study. Phage themselves are metabolically inert and their parasitism is largely dependent upon hijacking the host’s metabolic machinery and resources to replicate new virus particles. This is often deadly to host cells. One could hypothesize that facultative anaerobe cells under low oxygen possess an ample supply of energy metabolism to promote anabolism for the generation of macromolecules needed for virion replication and assembly.
Because time-kill kinetics provide insights over multiple phage infection cycles, we also characterized phage growth parameters, namely, adsorption constant, latent period, and burst size, under hypoxic and normoxic conditions. The lytic phage lifecycle begins with free-phage diffusion until attachment of the virion onto a specific host cell-surface structure. We found that 14% O2 significantly reduced phage PAK_P1 adsorption constant. The adsorption process is typically described by mass-action kinetics, which is assumed to be equally influenced by host density and adsorption rate (Shao and Wang, 2008). We demonstrated that phage PAK_P1 likely binds to the LPS in the outer membrane in P. aeruginosa. However, we cannot rule out hypoxia-induced alterations to LPS, which occur as a mechanism to evade recognition by mammalian defenses in vivo (Lam et al., 2011). The number of LPS monomers per bacterial cell appears to remain constant in different bacteria growth states (Walczak et al., 2012). On the contrary, one can assume that reduced bacterial motility under hypoxia might also influence phage adsorption (Schaible et al., 2017; Andre et al., 2022).
The latent period is defined by the timing of phage-induced host cell lysis, i.e., at the point when phage-progeny is released (Young, 2013). How phage PAK_P1 regulates lysis-timing remains to be determined, although we noticed that induced changes to the host cell under hypoxia did not affect the viral latent period and burst size. In contrast, several studies showed that the growth rate of bacteria influences phage growth parameters (Abedon et al., 2001; You et al., 2002; Golec et al., 2014; Nabergoj et al., 2018). A conclusion was that latent periods would tend to increase as host quality declines (Abedon et al., 2001). Our findings suggest that hypoxia did not diminish host quality to the extent of delayed lysis timing. Raya et al. (2011) showed that Escherichia coli under ambient and anaerobic growth conditions did not affect phage latent periods. This suggests that virulent phages precisely time the irreversible lysis of their host cells after infection independent of oxygen conditions.
The evolution of phage resistance was not influenced by the presence of the phage per se but rather the hypoxic conditions increased the adaptation tempo. Under normoxic conditions, phage PAK_P1 appeared to prevent bacterial growth in 25% of cocultures. In contrast, reducing O2 increased the pace of phage resistance, causing all phage–bacteria interactions to evolve resistance. Cellular metabolic rate is known to control mutation rates: mutations arise through unrepaired errors accrued during DNA replication and other damage-causing processes. Whole-genome sequencing determined that phage PAK_P1 was largely selected for mutations in genes related to LPS synthesis (galU, ssg, and wapH) under both normoxic and hypoxic conditions. For example, galU encodes a protein responsible for synthesizing UDP-glucose from UTP and glucose-1-phosphate during LPS outer core synthesis. Thus, modifications of the LPS core oligosaccharide were deemed the major mechanism for phage PAK_P1 resistance. The gram-negative bacterial LPS is a major component of the outer membrane that frequently plays a key role in pathogenesis. Bacterial adaptive changes including the modulation of LPS synthesis and structure are the conserved themes in infections, irrespective of the type of bacteria or the site of infection. In general, these changes result in immune system evasion, persisting inflammation and increased antimicrobial resistance (Priebe et al., 2004).
