
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 11 July 2022
Sec. Microbial Symbioses
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.904815
Bacterial and viral diseases in aquaculture result in severe production and economic losses. Among pathogenic bacteria, species belonging to the Vibrio genus are one of the most common and widespread disease-causing agents. Vibrio infections play a leading role in constraining the sustainable growth of the aquaculture sector worldwide and, consequently, are the target of manifold disease prevention strategies. During the early, larval stages of development, Vibrio species are a common cause of high mortality rates in reared fish and shellfish, circumstances under which the host organisms might be highly susceptible to disease preventive or treatment strategies such as vaccines and antibiotics use, respectively. Regardless of host developmental stage, Vibrio infections may occur suddenly and can lead to the loss of the entire population reared in a given aquaculture system. Furthermore, the frequency of Vibrio–associated diseases in humans is increasing globally and has been linked to anthropic activities, in particular human-driven climate change and intensive livestock production. In this context, here we cover the current knowledge of Vibrio infections in fish aquaculture, with a focus on the model species gilthead seabream (Sparus aurata), a highly valuable reared fish in the Mediterranean climatic zone. Molecular methods currently used for fast detection and identification of Vibrio pathogens and their antibiotic resistance profiles are addressed. Targeted therapeutic approaches are critically examined. They include vaccination, phage therapy and probiotics supplementation, which bear promise in supressing vibriosis in land-based fish rearing and in mitigating possible threats to human health and the environment. This literature review suggests that antibiotic resistance is increasing among Vibrio species, with the use of probiotics constituting a promising, sustainable approach to prevent Vibrio infections in aquaculture.
Global seafood production including fish, crustaceans, molluscs, and other aquatic animals but excluding aquatic mammals, reptiles, seaweeds, and other aquatic plants, was estimated to reach 179 million tonnes in 2018, with an approximate first sale value of 401 billion US$, being the aquaculture sector responsible for 250 billion US$. This corresponds to a worldwide production of 82.1 million tonnes derived from aquaculture practices compared with 96 million tonnes from wild captures (FAO, 2020) (Figure 1A). Totals of 22.2 million tonnes were used, in that year, for fish meal and fish oil production, while 156.4 million tonnes were used for human consumption, matching the demand for seafood production by the growing human population, which reached a record high of 20,5 kg per capita (FAO, 2020). Global fish production, including capture and aquaculture, both for human consumption and ancillary purposes, is still growing worldwide, mainly due to the contribution of the aquaculture sector (Figure 1A) (FAO, 1996, 2002, 2004, 2010, 2012, 2016, 2018, 2020). In fact, 52% of the fish biomass produced for human consumption currently derives from aquaculture activities (FAO, 2020). Global fish production is dominated by China (35%), closely followed by the remainder of the Asian continent (34%), Americas (14%), Europe (10%), Africa (7%) and Oceania (1%) (FAO, 2020).
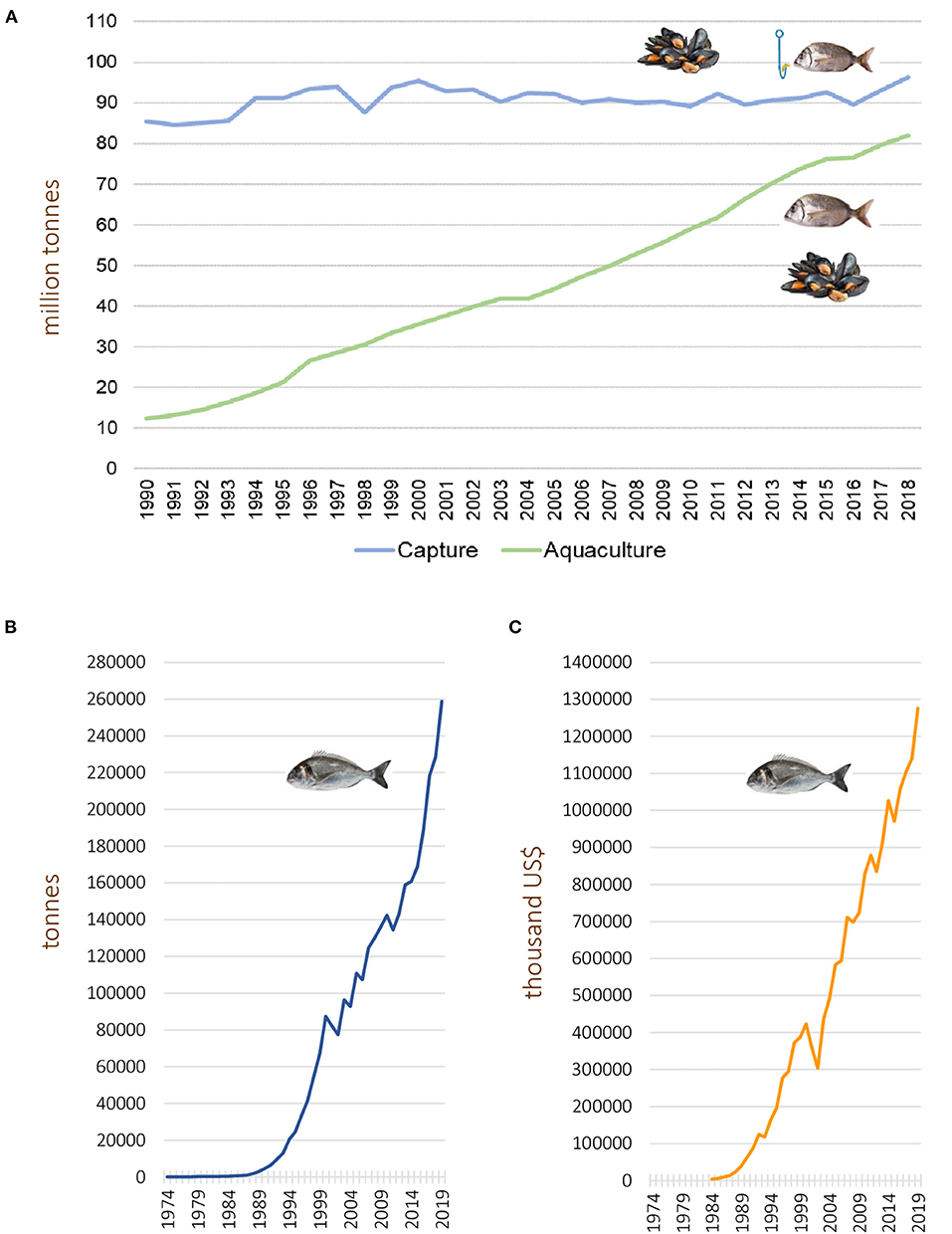
Figure 1. (A) Evolution of wild capture and aquaculture-based seafood production (million tonnes, live weight). The total seafood production is growing due to the increase in fish / shellfish biomass derived from the aquaculture sector, contrasting with near constant capture values. The data include fish, crustaceans, molluscs, and other cultured aquatic animals, and were retrieved from reports by the Food and Agriculture Organization of the United Nations (FAO) spanning the period (FAO, 1996, 2002, 2004, 2010, 2012, 2016, 2018, 2020). (B,C) show the worldwide gilthead seabream aquaculture production (in tonnes) (B) and their commercial value (thousand US$) (C). Data collected from FAO, query online, http://www.fao.org/fishery/statistics/global-aquaculture-production/query/en.
Given the obvious growth of the aquaculture industry and the emerging pressures it causes on human and environmental health, including the spread of bacterial diseases, this review covers alternative approaches to antibiotics and antimicrobials usage to suppress bacterial diseases in fish larvi- and aquaculture. Where appropriate, we place focus on studies of the model, cultured teleost fish gilthead seabream (Sparus aurata), an economically valuable reared species of relevance in Mediterranean countries. Our approach to pathogenicity in aquaculture emphasizes the opportunistic Vibrio species, highlighting environmental and public health concerns resulting from seafood vibriosis as well as human vibriosis acquired via seafood ingestion. In this context, the term vibriosis is herein defined as any sort of disease with clearly observable symptoms caused by Vibrio species on an animal host. PCR-based detection of virulence factors and mass spectrometry protocols used in the identification of Vibrio pathogens in fish and shellfish are thoroughly examined. Further emphasis is given to the existence of determinants of antibiotic resistance in Vibrio species present in commercial seafood products, given that they increase the risk of spread of antibiotic resistance genes from aquaculture to the consumer. New approaches for prophylaxis and treatment of vibriosis in fish relying on the management of pathobiomes and microbial communities in the aquaculture sector are then discussed, including the application of vaccines, bacteriophages, and probiotics to prevent bacterial disease proliferation. Particularly, we provide an overview of probiotics-based studies designed to supress Vibrio spp. across a broad range of host animals and aquaculture settings, portraying a solid body of work accumulated during the last 30 years which evidences great potential in the administration of probiotics for the control of vibriosis.
The marine perch-like fish Sparus aurata (Linnaeus, 1758), commonly known as gilthead seabream, is an economically valuable cultured species in southern European countries (Balebona et al., 1998b), ranking along with seabass as the most important fish species farmed in the Mediterranean zone (Firmino et al., 2019). World gilthead seabream aquaculture production, with regard to both quantity (tonnes) and value (thousand US$), has shown a consistent, continuous growth during the past three decades (Figures 1B,C).
Because of its plasticity and high amenability to rearing conditions, gilthead seabream can be cultured following extensive and semi-intensive methods in coastal ponds and lagoons. The extensive method relies partially on the species' natural migration and subsequent caught into fishing traps. Source juveniles obtained this way are, then, usually supplemented with additional juveniles reared in hatcheries by most of the modern stations employing the extensive method (FAO, 2021). A starting juvenile pool is, this way, seeded into a coastal lagoon, with juveniles (c. 45 DAH) weighting 2–3 g on average. Under this system, a juvenile achieves the first commercial size of 350 g in 20 months, with an average yield of 15–30 Kg/ha/yr and fish densities usually not exceeding 0.0025 kg/m3. Within semi-intensive rearing conditions, the increase of inputs derived from human activities (e.g., artificial feed and supplemental oxygen) results in a greater average production yield of 500–2,400 kg/ha/yr and higher fish densities of c. 1 kg/m3 (FAO, 2021).
Intensive rearing methods, in their turn, result in much higher yields in comparison with extensive and semi-intensive rearing methods. The densities of fish grown under this system, when raised in tanks receiving massive oxygen supply under optimal temperature conditions (18–26°C), are typically very high (15–45 kg/m3). In these circumstances, pre-fattened 5 g gilthead seabream may achieve the first commercial weight of 350 g in 1 year (FAO, 2021). Rearing of gilthead seabream in sea cages is a widely adopted methodology in the Mediterranean Sea, whereby reared fish densities may reach up to 10–15 kg/m3. Although intensive fish biomass production in sea cages is somewhat lower than that of land-based installations, the profits are much higher as there are no energy costs for pumping, aeration, or post-rearing water treatment (FAO, 2021). The main disadvantage is the absence of temperature control in sea cages and, consequently, the longer rearing period needed to reach the commercial size and to stock larger juveniles. Under this method, the larger, pre-fattened gilthead seabream (10 g) may take 1 year to reach the first commercial size of 350–400 g, while smaller seeding juveniles (5 g) achieve the same size in 16 months (FAO, 2021). Figure 2 lists the top ten gilthead seabream producing countries in the Mediterranean area. Interestingly, although not ranking among the three most producing countries in terms of quantity (tonnes/year), Italy (8,88 US$/kg), Portugal (7,59 US$/kg), Croatia (6,59 US$/kg), and Spain (5,73 US$/kg), in this order, were the countries presenting the highest commercial value of cultured gilthead seabream sold in the market.
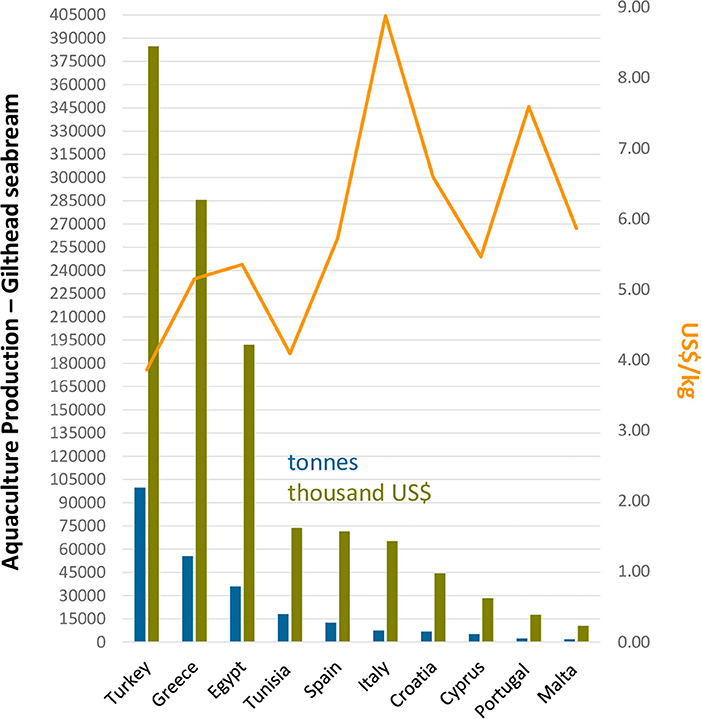
Figure 2. The top ten gilthead seabream producing countries in the Mediterranean zone. Colored bars represent quantity (tonnes) and commercial value (thousand US$/kg) of cultured gilthead seabream biomass produced for human consumption in 2019. The primary Y-axis represents quantity in tonnes, while the secondary axis represents the commercial value per kg of cultured gilthead seabream sold. Data collected from FAO, query online, http://www.fao.org/fishery/statistics/global-aquaculture-production/query/en.
Aquaculture facilities constitute a high-density species environment where the use of live feed, the stress and the animals' physical proximity increase the propagation of parasites and diseases (Guidi et al., 2018; Sanches-Fernandes et al., 2021a). Therefore, monitoring quality, safety, and microbiological indicators, across all production stages, will play a decisive role in the development of future, sustainable and cost-effective aquaculture practices (FAO, 2020). One main problem to be overcome is the fact that most fish species display very low survival rates during larval rearing, which can be partially attributed to the spread of bacterial diseases (Snoussi et al., 2008; Sanches-Fernandes et al., 2021a). In fact, bacterial diseases are responsible for mass stock mortalities in fish farms throughout Mediterranean waters, independently of the reared species and host developmental stage (Bordas et al., 1996; Akayli and Timur, 2002; Kahla-Nakbi et al., 2006), resulting in a significant production bottleneck. In this context, it is not only essential to move beyond traditional microbiological assessments of food items using more efficient methodologies, in particular next generation DNA sequencing, to ensure effective monitoring of microorganisms in animal and plant-derived foods and tissues (Lorenzo et al., 2018). It is also important to advance our understanding of the roles played by beneficial microorganisms in aquaculture facilities to effectively steer these built ecosystems toward a more environmentally friendly state, whereby disease proliferation and pollution can be mitigated using natural resources (Vadstein et al., 2013; Borges et al., 2021). Borges et al. (2021) have recently reviewed the diversity and properties of potentially beneficial microbes that occur in aquaculture facilities, including a vast diversity of Alphaproteobacteria species belonging to the Roseobacter clade (e.g., Phaeobacter inhibens) which rank as promising probiotic candidates to control bacterial diseases in these settings (see more in section Microbial-Based Strategies to Prevent Vibrio Diseases in Aquaculture).
In intensive larval rearing of commercial fish species, live feed provision is still mostly required, usually including rotifers (Brachionus spp.) as the first feed item provided, followed by brine shrimp Artemia sp. at nauplii and metanauplii developmental stages, according to the mouth size of the growing fish larvae (Pousão-Ferreira, 2009). Although strains of the genera Pseudomonas (Skjermo and Vadstein, 1993; Rombaut et al., 2001), Aeromonas (Dhert et al., 2001), Flavobacterium (Skjermo and Vadstein, 1993; Dhert et al., 2001; Rombaut et al., 2001), Marinomonas and Pseudoalteromonas (Rombaut et al., 2001) have been quite commonly found in cultured rotifers, Vibrio species were the dominant bacteria associated with these animals according to early, cultivation-dependent studies (Verdonck et al., 1997). Altogether, the ingestion of rotifers and Artemia is a potential mechanism of transport of various pathogens into fish larvae. In addition, intake of pathogens from water by fish larvae is a concern, even within recirculation aquaculture systems (RAS), which are regarded as the safest in terms of disease control (Vadstein et al., 2013). Therefore, in larviculture facilities which need to use high densities of both rotifers and Artemia as live feed to fish larvae, the high load of organic matter present in water increases the risk of proliferation of opportunistic pathogenic bacteria to the developing fish host (Verdonck et al., 1997; Rombaut et al., 2001; Prol-García et al., 2010; Haché and Plante, 2011; Asok et al., 2012; Vadstein et al., 2013; Interaminense et al., 2014). Section Vibrio species and Vibriosis in Aquaculture provides an overview of Vibrio spp. already reported in association with fish live feed.
The pathogenic load in fish larviculture stations is believed to be determined by the complexity and diversity of the microbial communities occurring in the microhabitats that constitute these multifaceted, man-made ecosystems, including the rearing water itself, particulate organic materials deriving from animal excretions and dietary foods, the fish host, and the live feed (Califano et al., 2017). As such, host-microbe, microbe-microbe, and microbe-environment interactive forces that prevail in each rearing setting are thought to determine the final state of aquaculture microbiomes across a theoretical symbiome–pathobiome continuum. As our ability to catalog the diversity and function of host-associated microbiomes in a cultivation-independent manner increases, the molecular mechanisms underpinning host colonization, persistence and disease development by opportunistic microorganisms are predicted to be revealed at a fast pace. It is relevant that the functional attributes of pathobiomes and symbiomes of fish larvae, juveniles and live feed are uncovered, so that disease control in aquaculture can be implemented in a consistent manner (Borges et al., 2021). Notably, there is currently a demand for the development of faster, more precise, and accurate molecular methods (see section Identification of Vibrio Pathogens in Aquaculture), beyond common rRNA gene amplicon sequencing, to better identify the microorganisms present in the pathobiome, opening new avenues to understand pathogenic mechanisms in aquaculture (Vayssier-Taussat et al., 2014).