Populations of pathogenic bacteria undergo bottlenecks during infection due to host conditions such as those imposed by the host immune system and physical properties of tissues including oxygen availability (Levin et al., 1999; Common and Westra, 2019; Garoff et al., 2020). The evolution of phage resistance under hypoxia was selected overwhelmingly for point mutations in galU, whereas evolution under normoxia was selected for LD excising up to 8% of the genome, including galU. That is, bottlenecks and varying selection levels imposed by the phage and the oxygen level favor a subset of genetically encoded phenotypes in the population, leading to the loss of genetic variants. Contrary to our prediction, bottlenecking worked to the advantage of the bacterium providing fitness advantages under hypoxia. LD also excised hmgA, which caused the overproduction of the red-brown pigment pyomelanin. hmgA converts homogentisic acid to 4-maleylacetoacetate during tyrosine catabolism to break down pyomelanin. Pyomelanin can improve pathogen resistance to oxidative stresses and increase iron uptake, which may bolster survival in harsh environments (Rodriguez-Rojas et al., 2009). However, pyomelanin-producing variants only account for 20–30% of P. aeruginosa isolates from CF airways (Mayer-Hamblett et al., 2014). In an oxygen-rich environment under normal physiological conditions, the lung mucosal surface is susceptible to conditions of oxygen deficiency or tissue hypoxia in patients with CF (Page et al., 2021).
While novel insights were gained into how hypoxia influences phage–bacteria dynamics, this study has some limitations. Since this study examined phage resistance under mild hypoxia, examining the phage–bacteria interactions under severe hypoxia may reveal different dynamics and evolutionary patterns. For example, Ceyssens et al. (2010) found that, although phage PEV2 was able to infect P. aeruginosa growing anaerobically, the phage latent period was significantly longer. Moreover, the phage–bacteria dynamics can vary across phage species and different host species. Finally, all our experiments were performed in the LB medium, which is a nutritionally rich medium but not a defined medium that mimics the human–host environment.
Antimicrobial resistance represents one of the few crises that unite global interests, concerns for human and animal health, and agriculture. ESKAPE pathogens represent the paradigm for antibiotic resistance, pathogenesis, and disease transmission in both the community and clinical settings. It is now well recognized that phages have the therapeutic potential for the treatment and management of ESKAPE pathogens (Roach et al., 2017; Dedrick et al., 2019; Luong et al., 2020b; Cano et al., 2021; Johri et al., 2021; Lebeaux et al., 2021). To make accurate therapeutic decisions, it may be important to determine phage susceptibility and resistance development under clinically relevant oxygen conditions. However, predator-prey dynamic studies are mostly conducted under standard laboratory conditions, although this is rarely how phage–bacteria interact in nature. P. aeruginosa is present in many healthcare settings, especially in the context of chronic wounds, respiratory support, or urinary tract devices, where hypoxia predisposes for persistence, immune evasion, and antimicrobial resistance. Examining the phage–bacteria interactions under microenvironments similar to those found in the human body may lead to new therapeutic interventions.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
ARS and ADS contributed to data acquisition. All authors contributed to the conception and design of the study, participated in data analysis and manuscript writing, read, and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Ryan Cook and Tiffany Luong for their comments on the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.905343/full#supplementary-material
Abedon, S. T., Herschler, T. D., and Stopar, D. (2001). Bacteriophage latent-period evolution as a response to resource availability. Appl. Environ. Microbiol. 67, 4233–4241. doi: 10.1128/AEM.67.9.4233-4241.2001
Andre, A. C., Laborde, M., and Marteyn, B. S. (2022). The battle for oxygen during bacterial and fungal infections. Trends Microbiol. 2022:002. doi: 10.1016/j.tim.2022.01.002
Bosi, E., Donati, B., Galardini, M., Brunetti, S., Sagot, M. F., Lio, P., et al. (2015). MeDuSa: a multi-draft based scaffolder. Bioinformatics 31, 2443–2451. doi: 10.1093/bioinformatics/btv171
Burki, T. K. (2021). Development of new antibacterial agents: a sense of urgency needed. Lancet Respir. Med. 9:e54.