Vibrio species are Gram-negative, asporogenous rods that are straight or curved, motile in aqueous environments usually by means of a single, polar flagellum (Kaysner et al., 2004). Vibrio spp. are mesophilic and chemoorganotrophic, possessing facultative fermentative metabolism (Kahla-Nakbi et al., 2007). They are ubiquitous inhabitants of aquatic environments including estuaries, marine coastal waters and sediments, and aquaculture settings (Balebona et al., 1998a; Thompson et al., 2004; Sarjito et al., 2009; Ringø, 2020). Except for V. cholerae and V. mimicus (Wong and Griffin, 2018), they are considered halophilic organisms (Wong and Griffin, 2018) commonly occurring at 30–35 ppt salinity although their aptitude to thrive in estuarine environments is also well-documented. The first Vibrio species described was Vibrio cholerae, in 1854, in the context of a study on cholera outbreaks in Florence (Thompson et al., 2004), but there are records of cholera-like diseases occurring in the times of Hippocrates (460–377 BC) (Blake, 1994). Currently, more than 130 species grouped in 14 clades in the Vibrio genus are recognized (Romalde et al., 2014; Huang et al., 2020), including commensal, mutualistic, and pathogenic species (Thompson et al., 2004).
The role of Vibrio spp. in marine organic carbon cycling (Romalde et al., 2014), particularly in coastal environments and marginal seas, has been underestimated (Zhang et al., 2018). Vibrio species are one of the best model marine heterotrophic bacterial groups, consuming several carbon compounds and growing with generation times as short as ~10 min (Zhang et al., 2018). They may represent about 60% of the total heterotrophic bacteria associated with aquatic organisms (Sonia and Lipton, 2012), being part of the normal microbiota of aquatic animals. Vibrio hosts are typically zooplankton, shellfish, crustaceans, benthic marine invertebrates such as sponges, corals and bryozoans, and fishes (Liu et al., 2016). Host-Vibrio relationships in nature may range from mutualistic through commensalistic to pathogenic (Liu et al., 2016). In general, Vibrio species proliferate well at warm temperatures, a condition that may favor their transition from commensal to pathogenic behavior. Environmental factors, including warming, have also been suggested to suppress fish immunity and increase their susceptibility to vibriosis (Haenen et al., 2014; El-Bouhy et al., 2016; El-Sayed et al., 2019).
In aquaculture, several Vibrio spp. are currently considered pathogens or opportunistic pathogens of reared finfish, shellfish, and shrimp (Liu et al., 2016). The most common Vibrionaceae spp. recorded in association with fish and shellfish diseases are V. anguillarum, V. ordalii, V. vulnificus, V. alginolyticus (Vera et al., 1991; Kahla-Nakbi et al., 2007; Korun and Timur, 2008), V. parahaemolyticus (Hamdan et al., 2016), Aliivibrio (formerly Vibrio; Urbanczyk et al., 2007) salmonicida, V. harveyi (Kahla-Nakbi et al., 2007; Korun and Timur, 2008), and V. tubiashii (Richards et al., 2014). Most of these species have been isolated both from reared and wild marine fish (Abdelaziz et al., 2017). Although V. cholerae is not referred to as a primary fish pathogen following Koch's postulates, it has been isolated from several freshwater and marine fish, which are considered a broad reservoir of V. cholerae strains that may cause infections in humans (Halpern and Izhaki, 2017). More recently, Devi et al. (2022) reported on a non-O1, non-O139 V. cholerae serotype (EMM1) capable of inducing high mortality in the freshwater species Labeo rohita, suggesting that V. cholerae strains other than the typical human pathogens shall be considered relevant aquatic pathogens as well. Vibriosis caused by the abovementioned species is the most common and devastating bacterial disease in fish larviculture and aquaculture, being a public health and economical concern affecting marine fishes, crustaceans, and bivalves worldwide (Balebona et al., 1998a; Sarjito et al., 2009; Ringø, 2020). The symptoms of vibriosis in fish are diverse, and include haemorrhagic septicaemias with extensive external skin lesions (haemorrhagic fins and ulcers), focal necrosis of some organs (liver, spleen, kidney), other tissue necrosis (Kahla-Nakbi et al., 2007), complete erosion of tail (Haldar et al., 2010), pale kidney, dark pigmentation, exophthalmic eyes, splenomegaly (Zorrilla et al., 2003), skeletal deformity (lordosis) (Abdel-Aziz et al., 2013), loss of appetite and lethargy (Korun and Timur, 2008).
Regarding the presence of Vibrio spp. in association with the live feed used for fish larviculture, several studies reported the isolation of the well-known causative agents of disease Vibrio alginolyticus (Yu et al., 1990), V. anguillarum (Dhert et al., 2001), V. parahaemolyticus (Balebona et al., 1998b) and V. rotiferianus (Gomez-Gil et al., 2003) from rotifers. The aquatic crustacean genus Artemia may also host Vibrio (Igarashi et al., 1989; Pousão-Ferreira, 2009), Pseudomonas (Igarashi et al., 1989) and Aeromonas (Interaminense et al., 2014) species. For instance, V. alginolyticus (Soto-Rodriguez et al., 2003; Interaminense et al., 2014), V. parahaemolyticus (Interaminense et al., 2014; Kumar et al., 2018), V. anguillarum (Campbell et al., 1993; Skjermo and Bergh, 2004), V. harveyi (Asok et al., 2012) and V. hispanicus (Gomez-Gil et al., 2004) have already been isolated from Artemia. Luminescent vibriosis (that is, vibriosis caused by luminescent Vibrio species) was as well-reported in Artemia and found to be caused mainly by V. harveyi and occasionally by V. splendidus (Soto-Rodriguez et al., 2003). Moreover, V. campbellii, frequently misidentified as V. harveyi in the past, was more recently found to be the etiological agent of luminescent vibriosis in shrimp hatcheries (Kumar et al., 2021).
Severe vibriosis in humans can be acquired by ingestion of contaminated water and raw or undercooked seafood (Wachsmuth et al., 1994; Finkelstein et al., 2002; Arab et al., 2020; Håkonsholm et al., 2020). Clinically, a few Vibrio species, despite their prevalently marine/estuarine origin, are able to elicit disease in humans. These include V. cholerae, V. parahaemolyticus, V. vulnificus (Wachsmuth et al., 1994; Finkelstein et al., 2002; Arab et al., 2020), V. alginolyticus (Gomathi et al., 2013; Citil et al., 2015), V. metschnikovii (Gomathi et al., 2013; Arab et al., 2020; Konechnyi et al., 2021), V. mimicus (Hernández-Robles et al., 2021), V. cincinnatiensis (Brayton et al., 1986), V. fluvialis (Ramamurthy et al., 2014; Kitaura et al., 2020), V. furnissi (Dalsgaard et al., 1997) and V. harveyi (Arab et al., 2020; Brehm et al., 2020). Well-documented symptoms of vibriosis in humans caused by Vibrio species which act as fish pathogens in aquaculture settings are highlighted below.
Vibrio alginolyticus had been frequently documented in early studies of gilthead seabream disease outbreaks in Mediterranean aquaculture (Balebona et al., 1998a). In humans, this bacterium was found to be associated with gastroenteritis in immunocompromised patients (Reina et al., 1995; Gomathi et al., 2013), causing extra-intestinal diseases (Gomez-Gil et al., 2003; Snoussi et al., 2008), wound infection, cellulitis, seawater-related otitis media (Abdel-Aziz et al., 2013; Gomathi et al., 2013), soft tissues and septicemia (Gomathi et al., 2013).
Vibrio parahaemolyticus is a well-known fish pathogen possessing a broad range of occurrence (Kumar et al., 2018). This bacterium was first recognized as a seafood borne pathogen to humans during an outbreak in 1950 in Osaka, Japan, involving 272 patients and causing the death of 20 people after the ingestion of Shirasu, a semi dried juvenile sardine (Aly et al., 2020). V. parahaemolyticus is the major food-borne pathogen worldwide (Bresee et al., 2002; Kawatsu et al., 2006), causing, after the ingestion of raw or undercooked seafood, acute dysentery and abdominal pain leading to diarrhea, nausea, vomiting, fever, chills, water-like stools, and an accentuated decrease of blood pressure leading to shock (Broberg et al., 2011; Siddique et al., 2021; Tan et al., 2021). In severe cases, patients become unconscious, with recurrent convulsions, becoming pale or cyanotic, eventually resulting in death. Antibiotic treatment and oral rehydration are the most common procedures to cure infections caused by V. parahaemolyticus. For individuals with critical physical or immunodeficiency diseases, the best practice to avoid severe illness is not to consume seafood at all (Wang et al., 2015). In the 21st century, increasing human disease outbreaks attributed to V. parahaemolyticus in Asia (Matsumoto et al., 2000), North America and Chile (Martinez-Urtaza et al., 2005), Europe namely France and Spain (Martinez-Urtaza et al., 2005; Quilici et al., 2005), Africa and Russia (Nair et al., 2007) have been described.
Vibrio vulnificus is highly pathogenic to humans (Snoussi et al., 2008). This bacterium causes epizootic outbreaks in seabream fish and can be transmitted to humans by ingestion, being a well-known cause of cellulitis and septicaemia in fishermen (Vinh et al., 2006). Besides, V. vulnificus is also able to infect the human host through an open cut or wound, in extreme cases resulting in necrotizing fasciitis, limb amputation and fatal septicaemia in susceptible individuals (Williams et al., 2014). The wound infections could start after the handling of infected fish and seafood, especially shellfish and after the practice of aquatic activities such as swimming (Hamdan et al., 2016; Baker-Austin and Oliver, 2018), being the consequences more severe when associated illnesses such as liver diseases, diabetes, and immune disorders are documented (Baker-Austin and Oliver, 2018). As usual among Vibrio spp., V. vulnificus possesses remarkable iron sequestration capabilities, meaning that the risk of infection is higher in humans with elevated iron levels (Wong and Griffin, 2018). More than 50% of primary septicaemia result in death within the first 72 h of hospitalization (Yun and Kim, 2018). Therefore, when there is a suspicion that the infection is caused by V. vulnificus, immediate and adequate antibiotic treatment and surgical interventions must be implemented. V. vulnificus is responsible for over 95% of deaths associated with seafood occurrences in the United States of America (Baker-Austin and Oliver, 2018). This is the highest fatality rate of any food-borne pathogen, which is in the range of category Biosafety Level 3 and 4 pathogens, namely anthrax, bubonic plague, Ebola, and Marburg fever (Baker-Austin and Oliver, 2018).
More recently, a few human infections caused by V. harveyi have as well been reported, underscoring the need of the public health sector to be aware of the possibility that wound infections caused by Vibrio species to humans may be becoming more likely to occur. With global warming, Vibrio-associated diseases will likely increase in the future (Brehm et al., 2020).
To avoid severe illness in humans caused by the ingestion of seafood contaminated with Vibrio species, thermal-based food processing such as low-temperature freezing (−18 or −24°C) or a 10 min high-temperature treatment (above 55°C) is common practice (Wang et al., 2015). Moreover, high-pressure processing and irradiation (using safety radioactive materials limits) are used to eliminate V. parahaemolyticus in oysters, keeping their original flavor (Wang et al., 2015). Notwithstanding the efficacy of hygiene measures employed in the preparation and processing of seafood for human consumption, devising novel, sustainable and green mechanisms of bacterial disease prevention in intensive fish farming holds promise in mitigating the impacts of the aquaculture industry on the environment and the risks posed to human health.
In 1997, the World Bank estimated that disease losses in aquaculture were worth US$3 billion per year, with Vibrio spp. having an important role in those losses (Laczka et al., 2014). Two decades later, estimates of disease losses duplicated (Stentiford et al., 2017). It has been suggested that all cultured marine fish around the world may, to varying degrees, host opportunistic vibrio species (Akayli and Timur, 2002), what does not necessarily imply that disease is always elicited nor that all vibrio species are pathogenic or will present pathogenic behavior. Yet multiple vibriosis outbreaks have been reported in several countries, infecting many fish species (Colorni et al., 1981; Akayli and Timur, 2002; Korun and Timur, 2008). In the case of the Mediterranean Sea, which is the primary habitat of gilthead seabream and a semi-closed water body, there is a limited rate of water exchange with open oceans. Aquatic pollution resulting from sewage, industrial effluents, crude oil refineries, and oil exploration affects the response of cultured fish to local environmental conditions (Guidetti et al., 2002). These factors also facilitate the invasion of bacterial pathogens (Vibrio, Streptococcus, Aeromonas, Pseudomonas) and parasites (nematodes, digeneans, acanthocephalans) into rearing systems. The ongoing chronic degradation of the Mediterranean Sea, thus, is considered to negatively impact the aquaculture industry in most of the North African coast (Eissa et al., 2017). For example, it has been suggested that the deterioration of water quality by sewage and agriculture discharges correlates with high prevalence of vibriosis in wild fish in the Mediterranean coast (Abdelaziz et al., 2017). Seawater exposed to higher anthropogenic pollution was found to display higher frequencies of Vibrio species, highlighting the importance of good manufacturing and hygiene practices to prevent and overcome fish vibriosis, even if innovative and “green” approaches were applied in industrial and domestic facilities (Abdelaziz et al., 2017; Arab et al., 2020).
The analysis of ten outbreaks involving Vibrio infections, affecting both cultured and wild gilthead seabream in the Mediterranean Sea (Table 1) revealed that V. alginolyticus was the Vibrio species most frequently isolated from gilthead seabream, followed by V. harveyi, V. splendidus, V. anguillarum, V. parahaemolyticus, and V. tubiashii. In these studies, Vibrio isolates were mainly recovered from seabream liver, spleen, and kidney, followed by external lesions and gills, but also from brain, eyes, gut, hepatopancreas, eroded tail and blood (Table 1). Interestingly, V. ichthyoenteri-like strains were isolated only from asymptomatic gilthead seabream individuals (Pujalte et al., 2003).
According to the Spanish outbreak (1990–1996) study performed by Balebona et al. (1998b), the species V. anguillarum, V. alginolyticus, V. harveyi, and V. splendidus were considered highly virulent for gilthead seabream by intraperitoneal inoculation, based on mean lethal dose (LD50) values between 104 and 106 CFU per g body weight. In a further disease outbreak study, V. alginolyticus and V. harveyi were identified as virulent to gilthead seabream with LD50 values between 105 and 106 CFU per g body weight (Kahla-Nakbi et al., 2007). In that study, V. alginolyticus and V. harveyi were isolated from the skin mucus of gilthead seabream, and no inhibitory effects of the skin mucus collected from gilthead seabream against those isolates was found. In fact, those Vibrio isolates showed remarkable serum resistance and were also able to adhere to skin mucus and grow using it as a nutrient source, suggesting high host colonization ability to eventually become an important infection risk.
Intriguingly, the etiological agents of human seafood-borne infections V. alginolyticus, V. cholera, and V. fluvialis were found in tissues of farmed gilthead seabream showing no disease symptoms (Arab et al., 2020), supporting the notion of a growing presence of the causing agent of cholera, V. cholerae, in farmed fish for human consumption (Halpern and Izhaki, 2017; Arab et al., 2020). Indeed, higher incidence of human pathogenic Vibrio species in coastal marine waters has been considered to result from climate change effects on the composition of marine microbial communities (Vezzulli et al., 2016). In this context, it is worth noting that vibriosis in cultured gilthead seabream has already been found to be induced by several factors such as transport stress, sudden temperature changes, low oxygen levels in water and handling procedures (Akayli and Timur, 2002). We posit that the trends observed in this review regarding gilthead seabream-Vibrio interactions are most likely applicable to a range of economically valuable fish species.
The continuous study of marine and estuarine microbiomes in coastal areas is of utmost relevance for a better understanding of long-term microbial community changes in highly productive ecosystems in the face of climate change. Such databases can guide the identification of beneficial microbes that can be used to mitigate the proliferation of opportunistic pathogens in immunocompromised hosts in built and open environments at large. Gilthead seabream and seabass are the main farmed species in the Mediterranean basin, and vibriosis was recently reported as the most common bacterial disease affecting these species (Muniesa et al., 2020). Based on the current literature, we argue that the occurrence of vibriosis in humans—and consequently the threats to human health posed by Vibrio species thriving in aquaculture settings—may be of a larger magnitude than previously thought. The frequency of human infections caused by estuarine and marine Vibrio spp. is likely to increase as ever-expanding intensive farming and global climate change synergistically interact to favor the proliferation of opportunistic microorganisms in livestock production systems (Reverter et al., 2020).
Species-level identification of members of the Vibrio genus, in an effective and standardized way, is necessary for a better bacteriological monitoring of farmed fish and the rearing environment within aquaculture facilities (Mustapha et al., 2013). As biochemical methods of identification often misidentify or are unsuccessful at the species level, molecular approaches must be implemented as common, accurate procedures for Vibrio species identification in seafood (Mustapha et al., 2013). However, it is important to note that, owing to the large genetic heterogeneity and fast diversification within Vibrio species, often 16S rRNA gene sequencing alone does not suffice for unequivocal identification of environmental strains at the species level. Bacterial species belonging to the Vibrio genus can differ in 16S rRNA gene nucleotide sequence from <1% up to 6% (Montieri et al., 2010). Particularly in the case of closely related species, the sequencing of a single marker gene may not be enough for precise taxon differentiation. For instance, V. parahaemolyticus and V. alginolyticus show quite similar biochemical properties (Mustapha et al., 2013) and are nearly identical with regards to 16S rRNA gene sequences (Montieri et al., 2010), prompting researchers to develop early DNA-based fingerprinting methods to discern between strains belonging to these species (Sadok et al., 2013). Indeed, V. alginolyticus was early designated V. parahaemolyticus biotype 2 (Aly et al., 2020), bearing testimony to the close phylogenetic relationship between these species.