Cano, E. J., Caflisch, K. M., Bollyky, P. L., Van Belleghem, J. D., Patel, R., Fackler, J., et al. (2021). Phage therapy for limb-threatening prosthetic knee Klebsiella pneumoniae infection: Case report and in vitro characterization of anti-biofilm activity. Clin. Infect Dis. 73, e144–e151. doi: 10.1093/cid/ciaa705
Carreau, A., El Hafny-Rahbi, B., Matejuk, A., Grillon, C., and Kieda, C. (2011). Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J. Cell Mol. Med. 15, 1239–1253. doi: 10.1111/j.1582-4934.2011.01258.x
CDC (2019). Antibiotic resistant threats in the United States 2019”. Centers for Disease Control and Prevention). Atlanta: CDC.
Ceyssens, P. J., Brabban, A., Rogge, L., Lewis, M. S., Pickard, D., Goulding, D., et al. (2010). Molecular and physiological analysis of three Pseudomonas aeruginosa phages belonging to the “N4-like viruses”. Virology 405, 26–30. doi: 10.1016/j.virol.2010.06.011
Common, J., and Westra, E. R. (2019). CRISPR evolution and bacteriophage persistence in the context of population bottlenecks. RNA Biol. 16, 588–594. doi: 10.1080/15476286.2019.1578608
Deatherage, D. E., and Barrick, J. E. (2014). Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188. doi: 10.1007/978-1-4939-0554-6_12
Dedrick, R. M., Guerrero-Bustamante, C. A., Garlena, R. A., Russell, D. A., Ford, K., Harris, K., et al. (2019). Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25, 730–733. doi: 10.1038/s41591-019-0437-z
Donovan, L., Welford, S. M., Haaga, J., Lamanna, J., and Strohl, K. P. (2010). Hypoxia–implications for pharmaceutical developments. Sleep Breath 14, 291–298. doi: 10.1007/s11325-010-0368-x
Forti, F., Roach, D. R., Cafora, M., Pasini, M. E., Horner, D. S., Fiscarelli, E. V., et al. (2018). Design of a broad-range bacteriophage cocktail that reduces Pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob. Agents Chemother. 2018:62. doi: 10.1128/AAC.02573-17
Garoff, L., Pietsch, F., Huseby, D. L., Lilja, T., Brandis, G., and Hughes, D. (2020). Population bottlenecks strongly influence the evolutionary trajectory to fluoroquinolone resistance in Escherichia coli. Mol. Biol. Evol. 37, 1637–1646. doi: 10.1093/molbev/msaa032
Gil-Marques, M. L., Pachon-Ibanez, M. E., Pachon, J., and Smani, Y. (2018). Effect of hypoxia on the pathogenesis of Acinetobacter baumannii and Pseudomonas aeruginosa in vitro and in murine experimental models of infection. Infect Immun. 86, e543–e518.
Golec, P., Karczewska-Golec, J., Los, M., and Wegrzyn, G. (2014). Bacteriophage T4 can produce progeny virions in extremely slowly growing Escherichia coli host: comparison of a mathematical model with the experimental data. FEMS Microbiol. Lett. 351, 156–161. doi: 10.1111/1574-6968.12372
Hatfull, G. F., Dedrick, R. M., and Schooley, R. T. (2022). Phage therapy for antibiotic-resistant bacterial infections. Annu. Rev. Med. 73, 197–211.
Hocquet, D., Petitjean, M., Rohmer, L., Valot, B., Kulasekara, H. D., Bedel, E., et al. (2016). Pyomelanin-producing Pseudomonas aeruginosa selected during chronic infections have a large chromosomal deletion which confers resistance to pyocins. Environ. Microbiol. 18, 3482–3493. doi: 10.1111/1462-2920.13336
Hodges, F. E., Sicheritz-Pontén, T., and Clokie, M. R. J. (2021). The effect of oxygen availability on bacteriophage infection: a review. Phage 2, 16–25. doi: 10.3390/v12101091
Hyman, P., and Abedon, S. T. (2009). Practical methods for determining phage growth parameters. Methods Mol. Biol. 501, 175–202.