Owing to the low discriminating power of highly conserved marker genes in distinguishing close Vibrio relatives, molecular identification based on species-specific markers and virulence genes have been considered adequate methods for species-level identification of Vibrio isolates (Mustapha et al., 2013). Because Vibrio spp. are symbiotic bacteria usually living in the intestine of aquatic species in a facultative way, genomic factors involved in the establishment of symbiosis and in the processes of host colonization and persistence may have evolved to confer adaptive advantage to species thriving in subtly different micro-niches. Several so-called “virulence factors”, such as enterotoxins, haemolysins, cytotoxins, proteases, lipases, phospholipases, siderophores, adhesive factors and/or haemagglutinins are produced by pathogenic species (Zhang and Austin, 2005). These traits allow Vibrio strains to adhere to the epithelial cells of fish juveniles, to break the first barrier of natural defense and to colonize all internal organs inducing vibriosis signs (Colorni et al., 1981; Paperna, 1984; Snoussi et al., 2008). It is important to note, however, that some of the abovementioned traits are common to several bacterial species and may likewise constitute adaptive features of mutualistic symbionts of fish (Borges et al., 2021). Overall, the use of genes coding for virulence or host-colonization factors as phylogenetic markers for the molecular detection of Vibrio species has gained increasing attention lately as nucleotide heterogeneities within such genes may reveal the adaptive behavior of different Vibrio species. A few PCR-based molecular identification systems of Vibrio species are listed in Table 2. These include protocols targeting genes coding for virulence factors which have been proved useful in discerning between closely related Vibrio species or in providing solid diagnosis of renowned pathogens, as reviewed more thoroughly in Supplementary File S1. For instance, several V. alginolyticus identification methods have been established based on the detection of hemolysin and collagenase encoding genes (Abdallah et al., 2011; Mustapha et al., 2013), and specific detection of V. parahaemolyticus and V. alginolyticus has been achieved through the exploration of nucleotide differences in genes encoding for the virulence regulatory proteins ToxR and ToxS (Abdallah et al., 2011; Aly et al., 2020). Also, a conserved virulence pathogenic island among Vibrio species has been exploited in the development of specific detection systems for the pathogen V. vulnificus (Table 2, see Supplementary File S1 for details).
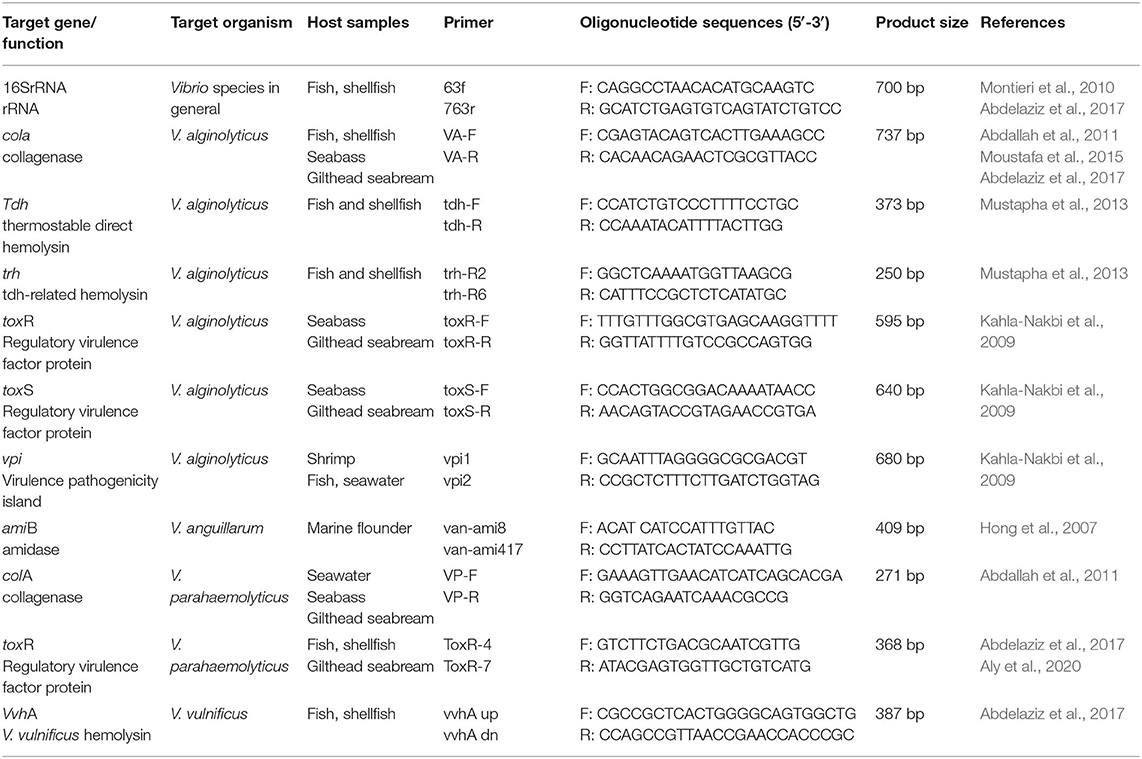
Table 2. Target genes, gene functions, and oligonucleotide primer sequences used for specific detection and identification of Vibrio species.
It has been reasoned that the use of gene-targeted molecular tools may facilitate prevention of an outbreak as they allow the identification of the potential pathogens present even in asymptomatic fish (Altinok and Kurt, 2004). Yet it is presumably challenging to implement multiple gene amplicon sequencing methods in routine diagnostics for each different pathogen. In this regard, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) is an alternative technique often used in the identification of Vibrio species that may show advantages over PCR-based detection of phylogenetic marker genes. Indeed, MALDI-TOF MS is not labor-intensive, does not require highly trained operators, and is suitable for the processing of many samples in an automated, rapid, and cost-effective way (Li et al., 2018; Mougin et al., 2020). However, for accurate identification of closely related Vibrio species using MALDI-TOF MS, database choice is crucial (as in the case of species identification using phylogenetic marker genes). For instance, Moussa et al. (2021) found that correct discrimination of isolates belonging to the species V. tubiashii/V. europaeus and V. owensii/ V. jasicida/V. campbellii could not be achieved using some of the commonly available databases for MALDI-TOF MS-based classification. However, successful identification of diverse Vibrio isolates was achieved by Mougin et al. (2020) through the combined use of the Luvibase and Bruker v.9.0.0.0 databases. Thus, to fully exploit the potential of MALDI-TOF MS in fast and accurate identification of Vibrio species in aquaculture facilities, continuous development of comprehensive databases that allow discrimination between closely related Vibrio species is fundamental.
In conclusion, the current tools for fast identification of Vibrio pathogens in aquaculture facilities, or cultured fish, usually rely on the use of toxin-encoding genes, or other alternative functional marker genes, in targeted, PCR-based approaches (Abdallah et al., 2009, 2011; Aly et al., 2020) as well as on mass spectrometry protocols which have been gaining momentum in recent years (Mougin et al., 2020; Moussa et al., 2021). The combination of highly specific molecular identification of Vibrio pathogens, either by means of gene-targeted or mass spectrometry approaches, and broad characterization of total microbial communities via high-throughput 16S rRNA gene sequencing, for instance, is likely to become an effective approach to accurately determine the presence of opportunistic/pathogenic bacteria in complex microbial communities inhabiting aquaculture facilities, which may include beneficial bacteria with the ability to supress the spread of pathogens present in the community. The steady development of well-curated databases in support of molecular diagnostic tools will play a decisive role in enabling (i) the identification of multiple pathogenic agents present in a sample including understudied organisms, such as the likely emerging pathogenic species V. chagasii (Sanches-Fernandes et al., 2021b) and V. jasicida (Sanches-Fernandes et al., 2021c); (ii) targeting the specific group of pathogenic agents typical of each facility in a straightforward manner (Stentiford, 2017). Given that each aquaculture facility is unique, with singular and distinct features (Stickney, 2016), these approaches shall be used in a complementary way and considered in a case-by-case manner.
Infections caused by drug-resistant pathogens are responsible for 700,000 annual deaths in aquaculture, estimated to reach 10 million deaths as of 2050 (O'Neill, 2015). Recent studies suggest that antibiotic resistant bacteria may not only emerge in the environment due to the use of antimicrobial agents but also due to the increase of local temperature (MacFadden et al., 2018; Reverter et al., 2020; Pepi and Focardi, 2021), since it can affect bacterial cell physiology and promote mutagenesis, allowing antibiotic resistance mutations to take place early (Pepi and Focardi, 2021). Accordingly, the occurrence of Vibrio outbreaks is commonly higher during spring and summer seasons (Aly et al., 2020). Nevertheless, V. anguillarum was referred to as the etiological agent of fish vibriosis in both warm and cold waters in aquaculture facilities (Lages et al., 2019). There is, however, evidence that Vibrio–associated diseases are increasing in a global manner because of climate change and human activities (Vezzulli et al., 2016). This highlights the urgent need for more effective actions to combat not only the indiscriminate use of antimicrobial agents but also global climate change and warming (Reverter et al., 2020; Pepi and Focardi, 2021). The development of multi-resistance traits among pathogenic Vibrio spp. has been reported steadily across several aquaculture stations worldwide (Scarano et al., 2014; Aly et al., 2020; Deng et al., 2020; Dutta et al., 2021; see Supplementary Table S1), and it may be reasonable to argue that this trend results from the synergistic effects of past and currently unsupervised antibiotics use and higher water temperatures.
The common practice and overuse of antibiotic administration for prophylactic reasons in aquaculture is an important factor to consider regarding the increase in transfer of antibiotic resistance genes to land animals and human pathogens (Costa et al., 2015). This is a global public health concern exacerbated by the fact that the increment in multiple antibiotic resistance is also observed in food-borne pathogens, opportunistic pathogens, and the commensal microbiota of animals for human consumption, resulting in antibiotic resistance in the human gastrointestinal tract (Nguyen et al., 2014). With regards to the most crucial players involved in horizontal gene transfer among Vibrio cells, along with plasmids, phages, transposons and integrons are also genomic islands and integrating conjugative elements (Rodríguez-Blanco et al., 2012; Costa et al., 2015). The inheritance of resistance traits is often acquired via conjugation of resistance plasmids (R-plasmids), which commonly contain genes encoding resistance to multiple antibiotics. R-plasmids have been reported for Vibrio capable of transferring drug resistance traits such as V. alginolyticus (Gomathi et al., 2013).
Resistance profiles of Vibrio species isolated from diseased gilthead seabream (including the most important Vibrio pathogens of fish and humans, such as V. alginolyticus, V. harveyi, V. parahaemolyticus and V. vulnificus) toward antibiotics frequently used in the aquaculture industry are summarized in Supplementary Table S1. This meta-analysis reveals that antibiotic resistance profiles may vary among strains of the same Vibrio species or across studies of the same species, which is the case of the data gathered for Vibrio alginolyticus and V. harveyi. Furthermore, we observed that most Vibrio species are sensitive to tetracycline, oxytetracycline, chloramphenicol, and florfenicol. All studies listed in Supplementary Table S1 were performed in gilthead seabream rearing facilities in the Mediterranean zone. Collectively, the data suggest a trend for increased antibiotic resistance among diverse Vibrionaceae species at the Mediterranean basin, possible to observe across time for species such as V. aestuarius (from 1998 to 2014), V. alginolyticus (from 1998 to 2014, where studies published in 2007 and 2008 were performed in the same region by the same research group), A. fischeri (from 1998 to 2003), V. harveyi (from 1998 to 2020), V. parahaemolyticus (from 2013 to 2020) and V. splendidus (from 1998 to 2003).
A more responsible and prudent use of antibiotics in the aquaculture sector is important as they are present in all production stages. In 2011, oxytetracycline was the antibiotic with the highest prescription for both prophylactic and therapeutic ends in aquaculture facilities, and also the one that is most of the times freely available (Bondad-Reantaso, 2018). There is currently no international uniformization regarding antibiotics usage approval, which are licensed by each country in accordance with their own legislation (Guidi et al., 2018). For an overview of antibiotics currently in use in aquaculture and existing policies among major producing countries, we refer the reader to the recent review by Lulijwa et al. (2020). We list some of the most frequently used antibiotics in the aquaculture sector and in the Mediterranean area in Supplementary Table S2, whereby those antibiotics approved for use in Norway, Italy, Brazil, and the United States are disclosed. We observed that Vibrio species are sensitive to several antibiotics licensed for use (Supplementary Table S1), including oxytetracycline and florfenicol. While this picture is congruent with the need of applying effective measures to deter vibriosis outbreaks, it simultaneously raises concerns regarding the development of broader multidrug resistance traits among Vibrio species. Perhaps as important as delineating which antimicrobials are permissible in what quantities and where, surveillance of the fate of antibiotics in the environment and seafood biomass is key to ensure adherence of farming stations to local/national policies. In this regard, it is worth noting that the concentration of permitted antibiotics in seafood biomass often exceeds maximum residual limits in most of the major producing countries (Lulijwa et al., 2020). This calls for an urgent up-scaling of surveillance capabilities for better traceability and follow-up of antibiotic use practices (Schar et al., 2020).
The fact that antibiotics have been usually applied for prophylactic, therapeutic, and metaphylactic purposes favors the loss of susceptibility among the target organisms, and hence an increasing trend of antibiotic resistance among Vibrio species is likely as suggested by the data present in this review. Although several antibiotics have been banned or subjected to strict regulations, particularly in industrialized countries, the legacy effects of their past and current indiscriminate use turn the development of multidrug resistance among bacterial pathogens an important and timely public health concern (Pepi and Focardi, 2021).
The negative impact of the over usage of antibiotics on farmed fish species and coastal environments worldwide urges the development of alternative methods to prevent disease proliferation and reduce ecosystem deterioration caused by emerging, multi-resistant opportunistic bacteria. The development of strategies that target bacterial pathogens based on the activation of the host's immune system (i.e., using vaccines), on biological interactions such as pathogen predation (i.e., phage therapy) and on competition/niche displacement among microbes or beneficial host-microbe interactions (e.g., using probiotics), are gaining increasing attention because they may offer a less hazardous alternative regarding the suppression of fish pathogens in aquaculture.
For an overview on the use of vaccines to prevent fish diseases, we refer the reader to the comprehensive reviews of Embregts and Forlenza (2016) and Ma et al. (2019), the latter on the promises and challenges of oral vaccine administration. In short, vaccination programmes are considered an efficient, pathogen-specific suppressive approach that is best employed for disease prevention among adult fish, especially when injection methods of antigen delivery are adopted (Embregts and Forlenza, 2016). Indeed, the implementation of efficient vaccination programmes in Norway during the nineties is nowadays considered a remarkable example of how alternative disease control methods can sharply reduce the use of antibiotics in intensive fish farming (Lulijwa et al., 2020). Presently, commercial Vibrio vaccines such as AquaVac™Vibromax™ (Wongtavatchai et al., 2010) and ALPHA JECT 3000 (PHARMAQ AS, Norway; see Torres-Corral et al., 2021 for an example of application) are available, which offer efficacy in vibriosis control in shrimp and finfish, respectively, under different administration methods, namely incubation of Artemia nauplii prior to shrimp feeding (AquaVacTMVibromaxTM; see Amatul-Samahah et al., 2020 for a review on vaccination of shrimp against vibriosis) and intraperitoneal injection of adult fish (AlphaJect 3000).
To improve fish wellbeing under vaccination programmes, oral administration methods using several modes of antigen encapsulation in delivery systems such as chitosan, alginates, and fish live feed such as Artemia and rotifers (bioencapsulation methods) have been attempted. However, improvements are still needed to ensure efficacy in antigen delivery in comparison with injection procedures. The main challenges of oral vaccine administration using encapsulation methods are to assure that vaccines reach the digestive tract of fish by ingestion, the maximum dosage allowed, which is dependent on the daily live feed intake, the time of exposure to be effective and the farmed fish tolerance to the vaccine (Embregts and Forlenza, 2016). The use of vaccines has moreover been considered not applicable to handle fish larvae and bivalves due to the lack of an adaptive immune system (Bentzon-Tilia et al., 2016), prompting researchers to consider alternative routes for disease prevention, such as the use of probiotics (see sub-section The Promise of Probiotics in Controlling Vibrio Diseases in Aquaculture).
Concerning phage therapy methods to prevent bacterial proliferation in aquaculture, the review by Richards (2014) covers pioneering studies on diverse bacteriophage-bacterial host systems and the efficacy of phage-based treatments to deter pathogens such as Aeromonas samonicida, Edwardsiella tarda, and V. harveyi, among others (Richards, 2014 and references therein). Like the vaccination approach, a key feature of phage therapy is its usual pathogen-specific nature, although the extent of specificity of the host-phage interaction may vary in case-dependent manner (Richards, 2014). In this regard, the use of phage mixtures has been considered a reasonable strategy to avoid the development of phage resistance by specific bacterial hosts while enabling the control of diverse pathogens (Richards, 2014). As for the use of vaccines and probiotics, phage dosage and delivery mode (immersion, oral via e.g., live feed ingestion, or injection) are crucial aspects for successful implementation of phage therapy (Richards, 2014; Soliman et al., 2019). To be cost-effective, phase dosage must be the lowest possible to induce bacterial infection with an associated, high phage replication rate (Soliman et al., 2019). Therefore, the use of lytic—instead of lysogenic—phages has been suggested as an imperative for the development of successful phage therapy methodologies (Richards, 2014). Since the ability to isolate and manipulate bacteriophages is strictly limited to the range of culturable bacterial hosts that can be captivated in the laboratory, and because the aquaculture pathobiome may include unculturable, or hard-to-culture, understudied bacteria, an intrinsic hurdle of the phage therapy approach relates with the development of novel methodologies leading to the control of bacterial populations for which no corresponding bacteriophages are known to date. Finally, large-scale application of phage therapy approaches in aquaculture shall be taken with caution, as concerns related with the environmental release of phages and its associated risks have been raised (Meaden and Koskella, 2013). The use of probiotics as a third, microbiome-based therapy approach to disease prevention in aquaculture is addressed below, as well as its likelihood to modulate aquaculture microbiomes toward a sustainable healthy state.
Probiotics can be defined as live bacterial species able to survive and thrive in the acidic gastric environment whose activity leads to a beneficial effect on the health of the host by re-establishing or improving the gut microbiota, when administered in adequate amounts (Zhou et al., 2020; Moroni et al., 2021), although less stringent definitions have been proposed (see Borges et al., 2021). Probiotics are ideally inoffensive and promote host fitness. A mandatory feature of commercially successful probiotics is their viability during storage and on/in the animal host. Their application may follow reasonably standardized and easy-to-implement methodology if commercial formulations are deployed (Abareethan and Amsath, 2015). Probiotics may be administered to the fish host through several mechanisms, including inoculation of the rearing water, of formulated foods or of the live feed (Verschuere et al., 2000). The use of non-pathogenic biological agents as probiotics can be highly advantageous as they may act successfully as anti-bacterial, anti-viral and anti-fungal agents (Chauhan and Singh, 2019), thus presenting the potential to increase reared fish health and rearing water quality globally (Abareethan and Amsath, 2015). This attractive way to face, prevent and combat disease among reared fishes requires (host) species-specific studies to be made on the advantages of probiotics application, as symbiotic bacteria may act as pathogenic or probiotic depending on the aquatic host species. To properly address the molecular mechanisms of action elicited by probiotics on the immune system of the host, if any, further research is needed (Hai, 2015).
Probiotics-based therapies for disease control, when applied to farmed fish, should ideally consider the use of strains native to the host. This ensures higher probabilities of effective host colonization and persistence by the probiotics in use, at the operational rearing conditions, ultimately promoting nutrient acquisition by the host and a safer environment to the reared species, humans, and surrounding ecosystems (Wanka et al., 2018; Borges et al., 2021). Several modes of action have been reported for effective probiotics. These comprise the biosynthesis of inhibitory compounds that avoid pathogen proliferation, amelioration of the host immune system, competition with pathogens for adhesion sites in the gut or for essential nutrients, and even improvement of rearing water quality (Verschuere et al., 2000; Pérez-Sánchez et al., 2014). Probiotics were also reported as a source of nutrients, fatty acids, and vitamins to the fish host, and as possessing the capacity to enhance the digestibility of foods by the host organism through modulation of the fish gut microbiome (Pérez-Sánchez et al., 2014; Borges et al., 2021). Best practices to evaluate the potential of novel probiotic organisms usually involve the performance of in vitro antagonism tests, exposing pathogens to potential probiotic strains or to extracellular products synthesized by them in liquid and/or solid medium. To determine the ability of a probiotic strain to prevent disease and epizootic outbreaks, it is necessary that in vivo tests are performed (Verschuere et al., 2000; Pérez-Sánchez et al., 2014).
The information regarding the use of probiotic approaches in farmed gilthead seabream is still very scarce and even absent concerning larviculture facilities. Promising clues to follow to evaluate the potential of probiotics to prevent, or even treat, vibriosis in gilthead seabream larvi- and aquaculture facilities are suggested from the information gathered in Table 3, which broadens our scope to list studies showing empirical evidence of the efficacy of probiotics use to supress vibriosis across a wide range of host organisms. Several benefits were identified in shellfish aquaculture associated with the use of diverse probiotics, namely Lactobacillus spp., Enterococcus spp., Bacillus spp., Aeromonas spp., Alteromonas spp., Arthrobacter spp., Bifidobacterium spp., Clostridium spp., Paenibacillus spp., Phaeobacter spp., Pseudoalteromonas spp., Pseudomonas spp., Rhodosporidium spp., Roseobacter spp., Streptomyces spp. and even Vibrio spp. (Ringø, 2020, Table 3). The main benefits include fish growth promotion, improved digestive capacity, inhibition of adherence and colonization of pathogenic bacteria in the digestive tract, gut microbiota modulation, and the improvement of hematological parameters and the immune response (Ringø, 2020). Curiously, a previous study referred to many avirulent V. alginolyticus strains that could be used as probiotics (Akayli et al., 2008). Vibrio alginolyticus was also reported to possess probiotic effects against Aeromonas salmonicida (Hoseinifar et al., 2018). The use of Bacillus cereus isolated from the intestine of shrimps Litopenaeus vannamei is an example of successful re-colonization of the host intestine at the post-larval stage, probably due to competitive exclusion via the secretion of antimicrobial substances, especially resulting in effective suppression of V. parahaemolyticus and V. harveyi, justifying Bacillus cereus use as probiotic bacterium in shrimp larviculture (Vidal et al., 2018). This is an example of probiotic screening from natural host microbiomes that can be successfully applied across several aquaculture systems. Several other Gram-positive and Gram-negative probiotic bacteria showing suppressive features against Vibrio spp. have already been identified (Table 3).
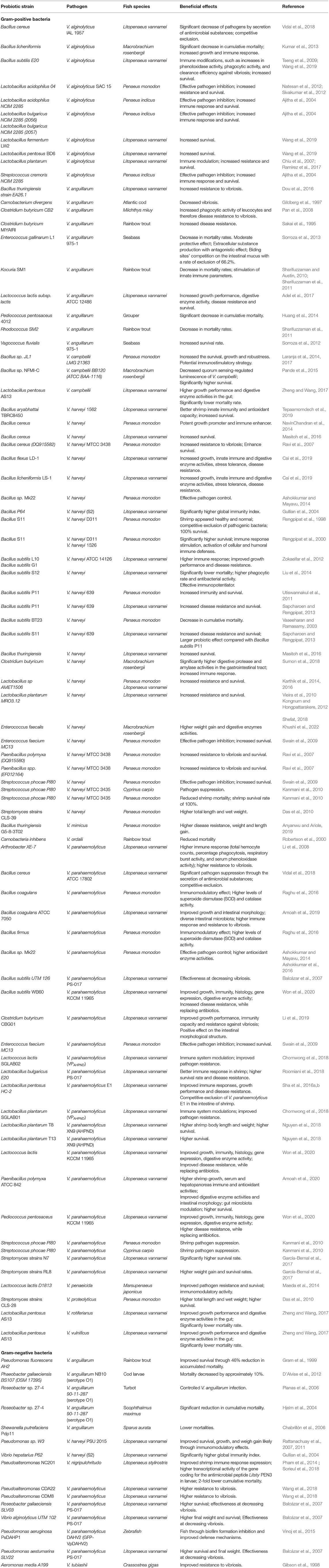
Table 3. Overview of the effects of probiotics against pathogenic Vibrio species in farmed fish and shrimp.
Antibiotic-producing bacterial probiotics such as V. hepatarius P62, Pseudomonas sp., Lactobacillus sp., Bacillus P64, along with yeast probiotics applied in shellfish and fish aquaculture have been usually selected from their natural environment (Cedeño and Rodríguez, 2006). The probiotic activity of Saccharomyces cerevisiae P13 against V. alginolyticus was demonstrated through the significant enhancement of survival rates of Pacific white shrimp Litopenaeus vannamei (Wang et al., 2019). Bacillus pumilus H2 could be very useful as an anti-Vibrio probiotic as it was shown to inhibit 29 different Vibrio strains (Gao et al., 2017). The anti-Vibrio compound was found to be amicoumacin, whose activity against Vibrio pathogens is based on the disruption of cell membranes, resulting in cell lysis. However, the minimum inhibitory concentration (MIC, expressed in μg/ml) of the purified anti-Vibrio compound amicoumacin A, isolated from Bacillus pumilus H2, was found to vary considerably depending on the Vibrio species/strain, from 0.5 μg/ml for V. vulnificus CZ-A2 and V. harveyi PH4 to 64 ug/ml for V. alginolyticus CGMCC 1.1607 and V. parahaemolyticus CGMCC 1.2164 (Gao et al., 2017).
Probiotic species able to inhibit diverse Vibrio strains or species in in situ experiments are scarcely documented. Future research should shed light on the potential use of Bacillus pumilus as a deterrent of multiple opportunistic species in aquaculture facilities. Tropodithietic acid (TDA) produced by Phaeobacter spp. can protect live feed, namely rotifers and Artemia, as well as turbot larvae and cod larvae against pathogenic Vibrio species (Rasmussen et al., 2018) such as V. anguillarum (D'Alvise et al., 2012). It was also found that the probiotic bacterium Phaeobacter inhibens strain S4Sm inhibited the growth of V. tubiashii and V. anguillarum in cultured oysters (Zhao et al., 2016). Furthermore, Phaeobacter inhibens antagonized V. anguillarum in cultures of copepod and in the copepod live feed Rhodomonas salina (Rasmussen et al., 2018), emerging as another candidate probiotic species with the ability to suppress multiple Vibrio species.
A multitude of readily culturable bacteria possessing potential probiotic features are currently available and well-described. These can be explored for the implementation of novel and effective methodologies of pathogen suppression, for example involving the development of multi-species probiotic inoculants or of smart delivery systems (e.g., using alginates) that may enhance the host colonization ability of probiotics. However, despite all the promising advances mentioned above, only three probiotic strains - the gut microbiota stabilizers Pediococcus acidilactici CNCM MA 18/5M and Pediococcus acidilactici CNCM I-4622 (bacteria), and the digestibility enhancer yeast Komagataella pastoris DSM 23036 - are authorized by the European Union to be used as live organisms in aquaculture facilities [European Commission 2021, Reg (EC) No 1831/2003]. In face of the acute challenges posed by increasing disease incidence in the aquaculture sector, and of the manifold possibilities of microbiome-based disease control revealed in the last three decades, it is reasonable to argue that time is ripe for advancing new legislation that is on par with current scientific knowledge regarding the use of environmentally safer, bio-based methodologies to deter bacterial diseases in this sector.
Microorganisms are major contributors to nutrient cycling and functioning within aquaculture facilities yet a fraction of the total microbiome thriving in these man-made is also responsible for disease and mortality affecting live feed, fish larvae, fish, and shellfish. The extent to which solutions to the pathogenicity problem within aquaculture facilities may be found in the naturally occurring aquaculture microbiome is a matter of current scientific debate. The effective implementation of protocols relying on the use of vaccines, phage therapy and probiotics holds promise in deterring pathogen proliferation in intensive fish farming. For instance, a wealth of Gram-positive and Gram-negative bacteria showing remarkable capacities to supress Vibrio pathogens or mitigate vibriosis symptoms in farmed fish and shellfish have been identified in the past 30 years. Nevertheless, to move beyond the proof-of-concept stage, such protocols still face technical and legal challenges that prevent their wide-range applicability at the production scale. Better understanding of microbiomes thriving in “healthy” and “diseased” hosts and aquaculture systems is key to instruct researchers in the pursuit of techniques leading to efficient microbiome manipulation and engineering toward safer rearing systems, eventually decreasing the need of using antibiotics and hazardous chemicals to control bacterial diseases in aquaculture.
The increasing pollution of coastal ecosystems caused by sewage and industrial effluent inputs, including oils, fertilizers, and heavy metals, may also result in a negative physiological response of reared fish, favoring the invasion of bacterial pathogens and parasites in rearing systems exposed to such pollutants. Therefore, proper environmental monitoring and ecosystem conservation are fundamental to prevent bacterial disease incidence in aquaculture. Aquaculture facilities may be in fact “hotspots” for gene transfer, as they contain dense and highly diverse bacterial communities whose structure and taxonomic composition result from the combination of current and past use of antibiotics, probiotics, prebiotics as well as other kinds of treatments or methods. In this context, it is important to discern the coding potential present in the mobile gene pool of Vibrio species and assess the intra- and interspecific transferability of these genes, as this bears implications to our understanding to the roles of Vibrio species as disease-causing agents and of the potential switch from commensal to pathogenic behavior based on processes of gene gain and loss in the Vibrio-associated plasmidome. Indeed, Bruto et al. (2017) revealed that pathogenicity of Vibrio crassostreae toward the cultured oyster species Crassostrea gigas is mediated by the acquisition of a large mobilizable plasmid. It is reasonable to argue that such processes mediate virulence of Vibrio pathogens of fish and may be promoted by high host densities in intensive rearing conditions.
Vibriosis is one of the most important diseases causing high mortality rates in the aquaculture industry. According to the meta-analysis discussed in this review, species such as V. alginolyticus and V. harveyi are among the main responsible for epizootic disease outbreaks causing economic losses in this sector, affecting several fish species, including the production of reared gilthead seabream in the Mediterranean zone. As the frequency of antibiotic- and multidrug-resistance Vibrio spp. is growing, constant surveillance and monitoring of antibiotic resistance and pollution must be assured to avoid the development of multi-resistant strains which may pose threats to both ecosystem and human health. Although certain Vibrio species from diseased farmed gilthead seabream were found to be sensitive to tetracycline, oxytetracycline, chloramphenicol and florfenicol, the development of alternative, cost-effective and sustainable pathogen suppression methods in aquaculture is encouraged from an environmental and a human health standpoint. Particularly worrisome is the current rise of human infections caused by environmental and seafood-associated Vibrio species, apparently influenced by climate-change drivers of microbial community assembly in coastal ecosystems, including intensive seafood farming systems.
The major goal of aquaculture production is supplying food for human consumption. Following several decades of heavy use of antimicrobial drugs and antibiotics to boost intensive fish rearing, current prophylaxis approaches that contribute to a more health-oriented management of aquaculture systems are being increasingly recommended to prevent or suppress epizootic disease outbreaks. They involve the use of less dangerous methods such as vaccines, immunostimulants and probiotics/microbiome therapy. This way, it is believed that healthier food for human consumption may be produced and bacterial resistance to antibiotics may be prevented or alleviated, thus reducing the transfer of acquired antibiotic resistance traits to human pathogens via mobile genetic elements.
GMMS-F, IS-C, and RC conceived and designed the study. GMMS-F performed the literature research, prepared figures, and tables. GMMS-F and RC interpreted the data and wrote the first manuscript draft. All authors read and revised the first manuscript draft and approved the final manuscript.
This study was supported by FCT—Fundação para a Ciência e a Tecnologia, I.P., through the research grants PTDC/MAR/112792/2009 and PTDC/BIA-MIC/31996/2017 and by the European Regional Development Fund (ERDF, Project # 031996, operational code ALG-01-0145-FEDER-031966) through the regional operational programs of Lisbon and Algarve, Portugal. This study was also financed by FCT in the scope of the projects UIDB/04565/2020 and UIDP/04565/2020 of the Research Unit Institute for Bioengineering and Biosciences—iBB, and the project LA/P/0140/2020 of the Associate Laboratory i4HB - Institute for Health and Bioeconomy.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.904815/full#supplementary-material
Abareethan, M., and Amsath, A. (2015). Characterization and evaluation of probiotic fish feed. Int. J. Pure Appl. Zool. 3, 148–153.
Abdallah, F. B., Bakhrouf, A., Ayed, A., and Kallel, H. (2009). Alterations of outer membrane proteins and virulence genes expression in gamma-irradiated Vibrio parahaemolyticus and Vibrio alginolyticus. Foodborne Pathog. Dis. 6, 1171–1176. doi: 10.1089/fpd.2009.0331
Abdallah, F. B., Ellafi, A., Lagha, R., Kallel, H., and Bakhrouf, A. (2011). Virulence gene expression, proteins secreted and morphological alterations of Vibrio parahaemolyticus and Vibrio alginolyticus in response to long-term starvation in seawater. Afr. J. Microbiol. Res. 5, 792–801. doi: 10.5897/AJMR10.653
Abdel-Aziz, M., Eissa, A. E., Hanna, M., and Okada, M. A. (2013). Identifying some pathogenic Vibrio/Photobacterium species during mass mortalities of cultured gilthead seabream (Sparus aurata) and European seabass (Dicentrarchus labrax) from some Egyptian coastal provinces. Int. J. Vet. Sci. Med. 1, 87–95. doi: 10.1016/j.ijvsm.2013.10.004
Abdelaziz, M., Ibrahem, M. D., Ibrahim, M. A., Abu-Elala, N. M., and Abdel-moneam, D. A. (2017). Monitoring of different vibrio species affecting marine fishes in Lake Qarun and Gulf of Suez: phenotypic and molecular characterization. Egypt. J. Aquat. Res. 43, 141–146. doi: 10.1016/j.ejar.2017.06.002
Adel, M., El-Sayed, A. F. M., Yeganeh, S., Dadar, M., and Giri, S. S. (2017). Effect of potential probiotic Lactococcus lactis subsp. lactis on growth performance, intestinal microbiota, digestive enzyme activities, and disease resistance of Litopenaeus vannamei. Probiot. Antimicrob. Prot. 9, 150–156. doi: 10.1007/s12602-016-9235-9
Ajitha, S., Sridhar, M., Sridhar, N., and Varghese, V. (2004). Probiotic effects of lactic acid bacteria against Vibrio Alginolyticus in Penaeus (Fenneropenaeus) indicus (H. Milne Edwards). Asian Fish. Sci. 17, 71–80. doi: 10.33997/j.afs.2004.17.1.008
Akayli, T., and Timur, G. (2002). Vibriosis in gilthead sea bream (Sparus aurata L.) in farms in the Aegean sea coast of Turkey. Turkish J. Fish. Aquat. Sci. 2, 89–91.