Johri, A. V., Johri, P., Hoyle, N., Pipia, L., Nadareishvili, L., and Nizharadze, D. (2021). Case report: chronic bacterial prostatitis treated with phage therapy after multiple failed antibiotic treatments. Front. Pharmacol. 12:692614. doi: 10.3389/fphar.2021.692614
Kropinski, A. M. (2009). Measurement of the rate of attachment of bacteriophage to cells. Methods Mol. Biol. 501, 151–155.
Labrie, S. J., Samson, J. E., and Moineau, S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327.
Lam, J. S., Taylor, V. L., Islam, S. T., Hao, Y., and Kocincova, D. (2011). Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front. Microbiol. 2:118.
Le, Q. T., Chen, E., Salim, A., Cao, H., Kong, C. S., Whyte, R., et al. (2006). An evaluation of tumor oxygenation and gene expression in patients with early stage non-small cell lung cancers. Clin. Cancer Res. 12, 1507–1514.
Lebeaux, D., Merabishvili, M., Caudron, E., Lannoy, D., Van Simaey, L., Duyvejonck, H., et al. (2021). A case of phage therapy against pandrug-resistant Achromobacter xylosoxidans in a 12-year-old lung-transplanted cystic fibrosis patient. Viruses 13:60. doi: 10.3390/v13010060
Levin, B. R., Lipsitch, M., and Bonhoeffer, S. (1999). Population biology, evolution, and infectious disease: convergence and synthesis. Science 283, 806–809.
Luong, T., Salabarria, A. C., and Roach, D. R. (2020b). Phage therapy in the resistance era: where do we stand and where are we going? Clin. Ther. 42, 1659–1680. doi: 10.1016/j.clinthera.2020.07.014
Luong, T., Salabarria, A. C., Edwards, R. A., and Roach, D. R. (2020a). Standardized bacteriophage purification for personalized phage therapy. Nat. Protoc. 15, 2867–2890.
Malhotra, S., Hayes, D. Jr., and Wozniak, D. J. (2019). Cystic fibrosis and Pseudomonas aeruginosa: The host-microbe interface. Clin. Microbiol. Rev. 32, e138–e118.
Mayer-Hamblett, N., Ramsey, B. W., Kulasekara, H. D., Wolter, D. J., Houston, L. S., Pope, C. E., et al. (2014). Pseudomonas aeruginosa phenotypes associated with eradication failure in children with cystic fibrosis. Clin. Infect Dis. 59, 624–631. doi: 10.1093/cid/ciu385
Mizuno, C. M., Luong, T., Cederstrom, R., Krupovic, M., Debarbieux, L., and Roach, D. R. (2020). Isolation and characterization of bacteriophages that infect Citrobacter rodentium, a model pathogen for intestinal diseases. Viruses 2020:12. doi: 10.3390/v12070737
Moradali, M. F., Ghods, S., and Rehm, B. H. (2017). Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell Infect Microbiol. 7:39. doi: 10.3389/fcimb.2017.00039
Nabergoj, D., Modic, P., and Podgornik, A. (2018). Effect of bacterial growth rate on bacteriophage population growth rate. Microbiologyopen 7:e00558.
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214.
Page, L. K., Staples, K. J., Spalluto, C. M., Watson, A., and Wilkinson, T. M. A. (2021). Influence of hypoxia on the epithelial-pathogen interactions in the lung: implications for respiratory disease. Front. Immunol. 12:653969. doi: 10.3389/fimmu.2021.653969
Pirnay, J. P., Verbeken, G., Ceyssens, P. J., Huys, I., De Vos, D., Ameloot, C., et al. (2018). The Magistral Phage. Viruses 10:64.
Priebe, G. P., Dean, C. R., Zaidi, T., Meluleni, G. J., Coleman, F. T., Coutinho, Y. S., et al. (2004). The galU Gene of Pseudomonas aeruginosa is required for corneal infection and efficient systemic spread following pneumonia but not for infection confined to the lung. Infect Immun. 72, 4224–4232. doi: 10.1128/IAI.72.7.4224-4232.2004
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A., and Korobeynikov, A. (2020). Using SPAdes De Novo Assembler. Curr. Protoc. Bioinformatics 70:e102.