Akayli, T., Timur, G., Aydemir, B., Kiziler, A. R., Coskun, O., Albayrak, G., et al. (2008). Characterization of Vibrio alginolyticus isolates from diseased cultured gilthead sea bream, Sparus aurata. Israel. J. Aquac. Bamidgeh. 60, 89–94. doi: 10.46989/001c.20487
Altinok, I., and Kurt, I. (2004). Molecular diagnosis of fish diseases: a review. Turk. J. Fish. Aquat. Sci. 3, 131–138.
Aly, S. M., Eisa, A. A., and Elbanna, N. I. (2020). Characterization of vibrio parahaemolyticus infection in gilthead seabream (Sparus aurata) cultured in Egypt. Egypt. J. Aquat. Biol. Fish. 24, 553–571. doi: 10.21608/ejabf.2020.76562
Amatul-Samahah, M. A., Wan Omar, W. H. H., Mohd Ikhsan, N. F., Amal Azmai, M. N., Zamri-Saad, M., and Ina-Salwany, M. Y. (2020). Vaccination trials against vibriosis in shrimp: a review. Aquac. Rep. 18, 100471. doi: 10.1016/j.aqrep.2020.100471
Amoah, K., Huang, Q.-C., Tan, B.-P., Zhang, S., Chi, S.-Y., Yang, Q.-H., et al. (2019). Dietary supplementation of probiotic Bacillus coagulans ATCC 7050, improves the growth performance, intestinal morphology, microflora, immune response, and disease confrontation of Pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 87, 796–808. doi: 10.1016/j.fsi.2019.02.029
Amoah, K., Huang, Q.-C., Dong, X.-H., Tan, B.-P., Zhang, S., Chi, S.-Y., et al. (2020). Paenibacillus polymyxa improves the growth, immune and antioxidant activity, intestinal health, and disease resistance in Litopenaeus vannamei challenged with Vibrio parahaemolyticus. Aquaculture 518, 734563. doi: 10.1016/j.aquaculture.2019.734563
Anyanwu, N. G., and Ariole, C. N. (2019). Probiotic potential of an indigenous marine Bacillus thuringiensis on shrimp (Penaeus monodon) culture infected with Vibrio mimicus. J. Appl. Sci. 19, 173–179. doi: 10.3923/jas.2019.173.179
Arab, S., Nalbone, L., Giarratana, F., and Berbar, A. (2020). Occurrence of Vibrio spp. along the algerian mediterranean coast in wild and farmed sparus aurata and dicentrarchus labrax. Vet. World 13, 1199–1208. doi: 10.14202/vetworld.2020.1199-1208
Ashokkumar, S., and Mayavu, P. (2014). Screening, identification and antagonistic activity of halo stable Bacillus sp. Mk22 used as probiotic in Penaeus monodon Fabricius, 1798. Afr. J. Food Sci. 8, 48–53. doi: 10.5897/AJFS2013.1048
Ashokkumar, S., Mayavu, P., and Kim, K. (2016). Growth enhancement of shrimp and reduction of shrimp infection by vibrio parahaemolyticus and white spot syndrome virus with dietary administration of bacillus sp. Mk22. Microbiol. Biotechnol. Lett. 44, 261–267. doi: 10.4014/mbl.1605.05001
Asok, A., Arshad, E., Jasmin, C., Somnath Pai, S., Bright Singh, I. S., Mohandas, A., et al. (2012). Reducing Vibrio load in Artemia nauplii using antimicrobial photodynamic therapy: a promising strategy to reduce antibiotic application in shrimp larviculture. Microb. Biotechnol. 5, 59–68. doi: 10.1111/j.1751-7915.2011.00297.x
Baker-Austin, C., and Oliver, J. D. (2018). Vibrio vulnificus: new insights into a deadly opportunistic pathogen. Environ. Microbiol. 20, 423–430. doi: 10.1111/1462-2920.13955
Balcázar, J. L., Rojas-Luna, T., and Cunningham, D. P. (2007). Effect of the addition of four potential probiotic strains on the survival of pacific white shrimp (Litopenaeus vannamei) following immersion challenge with Vibrio parahaemolyticus. J. Invertebr. Pathol. 96, 147–150. doi: 10.1016/j.jip.2007.04.008
Balebona, M. C., Andreu, M. J., Bordas, M. A., Zorilla, I., Moriñgo, M. A., and Borrego, J. J. (1998a). Pathogenicity of Vibrio alginolyticus for cultured gilt-head sea bream (Sparus aurata L.). Appl. Environ. Microbiol. 64, 4269–4275. doi: 10.1128/AEM.64.11.4269-4275.1998
Balebona, M. C., Zorrilla, I., Moriñigo, M. A., and Borrego, J. J. (1998b). Survey of bacterial pathologies affecting farmed gilt-head sea bream (Sparus aurata L.) in southwestern Spain from 1990 to 1996. Aquaculture 166, 19–35. doi: 10.1016/S0044-8486(98)00282-8
Bentzon-Tilia, M., Sonnenschein, E. C., and Gram, L. (2016). Monitoring and managing microbes in aquaculture – Towards a sustainable industry. Microb. Biotechnol. 9, 576–584. doi: 10.1111/1751-7915.12392
Blake, P. A. (1994). “Historical perspectives on pandemic cholera,” in Vibrio Cholerae and Cholera: Molecular to Global Perspectives, eds I. Kaye Wachsmuth, P. A. Blake, and Ø. Olsvik (Washington, DC: American Society for Microbiology), 291–295. doi: 10.1128/9781555818364.ch18
Bondad-Reantaso, M. G. (2018). “Country level implementation: FAO experience in Aquaculture, FAO Session 4: responsible and prudent use of veterinary antimicrobials: practical tools and experiences,” in 2nd OIE Global Conference on Antimicrobial Resistance and Prudent Use of Antimicrobial Agents in Animals, Putting Standards into Practice (Marrakesh).
Bordas, M. A., Balebona, M. C., Zorrilla, I., Borrego, J. J., and Moriñigo, M. A. (1996). Kinetics of adhesion of selected fish-pathogenic Vibrio strains to skin mucus of gilt-head sea bream (Sparus aurata L.). Appl. Environ. Microbiol. 62, 3650–3654. doi: 10.1128/aem.62.10.3650-3654.1996
Borges, N., Keller-Costa, T., Sanches-Fernandes, G. M. M., Louvado, A., Gomes, N. C. M., and Costa, R. (2021). Bacteriome structure, function, and probiotics in fish larviculture: the good, the bad, and the gaps. Annu. Rev. Anim. Biosci. 9, 1–30. doi: 10.1146/annurev-animal-062920-113114
Brayton, P. R., Bode, R. B., Colwell, R. R., MacDonell, M. T., Hall, H. L., Grimes, D. J., et al. (1986). Vibrio cincinnatiensis sp. nov., a new human pathogen. J. Clin. Microbiol. 23, 104–108. doi: 10.1128/jcm.23.1.104-108.1986
Brehm, T. T., Berneking, L., Rohde, H., Chistner, M., Schlickewei, C., Sena Martins, M., et al. (2020). Wound infection with Vibrio harveyi following a traumatic leg amputation after a motorboat propeller injury in Mallorca, Spain: a case report and review of literature. BMC Infect. Dis. 20, 1–7. doi: 10.1186/s12879-020-4789-2
Bresee, J. S., Widdowson, M. A., Monroe, S. S., and Glass, R. I. (2002). Foodborne viral gastroenteritis: challenges and opportunities. Clin. Infect. Dis. 35, 748–753. doi: 10.1086/342386
Broberg, C. A., Calder, T. J., and Orth, K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 13, 992–1001. doi: 10.1016/j.micinf.2011.06.013
Bruto, M., James, A., Petton, B., Labreuche, Y., Chenivesse, S., Alunno-Bruscia, M., et al. (2017). Vibrio crassostreae, a benign oyster colonizer turned into a pathogen after plasmid acquisition. ISME J. 11, 1043–1052. doi: 10.1038/ismej.2016.162
Cai, Y., Yuan, W., Wang, S., Guo, W., Li, A., Wu, Y., et al. (2019). In vitro screening of putative probiotics and their dual beneficial effects: To white shrimp (Litopenaeus vannamei) postlarvae and to the rearing water. Aquaculture 498, 61–71. doi: 10.1016/j.aquaculture.2018.08.024
Califano, G., Castanho, S., Soares, F., Ribeiro, L., Cox, C. J., Mata, L., et al. (2017). Molecular taxonomic profiling of bacterial communities in a gilthead seabream (Sparus aurata) hatchery. Front. Microbiol. 8, 1–16. doi: 10.3389/fmicb.2017.00204
Campbell, R., Adams, A., Tatner, M. F., Chair, M., and Sorgeloos, P. (1993). Uptake of Vibrio anguillarum vaccine by Artemia salina as a potential oral delivery system to fish fry. Fish Shellfish Immunol. 3, 451–459. doi: 10.1006/fsim.1993.1044
Cedeño, R., and Rodríguez, J. (2006). Uso de los probióticos vibrío hepatarius (p62) y bacillus sp. (p64) en el cultivo del camarón litopenaeus vannamei. Cenaim Informa, 134. Available online at: https://core.ac.uk/display/12398899 (accessed June 6, 2022).
Chabrillón, M., Arijo, S., Díaz-Rosales, P., Balebona, M. C., and Moriñigo, M. A. (2006). Interference of Listonella anguillarum with potential probiotic microorganisms isolated from farmed gilthead seabream (Sparus aurata L.). Aquac. Res. 37, 78–86. doi: 10.1111/j.1365-2109.2005.01400.x
Chauhan, A., and Singh, R. (2019). Probiotics in aquaculture: a promising emerging alternative approach. Symbiosis 77, 99–113. doi: 10.1007/s13199-018-0580-1
Chiu, C. H., Guu, Y. K., Liu, C. H., Pan, T. M., and Cheng, W. (2007). Immune responses and gene expression in white shrimp, Litopenaeus vannamei, induced by Lactobacillus plantarum. Fish Shellfish Immunol. 23, 364–377. doi: 10.1016/j.fsi.2006.11.010
Chomwong, S., Charoensapsri, W., Amparyup, P., and Tassanakajon, A. (2018). Two host gut-derived lactic acid bacteria activate the proPO system and increase resistance to an AHPND-causing strain of Vibrio parahaemolyticus in the shrimp Litopenaeus vannamei. Dev. Comp. Immunol. 89, 54–65. doi: 10.1016/j.dci.2018.08.002
Citil, B. E., Derin, S., Sankur, F., Sahan, M., and Citil, M. U. (2015). Vibrio alginolyticus associated chronic myringitis acquired in Mediterranean waters of Turkey. Case Rep. Infect. Dis. 2015, 1–3. doi: 10.1155/2015/187212
Colorni, A., Paperna, I., and Gordin, H. (1981). Bacterial infections in gilthead sea bream Sparus aurata cultured at Eilat. Aquaculture 23, 257–267. doi: 10.1016/0044-8486(81)90019-3
Costa, R. A., Araújo, R. L., Souza, O. V., and Vieira, R. H. S. D. F. (2015). Antibiotic-resistant vibrios in farmed shrimp. Biomed Res. Int. 2015, 1–5. doi: 10.1155/2015/505914
Dalsgaard, A., Glerup, P., Høybye, L. L., Paarup, A. M., Meza, R., Bernal, M., et al. (1997). Vibrio furnissii isolated from humans in Peru: a possible human pathogen? Epidemiol. Infect. 119, 143–149. doi: 10.1017/S095026889700798X
D'Alvise, P. W., Lillebø, S., Prol-Garcia, M. J., Wergeland, H. I., Nielsen, K. F., Bergh, Ø., et al. (2012). Phaeobacter gallaeciensis reduces vibrio anguillarum in cultures of microalgae and rotifers, and prevents vibriosis in cod larvae. PLoS ONE 7, 43996. doi: 10.1371/journal.pone.0043996
Das, S., Ward, L. R., and Chris Burke, C. (2010). Screening of marine Streptomyces spp. for potential use as probiotics in aquaculture. Aquaculture 305, 32–41. doi: 10.1016/j.aquaculture.2010.04.001
Deng, Y., Xu, L., Chen, H., Liu, S., Guo, Z., Cheng, C., et al. (2020). Prevalence, virulence genes, and antimicrobial resistance of Vibrio species isolated from diseased marine fish in South China. Sci. Rep. 10, 1–8. doi: 10.1038/s41598-020-71288-0
Devi, M. S., Paria, P., Kumar, V., Parida, P. K., Maurye, P., Behera, B. K., et al. (2022). Molecular identification and pathogenicity study of virulent Vibrio cholerae non O1/O139 serotype associated with mortality of farmed Labeo rohita (Hamilton, 1822), in India. Aquaculture 547, 737529. doi: 10.1016/j.aquaculture.2021.737529
Dhert, P., Rombaut, G., Suantika, G., and Sorgeloos, P. (2001). Advancement of rotifer culture and manipulation techniques in Europe. Aquaculture 200, 129–146. doi: 10.1016/S0044-8486(01)00697-4
Dou, C., Zuo, Z., Liu, Y., Zhang, Y., Geng, X., and Sun, J. (2016). Isolation and screening of digestive enzyme producing probiotics from intestine of Litopenaeus vannamei. J. Fish. China 40, 537–546.
Dutta, D., Kaushik, A., Kumar, D., and Bag, S. (2021). Foodborne pathogenic vibrios: antimicrobial resistance. Front. Microbiol. 12, 1–10. doi: 10.3389/fmicb.2021.638331
Eissa, A. E., Altakaly, M. B., Abolghait, S. K.;, Ismail, M. M., and Abumhara, A. (2017). Detection of the most common Vibrios affecting common pandora (Pagellus erythinus) from the coasts of Tripoli, Libya. J. Fish. Aquat. Sci. 12, 253–263. doi: 10.3923/jfas.2017.253.263
El-Bouhy, Z., El-Nobi, G., El-Murr, A. E., and El-Hakim, S. A. (2016). Study on vibriosis in Mugil Capito In El-Dakahlia And Damitta Governorates, Egypt. Abbassa Int. J. Aquat. 9, 19–35.
El-Sayed, M. E., Algammal, A. M., Abouel-Atta, M. E., Mabrok, M., and Emam, A. M. (2019). Pathogenicity, genetic typing, and antibiotic sensitivity of Vibrio alginolyticus isolated from Oreochromis niloticus and Tilapia zillii. Rev. Med. Vet. 170, 80–86.
Embregts, C. W. E., and Forlenza, M. (2016). Oral vaccination of fish: lessons from humans and veterinary species. Dev. Comp. Immunol. 64, 118–137. doi: 10.1016/j.dci.2016.03.024
FAO (1996). World Review of Fisheries and Aquaculture. Food and Agriculture Organization of the United Nations, Fisheries Department. Available online at: http://www.fao.org/3/w3265e/w3265e02.htm#b2-Trends (accessed June 6, 2022).
FAO (2002). The State of World Fisheries and Aquaculture 2002. Rome: Food and Agriculture Organization of the United Nations, Fisheries Department. Available online at: http://www.fao.org/3/y7300e/y7300e00.pdf (accessed June 6, 2022).
FAO (2004). The State of World Fisheries and Aquaculture 2004. Rome: Food and Agriculture Organization of the United Nations, Fisheries Department. Available online at: http://www.fao.org/3/y5600e/y5600e00.pdf (accessed June 6, 2022).
FAO (2010). The State of World Fisheries and Aquaculture 2010. Rome: Food and Agriculture Organization of the United Nations, Fisheries and Aquaculture Department. Available online at: http://www.fao.org/3/i1820e/i1820e.pdf (accessed June 6, 2022).
FAO (2012). The State of World Fisheries and Aquaculture 2012. Contributing to Food Security and Nutrition for All. Rome: Food and Agriculture Organization of the United Nations, Fisheries and Aquaculture Department. Available online at: http://www.fao.org/3/i2727e/i2727e.pdf (accessed June 6, 2022).
FAO (2016). The State of World Fisheries and Aquaculture 2016. Contributing to Food Security and Nutrition for All. Rome: Food and Agriculture Organization of the United Nations, Fisheries and Aquaculture Department. Available online at: http://www.fao.org/3/a-i5555e.pdf (accessed June 6, 2022).
FAO (2018). The State of World Fisheries and Aquaculture 2018. Meeting the Sustainable Development Goals. Rome: Food and Agriculture Organization of the United Nations, Fisheries and Aquaculture Department. Available online at: http://www.fao.org/3/i9540en/i9540en.pdf (accessed June 6, 2022).
FAO (2020). The State of World Fisheries and Aquaculture 2020. Sustainability in Action. Rome: Food and Agriculture Organization of the United Nations, Fisheries and Aquaculture Department. Available online at: http://www.fao.org/3/ca9229en/ca9229en.pdf (accessed June 6, 2022).
FAO (2021). “Sparus aurata. Cultured Aquatic Species Information Programme,” in Fisheries and Aquaculture Division, eds F. Colloca, and S. Cerasi (Rome). Available online at: https://www.fao.org/fishery/en/culturedspecies/sparus_aurata/en (accessed December 6, 2021).
Finkelstein, R., Edelstein, S., and Mahamid, G. (2002). Fulminant wound infections due to Vibrio vulnificus. Isr. Med. Assoc. J. 4, 654–655.