Raya, R. R., Oot, R. A., Moore-Maley, B., Wieland, S., Callaway, T. R., Kutter, E. M., et al. (2011). Naturally resident and exogenously applied T4-like and T5-like bacteriophages can reduce Escherichia coli O157:H7 levels in sheep guts. Bacteriophage 1, 15–24. doi: 10.4161/bact.1.1.14175
Roach, D. R., Leung, C. Y., Henry, M., Morello, E., Singh, D., Di Santo, J. P., et al. (2017). Synergy between the host immune system and bacteriophage Is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe 22:e34. doi: 10.1016/j.chom.2017.06.018
Rodriguez-Rojas, A., Mena, A., Martin, S., Borrell, N., Oliver, A., and Blazquez, J. (2009). Inactivation of the hmgA gene of Pseudomonas aeruginosa leads to pyomelanin hyperproduction, stress resistance and increased persistence in chronic lung infection. Microbiology 155, 1050–1057. doi: 10.1099/mic.0.024745-0
Schaible, B., Rodriguez, J., Garcia, A., Von Kriegsheim, A., Mcclean, S., Hickey, C., et al. (2017). Hypoxia reduces the pathogenicity of Pseudomonas aeruginosa by decreasing the expression of multiple virulence factors. J. Infect Dis. 215, 1459–1467. doi: 10.1093/infdis/jix139
Schaible, B., Taylor, C. T., and Schaffer, K. (2012). Hypoxia increases antibiotic resistance in Pseudomonas aeruginosa through altering the composition of multidrug efflux pumps. Antimicrob. Agents Chemother. 56, 2114–2118. doi: 10.1128/AAC.05574-11
Shao, Y., and Wang, I. N. (2008). Bacteriophage adsorption rate and optimal lysis time. Genetics 180, 471–482.
Shareefdeen, H., and Hynes, A. P. (2021). Does over a century of aerobic phage work provide a solid framework for the study of phages in the gut? Anaerobe 68:102319. doi: 10.1016/j.anaerobe.2021.102319
Sommer, M. O. A., Munck, C., Toft-Kehler, R. V., and Andersson, D. I. (2017). Prediction of antibiotic resistance: time for a new preclinical paradigm? Nat. Rev. Microbiol. 15, 689–696.
Walczak, J. J., Wang, L., Bardy, S. L., Feriancikova, L., Li, J., and Xu, S. (2012). The effects of starvation on the transport of Escherichia coli in saturated porous media are dependent on pH and ionic strength. Colloids Surf B Biointerfaces 90, 129–136. doi: 10.1016/j.colsurfb.2011.10.010
WHO (2017). Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. Geneva: World Health Organization.
Worlitzsch, D., Tarran, R., Ulrich, M., Schwab, U., Cekici, A., Meyer, K. C., et al. (2002). Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109, 317–325. doi: 10.1172/JCI13870
You, L., Suthers, P. F., and Yin, J. (2002). Effects of Escherichia coli physiology on growth of phage T7 in vivo and in silico. J. Bacteriol. 184, 1888–1894.
Keywords: bacteriophages, hypoxia, oxygen, phage resistance, pyomelanin, phage therapy, abiotic, galU
Citation: Schumann AR, Sue AD and Roach DR (2022) Hypoxia Increases the Tempo of Phage Resistance and Mutational Bottlenecking of Pseudomonas aeruginosa. Front. Microbiol. 13:905343. doi: 10.3389/fmicb.2022.905343
Received: 27 March 2022; Accepted: 16 June 2022;
Published: 01 August 2022.
Edited by:
Heejoon Myung, Hankuk University of Foreign Studies, South KoreaReviewed by:
Malgorzata Barbara Lobocka, Institute of Biochemistry and Biophysics (PAN), PolandCopyright © 2022 Schumann, Sue and Roach. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dwayne R. Roach, ZHdheW5lLnJvYWNoQHNkc3UuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.