Firmino, J., Furones, M. D., Andree, K. B., Sarasquete, C., Ortiz-Delgado, J. B., Asencio-Alcudia, G., et al. (2019). Contrasting outcomes of Vibrio harveyi pathogenicity in gilthead seabream, Sparus aurata and European seabass, Dicentrachus labrax. Aquaculture 511, 734210. doi: 10.1016/j.aquaculture.2019.734210
Gao, X. Y., Liu, Y., Miao, L. L., Li, E. W., Hou, T. T., and Liu, Z. P. (2017). Mechanism of anti-Vibrio activity of marine probiotic strain Bacillus pumilus H2, and characterization of the active substance. AMB Exp. 7, 23–32. doi: 10.1186/s13568-017-0323-3
García-Bernal, M., Medina-Marrero, R., Rodríguez-Jaramillo, C., Marrero-Chang, O., Campa-Córdova, Á. I., Medina-García, R., et al. (2017). Probiotic effect of Streptomyces spp. on shrimp (Litopenaeus vannamei) postlarvae challenged with Vibrio parahaemolyticus. Aquac. Int. 25, 927–939. doi: 10.1111/anu.12622
Gibson, L. F., Woodworth, J., and George, A. M. (1998). Probiotic activity of Aeromonas media on the Pacific oyster, Crassostrea gigas, when challenged with Vibrio tubiashii. Aquaculture 169, 111–120. doi: 10.1016/S0044-8486(98)00369-X
Gildberg, A., Mikkelsen, H., Sandaker, E., and Ring,ø, E. (1997). Probiotic effect of lactic acid bacteria in the feed on growth and survival of fry of Atlantic cod (Gadus morhua). Hydrobiologia 352, 279–285. doi: 10.1023/A:1003052111938
Gomathi, R. S., Vinothkumar, R., and Arunagiri, K. (2013). Isolation and Identification Vibrios from marine seafood samples. Int. J. Curr. Microbiol. Appl. Sci. 2, 36–43.
Gomez-Gil, B., Thompson, F. L., Thompson, C. C., Garcia-Gasca, A., Roque, A., and Swings, J. (2004). Vibrio hipanicus sp. nov., isolated from Artemia sp. and sea water in Spain. Int. J. Syst. Evol. Microbiol. 54, 261–265. doi: 10.1099/ijs.0.02775-0
Gomez-Gil, B., Thompson, F. L., Thompson, C. C., and Swings, J. (2003). Vibrio rotiferianus sp. nov., isolated from cultures of the rotifer Brachionus plicatilis. Int. J. Syst. Evol. Microbiol. 53, 239–243. doi: 10.1099/ijs.0.02430-0
Gram, L., Melchiorsen, J., Spanggaard, B., Huber, I., and Nielsen, T. F. (1999). Inhibition of Vibrio anguillarum by Pseudomonas fluorescens AH2, a possible probiotic treatment of fish. Appl. Environ. Microb. 65, 969–973. doi: 10.1128/AEM.65.3.969-973.1999
Guidetti, P., Fanelli, G., Fraschetti, S., Terlizzi, A., and Boero, F. (2002). Coastal fish indicate human-induced changes in the Mediterranean littoral. Mar. Environ. Res. 53, 77–94. doi: 10.1016/S0141-1136(01)00111-8
Guidi, L. R., Santos, F. A., Ribeiro, A. C. S. R., Fernandes, C., Silva, L. H. M., and Gloria, M. B. A. (2018). Quinolones and tetracyclines in aquaculture fish by a simple and rapid LC-MS/MS method. Food Chem. 245, 1232–1238. doi: 10.1016/j.foodchem.2017.11.094
Gullian, M., Thompson, F., and Rodriguez, J. (2004). Selection of probiotic bacteria and study of their immunostimulatory effect in Penaeus vannamei. Aquaculture 233, 1–14. doi: 10.1016/j.aquaculture.2003.09.013
Håkonsholm, F., Lunestad, B. T., Aguirre Sánchez, J. R., Martinez-Urtaza, J., Marathe, N. P., and Svanevik, C. S. (2020). Vibrios from the Norwegian marine environment: characterization of associated antibiotic resistance and virulence genes. Microbiologyopen 9, 1–19. doi: 10.1002/mbo3.1093
Haché, R., and Plante, S. (2011). The relationship between enrichment, fatty acid profiles and bacterial load in cultured rotifers (Brachionus plicatilis L-strain) and Artemia (Artemia salina strain Franciscana). Aquaculture 311, 201–208. doi: 10.1016/j.aquaculture.2010.11.034
Haenen, O. L. M., Van Zanten, E., Jansen, R., Roozenburg, I., Engelsma, M. Y., Dijkstra, A., et al. (2014). Vibrio vulnificus outbreaks in Dutch eel farms since 1996: Strain diversity and impact. Dis. Aquat. Organ. 108, 201–209. doi: 10.3354/dao02703
Hai, N. V. (2015). The use of probiotics in aquaculture. J. Appl. Microbiol. 119, 917–935. doi: 10.1111/jam.12886
Haldar, S., Maharajan, A., Chatterjee, S., Hunter, S. A., Chowdhury, N., Hinenoya, A., et al. (2010). Identification of Vibrio harveyi as a causative bacterium for a tail rot disease of sea bream Sparus aurata from research hatchery in Malta. Microbiol. Res. 165, 639–648. doi: 10.1016/j.micres.2009.12.001
Halpern, M., and Izhaki, I. (2017). Fish as hosts of Vibrio cholerae. Front. Microbiol. 8, 282. doi: 10.3389/fmicb.2017.00282
Hamdan, R. H., Peng, T. L., Ong, B. L., Suhana, M. Y. S., Hamid, N. H., Afifah, M. N. F., et al. (2016). “Antibiotics resistance of Vibrio spp. isolated from diseased seabass and tilapia in cage culture,” in Proceedings of International Seminar on Livestock Production and Veterinary Technology 2016, 554–560. doi: 10.14334/proc.intsem.lpvt-2016-p.554-560
Hernández-Robles, M. F., Natividad-Bonifacio, I., Álvarez-Contreras, A. K., Tercero-Alburo, J. J., Quiñones-Ramírez, E. I., and Vázquez-Salinas, C. (2021). Characterization of potential virulence factors of Vibrio mimicus isolated from fishery products and water. Int. J. Microbiol. 2021, 8397930. doi: 10.1155/2021/8397930
Hjelm, M., Bergh, Ø., Riaza, A., Nielsen, J., Melchiorsen, J., Jensen, S., et al. (2004). Selection and identification of autochthonous potential probiotic bacteria from turbot larvae (Scophthalmus maximus) rearing units. Syst. Appl. Microbiol. 27, 360–371. doi: 10.1078/0723-2020-00256
Hong, G. E., Kim, D. G., Bae, J. Y., Ahn, S. H., Bai, S. C., and Kong, I. S. (2007). Species-specific PCR detection of the fish pathogen, Vibrio anguillarum, using the amiB gene, which encodes N-acetylmuramoyl-L-alanine amidase. FEMS Microbiol. Lett. 269, 201–206. doi: 10.1111/j.1574-6968.2006.00618.x
Hoseinifar, S. H., Sun, Y. Z., Wang, A., and Zhou, Z. (2018). Probiotics as means of diseases control in aquaculture, a review of current knowledge and future perspectives. Front. Microbiol. 9, 1–18. doi: 10.3389/fmicb.2018.02429
Huang, J., Wu, Y. C., and Chi, S. C. (2014). Dietary supplementation of Pediococcus pentosaceus enhances innate immunity, physiological health and resistance to Vibrio anguillarum in orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 39, 196–205. doi: 10.1016/j.fsi.2014.05.003
Huang, Z., Yu, K., Fang, Y., Dai, H., Cai, H., Li, Z., et al. (2020). Comparative genomics and transcriptomics analyses reveal a unique environmental adaptability of Vibrio fujianensis. Microorganisms 8, 555–570. doi: 10.3390/microorganisms8040555
Igarashi, M. A., Sugita, H., and Deguchi, Y. (1989). Microflora associated with eggs and nauplii of Artemia salina. Nippon Suisan Gakkaishi 55, 2045–2045. doi: 10.2331/suisan.55.2045
Interaminense, J. A., Ferreira Calazans, N., do Valle, B. C., Lyra Vogeley, J., Peixoto, S., Soares, R., et al. (2014). Vibrio spp. control at brine shrimp, artemia, hatching and enrichment. J. World Aquac. Soc. 45, 65–74. doi: 10.1111/jwas.12096
Kahla-Nakbi, A., Ben Besbes, A., Bakhrouf, A., and Alcaide, E. (2007). Characterisation and virulence properties of Vibrio isolates from diseased gilthead sea bream (Sparus aurata) cultured in Tunisia. Bull. Eur. Assoc. Fish Pathol. 27, 90–99.
Kahla-Nakbi, A., Ben Chaieb, K., and Bakhrouf, A. (2009). Investigation of several virulence properties among Vibrio alginolyticus strains isolated from diseased cultured fish in Tunisia. Dis. Aquat. Organ. 86, 21–28. doi: 10.3354/dao02091
Kahla-Nakbi, A. B., Chaieb, K., Besbes, A., Zmantar, T., and Bakhrouf, A. (2006). Virulence and enterobacterial repetitive intergenic consensus PCR of Vibrio alginolyticus strains isolated from Tunisian cultured gilthead sea bream and sea bass outbreaks. Vet. Microbiol. 117, 321–327. doi: 10.1016/j.vetmic.2006.06.012
Kanmani, P., Satish Kumar, R., Yuvaraj, N., Paari, K. A., Pattukumar, V., and Arul, V. (2010). First identification of a novel probiotic bacterium Streptococcus phocae and its beneficial role in diseases control. J. Int. Dent. Med. Res. 3, 45–51.
Karthik, R., Jaffar Hussain, A., and Muthezhilan, R. (2014). Effectiveness of Lactobacillus sp (AMET1506) as probiotic against vibriosis in penaeus monodon and litopenaeus vannamei shrimp aquaculture. Biosci. Biotechnol. Res. Asia 11, 297–305. doi: 10.13005/bbra/1423
Karthik, R., Pushpam, A., Chelvan, Y., and Vanitha, M. (2016). Efficacy of probiotic and nitrifier bacterial consortium for the enhancement of Litopenaeus Vannamei aquaculture. Int. J. Vet. Sci. Res. 2, 001–006. doi: 10.17352/ijvsr.000006
Kawatsu, K., Ishibashi, M., and Tsukamoto, T. (2006). Development and evaluation of a rapid, simple, and sensitive immunochromatographic assay to detect thermostable direct hemolysin produced by Vibrio parahaemolyticus in enrichment cultures of stool specimens. J. Clin. Microbiol. 44, 1821–1827. doi: 10.1128/JCM.44.5.1821-1827.2006
Kaysner, C. A., and DePaola A. Jr Jones, J. (2004). Food and Drugs Administration: Bacteriological Analytical Manual, Methods for Specific Pathogens. Vibrio spp. U.S. Department of Health and Human Services. Available online at: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-9-vibrio (accessed June 6, 2022).
Khushi, S. S., Sumon, M. S., Ahmmed, M. K., Hasan Zilani, M. N., Ahmmed, F., Giteru, S. G., et al. (2022). Potential probiotic and health fostering effect of host gut-derived Enterococcus faecalis on freshwater prawn, Macrobrachium rosenbergii. Aquac. Fish. 7, 59–66. doi: 10.1016/j.aaf.2020.10.004
Kitaura, S., Okamoto, K., Wakabayashi, Y., Okada, Y., Okazaki, A., Ikeda, M., et al. (2020). Vibrio fluvialis liver abscess and bacteremia in a sashimi lover: a case report and review of the literature. Open Forum Infect. Dis. 7, 3–5. doi: 10.1093/ofid/ofaa212
Konechnyi, Y., Khorkavyi, Y., Ivanchuk, K., Kobza, I., Sekowska, A., and Korniychuk, O. (2021). Vibrio metschnikovii: current state of knowledge and discussion of recently identified clinical case. Clin. Case Rep. 9, 2236–2244. doi: 10.1002/ccr3.3999
Kongnum, K., and Hongpattarakere, T. (2012). Effect of Lactobacillus plantarum isolated from digestive tract of wild shrimp on growth and survival of white shrimp (Litopenaeus vannamei) challenged with Vibrio harveyi. Fish Shellfish Immunol. 32, 170–177. doi: 10.1016/j.fsi.2011.11.008
Korun, J., and Timur, G. (2008). Marine vibrios associated with diseased sea bass (Dicentrarchus labrax) in Turkey. J. Fish. Sci. 2, 66–76. doi: 10.3153/jfscom.2008007
Kumar, N. R., Raman, R. P., Jadhao, S. B., Brahmchari, R. K., Kumar, K., and Dash, G. (2013). Effect of dietary supplementation of Bacillus licheniformis on gut microbiota, growth and immune response in giant freshwater prawn, Macrobrachium rosenbergii (de Man, 1879). Aquac. Int. 21, 387–403. doi: 10.1007/s10499-012-9567-8
Kumar, S., Kumar, C. B., Rajendran, V., Abishaw, N., Anand, P. S. S., Kannapan, S., et al. (2021). Delineating virulence of Vibrio campbellii: a predominant luminescent bacterial pathogen in Indian shrimp hatcheries. Sci. Rep. 11, 1–16. doi: 10.1038/s41598-021-94961-4
Kumar, V., Baruah, K., Nguyen, D. V., Smagghe, G., Vossen, E., and Bossier, P. (2018). Phloroglucinol-mediated Hsp70 production in crustaceans: protection against Vibrio parahaemolyticus in Artemia franciscana and Macrobrachium rosenbergii. Front. Immunol. 9, 1091. doi: 10.3389/fimmu.2018.01091
Laczka, O. F., Labbate, M., Seymour, J. R., Bourne, D. G., Fielder, S. S., and Doblin, M. A. (2014). Surface immuno-functionalisation for the capture and detection of Vibrio species in the marine environment: a new management tool for industrial facilities. PLoS ONE 9, 1–9. doi: 10.1371/journal.pone.0108387
Lages, M. A., Balado, M., and Lemos, M. L. (2019). The expression of virulence factors in Vibrio anguillarum is dually regulated by iron levels and temperature. Front. Microbiol. 10, 1–11. doi: 10.3389/fmicb.2019.02335
Laranja, J., Amar, E., Ludevese, G., Niu, Y., Geaga, M. J., Schryver, D. P., et al. (2017). A probiotic Bacillus strain containing amorphous poly-beta-hydroxybutyrate (PHB) stimulates the innate immune response of Penaeus Monodon post-larvae. Fish Shellfish Immunol. 68, 202–210. doi: 10.1016/j.fsi.2017.07.023
Laranja, J. L. Q., Ludevese-Pascual, G. L., Amar, E. C., Sorgeloos, P., Bossier, P., and Schryver, P. D. (2014). Poly-β-hydroxybutyrate (PHB) accumulating Bacillus spp. improve the survival, growth and robustness of Penaeus monodon (Fabricius, 1798) post-larvae. Vet. Microbiol. 173, 310–317. doi: 10.1016/j.vetmic.2014.08.011
Li, H. D., Tian, X. L., and Dong, S. L. (2019). Growth performance, non-specific immunity, intestinal histology and disease resistance of Litopenaeus vannamei fed on a diet supplemented with live cells of Clostridium butyricum. Aquaculture 498, 470–481. doi: 10.1016/j.aquaculture.2018.09.003
Li, J., Tan, B., Mai, K., Ai, Q., Zhang, W., Liufu, Z., et al. (2008). Immune responses and resistance against Vibrio parahaemolyticus induced by probiotic bacterium Arthrobacter XE-7 in Pacific white shrimp, Litopenaeus vannamei. J. World Aquac. Soc. 39, 477–489. doi: 10.1111/j.1749-7345.2008.00188.x
Li, P., Xin, W., Xia, S., Luo, Y., Chen, Z., Jin, D., et al. (2018). MALDI-TOF mass spectrometry-based serotyping of V. parahaemolyticus isolated from the Zhejiang province of China. BMC Microbiol. 18, 1–10. doi: 10.1186/s12866-018-1328-z
Liu, H., Li, Z., Tan, B., Lao, Y., Duan, Z., Sun, W., et al. (2014). Isolation of a putative probiotic strain S12 and its effect on growth performance, non-specific immunity and disease-resistance of white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 41, 300–307. doi: 10.1016/j.fsi.2014.08.028
Liu, L., Ge, M., Zheng, X., Tao, Z., Zhou, S., and Wang, G. (2016). Investigation of Vibrio alginolyticus, V. harveyi, and V. parahaemolyticus in large yellow croaker, Pseudosciaena crocea (Richardson) reared in Xiangshan Bay, China. Aquac. Rep. 3, 220–224. doi: 10.1016/j.aqrep.2016.04.004
Lorenzo, J. M., Munekata, P. E., Dominguez, R., Pateiro, M., Saraiva, J. A., and Franco, D. (2018). “Main groups of microorganisms of relevance for food safety and stability: general aspects and overall description,” in Innovative Technologies for Food Preservation, eds F. J. Barba, A. S. Sant'Ana, V. Orlien, and M. Koubaa (London: Academic Press), 53–107. doi: 10.1016/B978-0-12-811031-7.00003-0
Lulijwa, R., Rupia, E. J., and Alfaro, A. C. (2020). Antibiotic use in aquaculture, policies and regulation, health and environmental risks: a review of the top 15 major producers. Rev. Aquacult. 12, 640–663. doi: 10.1111/raq.12344
Ma, J., Bruce, T. J., Jones, E. M., and Cain, K. D. (2019). A review of fish vaccine development strategies: conventional methods and modern biotechnological approaches. Microorganisms 7, 569. doi: 10.3390/microorganisms7110569
MacFadden, D. R., McGough, S. F., Fisman, D., Santillana, M., and Brownstein, J. S. (2018). Antibiotic resistance increases with local temperature. Nat. Clim. Chang. 8, 510–514. doi: 10.1038/s41558-018-0161-6
Maeda, M., Shibata, A., Biswas, G., Korenaga, H., Kono, T., Itami, T., et al. (2014). Isolation of lactic acid bacteria from kuruma shrimp (Marsupenaeus japonicus) intestine and assessment of immunomodulatory role of a selected strain as probiotic. Mar. Biotechnol 16, 181–192. doi: 10.1007/s10126-013-9532-1
Martinez-Urtaza, J., Simental, L., Velasco, D., DePaola, A., Ishibashi, M., Nakaguchi, Y., et al. (2005). Pandemic Vibrio parahaemolyticus O3:K6, Europe. Emerg Infect Dis. 11, 1319–1320. doi: 10.3201/eid1108.050322
Masitoh, M. M., Hariati, A. M., and Fadjar, M. (2016). Antimicrobial activity of Bacillus cereus and Bacillus thuringiensis on pathogenic Vibrio harveyi in Litopenaeus vannamei. J. Life Sci. Biomed. 6, 10–14.
Matsumoto, C., Okuda, J., Ishibashi, M., Iwanaga, M., Garg, P., Rammamurthy, T., et al. (2000). Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38, 578–585. doi: 10.1128/JCM.38.2.578-585.2000
Meaden, S., and Koskella, B. (2013). Exploring the risks of phage application in the environment. Front. Microbiol. 4, 1–8. doi: 10.3389/fmicb.2013.00358
Montieri, S., Suffredini, E., Ciccozzi, M., and Croci, L. (2010). Phylogenetic and evolutionary analysis of Vibrio parahaemolyticus and Vibrio alginolyticus isolates based on toxR gene sequence. New Microbiol. 33, 359–372.
Moroni, F., Naya-Català, F., Piazzon, M. C., Rimoldi, S., Calduch-Giner, J., Giardini, A., et al. (2021). The effects of nisin-producing Lactococcus lactis strain used as probiotic on gilthead sea bream (Sparus aurata) growth, gut microbiota, and transcriptional response. Front. Mar. Sci. 8, 659519. doi: 10.3389/fmars.2021.659519
Mougin, J., Flahaut, C., Roquigny, R., Bonnin-Jusserand, M., Grard, T., and Le Bris, C. (2020). Rapid identification of Vibrio species of the harveyi clade using MALDI-TOF MS profiling with main spectral profile database implemented with an in-house database: Luvibase. Front. Microbiol. 11, 586536. doi: 10.3389/fmicb.2020.586536
Moussa, M., Cauvin, E., Le Piouffle, A., Lucas, O., Bidault, A., Paillard, C., et al. (2021). A MALDI-TOF MS database for fast identification of Vibrio spp. potentially pathogenic to marine molluscs. Appl. Microbiol. Biotechnol. 105, 2527–2539. doi: 10.1007/s00253-021-11141-0
Moustafa, M., Eissa, A. E., Laila, A. M., Gaafar, A. Y., Abumourad, I. M. K., and Elgendy, M. Y. (2015). Investigations into the potential causes of mass kills in mari-cultured gilthead sea bream (Sparus aurata) at Northern Egypt. Res. J. Pharm. Biol. Chem. Sci. 6, 466–477.
Muniesa, A., Basurco, B., Aguilera, C., Furones, D., Reverté, C., Sanjuan-Vilaplana, A., et al. (2020). Mapping the knowledge of the main diseases affecting sea bass and sea bream in Mediterranean. Transbound. Emerg. Dis. 67, 1089–1100. doi: 10.1111/tbed.13482
Mustapha, S., Mustapha, E., and Nozha, C. (2013). Vibrio alginolyticus: an emerging pathogen of foodborne diseases. Int. J. Sci. Technol 2, 302–309.
Nair, G. B., Ramamurthy, T., Bhattacharya, S. K., Dutta, B., Takeda, Y., and Sack, D. A. (2007). Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20, 39–48. doi: 10.1128/CMR.00025-06
Natesan, S., Muthuraman, S., and Gopal, S. (2012). Probiotic effect of Lactobacillus acidophilus against vibriosis in juvenile shrimp (Penaeus monodon). African J. Biotechnol. 11, 15811–15818. doi: 10.5897/AJB12.1328
NavinChandran, M., Iyapparaj, P., Moovendhan, S., Ramasubburayan, R., Prakash, S., Immanuel, G., et al. (2014). Influence of probiotic bacterium Bacillus cereus isolated from the gut of wild shrimp Penaeus monodon in turn as a potent growth promoter and immune enhancer in P. monodon. Fish Shellfish Immunol. 36, 38–45. doi: 10.1016/j.fsi.2013.10.004
Nguyen, H. N. K., Van, T. T. H., Nguyen, H. T., Smooker, P. M., Shimeta, J., and Coloe, P. J. (2014). Molecular characterization of antibiotic resistance in Pseudomonas and Aeromonas isolates from catfish of the Mekong Delta, Vietnam. Vet. Microbiol. 171, 397–405. doi: 10.1016/j.vetmic.2014.01.028
Nguyen, T. T. G., Nguyen, T. C., Leelakriangsak, M., Pham, T. T., Pham, Q. H., Lueangthuwapranit, C., et al. (2018). Promotion of Lactobacillus plantarum on growth and resistance against acute hepatopancreatic necrosis disease pathogens in white-leg shrimp (Litopenaeus vannamei). Thai J. Vet. Med. 48, 19–28.
O'Neill, J. (2015). Antimicrobials in Agriculture and the Environment: Reducing Unnecessary Use and Waste. The Review on Antimicrobial Resistance. London: HM Government and the Wellcome Trust. Available online at: https://wellcomecollection.org/works/x88ast2u
Pan, X., Wu, T., Song, Z., Tang, H., and Zhao, Z. (2008). Immune responses and enhanced disease resistance in Chinese drum, Miichthys miiuy (Basilewsky), after oral administration of live or dead cells of Clostridium butyricum CB2. J. Fish. Dis. 31, 679–686. doi: 10.1111/j.1365-2761.2008.00955.x
Pande, G. S. J., Natrah, F. M. I., Flandez, A. V. B., Kumar, U., Niu, Y., Bossier, P., et al. (2015). Isolation of AHL-degrading bacteria from micro-algal cultures and their impact on algal growth and on virulence of Vibrio campbellii to prawn larvae. Appl. Microbiol. Biotechnol 99, 10805–10813. doi: 10.1007/s00253-015-6918-1
Paperna, I. (1984). “Review of diseases affecting cultured Sparus aurata and Dicentrarchus labrax,: in L'Aquaculture du Bar et des Sparidés, eds B. Barnabé, and R. Billard (Paris: INRA Publisher), 465–482.
Pepi, M., and Focardi, S. (2021). Antibiotic-resistant bacteria in aquaculture and climate change: A challenge for health in the Mediterranean area. Int. J. Environ. Res. Public Health 18. doi: 10.3390/ijerph18115723
Pérez-Sánchez, T., Ruiz-Zarzuela, I., de Blas, I., and Balcázar, J. L. (2014). Probiotics in aquaculture: a current assessment. Rev. Aquac. 6, 133–146. doi: 10.1111/raq.12033
Pham, D., Ansquer, D., Chevalier, A., Dauga, C., Peyramale, A., Wabete, N., et al. (2014). Selection and characterization of potential probiotic bacteria for Litopenaeus stylirostris shrimp hatcheries in New Caledonia. Aquaculture 432, 475–482. doi: 10.1016/j.aquaculture.2014.04.031
Planas, M., Pérez-Lorenzo, M., Hjelm, M., Gram, L., Uglenes Fiksdal, I., Bergh, Ø., et al. (2006). Probiotic effect in vivo of Roseobacter strain 27-4 against Vibrio (Listonella) anguillarum infections in turbot (Scophthalmus maximus L.) larvae. Aquaculture 255, 323–333. doi: 10.1016/j.aquaculture.2005.11.039
Pousão-Ferreira, P. M. (2009). Manual de cultivo e bioencapsulação da cadeia alimentar para a larvicultura de peixes marinhos. Ed. IPIMAR, 1–235. Available online at: https://www.ipma.pt/export/sites/ipma/bin/docs/publicacoes/pescas.mar/Manual_Cadeia__Alimentar_final.pdf (accessed June 6, 2022).
Prol-García, M. J., Planas, M., and Pintado, J. (2010). Different colonization and residence time of Listonella anguillarum and Vibrio splendidus in the rotifer Brachionus plicatilis determined by real-time PCR and DGGE. Aquaculture 302, 26–35. doi: 10.1016/j.aquaculture.2010.02.004
Pujalte, M. J., Sitjà-Bobadilla, A., Álvarez-Pellitero, P., and Garay, E. (2003). Carriage of potentially fish-pathogenic bacteria in Sparus aurata cultured in Mediterranean fish farms. Dis. Aquat. Organ. 54, 119–126. doi: 10.3354/dao054119
Quilici, M. L., Robert-Pillot, A., Picart, J., and Fournier, J. M. (2005). Pandemic vibrio parahaemolyticus o3:K6 spread, France [3]. Emerg. Infect. Dis. 11, 1148–1149. doi: 10.3201/eid1107.041008
Raghu, P., Rajikkannu, M., Baburajan, R., Deva, A., and Nandakumar, R. (2016). Effect of Bacillus coagulans and B. firmus incorporated probiotic diet on Superoxide dismutase activity and catalase activity in Penaeus monodon. World Sci. News 44, 224–235.
Ramamurthy, T., Chowdhury, G., Pazhani, G. P., and Shinoda, S. (2014). Vibrio fluvialis: an emerging human pathogen. Front. Microbiol. 5, 1–8. doi: 10.3389/fmicb.2014.00091
Ramírez, N. C. B., Rodrigues, M. S., Guimarães, A. M., Guertler, C., Rosa, J. R., Seiffert, W. Q., et al. (2017). Effect of dietary supplementation with butyrate and probiotic on the survival of Pacific white shrimp after challenge with Vibrio alginolyticus. Rev. Bras. Zootec. 46, 471–477. doi: 10.1590/s1806-92902017000600001
Rasmussen, B. B., Erner, K. E., Bentzon-Tilia, M., and Gram, L. (2018). Effect of TDA-producing Phaeobacter inhibens on the fish pathogen Vibrio anguillarum in non-axenic algae and copepod systems. Microb. Biotechnol. 11, 1070–1079. doi: 10.1111/1751-7915.13275
Rattanachuay, P., Kantachote, D., and Suntinanalert, P. (2007). Selection of proteolytic bacteria with ability to inhibit Vibrio harveyi during white shrimp (Litopenaeus vannamei) cultivation. Songklanakarin J. Sci. Technol. 29, 235–243.
Rattanachuay, P., Kantachote, D., Tantirungkij, M., Nitoda, T., and Kanzaki, H. (2011). Antivibrio compounds produced by Pseudomonas sp. W3: characterisation and assessment of their safety to shrimps. World J. Microbiol. Biotechnol. 27, 869–880. doi: 10.1007/s11274-010-0529-x
Ravi, A. V., Musthafa, K. S., Jegathammbal, G., Kathiresan, K., and Pandian, S. K. (2007). Screening and evaluation of probiotics as a biocontrol agent against pathogenic Vibrios in marine aquaculture. Lett. Appl. Microbiol. 45, 219–223. doi: 10.1111/j.1472-765X.2007.02180.x
Reina, J., Fernandez-Baca, V., and Lopez, A. (1995). Acute gastroenteritis caused by Vibrio alginolyticus in an immunocompetent patient. Clin Infect Dis. 21, 1044–1045. doi: 10.1093/clinids/21.4.1044
Rengpipat, S., Phianphak, W., Piyatiratitivorakul, S., and Menasveta, P. (1998). Effects of a probiotic bacterium on black tiger shrimp Penaeus monodon survival and growth. Aquaculture 167, 301–313. doi: 10.1016/S0044-8486(98)00305-6
Rengpipat, S., Rukpratanporn, S., Piyatiratitivorakul, S., and Menasaveta, P. (2000). Immunity enhancement in black tiger shrimp (Penaeus monodon) by a probiont bacterium (Bacillus S11). Aquaculture 191, 271–288. doi: 10.1016/S0044-8486(00)00440-3
Reverter, M., Sarter, S., Caruso, D., Avarre, J. C., Combe, M., Pepey, E., et al. (2020). Aquaculture at the crossroads of global warming and antimicrobial resistance. Nat. Commun. 11, 1–8. doi: 10.1038/s41467-020-15735-6
Richards, G. P. (2014). Bacteriophage remediation of bacterial pathogens in aquaculture: a review of the technology. Bacteriophage 4, e975540. doi: 10.4161/21597081.2014.975540
Richards, G. P., Needleman, D. S., Watson, M. A., and Bono, J. L. (2014). Complete genome sequence of the larval shellfish pathogen Vibrio tubiashii type strain ATCC 19109. Genome Announc. 2, 7–8. doi: 10.1128/genomeA.01252-14
Ringø, E. (2020). Probiotics in shellfish aquaculture. Aquac. Fish. 5, 1–27. doi: 10.1016/j.aaf.2019.12.001
Robertson, P. A. W., O'Dowd, C., Burrells, C., Williams, P., and Austin, B. (2000). Use of Carnobacterium sp. as a probiotic for Atlantic salmon (Salmo salar L.) and rainbow trout (Oncorhynchus mykiss, Walbaum). Aquaculture 185, 235–243. doi: 10.1016/S0044-8486(99)00349-X
Rodríguez-Blanco, A., Lemos, M. L., and Osorio, C. R. (2012). Integrating conjugative elements as vectors of antibiotic, mercury, and quaternary ammonium compound resistance in marine aquaculture environments. Antimicrob. Agents Chemother. 56, 2619–2626. doi: 10.1128/AAC.05997-11
Romalde, J. L., Diéguez, A. L., Lasa, A., and Balboa, S. (2014). New Vibrio species associated to molluscan microbiota: a review. Front. Microbiol. 4, 1–11. doi: 10.3389/fmicb.2013.00413
Rombaut, G., Suantika, G., Boon, N., Maertens, S., Dhert, P., Top, E., et al. (2001). Monitoring of the evolving diversity of the microbial community present in rotifer cultures. Aquaculture 198, 237–252. doi: 10.1016/S0044-8486(01)00594-4
Roomiani, L., Ahmadi, S., and Ghaeni, M. (2018). Immune response and disease resistance in the white shrimp, Litopenaeus vannamei induced by potential probiotic Lactobacillus bulgaricus. Ankara Univ. Vet. Fak. Derg. 65, 323–329. doi: 10.1501/Vetfak_0000002863
Sadok, K., Mejdi, S., Nourhen, S., et al. (2013). Phenotypic characterization and RAPD fingerprinting of Vibrio parahaemolyticus and Vibrio alginolyticus isolated during Tunisian fish farm outbreaks. Folia Microbiol. 58, 17–26. doi: 10.1007/s12223-012-0174-x
Sakai, M., Toshida, S., Atsuta, M., and Kobayashi, M. (1995). Enhancement of resistance to vibriosis in rainbow trout, Oncorhynchus mykiss (Walbaum), by oral administration of Clostridium butyricum bacterin. J. Fish Dis. 18, 187–190. doi: 10.1111/j.1365-2761.1995.tb00276.x
Sanches-Fernandes, G. M. M., Califano, G., Castanho, S., Soares, F., Ribeiro, L., Pousão-Ferreira, P., et al. (2021a). Effects of live feed manipulation with algal-derived antimicrobial metabolites on fish larvae microbiome assembly: a molecular-based assessment. Aquac. Res. 53, 1062–1083. doi: 10.1111/are.15648
Sanches-Fernandes, G. M. M., Califano, G., Keller-Costa, T., Castanho, S., Soares, F., Ribeiro, L., et al. (2021b). Draft genome sequence of Vibrio chagasii 18LP, isolated from gilthead seabream (Sparus aurata) larvae reared in aquaculture. Microbiol. Resour. Announc. 10, e0065821. doi: 10.1128/MRA.00658-21
Sanches-Fernandes, G. M. M., Califano, G., Keller-Costa, T., Castanho, S., Soares, F., Ribeiro, L., et al. (2021c). Draft genome sequence of Vibrio jasicida 20LP, an opportunistic bacterium isolated from fish larvae. Microbiol. Resour. Announc. 10, e0081321. doi: 10.1128/MRA.00813-21
Sapcharoen, P., and Rengpipat, S. (2013). Effects of the probiotic Bacillus subtilis (BP11 and BS11) on the growth and survival of Pacific white shrimp, Litopenaeus vannamei. Aquacult. Nutr. 19, 946–954. doi: 10.1111/anu.12040
Sarjito, R., Radjasa, O. K., Sabdono, A., Prayitno, S. B., and Hutabarat, S. (2009). Phylogenetic diversity of the causative agents of vibriosis associated with groupers fish from Karimunjawa Islands. Indonesia. Curr. Res. Bacteriol. 2, 14–21. doi: 10.3923/crb.2009.14.21
Scarano, C., Spanu, C., Ziino, G., Pedonese, F., Dalmasso, A., Spanu, V., et al. (2014). Antibiotic resistance of Vibrio species isolated from Sparus aurata reared in Italian mariculture. New Microbiol. 37, 329–337.
Schar, D., Klein, E. Y., Laxminarayan, R., Gilbert, M., and Van Boeckel, T. P. (2020). Global trends in antimicrobial use in aquaculture. Sci. Rep. 10, 1–9. doi: 10.1038/s41598-020-78849-3
Sha, Y., Liu, M., Wang, B., Jiang, K., Qi, C., and Wang, L. (2016a). Bacterial population in intestines of litopenaeus vannamei fed different probiotics or probiotic supernatant. J. Microbiol. Biotechnol. 26, 1736–1745. doi: 10.4014/jmb.1603.03078
Sha, Y., Wang, B., Liu, M., Jiang, K., and Wang, L. (2016b). Interaction between Lactobacillus pentosus HC-2 and Vibrio parahaemolyticus E1 in Litopenaeus vannamei in vivo and in vitro. Aquaculture 465, 117–123. doi: 10.1016/j.aquaculture.2016.09.007
Sharifuzzaman, S. M., Abbass, A., Tinsley, J. W., and Austin, B. (2011). Subcellular components of probiotics Kocuria SM1 and Rhodococcus SM2 induce protective immunity in rainbow trout (Oncorhynchus mykiss, Walbaum) against Vibrio anguillarum. Fish Shellfish Immunol. 30, 347–353. doi: 10.1016/j.fsi.2010.11.005
Sharifuzzaman, S. M., and Austin, B. (2010). Development of protection in rainbow trout (Oncorhynchus mykiss, Walbaum) to Vibrio anguillarum following use of the probiotic Kocuria SM1. Fish Shellfish Immunol. 29, 212–216. doi: 10.1016/j.fsi.2010.03.008
Shefat, S. H. T. (2018). Use of probiotics in shrimp aquaculture in bangladesh. Acta Sci. Microbiol. 1, 20–27.
Siddique, A. B., Moniruzzaman, M., Ali, S., Dewan, M. N., Islam, M. R., Islam, M. S., et al. (2021). Characterization of pathogenic Vibrio parahaemolyticus isolated from fish aquaculture of the southwest coastal area of Bangladesh. Front. Microbiol. 12, 635539. doi: 10.3389/fmicb.2021.635539
Sivakumar, N., Sundararaman, N., and Selvakumar, G. (2012). Probiotic effect of Lactobacillus acidophilus against vibriosis in juvenile shrimp (Penaeus monodon). African J. Biotechnol. 11, 15811–15818. doi: 10.5897/ajb12.1328
Skjermo, J., and Bergh, Ø. (2004). High-M alginate immunostimulation of Atlantic halibut (Hippoglossus hippoglossus L.) larvae using Artemia for delivery, increases resistance against vibriosis. Aquaculture 238, 107–113. doi: 10.1016/j.aquaculture.2004.05.038
Skjermo, J., and Vadstein, O. (1993). “Characterization of the bacterial flora of mass cultivated Brachionus plicatilis,” in Rotifer Symposium VI. Developments in Hydrobiology, eds J. J. Gilbert, E. Lubzens, and M. R. Miracle (Dordrecht: Springer), 83.
Snoussi, M., Hajlaoui, H., Noumi, E., Zanetti, S., and Bakhrouf, A. (2008). Phenotypic and genetic diversity of Vibrio alginolyticus strains recovered from juveniles and older Sparus aurata reared in a Tunisian marine farm. Ann. Microbiol. 58, 141–146. doi: 10.1007/BF03179458
Soliman, W. S., Shaapan, R. M., Mohamed, L. A., and Gayed, S. S. R. (2019). Recent biocontrol measures for fish bacterial diseases, in particular to probiotics, bio-encapsulated vaccines, and phage therapy. Open Vet. J. 9, 190–195. doi: 10.4314/ovj.v9i3.2
Sonia, G. A. S., and Lipton, A. P. (2012). Pathogenicity and antibiotic susceptibility of Vibrio species isolated from the captive-reared tropical marine ornamental blue damsel fish, Pomacentrus caeruleus (Quoy and Gaimard, 1825). Indian J. Mar. Sci. 41, 348–354.
Sorieul, L., Wabete, N., Ansquer, D., Mailliez, J. R., Pallud, M., Zhang, C., et al. (2018). Survival improvement conferred by the Pseudoalteromonas sp. NC201 probiotic in Litopenaeus stylirostris exposed to Vibrio nigripulchritudo infection and salinity stress. Aquaculture 495, 888–898. doi: 10.1016/j.aquaculture.2018.06.058
Sorroza, L., Padilla, D., Acosta, F., Román, L., Grasso, V., Vega, J., et al. (2012). Characterization of the probiotic strain Vagococcus fluvialis in the protection of European sea bass (Dicentrarchus labrax) against vibriosis by Vibrio anguillarum. Vet. Microbiol. 155, 369–373. doi: 10.1016/j.vetmic.2011.09.013
Sorroza, L., Real, F., Acosta, F., Acosta, B., Deniz, S., Roman, L., et al. (2013). A probiotic potential of Enterococcus gallinarum against Vibrio anguillarum infection, Fish Pathol. 48, 9–12. doi: 10.3147/jsfp.48.9
Soto-Rodriguez, S. A., Roque, A., Lizarraga-Partida, M. L., Guerra-Flores, A. L., and Gomez-Gil, B. (2003). Virulence of luminous vibrios to Artemia franciscana nauplii. Dis. Aquat. Organ. 53, 231–240. doi: 10.3354/dao053231
Stentiford, G. (2017). Solving the $6 Billion Per Year Global Aquaculture Disease Problem. Marine Science Blog Gov. Available online at: https://marinescience.blog.gov.uk/2017/02/02/solving-the-6-billion-per-year-global-aquaculture-disease-problem/ (accessed June 6, 2022).
Stentiford, G. D., Sritunyalucksana, K., Flegel, T. W., Williams, B. A. P., Withyachumnarnkul, B., Itsathitphaisarn, O., et al. (2017). New paradigms to help solve the global aquaculture disease crisis. PLoS Pathog. 13, 1–6. doi: 10.1371/journal.ppat.1006160
Sumon, M. S., Ahmmed, F., Khushi, S. S., Ahmmed, M. K., Rouf, M. A., Chisty, M. A. H., et al. (2018). Growth performance, digestive enzyme activity and immune response of Macrobrachium rosenbergii fed with probiotic Clostridium butyricum incorporated diets. J. King Saud Univ. Sci. 30, 21–28. doi: 10.1016/j.jksus.2016.11.003
Swain, S. M., Singh, C., and Arul, V. (2009). Inhibitory activity of probiotics Streptococcus phocae PI80 and Enterococcus faecium MC13 against vibriosis in shrimp Penaeus monodon. World J. Microbiol. Biotechnol. 25, 697–703. doi: 10.1007/s11274-008-9939-4
Tan, C. W., Rukayadi, Y., Hasan, H., Abdul-Mutalib, N. A., Jambari, N. N., Hara, H., et al. (2021). Isolation and characterization of six Vibrio parahaemolyticus lytic bacteriophages from seafood samples. Front. Microbiol. 12, 616548. doi: 10.3389/fmicb.2021.616548
Tepaamorndech, S., Chantarasakha, K., Kingcha, Y., Chaiyapechara, S., Phromson, M., Sriariyanun, M., et al. (2019). Effects of Bacillus aryabhattai TBRC8450 on vibriosis resistance and immune enhancement in Pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 86, 4–13. doi: 10.1016/j.fsi.2018.11.010
Thompson, F. L., Iida, T., and Swings, J. (2004). Biodiversity of Vibrios. Microbiol. Mol. Biol. Rev. 68, 403–431. doi: 10.1128/MMBR.68.3.403-431.2004
Torres-Corral, Y., Girons, A., González-Barreiro, O., Seoane, R., Riaza, A., and Santos, Y. (2021). Effect of bivalent vaccines against Vibrio anguillarum and Aeromonas salmonicida subspecies achromogenes on health and survival of turbot. Vaccines 9, 1–13. doi: 10.3390/vaccines9080906
Tseng, D. Y., Ho, P. L., Huang, S. Y., Cheng, S. C., Shiu, Y. L., et al. (2009). Enhancement of immunity and disease resistance in the white shrimp, Litopenaeus vannamei, by the probiotic, Bacillus subtilis E20. Fish Shellfish Immunol. 26, 339–344. doi: 10.1016/j.fsi.2008.12.003
Urbanczyk, H., Ast, J. C., Higgins, M. J., Carson, J., and Dunlap, P. V. (2007). Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb. nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov. Int. J. Syst. Evol. Microbiol. 57, 2823–2829. doi: 10.1099/ijs.0.65081-0
Utiswannakul, P., Sangchai, S., and Rengpipat, S. (2011). Enhanced growth of black tiger shrimp penaeus monodon by dietary supplementation with Bacillus (BP11) as a probiotic. J. Aquac. Res. Dev. S1, 1–9. doi: 10.4172/2155-9546.S1-006
Vadstein, O., Bergh, Ø., Gatesoupe, F. J., Galindo-Villegas, J., Mulero, V., Picchietti, S., et al. (2013). Microbiology and immunology of fish larvae. Rev. Aquac. 5, S1–S25. doi: 10.1111/j.1753-5131.2012.01082.x
Vaseeharan, B., and Ramasamy, P. (2003). Control of pathogenic Vibrio spp. by Bacillus subtilis BT23, a possible probiotic treatment for black tiger shrimp Penaeus monodon. Lett. Appl. Microbiol. 36, 83–87. doi: 10.1046/j.1472-765X.2003.01255.x
Vayssier-Taussat, M., Albina, E., Citti, C., Cosson, J. F., Jacques, M. A., Lebrun, M. H., et al. (2014). Shifting the paradigm from pathogens to pathobiome new concepts in the light of meta-omics. Front. Cell. Infect. Microbiol. 5, 1–7. doi: 10.3389/fcimb.2014.00029
Vera, P., Navas, J. I., and Fouz, B. (1991). First isolation of Vibrio damselae from sea bream (Sparus aurata). Bull. Eur. Assoc. Fish Pathol. 11, 112–113.
Verdonck, L., Grisez, L., Sweetman, E., Minkoff, G., Sorgeloos, P., Ollevier, F., et al. (1997). Vibrios associated with routine productions of Brachionus plicatilis. Aquaculture 149, 203–214. doi: 10.1016/S0044-8486(96)01451-2
Verschuere, L., Rombaut, G., Sorgeloos, P., and Verstraete, W. (2000). Probiotic bacteria as biological control agents in aquaculture. Microbiol. Mol. Biol. Rev. 64, 655–671. doi: 10.1128/MMBR.64.4.655-671.2000
Vezzulli, L., Grande, C., Reid, P. C., Hélaouët, P., Edwards, M., Höfle, M. G., et al. (2016). Climate influence on Vibrio and associated human diseases during the past half-century in the coastal North Atlantic. Proc. Natl. Acad. Sci. U. S. A. 113, E5062–E5071. doi: 10.1073/pnas.1609157113
Vidal, J., Aderaldo, M., Pessôa, M. N. D. C., Santos, F. L. D., Mendes, P. D. P., and Mendes, M. S. (2018). Probiotic potential of Bacillus cereus against Vibrio spp. in post-larvae shrimps. Rev. Caatinga 31, 495–503. doi: 10.1590/1983-21252018v31n226rc
Vieira, F. N., Buglione, C. C., Mouriño, J. P. L., Jatobá, A., Martins, M. L., Schleder, D. D., et al. (2010). Effect of probiotic supplemented diet on marine shrimp survival after challenge with Vibrio harveyi. Arq. Bras. Med. Vet. Zoot. 62, 631–638. doi: 10.1590/S0102-09352010000300019
Vinh, D. C., Mubareka, S., Fatoye, B., Plourde, P., and Orr, P. (2006). Vibrio vulnificus septicemia after handling Tilapia species fish: a Canadian case report and review. Can. J. Infect. Dis. Med. Microbiol. 17, 129–132. doi: 10.1155/2006/164681
Vinoj, G., Jayakumar, R., Chen, J. C., Withyachumnarnkul, B., Shanthi, S., and Vaseeharan, B. (2015). N-hexanoyl-L-homoserine lactone-degrading Pseudomonas aeruginosa PsDAHP1 protects zebrafish against Vibrio parahaemolyticus infection. Fish Shellfish Immunol. 42, 204–212. doi: 10.1016/j.fsi.2014.10.033
Wachsmuth, K., Olsvik, Ø., Evins, G. M., and Popovic, T. (1994). “Molecular epidemiology of cholera,” in Vibrio Cholerae and Cholera: Molecular to Global Perspectives, eds I. Kaye Wachsmuth, P. A. Blake, and Ø. Olsvik (Washington, DC: American Society for Microbiology), 357–370. doi: 10.1128/9781555818364.ch23
Wang, H., Wang, C., Tang, Y., Sun, B., Huang, J., and Song, X. (2018). Pseudoalteromonas probiotics as potential biocontrol agents improve the survival of Penaeus vannamei challenged with acute hepatopancreatic necrosis disease (AHPND)-causing Vibrio parahaemolyticus. Aquaculture 494, 30–36. doi: 10.1016/j.aquaculture.2018.05.020
Wang, R., Zhong, Y., Gu, X., Yuan, J., Saeed, A. F., and Wang, S. (2015). The pathogenesis, detection, and prevention of Vibrio parahaemolyticus. Front. Microbiol. 6, 1–13. doi: 10.3389/fmicb.2015.00144
Wang, Y.-C., Hu, S.-Y., Chiu, C.-S., and Liu, C.-H. (2019). Multiple-strain probiotics appear to be more effective in improving the growth performance and health status of white shrimp, Litopenaeus vannamei, than single probiotic strains. Fish Shellfish Immuno. 84, 1050–1058. doi: 10.1016/j.fsi.2018.11.017
Wanka, K. M., Damerau, T., Costas, B., Krueger, A., Schulz, C., and Wuertz, S. (2018). Isolation and characterization of native probiotics for fish farming. BMC Microbiol. 18, 1–13. doi: 10.1186/s12866-018-1260-2
Williams, T. C., Blackman, E. R., Morrison, S. S., Gibas, C. J., and Oliver, J. D. (2014). Transcriptome sequencing reveals the virulence and environmental genetic programs of vibrio vulnificus exposed to host and estuarine conditions. PLoS ONE 9, 1–27. doi: 10.1371/journal.pone.0114376
Won, S., Hamidoghli, A., Choi, W., Bae, J., Jang, W. J., Lee, S., et al. (2020). Evaluation of potential probiotics bacillus subtilis WB60, Pediococcus pentosaceus, and Lactococcus lactis on growth performance, immune response, gut histology and immune-related genes in whiteleg shrimp, Litopenaeus vannamei. Microorganisms 8, 1–15. doi: 10.3390/microorganisms8020281
Wong, K. K., and Griffin, P. M. (2018). “Other Vibrio species,” in Principles and Practices of Pediatric, Infectious Diseases, ed S. Long (Philadelphia, PA: Elsevier), 879–881. doi: 10.1016/B978-0-323-40181-4.00159-6
Wongtavatchai, J., López-Dóriga, M. V., and Francis, M. J. (2010). Effect of AquaVacTM VibromaxTM on size and health of post larva stage of pacific White shrimp Litopenaeus vannamei and black tiger shrimp Penaeus monodon. Aquaculture 308, 75–81. doi: 10.1016/j.aquaculture.2010.08.017
Yu, J. P., Hino, A., Noguchi, T., and Wakabayashi, H. (1990). Toxicity of Vibrio alginolyticus on the survival of the rotifer Brachionus plicatilis. Nippon Suisan Gakkaishi 56, 1455–1460. doi: 10.2331/suisan.56.1455
Yun, N. R., and Kim, D. M. (2018). Vibrio vulnificus infection: a persistent threat to public health. Korean J. Intern. Med. 33, 1070–1078. doi: 10.3904/kjim.2018.159
Zhang, X., Lin, H., Wang, X., and Austin, B. (2018). Significance of Vibrio species in the marine organic carbon cycle—A review. Sci. China Earth Sci. 61, 1357–1368. doi: 10.1007/s11430-017-9229-x
Zhang, X. H., and Austin, B. (2005). Haemolysins in Vibrio species. J. Appl. Microbiol. 98, 1011–1019. doi: 10.1111/j.1365-2672.2005.02583.x
Zhao, W., Dao, C., Karim, M., Gomez-Chiarri, M., Rowley, D., and Nelson, D. R. (2016). Contributions of tropodithietic acid and biofilm formation to the probiotic activity of Phaeobacter inhibens. BMC Microbiol. 16, 1–17. doi: 10.1186/s12866-015-0617-z
Zheng, C. N., and Wang, W. (2017). Effects of Lactobacillus pentosus on the growth performance, digestive enzyme and disease resistance of white shrimp, Litopenaeus vannamei (Boone, 1931). Aquac. Res. 48, 2767–2777. doi: 10.1111/are.13110
Zhou, Z., Chen, X., Sheng, H., Shen, X., Sun, X., Yan, Y., et al. (2020). Engineering probiotics as living diagnostics and therapeutics for improving human health. Microb. Cell Fact. 19, 1–12. doi: 10.1186/s12934-020-01318-z
Zokaeifar, H., Balcázar, J. L., Saad, C. R., Kamarudin, M. S., Sijam, K., Arshad, A., et al. (2012). Effects of Bacillus subtilis on the growth performance, digestive enzymes, immune gene expression and disease resistance of white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 33, 683–689. doi: 10.1016/j.fsi.2012.05.027
Keywords: biological control, fish larviculture, fish microbiome, host-microbe interactions, probiotics, Vibrio
Citation: Sanches-Fernandes GMM, Sá-Correia I and Costa R (2022) Vibriosis Outbreaks in Aquaculture: Addressing Environmental and Public Health Concerns and Preventive Therapies Using Gilthead Seabream Farming as a Model System. Front. Microbiol. 13:904815. doi: 10.3389/fmicb.2022.904815
Received: 25 March 2022; Accepted: 20 June 2022;
Published: 11 July 2022.
Edited by:
Malka Halpern, University of Haifa, IsraelReviewed by:
T. G. Sumithra, Indian Council of Agricultural Research (ICAR), IndiaCopyright © 2022 Sanches-Fernandes, Sá-Correia and Costa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rodrigo Costa, cm9kcmlnb3Njb3N0YUB0ZWNuaWNvLnVsaXNib2EucHQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.